Abstract
Objective
To investigate the prognostic and biologic significance of immune-related gene expression in high grade serous ovarian cancer (HGSOC).
Methods
Gene expression dependent survival analyses for a panel of immune related genes were evaluated in HGSOC utilizing The Cancer Genome Atlas (TCGA). Prognostic value of LCK was validated using IHC in an independent set of 72 HGSOC. Prognostic performance of LCK was compared to cytolytic score (CYT) using RNAseq across multiple tumor types. Differentially expressed genes in LCK high samples and gene ontology enrichment were analyzed.
Results
High pre-treatment LCK mRNA expression was found to be a strong predictor of survival in a set of 535 ovarian cancers. Patients with high LCK mRNA expression had a longer median progression free survival (PFS) of 29.4 months compared to 16.9 months in those without LCK high expression (p = 0.003), and longer median overall survival (OS) of 95.1 months versus 44.5 months (p = 0.001), which was confirmed in an independent cohort by IHC (p = 0.04). LCK expression was compared to CYT across tumor types available in the TCGA and was a significant predictor of prognosis in HGSOC where CYT was not predictive. Unexpectedly, LCK high samples also were enriched in numerous immunoglobulin-related and other B cell transcripts.
Conclusions
LCK is a better prognostic factor than CYT in ovarian cancer. In HGSOC, LCK high samples were characterized by higher expression of immunoglobulin and B-cell related genes suggesting that a cooperative interaction between tumor infiltrating T and B cells may correlate with better survival in this disease.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02385-x) contains supplementary material, which is available to authorized users.
Keywords: Ovarian cancer, Lymphocyte specific kinase, Biomarker, Cytolytic activity score, B lymphocyte
Introduction
Ovarian cancer is the leading cause of death from gynecologic malignancy, with over 22,000 cases per year in the United States and over 14,000 deaths [2]. The high mortality rate is due to the fact that the majority of ovarian cancer presents at advanced stage III/IV and has a high risk of recurrence despite initial response to traditional platinum based therapy. There is growing evidence to support a pivotal role of the immune system in the pathogenesis of cancer; in ovarian cancer and others the presence of high levels of tumor infiltrating lymphocytes (TILs) has been associated with improved progression free survival (PFS) and overall survival (OS) [3–9]. However, this impact is in the context of a complex interplay between multiple aspects of the tumor microenvironment, as T cell type, location, and tumor stromal factors have all been shown to modify survival rates [6, 10–14].
In the setting of this complexity, there is a need for reliable biomarker(s) with utility in prognostication and stratification of untreated ovarian cancers. One well published genomic prognostic feature is the cytolytic activity score (CYT), a quantitative measure of immune cytolytic activity based on transcript levels of perforin (PRF1) and granzyme A (GZMA) [15]. These two molecules reflect the central mechanism for cytotoxic lymphocyte killing; perforin is responsible for the creation of pores within the target cell membrane which then allow for the entry of granzymes that cleave caspases and induce apoptosis. CYT has been shown to be a useful metric of cyototoxic activation and subsequent improved survival in multiple other tumor types [15–18]. However, CYT captures only T lymphocyte activity and therefore may be limited in its representation of the immune microenvironment. Here our group reports on serial correlative studies within The Cancer Genome Atlas (TCGA) which demonstrated that high lymphocyte specific tyrosine kinase (LCK) expression is a better discriminator of PFS and OS than CYT not only in ovarian cancer, but also in many other cancer types. LCK is a canonical downstream T-cell receptor signaling molecule, but when transcriptional phenotype of high LCK expressing ovarian cancers was analyzed we noted the presence of a B-cell signature and chemokines, suggesting a positive prognostic effect when ovarian cancers are infiltrated by both T and B lymphocytes.
Materials and methods
TCGA data analysis
To explore the correlation between a variety of immune cell markers and clinical outcome, the high-grade serous ovarian cancer (HGSOC) provisional data set from The Cancer Genome Atlas was analyzed [19]. For mRNA expression analysis, Affymetrix U133 microarray data were used and only samples for which these data were available were included. Samples were divided into “high expression” and “non-high expression” groups using the CBioportal web interface, for the following markers: CD2, CD3E, CD3D, CD4, GZMA, PRF1, CD19, MS4A1 and LCK [20, 21] where high expression was defined as expression within the top 3% (1.86 SD). Gene expression and enrichment analyses were performed using BRB-ArrayTools (Version 4.5.1) developed by Dr. Richard Simon and the BRB-ArrayTools Development Team. Gene expression analysis was performed with p < 0.001 cutoff for significance to guard against false discovery due to multiple comparisons and at least twofold difference in the geometric mean of expression levels.
Subsequent analysis of RNA sequencing data was then performed across 30 tumor types available in the TCGA. The following tumor types (project code and n = sample size) were included: adrenocortical carcinoma (ACC, n = 92), bladder/urothelial (BLCA, n = 412), breast invasive carcinoma (BRCA, n = 1098), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, n = 307), cholangiocarcinoma (CHOL, n = 51), colon adenocarcinoma (COAD, n = 461), esophageal carcinoma (ESCA, n = 185), glioblastoma multiforme (GBM, n = 617), head and neck squamous cell carcinoma (HNSC, n = 528), kidney renal clear cell carcinoma (KIRC, n = 537), kidney renal papillary cell carcinoma (KIRP, n = 291), acute myeloid leukemia (LAML, n = 200), low grade glioma (LGG, n = 516), liver hepatocellular carcinoma (LIHC, n = 377), lung adenocarcinoma (LUAD, n = 585), lung squamous cell carcinoma (LUSC, n = 504), mesothelioma (MESO, n = 87), ovarian serous cystadenocarcinoma (OV, n = 608), pancreatic adenocarcinoma (PAAD, n = 185), pheochromocytoma and paraganglioma (PCPG, n = 179), prostate adenocarcinoma (PRAD, n = 500), rectum adenocarcinoma (READ, n = 172), sarcoma (SARC, n = 261), skin cutaneous melanoma (SKCM, n = 470), stomach adenocarcinoma (STAD, n = 443), testicular germ cell tumors (TGCT, n = 150), thyroid carcinoma (THCA, n = 507), uterine corpus endometrial carcinoma (UCEC, n = 560), uterine carcinosarcoma (UCS, n = 57), and uveal melanoma (UVM, n = 80). For this analysis in each cancer, the LCK high expressing population (the top 10%) was compared to the LCK-low population (bottom 10% in expression). This was compared to CYT which has been previously defined [15]. Briefly, total raw read counts per gene were converted to transcripts per million (TPM), which was calculated by dividing by the gene’s maximum transcript length to provide a coverage depth estimate and scaling to sum to a total depth of 1e6 per sample. CYT was calculated as the as the geometric mean of GZMA and PRF1 expression values in TPM, where similar high (top 10%) and low (bottom 10%) groups were compared.
Immunohistochemistry (IHC)
LCK protein expression was performed using immunohistochemistry on an independent cohort of 72 ovarian cancer samples using a commercially available anti-LCK antibody (HPA003494, Sigma-Aldrich). Additionally, CD8 and CD20 immunohistochemistry staining was performed in this cohort (CD20:SAB5600082, Sigma-Aldrich, CD8: CD8-4B11-L-CE, Leica Biosystems), and demographics and survival data was abstracted. All tumor tissue samples were resected from the primary tumor site of previously untreated HGSOC patients with stage 3 and 4 diseases. A semi-quantitative IHC score was assigned by pathology collaborators including a senior gynecologic pathologist. For scoring purposes tissue LCK + lymphocytes staining was as none (0, average of one or less LCK + lymphocyte), low (1, less than 10 LCK + lymphocytes), medium (2, greater than 10 but less than 40 LCK + lymphocytes), and high (3, greater than 40 LCK + lymphocytes or multiple germinal centers). The same cut offs were used for CD8 and CD20 positivity, and the counts were averaged over three fields for independent pathology samples. Samples were additionally investigated for presence of lymphoid aggregates and tertiary lymphoid structures (TLS). Given the difficulty in distinguishing lymphoid aggregate from true TLS due to potential for germinal center to be in an alternate plane than the section evaluated, lymphoid aggregates, defined as a rounded collection of lymphoid cells forming a mass outside of a lymph node, were coded as present or absent [22].
IHC was additionally performed across a range of benign and malignant serous neoplasms on a tissue microarray (TMA), where counts were averaged over the three cores. The TMA contained a spectrum of serous gynecological tissues, including normal fallopian tube epithelium obtained at the time of salpingo-oophorectomy for benign ovarian cystadenomas and high grade serous carcinomas. A total of 20 normal fallopian tube samples, 14 high-grade ovarian serous carcinoma tissues, and 13 benign serous cystadenomas were compared. Each tissue specimen was represented as three independent cores on the TMA.
Statistical analysis
Descriptive statistics (n, percent, mean, standard deviation) were calculated to summarize patient demographics. Cox regression and backwards stepwise regressions were performed to assess OS and PFS for immune-related genes and dichotomized CYT groups. Statistical analyses were performed using SAS 9.4 for Windows (SAS Institute Inc., Cary, NC). IHC score comparison was performed using the Mann–Whitney U test with p < 0.05 considered significant. Spearman correlations were performed to assess the strength of association of LCK, CD20, and CD8, and strength of correlation was assessed. Strength of correlations analysis performed using R version 3.4.1 package “cocor” [23].
Results
High LCK expression predicts improved survival in HGSOC
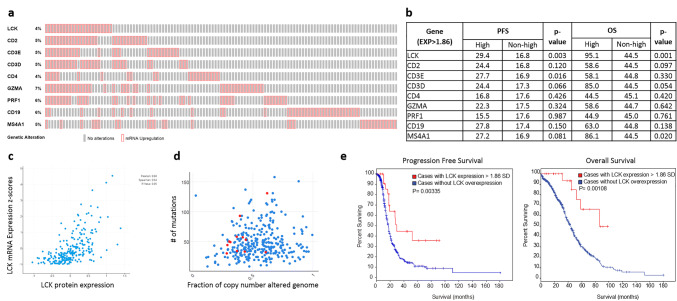
535 high-grade serous ovarian samples in the TCGA dataset were included using the cBioPortal platform, 520 of which had Affymetrix U133 microarray data available for mRNA analysis [19–21]. Analysis of the TCGA was performed investigating the upregulation of immune-related genes including CD3E, CD3D, CD2, CD4, Perforin 1 (PRF1), Granzyme A (GZMA), CD19, and CD20 (MS4A1) and LCK (Fig. 1a, b). Of note, CD8A data were unavailable within the TCGA microarray dataset. High LCK mRNA expression was present in 23 (4%) of all cases (Fig. 1a). Progression-free and overall survival data were collected for each of the above genes and compared in elevated and non-elevated samples. LCK was shown to have the strongest association with survival; patients with high LCK mRNA expression had a median progression free survival of 29.4 months, compared to 16.8 in those without high LCK expression (p = 0.003). Similarly, patients with high LCK had significantly longer overall survival than non-LCK high with median overall survival time of 95.1 months and 44.5 months respectively (p = 0.001) (Fig. 1e). As expected, LCK mRNA high samples also had significantly higher LCK protein levels as determined by reverse phase protein arrays (RPPA) (Fig. 1c). Only two other markers within the panel were statistically significantly associated with survival and were shown to have less dramatic prognostic differences. Specifically, high expression of B-cell marker CD20 (MS4A1) was associated with survival, with median PFS of 27.2 months (p = 0.08) and overall survival of 86.1 months (p = 0.02), while CD3E elevation had a significant association with PFS (p = 0.016) but was not associated with OS (p = 0.330). High expression of the other immune-related genes tested above was not associated with survival.
Fig. 1.
TCGA analysis of immune-related gene expression. a Altered gene expression samples: Total percentage of high-expressing samples demonstrated at left of serially tested immune-related genes in TCGA ovarian serous cystadenocarcinoma study (TCGA, provisional). Red boxes indicate sample with > 1.86 SD expression. b Kaplan–Meier analysis of progression-free survival and overall survival in gene-high as compared to not gene-high samples for respective immune related genes. c LCK mRNA expression levels (RNA Seq V2 RSEM) correlation with LCK protein expression (RPPA). d Mutation count and copy number alterations among total study tumors (blue) and LCK-high tumors (red). e Kaplan–Meier analysis of progression-free survival and overall survival in LCK high (red) tumors compared to non-LCK high (blue)
To examine if high LCK expression was simply a marker of high levels of tumor-infiltrating lymphocytes (TIL), we compared the levels of CD3 and TCR-related transcripts in LCK high samples. We also evaluated potential demographic, clinical, and pathological differences between LCK high and remaining samples (Table 1). The median age in the entire cohort was 59 years (30–89 years), and most patients were advanced stage (72.9% stage IIIC, 16.0% stage IV). No differences were detected between the two groups with respect to clinical characteristics, including age, race, ECOG performance status, clinical stage, and tumor grade. LCK expression was correlated with high expression of CD3 and TCR-related transcripts (Table 2), but as described above LCK had improved discriminatory prognostic ability than these markers alone.
Table 1.
Demographics by LCK expression level
| Total cohorta 520 |
LCK highb n = 23 |
Non-LCK high n = 497 | p value |
|---|---|---|---|
| Characteristic | |||
| Age (median) | 40–78 (58) | 30–89 (59) | 0.837 |
| ECOG performance | |||
| 0 | 4 | 69 | 0.633 |
| 1 | 3 | 72 | |
| 2 | 2 | 21 | |
| 3 | 0 | 4 | |
| Unknown | 14 | 331 | |
| Stage | |||
| I | 1 | 15 | 0.134 |
| II | 3 | 25 | |
| IIIA, B | 3 | 28 | |
| IIIC | 13 | 366 | |
| IV | 3 | 80 | |
| Unknown | 0 | 4 | |
| Grade | |||
| 1 | 0 | 5 | 0.552 |
| 2 | 3 | 61 | |
| 3 | 19 | 419 | |
| Unknown | 1 | 12 | |
| Race/ethnicity | |||
| Asian | 1 | 14 | 0.4696 |
| Black | 0 | 23 | |
| Hispanic | 1 | 7 | |
| White | 20 | 433 | |
| Other/unknown | 1 | 20 | |
a520 patients included for a total of 535 samples available
bLC- high: expression > 1.86SD within TCGA ovarian serous cystadenocarcinoma study (TCGA, provisional)
Table 2.
Gene expression and ontology enrichment in LCK-high samples
| Top Over Expressed Genes | ||||||
|---|---|---|---|---|---|---|
| Symbol | Name | LCK non-high (geometric mean of intensities) | LCK high (geometric mean of intensities) | Fold changea | Parametric p value | FDR |
| CXCL9 | Chemokine (C–X–C motif) ligand 9 | 55.65 | 870.35 | 15.640 | < 1e−07 | < 1e−07 |
| IGLC1 | Immunoglobulin lambda constant 1 (Mcg marker) | 186.45 | 1984.25 | 10.642 | < 1e−07 | < 1e−07 |
| IGHM | Immunoglobulin heavy constant mu | 26.53 | 221 | 8.330 | < 1e−07 | < 1e−07 |
| IGKC | Immunoglobulin kappa constant | 26.11 | 226.8 | 8.686 | < 1e−07 | < 1e−07 |
| JCHAIN | Joining chain of multimeric IgA and IgM | 25.76 | 217.56 | 8.446 | < 1e−07 | < 1e−07 |
| IGKC | Immunoglobulin kappa constant | 27.05 | 221.33 | 8.182 | < 1e−07 | < 1e−07 |
| CXCL13 | Chemokine (C–X–C motif) ligand 13 | 18.99 | 151.74 | 7.991 | < 1e−07 | < 1e−07 |
| IGHM | Immunoglobulin heavy constant mu | 16.18 | 113.07 | 6.988 | < 1e−07 | < 1e−07 |
| TRBC1 | T cell receptor beta constant 1 | 32.03 | 208.74 | 6.517 | < 1e−07 | < 1e−07 |
| IGLJ3 | Immunoglobulin lambda joining 3 | 23.3 | 154.25 | 6.620 | < 1e−07 | < 1e−07 |
| IGKC | Immunoglobulin kappa constant | 103.1 | 681.32 | 6.608 | < 1e−07 | < 1e−07 |
| CCL5 | Chemokine (C–C motif) ligand 5 | 27.65 | 175.54 | 6.349 | < 1e−07 | < 1e−07 |
| TRBC1 | T cell receptor beta constant 1 | 31.07 | 187.03 | 6.020 | < 1e−07 | < 1e−07 |
| IGLC1 | Immunoglobulin lambda constant 1 (Mcg marker) | 19.22 | 109.64 | 5.704 | < 1e−07 | < 1e−07 |
| CD2 | CD2 molecule | 24.16 | 129.36 | 5.354 | < 1e−07 | < 1e−07 |
| IGLJ3 | Immunoglobulin lambda joining 3 | 13.85 | 71.54 | 5.165 | < 1e−07 | < 1e−07 |
| CD8A | CD8a molecule | 16.17 | 82.27 | 5.088 | < 1e−07 | < 1e−e−07 |
| CD3D | CD3d molecule, delta (CD3-TCR complex) | 34.53 | 175.11 | 5.071 | < 1e−07 | < 1e−07 |
| IGLV1-44 | Immunoglobulin lambda variable 1-44 | 14.17 | 72.32 | 5.104 | < 1e−07 | < 1e−07 |
| GZMA | Granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) | 10.61 | 51.3 | 4.835 | < 1e−07 | < 1e−07 |
| PTPRC | Protein tyrosine phosphatase, receptor type, C | 36.38 | 174.69 | 4.802 | < 1e−07 | < 1e−07 |
| IGHD | Immunoglobulin heavy constant delta | 19.21 | 91.97 | 4.788 | < 1e−07 | < 1e−04 |
| ADAMDEC1 | ADAM-like, decysin 1 | 27.37 | 128.21 | 4.684 | < 1e−07 | < 1e−07 |
| IGLC1 | Immunoglobulin lambda constant 1 (Mcg marker) | 31.94 | 150.02 | 4.697 | < 1e−07 | < 1e−07 |
| LYZ | Lysozyme | 228.58 | 1099.09 | 4.808 | < 1e−07 | < 1e−07 |
| TRAC | T cell receptor alpha constant | 24.9 | 111.08 | 4.461 | < 1e−07 | < 1e−07 |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 156.2 | 697.99 | 4.469 | < 1e−07 | < 1e−07 |
| GZMB | Granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) | 17.16 | 74.13 | 4.320 | < 1e−07 | < 1e−07 |
| IGLJ3 | Immunoglobulin lambda joining 3 | 14.66 | 62.9 | 4.291 | < 1e−07 | < 1e−07 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 37.75 | 163.3 | 4.326 | < 1e−07 | < 1e−07 |
| CXCL11 | Chemokine (C–X–C motif) ligand 11 | 32.26 | 139 | 4.309 | < 1e−07 | < 1e−07 |
| CCL5 | Chemokine (C–C motif) ligand 5 | 49.09 | 207.14 | 4.220 | < 1e−07 | < 1e−07 |
| CXCL11 | Chemokine (C–X–C motif) ligand 11 | 21.48 | 90.56 | 4.216 | < 1e−07 | < 1e−07 |
| CXCL10 | Chemokine (C–X–C motif) ligand 10 | 393.84 | 1622.47 | 4.120 | 2.74E−05 | < 1e−07 |
| CD52 | CD52 molecule | 50.02 | 203.76 | 4.074 | < 1e−07 | < 1e−07 |
| PTPRC | Protein tyrosine phosphatase, receptor type, C | 74.72 | 304.33 | 4.073 | < 1e−07 | < 1e−07 |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 87.61 | 351.26 | 4.009 | < 1e−07 | < 1e−07 |
| Gene Ontology | ||||
|---|---|---|---|---|
| GO ID | GO term | Observed in selected subset | Expected in selected subset | Observed/expectedb |
| Cellular component | ||||
| GO:0042571 | Immunoglobulin complex, circulating | 7 | 0.15 | 46.41 |
| GO:0019814 | Immunoglobulin complex | 7 | 0.22 | 32.48 |
| GO:0042612 | MHC class I protein complex | 6 | 0.26 | 23.2 |
| GO:0061702 | z complex | 6 | 0.3 | 19.89 |
| GO:0042101 | T cell receptor complex | 6 | 0.39 | 15.47 |
| Molecular function | ||||
| GO:0032395 | MHC class II receptor activity | 7 | 0.17 | 41.73 |
| GO:0019957 | C–C chemokine binding | 5 | 0.17 | 29.8 |
| GO:0046977 | TAP binding | 6 | 0.22 | 26.82 |
| GO:0019865 | Immunoglobulin binding | 6 | 0.24 | 24.76 |
| GO:0004950 | Chemokine receptor activity | 8 | 0.34 | 23.84 |
| GO:0001637 | G-protein coupled chemoattractant receptor activity | 8 | 0.34 | 23.84 |
| GO:0023026 | MHC class II protein complex binding | 6 | 0.26 | 22.99 |
| GO:0019956 | Chemokine binding | 6 | 0.28 | 21.46 |
| GO:0045236 | CXCR chemokine receptor binding | 6 | 0.3 | 20.12 |
| GO:0023023 | MHC protein complex binding | 6 | 0.3 | 20.12 |
| Biological process | ||||
| GO:0002399 | MHC class II protein complex assembly | 5 | 0.15 | 32.44 |
| GO:0046113 | Nucleobase catabolic process | 5 | 0.18 | 27.8 |
| GO:0002396 | MHC protein complex assembly | 5 | 0.18 | 27.8 |
| GO:0010818 | T cell chemotaxis | 9 | 0.39 | 23.35 |
| GO:0002480 | Antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent | 5 | 0.23 | 21.62 |
| GO:0090026 | Positive regulation of monocyte chemotaxis | 7 | 0.36 | 19.46 |
| GO:0010819 | Regulation of T cell chemotaxis | 5 | 0.26 | 19.46 |
| GO:1901623 | Regulation of lymphocyte chemotaxis | 9 | 0.49 | 18.44 |
| GO:0036037 | CD8-positive, alpha–beta T cell activation | 5 | 0.31 | 16.22 |
aFold change < 4.0 are not reported
bObserved/expected < 15.0 not reported
Given the dramatic improvement in survival demonstrated in LCK-high samples, the influence of other established prognostic factors was tested in a multivariable model that included LCK status, age, race (white vs other), stage, grade, and ECOG status. Independent predictors of PFS included LCK status (p = 0.021, HR = 0.508) and race (p = 0.024, HR = 0.657). Additionally, LCK mRNA level was an independent predictor for OS (p = 0.001; HR = 0.315), as was race (p = 0.038; HR = 0.676) and age (p < 0.001; HR = 1.026).
High LCK does not correlate to increased mutation number
Non-synonymous somatic mutations in malignancies can lead to expression of “neo-epitopes” and hence increased potential immunogenicity; thus the relationship between LCK levels and number of somatic mutations in high-grade serous ovarian cancer samples was evaluated. High mutation load, as defined by mutation count > 100, was present in 18 out of 520 tumors with sequencing data available (3.5%). To determine a possible relationship between mutational load and LCK expression, the number of somatic mutations in LCK high samples was compared to that of non-LCK high tumors. This revealed no significant difference in mutation load or copy number alteration based on LCK expression status (Fig. 1d). In fact, in the LCK high samples, there was only one tumor with a mutation count greater than 100 (4.3% of the LCK high group).
LCK is a prognostic predictor in ovarian cancer and a subset of other malignancies where CYT is not prognostic
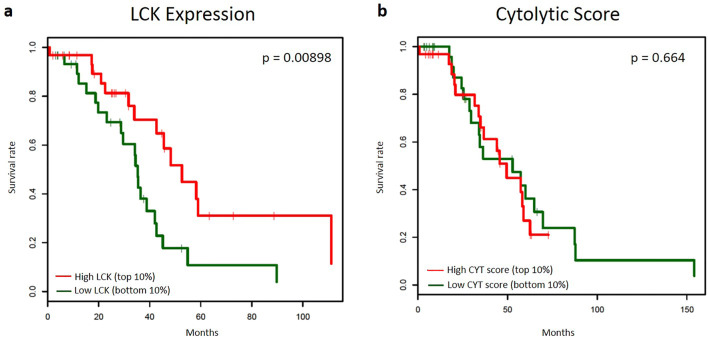
For this analysis, the definition of LCK high samples was liberalized (top 10%) and survival was compared to low LCK (bottom 10%) within the TCGA in order to reduce selection bias due to small numbers of LCK high/low cases. The median OS in the LCK high group was 52.6 months, as compared to 35.3 months in the LCK low group (p = 0.00898). Similar dichotomization of CYT, a measure of transcript levels of perforin (PRF1) and granzyme A (GZMA), was performed; samples were grouped by CYT score into highest and lowest 10%. CYT did not predict survival, with median OS was 49.4 and 52.8 months in high and low cohorts respectively (p = 0.664). Kaplan–Meier curves can be found in Fig. 2.
Fig. 2.
Kaplan–Meier analysis comparing the prognostic ability of LCK and CYT. a Kaplan–Meier analysis of overall survival in high LCK expression (top 10%, red) as compared to low LCK expression (bottom 10%, green). b Kaplan–Meier analysis of overall survival in high CYT score (top 10%, red) as compared to low CYT score (bottom 10%, green)
This analysis was then performed for 30 tumor types available in TCGA (Table 3). Of these 30 cancer types, CYT was a significant predictor of overall survival in five cancers including: breast invasive carcinoma (BRCA, p = 0.00293), cervical carcinoma (CESC, p = 0.0121), low-grade glioma (LGG, p = 0.0112), sarcoma (SARC, p = 0.0323), and cutaneous melanoma (SKCM, p = 0.00509). The LCK high group also had statistically significant improved survival in these subtypes (BRCA p = 0.0546, CESC p = 0.000748, LGG p = 0.0269, SARC p = 0.0166, and SKCM p = 0.0271). Interestingly, high LCK expression also had improved overall survival in an additional three cancer subtypes, namely ovary as described above, head and neck squamous carcinoma (HNSC, p = 0.0496), and uterine carcinosarcoma (UCS, p = 0.0358). Therefore, LCK was predictive of OS, including in a subset of three tumor types where CYT was not.
Table 3.
Survival analyses comparing the prognostic ability of LCK and CYT
| Cancer subtypea | LCK | Cytolytic Activity Score (CYT) | ||||
|---|---|---|---|---|---|---|
| Median OS bottom 10% (months) | Median OS top 10% (months) | p value | Median OS bottom 10% (months) | Median OS top 10% (months) | p value | |
| ACC | NA | NA | 0.818 | NA | NA | 0.99 |
| BLCA | NA | 94.3 | 0.254 | NA | NA | 0.506 |
| BRCA | 90.4 | 132 | 0.0546 | 84.5 | NA | 0.00293 |
| CESC | 19.4 | NA | 0.000748 | 136 | NA | 0.0121 |
| CHOL | 24.7 | NA | 0.87 | 9.03 | NA | 0.642 |
| COAD | NA | NA | 0.363 | NA | NA | 0.863 |
| ESCA | 42.1 | 26.1 | 0.93 | 26.1 | 16.1 | 0.617 |
| GBM | 13.2 | 12.5 | 0.623 | 13.2 | 10.6 | 0.295 |
| HNSC | 85.7 | 161.9 | 0.0496 | 28.7 | 58.7 | 0.109 |
| KIRC | NA | 66 | 0.497 | NA | 73 | 0.473 |
| KIRP | NA | NA | 0.232 | NA | 98 | 0.591 |
| LAML | 12.2 | 10.1 | 0.118 | 26.4 | 10.2 | 0.0838 |
| LGG | 63 | 63.8 | 0.0269 | 81.1 | 52.6 | 0.0112 |
| LIHC | NA | 54.1 | 0.865 | 59.7 | 56.2 | 0.763 |
| LUAD | 48.5 | 87.2 | 0.368 | 49.7 | 43.1 | 0.664 |
| LUSC | 74.1 | 56 | 0.603 | 74.1 | 61.9 | 0.918 |
| MESO | 17.6 | 13.8 | 0.584 | 25.2 | 13.8 | 0.959 |
| OV | 35.3 | 52.6 | 0.00898 | 52.8 | 49.4 | 0.664 |
| PAAD | NA | 23.4 | 0.687 | 21.7 | 50.1 | 0.973 |
| PCPG | NA | NA | 0.429 | NA | NA | 0.317 |
| PRAD | NA | NA | 0.304 | NA | NA | 0.893 |
| READ | NA | NA | 0.317 | NA | NA | 0.221 |
| SARC | 35.4 | NA | 0.0166 | 41.2 | NA | 0.0323 |
| SKCM | 54.3 | 164.3 | 0.0271 | 58.9 | 164.3 | 0.00509 |
| STAD | 58.2 | 22.3 | 0.857 | 73.2 | NA | 0.936 |
| TGCT | NA | NA | 0.317 | NA | NA | 0.289 |
| THCA | NA | NA | 0.631 | NA | NA | 0.659 |
| UCEC | NA | NA | 0.221 | NA | NA | 0.263 |
| UCS | 22.8 | 30.4 | 0.0358 | 31.6 | NA | 0.804 |
| UVM | NA | NA | 0.808 | NA | NA | 0.806 |
Median overall survival in high LCK expression and low LCK expression as compared to high and low CYT score. High and low groups are defined as top 10% and bottom 10% respectively
aThe following tumor types (project code and n = sample size) were included: adrenocortical carcinoma (ACC, n = 92), bladder/urothelial (BLCA, n = 412), breast invasive carcinoma (BRCA, n = 1098), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC, n = 307), cholangiocarcinoma (CHOL, n = 51), colon adenocarcinoma (COAD, n = 461), esophageal carcinoma (ESCA, n = 185), glioblastoma multiforme (GBM, n = 617), head and neck squamous cell carcinoma (HNSC, n = 528), kidney renal clear cell carcinoma (KIRC, n = 537), kidney renal papillary cell carcinoma (KIRP, n = 291), acute myeloid leukemia (LAML, n = 200), low grade glioma (LGG, n = 516), liver hepatocellular carcinoma (LIHC, n = 377), lung adenocarcinoma (LUAD, n = 585), lung squamous cell carcinoma (LUSC, n = 504), mesothelioma (MESO, n = 87), ovarian serous cystadenocarcinoma (OV, n = 608), pancreatic adenocarcinoma (PAAD, n = 185), pheochromocytoma and paraganglioma (PCPG, n = 179), prostate adenocarcinoma (PRAD, n = 500), rectum adenocarcinoma (READ, n = 172), sarcoma (SARC, n = 261), skin cutaneous melanoma (SKCM, n = 470), stomach adenocarcinoma (STAD, n = 443), testicular germ cell tumors (TGCT, n = 150), thyroid carcinoma (THCA, n = 507), uterine corpus endometrial carcinoma (UCEC, n = 560), uterine carcinosarcoma (UCS, n = 57), and uveal melanoma (UVM, n = 80)
LCK protein expression independently confirms impact on prognosis
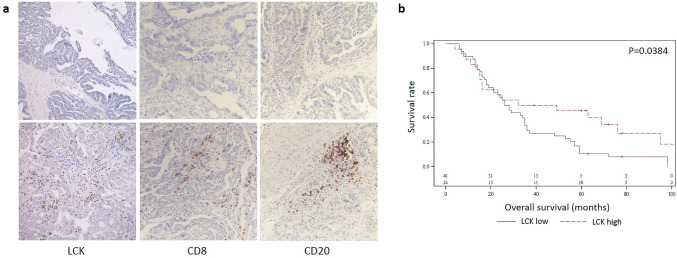
In order to determine if there was concordance between high LCK mRNA and protein expression, we investigated LCK protein levels in samples designated as LCK-high by mRNA expression in the TCGA cohort using reverse-phase protein arrays (RPPA). As expected, the LCK-high mRNA samples also expressed significantly higher levels of LCK protein (Fig. 1c). We also used an independent validation cohort of 72 high-grade serous ovarian cancer samples with available clinical data to compare LCK protein expression using IHC with CD8, and CD20 (markers of cytotoxic T lymphocytes and B-cells, respectively). Of the 72 samples, 24 (33.3%) were characterized as LCK-high by IHC scoring. This analysis confirmed that LCK expression was specific to tissue lymphocytes and that there was no confounding LCK expression by normal epithelial or by tumor cells. Furthermore, survival analysis revealed that only high LCK staining was statistically significantly correlated with overall survival, with median survival for high LCK staining of 40.5 months compared to 27.0 months (p = 0.04, Fig. 3). Neither LCK intensity nor LCK distribution (focal or diffuse) resulted in further stratification of the impact of LCK on survival.
Fig. 3.
LCK expression and survival analysis in an independent cohort. a Representative examples of varying LCK, CD8, CD20 expression by immunohistochemistry. Top row = low expression (from left to right: LCK, CD8, CD20). Bottom row = high expression (from left to right: LCK, CD8, CD20). b Kaplan–Meier analysis of overall survival in high LCK expression (red) as compared to low LCK expression (blue)
Transcriptional profile differs in LCK high samples
Given the prognostic importance of high LCK expression, we used the availability the U133 microarray data as part of the TCGA dataset to evaluate gene expression differences between LCK-high expressing (n = 23) and remaining samples (n = 496). This analysis revealed 291 differentially expressed transcripts (at a statistical cut-off of P < 0.001 and at least twofold change). As expected, LCK-high samples were characterized by higher expression of many transcripts associated with T cell function (Table 2). For example, CD2, CD3, TRBC1, GZMA, GZMB, TRAC, and several HLA class I and II transcripts were all significantly higher expressed in LCK-high samples. The greatest fold change was observed for Chemokine (CXC motif) ligand 9 (CXCL9, also known as chemokine induced by interferon γ (MIG)) with 15.64 higher expression level in the LCK-high samples. Given that LCK is a canonical T lymphocyte signaling molecule, it was surprising to find that many B lymphocyte/plasma cell-related transcripts including many immunoglobulin genes (e.g. IGHD, IGHM, IGKC, IGLJ3, IGLC1, and IGLV1-44) were also enriched in the LCK-high samples (Table 2). Interestingly, CXCL13 (also known as B lymphocyte chemoattractant (BLC)) was one of the chemokines enriched in LCK-high samples (7.7 fold).
We next performed gene ontology enrichment analysis (Table 2). This analysis confirmed that LCK-high samples were significantly enriched in B cell function and activity, as demonstrated by the highest observed-to-expected ratios in the “immunoglobulin complex circulating” gene ontology term (enrichment score: 46.41). In terms of molecular function, MHC II receptor (major histocompatibility complex) activity was most closely correlated with an enrichment score of 41.73, followed by C–C chemokine binding (29.8), and this was mirrored in the biologic process analysis where MHC class II protein complex assembly had the greatest enrichment (32.44, Table 2).
Given the enrichment of B-cell transcripts in LCK high samples, we also investigated the presence of tertiary lymphoid structures (TLS) in the independent cohort of 72 HGSOC samples. TLS ****represent transient colocalization of lymphoid cells in non-lymphoid tissues; the presence of TLS has been described in multiple solid tumor types and is felt to influence local and potentially systemic anti-cancer response. We found that LCK expression by IHC was moderately correlated with TLS (Spearman correlation: 0.53, p = < 0.0001). Proportional hazards regression analysis was performed including both TLS and LCK as predictors of OS, and both were significant independent predictors of survival (HRTLS = 4.1, p = 0.004, HRLCK = 3.8, p = 0.005). Finally, consistent with our mRNA expression analysis, there was moderate correlation between LCK, CD20, CD8 staining (Spearman correlation: LCK/CD8 = 0.465, LCK/CD20 = 0.416, CD8/CD20 = 0.382, all p value < 0.001). However, there was no evidence of any difference in strength of correlation between pairs of these markers (95% CI − 0.18–0.28 for LCK/CD8 vs LCK/CD20 and 0.31–0.14 for CD20/CD8 vs LCK/CD8).
Given the prognostic significance of LCK positive lymphocytes in HGSOC, we next sought to determine if the abundance of such lymphocytes differed between normal fallopian tube epithelium (tissue of origin for the vast majority of HGSOC), benign serous neoplasms, and HGSOC. LCK expression was evaluated by IHC in a TMA consisting of 20 normal Fallopian tube samples, 13 serous cystadenomas, and 14 HGSOC samples. We observed higher LCK expression in the malignant samples than in their benign counterparts (p = 0.023, Supplemental Fig. 1). However, LCK expressing lymphocytes were present (albeit at lower prevalence) among normal fallopian tube epithelium samples, suggesting a possible surveillance or a tissue resident function.
Discussion
The immunogenicity of EOC has been well documented, with extensive literature demonstrating the presence of tumor infiltrating lymphocytes in ovarian tumors and their prognostic significance [3–9]. However, the biological basis and the identification of reliable markers for this prognostic significance have proven elusive. The original publication of the ovarian cancer TCGA analysis identified an “immunoreactive” group as one of the four subtypes of high-grade serous ovarian cancer based on transcriptional profiling. However, there was no prognostic impact on survival associated with this immunoreactive subtype [19]. Recent publication reported a histotype-specific nature of immune infiltration and demonstrated that the magnitude of survival benefit in ovarian cancer was dose dependent on CD8 positive TILs [24, 25]. However, the use of TIL for clinical decision making currently remains in its early stage, and investigation into genomic markers has yielded mixed results.
The need for a robust, reproducible, and immune-related biomarker in HGSOC is further highlighted by the emerging data on immune checkpoint blockers resulting in response rates of 10–15% in heavily pretreated patients [15, 26–29]. Given the low response rates and significant toxicities of such therapies, studies aimed at identifying factors to provide more personalized prognostication for immune response in particular are of utmost importance. The use of PDL1 staining has emerged as a convenient and intuitive marker for prediction of response to immune checkpoint inhibitors, at least in some cancers. However, the predictive accuracy of this marker for ovarian cancer remains unknown. It is worth mentioning that the response rates to PD1/PDL1 targeting monoclonal antibodies is not appreciably higher in clinical trials that used PDL1 positivity by IHC as an eligibility criterion [28].
The current study demonstrates that high LCK expression identifies a small subset of high-grade serous ovarian cancers with better PFS and OS following treatment with standard frontline platinum-taxane adjuvant chemotherapy. Lymphocyte-specific kinase (LCK) is an attractive biomarker as it plays a central functional role in T-cell signaling. The T-cell receptor (TCR) is composed of an antigen recognition subunit (TCRαβ) as well as three signaling subunits (CD3) [30]. TCR-CD3 engagement with antigen induces phosphorylation by LCK, which then triggers downstream signaling cascades that lead to antigen specific T-cell immune response. Additionally, mice lacking LCK develop profound T cell deficiency [31]. Therefore, LCK is central to effective and specific T-cell response, including to tumor antigen. However, LCK is demonstrated herein to have greater discriminatory prognostic ability than previously validated metrics of T cell function alone such as CYT, which suggests it may capture additional facets of tumoral immune response such as B cell activity.
The impact of B cell infiltrates in ovarian malignancy is less clear than their T-cell counterparts, though they have been shown to similarly be associated with improved survival [13, 14, 32]. The role of B cells has been supported by prior analysis of the TCGA, which demonstrated improved survival with B-cell gene expression signatures in high-grade serous ovarian cancer [33]. The causality and mechanism of the herein reported correlation between LCK and B cell signatures remains to be determined. Prior literature suggests that B cells may induce the maturation of dendritic cells making them competent for T-cell activation, or preclinical studies demonstrate that depletion of B cells in a mouse model results in decreased expression of the degranulation marker CD107 on CD8+ T cells, suggesting impaired cytotoxic response [34, 35]. Interestingly, LCK has also been implicated in B-cell signaling at least in a minor but important B-cell subset, namely B-1 cells. These cells are found predominantly in peritoneal and pleural cavities, which are notably the primary location of ovarian cancer spread, and are characterized by deficient B-cell receptor (BCR) signaling [30, 31]. In future studies we plan to further investigate the potential prognostic significance of B1-cells and their LCK expression in HGSOC.
The limitations of the current research include small sample size, specifically due to the stringent criteria of top 3%; the low number of LCK high tumors within the TCGA limits the power of this analysis, specifically for gene enrichment and ontology. However, for all subsequent analyses, more liberal definitions of LCK-high tumors were used, including top 10% for comparison with CYT and pathologic criteria for IHC in the independent cohort. Therefore, the consistency of the association between LCK and survival lends strength to this conclusion. For the comparison to CYT, the high and low cohorts were defined arbitrarily, as has been done in other analyses; for example, significance of CYT in pancreas defined top decile and compared to bottom quartile resulting in a difference in significance level [18].
In summary, this study demonstrates that high LCK expression is associated with significantly longer survival than non-high LCK tumors and was found to be a more significant predictor of prognosis than the previously validated cytolytic activity score (CYT) across tumor types, including HGSOC. LCK-high samples demonstrated evidence of enriched B cell infiltration and function raising the possibility of that a cooperative interaction between tumor infiltrating T and B cells is correlated with better survival in this disease. Further research is needed to better elucidate the causality and mechanism of this correlation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- BCR
B cell receptor
- CYT
Cytolytic Activity Score
- GZMA
Granzyme A
- HGSOC
High grade serous ovarian cancer
- LCK
Lymphocyte specific tyrosine kinase
- MHC
Major histocompatibility complex
- PRF1
Perforin
- RPPA
Reverse phase protein array
- TCGA
The Cancer Genome Atlas
- TCR
T cell receptor
- TLS
Tertiary lymphoid structures
- TMA
Tissue microarray
- TPM
Transcripts per million
- CYT
Cytolytic Activity Score
- HGSOC
High grade serous ovarian cancer
- LCK
Lymphocyte specific tyrosine kinase
- TCGA
The Cancer Genome Atlas
- TLS
Tertiary lymphoid structures
- TMA
Tissue microarray
Author contributions
EH and AJ were the principle investigators. CP, SK and MHS performed immunohistochemistry and analysis. JR helped in TCGA analysis including comparison to CYT and related statistical analyses, while WP and PH aided in research question formulation and study design. SCM, TLY, QZ, MY contributed samples and support for analysis of independent cohort. EH wrote the manuscript, on which all co-authors commented.
Funding
This research was supported in part by the MD Anderson Cancer Center Support Grant (P30 CA016672), a T32 training grant for gynecologic oncology (CA101642; to K.H. Lu), and the Ovarian Cancer Research Program grants, Department of Defense (W81XWH-17-1-0126 and W81XWH-16-1-0038; to S.C. Mok).
Compliance with ethical standards:
Conflict of interest
The authors declare no potential conflicts of interest.
Ethical approval and ethical standards
Independent validation cohorts were enrolled on tissue and clinical data collection protocol approved by MD Anderson Cancer Center institutional review board (IRB, protocol #: LAB06-0412). All tissue included in the tissue microarray was obtained under an IRB approved protocol at the University of Virginia (protocol #:14461).
Informed consent
Because all information from the Cancer Genome Atlas is de-identified and publically available, informed consent by the study participants and approval of an ethics committee were unnecessary to perform this portion of the analyses in this study. All patients contributing tissue were enrolled under translational protocols as listed above and consent was obtained for the use of their specimens and data for research and for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hinchcliff EM, Paquette C, Roszik J, Kelting S, Stoler MH, Mok SC, Yeung T, Zhang Q, Yates M, Peng W (2019) Lymphocyte-specific protein tyrosine kinase expression predicts survival in ovarian high-grade serous carcinoma. In: Society for Gynecologic Oncology, Annual Meeting, Hawaii, March 2019
- 2.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A, Siegel RL. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi K, et al. Clonal expansion of T cells that are specific for autologous ovarian tumor among tumor-infiltrating T cells in humans1. Gynecol Oncol. 1999;74(1):86–92. doi: 10.1006/gyno.1999.5430. [DOI] [PubMed] [Google Scholar]
- 4.Ioannides CG, Freedman RS, Platsoucas CD, Rashed S, Kim YP. Cytotoxic T cell clones isolated from ovarian tumor-infiltrating lymphocytes recognize multiple antigenic epitopes on autologous tumor cells. J Immunol. 1991;146(5):1700–1707. [PubMed] [Google Scholar]
- 5.Peoples GE, Schoof DD, Andrews JV, Goedegebuure PS, Eberlein TJ. T-cell recognition of ovarian cancer. Surgery. 1993;114(2):227–234. [PubMed] [Google Scholar]
- 6.Preston CC, et al. The ratios of CD8+ T cells to CD4+ CD25+ FOXP3+ and FOXP3- T cells correlate with poor clinical outcome in human serous ovarian cancer. PLoS One. 2013;8(11):1–10. doi: 10.1371/journal.pone.0080063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato E, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci. 2005;102(51):18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webb JR, Milne K, Watson P, DeLeeuw RJ, Nelson BH. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker cd103 are associated with increased survival in high-grade serous ovarian cancer. Clin Cancer Res. 2014;20(2):434–444. doi: 10.1158/1078-0432.CCR-13-1877. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348(3):203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 10.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 11.Hwang WT, et al. Prognostic significance of tumor-infiltrating T-cells in ovarian cancer: a meta-analysis. Gynecol Oncol. 2012;124(2):192–198. doi: 10.1016/j.ygyno.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yildirim N, et al. Do tumor-infiltrating lymphocytes really indicate favorable prognosis in epithelial ovarian cancer? Eur J Obstet Gynecol Reprod Biol. 2017;215:55–61. doi: 10.1016/j.ejogrb.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Milne K, et al. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4(7):e6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bindea G, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25(8):2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balli D, Rech AJ, Stanger BZ, Vonderheide RH. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin Cancer Res. 2017;23(12):3129–3138. doi: 10.1158/1078-0432.CCR-16-2128. [DOI] [PubMed] [Google Scholar]
- 18.Roufas C, et al. The expression and prognostic impact of immune cytolytic activity-related markers in human malignancies: a comprehensive meta-analysis. Front Oncol. 2018;8:27. doi: 10.3389/fonc.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6(269):11 [DOI] [PMC free article] [PubMed]
- 22.Salgado R, et al. The evaluation of tumor-infiltrating lymphocytes (TILS) in breast cancer: recommendations by an International TILS Working Group 2014. Ann Oncol. 2015;26(2):259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diedenhofen B, Musch J. Cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One. 2015;10(3):e0121945. doi: 10.1371/journal.pone.0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goode EL, et al. Dose-response association of CD8+ tumor-infiltrating lymphocytes and survival time in high-grade serous ovarian cancer. JAMA Oncol. 2017;3(12):e173290. doi: 10.1001/jamaoncol.2017.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.James FR, et al. Association between tumour infiltrating lymphocytes, histotype and clinical outcome in epithelial ovarian cancer. BMC Cancer. 2017;17(1):657. doi: 10.1186/s12885-017-3585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma P, Allison JP. The future of immune checkpoint therapy. Science (80-) 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 27.Pakish JB, Jazaeri AA. Immunotherapy in gynecologic cancers: are we there yet. Curr Treat Options Oncol. 2017;18(10):59. doi: 10.1007/s11864-017-0504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varga A et al (2015) Antitumor activity and safety of pembrolizumab in patients (pts) with PD-L1 positive advanced ovarian cancer: Interim results from a phase Ib study. J Clin Oncol 33(15_suppl):5510
- 29.Brahmer JR, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat Rev Immunol. 2013;13(4):257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 31.Molina TJ, et al. Profound block in thymocyte development in mice lacking p56lck. Nature. 1992;357(6374):161–164. doi: 10.1038/357161a0. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen JS, et al. CD20+ tumor-infiltrating lymphocytes have an atypical CD27—memory phenotype and together with CD8+ T cells promote favorable prognosis in ovarian cancer. Clin Cancer Res. 2012;18(12):3281–3292. doi: 10.1158/1078-0432.CCR-12-0234. [DOI] [PubMed] [Google Scholar]
- 33.Iglesia MD, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20(14):3818–3829. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maddur MS, et al. Human B cells induce dendritic cell maturation and favour Th2 polarization by inducing OX-40 ligand. Nat Commun. 2014;5:4092. doi: 10.1038/ncomms5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiLillo DJ, Yanaba K, Tedder TF. B cells are required for optimal CD4+ and CD8+ T cell tumor immunity: therapeutic B cell depletion enhances B16 melanoma growth in mice. J Immunol. 2010;184(7):4006–4016. doi: 10.4049/jimmunol.0903009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.