Abstract
Measles virus (MV) isolated in B95a cells, a marmoset B-cell line, retains full pathogenicity for cynomolgus monkeys, while its derivative obtained by adaptation to the growth in Vero cells, a monkey kidney cell line, loses the pathogenic potential (F. Kobune, H. Sakata, and A. Sugiura, J. Virol. 64:700–705, 1990). Here, we show with a pair of strains, a fresh isolate (9301B) in B95a cells and its Vero cell-adapted form (9301V), that the in vivo attenuation parallels the decrease of replication and syncytium-inducing capabilities in the original B95a cells and that these in vitro phenotypes are attributable to impediment of transcription, which is already obvious at the level of primary transcription catalyzed by the virion-associated RNA polymerase. On the other hand, cell fusion assays detected no functional difference between the glycoproteins of the two viruses. Essentially the same transcriptional impediment with reduced syncytium induction following Vero cell adaptation was found with two other pairs of strains that had been similarly prepared. Nucleotide sequence comparison between the 9301B and 9301V viruses revealed that a few (at most five) amino acid changes, which sporadically took place in the polymerase (L and P proteins) and/or accessory V and C proteins, were responsible for the in vitro and in vivo attenuation through adaptation to growth in Vero cells.
Measles is a highly contagious disease characterized by a prodromal illness of fever, coryza, cough, and conjunctivitis followed by the appearance of a generalized maculopapular rash (for a review, see reference 9a). Despite the development of successful live attenuated vaccines, measles remains a major cause of mortality of children in developing countries and a cause of continuing outbreaks in industrialized nations (27).
Measles virus (MV), the etiologic agent, is an enveloped virus containing a nonsegmented, negative-strand (−) RNA as the genome. It is classified in the genus Morbillivirus of the family Paramyxoviridae. The viral genome of about 15 kb is organized starting with a short 3′ leader region, followed by the six genes encoding N (nucleocapsid), P (phospho), M (matrix), F (fusion), H (hemagglutinin), and L (large) proteins, and ending with a short 5′ trailer region. In addition to the P protein, the second gene expresses accessory genes V and C by a process known as cotranscriptional editing to insert a single nontemplated G residue and by using an overlapping frame, respectively. The genome is tightly associated with N subunits, forming the helical ribonucleoprotein complex (RNP). This RNP but not the naked RNA is the template for both transcription and replication. There is only a single promoter for the RNA polymerase (L-P) (10) at the 3′ end. By recognizing the stop (termination/polyadenylation) and restart signals at each gene boundary, the polymerase gives rise to leader RNA and each mRNA. After translation of these mRNAs and accumulation of the translation products, genome replication begins. In this step, the same polymerase copies the same RNP template but now somehow ignores the successive stop signals for the leader RNA and mRNAs and generates a full-length antigenomic positive-strand (+) RNP. The same polymerase enters the promoter at the 3′ end of (+) RNP to copy the genomic (−) RNP, which serves as the template for the next round of transcription and replication (for a review, see reference 16a).
Initiation of the infectious cycle begins with the binding of H glycoprotein on the envelope to the cell surface receptor on susceptible cells. This is followed by F glycoprotein-mediated fusion of the envelope with the cell membrane, delivering the viral content into the cytoplasm. The membrane cofactor protein or CD46 has been identified to be the specific receptor (4, 21, 22). However, some other molecules may be additionally involved in virus attachment to the cell surface or in the subsequent fusion process (5, 25). The final step of the virus life cycle is the assembly of viral components at the cell surface and the release of progeny by budding.
MV is most successfully isolated from peripheral blood leukocytes or respiratory secretions inoculated onto primary human and monkey kidney cells (6). Due to problems with animal use and possible contamination of primary cells with simian viruses, continuous monkey kidney cell lines (e.g., Vero and CV-1) are currently more frequently used for primary MV isolation. However, several blind passages in these cell lines are generally required before virus propagation and development of cytopathic effects (CPE) to detectable levels. More recently, an Epstein-Barr virus (EBV)-transformed marmoset B-lymphocyte line, B95-8 (18), and its derivatives have been found to be extremely susceptible to wild-type MV and to allow rapid isolation of the virus from clinical specimens (13). More importantly, MV isolated into one of the derivative lines, B95a, retained full pathogenicity for monkeys, whereas MV subsequently adapted to growth in Vero cells lost pathogenic potential (13, 14). These results suggest that Vero and related cell lines select a minor, nonpathogenic population in heterologous mixed populations (quasispecies) circulating in the body and that the virus must undergo further mutations and selections to become fully replicative in these cells. In this study, we compared a pair of MV strains, a fresh isolate in B95a cells and its derivative which had been adapted to full growth in Vero cells. The Vero cell-adapted virus was found to still be able to replicate in B95a cells, but its replication was significantly slower than that of the original B95a isolate. These two viruses were then compared for the entire genome sequence, functions of envelope glycoproteins, and gene expression and replication. The results suggested that MV attenuation through adaptation to growth in Vero cells could involve the impediment of transcription in the original B95a cells, which appeared to be caused by a few amino acid changes in the polymerase and/or the accessory V and C proteins.
MATERIALS AND METHODS
Viruses and cells.
A monkey kidney-derived cell line, Vero, and an EBV-transformed marmoset B-lymphocyte, B95a, were grown in Eagle’s minimal essential medium supplemented with 5% heat-inactivated (56°C for 30 min) fetal bovine serum (FBS) and RPMI 1640 supplemented with 10% FBS, respectively. The wild-type MV strain, 9301B, was isolated with B95a cells from throat swabs of children diagnosed with acute measles. The Vero cell-adapted strain, 9301V, was obtained by several blind passages of strain 9301B in Vero cells. Two additional pairs of fresh isolates in B95a cells and Vero cells strains 9303B and 9303V and strains 9403B and 9403V, were also obtained similarly.
Virus growth in cell cultures and experimental infection of monkeys.
The monolayer cultures of Vero cells and subconfluent B95a cells, both in 6-well cluster plates, were infected with viruses at different multiplicities of infection (MOIs). At various times postinfection (p.i.), the cells were scraped into culture medium. After three cycles of freezing and thawing, infectivity titers were determined by measuring the plaque titers in Vero cells or the 50% tissue culture infectious doses (TCID50) in B95a cells. The pair of viruses 9301B and 9301V were inoculated into cynomolgus monkeys subcutaneously and compared for the clinical manifestations and pathological changes as described previously (13).
Sequencing of the 9301B and 9301V genomes.
With the RNA isolation reagent RNAzol B (Biotex Laboratories, Houston, Tex.), the genome RNAs were extracted from 9301B and 9301V virions purified by sucrose gradient centrifugation from culture supernatants of 9301B-infected B95a cells and 9301V-infected Vero cells, respectively. Fifteen cDNA fragments (nucleotide positions 1 to 830, 82 to 1721, 1674 to 2480, 1777 to 3350, 3000 to 4462, 4261 to 4795, 5449 to 7130, 7000 to 8042, 7261 to 9144, 9001 to 10239, 10001 to 11240, 11197 to 12663, 12501 to 13900, 13857 to 14797, and 14755 to 15894) were amplified from these vRNA templates by reverse transcription-PCR with reverse transcriptase (Superscript II; Gibco-BRL, Gaithersburg, Md.), Taq DNA polymerase (Ex Taq; TaKaRa, Seta, Japan), and the 15 specific primer pairs. The cDNAs spanning the noncoding region between the M and F open reading frames (ORFs) were difficult to obtain, but this difficulty was overcome by using the Superscript One-Step reverse transcription-PCR system (Gibco-BRL). Paramyxovirus virions contain, in addition to the genomic (−) RNA, the antigenomic (+) RNA in different amounts (15). Therefore, the rapid amplification of cDNA ends (5′-RACE) system (Gibco-BRL) was used to sequence the 3′- and 5′-terminal regions including the leader and trailer sequences, respectively, with the genomic and antigenomic strands in the virion preparations as the templates. The detailed procedures for the above sequencing of the entire genomes and the data obtained will be published elsewhere.
Plasmid construction and fusion assay.
The F genes of 9301B and 9301V were identical in sequence, while their H genes differed by three nucleotides that were all nonsynonymous (see below). The cDNAs encoding the common (actually derived from 9301V) F protein, 9301B H protein, and 9301V H protein were inserted at the SmaI site just downstream of the T7 promoter in pGEM-3Z plasmids (Promega, Madison, Wis.). These recombinant plasmids were named pGEM-F, pGEM-H/9301B, and pGEM-H/9301V. Subconfluent B95a cells in 24-well cluster plate were infected with vaccinia virus expressing T7 polymerase, vTF7-3, a gift of B. Moss (9), at an input MOI of 2.0 PFU per cell. Then, plasmids pGEM-F(10 μg) and pGEM-H/9301B or pGEM-H/9301V (10 μg) were transfected individually or together into the VV-infected cells, with the aid of the lipofection reagent, DOSPER (Boehringer GmbH, Mannheim, Germany), as specified by the manufacturer. These B95a cells were incubated in RPMI 1640 with 10% FBS. At 24 h p.i., the cells were stained with Giemsa’s solution and then the number of nuclei within syncytia as a proportion of a total of 1,000 nuclei in several microscopic fields was counted. The fusion index (100 × the total number of nuclei within syncytia per 1,000 nuclei) was calculated by the formula of Ohgimoto et al. (23).
Detection of primary transcripts.
Subconfluent B95a cells in a 6-cm-diameter dish were infected with 9301B or 9301V at an input MOI of 0.02 TCID50 per cell in the presence of 100 μg of cycloheximide per ml so that the RNA transcription catalyzed by the virion-associated polymerase, but not genome replication or gene amplification, was allowed to occur. Total RNAs extracted from infected cells at 4, 8, and 11 h p.i. with the RNA isolation reagent RNAzol B were reverse transcribed into cDNAs with oligo(dT) primers and reverse transcriptase (Superscript II) at 42°C for 60 min. The cDNAs were amplified by PCR for 35 cycles of 1 min at 95°C, 2 min at 55°C, and 3 min at 72°C with Taq DNA polymerase. The primers used were 5′-1001CCTTGATGAACCTTTACCAG1020-3′ (sense for N gene), 5′-1680CTAGAAGATCACTGTCATTG1661-3′ (antisense for N gene), 5′-1778TCAACCATCCACTCCCACGA1797-3′ (sense for P gene), 5′-2480TCGGCAGTGCTGGCCCTACT2461-3′ (antisense for P gene), 5′-4001CAACCTGCTGGTGACCCTTA4020-3′ (sense for M gene), 5′-4462TTGCTGGGCACTACGGTCTA4443-3′ (antisense for M gene), 5′-6001ATCAATAATGAGCTGATACC6021-3′ (sense for F gene), 5′-7130GTTTCAAGAGTTGTAGAGGA7111-3′ (antisense for F gene), 5′-8501GGGGTCTTGTCTGTTGACCT8520-3′ (sense for H gene), 5′-9144AGATTGGTTCACTAGCAGCC9125-3′ (antisense for H gene), 5′-15001GGGATTTTGTTCAGGGATTT15020-3′ (sense for L gene), and 5′-15785TTAAT CCTTAATCAGGGCAC15766-3′ (antisense for L gene). The cDNA for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified as an internal control. The primer pair used for GAPDH was 5′-CAGCCGCATCTTCTTGTGC-3′ (sense) and 5′-CTCCTGGAAGATGGTGATGGG-3′ (antisense).
Detection of genome RNAs and mRNAs.
Total RNAs extracted from infected B95a cells at 16, 23 and 35 h p.i. were reverse transcribed into cDNAs with the specific sense primer for MV genome RNA, 5′-1ACCAAACAAAGTTGGGTAAG20-3′. The reverse transcriptase in the reaction mixture was heat denatured and then, using the same sense primer and an N gene-specific antisense primer, 5′-470GGTACCTCTTGATGCGAAGG451-3′, the synthesized first-strand cDNAs were amplified by PCR under the conditions described above. After every five cycles, amplified cDNAs were taken and analyzed by electrophoresis in 1.5% agarose–Tris-borate-EDTA (TBE) gels. At 16 and 24 h p.i., mRNAs were also extracted with the QuickProp Micro mRNA purification kit (Pharmacia, Uppsala, Sweden), electrophoresed in 0.9% agarose–formamide/morpholinepropanesulfonic acid (MOPS) gels, and capillary transferred onto Hybond-N filters (Amersham Pharmacia Biotech, Uppsala, Sweden) for Northern hybridization. They were hybridized with F and H gene-specific (NheI-PvuII fragment of pGEM-H/9301V) probes that had been labeled with [α-32P]dCTP with the Multiprime DNA-labeling system (Amersham Pharmacia Biotech) and with an [α-32P]GTP-labeled F gene-specific riboprobe made with SP6 RNA polymerase (Epicentre Technologies, Madison, Wis.) by using pGEM-F linearlized with EcoRV. The same filter was also hybridized with the [α-32P]GTP-labeled cellular GAPDH-specific riboprobe, which served as an internal control.
Western blotting.
Infected B95a cells were lysed and proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5% polyacrylamide), electrotransferred onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.), and immunoblotted with anti-Fcyt rabbit anti-peptide serum, anti-Hcyt rabbit anti-peptide serum (2) (gifts of R. Cattaneo), and anti-MV human polyclonal serum obtained from a healthy volunteer infected with MV vaccine strain Schwarz FF-8 (26), which was able to detect N protein clearly.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with accession no. AB012948 and AB012949.
RESULTS
Attenuation of MV through adaptation to growth in Vero cells.
The 9301B virus is a fresh isolate in B95a cells, which has been expanded to a sufficient titer for infection experiments only by several passages in the same cells. This virus was incapable of productively growing in Vero cells, but after 10 passages it became fully adapted to growth in Vero cells; this virus is named 9301V (Table 1).
TABLE 1.
Comparisons of 9301B and 9301V for their growth in Vero cells and pathogenicity for cynomolgus monkeysa
| Characteristic | 9301Bb | 9301Vb |
|---|---|---|
| Growth in Vero cells (TCID50/ml) | <10 | 1.7 × 106 |
| Symptom and histopathology in monkeys | ||
| Rash | ++ | − |
| Leukopenia | ++ | − |
| Body wt loss | ++ | − |
| Syncytium formation in lymphoid organsc | ++ | + |
Infections of Vero cells with 9301B and 9301V viruses were initiated at an input MOI of 0.2 TCID50/cell, and the titers on day 3 were assayed by measurement of TCID50 in B95a cells. Infections of monkeys were initiated with 105.5 TCID50 of the respective viruses, and symptoms and pathological changes were observed as described previously (13).
The degree of phenotype expression is shown by ++ (extensive), + (moderate), and − (not apparent).
Lymphoid organs include the spleen, thymus, and lymph nodes.
Table 1 summarizes the pathogenicity of the two viruses for cynomolgus monkeys. The wild-type 9301B virus produced maculopapular rashes typical of measles and caused lymphopenia and weight loss in a monkey after subcutaneous inoculation. Histopathologically, development of giant cells and necrosis in the entire thymus and other lymphoid tissues such as the spleen and lymph nodes were also evident. None of these extensive macroscopic and microscopic pathological changes was apparent with the 9301V virus, except for a moderate degree of syncytium formation in lymphoid tissues, which was limited in both number and size. These results confirm the previous view that measles virus becomes attenuated following adaptation to growth in Vero cells (13).
Growth comparison of 9301B and 9301V viruses in B95a cells.
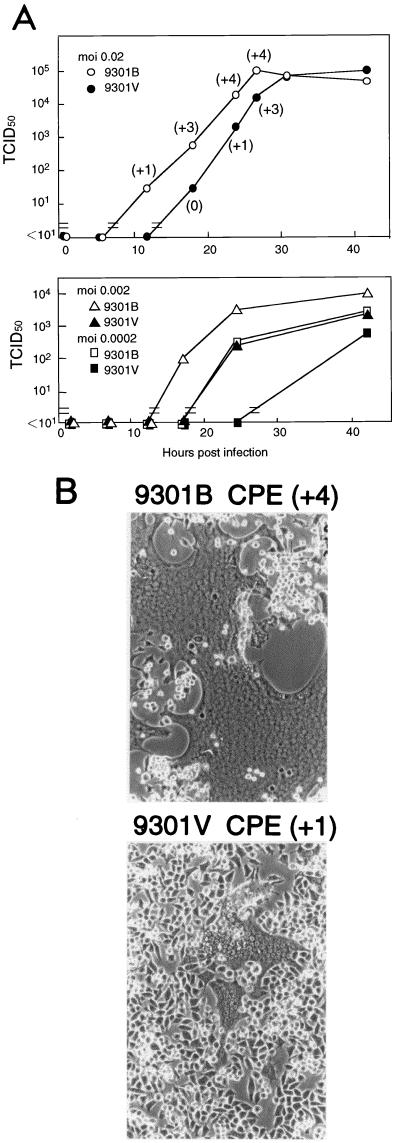
To learn how measles virus would replicate in the original B95a cells after adaptation to Vero cells, we compared the growth kinetics of 9301B and 9301V viruses in B95a cells. As shown in Fig. 1, the 9301V virus replicated significantly less fast than the 9301B virus following infections at three different MOIs, although the final titers were the same or nearly comparable between the two viruses. The 9301B virus caused severe CPE, indicated by extensive cell fusion (giant-cell formation), whereas no such strong CPE was found for the 9301V virus (Fig. 1). It was most probably due to severe CPE in the entire culture that 9301B virus growth appeared to be self-limiting, with the virus reaching final titers comparable to those of 9301V virus.
FIG. 1.
(A) Replication kinetics of 9301B and 9301V viruses in B95a cells. Infections were initiated with input MOIs of 0.02, 0.002, and 0.0002 TCID50 per cell. The degree of CPE is shown in parentheses as scores of +4 (cell fusion in the entire culture) to +1 (a few fusion foci in a microscopic field) and 0 (no apparent fusion). (B) Micrographs showing CPE manifested in 9301B- or 9301V-infected B95a cells 24 h after infection at a MOI of 0.02 per cell.
These results suggest that poor replication capability and low syncytium-inducing capacity in B95a cells are the in vitro correlates of reduced in vivo pathogenicity caused by adaptation of MV to Vero cells.
Nucleotide and amino acid sequence comparison between the 9301B and 9301V viruses.
Identification of nucleotide changes present throughout the entire genomes of 9301B and 9301V viruses would be a necessary first step toward revealing the basis underlying MV attenuation through adaptation to growth in Vero cells. Probably because of the tendency to form higher-order RNA structures, it was initially not easy to obtain cDNAs spanning the region between the M and F ORFs, but this difficulty was eventually overcome (see Materials and Methods), leading to sequencing of the entire genomes. The results revealed that the two genomes were identical in length (15,894 nucleotides) but differed by 8 nucleotides (Table 2). All these nucleotide changes were found in ORFs for the P, V, C, H, and L proteins. The other regions including the leader, trailer, transcription start and stop signals, intergenic sequences, and noncoding regions of all six genes were identical between the two. The pyrimidine-rich BB boxes, which were previously suggested to guide and facilitate RNA polymerase attachment (1), were also identical. Thus, we concluded that all the cis-acting elements for controlling viral transcription, translation, and replication were identical between the two viruses. All or some of the eight nonsilent changes would therefore be responsible for the above-described remarkable phenotypic changes caused by virus adaptation to growth in Vero cells.
TABLE 2.
Nucleotide and amino acid differences between the 9301B and 9301V viruses
| Gene | Nucleotide
|
Amino acid
|
||||
|---|---|---|---|---|---|---|
| Position | 9301B | 9301V | Position | 9301B | 9301V | |
| P/V/C | 1969 | G | A | 55 (P) | Ala | Thr |
| 55 (V) | Ala | Thr | ||||
| 2160 | A | T | 118 (P) | Glu | Asp | |
| 118 (V) | Glu | Asp | ||||
| 111 (C) | Lys | Met | ||||
| H | 7311 | A | G | 14 | Asp | Gly |
| 8538 | T | C | 423 | Leu | Pro | |
| 8906 | A | G | 546 | Ser | Gly | |
| L | 12976 | C | G | 1248 | Ala | Gly |
| 12977 | T | C | 1248 | Ala | Gly | |
| 14632 | A | G | 1800 | Glu | Gly | |
Functional comparisons of the 9301B and 9301V envelope glycoproteins.
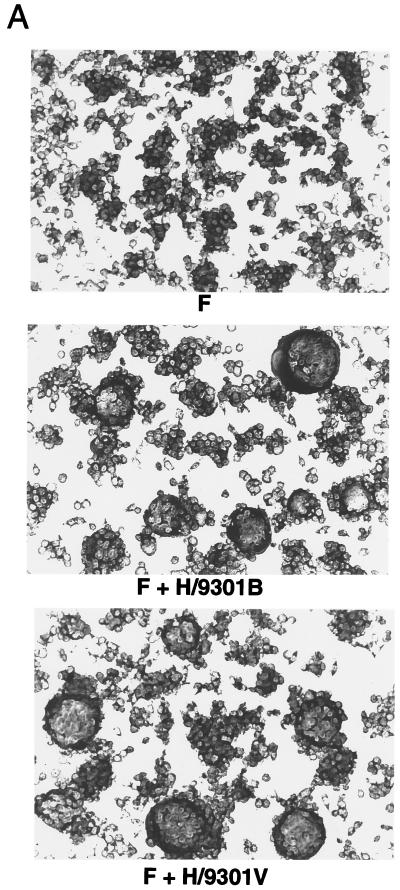
The H glycoprotein of MV is involved in receptor recognition, whereas the F protein is involved in the envelope-cell membrane fusion. Although the F protein is the direct mediator of cell fusion, coexpression of the receptor binding H protein is required for cell fusion (28). The remarkable differences in cell fusion capacity between the 9301B and 9301V viruses (Fig. 1) raised the possibility of structural and functional differences in one or both of their glycoproteins. Nucleotide sequencing revealed no difference in either coding or noncoding region of the F gene but identified three nucleotide changes, all nonsynonymous, in the H gene between the two viruses (Table 2). We examined whether the H proteins of 9301B and 9301V viruses similarly have a supporting function in cell fusion. Either of the H proteins was expressed in B95a cells along with the common F protein (actually derived from 9301V) from the respective transfected plasmids by vaccinia virus-encoded T7 polymerase.
As shown in Fig. 2, the H proteins derived from the 9301B and 9301V viruses were equally able to support cell fusion in the presence of F protein. Besides, although the 9301V virus replicated slowly in B95a cells (Fig. 1), the virus titer assayed in these cells (1.7 × 106 TCID50/ml) was comparable to that in Vero cells (2.1 × 106 TCID50/ml), to which the virus had been highly adapted. This comparable plating efficiency also suggested that the capacity of 9301V virus to bind to and penetrate into B95a cells was not impaired. So far, therefore, no functional impairment of glycoproteins has been found for the 9301V virus. Thus, the amino acid changes in the 9301V virus H protein appeared to be just incidental and irrelevant to measles virus adaptation to Vero cells. This notion was supported by the fact that there was no amino acid change in the H gene between another pair of strains (9403B and 9403V), which were similarly prepared.
FIG. 2.
Cell fusion-supporting activities of the H proteins derived from 9301B and 9301V. Subconfluent 24-well cluster plates of B95a cells were infected with vTF7-3 and transfected with pGEM-F alone or together with pGEM-H/9301B or pGEM-H/9301V. At 24 h p.i., the cells were stained with Giemsa (A) and fusion indices were determined as described in Materials and Methods (B).
Gene expression and replication.
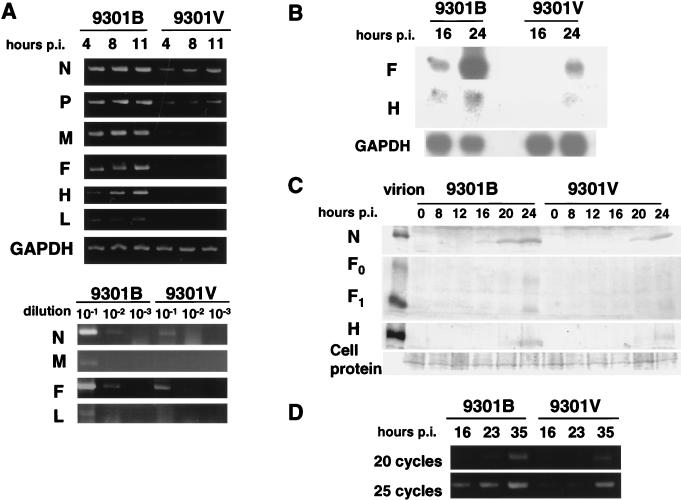
Paramyxovirus RNA polymerase consists of the catalytic L and cofactor P proteins (10). In addition, studies with Sendai virus demonstrated a significant contribution of the accessory V and C proteins to in vivo pathogenicity as well as in vitro replication (11, 12, 16). In addition, host cell factors appear to play roles (3, 17, 19, 20). The amino acid changes in these polymerase and accessory proteins between 9301B and 9301V prompted us to explore whether these two viruses would display differences at the steps of gene expression and replication in B95a cells. Infected cells were incubated in the presence of cycloheximide to block de novo protein synthesis and allow only the primary transcription catalyzed by the polymerase associated with the infecting viruses. At various time points, RNAs were extracted from the cells, the N, P, M, F, H, and L mRNAs were reverse transcribed, and the cDNAs were amplified by PCR (Fig. 3A, top). Compared with those of 9301B, the signals were much weaker or marginal for 9301V. The specificity of each band was confirmed by Southern blotting with specific DNA probes (data not shown). To be more quantitative, the RNAs at 4 h p.i. were diluted to 10−1 to 10−3 and the reverse-transcribed and PCR-amplified products were compared. Again, the specific products of 9301V infection were found to be significantly less abundant than those from 9301B infection when primers specific for N, M, F, and L (Fig. 3A, bottom), as well as for P and H (data not shown), were used. Amplified gene expression in the absence of cycloheximide in B95a cells was also compared between the two viruses by Northern hybridization. As represented by the results with F- and H-specific probes, the gene expression levels were considerably lower for 9301V than for 9301B (Fig. 3B). Western blotting further demonstrated that the levels of the gene products, i.e., the N, F, and H proteins, were all remarkably lower for 9301V than for 9301B (Fig. 3C). Finally, we compared the levels of intracellular genomic RNA between the two viruses. Paramyxovirus genomic RNA migrates in gels to the 50S position, but nonspecific aggregates of mRNAs also migrate to the same position. Therefore, we routinely use semiquantitative PCR, which amplifies a region from the 3′ terminus to an appropriate position within the N gene (11, 12, 16). Here, we amplified such a region of 470 nucleotides by using 20 and 25 cycles of PCR. It was clearly shown that the genome replication of 9301V was significantly impaired in B95a cells compared with that of 9301B (Fig. 3D).
FIG. 3.
Primary transcription (A), amplified gene expression (B), accumulation of gene products (C), and genome replication (D) during 9301B and 9301V virus infection in B95a cells. GAPDH expression was used as an internal control in the experiments in panels A and B. (A and D) PCR products; (B) Northern blot; (C) Western blot. RNAs extracted from infected cells at various times p.i. were directly reverse transcribed and PCR amplified with specific primers (A, top, and D), or the RNAs at 4 h p.i. were diluted to 10−1, 10−2, and 10−3 before being subjected to reverse transcription-PCR (A, bottom).
Studies with two other pairs of wild-type and Vero cell-adapted strains.
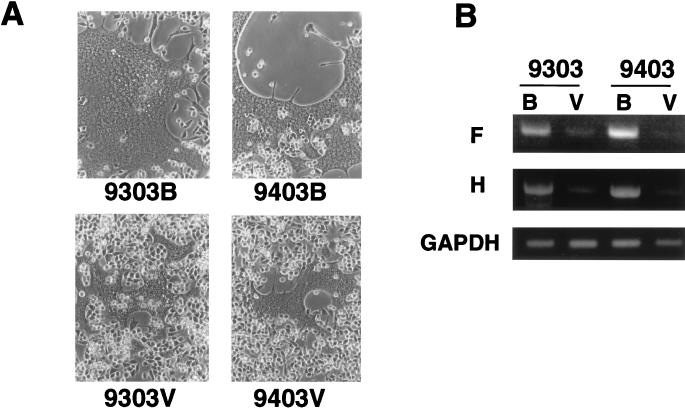
Essentially the same differences in fusion-inducing capacity (Fig. 4A) and replication capacity (data not shown) as were found between the 9301 pair were also observed between each new pair of 9303B-9303V and 9403B-9403V. Each pair of strains was then compared for primary transcription. It was again found that the Vero cell-adapted viruses were attenuated at the level of primary transcription (Fig. 4B). Thus, three independent trials of adaptation of fresh isolates to growth in Vero cells gave rise to very similar patterns of transcriptional attenuation in the original host, B95a cells.
FIG. 4.
Cell fusion 24 h after infection of B95a cells with 9303B, 9303V, 9403B, and 9403V (A) and RT-PCR detection of the primary transcripts of these viruses at 8 h p.i. in these cells (B).
DISCUSSION
Before the EBV-transformed marmoset B-lymphoblastoid cell line B95-8 and its derivatives such as B95a became available, monkey kidney cell lines (e.g., Vero and CV-1) were most frequently used for MV isolation and propagation (7, 8). However, spreading of the virus to a high titer in those monkey kidney lines has required at least several blind passages, indicating a highly selective pressure for the wild-type MV to become fully replicative in these cells. The viruses thus isolated can no longer produce clinical symptoms and pathological changes characteristic of measles in experimental animals (cynomolgus monkeys), as shown in the present study and previously (13). In contrast, the use of B95a and other related cell lines has allowed rapid isolation and propagation of fresh MV, suggesting that the selective pressure, if present, is marginal. Thus, the isolates closely represent the wild-type, pathogenic MV as a population and therefore have been readily able to reproduce the disease course of measles in monkeys, as shown here and previously (13, 14). MV adaptation to growth in Vero cells is accompanied by a decrease of its capacity to replicate and induce syncytia in B95a cells. Thus, these decreases are probably good in vitro markers for MV attenuation of in vivo pathogenicity. The purpose of our present study has been to decide the step(s) impeded in the life cycle of Vero cell-adapted, attenuated MV in the original B95a cells. For this, a fresh wild-type MV isolated in B95a cells (9301B) and its Vero cell-adapted strain (9301V) were systematically and comparatively analyzed for their genome sequences, glycoprotein structure and function, and gene expression and replication.
The nucleotide sequence comparison of the entire genomes revealed no change in the cis-acting regulatory regions for viral Zranscription and replication, including the leader, trailer, stop-restart, and intergenic sequences. Again, no change was found in noncoding regions of all six MV genes, which might be involved in translational regulation. The identified nucleotide changes were all within the ORFs and caused amino acid changes. These sequence data were highly informative in that the remarkable phenotypic differences both in vitro and in vivo were attributable to the changes of viral trans-acting but not cis-acting elements. While F protein amino acid sequences were identical between 9301B and 9301V, three amino acid differences were found in the H protein. The significance of these amino acid changes was studied by the fusion assay with either of the H proteins in combination with the common F protein. The results indicated that the amino acid changes led to no appreciable functional change but were neutral and hence provided no explanation for the reduced fusion activity during the course of 9301V infection in B95a cells. The functional integrity of 9301V H protein was further supported by the finding that this virus had the same plating efficiency (TCID50) on B95a and Vero cells. 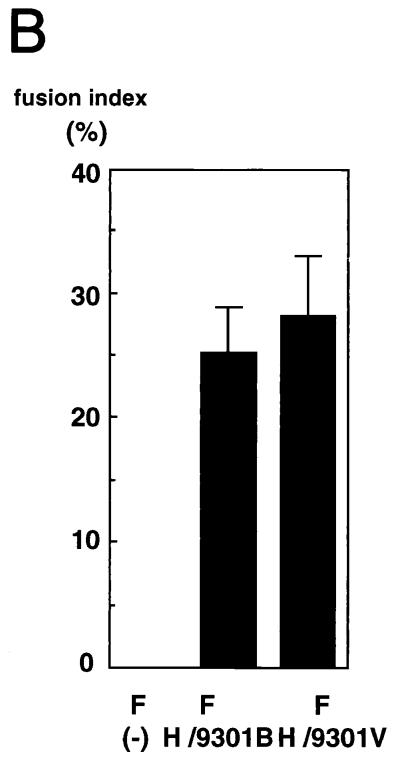
On the other hand, the gene transcription of 9301V virus in B95a cells was significantly impaired compared with that of the 9301B virus. This resulted in reduced levels of the gene products, including the F and H proteins, which in turn could clearly explain the reduced level of 9301V induced fusion in B95a cells. The impairment of transcription would be due to the amino acid changes in the L and P proteins constituting the polymerase complex (10). In view of the importance of the accessory V and C genes for Sendai virus transcription, replication, and pathogenicity (11, 12, 16), the amino acid changes in the V and C frames could also be relevant to the attenuated transcription. The facts that the primary transcription catalyzed by the virion-associated polymerase was already attenuated and that the V and C proteins were not present in the virions as major components (24) suggest that changes of P and L proteins would be more central to the attenuated phenotype of 9301V than those of V and C proteins. However the possibility of accessory-protein involvement, particularly of the C protein, remains open, because C protein knockout Sendai virus appeared to be less infectious (relative to the particle number, roughly represented by the hemagglutination units [16]). The genome replication of 9301V in B95a cells also appeared to be impaired. However, since any effect on transcription probably would decrease replication, it is not known whether the reduction of replication is a direct or an indirect result. Paramyxovirus transcription appears to be optimized by cellular cofactors (3, 17, 19, 20). The impaired transcription of the Vero-adapted virus in B95a cells could be due to the lack of such cofactors in cells or to reduced compatibility of the factors with the 9301V polymerase. Anyway, MV adaptation to growth in Vero cells seems to result in transcriptional attenuation in the original B95a cells, which could be caused by at most five amino acid changes sporadically found in the polymerase and accessory proteins. This attenuation so far appears to explain the retarded replication and reduced fusion activity in B95a cells, which are characteristic of Vero cell-adapted MV and are probably the primary cause of its reduced pathogenicity for the monkeys.
ACKNOWLEDGMENTS
We thank R. Cattaneo for providing anti-H and -F antibodies, M. Billeter for technical advice on the sequencing of the MV genome, and Y. Yogo, S. Ohgimoto, K. Takeuchi, K. Tanabayashi, K. Komase, M. Matsuda, and C. Moriya for helpful discussions. We are also grateful to the Human Genome Center (HGC) of our institution for providing the Genome Net services and databases.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture and the Ministry of Health and Welfare, Japan.
REFERENCES
- 1.Blumberg B M, Chan J, Udem S A. Function of paramyxiovirus 3′ and 5′ end sequences in theory and practice. In: Kingsbury D, editor. The paramyxioviruses. New York, N.Y: Plenum Press; 1991. pp. 235–247. [Google Scholar]
- 2.Cathomen T, Naim H Y, Cattaneo R. Measles virus with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72:1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das T, Mathur M, Gupta A K, Janssen G M C, Banerjee A K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 α β γ for its activity. Proc Natl Acad Sci USA. 1998;95:1449–1454. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorig R E, Marcil A, Chopra A, Richardson C D. The human CD46 molecule is a receptor for measles virus (Edmonston strain) Cell. 1993;75:295–305. doi: 10.1016/0092-8674(93)80071-l. [DOI] [PubMed] [Google Scholar]
- 5.Dunster L M, Schneider-Schaulies J, Loffler J, Lankes W, Schwartz-Albeiz R, Lottspeich F, ter Meulen V. Moesin: a cell membrane protein linked with susceptibility to measles virus infection. Virology. 1994;198:265–274. doi: 10.1006/viro.1994.1029. [DOI] [PubMed] [Google Scholar]
- 6.Enders J F, Peebles T C. Propagation in tissue culture of cytopathogenic agents from patients with measles. Proc Soc Exp Biol Med. 1954;86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 7.Enders J F, Peebles T C, McCarthy K, Milovanovic M, Mitus A, Holloway A. Measles virus: a summary of experiments concerned with isolation, properties, and behavior. Am J Public Health. 1957;47:275–282. doi: 10.2105/ajph.47.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enders J F. Measles virus: historical review, isolation and behavior in various systems. Am J Dis Child. 1962;103:282–287. [PubMed] [Google Scholar]
- 9.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Griffin D E, Bellini W J. Measles virus. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1267–1312. [Google Scholar]
- 10.Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 11.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobune F, Sakata H, Sugiura A. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J Virol. 1990;64:700–705. doi: 10.1128/jvi.64.2.700-705.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobune F, Takahashi H, Terao K, Ohkawa T, Ami Y, Suzaki Y, Nagata N, Sakata H, Yamanouchi K, Kai C. Nonhuman primate models of measles. Lab Anim Sci. 1996;46:315–320. [PubMed] [Google Scholar]
- 15.Kolakofsky D, Bruschi A. Antigenome in Sendai virions and Sendai virus-infected cells. Virology. 1975;66:185–191. doi: 10.1016/0042-6822(75)90189-0. [DOI] [PubMed] [Google Scholar]
- 16.Kurotani, A., K. Kiyotani, A. Kato, K. Mizumoto, T. Shioda, Y. Sakai, T. Yoshida, and Y. Nagai. Sendai virus C proteins are categorically nonessential proteins but silencing their expression severely impairs viral replication in vitro and totally attenuates pathogenicity in vivo. Genes Cells 3:111–124. [DOI] [PubMed]
- 16a.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 1177–1204. [Google Scholar]
- 17.Liston P, DiFlumeri C, Briedis D J. Protein interactions entered into by the measles virus P, V, and C proteins. Virus Res. 1995;38:241–259. doi: 10.1016/0168-1702(95)00067-z. [DOI] [PubMed] [Google Scholar]
- 18.Miller G, Shope T, Lisco H, Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci USA. 1972;69:383–389. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizumoto K, Muroya K, Takagi T, Omata-Yamada T, Shibuta H, Iwasaki K. Protein factors required for in vitro transcription of Sendai virus genome. J Biochem. 1995;117:527–534. doi: 10.1093/oxfordjournals.jbchem.a124740. [DOI] [PubMed] [Google Scholar]
- 20.Moyer S A, Baker S C, Horikami S M. Host cell proteins required for measles virus reproduction. J Gen Virol. 1990;71:775–783. doi: 10.1099/0022-1317-71-4-775. [DOI] [PubMed] [Google Scholar]
- 21.Naniche D, Wild T F, Rabourdin-Combe C, Gerlier D. Measles virus haemagglutinin induces down-regulation of gp57/67, a molecule involved in virus binding. J Gen Virol. 1993;74:1073–1079. doi: 10.1099/0022-1317-74-6-1073. [DOI] [PubMed] [Google Scholar]
- 22.Naniche D, Varior-Krishnan G, Cervoni F, Wild T F, Rossi B, Rabourdin-Comb C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohgimoto S, Tabata N, Suga S, Nishio M, Ohta H, Tsurudome M, Komada H, Kawano M, Watanabe N, Ito Y. Molecular characterization of fusion regulatory protein-1 (FRP-1) that induces multinucleated giant cell formation of monocytes and HIV gp160-mediated cell fusion. J Immunol. 1995;155:3585–3592. [PubMed] [Google Scholar]
- 24.Portner A, Gupta K C, Seyer J M, Beachey E H, Kingsbury D W. Localization and characterization of Sendai virus nonstractural C and C′ proteins by antibodies against synthetic peptides. Virus Res. 1986;6:109–121. doi: 10.1016/0168-1702(86)90043-2. [DOI] [PubMed] [Google Scholar]
- 25.Schneider-Schaulies J, Dunster L M, Schwartz-Albiez R, Krohne G, ter Meulen V. Physical association of moesin and CD46 as a receptor complex for measles virus. J Virol. 1995;69:2248–2256. doi: 10.1128/jvi.69.4.2248-2256.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz A J F. Preliminary tests of a highly attenuated measles vaccine. Am J Dis Child. 1962;103:216–219. doi: 10.1001/archpedi.1962.02080020398042. [DOI] [PubMed] [Google Scholar]
- 27.Weiss R. Measles battle loses potent weapon. Science. 1992;258:546–547. doi: 10.1126/science.1329205. [DOI] [PubMed] [Google Scholar]
- 28.Wild T F, Malvoisin E, Buckland R. Measles virus: both the haemagglutinin and fusion glycoproteins are required for fusion. J Gen Virol. 1991;72:439–442. doi: 10.1099/0022-1317-72-2-439. [DOI] [PubMed] [Google Scholar]