Abstract
Hepatic stellate cells (HSCs) are important stromal cells and pivotal mediators involved in the pathogenesis and immunosuppression of hepatocellular carcinoma (HCC). The liver has been demonstrated to be a site for accumulation of tumor-induced myeloid-derived suppressor cells (MDSCs). We previously reported that HSCs induced an increase in the number of MDSCs in HCC. However, how MDSCs are recruited in HCC remains largely unclear. In the present study, we found that HSC-conditioned medium (HSC-CM) induced bone marrow-derived cell and splenocyte migration, especially MDSC migration. Using chemokine-neutralizing antibodies and chemokine receptor inhibitors, we found that HSCs promoted MDSC migration through the SDF-1/CXCR4 axis. Subsequently, we used an orthotopic mouse liver tumor model to determine how HSCs mediated MDSC migration to HCC in vivo. The in vivo results indicated that pretreatment of MDSCs with a CXCR4 inhibitor or injection with SDF-1-knocked down HSCs inhibited MDSC migration to the spleen and liver of the tumor-bearing mice. Together, our findings indicate a central role for HSCs in MDSC migration mediated by the SDF-1/CXCR4 axis, thus revealing a potentially effective approach for modulating the tumor microenvironment by targeting HSCs in HCC.
Electronic supplementary material
The online version of this article (10.1007/s00262-019-02414-9) contains supplementary material, which is available to authorized users.
Keywords: Hepatic stellate cells, Myeloid-derived suppressor cells, Hepatocellular carcinoma, SDF-1, CXCR4, Migration
Introduction
Immune evasion, one of the hallmarks of cancer, is often achieved via recruitment of immunosuppressive cells, for example myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), to the tumor microenvironment. The liver, the largest solid organ, is enriched with blood and contains a large amount of innate and adaptive immune cells [1]. Published studies have demonstrated increased number of MDSCs in patients with hepatocellular carcinoma (HCC) and tumor-bearing mice; these MDSCs regulate immune suppression networks and then contribute to all aspects of tumor progression [2]. However, how MDSCs are recruited in HCC remains largely unclear.
MDSCs are a population of heterogeneous immature immunosuppressive myeloid cells, including myeloid progenitors and precursors of granulocytes, macrophages, and dendritic cells [3, 4], and have the notable ability to inhibit T cell function [5]. An increasing number of studies in in vivo systems such as tumor-bearing mice have demonstrated that diverse chemotactic factors, such as IL-8, IL -lβ, CCL2, CCL5, CCL12, CXCL12 (SDF-1), and CXCL5, produced by the tumor microenvironment stimulate the recruitment of MDSCs to tumors [6–9]. The liver has been demonstrated to be a site for tumor-induced MDSC accumulation [10, 11]. One study found that more MDSCs homed to the livers of tumor-bearing mice than to those of normal control mice [10].
In the last decade, the contribution of stromal cells to the immune escape has been extensively studied and is fulfilled by the expansion, recruitment, and activation of a variety of immunosuppressive cells in the tumor microenvironment [12]. As critical stromal cells in the liver, hepatic stellate cells (HSCs) are activated during liver injury, infection, or inflammation [13–16] and perform an immunomodulatory activity [17, 18]. Recent evidence suggests that activated HSCs can facilitate the differentiation of inflammatory monocytes into MDSCs through complement component 3 (C3) [19, 20] and in a CD44-dependent fashion [21]. In our previous studies, we reported that activated HSC cotransplatation induced more immunosuppressive Treg cells and MDSCs in the tumor microenvironment in a mouse liver tumor model [22, 23]. We further confirmed that HSCs induced MDSCs from bone marrow cells with upregulated immunosuppressive activity and that HSC-mediated MDSC expansion and HCC progression were impaired by suppression of HSC-derived PGE2 [24]. However, how HSCs influence MDSC mobilization and recruitment to the tumor microenvironment remains unclear.
In this study, we utilized MDSCs freshly isolated from tumor-bearing mice and investigated the relationship between HSCs and MDSC migration in vitro and in vivo. We demonstrated that HSCs regulate the migration of MDSCs through the SDF-1/CXCR4 axis.
Materials and methods
Cell lines and animals
The H22 murine HCC cell line was maintained in RPMI 1640 medium (HyClone, Logan, UT, USA), supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 U/mL streptomycin at 37 °C and 5% CO2. Eight-week-old male BALB/c (H-2d, haplotype) mice were used in this study and maintained under specific pathogen-free conditions in the animal center of Xiamen University.
HSC isolation and culture
HSCs were isolated from the livers of BALB/c mice as previously described [25]. HSC activation was assessed through α-SMA staining [22].
MDSC isolation
MDSCs were isolated from the spleens of tumor-bearing mice as previously described [26].
Knockdown of SDF-1 in HSCs
The shRNA retroviral plasmid containing a puromycin resistance gene was purchased from Clontech. Turbofect transfection reagent (Thermo Scientific, Waltham, MA, USA) was used for cell transfection and transfection efficiency was assessed by quantitative polymerase chain reaction. The primers for SDF-1 were 5ʹ-CTCTGCATCAGTGACGGTAAA-3ʹ (forward) and 5ʹ-CACAGTTTGGAGTGT TGAGGA-3ʹ (reverse).
Tumor inoculation
To study the effect of SDF-1 secreted by HSCs on MDSC mobilization, mice were given an intrahepatic injection of 1 × 106 H22 cells, a mixture of 1 × 106 H22 cells and 2 × 105 activated HSCs, or 2 × 105 SDF-1-knocked HSCs. Each group contained at least 5 mice.
To study the role of CXCR4 in MDSC migration in vivo, mice were given an intrahepatic injection with 1 × 106 H22 cells or 1 × 106 H22 cells plus 2 × 105 activated HSCs. Ten days after injection, MDSCs were pretreated with 10 μM AMD3100 (R&D Systems, Minneapolis, MN, USA), a CXCR4 inhibitor, for 2 h before transfer into the mice.
IVIS imaging of mice
To evaluate the effects of SDF-1 and CXCR4 on MDSC migration, purified MDSCs or CXCR4 inhibitor-pretreated MDSCs were incubated with 5 μM 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiR) (Invitrogen, Carlsbad, CA, USA) for 30 min, followed by intravenous (i.v.) injection of 3 × 106 cells/100 μL per mouse. The mice were scanned using an In Vivo Imaging System (IVIS) spectrum instrument (Lumina III, Caliper Life Sciences, Hopkinton, MA,USA) 24 h after the transfer of MDSCs. Data were analyzed using Living Image Software (Caliper Life Sciences, Hopkinton, MA,USA).
In vitro chemotaxis assay
MDSC migration was assessed using a 5.0-μm pore size transwell chamber (Costar, Cambridge, MA, USA) in triplicate experiments. First, 800 μL RPMI 1640 medium with 5% FBS and HSC-conditioned medium (HSC-CM) with 5% FBS were prepared, and one was added to the lower chamber. Then, 1 × 106/100 μL splenocytes, 5 × 105/100 μL bone marrow cells, or 5 × 105/100 μL purified MDSCs were seeded in the upper chamber. After 6 h, the migrated cells were collected to count the total cell number and then the ratio of cells of interest was analyzed using flow cytometry. The absolute number of indicated cells = the percentage of indicated cells × cell number in the lower chamber.
To explore the mechanism of HSC-promoted MDSC migration, HSC-CM was pre-incubated with 5 μg/mL neutralizing monoclonal antibodies against CCL2 or SDF-1 (R&D Systems), or MDSCs were pretreated with 10 μM CXCR4 inhibitor AMD3100 or CCR2 inhibitor RS102895 (R&D Systems) or 5 μg/mL CXCR2 antibody (R&D Systems) for 2 h before the migration assay.
HSC-CM chemokine array
To detect the chemokines secreted by HSCs, 10 mL HSC-CM was freeze dried and then re-dissolved in 1 mL ddH2O. The chemokines were detected using the Mouse Chemokine Array Kit (R&D Systems). The Mouse Chemokine Array coordinates are shown in Supplementary Table 1.
Flow cytometry analysis
The following monoclonal antibodies were used: APC-conjugated anti-CD11b, PE-conjugated anti-CD11c, anti-Ly6C, anti-Gr-1, anti-CXCR4, and FITC-conjugated anti-Ly6G antibodies (BD PharMingen, San Diego, CA, USA) and PE-conjugated anti-B220, FITC-conjugated anti-CD49b, and APC-conjugated anti-CD3 antibodies (eBioscience, San Diego, CA, USA). The appropriate isotype antibodies were used. The analysis was performed with a Gallio flow cytometer (Beckman Coulter, Miami, FL, USA), and the data were analyzed using FlowJo software.
Statistical analysis
Data are presented as the mean ± SD, and they were analyzed using SPSS software (version 22.0). Student’s t test was used for statistical analyses, and p < 0.05 was considered statistically significant.
Results
HSC-CM promoted MDSC migration in vitro
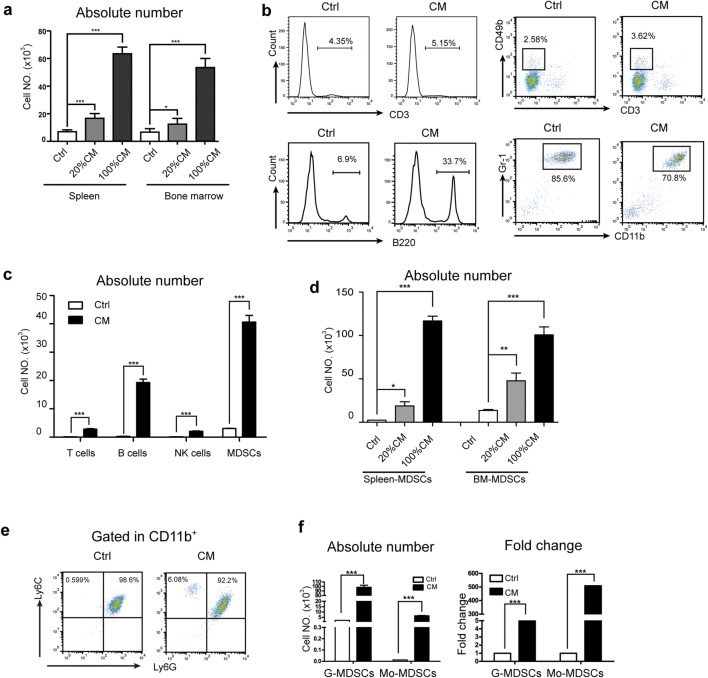
MDSCs are known to express a variety of chemokine receptors to enable their migration toward various cytokines and chemokines [27–30]. To explore the effect of HSCs on MDSC mobilization, first, we performed a transwell assay to evaluate the chemotactic activity of RPMI 1640 medium and HSC-CM. HSC-CM caused a dramatic increase in the absolute number of splenocytes and bone marrow-derived cells in a concentration-dependent manner (Fig. 1a). Then, we analyzed the percentage of the splenocyte subpopulation in the lower chamber of the transwell. As shown in Fig. 1b, both RPMI 1640 medium and HSC-CM chemoattracted CD3+, B220+, CD3−CD49+, and CD11b+Gr-1+ splenocytes. According to the splenocyte subpopulation ratios and cell numbers of migrated splenocytes, we calculated the absolute number of each splenocyte subpopulation. The results showed that HSC-CM chemoattracted many more splenocyte subpopulations than RPMI 1640 medium and that CD11b+Gr-1+ splenocytes represented the subpopulation that was attracted the most (Fig. 1c).
Fig. 1.
HSC-CM promoted MDSC migration in vitro. a The number of splenocytes and bone marrow cells that migrated to different concentration of HSC-CM. b–c The percentage (b) and absolute number (c) of CD3+, B220+, CD3−CD49+, and CD11b+Gr-1+ splenocytes in the lower chamber of the transwell. The absolute number was calculated as follows: number of indicated cells = the percentage of indicated cells × the cell number in the lower chamber. d–f HSC-CM induced MDSC migration. The cell number in the lower chamber was calculated (d). The ratio (e) and absolute number (f) of G-MDSCs and Mo-MDSCs in the lower chamber. The lower chamber contained HSC-CM with 5% FBS. Data are representative of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001
Next, CD11b+Gr-1+ MDSCs from the bone marrow and spleen of tumor-bearing mice were sorted, and the migration assay results showed that HSC-CM-mediated MDSC migration in a concentration-dependent manner as well (Fig. 1d). To determine whether MDSC subsets had different migration abilities toward HSC-CM, we analyzed the MDSC subsets in the lower chamber of the transwell by flow cytometry. As shown in Fig. 1e, the proportion of Ly6G+Ly6ClowCD11b+ G-MDSCs and Ly6G+Ly6ChighCD11b+ Mo-MDSCs in RPMI 1640 medium were 98.6% and 0.599%, respectively, whereas in HSC-CM, the proportions of G-MDSC and Mo-MDSCs were 92.2% and 6.08%, respectively. Indeed, HSC-CM induced a greater than fourfold and 500-fold increase in the absolute number of G-MDSCs and Mo-MDSCs, respectively, compared with RPMI 1640 medium (Fig. 1f).
HSCs promoted MDSC migration via CXCR4
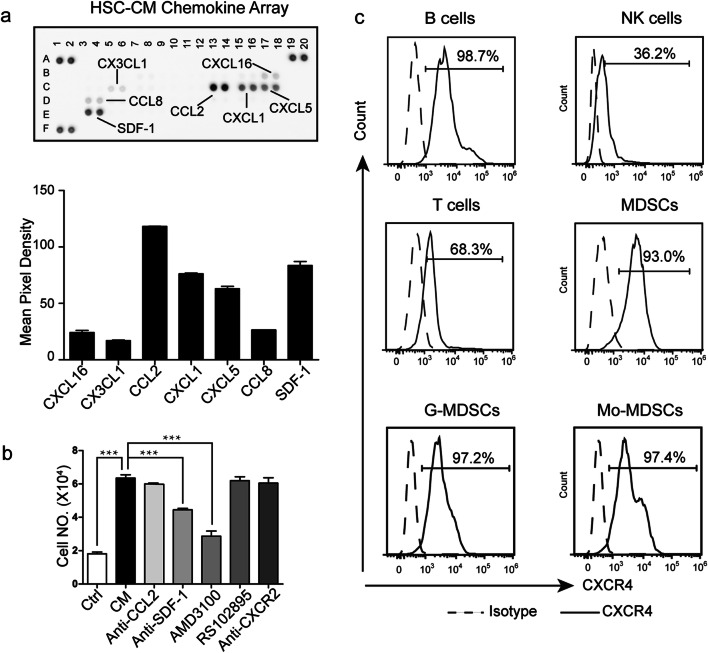
The mobilization of MDSCs is regulated by a complex chemokine–chemokine receptor axis. We first determined which major chemoattractants present in HSCs were responsible for the migration of MDSCs, and then, the chemokines present in the CM derived from the mouse HSCs were analyzed using a Mouse Chemokine Array. The chemokines detected are shown in Supplementary Table 1. The results revealed that HSCs secreted high levels of CCL2, CXCL1, CXCL5, and SDF-1 (Fig. 2a). The chemokine receptors for these chemokines are CCR2, CXCR2, and CXCR4. Moreover, we previously reported the spectrum of chemokine receptors in MDSC subsets, and CCR2, CXCR2, and CXCR4 were found to be highly expressed in MDSC subsets [31]. Based on these data, we postulated that CCL2/CCR2, CXCL1/CXCR2, CXCL5/CXCR2, and SDF-1/CXCR4 act as chemotactic axes for MDSC migration toward HSC-CM (Supplementary Fig. 1). To further confirm this hypothesis, specific inhibitors of CXCR2 and CXCR4 and neutralizing antibodies against CCL2, SDF-1, and CXCR2 were used in the migration assays. As expected, HSC-CM promoted more MDSC migration than RPMI 1640 medium (Fig. 2b). The CXCR4 inhibitor AMD3100 and the neutralizing antibody against SDF-1 suppressed the induction of MDSC chemotaxis toward HSC-CM, whereas the anti-CXCR2 antibody and CCR2 inhibitor RS102895 did not (Fig. 2b), which suggested that HSCs may promote MDSC migration through the SDF-1/CXCR4 axis. Because HSCs release factors that are also able to induce T cell, B cell, and NK cell migration to different extents, we investigated the CXCR4 expression of these cells. We found that the expression of CXCR4 in B cells, MDSCs, and MDSC subsets was much higher than that in T cells and NK cells (Fig. 2c). The CXCR4 expression level in these cells correlated with their efficiency of mobilization towards HSC-CM.
Fig. 2.
HSCs promoted MDSC migration via CXCR4. a The chemokines in HSC-CM were detected using a Mouse Chemokine Array. b The number of MDSCs in the lower chamber of the transwell after the indicated treatment. Ctrl: RPMI 1640 medium was added to the lower chamber; CM: HSC-CM was added to the lower chamber; Anti-CCL2/SDF-1/CXCR2: the CM was pretreated with CCL2 or SDF-1or CXCR2 neutralizing antibody 2 h before it was added to the lower chamber; AMD3100/RS102895: the MDSCs were pretreated with a CXCR4 or CCR2 inhibitor 2 h before migration; c Expression of CXCR4 on different splenocytes was detected by flow cytometry. *p < 0.05; **p < 0.01; ***p < 0.001
HSCs secreted SDF-1 to promote MDSC migration
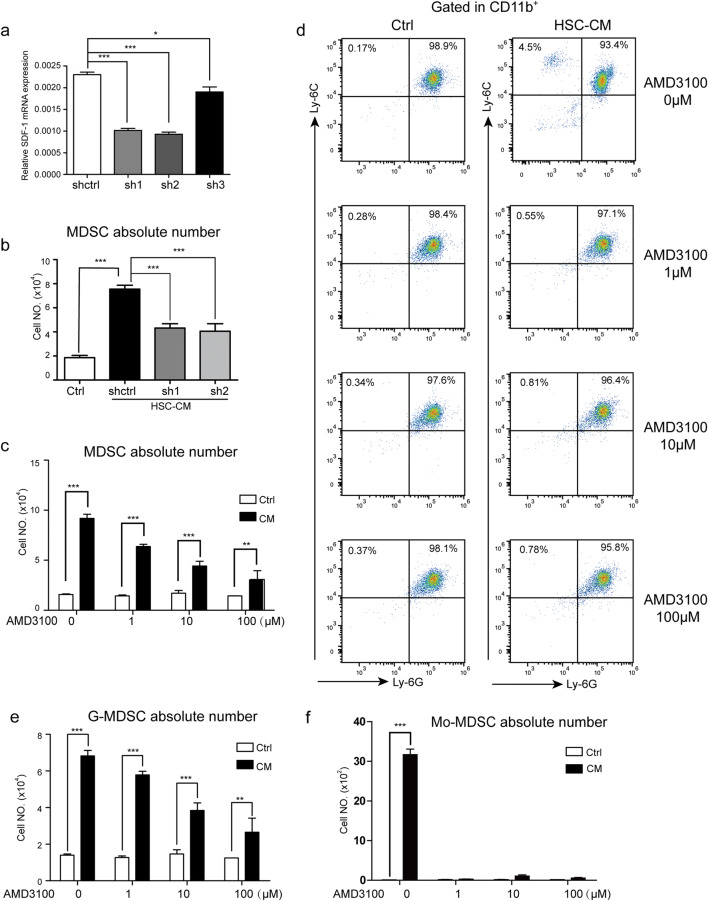
To further confirm the role of the SDF-1/CXCR4 axis in MDSC migration induced by HSCs, we used shRNA plasmids specific for SDF-1 to knockdown its expression level in HSCs and determine whether inhibiting SDF-1 would reduce the migration potential of MDSCs in response to HSCs. The sh1 and sh2 for SDF-1 were selected for further analysis (Fig. 3a). As shown in Fig. 3b, downregulation of SDF-1 significantly impaired the HSC-induced migration of MDSCs, which indicated that HSCs secreted SDF-1 to promote MDSC migration. MDSCs isolated from the spleens of tumor-bearing mice were treated with increasing concentrations of the CXCR4 inhibitor AMD3100 (1 µM, 10 µM, 100 µM) for 48 h, and cell migration was evaluated by counting the cell number in the lower chamber. As expected, AMD3100 inhibited MDSC migration in a dose-dependent manner (Fig. 3c). To further evaluate the effect of SDF-1/CXCR4 on MDSC subset migration, the ratio of G-MDSCs and Mo-MDSCs in the lower chamber was assessed by flow cytometry. The ratio of G-MDSCs and Mo-MDSCs and the cell number in the lower chamber (Fig. 3d) were used to calculate the absolute number of G-MDSCs and Mo-MDSCs, and the absolute numbers of both were reduced with AMD3100 treatment (Fig. 3e, f).
Fig. 3.
HSCs secreted SDF-1 to promote MDSC migration. a The SDF-1 mRNA level of HSCs was identified by real-time PCR. b The absolute number of MDSCs migrated to HSC-CM which collected from SDF-1-knocked down HSCs. c The effect of ADM3100 on MDSC migration induced by HSC-CM. d The ratios of MDSC subsets in the lower chamber of the transwell. e–f The absolute number of migrated G-MDSCs (e) and Mo-MDSCs (f) in the lower chamber. The absolute number of cells = the percentage of indicated cells × the cell number in the lower chamber
HSCs promoted MDSC migration to the liver and spleen via SDF-1/CXCR4 in vivo
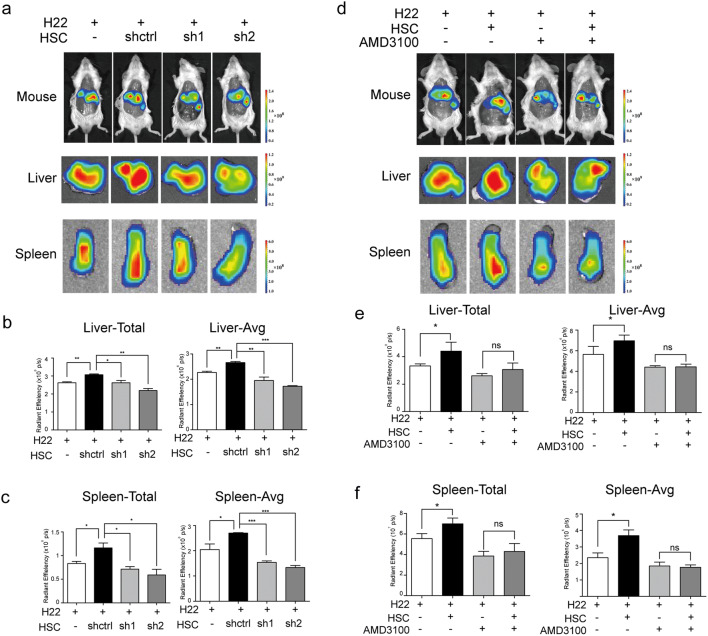
To assess the functional consequence of SDF-1 inducing MDSCs in vivo, an orthotopic mouse model of HCC was used in which mice were injected intra-hepatically with control or SDF-1-knocked down HSCs together with H22 cells or H22 cells alone. Ten days after the tumor model was established, the mice were inoculated with DiR-labeled MDSCs through the tail intravenous injection, and the signal was evaluated by IVIS. The fluorescence intensity of the inoculated MDSCs was observed 24 h after injection, and the livers and spleens of all tumor-bearing mice displayed high signal intensity (Fig. 4a). As shown in Fig. 4b and c, the quantitative fluorescence intensity data indicated an increase in the number of MDSCs in the resected livers and spleens from MDSC-infused mice bearing HSCs plus H22 cells compared with that in mice given H22 cells alone, indicating that HSCs induced MDSC migration to the tumor site and spleen. However, when co-transplanted with SDF-1-knocked down HSCs, the fluorescence intensity in the liver and spleen was much weaker than that in the HSC group, which suggested a pronounced reduction in the number of MDSCs in the SDF-1-knocked down HSC group. To further investigate whether the CXCR4-expressing cells were also recruited by HSCs in vivo, we performed the same animal experiment with DiR-labeled RAW 264.7, a mouse monocyte/macrophage cell line, which express CXCR4. The results shown in Supplementary Fig. 2 demonstrated that HSCs could recruit the CXCR4-expressing cells to livers via SDF-1.
Fig. 4.
HSCs promoted MDSC migration to the liver and spleen via SDF-1/CXCR4 in vivo. a Representative mice, livers, and spleens are shown. The mice were given an intrahepatic injection of 1 × 106 H22 cells, 1 × 106 H22 cells plus 2 × 105 activated shctrl HSCs (shctrl) or SDF-1-knocked down HSCs (sh1 or sh2). After the transfer of DiR-labeled MDSCs, the signal was evaluated by IVIS. b–c Total and average signal intensity in the livers (b) and spleens (c) of tumor-bearing mice are presented. d Representative mice, livers, and spleens. Mice were treated as indicated. MDSCs were pretreated with or without 10 µM AMD3100 before transfer of MDSCs and IVIS was performed 24 h later. e–f Total signal intensity and average signal intensity in the livers (e) and spleens (f) of tumor-bearing mice are presented. *p < 0.05, **p < 0.01
Next, we investigated the role of CXCR4 in MDSC migration, using a murine model in which H22 cells or HSCs plus H22 cells were inoculated orthotopically. Ten days later, MDSCs were pretreated with the CXCR4 inhibitor AMD3100 and then labeled with DiR and used to inoculate mice by intravenous injection in the tail. After 24 h, tumor-bearing mice were imaged by IVIS. As expected, MDSCs mainly accumulated in the spleen and tumor sites (Fig. 4d). Then, the livers and spleens were resected to assess the fluorescence intensity. The data showed that HSC co-transplantation clearly increased MDSC migration to the spleen and liver (Fig. 4e, f). However, when CXCR4 expression was inhibited by AMD3100 treatment, the effect of HSC on MDSC migration was attenuated. No significant difference in fluorescence intensity was observed between the HSCs and H22 groups when AMD3100 pretreated-MDSCs were used. Together, these results suggest that HSCs may secrete SDF-1, which can recruit MDSCs to the tumor microenvironment to exert a tumor-promoting effect.
Discussion
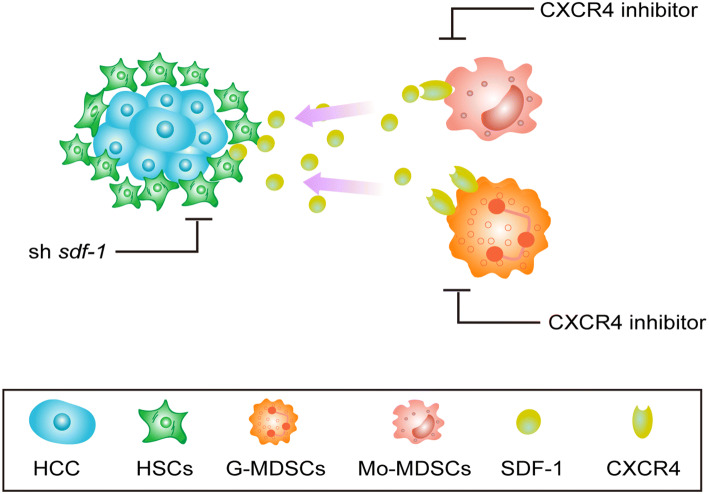
HSCs, the major stromal cells in the liver, are widely recognized as the key players in fibrogenesis. It has become increasingly clear that HSCs, in addition to their contribution to fibrogenesis, possess other pro-tumorigenic properties such as an immune-suppressive activity, resulting in them suppressing the immune microenvironment in HCC [22, 32]. Recently, there has been increasing interest in the cellular interactions between cancer cells and their microenvironment. HSCs, an important type of liver stromal cell, induce multiple types of immune inhibitory cells, leading to a complex suppressive microenvironment that not only modulates immune responses in the liver [33] but also plays vital roles in HCC progression [23, 34]. The molecular mechanisms that govern MDSC function and accumulation have been extensively explored. However, how HSCs induce MDSC recruitment to the HCC microenvironment remains largely unknown. Using an in situ HCC model in immune-competent mice, we showed that HSCs secreted SDF-1, which interacted with CXCR4 on MDSCs and promoted MDSC migration to the HCC microenvironment (Fig. 5). This work presents a novel mechanism of MDSC accumulation in the HCC microenvironment.
Fig. 5.
A schematic model depicting MDSC migration was induced by HSCs via SDF-1/CXCR4. In an orthotopic mouse model of HCC, activated HSC-derived SDF-1 induced MDSC migration to the HCC microenvironment via CXCR4. Knockdown of the expression of SDF-1 in HSCs or treatment of MDSCs with a CXCR4 inhibitor reduced the migration of MDSCs to the HCC microenvironment
Accumulating data have indicated the involvement of a variety of chemokine–chemokine receptors in MDSC recruitment to the tumor microenvironment. The CCL2/CCR2 and SDF-1/CXCR4 pathways have been shown to be essential for recruiting human MDSCs in breast, lung, and ovarian cancers [7, 35, 36]. However, the roles of HSCs in MDSC recruitment in HCC have rarely been addressed. HSCs are multifunctional fibroblast cells that can produce various cytokines. Activated HSCs produce large amounts of CCL2 but also secrete CXCL1, CXCL5, and SDF-1 (Fig. 2a). We previously reported that both G-MDSCs and Mo-MDSCs express high levels of CXCR4 and CXCR2 and that Mo-MDSCs express high levels of CCR2. Thus, CCL2/CCR2, CXCL1/CXCR2, CXCL5/CXCR2, and SDF-1/CXCR4 may function as a chemotactic axis for MDSC migration toward HSC-CM (Supplementary Fig. 1). The tumor histopathology and spectrum of chemokines in the tumor microenvironment determine the migration of particular MDSC subset [12]. It has been reported that CCL2 and its interaction with its receptors are involved in the recruitment of Mo-MDSCs in breast cancer and melanoma [37, 38]. A recent study showed that CCL2 recruits Ly-6C+ monocytic MDSCs through CCR2 within the glioma microenvironment [39], suggesting that MCP-1/CCR2 plays a vital role in inducing Mo-MDSC migration. However, the CCR2 inhibitor RS102895 did not attenuate MDSC migration toward HSC-CM, possibly because the total number of Mo-MDSCs was too low.
In rhabdomyosarcoma, CXCR2 deficiency can prevent CD11b+Ly6Ghi MDSC migration to the tumor [40], and CXCR2 loss was found to result in decreased MDSC infiltration into the colonic mucosa and tumor [41]. Additionally, CXCR2 ligands were shown to support G-MDSC migration to the tumor site [42]. In the current study, we found that anti-CXCR2 antibody did not suppress MDSC migration. Thus, the CXCL1/CXCR2 and CXCL5/CXCR2 axes do not seem to be involved in the HSC-mediated recruitment of MDSCs to the HCC environment. Meanwhile, both a CXCR4 inhibitor and SDF-1 neutralizing antibody exerted its inhibition on MDSC migration. Our data suggested that HSCs induced MDSCs through the SDF-1/CXCR4 axis, indicating that targeting MDSC recruitment is a novel therapeutic strategy for HCC.
As shown in Fig. 1c, CM generated by HSCs was able to recruit other immune cells, especially B220+ B cells. B cells were another key constituent for the adaptive immune system, however, the role of B cells on tumor occurence and development is not so extensively studied as MDSCs. Therefore, in this study, we mainly focus on the influence of HSCs on MDSC recruitment. It has been reported that HSCs could directly suppressed B cells via programmed death ligand 1 (PD-L1), which may contribute to the liver’s immune homeostasis maintained by HSCs [43]. Whether HSCs exert its effects on B cells to form a tumor-favoring environment requires further investigation.
Besides, CD3+ T cells and CD3−CD49+ NK cells, which are important in the innate immune response against tumorigenic cancer cells, were also recruited by HSC-CM. However, these cells were far fewer than MDSCs. Moreover, once MDSCs are attracted to the tumor microenvironment, they always affect the anti-tumor immune responses [44, 45], while the functioning of T, B, and NK cells is impaired [46]. The generation of oxidative stress, depletion of l-arginine and increasing of ROS and iNOS levels are the main mechanisms for MDSC-mediated lymphocyte suppression [47]. MDSCs impair B cell responses through IL-7 and STAT5 and suppress B cell proliferation in an arginase-dependent manner [48]. MDSCs from patients with cancer can inhibit FcR-mediated function and signal transduction in NK cells, in part through nitric oxide production [49]. HSCs also recruited RAW 264.7 cells in vivo (Supplementary Fig. 2), which further proved that HSCs could recruit the cells expressing CXCR4. These data demonstrated that the SDF-1/CXCR4 axis has an important role in tumor microenvironment to some extent. Therefore, in the tumor microenvironment, HSCs act as a mediator by attracting different types of immune cells to the tumor microenvironment and mediating the cross-talk between the immune cells. Whether HSC-CM induces changes in the gene expression pattern and behavior of these immune cells needs to be examined further.
In conclusion, HSCs are key regulators in HCC immunosuppression and elimination of HSCs might represent a potential effective treatment. Activated HSCs secrete SDF-1, which might mobilize and recruit MDSCs to promote tumor progression and immune evasion.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- C3
Complement component 3
- CCL/CCR
CC chemokine ligand/chemokine receptor
- CXCL/CXCR
CXC chemokine ligand/chemokine receptor
- DiR
1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide
- G-MDSCs
Granulocytic myeloid-derived suppressor cells
- HCC
Hepatocellular carcinoma
- HSC-CM
HSC conditioned medium
- HSCs
Hepatic stellate cells
- IL-1β
Interleukin-1 beta
- IL-8
Interleukin-8
- IVIS
In vivo imaging system
- MDSCs
Myeloid-derived suppressor cells
- Mo-MDSCs
Monocytic myeloid-derived suppressor cells
- PD-L1
Programmed death ligand 1
- RBC
Red blood cells
- SDF-1
Stromal cell-derived factor 1
Author contributions
YX performed most of the experiments, analyzed the data, and wrote the manuscript. FF, HJ, XZ, LH, and XY assisted with and/or performed some of the experiments and revised the manuscript. WZ contributed to the design of the experiments and analysis and interpretation of the data, and revised the manuscript.
Funding
This work was supported by grants from the National Nature Science Foundation of China (81602500 and 81572335); Outstanding Youth Science Research Personnel Training Plan of Fujian Province Colleges and Universities (2017); and Doctoral Start-up Foundation of Xiamen Medical College (K2016-07).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All animal experimental protocols were performed in compliance with the Guidelines for the Institutional Animal Care and Use Committee of Xiamen University. (the date of animal research approval: 2015-02-26).
Animal source
BALB/c (H-2d, haplotype) mice aged 8–12 weeks were purchased from the National Rodent Laboratory Animal Resources, Shanghai, China.
Cell authentication
Not applicable because murine primary HSCs and MDSCs and murine HCC cell line H22 cells were used, which are lacking of short tandem repeat data. The H22 cells were purchased from Shanghai Cell Bank, Chinese Academy of Sciences.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhang H, Li Z, Wang L, Tian G, Tian J, Yang Z, Cao G, Zhou H, Zhao L, Wu Z, Yin Z. Critical role of myeloid-derived suppressor cells in tumor-induced liver immune suppression through inhibition of NKT cell function. Front Immunol. 2017;8:129. doi: 10.3389/fimmu.2017.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoechst B, Voigtlaender T, Ormandy L, Gamrekelashvili J, Zhao F, Wedemeyer H, Lehner F, Manns MP, Greten TF, Korangy F. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology. 2009;50(3):799–807. doi: 10.1002/hep.23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid MC, Varner JA. Myeloid cells in tumor inflammation. Vasc Cell. 2012;4(1):14. doi: 10.1186/2045-824X-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Can Res. 2010;70(1):68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, Moses HL. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13(1):23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmid MC, Avraamides CJ, Foubert P, Shaked Y, Kang SW, Kerbel RS, Varner JA. Combined blockade of integrin-alpha4beta1 plus cytokines SDF-1alpha or IL-1beta potently inhibits tumor inflammation and growth. Cancer Res. 2011;71(22):6965–6975. doi: 10.1158/0008-5472.CAN-11-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, Fein M, Rasch MG, Tan YX, Qiu J, Park J, Sinha P, Bissell MJ, Frengen E, Werb Z, Egeblad M. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21(4):488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du R, Lu KV, Petritsch C, Liu P, Ganss R, Passegue E, Song H, Vandenberg S, Johnson RS, Werb Z, Bergers G. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13(3):206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilkovitch D, Lopez DM. The liver is a site for tumor-induced myeloid-derived suppressor cell accumulation and immunosuppression. Cancer Res. 2009;69(13):5514–5521. doi: 10.1158/0008-5472.CAN-08-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapanadze T, Gamrekelashvili J, Ma C, Chan C, Zhao F, Hewitt S, Zender L, Kapoor V, Felsher DW, Manns MP, Korangy F, Greten TF. Regulation of accumulation and function of myeloid derived suppressor cells in different murine models of hepatocellular carcinoma. J Hepatol. 2013;59(5):1007–1013. doi: 10.1016/j.jhep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umansky V, Sevko A. Tumor microenvironment and myeloid-derived suppressor cells. Cancer Microenviron. 2013;6(2):169–177. doi: 10.1007/s12307-012-0126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14(7):397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 14.Thompson AI, Conroy KP, Henderson NC. Hepatic stellate cells: central modulators of hepatic carcinogenesis. BMC gastroenterology. 2015;15:63. doi: 10.1186/s12876-015-0291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aimaiti Y, Jin X, Shao Y, Wang W, Li D. Hepatic stellate cells regulate hepatic progenitor cells differentiation via the TGF-beta1/Jagged1 signaling axis. J Cell Physiol. 2019;234(6):9283–9296. doi: 10.1002/jcp.27609. [DOI] [PubMed] [Google Scholar]
- 16.Ji F, Wang K, Zhang Y, Mao XL, Huang Q, Wang J, Ye L, Li Y. MiR-542-3p controls hepatic stellate cell activation and fibrosis via targeting BMP-7. J Cell Biochem. 2019;120(3):4573–4581. doi: 10.1002/jcb.27746. [DOI] [PubMed] [Google Scholar]
- 17.Yu MC, Chen CH, Liang X, Wang L, Gandhi CR, Fung JJ, Lu L, Qian S. Inhibition of T-cell responses by hepatic stellate cells via B7-H1-mediated T-cell apoptosis in mice. Hepatology. 2004;40(6):1312–1321. doi: 10.1002/hep.20488. [DOI] [PubMed] [Google Scholar]
- 18.Chen CH, Kuo LM, Chang Y, Wu W, Goldbach C, Ross MA, Stolz DB, Chen L, Fung JJ, Lu L, Qian S. In vivo immune modulatory activity of hepatic stellate cells in mice. Hepatology. 2006;44(5):1171–1181. doi: 10.1002/hep.21379. [DOI] [PubMed] [Google Scholar]
- 19.Chou HS, Hsieh CC, Yang HR, Wang L, Arakawa Y, Brown K, Wu Q, Lin F, Peters M, Fung JJ, Lu L, Qian S. Hepatic stellate cells regulate immune response by way of induction of myeloid suppressor cells in mice. Hepatology. 2011;53(3):1007–1019. doi: 10.1002/hep.24162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh CC, Chou HS, Yang HR, Lin F, Bhatt S, Qin J, Wang L, Fung JJ, Qian S, Lu L. The role of complement component 3 (C3) in differentiation of myeloid-derived suppressor cells. Blood. 2013;121(10):1760–1768. doi: 10.1182/blood-2012-06-440214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochst B, Schildberg FA, Sauerborn P, Gabel YA, Gevensleben H, Goltz D, Heukamp LC, Turler A, Ballmaier M, Gieseke F, Muller I, Kalff J, Kurts C, Knolle PA, Diehl L. Activated human hepatic stellate cells induce myeloid derived suppressor cells from peripheral blood monocytes in a CD44-dependent fashion. J Hepatol. 2013;59(3):528–535. doi: 10.1016/j.jhep.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W, Zhang L, Yin Z, Su W, Ren G, Zhou C, You J, Fan J, Wang X. Activated hepatic stellate cells promote hepatocellular carcinoma development in immunocompetent mice. Int J Cancer. 2011;129(11):2651–2661. doi: 10.1002/ijc.25920. [DOI] [PubMed] [Google Scholar]
- 23.Zhao W, Zhang L, Xu Y, Zhang Z, Ren G, Tang K, Kuang P, Zhao B, Yin Z, Wang X. Hepatic stellate cells promote tumor progression by enhancement of immunosuppressive cells in an orthotopic liver tumor mouse model. Lab Invest. 2014;94(2):182–191. doi: 10.1038/labinvest.2013.139. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Zhao W, Xu J, Li J, Hong Z, Yin Z, Wang X. Activated hepatic stellate cells promote liver cancer by induction of myeloid-derived suppressor cells through cyclooxygenase-2. Oncotarget. 2016;7(8):8866–8878. doi: 10.18632/oncotarget.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W, Su W, Kuang P, Zhang L, Liu J, Yin Z, Wang X. The role of hepatic stellate cells in the regulation of T-cell function and the promotion of hepatocellular carcinoma. Int J Oncol. 2012;41(2):457–464. doi: 10.3892/ijo.2012.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Zhao W, Wu D, Xu J, Lin S, Tang K, Yin Z, Wang X. Isolation of myeloid-derived suppressor cells subsets from spleens of orthotopic liver cancer-bearing mice by fluorescent-activated and magnetic-activated cell sorting: similarities and differences. Int J Clin Exp Pathol. 2014;7(11):7545–7553. [PMC free article] [PubMed] [Google Scholar]
- 27.Zollo M, Di Dato V, Spano D, De Martino D, Liguori L, Marino N, Vastolo V, Navas L, Garrone B, Mangano G, Biondi G, Guglielmotti A. Targeting monocyte chemotactic protein-1 synthesis with bindarit induces tumor regression in prostate and breast cancer animal models. Clin Exp Metastasis. 2012;29(6):585–601. doi: 10.1007/s10585-012-9473-5. [DOI] [PubMed] [Google Scholar]
- 28.Karin N, Razon H. The role of CCR28 in directing the mobilization and biological function of CD11b(+)Gr1(+)Ly6C(low) polymorphonuclear myeloid cells in cancer. Cancer Immunol Immunother CII. 2018;67(12):1949–1953. doi: 10.1007/s00262-018-2245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liepelt A, Tacke F. Stromal cell-derived factor-1 (SDF-1) as a target in liver diseases. Am J Physiol Gastrointest Liver Physiol. 2016;311(2):G203–209. doi: 10.1152/ajpgi.00193.2016. [DOI] [PubMed] [Google Scholar]
- 30.Shou D, Wen L, Song Z, Yin J, Sun Q, Gong W. Suppressive role of myeloid-derived suppressor cells (MDSCs) in the microenvironment of breast cancer and targeted immunotherapies. Oncotarget. 2016;7(39):64505–64511. doi: 10.18632/oncotarget.11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W, Xu Y, Xu J, Wu D, Zhao B, Yin Z, Wang X. Subsets of myeloid-derived suppressor cells in hepatocellular carcinoma express chemokines and chemokine receptors differentially. Int Immunopharmacol. 2015;26(2):314–321. doi: 10.1016/j.intimp.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Xia Y, Chen R, Ye SL, Sun R, Chen J, Zhao Y. Inhibition of T-cell responses by intratumoral hepatic stellate cells contribute to migration and invasion of hepatocellular carcinoma. Clin Exp Metastasis. 2011;28(7):661–674. doi: 10.1007/s10585-011-9399-3. [DOI] [PubMed] [Google Scholar]
- 33.Schildberg FA, Sharpe AH, Turley SJ. Hepatic immune regulation by stromal cells. Curr Opin Immunol. 2015;32:1–6. doi: 10.1016/j.coi.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Ji J, Eggert T, Budhu A, Forgues M, Takai A, Dang H, Ye Q, Lee JS, Kim JH, Greten TF, Wang XW. Hepatic stellate cell and monocyte interaction contributes to poor prognosis in hepatocellular carcinoma. Hepatology. 2015;62(2):481–495. doi: 10.1002/hep.27822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Damianakou C, Tsokas C. Clinical and technical characteristics of alginate hydrocolloid. Hellenika Stomatologika Chronika Hell Stomatol Ann. 1987;31(1):63–68. [PubMed] [Google Scholar]
- 36.Obermajer N, Muthuswamy R, Odunsi K, Edwards RP, Kalinski P. PGE(2)-induced CXCL12 production and CXCR36 expression controls the accumulation of human MDSCs in ovarian cancer environment. Can Res. 2011;71(24):7463–7470. doi: 10.1158/0008-5472.CAN-11-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umansky V, Blattner C, Gebhardt C, Utikal J. CCR38 in recruitment and activation of myeloid-derived suppressor cells in melanoma. Cancer Immunol Immunother CII. 2017;66(8):1015–1023. doi: 10.1007/s00262-017-1988-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, Kanojia D, Pituch KC, Qiao J, Pytel P, Han Y, Wu M, Zhang L, Horbinski CM, Ahmed AU, Lesniak MS. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 2016;76(19):5671–5682. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, Mackall CL. Disruption of CXCR1-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med. 2014;6(237):237ra267. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. CXCR41-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24(5):631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawanobori Y, Ueha S, Kurachi M, Shimaoka T, Talmadge JE, Abe J, Shono Y, Kitabatake M, Kakimi K, Mukaida N, Matsushima K. Chemokine-mediated rapid turnover of myeloid-derived suppressor cells in tumor-bearing mice. Blood. 2008;111(12):5457–5466. doi: 10.1182/blood-2008-01-136895. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Lu L, Qian S, Fung JJ, Lin F. Hepatic stellate cells directly inhibit B cells via programmed death-ligand 1. J Immunol. 2016;196(4):1617–1625. doi: 10.4049/jimmunol.1501737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostrand-Rosenberg S. Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother CII. 2010;59(10):1593–1600. doi: 10.1007/s00262-010-0855-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fruci D, Lo Monaco E, Cifaldi L, Locatelli F, Tremante E, Benevolo M, Giacomini P. T and NK cells: two sides of tumor immunoevasion. J Transl Med. 2013;11:30. doi: 10.1186/1479-5876-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pyzer AR, Cole L, Rosenblatt J, Avigan DE. Myeloid-derived suppressor cells as effectors of immune suppression in cancer. Int J Cancer. 2016;139(9):1915–1926. doi: 10.1002/ijc.30232. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Schafer CC, Hough KP, Tousif S, Duncan SR, Kearney JF, Ponnazhagan S, Hsu HC, Deshane JS. Myeloid-derived suppressor cells impair B cell responses in lung cancer through IL-7 and STAT5. J Immunol. 2018;201(1):278–295. doi: 10.4049/jimmunol.1701069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stiff A, Trikha P, Mundy-Bosse B, McMichael E, Mace TA, Benner B, Kendra K, Campbell A, Gautam S, Abood D, Landi I, Hsu V, Duggan M, Wesolowski R, Old M, Howard JH, Yu L, Stasik N, Olencki T, Muthusamy N, Tridandapani S, Byrd JC, Caligiuri M, Carson WE. Nitric oxide production by myeloid-derived suppressor cells plays a role in impairing Fc receptor-mediated natural killer cell function. Clin Cancer Res. 2018;24(8):1891–1904. doi: 10.1158/1078-0432.CCR-17-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.