Abstract
Altered numbers and functions of T cells have previously been demonstrated in chronic lymphocytic leukemia (CLL) patients. However, dynamics and specific T-cell subset alterations have not been studied in great detail. Therefore, we studied CLL blood lymphocyte subsets of individual patients in a longitudinal manner. Dynamic expansions of blood CD4+ and CD8+ T-cell numbers were consistently associated with a progressively increasing CLL leukemic compartment. Interestingly, the T-cell subset expansion over time was more pronounced in CD38+ CLL. Additionally, we performed gene expression profiling of CD3+ T cells of CLL patients and normal donors. Using gene set enrichment analysis, we found significant enrichment of genes with higher expression in CLL T cells within CD8+ effector memory and terminal effector T-cell gene signatures. In agreement with these data, we observed a marked expansion of phenotypic CD8+ effector memory T cells in CLL by flow cytometry. Moreover, we observed that increments of CD8+ effector memory T cells in human CLL and also mouse CLL (Eμ-TCL1 model) were due to an expansion of the inhibitory killer cell lectin-like receptor G1 (KLRG1) expressing cellular subset. Furthermore, higher plasma levels of the natural KLRG1 ligand E-cadherin were detected in CLL patients compared to normal donor controls. The predominance of KLRG1+ expression within CD8+ T cells in conjunction with increased systemic soluble E-cadherin might significantly contribute to CLL immune dysfunction and might additionally represent an important component of the CLL microenvironment.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-013-1473-z) contains supplementary material, which is available to authorized users.
Keywords: Chronic lymphocytic leukemia, Gene set enrichment analysis, CD8+ T cells, KLRG1
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most prevalent leukemia in Europe and the USA and is characterized by the accumulation of CD5+CD23+CD19+ clonal B cells [1]. The leukemic cells undergo spontaneous apoptosis in vitro, which implies that the interaction with the microenvironment is indispensable for leukemogenesis and disease maintenance.
Evidence suggests that abnormal T cells are part of the supporting microenvironment sustaining the malignant clone in vivo [2, 3]. In contrast to other malignancies, T-cell dysfunction coupled with T-cell expansion is a hallmark of CLL [4]. CLL T cells are characterized by molecular and functional abnormalities at the cellular level. A study by Gribben and colleagues detected defective pathways such as cytoskeleton formation and cell differentiation in CLL CD4+ and CD8+ T cells by expression profiling [5]. These investigations were complemented by functional studies, demonstrating defective immunological synapse formation in CLL [6]. Interestingly, these T-cell defects could be imposed on normal donor T cells by direct CLL co-culture. This finding was recapitulated in vivo with the Eμ-TCL1 murine CLL model [5–8].
We were able to show a dramatic decrease in T-cell telomere length especially in ZAP70+/CD38+ CLL patients. These data were indicative for extensive proliferation within the T-cell compartment of CLL patients [9]. However, due to the technical constraints of flow FISH analysis, this study was not able to discriminate between CD45RA+ naive and CD45RA+ terminally differentiated effector T cells.
At least in early CLL stage of disease, T cells appear not to be solely compliant to CLL cells: clonal T-cell expansion identified by preferential usage of a restricted TCR-BV gene usage was identified in CLL [10]. In vitro T-cell stimulation experiments with autologous B cells argued for the presence of leukemia-specific CD4+ and CD8+ T cells. However, not all expanded clones were able to recognize leukemic cells via their TCR [11].
In order to gain further insight into the emergence of T-cell abnormalities in CLL, we studied accessory lymphoid subsets of individual patients in a longitudinal fashion and correlated these with known prognostic factors. Moreover, we undertook gene expression profiling (GEP) analysis of CLL T cells compared to normal donor T cells and discovered a gene expression signature, which suggested the expansion of CD8+ effector memory T cells in CLL patients. This in silico observation was subsequently confirmed by flow cytometric analysis in CLL patients. Strikingly, we were able to observe in human as well as murine CLL that the expanded CD8+ effector memory T-cell subset was characterized by increased expression of the senescence mediating receptor KLRG1.
Materials and methods
Patients
Peripheral blood (PB) samples of 42 normal donors and 105 patients with CLL were analyzed after obtaining informed consent according to institutional guidelines, approved by the Ethics Commission of the University of Essen–Duisburg. The diagnosis of CLL required a persistent lymphocytosis of more than 5.0 × 109/l and a typical CD19+, CD20+, CD5+, CD23+, Ig light chain (κ or λ light chain) restricted immunophenotype as revealed by flow cytometry of PB. Furthermore, PB mononuclear cells (PBMC) were isolated using Lymphoprep (Invitrogen, Karlsruhe; Germany) density gradient centrifugation and were cryopreserved until further analysis. Longitudinal analysis of the accessory T-cell compartment (Fig. 1) was performed in 46 patients (median number of measurement time points per patient: 4 [range 2–10] over a median period of 16 months [range 3–80 months]). None of the patients included in the longitudinal analysis (Fig. 1) received therapy during the observation period.
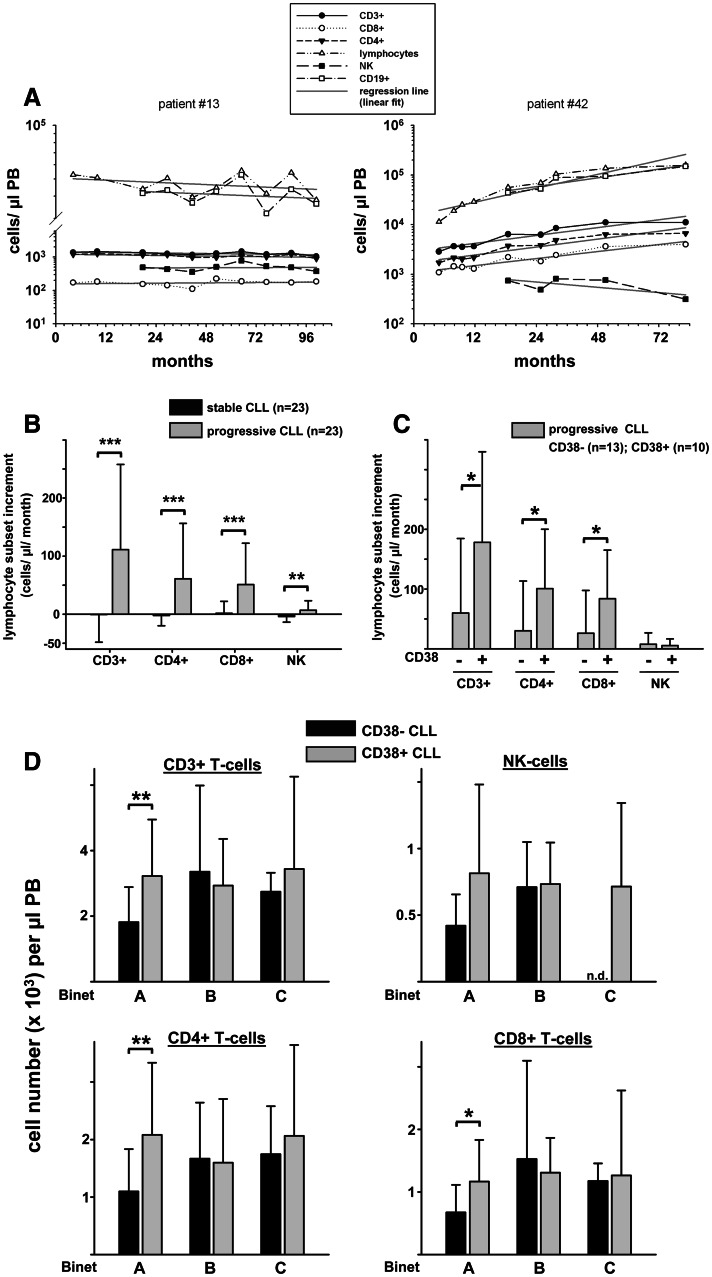
Fig. 1.
Longitudinal kinetics of the accessory T-cell compartment in relation to disease progression and CD38 status. a Levels of total CD3+, CD8+ and CD4+ T cells, NK cells (defined as CD56+CD16+ cells), absolute lymphocytes and CD19+ B cells of individual CLL patients (n = 46) were determined over time. The slope of the regression line was determined for each lymphocyte subset of every patient. An increment of ≥500 lymphocytes/μl per month was arbitrarily defined as progressive disease (n = 23 patients). An increment <500 lymphocytes/μl per month was regarded as stable disease (n = 23 patients). Representative graphs of a patient with stable (left) and a patient with progressive disease (right) are shown. b Comparison of lymphocyte subset increments of stable and progressive disease patients. c Lymphocyte subset increments per month of CD38+ are significantly higher than increments of CD38− progressive CLL patients. d Absolute numbers of peripheral blood accessory lymphocyte subsets including CD3+ total T cells, CD4+ T cells, CD8+ T cells and NK cells were analyzed according to Binet stage and CD38 expression status. Subgroups: Binet A, CD38−, n = 46; CD38+, n = 14; Binet B, CD38−, n = 14, CD38+, n = 12; Binet C, CD38−, n = 5; CD38+, n = 11. n.d., not determined; PB, peripheral blood; NK, natural killer cells. *P < 0.05; **P < 0.01; ***P < 0.001
Mice
TCL1 transgenic mice (C3H/B6 background) were provided by Dr. G. Fingerle-Rowson (Internal Medicine, Cologne, Germany) with kind permission of Dr. Carlo M. Croce (Ohio State University, Columbus, OH, USA). Mouse experiments were approved by the local institutional committee and were performed according to German animal legislation.
Gene expression profiling
CD3+ T cells were immunomagnetically isolated (MidiMacs, Miltenyi Biotec, Bergisch Gladbach, Germany) from the PB of normal donors (n = 8) and previously untreated patients with CLL (n = 25) resulting in a purity of >90 % as determined by flow cytometry. GEP was performed on total RNA extracted from 1 × 107 to 5 × 107 CD3+ cells (RNeasy Midi Kit, Qiagen, Hilden; Germany) using the Affymetrix U133A GeneChip platform (Affymetrix, Santa Clara, CA, USA) as described [12]. The raw experimental data are accessible through the Internet (http://www.ncbi.nlm.nih.gov/geo/; accession number GSE19147).
The analysis of gene expression data was performed with R Statistical Software version 2.8.1 (http://www.r-project.org) and extension packages of the Bioconductor platform (http://www.bioconductor.org). Affymetrix MAS 5.0 expression values and detection calls were computed using the affy package. Differentially expressed genes (DEG) between CLL and normal-donor-derived T-cell samples were identified with the significance analysis of microarrays (SAM) method [13] as implemented in the samr package. Only genes with “present” detection calls in at least 30 % across all arrays were considered for SAM analysis. The resulting list of DEG was further analyzed for associations with biological processes as defined in the Gene Ontology (GO) data base using the GOstats package. A hypergeometric probability was computed to assess whether the number of genes associated with a particular GO term was larger than expected. Gene set enrichment analysis (GSEA) to test for enrichment of our list of DEG in publicly available expression array data of human CD8+ T-cell subset signatures [14] was performed with a freely accessible online tool (Broad Institute, Cambridge, MA, USA). The normalized enrichment score (NES) was calculated for the highly expressed CLL T-cell gene set based on the relative position of the respective genes in the ranked list.
Flow cytometry
Absolute levels of CD4+ cells, CD8+ T cells, CD56+/16+ natural killer (NK) cells and CD19+ B cells in fresh EDTA whole PB samples were determined by four-color flow cytometry with the BD MultiTest CD3/CD4/CD8/CD45 and CD3/CD16 + CD56/CD45/CD19 Reagent kits (Becton–Dickinson, Heidelberg, Germany) and analyzed on a BD FACSCanto or FACSCalibur (Becton Dickonson; Table 1). In some cases, human and murine lymphocyte subpopulations were quantified using a two-platform assay by first staining samples with the respective antibodies and subsequently calculating absolute cell numbers on the basis of absolute lymphocyte counts (Coulter Counter FTKS or Z2 cell counter, Beckman Coulter, Krefeld, Germany). In primary plots, percentages of cells in a lymphocyte gate are displayed. CD8+ T-cell subsets of normal donor controls (n = 11; mean age 52 ± 15 years [±STD]) and CLL patients (n = 25; mean age 67 ± 11 years [±STD]) were analyzed with fluorescently labeled antibodies against CD8 (RPA-T8, BD Bioscience), CD45RA (HI100, BD Bioscience), CCR7 (150503, R&D Systems) and KLRG1 (clone 13A2 [15]). Murine lymphocyte subset analysis was performed with antibodies from BD Bioscience against CD3 (clone 145-2C11), CD5 (clone 53-7.3), CD8 (clone 53-6.7), CD19 (clone 1D3), CD44 (clone IM7) and CD62L (MEL-14). The antibody against KLRG1 (clone 2F1) and the corresponding Golden Syrian Hamster IgG isotype control were purchased from eBioscience (San Diego, CA, USA). Dead cells were excluded by forward and side scatter in addition to propidium iodide (PI) or 4′,6-diamidino-2-phenylindol (DAPI) positivity. Flow cytometric data were acquired on a FACSCalibur, FACSCanto or LSRII (Becton–Dickinson, Heidelberg, Germany), and analysis was performed with CellQuest, DiVa (Becton–Dickinson, San Jose, CA, USA) or FlowJo software (Treestar, San Carlos, CA, USA). Murine CD8+ T-cell subsets were defined by CD62L and CD44 expression [16–19].
Table 1.
Absolute numbers of peripheral blood lymphocyte subpopulations (per μl) of normal donors and CLL prognostic subgroups
| CD3 | CD8 | CD4 | CD4/CD8 | NK | CD19 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | |
| Normal donors (n = 31) | 1,548 | 459 | 552 | 278 | 990 | 307 | 2.18 | 1.16 | 337 | 138 | 267 | 109 |
| All patients (n = 105) | 2,559 | 1,802 | 1,012 | 882 | 1,501 | 1,056 | 2.15 | 3.0 | 580 | 412 | 47,534 | 47,221 |
| Binet (n = 102) | ||||||||||||
| A | 2,146 | 1,363 | 793 | 535 | 1,325 | 959 | 2.07 | 1.49 | 501 | 390 | 33,466 | 26,521 |
| B | 3,119 | 2,099 | 1,391 | 1,186 | 1,627 | 998 | 1.43 | 0.79 | 708 | 313 | 55,763 | 42,558 |
| C | 3,224 | 2,344 | 1,237 | 1,117 | 1,963 | 1,368 | 3.61 | 6.84 | 715 | 629 | 93,644 | 82,769 |
| P value | 0.0026 | 0.0017 | 0.028 | 0.15 | <0.01 | 0.023 | ||||||
| CD38 (n = 93) | ||||||||||||
| negative (n = 65) | 2,219 | 1,628 | 895 | 878 | 1,271 | 830 | 1.90 | 1.43 | 485 | 284 | 44,444 | 48,659 |
| positive (n = 38) | 3,163 | 1,968 | 1,223 | 872 | 1,905 | 1,284 | 2.58 | 4.53 | 753 | 539 | 55,609 | 45,892 |
| P value | <0.01 | 0.011 | 0.0053 | 0.76 | 0.037 | 0.18 | ||||||
| FISH (n = 94) | ||||||||||||
| del13 (n = 51) | 2,200 | 1,304 | 892 | 608 | 1,223 | 809 | 1.86 | 1.56 | 542 | 396 | 42,433 | 32,701 |
| No (n = 21) | 2,419 | 1,268 | 882 | 464 | 1,576 | 1,050 | 2.02 | 1.06 | 608 | 378 | 58,331 | 72,750 |
| del11 (n = 8) | 2,659 | 1,445 | 1,166 | 679 | 1,397 | 829 | 1.43 | 0.71 | 695 | 408 | 49,640 | 48,759 |
| Trisomy 12 (n = 11) | 4,580 | 3,680 | 1,904 | 2,011 | 2,587 | 1,609 | 2.1 | 1.4 | 645 | 230 | 61,596 | 66,416 |
| del17p (n = 3) | 3,406 | 342 | 691 | 497 | 2,682 | 669 | 11.17 | 15.1965 | 955 | 1,024 | 57,711 | 15,284 |
| P value | 0.014 | 0.35 | 0.0014 | 0.13 | 0.57 | 0.89 | ||||||
Statistical comparison between Binet and FISH subgroups was made with the Kruskal–Wallis test, between CD38 subgroups with the Mann–Whitney test
STD standard deviation
Soluble human epithelial cadherin enzyme-linked immunosorbent assay (ELISA)
Plasma was prepared from freshly collected heparinized peripheral blood samples and stored at −80 °C until analysis. Quantitative determination of soluble human epithelial cadherin (sE-cadherin) plasma levels was performed using a commercially available ELISA kit (R&D Systems, Wiesbaden-Nordenstadt, Germany) according to the manufacturer’s instructions.
Statistical analysis
Unless otherwise indicated, comparison of parameters between subgroups was made using the nonparametric Mann–Whitney U test or Kruskal–Wallis test for continuous variables. All mouse data were analyzed by Student’s t test. A P value <0.05 was regarded significant. All bar graphs represent mean ± standard deviation (STD).
Results
Expansion of accessory T cells is dependent on CLL progression and CD38 expression status
Absolute numbers of CD19+, CD3+, CD4+, CD8+ and CD16+CD56+ lymphocytes were assessed via flow cytometry in the PB of 105 patients with CLL (Table 1; clinical characteristics, Table S1). Consistent with previous work, clinical disease stages were characterized by significantly different numbers of PB T cells and NK cells. Patients with advanced disease generally exhibited higher cell numbers than Binet stage A patients. Furthermore, a significantly higher absolute number of T and NK cells was observed in patients expressing the CD38 surface antigen (>20 %) on their leukemic cells (Table 1). Unexpectedly, CLL patients with trisomy 12 exhibited particularly increased numbers of PB CD4+ T cells as compared to all other patients lacking this cytogenetic abnormality. Trisomy 12 patients also exhibited the greatest T-cell number variance, especially within the CD8+ T-cell compartment. Even though the number of cases analyzed with 17p deletion was limited, it is interesting to note that the CD8+ T-cell numbers within this group were comparatively low (Table 1).
Our finding that the absolute numbers of PB T- and NK-cell subsets are increased in Binet stage C patients as compared to early-stage CLL cases suggested that the expansion of accessory cells is related to disease progression (Table 1). To our knowledge, longitudinal data on accessory lymphocyte subsets within individual CLL patients have never been collected so far. Therefore, we sequentially quantified the numbers of PB T and NK cells in a subset of 46 patients. We calculated the slope (cellular increment/μl per month) of the regression line (linear fit) including all lymphocyte subset data of every individual patient (Fig. 1a). We found that the increment of CLL lymphocytes over time significantly correlated with the increment of CD3, CD4, CD8 and NK subsets (Table S2). Interestingly, T-cell increments did not correlate with NK-cell increments (Table S2), implying that NK-cell dynamics occur independently of T-cell dynamics in CLL.
During a mean observation period of 22 months, 23 of 46 patients showed evidence of disease progression [progressive disease (PD); arbitrarily defined as lymphocyte count increment ≥500 cells/μl per month; stable disease (SD) patients as lymphocyte count increment <500 cells/μl per month]. As expected, PD CLL was accompanied by increasing numbers of CD3+, CD4+, CD8+ and CD16+CD56+ cells. In contrast, this was not the case for SD CLL patients (Fig. 1b). The mean increment of accessory CD3+ T cells over time in PD patients was 111.3 cells/μl per month compared to −0.5 cells/μl per month in SD patients. This dramatic difference was also apparent in the CD8+ subset of T cells: the mean CD8+ T-cell increment over time in PD patients was more than 30-fold higher than in SD patients (Fig. 1b). Furthermore, the CD3+, CD4+ and CD8+ cellular increments in the PD patient cohort were significantly higher in CD38+ than in CD38− CLL (Fig. 1c). In light of the fact that lymphocyte increments between CD38+ (mean ± STD, 3,970 ± 2,811 cells/μl per month) and CD38− (3,135 ± 3,262 cells/μl per month) PD patients did not differ significantly (P = 0.31), the finding of differing accessory lymphocyte increments per time in CD38− and CD38+ disease is even more striking. This observation prompted us to reevaluate the absolute numbers of accessory lymphocytes in relation to both Binet stage and CD38 expression. This analysis revealed that the accelerated dynamics of CD38+ CLL T cells are restricted to Binet stage A patients (Fig. 1d). In summary, we demonstrated on the level of individual patients that an increasing leukemic burden is associated with the expansion of non-malignant lymphocyte populations.
Gene expression profiling and subsequent gene set enrichment analysis reveal an expanded CD8+ effector memory T-cell subset in CLL
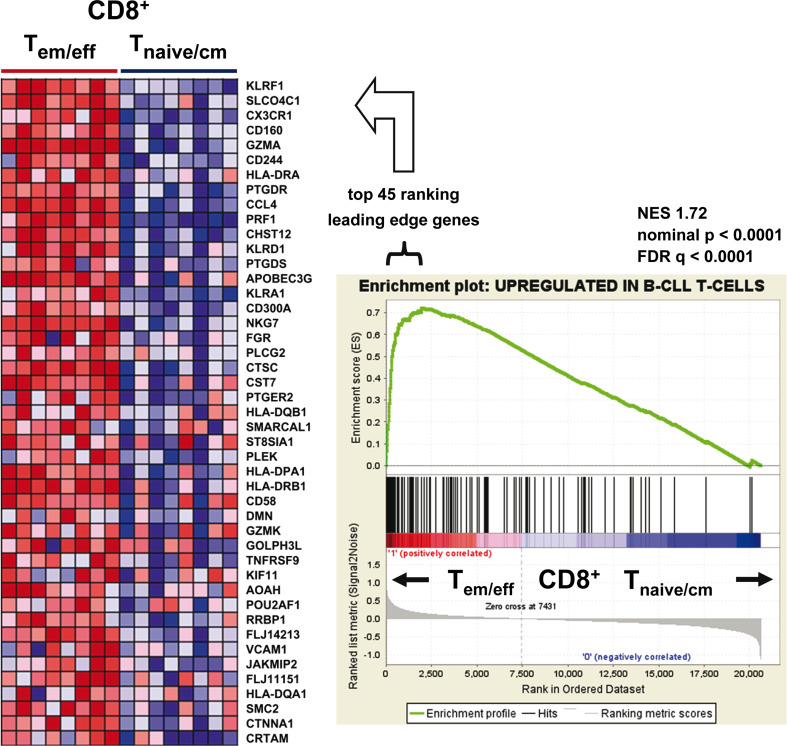
In order to gain mechanistic insight into the altered biology of CLL T cells, we compared the gene expression profiles of CLL T cells (n = 25 patients) with normal T cells (n = 8 normal donors). The SAM algorithm controlling the false discovery rate (FDR) at 1 % and using ±2.0 as fold change cut-off identified a list of 135 genes (defined by 150 probes) that had significantly higher expression and 11 genes (defined by 13 probes) that had significantly decreased expression in CLL T cells (Table S3). Notably, the genes with higher expression in CLL T cells included killer cell lectin-like receptor family members KLRA1, KLRC2, KLRD1 (CD94), KLRK1 and KLRF1 as well as the inhibitory receptors CD244 (NK-cell receptor 2B4), CD160 (NK-cell receptor BY55), CCL4, PRF1 (perforin 1) and CRTAM (class-I MHC-restricted T-cell-associated molecule). Because these receptors and genes are commonly expressed by CD8+ T cells exhibiting an effector memory phenotype [20–25], we hypothesized expansion of this subset in CLL. In agreement with this assumption, the list of genes with higher expression in CLL T cells was associated with biological processes implicated in immune response and antigen processing and presenting (Table S4). In order to verify our hypothesis of expanded CLL effector memory CD8+ T cells, we used gene set enrichment analysis (GSEA) to test for enrichment of our highly expressed CLL T-cell genes in a publicly available gene signatures derived from CD8+ effector and effector memory T cells [14]. To perform GSEA, the available data sets were grouped into effector memory (T em) and effector (T eff) versus naïve (T naïve) and central memory (T cm) CD8+ phenotype. Strikingly, the genes with higher expression in CLL T cells were enriched in the combined T em/T eff CD8+ T-cell gene signature (Fig. 2). The receptors KLRF1 (Nkp80), CD160, CD244, KLRD1 and KLRA1 were among the top ranking leading edge genes, suggesting an important role of these receptors in regulating CLL CD8+ T-cell function (Fig. 2). In conclusion, GEP analysis of CLL in comparison with normal donor T cells suggested expansion of CD8+ T em/T eff cells in CLL.
Fig. 2.
Genes with higher expression in CLL T cells are significantly enriched within gene signatures of CD8+ effector memory and terminal effector T cells. Gene set enrichment analysis (GSEA) showing enrichment of genes with higher expression in CLL T cells in published gene signatures of sorted normal donor CD8+ effector memory (T em)/terminal effector (T eff) T-cell populations (n = 8) in comparison with those of sorted CD8+ naive (T naive)/central memory (T cm) T-cell populations (n = 8) [14]. The GSEA enrichment plot (right) and the heat map of the top ranking leading edge genes (left) are shown. In the GSEA plot, the x-axis represents the ranked list of genes from the highest to the lowest probe signal between T em/eff versus T naive/cm CD8+ T-cell populations. The black lines relate to the position of genes with higher expression in CLL T cells. The height of the green enrichment curve illustrates the degree of enrichment. A strong correlation was observed between normal donor T em/eff genes and the CLL T-cell gene signature. In the heat map (left), signal intensities of leading edge genes (left) are shown by shades of red (upregulation) and blue (downregulation). NES normalized enrichment score; FDR false discovery rate
The CLL T-cell compartment is characterized by an expanded CD8+ effector memory subset
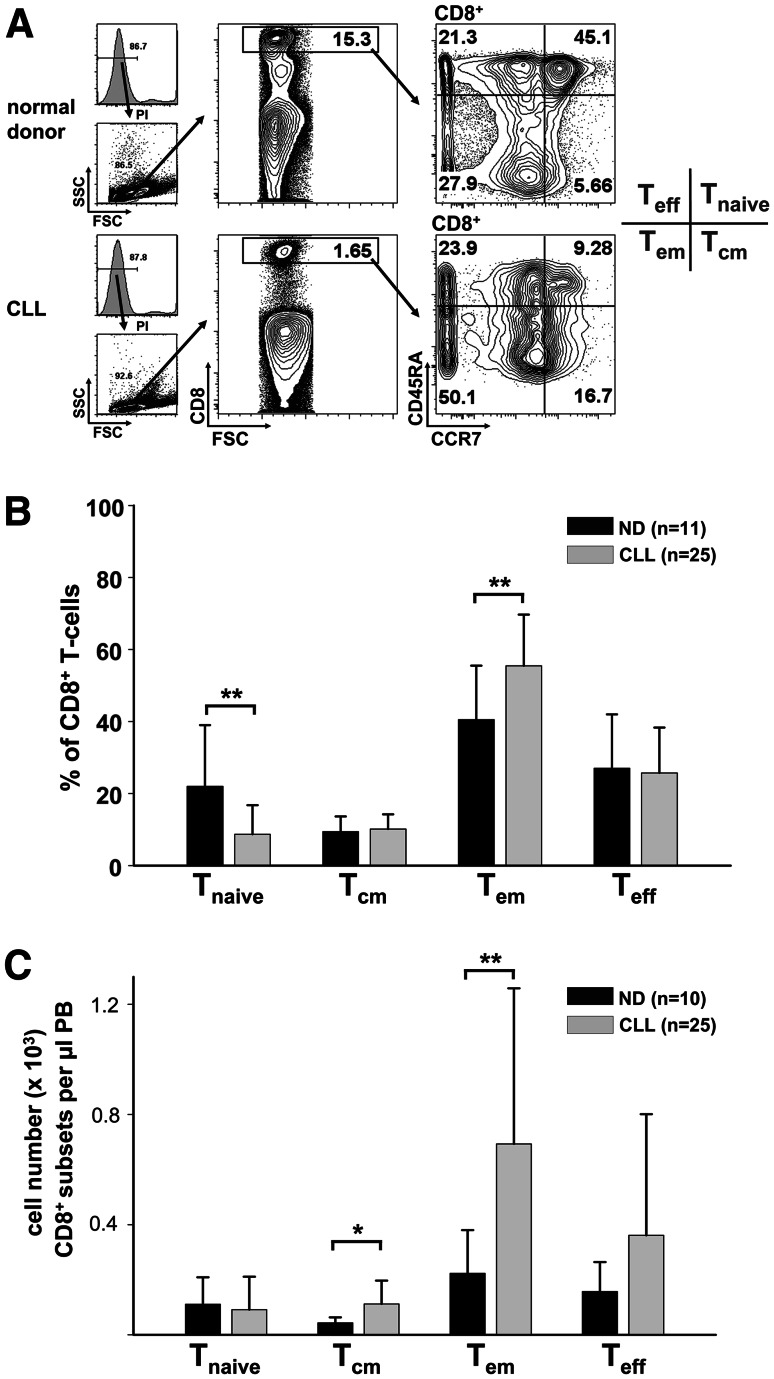
The CD8+ T-cell compartment can be subdivided into T naive (CCR7+CD45RA+), T cm (CCR7+CD45RA), T em (CCR7−CD45RA−) and T eff (CCR7−CD45RA+) cells by flow cytometric analysis [26]. In contrast to the CD8+ T naive and T cm subsets, CD8+ T em and T eff cells do not express lymph node homing molecules such as CD62L and CCR7. CD8+ T em and T eff cells directly populate inflammatory sites [27–30]. To confirm our GEP and GSEA data, we analyzed CLL and normal donor CD8+ T-cell subsets. Flow cytometric analysis revealed a relative shift of subsets within the CD8+ T-cell compartment. We observed a decreased proportion of CD8+ T naive cells and an increased proportion of CD8+ T em cells in the CLL cohort as compared to normal donor controls (Fig. 3a, b). However, when absolute cell numbers were calculated, it became evident that the increased number of overall CD8+ T cells in CLL was primarily attributable to the expansion of CD8+ T em cells (Fig. 3c). However, no significant difference of circulating CD8+ T naive cells between CLL and normal donors was noted (Fig. 3c). Taken together, these results confirmed our GEP-driven hypothesis of an expanded CD8+ T em compartment in CLL.
Fig. 3.
The CD8+ effector memory T-cell compartment of CLL patients is expanded. a Peripheral blood mononuclear cells were stained with antibodies against CD8, CD45RA, CCR7 and with propidium iodide (PI). Subsets of CD8+ T cells were defined as naive (T naive; CD45RA+, CCR7+), central memory (T cm; CD45RA−, CCR7+), effector memory (T em, CD45RA−, CCR7−) and terminal effector (T eff, CD45RA+,CCR7−) cells. Representative plots of normal donor (ND) and CLL samples are shown. b Quantification of the flow cytometric CD8+ T-cell subset analysis. The relative proportion of naive CD8+ T cells is decreased, while the relative proportion of CD8+ T em cells is increased. c Absolute cell numbers of CLL CD8+ T cm and T em cells were increased. FSC forward scatter; SSC side scatter; PB peripheral blood. *P < 0.05; **P < 0.01
The expansion of a senescent KLRG1+ cellular subset is responsible for increased numbers of CD8+ T cells in CLL
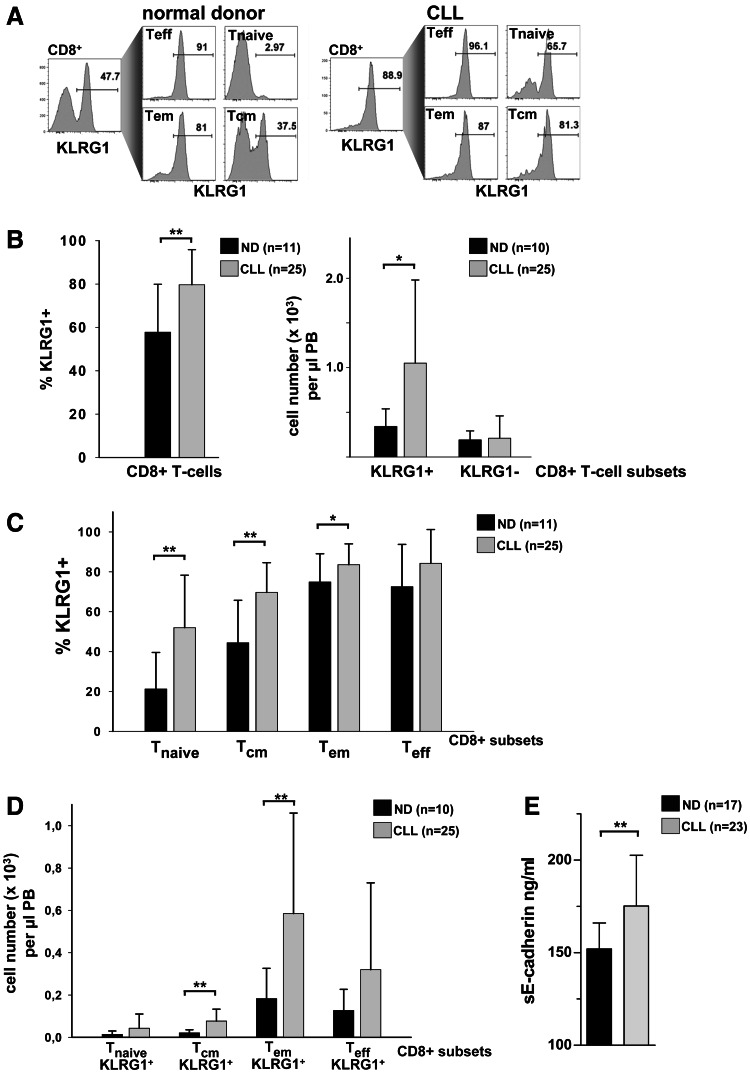
Genes found to be higher expressed in CLL T cells versus normal-donor-derived T cells included inhibitory receptors such as CD160 [20] and CD244 [21] (Fig. 2; Table S3). Importantly, the regulation of CD8+ T-cell exhaustion by various inhibitory receptors was shown to be non-redundant [31]. Another critical inhibitory receptor is killer cell lectin-like receptor G1 (KLRG1). This receptor was shown to mark CD8+ T cells, which have undergone extensive proliferation and lack replicative potential [15, 32–34]. Therefore, we examined KLRG1 surface expression on CD8+ T-cell subsets in CLL patients and in normal individuals (Fig. 4a). The proportion of cells expressing KLRG1 was found to be significantly higher in CLL CD8+ T cells compared to normal donor CD8+ T cells (Fig. 4b). When absolute cell numbers were calculated, it became evident that the elevation in the absolute number of overall CD8+ T cells in CLL was mainly attributed to the expansion of KLRG1+CD8+ T cells, while the number of KLRG1−CD8+ T cells did not differ significantly between the two groups (Fig. 4b). When analyzing CD8+ T-cell subsets for KLRG1 expression (Fig. 4a), we detected increased proportions of KLRG1+ cells in CD8+ T naive, T cm and T em subsets (Fig. 4c). However, only absolute KLRG1+CD8+ T cm and T em cell numbers were significantly increased in CLL patients (Fig. 4d). Comparable to our findings in CLL, Streeck et al. reported elevated KLRG1 expression of CD8+ T cells in patients with chronic progressive HIV infection. Additionally, the data of Streeck et al. [35] suggested that elevated plasma levels of the KLRG1 ligand, soluble E-cadherin (sE-cadherin), were responsible for inhibited effector functions of KLRG1+CD8+ T cells. We therefore also determined plasma sE-cadherin levels in our cohort of CLL patients. Strikingly, we detected significantly higher sE-cadherin levels in CLL patients compared to normal donor controls (Fig. 4e). In summary, the CLL T-cell compartment is characterized by elevated numbers of CD8+ T em cells expressing the inhibitory receptor KLRG1. Moreover, elevated CLL plasma sE-cadherin levels imply increased CD8+ T em functional inhibition via KLRG1 in CLL patients.
Fig. 4.
The high numbers of CD8+ T cells in CLL are due to an expansion of KLRG1+ cellular subsets. a KLRG1 expression within total CD8+ T cells and within CD8+ subsets according to the gating strategy depicted in Fig. 3 a. Representative plots of a normal donor and a CLL sample are shown. b The proportion of KLRG1+ cells within CD8+ T cells was compared between CLL patients and normal donor controls (left panel). Absolute numbers of KLRG1+CD8+ T cells were increased in the peripheral blood (PB) of CLL patients compared to normal donors (ND), while the numbers of KLRG1−CD8+ cells did not differ between the two groups (right panel). c Significantly increased proportions of KLRG1+ cells were found in CD8+ T naïve, T cm and T em cell subsets. d KLRG1+CD8+ T cm and T eff cell numbers were significantly increased in CLL patients compared to normal donors. e CLL plasma levels of soluble E-cadherin (sE-cadherin), ligand of KLRG1, are increased compared to normal donors. ND normal donor.*P < 0.05; **P < 0.01
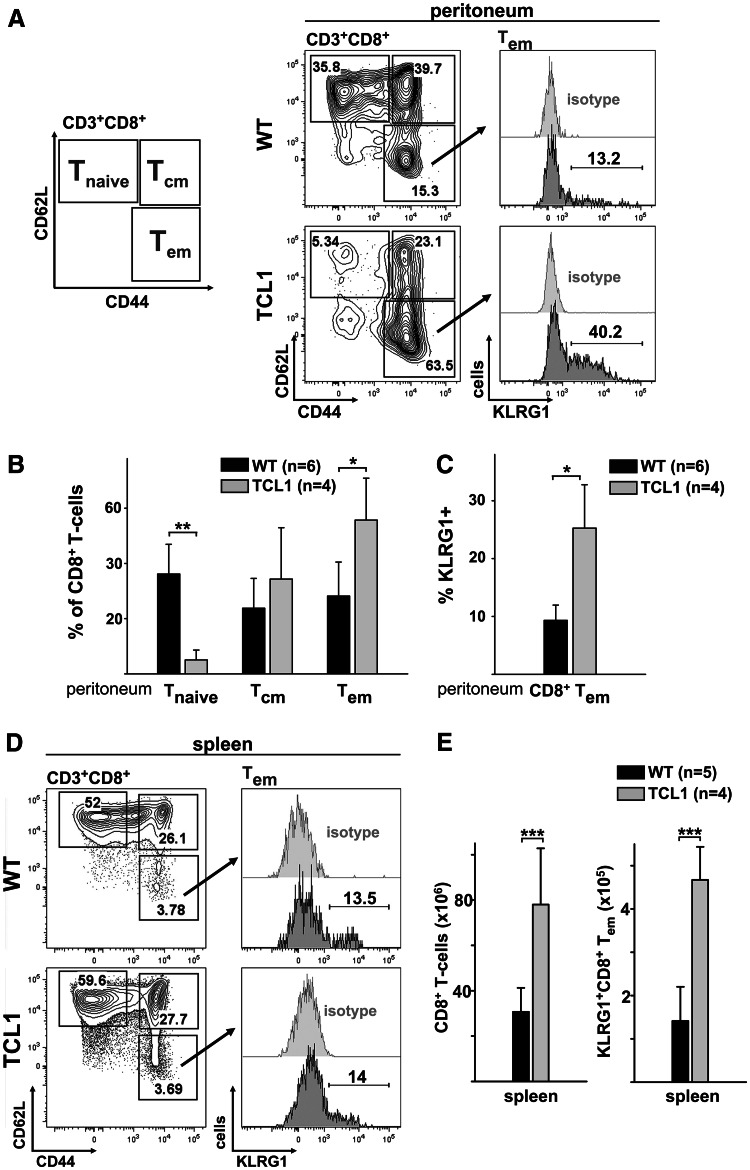
Murine CLL is characterized by expanded KLRG1+CD8+ effector memory T cells
In mice, the role of KLRG1 as a regulator as well as a marker of CD8+ T-cell senescence is well established [34, 36]. Therefore, we used a CLL murine model (Eμ-TCL1 [37]) to investigate whether the expansion of KLRG1+CD8+ T cells is also a hallmark of CLL in another mammalian system. Previous studies emphasized parallels between the TCL1 transgenic CLL mouse model and human CLL, which also included the non-malignant T-cell compartment [7, 8, 37]. TCL1 mice develop CLL at 8–12 months of age with preleukemic hyperplasia of CD5+ B cells developing as early as 2 and 4 months, respectively [37]. Since we observed CD8+ T-cell expansions already at early human CLL stages, we decided to analyze 7-month-old TCL1 mice. First, we confirmed CD5+CD19+ cell expansion in the peritoneal cavity and spleen of TCL1 transgenic mice (Figure S1A, B and C). Using a PCR for Ig heavy chain rearrangement, we were able to detect an emerging dominant clone in two of four TCL1 mice (Figure S1D). In parallel to the CD8+ T-cell subpopulations described for the human system, the murine CD8+ T-cell compartment can be subdivided into T naive, T cm and T em subsets (Fig. 5a). In concordance with our observations in human CLL PB, we detected a distinctly increased proportion of peritoneal CD8+ T em cells in TCL1 mice mainly at the expense of CD8+ T naive cells (Fig. 5a, b). Remarkably, the relative expansion of peritoneal CD8+ T em cells in TCL1 mice was also associated with an increased proportion of KLRG1-expressing cells (Fig. 5c). In contrast to the peritoneal cavity, no obvious shift within the CD8+ T-cell subpopulation was detectable in the spleen at this early disease stage (Fig. 5d). However, we observed a more than 2.5-fold increase in absolute splenic CD8+ T-cell numbers in TCL1 compared to wild-type mice (Fig. 5e). Again, in parallel to our observations in humans, numbers of KLRG1+CD8+ T em cells in the spleen were significantly increased in emerging mouse CLL (Fig. 5e). In synopsis, our human and murine data imply that expanded CLL CD8+ T cells are driven into a non-replication-competent KLRG1+ T em phenotype.
Fig. 5.
Peritoneal cavity and spleen CD8+ T cells of TCL1 transgenic mice are shifted toward a KLRG1+ effector memory phenotype. a Representative flow cytometric analysis of wild-type versus TCL1 peritoneal CD8+ T-cell subsets is displayed. Definition of murine CD8+ T-cell subsets: naïve (T naive: CD62L+, CD44low), central memory (T cm: CD62L+, CD44+) and effector memory (T em: CD62Llow, CD44+). Within the CD8+ T em compartment, KLRG1 expression was analyzed. b Compared to wild-type mice, the peritoneal CD8+ T-cell compartment of TCL1 transgenic mice was characterized by a decreased fraction of naive cells and an increased fraction of effector memory cells. c Within peritoneal TCL1 CD8+ T em compartment, the proportion of KLRG1+ cells was significantly increased. d Representative flow cytometric analysis of splenic CD8+ T-cell subsets. e TCL1 transgenic mice displayed a significant increase in CD8+ T-cell numbers (left). Significantly elevated KLRG1+CD8+ T em cell numbers were present in the spleens of TCL1 transgenic mice (right). *P < 0.05; **P < 0.01; ***P < 0.001. WT wild-type
Discussion
In this study, we initially combined clinical multicolor flow cytometry with microarray-based GEP analyses to investigate abnormalities in the T-cell compartment of CLL patients. Consistent with previous work, enumeration of PB lymphocyte subsets by flow cytometry revealed elevated numbers of circulating CD3+, CD4+, CD8+ and CD16+CD56+ lymphocytes depending on the disease stage: samples derived from patients with advanced disease contained higher numbers of these cells as compared to Binet A patients. This observation suggested that the expansion of non-malignant lymphocytes may be causally related to disease progression. In a previously unreported fashion, we further addressed this concept by sequentially characterizing the T-cell subsets and NK cells in a cohort of 46 patients over a mean time span of 22 months, showing that CLL progression was associated with an increase in accessory T cells. Remarkably, the mean increment of T cells over time in progressive CLL patients was more than 100-fold higher than in stable disease patients. Taken together, these results support the hypothesis that disease progression triggers an expansion of the CLL T-cell compartment in individual patients.
As another novel finding, we report that the absolute numbers of T and NK cells also correlated with the risk factor profile in that patients expressing the CD38 (>20 %) surface antigen showed higher circulating numbers of CD3+, CD4+, CD8+ T cells. In addition, the kinetics of T cell expansion appeared to differ in CD38− versus CD38+ patients in that the latter group responded more quickly showing significantly higher numbers of accessory T cells even in early Binet stage A. Strikingly, we found that in progressive CD38+ CLL patients, the mean increment of T cells over time was threefold higher than in progressive CD38− CLL patients. These data imply that expanding CD38+ leukemic cells are able to provoke a more dramatic accessory lymphocyte response than expanding CD38− leukemic cells.
Microarray-based GEP was used in a subset of 25 patients to further investigate the molecular mechanisms underlying the deregulation of the CLL T-cell compartment. Different from the study published by Gorgun et al. [5] who compared the transcriptomes of CD4+ and CD8+ T cells isolated from CLL patients and healthy blood donors, we employed immunomagnetically purified CD3+ T cells that were not further separated according to CD4 and CD8 expression. Comparative supervised analysis of normal donor- versus CLL-derived CD3+ T cells revealed a panel of 146 differentially expressed genes, the majority of which were found to be overexpressed in CLL T cells. A very recently published study used a similar approach comparing the gene expression profiles of PB CD3+ T cells of CLL, multiple myloma and normal donors [38]. As expected, numerous genes with high expression in CLL T cells including CX3CR1, CRTAM, KLRD1, KLRA1, TNFRSF9, CCL4 and GZMA identified by our GEP study were also found to be highly expressed in CLL T cells by Kiaii et al. [38]. In addition to the killer cell lectin-like receptors identified in our GEP analysis (KLRA1, KLRD1, KLRC2, KLRF1 and KLRK1), Kiaii et al. found KLRG1, KLRC3 and KLRC4 to be highly expressed by CLL T cells. The high CLL T-cell expression of killer cell lectin-like receptors might be part of the previously recognized broad upregulation of NK receptors by CLL T cells [39]. However, it remains unclear why the high expression of killer cell lectin-like receptors was not described by other CLL T-cell GEP studies [5].
We used GSEA to demonstrate that a previously published CD8+ T em/T eff T-cell signature [14] was significantly enriched within our differentially expressed CLL T-cell genes. The top leading edge gene of this analysis was KLRF1 (Fig. 2). Strikingly, we and others demonstrated that the KLRF1 ligand, CLEC2B (AICL), represents one of the top genes highly expressed in poor compared to good prognosis CLL [40, 41]. As it was shown that the KLRF1–CLEC2B interaction is capable of mediating mutual cellular activation [42], this molecular interface might represent a mechanism how CD8+ T em cells exert a propagating impact on CLL leukemic cells.
On the basis of these microarray data, we hypothesized that CLL cells not only cause an expansion of the accessory T cells but also induce a shift in their cellular composition toward a CD8+ T em phenotype. To test this hypothesis and to further validate our microarray data at the cellular level, we proceeded to perform a detailed flow cytometric analysis of the CLL CD8+ T-cell compartment. In line with the microarray data, this analysis revealed a significantly higher relative number of CD8+ T em cells and a lower relative number of CD8+ T naïve cells in the CLL cohort as compared to healthy controls. In order to test whether our observations in human CLL can be recapitulated in a murine transgenic CLL model, we studied the CD8+ T-cell compartment of 7-month-old TCL1 transgenic mice. We specifically studied how CD8+ T-cell subsets respond to arising CLL in the peritoneal cavity of TCL1 transgenic mice. Strikingly, we found that our observation of T em shifted CD8+ T cells in human CLL was phenocopied in the peritoneal cavity of TCL1 transgenic mice. Using a slightly different marker combination than our study, Hofbauer et al. [7] also described pronounced skewing toward a CD8+ T em phenotype in TCL1 transgenic mice.
The expansion of CD8+ T em cells in CLL supports a model in which the production of these cells is driven by the chronic stimulation provided by the leukemic cells. This cellular pathway parallels the increased production of CD8+ T em cells during chronic viral infection or repetitive vaccination [43, 44]. The relative expansion of the CLL CD8+ T em subset was previously noted in the context of a study by Davis and colleagues, which investigated the expression of Fc receptor-like 6 (FCRL6) protein on CLL cells and accessory lymphocyte subsets [45]. Our data corroborate the finding of a relative CD8+ T em cells expansion in CLL and extend this observation by demonstrating an absolute increase in these cells in the PB of CLL patients compared to normal donors. Greil and colleagues reported complementary data to our work for CD4+ T-cell subsets. Analogous to our study involving CD8+ T cells, a relative increase in CD4+ T em cells and a relative decrease in CD4+ T naive cells were reported comparing unmutated and mutated CLL. However, in contrast to our study, CLL CD4+ subsets were not compared with normal donor controls [46]. Assuming that absolute CD4+ T-cell numbers of CLL patients were increased compared to normal donor controls in the study by Greil and colleagues, one could conclude that CLL CD4+ and CD8+ T-cell subsets are skewed in a uniform direction toward a T em phenotype. This concordant behavior of CLL CD4+ and CD8+ T cells is further supported by our notion that an incremental CD4+ compartment correlates with an incremental CD8+ T-cell compartment in a significant manner (Table S2).
KLRG1 has been reported previously to be selectively expressed in murine and human CD8+ T em and T eff cells that have undergone extensive proliferation and lack replicative potential in the CD8 compartment [15, 33, 34]. Recent studies have shown that murine KLRG1 exerts a significantly weaker inhibitory signal than human KLRG1 [47, 48]. However, in CLL, our murine data recapitulate our human data of an expanded CLL CD8+ T em compartment with an increased proportion of KLRG1+ cells. The known ligands for KLRG1 are E-, N- and R-cadherin [49]. Comparable to our findings in CLL, Streeck et al. [35] demonstrated increased KLRG1+CD8+ T cells in chronic progressive HIV-1 patients. In parallel, these authors detected elevated levels of sE-cadherin plasma levels in these patients most likely caused by disrupted intestinal epithelium associated with HIV-1 infection. Strikingly, exposure to sE-cadherin in vitro impaired cytokine secretion and anti-viral response of KLRG1+CD8+ T cells. This study prompted us to study plasma sE-cadherin levels in our cohort of CLL patients, and indeed, we were able to detect significantly elevated sE-cadherin levels in CLL patients compared to normal donor controls. Therefore, we propose that systemically elevated sE-cadherin levels could contribute to increased functional inhibition of KLRG1+CD8+ T cells in CLL.
The expansion of CLL CD8+ T cells was previously attributed to chronic antigenic stimulation from viral pathogens such as CMV [50]. Furthermore, the CMV seroprevalence was shown to be higher in CLL patients than in healthy controls [51]. Therefore, we cannot rule out that CMV contributed the human CLL CD8+ T-cell alterations observed in our study. However, the validation of our findings in the CMV-free murine TCL1 system argues against this notion.
A number of previous studies support the concept of a particular role of CD8+ T cells for CLL biology. In particular, CLL CD8+ T cells were superior to CD4+ T cells in promoting survival of CLL cells in vitro [38]. Furthermore, when early-stage CLL CD8+ T-cell numbers were exceeding CD4+ T-cell numbers, this was associated with an inferior clinical course [52].
In summary, we show here that in CLL, an expansion of the peripheral leukemic cells is associated with a concomitant expansion of non-malignant peripheral T and NK lymphocytes. This implies that accessory lymphoid cells are expanding in response to leukemia progression and possibly contribute to the survival of CLL cells. Furthermore, we show that in human and rodent, CLL-expanding CD8+ T cells are driven into a senescent KLRG1+ T em phenotype. Additionally, inhibitory KLRG1 signaling in CD8+ T em cells might be potentiated by elevated CLL sE-cadherin levels. We argue for a model where expanded senescent CD8+ T em cells are an important component of the CLL supporting microenvironment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Ute Schmücker and Anja Führer for excellent technical assistance and Brigitte Fischer for help with compiling patient data. We are grateful to Carlo M. Croce for his permission to use Eμ-TCL1 transgenic mice. We thank Günter Fingerle-Rawson for providing Eμ-TCL1 transgenic mice. We are grateful to Peter Horn, Marc Seifert and Ralf Küppers for assistance organizing normal donor blood samples. This work was supported by a grant to J.D. from the Ministerium für Schule, Wissenschaft und Forschung des Landes Nordrhein-Westfalen. J.R.G. receives grant support by the Kompetenznetzwerk Stammzellforschung Nordrhein-Westfalen.
Conflict of interest
The authors declare no competing financial interest.
References
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R, Chum P, Yan XJ, Allen SL, Kolitz JE, Baskar S, Rader C, Mellstedt H, Rabbani H, Lee A, Gregersen PK, Rai KR, Chiorazzi N. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood. 2011;117(20):5463–5472. doi: 10.1182/blood-2010-12-324210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood. 2009;114(16):3367–3375. doi: 10.1182/blood-2009-06-225326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellstedt H, Choudhury A. T and B cells in B-chronic lymphocytic leukaemia: faust, Mephistopheles and the pact with the Devil. Cancer Immunol Immunother. 2006;55(2):210–220. doi: 10.1007/s00262-005-0675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorgun G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG. Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest. 2005;115(7):1797–1805. doi: 10.1172/JCI24176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, Byrd JC, Gribben JG. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008;118(7):2427–2437. doi: 10.1172/JCI35017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofbauer JP, Heyder C, Denk U, Kocher T, Holler C, Trapin D, Asslaber D, Tinhofer I, Greil R, Egle A. Development of CLL in the TCL1 transgenic mouse model is associated with severe skewing of the T-cell compartment homologous to human CLL. Leukemia. 2011;25(9):1452–1458. doi: 10.1038/leu.2011.111. [DOI] [PubMed] [Google Scholar]
- 8.Gorgun G, Ramsay AG, Holderried TA, Zahrieh D, Le Dieu R, Liu F, Quackenbush J, Croce CM, Gribben JG. Emu-TCL1 mice represent a model for immunotherapeutic reversal of chronic lymphocytic leukemia-induced T-cell dysfunction. Proc Natl Acad Sci USA. 2009;106(15):6250–6255. doi: 10.1073/pnas.0901166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth A, de Beer D, Nuckel H, Sellmann L, Duhrsen U, Durig J, Baerlocher GM. Significantly shorter telomeres in T-cells of patients with ZAP-70+/CD38+ chronic lymphocytic leukaemia. Br J Haematol. 2008;143(3):383–386. doi: 10.1111/j.1365-2141.2008.07363.x. [DOI] [PubMed] [Google Scholar]
- 10.Rezvany MR, Jeddi-Tehrani M, Osterborg A, Kimby E, Wigzell H, Mellstedt H. Oligoclonal TCRBV gene usage in B-cell chronic lymphocytic leukemia: major perturbations are preferentially seen within the CD4 T-cell subset. Blood. 1999;94(3):1063–1069. [PubMed] [Google Scholar]
- 11.Rezvany MR, Jeddi-Tehrani M, Wigzell H, Osterborg A, Mellstedt H. Leukemia-associated monoclonal and oligoclonal TCR-BV use in patients with B-cell chronic lymphocytic leukemia. Blood. 2003;101(3):1063–1070. doi: 10.1182/blood-2002-03-0746. [DOI] [PubMed] [Google Scholar]
- 12.Durig J, Nuckel H, Huttmann A, Kruse E, Holter T, Halfmeyer K, Fuhrer A, Rudolph R, Kalhori N, Nusch A, Deaglio S, Malavasi F, Moroy T, Klein-Hitpass L, Duhrsen U. Expression of ribosomal and translation-associated genes is correlated with a favorable clinical course in chronic lymphocytic leukemia. Blood. 2003;101(7):2748–2755. doi: 10.1182/blood-2002-09-2683. [DOI] [PubMed] [Google Scholar]
- 13.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willinger T, Freeman T, Hasegawa H, McMichael AJ, Callan MF. Molecular signatures distinguish human central memory from effector memory CD8 T cell subsets. J Immunol. 2005;175(9):5895–5903. doi: 10.4049/jimmunol.175.9.5895. [DOI] [PubMed] [Google Scholar]
- 15.Voehringer D, Koschella M, Pircher H. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1) Blood. 2002;100(10):3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]
- 16.Usherwood EJ, Hogan RJ, Crowther G, Surman SL, Hogg TL, Altman JD, Woodland DL. Functionally heterogeneous CD8+ T-cell memory is induced by Sendai virus infection of mice. J Virol. 1999;73(9):7278–7286. doi: 10.1128/jvi.73.9.7278-7286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gattinoni L, Klebanoff CA, Palmer DC, Wrzesinski C, Kerstann K, Yu Z, Finkelstein SE, Theoret MR, Rosenberg SA, Restifo NP. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J Clin Invest. 2005;115(6):1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gattinoni L, Zhong XS, Palmer DC, Ji Y, Hinrichs CS, Yu Z, Wrzesinski C, Boni A, Cassard L, Garvin LM, Paulos CM, Muranski P, Restifo NP. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15(7):808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts AD, Ely KH, Woodland DL. Differential contributions of central and effector memory T cells to recall responses. J Exp Med. 2005;202(1):123–133. doi: 10.1084/jem.20050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai G, Freeman GJ. The CD160, BTLA, LIGHT/HVEM pathway: a bidirectional switch regulating T-cell activation. Immunol Rev. 2009;229(1):244–258. doi: 10.1111/j.1600-065X.2009.00783.x. [DOI] [PubMed] [Google Scholar]
- 21.McNerney ME, Lee KM, Kumar V. 2B4 (CD244) is a non-MHC binding receptor with multiple functions on natural killer cells and CD8+ T cells. Mol Immunol. 2005;42(4):489–494. doi: 10.1016/j.molimm.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 22.Kuttruff S, Koch S, Kelp A, Pawelec G, Rammensee HG, Steinle A. NKp80 defines and stimulates a reactive subset of CD8 T cells. Blood. 2009;113(2):358–369. doi: 10.1182/blood-2008-03-145615. [DOI] [PubMed] [Google Scholar]
- 23.Tsujimura K, Obata Y, Matsudaira Y, Nishida K, Akatsuka Y, Ito Y, Demachi-Okamura A, Kuzushima K, Takahashi T. Characterization of murine CD160+ CD8+ T lymphocytes. Immunol Lett. 2006;106(1):48–56. doi: 10.1016/j.imlet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Arase N, Takeuchi A, Unno M, Hirano S, Yokosuka T, Arase H, Saito T. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int Immunol. 2005;17(9):1227–1237. doi: 10.1093/intimm/dxh299. [DOI] [PubMed] [Google Scholar]
- 25.Vivier E, Anfossi N. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat Rev Immunol. 2004;4(3):190–198. doi: 10.1038/nri1306. [DOI] [PubMed] [Google Scholar]
- 26.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291(5512):2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 28.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 29.Weninger W, Crowley MA, Manjunath N, von Andrian UH. Migratory properties of naive, effector, and memory CD8+ T cells. J Exp Med. 2001;194(7):953–966. doi: 10.1084/jem.194.7.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4(3):225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 31.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA, Wherry EJ. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henson SM, Franzese O, Macaulay R, Libri V, Azevedo RI, Kiani-Alikhan S, Plunkett FJ, Masters JE, Jackson S, Griffiths SJ, Pircher HP, Soares MV, Akbar AN. KLRG1 signaling induces defective Akt (ser473) phosphorylation and proliferative dysfunction of highly differentiated CD8+ T cells. Blood. 2009;113(26):6619–6628. doi: 10.1182/blood-2009-01-199588. [DOI] [PubMed] [Google Scholar]
- 33.Voehringer D, Blaser C, Brawand P, Raulet DH, Hanke T, Pircher H. Viral infections induce abundant numbers of senescent CD8 T cells. J Immunol. 2001;167(9):4838–4843. doi: 10.4049/jimmunol.167.9.4838. [DOI] [PubMed] [Google Scholar]
- 34.Heffner M, Fearon DT. Loss of T cell receptor-induced Bmi-1 in the KLRG1+ senescent CD8+ T lymphocyte. Proc Natl Acad Sci USA. 2007;104(33):13414–13419. doi: 10.1073/pnas.0706040104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Streeck H, Kwon DS, Pyo A, Flanders M, Chevalier MF, Law K, Julg B, Trocha K, Jolin JS, Anahtar MN, Lian J, Toth I, Brumme Z, Chang JJ, Caron T, Rodig SJ, Milner DA, Jr, Piechoka-Trocha A, Kaufmann DE, Walker BD, Altfeld M. Epithelial adhesion molecules can inhibit HIV-1-specific CD8+ T-cell functions. Blood. 2011;117(19):5112–5122. doi: 10.1182/blood-2010-12-321588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205(3):625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, Russo G, Hardy RR, Croce CM. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiaii S, Kokhaei P, Mozaffari F, Rossmann E, Pak F, Moshfegh A, Palma M, Hansson L, Mashayekhi K, Hojjat-Farsangi M, Osterborg A, Choudhury A, Mellstedt H. T cells from indolent CLL patients prevent apoptosis of leukemic B cells in vitro and have altered gene expression profile. Cancer Immunol Immunother. 2013;62(1):51–63. doi: 10.1007/s00262-012-1300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Junevik K, Werlenius O, Hasselblom S, Jacobsson S, Nilsson-Ehle H, Andersson PO. The expression of NK cell inhibitory receptors on cytotoxic T cells in B-cell chronic lymphocytic leukaemia (B-CLL) Ann Hematol. 2007;86(2):89–94. doi: 10.1007/s00277-006-0198-x. [DOI] [PubMed] [Google Scholar]
- 40.Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, Freedman A, Inghirami G, Cro L, Baldini L, Neri A, Califano A, La-Favera R. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194(11):1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttmann A, Klein-Hitpass L, Thomale J, Deenen R, Carpinteiro A, Nuckel H, Ebeling P, Fuhrer A, Edelmann J, Sellmann L, Duhrsen U, Durig J. Gene expression signatures separate B-cell chronic lymphocytic leukaemia prognostic subgroups defined by ZAP-70 and CD38 expression status. Leukemia. 2006;20(10):1774–1782. doi: 10.1038/sj.leu.2404363. [DOI] [PubMed] [Google Scholar]
- 42.Welte S, Kuttruff S, Waldhauer I, Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nat Immunol. 2006;7(12):1334–1342. doi: 10.1038/ni1402. [DOI] [PubMed] [Google Scholar]
- 43.Powell DJ, Jr, Rosenberg SA. Phenotypic and functional maturation of tumor antigen-reactive CD8+ T lymphocytes in patients undergoing multiple course peptide vaccination. J Immunother. 2004;27(1):36–47. doi: 10.1097/00002371-200401000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Champagne P, Ogg GS, King AS, Knabenhans C, Ellefsen K, Nobile M, Appay V, Rizzardi GP, Fleury S, Lipp M, Forster R, Rowland-Jones S, Sekaly RP, McMichael AJ, Pantaleo G. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410(6824):106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 45.Schreeder DM, Pan J, Li FJ, Vivier E, Davis RS. FCRL6 distinguishes mature cytotoxic lymphocytes and is upregulated in patients with B-cell chronic lymphocytic leukemia. Eur J Immunol. 2008;38(11):3159–3166. doi: 10.1002/eji.200838516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tinhofer I, Weiss L, Gassner F, Rubenzer G, Holler C, Greil R. Difference in the relative distribution of CD4+ T-cell subsets in B-CLL with mutated and unmutated immunoglobulin (Ig) VH genes: implication for the course of disease. J Immunother. 2009;32(3):302–309. doi: 10.1097/CJI.0b013e318197b5e4. [DOI] [PubMed] [Google Scholar]
- 47.Grundemann C, Schwartzkopff S, Koschella M, Schweier O, Peters C, Voehringer D, Pircher H. The NK receptor KLRG1 is dispensable for virus-induced NK and CD8+ T-cell differentiation and function in vivo. Eur J Immunol. 2010;40(5):1303–1314. doi: 10.1002/eji.200939771. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann M, Schweier O, Pircher H. Different inhibitory capacities of human and mouse KLRG1 are linked to distinct disulfide-mediated oligomerizations. Eur J Immunol. 2012;42(9):2484–2490. doi: 10.1002/eji.201142357. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Hofmann M, Wang Q, Teng L, Chlewicki LK, Pircher H, Mariuzza RA. Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition. Immunity. 2009;31(1):35–46. doi: 10.1016/j.immuni.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mackus WJ, Frakking FN, Grummels A, Gamadia LE, De Bree GJ, Hamann D, van Lier RA, van Oers MH. Expansion of CMV-specific CD8+CD45RA+CD27− T cells in B-cell chronic lymphocytic leukemia. Blood. 2003;102(3):1057–1063. doi: 10.1182/blood-2003-01-0182. [DOI] [PubMed] [Google Scholar]
- 51.Steininger C, Rassenti LZ, Vanura K, Eigenberger K, Jager U, Kipps TJ, Mannhalter C, Stilgenbauer S, Popow-Kraupp T. Relative seroprevalence of human herpes viruses in patients with chronic lymphocytic leukaemia. Eur J Clin Invest. 2009;39(6):497–506. doi: 10.1111/j.1365-2362.2009.02131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nunes C, Wong R, Mason M, Fegan C, Man S, Pepper C. Expansion of a CD8+PD-1+ replicative senescence phenotype in early stage CLL patients is associated with inverted CD4:CD8 ratios and disease progression. Clin Cancer Res. 2012;18(3):678–687. doi: 10.1158/1078-0432.CCR-11-2630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.