Abstract
Antigens encoded by genes of the LAGE family, including LAGE-1 and NY-ESO-1, are of interest for cancer immunotherapy because they are tumor-specific and shared by tumors of different histological types. Several clinical trials are in progress with NY-ESO-1 peptides, protein, recombinant poxviruses, and dendritic cells pulsed with peptides. In this study, CD8 T lymphocytes from an individual without cancer were stimulated with dendritic cells infected with a recombinant avian poxvirus encoding a complete LAGE-1 protein. A CTL clone was isolated that recognized a new LAGE-1 peptide, ELVRRILSR, which corresponds to position 103–111 of the protein sequence. It is presented by HLA-A6801 molecules. When tumor cells expressing LAGE-1 were transfected with HLA-A68, they were lysed by the CTL clone, indicating that the peptide is processed in tumor cells. These results indicate that the LAGE-1.A68 peptide can be used for antitumoral vaccination. We observed also that specific T cells could be detected in a blood sample with a high sensitivity by using an A68/LAGE-1 fluorescent multimer.
Keywords: HLA-A68, LAGE-1, Peptide, Poxvirus, SNP, Tumor
Introduction
LAGE-1 and NY-ESO-1/LAGE-2 belong to the “cancer-germline” gene families, like the MAGE, BAGE, and GAGE genes. These “cancer-germline” genes are expressed in many tumors and in male germline cells but are silent in all other normal tissues [5, 11, 12, 32]. Male germline cells do not express MHC class I and class II molecules and are therefore incapable of presenting antigens to T cells [17]. “Cancer-germline” genes therefore encode tumor-specific shared antigens, which have been used in therapeutic vaccination trials of cancer patients [7, 8, 10, 23].
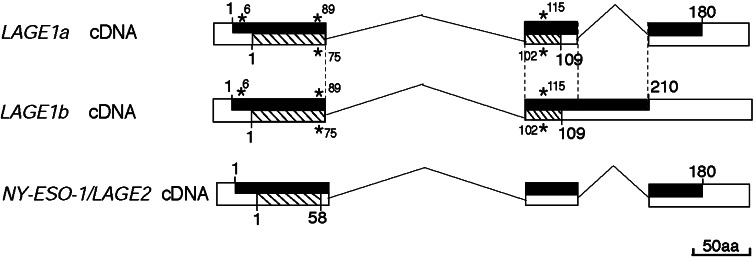
Two LAGE-1 transcripts have been described, namely, LAGE-1a and LAGE-1b (Fig. 1). LAGE-1b is incompletely spliced and codes for a putative protein of 210 aa, whereas the LAGE-1a gene product contains 180 aa [25]. An alternative reading frame is translated in both transcripts, producing a putative protein of 109 amino acids named CAMEL [1]. Expression of LAGE-1 was observed in many surgical tumor samples: 44% of bladder carcinomas, 33% of non-small-cell lung carcinomas, 29% of melanomas, 27% of head and neck cancers, and 25% of prostate adenocarcinomas [25]. LAGE-1 and NY-ESO-1/LAGE-2 are highly homologous proteins with up to 84% identity [9, 25].
Fig. 1.
Structure of the LAGE transcripts. Exons are represented as white boxes and spliced introns as broken lines. Open reading frames are represented as dark boxes for the major reading frame (ORF1), or as striped boxes for the second reading frame (ORF2). Amino acids are indicated with numbers. The protein encoded by ORF2 of LAGE-1 was named CAMEL [1]. A second reading frame was also reported for NY-ESO-1/LAGE-2 [27, 33]. The polymorphism of gene LAGE-1 was analyzed on RNA extracted from 17 tumor samples and 14 tumor cell lines obtained from both males (n=13) and females (n=16). The ORF contain three single nucleotide polymorphisms indicated by stars, namely Q6R (cag>cgg), Q89E (cag>gag) (F75L [ttc>ttg] in CAMEL), and P115P (cct>ccg) (L102R [ctc>cgc] in CAMEL). The three polymorphisms are linked with about 2/3 of the allele corresponding to the basic sequence (accession numbers AJ223023 and AJ223040). No polymorphism was observed in our series for NY-ESO-1/LAGE-2
Several peptides recognized by CD8+ CTL have been identified in NY-ESO-1/LAGE-2 and CAMEL proteins (Table 1). The specific CTL have been derived in vitro by stimulating lymphocytes of cancer patients either with autologous tumor cells or with antigen-presenting cells, which were pulsed with peptide or infected with an adenovirus containing a NY-ESO-1/LAGE-2-coding sequence. A few of these antigenic peptides are also encoded by LAGE-1 [1, 27].
Table 1.
Antigenic peptides encoded by LAGE genes and recognized by CD8+ T cells
| Gene | HLA | HLA frequency (%)a | Peptide | Position | Lymphocyte stimulation method | References | |
|---|---|---|---|---|---|---|---|
| Caucasians | Orientals | ||||||
| NY-ESO-1/LAGE-2 | A2 | 44 | 47 | SLLMWITQC | 157–165 | Autologous tumor cells | [6, 21, 27] |
| A2 | 44 | 47 | MLMAQEALAFL | ORF2, 1–11 | Autologous tumor cells | [1] | |
| A31 | 5 | 9 | ASGPGGGAPR | 53–62 | Autologous tumor cells | [33] | |
| A31 | 5 | 9 | LAAQERRVPR | ORF2, 18–27 | Autologous tumor cells | [33] | |
| B7 | 17 | 7 | APRGVRMAV | ORF2, 46–54 | Adenovirus-APC | [30] | |
| B35 | 20 | 10 | MPFATPMEA | 94–102 | Autologous tumor cells | [4] | |
| B51 | 12 | 13 | MPFATPMEA | 94–102 | Adenovirus-APC | [22] | |
| Cw3 | 24 | 37 | LAMPFATPM | 92–100 | Adenovirus-PBMCb | [15] | |
| Cw6 | 16 | 13 | ARGPESRLL | 80–88 | Adenovirus-PBMC | [15] | |
| LAGE-1 | A2 | 44 | 47 | MLMAQEALAFL | ORF2, 1–11 | Autologous tumor cellsc | [1] |
| A2 | 44 | 47 | SLLMWITQC | 157–165 | Peptided | [27] | |
| A31 | 5 | 9 | LAAQERRVPR | ORF2, 18–27 | Autologous tumor cells | [33] | |
a Based on Marsh et al. [26].
b Recombinant adenoviruses carrying the entire gene were used to infect the dendritic cells or PBMC that were subsequently used as stimulators.
c Antitumor CTL were obtained after repeated stimulation of blood lymphocytes with autologous tumor cells.
d Peptides chosen on the basis of consensus anchor residues were used to stimulate blood lymphocytes.
The identification of an additional set of antigenic peptides could be useful for the development of new vaccines to be made available for a large cohort of patients. It will also facilitate the design of vaccines comprising several antigens. To identify a new LAGE-1-derived peptide recognized by CTL on tumor cells, CD8+ T lymphocytes from a blood donor were stimulated with autologous dendritic cells (DC) infected with an avian poxvirus containing a complete LAGE-1b-coding sequence. As this requires the processing of the antigen by the DC, we surmised that the peptide that would be identified would also be processed in tumors that express LAGE-1.
Material and methods
Cell lines, recombinant viruses, cDNA clones, and reagents
The Epstein–Barr virus-transformed B (EBV-B) cell lines and tumor cell lines were cultured in IMDM (Invitrogen, Merelbeke, Belgium) supplemented with 10% fetal calf serum (Invitrogen). WEHI-164 clone 13 was cultured in DMEM (Invitrogen) supplemented with 5% fetal calf serum. COS-7 cells were maintained in H16 supplemented with 10% fetal calf serum and Hepes (10 mM). All the media were supplemented with 0.24 mM of L-asparagine, 0.55 mM of L-arginine, 1.5 mM of L-glutamine (AAG), 100 U/ml of penicillin and 100 mg/ml of streptomycin. Geneticin was purchased from Gibco BRL (Gaithersburg, MD, USA). Clonal line LB831-BLC/A68/c14 was obtained by transfecting the LB831-BLC cells with vector pcDNA3 carrying an HLA-A*68012 sequence using calcium phosphate. The transfectants were selected with 1 mg/ml of geneticin, and clones were derived from the geneticin-resistant cells. Aventis Pasteur (Lyon, France) provided the recombinant canarypoxvirus ALVAC- LAGE-1b, the vaccinia-LAGE-1b, and the parental vaccinia viruses. Retroviral vector LAGE-1b-CSM codes for a full-length LAGE-1b protein and a truncated form of the human low affinity nerve growth factor receptor (LNGFr). EBV-B cells transduced with LAGE-1b-CSM (LB1965-EBV/LAGE-1b) were obtained as described [34]. cDNA clones carrying truncated LAGE-1b sequences were inserted into vector pcDNA1/Amp (Invitrogen), cDNA encoding HLA-A*68012 and A*01 into pcDNA3 (Invitrogen), cDNA encoding HLA-Cw*0602 into pET3D–tag (Invitrogen), and cDNA encoding HLA-B*40012, B*5701 and Cw*0304 into pcDNA3.1/V5-His-TOPO (Invitrogen). Dr. M. Takenoyama, Department of Surgery, School of Medicine, University of Occupational and Environmental Health, Kitakyushu, Japan kindly provided the latter construct. Human recombinant IL-2 was purchased from Eurocetus (Amsterdam, The Netherlands), IL-7 from Genzyme (Cambridge, MA, USA), and GM-CSF from Schering-Plough (Brinny, Ireland). Human recombinant IL-4, IL-6, and IL-12 were produced in our laboratory.
Isolation of anti-LAGE-1 CTL
Peripheral blood mononuclear cells (PBMC) from HLA-A*6801 hemochromatosis patient LB1965 were isolated by Lymphoprep (Axis-shield PoCAS, Oslo, Norway) density gradient centrifugation. DC were obtained from adherent (60 min, 37°C) cells cultured for 6 days in RPMI 1640 supplemented with 10% FBS, 0.24 mM L-asparagine, 0.55 mM L-arginine, 1.5 mM L-glutamine, antibiotics (complete RPMI medium), IL-4 (200 U/ml), and GM-CSF (70 ng/ml) [28, 29]. CD8+ cells were purified from the non-adherent cells using magnetic microbeads coated with anti-CD8 antibodies (Miltenyi Biotech, Bergisch Gladbach, Germany), and the AutoMacs separator (Miltenyi Biotech). DC were incubated for 2 h at 37°C with ALVAC-LAGE-1b at a multiplicity of infection (MOI) of 30 and were washed. CD8+ T cells (1.5×105 cells/well) and infected DC (3×104 cells/well) were mixed in U-bottomed microwells in 200 μl of complete Iscove’s medium with IL-6 (1,000 U/ml) and IL-12 (10 ng/ml). On days 8 and 14, cultures were restimulated with infected DC in a medium supplemented with IL-2 (10 U/ml) and IL-7 (5 ng/ml). An aliquot of each microculture was tested for the presence of T cells with specific cytotoxicity against autologous EBV-B cells infected with vaccinia-LAGE-1b. Infection of autologous EBV-B cells was performed on 2×106 target cells for 2 h at an MOI of 20 in 200 μl of complete RPMI medium after virus sonication. Infected cells were washed, labeled with 100 μCi of Na(51 Cr)O4 during 1 h and washed again. Responder T cells were added at an E:T ratio of 40:1 in V-bottomed wells. Unlabeled K562 cells were also added (5×104 cells/well) to block natural killer activity. Individual microcultures were tested on each target in duplicate. The chromium release was measured after a 4 h incubation at 37°C. Positive microcultures were cloned by limiting dilution and stimulated with irradiated autologous EBV-B cells transduced with a retrovirus encoding LAGE-1b (10,000 cells/well), irradiated allogeneic LG2-EBV cells (20,000 cells/well) as feeder cells, IL-2 (50 U/ml), and IL-7 (5 ng/ml). The anti-LAGE-1b.A68 CTL clone LB1965-CTL-s10.E8, referred to below as clone E8, was kept in the culture and restimulated every 10 days with peptide-pulsed EBV-B cells, feeder cells, and cytokines.
Identification of the HLA presenting molecules
COS-7 cells (1.5×104) were distributed in flat-bottomed microwells and co-transfected using 1.5 μl of Lipofectamine (Invitrogen) with a LAGE-1b cDNA and with each of the sequences coding for the HLA molecules corresponding to the haplotype of the blood donor. Transfected cells were incubated for 24 h at 37°C and 8% CO2. A total of 2,000 CTL were added in the microwells containing the COS-7 transfectants in a total volume of 200 μl of complete IMDM supplemented with 25 U/ml of IL-2. After 20 h, the supernatant was collected and its TNF content was determined by testing its cytotoxic effect on WEHI-164 clone 13 cells in a MTT colorimetric assay [14, 18, 31].
Identification of the region containing the antigenic peptide
The LAGE-1b cDNA (clone 2) was previously retrieved from a cDNA library of LB373-MEL [25]. Different LAGE-1b fragments were produced by PCR that correspond to amino acids 1–210, 1–150, 1–118, and 1–108. The upper primer, 5′-CTCTCTgCCTCCgCATCC-3′, was located immediately downstream to a major transcription start of the LAGE-1 mRNA; the lower primers were 5′-gATCCACATCAACAagggAA-3′, 5′-CCCAACCCACCACCCTCA-3′, 5′-CCCTggTCgggggAgA-3′,and 5′-gATCCTgCggACCAgCTC-3′. The fragments were cloned into pcDNA3.1/V5-His-TOPO (Invitrogen). The HLA-A*68012 cDNA (80 ng) was transfected into COS-7 cells (1.5×104) together with each of the LAGE-1b cDNA fragments (80 ng). CTL (2,000 cells) were added to the transfected cells. The IFN-γ production was measured by ELISA after 20 h of coculture.
Peptide recognition assay
Peptides were synthesized on a solid phase using Fmoc for transient NH2-terminal protection and were characterized using mass spectrometry. All the peptides were 90% pure as indicated by analytic HPLC. Lyophilized peptides were dissolved at 2 mg/ml in 10 mM of acetic acid and 10% DMSO, and were stored at −20°C. The first screening by chromium release assay was performed with autologous EBV-B cells incubated with 16 amino acid long peptides at a concentration of 4–5 μM. Peptide-pulsed targets were tested for recognition by CTL at an E:T ratio of 5:1.
HLA-peptide fluorescent multimers
Recombinant HLA-A6801 molecules were folded in vitro with β2-microglobulin and peptide ELVRRILSR from LAGE-1 or MUM-3 peptide EAFIQPITR [3]. They were purified by gel filtration, biotinylated, and mixed as described [2] with Streptavidin-PE (Sigma, St. Louis, MT, USA) for the HLA-A68/ELVRRILSR multimer or streptavidin-APC (BD Biosciences) for the A68/EAFIQPITR control multimer [3]. For staining and sorting, cells from A*6801 melanoma patient DDHK2 were washed, resuspended at 107 cells/ml in Hank’s solution modified for flow cytometry [24] with 1% HS and incubated for 15 min at room temperature with HLA-A68 multimers loaded with LAGE-1 peptide (20 nM) or MUM-3 peptide (5 nM). Multimer-labeled cells (107 cells/80 μl) were incubated at 4°C with anti-PE microbeads (20 μl) according to the instructions of the manufacturer (Miltenyi Biotec), washed, and sorted through a separation column inserted to a magnet in an AUTOMACS at 0.5 ml/min (Miltenyi Biotec). The selected cells were distributed at 8,000 cells per well in U-bottomed microwells and cultured in 200 μl of IMDM supplemented with AAG, 10% human serum, IL-2 (100 U/ml), and IL-7 (5 ng/ml). They were stimulated on days 0 and 7 with irradiated (100 Gy) autologous cells incubated for 1 h with 4 μg/ml of peptide and washed. The autologous stimulator cells were obtained from the negative fraction of the magnetic sorting on day 0 and, on day 7, from cells amplified with 0.5 μg/ml of PHA and 100 U/ml of IL-2. Approximately 105 cells from each microculture were stained with multimer and analyzed by flow cytometry with a FACSCalibur (BD Biosciences) using the Cellquest software (BD Biosciences).
TCR analysis
Total RNA from T cell clones was extracted with the Tripure reagent (Roche Diagnostics). RNA was primed with oligo-dT and reverse-transcribed with MMLV-RT for 1 h at 42°C in 20 μl. cDNA (1/10) served as a template for PCR amplifications using panels of Vβ-specific upstream primers and one downstream Cβ primer. PCR products were purified and sequenced to obtain a complete identification of the CDR3 region.
Results and discussion
Isolation of an anti-LAGE-1 CTL clone
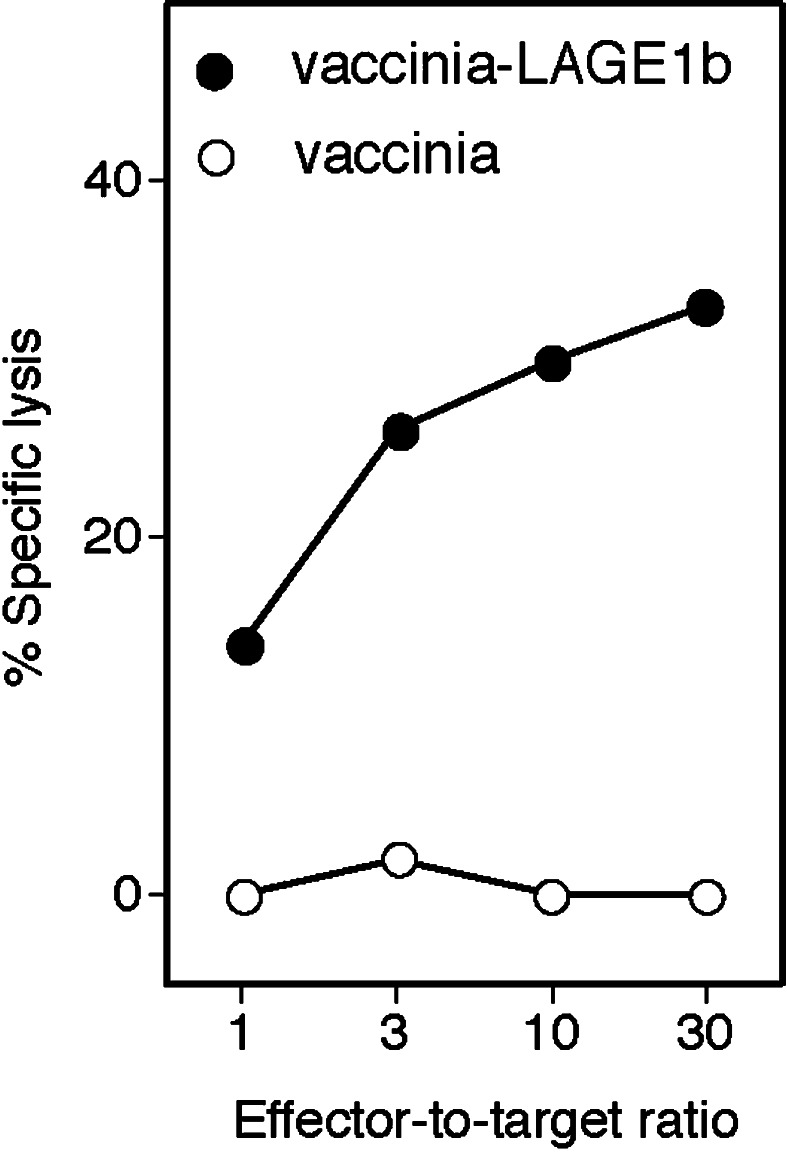
Monocyte-derived immature DC were obtained from a hemochromatosis patient without any evidence of cancer. DC were infected with an avian poxvirus carrying a LAGE-1b coding sequence (ALVAC-LAGE-1b) and used to stimulate autologous CD8+ T cells. Ninety-six microcultures were set up with 30,000 stimulator DC and 150,000 responder CD8+ T cells. Responder cells were restimulated once a week with autologous DC infected with ALVAC-LAGE-1b. Responder cells from each microculture were tested on day 21 for their lytic activity on autologous EBV-B cells infected with a vaccinia virus encoding LAGE-1b (vaccinia-LAGE-1b). An excess of unlabeled K562 target cells were added to quench the lytic activity of NK-like effectors. One microculture contained lymphocytes displaying anti-LAGE-1 reactivity. The lymphocytes were cloned by limiting dilution and stimulated with autologous EBV-B cells transduced with a retrovirus encoding LAGE-1b. CTL clones were obtained that specifically lysed EBV-B cells infected with vaccinia-LAGE-1b. Data obtained with representative clone E8 are shown in Fig. 2.
Fig. 2.
Lysis of EBV-B cells expressing LAGE-1b by CTL clone E8. LB1965-EBV-B cells were infected for 2 h with the indicated vaccinia vectors at multiplicity of infection of 20 and labeled for 1 h with 51 Cr. The targets were incubated with the autologous CTL at the indicated effector-to-target ratios and the chromium release was measured after 4 h. The TCRβ sequence of clone E8 is Vβ29-1*01, CSVE DG SNOPQHFG, Jβ1-5*01
The antigen recognized by CTL E8 is presented by HLA-A*6801 molecules
The blood donor was typed HLA-A*0101, A*68012, B*4014, B*5701, Cw*0304, and Cw*0602. To identify the HLA molecules that present the LAGE-1 peptide to CTL E8, COS-7 cells were co-transfected with a LAGE-1b cDNA and with each of the sequences coding for the HLA molecules corresponding to the haplotype of the blood donor. The transfected cells were tested for recognition by the CTL. Only the cells transfected with LAGE-1b and A*68012 stimulated the CTL to produce TNF (Fig. 3). COS-7 cells co-transfected with A68 and LAGE-1a, but not NY-ESO-1/LAGE-2, were recognized by the CTL (data not shown).
Fig. 3.
The LAGE-1 peptide is presented to CTL E8 by HLA-A68 molecules. Monkey COS-7 cells were transiently transfected with 80 ng of pcDNA1/Amp-LAGE-1b and 80 ng of expression vectors encoding each of the 6 HLA class I molecules of donor LB1965. After 24 h, 2,000 CTL were added to the transfected cells; 20 h later, TNF production was estimated by measuring the cytotoxicity of the supernatants on the TNF-sensitive WEHI-164 clone 13 cells. Experiments were performed in triplicate
Identification of the LAGE-1 antigenic peptide
To determine the region encoding the antigenic peptide, truncated cDNA fragments of LAGE-1b were cloned into an expression vector and co-transfected with an HLA-A68 construct into COS-7 cells (Fig. 4). Transfected cells were tested for their ability to stimulate CTL E8. The results indicated that CTL E8 recognized a peptide encoded by the region coding for amino acids 100–118 of open reading frame 1. This part of the protein does not contain any peptide carrying the two consensus anchor residues for HLA-A68, namely, V or T at position 2 and R or K at the C-terminus [16]. Several peptides with a R at the C-terminus were tested and ELVRRILSR appeared to be the optimal one (Fig. 4). Half maximal lysis of the EBV-B cells was obtained with 300 nM of the peptide. Peptide ELVRRILSR corresponds to position 103–111 of the LAGE-1a and LAGE-1b protein sequence. It is encoded by the different LAGE-1 alleles (Fig. 1) but not by the NY-ESO-1/LAGE-2 gene.
Fig. 4.
Identification of the LAGE-1 antigenic peptide. The LAGE-1b cDNA is shown in the upper part of the figure with exons appearing as open boxes, the spliced intron as kink, and the open reading frame as black boxes. Truncated LAGE-1b cDNA were co-transfected into COS-7 cells together with an HLA-A*68012 cDNA. The transfected cells were tested for recognition by the CTL. Several peptides encoded by this region were tested for recognition by CTL E8: HLA-A68+ chromium-labeled EBV-B cells were incubated for 15 min with each peptide at different concentrations, before addition of clone E8 at an E:T ratio of 5:1. Chromium release was measured after 4 h. The concentrations indicated in the figure are the concentrations during the 4 h of incubation
Before the construction of truncated cDNAs, we first tested CTL E8 for lysis and cytokine release upon contact with autologous EBV-B cells each incubated with a set of 74 peptides of 16 amino acids overlapping by 12 amino acids and covering the complete LAGE-1b protein sequence and the protein encoded by the alternative open reading frame. None of them was recognized by the CTL, including peptides SSPMEAELVRRILSRD and EAELVRRILSRDAAPL, both of which contained antigenic peptide ELVRRILSR (data not shown). We do not have any explanation but it has to be noted that the strategy of screening a peptide library was successful for all the antigenic peptides we have identified so far, with the exception of the MAGE-4.A2 peptide [13]. For the identification of the MAGE-4 peptide we also had to construct truncated cDNAs.
Lysis of HLA-A68 tumor cells expressing LAGE-1
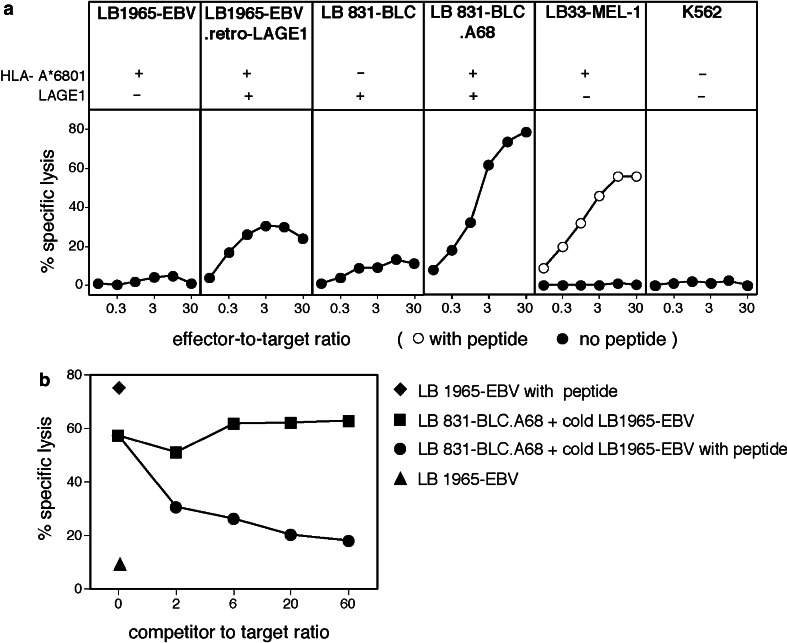
As we used DC expressing LAGE-1 to activate CTL E8, it was important to verify that tumor cells also process the LAGE-1.A68 antigen efficiently. As we had no appropriate tumor cell line, we transfected a LAGE-1+ bladder carcinoma line, LB831-BLC, with a vector carrying the A*6801 coding sequence and a sequence coding for the resistance to geneticin. Clonal populations were derived from the geneticin-resistant transfected cells. The A68+ cells were lysed by CTL clone E8, indicating that they processed the LAGE-1103–111 peptide (Fig. 5a). This lysis of LB831-BLC.A68 cells was quenched in the presence of an excess of unlabeled A68 EBV-B cells pulsed with peptide ELVRRILSR (Fig. 5b).
Fig. 5.
Lysis of tumor cell lines by CTL clone E8. a Cell line LB1965-EBV.retroLAGE-1b has been obtained by transduction of EBV-B cells with a retrovirus carrying a complete LAGE-1 coding sequence. LB831-BLC is a bladder carcinoma cell line. The A68-transfected cell line is a clonal population of cells stably transfected with an HLA-A*68012 cDNA. Target cells were 51Cr-labeled for 1 h, if indicated, pulsed for 15 min with 5 μM of peptide ELVRRILSR and incubated with CTL at various E:T ratios. The chromium release was measured after 4 h. b Unlabeled competitors were added before adding CTL at an E:T ratio of 5
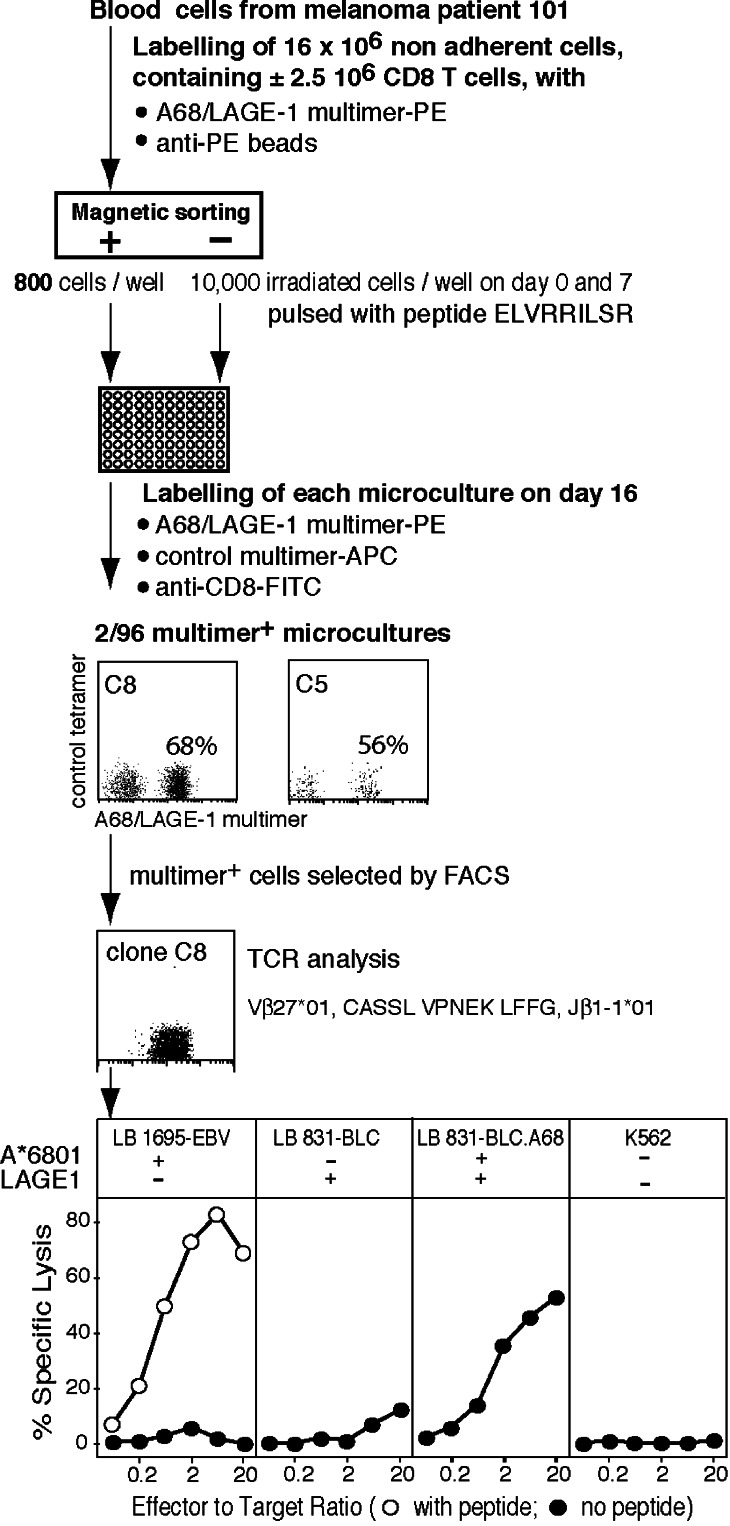
Isolation of anti-LAGE-1 CTL in a cancer patient using an A68/LAGE-1 fluorescent multimer
The identification of an antigenic peptide allows the use of HLA-peptide multimers to detect T cell responses in cancer patients. A fluorescent multimer was constructed by folding HLA-A6801 molecules with peptide ELVRRILSR. Sixteen million nonadherent blood cells (±2.5 million CD8 T cells) from an A*6801 melanoma patient, bearing a melanoma expressing LAGE-1, were incubated with A68/LAGE-1 multimers conjugated to phycoerythrin (PE) and anti-PE antibodies coupled to magnetic beads (Fig. 6). The multimer-positive cells selected by magnetic sorting were distributed in microwells and stimulated on days 0 and 7 with peptide-pulsed autologous cells. The microcultures were screened on day 16 for the presence of cells specifically labeled with multimers and two microcultures were positive, suggesting a frequency of CTL precursors of approximately 1 per million CD8 T cells (Fig. 6). We cannot exclude that the CTL precursors have been primed during the in vitro culture. A clonal population was obtained from one microculture. It lysed A68 cells pulsed with peptide ELVRRILSR and the bladder carcinoma cell line expressing LAGE-1 after transfection with HLA-A68.
Fig. 6.
Overview of the procedure to isolate anti-LAGE-1.A68 CTL with a fluorescent multimer from a blood sample of an A68 patient bearing a melanoma expressing LAGE-1. The clinical evolution of patient 101 was described in Zhang et al. [35]
This result validated the A68/LAGE-1 multimer as a tool for the monitoring of specific T cell responses in cancer patients. It also demonstrated that anti-LAGE-1.A68 CTL exist in different individuals with enough avidity to lyse tumor cells expressing LAGE-1. Therefore, we concluded that the new antigenic peptide, ELVRRILSR, could be used as target for therapeutic antitumoral vaccination of A68 patients carrying a tumor expressing LAGE-1. HLA-A68 molecules are expressed in 18% of blacks, 8% of Caucasians, and 3% of Orientals [26]. It has to be noted that 11 different HLA-A68 alleles have been identified encoding 9 different HLA molecules [26]. The binding specificity of the LAGE-1 peptide to the different alleles should be further analyzed; however, it is likely that most of the A68 molecules have an equivalent peptide anchor motif. Monitoring of T cell responses of cancer patients vaccinated with peptides should ideally be analyzed with tetramers corresponding precisely to their allele, because the contact sites between the T cell receptor and the HLA molecules could differ between different HLA alleles that can present the same peptide. Such a case has been encountered with B44: we isolated from a B*4402 individual an anti-MAGE-A3 CTL strictly restricted by B*4402 and from a B*4403 individual a CTL strictly restricted by B*4403 that recognized the same peptide [19, 20].
Acknowledgements
We thank Dr. Pierre Coulie for the critical reading and Mrs Nathalie Krack for the editorial assistance. This work was supported by the Belgian Programme on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming, and by a grant from the Fédération Belge contre le Cancer (Belgium).
References
- 1.Aarnoudse CA, van den Doel PB, Heemskerk B, Schrier PI. Interleukin-2-induced, melanoma-specific T cells recognize camel, an unexpected translation product of LAGE-1. Int J Cancer. 1999;82:442–448. doi: 10.1002/(SICI)1097-0215(19990730)82:3<442::AID-IJC19>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Altman JD, Moss PAH, Goulder PJR, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- 3.Baurain J-F, Colau D, van Baren N, Landry C, Martelange V, Vikkula M, Boon T, Coulie PG. High frequency of autologous anti-melanoma CTLs directed against an antigen generated by a point mutation in a new helicase gene. J Immunol. 2000;164:6057–6066. doi: 10.4049/jimmunol.164.11.6057. [DOI] [PubMed] [Google Scholar]
- 4.Benlalam H, Linard B, Guilloux Y, Moreau-Aubry A, Derré L, Diez E, Dreno B, Jotereau F, Labarrière N. Identification of five new HLA-B*3501-restricted epitopes derived from common melanoma-associated antigens, spontaneously recognized by tumor-infiltrating lymphocytes. J Immunol. 2003;171:6283–6289. doi: 10.4049/jimmunol.171.11.6283. [DOI] [PubMed] [Google Scholar]
- 5.Boël P, Wildmann C, Sensi M-L, Brasseur R, Renauld J-C, Coulie P, Boon T, van der Bruggen P. BAGE, a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2:167–175. doi: 10.1016/S1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen J-L, Dunbar PR, Gileadi U, Jäger E, Gnjatic S, Nagata Y, Stockert E, Panicali DL, Chen Y-T, Knuth A, Old LJ, Cerundolo V. Identification of NY-ESO-1 peptide analogues capable of improved stimulation of tumor-reactive CTL. J Immunol. 2000;165:948–955. doi: 10.4049/jimmunol.165.2.948. [DOI] [PubMed] [Google Scholar]
- 7.Chen Q, Jackson H, Parente P, Luke T, Rizkalla M, Tai TY, Zhu HC, Mifsud NA, Dimopoulos N, Masterman KA, Hopkins W, Goldie H, Maraskovsky E, Green S, Miloradovic L, McCluskey J, Old LJ, Davis ID, Cebon J, Chen W. Immunodominant CD4+ responses identified in a patient vaccinated with full-length NY-ESO-1 formulated with ISCOMATRIX adjuvant. Proc Natl Acad Sci USA. 2004;101:9363–9368. doi: 10.1073/pnas.0403271101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Q, Jackson H, Shackleton M, Parente P, Hopkins W, Sturrock S, MacGregor D, Maraskovsky E, Tai TY, Dimopoulos N, Masterman KA, Luke T, Davis ID, Chen W, Cebon J. Characterization of antigen-specific CD8+ T lymphocyte responses in skin and peripheral blood following intradermal peptide vaccination. Cancer Immun. 2005;5:5. [PubMed] [Google Scholar]
- 9.Chen Y-T, Scanlan MJ, Sahin U, Türeci Ö, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis ID, Chen W, Jackson H, Parente P, Shackleton M, Hopkins W, Chen Q, Dimopoulos N, Luke T, Murphy R, Scott AM, Maraskovsky E, McArthur G, MacGregor D, Sturrock S, Tai TY, Green S, Cuthbertson A, Maher D, Miloradovic L, Mitchell SV, Ritter G, Jungbluth AA, Chen YT, Gnjatic S, Hoffman EW, Old LJ, Cebon JS. Recombinant NY-ESO-1 protein with ISCOMATRIX adjuvant induces broad integrated antibody and CD4+ and CD8+ T cell responses in humans. Proc Natl Acad Sci USA. 2004;101:10697–10702. doi: 10.1073/pnas.0403572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Backer O, Arden KC, Boretti M, Vantomme V, De Smet C, Czekay S, Viars CS, De Plaen E, Brasseur F, Chomez P, Vanden Eynde B, Boon T, van der Bruggen P. Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res. 1999;59:3157–3165. [PubMed] [Google Scholar]
- 12.De Plaen E, Arden K, Traversari C, Gaforio JJ, Szikora J-P, De Smet C, Brasseur F, van der Bruggen P, Lethé B, Lurquin C, Brasseur R, Chomez P, De Backer O, Cavenee W, Boon T. Structure, chromosomal localization and expression of twelve genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 13.Duffour M-T, Chaux P, Lurquin C, Cornelis G, Boon T, van der Bruggen P. A MAGE-A4 peptide presented by HLA-A2 is recognized by cytolytic T lymphocytes. Eur J Immunol. 1999;29:3329–3337. doi: 10.1002/(SICI)1521-4141(199910)29:10<3329::AID-IMMU3329>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Espevik T, Nissen-Meyer J. A highly sensitive cell line, WEHI 164 clone 13, for measuring cytotoxic factor/tumor necrosis factor from human monocytes. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 15.Gnjatic S, Nagata Y, Jäger E, Stockert E, Shankara S, Roberts BL, Mazzara GP, Lee SY, Dunbar PR, Dupont B, Cerundolo V, Ritter G, Chen YT, Knuth A, Old LJ. Strategy for monitoring T cell responses to NY-ESO-1 in patients with any HLA class I allele. Proc Natl Acad Sci USA. 2000;97:10917–10922. doi: 10.1073/pnas.97.20.10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo H-C, Jardetzky TS, Garrett TPJ, Lane WS, Strominger JL, Wiley DC. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature. 1992;360:364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- 17.Haas GG, Jr, D’Cruz OJ, De Bault LE. Distribution of human leukocyte antigen-ABC and -D/DR antigens in the unfixed human testis. Am J Reprod Immunol Microbiol. 1988;18:47–51. doi: 10.1111/j.1600-0897.1988.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 18.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 19.Herman J, van der Bruggen P, Luescher I, Mandruzzato S, Romero P, Thonnard J, Fleischhauer K, Boon T, Coulie PG. A peptide encoded by the human gene MAGE-3 and presented by HLA-B44 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Immunogenetics. 1996;43:377–383. doi: 10.1007/BF02199806. [DOI] [PubMed] [Google Scholar]
- 20.Herman J, Jongeneel V, Kuznetsov D, Coulie PG. Differences in the recognition by CTL of peptides presented by the HLA-B*4402 and the HLA-B*4403 molecules which differ by a single amino acid. Tissue Antigens. 1999;53:111–121. doi: 10.1034/j.1399-0039.1999.530201.x. [DOI] [PubMed] [Google Scholar]
- 21.Jäger E, Chen Y-T, Drijfhout JW, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jäger E, Karbach J, Gnjatic S, Jäger D, Maeurer M, Atmaca A, Arand M, Skipper J, Stockert E, Chen Y-T, Old LJ, Knuth A. Identification of a naturally processed NY-ESO-1 peptide recognized by CD8+ T cells in the context of HLA-B51. Cancer Immun [serial online] 2002;2:12. [PubMed] [Google Scholar]
- 23.Khong HT, Yang JC, Topalian SL, Sherry RM, Mavroukakis SA, White DE, Rosenberg SA. Immunization of HLA-A*0201 and/or HLA-DPb1*04 patients with metastatic melanoma using epitopes from the NY-ESO-1 antigen. J Immunother. 2004;27:472–477. doi: 10.1097/00002371-200411000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann F, Marchand M, Hainaut P, Pouillart P, Sastre X, Ikeda H, Boon T, Coulie PG. Differences in the antigens recognized by cytolytic T cells on two successive metastases of a melanoma patient are consistent with immune selection. Eur J Immunol. 1995;25:340–347. doi: 10.1002/eji.1830250206. [DOI] [PubMed] [Google Scholar]
- 25.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. LAGE-1, a new gene with tumor specificity. Int J Cancer. 1998;76:903–908. doi: 10.1002/(SICI)1097-0215(19980610)76:6<903::AID-IJC22>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Marsh SGE, Parham P, Barber LD, editors. The HLA facts book. London: Academic; 2000. [Google Scholar]
- 27.Rimoldi D, Rubio-Godoy V, Dutoit V, Liénard D, Salvi S, Guillaume P, Speiser D, Stockert E, Spagnoli G, Servis C, Cerottini J-C, Lejeune F, Romero P, Valmori D. Efficient simultaneous presentation of NY-ESO-1/LAGE-1 primary and nonprimary open reading frame-derived CTL epitopes in melanoma. J Immunol. 2000;165:7253–7261. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]
- 28.Romani N, Gruner S, Brang D, Kämpgen E, Lenz A, Trockenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor a. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slager EH, van der Minne CE, Goudsmit J, van Oers JMM, Kostense S, Havenga MJE, Osanto S, Griffioen M. Induction of CAMEL/NY-ESO-ORF2-specific CD8+ T cells upon stimulation with dendritic cells infected with a modified Ad5 vector expressing a chimeric Ad5/35 fiber. Cancer Gene Ther. 2004;11:227–236. doi: 10.1038/sj.cgt.7700674. [DOI] [PubMed] [Google Scholar]
- 31.Traversari C, van der Bruggen P, Vanden Eynde B, Hainaut P, Lemoine C, Ohta N, Old L, Boon T. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 32.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Vanden Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 33.Wang R-F, Johnston SL, Zeng G, Topalian SL, Schwartzentruber DJ, Rosenberg SA. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J Immunol. 1998;161:3596–3606. [PubMed] [Google Scholar]
- 34.Zhang Y, Stroobant V, Russo V, Boon T, van der Bruggen P. A MAGE-A4 peptide presented by HLA-B37 is recognized on human tumors by cytolytic T lymphocytes. Tissue Antigens. 2002;60:365–371. doi: 10.1034/j.1399-0039.2002.600503.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Sun Z, Nicolay H, Meyer RG, Renkvist N, Stroobant V, Corthals J, Carrasco J, Eggermont AMM, Marchand M, Thielemans K, Wölfel T, Boon T, van der Bruggen P. Monitoring of anti-vaccine CD4 T cell frequencies in melanoma patients vaccinated with a MAGE-3 protein. J Immunol. 2005;174:2404–2411. doi: 10.4049/jimmunol.174.4.2404. [DOI] [PubMed] [Google Scholar]