Abstract
Previously we designed novel pseudotyped high-titer replication defective human immunodeficiency virus type 1 (HIV-1) vectors to deliver genes into nondividing cells (J. Reiser, G. Harmison, S. Kluepfel-Stahl, R. O. Brady, S. Karlsson, and M. Schubert, Proc. Natl. Acad. Sci. USA 93:15266–15271, 1996). Since then we have made several improvements with respect to the safety, flexibility, and efficiency of the vector system. A three-plasmid expression system is used to generate pseudotyped HIV-1 particles by transient transfection of human embryonic kidney 293T cells with a defective packaging construct, a plasmid coding for a heterologous envelope (Env) protein, and a vector construct harboring a reporter gene such as neo, ShlacZ (encoding a phleomycin resistance/β-galactosidase fusion protein), HSA (encoding mouse heat-stable antigen), or EGFP (encoding enhanced green fluorescent protein). The packaging constructs lack functional Vif, Vpr, and Vpu proteins and/or a large portion of the Env coding region as well as the 5′ and 3′ long terminal repeats, the Nef function, and the presumed packaging signal. Using G418 selection, we routinely obtained vector particles pseudotyped with the vesicular stomatitis virus G glycoprotein (VSV-G) with titers of up to 8 × 107 CFU/μg of p24, provided that a functional Tat coding region was present in the vector. Vector constructs lacking a functional Tat protein yielded titers of around 4 × 106 to 8 × 106 CFU/μg of p24. Packaging constructs with a mutation within the integrase (IN) core domain profoundly affected colony formation and expression of the reporter genes, indicating that a functional IN protein is required for efficient transduction. We explored the abilities of other Env proteins to allow formation of pseudotyped HIV-1 particles. The rabies virus and Mokola virus G proteins yielded high-titer infectious pseudotypes, while the human foamy virus Env protein did not. Using the improved vector system, we successfully transduced contact-inhibited primary human skin fibroblasts and postmitotic rat cerebellar neurons and cardiac myocytes, a process not affected by the lack of the accessory proteins.
Vectors based on oncoretroviruses such as Moloney murine leukemia virus (MoMLV) are useful to deliver therapeutic genes into primary cells in vitro and have also been applied in a number of gene marking and gene therapy trials with humans (13, 62). The principal advantages of retroviral vectors include the high efficiency of gene delivery, integration into the host cell genome, and high level of gene expression. One drawback of oncoretroviruses is their dependence on cell proliferation for completion of the life cycle (35, 56). Breakdown of the nuclear envelope that accompanies mitosis appears to be essential for the import of the viral preintegration complex into the nucleus and its integration into the genome of the host cell (26, 27, 51). In contrast, lentiviruses, including human immunodeficiency virus type 1 (HIV-1), differ fundamentally from oncoretroviruses in that they are independent of cell division for completion of their replicative cycle (58). This is an attractive feature in view of the need for vectors for nondividing cells such as neurons (14, 25). In common with all replication-competent retroviruses, the HIV-1 genome contains the gag, pol, and env coding regions, which encode the core proteins, the virion-associated enzymes, and the envelope (Env) glycoprotein, respectively, flanked by the long terminal repeats (LTRs). The LTRs include cis-acting sequences required for integration, transcription, and polyadenylation. HIV-1 also possesses regulatory functions encoded by the tat and rev genes as well as accessory genes that include vif, vpr, vpu, and nef, many of which are not required for virus replication in vitro (17).
Another advantage of HIV-1 and other retroviruses is the fact that they can be pseudotyped by the incorporation of heterologous glycoproteins, allowing an extension of the host range of such vectors beyond cells expressing CD4. Several studies have demonstrated that HIV-1 produced in cells infected with xenotropic murine leukemia virus (6, 30), amphotropic murine leukemia virus (9, 55), or herpes simplex virus (64) gave rise to phenotypically mixed virions with an expanded host range, suggesting that pseudotyped virions had formed. Additionally, phenotypic mixing of viral envelopes was shown to occur between HIV-1 and vesicular stomatitis virus (VSV) in coinfected cell cultures (64). Page et al. (40) showed that expression of amphotropic or ecotropic MoMLV Env glycoproteins in cells transfected with an HIV-1 vector construct produced virus capable of infecting both human and murine cells, and Landau et al. (23) demonstrated that HIV-1 efficiently incorporated the human T-cell leukemia virus type 1 Env. These observations were confirmed and extended by results showing that the VSV G glycoprotein (VSV-G) was efficiently incorporated into HIV-1 virions, with pseudotyped viral titers reaching 107 CFU/ml or higher (2, 38, 49).
A number of replication-defective HIV-1 vector systems have been described. With the original transient two-plasmid expression system (23, 40), titers of up to 2 × 105 CFU/ml were obtained on human osteosarcoma (HOS) cells. Several groups have designed transient three-component HIV-1 expression systems consisting of a packaging construct, a plasmid bearing the gene encoding gp160, and an expression vector carrying a reporter gene (5, 41, 42, 45, 50, 53, 59), thus reducing the likelihood of generating replication-competent virus. In general, these systems were quite inefficient, with titers of around 104 CFU/ml or below. A number of studies have dealt with the design of HIV-1-based packaging cell lines. While the initial vector titers were quite low (7), recent improvements which involve tetracycline-controlled HIV-1-based packaging constructs (63), different molecular clones (11, 44), or Rev-independent cell lines (57) suggest that the generation of high-titer HIV-1-based packaging cell lines will eventually be feasible. Improved three-component split packaging systems were recently described by Naldini et al. (38) and Kim et al. (21). In these systems, the viral particles were pseudotyped with the envelope of VSV. Titers of up to 9 × 105 transducing units per ml were obtained. In this report, we describe an improved and versatile high-titer three-plasmid-based packaging system and demonstrate its applicability for the efficient transduction of nondividing cells, including growth-arrested HOS cells, confluent primary human skin fibroblasts (HSFs), and postmitotic rat cardiac myocytes and cerebellar neurons.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were obtained through the AIDS Research and Reference Reagents Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), Bethesda, Md.: pHIVgpt and pHXB2-env from Kathleen Page and Dan Littman (40); pNL4-3 from Malcom Martin (1); and p210-13, p210-19, and p210-25 from Ronald Desrosiers (17). All nucleotides are numbered according to the work of Myers et al. (37). The HIV-neoΔ vector constructs were derived from the HXB2 molecular clone (48). They harbor deletions from the SpeI site (nucleotide 1506) to the EcoRI (nucleotide 5742), SalI (nucleotide 5784), and NdeI (nucleotide 6399) sites. The plasmids are referred to here as pHIV-neoΔE, pHIV-neoΔS, and pHIV-neoΔN, respectively. All three plasmids contain a truncated gp160 coding region with the sequences from the NdeI site at position 6399 to the BglII site at position 7611 deleted. They also contain a 168-bp fragment carrying the simian virus 40 (SV40) origin of replication (49). Plasmid HIV-neoΔTat (−) contains two consecutive termination codons after amino acid 10 within the 5′ tat exon. It is based on pTat(−)GV/4GSTm (19), which was kindly provided by K.-T. Jeang (NIAID). Plasmid NL-neo is based on the NL4-3 molecular clone and carries a deletion from the NsiI site (position 1246) to the BglII site at position 7611. A 1,169-bp fragment carrying the neo gene sequence and the SV40 early promoter derived from pBK-CMV (Stratagene) was inserted between the BamHI site (nucleotide 8464) and XhoI site (nucleotide 8886). Plasmid HIV-HSAΔE was derived from pHIV-HSA (49) by deleting the sequences between the SpeI (nucleotide 1506) and EcoRI (nucleotide 5742) sites. Plasmids HIV-ShlacZΔE and HIV-EGFPΔE are based on HIV-HSAΔE. The HSA (which encodes mouse heat-stable antigen) coding region was replaced with the ShlacZ sequence derived from pUT535 (CAYLA, Toulouse, France), which codes a for a bifunctional phleomycin/β-galactosidase fusion protein (3), or EGFP (which encodes enhanced green fluorescent protein) sequences derived from pEGFP-C1 (Clontech). All Env-encoding plasmids except pHXB2-env are based on pLTR-G (49). Plasmids encoding the rabies virus and Mokola virus G proteins (33) were kindly provided by Karl-Klaus Conzelmann (Federal Research Centre for Virus Disease of Animals, Tübingen, Germany). The human foamy virus (HFV) env coding region (16) was kindly provided by Axel Rethwilm (University of Würzburg, Würzburg, Germany), and the MoMLV 4070A Env-encoding plasmid (39) was from Alan Rein (National Cancer Institute, Frederick, Md.). In the C-Help packaging construct, the 5′ LTR was replaced with the human cytomegalovirus (CMV) immediate-early promoter (4). HIV-1 sequences from nucleotide 675 up to the ApaI site (nucleotide 2005) and sequences from the SalI site (nucleotide 5784) to the XhoI site (nucleotide 8886) were derived from the BH10 molecular clone (46). All other sequences were from the NL4-3 molecular clone. Plasmid C-Help carries a 1,212-bp env deletion (nucleotides 6399 to 7611) and an SV40 origin of replication (49). A 33-bp deletion harboring the putative packaging signal from nucleotides 756 to 789 between the 5′ major splice donor site and the beginning of the gag coding region was introduced. Sequences distal to the XhoI site (position 8886) were removed and replaced with the bovine growth hormone polyadenylation site. A helper construct with a mutated vpr coding region was made by Klenow fill-in of EcoRI-digested C-Help plasmid DNA. A helper plasmid encoding a defective IN (integrase) protein was designed by replacing the ApaI-SalI fragment (nucleotides 2005 to 5784) with the corresponding fragment from the D116N/7 molecular clone (15) (kindly provided by George Englund, NIAID). The C-Help Δvpr plasmid was constructed by replacing the ApaI-SalI fragment with the corresponding fragment from plasmid p210-19 (17). C-Help ΔvifΔvprΔvpu harbors sequences from p210-25 (ApaI-SalI fragment) and p210-13 (SalI-NdeI fragment) (17).
Plasmids pHIT60 and pHIT111 (54) were kindly provided by Alan Kingsman, Oxford University, Oxford, England). pCMV-G is based on pcDNA3.1/Zeo (Invitrogen) and harbors a 1.6-kb fragment encoding VSV-G. pG1-HSA is a MoMLV-based vector encoding mouse HSA.
Cells.
Human embryonic kidney 293T cells (12) were kindly provided by Warren Pear (Rockefeller University). HOS cells (CRL-1543), Rat-2 cells (CRL-1764), and primary HSFs (CRL-2072) were obtained from the American Type Culture Collection. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies Inc.) containing 10% heat-inactivated fetal bovine serum (FBS). The human H9 T-cell line was obtained from Robert Gallo (31) through the AIDS Research and Reference Reagent Program. The cells were grown in RPMI 1640 supplemented with 2 mM l-glutamine, 50 μg of gentamicin per ml, and 10% FBS. Neonatal ventricular myocytes were harvested from the hearts of 2- to 3-day-old Sprague-Dawley rats and cultured as described previously (46). Cerebellar granule cells from 8-day-old Sprague-Dawley rat pups (Taconic Farms) were prepared and cultured as described by Taniwaki et al. (60). The cells were plated in poly-l-lysine-coated 35-mm-diameter dishes. After 1 day in culture, cytosine arabinoside (final concentration, 10 μM) was added to the cells.
Virus production and infection.
For the preparation of HIV-1 pseudotypes, helper plasmid DNA (5 μg), Env plasmid DNA (5 μg), and vector plasmid DNA (5 μg) were cotransfected into subconfluent 293T cells by the calcium phosphate precipitation method (43). Approximately 2 × 106 cells were seeded into six-well plates 24 to 30 h prior to transfection. Chloroquine (25 μM, final concentration) was added to the cells immediately before transfection, and the medium was replaced with fresh DMEM–10% FBS (2 ml per well) 12 to 14 h later. MoMLV-based virus stocks were generated by transient cotransfection of pHIT60, pCMV-G, and pHIT111 or pG1-HSA, respectively. The virus stocks were harvested 60 to 65 h posttransfection and filtered through a 0.45-μm-pore-size filter, aliquoted, and subsequently frozen at −80°C. Target cells were infected in DMEM–10% FBS containing Polybrene (8 μg/ml) for 3 to 8 h. The medium was subsequently replaced with fresh DMEM–10% FBS or preconditioned medium for the cerebellar granule cells and heart ventricular myocytes. p24 assays were performed with a commercial kit (Cellular Products Inc.).
Analysis of transduced cells.
Cells expressing HSA were detached from the plate by using phosphate-buffered saline–2 mM EDTA and stained with a fluorescein isothiocyanate-labeled anti-HSA monoclonal antibody (Caltag) for 30 min on ice in Hanks’ balanced salt solution (Life Technologies) containing 2% FBS (Hanks’-FBS). The cells were washed twice with Hanks’-FBS, resuspended in 4% paraformaldehyde, and then subjected to fluorescence-activated cell sorting (FACS) analysis. Cells expressing EGFP were collected for FACS analysis as described above except that the paraformaldehyde step was omitted. Alternatively, they were analyzed by fluorescence microscopy. Cells expressing β-galactosidase were fixed and stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as described elsewhere (22). Rat cardiac myocytes were stained with the antimyosin monoclonal antibody MF 20 followed by rhodamine-labeled anti-mouse immunoglobulin G. Virus titers were determined by limiting dilution. Target cells were split into six-well plates the day prior to infection to give approximately 50% confluence at the time of infection. Infections were performed with serial dilutions of virus stock in a total of 0.5 ml of medium containing 8 μg of Polybrene per ml. After 3 to 6 h at 37°C, 2 ml of medium was added and the plates were incubated at 37°C for an additional 3 days. The medium was then aspirated, and 2 ml of medium supplemented with G418 (0.35 to 0.5 mg of active drug per ml; Life Technologies) was placed into each well. The medium was changed every 3 to 4 days, and the colonies were counted on day 10 or 14 after staining with crystal violet (0.2% in 20% ethanol). Alternatively, infected cells were trypsinized 3 days after infection and serially diluted into DMEM-FBS containing G418, and the colonies were stained 10 days later (23). The final yield of colonies was corrected for the increase in cell number between the time of infection and selection.
RESULTS
Design of an improved HIV-1-based vector system.
Our original two-component HIV-1-based vector system was composed of a vector construct and an independent Env-encoding component (49). The vector construct carried a deletion within the env coding region and harbored a reporter gene to be transferred to the target cells. It contained all sequences necessary for reverse transcription, vector integration, and expression of the reporter gene. In one of the vectors, an expression cassette consisting of the SV40 early promoter driving the bacterial neo gene was used. An additional vector contained the mouse HSA coding region as a reporter gene under the control of the human CMV immediate-early promoter. The formation of replication-competent HIV-1 was precluded because a substantial portion of the env coding region was missing in these vectors.
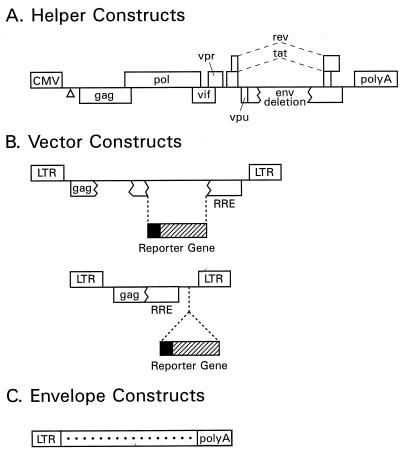
We have now improved this original vector system in terms of safety and flexibility. To minimize further the generation of replication-competent virus, the original two-component system was split into three components: a helper construct, a vector component, and an Env-encoding plasmid (Fig. 1). Our helper constructs (Fig. 1A) express the Gag, Pol, Tat, and Rev functions and, depending on the construct, retain functional vif, vpr, and vpu genes, but Nef is always absent. The 5′ LTR was replaced by the human CMV immediate-early promoter, and a heterologous polyadenylation signal was used instead of the 3′ LTR. All helper plasmids lack cis-acting sequences that have been implicated as important for efficient HIV-1 RNA packaging (10, 24, 32). The vector constructs (Fig. 1B) contain a reporter gene such as neo, HSA, ShlacZ, or EGFP. They also contain cis-acting sequences required for packaging, reverse transcription, and integration, including the 5′ and 3′ LTRs, and env-derived sequences encompassing the Rev response element (RRE). The expression cassette was placed either upstream or downstream of the RRE, depending on the design of the vector. The envelope constructs (Fig. 1C) provide a heterologous Env protein that leads to formation of HIV-1 pseudotypes. The Env proteins that were tested were those of VSV, rabies virus, Mokola virus, MoMLV, and HFV.
FIG. 1.
Components of the HIV-1 packaging system. (A) Helper constructs. The open triangle symbolizes a 33-bp deletion affecting the packaging signal between the 5′ splice donor site and the beginning of the gag sequence. Boxes interrupted by jagged lines contain partial deletions. (B) Transducing vector constructs. Top, vectors with expression cassette 5′ of the RRE; bottom, vectors with expression cassette 3′ of the RRE. (C) Env expression constructs.
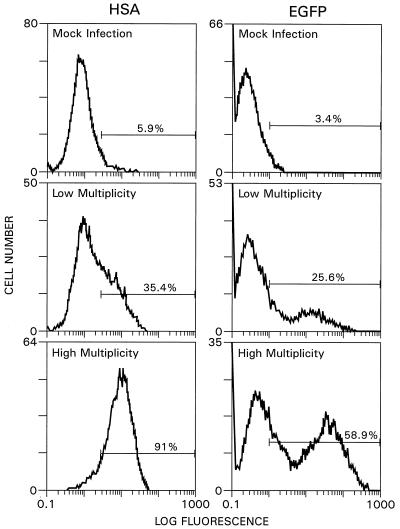
The ability of the newly designed vector system to mediate gene transfer was initially analyzed by infecting HOS cells with pseudotyped vectors carrying the HSA or EGFP reporter gene. Virus stocks were generated by transient cotransfection of 293T cells with a helper construct, together with the HIV-HSAΔE and HIV-EGFPΔE reporter vectors and a VSV-G-encoding plasmid. HOS cells were used primarily because they had previously been shown to be readily infectable by pseudotyped vectors (23, 49). To test for expression of the reporter genes, cells were collected 3 days after infection and processed for quantitative FACS analysis (Fig. 2). Depending on the multiplicity of infection used, up to 85% of the cells expressed HSA and up to 55% of the cells were EGFP positive (Fig. 2), indicating that the improved vector system was as efficient as our original two-component system in delivering the HSA reporter gene to HOS cells (49).
FIG. 2.
Efficiency of HSA and EGFP reporter gene expression in HOS cells. Cells were infected with HIV-HSAΔE or HIV-EGFPΔE pseudotypes and tested for expression of the reporter genes by FACS analysis. Approximately 5 × 105 cells were infected with 0.4 ml of a control virus supernatant for the mock infection; for the low- and high-multiplicity infections, 0.04 or 0.4 ml, respectively, of HIV-HSAΔE or HIV-EGFPΔE pseudotype stock were used. Infected cells were processed for FACS analysis 3 days later.
Effects of accessory proteins and IN on efficiency of the vector system.
To test the influence of the various accessory proteins and IN on the efficiency of the gene transfer system, virus stocks were generated by using a number of different helper constructs, together with the HIV-neoΔE or HIV-HSAΔE reporter vectors and a VSV-G-encoding plasmid. Virus stocks were harvested 60 h after transfection and subsequently used to infect HOS cells. Selection for growth in the presence of G418 allowed us to quantify the infection titer of each virus stock by counting G418-resistant colonies. The results presented in Table 1 show that pseudotype formation with the HIV-neoΔE vector was very efficient, with titers of the unconcentrated virus reaching 8.5 × 107 G418-resistant CFU/μg of p24. Colony formation was strictly dependent on the presence of a helper construct. In the absence of a helper construct, the titers were below the detection limit of the assay (Table 1). Also, a functional IN protein was necessary for efficient gene transfer. A helper construct with a defective IN core domain (C-Help IN) (15) yielded a G418 titer 3 orders of magnitude below the one obtained with helper constructs encoding a functional IN (Table 1), demonstrating that IN is required for efficient gene transfer to occur. To test the impact of the other accessory proteins on the formation of infectious pseudotypes, helper constructs lacking functional vif, vpu, and/or vpr coding regions were designed and tested. The results presented in Table 1 show that helper constructs with a mutated vpr coding region, carrying either a frameshift mutation at position 5743 (C-Help vpr−) or a 115-bp deletion (C-Help Δvpr), yielded vector particles that efficiently transduced HOS cells. In addition, C-Help constructs lacking the Vif, Vpr, and Vpu accessory proteins (C-Help ΔvifΔvprΔvpu) produced pseudotypes that efficiently delivered the neo reporter gene into HOS cells. We used quantitative FACS analysis to test in parallel the functionality and efficiency of the various helper constructs by monitoring the expression of the HSA reporter gene. HOS cells were stained 3 days after infection for cell surface-expressed HSA with fluorescein isothiocyanate-labeled anti-HSA antibody. No signal above background levels was seen with cells that had been infected with HIV-HSAΔE stocks previously prepared by using an IN-deficient helper construct (C-Help IN) (Table 1) or in the presence of 10 μM zidovudine (AZT) (36). The C-Help vpr−, C-Help Δvpr, and C-Help ΔvifΔvprΔvpu constructs produced pseudotypes that infected HOS cells as efficiently as virus stocks that had been prepared by using the C-Help construct producing intact Vif, Vpr, and Vpu, thus confirming the results obtained in assays using G418 selection. Taken together, these results indicate that Vif, Vpr, Vpu, and Nef are dispensable for infection of proliferating HOS cells.
TABLE 1.
Effect of packaging construct on pseudotype formationa
| Packaging construct | Functions missing | Virus titer (CFU/μg of p24)c | Relative HSA expressione (% of C-Help level) |
|---|---|---|---|
| None | NAb | <102d | <1 |
| C-Help | Ψ, Env, Nef, 5′ LTR, 3′ LTR | 5.8 × 107 | 100 |
| C-Help vpr− | Ψ, Vpr, Env, Nef, 5′ LTR, 3′ LTR | 5.0 × 107 | 153 |
| C-Help Δvpr | Ψ, Vpr, Env, Nef, 5′ LTR, 3′ LTR | 4.6 × 107 | 144 |
| C-Help ΔvifΔvprΔvpu | Ψ, Vif, Vpr, Vpu, Env, Nef, 5′ LTR, 3′ LTR | 8.5 × 107 | 109 |
| C-Help IN | Ψ, Int, Env, Nef, 5′ LTR, 3′ LTR | 5.3 × 104 | <1 |
Vector constructs used were HIV-neoΔE and HIV-HSAΔE; the Env construct used encoded VSV-G.
NA, not applicable.
Determined by endpoint dilution (n = 3). Cells were selected in medium containing G418. The colonies were stained 10 days later.
<102 indicates that no colonies were obtained above the detection limit of the assay.
Determined by FACS analysis (n = 3). <1 indicates signal at background level.
The presence of a functional tat coding region in the vector enhances pseudotype titers.
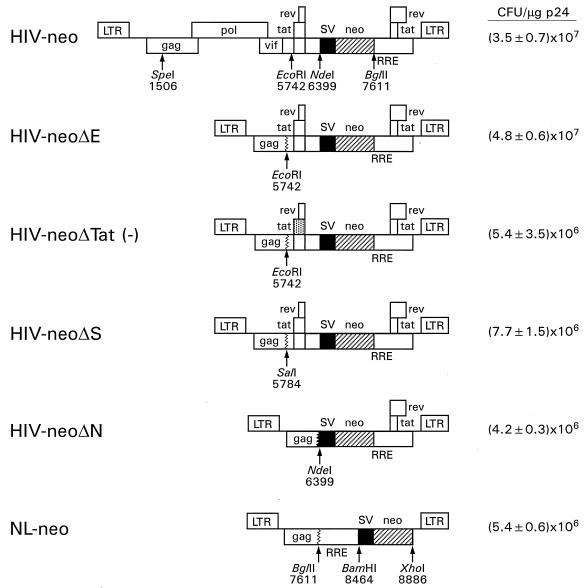
With a view toward constructing safe and efficient HIV-1-based gene transfer vectors, we designed constructs with deletions of various lengths affecting the gag, pol, vif, vpr, vpu, and 5′ tat and rev coding regions and tested the efficiency of formation of G418-resistant colonies. To do this, the previously described HIV-neo vector (49) was modified. A schematic diagram of this vector and the various deletion derivatives is shown in Fig. 3. In the HIV-neoΔE construct, sequences from the SpeI site (position 1506) up to the EcoRI site (position 5742) were deleted, thus completely eliminating the pol and vif genes and truncating the gag and vpr coding regions, but the 5′ tat and rev exons remain intact. In contrast, HIV-neoΔTat (−) contains a mutated tat coding region carrying two consecutive stop codons after amino acid 10, leading to a truncated version of Tat (19). However, since the rev coding region is unaltered, functional Rev protein is produced from this vector. In the HIV-neoΔS construct, the 5′ tat and rev exons are retained but the splice acceptor site at position 5777 is missing; in plasmid HIV-neoΔN, sequences up to the NdeI site at position 6399 were removed, thereby deleting the 5′ tat and rev exons and 178 nucleotides of the gp160 coding region. The efficiency of each of these HIV-neo constructs was tested by using the C-Help construct to produce pseudotyped virus stocks and testing the viruses on HOS cells. The efficiency of formation of G418-resistant colonies differed markedly among the various HIV-neo deletion constructs. While the HIV-neoΔE construct was as efficient as the original HIV-neo construct in generating G418-resistant colonies upon infection of HOS cells ([4.8 ± 0.6] × 107 versus [3.5 ± 0.7] × 107 CFU/μg of p24), the HIV-neoΔTat (−), HIV-neoΔS, and HIV-neoΔN derivatives gave six- to ninefold-reduced yields of G418-resistant colonies relative to that obtained with HIV-neoΔE. The NL-neo vector was constructed according to the design of Parolin et al. (41) and Naldini et al. (38), with the expression cassette located 3′ of the RRE. The titer obtained with this vector was comparable with the titers obtained with the other vector constructs lacking Tat. This finding suggests that the presence of Tat in the vector has a marked influence on the efficiency of formation of G418-resistant colonies.
FIG. 3.
Influence of Tat in vector construct on efficiency of formation of G418-resistant colonies. The top portion represents the original HIV-neo vector (49). The various deletion derivatives are shown below. The point-mutated 5′ tat exon in HIV-neoΔTat (−) is highlighted with dots. NL-neo carries the neo expression cassette 3′ of the RRE. The efficiency of colony formation (average ± standard deviation of the results of three to four independent experiments) is shown on the right for each vector construct.
Pseudotype formation using alternative Env glycoproteins.
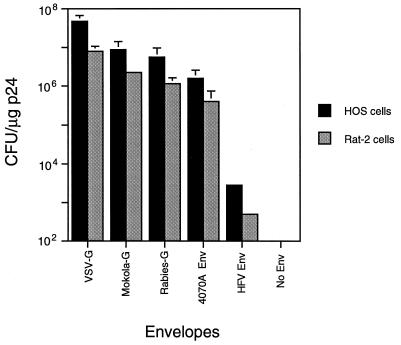
The capacity of HIV-1-based vectors to form pseudotypes with other Env proteins was investigated. The rabies virus G glycoprotein and the G protein of a related rhabdovirus, Mokola virus (Lyssavirus serotype 3), were tested, together with the Env protein of HFV and the amphotropic MoMLV 4070A Env, for the ability to yield infectious particles. All env coding regions were expressed under the control of the HIV-1 LTR. The normalized efficiency of formation of resistant colonies following packaging of the HIV-neoΔE construct and subsequent infection of HOS cells and Rat-2 cells was determined. Figure 4 shows that VSV-G yielded up to 5 × 107 CFU/μg of p24 on HOS cells and up to 8 × 106 CFU/μg of p24 on Rat-2 cells. Particles pseudotyped with the Mokola virus and rabies virus G glycoproteins, and the MoMLV 4070A Env, yielded infectious titers of up to 9 × 106 CFU/μg of p24 on HOS cells and up to 2 × 106 CFU/μg of p24 on Rat-2 cells. The HFV Env also led to formation of HIV-1 pseudotypes, but the titers obtained (2.8 × 103 CFU/μg of p24 on HOS cells and 2.5 × 102 CFU/μg of p24 on Rat-2 cells) were 4 orders of magnitude below those obtained with VSV-G. These results underscore the flexibility of the HIV-1 vector system to form infectious pseudotypes, but they show its limitation as far as the HFV Env is concerned.
FIG. 4.
Pseudotype formation using alternative Env proteins. HOS cells or Rat-2 cells were infected with HIV-neoΔE pseudotypes carrying different Env proteins. Cells were trypsinized 3 days after infection and serially diluted into DMEM-FBS containing G418 (0.35 mg of active drug per ml). Colonies were stained 10 to 14 days later. 4070A Env, amphotropic MoMLV Env. Error bars represent standard deviations.
Assay for the generation of replication-competent virus.
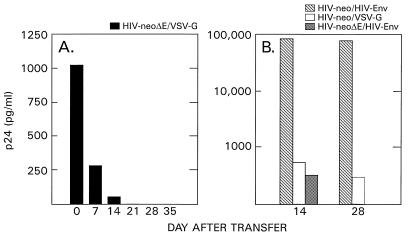
We next wished to determine if replication-competent virus that would subsequently be able to replicate in human T cells was produced during transient transfection. Two parallel cultures of the human H9 T-cell line (31) were infected with an HIV-neoΔE stock pseudotyped with VSV-G for 3 days and subsequently split 1:4. This procedure was repeated five more times over a period of 35 days, and the supernatants were assayed for the presence of p24. The data in Fig. 5A show that extracellular p24 levels decreased with increasing numbers of transfer and that final p24 concentrations reached background levels. A similar decrease in p24 levels was observed in cultures infected with HIV-neoΔE stocks harboring the HIV-1 HXB2 Env protein (Fig. 5B). This finding indicates that there was no substantial de novo production of p24-positive particles within the detection limits of the assay. However, cultures infected with HIV-neo stocks harboring the HIV-1 Env protein yielded extracellular p24 levels up to 80 ng/ml, and these levels remained high after five transfers (Fig. 5B). This finding is consistent with the view that replication-competent virus was emerging. HIV-neo vectors pseudotyped with VSV-G yielded p24 levels around 500 pg/ml after the third transfer and around 300 pg/ml after the fifth. Low but constant p24 levels were expected in this case, as the HIV-neo vector backbone contains a functional p24 coding region.
FIG. 5.
Detection of replication-competent virus. (A) Duplicate cultures of human H9 cells (0.9 × 106 to 1.2 × 106 cells per culture) were infected with 0.5 ml of HIV-neoΔE pseudotype stock. The cells were split six times at a ratio of 1:4 over a period of 35 days; p24 was assayed throughout the experiment. Average p24 levels are shown. (B) H9 cells were infected with HIV-neo and HIV-neoΔE stocks containing the HIV-1 Env protein (HIV-neo/HIV-Env and HIV-neoΔE/HIV-Env, respectively). HIV-neo stocks pseudotyped with VSV-G (HIV-neo/VSV-G) were run in parallel. The cells were split once a week, and p24 concentrations were determined. The p24 values observed at days 14 and 28 are shown.
Transduction of nondividing cells.
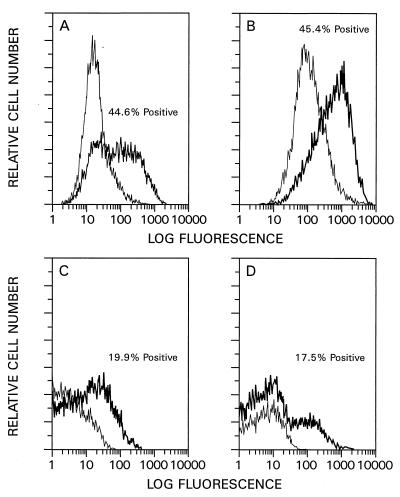
The ability of the newly designed HIV-1 vector system to mediate gene transfer into nondividing cells was analyzed by transducing growth-arrested HOS cells, contact-inhibited primary HSFs, postmitotic rat cardiac myocytes, and postmitotic rat cerebellar neurons. The relative efficiency of gene transfer into dividing and nondividing cells was investigated first. G2-arrested HOS cells were prepared by γ irradiation (26). Cell cycle analysis of the irradiated cells showed that up to 80% of the cells were in G2 (36). Such cells were subsequently infected with HIV-HSAΔE pseudotype stocks and analyzed by quantitative FACS analysis 3 days later. The numbers of HSA-positive cells were similar for irradiated (44.6% [Fig. 6B]) and nonirradiated (45.5% [Fig. 6A]) cells. Primary HSFs were growth arrested by being allowed to reach contact inhibition upon cultivation in medium containing 10% FBS for 3 weeks. Such cells have previously been shown to be highly enriched for populations in G0 and/or G1 (49, 61). Dividing control HSFs were prepared by subcultivation 2 days before infection. Dividing and nondividing HSFs were infected with HIV-EGFPΔE pseudotype stocks and analyzed by quantitative FACS analysis 3 days later. The fraction of EGFP-positive, dividing HSFs was 19.9% (Fig. 6C), while 17.5% of the contact-inhibited, nondividing HSFs (Fig. 6D) were EGFP positive, indicating that infection efficiency was independent of the proliferative status of the cells. Given that only about 20% of the infected HSFs were EGFP positive, a control infection was done with HOS cells in parallel. The results revealed that over 60% of these cells were EGFP positive, indicating that there are quantitative differences in the abilities of VSV-G-pseudotyped HIV-1-based vectors to infect primary cells versus established cell lines.
FIG. 6.
Relative infection efficiencies of dividing and nondividing cells. Nonirradiated (A) and irradiated (B) HOS cells were infected with pseudotyped HIV-HSAΔE vector stocks and subjected to FACS analysis 3 days later. Approximately 5 × 105 cells were plated into six-well plates and infected with 0.25 ml of virus stock. Dividing (C) and contact-inhibited (nondividing) (D) HSFs were infected with pseudotyped HIV-EGFPΔE vector stocks and subjected to FACS analysis 3 days after infection. Between 0.5 × 105 and 2.5 × 105 cells in six-well plates were infected with 0.25 ml of virus. HIV-neoΔE virus stocks were used as mock controls. Thick lines, HIV-HSAΔE- and HIV-EGFPΔE-infected cells; thin lines, mock-infected cells.
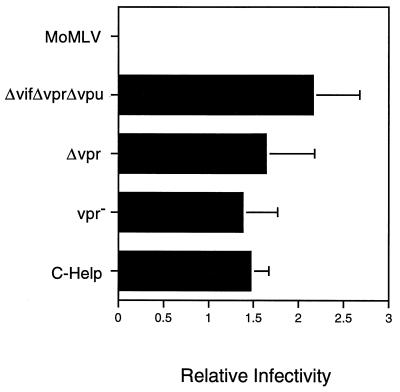
The influence of the Vpr, Vif, and Vpu accessory proteins on the efficiency of reporter gene transfer into G2-arrested HOS cells and contact-inhibited HSFs was determined next. HOS cells were irradiated (4,000 rads), subsequently infected with HIV-HSAΔE vector stocks, and processed for quantitative FACS analysis 3 days later. Subconfluent HOS cells were similarly infected and processed in parallel, and the ratios of the percentages of HSA-positive, nondividing (irradiated) HOS cells versus HSA-positive, dividing HOS cells were determined. These ratios varied only slightly, regardless of the C-Help construct used (Fig. 7). The MoMLV-derived G1-HSA vector stock served as a control. It did not transduce G2-arrested cells above background levels, indicating that the G2 block was effective. Contact-inhibited, nondividing HSFs were infected using HIV-EGFPΔE vector stocks and inspected by fluorescence microscopy 29 days later (Fig. 8). Vector stocks assembled from packaging constructs in which all accessory protein-encoding regions were absent were as efficient as the corresponding stocks assembled from wild-type constructs (compare Fig. 8B and A). A vector stock assembled from an IN-deficient helper was severely impaired (Fig. 8C). Taken together, these results show that the vif, vpr, and vpu genes in the helper construct are dispensable for infection of growth-arrested HOS cells and contact-inhibited HSFs.
FIG. 7.
Infection of growth-arrested HOS cells. Irradiated (4,000 rads) or nonirradiated HOS cells were infected with HIV-HSAΔE vector stocks previously prepared by using different helper constructs and subjected to FACS analysis 3 days later. Approximately 5 × 105 cells in six-well plates were infected with 0.25 ml of VSV-G-pseudotyped virus stocks. The relative infectivity represents the percentage of nondividing (irradiated) HSA-positive cells versus the percentage of dividing (nonirradiated) HSA-positive cells. The various helper constructs and the MoMLV-derived G1-HSA control vector stock are indicated. Error bars represent standard deviations.
FIG. 8.
Infection of contact-inhibited HSFs. (A to C) HSFs infected with HIV-EGFPΔE pseudotypes. (A) vif, vpr, and vpu genes present in helper construct; (B) all accessory protein-encoding genes absent in helper construct; (C) IN control; (D) mock infection.
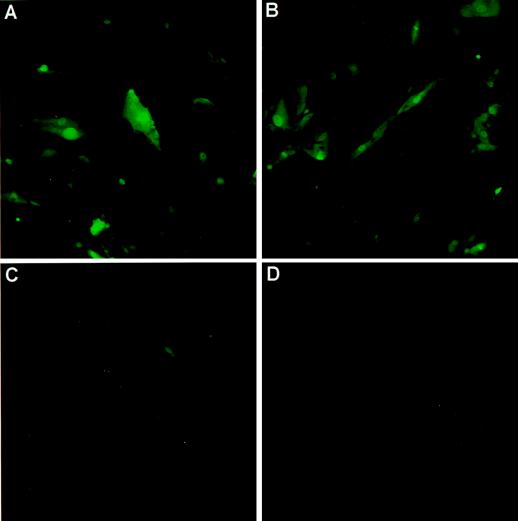
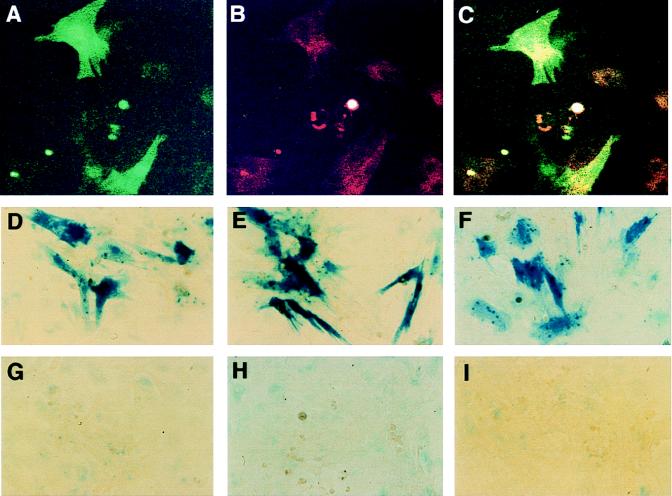
Primary neonatal rat ventricular myocytes prepared from the hearts of 2- to 3-day-old rats were prepared and infected 5 days later with HIV-EGFPΔE and HIV-ShlacZΔE pseudotypes or a pseudotyped MoMLV-lacZ control vector. The results presented in Fig. 9A to C show that myocytes had been successfully infected in that the EGFP-positive cells clearly overlapped the rhodamine-positive cells, labeled with a myosin-specific antibody; Fig. D to F show that none of the accessory proteins are required to generate virus that can infect such myocytes, as judged from the X-Gal staining; Fig. 9H (IN control) shows that the signals observed are not due to pseudotransduction. Moreover, the cells were no longer dividing at the time of infection, as judged from results for the MoMLV-lacZ virus control (Fig. 9G).
FIG. 9.
Infection of postmitotic rat cardiac myocytes. (A to C) Rat cardiac myocytes infected with HIV-EGFPΔE pseudotypes. (A) EGFP fluorescence; (B) myosin-specific rhodamine fluorescence; (C) combination of panels A and B. (D to I) Cardiac myocytes infected with HIV-ShlacZΔE. (D) vif, vpr, and vpu genes present in helper construct; (E) vif and vpu genes present in helper construct; (F) all accessory protein-encoding genes absent in helper construct; (G) MoMLV-lacZ vector; (H) IN control; (I) mock infection. Approximately 2 × 105 myocytes in six-well plates were infected with 0.3 to 0.5 ml of the various virus stocks. The cells were stained with X-Gal or processed for immunofluorescence 3 days after infection.
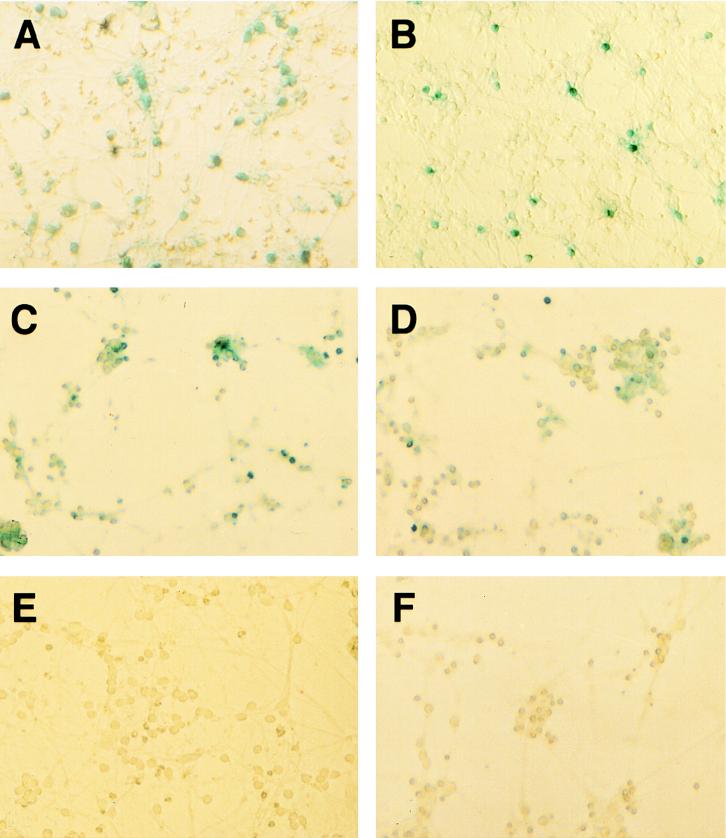
Cerebellar granule cells, also nondividing cells (60), were prepared from 8-day-old rat pups and infected 1 day later with HIV-ShlacZΔE and HIV-ShlacZΔN stocks pseudotyped with VSV-G. The HIV-ShlacZΔN vector is similar to the HIV-neoΔN vector because it lacks the 5′ tat and rev exons. Expression of the reporter gene was detected by β-galactosidase staining using the X-Gal substrate 3 days later. Up to 30% of the granule cells were X-Gal positive after overnight incubation (Fig. 10A and B). HIV-ShlacZΔE stocks previously prepared by using a vpr-deficient helper construct or helper constructs lacking all accessory proteins were as efficient as the corresponding stocks that had been prepared by using wild-type helper constructs (Fig. 10C and D). There was no X-Gal staining in granule cells that were infected in the presence of 10 μM AZT (Fig. 10F), suggesting that the staining is specific. MoMLV-based vectors encoding β-galactosidase did not produce signals above background levels (36).
FIG. 10.
Infection of postmitotic rat cerebellar granule cells. (A, C, D, and F) Rat cerebellar granule cells infected using HIV-ShlacZΔE pseudotypes; (B) rat cerebellar granule cells infected using HIV-ShlacZΔN pseudotypes; (C) vpr gene absent in helper construct; (D) all accessory protein-encoding genes absent in helper construct; (E) mock infection; (F) cells infected in the presence of 10 μM AZT. Approximately 1.6 × 105 granule cells in 35-mm-diameter dishes were infected with 0.3 ml of virus stock and stained with X-Gal 3 days later.
DISCUSSION
We have developed an efficient three-component packaging system to produce HIV-1 pseudotypes. The design of the system is based on the concept of split packaging systems that have been available for oncoretroviruses for over a decade (34). Transient three-component packaging systems that include the native HIV-1 gp160 have been available for some time (5, 45, 53, 50, 59), but their efficiency was generally quite low, with titers on the order of 103 to 104 transducing units per ml of supernatant. Naldini et al. (38) were the first to describe an HIV-1-based three-component system that involves heterologous Env proteins. Their pseudotype titers with VSV-G or the MoMLV amphotropic Env protein ranged from 1 × 105 to 4 × 105 transducing units per ml, based on β-galactosidase staining of transduced cells. The efficiency of our three-component system appears to be higher. The titers obtained with neo vectors were between 1 × 106 and 2 × 107 CFU/ml, depending on the vector construct used. Titers above 107 CFU/ml (up to 8 × 107 CFU/μg of p24) were routinely obtained provided that a functional Tat coding region was present in the vector. Vector constructs lacking a functional Tat protein typically yielded titers of around 106 CFU/ml (4 × 106 to 8 × 106 CFU/μg of p24).
The use of heterologous Env proteins such as VSV-G is assumed to preclude the formation of replication-competent HIV-1, and the separation of the helper functions from the vector functions further adds to the safety of the system. However, there is still substantial sequence overlap between the vector and helper constructs as far as gag sequences and sequences spanning the RRE are concerned. Theoretically, these regions of overlap could lead to recombinants that could reconstitute packageable helper constructs, and eliminating them will be mandatory in the long run to make safe HIV-1 vectors. Rev-independent HIV-1 helper constructs as described by Schneider et al. (52) and Srinivasakumar et al. (57) may be helpful in this respect. They may facilitate the design of packaging systems that lack RRE altogether, thus reducing the likelihood of recombination.
We have previously reported that functional vpr and nef coding regions are not required for generating high-titer HIV-1 vector stocks and for the subsequent transduction of proliferating and growth-arrested cells (49). The results presented here extend these findings and are consistent with the view that the Vif, Vpr, Vpu, and Nef functions are not required for the efficient transduction of proliferating and growth-arrested HOS cells in vitro. Also, the efficiency of transduction of postmitotic rat cardiac myocytes and cerebellar neurons was the same regardless of whether functional Vif, Vpr, or Vpu was present in the packaging construct. Some or all of the HIV-1 accessory proteins may be needed in other cell types to achieve efficient transduction. Zufferey et al. (65), Kafri et al. (20), and Kim et al. (21) recently described HIV-1 helper constructs in which several of the genes encoding accessory proteins have been deleted. Vector constructs packaged with these multiply deleted helper constructs retained the ability to transduce growth-arrested cells and monocyte-derived macrophages in culture and could efficiently deliver genes in vivo into muscle and adult neurons. Vpr and Vif were found to be required for efficient gene delivery into the liver (20).
In the study presented here, we compared different HIV-neo vector constructs for the ability to yield G418-resistant colonies. The HIV-neoΔE vector harbors intact 5′ tat and rev exons, while the HIV-neoΔTat (−) vector harbors a point-mutated 5′ tat exon and lacks the capacity to produce functional Tat. In the HIV-neoΔN construct, the 5′ tat and rev exons are missing. Compared to the HIV-neoΔTat (−) and HIV-neoΔN constructs, the HIV-neoΔE construct exhibited an approximately 6- to 10-fold-higher yield of G418-resistant colonies. This is probably due to the production of functional Tat by the HIV-neoΔE vector in infected target cells and concomitant improved neo gene expression from the Tat-regulated viral LTR, thus boosting the yield of G418-resistant colonies. This assumption is supported by the findings of Parolin et al. (42), who showed that Tat-driven reporter gene expression was substantially higher than expression of vectors that used heterologous internal promoter elements; the difference in expression was not due to improved packaging of the vector constructs. Although Tat-driven expression is very robust, the continuous production of Tat and Rev in the target cell may be detrimental, as Tat has been implicated in a number of effects besides its contribution to transcription, including cytotoxicity (8). However, for certain in vitro experiments, such as screening of cDNA expression libraries, the high efficiency of gene transfer may be advantageous and the presence of Tat in the vector construct may be desirable.
HIV-1 appears to be flexible in terms of forming pseudotypes, thereby allowing the expansion of its host range. In addition to VSV-G and the MoMLV amphotropic Env, rabies virus and Mokola virus G proteins formed pseudotypes, although the yield of G418-resistant colonies was highest with VSV-G pseudotypes. This difference may reflect, in part, receptor levels, pseudotype stability, and efficiency of pseudotype formation. Poor pseudotype formation involving the HFV Env may explain the low yield of G418-resistant colonies. This possibility is supported by the recent findings that the HFV Env cytoplasmic domain harbors an endoplasmic reticulum retention signal (18, 28), which may affect the proper assembly of HIV-1 pseudotypes at the plasma membrane.
The lacZ reporter gene can lead to pseudotransduction artifacts under conditions where highly concentrated VSV-G-pseudotyped MoMLV-derived vectors are used (29). We have ruled out pseudotransduction by including an IN control and by using AZT during infection. Also, because our titers are high, there is generally no need to concentrate the virus.
The three-component HIV-1 packaging system described here is efficient, robust, and safe. However, the design of efficient packaging cell lines is mandatory. The various constructs described here will be helpful in constructing such cell lines.
ACKNOWLEDGMENTS
We thank Kuan-Teh Jeang for critical reading of the manuscript and Kouji Horiba and Kazuyo Takeda for help with fluorescence microscopy. We are grateful to Manfred Schubert for making available his BL-2/BL-3 facility and to Delores Wilson for preparation of the neuron cultures.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkina R K, Walton R M, Chen M L, Li Q-X, Planelles V, Chen I S Y. High-efficiency gene transfer into CD34+ cells with a human immunodeficiency virus type 1-based retroviral vector pseudotyped with vesicular stomatitis virus envelope glycoprotein G. J Virol. 1996;70:2581–2585. doi: 10.1128/jvi.70.4.2581-2585.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron M, Reyes J P, Stassi D, Tiraby G. A selectable bifunctional β-galactosidase::phleomycin resistance fusion protein as a potential marker for eukaryotic cells. Gene. 1992;114:239–243. doi: 10.1016/0378-1119(92)90581-9. [DOI] [PubMed] [Google Scholar]
- 4.Boshart M, Weber F, Jahn G, Dorsch-Häsler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 5.Buchschacher G L, Jr, Panganiban A T. Human immunodeficiency virus vectors for inducible expression of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canivet M, Hoffman A D, Hardy D, Sernatinger J, Levy J A. Replication of HIV-1 in a wide variety of animal cells following phenotypic mixing with murine retroviruses. Virology. 1990;178:543–551. doi: 10.1016/0042-6822(90)90352-r. [DOI] [PubMed] [Google Scholar]
- 7.Carroll R, Lin J-T, Dacquel E J, Mosca J D, Burke D S, St. Louis D C. A human immunodeficiency virus type 1 (HIV-1)-based retroviral vector system utilizing stable HIV-1 packaging cell lines. J Virol. 1994;68:6047–6051. doi: 10.1128/jvi.68.9.6047-6051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang H K, Gallo R C, Ensoli B. Regulation of cellular gene expression and function by the human immunodeficiency virus type 1 Tat protein. J Biomed Sci. 1995;2:189–202. doi: 10.1007/BF02253380. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro B, Wehrly K, Maury W. Differential expression in human and mouse cells of human immunodeficiency virus pseudotyped by murine retroviruses. J Virol. 1990;64:4553–4557. doi: 10.1128/jvi.64.9.4553-4557.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbeau P, Kraus G, Wong-Staal F. Efficient gene transfer by a human immunodeficiency virus type 1 (HIV-1)-derived vector utilizing a stable packaging cell line. Proc Natl Acad Sci USA. 1996;93:14070–14075. doi: 10.1073/pnas.93.24.14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DuBridge R B, Tang P, Hsia H C, Phaik-Mooi L, Miller J H, Calos M P. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar C E. Gene transfer to hematopoietic stem cells: implications for gene therapy of human disease. Annu Rev Med. 1996;47:11–20. doi: 10.1146/annurev.med.47.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Emerman M. From curse to cure: HIV for gene therapy? Nat Biotechnol. 1996;14:943. doi: 10.1038/nbt0896-943. [DOI] [PubMed] [Google Scholar]
- 15.Englund G, Theodore T S, Freed E O, Engelman A, Martin M. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flügel R M, Rethwilm A, Maurer B, Darai G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987;6:2077–2084. doi: 10.1002/j.1460-2075.1987.tb02473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs J S, Regier D A, Desrosiers R C. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res Hum Retroviruses. 1994;10:343–350. doi: 10.1089/aid.1994.10.343. [DOI] [PubMed] [Google Scholar]
- 18.Goepfert P A, Shaw K L, Ritter G D, Jr, Mulligan M J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J Virol. 1997;71:778–784. doi: 10.1128/jvi.71.1.778-784.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang L-M, Joshi A, Wiley R, Orenstein J, Jeang K-T. Human immunodeficiency viruses regulated by alternative trans-activators: genetic evidence for a novel non-transcriptional function of Tat in virion infectivity. EMBO J. 1994;13:2886–2896. doi: 10.1002/j.1460-2075.1994.tb06583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kafri T, Blömer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 21.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landau N R, Page K, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lever A, Göttlinger H, Haseltine W, Sodroski J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J Virol. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lever A M L. HIV and other lentivirus-based vectors. Gene Ther. 1996;3:470–471. [PubMed] [Google Scholar]
- 26.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis P F, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindemann D, Bock M, Schweizer M, Rethwilm A. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J Virol. 1997;71:4815–4820. doi: 10.1128/jvi.71.6.4815-4820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M-L, Winther B L, Kay M A. Pseudotransduction of hepatocytes by using concentrated pseudotyped vesicular stomatitis virus G glycoprotein (VSV-G)–Moloney murine leukemia virus-derived retrovirus vectors: comparison of VSV-G and amphotropic vectors for hepatic gene transfer. J Virol. 1996;70:2497–2502. doi: 10.1128/jvi.70.4.2497-2502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusso P, Di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco S E, Kalyanaraman V S, Gallo R C. Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science. 1990;247:848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 31.Mann D L, O’Brien S J, Gilbert D A, Reid Y, Popovic M, Read-Conolle E, Gallo R, Gadzar A. Origin of the HIV-susceptible human CD4+ cell line H9. AIDS Res Hum Retroviruses. 1989;5:253–255. doi: 10.1089/aid.1989.5.253. [DOI] [PubMed] [Google Scholar]
- 32.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mebatsion T, Schnell M J, Conzelmann K-K. Mokola virus glycoprotein and chimeric proteins can replace rabies virus glycoprotein in the rescue of infectious defective rabies virus particles. J Virol. 1995;69:1444–1450. doi: 10.1128/jvi.69.3.1444-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller A D. Retrovirus packaging cells. Hum Gene Ther. 1990;1:5–14. doi: 10.1089/hum.1990.1.1-5. [DOI] [PubMed] [Google Scholar]
- 35.Miller D G, Adam M A, Miller A D. Gene transfer by retrovirus vectors occurs only in cells that are actively replicating at the time of infection. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki, H., and J. Reiser. Unpublished data.
- 37.Myers G, Foley B, Mellors J W, Korber B, Jeang K-T, Wain-Hobson S. Human retroviruses and AIDS 1996. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 38.Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 39.Ott D, Friedrich R, Rein A. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J Virol. 1990;64:757–766. doi: 10.1128/jvi.64.2.757-766.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page K A, Landau N R, Littman D R. Construction and use of human immunodeficiency virus vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parolin C, Dorfman T, Palú G, Göttlinger H, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parolin C, Taddeo B, Palú G, Sodroski J. Use of cis- and trans-acting viral regulatory sequences to improve expression of human immunodeficiency virus vectors in human lymphocytes. Virology. 1996;222:415–422. doi: 10.1006/viro.1996.0438. [DOI] [PubMed] [Google Scholar]
- 43.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poeschla E, Corbeau P, Wong-Staal F. Development of HIV vectors for anti-HIV gene therapy. Proc Natl Acad Sci USA. 1996;93:11395–11399. doi: 10.1073/pnas.93.21.11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poznansky M, Lever A, Bergeron L, Haseltine W, Sodroski J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. J Virol. 1991;65:532–536. doi: 10.1128/jvi.65.1.532-536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pracyk J B, Hegland D D, Tanaka K. Effect of a dominant negative ras on myocardial hypertrophy by using adenoviral-mediated gene transfer. Surgery. 1997;122:404–411. doi: 10.1016/s0039-6060(97)90033-7. [DOI] [PubMed] [Google Scholar]
- 47.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Jr, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N T, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 48.Ratner L, Fisher A, Jagodzinski L L, Mitsuya H, Liou R-S, Gallo R C, Wong-Staal F. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res Hum Retroviruses. 1987;3:57–63. doi: 10.1089/aid.1987.3.57. [DOI] [PubMed] [Google Scholar]
- 49.Reiser J, Harmison G, Kluepfel-Stahl S, Brady R O, Karlsson S, Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc Natl Acad Sci USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson J H, Kaye J F, Child L A, Lever A M L. Helper-free transfer of human immunodeficiency virus type 1 vectors. J Gen Virol. 1995;76:691–696. doi: 10.1099/0022-1317-76-3-691. [DOI] [PubMed] [Google Scholar]
- 51.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimada T, Fujii H, Mitsuya H, Nienhuis A W. Targeted and highly efficient gene transfer into CD4+ cells by a recombinant human immunodeficiency virus retroviral vector. J Clin Investig. 1991;88:1043–1047. doi: 10.1172/JCI115365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spector D H, Wade E, Wright D A, Koval V, Clark C, Jaquish D, Spector S A. Human immunodeficiency virus pseudotypes with expanded cellular and species tropism. J Virol. 1990;64:2298–2308. doi: 10.1128/jvi.64.5.2298-2308.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Springett G M, Moen R C, Anderson S, Blaese R M, Anderson W F. Infection efficiency of T lymphocytes with amphotropic retroviral vectors is cell cycle dependent. J Virol. 1989;63:3865–3869. doi: 10.1128/jvi.63.9.3865-3869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivasakumar N, Chazal N, Helga-Maria C, Prasad S, Hammarskjöld M-L, Rekosh D. The effect of viral regulatory protein expression on gene delivery by human immunodeficiency virus type 1 vectors produced in stable packaging cell lines. J Virol. 1997;71:5841–5848. doi: 10.1128/jvi.71.8.5841-5848.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevenson M. Portals of entry: uncovering HIV nuclear transport pathways. Trends Cell Biol. 1996;6:9–15. doi: 10.1016/0962-8924(96)81032-4. [DOI] [PubMed] [Google Scholar]
- 59.Strair R K, Medina D J, Nelson C J, Graubert T A, Mellors J W. Recombinant retroviral systems for the analysis of drug resistant HIV. Nucleic Acids Res. 1993;21:4836–4842. doi: 10.1093/nar/21.20.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taniwaki T, Becerra S P, Chader G J, Schwartz J P. Pigment epithelium-derived factor is a survival factor for cerebellar granule cells in culture. J Neurochem. 1995;64:2509–2517. doi: 10.1046/j.1471-4159.1995.64062509.x. [DOI] [PubMed] [Google Scholar]
- 61.Tobey R A, Valdez J G, Crissman H A. Synchronization of human diploid fibroblasts at multiple stages of the cell cycle. Exp Cell Res. 1988;179:400–416. doi: 10.1016/0014-4827(88)90279-0. [DOI] [PubMed] [Google Scholar]
- 62.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 63.Yu H, Rabson A B, Kaul M, Ron Y, Dougherty J P. Inducible human immunodeficiency virus type 1 packaging cell lines. J Virol. 1996;70:4530–4537. doi: 10.1128/jvi.70.7.4530-4537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu Z, Chen S S L, Huang A S. Phenotypic mixing between human immunodeficiency virus and vesicular stomatitis virus or herpes simplex virus. J Acquired Immune Defic Syndr. 1990;3:215–219. [PubMed] [Google Scholar]
- 65.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]