Abstract
Receptor for hyaluronan-mediated motility (RHAMM) is overexpressed in various tumors with high frequency, and was recently identified as an immunogenic antigen by serologic screening of cDNA expression libraries. In this study, we explored whether RHAMM is a potential target for dendritic cell (DC) immunotherapy. We constructed a plasmid for transduction of in vitro-transcribed mRNAs into DCs to efficiently transport the intracellular protein RHAMM into MHC class II compartments by adding a late endosomal/lysosomal sorting signal to the RHAMM gene. Immunization of mice with modified RHAMM mRNA-transfected DCs (DC/RHAMM) induced killing activity against RHAMM-positive tumor cells in splenocytes. To examine whether CD4+ and/or CD8+ T cells were required for this antitumor immunity, an anti-CD4 or anti-CD8 antibody was administered to mice after immunization with DC/RHAMM. Depletion of CD4+ T cells significantly diminished the induction of tumor cell-killing activity in splenocytes, whereas CD8+ T cell depletion had no effect. We then investigated the therapeutic effect of DC/RHAMM in a 3-day tumor model of EL4. DC/RHAMM was administered to mice on days 3, 7 and 10 after EL4 tumor inoculation. The treatment markedly inhibited tumor growth compared to control DCs. Moreover, antibody-mediated depletion of CD4+ T cells completely abrogated the therapeutic effect of DC/RHAMM, whereas depletion of CD8+ T cells had no effect. The results of this preclinical study indicate that DCs transfected with a modified RHAMM mRNA targeted to MHC class II compartments can induce CD4+ T cell-mediated antitumor activity in vivo.
Keywords: Dendritic cells, RHAMM, mRNA, CD4 T cells, MHC class II, Centrosomal protein
Introduction
Centrosome aberrations, which indicate an underlying deregulation of centrosome structure, duplication or segregation, have been found in a variety of cancers with high frequency [1, 2]. Probably connected with these events, there is an increasing evidence that some centrosomal proteins are overexpressed in cancer cells and may be related to cancer development [3–8]. These proteins could be of use not only as molecular tools for analyzing the mechanism for centrosome aberrations and chromosomal instability, but also as target antigens for cancer immunotherapy if they are immunogenic.
Recently, the genes encoding two centrosomal proteins, namely transforming acidic coiled-coil (TACC) protein and receptor for hyaluronan-mediated motility (RHAMM), were detected by serological screening of an expression library (SEREX) of cultured tumor cells or tumor tissues [9–11]. TACC transcripts, except for two alternatively spliced isoforms, were universally expressed in all normal tissues [9], whereas expression of full-length RHAMM mRNA in normal tissues was restricted to few organs including the thymus, placenta and testis [10]. Importantly, RHAMM has been found to be expressed in a wide variety of tumors. Its expression was revealed in breast cancer [12], colon cancer [13], multiple myeloma [14, 15] and acute myelogenous leukemia [16] by RT-PCR, and in breast [12, 17], stomach [18], urinary bladder [19] and endometrial [20] cancers by immunohistochemistry. Moreover, the expression of RHAMM was associated with metastasis, progression or prognosis in almost all of these malignancies [12, 13, 15, 17–20]. In addition to the centrosome, RHAMM distributes into multiple compartments including the cell surface, cytoskeleton, mitochondria and nucleus [21]. Especially, it should be noted that cell surface RHAMM–hyaluronan interactions regulate signaling through Ras and Ser [21]. It was shown that RHAMM antibodies, dominant-negative protein forms, soluble recombinant RHAMM protein and antisense RHAMM cDNA inhibit proliferation, motility and Ras-mediated transformation of immortalized fibroblasts in vitro [22]. It has therefore been suggested that RHAMM may be a good candidate as a tumor antigen for immunotherapy.
It is now evident that dendritic cell (DC) vaccination is a potent strategy for cancer immunotherapy in both pre-clinical and clinical settings [23, 24]. Various forms of tumor antigens have been applied to improve the induction efficacy of specific antitumor immune responses, including apoptotic tumor cells, tumor cell lysates, proteins, peptides and nucleic acids [25]. Among these, mRNA seems to be the most pertinent form for evaluating whether a new candidate gene identified by SEREX can be of use as an antigen for DC therapy, since it can easily be synthesized in vitro, transferred into DCs, and subjected to molecular engineering. Furthermore, mRNA is superior to cDNA in transfection efficiency and safety in vivo [26, 27], and also superior to synthetic peptides, since there is no need to determine that the peptides are presented to the immune system [26].
On the other hand, activation of CD4+ helper T cells is essential for optimal induction of cytotoxic T cells in DC therapy for cancer [28, 29]. It is known that exogenous antigens are taken up by DCs and processed in the endosomal pathway where resultant peptides are associated with MHC class II molecules leading to the induction of the CD4+ helper T cell response, whereas endogenous cytosolic and nuclear proteins are processed in a different manner, i.e. degradation by proteasomes, transportation of peptides into the endoplasmic reticulum, where they bind to MHC class I. RHAMM lacks a targeting sequence for the class II presentation pathway [30], thus making its presentation on class II unlikely or difficult.
In this study, we constructed a RHAMM mRNA targeted to MHC class II compartments and investigated whether DCs loaded with the modified mRNA (DC/RHAMM) could induce an antitumor immune response in vivo. Immunization of C57BL/6 mice with DC/RHAMM resulted in induction of tumor cell-killing activity in splenocytes, and DC/RHAMM exerted a remarkable therapeutic effect on tumors derived from subcutaneously inoculated EL4 cells. Of interest, this antitumor activity was mediated by CD4+ T cells, but not by CD8+ T cells.
Materials and methods
Animals and cell lines
Female C57BL/6 (H-2b) mice were purchased from Chiyoda Development Inc. (Tokyo, Japan) and bred at the Institute of Laboratory Animals, Yamaguchi University. For experimental purposes, mice of 6–12 weeks of age were used. Animal experiments were reviewed by the Committee of the Ethics on Animal Experiment in Yamaguchi University School of Medicine and carried out under the control of the Guideline for Animal Experiment in Yamaguchi University School of Medicine and the Low (No. 105) and Notification (No. 6) of the Government.
Murine cell lines including B16 melanoma, 9,10-dimetyl-1,2-benzanthracene induced EL4 lymphoma and YAC-1 lymphoma, and a human chronic leukemia cell line K562 were purchased from RIKEN BRC (Tsukuba, Japan). Murine KR158B brain tumor and colon 38 SL4 colon tumor were kindly donated by Dr. T. Jacks (Massachusetts Institute of Technology) and Dr. Tatsuro Irimura (Tokyo University), respectively. B16 and KR158B were maintained in complete media (CM, RPMI-1640 supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. All reagents were from SIGMA-Aldrich (Tokyo, Japan). EL4 and YAC1 were cultured in DMEM supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. Colon 38 SL4 was cultured in DMEM/F-12 1:1 mixture supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin. K562 was maintained in HAM F-12 supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine, 100 IU/ml penicillin and 100 μg/ml streptomycin.
Conventional reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was prepared from mouse normal tissues or tumor cell lines using TRIzol (Invitrogen, Carlbad, CA, USA) according to the manufacturer’s protocol. RNA (0.5 μg) was subjected to RT-PCR using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) according to the manufacturer’s protocol. The primers used were as follows; RHAMM forward primer, 5′-AAAGGCTAAGTTTTCTGAAGATGGT-3′; RHAMM reverse primer, 5′-CAAATCTTCTTCTAACTGGGCAATA-3′; TRP2 forward primer, 5′-TGGCACAGGTACCATCTGTTGT-3′; TRP2 reverse, 5′-TCCCCCGGGCTAGGCTTCCTCCGTG-3′; G3PDH forward primer, 5′-TCCACCACCCCTGTTGCTGTA-3′; G3PDH reverse, 5′-ACCACAGTCCATGCCATCAC-3′.
Plasmid constructs and in vitro transcription
For in vitro transcription, plasmids were cloned pSP64 vector (Promega, Madison, WI, USA). Tyrosinase-related protein-2 (TRP-2) signal sequence fragment (SS) and transmembrane-cytoplasmic domain (TM-Cyto) were amplified from TRP-2 cDNA by PCR (Ex Taq polymerase, Takara, Japan). The following primers were used: SS forward primer, 5′-AAAAAAGCTTATGGGCCTTGTGGGA-3′; SS reverse primer, 5′-AAAACTGCAGAGCCCGAGCTCTGAG-3′; TM-Cyto forward primer, 5′-CGGGATCCGTGGTCATTGGAATCCT-3′; TM-Cyto reverse primer, 5′-TCCCCCGGGCTAGGCTTCCTCCGTG-3′. PCR products were cloned as HindIII–PstI SS fragment and BamHI–SmaI TM-Cyto fragment into pSP64 to allow in vitro transcription under the control of a SP6 promoter (pSP64-cassette). To generate plasmid pSP64c-human RHAMM and pSP64c-mouse RHAMM, the fragment containing the human RHAMM and mouse RHAMM were amplified from K562 and B16 melanoma total RNA by RT-PCR (SuperScript One-step RT-PCR with Platinum Taq, Invitrogen), respectively. The following primers were used; human RHAMM forward primer, 5′-AAGTCGACATGTCCTTTCCTAAGGCGCCCTTAGG-3′; human RHAMM reverse primer, 5′-AAGCTAGCCTTCCATGATTCTTGACACTC-3′; mouse RHAMM forward primer, 5′-AAGTCGACATGTCCTTTCCTAAGGCGCCCCTGAA-3′; mouse RHAMM reverse primer, 5′-AATCTAGAGCAGCAGTTTGGGTTGCCT-3′. The products were cloned as a SalI–Nhe I human RHAMM fragment or a SalI–XbaI mouse RHAMM fragment into pSP64-cassette. The sequences were checked by the dideoxy methods. pSP64-TRP-2-enhanced green fluorescence protein (EGFP) was constructed as described previously [31]. For in vitro transcription, the plasmids were linearized and treated with proteinase K, followed by phenol–chloroform extraction and ethanol precipitation. In vitro transcription was carried out in a 100 μl reaction mix at 37°C for 2 h using mMESSAGE SP6 kit (Ambion, Austin, TX, USA) to generate 5′m7GpppG-capped in vitro transcribed mRNA. Purification of mRNA was performed by DNase I digestion followed by LiCl precipitation and 70% ethanol wash, in accordance with the manufacture’s instructions. mRNA quality was checked by agarose formaldehyde gel electrophoresis. RNA concentration was assayed by spectrophotometrical analysis at OD260.
DC generation
For DC culture, we used techniques described by Son et al. [32]. Briefly, femurs and tibiae were removed. The marrow was flushed with RPMI-1640 using a syringe with 26G needle and filtrated through a 70-μm cell strainer. Bone marrow cells were adjusted to 2×105 cells/ml in CM and plated on 100 mm dishes. They were cultured for up to 7 days in the presence of 1,000 U/ml of GM-CSF and 500 U/ml of IL-4 at 37°C, 5% CO2. On days 4 of culture, the same amount of cytokines was added to the cells. To isolate the DC population, the cells were collected and suspended in 5 ml of CM. The same volume of 14.5% (w/v) metrizamide in CM was underlaid and centrifugated at 1,200 g for 20 min at room temperature. After centrifugation, cells in the interface were collected and washed with RPMI-1640 three times and were used for subsequent RNA transfection. The expression of surface molecules on DC was analyzed under flow cytometry (Epics XL; Beckman Coulter Co., Miami, FL, USA). DC phenotypes showed CD11c+, CD40+, CD86+, H-2Kb+, I-Ab+ and DEC205+ (data not shown).
RNA transfection
mRNA transfection was performed using a commercially available reagent for RNA lipofection, TransMessenger Transfection Reagent (Qiagen), as described previously [33]. DCs were washed twice and resuspended in RPMI-1640. An amount of 1×106 DCs and 10 μg of mRNA were mixed and incubated for 3 h at 37°C. After transfection, cells were washed with PBS and subjected to the treatment of DC therapy.
Chloroquine inhibition assay
To assess the intracellular mechanisms of antigen processing, 50 μM chloroquine (Sigma-Aldrich) was added to the DCs immediately after mRNA transfection. The expression of EGFP was analyzed by flow cytometry. DCs were harvested at different time points after transfection, and treated with DAKO IntraStain (DAKO, Tokyo, Japan). EGFP was detected by antiEGFP monoclonal antibody through anti-mouse IgG-FITC as described previously [33].
Flow cytometry
Flow cytometry was performed for detection of MHC class I and class II molecules on EL4 and KR158B cells as described previously [34]. Briefly, the cells were cultured for 2 days with 10 ng/ml IFN-γ or medium control. The cells were harvested and incubated with FITC-conjugated anti-mouse MHC class I (H2-Kb, Caltag Lab., Burlingame, CA, USA) and anti-mouse class II (I-A/I-E, eBioscience, San Diego, CA, USA) monoclonal antibodies for 30 min on ice. After several washings with PBS, cells were analyzed with a flow cytometer (Epics XL; Beckman Coulter Co., Miami, FL, USA).
DC immunizations and 51Cr-release assay
All mice were injected intraperitonealy (i.p.) with 3×105 DCs once a week. After third immunization, mice were sacrificed and splenocytes were isolated. Splenocytes were subjected to 51Cr-release assay. For antibody depletion, 100 μl of anti-CD4 (GK1.5) or anti-CD8 (2.34) ascites [31] were injected i.p. into each mouse on 3 days prior to sacrifice.
Splenocytes were stimulated with DCs in vitro. On 4th day after stimulation in vitro, the standard 51Cr-release assay was performed as described previously [35]. KR158B cells were used as target cells in these experiments. The cells (1×106) were labeled with 100 μCi of [51Cr] sodium chromate (Perkin Elmer, Inc., Wellesley, MA, USA) for 1 h at 37°C and then washed to remove unincorporated isotope. The target (1×104) and effector cells (splenocytes) were added to each well of 96-well V-bottomed plates in a total volume of 200 μl of CM at a range of ratios and incubated for 4 h at 37°C. After incubation, the supernatant (100 μl) was harvested and counted in a Packard Cobra Quantum gamma counter (Packard Instrument Co., Meriden, CT, USA). The percentage of specific 51Cr release was calculated as follows: percentage of specific 51Cr release = (experimental release−spontaneous release)/(maximum release− spontaneous release)×100. Maximum or spontaneous release was determined from the supernatant of wells that contained 1% Triton X-100 or medium alone, respectively.
Tumor vaccination
For the 3-day EL4 tumor model, all mice were injected subcutaneously (s.c.) with 3×105 EL4 cells. Then, mice were injected with 3×105 mRNA-loaded DCs in the vicinity of the tumor inoculation site on days 3, 10 and 17. Tumor diameters were measured twice per week with a caliper and tumor volumes were calculated according to (A×B 2)/2 (A, the largest diameter; B, the smallest diameter).
For antibody depletion, ascites was obtained from hybridoma-bearing BALB/c nude mice and the mice were given i.p. transfers of the ascites (0.1 ml/mouse/transfer) biweekly from 3 days before to 24 days after EL4 tumor inoculation as described previously [31]. Depletion of CD4+ or CD8+ T cells was confirmed by analyzing circulating lymphocytes with flow cytometry. All experiments included 6 mice per group.
Statistical analysis
Two-tailed Student’s t test was used to determine the statistical significance of differences in tumor growth between treatment groups. A value of P<0.05 was considered significant. Statistical analyses were made using Stat View 5.0 software (Abacus Concepts, Calabasas, CA, USA).
Results
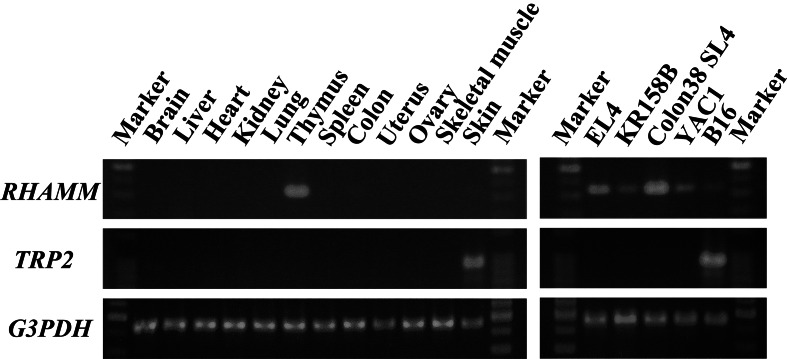
To clarify the tissue distribution of RHAMM in mice and its expression in mouse tumor cell lines, we examined RHAMM mRNA expression by conventional RT-PCR. Since the tyrosinase-related protein-2 (TRP-2) gene was utilized to construct the tumor antigen mRNA as described below, TRP-2 mRNA was simultaneously detected. In normal tissues, RHAMM mRNA was only detected in the thymus as shown in Fig. 1. In contrast, RHAMM mRNA was expressed in all 5 C57BL/6 tumor cell lines. TRP-2 mRNA was only detected in the skin and B16 melanoma (Fig. 1).
Fig. 1.
Expression of RHAMM mRNA in normal tissues and tumor cell lines. RT-PCR analysis was performed with primers specific for RHAMM or TRP-2. G3PDH was used as a control for equal template loading
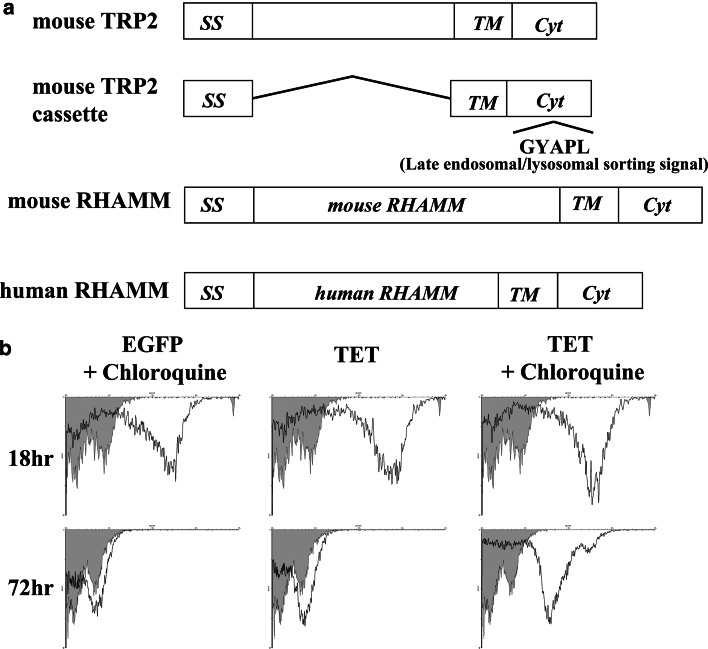
To efficiently transport RHAMM protein to MHC class II compartments, for eventual cross-presentation by both classes I and II on DCs in a cognate manner, the leader sequence and cytoplasmic domain containing a late endosomal/lysosomal sorting signal derived from the mouse TRP-2 gene were linked to the RHAMM gene at its N- and C-terminus, respectively (Fig. 2a). First, we investigated whether proteins translated from such mRNA constructs with a late endosomal/lysosomal sorting signal were really localized and degraded in the HLA-class II-processing compartments. For this purpose, enhanced green fluorescence protein (EGFP) and TRP2–EGFP mRNAs were used. TRP2–EGFP is a fusion mRNA in which EGFP was linked to the N-terminus region of TRP-2 at nucleotide position 169 as previously described [31]. After transfection of EGFP or TRP2–EGFP mRNA into DCs, the lysosomal degradation inhibitor chloroquine was added to the culture. EGFP expression by DCs cultured with or without chloroquine was monitored by flow cytometry. As shown in Fig. 2b, EGFP linked to the targeting signal (TRP2–EGFP) was almost completely degraded in non-treated DCs, whereas EGFP alone remained detectable in chloroquine-treated DCs at 72 h after transfection. Since the degradation of unlinked EGFP was not inhibited by chloroquine addition, these data suggest that the EGFP protein containing the TRP-2 targeting sequence is transported to and degraded in the MHC-class II-processing compartments.
Fig. 2.
Schematic representation and expression of constructed mRNAs. a Constructs used in this study. SS signal sequence, TM transmembrane domain, Cyt cytoplasmic domain. The mouse and human RHAMM genes were separately cloned into a mouse TRP-2 cassette plasmid. b Effect of chloroquine on EGFP expression in EGFP or TRP-2-EGFP mRNA-transfected DCs. Chloroquine was added to the DC culture and EGFP expression was analyzed by flow cytometry using an anti-EGFP antibody. Mouse IgG was used as a control antibody, as shown by the gray area
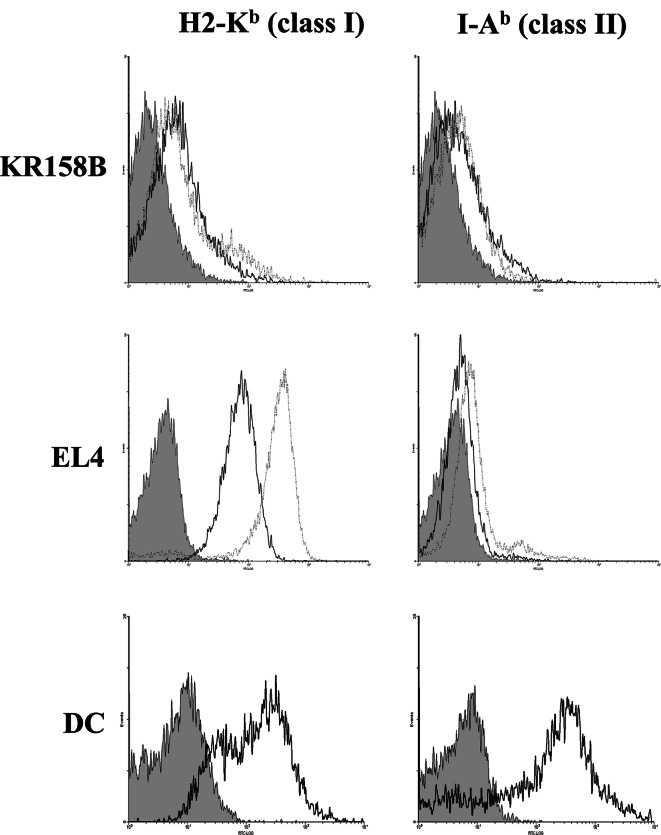
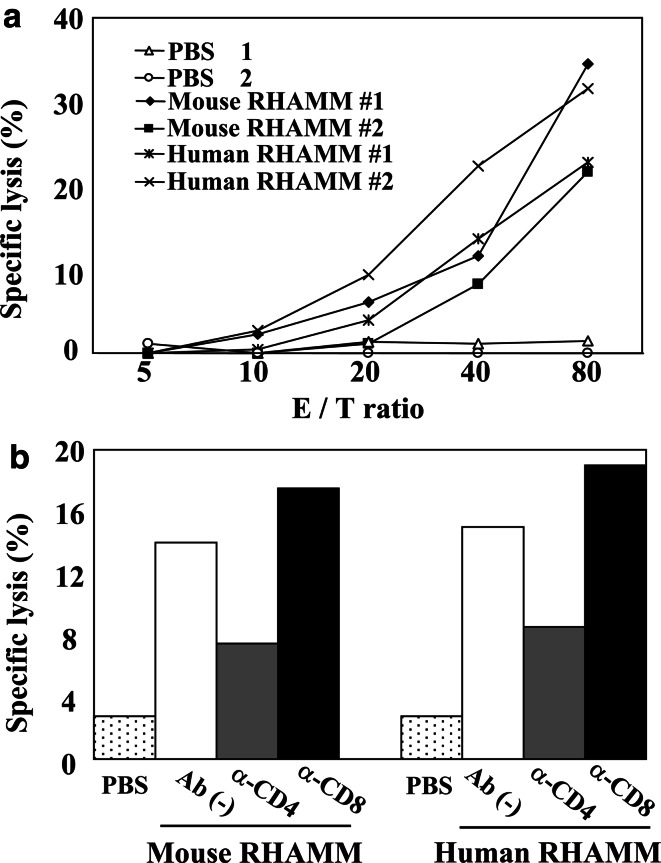
We then tested whether RHAMM mRNA-transfected DCs (DC/RHAMM) could induce an antitumor immune response in vivo. In addition to mouse RHAMM (mRHAMM) mRNA, human RHAMM (hRHAMM) mRNA was used as a xenogeneic antigen that could be expected to further enhance the immune response. DCs transfected with mRHAMM or hRHAMM mRNA were intraperitoneally (i.p) administered three times into C57BL/6 mice (3×105 DCs/injection). One week after the last immunization, the splenocytes were isolated and stimulated with DC/RHAMM in vitro, and then co-cultivated with 51Cr-labeled target KR158B cells at various E/T ratios to measure the specific killing. KR158B cells were shown to be weakly positive for both MHC class I and class II irrespective of IFN-γ treatment (Fig. 3). Those cells were therefore used for experiments without IFN-γ treatment. Splenocytes from mice immunized with DC/mRHAMM or DC/hRHAMM killed target cells in a dose-dependent manner compared to those from mice immunized with DCs without mRNA transfection (DC/PBS) (Fig. 4a). There was no significant difference in the induced killing activity between mouse and human mRNAs, suggesting that the immunogenicity of autologous RHAMM is as strong as that of the xenogeneic RHAMM.
Fig. 3.
Effect of IFN-γ treatment on the expression of MHC class I and class II. KR158B and EL4 cells were cultured for 2 days with 10 ng/ml IFN-γ (dotted line) or medium only (solid line). Cells were immunostained for MHC class I (H2-Kb) or class II (I-Ab). DCs cultured without IFN-γ were used as a positive control
Fig. 4.
In vivo induction of cytotoxic T cell responses. a Splenocytes taken from mice after immunization by DCs transfected with mouse or human RHAMM mRNA were restimulated with the same DCs in vitro for 4 days. The restimulated splenocytes were then co-cultured with the [51Cr] sodium chromate-labeled target KR158B cells for 4 h. Triplicate wells were averaged and the percentage of specific lysis was calculated as described in Materials and methods. The results of two independent experiments are shown as #1 and #2. PBS, mouse RHAMM or human RHAMM indicates immunization by DCs without mRNA, with mouse RHAMM mRNA or with human RHAMM mRNA, respectively. b Effect of CD4+ or CD8+ T cell depletion on the induction of cytotoxic activity in splenocytes by DC immunization was observed. Anti-CD4 or anti-CD8 antibodies were injected i.p. into each mouse on 3 days prior to sacrifice. Splenocytes were restimulated with DCs and subjected to 51Cr-release assay in the same manner as in a. The cytotoxicity assays were carried out at an E/T (effector to target) ratio of 80:1
To examine the requirement of CD4+ and/or CD8+ T cells for the antitumor activity induced by DC/RHAMM immunizations, CD4+ or CD8+ T cells were depleted in mice previously immunized with DC/RHAMM by administration of a specific anti-CD4 or anti-CD8 antibody. The antibodies were i.p. injected at 3 days prior to sacrifice. CD4+ T cell depletion reduced the antitumor acitivity of splenocytes to around 50%, whereas it was unaffected by CD8+ T cell depletion (Fig. 4b). These findings suggest that CD4+ T cells, but not CD8+ T cells, play an important role in this antitumor activity.
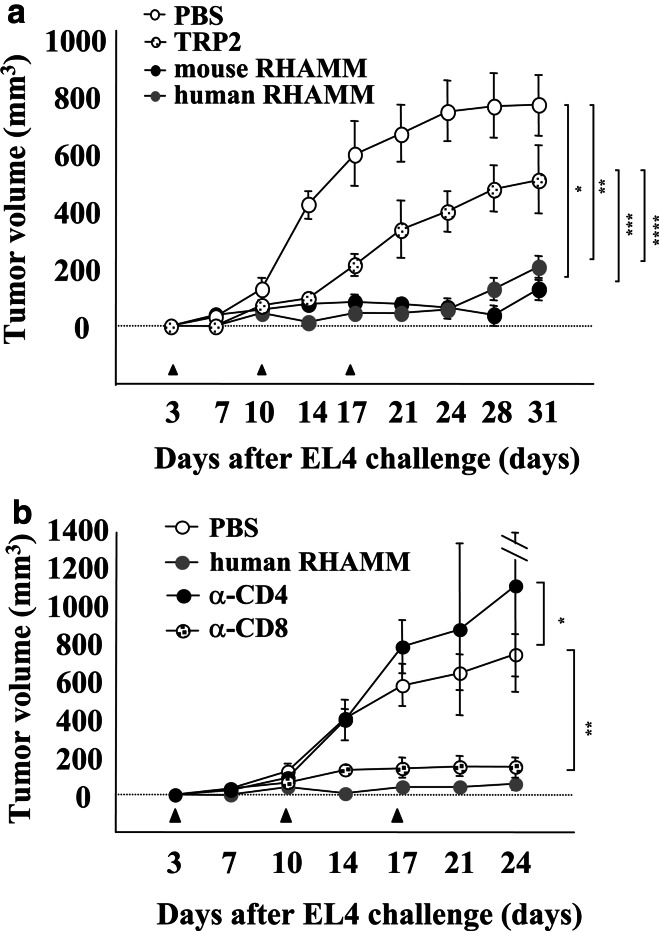
The in vivo antitumor effects of DC/RHAMM were evaluated to assess whether DC/RHAMM could be of use for immunotherapy. For this purpose, EL4 cells were used as targets, since they were clearly positive for MHC class I and negative for class II (Fig. 3). Three days after injection of EL4 cells (3×105 cells/mouse), the mice were immunized with 3 i.p. injections of DC/RHAMM. The tumor sizes were measured every 3–4 days. As shown in Fig. 5a, tumor growth was almost completely inhibited in mice immunized with DC/mRHAMM or DC/hRHAMM, in contrast to the lack of any therapeutic effect in mice immunized with DC/PBS. Although immunization with DC/TRP-2 conferred some degree of protection, a significant difference was observed between DC/TRP-2 and DC/RHAMM treatments (Fig. 5a).
Fig. 5.
Treatment of tumor-bearing mice with RHAMM mRNA-transfected DCs. a 3×105 EL4 cells were inoculated subcutaneously into the right flank of C57BL/6 mice. After 3, 10 and 17 days, 3×105 DC/PBS, DC/TRP-2, DC/mRHAMM or DC/hRHAMM were injected subcutaneously in the vicinity of the tumor inoculation site of mice. Tumor sizes were measured biweekly. A two-tailed Student’s t test was used to determine the statistical significance of differences in tumor growth among the treatment groups. *DC/PBS versus DC/mRHAMM (P<0.0001); **DC/PBS versus DC/hRHAMM (P<0.0001); ***DC/TRP2 versus DC/mRHAMM (P=0.0013); ****DC/TRP2 versus DC/hRHAMM (P=0.0008). b Effect of CD4+ or CD8+ T cell depletion on the efficacy of DC/RHAMM treatment. Mice were treated with an anti-CD4 or anti-CD8 antibody at 3 days before the tumor challenge, followed by two injections per week. 3×105 EL4 cells were inoculated subcutaneously into the right flank of the mice. After 3, 10 and 17 days, 3×105 DC/PBS or DC/hRHAMM were injected subcutaneously in the vicinity of the tumor inoculation site of mice. Tumor sizes were measured biweekly. A two-tailed Student’s t test was used to determine the statistical significance of differences in tumor growth among the treatment groups. *DC/PBS versus DC/hRHAMM + anti-CD4 (P=0.1177); **DC/PBS versus DC/hRHAMM + anti-CD8 (P=0.0006)
Depletion of CD4+ or CD8+ T cells was again performed to reveal their roles in the in vivo antitumor effects of DC/RHAMM. The anti-CD4 or anti-CD8 antibody was i.p. injected at 3 days before the tumor transplantation, followed by subsequent injections every 3–4 days. As shown in Fig. 5b, CD4+ T cell depletion completely abrogated the therapeutic effect of DC/hRHAMM, whereas CD8+ T cell depletion had no effect. These data further support the results shown in Fig. 4b.
Discussion
The distribution of RHAMM in normal human tissues was described in a recent report [10], but there is little data for that in normal mouse tissues. By RT-PCR examination, RHAMM mRNA showed a very restricted expression in mouse tissues (Fig. 1). Such an expression pattern is considered suitable for immunotherapy targeted against RHAMM in tumor cells. In contrast, all five tumor cell lines tested showed positive RHAMM expression (Fig. 1), consistent with a previous finding that RHAMM mRNA is ubiquitously expressed in murine tumor cells [36]. Considering that RHAMM has mitotic spindle-stabilizing functions [8], and oncogenic activity [22, 37], it is reasonable to anticipate that its expression would be limited to organs with increased cell proliferation such as the thymus, or transformed cells. In this context, further investigations into RHAMM expression in regenerative or inflammatory tissues will be required.
We constructed a modified mRNA joining the leader sequence and cytoplasmic domain containing a late endosomal/lysosomal sorting signal of TRP-2 to the N- and C-terminus of RHAMM, respectively, to transport the intracellular protein RHAMM to MHC class II compartments (Fig. 2a). This strategy has previously been demonstrated to be useful for efficient induction of a specific immune response by utilizing proteins with a late endosomal/lysosomal sorting signal, including lysosome-associated membrane protein [38, 39], DEC-205 [40] and an invariant chain [41]. The effect of the lysosomal degradation inhibitor chloroquine on the expression of EGFP after transduction of EGFP-TRP-2 mRNA, in which EGFP was linked to the N-terminal region of TRP-2 as described previously [31], was examined to assess whether the sorting signal of TRP-2 did actually target EGFP to late endosomes and lysosomes. The results revealed that EGFP expression was only found when chloroquine was added to the DC culture after EGFP-TRP-2 mRNA transfection (Fig. 2b).
Since the killing activity against the RHAMM-positive tumor cell line KR158B was easily detected in splenocytes after immunizing mice with DC/RHAMM, we next tried to confirm the induction of CD8+ cytotoxic T cells by in vivo depletion of these T cells with a specific antibody. Unexpectedly, one administration of the anti-CD8 antibody into mice after immunization had no effect on the antitumor activity of splenocytes, whereas similar administration of an anti-CD4 antibody reduced the killing activity to ~50% (Fig. 4a). These results suggest that CD4+ T cells, but not CD8+ T cells, play a major role in this antitumor immune response. The in vivo therapeutic effect of DC/RHAMM was also evident in a 3-day tumor model of EL4 (Fig. 5a). T cell depletion was again performed in this therapeutic model. At this time, an anti-CD4 or anti-CD8 antibody was injected into mice twice a week from 3 days before to 24 days after the tumor cell inoculation. The effect of DC/RHAMM was completely abolished by depletion of CD4+ T cells, whereas it remained unaffected by depletion of CD8+ T cells (Fig. 5b), further supporting the idea that CD4+ T cells, but not CD8+ T cells, could be involved in the induction of antitumor immunity by DC/RHAMM.
It is well known that CD4+ T cells are involved in the priming phase [42, 43] and maintenance of cytotoxic T cells [28], but their role in the effector phase of antitumor immunity remains unknown, despite the fact that CD4+ T cell-mediated tumor rejection has been found in various models [44–46]. Our present finding that the depletion of CD4+ T cells after DC immunization resulted in a significant reduction of the killing activity in splenocytes (Fig. 4b) also suggests that CD4+ T cells are a requisite for the effector phase. However, we observed that EL4 tumor cells did not express MHC class II molecules even after stimulation with interferon (IFN)-γ (Fig. 3). This observation means that CD4+ T cells are unable to recognize tumor cells and kill them on their own. It is known that CD4+ cytotoxic T cells induce Fas ligand-mediated apoptosis in Fas-positive target cells [47], but EL4 cells were previously shown to be resistant to Fas ligand-induced apoptosis due to FLIP overexpression [48]. It is therefore unlikely that CD4+ T cells are responsible for the killing activity of splenocytes induced by DC/RHAMM. Recent reports have suggested possible explanations for the effector mechanism of CD4+ T cell-mediated tumor cell rejection, i.e. the requirement of CD4+ T cells for the induction of natural killer cells by DCs [49] and inhibition of angiogenesis through IFN-γ production by T cells and IFN-γ receptor expression by non-hematopoietic cells [50]. In this context, it should be noted that DCs transfected with TRP-2 mRNA exerted only a slight inhibitory effect on the in vivo growth of EL4 tumors (Fig. 5a), suggesting that the ability of RHAMM to activate CD4+ T cells may be much higher than that of TRP-2. Another possible explanation is CD4+ T cell-mediated antibody production by DC immunization. Those antibodies may bind to RHAMM and inhibit the signal transduction for cell growth, since RHAMM has been shown to express on the cell surface and interact with hyaluronan, resulting in the signal regulation through Ras and Ser [21].
In addition to targeting the antigen to MHC class II compartments, we attempted an alternative strategy of enhancing the antitumor immune response with a xenogeneic, evolutionarily conserved antigen. Although the mechanism remains to be elucidated, these antigens have been revealed to be more effective at inducing an antitumor immune response than the corresponding autologous antigens in clinical trials as well as in animal models [51–54]. RHAMM is a well-conserved protein showing more than 80% amino acid sequence similarity between mice and humans [36]. The inducibility of the tumor cell-killing activity and the therapeutic effect on EL4 tumors were compared between DC/mRHAMM and DC/hRHAMM (Figs. 4, 5). Unexpectedly, there was no difference in the antitumor activities of mouse and human RHAMM. These data suggest that autologous RHAMM could have a potent immunogenicity equal to its xenogeneic counterpart, and this might be linked to a previous finding that centrosome proteins are intracellular targets of autoimmunity in both mice and humans [55, 56].
Collectively, the results of this study indicate that DCs loaded with RHAMM mRNA are able to induce antitumor immunity in vivo, and that this immunity is mediated by CD4+ T cells. Although further investigations into the mechanism of in vivo tumor suppression are required, DC therapy with RHAMM mRNA in clinical settings may be warranted, since RHAMM is able to induce antibody response in humans and expressed in a wide variety of cancers with high frequency.
Acknowledgements
This work was supported by a Grant-in-aid for Scientific Research on Priority Areas (C) (No. 12217097) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We would like to acknowledge the technical expertise of the DNA Core Facility at the Center for Gene Research, Yamaguchi University. We wish to thank Dr. Koji Kajiwara (Yamaguchi University) for supplying KR158B cells to us and for helpful discussion.
Abbreviations
- RHAMM
Receptor for hyaluronan-mediated motility
- DC
Dendritic cell
- TRP-2
Tyrosinase-related protein-2
- EGFP
Enhanced green fluorescence protein
References
- 1.Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 2.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression. Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 3.Charrasse S, Schroeder M, Gauthier-Roucviere C, Ango F, Cassimeris L, Gard DL, Larroque C. The TOGp protein is a new human microtubule-associated protein homorogous to the Xenopus XMAP215. J Cell Sci. 1998;111:1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- 4.Wells RA, Catzavelos C, Kamel-Reid S. Fusion of retinoic acid receptor alpha to NuMA, the nuclear mitotic apparatus protein, by a variant translocation in acute promyelocytic leukaemia. Nat Genet. 1997;17:109–113. doi: 10.1038/ng0997-109. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawamura K, Izumi H, Ma Z, Ikeda R, Moriyama M, Tanaka T, Nojima T, Levin LS, Fujikawa-Yamamoto K, Suzuki K, Fukasawa Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 2004;64:4800–4809. doi: 10.1158/0008-5472.CAN-03-3908. [DOI] [PubMed] [Google Scholar]
- 7.Neben K, Tews B, Wrobel G, Hahn M, Kokocinski F, Giesecke C, Krause U, Ho AD, Kramer A, Lichter P. Gene expression patterns in acute myeloid leukemia correlate with centrosome aberrations and numerical chromosome changes. Oncogene. 2004;23:2379–2384. doi: 10.1038/sj.onc.1207401. [DOI] [PubMed] [Google Scholar]
- 8.Maxwell AC, Keats JJ, Crainie M, Sun X, Yen T, Shibuya E, Hendzel M, Chan G, Pilarski LM. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol Biol Cell. 2003;14:2262–2276. doi: 10.1091/mbc.E02-07-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Line A, Slucka Z, Stengrevics A, Li G, Rees RC. Altered splicing pattern of TACC1 mRNA in gastric cancer. Cancer Genet Cytogenet. 2002;139:78–83. doi: 10.1016/S0165-4608(02)00607-6. [DOI] [PubMed] [Google Scholar]
- 10.Greiner J, Ringhoffer M, Taniguchi M, Schmitt A, Kirchner D, Krahn G, Heilmann V, Gschwend J, Bergmann L, Dohner H, Schmitt M. Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol. 2002;30:1029–1035. doi: 10.1016/S0301-472X(02)00874-3. [DOI] [PubMed] [Google Scholar]
- 11.Greiner J, Ringhoffer M, Taniguchi M, Hauser T, Schmitt A, Dohner H, Schimitt M. Characterization of several leukemia-associated antigens inducing humoral immune responses in acute and chronic myeloid leukemia. Int J Cancer. 2003;106:224–231. doi: 10.1002/ijc.11200. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Thor AD, Moore DH, II, Zhao Y, Kerschmann R, Stern R, Watson PH, Turley EA. The overexpression of RHAMM, a hyalurinan-binding protein that regulates ras signaling, correlates with overexpression of mitogen-activated protein kinase and is a significant parameter in breast cancer progression. Clin Cancer Res. 1998;4:567–576. [PubMed] [Google Scholar]
- 13.Yamada Y, Itano N, Narimatsu H, Kudo T, Hirohashi S, Ochiai A, Niimi A, Ueda M, Kimata K. Receptor for hyaluronan-mediated motility and CD44 expressions in colon cancer assessed by quantitative analysis using real-time reverse transcriptase-polymerase chain reaction. Jpn J Cancer Res. 1999;90:987–992. doi: 10.1111/j.1349-7006.1999.tb00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crainie M, Belch AR, Mant MJ, Pilarski LM. Overexpression of the receptor for hyaluronan-mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: identification of three distinct RHAMM variants. Blood. 1999;93:1684–1696. [PubMed] [Google Scholar]
- 15.Maxwell CA, Rasmussen E, Zhan F, Keats JJ, Adamia S, Strachan E, Crainie M, Walker R, Belch AR, Pilarski LM, Barlogie B, Jr, Shaughnessy J, Reiman T. RHAMM expression and isoform balance predict aggressive disease and poor survival in multiple myeloma. Blood. 2004;104:1151–1158. doi: 10.1182/blood-2003-11-4079. [DOI] [PubMed] [Google Scholar]
- 16.Greiner J, Ringhoffer M, Taniguchi M, Li L, Schmitt A, Shiku H, Dohner H, Schmitt M. mRNA expression of leukemia-associated antigens in patients with acute myeloid leukemia for the development of specific immunotherapies. Int J Cancer. 2004;108:704–711. doi: 10.1002/ijc.11623. [DOI] [PubMed] [Google Scholar]
- 17.Assman V, Gillett CE, Poulsom R, Ryder K, Hart IR, Hanby AD. The pattern of expression of the microtubule-binding protein RHAMM/IHABP in mammary carcinoma suggests a role in the invasive behaviour of tumor cells. J Pathol. 2001;195:191–196. doi: 10.1002/path.941. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Guo L, Li JW, Liu N, Qi R, Liu J. Expression of hyaluronan receptors CD44 and RHAMM in stomach cancers: relevance with tumor progression. Int J Oncol. 2000;17:927–932. [PubMed] [Google Scholar]
- 19.Kong QY, Liu J, Chen XY, Wang XW, Sun Y, Li H. Differntiall expression patterns of hyaluronan receptors CD44 and RHAMM in transitional cell carcinomas of urinary bladder. Oncol Rep. 2003;10:51–55. [PubMed] [Google Scholar]
- 20.Rein DT, Roehrig K, Schondorf T, Lazar A, Fleisch M, Niederacher D, Bender HG, Dall P. Expression of the hyaluronan receptor RHAMM in endometrial carcinomas suggests a role in tumor progression and metastasis. J Cancer Res Clin Oncol. 2003;129:161–164. doi: 10.1007/s00432-003-0415-0. [DOI] [PubMed] [Google Scholar]
- 21.Turley EA, Noble PW, Bourguignon LY. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–4592. doi: 10.1074/jbc.R100038200. [DOI] [PubMed] [Google Scholar]
- 22.Tolg C, Poon R, Fodde R, Turley EA, Alman BA. Genetic deletion of receptor for hyaluronan-mediated motility (Rhamm) attenuates the formation of aggressive fibromatosis (desmoid tumor) Oncogene. 2003;22:6873–6882. doi: 10.1038/sj.onc.1206811. [DOI] [PubMed] [Google Scholar]
- 23.Paczesny S, Ueno H, Fay J, Banchereau J, Palucka AK. Dendritic cells as a vectors for immunotherapy of cancer. Semin Cancer Biol. 2003;13:439–447. doi: 10.1016/j.semcancer.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Yannelli JR, Wroblewski JM. On the road to a tumor cell vaccine: 20 years of cellular immunotherapy. Vaccine. 2004;23:97–113. doi: 10.1016/j.vaccine.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt T, Ziske C, Marten A, Endres S, Teimann K, Schmitz V, Gorschluter M, Schneider C, Sauerbruch T, Schmidt-Wolf IG. Intratumoral immunization with tumor RNA-pulsed dendritic cells confers antitumor immunity in a C57BL/6 pancreatic murine tumor model. Cancer Res. 2003;63:8962–8967. [PubMed] [Google Scholar]
- 26.Mitchell DA, Nair SK. RNA-transfected dendritic cells in cancer immunotherapy. J Clin Invest. 2000;106:1065–1069. doi: 10.1172/JCI11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponsaerts P, van Tendeloo VF, Berneman ZN. Cancer immunotherapy using RNA-loaded dendritic cells. Clin Exp Immunol. 2003;134:378–384. doi: 10.1046/j.1365-2249.2003.02286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ossendorp F, Mengede E, Camps M, Filius R, Melief CJ. Specific T helper cell requirement for optimal induction of cytotoxic T lymphocytes against major histocompatibility complex class II negative tumors. J Exp Med. 1998;187:693–702. doi: 10.1084/jem.187.5.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuler-Thurner B, Schultz ES, Berger TG, Weinlich G, Ebner S, Woerl P, Bender A, Feuerstein B, Fritsch PO, Romani N, Schuler G. Rapid induction of tumor-specific type 1 T helper cells I metastatic melanoma patients by vaccination with mature, cryopreserved, peptide-loaded monocyte-derived dendritic cells. J Exp Med. 2002;195:1279–1288. doi: 10.1084/jem.20012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Assmann V, Marshall JF, Fieber C, Hofmann M, Hart IR. The human hyaluronan receptor RHAMM is expressed as an intracellular protein ini breast cancer cells. J Cell Sci. 1998;111:1685–1694. doi: 10.1242/jcs.111.12.1685. [DOI] [PubMed] [Google Scholar]
- 31.Fukui M, Nakano-Hashimoto T, Okano K, Maruta Y, Suehiro Y, Hamanaka Y, Yamashita H, Imai K, Hinoda Y. Therapeutic effect of dendritic cells loaded with a fusion mRNA encoding tyrosinase-related protein 2 and enhanced green fluorescence protein on B16 melanoma. Tumour Biol. 2004;25:252–257. doi: 10.1159/000081388. [DOI] [PubMed] [Google Scholar]
- 32.Son YI, Egawa S, Tatsumi T, Redlinger RE, Jr, Kalinski K, Kanto T. A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J Immunol Methods. 2002;262:145–157. doi: 10.1016/S0022-1759(02)00013-3. [DOI] [PubMed] [Google Scholar]
- 33.Okano K, Fukui M, Suehiro Y, Hamanaka Y, Imai K, Hinoda Y. Evaluation of an mRNA lipofection procedure for human dendritic cells and induction of cytotoxic T lymphocytes against enhanced green fluorescence protein. Tumour Biol. 2003;24:317–324. doi: 10.1159/000076464. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi T, Makiguchi Y, Hinoda Y, Kakiuchi H, Nakagawa N, Imai K, Yachi A. Expression of MUC1 on myeloma cells and induction of HLA-unrestricted CTL against MUC1 from a multiple myeloma patient. J Immunol. 1994;153:2102–2109. [PubMed] [Google Scholar]
- 35.Adachi T, Hinoda Y, Nishimori I, Adachi M, Imai K. Increased sensitivity of gastric cancer cells to natural killer and lymphokine-activated killer cells by antisense suppression of N-acetylgalactosaminyltransferase. J Immunol. 1997;159:2645–2651. [PubMed] [Google Scholar]
- 36.Hofmann M, Fieber C, Assmann V, Gottllcher M, Sleeman J, Plug R, Howells N, von Stein O, Ponta H, Herrlich P. Identification of IHABP, a 95 kDa intracellular hyaluronate binding protein. J Cell Sci. 1998;111:1673–1684. doi: 10.1242/jcs.111.12.1673. [DOI] [PubMed] [Google Scholar]
- 37.Hall CL, Yang B, Yang X, Zhang S, Turley M, Samuel S, Lange LA, Wang C, Curpen GD, Savani RC, Greenberg AH, Turley EA. Overexpression of the hyaluronan receptor RHAMM is transforming and is also required for H-ras transformation. Cell. 1995;82:19–26. doi: 10.1016/0092-8674(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 38.Wu TC, Guarnieri FG, Staveley-O’Carroll KF, Viscidi RP, Levitsky HI, Hedrick L, Cho KR, August JT, Pardoll DM. Engineering an intracellular pathway for major histocompatibility complex class II presentation of antigens. Proc Natl Acad Sci USA. 1995;92:11671–11675. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su Z, Vieweg J, Weizer AZ, Dahm P, Yancey D, Turaga V, Higgins J, Boczkowski D, Gilboa E, Dannull J. Enhanced induction of telomerase-specific CD4 (+) T cells using dendritic cells transfected ith RNA encoding a chimeric gene product. Cancer Res. 2002;62:5041–5048. [PubMed] [Google Scholar]
- 40.Mahnke K, Guo M, Lee S, Sepulveda H, Swain SL, Nussenzweig M, Steiman RM. The dendritic cell receptor for endocytosis, DEC-205, can recycle and enhance antigen presentation via major histocompaibility complex class II-positive lysosomal compartments. J Cell Biol. 2000;151:673–684. doi: 10.1083/jcb.151.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonehill A, Heirman C, Tuyaerts S, Michiels A, Zhang Y, van der Bruggen P, Thielemans K. Efficient presentation f known HLA class II-restricted MAGE-A3 epitope by dendritic cells electroporated with messenger RNA encoding an invariant chain with genentic exchange of class II-associated invariant chain peptide. Cancer Res. 2003;63:5587–5594. [PubMed] [Google Scholar]
- 42.Bannerji R, Arroyo CD, Cordon-Cardo C, Gilboa E. The role of IL-2 secreted from genetically modified tumor cells in the establishment of antitumor immunity. J Immunol. 1994;152:2324–2332. [PubMed] [Google Scholar]
- 43.Grohmann U, Fioretti MC, Bianchi R, Belladonna ML, Ayroldi E, Surace D, Silla S, Puccetti P. Dendritic cells, interleukin 12, and CD4+ lymphocytes in the initiation of class I-restricted reactivity to a tumor/self peptide. Crit Rev Immunol. 1998;18:87–98. doi: 10.1615/critrevimmunol.v18.i1-2.100. [DOI] [PubMed] [Google Scholar]
- 44.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4+ T cells in the antitumor immune response. J Exp Med. 1998;188:2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mumberg D, Monach PA, Wanderling S, Philip M, Toledano AY, Schreiber RD, Schreiber H. CD4+ T cells eliminate MHC class II-negative cancer cells in vivo by indirect effects of IFN-γ. Proc Natl Acad Sci USA. 1999;96:8633–8638. doi: 10.1073/pnas.96.15.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schurmans LR, Diehl L, den Boer AT, Sutmuller RP, Boonman ZF, Medema JM, van der Voort EI, Laman J, Melief CJ, Jager MJ, Toes RE. Rejection of intraocular tumors by CD4+ T cells without induction of phthisis. J Immunol. 2001;167:5832–5837. doi: 10.4049/jimmunol.167.10.5832. [DOI] [PubMed] [Google Scholar]
- 47.Hahn S, Erb P. The immunomodulatory role of CD4-positive cytotoxic T-lymphocytes in health and disease. Int Rev Immunol. 1999;18:449–464. doi: 10.3109/08830189909088493. [DOI] [PubMed] [Google Scholar]
- 48.Kataoka T, Ito M, Budd RC, Tschopp J, Nagai K. Expression level of c-FLIP versus Fas determines susceptibility to Fas ligand-induced cell death in murine thymoma EL-4 cells. Exp Cell Res. 2002;273:256–264. doi: 10.1006/excr.2001.5438. [DOI] [PubMed] [Google Scholar]
- 49.van den Broeke LT, Daschbach E, Thomas EK, Andringa G, Berzofsky JA. Dendritic cell-induced activation of adaptive and innate antitumor immunity. J Immunol. 2003;171:5842–5852. doi: 10.4049/jimmunol.171.11.5842. [DOI] [PubMed] [Google Scholar]
- 50.Qin Z, Blankenstein T. CD4+ T cell-mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN-γ receptor expression by nonhematopoietic cells. Immunity. 2000;12:677–686. doi: 10.1016/S1074-7613(00)80218-6. [DOI] [PubMed] [Google Scholar]
- 51.Naftzger C, Takechi Y, Kohda H, Hara I, Vijayasaradhi S, Houghton AN. Immune response to a differentiation antigen induced by altered antigen: a study of tumor rejection and autoimmunity. Proc Natl Acad Sci USA. 1996;93:14809–14814. doi: 10.1073/pnas.93.25.14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steitz J, Bruck J, Steinbrink K, Fnk A, Knop J, Tuting T. Genetic immunization of mice with human tyrosinase-related protein 2: implications for the immunotherapy of melanoma. Int J Cancer. 2000;86:89–94. doi: 10.1002/(SICI)1097-0215(20000401)86:1<89::AID-IJC14>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 53.Fong L, Brockstedt D, Benike C, Breen JK, Strang G, Ruegg CL, Engleman EG. Dendritic cell-based xenoantigen vaccination for prostate cancer immunotherapy. J Immunol. 2001;167:7150–7156. doi: 10.4049/jimmunol.167.12.7150. [DOI] [PubMed] [Google Scholar]
- 54.Bergman PJ, McKnight J, Novosad A, Charney S, Farrelly J, Craft D, Wulderk M, Jeffers Y, Sadelain M, Hohenhause AE, Segal N, Gregor P, Engelhorn M, Riviere I, Houghton AN, Wolchok JD. Long-term survival of dogs with advanced malignant melanoma after DNA vaccination with xenogeneic human tyrosinase: a phase I trial. Clin Cancer Res. 2003;9:1284–1290. [PubMed] [Google Scholar]
- 55.Gavanescu I, Pihan G, Halilovic E, Szomolanyi-Tsuda E, Welsh RM, Doxsey S. Mycoplasma infection induces a scleroderma-like centrosome autoantibody response in mice. Clin Exp Immunol. 2004;137:288–297. doi: 10.1111/j.1365-2249.2004.02535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gavanescu I, Vazquez-Abad D, McCauley J, Senecal J-L, Doxsey S. Centrosome proteins: a major class of autoantigens in scleroderma. J Clin Immunol. 1999;19:166–171. doi: 10.1023/A:1020551610319. [DOI] [PubMed] [Google Scholar]