Abstract
NY-ESO-1 is frequently expressed in epithelial ovarian cancer (EOC) and elicits spontaneous humoral and cellular immune responses in a proportion of EOC patients. The identification of NY-ESO-1 peptide epitopes with dual HLA-class I and class II specificities might be useful in vaccination strategies for generating cognate CD4+ T cell help to augment CD8+ T cell responses. Here, we describe two novel NY-ESO-1-derived MHC class I epitopes from EOC patients with spontaneous humoral immune response to NY-ESO-1. CD8+ T cells derived from NY-ESO-1 seropositive EOC patients were presensitized with a recombinant adenovirus encoding NY-ESO-1or pooled overlapping peptides. These epitopes, ESO127–136 presented by HLA-A68 molecule, and ESO127–135 restricted by HLA-Cw15 allele, are located within ESO119–143, a promiscuous HLA-class II region containing epitopes that bind to multiple HLA-DR alleles. The novel epitopes were naturally processed by APC or naturally presented by tumor cell lines. In addition, these epitopes induced NY-ESO-1-specific CTL in NY-ESO-1 seropositive EOC patients. Together, the results indicate that ESO119–143 epitope has dual HLA classes I and II specificities, and represents a potential vaccine candidate in a large number of cancer patients.
Keywords: Epithelial ovarian cancer, Tumor immunity, NY-ESO-1, CD8, Epitope, MHC class I
Introduction
NY-ESO-1 belongs to the family of cancer-testis (CT) antigens, and was initially defined by serological analysis of recombinant cDNA expression libraries (SEREX) in esophageal cancer [7]. The defining characteristics of CT antigens are high levels of expression in adult male germ cells, but not in normal adult tissues, and aberrant expression in a variable proportion of a wide range of different cancer types [23]. Among CT antigens, NY-ESO-1 is one of the most spontaneously immunogenic, eliciting both cellular and humoral immune responses in a high proportion of patients with advanced NY-ESO-1-expressing tumors [9, 15, 26]. Several NY-ESO-1 MHC class I- and MHC class II-restricted epitopes have been characterized (http://www.cancerimmunity.org/peptidedatabase/tumorspecific.htm) and additional epitopes are still being identified. Some of these immunogenic NY-ESO-1-derived peptides have been investigated in a number of clinical trials [2, 14]. In general, while peptide vaccination with NY-ESO-1 and other antigens elicit tumor antigen-specific T cell responses, the clinical efficacy has been limited in immunized patients, probably because of lack of recognition of naturally processed antigen [8, 10]. Emerging evidence suggests that one of the critical factors for effective peptide vaccination is the identification of immunodominant regions of a tumor antigen with dual MHC classes I and II specificities [35]. Immunodominance is a phenomenon whereby the immune system focuses on a few abundant highly immunogenic peptides that bind to CD8+ and CD4+ T cells [6, 31, 32]. Thus, identification of such regions could be important for focusing peptide vaccination in the direction of these highly immunogenic epitopes.
Previously, we reported frequent expression of NY-ESO-1 in epithelial ovarian cancer (EOC) patients and evidence of spontaneous humoral and cellular immune responses to NY-ESO-1 in a high proportion of the patients [18, 21]. These findings suggest that NY-ESO-1 is a promising target for specific immunotherapy of EOC. While several immunodominant NY-ESO-1-specific HLA class I and class II epitopes have been identified in melanoma patients with spontaneous immune responses to NY-ESO-1 [13, 30], it is unknown whether CD8+ and CD4+ T cells from EOC patients recognize immunodominant NY-ESO-1 epitopes. In this study, we report the successful identification of two novel MHC class I epitopes of NY-ESO-1 from ovarian cancer patients with spontaneous humoral immune response to NY-ESO-1. They are both located in the NY-ESO-1119–143 sequence which is an HLA-class II rich region, containing epitopes binding to multiple HLA-DR alleles [33, 34]. We defined the minimal epitopes that are naturally processed by APC or naturally presented by tumor. Moreover, the epitopes are capable of inducing NY-ESO-1-specific CTL from PBMC of NY-ESO-1 seropositive EOC patients. These findings indicate that the ESO119–143 sequence comprises several naturally occurring HLA class I and class II epitopes, and a candidate for inducing tumor-specific CD4+ and CD8+ T cells in a large number of cancer patients.
Materials and methods
Patients
Tissue specimens and peripheral blood mononuclear cells (PBMCs) were obtained from patients undergoing debulking surgery for EOC at the Roswell Park Cancer Institute, Buffalo, NY. All specimens were collected under an approved protocol from the Institutional Review Board (IRB). Expression of NY-ESO-1 and/or LAGE-1 was detected in tumors by RT-PCR and/or immunohistochemistry, as previously described [18].
Peptides and multimer
Peptides (>90% purity) were synthesized according to good manufacturing practice guidelines (Multiple Peptide Systems, San Diego, CA, USA) and formulated (2 mg/ml in 100% DMSO) by the Biological Production Facility, Lausanne, Switzerland. Phycoerythrin (PE)-labeled HLA-A*0201/peptide multimer were assembled with NY-ESO-1-derived peptides 157–165, as described previously [11].
Molecular typing of HLA molecules
HLA-class I typing was performed at the HLA typing laboratory of the Roswell Park Cancer Institute using sequence-specific primer pairs obtained from Genovision [1]. Table 1 indicates HLA typing of patients, Epstein-Barr virus-transformed B lymphocyte (EBV-B) cell lines and tumor cell lines used in this study.
Table 1.
HLA molecule and NY-ESO-1 expression of the cells used in this report
| Cell line | HLA-A | HLA-B | HLA-C | Serum Ab | ESO | |||
|---|---|---|---|---|---|---|---|---|
| Patient B01 | 02 | 68 | 39 | 51 | 05 | 12 | + | |
| Patient B02 | 02 | 44 | 51 | 05 | 15 | + | ||
| Patient B03 | 33 | 68 | 15 | 8101 | 0202 | 18 | + | |
| Patient B04 | 11 | 68 | 44 | 52 | 07 | 12 | − | |
| Patient B05 | 11 | 68 | 06 | 07 | − | |||
| Patient B06 | 26 | 29 | 44 | 51 | 16 | 15 | + | |
| B07 T-APC | 01 | 25 | 18 | 37 | 06 | 12 | ||
| B08 EBV-B | 0201 | 0101 | 1801 | 3701 | 0602 | 0701 | ||
| B09 EBV-B | 0301 | 2402 | 35 | 4402 | 05 | 04 | ||
| SK-OV-3 | 0301 | 6801 | 1801 | 3501 | 0401 | 0501 | + | |
| SK-MEL-29 | 0201 | 6801 | 4402 | 4501 | 0501 | 0602 | − | |
| SK-MEL-128 | 0101 | 2402 | 4402 | 5101 | 0102 | 0501 | + | |
| SK-MEL-37 | 0201 | 1101 | 1501 | 5601 | 0102 | + | ||
Bold number indicates HLA molecule matched with Patient B01. The bold underline indicates HLA molecule matched with both Patients B01 and B02
Serum Ab (+) NY-ESO-1 seropositive patient
Serum Ab (−) NY-ESO-1 seronegative patient
ESO (+) NY-ESO-1 expressing cell line
ESO (+) NY-ESO-1 nonexpressing cell line
NY-ESO-1 serum antibody
NY-ESO-1-specific antibodies were measured in the serum by ELISA analysis, as described previously [18, 21].
In vitro stimulation of PBMCs
PBMCs were collected using a Ficoll gradient and were frozen in 90% fetal calf serum (FCS; Biofluid Inc., Gaithersburg, MD, USA) and 10% DMSO in liquid nitrogen until use. NY-ESO-1-specific CD8+ cells were elicited as described previously [11]. Briefly, CD8+ T lymphocytes were separated from PBMCs using magnetic beads (Dynabeads, Dynal, Oslo, Norway) and stimulated with irradiated autologous CD4/CD8-depleted PBMCs, infected with 1,000 pfu/cell adenoviral-NY-ESO-1 constructs (Ade-ESO) for patient B01; or pulsed with 2 μM pooled peptides (seventeen 20–25-mer overlapping peptides) for patient B02. The cells were cultured in complete RPMI medium [RPMI 1640 (Invitrogen Inc., Rockville, MO, USA) supplemented with 10% human AB serum (NABI, Boca Raton, FL, USA), l-glutamine (2 mM), penicillin (100 units/ml), streptomycin (100 μg/ml), 1% non-essential amino acids] in the presence of rhIL-2 (10 IU/ml, Roche Molecular Biochemicals, Indianapolis, IN, USA).
Target cells
The activated T cell APC (T-APC) as target cells were generated from a fraction of CD4+ T cells by stimulated with 10 μg/ml PHA (HA15, Murex Diagnostics, Dartford, UK). For HLA-restriction analysis, partially HLA-matched allogeneic EBV-B cells or T-APC were pulsed with 10 μM peptide in X-VIVO-15 medium overnight. In some experiments, target cells were incubated with graded concentration of peptides (10−5 to 10−11 M).
Intracellular staining
Presensitized CD8+ T cells were incubated with peptide-pulsed APC in the presence of anti-CD28 (1 μg/ml) and anti-CD49d (1 μg/ml), FITC-labeled CD107a and CD107b mAbs for 2 h. Brefeldin-A (BFA) and monensin (Sigma, St Louis, MO, USA) were added to the samples and the cells were incubated for an additional 4 h. After that, cells were stained with tricolor (TC)-labeled CD8 mAb and/or PE-labeled HLA-A2/ESO157–165 multimer, fixed, and stained intracellular cytokine with allophycocyanin-labeled IFN-γ mAb and/or PE-labeled tumor necrosis factor (TNF)-α, IL-2, IL-4, IL-5 or IL-10 mAbs in permeabilizing solution (CALTAG, Burlingame, CA, USA) containing normal mouse IgG (CALTAG) at room temperature for 30 min. Results were analyzed by flow cytometry by gating on CD8+ lymphocytes. All monoclonal antibodies were obtained from BD Pharmingen (San Diego, CA, USA).
ELISPOT assay
The number of IFN-γ secreting antigen-specific T cells was assessed by ELISPOT assays as described previously [15]. Briefly, flat-bottomed, 96-well nitrocellulose plates (Millititer; Millipore) were coated with IFN-γ mAb (2 μg/ml, 1-D1K; MABTECH, Stockholm, Sweden). CD8+ T cells and target cells were added to each well and incubated in plain RPMI 1640 medium, following blocking plates with complete RPMI medium. After 22–24 h of incubation, spots were developed using biotin-labeled IFN-γ mAb (0.2 μg/ml, 7-B6-1-biotin; MABTECH), streptavidin-alkaline phosphatase (1 μg/ml; MABTECH) and 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Sigma). The dark-violet spots were counted by an automated ELISPOT reader (Zeiss).
Cell lines
Established human melanoma cell lines were obtained from the cell bank maintained at the New York Branch of the Ludwig Institute. Melanoma lines; SK-MEL-29 (HLA-A68+ve; NY-ESO-1−ve), SK-MEL-37 (HLA-A2+ve; NY-ESO-1+ve) and SK-MEL-128, ovarian cancer cell line; SK-OV-3 (HLA-A68+ve; NY-ESO-1+ve) were maintained in RPMI 1640 (Invitrogen Inc., Rockville, MD, USA) supplemented with 10% FCS (Table 1).
Cloning of peptide-specific CD8+ cells
For establishing antigen-specific CD107 expressing CTL clone, CD8+ cells were stimulated with autologous T-APC pulsed with ESO119–143 peptide in the presence of anti-CD28, anti-CD49d, FITC-conjugated anti-CD107a/b mAbs for 1 h, and incubated in the presence of BFA and monensin for additional 4 h, and then, cells were stained with TC-labeled anti-CD8+ mAb. CD8+CD107a/b+ cells were sorted by FACSAria. Isolated ESO119–143 specific CD8+ cells were cloned by limiting dilution, and expanded in 10% FCS-RPMI 1640 medium in the presence of 10 μg/ml PHA, allogeneic irradiated feeder cells and 50 IU/ml rhIL-2. Antigen-specificity of CTL clones were screened by IFN-γ ELISPOT assay against autologous T-APC pulsed with or without ESO119–143 peptide.
Results
Immunodominant region of NY-ESO-1-specific CD8+ T cells in EOC patient
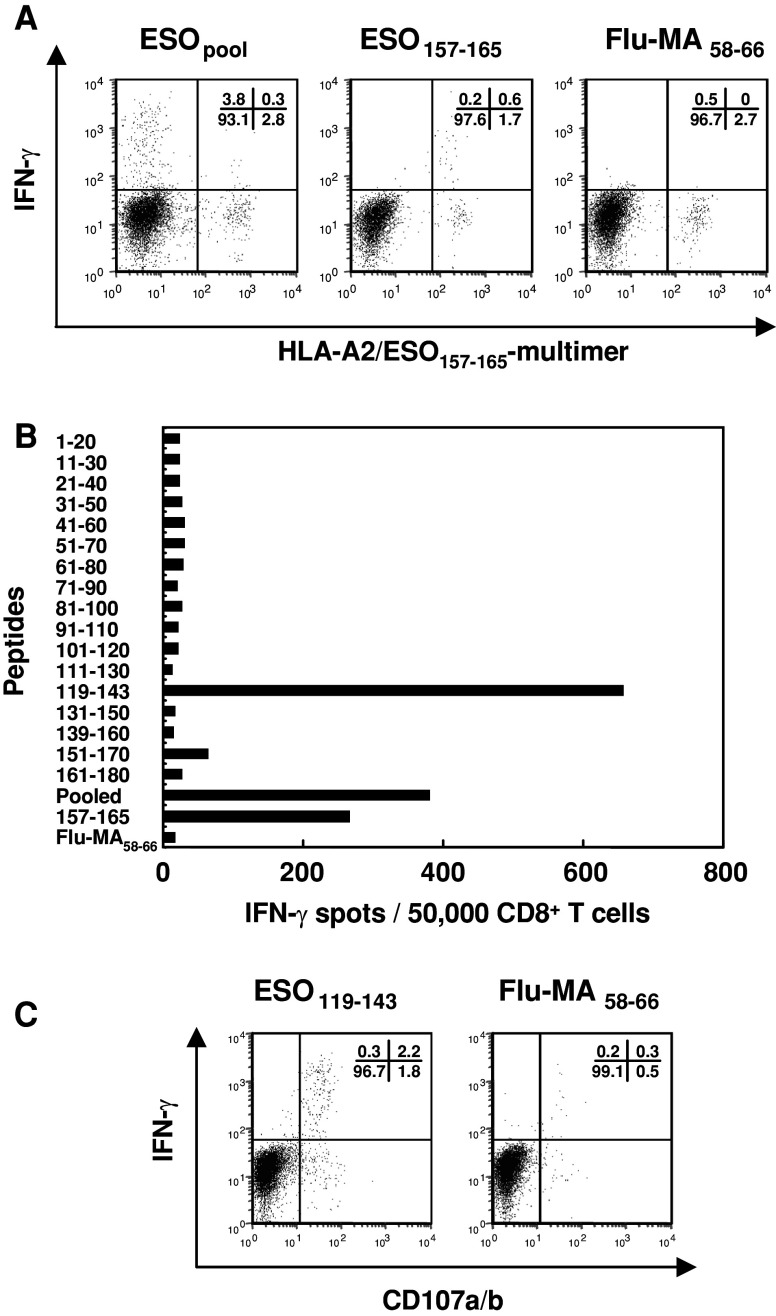
Previously, we reported that the presence of serum antibodies to NY-ESO-1 was tightly correlated with the presence of antigen-specific CD4+ and CD8+ T cell responses in cancer patients [9, 11]. Since patients B01 and B02 were NY-ESO-1 seropositive and HLA-A2+ve (Table 1), we predicted that these patients could have immunodominant CD8+ T cell responses to the HLA-A2 restricted peptide epitope, ESO157–165 [11]. Thus, to examine the response of patient B01, CD8+ cells were presensitized with NY-ESO-1 recombinant adenovirus (Ade-ESO) infected APC and tested for reactivity against target cells pulsed with pooled (seventeen 20–25-mer overlapping peptides), ESO157–165 or irrelevant Flu-MA58–66 (HLA-A2 epitope of influenza matrix protein) peptides. As shown in Fig. 1a, although the HLA-A2/ESO157–165 multimer positive population produced IFN-γ, there was a higher frequency of multimer-negative CD8+ cells that produced IFN-γ upon stimulation with pooled peptides. To identify the immunodominant region, the CD8+ cells were screened by ELISPOT assay using 17 individual overlapping peptides, i.e. components of the pooled peptides. Clearly, CD8+ T cell response was detected to ESO119–143 peptide and ESO157–165 but not to other ESO peptides (Fig. 1b). This response to ESO119–143 was confirmed by intracellular staining for IFN-γ and CD107a/b (Fig. 1c). CD107 expression has been shown to be indicative of CTL activity in previous studies [4, 22]. As shown in Fig. 1c, most of the ESO119–143 peptide specific-CD8+ cells were doubly positive for IFN-γ and CD107a/b or single positive of CD107a/b.
Fig. 1.
Immunodominant CD8+ T cell response in naturally occurring NY-ESO-1-seropositive EOC patient. CD8+ cells were co-cultured with Ade-ESO infected APC for 14 days. a IFN-γ production in HLA-A2/ESO157–165 multimer positive or negative CD8+ cells was analyzed against autologous T-APC pulsed with pooled peptides, ESO157–165 or Flu-MA58–66 by intracellular staining. b The number of IFN-γ producing CD8+ cells was tested by ELISPOT assay against autologous T-APC pulsed with 17 overlapping NY-ESO-1 20–25-mer peptides, ESO157–165 or Flu-MA58–66 peptides. c CD107 and IFN-γ expression of CD8+ cells were examined by intracellular staining upon stimulation with ESO119–143 or Flu-MA58–66 peptides
Identification of HLA restriction of NY-ESO-1-specific CD8+ cells
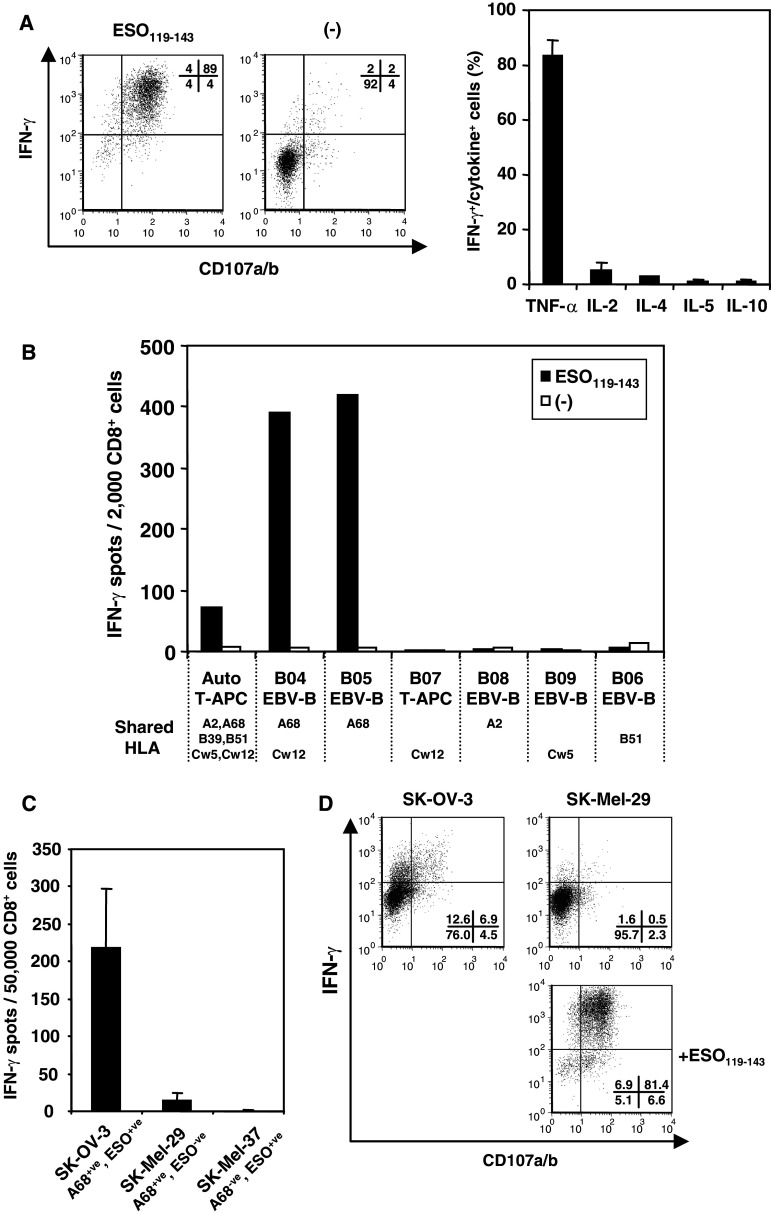
To further characterize ESO119–143-specific CD8+ T cell responses, we sorted CD107a/b expressing cells after peptide stimulation and generated a CTL clone. The B01 clone showed peptide specific-IFN-γ and CD107 expression, and the cells secreted high amounts of TNF-α but little or no production of other cytokines (Fig. 2a). To define the restricting HLA molecule by B01 clone, we examined their ability to respond to partially HLA-matched EBV-B, T-APC or melanoma cell lines pulsed with or without ESO119–143 peptide. As shown in Fig. 2b, only HLA-A68+ve target cells were able to present the peptide to B01 clone. To test this finding for tumor recognition, we used tumor cell lines that were HLA-A68+ve and were either NY-ESO-1 positive (SK-OV-3) or negative (SK-MEL-29). The ability of B01 clone to recognize the tumor cell lines was evaluated in ELISPOT assay (Fig. 2c). The B02 clone demonstrated specific response to SK-OV-3 but not SK-MEL-29 or HLA-A68−ve NY-ESO-1+ve SK-MEL-37 tumor cell lines. Furthermore, CD107 and IFN-γ expression was determined in the cloned cells by intracellular staining (Fig. 2d). To enhance the reactivity of B01 clone, the cells were treated with IL-2, IL-15 and IL-12 for 7 days, and then CD107 expression was examined by intracellular staining. Both CD107 expressing and IFN-γ producing cells were detected against SK-OV-3 and ESO119–143-pulsed SK-MEL-29 but not peptide-unpulsed SK-MEL-29. These results indicated that the CTL clone recognizing ESO119–143 peptide was restricted by HLA-A68 and the HLA-A68-restricted epitope was naturally presented by tumor cell line.
Fig. 2.
Determination of HLA restriction for CTL clone cells obtained from B01 patient. a IFN-γ and CD107 expression by B01 clone was examined in intracellular staining against autologous T-APC pulsed with or without ESO119–143 (left). Various cytokine productions were examined by intracellular staining using anti-TNF-α, anti-IL-2, anti-IL-4, anti-IL-5 and anti-IL-10 mAbs, respectively (right). b B01 clone was stimulated with partially HLA-matched T-APC or EBV-B cells pulsed with or without ESO119–143 peptide. IFN-γ productions from CTL clone were examined by ELISPOT assay. Only shared HLA molecules between the patient and other APCs are represented under the cell names. c The response of B01 clone against HLA-matched tumor cell lines were tested by ELISPOT assay. SK-OV-3 and SK-MEL-37 but not SK-MEL-29 express NY-ESO-1 antigen. d IFN-γ and CD107 expression of B01 clone against HLA-A68+ve tumor cell lines were tested by intracellular staining. For this assay, CTL clone was cultured under the combination of 10 U/ml IL-2, 10 ng/ml IL-15 and 10 U/ml IL-12 for 5 days, and then, the response against tumor cell lines were tested
The novel HLA-A68-restricted epitope was naturally presented by a tumor cell line
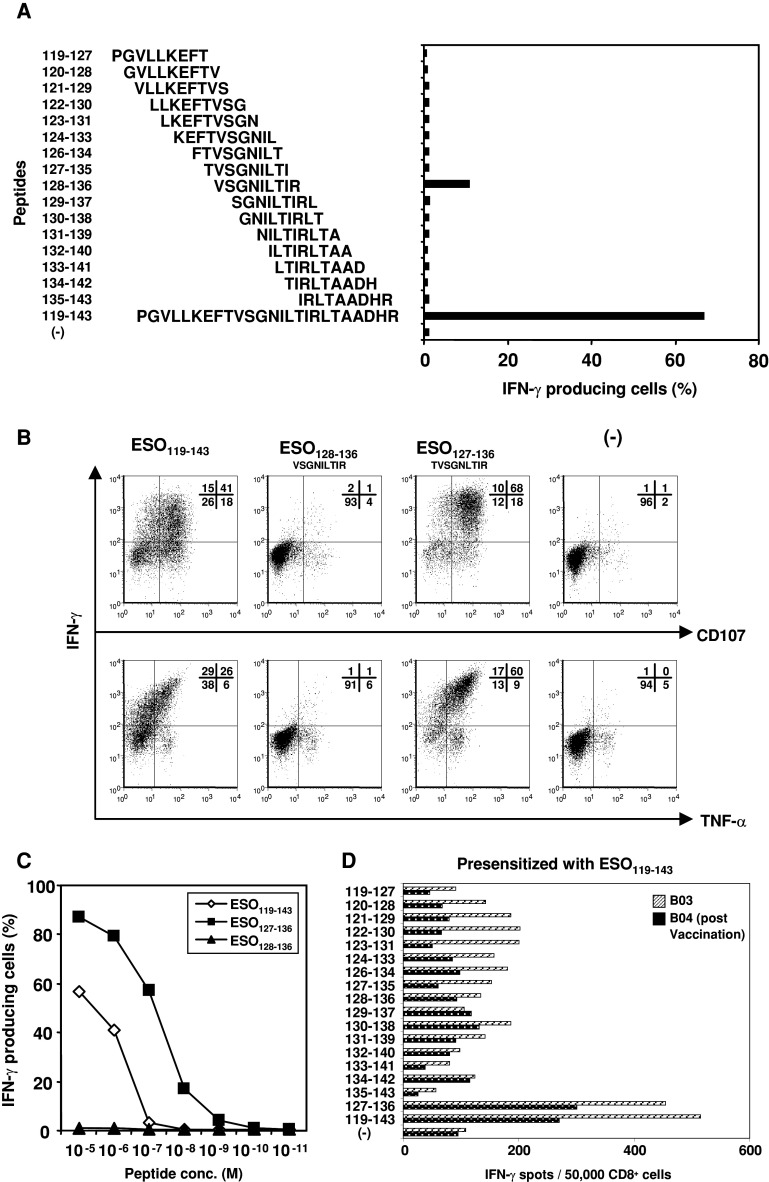
To more precisely define the epitope recognized by B01 clone, we synthesized and tested the response of the B01 clone against sets of overlapping 9- or 10-mer peptides that cover the sequence of NY-ESO-1 119–143. First, we tested the reactivity of the cloned cells to 9- or 10-mer overlapping peptides covering the ESO119–143 sequence. The B02 clone produced IFN-γ when stimulated with ESO128–136 peptide, but the response was significantly lower than that against the ESO119–143 peptide (Fig. 3a). Therefore, we searched the peptide sequence in ESO119–143 using a predictive algorithm for their dissociation rate with HLA-A68/allele [HLA Peptide Binding Predictions* (BIMAS) http://bimas.dcrt.nih.gov/molbio/hla_bind/]. The predictive binding score of ESO127–136 sequence was significantly higher than that of 9-mer, ESO128–136 peptide (200.00 vs. 30.00). Indeed, when we tested the response of B01 CTL clone against ESO127–136, the cells showed stronger fluorescence intensity of IFN-γ and higher double positive population of IFN-γ/TNF-α and IFN-γ/CD107 to the peptide than the reactivity against ESO128–136 (Fig. 3b), and the T cells were able to recognize peptide concentration as low as 10−8 M (Fig. 3c). Finally, we tested whether peptide-specific CD8+ cells could be induced by presensitizing with ESO119–143 or ESO127–136 from HLA-A68+ve EOC seropositive patients (patient B03). The CD8+ cells presensitized with ESO119–143 peptide induced IFN-γ production against both ESO119–143 and ESO127–136 peptide but not other peptides (Fig. 3d). Interestingly, antigen-specific HLA-A68 restricted CD8+ T cells were also detectable in a patient (#B04) enrolled in an on-going trial consisting of heterologous prime-boost vaccination with recombinant vaccinia-NY-ESO-1 and recombinant fowlpox-NY-ESO-1. However, presensitization with ESO127–136 peptide was not able to induce NY-ESO-1-specific CD8+ T cells from both HLA-A68+ve patients.
Fig. 3.
Identification of the optimal epitope within ESO119–143 recognized by HLA-A68-restricted B01 clone. a B01 clone was stimulated with B05-EBV-B cells pulsed with overlapping 9- or 10-mer peptides between ESO119–143, and cytoplasmic IFN-γ productions were determined by intracellular staining. b B01 clone was cocultured with autologous T-APC pulsed with the indicated peptides, and CD107 expression and IFN-γ and TNF-α productions were examined by intracellular staining. c The affinity to peptides were analyzed by intracellular staining against autologous T-APC pulsed with the different concentration of ESO119–143, ESO128–136 and ESO127–136 peptides. d CD8+ cells from HLA-A68+ve patients who was NY-ESO-1 seropositive (patient B03), and was seronegative but vaccinated with recombinant fowlpox-NY-ESO-1 and recombinant vaccinia-NY-ESO-1 (patient B04) were presensitized with ESO119–143 peptide for 14 days, and IFN-γ secretion were determined by ELISPOT assay against the indicated peptide pulsed autologous T-APC
Shared ESO-specific immunodominant region in another HLA-A2+ve EOC patient
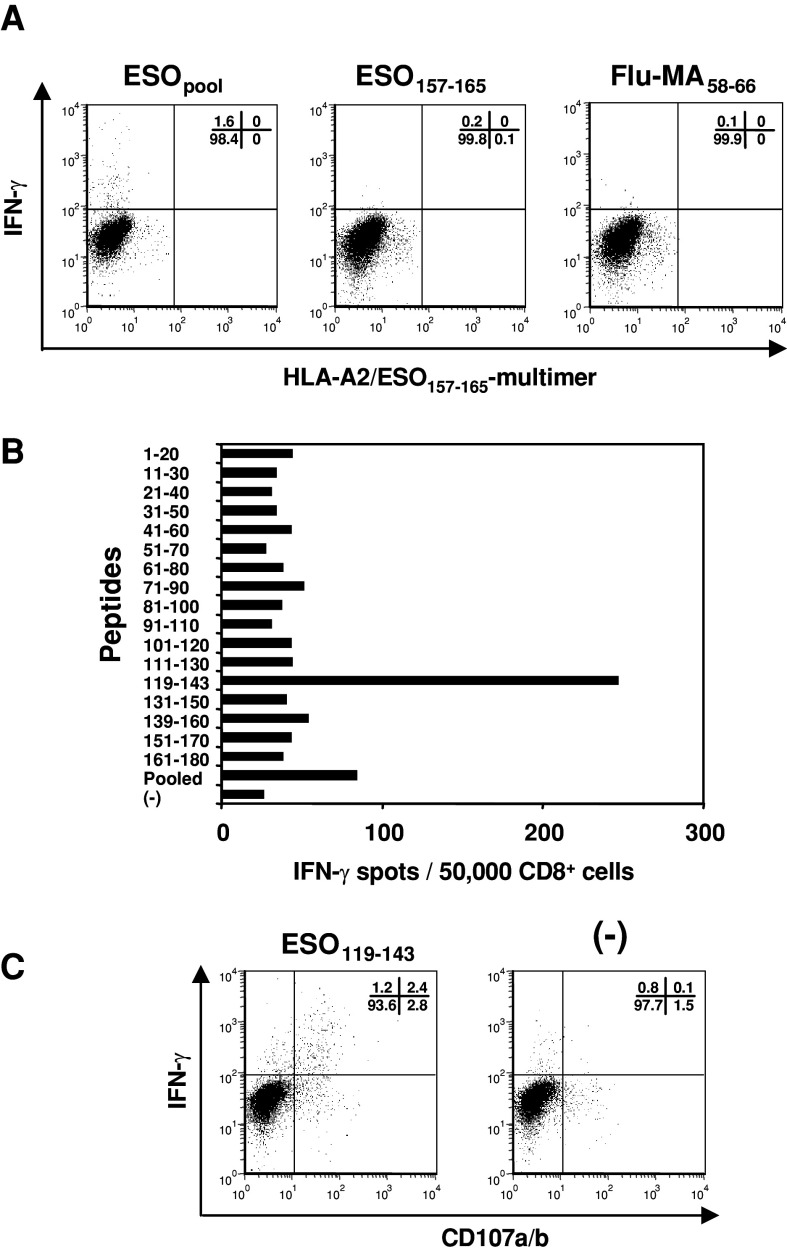
We next investigated for the presence of immunodominant CTL epitopes in another HLA-A2+ve and NY-ESO-1 seropositive EOC patient (B02) by presensitizing CD8+ cells with pooled overlapping NY-ESO-1 peptides. As shown in Fig. 4a, there were no detectable HLA-A2/ESO157–165-multimer reactive CD8+ T cells. In contrast, the multimer-negative population showed IFN-γ production following stimulation with pooled overlapping peptides. Interestingly, although the patient was HLA-A68−ve, the immunodominant epitopes of CD8+ cells were ESO119–143 (Fig. 4b, Table 1). Furthermore, by stimulating with ESO119–143 peptide, CD8+ cells showed high expression of IFN-γ and CD107 (Fig. 4c). These findings indicated that there was another CD8+ T cell epitope that was not restricted by HLA-A68, within the ESO119–143 sequence.
Fig. 4.
Detection of immunodominant CD8+ T cell response in HLA-A2+ve EOC patient with spontaneous antibody response to NY-ESO-1. CD8+ cells were presensitized with pooled NY-ESO-1 overlapping peptides for 14 days. a CD8+ cells were analyzed for reactivity against autologous T-APC pulsed with pooled peptides, ESO157–165 or Flu-MA58–66 peptide by intracellular staining. b The responses of CD8+ cells were tested by IFN-γ ELISPOT assay against autologous T-APC pulsed with 17 overlapping NY-ESO-1 20–25-mer peptides. c CD107 and IFN-γ expression of CD8+ cells was examined by intracellular staining upon stimulation with autologous T-APC pulsed with or without ESO119–143
Determination of the minimal epitope recognized by B02 clone
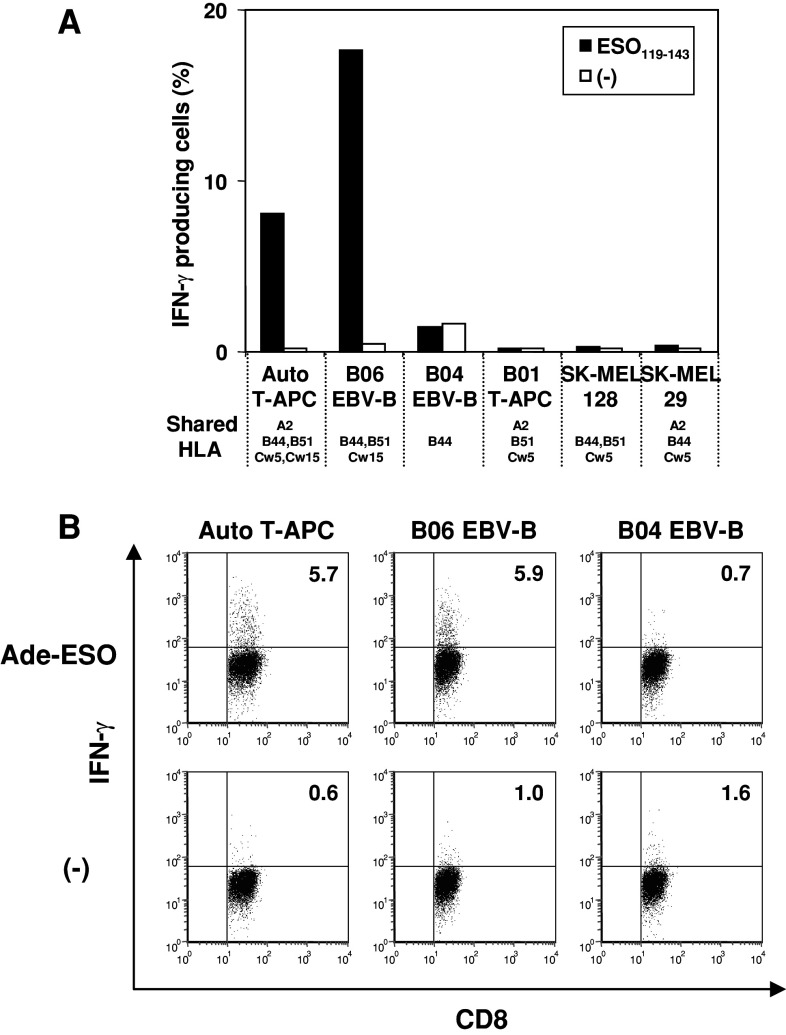
To characterize NY-ESO-1 specific CD8+ cells of patient B02, we established another CTL clone (named B02 clone), as described above. Established B02 clone produced IFN-γ and TNF-α but not other Th2 cytokines such as IL-4, IL-5 and IL-10 in response to ESO119–143 (data not shown). To define the restricting HLA molecule by B02 clone, we examined their ability to respond to partially HLA-matched EBV-B, T-APC or melanoma cell lines pulsed with or without ESO119–143 peptide. As shown in Fig. 5a, B02 clone specifically recognized HLA-Cw15+ve target cells, whereas there was no response to other shared alleles. Moreover, B02 clone produced IFN-γ in response to Ade-ESO-infected autologous T-APC and HLA-Cw15+ve B06-EBV-B cells, but not HLA-Cw15−ve B04-EBV-B cells (Fig. 5b). Together, this data indicate that the B02 CD8+ T cell clone recognized naturally processed NY-ESO-1 antigen restricted by HLA-Cw15.
Fig. 5.
Analyses of HLA restriction of CTL clone from patient B02 in response to ESO119–143. a B02 clone was stimulated with partially HLA-matched T-APC, EBV-B or melanoma cell lines pulsed with or without ESO119–143 peptide. IFN-γ production from the clone was examined by intracellular staining. b The response of B02 clone was detected by intracellular staining against Ade-ESO infected autologous T-APC or partially HLA-matched EBV-B cells. Only shared HLA molecules between the patient and other APC are represented under the cell names
Identification of the optimal epitope restricted by HLA-Cw15
We then performed studies as previously described (Fig. 3) to identify the optimal HLA-Cw15-restricted NY-ESO-1 epitope recognized by the B02 clone. B02 cells showed a stronger IFN-γ production in response to ESO127–135 and ESO128–136 compared to ESO119–143 (Fig. 6a). Since B02 clone showed reactivity against both ESO127–135 and ESO128–136 peptides, we tested additional 10-mer ESO127–136 peptide covering both ESO127–135 and ESO128–136. As shown in Fig. 6b, B02 clone expressed more CD107 and produced higher amounts of IFN-γ and TNF-α upon stimulation with ESO127–135 as compared with ESO127–136 and ESO128–136 peptides. Serial dilutions of the peptides confirmed that ESO127–135 was the minimal epitope within ESO119–143 sequence because B02 cells were able to recognize the peptide at a concentration of 1 nM and the recognition decreased dramatically as the peptide sequence was shifted or extended by a single amino acid (Fig. 6c). In addition, the ESO127–135 peptide was capable of inducing peptide-specific CD8+ T cells upon in vitro stimulation of PBMCs from HLA-Cw15+ve NY-ESO-1 seropositive ovarian cancer patients, B02 and B06 (Fig. 6d). These results indicated that ESO127–135 was the minimal and optimal epitope recognized by HLA-Cw15-restricted CD8+ T cells.
Fig. 6.
Identification of the minimal epitope of ESO119–143 recognized by HLA-Cw15-restricted B02 clone. a B02 clone were stimulated with autologous T-APC pulsed with overlapping 9- or 10-mer peptides in ESO119–143, and cytoplasmic IFN-γ productions were determined by intracellular staining. b B02 clone was cocultured with autologous T-APC pulsed with the indicated peptides, and CD107 expression and IFN-γ and TNF-α productions were examined by intracellular staining. c The affinity to peptides were analyzed in ELISPOT assay against autologous T-APC pulsed with the different concentration of ESO119–143, ESO127–135, ESO128–136 and ESO127–136 peptides. d CD8+ cells in NY-ESO-1 seropositive and HLA-Cw15+ve EOC patients were presensitized with ESO127–135 peptide for 20 days, and IFN-γ and CD107 expression was examined by intracellular staining against the indicated peptide-pulsed autologous T-APC
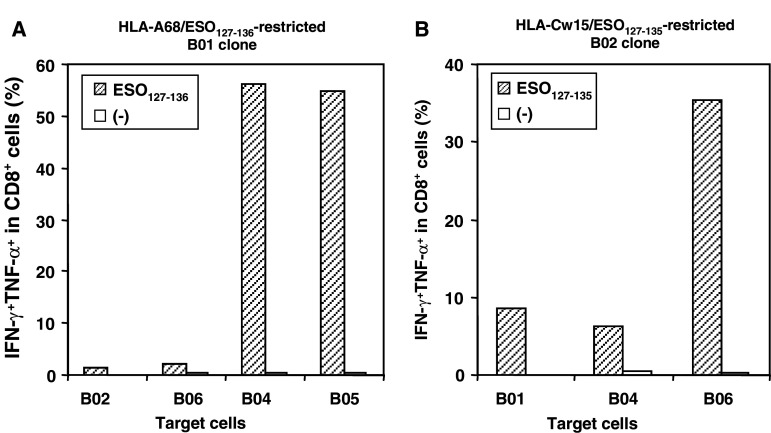
Since these two novel epitopes differ by only one amino acid, we ascertained that the CD8+ clones are not cross-reactive against the peptide presented by the other’s cognate APC. HLA-A68-restricted CD8+ cells did not recognize the epitope presented on HLA-Cw15+ve APC, and HLA-Cw15-restricted CTL clones did not cross-react with ESO127–135 presented by HLA-A68+ve APC (Fig. 7). These findings support our conclusion that the optimal sequence for HLA-A68-restricted NY-ESO-1 was ESO127–136, and the one for HLA-Cw15-specific NY-ESO-1 epitope was ESO127–135.
Fig. 7.
CTL clone cells did not recognize the epitope presented on the different HLA-expressing APCs. a IFN-γ/TNF-α productions from B01 clone were examined by intracellular staining against HLA-A68+ve or HLA-Cw15+ve APCs in the presence or absence of ESO127–136 peptide. b HLA-Cw15/ESO127–135-specific B02 clone was stimulated with the indicated APCs pulsed with or without ESO127–135 peptide and IFN-γ/TNF-α production were analyzed by intracellular staining
Discussion
In the present study, we have identified two novel NY-ESO-1 HLA-class I epitopes within the ESO119–143 sequence, recognized by CD8+ T cells of NY-ESO-1 seropositive EOC patients. These epitopes were naturally presented by tumor cell lines as well as APC. Importantly, the epitopes were able to induce antigen-specific CD8+ T cells with effector function following in vitro presensitization. Originally, ESO119–143 peptide was identified as a promiscuous HLA-class II epitope, which binds to several HLA-DR molecules (e.g. HLA-DRB1*0101, DRB1*0401, DRB1*0701, DRB1*1101, DRB1*1501, DRB3*0101, DRB4*0101 and DRB5*0101). In addition, the peptide induces tumor-reactive CD4+ T cells in vitro [33, 34]. The HLA-DRB1 molecules which bind the ESO119–143 are expressed by 46% of the American–Caucasian population and other HLA molecules (HLA-DRB3*0101, DRB4*0101 and DRB5*0101) are expressed with high frequency in the Caucasian population [33]. Altogether, ESO119–143 peptide is a promiscuous peptide sequence that binds to HLA-DR molecules expressed by 91.6% of the American–Caucasian population [33]. In addition, this peptide is a pan MHC-class II epitope that binds to multiple HLA-DR and HLA-DP4 molecules and stimulate antigen-specific CD4+ T cells in the context of these molecules [5, 16, 33]. Moreover, Ohkuri et al. [20] recently reported that additional HLA-DR molecules are capable of binding ESO119–143 sequence. We were able to induce ESO119–143 specific CD4+ cells by presensitizing CD4+ cells with ESO119–143 peptide or pooled peptides from patients B02 (DRB3*0101) and B06 (DRB1*0401) (data not shown). In previous reports, two HLA-class I epitopes restricted by HLA-A66 and HLA-Cw3 molecules were described in the ESO119–143 sequence [13, 30]. Taken together with the novel epitopes found in the present study, HLA class I restriction of ESO119–143 peptide could potentially occur in approximately 27–48% of Caucasians (HLA-A68: 4–12%; HLA-Cw15: 3–7%; HLA-Cw3: 20–28%; HLA-A66: 0–1%), and 41–55% of Blacks (HLA-A68: 16–24%; HLA-Cw15: 4–6%; HLA-Cw3: 18%; HLA-A66: 3–7%) (http://www.allelefrequencies.net/test/default1.asp). In addition, ESO119–143 was shown to be an immunodominant HLA-class I peptide that is efficiently processed by proteasomes in a study of melanoma patients [30]. Therefore, ESO119–143 is an immunodominant NY-ESO-1 derived peptide that is likely to elicit vaccine induced CD4+ and CD8+ T cells in a high frequency of Caucasian and non-Caucasian populations.
Sun et al. [28] found that the HLA-A68-restricted LAGE-1 epitope corresponds to residues 103–111 of the protein sequence. It is well known that LAGE-1 and NY-ESO-1/LAGE-2 are highly homologous proteins with up to 84% identity [17], and several epitopes such as HLA-A2/SLLMWITQC (157–165) are the same sequences. However, the residues 103–111 contain four amino acids that are different between NY-ESO-1 (ELARRSLAQ) and LAGE-1 (ELVRRILSR). Moreover, Sun et al. [28] showed that COS-7 cells co-transfected with HLA-A68 and NY-ESO-1 were not recognized by HLA-A68/LAGE-1103–111-specific CTL cells. Similarly, there are two different amino acids in residues 127–136; the sequence of NY-ESO-1 is TVSGNILTIR and that of LAGE-1 is TVSGNLLFIR, and ESO127–136 but not LAGE-1127–136 contains a potential phosphorylation site for protein kinase C (Thr 134). Thus, for HLA-A68, it is not surprising that the HLA-class I epitope is not the same for NY-ESO-1 and LAGE-1.
The HLA-Cw15-restricted 9-mer epitope, ESO127–135, was able to expand antigen-specific CD8+ cells from HLA-Cw15+ve EOC patients (Fig. 6d). In contrast, although we were able to detect HLA-A68 restricted antigen-specific CD8+ cells by presensitizing with ESO119–143 peptide (Fig. 3d), ESO127–136 peptide stimulation did not expand NY-ESO-1-specific CD8+ T cell precursors from HLA-A68+ve patient’s PBMC. While this observation could be due to differences in peptide stability, we found that stability of ESO127–136 was almost similar to ESO119–143 peptide (data not shown). In our previous study, we showed that HLA-A2/ESO157–165-specific CD8+ T cells could be detected at higher frequency by stimulating with ESO157–170 peptide, rather than ESO157–165 peptide [19]. In addition, compared with HLA-Cw15/ESO127–135, the avidity of the HLA-A68 restricted clone for ESO119–143 and ESO127–136 was not significantly different. These findings suggest that when peptide avidity is not remarkably different, the longer peptide (ESO119–143) containing the minimal epitope might induce antigen-specific CD8+ cells more efficiently than the minimal epitope (ESO127–136) alone.
Peptide vaccines remain attractive candidates for clinical use because of their ease of production and minimal risk of side effects. While the majority of cancer vaccine trials have focused on eliciting antigen-specific CD8+ effector T cells, it is now generally accepted that tumor-specific CD4+ cells also play an important role in tumor rejection [3, 12, 29]. The multiple roles of antigen-specific CD4+ T cells include the provision of help to antigen-specific CD8+ T cells during the primary and secondary immune responses, direct cytolysis, and activation of B cells for production of tumor antigen-specific antibodies [24, 25, 27]. Thus, identifying immunodominant epitopes capable of eliciting integrated CD4+ and CD8+ T cell responses will be critical for enhancing the efficacy of peptide vaccines. The results of our recent clinical study using ESO157–170 peptide of dual HLA-class I and II specificities underlines the role of CD4+ T cells in augmenting expansion of functional and long-lasting CD8+ T cells with capacity to recognize tumor targets [19]. Taken together, the present study strongly support ESO119–143 peptide as a vaccine candidate for the induction of tumor-reactive CD4+ and CD8+ T cells in ovarian cancer patients.
Acknowledgments
This work was supported by Cancer Research Institute/Ludwig Institute for Cancer Research Cancer Vaccine Collaborative Grant; and by Anna-Marie Kellen Clinical Investigator Award of the Cancer Research Institute, NY (K. O.).
References
- 1.Aldener-Cannava A, Olerup O. HLA-DPB1 typing by polymerase chain reaction amplification with sequence-specific primers. Tissue Antigens. 2001;7:287–299. doi: 10.1034/j.1399-0039.2001.057004287.x. [DOI] [PubMed] [Google Scholar]
- 2.Baumgaertner P, Rufer N, Devevre E, Derre L, Rimoldi D, Geldhof C, Voelter V, Liénard D, Romero P, Speiser DE. Ex vivo detectable human CD8 T-cell responses to cancer-testis antigens. Cancer Res. 2006;66:1912–1916. doi: 10.1158/0008-5472.CAN-05-3793. [DOI] [PubMed] [Google Scholar]
- 3.Baxevanis CN, Voutsas IF, Tsitsilonis OE, Gritzapis AD, Sotiriadou R, Papamichail M. Tumor-specific CD4+ T lymphocytes from cancer patients are required for optimal induction of cytotoxic T cells against the autologous tumor. J Immunol. 2000;164:3902–3912. doi: 10.4049/jimmunol.164.7.3902. [DOI] [PubMed] [Google Scholar]
- 4.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/S0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 5.Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, Gahery-Ségard H, Guillet JG, Ménez A, Georges B, Maillère B. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol. 2002;169:6928–6934. doi: 10.4049/jimmunol.169.12.6928. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, McCluskey J. Immunodominance and immunodomination: critical factors in developing effective CD8+ T-cell-based cancer vaccines. Adv Cancer Res. 2006;95:203–247. doi: 10.1016/S0065-230X(06)95006-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen YT, Scanlan MJ, Sahin U, Türeci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutoit V, Taub RN, Papadopoulos KP, Talbot S, Keohan ML, Brehm M, Gnjatic S, Harris PE, Bisikirska B, Guillaume P, Cerottini JC, Hesdorffer CS, Old LJ, Valmori D. Multiepitope CD8+ T cell response to a NY-ESO-1 peptide vaccine results in imprecise tumor targeting. J Clin Invest. 2002;110:1813–1022. doi: 10.1172/JCI200216428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnjatic S, Atanackovic D, Jäger E, Matsuo M, Selvakumar A, Altorki NK, Maki RG, Dupont B, Ritter G, Chen YT, Knuth A, Old LJ. Survey of naturally occurring CD4+ T cell responses against NY-ESO-1 in cancer patients: correlation with antibody responses. Proc Natl Acad Sci USA. 2003;100:8862–8867. doi: 10.1073/pnas.1133324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gnjatic S, Jäger E, Chen W, Altorki NK, Matsuo M, Lee SY, Chen Q, Nagata Y, Atanackovic D, Chen YT, Ritter G, Cebon J, Knuth A, Old LJ. CD8+ T cell responses against a dominant cryptic HLA-A2 epitope after NY-ESO-1 peptide immunization of cancer patients. Proc Natl Acad Sci USA. 2002;99:11813–11818. doi: 10.1073/pnas.142417699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnjatic S, Nagata Y, Jager E, Stockert E, Shankara S, Roberts BL, Mazzara GP, Lee SY, Dunbar PR, Dupont B, Cerundolo V, Ritter G, Chen YT, Knuth A, Old LJ. Strategy for monitoring T cell responses to NY-ESO-1 in patients with any HLA class I allele. Proc Natl Acad Sci USA. 2000;97:10917–10922. doi: 10.1073/pnas.97.20.10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung K, Hayashi R, Lafond-Walker A, Lowenstein C, Pardoll D, Levitsky H. The central role of CD4+ T cells in the antitumor immune response. J Exp Med. 1998;188:2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson H, Dimopoulos N, Mifsud NA, Tai TY, Chen Q, Svobodova S, Browning J, Luescher I, Stockert L, Old LJ, Davis ID, Cebon J, Chen W. Striking immunodominance hierarchy of naturally occurring CD8+ and CD4+ T cell responses to tumor antigen NY-ESO-1. J Immunol. 2006;176:5908–5917. doi: 10.4049/jimmunol.176.10.5908. [DOI] [PubMed] [Google Scholar]
- 14.Jäger E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jäger E, Nagata Y, Gnjatic S, Wada H, Stockert E, Karbach J, Dunbar PR, Lee SY, Jungbluth A, Jäger D, Arand M, Ritter G, Cerundolo V, Dupont B, Chen YT, Old LJ, Knuth A. Monitoring CD8 T cell responses to NY-ESO-1: correlation of humoral and cellular immune responses. Proc Natl Acad Sci USA. 2000;97:4760–4765. doi: 10.1073/pnas.97.9.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudela P, Janjic B, Fourcade J, Castelli F, Andrade P, Kirkwood JM, El-Hefnawy T, Amicosante M, Maillere B, Zarour HM. Cross-reactive CD4+ T cells against one immunodominant tumor-derived epitope in melanoma patients. J Immunol. 2007;179:7932–7940. doi: 10.4049/jimmunol.179.11.7932. [DOI] [PubMed] [Google Scholar]
- 17.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. LAGE-1, a new gene with tumor specificity. Int J Cancer. 1998;76:903–908. doi: 10.1002/(SICI)1097-0215(19980610)76:6<903::AID-IJC22>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Odunsi K, Jungbluth AA, Stockert E, Qian F, Gnjatic S, Tammela J, Intengan M, Beck A, Keitz B, Santiago D, Williamson B, Scanlan MJ, Ritter G, Chen YT, Driscoll D, Sood A, Lele S, Old LJ. NY-ESO-1 and LAGE-1 cancer-testis antigens are potential targets for immunotherapy in epithelial ovarian cancer. Cancer Res. 2003;63:6076–6083. [PubMed] [Google Scholar]
- 19.Odunsi K, Qian F, Matsuzaki J, Mhawech-Fauceglia P, Andrews C, Hoffman EW, Pan L, Ritter G, Villella J, Thomas B, Rodabaugh K, Lele S, Shrikant P, Old LJ, Gnjatic S. Vaccination with an NY-ESO-1 peptide of dual HLA class I and II specificities plus incomplete Freund adjuvant induces simultaneous humoral, CD4+ and CD8+ T-cell responses in ovarian cancer patients. Proc Natl Acad Sci USA. 2007;104:12837–12842. doi: 10.1073/pnas.0703342104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohkuri T, Sato M, Abe H, Tsuji K, Yamagishi Y, Ikeda H, Matsubara N, Kitamura H, Nishimura T. Identification of a novel NY-ESO-1 promiscuous helper epitope presented by multiple MHC class II molecules found frequently in the Japanese population. Cancer Sci. 2007;98:1092–1098. doi: 10.1111/j.1349-7006.2007.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian F, Gnjatic S, Jäger E, Santiago D, Jungbluth A, Grande C, Schneider S, Keitz B, Driscoll D, Ritter G, Lele S, Sood A, Old LJ, Odunsi K. Th1/Th2 CD4+ T cell responses against NY-ESO-1 in HLA-DPB1*0401/0402 patients with epithelial ovarian cancer. Cancer Immun. 2004;4:12. [PubMed] [Google Scholar]
- 22.Rubio V, Stuge TB, Singh N, Betts MR, Weber JS, Roederer M, Lee PP. Ex vivo identification, isolation and analysis of tumor-cytolytic T cells. Nat Med. 2003;9:1377–1382. doi: 10.1038/nm942. [DOI] [PubMed] [Google Scholar]
- 23.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 24.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 25.Smith CM, Wilson NS, Waithman J, Villadangos JA, Carbone FR, Heath WR, Belz GT. Cognate CD4+ T cell licensing of dendritic cells in CD8+ T cell immunity. Nat Immunol. 2004;5:1143–1148. doi: 10.1038/ni1129. [DOI] [PubMed] [Google Scholar]
- 26.Stockert E, Jäger E, Chen YT, Scanlan MJ, Gout I, Karbach J, Arand M, Knuth A, Old LJ. A survey of the humoral immune response of cancer patients to a panel of human tumor antigens. J Exp Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Z, Lethé B, Zhang Y, Russo V, Colau D, Stroobant V, Boon T, van der Bruggen P. A new LAGE-1 peptide recognized by cytolytic T lymphocytes on HLA-A68 tumors. Cancer Immunol Immunother. 2006;55:644–652. doi: 10.1007/s00262-005-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toes RE, Ossendorp F, Offringa R, Melief CJ. CD4 T cells and their role in antitumor immune responses. J Exp Med. 1999;189:753–756. doi: 10.1084/jem.189.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valmori D, Lévy F, Godefroy E, Scotto L, Souleimanian NE, Karbach J, Tosello V, Hesdorffer CS, Old LJ, Jager E, Ayyoub M. Epitope clustering in regions undergoing efficient proteasomal processing defines immunodominant CTL regions of a tumor antigen. Clin Immunol. 2007;122:163–172. doi: 10.1016/j.clim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 32.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533–543. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Zarour HM, Maillere B, Brusic V, Coval K, Williams E, Pouvelle-Moratille S, Castelli F, Land S, Bennouna J, Logan T, Kirkwood JM. NY-ESO-1 119–143 is a promiscuous major histocompatibility complex class II T-helper epitope recognized by Th1- and Th2-type tumor-reactive CD4+ T cells. Cancer Res. 2002;62:213–218. [PubMed] [Google Scholar]
- 34.Zarour HM, Storkus WJ, Brusic V, Williams E, Kirkwood JM. NY-ESO-1 encodes DRB1*0401-restricted epitopes recognized by melanoma-reactive CD4+ T cells. Cancer Res. 2000;60:4946–4952. [PubMed] [Google Scholar]
- 35.Zeng G, Wang X, Robbins PF, Rosenberg SA, Wang RF. CD4+ T cell recognition of MHC class II-restricted epitopes from NY-ESO-1 presented by a prevalent HLA DP4 allele: association with NY-ESO-1 antibody production. Proc Natl Acad Sci USA. 2001;98:3964–3969. doi: 10.1073/pnas.061507398. [DOI] [PMC free article] [PubMed] [Google Scholar]