Abstract
Neoplastic transformation is a multistep process in which gene products of specific regulatory pathways are involved at each stage. Identification of these overexpressed or mutated gene products provides an unprecedented opportunity to address the immune system against defined antigens and eliminate transformed cells. Mice transgenic for these oncogenes (e.g. HER-2/neu, a prototype of deregulated oncogenic protein kinase membrane receptors) are ideal experimental models for assessing the potential of active immunization. The demonstration that vaccines can cure HER-2/neu transplantable tumors, prevent their onset and delay the progression of preneoplastic lesions in mice at risk suggests that efficient immunological inhibition of HER-2/neu carcinogenesis can be achieved by specific vaccination. To further explore this issue, halting of tumor progression in the mammary glands of BALB-neuT mice with two immunization protocols in two laboratories has been studied independently by DNA microarray analysis. Combination of the two sets of results revealed a clear correlation between them when the tumor mass was titrated by transcription profiling. It was also clear that both protocols induced a strong, polyclonal antibody response and halted tumor growth at a condition very similar to that at which the vaccination began. Differences in the expression profiles were mainly related to the expression levels of a few chemokines and T-cell-specific genes that may be in some way correlated with the efficacy of the vaccination. Last, combination of the expression data with the protection results indicated that chronic vaccination is needed to maintain an active IFN-γ-mediated response in the mammary gland.
Keywords: HER-2/neu, Tumor prevention, Mammary carcinogenesis, Cancer vaccines, Microarray analysis, Transcriptional profiling
Introduction
Neoplastic transformation is a multistep process in which gene products of specific regulatory pathways are involved at each stage. It may thus be supposed that a given stage can be influenced or even halted by interfering with its pathway/component. Identification of these distinct overexpressed or mutated gene products provides an unprecedented opportunity to address the immune system against defined antigens and eliminate transformed cells [10]. Mice transgenic for oncogenes are ideal experimental models for assessing the potential of active immunization [11, 23]. Those transgenic for the HER-2/neu oncogene are extensively studied because HER-2/neu is a prototype of the deregulated oncogenic protein kinase membrane receptors directly involved in cell carcinogenesis and markedly overexpressed by early neoplastic lesions. In addition, its amplification and consequent overexpression are associated with tumorigenesis in the breast, ovaries, brain and prostate, and an aggressive course [1, 13].
Vaccines can cure HER-2/neu transplantable tumors by preventing their onset and delaying the progression of preneoplastic alterations to invasive carcinomas [32]. We have used HER-2/neu mammary carcinogenesis in BALB-neuT mice, one of the most aggressive models [3, 32], to show that its efficient immunological inhibition can be achieved in this way by vaccination [21, 25–28]. This protection always stems from the concurrence of IFN-γ release and specific antibodies against the oncogene’s protein product p185neu. T-cell cytotoxicity, on the other hand, appears of only marginal importance [4, 5, 8, 21, 25–28] despite its proven association with the response against transplantable tumors. This paper describes our further investigation of the role of antibodies in this context through the employment of DNA microarray analysis to study the outcome of two immunization protocols independently in two laboratories.
Combined allogeneic tumor cell vaccinations halt HER-2/neu carcinogenesis in BALB-neuT mice
Two vaccination protocols halt mammary carcinogenesis in BALB-neuT mice (H-2d) [21, 26] transgenic for the activated transforming rat HER-2/neu oncogene under the transcriptional control of the mouse mammary tumor virus promoter [19]. These mice are one of the most aggressive models of mammary carcinogenesis, since p185neu is already overexpressed on the cell surface of the rudimentary mammary glands of 3-week-old females. All ten glands inexorably develop invasive and metastasizing carcinomas through a well-established and almost synchronous progression [3, 5, 21, 26]. Lollini et al. [21] observed that a combination of MHC allogeneic (H-2q) tumor cells expressing p185neu and systemic recombinant interleukin (IL-) 12 administered to healthy, 6-week-old BALB-neuT mice reduced tumor incidence by 90–100% and more than doubled life expectancy. Almost the same protection was provided by a single-cell vaccine composed of H-2q p185neu-positive tumor cells gene-engineered to release IL-12 [8]. Cavallo et al. [26] have shown that vaccination of 10-week-old mice bearing multiple in situ carcinomas with DNA plasmids coding for the extracellular and transmembrane domain of p185neu, followed by a boost with p185neu-positive H-2q tumor cells gene-engineered to secrete IFN-γ, keeps 48% of mice tumor free until week 32.
In both studies, cytokine release and antibody production were the most prominent immune mechanisms, whereas T-cell cytotoxicity was absent, and high serum titers of anti-p185neu antibodies were found. Since these antibodies were mainly IgG2a and to a lesser extent IgG2b, whereas IgG3 was a marginal component, vaccination may be supposed to activate T cells producing IFN-γ, the primary switch factor for IgG2a. The gene expression patterns of the mammary glands of these mice have now been investigated independently in the two laboratories [26; Astolfi et al., unpublished results] to determine the mechanisms through which protection is provided. The results are compared in this paper.
Cross-study validation
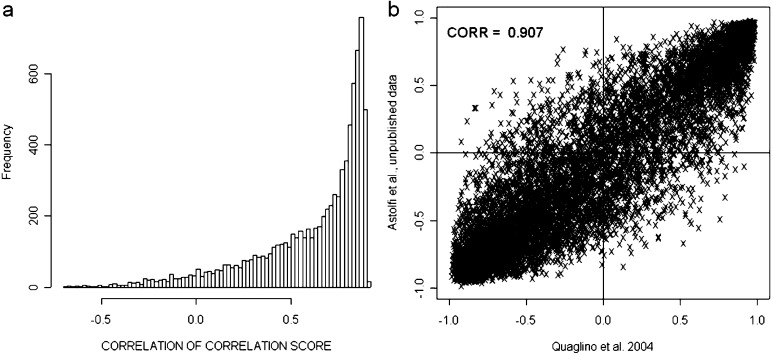
Integration and cross-study validation of the gene expression profiles were hampered by the use of distinct microarray platforms, sample sets, protocols, and analytical approaches. Parmigiani et al. [24] have devised an integrative correlation (IC) coefficient for quantification of the extent to which independent studies can be reliably analyzed together for the systematic comparison of microarray profiles. This approach has been used for cross-study validation of the data of Cavallo [26] and Lollini [Astolfi, unpublished results]. All pairwise correlations (Pearson correlation coefficient) of gene expression across samples within individual projects were calculated and the reproducibility of the results was defined without relying on direct comparison of expression across platforms. Overall reproducibility was summarized by correlations of IC coefficients equal to 0.907 and it was also evaluated gene by gene (Fig. 1). Both overall and gene-by-gene reproducibility revealed the striking similarity of the two studies and hence allowed comparison of their data.
Fig. 1.
Cross-study validation of data generated by Quaglino et al. [26] and De Giovanni et al. [8; Astolfi, unpublished results] reveals significant agreement of the gene expression patterns in these two independent studies. Cross-experiment consistency of expression patterns was evaluated with an integrative correlation (IC) coefficient that quantifies cross-study reproducibility without relying on direct assimilation of expression measurements across experiments. a Histogram of the empirical distribution of the gene-specific correlation of correlations. b Scatterplot of the pairwise correlation comparison within the probe sets in the two datasets.
Titration of the tumor by transcriptional profiling
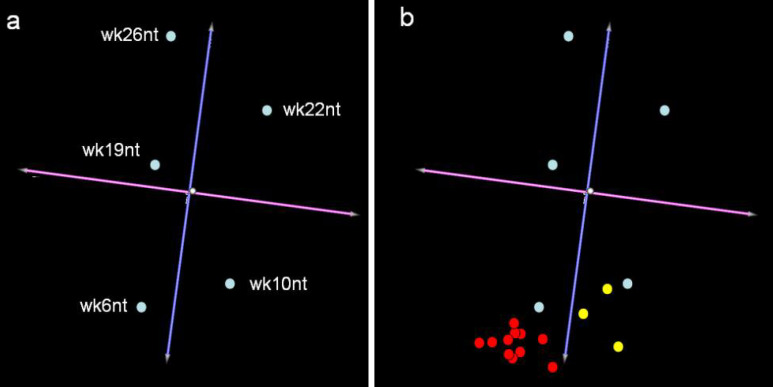
The number of neoplastic cells in the mammary tissue of BALB-neuT mice increases constantly and by 22 weeks of age a tumor mass becomes palpable in all their mammary glands. Quaglino et al. [26] used Affymetrix MGU74Av2 genechips (Affymetrix Inc., Santa Clara, CA) to profile the gene expression in the glands of non-treated 10-week-old (wk10nt) and 22-week-old (wk22nt) mice, De Giovanni et al. [8] employed them to study gene expression changes in the glands from 6-week-old (wk6nt), 19-week-old (wk19nt), and 26-week-old (wk26nt) mice. Despite their slightly different approaches (Cavallo uses experimental replicates made of pools of animals, while Lollini uses multiple animals, etc.), the studies display a good IC coefficient and can be compared [24]. Virtual two-dye experiments [14] were generated by comparing the gene expression linked to HER-2/neu neoplastic alteration in mammary glands from 6- to 26-week-old BALB-neuT mice with that linked to the hormonal hyperplasia of those from 2-weeks-pregnant (wk2pgr) wild-type BALB/c mice. The replicates were considered individually to take account of their biological differences. The results were evaluated by PCA (principal component analysis). This technique represents the global gene expression changes between the samples as coordinates in a multidimensional space in which the axes are called the “principal components”. Since the first components account for the greatest part of the variability within the dataset, graphical representation of the first three components only (Fig. 2a) will catch the differences between the samples. This tridimensional representation groups mice in function of their age and hence in function of the increase in the tumor mass in their mammary glands, since 91.3% of the total variance is distributed around the blue axis (Fig. 2a). Less than 5% of the total variance is distributed around the purple axis (Fig. 2a). This shows that the variance in function of the laboratory in which the experiments were performed (lab effect) is of little weight. Moreover, it was also found that the lab effect itself is due to a very small number of genes related to the different ways in which the samples were collected (data not shown).
Fig. 2.
Principal component analysis of the transcriptional profiling of gene expression linked to HER-2/neu neoplastic alteration and anti-HER-2/neu vaccination. a The panel shows the PCA experiment clustering generated using TMEV (http://www.tigr.org). Each light blue dot relates to the average of the experimental replicates for each gene in each experiment. Even though the data have been generated by two independent laboratories with slightly different approaches, the variance accounting for the differences between the groups is related to age, as the total variance is distributed around the blue axis. b Yellow dots showing transcriptional profiling at week 22 following vaccination at week 10 reveals that growth of the tumor mass is halted at week 10 (Cavallo’s laboratory). Since the profiles at week 15 and week 26 following vaccination at week 6 (red dots Lollini’s laboratory) resemble that of wk6nt mice, it is clear that vaccination time determines the stage at which tumor progression is halted.
A major open question in the field of microarray studies is whether the analysis should be performed on a pure cell population obtained, for example, by microdissection, or on whole tissues. This may depend on the tissue, organ or biological process involved. In the case of mammary carcinogenesis, we firmly believe that whole-gland analysis is the only meaningful way to study gene expression profiles. Both normal and neoplastic glands are complex microenvironments containing many cell types that co-ordinately change their expression profiles during physiological (e.g. pregnancy, lactation, involution) and pathological (e.g. carcinogenesis, inflammation, fibrosis/desmoplasia) processes [6, 9, 29]. Analysis of a single cell type invariably leads to the loss of a global functional perspective. The classic metaphor of the blind men trying to understand what an elephant is just by touching its legs, trunk, or tail [2] is a particularly appropriate illustration of this situation (Fig. 3).
Fig. 3.
“The blind men and the elephant”: poem by John Godfrey Saxe.
Even if attention is solely devoted to mammary epithelial cells, one is faced with a daunting prospect. It is now clear that these cells are of several types (or in several differentiation states) [31]. Carcinogenesis as opposed to normal development presents the same difficulties. The definition of stem cell markers (albeit imperfect) has demonstrated that the proportion of cancer stem cells in transgenic model systems varies considerably and is apparently unrelated to tumor aggressiveness. In the mammary carcinomas of HER-2/neu transgenic mice of another line, for example, this proportion was as low as 6% [16]. One may thus ask whether microarray analysis should be restricted to stem cells rather than the whole tumor. We studied whole glands because we believe that both carcinogenesis and immune responses affect the gland as a whole, and hence that microdissection would be meaningless. The only single structure considered was the inguinal lymph node that sits in the middle of the fourth pair of glands (Fig. 4). In a few samples, we assessed the gene expression profiles of glands with and without this node to compare the changes that take place in the mammary tissue and in the node.
Fig. 4.
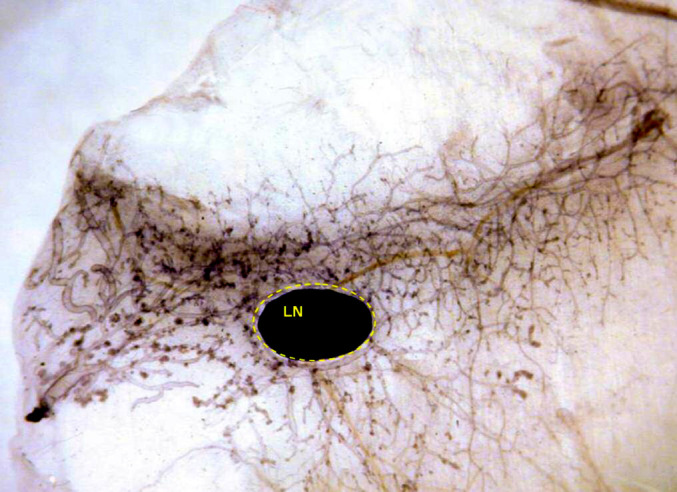
Whole mount of the fourth mammary gland of a 26-week-old BALB-neuT mouse vaccinated with allogeneic mammary carcinoma cells expressing HER-2/neu and engineered to produce IL-12. The black dot is the lymph node (LN). The gland is free from carcinoma and displays only some hyperplastic foci in the end buds.
Effects of vaccinations on the gene expression profile of the mammary tissue
In Cavallo’s study, BALB-neuT mice bearing multiple in situ carcinomas were primed at weeks 10 and 12 with plasmids coding for the transmembrane and extracellular domains of the p185neu. One week later, they were boosted with p185neu-positive H-2q cells gene-engineered to release IFN-γ [26]. This prime-and-boost (pb) method ensured sustained control of the progression of HER-2/neu precancerous lesions. Transcriptome expression profiling of mammary glands from 22-week-old primed and boosted mice (wk22pb) and the whole-mount findings show that vaccination instantly freezes the progression of preneoplastic lesions [26]. In Lollini’s study, 6-week-old mice received a 2-weeks course of four twice-weekly vaccinations with H-2q cells expressing the p185neu and engineered to secrete IL-12. After 2 weeks of rest, this course was repeated until mice were killed or were 1 year old. This vaccination was called “Triplex” from its three immunological stimuli (p185neu, allogeneic MHC and IL-12). Its effects were analyzed at weeks 15 and 26. Two distinct p185neu-positive H-2q cell lines were transduced with IL-12 and used for vaccination (vaxA and vaxB). These two genetically engineered vaccines produced different amounts of IL-12: 50±12 ng/ml/106 cells/72 h for vaxA and 285±87 for vaxB. Both yielded the same results and protected up to 90% of the mice up to one year of age [8]. The whole-mount analysis and the transcriptome expression profiling of glands from vaccinated mice resemble those of a wk6nt mouse (unpublished observations). Integration of the PCA results and vaccination data (Fig. 2b) showed that vaccinations halt tumor growth in a condition very similar to that at the moment of their commencement (week 6 for vaxA and vaxB and week 10 for prime and boost), as illustrated by the transcription profiles, though some differences are evident. To identify the genes changed by vaccination, we compared mice vaccinated at different time points with control untreated mice at 10 weeks of age for primed and boosted (pb) and 6 weeks of age for Triplex (vaxA and vaxB) mice. These comparisons (wk22pb vs. wk10nt, wk15vaxA/B vs. wk6nt and wk26vaxA/B vs. wk6nt) were made with the significance analysis of microarrays (SAM) software [35]. Only genes that changed at least twofold were selected and a similar error rate was imposed for each comparison. The genes expressed differently in vaccinated and nt mice at the commencement of vaccination were: 42, 100 and 94 in wk22pb, wk15vaxA/B and wk26vaxA/B, respectively. These three groups together yielded 134 probe sets linked to gene bank annotations (locus link Id, Chromosomal location, gene description, etc.). A binomial test for proportionality disclosed strong enrichment of probe sets located in chromosome 6 (P value <2.2x10−16). Nearly all the probe sets in this chromosome were up-modulated in the vaccinated mice. However, this enrichment was not related to specific amplification of a chromosome 6 locus, but to over-representation of a limited number of locus links related to Ig κ chains (see Table 1) in the MGU74Av2 Affymetrix genechip. Over-represented probes clearly hamper microarray data interpretation, since they naturally tend to form a cluster on their own being the results similar for every probe of the same gene. This leads to a loss of information on the relationship of the over-represented gene to all the other genes, and its biological significance within a gene list may be overestimated. In immunological terms, this is particularly true for Ig chain or T-cell receptor genes on account of the homology between the chains and members of the family, while the long time that has elapsed since their discovery has resulted in many independent annotations of the same sequence. One solution (adopted in our study) is to hive off the cluster of homologous probes. Alternatively one could create a virtual gene by averaging the signals of many probes.
Table 1.
Probe sets found differentially expressed in vaccinated as opposed to untreated mice
| Probe Set IDab | Cluster in Fig. 5 | Chromosomal location | Locus link | Description | Function |
|---|---|---|---|---|---|
| 160353_i_at | c | 1 E4 | 17164 | MAP kinase-activated protein kinase 2 | Signal transduction |
| 102906_at | b | 11 B1.2 | 21822 | T-cell specific GTPase | Signal transduction |
| 98349_at | c | 13 D2.2 | 16195 | Interleukin 6 signal transducer | Signal transduction |
| 104750_at | d | 11 B1.2 | 18356 | Interferon gamma hitinos protein | Signal transduction |
| 160933_at | d | 11 32.0 cM | 16145 | Interferon gamma induced GTPase | Signal transduction |
| 97409_at | d | 11 B1.2 | 15944 | Interferon protein 1 | Signal transduction |
| 102259_at | c | 5 75.0 cM | 22628 | 3-Monooxgenase/tryptophan 5-monooxgenase activation protein | Signal transduction |
| 97227_at | c | 5 82.0 cM | 14673 | Guanine nucleotide binding protein, alpha 12 | Signal transduction |
| 101465_at | d | 1 25.9 cM | 20846 | Signal transducer, activator of transcription 1 | Signal transduction |
| 100368_at | c | 4 E1 | 12371 | Caspase 9 | Apoptosis |
| 101845_s_at | d | 1 53.1 cM | 114564 | Kappa light chain variable region mRNA | Immunoglobulin |
| 100362_f_at | e | – | – | Ig V(H)II gene H8 | Immunoglobulin |
| 100376_f_at | e | – | – | Heavy chain variable region precursor gene | Immunoglobulin |
| 100583_at | e | 12 59.0 cM | 16061 | IgH chain gene | Immunoglobulin |
| 101718_f_at | e | – | 213684 | Similar to Ig light chain variable region | Immunoglobulin |
| 101743_f_at | e | 12 59.0 cM | 16061 | Ig heavy chain (J558 family) | Immunoglobulin |
| 101870_at | e | 12 58.0 cM | 16017 | Ig heavy chain 4 (serum IgG1) | Immunoglobulin |
| 101871_f_at | e | 12 F1 | 382657 | Similar to anti-human CD19 moAb 4G7 Ig gamma1 heavy chain | Immunoglobulin |
| 102372_at | e | 5 E1 | 16069 | Ig joining chain | Immunoglobulin |
| 102823_at | e | 12 F1 | 380794 | Similar to Ig gamma-2b chain membrane | Immunoglobulin |
| 102824_g_at | e | 12 F1 | 380794 | Similar to Ig gamma-2b chain membrane | Immunoglobulin |
| 102843_s_at | e | 12 58.0 cM | 16014 | Ig heavy chain 4 (serum IgG1) | Immunoglobulin |
| 93638_s_at | e | 16 13.0 cM | 16142 | Ig lambda chain, variable 1 | Immunoglobulin |
| 93904_f_at | e | – | – | Ig heavy chain VDJ region gene | Immunoglobulin |
| 93927_f_at | e | 12 59.0 cM | 16061 | Ig heavy chain 4 (serum IgG1) | Immunoglobulin |
| 97008_f_at | e | – | – | Ig variable region heavy chain precursor | Immunoglobulin |
| 97563_f_at | e | 12 | – | Ig heavy chain 1 (serum IgG2a) | Immunoglobulin |
| 97574_f_at | e | – | – | Ig heavy chain variable region precursor | Immunoglobulin |
| 97575_f_at | e | – | – | Ig heavy chain variable region precursor | Immunoglobulin |
| 97576_f_at | e | 12 | – | Ig heavy chain 4 | Immunoglobulin |
| 98765_f_at | e | – | – | Ig heavy chain V region mRNA | Immunoglobulin |
| 99420_at | e | 12 59.0 cM | 16059 | Ig heavy chain (V7183 family) | Immunoglobulin |
| 102904_at | b | 17 18.7 cM | 14968 | Histocompatibility 2, class II antigen E alpha | Immune response |
| 94285_at | d | 17 18.66 cM | 14969 | Histocompatibility 2, class II antigen E beta | Immune response |
| 103202_at | d | 3 H1 | 55932 | Guanylate nucleotide binding protein 3 | Immune response |
| 104597_at | d | 3 67.4 cM | 14469 | Guanylate nucleotide binding protein 2 | Immune response |
| 95974_at | d | 3 67.4 cM | 14468 | Guanylate nucleotide binding protein 1 | Immune response |
| 161968_f_at | b | 9 | – | Chemokine (C-C motif) receptor 5 | Inflammatory response |
| 92459_at | b | 11 47.0 cM | 20307 | Chemokine (C-C motif) ligand 8 | Inflammatory response |
| 101436_at | d | 5 53.0 cM | 17329 | Chemokine (C-X-C motif) ligand 9 | Inflammatory response |
| 102719_f_at | d | 9 72.0 cM | 12774 | Chemokine (C-C motif) receptor 5 | Inflammatory response |
| 101028_i_at | c | 2 64.0 cM | 11464 | Actin, alpha | Structural molecule activity |
| 101080_at | c | 2 18.0 cM | 12831 | Procollagen, type V, alpha 1 | Structural molecule activity |
| 101095_at | c | 4 D3-E1 | 17150 | Microfibrillar-associated protein 2 | Structural molecule activity |
| 102990_at | c | 1 21.1 cM | 12825 | Procollagen, type III, alpha 1 | Structural molecule activity |
| 103709_at | c | 11 56.0 cM | 12842 | Procollagen, type I, alpha 1 | Structural molecule activity |
| 92207_at | c | 5 75.0 cM | 13717 | Elastin | Structural molecule activity |
| 92836_at | c | 5 75.0 cM | 13717 | Elastin | Structural molecule activity |
| 93517_at | c | 10 41.1 cM | 12834 | Procollagen, type VI, alpha 2 | Structural molecule activity |
| 94305_at | c | 11 56.0 cM | 12842 | Procollagen, type I, alpha 1 | Structural molecule activity |
| 99367_at | c | 2 H1 | 13589 | Microtubule-associated protein, RP/EB family, member 1 | Structural molecule activity |
| 92559_at | d | 3 50.8 cM | 22329 | Vascular cell adhesion hitinos 1 | Structural molecule activity |
| 100463_at | a | 5 44.93 cM | 12993 | Casein gamma | Transport |
| 96030_at | a | 5 44.9 cM | 12990 | Casein alpha | Transport |
| 99130_at | a | 5 44.91 cM | 12991 | Casein beta | Transport |
| 102704_at | c | 18 6.0 cM | 11829 | Aquaporin 4 | Transport |
| 102708_at | a | 13 12.0 cM | 12231 | Butyrophilin, subfamily 1, member A1 | Transport |
| 160949_at | c | 14 B | 26430 | Poly (ADP-ribose) glycohydrolase | DNA damage responses |
| 92833_at | b | 10 51.0 cM | 15109 | Histidine ammonia lyase | Biosynthesis |
| 160617_at | c | 7 B5 | 50794 | Kruppel-like factor 13 | Transcription |
| 103926_at | c | 16 A1 | 208643 | Ribosomal protein L24 | Translation |
| 101139_r_at | c | 5 E1 | 17830 | Mucin 10 | Biological-process unknown |
| 102860_at | d | 12 E | 20715 | Serine (or cysteine) proteinase inhibitor, clade A, member 3G | Biological-process unknown |
| 93085_at | d | 17 18.59 cM | 16912 | Proteosome subunit, beta type 9 (large multifunctional protease 2) | Biological-process unknown |
| 102931_at | a | 11 0.5 cM | 22373 | Whey acidic protein (WAP) | Biological-process unknown |
| 93028_at | c | 7 69.03 cM | 14955 | H19 fetal liver mRNA | Biological-process unknown |
| 103033_at | b | 17 18.83 cM | 20567 | Sex-limited protein | Biological-process unknown |
| 99126_at | c | X | – | Inactive X specific transcripts | Biological-process unknown |
| 99850_at | b | 15 | – | RIKEN cDNA 4432409D24 gene | Biological-process unknown |
| 101518_at | c | 11 B5 | 55978 | RIKEN cDNA 0610009H04 gene | Biological-process unknown |
| 103905_at | c | 9 C | 76459 | RIKEN cDNA 2310047E01 gene | Biological-process unknown |
| 100944_at | b | 19 | – | Expressed sequenze AW112010 | Biological-process unknown |
| 103963_f_at | d | 18 E1 | 60440 | Expressed sequenze AW111922 | Biological-process unknown |
| 96764_at | d | 18 E1 | 60440 | Expressed sequence AW111922 | Biological-process unknown |
| 98410_at | d | 11 B1.3 | 54396 | Expressed sequenze AI481100 | Biological-process unknown |
| 102721_at | e | 12 F1 | 380795 | EST | Biological-process unknown |
| 102722_g_at | e | 12 F1 | 380795 | EST | Biological-process unknown |
| 160173_at | c | – | – | EST (acc: AI852838) | Biological-process unknown |
| 100880_at | d | – | – | EST (acc: AA816121) | Biological-process unknown |
aExpression data related to the present probe sets can be requested to the authors
b34/37 probe sets found over-represented in chromosome 6 were linked to Ig k light chain genes: 21 probe sets were linked to variable region (V) 28, 3 to V8, 1 to V1, 1 to V20, 1 to V21, and the others were simply annotated as Ig k genes. Probe sets are grouped on the basis of their putative function. Functional groups were inferred by probesets Gene Ontology (http://www.geneontology.org) annotation or by GeneRIF (http://www.ncbi.nih.gov/entrez/query.fcgi?db=gene) annotation
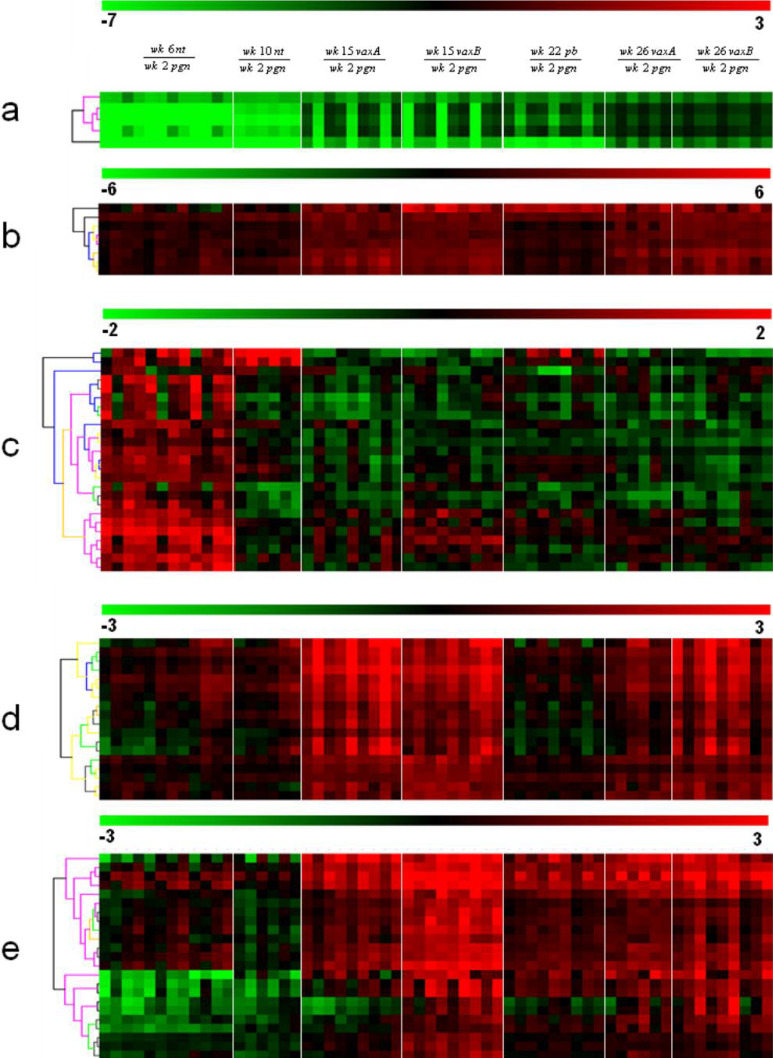
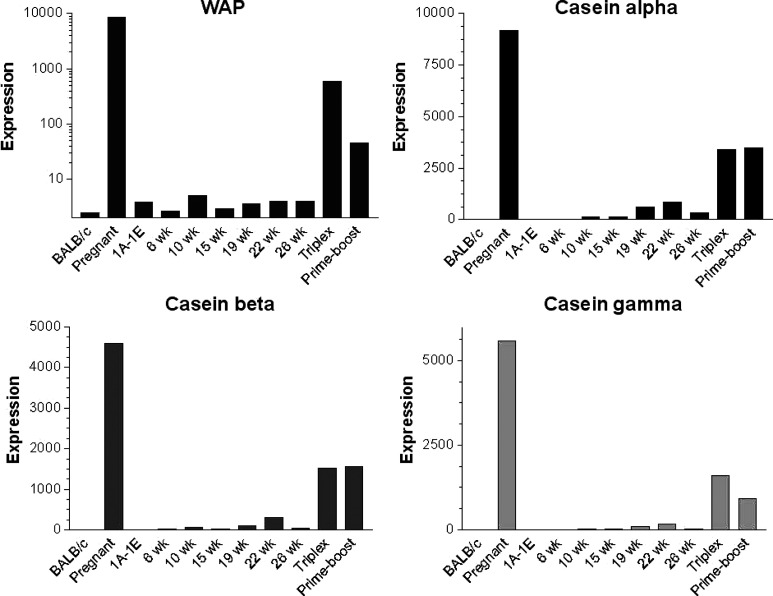
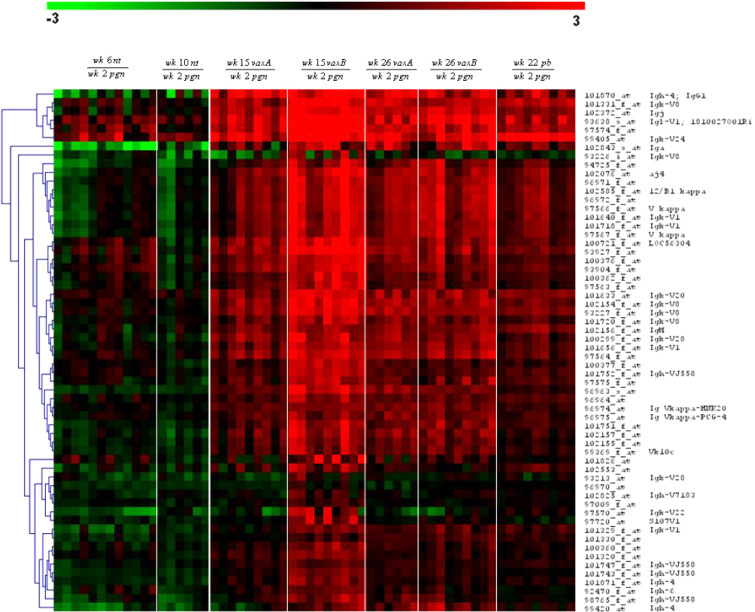
Hierarchical clustering of the remaining 97 probe sets (Table 1) was undertaken to group the genes and the samples into clusters with a similar expression profile. This yielded five gene clusters characterizing the components of the experimental groups (Fig. 5). The differential expression levels in wk15vaxA/B and wk26vaxA/B are much higher than those in wk22pb (Fig. 5, clusters b, d). There may thus be a direct correlation between the expression levels of the genes identified by the probe sets in these clusters and the efficacy of vaccination. The procedure of Lollini et al. kept all mice tumor-free until 1 year of age [8]. pb vaccination was less effective [26]. It delayed the development of palpable tumors and kept 48% of mice tumor-free until week 32 [26], while the follow-up showed that it significantly delayed the first tumor appearance and more than doubled disease-free survival, though all mice developed one or more tumors by week 52 (Cavallo, unpublished observation). It may be supposed that the two procedures halt tumor progression via different pathways, though in the procedure of Lollini et al. vaccinations were repeated, whereas pb vaccination was performed only once [26]. As in other tumor systems, halting of tumor progression in pb mice may coincide with a definitive cure. However, the activated rat HER-2/neu oncogene is embedded in the genome of BALB-neuT mice and continuous neoplastic stem cell production is likely. This makes the tumor-halting condition a dynamic result between p185neu cell onset and their immunological clearance. Many interfering factors may tip the scales and restart tumor growth as the immune response fades with age, whereas chronic vaccination may maintain a lifelong high response. Repeated DNA vaccination with the plasmids coding for the transmembrane and extracellular domains of p185neu keep all BALB-neuT mice free from tumors up to 52 weeks [25]. This is an indication that the differences between the two studies are mainly related to the fact that at week 22 some of the mRNA of genes transcribed during the early phase of the immune response have already decayed, whereas they are constantly regenerated by repeated vaccination. The five clusters are illustrated in Fig. 5. Cluster “a” groups genes strongly down-regulated in the mammary glands of wk6nt and wk10nt mice. It is characterized by the presence of three isoforms of casein (α, β, γ) and whey acidic protein (WAP), which are highly induced in mammary glands from pregnant and vaccinated mice. Induction of genes related to late pregnancy and lactation in the glands of vaccinated virgin mice is an unexpected finding that emerged from both sets of data. Raw expression levels are shown in more detail in Fig. 6, which also includes results from additional microarray experiments with non-transgenic mammary glands of BALB/c mice and with in vitro cultured N202.1A and N202.1E cell lines [22]. The results clearly show that the induction of genes related to late pregnancy and lactation was caused by vaccination and unrelated to tumor growth. The WAP was only expressed in the glands of pregnant mice and those in the two vaccination groups. Expression of casein genes was detectable in late progression samples (19–26 weeks of age), but the levels induced by vaccination in tumor-free glands were one order of magnitude higher than those found in tumors. The glands of vaccinated mice expressed a signature normally induced by pregnancy and lactation, rather than by carcinoma growth and progression. The strong IFN-γ response induced by vaccination may well be involved in the induction of this group of genes. In late pregnancy and lactation, WAP and caseins are induced by prolactin acting via Stat5 [15]. The ubiquitous effects of IFN-γ are mediated by Stat1 activation, but in the breast and a few other tissues IFN-γ also activates Stat5 [34]. Induction of WAP and casein γ was higher in Triplex than in the pb mice, in parallel with the higher induction of all other IFN-γ-sensitive genes. Prolactin-like activity triggered by IFN-γ in the mammary gland is a possibility that deserves further investigation to determine the mechanism involved and its potential biological consequences.
Fig. 5.
Hierarchical clustering of probe sets differentially expressed in vaccinated as opposed to nt mice. Probe sets were converted in virtual two-dye experiments by comparing all replicates of each group with the wk2pgr replicates. Virtual two-dye experiments of wk6nt, wk10nt, wk15vax, wk22pb and wk26vax, grouped by hierachical clustering, yielded five gene clusters characterizing the experimental groups. a Cluster strongly down-regulated in wk6nt and wk10nt mice, and encompassing three casein isoforms and WAP. b Cluster mainly up-modulated in Triplex-vaccinated mice and encompassing two chemokine genes, two MHC II antigens and one T-cell-specific GTPase. c Cluster with many “stromal” genes (protocollagens, actins and elastins) mainly expressed in wk6nt mice. d Cluster containing IFN-induced genes, up-modulated almost solely in triplex-vaccinated mice. e Cluster containing only Ig chain genes.
Fig. 6.
Level of expression measured with microarrays of WAP and alpha, beta and gamma casein in mammary glands from non-transgenic BALB/c mice, pregnant mice, cell lines derived from tumors in HER-2/neu transgenic mice (1A-1E), mammary glands from HER-2/neu transgenic nt mice at different time points, and from vaccinated mice (Triplex, Prime-boost). Both vaccinations increase the level of expression of these genes, which are normally related to pregnancy and lactation.
Cluster “b” contains genes prevalently up-modulated in vaccinated mice, particularly in wk15vaxA/vaxB and wk26vaxA/vaxB compared to wk22pb. Chemokine receptor 5 (CCR5), chemokine ligand 8 (CCL8) and a T-cell-specific GTPase (TGTP) are enclosed in this cluster, together with MHC II antigens. Both CCR5 and CCL8 are involved in inflammatory responses. Expression of CCR5 has been detected in a promyeloblastic cell line, suggesting that it plays a role in granulocyte lineage proliferation and differentiation [30], while CCL8 recruits leukocytes to sites of inflammation and contributes to tumor-associated leukocyte infiltration [37].
Cluster “c” mainly contains “stromal” genes such as elastin, collagen and IL-6. These genes are down-modulated during tumor progression, as the epithelial component overcomes all other cell types in the mammary tissue. Their low expression in all the vaccinated samples suggests remodeling of the mammary stroma due to age or perhaps due to the immune response induced by vaccination. However, this remodeling has not yet been established morphologically and further investigation of this discrepancy is needed.
Of the other genes in cluster “c”, caspase-9 is up-modulated in vaccinated mice. It is processed by caspase APAF1 [36] as one of the earliest steps in the caspase activation cascade. Mapkapk2 kinase [33], also present in this cluster, is involved in many cellular processes, including stress and inflammatory responses, nuclear export [18], gene expression regulation and cell proliferation [38]. Furthermore, the RAC1–MKK3–P38–Mapkapk2 pathway promotes urokinase plasminogen activator mRNA stability in invasive breast cancer cells [12].
Cluster “d” contains IFN-induced genes (Stat1, Ifi1, Ifi47, Igtp, Gbp1-3, CXCL9, PSMB9) that are only up-modulated strongly in vaxA and vaxB. Igtp is a Stat1-dependent gene [7]. Its role in cell survival relies on activation of PI3-K/Akt, inactivation of GSK-3 and suppression of caspase-9 and caspase-3 [39]. The genes in this cluster are early-response genes that are probably already decayed in the pb experiments. The serum IFN-γ burst occurs between day 1 and day 3 after vaccination with IL-12-transduced cells, and plasma levels return to the baseline by the 4th day [8]. The IFN-γ-mediated response may be short-lived also in the mammary epithelium, particularly since these IFN-γ-induced genes are much more up-regulated in wk15vaxA/B than in wk26vaxA/B mice. The 15th week, in fact, corresponds to the 2nd week of injection in the third course, whereas at 26 weeks mice are at the end of a 2-week pause before the sixth course. So we can conclude that chronic vaccination is necessary to maintain an active IFN-γ-mediated response in the mammary gland, which is probably necessary (but not sufficient) for long-term protection from tumor onset, since only chronic boosting of this response can prevent tumor development for a long time [8, 21, 25]. Cluster “d” also comprises VCAM1, which is a member of the Ig superfamily and encodes a cell surface sialoglycoprotein expressed by cytokine-activated endothelium [20].
Cluster “e” contains only Ig genes heavily up-regulated in vaccinated as opposed to untreated mice. To see whether antibody-producing B cells are mainly located in the lymph node, or are also present in the mammary tissue, we analyzed the differences in gene expression between whole and lymph-node-deprived mammary glands. Close study of the Ig-coding genes showed that expression of the vast majority of Ig increased in vaccinated compared with nt mice, though this increase was almost completely abolished when glands were collected without the lymph node (data not shown).
Figure 7 illustrates the Ig chain genes induced by Triplex vaccination. pb vaccination increases Ig expression, but its induction of Ig genes is lower than that of Triplex vaccination because the lymph node was not analyzed.
Fig. 7.
Virtual two-dye experiments showing Ig chain genes in wk6nt, wk10nt, wk15vax, wk26vax and wk22pb with respect to wk2pgn grouped by hierarchical clustering (Euclidean distance, average linkage). All the vaccinated mice show up-regulation of immunoglobulin genes with respect to nt mice. Triplex-vaccinated mice (wk15vax, wk26vax) whose mammary glands were collected with the lymph node display a higher Ig gene induction than wk22pb mice whose lymph node was not analyzed. This demonstrates that the B-cell response resides mainly in the lymph node rather than in the surrounding mammary tissue.
This suggests that the B-cell-specific response resides almost entirely in the lymph node. Some probes recognizing antibodies were not expressed or were decreased in our samples. They can be divided into three groups: (a) antibody chains known to be expressed in the bone marrow by pre-B cells (e.g. λ5 chain), but not in the periphery. Their absence in our gland samples was expected. (b) Probes related to primary antibody responses, e.g. IgM and IgD chains. A decrease of these antibodies may be expected in hyperimmune mice. (c) Probes that should in principle be highly expressed in our vaccinated mice (Igh-4, Igh-VJ558, Igl-V1, Igl-5). It may be supposed that their small number reflects the presence of “dead” probes or wrong DNA sequences, since some Affymetrix probe sets do not match the corresponding reference sequence (RefSeq) [17], (the curated, non-redundant sequence for every transcript). A small number of wrong probes may be expected in early chip releases. Studies of the kind described here can help in pinpointing these probes and excluding them from further releases.
Conclusions
If the experimental design and the biological part of the microarray analysis are carried out accurately, the results produced (using the same animal model and technological platform) by laboratories with slightly different designs, but addressing the same biological question, are strikingly comparable. This general observation shows that microarray analysis is now mature and ensures a good level of inter-laboratory reproducibility.
A modern retelling of the fable of the blind men and the elephant [2] ends with the blind men discovering that the elephant is also a means of transport that can carry them effortlessly. In a similar vein, we can conclude that microarrays per se cannot restore the sight of blind cancer immunologists, but the combination of conventional immunological and pathological studies with gene expression profiles significantly furthers our understanding of tumor immunology and leads to better appreciation of the complex network of immune and non-immune interactions triggered by cancer vaccines. A scientist working with microarrays is deprived of the intelligent eye of the experienced pathologist. If he can manage to identify elephants with some accuracy, what is the advantage of his blindness? One wonders, indeed, whether microarrays can really yield any worthwhile new and valuable information. Do the results they provide primarily corroborate what can be found by integrated pathological and immunological analysis? We believe that the prime advantage of the microarray technology is that it probes a large array of genes, most of which are absolutely meaningless for the experimenter. Moreover, the substantial results it provides, in addition to confirming those offered by more conventional techniques, will in most cases be totally unexpected or difficult to predict. In our study, for example, casein could have been detected histochemically, but who would have thought of looking for it? Everything can be seen histochemically, except that which is not being sought, whereas this is rendered evident by microarrays. Wisdom will stem from the ability to determine whether the picture is that of a gnat viewed from a millimetre away or an elephant viewed from a mile.
Acknowledgements
This work was supported by the Italian Association for Cancer Research (AIRC), the Italian Foundation for Cancer Research (FIRC), the Italian Ministry for Education, University and Research, and the University of Bologna. Annalisa Astolfi is the recipient of FIRC fellowship.
References
- 1.Alroy I, Yardeen Y. The ErbB signaling network in embryogenesis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 1997;410:83–86. doi: 10.1016/S0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 2.Backstein K. The blind men and the elephant. New York: Scholastic; 1992. [Google Scholar]
- 3.Boggio K, Nicoletti G, Di Carlo E, Cavallo F, Landuzzi L, Melani C, Giovarelli M, Rossi I, Nanni P, De Giovanni C, Bouchard P, Wolf S, Modesti A, Musiani P, Lollini PL, Colombo MP, Forni G. Interleukin-12 mediated prevention of spontaneous mammary adenocarcinomas in two lines of Her-2/neu transgenic mice. J Exp Med. 1998;188:589. doi: 10.1084/jem.188.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calogero RA, Musiani P, Forni G, Cavallo F. Toward a long-lasting immune prevention of HER2 mammary carcinomas: directions from transgenic mice. Cell Cycle. 2004;3:704–706. [PubMed] [Google Scholar]
- 5.Cappello P, Triebel F, Iezzi M, Caorsi C, Quaglino E, Nanni P, Amici A, Di Carlo E, Musiani P, Giovarelli M, Forni G. LAG-3 enables DNA vaccination to persistently prevent mammary carcinogenesis in HER-2/neu transgenic BALB/c mice. Cancer Res. 2003;63:2518–2525. [PubMed] [Google Scholar]
- 6.Clarkson RW, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6:R92–R109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collazo CM, Yap GS, Hieny S, Caspar P, Feng CG, Taylor GA, Sher A. The function of gamma interferon-inducible GTP-binding protein IGTP in host resistance to Toxoplasma gondii is Stat1 dependent and requires expression in both hematopoietic and nonhematopoietic cellular compartments. Infect Immun. 2002;70:6933–6939. doi: 10.1128/IAI.70.12.6933-6939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Giovanni C, Nicoletti G, Landuzzi L, Astolfi A, Croci S, Comes A, Ferrini S, Meazza R, Iezzi M, Di Carlo E, Musiani P, Cavallo F, Nanni P, Lollini PL. Immunoprevention of HER-2/neu transgenic mammary carcinoma through an interleukin 12-engineered allogeneic cell vaccine. Cancer Res. 2004;64:4001–4009. doi: 10.1158/0008-5472.CAN-03-2984. [DOI] [PubMed] [Google Scholar]
- 9.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, Simon R, Liu ET, Green JE. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci U S A. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn OJ. Cancer vaccines: between the idea and the reality. Nat Rev Immunol. 2003;3:630–641. doi: 10.1038/nri1150. [DOI] [PubMed] [Google Scholar]
- 11.Finn OJ, Forni G. Prophylactic cancer vaccines. Curr Opin Immunol. 2002;14:172–177. doi: 10.1016/s0952-7915(02)00317-5. [DOI] [PubMed] [Google Scholar]
- 12.Han Q, Leng J, Bian D, Mahanivong C, Carpenter KA, Pan ZK, Han J, Huang S. Rac1-MKK3-p38-MAPKAPK2 pathway promotes urokinase plasminogen activator mRNA stability in invasive breast cancer cells. J Biol Chem. 2002;277:48379–48385. doi: 10.1074/jbc.M209542200. [DOI] [PubMed] [Google Scholar]
- 13.Hynes NE, Stern DF. The biology of erbB-2/neu/HER-2 and its role in cancer. Biochim Biophys Acta. 1994;1198:165–184. doi: 10.1016/0304-419X(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 14.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol. 2002;27:125–132. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Rosen JM. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol Cell Biol. 1995;15:2063–2070. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, Tan LK, Rosen JM, Varmus HE. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mecham BH, Wetmore DZ, Szallasi Z, Sadovsky Y, Kohane I, Mariani TJ. Increased measurement accuracy for sequence-verified microarray probes. Physiol Genomics. 2004;18:308–315. doi: 10.1152/physiolgenomics.00066.2004. [DOI] [PubMed] [Google Scholar]
- 18.Meng W, Swenson LL, Fitzgibbon MJ, Hayakawa K, Ter Haar E, Behrens AE, Fulghum JR, Lippke JA. Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J Biol Chem. 2002;277:37401–37405. doi: 10.1074/jbc.C200418200. [DOI] [PubMed] [Google Scholar]
- 19.Muller WJ, Sinn E, Wallance R, Pattengale PK, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 20.Nakao S, Kuwano T, Ishibashi T, Kuwano M, Ono M. Synergistic effect of TNF-alpha in soluble VCAM-1-induced angiogenesis through alpha 4 integrins. J Immunol. 2003;170:5704–5711. doi: 10.4049/jimmunol.170.11.5704. [DOI] [PubMed] [Google Scholar]
- 21.Nanni P, Nicoletti G, De Giovanni C, Landuzzi L, Di Carlo E, Cavallo F, Pupa SM, Rossi I, Colombo MP, Ricci C, Astolfi A, Musiani P, Forni G, Lollini P-L. Combined allogeneic tumor cell vaccination and systemic IL-12 prevents mammary carcinogenesis in HER-2/neu transgenic mice. J Exp Med. 2001;194:1195–1205. doi: 10.1084/jem.194.9.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanni P, Pupa SM, Nicoletti G, De Giovanni C, Landuzzi L, Rossi I, Astolfi A, Ricci C, De De Vecchi R, Invernizzi AM, Di Carlo E, Musiani P, Forni G, Menard S, Lollini PL. p185(neu) protein is required for tumor and anchorage-independent growth, not for cell proliferation of transgenic mammary carcinoma. Int J Cancer. 2000;87:186–194. doi: 10.1002/1097-0215(20000715)87:2<186::AID-IJC5>3.3.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 23.Ostrand-Rosenberg S. Animal models of tumor immunity, immunotherapy and cancer vaccines. Curr Opin Immunol. 2004;14:172–177. doi: 10.1016/j.coi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Parmigiani G, Garrett-Mayer ES, Anbazhagan R, Gabrielson E. A cross-study comparison of gene expression studies for the molecular classification of lung cancer. Clin Cancer Res. 2004;10:2922–2927. doi: 10.1158/1078-0432.ccr-03-0490. [DOI] [PubMed] [Google Scholar]
- 25.Quaglino E, Iezzi M, Mastini C, Amici A, Pericle F, Di Carlo E, Pupa SM, De Giovanni C, Spadaro M, Curcio C, Lollini PL, Musiani P, Forni G, Cavallo F. Electroporated DNA vaccine clears away multifocal mammary carcinomas in Her-2/neu transgenic mice. Cancer Res. 2004;64:2858–2864. doi: 10.1158/0008-5472.can-03-2962. [DOI] [PubMed] [Google Scholar]
- 26.Quaglino E, Rolla S, Iezzi M, Spadaro M, Musiani P, De Giovanni C, Lollini PL, Lanzardo S, Forni G, Sanges R, Crispi S, De Luca P, Calogero R, Cavallo F. Concordant morphologic and gene expression data show that a vaccine halts HER-2/neu preneoplastic lesions. J Clin Invest. 2004;113:709–717. doi: 10.1172/JCI200419850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rovero S, Amici A, Carlo ED, Bei R, Nanni P, Quaglino E, Porcedda P, Boggio K, Smorlesi A, Lollini PL, Landuzzi L, Colombo MP, Giovarelli M, Musiani P, Forni G. DNA vaccination against rat her-2/Neu p185 more effectively inhibits carcinogenesis than transplantable carcinomas in transgenic BALB/c mice. J Immunol. 2000;165:5133–5142. doi: 10.4049/jimmunol.165.9.5133. [DOI] [PubMed] [Google Scholar]
- 28.Rovero S, Boggio K, Di Carlo E, Amici A, Quaglino E, Porcedda P, Musiani P, Forni G. Insertion of the DNA for the 163–171 peptide of IL1β enables a DNA vaccine encoding p185 (neu) to inhibit mammary carcinogenesis in Her-2/neu transgenic BALB/c mice. Gene Ther. 2001;8:447–452. doi: 10.1038/sj.gt.3301416. [DOI] [PubMed] [Google Scholar]
- 29.Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia. 2003;8:287–307. doi: 10.1023/B:JOMG.0000010030.73983.57. [DOI] [PubMed] [Google Scholar]
- 30.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 31.Smalley M, Ashworth A. Stem cells and breast cancer: a field in transit. Nat Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- 32.Spadaro M, Lanzardo S, Curcio C, Forni G, Cavallo F. Immunological inhibition of carcinogenesis. Cancer Immunol Immunother. 2004;53:204–216. doi: 10.1007/s00262-003-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokoe D, Caudwell B, Cohen PTW, Cohen P. The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem J. 1993;296:843–849. doi: 10.1042/bj2960843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tourkine N, Schindler C, Larose M, Houdebine LM. Activation of STAT factors by prolactin, interferon-gamma, growth hormones, and a tyrosine phosphatase inhibitor in rabbit primary mammary epithelial cells. J Biol Chem. 1995;270:20952–20961. doi: 10.1074/jbc.270.36.20952. [DOI] [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Twiddy D, Brown DG, Adrain C, Jukes R, Martin SJ, Cohen GM, MacFarlane M, Cain K. Pro-apoptotic proteins released from the mitochondria regulate the protein composition and caspase-processing activity of the native Apaf-1/caspase-9 apoptosome complex. J Biol Chem. 2004;279:19665–19682. doi: 10.1074/jbc.M311388200. [DOI] [PubMed] [Google Scholar]
- 37.Van Coillie E, Fiten P, Nomiyama H, Sakaki Y, Miura R, Yoshie O, Van Damme J, Opdenakker G. The human MCP-2 gene (SCYA8): cloning, sequence analysis, tissue expression, and assignment to the CC chemokine gene contig on chromosome 17q11.2. Genomics. 1997;40:323–331. doi: 10.1006/geno.1996.4594. [DOI] [PubMed] [Google Scholar]
- 38.Yannoni YM, Gaestel M, Lin LL. P66(ShcA) interacts with MAPKAP kinase 2 and regulates its activity. FEBS Lett. 2004;564:205–211. doi: 10.1016/S0014-5793(04)00351-5. [DOI] [PubMed] [Google Scholar]
- 39.Zhang HM, Yuan J, Cheung P, Luo H, Yanagawa B, Chau D, Stephan-Tozy N, Wong BW, Zhang J, Wilson JE, McManus BM, Yang D. Overexpression of interferon-gamma-inducible GTPase inhibits coxsackievirus B3-induced apoptosis through the activation of the phosphatidylinositol 3-kinase/Akt pathway and inhibition of viral replication. J Biol Chem. 2003;278:33011–33019. doi: 10.1074/jbc.M305352200. [DOI] [PubMed] [Google Scholar]