Abstract
Background
Metabolic dysfunction-associated steatotic liver disease (MASLD) is a common complication of obesity and, in severe cases, progresses to metabolic dysfunction-associated steatohepatitis (MASH). Small heterodimer partner (SHP) is an orphan member of the nuclear receptor superfamily and regulates metabolism and inflammation in the liver via a variety of pathways. In this study, we investigate the molecular foundation of MASH progression in mice with hepatic SHP deletion and explore possible therapeutic means to reduce MASH.
Methods
Hepatic SHP knockout mice (SHPΔhep) and their wild-type littermates (SHPfl/fl) of both sexes were fed a fructose diet for 14 weeks and subjected to an oral glucose tolerance test. Then, plasma lipids were determined, and liver lipid metabolism and inflammation pathways were analyzed with immunoblotting, RNAseq, and qPCR assays. To explore possible therapeutic intersections of SHP and inflammatory pathways, SHPΔhep mice were reconstituted with bone marrow lacking interferon γ (IFNγ−/−) to suppress inflammation.
Results
Hepatic deletion of SHP in mice fed a fructose diet decreased liver fat and increased proteins for fatty acid oxidation and liver lipid uptake, including UCP1, CPT1α, ACDAM, and SRBI. Despite lower liver fat, hepatic SHP deletion increased liver inflammatory F4/80+ cells and mRNA levels of inflammatory cytokines (IL-12, IL-6, Ccl2, and IFNγ) in both sexes and elevated endoplasmic reticulum stress markers of Cox2 and CHOP in female mice. Liver bulk RNAseq data showed upregulation of genes whose protein products regulate lipid transport, fatty acid oxidation, and inflammation in SHPΔhep mice. The increased inflammation and fibrosis in SHPΔhep mice were corrected with bone marrow-derived IFNγ−/− myeloid cell transplantation.
Conclusion
Hepatic deletion of SHP improves fatty liver but worsens hepatic inflammation possibly by driving excess fatty acid oxidation, which is corrected by deletion of IFNγ specifically in myeloid cells. This suggests that hepatic SHP limits fatty acid oxidation during fructose diet feeding but, in doing so, prevents pro-MASH pathways. The IFNγ-mediated inflammation in myeloid cells appears to be a potential therapeutic target to suppress MASH.
Keywords: Fatty liver (MASLD/MASH), Inflammation and fibrosis, Fructose diet, Bulk RNAseq, Fatty acid oxidation, Interferon gamma
Highlights
-
•
Hepatic SHP is required to limit lipotoxicity under nutrient excess conditions.
-
•
Hepatic SHP is required to suppress mRNA levels of genes whose products may promote liver inflammation.
-
•
Blocking IFNγ in myeloid cells suppresses inflammation and fibrosis in MASH liver.
-
•
Sex differences in MASH transition with a fructose diet.
1. Introduction
Metabolic dysfunction-associated steatotic liver disease (MASLD) is highly prevalent worldwide and impacts about 25% of American adults [1,2]. MASLD is closely associated with the features of metabolic syndrome including insulin resistance and hyperlipidemia under the condition of nutrient excess. Nutrition excess increases lipogenesis and causes lipotoxicity through multiple metabolic pathways. First, increased delivery of free fatty acids (FFAs) from adipose to the liver may promote the expression of inflammatory genes by acting as ligand activators for transcription factors or by modifying membrane lipid draft functions [3]. In response to the elevated FFA flux to the liver, both hepatic fatty acid oxidation and triacylglycerol (TG) biosynthesis can be adaptively increased [1,2]. Second, fatty acid oxidation overload may lead to the accumulation of reactive oxidative species, which causes hepatocellular injury and inflammation [1,2]. Third, the liver content of diacylglycerol (DAG), an intermediate product for TG synthesis, may rise and promote chronic inflammation and insulin resistance [4]. Sustained inflammation drives the transition of MASLD to metabolic dysfunction-associated steatohepatitis (MASH), which is characterized by inflammation and hepatocellular injury with or without fibrosis [1]. A key aspect in the transition of MASLD to MASH is increased inflammation [2]. About 10% of MASH cases in clinics are not associated with excess liver fat accumulation, indicating the significance of inflammation in the transition of MASH [1]. About one-third of MASH cases develop into more severe pathology situations of cirrhosis, cancer, and liver failure [1], contributing to the high mortality risk associated with MASH [2,5]. Unfortunately, there are not any FDA-approved drugs to treat this severe stage of chronic liver disease.
Small heterodimer partner (SHP, Nr0b2 gene) is a nuclear receptor and acts as a transcriptional repressor to regulate a diverse range of biological processes [6]. SHP regulates sex hormonal signaling pathways by interacting with androgen receptors (AR) and estrogen receptors [7,8]. We previously demonstrated that liver SHP contributes to sex differences in blood TG levels in an AR-dependent manner [9]. SHP suppresses virus-mediated infection by repressing the transcription of type I interferon (IFN) [10]. In the liver, SHP actively regulates the expression of cytochrome P450 (CYP) genes, which are important regulators in liver inflammation and lipid metabolism via monooxygenase and epoxygenase pathways [[11], [12], [13]]. SHP inhibits bile acid biosynthesis by suppressing Cyp7a1 and/or Cyp8b1 to maintain liver cholesterol and bile acid homeostasis [12,13]. SHP suppresses liver glucose production by inhibiting the gene expression of glucose-6-phosphatase via interaction with hepatocyte nuclear factors (HNFs) [14,15]. In hyperlipidemic mice, global deletion of SHP reduces the expression of inflammatory genes in the liver with inconsistent impact on liver lipid content and hyperlipidemia [[16], [17], [18]]. In mice lacking hepatic bile acid sensor farnesoid X receptor (FXR), the mRNA level of Nr0b2 inversely correlates with the expression of inflammatory genes [19]. While overexpression of hepatic SHP may lead to liver steatosis in mice [11], hepatic deletion of SHP decreases liver lipid content but increases liver inflammation [20]. The underlying mechanisms for SHP-mediated metabolic pathways and their impact on hepatic inflammation in nutrient-excess conditions remain unclear.
In this study, we investigate the metabolic and inflammatory pathways in mice with hepatic SHP deletion after a fructose diet feeding. To further study the role of inflammation in MASH transition, inflammation pathways were suppressed in SHPΔhep mice by transplanting interferon γ deficient (IFNγ−/−) bone marrow following lethal irradiation. We show that overlaying the deletion of IFNγ in myeloid cells suppressed liver inflammation and fibrogenesis.
2. Materials and methods
2.1. Animals
All mouse experiments were approved under the Vanderbilt University Institutional Animal Care and Use Committee. Mice were housed in 12-hour light/dark cycles in temperature and humidity-controlled facilities with ad-libitum access to a chow diet and water. Mice with homozygous floxed Nr0b2 allele were crossed with mice with Cre recombinase under the control of an albumin promoter to generate hepatocyte-specific knockout of SHP (SHPΔhep), which we have described previously [21]. Floxed littermates lacking Cre were wild-type controls (SHPfl/fl) in all experiments. All strains were backcrossed at least 10 generations onto the C57BL/6 background. Genotyping was done from ear punch DNA as we described before [22]. Mice were fed 20% fructose water with a chow diet at 10 ± 1-week old for 14 weeks.
2.2. Lipid and enzyme activity analysis
Plasma triacylglycerol (TG) and cholesterol levels were measured using colorimetric kits (Infinity) as we reported previously [21]. Lipoproteins were separated by fast-performance liquid chromatography (FPLC, Superose 6 column, GE Healthcare) from pooled plasma samples (100 μl total pooled plasma used).
Liver TG and cholesterol contents were determined by the Vanderbilt Hormone Assay Core as we described before [23]. Briefly, 50–100 mg liver was Folch-extracted and separated by thin layer chromatography, which was then analyzed by gas chromatography with internal standards used to control extraction efficiency.
Blood aspartate aminotransferase (AST) and alanine transaminase (ALT) activities were measured using the colorimetric activity assay kits from Cayman following the manufactory protocol (Cat# 701640 and 700260).
2.3. Oral glucose tolerance test (OGTT)
Mice were fasted at 7 AM and an OGTT was performed after 5 h of fasting. Mice were gavaged with 20% w/v dextrose (final 2 g/kg body weight), and tail-vein blood glucose was measured via a glucometer (Accu-Chek, Accu-Chek Aviva Plus Meter) at 10 min before gavage (−10 min) and 5, 10, 15, 30, 45, 60, 90, and 120 min after gavage. Plasma insulin was measured using the Crystal Chem Ultrasensitive Mouse Insulin ELISA Kit (catalog # 90080).
2.4. Overlay the IFNγ−/− myeloid cells in SHPΔhep mice
A second cohort of mice were used for targeting IFNγ-mediated inflammation in immune cells in SHPΔhep mice with a fructose diet. When starting the fructose diet, 10-week-old SHPΔhep mice were reconstituted with bone marrow from IFNγ null mice following lethal irradiation (900 rad). SHPΔhep mice received bone marrow from SHPΔhep mice were used as the controls for bone marrow transplant procedure; SHPfl/fl mice received bone marrow from SHPfl/fl mice were also included as isogenic controls (Figure 2A). All the mice were fed 20% fructose water with chow diet for 14 weeks followed by OGTT; mice were sacrificed after 5 h fasting and tissues and blood were collected for investigation of metabolic changes (Figure 2A).
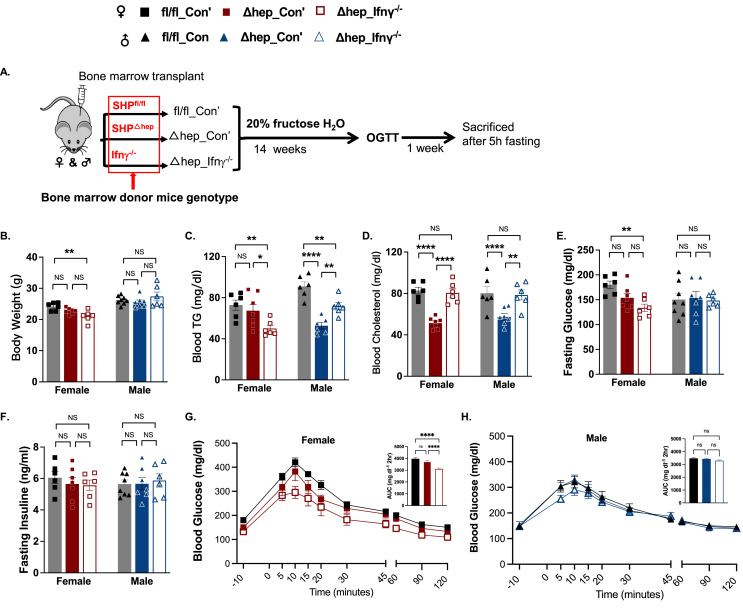
Figure 2.
Overlaying IFNγ−/−myeloid cells reversed blood cholesterol in SHPΔhepmice. A: Schematic of the study design for overlaying bone marrow-derived IFNγ−/− myeloid cells in mice with hepatic SHP deletion. After lethal irradiation, SHPΔhep mice were reconstituted with bone marrow from IFNγ−/− mice (Δhep_Ifnγ−/−) to suppress liver inflammation when starting the fructose diet. SHPfl/fl and SHPΔhep mice reconstituted with bone marrow from donors of respective genotypes were included (fl/fl_Con’ and Δhep_Con’) as isogenic controls. B: Body weight was not affected by the bone marrow transplant. C: Overlaying IFNγ−/− bone marrow reduced fasting blood TG in female mice but increased fasting TG in male mice. D: Fasting blood cholesterol was decreased by the hepatic deletion of SHP and was reversed by overlaying IFNγ−/− myeloid cells in both sexes. E. Changes in fasting glucose levels were similar to changes in body weight. F. Fasting insulin levels were not significantly different between groups in either sex. G-H: Overlaying IFNγ−/− myeloid cells improved OGTT only in female mice (G) but not in male mice (H). n = 6, statistical analysis was performed with 2-way ANOVA with Tukey's multiple comparisons. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗∗, p < 0.0001.
2.5. Liver RNAseq and qPCR
Tissue samples were flash-frozen in liquid nitrogen immediately when mice were sacrificed and stored at −80 °C until later use. Liver tissue (25 mg) was bead homogenized, and total RNA was isolated according to manufacturer instructions (Direct-zol RNA Miniprep Kit, Zymo Research, Irvine, CA, USA). For qPCR: complementary DNA was synthesized from 1 μg of RNA (iScript, Bio-Rad, Hercules, CA, USA). Quantitative PCR was performed in duplicate using TaqMan™ primers (supplemental Table 1) and reagents. For RNAseq, samples of DNase-treated RNA (∼80 ng/μL) were submitted to the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core laboratory for Next-Gen sequencing. A total of 30 samples were sequenced at multiplex Paired-End 150 bp on the Illumina NovaSeq 6000 and submitted for RNAseq. The quality control (QC) was evaluated at different levels including RNA quality, raw read data, alignment, and gene expression. Raw RNA-seq paired-ends were mapped to the mouse reference genome mm10 using STAR 2.7.3 [24]. Raw read counts were calculated by FeatureCount [25] and used for downstream analysis. Differential gene expression and functional enrichment analyses were performed in R (version 3.6) with several Bioconductor packages including DESeq2 [26] and clusterProfiler [27]. Using sample-to-sample PCA analysis, six outliers were excluded for downstream analyses. Comparisons were made with 24 samples by including sex (male and female) and genotypes (fl/fl_Con’, Δhep_Con’, and Δhep_Ifnγ−/−) as variables. Results for specific comparison were extracted by combinations of coefficients with DESeq2. A 5% false discovery rate threshold was used for significant results filtering. The ranking gene lists based on the statistics of differential expression analyses were used for IPA [28] analysis (QIAGEN Inc., https://digitalinsights.qiagen.com/IPA) for further pathway enrichment digestion.
2.6. Liver tissue F4/80 and Masson's trichrome staining
Sections of liver tissue collected from the left lobe of each mouse were fixed in either formalin (10% neutral buffered formalin) or Tissue-Tek OCT Compound (Tissue Tek OCT Compound, Sakura Finetek, USA). Samples were sent to the Vanderbilt University Translational Pathology Shared Resource (TPSR) Core, where they were sectioned into 5 μm slices and stained with Masson's Trichrome and F4/80 antibody. These stained samples were placed onto slides for imaging with a light microscope (AmScope). Six images were taken for every mouse for each stain and the images were analyzed using ImageJ. Macros were written on ImageJ to quantify the density of macrophage staining or Masson's Trichrome staining area in each image.
2.7. Western blotting
Immunoblotting with liver tissues was performed as we reported previously [23,29]. The primary antibody for SR-BI (NB400104) was from Novus. CD36 polyclonal antibody (PA1-16813) was from Invitrogen. The primary antibodies for AKT (9272), pAKT (4060), pACC (11818), ACC (3676), FASN (3189), SCD1 (2794), CHOP (L63F7, 2895), AMPKβ (4150), pAMPKβ (4186), pAMPKα (2531S), ERK (9107), pERK (4370), and STAT5 (94205) were from Cell Signaling. The following primary antibodies were all from Abcam: anti-LDLr (ab52818), anti-UCP1 (ab10983), anti-PGC1α (ab54481), anti-PPARα (ab8934), anti-CPT1α (ab128568), anti-ACADM (ab110296). Anti-Cox1 (sc-19998) and -Cox2 (sc-514489) antibodies were from Santa Cruz. Liver β-actin was detected with either rabbit anti-actin (I-19) antibody (sc-1616) from Santa Cruz or with mouse anti-human actin (MCA5775GA) from Bio-Rad. Secondary antibodies were from LI-COR biotech (Lincoln, NE). All primary antibodies were diluted at 1:1000 and incubated at 4 °C overnight. All secondary antibodies were diluted at 1:10,000 and were incubated at room temperature for 1 h. Images were acquired using an LI-COR Odyssey infrared imaging system (LI-COR Biotechnology). Blot densities were quantified with Image J.
2.8. Statistical analysis
Data are summarized using the mean and standard error of the mean. Statistical differences were analyzed by 1-way ANOVA with Bonferroni post-hoc comparisons or 2-way ANOVA with Tukey's multiple comparison test as indicated in each figure legend. Repeated measures 1-way ANOVA with Bonferroni post test comparison was used to statistically analyze blood glucose levels during OGTT. P-values <0.05 were considered statistically significant. Statistical analysis of RNA-seq datasets is indicated above.
3. Results
3.1. Hepatic deletion of SHP lowered blood lipids but promoted liver inflammation
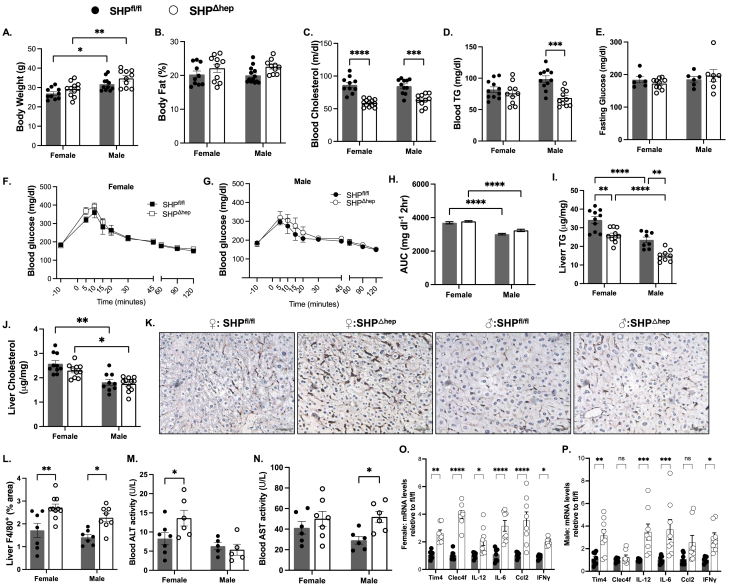
To induce fatty liver, male and female hepatic SHP knockout (SHPΔhep) mice and their wild-type littermates (SHPfl/fl) were fed a diet of 20% fructose water ad libitum for 14 weeks. Hepatic deletion of SHP did not change body weight or body composition in either sex, and body weight was generally higher in male than female mice (Figure 1A–B). Hepatic deletion of SHP lowered fasting blood cholesterol in both sexes and lowered the fasting blood triacylglycerol (TG) level only in male SHPΔhep mice (Figure 1C–D). Hepatic SHP deletion did not change fasting glucose levels, glucose tolerance, or liver cholesterol content but decreased liver TG content in both sexes (Figure 1E–J). Glucose tolerance was better in male mice, which was associated with a lower liver TG content compared to female mice (Figure 1F–I).
Figure 1.
Hepatic deletion of SHP with a fructose diet decreased blood lipids but promoted liver inflammation. Female and male hepatic SHP knockout mice (SHPΔhep) and their wild-type littermates (SHPfl/fl) were fed a fructose diet for 14 weeks. A: The body weight was not different between phenotypes and was higher in male than female mice. B: The body fat composition was not different between genotypes or sexes. C: The fasting blood cholesterol level was decreased in SHPΔhep mice in both sexes. D: The fasting blood TG levels were only decreased in male SHPΔhep mice. E: Fasting blood glucose level was not affected by the hepatic deletion of SHP in either sex. F–H: There was no difference in glucose tolerance between genotypes in both sexes, and male mice presented better glucose tolerance than female mice. The area under the curve (AUC) for blood glucose in F and G panels was analyzed with Prism by group and plotted with mean ± SD with n numbers in H. I: Hepatic deletion of SHP decreased liver TG content in both sexes, and liver TG was lower in male than female mice. J: Hepatic deletion of SHP did not change liver cholesterol content in both sexes, and liver cholesterol was lower in male than female mice. K-L: Representative images of immunostaining for liver F4/80+ cells from each group are shown in K and the quantification is shown in L. M: Blood ALT activity was increased in female SHPΔhep mice. N: Blood AST activity was increased in male SHPΔhep mice. O–P: mRNA levels of liver inflammatory cytokines were respectively increased in female (O) and male (P) SHPΔhep mice (n = 6∼11). Statistical analysis was performed with 2-way ANOVA (Tukey's multiple comparisons); ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
Despite lower liver TG content, hepatic deletion of SHP increased liver inflammation. Inflammatory F4/80+ cell numbers in the liver were larger in SHPΔhep than SHPfl/fl mice (Figure 1K–L), which was associated with increased mRNA levels of Timd4 and Clec4f (Figure 1O–P). Both Tim4 and Clec4f are specifically expressed in F4/80+ Kupffer cells in the liver [30]. In line with these changes, blood alanine transaminase (ALT) activity in female SHPΔhep mice (Figure 1M) and increased aspartate aminotransferase (AST) activity in male SHPΔhep mice (Figure 1N). The liver collagen content determined by Mason's Trichrome Blue staining did not differ between genotypes or sexes (data not shown). The mRNA level of the inflammatory cytokine interferon γ (IFNγ) was increased in SHPΔhep mice in both sexes, which was associated with increased mRNA levels for inflammatory cytokines of interleukin 12 (IL-12), IL-6, and chemokine CC-motif ligand 2 (Ccl2), in either one or both sexes (Figure 1O–P).
3.2. The ablation of IFNγ−/− in bone marrow-derived myeloid cells suppressed liver inflammation and fibrosis in SHPΔhep mice
To test whether liver inflammation induced by the hepatic SHP deletion with a fructose diet could be reduced by suppressing IFNγ-mediated pathways in myeloid cells, SHPΔhep mice were reconstituted with bone marrow either from IFNγ−/− mice (Δhep_ Ifnγ−/−) or from SHPΔhep mice (Δhep_Con’, isogenic controls) following exposure to a lethal dose of irradiation, and then mice were fed a fructose diet for 14 weeks (Figure 2A). The wild type SHPfl/fl littermates that were reconstituted with bone marrow from the same genotype donors were used as “sham” controls (fl/fl_Con’) for the bone marrow transplant procedure (Figure 2A). Similar to the metabolic changes presented with the 1st cohort mice without bone marrow transplant (Figure 1), body weight (Figure 2B) and body composition (data not shown) were not influenced by hepatic SHP deletion. Blood cholesterol levels were lower in Δhep_Con’ than fl/fl_Con’ in both sexes and blood TG levels were decreased only in male Δhep_Con’ mice compared to fl/fl_Con’ controls (Figure 2C–D). Changes in blood cholesterol and TG distribution in lipoproteins by the SHP deletion were also similar between the two cohort mice (Suppl. Figs. 1 and 2). These results indicate that the bone marrow transplant procedure did not significantly impact the phenotype of hepatic SHP deletion with fructose-diet feeding.
Overlaying bone marrow-derived IFNγ−/− myeloid cells did not change body weight compared to Δhep_Con’ mice in either sex (Figure 2B). In comparison to Δhep_Con’ mice, overlying IFNγ−/− myeloid cells reduced blood TG levels in females but increased blood TG levels in males. Overlying IFNγ−/− myeloid cells increased blood cholesterol levels compared to Δhep_ Con’ mice in both sexes (Figure 2C–D). Fasting glucose levels were not different between fl/fl_Con’ and Δhep_Con’ mice, and overlaying IFNγ−/− myeloid cells did not significantly change fasting glucose in comparison to Δhep_Con’ mice in either sex (Figure 2E). Fasting insulin levels were not impacted either by the deletion of hepatic SHP or by overlying IFNγ−/− myeloid cells (Figure 2F). Glucose tolerance was not altered by the hepatic SHP deletion (Figure 2G and H). The ablation of IFNγ in myeloid cells improved glucose tolerance only in female mice but not in male mice (Figure 2G and H).
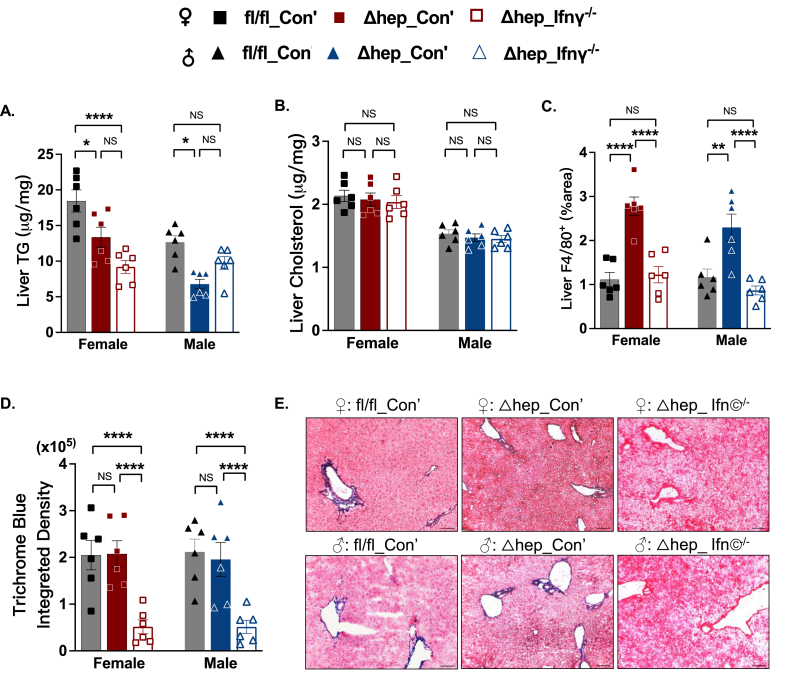
Liver TG content was reduced in Δhep_Con’ mice in both sexes, and overlaying IFNγ−/− myeloid cells did not significantly affect liver TG content compared to Δhep_Con’ in either sex (Figure 3A). Liver cholesterol content was not altered either by the deletion of hepatic SHP or by overlying IFNγ−/− myeloid cells (Figure 3B). Also similar to the cohort mice without bone marrow transplant, liver F4/80+ cell number was higher in Δhep_Con’ mice than in fl/fl_Con’ mice, and overlaying IFNγ−/− myeloid cells reduced liver F4/80+ cell number in both sexes (Figure 3C). Furthermore, overlaying IFNγ−/− myeloid cells decreased collagen content in Δhep_Ifnγ−/− mice in both sexes (Figure 3D and E), demonstrating that myeloid IFNγ plays a key role in the inflammatory response in SHPΔhep mice.
Figure 3.
Overlaying IFNγ−/−myeloid cells improved the MASH liver in SHPΔhepmice. A: Liver TG content was lower in Δhep_Con’ than fl/fl_Con’ mice and was not significantly changed by the ablation of IFNγ in myeloid cells in either sex. B: Liver cholesterol content was not different between genotypes and treatments in both sexes. C. Liver F4/80+ cells were increased by the hepatic deletion of SHP and were reduced by the ablation of IFNγ in myeloid cells in both sexes. D. Quantification of Masson's trichrome blue staining showed that liver collagen content was not changed by the hepatic deletion of SHP and was suppressed by overlaying IFNγ−/− myeloid cells in both sexes. E. Representative images of Masson's trichrome blue staining from different groups (n = 6, scale bar in each image equals 100 um). Statistical analysis was performed with 2-way ANOVA with Tukey's multiple comparisons. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗∗, p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Pathways for liver lipid uptake and fatty acid oxidation were upregulated with hepatic SHP deletion, which was partially reversed by overlaying IFNγ−/− myeloid cells
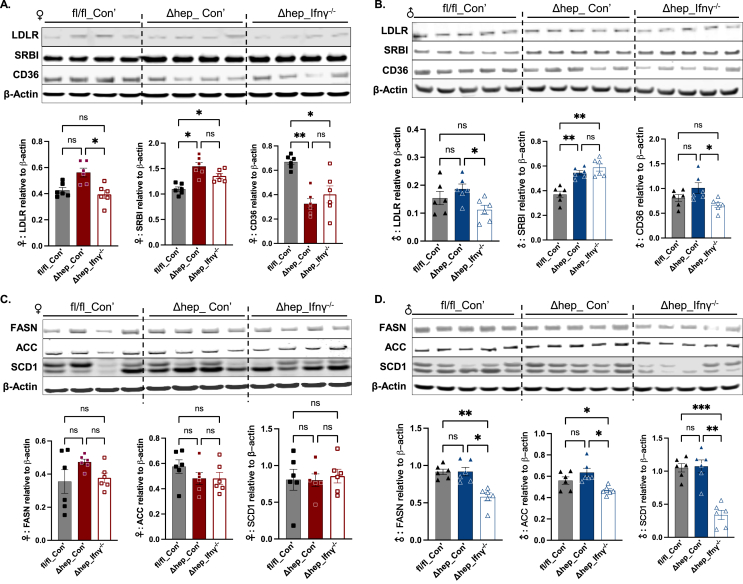
To better understand mechanisms for the modification of blood and liver lipids by hepatic SHP deletion, we investigated lipid metabolic pathways in the liver. While there was no significant difference in low-density lipoprotein receptor (LDLR) protein amounts between fl/fl_Con’ and Δhep_Con’ mice, LDLR protein content was slightly reduced by overlaying IFNγ−/− myeloid cells (Δhep_ Ifnγ−/− mice) in both sexes (Figure 4A and B). The protein level of scavenger receptor class B type I (SRBI), a receptor for HDL-cholesterol uptake, was increased by the hepatic SHP deletion in Δhep_Con’ mice and was not significantly influenced by overlaying IFNγ−/− myeloid cells in either sex (Figure 4A and B). The protein level of CD36 was decreased by the hepatic SHP deletion only in female mice and was maintained at lower levels in Δhep_Ifnγ−/− mice in both sexes (Figure 4A and B). These results indicate that increased SRBI may contribute to decreased blood cholesterol levels with hepatic SHP deletion. Reduced LDLR by the ablation of myeloid IFNγ may drive the corrected blood cholesterol levels in Δhep_ Ifnγ−/− mice in comparison to Δhep_Con’ mice.
Figure 4.
Changes in liver proteins in lipid uptake and lipogenesis pathways by hepatic deletion of SHP and by overlaying IFNγ−/−myeloid cells. Western blots with liver proteins of lipid uptake receptors are shown in A for female mice and in B for male mice. Western blots with liver proteins in lipogenesis are shown in C for female mice and in D for male mice. Quantification of blots is shown underneath the respective blots in each panel (n = 6). Statistical analysis was performed with a Student's t-test for each protein. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
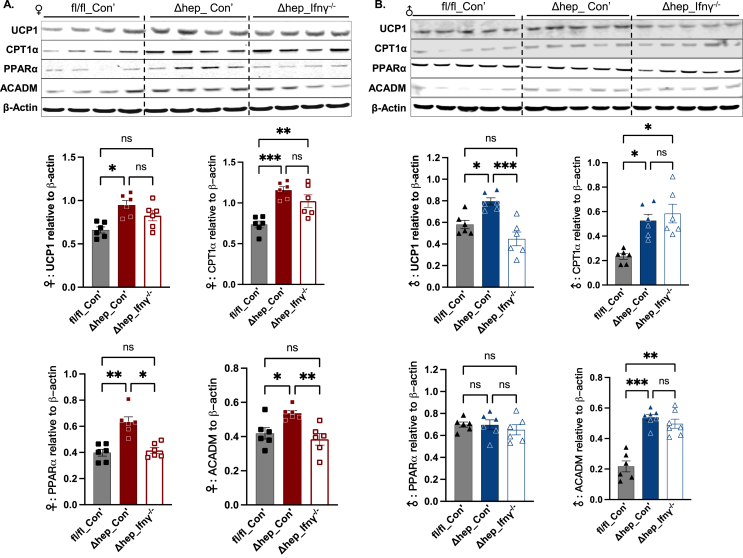
Proteins from lipogenesis pathways, such as fatty acid synthesis (FASN), acetyl-CoA carboxylase (ACC), and stearoyl CoA desaturase 1 (SCD1), were not influenced by the hepatic SHP deletion in either sex (Figure 4C and D). Phosphorylation of ACC (pACC) was also not significantly different between genotypes in either sex (Suppl. Fig. 3). Carnitine palmitoyl transferase 1α (CPT1α) regulates β-oxidation by limiting fatty acid transport to mitochondria [31]. Protein amounts of CPT1α and medium-chain acyl-CoA dehydrogenase (ACADM), a rate-limiting enzyme in β-oxidation process, were increased in mice with the hepatic SHP deletion in both sexes (Figure 5). Consistent with this, the protein level of uncoupling protein 1 (UCP1) was increased in both sexes while proteins for peroxisome proliferator-activated receptor α (PPARα) were only increased in female mice (Figure 5A), indicating that fatty acid oxidation was increased by the hepatic SHP deletion. Interestingly, overlaying IFNγ−/− myeloid cells decreased lipogenic proteins of FASN and SCD1 in male Δhep_Ifnγ−/− mice (Figure 4D) and suppressed proteins governing fatty acid β-oxidation in Δhep_Ifnγ−/− mice in a sex-specific manner (ACDAM, PPARα in female mice and UCP1 in male mice, Figure 5). Phosphorylation of AMPKα was not influenced by either hepatic SHP deletion or overlaying IFNγ−/− myeloid cells in either sex, however, total liver AMPKα was not detectable with available antibodies (Suppl. Fig. 4. A and B). In addition, neither the protein content of AMP-activated protein kinase β (AMPKβ) nor its phosphorylation (pAMPKβ) was different between genotype and bone marrow transplant groups (Suppl. Fig. 4, A and B). These results indicate that the hepatic SHP deletion promotes fatty acid oxidation independent of AMPK pathways in both sexes. Overlaying IFNγ−/− myeloid cells suppressed proteins involved in lipogenesis in male mice and decreased proteins involved in fatty acid oxidation in both sexes with a sex-specific pattern for certain proteins (Figure 4, Figure 5).
Figure 5.
Changes in proteins in fatty acid oxidation pathways by hepatic deletion of SHP and by overlaying IFNγ−/−myeloid cells. Western blots of UCP1, CPT1α, PPARα, and ACADM are shown in A for female and in B for male mice. Quantification of blots is shown underneath the respective blots in each panel (n = 6). Statistical analysis was performed with a Student's t-test for each protein. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001.
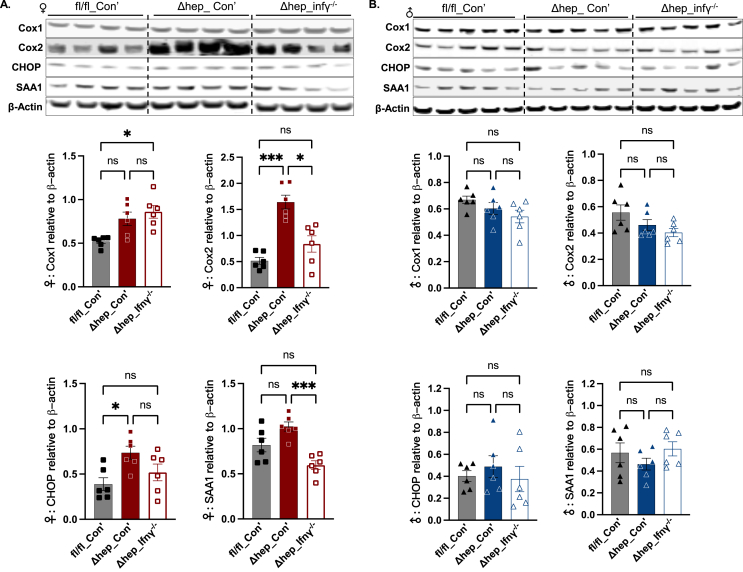
3.4. Protein contents for cyclooxygenase 2 (Cox2) and CCAAT-enhancer-binding protein homologous protein (CHOP) were increased by the hepatic SHP deletion in female mice
To further understand the association between inflammation and the upregulated lipid metabolism by hepatic SHP deletion with a fructose diet, we examined protein levels of Cox1 and Cox2 in the liver. The activities of Cox enzymes are associated with UCP1 and fatty acid oxidation in adipose tissue [32]. Cox enzymes play a critical role in the conversion of long-chain fatty acids to bioactive metabolites, such as prostaglandins, eicosanoids, and ceramides, and Cox2 is well known for its correlation with inflammation and cell death in multiple tissues [33,34]. In female mice, Cox2 protein content was increased by the hepatic SHP deletion and was decreased by overlaying IFNγ−/− myeloid cells while Cox1 protein level was not changed either by the SHP deletion or overlaying myeloid IFNγ−/− (Figure 6A). Associated with the elevated levels of Cox2 and UCP1, the protein content of CHOP, an endoplasmic reticulum (ER) stress marker, was also increased by the hepatic SHP deletion in female mice (Figure 6A). The liver protein content of acute response protein serum amyloid A1 (SAA1) was not impacted by the SHP deletion but was decreased by overlaying IFNγ−/− myeloid cells in female Δhep_Ifnγ−/− mice (Figure 6A). In male mice, liver Cox1, Cox2, CHOP, and SAA1 proteins were not significantly different between groups (Figure 6B). In response to inflammation, extracellular signal-related kinases 1 and 2 (ERK1/2) phosphorylate c-Fos for inflammatory cytokine production [35]. Protein amounts of ERK1/2 and their phosphorylation status were not significantly changed by the hepatic SHP deletion or by overlaying IFNγ−/− myeloid cells in either sex (Suppl. Fig. 4, C and D). These results suggest that Cox2-related inflammation is induced by hepatic SHP deletion and corrected by the ablation of IFNγ in myeloid cells in female mice, which is independent of ERK1/2.
Figure 6.
The abundance of liver proteins involved in inflammation and endoplasmic reticulum stress. Western blots of representative proteins in inflammation and endoplasmic reticulum stress (A for female and B for male mice). Quantification of blots is shown underneath the respective blots in each panel (n = 6). Statistical analysis was performed with a Student's t-test for each protein. ∗, p < 0.05; ∗∗∗, p < 0.001.
3.5. Liver transcriptome analysis for metabolic and inflammatory pathways in mice with hepatic SHP deletion
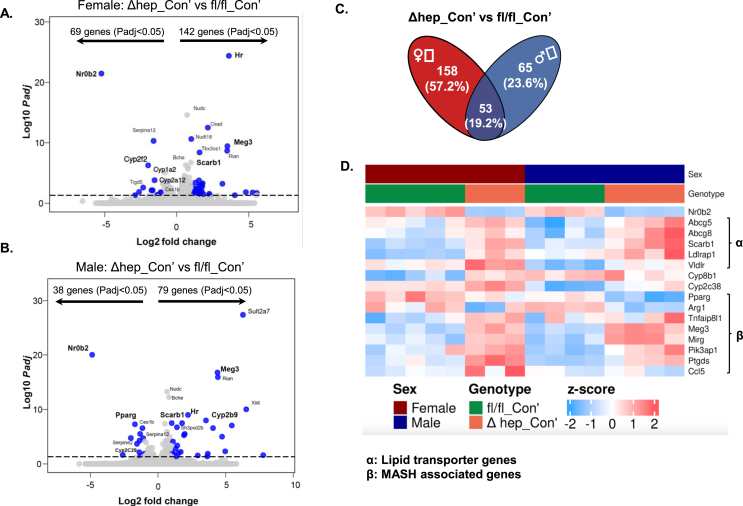
To further understand the metabolic changes regulated by hepatic SHP, we performed bulk RNAseq analysis with liver samples from fl/fl_Con’ and Δhep_Con’ mice (Figure 2A). Hepatic SHP deletion significantly (Padj<0.05) modified mRNA levels of 211 transcripts in female mice with 69 transcripts downregulated and 142 transcripts upregulated (Figure 7A). Hepatic SHP deletion significantly changed mRNA levels of 117 transcripts in male mice with 38 transcripts downregulated and 79 transcripts upregulated (Figure 7B). The commonly changed 53 transcripts by the SHP deletion in both sexes were involved in multiple gene ontology (GO) items including monooxygenase, steroid hydroxylase, arachidonic acid monooxygenase, and aromatase activities and bile secretion pathway from Kyoto encyclopedia of genes and genomes (KEGG) database (Figure 7C, Suppl. Fig. 7).
Figure 7.
Bulk RNAseq analysis of liver transcripts that were modified by hepatic deletion of SHP with a fructose diet. A-B. Volcano plots of transcripts changed in the liver in Δhep_Con’ vs fl/fl_Con’ mice in female (A) and male (B) mice. C: The Venn graph of the number of transcripts changed by hepatic deletion of SHP in female and male mice. D. A heatmap of genes whose products are involved in lipid metabolism, inflammation, and fibrogenesis with Padj < 0.05 in at least one sex, additional genes are shown in Supplemental Figure 5.
Associated with the deletion of hepatic SHP (N0b2 gene), the mRNA levels of cholesterol transporters (Abcg5/8, Suppl. Table 2) and lipid uptake receptors (Scarb1, Ldlrap1, and Vldlr) were upregulated in both sexes (Figure 7D, Suppl. Fig. 5A). This result is in line with the changes in the protein amounts for SRBI in mice without hepatic SHP (Figure 4). Peroxisome proliferator-activated receptor γ (PPARγ, Pparg gene) along with SHP regulates inflammation resolution in the liver with cholestasis [36]. Pparg mRNA was decreased by hepatic SHP deletion, which was associated with increased mRNAs for Tnfaip8l1, Meg3, and Mirg (Figure 7D). These genes have been reported to be associated with MASH transition [[37], [38], [39]]. In addition, reduction of Pparg expression initiates hepatic stellate cell activation [40]. Associated with the downregulation of Pparg, transcripts for multiple hepatic stellate cell activation markers such as Cd9 and Pik3ap1 were also upregulated (Suppl. Fig. 5C). In the condition of inflammation, Ccl5 recruits immune cells and activates cytokine production in the inflammatory sites to promote liver injury and fibrogenesis [41]. The mRNA level of Ccl5 was significantly increased by hepatic SHP deletion in female mice (Figure 7D). These data suggest that hepatic SHP is required to suppress the expression of genes whose products promote MASH progress.
3.6. Overlaying bone marrow-derived IFNγ−/− myeloid cells suppressed pro-MASH pathways
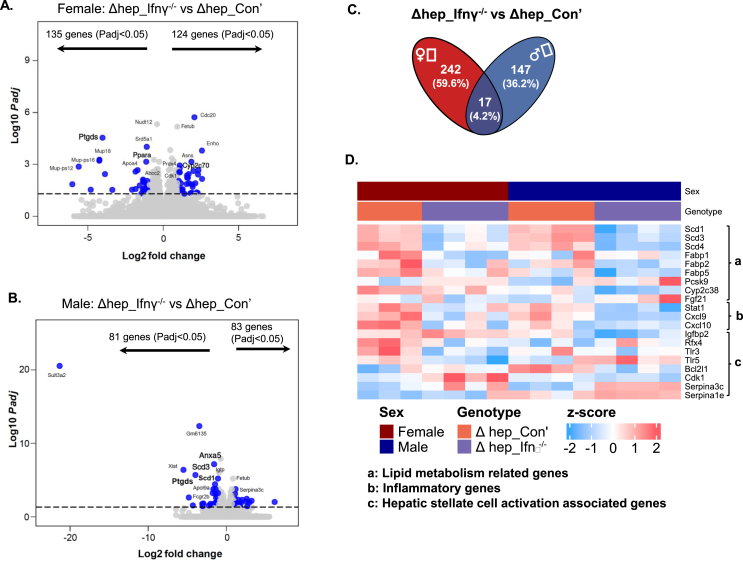
To better understand the mechanisms for the suppression of liver inflammation and fibrogenesis by bone marrow-derived IFNγ−/− myeloid cells, we performed bulk RNAseq analysis with liver samples from hepatic SHP knockout mice overlaid with IFNγ−/− bone marrow (Figure 2A). The gene expression regulated by overlaying IFNγ−/− myeloid cells diverged between sexes. A total of 259 transcripts were modified in female mice, and 164 transcripts were modified in male mice, with only 17 transcripts commonly changed in both sexes (Figure 8A–C). Of note, a large portion of transcripts modified by overlaying IFNγ−/− myeloid cells were in inflammation and fibrogenesis pathways (Suppl. Fig. 6). In both sexes, overlaying IFNγ−/− myeloid cells downregulated pro-inflammation transcripts of Stat1 and Cxcl9/10, and upregulated anti-inflammation transcripts including Apom (Figure 7D; Suppl. Fig. 5). Of note, it is known that Ptgds and Cxcl9/10 are associated with hepatic stellate cell activation [42,43]. Overlaying IFNγ−/− myeloid cells suppressed fibrosis in a sex-specific manner. Regarding hepatic stellate cells activation genes [40,44], Igfbp2, Bcl2l1, and Cdk1 were downregulated in male Δhep_Ifnγ−/− mice while Rfx4 and Tlr3/5 were downregulated in female Δhep_Ifnγ−/− mice (Figure 8D). Recent studies showed that the upregulation of Serpina1e and Serpina1b were associated with the de-activation of hepatic stellate cells [44]. The expression of Serpina1e and Serpina3C were both upregulated in male Δhep_Ifnγ−/− mice and the expression of Serpina3c was upregulated in female Δhep_Ifnγ−/− mice (Figure 8D). Furthermore, transcripts for inflammatory and fibrogenesis proteins including Il15ra, Iglv1, Ttc9, Ccl200, and Cxcl15 were modified only in female Δhep_Ifnγ−/− mice, while transcripts for fibrogenesis including Rab36/13, Lcn2, and Atf4 were changed only in male Δhep_Ifnγ−/− mice (Suppl. Fig. 6). Analysis with GO and KEGG enrichment showed that the ablation of IFNγ in myeloid cells suppressed inflammation and fibrogenesis through different pathways between sexes (Suppl. Fig. 7). IPA analysis revealed that overlaying IFNγ−/− myeloid cells suppressed interferon regulatory factors (IRFs) and IFNα2 as well as PPARα- and PPARγ-mediated pathways in female Δhep_Ifnγ−/− mice, and suppressed IRFs and IFNs (α2, β1, and λ1) as well as STAT1-mediated pathways in male Δhep_Ifnγ−/− mice (Suppl. Fig. 8, C and D).
Figure 8.
Bulk RNAseq analysis of liver transcripts that were modified by the overlay of IFNγ−/−myeloid cells. A-B: Volcano plots of transcripts changed in the liver in Δhep_Ifnγ−/− vs Δhep_Con’ mice in female (A) and male (B) mice. C. The Venn graph of the number of transcripts changed by overlaying IFNγ−/− myeloid cells in female and male mice. D. A heatmap of transcripts which products are involved in lipid metabolism, inflammation, and stellate cell activation markers with Padj<0.05 in at least one sex, additional genes are shown in Supplemental Figure 6.
Interestingly, overlaying IFNγ−/− myeloid cells not only modified transcripts for genes whose products are part of inflammation pathways but those for lipid metabolic pathways in both sexes (Figure 8D and Supple. Fig. 6). The changes in the expression of apolipoprotein transcripts (Apoa1/4, Apom, Apob, Apol9a/b) may have contributed to the elevation in HDL-cholesterol in Δhep_Ifnγ−/− mice (Suppl. Fig. 2 and Suppl. Fig. 6). HDL particles are known to perform important anti-inflammatory functions in metabolic disorder conditions [45]. Furthermore, overlaying IFNγ−/− myeloid cells reduced the expression of lipogenic genes (Scd1/3/4) and fatty acid binding protein (Fabp1/2/5) genes in both sexes (Figure 8D). Moreover, overlaying IFNγ−/− myeloid cells increased the expression of Pcsk9, which may have caused the lower protein level of LDLR in Δhep_Ifnγ−/− mice (Figure 4, Figure 8C). These results indicate that the changes in lipid metabolism may potentially contribute to the suppression of inflammation by the ablation of IFNγ in myeloid cells.
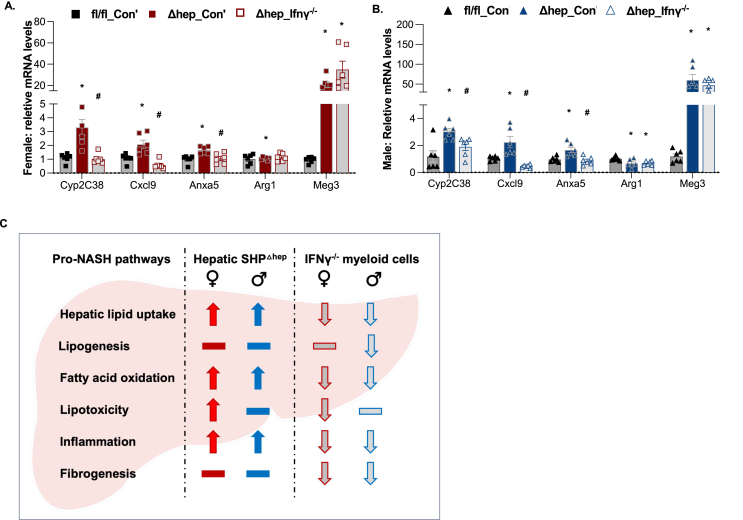
We next verified the gene expression with qPCR for several key transcripts that were indicated by the RNAseq analysis. The mRNA of Cyp2c38 was increased by hepatic SHP deletion and was reversed by overlaying IFNγ−/− myeloid cells in Δhep_Ifnγ−/− mice in both sexes (Figure 8A–B). Regulation of Cxlcl9 and Anxa5 by the SHP deletion and overlying IFNγ−/− myeloid cells fell in the same pattern (Figure 9A–B). The expression of maternally expressed gene 3 (Meg3), a long non-coding RNA, has been reported to be closely associated with MASH transition in mice and humans [39]. The transcript of Meg3 was dramatically increased by hepatic SHP deletion and was maintained at high levels in Δhep_Ifnγ−/− mice in both sexes (Figure 7, Figure 9, A-B). The analysis of RNAseq data shows that overlaying IFNγ−/− myeloid cells reduced the expression of the transcripts in inflammation pathways in both sexes but in a sex-specific manner for certain genes.
Figure 9.
Validation of the gene expression with qPCR and proposed metabolic pathways in the liver regulated hepatic SHP deletion and overlaying IFNγ−/−myeloid cells. A-B: Expression of selective transcripts indicated from bulk RNAseq analysis was validated with qPCR analysis (n = 6). Statistical analysis was performed with 2-way ANOVA with multiple comparisons. ∗, p < 0.05 for significant differences between Δhep_Con’ and fl/fl_Con’; #, p < 0.05 for significant differences between Δhep_Ifnγ−/− and Δhep_Con’. The expression of genes in fl/fl_Con’ was used as the control. C. Summary for the pro-MASH pathways changed by hepatic SHP deletion and overlaying bone marrow-derived IFNγ−/− myeloid cells in female and male mice. Pathways that were up- or down-regulated were denoted by the direction of the arrowheads. Pathways not found significantly to be changed in the current study were denoted by horizontal lines.
4. Discussion
We show that hepatic SHP deletion promoted pathways for liver lipid uptake and hepatic fatty acid oxidation, which was associated with reduced blood cholesterol and liver TG content in both sexes (Figure 9C). Despite decreasing liver fat content, hepatic SHP deletion increased liver inflammation by modifying the expression of a variety of genes including cytokine genes and PPARγ (Figure 9C). In addition, hepatic SHP deletion with a fructose diet also promoted hepatic inflammation by increasing ER stress, which was evidenced by elevated protein amounts of Cox2 and CHOP in female mice. Overlaying IFNγ−/− myeloid cells suppressed inflammation and fibrogenesis (Figure 9C) by modifying mRNA transcripts related to pathways for MASH transition and limited lipotoxicity by reducing mRNAs of apolipoproteins and proteins in lipogenesis and fatty acid oxidation pathways.
Our studies suggest that hepatic SHP decreases liver lipid uptake and cholesterol transport pathways and limits lipid flux to the liver. In hepatic SHP knockout mice, the mRNA levels of hepatic lipid receptors (Srarb1 and Vldlr) and cholesterol transporters (Abcg5/8) were upregulated (Figure 7D). In line with this, the increased liver lipid uptake associated with the downregulation of hepatic SHP has been reported by different groups [9,16,20]. It has been reported that the bile acid pool size in the liver can be enlarged in SHP knockout mice due to the lack of suppression of Cyp7a1 and/or Cyp8b1 expression [9,13,20,46]. Bile acids are the major endogenous activating ligands for FXR transcriptional activity [47], while Srarb1 [48,49], Abcg5 and Abcg8 [48], and Vldlr [50] are FXR target genes. In the current study, Cyp8b1 expression was increased by the deletion of SHP in female mice (Suppl. Fig. 5B), suggesting that FXR-regulated pathways may play a role in upregulating liver lipid uptake by hepatic SHP deletion. Although neither Cyp8b1 nor Cyp7a1 expression was increased by the SHP deletion in male mice, the bile acid secretion pathways were enriched as demonstrated by IPA analysis of the RNAseq data (Suppl. Fig. 8, A and C). These data indicate additional mechanisms for the upregulation of liver lipid uptake existing in the liver with hepatic SHP deletion, which might be predominant in males compared to females.
Our study and published data together suggest that hepatic SHP limits diet-induced lipotoxicity from the overload of fatty acid oxidation. Chronic exposure to dietary fructose drives MASLD by contributing to lipogenic substrate overflow and lipid accumulation in hepatocytes [51]. In our study, despite increased pathways for lipid uptake, liver TG content was lower in mice with hepatic SHP deletion than in their wild-type littermates. This suggests fatty acid oxidation was overloaded in the liver with SHP deletion after high fructose feeding. We show that the protein amounts of several key enzymes in fatty acid oxidation pathways, such as CPT1α, ACDAM, and UCP1, were increased in the liver with SHP deletion in both sexes while the key enzymes in lipogenesis, such as FASN and ACC, were unchanged (Figure 5). Under physiological conditions, PPARα is activated by unsaturated free fatty acids and their metabolites during fasting to upregulate the expression of enzymes in fatty acid oxidation pathways [52]. With hepatic SHP deletion, the protein amount of PPARα was increased in female mice (Figure 5) and PPARα-mediated pathways were predicted to be upregulated in male mice (Suppl. Fig. 8C). Bile acids may upregulate PPARα expression by activating FXR in human hepatocytes [52,53]. However, this mechanism has not been reported in mice. Activities of monooxygenase- and arachidonic acid epoxygenase-mediated pathways were enriched in the liver with SHP deletion in both sexes (Suppl. Fig. 7). Thus, it is possible that, with increased lipid flux in the liver associated with a fructose diet, metabolites of long-chain fatty acids derived from cytochrome P450 (CYP) monooxygenase- and epoxygenase-mediated pathways may have acted as a ligand for the transactivation of PPARα. Of note, the overload of fatty acid oxidation may increase ER stress and lead to inflammation. Therefore, SHP is likely required for inflammation resolution by limiting lipid flux in addition to regulating the expression of genes in inflammation pathways under a pro-MASH condition.
Our study suggests that SHP suppresses inflammation pathways in the early stages of MASH. Despite reduced liver lipid content, liver F4/80+ cells were increased in mice with hepatic SHP deletion, which was associated with the upregulation of inflammatory cytokines and increased ALT or AST activity. Consistent with our observation, preclinical and clinical studies showed that downregulation of hepatic SHP was associated with increased inflammation and fibrosis in the liver [20,54,55]. In human liver samples from MASH patients, the reduction of liver SHP mRNA was associated with increased expression of genes in inflammation and fibrogenesis pathways [54]. SHP could suppress inflammation by inhibiting the expression of Ccl2, TNFα, IL-1β, and IL-6 and the activity of NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome via multiple pathways [52,56,57]. PPARγ-mediated pathways suppress inflammation by upregulating the expression of anti-inflammatory genes and downregulating pro-inflammatory mediators [58,59]. Consistent with our observation, the downregulation of PPARγ expression has been reported in SHP knockout mice [60]. Moreover, inflammation can also be induced by metabolites of long-chain fatty acids associated with a fructose diet [61] or by liver injury associated with high ALT and AST activity [57] in mice with hepatic SHP deletion. Despite the mounting evidence showing the protective roles of hepatic SHP, Lee et al. reported that hepatic SHP deletion protected mice from MASH after a long-term WD (Western-type diet) or an MCD (methionine-choline deficient diet) in apoE knockout mice [62]. The approaches of long-term WD diet or the MCD in apoE knockout mice model the advanced MASH subgroup, which represents injury-associated fibrogenesis rather than metabolic dysfunction-associated inflammation and fibrogenesis. SHP is involved in a broad spectrum of biological functions, including metabolism, inflammation, cell apoptosis and replication, etc. It is possible that during the late stages of MASH when the metabolic context changes, the dominant SHP-mediated pathways might be altered compared to earlier stages studied here. Thus, results from our studies and others indicate that hepatic SHP is required to suppress MASH at its early stages.
Hepatic SHP has a stronger effect on suppressing markers of diet-induced ER stress pathways in females than in males. Lipotoxicity from increased fatty acid flux and oxidation may cause ER stress and damage mitochondria DNA, leading to cell death and inflammatory infiltration [3]. With a fructose diet, liver ER stress associated with impaired glucose tolerance was more pronounced in female than male mice (Figure 1F–H). The network of pathways analyzed with IPA showed increased lipid metabolism, including lipid oxidation and cholesterol transport, in female mice compared to male mice (Suppl. Fig. 9). Furthermore, female mice consistently exhibited higher expression of Sult3a1 and Sult3a2 (Suppl. Fig. 5C and Suppl. Table 2), products of both genes play an important role in bile acid (BA) catabolism [63], indicating the BA-FXR activated fatty acid oxidation may be more vigorous and lipotoxicity may be more severe in female mice. Thus, SHP-mediated suppression of inflammation may play a stronger role in MASH transition in females than in males.
With our bone marrow transplant approach, we found that blocking IFNγ pathways in myeloid cells inhibits MASH progression by suppressing CXCL9/10 expression for fibrogenesis and modifying genes in inflammatory pathways in a sex-specific manner. Recent studies showed that activation of macrophages occurs at the onset of MASLD, starting as early as one week after feeding a Western-type diet [37]. We performed bone marrow transplants using IFNγ−/− mice as donors to suppress inflammation pathways in myeloid cells in mice with hepatic SHP deletion. CXCL9 and CXCL10 are primarily induced by IFNγ or its downstream chemokines [42,64,65]. Clinical studies showed that blood CXCL9 and CXCL10 levels are positively associated with fibrosis in multiple organs [42]. STAT1 is currently considered a negative regulator for hepatic fibrosis and is primarily activated by IFNγ [66]. Thus, the downregulation of STAT1 in the mouse liver after the IFNγ−/− bone marrow transplant is likely a subsequent response to the ablation of IFNγ in myeloid cells. Surprisingly, in addition to suppressing liver inflammation, inhibiting IFNγ-mediated inflammation in myeloid cells also reduced the mRNA levels of lipogenic genes (Scd1/3/4 for both sexes) and proteins in fatty acid oxidation pathways (ACDAM in female mice and CPT1α in male mice). The risk for cardiovascular disease events is increased in patients with chronic inflammation such as SLE (systemic lupus erythematosus). Future studies are required to define if pathways modified by anti-inflammation regimes for SLE patients would influence atherosclerosis risk.
Our study demonstrates that hepatic SHP is required to suppress gene expression in inflammation pathways and limit lipotoxicity associated with diet-induced metabolic dysfunction. We show proof-of-principle that suppression of inflammatory pathways in myeloid cells may be effective in treating MASH. The increased fatty acid oxidation associated with obesity is an adaptive response to limit fatty liver, however, our studies support that this excess fatty acid oxidation could be a driver of inflammation that promotes the transition to MASH. Additionally, our RNAseq data provide a resource for future studies regarding sex differences in the development and treatments for MASH.
Funding sources
The NIA (K01AG077038) provided support to LZ. The Department of Veterans Affairs (BX002223) and NIH (R01DK109102, R01HL144846) provided support to JMS. NIH (R01DK109102, R01HL144846) also provided support to YW and XZ. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CRediT authorship contribution statement
Lin Zhu: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Bridget Litts: Data curation, Investigation, Methodology, Writing – review & editing. Yu Wang: Writing – review & editing, Software, Data curation, Investigation, Methodology. Jeffrey A. Rein: Writing – review & editing, Methodology, Investigation, Data curation. Cassandra L. Atzrodt: Writing – review & editing, Data curation. Sivaprakasam Chinnarasu: Writing – review & editing, Data curation. Julia An: Writing – review & editing, Methodology, Data curation. Ariel S. Thorson: Writing – review & editing, Data curation. Yaomin Xu: Writing – review & editing, Supervision, Software, Resources, Project administration, Investigation, Funding acquisition, Formal analysis, Data curation. John M. Stafford: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Resources, Supervision, Visualization, Writing – review & editing.
Declaration of competing interest
None.
Acknowledgments
The authors acknowledge the helpful assistance of the Vanderbilt Hormone Assay Core (supported by NIH grant DK020593 to the Vanderbilt Diabetes Research Center). We also acknowledge excellent support of the Vanderbilt Mouse Metabolic Phenotyping Core (supported by NIH grant DK59637). We additionally recognize the support of the Vanderbilt Diabetes Research Center (P30DK020593) and the Vanderbilt Digestive Disease Research Center (P30DK058404).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2024.101932.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Schuster S., Cabrera D., Arrese M., Feldstein A.E. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol. 2018;15(6):349–364. doi: 10.1038/s41575-018-0009-6. [DOI] [PubMed] [Google Scholar]
- 2.Hardy T., Oakley F., Anstee Q.M., Day C.P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annu Rev Pathol. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. [DOI] [PubMed] [Google Scholar]
- 3.Rada P., Gonzalez-Rodriguez A., Garcia-Monzon C., Valverde A.M. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis. 2020;11(9):802. doi: 10.1038/s41419-020-03003-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jornayvaz F.R., Shulman G.I. Diacylglycerol activation of protein kinase Cepsilon and hepatic insulin resistance. Cell Metabol. 2012;15(5):574–584. doi: 10.1016/j.cmet.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekstedt M., Franzen L.E., Mathiesen U.L., Thorelius L., Holmqvist M., Bodemar G., et al. Long-term follow-up of patients with NAFLD and elevated liver enzymes. Hepatology. 2006;44(4):865–873. doi: 10.1002/hep.21327. [DOI] [PubMed] [Google Scholar]
- 6.Wu J., Lin T., Gao Y., Li X., Yang C., Zhang K., et al. Long noncoding RNA ZFAS1 suppresses osteogenic differentiation of bone marrow-derived mesenchymal stem cells by upregulating miR-499-EPHA5 axis. Mol Cell Endocrinol. 2022;539 doi: 10.1016/j.mce.2021.111490. [DOI] [PubMed] [Google Scholar]
- 7.Gobinet J., Auzou G., Nicolas J.C., Sultan C., Jalaguier S. Characterization of the interaction between androgen receptor and a new transcriptional inhibitor. SHP. Biochemistry. 2001;40(50):15369–15377. doi: 10.1021/bi011384o. [DOI] [PubMed] [Google Scholar]
- 8.Johansson L., Thomsen J.S., Damdimopoulos A.E., Spyrou G., Gustafsson J.A., Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J Biol Chem. 1999;274(1):345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 9.Palmisano B.T., Zhu L., Litts B., Burman A., Yu S., Neuman J.C., et al. Hepatocyte small heterodimer partner mediates sex-specific effects on triglyceride metabolism via androgen receptor in male mice. Metabolites. 2021;11(5) doi: 10.3390/metabo11050330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J.H., Yoon J.E., Nikapitiya C., Kim T.H., Uddin M.B., Lee H.C., et al. Small heterodimer partner controls the virus-mediated antiviral immune response by targeting CREB-binding protein in the nucleus. Cell Rep. 2019;27(7):2105–2118 e2105. doi: 10.1016/j.celrep.2019.04.071. [DOI] [PubMed] [Google Scholar]
- 11.Boulias K., Katrakili N., Bamberg K., Underhill P., Greenfield A., Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24(14):2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu T.T., Makishima M., Repa J.J., Schoonjans K., Kerr T.A., Auwerx J., et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6(3):507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 13.Goodwin B., Jones S.A., Price R.R., Watson M.A., McKee D.D., Moore L.B., et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y.S., Kim D.K., Kim Y.D., Park K.C., Shong M., Seong H.A., et al. Orphan nuclear receptor SHP interacts with and represses hepatocyte nuclear factor-6 (HNF-6) transactivation. Biochem J. 2008;413(3):559–569. doi: 10.1042/BJ20071637. [DOI] [PubMed] [Google Scholar]
- 15.Yamagata K., Daitoku H., Shimamoto Y., Matsuzaki H., Hirota K., Ishida J., et al. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J Biol Chem. 2004;279(22):23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 16.Hartman H.B., Lai K., Evans M.J. Loss of small heterodimer partner expression in the liver protects against dyslipidemia. J Lipid Res. 2009;50(2):193–203. doi: 10.1194/jlr.M800323-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Mifflin R., Park J.E., Lee M., Jena P.K., Wan Y.Y., Barton H.A., et al. Microbial products linked to steatohepatitis are reduced by deletion of nuclear hormone receptor SHP in mice. J Lipid Res. 2023 doi: 10.1016/j.jlr.2023.100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J., Iqbal J., Saha P.K., Liu J., Chan L., Hussain M.M., et al. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46(1):147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 19.Renga B., Mencarelli A., Migliorati M., Cipriani S., D'Amore C., Distrutti E., et al. SHP-dependent and -independent induction of peroxisome proliferator-activated receptor-gamma by the bile acid sensor farnesoid X receptor counter-regulates the pro-inflammatory phenotype of liver myofibroblasts. Inflamm Res. 2011;60(6):577–587. doi: 10.1007/s00011-010-0306-1. [DOI] [PubMed] [Google Scholar]
- 20.Magee N., Zou A., Ghosh P., Ahamed F., Delker D., Zhang Y. Disruption of hepatic small heterodimer partner induces dissociation of steatosis and inflammation in experimental nonalcoholic steatohepatitis. J Biol Chem. 2020;295(4):994–1008. doi: 10.1074/jbc.RA119.010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmisano B.T., Le T.D., Zhu L., Lee Y.K., Stafford J.M. Cholesteryl ester transfer protein alters liver and plasma triglyceride metabolism through two liver networks in female mice. J Lipid Res. 2016;57(8):1541–1551. doi: 10.1194/jlr.M069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmisano B.T., Anozie U., Yu S., Neuman J.C., Zhu L., Edington E.M., et al. Cholesteryl ester transfer protein impairs triglyceride clearance via androgen receptor in male mice. Lipids. 2021;56(1):17–29. doi: 10.1002/lipd.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu L., Luu T., Emfinger C.H., Parks B.A., Shi J., Trefts E., et al. CETP inhibition improves HDL function but leads to fatty liver and insulin resistance in CETP-expressing transgenic mice on a high-fat diet. Diabetes. 2018;67(12):2494–2506. doi: 10.2337/db18-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 26.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu L., An J., Chinnarasu S., Luu T., Pettway Y.D., Fahey K., et al. Expressing the human cholesteryl ester transfer protein minigene improves diet-induced fatty liver and insulin resistance in female mice. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.799096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bleriot C., Ginhoux F. Understanding the heterogeneity of resident liver macrophages. Front Immunol. 2019;10:2694. doi: 10.3389/fimmu.2019.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreurs M., Kuipers F., van der Leij F.R. Regulatory enzymes of mitochondrial beta-oxidation as targets for treatment of the metabolic syndrome. Obes Rev. 2010;11(5):380–388. doi: 10.1111/j.1467-789X.2009.00642.x. [DOI] [PubMed] [Google Scholar]
- 32.Banhos Danneskiold-Samsoe N., Sonne S.B., Larsen J.M., Hansen A.N., Fjaere E., Isidor M.S., et al. Overexpression of cyclooxygenase-2 in adipocytes reduces fat accumulation in inguinal white adipose tissue and hepatic steatosis in high-fat fed mice. Sci Rep. 2019;9(1):8979. doi: 10.1038/s41598-019-45062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amior L., Srivastava R., Nano R., Bertuzzi F., Melloul D. The role of Cox-2 and prostaglandin E(2) receptor EP3 in pancreatic beta-cell death. Faseb J. 2019;33(4):4975–4986. doi: 10.1096/fj.201801823R. [DOI] [PubMed] [Google Scholar]
- 34.Linton M.F., Fazio S. Cyclooxygenase-2 and inflammation in atherosclerosis. Curr Opin Pharmacol. 2004;4(2):116–123. doi: 10.1016/j.coph.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Lucas R.M., Luo L., Stow J.L. ERK1/2 in immune signalling. Biochem Soc Trans. 2022;50(5):1341–1352. doi: 10.1042/BST20220271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park Y.J., Qatanani M., Chua S.S., LaRey J.L., Johnson S.A., Watanabe M., et al. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008;47(5):1578–1586. doi: 10.1002/hep.22196. [DOI] [PubMed] [Google Scholar]
- 37.Padiadpu J., Spooner M.H., Li Z., Newman N., Apperson D.K., Lohr C.V., et al. Early transcriptome changes associated with western diet induced NASH in Ldlr-/- mice points to activation of hepatic macrophages and an acute phase response. Front Nutr. 2023;10(Sec. Nutrigenomics) doi: 10.3389/fnut.2023.1147602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todoric J., Di Caro G., Reibe S., Henstridge D.C., Green C.R., Vrbanac A., et al. Fructose stimulated de novo lipogenesis is promoted by inflammation. Nat Metab. 2020;2(10):1034–1045. doi: 10.1038/s42255-020-0261-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J., Nagy L.E., Wang L. The long and the small collide: LncRNAs and small heterodimer partner (SHP) in liver disease. Mol Cell Endocrinol. 2021;528 doi: 10.1016/j.mce.2021.111262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Xu J., Rosenthal S., Zhang L.J., McCubbin R., Meshgin N., et al. Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology. 2020;158(6):1728–1744 e1714. doi: 10.1053/j.gastro.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohs A., Kuttkat N., Reissing J., Zimmermann H.W., Sonntag R., Proudfoot A., et al. Functional role of CCL5/RANTES for HCC progression during chronic liver disease. J Hepatol. 2017;66(4):743–753. doi: 10.1016/j.jhep.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Arger N.K., Ho M.E., Allen I.E., Benn B.S., Woodruff P.G., Koth L.L. CXCL9 and CXCL10 are differentially associated with systemic organ involvement and pulmonary disease severity in sarcoidosis. Respir Med. 2020;161 doi: 10.1016/j.rmed.2019.105822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao R., Wang J., He X., Wang T., Zhou L., Ren Z., et al. Comprehensive analysis of endoplasmic reticulum-related and secretome gene expression profiles in the progression of non-alcoholic fatty liver disease. Front Endocrinol. 2022;13 doi: 10.3389/fendo.2022.967016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mannaerts I., Schroyen B., Verhulst S., Van Lommel L., Schuit F., Nyssen M., et al. Gene expression profiling of early hepatic stellate cell activation reveals a role for Igfbp3 in cell migration. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0084071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaji H. High-density lipoproteins and the immune system. J Lipids. 2013;2013 doi: 10.1155/2013/684903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw R.P.H., Kolyvas P., Dang N., Hyon A., Bailey K., Anakk S. Loss of hepatic small heterodimer partner elevates ileal bile acids and alters cell cycle-related genes in male mice. Endocrinology. 2022;163(6) doi: 10.1210/endocr/bqac052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiao Y., Lu Y., Li X.Y. Farnesoid X receptor: a master regulator of hepatic triglyceride and glucose homeostasis. Acta Pharmacol Sin. 2015;36(1):44–50. doi: 10.1038/aps.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lambert G., Amar M.J., Guo G., Brewer H.B., Jr., Gonzalez F.J., Sinal C.J. The farnesoid X-receptor is an essential regulator of cholesterol homeostasis. J Biol Chem. 2003;278(4):2563–2570. doi: 10.1074/jbc.M209525200. [DOI] [PubMed] [Google Scholar]
- 49.Dong B., Singh A.B., Guo G.L., Young M., Liu J. Activation of FXR by obeticholic acid induces hepatic gene expression of SR-BI through a novel mechanism of transcriptional synergy with the nuclear receptor LXR. Int J Mol Med. 2019;43(5):1927–1938. doi: 10.3892/ijmm.2019.4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirvent A., Claudel T., Martin G., Brozek J., Kosykh V., Darteil R., et al. The farnesoid X receptor induces very low density lipoprotein receptor gene expression. FEBS Lett. 2004;566(1–3):173–177. doi: 10.1016/j.febslet.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 51.Yu S., Li C., Ji G., Zhang L. The contribution of dietary fructose to non-alcoholic fatty liver disease. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.783393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Hagedorn C.H., Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812(8):893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pineda Torra I., Claudel T., Duval C., Kosykh V., Fruchart J.C., Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol Endocrinol. 2003;17(2):259–272. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., Xu N., Xu J., Kong B., Copple B., Guo G.L., et al. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr-1/SHP/EID1 network. Hepatology. 2014;60(3):919–930. doi: 10.1002/hep.27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J., Li Y., Chen W.D., Xu Y., Yin L., Ge X., et al. Hepatic carboxylesterase 1 is essential for both normal and farnesoid X receptor-controlled lipid homeostasis. Hepatology. 2014;59(5):1761–1771. doi: 10.1002/hep.26714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou A., Magee N., Deng F., Lehn S., Zhong C., Zhang Y. Hepatocyte nuclear receptor SHP suppresses inflammation and fibrosis in a mouse model of nonalcoholic steatohepatitis. J Biol Chem. 2018;293(22):8656–8671. doi: 10.1074/jbc.RA117.001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuk J.M., Shin D.M., Lee H.M., Kim J.J., Kim S.W., Jin H.S., et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat Immunol. 2011;12(8):742–751. doi: 10.1038/ni.2064. [DOI] [PubMed] [Google Scholar]
- 58.Bouhlel M.A., Derudas B., Rigamonti E., Dievart R., Brozek J., Haulon S., et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metabol. 2007;6(2):137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Kapadia R., Yi J.H., Vemuganti R. Mechanisms of anti-inflammatory and neuroprotective actions of PPAR-gamma agonists. Front Biosci. 2008;13:1813–1826. doi: 10.2741/2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohn J.H., Hwang J.Y., Moon M.K., Ahn H.Y., Kim H.H., Koo Y.D., et al. Small heterodimer partner (SHP) deficiency protects myocardia from lipid accumulation in high fat diet-fed mice. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0186021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calder P.C. Long-chain fatty acids and inflammation. Proc Nutr Soc. 2012;71(2):284–289. doi: 10.1017/S0029665112000067. [DOI] [PubMed] [Google Scholar]
- 62.Lee Y.K., Park J.E., Lee M., Mifflin R., Xu Y., Novak R., et al. Deletion of hepatic small heterodimer partner ameliorates development of nonalcoholic steatohepatitis in mice. J Lipid Res. 2023;64(11) doi: 10.1016/j.jlr.2023.100454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Runge-Morris M., Kocarek T.A., Falany C.N. Regulation of the cytosolic sulfotransferases by nuclear receptors. Drug Metab Rev. 2013;45(1):15–33. doi: 10.3109/03602532.2012.748794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Proost P., Verpoest S., Van de Borne K., Schutyser E., Struyf S., Put W., et al. Synergistic induction of CXCL9 and CXCL11 by Toll-like receptor ligands and interferon-gamma in fibroblasts correlates with elevated levels of CXCR3 ligands in septic arthritis synovial fluids. J Leukoc Biol. 2004;75(5):777–784. doi: 10.1189/jlb.1003524. [DOI] [PubMed] [Google Scholar]
- 65.Proost P., Vynckier A.K., Mahieu F., Put W., Grillet B., Struyf S., et al. Microbial Toll-like receptor ligands differentially regulate CXCL10/IP-10 expression in fibroblasts and mononuclear leukocytes in synergy with IFN-gamma and provide a mechanism for enhanced synovial chemokine levels in septic arthritis. Eur J Immunol. 2003;33(11):3146–3153. doi: 10.1002/eji.200324136. [DOI] [PubMed] [Google Scholar]
- 66.Liu J., Wang F., Luo F. The role of JAK/STAT pathway in fibrotic diseases: molecular and cellular mechanisms. Biomolecules. 2023;13(1) doi: 10.3390/biom13010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.