Abstract
There is a long-standing debate about the magnitude of the contribution of gene-environment interactions to phenotypic variations of complex traits owing to the low statistical power and few reported interactions to date. To address this issue, the Gene-Lifestyle Interactions Working Group within the Cohorts for Heart and Aging Research in Genetic Epidemiology Consortium has been spearheading efforts to investigate G × E in large and diverse samples through meta-analysis. Here, we present a powerful new approach to screen for interactions across the genome, an approach that shares substantial similarity to the Mendelian randomization framework. We identify and confirm 5 loci (6 independent signals) interacted with either cigarette smoking or alcohol consumption for serum lipids, and empirically demonstrate that interaction and mediation are the major contributors to genetic effect size heterogeneity across populations. The estimated lower bound of the interaction and environmentally mediated heritability is significant (P < 0.02) for low-density lipoprotein cholesterol and triglycerides in Cross-Population data. Our study improves the understanding of the genetic architecture and environmental contributions to complex traits.
Subject terms: Genetic association study, Risk factors, Statistical methods
Here, the authors report 5 loci interacting with smoking/alcohol for serum lipids using a new method akin to Mendelian randomization. They unveil significant heritability through gene-environment interaction and mediation, enhancing understanding of complex trait genetics.
Introduction
Current genome-wide association studies (GWAS) focus on detecting genetic variants that lead to different phenotype means across genotype groups1,2, and have identified a large number of genetic loci that, in some cases, explain large proportions of the trait’s SNP-heritability3–5. While it is commonly agreed that complex traits are influenced by genetic and environmental factors and their interactions6–9, there is a long-standing disagreement about the magnitude of the contribution to heritability because of different theoretical models and assumptions10,11. As pointed out in ref. 12, arbitrarily defined parameterizations of genetic effects with non-additive gene actions may explain the same degree of genetic variation as the currently prevailing additive model. Thus, while using additive genetic models such as polygenic risk scores to predict individual quantitative or qualitative phenotypes has become standard5, these models may not be fully informative in understanding genetic architecture.
Interactions are often studied secondarily in comparison to additive variance, whose advantage is to explain most of the correlations among relatives and fit natural selection model well10,13. Theoretical studies have demonstrated that a significant portion of variance can be explained by an additive model even when the genetic contribution to a phenotype is purely through 14. This limitation is one of the key factors explaining the low power of approaches modeling interactions conditional on additive variance. As a result, studies focusing on detecting at the genome-wide level are seldom considered as primary analyses. By contrast, the joint evidence of main genetic and effects, in addition to the alone, is tested in the Gene-Lifestyle Interactions (GLI) Working Group within the Cohorts for Heart and Aging Research in Genetic Epidemiology Consortium (CHARGE)9,15, where only a modest number of genetic loci have been identified through testing for alone16–19. The joint test limits our ability to determine to what degree the currently identified loci reflect evidence for contribution, making it difficult to understand the precise interplay between genetic and environmental factors.
Concurrently, Mendelian randomization (MR) has been developed and widely applied to study causal relationships between risk exposures and outcomes in the post-GWAS era20,21. Although an MR approach has been used to explain 22, the underlying connection between testing pleiotropic variants in the MR framework and the detection of is currently unclear. Here, we conceptually connect with MR framework, illuminate their similarities and demonstrate that the test of horizontal pleiotropy in MR23,24 can be used for detecting . Based on this principle, one can identify using existing available GWAS and GWIS summary statistics. We applied this approach to the summary statistics from the Global Lipids Genetics Consortium study (GLGC, n = 1.65 M)3 and the summary statistics in the interaction analysis with cigarette smoking and alcohol drinking in the CHARGE GLI working group17, with replication using direct interaction tests performed in the UK Biobank (UKBB) data. Although the UKBB data accounted for about one third of sample in the GLGC consortium, theoretical work suggests that such replication is statistically independent (Supplementary Note)
Results
Testing and mediation based on Mendelian randomization (MR)
Traditionally a genome-wide interaction study (GWIS) with an environmental exposure on a quantitative trait Y is modeled through a linear regression:
| 1 |
where and correspond to the ‘main’ effect of (in the presence of ), the main effect of and the interaction effect of , respectively, and is a random noise. Here , , and represent a genotype value, environmental factor, and their interaction respectively. For simplicity, we do not include any covariates, but it will not affect the general conclusion. The interaction effect is evaluated by the direct test statistic , where refers the estimate from the regression model (1). Theoretical work indicates that the test statistics for the main effect and the interaction effect are correlated, with the correlation coefficient equal to , where and are the mean and variance of the environmental factor in the data14. However, the power of the direct test is usually low because of the collinearity between and which induces a covariance between the estimates of and . This covariance produces uncertainty (i.e. larger standard error) which by itself reduces power for testing either or , even if the underlying true model includes alone (i.e., and )10,14.
In practice, a GWAS is routinely conducted first when studying the genetic contribution to a trait, which is typically done through a linear regression model without including environmental factors, i.e.,
| 2 |
where we refer to as the ‘marginal’ effect from a GWAS (in the absence of ) to differentiate from the main effect in model (1). We show that , where is the mediation contribution of through , , , and represent the environmental mean, standard deviation, and genotype standard deviation in GWAS data, respectively, suggesting that testing the hypothesis H0: α–β1 = 0 for the difference between the marginal and main effects is equivalent to testing for the combined effect of and mediation, and further reduces to testing for the when and are independent (i.e., Supplementary Note). This hypothesis can be tested by the statistic , where , , and their corresponding standard errors are estimated from the GWAS and GWIS analyses, respectively. In fact, and are equivalent when GWAS and GWIS are performed in the same data. We verified this property using real data analysis in the GLI studies17, from which the summary statistics of the marginal, main, and interaction effects are available and the marginal effect was obtained after adjusting for . We observed that the correlation between the statistics of the and the direct test is 0.98 for LDL and current smoking (Supplementary Fig. 5). However, GWAS is often performed in a much larger sample than the GWIS because of data availability. The environmental exposure may have different distributions in cohorts for conducting GWAS and GWIS (i.e., different mean and variance). Furthermore, models (1) and (2) are likely to be performed by two different groups of investigators, which will bring variation across studies in trait definitions, trait measurement procedures, quality control procedures, and covariates. Moreover, the summary statistics are obtained through meta-analyses in both GWAS and GWIS analyses, which can bring additional variation and confounding factors, including population stratification and cryptic relatedness, leading to a potentially invalid comparison between the marginal and main effects. In fact, it has been reported that the confounding of population stratification is not sufficiently corrected in large GWAS25,26. Therefore, directly using to screen the genome can be biased even for testing the combined contribution of interaction and mediation.
To overcome this bias, we note that the marginal effect estimate and the main effect estimate have a linear relationship,
| 3 |
where reflects the contribution of main effect to marginal effect, which converges to 1 when GWAS and GWIS are conducted using homogeneous measurements of phenotypes and environments (“Methods”). The genetic variants with no and no mediation will fall on the regression line but the variants with or mediation will depart from this line. We do not expect this pattern to be systematically impacted by the variation across studies. Therefore, we search the genetic variants that depart from this regression line to test the combined effect of and mediation, providing can be correctly estimated. This idea is conceptually the same as the MR framework when we introduce a pseudo exposure , representing a polygenic score of the trait (Fig. 1). We do not need to construct this pseudo exposure in our analysis because we work directly on the summary statistics under the MR framework. We then estimate the causal effect of the pseudo exposure on trait in the MR framework and the effect or mediation through is tested in the same way as testing for horizontally pleiotropic variants23. In doing so, we first select a set of independent variants associated with trait and perform the inverse variance weighted analysis to estimate θ, . Second, we test the or mediation of a genetic variant through by the statistic . This test can be performed by the iterative Mendelian randomization and pleiotropy (IMRP) approach23,27. The statistic is an asymptotically unbiased test for testing the combined effect of and mediation through (Supplementary Note).
Fig. 1. Illumination of Mendelian randomization and G × E.
A Left panel: the path diagram of the MR, where U refers to all confounders. Genetic variants (G) contributing to outcome Y through mediation of exposure X are often selected as the valid genetic instrumental variables (black paths). Genetic variants contributing to Y through both black and red paths independently are horizontal pleiotropic variants. Genetic variants contributing to Y through confounders (U) are invalid instrumental variables and need be blocked (x). Right panel: a scatter plot of effect sizes of genetic instrumental variants for an exposure and an outcome. Each + corresponds to the 95% confidence intervals of the exposure effect size (horizontal line segment) and the outcome effect size (vertical line segment). The horizontal pleiotropic variants (red +) depart from the regression line and can be separated from the variants with no pleiotropic effect (blue +). B Left panel: the framework, with the goal of testing . Instead of an explicit exposure, we create a pseudo exposure , which can be viewed as a polygenic score for trait Y based on marginal effect sizes. However, our analysis does not require estimating this pseudo exposure. The genetic variants associated with the pseudo exposure but not through either the environment E or are valid instrumental variables. The genetic variants interacting with E can be viewed the same as horizontally pleiotropic variants in the MR framework. Genetic variants associated with Y via mediation through E can contribute to both the pseudo exposure and Y, and thus have similar effects as and cannot be distinguished from . Thus, testing the combined effect of interaction and mediation is conceptually equivalent with testing the horizontally pleiotropic effect in the MR framework. Right panel: a scatter plot of genetic variants for GWIS main effects and GWAS marginal effects. Each + corresponds to the 95% confidence intervals of the GWIS main effect size (horizontal line segment) and the GWAS marginal effect size (vertical line segment). Like the horizontal pleiotropic variants in the MR framework, variants (red +) depart from the regression line and can be separated from variants with no assuming no mediation.
Two-step procedure for testing
Note that likely tests for the combined effect of and mediation unless and are independent (i.e., ). To test for , we propose a two-step procedure by using to screen the whole genome and then performing on the variants surviving the screen. We set the significance level at 5 × 10−8 for the first step ( test), and the significance level at 0.05/X for the step 2 test, where X is the number of independent significant variants in the first step/test. This two-step procedure can increase power at the screening step when there is interaction and mediation and increases power at the direct testing step by substantially reducing the multiple comparison burden. and are not independent (Supplementary Note), and therefore, the variants detected by the two-step procedure could still reflect the contribution of mediation and , and it is necessary for further replication by performing in independent data. To mitigate this problem, we can exclude the variants identified through GWAS of , which could represent large mediation effect.
Type I error rate and power of and the two-step procedure
We first performed a series of simulations to investigate the type-I error rate and power of in the absence of mediation. In simulations we observed that () is close to 1 and the estimate converges to 1 when sample size increases, which is expected by theoretical prediction (Fig. 2A and Supplementary Fig. 6a). The direct estimate of the interaction effect as well as of is also unbiased (Fig. 2B, Supplementary Fig. 6b), although the standard error of is affected by the environmental means in GWAS and GWIS. When no mediation is present, the type-I error rates for both and the direct test are well controlled (Fig. 2C and Supplementary Fig. 6c)). The power of depends on multiple parameters, including and allele frequency in GWAS and GWIS and is less powerful than when the environmental mean in GWAS is lower (Fig. 2D and Supplementary Fig. 6d). Additional simulations for the estimates of , interaction effect , type-I error rate and power are presented in Supplementary Figs. 7–9.
Fig. 2. Simulation performance of TMR_GXE and the two-step procedure.
A–D No medication was present. The simulation details were described in “Methods”. A Box plots of in simulations under different environments in GWAs data. The top and bottom edges of the box plots represent the 25th and 75th percentiles of , and the horizontal middle line represents the 50th percentile. The vertical bars extend from the 25th (or 75th) percentile of to the minimum (or maximum) value of simulated data. is close to 1 as expected. B Box plots of the direct estimate of in GWIS (top panel) and by through MR- analysis (bottom panel). The box plots are interpreted the same as in (A) accordingly. Both the estimates of and that by are unbiased. Here s = −1 refers to the scenario when the main effect and interaction effect have opposite effect directions; s = 0 refers to no main effect; and s = 1 refers to the scenario when the main effect and interaction effect have the same effect direction. C Type I error rate comparison between and the direct test for different main and interaction effect directions. Both and the direct test maintain the type I error rate well. D Power comparison between and the direct test for different main and interaction effect directions. E, F 20 variants were tested when mediation was present or not. The simulation details were described in ”Methods”. E Type I error comparison for , and two-step procedure. The dash lines represent the 95% confidence interval. F Power comparison for , and two-step procedure.
We next investigated the performance of , and the two-step procedure when mediation is present and multiple variants are tested. We generated 20 independent variants with one variant having mediation, interaction, or both. All three tests have well controlled type I error rates when mediation is absent (Fig. 2E and Supplementary Fig. 10A). When mediation is present, the type-I error rate was still well controlled, although inflation can be observed for the two-step test and when contributes to 5% of the outcome variation and the samples between GWAS and GWIS are completely overlapped (Supplementary Fig. 10B, C). This inflation was caused by mediation and quickly disappeared when the overlapping rate between GWAS and GWIS subjects was reduced. The statistical power of and the two-step procedure for testing was much more improved than when mediation was present (Fig. 2F and Supplementary Fig. 10D–F).
Identifying gene-smoking and gene-alcohol drinking interactions to serum lipids
We applied the two-step procedure to search for genetic variants interacting with cigarette smoking and alcohol drinking for serum lipids, using the summary statistics of high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG) from the GLGC (n = 1.65 M) and the CHARGE GLI (n = 134 K). To mitigate the effects of mediation through cigarette smoking or alcohol drinking, we excluded all loci with P-value < 5 × 10−7 reported in the early GWAS of cigarette smoking status or alcohol drinking28, which represent relatively large effect sizes of variants on cigarette smoking and alcohol drinking. We observed that ranged from 0.92−1.33, 0.95−1.62, 0.83−1.25, 0.87−1.37, and 0.95–1.28 for European, African, Asian, Hispanic, and Cross-population data, respectively (Supplementary Data S1). The departure of from 1 suggests that the phenotype treatments, analysis protocols, and corrections for population structure were not identical between the GLGC and CHARGE GLI consortiums. For example, CHARGE GLI performed a natural logarithmic transformation to the lipid measurements, whereas GLGC further performed an inverse normal transformation. The number of principal components (PCs) for correcting populations was also different between GLGC and CHARGE GLI. Despite these discrepancies, we did not observe an inflation for , with the genomic control values being close to 1 (range 0.93–1.05, Supplementary Data S2).
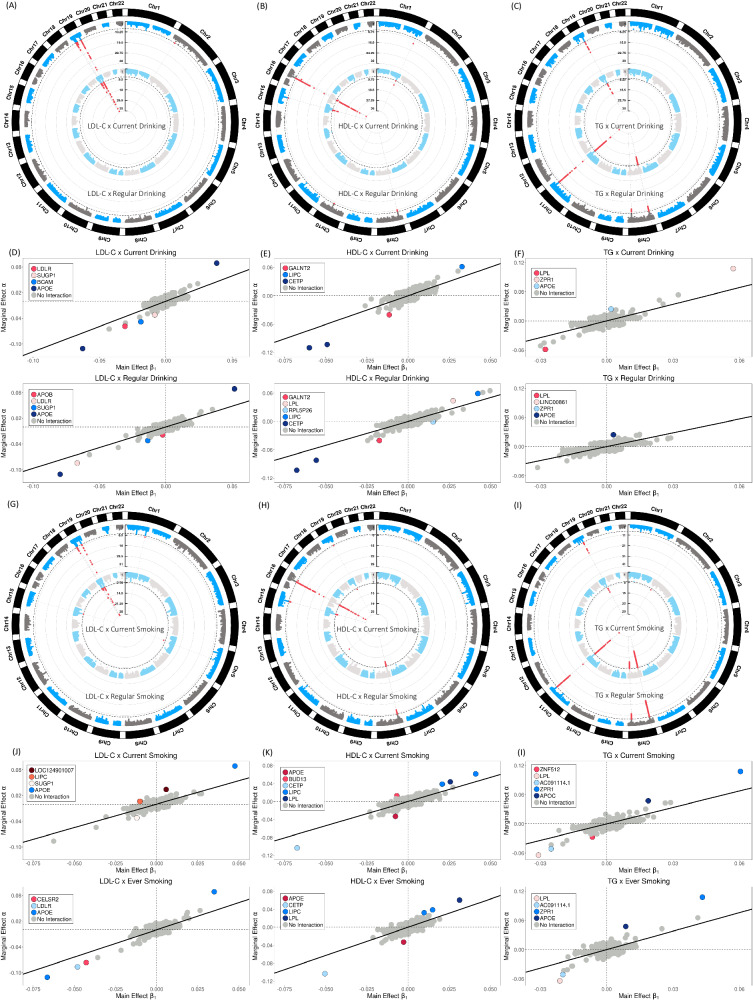
Using to screen the genome, we observed 15 genome-wide significant loci consisting of 17 independent signals (P < 5 × 10−8, ), including 4 and 5 loci for LDL-C, 7 and 5 loci for HDL-C, and 5 and 6 loci for TG, interacting with cigarette smoking and alcohol drinking or mediating through them, respectively (Fig. 3A–C, G–I, Supplementary Data S3a). All but 3 loci have been reported to be associated with either cigarette smoking or alcohol drinking in the recent largest GWAS study with over 3 million samples29, suggesting the contribution of both and mediation. Since we already excluded the cigarette smoking and alcohol drinking variants identified from a relatively smaller study28, these detected variants should represent modest mediation effects. Locus-specific plots of all significantly associated loci are presented in Supplementary Fig. 11, which suggest that multiple protein-coding genes are present in these loci. Strikingly, all the loci have previously been mapped to lipids traits except RPL5P26 on chromosome 10. The or mediation loci are clearly departing from most of the lipids-associated variants (Fig. 3D–F, J–L). The population-specific results are presented in Supplementary Fig. 12, which are also consistent with the Cross-population results, although the main contribution comes from the European population.
Fig. 3. Manhattan plots, marginal and main effect size comparisons.
The circle Manhattan plots of gene × alcohol drinking interactions for A LDL-C; B HDL-C; and C TG, respectively. The genome-wide significant loci are presented in red dots. The marginal and main effect sizes corresponding to alcohol drinking for D LDL-C; E HDL-C, and F TG, respectively. The colored circles represent the genome-wide significant loci and gray circles represent insignificant loci by test. The circle Manhattan plots of gene × cigarette smoking interactions for G LDL-C; H HDL-C; and I TG, respectively. The marginal and main effect sizes corresponding to cigarette smoking for J LDL-C; K HDL-C, and L TG, respectively.
By applying the two-step procedure, we observed that 8 of the 17 independent signals are significant when using the direct test after correcting for the 17 tests and 4 environmental factors (Table 1, P < 7.35 × 10−4). In comparison to the direct test in GWIS, the two-step procedure identified more signals for each of the three lipid traits and four environmental factors (Supplementary Data S3b). This provides additional support for the enhanced statistical power of the two-step procedure. The tissue enrichment analysis using the GWAS-based pathway analysis tools, MAGMA30 and FUMA31, suggest that these loci are enriched in liver, hippocampus, small intestine, and stomach tissues (Supplementary Fig. 13). Multiple loci were colocalized with expression quantitative trait loci (eQTLs) in the corresponding liver, lung, and blood tissues in the genotype-tissue expression database (GTEx)32 (Supplementary Fig. 14).
Table 1.
Interaction loci screened by and followed by the direct test in GLI (two-step test) and replicated by the direct test in UK Biobank
| Mapping Gene | CHR: BP | Lead SNP | Environmental factor | Lipid traits | MR_GxE test P-value | GLI direct test P-value | UKBB direct test P-value | Combined GLI and UKBB direct test P-value |
|---|---|---|---|---|---|---|---|---|
| Signals identified by (P < 5E−08), by (P < 7.35E−04) and replicated by in UKBB (P < 1.56E−3) or combined GLI and UKBB P < 5E−8 | ||||||||
| BUD13* | 11:116637146 | rs12294259 | Regular Drinking | TG | 2.47E−18 | 3.61E−06a | 1.97E−04a | 2.14E−08 |
| 11:116657561 | rs3741298 | Current Smoking | TG | 2.80E−13 | 1.16E−10a | 4.24E−01a | 6.99E−10 | |
| CETP | 16:57000696 | rs8045855 | Current Drinking | HDL-C | 6.12E−24 | 1.85E−07a | 4.97E−07a | 4.05E−12 |
| 16:57006829 | rs289713 | Regular Drinking | HDL-C | 5.01E−19 | 3.63E−07a | 3.16E−06a | 4.62E−11 | |
| BCAM* | 19:45392254 | rs6857 | Regular Drinking | LDL-C | 4.02E−12 | 1.28E−06a | 2.95E−04a | 8.57E−09 |
|
NECTIN2* TOMM40 APOE APCO1 |
19:45422946 | rs4420638 | Regular Drinking | LDL-C | 6.55E−36 | 4.41E−05a | 1.95E−06a | 2.08E−09 |
| LPL* | 8:19830170 | rs1569209 | Current Smoking | TG | 4.77E−10 | 1.01E−13b | 3.49E−02b | 1.04E−13 |
| SMARCA4 | 19:11191677 | rs10402112 | Regular Drink | LDL-C | 1.85E−15 | 5.75E−04a | 9.04E−04a | 8.04E−06 |
| Signals identified by (P < 5E−08) and by (P < 7.35E−04) but failed in UKBB replication | ||||||||
| RPL5P26* | 10:71533084 | rs11591480 | Regular Drinking | HDL-C | 3.34E−08 | 1.11E−04a | 5.23E−02a | 8.69E−05 |
| ZPR1** | 11:116662579 | rs651821 | Ever Smoking | TG | 7.34E−17 | 3.44E−05a | 6.87E−01a | 1.73E−04 |
The P-values of and TDirect are two-sided P-values based on Z-scores. The P-values in the last column (combined GLI and UKBB direct Test P-value) are calculated from a chi-square test with 4 degrees of freedom. All P-values were not adjusted for multiple comparisons. The bold P-values represent significant variants after adjusting for multiple comparisons.
*The locus has been reported to be associated with cigarette smoking.
**The locus has been reported to be associated with both cigarette smoking and alcohol drinking.
aThe interaction effect direction is the same in GLI and UKBB. Detailed effect sizes and standard errors are presented in Supplementary Data S3a.
bThe interaction effect direction is opposite in GLI and UKBB. Detailed effect sizes and standard errors are presented in Supplementary Data S3a.
Independent replication
We next attempted to replicate the evidence for these 8 independent signals in the UKBB. Although the UKBB data accounted for about one third of samples in the GLGC consortium, the direct test statistic calculated in the UKBB is independent of , so are the test statistics calculated in UKBB and CHARGE GLI, thus qualifying this as an independent replication (Supplementary Note). Six of the 8 signals are replicated in the UKBB after adjusting for 32 tests (P < 1.56 × 10−3), and 5 of them were genome-wide significant by the test in combined CHARGE GLI and UK Biobank data (Table 1). All 8 independent signals have the same interaction direction in CHARGE GLI and UKBB except LPL, which is not significant in UKBB (Supplementary Data S3a). The CETP and SMARCA4 loci are the only two loci with no reported mediation evidence through either cigarette smoking or alcohol drinking.
We now aim to understand if the interaction evidence observed in this study has an alternative explanation33 because of linkage disequilibrium (LD) with a variant which has causal effect on cigarette smoking or alcohol drinking. To examine this, we searched if there exists a variant(s) at each of the loci in Table 1 explaining the observed interaction evidence in the UKBB. However, we did not observe such variants (Supplementary Fig. 15), suggesting that the interaction evidence presented in Table 1 is genuine. In total, we identified 5 loci consisting of 6 independent signals that have evidence of interaction with either cigarette smoking or alcohol drinking.
interaction and mediation to SNP heritability
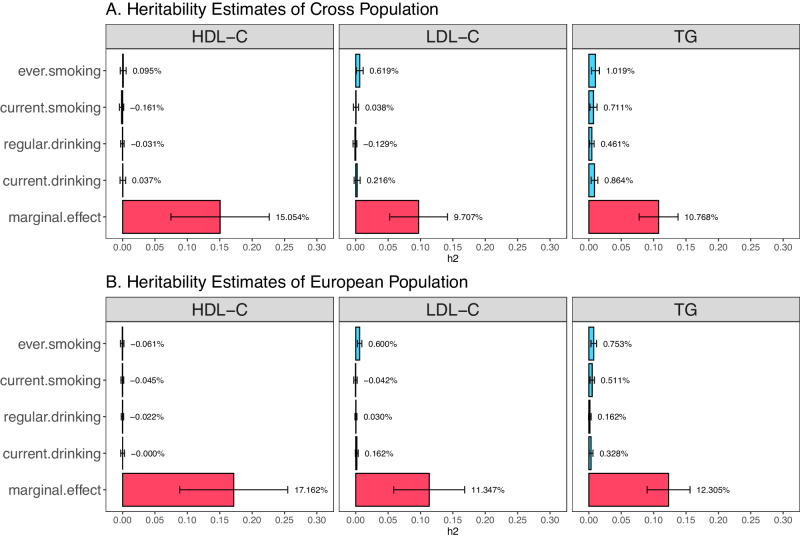
Since refers to the combined interaction and mediation contribution to the marginal effect, we use to estimate the heritability contributed by the interaction and mediation through the LD score (LDSC) regression34. Note that this heritability is a lower bound of the phenotype variance contributed by the and mediation through E and is a part of the heritability estimated through the marginal effect, which is often referred to as the SNP-heritability in GWAS. In both Cross-Population (Fig. 4A) and European population (Fig. 4B), we observed significant interaction and mediation heritability (P < 0.03) with ever cigarette smoking for LDL-C, and alcohol consumption or cigarette smoking for TG, suggesting that the heritability estimates based on marginal effects also include significant contributions from and mediation through the corresponding environment factors (Supplementary Data S4).
Fig. 4. The estimated heritability of HDL-C, LDL-C, and TG using LDSC regression.
A Cross-Population. B European population. X-axis represents heritability in percentage. Y-axis represents the corresponding heritability estimated in percentage (marginal.effect: marginal effect heritability; current.drinking: gene and current drinking interaction effect heritability; regular.drinking: gene and regular drinking interaction effect heritability; current.smoking: gene and current smoking interaction effect heritability; ever.smoking: gene and ever smoking interaction effect heritability). Marginal effect heritability refers to the heritability estimated through the marginal effect , and interaction effect heritability refers to the heritability estimated through . The percentage number displayed on the right side of each bar represents the estimated heritability, and the corresponding 95% confidence interval shown as horizontal error bars. For the marginal effect analysis, the sample size is 1.65 M and 1.32 M for cross-population and European population analysis, respectively. For the interaction effect analysis, the sample size is 134 K and 80 K for cross-population and European population analysis, respectively.
interaction and mediation to heterogeneity of genetic effect sizes across populations
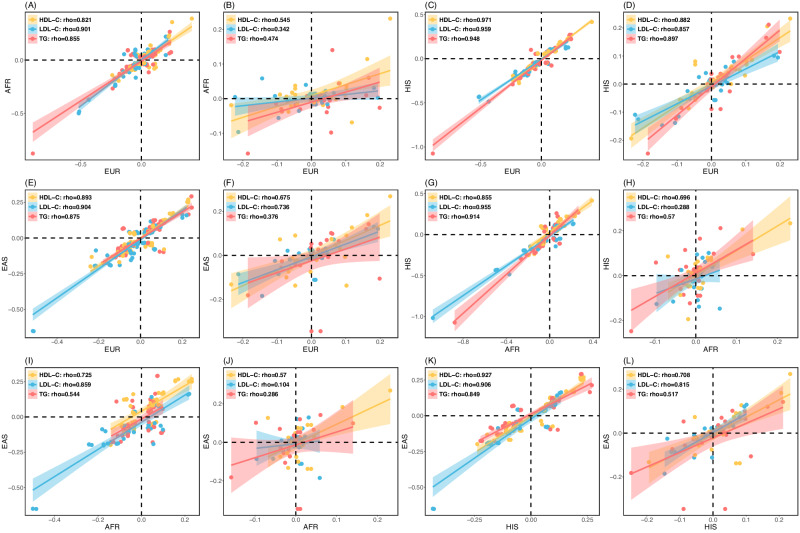
As noted in Eq. (3), the marginal effect estimate of a genetic variant in GWAS consists of the and mediation contribution when the and mediation occur. Because of the environment heterogeneity across populations, we expected that the marginal effect sizes of the variants will be less correlated across populations for the variants with than without interaction or mediation. We calculated the marginal effect size correlations between European, African, Hispanics, and Eastern Asian for these variants reported in Graham et al3 after excluding the variants in Supplementary Data S3a where their interactions or mediations were observed in this study. Similarly, we calculated the marginal effect size correlations for the variants in Supplementary Data S3a. We compared the correlation and observed a median of 24.4% drop of the cross-population correlation coefficient (Fig. 5), strongly support that interactions or mediations contribute to the marginal effect size heterogeneity across populations.
Fig. 5. Cross-population comparison of the LDL-C, HDL-C, and TG marginal effect sizes of the variants reported in Graham et al.3.
A EUR vs AFR, no interaction or mediation. B EUR vs AFR, interaction or mediation. C EUR vs HIS, no interaction or mediation. D EUR vs HIS, interaction or mediation. E EUR vs EAS, no interaction or mediation. F EUR vs EAS, interaction or mediation. G AFR vs HIS, no interaction or mediation. H AFR vs HIS, interaction or mediation. I AFR vs EAS, no interaction or mediation. J AFR vs EAS, interaction or mediation. K HIS vs EAS, no interaction or mediation. L HIS vs EAS, interaction or mediation. The variants with no interaction or mediation are those not included in Supplementary Data S3a. The variants with interaction or mediation are those in Supplementary Data S3a. We only included independent variants. The shadow error bands represent the 95% confidence intervals. Clearly the variants without interactions or mediations have substantially larger cross-population correlations than the variants with interactions or mediations, suggesting that interactions or mediations contribute the marginal effect size heterogeneity across populations. (European (EUR), African (AFR), Hispanics (HIS), Eastern Asian (EAS)).
Discussion
In this study, we utilize marginal effects from GWAS to search for . We conceptually demonstrated the deep connection between detecting and MR for causal inference. Although tests for the combined effect of and mediation, the two-step procedure of followed by in fact tests for , and its statistical power is much improved because of the following reasons: (1) the step 1 can increase power when a genetic variant has a mediation effect through the environmental factor. In this case, we expect a larger difference between the marginal effect and the main effect (Eq. (3)) than no mediation. (2) The difference between the marginal and main effect can further increase when the environmental distributions between GWAS and GWIS cohorts are different (Eq. (3) and Fig. 1D). (3) At the two-step procedure, multiple comparison burden is significantly alleviated because only significant variants survived at step 1 need to be examined. As demonstrated in this study, the two-step procedure identified 8 independent signals in comparing with two by the direct test in GWIS. This is also consistent with when comparing with the direct test in GWIS, the two-step procedure identified more signals for each of the three lipid traits and four environmental factors (Supplementary Data S3b). Detecting using direct tests can be biased by unmeasured confounders due to omitting covariates in the regression models35, but the two-step procedure is robust because is not affected by confounders such as population structure. Considering the advantages of the two-step procedure, we view it as a complement rather than a replacement of the direct test. This perspective arises from the fact that the two-step test necessitates additional GWAS summary statistics and may be less powerful than the direct test in some situations (Fig. 1D).
Our study demonstrated that the current heritability estimates based on marginal effects could also include contributions from and mediation through the corresponding environment factors (Fig. 4 and Supplementary Data S4). We excluded cigarette smoking- or alcohol drinking-associated variants identified from a large cigarette smoking and alcohol consumption GWAS of 1.2 million individuals28 in our analysis, which mitigates the potential mediation contribution in the analysis. However, among the 15 loci identified by , only three were not reported in the much larger recent cigarette smoking and alcohol consumption GWAS of 3.4 million individuals, suggesting mediation through cigarette smoking and/or alcohol consumption is still present but with modest effects. Among the six variants identified, 4 are associated with either cigarette smoking or alcohol drinking, suggesting that the variants are also likely to be mediated through E and the mediation improves power to detect . Furthermore, we demonstrated that the current SNP heritability estimates based on marginal effects could also include significant contributions from and mediation through the corresponding environment factors for LDL-C and TG (Fig. 4 and Supplementary Data S4). We did not observe significant contributions from and mediation to heritability for HDL-C, potentially attributable to the relatively small sample sizes in our GWIS. Since the LDSC regression34 cannot be used to estimate heritability, our estimates reflect the low bound of the interaction and environmentally mediated heritability. We therefore suggest that the current SNP heritability estimates based on the marginal genetic effects be called marginal SNP heritability, to differentiate it from narrow-sense heritability36 that is defined by additive genetic actions without the inclusion of or mediation contributions. We believe this differentiation is important for correctly interpreting the current heritability estimates and understanding the genetic architecture of complex traits.
The 5 (6 independent signals) replicated loci interacting with cigarette smoking and alcohol consumption contain genes that are enriched in liver tissue, possibly reflecting the effect of alcohol drinking on aspartate amino transferase, alanine aminotransferase and γ-glutamyl transferase activities via the actions of numerous ingredients that alter the activities of enzymes found in the liver37. Among them, the interaction between alcohol consumption and cholesteryl ester transfer protein (CETP) has been reported for HDL-C and coronary artery disease38–40. The interaction between alcohol consumption and APOE on LDL-C has also been reported in a Mediterranean Spanish population41, while the interactions between APOA5 and cigarette smoking and alcohol drinking status associated with elevated TG and reduced HDL-C were observed in the Chinese and Korean populations42,43. However, our study is the only well-powered study demonstrating significant evidence at the genome-wide level and the interaction loci are replicable. SMARCA4 was reported to be associated with LDL-C in the lipids GWAS in Africans44 but not in the recent largest lipids GWAS which is predominantly European ancestry3. Overall, the marginal effect sizes of the variants are less correlated across populations for the variants with than without interaction or mediation (Fig. 5), empirically verified that and mediation contribute to marginal effect differences across different populations45. We expect that including interactions should improve polygenetic risk score prediction across populations.
It is well known that causal effect estimate in MR framework can be biased when the three IV assumptions are violated. However, our goal is to detect rather than to estimate the causal effect. Detecting based on MR is less likely to be biased for these reasons: (1) the effect sizes of IVs on the pseudo exposure are all highly significant in GWAS, which represent strong IVs. (2) It is less likely to have a confounding effect between a trait and its pseudo exposure, i.e., a polygenic score. (3) The iterative Mendelian randomization and pleiotropy test is a powerful method to detect pleiotropy when the two IV conditions are satisfied23, in particular, it is expected that most of the IVs are not interacted with E. (4) Although the causal effect estimate can be affected if population structure is not well corrected, testing by is not. The reason is that can be viewed as a weighted linear regression of the effect size of GWAS on the effect size of GWIS followed by searching the outlies of variants departing from the regression line. While the regression line (equivalent to causal effect estimate in MR) can be affected by population structure, the outlie detection is not.
In summary, our approach is powerful and able to detect genetic loci interacting with environments that account for significant phenotypic variability. Our findings indicate that the contribution of in lipids is not ignorable. Our study only focuses on the interactions of genes with cigarette smoking and alcohol consumption in lipids. The cumulative interaction contribution with many environmental factors can even be greater. Detecting individual genetic loci with environmental interactions facilitates a better understanding of the genetic architecture of complex traits and can improve phenotype prediction.
Methods
Summary statistics data
The marginal summary statistics of high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG) from the Global Lipids Genetics Consortium study (GLGC, n = 1.65 M)3 were downloaded at http://csg.sph.umich.edu/willer/public/glgc-lipids2021.
GLGC consists of GWAS results from 1.65 M subjects representing five genetic ancestry groups: European (N = 1.32 M); African or admixed African (N = 99 K); East Asian (N = 146 K); Hispanic (N = 48 K); and South Asian (N = 41 K). We did not perform South Asian specific analysis because there was no corresponding GWIS in the Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) consortium. The GWIS summary statistics from CHARGE gene-lifestyle (GLI) working group in this study are available via dbGaP (accession number phs000930). The CHARGE GWIS consists of 60 GWIS summary datasets: (LDL-C, HDL-C, and TG)-current smoking, (LDL-C, HDL-C, and TG)-ever smoking, (LDL-C, HDL-C, and TG)-current alcohol drinking, and (LDL-C, HDL-C, and TG)-regular alcohol drinking, for European, African or Admixed African, East Asian, Hispanic and multi-ancestry.
QCs for performing analysis
To perform MR analysis, we aligned the GWAS summary statistics HDL-C, LDL-C, and TG from the GLGL with the corresponding GWIS summary statistics from the CHARGE gene-lifestyle consortium. We flipped the effect size if the corresponding reference allele did not match. We dropped a genetic variant if the two alleles were either {A, T} or {C, G}. We also excluded any variants with minor allele frequency (MAF) difference larger than 0.15 between GWAS and GWIS study. If multiple variants fall on the same chromosome position, we required the matched variants with MAF difference less than 0.01. We further excluded any variants with the effective sample size in GLGC trans-ethics or European less than 100 K and the other populations (African, Hispanic, East Asian) less than 30 K. To reduce the effect by mediations through the smoking and alcohol drinking, we excluded all loci with P-value < 5E−7 identified by the GWAS of smoking status or alcohol drinking28.
analysis
To perform , we applied the Mendelian randomization (MR) software IMRP23 to estimate the causal effect by considering the main effect sizes from the GWIS of the CHARGE gene-lifestyle consortium as the exposure effects, and the marginal effects from the GLGC as the outcome effects, respectively. To identify instrument variables, we first selected all the variants with the P-value < 5E−8 after GC-correction in the GLGC, and then pruned them using the window size 500 KB and value 0.1 by the Plink software46. We standardized the effect sizes as in27. IMRP requires the input of the correlation coefficient to account for the effect of sample overlapping between GWAS and GWIS cohorts and this correlation was calculated based on the unsignificant variants (P-value > 0.05) across the genome. After estimate the causal effect, we performed which is equivalent to the pleiotropy test in the IMRP, to all the genetic variants across the genome.
Independent locus definition
Independent loci were defined as the regions within 1 Mb of the most significant variants by the test. Independent signals were defined as the variants in a locus with < 0.1. The 1000 G data was used as the reference genetic data for LD calculation.
Choosing independent variants for replication in UK Biobank
By applying , we observed that 15 genome-wide significant loci consisting of 17 independent signals (P-value < 5E−8), including 4 and 5 loci for LDL-C, 7 and 5 loci for HDL-C, and 5 and 6 loci for TG, interacted with alcohol drinking and cigarette smoking, respectively (Supplementary Data S3a). At a locus with the significant (P-value < 5E−8) for a lipid trait (LDL-C, HDL-C, or TG) and environment (smoking or alcohol drinking), we searched the variant with the smallest P-value of the direct test among the significant variants by the . The variants with P-value < 7.35E−4, which correct for the 17 tests and 4 environmental factors, were considered as significant for interaction (two-step procedure). We obtained 8 independent variants in 6 loci among these 17 independent signals survived the threshold P-value = 7.35E−4 and these variants were further tested for the replication of the interaction effects in UK Biobank using test.
LD score regression
We applied the LD score regression to estimate heritability contributed by interaction and mediation through the environment factor . We estimated heritability by combining all chromosomes rather than chromosome specifically. We used the R package bigsnpr47 to estimate LD scores in the corresponding populations from 1000 G reference data with default settings.
Functional mapping and annotation
We performed overall enrichment tests using the residual as the effect size and as the corresponding standard error. We used MAGMA30 (Multi-marker Analysis of GenoMic Annotation) and DEPICT48 (Data-driven Expression Prioritized Integration for Complex Traits) to identify tissues and cells that are highly expressed at genes within the loci. We also used DEPICT to test for enrichment in gene sets associated with gene ontology (GO) ontologies, mouse knockout phenotypes and protein-protein interaction networks. In addition, we reported significant enrichments with a false discovery rate 0.05. Analysis was done using the online platform FUMA GWAS.
Colocalization
We performed colocalization analysis by using the software ezQTL (https://analysistools.cancer.gov/ezqtl/#/home). We chose the public genotype-tissue expression (GTEx) v7 with eQTL32 as the QTL data and chose the public European reference panels for calculating the LD data. We performed colocalization analysis between GWIS and QTL results within a locus using eCAVIAR (eQTL and GWAS Causal Variant Identification in Associated Regions)49, where the Colocalization Posterior Probability (CLPP) is used to describe the significance level of colocalization. We only recorded colocalization with CLPP > 0.01, as suggested by the authors of eCAVIAR.
UK Biobank individual level data for replication
The UK Biobank (UKBB)50 individual-level data used for replications were available through Application ID: 81097. Quality Controls Participants in the UKBB were genotyped using a custom Affymetrix UK Biobank Axiom array51. Genotypes were imputed by the UKBB using the Haplotype Reference Consortium reference panel52 with imputation value greater than 0.3. Related individuals with pairwise kinship coefficient greater than 0.0884 (suggested by UKBB) were removed from analysis, resulting in N = 445,424 individuals of European, African, and East Asian ancestries. The principal components were calculated by UKBB with genotype data within each ancestry to account for population structure. These data were independent of GLI cohorts and consisted of European, African, and Asian individuals (race determined using UKBB field ID 21000-0.0) in UKBB who were unrelated (genetic kinship coefficient less than 0.0884; 22021-0.0). Linear regression model (1) in main text was performed. Covariates included age at assessment (21003-0.0), age2, sex (31-0.0), the first 10 PCs (22009-0.1 to 22009-0.10), and environment exposure, a genetic variant and their interaction. Environmental exposures included ever/never smoking status (20116-0.0), current/non-current smoking status (20116-0.0), and alcohol intake frequency (1558-0.0).
Analogous to the analysis in ref. 17, HDL-C (30760-0.0) and TG (30870-0.0) measurements were natural log transformed and LDL-C measurements (30780-0.0) were converted from mmol/L to mg/dl then multiplied by a factor of 0.7 if there was a history of lipid-lowering medication (6177-0.0) present. LDL-C measurements were therefore considered no medication if there were missing values for medication history. This introduced missing values in LDL-C for 248,419 individuals.
Theoretical properties of
In MR analysis, the instrumental variables are independent and are genome-wide significant variants selected from GWAS. Let and , , be the corresponding effect size estimates in GWIS (model (1) and GWAS (model 2)) for the m instrument variables.
The causal effect of the inverse variance weighted (IVW) is estimated by
| 4 |
It is much simply to work on by standardizing the IVs and this procedure does not change the conclusion. Thus, we let , j = 1 …, m, in both GWAS and GWIS data. Further, we let the phenotype residue variance By equation (S15) in Supplementary Note, we have
Since only the variants without either interaction or mediation are valid in the MR analysis, we assume (no mediation) and (no interaction). We have
| 5 |
By applying the Slutsky’s theorem, and let , we have:
| 6 |
Because , is the average phenotypic variance accounted by an IV. Define , we have:
| 7 |
which converges to 1 when n1 and n2→∞. However, when is small (weak instrument in MR analysis), the converge of to 1 is slow. We also note that .
Simulation settings without medication contribution (Fig. 2A–D, Supplementary Figs. 6–9)
For ith individual, we generated m = 102 number of independent variants through for by , where pj∼unifom (0.05,0.5). We standardized genotypes by For the environment factor in the GWAS model, we generated . For the environment factor in the GWIS model, we generated . For the samples overlapped between the GWAS and GWIS, we generated their environment values through We varied the values of , and the proportion of overlapped samples.
The main effect size of the jth variant was generate by where is the trait variance accounted for by the IVs. For the first variant, we added its interaction effect with E. The phenotype by generated by
| 8 |
where . The causal effect was estimated using the last 100 variants as the IVs. The power and type I error rate for and were calculated based on the first and second variants, respectively.
Simulation settings without medication contribution (Fig. 2E–F), Supplementary Fig. 10
We generated independent variants by and standardized it but without mean correction. We simulated environment E according to mediation present or not present. If no mediation, is generated from . If there is mediation, , or contributes 0.25% variation of E. The phenotype is generated according to the following models:
No mediation and no interaction: , where
Mediation but no interaction: , where .
Mediation and interaction: , where .
We let take values of 1 and . We also simulated data with environment mean 0.5 (Supplementary Fig. 10). We first simulated subjects for GWIS cohort (or main effect estimation). The sample size for marginal effect estimation varied from to 300,000, with the 20,000 subjects in GWIS cohort was always included. For the non-overlapped subjects, we let the environment mean to be 1.5 times of the environment mean in GWIS cohort. The type I error and power for and were calculate by correcting for 20 tests using the Bonferroni correction. For the two-step procedure, we first applied and Bonferroni correction. The variants survived after were further tested by and Bonferroni correction was further applied.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by grant R01HG011052 and R01HG01152-03S1 (to X.Z.) from the National Human Genome Research Institute (NHGRI) and R01HL118305 and R01HL156991 (to D.R.) from the National Heart, Lung and Blood Institute. This work was also supported in part by the Intramural Research Program of NHGRI through the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is also supported by the National Institute of Diabetes and Digestive and Kidney Diseases and the Office of the Director of the National Institutes of Health (Z01HG200362 to A.R.B.).
Author contributions
X.Z. conceived and designed the study. X.Z., Y.Y., N.L., and G.L. performed analysis. X.Z. drafted the initial manuscript with inputs from others. X.Z., Y.Y., N.L., G.L., A.R.B., P.S.d.V., M.B., A.C.M, C.N.R., W.J.G., D.R., and H.A. critically revised and approved the manuscript.
Peer review
Peer review information
Nature Communications thanks Wei Pan and Doug Speed for their contribution to the peer review of this work. A peer review file is available.
Data availability
The marginal summary statistics of HDL-C, LDL-C, and TG from the Global Lipids Genetics Consortium study (GLGC, n = 1.65 M) were downloaded at http://csg.sph.umich.edu/willer/public/glgc-lipids2021. GLGC consists of GWAS results from 1.65 M subjects representing five genetic ancestry groups: European (N = 1.32 M); African or admixed African (N = 99k); East Asian (N = 146k); Hispanic (N = 48k); and South Asian (N = 41k). We did not perform South Asian specific analysis because there was no corresponding GWIS in the Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) consortium. The GWIS summary statistics from CHARGE gene-lifestyle (GLI) working group in this study are available via dbGaP (accession number phs000930). The UKBB individual-level data for replications were available through Application ID: 81097.
Code availability
test was used the software IMRP which is available in the Github repository with the following link, https://github.com/XiaofengZhuCase/IMRP23. Heritability analysis was performed by Bigsnpr, https://privefl.github.io/bigsnpr/reference/snp_ld_scores.html47 and LDSC regression, https://github.com/bulik/ldsc47. FUMA: https://fuma.ctglab.nl/31. Software ezQTL: https://analysistools.cancer.gov/ezqtl/#/home. MAGMA: https://ctg.cncr.nl/software/magma30. DEPICT: https://github.com/perslab/depict48. Plink: https://www.cog-genomics.org/plink2/46. The codes for performing simulations and analyzing interaction of lipids data were deposited in the Zenodo database53.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-47806-3.
References
- 1.Manolio TA. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 2.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6:109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 3.Graham SE, et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yengo L, et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610:704–712. doi: 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdellaoui A, Yengo L, Verweij KJH, Visscher PM. 15 years of GWAS discovery: realizing the promise. Am. J. Hum. Genet. 2023;110:179–194. doi: 10.1016/j.ajhg.2022.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter DJ. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 7.Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Elston RC, Zhu X. The meaning of interaction. Hum. Hered. 2010;70:269–277. doi: 10.1159/000321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao DC, et al. Multiancestry study of gene-lifestyle interactions for cardiovascular traits in 610 475 individuals from 124 cohorts: design and rationale. Circ. Cardiovasc Genet. 2017;10:e001649. doi: 10.1161/CIRCGENETICS.116.001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4:e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: genetic interactions create phantom heritability. Proc. Natl Acad. Sci. USA. 2012;109:1193–1198. doi: 10.1073/pnas.1119675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang W, Mackay TF. The genetic architecture of quantitative traits cannot be inferred from variance component analysis. PLoS Genet. 2016;12:e1006421. doi: 10.1371/journal.pgen.1006421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow JF. On epistasis: why it is unimportant in polygenic directional selection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1241–1244. doi: 10.1098/rstb.2009.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aschard H. A perspective on interaction effects in genetic association studies. Genet. Epidemiol. 2016;40:678–688. doi: 10.1002/gepi.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laville V, et al. Gene-lifestyle interactions in the genomics of human complex traits. Eur. J. Hum. Genet. 2022;30:730–739. doi: 10.1038/s41431-022-01045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sung YJ, et al. A large-scale multi-ancestry genome-wide study accounting for smoking behavior identifies multiple significant loci for blood pressure. Am. J. Hum. Genet. 2018;102:375–400. doi: 10.1016/j.ajhg.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentley AR, et al. Multi-ancestry genome-wide gene-smoking interaction study of 387,272 individuals identifies new loci associated with serum lipids. Nat. Genet. 2019;51:636–648. doi: 10.1038/s41588-019-0378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vries PS, et al. Multiancestry genome-wide association study of lipid levels incorporating gene-alcohol interactions. Am. J. Epidemiol. 2019;188:1033–1054. doi: 10.1093/aje/kwz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kilpelainen TO, et al. Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat. Commun. 2019;10:376. doi: 10.1038/s41467-018-08008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith GD, Ebrahim S. Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 21.Smith GD, et al. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gage SH, Davey Smith G, Ware JJ, Flint J, Munafo MR. G = E: what GWAS can tell us about the environment. PLoS Genet. 2016;12:e1005765. doi: 10.1371/journal.pgen.1005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Li X, Xu R, Wang T. An iterative approach to detect pleiotropy and perform Mendelian Randomization analysis using GWAS summary statistics. Bioinformatics. 2021;37:1390–1400. doi: 10.1093/bioinformatics/btaa985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg JJ, et al. Reduced signal for polygenic adaptation of height in UK Biobank. Elife. 2019;8:e39725. doi: 10.7554/eLife.39725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sohail M, et al. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. Elife. 2019;8:e39702. doi: 10.7554/eLife.39702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Zhu L, Wang H, Cooper RS, Chakravarti A. Genome-wide pleiotropy analysis identifies novel blood pressure variants and improves its polygenic risk scores. Genet Epidemiol. 2022;46:105–121. doi: 10.1002/gepi.22440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M, et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saunders, G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature, 612, 720–724 (2022). [DOI] [PMC free article] [PubMed]
- 30.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium GT, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood AR, et al. Another explanation for apparent epistasis. Nature. 2014;514:E3–E5. doi: 10.1038/nature13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller MC. Gene x environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol. Psychiatry. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics (Longman, 1996).
- 37.Wannamethee SG, Shaper AG. Cigarette smoking and serum liver enzymes: the role of alcohol and inflammation. Ann. Clin. Biochem. 2010;47:321–326. doi: 10.1258/acb.2010.009303. [DOI] [PubMed] [Google Scholar]
- 38.Fumeron F, et al. Alcohol intake modulates the effect of a polymorphism of the cholesteryl ester transfer protein gene on plasma high density lipoprotein and the risk of myocardial infarction. J. Clin. Invest. 1995;96:1664–1671. doi: 10.1172/JCI118207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dachet C, Poirier O, Cambien F, Chapman J, Rouis M. New functional promoter polymorphism, CETP/-629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb. Vasc. Biol. 2000;20:507–515. doi: 10.1161/01.ATV.20.2.507. [DOI] [PubMed] [Google Scholar]
- 40.Williams PT. Quantile-dependent expressivity and gene-lifestyle interactions involving high-density lipoprotein cholesterol. Lifestyle Genom. 2021;14:1–19. doi: 10.1159/000511421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corella D, et al. Environmental factors modulate the effect of the APOE genetic polymorphism on plasma lipid concentrations: ecogenetic studies in a Mediterranean Spanish population. Metabolism. 2001;50:936–944. doi: 10.1053/meta.2001.24867. [DOI] [PubMed] [Google Scholar]
- 42.Lin E, et al. Association and interaction of APOA5, BUD13, CETP, LIPA and health-related behavior with metabolic syndrome in a Taiwanese population. Sci. Rep. 2016;6:36830. doi: 10.1038/srep36830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S, Kang S. Alcohol, carbohydrate, and calcium intakes and smoking interactions with APOA5 rs662799 and rs2266788 were associated with elevated plasma triglyceride concentrations in a cross-sectional study of Korean adults. J. Acad. Nutr. Diet. 2020;120:1318–1329 e1. doi: 10.1016/j.jand.2020.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Bentley AR, et al. GWAS in Africans identifies novel lipids loci and demonstrates heterogenous association within Africa. Hum. Mol. Genet. 2021;30:2205–2214. doi: 10.1093/hmg/ddab174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel RA, et al. Genetic interactions drive heterogeneity in causal variant effect sizes for gene expression and complex traits. Am. J. Hum. Genet. 2022;109:1286–1297. doi: 10.1016/j.ajhg.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prive F, Aschard H, Ziyatdinov A, Blum MGB. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics. 2018;34:2781–2787. doi: 10.1093/bioinformatics/bty185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hormozdiari F, et al. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, X. et al. Code for the manuscript A new approach to identify gene-environment interactions and reveal new biological insight in complex traits. https://zenodo.org/records/10815731 (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The marginal summary statistics of HDL-C, LDL-C, and TG from the Global Lipids Genetics Consortium study (GLGC, n = 1.65 M) were downloaded at http://csg.sph.umich.edu/willer/public/glgc-lipids2021. GLGC consists of GWAS results from 1.65 M subjects representing five genetic ancestry groups: European (N = 1.32 M); African or admixed African (N = 99k); East Asian (N = 146k); Hispanic (N = 48k); and South Asian (N = 41k). We did not perform South Asian specific analysis because there was no corresponding GWIS in the Cohorts for Heart and Aging Research in Genetic Epidemiology (CHARGE) consortium. The GWIS summary statistics from CHARGE gene-lifestyle (GLI) working group in this study are available via dbGaP (accession number phs000930). The UKBB individual-level data for replications were available through Application ID: 81097.
test was used the software IMRP which is available in the Github repository with the following link, https://github.com/XiaofengZhuCase/IMRP23. Heritability analysis was performed by Bigsnpr, https://privefl.github.io/bigsnpr/reference/snp_ld_scores.html47 and LDSC regression, https://github.com/bulik/ldsc47. FUMA: https://fuma.ctglab.nl/31. Software ezQTL: https://analysistools.cancer.gov/ezqtl/#/home. MAGMA: https://ctg.cncr.nl/software/magma30. DEPICT: https://github.com/perslab/depict48. Plink: https://www.cog-genomics.org/plink2/46. The codes for performing simulations and analyzing interaction of lipids data were deposited in the Zenodo database53.