Abstract
Reverse transcriptase (RT) plays a critical role in retrovirus replication, directing the synthesis of a double- stranded DNA copy of the viral RNA genome. We have previously described a mutant RT of the Moloney murine leukemia virus in which F155 was replaced by valine, and we demonstrated that this substitution allowed the enzyme to incorporate ribonucleotides to form RNA while still retaining its normal ability to incorporate deoxyribonucleotides to form DNA. When introduced into the viral genome, this mutation rendered the virus incapable of replication. Characterization of the mutant virus revealed that the enzyme was still active and able to synthesize minus-strand strong stop DNA and some longer products but failed to make full-length minus-strand DNA. We propose that the failure of the enzyme to complete DNA synthesis in vivo resulted from its ability to incorporate ribonucleotides into the products, which served as inhibitors for DNA synthesis. We also tested seven other amino acid residues for their abilities to substitute for F155 in virus replication; of these, only tyrosine could support virus replication. In an attempt to select for second-site suppressor mutations, the F155V mutant was subjected to random mutagenesis and was used as a parent for the isolation of revertant viruses. Two independent revertants were found to have changed the valine residue at position 155 back to the wild- type phenylalanine. These results suggest that an aromatic ring at this position is important for virus replication.
Reverse transcriptase (RT) plays a defining role in the retrovirus life cycle (2, 26): the enzyme is responsible for the synthesis of a double-stranded DNA copy of the single-stranded RNA genome, which is inserted into the host genome to establish the integrated provirus. RT performs this task with three catalytic activities: RNA- and DNA-directed DNA polymerase activities and RNase H activity, which degrades RNA in an RNA-DNA hybrid. The process of reverse transcription begins with the extension of a tRNA primer using the RNA viral genome as a template to synthesize a short DNA intermediate product, minus-strand strong stop DNA, which corresponds to the sequence from the primer binding sequence on the genomic RNA to the 5′ end of the genome (25). The minus-strand strong stop DNA is then translocated to the 3′ end of the RNA genome and extended to give rise to long minus-strand DNA products; as synthesis proceeds, the RNA genome is degraded by the RNase H activity of RT. A specific RNA product, the polypurine tract (PPT), serves as a primer for the initiation of the plus-strand DNA, forming a plus-strand strong stop DNA. This intermediate can anneal to the 3′ end of its template, and further elongation of both plus- and minus-strand DNAs can then proceed. The final product is a long double-stranded DNA copy of the genome capable of integration into the host genome.
The bulk of the DNA synthesis mediated by RT takes place in a protein complex in the cytoplasm of the recently infected cell. The intracellular nucleotide pools contain much higher levels of ribonucleoside triphosphates (rNTPs) than of deoxyribonucleoside triphosphates (12), and thus to synthesize DNA efficiently the RT enzyme must have high selectivity for deoxyribonucleotides. Like other DNA polymerases, RT exhibits such selectivity (27). We have previously described a mutant Moloney murine leukemia virus (M-MuLV) RT in which F155 was changed to valine (8). Examination of this mutant enzyme (RT-F155V) isolated after expression in Escherichia coli demonstrated that it could act either as an RNA polymerase or as a DNA polymerase. In contrast to the wild-type RT (RT-WT), RT-F155V showed a dramatically increased affinity for ribonucleotides, comparable to the affinity for deoxyribonucleotides. The Vmax for the incorporation of ribonucleotides, however, remained almost the same as that for RT-WT, approximately 100-fold lower than the Vmax for the incorporation of deoxyribonucleotides. The low Vmax was a result of both slow incorporation of ribonucleotides and slow extension of primers with incorporated ribonucleotides. In contrast, the affinity and Vmax of RT- F155V for deoxyribonucleotides were almost the same as those for RT-WT.
In this report, we describe the replication-defective phenotype of mutant viruses containing this same mutation. Characterization of the enzymatic activities of RT-F155V in virions and monitoring the synthesis of reverse transcription intermediates suggest a basis for the failure of the virus to replicate.
MATERIALS AND METHODS
Plasmid construction.
The following plasmid constructs were used in this report: pNCA-WT, carrying a complete infectious M-MuLV proviral DNA (6); pNCA-WT-H, equivalent to pNCA-WT with a point mutation (D524N) inactivating RNase H (4); pNCA-F155V, carrying the F155V mutation in the polymerase domain; pNCA-F155V-H, a provirus with both mutations; pNCS-WT, a version of pNCA-WT with a simian virus 40 (SV40) replication origin in the vector to allow high-level expression in 293T cells; pNCS-F155V, a pNCS version of pNCA-F155V; pNCS-F155V-H, a pNCS version of pNCA-F155V-H; and RT-WT-His, a plasmid to express 6× His-tagged RT-WT. RT-WT-His was constructed by replacing the BglII-XbaI sequence (nucleotides [nt] 1847 to 2013) in pRT-30-2 (encoding the RT-WT) (24) with a PCR-derived fragment of the corresponding region in which 18 nt encoding six histidine residues were introduced at the COOH end of RT. pNCA-F155V, pNCA-F155V-H, and RT-F155V-His were generated by using the BclI-SalI fragment (nt 143 to 1108) from RT-F155V-H (8) to replace the same sequence in pNCA-WT, pNCA-WT-H, and RT-WT-His, respectively. To generate pNCS-WT, a blunt-end fragment containing the SV40 origin from pSV2neo was cloned into pNCA-WT after digestion by EcoRI and treatment with the Klenow fragment to form blunt ends. pNCS-WT-H, pNCS-F155V, and pNCS-F155V-H were constructed in the same way. Mutants RT-F155A, RT-F155H, RT-F155I, RT-F155L, RT-F155M, RT-F155W, and RT-F155Y were constructed by the same PCR strategy as described previously (8). The mutations were then introduced into pNCA as described above.
Cells and transfection.
NIH 3T3 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% bovine calf serum. Viral spread assay in NIH 3T3 cells has been previously described (10). In the revertant screening experiment, each subpool of mutagenized DNA was introduced into 2 × 106 NIH 3T3 cells by Lipofectamine-mediated DNA transfection following the manufacturer’s instructions (Gibco-BRL). 293T cells were maintained in DMEM supplemented with 10% fetal bovine serum. To produce viruses, the pNCS versions of the proviral DNAs were introduced into these cells by standard calcium phosphate transfection for 8 h. To minimize ribonucleotide contamination in the virus stock, 24 h after transfection fresh medium containing 10% dialyzed fetal bovine serum (Gibco-BRL) was used to wash and maintain the cells. Twenty-four hours later, the virus-containing culture supernatants were harvested and cleared through a 0.45-μm filter. Rat2 cells were maintained in DMEM supplemented with 10% bovine calf serum. All acute infections were performed in the presence of 8 μg of Polybrene per ml for 2 h.
Protein purification.
Recombinant RT enzymes for use in homopolymer assays were expressed in E. coli DH5α and were partially purified with DE52 resin as previously described (8). Recombinant enzymes for use in the processivity assay were more carefully prepared to exclude any contaminating bacterial DNA polymerase activity. RT-WT-His and RT-F155V-His were introduced into C2110, a bacterial strain deficient in DNA polymerase I activity (21). Due to the inability of the plasmids to replicate in this strain, only integration of the plasmid sequence into the bacterial genome could lead to RT expression. Potential transformants were selected for ampicillin resistance and then screened for expression of RT by Western blotting with anti M-MuLV-RT antiserum (2). Constitutively expressed RT enzymes were purified to near homogeneity, as shown by Coomassie staining, by means of Ni-nitrilotriacetic acid agarose batch separation following the manufacturer’s instructions (Qiagen). In parallel, the purification of an inactive RT (RT-YMNN, double mutation in the YMDD active site [7, 15]) through the same procedure resulted in no DNA polymerase activity, further demonstrating the lack of contaminating polymerase activity in the purified products.
Endogenous RT reactions.
Standard methods for endogenous RT reactions have been described elsewhere (23). Some minor modifications were made in our experiments. To detect minus-strand strong stop DNA, the reaction was carried out for 2 h under an RNase-free condition and treatment of the products with NaOH was omitted to protect ribonucleotide-containing products. To detect incorporation of ribonucleotides into minus-strand strong stop DNA, [α-32P] rUTP instead of [α-32P]dTTP was included in the substrates (for better detection, fivefold more [α-32P]rUTP than [α-32P]dTTP was used).
Processivity assay.
RNA template-primer complex was prepared as previously described (8). Template-primer (at a concentration of 100 nM primer termini) was preincubated with purified recombinant RT enzyme (150 nM) at 37°C for 10 min in standard RT reaction buffer (8) containing 10 mM MgCl2. The primer extension reaction was initiated by the addition of 500 μM each deoxynucleoside triphosphate (dNTP), in the absence or presence of a huge excess of poly(rA) · oligo(dT)12–18 (10 μM as primer termini) as a competitor trap for the enzyme. Thirty minutes later, the reaction was terminated by the addition of EDTA, and the products were resolved by electrophoresis on a 15% polyacrylamide denaturing gel, followed by autoradiography.
DNA mutagenesis.
Two independent procedures were used to create a pool of randomly mutagenized DNAs. Method 1 was as follows. Two fragments of the RT-F155V sequence—one from the BclI site (nt 143) to the SalI site (nt 1108), and the other from the SalI site (nt 1108) to the HindIII site (in the beginning of the integrase sequence)—were separately mutagenized by PCR. The fragments were first amplified from pNCA-F155V with native Taq polymerase (Perkin-Elmer) under standard conditions for 25 cycles (95°C for 1 min, 60°C for 2 min, and 72°C for 3 min for each cycle). Twenty microliters of the product was used as a template in a 100-μl reaction mixture for further amplification under mutagenesis PCR conditions (14) for 25 cycles (95°C for 1 min, 42°C for 2 min, and 72°C for 3 min for each cycle). The products were digested with the appropriate restriction enzymes and used to replace the corresponding fragment in the pNCA-F155V parent.
Method 2 was as follows. Plasmid pNCA-F155V was used to transform XL1-red cells following the instructions of the manufacturer (Stratagene). Approximately 2,000 colonies were pooled and grown in 5 ml of LB medium. A 100-μl volume of this culture was grown for 1 more day in 5 ml of LB medium. DNA extracted from these two cultures was used to transform ElectroMax DH10B cells (Gibco-BRL). Approximately 5 million colonies were recovered from the first DNA sample, and approximately 2 million colonies were recovered from the DNA subjected to the extended culture period. Large-scale DNA preparations were made from the transformed DH10B cells and were used for transfection into NIH 3T3 cells. In parallel, pUC18 plasmid provided by the manufacturer was also mutagenized to monitor mutagenesis efficiency.
RESULTS
Test of viral spread of M-MuLV harboring the F155V mutation.
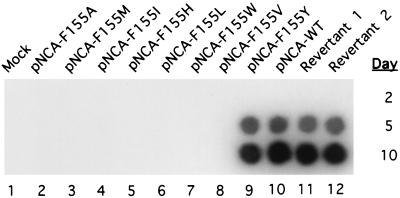
Previous experiments showed that substitution of F155 of M-MuLV-RT with valine rendered the enzyme capable of incorporating ribonucleotides into the products while the DNA polymerase activity remained unchanged (8). To analyze the effect of this F155V mutation on viral replication, the mutation was introduced into the provirus genome and the resulting DNA was then analyzed for infectivity. In one method, the mutant provirus DNA was introduced into NIH 3T3 cells by transient transfection and viral spread was monitored by assaying for RT activity in the supernatant. If the mutant RT was present, it would be detected under standard assay conditions (8) (see Fig. 2). RT activity could be easily detected from pNCA-WT-transfected cells 5 days after transfection (Fig. 1, column 10), demonstrating its ability to replicate. In contrast, pNCA-F155V-transfected cells failed to display any RT activity even 30 days after transfection (Fig. 1, column 8; data not shown).
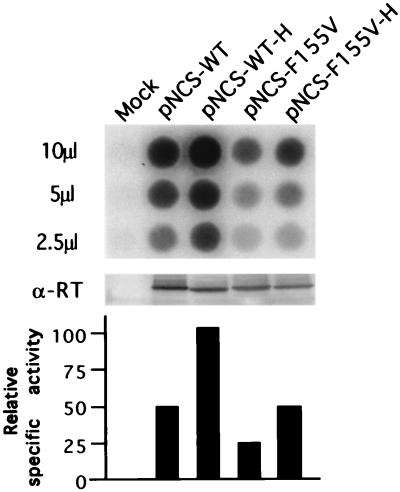
FIG. 2.
Comparison of mutant and wild-type viruses for virion-associated RT protein and RT activities. 293T cells were transiently transfected with the indicated proviral DNAs on a plasmid containing the SV40 origin of replication. Mock, no DNA; pNCS-WT, wild-type viral DNA; pNCS-WT-H, RNase H-deficient mutant; pNCS-F155V, the substitution mutant; and pNCS-F155V-H, the double mutant. Culture supernatants were harvested 48 h after transfection, and various amounts of the supernatant, as indicated, were analyzed for RT activity by homopolymer RT assay (upper panel). The virions were pelleted through a 25% sucrose cushion and then analyzed by Western blotting for RT levels with a polyclonal anti-RT antiserum (middle panel). Relative specific activities for each enzyme were calculated (lower panel).
FIG. 1.
Test of replication of viral RT mutants. Proviral DNAs encoding RTs with various amino acid substitutions of Y155 were analyzed for replication ability by DEAE dextran-mediated transient transfection of NIH 3T3 cells. Mock, mock transfection; WT, wild-type DNA; F, phenylalanine; A, alanine; M, methionine; I, isoleucine; H, histidine; L, leucine; W, tryptophan; V, valine; and Y, tyrosine. Revertant 1 and 2, revertants isolated from the F155V parent. Culture supernatants were harvested at 2, 5, and 10 days after transfection and analyzed for RT activity by the homopolymer RT assay as a measure of viral spread.
In the second method, viruses were first produced by transient transfection of 293T cells by the provirus DNAs and were then collected from the culture supernatants and used to infect NIH 3T3 cells. The spread of infectious virus in the 3T3 cells results in the appearance of RT activity in the culture supernatants. The mutant F155V virus failed to induce any detectable RT activity even after 30 days, whereas wild-type virus induced detectable RT activity 2 days postinfection (data not shown). These results clearly established that virus harboring the F155V mutation could not replicate.
Analysis of virion-associated RT.
As a first step toward understanding why the F155V virus could not replicate, we analyzed the enzymatic activity of the RT-F155V enzyme present in virions to test whether the enzyme was still active. We purified virions from the culture supernatants after transfection of 293T cells, which can release high levels of virions even from replication-defective genomes. Both RNase H-defective and wild-type versions were utilized to rule out any possible degradation of ribonucleotide-containing products by RNase H. Virions collected from the supernatants were examined by Western blotting to estimate the protein levels of RT (3). The relative DNA polymerase activities of mutant RTs and RT-WT in the virions were measured by standard exogenous assays on homopolymer template-primers. With either the RNase H-deficient version or the RNase H wild-type version, the level of the pNCS-F155V-RT as judged by protein was about twofold less than that of the pNCS-WT-RT (Fig. 2, middle panel; compare lanes 2 and 4 and lanes 3 and 5). The mutant enzyme consistently displayed an approximately fourfold-lower level of DNA polymerase activity compared to that of pNCS-WT (Fig. 2, upper panel; compare columns 2 and 3 and columns 4 and 5), with either crude (Fig. 2) or purified (data not shown) virions. Therefore, when normalized to the amount of protein, pNCS-F155V exhibited specific activity approximately twofold less than that of pNCS-WT, with either the RNase H mutant or the wild-type version (Fig. 2, lower panel). In the subsequent experiments, the same amounts of virions (based on the Western blot result) were used for comparisons. Interestingly, the viral RNase H-active enzymes consistently displayed activity approximately twofold less than those of the RNase H-deficient ones (Fig. 2, upper panel; compare columns 2 and 3 and columns 4 and 5), suggesting that the point mutations in RNase H might slightly enhance the polymerase activity of RT in virus.
Analysis of the synthesis of preintegrative viral DNA in cells.
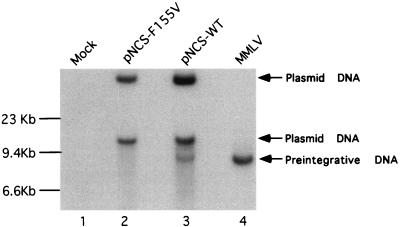
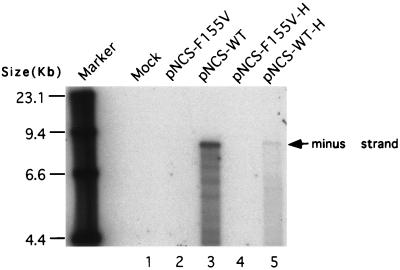
The above results suggested that the F155V mutant RT showed substantial DNA polymerase activity in the virion but was still unable to support virus replication. To test whether the mutant could direct the synthesis of any DNA in vivo, virus preparations were collected from transfected 293T cells and used to infect NIH 3T3 cells. The low-molecular-weight preintegrative viral DNA was harvested by the Hirt method (11) and was analyzed by Southern blot with a labeled viral DNA as probe. In wild-type virus-infected cells, the viral DNA was easily detectable, whereas in F155V virus-infected cells the viral DNA was absent (Fig. 3), indicating the inability of F155V virus to synthesize full-length viral DNA.
FIG. 3.
Detection of viral DNA in wild-type and mutant virus-infected NIH 3T3 cells by Southern blotting. Supernatants harvested from transfected 293T cells were used to acutely infect NIH 3T3 cells. Twelve hours after infection, low-molecular-weight DNA was isolated, and resolved by electrophoresis, and viral DNA was detected by Southern blotting with a viral DNA probe. MMLV, virus collected from a stable NIH 3T3 cell line producing wild-type M-MuLV. Note that two forms of the plasmid DNAs used to transfect the 293T cells were carried over in the viral supernatants to the NIH 3T3 cells and detected in the analysis. The migration positions of the double-stranded viral DNA (8.8 kb) and plasmid DNA are indicated.
Endogenous reaction: DNA synthesis in virions.
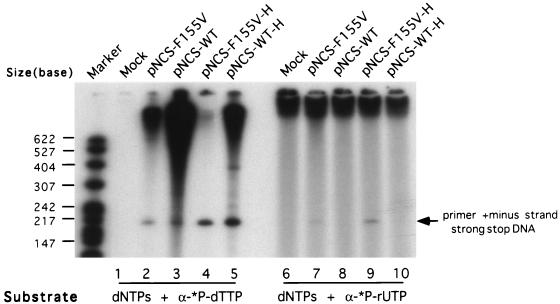
To define the step at which reverse transcription was blocked, virions were purified and incubated in vitro with nucleotides to monitor the synthesis of reverse transcription intermediates on the viral DNA template (the endogenous reaction) (9, 19, 20). The reactions were performed for only 2 h to monitor the synthesis of early products. When incubated with labeled deoxyribonucleotides, minus-strand strong stop DNA and some longer products were synthesized in all of the virions (Fig. 4, lanes 1 to 5), although the level of DNA made by RT-F155V was much lower than that made by RT-WT (compare lanes 2 and 3 and lanes 4 and 5). When the mutant RT-F155V was incubated with a labeled ribonucleotide, it incorporated rUTP into the strong stop DNA, while wild-type virus failed to do so (Fig. 4, lanes 6 to 10). These results suggest that the virion-associated RT was similar to the recombinant enzyme in its altered selectivity for nucleotides. The endogenous reaction was then extended to 12 h to address whether the products seen above could be extended to full-length minus-strand DNA. As shown in Fig. 5, wild-type virions were able to form full-length minus-strand DNA at high levels; as previously reported, RTs lacking the RNase H activity showed profoundly reduced levels of long products, due to their inability to efficiently mediate strong stop DNA translocation (4). Full-length DNA was not detectable in F155V virions. Therefore, even though the F155V mutant enzyme was still enzymatically active and able to synthesize minus-strand strong stop DNA and some longer products, it failed to complete minus-strand DNA synthesis, in agreement with the absence of preintegrative DNA in F155V virus-infected cells.
FIG. 4.
Endogenous RT assay of products generated in wild-type and mutant virions collected from transfected 293T cells. Either [α-32P]dTTP (lanes 1 to 5) or [α-32P]rUTP (lanes 6 to 10) was included with all four dNTP substrates. The migration position of minus-strand strong stop DNA covalently linked to the tRNA primer is indicated.
FIG. 5.
Endogenous assay to detect full-length minus-strand DNA in wild-type and mutant virions. DNA products were labeled by [α-32P]dTTP and resolved on a 0.8% agarose gel, followed by autoradiography. The migration position of full-length minus-strand DNA is indicated.
Processivity analysis of RT-WT and mutant RTs.
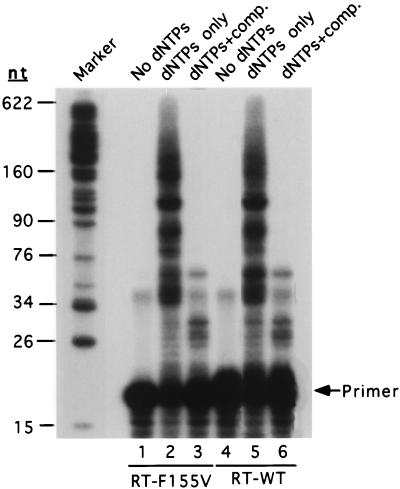
One possible explanation for the inability of the mutant enzyme to complete minus-strand DNA synthesis was that the processivity of the mutant enzyme, which is an indicator of the ability of an enzyme to continuously synthesize long products, was diminished. We addressed this question by directly comparing the processivities of mutant and wild-type recombinant enzymes. To mimic the conditions under which RT carries out DNA synthesis in virions, purified RNase H-proficient enzymes and an RNA template were used. Reactions were performed on a defined template, with or without competitor template added. RT-F155V showed no difference from RT- WT in processivity, with or without template-primer competitors (Fig. 6). The failure of RT-F155V to make full-length minus-strand DNA in virions could not simply be accounted for based on a reduced processivity.
FIG. 6.
Processivity analysis of RT-WT and mutant RTs. 32P-end-labeled primer was extended by purified recombinant RT-F155V (lanes 2 to 4) or RT- WT with a 320-nt-long RNA as the template. No dNTPs, no dNTP substrates added to the reaction; dNTPs only, 500 μM each dNTP added to the reaction; dNTPs + comp., dNTPs substrates, together with excess template-primer competitor, poly(rA) · oligo(dT)12–18, added to the reaction to trap enzymes that dissociate from the elongation complex. The migration position of labeled primer is indicated.
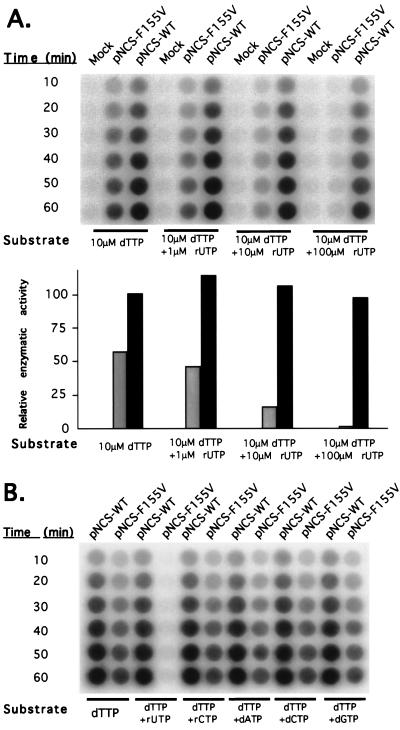
Inhibition of the enzymatic activity of RT-F155V by ribonucleotides.
Another possible explanation for the failure of the F155V virus to make full-length viral DNA was the selective inhibition of the mutant RT-F155V by ribonucleotides. Since RT-F155V demonstrated similar affinities for ribonucleotides and deoxyribonucleotides but much less polymerase activity with ribonucleotides than with deoxyribonucleotides as substrates (8), ribonucleotides could serve directly as inhibitors. Further, ribonucleotides might be incorporated into the product and serve as inhibitors for the subsequent extension of the product. To test these ideas, assays were performed with virion-derived RT acting on a homopolymer template primer, with or without exogenous rUTP added. When only 10 μM dTTP was provided, the mutant enzyme was marginally inhibited by 1 μM rUTP, was dramatically inhibited by 10 μM rUTP, and was completely blocked by the addition of 100 μM rUTP (Fig. 7A). In contrast, the addition of rUTP to the wild-type virus reaction mixture, even at 100 μM, had no effect. Furthermore, the addition of other NTPs, including rCTP, dATP, dCTP, and dGTP, had no such inhibitory effect (Fig. 7B), indicating that such inhibition is base pairing dependent. The same phenomenon was also observed with the RNase H-deficient versions of the enzymes (data not shown), indicating that inhibition occurred on the DNA synthesis level and not through degradation of the product by RNase H. The inhibitory effect of ribonucleotides on DNA synthesis by RT-F155V could account, at least in part, for the enzyme’s inability to make full-length DNA in vivo. The consequences of such inhibition would be magnified in reverse transcription of the genome, for which completion of long DNA products is required.
FIG. 7.
Inhibition of the enzymatic activity of RT-F155V by ribonucleotide rUTP. (A) Supernatants from transfected 293T cells were analyzed for RT activity in the homopolymer RT assay with poly(rA) · oligo(dT) as the template-primer, in the presence of various concentrations of rUTP as inhibitors as indicated. Aliquots were removed at the indicated time points and spotted on DEAE paper. The paper was then washed and exposed to either X-ray film for autoradiography (upper panel) or PhosphoImager analysis for quantitation. The relative enzymatic activities of RT-WT and mutant RTs were plotted and normalized to the wild-type enzyme without inhibitor (lower panel). Gray bars, RT-F155V; black bars, RT-WT. (B) Culture supernatants containing either wild-type or F155V virions were analyzed for RT activity on poly(rA) · oligo(dT)12–18 as the template-primer, with labeled dTTP as the substrate, in the presence of different unlabeled NTPs as indicated. Reactions were performed with dTTP at 10 μM and with each inhibitor at 100 μM.
Test of more mutants for virus replication.
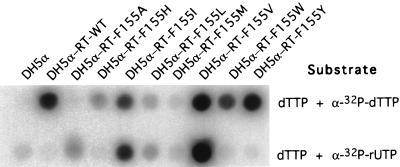
To further investigate the importance of F155 of M-MuLV-RT in retrovirus replication, different point mutations were generated at this position in the viral genome. These mutations can be roughly divided into two groups as follows: in F155A, F155I, F155L, and F155M, the phenylalanine residue was replaced by a relatively smaller residue; and in F155H, F155W, and F155Y, the phenylalanine was replaced by similarly bulky residues. The mutant proviral DNAs were introduced into NIH 3T3 cells, and the cultures were analyzed for viral spread as described before. As shown in Fig. 1, only mutant pNCA-F155Y was replication competent.
To understand why the majority of these mutants failed to replicate, the mutations were also introduced into a plasmid construct expressing high levels of recombinant RT in E. coli (24). The proteins were expressed, partially purified, and tested for their DNA polymerase activities and abilities to incorporate ribonucleotides into the products on a homopolymer substrate (Fig. 8). The activities of RT-F155V and RT-F155Y were as observed previously. Both were as active as RT-WT with a deoxyribonucleotide substrate; RT-F155V was also active with the ribonucleotide substrate, whereas RT-F155Y remained inactive like the wild-type enzyme. In accord with the viral replication phenotypes, mutants RT-F155A and RT- F155M were totally inactive. Mutants RT-F155H and RT-F155L showed low activity, and RT-F155W was somewhat more active but much less so than the wild-type enzyme. Interestingly, RT-F155I exhibited activities similar to those of RT- F155V, including both DNA and RNA polymerase activities, although the overall enzymatic activity was lower.
FIG. 8.
Enzymatic activities of mutant RT and RT-WT enzymes with deoxyribonucleotides or ribonucleotides as substrates. Recombinant enzymes were partially purified and quantitated by Western blotting so that the same amount of RT protein was used in each reaction. Reactions were performed for 5 min with poly(rA) · oligo(dT) as the template-primer with substrates as indicated.
The search for a suppressor mutation.
Mutant F155V shows normal DNA polymerase activity in vitro but is clearly defective at reverse transcription in vivo. To determine the range of mechanisms by which this RT could become functional, we attempted to recover a second-site mutation in the RT which could suppress the phenotype of the F155V mutation and restore normal retrovirus replication. Random mutations were introduced into pNCA-F155V, and pools of the mutagenized plasmids were then transiently transfected into NIH 3T3 cells. The cultures were passaged to allow viral spread until any replication-competent virus could expand to most of the cells, as indicated by RT activity in the supernatant.
Two methods were used to mutagenize pNCA-F155V (see Materials and Methods). Two separate pools of mutants were generated with a PCR mutagenesis method; one was with mutations in the fragment from the BclI site to the SalI site (pool 1), and the other was from the SalI site to the HindIII site (pool 2). In each pool, more than 1 million independent bacterial transformants were obtained. Five randomly picked clones were sequenced to estimate the mutation rate as approximately one mutation per 1,000 bases. Two pools of mutants were also generated with a bacterial mutator (XL1-red) method: approximately 5 million independent transformants were obtained after an overnight culture period of the mutator strain (pool 3), and approximately 2 million transformants were obtained from a 2-day culture (pool 4), in which the plasmid was more heavily mutagenized. Each pool was divided into 10 subpools, which were used to transfect NIH 3T3 cells (approximately 2 million cells for each subpool) to select for replication-competent virus. The success of transfection was indicated by the transient appearance of RT activity in the supernatant 2 days posttransfection. This RT signal, resulting from the transient expression of the transfected DNA, disappeared a few days later because of the replication inability of the transfected DNA. Four weeks after transfection with the mutagenized DNAs, two cultures became RT positive (one from pool 1 and the other from pool 3); all 10 cultures from pools 2 and 4 remained negative.
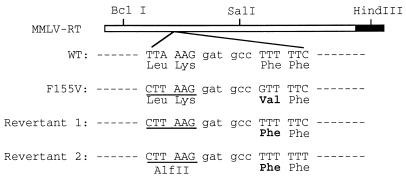
The viruses from these two revertant cultures were used to infect Rat2 cells, and low-molecular-weight DNA was collected 12 h after infection. A portion of the pol gene was amplified by PCR and cloned into pNCA for confirmation of the replication competence (Fig. 1, columns 11 and 12) and for sequence analysis. Two independent clones for each revertant were sequenced to ensure that any mutations detected were not introduced during PCR amplification of the viral DNA. The sequences of the two revertants are shown in Fig. 9, aligned with the sequences of RT-WT and RT-155V. During construction of RT-F155V, a new restriction site was introduced near the F155V mutation but not affecting the encoded protein. This restriction site now served as a hallmark for the original mutation. In both revertants, this hallmark was retained, indicating that the revertants were not derived from any contamination by wild-type virus. In both revertants, the mutant V155 was changed back to wild-type sequence, i.e., F155. It is worth noting that the two revertants are clearly independent because the codons for the adjacent residue, F156, are different (TTC, the same as the wild-type, for one, and TTT for the other). No other change was found in the fragment which fully restored virus replication to the F155V mutant. Thus, no second-site suppression mutations were present in these clones. We did not recover a revertant in which V155 was changed to Y155, which would render the virus replication competent (Fig. 1, column 10). The amino acid change from valine to phenylalanine requires only one nucleotide change (from GTT to TTT), whereas the change from valine to tyrosine requires at least a two-nucleotide change (from GTT to TAT). The failure to recover the Y155 revertant implies that two nucleotides change events were rare in our assay (see below for further discussion). Other possible codons that could be generated from GTT (codon for V155) by the introduction of one nucleotide mutation include ATT (I), CTT (L), GAT (D), GGT (G), and GCT (A). The absence of these residues in the revertants was consistent with the observation that these residues rendered the virus incapable of replication (Fig. 1).
FIG. 9.
Sequence comparison of the revertants, RT-WT and the original F155V mutant. The restriction sites used in the random mutagenesis are shown on the schematic sequences of RT (open bar) and part of integrase (closed bar). The AflII restriction site, which was generated by silent mutations during the construction of RT-F155V, is underlined.
DISCUSSION
In this report, we demonstrate that although the F155V mutant of RT is exceedingly similar to RT-WT in its enzymatic properties, it could not support virus replication. Characterization of the mutant virions revealed that the mutant RT was incorporated into virions and showed normal DNA polymerase activity and processivity. As expected from studies of the recombinant enzyme, the mutant virions could incorporate ribonucleotides into products in the endogenous reaction. We suggest that this incorporation of ribonucleotides may be at least one of the reasons that the virus is replication defective. The addition of modest levels of ribonucleotides to reactions performed with homopolymer templates did indeed inhibit DNA synthesis; similar inhibition by ribonucleotides may explain why the F155V virus could not make full-length viral DNA in vivo. The levels of ribonucleotides inside the cell are at least 10-fold higher than the levels of deoxyribonucleotides (12), which is more than sufficient to block the mutant enzyme. These results suggest that the wild-type enzyme has evolved a high selectivity for deoxyribonucleotides, in part, to prevent this inhibition.
We noted that the mutant RT was also defective at forming long DNAs in the endogenous reaction even when only deoxyribonucleotides were provided to the virions, suggesting that there may be additional problems. Although no ribonucleotides were intentionally added in the endogenous reaction, small levels of ribonucleotides may well be present in the virions or in the deoxyribonucleotides supplied to the enzyme. These low levels may have allowed for the synthesis of minus-strand strong stop DNA and some elongated products, at reduced yields, but they may very potently block long products. The inhibitory effect of ribonucleotides on DNA synthesis by RT-F155V could also explain why the virion-associated enzyme was roughly twofold less active than the recombinant enzyme. Unfortunately, it would be very difficult to distinguish this possibility from an intrinsic lower enzymatic activity of viral RT-F155V.
The corresponding tyrosine residue in human immunodeficiency virus type 1 RT (HIV-1-RT), Y115, has been extensively studied by mutational analysis (5, 13, 16, 17). In this enzyme, the effects of these mutations are somewhat different. Consistent with the computer-modeling data suggesting that this residue may interact directly with the base of the incoming dNTP (18, 22), the substitution of Y115 of HIV-1-RT with other residues strongly affected the enzyme’s affinity for dNTP substrates and the fidelity of DNA synthesis (16, 17). Thus, Y115 mutations in HIV-1-RT had a major effect on the base pairing of the incoming nucleotide with the template. Unfortunately, whether the mutant HIV-1-RT-Y115V is able to incorporate ribonucleotides into the product has not been reported. However, the substitution of phenylalanine with valine in M- MuLV- RT does not affect the fidelity of the enzyme (7a) or the affinity of the enzyme for the substrate dTTP (8), suggesting that there was little or no effect of the mutation on base pairing. Even with respect to inhibition by rNTPs, stringent base pairing fidelity is apparently conserved and required for inhibitory activity (Fig. 7B). This difference between M- MuLV- RT and HIV-1-RT in their responses to alteration of the corresponding tyrosine suggests that in different contexts, the two residues play somewhat different roles. This idea is further supported by a recent mutational analysis of the Klenow fragment of DNA polymerase I (1). In DNA polymerase I, the residue corresponding to F155 of M-MuLV-RT in the primary sequence is a glutamate, E710. The substitution of this residue with a smaller one also resulted in a decrease in the enzyme’s ability to discriminate against ribonucleotides. However, the effect of the substitution in this case was mainly on the rate constant, kcat, for incorporation of rNTPs, rather than on the Km for the binding of the substrate (1). Thus, even when the consequences of homologous mutations in two enzymes appear to be similar, detailed examination can show that the mutations may mediate their respective effects on the enzymes through distinct mechanisms.
In this report, we also tested other amino acid residues for their abilities to substitute for F155 in virus replication. Only tyrosine was functional. The substitution of F155 with seven relatively conservative residues failed to restore infectivity, suggesting the importance of an aromatic residue at this position for virus replication. Such importance was further supported by our failure to find a second-site suppressor.
The mutagenesis and selection methods used here for the recovery of revertant viruses were effective, since we did isolate two independent revertants from the defective parent. However, the procedures might not be powerful enough to recover all of the possible single-site revertants that would render the virus replication competent, as indicated by the absence in our screening of reversion from V155 to Y155. It is possible that a second-site mutation that could suppress the F155V mutation requires a change of more than 1 nt. The mutation frequency was estimated to be approximately one mutation per 1 kb; the probability of finding one particular amino acid change at a particular position is thus approximately 1 of 1,000 independent clones, and the probability of finding two particular amino acid changes at particular positions would be roughly 1 in 106. It is difficult to recover mutants from our pools of mutagenized DNAs, as indicated by our failure to find the V155Y revertant, that would require at least a 2-nt change (from GTT to TAT or TAC). Increasing the mutagenesis efficiency might not help, as suggested by the failure to find any revertants from pool 4, which should harbor more mutations than pool 3 (see Materials and Methods). The heavy mutagenesis may have introduced more lethal mutations that prevented the recovery of viable revertants.
It is interesting to note that revertant 2 contained a linked nucleotide change in the adjacent codon, which did not alter the residue F156 and thus was unselected. The appearance of this mutation suggests that mutagenesis by the mutator polymerase in E. coli may have altered two residues during patch repair synthesis of a small stretch of continuous bases. The frequency of unlinked pairs of mutations might be lower, and mutants arising from these events might be correspondingly harder to recover.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant CA 30488 from the National Cancer Institute. G.G. is an Associate and S.P.G. is an Investigator of the Howard Hughes Medical Institute.
We thank Eran Bacharach and Jason Gonsky for helpful discussion and Kenia delos Santos and Sharon Boast for technical support during the course of the work.
REFERENCES
- 1.Astatke M, Ng K, Grindley N D F, Joyce C M. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc Natl Acad Sci USA. 1998;95:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 3.Blain S W, Goff S P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase: mutants with altered substrate specificities. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 4.Blain S W, Goff S P. Effects on DNA synthesis and translocation caused by mutations in the RNase H domain of Moloney murine leukemia virus reverse transcriptase. J Virol. 1995;69:4440–4452. doi: 10.1128/jvi.69.7.4440-4452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer P L, Ferris A L, Hughes S H. Mutational analysis of the fingers domain of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1992;66:7533–7537. doi: 10.1128/jvi.66.12.7533-7537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colicelli J, Goff S P. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle R F, Feng D-F, Johnson M S, McClure M A. Origin and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 7a.Gao, G., and S. P. Goff. Unpublished data.
- 8.Gao G, Orlova M, Georgiadis M M, Hendrickson W A, Goff S P. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc Natl Acad Sci USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilboa E, Mitra S W, Goff S P, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 10.Goff S P, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 12.Kornberg A, Baker T A. DNA replication. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 13.Larder B A, Kemp S D, Purifoy D J M. Infectious potential of human immunodeficiency virus type 1 reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci USA. 1989;86:4803–4807. doi: 10.1073/pnas.86.13.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 15.Lowe D M, Parmar V, Kemp S D, Larder B A. Mutational analysis of two conserved sequence motifs in HIV-1 reverse transcriptase. FEBS Lett. 1991;282:231–234. doi: 10.1016/0014-5793(91)80484-k. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Hernandez A M, Domingo E, Menendez-Arias L. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 1996;15:4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Hernandez A M, Gutierrez-Rivas M, Domingo E, Menendez-Arias L. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 1997;25:1383–1389. doi: 10.1093/nar/25.7.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel P H, Jacobo-Molina A, Ding J, Tantillo C, Clark A D J, Raag R, Nanni R G, Hughes S H, Arnold E. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 19.Rothenberg E, Baltimore D. Synthesis of long representative DNA copies of the murine RNA tumor virus genome. J Virol. 1976;17:168–174. doi: 10.1128/jvi.17.1.168-174.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothenberg E, Smotkin D, Baltimore D, Weinberg R A. In vitro synthesis of infectious DNA of murine leukemia virus. Nature. 1977;269:122–126. doi: 10.1038/269122a0. [DOI] [PubMed] [Google Scholar]
- 21.Tanese N, Roth M, Goff S P. Expression of enzymatically active reverse transcriptase in E. coli. Proc Natl Acad Sci USA. 1985;82:4944–4948. doi: 10.1073/pnas.82.15.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 23.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 24.Telesnitsky A, Blain S W, Goff S P. Defects in Moloney murine leukemia virus replication caused by a reverse transcriptase mutation modeled on the structure of Escherichia coli RNase H. J Virol. 1992;66:615–622. doi: 10.1128/jvi.66.2.615-622.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA, 121–160. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 26.Temin H, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 27.Verma I M. Studies on reverse transcriptase of RNA tumor viruses. I. Localization of thermolabile DNA polymerase and RNase H activities on one polypeptide. J Virol. 1975;15:121–126. doi: 10.1128/jvi.15.1.121-126.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]