Abstract
The site-specific integration of wild-type adeno-associated virus (wtAAV) into the human genome is a very attractive feature for the development of AAV-based gene therapy vectors. However, knowledge about integration of wtAAV, as well as currently configured recombinant AAV (rAAV) vectors, is limited. By using a modified Alu-PCR technique to amplify and sequence the vector-cellular junctions, we provide the first direct evidence both in vitro and in vivo of rAAV-mediated transgene integration in several types of nondividing cells, including neurons. This novel technique will be highly useful for further delineating the mechanisms underlying AAV-mediated integration, including issues of frequency, site preference, and DNA rearrangement in human as well as animal cells. Results from these studies should be beneficial for the development of the next generation of gene delivery vectors.
Efficient gene transfer and stable transgene expression are two key features required for effective human gene therapy for congenital as well as many acquired disorders. Adeno-associated virus (AAV)-derived vectors are promising candidates for gene therapy because of their ability to mediate highly efficient gene transfer into several clinically relevant nondividing cell types, including neurons (7, 21, 28) and muscle cells (9, 13, 15, 22, 42), without inducing pathogenic or inflammatory side effects. Long-term expression of transgenes after AAV vector transduction into these tissues has also been documented (13, 15, 28, 42). One possible mechanism of AAV transgene stability is integration into the genomes of the transduced cells.
AAV is a human parvovirus with a 4.7-kb DNA genome containing two open reading frames, which encode the viral regulatory (rep) and structural (cap) proteins under the control of the p5, p19, and p40 promoters (25). The wild-type AAV (wtAAV) is able to integrate site specifically into the human host genome, at a particular locus (AAVS1) in chromosome 19 (16, 20, 34). This specific integration feature of AAV is believed to be mediated through its inverted terminal repeat (ITR) and Rep68 or Rep78 protein (4, 11, 27, 38). Rep-containing AAV vectors retain this specific integration feature, as demonstrated by our group (40) and others (1, 35). However, the rep gene is undesirable for engineering AAV-transducing vectors, due to its strong inhibitory effects on gene expression and cell growth (2). Recently, several groups (8, 14, 30, 33) demonstrated that rep− AAV vectors could also induce DNA integration in immortalized cell lines including 293 cells, although not in a site-specific fashion. However, there has been no fully convincing and direct evidence confirming the integration of AAV recombinants in nondividing cells. Our previous observation of a progressively increasing PCR-amplified transgene-specific signal in high-molecular-weight DNA samples from transduced human NT neurons was shadowed by the possibility of episomal replication and/or single-to-double-strand conversion of the AAV vector (7). Even recently reported Southern analysis data, claimed as evidence of AAV-mediated integration in nondividing human CD34+ hematopoietic progenitor cells (10) and mouse muscle cells (42), could not rule out possible contributions from undefined concatemers of episomal forms of the vectors. Furthermore, the requirement for large amounts of genomic DNA renders this technique problematic for addressing the integration issue in small samples of cells or tissues both in vitro and in vivo. In support of AAV vector integration in mouse muscle cells, Fisher et al. (9) recently reported other lines of evidence, including PCR detection of AAV vector persistence as head-to-tail concatemers, which are frequently found in the integrated stage. However, it is not clear whether episomal forms of AAV or its derivatives exist with this head-to-tail structure, and thus the possibility of episomal contamination again could not be absolutely excluded. PCR amplification with one primer targeting the chromosome 19 AAVS1 region is also not applicable, due to the fact that rep− AAV vectors do not appear to integrate with the same site-specific pattern as the wild-type virus. Thus, none of the current methodologies is able to comprehensively characterize the frequency of integration or address the possibility of AAV-mediated rearrangement in the integration site (22) in nondividing populations, especially in an in vivo setting.
In seeking a reliable method to analyze AAV-mediated integration in neural tissues, an advanced Alu-PCR approach (24) attracted our attention. As the major family of short interspersed repeat DNAs in humans, Alu sequences are present at the level of 9 × 105 copies per genome (26). They have been well characterized as 7SL RNA dimeric retropseudogenes, about 300 bp in length (36, 37). The high frequency of Alu sequences, appearing on average every 4 kb in the human genome, in combination with PCR techniques which can amplify long DNA fragments should in theory allow identification of any integrated exogenous DNA. One of the obvious advantages of an Alu-PCR technique is its ability to discriminate against episomal forms of AAV vectors by employing one primer specifically targeting the Alu sequences in the human genome. Furthermore, it has been shown that an Alu-equivalent sequence, B1, is present in the rodent genome at a similar frequency. Since the rodent is one of the most frequently used animal models for in vivo gene transfer studies, it is important to determine whether these AAV vectors, derived from a human virus, integrate into rodent tissues in vivo. We now report a successful methodology applying Alu-PCR and B1-PCR to identify rep− AAV vector-mediated DNA integration in nondividing cells of human origin in vitro and rodent in vivo, respectively. Our findings demonstrate stable AAV vector integration occurring in several highly susceptible targets.
MATERIALS AND METHODS
CMVβ-gal AAV vectors.
Construction and packaging of a recombinant AAV (rAAV) plasmid, CMVβ-gal (previously named AAVβ-gal), have been described in detail previously (7). Briefly, semiconfluent 293 cells were infected with adenovirus type 5 (Ad5; provided by N. Muzyczka, University of Florida) and then cotransfected with 18 μg of CMVβ-gal vector plasmid and 6 μg of pAd8 helper plasmid (provided by R. J. Samulski, University of North Carolina), using a standard calcium phosphate precipitation method. Three days posttransfection, cells were harvested and subjected to four freeze-thaw cycles to release the cell-associated virions. Contaminating Ad5 was inactivated by heating to 56°C for 30 min, and any remaining plasmid DNA was removed by treatment with 25 U of RNase-free DNase per ml at 37°C for 30 min. In some experiments, these AAV preparations were further purified by two rounds of ultracentrifugation on a CsCl gradient (1.38 g/ml). Packaged CMVβ-gal viral stocks were titered by the infectious center assay, a modification of the method described by McLaughlin et al. (23). Briefly, 50,000 293 cells were infected with serially diluted AAV stocks (1:1 to 1:105) together with wtAAV and Ad5, both at a multiplicity of infection (MOI) of 5. After 30 h, cells were resuspended in 25 mM EDTA–phosphate-buffered saline and then trapped on a nylon membrane by vacuum filtration. Total DNA was fixed and then hybridized with a specific 32P-labeled β-galactosidase probe. The numbers of labeled dots representing the number of infected cells containing the actively replicated CMVβ-gal were counted and plotted to determine the infectious units (IU) per ml. The titers of our packaged viral lysates were usually in the range of 108 IU/ml.
Cells.
293 cells were maintained in Eagle’s minimal essential medium containing 10% fetal bovine serum (FBS) and penicillin-streptomycin (pen/strep). The method for preparing human NT neurons was described previously (39). Briefly, the precursor NT2 cells were treated with 10 μM retinoic acid for 4 to 5 weeks and then replated at low density (1:6). One to two days later, top layers containing differentiated cells were mechanically dislodged and replated in culture dishes precoated with 0.01% poly-d-lysine–1:20 Matrigel (Collaborative Research). Cells were cultured in Dulbecco modified Eagle essential medium–10% FBS–pen/strep plus mitotic inhibitors for 3 weeks. The enriched and terminally differentiated neurons were maintained in Opti-MEM 1–5% FBS–pen/strep. Human primary alveolar macrophages from bronchoalveolar lavages were cultured in RPMI 1640–10% FBS–pen/strep. Transduction was performed by incubating cells with packaged CMVβ-gal vector at an MOI of 5 in a limited amount of medium without serum for 90 min at 37°C. Cells were then cultured in appropriate media with serum for another week.
Animals.
Adult male Sprague-Dawley rats (weighing 250 to 270 g; Harlan Sprague-Dawley, Inc., Indianapolis, Ind.) were maintained in well-ventilated rooms at 24°C, 50% relative humidity, and a 12-h/12-h daily light cycle. Purina Rat Chow and tap water were given ad libitum. Intracerebroventricular injection of AAV vectors was performed as previously described (29). Briefly, animals were anesthetized with pentobarbital (65 mg/kg of body weight intraperitoneally). Five microliters of CMVβ-gal vector (5 × 108 IU/ml) or artificial cerebrospinal fluid was injected into the left lateral ventricle on the coordinates of A-P (1 mm behind the bregma), D-V (5 mm from the skull), and Lat (1.2 mm). Two weeks postinjection, animals were euthanized and their brain tissues were collected for DNA isolation.
Alu-PCR and Southern hybridization.
High-molecular-weight DNA was extracted from cultured cells or brain tissues 1 to 2 weeks after CMVβ-gal transduction, using Puregene DNA isolation kits (Gentra Systems, Research Triangle Park, N.C.); 100-ng DNA samples were subjected to Alu- or B1-PCR.
dUTP-containing primers used in the first 10 PCR cycles included one B1, one CMV, and two Alu primers. The Alu5 (Fig. 1) and Alu3 primers were complementary to the 5′ and 3′ regions of the Alu consensus sequence, respectively. They were designed according to the description of Minami et al. (previously named A5 and A3 [24]) with their 5′ ends linked to a tag sequence. The B1-5 primer (CAGUGCCAAGUGUUUGCUGACGCCAAGUGCUGGGAUUAAAGG) included a sequence covering the 5′ region of the B1 consensus sequence (5) and the same tag (underlined nucleotides) used in the Alu primers. C1 (AUUAUUGACGUCAAUGGGCGGGGGUCGUUG) was a CMV-specific primer (Fig. 1). The GeneAmp XL PCR kit and AmpliWax PCR Gem (both from Perkin-Elmer/Applied Biosystems, Foster City, Calif.) were used in combination with the hot-start technique in order to achieve the optimal DNA amplification. The rTth DNA polymerase-XL (for extra-long [XL] amplification), the optimized enzyme used in the kit, contains both polymerase and proofreading activities and allows XL PCR amplification of up to 22 kb from human genomic DNA. The first 10 cycles were conducted according to the manufacturer’s instructions. Different amounts and combinations of either 5′ or 3′ primer ranging from 10 to 100 pmol were tested empirically. Following 10 2-step cycles (94°C for 30 s and 61°C for 6 min), each sample was incubated with 1 U of uracil DNA glycosylase at 37°C for 30 min and then heated for 10 min at 94°C to break DNA strands at apurinic dUTP sites.
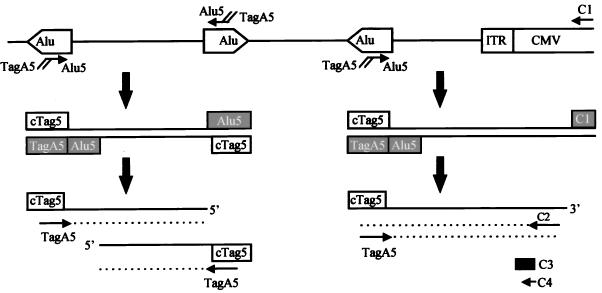
FIG. 1.
Schematic of Alu-PCR. Human genomic DNA from CMVβ-gal-transduced cells was amplified by using the first set of primers: Alu5, an Alu-specific primer containing a tag; and C1, a CMV-specific primer. After an initial 10 cycles of PCR, the first set of primers was destroyed by uracil DNA glycosylase-induced nicks at dUTP (filled box). cTag5 (open box) represents a sequence complementary to the tag sequence on the newly synthesized strand. These PCR products were further amplified by using an internal primer, C2, for the CMV sequence plus TagA5, containing 16 nucleotides of Tag sequence and 6 nucleotides of Alu5 sequence. C3 was used as a specific CMV oligonucleotide probe for Southern hybridization. C4 and TagA5 were used to further amplify the above Alu-PCR products in order to obtain discrete bands for direct sequencing.
These initial PCR products were further amplified by using an internal primer, C2 (Fig. 1), corresponding to the CMV sequence (GGCGGGCCATTTACCGTAAGTTATGT), in combination with one of the following tag primers. TagA5 (or Tag5) (24) and TagA3 (or Tag3) (24) were so designated because they contained mainly the tag sequence but also a small portion (six nucleotides) of 5′ and 3′ regions of the Alu repeat, respectively. TagB (CAAGTGTTTGCTGACGCCAAGT) had the same tag sequence with an additional six nucleotides derived from the B1 repeat. After addition of the second pair of primers (the Tag and CMV nested primers), the “touchdown” PCR technique was used for further amplification. This specific technique was developed to minimize mispriming-induced nonspecific PCR amplification, especially when a complex genome is used as a template (6). It started with a denaturation step at 94°C for 30 s, followed by an annealing step for 30 s with the temperature decreasing 1°C every second cycle from 65 to 55°C and then an extension step at 72°C for 5 min; 20 cycles at the final touchdown temperature (55°C) were followed by a final extension at 72°C for 8 min.
Aliquots of 25 to 35 μl of each reaction product (out of a 100-μl total) were fractionated on a 1% agarose gel, transferred to nylon membrane, and then hybridized with a 32P-end-labeled CMV-specific oligonucleotide probe (C3, ACGGGGTCATTAGTTCATAGCCCATATATGGAGTTCCGCG [Fig. 1]) at 64°C overnight. Membranes were stringently washed and then subjected to autoradiography. All of the primers targeting the AAV vector as well as the specific probe were in the CMV region and at least 30 bp away from the AAV ITR, which has been reported to undergo frequent rearrangement (19) when integrated into the host genome.
Nested PCR, cloning, and sequencing.
One microliter of the above PCR products from CMVβ-gal-transduced cells was subjected to another run of touchdown PCR amplification using a nested CMV primer, C4 (AACCCAAGCTTGCGGAACTCCATATATGG [Fig. 1]), in combination with TagA5, both at 10 pmol. Ten microliters of the amplification product was gel fractionated, transferred to a membrane, and then hybridized with the C3 probe. The rest of the reaction was fractionated in a 0.8% agarose gel. To reduce the background of nonspecific genomic DNA and to enhance the amplification of signals, the CMV-specific bands were extracted and then subjected to another round of nested PCR using a pair of internal primers: A5N (AAAAAGAATTCAGTGTTTGCTGACGCCAA) and C5N (ACACCAAGCTTATATATGGGCTATGAACT). The addition of the EcoRI and HindIII sites in A5N and C5N, respectively, was to facilitate cloning of the PCR fragments into plasmids. These PCR products were gel fractionated. The most intense fragment was then extracted and cloned into pGEM11Z (Promega, Madison, Wis.). In other cases, the original Alu-PCR products were subjected to three rounds of nested PCR, and the end products were then cloned directly into the pCR 2.1 vector according to the manufacturer’s instructions (Original TA Cloning kit; Invitrogen Co., Carlsbad, Calif.). Purified plasmid DNA containing the PCR fragment was then subjected to dideoxy-chain termination sequencing.
RESULTS
Development of Alu-PCR to study AAV integration.
As a first test of whether an Alu-PCR technique was applicable for investigation of AAV vector-mediated integration in human cells, we applied this method in human 293 cells, since AAV vector integration is already well documented by fluorescence in situ hybridization in this cell line (8). rAAV vector-mediated gene transfer into this cell type is highly efficient. The result of a typical CMVβ-gal transduction into 293 cells is shown in Fig. 2.
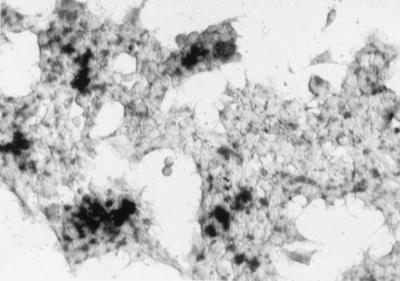
FIG. 2.
5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) histochemical staining of 293 cells 2 days after transduction with the CMVβ-gal vector at an MOI of 15.
The schematic drawing in Fig. 1 outlines the basic strategy of the Alu-PCR technique. Chromosomal DNA from transduced cells was subjected to two rounds of PCR amplification, with the first pair of primers specifically targeting the 5′ end of the human Alu sequence (Alu5) and the CMV immediate-early (IE) promoter (C1) in our CMVβ-gal AAV vector. Two unique features were built into this pair of primers: (i) they were both made with dUTP instead of dTTP, and (ii) a tag sequence was introduced at the 5′ end of the Alu5 primer (Fig. 1). These primers were then destroyed by uracil DNA glycosylase-induced cleavage at the uracil residues after the first 10 PCR cycles. The amplified products, encompassing fragments between integrated vector sequences and nearby Alu sites, were then further amplified by using a second pair of primers targeting the tag and a nested CMV sequence (C2 [Fig. 1]). Further amplification of any initial PCR products formed between two Alu sequences was limited due to the presence of only one homologous primer (TagA5) (Fig. 1, left array). A touchdown protocol (6, 24) was applied for the second part of the PCR amplification to further enhance sensitivity and specificity by gradually decreasing the annealing temperature to minimize products resulting from misprimed amplification. Various ratios of Alu5 to C1, the first pair of primers, were tested in order to obtain the optimal conditions for PCR amplification. Other optimal cycling conditions, such as the concentration levels of magnesium and the annealing temperature in the first 10 cycles, etc., were also determined empirically. All samples having different first primer sets were then treated equally for the rest of the procedure.
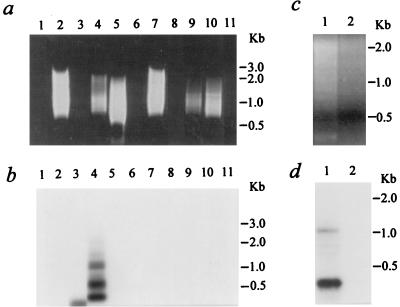
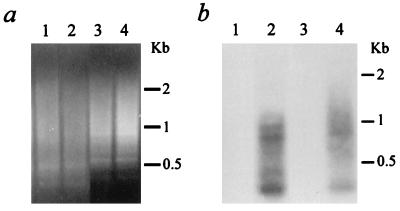
We further modified the original method of Minami et al. (24) by using rTth DNA polymerase-XL (Perkin-Elmer/Applied Biosystems), an optimized enzyme containing both polymerase and proofreading activities and allowing XL PCR amplification of up to 22 kb from human genomic DNA. Other modifications included using a lower concentration of magnesium (1.1 mM) and two-step PCR cycling. Together, these changes resulted in dramatically different amplification efficiencies between our Alu-PCR and that of Minami et al. when using the first pair of primers at a ratio of 10:100 pmol. With this ratio, we could not detect any visible PCR products on an ethidium bromide-stained gel (Fig. 3a). However, other ratios, including 10 pmol of Alu5 primer only and Alu5 plus C1 at 10:10 or 100:10, all gave some smeared bands with sizes of up to 3 kb visible on an ethidium bromide-stained gel (Fig. 3a). The larger fragment sizes that we were able to amplify are close to the average distance, 4 kb, between copies of Alu in the human genome. Specific signals detected by a 32P-labeled CMV oligonucleotide probe (C3 [Fig. 1]) were found in the sample from the CMVβ-gal-transduced cells that was amplified by using the Alu5-C1 combination at a 10:10 ratio (Fig. 3b, lane 4). These six specific bands were 2.1, 1.6, 1.2, 1.1, 0.52, and 0.4 kb in size. In contrast, a similar but not identical smeared pattern of PCR products from the control cells (Fig. 3a, lane 9), likely due to amplifications of adjacent Alu sequences located in opposite orientations, lacked any detectable specific hybridization signals (Fig. 3b, lane 9). Although no PCR products were visible in a sample with 100 pmol of the C1 primer alone by ethidium bromide staining (Fig. 3a, lanes 3 and 8), a 270-bp band was detected by the C3 probe (Fig. 3b, lane 3). This specific signal might be the result of amplification of head-to-head tandem concatemers of the rAAV, which could be present in either integrated or episomal forms. Based on the fact that this 270-bp fragment was not found in the same DNA sample subjected to Alu-PCR with both the cellular (Alu5) and vector (C1) primers (Fig. 3b, lane 4), we hypothesize that the presence of the two primers together may inhibit amplification of any vector-vector templates under the conditions used.
FIG. 3.
Alu-PCR analysis on 293 cell samples. (a and b) Cells were either untreated or transduced with a crude preparation of CMVβ-gal. (a) Aliquots of 25 μl of PCR products were loaded in a 1% agarose gel containing ethidium bromide. Lane 1, control sample without a genomic DNA template; lanes 2 to 6, 100 ng of genomic DNA isolated 8 days after transduction of the CMVβ-gal vector into 293 cells; lanes 7 to 11, 100 ng DNA from control 293 cells; lanes 2 and 7, 10 pmol of Alu5 primer alone for the first 10 cycles; lanes 3 and 8, 10 pmol of C1 primer alone; lanes 1, 4, and 9, Alu5-C1 at 10 pmol: 10 pmol; lanes 5 and 10, Alu5-C1 at 100:10; lanes 6 and 11, Alu5-C1 at 10:100. (b) The gel shown in panel a subjected to Southern blot analysis. PCR-amplified products on a nylon membrane were hybridized with a 32P-end-labeled specific CMV oligonucleotide probe (C3). (c and d) Alu-PCR amplification from 100 ng of genomic DNA isolated 8 days after transduction with CsCl gradient-purified CMVβ-gal. (c) Aliquots of 25 μl of PCR products loaded in an agarose gel. Lane 1, Alu5-C1 at 10 pmol: 10 pmol; lane 2, C1 primer alone. (d) The gel shown in panel c subjected to Southern blot analysis.
The Alu-PCR and Southern hybridization were performed at least five times on the same DNA sample of 293 cells treated with the crude preparation of CMVβ-gal. A pattern of C3-detected bands similar to that shown in Fig. 3 was consistently and repeatably detected. One or two bands were absent in some instances, while one or two additional bands were evident in others. These differences were likely due to variations in the performance of the Alu-PCR amplification.
To minimize the possible effects of contaminating Ad5 or its proteins on the integration of rAAV, we conducted the same analyses on a genomic DNA sample isolated from 293 cells following transduction with a CsCl gradient-purified CMVβ-gal viral stock at an MOI of 5 (Fig. 3c and d). Similar to what we observed in the samples from 293 cells treated with a crude preparation of the rAAV, multiple bands (1.2, 0.8, 0.6, and 0.3 kb in size) were detected by the C3 probe (Fig. 3d, lane 1). No specific signals were detected when the same sample was subjected to Alu-PCR with only the CMV (vector) primer (Fig. 3d, lane 2).
To further exclude the possibility that nonspecific amplification from episomal forms of the AAV vector contributed to these signals, we performed the same Alu-PCR analysis on either 0.1 or 1 ng of the CMVβ-gal plasmid, mixed with chromosomal DNA from control 293 cells (data not shown). The absence of specific hybridization signals in these samples confirmed that the Alu-PCR technique is specific in distinguishing integrated chromosomal copies of the transgene from episomal plasmids. These findings indicated that this Alu-PCR technique was successful in our initial trial to identify AAV-mediated integration in 293 cells.
Further evaluation of Alu-PCR as a significant and reliable tool for studies of AAV vector-mediated integration was accomplished by successful sequencing of vector-cellular junctions. As described in Materials and Methods, Alu-PCR products derived from CMVβ-gal-transduced 293 cells were subjected to several rounds of touchdown PCR amplification using nested primers. The discrete bands were purified by gel fractionation and inserted into plasmids. Three clones, containing PCR fragments ranging from 400 to 1,300 bp in size, were either fully or partially sequenced. Detailed data for one representative clone containing the 0.4-kb fragment pictured in Fig. 3b, lane 4, are shown in Fig. 4. The 362 nucleotides depicted in Fig. 4 revealed sequences present in the CMVβ-gal vector, as well as other sequences which were not vector derived. The vector sequence included 25 nucleotides from the 5′ end of the CMV IE promoter and, most importantly, the 5′ end of the AAV ITR covering the intact D, A′, and B′ regions. The B, C, C′, and A regions in the 5′ end of the ITR (3, 25) were deleted. The presumptive cellular sequences included a 73-nucleotide stretch in the middle portion (dash-boxed in Fig. 4a) possessing 64% homology with the human sequence from PAC 448E20 on chromosome Xq26.1, based on an advanced BLAST search conducted through the National Center for Biotechnology Information service. Both vector (including AAV ITR) and cellular sequences were present in all three clones, demonstrating that this Alu-PCR technique can amplify junction fragments between integrated AAV vectors and the flanking chromosomal DNA.
FIG. 4.
Sequence of a vector-cellular junction identified by Alu-PCR from 293 cells transduced with the CMVβ-gal rAAV vector. (a) The vector portion of the junction includes the 5′ end of the CMV IE promoter (open box) and some AAV sequences (underlined) containing a partial AAV ITR. The cellular sequence contains a fragment with 64% homology to a human DNA sequence on chromosome Xq26.1 (dashed box), which is flanked by unidentified sequences not present in the CMVβ-gal vector (unmarked). The actual sequence in the shaded box is shown in panel b. The arrows in panels a and b point to the site of the vector-cellular junction.
AAV vector integration in nondividing cells in vitro.
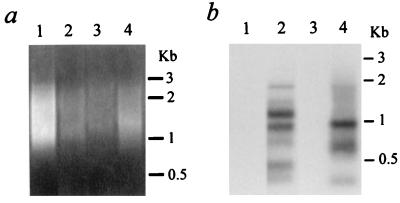
The same Alu-PCR technique that we used successfully to detect AAV integration in 293 cells was applied next in several types of nondividing human cells. One of the cell types chosen was human NT neurons, because our group and others have noted dramatic gene transfer efficiencies mediated by AAV vectors into neuron populations, both in vitro and in vivo (7, 21, 28). Other reasons for selecting this particular cell system for in vitro integration studies included its human origin, our previous successes using this line (7), and the fact that it represents a highly pure population of nondividing cells exhibiting typical neuronal features (39). NT neurons at 4 weeks of age were transduced with the packaged CMVβ-gal vector at an MOI of 3. One week later, chromosomal DNA was isolated and subjected to Alu-PCR amplification using the Alu5 and C1 primers. About one-third of the resulting PCR products were gel fractionated and subjected to hybridization with the C3 probe labeled with 32P. A smeared pattern of amplified products was visible in sample lanes from both untreated and transduced NT neurons (Fig. 5a). However, specific signals detected by the C3 probe were found only in the sample from the CMVβ-gal-transduced cells (Fig. 5b). Consistent with the results from our previous study on 293 cells, multiple bands (1.9, 1.4, 1.1, 0.9, 0.6, 0.4, and 0.3 kb) were found in the transduced NT neuronal samples.
FIG. 5.
Alu-PCR analysis using an Alu5 primer. (a) Aliquots of 33 μl of Alu-PCR products from 100 ng of genomic DNA were loaded on a 1% agarose gel containing ethidium bromide. Lane 1, control human NT neurons; lane 2, NT neurons transduced with CMVβ-gal for 7 days; lane 3, control human alveolar macrophages; lane 4, transduced alveolar macrophages. (b) The gel shown in panel a subjected to Southern blot analysis with a 32P-end-labeled specific CMV oligonucleotide probe (C3).
As in the case of vector-treated 293 cells, confirmation that fragments being amplified by the Alu-PCR technique represented AAV vector-mediated integration events in the nondividing human NT neurons was obtained by successful sequencing of vector-cellular junctions. Following several rounds of nested PCR, the end products of amplification were subjected directly to TA cloning. Three clones, containing PCR fragments ranging from approximately 200 to 800 bp in size, were either fully or partially sequenced. Both vector and cellular sequences were present in all three clones. Detailed data for one representative clone containing the 0.3-kb fragment pictured in Fig. 5b, lane 2, are shown in Fig. 6. The 272 nucleotides depicted in Fig. 6 contained both vector-derived and non-vector-derived sequences. The vector sequence included 46 nucleotides from the 5′ end of the CMV IE promoter and the 5′ end of the AAV ITR covering the intact D, A′, and C′ regions. The B, B′, C, and A regions in the 5′ end of the ITR (3, 25) were deleted. The presumptive cellular sequences included an 88-nucleotide stretch (dash-underlined in Fig. 6a) possessing 98% homology with a sequence on human chromosome segment 9q34, based on an advanced BLAST search. In addition to the minor deletion of the ITR sequence in this junction, substantial vector rearrangement was also evident in one of the other clones (data not shown).
FIG. 6.
Sequence of a vector-cellular junction identified by Alu-PCR from human NT neurons transduced with the CMVβ-gal rAAV vector. (a) The vector portion of the junction includes the 5′ end of the CMV IE promoter (open box) and some AAV sequences (underlined) containing a partial AAV ITR. The cellular sequence includes a fragment with 98% homology to a human DNA sequence on chromosome 9q34 (dashed underlined). The actual sequence in the shaded box is shown in panel b. The arrows in panels a and b point to the site of the vector-cellular junction.
A very different type of nondividing human cell was applied at the same time to this AAV integration study, using the Alu-PCR method. Primary human alveolar macrophages were chosen because, like neurons, they appear to be highly susceptible to AAV-mediated transduction (12). Macrophages were isolated by bronchoalveolar lavage and cultured for 1 week before CMVβ-gal exposure. About 1 week after transduction, high-molecular-weight DNA extracted from either treated or control cells was subjected to Alu-PCR and Southern hybridization. Similar results were obtained as with the neuronal cells, with multiple transgene-specific signals (1.0, 0.6, and 0.3 kb) apparent only in the chromosomal DNA sample from cells transduced with the CMVβ-gal vector (Fig. 5).
It is noteworthy that the above Alu-PCR analyses were performed at least three times and the results were consistent within the same DNA sample from either the NT neurons or the alveolar macrophages. No specific signals were detected by the C3 probe in either cell type when the PCR was performed with only the vector primer (C1) (data not shown).
To further confirm the validity of these findings, an alternative primer, Alu3, specifically targeting the 3′ end of the human Alu sequence, was used in place of Alu5 in the first 10 cycles of Alu-PCR amplification. Specific C3 probe-labeled bands (ranging from 0.3 to 0.9 kb) were still detected in CMVβ-gal-transduced NT neurons or alveolar macrophages but not in either type of control cell (Fig. 7). Together, these results confirmed that Alu-PCR is a viable technique for studies of AAV vector-mediated integration and demonstrated that rep− AAV vectors can integrate into the genomes of at least some types of nondividing cells.
FIG. 7.
Alu-PCR analysis using an Alu3 primer. (a) Aliquots of 33 μl of Alu-PCR products from 100 ng of genomic DNA were loaded on a 1% agarose gel containing ethidium bromide. Lane 1, control human NT neurons; lane 2, NT neurons transduced with CMVβ-gal for 7 days; lane 3, control human alveolar macrophages; lane 4, transduced alveolar macrophages. (b) The gel shown in panel a subjected to Southern blot analysis with a 32P-end-labeled specific CMV oligonucleotide probe (C3).
AAV vector integration in rat brain in vivo.
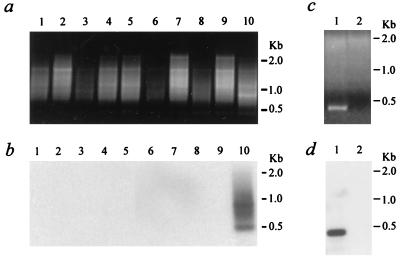
Based on our success using Alu-PCR to detect integration of an AAV transgene cassette in nondividing human NT neurons, we applied an Alu-PCR equivalent, B1-PCR, to brain samples from rats exposed to the same CMVβ-gal AAV vector. Two weeks after injection of 5 μl of crude preparation of CMVβ-gal viral stock (5 × 105 IU, titered by infectious center assay) into the left lateral ventricle, genomic DNA from different brain regions was isolated separately and subjected to CMV-B1-PCR analysis. B1-5 is a primer covering the 5′ region of the B1 consensus sequence (5), and its 5′ end is linked to the same tag sequence used in the Alu primer (Fig. 1). Both B1-5 and the CMV-specific primer C1 (Fig. 1) were used at 10 pmol for the first 10 cycles of PCR amplification. The rest of the procedures, including treatment with uracil DNA glycosylase, touchdown PCR, gel fractionation, and Southern blot hybridization with the CMV-specific oligonucleotide probe, were carried out just as performed in the above Alu-PCR studies. Smeared B1-PCR products were visible in samples from both control and vector-injected rats in all of the sampled brain regions, including the frontal cerebral cortex, cerebellum, striatum, hypothalamus, and brainstem (Fig. 8a). However, only the hypothalamic DNA sample from the CMVβ-gal-injected animal had specific signals (0.9, 0.7, and 0.5 kb) reactive with the CMV probe (Fig. 8b).
FIG. 8.
B1-PCR analysis of rat brain samples. (a) Aliquots of 30 μl of B1-PCR products from 100 ng of genomic DNA were loaded on a 1% agarose gel containing ethidium bromide. Lanes 1 to 5, brain samples from a rat 2 weeks after injection with artificial CSF; lanes 6 to 10, samples from a rat injected with 5 × 105 IU of a crude preparation of the CMVβ-gal AAV vector; lanes 1 and 6, cerebral cortex; lanes 2 and 7, cerebellum; lanes 3 and 8, striatum; lanes 4 and 9, brainstem; lanes 5 and 10, hypothalamus. (b) The gel shown in panel a subjected to Southern blot analysis with a 32P-end-labeled specific CMV oligonucleotide probe (C3). (c and d) Alu-PCR amplification from 100 ng of hypothalamic genomic DNA isolated 2 weeks after injection with the CsCl gradient-purified CMVβ-gal. (c) Aliquots of 40 μl of PCR products were loaded in an agarose gel. Lane 1, B1-5-C1 at 10 pmol: 10 pmol; lane 2, C1 primer alone. (d) The gel shown in panel c subjected to Southern blot analysis.
The B1-PCR and Southern hybridization were also performed on genomic DNA samples from rat brain injected with CsCl gradient-purified CMVβ-gal vector. One dominant fragment (0.4 kb) as well as two faint bands (0.8 and 0.2 kb) were observed in the hypothalamic sample (Fig. 8c and d). Similar to the earlier studies on genomic DNA samples from NT neurons and alveolar macrophages, no specific signals were detected by the C3 probe when the same DNA samples were subjected to PCR analysis using only the CMV primer (Fig. 8d).
DISCUSSION
A PCR strategy targeting Alu sequences has been used previously to examine cellular sequences flanking integrated hepatitis B virus in human hepatoma cells (24). We have now successfully adapted an Alu-PCR approach and an equivalent B1-PCR technique to directly identify AAV vector-mediated integration in nondividing cells of either human or rat origin. The samples included a rapidly dividing human cell line, terminally differentiated nondividing human NT neurons, primary human lung alveolar macrophages, and rat brain tissues. Findings from the dividing cells were in good agreement with those of other groups (8, 12, 30, 33), including observations using the fluorescence in situ hybridization method to analyze metaphase spreads. The reliability of this Alu-PCR technique in combination with Southern hybridization was confirmed by our observations of consistent patterns of specific signals in repeated amplifications from the same DNA sample. Most importantly, the successful sequencing of vector-cellular junctions identified by this new technique will enable further examination of the integration events of rAAV vectors in primary, nondividing cells or tissues.
Multiple transgene-specific bands were detected from each type of cells or tissues exposed to the packaged CMVβ-gal vector. It is possible that two signals represent different amplified products targeting the same AAV integration locus but flanked by different nearby Alu sequences. However, the chances that all the specific bands in a given sample were derived from a single integration site are very slim. It is unlikely that Alu sequences reside in such a high frequency in the genome, e.g., seven copies within a 1.8-kb span in the case of the transduced NT neurons. In support of this view, although only a small number of amplified junctions were sequenced, no site duplications were noted. It also cannot be ruled out that some AAV integration sites are undetectable by this Alu-PCR method. These would include sites located either too far from an Alu sequence (26) or near a particular Alu element with a sequence too divergent from the consensus and therefore unable to anneal efficiently with the Alu primer. Nevertheless, the detection of relatively small numbers of specific bands suggests that rep− AAV vector-mediated integration is not entirely random; otherwise, a greater smearing of specific signals might be expected in the amplified chromosomal DNA samples from the mixed cell populations. Alternatively, many integration sites may be present but accompanied by a significant degree of bias toward a few select sites. It is possible that the junctions amplified by Alu-PCR are drawn primarily from these most preferred sites. It is also noteworthy that neurons and alveolar macrophages represent cell types known to be highly susceptible to stable AAV-mediated gene transfer. The detection of integrated transgenes in these lineages cannot be assumed to be characteristic of every primary cell type following exposure to rAAV.
It is well known that Rep protein provided in trans is critical for wtAAV-mediated site-specific integration into human chromosome 19. Although the rep gene cassette is not encoded by the rAAV vectors used, there are two possible sources of Rep in the rAAV stocks. The first one is contamination by wtAAV-like particles in the rAAV preparation. Using an infectious center assay, we detected very low levels of wtAAV-like particles, 2 IU rather than the 105 IU of rAAV, in our crude preparations of rAAV. This contamination was not introduced from the helper Ad5 viral stocks, since no positive signals were detected in assays using a rep-specific DNA probe in a cell sample treated with Ad5 alone (41a). Kube et al. (17) reported similar findings for studies using CsCl gradient-purified rAAV stocks and hypothesized that “wild-type AAV-like particles can be generated by recombination events involving the recombinant AAV plasmid and the AAV helper plasmid, pAAV/Ad, during production of recombinant AAV.” A second potential source of Rep could be Rep proteins attached to rAAV virions. Recently, two groups have reported that functional Rep proteins are tightly associated with and even encapsidated within wtAAV and rAAV virions (17, 31, 32). The role of these associated proteins in rAAV-mediated nonspecific integration remains to be defined. However, our results from sequencing several amplified vector junctions support the view that the strong affinity of wtAAV for a small region of human chromosome 19 is not mimicked by the rAAV vectors used here.
One of the main issues to be addressed in our present in vivo study was whether this type of human viral vector could mediate transgene integration in a rodent species. In other studies using the CMVβ-gal vector, we have noted little or no transgene expression in brain regions following intraventricular injection of the vector. However, using an otherwise isogenic AAV vector, AVPβ-gal, in which the transgene is driven by a native neurohormone promoter, we have detected significant amounts of gene expression, principally in the hypothalamus, although with weaker staining in a few other brain regions (41). Recent studies by other groups (21, 28) have demonstrated stable AAV transduction for at least 3 months in the rat or mouse central nervous system (CNS) in vivo, yet little is known about the mechanism(s) underlying the stability of AAV vectors in neuronal tissues. AAV-mediated integration is one possibility. Another key issue was whether this Alu-PCR methodology would be sufficiently sensitive to detect integrated AAV transgenes in brain cells. This is a critical question, especially for the brain, into which only small amounts of vector may be injected. Restricted diffusion of AAV vectors in brain tissue may also limit the numbers of cells in any given brain region harboring an AAV vector.
We therefore developed B1-PCR, an Alu-PCR equivalent, to examine AAV vector integration in the rat brain. By using the CMVβ-gal vector, we were able to take advantage of the same basic methodology that we had established and optimized for human cells. Two weeks after injection of the packaged CMVβ-gal into the lateral ventricle, transgene-specific signals were detected in the high-molecular-weight DNA sample from the hypothalamus but not from other regions including the frontal cerebral cortex, cerebellum, striatum, and brainstem. This result was not surprising since intraventricularly injected AAV vectors can easily reach the hypothalamus, due to the close proximity of this structure to the ventricular system; thus, a considerable amount of virus was likely delivered into hypothalamic tissue compared to the other sampled regions. This finding was consistent with the pattern of vector expression detected in our earlier histochemical study using the AVPβ-gal vector. The results also support a previous report by McCown et al. (21), suggesting that low levels of expression from an AAV-delivered transgene in the brain were due to inactivation of the CMV IE promoter rather than to the absence of the gene cassette. Most importantly, our B1-PCR data indicate for the first time that a human AAV-derived vector can integrate into the genomes of brain cells in a rodent species and confirm the ability of the B1-PCR technique to detect limited integration signals in vivo.
Several features of Alu-PCR analysis would appear to render it more attractive for the study of AAV vector integration in nondividing cells than other methodologies such as Southern blot analysis (10, 42) or PCR amplification of AAV head-to-tail junctions (9). Alu-PCR directly targets the vector-cellular junction sequences, which is not possible with alternative methods, and therefore avoids the attendant problems in interpretation resulting from episomal contamination. In addition, the combination of nested PCR and sequencing provides a relatively simple way to further examine the integration sites of AAV vectors in vivo and to elucidate possible sequence rearrangements near the junction site in both cellular and vector regions. By targeting a transgene possessing an endogenous counterpart present at a low copy number, it should also be possible to determine the actual AAV integration frequency by using quantitative Alu-PCR to compare intensities between endogenous and exogenous signals.
As the only known mammalian virus which can integrate into the human genome at a specific site (18), AAV possesses uniquely attractive features for certain applications of gene therapy. The demonstration of vector integration in treated brain and neuronal cell samples provides strong encouragement for further development of the AAV system for application in the CNS. Further characterization of this event in rats should be beneficial to the broad usage of the rodent as an animal model for investigating AAV-mediated gene delivery into the CNS. The Alu-PCR method described here will be a useful tool for further dissecting the mechanisms underlying this integration and may provide insights into designing strategies for the next generation of AAV vectors for human gene therapy.
ACKNOWLEDGMENTS
We thank Henry Koziel (Division of Pulmonary Medicine, Beth Israel Deaconess Medical Center) for providing human lung alveolar macrophages and Jerome E. Groopman, Yongjia Yu, and In-Woo Park for helpful discussions. We also thank Xianglin Ren for technical assistance and Janet Delahanty for editing the manuscript, as well as Evelyn Gould and Nancy DesRosiers for assistance in preparing the figures.
This work was supported by NIH grants P01 HL43510-06 and RO1 HL44846 and by an award to E.F.T. from the Concerned Parents for AIDS Research.
REFERENCES
- 1.Balagué C, Kalla M, Zhang W-W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns K I. Parvovirus replication. Microbiol Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I, Linden R M. The cryptic life style of adeno-associated virus. Bioessays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 4.Chiorini J A, Wiener S M, Owens R A, Kyöstiö S R M, Kotin R M, Safer B. Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J Virol. 1994;68:7448–7457. doi: 10.1128/jvi.68.11.7448-7457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Dunnen J T, Schoenmakers J G. Consensus sequences of the Rattus norvegicus B1 and B2 repeats. Nucleic Acids Res. 1987;15:2772. doi: 10.1093/nar/15.6.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du B, Wu P, Boldt-Houle D M, Terwilliger E F. Efficient transduction of human neurons with an adeno-associated virus vector. Gene Ther. 1996;3:254–261. [PubMed] [Google Scholar]
- 8.Duan D, Fisher K J, Burda J F, Engelhardt J F. Structural and functional heterogeneity of integrated recombinant AAV genomes. Virus Res. 1997;48:41–56. doi: 10.1016/s0168-1702(96)01425-6. [DOI] [PubMed] [Google Scholar]
- 9.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 10.Fisher-Adams G, Wong K K, Jr, Podsakoff G, Forman S J, Chatterjee S. Integration of adeno-associated virus vectors in CD34+ human hematopoietic progenitor cells after transduction. Blood. 1996;88:492–504. [PubMed] [Google Scholar]
- 11.Flotte T R, Carter B J. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 12.Inouye R T, Du B, Boldt-Houle D, Ferrante A, Park I-W, Hammer S M, Duan L, Groopman J E, Pomerantz R J, Terwilliger E F. Potent inhibition of human immunodeficiency virus type 1 in primary T cells and alveolar macrophages by a combination anti-Rev strategy delivered in an adeno-associated virus vector. J Virol. 1997;71:4071–4078. doi: 10.1128/jvi.71.5.4071-4078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaplitt M G, Xiao X, Samulski R J, Li J, Ojamaa K, Klein I L, Makimura H, Kaplitt M J, Strumpf R K, Diethrich E B. Long-term gene transfer in porcine myocardium after coronary infusion of an adeno-associated virus vector. Ann Thorac Surg. 1996;62:1669–1676. doi: 10.1016/s0003-4975(96)00946-0. [DOI] [PubMed] [Google Scholar]
- 14.Kearns W G. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 15.Kessler P D, Podsakoff G M, Chen X, McQuiston S A, Colosi P C, Matelis L A, Kurtzman G J, Byrne B J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci USA. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotin R M, Linden R M, Berns K I. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kube D M, Ponnazhagan S, Srivastava A. Encapsidation of adeno-associated virus type 2 Rep proteins in wild-type and recombinant progeny virions: Rep-mediated growth inhibition of primary human cells. J Virol. 1997;71:7361–7371. doi: 10.1128/jvi.71.10.7361-7371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linden R M, Berns K I. Site-specific integration by adeno-associated virus: a basis for a potential gene therapy vector. Gene Ther. 1997;4:4–5. doi: 10.1038/sj.gt.3300357. [DOI] [PubMed] [Google Scholar]
- 19.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCown T J, Xiao X, Li J, Breese G R, Samulski R J. Differential and persistent expression patterns of CNS gene transfer by an adeno-associated virus (AAV) vector. Brain Res. 1996;713:99–107. doi: 10.1016/0006-8993(95)01488-8. [DOI] [PubMed] [Google Scholar]
- 22.McKeon C, Samulski R J. NIDDK workshop on AAV vectors: gene transfer into quiescent cells. Hum Gene Ther. 1996;7:1615–1619. doi: 10.1089/hum.1996.7.13-1615. [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minami M, Poussin K, Brechot C, Paterlini P. A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics. 1995;29:403–408. doi: 10.1006/geno.1995.9004. [DOI] [PubMed] [Google Scholar]
- 25.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–131. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 26.Nelson D L, Ledbetter S A, Corbo L, Victoria M F, Ramirez-Solis R, Webster T D, Ledbetter D H, Caskey C T. Alu polymerase chain reaction: a method for rapid isolation of human-specific sequences from complex DNA sources. Proc Natl Acad Sci USA. 1989;86:6686–6690. doi: 10.1073/pnas.86.17.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens R A, Weitzman M D, Kyöstiö S R M, Carter B. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep proteins. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peel A L, Zolotukhin X, Schrimsher G W, Muzyczka N, Reier P J. Efficient transduction of green fluorescent protein in spinal cord neurons using adeno-associated virus vectors containing cell-type specific promoter. Gene Ther. 1997;4:16–24. doi: 10.1038/sj.gt.3300358. [DOI] [PubMed] [Google Scholar]
- 29.Phillips M I, Mohuczy-Dominiak D, Coffey M, Galli S M, Kimura B, Wu P, Zelles T. Prolonged reduction of high blood pressure with an in vivo, nonpathogenic, adeno-associated viral vector delivery of AT1-R mRNA antisense. Hypertension. 1997;29:374–380. doi: 10.1161/01.hyp.29.1.374. [DOI] [PubMed] [Google Scholar]
- 30.Ponnazhagan S, Erikson D, Kearns W G, Zhou S Z, Nahreini P, Wang X-S, Srivastava A. Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum Gene Ther. 1997;8:275–284. doi: 10.1089/hum.1997.8.3-275. [DOI] [PubMed] [Google Scholar]
- 31.Prasad K M, Trempe J P. The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology. 1995;214:360–370. doi: 10.1006/viro.1995.0045. [DOI] [PubMed] [Google Scholar]
- 32.Prasad K M, Zhou C, Trempe J P. The adeno-associated virus Rep78 protein is covalently linked to viral DNA in a preformed virion. Virology. 1995;214:360–370. doi: 10.1006/viro.1995.0045. [DOI] [PubMed] [Google Scholar]
- 33.Rutledge E A, Russell D W. Adeno-associated virus vector integration junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shelling A N, Smith M G. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 36.Singer M F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- 37.Weiner A M, Deininger P L, Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- 38.Weitzman M D, Kyöstiö S R M, Kotin R M, Owens R A. Adeno-associated virus (AAV) rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu P, Price P, Du B, Hatch W C, Terwilliger E F. Direct cytotoxicity of HIV-1 envelope protein gp120 on human NT neurons. NeuroReport. 1996;7:1045–1049. doi: 10.1097/00001756-199604100-00018. [DOI] [PubMed] [Google Scholar]
- 40.Wu P, Ziska D, Bonnell M, Grouzmann E, Millard W J, Meyer E M. Differential neuropeptide Y gene expression in post-mitotic versus dividing neuroblastoma cells driven by an adeno-associated virus vector. Mol Brain Res. 1994;24:27–33. doi: 10.1016/0169-328x(94)90114-7. [DOI] [PubMed] [Google Scholar]
- 41.Wu P, Du B, Phillips M I, Terwilliger E F. Efficient and long-term gene delivery into rat brain using adeno-associated viral vectors, abstr. 1450. In Abstracts of the 27th Annual Meeting of the Society for Neuroscience 1997. New Orleans, La: Society for Neuroscience; 1997. [Google Scholar]
- 41a.Wu, P., et al. Unpublished data.
- 42.Xiao X, Li J, Samulski R J. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]