Abstract
MeCP2 is a general regulator of transcription involved in the repression/activation of genes depending on the local epigenetic context. It acts as a chromatin regulator and binds with exquisite specificity to gene promoters. The set of epigenetic marks recognized by MeCP2 has been already established (mainly, cytosine modifications in CpG and CpA), as well as many of the constituents of its interactome. We unveil a new set of interactions for MeCP2 with the four canonical nucleosomal histones. MeCP2 interacts with high affinity with H2A, H2B, H3 and H4. In addition, Rett syndrome associated mutations in MeCP2 and histone epigenetic marks modulate these interactions. Given the abundance and the structural/functional relevance of histones and their involvement in epigenetic regulation, this new set of interactions and its modulating elements provide a new addition to the ‘alphabet’ for this epigenetic reader.
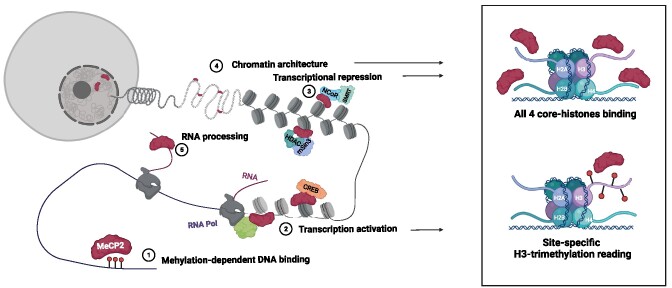
Graphical Abstract
Graphical Abstract.
Introduction
Methyl-CpG binding protein 2 (MeCP2) is a transcriptional regulator involved in early stages of neuronal development, differentiation, maturation, and control of synaptic plasticity (1). MeCP2 exhibits precise dsDNA interaction at specific promoters, such as BDNF (2). The ability to repress or activate gene transcription depends on the existing epigenetic marks in the promoter and the recruitment of specific repressor or activator partners. In addition, MeCP2 can also extensively bind to chromatin acting as a chromatin architectural factor (3–6).
The ability of MeCP2 to interact with many various biological partners (e.g. nucleic acids, proteins) and its multifunctional activity as an important hub within gene transcription regulation networks, regulated by post-translational modifications and allosteric interdomain interactions, rely on the modular, dynamic and adaptive structure of MeCP2 (7,8). Still, recent evidence suggests a primary role for MeCP2 in recruiting co-repressor complexes to methylated sites in the genome, resulting in dampening of gene expression (9).
Dysregulation of MeCP2 leads to disease (10–12). Deleterious mutations in MeCP2 causing activity loss or deficiency are associated with Rett syndrome (RTT), a rare neurodevelopmental disorder mainly affecting girls with an incidence of 1 in 10000 live births. RTT is characterized by a broadly varying clinical phenotype, sharing many features observed in neurological autistic diseases. On the other hand, MECP2 duplication causes MeCP2 overdosage resulting in MECP2 duplication syndrome (MDS), another even rarer disorder that affects males. Interestingly, MDS shares some phenotypic features with RTT, such as severe intellectual disability and impaired motor function.
MeCP2 is an intrinsically disordered protein (IDP) organized into six domains: N-terminal domain (NTD), methyl binding domain (MBD), intervening domain (ID), transcriptional repression domain (TRD), C-terminal domain α (CTDα), and C-terminal domain β (CTDβ) (4,13). About 60% of the polypeptide chain lacks well-defined secondary/tertiary structure. Flexible, highly dynamic regions facilitate structural rearrangements necessary for allosteric interdomain regulation and for exposing different motifs to interact with different biological partners. Interactions in IDPs are often characterized by a moderate-to-low binding affinity and a transient nature, due to the energetic penalty stemming from the conformational change coupled to the binding (14,15).
The most important domains are MBD, initially associated with binding to DNA containing methylated CpG dinucleotides, and TRD, associated with the repression of gene transcription (16,17). However, it has been shown that other domains (ID, CTDα and CTDβ) are directly or indirectly involved in DNA interaction independent of methylation (18,19).
We have studied the interaction of several MeCP2 constructs (MBD, NTD-MBD, NTD-MBD-ID, NTD-MBD-ID-TRD, NTD-MBD-ID-TRD-CTDα and NTD-MBD-ID-TRD-CTDα-CTDβ) with unmethylated, methylated, and hydroxymethylated DNA (20–25). In addition, we have studied the effect of two primary RTT-associated mutations: R106W and R133C. In summary, we can state: (i) NTD-MBD-ID is more appropriate for studying DNA interaction than the isolated MBD; (ii) the presence of the disordered flanking domains for MBD increases considerably the affinity for DNA and ID provides a second DNA binding site; (iii) the MBD flanking domains increase the structural stability of MBD; (iv) methylation of CpG moderately increases DNA affinity compared to unmethylated CpG, while hydroxymethylation induces an intermediate effect; (v) RTT-associated mutations far from the DNA binding interface (e.g. R106W) may show a more deleterious effect than those located in the DNA binding interface (e.g. R133C), through an indirect conformational rearrangement, in agreement with the observed clinical burden; and vi) hydroxymethylation in CpG specifically affects R133C, which could be associated to the onset and time evolution of RTT.
There are many (completely or partially) unexplained issues related to MeCP2 function, including: (i) the limited ability to discriminate modified cytosines in CpG dinucleotides in vitro is far from being biologically significant to explain its preferential distribution tracking the density of heterochromatin foci enriched with 5-methyl-cytosine (7,26,27); (ii) the mechanisms by which MeCP2 can both activate and deactivate the transcription of hundreds of genes, depending on the context (e.g. decreasing transcriptional noise and adapting gene expression patterns to different physiological or environmentally-induced conditions) (28,29) and (iii) the ability of MeCP2 to simultaneously exhibit methylation-dependent specific binding to certain target gene promoter locations and genome-wide methylation-independent unspecific binding to heterochromatin (30–32). In this work, we present experimental evidence for a new set of protein–protein interactions involving MeCP2 that may shed light on these issues.
MeCP2 interactome comprises dsDNA (and other nucleic acids, such as ssDNA and ssRNA), and many proteins: ATRX (33), histone H3 lysine 9 methyltransferase activity (34), c-Ski (35), DNMT1 (36), CoRest (37), LANA (38), PU1 (39), splicing factors (40), Y box-binding protein 1 (41) and Brm (42). Then, it would be no surprise that other key interactions remain to be unveiled. The function of MeCP2 will be the result of the combinatorial binding of these biological partners, ultimately governed by their effective intracellular concentrations and interaction affinities, which will depend on the cellular type and its developmental or metabolic stage.
MeCP2 function is interconnected to histones in various ways. For example, although MeCP2 interaction with chromatin seem to be guided by epigenetic marks, MeCP2 is expressed at nearly histone-octamer levels in cortical neurons (31) and it might compete with H1 for common binding sites (3,43), although more recent findings suggest MeCP2 binding to chromatin might be independent from H1 (44). In addition, MeCP2 may recruit elements that modify histones by adding epigenetic marks to modulate chromatin accessibility for gene transcription and DNA repair, among other processes. Histones are highly basic proteins, abundant in lysine and arginine residues. One of the most common modifications is the methylation of lysine, which can affect how other protein, such as transcription factors, interact with the nucleosomes. The four nucleosomal histones are relatively similar in structure and are highly conserved throughout evolution, all of them featuring a ‘helix-turn-helix-turn-helix’ motif. They also have disordered tails protruding from the nucleosome and containing the main spots for stimulus-dependent posttranslational modifications. These tails serve as crucial regulatory elements that modulate the attachment of structural and functional elements to the chromatin. In fact, many proteins can recognize lysine methylation in H3 and H4 with exquisite sensitivity, and the capability to differentiate mono-, di- and tri-methyl lysine. Histone trimethylation at lysines may be associated with activated or repressed genes depending on the context (45,46).
Since MeCP2 binds with high efficiency to chromatin at nucleosomal sites, it is possible that this protein interacts not only with dsDNA, but also with other nucleosomal elements. That is the hypothesis guiding this study. We present a biophysical study of the interaction of MeCP2 with the four nucleosomal histones: H2A, H2B, H3 and H4, which we believe must be included within the MeCP2 interactome. Given the high concentration of these biological partners, and their considerable interaction affinity with MeCP2, they may significantly contribute to various MeCP2 functions (e.g. chromatin organization and substitution of histone H1 as nucleosomal linker element) and their regulation. Furthermore, epigenetic modifications in histones (e.g. lysine methylation) may represent an additional layer of MeCP2 regulation and a new addition to the ‘alphabet’ of this epigenetic reader. We also provide information on the effect of Rett syndrome-associated mutations and lysine trimethylation on these MeCP2-histone interactions.
Materials and methods
MeCP2 expression and purification
The MeCP2 variants (corresponding to isoform E2) were expressed using pET30b plasmid as expression vector for BL21 (DE3) Star E. coli strain. The sequences contained an N-terminal polyhistidine-tag for purification by immobilized metal ion affinity chromatography (IMAC). Cultures were grown in 150 ml of LB/kanamycin (50 μg/ml) media at 37°C overnight. Then, 4 l of LB/kanamycin (25 μg/ml) were inoculated (1:100 dilution) and incubated under the same conditions until reaching an OD (at a wavelength of 600 nm) of 0.6. Protein expression was induced with 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) at 18°C overnight. Cells were ruptured by sonication in ice and benzonase (Merck-Millipore, Madrid, Spain) was added (20 U/ml) to degrade nucleic acids.
Proteins were purified in a HiTrap TALON column (GE-Healthcare Life Sciences, Barcelona, Spain) with two washing steps: buffer sodium phosphate 50 mM, pH 7, NaCl 300 mM, and in buffer sodium phosphate 50 mM, pH 7, NaCl 800 mM (to remove potential DNA contamination from the protein), before an imidazole 10–150 mM elution gradient. Purity was checked by SDS-PAGE. The polyhistidine-tag was removed by GST-tagged PreScission Protease cleavage in buffer Tris–HCl 50 mM, pH 7.5, NaCl 150 mM, at 4 °C for 4 h. The final step consisted of a combination of two affinity chromatographic steps: removal of the polyhistidine-tag, or the unprocessed protein, using a HiTrap TALON column (GE-Healthcare Life Sciences, Barcelona, Spain) and removal of the GST-tagged PreScission Protease using a GST TALON column (GE-Healthcare Life Sciences, Barcelona, Spain). Purity and homogeneity were checked by SDS-PAGE. The proteins were stored in buffer Tris 50 mM, pH 7.0 at −80°C. Potential DNA contamination was always assessed by the ratio of UV absorption at 260 nm versus absorption at 280 nm. Extinction coefficients at 280 nm of 11 460, 13 075 and 16 960 M−1 cm−1 were employed for wild-type/R133C MBD, full-length MeCP2, and R106W MBD, respectively.
Histones expression and purification
All the histones were expressed using pET11a plasmid as expression vector for BL21 (DE3) Star E. coli strain. Cultures were grown in 150 ml of LB/ampicillin (100 μg/ml) media at 37°C overnight. Then, 4 l of LB/ampicillin (25 μg/ml) were inoculated (1:100 dilution) and incubated under the same conditions until reaching an OD (at a wavelength of 600 nm) of 0.4. Protein expression was induced with 1 mM IPTG at 37°C for 2–4 h. Cells were ruptured by sonication in ice and benzonase (Merck-Millipore, Madrid, Spain) was added (20 U/ml) to degrade nucleic acids.
The extraction and purification protocol was adapted from (47). Briefly, the cellular pellet was resuspended in 10 ml SA10X buffer, containing sodium acetate 400 mM pH 5.2, EDTA 10 mM pH 8, methionine 100 mM, and 18 g of urea powder, 0.58 mg of NaCl were added, and completed up to a 50 ml final volume containing β-mercaptoethanol 5 mM. After sonication, the filtered supernatant was centrifuged to remove insoluble cellular debris.
Proteins were purified by ion exchange chromatography, using a column tandem Sepharose Q–Sepharose SP (top–bottom) (GE-Healthcare Life Sciences, Barcelona, Spain) pre-equilibrated in buffer SAU200 (sodium acetate 40 mM, pH 5.2, urea 6 M, EDTA 1 mM pH 8, β-mercaptoethanol 5 mM, methionine 10 mM, NaCl 200 mM). After a 2 ml/min injection, SP column was removed and the protein was eluted from Q column using a gradient with elution Buffer (sodium acetate 40 mM, pH 5.2, urea 6 M, EDTA 1 mM pH 8, β-mercaptoethanol 5 mM, methionine 10 mM, NaCl 1 M. Refolding of the protein was accomplished by dialyzing the protein-containing fractions with deionized water, followed by lyophilization for storage. Purity and homogeneity were checked by SDS-PAGE. Potential DNA contamination was always assessed by the ratio of UV absorption at 260 nm versus absorption at 280 nm. The extinction coefficients respectively at 280 nm used for protein concentration determination are 4050, 6070, 4040 and 5400 M−1 cm−1 for H2A, H2B, H3 (wild-type and variants) and H4, respectively.
Histone 3 trimethylation
H3 variants in which K4, K9, K27 and K36 were trimethylated were prepared according to a standard procedure (48). The procedure consists in a chemical approach to produce analogs of methyl-lysine into recombinant proteins, efficiently generating protein variants in which the site and degree of methylation can be specified. Briefly, the cysteine naturally present in H3 (C111) was mutated to alanine, and lysine-to-cysteine substitution H3 mutants were obtained at particular locations (K4C, K9C, K27C and K36C). Because H3 lacks additional cysteine residues, it is possible to target trimethylation to specific sites. The expression and purification protocol for H3 K-to-C mutants was the same as that for wild-type H3. Then, 5–10 mg of lyophilized mutant H3 was suspended in 980 μl of alkylation buffer (HEPES 1 M pH 7.8, guanidinium chloride 4 M, d/l-methionine 10 mM), and 20 μl of dithiothreitol (DTT) 1 M was added, incubating at 37°C for 1 h. After that, 100 mg of (2-bromoethyl)trimethylammonium bromide was added, and incubated at 50°C for 2.5 h. Another 10 μl of DTT 1 M was added, and incubated for another 2.5 h at the same temperature. Alkylation reaction was quenched by adding 50 μl of β-mercaptoethanol. Refolding was accomplished using a PD-10 column for buffer exchange to deionized water. Trimethylation was assessed by MALDI-TOF mass spectrometry.
H3-related peptides
Peptides derived from H3 were purchased from NZYTech (Lisbon, Portugal). They correspond to the K27 region TKAARKSAPAT, where the trimethylated K27 is the second lysine in the sequence. The N-terminus was acetylated and the C-terminus was amidated to reduce end-effects and mimic the natural fragment (e.g. electrostatic charges) within the protein structure.
Double-stranded DNA
HPLC-purified 45-base single-stranded DNA (ssDNA) oligomers corresponding to the promoter IV of the mouse brain-derived neurotrophic factor (BDNF) gene were obtained from Integrated DNA Technologies (Leuven, Belgium): (forward) 5′-GCCATGCCCTGGAACGGAACTCTCCTAATAAAAGATGTATCATTT-3′; and (reverse) 5′-AAATGATACATCTTTTATTAGGAGAGTTCCGTTCCAGGGCATGGC-3′.
The DNA fragments were purchased as ssDNA oligonucleotides and they were subsequently annealed to obtain 45-bp double-stranded DNA (dsDNA). They were dissolved to obtain a 0.5 mM ssDNA solution for each oligonucleotide. Then, they were mixed at an equimolar ratio and were annealed using a Stratagene Mx3005P qPCR real-time thermal cycler (Agilent Technologies, Santa Clara, CA, USA). The thermal annealing profile consisted of four steps: (i) equilibration at 25°C for 30 s; (ii) heating ramp up to 99°C; (iii) equilibration at 99°C for 60 s and (iv) 3-h cooling process down to 25°C at a rate of 1°C/180 s.
Circular dichroism (CD)
Circular dichroism spectra were recorded in a thermostated Chirascan spectrometer (Applied Photophysics, Leatherhead, UK) using a 0.1 cm path-length quartz cuvette (Hellma Analytics, Müllheim, Germany) with a bandwidth of 1 nm, a spectral resolution of 0.5 nm, and a response time of 5 s. Temperature was controlled by a Peltier unit and monitored using a temperature probe. The assays were performed in the far-UV range (190–260 nm). Protein concentration was set at 7 μM for MeCP2 and H3 variants. Experiments were performed in buffer Tris 50 mM, pH 7, at 20°C.
Individual spectra of MeCP2 variants and H3 were recorded, as well as their complexes, employing the same concentrations for the interacting molecules. All experiments were performed in triplicate. Far-UV spectra contain information about the secondary structure content in a protein, while near-UV spectra reflect the asymmetric microenvironment of aromatic residues when that protein has some tertiary structure. Comparing the spectra of individual proteins, the spectrum of the complex, and the additive effect of the individual spectra, provided direct evidence for the interaction: when the additive effect does not coincide with the spectrum of the complex, the two protein interacts. No normalization (concentration, number of residues) was performed because the spectra correspond in many cases to protein mixes, and the raw CD data (ellipticity in millidegrees) is reported.
Isothermal titration calorimetry (ITC)
The interaction between the MeCP2 variants and histones was characterized using an Auto-iTC200 microcalorimeter (MicroCal, Malvern-Panalytical, Malvern, UK). MeCP2 variant in the calorimetric cell at 2–5 μM was titrated with each histone solution at 25–50 μM, in buffer Tris 50 mM, pH 7, at 20°C. In the case of the titrations with the H3-derived peptides, MeCP2 at 50 μM was titrated with each peptide solution at 500 μM. A sequence of 2 μl-injections of titrant solution every 150 s was programmed and the stirring speed was set to 750 rpm. All assays were performed in duplicate or triplicate, providing representative results. The association constant, Ka, and the enthalpy of binding, ΔH, were estimated through non-linear regression of the experimental data employing a single ligand binding site model (1:1 stoichiometry) or two ligand binding sites (1:2 stoichiometry) implemented in Origin (OriginLab, Northampton, MA). Importantly, the background injection heat effect was accounted for including an adjustable parameter in the fitting routine. The dissociation constant Kd was calculated as the inverse of the association constant.
Ternary experiments, corresponding to the interaction of MecP2 variants with H3 in the presence of dsDNA or interaction of MeCP2 variants with dsDNA in the presence of H3, were performed employing the same experimental protocol but premixing MeCP2 variants with H3 or dsDNA in the calorimetric cell and injecting dsDNA or H3 from the syringe.
Nuclei isolation, protein fractionation and analysis
Nuclei from whole mouse brains previously frozen at –80°C were prepared as described in (49). The nuclear pellet was homogenized with 0.65 N HCl using a Dounce homogenizer. The sample was centrifuged at 13 000 g for 10 min at 4°C. Then, 6 volumes of acetone were added to the supernatant and, after thorough mixing, the mixture was incubated at –20°C overnight to precipitate nuclear proteins. The protein precipitate was processed as described in (50) and was subsequently fractionated by reverse phase HPLC (50). Proteins were eluted using a 0–60% acetonitrile gradient in the presence of 0.1% TFA for 60 min. Fractions were collected at 1 min interval and analyzed by SDS-PAGE (50). Western blotting was carried out as described elsewhere (25) using a MeCP2 antibody (Sigma Aldrich; M9317: 1:8000 dilution). Images were analyzed with Li-Cor Odyssey (Li-Cor Biosciences Lincoln, NE, USA) and Li-Cor Image Studio Lite 5.2.5 software. Assays were performed replicated four times, providing representative results.
Results
MeCP2 interacts with H3
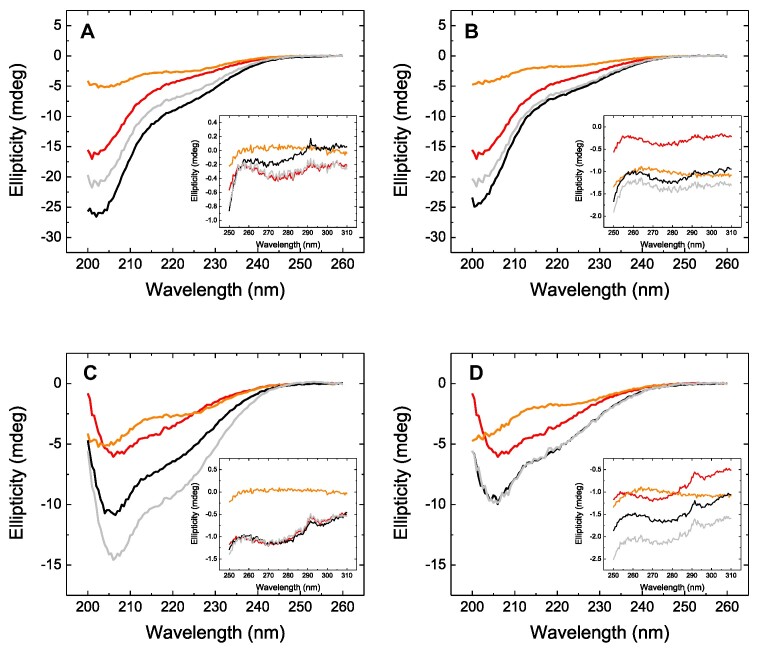
First, the interaction of MeCP2 was tested by CD. As it can be seen in Figure 1, the far-UV spectra of the complexes did not coincide with the addition of the individual spectra, indicating that an interaction was taking place. A similar result could be observed in the near-UV spectra, but the interpretation is more ambiguous. The results for MBD also suggested a clear interaction. Interestingly, the spectrum of the MeCP2-H3 complex showed a larger ellipticity than the additive effect of the individual spectra, whereas the spectrum of the MBD-H3 complex showed a smaller ellipticity than the additive effect of the individual spectra. This might suggest some kind of additional structure loss for the interaction with the isolated domain, and may also be related to the different protein size (full-length versus isolated domain) and the different interaction stoichiometry (1:2 versus 1:1), as mentioned below. An important observation was the reduction in the difference between the spectrum of the complex and the additive spectrum for both MeCP2 variants when mixed with the H3K27me3, more evident for MBD. This might suggest that trimethylation of K27 reduced the affinity of the interaction.
Figure 1.
Circular dichroism spectra. Far-UV circular dichroism spectra for individual proteins (H3: orange, MeCP2 variant: red), and the complex (black). The additive effect of the individual spectra is shown in gray. The spectra for full-length MeCP2 (A and B) are shown, as well as the spectra for MBD (C and D). The interaction of the MeCP2 variants was tested with H3 (A and C) and H3K27me3 (B and D). The corresponding near-UV spectra are shown as inset accompanying the far-UV spectra.
Full-length MeCP2 contains two binding sites for H3
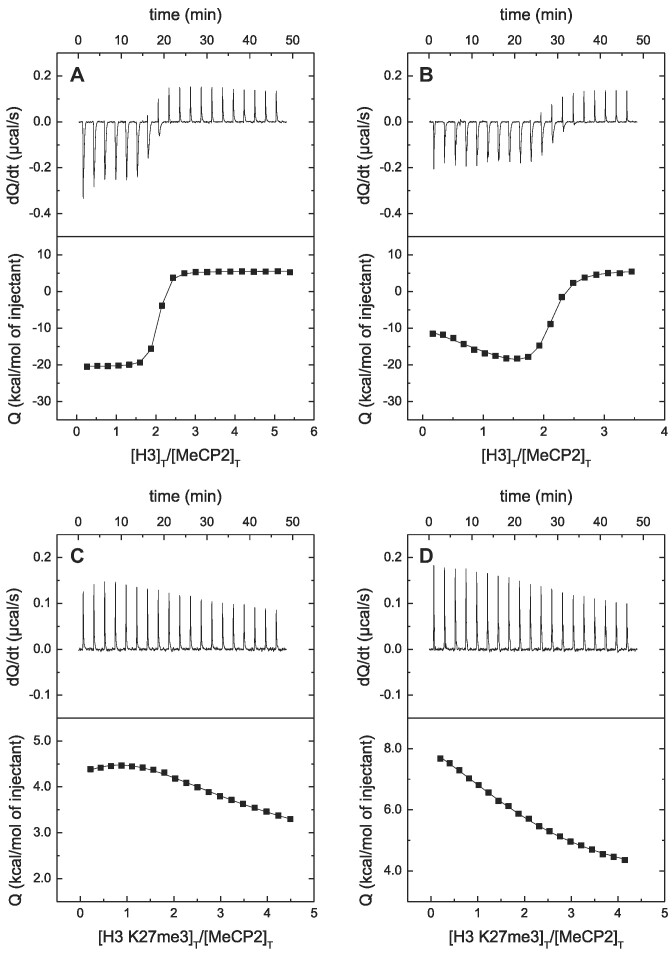
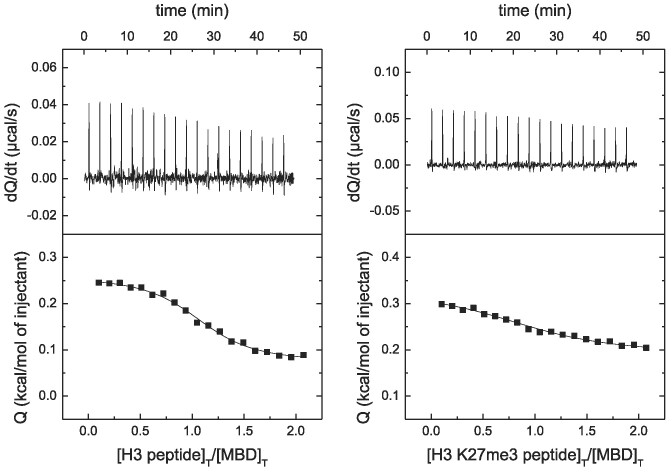
Direct evidence of the interaction (target engagement) of MeCP2 with H3 was obtained by ITC, which is considered the gold standard for affinity determination in biomolecular interactions. As shown in Figure 2, MeCP2 interacted with H3 with high affinity (dissociation constant in the nanomolar range). MeCP2 contained two binding sites for H3 with similar affinities at low ionic strength, but with slightly different affinities at physiological ionic strength (see Table 1). Increase in ionic strength elicited a small reduction in affinity, with no modification in the stoichiometry. Interestingly, trimethylation of K27 largely reduced the affinity of the interaction, in agreement with the interpretation of the far-UV CD spectra.
Figure 2.
MeCP2 interaction with H3 by ITC. Calorimetric titrations of full-length MeCP2 interacting with H3 (A and B) and with H3K27me3 (C and D). The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a two-binding site model.
Table 1.
MeCP2 interacts with H3
| K a (M−1) | K d (μM) | ΔH (kcal/mol) | |||
|---|---|---|---|---|---|
| MeCP2 | H3 | NaCl 20 mM | 3.6 × 107 | 0.028 | –26.0 |
| NaCl 150 mM | 1.9 × 107 | 0.053 | 1.5 | ||
| 1.1 × 107 | 0.093 | –48.5 | |||
| MeCP2 | H3 K27me3 | NaCl 20 mM | 2.1 × 105 | 4.7 | 1.0 |
| 3.2 × 104 | 32 | 24.3 | |||
| NaCl 150 mM | 1.7 × 105 | 5.8 | 6.8 |
Binding parameters for MeCP2 (full length) interacting with H3 and H3 K27me3 at pH 7, 20°C
MBD interacts with the four canonical nucleosomal histones
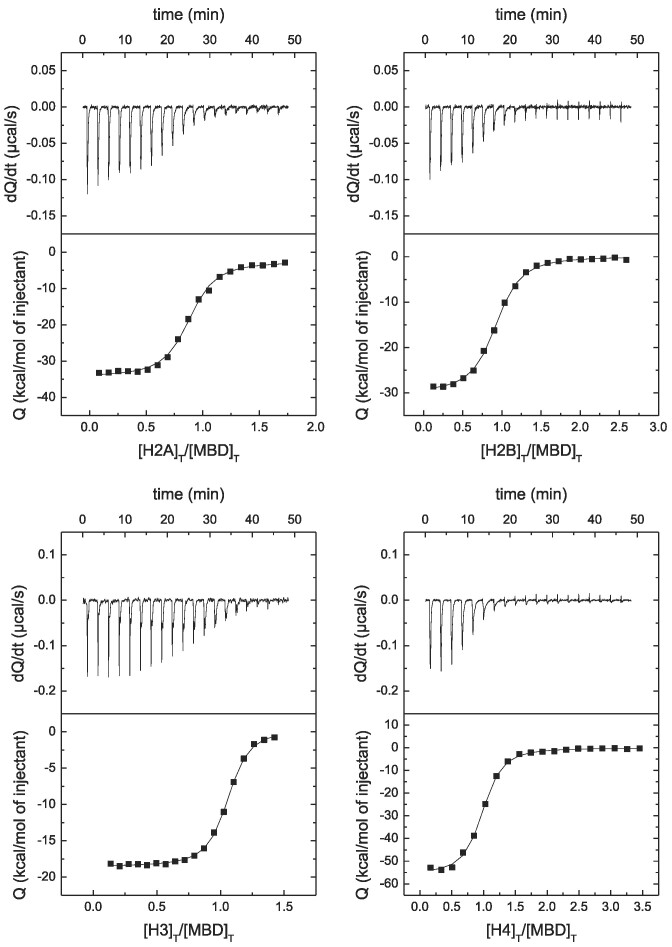
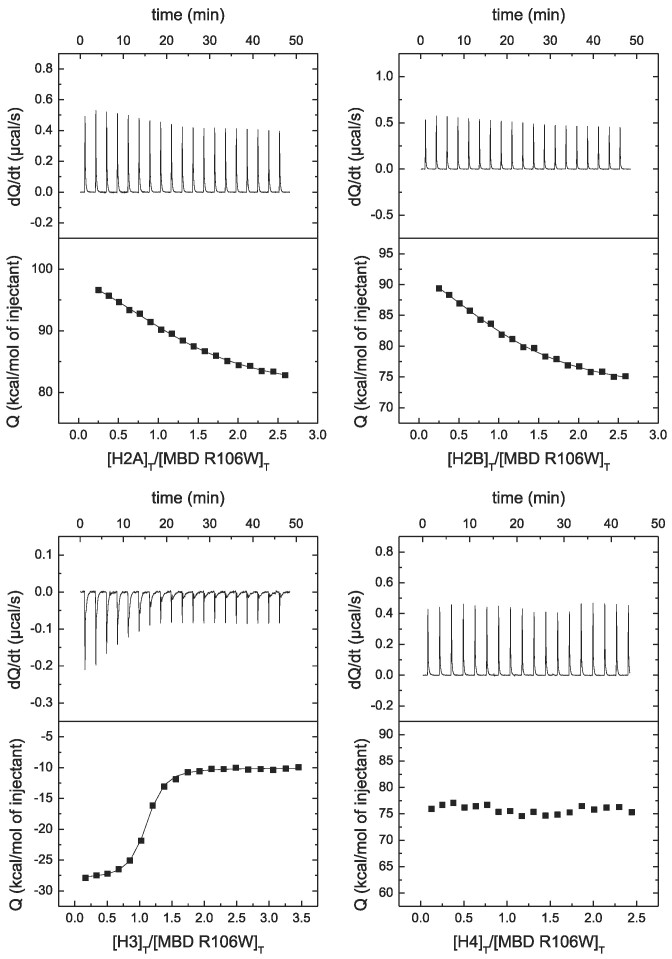
Although MeCP2 contains six domains, all important for its different functions, MBD is one of the key domains, together with TRD/NID. We evaluated the interaction of the four nucleosomal histones: H2A, H2B, H3, and H4. MBD was able to interact with the four histones with high affinity (see Figure 3 and Table 2).
Figure 3.
MBD interaction with canonical histones by ITC. Calorimetric titrations of MBD interacting with H2A, H2B, H3, and H4. The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a single binding site model.
Table 2.
MBD interacts with the four nucleosomal histones. Binding parameters for MBD (WT and Rett-associated variants) interacting with H2A, H2B, H3, and H4 at pH 7, 20°C
| K a (M−1) | K d (μM) | ΔH (kcal/mol) | ||
|---|---|---|---|---|
| MBD | H2A | 2.2 × 107 | 0.046 | –31.9 |
| H2B | 1.8 × 107 | 0.057 | –30.2 | |
| H3 | 2.0 × 107 | 0.050 | –19.0 | |
| H4 | 2.7 × 107 | 0.037 | –56.0 | |
| MBD R106W | H2A | 1.0 × 106 | 1.0 | 25.4 |
| H2B | 8.9 × 105 | 1.1 | 29.0 | |
| H3 | 1.9 × 107 | 0.052 | –18.1 | |
| H4 | – | – | – | |
| MBD R133C | H2A | 3.0 × 107 | 0.034 | –42.0 |
| H2B | 1.2 × 107 | 0.083 | –28.3 | |
| H3 | 2.5 × 107 | 0.040 | –26.2 | |
| H4 | 1.8 × 107 | 0.056 | –39.4 |
Interestingly, the stoichiometry of the interaction was 1:1 and the high affinity was due to a strongly exothermic interaction. The highest affinity corresponded to H4 (dissociation constant of 37 nM), while the weakest interaction corresponded to H2B (dissociation constant of 57 nM).
Rett-syndrome associated mutations differentially affect the interaction with histones
Substitutions R106W and R133C, located in MBD, are two major mutations linked to Rett syndrome. While R133C is associated with a mild Rett phenotype, R106W is associated with a severe phenotype. Interestingly, R133C is located within the dsDNA binding interface, whereas R106W is within the folded core of MBD, likely distorting the interaction with dsDNA and many other biological partners. This underlines the importance of MeCP2 interactions with other biomolecules, not only dsDNA.
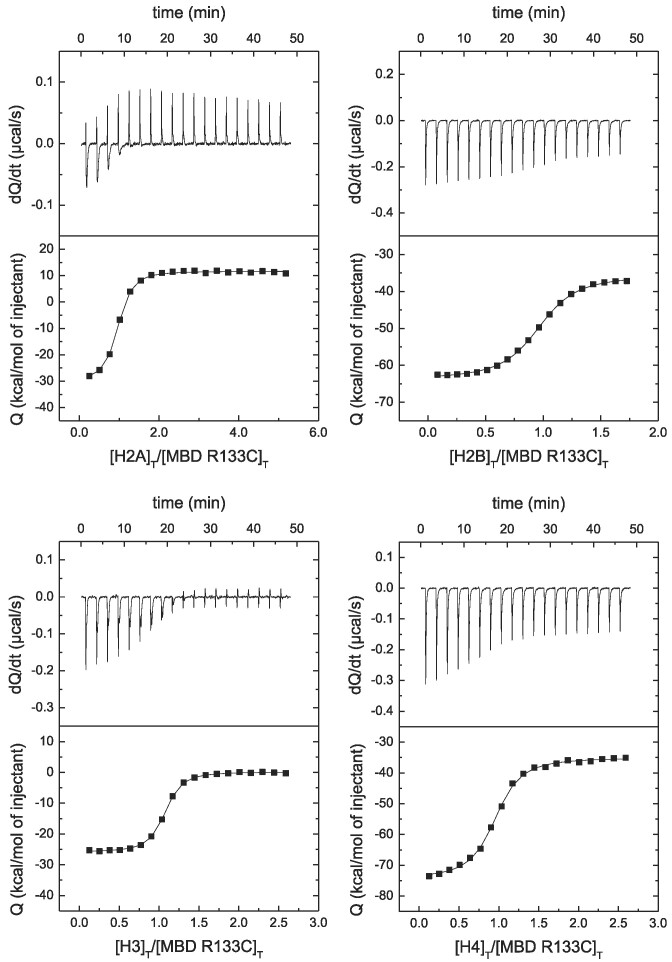
According to the results (see Figure 4 and Table 2), R106W lowers the affinity for H2A, H2B and H4, but does not affect the interaction with H3. Strikingly, R106W abolishes the interaction with H4. On the contrary, R133C does not affect the interaction with the four histones (see Figure 5 and Table 2). This is an example of how Rett syndrome associated mutations may impact differentially the interaction of MeCP2 with a given biological partner.
Figure 4.
MBD R106W interaction with canonical histones by ITC. Calorimetric titrations of MBD R106W interacting with H2A, H2B, H3 and H4. The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a single binding site model.
Figure 5.
MBD R133C interaction with canonical histones by ITC. Calorimetric titrations of MBD R133C interacting with H2A, H2B, H3 and H4. The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a single binding site model.
The interaction of MeCP2 with histones might account for its unusual rpHPLC fractionation
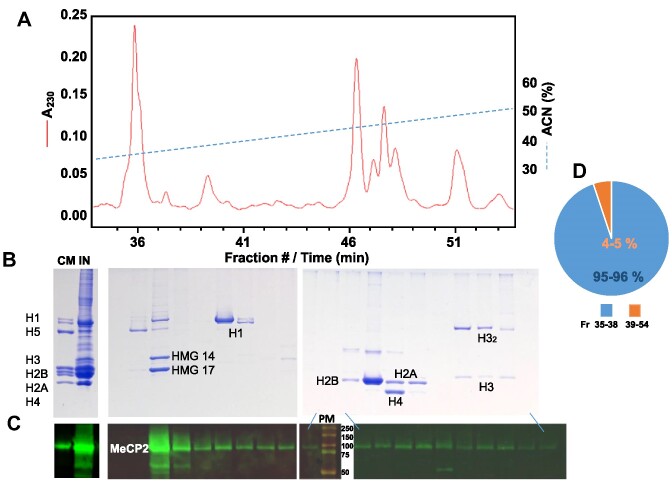
Histones can be readily fractionated by reverse phase HPLC (50–52). However, rpHPLC fractionation of MeCP2 had proven to be more complex as a result of its co-elution with the histone fractions (Figure 6). When the elution of proteins from the column is carried out using an acetonitrile gradient (Figure 6A), most of MeCP2 (>80%) co-elutes with the high mobility group (HMG) proteins, in particular HMG-14/17 (HMGN1/2), and the rest co-elutes with the histone fractions. Although over the years an explanation remained elusive, such an unusual pattern of elution could potentially be explained by the propensity of MeCP2 to interact with histones described above.
Figure 6.
HPLC analysis of MeCP2 and histones elution. A fraction of MeCP2 elutes with histones during reverse phase HPLC fractionation. (A) Elution profile of an HCl extract from mouse brain nuclei. A230, absorbance at 230 nm wavelength; ACN, acetonitrile. (B) SDS-PAGE of the fractions shown in (A). CM, chicken marker; IN, input sample. (C) Western blot analysis of the fractions shown in (B) using a MeCP2 antibody. PM, protein marker (the molecular masses in kDa are indicated). (D) Pie chart of the MeCP2 based on the immunofluorescence quantification of the bands shown in (C).
Trimethylation in H3 lysines differentially affects the interaction with H3
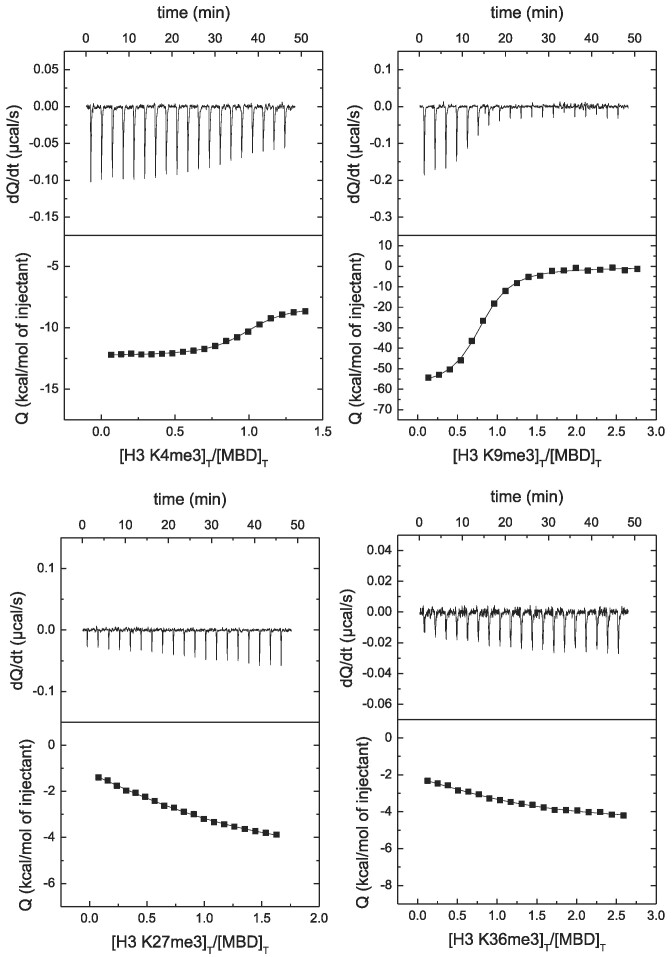
Lysine trimethylation in H3 has important biological consequences depending on the position of such lysine. Then, it was important to assess the impact of those epigenetic marks on the interaction of MBD with H3. Trimethylation induced very small changes in the structure of H3 (see Supplementary Figure S1 as an example for H3K27me3). It can be observed in Figure 7 that trimethylation in K4 and K9 affects very little the interaction of MBD with H3, while trimethylation in K27 and K36 considerably lowers the affinity (see Table 3).
Figure 7.
MBD interaction with H3 trimethylated at different lysine positions by ITC. Calorimetric titrations of MBD interacting with H3 trimethylated at K4, K9, K27 and K36. The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a single binding site model.
Table 3.
MBD interacts with the lysine-trimethylated H3 variants
| K a (M−1) | K d (μM) | ΔH (kcal/mol) | ||
|---|---|---|---|---|
| MBD | H3 | 2.0 × 107 | 0.050 | –19.0 |
| H3 K4me3 | 8.4 × 106 | 0.12 | –4.1 | |
| H3 K9me3 | 1.3 × 107 | 0.075 | –59.0 | |
| H3 K27me3 | 1.7 × 105 | 5.9 | 7.0 | |
| H3 K36me3 | 3.3 × 105 | 3.1 | 4.0 | |
| MBD R106W | H3 | 1.9 × 107 | 0.052 | –18.1 |
| H3 K4me3 | 3.2 × 106 | 0.32 | –15.5 | |
| H3 K9me3 | 2.0 × 107 | 0.051 | –51.9 | |
| H3 K27me3 | 1.6 × 105 | 6.2 | 15.3 | |
| H3 K36me3 | 6.1 × 104 | 16 | 7.7 | |
| MBD R133C | H3 | 2.5 × 107 | 0.040 | –26.2 |
| H3 K4me3 | 3.5 × 106 | 0.29 | –12.8 | |
| H3 K9me3 | 2.2 × 107 | 0.046 | –51.1 | |
| H3 K27me3 | 1.3 × 105 | 7.9 | 10.7 | |
| H3 K36me3 | 6.7 × 105 | 1.5 | –3.7 |
Binding parameters for MBD (WT and Rett-associated variants) interacting with H3 trimethylated at K4, K9, K27 and K36 at pH 7, 20°C.
The interplay between Rett syndrome linked mutations in MBD and lysine trimethylations in H3 was further assessed (see Supplementary Figures S2 and S3, and Table 3). Both R106W and R133C did not affect the interaction with trimethylated H3 variants differentially compared to wild-type MBD.
Conformational constraints in H3 modulate the impact of H3K27me3 on the interaction
Two peptides spanning 11 residues and centered on K27 of H3 were synthesized, one of them with K27 trimethylated, and their interaction with MBD was evaluated. According to the experimental results (see Figure 8), the affinity of interaction was similar, with a minor effect of the lysine trimethylation (dissociation constant of 3.4 μM for wild-type peptide and 12 μM for trimethylated peptide), much smaller fold-change in dissociation constant than that observed for trimethylation of full-length H3. In addition, the wild-type peptide interacted with much less affinity than the full-length H3. These results prove that MBD binds to H3 at the histone tail close to the folded core, but only partially, as some contacts would be established between the MBD and the H3-core to improve not only the affinity of the interaction but also the methylation reading capability.
Figure 8.
MBD interaction with H3-derived peptides by ITC. Calorimetric titrations of MBD interacting with H3-derived peptides: wild-type and trimethylated at K27. The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a single binding site model.
These results also suggest that, considering that the peptides are quite flexible in solution, even though the H3 tail is fully disordered in solution, it may still maintain some residual structural propensity in such a way that the K27 trimethylation effect on the interaction with MBD is different in the full-length protein and in the 11-residue peptide. In fact, according to the AlphaFold database (53,54) the region around K27 might transiently adopt residual α-helical structure which could affect the conformational propensity around K27.
Ternary interactions in MBD: dsDNA and H3
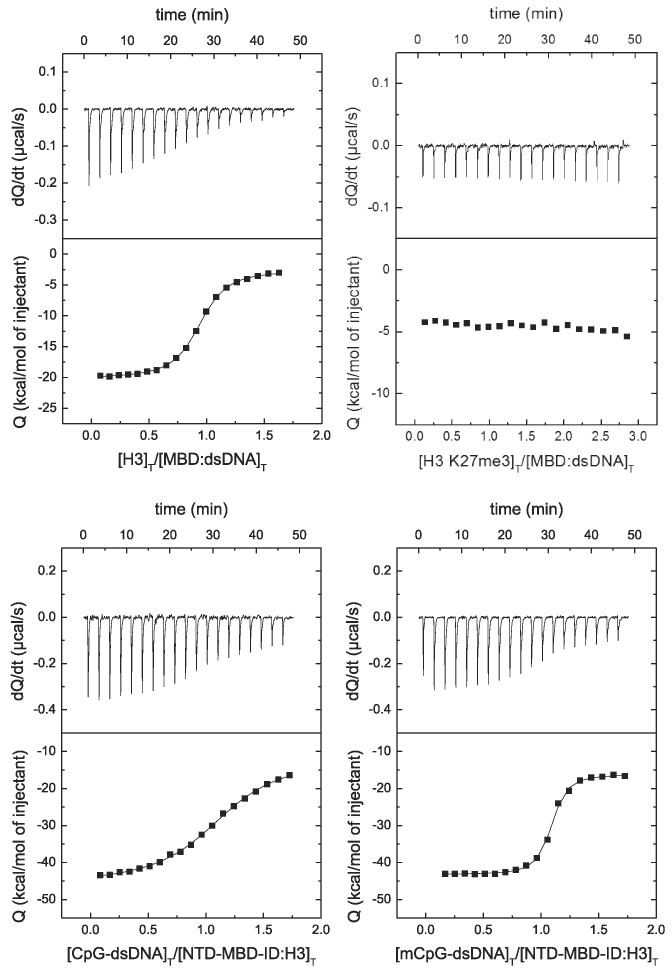
Because MeCP2 is a multifunctional protein able to interact with many different biological partners and establish high-order interaction complexes (e.g. binding to methylated promoters and recruit repressor elements), we wanted to further assess the interplay of the interaction of MBD with H3 and dsDNA focusing on the potential impact of K27 trimethylation. As shown in Figure 9, H3 interacted with the MBD:dsDNA (dissociation constant of 0.10 μM), but trimethylation in K27 hampered such interaction.
Figure 9.
MBD interaction with H3 and dsDNA by ITC. (Upper plots) Calorimetric titrations of H3 interacting with the binary complex MBD:dsDNA, observing the impact of K27 trimethylation on such interaction. (Lower plots) Calorimetric titrations of (unmethylated and methylated) dsDNA interacting with the binary complex NTD-MBD-ID:H3, observing the impact of DNA methylation on such interaction. The upper panels show the thermograms (thermal power as a function of time to maintain the same temperature in the sample cell with respect to the reference cell), and the lower panels show the binding isotherms (ligand-normalized heat effect per injection as a function of the molar ratio in the sample cell). The continuous lines correspond to the non-linear least-squares fitting according to a single binding site model.
It has been established that MeCP2 is able to discriminate between symmetrically CpG-methylated dsDNA and unmethylated dsDNA in vivo (55). However, in vivo and in vitro experiments have provided controversial results, some of them suggesting that MeCP2 (full-length and MBD) interact with methylated and unmethylated with comparable affinities, resulting in a very limited cytosine-methylation discrimination ability (24,56). This suggests that additional intrinsic or extrinsic factors may come into play (55). As observed in Figure 9, the interaction of H3 with NTD-MB-ID (construction containing the MBD flanked by two disordered domains, as a better scaffold and minimal construction for dsDNA studies with MeCP2) elicits a 20-fold increase in affinity when interacting with methylated dsDNA compared to unmethylated dsDNA (dissociation constant of 410 nM compared to 23 nM). Previously, we reported a mere 3-fold increase in DNA binding affinity induced by cytosine methylation (24). Therefore, the interaction with H3 considerably improved the capability to discriminate between unmethylated and methylated dsDNA.
Discussion
Considering MeCP2 capability to interact extensively with chromatin at the level of the nucleosome, MeCP2 function as a scaffolding protein replacing H1 at the nucleosomal linker, the presence of long disordered histone tails protruding from the nucleosomes acting as epigenetic hot-spots modulating the accessibility of many proteins to the DNA, and the promiscuous ability of MeCP2 to interact with a vast array of biological partners, some kind of interaction between MeCP2 and nucleosomal histones would not be surprising. Following this hypothesis, in this work we provide direct experimental evidence for the interaction of MeCP2 and MBD with the four canonical histones which might account for the unusual HPLC elution of MeCP2 (Figure 6).
Although the initial evidence of MeCP2 interaction with histones provided here involved the entire protein, most of the experiments have been performed with the MBD. Working with the MBD may have some limitations, as we have reported previously. However, MBD together with the TRD/NID represent a minimal MeCP2 construct containing the main necessary and sufficient elements for MeCP2 function (57,58). The discrepancy between a minimal peptide and the diverse functionality of a protein with six domains may be explained considering that the main function of MeCP2 is to mediate transcriptional repression via NCoR complex recruitment to methylated DNA. Therefore, we studied the potential interaction of the isolated MBD with histones, and we have provided direct evidence for the existence of a histone binding site in that domain. Since full-length MeCP2 contains two binding sites, the second one is likely located in TRD/NID.
Applying biophysical techniques (CD and ITC) we have provided clear and direct evidence for the interaction of MeCP2 with the four nucleosomal histones. Nowadays, ITC is considered the gold standard for studying biomolecular interactions (59,60). MBD interacts with the four canonical nucleosomal histones with similar affinity: dissociation constant in the 30–60 nM range. This is not completely unexpected taking into account the structural and functional similarity between histones. Then, the MBD-histone interaction exhibits low specificity (within the histone protein family), but, overall, in a broader sense, it may be considered to exhibit high specificity within the full MeCP2 interactome because of their high affinity towards MeCP2 and their abundance, considering the large size of MeCP2 interactome (61). In fact, from the point of view of interactions, some of the main differences between globular proteins and IDPs are their adaptability and dynamics, and the size of their interactomes (62). Therefore, the interaction specificity is usually much lower for IDPs, but this is not due to lower affinities, but to larger interactomes. In any case, the interaction specificity for a given protein must be accounted for by considering its interactome, its affinity for each partner, and the concentration of each partner.
Binding specificity for a certain protein quantifies the ability to establish different interactions while being selective and positively discriminate some particular interaction(s). High binding specificity implies there are only a few interactions that are quantitatively relevant in a given context; on the contrary, low binding specificity means that all potential interactions are more or less equally relevant. The most relevant interaction in quantitative terms regarding specificity will be that with a large concentration for the binding partner and with a high affinity for such partner. There might be some interactions that are not biologically relevant, despite their high affinity, because the binding partner may be present at a very low concentration, and, on the contrary, some interactions that are biologically relevant, despite their low affinity, because the binding partner is present at a sufficiently large concentration. In the case of MeCP2 interacting with histones, both factors, partner abundance and affinity, are considerably high.
Because possible buffer contributions to the overall enthalpies have not been estimated, not much information can be inferred from them (see below). However, provided that the observed apparent enthalpies are very negative (from –19 to –56 kcal/mol), the interaction of MBD with histones must be enthalpically driven with a very favorable enthalpic contribution (i.e. strongly exothermic) accompanied by a large entropic penalty, probably arising from mobility restriction, or even partial refolding, of some disordered regions upon binding, as observed in the circular dichroism spectra of the complexes, which overwhelms the favorable entropic contribution from dehydration of the binding interfaces. Strong enthalpic contributions are common in protein-protein interactions involving IDPs (63,64). In the cases where a considerable reduction of binding affinity was observed (e.g. the impact of R106W on the interaction with H2A and H2B), such affinity reduction was accompanied by a less favorable or even very unfavorable interaction enthalpy.
The experiments have been performed under non-physiological interactions: buffer Tris 50 mM, pH 7, NaCl 20 mM. It is well-known that Tris is a buffer that may distort the results in ITC, often yielding apparent enthalpies and entropies of interaction much different from their intrinsic values. This is due to its large ionization enthalpy and potential changes in pKa’s of ionizable groups elicited by the formation of the protein-protein complex resulting in a net proton exchange between the complex and the bulk solution. Thus, there could be a significant contribution of the buffer (de)protonation to the observed apparent enthalpy. However, the goal was to prove, beyond doubt, the in vitro interactions reported in this work, and, fortunately, using Tris as buffer will not affect the interaction affinity. In addition, low ionic strength may also affect the interaction strength and specificity, and this is even more important when polar and charged functional groups are expected to be involved in the intermolecular interactions, as it is the case of interactions between intrinsically disordered proteins. Nevertheless, the experiments at physiological ionic strength showed that, although the interaction affinity may be lower than that a low ionic strength (a dissociation constant in the micromolar range, compared to a dissociation constant in the nanomolar range), there is still sufficient affinity to be relevant regarding the specificity of MeCP2 interactions.
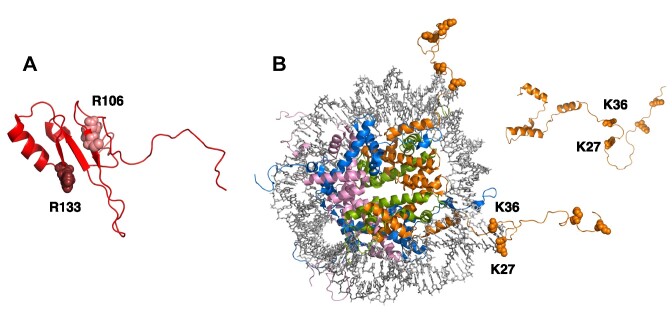
The R106W mutation corresponds to the substitution of a polar residue in the folding core of MBD by a bulkier and non-polar residue. On the other hand, the mutation R133C corresponds to the substitution of a polar residue in the DNA binding interface by a smaller and non-polar residue (see Figure 10). MBD is a small, malleable domain that may efficiently adapt to many aminoacid substitutions. But, considering the impact of R106W and R133C on the interaction affinity of MBD with the different histones (large reduction in affinity induced by R106W, compared to negligible effect of R133C), it is reasonable to conclude that the binding spot for histones in MBD is located close to arginine 106, behind the DNA binding site. The R106W substitution also considerably affects the interaction with dsDNA, and this mutation is associated with a high severity score in Rett syndrome. The findings reported here may help to explain this effect. The fact that the histone binding site in MBD is far from the DNA binding interface suggests that this domain contains two functional binding sites that may work cooperatively in an allosteric fashion and that may be regulated by further modifications (e.g. methylation) in the binding partners.
Figure 10.
Structural models for MBD and H3. (A) AlphaFold model for the structure of MBD (UniProt: A0A075BPH5) showing the location of the two Rett syndrome associated mutations studied in this work: R106 (light red) and R133 (dark red). (B) Structure of the nucleosome (PDB code: 1kx5), showing the histone octamer as cartoon model (histones in different colors, H3 in orange) and the dsDNA as stick model (grey). The AlphaFold model for the isolated histone H3 (UniProt: B4E380) is also shown as an inset. Lysines K4, K9, K27 and K36 are shown as sphere model. Lysines K27 and K36 are located close to the nucleosomal dsDNA.
An additional piece of valuable information comes from the ternary interaction experiments: the dissociation constant for H3 interacting with MBD is 0.1 μM, whereas the dissociation constant for H3 interacting with MBD bound to dsDNA is 0.05 μM, revealing a 2-fold reduction in affinity due to the presence of dsDNA. These results suggest that H3 and dsDNA do not interact at the same binding site in MeCP2 and their interaction is not reciprocally competitive. If the binding of H3 and dsDNA with MBD were competitive (i.e. binding to the same site in MBD or coupled through an excluding conformational rearrangement), there would be a 23-fold reduction in affinity caused by the presence of dsDNA (whose interaction with MBD is characterized by a dissociation constant of 0.45 μM).
Trimethylation in K4 and K9 hardly affects the interaction affinity between MBD and histones, but trimethylation in K27 and K36 reduces considerably the interaction affinity. Then, it is reasonable to conclude that the region of the disordered histone tail close to the nucleosome represents the binding spot for MeCP2 (see Figure 10). This is in agreement with MeCP2 acting as pseudo-H1 as a nucleosomal linker. And this way MeCP2 would be close to dsDNA and histones, involved in ternary (or higher-order) interactions.
As trimethylation in K9 and K27 is often associated with transcriptional repression, contrary to trimethylation in K4 and K36, functional correlation with MeCP2 binding (a well-known repression mediator) would be expected: MeCP2 binding to histone marks for repression and reduced binding to histone marks for activation. Instead, the MBD binding selectivity regarding DNA methylation does not correlate with the binding selectivity regarding histone methylation, which contributes to the idea that epigenetic regulation is not an on-off switch, but the multifactorial effect resulting from the fine tune of many disparate, diverging, opposing signals pushing towards different directions depending on the molecular context, with MeCP2 acting as a transcriptional regulator, rather than a mere transcriptional repressor (65).
It has been reported before that MeCP2 specifically binds to nucleosomes in which K27 is trimethylated, with some kind of DNA methylation dependence as a reinforcing mechanism towards transcriptional repression (49,66). Although we have observed a similar result with the NTD-MBD-ID construction (Figure 9), the affinity of interaction of MBD with H3 decreases when K27 is trimethylated (Figure 9). The most parsimonious explanation for the potential interaction of MeCP2 with H3K27me3 is that the sites of methylation recognition are the same for DNA CpG and histone methylation (67). However, the binding site of MeCP2 to H3K27me3, other than being located in MBD, has never been experimentally determined. In such instance, and considering that H3K27me3 alters the secondary structure of that region of the tail, it would then not be surprising that such alteration perturbs MBD binding at this important site of generic H3 binding. An alternative explanation could be that, in vivo within the nucleus, the interaction of MeCP2 with H3K27me3 is mediated through an intermediary protein interacting with MeCP2 such as HP1 (68). Indeed, it has been shown that H3K9me3 cooperates with H3K27me3 and Polycomb repressive complex2 (PRC2) to anchor HP1α to chromatin (69). However, a direct comparison of previous results is not straightforward because we did most of the experiments with isolated MBD and purified proteins in solution, which is a different scenario compared to MeCP2 interacting with nucleosomes within the cell, which may add additional intrinsic or extrinsic elements/factors (e.g. MeCP2 domains or other molecules) mediating such interaction, and further work is required to evaluate such cooperativity effect. Moreover, in vivo there will be additional factors (intrinsic from MeCP2 itself, and extrinsic from chromatin) that could mediate meCP2 recognition of specific elements. We may speculate on the impact of high ionic strength on the interactions of MBD and MeCP2 with histones, but we do not have information currently and this is part of ongoing work. The ionic strength is another factor whose influence might explain why MeCP2 is specifically associated to H3K27me3-containing nucleosomes in vivo, while it is observed an affinity reduction caused by H3K27me3 in vitro. However, the scenario in vivo is considerably different: histones forming a nucleosome, containing nucleosomal DNA and DNA linker, and very likely some other chromatin elements.
The interaction between MBD and histones represent a paradigmatic example of interaction between two intrinsically disordered proteins. The interaction is established between the disordered tail of histones, close to their folded core, and the folded core in MBD. However, there is a possibility that other disordered regions of MBD (about 60% of MBD remains unstructured) also participate in this interaction. The binding affinities for MBD interacting with the different histones are quite similar (dissociation constant in the 30–60 nM range), but the observed apparent interaction enthalpies are markedly different. Not much information can be extracted from the differences in interaction enthalpies since possible buffer contributions to the overall enthalpies have not been estimated.
Nucleosomal histones are present in the nucleus at a very high concentration, and their estimated interaction affinity with MeCP2 is considerable (in the nanomolar range). Rett syndrome associated mutations, which affect differentially to the interaction of MeCP2 with DNA, also affect differentially to the interaction of MeCP2 with histones. In addition, lysine trimethylation, an important epigenetic mark for gene transcription/repression, also affects differentially the interaction of MeCP2 with histones, and it shows some reciprocal effects with Rett-associated mutations. Therefore, the relevance of these newly reported interactions relies on their abundance and their potential different layers for regulation (e.g. post-translational modifications in histones).
These findings open a new level of direct regulation for MeCP2 that may shed light on some controversial issues in MeCP2 physiology and pathology. Thus, for MeCP2 function, not only the methyl-cytosine density in promoters is a key regulatory element, but also different histone modifications may play a fundamental role in MeCP2 function. From these results, which extend MeCP2 proteome and the ‘library’ for this epigenetic reader, many questions remain to be answered: How do the two histone binding sites on MeCP2 interplay and cooperate in MeCP2 function? Which other histone modifications might be relevant for MeCP2 function? Do other non-canonical histones interact with MeCP2? How does MeCP2 establish ternary (or higher order) complexes with DNA and histones in nucleosomes? Which are the functional (physiological and pathological) consequences in vivo? Further work, beyond the scope of this manuscript, is required to answer these questions.
MeCP2 is a multifaceted protein involved in many key cellular events. MeCP2 shows a gene-dependent regulatory function linked to the specific identification of methyl-cytosines (mainly, CpG and CpA) at promoters, but MeCP2 also recognizes other epigenetic marks such as hydroxymethyl-, formyl- and carboxy-cytosine (70). On the other hand, MeCP2 shows an apparently unspecific, global chromatin binding ability, which is at times reminiscent of the function of histone H1, likely linked to its chromatin architectural functions. Applying biophysical techniques (spectroscopy, calorimetry, and chromatography) we have provided unequivocal evidence for the interaction of MeCP2 with the four nucleosomal histones, revealing a new set of interactions for MeCP2 and extending its interactome to core histones. In summary, MeCP2 interacts with high affinity with H2A, H2B, H3, and H4. Moreover, Rett syndrome associated mutations in MeCP2 (e.g. R106W and R133C) and epigenetic marks in histones (e.g. lysine trimethylation) modulate these interactions. Given the abundance and the structural/functional relevance of these proteins, and their involvement in epigenetic regulation, this new set of interactions and its modulating elements provide a new addition to the ‘alphabet’ for this epigenetic reader.
Supplementary Material
Acknowledgements
Authors would like to acknowledge the use of Scientific and Technical Services of the Biomedical Research Center of Aragon (CIBA, IACS – Universidad de Zaragoza) for the MALDI-TOF mass spectrometry protein analysis. Graphical Abstract was created with BioRender.com.
Contributor Information
David Ortega-Alarcon, Institute of Biocomputation and Physics of Complex Systems (BIFI), Joint Unit GBsC-CSIC-BIFI, Universidad de Zaragoza, 50018 Zaragoza, Spain; Instituto de Investigación Sanitaria Aragón (IIS Aragón), 50009 Zaragoza, Spain.
Rafael Claveria-Gimeno, Certest Biotec S.L., 50840 Zaragoza, Spain.
Sonia Vega, Institute of Biocomputation and Physics of Complex Systems (BIFI), Joint Unit GBsC-CSIC-BIFI, Universidad de Zaragoza, 50018 Zaragoza, Spain.
Ladan Kalani, Department of Biochemistry and Microbiology, University of Victoria, Victoria, BCV8W 2Y2, Canada.
Olga C Jorge-Torres, Josep Carreras Leukaemia Research Institute (IJC), 08916 Badalona, Barcelona, Spain.
Manel Esteller, Josep Carreras Leukaemia Research Institute (IJC), 08916 Badalona, Barcelona, Spain; Centro de Investigación Biomédica en Red Cáncer (CIBERONC), 28029 Madrid, Spain; Institucio Catalana de Recerca i Estudis Avançats (ICREA), 08010 Barcelona, Spain; Physiological Sciences Department, School of Medicine and Health Sciences, University of Barcelona (UB), 08907 l’Hospitalet de Llobregat, Barcelona, Spain.
Juan Ausio, Department of Biochemistry and Microbiology, University of Victoria, Victoria, BCV8W 2Y2, Canada.
Olga Abian, Institute of Biocomputation and Physics of Complex Systems (BIFI), Joint Unit GBsC-CSIC-BIFI, Universidad de Zaragoza, 50018 Zaragoza, Spain; Instituto de Investigación Sanitaria Aragón (IIS Aragón), 50009 Zaragoza, Spain; Centro de Investigación Biomédica en Red en el Área Temática de Enfermedades Hepáticas y Digestivas (CIBERehd), 28029 Madrid, Spain; Departamento de Bioquímica y Biología Molecular y Celular, Universidad de Zaragoza, 50009 Zaragoza, Spain.
Adrian Velazquez-Campoy, Institute of Biocomputation and Physics of Complex Systems (BIFI), Joint Unit GBsC-CSIC-BIFI, Universidad de Zaragoza, 50018 Zaragoza, Spain; Instituto de Investigación Sanitaria Aragón (IIS Aragón), 50009 Zaragoza, Spain; Centro de Investigación Biomédica en Red en el Área Temática de Enfermedades Hepáticas y Digestivas (CIBERehd), 28029 Madrid, Spain; Departamento de Bioquímica y Biología Molecular y Celular, Universidad de Zaragoza, 50009 Zaragoza, Spain.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Ministerio de Economía y Competitividad and European Regional Development Fund (MCIU/AEI/FEDER, EU) [BFU2016-78232-P to A.V.C., BES-2017-080739 to D.O.A.]; Ministerio de Ciencia e Innovación MCIN/AEI/10.13039/501100011033/ and ‘ERDF A way of Making Europe’ [PID2021-127296OB-I00 to A.V.C.]; Fondo de Investigaciones Sanitarias from Instituto de Salud Carlos III and European Union (ERDF/ESF, ‘Investing in your future’) [PI21/00394 to O.A.]; Spanish Ministry of Science and Innovation with funds from the European Union NextGenerationEU [PRTR-C17.I1]; Autonomous Community of Aragón within the framework of the Biotechnology Plan Applied to Health (LA3); Gobierno de Aragón [Protein Targets and Bioactive Compounds Group E45_23R to A.V.C., and Digestive Pathology Group B25_23R to O.A.]; Centro de Investigación Biomédica en Red en Enfermedades Hepáticas y Digestivas (CIBERehd). Funding for open access charge: Ministerio de Ciencia e Innovación MCIN/AEI/10.13039/501100011033/ and ‘ERDF A way of Making Europe’ [PID2021-127296OB-I00].
Conflict of interest statement. None declared.
References
- 1. Ausio A., Martinez de Paz A., Esteller M. MeCP2: the long trip from a chromatin protein to neurological disorders. Trends Mol. Med. 2014; 20:487–498. [DOI] [PubMed] [Google Scholar]
- 2. Bach S., Ryan N.M., Guasoni P., Corvin A.P., El-Nemr R.A., Khan D., Sanfeliu A., Tropea D. Methyl-CpG-binding protein 2 mediates overlapping mechanisms across brain disorders. Sci. Rep. 2020; 10:22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghosh R.P., Horowitz-Scherer R.A., Nikitina T., Shlyakhtenko L.S., Woodcock C.L. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol. Cell. Biol. 2010; 30:4656–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Adkins N.L., Georgel P.T. MeCP2: structure and function. Biochem. Cell Biol. 2011; 89:1–11. [DOI] [PubMed] [Google Scholar]
- 5. Nan X., Campoy F.J., Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997; 88:471–481. [DOI] [PubMed] [Google Scholar]
- 6. Hansen J.C., Ghosh R.P., Woodcock C.L. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010; 62:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guy J., Cheval H., Selfridge J., Bird A. The role of MeCP2 in the brain. Annu. Rev. Cell Dev. Biol. 2011; 27:631–652. [DOI] [PubMed] [Google Scholar]
- 8. Claveria-Gimeno R., Abian O., Velazquez-Campoy A., Ausio J. MeCP2… Nature's wonder protein or medicine's most feared one?. Curr. Genet. Med. Rep. 2016; 4:180–194. [Google Scholar]
- 9. Tillotson R., Bird A. The molecular basis of MeCP2 function in the brain. J. Mol. Biol. 2020; 432:1602–1623. [DOI] [PubMed] [Google Scholar]
- 10. Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999; 23:185–188. [DOI] [PubMed] [Google Scholar]
- 11. Lorenz J., Neul M.D. The relationship of Rett syndrome and MeCP2 disorders to autism. Dialogues Clin. Neurosci. 2014; 14:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sandweiss A.J., Brandt V.L., Zoghbi H.Y. Advances in understanding of Rett syndrome and MECP2 duplication syndrome: prospects for future therapies. Lancet Neurol. 2020; 19:689–698. [DOI] [PubMed] [Google Scholar]
- 13. Hite K.C., Adams V.H., Hansen J.C. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009; 87:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hadži S., Lah J. The free energy folding penalty accompanying binding of intrinsically disordered α-helical motifs. Protein Sci. 2022; 31:e4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morris O.M., Torpey J.H., Isaacson R.L. Intrinsically disordered proteins: modes of binding with emphasis on disordered domains. Open Biol. 2021; 11:210222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nan X., Meehan R.R., Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993; 21:4886–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adam H., Georgel P.T. CHD proteins: a diverse family with strong ties. Biochem. Cell Biol. 2007; 85:463–476. [DOI] [PubMed] [Google Scholar]
- 18. Hite K.C., Adams V.H., Hansen J.C. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009; 87:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh R.P., Nikitina T., Horowitz-Scherer R.A., Gierasch L.M., Uversky V.N., Hite K., Hansen J.C., Woodcock C.L. Unique physical properties and interactions of the domains of methylated DNA binding protein 2. Biochemistry. 2010; 49:4395–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ortega-Alarcon D., Claveria-Gimeno R., Vega S., Jorge-Torres O.C., Esteller M., Abian O., Velazquez-Campoy A. Unexpected thermodynamic signature for the interaction of hydroxymethylated DNA with MeCP2. Int. J. Biol. Macromol. 2023; 232:123373. [DOI] [PubMed] [Google Scholar]
- 21. Ortega-Alarcon D., Claveria-Gimeno R., Vega S., Jorge-Torres O.C., Esteller M., Abian O., Velazquez-Campoy A. Stabilization effect of intrinsically disordered regions on multidomain proteins: the case of the methyl-CpG protein 2, MeCP2. Biomolecules. 2021; 11:1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ortega-Alarcon D., Claveria-Gimeno R., Vega S., Jorge-Torres O.C., Esteller M., Abian O., Velazquez-Campoy A. Influence of the disordered domain structure of MeCP2 on its structural stability and dsDNA interaction. Int. J. Biol. Macromol. 2021; 175:58–66. [DOI] [PubMed] [Google Scholar]
- 23. Ortega-Alarcon D., Claveria-Gimeno R., Vega S., Jorge-Torres O.C., Esteller M., Abian O., Velazquez-Campoy A. Molecular context-dependent effects induced by Rett syndrome-associated mutations in MeCP2. Biomolecules. 2020; 10:1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Claveria-Gimeno R., Lanuza P.M., Morales-Chueca I., Jorge-Torres O.C., Vega S., Abian O., Esteller M., Velazquez-Campoy A. The intervening domain from MeCP2 enhances the DNA affinity of the methyl binding domain and provides an independent DNA interaction site. Sci. Rep. 2017; 7:41635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez de Paz A., Khajavi L., Martin H., Claveria-Gimeno R., Tom Dieck S., Cheema M.S., Sanchez-Mut J.V., Moksa M.M., Carles A., Brodie N.I. et al. MeCP2-E1 isoform is a dynamically expressed, weakly DNA-bound protein with different protein and DNA interactions compared to MeCP2-E2. Epigenetics Chromatin. 2019; 12:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ballestar E., Yusufzai T.M., Wolffe A.P. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry. 2000; 39:7100–7106. [DOI] [PubMed] [Google Scholar]
- 27. Fraga M.F., Ballestar E., Montoya G., Taysavang P., Wade P.A., Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003; 31:1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh J., Saxena A., Christodoulou J., Ravine D. MeCP2 genomic structure and function. Nucleic Acids Res. 2008; 36:6035–6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klose R.J., Bird A.P. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006; 32:89–97. [DOI] [PubMed] [Google Scholar]
- 30. Georgel P.T., Horowitz-Scherer R.A., Adkins N., Woodcock C.L., Wade P.A., Hansen J.C. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003; 278:32181–32188. [DOI] [PubMed] [Google Scholar]
- 31. Skene P.J., Illingworth R.S., Webb S., Kerr A.R., James K.D., Turner D.J., Andrews R., Bird A.P. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010; 37:457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cohen S., Gabel H.W., Hemberg M., Hutchinson A.N., Sadacca L.A., Ebert D.H., Harmin D.A., Greenberg R.S., Verdine V.K., Zhou Z. et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011; 72:72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nan X., Hou J., Maclean A., Nasir J., Lafuente M.J., Shu X., Kriaucionis S., Bird A. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:2709–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fuks F., Hurd P.J., Wolf D., Nan X., Bird A.P., Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003; 278:4035–4040. [DOI] [PubMed] [Google Scholar]
- 35. Kokura K., Kaul S.C., Wadhwa R., Nomura T., Khan M.M., Shinagawa T., Yasukawa T., Colmenares C., Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. J. Biol. Chem. 2001; 276:34115–34121. [DOI] [PubMed] [Google Scholar]
- 36. Kimura H., Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 2003; 278:4806–4812. [DOI] [PubMed] [Google Scholar]
- 37. Lunyak V.V., Burgess R., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G. et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002; 298:1747–1752. [DOI] [PubMed] [Google Scholar]
- 38. Krithivas A., Fujimuro M., Weidner M., Young D.B., Hayward S.D. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 2002; 76:11596–11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki M., Yamada T., Kihara-Negishi F., Sakurai T., Oikawa T. Direct association between PU.1 and MeCP2 that recruits mSin3A-HDAC complex for PU.1-mediated transcriptional repression. Oncogene. 2003; 22:8688–8698. [DOI] [PubMed] [Google Scholar]
- 40. Buschdorf J.P., Strätling W.H. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J. Mol. Med. (Berl.). 2004; 82:135–143. [DOI] [PubMed] [Google Scholar]
- 41. Young J.I., Hong E.P., Castle J.C., Crespo-Barreto J., Bowman A.B., Rose M.F., Kang D., Richman R., Johnson J.M., Berget S. et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17551–17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harikrishnan K.N., Chow M.Z., Baker E.K., Pal S., Bassal S., Brasacchio D., Wang L., Craig J.M., Jones P.L., Sif S. et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005; 37:254–264. [DOI] [PubMed] [Google Scholar]
- 43. Ishibashi T., Thambirajah A.A., Ausio J. MeCP2 preferentially binds to methylated linker DNA in the absence of the terminal tail of histone H3 and independently of histone acetylation. FEBS Lett. 2008; 582:1157–1162. [DOI] [PubMed] [Google Scholar]
- 44. Ito-Ishida A., Yamalanchili H.K., Shao Y., Baker S.A., Heckman L.D., Lavery L.A., Kim J.Y., Lombardi L.M., Sun Y., Liu Z. et al. Genome-wide distribution of linker histone H1.0 is independent of MeCP2. Nat. Neurosci. 2018; 21:794–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Black J.C., Van Rechem C., Whetstine J.R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012; 48:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005; 6:838–849. [DOI] [PubMed] [Google Scholar]
- 47. Klinker H., Haas C., Harrer N., Becker P.B., Mueller-Planitz F. Rapid purification of recombinant histones. PLoS One. 2014; 9:e104029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Simon M.D., Chu F., Racki L.R., de la Cruz C.C., Burlingame A.L., Panning B., Narlikar G.J., Shokat K.M. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007; 128:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thambirajah A.A., Ng M.K., Frehlick L.J., Li A., Serpa J.J., Petrotchenko E.V., Silva-Moreno B., Missiaen K.K., Borchers C.H., Adam Hall J. et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2012; 40:2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheema M.S., Ausio J. Analytical ultracentrifuge analysis of nucleosomes assembled from recombinant, acid-extracted, HPLC-purified histones. Methods Mol. Biol. 2017; 1528:75–95. [DOI] [PubMed] [Google Scholar]
- 51. Moore S.C., Rice P., Iskandar M., Ausio J. Reconstitution of native-like nucleosome core particles from reversed-phase-HPLC-fractionated histones. Biochem. J. 1997; 328:409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ausio J., Moore S.C. Reconstitution of chromatin complexes from high-performance liquid chromatography-purified histones. Methods. 1998; 15:333–342. [DOI] [PubMed] [Google Scholar]
- 53. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klose R.J., Sarraf S.A., Schmiedeberg L., McDermott S.M., Stancheva I., Bird A.P. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell. 2005; 19:667–678. [DOI] [PubMed] [Google Scholar]
- 56. Connelly J.C., Cholewa-Waclaw J., Webb S., Steccanella V., Waclaw B., Bird A. Absence of MeCP2 binding to non-methylated GT-rich sequences in vivo. Nucleic Acids Res. 2020; 48:3542–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Good K.V., Vincent J.B., Ausio J. MeCP2: the genetic driver of Rett syndrome epigenetics. Front. Genet. 2021; 12:620859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tillotson R., Selfridge J., Koerner M.V., Gadalla K.K.E., Guy J., De Sousa D., Hector R.D., Cobb S.R., Bird A. Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature. 2017; 550:398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Freire E., Mayorga O.L., Straume M. Isothermal titration calorimetry. Anal. Chem. 1990; 62:950A–959A. [Google Scholar]
- 60. Bastos M., Abian O., Johnson C.M., Ferreira da Silva F., Vega S., Jimenez-Alesanco A., Ortega-Alarcon D., Velazquez-Campoy A. Isothermal titration calorimetry. Nat. Rev. Methods Primers. 2023; 3:17. [Google Scholar]
- 61. Teilum K., Olsen J.G., Kragelund B.B. On the specificity of protein-protein interactions in the context of disorder. Biochem. J. 2021; 478:2035–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Perovic V., Sumonja N., Marsh L.A., Radovanovic S., Vukicevic M., Roberts S.G.E., Veljkovic N. IDPpi: protein-protein interaction analyses of human intrinsically disordered proteins. Sci. Rep. 2018; 8:10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zavrtanik U., Hadži S., Lah J. Unraveling the thermodynamics of ultra-tight binding of intrinsically disordered proteins. Front. Mol. Biosci. 2021; 8:726824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hadži S., Lah J. The free energy folding penalty accompanying binding of intrinsically disordered α-helical motifs. Protein Sci. 2022; 31:e4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., Zoghbi H.Y. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008; 320:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee W., Kim J., Yun J.M., Ohn T., Gong Q. MeCP2 regulates gene expression through recognition of H3K27me3. Nat. Commun. 2020; 11:3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vincent J.B., Ausio J. MeCP2: latest insights fundamentally change our understanding of its interactions with chromatin and its functional attributes. Bioessays. 2021; 43:e2000281. [DOI] [PubMed] [Google Scholar]
- 68. Della Ragione F., Vacca M., Fioriniello S., Pepe G., D’Esposito M. MECP2, a multi-talented modulator of chromatin architecture. Brief. Funct. Genomics. 2016; 15:420–431. [DOI] [PubMed] [Google Scholar]
- 69. Boros J., Arnoult N., Stroobant V., Collet J.F., Decottignies A. Polycomb repressive complex 2 and H3K27me3 cooperate with H3K9 methylation to maintain heterochromatin protein 1α at chromatin. Mol. Cell. Biol. 2014; 34:3662–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang Y., Kucukkal T.G., Li J., Alexov E., Cao W. Binding analysis of methyl-CpG binding domain of MeCP2 and Rett syndrome mutations. ACS Chem. Biol. 2016; 11:2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.