Abstract
The study aims at developing a neural network model to improve the performance of Human Phenotype Ontology (HPO) concept recognition tools. We used the terms, definitions, and comments about the phenotypic concepts in the HPO database to train our model. The document to be analyzed is first split into sentences and annotated with a base method to generate candidate concepts. The sentences, along with the candidate concepts, are then fed into the pre-trained model for re-ranking. Our model comprises the pre-trained BlueBERT and a feature selection module, followed by a contrastive loss. We re-ranked the results generated by three robust HPO annotation tools and compared the performance against most of the existing approaches. The experimental results show that our model can improve the performance of the existing methods. Significantly, it boosted 3.0% and 5.6% in F1 score on the two evaluated datasets compared with the base methods. It removed more than 80% of the false positives predicted by the base methods, resulting in up to 18% improvement in precision. Our model utilizes the descriptive data in the ontology and the contextual information in the sentences for re-ranking. The results indicate that the additional information and the re-ranking model can significantly enhance the precision of HPO concept recognition compared with the base method.
1. Introduction
Deep phenotyping has been identified as an efficient way to better describe the observable abnormalities of diseases [1]. Phenotypic concept recognition plays a vital role in this process. HPO [2] is a controlled vocabulary that has been widely used for computational deep phenotyping and precision medicine [3]. HPO has been adopted as a standardized terminology in some phenotyping analysis platforms for clinical practice (e.g., PhenoTips [4], PhenoDB [5]). To capture the comprehensive phenotypic information of patients, clinicians use these platforms to search and select the most appropriate HPO terms for characterizing patients’ symptoms.
The ontology repository with the data version of 2021-02-08 contains 15,783 terms that characterize the phenotypic concepts in human disease. The terms are connected via semantic links representing the hierarchical relations among those concepts. The ontology database also includes cross-links to other knowledge databases, such as Online Mendelian Inheritance in Man (OMIM) [6], Unified Medical Language System (UMLS) [7], and Systematized Nomenclature of Medicine-Clinical Terms (SNOMED-CT) [8]. Each term’s detailed definition and evidence make HPO valuable for precise and dependable phenotype analysis. HPO is not widely employed in the literature, case reports, or most electronic health record (EHR) data to describe phenotypic information. Free text makes it challenging to conduct comparative analyses of phenotypes among these data sources or with new curated patient records, using HPO terms. Therefore, automatic recognition of HPO concepts from free text would be helpful for phenotype-based analysis.
The existing HPO concept recognition approaches are developed using different techniques. OBO annotator [9], NCBO annotator [10], ClinPhen [11], and MetaMap [12] adopt dictionary-based and traditional NLP methods to solve the above-noted problem. Machine-learning approaches also have been applied to this problem for performance improvement [13], but high-quality annotated datasets are limited. Arbabi et al. presented a neural dictionary model, termed Neural Concept Recognizer (NeuralCR), for HPO and SNOMED-CT concept recognition [14]. Other tools for HPO recognition are available via online platforms or APIs such as Doc2hpo [15], Monarch Initiative Annotator1 [16], and Track.Health2. These existing HPO annotation tools are built on the literal terms in the ontology database and the datasets with limited coverage of concept curation.
In our previous study [17], we built PhenoTagger for HPO concept annotation, combining dictionary-based and weakly supervised machine-learning techniques to obtain better recall, outperforming the existing machine-learning approaches. Filtering the false positives would be a straightforward solution to further improve overall performance, especially for those downstream tasks that require better precision or focus on a specific scope of phenotypic abnormality.
This study proposes a re-ranking model pre-trained on the HPO database to filter the results retrieved from the dictionary-based or machine-learning methods. The model utilizes the informative text in the ontology and employs a deep-learning approach to boost overall performance. To construct a pre-training dataset, many textual descriptions of the corresponding HPO concepts are extracted from the HPO ontology database. A pre-built language model is used to obtain the contextual information from the input text, and a novel feature selection component for deep-learning models is proposed to refine the context. Significant improvements as compared to existing methods are observed in the experimental results. The contribution of our proposed feature selection module also is validated in the exclusion experiment.
2. Methodology
2.1. Dataset
BiolarkGSC+
The Bio-LarK gold-standard corpus (GSC) [18] is broadly used in building and testing HPO concept recognition systems. The Bio-LarK CR system, NCBO Annotator [10], and OBO Annotator [9] have been evaluated on this dataset for comparison. The GSC dataset is then fixed in terms of HPO entity inconsistencies and referred to as BiolarkGSC+ [19]. These inconsistencies originate from similar concept mentions that were curated differently in various locations of the corpus. The updated version contains adjusted annotations that diminish the confusion. It enhances the quality of the annotations and reduces the errors of the machine-learning-based annotators.
This dataset comprises 228 abstracts manually annotated with HPO concepts on textual spans (mention level). There are 497 unique HPO terms located at the subtrees of “Phenotypic abnormality” (HP:0000118) and “Mode of inheritance” (HP:0000005). As shown in Supplementary Table S1, each document in the corpus has roughly seven sentences and 149 tokens on average, implying that it can fit into memory for most machine-learning models. The number of labels varies from 1 to 54, indicating that the distribution of annotations over all the documents is sparse.
COPD-HPO
Besides the updated abstract-level corpus GSC+, we also employed another corpus containing phenotypic annotations, COPD [20]. It consists of 30 full-text articles that focus on Chronic obstructive pulmonary disease (COPD) phenotypes. The full-text articles are split into paragraphs in advance, and the annotations are given for each paragraph to form a sample. The named entity annotations in the COPD corpus are linked to UMLS concept identifiers (CUIs). We map the CUIs to HPO ids through the cross-links provided in the HPO database, named COPD-HPO in this paper. As shown in Fig. 1, since this dataset only focuses on the phenotypes related to a specific disease and not every CUI can be mapped to a corresponding HPO concept, the documents contain fewer labels than those in GSC+. Because the COPD-HPO corpus is not intuitively designed for HPO annotation evaluation, we regard it as a silver standard corpus that serves as a secondary dataset for benchmarking.
Fig. 1.
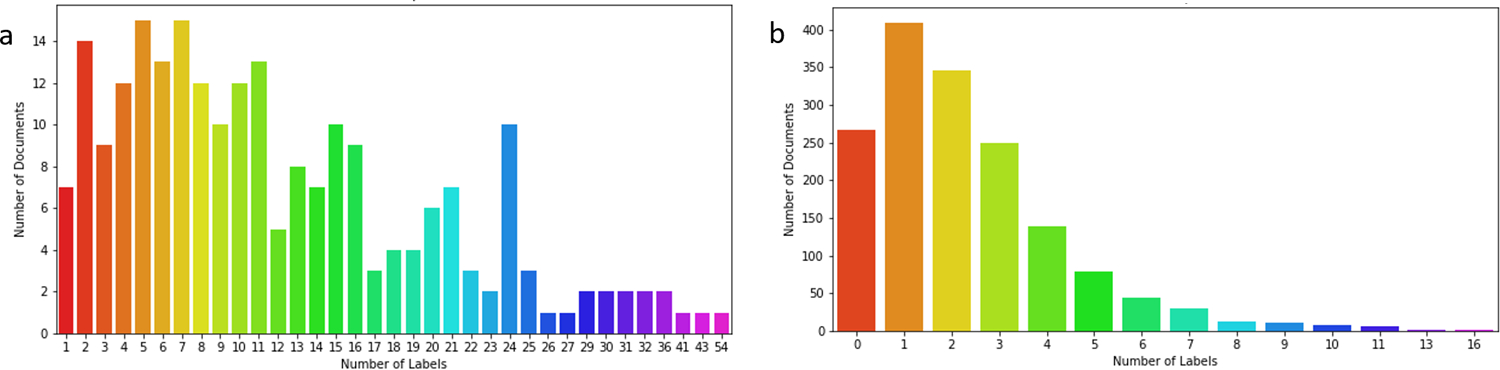
Distributions of labels for (a) BiolarkGSC+ and (b) COPD-HPO datasets
In practice, most of the clinical notes are annotated at the document level. To align with those practical scenarios, we evaluate all the approaches only at the document level. Document classification and named entity recognition are two common tasks in NLP. However, formalizing HPO recognition as either one of these two tasks is not appropriate. It can be directly observed from Fig. 1 that documents in BiolarkGSC+ are annotated with more HPO labels than those in COPD-HPO on average. In other words, it would be a challenge to formalize the HPO annotation task as a multi-label classification problem due to the number and distribution of labels. Considering the HPO annotation task as a named entity recognition problem seems more reasonable. Document-level annotations, however, are adequate for downstream tasks. In addition, mention-level curations are immensely expensive and time-consuming. Currently, corpora with mention-level annotations of HPO terms are limited for training a supervised NER model. Therefore, we formalized the problem as a document-concept pair similarity task with additional pre-processing and post-processing steps. Our model addresses the problem of insufficient curated data by leveraging the materials in the HPO database. The pre-training process does not need any datasets with manual curations of HPO labels, while the optional fine-tuning step requires only sentence- or mention-level annotations.
2.2. Problem formalization and transformation
As depicted in Table S1 about the data statistics, the machine-learning models may not work well in the multi-label classification problem in BiolarkGSC+ due to the sparse distribution of the labels. A conventional approach to address this is problem transformation for multi-label classification [21]. Although the transformation will yield multiple classifiers, training more than 10,000 classifiers for HPO concept recognition is inappropriate when using a language model. Based on the above observations, we propose a framework that can improve the HPO annotator’s performance, specifically in terms of precision, by leveraging the language model. We formalize the concept recognition as a sentence similarity prediction problem. First, we derive the corresponding label names and definitions from the ontology database for the HPO IDs. For each document annotated with HPO IDs, we transform the document into pairs of records, . If the document contains multiple long sentences that cannot fit into the models, we further split the document into sentences, treating each sentence as a document that is then paired with the annotated labels. This splitting step is only required when the text is too long to be handled by the model and the annotations are sentence- or mention-level. Adopting a new model that accepts an arbitrary input length is an alternative solution for this issue. After that, we link the label names and the definitions of the HPO IDs to these records. The objective is to predict whether the document contains the HPO label names or has a similar meaning based on its definition. It is not feasible to iterate over all pairs of documents and HPO IDs. Accordingly, we adopt a base method to retrieve a candidate set of labels before constructing pairwise records. Based on the benchmarking results, we employed three robust methods, text annotator from Monarch Initiative, MetaMap, and the state-of-the-art HPO annotator PhenoTagger [17], to annotate the corpora with HPO concepts. The resulting candidate labels for each document were transformed into pairs of text and labels and fed into the trained re-ranking model. The final re-ranking model acts as a filter to diminish false-positive predictions.
The example in Fig. 2 demonstrates how we re-rank the results of the HPO annotator from Monarch Initiative. The predictions of a base method may contain irrelevant concepts (colored in pink). In this example, the terms “distal” and “proximal” can fully match the concepts under the sub-tree of clinical modifier. However, they better fit the longer text span according to the context of the sentence. Our model takes as input both the text and the predictions of the base method, and then determines if the predicted concepts fit the context.
Fig. 2.

An example of HPO concept re-ranking. The predictions are generated using Monarch Initiative Annotator and then re-ranked for more accurate results. The concepts in bold are ground truth annotations, whose corresponding text is highlighted in yellow. The problem is to filter out the false positives colored in pink and keep the true positives.
2.3. Re-ranking model
BlueBERT language model
We illustrate the workflow and architecture of our proposed re-ranking model in Fig. 3. The flow chart on the left side demonstrates the processes of prediction. The model on the right side is pre-trained on the informative content extracted from the ontology database before re-ranking. The parts before the linear transformation module in the re-ranking model can be divided into two streams. The left stream is a conventional sentence-similarity prediction workflow that employs BlueBERT, a BERT model pre-trained on PubMed abstracts, and MIMIC-III clinical notes [22]. The model takes a text pair as input, where the first one is the sentences in the documents, and the second one is the label names or definitions of the paired HPO term. It encodes the text pairs to a tensor with dimension , which depends on the hyper-parameters of the BlueBERT model.
Fig. 3.
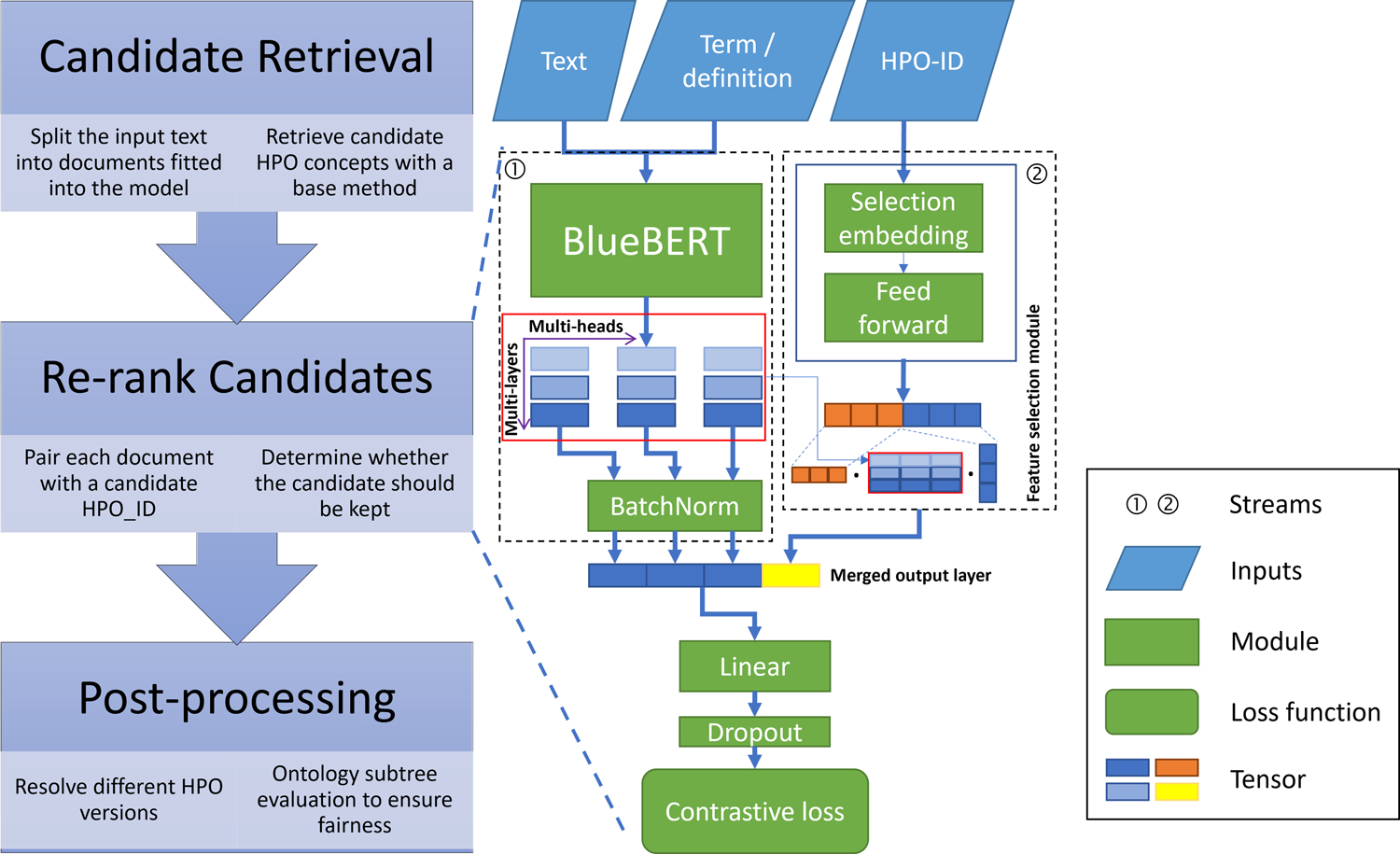
Architecture of the re-ranking model. Pairs of document and candidate HPO terms are inputted into the model. The text in the document and HPO term (or term definition) are merged into the BlueBERT model, while the HPO IDs are fed into the selection-embedding module. The outputs of these two streams are concatenated before the linear transformation module.
Feature selection module
Commonly, the output of the last layer (colored in blue in Fig. 3) in the BlueBERT model will be used as input of a linear transformation module. However, different layers of the output encode the granularity of the context, where the layers close to the textual tokens are more likely to represent word embeddings. In contrast, those close to the loss function imply features for the task labels. The strategy of selecting the best output layers varies for different labels, and it is time-consuming to exploit the performance of all layers [23]. The HPO annotation problem that we formalized also can be regarded as a multiple binary classification problem for different HPO terms. Therefore, we developed a feature selection module that utilizes the optimum choice of output from the language model for each label. As shown in the right stream in Fig. 3, we employ a selection-embedding module for the HPO concepts. After pre-training, this module’s parameters will be fixed, which means that we encode all the HPO IDs into a continuous space that represents each unique HPO concept. A feed-forward module is employed to encode the trained embeddings into weight vectors used to calculate weighted sums on BlueBERT’s multi-layer outputs. The resulting output (colored in yellow in Fig. 3) is concatenated to the output of the last layer of BlueBERT.
Contrastive loss
We append a batch normalization module [24] after the BlueBERT model’s output to accelerate the training process. After the merged output tensor is applied to the linear module, we append a dropout module to prevent overfitting. Last, we adopted a pairwise ranking loss, contrastive loss [25], to optimize the predicted similarity scores for the input pairs of text. Assume the ground truth label and predicted label for document is and , the contrastive loss of the model on sample is , where the parameter m is set to 2 normally. We also employ an early-stopping strategy to terminate the training process when the loss value no longer decreases. The detailed hyperparameters of our proposed model are shown in Supplementary Table S2.
2.4. Weakly supervised pre-training
Due to the lack of sufficient annotated corpora for HPO concept recognition, we use the intrinsic data of HPO database for weakly supervised pre-training. In addition to term names, the HPO database contains multiple types of textual data (e.g., synonyms, definitions, comments) for most concepts that can be used for pre-training. The labels are first obtained from the associations between the concepts and the textual content provided in the HPO ontology database. We observe that those textual records either imply or explicitly contain the corresponding concept terms, given that the associations between the text and concepts are extracted from the ontology database. Then, a group of randomly selected non-ancestor concepts and the correct concept are paired with each record. The objective of this pre-training task is to predict whether the textual records imply the paired concept terms. The textual data embedded, involving almost all the HPO labels in the ontology database, are used to initialize the model. Consequently, the parameters of the pre-trained model are shared with the downstream tasks.
To derive a robust model that can perform concept recognition on the complete set of HPO terms, we fed the model with different types of ontology content for pre-training. The textual content is paired with the corresponding concept terms as input. The version of the ontology database we used in our experiments contains definitions of about 11,084 of the 15,783 HPO concepts. We adopted the synonyms of the remaining 4,699 concepts to represent their meanings. In addition to definitions and synonyms, the database consists of many comment notes that can be organized as a corpus in which the notes describe the corresponding HPO concept. Cross-links with other databases also contain similar notes that can be merged into this corpus. We constructed a corpus with all the textual data that we extracted from the ontology database, including concept terms, definitions, comments, and synonyms for pre-training. As for the negative samples of a specific label annotated in each document, we used the Damerau-Levenshtein distance [26] to find similar terms from the HPO database, except the ancestors and descendants of a specific HPO ID. The negative samples are then randomly selected from a large set of candidates. The definitions and comments of those negative labels are paired with the concept terms, as with the positive samples. We used synonyms instead for those concepts without definitions or comments in the ontology database. The negative samples can help our model discriminate against the positive samples from a large number of negatives. To handle this imbalanced dataset, we applied a label-based weighting scheme to the loss function.
2.5. Fine-tuning
After pre-training, we fine-tuned the model using a target dataset (e.g., BiolarkGSC+ and COPD-HPO). This is an optional step because the pre-training corpus already had covered all the available HPO concepts, and the pre-trained model can work on most HPO recognition tasks. If better performance is desired and a portion of the target dataset has been annotated, however, the fine-tuning step is helpful to further improve predictive performance. Nevertheless, some studies focus on only a specific type of disease or a set of phenotypes, in which the existing methods will retrieve irrelevant concepts that appear in the text. Re-ranking approaches can better resolve this problem by fine-tuning the model on the target dataset. We evaluated the difference in performance when using this optional fine-tuning step, as shown in the Results section below.
2.6. Evaluation
Because some existing methods have different behaviors for HPO concept extraction, a consolidated post-processing procedure was applied to all compared methods to ensure fairness. The first and most crucial step is the unification of concept IDs among different HPO releases. We obtained the mapping between each deprecated alternative ID and its primary concept ID from the ontology database and applied this mapping to the datasets and predictions. Another crucial step is to evaluate the model on a specific subtree of the HPO hierarchical structure to ensure fairness due to the limitations of some existing methods (e.g., OBO annotator works on concepts under HP:0000118 “phenotypic abnormality”). This step, however, is not necessary for a real-world application unless the task is required to focus on a specific subtree of the ontology. As a result of evaluating our model on a subset of labels, the original data records may have empty lists of annotated concepts. Calculating some metrics may encounter a zero-divisor problem if the prediction is also an empty list of concepts. We addressed the null label issue by adding a dummy label to both the gold-standard annotations and the record predictions and adopting the original metrics during the evaluation.
3. Results
3.1. Experimental setting
We implemented our model using PyTorch3 and trained it on four Nvidia Tesla V100 GPUs. We wrote custom wrapper code for each comparison method to ensure consistency between all inputs and outputs. From the perspective of practicability, the document-level annotations are sufficient for the downstream analysis. Therefore, we adopted macro average [27] document-level metrics to evaluate all the comparison approaches’ performance.
We compared our approaches with the existing methods on BiolarkGSC+ and COPD-HPO. We evaluated our model on the whole target datasets when it is pre-trained only on the corpus generated from the ontology database. We compared our method to others by not training on the target datasets. We built three versions of our approach adopting different base methods, Monarch Initiative annotator (MNI), MetaMap (MM), and PhenoTagger (PT), reported as MNIRerank, MMRerank, and PTRerank, respectively, because they achieved the most robust performance in the benchmark experiment. PhenoTagger and MetaMap have the highest recall on BiolarkGSC+ and COPD-HPO, respectively. As shown in Table 1, our re-ranking model sacrifices a little recall for a significant increase in precision. Compared with the original method, PTRerank, MMRerank, and MNIRerank achieved 3%, 1.9%, and 1.1% F1-score improvement on BiolarkGSC+, while obtaining 9.4%, 9.6%, 2.4% F1-score improvement on COPD-HPO. Precision and recall play an essential role in deep phenotyping. We make full use of the dictionary-based methods to retrieve as many relevant candidate labels as possible, then employ a model with deep-language understanding to filter out irrelevant labels. As we generate plenty of negative samples for training, a label-based weighting scheme is applied. Namely, we penalize false negatives more than false positives in the loss function. That is why the recall of the base method and our re-ranking method are so similar.
Table 1.
Performance comparison on BiolarkGSC+ and COPD-HPO
| BiolarkGSC+ | COPD-HPO | |||||
|---|---|---|---|---|---|---|
| Method/Metric | Precision | Recall | F1-score | Precision | Recall | F1-score |
| OBO Anotator [9] | 0.810 | 0.568 | 0.668 | 0.318 | 0.282 | 0.299 |
| NCBO [10] | 0.777 | 0.521 | 0.624 | 0.756 | 0.763 | 0.760 |
| MonarchInitiative [16] | 0.751 | 0.608 | 0.672 | 0.741 | 0.747 | 0.744 |
| Doc2hpo-Ensemble [15] | 0.754 | 0.608 | 0.673 | 0.779 | 0.755 | 0.767 |
| MetaMap [12] | 0.707 | 0.599 | 0.649 | 0.640 | 0.781 | 0.704 |
| Clinphen [11] | 0.590 | 0.418 | 0.489 | 0.377 | 0.328 | 0.351 |
| NeuralCR [14] | 0.736 | 0.610 | 0.667 | 0.543 | 0.719 | 0.619 |
| TrackHealth | 0.757 | 0.595 | 0.666 | 0.719 | 0.669 | 0.693 |
| PhenoTagger [17] | 0.720 | 0.760 | 0.740 | 0.623 | 0.820 | 0.708 |
| MMRerank | 0.754 | 0.599 | 0.668 | 0.822 | 0.779 | 0.800 |
| MNIRerank | 0.789 | 0.603 | 0.683 | 0.802 | 0.736 | 0.768 |
| PTRerank | 0.843 | 0.708 | 0.770 | 0.836 | 0.771 | 0.802 |
Note. MMRerank, MNIRerank, and PTRerank represent the re-ranking models based on MetaMap, MonarchInitiative methods, and PhenoTagger. The digits in bold indicate the best scores in terms of the corresponding metrics.
We also conducted experiments to fine-tune the model on the target datasets. We split the dataset into 80:20 samples for training and testing, respectively. We recalculated the performance of the existing methods on the testing set of the dataset. The training sets were then fitted into our pre-trained re-ranking model before evaluating it on the testing set. As depicted in Supplementary Table S3, the performance improvement is consistent with evaluations on the whole dataset. The re-ranking model boosted the F1-score at most by 8.1% and 7.6% on the testing set of BiolarkGSC and COPD-HPO, respectively.
As shown in Supplementary Table S4, we conducted the same experiments without post-processing applied to all the methods on the whole dataset to highlight the importance of post-processing steps. To align with the results in our previous study [17], we also ran the experiments on the pre-defined subset of BiolarkGSC+. As illustrated in Supplementary Table S5, the results are mainly consistent with those conducted on the whole dataset. The re-ranking results demonstrate that our model can fit both dictionary-based and machine-learning approaches for performance boosting. Details are provided in Appendix A.
3.2. Excluding selection embedding
The selection embedding shown in Stream 2 of Fig. 3 plays an essential role in our proposed method. The multi-layer multi-head transformer-based model captures different types and levels of context, represented over all the layers and heads. Each HPO concept has a different focus in this context. The selection embedding encodes the focus for each HPO concept and helps the model make the optimum choice of context according to the observed patterns. We conducted additional experiments to verify the effectiveness of the selection embedding. We excluded Stream 2 of Fig. 3 and repeated the experiments for our re-ranking model. As tabulated in Table 2, the re-ranking model without selection embedding slightly improved the precision but resulted in a more significant decrease in the recall, resulting in lower overall performance.
Table 2.
Performance of the re-ranking model without the selection embedding component
| Method | Precision | Recall | F1-score | Precision | Recall | F1-score |
|---|---|---|---|---|---|---|
| BiolarkGSC+ | COPD-HPO | |||||
| MMRerank w/o SE | 0.605(−20%) | 0.460(−23%) | 0.523(−22%) | 0.463(−44%) | 0.615(−21%) | 0.528(−34%) |
| MNIRerank w/o SE | 0.567(−28%) | 0.483(−20%) | 0.522(−24%) | 0.445(−45%) | 0.564(−23%) | 0.497(−35%) |
| PTRerank w/o SE | 0.753(−11%) | 0.407(−43%) | 0.528(−31%) | 0.683(−18%) | 0.506(−34%) | 0.581(−28%) |
| BiolarkGSC+ Testing Set | COPD-HPO Testing Set | |||||
| MMRerank w/o SE | 0.581(−27%) | 0.377(−7%) | 0.457(−15%) | 0.478(−33%) | 0.623(−4%) | 0.541(−20%) |
| MNIRerank w/o SE | 0.714(−14%) | 0.349(−21%) | 0.468(−19%) | 0.658(−1%) | 0.590(−1%) | 0.622(−1%) |
| PTRerank w/o SE | 0.580(−19%) | 0.218(−50%) | 0.317(−41%) | 0.637(−10%) | 0.558(−21%) | 0.595(−15%) |
Note. w/o SE represents without the selection embedding component. Each percentage number in the brackets is the change compared with the sampe approach with selection embedding.
3.3. Detailed comparison of predictions
To further illustrate how our proposed method works, we compared the prediction results of the base approaches, our re-ranking methods, and the re-ranking methods without selection embedding. The Venn diagram in Fig. 4 demonstrates the number of overlapping HPO terms predicted by the compared methods and lists the number of true positives in brackets. Our re-ranking model filters out most of the false positives produced by the base approaches. The Venn diagram counts consider only the HPO term predicted in all documents. The number of false positives can be calculated by subtracting the numbers from the ones in parenthesis for each part of the Venn diagrams. PTRerank, MMRerank, and MNIRerank eliminate 83.2%, 85.7%, and 84.7%, respectively, of the false positives on BiolarkGSC+. However, the models without the selection embedding module kept many false positives, leading to poorer re-ranking performance.
Fig. 4.
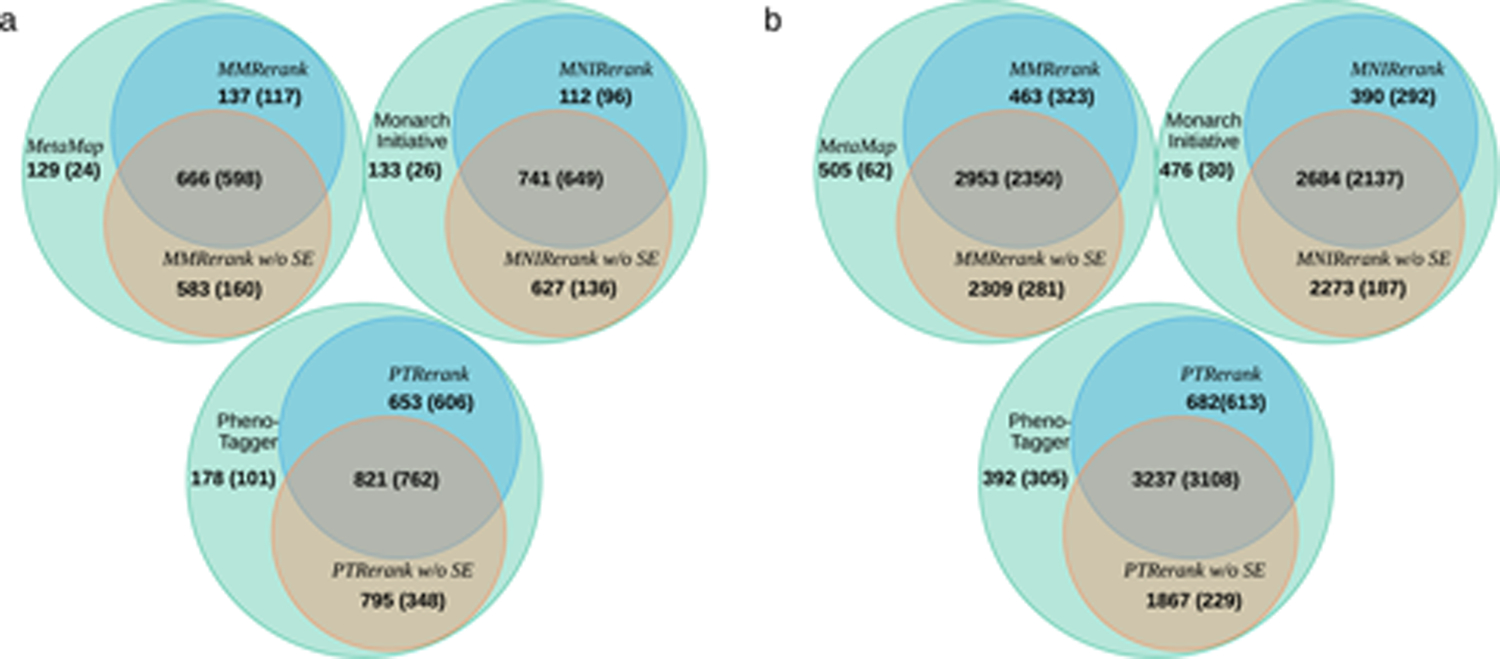
Venn diagram of the prediction results among the base approaches (MetaMap, MonarchInitiative, PhenoTagger), the re-ranking approaches (MMRerank, MNIRerank, PTRerank), and the re-ranking without selection embedding (without SE) approaches (MMRerank without SE, MNIRerank without SE, PTRerank without SE) on (a) BiolarkgGSC+ and (b) COPD-HPO. The digits outside parentheses represent the number of predicted labels, and the ones in parentheses indicate the true positive among the predictions.
Another finding is that the total number of concepts contained in COPD-HPO is much larger than that in BioLarkGSC+, which is caused by the large difference in the number of samples in both datasets. Along with the fact that fewer labels are contained in COPD-HPO, it leads to larger sparsity of the label distribution in BioLarkGSC+. As a result, the recall of most of the methods on BioLarkGSC+ is lower than that on COPD-HPO. The difference between BioLarkGSC+ and COPD-HPO reflects the complexity in the real-world applications and the challenge in HPO concept recognition. Our approach addresses this issue by problem transformation and pre-training on the ontology.
3.4. Error analysis
We randomly pick some samples from the categories of true positives that were wrongly filtered out (false negatives) and the false positives that were wrongly kept in by our proposed re-ranking model (jointly predicted by PTRerank, MMRerank, and MNIRerank) on BiolarkGSC+ for error analysis. The re-ranking model might fail to recognize the completed different expressions of these HPO concepts. For example, the sentence, “The analysis of a de novo 8q12.2-q21.2 deletion led to the identification of a proposed previously undescribed contiguous gene syndrome consisting of Branchio-Oto-Renal (BOR) syndrome, Duane syndrome, hydrocephalus and trapeze aplasia.” in document PMID:7849713 contains the HPO concept HP:0004253 “Absent trapezium”, mentioned as “trapeze aplasia”. The base approach finds this concept by string matching, but the re-ranking model filters it out according to the context. This is because this concept term is completely different from the mentions, and it has limited information in the ontology including the definition and synonyms. The model wrongly filtered out this concept during the re-ranking step. However, if the information is sufficient for the model to learn the alternative expressions, it could help prevent this type of error. For instance, the sentence “All affected individuals had shortness principally affecting the second and fifth phalanges and first metacarpal.” in document PMID:9024575 is predicted with HPO:0010692 “2–5 finger syndactyly”. However, this concept emphasizes the fusion of fingers but it is not reflected in the context, which is then filtered by our re-ranking model.
Another example is that the re-ranking model may not be able to discriminate against variant expressions with few common words. “These features include severe mental retardation, epileptic seizures, easily provoked and prolonged paroxysms of laughter, atactic jerky movements, hypotonia, large mandible with prognathia, and 2–3 cps spike and wave activity in the EEG” in PMID:7450780 contains concept HP:0000303 “Mandibular prognathia” but expressed as “large mandible with prognathia” in the paragraph. The relation between these two phrases is not captured by the re-ranking model, which leads to a false negative. For instance, the base approach detects concept HP:0100272 “Branchial sinus” in the sentence, “The earpits-deafness syndrome is an autosomal dominant disorder in which affected individuals may have sensorineural, conductive or mixed hearing loss, preauricular pits, structural defects of the outer, middle and inner ear, lacrimal duct stenosis, branchial fistulas or cysts of the second branchial arch, and renal anomalies ranging from mild hypoplasia to complete absence” of PMID:6964893. The re-ranking method and the base approach think that “branchial fistulas or cysts” has the identical meaning of “branchial sinus.” Another possibility is that the dataset has confusing gold-standard annotations. The sentence, “Tongue thrusting is common,” in PMID:2466440 should match concept HP:0100703 “Tongue thrusting.” The gold-standard annotations, however, indicate it as concept HP:0000182 “Movement abnormality of the tongue.” The curator of the gold-standard annotations has a different understanding of the phenotype contained in this sentence. Given the limited training samples for this particular concept, it is difficult for the algorithm to learn this pattern.
In addition to prediction errors, annotation bias is inevitable in manually curated datasets that would affect the evaluation results. Some of the false positives identified by algorithms might be “true positives” that curators miss. Therefore, this bias might affect our approach since the re-ranking model is designed to filter out the actual false positives. We sampled 10% and 3% of the records from BioLarkGSC+ and COPD-HPO, which we manually reviewed to estimate the error rate of each approach. We then calculated the calibrated precisions by for each document and obtained the overall results using macro averaging. As shown in Supplementary Table S6, the improvement of precision by our re-ranking approaches is slightly decreased, but the ranking of precisions is consistent with the original results. It reveals that the evaluation results are still meaningful since the pre-training step of our re-ranking model does not heavily rely on those manually curated datasets which might contain annotation bias.
4. Discussion
We have presented a benchmarking experiment on the BiolarkGSC+ dataset over several existing approaches and our approach. Our method, which re-ranks results derived from other dictionary-based approaches, significantly improves precision (from 3.8% to 12.3% on BiolarkGSC+ and 1.82% to 21.3% on COPD-HPO, as seen in Table 1) at the cost of a minor reduction in recall compared to the base approach. The existing methods rely mainly on dictionary data instead of the complete ontology information for HPO concept annotation, resulting in more false positives. Our proposed re-ranking method acts as a filter to help eliminate irrelevant results according to context. We pair each sentence and candidate HPO term together to form a textual pair instance, which can be fed into a language model for improved classification.
The method presented here does not require mention-level annotations; instead, it only utilizes the description data in the ontology database for model training. It addresses the problem of limited annotated resources in terms of label coverage and annotation level. It also can be fine-tuned to a specific dataset for better performance when provided with sentence- or abstract-level annotations for training. However, identifying a base approach with high recall is a limitation of our proposed method. Our benchmark experiments revealed that PhenoTagger, MetaMap, and the annotator from the Monarch Initiative team worked consistently well on BiolarkGSC+ and COPD-HPO. They yielded higher recall, which implies that they are more suitable to be employed in our re-ranking model.
5. Conclusion
In this study, we conducted a comprehensive study on HPO concept recognition, and evaluated most existing approaches on a gold-standard dataset as well as a large corpus focuses on a specific disease. The benchmark results indicated that those approaches with higher recall have room for improvement in precision. Precision becomes more important when the text contains a number of irrelevant concepts but the annotations only focus on specific diseases. This is common in clinical notes when patients have multiple visits and tests. To achieve this goal, we proposed a re-ranking model that can refine the results of an existing base approach. We formalized the HPO concept annotation task as a multi-label classification problem and transformed the problem into a sentence-concept similarity prediction. The problem transformation intends to build a weakly supervised model that employs knowledge from the HPO database. The model was fed with the textual information in the HPO database, including terms, synonyms, definitions, notes, and comments. The objective of training the model is to learn whether a sentence contains a specific HPO concept. The re-ranking approach was shown to work well with base algorithms that demonstrate high recall and retrieve as many true positives as possible. The re-ranking model improves precision by filtering out most retrieved false positives, resulting in a significant improvement in the F1-score. A clear advantage of using our re-ranking model is that it does not need mention-level annotations for training and can be fine-tuned on specific datasets by providing corresponding sentence- or abstract-level curations. The relied-upon base approach may limit the power of our model in terms of recall. However, our general re-ranking method can be paired with future approaches to obtain a better overall recall. It ensures that our method remains flexible for use with different types of datasets. Future work in this area would involve developing a more accurate HPO annotator for documents with arbitrary lengths and considering correlations among the predicted concepts. Negation detection and clinical modifier recognition of phenotype [28] are also important for real-world applications. Our clinical NLP tool NegBio [29] would be helpful in handling the negation problem for practical usage. They could further improve the accuracy and refine the granularity of the extracted phenotypic information, which is worth exploring in a future study.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Footnotes
Availability
The source code of the re-ranking model, the benchmarking scripts, and the pre-processed datasets are available at https://github.com/ncbi-nlp/PhenoRerank.
References
- [1].Robinson PN. Deep phenotyping for precision medicine. Hum Mutat 2012;33:777–80. 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- [2].Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res 2017;45:D865–76. 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Robinson PN, Mungall CJ, Haendel M. Capturing phenotypes for precision medicine. Mol Case Stud 2015;1:a000372. 10.1101/mcs.a000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Girdea M, Dumitriu S, Fiume M, Bowdin S, Boycott KM, Chénier S, et al. PhenoTips: Patient phenotyping software for clinical and research use. Hum Mutat 2013;34:1057–65. 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- [5].Hamosh A, Sobreira N, Hoover-Fong J, Sutton VR, Boehm C, Schiettecatte F, et al. PhenoDB: A New Web-Based Tool for the Collection, Storage, and Analysis of Phenotypic Features. Hum Mutat 2013;34:n/a–n/a. 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Amberger JS, Bocchini CA, Scott AF, Hamosh A. Omim. org: leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res 2019;47:D1038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bodenreider O The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lee D, de Keizer N, Lau F, Cornet R. Literature review of SNOMED CT use. J Am Med Informatics Assoc 2014;21:e11. 10.1136/amiajnl-2013-001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Taboada M, Rodríguez H, Martínez D, Pardo M, Sobrido MJ. Automated semantic annotation of rare disease cases: a case study. Database 2014;2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Whetzel PL. NCBO Technology: Powering semantically aware applications. J Biomed Semantics 2013;4:S8. 10.1186/2041-1480-4-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Deisseroth CA, Birgmeier J, Bodle EE, Kohler JN, Matalon DR, Nazarenko Y, et al. ClinPhen extracts and prioritizes patient phenotypes directly from medical records to expedite genetic disease diagnosis. Genet Med 2019;21. 10.1038/s41436-018-0381-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aronson AR, Lang F-MM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229–36. 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Huang M-S, Lai P-T, Lin P-Y, You Y-T, Tsai RT-H, Hsu W-L. Biomedical named entity recognition and linking datasets: survey and our recent development. Brief Bioinform 2020. [DOI] [PubMed] [Google Scholar]
- [14].Arbabi A, Adams DR, Fidler S, Brudno M. Identifying clinical terms in medical text using ontology-guided machine learning. J Med Internet Res 2019;21:e12596. 10.2196/12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu C, Peres Kury FS, Li Z, Ta C, Wang K, Weng C. Doc2Hpo: a web application for efficient and accurate HPO concept curation. Nucleic Acids Res 2019;47:W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shefchek KA, Harris NL, Gargano M, Matentzoglu N, Unni D, Brush M, et al. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 2020;48:D704–15. 10.1093/nar/gkz997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Luo L, Yan S, Lai P-T, Veltri D, Oler A, Xirasagar S, et al. PhenoTagger: A Hybrid Method for Phenotype Concept Recognition using Human Phenotype Ontology. ArXiv Prepr ArXiv200908478 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Groza T, Kohler S, Doelken S, Collier N, Oellrich A, Smedley D, et al. Automatic concept recognition using the Human Phenotype Ontology reference and test suite corpora. Database 2015;2015:bav005–bav005. 10.1093/database/bav005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lobo M, Lamurias A, Couto FM. Identifying human phenotype terms by combining machine learning and validation rules. Biomed Res Int 2017;2017:1–8. 10.1155/2017/8565739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ju M, Short AD, Thompson P, Bakerly ND, Gkoutos GV, Tsaprouni L, et al. Annotating and detecting phenotypic information for chronic obstructive pulmonary disease. JAMIA Open 2019;2:261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tsoumakas G, Katakis I. Multi-label classification: An overview. Int J Data Warehous Min 2007;3:1–13. [Google Scholar]
- [22].Peng Y, Yan S, Lu Z. Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets 2019:58–65.
- [23].Huang H-Y, Zhu C, Shen Y, Chen W. FusionNet: Fusing via Fully-Aware Attention with Application to Machine Comprehension. 6th Int Conf Learn Represent ICLR 2018 - Conf Track Proc 2017. [Google Scholar]
- [24].Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. 32nd Int. Conf. Mach. Learn. ICML 2015, vol. 1, International Machine Learning Society (IMLS); 2015, p. 448–56. [Google Scholar]
- [25].Hadsell R, Chopra S, LeCun Y. Dimensionality reduction by learning an invariant mapping. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit, vol. 2, 2006, p. 1735–42. 10.1109/CVPR.2006.100. [DOI] [Google Scholar]
- [26].Brill E, Moore RC. An improved error model for noisy channel spelling correction. Proc. 38th Annu. Meet. Assoc. Comput. Linguist., 2000, p. 286–93. [Google Scholar]
- [27].Van Asch V. Macro-and micro-averaged evaluation measures. Belgium: CLiPS 2013;49. [Google Scholar]
- [28].Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine J-P, et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res 2019;47:D1018–27. 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Peng Y, Wang X, Lu L, Bagheri M, Summers R, Lu Z. NegBio: a high-performance tool for negation and uncertainty detection in radiology reports. AMIA Summits Transl Sci Proc 2018;2018:188. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source code of the re-ranking model, the benchmarking scripts, and the pre-processed datasets are available at https://github.com/ncbi-nlp/PhenoRerank.


