Abstract
Formation of protein complexes is crucial to most biological functions. The cellular mechanisms governing protein complex biogenesis are not yet well understood, but some principles of co-translational and post-translational assembly are beginning to emerge. In bacteria, this process is favored by operons encoding subunits of protein complexes. Eukaryotic cells do not have polycistronic mRNAs raising the question of how they orchestrate the encounter of unassembled subunits. Here we review the constraints and mechanisms governing eukaryotic co- and post-translational protein folding and assembly, including the influence of elongation rate on nascent chain targeting, folding, and chaperone interactions. Recent evidence shows that mRNAs encoding subunits of oligomeric assemblies can undergo localized translation and form cytoplasmic condensates that might facilitate the assembly of protein complexes. Understanding the interplay between localized mRNA translation and co-translational proteostasis will be critical to define protein complex assembly in vivo.
Keywords: Protein folding, Translation elongation, Cotranslational folding, Chaperones, Hsp70, Chaperonin, TRiC, Organelle targeting, NAC, Protein Complex Assembly, RNA granules, Nascent polypeptide
1. Introduction
How proteins acquire their unique three-dimensional structure is one of the fundamental questions in biology. While biophysical experiments showed that the information to reach the native structure is encoded in the amino acid sequence1, the last 20 years have demonstrated that protein folding in vivo is governed by biological constraints. Eukaryotic proteins are structurally diverse: some have complex domain folds, others are multidomain proteins, while others work as part of multi-subunit protein complexes2. In many, if not most, protein complexes, completing polypeptide folding is dependent on binding to oligomeric partner proteins. In order to orchestrate these processes within the densely crowded eukaryotic cytosol, cells have evolved different molecular pathways, collectively termed the Proteostasis Network (PN; Figure 1). The proteostasis network comprises around 2,000 proteins in human cells3. These factors survey protein conformation and fate from the moment they are synthesized on ribosomes through their final degradation by cellular clearance pathways. During biogenesis, the PN assists co-translational folding, protein trafficking to organelles -such as the Endoplasmic Reticulum (ER) and mitochondria-, and when correct folding and assembly are impaired, facilitates spatial sequestration and degradation of misfolded proteins4,5, avoiding the accumulation of toxic or aggregation-prone protein species6.
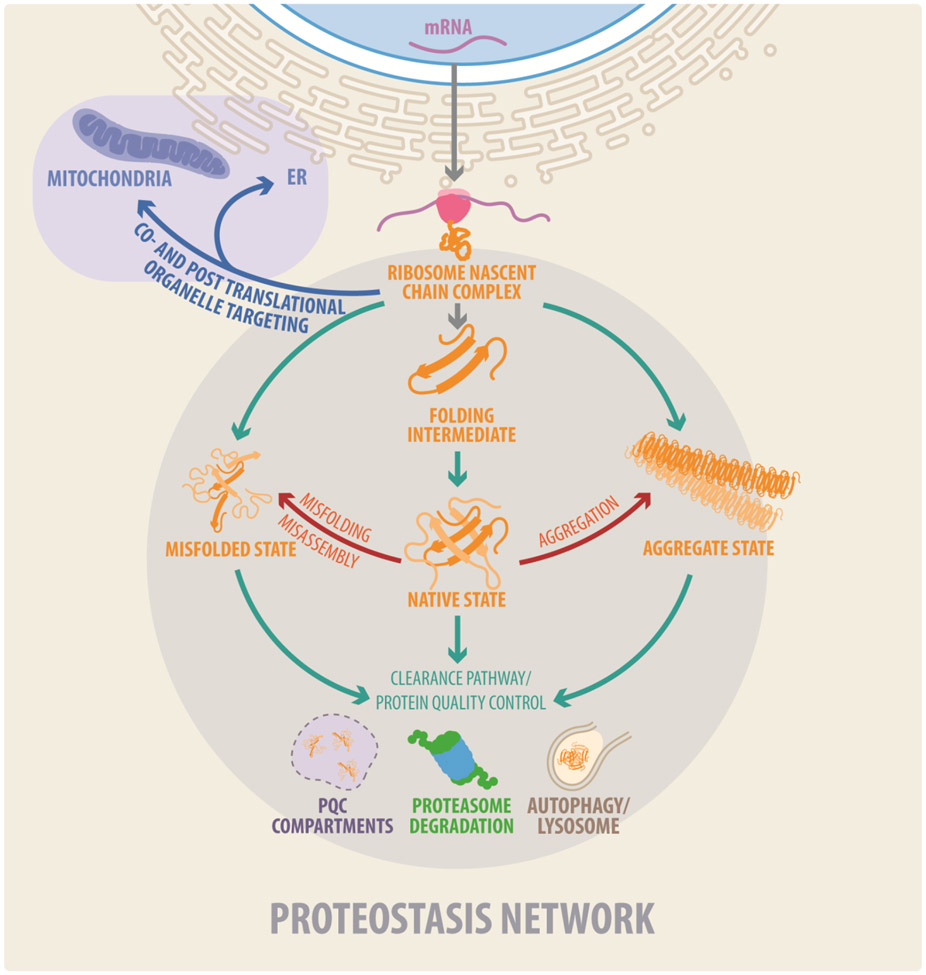
Figure 1. The Proteostasis Network.
Proteostasis (protein homeostasis) is required to maintain a functional proteome. It results from the integration of cellular processes that mediate protein synthesis, protein folding, targeting to organelles, and protein quality control (PQC) and clearance pathways. The proteostasis network consists of factors that together promote these processes, including the translation machinery, molecular chaperones, the ubiquitin proteasome system (UPS), and autophagy–lysosomal processes. Misfolded and aggregated proteins are detected by quality control mechanisms and subjected to the ubiquitin–proteasome system and the autophagosomal–lysosomal machinery to protect cells against accumulating abnormal proteins.
Recent advances illuminate how the PN facilitates co-and post-translational folding of monomeric proteins and assembly of protein complexes7,8. Here we review the principles and complex pathways promoting protein biogenesis and summarize our current understanding of the factors assisting these processes in the eukaryotic cell. Sections 2-5 reviews the roles of specific chaperones and targeting factors that associate with nascent and newly translated polypeptide in promoting de novo protein folding and targeting in the cell. Section 6 summarized the increasingly clear coordination between the fine-tuning of translation elongation rate and cotranslational protein folding, chaperone binding and targeting. Finally, Sections 7-9 discuss emerging ideas for how different protein subunits of oligomeric complexes may come together to form fully assembled protein complexes. Section 7 overviews the role of translation in orchestrating the encounter of subunits of protein complexes, while Sections 8 and 9 discuss the role of localized translation and RNA granules and condensates in the organization of assembly processes in the crowded cellular milieu of the eukaryotic cell.
2. Ribosome-nascent chain associated factors during protein biogenesis
The journey of every protein starts at the ribosome, where messenger RNA (mRNA) is decoded into a polypeptide (see Table 1 for List of Abbreviations). The elongating nascent protein is protected from cytosol inside the ribosome exit tunnel until it reaches ~30~40 amino acids9. Once a nascent protein emerges from the ribosomal exit tunnel, it encounters many ribosome-associated nascent chain-binding proteins, including N-terminal modification enzymes, co-translationally acting chaperones, and membrane targeting factors (Fig. 2, A and Table 2)9,10 These ribosome interactors often have overlapping binding sites on the ribosome, close to the polypeptide exit site (Fig.2, B), suggesting that their functions are highly coordinated and temporally regulated. Together, these ribosome interactors ensure proper folding and biogenesis of nascent proteins as discussed below (refer to Ref 10 for a comprehensive review).
Table 1.-.
List of abbreviations
| Abbreviation | Definition |
|---|---|
| 3′-UTR | 3′-untranslated region |
| CoFe | Core Fermentation |
| cryo-EM | cryogenic electron microscopy |
| DiSP | Selective Disome Profiling |
| DN | Dominant-negative |
| ER | Endoplasmic Reticulum |
| ER | Endoplasmic Reticulum |
| GET | Guided-entry of tail-anchored proteins pathway |
| GFP | Green fluorescent Protein |
| Hsp70 | Heat Shock protein 70 |
| MAP | Methionine aminopeptidases |
| mRNA | messenger RNA |
| mRNPs | messenger ribonucleoprotein particles |
| MTS | N-terminal mitochondrial targeting sequence |
| NAC | Nascent polypeptide associated complex |
| NAT | N-acetyltransferase |
| NPC | Nuclear Pore Complexes |
| ORFs | Open Reading Frames |
| PFD | Prefoldin |
| PN | Proteostasis Network |
| RAC | Ribosome-Associated complex |
| RBPs | RNA Binding Proteins |
| RNA | Rubonucleic acid |
| RNC | Ribosome-Nascent Chain Complex |
| RQC | Ribosome Quality Control |
| RT-qPCR | Reverse transcription quantitative real-time PCR |
| SeRP | Selective Ribosome Profiling |
| SGs | Stress granules |
| SR | SRP receptor |
| SRP | Signal Recognition Particle |
| SRPR | SRP receptor |
| SS | N-terminal signal sequences |
| TA | tail-anchored |
| TCP-seq | Selective Translation Complex profiling |
| TF | Trigger factor |
| TMDs | Transmembrane domains |
| TRiC | T-complex protein ring complex |
| tRNA | Transfer RNA |
| UPS | Ubiquitin-Proteasome System |
Figure 2. Overview of factors involved in co-translational protein biogenesis.
A.- Overview of protein biogenesis pathways. Translation initiates on methionine. The nascent chains are directed to their fate by co-translational interactors. (arrow SRP) SRP recognizes signal sequences (Sig. Seq.) or transmembrane domains (TMD) emerging out of ribosome exit tunnel in cooperation with early acting NAC chaperone. SRP delivers the translating complex to to Sec61p on ER through interactions with SR. (arrow GET) Get3 (TRC40) captures tail-anchored proteins that have TMD at the C-terminus and delivers to Get1/2 on the ER. Other players in this pathway are not shown for simplicity. (arrow down) Initiator methionine is removed by MetAP and/or acetylated by NATs. Co-translational chaperones RAC, Ssb (Hsp70), and NAC protect and fold nascent protein elongating on the ribosome. As the nascent chain gets longer, it forms secondary structures and further recruits TRiC and/or Prefoldin to reach near-native structures. Translation terminates and nascent protein reaches its native structure. Targeting to Mitochondria can happen both post- or co-translationally B.- View of ribosome exit tunnel highlighting major binding sites of ribosome interactors. While this figure indicates major binding sites for each ribosome interactors, the actual region occupied by each interactor is much larger. Therefore, simultaneous ribosome binding of these interactors may not be possible even if major binding sites are not overlapping.
Table 2.-.
Chaperones and enzymes involved in protein biogenesis
| Protein | Function | References |
|---|---|---|
| Methionine Aminopeptidase (MAP) | Co-translational cleavage of N-terminal initiator methionine on nascent polypeptide | 10, 17 |
| N-terminal Acetylase (NAT) | Co-translational acetylation of N-terminus of nascent polypeptide | 10, 11 |
| Ribosome-associated complex (RAC) | Heterodimer of J-domain/Hsp70 [Zuo1/Ssz1(Yeast) MPP11/Hsp70L1(Mammals)]. Chaperones nascent polypeptide and recruits Ssb/Hsp70 to ribosomes | 10, 17, 28, 31 |
| Hsp70 (Ssb) | ATP-dependent co- and post-translational protein folding. Cooperates with J-domain proteins and NEFs | 10, 17, 26, 27, 30 |
| Nascent-polypeptide Associated Complex (NAC) | Enhances fidelity of protein localization. Protects nascent polypeptide from misfolding and aggregation. | 10, 17, 21, 22, 23 |
| Prefoldin (PFD) | Protects and relays nascent proteins to TRiC. Also called GIMc | 34, 35, 36, 37 |
| TCP-1 Ring Complex (TRiC) | Forms an enclosed chamber providing optimal folding environment for proteins that are difficult to fold in cytosolic environment. Also called CCT | 36, 37, 38 |
| Signal Recognition Particle (SRP) | Co-translational delivery of ribosomes translating ER/Secretory proteins to ER | 39, 40, 43 |
| Guided-entry of Tail-anchored proteins (GET) | Post-translational delivery of tail-anchored proteins to ER | 39, 42 |
3. N-terminal modification enzymes
While translation almost invariably starts at a methionine codon, the N-termini of the cellular proteome are diverse, indicating that N-terminal methionines are removed from proteins during biogenesis10. Roughly 70-90% of cytosolic proteins in eukaryotes have their initiator methionine removed by Methionine Aminopeptidases (MAPs). Another significant modification that affects ~80% of proteins is N-terminal acetylation, mediated by N-acetyltransferases (NATs)11. These enzymes bind ribosomes co-translationally as soon as nascent protein emerges out of ribosome exit tunnel12,13. N-terminal modification is central for protein-protein interactions and protein folding for many proteins11, and thus is an essential step in proper biogenesis and complex assembly. N-terminal modifications are also proposed to serve as degrons that help maintain protein levels and stoichiometries14 although recent studies found a poor correlation between N-acetylation and protein stability15.
4. Co-translational chaperone recruitment to nascent polypeptides
Nascent polypeptides emerging from the ribosome are highly vulnerable to misfolding and aggregation as they have not reached their stable native structure, have not found their interacting partners, and thus expose many potential interaction surfaces9,16. In eukaryotes, a diverse set of chaperones are recruited to translating ribosomes to protect nascent proteins from misfolding 10,17 and function cooperatively to facilitate nascent protein folding and assist co- or post-translational complex assembly8,18 (Fig. 2).
4.1. Nascent polypeptide associated complex (NAC)
NAC is an abundant heterodimeric chaperone composed of two related subunits called alpha- and beta-NAC19. It is present at nearly equal concentration to ribosomes and thus every ribosome is thought to be bound by NAC19. Although much evidence suggests the importance of NAC in protein biogenesis17, its precise function on the ribosome is poorly understood. Most research on NAC function has focused on events after nascent proteins emerge out of ribosome exit tunnel. However, a recent Cryogenic Electron Microscopy (cryo-EM) structure of NAC-bound to the 60S large ribosomal subunit unexpectedly showed the disordered N-terminus of the beta-NAC subunit inserts into the ribosome exit tunnel20. This suggests that NAC may interact with (at least some) nascent proteins while still in the tunnel, even before their exposure to the cytosol. The functional consequence of this early recognition is still unclear, but this finding raises obvious questions on how N-terminal modification enzymes or other chaperones access the nascent chain if they contact NAC at this early stage. In addition to chaperone activity, perhaps the best-studied function of NAC is its interplay with the Signal Recognition Particle (SRP), where it enhances the specificity of membrane targeting as discussed in detail below21-23. Notably, a recent proteomic study of NAC interactors in P. torridus identified some elongation factors (eEF1A and eIF5A)24. As translation elongation rate can profoundly affect co-translational folding and assembly (see Section 6 below), these results open the intriguing possibility that NAC function may also regulate co-translational maturation by affecting translation elongation rate.
4.2. Co-translational Hsp70 action: Ribosome-associated complex (RAC) and Hsp70 (Ssb)
Ribosome-associated complex (RAC) is another chaperone system that cooperates with ribosome-bound Hsp70 (Ssb in yeast, Table 2)10,17. RAC is evolutionarily conserved from yeast to human, and consists of a heterodimer formed by a J-domain containing protein, called Zuo1 in yeast, and an Hsp70 called Ssz1 in yeast (see Tabble 2) 10,17. In yeast, RAC was shown to function primarily to recruit Ssb/Hsp70 to the ribosome via its J-domain and modulate Hsp70 activity to fold nascent proteins25-27. A recent study shows a direct crosslink between nascent polypeptides and RAC, providing evidence for possible substrate relay mechanism from RAC to Hsp7028. Such a mechanism would better protect the nascent chain from misfolding by minimizing exposure to the cytosolic environment.
Selective Ribosome Profiling (SeRP, See Sidebar) methods29 enabled interrogation of the substrate pool of yeast Hsp70 Ssb, and precise timing of its engagement to translating ribosomes. Sevral recent studies consistently showed Ssb has an extensive substrate pool encompassing ~80% of cytosolic and nuclear proteins and a smaller fraction of membrane proteins26,27,30. Hsp70/Ssb generally recognizes linear stretches of hydrophobic residues flanked by positively charged motifs27,30 occuring in regions with high beta-sheet propensity30. Presumably, early recognition by Hsp70 minimizes competition with other chaperones and reduces exposure of aggregation-sensitive nascent polypeptide regions to the cytosolic environment. Importantly, these binding events ensure domain-wise folding and subsequent co-translational complex assembly. Despite much progress on defining co-translational Hsp70/Ssb-substrate interactions, the field has struggled to get any structural information on how Hsp70/Ssb binds to the ribosome. Partly explaining this difficulty, a recent crosslinking study uncovered the dynamic nature of Ssb-ribosome interaction, where it cycles between RAC and ribosome depending on the nucleotide state31. This result is consistent with a substrate relay model that couples ATPase cycle of Ssb to co-translational folding. RAC transfers nascent chains to Ssb, and also promotes ATP hydrolysis, stabilizing the substrate-bound ADP-state of Ssb/Hsp7026, 28 . The ATPase cycle of Hsp70/Ssb is completed by another chaperone, SSE/Hsp110, which acts to promote nucleotide exchange by Hsp70 after its dissociation from the ribosome, and thus promote release, and presumably folding, of the bound polypeptide 26,32.
Sidebar: SELECTIVE RIBOSOME PROFILING (SeRP).
Ribosomes function as integration hubs at an interface between translation and folding, coordinating the activity of chaperones, enzymes, and membrane targeting. These factors co-translationally interact with specific ribosome-nascent chain complexes (RNCs). The Selective Ribosome Profiling (SeRP) method (discussed here in 4.2, 4.3, and 7.2), is a powerful tool to capture the interaction of molecular chaperones with nascent chains and ribosomes along the process of translation elongation in vivo. In addition, it has been used to study the interaction of fully folded protein complex subunits with nascent interacting subunits. SeRP allows the identification and description of the mode of action of the co-translational interactions of nascent-chains at near-codon resolution by deep sequencing the ribosome-protected mRNA fragments from the ribosome subpopulation engaged by the factor of interest.
4.3. Co-translational chaperonin action: Prefoldin and T-complex protein ring complex (TRiC)
The hetero-oligomeric chaperones Prefoldin (PFD) and TRiC/CCT function both co- and post-translationally to promote protein folding (Table 2). Both TRiC and PFD interact with translating ribosomes33 but yt is not known whether PFD and TRiC have direct binding sites on the ribosome or whether they are recruited by upstream factors to bind nascent proteins emerging from ribosomes. The substrate spectrum of PFD has not been defined but it was shown to bind nascent actin and tubulin34,35. While PFD is thought to function co-translationally to relay nascent proteins to TRiC5,16,36,37, it may have a more general chaperone function independent of TRiC. Many studies show PFD has a general protective role preventing protein aggregation and suppressing toxicity of neurodegenerative disease-linked proteins (reviewed in 35). However, the extent to which PFD contributes to co-translational folding is unclear.
TRiC has a clear role in co-translational folding of a subset of approximately 10 % of the proteome, including many topologically complex proteins38. Recent work employing SeRP identified ~500 proteins that engage TRiC co-translationally in yeast30. These were predominantly cytosolic and nuclear proteins with complex folding intermediates, which are likely difficult to fold. TRiC engaged these substrates once nearly complete domains have emerged out of the ribosomal exit site; further comparison with Hsp70/Ssb cotranslational binding sites indicated that TRiC associates with nascent chains and functions after Hsp70 has bound, thus providing a bridge between co- and post-translational folding events30.
5. Targeting to membrane-bound organelles
While most proteins are translated in the cytosol, some nascent proteins need to localize correctly to membrane-bound organelles for their function. As discussed below, protein delivery to mitochondria is poorly understood but at least three pathways are known for nascent protein delivery to the ER (Table 2; reviewed in39): The Signal Recognition Particle or SRP pathway (reviewed in40), the Snd pathway41, and the Guided-entry of tail-anchored proteins or GET pathway (reviewed in42). In general, these pathways recognize different placements of hydrophobic targeting sequences along with features in the mRNA transcript. SRP recognizes hydrophobic sequences towards the N-terminus as well as transmembrane domains40, while the GET pathway recognizes hydrophobic sequences at the very C-terminus42. The Snd pathway tends to recognize hydrophobic sequences in the middle of the protein41, but the mechanistic details of the Snd pathway are still unclear. Together, these three pathways theoretically capture and correctly target most ER destined nascent proteins.
5.1. Signal Recognition Particle (SRP)
SRP is responsible for the co-translational delivery of ~30% of the proteome to the ER. It recognizes ribosomes translating N-terminal signal sequences (SS) or transmembrane domains (TMDs) and delivers them to the ER40,43. Recently, a subset of ribosome-nascent chain complexes was found to recruit SRP before the SS is exposed to the cytosol, suggesting an early “preloading” mechanism of SRP recognition acting in a pioneer round of translation to pre-select mRNAs destined to the ER44. Given that many nascent chain interacting factors bind near the ribosomal exit site, this is an attractive mechanism to bypass the competition with other factors and maintain specificity of ER targeting. Since the SRP receptor (SR) at the ER membrane depends on a ribosome-exposed SS45,46 for ribosome-nascent chain-SRP recruitment and subsequent conformational changes leading to translocation, the “preloading” mechanism subsequently recruits SRP receptors once the SS or TMDs emerged from the ribosome.
A prominent example of the interplay between ribosome bound nascent-chain associated factors can be found between SRP and NAC. NAC is not essential in yeast, but the absence of NAC is embryonically lethal in a number of metazoans, and its depletion induces an unfolded protein response in the ER22,47. This is attributed mainly to the mistargeting of cytosolic proteins to the ER by SRP, indicating that NAC modulates SRP specificity by preventing non-ER destined proteins from entering SRP pathway21,22. Due to partially overlapping binding sites of SRP and NAC on the ribosome, it has been hypothesized that NAC exerts this effect by preventing SRP binding to ribosomes translating non-ER destined proteins. Confirming this hypothesis, a recent study showed how NAC occupies the SRP binding site on most ribosomes to reduce SRP binding. However, the NAC-ribosome interaction is destabilized by emerging hydrophobic signal sequences to allow SRP access to ER-destined ribosomes. Furthermore, a domain of NAC interacts with SRP to promote SRP recruitment to the ribosome, emphasizing an active role of NAC during this process. These results highlight diverse and context-dependent roles of NAC and expands its role as an active ‘gatekeeper’ of translating ribosomes48. Adding additional layer of regulation, another recent study shows that NAC also prevents SRP from attaining a conformation conducive for SR recruitment for non-ER destined ribosomes, which results in reduced mis-targeting to ER23. Thus, NAC enhances specificity of the SRP pathway at multiple levels; enhancing cognate SRP-ribosome interactions and preventing non-cognate membrane targeting during SRP-SR interaction. These results parallel findings in bacteria between Trigger Factor (TF) and SRP, suggesting common principles acting to improve membrane targeting specificity across different kingdoms of life49. With so many factors crowding the ribosome exit site, additional layers of regulations through their interplay are likely yet to be uncovered.
5.2. Guided-entry of tail-anchored proteins (GET)
Tail-anchored (TA) proteins have their hydrophobic transmembrane domains at the very C-terminus. These proteins escape the SRP pathway as translation terminates before the transmembrane domains emerge from the ribosome, making it necessary for an alternative pathway. Guided-entry of tail-anchored protein (GET) pathway captures TA proteins post-translationally and delivers them to the ER42. Although the GET pathway mainly functions in a post-translational manner, recent studies show multiple GET pathway components binding to the ribosome50,51. This is somewhat expected as initial TA capture needs to be highly coordinated with translation to prevent transmembrane domain aggregation. Nonetheless, these results show that the GET pathway is not entirely post-translational and adds additional factors to the already-crowded ribosomal exit tunnel. The functions of the GET proteins likely extend to general protein homeostasis functions, such as preventing misfolding and ubiquitination51. Therefore, this pathway could, in theory, interact with other aggregation-prone proteins in the cytosol and play an important yet to be defined role in maintaining proteostasis.
5.3. Targeting to the mitochondria
Protein targeting to mitochondria faces several challenges, as some mitochondrial proteins must transverse one or both membranes inside the mitochondria, while other must assemble into large complexes containing soluble and membrane subunits synthesized in both the cytosol and the mitchondria52. While the mechanisms of mitochondrial protein targeting are not fully understood, there are examples of pre-, co-, and post-translational targeting. The evidence for pre-translational mitochondrial targeting of precursors comes from the observation that mRNAs associate with the cytoplasmic mitochondrial surface53. A subpopulation of mRNAs localizes to mitochondria through their 3′-untranslated region (3′-UTR, see Table 1 for abbreviations), which is recognized by specific RNA Binding Proteins (RBPs) at the Outer Mitochondrial Membrane (OMM)54. In yeast Puf3 is responsible for mitochondrial mRNA recruitment55, but the functionally equivalent RBPs in mammalian cells are not yet known56.
The presence of translating ribosomes on the surface of mitochondria is also indicative of a co-translational targeting model53,57. Co-translational targeting to the mitochondrial membrane is enhanced by longer ORFs and ribosome elongation slowdowns. These features may increase mitochondrial mRNA localization by extending the time-window available to nascent chains exposing an N-terminal mitochondrial targeting sequence (MTS) to associate with translocases on OMM58, thus bringing translating ribosomes to mitochondria59. NAC is proposed to direct Ribosome-Nascent Chain Complexes (RNC) to the vicinity of the mitochondria60, 61. By occupying rthe ibosome binding sites of SRP and Sec61 translocon, NAC may also prevent non-specific interactions between RNC and SRP or Sec61 enhancing mitochondrial targeting22.
Most of the yeast mitochondrial proteome is post-translationally targeted to mitochondria57. Many co- and post-translational acting chaperones, including cytosolic chaperones such as Hsp70, Hsp40 (a J-domain protein), and Hsp90 maintain the import-competent conformation of the newly translated mitochondrial precursors by preventing misfolding and aggregation. Chaperones such as Hsp70 and Hsp90 also ensure proteins reach import sites via specific interaction with the TOM receptors62. Although numerous cytosolic factors contribute to the successful targeting of mitochondrial precursor proteins to the organelle, we lack a detailed mechanistic understanding of their function. It is unclear whether and how many additional factors or pathways will be discovered in the future.
6. Role of translation elongation rate in co-translational proteostasis
Nascent polypeptides can begin to fold co-translationally63. The ribosomal exit tunnel constrains ~30-40 amino acids of the nascent polypeptide, most likely in a largely extended state, but secondary structures can form once the polypeptide reaches the vestibule near the ribosomal exit site64. Once the nascent polypeptide emerges out of the exit tunnel it can form folded intermediates and can interact with chaperones and targeting factors as described above. It has become increasingly clear that these co-translational proteostasis processes are tightly attuned to translation elongation kinetics. In general, as translation elongation rate (~4-6 amino acids per second) is much slower than formation of folded protein structures (milliseconds), elongation speed sets an upper limit for co-translational folding65. At the mRNA level, the speed of translation elongation is non-uniform across the mRNAs with local slowdowns and speedups present66. Local elongation slowdowns may serve to allow nascent proteins time to form specific folding intermediates and circumvent kinetic traps by preventing premature tertiary interactions65. Transient translation elongation slowdowns can also facilitate the recruitment of ribosome-associated factors required for folding and membrane targeting of the nascent chain (Figure 3).
Figure 3. Local slowdowns and speedups determine the fate of nascent proteins.
Local translation rates are controlled by several upstream determinants such as tRNA abundance (codon optimality), codon usage and context, mRNA secondary structure, and protein sequence. Local speedups during translation allows faster synthesis of protein domains that enhance translation fidelity and efficiency. On the other hand, local slowdowns are strategically incorporated at different positions along the transcript to facilitate various downstream functional consequences for co-translational proteostasis. For example, slowdowns after TMD allows more time for SRP to recognize and deliver translating ribosomes to the membrane. Similarly, slowdowns are coordinated with chaperone binding to enhance co-translational folding. Stronger slowdowns can cause stalls in translation that recruit ribosome-quality control machinery along with DEAD-box helicase Dhh1 to degrade nascent protein and defective mRNAs.
The precise mechanisms and factors regulating local elongation rate are not fully defined, but it is clear that codon usage plays an important role, incorporating factors such as tRNA abundance, tRNA selection, and modification. The balance between the codon and the cognate-tRNA causes variable decoding rates which influence local translation elongation rates67. Codon optimality can be understood as a scale that reflects the balance between the availability of the decoding tRNA and the demand of tRNA usage by translating ribosomes. Optimal codons are usually codons that can be efficiently decoded by abundant tRNAs. Conversely, nonoptimal codons tend to be rare codons decoded through wobble interactions or less abundant tRNAs68. As the same amino acid residues can be encoded by synonymous codons decoded at very different rates and efficiencies, the choice of codon in a sequence may significantly affect the fate of the nascent chain. Previously thought to be “silent”, synonymous codon usage within specific mRNAs regions appears to fine-tune co-translational protein folding and nascent chain interactions66,69.
The biophysical properties of nascent chains also influence translation elongation70,71. This is the case of polybasic stretches, which are proposed to slow decoding due to their interaction with the ribosomal tunnel72. Prolines, which have an atypical side-chain geometry that slows the peptide-transfer reaction, also cause ribosome slowdowns. Consecutive polyproline stretches can cause stalling during translation73, leading to destabilization of the peptidyl-tRNA in the P site and the nascent chain in ribosome exit tunnel72. Ribosomes stalled on polyproline stretches are recognized by eIF5A (EF-P in bacteria), an elongation factor that binds to the E-site of the ribosome to alleviate the ribosome stall and promote elongation73-75.
Recent studies show that local slowdown of translation elongation encoded by stretches of rare codons or sequences rich in positively charged amino acids or proline residues can facilitate binding of SRP to RNCs to enhance co-translational targeting of secretory proteins to the ER44,76,77. Similarly, mutations that impair tRNA modification also influence elongation rate by affecting decoding efficiency, causing codon usage–dependent changes in protein synthesis78,79. Recent studies indicate that codon choice is an important evolutionary constraint in the evolution of co-translational folding80. Indeed, patterns of codon optimality that correlate with specific secondary structure elements in the nascent chain are conserved across eukaryotic genomes, indicating evolutionary pressure to optimize the speed of protein elongation to facilitate binding of molecular chaperones and co-translational folding81.
While local slowdowns in elongation promote correct proteostasis, prolonged ribosome stalling is deleterious and triggers protein degradation and mRNA decay pathways82. Stalled translation elongation events can lead to ribosome collisions, which are recognized by Ribosome Quality Control (RQC) pathways to effectively targets the stalled mRNA and nascent chain for clearance. Several factors that recognize ribosome collisions have been identified, including E3 ubiquitin ligases that modify the stalled ribosomes. These then orchestrate a set of events that target mRNA for efficient no-go decay and nascent proteins for proteasomal degradation. Defective mRNAs are degraded via mRNA no-go decay83. The RQC pathway82,84, also senses ribosome-stalled polypeptides and engages Ubiquitin-Proteasome system (UPS) components for degradation of the translated nascent protein chain. Failure of the RQC machinery leads to protein aggregation, cellular toxicity, and ageing85-87.
An important unanswered question is how the determinants and frequency of functional ribosomal pausing are coordinated to prevent deleterious ribosomal stalling. Ribosome profiling approaches that sequence RNA footprints protected by RNAse-resistant ribosome pairs, presumably indicative of stalled ribosomes, are beginning to provide some insights into this question. For instance, a technique called Disome Profiling88-90 showed that collisions are widespread on endogenous mRNAs in yeast and in mouse liver cells 89,91. Such programmed ribosome queuing may capture functional elongation slowdowns, besides RQC activation. A recent study identified collisions near the SS peptide coding regions, which may arise from ribosome collisions that help co-translational targeting of RNCs to the ER. Additionally, these experiments identify pause sites disome peaks in mRNA regions correlating with structural features of the final protein or encoding unstructured segments of some proteins, further pointing to the potential involvement of collisions in modulating co-translational folding89.
7. From folding of monomers to assembly of oligomers in the cell
A significant percentage of the functional proteome corresponds to oligomeric complexes formed by either identical or different subunits – known as homomers and heteromers, respectively. Protein complexes need to strictly maintain their functional stoichiometry, an aspect thought to be regulated at the translational level as a core principle essential to protein homeostasis92. An emerging question to understand protein biogenesis is how the constraints and mechanisms described above facilitate protein complex assembly. Formation of oligomeric complexes usually follows ordered assembly pathways93.
In principle, oligomeric assembly could occur post-translationally by random encounter of (quasi) folded or chaperone-bound subunits94. Indeed, chaperones can maintain fully synthesized subunits in an assembly competent state for post-translational assembly95. Many of the substrates of chaperonin TRiC are subunits of oligomeric assemblies, biochemical evidence shows the fully made subunits are only released from TRiC in the presence of their oligomeric partners30,38. Additionally, depending on the geometry of the complex, the oligomeric partners could begin to associate co-translationally7. Based on current evidence, it appears that both mechanisms exist to mediate assembly of protein complexes and maintain the cellular proteome. Below we discuss different mechanisms of co-translational assembly presented in the literature (Figure 4).
Figure 4. Diagram depicting major mechanisms of co-translational complex assembly.
Co- post-translational assembly (upper) in bacterial (left) or eukaryotic (right) cells can mediate formation of both homomeric and heteromeric complexes. The Co-Post assembly mechanism involves the interaction of one fully synthetized and folded subunit, diffusing protein with the nascent, polysome-bound that are concurrently translated from the same (cis) or different mRNAs (trans).
Co- Co-translational assembly (lower) in bacterial and eukaryotic cells can mediate formation of both homomeric and heteromeric complexes. The Co-Co-translational assembly mechanism involves the direct interaction of two nascent subunits emerging from proximal ribosomes translating the same (cis) or different mRNAs (trans).
Both mechanisms of assembly are facilitated by operons in bacteria and by different cellular condensates in eukaryotes.
Post-translational assembly: Following translation, encounter of (quasi) folded subunits in the cellular milieu.
7.1. Translation-coupled protein complex assembly
Linking translation to assembly has a number of advantages: it can impede nonproductive interactions of nascent chains that might lead to aggregation96, and could orchestrate folding and subsequent assembly of individual subunits in proximity to chaperones to ensure the fulfillment of strict sequential assembly pathways96. Nevertheless, a co-translational mechanism in the context of the cell would require proximity between their respective RNCs. Initial studies demonstrated co-translational assembly for β-glucosidase in yeast97, and penicillinase in Bacillucereus98,which exhibited enzymatic activity as nascent chains. Among these lines, one of the first pieces of evidence that indirectly demonstrated translation-associated complex assembly was the co-migration on a sucrose gradient of the E. coli active form of the 16-mer homooligomeric enzyme β-galactosidase99. Additionally, Fulton et al. observed polyribosomes attached to the cytoskeleton suggesting some cytoskeletal proteins are assembled during translation100. Supporting co-translational assembly of myosin heavy chains, 80% of RNCs encoding nascent myosin heavy chains associated with the cytoskeleton in developing cultured skeletal muscle cells101. The concept of translation-associated assembly received its strongest support in a study of co-translational assembly of the σ1 reovirus cell attachment homotrimer, which demonstrated that trimerization of nascent σ1 chains occurs on vicinal nascent chains after the N-terminal trimerization domain emerged from the ribosome102.
7.2. Sequential Assembly of Protein Complexes or co-post translational
One reported mechanisms for the translation-dependent assembly of protein complexes involves one free fully translated subunit and at least one ribosome-associated nascent polypeptide: a strategy known as sequential96 or “co-post” translational assembly7,8. This mechanism might serve the formation of either homo and heteromers, and it could take place either in cis (i.e. polypeptides translated in the same polysome) or in trans (i.e. polypeptides translated in different polysomes), depending on the number of mRNA molecules being translated at any given time (Figure 4). In this mechanism a recently completed subunit, either folded or chaperone-bound) can interact with nascent chains being synthesized by downstream ribosomes within the same mRNA molecule or with ribosome-bound nascent polypeptides produced by another molecule of mRNA -encoding either the same or another subunit.
In eukaryotes, the co-post assembly mechanism for multi-subunit protein complexes was first demonstrated by Duncan and Mata, which showed that translating mRNAs for diverse proteins engage their fully made oligomeric partners proteins7. A “co-post” sequential mechanism for co-translational assembly for heteromeric protein complexes was subsequently also observed in prokaryotes103 for a bicistronic mRNA encoding heterodimeric LuxAB luciferase. The arrangement of the luciferase subunits LuxA and LuxB side by side into an operon enables their colocalized translation and assembly into an active enzyme complex. The initially translated and completed subunit LuxA remains protected by the bacterial trigger factor (TF) and engages nascent LuxB in vivo only when the N-terminal dimer interface of LuxB emerges from the ribosome103. Interestingly, the authors observed that the chance of productive encounter of LuxA with nascent LuxB for luciferase assembly diminished when subunits were translated from mRNAs produced by genes integrated distant with each other on the bacterial chromosome. Therefore, the subcellular proximity of encoding mRNAs of complex subunits is relevant and needed for assembly efficiency in the bacterial cell. In this case, the ordered arrangement of genes in a bacterial operon appears to have evolved to promote their co-localized synthesis and oligomeric association in the context of translation104. For eukaryotic cells lacking polycistronic mRNAs, the determinants of co-post translational assembly of protein complexes must be different, as operons are absent and the encoding genes localize into different loci in the genome103.
Insight into eukaryotic co- post- translational assembly was obtained using SeRP which showed that nine out of 12 well-characterized yeast heterocomplexes have a directional assembly mode. The three that did not were chaperone-dependent and in one case presumably bidirectional (i.e., phosphofructokinase)8. Moreover, when one of the subunits was deleted, C-terminally GFP tagged knock-in strains for the corresponding partner proteins formed cytoplasmic aggregates. A recent model proposes that elongating nascent chains expose their domains sequentially to interact with their already-synthesized partner proteins96. Oligomeric assembly is proposed to be regulated by molecular chaperones, electrostatic interactions between nascent chains and the ribosome surface105,106 and elongation rates107,108 . In particular, ribosome pausing and collisions are proposed to facilitate the encounter of nascent chains to produce heteromers in yeast109, and homomers in U2O2 and HEK-293T cells18. In these cases, these “functional” ribosome collisions appear to escape ribosomal quality control mechanisms. In principle, the subcellular availability of the interacting subunits around the translating ribosomes seems to be the most relevant requirement for co-translational assembly. Future studies will elucidate how the cell controls and integrates co-translational assembly in the crowded cellular milieu, depending on the strategy and mechanism employed to form any given multi-subunit complex machine. Current insights into these mechanisms are discussed below.
Further evidence has also shown a sequential co-post translational assembly mechanism for the assembly of specific hetero-oligomeric complexes in eukaryotes. For instance, using a modified version of SeRP named Selective Translation Complex profiling (TCP-seq), the authors could demonstrate a multidirectional mechanism of co- post-translational assembly of the translation initiation factor 3, eIF3110. In yeast, the SET1C/COMPASS histone methyltransferase - a hetero octameric complex - is assembled on nascent Set1 via a subcomplex named SET1RC (SET1 mRNA associated complex, containing SET1 mRNA, Set1, Swd1, Spp1, and Shg1)111. Set1p did not interact with SET1 mRNA when expressed from an independent transcript or forced to localize in the nucleus. Similarly, this interaction was lost under EDTA and puromycin treatment, indicating that SET1 mRNA binding by SET1RC was dependent on translation and likely by engaging nascent Set1 for assembly111.
More recently, the Tora lab has shown that the position of the heterodimerization domain in a protein (either N or C terminally located) guides the mechanism of translation-dependent assembly in mammalian nuclear multi-subunit complexes112. Among three different dimerization subcomplexes, TAF8-TAF10 and TAF1-TBP engaged sequential co-translational assembly. Using immunoprecipitation of N-terminally and C-terminally tagged versions of the subunits followed by RT-qPCR, the authors concluded that fully made TAF10 engages nascent TAF8 for assembly and that the translation, folding, and protein abundance of TAF8 is impaired in the absence of TAF10. The other subcomplex, TAF6-TAF9, engaged in simultaneous96 or “co-co” translational assembly18, a mechanism that involves association between more than one nascent chain113.
Similar conclusions were reached by a recent study of the biogenesis of Nuclear Pore Complexes (NPC) in yeast, where co-post assembly and co-co assembly were observed114. While nascent Nup1 and Nup2 are recognized by karyopherins and recruited to the NPC where they can complete translation in its proximity, other fully made subunits - like Nup82 can recruit nascent Nup159, Nup116, and Gle2 in a co-post assembly fashion114. Complementing this study, a second report showed some proteins of the NPC have a moonlighting function as part of multiple complexes. These proteins engage in co-post translational assembly in one but not in all possible pathways, suggesting that co-translational assembly is used to seed ordered pathways, avoiding potential harmful or non-functional outcomes115.
7.3. Simultaneous Assembly of Protein Complexes or co-co translational.
7.3.1. Co-translational assembly in polypeptides translated by two or more mRNAs
Co-translational protein assembly could also take place by interaction of more than one nascent subunit. This assembly mode could occur either in cis -if the nascent chains are produced from one discrete mRNA molecule in a polysome- or in trans when two or more different mRNAs are involved, encoding the same or different subunits18. While the existence of operons enables the simultaneous co-co assembly in bacteria of heteromers, the formation of hetero complexes in the eukaryotic cell would take place in trans whereby two different nascent chains should find each other in the cytoplasm. A growing body of evidence suggests that this may be a widespread mechanism of assembly109,112,116-118, however mechanistic details awaist further studies, particularly to distinguish nascent and mature proteins 117,119. Single-molecule FISH experiments showing significant proximity of mRNAs encoding interacting polypeptides in vivo support this model, as in the case of the TAF6 and TAF9 mRNAs encoding subunits for nuclear complexes112, the co-translational association of mRNAs encoding subunits of ion channels118, the glycolytic enzymes in the Core Fermentation (CoFe) granules116, some subunits of translation factors110,120, the proteasomal subunits RPT1 and RPT2109, and the previously discussed example of Nup1 and Nup2 being co-translated on the NPC114.
Evidence of co-localization of mRNAs encoding subunits of complexes, as well as the co-precipitation of fully made subunits with their corresponding partner mRNAs, cannot rule out a potential sequential mechanism of assembly. A recent study used a synthetic “complex” to demonstrate experimentally the formation of oligomeric complex assembly factories in yeast, in which the nascent chains of one of the subunits sufficed to co-translationally localize its encoding mRNA to the condensates117. This study indicates that formation of cellular condensates in the cell might bring together fully made and/or nascent proteins to facilitate protein complex formation locally, similarly to the recently suggested working model on RBPs for post and co-post translational assembly of protein complexes in the cell121.
7.3.2. Co-translational assembly comprising vicinal nascent chains in the polysome
Various studies have demonstrated the existence of a cis mechanism for assembly of homomers. Initial pulse-chase studies in U-138 M human glioma cells suggested a co-translational mechanism of assembly for the tenascin hexabrachion, an extracellular matrix homohexamer that modulates cell adhesion. The authors observed that full-length tenascin was always hexameric, and no monomers, dimers or trimers were observed during the chase. The absence of assembly intermediates suggested nascent chains could be associating prior to completion of translation, driven by the amino-terminal location of the assembly domain122. Similarly, in-vitro translation studies investigating the trimerization of the σ1 reovirus attachment protein demonstrated co-translational assembly of the triple N-terminal α-helix coiled coil helps the C-terminal trimeric globular head to assemble post-translationally102. A more recent study showed assembly of the large ribonucleoprotein vault particle follows a so-called “polyribosome templating” mode: the ribosome serves as a 3D nanoprinter that directs the ordered assembly of this homopolymer from the same polysome. In detail, two adjacent N-terminal domains dimerize during translation, and when the disome approaches termination, the dimers start stacking successively to fully form the complete particle. Interestingly, Electron microscopy (EM) experiments following in vitro translation showed direct proximity of the vault particles to polysomes. Moreover, when co-expressing different mutant variants, the authors detect particles containing uniquely one protein variant, further supporting a co-co translational mechanism of assembly123. Therefore, it is reasonable to think that a co-translational mechanism of assembly in cis might has evolved to preclude the deleterious effect of specific mutations.
A co-co mechanism of assembly could be relevant to genetic disorders with dominant-negative (DN) phenotypes, which usually affect genes that encode homomers124,125. p53 is a tetra-homomeric protein known as the “guardian of the genome” that induces the expression of genes involved in cell death/apoptosis, DNA surveillance pathways, and cell cycle arrest, as an attempt to counteract the damage imposed by different types of cellular stress126. Mutations of only one allele of p53 are sufficient to drive cancer progression suggesting a DN effect of p53 mutations inactivating WT p53 in the tetramer. In vitro translation studies report p53 dimerizes co-translationally from the same polysome, whereas tetramerization occurs post-translationally between two already-formed dimers. The simultaneous expression of wild type and mutant p53 caused the generation of heterotetrameric species of p53. These hetero-tetramers bound poorly to DNA, presumably due to the lack of dimer-dimer cooperativity; thus suggesting a mechanism underlying pathologies where p53 function is impaired127.
Another example of co-translational assembly potentially relevant to disease is the case of NF-κB, a major immunological modulator. The NF-κB transcription factor is a heterodimeric complex formed by p50 and RelA subunits harboring a transactivation domain. The NFKB1 gene encodes for NF-κB isoforms, p50 and the longer p105 protein (a RelA protein). Interestingly, NF-κB assembles in two steps: an initial co-co translational assembly in cis of vicinal N-terminal domains and later, a co-translational proteasomal-directed degradation of one of the subunits128,129. During translation, two adjacent N-terminal Rel homology domains (RHD) of ~300-residues dimerize co- co-translationally, allowing the degradation of only the downstream C-terminal fraction one subunit producing p50-p105 heterodimers129. This mechanism suggests p50 is under the control of p105 during biogenesis, precluding an early activation of immunological responses within the cell.
Selective Disome Profiling, DiSP18 was recently used to map at a codon level resolution the exact moment at which neighboring nascent polypeptides dimerized co-translationally yielding a proteome-wide picture of simultaneous co-co translational assembly in human cells. The authors suggest that ~32% of the total proteome engage in co-co translational assembly in cis, but did not observe association of nascent proteins encoded by different mRNAs. This study replicated previous findings for homocomplexes, i.e., NFκB129, but not for heterocomplexes like TAF6 -TAF9112 and the proteasome109. Recapitulation of co-translational dimerization of LMNA and DCTN1 in bacteriasuggest this process is intrinsic to the nascent chains, in contrast with findings suggesting cotranslational assembly of the proteasome and other complexes114 rely of specialized factors. Further studies are needed to clarify the mechanisms and factors governing co-co complex assembly.
8. Localized translation and Mechanisms of subcellular Localization of mRNAs
Localizing mRNAs to specific sites in the cell is proposed to ensure the spatially restricted production of proteins on site56,130. One of the best examples of localized translation occurs in yeast, where the ASH1 mRNA localizes to the distal bud tip to trigger the mating-type switching in the daughter cell131. The mechanisms that regulate localization and local translation of ASH1 are finely tuned. A complex called the “locasome” functions as an RNA-protein motor adaptor that assists localization132. Here, the She2p is an RNA binding protein that interacts with RNA elements, and to She3p, bridging ASH1 to the heavy myosin motor protein Myo4p located at the bud tip133. The regulated translation of ASH1 mRNA relies on cis-acting localization motifs acting as a ZIP code within the coding sequence; the association with Puf6p via UUGU elements in the 3’UTR of ASH1 and a translation-arresting complex formed by the protein Khd1 that interacts with an mRNA localization element (called E1) with the C-terminal domain of the translation initiation factor eIF4G1134. Upon arrival of ASH1 to the bud tip, Yck1p phosphorylates Khd1 disrupting the Khd1- eIF4G1 complex that initiates translation at the correct location134.
Similar examples of coordinated regulation of mRNA localization and translation have been reported in human cells, where mRNAs need to travel long distances to reach their final destination135. Single-molecule in situ hybridization (smFISH) experiments in human neurons have shown that both β-actin mRNA and ribosomes travel long distances to dendrites to provide a mechanism to control the space and time at which the mRNA is translated136. Similarly, β-actin mRNA localizes to the leading edge of human fibroblasts through binding of zip-code binding protein 1 (ZBP1) to the actin 3’UTR130. Similarly, all seven mRNAs encoding the actin-polymerization nucleator hetero complex Arp2/3 also localize to the leading edge of cells presumably to facilitate its assembly either co- or peri-translationally137.
Membrane-bound organelles -like the ER- are also hubs for localized translation of functionally related mRNAs to facilitate protein-complex assembly likely via RNC targeting to the ER 39,138. For instance, the tetrameric voltage-gated potassium channel (Kv) assemble in trans while the nascent interacting T1 domains are emerging from the ribosomes and have just translocated the membrane of the ER139. The ER membrane itself may provide a platform for RNC proximity or specific factors may facilitate co-localization of mRNAs. For instance, the kinase with no lysine 1 (WNK1) has been described as an assembly factor for the human ER membrane protein complex (EMC) acting to prevent non-specific interactions and cotranslational misfolding of the complex subunits140.
9. Compartment-specific protein complex assembly in eukaryotic cells
Since eukaryotic cells do not have operons, formation of messenger ribonucleoprotein particles (mRNPs) could orchestrate the localization of specific mRNAs, RBPs, and chaperones to facilitate protein production and assembly. These would be different from the well-defined RNA granules formed under stress, i.e., P-bodies and Stress granules (SGs) which are not translation-active (reviewed in 141) (Figure 5). In principle, the likelihood of two fully folded interacting subunits to find each other post-translationally depends on the distance between their encoding mRNAs and their degradation rates. Mathematical stochastic models show that the average rate of encounter would be slow, and therefore cellular co-localization of mRNAs could facilitate protein interactions and complex formation142. Nevertheless, experimental evidence has been scarce, and just few examples have started of emerge.
Figure 5. RNA granules and cytoplasmic condensates in the eukaryotic cell.
A. Following stress, mRNAs partition to P-bodies and stress granules where they are degraded or stored, respectively. Assemblysome: Following arsenite stress, mRNAs encoding proteasomal subunits Rpt1 and Rpt2 co-localize in granules enriched in Not1p/CNOT1 proving a protected environment. Under unstressed conditions we find: Transperons: The chromosomal rearrangement and concerted transcription of genes encoding functionally related mRNAs form cytoplasmic condensates. Translation factory: Translation competent cytoplasmic condensate that harbor homotypic or heterotypic mRNAs. TIS granule: The RBP TIS11B forms an intricated condensate intertwined with the ER that facilitates 3’UTR-mediated protein-protein interactions and protein complex formation. SRP preloading: SRP preloads specific ribosomes translating mRNAs before any SS or TMTs domain has emerged from the ribosome. Once in the ER membrane, a pioneering round of translation is complete, further rounds of translation initiation and elongation occur. B. RBPs serve as a hub for protein complex assembly. Homotypic interactions within RBPs form the condensate, and heterotypic interactions allow interactions of fully folded proteins that enrich locally in the condensate (Chen and Mayr, 2021) C. Models of co-co assembly of heteromers in the eukaryotic cell within condensates harboring mRNAs encoding protein complex subunits: A cytoplasmic condensate favor protein complex assembly peri-translationally from where fully assembled complexes are ejected into the cytoplasm. Alternately, mRNAs could find each other in the cytoplasm in a pioneer round via nascent chain interactions facilitating the assembly of the granule. Further rounds of translation would facilitate the synthesis of new protein complexes locally. D. Model of co-co assemble of homomers in the eukaryotic cell: independent and homotypic mRNA condensates facilitate the assembly of homomers. In the diagram, ENO1 and ENO2 independent yeast granules presumably would impede the formation of heteromers by spatially sequestration of mRNAs in independent granules (Morales-Polanco et al., 2021)
9.1. Translation factories
A recent dual protein-mRNA localization screen on 523 human cells lines expressing GFP-tagged proteins showed mRNAs sometimes have a specific localization in the cell that unlike yeast ASH1 mRNA, is determined co-translationally. This study observed that four mRNAs frequently formed cytoplasmic granules distinct from P-bodies and SGs, which they termed “translation factories” (Figure 5, A, unstressed cell). Their function on cotranslational protein folding and assembly was not evaluated143. Similar translation competent condensates have also been described in yeast, and proposed to serve as seeds of P-bodies following glucose starvation116,120,144.
Recently, SunTag method was used to visualize and quantify the location, kinetics, and timing of the translation of single mRNAs in living fruit fly embryos145. The authors observed a remarkable spatial heterogeneity, revealing the existence of mRNA containing translation factories. Similarly, mRNAs encoding glycolytic enzymes were shown to co-localize in cytoplasmic granules known as CoFe or “Core fermentation granules”, where they are actively translated116. These newly described translation factories represent a site of active “co-translation” of mRNAs where presumably chaperones and other translational regulators might interact. In principle, these translation factories may serve as a hub for mRNA translation, chaperone binding, and co- and post-translational assembly. Newly synthesized proteins would then diffuse to the cytoplasm, as supported by fluorescent recovery after photobleaching (FRAP) and microscopy experiments143,144 (Figure 5, A). However, the dynamics and functional relevance of these factories in protein complex assembly remain to be determined.
Finally, mRNAs have the inherent ability to form homotypic assemblies -independent of its sequence-, principally under Translation factories containing only one isoform encoding mRNA may be formed by molecular crowding conditions similar to what is observed for other RNA protein granules146, to favor homocomplex formation, presumably as in the case of ENO1- and ENO2-containing RNA glycolytic granules in yeast (Figure 5, D)116. Future studies should determine if RBPs, mRNA export, and splicing factors have evolved to coordinate the interdependent processes of transcription, splicing, mRNA maturation, post-transcriptional processing, translation, folding, and complex assembly147.
9.2. TIS granules, RBPs condensates and Assemblysomes
Another type of cellular condensates with a role in complex assembly is proposed to form by homotypic interactions of high valency RBPs. Here, the heterotypic interactions of these RBPs would recruit mRNAs and interactors to the condensate to favor protein complex assembly (Figure 5, B)121. Once assembly is fulfilled, the RBPs remain in the condensate due to their high valency, while the assembled protein complex leaves the condensate. Recently, a highly expressed RBP called TIS11B was shown to form condensates, named TIS granules, that interact closely with the ER, giving rise to a so-called TIGER domain- a separate compartment from the cytoplasm148. This compartment places mRNAs in TIS granules on the ER membrane in a 3’UTR dependent manner to amplify surface expression of protein complexes, including CD47 and PD-L1.
A phase-separated condensate named “assemblysome” is proposed to facilitate the co co-translational assembly in trans of proteasomal sub-units Rpt1 and Rpt2 in coordination with changes of ribosome speed following arsenite stress in yeast and human cells109 (Figure 5, stressed cell). Detailed fluorescent microscopy experiments in human cells show that both RPT1 and RPT2 mRNAs co-localize with CNOT1, and the authors propose that these granules might limit new rounds of translation initiation, avoiding the activation of ribosome collisions and therefore the activation of RQC, giving time for partner subunits to associate109. It is however unclear how these stuctures differ from translation factories present in unstressed cells114,149,150.
In sum, localized translation may be mediated by RBPs, by the nascent chains117 or by elements in the mRNA, such as the 3’UTR for SRP pre-recruitment to secretory RNCs in yeast44 (Figure 5, A). Hence, different features might contribute to ensure post- and co-translational protein complex assembly in the cell (Figure 5, C).
9.3. Transperons, chromosomal re-arrangement and eukaryotic operons
To further understand the mechanisms driving co-localization of mRNAs encoding oligomeric complex subunits, a recent study used RNA immunoprecipitation and sequencing in unstressed yeast to suggest the existence of “transperons” (Figure 5, A unstressed cell): discrete condensates that co-localize mRNAs encoding proteins with diverse physiological functions, i.e., heat shock proteins, mitochondrial outer membrane proteins, and mating type proteins151. Using chromatin capture and allele tagging, the authors revealed that the assembly of these condensates might result from co-regulated transcription and chromosomal re-arrangement mediated by histone H4. No evidence of transperon formation was detected for well-known co-post translational assembly candidates8, nor for mRNAs in translation factories like CoFe granules116.
In addition, recent experiments using single-cell RNAseq provide evidence that “transcription factories” gather regions of different chromosomes to coordinate transcription of mRNAs encoding subunits of protein complex152. RBPs could serve as to connect transcription, nuclear export, and coordinated translation in the cytoplasm153. For instance, yeast mRNA export factors Yra1p and Mex67p bind functionally related mRNAs by direct association with specific transcription factors (i.e., Abf1p TF binds Yra1p but not Mex67p). These mRNA export factors could coordinate the nucleocytoplasmic export of transcriptionally co-regulated classes of mRNAs, apparently via interactions with transcription complexes in yeast154 as well as mammalian cells and plants155.
10. Conclusions and future perspectives
Defining how the cell coordinates protein translation and biogenesis raises many questions regarding mechanisms directing folding and assembly of protein complexes, including the contribution of spatially localized translation, the role of chaperones and how translational kinetics regulates these processes156. The identification of condensates that co-localize mRNAs encoding protein-complex subunits partners represents an excellent opportunity to investigate the functional and cellular importance of this mechanism of protein complex assembly, and in particular if it facilitates the encounter of related mRNAs prior to the first round of assembly.
The role of ribosome elongation rate and stalling should also be clarified, including its role in promoting co-translational encounters between two nascent chains encoded by different mRNAs. Since ribosome slowdowns and collisions may be beneficial to drive co-translational assembly, translation within condensates might provide a protected environment where they can escape RQC. Surprisingly, a new quality control pathway named dimerization quality control (DQC) monitors and eliminates nonfunctional dimer complexes157. Clearly this is an exciting area of biology, which will yield many conceptual advances and that has many important implications for biotechnological and therapeutic potential.
Summary Points:
1.- As nascent proteins are translated, NAC may contact nascent chains before they emerge from the ribosome exit tunnel. Subsequent recruitments of ATP-dependent Hsp70 allows local folding, but some proteins require subsequent addition of TRiC for domain-wise folding to reach the native structure.
2.- Elongation rate is locally variable and regulated with slowdowns and speedups strategically encoded in the mRNA sequence to promote chaperone binding, membrane targeting, and complex assembly.
3.- An increasing body of evidence points to widespread translation-dependent mechanisms that coordinate assembly of protein complexes. Elongation rate and structural topologies of interacting subunits determine the mechanism of co-translational assembly. In “co- post-translational assembly a fully made subunit binds its nascent-protein partner while in co-co translational assembly, two nascent chains can dimerize simultaneously. Adjacent nascent chains on the same polysome might facilitate the assembly of homomeric proteins, while the interaction of nascent chains in trans would favor heteromeric protein complex formation.
4.- Newly described membrane-less organelles suggest cellular mechanisms for co-translational assembly in eukaryotic cells, that involve formation of translation-active condensates and/or “translation factories” to provide a platform for complex assembly.
Future Issues:
How are interactions of the plethora of ribosome-associated chaperones and nascent chain binding factors coordinated? Do chaperones modulate aspects of translation elongation favoring the folding of nascent proteins?
How specific are the mechanisms of co-translational assembly? For a given complex: are there unique or parallel models of assembly?
How pervasive is the co-localization of mRNAs encoding subunits of oligomeric complexes? What are the mechanisms directing their co-localization?
Are translation factories, assemblysomes and TIS granules the same type of condensates? How do their interactomes differ?
How does ribosome and polysome structure help orchestrate elongation, folding and assembly of protein complexes.
Acknowledgements:
The authors would like to apologize to scientists and colleagues whose work could not be cited due to space limitations. We thank Felipe G. Serrano (https://illustrative-science.com) for help with illustrations, and members of the Frydman lab for their fruitful comments on the text and figures.
Funding:
F.M.-P. was supported by The Pew Trusts in the Biomedical Sciences postdoctoral award 00034104, and J.H.L. was supported by NIH/NIA postdoctoral grant T32AG000266. Work in the Frydman lab is supported by National Institutes of Health grants GM56433 and AG054407.
Terms and Definitions:
- Domain Fold
Section of a polypeptide chain able to fold stably and independently from the entire protein
- cryo-EM
A technique used to study the macromolecular structure of proteins using cryogenic temperatures on samples on a liquid media.
- Signal Sequence/SS
A short peptide of 15-30 aminoacids present at the N-terminal of most proteins to be targeted to the secretory pathway.
- Transmembrane domain/TMD
A usually hydrophobic membrane-spanning protein domain
- Tail-anchored protein
membrane proteins thar are targeted to the ER by the GET and the (TMD)-recognition complex pathway
- UTR
Untranslated region of mRNAs either at the 5’ and the 3’.
- RNA binding Protein/RBP
Proteins with high valency that bind single and double stranded RNA molecules and form ribonucleoprotein complexes.
- OMM
Outer mitochondrial membrane that contains many porins that favor trafficking with the cellular cytoplasm.
- Ribosome nascent-chain complex/RNC
Refers to the group of biomolecules that accompany the ribosome and the newly synthesizing polypeptide during translation.
- Disome profiling
A variant of the ribosome profiling technique that maps the 60 nt footprints on mRNAs covered by two vicinal ribosomes during translation.
- Operon
A bacterial loci or DNA unit that harbors a group of functionally related genes under the control of a unique promotor. Transcription of operons usually derives in polycistronic mRNAs.
- qPCR
Quantitative Polymerase Chain Reaction, a technology used to measure amounts of a certain DNA molecule.
- Single Molecule FISH
A modern fluorescence in situ hybridization technique that uses a set of multiple probes tagging the whole body of a particular RNA facilitating the visualization of single molecules in cells.
- Dominant negative mutation
A type of mutation in which its expression of a mutant allele precludes the function of the native allele
- Locasome
The ASH1 mRNA transport machinery which comprises She2p and She3p for She1p/Myo4p myosin mediated cellular mobilization.
- Suntag Method
A fluorescent microscopy technique using epitopes on nascent chains (Suntag) which are tether by single chain variable fragments, allowing observation and monitoring of nascent proteins in vivo.
- DQC
Dimerization quality control. A newly described quality control pathway that surveillance and degrades nonfunctional dimer complexes.
- Homomer
protein complex formed by two or more identical subunits
- Heteromer
protein complex formed by nonidentical subunits
Footnotes
Declaration of interests: The authors declare no competing interests
References
- 1.Anfinsen CB Principles that govern the folding of protein chains. Science (80-. ). 181, 223–230 (1973). [DOI] [PubMed] [Google Scholar]
- 2.Netzer WJ & Hartl FU Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388, 343–349 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Klaips CL, Jayaraj GG & Hartl FU Pathways of cellular proteostasis in aging and disease. J Cell Biol 217, 51–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sontag EM, Samant RS & Frydman J Mechanisms and Functions of Spatial Protein Quality Control. Annu Rev Biochem 86, 97–122 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Balchin D, Hayer-Hartl M & Hartl FU In vivo aspects of protein folding and quality control. Science (80-. ). 353, (2016). [DOI] [PubMed] [Google Scholar]
- 6.Chiti F & Dobson CM Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu Rev Biochem 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 7. Duncan CDS & Mata J Widespread cotranslational formation of protein complexes. PLoS Genet. 7, e1002398 (2011). This is the very first publication showing that co-post translational assembly is widespread in eukaryotes
- 8. Shiber A. et al. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272 (2018). Here the authors used C-terminally GFP tagged subunits of pre-selected protein complexes to investigate co-post-translational assembly in yeast using seRP
- 9.Cassaignau AME, Cabrita LD & Christodoulou J How Does the Ribosome Fold the Proteome? Annu. Rev. Biochem 89, 389–415 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Kramer G, Shiber A & Bukau B Mechanisms of Cotranslational Maturation of Newly Synthesized Proteins. Annu. Rev. Biochem 88, 337–364 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Aksnes H, Ree R & Arnesen T Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases. Mol. Cell 73, 1097–1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knorr AG et al. Ribosome–NatA architecture reveals that rRNA expansion segments coordinate N-terminal acetylation. Nat. Struct. Mol. Biol 26, 35–39 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Yang C-I, Hsieh H-H & Shan S Timing and specificity of cotranslational nascent protein modification in bacteria. Proc. Natl. Acad. Sci 116, 23050–23060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shemorry A, Hwang C-S & Varshavsky A Control of Protein Quality and Stoichiometries by N-Terminal Acetylation and the N-End Rule Pathway. Mol. Cell 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedrich UA et al. N$α$-terminal acetylation of proteins by NatA and NatB serves distinct physiological roles in Saccharomyces cerevisiae. Cell Rep. 34, 108711 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Hartl FU, Bracher A & Hayer-Hartl M Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Deuerling E, Gamerdinger M & Kreft SG Chaperone Interactions at the Ribosome. Cold Spring Harb. Perspect. Biol 11, a033977 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bertolini M. et al. Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. Science (80-. ). 371, 57–64 (2021). Using a new technique named DiSP the authors show that some adjacent nascent proteins dimerize co-co- translationally to favor homomer assembly
- 19.Raue U, Oellerer S & Rospert S Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem 282, 7809–7816 (2007). [DOI] [PubMed] [Google Scholar]
- 20. Gamerdinger M. et al. Early Scanning of Nascent Polypeptides inside the Ribosomal Tunnel by NAC. Mol. Cell 75, 996–1006.e8 (2019). This paper provides the first structural information on how NAC interacts with the ribosome and describes an unexpected feature where NAC protrudes into the ribosome exit tunnel which imply early interaction between NAC and nascent chain
- 21.del Alamo M. et al. Defining the Specificity of Cotranslationally Acting Chaperones by Systematic Analysis of mRNAs Associated with Ribosome-Nascent Chain Complexes. PLoS Biol. 9, e1001100 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamerdinger M, Hanebuth MA, Frickey T & Deuerling E The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science (80-. ). 348, 201–207 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Hsieh H-H, Lee JH, Chandrasekar S & Shan S A ribosome-associated chaperone enables substrate triage in a cotranslational protein targeting complex. Nat. Commun 11, 5840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singhal N. et al. Biophysical and Biochemical Characterization of Nascent Polypeptide-Associated Complex of Picrophilus torridus and Elucidation of Its Interacting Partners. Front. Microbiol 11, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koplin A. et al. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol 189, 57–68 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willmund F. et al. The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Döring K. et al. Profiling Ssb-Nascent Chain Interactions Reveals Principles of Hsp70-Assisted Folding. Cell 170, 298–311.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y. et al. The ribosome-associated complex RAC serves in a relay that directs nascent chains to Ssb. Nat. Commun 11, 1504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galmozzi CV, Merker D, Friedrich UA, Döring K & Kramer G Selective ribosome profiling to study interactions of translating ribosomes in yeast. Nature Protocols vol. 14 (Springer; US, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stein KC, Kriel A & Frydman J Nascent polypeptide domain topology and elongation rate direct the cotranslational hierarchy of Hsp70 and TRiC/CCT. Mol. Cell 75, 1117–1130 (2019). Through SeRP, the authors show how Hsp70 and TRiC/CCT cooperatively work to fold nascent proteins on the ribosome.
- 31. Lee K. et al. Pathway of Hsp70 interactions at the ribosome. Nat. Commun. 12, 5666 (2021). This paper identified two modes of interactions between ribosome and Hsp70 that couples ATPase cycle of Hsp70 with ribosome interactions
- 32.Yam AY-W, Albanèse V, Lin H-TJ & Frydman J Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem 280, 41252–41261 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Albanèse V, Yam AYW, Baughman J, Parnot C & Frydman J Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124, 75–88 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Hansen WJ, Cowan NJ & Welch WJ Prefoldin–Nascent Chain Complexes in the Folding of Cytoskeletal Proteins. J. Cell Biol 145, 265–277 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahlan M, Zako T & Yohda M Prefoldin, a jellyfish-like molecular chaperone: functional cooperation with a group II chaperonin and beyond. Biophys. Rev 10, 339–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartl FU & Hayer-Hartl M Molecular Chaperones in the Cytosol: from Nascent Chain to Folded Protein. Science (80-. ). 295, 1852–1858 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Gestaut D. et al. The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis. Cell 177, 751–765.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yam AY et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol 15, 1255–1262 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegde RS & Keenan RJ The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol 1–18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X & Shan S Fidelity of Cotranslational Protein Targeting by the Signal Recognition Particle. Annu. Rev. Biophys 43, 381–408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aviram N. et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 540, 134–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan S. Guiding tail-anchored membrane proteins to the endoplasmic reticulum in a chaperone cascade. J. Biol. Chem 294, 16577–16586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akopian D, Shen K, Zhang X & Shan S Signal Recognition Particle: An Essential Protein-Targeting Machine. Annu. Rev. Biochem 82, 693–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chartron JW, Hunt KCL & Frydman J Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536, 224–228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JH et al. Sequential activation of human signal recognition particle by the ribosome and signal sequence drives efficient protein targeting. Proc. Natl. Acad. Sci 115, E5487–E5496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH et al. Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER. Sci. Adv 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murayama E. et al. NACA deficiency reveals the crucial role of somite-derived stromal cells in haematopoietic niche formation. Nat. Commun 6, 8375 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Jomaa A. et al. Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting. Science (80-. ). 375, 839–844 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ariosa A, Lee JH, Wang S, Saraogi I & Shan S Regulation by a chaperone improves substrate selectivity during cotranslational protein targeting. Proc. Natl. Acad. Sci 112, E3169–E3178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y. et al. Ribosome-bound Get4/5 facilitates the capture of tail-anchored proteins by Sgt2 in yeast. Nat. Commun 12, 782 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leznicki P & High S sgta associates with nascent membrane protein precursors. EMBO Rep. 21, e48835 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Couvillion MT, Soto IC, Shipkovenska G & Churchman LS Synchronized mitochondrial and cytosolic translation programs. Nature 533, 499–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kellems RE & Butow RA Cytoplasmic-type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J Biol Chem 247, 8043–8050 (1972). [PubMed] [Google Scholar]
- 54.Marc P. et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3, 159–164 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saint-Georges Y. et al. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One 3, e2293 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fazal FM et al. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell 178, 473–490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams CC, Jan CH & Weissman JS Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science (80-. ). 346, 748–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsuboi T. et al. Mitochondrial volume fraction and translation duration impact mitochondrial mRNA localization and protein synthesis. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eliyahu E. et al. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol. Cell. Biol 30, 284–294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiedmann B, Sakai H, Davis TA & Wiedmann M A protein complex required for signal-sequence-specific sorting and translocation. Nature 370, 434–440 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Lesnik C, Cohen Y, Atir-Lande A, Schuldiner M & Arava Y OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat. Commun 5, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young JC, Hoogenraad NJ & Hartl FU Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Cabrita LD, Dobson CM & Christodoulou J Protein folding on the ribosome. Curr Opin Struct Biol 20, 33–45 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Lu J & Deutsch C Folding zones inside the ribosomal exit tunnel. Nat Struct Mol Biol 12, 1123–1129 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Kaiser CM & Liu K Folding up and Moving on-Nascent Protein Folding on the Ribosome. J Mol Biol 430, 4580–4591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chaney JL & Clark PL Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys 44, 143–166 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Letzring DP, Dean KM & Grayhack EJ Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16, 2516–2528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanson G & Coller J Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol 19, 20–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai CJ et al. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383, 281–291 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brule CE & Grayhack EJ Synonymous codons: choose wisely for expression. Trends Genet. 33, 283–297 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seidelt B. et al. Structural insight into nascent polypeptide chain–mediated translational stalling. Science (80-. ). 326, 1412–1415 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huter P. et al. Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P. Mol Cell 68, 515–527.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Gutierrez E. et al. eif5A promotes translation of polyproline motifs. Mol. Cell (2013) doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Doerfel LK et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science (80-. ). 339, 85–88 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Schuller AP, Wu CCC, Dever TE, Buskirk AR & Green R eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 66, 194–205.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pechmann S & Frydman J Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20, 237–243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pechmann S, Chartron JW & Frydman J Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo. Nat Struct Mol Biol 21, 1100–1105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lyu X. et al. Adaptation of codon usage to tRNA I34 modification controls translation kinetics and proteome landscape. PLoS Genet 16, e1008836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rapino F. et al. Wobble tRNA modification and hydrophilic amino acid patterns dictate protein fate. Nat Commun 12, 2170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Geller R, Pechmann S, Acevedo A, Andino R & Frydman J Hsp90 shapes protein and RNA evolution to balance trade-offs between protein stability and aggregation. Nat. Commun 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pechmann S, Willmund F & Frydman J The ribosome as a hub for protein quality control. Mol. Cell 49, 411–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Joazeiro CAP Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu Rev Cell Dev Biol 33, 343–368 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Simms CL, Yan LL & Zaher HS Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol Cell 68, 361–373.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brandman O & Hegde RS Ribosome-associated protein quality control. Nat. Struct. Mol. Biol 23, 7–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yonashiro R. et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. Elife 5, e11794 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Izawa T, Park SH, Zhao L, Hartl FU & Neupert W Cytosolic Protein Vms1 Links Ribosome Quality Control to Mitochondrial and Cellular Homeostasis. Cell 171, 890–903.e18 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Stein KC, Morales-Polanco F, van der Lienden J, Rainbolt TK & Frydman J Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature 1–6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao T. et al. Disome-seq reveals widespread ribosome collisions that promote cotranslational protein folding. Genome Biol. 22, 1–35 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arpat AB et al. Transcriptome-wide sites of collided ribosomes reveal principles of translational pausing. Genome Res. 30, 985–999 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Han P. et al. Genome-wide survey of ribosome collision. Cell Rep. 31, 107610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Meydan S & Guydosh NR Disome and trisome profiling reveal genome-wide targets of ribosome quality control. Mol. Cell 79, 588–602 (2020). By profiling disomes and trisomes, the authors show widespread ribosome collisions on endogenous mRNAs on normal conditions, which may play functional roles on cotranslational protein folding without inducing ribosomal quality control
- 92.Taggart JC & Li G-W Production of protein-complex components is stoichiometric and lacks general feedback regulation in eukaryotes. Cell Syst. 7, 580–589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marsh JA et al. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell 153, 461–470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hemmingsen SM et al. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333, 330–334 (1988). [DOI] [PubMed] [Google Scholar]
- 95.Minton AP Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol 10, 34–39 (2000). [DOI] [PubMed] [Google Scholar]
- 96.Schwarz A & Beck M The benefits of cotranslational assembly: a structural perspective. Trends Cell Biol. 29, 791–803 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Kihara HK, Hu ASL & Halvorson HO The identification of a ribosomal-bound β-glucosidase. Proc. Natl. Acad. Sci. U. S. A 47, 489 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Duerksen JD & O’Connor ML The demonstration of ribosome-bound penicillinase in Bacilluscereus. Biochem. Biophys. Res. Commun 10, 34–39 (1963). [Google Scholar]
- 99.Zipser D. Studies on the ribosome-bound β-galactosidase of Escherichia coli. J. Mol. Biol 7, 739–751 (1963). [DOI] [PubMed] [Google Scholar]
- 100.Fulton AB, Wan KM & Penman S The spatial distribution of polyribosomes in 3T3 cells and the associated assembly of proteins into the skeletal framework. Cell 20, 849–857 (1980). [DOI] [PubMed] [Google Scholar]
- 101.Isaacs WB & Fulton AB Cotranslational assembly of myosin heavy chain in developing cultured skeletal muscle. Proc. Natl. Acad. Sci 84, 6174–6178 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gilmore R, Coffey MC, Leone G, McLure K & Lee PW Co-translational trimerization of the reovirus cell attachment protein. EMBO J. 15, 2651–2658 (1996). [PMC free article] [PubMed] [Google Scholar]
- 103.Shieh Y-W et al. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science (80-. ). 350, 678–680 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Wells JN, Bergendahl LT & Marsh JA Operon gene order is optimized for ordered protein complex assembly. Cell Rep. 14, 679–685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cassaignau AME et al. Interactions between nascent proteins and the ribosome surface inhibit co-translational folding. Nat. Chem 1–7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Marino J, Buholzer KJ, Zosel F, Nettels D & Schuler B Charge interactions can dominate coupled folding and binding on the ribosome. Biophys. J 115, 996–1006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang L, Paakkarinen V, van Wijk KJ & Aro E-M Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem 274, 16062–16067 (1999). [DOI] [PubMed] [Google Scholar]
- 108.Zhang L, Paakkarinen V, van Wijk KJ & Aro E-M Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12, 1769–1781 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Panasenko OO et al. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct. Mol. Biol 26, 110–120 (2019). Here the authors show that the proteasomal subunits Rpt1 anf Rpt2 assemble co-co-translationally in trans within cytosplasmic granules termed “assemblysomes”
- 110.Wagner S. et al. Selective translation complex profiling reveals staged initiation and co-translational assembly of initiation factor complexes. Mol. Cell 79, 546–560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Halbach A. et al. Cotranslational assembly of the yeast SET1C histone methyltransferase complex. EMBO J. 28, 2959–2970 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kamenova I. et al. Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun 10, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Umlauf D. et al. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 126, 2656–2667 (2013). [DOI] [PubMed] [Google Scholar]
- 114.Lautier O. et al. Co-translational assembly and localized translation of nucleoporins in nuclear pore complex biogenesis. Mol. Cell 81, 2417–2427 (2021). [DOI] [PubMed] [Google Scholar]
- 115.Seidel M. et al. Co-translational assembly counteracts promiscuous interactions. bioRxiv (2021). [Google Scholar]
- 116. Morales-Polanco F. et al. Core Fermentation (CoFe) granules focus coordinated glycolytic mRNA localization and translation to fuel glucose fermentation. IScience 24, 102069 (2021). The authors show that mRNAs encodying glycolitic enzymes can co-localize in translationally active RNA granules: translation factories for the effective production of highly expressed proteins.
- 117.Heidenreich M. et al. Designer protein assemblies with tunable phase diagrams in living cells. Nat. Chem. Biol 16, 939–945 (2020). [DOI] [PubMed] [Google Scholar]
- 118.Liu F, Jones DK, de Lange WJ & Robertson GA Cotranslational association of mRNA encoding subunits of heteromeric ion channels. Proc. Natl. Acad. Sci 113, 4859–4864 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao N. et al. A genetically encoded probe for imaging nascent and mature HA-tagged proteins in vivo. Nat. Commun 10, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pizzinga M. et al. Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol 218, 1564–1581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen X & Mayr C A working model for condensate RNA-binding proteins as matchmakers for protein complex assembly. RNA rna-078995 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Redick SD & Schwarzbauer JE Rapid intracellular assembly of tenascin hexabrachions suggests a novel cotranslational process. J. Cell Sci 108, 1761–1769 (1995). [DOI] [PubMed] [Google Scholar]
- 123.Mrazek J. et al. Polyribosomes are molecular 3D nanoprinters that orchestrate the assembly of vault particles. ACS Nano 8, 11552–11559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Veitia RA Exploring the molecular etiology of dominant-negative mutations. Plant Cell 19, 3843–3851 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Natan E, Wells JN, Teichmann SA & Marsh JA Regulation, evolution and consequences of cotranslational protein complex assembly. Curr. Opin. Struct. Biol 42, 90–97 (2017). [DOI] [PubMed] [Google Scholar]
- 126.Niazi S, Purohit M & Niazi JH Role of p53 circuitry in tumorigenesis: a brief review. Eur. J. Med. Chem 158, 7–24 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Nicholls CD, McLure KG, Shields MA & Lee PWK Biogenesis of p53 Involves Cotranslational Dimerization of Monomers and Posttranslational Dimerization of Dimers: implications on the dominant negative effect. J. Biol. Chem 277, 12937–12945 (2002). [DOI] [PubMed] [Google Scholar]
- 128.Lin L, DeMartino GN & Greene WC Cotranslational biogenesis of NF-κB p50 by the 26S proteasome. Cell 92, 819–828 (1998). [DOI] [PubMed] [Google Scholar]
- 129.Lin L, DeMartino GN & Greene WC Cotranslational dimerization of the Rel homology domain of NF-κB1 generates p50–p105 heterodimers and is required for effective p50 production. EMBO J. 19, 4712–4722 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Das S, Vera M, Gandin V, Singer RH & Tutucci E Intracellular mRNA transport and localized translation. Nat. Rev. Mol. Cell Biol 22, 483–504 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sil A & Herskowitz I Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell 84, 711–722 (1996). [DOI] [PubMed] [Google Scholar]
- 132.Böhl F, Kruse C, Frank A, Ferring D & Jansen R She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 19, 5514–5524 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chartrand P, Meng XH, Huttelmaier S, Donato D & Singer RH Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell 10, 1319–1330 (2002). [DOI] [PubMed] [Google Scholar]
- 134.Paquin N. et al. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26, 795–809 (2007). [DOI] [PubMed] [Google Scholar]
- 135.Yoon YJ et al. Glutamate-induced RNA localization and translation in neurons. Proc. Natl. Acad. Sci 113, E6877–E6886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Buxbaum AR, Wu B & Singer RH Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science (80-. ). 343, 419–422 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mingle LA et al. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci 118, 2425–2433 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shurtleff MJ et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife 7, e37018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu J, Robinson JM, Edwards D & Deutsch C T1– T1 interactions occur in ER membranes while nascent Kv peptides are still attached to ribosomes. Biochemistry 40, 10934–10946 (2001). [DOI] [PubMed] [Google Scholar]
- 140.Pleiner T. et al. WNK1 is an assembly factor for the human ER membrane protein complex. Mol. Cell (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ivanov P, Kedersha N & Anderson P Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol 11, a032813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Batada NN, Shepp LA & Siegmund DO Stochastic model of protein–protein interaction: Why signaling proteins need to be colocalized. Proc. Natl. Acad. Sci 101, 6445–6449 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chouaib R. et al. A dual protein-mrna localization screen reveals compartmentalized translation and widespread co-translational rna targeting. Dev. Cell 54, 773–791 (2020). [DOI] [PubMed] [Google Scholar]
- 144.Lui J. et al. Granules Harboring Translationally Active mRNAs Provide a Platform for P-Body Formation following Stress. Cell Rep. 9, 944–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dufourt J. et al. Imaging translation dynamics in live embryos reveals spatial heterogeneities. Science (80-. ). 372, 840–844 (2021). [DOI] [PubMed] [Google Scholar]
- 146.Trcek T. et al. Sequence-independent self-assembly of germ granule mRNAs into homotypic clusters. Mol. Cell 78, 941–950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hyjek-Składanowska M. et al. Core spliceosomal Sm proteins as constituents of cytoplasmic mRNPs in plants. Plant J. 103, 1155–1173 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Ma W & Mayr C A membraneless organelle associated with the endoplasmic reticulum enables 3′ UTR-mediated protein-protein interactions. Cell 175, 1492–1506 (2018). The authors show that the RBP TIS11B forms a condensate that facilitates protein complex assembly either post-translationally or co post-translationally.
- 149.Chang L, Shav-Tal Y, Trcek T, Singer RH & Goldman RD Assembling an intermediate filament network by dynamic cotranslation. J. Cell Biol 172, 747–758 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hampoelz B. et al. Nuclear pores assemble from nucleoporin condensates during oogenesis. Cell 179, 671–686 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Nair RR et al. Multiplexed mRNA assembly into ribonucleoprotein particles plays an operon-like role in the control of yeast cell physiology. Elife 10, e66050 (2021). This paper shows that functionally related mRNAs can arrange in condensates at the moment they are being transcribed.
- 152.Tarbier M. et al. Nuclear gene proximity and protein interactions shape transcript covariations in mammalian single cells. Nat. Commun 11, 1–12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Keene JD RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet 8, 533–543 (2007). [DOI] [PubMed] [Google Scholar]
- 154.Hieronymus H & Silver PA Genome-wide analysis of RNA–protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet 33, 155 (2003). [DOI] [PubMed] [Google Scholar]
- 155.Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA & Carmo-Fonseca M Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol. 7, 1–14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Juszkiewicz S & Hegde RS Quality control of orphaned proteins. Mol. Cell 71, 443–457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Herhaus L & Dikic I Dimerization quality control via ubiquitylation. Science (80-. ). 362, 151–152 (2018). [DOI] [PubMed] [Google Scholar]