Abstract
Individuals who are homozygous for the 32-bp deletion in the gene coding for the chemokine receptor and major human immunodeficiency virus type 1 (HIV-1) coreceptor CCR5 (CCR5 −/−) lack functional cell surface CCR5 molecules and are relatively resistant to HIV-1 infection. HIV-1 infection in CCR5 −/− individuals, although rare, has been increasingly documented. We now report that the viral quasispecies from one such individual throughout disease is homogenous, T cell line tropic, and phenotypically syncytium inducing (SI); exclusively uses CXCR4; and replicates well in CCR5 −/− primary T cells. The recently discovered coreceptors BOB and Bonzo are not used. Although early and persistent SI variants have been described in longitudinal studies, this is the first demonstration of exclusive and persistent CXCR4 usage. With the caveat that the earliest viruses available from this subject were from approximately 4 years following primary infection, these data suggest that HIV-1 infection can be mediated and persistently maintained by viruses which exclusively utilize CXCR4. The lack of evolution toward the available minor coreceptors in this subject underscores the dominant biological roles of the major coreceptors CCR5 and CXCR4. This and two similar subjects (R. Biti, R. Ffrench, J. Young, B. Bennetts, G. Stewart, and T. Liang, Nat. Med. 3:252–253, 1997; I. Theodoreu, L. Meyer, M. Magierowska, C. Katlama, and C. Rouzioux, Lancet 349:1219–1220, 1997) showed relatively rapid CD4+ T-cell declines despite average or low initial viral RNA load. Since viruses which use CXCR4 exclusively cannot infect macrophages, these data have implications for the relative infection of the T-cell compartment versus the macrophage compartment in vivo and for the development of CCR5-based therapeutics.
Cellular entry of human immunodeficiency virus type 1 (HIV-1) requires binding both to CD4 (14, 33, 40) and to one of the seven transmembrane G-protein-coupled chemokine receptors recently discovered to act as coreceptors (2, 6, 8, 11, 16, 19, 20, 24, 46). Viruses able to infect cultured T-cell lines (T tropic) are syncytium inducing (SI), are frequently found in late-stage HIV disease, and utilize the chemokine receptor CXCR4; macrophage-tropic (M-tropic) viruses are non-SI (NSI) in T-cell lines, are found throughout disease, and utilize CCR5 (2, 6, 8, 11, 16, 19, 20, 24, 46). Two other chemokine receptors, CCR2B and CCR3, function as minor HIV-1 entry coreceptors (11, 19, 48), with CCR3 likely playing a role in central nervous system HIV-1 infection (27). Recently, two seven-transmembrane receptors with extensive sequence homology to CCR5 and CXCR4—Bonzo (3, 17) and BOB (17, 22)—have been shown to mediate entry of simian immunodeficiency virus (SIV), as well as some M-tropic HIV-1 and HIV-2 strains. Another seven-transmembrane receptor, GPR1, mediates the entry of SIV but not HIV-1 (22). The CC chemokines RANTES, MIP-1α, and MIP-1β are natural ligands for CCR5 (49, 51), and the CXC chemokine stromal-cell-derived factor 1 (SDF-1) is the only known natural ligand for CXCR4 (8, 46, 49, 51). Ligand binding to both receptors is associated with G-protein-coupled signal transduction and leukocyte chemoattraction (8, 46, 49, 51), as well as partial viral-entry antagonism (2, 8, 11–13, 16, 20, 29, 46, 58). Viral entry and signal transduction are separable in vitro functions for CCR5 (4, 23, 26), but the two may be biologically related to viral pathogenesis in vivo.
Most viral isolates recovered during primary and early chronic infection are NSI regardless of the route of infection (56, 59). Evolution of coreceptor use from CCR5 to CXCR4 is coincident with viral phenotypic switch from NSI to SI and progression to AIDS in approximately half of all HIV-1-seropositive subjects (31, 32, 34, 43, 54). A 32-bp inactivating deletion in CCR5 (CCR5 Δ32) common to northern European populations (41) has been associated with delayed disease progression in heterozygotes (15, 18, 21, 28, 43, 50, 60) and especially in those harboring NSI virus (18, 43). Subjects homozygous for CCR5 Δ32 (CCR5 −/−) are at a sharply reduced risk for virus transmission (15, 21, 28, 38, 43, 52, 60). However, reports of HIV-1 infected CCR5 −/− individuals, by our group and others, demonstrate that this risk reduction is finite (5, 7, 47, 55). We now report the viral phenotype, replication kinetics, macrophage tropism, and coreceptor usage of viruses derived early and late in disease from an HIV-1-infected CCR5 −/− subject.
MATERIALS AND METHODS
Case history and specimen availability.
Details of the clinical history have been reported previously (47). Briefly, patient UNC116 was born in 1969 with severe hemophilia A and received over 500,000 U of non-heat-treated factor VIII concentrates between 1978 and 1984. He had no other HIV-1 transmission risk factors. UNC116 was enrolled in the Multicenter Hemophilia Cohort Study (MHCS) with estimated HIV-1 infection in January 1982 and serologic diagnosis in 1985. His time to a CD4+ T-cell count of less than 200 cells/μl (4.4 years) and progression to AIDS (13.7 years) were at the 7th and 42nd percentiles, respectively, for the MHCS. The serum HIV-1 RNA concentration of 3.7 log10 copies/ml in 1986 was median for the MHCS. We previously showed that multiple peripheral blood mononuclear cell (PBMC) DNA samples from UNC116 were CCR5 −/− (47). The subject’s parents were CCR5 −/− and CCR5 +/− with confirmation of lineage by microsatellite DNA typing. Serum and plasma samples were available throughout this subject’s course, but PBMC samples were available only at later time points.
HIV-1 molecular clones and isolates.
The following infectious molecular clones and viral isolates were obtained through the AIDS Research and Reference Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: pYU-2 from Beatrice Hahn and George Shaw; pNL4-3 from Malcolm Martin; HIV-1ADA-M from Howard Gendelman; HIV-1BaL from Suzanne Gartner, Mikulas Popovic and Robert Gallo; HIV-1SF162 from Jay Levy; and HIV-189.6 from Ronald Collman. HIV-1 and HIV-2 infectious molecular clones LAI and ROD, respectively, were obtained from Keith Peden. A molecular clone of JR-CSF was a gift of Irvin S. Y. Chen, and subtype E primary isolates CM235 and CM240 were collected by the Walter Reed Army Institute of Research. pNL4-3 is an infectious molecular clone of an SI, T-tropic virus (1). pYU-2 is an NSI, M-tropic infectious molecular clone (37) which uses the coreceptors CCR5 and CCR3 (11, 37). Both clones belong to genetic subtype B. HIV-1ADA-M, HIV-1BaL, HIV-1JR-CSF, HIV-1CM235, HIV-1CM240, and HIV-1SF162 are NSI viruses which exclusively use CCR5 (17; also, data not shown), while HIV-189.6 is an SI, dual-tropic strain which uses CCR5, CXCR4, CCR3, and CCR2B (19). Plasmids were propagated in Escherichia coli DH5α and purified for transfection and PCR experiments by using Qiagen columns (Qiagen, Inc., Chatsworth, Calif.).
Amplification and DNA sequencing of env V1 through V3 cDNA.
Separate 100-μl aliquots of serum drawn from the subject in December 1985 and June 1986 were mixed with 900 μl of RNAzol (Tel-Test, Inc., Friendswood, Tex.) prior to the addition of 10 μg of an E. coli rRNA carrier (Boehringer Mannheim, Indianapolis, Ind.) and RNA purification as recommended by the manufacturer. cDNA fragments representing HIV-1 env domains V1 through V3 were subsequently amplified, cloned, and sequenced as described previously (42). Control amplifications in the absence of template were uniformly negative.
Viral RNA was isolated from a second 100-μl aliquot of serum drawn in June 1986 by using a Qiagen viral RNA isolation kit. The RNA was eluted with 50 μl of H2O. Reverse transcriptase PCR (RT-PCR) was performed with primers V3L2 (5′-ACTCAACTGCTGTTAAATGG-3′) and V3R1 (5′-CACTTCTCCAATTGTCCCTCA-3′) to produce an approximately 650-bp product spanning V3 to V5. The reaction conditions were as follows. Ten-microliter RT reaction mixtures consisted of 5 μl of viral RNA, 1× PCR buffer (50 mM KCl, 10 mM Tris [pH 8.3]), 5 mM MgCl2, 1 mM each deoxynucleoside triphosphate (United States Biochemicals, Inc.), 7.5 pmol of primer V3R1, 10 U of RNase inhibitor (Perkin-Elmer), and 25 U of Moloney murine leukemia virus (MLV) RT (Perkin-Elmer). Reverse transcription was done at 42°C for 30 min, followed by 10 min at 99°C to inactivate the RT. Forty microliters of a PCR master mix (1× PCR buffer, 2.5 mM MgCl2, 7.5 pmol of primer V3L2, 1.25 U of Perkin-Elmer AmpliTaq polymerase) was then added to the RT reaction mixture. PCR was carried out in a Stratagene Gradient-40 Robocycler with a program of 96°C for 3 min, followed by 40 cycles of 96°C for 45 s, 52°C for 45 s, and 72°C for 1 min. The RT-PCR product was cloned into the pT7Blue vector (Novagen) and propagated in DH5α. Clones were screened by amplifying the 141-bp V3 fragment by colony PCR (36) with primers V3L4 and V3R5 and analyzing the colony PCR products by the V3 heteroduplex tracking assay (V3-HTA) as described elsewhere (45). The clones were sequenced with an ABI PRISM dye terminator kit (Perkin-Elmer). cDNA amplicons representing regions V1 through V3 and V3 through V5 were obtained in two separate laboratories. These sequences were both unique to the laboratories where they were generated and highly related to each other, mitigating the possibility of contamination.
V3-HTA.
RT-PCR of the V3 region was performed as described elsewhere (45) to generate a 141-bp product. The reaction conditions for this RT-PCR were the same as above, except that primers V3L4 and V3R5 were used. Cycling conditions were as described above with an annealing temperature of 51°C and an extension time of 45 s. The PCR product described above was analyzed by V3-HTA (45) to determine whether the plasma sample contained SI-like variants and whether it was homogeneous. Heteroduplexes were formed between the RT-PCR product and a 35S-labeled probe which contains a V3 sequence nearly identical to the subtype B consensus from the JR-FL molecular clone (35). The heteroduplexes were separated on a 12% polyacrylamide gel as described elsewhere (45).
Construction of isogenic recombinant provirus.
HXB2 proviral clones chimeric in the V3 region of env were generated from a cloned serum cDNA sequence as previously described (39) except that the V3 fragments were generated by PCR and not by overlapping oligonucleotide synthesis (Fig. 1a). HIV clones chimeric in domains V3 through V5 were generated as follows. A recombinant env subclone (pKMJ) was constructed for convenient replacement of a 560-bp BglII-BglII fragment spanning V3 to V5 in JR-FL. The pKMJ vector was constructed by subcloning the 3.1-kb EcoRI-XhoI fragment of the pNL4-3 proviral clone (1) into pET21a (Novagen), in which the BglII site had been destroyed by Klenow fill-in. The 2.9-kb SacI-XhoI fragment was then replaced with the same region from the pUC112-1 subclone of JR-FL. This subclone has only two BglII sites which flank V3 to V5, which allows direct exchange of V3 to V5 sequences. Two of the 650-bp clones (1-7 and 5-3) generated from the June 1986 sample were chosen for insertion into a recombinant infectious clone. The V3 to V5 sequences were reamplified from clones 1-7 and 5-3 with primers 5′BGL (5′-ATTAGATCTGAAAATTTCACGGAC-3′) and 3′BGL (5′-TCCTCCTCCAGGTCTGAAGATCTC-3′). Reaction mixtures contained 1× PCR buffer, 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 5 pmol of each primer, 1 ng of plasmid, and 2.5 U of Gibco BRL Taq polymerase, in a final volume of 50 μl. Cycling conditions were as described above. The products were ligated into the pT7Blue vector, and clones with inserts were sequenced by using the ABI PRISM dye terminator kit. The BglII insert from one clone of each sequence (1-7 and 5-3) was ligated into the pKMJ vector to replace the JR-FL V3 to V5 sequence. Control recombinant clones were constructed by inserting the BglII-BglII fragments from the pYU-2 and pNL4-3 molecular clones into pKMJ. The EcoRI-XhoI fragment from each subclone was then ligated into pNL4-3 to make infectious proviral clones (Fig. 1b). Recombinant molecular-clone DNA was prepared from E. coli STBL2 (Gibco BRL) by using a Qiagen Plasmid Maxi kit. The inserts of all isogenic recombinant proviral constructions were confirmed by DNA sequence analysis.
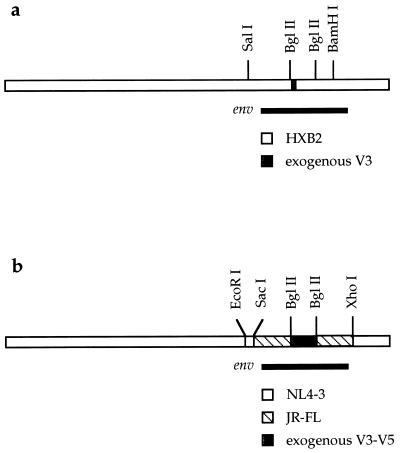
FIG. 1.
Construction of isogenic recombinant proviruses. (a) Exogenous V3 region env sequences were inserted into a portion of the HIV-1HXB2 env gene contained within the SalI-BamHI fragment and then inserted into the HIV-1HXB2 proviral background. (b) Exogenous sequences V3 through V5 contained by BglII sites were inserted into the HIV-1JR-FL background bounded by SacI and XhoI sites and then inserted into the HIV-1NL4-3 proviral background.
Preparation of viral stocks from molecular clones.
Twenty-microgram amounts of proviral clones were transfected into 1.5 × 107 COS-1 cells by electroporation (0.22 kV, 960 μF) and subsequently maintained in a humidified, 5% CO2 environment at 37°C for 48 h in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) (PAA Laboratories, Inc., Newport Beach, Calif.), 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine. Supernatants were saved, and 3 ml of fresh medium was added to each well for an additional 24 h. Supernatant fluids were then cleared by centrifugation at 300 × g, filtered through 0.22-μm syringe filters (Millipore, Bedford, Mass.), and monitored for p24 antigen production by HIV-1 core antigen enzyme-linked immunosorbent assay (Coulter, Hialeah, Fla.). Approximately 2 × 106 phytohemagglutinin (PHA)-stimulated seronegative donor PBMC were exposed to 1 ml of the p24 antigen-positive COS-1 cell supernatant. After overnight infection, cells were washed and maintained in culture medium (RPMI 1640 supplemented with 15% FCS, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 2 mM l-glutamine) containing 20 U of human recombinant interleukin 2 (IL-2). Virus replication was monitored by p24 antigen production, and culture supernatant harvested at peak p24 antigen production was used to measure the 50% tissue culture infective dose (TCID50). Stocks for HIV-1 isolates LAI and JR-CSF, HIV-2 isolate ROD, and MLV-pseudotyped HIV-1 were prepared as described elsewhere (17).
Twenty-microgram samples of proviral DNA of the June 1986 chimeric clones (1-7 and 5-3) and the YU-2 and NL4-3 control chimeric clones were used in calcium phosphate transfection of HeLa cells as previously described (10). Supernatants were harvested, and virus production was quantitated by RT assay (9). Virus stocks were produced by infection of uninfected PHA-stimulated PBMC.
Recovery of plasma virus.
Frozen plasma from June 1994 was thawed at 37°C, and 100 μl was added to triplicate wells of a 96-well microtiter plate. An aliquot of 5 × 104 PHA-stimulated PBMC was added to each well and incubated overnight. Cells were washed and maintained in IL-2-supplemented culture medium. One-half of the culture supernatant was harvested and replaced every 2 to 3 days for 20 days. Culture supernatant from one p24 antigen-positive well was used to prepare an expanded virus stock.
PBMC replication kinetics.
In triplicate wells of a 96-well microtiter plate, 1.5 × 105 PHA-stimulated PBMC per well were infected overnight with ∼100 TCID50 of each virus stock. Cells were washed and maintained in IL-2-supplemented culture medium. One-half of the culture supernatant was harvested and replaced every other day for 20 days. Culture supernatant from triplicate wells was pooled, and p24 antigen was measured in batch at the end of the time course.
Macrophage replication kinetics.
Macrophages were prepared from PBMC of HIV-1-seronegative donors by overnight adherence to plastic flasks precoated with heat-inactivated normal human serum. Nonadherent cells were removed by vigorous washing with culture medium, and macrophages were detached with cold Ca2+- and Mg2+-free phosphate-buffered saline (PBS) as described elsewhere (25). Macrophages were then seeded at 105 cells per well in flat-bottom 96-well microtiter plates and maintained in culture medium supplemented with 10% normal human serum for 7 days prior to infection. To evaluate virus growth kinetics, cells in triplicate wells were exposed overnight to ∼400 TCID50 of virus stock and washed thoroughly, and culture supernatant p24 antigen was monitored every 3 to 4 days for 30 days. M-tropic (ADA-M, SF162, and 89.6) and T-tropic (MN) viruses were propagated in PHA-stimulated PBMC for use as positive and negative macrophage infectivity controls, respectively.
MT-2 phenotyping assay.
The ability of the PBMC-derived viruses to induce syncytia in MT-2 cells was evaluated by using the AIDS Clinical Trials Group consensus protocol (30). MT-2 cells were exposed to 200 TCID50 of each virus in duplicate wells. Viruses were judged to be of the SI phenotype if three or more syncytia per low-magnification (100×) field were observed during a 21-day observation period.
Coreceptor usage in GHOST cells.
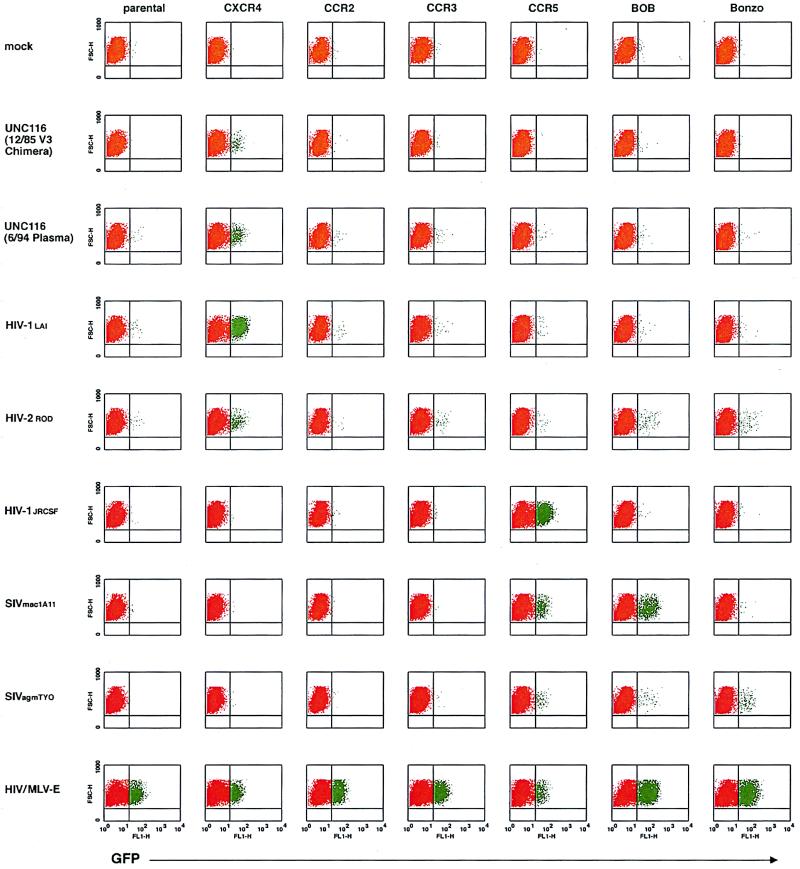
Virus coreceptor preferences were assayed by single-round infection using a novel indicator cell panel (32a). Human osteosarcoma (HOS) cells were stably transduced with human CD4 encoded on the MLV vector pMV7neo. HOS.T4 cells were next stably cotransfected with pCMV-hph and a reporter construct encoding humanized green fluorescent protein (GFP) under the control of the HIV-2 long terminal repeat (LTR). GFP-HOS.T4 (GHOST) clones were screened for their ability to induce GFP after transduction with the HIV-1 tat gene encoded on an MLV vector. One clone which demonstrated a high level of Tat-dependent activated transcription of GFP was chosen and subsequently stably transduced with the MLV vector pBABE-puro (44), encoding various chemokine receptor or HIV/SIV coreceptor genes. GHOST cells were maintained in DMEM supplemented with 10% FCS, 500 μg of G418 per ml, and 100 μg of hygromycin per ml. GHOST cells expressing coreceptor genes were additionally supplemented with 1 μg of puromycin per ml. In addition to the parental line which served as a background control, 10 other GHOST cell lines expressing human CCR1, CCR2B, CCR3, CCR4, CCR5, CXCR4, GPR1, BOB/GPR15, or Bonzo/STRL33 were used in analyses.
To determine virus coreceptor usage, 2 × 104 cells were plated per well of 12-well dishes the night prior to infection. The next day, equivalent amounts of virus stocks were applied in the presence of 10 μg of Polybrene per ml onto cells for 2 h, after which virus-containing medium was replaced with growth medium. Two days postinfection, cells were detached from plates with PBS–0.02% EDTA, fixed in 2% paraformaldehyde–PBS for 1 h, washed twice with PBS, and resuspended in PBS–2% FCS–0.2% sodium azide. Cells were then analyzed for humanized-GFP expression on a Becton Dickinson FACScan using CellQuest software. For each infection, the percentage of FL1-positive cells in mock-infected GHOST cells was subtracted from that of virally infected GHOST cells. The percentage of FL1-positive virally infected parental cells was then subtracted from that of all other GHOST cell infections to control for low-level, endogenous CXCR4 expression. Finally, these corrected percentages of FL1-positive cells were normalized for relative cell line GFP production reflected by parallel infections with amphotropic-MLV envelope (MLV-E)-pseudotyped HIV-1 (which enters GHOST cells independently of CD4 or coreceptor utilization) by multiplying them by the ratio of the mean GHOST MLV-E percent FL1 positives to coreceptor-specific MLV-E percent FL1 positives.
Coreceptor usage in HOS.CD4.coreceptor cells (p24 assay).
Virus coreceptor entry was evaluated by using HOS cells transduced with CD4 alone or in combination with CCR1, CCR2B, CCR3, CCR4, CCR5, or CXCR4 (generously provided by N. Landau, Aaron Diamond AIDS Research Center). HOS cells (maintained in DMEM supplemented with 10% FCS, 100 U of penicillin per ml, 100 μg of streptomycin per ml, 2 mM l-glutamine, and 1 μg of puromycin per ml) were seeded at 5 × 103 per well in flat-bottom 96-well microtiter plates. In triplicate wells, HOS cells were exposed overnight to 500 to 1,000 TCID50 of virus, and gently washed, and p24 antigen in the culture supernatant was measured on days 4 and 7. p24 production in HOS.CD4 wells was subtracted from that in HOS.CD4.coreceptor wells to account for low-level CXCR4 expression in these cells.
Nucleotide sequence accession numbers.
Nucleotide sequences V1 to V3 and V3 to V5 derived from patient UNC116 have been assigned GenBank accession no. AF034375 to AF034385.
RESULTS
HIV-1 V3 sequences from patient UNC116 are homogenous and are SI by both genotype and phenotype.
We previously showed that HIV-1 env sequences amplified from clinical samples obtained from patient UNC116 between 1985 and 1992 were of the SI genotype based on the presence of positively charged amino acids in the deduced amino acid sequence of the V3 loop region (47) and contained minimal sequence diversity (47; also, data not shown). The presence of the SI genotype and homogeneity were confirmed at the viral cDNA population level by the newly described V3-HTA (45) (Fig. 2). V3 sequences were amplified by RT-PCR from viral RNA isolated from a June 1986 serum sample from subject UNC116. Heteroduplexes were formed between these RT-PCR products and a radioactively labeled probe containing the near consensus V3 sequence for subtype B viruses. After resolution of the heteroduplexes, a single dominant band was seen on the autoradiograph, indicating limited V3 quasispecies diversity in the 1986 serum sample. The intermediate mobility of the heteroduplexes from UNC116, compared with the faster NSI control YU-2 and the slower SI control NL4-3 (whose envelope sequences are derived from HIV-1LAI [1]), indicated that the sequences had clustering of mutations that has been associated with the SI genotype (45). Nucleotide sequencing of molecular clones containing env cDNA sequences V1 to V3 and V3 to V5 from the December 1985 and June 1986 samples revealed a range of interclone divergence of 2.1 to 5.7%, which rose to only 7.5 to 9.9% by inclusion of cDNA sequences derived from a June 1994 plasma sample; all clones were of the SI genotype and were highly associated phylogenetically (data not shown; the sequences have been deposited in GenBank).
FIG. 2.
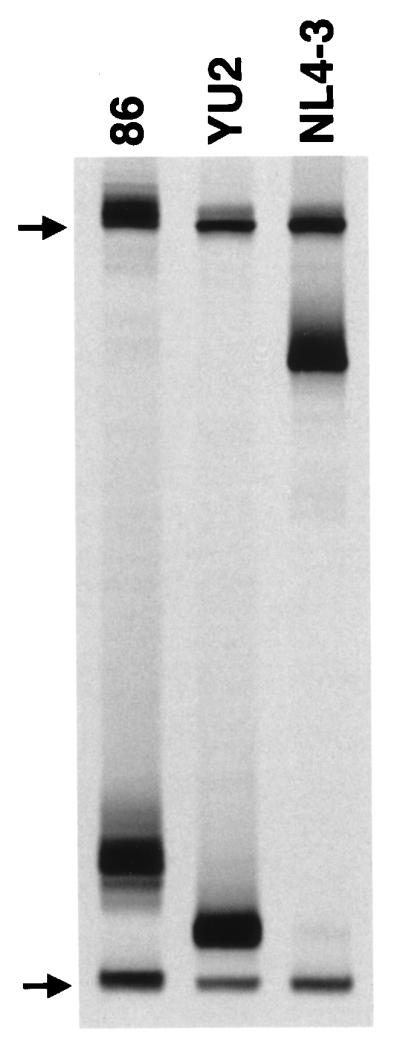
Analysis by V3-HTA. V3 loop RT-PCR products from the June 1986 serum sample from patient UNC116 (lane 86) and control YU-2 and NL4-3 recombinant proviral clones were annealed with the JR-FL V3 probe that was 35S labeled on one strand. The heteroduplexes were resolved on a native polyacrylamide gel and visualized by autoradiography. Only the bottom half of the autoradiograph is shown. The positions of the single-stranded probe (upper arrow) and double-stranded probe homoduplexes (lower arrow) are indicated. Heteroduplexes in each lane resolved between the single-stranded probe and probe homoduplexes.
The earliest available specimens for UNC116 consisted of small volumes of serum that had been stored under nonideal conditions for successful viral culture. Rather than attempt viral culture of this serum, we constructed isogenic recombinant proviruses containing exogenous sequences of either V3 or V3 through V5 (Fig. 1) derived from UNC116 env cDNAs as described in Materials and Methods. The predominant V3 sequence in the December 1985 serum sample was put into the HXB2 proviral clone to create the 12/85 chimera (Fig. 1a). The complete HXB2 proviral clone was used as a control for this recombinant. The 6/86 chimeras were made by putting two different V3 to V5 sequences (1-7 and 5-3) derived from the June 1986 serum sample into a JR-FL env gene within an NL4-3 proviral clone (Fig. 1b). Each of these V3 to V5 sequences had the same V3 sequence as that used in the 12/85 recombinant virus. Control recombinant viruses were constructed by placing the V3 to V5 sequences from the YU-2 (NSI control) and NL4-3 (SI control) proviral clones into the same NL4-3/JR-FL recombinant provirus. A viral isolate from late in disease was obtained from a June 1994 plasma specimen with high viral load.
The three early (12/85 and 6/86) viral chimeras, the late (6/94) plasma viral isolate, and the control viruses were tested for biological phenotype in the MT-2 cell assay. All of the viruses except the YU-2 recombinant control virus were phenotypically SI (Table 1 and data not shown).
TABLE 1.
Biological phenotype and coreceptor usage of experimental and control viral stocks
| Virus (phenotypea) | % GHOST cell infectionb
|
|||||
|---|---|---|---|---|---|---|
| CXCR4 | CCR2B | CCR3 | CCR5 | BOB | Bonzo | |
| 12/85 V3 chimerac (SI) | 100.0 | 0 | 0 | 0 | 0 | 0 |
| 6/94 plasma isolate (SI) | 100.0 | 0 | 0 | 0 | 0 | 0 |
| HIV-1LAI (SI) | 100.0 | 0 | 0 | 0 | 0 | 0 |
| HIV-2ROD (SI) | 85.5 | 0 | 2.7 | 0 | 6.8 | 5.0 |
| HIV-1JRCSF (NSI) | 0 | 0 | 0 | 100 | 0 | 0 |
| SIVmac1A11 | 0 | 0 | 0 | 71.2 | 28.8 | 0 |
| SIVagmTYO | 0 | 0 | 0 | 75.7 | 6.1 | 18.2 |
Determined by the ACTG consensus MT-2 cell assay.
Normalized to MLV-E-pseudotyped HIV-1 infection as described in Materials and Methods.
The 6/86 V3-V5 chimera stock was not tested in the GHOST cell assay. This SI stock was tested in HOS.CD4.coreceptor cells with p24 readout and found to exclusively use CXCR4 (data not shown).
The viral quasispecies displays exclusive and persistent CXCR4 coreceptor utilization.
Equivalent amounts of the viral stocks were used to infect GHOST cells to determine relative coreceptor utilization in a single-step infection assay. This HOS cell line is stably transduced with human CD4, GFP under the control of the Tat-sensitive HIV-2 promoter, and specific human coreceptor cDNAs. Infection of these GHOST cells by HIV-1 results in the production of GFP that can be detected by flow cytometry less than 48 h postinfection. Whereas mock infection of these cells results in virtually no signal above background (red in Fig. 3), infection with CD4/coreceptor-independent amphotropic-MLV-E-pseudotyped HIV-1 virions results in 5 to 32% of these cells becoming positive for GFP production (green in Fig. 3). Inspection of the fluorescence-activated cell sorter profiles in Fig. 3 and the relative coreceptor utilization in Table 1 for the 12/85 viral chimera and 6/94 plasma isolate show CXCR4 use to the exclusion of the other known entry coreceptors (CCR2B, CCR3, CCR5, BOB, and Bonzo). CCR1, CCR4, and GPR1 were also tested and were not used by these stocks (data not shown). The SI control virus HIV-1LAI exclusively used CXCR4, whereas the NSI control virus HIV-1JR-CSF exclusively used CCR5. HIV-2ROD used primarily CXCR4 but also CCR3, BOB, and Bonzo; SIVmac1A11 used CCR5 and BOB; and SIVagmTYO used CCR5, BOB, and Bonzo. Exclusive CXCR4 usage was confirmed for the 12/85 and 6/86 viral chimeras and 6/94 plasma isolate in a HOS.CD4.coreceptor assay using p24 antigen readout, in which CXCR4, CCR1, CCR2B, CCR3, CCR4, and CCR5 were tested (data not shown). Lower viral titers for the 6/86 viral chimeras compared with the 12/85 chimeras or 6/94 plasma isolate obviated coreceptor typing with GHOST cells, whose readout is 2 days. HOS .CD4 .coreceptor typing with p24 antigen readout at 7 days afforded greater sensitivity for viruses with delayed replication kinetics.
FIG. 3.
HIV-1 entry coreceptor typing of viral stocks in GHOST cells. HOS cells nonpermissive for HIV-1 infections were transduced with human CD4 and the GFP under the control of the HIV-2 LTR to generate parental GHOST cells. These cells were then individually transduced with the HIV-1 entry coreceptor cDNAs indicated at the top. HIV-1 entry and subsequent tat gene expression transactivates the HIV-2 LTR and upregulates the expression of GFP. Following infection of parental and coreceptor-transduced cells with equivalent amounts of specific viral stocks (indicated at the left), the cells were fixed and analyzed by flow cytometry. For each fluorescence-activated cell sorter profile, linear forward side scatter is plotted against log fluorescence intensity for GFP in the FL1 window. Basal-level production of GFP is red; transactivated GFP expression is green.
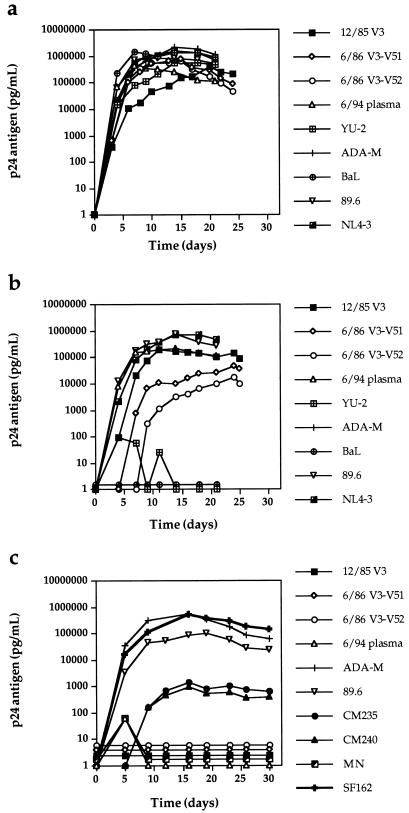
The viral quasispecies replicates in CCR5 +/+ and CCR5 −/− PBMC but not in macrophages.
The early viral chimeras and late plasma isolate from UNC116 displayed typical replication kinetics in CCR5 +/+ PBMC compared with virus derived from the M-tropic molecular clone pYU-2, virus derived from the T-tropic molecular clone pNL4-3, the dual-tropic isolate 89.6, and the NSI isolates ADA-M and BaL (Fig. 4a). The 12/85 chimera and the 89.6 and NL4-3 viruses also replicated well in seronegative CCR5 −/− PBMC, while YU-2, ADA-M, and BaL did not (Fig. 4b). The replication of the 6/86 V3-V5 viral chimeras was delayed in these cells. This observation could reflect stochastic selection of relatively less fit members of the quasispecies or the reduced activity of a chimeric protein or involve subtle host cell differences between the CCR5 +/+ and −/− PBMC.
FIG. 4.
Replication kinetics of viral stocks in CCR5 +/+ (a) and CCR5 −/− (b) PBMC and CCR5 +/+ macrophages (c). One hundred and 500 TCID50 of PBMC-derived viral stocks were used to infect PHA- and IL-2-stimulated PBMC and monocyte-derived macrophages, respectively. Virus replication was monitored by supernatant p24 antigen production.
To rule out the use of as yet undiscovered coreceptors on primary macrophages, the replication kinetics of the UNC116 and control viruses were studied in CCR5 +/+ primary macrophages (Fig. 4c). All of the UNC116 viruses and the SI control HIV-1MN failed to replicate in macrophages, but the NSI primary isolates CM235, CM240, SF162, and ADA-M, as well as the dual-tropic 89.6, replicated to various degrees in primary macrophages. Thus, if the UNC116 viruses are capable of using as yet undiscovered HIV-1 entry coreceptors, these putative receptors are not expressed in primary macrophages under culture conditions.
DISCUSSION
Given the importance of CCR5 coreceptor utilization in transmission and early HIV-1 disease, the virology of HIV-1-infected CCR5 −/− individuals provides an important insight into HIV-1 pathogenesis. We now report that the viral quasispecies from one such individual is persistently homogenous, SI, and T tropic and exclusively uses the HIV-1 entry coreceptor CXCR4 during the observation period. Although early and persistent SI variants (32, 34) or evolution to exclusive CXCR4 usage (53) has been previously found in longitudinal cohorts, UNC116 represents the first demonstration of exclusive and persistent CXCR4 usage in an HIV-1-infected individual, albeit with the important caveat that the earliest viral sampling in this study occurred approximately 4 years postinfection.
As most other primary viruses reported to date can use CCR5 for cell entry, we speculate that patient UNC116 was initially exposed to a swarm containing viruses capable of using both CCR5 and CXCR4 but only viruses with CXCR4 entry capacity were propagated in the context of the host environment. The quasispecies did not expand to use “minor” coreceptors such as CCR2B, CCR3, BOB, Bonzo, or GPR1, a finding consistent with the dominant roles of CCR5 and CXCR4 as HIV-1 entry coreceptors. As virus from UNC116 was first sampled roughly 4 years postinfection, it is not clear when selection for exclusive CXCR4 use occurred, but our data suggest that virus with CXCR4-restricted coreceptor utilization was present during most, if not all, of the period of chronic HIV infection. As these observations are based on a single patient, it will be critical to learn if they apply to other CCR5 −/− seropositive individuals. Within this context, it may be notable that virus from a laboratory worker initially infected with the CXCR4-exclusive, T-tropic strain HIV-1IIIB was subsequently recovered in both autologous macrophage culture and donor PBMC. The PBMC-derived virus was capable of passage into the human T-cell line H9 (57), indicative of the utilization of both CCR5 and CXCR4. Together, these reports suggest that HIV-1 quasispecies may evolve to fit the CCR5 environment of the host.
UNC116 V3 sequences embedded in the env background of the exclusive CXCR4-using virus HIV-1HXB2 and sequences V3 to V5 embedded in the env background of the exclusive CCR5-using virus HIV-1JR-FL resulted in viral chimeras restricted to CXCR4. Although we cannot formally exclude the possibility that sequences outside the V3 region contributed to coreceptor selectivity, this finding confirms previous data showing the strong influence of the V3 domain upon CCR5 and CXCR4 usage (13).
SI variant, CXCR4-using HIV-1 quasispecies are associated with increased in vivo virulence in cohort studies in which the presence of CCR5 −/− infected individuals is either absent or undocumented but is presumably very rare (31, 32, 34, 43, 54). The increased pathogenicity of such strains appears to be independent of the viral load (31). In agreement with those observations, UNC116 and two similar patients with estimable dates of infection (7, 55) showed relatively rapid CD4+ T-cell decline despite average to low early viral RNA load. Because case reports of HIV-1-infected patients with the CCR5 −/− genotype are few and potentially biased toward those with a better clinical course, available data are insufficient to predict the prognosis of such persons. However, these reports provide no evidence that, once infected with HIV-1, CCR5 −/− patients have a better prognosis than other persons.
We speculate that HIV-1 infection in CCR5 −/− individuals involves the persistent and dominant (if not exclusive) use of the coreceptor CXCR4 with cell-free infection largely confined to the T-cell (not macrophage) compartment. Infection of macrophages in such individuals may occur through cell-cell fusion events, although this could limit the ability of macrophages to serve as a reservoir of viral replication. Limited data suggest that HIV-1 quasispecies may evolve in response to the host’s CCR5 environment and that HIV-1-infected CCR5 −/− patients have no better prognosis than other infected persons. These points should be carefully considered in the design of therapeutic modalities such as blockade of CCR5 by small molecules or bone marrow transplantation with CCR5 −/− stem cells.
ACKNOWLEDGMENTS
We thank Deborah Birx, Suzanne Gartner, James Goedert, Mika Popovic, Merlin Robb, and Steven Wolinsky for helpful discussions. We also thank Irvin Chen for providing the HIV-1JR-FL subclone pUC112-1 and Malcolm Martin for providing the pNL4-3 clone.
This work was supported in part by National Cancer Institute contract NO1-CP-85649 with Research Triangle Institute, U.S. Army Medical Research and Materiel Command contract DAMD17-94-J-4430, and cooperative agreement DAMD17-93-V-3004 between the U.S. Army Medical Research and Materiel Command and the Henry M. Jackson Foundation for the Advancement of Military Medicine. J.A.E.N. was supported by postdoctoral training grant T32-CA-09156 from the National Cancer Institute. V.N.K. was supported by a postdoctoral fellowship from the Damon Runyon Walter Winchell Foundation. D.R.L. is supported by grants from the NIH and is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Liao F, Berger E A, Farber J M, Peden K W C. A new SIV co-receptor, STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 4.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 5.Balotta C, Bagnarelli P, Violin M, Ridolfo A L, Zhou D, Berlusconi A, Corvasce S, Corbellino M, Clementi M, Clerici M, Moroni M, Galli M. Homozygous Δ32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS. 1997;11:F67–F71. doi: 10.1097/00002030-199710000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Berson J F, Long D, Doranz G J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biti R, Ffrench R, Young J, Bennetts B, Stewart G, Liang T. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat Med. 1997;3:252–253. doi: 10.1038/nm0397-252. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Buckheit R W, Swanstrom R. Characterization of an HIV-1 isolate displaying an apparent absence of virion-associated reverse transcriptase activity. AIDS Res Hum Retroviruses. 1991;7:295–302. doi: 10.1089/aid.1991.7.295. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 12.Cocchi F, De Vico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, De Vico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 14.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J Hemophilia Growth and Development Study; Multicenter AIDS Cohort Study; Multicenter Hemophilia Cohort Study; San Francisco City Cohort; ALIVE Study. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 16.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 17.Deng H, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 18.de Roda Husman A-M, Koot M, Cornelissen M, Keet I P M, Brouwer M, Broersen S, Bakker M, Roos M T L, Prins M, de Wolf F, Coutinho R A, Miedema F, Goudsmit J, Schuitemaker H. The association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127:882–890. doi: 10.7326/0003-4819-127-10-199711150-00004. [DOI] [PubMed] [Google Scholar]
- 19.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 20.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 21.Eugen-Olsen J, Iversen A K N, Garred P, Koppelhus U, Pedersen C, Benfield T L, Sorensen A M, Katzenstein T, Dickmeiss E, Gerstoft J, Skinhoj P, Svejgaard A, Nielsen J O, Hoffman B. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS. 1997;11:305–310. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- 22.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farzan M, Choe H, Martin K A, Sun Y, Sidelko M, Mackay C R, Gerard N P, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y, Broder C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 25.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 26.Gosling J, Monteclaro F S, Atchinson R E, Arai H, Tsou C-L, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 29.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Sensitivity to inhibition by beta-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Japour A J, Fiscus S A, Arduino J-M, Mayers D L, Reichelderfer P S, Kuritzkes D R. Standardized microtiter assay for determination of syncytium-inducing phenotypes of clinical human immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurriaans S, Van Gemen B, Weverling G J, Van Strijp D, Nara P, Coutinho R, Koot M, Schuitemaker H, Goudsmit J. The natural history of HIV-1 infection: virus load and virus phenotype independent determinants of clinical course? Virology. 1994;204:223–233. doi: 10.1006/viro.1994.1526. [DOI] [PubMed] [Google Scholar]
- 32.Karlsson A, Parsmyr K, Aperia K, Sandstrom E, Fenyo E M, Albert J. MT-2 cell tropism of human immunodeficiency virus type 1 isolates as a marker for response to treatment and development of drug resistance. J Infect Dis. 1994;170:1367–1375. doi: 10.1093/infdis/170.6.1367. [DOI] [PubMed] [Google Scholar]
- 32a.Kewal Ramani, V. N., B. Volsky, and D. R. Littman. Unpublished data.
- 33.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 34.Koot M, Keet I P M, Vos A H V, de Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 35.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 36.Lee A B, Cooper T A. Improved direct PCR screen for bacterial colonies: wooden toothpicks inhibit PCR amplification. BioTechniques. 1995;18:225–226. [PubMed] [Google Scholar]
- 37.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 39.Lusso P, Cocchi F, Balotta C, Markham P D, Louie A, Farci P, Pal R, Gallo R C, Reitz M S., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 41.Martinson J J, Chapman N H, Rees D C, Liu Y-T, Clegg J B. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet. 1997;16:100–103. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- 42.Michael N L, Chang G, Ehrenberg P K, Vahey M T, Redfield R R. HIV-1 proviral genotypes from the peripheral blood mononuclear cells of an infected patient are differentially represented in expressed sequences. J Acquired Immune Defic Syndr. 1993;6:1073–1085. [PubMed] [Google Scholar]
- 43.Michael N L, Chang G, Louie L G, Mascola J R, Dondero D, Birx D L, Sheppard H W. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 44.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legier D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTER/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 47.O’Brien T R, Winkler C, Dean M, Nelson J A, Carrington M, Michael N L, White G C., II HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet. 1997;349:1219. doi: 10.1016/s0140-6736(97)24017-1. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 48.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H-H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raport C J, Gosling J, Schweickart V L, Gray P W, Charo I F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1β, and MIP-1α. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 50.Rappaport J, Cho Y Y, Hendel H, Schwartz E J, Schachter F, Zagury J F. 32 bp CCR-5 gene deletion and resistance to fast progression in HIV-1 infected heterozygotes. Lancet. 1997;349:922–923. doi: 10.1016/S0140-6736(05)62697-9. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 51.Sampson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 52.Sampson M, Libert F, Doranz B L, Rucker J, Liesnard C, Farber C-M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, George M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 53.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnatt M S, Plebani A, Siccardi A G, Littman D L, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 54.Sheppard H W, Lang W, Ascher M S, Vittinghoff E, Winkelstein W. The characterization of non-progressors: long-term HIV-1 infection with stable CD4+ T-cell levels. AIDS. 1993;7:1159–1166. [PubMed] [Google Scholar]
- 55.Theodorou I, Meyer L, Magierowska M, Katlama C, Rouzioux C. HIV-1 infection in an individual homozygous for CCR5 delta 32: Seroco study group. Lancet. 1997;349:1219–1220. . (Letter.) [PubMed] [Google Scholar]
- 56.van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss S H, Goedert J J, Gartner S, Popovic M, Waters D, Markham P, di Marzo Veronese F, Gail M H, Barkley W E, Gibbons J, Gill F A, Leuther M, Verpmese F M, Shaw G M, Gallo R C, Blattner W A. Risk of human immunodeficiency virus (HIV-1) infection among laboratory workers. Science. 1988;239:68–71. doi: 10.1126/science.3336776. [DOI] [PubMed] [Google Scholar]
- 58.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR levels and expression pattern correlate with infectibility by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 60.Zimmerman P A, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P E, Kumaraswami V, Giorgi J V, Detels R, Hunter J, Chopek M, Berger E A, Fauci A S, Nutman T B, Murphy P M. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]