Abstract
The development of complex eukaryotic organisms can be viewed as the selective expression of distinct fractions of the genome in different organs or tissue types in response to developmental and environmental cues. Here, we generated a genome expression atlas of 18 organ or tissue types representing the life cycle of Arabidopsis (Arabidopsis thaliana). We showed that each organ or tissue type had a defining genome expression pattern and that the degree to which organs share expression profiles is highly correlated with the biological relationship of organ types. Further, distinct fractions of the genome exhibited expression changes in response to environmental light among the three seedling organs, despite the fact that they share the same photoperception and transduction systems. A significant fraction of the genes in the Arabidopsis genome is organized into chromatin domains exhibiting coregulated expression patterns in response to developmental or environmental signals. The knowledge of organ-specific expression patterns and their response to the changing environment provides a foundation for dissecting the molecular processes underlying development.
All complex eukaryotic organisms, including mammals and higher plants, consist of multiple organ and tissue types. The organ and tissue types for a given organism are generated during its life cycle through a temporally and spatially regulated process of selective expression of specific fractions of the same genome in different cells (Meyerowitz, 2002). Therefore, a long-sought objective of developmental biology has been to define the subset of genes expressed and their relative abundance for each organ or tissue type. Higher plants possess a relatively simple developmental process, with only three nonreproductive organ systems and fewer than 25 major tissue and cell types (Eeau, 1977), thereby providing a good model for defining the organ- and tissue-specific genome expression patterns during development. Early studies using an RNA-excess/single-copy DNA hybridization strategy have revealed much about the mRNA complexity in six selected organ types of a tetraploid tobacco (Nicotiana tabacum) strain (Goldberg, 1988; Goldberg and Barker, 1989). These studies suggested that about 24,000 to 27,000 average-sized mRNA species were present in leaf, root, stem, petal, anther, and ovary (Goldberg et al., 1978; Kamalay and Goldberg, 1980). However, it provided little information toward the identities and relative abundance of the individual mRNA species expressed in each organ type. Other approaches to determining the gene expression pattern include RNA blot and in situ hybridization. These later approaches can give detailed information regarding when and where the detected gene is expressed. However, due to the labor intensive nature of these approaches, they have been applied to only a small number of genes (Barker et al., 1988; Yanofsky et al., 1990).
DNA microarrays can measure the individual transcript level of tens of thousands of genes simultaneously, thus providing a high throughput means to analyze gene expression levels on a larger scale (Schena et al., 1995; Chu et al., 1998). For instance, Zhu et al. (2001) used a partial-genome array to study the gene expression profiles from six organs and identified some interesting features. Wellmer et al. (2004) analyzed the gene expression profiles of inflorescences from wild type and several floral-homeotic mutants using a whole-genome oligo array and identified genes specifically or predominantly expressed in one type of floral organ within flowers. The complete sequence of the Arabidopsis (Arabidopsis thaliana) genome provides the means to design a microarray with essentially all known and predicted genes in the genome, which can be used to assay the expression of all the genes at once. For those genes that have been defined solely by prediction, a whole-genome expression analysis will provide a confirmation of expression as well. In recent microarray analyses, the expression profiles of the Arabidopsis genome from seedling, flower, root, and cultured cells were compared, which provided confirmation for many predicted genes, as well as led to discovery of new genes (Birnbaum et al., 2003; Yamada et al., 2003).
In this study, a 70-mer oligo microarray that covers 25,676 unique known and predicted genes of Arabidopsis (Fig. 1A) was used to profile their expression level from 18 representative organ or tissue types throughout the life cycle of Arabidopsis. In addition, we also examined the light-responsive expression of the Arabidopsis genome among the three organs of seedlings. Several interesting insights on the regulation of genome expression during Arabidopsis development and its response to light were observed. In the process, it was observed that a significant fraction of adjacent genes are organized into chromatin domains with similar expression patterns.
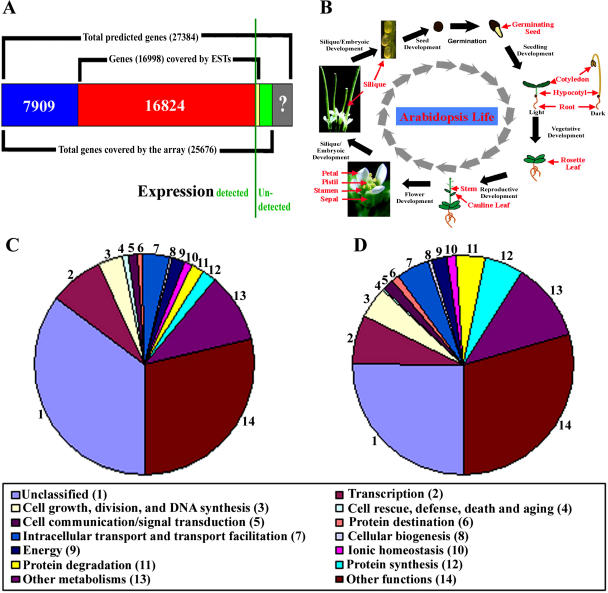
Figure 1.
Experimental expression analysis of known and predicted genes in the Arabidopsis genome. A, Summary of EST/mRNA-supported and purely predicted genes in the Arabidopsis genome (as of March 20, 2003) and its coverage by the 70-mer oligo microarray used in this study. The relative proportions of the genes either expression confirmed or not in our analysis are separated by a green vertical line. B, Diagram of organ sample collection during the Arabidopsis life cycle. C and D, GO categories of genes expressed in at least one organ or in cultured cells (C) and in all collected organs and cultured cells (D).
RESULTS
Analysis of Arabidopsis Representative Organ Transcriptomes Supports Expression of Most Known and Predicted Genes
During the life cycle of Arabidopsis, vegetative (root, stem, and leaf) and reproductive (petal, sepal, stamen, pistil, silique, and seed) organs are formed, and individual organs are specialized to carry out specific biological functions. We selected samples from 17 representative organs throughout the life cycle of Arabidopsis (Fig. 1B). We also included Arabidopsis suspension cultured cells as a common control, which allowed us to estimate the relative expression abundance for each transcript in different organs. We refer to this relative abundance of individual genes in each organ as the gene's expression level.
We first estimated the number of known and predicted genes for which expression can be detected experimentally. Among the known and predicted genes covered by the array, the expression for 24,733 (96%) out of 25,676 can be detected in at least one of the 17 organs or cultured cells under our experimental conditions (Fig. 1A). To assess our results, we examined the expression of those genes that have an available expressed sequence tag (EST) match. There are 16,998 unique annotated genes represented in this microarray that have available EST matches, and 16,824 (99%) of them showed detectable expression in at least one of 17 organs or cultured cells (Fig. 1A). This result indicates that our detection of gene expression is adequately sensitive and that the vast majority of known and predicted genes are expressed during Arabidopsis development. Our results further showed that the majority (7,909 genes, or 89%) of the computer-predicted genes (a total of 8,852 genes represented by this microarray) that lacked prior confirmation are expressed during Arabidopsis development, which validates the notion that they correspond to real genes.
Annotation of genes using gene ontology (GO) functional categories assigns functions to genes with a dynamic and controlled vocabulary (Gene Ontology Consortium, 2000). We functionally classified all expressed genes using GOslim terms from The Arabidopsis Information Resource (TAIR) annotation (Rhee et al., 2003). The functional classifications for genes that were expressed in either one or more organs, and in all organs are shown in Figure 1, C and D, respectively. In total, 4,070 (16%) out of the 24,733 expressed genes were expressed in all 17 organs and culture cells. This group of commonly expressed genes is likely to be essential for fundamental cellular processes and may be regarded as housekeeping genes in Arabidopsis. Indeed, GO-based functional classification indicates that this group of commonly expressed housekeeping genes were enriched for genes encoding proteins involved in essential cellular processes such as protein synthesis, protein degradation, protein destination, energy metabolism, cell growth, cell division, and DNA synthesis (Fig. 1, C and D).
Different Proportions of the Genome Are Expressed in Representative Organs
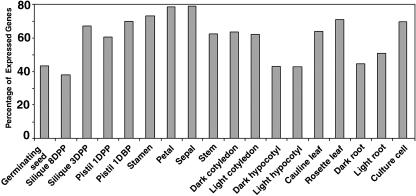
Examination of the fractions of the genome expressed in each organ type revealed that the percentage of expressed genes varies from organ to organ. Over 70% of the total genes examined were expressed in stamen, petal, rosette leaf, and sepal, while about 40% of the total genes were expressed in root, hypocotyl, germinating seed, and late-stage silique (Fig. 2). These numbers are consistent with previous results from a tetraploid tobacco (Nicotiana tabacum), in which leaf and petal had the highest expressed mRNA species among the six organs examined (Goldberg et al., 1978; Kamalay and Goldberg, 1980). However, the higher absolute number of expressed mRNA species in their estimation is possibly due to the tetraploid nature of the tobacco strain they used. It is interesting to note that approximately 70% of the total genes were expressed in cultured cells (Fig. 2), placing them among the organ or tissue types with the highest percentage of expressed genes.
Figure 2.
The percentage of the genome expressed in different organs and cultured cells. DBP, Days before pollination.
Relatedness of Genome Expression Correlates with Developmental Relationship of Organs in Arabidopsis
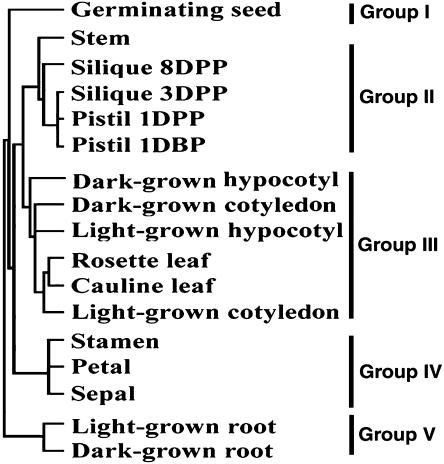
We next used the overall genome expression profiles of individual organs relative to cultured cells to examine the relatedness of the genome expression changes across the selected organs based on the average linkage clustering with correlation distance (Eisen et al., 1998). As shown in Figure 3, it is apparent that different leaf types (cotyledon, cauline leaf, and rosette leaf) have similar genome expression profiles and fall into one general clade. Similarly, pistil and silique had similar genome expression profiles, which were more closely related to that of stem. The three outer-whorl organs of flower (stamen, petal, and sepal) had similar expression patterns and formed a clade that was significantly diverged from the leaf clade, although those floral organs evolved from leaves. Both light- and dark-grown roots showed similar genome expression profiles that were significantly diverged from those of the other organ systems. On the other hand, germinating seed had a unique genome expression profile and formed a branch of its own. These results firmly indicate that developmentally related organs have similar genome expression profiles that distinguish them from those in other organs.
Figure 3.
Relatedness of the genome expression patterns across selected organs and distribution of organ-specific expressed genes. An average-linkage clustering with correlation distance analysis of overall relatedness for expression ratios from selected organ versus cultured-cell pairs.
Organ-Enriched Expression of the Arabidopsis Genome
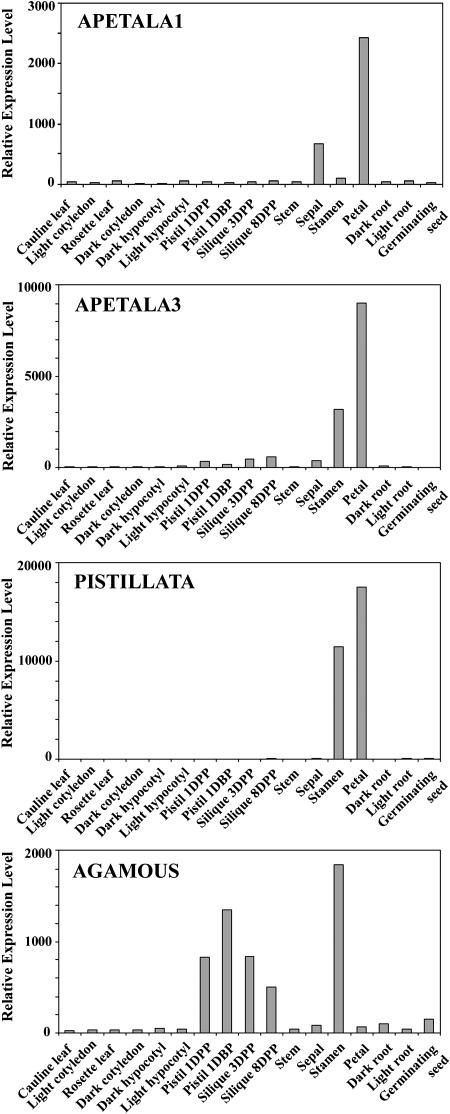
We designated genes as organ enriched if they fulfilled the two criteria: (1) showing differential expression (P < 0.05) based on the F test in a group of samples; and (2) their expression levels were at least 2-fold higher than that in any other nonhomologous organs. With these criteria, the expression of 699 (2.7%), 747 (2.9%), 762 (3.0%), 805 (3.2%), 827 (3.2%), 143 (0.5%), 89 (0.4%), 317 (1.2%), 36 (0.1%), and 187 (0.7%) genes were found to be enriched in germinating seed, rosette leaf, root, stamen, petal, sepal, silique, pistil, hypocotyl, and stem, respectively (Fig. 4A; Supplemental Table I). Most of the specific functional groups of genes followed similar trends, as shown by the transcription factor genes (Fig. 4A). We further analyzed the organ-enriched genes within several organ groups. For example, 731 (2.9%), 669 (2.7%), 261 (1.0%), and 143 (0.6%) genes showed specifically high expression in pistil 1 d before pollination, 1, 3, and 8 d after pollination, respectively (Fig. 4B; Supplemental Table II). There were 628 (2.5%), 155 (0.6%), and 778 (3.1%) genes whose expressions were enriched in rosette leaf, cauline leaf, and light-grown cotyledon, respectively (Fig. 4C; Supplemental Table III). Among the three floral organs, the expressions of 558 (2.1%), 1,710 (6.6%), and 1,282 (5.0%) genes were enriched in sepal, petal, and stamen, respectively (Fig. 4D; Supplemental Table IV). These organ-specific expression data are consistent with previously documented expression patterns of known genes. For example, four well-characterized floral pattern determination genes (AP1, AP3, PI, and AG) exhibited expression patterns among floral organs (Fig. 5) as previously reported (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994). In addition, CPC and GL2 are enriched in root (Masucci et al., 1996; Wada et al., 1997), CCA1 has the highest expression level in leaf (Wang et al., 1997), and BELL1 shows the highest expression level in pistil (Reiser et al., 1995). Together, these results suggest that the whole-genome microarray is a valid approach to determine specific gene expression patterns that may reveal the function of the corresponding genes.
Figure 4.
Organ-specific expressed genes. A, Distribution of total organ-specific expressed genes and transcription factor (TF) genes (Jiao et al., 2003) among all representative organ types. B, Distribution of total organ-specific expressed genes and TF genes at distinct phases of pistils and siliques. DBP, Days before pollination. C, Distribution of total organ-specific expressed genes and TF genes within the leaf organ group (cotyledon, rosette leaf, and cauline leaf). D, Distribution of total organ-specific expressed genes and TF genes among sepal, petal, and stamen of flower.
Figure 5.
The relative expression levels of the four well-characterized floral pattern determination genes, Apetala 1 (AP1), Apetala 3 (AP3), Pistillata (PI), and Agamous (AG) among the 17 Arabidopsis organs as determined by our microarray analysis. The observed expression patterns from our whole-genome microarray studies consistent with the whorl 1- and 2-specific expression of AP1, whorl 2- and 3-specific expression of AP3 and PI, and whorl 3- and 4-specific expression of AG as previously reported (Yanofsky, et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994).
Distinct Portions of the Genome Respond to Light Regulation in the Three Arabidopsis Seedling Organs
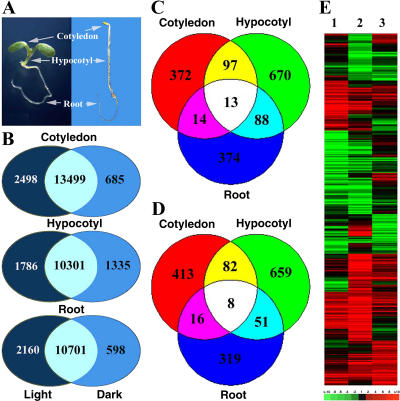
Arabidopsis seedling development is dramatically regulated by light. The three seedling organs (root, hypocotyl, and cotyledon) exhibit distinct developmental responses to light (Fig. 6A), even though the same photoperception and signaling systems seem to operate in all three organs (Cashmore et al., 1999; Quail, 2002). As an initial step to understand this regulatory mechanism, the genome expression profiles of light- and dark-grown cotyledon, hypocotyl, and root organs were examined and compared. We noted that for each of the three seedling organs, largely overlapping sets of genes were expressed in both dark- and light-grown conditions (Fig. 6B; Supplemental Table V). However, 1,015 (4.0%), 1,668 (6.5%), and 883 (3.5%) of the genes showed differential expression (at least 2-fold and P < 0.05; see “Materials and Methods”) between light- and dark-grown cotyledon, hypocotyl, and root, respectively (Fig. 6, C and D; Supplemental Tables VI and VII). Careful examination of those genes that are differentially expressed in response to light indicates only small overlaps among cotyledon, hypocotyl, and root (Fig. 6, C and D; Supplemental Tables VI and VII), with a small number of genes even oppositely regulated by light in different organs (Fig. 6E; Supplemental Table VIII). Clearly, the light signal, perceived and transduced by the same photoperception and transduction systems, results in distinct expression changes in the genome in different organs.
Figure 6.
The three seedling organs exhibited distinct genome expression changes under light. A, Morphological comparison of continuous white-light- and dark-grown Arabidopsis seedlings, with the three organs marked. B, Diagrams illustrating both the overlapping and uniquely expressed gene numbers between white-light- and dark-grown seedling organ pairs. The numbers in overlapping areas show the shared gene number between the light- and dark-grown organ pairs, while the numbers in nonoverlapping areas show the unique gene number in light- or dark-growth conditions. C and D, Venn diagrams of the differentially expressed genes that exhibited significant (at least 2-fold with P < 0.05) up-regulation (C) or down-regulation (D) for each of the three organ pairs. The numbers in the overlapping areas indicate the shared number of genes that exhibited 2-fold or greater differential expression in either two or three organ pairs, while those numbers in nonoverlapping areas indicated organ-specifically regulated genes. E, Hierarchical clustering display of white-light-regulated genome expression among the three seedling organs. Any gene that exhibited light-regulated expression (at least 2-fold with P < 0.05) in at least one of the three organs was selected for analysis. Lane 1, Expression ratios of white-light- and dark-grown cotyledon. Lane 2, Expression ratios of white-light- and dark-grown hypocotyl. Lane 3, Expression ratios of white-light- and dark-grown root. The color scale is shown at the bottom of the cluster.
Organ-Specific Light Regulation of Metabolic Pathways in Arabidopsis Seedlings
In a previous study using whole seedlings, more than 26 metabolic and regulatory pathways were found to be regulated by light in Arabidopsis (Ma et al., 2001). Our genome expression profiles from the three seedling organs showed distinct light regulation patterns, with only a small overlap of light-regulated genes (Fig. 6). Thus, possible organ specificity in the light regulation of those metabolic pathways among the three organs was examined. Some of the metabolic pathways were regulated by light in all three organs. For example, the genes encoding the anthocyanin-biosynthetic pathways (Fig. 7A) and the jasmonic acid biosynthetic and response pathways (Fig. 7B) were significantly induced (more than 2-fold and P < 0.05) by light in all three organs. The degrees of light responsiveness in those three organs differ significantly. The responsiveness of the jasmonate biosynthetic and responsive pathways to light might indicate that jasmonate plays a role in light regulation of seedling development. Some other pathways were found to be regulated by light in cotyledon and hypocotyl, but not in root. For example, the genes encoding proteins involved in the photosynthetic light reaction, carbon metabolism, and photorespiration pathways were induced by light in both cotyledon and hypocotyl, but not in root (Fig. 7C). On the other hand, the ethylene biosynthetic pathway was repressed by light in both cotyledon and hypocotyl, but not in root (Fig. 7D).
Figure 7.
Light-regulated expression profiles of 10 representative genes from selected metabolic and regulatory pathways in the cotyledon, hypocotyl, and root of Arabidopsis seedling. See legend of Figure 6 for further details.
Many pathways are subjected to light regulation in only one of the three organs. For example, the starch and Suc biosynthetic pathways were up-regulated by light only in cotyledon (Fig. 7E). The genes encoding light repressible receptor-like protein kinase and several other receptor-like kinases were repressed only in root (Fig. 7F).
Many Gene Family Members Exhibit Distinct Light or Organ Regulation Patterns in Arabidopsis
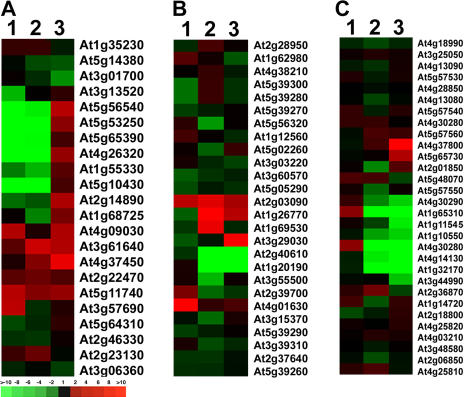
Large portions of genes in the Arabidopsis genome are classified into gene families based on their sequence homologies (Arabidopsis Genome Initiative, 2000). Thus, we examined light regulation profiles for the gene family members among the three seedling organs. It is evident that the light regulation and organ-specific expression patterns for gene members in the same family are not identical. To illustrate this point clearly, we chose three medium-size gene families, arabinogalactan protein (AGP), expansin, and xyloglucan endotransglucosylase (XTH) families, for hierarchical clustering analysis using the average linkage method. It should be pointed out that expansin and XTH families are both involved in cell elongation and growth (Campbell and Braam 1999; Li et al., 2003). As shown in Figure 8A, the light regulated expression of 22 AGP gene members is quite diverse. Among these 22 genes, nine family members were repressed by light in cotyledon, five gene members were induced by light in cotyledon, and the remaining eight gene members were not light regulated in cotyledon (Fig. 8A). Among the three seedling organs (Fig. 8A), several AGP gene members had the same or similar light regulation pattern(s) among the three organs (e.g. At5g11740); some gene members had similar light regulation patterns in two organs (e.g. At5g56540 and At4g09030), while other family members showed a distinct light regulation pattern among the three organs (e.g. At3g13520 and At1g68725). Similar, or more evident, diversity in their light-regulated and organ-specific expression of the other two gene families (expansin and XTH families) was observed (Fig. 8, B and C).
Figure 8.
Hierarchical clustering display of expression ratios from light- versus dark-grown seedling organs for three representative gene families. A, AGP family; B, expansin family; C, XTH family. The three lanes in each cluster are: 1, light-grown cotyledon versus dark-grown cotyledon; 2, light-grown hypocotyl versus dark-grown hypocotyl; and 3, light-grown root versus dark-grown root. The color scale is shown at the bottom of cluster A.
We further examined the gene expression patterns for different gene families among all 17 organs examined. We found that the organ-specific expression pattern for gene members in the same family is not identical in all 17 organs. Again, we used the above-mentioned three gene families to do cluster analyses. Because we compared each organ with cultured cells to obtain the genome expression profile for each organ, we used the expression ratio of organ and cultured cells to show the gene expression patterns among all 17 organs. As shown in Figure 9, different members in the gene family had distinct expression patterns in the same organ. Further, organ-specific regulation of a gene family was also distinct among organs, while similar organ types, for example, rosette and cauline leaf, pistil and silique, tended to show similar expression patterns (Fig. 9, A–C). Such diversified expression patterns for members of the same gene family would be consistent with the notion that different members in a gene family might have evolved distinct functions.
Figure 9.
Hierarchical clustering display of expression ratios from organ versus cultured cell for three representative gene families. A, AGP family; B, expansin family; C, XTH family. The 17 lanes in each cluster are: 1, light-grown cotyledon versus cultured cell; 2, dark-grown cotyledon versus cultured cell; 3, light-grown hypocotyl versus cultured cell; 4, dark-grown hypocotyl versus cultured cell; 5, rosette leaf versus cultured cell; 6, cauline leaf versus cultured cell; 7, stem versus cultured cell; 8, sepal versus cultured cell; 9, petal versus cultured cell; 10, stamen versus cultured cell; 11, pistil 1DBP versus cultured cell; 12, pistil 1DPP versus cultured cell; 13, silique 3DPP versus cultured cell; 14, silique 8DPP versus cultured cell; 15, germinated seed versus cultured cell; 16, light-grown root versus cultured cell; and 17, dark-grown root versus cultured cell. The color scale is shown at the bottom of cluster C.
Coregulation of Gene Expression Patterns in the Arabidopsis Genome
Recent studies from several species suggest that a significant fraction of those organisms' genomes may be organized into chromatin domains that contain a number of adjacent genes whose expressions are coordinately regulated (Cohen et al., 2000; Caron et al., 2001; Lercher et al., 2002; Spellman and Rubin, 2002; Birnbaum et al., 2003). For example, in budding yeast (Saccharomyces cerevisiae), pairs or triplets of adjacent genes displayed similar expression patterns (Cohen et al., 2000), whereas groups of 10 to 30 adjacent genes showed similar expression patterns in Drosophila (Spellman and Rubin, 2002). To further examine whether the Arabidopsis genome also contains these chromatin domains with coregulated adjacent genes, we used the organ expression data sets to calculate the coregulated gene clusters (with a block size of 10 genes) based on their physical positions on the chromosome (Spellman and Rubin, 2002). We also considered the fact that the Arabidopsis genome contains many tandemly repeated genes and therefore excluded these from the fraction of coregulated genes during our calculation. This analysis indicated that the chromatin domains of coregulated genes are evident in the Arabidopsis genome, possibly accounting for over 12% of the Arabidopsis genome in one calculation (Table I). This estimated fraction of the genome organized into these coregulated chromatin domains in the Arabidopsis genome is similar to the reported 20% for Drosophila (Spellman and Rubin, 2002).
Table I.
The total number of unique genes identified as being within the coregulated chromatin domains as genes ordered in their native chromosomal locations (ordered genes), compared to the situation where all genes are randomized in their relative positions (randomized genes)
The organ-specific gene expression data from 17 organs versus tissue types were used in the calculation, with a chromatin domain window size of 10 genes. Calculation of the chromatin domains is based on a recently reported method (Spellman and Rubin, 2002).
| Significance (P Value) | Ordered Genes | Randomized Genes |
|---|---|---|
| 0.001 | 1,415 | 135 |
| 0.005 | 2,425 | 512 |
| 0.01 | 3,262 | 1,038 |
DISCUSSION
In this study, we provide an organ-specific genome expression atlas during Arabidopsis development by analyzing the genome expression profile of individual representative organs using a 70-mer oligomer microarray. This analysis provides experimental evidence for a large fraction of those predicted genes in the Arabidopsis genome (Fig. 1). In addition, we observed that different organs expressed distinct sets of genes from the Arabidopsis genome (Figs. 2 and 4), and only about 16% of the total genes are expressed in all examined organs (Fig. 1). Based on those organ-specific genome expression profiles, we further demonstrated that the relatedness of the organ types based on development is reflected by their whole-genome expression patterns as well (Fig. 3). These results support the conclusion that the genome expression patterns are defining characteristics of Arabidopsis organs during development. Therefore, defining the exact genome expression pattern (the abundance of each individual expressed gene) of each organ will provide us with much-needed information in understanding the developmental characteristics of each organ type. Further, the comprehensive genome expression information made available by this and other related works, together with genome-wide insertional mutagenesis (Alonso et al., 2003), should provide a foundation toward description of the molecular mechanism leading to the observed genome expression pattern changes during Arabidopsis development.
Development in plants is often reprogrammed by environmental signals. For example, light is one of most important environmental signals for controlling plant growth and development (Kendrick and Kronenberg, 1994; Deng and Quail, 1999; Neff et al., 2000). Plants undergo dramatic changes in developmental patterns depending on the presence or absence of light in the growth environment. When grown in the dark, an Arabidopsis seedling develops with a long hypocotyl, unopened small cotyledons with an apical hook, and short root, whereas the light-grown seedling exhibits a photomorphogenic phenotype with a short hypocotyl, open cotyledons without an apical hook, and long strong root (Kendrick and Kronenberg, 1994; Deng and Quail, 1999; Neff et al., 2000; Fig. 6A). For light control of plant development, it is generally assumed that photoreceptors perceive and interpret incident light and transduce the signals to modulate light-responsive nuclear genes, which direct appropriate growth and developmental responses (Ma et al., 2001; Tepperman et al., 2001; Quail, 2002). Our previous studies using an EST-based microarray suggested that a significant fraction of expressed genes in the Arabidopsis genome are controlled by light in the whole seedling and at later developmental stages (Ma et al., 2001, 2003). In this study, we further checked the light regulation of the Arabidopsis genome expression in different organ types of Arabidopsis seedlings. We found that about 3.5% of the Arabidopsis genome is controlled by light in the root, while light regulation exists for a larger proportion of the genome in both the cotyledon and hypocotyl (Fig. 6, C and D). This is consistent with the observation that roots also expressed a high level of photoreceptors (Somers and Quail, 1995) and the contrasting root phenotypes grown under dark and light conditions (Fig. 6A). However, we cannot rule out the possibility that a fraction of the observed light regulated genes may be caused through secondary responses to the possible stress conditions under the light- or dark-growth conditions for Arabidopsis seedlings.
An interesting feature is that only a small set of light-regulated genes are shared among the three seedling organ types. Some of the genes that differ among the three organ types even show opposite light regulation patterns (e.g. light induces a gene expression in cotyledon, while the same gene is repressed by light in root or hypocotyl; Fig. 6, C–E). These results suggest that the light signal triggers expression changes in distinct target genes in different organs or in some instances distinct responses of the same target genes. Therefore, although light signals are perceived by the same photoreceptors and may be transduced by the same transduction pathways, distinct changes in genome expression occur in different seedling organ types. The mechanism responsible for light regulation in different organ types is not yet clear.
Recent results from human (Caron et al., 2001; Lercher et al., 2002), Drosophila (Spellman and Rubin, 2002), Arabidopsis (Birnbaum et al., 2003; this study), and yeast (Cohen et al., 2000) suggest that the regulation of genome expression involves coordinated regulation of adjacent genes in chromosomal regions. Our results further suggest that approximately 12% of the Arabidopsis genome shows a coregulation expression pattern (Table I). However, the mechanism for this coregulated expression pattern in the genome is not clear yet. One reasonable possibility is the involvement of chromatin modification mechanism in these coregulated neighbor genes. As histone proteins in the nucleosomes around a given gene are modified (e.g. acetylation) by chromatin remodeling mediators according to a given signal, the chromatin domain is opened not only for the gene it is binding directly, but also for those neighbor genes within the whole chromatin domain. Genome-wide analysis of histone acetylation or methylation patterns in representative Arabidopsis organs or in response to the given signals may provide evidence for this prediction.
MATERIALS AND METHODS
Plant Materials
The wild-type Arabidopsis (Arabidopsis thaliana) used in this study was the Columbia ecotype. Surface sterilization, cold treatment of the seed, and seedling growth were performed as described previously (Ma et al., 2001). The germinating seeds were collected after the seeds were planted on growth medium agar plates containing 1% Suc and grown under continuous white light (150 μmol m−2 s−1) for 48 h at 22°C. Arabidopsis seedlings used in this study were 6 d old. The seedlings were planted on agar plates containing growth medium with 1% Suc and grown at 22°C in continuous white light (150 μmol m−2 s−1) or darkness. The cotyledon, hypocotyl, and root were collected from the same seedling, respectively. The rosette leaf was collected from 3-week-old plants grown at 22°C under continuous white light (150 μmol m−2 s−1), with the root, bolt, and senescing leaves removed. Adult Arabidopsis plants were grown in a walk-in Environmental Growth Chamber (EGC, Chagrin Falls, OH) at 22°C under continuous white light (250 μmol m−2 s−1). The cauline leaf and stem were collected from 4-week-old adult plants, respectively. The floral organs were collected from the mature flowers of adult plants at flower stage 14 (Bowman, 1994). The silique was collected from adult plants 3 or 8 d after pollination. Suspension culture cells were prepared starting from seeds as described by Martinez-Zapater and Salinas (1998). The cultured cells used for RNA isolation were collected at the logarithmic growth phase.
Oligo Microarray
The 70-mer oligo set for the Arabidopsis genome was designed and synthesized by Qiagen/Operon (http://oligos.qiagen.com/arrays/omad.php) based on the Arabidopsis genome information available on February 20, 2002. The oligos were purchased from Qiagen (Valenica, CA) and printed onto polylysine coated microscope slides in the DNA microarray laboratory at Yale University (http://info.med.yale.edu/wmkeck/dna_arrays.htm). There were 26,090 unique oligos, and 12 distinct negative control oligos. Each negative control oligo was printed 16 times at well-spaced locations on each slide. Thus, each slide included a total of 26,090 oligo spots and 192 negative control spots. The negative controls were positioned all over the slide to avoid potential errors caused by spatial effects. These negative controls do not have a match in the genome sequence.
RNA Isolation, Probe Labeling, and Hybridization
Total RNA was extracted from the above-mentioned organs using the Qiagen RNeasy Plant Mini prep kit. RNA preparations from two to four independent biological samples for each test were made and used for probe synthesis. Thus, each experiment produced two to four biological replicate data sets. Total RNA (50 μg) was first labeled with aminoallyl-dUTP (aa-dUTP; Sigma, St. Louis) by direct incorporation of aa-dUTP during reverse transcription, as described previously (Ma et al., 2002). The purified probe was further labeled with fluorescent dye by conjugating aa-dUTP and monofunctional Cy-3 or Cy-5 (Amersham Pharmacia Biotech, Piscataway, NJ). The dye-labeled probe was purified from the unincorporated dye molecules by washing three times through a Microcon YM-30 filter (Millipore, Bedford, MA). The purified labeled probes from specific organ versus culture cell control pairs or dark versus light organ pairs were combined to hybridize the microarray slide for 12 to 16 h at 42°C (Ma et al., 2001). Except petal (two replicates) and pistil of 1 d postpollination (1DPP, four replicates), there were three biological replicates used for all other organ types, with one quality data set from each replicate.
Coverage of Oligo Set for Arabidopsis Genes and EST Clones
We first blasted the sequences of 26,090 oligos individually against Munich Information Center for Protein Sequences (MIPS) Arabidopsis gene annotation (March 20, 2003 version) to associate oligos with gene IDs (e.g. AT3G20980) if they matched to the annotated genes. We used the criterion that the matching identity between oligo sequence and genome sequence should be 70% (or at least matching 49 out of 70 nucleotides) or higher, to define the match between oligo and gene locus. In fact, the vast majority of oligos (97%) were mapped to the chromosomal genes with more than 90% identity. For oligos with no match in the MIPS annotated gene sequences, we blasted them against The Institute for Genomic Research (TIGR) annotation (July 31, 2002 version) downloaded from TAIR (ftp://ftp.arabidopsis.org/home/tair/Sequences/blast_datasets/). More corresponding gene loci were obtained, and their locus IDs were assigned to those oligos. In total, we are able to assign 25,822 oligos that represent 25,676 unique locus IDs (genes). Among them, the majority (94%) of the oligos had a single match to a unique gene locus. A small fraction of the oligos (6%, 1,553) fall into the following two categories: one oligo matches two or more unique genes, or one unique gene matches more than one oligo (with the 70% identity cutoff). In the former case, we assigned the oligo with multiple locus IDs to cover all possible genes that may contribute to the detected expression signal. While in the latter case, as multiple oligos assigned the same locus ID, the median intensity of those oligo spots was taken as its expression level.
To define the number of genes covered by the oligo array that have EST hits, we joined together the information in ESTtoAT and mRNAtoAT at TAIR (ftp://ftp.arabidopsis.org/home/tair/Sequences/) and ESTmatchingtoAT at MIPS (http://mips.gsf.de/proj/thal/; downloaded on July 8, 2003) to obtain the unique locus numbers with at least one EST or mRNA hit from the above three resources. This analysis resulted in the number (16,998) of the genes from the above 25,676 unique locus IDs that have at least one EST or mRNA hit.
GO annotation for all gene models were downloaded from TAIR (Rhee et al., 2003; ftp://ftp.arabidopsis.org/home/tair/Genes/Gene_Ontology/). We functionally classified all genes from GO annotation using GOslim terms. We followed the TAIR April 14, 2003 version GO annotation. We also updated the annotation for (basic helix-hoop-helix) transcription factor family according a recent report (Toledo-Ortiz et al., 2003).
Data Normalization and Determination of Expression
Spot intensities were quantified using Axon GenePix Pro 3.0 image analysis software. The net intensities for each channel and channel ratios were measured using the GenePix Pro 3.0 median of intensity or ratio method. Replicates were normalized first to remove artifacts due to experimental variations using custom-designed programs (http://bioinformatics.med.yale.edu/software.html). Then normalization based on median of intensities was performed among all the experiments. We followed a commonly used strategy (Rinn et al., 2003) to define whether a gene is expressed or not with minor adjustment. First, we stipulated that the normalized intensity of an expressed gene (spot) has to be higher than the intensity value at the 90% of the normalized median intensities of 192 negative control oligos. Second, we consider that the expression of a gene is detectable only if the majority (two out of two, at least two out of three, and at least three out of four) of the corresponding spots from multiple experiments showed experimentally detectable expression as defined in the first criterion. Third, those spots that exhibited a large difference between replicates were defined as outliers and eliminated from further analysis.
Identification of Differential Expression
To identify differentially expressed genes among organ groups (Fig. 4), we fitted normalized replicate intensities of all organs together with a cultured-cell control into an ANOVA model. For each data set, the gene intensities of both organ and reference cultured cell were used. The model is given by yijkl = μ + Ai + Dj + ADij + Gl + VGkl + DGjl + AGil + ɛijkl, where yijkl denotes the logarithm transformed signal for gene l on slide i labeled with dye j of sample k. The overall mean effect was represented by μ; A, D, and G represented main effects from array, dye, and gene. The interaction terms AD, VG, DG, and AG represented array by dye, sample (variation) by gene, dye by gene, and array by gene. The random error was denoted by ɛijkl. We were interested in the term VG. The above ANOVA was performed on each spot using MAANOVA for R with F statistics computed on the James-Stein shrinkage estimates of the error variance (Cui and Churchill, 2003; Wu et al., 2003). We selected organ-enriched genes with a P value <0.05 in the above F test, and if the expression intensities in one organ were 2-fold, or more, higher than in other organs in the same comparison group.
Similar statistical analyses were performed for the identification of light regulated genes. A student's t test was performed for each gene between light-growth condition and dark-growth condition. We consider genes with a P value <0.05 as light regulated. To reduce the occurrence of false positives, we applied an additional 2-fold expression changed for some subsequent analysis.
Estimation of Average Intensities for Each Gene (Spot)
For other analyses in this work, normalized intensities were averaged among all replicates of the same sample to obtain a single statistic, which is considered a relative expression for a gene. Similarly, we also used a single statistic for ratios for each gene. We calculated the expression ratio for a sample pair only when at least one channel showed experimentally detectable expression as defined above.
Calculation of Chromatin Domains with Coregulated Adjacent Genes
We used the method reported by Spellman and Rubin (2002) to calculate the coregulated adjacent genes. For each given block size, we calculated the average correlation coefficient among the gene expression data. The tandem repeat genes were excluded from our analysis. The value was compared to the values from 10,000 sets of randomly selected genes of the same number of genes to calculate the P value. We carried out such analysis for the block sizes of 2, 4, 6, 8, 10, 15, 20, 25, and 30 genes and found that the block size of 10 represented the major block size. We then calculated the number of genes showing coexpression at P values 0.001, 0.005, and 0.01 with the block size of 10 genes.
All the microarray data described in this study were deposited into the NCBI GEO database (accession no. GSE 1599).
Acknowledgments
We thank Mr. Matthew Holford for assisting with the Java programming, Elizabeth Strickland, Jessica Habashi, and Lei Li for reading and commenting on this manuscript, and the Yale DNA microarray laboratory of the Keck Biological Resource Center for the production of the microarray used in this study (http://info.med.yale.edu/wmkeck/dna_arrays.htm).
This work was supported by the National Science Foundation of China (strategic international corporation project grant no. 30221120261), by the National Institutes of Health (grant nos. GM–47850 to X.W.D. and GM59507 to H.Z.), and by the National Science Foundation (grant no. DMS 0241160). L.M. is a long-term postdoctoral fellow of the Human Frontier Science Program.
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk K, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barker J, Harada JJ, Goldberg RB (1988) Cellular localization of soybean storage protein mRNA in transformed tobacco seeds. Proc Natl Acad Sci USA 85: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bowman J (1994) Arabidopsis: An Atlas of Morphology and Development. Springer-Verlag, New York
- Campbell P, Braam J (1999) Xyloglucan endotransglycosylases: diversity of genes, enzymes and potential wall-modifying functions. Trends Plant Sci 9: 361–366 [DOI] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, et al (2001) The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291: 1289–1292 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I (1998) The transcriptional program of sporulation in budding yeast. Science 282: 699–705 [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM (2000) A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet 26: 183–186 [DOI] [PubMed] [Google Scholar]
- Cui X, Churchill GA (2003) Statistical tests for differential expression in cDNA microarray experiments. Genome Biol 4: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Quail PH (1999) Signalling in light-controlled development. Semin Cell Dev Biol 10: 121–129 [DOI] [PubMed] [Google Scholar]
- Eeau K (1977) Anatomy of Seed Plants, Ed 2. John Wiley & Sons, New York
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium (2000) Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB (1988) Plants: novel developmental processes. Science 240: 1460–1467 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ (1989) Regulation of gene expression during plant embryogenesis. Cell 56: 149–160 [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Kamalay JC, Timberlake WE (1978) Sequence complexity of nuclear and polysomal RNA in leaves of the tobacco plant. Cell 14: 123–131 [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM (1994) Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev 8: 1548–1560 [DOI] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68: 683–697 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Yang H, Ma L, Sun N, Yu H, Liu T, Gao Y, Gu H, Chen Z, Wada M, et al (2003) A genome-wide analysis of blue-light regulation of Arabidopsis transcription factor gene expression during seedling development. Plant Physiol 133: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamalay JC, Goldberg RB (1980) Regulation of structural gene expression in tobacco. Cell 19: 935–946 [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM (1994) Photomorphogenesis in Plants, Ed 2. Kluwer Academic Publishers, Dordrecht, The Netherlands
- Lercher MJ, Urrutia AO, Hurst LD (2002) Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet 31: 180–183 [DOI] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6: 603–610 [DOI] [PubMed] [Google Scholar]
- Ma L, Gao Y, Qu L, Chen Z, Li J, Zhao H, Deng XW (2002) Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14: 2383–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Zhao H, Deng XW (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130: 969–981 [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Salinas J (1998) Arabidopsis Protocols. Humana Press, Totowa, NJ, pp 27–30
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MW, Schiefelbein JW (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM (2002) Plants compared to animals: the broadest comparative study of development. Science 295: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Reiser L, Modrusan Z, Margossian L, Samach A, Ohad N, Haughn GW, Fischer RL (1995) The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83: 735–742 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S, Harrison PM, Nelson FK, Miller P, Gerstein M, et al (2003) The transcriptional activity of human Chromosome 22. Genes Dev 17: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Somers DE, Quail PH (1995) Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis. Plant J 7: 413–427 [DOI] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM (2002) Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol 1: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH (2001) Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Wang Z, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM (1997) A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9: 491–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM (2004) Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kerr K, Cui X, Churchill GA (2003) MAANOVA: a software package for the analysis of spotted cDNA microarray experiments. In G Parmigiani, ES Garett, RA Irizarry, SL Zeger, eds, The Analysis of Gene Expression Data: Methods and Software. Springer-Verlag, Heidelberg, Germany, pp 313–341
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Feldmann GN, Meyerowitz EM (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Zhu T, Buwworth P, Han B, Brown D, Chang HS, Zou G, Wang X (2001) Toward elucidating the global gene expression patterns of developing Arabidopsis: parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant Physiol Biochem 39: 221–242 [Google Scholar]