Abstract
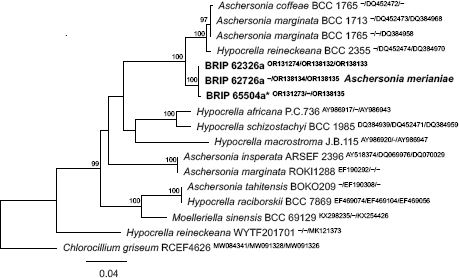
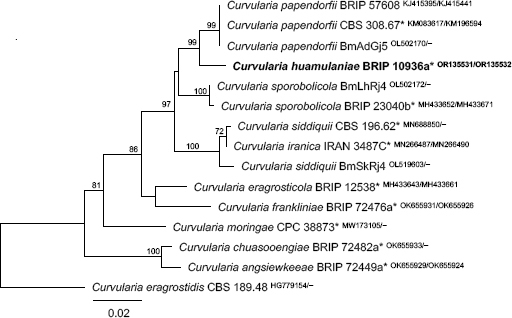
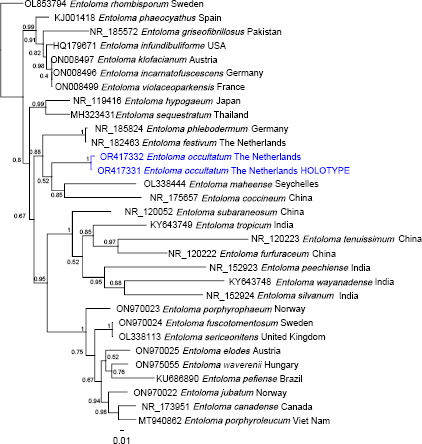
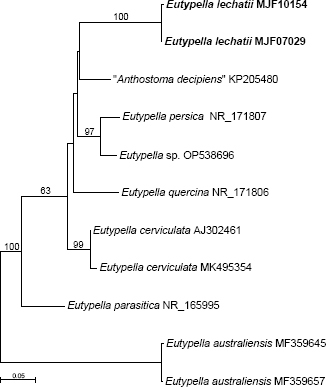
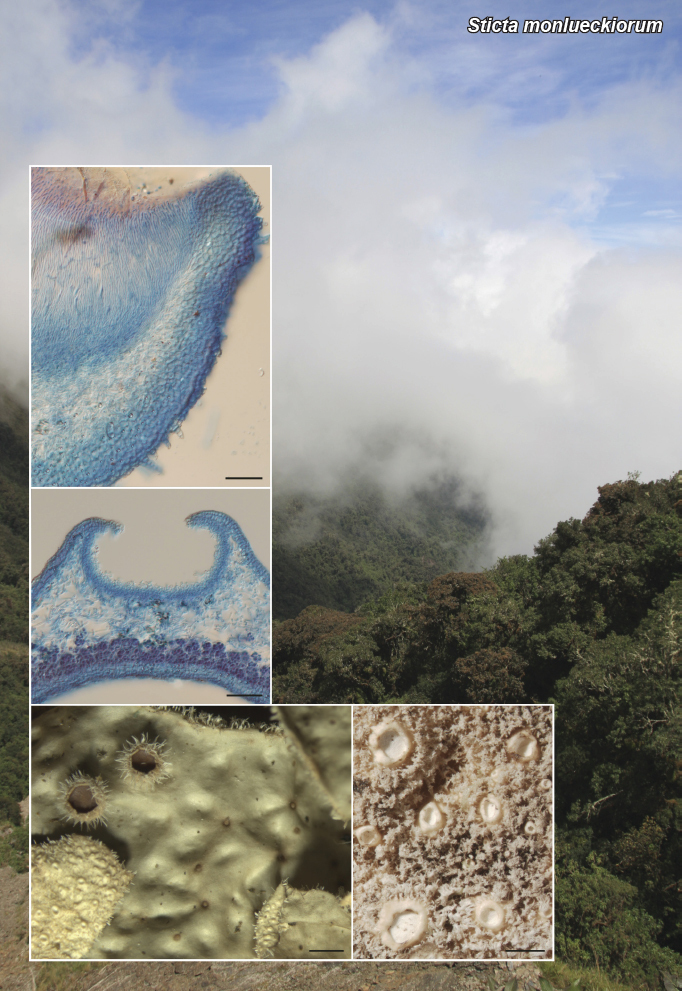
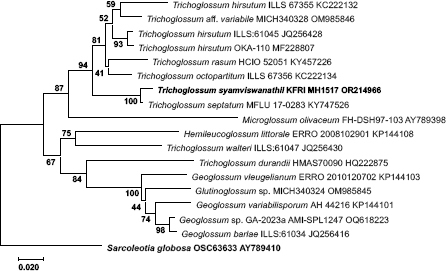
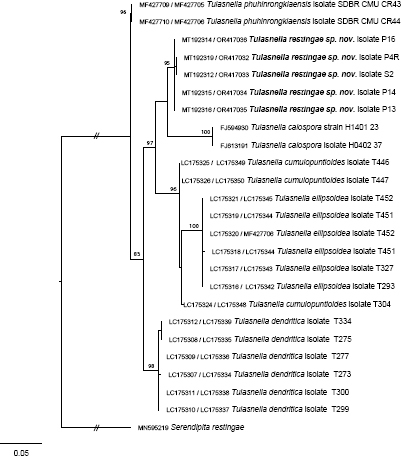
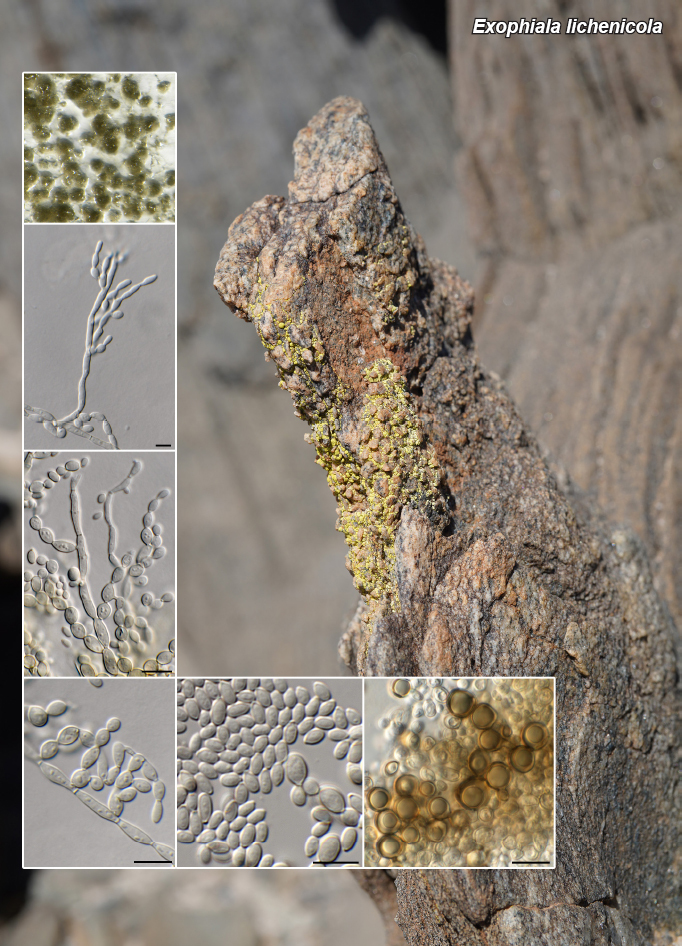
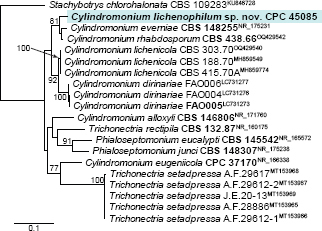
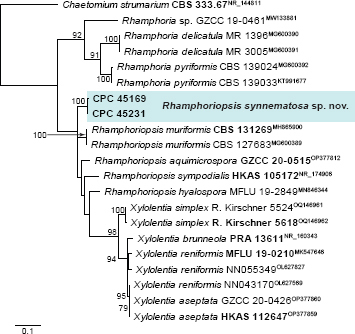
Novel species of fungi described in this study include those from various countries as follows: Argentina, Neocamarosporium halophilum in leaf spots of Atriplex undulata. Australia, Aschersonia merianiae on scale insect (Coccoidea), Curvularia huamulaniae isolated from air, Hevansia mainiae on dead spider, Ophiocordyceps poecilometigena on Poecilometis sp. Bolivia, Lecanora menthoides on sandstone, in open semi-desert montane areas, Sticta monlueckiorum corticolous in a forest, Trichonectria epimegalosporae on apothecia of corticolous Megalospora sulphurata var. sulphurata, Trichonectria puncteliae on the thallus of Punctelia borreri. Brazil, Catenomargarita pseudocercosporicola (incl. Catenomargarita gen. nov.) hyperparasitic on Pseudocercospora fijiensis on leaves of Musa acuminata, Tulasnella restingae on protocorms and roots of Epidendrum fulgens. Bulgaria, Anthracoidea umbrosae on Carex spp. Croatia, Hymenoscyphus radicis from surface-sterilised, asymptomatic roots of Microthlaspi erraticum, Orbilia multiserpentina on wood of decorticated branches of Quercus pubescens. France, Calosporella punctatispora on dead corticated twigs of Aceropalus. French West Indies (Martinique), Eutypella lechatii on dead corticated palm stem. Germany, Arrhenia alcalinophila on loamy soil. Iceland, Cistella blauvikensis on dead grass (Poaceae). India, Fulvifomes maritimus on living Peltophorum pterocarpum, Fulvifomes natarajanii on dead wood of Prosopis juliflora, Fulvifomes subazonatus on trunk of Azadirachta indica, Macrolepiota bharadwajii on moist soil near the forest, Narcissea delicata on decaying elephant dung, Paramyrothecium indicum on living leaves of Hibiscus hispidissimus, Trichoglossum syamviswanathii on moist soil near the base of a bamboo plantation. Iran, Vacuiphoma astragalicola from stem canker of Astragalus sarcocolla. Malaysia, Neoeriomycopsis fissistigmae (incl. Neoeriomycopsidaceae fam. nov.) on leaf spots on flower Fissistigma sp. Namibia, Exophiala lichenicola lichenicolous on Acarospora cf. luederitzensis. Netherlands, Entoloma occultatum on soil, Extremus caricis on dead leaves of Carex sp., Inocybe pseudomytiliodora on loamy soil. Norway, Inocybe guldeniae on calcareous soil, Inocybe rupestroides on gravelly soil. Pakistan, Hymenagaricus brunneodiscus on soil. Philippines, Ophiocordyceps philippinensis parasitic on Asilus sp. Poland, Hawksworthiomyces ciconiae isolated from Ciconia ciconia nest, Plectosphaerella vigrensis from leaf spots on Impatiens noli-tangere, Xenoramularia epitaxicola from sooty mould community on Taxus baccata. Portugal, Inocybe dagamae on clay soil. Saudi Arabia, Diaporthe jazanensis on branches of Coffea arabica. South Africa, Alternaria moraeae on dead leaves of Moraea sp., Bonitomyces buffels-kloofinus (incl. Bonitomyces gen. nov.) on dead twigs of unknown tree, Constrictochalara koukolii on living leaves of Itea rhamnoides colonised by a Meliola sp., Cylindromonium lichenophilum on Parmelina tiliacea, Gamszarella buffelskloofina (incl. Gamszarella gen. nov.) on dead insect, Isthmosporiella africana (incl. Isthmosporiella gen. nov.) on dead twigs of unknown tree, Nothoeucasphaeria buffelskloofina (incl. Nothoeucasphaeria gen. nov.), on dead twigs of unknown tree, Nothomicrothyrium beaucarneae (incl. Nothomicrothyrium gen. nov.) on dead leaves of Beaucarnea stricta, Paramycosphaerella proteae on living leaves of Protea caffra, Querciphoma foliicola on leaf litter, Rachicladosporium conostomii on dead twigs of Conostomium natalense var. glabrum, Rhamphoriopsis synnematosa on dead twig of unknown tree, Waltergamsia mpumalanga on dead leaves of unknown tree. Spain, Amanita fulvogrisea on limestone soil, in mixed forest, Amanita herculis in open Quercus forest, Vuilleminia beltraniae on Cistus symphytifolius. Sweden, Pachyella pulchella on decaying wood on sand-silt riverbank. Thailand, Deniquelata cassiae on dead stem of Cassia fistula, Stomiopeltis thailandica on dead twigs of Magnolia champaca. Ukraine, Circinaria podoliana on natural limestone outcrops, Neonematogonum carpinicola (incl. Neonematogonum gen. nov.) on dead branches of Carpinus betulus. USA, Exophiala wilsonii water from cooling tower, Hygrophorus aesculeticola on soil in mixed forest, and Neocelosporium aereum from air in a house attic. Morphological and culture characteristics are supported by DNA barcodes.
Citation: Crous PW, Costa MM, Kandemir H, et al. 2023. Fungal Planet description sheets: 1550–1613. Persoonia 51: 280–417. doi: 10.3767/persoonia.2023.51.08.
Keywords: ITS nrDNA barcodes, LSU, new taxa, systematics
Acknowledgments
The work of P.W. Crous and colleagues benefitted from funding by the European Union’s Horizon 2020 research and innovation program (RISE) under the Marie Sklodowska-Curie grant agreement No. 101008129, project acronym ‘Mycobiomics’, and the Dutch NWO Roadmap grant agreement No. 2020/ENW/00901156, project ‘Netherlands Infrastructure for Ecosystem and Biodiversity Analysis – Authoritative and Rapid Identification System for Essential biodiversity information’ (acronym NIEBA-ARISE). M. Plaza and T. Illescas are grateful to the Junta de Andalucía for facilitating their vehicle access to areas of special protection; and to both their daughters, C. Plaza and M. Illescas, for revising the English text, and the Asociación Botánica y Micológica de Jaén, for funding part of the DNA sequences included in this study. The study of T.T. Denchev & C.M. Denchev was supported by the Bulgarian National Science Fund (Grant no. KP-06-N51/10/16.11.2021). K. Karasungur is thanked for technical help with the Austrian material of Arrhenia sequenced within the Austrian Barcode of Life project, supported by the Austrian Federal Ministry of Education, Science and Research. Y.P. Tan and colleagues acknowledge The Australian Biological Resources Study for funding. F.A. Custódio and O.L. Pereira are thankful to the Coordenação de Aperfeiçoamento de Pessoal de Níve Superior, Brazil (CAPES), finance code 001, Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), and Conselho Nacional de Desenvolvimiento Científico e Tecnológico (CNPq) for their financial support. The authors acknowledge K. Bensch for the help with the Latin name of the new fungus. A. Miller would like to thank the Roy J. Carver Biotechnology Center at the University of Illinois for Sanger sequencing. J. Fournier is thankful to R. Courtecuisse (Lille University) for having organised field trips to Guadeloupe and Martinique from 2003 to 2016 and special thanks go to P.-A. Moreau who attended several of these forays and shared his image of C. Lechat at La Caravelle. V. Vasan, N. Chellappan, E. Arumugam, R. Murugadoss and M. Kaliyaperumal thank the Director, Centre of Advanced Studies in Botany, University of Madras for the laboratory facilities. M. Kaliyaperumal and E. Arumugam acknowledge the Tamil Nadu State Council for Higher Education (RGP/2019-20/MU/HECP-0040) for providing financial aid. The authors acknowledge C. Janagar Dhas, Centre of Advanced Studies in Botany, University of Madras for assistance with photography. R. Murugadoss would like to acknowledge CSIR – Junior Research Fellowship (JRF-NET), New Delhi, India (09/0115(13300)/2022-EMR-I) for the financial assistance. M. Kaliyaperumal and co-authors thank Dr. J. Bhat, for his suggestion with the nomenclature. M. Kaliyaperumal, E. Arumugam and S. Gunaseelan would like to thank Prof. N. Mathivanan, Director, Centre for Advanced Studies in Botany, University of Madras, Chennai, for providing laboratory facilities. The study of R. Jankowiak was funded by the Ministry of Science and Higher Education of the Republic of Poland (SUB/040013/ D019) and by the National Science Centre, Poland (contract no. 2021/41/B/ NZ8/03456). R.G. Thorn and co-authors thank the Walpole Island First Nation for providing permission to search for and collect Hygrophorus aesculeticola at Bkejwanong. Participation of M.E. Smith was supported by the US National Science foundation grant DEB-2106130 and NIFA-USDA award FLA-PLP-005289. Y. Lamoureux is acknowledged for microscopical work on his collection of Hygrophorus paludosoides. G. Delgado thanks W. Colbert and S. Ward (Eurofins Built Environment) for the provision of laboratory facilities. J.G. Maciá-Vicente acknowledges the support of the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) of the state of Hesse within the framework of the Cluster for Integrative Fungal Research (IPF) of Goethe University Frankfurt. Both authors thank H.-O. Baral for providing sequence data and his feedback on the phylogeny of Hymenoscyphus. V. Fachada and co-authors would like to thank C. Vieira (MHNC-UP) for managing the voucher collections in the PO herbarium. We are grateful for the valuable assistance provided by K. Bensch and S. Pennycook in determining the correct taxonomic nomenclature. E. Larsson acknowledges the Swedish Taxonomy Initiative, SLU Artdatabanken, Uppsala, Sweden. G.M. Jansen is grateful to J. Vauras for making collections from TUR available and for his valuable ideas about neighbouring species, to M. Gotink, M. van der Vegte and M. Plekkenpol for their collections and their help in financing the sequencing, to D. Bandini for supplying a gill of the isotype of Inocybe mytiliodora for sequencing, to P.B. Matheny for making his description and sequence of his collection PBM1572 available and for sharing his ideas for rooting the tree, and to the Ger van Zaanen Fund for financing the sequencing of L4343844. The description of Inocybe pseudomytiliodora was critically reviewed by J. Vauras and P.B. Matheny, and their valuable suggestions and improvements are gratefully acknowledged. E. Mazur acknowledges the staff of the Herbario Nacional de Bolivia, Instituto de Ecología, Universidad Mayor de San Andrés, La Paz, for their generous long-term cooperation with lichenologists in the W. Szafer Institute of Botany, Polish Academy of Sciences, the collectors: A. Flakus, K. Wilk and B. Cykowska for making their material available for the study, and L. Sliwa who supervised her doctoral thesis. E. Mazur was supported by statutory funds from the W. Szafer Institute of Botany, Polish Academy of Sciences, and the National Science Centre, Poland, project 2016/21/B/NZ8/02463. A.R. Podile acknowledges the Department of Science and Technology, Govt. of India for the award of the JC Bose Fellowship (Grant No. JCB/2017/000053) and the Ministry of Education, Govt. of India and Institution of Excellence Directorate, University of Hyderabad for the award of the project (Grant No.UOH-IOE-RC3-21-065). S. Mahadevakumar thanks the Director, KSCSTE - Kerala Forest Research Institute & Head of Office Botanical Survey of India, Andaman and Nicobar Regional Centre, Port Blair, M. Madappa, Department of Studies in Botany, University of Mysore and K.T. Mufeeda Forest Patho-logy Department, Kerala Forest Research Institute, Peechi, Thrissur for their kind support and technical assistance. K.G.G. Ganga acknowledges support from the University Grants Commission (UGC), India, in the form of a UGC research fellowship (Ref No. 20/12/2015(ii) EU-V). K.G.G. Ganga and co-authors also thank the authorities of the University of Calicut for providing facilities for this study. R.M. Sanchez and M.V. Bianchinotti acknowledge the National Council for Scientific and Technical Research (CONICET) for providing the funds for this research (PIP11220130100280CO). D.A. Acal would like to thank the Department of Science and Technology (DOST-SEI), Republic of Philippines for the financial support during the collection of material and the Department of Environmental and Natural Resources of Biodiversity Management Bureau (DENR-BMB), Republic of Philippines for the issuance of Wildlife Gratuitous Permits (no. 319). F. Fuljer was funded by the Operational Program of Integrated Infrastructure, co-financed with the European Fund for Regional Development (EFRD) ITMS2014+313021W683: ‘DNA barcoding of Slovakia (SK-BOL), as a part of international initiative International Barcode of Life (iBOL)’. The authors would also like to thank I. Kušan and N. Matočec for conveying valuable information about bioclimatic zones of localities. K. Hansen is thankful to Juan Carlos Zamora, Ibai Olariaga and Timo Kosonen for their help on field, microscopic and molecular work, to Henning Knudsen for nomenclatural advice, Donald H. Pfister for discussions on the new species, and to C, CUP, NICE and UPS for arranging loans of specimens, and the Swedish Taxonomy Initiative, SLU Artdatabanken, Sweden for providing funding for this research. S. Kumar and co-authors are grateful to the Director, KSCSTE-Kerala Forest Research Institute, Peechi for providing library and laboratory facilities. They also acknowledge the Science & Engineering Research Board (SERB), Department of Science & Technology (DST), Govt. of India for financial support (CRG/2019/005014). The research of K. Patejuk, W. Pusz and A. Baturo-Cieśniewska (in part) was funded by the Forest Fund under the agreement concluded between the State Forests National Forest Holding and the Wigry National Park (contracts no. EZ.0290.1.21.2021 and EZ.0290.1.21.2022). E.A. Ossowska and co-authors are grateful to the members of Herbario Nacional de Bolivia, Instituto of Herbario Nacional de Bolivia, Instituto de Ecología, Universidad Mayor de San Andrés, La Paz, for the generous cooperation and M. Kukwa for help with the description. This research received support from the SYNTHESYS Project (DE-TAF-8180) http://www.synthesys.info/ which is financed by European Community Research Infrastructure Action under the FP7 ‘Capacities’ Programme and University of Gdansk, granted to EAO. V. Darmostuk and co-authors acknowledge our colleagues and all staff of the Herbario Nacional de Bolivia, Instituto de Ecología, Universidad Mayor de San Andrés, La Paz, for their generous long-term cooperation. They would also like to thank the SERNAP (http://sernap.gob.bo), and all protected areas staff, for providing permits for scientific studies, as well as their assistance and logistical support during the field works. This research was financially supported by the National Science Centre (NCN) in Poland (grant number DEC-2013/11/D/NZ8/03274). The study of P. Eis-vand and M. Mehrabi-Koushki was financially supported by grant (SCU. AP1401.294) from the Research Council of Shahid Chamran University of Ahvaz. Financial support to M. Dueñas and colleagues was provided by Plan Nacional I+D+I project no. CGL2009-07231 and Ref. 202030E059. They also acknowledge M. Glenn (Seton Hall University, USA) for her kind English revision, and M. Ghobad-Nejhad for sending the ITS nrDNA alignment with sequences published in Ghobad-Nejhad & Duhem (2014). The study of M. Piątek and co-authors was funded by the National Science Centre, Poland, under the project 2017/27/B/NZ9/02902. V. Hubka was supported by a Czech Academy of Sciences Long-term Research Development Project (RVO: 61388971). A. Ismail and co-authors thank the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia, for supporting this research for work through grant number (GRANT 4597). M. Thines acknowledges funding by the LOEWE excellence initiative of the government of Hesse, in the framework of the Centre for Translational Biodiversity Genomics (TBG).
REFERENCES
- Aalto M. 1974. Amanita magnivolvata sp. nova (Agaricales). Karstenia 14: 93–96. [Google Scholar]
- Acharius E. 1810. Lichenographia Universal is. Frid. Danckwerts., Gottingen. [Google Scholar]
- Ajitomi A, Takushi T, Sato T, et al. 2017. First report of flyspeck of mango caused by Stomiopeltis sp. in Japan. Journal of General Plant Pathology 83: 299–303. [Google Scholar]
- Alidadi A, Kowsari M, Javan-Nikkham M, et al. 2019. Deniquelata quercina sp. nov.; a new endophyte species from Persian oak in Iran. Phytotaxa 405: 187–194. [Google Scholar]
- Almaraz T. 2002. Bases corológicas de Flora Micológica Ibérica. Numeros 1766–1932. In: Pando F, Hernández JC. (eds), Cuadernos de trabajo de Flora Micológica Ibérica 17: 1–124. [Google Scholar]
- Amandeep K, Atri NS, Munruchi K. 2015. Taxonomic study on the coprophilous mushrooms from Punjab, India: new records of family Agaricaceae. Current Research in Environmental & Applied Mycology 5: 27–45. [Google Scholar]
- Ariyawansa HA, Maharachchikumbura SS, Karunarathne SC, et al. 2013. Deniquelata barringtoniae gen. et sp. nov., associated with leaf spots of Barringtonia asiatica. Phytotaxa 105: 11–20. [Google Scholar]
- Ariyawansa HA, Thambugala KM, Manamgoda DS, et al. 2015. Towards a natural classification and backbone tree for Pleosporaceae. Fungal Diversity 71: 85–139. [Google Scholar]
- Armand A, Bundhun D, Bhunjun CS, et al. 2023. Paramyrothecium amorphophalli sp. nov., a causal agent of leaf blight on elephant foot yam in northern Thailand. Current Research in Environmental & Applied Mycology 13: 57–67. [Google Scholar]
- Arnolds E. 1990. Genus Hygrophorus. In: Bas C, Kuyper TW, Noordeloos ME, et al. (eds), Flora Agaricina Neerlandica 2: 115–133. Balkema, Lisse. [Google Scholar]
- Arup U, Ekman S, Grube M, et al. 2007. The sister group relation of Parmeliaceae (Lecanorales, Ascomycota). Mycologia 99: 42–49. [DOI] [PubMed] [Google Scholar]
- Aveskamp MM, De Gruyter J, Woudenberg JHC, et al. 2010. Highlights of the Didymellaceae: a polyphasic approach to characterise Phoma and related pleosporalean genera. Studies in Mycology 65: 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandini D, Huijser H. 2017. Vezelkoppen (Inocybe) van Ameland, Inocybegriseotarda Poirier. Coolia 60: 243–247. [Google Scholar]
- Bandini D, Oertel B, Eberhardt U. 2021. A fresh outlook on the smooth-spored species of Inocybe: type studies and 18 new species. Mycological Progress 20: 1019–1114. [Google Scholar]
- Baral HO, Weber E, Marson G. 2020. Monograph of Orbiliomycetes (Ascomycota) based on vital taxonomy. Part I + II. National Museum of Natural History Luxembourg. [Google Scholar]
- Bas C. 1969. Morphology and subdivision of Amanita and a monograph of its section Lepidella. Persoonia 5: 285–579. [Google Scholar]
- Bas C, Kuyper TW, Noordeloos ME, et al. 1995. Flora Agaricina Neerlandica 3. Critical monographs on families of agarics and boleti occurring in the Netherlands. Balkema, Rotterdam Brookfield. [Google Scholar]
- Bellanger JM, Lebeuf R, Sesli E, et al. 2021. Hygrophorus sect. Olivaceoumbrini: new boundaries, extended biogeography and unexpected diversity unravelled by transatlantic studies. Persoonia 46: 272–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkeley MJ, Broome CE. 1873. ‘1875. ’. Enumeration of the Fungi of Ceylon. Part II, containing the remainder of the Hymenomycetes, with the remaining established tribes of Fungi. Botanical Journal of the Linnean Society 14: 29–140. [Google Scholar]
- Bizio E, Ferisin G, Dovana F. 2017. Note sul campo di variabilità di Inocybe griseotarda. Rivista di Micologia 60: 59–70. [Google Scholar]
- Błońska E, Lasota J, Jankowiak R, et al. 2021. Biological and physicochemical properties of the nests of White Stork Ciconia ciconia reveal soil entirely formed, modified and maintained by birds. Science of the Total Environment 763: 43020. [DOI] [PubMed] [Google Scholar]
- Boldt-Burisch K, Douanla-Meli C. 2023. First report of Querciphoma minuta causing branch and stem canker in Platanus × hispanica in Germany. New Disease Reports 47: e12153. [Google Scholar]
- Bon M. 1978. Amanitopsis pachyvolvata. Documents Mycologiques 8: 36. [Google Scholar]
- Bon M. 1990. Flore mycologique du littoral - 05 - Inocybe. Documents Mycologiques 20: 61–66. [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, et al. 2014. BEAST 2: A software platform for bayesian evolutionary analysis. PLoS Computational Biology 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresadola G. 1903. Fungi polonici. Annales Mycologici 1: 97–131. [Google Scholar]
- Calduch M, Gené J, Guarro J, et al. 2002. Hyphomycetes from Nigerian Rain Forests. Mycologia 94: 127–135. [PubMed] [Google Scholar]
- Candusso M. 1997. Fungi Europaei 6: Hygrophorus s.l. Edizioni Candusso, Alassio, Italia. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chater AO. 1980. Carex L. In: Tutin TG, Heywood VH, Burges NA, et al. (eds), Flora Europaea. Vol. 5: 290–323. Cambridge University Press, Cambridge. [Google Scholar]
- Chernomor O, Von Haeseler A, Quang Minh B. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65: 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhetri PK, Pradhan BK, Chhetri B. 2019. First record of Ophiocordyceps dipterigena Berk. & Broome (Ophiocordycipitaceae) in the Himalayas. National Academy Science Letters 43: 367–369. [Google Scholar]
- Clemençon H. 1982. European Omphalinoid Tricholomataceae. Zeitschrift für Mykologie 48: 195–237. [Google Scholar]
- Coker TLR, Rozsypálek J, Edwards A, et al. 2019. Estimating mortality rates of European ash (Fraxinus excelsior) under the ash dieback (Hymenoscyphus fraxineus) epidemic. Plants, People, Planet 1: 48–58. [Google Scholar]
- Consiglio G, Contu M. 1999. Amanita dryophila (Amanitaceae) spec. nov. and the species of the section Vaginatae with a semifriable universal veil and ellipsoid spores. Persoonia 17: 287–190. [Google Scholar]
- Contu M. 1984. Novitates (2). Documents Mycologiques 14: 26. [Google Scholar]
- Contu M. 1999. Appunti sul genere Amanita IX Nuove specie e studi tassonomico-nomenclaturali nella sezione Vaginatae. Bollettino dell’Associazione Micologica ed Ecologica Romana 46: 3–22. [Google Scholar]
- Crous PW, Akulov A, Balashov S, et al. 2023. New and interesting fungi. 6. Fungal Systematics and Evolution 11: 109–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. 2020. Fungal Planet description sheets: 1112-1181. Persoonia 45: 251–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Cowan DA, Maggs-Kölling G, et al. 2021a. Fungal Planet description sheets: 1182-1283. Persoonia 46: 313–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ. 2017. The genera of fungi - G4: Camarosporium and Dothiora. IMA Fungus 8: 131–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Shivas RG, et al. 2011. Fungal Planet description sheets: 69–91. Persoonia 26: 108–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Hernández-Restrepo M, Schumacher RK, et al. 2021b. New and interesting fungi. 4. Fungal Systematics and Evolution 7: 255–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Luangsa-ard JJ, Wingfield MJ, et al. 2018a. Fungal Planet description sheets: 785–867. Persoonia 41: 238–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Mohammed C, Glen M, et al. 2007. Eucalyptus microfungi known from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Diversity 25: 19–36. [Google Scholar]
- Crous PW, Osieck ER, Jurjević Ž, et al. 2021c. Fungal Planet description sheets: 1284–1382. Persoonia 47: 178–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schoch CL, Hyde KD, et al. 2009. Phylogenetic lineages in the Capnodiales. Studies in Mycology 64: 17–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Schumacher RK, Wingfield MJ, et al. 2018b. New and interesting fungi. 1. Fungal Systematics and Evolution 1: 169–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Shivas RG, Quaedvlieg W, et al. 2014. Fungal Planet description sheets: 214–280. Persoonia 32: 184–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, et al. 2016. Fungal Planet description sheets: 469–557. Persoonia 37: 218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, et al. 2013. Fungal Planet description sheets: 154–213. Persoonia 31: 188–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, et al. 2015. Fungal Planet description sheets: 320–370. Persoonia 34: 167–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, et al. 2019. Fungal Planet description sheets: 951–1041. Persoonia 43: 223–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Posada D, Kozlov AM, et al. 2020. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37: 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, et al. 2012. jModel test 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EA, Yakovchenko LS, Hollinger J, et al. 2021. The new genus Pulvinora (Lecanoraceae) for species of the ‘Lecanora pringlei’ group, including the new species Pulvinora stereothallina. Bryologist 124: 242–256. [Google Scholar]
- Dayarathne MC, Jones EBG, Maharachchikumbura SSN, et al. 2020. Morphomolecular characterization of microfungi associated with marine based habitats. Mycosphere 11: 1–188. [Google Scholar]
- De Beer ZW, Marincowitz S, Duong TA, et al. 2016. Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biology 120: 1323–1340. [DOI] [PubMed] [Google Scholar]
- Denchev CM. 2001. Class Ustomycetes (Orders Tilletiales, Ustilaginales, and Graphiolales). In: Fakirova V. (ed.), Fungi of Bulgaria. Vol. 4: 1–286. Editio Academica ‘Prof. Marin Drinov’ & Editio Pensoft, Sofia. [Google Scholar]
- Denchev TT, Denchev CM, Begerow D, et al. 2023. Anthracoidea obtusatae (Anthracoideaceae, Ustilaginales), a new smut fungus on Carex obtusata (Cyperaceae) from Central Asia. Phytotaxa 595: 139–148. [Google Scholar]
- Denchev TT, Denchev CM, Koopman J, et al. 2021. Host specialization and molecular evidence support a distinct species of smut fungus, Anthracoidea hallerianae (Anthracoideaceae), on Carex halleriana (Cyperaceae). Willdenowia 51: 57–67. [Google Scholar]
- Denchev TT, Denchev CM, Michikawa M, et al. 2013. The genus Anthracoidea (Anthracoideaceae) in Japan and some adjacent regions. Mycobiota 2: 1–125. [Google Scholar]
- Devadatha B, Sarma VV, Ariyawansa HA, et al. 2018. Deniquelata vittalii sp. nov., a novel Indian saprobic marine fungus on Suaeda monoica and two new records of marine fungi from Muthupet mangroves, East coast of India. Mycosphere 9: 565–582. [Google Scholar]
- Dos Santos LA, Aptroot A, Lücking R, et al. 2023. Lecanora s.lat. (Ascomycota, Lecanoraceae) in Brazil: DNA barcoding coupled with phenotype characters reveals numerous novel species. Journal of Fungi 9: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Cao TX, Wu YD, et al. 2021. Two new species of Hymenochaetaceae on Dracaena cambodiana from tropical China. MycoKeys 80: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edler D, Klein J, Antonelli A, et al. 2021. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12: 373–377. [Google Scholar]
- Edler F, Miccoli I, Pfnur H, et al. 2019. Space charge layer effects in silicon studied by in situ surface transport. Journal of Physics: Condensed Matter 31: 1–8. [DOI] [PubMed] [Google Scholar]
- Egidi E, De Hoog GS, Isola D. et al. 2014. Phylogeny and taxonomy of meristematic rock-inhabiting black fungi in the Dothideomycetes based on multi-locus phylogenies. Fungal Diversity 65: 127–165. [Google Scholar]
- Egorova TV. 1999. The sedges (Carex L.) of Russia and adjacent states (within the limits of the former USSR). Saint Petersburg State Chemical-Pharmaceutical Academy, Saint Petersburg, Russia & Missouri Botanical Garden Press, St. Louis, USA. [Google Scholar]
- Ekanayaka AH, Hyde KD, Jones EBG, et al. 2017. A new species of Trichoglossum (Geoglossales, Ascomycota) from Thailand. Phytotaxa 316: 11–170. [Google Scholar]
- Enderle M, Krieglsteiner GJ, Bender H. 1986. Studien zur Gattung Coprinus (Pers.: Fr.) SF Gray in der Bundesrepublik Deutschland. III. Zeitschrift für Mykologie 52: 101–132. [Google Scholar]
- Etayo J. 2001. Hongos liquenícolas de Ecuador. I. Dos especies nuevas del orden Hypocreales (Ascomycota): Pronectria parmotrematis y Trichonectria leptogiicola. Anales del Jardín Botánico de Madrid 58: 219–222. [Google Scholar]
- Etayo J, Sancho LG. 2008. Hongos liquenícolas del Sur de Sudamérica, especialmente de Isla Navarino (Chile). Bibliotheca Lichenologica 98: 1–302. [Google Scholar]
- Etayo J, Van den Boom PPG. 2005. Contribution to the lichen flora of the Canary Islands. VIII. Some lichenicolous fungi. Nova Hedwigia 81: 157–162. [Google Scholar]
- Fathima M, Usman M, Khalid AN. 2023. Fulvifomes aurantiacus sp. nov. (Basidiomycota; Hymenochaetaceae) from Pakistan. Phytotaxa 599: 078–088. [Google Scholar]
- Favre J. 1955. Les champignons supérieurs de la zone alpine du Parc National Suisse. Ergebnisse der Wissensnschaftlichen Untersuchungen des Schweizerrischen National Parks 5: 1–212. [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Ferrari E. 2010. Inocybe dai litorali alla zona alpina. Fungi Non Delineati Pars LIV–LV: 1–212. [Google Scholar]
- Fletcher A, Purvis OW, Coppins BJ. 2009. Aspicilia A. Massal. In: Smith CW, Aptroot A, Coppins BJ, et al. (eds), The lichens of Great Britain and Ireland: 181–188. The British Lichen Society, London. [Google Scholar]
- Fournier J, Lechat C. 2011. Eutypella phaeospora, a new species on Chenopodiaceae. Mycotaxon 118: 441–446. [Google Scholar]
- Fraiture A. 1993. Les Amanitopsis d’Europe (genre Amanita, Agaricales, Fungi) Synthèse critique de la littérature. Opera Botanica Belgica 5: 1–128. [Google Scholar]
- Friedrich RC, Shrestha B, Salvador-Montoya CA, et al. 2018. Ophiocordyceps neonutans sp. nov., a new neotropical species from O. nutans complex (Ophiocordycipitaceae, Ascomycota). Phytotaxa 344: 215–27. [Google Scholar]
- Fujimori S, Abe JP, Okane I, et al. 2019. Three new species in the genus Tulasnella isolated from orchid mycorrhiza of Spiranthes sinensis var. amoena (Orchidaceae). Mycoscience 60: 71–81. [Google Scholar]
- Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 12: 543–548. [DOI] [PubMed] [Google Scholar]
- Gams W. 1971. Cephalosporium-artige Schimmelpilze (Hyphomycetes). Fischer Publishing, Stuttgart, Germany. [Google Scholar]
- Gams W, Holubová-Jechová V. 1976. Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Studies in Mycology 13: 1–99. [Google Scholar]
- Gams W, Stielow B, Gräfenhan T, et al. 2019. The ascomycete genus Niesslia and associated monocillium-like anamorphs. Mycological Progress 18: 5–76. [Google Scholar]
- Gams W, Zare R. 2001. A revision of Verticillium sect. Prostrata. III. Generic classification. Nova Hedwigia 72: 47–55. [Google Scholar]
- Gao H, Yin J, Li Y. et al. 2022. Introducing Querciphoma styphnolobii sp. nov., the first sexual morph of Querciphoma (Leptosphaeriaceae, Pleosporales). Phytotaxa 555: 279–290. [Google Scholar]
- Ghobad-Nejhad M, Duhem B. 2014. Novelties in the Corticiales: Vuillemina nilsii sp. nov. and Dendrominia gen. nov. (Basidiomycota). Mycological Progress 13: 1–11. [Google Scholar]
- Gibbs T. 1908. A new Coprinus. The Naturalist 614: 100. [Google Scholar]
- Giraldo A, Crous PW. 2019. Inside Plectosphaerellaceae. Studies in Mycology 92: 227–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo A, Hernández-Restrepo M, Crous PW. 2019. New plectosphaerellaceous species from Dutch garden soil. Mycological Progress 18: 1135–1154. [Google Scholar]
- Glenn MG, Gomez-Bolea A, Orsi EV. 1997. Effects of thallus damage on interactions of lichens with non-lichenized fungi under natural and laboratory conditions. The Lichenologist 29: 51–65. [Google Scholar]
- Glynou K, Ali T, Buch A-K, et al. 2016. The local environment determines the assembly of root endophytic fungi at a continental scale. Environmental Microbiology 18: 2418–2434. [DOI] [PubMed] [Google Scholar]
- Glynou K, Thines M, Maciá-Vicente JG. 2018. Host species identity in annual Brassicaceae has a limited effect on the assembly of root-endophytic fungal communities. Plant Ecology & Diversity 11: 569–580. [Google Scholar]
- Goettel MS, Koike M, Kim JJ, et al. 2008. Potential of Lecanicillium spp. for management of insects, nematodes and plant diseases. Journal of Invertebrate Pathology 98: 256–261. [DOI] [PubMed] [Google Scholar]
- Gomes RR, Glienke C, Videira SI, et al. 2013. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 31: 1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves MFM, Aleixo A, Vicente TFL, et al. 2019. Three new species of Neocamarosporium isolated from saline environments: N. aestuarinum sp. nov., N. endophyticum sp. nov. and N. halimiones sp. nov. Mycosphere 10: 608–621. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Grube M, Baloch E, Arup U. 2004. A phylogenetic study of the Lecanora rupicola group (Lecanoraceae, Ascomycota). Mycological Research 108: 506–514. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hagestad OC, Hou L, Andersen JH, et al. 2021. Genomic characterization of three marine fungi, including Emericellopsis atlantica sp. nov. with signatures of a generalist lifestyle and marine biomass degradation. IMA Fungus 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hanss JM, Moreau PA. 2020. Une révision des Amanites «vaginées» (Amanita sect. Vaginatae) en Europe, 1re partie: quelques Amanites argentées. Bulletin de la Société Mycologique de France 133: 67–141. [Google Scholar]
- Hattori T, Ota Y, Sotome K. 2022. Two new species of Fulvifomes (Basidiomycota, Hymenochaetaceae) on threatened or near threatened tree species in Japan. Mycoscience 63: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Sakayaroj J, Jones EBG, et al. 2014. Three species of Fulvifomes (Basidiomycota, Hymenochaetales) associated with rots on mangrove tree Xylocarpus granatum in Thailand. Mycoscience 55: 344–354. [Google Scholar]
- Hendrichs M, Begerow D, Bauer R, et al. 2005. The genus Anthracoidea (Basidiomycota, Ustilaginales): a molecular phylogenetic approach using LSU rDNA sequences. Mycological Research 109: 31–40. [DOI] [PubMed] [Google Scholar]
- Heredia G, Li DW, Wendt L, et al. 2020. Natonodosa speciosa gen. et sp. nov. and rediscovery of Poroisariopsis inornata: neotropical anamorphic fungi in Xylariales. Mycological Progress 19: 15–30. [Google Scholar]
- Hesler LR, Smith AH. 1963. North American species of Hygrophorus. University of Tennessee Press, Knoxville. [Google Scholar]
- Hietala AM, Agan A, Nagy NE, et al. 2022. The native Hymenoscyphus albidus and the invasive Hymenoscyphus fraxineus are similar in their necrotrophic growth phase in ash leaves. Frontiers in Microbiology 13: 892051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobart C, Henrici A. 2011. Inocybe mytiliodora new to Britain. Field Mycology 12: 77–80. [Google Scholar]
- Hongsanan S, Hyde KD, Phookamsak R, et al. 2020. Refined families of Dothideomycetes: orders and families incertae sedis in Dothideomycetes. Fungal Diversity 105: 17–318. [Google Scholar]
- Hosen MI, Song ZP, Gates G, et al. 2017. Two new species of Xanthagaricus and some notes on Heinemannomyces from Asia. Mycokeys 28: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosen MI, Song ZP, Gates G, et al. 2018. Xanthagaricus caeruleus, a new species with ink-blue lamellae from southeast China. Mycoscience 59: 188–192. [Google Scholar]
- Hosen MI, Yang ZL. 2013. Coniolepiota spongodes (Agaricaceae, Basidiomycota) in Bangladesh and China. Mycotaxon 124: 341–347. [Google Scholar]
- Hosoya T, Maruyama K. 2004. Pachyella globispora sp. nov. (Pezizaceae) from Japan. Mycoscience 45: 112–115. [Google Scholar]
- Hou LW, Giraldo A, Groenewald JZ, et al. 2023. Redisposition of acremonium-like fungi in Hypocreales. Studies in Mycology 105: 23–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou LW, Groenewald JZ, Pfenning LH, et al. 2020. The phoma-like dilemma. Studies in Mycology 96: 309–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Hussain S, Afshan NS, Ahmad H, et al. 2018. Xanthagaricus pakistanicus sp. nov. (Agaricaceae), the first report of the genus from Pakistan. Turkish Journal Botany 42: 123–133. [Google Scholar]
- Hyde KD, Chaiwan N, Norphanphoun C, et al. 2018. Mycosphere notes: 169–224. Mycosphere 9: 271–430. [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ, et al. 2020. The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 103: 219–271. [Google Scholar]
- Illescas T. 2022. Amanita dryophila f. grisea, f. nov., una forma no descrita de una Amanita poco conocida. Boletín Informativo de la Sociedad Micológica Extremeña 22: 13–19. [Google Scholar]
- Illescas T, Plaza M. 2022. Amanita calida sp. nov., una nueva especie europea de Amanita sect. Vaginatae. Fungi Iberici 2: 41–54. [Google Scholar]
- Jaklitsch WM, Voglmayr H. 2012. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales). Fungal Diversity 52: 75–98. [Google Scholar]
- Jayasiri SC, Hyde KD, Jones EBG, et al. 2019. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10: 1–186. [Google Scholar]
- Jayawardena RS, Hyde KD, Wang S, et al. 2022. Fungal diversity notes 1512–1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 117: 1–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji XH, Wu F, Dai YC, et al. 2017. Two new species of Fulvifomes (Hymenochaetales, Basidiomycota) from America. MycoKeys 22: 1–13. [Google Scholar]
- Johnston PR, Quijada L, Smith CA, et al. 2019. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. 2017. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatygin IV, Simonyan SA. 1985. Species Ustilaginalium pro URSS novae et rarae in Armenia et Georgia inventae. Novosti Sistematiki Nizshikh Rastenii 22: 119–121. [In Russian.] [Google Scholar]
- Kärcher R. 1988. Une variété nouvelle d’Amanitopsis sous Fagaceae: Amanita vaginata (Bull. : Fr.) Quél. var. elongata v. nov. Documents Mycologiques 19: 53–55. [Google Scholar]
- Kärcher R, Contu M. 1999. Amanita praelongipes Kärcher & Contu, nom. nov. e un parametron micromorfologico rilevante per la taxificazione nel genere Amanita sect. Vaginatae. Bollettino Gruppo Micologico G. Bresadola 42: 351–359. [Google Scholar]
- Katoh K, Misawa K, Kuma K, et al. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26: 1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirle MR, Hemmes DE, Desjardin DE. 2004. Agaricales of the Hawaiian Islands. 8. Agaricaceae: Coprinus and Podaxis; Psathyrellaceae: Coprinopsis, Coprinellus and Parasola. Fungal Diversity 15: 33–124. [Google Scholar]
- Kelly KL, Judd DB. 1976. COLOR. Universal language and dictionary of names. National Bureau of Standards, Special Publication; 440. [Google Scholar]
- Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, et al. 2017. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8: 335–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khao-ngam S, Mongkolsamrit N, Rungjjndamai W, et al. 2021. Ophiocordyceps asiana and Ophiocordyceps tessaratomidarum (Ophiocordycipitaceae, Hypocreales), two new species on stink bugs from Thailand. Mycological Progress 20: 341–353. [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kirika P, Parnmen S, Lumbsch T. 2012. Two new species of Lecanora sensu stricto (Lecanoraceae, Ascomycota) from east Africa. MycoKeys 3: 37–47. [Google Scholar]
- Kondratyuk S, Popova LP, Khodosovtsev OY, et al. 2021. The fourth checklist of Ukrainian lichen-forming and lichenicolous fungi with analysis of current additions. Acta Botanica Hungarica 63: 97–163. [Google Scholar]
- Kornerup A, Wanscher JH. 1961. Methuen handbook of colour, 1st edn. Eyre Methuen, London. [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour, 3rd ed. Eyre Methuen, London. [Google Scholar]
- Kosonen T, Huhtinen S, Hansen K. 2021. Taxonomy and systematics of Hyaloscyphaceae and Arachnopezizaceae. Persoonia 46: 26–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukol O. 2012. A new species of Infundichalara from pine litter. Mycotaxon 120: 343–352. [Google Scholar]
- Kozlov AM, Darriba D, Flouri T, et al. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen R. 2011. Pachyella hydrophila (Pezizales) – the first finding in Europe. Agarica 30: 97–102. [Google Scholar]
- Kück P, Meusemann K. 2010. FASconCAT: Convenient handling of data matrices. Molecular Phylogenetics and Evolution 56: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Kühner R. 1988. Diagnoses de quelques nouveaux Inocybes récoltés en zone alpine de la Vanoise (Alpes françaises). Documents Mycologiques 19: 1–27. [Google Scholar]
- Kühner R, Lamoure D. 1972. Agaricales de la zone alpine. Pleurotacées. Le Botaniste 55: 7–37. [Google Scholar]
- Kukkonen I. 1963. Taxonomic studies on the genus Anthracoidea (Ustilaginales). Annales Botanici Societatis Zoologicae Botanicae Fennicae ‘Vanamo’ 34: 1–122. [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumla J, Suwannarach N, Sringernyuang K, et al. 2018. Xanthagaricus thailandensis sp. nov. (Agaricales, Basidiomycota), from northern Thailand. Phytotaxa 348: 109–117. [Google Scholar]
- Kumla J, Suwannarach N, Wannathes N. 2021. Hymenagaricus saisamornae sp. nov. (Agaricales, Basidiomycota) from Northern Thailand. Chiang Mai Journal of Science 48: 827–836. [Google Scholar]
- Kuyper TW. 1986. A revision of the genus Inocybe in Europe 1. Subgenus Inosperma and the smooth-spored species of subgenus Inocybe. Persoonia Supplement 3: 1–247. [Google Scholar]
- Lanfear R, Calcott B, Ho SYW, et al. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, et al. 2016. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. [DOI] [PubMed] [Google Scholar]
- Lawrence DP, Gannibal PB, Peever TL, et al. 2013. The sections of Alternaria: Formalizing species-groups concepts. Mycologia 105: 530–546. [DOI] [PubMed] [Google Scholar]
- Lebeuf R, Alexandrova AV, Cerna-Mendoza A, et al. 2021. Fungal Systematics and Evolution: FUSE 8. Sydowia 74: 193–249. [Google Scholar]
- Loizides M, Bellanger JM, Yiangou Y. et al. 2018. Preliminary phylogenetic investigations into the genus Amanita (Agaricales) in Cyprus, with a review of previous records and poisoning incidents. Documents Mycologiques 37: 201–218. [Google Scholar]
- Lu L, Karunarathna SC, Dai DQ, et al. 2022. Description of four novel species in Pleosporales associated with coffee in Yunnan, China. Journal of Fungi 8: 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig E. 2001. Pilzkompendium Band 1. Beschreibungen. IHW Verlag. Eching. [Google Scholar]
- Lumbsch HT, Elix J. 2004. Lecanora. Flora Australia 56A: 10. [Google Scholar]
- Lumyong S. 2020. Xanthagaricus siamensis sp. nov. (Agaricaceae), a new species with dull green lamellae from northern Thailand. Phytotaxa 437: 14–22. [Google Scholar]
- Luo H, Tao YQ, Fan XY, et al. 2018. Identification and characterization of Alternaria iridiaustralis causing leaf spot on Iris ensata in China. Mycobiology 46: 168–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzoni FM. 1997. Phylogeny of lichen- and non-lichen forming omphalinoid mushrooms and the utility for testing for combinability among multiple data sets. Systematic Biology 46: 373–406. [DOI] [PubMed] [Google Scholar]
- Magain N, Sérusiaux E. 2015. Dismantling the treasured flagship lichen Sticta fuliginosa (Peltigerales) into four species in Western Europe. Mycological Progress 14: e97. [Google Scholar]
- Malícek J, Berger F, Palice Z, et al. 2017. Corticolous sorediate Lecanora species (Lecanoraceae, Ascomycota) containing atranorin in Europe. The Lichenologist 49: 431–455. [Google Scholar]
- Mark K, Cornejo C, Keller C, et al. 2016. Barcoding lichen-forming fungi using 454 pyrosequencing is challenged by artifactual and biological sequence variation. Genome 59: 685–704. [DOI] [PubMed] [Google Scholar]
- Martinez M, Salvador-Montoya CA, De Errasti A, et al. 2023. Fulvifomes wrightii (Hymenochaetales), a new species related to F. robiniae from Argentina and Paraguay. Fungal Systematics and Evolution 12: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield DA, Karakaya A, Batzer JC, et al. 2013. Diversity of sooty blotch and flyspeck fungi from apples in northeastern Turkey. European Journal of Plant Pathology 135: 805–815. [Google Scholar]
- Medeiros ID, Mazur E, Miadlikowska J, et al. 2021. Turnover of lecanoroid mycobionts and their Trebouxia photobionts along an elevation gradient in Bolivia highlights the role of environment in structuring the lichen symbiosis. Frontiers in Microbiology 12: 774839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Printzen C. 2000. Proposal for a standardized nomenclature and characterization of insoluble lichen pigments. The Lichenologist 32: 571–583. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA: 1–8. [Google Scholar]
- Minh BQ, Nguyen MA, Von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30: 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. 2020. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncada B. 2012. El género Sticta (Schreb.) Ach. en Colombia, Taxonomía, Ecogeografía e Importancia. Doctoral thesis, Universidad Nacional de Colombia, Bogotá. [Google Scholar]
- Mongkolsamrit S, Noisripoom W, Tasanathai K, et al. 2022. Comprehensive treatise of Hevansia and three new genera Jenniferia, Parahevansia and Polystromomyces on spiders in Cordycipitaceae from Thailand. MycoKeys 91: 113–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsell A. 2009. Munsell book of soil color charts 2009 revision. X-Rite, Incorporated. Grand Rapids. Michigan. [Google Scholar]
- Munsell AH. 1919. A color notation. Munsell Color Company. [Google Scholar]
- Neville P, Poumarat S. 2009. Quelques espèces nouvelles ou mal délimitées d'Amanita de la sous-section Vaginatinae. 1er complément à Amaniteae, Fungi Europaei 9. Fungi non Delineati 51–52: 1–197. [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, et al. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa J, Nakashima C. 2020. Japanese species of Alternaria and their species boundaries based on host range. Fungal Systematics and Evolution 5: 197–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordin A, Savic S, Tibell L. 2010. Phylogeny and taxonomy of Aspicilia and Megasporaceae. Mycologia 102: 1339–1349. [DOI] [PubMed] [Google Scholar]
- Ohmaki A, Okane I, Crous PW, et al. 2023. Cylindromonium dirinariae sp. nov. (Ascomycota, Hypocreales), a new nectrioid lichenicolous species on Dirinaria applanata in Japan. Fungal Systematics and Evolution 11: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olou BA, Ordynets A, Langer E. 2019. First new species of Fulvifomes (Hymenochaetales, Basidiomycota) from tropical Africa. Mycological Progress 18: 1383–1393. [Google Scholar]
- Ortega A, Contu M. 2003. Sobre algunas especies interesantes del género Amanita Sección Vaginatae en Andalucía (España). Revista Catalana de Micología 25: 71–77. [Google Scholar]
- Orton PD, Watling R. 1979. British Fungus Flora, Agarics and Boleti. 2 Coprinaceae part 1: Coprinus. Her Majesty’s Stationery Office. Edinburgh. [Google Scholar]
- Ossowska EA. 2021. First records of Sticta weigelii s.str. from Bolivia confirmed by molecular data. Folia Cryptogamica Estonica 58: 65–72. [Google Scholar]
- Ossowska EA, Kosecka M, Jaskólska J, et al. 2022b. Two taxa of the genus Sticta (Peltigerales, Ascomycota), S. andina and S. scabrosa subsp. scabrosa, new to Bolivia confirmed by molecular data. Plant and Fungal Systematics 67: 45–54. [Google Scholar]
- Ossowska EA, Moncada B, Kukwa M, et al. 2022a. New species of Sticta (lichenised Ascomycota, lobarioid Peltigeraceae) from Bolivia suggest a high level of endemism in the Central Andes. MycoKeys 92: 131–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owe-Larsson B, Nordin A, Tibell L. 2007. Aspicilia. In: Nash TH, III, Ryan BD, Diederich P. et al. (eds), Lichen flora of the Greater Sonoran Desert Region, Vol. 3: 61–108. Tempe, Lichens Unlimited, Arizona State University. [Google Scholar]
- Papizadeh M, Wijayawardene NN, Amoozegar MA, et al. 2018. Neocamarosporium jorjanensis, N. persepolisi, and N. solicola spp. nov. (Neocamarosporiaceae, Pleosporales) isolated from saline lakes of Iran indicate de possible halotolerant nature for the genus. Mycological Progress 17: 661–679. [Google Scholar]
- Papong K, Boonpragob K, Parnmen S, et al. 2012. Molecular phylogenetic studies on tropical species of Lecanora sensu stricto (Lecanoraceae, Ascomycota). Nova Hedwigia 96: 1–13. [Google Scholar]
- Perera RH, Hyde KD, Jones EBG, et al. 2023. Profile of Bionectriaceae, Calcarisporiaceae, Hypocreaceae, Nectriaceae, Tilachlidiaceae, Ijuhyaceae fam. nov., Stromatonectriaceae fam. nov. and Xanthonectriaceae fam. nov. Fungal Diversity 118: 95–271. [Google Scholar]
- Persoon CH. 1828. Mycologia Europaea 3. Palmius, Erlangen. [Google Scholar]
- Petch T. 1931. Notes on entomogenous fungi. Transactions of the British Mycological Society 16: 55–75. [Google Scholar]
- Pfister DH. 1973. The psilopezioid fungi. IV. The genus Pachyella (Pezizales). Canadian Journal of Botany 51: 2009–2023. [Google Scholar]
- Pfister DH. 1979. Type studies in the genus Peziza. VI. Species described by C.H. Peck. Mycotaxon 8: 333–338. [Google Scholar]
- Pfister DH. 1995. The psilopezioid fungi IX. Pachyella habrospora, a new species from Brazil. Mycotaxon 54: 393–396. [Google Scholar]
- Pfister DH, Candoussau F. 1981. The psilopezioid fungi. VIII. Additions to the genus Pachyella. Mycotaxon 13: 457–464. [Google Scholar]
- Pfister DH, Matocec N, Kušan I. 2008. ‘2009. ’. Integrated studies in the classification of the Pezizaceae. Re-evaluation of the genus Pachyella with a new segregate genus Adelphella. Mycologia Montenegrina 11: 7–17. [Google Scholar]
- Piątek M, Lutz M, Nobis M, et al. 2015. Phylogeny and morphology of Anthracoidea pamiroalaica sp. nov. infecting the endemic sedge Carex koshewnikowii in the Pamir Alai Mts (Tajikistan). Mycological Progress 14: 120. [Google Scholar]
- Poirier J. 2002. Notes sur le genre Inocybe - 1. Documents Mycologiques 31: 3–13. [Google Scholar]
- Prematunga C, Boonmee S, Jones EBG, et al. 2023. Neocamarosporium aquaticum (Neocamarosporiaceae, Dothideomycetes), a novel fungus from salt marsh habitats. Botanica Marina 66: 271–279. [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: Computing large minimum-evolution trees with profiles instead of a distance matrix. Molecular Biology and Evolution 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: Approximately maximum-likelihood trees for large alignments. PLoS ONE 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusz W, Patejuk K, Kaczmarek-Pieńczewska A. 2022. Mycobiota of infected organs of small balsam (Impatiens parviflora DC.) seeds in Wigry National Park. Progress in Plant Protection 62: 37–43. [Google Scholar]
- Qiao M, Zheng H, Guo J-S, et al. 2021. Two new asexual genera and six new asexual species in the family Microthyriaceae (Dothideomycetes, Ascomycota) from China. MycoKeys 85: 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Y, Deng S, Prenafeta-Boldủ FX. et al. 2023. The origin of human pathogenicity and biological interactions in Chaetothyriales. Fungal Diversity 10.1007/s13225-023-00518-3. [DOI]
- Quan Y, Muggia L, Moreno LF, et al. 2020. A re-evaluation of the Chaetothyriales using criteria of comparative biology. Fungal Diversity 103: 47–85. [Google Scholar]
- Quélet L. 1886. Enchiridion Fungorum in Europa media et praesertim in Gallia Vigentium.
- Quijada L, Huhtinen S, Beltrán-Tejera E. 2015. Studies in Hyaloscyphaceae associated with major vegetation types in the Canary Islands I: Cistella and Hyphodiscus. Willdenowia 45: 131–146. [Google Scholar]
- Rafiqi M, Kosawang C, Peers JA, et al. 2023. Endophytic fungi related to the ash dieback causal agent encode signatures of pathogenicity on European ash. IMA Fungus 14: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. 2009. FigTree ver. 1.4.3. http://tree.bio.ed.ac.uk/software/figtree [accessed 4 Oct. 2016].
- Rannala B, Yang Z. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. [DOI] [PubMed] [Google Scholar]
- Rappaz F. 1987. Taxonomie et nomenclature des Diatrypacées à asques octosporus. Mycologia Helvetica 2: 285–648. [Google Scholar]
- Rappaz F. 1992. Anthostoma decipiens et sa position systématique. Mycologia Helvetica 5: 21–32. [Google Scholar]
- Rashmi M, Kushveer JS, Sarma VV. 2019. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10: 798–1079. [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. CMI and British Mycological Society. Kew, Surrey, UK. [Google Scholar]
- Réblová M, Gams W, Štěpánek V. 2011. The new hyphomycete genera Brachyalara and Infundichalara, the similar Exochalara and species of ‘Phialophora sect. Catenulatae’ (Leotiomycetes). Fungal Diversity 46: 67–86. [Google Scholar]
- Réblová M, Štěpánek V. 2018. Introducing the Rhamphoriaceae, fam. nov. (Sordariomycetes), two new genera, and new life histories for taxa with Phaeoisaria- and Idriella-like anamorphs. Mycologia 110: 750–770. [DOI] [PubMed] [Google Scholar]
- Rico VJ. 1999. Aspicilia crespiana, a new lichen species from southern Europe. Lichenologist 31: 129–139. [Google Scholar]
- Ridgway R. 1912. Color standards and color nomenclature. Ridgway, Washington, DC. [Google Scholar]
- Roalson EH, Jiménez-Mejías P, Hipp AL, et al. 2021. A framework infrageneric classification of Carex (Cyperaceae) and its organizing principles. Journal of Systematics and Evolution 59: 726–762. [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, et al. 2012. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematics Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman AY. 1983. The phragmosporous species of Nectria and related genera. Mycological Papers 150: 1–164. [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, et al. 1999. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Studies in Mycology 42: 1–248. [Google Scholar]
- Roux C, Bertrand M, Nordin A. 2016. Aspicilia serenensis Cl. Roux et M. Bertrand sp. nov., espèce nouvelle de lichen (groupe d’A. calcarea, Megasporaceae). Bulletin de la Societe Linneenne de Provence 67: 165–182. [Google Scholar]
- Ryberg M, Larsson E, Jacobsson S. 2010. An evolutionary perspective on morphology and ecological characters in the mushroom family Inocybaceae (Agaricomycotina, Fungi). Molecular Phylogenetics and Evolution 55: 431–442. [DOI] [PubMed] [Google Scholar]
- Ryvarden L. 2004. Neotropical polypores Part 1 – Introduction, Ganodermataceae and Hymenochaetaceae. Synopsis Fungorum 19: 1–227, Fungiflora, Oslo, Norway. [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Salgado-Neto G, Sanz-Veiga PA, Braz Vaz MA. 2018. First record of Ophiocordyceps dipterigena (Ascomycota: Hypocreales: Ophiocordycipitaceae) infecting adults of Melanagromyza sojae (Diptera: Agromyzidae) in Brazil. Ciência Rural 48.
- Salvador-Montoya CA, Popoff OF, Reck M, et al. 2018. Taxonomic delimitation of Fulvifomes robiniae (Hymenochaetales, Basidiomycota) and related species in America: F. squamosus sp. nov. Plant Systematics and Evolution 304: 445–459. [Google Scholar]
- Samson RA, Brady BL. 1982. Akanthomyces novoguineensis sp. nov. Transactions of the British Mycological Society 79: 571–572. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert K, Morgan-Jones G, Gams W, et al. 2011. The genera of Hyphomycetes. CBS Biodiversity Series no. 9: 1-997. CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands. [Google Scholar]
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. [Google Scholar]
- Simmons EG. 2007. Alternaria. An identification manual. CBS Biodiversity Series 6. CBS Fungal Biodiversity Centre, Utrecht, The Netherlands. [Google Scholar]
- Sohrabi M, Stenroos S, Myllys L, et al. 2013. Phylogeny and taxonomy of the ‘manna lichens’. Mycological Progress 12: 231–269. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangl J. 1989. Die Gattung Inocybe in Bayern. Hoppea 46: 5–388. [Google Scholar]
- Sung G, Hywel-Jones NL, Sung J-M, et al. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP: Phylogenetic AnalysisUusing Parsimony (and other methods). Version 4.0b10. Sunderland, MA, Sinauer Associates. [Google Scholar]
- Swofford DL. 2003. PAUP* 4.0b10. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland, MA, USA. [Google Scholar]
- Syed MF, Saba M, Chattha SM, et al. 2023. Hymenagaricus pakistanicus (Agaricaceae, Agaricales), a new species from Pakistan based on morphological and molecular evidence. Phytotaxa 594: 292–300. [Google Scholar]
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: Molecular evolutionary genetics analysis version 11. Molecular Biology and Evolution 38: 3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Bishop-Hurley SL, Shivas RG, et al. 2022. Fungal Planet description sheets: 1436-1477. Persoonia 49: 261–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YP, Shivas R.G. 2023. Index of Australian Fungi 8: 1–14. [Google Scholar]
- Tan YP, Shivas RG. 2022. Nomenclatural novelties. Index of Australian Fungi 1: 1–18. [Google Scholar]
- Tasanathai K, Noisripoom W, Chaitika T, et al. 2019. Phylogenetic and morphological classification of Ophiocordyceps species on termites from Thailand. MycoKeys 56: 101–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchoumi JMT, Coetzee MPA, Rajchenberg M, et al. 2020. Poroid Hymenochaetaceae associated with trees showing wood-rot symptoms in the Garden Route National Park of South Africa. Mycologia 112: 722–741. [DOI] [PubMed] [Google Scholar]
- Tibuhwa DD, Mwanga ZN. 2014. A comprehensive study on Agaricus-like mushrooms from Mwalimu JK Nyerere Mlimani Campus. Tanzania Journal of Biology, Agriculture and Healthcare 4: 70–78. [Google Scholar]
- Torres-Garcia D, García D, Réblová M, et al. 2023. Diversity and novel lineages of black yeasts in Chaetothyriales from freshwater sediments in Spain. Persoonia 51: 194–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, Von Haeseler A, et al. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1): W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimbach J. 1990. Pachyella lazzariana, espèce subalpine nouvelle. Rivista di Micologia 33: 341–345. [Google Scholar]
- Tulloss RE. 1994. Type studies in Amanita section Vaginatae I: Some taxa described in this Century (studies 1-23) with notes on description of spores and refractive hyphae in Amanita. Mycotaxon 52: 305–396. [Google Scholar]
- Tulloss RE, Yang ZL. 2023. Amanitaceae studies. http://www.amanitaceae.org [accessed 5 Aug. 2023].
- Uljé CB. 2005. Coprinus. In: Noordeloos ME, Kuyper TW, Vellinga EC. (eds), Flora agaricina neerlandica 6: 22–109. Taylor & Francis, Boca Raton. [Google Scholar]
- Valenzuela-Lopez N, Cano-Lira JF, Guarro J, et al. 2018. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Studies in Mycology 90: 1–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Aa HA. 1967. A new species of Curvularia. Persoonia 5: 45–46. [Google Scholar]
- Vandepol N, Liber J, Desirò A. et al. 2020. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Diversity 104: 267–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vánky K. 1979. Species concept in Anthracoidea (Ustilaginales) and some new species. Botaniska Notiser 132: 221–231. [Google Scholar]
- Vánky K. 1985. Carpathian Ustilaginales. Symbolae Botanicae Upsalienses 24: 1–309. [Google Scholar]
- Vánky K. 1994. European smut fungi. Fischer Verlag, Stuttgart, Jena, New York. [Google Scholar]
- Vánky K. 2011. Smut fungi of the world. APS Press, St. Paul, Minnesota, USA. [Google Scholar]
- Vellinga EC, Sysouphanthong P, Hyde KD. 2011. The family Agaricaceae: phylogenies and two new white-spored genera. Mycologia 103: 494–509. [DOI] [PubMed] [Google Scholar]
- Videira SIR, Groenewald JZ, Braun U, et al. 2016. All that glitters is not Ramularia. Studies in Mycology 83: 49–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videira SIR, Groenewald JZ, Nakashima C, et al. 2017. Mycosphaerellaceae – chaos or clarity? Studies in Mycology 87: 257–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzini A, Zotti M, Traverso M, et al. 2016. Variability, host range, delimitation and neotypification of Amanita simulans (Amanita section Vaginatae): collections associated with Helianthemum grasslands, and epitypification of A. lividopallescens. Phytotaxa 280: 1–22. [Google Scholar]
- Voglmayr H, Jaklitsch WM. 2014. Stilbosporaceae resurrected: generic reclassification and speciation. Persoonia 33: 61–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voitk A, Saar I, Lücking R, et al. 2020. Surprising morphological, ecological and ITS sequence diversity in the Arrhenia acerosa complex (Basidiomycota: Agaricales: Hygrophoraceae). Sydowia 73: 133–162. [Google Scholar]
- Wanasinghe DN, Hyde KD, Jeewon R, et al. 2017. Phylogenetic revision of Camarosporium (Pleosporinae, Dothideomycetes) and allied genera. Studies in Mycology 87: 207–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling R. 1998. Heinemannomyces: a new lazuline-spored agaric genus from South East Asia. Belgian Journal of Botany 131: 133–138. [Google Scholar]
- Widhelm TJ, Bertoletti FR, Asztalos MJ, et al. 2018. Oligocene origin and drivers of diversification in the genus Sticta (Lobariaceae, Ascomycota). Molecular Phylogenetics and Evolution 126: 58–73. [DOI] [PubMed] [Google Scholar]
- Wirth V, Hauck M, Schultz M. 2013. Die Flechten Deutschlands. Eugen Ulmer KG, Stuttgart, 2 Band. [Google Scholar]
- Woudenberg JHC, Groenewald JZ, Binder M, et al. 2013. Alternaria redefined. Studies in Mycology 75: 171–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhou LW, Vlasák J, et al. 2022. Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Diversity 113: 1–192. [Google Scholar]
- Wu W, Diao Y. 2023. The chalara-like anamorphs of Leotiomycetes. Fungal Diversity 119: 213–490. [Google Scholar]
- Xiao Y-P, Hongsanan S, Hyde KD, et al. 2019. Two new entomopathogenic species of Ophiocordyceps in Thailand. MycoKeys 47: 53–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XQ, Ma SY, Peng ZX, et al. 2021. Diversity of Plectosphaerella within aquatic plants from southwest China, with P. endophytica and P. sichuanensis spp. nov. MycoKeys 80: 57–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlic I, et al. 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20: 348–355. [DOI] [PubMed] [Google Scholar]
- Zhang ZF, Zhou SY, Eurwilaichitr L. et al. 2021. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Diversity 106: 29–136. [Google Scholar]
- Zhang ZY, Chen WH, Zou X, et al. 2019. Phylogeny and taxonomy of two new Plectosphaerella (Plectosphaerellaceae, Glomerellales) species from China. MycoKeys 57: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Karunarathna S, Raspé O, et al. 2011. Major clades in tropical Agaricus. Fungal Diversity 51: 279–296. [Google Scholar]
- Zhao RL, Desjardin DE, Soytong K, et al. 2010. A monograph of Micropsalliota in Northern Thailand based on morphological and molecular data. Fungal Diversity 45: 33–79. [Google Scholar]
- Zhao X, Leavitt SD, Zhao ZT, et al. 2016. Towards a revised generic classification of lecanoroid lichens (Lecanoraceae, Ascomycota) based on molecular, morphological and chemical evidence. Fungal Diversity 78: 293–304. [Google Scholar]
- Zheng HF, Huang FC, Liu B, et al. 2021. Fulvifomes nonggangensis and F. tubogeneratus (Hymenochaetales, Basidiomycota): Two new species from Southern China based on morphological and molecular evidence. Mycobiology 49: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LW. 2014. Fulvifomes hainanensis sp. nov. and F. indicus comb. nov. (Hymenochaetales, Basidiomycota) evidenced by a combination of morphology and phylogeny. Mycoscience 55: 70–77. [Google Scholar]
- Zhou LW. 2015. Fulvifomes imbricatus and F. thailandicus (Hymenochaetales, Basidiomycota): Two new species from Thailand based on morphological and molecular evidence. Mycological Progress 14: 89. [Google Scholar]
- Zwetko P, Blanz P. 2004. Die Brandpilze Österreichs. Doassansiales, Entorrhizales, Entylomatales, Georgefischeriales, Microbotryales, Tilletiales, Urocystales, Ustilaginales. In: Ehrendorfer F. (ed.), Catalogus Florae Austriae 3(3), Biosystematics and Ecology Series 21. Verlag der Österreichischen Akademie der Wissenschaften, Vienna, Austria. [Google Scholar]