Summary
Exosomes are a type of extracellular vesicle (EV) with diameters of 30–150 nm secreted by most of the cells into the extracellular spaces and can alter the microenvironment through cell-to-cell interactions by fusion with the plasma membrane and subsequent endocytosis and release of the cargo. Because of their biocompatibility, low toxicity and immunogenicity, permeability (even through the blood–brain barrier (BBB)), stability in biological fluids, and ability to accumulate in the lesions with higher specificity, investigators have started making designer’s exosomes or engineered exosomes to carry biologically active protein on the surface or inside the exosomes as well as using exosomes to carry drugs, micro RNA, and other products to the site of interest. In this review, we have discussed biogenesis, markers, and contents of various exosomes including exosomes of immune cells. We have also discussed the current methods of making engineered and designer’s exosomes as well as the use of engineered exosomes targeting different immune cells in the tumors, stroke, as well as at peripheral blood. Genetic engineering and customizing exosomes create an unlimited opportunity to use in diagnosis and treatment. Very little use has been discovered, and we are far away to reach its limits.
Keywords: engineered exosomes, designer’s exosomes, manipulation of biogenesis, exosomes, and immune cells, separation of exosomes, therapeutic exosomes
1 |. INTRODUCTION
Exosomes are a type of extracellular vesicle (EV) with diameters of 30–150 nm secreted by most of the cells into the extracellular spaces and can alter the microenvironment through cell-to-cell interactions by fusion with the plasma membrane and subsequent endocytosis and release of their cargo.1–6 Irrespective of the origin of parent cells, exosomes share common features such as certain tetraspanins (CD9, CD63, and CD81), heat shock proteins (HSP 60, Hsp 70, and Hsp 90), biogenesis-related proteins (Alix and TSG 101), membrane transport and fusion proteins (GTPases, annexins, and Rab proteins), nucleic acids (mRNA, miRNA, and long noncoding RNAs and DNAs), and lipids (cholesterol and ceramide).2,7,8 Because of their biocompatibility, low toxicity and immunogenicity, permeability (even through the blood–brain barrier (BBB)), stability in biological fluids, and ability to accumulate in the lesions with higher specificity,9–15 investigators have started making designer’s exosomes or engineered exosomes to carry biologically active protein on the surface or inside the exosomes as well as using exosomes to carry drugs, micro RNA, and other products to the site of interest.11,16–19
When searched in PubMed using the term exosomes, there were only 84 publications between 1950 and 2000. However, using the same search word there were 5001 publications in 2021, and 1402 are published as review articles. When the search term is used as “exosomes in immunology” PubMed produce 581 publications in 2021 including 215 review articles. A total of 123 publications since 2010 mentioned the term “engineered exosomes” in the title or the abstract and only 21 publications dealt with engineered exosomes in immunology since 2010. Among them, only 6 review articles discuss the application of engineered exosomes in immunotherapy.16,20–24 It is obvious that engineered exosomes in the field of immunotherapy are still in infancy and untapped.
Recently our laboratory has achieved a few milestones in exosome technology: (1) we developed a platform to make engineered exosomes using nontumorous HEK293 cells that carry and express specific cell-targeting peptides to detect specific cells in vivo when administered intravenously; (2) we used these engineered exosomes as a therapeutic probe to deplete specific cells in the body; (3) we optimized the methods to collect a uniform-sized large amount of exosomes from different cells using a combination of size exclusion and centrifugal filters in shortest possible time; (4) we showed differential biodistribution of exosomes collected from different cells in tumor-bearing animals using clinically relevant single-photon emission computed tomography (SPECT).25,26 In this review article, we will revisit the current version of the biogenesis of exosomes using tumorous and nontumorous cells, how to manipulate the biogenesis mechanism to make engineered exosomes to express protein or RNA of interest in the exosomes and how to make designer’s exosomes to carry nanoparticles, micro RNA, chemotherapeutics, and others. All possible biogenesis of engineered exosomes and their applications will be around the subject matters of immunotherapy, especially targeting tumor microenvironment (TME).
2 |. CURRENT VIEW OF BIOGENESIS OF EXOSOMES
The biogenesis of exosomes starts from the process of plasma membrane invagination, the formation of early and late endosomes, the formation of the multivesicular body (MVB), the generation of exosomes as intraluminal vesicles (ILVs), and the secretion of the ILVs as exosomes extracellularly.27 MVB is composed of ILV particles of different sizes, which range from a few nanometers (nm) to micrometers (μm). The common consensus is that size of the exosomes ranges from 30 to 150 nm.28 The biogenesis of exosomes is a highly regulated process and involves many steps and proteins.29 First ubiquitin-binding protein Golgi-Localized γ-Ear-Containing ARF-Binding (GGA), Vps27/Hse1, and clathrin form an endosomal clathrin coat, which acts as a cargo loading site for ESCRT machinery. The ESCRTs ESCRT-0, -I, -II, -III, and Vps4 then form the multivesicular body. Interestingly, the ESCRTs are also involved in the invagination of multivesicular body and formation of ILVs7 where ESCRT III takes part in scission1 of ILVs in the lumen. Along with ESCRTs syndecan, ceramide and tetraspanins are involved in ILVs biogenesis.30 Several ESCRT and related proteins including HRS, STAM1, TSG101, ALIX, and VPS4 are involved in MVB docking31 with membrane and SNAREs are responsible for fusion of MVB with membrane and release of ILVs.32 In contrast to exosomes, microvesicles, another type of EV sizing 100–1000 nm are generated by direct outward budding of the plasma membrane with the help of several GTPases. Figure 1 shows the current view of exosome biogenesis.
FIGURE 1.
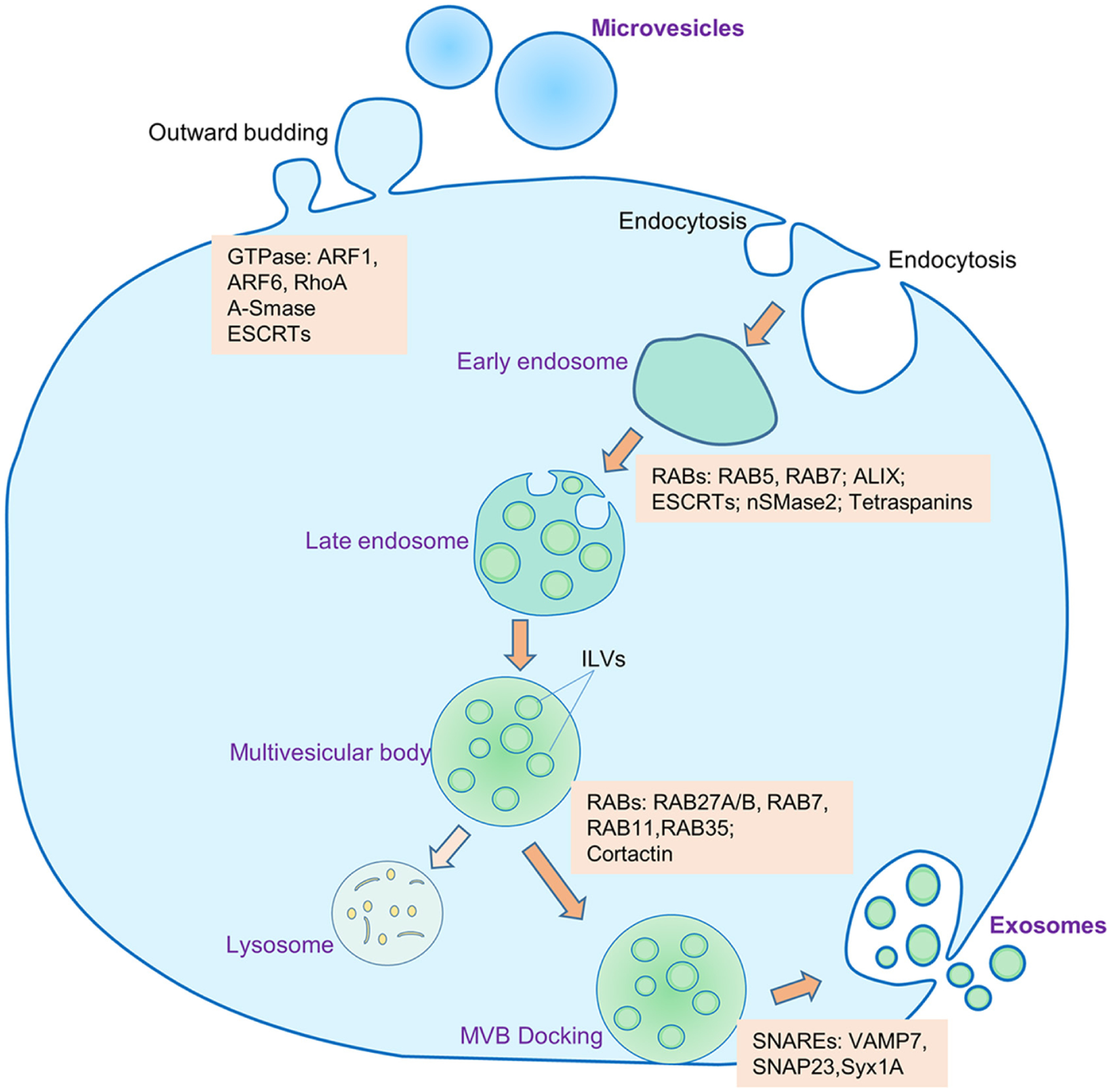
Biogenesis and secretion of exosomes and microvesicles. Microvesicles are generated by outward budding of the plasma membrane with the help of several GTPases. The process of biogenesis and release of exosomes into the extracellular space encompasses several distinct steps: (1) invagination of plasma membrane and formation of early endosomes, (2) inward protrusion of early endosomal membrane to generate late endosomes, (3) formation of multivesicular bodies (MVBs) that contain intraluminal vesicles (ILVs), (4) docking of the MVBs to the cellular plasma membrane, (5) exocytosis of the exosomes into the extracellular milieu. Some of the MVBs may go into lysosomal degradation. Several molecules are involved in the biogenesis and release of microvesicles and exosomes. ESCRT, endosome sorting complex required for transport; RAB, RAS-related protein; ALIX, ALG-2 interacting protein X; nSMase2, neutral sphingomyelinase 2; SNARE, soluble NSF attachment protein receptor; VAMP7, vesicle-associated membrane protein 7; SNAP23, synaptosomal-associated protein 23; Syx1A, syntaxin 1A; ARF, ADP, ribosylation factor; RohA, Ras homolog family member A; A-SMase, acid sphingomyelinase
2.1 |. Importance of tetraspanins and their manipulation for biogenesis
EVs are secreted by all types of cells. Among the EVs, exosomes contain a specific amount and types of components based on the cell of origin. Alongside genetic materials and lipids, proteins are one of the major components in the exosomes. Exosomes show protein heterogeneity because the parent cells are secreted from having different types of protein contents. One of the large protein families present on the surface of exosomes is tetraspanins. Exosomes have their tetraspanin-enriched microdomains (TEMs) and form a cluster on the surface. By their cluster, they can interact with numerous signaling molecules.33,34 Almost all exosomes have three major types of tetraspanins CD63, CD9, and CD81, which are also being used as exosome markers. Tetraspanins are involved in exosome biogenesis processes and sorting cargo of the exosomes. Tetraspanins are also involved in the attachment with the target cell as well as in antigen presentation.35 They also regulate cellular motility and migration and have shown their role in the metastasis of tumors.36 CD63 interacts by its C terminal with protein complex and attaches the exosomes with membrane to clathrin-dependent pathways.37 CD9 marker is not specific for endosomes small vesicles (like exosomes) because the presence of this marker in large vesicles was also noted.38 CD9 transfer from the endoplasmic reticulum to Golgi in B cell has the involvement of CD81.39 CD9 and CD81 have been shown to interact with G proteins.40 Additional to these, tetraspanins have different other functions.
2.2 |. Manipulation biogenesis to control the contents of the exosomes in the lumen and on its surface
To meet up the protein deficiency and dysfunction, overexpression of the target protein is a way to increase the protein content of exosomes.41 In this process, certain proteins in the donor cells are overexpressed and that overexpressed protein goes to exosomes by their normal sorting. Excessive protein-containing exosomes are released from cells and can be collected for using further studies or therapies. The downside of this process is possible cytotoxicity and the proliferation inhibition of donor cells. An alternative approach to this process is using ubiquitin. Ubiquitin is one of the most abundant proteins.42 By using Ubiquitin, a target protein can be expressed 10-fold higher than normal by conjugating the target protein in the C terminal of ubiquitin, which has been shown in HEK 293 cell.43 In exosomes, MHC-II β-chain cytoplasmic tail ubiquitination turns them to be sorted, therefore this ubiquitination platform could be used to package cargo protein in exosomes.44
2.3 |. Biogenesis of exosomes in immune cells
Similar to other cells, immune cells also release exosomes abundantly that carry membranous, cytosolic, and even nuclear molecules (DNA, RNA) characteristic of the cells of origin. Given the numerous types of immune cells, exosomes derived from immune cells play crucial and complex physiological and pathological roles within the already complex immune system. Functional molecules of exosomes, derived from various immune cells and their effects are summarized in Figure 2.
FIGURE 2.

Functional molecules in the exosomes released from different immune cells
2.3.1 |. Macrophages
Macrophages are innate immune cells, which exert diverse functions through their secreted exosomes and are shown to be involved in the progression of the disease by their bioactive molecules.45,46 The biogenesis of exosomes in macrophages is the same as in other cell types. Different studies have confirmed that the exosome contents of macrophages and the surface proteins are secretory cell-specific.47 As macrophages are of three types, M0 (nonpolarized), M1, and M2 (polarized), there are three types of macrophage-derived exosomes, and different studies investigated the role of M0, M1, and M2 macrophage-derived exosomes.48–50 Macrophage-derived exosomes are shown to exert their effects in different pathological conditions by activating different gene signaling pathways; mostly for progression and metastasis.51
The main content of macrophage exosomes are miRNAs, long noncoding RNAs (lncRNAs), and proteins.52 Some miRNAs are found in higher levels in M2 macrophage-derived exosomes than in M1 macrophage-derived exosomes.53 Among the major miRNA in macrophage-derived exosomes for cancer progression, drug resistance and cancer inhibitions are miR-29a, miR-92a-2, miR-95, miR-125a/b, miR-142, miR-21, miR-155, miR-7, and miR-146a. The major proteins present in the macrophage-derived exosome are ApoE, IL-6, and AMAD15.53 Other components in macrophage exosomes are mRNA, tRNA, and ribosome but there is no evidence of active DNA.54,55
2.3.2 |. Myeloid-derived suppressor cells
MDSCs are myeloid heterogeneous cells grouped in an immature state. The major two subgroups of MDSC are monocytic MDSC (M-MDSC) and granulocytic MDSC (G-MDSC), which are differentiated based on Ly6C high (M-MDSC) or Ly6G high (G-MDSC). Annexins, tetraspanins, cytoskeletal proteins, and heat shock proteins (HSPs) are common in exosomes released by MDSC, which are similar to other cell-derived exosomes. Tetraspanins (including CD9, CD177), Hsp70, Hsp90α, Hsp90β, Alix, and the ESCRT complex are characteristic proteins of exosome biogenesis and cargo sorting are also present in MDSC exosomes.56 Some pro-inflammatory proteins S100A8/9, CD47, and thrombospondin-1 as well as platelet factor-4 are also enriched in exosomes are also enriched in MDSC-derived exosomes. MDSC-derived exosomes contain abundant ubiquitinated proteins such as the ubiquitinated histones, and the nonhistone nuclear protein high mobility group box (HMBG).57 Transforming growth factor-β1(TGF-β1) is 4.3 times higher in MDSC-derived exosomes than in the MDSC cell.58 Cancer progression miRNAs are abundant in exosomes derived from MDSC, which are mainly miR-146a, miR-146b, miR-155, miR-125b, miR-100, let-7e, miR-125a, and miR-99b.59
2.3.3 |. T cells
Like many other cells, T cells also release exosomes but very few studies have been conducted about the T cell exosome biogenesis. T cell released exosomes mostly showed the immune modulation function.60–62 Cytotoxic T cells release the lethal protein perforin as well as granzymes to the target cell through exosomes.63 Even apoptosis changes the protein content of T cell released exosomes when compared between an activated T cell released exosome and apoptotic T cell released exosome.64,65 One of the components of T cell exosomes is FasL, secreted as “lethal exosomes” following activation-induced fusion of the MVB with the plasma membrane.65 Along with FasL, APO2 ligand (APO2L)/TNF-related apoptosis-inducing ligand (TRAIL) has also been found in T cell exosomes.66 A study demonstrated that T cell exosomes express thrombospondin-1 receptor CD47 and it regulates endothelial cell responses to vascular endothelial growth factor (VEGF).67
2.3.4 |. Dendritic cells
Dendritic cells (DCs) are regarded as specialized and most potent antigen-presenting cells (APC) mediating crucial functions in innate and adaptive immune responses. They can efficiently process and present antigens followed by triggering the proliferation, activation, and differentiation of naive T cells.68 Research has shown that like DCs, exosomes released from activated DC also express MHCII complex and T cell co-stimulatory molecules and are involved in antigen presentation.69,70 It has been found that exosomes derived from mature DCs contain CCR7, a chemokine receptor that directs mature DCs to peripheral lymphoid tissues which also analogously regulates the increased accumulation of these exosomes in the spleen and inflammatory responses upon injection in mice.71 Although DC-derived exosomes can activate T cells through stable interactions with TCR complexes, the extent of the activation depends on DC developmental stage. Generally, T cells are more efficiently activated by mature DCs than immature DCs, and mature DCs release exosomes to facilitate immune-stimulatory responses, whereas immature DC exosomes exhibit a potent immune-suppressive response.72,73 Immunosuppressive molecules, such as TGF-β, NKG2D, and death ligand FasL expressed by immature DCs following response to tumors, can inhibit natural killer (NK) cells, macrophages, and neutrophils.74,75 Furthermore, DC-derived exosomes expressing HLA-B associated transcript-3 (BAT3) bound to NKp30 receptor in NK cells and stimulate the secretion of TNF-α and IFN-γ.76 Through IL-15Rα and NKG2D, DC-derived exosomes also enhanced NK cell proliferation and activation.20
3 |. CURRENT METHODS TO DIFFERENTIATE EXOSOMES VS EV PARTICLES
EVs are divided into three main classes77: Exosomes, Microvesicles (also known as microparticles or ectosomes), and apoptotic bodies. Exosomes are produced within the endosomal network as MVB, which are released upon fusion with the plasma membrane. Exosomes are identified by specific markers, for example, Alix, tetraspanin. These markers denote their specific endocytic origins and a combination of the markers is preferred. Microvesicles are formed by outward budding and fission of the plasma membrane. Apoptotic bodies are released as blebs of cells undergoing apoptosis. Characteristics and main differences between different EVs are shown in Table 1.
TABLE 1.
| Traits | Exosomes | Microvesicles | Apoptotic bodies |
|---|---|---|---|
| Biogenesis | Endosomal origin and exocytosis | Outward budding of the plasma membrane | Outward blebbing and fragmentation of the plasma membrane |
| Release time | Ten minutes or more | Few seconds | – |
| Pathways | ESCRT-dependent Tetraspanin-dependent Ceramide-dependent Stimuli-dependent | Ca2+-dependent Stimuli- and cell-dependent | Apoptosis-related |
| Size | 30–150 nm | 100–1000 nm | 1000–5000 nm |
| Appearance-electron microscopy | Spheroid/cup shape | Irregular and electron-dense | Heterogeneous |
| Density | 1.13–1.19 g/ml | 1.04–1.07 g/ml | 1.16–1.28 g/ml |
| Isolation method | ultracentrifugation, ultrafiltration, precipitation, size exclusion chromatography, immunoaffinity capture-based, microfluidics-based, polymer-based, etc. | Ultracentrifugation | No standardized protocol |
| Content | Proteins, nucleic acids, lipids, and metabolites | Proteins, nucleic acids, lipids, and metabolites | DNA fragments and histone, chromatin remnants, cell organelles, cytosol portions, degraded proteins |
| Typical constituent proteins | Tetraspanins (CD9, CD63, CD81), ESCRT proteins (Alix, TSG101), Integrins (−α, −β), heat shock proteins (HSP90, HSP70) | Anexin V, Flotillin-2, Selectins, Integrins, CD40 ligand, metalloproteinase | Anexin V, DNA, histones |
| Function | Cell-cell communication | Cell-cell communication | Product of programmed cell death. Removal of unwanted cells |
3.1 |. Specific markers of exosomes
Several exosomal proteins have been identified and are generally been used as exosome markers. A summary of common exosomal protein markers, their location in the exosomes, and collection and detection methods of these proteins is shown in Table 2 and Figure 3.
TABLE 2.
Exosome protein markers
| Exosome markers | Location | Collection and detection methods | References |
|---|---|---|---|
| Alix (PDCD6IP) | Cytoplasm | Ultracentrifugation | 82,83 |
| CD9 | Plasma membrane | Immunohistochemistry | 84–86 |
| CD24 | Plasma | Ultracentrifugation; Immunohistochemistry | 87,88 |
| CD63 | Plasma membrane and cytosol | Immunohistochemistry | 86,89,90 |
| CD81 | Plasma membrane | Immunohistochemistry | 84 |
| CPNE3 | Plasma | Ultracentrifugation; Immunohistochemistry | 91 |
| EDIL3 | Plasma | Ultracentrifugation | 87,92 |
| Exo-PD-L1 | Serum | Immunohistochemistry | 93 |
| Fibronectin | Plasma | Ultracentrifugation | 87,94 |
| FLOT1 | Plasma membrane | Ultracentrifugation, immunoblotting | 95,96 |
| HSP70 | Serum, plasma membrane | Ultracentrifugation | 97 |
| TEX | Plasma | Density gradient ultracentrifugation; size exclusion chromatography (SEC); Differential centrifugation; Ultrafiltration | 98 |
| TfR | Perinuclear or plasma membrane | Ultracentrifugation; Size-exclusion chromatography (SEC) | 99,100 |
| TSG101 | Cytoplasm | Ultracentrifugation, immunoblotting | 101,102 |
Abbreviations: CPNE3, Copine III; EDIL3, EGF-Like Repeats and Discoidin I-Like Domains Protein 3; Exo-PD-L1, Exosomal programmed cell-death ligand 1; FLOT1, flotillin 1; PDCD6IP, Programmed cell death 6-interacting protein; TEX, Tumor-Derived Exosomes; TfR, Transferrin receptor; TSG101, Tumor susceptibility gene 101.
FIGURE 3.
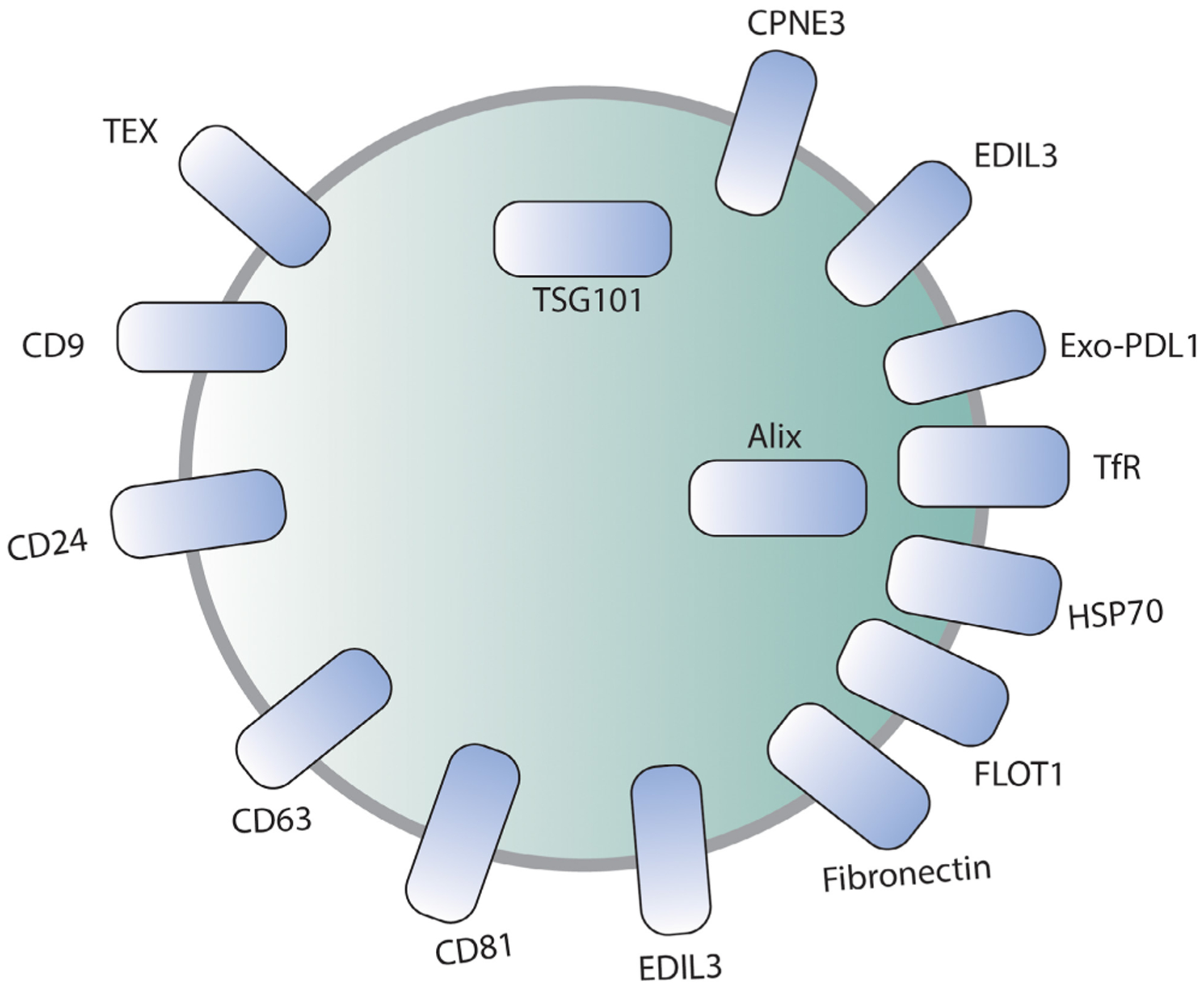
Schematic diagram showing common exosomal markers
Alix (or PDCD6IP, also known as Programmed cell death 6-interacting protein) regulates the endolysosomal system and regulates neuronal death as demonstrated by the upregulation of Alix in degenerating hippocampal neurons after epileptic seizures.103
EDIL 3 (or EGF Like Repeats and Discoidin I-Like Domains Protein 3, also known as Developmental Endothelial Locus 1) is a pro-angiogenic factor and a regulator of endothelial cell adhesion and migration.104 It is an extracellular matrix protein that contains 3 EGF-like domains. One of the domains contains an RGD (Arg–Gly–Asp) motif, which facilitates its interaction with integrins.105
HSP70 (heat shock proteins) are membrane-bound and extracellularly located proteins that maintain protein homeostasis as a chaperone in the cytosol. It also has cytoprotective effects. Since the synthesis of HSPs is induced by stress, heat, and other chemical and mechanical stimuli, a variety of HSPs (namely HSP70 and HSP90) have been frequently found in the plasma membrane of the tumor cells. HSPs are isolated by ultracentrifugation.97
Several isolation methods have been developed to detect exosomes but the combination of methods yields the best results. Exosome markers like PDCD6IP (Programmed cell death 6-interacting protein, also known as Alix), CD24, CPNE3, EDIL3, Fibronectin, FLOT1, HSP70, TEX, TfR, and TSG101 can be detected by ultracentrifugation.82,83 Immunohistochemistry detects exosomes like CD9, CD24, CD63, CPNE3, Exo-PD-L1, and CD81.86
TEX (or tumor-derived exosomes) are ubiquitously present in the plasma and TME in all body fluids of cancer patients.106 These exosomes facilitate immune-regulatory activities.98
Pineles et al. (2022) conducted an observational cohort study on term/near-term neonates undergoing therapeutic hypothermia (TH) for hypoxic-ischemic encephalopathy (HIE), where they purified CNS exosomes from serum using several established methods. In this study, the researchers concluded that CNS exosome cargo acts as biomarkers that correspond with the severity of brain injury, response to TH, and quantify pharmacological response to neuroactive therapeutic/adjuvant agents. Synaptopodin (SYNPO) is a protein contained within the neonatal CNS exosomes and is specific to HIE.107
3.2 |. Specific markers for immune cell-derived exosomes
Various cellular components take part in the formation of both the innate and adaptive components of the immune system. Among the several biological functions of exosomes on immune systems, the most significant ones are immunomodulation including immune suppression and various anti-inflammatory processes; cell-to-cell communication including antigen presentation, NK cell, and T cell activation.108
Among all the immune-cell-derived exosomes, DC-derived exosomes are the most vital as they exist in multiple populations, and effectively initiate the antigen-specific immune response by efficient activation and proliferation of T cells, thus promoting immunity. A combination of various cell markers is used to identify the DCs. Exosomes derived from DCs have an essential role in several diseases, including autoimmune diseases, cardiovascular diseases for example, acute MI, or transplant medicine.109 Leone et al. demonstrated that DCs are identified by CD107a/LAMP-1 (lysosome-associated membrane protein-1) and CD107b/LAMP-2 (lysosome-associated membrane protein-2) that are present on the surface of DCs.110 APC-derived exosomes originate from inward invagination of the internal vesicles of the MHC class II compartment (MIIC). Immunoelectron microscopy of B cells and DCs demonstrates that MVE (multivesicular endosomes) limiting membranes fuse with the plasma membranes and the internal vesicles within the MVE express MIIC-specific markers LAMP-1, MHC-II, CD63 and CD82.111 DC-derived exosomes stimulate the proliferation of allogeneic lymphocytes. On the other hand, APC-derived exosomes express MHC-II and stimulate T cells.112 DC-derived exosomes that express MHC-I and CD86 can effectively generate CD8+ T cell response against tumors.69 The long-term culture method which supports the production of myeloid-like and immature myeloid DC,113 both lack expression of MHC-II or CD40 but myeloid-like DC expresses CD11c, CD11b, CD80, CD86, and immature DC expresses FcγII/IIIR.114
Macrophage-derived exosomes are of monocytic lineage. These exosomes participate in immune response after cardiac injury following MI or other cardiac injuries through the recruitment of other macrophage components. Following MI (or cardiac injury), for the first few days, the M1 macrophage peaks, then macrophages shift from M1 to M2. This shift signifies the pro-inflammatory and pro-phagocytic response of M1 macrophage and the anti-inflammatory response of M2.115 Notable microRNA contained within the exosomes taking part in this process are miR-155, miR-19, miR-21, miR-146, and miR-223. Of note, these miR-NAs inhibit fibroblast proliferation and stimulate inflammation, which in turn creates a pro-inflammatory environment in cardiac muscles. Detecting these miRNAs in macrophage-derived exosomes can provide a significant clinical understanding of myocardial diseases.116
B-cell-derived exosomes contain MHC-II complexes. Schroeder et al demonstrated that in HNSCC (head neck squamous cell carcinoma) involving B cells PD-1, CTLA, LAG3, and CD137 are increased in some patients.117 PD-1 expression decreases BCR signaling, and subsets of PD-1 may also be found to be elevated in hepatocellular cancer and thyroid cancer.91 CTLA4 expression, which is associated with inhibitory effects on immunoglobulin production, is reported to be elevated in B cell malignancies and malignant melanoma,118 LAG3 (CD223) is a “checkpoint receptor” that regulates TCR signaling and function.119 CD137, expressed on activated B cells in peripheral blood and on tonsillar B cells, in turn, enhances B cell proliferation, improves survival, and induces secretion of TNF-α & -β.120 Mature B cells express CD39 (“B cell activation marker”) and CD73 on their surface. CD39 and CD73 are considered “immunological switches,” that shift from pro- to anti-inflammatory activity in the cells, create an immunocompromised environment, and contribute to the progression of cancer.121
T cell-derived exosomes are determined by their surface markers, CD3, CD4, CD8, CD27, and CD28. Loss of CD27/CD28 has been associated with suppressive function and cancer cells maintain their proliferative capacity.61 Wahlgren et al.122 showed that exosomes from IL-2, anti-CD3, and anti-CD28 stimulated T cells to express CD9, CD63, and CD81 markers on their surface. These exosomes carry RANTES (CCL5) which promotes cytotoxic response.
4 |. CURRENT METHODS OF SEPARATION/COLLECTION OF EXOSOMES
The most commonly used methods of exosome isolation are ultracentrifugation and precipitation. The gold standard for exosome isolation is ultracentrifugation. Precipitation is another most common method for exosome isolation from plasma. Coughlan et al. (2020) used ExoQuick® ULTRA EV Isolation Kit for Serum and Plasma (Systems Biosciences) for precipitation of exosomes due to the ease of extracellular vesicle preparation, significantly depleted number of both IgGs or albumin, and relative enrichment of exosomes based on Nanoparticle Tracking Analysis (NTA) assessment of size and concentration.123 Ultracentrifugation method produces highly enriched EVs but it is low-throughput and specific infrastructure (i.e., ultracentrifuge) and expertise is required to be performed correctly.123 Precipitation methods are significantly faster than ultracentrifugation methods and they prepare higher concentrations of exosomes. It also produces extracellular vesicles that have a significantly low number of both IgGs and albumin. A schematic summary of the processes involved in different exosome isolation techniques is shown in Figure 4.
FIGURE 4.
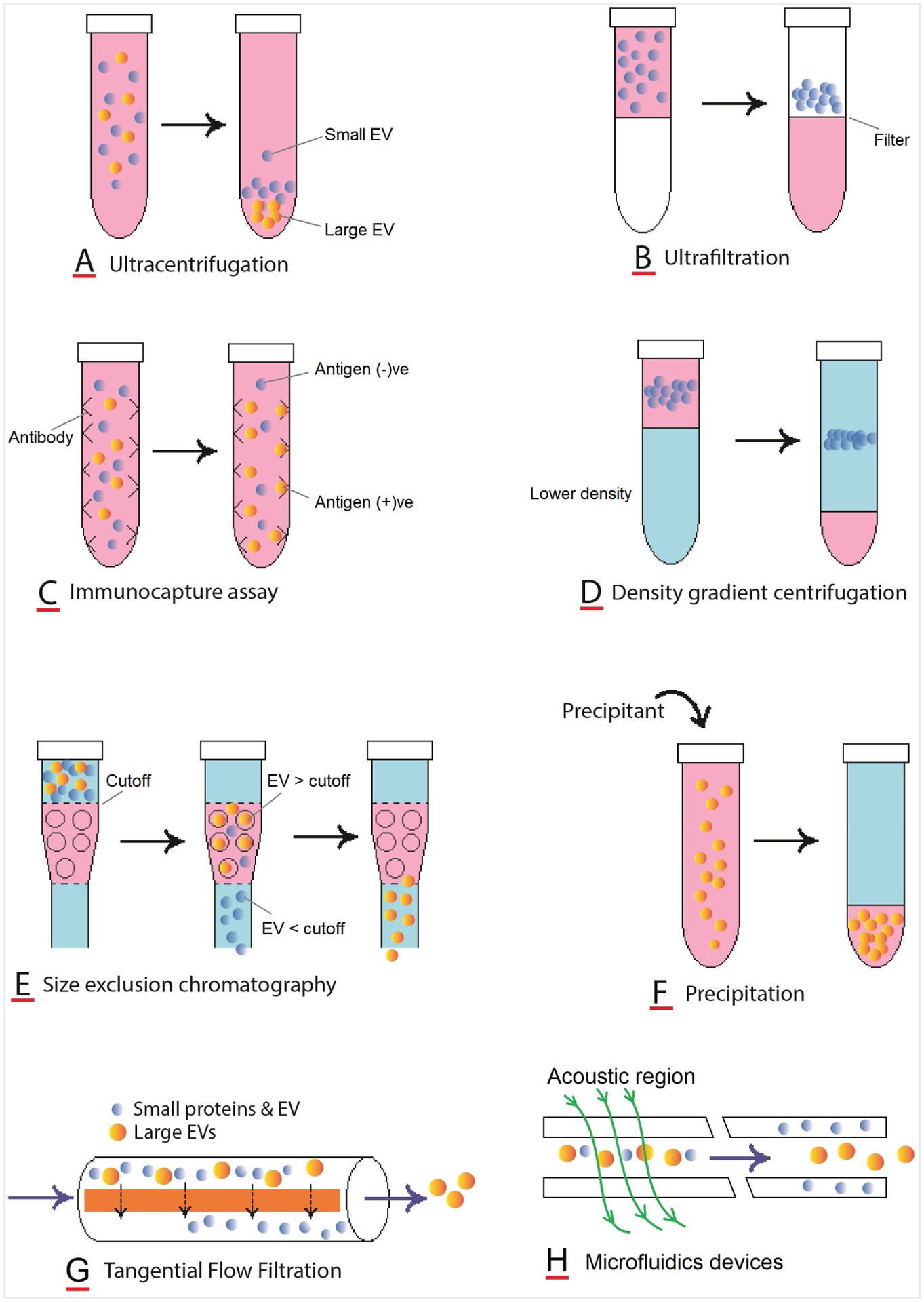
Schematic presentation of processes involved in different techniques of exosome isolation
For isolating exosomes, several techniques have been developed by exploiting a particular trait, such as the size, density, and surface markers of exosomes. However, each of these techniques comes with its own limitations which must be addressed for downstream applications. The advantages and disadvantages of commonly applied methods are shown in Table 3.
TABLE 3.
Exosome isolation methods
| Advantages | Disadvantages | References | |
|---|---|---|---|
| Ultracentrifugation (Most commonly used method) [Differential centrifugation (DC)] |
|
|
87,111,123–127 |
| Precipitation |
|
|
123–125,128 |
| Size exclusion chromatography (SEC) |
|
|
124,125 |
| Ultrafiltration (UF) |
|
|
87,124,125,129–131 |
| Immuno-capture assays (ICA) |
|
|
87,124,125,132,133 |
| Density gradient centrifugation (DGC) |
|
|
87,111,134,135 |
| Polymer-based precipitation |
|
|
136 |
| Microfluidics |
|
|
124,137,138 |
| Tangential flow filtration |
|
|
139–142 |
| Commercial kits miRCURY ExoQuick TEIR |
|
143 |
Abbreviations: UC, Ultracentrifugation; TEIR, Total Exosome Isolation Reagent.
While the precipitation method provides the most effective exosome isolation (~90%), it takes a long time to achieve exosomes via this method. On the other hand, differential centrifugation takes less time (~9 hours) but the EV yield is variable, sometimes as low as 2%. AF4 (Asymmetric flow field-flow fractionation) process takes only 1 hour but its sample preparation may take up to 3 days. Newer, methods are quick, easier to detect, and can be commercially used.
Helwa et al.143 compared different exosome extraction methods. They used 6 different volumes of human serum samples versus commercial serum samples from human donors and concluded that even with limited amounts of biological samples, commercial kits miRCURY, ExoQuick, and TEIR are suitable alternatives to ultracentrifugation. Also, exosomes isolated by these techniques and serum volumes had similar zeta potentials to previous studies. In this study, the NTA results showed that all isolation techniques produced exosomes within the expected size range (30–150 nm).
Additionally, exosome isolation methods can be categorized based on their recovery time and assay time (Tables 4 and 5).
TABLE 4.
Categorizing isolation methods based on EV recovery
TABLE 5.
Categorizing isolation methods based on assay time
Newer methods have been developed that aim for better recovery and specificity. These include:
Acoustics (or acoustic-based separation methods)144: This exosome separation method uses acoustic frequency (as high as ~40 MHz) through a series of cell-removal and exosome-isolation modules and can separate particles based on their physical properties such as size difference, and acoustic contrast factors.145 Current methods are only based on biological fluids (e.g., undiluted blood samples). This method requires specialized equipment and significant time owing to its preprocessing of liquid samples.146
Alternating current electrophoretic147: Another rapid exosome isolation technique is alternating current electrokinetic (ACE) microarray that has been shown to rapidly isolate and recover glioblastoma exosomes from undiluted human plasma samples.147 This method requires a small plasma sample and can take up to 15 minutes to isolate exosomes. This method is used to isolate various sample types including undiluted blood, plasma, serum, high-molecular-weight DNA, viruses from high conductance buffers, and drug delivery nanoparticles. The principle of this method is based on creating an alternating current (AC) electric field by generating a dielectrophoretic (DEP) separation force generated by the ACE microarray.148
Field-flow fractionation (FFF): Field-flow fractionation is a chromatography-like separation technique that is based on the principle of fractionation of macromolecules, colloids, and particles. A laminar flow of liquid between two walls is pushed by an external field force.149 It is a rarely used method of exosome separation.150
Asymmetric flow field-flow fractionation (AFFF, A4F, or AF4): The principles of AF4 isolation methods are based on the techniques of “field-flow fraction (FFF)” which was developed in 1966 by Giddings.151 The AF4 instruments are commercially available and require minimal expertise (requires only basic knowledge of software) and can separate exomeres from other exosome subpopulations. Although the AF4 fractionation step takes only one hour, the total steps from cell culture to exosome/exomere isolation from the conditioned media by ultracentrifugation can take approximately three days. Although this method certainly has some major advantages, the significant drawbacks this method possesses are its inability to handle large samples and its inadequate separation of exosomes based on their sizes.29
Deterministic lateral displacement (DLD) arrays: DLD is a passive microfluidic technique that separates particles based on their size, shape, deformability, and charge. A flat microfluidic channel is filled with a regular array of micropillar obstacles, which creates a periodic flow pattern in a “zigzag” manner, creating the potential for the separation of both cellular and nanoparticles. It is a low-cost separation method.152
(Moved up) Field-free viscoelastic flow: This method is based on the principle that particle migration is caused by size-dependent elastic forces in a viscoelastic medium. This method is more precise than other microfluidics techniques because it is possible to separate particles of submicrometer diameter from a very small volume of samples.153
Fluorescence-activated sorting (especially for larger EVs including large apoptotic bodies and large oncosomes): This sorting method separates specific cell populations by phenotypes that can be detected by flow cytometry. This method is best for characterizing a single cell population without being contaminated by other cell populations.154
High-throughput/high-pressure methods such as fast protein/high-performance liquid chromatography (FPLC/ HPLC) that involve some form of chromatography
Hydrostatic filtration dialysis: Musante et al.155 demonstrated that urine exosomal vesicles can be effectively isolated by hydration pressure pushed through a dialysis membrane and samples passing through a dialysis membrane of 1000 kDa molecular weight cut-off are separated based on their sizes.
Ion exchange chromatography (IEX): It is a chromatographic separation method that separates molecules based on the net charge on the surface of the proteins. Depending on the ion, IEX is divided into 2 types, cation IEX and anion IEX. Since different proteins have different charges on their surface, this method of separation can easily isolate based on even the tiniest ion change on the surface of the proteins.156
Microfiltration: In order to isolate urinary exosomes, microfiltration methods are developed that uses a hydrophilized, commercially available membrane. This method can isolate LMW proteins from HMW proteins irrespective of the abundance of proteins in the cell sample population.157
Column-based separation protocols yield exosomes with high purity but they produce diluted exosomes and this process is time-consuming. This method of separation involves size exclusion chromatography.158,159
4.1 |. Importance of heterogeneity of exosomes
Exosomes are a heterogeneous group of EVs and their heterogeneity is due to their varied size, constituents, function, and cellular origin, which adds complexity to their characterization. Such diversity is likely because of the limiting membrane of MVBs during ILV formation or differences in molecular routes partaken during exosome biogenesis.160 This heterogeneity leads to differential exosome qualitative and quantitative content which in turn produces miscellaneous exosome subpopulations that are distinct in both their biophysical properties and composition. Generally, we can separate exosomes based on their sizes. Large exosomes (Exo-L) are 90–150 nm; small exosomes (Exo-S) are 60–80 nm in size, and the smallest exosomes are exomeres that are 30–35 nm in size. The exomeres are only recently discovered using asymmetric flow field-flow fractionation. Their study showed that exomeres can transfer functional cargo. In this study, AREG-containing exomeres and exosomes elicited prolonged EGFR effect to modulate EGFR trafficking in intestines, and significantly enhanced the growth of colonic tumor organoids. The increased activity of nanoparticle AREG elicited effects at 1:1,000th of the concentration of rAREG.29,161 Furthermore, separation with density centrifugation exosomes can be classified as high and low-density exosomes.78
Lee, Sang-Soo, et al. identified a new group of EV in the P200 vesicles that were smaller than exosomes in size. Exosomes and the P200 vesicles are found in CM (conditioned medium) of human cell lines. These involve a different biogenesis pathway that is independent of the endocytic pathway. While exosome markers (e.g., Hsp70, TSG101, and CD63) are present in both P100 and P200 vesicle types, the CD81 exosome marker is not detected in the smaller EVs. The addition of the P200 vesicles to human cell cultures enhanced exosome production and cell proliferation.162
5 |. METHODS OF ENGINEERING EXOSOMES USING DNA TECHNOLOGY
As the research enlightened exosomes’ stability, low immunogenicity, and permeability in the body, the idea of using exosomes as a diagnostic and therapeutic tool has emerged. Genetic engineering became a major tool for generating modified exosomes. These engineering processes served to display a peptide/protein on the surface as a cargo or targeting sequence, load cargo into exosomes, and escape micropinocytosis in the circulation.
5.1 |. Methods of designing exosomes to carry payload outside the exosomes
In one of the earliest studies of exosome engineering, Delcayre et al. reported that the lactadherin protein binds to exosome lipids with its C1C2 domain and presents on the exosome surface. They showed that engineered fusion proteins with C1C2 domains were presented in the exosomes and called this Exosome Display Technology.163 Another group used a similar strategy, engineered lactadherin with Gaussia luciferase, and overexpressed this construct in B16-BL6 cells. Following the exosome isolation and intravenous injection into mice, they could track exosomes in mice with bioluminescence imaging.164 Gassart et al. utilized the cytosolic domain of TM Env protein from the bovine leukemia virus and fused it with the CD8 ectodomain. Expression of this construct resulted in a CD8 enrichment in exosomes.165
LAMP2b is another useful exosome membrane protein expressed in murine exosomes76 and widely engineered to present polypeptides in exosome surfaces. Inserting a polypeptide following its N terminal signal peptide results in the expression of the poly-peptide fused with Lamp2b protein and presentation on the surface of exosomes. Alverez-Erviti et al.12 fused the neuron-specific peptide RVG to the LAMP2b DNA sequence and generated engineered exosomes with RVG peptides to target neurons. After loading exosomes with siRNA by electroporation, they observed a significant uptake of exosomes to the brain of wild-type mice, which resulted in specific knockdown of BACE1, a target in Alzheimer’s disease, in mRNA and protein level. Bellavia et al.166 utilized the Interleukin-3 fragment fused LAMP2b to target chronic myelogenous leukemia cells. By loading engineered exosomes with Imatinib or siRNA against BCR-ABL, they could inhibit the growth of CML cells in vitro and in vivo.
Stickney et al.167 investigated the use of exosomal surface proteins as an anchor for fluorescent proteins and demonstrated the feasibility of CD63, CD9, and CD81 fusions with RFP. They also showed the possibility of presenting fluorescent protein either in the lumen or at the surface, depending on the location of the inserted fluorescent protein in the CD63 sequence. Besides well-known exosomal surface proteins, Ohno et al. presented GE11 peptide on exosome surface by genetic engineering of platelet-derived growth factor receptor in HEK293 cells. By inserting Let-7a miRNA into modified exosomes with liposomes, they successfully targeted EGFR-expressing cancer cells in RAG2−/− mice and inhibited tumor growth.168 Curley et al.169 also investigated the topology of CD63, exosomal membrane protein, to optimize engineering exosomes to use delivering proteins and peptides.
Dooley et al.170 conducted a comprehensive study to identify exosomal proteins to carry proteins/peptides on the surface and inside the exosomes. Apart from previous studies, they conjugated GFP to candidate proteins to make this study a functional assay with ELISA and flow cytometry. After optimizing exosomes to work, followed by proteomics, they identified Prostaglandin F2 receptor negative regulator protein, PTGFRN, a previously unreported scaffold protein, to efficiently present GFP on the exosome’s surface. Finally, they completed the study with optimization of truncated PTGFRN, which has the potential to become a standard of exosome modification. The same group used PTGFRN as a scaffold to carry IL12 on the surface of the exosome, generating engineered exosome exoIL12. Intra-tumoral injections of exoIL12 showed greater antitumor activity than recombinant IL12 in the MC38 tumor model in mice. exoIL12 also demonstrated one typical advantage that is expected from engineered exosome treatment, compared to recombinant protein counterparts, prolonged half-life/retention. The complete response to exoIL12 at a rate of 63% compared to 0% at recombinant IL12 shows exosomes have the potential to bring many protein-based therapies into the clinic.171
Gao et al.172 developed a novel method to use exosomes for targeting and therapeutic purposes. They identified the CP05 peptide as a CD63 ligand using the phage display technology. By conjugating CP05 with different targeting peptides (M12 for muscle, RVG for neuron, SP94 for hepatocellular carcinoma), they achieved specific targeting of exosomes to target tissue. Furthermore, they showed dual-labeling with neuron-specific NP41 peptide and fluorescein isothiocyanate fluorescent marker, allowing feasible tracking and detecting of specific cells. Also using the amide linker, they conjugated antisense oligonucleotides for exon skipping therapy in Duchene muscular dystrophy in the mouse model and demonstrated an enhanced dystrophin expression. This approach could have great translational potential since it allows for modification of native exosomes of the organism and involves minimum disturbance. For example, it would be possible to collect exosomes from patients, label them with CP05-conjugated proteins or peptides, and give them back for diagnostic, and therapeutic purposes.
Other than genetic engineering of membrane proteins to load cargo on the surface, another method to conjugate peptides into the exosome membrane is click chemistry. Jia et al.173 modified the exosome membrane by inserting (1-Ethyl-3-[3-dimethylaminoprop yl]-carbodiimide hydrochloride-N-Hydroxysuccinimide (EDC-NHS) and attaching a neuropilin-targeting RGE peptide to target glioma. With the addition of curcumin and super paramagnetic iron oxide nanoparticle (SPION) into the exosome by electroporation, they showed SPION-labeled exosomes enriched in glioma on magnetic resonance imaging (MRI). The tumor volume is decreased and survival increased in the mouse glioma model. Kim et al.174 also used 1,2-Distearoyl-sn-glycero-3-phosphorylethanolamine (DSPE) and polyethylene glycol to incorporate AA ligand (which has a high affinity for sigma receptors in nonsmall cell lung carcinoma) into macrophage-derived exosomes. After loading paclitaxel into exosomes by sonication, they demonstrate that modified exosomes specifically target the pulmonary metastasis of the Lewis lung carcinoma mouse model and improve survival. Choi et al.175 modified exosomes by mannose-conjugated polyethylene glycol modification of exosomal membrane to target DCs. To increase immune response, monophosphoryl lipid A (adjuvant) loaded into exosomes in the presence of DMSO, and they managed to target DCs specifically and increased inflammatory cytokines TNF-α and IL-6. Figure 5 shows currently available methods to display protein on exosome surface.
FIGURE 5.

Current methods to display a cargo on the surface of exosomes. PDGFR, Platelet-derived growth factor receptor; PTGFRN, Prostaglandin F2 Receptor Inhibitor; BLV, Bovine Leukemia Virus; LAMP2b, Lysosome-associated membrane protein 2; NGFR, Nerve Growth Factor Receptor; DSPE, 1,2-Distearoyl-snglycero-3-phosphorylethanolamine; DMPE, 1,2-Dimyristoyl-sn-glycero-3-phosphoethanolamine; DOPE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; PEG, Polyethylene glycol; EDC-NHS, (1-Ethyl-3-[3-dimethylami- nopropyl]-carbodiimide hydrochloride – N-Hydroxysuccinimide; N3, azide radical
5.2 |. Methods to load cargo into exosomes
Exosome lumen can be used to carry protein and nucleic acid cargoes, as the content can travel without risk of degradation or unintended interaction. In one approach, Lai et al.176 genetically engineered cells to express nuclear localization signal (NLS) fused carrier protein (GFP) with MS2 coat protein (MS2CP) that would work as a dock inside the exosome. They also expressed a reporter mRNA with MS2 binding site (MBS), which will bind to MS2CP of docking protein. As a result, exosomes collected from these cells contained carrier protein with attached mRNA, fused with their MS2CP and MBS domains, respectively. Another docking approach utilized by Yim et al.177 who used the interaction between photoreceptor cryptochrome 2 (CRY2) and CIBN, truncated version of CRY-interaction basic-loop-helix 1 protein in a technique they called “EXPLOR.” by genetically integrating CIBN into the luminal side of CD9 and CRY2 into cargo protein, they were able photo-activate docking of cargo protein to CD9 through CRY2 and CIBN interaction and generated exosomes filled with cargo protein in the presence of blue light. They successfully delivered Cre-carrying exosomes into the brain of lox EYFP transgenic mice and demonstrated the expression of EYFP proteins in vivo. Further, this group used the same experimental design to introduce super-repressor IκB (srIκB), an engineered protein without phosphorylation sites, which inhibits translocation of nuclear factor κB into the nucleus to prevent sepsis. After generating engineered exosomes in HEK293T cells, they have shown that local injection of engineered exosomes significantly reduced inflammatory response and mortality in the septic mouse model.178
Dooley et al.,170 who identified PTGFRN protein to carry cargoes on exosome surfaces, also studied proteins to carry cargo inside the exosomes. They identified BASP1 as associated with the inner leaf-let of membranes. Further optimization with truncation of BASP1 identified eight amino acid peptides efficiently load GFP into exosomes, comparable to full-length BASP2 protein. Furthermore, an ovalbumin-loaded exosome, exoOVA, successfully induced IFNγ and OVA-reactive CD8 T cells much more efficiently than Ovalbumin alone. This also indicates the advantage of using engineered exosomes over recombinant protein counterparts.
Sterzenbach et al.17 reported that the late-domain pathway could be used to load molecules into exosomes. They fused the WW tag into Cre recombinase, which is recognized by late-domain containing protein Ndfip1, ubiquitylated, and subsequently loaded into exosomes. Upon nasal administration of these engineered exosomes, exosomes were taken up by floxed reporter cells, resulting in tdTomato expression, indicating functional delivery of proteins. They also found that proteinase K treatment did not diminish WW-Cre protein in the absence of Triton X-100, showing the cargo protein is located inside the exosome. We also employed this approach in our lab and found that WW tagged Neuroglobins enriched in exosomes (unpublished).
It might be argued that protein loading techniques into exosomes with physical force and disruption may damage the exosome membrane and cause content loss. Busatto et al.179 have used cationic amphiphilic molecules, which can penetrate membranes, to load proteins inside exosome.
There are several methods developed to load nucleic acid into the exosomes. Li et al.180 employed Human Antigen R (HuR), an RNA binding protein, into the luminal surface of exosomal membrane protein CD9 and loaded exosomes with specific miRNA, which bound to HuR through Adenylate-uridylate-rich elements (AU-rich elements). As a result, after successfully delivering engineered exosomes to target cells, they reduced the target protein expression in vivo and in vitro. They also delivered CRISPR/dCas9 system in vivo by adding AU-rich elements to dCas9 mRNA and repressed C/ebpα expression. Kojima et al.181 developed an RNA packaging device using archaeal ribosomal protein L7Ae that binds to the C/Dbox RNA structure. They conjugated L7Ae into the C terminus of CD63 to place inside exosomes and inserted the C/Dbox region in the 3′-untranslated region of reporter gene coding nanoluc bioluminescence reporter protein. Along with RVG targeting peptide attached to exosome in LAMP2b exosomal membrane protein, they demonstrated that exosomes targeted the brain and delivered their mRNA, and detected luminescence in target cells. Figure 6 shows genetic engineering and physical methods to load cargo into exosomes.
FIGURE 6.

Methods to load cargo inside exosomes. (A) Genetic engineering methods to load exosomes with protein and nucleic acid. MS2CP, MS2 coat protein; MBS, MS2 binding site; CRY2, cryptochrome 2; CIBN, truncated version of CRY-interaction basicloop- helix 1 protein; NLS, nuclear localization signal; BASP1, Brain Abundant Membrane Attached Signal Protein 1; HuR, Human Antigen R. (B) Physical methods to load proteins and nucleic acids into exosomes
5.3 |. Immunological use of engineered exosomes
Huang et al. modified HELA cells by overexpression α-Lactalbumin (α-LA), a breast-specific protein expressed in human breast cancers, and collected α-LA-enriched exosomes. After loading TLR3 agonist Hiltonol and immunogenic cell death inducer human neutrophil elastase, they treated mouse breast tumor models with this exosome. They found an increased accumulation of DCs and CD8 T cells in the tumor and reduced tumor size in MDA-MB-231 tumor-bearing mice.182
Antigen-presenting features of DCs are key in inducing the immune response. Dendritic cell exosomes have the potential to induce the immune system. Hong et al.183 modified dendritic cell-derived exosome to utilize MHC-I molecule on the surface of exosome by integrating respiratory syncytial virus antigen. Despite the failure to activate CD8 T cells in vivo, it is in vitro success proves that it works but needs further optimization. In another effort, Kim et al.184 genetically engineered K562 cells by overexpressing human leukocyte antigen-A2 and costimulatory molecules CD80, CD83, and 41BBL to use exosomes to stimulate antigen-specific CD8 T cells. This effort to overcome the inherent difficulty of exosome generation in DCs for the same purpose proved successful in cell culture by having a comparable CD8 stimulation. This approach could be used for adoptive cell therapies. In another attempt to employ exosomes in immune system activation, Morishita et al.185 developed a lactadherin with streptavidin fusion protein and genetically engineered murine B16-BL6 melanoma cells. After collecting modified exosomes, they incubated exosomes with biotinylated CpG DNA (innate immune response activators) and labeled these exosomes using streptavidin on the exosome surface. They reported these engineered exosomes activated DC2.4 cells and enhanced their tumor antigen presentation.
Exosomes, through engineering surface proteins, can turn into immunological reagents. Hartman et al.186 used the C1C2 domain of lactadherin to present carcinoembryonic antigen (CEA) and human epidermal growth factor receptor 2 (HER2) tumor-associated antigens on exosome surface to antigen-presenting cells to enhance the anti-tumor immune response. By fusing CEA and HER2 to the C1C2 domain, they expressed these antigens on the exosome surface and enhanced T and B cell responses. Shi et al.187 turned exosomes into immunological mediators by anchoring anti-CD3 and anti-HER2 into the exosomal membrane by PDGFR. By dual targeting T cells and HER2-expressing cancer cells, the directed T cell demonstrated anti-tumor activity in the mouse breast cancer model. We used LAMP2b protein to display CD206-targeting peptide to target M2 type of macrophages and Fc fragment of mouse IgG2b to bind natural killer cells, aiming antibody-dependent cellular cytotoxicity (ADCC) to eliminate M2 type macrophages. Our engineered exosomes specifically targeted M2 macrophages and significantly reduced CD206+ cells in vivo. Further treatment with engineered exosomes reduced tumor growth and prevented early metastasis in 4T1 tumors in mice.26 Figure 7 shows the immunological use of engineered exosomes.
FIGURE 7.

Immunological use of engineered exosomes. CEA, carcinoembryonic antigen; HER2, human epidermal growth factor receptor 2; RSV, respiratory syncytial virus; IL6ST, Interleukin 6 Cytokine Family Signal Transducer, mIgG2b, mouse immunoglobulin G 2b
Fan et al. followed a hybrid approach in engineering exosomes for immunotherapy. First, they collected ovalbumin-induced dendritic cell exosomes, which already express MHC and CD86, needed for T cell activation. Then, they enriched the exosome membrane with anti-CD3 and anti-EGFR receptors to bind T cells and B16-OVA tumor cells, respectively, and induce cytotoxicity by bringing them into the vicinity. Engineered exosome treatment resulted in an immune response augmented with a PD-L1 inhibitor, decreased tumor size, and increased survival in the B-16 OVA tumor model in mice.188
In the TME, one commonly studied phenomenon is the M1 and M2 macrophages and their pro-inflammatory and immunosuppressive roles, respectively. Gunassekaran et al.189 engineered M1-derived exosomes to deliver siRNA and miRNA to M2 type of macrophages to induce M2 to M1 polarization. To achieve this, M1 exosomes were transfected with miR-5aa-3p and NK-κB siRNA. To target M2 macrophages, IL4R-binding peptide attached to exosome membrane using DOPE-PEG amine. The engineered exosomes achieved M2 to M1 polarization and reduced the tumor volume in the 4T1 mouse breast tumor model. Engineered exosomes could also modify the immune system through their displayed proteins. Conceição et al.190 engineered exosomes as a decoy for interleukin 6 (IL-6), a key mediator of inflammation in skeletal muscle, to inhibit the IL-6 trans-signaling pathway and inflammation. They found engineered exosomes reduced STAT3 signaling, which indicates the inhibition of the inflammation and shows it can be used in Duchenne treatment to reduce muscle wasting as an alternative to anti-inflammatories. Duong et al.191 have engineered exosomes by presenting the TNFα receptor on their surface to antagonize TNFα and prevent inflammation in vitro model.
5.4 |. Engineered exosomes in metastasis
Tumor exosomes indicate the immune status and play a significant role in metastasis. In an indepth study, Chen et al.192 found that PD-L1, which suppresses immune response against the tumor, on exosomes was abundant in metastatic melanoma compared to healthy donors. Exosomal PD-L1 was found to inhibit CD8 T cells and correlated with poor Pembrolizumab (antibody against PD-L1 receptor, PD-1) response. This indicates that tumors use exosomes as “decoy” to overcome immune responses.
One of the central concepts in cancer metastasis research is the “seed and soil” hypothesis. Suetsugu et al.193 tagged CD63 exosome membrane protein with GFP in tumor cells and tracked the exosome traffic in mouse breast cancer models. They were able to track tumor-derived exosomes in organs and tumor-associated cells in the circulation and demonstrated the use of exosome tracking in investigating cancer metastasis. This approach would be particularly useful in metastasis research by tracking exosome traffic from cancer cells. Pucci et al.194 adopted a methodical approach to investigating tumor cell communications by modifying melanoma to express luciferase. They found luciferase activity in tumor-draining lymph nodes and identified CD169+ macrophages as a tumor suppressor that prevents tumor exosome spread. This study proves that the exosome study holds a great potential to understand metastasis. Pucci’s group further engineered exosomes to express bacterial Sortase A on PDGFR or dNGFR membrane proteins, which transfers substrate peptides (e.g., biotin-containing peptides) to N terminal glycine of surface proteins. Compared to employing GFP-labeled exosome, this method showed 10–100 fold increased sensitivity in detecting exosome-target cell interaction and a promising strategy to study specific exosome-cell interactions.195
5.5 |. Engineered exosomes in various diseases
Organ and tissue-specific exosome delivery are achieved by also physical forces. Lee et al. loaded mesenchymal stem cell-derived exosomes with iron oxide nanoparticles by supplying them in cell culture to increase the delivery to target organs. By implanting a magnet next to the heart, they achieved an increased delivery into the infarcted myocardium.196 In another concept study, researchers labeled exosomes through their transferrin receptor using superparamagnetic iron oxide nanoparticles conjugated to transferrin with the help of carboxylated chitosan. Exosomes are loaded with BAY55–9837 peptide for type 2 diabetes mellitus treatment and targeted to pancreatic islet cells using magnets to attract SPION labeled exosomes. They observed a significant increase in delivery of exosomes and alleviation of hyperglycemia in db/db diabetic mice.197 Mizuta et al.198 also used magnetic nanogels to increase the delivery of exosomes. After hybridizing exosomes with magnetic nanogels, magnetic force significantly improved the delivery of exosomes to cells in culture.
Liu et al. utilized the intrinsic feature of ferritin use as an MRI contrast reagent and engineered exosomes to carry ferritin in modified lactadherin transmembrane protein on the exosome surface. With this, they were able to use exosomes as MRI contrast reagents without the need for further labeling.199 Furthermore, with further modification to target specific cells, engineered exosomes could be used to image cells or tissue in the body.
Maguire et al. found that Adeno-associated virus (AAV) generating cells also release the virus in exosomes, and called these “vexosomes.” They found vexosomes have outperformed AAV alone in transfecting the cells. Further modification of the exosome membrane with biotin attachment and magnetic bead labeling, followed by attraction with magnets in cell culture further improved the transfection efficiency.200 Maguire’s group further explored the exosome-associated AAV gene delivery/therapy in the mouse. They found the same level of exosome-associated AAV delivery in blood, but lesser performance, still comparable in the spleen, lymph node, and liver compared to conventional AAV.201 With the use of engineered exosome-associated AAV to target specific cells, exosomes would be a new and more effective method to be used in gene delivery and therapy.
Jhan et al.202 fused exosomes with synthetic lipids to increase the number of vesicles and increased the vesicle amount 6–43 fold. Their siRNA cargo loading and delivery were successful. Sato et al.203 used freeze-thaw cycles to fuse functional lipids with exosome membrane. Although these processes have the advantage of increasing quantity and modifying membranes, many cargo proteins might be lost, and surface proteins lessened through the process.
Membrane receptors are major drug targets, and molecular assays in protein’s native conformation are crucial in biotechnology and clinical research. Desplantes et al.204 engineered exosomes to study multiple membrane proteins by directing membrane proteins to exosome membranes by conjugating patented “DCTM” peptides.
Exosomes are subject to elimination via multiple mechanisms in circulation, and there are various studies to prolong exosome half-life in the organism (Figure 8). Hung et al.205 observed an in-sufficient peptide presentation in engineered LAMP2b of exosomes. They hypothesized that glycosylation would protect these peptides and demonstrated that glycosylation protects peptides in LAMP2b and enhances the delivery of exosomes to recipient cells. Kamerkar et al.206 found that CD47, a ligand for signal regulatory protein (SIRPα), is crucial in protecting exosomes from micropinocytosis, the presence of CD47 on the surface protects exosomes from phagocytosis. They also showed loading exosomes with KrasG12D shRNA, which targets a common mutation in pancreatic cancer, proved to suppress pancreatic tumor growth and metastasis in mice in the presence of CD47. Another group investigated different aspect and use of CD47 interaction. Koh et al.207 stated that CD47 is present in most tumors, making the tumors immune to phagocytosis. They overexpressed SIRPα in HEK293 cells and generated SIRPα-enriched exosomes using pDisplay. By saturating all CD47 (don’t eat me) receptors of tumors with these engineered exosomes, they showed a significant decrease in tumor volume of CT26.CL25 in immunocompetent BALB/c mice, but not in HT29 in BALB/c nude mice, indicates T cell immunity may be required for effective treatment in CD47 blockade. In another attempt to prevent the elimination of exosomes, Lathwal et al.208 used cholesterol-modified DNA tethers and complementary DNA block copolymers to enhance the stability of exosomes. They found modified exosomes have fourfold higher blood circulation time. The methods to extend exosome half-life in the organism are described in Figure 8.
FIGURE 8.
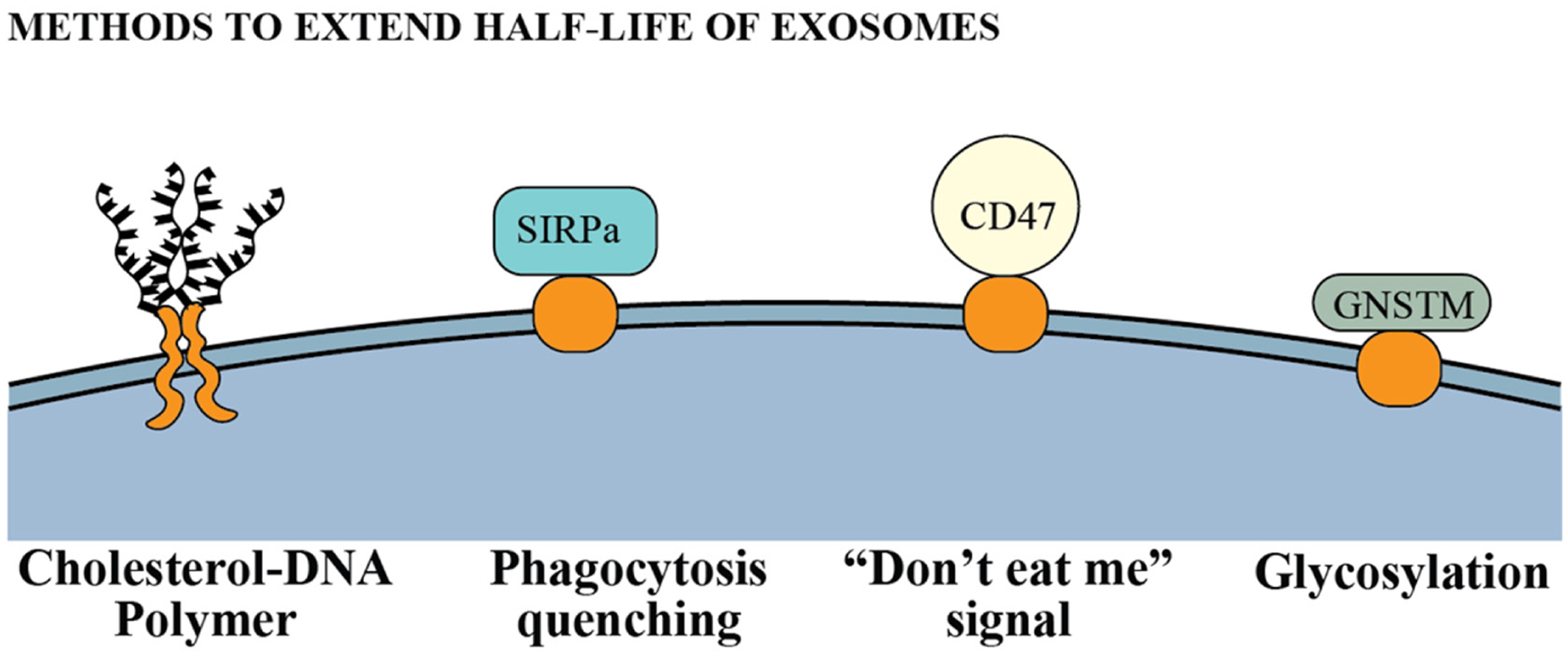
Engineering methods to extend half-life of exosomes in circulation. SIRPα, signal regulatory protein; GNSTM, glycosylation motif
6 |. USE OF ENGINEERED EXOSOMES FOR TARGETING SPECIFIC CELLS IN VIVO
Based on the above description and details of the engineering method of exosomes using DNA technology, it should be obvious to the readers that engineered exosomes could be the next nanotechnology that would be widely used to target specific cells in vivo not only to determine the distribution of specific cells and enhance the functional status of specific cells but also to target and deplete the specific cells. Irrespective of the origin or parent cells, exosomes share common features such as certain tetraspanins (CD9, CD63, and CD81), heat shock proteins (HSP 60, Hsp 70, and Hsp 90), biogenesis-related proteins (Alix and TSG 101), membrane transport and fusion proteins (GTPases, annexins, and Rab proteins), nucleic acids (mRNA, miRNA, and long noncoding RNAs and DNAs), and lipids (cholesterol and ceramide).2,7,8 Investigators have started making engineered exosomes to carry biologically active protein on the surface or inside the exosomes and using exosomes to carry drugs to the site of interest.11,16–19 Recently, our laboratory has achieved a few milestones in exosome technology.25,26 Our laboratory is heavily engaged in the investigations of the TME and microenvironment of cerebrovascular diseases (CVD). We are working on determining the roles of myeloid cells, especially MDSC and immune suppressive M2 macrophages in the TME, and the roles of neutrophils on the exacerbation of edema in stroke or their roles in tumors following therapies. The following paragraphs will detail the methods and possible utility of immune cell-specific engineered exosomes that can be used to target and deplete cells in the TME or CVD.
6.1 |. Engineered exosomes to target M2-macrophages
Depending on the stimuli, macrophages undergo a series of functional reprogramming as described by two different polarization states, known as M1 (classically activated) and M2 (alternatively activated).209,210 Phenotypically, M1 macrophages express high levels of major histocompatibility complex class II (MHC II), the CD68 marker, and co-stimulatory molecules CD80 and CD86. On the other hand, M2 macrophages express high levels of MHC II, CD163, CD206/MRC1, Arg-1 (mouse only), and others. M2-polarized macrophages are induced by IL-4, IL-13, IL-21, and IL-33 cytokines.211,212 M2 macrophages release high levels of IL-10, transforming growth factor-beta (TGF-β), and low levels of IL-12 and IL-23 (type 2 cytokines). M2 macrophages also produce CCL-17, CCL-22, and CCL-24 chemokines that regulate the recruitment of Tregs, Th2, eosinophils, and basophils (type-2 pathway) in tumors.213,214 The Th2 response is associated with the anti-inflammatory and immunosuppressive microenvironment. CD206, also known as mannose receptor (MR), is a 175 kD type-I membrane protein and is expressed predominantly by alternatively activated M2 macrophages and resident tissue macrophages mostly in the lungs, spleen, and liver.215 It functions in endocytosis and phagocytosis and plays an important role in immune homeostasis by scavenging unwanted mannoglycoproteins.216 Alternately activated M2 macrophages are known to be associated with therapy-resistant, metastasis, and poor survival in different malignant tumors.217–219 Figure 9 shows an increased number of M2 macrophages in metastatic breast cancer and there is poor overall survival and disease-free survival in tumors showing a higher number of MRC1.
FIGURE 9.
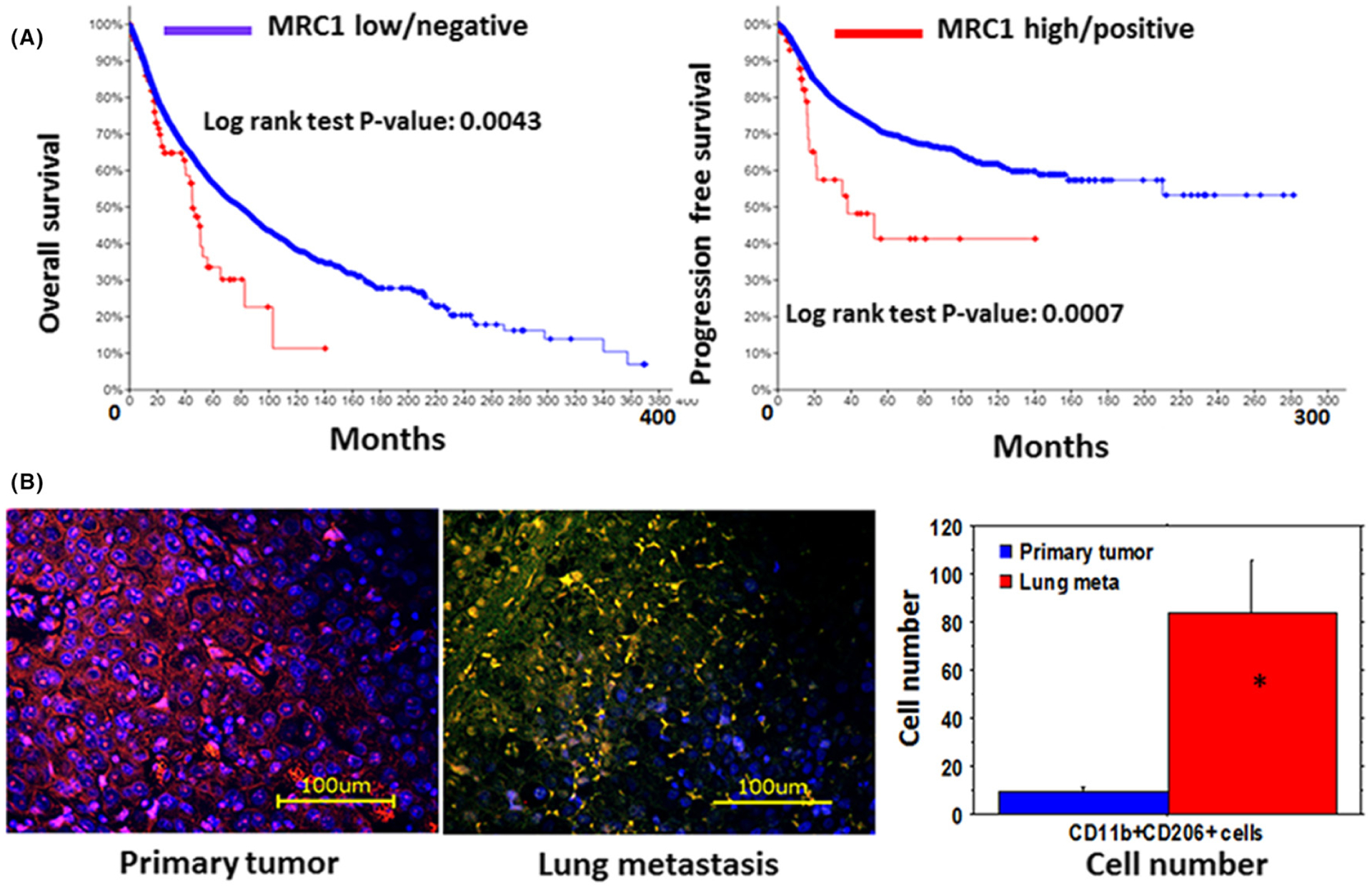
(A) Disease-free and overall survival of patients with different cancers expressing mannose receptor (MRC1) in the tumor tissues (TCGA data). (B) Increased number of CD206+/CD11b+ cells in lung metastasis (middle panel, yellow cells) from breast cancer compared to that in the primary tumor (left panel). Quantitative analysis showed a significantly increased number of CD11b+CD206+ cells. The samples are from multiple patients and randomly selected histochemical sections (n = 6). * = P < 0.01
In recent years, investigators have identified a peptide sequence CSPGAKVRC or its linear form CSPGAK that binds specifically to CD206+ M2 macrophages in the tumors and sentinel lymph nodes in different tumor models.220,221 It is to note that the linear form of this peptide CSPGAK also binds to human M2 macrophages.221 We have developed engineered exosomes that carry these peptides and precisely detected M2 macrophages both in vitro and in vivo and showed our results in recent publications.26,222 We have used nontumorous cells (HEK-293 cells) to develop the engineered endosome carrying M2-macrophages targeting peptides as well as the Fc-portion of mouse IgG2b (Fc-mIgG2b) on the surface to target and deplete alternatively activated immunosuppressive CD206+ M2 macrophages in vivo through ADCC and apply these engineered exosomes to alter immunosuppressive TME to enhance the effect of different therapies (including immunotherapy) to decrease tumor burden and improve survival. Figure 10 shows the vector design and mechanisms of action of the engineered exosomes to initiate ADCC to kill targeted cells.
FIGURE 10.
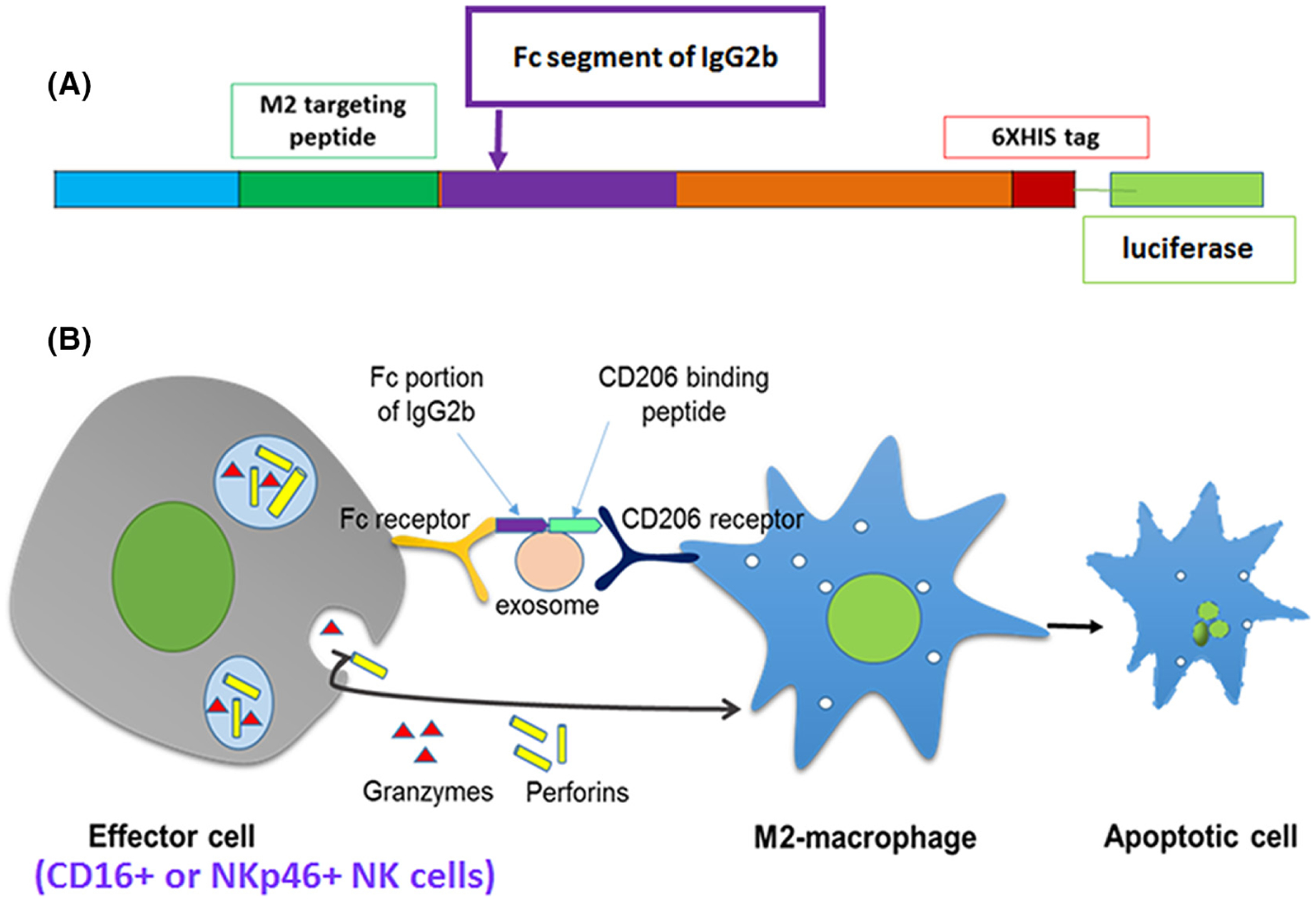
(A) Vector design to express M2 targeting peptide and Fc-mIgG2b on exosomes. (B) Cartoon to show the mechanisms of ADCC through engineered exosomes
6.2 |. The exosome is a better vehicle to enhance antibody-dependent cellular cytotoxicity
ADCC is a nonphagocytic mechanism by which most NK cells (effector cells) can kill antibody-coated target cells in the absence of complement and without major histocompatibility complex (MHC).223 Targeted therapy utilizing monoclonal antibodies (mAbs) has instituted immunotherapy as a robust new tool to fight against cancer and other noncancerous disorders, such as cryoglobulinemia, Wegener’s granulomatosis, and bullous pemphigoid.224,225 As mAb therapy has revolutionized immunotherapies, ADCC has become more applicable in a clinical context. Clinical trials have demonstrated that many mAbs perform somewhat by eliciting ADCC.226 Antibodies serve as a bridge between Fc-receptors on the effector cell and the target antigen on the cell that is to be killed. Crosslinking of receptors in both effector cells and target cells is required for triggering the cytotoxic event. ADCC occurs through various pathways, including (a) release of cytotoxic granules; (b) TNF family death receptors signaling; (c) release of pro-inflammatory cytokines, such as IFN-γ.227 Both the uptake of perforin and granzymes by target cells and TNF family death receptor signaling induce target cell apoptosis,228 while effector cell-released IFN-γ actuates nearby immune cells to stimulate antigen presentation and adaptive immune responses.229 Our goal is to target the Fc gamma-receptor (FcγR)-based platform to deplete M2 macrophages (Figure 10). We have identified the sequence of the Fc-mIgG2b that triggers FcγR-mediated phagocytosis and cytotoxicity230 and recently we have reported the utility of engineered exosomes as imaging and therapeutic probes.26 It is to note that we have also identified the sequence Fc portion of human IgG that triggers FcγR-mediated phagocytosis and cytotoxicity for designing human M2 macrophages targeting engineered exosomes.
Because of the cellular origin, exosomes show enhanced permeability even through the intact BBB, which is an advantage over synthetic nanoparticles.231–234 Exosomes are also shown to utilize enhanced permeability and retention (EPR) effects.234,235 Due to higher stability in biological fluids and enhanced permeability, exosomes are better for targeted delivery of therapeutic payloads.231–234,236 Investigators have used either synthetic nanoparticles or fusion protein to deliver Fc-IgG2b to initiate ADCC but because of the rigid body, synthetic nanoparticles rely most on the ERP effect and reports are showing a lack of ADCC following tagging with gold nanoparticles.237–241 Moreover, due to a size-dependent manner, synthetic nanoparticles can be cleared by the kidneys or reticuloendothelial system, even with targeting moieties.242–244 On the other hand, fusion protein-based ADCC did not show promise due to rapid clearance and nonspecific bindings.245–248 Antibody-mediated ADCC also depends on the antibody design with intact Fc-portion and specific attachment to the target cells.249 Most of the antibodies that are used to initiate ADCC are monoclonal.250 We postulate that engineered exosomes developed in nontumorous cells, HEK293, will be a better choice to carry therapeutic payloads to enhance ADCC.
6.3 |. Engineered exosomes to target Myeloid-derived suppressor cells or CSF1R+ myeloid cells
Our decade-long investigations and investigations by others proved that bone marrow-derived progenitor cells (BMDPC) influence the TME tremendously causing dynamic changes from inflammatory to the immunosuppressive milieu, neovascularization, recurrence, local invasion, and distal metastasis.251–258 These dynamic changes are pronounced due to mobilization and accumulation of BMDPC following different therapies including radiation, chemo, and antiangiogenic causing therapy resistance.253,254,256,259–261 Based on the status of the microenvironment such as inflammatory vs immunosuppressive, the treatment effects differ significantly and the recent addition of immunotherapy also becomes noneffective in the solid tumors.255,256,262,263 Recently, we have pointed out the involvement of myeloid cells in the development of therapy resistance and recurrence of different tumors.255,256,264,265 In our recent publications, we have used small molecular agents that inhibit CSF1R tyrosine kinase and showed the retardation of growth of GBM and breast cancers, which was corroborated with animal models where all CSF1R+ cells were conditionally depleted.253,256,266,267 Therefore, using DNA engineering technology we can make exosomes to carry CSF1R targeting peptides and use payloads for depleting the myeloid cells at different stages of TME status. We have already identified a truncated version (peptide sequences from 36 aa to 147 aa) of CSF1 protein, which showed 100% sensitivity to react with CSF1R and made vectors for making engineered exosomes. We used a similar platform as shown in Figure 10 to make the vector and engineered exosomes. We are also in preparation to make targeting exosomes without inserting Fc-mIgG2b to see the distribution of CSF1R+ cells in the TME at different stages of TME following therapy. We stipulate that our engineered exosomes to target and deplete CSF1R+ cells along with established immunotherapy (anti-PD1) will show synergistic effects.
6.4 |. Engineered exosomes to target neutrophils
Each year more than 795 000 people in the United States have a stroke and it kills about 140 000 people, placing a $34 billion annual economic burden on society.268 Though major advances in our understanding of cerebral ischemia have been made, the need for novel effective therapies remains imperative. Unfortunately, the success of different therapies is highly variable, and none can be employed early before significant vascular pathology and damage to the brain have occurred. Activated neutrophils have pivotal roles in acute ischemic brain injury, atherosclerosis, and thrombus formation.269 Neutrophils are the most abundant polymorph nuclear (PMN) white blood cells (WBCs) in the blood and make up part of the innate immune system. Neutrophils are an essential part of the inflammatory cascade, being the first cell type to migrate from the bloodstream to the site of inflammation.270,271 Following recruitment, neutrophils get activated and subsequently express adhesion molecules and release reactive oxygen species, cytokines/chemokines, and proteolytic enzymes causing damage to the tissues.272,273 Infiltration of neutrophils in the ischemia-reperfusion stroke area occurs early, at the same time as brain injury. This increased accumulation of neutrophils is associated with stroke severity,274 infarct volume,275 and worse functional outcomes.276 Several studies have started to evaluate broadly targeting anti-neutrophil treatments to minimize stroke injury and to improve stroke outcomes.269,277,278
Our laboratory studies in male B6 mice (10–12 months old) (Figure 11) show the mobilization of neutrophils (Ly6G+), natural killer (NK) cells (NKp46+CD3−), macrophages (F4/80), and M1 macrophages (CD80) in the peripheral blood and at the sites of a stroke at 3, 24, and 72 hours after stroke.
FIGURE 11.
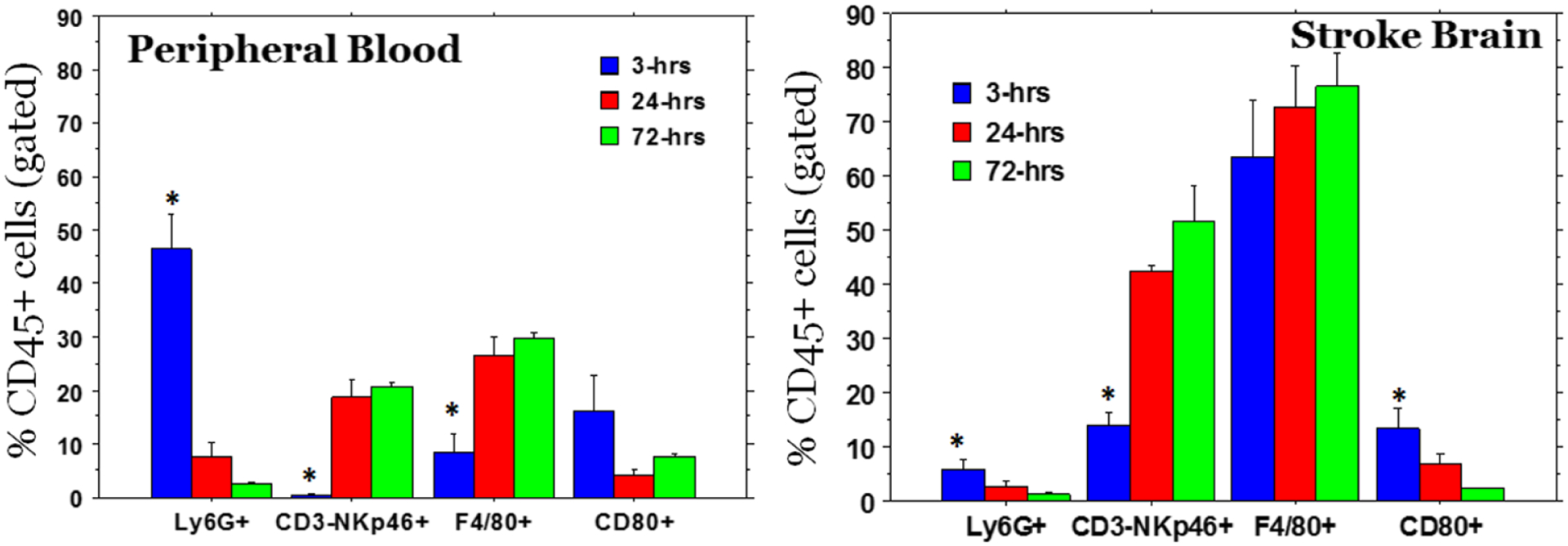
Mobilization of neutrophils and M1 macrophages in the peripheral blood and in the stroke area was observed as early as 3 hrs. Whereas other cell types such as NK cells and macrophages (F4/80+, which also contain M2 type macrophages) gradually increased in the stroke areas. Following collection of peripheral blood from each stroke animal, animals were euthanized and perfused with ice cold PBS and the brain tissues from stroke area were collected and single cell suspensions were made for flow cytometry. * = significant differences
Recent studies also pointed out the involvement of tumor-associated neutrophils (TAN) and tumor-associated macrophages (TAM) in maintaining the inflammatory or immunosuppressive TME that dictates the effect of therapies.217–219,279–286 Neutrophils are the most abundant polymorph nuclear (PMN) cells available in the peripheral blood and early accumulated tumor-associated cells following therapies that make the inflammatory milieu.280,286–290 However, based on the tumor cell-secreted cytokines and chemokines due to therapy insults, tumor-associated TAN polarized into N1 (CD11b+Ly6G+CD206-TNF-α+) and N2 (CD11b+Ly6G+CD206+IL10+) phenotypes.
Neutrophil migration to sites of inflammation and subsequent activation and multiple functions are highly regulated and orchestrated processes that are controlled by interactions between numerous receptors and their cognate ligands. FPRs are G protein-coupled receptors that transduce chemotactic signals in phagocytes and mediate host-defense as well as inflammatory responses including cell adhesion, directed migration, granule release, and superoxide production.291 Although there are a few ligands that are an agonist for FPRs, we cannot utilize those for targeting neutrophils because they may stimulate the neutrophils for hyper-functioning. Chemotaxis inhibitory protein of S. aureus is a native protein and part of it is FTFEPF, which shows FPR (specially FPR1) antagonistic activity.292 A coronavirus 229E-derived 12-mer peptide (ETYIKPWWVWL) was identified as a potent antagonist of FPR1 with a Ki of 230 nM.293 Investigators have pointed out a lower survival probability if FPR1 is highly expressed in breast cancer patients (Figure 12). We have used our platform (vector design, Figure 10) to make engineered exosomes to target and deplete activated neutrophils at the lesions (stroke or tumors) and in the peripheral blood. Our initial studies showed decreased number of neutrophils in the stroke areas following IV administration of the engineered exosomes.
FIGURE 12.

Survival probability in BRCA+ breast cancer patients expressing FPR1. (TCGA data from UACLAN)
7 |. KEY TAKEAWAYS
Genetic engineering and customizing exosomes create an unlimited opportunity to use in diagnosis and treatment. Very little use has been discovered, and we are far away to reach its limits.
Exosomes, in a sense, work like hormones and transfer messages between cells. They have, along with potentially bigger extracellular vesicles, the potential to revolutionize cancer metastasis research and expand our understanding of it.
Because the human body already has exosomes, making use of their own exosomes after isolation and extracorporal modifications with treatment/diagnosis approaches that are already in use could fasten the entering of exosomes into the clinic. For example, after collecting patients’ exosomes, they could be loaded with an Alzheimer’s drug that has poor blood–brain barrier permeability. And at the same time, exosomes could be tagged with neuron-specific peptides to increase the permeability and targeting capabilities.
Because exosomes carry proteins, lipids, nucleic acid, and metabolites from their parental cells, it should be considered a transfusion/ transplant. Until the safety of extra-corporal sourced exosomes, its clinical use will not be possible. Once the safety of these extra-corporal exosomes is proven, we could see many different uses of it in near future. Another way to overcome this safety concern is to collect and modify exosomes with already approved treatment modalities, which could be more safe and potential translate into the clinic in a short time.
ACKNOWLEDGEMENTS
The study was funded by American Heart Associate (AHA) grant 19TPA34850076 and part of the Georgia Cancer Center start-up fund and NIH grant R01NS110378 to ASA.
Footnotes
CONFLICT OF INTEREST
None declared.
DATA AVAILABILITY STATEMENT
Data derived from public domain resources. Data available on request from the authors.
REFERENCES
- 1.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Corbett AL, Taatizadeh E, et al. Challenges and opportunities in exosome research—Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3(1):011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. [DOI] [PubMed] [Google Scholar]
- 4.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8(5):470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113(Pt 19):3365–3374. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W, Peng P, Shen K. Role of exosome shuttle RNA in cell-to-cell communication. Acta Acad Med Sinicae. 2016;38(4):480–483. [DOI] [PubMed] [Google Scholar]
- 7.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255–289. [DOI] [PubMed] [Google Scholar]
- 8.Kalluri R The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ELA S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12(5):347–357. [DOI] [PubMed] [Google Scholar]
- 10.Lener T, Gimona M, Aigner L, et al. Applying extracellular vesicles based therapeutics in clinical trials – an ISEV position paper. J Extracell Vesicles. 2015;4:30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang XC, Gao JQ. Exosomes as novel biocarriers for gene and drug delivery. Int J Pharm. 2017;521(1–2):167–175. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. [DOI] [PubMed] [Google Scholar]
- 13.El Andaloussi S, Lakhal S, Mager I, Wood MJ. Exosomes for targeted siRNA delivery across biological barriers. Adv Drug Deliv Rev. 2013;65(3):391–397. [DOI] [PubMed] [Google Scholar]
- 14.El-Andaloussi S, Lee Y, Lakhal-Littleton S, et al. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7(12):2112–2126. [DOI] [PubMed] [Google Scholar]
- 15.Mashouri L, Yousefi H, Aref AR, Ahadi Am, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer 2019;18(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellavia D, Raimondi L, Costa V, et al. Engineered exosomes: a new promise for the management of musculoskeletal diseases. Biochim Biophys Acta Gen Subj. 2018;1862(9):1893–1901. [DOI] [PubMed] [Google Scholar]
- 17.Sterzenbach U, Putz U, Low L-H, Silke J, Tan S-S, Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol Ther. 2017;25(6):1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38:754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian T, Zhang H-X, He C-P, et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. [DOI] [PubMed] [Google Scholar]
- 20.Kugeratski FG, Kalluri R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021;288(1):10–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taghikhani A, Farzaneh F, Sharifzad F, Mardpour S, Ebrahimi M, Hassan ZM. Engineered tumor-derived extracellular vesicles: potentials in cancer immunotherapy. Front Immunol. 2020;11:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawikova I, Askenase PW. Diagnostic and therapeutic potentials of exosomes in CNS diseases. Brain Res. 2015;1617:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pi YN, Xia BR, Jin MZ, Jin WL, Lou G. Exosomes: powerful weapon for cancer nano-immunoengineering. Biochem Pharmacol. 2021;186:114487. [DOI] [PubMed] [Google Scholar]
- 24.Tan A, De La Peña H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomed. 2010;5:889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashid MH, Borin TF, Ara R, et al. Differential in vivo biodistribution of 131I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomedicine. 2019;21:102072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid MH, Borin TF, Ara R, Alptekin A, Liu YT, Arbab AS. Generation of novel diagnostic and therapeutic exosomes to detect and deplete protumorigenic M2 macrophages. Adv Ther-Germany. 2020;3(7):1900209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36(3):301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cell. 2019;8(7):727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Higginbotham JN, Jeppesen DK, et al. Transfer of functional cargo in exomeres. Cell Rep. 2019;27(3):940–954.e946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teng F, Fussenegger M. Shedding light on extracellular vesicle biogenesis and bioengineering. Adv Sci. 2020;8(1):2003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–5565. [DOI] [PubMed] [Google Scholar]
- 32.Han J, Pluhackova K, Bockmann RA. The multifaceted role of SNARE proteins in membrane fusion. Front Physiol. 2017;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420(2):133–154. [DOI] [PubMed] [Google Scholar]
- 34.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. [DOI] [PubMed] [Google Scholar]
- 35.Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JC, Bégin LR, Bérubé NG, et al. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin Cancer Res. 2007;13(8): 2354–2361. [DOI] [PubMed] [Google Scholar]
- 37.Rous BA, Reaves BJ, Ihrke G, et al. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13(3):1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bobrie A, Colombo M, Krumeich S, Raposo G, Thery C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J Extracell Vesicles. 2012;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Susa KJ, Seegar TC, Blacklow SC, Kruse AC. A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking. eLife. 2020;9:e52337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little KD, Hemler ME, Stipp CS. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol Biol Cell. 2004;15(5):2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9(4):1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26(4):399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen W, Huang Y, Han J, et al. Immunomodulatory effects of mesenchymal stromal cells-derived exosome. Immunol Res. 2016;64(4):831–840. [DOI] [PubMed] [Google Scholar]
- 44.Gauvreau ME, Côté MH, Bourgeois-Daigneault MC, et al. Sorting of MHC class II molecules into exosomes through a ubiquitin-independent pathway. Traffic. 2009;10(10):1518–1527. [DOI] [PubMed] [Google Scholar]
- 45.Wang M, Su Z, Amoah BP. Crosstalk among colon cancer-derived exosomes, fibroblast-derived exosomes, and macrophage phenotypes in colon cancer metastasis. Int Immunopharmacol. 2020;81:106298. [DOI] [PubMed] [Google Scholar]
- 46.Wang R, Ji Q, Meng C, et al. Role of gingival mesenchymal stem cell exosomes in macrophage polarization under inflammatory conditions. Int Immunopharmacol. 2020;81:106030. [DOI] [PubMed] [Google Scholar]
- 47.Rana S, Yue S, Stadel D, Zöller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012;44(9):1574–1584. [DOI] [PubMed] [Google Scholar]
- 48.Xia Y, He XT, Xu XY, Tian BM, An Y, Chen FM. Exosomes derived from M0, M1 and M2 macrophages exert distinct influences on the proliferation and differentiation of mesenchymal stem cells. PeerJ. 2020;8:e8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H, Wang SY, Kwak G, Yang Y, Kwon IC, Kim SH. Exosome-guided phenotypic switch of M1 to M2 macrophages for cutaneous wound healing. Adv Sci. 2019;6(20):1900513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bardi GT, Smith MA, Hood JL. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine. 2018;105:63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker ND, Elias M, Guiro K, et al. Exosomes from differentially activated macrophages influence dormancy or resurgence of breast cancer cells within bone marrow stroma. Cell Death Dis. 2019;10(2):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Si L, Bai J, Fu H, Qiu H, Guo R. The functions and potential roles of extracellular vesicle noncoding RNAs in gynecological malignancies. Cell Death Dis. 2021;7(1):258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binenbaum Y, Fridman E, Yaari Z, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78(18):5287–5299. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Xu X, Su X. Noncoding RNAs in cancer immunity: functions, regulatory mechanisms, and clinical application. Mol Cancer. 2020;19(1):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76(11):2059–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen M, Chen J, Huang W, et al. Exosomes from human induced pluripotent stem cells derived mesenchymal stem cells improved myocardial injury caused by severe acute pancreatitis through activating Akt/Nrf2/HO-1 axis. Cell Cycle. 2022;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams KR, Chauhan S, Patel DB, et al. Ubiquitin conjugation probed by inflammation in myeloid-derived suppressor cell extracellular vesicles. J Proteome Res. 2018;17(1):315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geis-Asteggiante L, Belew AT, Clements VK, et al. Differential content of proteins, mRNAs, and miRNAs suggests that MDSC and their exosomes may mediate distinct immune suppressive functions. J Proteome Res. 2018;17(1):486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daveri E, Vergani E, Shahaj E, et al. microRNAs shape myeloid cell-mediated resistance to cancer immunotherapy. Front Immunol. 2020;11:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segura E, Amigorena S, Théry C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood Cells Mol Dis. 2005;35(2):89–93. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H, Xie Y, Li W, Chibbar R, Xiong S, Xiang J. CD4(+) T cell-released exosomes inhibit CD8(+) cytotoxic T-lymphocyte responses and antitumor immunity. Cell Mol Immunol. 2011;8(1): 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peters PJ, Borst J, Oorschot V, et al. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J Exp Med. 1991;173(5):1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tucher C, Bode K, Schiller P, et al. Extracellular vesicle subtypes released from activated or apoptotic T-lymphocytes carry a specific and stimulus-dependent protein cargo. Front Immunol. 2018;9:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mazzeo C, Calvo V, Alonso R, Mérida I, Izquierdo M. Protein kinase D1/2 is involved in the maturation of multivesicular bodies and secretion of exosomes in T and B lymphocytes. Cell Death Differ. 2016;23(1):99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martínez-Lorenzo MJ, Anel A, Gamen S, et al. Activated human T cells release bioactive Fas ligand and APO2 ligand in microvesicles. J Immunol. 1999;163(3):1274–1281. [PubMed] [Google Scholar]
- 67.Kaur S, Singh SP, Elkahloun AG, Wu W, Abu-Asab MS, Roberts DD. CD47-dependent immunomodulatory and angiogenic activities of extracellular vesicles produced by T cells. Matrix Biol. 2014;37:49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mellman I Dendritic cells: master regulators of the immune response. Cancer Immunol Res. 2013;1(3):145–149. [DOI] [PubMed] [Google Scholar]
- 69.Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4(5):594–600. [DOI] [PubMed] [Google Scholar]
- 70.Lindenbergh MFS, Stoorvogel W. Antigen presentation by extra-cellular vesicles from professional antigen-presenting cells. Annu Rev Immunol. 2018;36:435–459. [DOI] [PubMed] [Google Scholar]
- 71.Wei G, Jie Y, Haibo L, et al. Dendritic cells derived exosomes migration to spleen and induction of inflammation are regulated by CCR7. Sci Rep. 2017;7:42996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelissier Vatter FA, Cioffi M, Hanna SJ, et al. Extracellular vesicle- and particle-mediated communication shapes innate and adaptive immune responses. J Exp Med. 2021;218(8):e20202579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seo N Exosome-mediated immune regulation and its clinical application. Trends Immunotherapy. 2020;4(1):36–41. [Google Scholar]
- 74.Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. 2019;50(4):924–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim SH, Bianco N, Menon R, et al. Exosomes derived from genetically modified DC expressing FasL are anti-inflammatory and immunosuppressive. Mol Ther. 2006;13(2):289–300. [DOI] [PubMed] [Google Scholar]
- 76.Simhadri VR, Reiners KS, Hansen HP, et al. Dendritic cells release HLA-B-associated transcript-3 positive exosomes to regulate natural killer function. PLoS One. 2008;3(10):e3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yáñez-Mó M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willms E, Johansson HJ, Mäger I, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Borges FT, Reis LA, Schor N. Extracellular vesicles: structure, function, and potential clinical uses in renal diseases. Braz J Med Biol Res. 2013;46(10):824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ståhl A-L, Johansson K, Mossberg M, Kahn R, Karpman D. Exosomes and microvesicles in normal physiology, pathophysiology, and renal diseases. Pediatr Nephrol. 2019;34(1):11–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. [DOI] [PubMed] [Google Scholar]
- 82.Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J Cell Sci. 2005;118(Pt 12):2625–2635. [DOI] [PubMed] [Google Scholar]
- 83.Nakamichi E, Sakakura H, Mii S, et al. Detection of serum/salivary exosomal Alix in patients with oral squamous cell carcinoma. Oral Dis. 2021;27(3):439–447. [DOI] [PubMed] [Google Scholar]
- 84.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol. 2007;177(2):329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyado K, Yamada G, Yamada S, et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287(5451):321–324. [DOI] [PubMed] [Google Scholar]
- 86.Khushman M, Bhardwaj A, Patel GK, Laurini JA, et al. Exosomal markers (CD63 and CD9) expression pattern using immunohistochemistry in resected malignant and nonmalignant pancreatic specimens. Pancreas 2017, 46, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boriachek K, Islam MN, Möller A, et al. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14(6):1702153. [DOI] [PubMed] [Google Scholar]
- 88.Keller S, Rupp C, Stoeck A, et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72(9):1095–1102. [DOI] [PubMed] [Google Scholar]
- 89.Nieuwenhuis HK, van Oosterhout JJ, Rozemuller E, et al. Studies with a monoclonal antibody against activated platelets: Evidence that a secreted 53,000-molecular weight lysosome-like granule protein is exposed on the surface of activated platelets in the circulation. Blood. 1987;70(3):838–845. [PubMed] [Google Scholar]
- 90.Toothill VJ, Van Mourik JA, Niewenhuis HK, et al. Characterization of the enhanced adhesion of neutrophil leukocytes to thrombin-stimulated endothelial cells. J Immunol. 1990;145(1):283–291. [PubMed] [Google Scholar]
- 91.Sun B, Li Y, Zhou Y, et al. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234(2):1416–1425. [DOI] [PubMed] [Google Scholar]
- 92.Beckham Carla J, Olsen J, Yin P-N, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol. 2014;192(2):583–592. [DOI] [PubMed] [Google Scholar]
- 93.Li C, Li C, Zhi C, et al. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J Transl Med. 2019;17(1):355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem. 2016;291(4):1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skottvoll FS, Berg HE, Bjørseth K, et al. Ultracentrifugation versus kit exosome isolation: nanoLC–MS and other tools reveal similar performance biomarkers, but also contaminations. Fut Sci. 2019;5(4):FSO359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaswani K, Mitchell MD, Holland OJ, et al. A method for the isolation of exosomes from human and bovine milk. J Nutr Metabol. 2019;2019:5764740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Multhoff G Heat shock protein 70 (Hsp70): membrane location, export and immunological relevance. Methods. 2007;43(3):229–237. [DOI] [PubMed] [Google Scholar]
- 98.Whiteside TL, Diergaarde B, Hong C-S. Tumor-derived exosomes (TEX) and their role in immuno-oncology. Int J Mol Sci. 2021;22(12):6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gross CN, Irrinki A, Feder JN, Enns CA. Co-trafficking of HFE, a nonclassical major histocompatibility complex class I protein, with the transferrin receptor implies a role in intracellular iron regulation. J Biol Chem. 1998;273(34):22068–22074. [DOI] [PubMed] [Google Scholar]
- 100.Díaz-Varela M, de Menezes-Neto A, Perez-Zsolt D, et al. Proteomics study of human cord blood reticulocyte-derived exosomes. Sci Rep. 2018;8(1):14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Welsch S, Habermann A, Jäger S, et al. Ultrastructural analysis of ESCRT proteins suggests a role for endosome-associated tubular-vesicular membranes in ESCRT function. Traffic. 2006;7(11):1551–1566. [DOI] [PubMed] [Google Scholar]
- 102.Fernandez-Llama P, Khositseth S, Gonzales PA, Star RA, Pisitkun T, Knepper MA. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77(8):736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trioulier Y, Torch S, Blot B, et al. Alix, a protein regulating endosomal trafficking, is involved in neuronal death*. J Biol Chem. 2004;279(3):2046–2052. [DOI] [PubMed] [Google Scholar]
- 104.Aoki M, Kanamori M, Ohmori K, et al. Expression of developmentally regulated endothelial cell locus 1 was induced by tumor-derived factors including VEGF. Biochem Biophys Res Commun 2005;333(3):990–995. [DOI] [PubMed] [Google Scholar]
- 105.Hidai C, Kawana M, Kitano H, Kokubun S. Discoidin domain of Del1 protein contributes to its deposition in the extracellular matrix. Cell Tissue Res. 2007;330(1):83–95. [DOI] [PubMed] [Google Scholar]
- 106.Muller L, Simms P, Hong C-S, et al. Human tumor-derived exosomes (TEX) regulate Treg functions via cell surface signaling rather than uptake mechanisms. OncoImmunology. 2017;6(8):e1261243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pineles B, Mani A, Sura L, et al. Neuronal exosome proteins: novel biomarkers for predicting neonatal response to therapeutic hypothermia. Arch Dis Child Fetal Neonatal Ed. 2022;107(1):60. [DOI] [PubMed] [Google Scholar]
- 108.Tavasolian F, Hosseini AZ, Rashidi M, et al. The impact of immune cell-derived exosomes on immune response initiation and immune system function. Curr Pharm Des. 2021;27(2):197–205. [DOI] [PubMed] [Google Scholar]
- 109.Yin W, Ouyang S, Luo Z, et al. Immature exosomes derived from microRNA-146a overexpressing dendritic cells act as antigen-specific therapy for myasthenia gravis. Inflammation. 2017;40(4):1460–1473. [DOI] [PubMed] [Google Scholar]
- 110.Leone DA, Peschel A, Brown M, et al. Surface LAMP-2 is an endocytic receptor that diverts antigen internalized by human dendritic cells into highly immunogenic exosomes. J Immunol. 2017;199(2):531. [DOI] [PubMed] [Google Scholar]
- 111.Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zitvogel L, Fernandez N, Lozier A, et al. Dendritic cells or their exosomes are effective biotherapies of cancer. Eur J Cancer. 1999;35(Suppl 3):S36–S38. [DOI] [PubMed] [Google Scholar]
- 113.Ni K, O’Neill HC. Long-term stromal cultures produce dendritic-like cells. Br J Haematol. 1997;97(4):710–725. [DOI] [PubMed] [Google Scholar]
- 114.Quah B, Ni K, O’Neill HC. In vitro hematopoiesis produces a distinct class of immature dendritic cells from spleen progenitors with limited T cell stimulation capacity. Int Immunol. 2004;16(4):567–577. [DOI] [PubMed] [Google Scholar]
- 115.Mongue-Din H, Patel AS, Looi YH, et al. NADPH oxidase-4 driven cardiac macrophage polarization protects against myocardial infarction-induced remodeling. JACC Basic Transl Sci. 2017;2(6):688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Diehl P, Fricke A, Sander L, et al. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93(4):633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schroeder JC, Puntigam L, Hofmann L, et al. Circulating exosomes inhibit B cell proliferation and activity. Cancer. 2020;12(8):2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Do P, Beckwith KA, Cheney C, et al. Leukemic B cell CTLA-4 suppresses costimulation of T cells. J Immunol. 2019;202(9):2806–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andrews LP, Marciscano AE, Drake CG, Vignali DAA. LAG3 (CD223) as a cancer immunotherapy target. Immunol Rev. 2017;276(1):80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang X, Voskens CJ, Sallin M, et al. CD137 promotes proliferation and survival of human B cells. J Immunol. 2010;184(2):787–795. [DOI] [PubMed] [Google Scholar]
- 121.Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013;19(6):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wahlgren J, Karlson TDL, Glader P, Telemo E, Valadi H. Activated human T cells secrete exosomes that participate in IL-2 mediated immune response signaling. PLoS One. 2012;7(11):e49723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coughlan C, Bruce KD, Burgy O, et al. Exosome isolation by ultracentrifugation and precipitation and techniques for downstream analyses. Curr Protoc Cell Biol. 2020;88(1):e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ayala-Mar S, Donoso-Quezada J, Gallo-Villanueva RC, Perez-Gonzalez VH, González-Valdez J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis. 2019;40(23–24):3036–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Coumans FAW, Brisson AR, Buzas EI, et al. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120(10):1632–1648. [DOI] [PubMed] [Google Scholar]
- 126.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235–246. [DOI] [PubMed] [Google Scholar]
- 127.Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracellular Vesicles. 2013;2(1):20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim J, Shin H, Kim J, Kim J, Park J. Isolation of high-purity extracellular vesicles by extracting proteins using aqueous two-phase system. PLoS One. 2015;10(6):e0129760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alvarez ML, Khosroheidari M, Kanchi Ravi R, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82(9):1024–1032. [DOI] [PubMed] [Google Scholar]
- 130.Quintana JF, Makepeace BL, Babayan SA, et al. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit Vectors. 2015;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheruvanky A, Zhou H, Pisitkun T, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol. 2007;292(5):F165 7–F1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. 2015;4:26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oksvold MP, Neurauter A, Pedersen KW. Magnetic bead-based isolation of exosomes. Meth Molec Biol. 2015;1218:465–481. [DOI] [PubMed] [Google Scholar]
- 134.Gonzales PA, Zhou H, Pisitkun T, et al. Isolation and purification of exosomes in urine. Meth Molec Biol. 2010;641:89–99. [DOI] [PubMed] [Google Scholar]
- 135.Poliakov A, Spilman M, Dokland T, Amling CL, Mobley JA. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate. 2009;69(2):159–167. [DOI] [PubMed] [Google Scholar]
- 136.Lane RE, Korbie D, Anderson W, Vaidyanathan R, Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci Rep. 2015;5:7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bhagat AAS, Kuntaegowdanahalli SS, Papautsky I. Continuous particle separation in spiral microchannels using dean flows and differential migration. Lab Chip. 2008;8(11):1906–1914. [DOI] [PubMed] [Google Scholar]
- 138.Winkleman A, Perez-Castillejos R, Gudiksen KL, Phillips ST, Prentiss M, Whitesides GM. Density-based diamagnetic separation: devices for detecting binding events and for collecting unlabeled diamagnetic particles in paramagnetic solutions. Anal Chem. 2007;79(17):6542–6550. [DOI] [PubMed] [Google Scholar]
- 139.Watson DC, Yung BC, Bergamaschi C, et al. Scalable, cGMP-compatible purification of extracellular vesicles carrying bioactive human heterodimeric IL-15/lactadherin complexes. J Extracell Vesicles. 2018;7(1):1442088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Grimm KM, Trigona WL, Heidecker GJ, et al. An enhanced and scalable process for the purification of SIV gag-specific MHC tetramer. Protein Expr Purif. 2001;23(2):270–281. [DOI] [PubMed] [Google Scholar]
- 141.Dizon-Maspat J, Bourret J, D’Agostini A, Li F. Single pass tangential flow filtration to debottleneck downstream processing for therapeutic antibody production. Biotechnol Bioeng. 2012;109(4):962–970. [DOI] [PubMed] [Google Scholar]
- 142.Busatto S, Vilanilam G, Ticer T, et al. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cell. 2018;7(12):273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Helwa I, Cai J, Drewry MD, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017;12(1): e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee K, Shao H, Weissleder R, Lee H. Acoustic purification of extracellular microvesicles. ACS Nano. 2015;9(3):2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li M, Rai AJ, DeCastro GJ, et al. An optimized procedure for exosome isolation and analysis using serum samples: Application to cancer biomarker discovery. Methods. 2015;87:26–30. [DOI] [PubMed] [Google Scholar]
- 146.Wu M, Ouyang Y, Wang Z, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci USA. 2017;114(40):10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ibsen SD, Wright J, Lewis JM, et al. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano. 2017;11(7):6641–6651. [DOI] [PubMed] [Google Scholar]
- 148.Ramos A, Morgan H, Green NG, Castellanos A. Ac electrokinetics: a review of forces in microelectrode structures. J Phys D Appl Phys. 1998;31(18):2338–2353. [Google Scholar]
- 149.Schimpf ME. POLYMERS | field flow fractionation. In: Wilson ID, ed. Encyclopedia of Separation Science. Academic Press; 2000:3906–3915. [Google Scholar]
- 150.Petersen KE, Shiri F, White T, et al. Exosome isolation: cyclical electrical field flow fractionation in low-ionic-strength fluids. Anal Chem. 2018;90(21):12783–12790. [DOI] [PubMed] [Google Scholar]
- 151.Giddings JC. A new separation concept based on a coupling of concentration and flow nonuniformities. Sep Sci. 1966;1(1):123–125. [Google Scholar]
- 152.Hochstetter A, Vernekar R, Austin RH, et al. Deterministic lateral displacement: challenges and perspectives. ACS Nano. 2020;14(9):10784–10795. [DOI] [PubMed] [Google Scholar]
- 153.Liu C, Ding B, Xue C, Tian Y, Hu G, Sun J. Sheathless focusing and separation of diverse nanoparticles in viscoelastic solutions with minimized shear thinning. Anal Chem. 2016;88(24):12547–12553. [DOI] [PubMed] [Google Scholar]
- 154.Liao J, Liu R, Shi YJ, Yin LH, Pu YP. Exosome-shuttling microR-NA-21 promotes cell migration and invasion-targeting PDCD4 in esophageal cancer. Int J Oncol. 2016;48(6):2567–2579. [DOI] [PubMed] [Google Scholar]
- 155.Musante L, Tataruch D, Gu D, et al. A simplified method to recover urinary vesicles for clinical applications and sample banking. Sci Rep. 2014;4(1):7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kadakeri S, Arul MR, Bordett R, Duraisamy N, Naik H, Rudraiah S. 6 – Protein synthesis and characterization. In: Wei G, Kumbar SG, eds. Artificial Protein and Peptide Nanofibers. Woodhead Publishing; 2020:121–161. [Google Scholar]
- 157.Merchant ML, Powell DW, Wilkey DW, et al. Microfiltration isolation of human urinary exosomes for characterization by MS. PROTEOMICS – Clin Appl. 2010;4(1):84–96. [DOI] [PubMed] [Google Scholar]
- 158.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Welton JL, Webber JP, Botos LA, Jones M, Clayton A. Ready-made chromatography columns for extracellular vesicle isolation from plasma. J Extracell Vesicles. 2015;4:27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee S-S, Won J-H, Lim GJ, et al. A novel population of extracellular vesicles smaller than exosomes promotes cell proliferation. Cell Commun Signal. 2019;17(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Delcayre A, Estelles A, Sperinde J, et al. Exosome Display technology: applications to the development of new diagnostics and therapeutics. Blood Cells Mol Dis. 2005;35(2):158–168. [DOI] [PubMed] [Google Scholar]
- 164.Takahashi Y, Nishikawa M, Shinotsuka H, et al. Visualization and in vivo tracking of the exosomes of murine melanoma B16-BL6 cells in mice after intravenous injection. J Biotechnol. 2013;165(2):77–84. [DOI] [PubMed] [Google Scholar]
- 165.De Gassart A, Trentin B, Martin M, et al. Exosomal sorting of the cytoplasmic domain of bovine leukemia virus TM Env protein. Cell Biol Int. 2009;33(1):36–48. [DOI] [PubMed] [Google Scholar]
- 166.Bellavia D, Raimondo S, Calabrese G, et al. Interleukin 3-receptor targeted exosomes inhibit in vitro and in vivo Chronic Myelogenous Leukemia cell growth. Theranostics. 2017;7(5):1333–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stickney Z, Losacco J, McDevitt S, Zhang Z, Lu B. Development of exosome surface display technology in living human cells. Biochem Biophys Res Commun. 2016;472(1):53–59. [DOI] [PubMed] [Google Scholar]
- 168.Ohno S, Takanashi M, Sudo K, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Curley N, Levy D, Do MA, et al. Sequential deletion of CD63 identifies topologically distinct scaffolds for surface engineering of exosomes in living human cells. Nanoscale. 2020;12(22):12014–12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dooley K, McConnell RE, Xu K, et al. A versatile platform for generating engineered extracellular vesicles with defined therapeutic properties. Mol Ther. 2021;29(5):1729–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lewis ND, Sia CL, Kirwin K, et al. Exosome surface display of IL12 results in tumor-retained pharmacology with superior potency and limited systemic exposure compared with recombinant IL12. Mol Cancer Ther. 2021;20(3):523–534. [DOI] [PubMed] [Google Scholar]
- 172.Gao X, Ran N, Dong X, et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci Transl Med. 2018;10(444):eaat0195. [DOI] [PubMed] [Google Scholar]
- 173.Jia G, Han Y, An Y, et al. NRP-1 targeted and cargo-loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. [DOI] [PubMed] [Google Scholar]
- 174.Kim MS, Haney MJ, Zhao Y, et al. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: in vitro and in vivo evaluations. Nanomedicine. 2018;14(1):195–204. [DOI] [PubMed] [Google Scholar]
- 175.Choi ES, Song J, Kang YY, Mok H. Mannose-modified serum exosomes for the elevated uptake to murine dendritic cells and lymphatic accumulation. Macromol Biosci. 2019;19(7):1900042. [DOI] [PubMed] [Google Scholar]
- 176.Lai CP, Kim EY, Badr CE, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6(1):7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yim N, Ryu SW, Choi K, et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat Commun. 2016;7:12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Choi H, Kim Y, Mirzaaghasi A, et al. Exosome-based delivery of super-repressor IκBα relieves sepsis-associated organ damage and mortality. Science Advances 2020;6(15):eaaz6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Busatto S, Iannotta D, Walker SA, Di Marzio L, Wolfram J. A simple and quick method for loading proteins in extracellular vesicles. Pharmaceuticals. 2021;14(4):356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li Z, Zhou X, Wei M, et al. In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019;19(1):19–28. [DOI] [PubMed] [Google Scholar]
- 181.Kojima R, Bojar D, Rizzi G, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat Commun. 2018;9(1):1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang L, Rong Y, Tang X, et al. Engineered exosomes as an in situ DC-primed vaccine to boost antitumor immunity in breast cancer. Mol Cancer. 2022;21(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hong S, Ruan S, Greenberg Z, He M, McGill JL. Development of surface engineered antigenic exosomes as vaccines for respiratory syncytial virus. Sci Rep. 2021;11(1):21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kim S, Sohn HJ, Lee HJ, et al. Use of engineered exosomes expressing HLA and costimulatory molecules to generate antigen-specific CD8+ T cells for adoptive cell therapy. J Immunotherapy. 2017;40(3):83–93. [DOI] [PubMed] [Google Scholar]
- 185.Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y. Exosome-based tumor antigens-adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA. Biomaterials. 2016;111:55–65. [DOI] [PubMed] [Google Scholar]
- 186.Hartman ZC, Wei J, Glass OK, et al. Increasing vaccine potency through exosome antigen targeting. Vaccine. 2011;29(50):9361–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shi X, Cheng Q, Hou T, et al. Genetically engineered cell-derived nanoparticles for targeted breast cancer immunotherapy. Mol Ther. 2020;28(2):536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fan M, Liu H, Yan H, et al. A CAR T-inspiring platform based on antibody-engineered exosomes from antigen-feeding dendritic cells for precise solid tumor therapy. Biomaterials. 2022;282: 121424. [DOI] [PubMed] [Google Scholar]
- 189.Gunassekaran GR, Poongkavithai Vadevoo SM, Baek MC, Lee B. M1 macrophage exosomes engineered to foster M1 polarization and target the IL-4 receptor inhibit tumor growth by reprogramming tumor-associated macrophages into M1-like macrophages. Biomaterials. 2021;278:121137. [DOI] [PubMed] [Google Scholar]
- 190.Conceição M, Forcina L, Wiklander OPB, et al. Engineered extracellular vesicle decoy receptor-mediated modulation of the IL6 trans-signalling pathway in muscle. Biomaterials. 2021;266: 120435. [DOI] [PubMed] [Google Scholar]
- 191.Duong N, Curley K, Brown A, et al. Decoy exosomes as a novel biologic reagent to antagonize inflammation. Int J Nanomedicine. 2019;14:3413–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65(3):383–390. [DOI] [PubMed] [Google Scholar]
- 194.Pucci F, Garris C, Lai CP, et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science. 2016;352(6282):242–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hamilton N, Claudio NM, Armstrong RJ, Pucci F. Cell Surface Labeling by Engineered Extracellular Vesicles. Advanced biosystems. 2020;4(12):e2000007. [DOI] [PubMed] [Google Scholar]
- 196.Lee JR, Park BW, Kim J, et al. Nanovesicles derived from iron oxide nanoparticles-incorporated mesenchymal stem cells for cardiac repair. Sci Adv. 2020;6(18):eaaz0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhuang M, Du D, Pu L, et al. SPION-decorated exosome delivered BAY55–9837 targeting the pancreas through magnetism to improve the blood GLC response. Small. 2019;15(52):e1903135. [DOI] [PubMed] [Google Scholar]
- 198.Mizuta R, Sasaki Y, Kawasaki R, et al. Magnetically navigated intra-cellular delivery of extracellular vesicles using amphiphilic nanogels. Bioconjug Chem. 2019;30(8):2150–2155. [DOI] [PubMed] [Google Scholar]
- 199.Liu T, Zhu Y, Zhao R, Wei X, Xin X. Visualization of exosomes from mesenchymal stem cells in vivo by magnetic resonance imaging. Magn Reson Imaging. 2020;68:75–82. [DOI] [PubMed] [Google Scholar]
- 200.Maguire CA, Balaj L, Sivaraman S, et al. Microvesicle-associated AAV vector as a novel gene delivery system. Mol Ther. 2012;20(5):960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Breuer CB, Hanlon KS, Natasan JS, et al. In vivo engineering of lymphocytes after systemic exosome-associated AAV delivery. Sci Rep. 2020;10(1):4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jhan YY, Prasca-Chamorro D, Palou Zuniga G, et al. Engineered extracellular vesicles with synthetic lipids via membrane fusion to establish efficient gene delivery. Int J Pharm. 2020;573:118802. [DOI] [PubMed] [Google Scholar]
- 203.Sato YT, Umezaki K, Sawada S, et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep. 2016;6(1):21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Desplantes R, Lévêque C, Muller B, et al. Affinity biosensors using recombinant native membrane proteins displayed on exosomes: application to botulinum neurotoxin B receptor. Sci Rep. 2017;7(1):1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hung ME, Leonard JN. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290(13):8166–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Koh E, Lee EJ, Nam G-H, et al. Exosome-SIRPα, a CD47 blockade increases cancer cell phagocytosis. Biomaterials. 2017;121:121–129. [DOI] [PubMed] [Google Scholar]
- 208.Lathwal S, Yerneni SS, Boye S, et al. Engineering exosome polymer hybrids by atom transfer radical polymerization. Proc Natl Acad Sci USA. 2021;118(2):e2020241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122(3):787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mantovani A, Sozzani S, Locati M, et al. Infiltration of tumours by macrophages and dendritic cells: tumour-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Novartis Found Symp. 2004;256:137–145. discussion 146–138, 259–169. [PubMed] [Google Scholar]
- 211.Pesce J, Kaviratne M, Ramalingam TR, et al. The IL-21 receptor augments Th2 effector function and alternative macrophage activation. J Clin Invest. 2006;116(7):2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kurowska-Stolarska M, Stolarski B, Kewin P, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183(10):6469–6477. [DOI] [PubMed] [Google Scholar]
- 213.Germano G, Allavena P, Mantovani A. Cytokines as a key component of cancer-related inflammation. Cytokine. 2008;43(3):374–379. [DOI] [PubMed] [Google Scholar]
- 214.Sica A, Allavena P, Mantovani A. Cancer related inflammation: the macrophage connection. Cancer Lett. 2008;267(2):204–215. [DOI] [PubMed] [Google Scholar]
- 215.Gordon S, Plüddemann A. Tissue macrophages: heterogeneity and functions. BMC Biol. 2017;15(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Azad AK, Rajaram MVS, Schlesinger LS. Exploitation of the macro-phage mannose receptor (CD206) in infectious disease diagnostics and therapeutics. J Cytol Mol Biol. 2014;1(1):1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Qiu S-Q, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, Schröder CP. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat Rev. 2018;70:178–189. [DOI] [PubMed] [Google Scholar]
- 218.DeNardo DG, Brennan DJ, Rexhepaj E, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shree T, Olson OC, Elie BT, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25(23):2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Scodeller P, Simón-Gracia L, Kopanchuk S, et al. Precision targeting of tumor macrophages with a CD206 binding peptide. Sci Rep. 2017;7(1):14655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Asciutto EK, Kopanchuk S, Lepland A, et al. Phage-display-derived peptide binds to human CD206 and modeling reveals a new binding site on the receptor. J Phys Chem B. 2019;123(9):1973–1982. [DOI] [PubMed] [Google Scholar]
- 222.Rashid MH, Borin TF, Ara R, et al. CD206 positive M2-macrophage targeting engineered exosomes as a potential diagnostic and therapeutic tool. Cancer Res. 2019;79(13_Supplement):1139. [Google Scholar]
- 223.Teillaud JL. Antibody-dependent Cellular Cytotoxicity (ADCC). In: eLS. Chichester: John Wiley & Sons, Ltd; 2006:1–8. [Google Scholar]
- 224.Polansky M, Eisenstadt R, DeGrazia T, Zhao X, Liu Y, Feldman R. Rituximab therapy in patients with bullous pemphigoid: a retrospective study of 20 patients. J Am Acad Dermatol. 2019;81(1):179–186. [DOI] [PubMed] [Google Scholar]
- 225.Sneller MC. Rituximab and Wegener’s granulomatosis: Are B cells a target in vasculitis treatment? Arthritis Rheum. 2005;52(1):1–5. [DOI] [PubMed] [Google Scholar]
- 226.Aldeghaither DS, Zahavi DJ, Murray JC, et al. A mechanism of resistance to antibody-targeted immune attack. Cancer Immunol Res. 2019;7(2):230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang W, Erbe AK, Hank JA, Morris ZS, Sondel PM. NK cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front Immunol. 2015;6:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Smyth MJ, Cretney E, Kelly JM, et al. Activation of NK cell cytotoxicity. Mol Immunol. 2005;42(4):501–510. [DOI] [PubMed] [Google Scholar]
- 229.Srivastava RM, Lee SC, Andrade Filho PA, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin Cancer Res. 2013;19(7):1858–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Qin H, Lerman B, Sakamaki I, et al. Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice. Nat Med. 2014;20(6):676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Srivastava A, Filant J, Moxley KM, Sood A, McMeekin S, Ramesh R. Exosomes: a role for naturally occurring nanovesicles in cancer growth, diagnosis and treatment. Curr Gene Ther. 2015;15(2):182–192. [DOI] [PubMed] [Google Scholar]
- 232.Haqqani AS, Delaney CE, Tremblay TL, Sodja C, Sandhu JK, Stanimirovic DB. Method for isolation and molecular characterization of extracellular microvesicles released from brain endothelial cells. Fluids Barriers CNS. 2013;10(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lakhal S, Wood MJ. Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. BioEssays. 2011;33(10):737–741. [DOI] [PubMed] [Google Scholar]
- 234.Sun D, Zhuang X, Zhang S, et al. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv Drug Deliv Rev. 2013;65(3):342–347. [DOI] [PubMed] [Google Scholar]
- 235.Kibria G, Ramos EK, Wan Y, Gius DR, Liu H. Exosomes as a drug delivery system in cancer therapy: potential and challenges. Mol Pharm. 2018;15(9):3625–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. 2015;9(3–4):358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Rückert R, Herz U, Paus R, et al. IL-15-IgG2b fusion protein accelerates and enhances a Th2 but not a Th1 immune response in vivo, while IL-2-IgG2b fusion protein inhibits both. Eur J Immunol. 1998;28(10):3312–3320. [DOI] [PubMed] [Google Scholar]
- 238.Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: new developments and future perspectives. EMBO Mol Med. 2012;4(10):1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ahmed M, Pan DW, Davis ME. Lack of in vivo antibody dependent cellular cytotoxicity with antibody containing gold nanoparticles. Bioconjug Chem. 2015;26(5):812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Leon PE, He W, Mullarkey CE, et al. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci. 2016;113(40):E5944–E5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Sioud M, Westby P, Olsen JKE, Mobergslien A. Generation of new peptide-Fc fusion proteins that mediate antibody-dependent cellular cytotoxicity against different types of cancer cells. Mol Ther Methods Clin Dev. 2015;2:15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Han SG, Lee JS, Ahn K, et al. Size-dependent clearance of gold nanoparticles from lungs of Sprague–Dawley rats after short-term inhalation exposure. Arch Toxicol. 2015;89(7): 1083–1094. [DOI] [PubMed] [Google Scholar]
- 243.Hoshyar N, Gray S, Han H, Bao G. The effect of nanoparticle size on in vivo pharmacokinetics and cellular interaction. Nanomedicine. 2016;11(6):673–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5(4):505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Arora T, Padaki R, Liu L, et al. Differences in binding and effector functions between classes of TNF antagonists. Cytokine. 2009;45(2):124–131. [DOI] [PubMed] [Google Scholar]
- 246.Gillies SD, Young D, Lo KM, Roberts S. Biological activity and in vivo clearance of antitumor antibody/cytokine fusion proteins. Bioconjug Chem. 1993;4(3):230–235. [DOI] [PubMed] [Google Scholar]
- 247.Wu B, Sun Y-N. Pharmacokinetics of peptide–Fc fusion proteins. J Pharm Sci. 2014;103(1):53–64. [DOI] [PubMed] [Google Scholar]
- 248.Datta-Mannan A, Lu J, Witcher DR, Leung D, Tang Y, Wroblewski VJ. The interplay of non-specific binding, target-mediated clearance and FcRn interactions on the pharmacokinetics of humanized antibodies. MAbs. 2015;7(6):1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zahavi D, AlDeghaither D, O’Connell A, Weiner LM. Enhancing antibody-dependent cell-mediated cytotoxicity: a strategy for improving antibody-based immunotherapy. Antibody Therapeutics. 2018;1(1):7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gül N, van Egmond M. Antibody-dependent phagocytosis of tumor cells by macrophages: a potent effector mechanism of monoclonal antibody therapy of cancer. Cancer Res. 2015;75(23):5008–5013. [DOI] [PubMed] [Google Scholar]
- 251.Rashid MH, Borin TF, Ara R, et al. Critical immunosuppressive effect of MDSCderived exosomes in the tumor microenvironment. Oncol Rep. 2021;45(3):1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Angara K, Arbab AS. Chapter 16 – Mechanisms of glioblastoma resistance to antiangiogenic agents and reversal approaches. In: Paulmurugan R, Massoud TF, eds. Glioblastoma Resistance to Chemotherapy: Molecular Mechanisms and Innovative Reversal Strategies. Vol 15. Academic Press; 2021:429–452. [Google Scholar]
- 253.Ali S, Borin TF, Piranlioglu R, et al. Changes in the tumor microenvironment and outcome for TME-targeting therapy in glioblastoma: a pilot study. PLoS One. 2021;16(2):e0246646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Achyut BR, Shankar A, Iskander ASM, et al. Chimeric Mouse model to track the migration of bone marrow derived cells in glioblastoma following anti-angiogenic treatments. Cancer Biol Ther. 2016;17(3):280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Achyut BR, Arbab AS. Myeloid cell signatures in tumor microenvironment predicts therapeutic response in cancer. Onco Targets Ther. 2016;9:1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Achyut BR, Shankar A, Iskander AS, et al. Bone marrow derived myeloid cells orchestrate antiangiogenic resistance in glioblastoma through coordinated molecular networks. Cancer Lett. 2015;369(2):416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Arbab AS. Activation of alternative pathways of angiogenesis and involvement of stem cells following anti-angiogenesis treatment in glioma. Histol Histopathol. 2012;27(5):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ali MM, Janic B, Babajani-Feremi A, et al. Changes in vascular permeability and expression of different angiogenic factors following anti-angiogenic treatment in rat glioma. PLoS One. 2010;5(1):e8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Scarlett CJ. Contribution of bone marrow derived cells to the pancreatic tumor microenvironment. Front Physiol. 2013;4:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Vorontsova A, Kan T, Raviv Z, Shaked Y. The dichotomous role of bone marrow derived cells in the chemotherapy-treated tumor microenvironment. J Clin Med. 2020;9(12):3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Hirata E, Sahai E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb Perspect Med. 2017;7(7):a026781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Denis M, Duruisseaux M, Brevet M, Dumontet C. How can immune checkpoint inhibitors cause hyperprogression in solid tumors? Front Immunol. 2020;11:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Liu J, Chen Z, Li Y, Zhao W, Wu J, Zhang Z. PD-1/PD-L1 checkpoint inhibitors in tumor immunotherapy. Front Pharmacol. 2021;12(2339):731798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Achyut BR, Arbab AS. Myeloid derived suppressor cells: fuel the fire. Biochem Physiol. 2014;3(3):e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Achyut BR, Arbab AS. Taming immune suppressor: application of myeloid-derived suppressor cells in anti-cancer gene therapy. Transl Cancer Res. 2017;6:S160–S162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Borin TF, Angara K, Rashid M, et al. CSF-1R inhibitor prevented pre-metastatic lung niches in metastatic mammary tumor. Cancer Res. 2017;77(13_Supplement):1043. [Google Scholar]
- 267.Khan M, Hasan MM, Barnett A, et al. Co-axial electrospraying of injectable multi-cancer drugs nanocapsules with polymer shells for targeting aggressive breast cancers. Cancer Nanotechnol. 2022;13(1):6. [Google Scholar]
- 268.Yang Q, Tong X, Schieb L, et al. Vital signs: recent trends in stroke death rates – United States, 2000–2015. MMWR Morb Mortal Wkly Rep. 2017;66(35):933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Jickling GC, Liu D, Ander BP, Stamova B, Zhan X, Sharp FR. Targeting neutrophils in ischemic stroke: translational insights from experimental studies. J Cereb Blood Flow Metab. 2015;35(6):888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Downing LJ, Strieter RM, Kadell AM, et al. Neutrophils are the initial cell type identified in deep venous thrombosis induced vein wall inflammation. ASAIO J. 1996;42(5):M677–M682. [DOI] [PubMed] [Google Scholar]
- 271.Schabitz WR, Minnerup J. Neutrophils in acute stroke pathophysiology. Stroke. 2019;50(3):e44–e45. [DOI] [PubMed] [Google Scholar]
- 272.Jasper AE, McIver WJ, Sapey E, Walton GM. Understanding the role of neutrophils in chronic inflammatory airway disease. F1000Res. 2019;8:1–17. F1000 Faculty Rev-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Aronowski J, Roy-O’Reilly MA. Neutrophils, the felons of the brain. Stroke. 2019;50(3):e42–e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kim J, Song TJ, Park JH, et al. Different prognostic value of white blood cell subtypes in patients with acute cerebral infarction. Atherosclerosis. 2012;222(2):464–467. [DOI] [PubMed] [Google Scholar]
- 275.Buck BH, Liebeskind DS, Saver JL, et al. Early neutrophilia is associated with volume of ischemic tissue in acute stroke. Stroke. 2008;39(2):355–360. [DOI] [PubMed] [Google Scholar]
- 276.Kumar AD, Boehme AK, Siegler JE, Gillette M, Albright KC, Martin-Schild S. Leukocytosis in patients with neurologic deterioration after acute ischemic stroke is associated with poor outcomes. J Stroke Cerebrovasc Dis. 2013;22(7):e111–e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hayakawa K, Mishima K, Irie K, et al. Cannabidiol prevents a post-ischemic injury progressively induced by cerebral ischemia via a high-mobility group box1-inhibiting mechanism. Neuropharmacology. 2008;55(8):1280–1286. [DOI] [PubMed] [Google Scholar]
- 278.Chou WH, Choi DS, Zhang H, et al. Neutrophil protein kinase Cdelta as a mediator of stroke-reperfusion injury. J Clin Invest. 2004;114(1):49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Lecot P, Sarabi M, Pereira Abrantes M, et al. Neutrophil het-erogeneity in cancer: from biology to therapies. Front Immunol. 2019;10:2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Long W, Chen J, Gao C, Lin Z, Xie X, Dai H. Brief review on the roles of neutrophils in cancer development. J Leukoc Biol. 2021;109(2):407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wu L, Saxena S, Awaji M, Singh RK. Tumor-associated neutrophils in cancer: going pro. Cancer. 2019;11(4):564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Keibel A, Singh V, Sharma MC. Inflammation, microenvironment, and the immune system in cancer progression. Curr Pharm Des. 2009;15(17):1949–1955. [DOI] [PubMed] [Google Scholar]
- 283.Santana Carrero RM, Beceren-Braun F, Rivas SC, et al. IL-15 is a component of the inflammatory milieu in the tumor microenvironment promoting antitumor responses. Proc Natl Acad Sci. 2019;116(2):599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Chen X, Gao A, Zhang F, et al. ILT4 inhibition prevents TAM- and dysfunctional T cell-mediated immunosuppression and enhances the efficacy of anti-PD-L1 therapy in NSCLC with EGFR activation. Theranostics. 2021;11(7):3392–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Eisinger S, Sarhan D, Boura VF, et al. Targeting a scavenger receptor on tumor-associated macrophages activates tumor cell killing by natural killer cells. Proc Natl Acad Sci U S A. 2020;117(50):32005–32016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Rosales C Neutrophil: a cell with many roles in inflammation or several cell types? Front Physiol. 2018;9:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Shinde-Jadhav S, Mansure JJ, Rayes RF, et al. Role of neutrophil extracellular traps in radiation resistance of invasive bladder cancer. Nat Commun. 2021;12(1):2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hedrick CC, Malanchi I. Neutrophils in cancer: heterogeneous and multifaceted. Nat Rev Immunol. 2021;22(3):173–187. [DOI] [PubMed] [Google Scholar]
- 289.Wisdom AJ, Hong CS, Lin AJ, et al. Neutrophils promote tumor resistance to radiation therapy. Proc Natl Acad Sci. 2019;116(37):18584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Masucci MT, Minopoli M, Carriero MV. Tumor associated neutrophils. their role in tumorigenesis, metastasis, prognosis and therapy. Front Oncol. 2019;9:1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.He H-Q, Ye RD. The formyl peptide receptors: diversity of ligands and mechanism for recognition. Molecules. 2017;22(3):455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Haas PJ, de Haas CJ, Kleibeuker W, et al. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. J Immunol. 2004;173(9):5704–5711. [DOI] [PubMed] [Google Scholar]
- 293.Mills JS. Peptides derived from HIV-1, HIV-2, Ebola virus, SARS coronavirus and coronavirus 229E exhibit high affinity binding to the formyl peptide receptor. Biochim Biophys Acta. 2006;1762(7): 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data derived from public domain resources. Data available on request from the authors.


