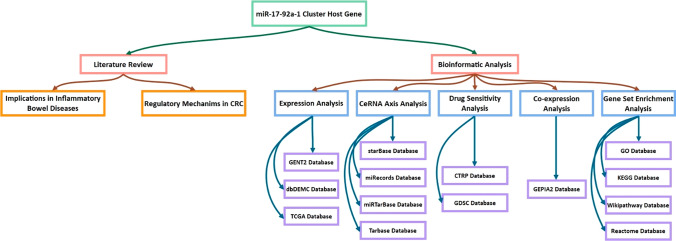
Abstract
Colorectal cancer (CRC), recognized among the five most prevalent malignancies and most deadly cancers, manifests multifactorial influences stemming from environmental exposures, dietary patterns, age, and genetic predisposition. Although substantial progress has been made in comprehending the etiology of CRC, the precise genetic components driving its pathogenesis remain incompletely elucidated. Within the expansive repertoire of non-coding RNAs, particular focus has centered on the miR-17-92a-1 cluster host gene (MIR17HG) and its associated miRNAs, which actively participate in diverse cellular processes and frequently exhibit heightened expression in various solid tumors, notably CRC. Therefore, the primary objective of this research is to undertake an extensive inquiry into the regulatory mechanisms, structural features, functional attributes, and potential diagnostic and therapeutic implications associated with this cluster in CRC. Furthermore, the intricate interplay between this cluster and the development and progression of CRC will be explored. Our findings underscore the upregulation of the miR-17-92a-1 cluster host gene (MIR17HG) and its associated miRNAs in CRC compared to normal tissues, thus implying their profound involvement in the progression of CRC. Collectively, these molecules are implicated in critical oncogenic processes, encompassing metastatic activity, regulation of apoptotic pathways, cellular proliferation, and drug resistance. Consequently, these findings shed illuminating insights into the potential of MIR17HG and its associated miRNAs as promising targets for therapeutic interventions in the management of CRC.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-024-01331-1.
Keywords: MIR17HG, miR-17-92a-1 cluster host gene, Colorectal cancer, Non-coding RNA, miRNA
Introduction
Colorectal cancer (CRC) stands as a highly prevalent global affliction, ranking as the third most common cancer worldwide [1]. In terms of mortality, CRC assumes the grim position of being the second most lethal malignancy [1, 2]. Its incidence is markedly impacted by an interplay of environmental factors, lifestyle choices, dietary habits, age, and genetic predispositions [3]. While the full etiology of this disease remains elusive, substantial evidence has substantiated the pivotal role of genetic factors in its pathogenesis [4, 5].
Exceeding 75% of the human genome is subject to transcription; however, the majority of these transcripts do not undergo protein translation and are classified as non-coding RNAs [6]. Prominent categories of non-coding RNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) [6, 7] have been elucidated for their pivotal roles in a multitude of diseases, especially diverse types of cancer [8]. These RNA species exhibit distinct attributes in terms of length, structure, and functionality. For instance, miRNAs typically comprise approximately 22 nucleotides, whereas lncRNAs are predominantly longer than 200 nucleotides [9]. Additionally, in recent years, the utilization of non-coding RNAs as non-invasive diagnostic biomarkers has gained significant popularity [10, 11]. Hundreds of microRNA clusters have been recognized within the human genome, including the miR17-92a-1 cluster host gene, also known as MIR17HG [12]. The miR-17–92 cluster host gene, located on human chromosome 13q31.3, is a gene cluster with a polycistronic promoter that gives rise to mature miRNAs: hsa-miR-17, hsa-miR-18a, hsa-miR-19a, hsa-miR-19b-1, hsa-miR-20a, and hsa-miR-92a. This cluster contributes to the development of malignancies and has considerable overexpression in a variety of solid tumors [13]. Mogilyansky et al., in their study, show that the miR-18a and miR-20a are overexpressed in CRC [14]. Alternatively, XU et al. explored MIR17HG as a pivotal BLNK inhibitor, revealing that BLNK suppression enhances metastatic potential and tumorigenesis in CRC [15]. Additionally, Cellura and colleagues demonstrated that miR-19, through its inhibitory effects on transglutaminase 2, fosters augmented invasion and metastasis in CRC [16]. However, the generalizability of many published papers on this issue is limited, and such expositions are far from satisfactory due to their failure to undertake a thorough exploration into the regulatory mechanism, structure, function, and potential therapeutic implications of this specific cluster in CRC. Therefore, this study aims to comprehensively investigate the miR-17-92a-1 cluster host gene and its intricate regulatory relationship with the development and progression of CRC.
miR-17-92a-1 cluster host gene overview
The miR-17-92a-1 cluster host gene (MIR17HG) is situated at 13q31.3, position 91,347,820 to 91,354,575, within the genomic sequence with accession number NC_000013.11 (GRCh38.p14 primary assembly). This gene has two linear long non-coding RNA variants called MIR17HG transcript variants 1 and 2, which have lengths of 5,018 bp (NR_027350.1) and 927 bp (NR_027349.1), respectively. MIR17HG, on the other hand, serves as a host gene for six microRNAs (hsa-miR-17, hsa-miR-18a, hsa-miR-19a, hsa-miR-19b-1, hsa-miR-20a, and hsa-miR-92a-1) (https://www.ncbi.nlm.nih.gov/gene/407975). Based on the NCBI database, we have added more information regarding the MIR17HG-derived RNAs as follows:
MIR17HG LncRNA
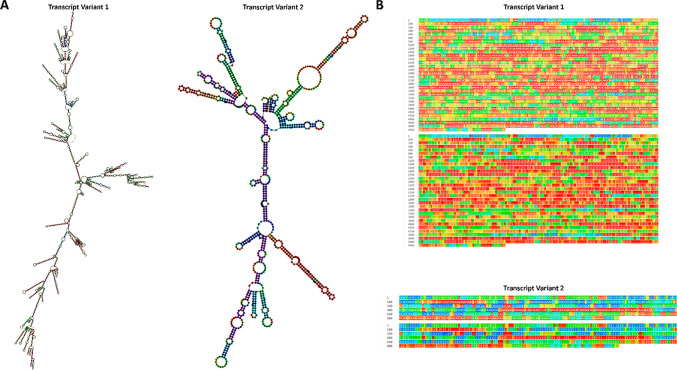
The optimal secondary structure of transcript variants 1 and 2 for the non-coding RNA MIR17HG was analyzed using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) [17, 18]. The structures were drawn in both minimum free energy (MFE) and dot-bracket notation. The MFE structure for variant 1 was calculated to be -1581.60 kcal/mol, whereas that for variant 2 was -262.50 kcal/mol. The results suggest that the secondary structure of the MIR17HG transcript variants 1 and 2 is highly stable with low MFE values, indicating their potential biological significance, the details in Fig. 1A–B.
Fig. 1.
The optimal minimum free energy (MFE) construction A and dot-bracket representation B related to two ncRNA-derived MIR17HG transcript variants. The results showed that MIR17HG transcript variant 1 had a MFE of 1398.30 kcal/mol, while MIR17HG transcript variant 2 had a MFE of − 262.50 kcal/mol
MIR17HG -derived miRNAs
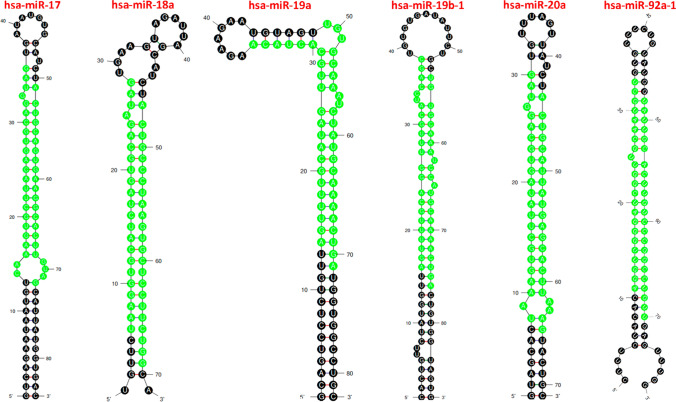
We utilized the mfold web server (http://unafold.rna.albany.edu/?q=node/60) [19] to visualize the optimized secondary structures and the precise location of MIR17HG-related mature miRNAs (identified by green highlights) derived from their precursor (pre-miRNAs). This analysis allows us to obtain a clear and in-depth understanding of the miRNA’s location and structure (Fig. 2). Table 1 presents comprehensive attributes of six pre-miRNAs that are linked with the miR-17-92a-1 cluster host gene, namely hsa-miR-17, hsa-miR-18a, hsa-miR-19a, hsa-miR-19b-1, hsa-miR-20a, and hsa-miR-92a-1.
Fig. 2.
The optimized secondary structure of the MIR17HG-related miRNAs, which includes hsa-miR-17 (ΔG = − 34.30), hsa-miR-18a (ΔG = − 22.00), hsa-miR-19a (ΔG = − 39.10), hsa-miR-19b-1 (ΔG = − 38.40), hsa-miR-20a (ΔG =− 31.00), and hsa-miR-92a-1 (ΔG = − 35.70) stem-loop precursor miRNAs (pre-miRNAs). The green highlighting in the structure indicates the position where the mature miRNAs originate
Table 1.
The fundamental details of six microRNAs that originate from the miR17HG
| miRNAs | miRBase Accession Number | Minimum Free Energy (MFE) kCal/mol | Mature miRNA | Mature miRNA sequence |
|---|---|---|---|---|
| hsa-miR-17 | MI0000071 | − 34.30 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAG |
| hsa-miR-17-3p | ACUGCAGUGAAGGCACUUGUAG | |||
| hsa-miR-18a | MI0000072 | − 22.00 | hsa-miR-18a-5p | UAAGGUGCAUCUAGUGCAGAUAG |
| hsa-miR-18a-3p | ACUGCCCUAAGUGCUCCUUCUGG | |||
| hsa-miR-19a | MI0000073 | − 39.10 | hsa-miR-19a-5p | AGUUUUGCAUAGUUGCACUACA |
| hsa-miR-19a-3p | UGUGCAAAUCUAUGCAAAACUGA | |||
| hsa-miR-19b-1 | MI0000074 | − 38.40 | hsa-miR-19b-1-5p | AGUUUUGCAGGUUUGCAUCCAGC |
| hsa-miR-19b-1-3p | UGUGCAAAUCCAUGCAAAACUGA | |||
| hsa-miR-20a | MI0000076 | − 31.00 | hsa-miR-20a-5p | UAAAGUGCUUAUAGUGCAGGUAG |
| hsa-miR-20a-3p | ACUGCAUUAUGAGCACUUAAAG | |||
| hsa-miR-92a-1 | MI0000093 | − 35.70 | hsa-miR-92a-1-5p | AGGUUGGGAUCGGUUGCAAUGCU |
| hsa-miR-92a-3p | UAUUGCACUUGUCCCGGCCUGU |
MIR17HG and miRNAs expression analysis
The clinical data of Colon Adenocarcinoma (COAD) were obtained from The cancer genome atlas (TCGA) database (https://portal.gdc.cancer.gov/repository) [20], specifically to analyze RNA-sequencing (RNA-seq) and microRNA-sequencing (miRNA-seq) data. The inclusion criteria were as follows: (1) verification of a histopathological diagnosis of COAD; (2) confirmation of the primary tumor site as the colon; (3) categorization of the disease type as adenoma or adenocarcinoma. Cases lacking complete demographic information, including gender and vital status, as well as those lacking comprehensive RNA-seq and miRNA-seq transcriptome profiling data, were systematically excluded. Ultimately, our study encompassed 380 COAD patients for RNA-seq data analysis and 369 COAD patients for miRNA-seq data analysis. The clinical characteristics of the patients are summarized in Table 2.
Table 2.
Demographic data of COAD patients
| RNA-seq data analysis | miRNA-seq data analysis | |||
|---|---|---|---|---|
| Characteristics | Number | Frequency (%) | Number | Frequency (%) |
| Gender | ||||
| Male | 202 | 53.1 | 194 | 52.5 |
| Female | 178 | 46.9 | 175 | 47.5 |
| Age (year) (mean (SD)) | 67 (12.7) | 66.9 (12.8) | ||
| < 69 | 189 | 49.7 | 184 | 49.8 |
| ≥ 69 | 191 | 50.3 | 185 | 50.2 |
| Vital status | ||||
| Alive | 301 | 79.2 | 290 | 78.5 |
| Dead | 79 | 20.8 | 79 | 21.5 |
The corresponding high-level data from RNA-Seq and miR-Seq were acquired using TCGA database. Raw read counts from RNA-Seq and miR-Seq datasets underwent normalization using the Trimmed Mean of M values (TMM) method. Differential expression analysis was conducted to identify differentially expressed long non-coding RNAs (DElncRNAs) and microRNAs (DEmiRNAs) between healthy solid tissues and primary malignancies. This analysis was executed using The “limma” package [21, 22] and Voom statistical analysis [23] within the R programming software (R-4.2.1, 64-bit, https://www.r-project.org/) [24], alongside RStudio Desktop version 2022.7.0.548 [25]. Subsequently, the miRNAmeConverter package [26] was employed to find the latest names of final DEmiRNAs from the miRbase version 22 database [27–32]. Furthermore, we employed GENT2, a platform designed for the exploration of gene expression patterns across normal and tumor tissues, to validate the DElncRNAs identified using Affymetrix microarray data from the GPL570 platform [33]. To validate the DEmiRNAs, we utilized microarray data from the GSE53592, GSE35834, and GSE18392 data series, sourced from the dbDEMC version 3.0 database, a comprehensive repository of differentially expressed miRNAs in human cancers [34]. Data filtering criteria for RNA/miRNA-seq and microarray data included a requirement for |log2 Fold Change (FC)|> 1, as well as a significance threshold based on P-value and adjusted P-value (false discovery rate (FDR)) < 0.01.
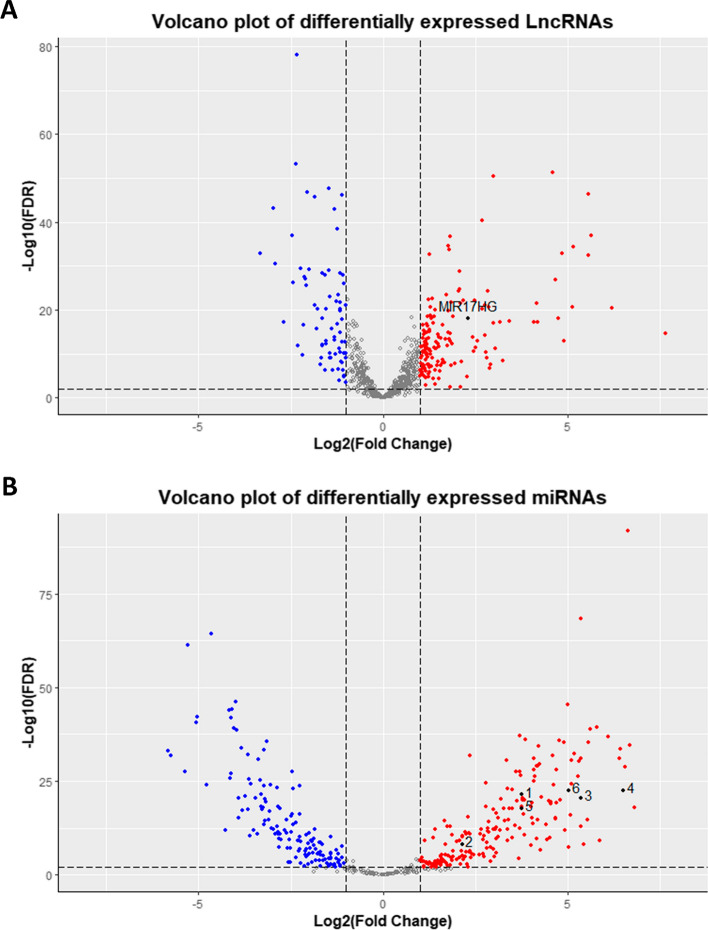
The RNA-seq and microarray analysis results revealed that MIR17HG exhibits significant overexpression in tumor tissues compared to their normal (Table 3, Fig. 3A). Alternatively, while the expression profiles of hsa-miR-18a-3p, hsa-miR-20a-3p, and hsa-miR-92a-3p appeared inconsistent between miRNA-seq and microarray analyses, the other MIR17HG-derived miRNAs, including hsa-miR-17-5p, hsa-miR-17-3p, hsa-miR-18a-5p, hsa-miR-19a-3p, hsa-miR-19b-1-5p, and hsa-miR-20a-5p, collectively demonstrated an upregulation pattern in patients diagnosed with COAD (Table 4, Fig. 3B).
Table 3.
The MIR17HG RNA-seq and microarray data analysis
| TCGA database | GENT2 database | Expression pattern | |||
|---|---|---|---|---|---|
| LncRNA | LogFC | FDR | LogFC | P-value | |
| MIR17HG | 2.28 | 0.00*** | 1.047 | 0.00*** | Up-regulation |
*** < 0.001
Fig. 3.
Volcano plots of the DELncRNAs and DEmiRNAs from RNA/miRNA-seq data. A The volcano plot depicts the DELncRNAs, with MIR17HG highlighted in black. B Volcano plot of DEmiRNAs denoting hsa-miR-17-5p, hsa-miR-17-3p, hsa-miR-18a-5p, hsa-miR-19a-3p, hsa-miR-19b-1-5p, and hsa-miR-20a-5p as data points 1 to 6, respectively. Over-expressed genes are represented in red, while under-expressed ones are displayed in Blue
Table 4.
The MIR17HG-derived miRNAs miRNA-seq and microarray data analysis
| TCGA database | dbDEMC database | Expression Pattern | ||||
|---|---|---|---|---|---|---|
| miRNA | LogFC | FDR | LogFC | Adjusted P-value | Data series | |
| hsa-miR-17-5p | 3.75 | 0.00*** | 1.15 | 0.00*** | GSE35834 | Upregulation |
| hsa-miR-17-3p | 2.13 | 0.00*** | 0.98 | 0.00*** | GSE18392 | Upregulation |
| hsa-miR-18a-5p | 5.36 | 0.00*** | 1.13 | 0.003 | GSE53592 | Upregulation |
| hsa-miR-18a-3p | − 1.31 | 0.002 | 0.52 | 0.00*** | GSE18392 | Contradiction |
| hsa-miR-19a-5p | – | NS | – | NS | – | – |
| hsa-miR-19a-3p | 6.51 | 0.00*** | 0.68 | 0.00*** | GSE18392 | Upregulation |
| hsa-miR-19b-1-5p* | 3.75 | 0.00*** | 0.45 | 0.00*** | GSE35834 | Upregulation |
| hsa-miR-19b-1-3p | – | NS | – | NS | – | – |
| hsa-miR-20a-5p | 5.03 | 0.00*** | 0.62 | 0.00*** | GSE35834 | Upregulation |
| hsa-miR-20a-3p | 4.66 | 0.00*** | – | NS | – | – |
| hsa-miR-92a-1-5p | – | NS | – | NS | – | – |
| hsa-miR-92a-3p | − 2.03 | 0.00*** | 0.20 | 0.00*** | GSE18392 | Contradiction |
*hsa-miR-19b-1 in the GSE35834 was considered as the hsa-miR-19b-1-5p; ***: < 0.001; NS: not significant with P-value and adjusted P-value (false discovery rate (FDR)) threshold < 0.01.
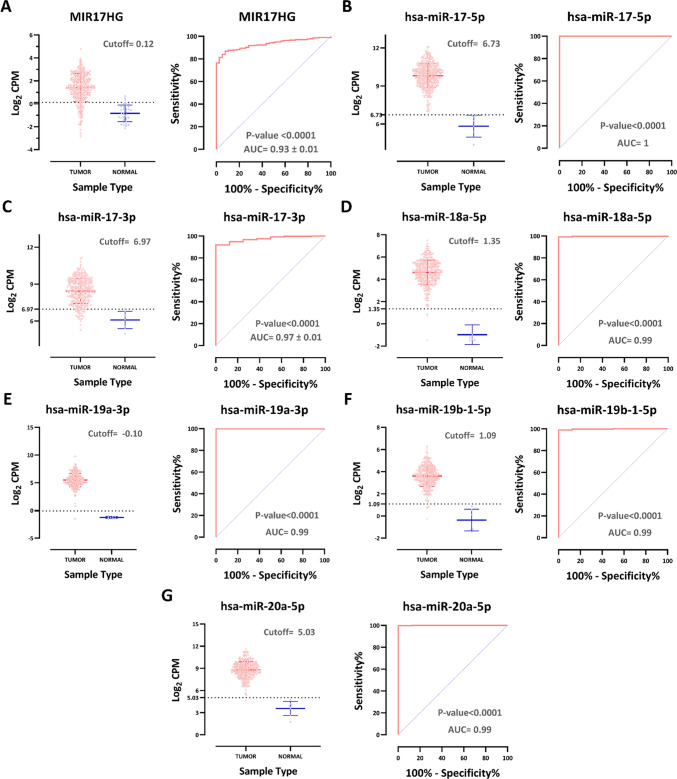
The diagnostic potential of MIR17HG and its associated definitive upregulated miRNAs in COAD was assessed through receiver operating characteristic (ROC) analysis from RNA/miRNA-seq data, which yielded metrics including the area under the curve (AUC) and a 95 percent confidence interval (CI). These analyses were performed using GraphPad Prism 9.5.1, with statistical significance defined as a P-value < 0.01.
Our analysis revealed an impressive AUC value of 0.93 for MIR17HG (P-value < 0.0001, Fig. 4A). Furthermore, the AUC values of other microRNAs, including hsa-miR-17-5p, hsa-miR-17-3p, hsa-miR-18a-5p, hsa-miR-19a-3p, hsa-miR-19b-1-5p, and hsa-miR-20a-5p, were also evaluated. Remarkably, these microRNAs exhibited high AUC values of 1, 0.97, 0.99, 0.99, 0.99, and 0.99, respectively (P-values < 0.0001, Figs. 4B–G). This robust performance underscores the strong discriminatory power of MIR17HG and its associated miRNAs to distinguish COAD from normal tissues. Additionally, Fig. 4 serves as a pivotal resource by delineating the optimal cutoff values for distinguishing between tumor and normal tissue for each of these non-coding RNAs. Such delineation further enhances the chance of clinical utility of these ncRNAs by providing precise guidelines for their application in COAD. Further studies and validation are warranted to affirm their clinical utility.
Fig. 4.
MIR17HG and its derived miRNAs diagnostic potential analysis. A–G Significantly increased expression of lncRNA MIR17HG and its derived miRNAs can distinguish tumor from normal tissue with different cutoff values and high specificity and sensitivity (P-value < 0.0001)
MIR17HG/miRNA/mRNA CeRNA axis
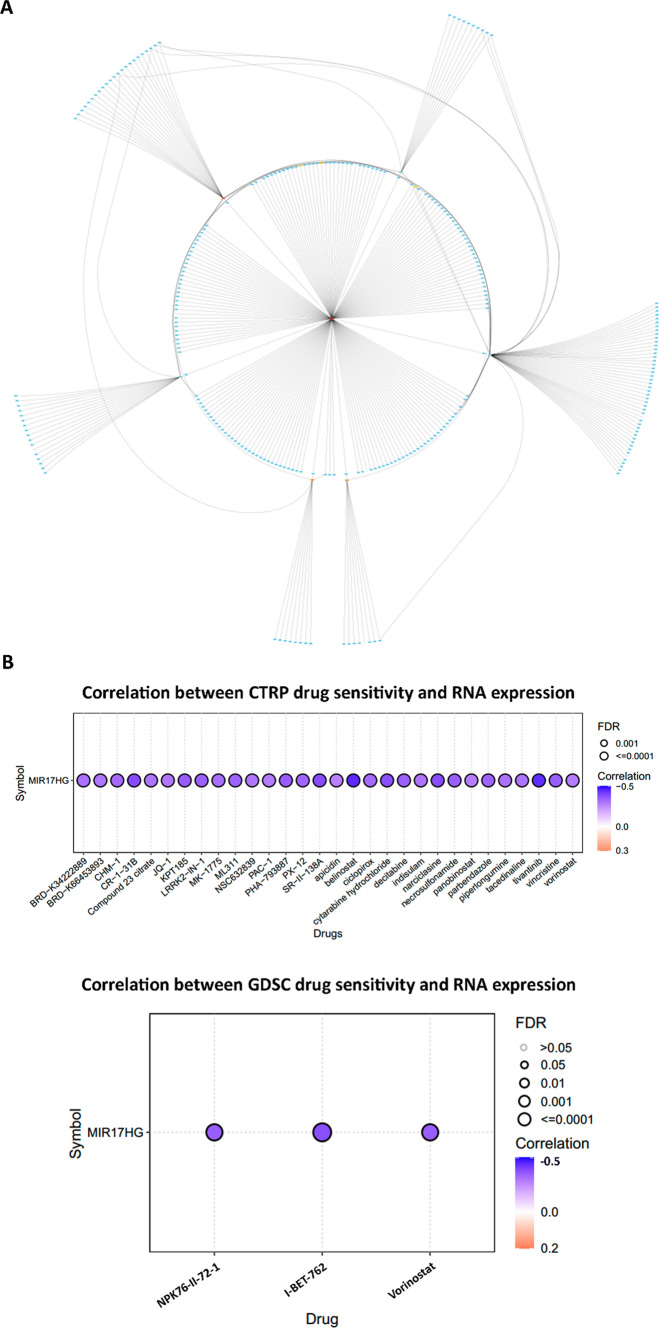
To elucidate the regulatory role of MIR17HG, we established a Competing Endogenous RNA (CeRNA) axis. The MIR17HG-associated miRNAs were identified by applying a stringent criterion, setting the CLIP region P-value to ≤ 0.01 utilizing the starBase v2.0 database (https://rnasysu.com/encori/index.php) [35, 36]. To harmonize the nomenclature with the latest miRNA standards, the miRNAs retrieved from starBase were subjected to the miRNAmeConverter package [26] aligning with miRbase version 22 [27–32]. Subsequently, we got an intersection between the down-regulated miRNAs from our TCGA miRNA-seq data analysis (LogFC < -1 and FDR < 0.01) and starBase miRNAs with the Venny 2.1 web tool (https://bioinfogp.cnb.csic.es/tools/venny/) [37]. Validated target mRNAs were identified using the multiMIR package, referencing data from miRecords, miRTarBase, and Tarbase databases [38]. Similarly, an intersection analysis was performed to align up-regulated mRNAs derived from our TCGA RNA-seq data analysis (with LogFC > 1 and FDR < 0.01) with the mRNAs from the multiMIR package, again utilizing the Venny 2.1 web tool. Data analysis was carried out using the R programming software (version R-4.2.1, 64-bit, https://www.r-project.org/) [24] in conjunction with RStudio Desktop (version 2022.7.0.548) [25]. Ultimately, the Cytoscape software (version 3.9.1) [39] was employed to construct the MIR17HG-miRNAs-mRNAs CeRNA axis, with the aid of the CytoHubba app [40] to identify central nodes within the network.
Consequently, we have identified the top ten molecules according to the maximal clique centrality (MCC) ranking approach within the regulatory network of MIR17HG. Specifically, hsa-miR-214-3p, hsa-miR-140-3p, hsa-miR-501-3p, hsa-miR-1287-5p, hsa-miR-28-3p, hsa-miR-323a-3p, hsa-miR-664b-3p, CDCA4, NCAPG2, and MIR17HG were ranked with the highest MCC scores (Fig. 5A and Table 5). Collectively, these findings introduce the MIR17HG network as a central regulatory axis in the regulation of MIR17HG-mediated cellular processes in the context of COAD. However, it is crucial to emphasize that further experimental investigations are warranted to validate the intricate interactions among these nodes and MIR17HG, thereby enhancing our comprehension of the underlying molecular mechanisms governing COAD pathobiology.
Fig. 5.
A The CeRNA network of the MIR17HG in COAD. B The drug sensitivity analysis for the MIR17HG gene using data from the GSCA database, showing the correlation between MIR17HG gene expression and drug sensitivity. A negative correlation indicates that elevated expression of the gene is associated with decreased drug sensitivity or potential resistance, while a positive correlation implies the opposite
Table 5.
Top 10 nodes in MIR17HG-miRNAs-mRNAs network ranked by MCC method
| Rank | Node Name | Score |
|---|---|---|
| 1 | hsa-miR-214-3p | 175 |
| 2 | hsa-miR-140-3p | 72 |
| 3 | hsa-miR-501-3p | 34 |
| 4 | hsa-miR-1287-5p | 25 |
| 5 | hsa-miR-28-3p | 16 |
| 6 | hsa-miR-323a-3p | 12 |
| 7 | hsa-miR-664b-3p | 10 |
| 8 | MIR17HG | 7 |
| 9 | CDCA4 | 4 |
| 9 | NCAPG2 | 4 |
MIR17HG drug sensitivity analysis
To investigate the potential utility of aberrant MIR17HG gene expression as a clinical response indicator and its suitability as a candidate predictive biomarker for drug screening, we conducted a drug sensitivity analysis utilizing the Gene Set Cancer Analysis (GSCA) database (http://bioinfo.life.hust.edu.cn/GSCA/#/) [41]. This database compiles the IC50 of several small molecules in human pan-cancer cell lines and its corresponding gene expression from two primary sources, the Cancer therapeutics response portal (CTRP) and Genomics of Drug Sensitivity in Cancer (GDSC) databases [42–47]. The analysis involves employing Pearson correlation to assess the relationship between gene expression and drug sensitivity, and the derived P-values are subjected to FDR adjustment. A negative correlation signifies that increased expression of the gene of interest corresponds to reduced drug sensitivity or potential resistance, and vice versa. While the outcomes from the GDSC database have suggested that cancer patients exhibiting high expression levels of the MIR17HG gene tend to display resistance to I-BET-762, NPK76-II-72–1, and Vorinostat, the CTRP results have further extended our understanding. The CTRP data have illuminated that elevated MIR17HG gene expression is likely associated with resistance to additional compounds, namely Belinostat, Tivantinib, and CR-1-31B. For a comprehensive overview of these correlations between MIR17HG expression and drug sensitivity, we refer to Fig. 5B, which visually encapsulates the intricate relationship between MIR17HG and drug responses. These findings collectively highlight the potential clinical significance of MIR17HG expression as an indicator of drug resistance in the context of cancer treatment. Given the absence of dedicated databases or datasets exclusively tailored for CRC, we have leveraged this comprehensive database to investigate the potential correlation between MIR17HG expression and drug sensitivity at the pan-cancer level. Nevertheless, it is essential to acknowledge the need for additional validation specifically within the context of CRC, as a means to address this particular limitation and enhance the robustness of our findings.
MIR17HG co-expression analysis
The “Similar Gene Detection” module of GEPIA2, a gene expression profiling interactive analysis database (http://gepia2.cancer-pku.cn/#index) [48] was employed to identify a set of 500 genes exhibiting transcription profiles most closely resembling that of MIR17HG, drawing from the TCGA-COAD project. Only genes displaying a Pearson’s correlation coefficient ≥ 0.4 were considered for inclusion in this analysis. Subsequently, we assessed the expression patterns of the identified genes using our RNA-seq data and specifically selected those protein-coding genes that displayed significant differential expression (FDR < 0.01, LogFC > 1). Consequently, our focus was directed solely toward those protein-coding genes exhibiting a positive correlation with MIR17HG and concurrently displaying an upregulated expression pattern in COAD for subsequent analyses (63 protein-coding genes that are shown in supplementary material 1).
MIR17HG gene set enrichment analysis
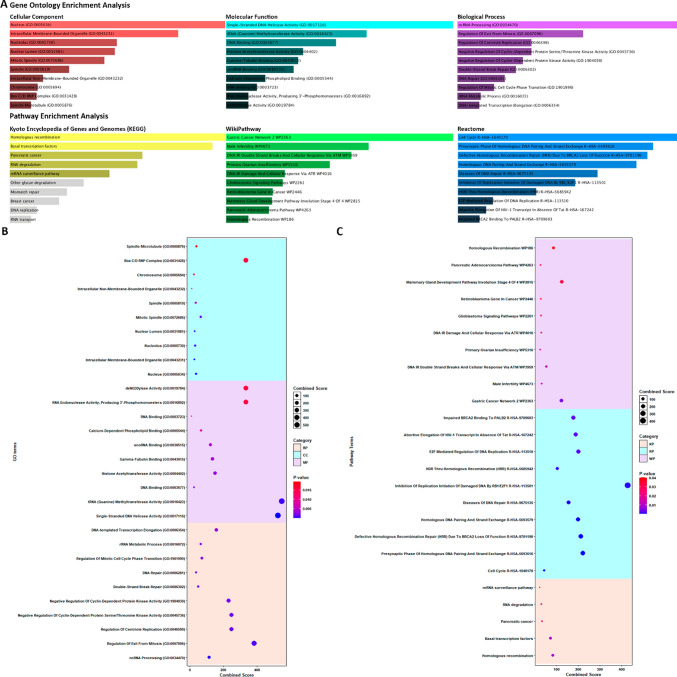
We conducted an enrichment analysis on the set of 63 mentioned mRNAs to elucidate the potential functional roles of protein-coding genes that exhibit co-expression with MIR17HG. This analysis encompassed gene ontology (GO) enrichment assessments across cellular component, molecular function, and biological process categories. Additionally, pathway enrichment analysis was performed, encompassing the Kyoto Encyclopedia of Genes and Genomes (KEGG), Wikipathway, and Reactome databases. The Enrichr tool (https://maayanlab.cloud/Enrichr/) was employed for these enrichment analyses, utilizing a significance threshold set at a P-value below 0.05 [49–51]. Furthermore, to visually represent the results of the GO and pathway enrichment analysis, dot plots were created. This was accomplished using the ggplot2 package [52] within the R programming software (version R-4.2.1, 64 bit, https://www.r-project.org/) [24] and RStudio Desktop (version 2022.7.0.548) [25]).
The findings from our analysis reveal significant involvement of the identified genes in a variety of crucial cellular processes. Notably, these genes play key roles in non-coding RNA (ncRNA) processing, the regulation of exit from the mitotic phase, and the control of centriole replication, highlighting their importance in fundamental biological mechanisms. Furthermore, our investigation unveiled that a substantial proportion of these genes exhibit enrichment in various molecular activities, including single-stranded DNA helicase activity, tRNA (Guanine) methyltransferase activity, as well as DNA and RNA binding. Additionally, some of these genes were associated with histone acetyltransferase activity, underscoring their potential influence on chromatin modification. In terms of cellular localization, our results indicate that these genes are predominantly situated within the cellular nucleus, with a noteworthy presence in the nucleolus. This subcellular distribution further emphasizes their essential roles in nuclear processes and gene regulation. Moreover, our analysis extends beyond functional annotations and encompasses insights into pathway involvement. We have identified prominent pathways based on comprehensive databases such as KEGG, Wikipathway, and Reactome. These pathways include Homologous recombination, basal transcription factors, DNA double-strand break repair and cellular responses via ATM (ataxia-telangiectasia mutated), and gastric cancer network pathways. These pathway associations shed light on the potential implications of the identified genes in various biological contexts (Fig. 6).
Fig. 6.
The MIR17HG-related gene set enrichment analysis. The GO enrichment analysis and pathway enrichment analysis from the Enrichr tool were visualized by bar plot A and dot plots B, C, highlighting statistically significant enrichments with a significance threshold set at P-value < 0.05
Regulatory mechanisms of MIR17HG and its derived ncRNAs
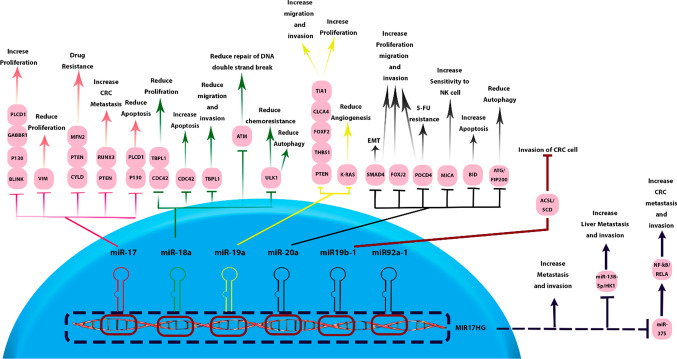
The MIR17HG exerts multifaceted functions that are pivotal in normal developmental biology as well as in the pathogenesis of cancer. In the context of carcinogenesis, MIR17HG demonstrates a shift towards oncogenic activities. It actively promotes cellular proliferation, fosters a milieu conducive to angiogenesis, and inhibits cellular differentiation—all hallmarks of cancer progression. Furthermore, it contributes to the sustenance of cell viability, thereby potentially aiding in the persistent survival of malignant cells. These actions position MIR17HG as a crucial regulator within the cellular microenvironment [53]. Therefore, the specific mechanisms of MIR17HG and its associated miRNAs are delineated, elucidating their roles in the intricate network of gene regulation within oncogenic outcomes (summarized in Table 6 and Fig. 7).
Table 6.
Regulatory Networks of MIR17HG and its associated ncRNAs in CRC oncogenesis
| LncRNA/miRNA | Downstream target | Outcomes in CRC cells | Cell line | Refs. |
|---|---|---|---|---|
| MIR17HG | hsa-miR-375/NF-κB/RELA | Enhance invasion and metastasis of CRC cells | HCT116, SW620 | [15] |
| hsa-miR-138-5p/HK1 | Enhance invasion of CRC cells / Increase liver metastasis | SW480, RKO | [54] | |
| hsa-miR-17 | MFN2 | Resistance to 5-FU | HCT116 | [55] |
| BLINK | Enhance migration and invasion of CRC cells | SW620, HCT116 | [15] | |
| PTEN | Metastatic behavior and multiple drug resistance | SW480, HCT116 | [56] | |
| RUNX3 | Enhance CRC metastasis | SW480, HCT116 | [57] | |
| CYLD | Resistance to 5-FU | T84 | [58] | |
| VIM | Decrease migration and invasion of CRC cells | LoVo, HT29 | [59] | |
| P130 | Increase proliferation and invasion / Decrease apoptosis | SW480, LoVo | [60] | |
| PLCD1 | Reduce apoptosis, and increase invasion and migration of CRC cells | LoVo, HCT116 | [61] | |
| GABBR1 | Promote invasion and proliferation of CRC cells | HCT116, HT29 | [62] | |
| hsa-miR-19a | TIA1 | Increase CRC cell invasion and proliferation | SW480 | [63] |
| CLCA4 | Promote invasion, proliferation, and migration of CRC cells | CaCo2, SW480 | [64] | |
| FOXF2 | Increase CRC cell proliferation and migration | HCT116 | [65] | |
| THBS1 | Increase viability and migration of CRC cells | SW480 | [66] | |
| PTEN | Enhance CRC cell migration and invasion | HCT116, SW480 | [67] | |
| KRAS | Decrease angiogenesis of CRC cells | HCT116 | [68] | |
| hsa-miR-19b-1 | ACSL/SCD | Impede invasion of CRC cells | SW620 | [69] |
| hsa-miR-18a | CDC42 | Reduce proliferation, and migration /Enhance apoptosis of CRC cells | HCT116, LIM1215 | [70] |
| TBPL1 | Suppress growth, migration, and invasion of CRC cells | HCT116, SW620 | [71] | |
| ULK1 | Reduce autophagy activation/ Reduce Chemoresistance in CRC cells | HCT116 | [72] | |
| ATM | Reduce DNA double-strand breaks repair | HCT116 | [73] | |
| hsa-miR-20a | BID | Reduce apoptosis of CRC cells | SW480 | [74] |
| MICA | Reduce CRC cell sensitivity to NK cells | SW480, HCT116 | [75] | |
| FOXJ2 | Increase proliferation, invasion, and migration of CRC cells | HCT116 | [76] | |
| SMAD4 | Induce EMT / increase proliferation and invasion of CRC cells | SW480 | [77] | |
| POCD4 | Increase CRC cells proliferation / Resistance to 5-FU | SW480, HCT116 | [78] | |
| ATG5/FIP200 | Inhibit the autophagic response initiated by hypoxia | LoVo, SW480 | [79] |
Fig. 7.
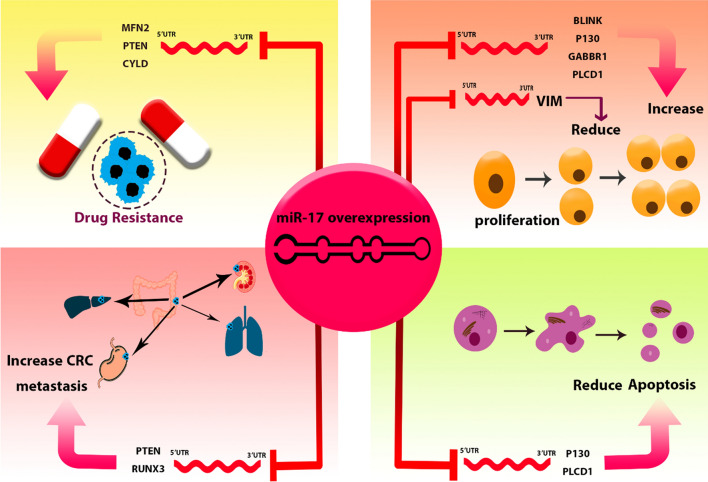
Regulatory mechanisms of MIR17HG and its associated miRNAs in CRC. MIR17HG and its derived miRNAs exert comprehensive regulatory mechanisms in CRC cells by targeting a diverse array of mRNAs. This cluster is recognized as a pivotal factor in the progression of CRC
MIR17HG
The long non-coding RNA MIR17HG enhances the metastatic and invasive capabilities of SW620 and HCT116 CRC cell lines. It functions as a CeRNA and modulates the expression of NF-κB/RELA by downregulation of miR-375 [15]. It enhances glycolysis activity in CRC cells through the regulation of the miR-138-5p/HK1 axis. Alternatively, overexpression of MIR17HG significantly increases the invasive potential of SW480 and RKO cell lines. Transplantation of these modified cell lines into nude mice increases the incidence of liver metastasis lesions [54].
hsa-miR-17
Studies conducted in the HCT116 cell line have provided insights into the regulatory role of miR-17-5p in colorectal cancer. This microRNA suppresses the expression of MFN2, leading to reduced mitochondrial fusion, increased mitochondrial fission, and enhanced mitophagy. These dynamic changes in mitochondrial behavior contribute to the development of resistance to 5-FU chemotherapy. Notably, these findings have been consistently replicated in mouse models, emphasizing their significance [55]. Furthermore, miR-17-5p directly targets BLINK mRNA and reduces its expression in SW620 and HCT116 cell lines, resulting in an enhanced capacity for invasion and migration in CRC cells [15]. Elevated expression of miR-17-5p in SW480 and COLO205 cell lines is associated with enhanced metastatic behavior and drug resistance. This outcome is attributed to the direct targeting of PTEN by miR-17-5p, with the down-regulation of PTEN playing a pivotal role in the promotion of metastasis and the acquisition of multiple drug resistance [56]. Additionally, using SW480 and HCT-116 cell lines, it was observed that miR-17-5p, by targeting RUNX3, activates the TGF-β signaling pathway and enhances colorectal cancer metastasis [57]. Pan et al. have indicated that miR-17 inhibits CYLD expression, thereby contributing to 5-FU resistance in the T84 cell line [58]. In primary colorectal cancer tissues, miR-17-5p is downregulated in the presence of metastasis compared to non-metastasis CRC, and it directly interacts with VIM mRNA. Increasing miR-17-5p expression reduces vimentin levels, thereby mitigating cell migration and invasion in LoVo and HT29 cells. In summary, miR-17-5p exerts a regulatory influence on vimentin expression, thereby impacting CRC metastasis [59]. Another study involving SW480 and LoVo cell lines revealed that miR-17-5p specifically targets P130, leading to the activation of the Wnt/β-catenin pathway and driving the progression of CRC by increasing the proliferation, invasion and migration, and decreasing apoptosis of CRC cells [60]. Alternatively, experiments carried out on CRC cell lines, specifically LoVo and HCT116, have revealed that the upregulation of miR-17-3p results in a decrease of apoptosis and a simultaneous enhancement of invasion, migration, and proliferation in CRC cells. These functional changes are attributed to the regulatory interaction between miR-17-3p and PLCD1 [61]. Zhang et al. demonstrated that exosomes derived from cancer-associated fibroblasts exhibit high expression of miR-17-5p compared to normal fibroblasts. miR-17 also exhibits a targeted binding to the 3'UTR of GABBR1 mRNA, resulting in heightened invasion and proliferation in HCT116 and HT-29 cell lines [62]. Generally, the oncogenic potential of hsa-miR-17 in CRC is summarized in Fig. 8.
Fig. 8.
The oncogenic potential of hsa-miR-17 in CRC. miR-17 regulates various cellular processes, including cell proliferation and apoptosis. Its overexpression results in elevated drug resistance and metastasis in CRC cell lines
hsa-miR-19a / hsa-miR-19b-1
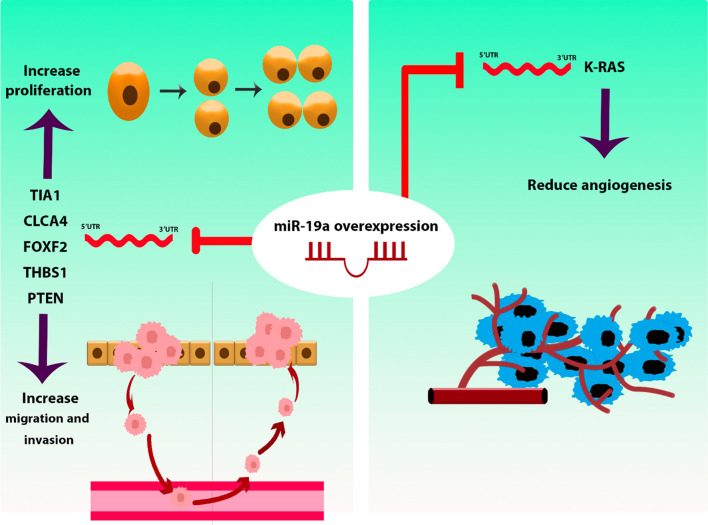
miR-19a, via its interaction with TIA1, facilitates the promotion of migration and proliferation in SW480 CRC cells. Consistently, xenograft mouse models corroborated this observed effect [63]. The studies on CaCo2 and SW480 CRC cells established that miR-19a overexpression significantly increased the proliferation, invasion, and migration of CRC cells by under-expressing CLCA4. Furthermore, it was demonstrated that the miR-19a/CLCA4 axis plays a regulatory role in the PI3K/AKT pathway in these cell lines [64]. Alternatively, miR-19a-3p fosters proliferation and migration in HCT116 CRC cells by inhibiting the expression of FOXF2 [65]. Inhibiting the expression of miR-19a-3p can result in the elevation of the FOXF2-associated Wnt/β-catenin signaling pathway. This, in turn, impacts the epithelial-mesenchymal transition (EMT), cell proliferation, invasion, and cellular migration in both HT29 and HCT116 cells [80]. In their study, Yin and colleagues investigated the regulatory role of miR-19a in SW480 CRC cell lines. miR-19a was found to exert control over the viability, invasiveness, and migratory properties of CRC cells by directly interacting with THBS1. Notably, the application of a miR-19a inhibitor effectively mitigated the malignant characteristics of CRC cells, concomitantly leading to the down-regulation of matrix metallopeptidase 9 (MMP-9) and vascular endothelial growth factor C (VEGFC) expression [66]. In hypoxic conditions, miR-19a initiates the activation of the PI3K/AKT pathway, leading to an enhancement in cell migration and invasion within HCT116 and SW480 CRC cells. This functional modulation is primarily attributed to the targeting of PTEN mRNA by miR-19a [67]. In contrast to the prevailing consensus that regards miR-19a as an oncogenic factor, the findings by Chen et al. indicated that elevated miR-19a expression led to the inhibition of KRAS expression and subsequently curtailed angiogenesis in HCT116 cells. Importantly, this effect was consistently evident in a mouse model as well [68]. Figure 9 delineates a compendium of the regulatory mechanisms pertinent to hsa-miR-19a.
Fig. 9.
hsa-miR-19a regulatory mechanisms in CRC. miR-19a plays a pivotal role in controlling cell proliferation, invasion, and migration by targeting multiple mRNAs. Additionally, it regulates angiogenesis in CRC cells
miR-19b-1 assumes a significant role as a central regulator of genes associated with lipid metabolism, specifically focusing on ACSL/SCD. miR-19b-1 directly oversees the pro-tumorigenic axis involving ACSL/SCD and exhibits the capability to impede invasion in SW620 and LoVo CRC cells. Importantly, it should be highlighted that diminished expression of miR-19b-1 is associated with a reduced survival rate among CRC patients, implying the potential involvement of ACSL/SCD in patient relapse [69].
hsa-miR-18a
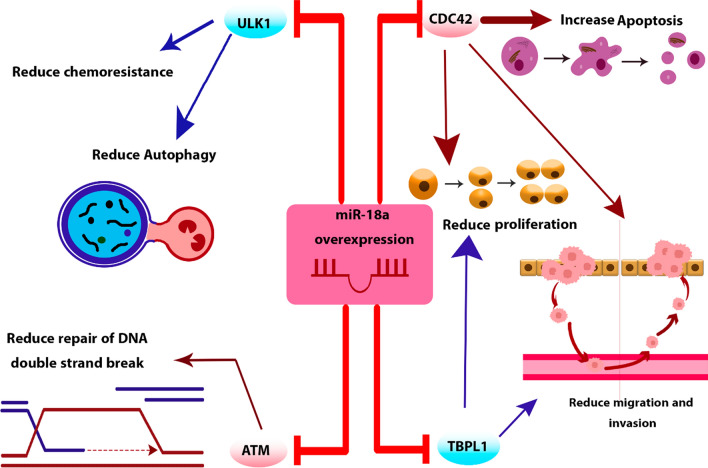
Mir-18a exhibits direct binding to the 3’ UTR of CDC42, a pivotal mediator within the PI3K pathway. Research conducted on HCT116 and LIM1215 CRC cell lines unveiled that miR-18a significantly reduces cell proliferation and migration while simultaneously enhancing apoptosis and the efficacy of pro-apoptotic agents [70]. Furthermore, it has been revealed that miR-18a exerts inhibitory effects on the growth, invasiveness, and migratory capabilities of HCT116 and SW620 colorectal cancer (CRC) cell lines. This regulatory influence is achieved through the specific targeting of TBPL1 by miR-18a [71]. Vu et al. brought to light the potential role of miR-18a in Fusobacterium nucleatum-mediated chemoresistance among CRC patients. Their research elucidated that miR-18a functions in the regulation of autophagy within HCT116 cells by suppressing ULK1 mRNA. Furthermore, their findings indicated that individuals with recurrent CRC exhibited elevated Fusobacterium nucleatum levels and diminished miR-18a expression when compared to patients who did not experience recurrence [72]. Moreover, in another study, it was demonstrated that miR-18a establishes a direct binding interaction with ATM mRNA. This interaction results in a reduction of DNA double-strand break repair within HCT116 CRC cells [73]. The different functions of hsa-miR-18a in CRC are delineated in Fig. 10.
Fig. 10.
The various functions of Hsa-miR-18a in CRC. miR-18a regulates cell proliferation, invasion, migration, apoptosis, and autophagy. Additionally, it reduces chemoresistance and controls DNA double-stranded break repair in CRC cells
hsa-miR-20a
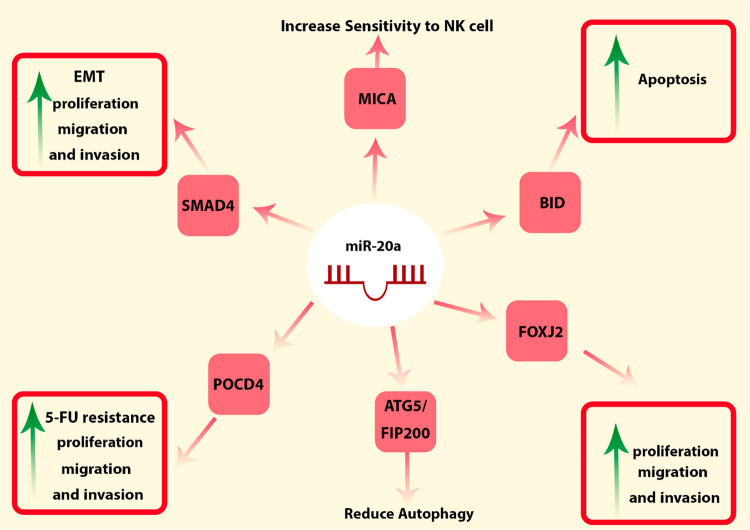
Research conducted on SW480 cells revealed that miR-20a plays a role in the regulation of apoptosis by specifically targeting BID mRNA, a member of the pro-apoptotic gene family within BCL-2 [74]. miR-20a also influences the responsiveness of SW480 and HCT116 CRC cells to NK cells through its targeting of MICA [75]. miR-20a serves as a direct regulator of Foxj2, with experimental findings indicating that heightened miR-20a expression leads to a notable escalation in the proliferation, invasion, and migration of HCT116 CRC cells. This functional modulation is primarily attributed to the suppressive action of miR-20a on Foxj2 mRNA, underscoring its pivotal role in these cellular responses [76]. Zhang et al. indicated that miR-20a induced EMT and took part in the regulation of migration and invasion in SW480 cells, primarily by suppressing the expression of SMAD4 [77]. Moreover, It also directly binds to 3' UTR of POCD4 mRNA, resulting in the promotion of cell proliferation and resistance to 5-FU in SW480 and HCT116 CRC cell lines [78]. In another study, miR-20a displayed substantial downregulation compared to non-hypoxic conditions in colon cancer cells when subjected to hypoxic conditions. This reduction in miR-20a expression was found to impede the autophagic response triggered by hypoxia. Notably, miR-20a achieves this inhibitory effect by directly targeting several key regulators of autophagy, including ATG5 and FIP200, within LoVo and SW480 cell lines [79]. Figure 11 encapsulates the regulatory roles of miR-20a within CRC.
Fig. 11.
The regulatory roles of hsa-miR-20a in CRC. miR-20a controls cell proliferation, invasion, migration, autophagy, and apoptosis. This microRNA increases the CRC cells sensitivity to NK cells and also extends the resistance of these cells to 5-FU. Additionally, miR-20a facilitates EMT in CRC cells
Inflammatory bowel diseases and MIR17HG-derived ncRNAs
Inflammatory bowel diseases (IBDs) represent persistent intestinal disorders, commonly classified into two principal subtypes: Crohn’s disease (CD) and ulcerative colitis (UC) [81]. Recent investigations have brought to light the intricate role of MIR17HG-derived ncRNAs in the pathogenesis and control of essential molecular pathways associated with IBDs, several of which are elucidated below:
Wang et al. revealed that miR-20a-5p exhibited the most expression reduction in patients with CD compared to healthy controls. Their intervention, utilizing microspheres composed of poly (lactic-co-glycolic acid) and loaded with miR-20a-5p, led to remarkable enhancements in colitis, a decrease in mucosal inflammation, and an enhancement in the function of the epithelial barrier in mouse models of CD [82]. In a separate study, it was demonstrated that within the unaffected mucosal tissues of patients with CD, the expression of miR-18a and mir-20a was elevated in comparison to the control group [83]. Chen et al. identified a significant reduction in the expression of miR-19a and a remarkable increase in TNF-α within human colon tissues afflicted with UC [84]. However, findings from a study by Schaefer et al. indicated a significant elevation in miR-19a levels in both UC colon biopsies and blood samples compared to those from normal controls [85]. The serum levels of miR-19a-3p and miR-19b-3p were found to be lower in CD patients exhibiting a stricture phenotype compared to control CD patients. This analysis in patients with a 4-year follow-up period provided support for the hypothesis that diminished levels of miR-19a-3p and miR-19b-3p precede the development of strictures phenotype [86]. An examination conducted on IL-10 knockout mice revealed that the expression levels of ten miRNAs, including miR-19a, exhibited elevation in both the colon tissues and peripheral blood leukocytes compared to healthy control mice [87]. Additionally, there is supporting evidence indicating a significant upregulation of circulating miR-19b in individuals with IBD when compared to the control group [88].
Patients diagnosed with UC and CD encounter a substantial long-term concern, notably an elevated susceptibility to the development of CRC. In the context of CD, a noteworthy pattern emerges during the transition from non-neoplastic tissue to dysplasia, with miR-17 showing an upregulation. However, as the disease progression advances from dysplasia to full-blown cancer, a subsequent downregulation of miR-17 is observed [89]. This dynamic miR-17 expression profile signifies its intricate involvement in the multi-step process of CRC development within the context of CD, emphasizing the importance of further investigations into its regulatory mechanisms and potential implications for disease management.
Conclusion and future prospect
The data presented in this study underscore the miR-17-92a-1 cluster host gene (MIR17HG) and its associated miRNAs upregulation in CRC, suggesting their significant involvement in the development and progression of CRC. They are implicated in critical oncogenic processes, including metastatic activity, apoptosis regulation, cell proliferation, and drug resistance. These findings shed light on the potential of MIR17HG and its associated miRNAs as therapeutic targets in CRC.
Future research should prioritize comprehensive investigations utilizing more human clinical samples to validate the oncogenic mechanisms of the miR-17-92a-1 cluster host gene and its associated miRNAs in CRC. Longitudinal studies would be particularly beneficial to trace miRNA expression over the cancer progression timeline. Additionally, functional studies aimed at exploring their background regulation by other molecular players in the CRC milieu could illuminate synergistic targets for intervention. Finally, developing advanced therapeutic strategies, including miRNA mimics or inhibitors, could pave the way for improved personalized treatment modalities for CRC patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- CRC
Colorectal cancer
- miRNA
MicroRNA
- lncRNA
Long non-coding RNA
- circRNA
Circular RNAs
- MFE
Minimum free energy
- COAD
Colon adenocarcinoma
- TCGA
The cancer genome atlas
- RNA-seq
RNA-sequencing
- miRNA-seq
MicroRNA-sequencing
- TMM
Mean of M values
- DElncRNA
Differentially expressed long non-coding RNA
- DEmiRNA
Differentially expressed microRNAs
- FC
Fold change
- FDR
False discovery rate
- ROC
Receiver operating characteristic
- AUC
Area under the curve
- CI
Confidence interval
- CeRNA
Competing endogenous RNA
- MCC
Maximal clique centrality
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GSCA
Gene set cancer analysis
- CTRP
Cancer therapeutics response portal
- GDSC
Genomics of drug sensitivity in cancer
- GO
Gene ontology
- ncRNA
Non-coding RNA
- ATM
Ataxia-telangiectasia mutated
- MMP-9
Matrix metallopeptidase 9
- VEGFC
Vascular endothelial growth factor C
- IBD
Inflammatory bowel disease
- CD
Crohn’s disease
- UC
Ulcerative colitis
Authors contributions
AMK: conceptualization, bioinformatic analysis, literature review, and writing article. SM: conceptualization, literature review, and writing article. AR: literature review and writing article. BHMH: writing article. BB: writing article. PM: supervision, edit, and review.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
The data used for this research are available from the corresponding author upon request.
Declarations
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, et al. Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr Drug Targets. 2021;22(9):998–1009. [DOI] [PubMed] [Google Scholar]
- 3.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–80. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham C, Dunlop MG. Genetics of colorectal cancer. Br Med Bull. 1994;50(3):640–55. [DOI] [PubMed] [Google Scholar]
- 5.Hull R, Francies FZ, Oyomno M, Dlamini Z. Colorectal cancer genetics, incidence and risk factors in search for targeted therapies. Cancer Manag Res. 2020;12:9869–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65(4):625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Xu Z, Liu D. Small non-coding RNA and colorectal cancer. J Cell Mol Med. 2019;23(5):3050–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(suppl_1):R17–29. [DOI] [PubMed] [Google Scholar]
- 10.Zhang R, Zeng Y, Deng JL. Long non-coding RNA H19: a potential biomarker and therapeutic target in human malignant tumors. Clin Exp Med. 2023;23(5):1425–40. [DOI] [PubMed] [Google Scholar]
- 11.Zhu M, Li X, Zhu S, Li P, Min L, Zhang S. Long non-coding RNA BLACAT1, a novel promising biomarker and regulator of human cancers. Biomed Pharmacother. 2020;132:110808. [DOI] [PubMed] [Google Scholar]
- 12.Pidíková P, Herichová I. miRNA clusters with up-regulated expression in colorectal cancer. Cancers. 2021;13(12):2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Li Y, Qi P, Ma Z. Biology of MiR-17-92 cluster and its progress in lung cancer. Int J Med Sci. 2018;15(13):1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20(12):1603–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Meng Q, Li X, Yang H, Xu J, Gao N, et al. Long noncoding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Can Res. 2019;79(19):4882–95. [DOI] [PubMed] [Google Scholar]
- 16.Cellura D, Pickard K, Quaratino S, Parker H, Strefford JC, Thomas GJ, et al. miR-19-mediated inhibition of transglutaminase-2 leads to enhanced invasion and metastasis in colorectal cancer. Mol Cancer Res. 2015;13(7):1095–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz R, Bernhart SH, Hönerzu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10(2):946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.R core team (2022). R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. URL https://www.R-project.org/.
- 25.RStudio Team (2022). RStudio: integrated development environment for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com/.
- 26.Haunsberger SJ, Connolly NM, Prehn JH. miRNAmeConverter: an R/bioconductor package for translating mature miRNA names to different miRBase versions. Bioinformatics. 2017;33(4):592–3. [DOI] [PubMed] [Google Scholar]
- 27.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32(suppl1):D109–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S-J, Yoon B-H, Kim S-K, Kim S-Y. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12(5):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu F, Wang Y, Ling Y, Zhou C, Wang H, Teschendorff AE, et al. dbDEMC 3.0: functional exploration of differentially expressed miRNAs in cancers of human and model organisms. Genom Proteom Bioinform. 2022;20(3):446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou KR, Huang JH, Liu S, Li B, Liu SR, Zheng WJ, et al. An encyclopedia of RNA interactomes in ENCORI.
- 36.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(1):D92–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveros, JC (2007–2015) Venny. An interactive tool for comparing lists with Venn's diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
- 38.Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, et al. The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42(17):e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin C-H, Chen S-H, Wu H-H, Ho C-W, Ko M-T, Lin C-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(4):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34(21):3771–2. [DOI] [PubMed] [Google Scholar]
- 42.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12(2):109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, et al. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5(11):1210–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012;41(D1):D955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166(3):740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie Z, Bailey A, Kuleshov MV, Clarke DJ, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with enrichr. Current protocols. 2021;1(3): e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 53.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42(8):1348–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao S, Guan B, Mi Y, Shi D, Wei P, Gu Y, et al. LncRNA MIR17HG promotes colorectal cancer liver metastasis by mediating a glycolysis-associated positive feedback circuit. Oncogene. 2021;40(28):4709–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun K, Chen L, Li Y, Huang B, Yan Q, Wu C, et al. METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer. Cell Death Dis. 2023;14(2):148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang L, Li H, Wang L, Hu J, Jin T, Wang J, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, Wang S, Lai Q, Fang Y, Wu C, Liu Y, et al. Cancer-associated fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive feedback loop. Cancer Lett. 2020;491:22–35. [DOI] [PubMed] [Google Scholar]
- 58.Pan S, Bao D, Li Y, Liu D, Quan S, Wang R. SOX4 induces drug resistance of colorectal cancer cells by downregulating CYLD through transcriptional activation of microRNA-17. J Biochem Mol Toxicol. 2022;36(1):e22910. [DOI] [PubMed] [Google Scholar]
- 59.Kim TW, Lee YS, Yun NH, Shin CH, Hong HK, Kim HH, et al. MicroRNA-17-5p regulates EMT by targeting vimentin in colorectal cancer. Br J Cancer. 2020;123(7):1123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma Y, Zhang P, Wang F, Zhang H, Yang Y, Shi C, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3(1):1291. [DOI] [PubMed] [Google Scholar]
- 61.Ji J, Fu J. MiR-17-3p Facilitates aggressive cell phenotypes in colon cancer by targeting PLCD1 through affecting KIF14. Appl Biochem Biotechnol. 2023;195(3):1723–35. [DOI] [PubMed] [Google Scholar]
- 62.Longqiu Y, Pengcheng L, Xuejie F, Peng Z. A mi RNA s panel promotes the proliferation and invasion of colorectal cancer cells by targeting GABBR 1. Cancer Med. 2016;5(8):2022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S, et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Huang B. miR-19a targets CLCA4 to regulate the proliferation, migration, and invasion of colorectal cancer cells. Eur J Histochem. 2022;66(1):3381. [DOI] [PMC free article] [PubMed]
- 65.Dai W, Zeng W, Lee D. lncRNA MCM3AP-AS1 inhibits the progression of colorectal cancer via the miR-19a-3p/FOXF2 axis. J Gene Med. 2021;23(3):e3306. [DOI] [PubMed] [Google Scholar]
- 66.Yin Q, Wang P-P, Peng R, Zhou H. MiR-19a enhances cell proliferation, migration, and invasiveness through enhancing lymphangiogenesis by targeting thrombospondin-1 in colorectal cancer. Biochem Cell Biol. 2019;97(6):731–9. [DOI] [PubMed] [Google Scholar]
- 67.Tang Y, Weng X, Liu C, Li X, Chen C. Hypoxia enhances activity and malignant behaviors of colorectal cancer cells through the stat3/microrna-19a/pten/pi3k/akt axis. Anal Cell Pathol. 2021;2021:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen M, Lin M, Wang X. Overexpression of miR-19a inhibits colorectal cancer angiogenesis by suppressing KRAS expression. Oncol Rep. 2018;39(2):619–26. [DOI] [PubMed] [Google Scholar]
- 69.Cruz-Gil S, Sanchez-Martinez R, de Cedron MG, Martin-Hernandez R, Vargas T, Molina S, et al. Targeting the lipid metabolic axis ACSL/SCD in colorectal cancer progression by therapeutic miRNAs: miR-19b-1 role. J Lipid Res. 2018;59(1):14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Humphreys KJ, McKinnon RA, Michael MZ. miR-18a inhibits CDC42 and plays a tumour suppressor role in colorectal cancer cells. PLoS ONE. 2014;9(11):e112288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu G, Liu Y, Yang Z, Wang J, Li D, Zhang X. Tumor suppressor microRNA-18a regulates tumor proliferation and invasion by targeting TBPL1 in colorectal cancer cells. Mol Med Rep. 2015;12(5):7643–8. [DOI] [PubMed] [Google Scholar]
- 72.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu C-W, Dong Y-J, Liang Q-Y, He X-Q, Ng SS, Chan FK, et al. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS ONE. 2013;8(2):e57036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang G, Chen X, Cai Y, Wang X, Xing C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncol Rep. 2017;37(1):571–8. [DOI] [PubMed] [Google Scholar]
- 75.Tang S, Fu H, Xu Q, Zhou Y. miR-20a regulates sensitivity of colorectal cancer cells to NK cells by targeting MICA. Biosci Rep. 2019;39(7):BSR20180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiang Y, Feng L, Wang G, Liu J, Zhang J, Xiang L, et al. miR-20a/Foxj2 axis mediates growth and metastasis of colorectal cancer cells as identified by integrated analysis. Med Sci Monitor Int Med J Exp Clin Res. 2020;26:e923559–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang GJ, Li Y, Zhou H, Xiao HX, Zhou T. miR-20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasis. Mol Med Rep. 2014;10(1):283–91. [DOI] [PubMed] [Google Scholar]
- 78.Jiang Z, Li L, Hou Z, Liu W, Wang H, Zhou T, et al. LncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer. Cell Signal. 2020;66:109483. [DOI] [PubMed] [Google Scholar]
- 79.Che J, Wang W, Huang Y, Zhang L, Zhao J, Zhang P, et al. miR-20a inhibits hypoxia-induced autophagy by targeting ATG5/FIP200 in colorectal cancer. Mol Carcinog. 2019;58(7):1234–47. [DOI] [PubMed] [Google Scholar]
- 80.Yu F-B, Sheng J, Yu J-M, Liu J-H, Qin X-X, Mou B. MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J Gastroenterol. 2020;26(6):627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang JT. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383(27):2652–64. [DOI] [PubMed] [Google Scholar]
- 82.Wang H, Xing H, Xia Y, Zhou Y, Zhou J, Li L, et al. PLGA microspheres carrying miR-20a-5p improved intestinal epithelial barrier function in patients with Crohn’s disease through STAT3-mediated inhibition of Th17 differentiation. Int Immunopharmacol. 2022;110:109025. [DOI] [PubMed] [Google Scholar]
- 83.Béres NJ, Kiss Z, Sztupinszki Z, Lendvai G, Arató A, Sziksz E, et al. Altered mucosal expression of microRNAs in pediatric patients with inflammatory bowel disease. Dig Liver Dis. 2017;49(4):378–87. [DOI] [PubMed] [Google Scholar]
- 84.Chen B, She S, Li D, Liu Z, Yang X, Zeng Z, et al. Role of miR-19a targeting TNF-α in mediating ulcerative colitis. Scand J Gastroenterol. 2013;48(7):815–24. [DOI] [PubMed] [Google Scholar]
- 85.Schaefer JS, Attumi T, Opekun AR, Abraham B, Hou J, Shelby H, et al. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015;16:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lewis A, Mehta S, Hanna LN, Rogalski LA, Jeffery R, Nijhuis A, et al. Low serum levels of microRNA-19 are associated with a stricturing Crohn’s disease phenotype. Inflamm Bowel Dis. 2015;21(8):1926–34. [DOI] [PubMed] [Google Scholar]
- 87.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. J Immunol. 2011;187(11):5834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zekri AR, Youssef AS, Lotfy MM, Gabr R, Ahmed OS, Nassar A, Hussein N, Omran D, Medhat E, Eid S, Hussein MM. Circulating serum miRNAs as diagnostic markers for colorectal cancer. PLoS ONE. 2016;11(5):e0154130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanaan Z, Rai SN, Eichenberger MR, Barnes C, Dworkin AM, Weller C, et al. Differential microRNA expression tracks neoplastic progression in inflammatory bowel disease-associated colorectal cancer. Hum Mutat. 2012;33(3):551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used for this research are available from the corresponding author upon request.