Abstract
Purpose
Genome-wide association studies have recently uncovered many loci associated with variation in intraocular pressure (IOP). Artificial intelligence (AI) can be used to interrogate the effect of specific genetic knockouts on the morphology of trabecular meshwork cells (TMCs) and thus, IOP regulation.
Design
Experimental study.
Subjects
Primary TMCs collected from human donors.
Methods
Sixty-two genes at 55 loci associated with IOP variation were knocked out in primary TMC lines. All cells underwent high-throughput microscopy imaging after being stained with a 5-channel fluorescent cell staining protocol. A convolutional neural network was trained to distinguish between gene knockout and normal control cell images. The area under the receiver operator curve (AUC) metric was used to quantify morphological variation in gene knockouts to identify potential pathological perturbations.
Main Outcome Measures
Degree of morphological variation as measured by deep learning algorithm accuracy of differentiation from normal controls.
Results
Cells where LTBP2 or BCAS3 had been perturbed demonstrated the greatest morphological variation from normal TMCs (AUC 0.851, standard deviation [SD] 0.030; and AUC 0.845, SD 0.020, respectively). Of 7 multigene loci, 5 had statistically significant differences in AUC (P < 0.05) between genes, allowing for pathological gene prioritization. The mitochondrial channel most frequently showed the greatest degree of morphological variation (33.9% of cell lines).
Conclusions
We demonstrate a robust method for functionally interrogating genome-wide association signals using high-throughput microscopy and AI. Genetic variations inducing marked morphological variation can be readily identified, allowing for the gene-based dissection of loci associated with complex traits.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found in the Footnotes and Disclosures at the end of this article.
Keywords: Glaucoma, Genetics, CRISPR, Transcriptomics, Morphological profiling
Primary open-angle glaucoma (POAG) is a blinding disease characterized by progressive degeneration of the optic nerve and retinal nerve fiber layer.1,2 Primary open-angle glaucoma is one of the leading causes of blindness globally.3 Whilst the precise pathophysiology of glaucoma is unknown, the most important modifiable risk factor is raised intraocular pressure (IOP).1,4 Raised IOP in POAG is primarily caused by dysfunctional aqueous humor drainage through the trabecular meshwork.1 Family heritage studies and genome-wide association studies (GWASs) have demonstrated a genetic contribution to trabecular meshwork dysfunction in POAG; however, the exact cellular and genetic processes involved remain unknown.1 Current treatments for POAG focus on reducing IOP by decreasing the production of aqueous humor or increasing outflow, with medications, or through the use of pressure-lowering surgery. However, there is currently no definitive cure for all patients with POAG.5 For novel pressure-lowering treatments to be developed, the pathophysiology of raised IOP in POAG must be understood and molecular pathways for this vision threatening disease uncovered.
Previous research has implicated a number of genes that contribute to POAG development and variation in IOP.1,6 Linkage analysis identified variants in the MYOC gene as being strongly associated with POAG.7, 8, 9 Disease-causing mutations in this gene have been shown to cause accumulation of a misfolded protein (myocilin), resulting in endoplasmic reticulum stress in trabecular meshwork cells (TMCs) and a subsequent rise in IOP.6 Genome-wide association studies have identified numerous genetic variants associated with raised IOP, many of which have also been associated with POAG.10,11 However, further investigation into these genetic variants is required to identify which individual genes may be affected by these variants and, thus, what cellular mechanisms may be involved. The ongoing development of artificial intelligence (AI) and deep learning tools such as convolutional neural networks (CNNs) provides a unique opportunity to investigate the genes of interest highlighted in GWAS and their effect on single cell morphology.
Deep learning is a rapidly advancing field of machine learning that relies on neural networks to learn abstract representations of data. A CNN is a specialized deep learning model designed to learn features of image data. In supervised learning, the original images are labeled, allowing CNNs to learn the correct representation for a given label. Given the effectiveness of CNNs at image classification,12 they have been extensively used in the analysis of cellular morphology, which is relevant in many domains of biology and medicine such as phenotype analysis,13,14 drug screening,15,16 and cell sorting.17,18
This study aimed to train a CNN to distinguish between primary TMCs that had specific genes from selected IOP-associated loci,10,11 knocked out using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas, and control TMCs transfected with nontargeting guide RNAs. The accuracy, as measured by the area under the receiver operator curve (AUC) metric, was used to quantify variation in morphological profiles between target gene knockouts and control cells. This high-throughput approach uncovered genes at IOP loci, which, when perturbed, lead to marked variation in TMC morphology.
Methods
Cell Culture and Passaging
Primary TMCs were collected from a 58-year-old donor through the Lions Eye Donation service (Human Research Ethics Committee of the Royal Victorian Eye and Ear Hospital - reference number 13-1151H). Cells were cultured in Dulbecco’s Minimal Essential Medium (Gibco, 11965118) with 10% fetal bovine serum (Gibco, 16000044) and 0.5% antibiotic-antimycotic (Gibco, 15240-062) (herein referred to as “culture medium”) at 37°C with 5% CO2. Cells were passaged by removing the culture medium and washing twice with phosphate buffered saline (Gibco, 14190144). Trypsin 0.25% diluted in phosphate buffered saline (Gibco, 25200056) was then added and the cells were incubated for 3 minutes at 37°C with 5% CO2. The trypsin was deactivated with cell culture medium and cells were then aspirated into tubes and centrifuged at 1000 rpm for 5 minutes. The supernatant was aspirated and the cell pellet was resuspended in culture medium before being plated at the desired ratio for ongoing culture. All TMCs were cultured in tissue-culture treated polystyrene plates (Corning, 3516, 3524). Prior to use, our primary TMCs were characterized as previously described.19 In brief, we verified phagocytosis and expression of CAV1 and TIMP3 by immunostaining, as well as MYOC induction by dexamethasone exposure (Fig S1). Cell lines were tested for mycoplasma on an alternate weeks using the PCR Mycoplasma Test Kit (PromoKine, PK-CA91-1096). The primary TMCs were regarded as passage zero and were seeded at passage 3 before undergoing respective cell painting immunohistochemistry or RNA-sequencing protocols at passage 4.
Cell Transfection and CRISPR Gene Knockout
A total of 67 TMC lines were generated using a library of 124 targeting single guide RNAs (sgRNAs) (2 for each target gene), together with 10 nontargeting sgRNAs as negative controls. Single guide RNAs were designed using GUIDES20 and are displayed in Table S1. Following synthesis, sgRNAs were cloned into a novel construct that had previously been developed for the pooled single-cell RNA sequence profiling of primary cells (CROPseq-Guide-pEFS-SpCas9-p2a-puro; Addgene: #99248).21 The lentivirus was then packaged by transfecting human embryonic kidney 293FT cells with pCMV delta 8.91, pMDG, and the recombinant plasmid via lipofectamine 2000. Lentivirus was chosen as the optimal viral vector due to its large size of ∼8.5 kB allowing sgRNA, Cas9, and puromycin resistance genes to be packaged into 1 viral vector.22
P1 primary TMCs were transfected with 50 μl of lentiviral plasmid to give a multiplicity of infection of approximately 3, and each CRISPR/Cas9/sgRNA/puromycin plasmid in an arrayed format. Individually cloned CRISPR/Cas9/sgRNA/puromycin plasmids were separately added to 450 μl of TMCs in culture mixed with 1:100 lentiblast (OZ Bioscience, LB01500) in 24 well plates. Each well was seeded with approximately 3.0 × 104 cells. Cell cultures were incubated for 3 days before 1 μg/ml puromycin selection occurred over 4 days. Transfected TMCs underwent standard cell passaging and were then resuspended in 100 μl to 500 μl Dulbecco’s Minimal Essential Medium depending on initial cell density. Initial cell density was qualitatively checked with brightfield microscopy before seeding. The predicted on-target editing efficiency for each sgRNA was generated for each sgRNA (Table S1). The mRNA expression of each gene knockout can be quantified from RNA sequencing data; however, while CRISPR introduces indels into the targeted sequence, the transcription of mRNA for each target gene still occurs. To investigate the efficacy of these CRISPR-constructs we compared the targeted gene transcript in each knockout line to that of the nontargeting control cells. Overall, 25 of the target cells had lower transcript counts compared with the controls at the Bonferroni corrected level (P < 0.0008; Fig S2), which is reassuring given that the transcripts would be transcribed though susceptible to nonsense mediated decay.
Cell Painting and Imaging Protocols
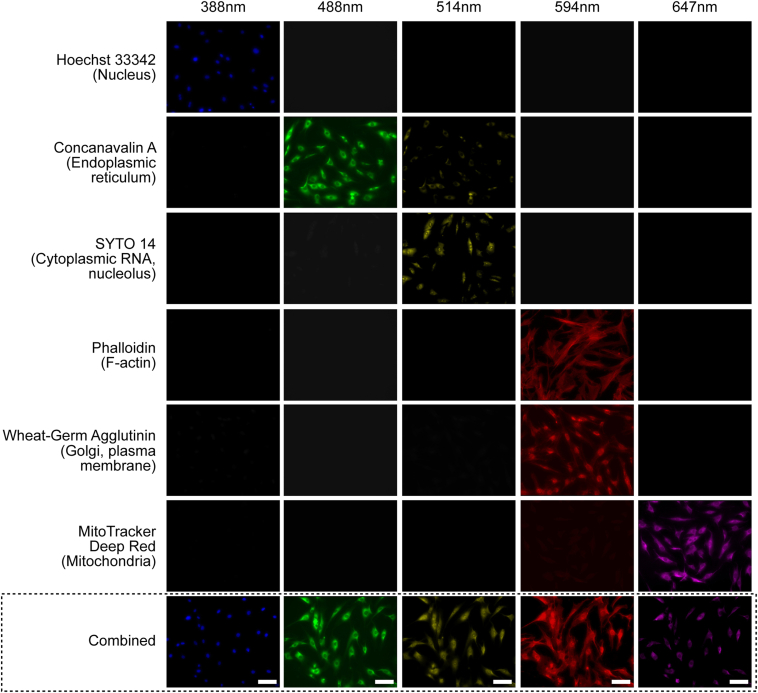
Cells were seeded at random in triplicates across 96 well plates at a density of 4.0 × 103 cells per well using a Beckman Coulter MoFlo Astrios EQ fluorescence-activated cell sorter to ensure an equal distribution of cells. The Cell Painting protocol as described by Bray et al23 was then followed. Briefly, TMCs were incubated in culture medium containing 500 nM Mitotracker (Invitrogen, M22436) and 30 μg/ml wheat germ agglutinin Alexa594 conjugate (Invitrogen, W11262) for 30 minutes at 37°C. Then TMCs were fixed with 4% paraformaldehyde at room temperature for 20 minutes and washed with 150 μl of Hanks' balanced salt solution (HBSS) (Gibco, 14025134). Next, TMCs were permeabilized with 0.1% solution of Triton X-100 (Sigma, T8787) for 20 minutes and washed with 150 μl HBSS twice. Lastly, TMCs were incubated with HBSS staining solution containing 1% bovine serum albumin (Merck, A8806), 50 μg/ml ConcanavalinA (Invitrogen, C11252), 3 μM Syto14 (Invitrogen, S7576), 5 μg/ml Hoechst (Invitrogen, H3570), and 1 unit/ml Phalloidin (Invitrogen, A12381) for 30 minutes at room temperature. Trabecular meshwork cells were washed 3 times with HBSS without final aspiration and then sealed with parafilm. All 96 well plates were kept at 4°C in the dark before imaging. Then the TMCs were imaged with high content microscopy taken at 20× magnification across 5 fluorescent channels on a Zeiss CellDiscoverer7 as outlined in Table 1. Images were autofocused using the definite focus strategy (a set focus point for each image) at 25 sites per well as shown in Figure 1.
Table 1.
Cell Painting Reagents, Fluorescent Channels, and Associated Cellular Organelles
| Cell Painting Reagent | Fluorescent Channel | Excitation Filter (nm) | Emission Filter (nm) | Organelles |
|---|---|---|---|---|
| Hoechst 33342 | DAPI | 387/11 | 417–477 | Nucleus |
| Concanavalin A/Alexa Fluor 488 conjugate | EGFP | 472/30 | 503–538 | Endoplasmic reticulum |
| SYTO 14 Green Fluorescent Nucleic Acid stain | AF514 | 531/40 | 573–613 | Cytoplasmic RNA, nucleolus |
| Phalloidin/AlexaFluor 568 WGA/AlexaFluor 555 conjugate |
AF594 | 581/609 (phalloidin) 590/617 (WGA) |
622–662 | F-actin, golgi complex, cell membrane |
| MitoTracker deep red | AF647 | 628/40 | 672–712 | Mitochondria |
WGA = wheat germ agglutinin.
The Cell Painting protocol was designed to allow a maximum number of cellular organelles to be visualized with minimal overlap of fluorescent channels.
Figure 1.
Cell Painting of trabecular meshwork cells (TMCs). Example images of TMCs stained with the Cell Painting protocol in which 6 fluorophores are imaged over 5 channels to identify 8 distinct intracellular organelles for morphological profiling. Each row shows different cells stained with the single fluorophore, or 1 group with all fluorophores combined (bottom row); columns indicate excitation wavelengths. Single channel testing shows minimal overlap across channels except for the phalloidin and wheat germ agglutinin stains which are analyzed together. This ensures that only a single stain will fluoresce when exposed to a particular wavelength of light. This figure shows whether a single stain would contaminate other emission channels and whether the signal of the light emission channel was dominated by the dyes we selected. Scale bar 50 μm.
Image Preprocessing and Quality Control
All images were separated into multiple single-cell images using the “Save Cropped Objects” function in CellProfiler (version 3.1.9, Broad Institute, Massachusetts Institute of Technology).24,25 This was undertaken to ensure that single-cell morphology was the only feature of the image, and classification was not influenced by overall cell confluency. An image quality filter was then applied using CellProfiler, which flagged any low-quality images that may contain artefacts or were inadequate for analysis, and these were subsequently removed. CellProfiler analysis data was used to calculate Spearman's rank correlation of individual cells for all cell lines. Noncorrelated cells from each line were then removed by setting a Spearman correlation cutoff value of 0.15 to reduce well-to-well and batch-to-batch variation.
CNN Architecture, Training, and Evaluation
The CNN architecture is outlined in Table S2 and accessible via GitHub. We sought to randomly select 3000 cell images from control and gene knockout groups to allocate into training (80%), validation (10%), and testing (10%) sets. A separate CNN was trained for each fluorescent channel of each gene across 5 replicates (each with a different random seed to create individual datasets). Training was conducted for 100 epochs, with the model being saved at each epoch. An Adam optimizer was used with a learning rate of 0.0001. For evaluation, the best performing model of the 100 epochs as per the loss function was selected and evaluated on the test set. Testing was performed by training a network which sought to distinguish control images from gene knockouts and the AUC metric was used to quantify CNN performance and thus, the degree of morphological variation induced by genetic variations. The highest performing models were all selected prior to reaching 100 epochs where model overfitting began to reduce model accuracy.
Results
Image Filtering and Data Split
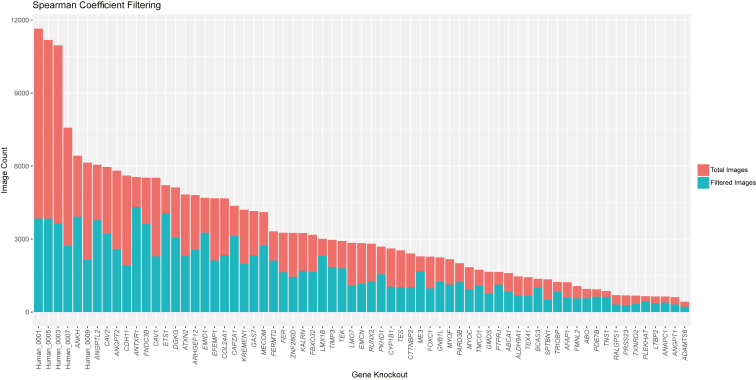
Filtering using CellProfiler and by Spearman correlation reduced the total dataset size from 225 095 images per channel to 114 830 images per channel, yielding a total of 574 150 images for analysis. The proportion of images removed via Spearman filtering varied across groups from 22.1% (ANTXR1) to 70.0% (nontargeting group 1). The 5 nontargeting control lines had the greatest proportion of images removed via Spearman filtering as shown in Figure 2. The total number of cell images after filtering ranged from 221 (ADAMTS6) to 4323 (ANTXR1). This intergroup variability was balanced during training with image rotation data augmentation (0, 90, 180, 270, with or without horizontal mirroring) to reach 3000 images per group. When 3000 were not obtained using data augmentation, the control group numbers were reduced to match and maintain a 50:50 balanced split between knockout and control images. This occurred in only 6 knockout cell lines (ADAMTS6, PRSS23, RALGPS1, ANGPT1, TXNRD2, and LTBP2) which had 1768, 2296, 2488, 2520, 2688, and 2872 images, respectively. A random selection of nontargeting control images was then selected to produce a balanced dataset of gene knockout and nontargeting control images. The same nontargeting images were chosen for each knockout comparison. The dataset was split into training (80%), validation (10%), and testing (10%) sets.
Figure 2.
Total number of images for each arrayed cell line following Spearman correlation filtering. Images were removed from the dataset if the Spearman correlation was > 0.15 in order to improve the quality of the dataset and reduce the effect of well-to-well and batch-to-batch variation. Ultimately, the percentage of cells removed ranged from 67% (control line 1) to 22% (ANTXR1).
Overall Morphological Variation Induced by Genetic Knockouts
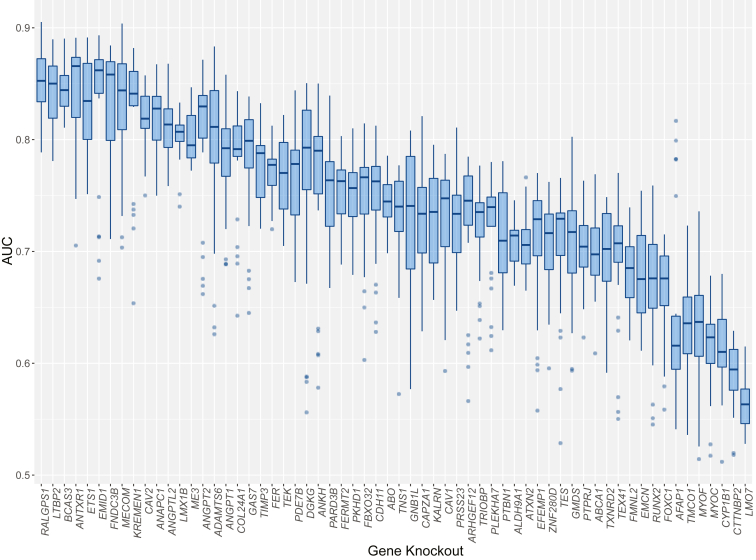
The AUC metric was used to assess the ability of the CNN to distinguish genetic knockout lines from nontargeting control lines, thereby providing a quantifiable value of morphological variation induced by gene knockouts. The mean AUC of 5 replicates across 5 channels was calculated to produce an overall AUC for each target gene. Knockout of RALGPS1 produced the most morphologically distinct TMCs (AUC 0.851, standard deviation [SD] 0.030); however, the RALGPS1 cell line was not significantly knocked down by the CRISPR system (P = 0.605) as seen in Fig S2. The second most morphologically distinct was LTBP2 (AUC 0.846, SD 0.029), followed by BCAS3 (AUC 0.845, SD 0.020). Both LTBP2 and BCAS3 had statistically significant gene knockout efficacy (P = 2.22 × 10−16 and 2.29 × 10−3). The overall AUCs ranged from 0.564 (LMO7) to the most distinguishable at 0.851 (RALGPS1) as displayed in Figure 3.
Figure 3.
Mean convolutional neural network (CNN) area under the receiver operator curve (AUC) scores for each gene knockout cell line. The mean AUC score when training a CNN to distinguish between gene knockout cell lines and nontargeting control cell lines. A higher AUC indicates a more distinct morphological variation induced by a particular gene knockout. The gene knockouts are in decreasing order of mean AUC across all organelles. The bars represent the median AUC with upper and lower quartile boxes. Outliers are displayed as single dots.
Morphological Variation Induced in Individual Organelles
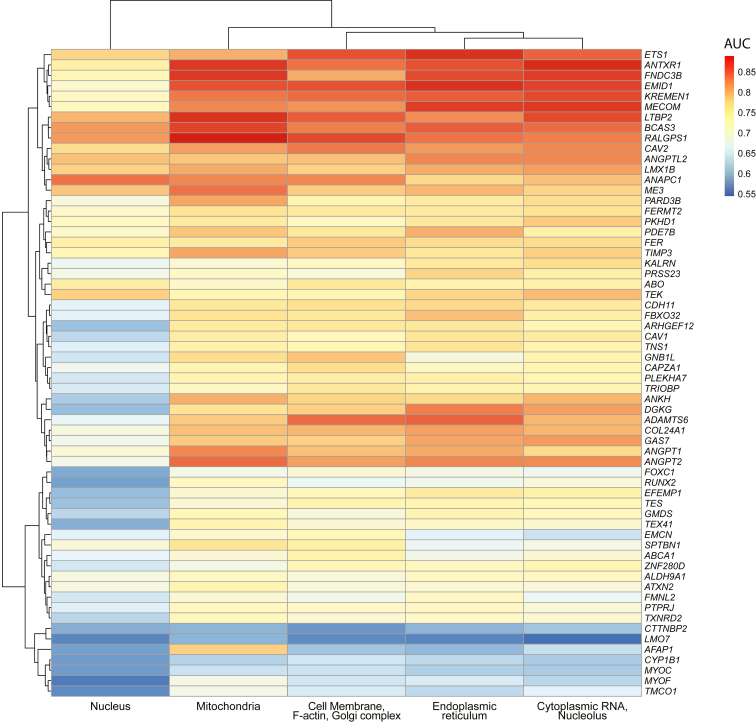
Twenty-one (33.9%) gene knockout groups had greater morphological distinction in the mitochondrial channel (mean AUC 0.760 of all cell lines, SD 0.070) compared with other organelles, illustrating that mitochondrial variation occurs in a large proportion of the gene knockouts. The relative AUC of each gene across all organelles is shown in Figure 4. Endoplasmic reticulum showed the next greatest morphological variation evident in 16 (25.8%) of the gene knockout lines (mean AUC 0.756, SD 0.079). The F-actin/cell membrane/Golgi body channel showed the highest morphological variation in 13 (20.9%) gene knockout lines (mean AUC 0.751, SD 0.073), followed by 11 (17.7%) knockout lines in the cytoplasmic RNA/nucleolus channels (mean AUC 0.753, SD 0.078). Finally, only the ANAPC1 knockout showed morphological variation most in the nucleus (mean AUC 0.677, SD 0.079).
Figure 4.
Gene knockout cell line area under the receiver operator curve (AUC) for each organelle. Heatmap of the morphological variation (AUC) across individual fluorescent channels for each gene knockout. Red shading indicates a higher degree of morphological variation as indicated by a higher AUC.
Gene Prioritization
Finally, we used the trained CNN AUC metrics to investigate TMC morphological variation for genes at multigene loci.10,11 Table 2 displays the AUC (knockout of target gene compared to nontargeting control) for 15 genes at overlapping IOP associated loci. These analyses prioritized 4 gene knockouts (ALDH9A1, CAV2, ME3, and RALGPS1) at these loci, which resulted in greater morphological variation than knockout of their neighboring gene counterparts. However, when considering gene knockdown efficacy, of these 4 targets only ALDH9A1 and CAV2 had statistically significant changes in respective gene expression (P = 3.47 ×10−2 and 2.22 ×10−16, respectively). Knockout of genes at 2 multigene loci (EMID1-KREMEN1 and GNB1L-TXNRD2) generated TMCs that were morphologically similar and thus could not be resolved. KREMEN1 had a statistically significant knockdown effect (P = 1.84 ×10−11) while the remaining 3 had no significant change in gene expression, which may explain why these multigene loci could not be resolved.
Table 2.
Comparison of Convolutional Neural Networks to Morphologically Distinguish Trabecular Meshwork Cells With Knockout of Genes at Overlapping IOP-Associated Loci10
| Top IOP GWAS SNP | Overlapping Genes (Mean AUC) | P Value |
|---|---|---|
| rs7518099 |
ALDH9A1 (AUC 0.709, SD 1.93e-02) TMCO1 (AUC 0.634, SD 4.76e-02) |
7.78 × 10−05 |
| rs11795066 |
RALGPS1 (AUC 0.851, SD 3.05e-02) ANGPTL2 (AUC 0.811, SD 2.50e-02) |
4.12 × 10−04 |
| rs6478746 |
LMX1B (AUC 0.803, SD 2.03e-02) RALGPS1 (AUC 0.851, SD 3.12e-02) |
5.5 × 10−06 |
| rs10281637 rs55892100 |
CAV1 (AUC 0.726, SD 5.53e-02) CAV2 (AUC 0.817, SD 2.71e-02) TES (AUC 0.704, SD 5.79e-02) |
4.49 × 10−01 (CAV1 vs. TES) 3.00 × 10−03 (CAV2 vs. TES) 4.00 × 10−03 (CAV1 vs. CAV2) |
| rs9608740 |
EMID1 (AUC 0.834, SD 6.50e-02) KREMEN1 (AUC 0.824, SD 5.70e-02) |
5.73 × 10−01 |
| rs8141433 |
GNB1L (AUC 0.729, SD 5.97e-02) TXNRD2 (AUC 0.695, SD 4.47e-02) |
3.75 × 10−01 |
| rs746491 |
ME3 (AUC 0.803, SD 2.45e-02) PRSS23 (AUC 0.725, SD 4.25e-02) |
3.47 × 10−04 |
AUC = area under the receiver operator curve; GWAS = genome-wide association studies; IOP = intraocular pressure; SD = standard deviation.
The mean AUC across all fluorescent channels of target knockouts vs. nontargeting control cells was compared for genes at the same locus. A higher AUC indicates a larger degree of morphological variation compared with normal control cells. This allows for prioritization of overlapping genes at given loci.
Discussion
There has been a shift in recent years towards using high-throughput profiling to undertake large-scale studies investigating the cellular basis of disease. This shift has been accelerated by advancements in computational technology and AI as a method of rapidly analyzing large, complex datasets. In this study, we utilized a CNN to perform a high-throughput morphological analysis of genetic variations associated with IOP variation in primary human TMCs. By training the CNN to distinguish gene knockout cells from healthy control cells, we could use the AUC as a metric to quantify differences in cellular morphology induced by various genetic variations. Therefore, the AUC can be used to identify which variations invoke a greater degree of morphological change and thus, which are more likely to be involved in IOP dysregulation and the pathogenesis of POAG.
This study highlights the complex genetic basis of POAG, and has clinical relevance in the development of new therapeutics to treat this vision-threatening disease. If the precise pathophysiology of POAG can be understood at the cellular level, new drug targets may be uncovered. Further, the characterization of gene-based perturbations in TMCs is an important first step in the high-throughput screening of TMC modulators. Herein, we have described an AI framework for the large-scale profiling of TMCs.
Of the genes known to cause primary congenital glaucoma or anterior segment dysgenesis, LTBP2 and TEK showed marked differentiation from normal control morphology. The LTBP2 knockout cell line was readily distinguished from normal control TMCs (AUC 0.846) with the greatest degree of difference occurring in mitochondrial morphology, indicating that LTBP2 may play a role in mitochondrial function. LTBP2 encodes for latent transforming growth factor beta binding protein 2, which is an extracellular matrix protein associated with fibrillin-1 containing microfibrils and is hypothesized to modulate extracellular matrix production.26 Variations in LTBP2 have been previously associated with primary congenital glaucoma, microspherophakia, megalocornea, and Weill-Marchesani syndrome.26, 27, 28, 29 A previous study has identified that LTBP2 knockout may contribute to the development of POAG via dysregulation of the extracellular matrix, a crucial component of the trabecular meshwork.30 Studies looking at dilated cardiomyopathy and right ventricular failure have also implicated LTBP2 function in fibrosis regulation which may indicate a role in the pathogenesis of trabecular meshwork dysfunction.31,32 Interestingly, although LTBP2 encodes for an extracellular protein, we demonstrated a distinct mitochondrial morphology in LTBP2 knockout cell lines for which we speculated that LTBP2 may lead to oxidative stress on mitochondria either directly or via changes in gene expression involved in TGFβ and bone morphogenetic protein signaling pathway that may affect mitochondrial function.
The TEK knockout cell line also showed significant differentiation (AUC 0.768) most prominent in the cytoplasmic RNA and nucleolus channel. Variations in TEK have been associated with raised IOP and congenital glaucoma primarily due to disruption of Schlemm’s canal, indicating a potential interaction with ANGPT1 in the development of glaucoma.33, 34, 35, 36 However, it must be noted that the knockdown efficacy of TEK in this study was insignificant (P = 0.858), indicating that the learned morphological variation may be due to a confounding factor learned by the deep learning system. MYOC, CYP1B1, GMDS, and FOXC1 knockouts resulted in only mild differentiation from control TMC morphology (AUCs of 0.615, 0.612, 0.704, and 0.665, respectively) despite an association with glaucoma and anterior segment dysgenesis;7,37, 38, 39, 40 however, these genes were seen to have low knockout efficiency which may explain the limited morphological change. Additionally, these gene knockouts may not invoke significant morphological variation as they are primarily involved with trabecular meshwork development rather than their homeostatic maintenance.41 Furthermore, some gene mutations associated with congenital glaucoma are gain-of-function mutations, and may not show significant change when knocked out. Another reason for not seeing change in cellular morphology is that these genes may primarily act extracellularly, such as MYOC, which has been shown to demonstrate accumulation of extracellular products in specific mutations.42
Overall, the mitochondrial channel most frequently displayed the greatest degree of differentiation (33.9% of all cell lines). This supports previous work, where TMCs in POAG have been shown to demonstrate mitochondrial dysfunction resulting in sensitivity to calcium stress.43 The endoplasmic reticulum channel also showed the most morphological variation in a large proportion of cell lines (25.8%), which is in keeping with many studies that have highlighted a link between glaucoma and endoplasmic reticulum stress.44, 45, 46
This work introduced a novel method for prioritizing genes at overlapping loci identified in GWAS using CNN analysis.10,11 The results show that ALDH9A1 and CAV2 show statistically greater differentiation from control cells than the respectively associated gene at the same locus. Studies have previously associated POAG with genetic variants at the intergenomic region of TMCO1 and ALDH9A1.47, 48, 49 The results of this study point toward ALDH9A1 being the implicated gene in POAG due to inducing a greater degree of morphological change compared with TMCO1 (P = 7.78 × 10−05). The mitochondrial channel in ALDH9A1 displayed the greatest degree of differentiation, highlighting the potential role of mitochondrial dysfunction in ALDH9A1 interruption in POAG. This is supported by the role of ALDH9A1 in carnitine synthesis, which takes place in the mitochondrial matrix.50 There have also been numerous studies illustrating an association between POAG and variations at the intergenomic region of CAV1 and CAV2.51, 52, 53, 54 This analysis prioritized CAV2 as a potential causative gene, with a higher degree of morphological change from control cells than CAV1 (P = 4.00 ×10−03). The CAV2 knockout cell line displayed the most prominent changes in the F-actin, Golgi complex, and cell membrane fluorescent channel. Supporting this, previous studies have highlighted the interaction between CAV2 and the Golgi complex.55, 56, 57 The remaining genes at overlapping loci (EMID1 vs. KREMEN1 and GNB1L vs. TXNRD2) showed no statistically significant differences in morphology as well as limited gene knockdown efficiency. They will require further investigation to prioritize which of these may be the causative gene.
A further application of AI-based analysis of single cell morphology is to predict gene expression as demonstrated in prior studies. For example, Chlis et al58 developed a machine learning model to predict gene expression of human mononuclear blood cells and mouse myeloid progenitor cells based on cellular morphology. Our study further highlights the complex interaction between cell morphology and gene expression and the opportunity that AI poses as a means of analyzing the large amounts of data produced. Further investigation into this field could uncover the genetic drivers behind pathological changes in morphology that drive disease processes and allow for identification of novel therapeutic targets.58,59
One of the main limitations of this study lies in the intrinsic difficulty in interpreting the decision-making process of CNNs. This means it can be difficult to establish if morphological features learned by the CNN are truly pathological or simply due to systematic bias. For example, if wells had lower cell density, the cells may grow to a larger size, and as such cell size may inadvertently influence the decision-making of the CNN. Certain gene knockouts may invoke cell death, which may account for lower cell numbers in particular cell lines as illustrated in Figure 2. A potential solution to this is to utilize attention-based CNN models which highlight areas of interest within the image used for decision-making.60 This may reveal which cell features are responsible for morphological variation and if features such as cell size or cell density are contributing. In addition, alternative functional systems, such as animal models, could be used to validate our findings. A further limitation of this study is that TMC cell lines were generated from a single donor which may reduce the generalizability of these results to the general population. Of course, carrying out such a study with larger donor numbers would provide a more robust dataset and increase generalizability. As well as this, it was noted that a number of cell lines had limited gene knockout efficacy as demonstrated by insignificant variations in gene expression (Fig S2). Nevertheless, this study does provide the foundations for similar larger scale studies to follow with greater donor numbers. Finally, given that we applied the Cell Painting protocol as described by Bray et al,23 which combines wheat germ agglutinin and phalloidin into the same fluorescent channel (Fig 1), it was challenging for us to distinguish actin cytoskeleton from Golgi apparatus and cell membrane features. Future studies may be able to expand on the use of Cell Painting by utilizing phalloidin alone in a separate assay to assess cytoskeletal changes that have been proposed to contribute to POAG.
In summary, this study used a powerful approach to quantify morphological change induced by genetic variations associated with POAG. RALGPS1 produced the greatest morphological variation. In addition, we could prioritize genes at overlapping loci identified to have an association with IOP. However, there are some limitations due to the difficulty in removing systematic bias from the methodology. This bias may result in the CNN learning features that are not directly associated with IOP physiology. This study highlights a new avenue for utilizing CNNs trained on single-cell morphology to further interpret the results of GWASs.
Acknowledgments
D.A.M., J.E.C., J.E.P, S.M., and A.W.H. are supported by the Australian National Health and Medical Research Council (NHMRC) Fellowships. X.H. was supported by the University of Queensland Research Training Scholarship and QIMR Berghofer Medical Research Institute PhD Top Up Scholarship. The authors thank for funding from Australian Vision Research; a NHMRC Program grant (1150144), Partnership grant (1132454) and the Clifford Craig Foundation, Australia.
Manuscript no. XOPS-D-23-00180.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures: D.A.M: Financial support – Australian National Health and Medical Research Council Fellowship, Australian National Health and Medical Research Council Partnership Grant, and Australian National Health and Medical Research Council Program Grant as payment to institution.
A.L.C.: Financial support – Clifford Craig Foundation as payment to institution.
J.E.P.: Financial support – Australian National Health and Medical Research Council Fellowship and Australian Vision Research as payment to institution.
J.E.C. and S.M.: Financial support – Australian National Health and Medical Research Council Fellowship and Australian National Health and Medical Research Council Program Grant as payment to institution.
A.H.: Financial support – Australian National Health and Medical Research Council Fellowship, Australian Vision Research, Australian National Health and Medical Research Council Program Grant, and Australian National Health and Medical Research Council Partnership Grant as payment to institution.
HUMAN SUBJECTS: No human subjects were included in this study. Primary trabecular meshwork cells were collected from donors through the Lions Eye Donation service. The Human Research Ethics Committee of the Royal Victorian Eye and Ear Hospital approved the study. All research adhered to the tenets of the Declaration of Helsinki.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Greatbatch, Lu, Powell, Craig, MacGregor, Hewitt
Data collection: Greatbatch, Lu, Hung, Wing, Liang, Zhou, Siggs, Cook, Craig
Analysis and interpretation: Greatbatch, Lu, Hung, Tran, Wing, Han, Siggs, Mackey, Liu, Cook, Powell, MacGregor, Hewitt
Obtained funding: Powell, Craig, MacGregor, Hewitt
Overall responsibility: Greatbatch, Lu, Powell, Craig, MacGregor, Hewitt
Supplementary Data
References
- 1.Weinreb R.N., Leung C.K.S., Crowston J.G., et al. Primary open-angle glaucoma. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.67. [DOI] [PubMed] [Google Scholar]
- 2.Weinreb R.N., Aung T., Medeiros F.A. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2019 Blindness and Vision Impairment Collaborators. Vision Loss Expert Group of the Global Burden of Disease Study Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the right to sight: an analysis for the Global Burden of Disease Study. Lancet Glob Health. 2021;9:e144–e160. doi: 10.1016/S2214-109X(20)30489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lusthaus J., Goldberg I. Current management of glaucoma. Med J Aust. 2019;210:180–187. doi: 10.5694/mja2.50020. [DOI] [PubMed] [Google Scholar]
- 5.Beidoe G., Mousa S.A. Current primary open-angle glaucoma treatments and future directions. Clin Ophthalmol. 2012;6:1699–1707. doi: 10.2147/OPTH.S32933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ojha P., Wiggs J.L., Pasquale L.R. The genetics of intraocular pressure. Semin Ophthalmol. 2013;28:301–305. doi: 10.3109/08820538.2013.825291. [DOI] [PubMed] [Google Scholar]
- 7.Stone E.M., Fingert J.H., Alward W.L., et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 8.Hewitt A.W., Mackey D.A., Craig J.E. Myocilin allele-specific glaucoma phenotype database. Hum Mutat. 2008;29:207–211. doi: 10.1002/humu.20634. [DOI] [PubMed] [Google Scholar]
- 9.Burdon K.P., Graham P., Hadler J., et al. Specifications of the ACMG/AMP variant curation guidelines for myocilin: recommendations from the clingen glaucoma expert panel. Hum Mutat. 2022;43:2170–2186. doi: 10.1002/humu.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacGregor S., Ong J.-S., An J., et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018;50:1067–1071. doi: 10.1038/s41588-018-0176-y. [DOI] [PubMed] [Google Scholar]
- 11.Khawaja A.P., Cooke Bailey J.N., Wareham N.J., et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat Genet. 2018;50:778–782. doi: 10.1038/s41588-018-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. Vol. 25. Curran Associates, Inc; New York: 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 13.Oei R.W., Hou G., Liu F., et al. Convolutional neural network for cell classification using microscope images of intracellular actin networks. PLoS One. 2019;14 doi: 10.1371/journal.pone.0213626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kensert A., Harrison P.J., Spjuth O. Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS Discov. 2019;24:466–475. doi: 10.1177/2472555218818756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H., Engkvist O., Wang Y., et al. The rise of deep learning in drug discovery. Drug Discov Today. 2018;23:1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 16.Gibson C.C., Zhu W., Davis C.T., et al. Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation. 2015;131:289–299. doi: 10.1161/CIRCULATIONAHA.114.010403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitta N., Sugimura T., Isozaki A., et al. Intelligent image-activated cell sorting. Cell. 2018;175:266–276.e13. doi: 10.1016/j.cell.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Ota S., Horisaki R., Kawamura Y., et al. Ghost cytometry. Science. 2018;360:1246–1251. doi: 10.1126/science.aan0096. [DOI] [PubMed] [Google Scholar]
- 19.Keller K.E., Bhattacharya S.K., Borrás T., et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp Eye Res. 2018;171:164–173. doi: 10.1016/j.exer.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meier J.A., Zhang F., Sanjana N.E. GUIDES: sgRNA design for loss-of-function screens. Nat Methods. 2017;14:831–832. doi: 10.1038/nmeth.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datlinger P., Rendeiro A.F., Schmidl C., et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Methods. 2017;14:297–301. doi: 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anon Lentiviral guide. Addgene. https://www.addgene.org/guides/lentivirus/
- 23.Bray M.-A., Singh S., Han H., et al. Cell painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc. 2016;11:1757–1774. doi: 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter A.E., Jones T.R., Lamprecht M.R., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McQuin C., Goodman A., Chernyshev V., et al. CellProfiler 3.0: next-generation image processing for biology. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haji-Seyed-Javadi R., Jelodari-Mamaghani S., Paylakhi S.H., et al. LTBP2 mutations cause Weill-Marchesani and Weill-Marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum Mutat. 2012;33:1182–1187. doi: 10.1002/humu.22105. [DOI] [PubMed] [Google Scholar]
- 27.Désir J., Sznajer Y., Depasse F., et al. LTBP2 null mutations in an autosomal recessive ocular syndrome with megalocornea, spherophakia, and secondary glaucoma. Eur J Hum Genet. 2010;18:761–767. doi: 10.1038/ejhg.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali M., McKibbin M., Booth A., et al. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84:664–671. doi: 10.1016/j.ajhg.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A., Duvvari M.R., Prabhakaran V.C., et al. A homozygous mutation in LTBP2 causes isolated microspherophakia. Hum Genet. 2010;128:365–371. doi: 10.1007/s00439-010-0858-8. [DOI] [PubMed] [Google Scholar]
- 30.Suri F., Yazdani S., Elahi E. LTBP2 knockdown and oxidative stress affect glaucoma features including TGFβ pathways, ECM genes expression and apoptosis in trabecular meshwork cells. Gene. 2018;673:70–81. doi: 10.1016/j.gene.2018.06.038. [DOI] [PubMed] [Google Scholar]
- 31.Pang X.-F., Lin X., Du J.-J., Zeng D.-Y. LTBP2 knockdown by siRNA reverses myocardial oxidative stress injury, fibrosis and remodelling during dilated cardiomyopathy. Acta Physiol. 2020;228 doi: 10.1111/apha.13377. [DOI] [PubMed] [Google Scholar]
- 32.Potus F., Hindmarch C.C.T., Dunham-Snary K.J., et al. Transcriptomic signature of right ventricular failure in experimental pulmonary arterial hypertension: deep sequencing demonstrates mitochondrial, fibrotic, inflammatory and angiogenic abnormalities. Int J Mol Sci. 2018;19:2730. doi: 10.3390/ijms19092730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J., Park D.-Y., Bae H., et al. Impaired angiopoietin/Tie2 signaling compromises Schlemm's canal integrity and induces glaucoma. J Clin Invest. 2017;127:3877–3896. doi: 10.1172/JCI94668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson B.R., Souma T., Tompson S.W., et al. Angiopoietin-1 is required for Schlemm’s canal development in mice and humans. J Clin Invest. 2017;127:4421–4436. doi: 10.1172/JCI95545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabra M., Zhang W., Rathi S., et al. Angiopoietin receptor TEK interacts with CYP1B1 in primary congenital glaucoma. Hum Genet. 2017;136:941–949. doi: 10.1007/s00439-017-1823-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souma T., Tompson S.W., Thomson B.R., et al. Angiopoietin receptor TEK mutations underlie primary congenital glaucoma with variable expressivity. J Clin Invest. 2016;126:2575–2587. doi: 10.1172/JCI85830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaur K., Reddy A.B.M., Mukhopadhyay A., et al. Myocilin gene implicated in primary congenital glaucoma. Clin Genet. 2005;67:335–340. doi: 10.1111/j.1399-0004.2005.00411.x. [DOI] [PubMed] [Google Scholar]
- 38.Pasutto F., Chavarria-Soley G., Mardin C.Y., et al. Heterozygous loss-of-function variants in CYP1B1 predispose to primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2010;51:249–254. doi: 10.1167/iovs.09-3880. [DOI] [PubMed] [Google Scholar]
- 39.Gharahkhani P., Burdon K.P., Fogarty R., et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46:1120–1125. doi: 10.1038/ng.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siggs O.M., Souzeau E., Pasutto F., et al. Prevalence of FOXC1 variants in individuals with a suspected diagnosis of primary congenital glaucoma. JAMA Ophthalmol. 2019;137:348–355. doi: 10.1001/jamaophthalmol.2018.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauthier A.C., Wiggs J.L. Childhood glaucoma genes and phenotypes: focus on FOXC1 mutations causing anterior segment dysgenesis and hearing loss. Exp Eye Res. 2020;190 doi: 10.1016/j.exer.2019.107893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueda J., Wentz-Hunter K., Yue B.Y.J.T. Distribution of myocilin and extracellular matrix components in the juxtacanalicular tissue of human eyes. Invest Ophthalmol Vis Sci. 2002;43:1068–1076. [PubMed] [Google Scholar]
- 43.He Y., Ge J., Tombran-Tink J. Mitochondrial defects and dysfunction in calcium regulation in glaucomatous trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2008;49:4912–4922. doi: 10.1167/iovs.08-2192. [DOI] [PubMed] [Google Scholar]
- 44.Peters J.C., Bhattacharya S., Clark A.F., Zode G.S. Increased endoplasmic reticulum stress in human glaucomatous trabecular meshwork cells and tissues. Invest Ophthalmol Vis Sci. 2015;56:3860–3868. doi: 10.1167/iovs.14-16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anholt R.R.H., Carbone M.A. A molecular mechanism for glaucoma: endoplasmic reticulum stress and the unfolded protein response. Trends Mol Med. 2013;19:586–593. doi: 10.1016/j.molmed.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z., Ge Y., Pease M., et al. Role of endoplasmic reticulum stress in retinal ganglion cell death in glaucoma and optic nerve injury. Invest Ophthalmol Vis Sci. 2010;51:5812. [Google Scholar]
- 47.Verkuil L., Danford I., Pistilli M., et al. SNP located in an AluJb repeat downstream of TMCO1, rs4657473, is protective for POAG in African Americans. Br J Ophthalmol. 2019;103:1530–1536. doi: 10.1136/bjophthalmol-2018-313086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burdon K.P., Macgregor S., Hewitt A.W., et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43:574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 49.Hysi P.G., Cheng C.-Y., Springelkamp H., et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46:1126–1130. doi: 10.1038/ng.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Končitíková R., Vigouroux A., Kopečná M., et al. Kinetic and structural analysis of human ALDH9A1. Biosci Rep. 2019;39 doi: 10.1042/BSR20190558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiggs J.L., Kang J.H., Yaspan B.L., et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet. 2011;20:4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen F., Klein A.P., Klein B.E.K., et al. Exome array analysis identifies CAV1/CAV2 as a susceptibility locus for intraocular pressure. Invest Ophthalmol Vis Sci. 2014;56:544–551. doi: 10.1167/iovs.14-15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L.J., Tam P.O.S., Pang C.P. Revisit the association of CAV1/CAV2 with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2013;54:6236. [Google Scholar]
- 54.Loomis S.J., Kang J.H., Weinreb R.N., et al. Association of CAV1/CAV2 genomic variants with primary open-angle glaucoma overall and by gender and pattern of visual field loss. Ophthalmology. 2014;121:508–516. doi: 10.1016/j.ophtha.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mora R., Bonilha V.L., Marmorstein A., et al. Caveolin-2 localizes to the golgi complex but redistributes to plasma membrane, caveolae, and rafts when co-expressed with caveolin-1. J Biol Chem. 1999;274:25708–25717. doi: 10.1074/jbc.274.36.25708. [DOI] [PubMed] [Google Scholar]
- 56.Aga M., Bradley J.M., Wanchu R., et al. Differential effects of caveolin-1 and -2 knockdown on aqueous outflow and altered extracellular matrix turnover in caveolin-silenced trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2014;55:5497–5509. doi: 10.1167/iovs.14-14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parolini I., Sargiacomo M., Galbiati F., et al. Expression of caveolin-1 is required for the transport of caveolin-2 to the plasma membrane. Retention of caveolin-2 at the level of the Golgi complex. J Biol Chem. 1999;274:25718–25725. doi: 10.1074/jbc.274.36.25718. [DOI] [PubMed] [Google Scholar]
- 58.Chlis N.-K., Rausch L., Brocker T., et al. Predicting single-cell gene expression profiles of imaging flow cytometry data with machine learning. Nucleic Acids Res. 2020;48:11335–11346. doi: 10.1093/nar/gkaa926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cutiongco M.F.A., Jensen B.S., Reynolds P.M., Gadegaard N. Predicting gene expression using morphological cell responses to nanotopography. Nat Commun. 2020;11:1384. doi: 10.1038/s41467-020-15114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S., Ren S., Wang G., et al. Interpretable CNN-multilevel attention transformer for rapid recognition of pneumonia from chest X-ray images. IEEE J Biomed Health Inform. 2024;28:753–764. doi: 10.1109/JBHI.2023.3247949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.