Abstract
Objective
Genomics is rapidly changing treatment paradigms for cancers, obligating oncologists to have good genomics knowledge. Through this survey, we aimed to assess the current understanding of cancer genomics among UK oncologists.
Methods
We conducted a web-based nation-wide self-assessment survey of the cancer genomics knowledge of UK clinical and medical oncology trainees and consultants.
Results
In total, 150 oncologists (81 consultants and 69 trainees) responded, representing 10% of UK oncologists.
Formal training in genomics had not been received by 38.7% of oncologists and 92.7% identified a need for additional genomics training.
In total, 71.3% self-reported to have good knowledge of defining somatic and germline mutations, falling to 35.3% for understanding principles of gene expression and regulation. Knowledge of cancer-predisposing syndromes was highest for Lynch syndrome (40.7% good knowledge) and lowest for multiple endocrine neoplasia (14.0% good knowledge).
Overall, 49.0% of respondents had consented patients for germline testing, but 80.7% reported a lack of training in genetic counselling.
Conclusion
Large knowledge gaps have been identified through this survey, highlighting the need for incorporation of improved formal training in cancer genomics for consultants and trainees, with an aim to equip oncologists for advances in clinical practice and to take up genetic mainstreaming confidently.
KEYWORDS: germline genetics, somatic genetics, oncology, genomics, mainstreaming
Introduction
The completion of the Human Genome Project in 2003 marked a milestone in our understanding of human genetics. However, it was the technology of genome sequencing that developed alongside it that provided a detailed understanding of both human and human cancer genetics.1 The identification of key somatic driver mutations in some cancers has enabled the development of therapeutic agents that specifically target the aberrant protein product, increasing effectiveness and reducing toxicity. Simultaneously, the rapid fall in DNA-sequencing costs secondary to the development of next generation sequencing (NGS) techniques has made routine testing of tumour samples for specific mutations a viable option.2
The same technological advances have also driven a dramatic expansion in testing for inherited variations in cancer susceptibility genes (CSGs).3, 4 With NGS, testing for germline variants of CSGs could be included as part of the diagnostic workup for selected patients with breast, ovarian or colorectal cancers, and when germline CSG mutations are implicated by analysis of tumour material.5, 6
It is estimated that 3% of cancers arise on a background of germline mutations in CSGs.7 Until recently, knowledge of a CSG mutation mainly benefited healthy relatives of patients with cancer, through genetic screening and appropriate clinical interventions for relatives carrying the CSG mutation. The advent of therapies targeted to germline CSG mutations, and recognition that some non-targeted systemic therapies have differing effects in CSG-mutation carriers, means that there is now a potential benefit from determining CSG mutation status at cancer diagnosis. Examples include immunotherapy in Lynch syndrome and carboplatin or PARP inhibitors in patients with BRCA1/2-mutated breast cancer.8, 9, 10, 11, 12, 13, 14, 15
An ‘oncogenetic’ model of CSG testing, whereby testing of patients with cancer can be performed via the cancer team, with support as required from clinical genetics, has now been established at a few cancer centres. This is frequently called ‘mainstreaming’ and examples include the Birmingham Women's and Children's Hospital ‘Generate’ training16 and the Royal Marsden Hospital (RMH) ‘Mainstreaming Cancer Genetics programme’.17, 18
There are 62 UK cancer centres and 23 regional genetics centres,19 highlighting a potential disparity in interaction between oncology and genetics between sites with and without on-site clinical genetic services.20 Genomic testing is delivered by the National Genomic Medicine Service in England through seven Genomic Laboratory Hubs (GLHs), which provide regional testing services. These are situated with a lead provider for the seven regions aligned to a regional genetics service.19 In the devolved nations, these are delivered through one or two genomic laboratories attached to regional genetics centres in the main cities (ie Belfast, Glasgow, Edinburgh and Cardiff). Turnaround time is dependent on the type of test and clinical urgency. It can range from 2–4 weeks, for urgent cancer gene testing, to 6–12 months, for large cancer gene panels or whole-genome sequencing in the routine setting.
It is increasingly acknowledged that clinical genetics departments are not able to meet the current and future demand for germline genetic testing. The integration of genetics into ‘mainstream’ specialties is broadly agreed as the optimum way for healthcare services to evolve21 and improve services with a limited workforce, but requires the clinician to be sufficiently versed and confident in genomics. The rapidly increasing complexity of genomic data highlights the obligation of healthcare professionals to ensure that patients understand the issues and options around genetic testing.22
In 2015, the Association of Cancer Physicians (ACP) established an oncogenetic training working party (OTWP) to standardise and enhance the training of medical oncologists in oncogenetics, resulting in the inclusion of a new genomics section in the 2017 medical oncology training curriculum. However, the current level of genetics knowledge within the oncology community is unknown.
A 2019 survey of gastroenterology trainees regarding mainstreaming highlighted that most trainees felt ill-equipped to practice genomic and personalised medicine as consultants.23 In addition, a previous survey of breast cancer specialists described a lack of knowledge in interpreting and communicating variants of uncertain significance (VUS).24 These issues might also be present in oncology, but no comprehensive survey is available.
Therefore, a training needs assessment was designed and disseminated in collaboration with the Cancer Research UK (CRUK)-funded CanGene-Canvar (CGCV) programme to investigate the current level of knowledge and experience of cancer genetics within the oncology workforce. This information will help improve our understanding of the need for further training of oncology clinicians, alongside informing development of educational resources, to help underpin future expansions in genetic testing and mainstreaming access.
Methods
Survey design and dissemination
We conducted a web-based nation-wide survey using the SurveyMonkey® platform25 between September and December 2021, targeting all UK oncology specialty trainees and consultants. An overview of the survey questionnaire is shown in the supplementary material online (supplementary material S1). The survey comprised 36 questions, including yes/no questions (n=10), multiple choice questions (n=3), visual analogue rating scales (Likert scales) (n=19) and free-text entries. The rating scales were later grouped into poor (0–3), some (4–7) and good knowledge (8–10) categories. Each survey also collected basic demographic data and details regarding current speciality, position and level of education. A separate questionnaire was developed for consultants and trainees. All questions regarding the genetics survey were the same for the consultant/trainee questionnaires apart from question 29, which asked for an opinion on trainees and consultants, respectively. Questions were split into five major topics: previous training received in genomics; the basics of genomics; high-risk cancer predisposition syndromes; knowledge of the local clinical genetics service; interpretation and communication of test results; and requirements for training. The questionnaire was reviewed and piloted in three cancer genetics centres (University Hospital Southampton, RMH and St George's London) and iteratively modified based on feedback to ensure completeness and ease of use.
Invitations to the survey were sent via email and social media to all clinical and medical oncology trainees and consultants via the contact lists for medical and clinical oncologists from the Association of Cancer Physicians (ACP) and Royal College of Radiologists (RCR), respectively. This included trainees in medical and clinical oncology with a national training number, including those currently out of programme, and those in the locum appointment to service, academic clinical fellow and academic clinical lecturer positions. Consultant posts included academic and NHS posts and associate specialists. Assuming that 75% of oncologists are members of these bodies, a total of ∼1,100 oncologists were contacted.20, 26
Continuous variables were analysed using the Mann–Whitney (MW) test for comparison of two variables, and Kruskal–Wallis (KW) test for three variables and more; all p-values are two-sided. p≤0.05 was counted as significant for the KW test. The Bonferroni correction method was used to reduce the type 1 error rate resulting from multiple MW comparisons. Using the standard level of significance for a single MW test of p≤0.05, a corrected MW p-value of ≤0.01 was required for significance (five comparisons).
UK Research Ethics Committee and NHS Research Authority regulatory permission were not required because no patients were recruited and no personal identification information was stored.
Patient and public involvement
Patients and the public were not involved in our work.
Results
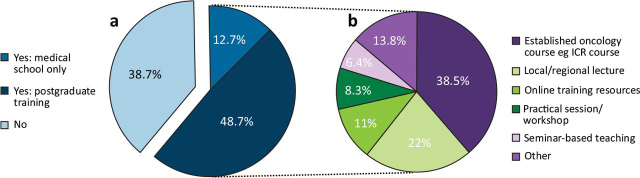
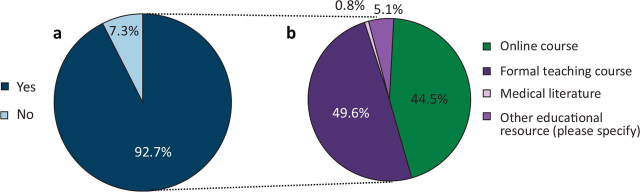
We received 150 responses, representing ∼10% of oncologists across the UK.20, 26 This comprised 81 consultant responses, including 12 with an academic appointment, and 69 trainee responses, including 10 with an academic position. Of the respondents, 68.7% had been involved in a clinical trial incorporating genomic stratification and 61.3% had received some form of training in genetics (63.8% of trainees and 59.3% of consultants), with 12.7% having received training in medical school and 48.7% during postgraduate training. The format of genetics training was variable and included: an established course (38.5%); lectures (22.0%); online training resources (11.0%); workshop (8.3%); seminars (6.4%); and other forms (13.8%) (Fig 1).
Fig 1.
Previous training received in cancer genomics and the different formats of teaching. (a) Has specific training in cancer genomics been completed? (b) Format of teaching. ICR = Institute for Cancer Research.
Basics of genomics
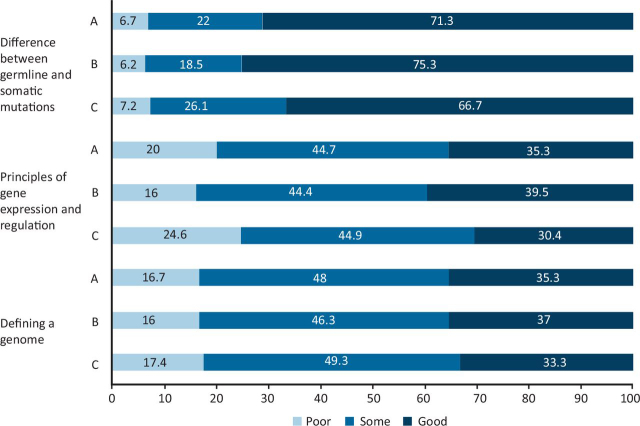
Approximately three-quarters (71.3%) of the respondents reported good knowledge of defining the difference between germline and somatic mutations. Just over one-third of respondents (35.3%) were confident in defining a genome and 35.3% had good knowledge of the principles of gene expression and regulation (Fig 2).
Fig 2.
Respondents' perception of basics of genomics. Total respondents: 150. A = all; B = consultants; C = trainees.
High-risk cancer predisposition syndromes
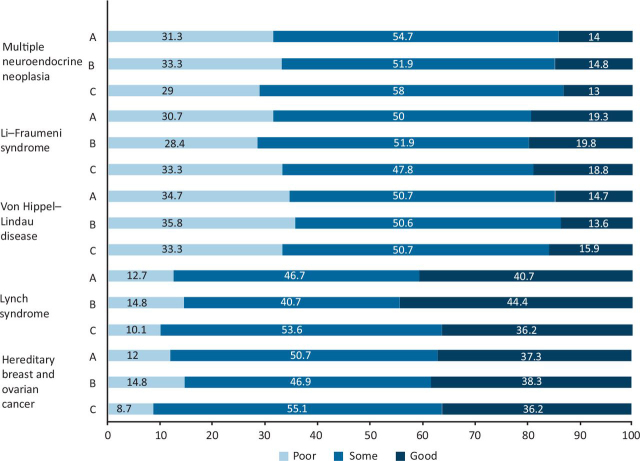
Overall, there was a significant difference in respondents' knowledge of the different patterns of malignancies and underlying gene mutations associated with cancer predisposition syndromes (KW p<0.00001). Respondents had the best knowledge of Lynch syndrome (40.7% reporting good knowledge), followed by hereditary breast and ovarian cancer syndrome (37.3% reporting good knowledge). Few respondents had good knowledge of the rarer syndromes: Von Hippel–Lindau (14.7%); Li–Fraumeni (19.3%); and multiple endocrine neoplasia (MEN) syndromes (14.0%) (Fig 3).
Fig 3.
Describing the pattern of malignancies and underlying gene mutations associated with various high-risk cancer predisposition syndromes. A = all; B = consultants; C = trainees.
Referring a patient to clinical genetics service
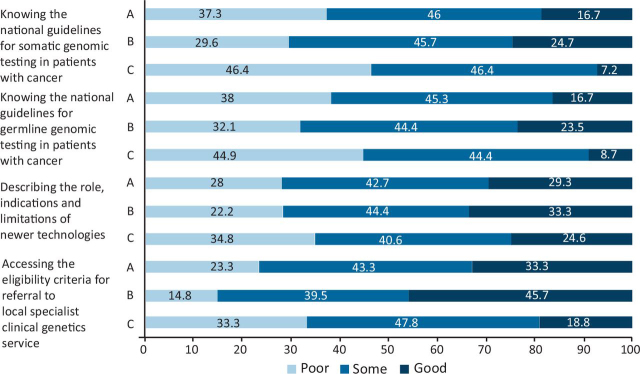
In total, 29.3% of respondents had good knowledge of describing the role, indications and limitations of newer genetic testing technologies. Of respondents, 16.7% reported good knowledge of current national guidelines for somatic testing of patients with cancer, and similarly 16.7% for the national guidelines for germline testing. One-third (33.3%) of respondents had good knowledge of how to access eligibility criteria for a formal referral of a patient to their local specialist clinical genetics service. with better knowledge for consultants versus trainees (45.7% consultant versus 18.8% trainee good knowledge; MW<0.00001) (Fig 4).
Fig 4.
Respondents' knowledge of local clinical genetics services. A = all; B = consultants; C = trainees.
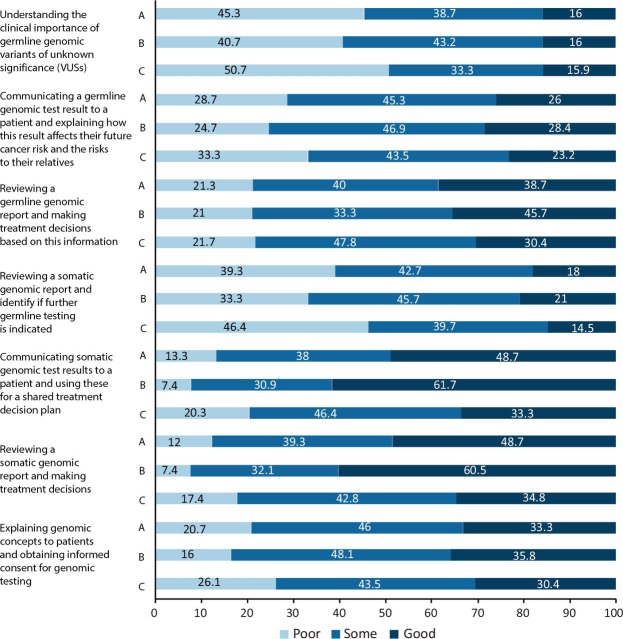
Interpreting and communicating test results
Only 16.0% of respondents had good knowledge of the clinical importance of germline VUS. Respondents had better knowledge of the communication of somatic versus germline results (overall 48.7 versus 26.0% good knowledge, respectively; MW p<0.00001), but had more similar knowledge of making treatment decisions acting on somatic versus germline reports (overall 48.7% versus 38.7% good knowledge, respectively; MW p=0.023). There was a significant difference in knowledge between consultants and trainees for ‘communication of somatic genomic test results and using them for a shared treatment plan’ (61.7% consultant versus 33.3% trainee good knowledge; MW p=0.0004) and ‘using somatic genomic reports for making treatment decision’ (60.5% consultant versus 34.8% trainee good knowledge; MW p=0.0002). Few respondents had good knowledge of how to identify patients in potential need of germline testing based on a somatic report (18.0%). One-third (33.3%) of respondents had good knowledge of how to explain genomic concepts to patients and to obtain informed consent for genomic testing (Fig 5).
Fig 5.
Respondents' knowledge of interpreting and communicating results. A = all; B = consultants; C = trainees.
Need for specific training in cancer genomics
Almost half of the respondents (49.0%) reported to have consented at least one patient with cancer for germline testing for mutations in a cancer susceptibility gene, but 80.7% had not received training in counselling patients. Concerns regarding the adoption of genomic science and whole-genome sequencing into routine clinical practice were expressed by 57.3% of respondents. In addition, 92.7% stated a need for further specific training in cancer genomics, with most respondents opting for formal teaching courses (45.3%) followed by online courses (40.7%) (Fig 6).
Fig 6.
Reported need for specific training in cancer genomics. (a) Do oncologists feel the need for further training? (b) Preferred format for delivery of training.
Discussion
This is the largest survey of oncologists to date that has assessed the extent of knowledge and training in cancer genomics received by UK oncologists. Although it is reassuring that 61.3% of the respondents had received training in some form, the lack of training in 38.7% of respondents is concerning. There was marked variability in the genomics training experience received by oncologists in the UK, with 38.5% having undertaken established courses and the rest lectures, online training resources, seminars and workshops. Given that this survey was undertaken after the start of the coronavirus 2019 (COVID-19) pandemic, this would have affected opportunities for face-to-face teaching and might have contributed to the diversity of genomics training experience.
This survey identifies some significant knowledge gaps in the understanding of genomic principles among oncologists in the UK. Although 71.3% of respondents felt confident to define the difference between somatic and germline mutations, it is concerning that 28.7% of respondents were not confident, because this is core knowledge for oncology doctors, who increasingly use both somatic and germline test results to select treatments. In addition, only 35.3% had good knowledge of defining a genome or the principles of gene expression and regulation, indicating the need for additional genetics education.
Significant variance was also identified in the respondents' knowledge of the cancer patterns in high-risk cancer predisposing genetic syndromes, ranging from 40.7% of respondents acknowledging good knowledge of the relatively common Lynch syndrome, to 14% for rarer syndromes, such as MEN (KW p<0.00001). Given that oncologists frequently specialise in one or two cancer types, this variance in knowledge of CSGs might occur because of consultant oncologists being most familiar with the CSGs related to their cancer types, although some, such as Li-Fraumeni, can cause a range of primary cancers.
There is also a clear need for better liaison between the oncology and genetics departments, evidenced by lack of awareness of how to access the eligibility criteria for a formal referral to a local clinical genetics service (33.3% good knowledge, although better for consultants versus trainees) and poor knowledge of both the national guidelines for somatic (16.7% good knowledge) and germline (16.7% good knowledge) testing for cancers.
Although 49.0% of respondents have consented patients for germline testing for mutations in a cancer susceptibility gene, only 33.3.% reported good knowledge of how to take informed consent for genetic testing and 80.7% acknowledged lack of training in counselling patients. In addition, only 38.7% had good knowledge of interpretation of these results (although consultant knowledge was better than of trainees), and only 16.0% had good knowledge of how to interpret a VUS. This demonstrates that many oncologists feel that they have received insufficient training to undertake or interpret genetic testing and are ill-equipped to adapt to the recent advances in cancer management, which are increasingly targeting CSG mutations.
Given that genomic testing is being increasingly included into the pathway of cancer diagnosis to ultimately guide treatment decisions for a whole array of cancers, it is acknowledged that clinical genetics departments will not be able to meet the increasing demand of testing and oncologists would be required to step up in the form of mainstreaming of genomics. The lack of knowledge highlighted in this study indicates that a significant amount of work is required to bring oncologist knowledge of genetics up to the required level.
It is not necessary for all oncologists to have expert knowledge of all areas of cancer genomics. The three-tiered approach to oncogenetics training recommended by the ACP OTWP27 envisages three levels of expertise:
-
•
For all medical oncologists: a comprehensive understanding of basic genomics, including limitations of current technology, key differences between somatic and germline mutations, and principles of stratified cancer medicine.
-
•
Practical experience in cancer genetics clinics for those who are interested, with the aim of providing medical oncologists who will be able to offer enhanced germline genetic advice in a multidisciplinary team (MDT) meeting setting.
-
•
Advanced experience in oncogenetics provided by formal post-completion of speciality training cancer genetics fellowships at a small number of tertiary centres for oncologists who have a specialist interest in oncogenetics. It is envisaged that, in the future, there should be at least one medical oncology consultant at each tertiary centre with this level of experience and that these oncologists would have regular participation in molecular tumour boards and genetics MDTs.
However, this survey identifies that many oncologists lack the basic levels of genetics knowledge. Without this knowledge, it is not possible for these doctors to provide mainstreaming or to safely interpret somatic or germline genetic information, indicating an urgent need to improve genetics education to ensure the optimal management of patients.
This study is the largest study on genomics knowledge within oncology to date, but represents a response of ∼10% of all oncologists,20, 26 which is a small proportion of all oncologists in the UK. We have no significant reason to believe that this is a non-representative sample, given the similar responses from trainees and consultants. However, a potential weakness of this study is that oncologists with some interest in genetics and genomics might be more, or indeed less, likely to respond to the survey and, therefore, this study might over or underestimate the current level of knowledge of oncologists to some degree.
Education and training opportunities in cancer genomics
Several existing free-at-point-of-access national educational initiatives exist to support oncologists wanting to upskill in cancer genomics.28, 29, 30, 31 The Genomics Education Programme (GEP) within Health Education England (HEE) have a suite of multi-media online materials on the basics of genomics and, more specifically, on cancer genomics, which are freely accessible to all.28 Educational materials range from short 2–10-min articles to more in-depth online modules. The GeNotes platform29 developed by GEP has launched oncology-specific genomics training. This resource is freely available online and designed to meet the needs of busy clinicians both at point of need with information to enable clinicians to undertake genetic testing in clinic and prospectively with case studies and more in-depth educational materials.
The educational arm of the CRUK CGCV research programme has developed high-level Massive Open Online Courses (MOOCs) in variant interpretation for cancer susceptibility genes30 and will be embedding oncology-specific guidelines for cancer variant interpretation within the GeNotes platform. Established synchronous virtual courses in cancer genomics are also available within the NHS.31 In addition, the CGCV Cancer Variant Interpretation Group (CanVIG-UK) have developed cancer susceptibility gene templates, which have been adopted across Genomic Laboratory Hubs in the UK, as well as gene-specific variant interpretation guidelines adapting the American College of Medical Genetics and Genomics framework.32, 33 The increase in somatic testing and requirement to establish Genomic Tumour Advisory Boards (GTABs) for interpretation and patient management, will also provide more workplace-based training opportunities.
Given that genetics is a rapidly changing area of medicine, it is important that clinicians engaged in genetic testing keep their knowledge up to date. Therefore, appropriate training courses should be repeated regularly to ensure maintenance of an appropriate knowledge base, further increasing the need for up-to-date freely accessible online courses.
Conclusion
Through this survey the oncology fraternity have reflected, identified gaps in their knowledge and highlighted their concerns regarding the adoption of genomics and whole genome sequencing into routine clinical practice. Although mainstreaming into clinical practice has been proposed to be the best way to optimise healthcare services to keep up with the increasing demand for germline genetic testing, many oncologists have insufficient knowledge to prepare them for this new role and most (92.7%) have highlighted the need for additional training in cancer genomics. We have highlighted several online training opportunities in cancer genetics for oncologists, but to fully address this educational need, oncology professional and educational bodies will need to work closely with clinical genetics to ensure oncologists at all levels are provided with appropriate training opportunities, thereby improving the service offered to patients.
Summary
What is known?
Oncologists require good genomics knowledge to select appropriate treatments. This knowledge also allows mainstream testing of appropriate patients for CSGs.
What is the question?
What is the current genomics knowledge of UK oncologists? This project carried out a survey of UK oncologists to define the current degree of genomics knowledge of UK oncologists.
What was found?
This survey highlights significant gaps in knowledge of both germline and somatic genetics; 92.7% of respondents identified a need for additional cancer genomics training.
Implication for practice
Professional and educational bodies will need to improve genetics training to enable oncologists to manage patients appropriately.
Supplementary material
Additional supplementary material may be found in the online version of this article at www.rcpjournals.org/clinmedicine.
S1. An overview of the survey questionnaire.
References
- 1.Schloss JA, Gibbs RA, Makhijani VB, Marziali A. Cultivating DNA Sequencing Technology After the Human Genome Project. Ann Rev Genomics Hum Genet. 2020;21:117–138. doi: 10.1146/annurev-genom-111919-082433. [DOI] [PubMed] [Google Scholar]
- 2.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 3.Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics. 2016;107:1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahassi el M, Stambrook PJ. Next-generation sequencing technologies: breaking the sound barrier of human genetics. Mutagenesis. 2014;29:303–310. doi: 10.1093/mutage/geu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng DT, Prasad M, Chekaluk Y, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10:33. doi: 10.1186/s12920-017-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt KT, Chau CH, Price DK, Figg WD. Precision oncology medicine: the clinical relevance of patient-specific biomarkers used to optimize cancer treatment. J Clin Pharmacol. 2016;56:1484–1499. doi: 10.1002/jcph.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans JP, Green RC. Direct to consumer genetic testing: Avoiding a culture war. Genet Med. 2009;11:568–569. doi: 10.1097/GIM.0b013e3181afbaed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 10.Turner NC, Tutt AN. Platinum chemotherapy for BRCA1-related breast cancer: do we need more evidence? Br Cancer Res. 2012;14:115. doi: 10.1186/bcr3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT, Kim TW, Van Cutsem E, et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J Clin Oncol. 2020;38:11–19. doi: 10.1200/JCO.19.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz LA, Jr, Shiu KK, Kim TW, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high or mismatch repair-deficient metastatic colorectal cancer (KEYNOTE-177): final analysis of a randomised, open-label, phase 3 study. Lancet Oncol. 2022;23:659–670. doi: 10.1016/S1470-2045(22)00197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tutt ANJ, Garber JE, Kaufman B, et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-Mutated Breast Cancer. N Engl J Med. 2021;384:2394–2405. doi: 10.1056/NEJMoa2105215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 16.16 Birmingham Women's and Children's NHS Foundation Trust. Information for professionals: Genetics. https://bwc.nhs.uk/information-for-professionals-genetics/.
- 17.Rahman N. Mainstreaming genetic testing of cancer predisposition genes. Clin Med. 2014;14:436–439. doi: 10.7861/clinmedicine.14-4-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp Z, Turnbull A, Yost S, et al. Evaluation of cancer-based criteria for use in mainstream BRCA1 and BRCA2 genetic testing in patients with breast cancer. JAMA Netw Open. 2019;2:e194428. doi: 10.1001/jamanetworkopen.2019.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.19 Genetic Alliance,. NHS Genetic Services in the UK 2020. https://geneticalliance.org.uk/information/service-and-testing/nhs-genetic-services-in-the-uk-2/.
- 20.Royal College of Radiologists . RCR; 2018. Clinical Oncology - UK workforce census report 2018.www.rcr.ac.uk/system/files/publication/field_publication_files/bfco192-co-workforce-census-2018.pdf [Google Scholar]
- 21.Independent Cancer Taskforce . NHS England; 2016. Achieving world-class cancer outcomes: A strategy for England 2015-2020.www.england.nhs.uk/publication/achieving-world-class-cancer-outcomes-a-strategy-for-england-2015-2020 . [Google Scholar]
- 22.Stacey D, Légaré F, Lewis K, et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst Rev. 2017;4:CD001431. doi: 10.1002/14651858.CD001431.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al Bakir I, Sebepos-Rogers GM, Burton H, Monahan KJ. Mainstreaming of genomic medicine in gastroenterology, present and future: a nationwide survey of UK gastroenterology trainees. BMJ Open. 2019;9:e030505. doi: 10.1136/bmjopen-2019-030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eccles BK, Copson E, Maishman T, Abraham JE, Eccles DM. Understanding of BRCA VUS genetic results by breast cancer specialists. BMC Cancer. 2015;15:936. doi: 10.1186/s12885-015-1934-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.25 SurveyMonkey: Momentive Inc.; 2022.
- 26.Association of Cancer Physicians . ACP; 2019. ACP medical oncology training survey report 2019.www.theacp.org.uk/members/trainees . [Google Scholar]
- 27.Baird R, Banks I, Cameron D, et al. An Association of Cancer Physicians' strategy for improving services and outcomes for cancer patients. Ecancermedicalscience. 2016;10:608. doi: 10.3332/ecancer.2016.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.28 Health Education England,. Genomics Education Programme. www.genomicseducation.hee.nhs.uk/.
- 29.29 Health Education England,. GeNotes: genomic notes for clinicians. www.genomicseducation.hee.nhs.uk/about-us/genotes-genomic-notes-for-clinicians/.
- 30.30 St George's University of London,. Interpreting genomic variation: inherited cancer susceptibility. www.futurelearn.com/courses/interpreting-genomic-variation-inherited-cancer-susceptibility.
- 31.31 Guy's and St Thomas' NHS Foundation Trust,. Virtual Cancer Genetics Course 2022. www.guysandstthomasevents.co.uk/courses/virtual-cancer-genetics-course-2022/.
- 32.Ellard S, Baple EL, Callaway A, et al. ACGS; 2020. UK Practice Guidelines for variant classification. [Google Scholar]
- 33.CanVIG-UK CanVIG-UK Resource Templates. 2022 [Google Scholar]