Abstract
Simple Summary
Cancer is a heterogeneous disease characterized by uncontrollable abnormal cell growth. KRAS plays a crucial role in cell proliferation and differentiation. KRAS mutations are frequently found in many types of cancer, including non-small-cell lung cancer. However, targeting this oncogene has been a real challenge for many years. Recent advances in KRAS targeting have changed the current therapeutic landscape. In this review, we summarize the therapeutic strategies for KRAS-mutated NSCLC with encouraging results and restrictions. Moreover, we present possible combination therapies aiming to achieve individualized treatment.
Abstract
Kirsten rat sarcoma virus (KRAS) is the most frequently found oncogene in human cancers, including non-small-cell lung cancer (NSCLC). For many years, KRAS was considered “undruggable” due to its structure and difficult targeting. However, the discovery of the switch II region in the KRAS-G12C-mutated protein has changed the therapeutic landscape with the design and development of novel direct KRAS-G12C inhibitors. Sotorasib and adagrasib are FDA-approved targeted agents for pre-treated patients with KRAS-G12C-mutated NSCLC. Despite promising results, the efficacy of these novel inhibitors is limited by mechanisms of resistance. Ongoing studies are evaluating combination strategies for overcoming resistance. In this review, we summarize the biology of the KRAS protein and the characteristics of KRAS mutations. We then present current and emerging therapeutic approaches for targeting KRAS mutation subtypes intending to provide individualized treatment for lung cancer harboring this challenging driver mutation.
Keywords: KRAS, NSCLC, G12C mutation, undruggable, sotorasib, adagrasib, mechanisms of resistance, emerging therapies
1. Introduction
For several decades, lung cancer has been the deadliest form of cancer worldwide. The majority of lung cancer cases (approximately 85%) belong to the non-small-cell lung cancer type, especially the adenocarcinoma histological subtype [1,2]. In recent years, the introduction of targeted therapies with tyrosine kinase inhibitors (TKIs) has significantly changed the therapeutic management of NSCLC [3,4]. Molecular characterization of the tumor at baseline gives us the opportunity to identify those patients with oncogenic alterations and offer them the optimal therapeutic strategy, thus improving their survival and quality of life [5,6]. Among all the genetic alterations found in NSCLC, Kirsten rat sarcoma virus (KRAS) is the most common oncogene that serves as a driver mutation in approximately 30% of all cases [7,8]. For many years, the KRAS protein was considered an “undruggable” target due to the failure of many therapeutic approaches [9]. However, recently, promising KRAS inhibitors were designed, leading to a new era of effective KRAS-targeting agents [10]. In this review, we provide an overview of the structure and function of the KRAS protein and discuss the emerging therapies targeting KRAS, shedding light on future perspectives and restrictions.
2. KRAS Biology
2.1. Structure and Signaling Pathways
The Kirsten rat sarcoma virus (KRAS) gene was first discovered in 1982 and belongs to the RAS gene family along with the Harvey rat sarcoma virus (HRAS) and the neuroblastoma rat sarcoma virus (NRAS) [11]. It is located in the short arm of chromosome 12 (12p), and it encodes two alternative spliced variants: KRAS 4A and KRAS 4B [12,13].
Structurally, KRAS protein is composed of two main regions: the catalytic domain (G-domain) and the C-terminal hypervariable region (HVR). The G-domain consists of the P-loop and switch regions (I and II) and is responsible for the interaction and binding with regulatory proteins. It enables the GTP function of the KRAS protein. HVR is required to bind the protein to the cell membrane, a necessary step for it to be activated [14,15,16].
Under normal conditions, KRAS functions as a GTPase protein that relays extracellular signals to the nucleus, leading to the activation of several intracellular pathways that regulate proliferation, differentiation, survival, and apoptosis [17,18]. GTP and GDP molecules are required for the activation and inactivation of KRAS. It acts like a molecular switch that is “turned on” (activated) when bound to GTP and “turned off” (inactivated) when bound to GDP [17,19]. The alteration between the two function states of KRAS is mediated by regulatory proteins. Guanine nucleotide-exchange factors (GEFs), including Son of Sevenless (SOS 1), promote the GTP-bound form of KRAS by the release of GDP and GTPase-activating proteins (GAPs), such as neurofibromin 1 (NF1), hydrolyze GTP to GDP, thus deactivating the protein [20,21,22]. In basal conditions, KRAS remains bound to the GDP in an inactivated form. When extracellular stimuli bind and activate the receptor tyrosine kinase (RTK) on the cell membrane, SOS 1 promotes the substitution of GDP to GTP with a conformational change in the switch I and II regions [10,13,23]. The activation of KRAS is also mediated by the SHP2 protein, which interacts with the RTK and SOS 1 [24,25]. The GTP-bound KRAS stimulates multiple downstream pathways, including the mitogen-activated protein kinase (MARK) pathway (RAF–MEK–ERK), the phosphatidylinositol 3-kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR) pathway and the RAS-like (RAL) pathway [14]. When a mutation in KRAS occurs, it becomes an oncogene. GAP is hindered from binding to KRAS, blocking GTP hydrolysis and maintaining the protein in an active state. The constitutive stimulation of the downstream signaling cascades leads to oncogenic cell proliferation and tumorigenesis [26,27].
2.2. Cooperation of KRAS and WNT Pathways
According to studies, KRAS and WNT/β-catenin pathways share a significant connection leading to cancer development and progression, especially colorectal cancer (CRC). Gene mutations such as APC and KRAS mutations are frequently detected in CRC, and their interactions promote tumorigenesis. The loss of APC causes the stabilization of β-catenin and KRAS. The cooperation of WNT/β-catenin and KRAS-ERK pathways is associated with tumor growth and treatment resistance. Consequently, this intriguing crosstalk stands as a potential therapeutic avenue for cancer treatment [28,29,30,31].
2.3. Mutations
KRAS is the most common driver mutation in NSCLC, discovered in approximately 30% of all cases [11]. The majority of alterations are located in codon 12 (90%), followed by codons 13 and 61. The KRAS-G12C mutation, characterized as a replacement of glycine with cysteine at codon 12, is the prevailing KRAS mutation (40% of KRAS mutations) found in 13% of all NSCLC cases [8,32]. Substitution of glycine with valine (KRAS G12V) and substitution of glycine with aspartic acid (KRAS G12D) are less frequent alterations [33]. KRAS mutations have been associated with characteristics such as the female sex, adenocarcinoma histology, and smoking habits [34,35]. However, this depends on the specific KRAS mutation subtype. So, among smokers, it is more likely to discover the G12C or G12V mutations, whereas G12D is usually found in patients who do not smoke [32]. Moreover, the incidence of KRAS alterations is higher in Western countries compared with Asian countries [36,37]. Although the prognostic role of KRAS mutations remains a matter of debate, some studies have associated patients harboring these mutations with lower progression-free survival (PFS) and overall survival (OS) [38,39]. However, these survival outcomes need to be confirmed in clinical trials.
2.4. Comutations
KRAS mutations may be mutually exclusive with oncogenic drivers such as EGFR and ALK, but they often coexist with mutations in other genes [40,41]. The most common concurrent molecular alterations are TP53, STK11, and KEAP1. KRAS and its comutations are categorized into three clusters: (1) KRAS with TP53 mutation (KP group), (2) KRAS with STK11/LKB1 mutation including the KEAP1 mutation (KL group), and (3) KRAS with inactivation of CDKN2A/B (KC group) [42]. According to studies, STK11 and KEAP1 mutations are negative prognostic factors and forecast impaired OS [43]. Furthermore, some comutations have been associated with an altered immunogenic profile. Skoulidis et al. elucidated the lower expression of PD-L1 and deficient tumor microenvironment when the STK11 comutation is present and the higher levels of tumor-infiltrating lymphocytes (TILs) and PD-L1 when the TP53 mutation coexists [42,44,45]. All the above data propose the key role of concomitant mutations on the heterogeneity of KRAS-mutated tumors and their impact on different therapeutic strategies [46].
2.5. Why Was the KRAS Mutation Undruggable?
For many years, all efforts of therapeutic targeting KRAS mutations had been unsuccessful, leading to its characterization as an “undruggable” target. The main reasons making this process difficult were the structure and complexity of KRAS pathways [47,48]. KRAS protein consists of a highly smooth surface and a unique GTP binding site. The absence of available binding pockets, along with the protein’s high affinity for GTP, have made the development of effective targeted agents a challenge [49]. However, the discovery of the switch II pocket on the KRAS-G12C-mutated protein by Shokat lab in 2013 paved the way for the design of agents that selectively bind to the cysteine residues [50]. Even though many molecules have been developed since then, only two of them, sotorasib (AMG510) and adagrasib (MRTX849), have demonstrated clinical efficacy and received FDA approval [10,13].
3. Direct Targeting of KRAS
3.1. Sotorasib (AMG510)
Sotorasib is the first clinically effective selective inhibitor of KRAS-G12C mutation that gained FDA approval [51]. Lumakras (Sotorasib) is an oral small molecule that covalently binds to the cysteine in the switch II pocket, blocking the interaction between KRAS and SOS 1 and locking KRAS in its inactive GDP-bound state [50]. CodeBreak 100 was a phase I/II study that evaluated the safety, pharmacokinetics, and efficacy of sotorasib in patients with locally advanced or metastatic KRAS-G12C tumors who had already received previous lines of therapy [52,53]. In the phase I trial, safety was the primary endpoint. After 11.7 months of median follow-up, the median PFS was 6.3 months. The dose of 960 mg daily was set as the dose for the expansion cohort. The most frequently reported treatment-related adverse events (TRAEs) were diarrhea, fatigue, nausea, vomiting, and abnormal levels of aminotransferases [52]. In phase II of the CodeBreak 100 trial, the patients enrolled had progressed after platinum-based chemotherapy and immunotherapy alone or combined. After 15.3 months of follow-up, the median PFS and OS were identified as 6.8 months and 12.5 months, respectively. The TRAEs reported were similar to the ones in the phase I trial and included gastrointestinal AEs and fatigue. In an exploratory analysis of the phase II trial, Skoulidis et al. investigated the relation between comutation subgroups and the efficacy of sotorasib treatment. Patients with STK11 mutations and wild-type KEAP1 demonstrated better response (50%) in comparison with mutated KEAP1 and wild-type STK11 (14%) [53]. Moreover, according to a post hoc analysis, Lumakras showed intracranial efficacy in brain metastases [54,55]. Based on the promising results of CodeBreak 100, sotorasib received accelerated approval from the FDA in May 2021 for patients with KRAS-G12C-mutated NSCLC who had been treated with at least one prior line of anticancer therapy [56].
Additional studies were designed to evaluate sotorasib as monotherapy or in combination with other targeted agents, immunotherapy, and chemotherapy. CodeBreak 200 is a global, randomized, open-label phase III trial aiming to assess the activity of sotorasib versus docetaxel in the second line for patients with advanced KRAS-mutated NSCLC. Preliminary data were published at ESMO 2022. The KRAS-G12C inhibitor was associated with a better PFS and safety profile compared to docetaxel but showed no difference in terms of OS [57,58,59].
In another phase II study, known as CodeBreak 201 (NCT04933695), sotorasib is being evaluated as a first-line therapy in patients with KRAS-mutant NSCLC with the following criteria: (1). stage IV, (2). PD-L1 TPS score < 1%, and/or (3). comutated STK11 [52]. Finally, CodeBreak 101 (NCT041185883) is an ongoing phase Ib/II study testing the efficacy and safety of sotorasib in combination with targeted molecules, such as EGFR, SHP2, mTOR, and CDK inhibitors, but also with chemotherapy and immune checkpoint inhibitors (ICIs) in advanced KRAS-G12C-mutated lung cancer [60].
3.2. Adagrasib (MRTX849)
Adagrasib is the second direct KRAS-G12C inhibitor that demonstrated sufficient clinical activity. The mechanism of action is similar to sotorasib and is based on the irreversible bind to the protein, trapping it in its inactive GDP-bound state. Interestingly, adagrasib has shown superiority in terms of pharmacokinetic properties such as long half-life (approximately 24 h), elevated oral bioavailability, and extensive tissue distribution. In addition, adagrasib remains efficient even at low concentrations and is highly selective for the KRAS-G12C mutation [61].
KRYSTAL-1 was the phase I/Ib study that evaluated the safety and efficacy of adagrasib in patients with KRAS-G12C-mutated NSCLC [62,63,64]. After 19.6 months of follow-up, the median PFS was 11.1 months. The recommended dose for phase II (RP2D) was identified as 600 mg twice daily. The most commonly experienced treatment-related adverse (TRAEs) were nausea, diarrhea, vomiting, and fatigue [62]. In the phase II expansion cohort of KRYSTAL-1, the patients enrolled were already treated with chemotherapy and/or immunotherapy in previous lines. They received 600 mg of adagrasib per os twice a day, and after follow-up, the reported mPFS and mOS were 6.5 months and 12.6 months, respectively. The safety profile included gastrointestinal AEs (diarrhea, nausea, vomiting), fatigue, and increased levels of aminotransferases (ALT and AST). Moreover, in patients with stable, previously treated brain metastases, the intracranial confirmed response rate (ORR) was 33.3%. As for comutations, KEAP1 was associated with worse responses [65,66]. Based on the aforementioned results, adagrasib obtained FDA approval for patients with previously treated KRAS-G12C mutation [67].
Ongoing clinical trials aim to investigate novel treatment approaches with the combination of adagrasib with other anticancer drugs. The KRYSTAL-12 (NCT04685135) phase III trial tests the clinical activity of adagrasib versus docetaxel in pretreated patients [68,69]. Another phase II/III study (KRYSTAL-7) uses adagrasib with pembrolizumab in the first line and evaluates the efficacy in three subgroups: 1. PD-L1 TPS < 1%—combination, 2. PD-L1 TPS < 1%—adagrasib monotherapy, and 3. PD-L1 TPS > 1%—combination (NCT04613596) [70]. Finally, the associations between the KRAS inhibitor and other agents, including BI 1701963 (SOS inhibitor), TNO155 (SHP2 inhibitor), and palbociclib (CDK4/6 inhibitor), are being investigated in the studies KRYSTAL-14 (NCT04975256), KRYSTAL-2 (NCT04330664), and KRYSTAL-16 (NCT05178888), respectively.
3.3. Novel Direct Inhibitors of KRAS G12C
Many phase I/II clinical trials are ongoing for the evaluation of newly designed KRAS-G12C inhibitors.
GDC-6036 is a novel drug that is being tested in a phase I trial (NCT04449874) as monotherapy or in combination with other molecules, including bevacizumab, atezolizumab, and cetuximab in patients with KRAS-mutated tumors. According to the preliminary results presented at the 2022 World Congress on Lung Cancer (WCLC), the ORR was 46%. As for treatment-related adverse events (TRAEs), the most commonly reported were nausea, diarrhea, vomiting, fatigue, elevated liver enzymes (ALT and AST), and reduced appetite [71,72].
Another novel molecule that selectively inhibits KRAS-G12C mutation is D-1553. In the phase I study (NCT04585035), where pretreated patients were enrolled, it demonstrated an ORR of 40.4% [73].
Moreover, JDQ443 is currently being tested in a phase I/II trial (NCT04699188) as monotherapy and in combination with TNO155 (SHP2 inhibitor) or tislelizumab (antiPD1 antibody). Finally, JAB-21822, LY3537982 (NCT04956640), RMC6291 (NCT05462717), and multiple novel agents have entered clinical trials and are currently under investigation (Table 1).
Table 1.
Direct KRAS-G12C inhibitors.
| Trial | Phase | KRAS Inhibitor |
Combinations | Status/Outcomes | References |
|---|---|---|---|---|---|
| CodeBreak 100 NCT03600883 |
I/II | sotorasib | monotherapy | mPFS 6.8 months mOS 12.5 months FDA Approval |
[52,53,56] |
| CodeBreak 200 NCT04303780 |
III | sotorasib | monotherapy vs. docetaxel | mPFS 5.6 months vs. 4.5 months (HR 0.66) |
[57,58,59] |
| CodeBreak 201 NCT04933695 |
II | sotorasib | monotherapy first line (st. IV, TPS <1% and/or comut. STK11) |
ongoing | [52] |
| CodeBreak 101 NCT04185883 |
Ib/II | sotorasib | EGFR inh, SHP2 inh, mTOR inh, CDK inh. Chemotherapy, ICIs, etc. |
Ongoing | [60] |
| KRYSTAL-1 NCT03785249 |
I/II | adagrasib | monotherapy/afatinib, pembrolizumab, cetuximab | mPFS 6.5 months mOS 12.6 months FDA Approval |
[62,63,64,67] |
| KRYSTAL-12 NCT04685135 |
III | adagrasib | monotherapy vs. docetaxel | ongoing | [68,69] |
| KRYSTAL-7 NCT04613596 |
II/III | adagrasib | pembrolizumab/pembrolizumab + adagrasib vs. pembrolizumab + chemotherapy (first line) | ongoing | [70] |
| KRYSTAL-14 NCT04975256 |
I/Ib | adagrasib | BI 1701963 (SOS inhibitor) | completed | |
| KRYSTAL-2 NCT04330664 |
I/II | adagrasib | TNO155 (SHP2 inhibitor) |
ongoing | |
| KRYSTAL-16 NCT05178888 |
I/Ib | adagrasib | palbociclib (CDK4/6 inh.) |
ongoing | |
| NCT04449874 | I | GDC-6036 | monotherapy/ bevacizumab, atezolizumab, cetuximab, erlotinib, etc. |
ongoing | [71,72] |
| NCT04585035 | I/II | D-1553 | monotherapy/other agents | ongoing | [73] |
| KontRASt-01 NCT04699188 |
I/II | JDQ443 | monotherapy/ TNO155, tislelizumab |
ongoing | |
| NCT05009329 | I/II | JAB-21822 | monotherapy | ongoing | |
| NCT04956640 | I | LY3537982 | monotherapy/ abemaciclib, cetuximab, pembrolizumab, chemotherapy, etc. |
ongoing | |
| NCT05462717 | I/Ib | RMC-6291 | monotherapy | ongoing |
4. Resistance to KRAS-G12C Inhibitors
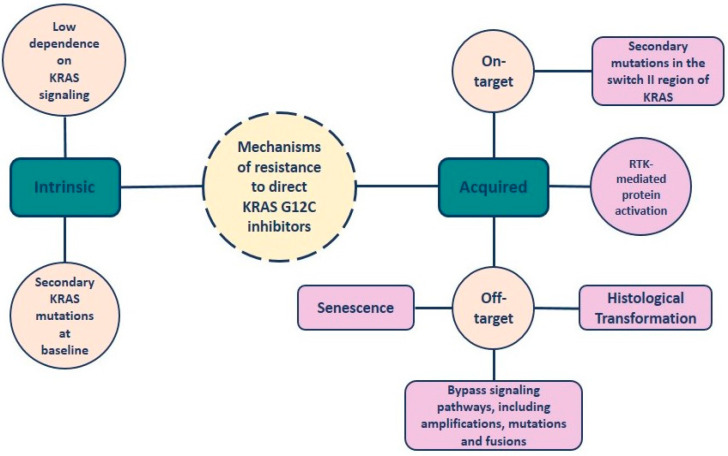
Despite the aforementioned encouraging clinical outcomes that highlight the efficacy of novel KRAS-G12C inhibitors, the possibility of patients demonstrating disease progression is high. This can be attributed to a variety of resistance mechanisms that are divided into two types: intrinsic and acquired mechanisms of resistance.
Regarding intrinsic resistance, one of the main reasons for the limited response to KRAS inhibitors is the low dependence on KRAS signaling. The RAS protein regulates multiple downstream pathways that, in some cases, can be activated even after KRAS knockdown [74,75]. Specifically, the PI3K pathway can be stimulated even in the presence of KRAS inhibitors [76]. Moreover, secondary KRAS mutations at baseline result in a heterogeneous response to direct inhibitors of KRAS-G12C mutation [77].
As for the acquired type of resistance, two mechanisms have been identified: on-target and off-target [78]. The on-target mutations impact the switch II region, which is essential for successful drug binding. KRAS G12D/R/V and other secondary mutations have been reported in accordance with the inhibitor that was used for treatment. KRAS G13D has been associated with sotorasib-induced resistance, and KRAS Q99L with adagrasib-induced resistance. Y96D has been observed after exposure to either one of them [79,80]. RM-018 is a novel inhibitor of the KRAS GTP-bound (active) form, effective in overcoming the secondary KRAS-G12C/Y96D mutations [81]. Furthermore, resistance to KRAS-G12C inhibition can arise due to off-target mechanisms. Multiple bypass signaling pathways have been identified to date, including MET amplification; mutations activating NRAS, BRAF, MAP2K1, and RET; fusions of ALK, RET, BRAF, RAF1, and FGFR3; and loss-of-function of PTEN and NF1 [82]. Another way for acquiring resistance is epithelial-to-mesenchymal transformation (EMT) induced by the activation of the PI3K pathway [40]. In some cases, a histological transformation of lung adenocarcinoma to squamous cell carcinoma can be observed [82]. In addition, senescence resulting from the activation of aurora kinase signaling can induce resistance [10].
Moreover, the inhibition of KRAS and, subsequently, the block of the MAPK pathway can cause the synthesis of a novel KRAS-G12C protein, which remains active by the receptor tyrosine kinase (RTK), even if an inhibitor is present [83].
Finally, the activation of wild-type RAS (HRAS and NRAS) mediated by RTKs is supposed to be another mechanism of resistance to direct KRAS inhibitors (Figure 1) [75,84].
Figure 1.
Resistance mechanisms to direct KRAS-G12C inhibition.
In conclusion, an abundance of intrinsic and acquired mechanisms are responsible for the progression of disease in patients harboring KRAS-G12C mutation and treated with direct inhibitors. However, many studies still need to be conducted in order for us to understand those mechanisms fully and develop the optimal treatment strategies to overcome them. For this purpose, efforts to combine KRAS inhibitors with targeted agents depending on the type of resistance are currently being made, and results are eagerly expected [85,86].
5. Indirect Inhibition of KRAS
As mentioned before, the development of resistance has overshadowed the encouraging clinical efficacy of novel direct KRAS-G12C inhibitors. The combination of inhibitors with targeted agents of the upstream, downstream, or other important signaling pathways is a promising therapeutic approach and a potential solution to this predicament [85,86].
5.1. Inhibitors of the Upstream Signaling Pathway
EGFR inhibitors
RTKs, such as EGFR, can induce the rebound activation of KRAS and its downstream pathways as a result of the inhibition of KRAS. EGFR inhibitors, in combination with KRAS-G12C inhibitors, can possibly overcome the resistance mechanism [61].
Combinations are already being tested in studies, including CodeBreak 101 (sotorasib with a pan-ErbB inhibitor) and a phase I trial (GDC-6036 with erlotinib—NCT04449874).
SHP2 and SOS1 inhibitors
Other promising target proteins for the indirect inhibition of KRAS are SHP2 and SOS1. These molecules are essential for the activation of KRAS as part of the upstream pathway [87]. SHP2 inhibitors, including RMC-4630 and TN0155, have been designed and are being evaluated in studies as monotherapy or in combination with KRAS inhibitors (NCT03634982, NCT03114319, NCT04185883). Similarly, BI 1701963 is a pan-KRAS SOS1 inhibitor under investigation in a phase I trial as a single agent or in association with trametinib (MEK inhibitor) (NCT04111458).
5.2. Inhibitors of the Downstream Signaling Pathway
MAPK pathway (RAF/MEK/ERK)
Regarding the MAPK pathway, the GTP-bound KRAS protein activates RAF, which stimulates the phosphorylation of MEK and then ERK, leading to the activation of transcription factors [88,89,90,91]. RAF inhibitors (belvarafenib) and MEK inhibitors (binimetinib, cobimetinib) are being tested for the indirect inhibition of KRAS (NCT03284502, NCT01859026, NCT01986166). Also, RAF–MEK inhibitor VS-6766 is being used as monotherapy or in combination with defactinib (FAK inhibitor) [92], sotorasib, or adagrasib (KRAS-G12C inhibitors) in ongoing trials (NCT05074810, NCT05375994). The resistance induced by the treatment with RAF or MEK inhibitors led to the development and evaluation of ERK inhibitors (JSI-1187-01) (NCT04418167). The combination of a RAF/MEK inhibitor with a FAK inhibitor and the combinations of MEK and PI3K/mTOR/AKT inhibitors are potent therapeutic strategies targeting the downstream signaling pathway of KRAS [93].
PI3K/AKT/mTOR pathway
The PI3K/AKT/mTOR is another well-known downstream pathway activated by KRAS, which contains potential targets for effective inhibition. PI3K inhibitors (BKM120, serabelisib—NCT04073680, XL147), an AKT inhibitor (TAS0612—NCT04586270), and an mTOR inhibitor (A2D2014—NCT02583542) were designed and are being investigated in phase I/II studies [94,95,96]. In addition, the combination of PI3K inhibitors with ARS1620 (second-generation KRAS-G12C inhibitor) has demonstrated efficacy in preclinical studies [97].
5.3. FAK Inhibitors
Focal adhesion kinase (FAK) is a non-receptor tyrosine kinase that can cause KRAS mutant cell death if inhibited [98]. The FAK inhibitor defactinib was evaluated in a phase II study (NCT01951690) in previously treated patients showing average clinical activity [99].
5.4. DDR1 Inhibitors
Discoidin domain receptor 1 is being evaluated as a novel biomarker and therapeutic target in KRAS mutant NSCLC. DDR1 is a receptor tyrosine kinase with a significant role in cell proliferation and differentiation. According to studies, the upregulation of DDR1 in lung cancer is associated with lymph node metastasis, increased invasiveness, and poor prognosis. Moreover, the DDR1–BCR–ABL axis is strongly correlated with the KRAS PI3K–AKT–mTOR axis. The combinational targeting of DDR1, BCR-ABL, and EGFR-ERBB2 is a promising therapeutic approach with future perspectives [28,100,101].
5.5. YB-1 Inhibitors
Y-box-binding protein 1 (YB-1) serves as an oncoprotein that plays a crucial role in cancer cell proliferation, stemness, DNA repair, and chemotherapy resistance. YB-1 is activated by the PI3K/AKT and MAPK/ERK pathways, with phosphorylation at S102 mediated by the p90 ribosomal S6 kinase (RSK). RSK inhibitors, such as LJI308, were designed; however, their long-term use leads to AKT reactivation. As a result, the combination of AKT and RSK inhibitors can be a possible solution and an effective therapeutic strategy [102,103].
5.6. HSP90 Inhibitors
Heat shock protein 90 (HSP90) is a chaperone responsible for protein stabilization, adjustment to heat, and cell proliferation [104]. Ganetespib, an HSP90 inhibitor, was efficient in blocking tumorigenesis in a phase II study [105]; however, its clinical activity was inadequate in the phase III study (GALAXY II) with docetaxel in KRAS-mutated NSCLC [106]. A novel HSP90 inhibitor, SNX-5422, was associated with promising results in a phase I study with chemotherapy (Table 2) [107].
Table 2.
Indirect KRAS-G12C inhibitors.
| Trial | Phase | KRAS Inhibitor | Combinations | Status/ Outcomes |
References |
|---|---|---|---|---|---|
| NCT03634982 | I | RMC-4630 (SHP2 Inhibitor) |
monotherapy | completed | [87] |
| NCT03114319 | I | TNO155 (SHP2 Inhibitor) |
monotherapy | ongoing | [87] |
| NCT04111458 | I | BI 1701963 (SOS1 Inhibitor) | monotherapy/ trametinib (MEK Inh.) |
ongoing | [87] |
| NCT03284502 | I | belvarafenib (RAF Inh.) | cobimetinib, cetuximab |
ongoing | [88,89,90,91] |
| NCT01859026 | I | binimetinib (MEK Inh.) | erlotinib | completed | [88,89,90,91] |
| NCT01986166 | I | cobimetinib (MEK Inh.) | duligotuzumab (EGFR/HER3 Inh.) |
completed | [88,89,90,91] |
| RAMP203 NCT05074810 | I/II | Avutometinib [VS-6766] (RAF–MEK Inhibitor) |
sotorasib | ongoing | [92] |
| RAMP204 NCT05375994 | I/II | Avutometinib [VS-6766] (RAF–MEK Inhibitor) |
adagrasib | ongoing | [92] |
| NCT04418167 | I | JSI-1187-01 (ERK Inhibitor) |
monotherapy/ dabrafenib (BRAF Inh.) |
ongoing | |
| NCT04073680 | I/II | serabelisib (PI3K Inhibitor) |
canagliflozin (SGLT2 Inh.) | completed | [94,95,96] |
| NCT04586270 | I | TAS0612 (AKT Inhibitor) |
monotherapy | ongoing | [94,95,96] |
| TORCMEK NCT02583542 | I/II | AZD2014 (mTOR Inhibitor) |
selumetinib (MEK Inh.) | completed | [94,95,96] |
| NCT01951690 | II | defactinib (FAK Inhibitor) |
monotherapy | completed | [99] |
| GALAXY 2 NCT01798485 | III | ganetespib (HSP90 Inhibitor) |
docetaxel vs. docetaxel alone | terminated (futility) |
[106] |
| NCT01892046 | I | SNX-5422 (HSP90 Inhibitor) |
chemotherapy | completed | [107] |
5.7. CDK 4/6 Inhibitors
Cyclin-dependent kinases (CDKs) 4/6 are regulatory agents of the cell cycle, and their activation is connected with the KRAS pathway [108]. Although monotherapy abemaciclib or palbociclib in KRAS-mutant NSCLC was ineffective [109,110], the combination of CDK 4/6 inhibitors with KRAS direct inhibitors seems promising according to preclinical data [74]. Clinical trials are ongoing (CodeBreak 101—sotorasib with palbociclib and KRYSTAL 16—adagrasib with palbociclib—NCT04185883).
6. Platinum-Based Chemotherapy in KRAS-Mutated NSCLC
Despite the tremendous progression made in targeting the KRAS mutation in recent years, platinum-based chemotherapy remains the standard of care for patients with KRAS-mutant NSCLC. But how does the KRAS oncogene influence the response to platinum-based chemotherapy? The data are scarce. However, several studies conclude that KRAS mutation is associated with shorter PFS or OS when patients are treated with traditional chemotherapy. Moreover, among KRAS mutation subtypes, the KRAS G12V is associated with the worst PFS. As a result, KRAS oncogene is a negative predictive factor of PFS in patients who receive first-line platinum-based chemotherapy, but treatment responses differ among KRAS mutation subtypes [111,112].
7. Immunotherapy in KRAS-Mutated NSCLC
In recent years, the advent of immune checkpoint inhibitors (ICIs) has significantly altered the therapeutic approach of NSCLC [113,114,115,116]. According to preclinical data, patients harboring the KRAS mutation can benefit from monoclonal antibodies targeting PD1 and PD-L1. KRAS-mutated lung cancer has been associated with smoking habits and, consequently, an inflammatory tumor microenvironment (TME) and high tumor mutation burden (TMB) [117,118]. Moreover, CD8+ tumor-infiltrating lymphocytes (TILS) and PD-L1 expression are increased when the KRAS mutation is present [119,120,121,122].
The efficacy of immunotherapy in KRAS-mutated NSCLC is also evident in clinical trials. In the CheckMate 057 trial, the KRAS mutation was linked with higher OS when comparing nivolumab (anti-PD-1 antibody) with chemotherapy (HR 0.52, 95% CI: 0.29–0.95) [113]. In addition, in the phase III OAK study, the benefit of atezolizumab was observed in KRAS-mutated patients (mOS: HR = 0.71 [95% CI: 0.38–1.35]) [115]. Similarly, based on the results of the KEYNOTE-042 study (mOS KRASm vs. wt: 21.1 vs. 13.6 months, p = 0.03) and the IMMUNOTARGET study (mPFS: 3.2 months and mOS 13.5 months), immunotherapy can be therapeutically exploited in KRAS mutations [123,124].
However, it is under consideration whether KRAS point mutations or co-occurring mutations influence the efficacy of ICIs. KRAS-G12D-mutated tumors have been associated with decreased levels of PD-L1, CD8+ TILS, and TMB. These characteristics suggest a lower response to immunotherapy in KRAS-G12D mutation [125,126]. On the contrary, other studies do not verify different results between KRAS mutations [122]. Furthermore, as mentioned before, STK11 or KEAP1 comutations have been linked with decreased PD-L1 and TILS levels, resulting in impaired activity of ICIs [44,127,128]. On the other hand, tumors harboring TP53 comutation benefit more from immunotherapy due to increased expression of PD-L1 [128,129,130]. More studies need to be conducted in order to evaluate the aforementioned outcomes. Among the novel therapeutic approaches targeting the KRAS mutation, the combination of KRAS-G12C inhibitors with immunotherapy seems promising. In preclinical studies, sotorasib induced an inflammatory tumor microenvironment and enhanced the immune system [74]. Similar results were observed in tumors treated with adagrasib [131]. The synergistic effect of KRAS-G12C inhibitors and ICIs is currently being evaluated in phase I/II studies, including the CodeBreak 100 and 101 trials (sotorasib with PD-1/PD-L1 monoclonal antibodies) [132] and the KRYSTAL-7 study (adagrasib with pembrolizumab) [133].
8. KRAS-G12V and -G12D Mutations
KRAS-G12V mutation, where glycine is substituted with valine at position 12, is observed in 21% of patients with KRAS-mutated tumors in the lung, especially smokers [134]. This mutation has been associated with impaired OS and increased recurrence rate [135]. Regarding the treatment strategies, chemotherapeutic agents with higher efficacy in KRAS-G12V mutation are taxanes and cisplatin [136,137]. In addition, immunotherapy is another potential approach due to the increased expression of PD-L1 [138]. Emerging therapies with KRAS-G12V inhibitors, including JAB23000, are currently under investigation [87,139].
KRAS-G12D mutation, where glycine is replaced with aspartic acid at position 12, has been reported in 15% of KRAS-mutated cases and is usually observed in nonsmokers [134]. As mentioned before, KRAS G12D has been linked with a lower level of PD-L1 protein and immune suppression, resulting in decreased activity of ICIs [140]. As for the direct inhibition of the G12D allele, MRTX1133 is a novel molecule targeting both active and inactive states of the protein and has shown promising activity in preclinical studies. Recently, the US FDA approved the initiation of a phase I study for the evaluation of this first-in-class inhibitor [141].
9. Other Emerging Therapies
Except for the conventional inhibitors directly targeting the KRAS-G12C protein or molecules of the upstream or downstream KRAS pathways, there is a need for new treatment strategies, including PROTACs, pan-KRAS inhibitors, and agents targeting metabolic rewiring processes and regulatory molecules of the immune system. Proteolysis-targeting chimeras (PROTACs) are emerging targeted agents that degrade the protein of interest using cellular proteasomal degradation mechanisms [142]. KRAS-G12C PROTACs, such as LC-2, have been designed to exploit KRAS inhibitors (MRTX849 in the case of LC-2) for the degradation of KRAS-G12C protein [143].
Another novel therapeutic approach targeting multiple KRAS mutations (G12D, G12V, G13D, and G12C) is an mRNA-based cancer vaccine. V941 is being tested in a phase I study (NCT03948763) as a single agent or in combination with pembrolizumab in three different types of cancer, including KRAS-mutated NSCLC [144].
Regarding the metabolic rewiring of cancer cells, autophagy plays a crucial role in cell homeostasis. Autophagy inhibition is a potential therapeutic strategy aiming to achieve cancer cell death [145]. Hydroxychloroquine is an autophagy inhibitor that is currently being evaluated in a study along with MEK-inhibitor binimetinib for the treatment of patients with KRAS-mutated lung cancer (NCT04735068).
Moreover, the KRAS oncogene influences metabolism and especially upregulates the expression of fatty acid synthase (FASN), the enzyme of de novo lipogenesis, in order to prevent lipid peroxidation and, consequently, ferroptosis. Ferroptosis is an oxidative-stress-dependent type of programmed cell death. According to these data, the induction of ferroptosis could be an effective antitumor strategy. TVB-2640 is a FASN inhibitor that is being studied in a phase II trial (NCT03808558) with promising results [146].
Furthermore, the intriguing relationship between KRAS and miRNA needs to be explored. Physiologically, miRNAs are responsible for post-transcriptional gene regulation through interaction with target mRNAs. However, KRAS mutations tend to misregulate the miRNA regulatory pathway in multiple ways, including miRNA biosynthesis, expression, and function. As a result, the miRNA machinery can be therapeutically exploited in KRAS-mutated cancers. Studies need to be conducted in order to navigate novel strategies, including simultaneous targeting of KRAS and miRNA pathways [147].
Finally, the remodeling of the tumor microenvironment (TME) by cancer cells is another key to the achievement of effective targeted therapy [148]. According to studies, cytokines IL-1β and IL-6 have been associated with cancer cell proliferation due to signal transduction through multiple pathways [149,150,151]. Several trials are ongoing for the evaluation of interleukin inhibitors. Canakinumab is an anti-IL-1β agent under investigation as a monotherapy (NCT03447769) or in combination with immunotherapy (pembrolizumab—NCT03968419 and durvalumab—NCT04905316) or chemotherapy (docetaxel—NCT03626545). Similarly, tecilizumab (IL-6 inhibitor) is being tested in association with ICIs (atezolizumab—NCT04691817 and nivolumab/ipilimumab—NCT04940299) for the treatment of NSCLC, including tumors harboring the KRAS mutation (Table 3 and Table 4).
Table 3.
Novel treatment strategies.
| Trial | Phase | KRAS Inhibitor | Combinations | Status/ Outcomes |
References |
|---|---|---|---|---|---|
| NCT05737706 | I/II | MRTX1133 (KRAS-G12D Inh.) |
monotherapy | ongoing | [141] |
| V941-001 NCT03948763 | I | V941 (mRNA cancer vaccine) |
monotherapy/pembrolizumab | completed | [144] |
| NCT04735068 | II | hydroxychloroquine (autophagy inhibitor) |
binimetinib (MEK Inh.) |
ongoing | [145] |
| CANOPY-N NCT03968419 | II | canakinumab (anti-IL-1β agent) |
monotherapy/pembrolizumab | terminated (low enrollment) |
[149,150,151] |
| CHORUS NCT04905316 | II | canakinumab (anti-IL-1β agent) |
durvalumab, chemoradiation | ongoing | [149,150,151] |
| CANOPY-2 NCT03626545 | III | canakinumab (anti-IL-1β agent) |
docetaxel vs. placebo + docetaxel | terminated (lack of efficacy) |
[149,150,151] |
| NCT04691817 | I/II | tecilizumab (IL-6 Inhibitor) | atezolizumab | ongoing | [149,150,151] |
| NCT04940299 | II | tecilizumab (IL-6 Inhibitor) |
nivolumab—ipilimumab | ongoing | [149,150,151] |
Table 4.
Indirect actionable pathways and targets against KRAS-mutated NSCLC.
| Pathway | Target | Inhibitor | References |
|---|---|---|---|
| Upstream signaling pathway | EGFR | erlotinib | [61] |
| SHP2 | RMC-4630 TNO-155 |
[87] | |
| SOS 1 | BI 1701963 | [87] | |
| Downstream signaling pathway MAPK pathway (RAF–MEK–ERK) |
RAF | belvarafenib | [88,89,90,91] |
| MEK RAF–MEK |
binimetinib cobimetinib VS-6766 |
[88,89,90,91] [92] |
|
| ERK | JSI-1187-01 | ||
| Downstream signaling pathway PI3K–AKT–mTOR pathway |
PI3K |
BKM120 serabelisib XL147 |
[94,95,96] |
| AKT | TAS0612 | [94,95,96] | |
| mTOR | AZD2014 | [94,95,96] | |
| Other pathways | FAK | defactinib | [99] |
| HSP90 | ganetespib SNX-5422 |
[105,106] [107] |
|
| CDK 4/6 | palbociclib | [74,108,109,110] | |
| DDR1 | |||
| YB-1—RSK | LJI308 | ||
| Immune system | ICIs | nivolumab atezolizumab |
[113] [115] |
| Metabolic rewiring | autophagy | hydroxychloroquine | [145] |
| FASN | TVB-2640 | [146] | |
| Tumor microenvironment (TME) |
IL-1β IL-6 |
canakinumab tecilizumab |
[148,149,150,151] |
10. Conclusions
The thought of KRAS as “undruggable” for many years multiplied the efforts for the development of effective targeted therapies. Sotorasib and adagrasib were the first approved direct KRAS-G12C inhibitors, altering the therapeutic landscape of KRAS. However, the emergence of intrinsic or acquired resistance mechanisms limits the duration of activity and efficacy of the novel KRAS-G12C inhibitors. Many ongoing studies are evaluating combination approaches in order to overcome resistance. Moreover, the heterogeneity and complexity of KRAS-mutated tumors are the main reasons why they exhibit different sensitivity to different therapies. According to studies, KRAS mutation subtypes, as well as comutations, have a significant impact on the efficacy of immunotherapy. Molecular profiling of tumors at baseline is an emerging need in order to determine the optimal therapeutic regimen and potential sequencing of therapies. In addition, results of novel strategies, including cancer vaccines, PROTACs, and pan-KRAS inhibitors, are eagerly awaited so as to be introduced in clinical practice. In conclusion, KRAS remains one of the most attractive target mutations in NSCLC, and personalized therapy seems to be the key to the achievement of successful and durable treatment in the future.
Author Contributions
Conceptualization, A.K. and E.K.; writing—original draft preparation, A.K.; writing—review and editing, E.K. and O.F.; supervision, K.S. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ganti A.K., Klein A.B., Cotarla I., Seal B., Chou E. Update of Incidence, Prevalence, Survival, and Initial Treatment in Patients With Non–Small Cell Lung Cancer in the US. JAMA Oncol. 2021;7:1824–1832. doi: 10.1001/jamaoncol.2021.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thai A.A., Solomon B.J., Sequist L.V., Gainor J.F., Heist R.S. Lung Cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 3.Gomatou G., Syrigos N., Kotteas E. Osimertinib Resistance: Molecular Mechanisms and Emerging Treatment Options. Cancers. 2023;15:841. doi: 10.3390/cancers15030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsamis I., Gomatou G., Chachali S.P., Trontzas I.P., Patriarcheas V., Panagiotou E., Kotteas E. BRAF/MEK inhibition in NSCLC: Mechanisms of resistance and how to overcome it. Clin. Transl. Oncol. 2023;25:10–20. doi: 10.1007/s12094-022-02849-0. [DOI] [PubMed] [Google Scholar]
- 5.Lindeman N.I., Cagle P.T., Aisner D.L., Arcila M.E., Beasley M.B., Bernicker E.H., Colasacco C., Dacic S., Hirsch F.R., Kerr K., et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Mol. Diagn. 2018;20:129–159. doi: 10.1016/j.jmoldx.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Ho C., Tong K.M., Ramsden K., Ionescu D., Laskin J. Histologic classification of non-small-cell lung cancer over time: Reducing the rates of not-otherwise-specified. Curr. Oncol. 2015;22:164–170. doi: 10.3747/co.22.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prior I.A., Hood F.E., Hartley J.L. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biernacka A., Tsongalis P.D., Peterson J.D., de Abreu F.B., Black C.C., Gutmann E.J., Liu X., Tafe L.J., Amos C.I., Tsongalis G.J. The potential utility of re-mining results of somatic mutation testing: KRAS status in lung adenocarcinoma. Cancer Genet. 2016;209:195–198. doi: 10.1016/j.cancergen.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L., Guo Z., Wang F., Fu L. KRAS Mutation: From Undruggable to Druggable in Cancer. Signal Transduct. Target. Ther. 2021;6:386. doi: 10.1038/s41392-021-00780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox A.D., Der C.J. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang E.H., Gonda M.A., Ellis R.W., Scolnick E.M., Lowy D.R. Human genome contains four genes homologous to transforminggenes of Harvey and Kirsten murine sarcoma viruses. Proc. Natl. Acad. Sci. USA. 1982;79:4848–4852. doi: 10.1073/pnas.79.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore A.R., Rosenberg S.C., McCormick F., Malek S. RAS targeted therapies: Is the undruggable drugged? Nat Rev Drug Discov. 2020;19:533–552. doi: 10.1038/s41573-020-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock J.F. Ras Proteins: Different Signals from Different Locations. Nat. Rev. Mol. Cell Biol. 2003;4:373–385. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 15.Westcott P.M.K., To M.D. The Genetics and Biology of KRAS in Lung Cancer. Chin. J. Cancer. 2013;32:63–70. doi: 10.5732/cjc.012.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall B.E., Bar-Sagi D., Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc. Natl. Acad. Sci. USA. 2002;99:12138–12142. doi: 10.1073/pnas.192453199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malumbres M., Barbacid M. RAS oncogenes: The first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 18.Tímár J. The clinical relevance of KRAS gene mutation in non-small-cell lung cancer. Curr. Opin. Oncol. 2014;26:138–144. doi: 10.1097/CCO.0000000000000051. [DOI] [PubMed] [Google Scholar]
- 19.Barbacid M. ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 20.Pantsar T. The Current Understanding of KRAS Protein Structure and Dynamics. Comput. Struct. Biotechnol. J. 2019;18:189–198. doi: 10.1016/j.csbj.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.PNAS Drugging an Undruggable Pocket on KRAS. [(accessed on 26 February 2024)]. Available online: https://www.pnas.org/doi/10.1073/pnas.1904529116.
- 22.Maertens O., Cichowski K. An Expanding Role for RAS GTPase Activating Proteins (RAS GAPs) in Cancer. Adv. Biol. Regul. 2014;55:1–14. doi: 10.1016/j.jbior.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Drosten M., Barbacid M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell. 2020;37:543–550. doi: 10.1016/j.ccell.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Dance M., Montagner A., Salles J.-P., Yart A., Raynal P. The Molecular Functions of Shp2 in the Ras/Mitogen-Activated Protein Kinase (ERK1/2) Pathway. Cell. Signal. 2008;20:453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Mainardi S., Mulero-Sánchez A., Prahallad A., Germano G., Bosma A., Krimpenfort P., Lieftink C., Steinberg J.D., de Wit N., Gonçalves-Ribeiro S., et al. SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat. Med. 2018;24:961–967. doi: 10.1038/s41591-018-0023-9. [DOI] [PubMed] [Google Scholar]
- 26.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren G.W., Cummings K.M. Tobacco and lung cancer: Risks, trends, and outcomes in patients with cancer. Am. Soc. Clin. Oncol. Educ. Book. 2013;33:359–364. doi: 10.14694/EdBook_AM.2013.33.359. [DOI] [PubMed] [Google Scholar]
- 28.Gupta K., Jones J.C., Farias V.D.A., Mackeyev Y., Singh P.K., Quiñones-Hinojosa A., Krishnan S. Identification of Synergistic Drug Combinations to Target KRAS-Driven Chemoradioresistant Cancers Utilizing Tumoroid Models of Colorectal Adenocarcinoma and Recurrent Glioblastoma. Front. Oncol. 2022;12:840241. doi: 10.3389/fonc.2022.840241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong C.C., Xu J., Bian X., Wu J.L., Kang W., Qian Y., Li W., Chen H., Gou H., Liu D., et al. In Colorectal Cancer Cells With Mutant KRAS, SLC25A22-Mediated Glutaminolysis Reduces DNA Demethylation to Increase WNT Signaling, Stemness, and Drug Resistance. Gastroenterology. 2020;159:2163–2180.e6. doi: 10.1053/j.gastro.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 30.Hu J., Zhang Z., Zhao L., Li L., Zuo W., Han W.Z.A.L. High Expression of RAD51 Promotes DNA Damage Repair and Survival in KRAS-Mutant Lung Cancer Cells. BMB Rep. 2019;52:151–156. doi: 10.5483/BMBRep.2019.52.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L., Shen C., Estrada-Bernal A., Robb R., Chatterjee M., Sebastian N., Webb A., Mo X., Chen W., Krishnan S., et al. Oncogenic KRAS Drives Radioresistance Through Upregulation of NRF2- 53BP1-Mediated non-Homologous End-Joining Repair. Nucleic Acids Res. 2021;49:11067–11082. doi: 10.1093/nar/gkab871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogan S., Shen R., Ang D.C., Johnson M.L., D’Angelo S.P., Paik P.K., Brzostowski E.B., Riely G.J., Kris M.G., Zakowski M.F., et al. Molecular epidemiology of EGFR and KRAS mutations in 3026 lung adenocarcinomas: Higher susceptibility of women to smoking-related KRAS mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herdeis L., Gerlach D., McConnell D.B., Kessler D. Stopping the beating heart of cancer: KRAS reviewed. Curr. Opin. Struct. Biol. 2021;71:136–147. doi: 10.1016/j.sbi.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Judd J., Abdel Karim N., Khan H., Naqash A.R., Baca Y., Xiu J., VanderWalde A.M., Mamdani H., Raez L.E., Nagasaka M., et al. Characterization of KRAS Mutation Subtypes in Non–Small Cell Lung Cancer. Mol. Cancer Ther. 2021;20:2577–2584. doi: 10.1158/1535-7163.MCT-21-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reck M., Carbone D.P., Garassino M., Barlesi F. Targeting KRAS in Non-Small-Cell Lung Cancer: Recent Progress and New Approaches. Ann. Oncol. 2021;32:1101–1110. doi: 10.1016/j.annonc.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 36.El Osta B., Behera M., Kim S., Berry L.D., Sica G., Pillai R.N., Owonikoko T.K., Kris M.G., Johnson B.E., Kwiatkowski D.J., et al. Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: The lung cancer mutation consortium experience. J. Thorac. Oncol. 2019;14:876–889. doi: 10.1016/j.jtho.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dearden S., Stevens J., Wu Y.L., Blowers D. Mutation incidence and coincidence in non small-cell lung cancer: Meta-analyses by ethnicity and histology (mutMap) Ann. Oncol. 2013;24:2371–2376. doi: 10.1093/annonc/mdt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H.A., Sima C.S., Shen R., Kass S., Gainor J., Shaw A., Hames M., Iams W., Aston J., Lovly C.M., et al. Prognostic Impact of KRAS Mutation Subtypes in 677 Patients with Metastatic Lung Adenocarcinomas. J. Thorac. Oncol. 2015;10:431–437. doi: 10.1097/JTO.0000000000000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goulding R.E., Chenoweth M., Carter G.C., Boye M.E., Sheffield K.M., John W.J., Leusch M.S., Muehlenbein C.E., Li L., Jen M.-H., et al. KRAS Mutation as a Prognostic Factor and Predictive Factor in Advanced/Metastatic Non-Small Cell Lung Cancer: A Systematic Literature Review and Meta-Analysis. Cancer Treat. Res. Commun. 2020;24:100200. doi: 10.1016/j.ctarc.2020.100200. [DOI] [PubMed] [Google Scholar]
- 40.Adachi Y., Ito K., Hayashi Y., Kimura R., Tan T.Z., Yamaguchi R., Ebi H. Epithelial-to-Mesenchymal transition is a cause of both intrinsic and acquired resistance to KRAS G12C inhibitor in KRAS G12C-mutant non-small cell lung cancer. Clin. Cancer Res. 2020;26:5962–5973. doi: 10.1158/1078-0432.CCR-20-2077. [DOI] [PubMed] [Google Scholar]
- 41.West H.J., McCleland M., Cappuzzo F., Reck M., Mok T.S., Jotte R.M., Nishio M., Kim E., Morris S., Zou W., et al. Clinical efficacy of atezolizumab plus bevacizumab and chemotherapy in KRAS-mutated non-small cell lung cancer with STK11, KEAP1, or TP53 comutations: Subgroup results from the phase III IMpower150 trial. J. Immunother. Cancer. 2022;10:e003027. doi: 10.1136/jitc-2021-003027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoulidis F., Byers L.A., Diao L., Papadimitrakopoulou V.A., Tong P., Izzo J., Behrens C., Kadara H., Parra E.R., Canales J.R., et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arbour K.C., Jordan E., Kim H.R., Dienstag J., Yu H.A., Sanchez-Vega F., Lito P., Berger M., Solit D.B., Hellmann M., et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skoulidis F., Goldberg M.E., Greenawalt D.M., Hellmann M.D., Awad M.M., Gainor J.F., Schrock A.B., Hartmaier R.J., Trabucco S.E., Gay L., et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marinelli D., Mazzotta M., Scalera S., Terrenato I., Sperati F., D’Ambrosio L., Pallocca M., Corleone G., Krasniqi E., Pizzuti L., et al. KEAP1-driven co-mutations in lung adenocarcinoma unresponsive to immunotherapy despite high tumor mutational burden. Ann. Oncol. 2020;31:1746–1754. doi: 10.1016/j.annonc.2020.08.2105. [DOI] [PubMed] [Google Scholar]
- 46.Skoulidis F., Heymach J.V. Co-occurring genomic alterations in non-small cell lung cancer biology and therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fell J.B., Fischer J.P., Baer B.R., Blake J.F., Bouhana K., Briere D.M., Brown K.D., Burgess L.E., Burns A.C., Burkard M.R., et al. Identification of the clinical development candidate MRTX849, a covalent KRAS(G12C) inhibitor for the treatment of cancer. J. Med. Chem. 2020;63:6679–6693. doi: 10.1021/acs.jmedchem.9b02052. [DOI] [PubMed] [Google Scholar]
- 48.Goebel L., Müller M.P., Goody R.S., Rauh D. KRasG12C inhibitors in clinical trials: A short historical perspective. RSC Med. Chem. 2020;11:760–770. doi: 10.1039/D0MD00096E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwan A.K., Piazza G.A., Keeton A.B., Leite C.A. The Path to the Clinic: A Comprehensive Review on Direct KRASG12C Inhibitors. J. Exp. Clin. Cancer Res. 2022;41:27. doi: 10.1186/s13046-021-02225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.FDA FDA Approves First Targeted Therapy for Lung Cancer Mutation Previously Considered Resistant to Drug Therapy. [(accessed on 26 February 2024)]; Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-lung-cancer-mutation-previously-considered-resistant-drug.
- 52.Hong D.S., Fakih M.G., Strickler J.H., Desai J., Durm G.A., Shapiro G.I., Falchook G.S., Price T.J., Sacher A., Denlinger C.S., et al. KRAS G12C Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skoulidis F., Li B.T., Dy G.K., Price T.J., Falchook G.S., Wolf J., Italiano A., Schuler M., Borghaei H., Barlesi F., et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramalingam S., Skoulidis F., Govindan R., Velcheti V., Li B., Besse B., Dy G., Kim D., Schuler M., Vincent M., et al. P52.03 Efficacy of Sotorasib in KRAS p.G12C-Mutated NSCLC with Stable Brain Metastases: A Post-Hoc Analysis of CodeBreak 100. J. Thorac. Oncol. 2021;16:S1123. doi: 10.1016/j.jtho.2021.08.547. [DOI] [Google Scholar]
- 55.Skoulidis F., Schuler M., Wolf J., Barlesi F., Price T., Dy G., Govindan R., Borghaei H., Falchook G., Li B., et al. Genomic profiles and potential determinants of response and resistance in KRAS p/G12C-mutated NSCLC treated with sotorasib. J. Thorac. Oncol. 2020;16:S929–S930. doi: 10.1016/j.jtho.2021.08.184. [DOI] [Google Scholar]
- 56.Nakajima E.C., Drezner N., Li X., Mishra-Kalyani P.S., Liu Y., Zhao H., Bi Y., Liu J., Rahman A., Wearne E., et al. FDA Approval Summary: Sotorasib for KRAS G12C-Mutated Metastatic NSCLC. Clin. Cancer Res. 2021;28:1482–1486. doi: 10.1158/1078-0432.CCR-21-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reck M., Spira A., Besse B., Wolf J., Skoulidis F., Borghaei H., Goto K., Park K., Griesinger F., Felip E., et al. CodeBreak 200: A phase III multicenter study of sotorasib (AMG 510), a KRAS (G12C) inhibitor, versus docetaxel in patients with previously treated advanced non small cell lung cancer (NSCLC) harboring KRAS p. G12C mutation. Ann. Oncol. 2020;31:S894–S895. doi: 10.1016/j.annonc.2020.08.1730. [DOI] [Google Scholar]
- 58.Johnson M.L., De Langen J., Waterhouse D.M., Mazieres J., Dingemans A.C., Mountzios G., Pless M., Wolf J., Schuler M., Lena H., et al. Sotorasib versus Docetaxel for Previously Treated Non-Small Cell Lung Cancer with KRASG12C Mutation: CodeBreaK 200 Phase III Study. ESMO Congress 2022, LBA10. [(accessed on 26 February 2024)]. Available online: https://dailyreporter.esmo.org/esmo-congress-2022/news/sotorasib-improves-pfs-versus-docetaxel-in-patients-with-pre-treated-kras-g12c-mutated-nsclc.
- 59.Lumakras®/Lumykras®(Sotorasib) Demonstrates Superior Progression-Free Survival over Docetaxel in First Positive Phase 3 Trial of a Kras G12c Inhibitor in Non-Small Cell Lung Cancer. [(accessed on 26 February 2024)]. Available online: https://www.prnewswire.com/news-releases/lumakraslumykras-sotorasib-demonstrates-superior-progression-free-survival-over-docetaxel-in-first-positive-phase-3-trial-of-a-kras-g12c-inhibitor-in-non-small-cell-lung-cancer-301621632.html.
- 60.Hong D.S., Strickler J.H., Fakih M., Falchook G.S., Li B.T., Durm G.A., Burns T.F., Ramalingam S.S., Goldberg S.B., Frank R.C., et al. Trial in progress: A phase 1b study of sotorasib, a specific and irreversible KRAS G12C inhibitor, as monotherapy in non-small cell lung cancer (NSCLC) with brain metastasis and in combination with other anticancer therapies in advanced solid tumors (CodeBreaK 101) J. Clin. Oncol. 2021;39:TPS2669. doi: 10.1200/JCO.2021.39.15_suppl.TPS2669. [DOI] [Google Scholar]
- 61.Hallin J., Engstrom L.D., Hargis L., Calinisan A., Aranda R., Briere D.M., Sudhakar N., Bowcut V., Baer B.R., Ballard J.A., et al. The KRAS G12C Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ou S.-H.I., Jänne P.A., Leal T.A., Rybkin I.I., Sabari J.K., Barve M.A., Bazhenova L.A., Johnson M.L., Velastegui K.L., Cilliers C., et al. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients With Advanced KRASG12C Solid Tumors (KRYSTAL1) J. Clin. Oncol. 2022;40:2530–2538. doi: 10.1200/JCO.21.02752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papadopoulos K.P., Ou S.H.I., Johnson M.L., Christensen J., Velastegui K., Potvin D., Faltaos D., Chao R.C. A phase I/II multiple expansion cohort trial of MRTX849 in patients with advanced solid tumors with KRAS G12C mutation. J. Clin. Oncol. 2019;37:TPS3161. doi: 10.1200/JCO.2019.37.15_suppl.TPS3161. [DOI] [Google Scholar]
- 64.Jänne P.A., Papadopoulos K., Rybkin I., Johnson M. A Phase 1 Clinical Trial Evaluating the Pharmacokinetics (PK), Safety, and Clinical Activity of MRTX849, a Mutant-Selective Small Molecule KRAS G12C Inhibitor, in Advanced Solid Tumors. [(accessed on 26 February 2024)]. Available online: https://www.mirati.com/wp-content/uploads/AACR-NCI-EORTC-Clinical-Data-Presentation_Janne_October-2019-1-1.pdf.
- 65.Jänne P.A., Riely G.J., Gadgeel S.M., Heist R.S., Ou S.I., Pacheco J.M., Johnson M.L., Sabari J.K., Leventakos K., Yau E., et al. Adagrasib in non-small-cell lung cancer harboring a KRASG12C mutation. N. Engl. J. Med. 2022;387:120–131. doi: 10.1056/NEJMoa2204619. [DOI] [PubMed] [Google Scholar]
- 66.Spira A.I., Riely G.J., Gadgeel S.M., Heist R.S., Ou S.H.I., Pacheco J.M., Johnson M.L., Sabari J.K., Leventakos K., Yau E., et al. KRYSTAL-1: Activity and safety of adagrasib (MRTX849) in patients with advanced/metastatic non–small cell lung cancer (NSCLC) harboring a KRASG12C mutation. J. Clin. Oncol. 2022;40:9002. doi: 10.1200/JCO.2022.40.16_suppl.9002. [DOI] [Google Scholar]
- 67.New Drug Application for Adagrasib Accepted by FDA for KRAS G12C+ NSCLC. [(accessed on 26 February 2024)]. Available online: https://www.cancernetwork.com/view/new-drug-application-for-adagrasib-accepted-by-fda-for-kras-g12c-nsclc.
- 68.Mirati Therapeutics Inc. A Randomized Phase 3 Study of MRTX849 versus Docetaxel in Patients with Previously Treated Non-Small Cell Lung Cancer with KRAS G12C Mutation. [(accessed on 26 February 2024)];2022 Available online: https://clinicaltrials.gov/ct2/show/NCT04685135.
- 69.Mok T.S.K., Lawler W.E., Shum M.K., Dakhil S.R., Spira A.I., Barlesi F., Reck M., Garassino M.C., Spigel D.R., Alvarez D., et al. KRYSTAL-12: A randomized phase 3 study of adagrasib (MRTX849) versus docetaxel in patients (pts) with previously treated non-small-cell lung cancer (NSCLC) with KRASG12C mutation. J. Clin. Oncol. 2021;39:TPS9129. doi: 10.1200/JCO.2021.39.15_suppl.TPS9129. [DOI] [Google Scholar]
- 70.Riely G.J., Ou S.I., Rybkin I., Spira A., Papadopoulos K., Sabari J., Johnson M., Heist R., Bazhenova L., Barve M., et al. 99O_PR KRYSTAL-1: Activity and preliminary pharmacodynamic (PD) analysis of adagrasib (MRTX849) in patients (Pts) with advanced non–small cell lung cancer (NSCLC) harboring KRASG12C mutation. J Thoracic Oncol. 2021;16:S751–S752. doi: 10.1016/S1556-0864(21)01941-9. [DOI] [Google Scholar]
- 71.Evidence of Antitumor Effect with GDC-6036 Monotherapy in KRAS G12C+ NSCLC Revealed at 2022 WCLC. [(accessed on 26 February 2024)]. Available online: https://www.cancernetwork.com/view/evidence-of-antitumor-effect-with-gdc-6036-monotherapy-in-kras-g12c-nsclc-revealed-at-2022-wclc.
- 72.Sacher A., Patel M.R., Miller W.H., Desai J., Garralda E., Bowyer S., Kim T., De Miguel M., Falcon A., Krebs M., et al. OA03.04 Phase IA Study to Evaluate GDC-6036 Monotherapy in Patients with Non-small Cell Lung Cancer (NSCLC) with KRAS G12C Mutation. J. Thorac. Oncol. 2022;17:S8–S9. doi: 10.1016/j.jtho.2022.07.023. [DOI] [Google Scholar]
- 73.Luo J., Ostrem J., Pellini B., Imbody D., Stern Y., Solanki H.S., Haura E.B., Villaruz L.C. Overcoming KRAS-mutant lung cancer. Am. Soc. Clin. Oncol. Educ. Book. 2022;41:1–11. doi: 10.1200/EDBK_360354. [DOI] [PubMed] [Google Scholar]
- 74.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 75.Jiao D., Yang S. Overcoming resistance to drugs targeting KRASG12C mutation. Innovation. 2020;1:100035. doi: 10.1016/j.xinn.2020.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Janku F., Yap T.A., Meric-Bernstam F. Targeting the PI3K pathway in cancer: Are we making headway? Nat. Rev. Clin. Oncol. 2018;15:273–291. doi: 10.1038/nrclinonc.2018.28. [DOI] [PubMed] [Google Scholar]
- 77.Kordiak J., Szemraj J., Grabska-Kobylecka I., Bialasiewicz P., Braun M., Kordek R., Nowak D. Intratumor heterogeneity and tissue distribution of KRAS mutation in non-small cell lung cancer: Implications for detection of mutated KRAS oncogene in exhaled breath condensate. J. Cancer. Res. Clin. Oncol. 2019;145:241–251. doi: 10.1007/s00432-018-2779-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blaquier J.B., Cardona A.F., Recondo G. Resistance to KRAS (G12C) Inhibitors in Non-Small Cell Lung Cancer. Front. Oncol. 2021;11:787585. doi: 10.3389/fonc.2021.787585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Désage A.-L., Léonce C., Swalduz A., Ortiz-Cuaran S. Targeting KRAS Mutant in Non-Small Cell Lung Cancer: Novel Insights Into Therapeutic Strategies. Front. Oncol. 2022;12:796832. doi: 10.3389/fonc.2022.796832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koga T., Suda K., Fujino T., Ohara S., Hamada A., Nishino M., Chiba M., Shimoji M., Takemoto T., Arita T., et al. KRAS Secondary Mutations That Confer Acquired Resistance to KRAS G12C Inhibitors, Sotorasib and Adagrasib, and Overcoming Strategies: Insights From In Vitro Experiments. J. Thorac. Oncol. 2021;16:1321–1332. doi: 10.1016/j.jtho.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 81.Tanaka N., Lin J.J., Li C., Ryan M.B., Zhang J., Kiedrowski L.A., Michel A.G., Syed M.U., Fella K.A., Sakhi M., et al. Clinical acquired resistance to KRASG12C inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11:1913–1922. doi: 10.1158/2159-8290.CD-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Awad M.M., Liu S., Rybkin I.I., Arbour K.C., Dilly J., Zhu V.W., Johnson M.L., Heist R.S., Patil T., Riely G.J., et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xue J.Y., Zhao Y., Aronowitz J., Mai T.T., Vides A., Qeriqi B., Kim D., Li C., de Stanchina E., Mazutis L., et al. Rapid nonuniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature. 2020;577:421–425. doi: 10.1038/s41586-019-1884-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ryan M.B., Fece de la Cruz F., Phat S., Myers D.T., Wong E., Shahzade H.A., Hong C.B., Corcoran R.B. Vertical pathway inhibition overcomes adaptive feedback resistance to KRAS(G12C) inhibition. Clin. Cancer Res. 2020;26:1633–1643. doi: 10.1158/1078-0432.CCR-19-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim D., Xue J.Y., Lito P. Targeting KRAS (G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell. 2020;183:850–859. doi: 10.1016/j.cell.2020.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosell R., Aguilar A., Pedraz C., Chaib I. KRAS inhibitors, approved. Nat. Cancer. 2021;2:1254–1256. doi: 10.1038/s43018-021-00289-3. [DOI] [PubMed] [Google Scholar]
- 87.Nagasaka M., Potugari B., Nguyen A., Sukari A., Azmi A.S., Ou S.-H.I. KRAS Inhibitors- yes but what next? Direct targeting of KRAS vaccines, adoptive T cell therapy and beyond. Cancer Treat. Rev. 2021;101:102309. doi: 10.1016/j.ctrv.2021.102309. [DOI] [PubMed] [Google Scholar]
- 88.Knight T., Irving J.A. Ras/Raf/MEK/ERK Pathway Activation in Childhood Acute Lymphoblastic Leukemia and Its Therapeutic Targeting. Front. Oncol. 2014;4:160. doi: 10.3389/fonc.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Santarpia L., Lippman S.M., El-Naggar A.K. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets. 2012;16:103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cseh B., Doma E., Baccarini M. “RAF” neighborhood: Protein-protein interaction in the Raf/Mek/Erk pathway. FEBS Lett. 2014;588:2398–2406. doi: 10.1016/j.febslet.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rauen K.A. The RASopathies. Annu. Rev. Genom. Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Capelletto E., Bironzo P., Denis L., Koustenis A., Bungaro M., Novello S. Single agent VS6766 or VS-6766 plus defactinib in KRAS-mutant nonsmall-cell lung cancer: The RAMP-202 phase II trial. Future Oncol. 2022;18:1907–1915. doi: 10.2217/fon-2021-1582. [DOI] [PubMed] [Google Scholar]
- 93.Cucurull M., Notario L., Sanchez-Cespedes M., Hierro C., Estival A., Carcereny E., Saigí M. Targeting KRAS in Lung Cancer Beyond KRAS G12C Inhibitors: The Immune Regulatory Role of KRAS and Novel Therapeutic Strategies. Front. Oncol. 2022;11:793121. doi: 10.3389/fonc.2021.793121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bendell J.C., Rodon J., Burris H.A., de Jonge M., Verweij J., Birle D., Demanse D., De Buck S.S., Ru Q.C., Peters M., et al. Phase I, dose escalation study of BKM120, an oral pan Class I PI3K inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2012;30:282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 95.Ding X., Morrison G., Dean B., Hop C., Tobler L., Percey S., Meng M., Reuschel S., West D., Holden S., et al. A solid phase extraction-liquid chromatographic-tandem mass spectrometry method for determination of concentrations of GDC-0941, a small molecule class I phosphatidylinositide 3-kinase inhibitor, to support clinical development. J. Pharm. Biomed. Anal. 2012;61:1–7. doi: 10.1016/j.jpba.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 96.Shapiro G.I., Rodon J., Bedell C., Kwak E.L., Baselga J., Braña I., Pandya S.S., Scheffold C., Laird A.D., Nguyen L.T., et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of SAR245408 (XL147), an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 2014;20:233–245. doi: 10.1158/1078-0432.CCR-13-1777. [DOI] [PubMed] [Google Scholar]
- 97.Misale S., Fatherree J.P., Cortez E., Li C., Bilton S., Timonina D., Myers D.T., Lee D., Gomez-Caraballo M., Greenberg M., et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin. Cancer Res. 2019;25:796–807. doi: 10.1158/1078-0432.CCR-18-0368. [DOI] [PubMed] [Google Scholar]
- 98.Konstantinidou G., Ramadori G., Torti F., Kangasniemi K., Ramirez R.E., Cai Y., Behrens C., Dellinger M.T., Brekken R.A., Wistuba I.I., et al. RHOAFAK is a required signaling axis for the maintenance of KRAS-driven lung adenocarcinomas. Cancer Discov. 2013;3:444–457. doi: 10.1158/2159-8290.CD-12-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gerber D.E., Camidge D.R., Morgensztern D., Cetnar J., Kelly R.J., Ramalingam S.S., Spigel D.R., Jeong W., Scaglioni P.P., Zhang S., et al. Phase 2 study of the focal adhesion kinase inhibitor defactinib (VS-6063) in previously treated advanced KRAS mutant non-small cell lung cancer. Lung Cancer. 2020;139:60–67. doi: 10.1016/j.lungcan.2019.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altaf R., Ilyas U., Ma A., Shi M. Identification and validation of differentially expressed genes for targeted therapy in NSCLC using integrated bioinformatics analysis. Front. Oncol. 2023;13:1206768. doi: 10.3389/fonc.2023.1206768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang S.H., Baek H.A., Lee H.J., Park H.S., Jang K.Y., Kang M.J., Lee D.G., Lee Y.C., Moon W.S., Chung M.J. Discoidin domain receptor 1 is associated with poor prognosis of non-small cell lung carcinomas. Oncol. Rep. 2010;24:311–319. doi: 10.3892/or_00000861. [DOI] [PubMed] [Google Scholar]
- 102.Khozooei S., Veerappan S., Toulany M. YB-1 activating cascades as potential targets in KRAS-mutated tumors. Strahlenther Onkol. 2023;199:1110–1127. doi: 10.1007/s00066-023-02092-8. [DOI] [PubMed] [Google Scholar]
- 103.Maier E., Attenberger F., Tiwari A., Lettau K., Rebholz S. Dual Targeting of Y-Box Binding Protein-1 and Akt Inhibits Proliferation and Enhances the Chemosensitivity of Colorectal Cancer Cells. Cancers. 2019;11:562. doi: 10.3390/cancers11040562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zuehlke A.D., Moses M.A., Neckers L. Heat shock protein 90: Its inhibition and function. Philos. Trans. R Soc. Lond. B Biol. Sci. 2018;373:20160527. doi: 10.1098/rstb.2016.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Socinski M.A., Goldman J., El-Hariry I., Koczywas M., Vukovic V., Horn L., Paschold E., Salgia R., West H., Sequist L.V., et al. A multicenter phase II study of ganetespib monotherapy in patients with genotypically defined advanced non-small cell lung cancer. Clin. Cancer Res. 2013;19:3068–3077. doi: 10.1158/1078-0432.CCR-12-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pillai R.N., Fennell D.A., Kovcin V., Ciuleanu T.E., Ramlau R., Kowalski D., Schenker M., Yalcin I., Teofilovici F., Vukovic V.M., et al. Randomized Phase III Study of Ganetespib, a Heat Shock Protein 90 Inhibitor, With Docetaxel Versus Docetaxel in Advanced Non-Small Cell Lung Cancer (GALAXY-2) J. Clin. Oncol. 2020;38:613–622. doi: 10.1200/JCO.19.00816. [DOI] [PubMed] [Google Scholar]
- 107.Gutierrez M., Guo R., Giaccone G., Liu S.V., Hao Z., Hilton C., Hinson J.M., Jr., Kris M.G., Orlemans E.O., Drilon A. Phase 1 multicenter study of the HSP90 inhibitor SNX-5422 plus carboplatin and paclitaxel in patients with lung cancers. Lung Cancer. 2021;162:23–28. doi: 10.1016/j.lungcan.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Puyol M., Martín A., Dubus P., Mulero F., Pizcueta P., Khan G., Guerra C., Santamaría D., Barbacid M. A synthetic lethal interaction between K-Ras oncogenes and Cdk4 unveils a therapeutic strategy for non-small cell lung carcinoma. Cancer Cell. 2010;18:63–73. doi: 10.1016/j.ccr.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 109.Goldman J.W., Mazieres J., Barlesi F., Dragnev K.H., Koczywas M., Göskel T., Cortot A.B., Girard N., Wesseler C., Bischoff H., et al. A randomized phase III study of abemaciclib versus erlotinib in patients with stage IV non-small cell lung cancer with a detectable KRAS mutation who failed prior platinum-based therapy: JUNIPER. Front. Oncol. 2020;10:578756. doi: 10.3389/fonc.2020.578756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Passiglia F., Cappuzzo F., Alabiso O., Bettini A.C., Bidoli P., Chiari R., Defferrari C., Delmonte A., Finocchiaro G., Francini G., et al. Efficacy of nivolumab in pre-treated nonsmall-cell lung cancer patients harbouring KRAS mutations. Br. J. Cancer. 2019;120:57–62. doi: 10.1038/s41416-018-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou H., Dai Y., Zhu L., Wang C., Fei X. Poor response to platinum-based chemotherapy is associated with KRAS mutation and concomitant low expression of BRAC1 and TYMS in NSCLC. J. Inter. Med. Res. 2016;44:89–98. doi: 10.1177/0300060515607383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jia Y., Jiang T., Li X., Zhao C., Zhang L. Characterization of distinct types of KRAS mutation and its impact on first line platinum based chemotherapy in Chinese patients with advanced non small cell lung cancer. Oncol. Let. 2017;14:6525–6532. doi: 10.3892/ol.2017.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borghaei H., Paz-Ares L., Horn L., Spigel D.R., Steins M., Ready N.E., Chow L.Q., Vokes E.E., Felip E., Holgado E., et al. Nivolumab versus docetaxel in advanced nonsquamous nonsmall-cell lung cancer. N. Engl. J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brahmer J., Reckamp K.L., Baas P., Crinò L., Eberhardt W.E.E., Poddubskaya E., Antonia S., Pluzanski A., Vokes E.E., Holgado E., et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015;373:123–135. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rittmeyer A., Barlesi F., Waterkamp D., Park K., Ciardiello F., von Pawel J., Gadgeel S.M., Hida T., Kowalski D.M., Dols M.C., et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicenter randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Herbst R.S., Baas P., Kim D.W., Felip E., Pérez-Gracia J.L., Han J.Y., Molina J., Kim J.H., Arvis C.D., Ahn M.J., et al. Pembrolizumab versus docetaxel for previously treated, PD-L1positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 117.Spigel D.R., Schrock A.B., Fabrizio D., Frampton G.M., Sun J., He J., Gowen K., Johnson M.L., Bauer T.M., Kalemkerian G.P., et al. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J. Clin. Oncol. 2016;34:9017. doi: 10.1200/JCO.2016.34.15_suppl.9017. [DOI] [Google Scholar]
- 118.Kadara H., Choi M., Zhang J., Parra E.R., Rodriguez-Canales J., Gaffney S.G., Zhao Z., Behrens C., Fujimoto J., Chow C., et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann. Oncol. 2017;28:75–82. doi: 10.1093/annonc/mdw436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen N., Fang W., Lin Z., Peng P., Wang J., Zhan J., Hong S., Huang J., Liu L., Sheng J., et al. KRAS Mutation-Induced Upregulation of PD-L1 Mediates Immune Escape in Human Lung Adenocarcinoma. Cancer Immunol. Immunother. 2017;66:1175–1187. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu C., Zheng S., Jin R., Wang X., Wang F., Zang R., Xu H., Lu Z., Huang J., Lei Y., et al. The Superior Efficacy of Anti-PD1/PD-L1 Immunotherapy in KRAS-Mutant Non-Small Cell Lung Cancer That Correlates with an Inflammatory Phenotype and Increased Immunogenicity. Cancer Lett. 2020;470:95–105. doi: 10.1016/j.canlet.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 121.D’Incecco A., Andreozzi M., Ludovini V., Rossi E., Capodanno A., Landi L., Tibaldi C., Minuti G., Salvini J., Coppi E., et al. PD-1 and PD-L1 Expression in Molecularly Selected Non-Small-Cell Lung Cancer Patients. Br. J. Cancer. 2015;112:95–102. doi: 10.1038/bjc.2014.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salem M.E., El-Refai S.M., Sha W., Puccini A., Grothey A., George T.J., Hwang J.J., O’Neil B., Barrett A.S., Kadakia K.C., et al. Landscape of KRASG12C, Associated Genomic Alterations, and Interrelation With Immuno-Oncology Biomarkers in KRASMutated Cancers. JCO Precis. Oncol. 2022;6:e2100245. doi: 10.1200/PO.21.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Herbst R., Lopes G., Kowalski D., Kasahara K., Wu Y.-L., De Castro G., Cho B., Turna H., Cristescu R., Aurora-Garg D., et al. LBA4 Association of KRAS mutational status with response to pembrolizumab monotherapy given as first-line therapy for PD-L1-positive advanced non-squamous NSCLC in Keynote-042. Ann. Oncol. 2019;30:xi63–xi64. doi: 10.1093/annonc/mdz453.001. [DOI] [Google Scholar]
- 124.Mazieres J., Drilon A., Lusque A., Mhanna L., Cortot A., Mezquita L., Thai A.A., Mascaux C., Couraud S., Veillon R., et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: Results from the IMMUNOTARGET registry. Ann. Oncol. 2019;30:1321–1328. doi: 10.1093/annonc/mdz167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu C., Zheng S., Wang Z., Wang S., Wang X., Yang L., Xu H., Cao Z., Feng X., Xue Q., et al. KRAS-G12D Mutation Drives Immune Suppression and the Primary Resistance of Anti-PD-1/PD-L1 Immunotherapy in Non-Small Cell Lung Cancer. Cancer Commun. 2022;42:828–847. doi: 10.1002/cac2.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ricciuti B., Alessi J.V., Elkrief A., Wang X., Cortellini A., Li Y.Y., Vaz V.R., Gupta H., Pecci F., Barrichello A., et al. Dissecting the Clinicopathologic, Genomic, and Immunophenotypic Correlates of KRASG12D-Mutated Non-Small-Cell Lung Cancer. Ann. Oncol. 2022;33:1029–1040. doi: 10.1016/j.annonc.2022.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheng W., Xu B., Zhang H., Fang S. Lung Adenocarcinoma Patients with KEAP1 Mutation Harboring Low Immune Cell Infiltration and Low Activity of Immune Environment. Thorac. Cancer. 2021;12:2458–2467. doi: 10.1111/1759-7714.14089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ricciuti B., Arbour K.C., Lin J.J., Vajdi A., Vokes N., Hong L., Zhang J., Tolstorukov M.Y., Li Y.Y., Spurr L.F., et al. Diminished Efficacy of Programmed Death-(Ligand)1 Inhibition in STK11- and KEAP1-Mutant Lung Adenocarcinoma Is Affected by KRAS Mutation Status. J. Thorac. Oncol. 2022;17:399–410. doi: 10.1016/j.jtho.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dong Z.-Y., Zhong W.-Z., Zhang X.-C., Su J., Xie Z., Liu S.-Y., Tu H.-Y., Chen H.-J., Sun Y.L., Zhou Q., et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 130.Assoun S., Theou-Anton N., Nguenang M., Cazes A., Danel C., Abbar B., Pluvy J., Gounant V., Khalil A., Namour C., et al. Association of TP53 Mutations with Response and Longer Survival under Immune Checkpoint Inhibitors in Advanced Non Small-Cell Lung Cancer. Lung Cancer. 2019;132:65–71. doi: 10.1016/j.lungcan.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 131.Briere D.M., Li S., Calinisan A., Sudhakar N., Aranda R., Hargis L., Peng D.H., Deng J., Engstrom L.D., Hallin J., et al. The KRAS (G12C) Inhibitor MRTX849 Reconditions the Tumor Immune Microenvironment and Sensitizes Tumors to Checkpoint Inhibitor Therapy. Mol. Cancer Ther. 2021;20:975–985. doi: 10.1158/1535-7163.MCT-20-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li B.T., Falchook G.S., Durm G.A., Burns T.F., Skoulidis F., Ramalingam S.S., Spira A., Bestvina C.M., Goldberg S.B., Veluswamy R., et al. OA03.06 CodeBreaK 100/101: First Report of Safety/Efficacy of Sotorasib in Combination with Pembrolizumab or Atezolizumab in Advanced KRAS p.G12C NSCLC. J. Thorac. Oncol. 2022;17:S10–S11. doi: 10.1016/j.jtho.2022.07.025. [DOI] [Google Scholar]
- 133.Mirati Therapeutics Inc. A Phase 2 Trial of MRTX849 Monotherapy and in Combination with Pembrolizumab in Patients with Advanced Non-Small Cell Lung Cancer with KRAS G12C Mutation. [(accessed on 26 February 2024)]; Available online: https://clinicaltrials.gov/ct2/show/NCT04613596.
- 134.Xie M., Xu X., Fan Y. KRAS-mutant non-small cell lung cancer: An emerging promisingly treatable subgroup. Front. Oncol. 2021;11:672612. doi: 10.3389/fonc.2021.672612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Renaud S., Seitlinger J., Falcoz P.E., Schaeffer M., Voegeli A.C., Legrain M., Beau-Faller M., Massard G. Specific KRAS amino acid substitutions and EGFR mutations predict site-specific recurrence and metastasis following non-small-cell lung cancer surgery. Br. J. Cancer. 2016;115:346–353. doi: 10.1038/bjc.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mellema W.W., Masen-Poos L., Smit E.F., Hendriks L.E., Aerts J.G., Termers A., Goosens M.J., Smit H.J., Heuvel M.M.v.D., van der Wekken A.J., et al. Comparison of clinical outcome after first-line platinum based chemotherapy in different types of KRAS mutated advanced non-small-cell lung cancer. Lung Cancer. 2015;90:249–254. doi: 10.1016/j.lungcan.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 137.Garassino M.C., Marabese M., Rusconi P., Rulli E., Martelli O., Farina G., Scanni A., Broggini M. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in nonsmall-cell lung cancer. Ann. Oncol. 2011;22:235–237. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 138.Pan L.N., Ma Y.F., Li Z., Hu J.A., Xu Z.H. KRAS G12V mutation upregulates PD-L1 expression via TGF-β/EMT signaling pathway in human nonsmall-cell lung cancer. Cell Biol. Int. 2021;45:795–803. doi: 10.1002/cbin.11524. [DOI] [PubMed] [Google Scholar]
- 139.Koltun E., Cregg J., Rice M.A., Whalen D.M., Freilich R., Jiang J., Hansen R., Bermingham A., Knox J.E., Dinglasan J., et al. Abstract 1260: First-in-Class, Orally Bioavailable KRASG12V(ON) Tri-Complex Inhibitors, as Single Agents and in Combinations, Drive Profound Anti-Tumor Activity in Preclinical Models of KRASG12V Mutant Cancers. Cancer Res. 2021;81:1260. doi: 10.1158/1538-7445.AM2021-1260. [DOI] [Google Scholar]
- 140.Gao G., Liao W., Ma Q., Zhang B., Chen Y., Wang Y. KRAS G12D mutation predicts lower TMB and drives immune suppression in lung adenocarcinoma. Lung Cancer. 2020;149:41–45. doi: 10.1016/j.lungcan.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 141.Wang X., Allen S., Blake J.F., Bowcut V., Briere D.M., Calinisan A., Dahlke J.R., Fell J.B., Fischer J.P., Gunn R.J., et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS (G12D) Inhibitor. J. Med. Chem. 2022;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 142.Békés M., Langley D.R., Crews C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022;21:181–200. doi: 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bond M.J., Chu L., Nalawansha D.A., Li K., Crews C.M. Targeted degradation of oncogenic KRAS G12C by VHL-recruiting PROTACs. ACS Cent. Sci. 2020;6:1367–1375. doi: 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.A Study of mRNA-5671/V941 as Monotherapy and in Combination with Pembrolizumab (V941-001) [(accessed on 26 February 2024)]; Available online: https://classic.clinicaltrials.gov/ct2/show/NCT03948763.
- 145.Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M., Karantza V., et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bartolacci C., Andreani C., Vale G., Berto S., Melegari M. Targeting de novo lipogenesis and the Lands cycle induces ferroptosis in KRAS-mutant lung cancer. Nat. Comm. 2022;13:4327. doi: 10.1038/s41467-022-31963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bortoletto A.S., Parchem R.J. KRAS Hijacks the miRNA Regulatory Pathway in Cancer. Cancer Res. 2023;83:1563–1572. doi: 10.1158/0008-5472.CAN-23-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 149.McLoed A.G., Sherrill T.P., Cheng D.S., Han W., Saxon J.A., Gleaves L.A., Wu P., Polosukhin V.V., Karin M., Yull F.E., et al. Neutrophil-Derived IL-1β Impairs the Efficacy of NF-κB Inhibitors against Lung Cancer. Cell Rep. 2016;16:120–132. doi: 10.1016/j.celrep.2016.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Müller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/bj20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Deng S., Clowers M.J., Velasco W.V., Ramos-Castaneda M., Moghaddam S.J. Understanding the Complexity of the Tumor Microenvironment in K-ras Mutant Lung Cancer: Finding an Alternative Path to Prevention and Treatment. Front. Oncol. 2019;9:1556. doi: 10.3389/fonc.2019.01556. [DOI] [PMC free article] [PubMed] [Google Scholar]