Abstract
Herpes simplex virus genes are predominantly intronless. We identified cis-acting elements in the intronless herpes simplex virus type 1 thymidine kinase (TK) gene that facilitate intron-independent gene expression. TK sequences functionally replaced the hepatitis B virus (HBV) posttranscriptional regulatory element (PRE) by inducing the expression of the intronless HBV surface message. TK also activated the pDM138 assay by inducing the cytoplasmic accumulation of intron-containing RNA. Multiple cis-acting RNA sequences, or subelements, that induce cytoplasmic localization of unspliced RNA were mapped within the TK gene. The presence of multiple RNA subelements within the TK gene is reminiscent of the multiple subelements in the HBV PRE required for the cytoplasmic accumulation of intronless HBV RNAs. Similar to HBV PRE subelements, duplication of a single TK subelement resulted in greater-than-additive increases in activity. A reporter chimera containing a single TK subelement juxtaposed to an HBV PRE subelement demonstrated a commensurate increase in activity. These results suggest that viral intronless genes utilize a similar strategy for intron-independent gene expression that requires multiple cis-acting RNA signals. Furthermore, like HBV PRE-containing RNA, TK cytoplasmic localization is not sensitive to leptomycin B, a drug that inhibits the export of proteins containing nuclear export signals. From this, we conclude that proteins that bind TK and facilitate its cytoplasmic accumulation do not travel through a CRM1-dependent RNA transport pathway.
The study of intron-dependent gene expression has, in large part, been inseparable from that of intron-independent gene expression. Several groups that investigated the intron dependence of β-globin gene expression also studied the intron independence of the thymidine kinase (TK) gene from herpes simplex virus type 1 (HSV-1) (3, 10). They concluded that β-globin gene expression absolutely requires the presence of an intron but TK expression is intron independent. In fact, the TK gene induced the cytoplasmic accumulation of unspliced RNA when placed 5′, but not when placed 3′, of a β-globin intron (10). The ability to generate cytoplasmic intron-containing RNA in an orientation-dependent manner suggested that TK may utilize an intronless RNA-processing pathway, distinct from that utilized by intron-containing messages, that lacked splicing. The groups concluded that sequences within the TK gene may be responsible for intron-independent gene expression, yet none of the groups mapped any activity to a specific region. A report by Liu and Mertz was the first to precisely map a positive cis-acting RNA sequence within TK that enables intron-independent expression of intronless β-globin (18). The 119-nucleotide sequence, designated a pre-mRNA-processing enhancer (PPE), was shown to interact with heterogeneous nuclear ribonucleoprotein (hnRNP) L from nuclear extracts in a sequence-specific manner. The PPE was postulated to increase the efficiency of intronless mRNA expression posttranscriptionally.
Besides the TK PPE, the only other known viral cis-acting RNA sequences responsible for the cytoplasmic accumulation of intronless RNAs are the hepatitis B virus (HBV) and woodchuck hepatitis virus posttranscriptional regulatory elements, HBV PRE and WPRE, respectively (5, 14, 16, 17). The HBV PRE has a bipartite structure composed of two subelements, α and β, while the WPRE is composed of three subelements, α, β, and γ (4, 5). Similar to the PPE, the PREs function independently of any virus-encoded proteins.
We investigated whether the TK gene and the PREs function similarly. The PPE of TK seemed a likely candidate for a cis-acting RNA element homologous to a PRE subelement. Liu and Mertz alluded to the possibility that more than one PPE is present within the TK gene. Furthermore, if there were multiple PPEs within the TK gene, we hypothesized that they would act cooperatively in facilitating cytoplasmic localization of unspliced RNA similar to PRE subelements (4, 5). By studying cis-acting RNA sequences within TK, we sought to determine whether the TK gene contains positive cis-acting sequences besides the PPE element capable of facilitating cytoplasmic accumulation of intronless RNA and whether the TK sequences behave similarly to PRE subelements.
MATERIALS AND METHODS
Plasmids.
The chloramphenicol acetyltransferase (CAT) reporter plasmid pDM138 (henceforth referred to as 138) has been previously reported (13). The TK gene fragments tested in this study were amplified by 40 cycles of PCR with an HSV TK (strain CL101) plasmid clone as a DNA template. To construct the TK reporter derivatives, 34-base oligonucleotides were synthesized and used to PCR amplify the fragment of interest from the TK DNA template. The oligonucleotides contained either one of two possible 5′ sequences (GCGCTCGAGAATCGAT or GCGGGATCCATCGAT), followed by 18 bases of the TK sequence. All TK nucleotides are numbered relative to the transcription initiation site (27). The PCR products were purified on a 2% agarose gel, digested with ClaI or BamHI, and ligated into either the ClaI or the BglII site of 138. The TK119LSO mutation was generated by PCR mutagenesis. The cytomegalovirus (CMV) surface gene expression construct HSAg (HBV surface antigen) was synthesized by amplifying nucleotides (nt) 135 to 1685 by utilizing HBV (accession no. D00329) DNA as the template. The amplified fragment was digested with SacI and BglII and ligated into a SacI-BglII-digested CMV expression construct. The HBV PRE was then removed from HSAg by digestion with EcoRV, and the vector was religated to yield the ΔR5 surface expression vector. TK(60-1242), TK(641-841), and WPRE PCR-amplified fragments were gel purified, digested with ClaI, and then ligated into the ClaI site of ΔR5.
Cell culture and transfections.
293 and CV-1 cells were grown at 37°C (5% CO2) in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. For transfection, a confluent 10-cm-diameter plate was split 1:250 into the wells of a 24-well plate approximately 24 h before transfection. For CAT assays, 293 cells were transfected in triplicate with 80 ng of the reporter plasmid, 10 ng of the β-galactosidase expression vector (pCH110; Pharmacia), and 310 ng of pUC118 via the CaPO4 method. 293 cells were transfected by adding the DNA-CaPO4 mixture to the medium. The medium was changed 16 h after transfection. Cells were harvested 36 to 48 h after transfection with Reporter Lysis Buffer (Promega), cellular debris was pelleted, lysates were normalized for transfection efficiency with β-galactosidase, and normalized amounts of lysate were used for CAT assays as previously described (4). For CAT assays utilizing leptomycin B (LMB), CaPO4 transfections were done as mentioned above, except that the precipitates were divided in half and added to cells that would be treated with and without LMB. At 18 h posttransfection, the medium on the cells was changed with fresh medium containing 5 nM LMB or medium alone. The cells were harvested 24 h later in Reporter Lysis Buffer and processed as described above. For HBV surface gene expression assays, CV-1 cells were transfected in duplicate via the CaPO4 method with 25 μg of surface expression reporter and 5 μg of CMV secreted alkaline phosphatase reporter. To prepare CV-1 cells for transfection, a confluent 10-cm-diameter plate was split 1:8 into 10-cm-diameter plates 24 h prior to transfection. To transfect CV-1 cells, the medium was removed and the DNA-CaPO4 mixture was added directly to the cells. After 10 min, 6 ml of medium was placed onto the cells. The medium was changed approximately 16 h after transfection. The medium was harvested 48 h later and assayed for HSAg.
CAT assay.
293 cells were resuspended in 200 μl of Reporter Lysis Buffer (Promega). The lysates were spun briefly to pellet insoluble debris. An aliquot of each lysate was assayed for β-galactosidase activity, which was then used to normalize each lysate for transfection efficiency. The normalized lysates were equalized with Reporter Lysis Buffer and incubated at 37°C for 1 h to several h with 1.5-nCi/ml [14C]chloramphenicol (50 to 60 mCi/mmol) and 1 mM acetyl coenzyme A in 50-μl volumes. The substrate and products were resolved by thin-layer chromatography and quantitated by a PhosphorImager (Molecular Dynamics). The mean percent acetylation was calculated from the percent conversion of each repeated point. The standard error of the mean was calculated by dividing the standard deviation of a datum set by the square root of the number of samples within the datum set. All experimental points were assayed in triplicate, each experiment was repeated at least twice, and representative results are presented.
Surface gene expression assay.
The medium from duplicate transfections was assayed for secreted alkaline phosphatase, which was then used to normalize each supernatant for transfection efficiency. Normalized amounts of supernatant were then assayed for the presence of HSAg with the Ausria II kit (Abbott Laboratories), and the HSAg was quantitated in a gamma counter. All experimental points were assayed in duplicate, each experiment was repeated at least twice, and representative results are presented.
RNA analysis.
For Northern blot analysis, 293 cells were transiently transfected with 10 μg of 138TK and 10 μg of 138RRE. A 1-μg sample of β-galactosidase reporter was included in each transfection. Two DNA-CaPO4 precipitates were prepared for each reporter and used to transfect two plates (10-cm diameter) each. This was done to achieve similar transfection efficiencies for all reporters. The cells were harvested and lysed in cytoplasmic lysis buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 0.1 mM EDTA, 20% glycerol, 0.5% Nonidet P-40). The lysed cells were spun at 8,000 × g, and the supernatant was recovered and spun for an additional 5 min at 14,000 × g. The supernatant was then transferred to 1 ml of RNA Stat-50LS (Tel-Test). The nuclear pellet from the first spin was resuspended in 1 ml of cytoplasmic lysis buffer and spun at 8,000 × g for 3 min. The supernatant was discarded, and the pellet was resuspended in 800 μl of nuclear buffer (10 mM Tris [pH 8.4], 1.5 mM MgCl2, 140 mM NaCl, 20% glycerol). The sample was centrifuged at 8,000 × g, and the supernatant was discarded. The pellet was resuspended in 300 μl of nuclear buffer and lysed with 1 ml of RNA Stat-50LS. The manufacturer’s RNA Stat-50 protocol was followed. After RNA purification, the samples were treated with DNase for 15 min at 37°C. Nuclear (5 μg) and cytoplasmic (10 μg) RNAs were run on a 1% agarose–formaldehyde gel and transferred to a nylon membrane (Stratagene). The Northern blot was prehybridized in QuikHyb (Stratagene) for 1 h at 65°C and then hybridized with 32P-labeled CAT and glyceraldehyde-3-phosphate dehydrogenase probes for 2 h at 65°C. The blot was washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and then in 0.1× SSC at 65°C and exposed to X-ray film, and an autoradiograph was obtained.
RESULTS
The TK gene can functionally replace the HBV PRE in an HSAg expression assay.
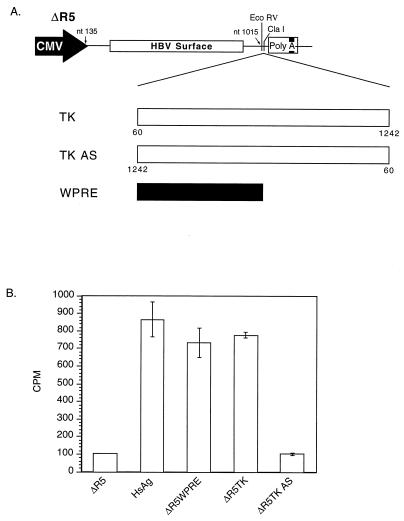
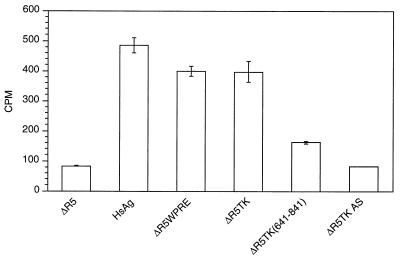
To assess whether the TK gene can functionally replace the HBV PRE, we utilized an intronless HSAg expression construct that is dependent on the HBV PRE for cytoplasmic localization of surface RNA (16). Previous studies have demonstrated that efficient HBV surface gene expression requires the PRE (16). We used an HSAg construct, ΔR5, with HBV PRE deleted to construct the surface reporter vectors (5). WPRE and TK nt 60 to 1242 (relative to the transcription start site [27]) were inserted into ΔR5 in the sense and antisense orientations (Fig. 1A) and transfected into CV-1 cells, and the supernatants were assayed for surface protein 48 h after transfection. The results in Fig. 1B demonstrate that ΔR5TK induced surface protein expression to levels similar to those of the positive HSAg control and ΔR5WPRE. The ΔR5 reporter with the TK gene in the antisense orientation had no increase in activity over that of the ΔR5 vector alone. We concluded that the TK gene can functionally replace PREs and induce cytoplasmic localization of intronless surface transcripts in an orientation-dependent manner.
FIG. 1.
(A) Schematic illustration of HBV surface vector ΔR5 and derivative reporter constructs containing the WPRE and the TK gene in the sense and antisense (AS) orientations. (B) Surface protein assay demonstrating the effect of TK gene sequences on surface protein expression. The TK gene induces surface gene expression to levels comparable to those produced by WPRE and the HSAg control which contains the HBV PRE. The TK gene in the antisense orientation has no effect over that of empty vector ΔR5.
The TK gene can localize unspliced reporter RNAs to the cytoplasm.
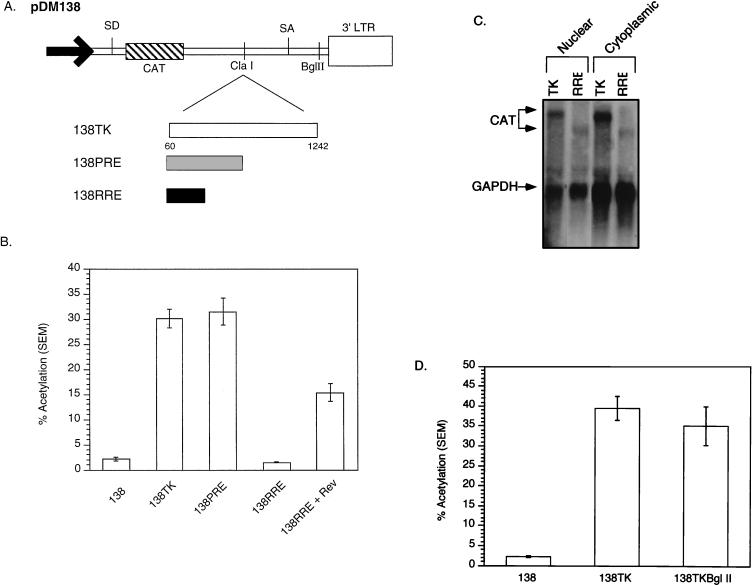
To delineate cis-acting RNA sequences within the TK gene, we employed a well-characterized reporter system used to map PRE subelements (4, 5) and the RNA export elements of complex retroviruses (12, 13, 19). The system utilizes pDM138, a plasmid reporter derived from the second intron of human immunodeficiency virus type 1 (HIV-1) in which the HIV env gene is replaced with the CAT gene (13). When pDM138 is transiently transfected, the CAT gene is translated only if unspliced RNAs accumulate in the cytoplasm. Export of unspliced HIV-1 Rev response element (RRE)-containing RNA depends on the presence of HIV-1 Rev.
To generate reporter plasmids, TK fragments were inserted into 138. TK gene nt 60 to 1242 were inserted into 138 in the sense orientation (Fig. 2A). The resulting reporter, 138TK, was transiently transfected into 293 cells, which were subsequently assayed for CAT activity. To compare the relative strength of the TK gene with regard to that of known cis-acting RNA elements, we also transfected 138 constructs containing the HBV PRE (138PRE) (4) and the HIV-1 RRE (138RRE) with pRSVRev. The results in Fig. 2B demonstrate that 138TK induces CAT activity comparably to 138PRE. The HBV PRE functions more efficiently than RRE/Rev due to the activation of PRE enhancer I in 293 cells. The 138 reporter with the TK gene in the antisense orientation did not induce any significant CAT activity (data not shown).
FIG. 2.
(A) Schematic illustration of the pDM138 constructs containing the TK gene, the HBV PRE, and the RRE. Base pairs are numbered from the transcription start site (27). SD, splice donor; SA, splice acceptor; LTR, long terminal repeat. (B) pDM138 CAT assay demonstrating induction of the cytoplasmic localization of unspliced, intron-containing RNA by TK gene sequences. The TK gene induces CAT expression to levels comparable to those induced by the HBV PRE. (C) Northern blot analysis of nuclear and cytoplasmic RNAs from 293 cells transiently transfected with 138TK and 138RRE. The blot was hybridized with a probe that detects CAT-containing RNAs. Both the TK- and RRE-containing CAT RNAs are present in the nucleus and cytoplasm at the predicted sizes for full-length, unspliced RNAs. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) pDM138 CAT assay demonstrating the ability of TK gene sequences to induce cytoplasmic localization of intron-containing RNA when positioned outside the 138 intron. The TK gene induces CAT expression to similar levels when positioned outside or within the intron. SEM, standard error of the mean.
To verify that 138TK CAT activity was due to the appearance of unspliced RNA in the cytoplasm, a Northern blot was performed on nuclear and cytoplasmic RNAs isolated from 293 cells transiently transfected with 138TK and 138RRE. Figure 2C illustrates that CAT-containing transcripts in the nucleus and cytoplasm derived from 138TK are unspliced.
TK can function within the intron or exon of unspliced RNA.
The HBV PRE is position independent, since it induces cytoplasmic accumulation of unspliced RNA when placed within the intron or exon of 138 (4). To determine whether TK is also position independent, TK was inserted into the BglII site within the 3′ exon of 138 (Fig. 2A). 138, 138TK, and 138TKBglII were transiently transfected into 293 cells, which were subsequently assayed for CAT activity. The results in Fig. 2D demonstrate that, similar to the HBV PRE, 138TK reporters induced CAT activity to similar levels whether positioned within the intron or the exon. The position independence of the TK gene in 138 further supports the hypothesis that sequences within the TK gene that facilitate cytoplasmic accumulation are functionally similar to the HBV PRE.
Deletion analysis of the TK gene.
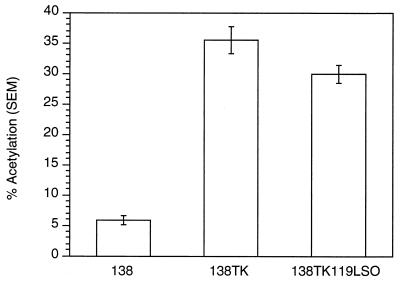
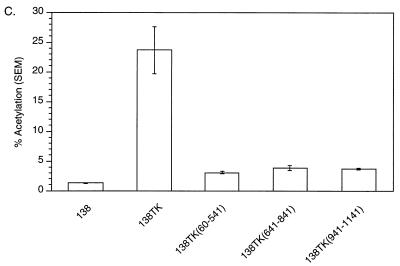
To test whether the PPE functioned as a cis-acting minimal element in the CAT assay, we constructed a 138TK reporter with a mutation in the PPE (119LSO) reported to knock out its function (18). This reporter (138TK119LSO), 138TK, and 138 were transiently transfected into 293 cells, which were then assayed for CAT activity. The 138TK119LSO reporter induced CAT activity 84% compared to 138TK (Fig. 3). A small but significant decrease in CAT activity was observed in the absence of PPE function. However, the large amount of remaining CAT activity in 138TK119LSO is evidence that the PPE is not the sole cis-acting RNA sequence within the TK gene and that other elements exist.
FIG. 3.
pDM138 CAT assay demonstrating the effect a PPE mutation has on the level of TK-induced CAT activity. The reporter containing the PPE-mutated TK gene, 138TK119LSO, has reduced activity (84%) compared to 138TK. SEM, standard error of the mean.
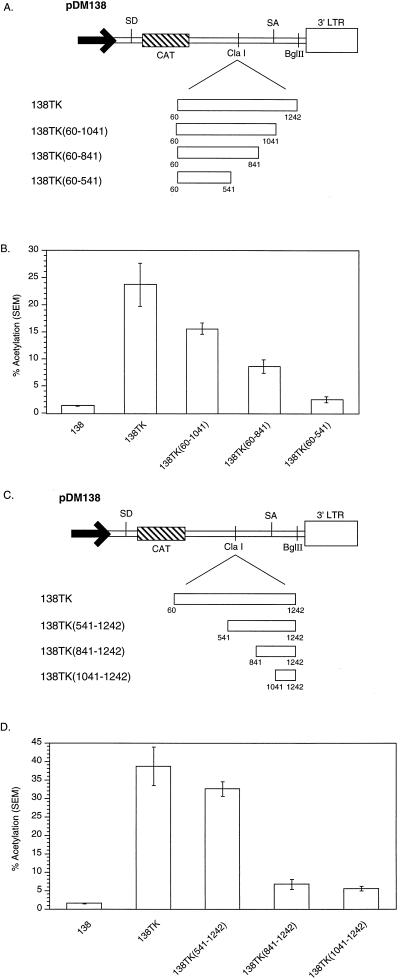
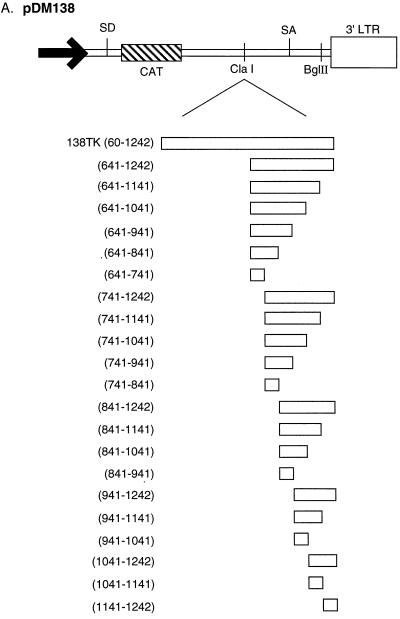
Several 5′ and 3′ deletions of the TK gene were cloned into 138 (Fig. 4A and C). These TK deletion constructs and 138 were transiently transfected into 293 cells, and they were assayed for CAT activity. The TK gene fragment including nt 60 to 541 contains the PPE (nt 361 to 479). As can be seen in Fig. 4B, 138TK(60-541) induced CAT activity 10% compared to 138TK. The larger 3′ deletions, 138TK(60-841) and 138TK(60-1041), induced CAT activity 36 and 66%, respectively, compared to 138TK. The low-level CAT activity observed with the PPE-containing reporter, 138TK(60-541), supports the conclusion reached by others that the PPE is a minimal activity element (18). Furthermore, the stepwise decrease in CAT activity produced by the 3′ deletions suggest that there are additional cis-acting RNA sequences within the TK gene, 3′ of the PPE, responsible for cytoplasmic localization of unspliced RNA.
FIG. 4.
(A) Schematic illustration of the pDM138 constructs containing the TK gene and several 3′ deletions. (B) pDM138 CAT assay demonstrating the stepwise decrease in activity of the 3′ TK deletions. (C) Schematic illustration of the pDM138 constructs containing the TK gene and several 5′ deletions. (D) pDM138 CAT assay demonstrating the stepwise decrease in activity of the 5′ TK deletions. For definitions of abbreviations, see the legend to Fig. 2.
The 5′ TK deletion construct 138TK(541-1242) induced CAT activity 80% compared to 138TK (Fig. 4D). Deletions 3′ of nt 541 led to large decreases in activity. 138TK(841-1242) and 138TK(1041-1242) retained only 15% of the CAT activity of 138TK. Similar to the results obtained with 138TK119LSO (Fig. 3), a significant decrease in CAT activity was caused by deleting the PPE. These results further support the conclusion that the PPE is a minimal activity element. Conversely, the large amount of activity induced by 138TK(541-1242) and the stepwise decrease in CAT activity caused by the 5′ deletions indicate that there are additional cis-acting RNA elements within this region, a conclusion also reached with the 3′ deletions. Therefore, extensive deletion analysis was performed to map the remaining minimal cis-acting RNA elements in the TK gene.
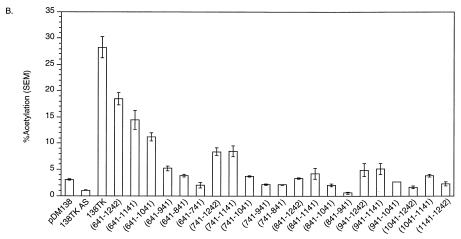
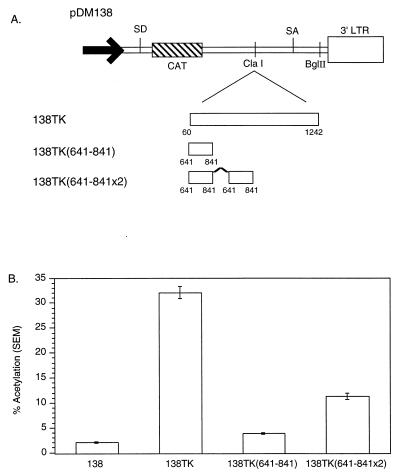
The TK gene contains three cis-acting subelements.
TK deletion analysis localized the cis-acting sequences to three regions in the gene. One cis-acting sequence maps to TK nt 60 to 541 and contains the PPE (Fig. 4B and data not shown). The other two previously unreported elements map to TK nt 641 to 841 and 941 to 1141 (Fig. 5B). Several similar experiments were performed to confirm the minimal activity of the TK subelements (data not shown). Each region separately induced a minimal amount of CAT activity in the 138 CAT assay, i.e., two- to threefold activation compared to 138 (Fig. 5C). Sequences that induce minimal CAT activity are defined as cis-acting RNA subelements. Activities similar to those reported here for TK subelements have been reported for HBV PRE and WPRE subelements (4, 5).
FIG. 5.
(A) Schematic illustration of the pDM138 constructs containing several TK internal deletions. (B) pDM138 CAT assay demonstrating the activity of several TK internal deletions used to map the remaining cis-acting elements in TK. 138TK AS contains TK in the antisense orientation. (C) pDM138 CAT assay demonstrating the activity of the three TK minimal cis-acting RNA subelements mapped by deletion analysis. For definitions of abbreviations, see the legend to Fig. 2.
To test whether a single TK gene subelement defined by the 138 assay would induce intronless surface RNA expression, we constructed a ΔR5 reporter that contained TK gene nt 641 to 841. This construct, ΔR5TK(641-841), was transiently transfected into CV-1 cells and then assayed for surface gene expression. As can be seen in Fig. 6, a single TK gene subelement induced surface gene expression 41% compared to ΔR5TK.
FIG. 6.
HBV surface gene expression assay demonstrating that one TK gene subelement, nt 641 to 841, is sufficient to induce intronless surface gene expression.
Duplications of TK gene subelements exhibit increased activity.
Donello et al. demonstrated that duplication of either HBV PRE α or β subelements resulted in an element that is at least twice as active as a single subelement (4). To examine whether TK gene subelements function similarly, TK nt 641 to 841 were duplicated and inserted into the 138 reporter (Fig. 7A). This 138TK(641-841x2) construct, 138TK(641-841), and 138 were transiently transfected into 293 cells, and the cells were assayed for CAT activity. The duplication of the TK(641-841) subelement increased CAT activity twice more than as much as a single subelement (Fig. 7B). While 138TK(641-841) induced 12% of the CAT activity induced by 138TK, 138TK(641-841x2) induced 35% of that induced by 138TK. Duplications of the PPE were also observed to increase reporter function in a greater-than-additive manner (18).
FIG. 7.
(A) Schematic illustration of the TK(641-841) subelement duplication construct in pDM138. (B) pDM138 CAT assay demonstrating that the TK(641-841) subelement duplication, TK(641-841x2), induces more than twice the CAT activity of a single subelement. For definitions of abbreviations, see the legend to Fig. 2.
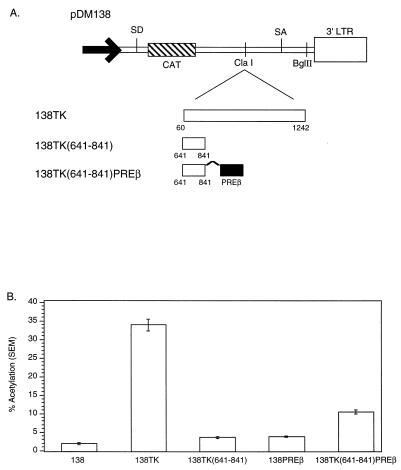
Chimeric constructs containing TK and HBV PRE subelements exhibit increased activity.
When duplicated, TK nt 641 to 841 behaved like HBV PRE subelements, suggesting that they are functionally similar. We hypothesized that if these different subelements are functionally similar, reporters containing multiple subelements from different intronless genes may induce activity similarly to reporters containing subelement duplications. To test this hypothesis, we created chimeric reporters containing an HBV PRE subelement juxtaposed to a TK subelement.
The HBV PREβ subelement was inserted 3′ of the TK nt 641 to 841 subelement (Fig. 8A) in 138. This chimeric construct [138TK(641-841)PREβ, 138TK], 138TK(641-841), 138PREβ, and 138 were transiently transfected into 293 cells, and the cells were assayed for CAT activity. The results shown in Fig. 8B demonstrate that (i) 138TK(641-841) and 138PREβ induced CAT activity to similar levels, i.e., 6% of that of 138TK, and (ii) when juxtaposed, TK 641-841 and HBV PREβ induced an increase in CAT activity that was greater than the additive effects of the separate subelements. 138TK(641-841)PREβ induced activity that was 25% of that of 138TK. The increase in activity of the chimeric subelement reporter suggests that HBV PRE and TK subelements are functionally similar.
FIG. 8.
(A) Schematic illustration of the TK(641-841) subelement and HBV PREβ subelement chimeric construct in pDM138. (B) pDM138 CAT assay demonstrating that the TK(641-841)PREβ chimeric construct induces CAT activity greater than the sum of the activities induced by individual subelements. For definitions of abbreviations, see the legend to Fig. 2.
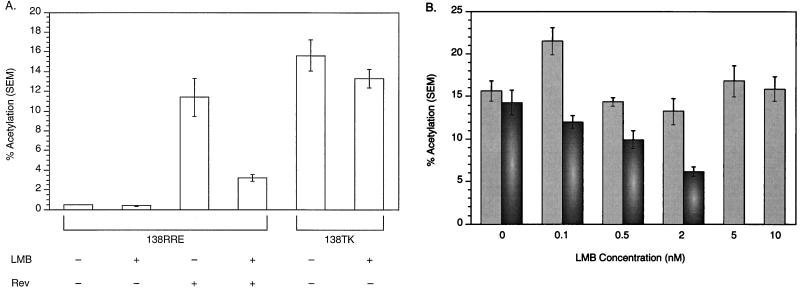
TK function is not CRM1 dependent.
The hnRNP L-PPE interaction was postulated to possibly function analogously to Rev export of RRE-containing RNA (18). Rev export depends on the presence of a leucine-rich nuclear export signal (NES; reviewed in reference 11). If TK RNA was actively exported through an NES-dependent pathway, like Rev, the cytoplasmic accumulation of TK RNA should be inhibited by LMB. LMB has been shown to block Rev and other NES-containing protein export (9, 20, 25, 28) by binding the NES receptor, CRM1 (exportin 1), thereby preventing the NES-exportin 1-RanGTP complex from assembling (8). LMB specifically blocks NES-dependent RNA export but does not block the mRNA or tRNA export pathway (8). Work in this laboratory has demonstrated that LMB does not inhibit cytoplasmic accumulation of Mason-Pfizer monkey virus constitutive transport element (CTE) (2, 6, 7, 26, 30)- or PRE (21)-containing RNA. We transiently transfected 293 cells with 138TK and 138RRE with pRSVRev in the presence and absence of LMB. Figure 9A demonstrates that while LMB inhibited Rev-induced CAT activity of RRE-containing RNA, LMB had no significant effect on 138TK CAT activity. To rule out the possibility that the difference in LMB sensitivity between RRE-containing RNA and TK RNA was dependent on the concentration of LMB used (5 nM), we performed transient transfections and CAT assays on 138RRE with pRSVRev and 138TK, in which the LMB concentration was titrated over a broad range. As can be seen in Fig. 9B, while Rev-dependent gene expression was maximally inhibited at 2 nM LMB, TK RNA expression was not significantly decreased over the range of LMB concentrations. PRE-containing RNA included in the titration experiments was also not affected by LMB as previously reported (data not shown). We conclude that, like the PRE and the CTE, TK RNA travels through a CRM1-independent RNA-processing pathway.
FIG. 9.
(A) Effect of LMB on TK-induced expression of intron-containing RNA in the pDM138 CAT assay. 138RRE was transfected with and without pRSVRev in the presence and absence of LMB. 138TK was transfected in the presence and absence of LMB. LMB had no significant effect on TK-induced CAT expression, while LMB inhibited Rev-induced CAT expression 72%. (B) Dose-response curve of LMB effects on Rev-dependent and TK-mediated gene expression in the CAT assay. 138TK (gray) and 138RRE and pRSVRev (dark gray) were transfected in the presence of increasing concentrations of LMB. As reported in reference 21, Rev-dependent CAT expression decreased until maximally inhibited at 2 nM LMB. TK-mediated CAT expression was not adversely affected between 0 and 10 nM LMB. SEM, standard error of the mean.
DISCUSSION
There has been much research regarding the role of introns in gene expression. Several reports support the hypothesis that introns, including β-globin introns, increase 3′-end formation and accumulation of polyadenylated RNA posttranscriptionally (3, 10, 15, 29). Similarly, several groups have demonstrated that TK sequences eliminate the intron dependence of intronless β-globin expression and facilitate the cytoplasmic accumulation of polyadenylated β-globin–TK hybrid RNA posttranscriptionally (1, 10, 18). It appears that TK sequences induce proper processing and cytoplasmic localization of the intronless hybrid RNA, presumably as it would for itself, in the absence of splicing. These observations suggest that a pathway or mechanism divergent from that used to process intron-containing RNAs may exist in order to process intronless RNAs. From transcription to export, these putative RNA-processing pathways may act in parallel at some steps in processing, while converging at others, including 3′ cleavage and polyadenylation.
Interestingly, TK sequences present upstream but not downstream of a β-globin intron suppress splicing of the intron and transport the intron-containing transcript to the cytoplasm (10). We also observed that TK induces cytoplasmic expression of intron-containing CAT RNA. It seems that cis-acting TK sequences are capable of driving intron-containing transcripts through a processing pathway that lacks splicing when they are upstream of or within an intron. However, when TK is 3′ of the 138 intron, it still induces expression of intron-containing RNA (Fig. 2D), contradictory to the report by Greenspan and Weissman (10). This discrepancy in TK effectiveness when it is 3′ of an intron likely depends on the efficiency of the splicing intron. The intronic and positional variables that affect the ability of TK sequences to override splicing suggest that there is a posttranscriptional competition between TK sequences attempting to process through a pathway devoid of splicing and the more conventional splicing pathway.
Our results support a hypothesis put forth by Greenspan and Weissman, who suggested that a gene which has evolved to express unspliced RNA may contain multiple regions that direct nuclear export of the transcript without splicing (10). We favor an alternative hypothesis in which the RNA subelements of the TK gene are bound by cellular proteins that increase the efficiency of RNA processing in the absence of splicing. In light of evidence that introns increase the efficiency of 3′ processing, and since TK mRNA lacks introns, we speculate that TK subelements may induce efficient 3′-end formation. TK subelements are capable of inducing the expression of intronless mRNAs like ΔR5 and β-globin. These mRNAs apparently lack some signals, like the PRE or an intron, required for expression. Additionally, the TK subelements are, in some instances, capable of overriding splicing and inducing the expression of intron-containing mRNAs. In these cases, despite the presence of an intron, TK subelements may induce proper processing of the mRNA without splicing, allowing it to be released from the nuclear retention normally associated with introns.
Donello et al. concluded that there is a direct correlation between the number of PRE subelements a 138WPRE reporter contains and the amount of CAT activity it induces (5). We observed the same correlation with TK subelements in the 138TK deletion analysis and the duplication of subelements (Fig. 4 and 7). It appears that the more TK or PRE subelements an intron-containing mRNA has, the more likely it is to be processed without being spliced. We speculate that the same correlation exists in the intronless context of the HSAg assay. One TK subelement induced a small amount of surface protein expression, while the entire TK gene induced 2.5 times as much activity (Fig. 6).
To address the possibility that cellular proteins bind TK subelements and induce nuclear export of RNA similar to Rev-induced export of RRE-containing RNA, we performed 138 CAT assays with 138TK in the presence and absence of LMB. Our results demonstrated that, similar to that of the PRE and the CTE (21–23), TK cytoplasmic localization was not analogous to the CRM1-dependent, Rev-induced nuclear export of RRE-containing RNA (Fig. 9). It is interesting that the CTE, the PRE, and the TK gene all utilize cellular factors and a CRM-1-independent pathway for cytoplasmic localization.
Since hnRNP L is not reported to have a leucine-rich NES, and hence may travel through a CRM1-independent pathway, these results do not rule out the putative role hnRNP L has in binding the PPE and inducing cytoplasmic localization. This finding is striking in light of a report claiming that ICP27 binds TK mRNA and facilitates its expression through an NES-dependent pathway (24). The discrepancies in TK mRNA pathway usage are most likely due to in vivo conditions that vary between HSV infections and the transient transfections that we and others have done to study TK expression. It is plausible that HSV has evolved a protein, similar to HIV-1 Rev, that induces the expression of several of its intronless mRNAs through an NES-dependent pathway at a time during HSV infection when the majority of cellular transcripts are not being expressed. However, in the absence of HSV infection and ICP27, TK mRNA is still efficiently expressed in transient transfections. Perhaps TK RNA expression proceeds through an intronless mRNA pathway early in HSV infection and then proceeds through an ICP27/CRM1-dependent pathway during late stages of infection. Experiments are under way in our laboratory that address the effect ICP27 has on TK expression in the 138 and HSAg assays. We are also conducting experiments to identify trans-acting factors, other than hnRNP L, that bind TK subelements.
In either the intronless HSAg assay or the intron-containing 138 assay, the entire TK gene functions as well as the HBV PRE. We have demonstrated that the coding sequence of the naturally intronless TK gene possesses three cis-acting RNA subelements. A single TK subelement increases RNA expression of intronless, as well as intron-containing, transcripts. Additionally, TK and HBV PRE subelements function cooperatively when duplicated and in chimeric reporters. The data support the hypothesis that TK and HBV PRE subelements are functionally similar and utilize the same RNA-processing pathway. We hypothesize that TK RNA travels through the mRNA pathway without splicing under normal intronless conditions. However, as has been shown by this laboratory and others, in some instances, TK subelements are strong enough to override splicing to localize intron-containing RNA to the cytoplasm. This type of posttranscriptional regulation in an intronless transcript has only been described previously in conjunction with the HBV PRE and the WPRE (4, 5). From these results, we hypothesize that the TK gene and the genes containing PREs are the first members of an emerging family of intronless viral genes that utilize multiple cis-acting sequences to facilitate their cytoplasmic localization. It remains to be determined at which stages of mRNA processing and export the PRE and TK subelements exert their effects. With multiple subelements and multiple steps in mRNA processing, there exists the opportunity for both specialization and redundancy of function for the subelements.
ACKNOWLEDGMENTS
We thank John Donello, Matthew Harris, Katja Straesser, and Mario McLean for assistance and helpful discussions. We thank Allison Bocksruker for help in preparing the manuscript.
This work was supported by National Institutes of Health grant AI35477 (T.J.H.). Glen Otero was supported by a Ford Foundation postdoctoral fellowship. T.J.H. is supported in part by the Gene and Ruth Posner Foundation.
REFERENCES
- 1.Bordonaro M, Nordstrom J L. Different mechanisms are responsible for the low accumulation of transcripts from intronless and 3′ splice site deleted genes. Biochem Biophys Res Commun. 1994;203:128–132. doi: 10.1006/bbrc.1994.2158. [DOI] [PubMed] [Google Scholar]
- 2.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjold M-L. A small element from Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchman A R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donello J E, Beeche A A, Smith G J, Lucero G R, Hope T J. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donello J E, Loeb J E, Hope T J. The woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst R K, Bray M, Rekosh D, Hammarskjöld M-L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst R K, Bray M, Rekosh D, Hammarskjöld M-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. . [Comments.] [DOI] [PubMed] [Google Scholar]
- 9.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 10.Greenspan D S, Weissman S M. Synthesis of predominantly unspliced cytoplasmic RNAs by chimeric herpes simplex virus type 1 thymidine kinase–human β-globin genes. Mol Cell Biol. 1985;5:1894–1900. doi: 10.1128/mcb.5.8.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hope T J. Viral RNA export. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 12.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type 1 Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hope T J, Huang X, McDonald D, Parslow T. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang M T F, Gorman C M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Z-M, Yen T S B. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z-M, Yen T S B. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Mertz J E. hnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 19.Mancuso V A, Hope T J, Zhu L, Derse D, Phillips T, Parslow T G. Posttranscriptional effector domains in the Rev proteins of feline immunodeficiency virus and equine infectious anemia virus. J Virol. 1994;68:1998–2001. doi: 10.1128/jvi.68.3.1998-2001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 21.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasquinelli A, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M-L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 CTE, unlike the HIV-1 RRE, utilises factors required for the export of cellular mRNAs. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 24.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A, Wang J Y J, Hope T J. Nuclear-cytoplasmic shuttling of c-abl tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner M J, Sharp J A, Summers W C. Nucleotide sequence of the thymidine kinase gene of herpes simplex virus type 1. Proc Natl Acad Sci USA. 1981;78:1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 29.Yu X-M, Gelembiuk G W, Wang C-Y, Ryu W-S, Mertz J E. Expression from herpesvirus promoters does not relieve the intron requirement for cytoplasmic accumulation of human β-globin mRNA. Nucleic Acids Res. 1991;19:7231–7234. doi: 10.1093/nar/19.25.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]