Abstract
Thaumatin-like proteins (TLPs) comprise a complex and evolutionarily conserved protein family that participates in host defense and several developmental processes in plants, fungi, and animals. Importantly, TLPs are plant host defense proteins that belong to pathogenesis-related family 5 (PR-5), and growing evidence has demonstrated that they are involved in resistance to a variety of fungal diseases in many crop plants, particularly legumes. Nonetheless, the roles and underlying mechanisms of the TLP family in legumes remain unclear. The present review summarizes recent advances related to the classification, structure, and host resistance of legume TLPs to biotic and abiotic stresses; analyzes and predicts possible protein–protein interactions; and presents their roles in phytohormone response, root nodule formation, and symbiosis. The characteristics of TLPs provide them with broad prospects for plant breeding and other uses. Searching for legume TLP genetic resources and functional genes, and further research on their precise function mechanisms are necessary.
Keywords: legume, TLP, PR-5, biotic stress, abiotic stress
1. Introduction
Given their sessile lifestyle, plants have developed a highly complex defense system against different threats, including biotic and abiotic stresses [1]. Global environmental deterioration and pathogen invasions can cause enormous harm to agricultural production. In both animals and plants, several fungal and fungal-like diseases have caused some of the most severe die-offs and extinctions ever witnessed in wild species, and this is jeopardizing food security [2].
Plant pathogenic fungi attack a wide range of crops, but controlling fungal diseases with fungicides is cost intensive and comes with detrimental effects on the environment [3]. Fungal infection threats can be seen across many sources of data, including a high risk of biodiversity loss, host extinction, and low food yield and quality [2]. When plants perceive a pathogenic infection, pathogenesis-related (PR) protein genes are significantly upregulated, acting as the first line of plant defense [1,2]. A recent study showed that 19 classes of PR proteins can be distinguished based on structural similarity and functional activity [1,4], of which, thaumatin-like proteins (TLPs)—the homologies of sweet-tasting thaumatin isolated from the plant Thaumatococcus daniellii—belong to the PR-5 family [5,6]. Growing evidence shows that TLPs are involved in resistance to a variety of fungal diseases in many species, such as Gossypium hirsutum [7], Gossypium barbadense [8], Solanum lycopersicum [4,9], Phyllostachys edulis [10], Ganoderma lingzhi [11], Triticum aestivum L. [12,13], Carya cathayensis [14], Fragaria ananassa [15], Allium sativum L. [16], Pinus radiata [17], Lentinula edodes [18], Camellia sinensis [19], Cucumis melo L. [20], Avena nuda [5], Musa acuminate [21], Manihot esculenta [22], Vitis amurensis [23], Populus szechuanica [24], Camellia sinensis [25], Piper colubrinum [26], Cynanchum komarovii [27], Castanea sativa [28], Actinidia chinensis [29], and Lyophyllum shimeji [30].
Plant TLPs also play diverse roles in abiotic stresses such as drought [31], freezing [32], and salinity [33,34]. Computational approaches such as cis-regulatory analysis, promoter analysis, and co-expressed gene analysis have revealed that TLPs play diverse roles in abiotic stresses [35]. The transcript levels of TLPs are significantly altered under different stress conditions. These findings reveal the functional diversity of TLPs throughout the evolution of genes [36].
Twelve thousand years ago, grain legumes played a crucial role in the development of Neolithic agriculture [37]. Legumes are a globally important crop for food, oil, forage, and bioactive components, and are the main source of edible vegetable protein and oil [38,39,40]. Several phytopathogenic diseases are known to limit legume productivity [41,42]. For example, soybean rust [43], soybean stay-green disease [44], soybean red leaf blotch disease [45], pea blight disease [45], peanut rust [46], peanut leaf spot [47], faba bean root rot [48], and other legume pathogenic diseases are caused by multiple pathogens and are responsible for severe yield loss. Reports have also demonstrated that the productivity of legume crops such as soybeans, chickpeas, and peanuts has been inhibited by different abiotic stresses [49,50,51].
Legumes are some of the most important food crops, and as such, studying biotic and abiotic stresses in the TLP families is worthwhile. This review provides a comprehensive discussion of the research progress regarding TLPs in leguminous plants.
2. Classification and Characteristics of TLPs
TLPs are widely distributed in plants, animals, and fungi [52]. In plants, the thaumatin-like PR-5 family includes PR-5 proteins; OLPs (osmotin-like proteins); PR-like rash; the PR5-like protein kinase receptor; and permatins such as zeamation in maize, hordomatin in barley, and avematin in oats [27,53]. Despite their significant diversity in plants, the amino acid sequences of TLPs have a well-defined thaumatin family signature (PS00316): G-x-[GF]-x-C-x-T-[GA]-d-C-x(1,2)-[GQ]-x(2,3)-C [28,33,54]. Furthermore, promoter analysis has shown that TLPs contain five conserved TFBSs (transcription factor binding sites) called ASRC, CCAF, L1BX, NCS1, and WBXF [55]. Specifically, ASRC and WBXF are responsible for pathogen defense, CCAF is associated with the circadian clock, L1BX is a homeodomain protein recognition motif, and NCS1 is a nodulin consensus sequence.
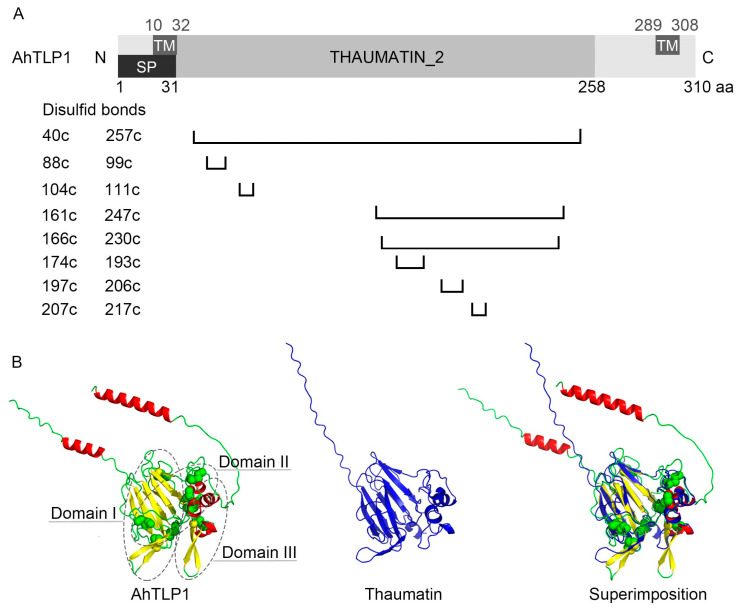
Furthermore, based on their molecular mass, TLPs can be divided into two types. Large (L)-type TLPs range from 21 to 26 kD and contain 16 conserved cysteine residues; most TLPs belong to this type, including TLP-1 to -5 in Medicago truncatula [56], Rj4 in Glycine max [57], AdTLP in Arachis diogoi [58], and CdTLP in Cassia didymobotrya [59]. Small (S)-type TLPs have molecular masses ranging from 16 to 17 kD and have only 10 cysteines in conserved positions because of a peptide deletion [28]. All plant TLPs have similar 3D structures within three domains: domain I, containing 11 stranded β-sheets organized as a β-barrel, forming the protein core; domain II, containing an α-helix and a set of disulfide-rich loops; and domain III, containing a β-hairpin and a coil motif, both maintained by a disulfide bond [31]. There is a cleft structure between domains I and II [60], and each domain is stabilized by at least one disulfide bridge connected by up to 16 cysteine residues with a conserved spatial distribution throughout the protein [61]. Below, AhTLP1 (Arachis hypogaea TLP1) is used as an example to exhibit the general legume TLP structure (Figure 1).
Figure 1.
The protein structure of AhTLP1. (A) The protein structure and eight disulfide bonds of AhTLP (http://www.ebi.ac.uk./interpro/, accessed on 28 March 2024; http://prosite.expasy.org/, accessed on 28 March 2024). TM: transmembrane helix; SP: signal peptide; N: N-terminal; C: C-terminal; c: conserved cysteine involved in a disulfide bond. (B) Three-dimensional structure of AhTLP1 and thaumatin. The TLP protein family contains three conserved domains: domain I, containing 11 stranded β-sheets organized as a β-barrel, forming the protein core; domain II, containing an α-helix and a set of disulfide-rich loops (green balls); and domain III, containing a β-hairpin and a coil motif, both maintained by a disulfide bond. This image was generated with PyMOL Molecular Graphics System version 2.2.0 (Schrödinger, LLC, https://pymol.org).
In addition to similar core domains, one domain is often responsible for localization at either end of the TLP. The N-terminal signal peptide always targets mature proteins in the secretory pathway. For instance, CkTLP in Cynanchum komarovii [27] and AdTLP [58] are located in the extracellular space or cell wall. However, the C-terminal peptide diversifies the subcellular localization of TLPs. For example, the C-terminal propeptide of tobacco osmotin is responsible for its vacuolar localization [62]. Rj4 is presumed to be localized in cell membranes because of a unique transmembrane domain in its C-terminus [57].
3. Biological Functions of TLPs in Legumes
TLPs in plants have various biological functions including host–pathogen interactions, stress tolerance, and cell-signaling transduction. They mostly participate in responses to biotic and abiotic stresses [48]. Therefore, it is believed that there are many plant TLPs, and several reports confirm this belief. In total, there are 106 TLPs in Zea mays, 97 TLPs in Gossypium hirsutum, 78 TLPs in Oryza sativa, 66 TLPs in Nicotiana tabacum, and 51 TLPs in Arabidopsis thaliana [48]. In this regard, research on legumes is still quite limited. The legume genome is extremely complex, and several TLPs have distinct differences. For example, 56 TLPs have been found in Medicago truncatula, and only a few MtTLPs have been well studied. However, although research is lacking, a recent study suggests that MtTLPs exhibit high antifungal activity [41].
3.1. Legume TLPs in Response to Biotic Stresses
Legume TLPs play important roles in plant defense against various biotic stresses. In 1999, a legume TLP was first isolated from the French bean (Phaseolus vulgaris L.), and it exerted antifungal activity against Fusarium oxysporum, Pleurotus ostreatus, and Coprinus comatus [63]. In recent years, more and more TLPs isolated from plants have been proven to have potential antifungal activities [28]. In legumes, the CdTLP protein significantly inhibits fungal strains such as Candida albicans, Candida krusei, and Candida parapsilosis [59]. AdTLP has strong antifungal activity, and transgenic tobacco plants expressing AdTLP have shown enhanced resistance to the soil-borne pathogen Rhizoctonia solani [58]. TLP transcript accumulation was detected after inoculating Medicago truncatula with Colletotrichum trifolii and Erysiphe pisi [64], and recent experimental evidence shows that MtTLP1-5 have strong in vitro antifungal activities against Rhizoctonia solani, Alternaria alternata, Fusarium graminearum, Fusarium solani, Verticillium sp., and Phytophthora spp.; all five MtTLPs reduce the viability of fungal hyphae and significantly reduce A. alternata spore germination [56]. Chickpea (Cicer arietinum) cells have been shown to experience a rapid response in their activated defense-related genes, including a TLP gene, after treatment with a Pmg (Phytophthora megasperma) elicitor and cantharidin [65]. In addition, bulked segregant analysis (BSA) has shown that chickpeas also express TLPs for Fusarium oxysporum resistance [66]. The roles of TLPs in different legumes are summarized in Table 1.
Some TLPs have glucan-binding and glucanase activity, inhibition activity similar to xylanase, α-amylase, or trypsin. Therefore, they have the potential to weaken the cell walls of fungi and interfere with the metabolic processes of pathogens or even affect the digestive ability of insects [28,67,68]. As mentioned above, TLPs contain a conserved motif, and most plant TLPs are the L-type, with 16 highly conserved cysteine residues. Significantly, the 3D structures of TLPs consist of three domains, of which, domains I and II possess a special cleft structure that is crucial for receptor binding and antifungal activity [60]. Specifically, the cleft between domains I and II is made up of five evolutionarily conserved amino acids (arginine, glutamic acid, and three aspartic acid residues) and provides an acidic environment for ligand/receptor binding [61]. Further analysis of this protein superfamily is still necessary to reveal its underlying pathogen-resistance mechanism.
PR proteins accumulate during the plant’s inducible immunity process under pathogenic infection conditions [69]. Pathogens attack plants and trigger innate immune responses, including pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) [70]. Recently, research has shown that host-defense peptides (HDPs) have both direct antimicrobial and immunomodulatory activity, thus protecting multicellular eukaryotes from infections. Importantly, two TLPs—sweet potato LbACP (Lpomea batatas anti-cancer peptide) and European plum (Prunus domestica L.) PdPR5-1—were recently discovered to be HDPs, and they may provide us with valuable tools for developing phytosanitary products [71]. Regarding legumes, the peanut gene TLP1b is a candidate resistant gene that may be used to impart an immune response to Aspergillus flavus infections [72]. However, little is known about legume TLPs in terms of plant immune responses. Future research should clarify this subject.
3.2. Legume TLPs in Response to Abiotic Stresses
Legume TLPs also protect plants from various abiotic stresses besides biotic invasions. Research has reported that two TLPs in the faba bean (Vicia faba L.), VfTLP4-3 and VfTLP5, play important roles in mediating the drought response. These two genes are significantly upregulated under drought conditions, and confer drought resistance and higher POD activity when ectopically expressed in tobacco [36]. Soybean osmotins such as GmOLPa, GmOLPb, and P21e are involved in high-salt stress [33,34]. GmOLPa-like and P21-like osmotins play important roles in drought tolerance; the former shows higher expression levels in roots, and both are also expressed in nodules. Their highest expression levels can be found in the leaves or roots of the drought-tolerant cultivar [31]. The TLP1b gene can be induced by wounding the Caesalpinioideae Senna tora [73]. The AHCSP33 cold shock protein is a TLP homologous protein secreted into the leaf apoplasts of peanuts (Arachis hypogaea) during low-temperature exposure. AHCSP33 acts as a cryoprotecting protein and can prevent the freeze-induced denaturation of L-lactate dehydrogenase (LDH) [32]. In addition, a rare evergreen broad-leaved leguminous shrub, Ammopiptanthus nanus, expresses large quantities of TLPs to resist low-temperature stress in inhospitable desert areas. Most AnTLP genes contain multiple cis-acting elements in promoter regions related to the environmental stress response, and when heterologously expressed in Escherichia coli, yeast cells, and tobacco leaves, a cold-induced AnTLP13 can better enhance low-temperature stress tolerance compared with control cells or seedlings [74]. Moreover, a study on cowpeas (Vigna unguiculata) with a supply of excess manganese (Mn) showed a significant increase in soluble apoplastic proteins, including TLPs, in the apoplastic washing fluid of leaf tissue, suggesting that these proteins have specific physiological roles in response to Mn stress [75]. The roles of TLPs resistant to abiotic stress in different legumes are summarized in Table 1.
Table 1.
The function of legume TLPs in response to stresses.
| Name | Sources | Function/Biological Role | References |
|---|---|---|---|
| TLP | Phaseolus vulgaris | Antifungal activity | [63] |
| CdTLP | Cassia didymobotrya | Antifungal activity | [59] |
| AdTLP | Arachis diogoi | Antifungal activity | [58] |
| MtTLP | Medicago truncatula | Antifungal activity | [56,64] |
| TLP | Cicer arietinum | Phytoalexin response | [65,66] |
| VfTLP4-3, VfTLP5 | Vicia faba | Drought response | [36] |
| StTLP1b | Senna tora | Wound response | [73] |
| AHCSP33 | Arachis hypogaea | Cold response | [32] |
| AnTLPs | Ammopiptanthus nanus | Cold response | [74] |
| GmOLPa | Glycine max | Salt response | [33,34] |
| GmOLPb | Glycine max | Salt response | [33,34] |
| TLP | Vigna unguiculata | Manganese toxicity response | [75] |
| TLP1b | Arachis hypogaea | Immune response | [72] |
| GmOLPa-like and P21-like osmotins | Glycine max | Drought response | [31] |
3.3. The Role of TLPs in Phytohormone Responses
Several expression analyses have shown that hormone treatments have an induction effect on legume TLP gene transcript levels, suggesting that TLPs may have a role in different hormone responses with significant crosstalk [34,58,76,77]. Cis-acting elements are commonly identified in the promoter region of TLP genes, including motifs controlling phytohormone responses such as auxin, abscisic acid (ABA), ethylene (ET), salicylic acid (SA), jasmonic acid (JA), and gibberellic acid (GA) [74,78,79]. However, there have been few reports on how TLPs act in hormone signal transduction pathways, and there are even fewer regarding legumes. One study showed that two osmotin-like proteins bind cytokinin and its analogs, so they are called cytokinin-binding proteins [80]. It has been suggested that TLPs might be involved in cytokinin transportation partly because of their extracellular locations [58,74].
SA and JA are two important defense hormones in plants. SA is generally involved in defense responses against (hemi-)biotrophic pathogens, while JA is associated with defense responses against necrotrophic pathogens and herbivorous insects; therefore, the SA and JA defense pathways are usually antagonistic [78,79]. PR5 is activated as an SA response gene during the plant–pathogen interaction process [81]. In leguminous TLP genes, SA and JA are spatiotemporally specific to their induction location, and even the same gene usually has opposite expression patterns in a single tissue when faced with these two hormonal stimuli. In soybeans, SA stimulation induces two acidic PR-5 genes, GmOLPa and P21e, notably increasing GmOLPa levels in the lower leaves. MeJA markedly induces not only neutral GmOLPb but also P21e in these leaves, but GmOLPa transcription levels will then be lower [33]. PR-5 genes are SA-responsive genes and respond more strongly to SA treatments than JA treatments in Medicago truncatula, even though they have similar expression patterns in response to both hormones [82]. TLP1bs in Senna tora are marker genes for SA signaling and can be induced via wounds: they appear after 6 h and peak after 24 h. However, JA biosynthesis genes can be strongly induced after 1 h and decrease after 3 h, which is unlike that observed in undamaged leaves. This suggests that JA can affect TLP1b transcription [73]. Generally, TLP genes are more active in the SA pathway, and their activities can be affected by protein surface pH levels and other changes. However, more evidence is needed to support these hypotheses (Figure 2).
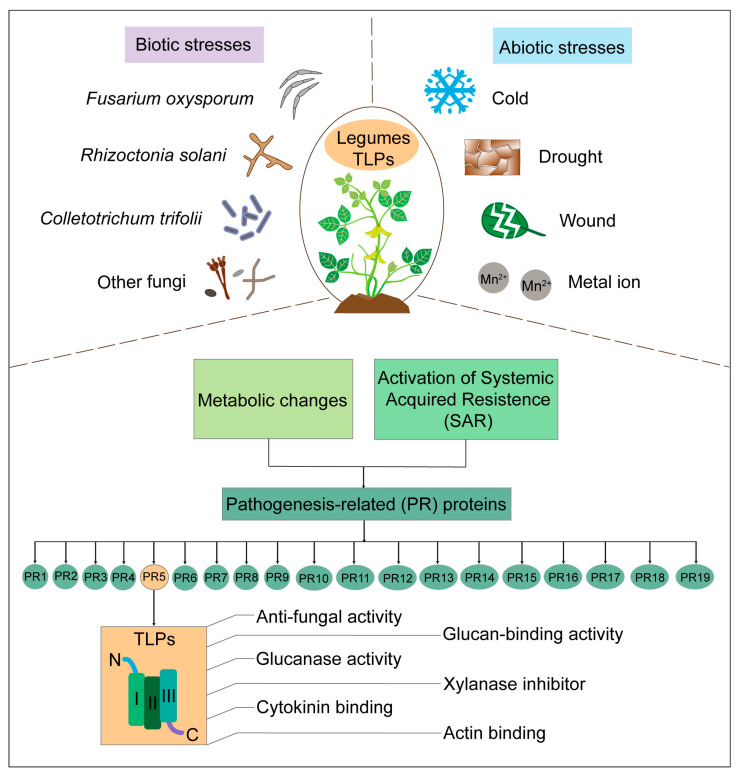
Figure 2.
Plant defense mechanisms of TLPs. Plants respond to biotic and abiotic stresses through several pathways. Under stress conditions, metabolic changes and systemic acquired resistance (SAR) are quickly stimulated and initiate the expression of PR proteins, including PR5. TLPs are PR5 members, multifunctional proteins that exhibit antifungal activity, glucan-binding activity, glucanase activity, xylanase inhibitor abilities, cytokinin-binding abilities, and actin-binding abilities. TLPs contain three domains: I, II, and III. N: N-terminal, C: C-terminal.
3.4. The Role of TLPs in Leguminous Nodulation and Symbiosis
Nitrogen is a component of many important compounds in plants, and it plays a crucial role in the synthesis of proteins, nucleic acids, and chlorophylls. However, most plants are unable to directly use atmospheric nitrogen gas. Uniquely, legumes with special metabolic pathways can form root nodules with nitrogen-fixing bacteria to secure nitrogen for growth and development [83,84]. Soybean nodulation is controlled by several host genes referred to as Rj (rj) genes; these include the Rj4 gene, which encodes a thaumatin-like protein. Rj4 is constitutively expressed in roots, including in root nodules, so it is very likely that Rj4 is involved in gene-for-gene resistance against specific Bradyrhizobium strains. These strains are highly competitive for nodulation but have low nitrogen fixation efficiency; thus, cultivars harboring an Rj4 allele are considered favorable. It is also possible that Rj4 interacts with rhizobial surface polysaccharides given its glucan-binding and glucanase activities [57]. The nonmycorrhizal legume white lupin (Lupinus albus) prevents mycorrhiza infection, and a TLP peptide has been identified from its extracellular protein components [85]. TLPs have also been detected in the intercellular fluid and different tissues of healthy white lupins [86] and in the intercellular washing fluid of chickpea leaves [87], suggesting that these proteins may have inhibitory effects on symbiosis in addition to their anti-pathogenic functions.
In one study investigating short-term dynamics in proteomic patterns using continuous sampling, extrafloral nectar produced by an obligate ant plant (Acacia cornigera) showed that anabolism involving TLP accumulated in the plant’s nectary directly before secretion, and it diminished quantitatively after the daily secretion process [88]. This plant is inhabited by mutualistic ants, and large quantities of TLPs in the biochemically complex components secreted by its nectary may play a role in its symbiotic relationship with the insects.
3.5. Prediction of Protein–Protein Interactions in Legume TLPs
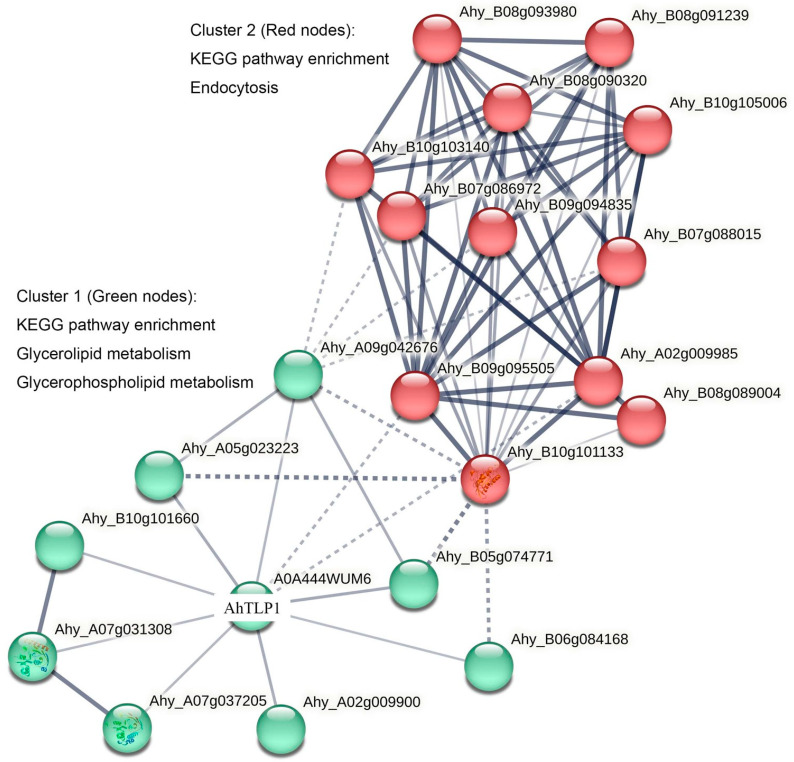
Protein–protein interactions in legume TLPs and the metabolic pathways they participate in may reflect their biological functions and potential roles. To demonstrate this, we analyzed protein–protein interactions between a TLP protein from Arachis hypogaea, AhTLP1 (NCBI accession number: XP_025651708.1) and two soybean TLP proteins, namely GmOLPa (NCBI accession number: NP_001236405.2) and GmOLPb (NCBI accession number: NP_001235877.1), using the STRING database (https://cn.string-db.org, accessed on 29 January 2024).
By analyzing the results using association rules and k-means clustering, we found that the interaction network of AhTLP1 can be divided into two clusters (Figure 3, Table 2). AhTLP1 may physically interact with 10 proteins, including the TYR_PHOSPHATASE_2 domain-containing protein, protein phosphatase 2C, the cyclic nucleotide-binding/kinase domain-containing protein, the PlsC domain-containing protein, 1-acyl-sn-glycerol-3-phosphate acyltransferase, 3-deoxy-d-manno-octulosonic-acid transferase, E3 ubiquitin–protein ligase, and two uncharacterized proteins. Functional association analyses showed that AhTLP1 may indirectly interact with another 10 proteins, including Rac-like GTP-binding protein RAC13, ribonucleoside–diphosphate reductase subunit beta, Clathrin heavy chain, the beta-adaptin-like protein, the FYVE-type domain-containing protein, the subunit of adaptor protein complex 2, and the beta-adaptin-like protein.
Figure 3.
Protein–protein interaction predictions for Arachis hypogaea TLP1 analyzed using STRING. A0A444WUM6: AhTLP1, Arachis hypogaea TLP1; KEGG: Kyoto Encyclopedia of Genes and Genomes. The thickness of the line indicates the degree of confidence prediction of the interaction.
Table 2.
The AhTLP1 protein–protein interacting network information.
| Interactions | Predicted Interaction Partners | Protein Accessions |
|---|---|---|
| Direct interaction with AhTLP1 | TYR_PHOSPHATASE_2 domain-containing protein | Ahy_A09g042676 |
| Protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein | Ahy_A05g023223, Ahy_B05g074771 | |
| PlsC domain-containing protein | Ahy_B10g101660, Ahy_A07g037205 | |
| 1-acyl-sn-glycerol-3-phosphate acyltransferase | Ahy_A07g031308 | |
| 3-deoxy-d-manno-octulosonic-acid transferase | Ahy_A02g009900 | |
| E3 ubiquitin–protein ligase | Ahy_B06g084168 | |
| Uncharacterized protein | Ahy_A02g009985, Ahy_B09g095505 | |
| Indirect interaction with AhTLP1 | Rac-like GTP-binding protein RAC13 | Ahy_B10g101133 |
| Ribonucleoside–diphosphate reductase subunit beta | Ahy_B08g089004 | |
| Clathrin heavy chain | Ahy_B07g088015, Ahy_B09g094835, Ahy_B07g086972, Ahy_B10g103140 | |
| Beta-adaptin-like protein | Ahy_B10g105006 | |
| FYVE-type domain-containing protein | Ahy_B08g090320 | |
| Subunit of the adaptor protein complex 2 | Ahy_B08g093980 | |
| Beta-adaptin-like protein | Ahy_B08g091239 |
KEGG pathway analysis has shown that the proteins of cluster 1 mainly participate in glycerolipid metabolism and glycerophospholipid metabolism because of the following two proteins: the PlsC domain-containing protein 1-acyl-sn-glycerol-3-phosphate acyltransferase (which is involved in the formation of phosphatidic acid), a precursor of various membrane phospholipids (PLs); and 3-deoxy-d-manno-octulosonic-acid transferase (involved in the biosynthesis of lipid A), a phosphorylated glycolipid that anchors the lipopolysaccharide to the outer membrane of the cell in bacteria [89]. The TYR_PHOSPHATASE_2 domain-containing protein is an enzyme responsible for dephosphorylating phosphor-Tyr from proteins [90]. Protein phosphatase type 2C functions in the ABA signaling pathway, which is one of the major signal transduction pathways in abiotic stress responses [91]. E3 ubiquitin–protein ligase transfers ubiquitin and ubiquitin-like proteins through an E2 enzyme to a target substrate, which is the final step of ubiquitination [92]. The AhTLP1’s interaction network indicates its potential role in different cellular processes. In addition, KEGG pathway analysis of cluster 2 has shown that there are 12 proteins involved in the endocytosis pathway. Importantly, one study showed that the thaumatin-like protein CalA in Aspergillus fumigatus assists this fungal pathogen in invading pulmonary epithelial cells and vascular endothelial cells by inducing its own endocytosis [93]. The above research shows that TLPs may play a role in the endocytosis pathway.
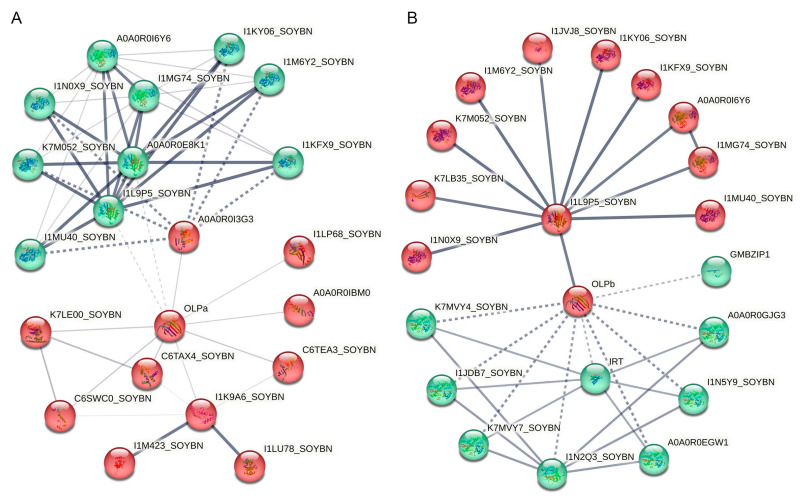
For soybean TLP proteins, analyses of GmOLPa and GmOLPb protein–protein interactions conducted on the STRING database have shown that GmOLPa is involved in sucrose biosynthetic process peroxisome organization and the carbohydrate metabolic process (Figure 4A, Table 3). Furthermore, GmOLPb is involved in the sucrose biosynthetic process, defense responses to other organisms, the carbohydrate metabolic process, responses to biotic stimuli, and defense responses (Figure 4B, Table 4).
Figure 4.
Protein–protein interaction predictions for Glycine max GmOLPa and GmOLPb analyzed using STRING. (A) STRING analysis of GmOLPa; (B) STRING analysis of GmOLPb. The thickness of the line indicates the degree of confidence prediction of the interaction.
Table 3.
GmOLPa protein–protein interaction network information.
| Interactions | Predicted Interaction Partners | Protein Accessions |
|---|---|---|
| Direct interaction with GmOLPa | Sucrose–phosphatase 2 | A0A0R0I3G3, A0A0R0E8K1 |
| Bet_v_1 domain-containing protein | I1LP68_SOYBN | |
| Regulatory protein NPR1 | A0A0R0IBM0 | |
| Sucrose–phosphatase 1 | I1L9P5_SOYBN | |
| Ribonucleoprotein | C6TEA3_SOYBN | |
| UBIQUITIN_CONJUGAT_2 domain-containing protein | I1K9A6_SOYBN | |
| Calmodulin-like protein 1 | C6SWC0_SOYBN, C6TAX4_SOYBN | |
| ABC transporter family protein | K7LE00_SOYBN | |
| Indirect interaction with GmOLPa | Glycosyl hydrolase 31 family | I1MG74_SOYBN, A0A0R0I6Y6 |
| Sucrose–phosphate synthase 2 | I1KY06_SOYBN, I1M6Y2_SOYBN, I1N0X9_SOYBN, I1KFX9_SOYBN, K7M052_SOYBN, I1MU40_SOYBN | |
| Peroxisome biogenesis protein | I1M423_SOYBN, I1LU78_SOYBN |
Table 4.
GmOLPb protein–protein interaction network information.
| Interactions | Predicted Interaction Partners | Protein Accessions |
|---|---|---|
| Direct interaction with GmOLPb | Cytochrome P450 family protein | K7MVY7_SOYBN, A0A0R0GJG3, I1N5Y9_SOYBN, K7MVY4_SOYBN, A0A0R0EGW1, I1JDB7_SOYBN |
| Trypsin inhibitor A | I1N2Q3_SOYBN | |
| Fe2+/Zn2+ regulated transporter | IRT | |
| Sucrose–phosphatase 1 | I1L9P5_SOYBN | |
| bZIP transcription factor | GmbZIP1 | |
| Indirect interaction with GmOLPb | Tim44 domain-containing protein | I1JVJ8_SOYBN, K7LB35_SOYBN |
| Sucrose–phosphate synthase 2 | I1KY06_SOYBN, I1KFX9_SOYBN, I1M6Y2_SOYBN, K7M052_SOYBN, I1N0X9_SOYBN, I1MU40_SOYBN | |
| Glycosyl hydrolase 31 family | A0A0R0I6Y6, I1MG74_SOYBN |
Specifically, our protein–protein interaction analysis revealed that GmOLPa interacts with sucrose–phosphatase 2, Bet_v_1 domain-containing protein, regulatory protein NPR1, sucrose–phosphatase 1, ribonucleoprotein, the UBIQUITIN_CONJUGAT_2 domain-containing protein, calmodulin-like protein 1, and the ABC transporter family protein. Functional association analyses showed that GmOLPa may indirectly interact with another 10 proteins, including the glycosyl hydrolase 31 family, sucrose–phosphate synthase 2, and the peroxisome biogenesis proteins. Moreover, our protein–protein interaction analysis revealed that GmOLPb interacts with cytochrome P450 family proteins, trypsin inhibitor A, the Fe2+/Zn2+-regulated transporter, sucrose–phosphatase 1, and the bZIP transcription factor. The functional association analysis also showed that GmOLPb may indirectly interact with another 10 proteins, including Tim44 domain-containing proteins, sucrose–phosphate synthase 2, and the glycosyl hydrolase 31 family. Most importantly, the nonexpressor of pathogenesis-related genes 1 (NPR1) is a master regulator of SA-mediated SAR, a broad-spectrum disease-resistance mechanism in plants [94]. Basic leucine zipper (bZIP) genes encode transcription factors (TFs) that control important biochemical and physiological processes, including various kinds of abiotic and biotic stress responses, such as salt responses, drought responses, and pathogen responses in soybean [95,96,97]. This might explain why GmOLPs participate in high-salt and drought responses.
The above research shows that legume TLPs may be involved in multiple metabolic pathways. However, the proteins that interact with TLPs predicted by STRING must be confirmed through experimental verification to comprehensively explore their potential functions.
4. Potential Biotechnological Applications for Legume TLPs
The antifungal activity and stability of TLPs provide them with broad prospects for agricultural applications in the pathogen-resistance field. Highly conserved cysteine residues with disulfide bridge structures in most plant TLPs provide stabilization under extreme thermal and pH conditions, as well as resistance to protease degradation [98,99]. In addition, TLPs also protect plants from abiotic stresses, including cold, salinity, and drought [28,100]. These characteristics make TLPs a good source for breeding and plant transformation aimed at producing better performance under biotic and abiotic stresses.
Research shows that TLPs may play a role in plant herbicide resistance. Transcriptomic studies of chickpeas using two herbicide-susceptible and -tolerant genotypes exposed to imidazoline (Imazethapyr) have revealed that gene encoding for thaumatin-like protein-1 shows a five-fold change in differential expression as an effect of herbicide on tolerant plants, indicating that this TLP can be used for further investigation and association application studies [101].
Legume TLPs may have wide applications in the human food and pharmaceutical industries. Antifungal proteins (including TLPs) isolated from the seeds of legume plants have shown inhibitory activity against enzymes that are essential to the life cycle of human immunodeficiency virus type 1 (HIV-1); for instance, French bean TLPs have been found to potently inhibit HIV-1 integrase and reverse transcriptase, as well as low HIV-1 protease inhibitory activity [102]. Thaumatin has been approved and commercialized as a safe sweetener and flavor enhancer in food because it is 1600 times sweeter than sucrose on a weight-to-weight basis [103,104]. As homologies of thaumatin, TLPs can be developed as natural flavor modifiers or enhancers, replacing synthetic sweeteners. TLP genes can also be transferred to vegetables or fruit crops as flavor enhancers [28,105,106]. Exploring the pharmaceutical and sweet proteins in edible legume plants and their potential applications is a worthwhile research direction.
5. Perspectives
TLPs are a group of proteins with broad application prospects, but corresponding in-depth research on this family in leguminous plants remains quite limited. Many varieties of leguminous plants are edible and can be used for chemical industry purposes. Moreover, recombinant TLP products can be expressed and refined in different bioreactors, such as bacteria, fungi, and other plants, on account of the antifungal activity of TLPs. Furthermore, transgenic plants with TLPs have displayed increased activity under biotic and abiotic stresses. TLPs are valuable candidates for plant breeding. Further research on the precise mechanisms underlying legume TLP protein regulation, localization, and function is necessary, especially regarding whether TLPs can degrade fungal glucan, as well as whether they possess pathogen-associated molecular patterns for cell surface pattern recognition receptors, which can trigger plant immune responses in extracellular regions.
Author Contributions
All authors contributed to the conceptualization of this study. Data collection was performed by L.F. and Y.L. The first draft of the manuscript was written by L.F., Y.L. and S.W. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing does not apply to this article.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by the National Natural Science Foundation of China, grant number 31171625; the Science and Technology Planning Project of Guangdong Province, China; grant number 2020A1414040005; and the Guangdong Basic and Applied Basic Research Foundation, China, grant number 2021A1515110341.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Dos Santos C., Franco O.L. Pathogenesis-Related Proteins (PRs) with Enzyme Activity Activating Plant Defense Responses. Plants. 2023;12:2226. doi: 10.3390/plants12112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher M.C., Henk D.A., Briggs C.J., Brownstein J.S., Madoff L.C., McCraw S.L., Gurr S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kou Y., Naqvi N.I. Surface sensing and signaling networks in plant pathogenic fungi. Semin. Cell Dev. Biol. 2016;57:84–92. doi: 10.1016/j.semcdb.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Li X., Xu B., Xu J., Li Z., Jiang C., Zhou Y., Yang Z., Deng M., Lv J., Zhao K. Tomato-Thaumatin-like Protein Genes Solyc08g080660 and Solyc08g080670 Confer Resistance to Five Soil-Borne Diseases by Enhancing beta-1,3-Glucanase Activity. Genes. 2023;14:1622. doi: 10.3390/genes14081622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Han D., Shi Y. Gene Cloning, Expression, and Antifungal Activities of Permatin from Naked Oat (Avena nuda) Probiotics Antimicrob. Proteins. 2019;11:299–309. doi: 10.1007/s12602-018-9422-y. [DOI] [PubMed] [Google Scholar]
- 6.Nawrot R., Musidlak O., Barylski J., Nowicki G., Baldysz S., Czerwoniec A., Gozdzicka-Jozefiak A. Characterization and expression of a novel thaumatin-like protein (CcTLP1) from papaveraceous plant Corydalis cava. Int. J. Biol. Macromol. 2021;189:678–689. doi: 10.1016/j.ijbiomac.2021.08.067. [DOI] [PubMed] [Google Scholar]
- 7.Zhou H., Xie Y., Jiang Y., Nadeem H., Wang Y., Yang N., Zhu H., Tang C. GhTLP1, a thaumatin-like protein 1, improves Verticillium wilt resistance in cotton via JA, ABA and MAPK signaling pathway-plant pathways. Int. J. Biol. Macromol. 2023;253:127388. doi: 10.1016/j.ijbiomac.2023.127388. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Chen W., Sang X., Wang T., Gong H., Zhao Y., Zhao P., Wang H. Genome-Wide Identification of the Thaumatin-like Protein Family Genes in Gossypium barbadense and Analysis of Their Responses to Verticillium dahliae Infection. Plants. 2021;10:2647. doi: 10.3390/plants10122647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu H., Deng M., Yang Z., Mao L., Jiang S., Yue Y., Zhao K. Two Tomato (Solanum lycopersicum) Thaumatin-Like Protein Genes Confer Enhanced Resistance to Late Blight (Phytophthora infestans) Phytopathology. 2021;111:1790–1799. doi: 10.1094/PHYTO-06-20-0237-R. [DOI] [PubMed] [Google Scholar]
- 10.Gu Y., Yu H., He S., Zhang P., Ma X. Genome-Wide Identification and Characterization of the TLP Gene Family in Phyllostachys edulis and Association with Witches’ Broom Disease Resistance in Bamboo. Int. J. Mol. Sci. 2023;24:10257. doi: 10.3390/ijms241210257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T., Li X., Zhang C., Xu J. Transcriptome analysis of Ganoderma lingzhi (Agaricomycetes) response to Trichoderma hengshanicum infection. Front. Microbiol. 2023;14:1131599. doi: 10.3389/fmicb.2023.1131599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gul S., Hussain A., Ali Q., Alam I., Alshegaihi R.M., Meng Q., Zaman W., Manghwar H., Munis M.F.H. Hydropriming and Osmotic Priming Induce Resistance against Aspergillus niger in Wheat (Triticum aestivum L.) by Activating beta-1,3-glucanase, Chitinase, and Thaumatin-like Protein Genes. Life. 2022;12:2061. doi: 10.3390/life12122061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Tang C., Deng L., Cai G., Liu X., Liu B., Han Q., Buchenauer H., Wei G., Han D., et al. Characterization of a pathogenesis-related thaumatin-like protein gene TaPR5 from wheat induced by stripe rust fungus. Physiol. Plant. 2010;139:27–38. doi: 10.1111/j.1399-3054.2009.01338.x. [DOI] [PubMed] [Google Scholar]
- 14.Li P., Xu Y., Wang K., Guo W., Gu Y., Lyu S., Huang J., Lin H., Huang C., Xu Z., et al. Genome-Wide Identification of TLP Gene Family and Their Roles in Carya cathayensis Sarg in Response to Botryosphaeria dothidea. Front. Plant Sci. 2022;13:849043. doi: 10.3389/fpls.2022.849043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Miao L., Yang X., Jiang G. Genome-wide characterization and expression of the TLP gene family associated with Colletotrichum gloeosporioides inoculation in Fragaria x ananassa. PeerJ. 2022;10:e12979. doi: 10.7717/peerj.12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anisimova O.K., Kochieva E.Z., Shchennikova A.V., Filyushin M.A. Thaumatin-like Protein (TLP) Genes in Garlic (Allium sativum L.): Genome-Wide Identification, Characterization, and Expression in Response to Fusarium proliferatum Infection. Plants. 2022;11:748. doi: 10.3390/plants11060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aloi F., Zamora-Ballesteros C., Martin-Garcia J., Diez J.J., Cacciola S.O. Co-Infections by Fusarium circinatum and Phytophthora spp. on Pinus radiata: Complex Phenotypic and Molecular Interactions. Plants. 2021;10:1976. doi: 10.3390/plants10101976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma X., Fan X., Wang G., Xu R., Yan L., Zhou Y., Gong Y., Xiao Y., Bian Y. Enhanced Expression of Thaumatin-like Protein Gene (LeTLP1) Endows Resistance to Trichoderma atroviride in Lentinula edodes. Life. 2021;11:863. doi: 10.3390/life11080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S., Liu L., Mi X., Zhao S., An Y., Xia X., Guo R., Wei C. Multi-omics analysis to visualize the dynamic roles of defense genes in the response of tea plants to gray blight. Plant J. 2021;106:862–875. doi: 10.1111/tpj.15203. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Cui J., Zhou X., Luan Y., Luan F. Genome-wide identification, characterization and expression analysis of the TLP gene family in melon (Cucumis melo L.) Genomics. 2020;112:2499–2509. doi: 10.1016/j.ygeno.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Jiao W., Li X., Zhao H., Cao J., Jiang W. Antifungal Activity of an Abundant Thaumatin-Like Protein from Banana against Penicillium expansum, and Its Possible Mechanisms of Action. Molecules. 2018;23:1442. doi: 10.3390/molecules23061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odeny Ojola P., Nyaboga E.N., Njiru P.N., Orinda G. Overexpression of rice thaumatin-like protein (Ostlp) gene in transgenic cassava results in enhanced tolerance to Colletotrichum gloeosporioides f. sp. manihotis. J. Genet. Eng. Biotechnol. 2018;16:125–131. doi: 10.1016/j.jgeb.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He R., Wu J., Zhang Y., Aguero C.B., Li X., Liu S., Wang C., Walker M.A., Lu J. Overexpression of a thaumatin-like protein gene from Vitis amurensis improves downy mildew resistance in Vitis vinifera grapevine. Protoplasma. 2017;254:1579–1589. doi: 10.1007/s00709-016-1047-y. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z.J., Cao Z.M., Yu Z.D., Yu D. Cloning and characterization of defense-related genes from Populus szechuanica infected with rust fungus Melampsora larici-populina. Genet. Mol. Res. 2016;15:1–15. doi: 10.4238/gmr.15017314. [DOI] [PubMed] [Google Scholar]
- 25.Acharya K., Pal A.K., Gulati A., Kumar S., Singh A.K., Ahuja P.S. Overexpression of Camellia sinensis thaumatin-like protein, CsTLP in potato confers enhanced resistance to Macrophomina phaseolina and Phytophthora infestans infection. Mol. Biotechnol. 2013;54:609–622. doi: 10.1007/s12033-012-9603-y. [DOI] [PubMed] [Google Scholar]
- 26.Mani T., Sivakumar K.C., Manjula S. Expression and functional analysis of two osmotin (PR5) isoforms with differential antifungal activity from Piper colubrinum: Prediction of structure-function relationship by bioinformatics approach. Mol. Biotechnol. 2012;52:251–261. doi: 10.1007/s12033-011-9489-0. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q., Li F., Zhang X., Zhang Y., Hou Y., Zhang S., Wu Z. Purification and characterization of a CkTLP protein from Cynanchum komarovii seeds that confers antifungal activity. PLoS ONE. 2011;6:e16930. doi: 10.1371/journal.pone.0016930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J.J., Sturrock R., Ekramoddoullah A.K. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 2010;29:419–436. doi: 10.1007/s00299-010-0826-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang H., Ng T.B. Isolation of an antifungal thaumatin-like protein from kiwi fruits. Phytochemistry. 2002;61:1–6. doi: 10.1016/S0031-9422(02)00144-9. [DOI] [PubMed] [Google Scholar]
- 30.Lam S.K., Ng T.B. First simultaneous isolation of a ribosome inactivating protein and an antifungal protein from a mushroom (Lyophyllum shimeji) together with evidence for synergism of their antifungal effects. Arch. Biochem. Biophys. 2001;393:271–280. doi: 10.1006/abbi.2001.2506. [DOI] [PubMed] [Google Scholar]
- 31.Faillace G.R., Caruso P.B., Timmers L., Favero D., Guzman F.L., Rechenmacher C., de Oliveira-Busatto L.A., de Souza O.N., Bredemeier C., Bodanese-Zanettini M.H. Molecular Characterisation of Soybean Osmotins and Their Involvement in Drought Stress Response. Front. Genet. 2021;12:632685. doi: 10.3389/fgene.2021.632685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dave R.S., Mitra R.K. A low temperature induced apoplastic protein isolated from Arachis hypogaea. Phytochemistry. 1998;49:2207–2213. doi: 10.1016/S0031-9422(98)00372-0. [DOI] [PubMed] [Google Scholar]
- 33.Tachi H., Fukuda-Yamada K., Kojima T., Shiraiwa M., Takahara H. Molecular characterization of a novel soybean gene encoding a neutral PR-5 protein induced by high-salt stress. Plant Physiol. Biochem. 2009;47:73–79. doi: 10.1016/j.plaphy.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 34.Onishi M., Tachi H., Kojima T., Shiraiwa M., Takahara H. Molecular cloning and characterization of a novel salt-inducible gene encoding an acidic isoform of PR-5 protein in soybean (Glycine max [L.] Merr.) Plant Physiol. Biochem. 2006;44:574–580. doi: 10.1016/j.plaphy.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Kumar D., Kirti P.B. The genus Arachis: An excellent resource for studies on differential gene expression for stress tolerance. Front. Plant Sci. 2023;14:1275854. doi: 10.3389/fpls.2023.1275854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Yang X., Zhang J., Huang L., Shi Z., Tian Z., Sha A., Lu G. Thaumatin-like protein family genes VfTLP4-3 and VfTLP5 are critical for faba bean’s response to drought stress at the seedling stage. Plant Physiol. Biochem. 2023;206:108243. doi: 10.1016/j.plaphy.2023.108243. [DOI] [PubMed] [Google Scholar]
- 37.Bohra A., Tiwari A., Kaur P., Ganie S.A., Raza A., Roorkiwal M., Mir R.R., Fernie A.R., Smykal P., Varshney R.K. The Key to the Future Lies in the Past: Insights from Grain Legume Domestication and Improvement Should Inform Future Breeding Strategies. Plant Cell Physiol. 2022;63:1554–1572. doi: 10.1093/pcp/pcac086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo B., Sun L., Jiang S., Ren H., Sun R., Wei Z., Hong H., Luan X., Wang J., Wang X., et al. Soybean genetic resources contributing to sustainable protein production. Theor. Appl. Genet. 2022;135:4095–4121. doi: 10.1007/s00122-022-04222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanthakumar P., Klepacka J., Bains A., Chawla P., Dhull S.B., Najda A. The Current Situation of Pea Protein and Its Application in the Food Industry. Molecules. 2022;27:5354. doi: 10.3390/molecules27165354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akram N.A., Shafiq F., Ashraf M. Peanut (Arachis hypogaea L.): A Prospective Legume Crop to Offer Multiple Health Benefits Under Changing Climate. Compr. Rev. Food Sci. Food Saf. 2018;17:1325–1338. doi: 10.1111/1541-4337.12383. [DOI] [PubMed] [Google Scholar]
- 41.Mondal S., Badigannavar A.M. Mapping of a dominant rust resistance gene revealed two R genes around the major Rust_QTL in cultivated peanut (Arachis hypogaea L.) Theor. Appl. Genet. 2018;131:1671–1681. doi: 10.1007/s00122-018-3106-6. [DOI] [PubMed] [Google Scholar]
- 42.Yang Y., Chen T., Dai X., Yang D., Wu Y., Chen H., Zheng Y., Zhi Q., Wan X., Tan X. Comparative transcriptome analysis revealed molecular mechanisms of peanut leaves responding to Ralstonia solanacearum and its type III secretion system mutant. Front. Microbiol. 2022;13:998817. doi: 10.3389/fmicb.2022.998817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S., Smith J.R. Phenotypic Evaluation of Soybean Genotypes for Their Reaction to a Mississippi Isolate of Phakopsora pachyrhizi Causing Soybean Rust. Plants. 2023;12:1797. doi: 10.3390/plants12091797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Wang M., Wang L., Feng H., He X., Chang S., Wang D., Wang L., Yang J., An G., et al. Whole-plant microbiome profiling reveals a novel geminivirus associated with soybean stay-green disease. Plant Biotechnol. J. 2022;20:2159–2173. doi: 10.1111/pbi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukanda M.M., Dramadri I.O., Adjei E.A., Badji A., Arusei P., Gitonga H.W., Wasswa P., Edema R., Ochwo-Ssemakula M., Tukamuhabwa P., et al. Genome-Wide Association Analysis for Resistance to Coniothyrium glycines Causing Red Leaf Blotch Disease in Soybean. Genes. 2023;14:1271. doi: 10.3390/genes14061271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You Y., Liao J., He Z., Khurshid M., Wang C., Zhang Z., Mao J., Xia Y. Effects of Peanut Rust Disease (Puccinia arachidis Speg.) on Agricultural Production: Current Control Strategies and Progress in Breeding for Resistance. Genes. 2024;15:102. doi: 10.3390/genes15010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzales M., Kemerait R., Jr., Bertioli D., Leal-Bertioli S. Strong Resistance to Early and Late Leaf Spot in Peanut-Compatible Wild-Derived Induced Allotetraploids. Plant Dis. 2023;107:335–343. doi: 10.1094/PDIS-03-22-0721-RE. [DOI] [PubMed] [Google Scholar]
- 48.Yu H., Yang F., Hu C., Yang X., Zheng A., Wang Y., Tang Y., He Y., Lv M. Production status and research advancement on root rot disease of faba bean (Vicia faba L.) in China. Front. Plant Sci. 2023;14:1165658. doi: 10.3389/fpls.2023.1165658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumari P., Rastogi A., Yadav S. Effects of Heat stress and molecular mitigation approaches in orphan legume, Chickpea. Mol. Biol. Rep. 2020;47:4659–4670. doi: 10.1007/s11033-020-05358-x. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y., Cao Y. GmWRKY17-mediated transcriptional regulation of GmDREB1D and GmABA2 controls drought tolerance in soybean. Plant Mol. Biol. 2023;113:157–170. doi: 10.1007/s11103-023-01380-2. [DOI] [PubMed] [Google Scholar]
- 51.Banavath J.N., Chakradhar T., Pandit V., Konduru S., Guduru K.K., Akila C.S., Podha S., Puli C.O.R. Stress Inducible Overexpression of AtHDG11 Leads to Improved Drought and Salt Stress Tolerance in Peanut (Arachis hypogaea L.) Front. Chem. 2018;6:34. doi: 10.3389/fchem.2018.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J.P., Su X.H. Patterns of molecular evolution and predicted function in thaumatin-like proteins of Populus trichocarpa. Planta. 2010;232:949–962. doi: 10.1007/s00425-010-1218-6. [DOI] [PubMed] [Google Scholar]
- 53.van Loon L.C., Rep M., Pieterse C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 54.Jami S.K., Swathi Anuradha T., Guruprasad L., Kirti P.B. Molecular, biochemical and structural characterization of osmotin-like protein from black nightshade (Solanum nigrum) J. Plant Physiol. 2007;164:238–252. doi: 10.1016/j.jplph.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Deihimi T., Niazi A., Ebrahimi M., Kajbaf K., Fanaee S., Bakhtiarizadeh M.R., Ebrahimie E. Finding the undiscovered roles of genes: An approach using mutual ranking of coexpressed genes and promoter architecture-case study: Dual roles of thaumatin like proteins in biotic and abiotic stresses. Springerplus. 2012;1:30. doi: 10.1186/2193-1801-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saeidi M., Zareie R. Prediction, isolation, overexpression and antifungal activity analysis of Medicago truncatula var. truncatula putative thaumatin like proteins (TLP-1, -2, -3, -4 and -5) Turk. J. Biol. 2020;44:176–187. doi: 10.3906/biy-1912-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi M., Shiro S., Kanamori H., Mori-Hosokawa S., Sasaki-Yamagata H., Sayama T., Nishioka M., Takahashi M., Ishimoto M., Katayose Y., et al. A thaumatin-like protein, Rj4, controls nodule symbiotic specificity in soybean. Plant Cell Physiol. 2014;55:1679–1689. doi: 10.1093/pcp/pcu099. [DOI] [PubMed] [Google Scholar]
- 58.Singh N.K., Kumar K.R., Kumar D., Shukla P., Kirti P.B. Characterization of a pathogen induced thaumatin-like protein gene AdTLP from Arachis diogoi, a wild peanut. PLoS ONE. 2013;8:e83963. doi: 10.1371/journal.pone.0083963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vitali A., Pacini L., Bordi E., De Mori P., Pucillo L., Maras B., Botta B., Brancaccio A., Giardina B. Purification and characterization of an antifungal thaumatin-like protein from Cassia didymobotrya cell culture. Plant Physiol. Biochem. 2006;44:604–610. doi: 10.1016/j.plaphy.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Ghosh R., Chakrabarti C. Crystal structure analysis of NP24-I: A thaumatin-like protein. Planta. 2008;228:883–890. doi: 10.1007/s00425-008-0790-5. [DOI] [PubMed] [Google Scholar]
- 61.Min K., Ha S.C., Hasegawa P.M., Bressan R.A., Yun D.J., Kim K.K. Crystal structure of osmotin, a plant antifungal protein. Proteins. 2004;54:170–173. doi: 10.1002/prot.10571. [DOI] [PubMed] [Google Scholar]
- 62.Melchers L.S., Sela-Buurlage M.B., Vloemans S.A., Woloshuk C.P., Van Roekel J.S., Pen J., van den Elzen P.J., Cornelissen B.J. Extracellular targeting of the vacuolar tobacco proteins AP24, chitinase and beta-1,3-glucanase in transgenic plants. Plant Mol. Biol. 1993;21:583–593. doi: 10.1007/BF00014542. [DOI] [PubMed] [Google Scholar]
- 63.Ye X.Y., Wang H.X., Ng T.B. First chromatographic isolation of an antifungal thaumatin-like protein from French bean legumes and demonstration of its antifungal activity. Biochem. Biophys. Res. Commun. 1999;263:130–134. doi: 10.1006/bbrc.1999.1166. [DOI] [PubMed] [Google Scholar]
- 64.Samac D.A., Penuela S., Schnurr J.A., Hunt E.N., Foster-Hartnett D., Vandenbosch K.A., Gantt J.S. Expression of coordinately regulated defence response genes and analysis of their role in disease resistance in Medicago truncatula. Mol. Plant Pathol. 2011;12:786–798. doi: 10.1111/j.1364-3703.2011.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otte O., Pachten A., Hein F., Barz W. Early elicitor-induced events in chickpea cells: Functional links between oxidative burst, sequential occurrence of extracellular alkalinisation and acidification, K+/H+ exchange and defence-related gene activation. Z. Naturforsch. C J. Biosci. 2001;56:65–76. doi: 10.1515/znc-2001-1-212. [DOI] [PubMed] [Google Scholar]
- 66.Benko-Iseppon A.M., Winter P., Huettel B., Staginnus C., Muehlbauer F.J., Kahl G. Molecular markers closely linked to fusarium resistance genes in chickpea show significant alignments to pathogenesis-related genes located on Arabidopsis chromosomes 1 and 5. Theor. Appl. Genet. 2003;107:379–386. doi: 10.1007/s00122-003-1260-x. [DOI] [PubMed] [Google Scholar]
- 67.Trudel J., Grenier J., Potvin C., Asselin A. Several thaumatin-like proteins bind to beta-1,3-glucans. Plant Physiol. 1998;118:1431–1438. doi: 10.1104/pp.118.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grenier J., Potvin C., Trudel J., Asselin A. Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. Plant J. 1999;19:473–480. doi: 10.1046/j.1365-313X.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 69.Petre B., Major I., Rouhier N., Duplessis S. Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biol. 2011;11:33. doi: 10.1186/1471-2229-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan M., Ngou B.P.M., Ding P., Xin X.F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021;62:102030. doi: 10.1016/j.pbi.2021.102030. [DOI] [PubMed] [Google Scholar]
- 71.Petre B. Toward the Discovery of Host-Defense Peptides in Plants. Front. Immunol. 2020;11:1825. doi: 10.3389/fimmu.2020.01825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jayaprakash A., Roy A., Thanmalagan R.R., Arunachalam A., Ptv L. Immune response gene coexpression network analysis of Arachis hypogaea infected with Aspergillus flavus. Genomics. 2021;113:2977–2988. doi: 10.1016/j.ygeno.2021.06.027. [DOI] [PubMed] [Google Scholar]
- 73.Kang J.N., Lee W.H., Won S.Y., Chang S., Hong J.P., Oh T.J., Lee S.M., Kang S.H. Systemic Expression of Genes Involved in the Plant Defense Response Induced by Wounding in Senna tora. Int. J. Mol. Sci. 2021;22:10073. doi: 10.3390/ijms221810073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Q., Sui X., Wang Y., Zhu M., Zhou Y., Gao F. Genome-Wide Analyses of Thaumatin-like Protein Family Genes Reveal the Involvement in the Response to Low-Temperature Stress in Ammopiptanthus nanus. Int. J. Mol. Sci. 2023;24:2209. doi: 10.3390/ijms24032209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fecht-Christoffers M.M., Braun H.P., Lemaitre-Guillier C., VanDorsselaer A., Horst W.J. Effect of manganese toxicity on the proteome of the leaf apoplast in cowpea. Plant Physiol. 2003;133:1935–1946. doi: 10.1104/pp.103.029215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Del Campillo E., Lewis L.N. Identification and kinetics of accumulation of proteins induced by ethylene in bean abscission zones. Plant Physiol. 1992;98:955–961. doi: 10.1104/pp.98.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Piromyou P., Nguyen H.P., Songwattana P., Boonchuen P., Teamtisong K., Tittabutr P., Boonkerd N., Alisha Tantasawat P., Gottfert M., Okazaki S., et al. The Bradyrhizobium diazoefficiens type III effector NopE modulates the regulation of plant hormones towards nodulation in Vigna radiata. Sci. Rep. 2021;11:16604. doi: 10.1038/s41598-021-95925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bari R., Jones J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009;69:473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 79.Wang L., Xu Z., Yin W., Xu K., Wang S., Shang Q., Sa W., Liang J., Wang L. Genome-wide analysis of the Thaumatin-like gene family in Qingke (Hordeum vulgare L. var. nudum) uncovers candidates involved in plant defense against biotic and abiotic stresses. Front. Plant Sci. 2022;13:912296. doi: 10.3389/fpls.2022.912296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kobayashi K., Fukuda M., Igarashi D., Sunaoshi M. Cytokinin-binding proteins from tobacco callus share homology with osmotin-like protein and an endochitinase. Plant Cell Physiol. 2000;41:148–157. doi: 10.1093/pcp/41.2.148. [DOI] [PubMed] [Google Scholar]
- 81.Ding L.N., Li Y.T., Wu Y.Z., Li T., Geng R., Cao J., Zhang W., Tan X.L. Plant Disease Resistance-Related Signaling Pathways: Recent Progress and Future Prospects. Int. J. Mol. Sci. 2022;23:16200. doi: 10.3390/ijms232416200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamchi A., Ben C., Rossignol M., Zareie S.R., Mirlohi A., Sayed-Tabatabaei B.E., Pichereaux C., Sarrafi A., Rickauer M., Gentzbittel L. Proteomics analysis of Medicago truncatula response to infection by the phytopathogenic bacterium Ralstonia solanacearum points to jasmonate and salicylate defence pathways. Cell. Microbiol. 2018;20:e12796. doi: 10.1111/cmi.12796. [DOI] [PubMed] [Google Scholar]
- 83.Yang J., Lan L., Jin Y., Yu N., Wang D., Wang E. Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 2022;64:244–267. doi: 10.1111/jipb.13207. [DOI] [PubMed] [Google Scholar]
- 84.Berrabah F., Ratet P., Gourion B. Legume Nodules: Massive Infection in the Absence of Defense Induction. Mol. Plant Microbe Interact. 2019;32:35–44. doi: 10.1094/MPMI-07-18-0205-FI. [DOI] [PubMed] [Google Scholar]
- 85.Liao C., Hochholdinger F., Li C. Comparative analyses of three legume species reveals conserved and unique root extracellular proteins. Proteomics. 2012;12:3219–3228. doi: 10.1002/pmic.201100629. [DOI] [PubMed] [Google Scholar]
- 86.Regalado A.P., Ricardo C.P. Study of the intercellular fluid of healthy Lupinus albus organs. Presence of a chitinase and a thaumatin-like protein. Plant Physiol. 1996;110:227–232. doi: 10.1104/pp.110.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanselle T., Ichinoseb Y., Barz W. Biochemical and molecular biological studies on infection (Ascochyta rabiei)-induced thaumatin-like proteins from chickpea plants (Cicer arietinum L.) Z. Naturforsch. C J. Biosci. 2001;56:1095–1107. doi: 10.1515/znc-2001-11-1229. [DOI] [PubMed] [Google Scholar]
- 88.Orona-Tamayo D., Wielsch N., Escalante-Perez M., Svatos A., Molina-Torres J., Muck A., Ramirez-Chavez E., Adame-Alvarez R.M., Heil M. Short-term proteomic dynamics reveal metabolic factory for active extrafloral nectar secretion by Acacia cornigera ant-plants. Plant J. 2013;73:546–554. doi: 10.1111/tpj.12052. [DOI] [PubMed] [Google Scholar]
- 89.Toyotake Y., Cho H.N., Kawamoto J., Kurihara T. A novel 1-acyl-sn-glycerol-3-phosphate O-acyltransferase homolog for the synthesis of membrane phospholipids with a branched-chain fatty acyl group in Shewanella livingstonensis Ac10. Biochem. Biophys. Res. Commun. 2018;500:704–709. doi: 10.1016/j.bbrc.2018.04.140. [DOI] [PubMed] [Google Scholar]
- 90.Alonso A., Pulido R. The extended human PTPome: A growing tyrosine phosphatase family. FEBS J. 2016;283:2197–2201. doi: 10.1111/febs.13748. [DOI] [PubMed] [Google Scholar]
- 91.Chuong N.N., Hoang X.L.T., Nghia D.H.T., Dai T.N.T., Thi V.L., Thao N.P. Protein Phosphatase Type 2C Functions in Phytohormone-Dependent Pathways and in Plant Responses to Abiotic Stresses. Curr. Protein Pept. Sci. 2021;22:430–440. doi: 10.2174/1389203722666210322144442. [DOI] [PubMed] [Google Scholar]
- 92.Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 93.Liu H., Lee M.J., Solis N.V., Phan Q.T., Swidergall M., Ralph B., Ibrahim A.S., Sheppard D.C., Filler S.G. Aspergillus fumigatus CalA binds to integrin α5β1 and mediates host cell invasion. Nat. Microbiol. 2016;2:16211. doi: 10.1038/nmicrobiol.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun Y., Detchemendy T.W., Pajerowska-Mukhtar K.M., Mukhtar M.S. NPR1 in JazzSet with Pathogen Effectors. Trends Plant Sci. 2018;23:469–472. doi: 10.1016/j.tplants.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 95.Zhang M., Liu Y., Cai H., Guo M., Chai M., She Z., Ye L., Cheng Y., Wang B., Qin Y. The bZIP Transcription Factor GmbZIP15 Negatively Regulates Salt- and Drought-Stress Responses in Soybean. Int. J. Mol. Sci. 2020;21:7778. doi: 10.3390/ijms21207778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sornaraj P., Luang S., Lopato S., Hrmova M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta. 2016;1860:46–56. doi: 10.1016/j.bbagen.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 97.Zhang M., Liu Y., Li Z., She Z., Chai M., Aslam M., He Q., Huang Y., Chen F., Chen H., et al. The bZIP transcription factor GmbZIP15 facilitates resistance against Sclerotinia sclerotiorum and Phytophthora sojae infection in soybean. Iscience. 2021;24:102642. doi: 10.1016/j.isci.2021.102642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fierens E., Gebruers K., Voet A.R., De Maeyer M., Courtin C.M., Delcour J.A. Biochemical and structural characterization of TLXI, the Triticum aestivum L. thaumatin-like xylanase inhibitor. J. Enzym. Inhib. Med. Chem. 2009;24:646–654. doi: 10.1080/14756360802321831. [DOI] [PubMed] [Google Scholar]
- 99.Smole U., Bublin M., Radauer C., Ebner C., Breiteneder H. Mal d 2, the thaumatin-like allergen from apple, is highly resistant to gastrointestinal digestion and thermal processing. Int. Arch. Allergy Immunol. 2008;147:289–298. doi: 10.1159/000144036. [DOI] [PubMed] [Google Scholar]
- 100.He L., Li L., Zhu Y., Pan Y., Zhang X., Han X., Li M., Chen C., Li H., Wang C. BolTLP1, a Thaumatin-like Protein Gene, Confers Tolerance to Salt and Drought Stresses in Broccoli (Brassica oleracea L. var. Italica) Int. J. Mol. Sci. 2021;22:11132. doi: 10.3390/ijms222011132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iquebal M.A., Soren K.R., Gangwar P., Shanmugavadivel P.S., Aravind K., Singla D., Jaiswal S., Jasrotia R.S., Chaturvedi S.K., Singh N.P., et al. Discovery of Putative Herbicide Resistance Genes and Its Regulatory Network in Chickpea Using Transcriptome Sequencing. Front. Plant Sci. 2017;8:958. doi: 10.3389/fpls.2017.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ng T.B., Au T.K., Lam T.L., Ye X.Y., Wan D.C. Inhibitory effects of antifungal proteins on human immunodeficiency virus type 1 reverse transcriptase, protease and integrase. Life Sci. 2002;70:927–935. doi: 10.1016/S0024-3205(01)01458-8. [DOI] [PubMed] [Google Scholar]
- 103.Zemanek E.C., Wasserman B.P. Issues and advances in the use of transgenic organisms for the production of thaumatin, the intensely sweet protein from Thaumatococcus danielli. Crit. Rev. Food Sci. Nutr. 1995;35:455–466. doi: 10.1080/10408399509527709. [DOI] [PubMed] [Google Scholar]
- 104.Faus I. Recent developments in the characterization and biotechnological production of sweet-tasting proteins. Appl. Microbiol. Biotechnol. 2000;53:145–151. doi: 10.1007/s002530050001. [DOI] [PubMed] [Google Scholar]
- 105.Bartoszewski G., Niedziela A., Szwacka M., Szczytt K.N. Modification of tomato taste in transgenic plants carrying a thaumatin gene from Thaumatococcus Daniellii Benth. Plant Breed. 2003;122:347–351. doi: 10.1046/j.1439-0523.2003.00864.x. [DOI] [Google Scholar]
- 106.Masuda T., Kitabatake N. Developments in biotechnological production of sweet proteins. J. Biosci. Bioeng. 2006;102:375–389. doi: 10.1263/jbb.102.375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing does not apply to this article.