Abstract
A set of 18 plasmid subclones of the Autographa californica nuclear polyhedrosis virus genome, each containing an identified late expression factor gene (lef), supports expression from a late viral promoter in transient expression assays in the SF-21 cell line derived from Spodoptera frugiperda. We have constructed a further set of plasmids in which each lef open reading frame (ORF) is controlled by the Drosophila melanogaster heat shock protein 70 (hsp70) promoter and epitope tagged. Failure of this set of plasmids to support transient late gene expression, and the inability of the p47 ORF to replace the p47-containing plasmid supplied in the lef plasmid library, led to the identification of a 19th late expression factor gene (lef-12) located adjacent to the p47 gene. The sequence of lef-12 is predicted to encode a protein of 21 kDa with no homology to any previously identified protein. The set of 19 hsp70-controlled lef ORFs (HSEpiHis lef library) supports transient expression from a late viral promoter. lef-12 did not affect expression from an early baculovirus promoter. In TN-368 cells, which are also permissive for virus replication, lef-12 provided a stimulatory effect but did not appear to be essential.
Genes involved in regulating late gene expression of the baculovirus Autographa californica nuclear polyhedrosis virus (AcMNPV) have been identified primarily by using a transient expression assay which is based on activation of a reporter gene under control of a late viral promoter by cotransfection with a combination of viral genes (38, 41). In addition to the reporter gene, the reporter plasmid used in this assay contains an AcMNPV homologous repeat (hr) region which serves as an origin of DNA replication (21, 22, 28, 43). When the reporter plasmid is cotransfected into cells in the presence of 12 overlapping clones (designated the genomic library) which collectively represent the AcMNPV genome, the levels of reporter gene expression are approximately 1,000-fold higher than those observed in the absence of viral sequences (41). Removal of certain clones from this genomic library results in greater than 20-fold decreases in transactivation levels. By replacing individual or overlapping sets of clones from the genomic library with subclones capable of supplying the transactivating activity, a set of 18 plasmids which support transient late gene expression in the IPBL-SF-21 (SF-21) cell line derived from the fall armyworm Spodoptera frugiperda was identified (23, 27, 33, 38–42, 50, 51). This set of 18 plasmids, each containing an identified late expression factor gene (lef), is designated the lef library. Very late gene expression, which is required for occlusion body formation, additionally requires the function of the very late expression factor 1 gene (vlf-1) product, a polypeptide with sequence motifs characteristic of a family of integrases/resolvases (30, 50).
A subset of lef genes, ie-1, ie-2, lef-1, lef-2, lef-3, p143, dnapol, p35, and lef-7, are required for optimal hr-dependent plasmid DNA replication in SF-21 cells and are therefore known as replication lef genes (20, 28). The need for replication lef genes demonstrates the dependence of late gene expression on DNA replication or reporter plasmid stability in the transient expression system (20, 28). Expression of late and very late baculovirus genes also depends on viral DNA replication during infection (25, 46, 49). IE-1 transactivates early gene expression and may be involved in transactivating lef gene expression (15, 16, 34). However, it also recognizes and binds imperfect palindromes within hr regions and may therefore be directly involved in DNA replication (8, 14, 47). IE-2 has two known functions: transactivating early gene expression (4, 5) and blocking cell cycle progression (44). IE-2 is not required for transient late gene expression in TN-368 cells (26) derived from Trichoplusia ni, and its role in the SF-21 assays is not clear. LEF-1 contains three sequence motifs which are conserved in DNA primases, suggesting that LEF-1 may be a baculovirus primase (2, 11). LEF-2 is known to be an essential gene for AcMNPV replication, and some mutant alleles display a delay in late gene expression and a defect in very late gene expression (31). LEF-3 is a single-stranded DNA binding protein (17). p143 and dnapol encode polypeptides with sequence similarities to DNA helicases and DNA polymerases, respectively. p35 is a general caspase inhibitor which is required to block virus-induced apoptosis in SF-21 cells (3, 9), and its role in this assay may be to stabilize the reporter plasmid from nucleolytic degradation (50). LEF-7, like IE-2 and p35, has little or no influence on plasmid DNA replication or stability in TN-368 cells, suggesting cell line-specific or host-specific factors are required for hr-dependent DNA replication or stability (26). In the cases of p35 and lef-7, such host or cell line specificity has been confirmed by analysis of mutant virus phenotypes (7, 9). An additional replicative lef, hcf-1, is required for optimal hr-dependent DNA replication and transient late gene expression in TN-368 cells (26), and virus mutants with deletions in hcf-1 show delayed DNA replication and defective late gene expression in TN-368 cells (29).
The remaining nine lef genes, termed transcription-specific lef genes, affect the steady-state levels of reporter gene transcripts but not plasmid DNA and are thus likely to be involved in transcription or RNA processing and stability (28). Two of these genes (lef-8 and lef-9) are predicted to be components of a viral RNA polymerase, based on amino acid sequence motifs that are conserved in prokaryotic and eukaryotic RNA polymerases (27, 42), and lef-6 may have a sequence motif related to one found in vaccinia virus RNA polymerase (39). The roles of p47 and LEF-4 in late gene transcription were identified by analysis of conditional lethal mutants of AcMNPV (6, 37). These mutants synthesize viral DNA at the nonpermissive temperature but are defective in late and very late gene expression. The remaining transcription-specific genes, lef-5, 39k, lef-10, and lef-11, appear to have no significant homology to genes in existing sequence databases.
Plasmids of the lef library supply the 18 lef genes, but because of additional flanking sequences included during subcloning, the lef library collectively contains approximately one-third of the AcMNPV genome. To define the role of each lef gene more thoroughly, it was of interest to construct a library of just the open reading frames (ORFs) of the lef genes. Furthermore, the genes comprising the lef library are all under the control of their original promoters, and at least some of them are known to be transactivated by IE-1 and/or IE-2 (5, 15, 24, 33–35). Therefore, we constructed a set of plasmids, each containing only a single lef ORF, fused to epitope and His6 tags and placed under constitutive promoter control. This collection of plasmids is designated the HSEpiHis lef library. In the process of constructing the HSEpiHis lef library, we discovered a previously unidentified lef, lef-12, on the plasmid subclone p47. The p47 plasmid is subsequently referred to as p47* since it contains two lef genes (p47 and lef-12) which are required for late and very late gene expression. We also found that all 19 lef genes are necessary for transient late gene expression in SF-21 cells and that collectively these genes are sufficient to support late gene expression at levels that are 30% or higher than that observed in the presence of the entire genome.
MATERIALS AND METHODS
Cells.
S. frugiperda SF-21 (53) and T. ni TN-368 (19) cells were grown at 27°C in TC-100 medium (Life Technologies, Inc., Gaithersburg, Md.) containing 10% fetal bovine serum and 0.26% tryptose broth (36).
Plasmid constructs.
The previously described reporter plasmids pETCAThr5 (41), pCAPCAT (49), and phcwt (45) contain the early ETL, late vp39, and very late polh promoters, respectively, controlling the reporter gene, which encodes chloramphenicol acetyltransferase (cat). The construction of pHSEpip35VI+ has been described elsewhere (48).
The lef library and overlapping AcMNPV clone library used in these experiments (Table 1) have been described elsewhere (41).
TABLE 1.
Oligonucleotides and plasmid templates used to construct the HSEpiHis lef genes
| Primer | Sequencea | GenBank coordinatesb | Template | Reference |
|---|---|---|---|---|
| NIE1 | 5′-GAAGATCTACGCAAATTAATTTTAACGC-3′ | 127201–127220 | pPS-IE1/HC | 41 |
| CIE1 | 5′-TTTTCCCCCCGGGTCGCCAACTCCCATTGTTAT-3′ | 129048–129067 | pPS-IE1/HC | 41 |
| N2IE2 | 5′-CGCGGATCCAGTCGCCAAATCAACGCCGCC-3′ | 132060–132080 | pBSP-PstNA | 41 |
| CIE2 | 5′-AAGGAAAAAAGCGGCCGCTGGATGTACCGCTAACCAAA-3′ | 130750–130731 | pBSP-PstNA | 41 |
| NLF2noIM | 5′-GAAGATCTGCGAATGCATCGTATAACGT-3′ | 3092–3111 | pBE42-630 | 41 |
| CLEF2 | 5′-TTTTCCCCCCGGGATTTATAATTGTTTTATTAT-3′ | 3722–3741 | pBE42-630 | 41 |
| NLEF1 | 5′-GAAGATCTTTAGTGTGCAATTATACGCA-3′ | 11291–11310 | pBS BCNE | 38 |
| CLEF1 | 5′-TTTTCCCCCCGGGTTGCATTTGAATGAGTCCCA-3′ | 10463–10483 | pBS BCNE | 38 |
| NLEF6 | 5′-GAAGATCTAACATGGTGTTCAACGTGTACTAC-3′ | 23465–23485 | pAcIAP-PstI/NsiI | 39 |
| CLEF6 | 5′-TACTCCCGGGGTTTTATTGTTTTTCTAATACATTC-3′ | 23965–23989 | pAcIAP-PstI/NsiI | 39 |
| N39K | 5′-GAAGATCTAACATGGTAAACGTGCCGGAG-3′ | 30052–30072 | pNspAfl | 51 |
| C39Klong | 5′-TTTCCCCCCGGGTTAATCTGACATATTTGTAT-3′ | 29242–29261 | pNspAfl | 51 |
| NLEF11 | 5′-GAAGATCTAACATGCCCCCCAAAAATTGCAC-3′ | 30382–30401 | pHindIIIR | 51 |
| CLEF11 | 5′-ATATCCCGGGTGTTGCTCCGGCACGTTTAC-3′ | 30047–30066 | pHindIIIR | 51 |
| N2P47 | 5′-GAAGATCTTTTGTCACCCGGTTGGAGCACA-3′ | 33358–33379 | p47* | 51 |
| CP47 | 5′-ATATCCCGGGTTTGTCCATGATTGGCTCAG-3′ | 32161–32180 | p47* | 51 |
| NLEF12 | 5′-GAAGATCTACTATGGAAAATAACGCGGAATTCA-3′ | 33384–33408 | p47* | 51 |
| CLEF12 | 5′-TTTTCCCCCCGGGTATTGCGCGCTCGTAATCTA-3′ | 33954–33973 | p47* | 51 |
| N2LEF8 | 5′-GAAGATCTACGGACGTGGTTCAAGATTT-3′ | 43131–43150 | pBS-RI-M | 42 |
| CLEF8LONG | 5′-TTTCCCCCCGGGCAGTGATTCTAATTGCAGCTGC-3′ | 40420–40441 | pBS-RI-M | 42 |
| N2LEF10 | 5′-GAAGATCTACGAACGTATGGTTCGCGACGG-3′ | 45131–45152 | pPstHIME0.5 | 27 |
| CLEF101g | 5′-TTTCCCCCCGGGAATAATTGTTACGTGGACGCG-3′ | 45352–45372 | pPstHIME0.5 | 27 |
| N2LEF9 | 5′-GAAGATCTTTTTCTTTTTTGGATAAAACTCC-3′ | 49265–49286 | pPstHISB2.35 | 27 |
| CLEF9LONG | 5′-TTTCCCCCCGGGTTATCATTCAATGAACATGTCG-3′ | 50716–50737 | pPstHISB2.35 | 27 |
| NDNAp | 5′-GAAGATCTAAAATATATCCTTACAATGA-3′ | 55261–55280 | pDNAp | 27 |
| CDNAp | 5′-AAGGAAAAAAGCGGCCGCAACCGCTGTCGTAATCTTGG-3′ | 52227–52246 | pDNAp | 27 |
| N2LEF3 | 5′-CGCGGATCCGCGACCAAAAGATCTTTGTC-3′ | 58856–58875 | pSDEM2 | 23 |
| C2LEF3 | 5′-TTTTCCCCCCGGGTTGCAGATCAGGCTGTCAAA-3′ | 57534–57553 | pSDEM2 | 23 |
| 5′ hcflbgl | 5′-GAAGATCTGATTCGCTAGCCAATTTGTGCTTG-3′ | 60113–60136 | pXABgE3.6 | 26 |
| 3′ hcflpspa | 5′-TTTTCCCCCCGGGTCTAGCGACATGTTTGCC-3′ | 61075–61092 | pXABgE3.6 | 26 |
| NVLF1 | 5′-GAAGATCTAACGGTTTTAATGTTCGCAA-3′ | 64930–64949 | pXA-7 | 30 |
| CVLF1 | 5′-TTTCCCCCCGGGCCGGGCTGACGATAATAAAC-3′ | 63677–63696 | pXA-7 | 30 |
| NLEF4 | 5′-GAAGATCTAACATGGACTACGGCGATTTTG-3′ | 76596–76614 | p86D5B | 40 |
| CLEF4 | 5′-TATCCCGGGGCTTTAATTTGGCACGATTC-3′ | 77974–77992 | p86D5B | 40 |
| p143N | 5′-GAAGATCTATTGACAACATTTTACAATT-3′ | 84337–84356 | pBR322-RI-D/SmaIΔORF6 | 40 |
| p143C | 5′-TTTTCCCCCCGGGATTGTGTGTTTGATCGACCC-3′ | 80460–80479 | pBR322-RI-D/SmaIΔORF6 | 40 |
| NLEF5 | 5′-GAAGATCTAACATGTCGTTTGATGATG-3′ | 85918–85933 | pH3H/SH/ORF6 | 40 |
| CLEF5 | 5′-TATCCCGGGCTATTAACAACCAGACATTCC-3′ | 86698–86718 | pH3H/SH/ORF6 | 40 |
| NLEF7 | 5′-GAAGATCTTCGAGCGTTACAAAGCGCCC-3′ | 105211–105230 | pBSXBglII | 33 |
| CLEF7 | 5′-TTTTCCCCCCGGGCGCCACCGTTCTCTAACATT-3′ | 104487–104506 | pBSXBglII | 33 |
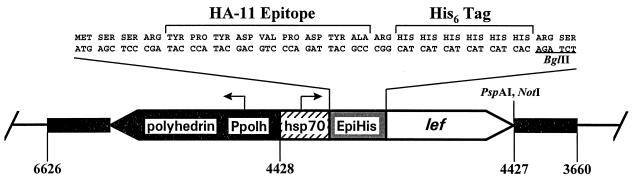
Plasmid pHSEpiHisVI+ was constructed by digesting pHSEpiOpIAPVI+ (18) with XmaI and NotI to remove the OpIAP ORF and inserting overlapping oligonucleotides with XmaI and NotI cohesive ends encoding a His6 tag and BglII, PspAI, and NotI sites. A schematic diagram of pHSEpiHisVI+ with a generic lef inserted in the BglII and PspAI sites is shown in Fig. 1. In the name pHSEpiHisVI+, “HS” refers to the Drosophila melanogaster heat shock protein 70 (hsp70) promoter (52), “Epi” refers to the HA.11 epitope (12), and “His” refers to the six-histidine tag fused to the N terminus of the lef gene cloned into the vector; “VI+” indicates that the vector contains the polyhedrin gene and sequences flanking this gene which allow selectable homologous recombination into the baculovirus genome for expression in future studies.
FIG. 1.
Schematic diagram of pHSEpiHisVI+. Arrows indicate the direction of transcription of the polyhedrin gene and D. melanogaster hsp70 gene promoters (Ppolh and hsp70, respectively). “EpiHis” refers to the HA.11 epitope and His6 tag which are fused to the N terminus of each lef. Only the BglII, PspAI, and NotI sites of the multiple cloning site are shown. The orientation of a generic lef inserted between the BglII and PspAI sites is indicated by an open arrow. Black portions indicate the polyhedrin gene and flanking viral sequences. The limits of the viral sequences in this vector are indicated by the numbers at the bottom (1) (GenBank accession no. L22858). The pUC8 sequences are represented by the single black line.
To construct the HSEpiHis lef library, the ORF for each lef was amplified with Pfu DNA polymerase (Stratagene, La Jolla, Calif.), using the primers and plasmid templates of the lef library listed in Table 1. The N2LEF9 primer corresponded to the second ATG of the lef-9 ORF. PCR products were digested with the appropriate restriction enzymes (sites underlined in Table 1) and gel purified prior to their insertion into pHSEpiHisVI+. The 5′ end of each lef PCR product was ligated to the BglII site of the pHSEpiHisVI+ multiple cloning site (Fig. 1).
Construction of lef-12 frameshift mutants.
The AcMNPV plasmid subclone p47* was digested with EcoRI; the ends were filled in with T4 polymerase and religated to produce plasmid p47*/EcoRI fs. The mutation in this plasmid was confirmed by sequencing, and the mutant gene is predicted to produce a 73-amino-acid peptide containing the first 9 amino acids of lef-12. To construct p47*/ApaI fs, p47* was digested with ApaI, and the ends were removed by using T4 DNA polymerase and religated. This plasmid, which was sequenced and found to contain a 5-bp deletion in the lef-12 ORF, is predicted to produce a 106-amino-acid peptide that contains the first 95 amino acids of lef-12. The same mutations were made in the plasmid containing the hsp70-promoted lef-12 ORF (pHSEpiHis lef-12) and confirmed by sequencing; the resultant plasmids were called pHSEpiHis lef-12/EcoRI fs and pHSEpiHis lef-12/ApaI fs.
Transfections and transient expression assays.
SF-21 or TN-368 cells were transfected by using Lipofectin reagent (Life Technologies). Cells were transfected with 2.0 μg of reporter plasmid and either 0.5 μg of each of the clones of the AcMNPV genomic library or 1.0 μg of each plasmid of the lef library or HSEpiHis lef library.
Cells were collected at 24, 48, and 72 h after transfection for samples containing pETCAThr5, pCAPCAT, and phcwt, respectively, and lysates were assayed for CAT activity (13), using 1/10 of the lysate or dilutions thereof for quantitation purposes. Quantitations of CAT assays were done directly on the thin-layer chromatography plates with a PhosphorImager 4000 (Molecular Dynamics, Sunnyvale, Calif.).
DNA replication assays.
The method has been described previously (28). Briefly, 1.8 × 106 SF-21 cells were cotransfected with pCAPCAT and clones of the AcMNPV genomic library or the HSEpiHis lef library. DNA from each sample was digested with BglII and DpnI, electrophoresed through a 0.7% agarose gel, and transferred to Zeta-probe nylon membranes (Bio-Rad, Richmond, Calif.). Membrane-bound DNA was hybridized to a [32P]dCTP nick-translated BglII-KpnI fragment of pCAPCAT containing the cat ORF. The relative levels of DNA replication were determined by using a PhosphorImager 4000.
Immunoblotting.
SF-21 cells (5.4 × 105) were transfected with 2 μg of each HSEpiHis lef as described above. At 14 h after transfection, the cells were heat shocked for 30 min at 42°C. At 2 h after heat shock treatment, cells were lysed in sodium dodecyl sulfate (SDS) sample buffer, and equal volumes representing the same cell number were resolved on an SDS–10 to 18% gradient polyacrylamide gel. The resolved proteins were then blotted onto polyvinylidene fluoride membranes (Millipore, Bedford, Mass.). The membranes were blocked and then incubated with anti-HA.11 mouse immunoglobulin G (BAbCO, Richmond, Calif.) followed by anti-mouse immunoglobulin G conjugated to horseradish peroxidase. Immunoreactive proteins were visualized with the enhanced chemiluminescence Western blotting system (Amersham Life Science Inc., Arlington Heights, Ill.).
RESULTS
Construction of the HSEpiHis lef library.
We constructed a lef library (HSEpiHis lef library) in which all of the individual lef ORFs were placed under D. melanogaster hsp70 promoter control within plasmid pHSEpiHisVI+ (Fig. 1). The hsp70 promoter was chosen for expression of lef genes since earlier studies have shown that the promoter is constitutively expressed in the absence of heat shock in SF-21 and TN-368 cells (32) but can be strongly induced with heat shock treatment (10).
ORF41 (lef-12) is involved in late gene expression in SF-21 cells.
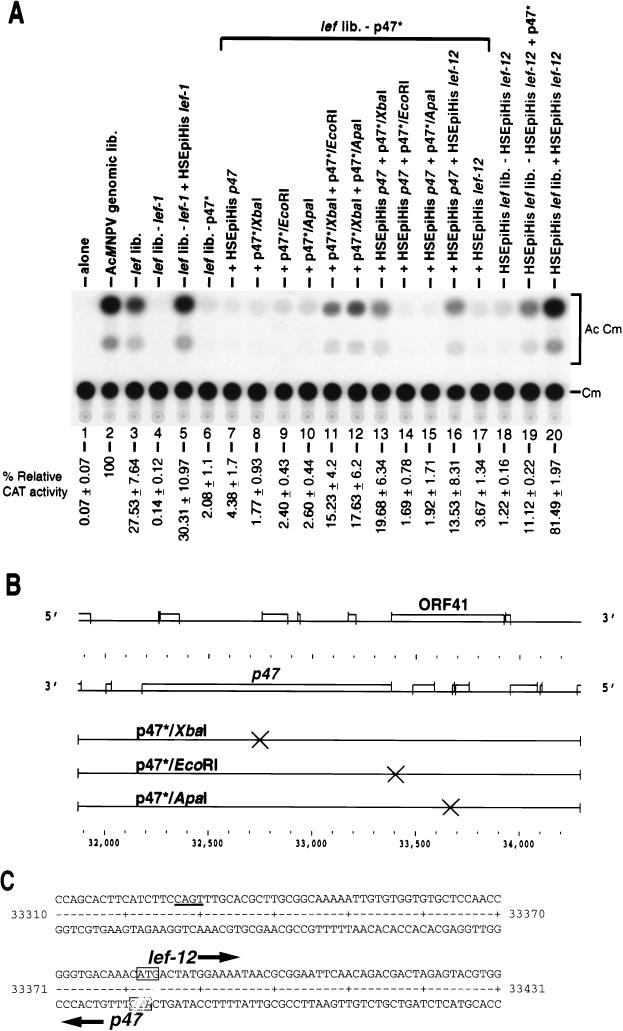
CAT gene expression from the late promoter of reporter plasmid pCAPCAT is activated over 100-fold by cotransfection with the set of genomic clones representing the entire AcMNPV genome (Fig. 2A; compare lanes 1 and 2). Similarly, addition of the 18 clones constituting the lef library stimulates expression over 100-fold (lane 3). The level of expression from the lef library is three- to fourfold lower than that from the genomic library in this experiment. In other experiments using other preparations of plasmids, the two libraries can be virtually equivalent (28), although the general trend is for expression from the lef library to be slightly lower than that from the genomic library. Since we define lef genes as those genes which provide at least a 10-fold stimulation to late reporter gene expression, the lef library appears to contain all the necessary lef genes for late gene expression but may lack genes which can stimulate expression mildly. Deletion of any one of the lef clones from the library decreases expression by 10-fold or more (e.g., lef-1 [lane 4]) (28).
FIG. 2.
Involvement of ORF 41 (lef-12) in late gene expression. (A) Transient expression assays showing the levels of CAT activity from SF-21 cells transfected with the late reporter plasmid pCAPCAT alone (lane 1) or cotransfected with pCAPCAT and the complete AcMNPV genomic library (lane 2), the lef library (lane 3), the lef library with lef-1 omitted (lanes 4 and 5), the lef library with p47* omitted (lanes 6 through 17), or the HSEpiHis lef library (lanes 18 through 20). Additional plasmid clones or clones missing from cotransfections are shown above each lane. Lanes 4 and 6 contained the empty vector plasmid pHSEpiHisVI+ to maintain DNA concentrations equivalent to those in lanes 5 and 7. The acetylated chloramphenicol products (Ac Cm) and unacetylated substrate (Cm) are indicated on the right. Relative CAT activities are shown below each lane. (B) ORFs and frameshift constructs of p47*. The ORF diagram was generated by the FRAMES program of the Wisconsin Package, version 8.1 (Genetics Computer Group, Inc., Madison, Wis.). Frameshift mutations introduced into p47* (×) are indicated on the XbaI, EcoRI, and ApaI sites. Plasmid p47*/XbaI has been previously described (51). Numbers at the bottom refer to sequence coordinates for GenBank accession no. L22858 (1). (C) Nucleotide sequence of the p47 and lef-12 overlapping N-terminal regions. Initiating methionine codons are boxed. Large arrows show the direction of the lef-12 and p47 ORFs. A possible initiation site for lef-12 transcription is underlined. Numbers at the sides refer to coordinates for GenBank accession no. L22858 (1).
Each plasmid of the HSEpiHis lef library was tested for its ability to substitute in transient expression assays for its counterpart in the lef library. A representative example of how each HSEpiHis lef was tested is shown for HSEpiHis lef-1, which restored the relative CAT activity to levels observed with the lef library containing lef-1 (Fig. 2A, lanes 3 through 5). Unlike the other 17 HSEpiHis lef genes which could substitute for their counterpart in the lef library in the transient assay, HSEpiHis p47 did not restore CAT expression to the level observed with the lef library containing p47* (lanes 6 and 7). Therefore, a set of frameshift mutants of p47* (Fig. 2B) was used to investigate the possibility that p47* contained an unidentified lef (Fig. 2A, lanes 8 through 15). In the absence of the p47* plasmid but in the presence of HSEpiHisp47, the addition of p47*/XbaI fs, which is defective in p47 function (51), restored CAT activity to levels similar to those observed with the lef library (lane 13), suggesting that p47* contains a second element required for transient late gene expression. The p47* plasmid, formerly known as p47, contains 911 bp upstream and 307 bp downstream of the p47 ORF (Fig. 2B). One other ORF, ORF41, is present on p47*; this ORF has the potential to encode a polypeptide of at least 50 amino acids and could supply lef function in the assay. In the absence of p47*, frameshift mutants of ORF41, p47*/EcoRI, and p47*/ApaI did not restore expression of the reporter plasmid (Fig. 2A, lanes 9 and 10) indicating that ORF41 had transactivating activity and/or that the EcoRI and ApaI mutations interfered with the expression of p47. The possibility that the frameshift mutations in ORF41 were interfering with p47 expression was excluded since in the absence of p47*, both p47*/EcoRI and p47*/ApaI were able to restore CAT activity when used in combination with p47*/XbaI (which is defective in p47) and thus were able to supply p47 activity (lanes 11 and 12). In addition, ORF41 was cloned into pHSEpiHisVI+ and tested in the lef assay in combination with HSEpiHis p47 (lane 16) or without the p47 ORF (lane 17). CAT activity was partially restored when both the p47 and ORF41 coding sequences were expressed constitutively in the context of the lef library (lane 16). In the context of the HSEpiHis lef library, the coding sequence of ORF41, subsequently known as lef-12, transactivated expression from the vp39 promoter significantly and gave levels of CAT activity 80% of those seen for the AcMNPV genomic library (lanes 18 through 20). It is possible that lef-12 down regulates one or more lef promoters while transactivating the vp39 promoter, and thus its effect is most easily observed in the context of the HSEpiHis lef library.
Examination of the sequence of p47* revealed that the translational initiation codons for the p47 ORF and the lef-12 ORF divergently overlap by two nucleotides, based on the predicted ORFs described by Ayres et al. (1) (Fig. 2C).
Immunoblot detection of each HSEpiHis lef.
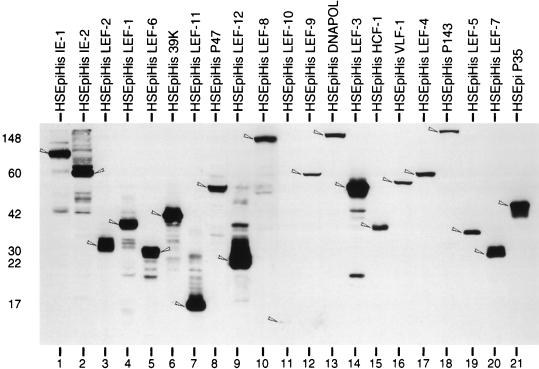
To test the expression of each HSEpiHis lef, plasmids were transfected individually into SF-21 cells; the cells were subsequently heat shocked, and the cell lysates were subjected to immunoblot analysis (Fig. 3). Predicted molecular masses of the HSEpiHis proteins ranged from 11.1 kDa for HSEpiHis lef-10 (Fig. 3, lane 11) to 146 kDa for HSEpiHis p143 (lane 18). Although equal portions of cell lysates representing equal numbers of cells were loaded on the gel for each HSEpiHis lef gene, expression levels varied widely. HSEpiHis lef-10 (Fig. 3, lane 11) reproducibly showed the lowest level of expression among the HSEpiHis lef genes. In contrast, HSEpiHis lef-3 (lane 14) and HSEpiHis lef-12 (lane 9) reproducibly showed the highest levels of expression and were easily detectable.
FIG. 3.
Immunoblot of HSEpiHis lef genes expressed in transfected cells. SF-21 cells were transfected with 2 μg of each HSEpiHis lef, heat shocked for 30 min at 18 h posttransfection, and harvested 2 h after heat shock. Equal amounts of total cell lysates were analyzed on an SDS–10 to 18% gradient polyacrylamide gel followed by immunoblot analysis using an anti-hemagglutinin monoclonal antibody. Arrowheads indicate the bands corresponding to the predicted molecular weight of each HSEpiHis LEF. Positions of molecular weight standards (in thousands) are shown on the left.
lef-12 is involved in late gene expression in the context of the overlapping genomic library in SF-21 cells.
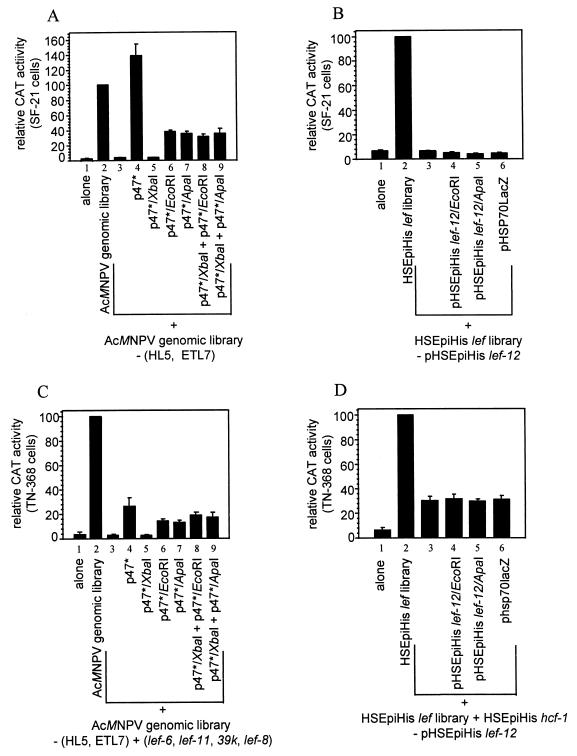
Since p47 was originally identified as the only late expression factor within p47* responsible for transient late gene expression in the context of the AcMNPV genomic library, we reexamined the roles of both p47 and lef-12 in this context. The genes for lef-12 and p47 are located on two adjacent overlapping genomic clones, HL5 (starting at bp 22060 and ending at bp 38451) and ETL7 (starting at bp 29032 and ending at bp 45037) (1). In addition to p47 and lef-12, HL5 and ETL7 contain copies of the genes for late expression factors lef-6, 39k, lef-11, and lef-8. However, unlike p47 and lef-12, one or more copies of each of these lef genes are present on other library clones. To investigate the influence of lef-12 in transient late gene expression assays with the AcMNPV genomic library, we removed HL5 and ETL7 and replaced them with plasmids containing intact or frameshift versions of lef-12 and p47 (Fig. 4A). Omission of HL5 and ETL7 from the AcMNPV library reduced CAT gene expression to background levels (Fig. 4A; compare lanes 2 and 3). The addition of p47*, which contains both p47 and lef-12, increased CAT expression levels to 140% of that seen using the AcMNPV genomic library (lane 4); thus, sufficient quantities of lef-6, 39k, lef-11 and lef-8 are expressed from genes located on other genomic clones, and p47* was able to provide the remaining lef genes found in the AcMNPV genomic library. Substitution of p47* with a plasmid containing a frameshift mutant of p47 (p47*/XbaI) reduced CAT gene expression to background levels (lane 5) and supports the previous finding that p47 is required for optimal late gene expression (51). Substituting p47* with clones containing intact p47 and frameshifts of lef-12 (p47*/EcoRI and p47*/ApaI) reduced CAT gene expression to approximately 28% of that seen with wild-type lef-12 (compare lanes 6 and 7 with lane 4) and to about 40% of that seen with the complete genomic library (compare lanes 6 and 7 with lane 2). This finding indicated that lef-12 has a role in late gene expression in transient assays using the genomic library but does not appear to be required in this context.
FIG. 4.
Requirement for lef-12 in transient late gene expression assays. SF-21 cells (A and B) or TN-368 cells (C and D) were transfected with the AcMNPV genomic library (A and C) or the HSEpiHis lef library (B and D). Library clones supplied or omitted from the complete libraries are indicated below each graph. In panel C, plasmid clones pAcIAP-PstI/NsiI and pBS-RI-M were added to supply lef-6 and lef-8, respectively, and plasmid pFspAfl (51) was added to supply both 39k and lef-11. Plasmids supplying wild-type p47 or lef-12 or corresponding frameshift mutations are shown below the lanes. CAT activities are reported relative to those of the full libraries; data represent the mean of at least three independent experiments, and bars represent the standard error.
In assays using the AcMNPV genomic library, substitution of clones HL5 and ETL7 with two plasmids, p47*/XbaI, the plasmid containing a mutant p47 but an intact lef-12, and either p47*/EcoRI or p47*/ApaI, both of which contain a mutated lef-12 and an intact p47, did not give high levels of CAT expression (Fig. 4A; compare lanes 8 and 9 with lanes 6 and 7). The reason for the lack of complementation between these plasmids in this assay is not clear.
As demonstrated in Fig. 2A, lef-12 was required for optimal late gene expression in transient assays using a set of 19 plasmids in the context of the HSEpiHis lef library. Removal of pHSEpiHis lef-12 or substitution with a pHSEpiHisVI+-based plasmid containing a frameshift of lef-12 or a lacZ gene reduced CAT gene expression to background levels (Fig. 4B; compare lanes 3 to 6 with lane 2), confirming that lef-12 is a late expression factor in this context.
LEF-12 has a stimulatory role on late gene expression in TN-368 cells in the context of the HSEpiHis lef library.
In transient late expression assays of TN-368 cells, when clones HL5 and ETL7 were removed from the genomic library and p47 and lef-12 were provided by the addition of p47*, CAT expression levels were only 15 to 20% of that seen for the complete AcMNPV genomic library (data not shown). To eliminate the possibility that the reduced copy numbers of the other lef genes present on HL5 and ETL7 limited CAT expression, the assays were supplemented with plasmids containing lef-6, 39k, lef-11, and lef-8. The level of CAT gene expression increased marginally to about 25 to 30% of that seen with the complete genomic library (Fig. 4A, lane 4). The reason why p47* cannot fully substitute for HL5 and ETL7 in TN-368 cells is not known. Substituting a frameshift mutant p47 in this system resulted in background levels of CAT expression (Fig. 4C; compare lane 5 with lane 1), confirming a role for p47 in late gene expression in transient assays in TN-368 cells. Lack of a functioning lef-12 gene caused only a 40 to 50% reduction in the levels of CAT gene expression (compare lanes 6 and 7 with lane 4) and suggested that lef-12 may play only a stimulatory role in TN-368 cells. As in SF-21 cells, addition of two plasmids each containing a frameshift in p47 or lef-12 did not further augment expression (lanes 8 and 9).
In TN-368 cells, removal of pHSEpiHis lef-12 from the HSEpiHis lef library or replacement with a construct containing a pHSEpiHisVI+-based frameshifted lef-12 gene or lacZ gene reduced CAT gene expression by a factor of 3 (Fig. 4D; compare lanes 3 to 5 with lane 2). This contrasts with the 10-fold reduction seen in SF-21 cells and suggests that lef-12 has a stimulatory rather than an essential role in late transient gene expression in TN-368 cells.
lef-12 is involved in late and very late but not early gene expression.
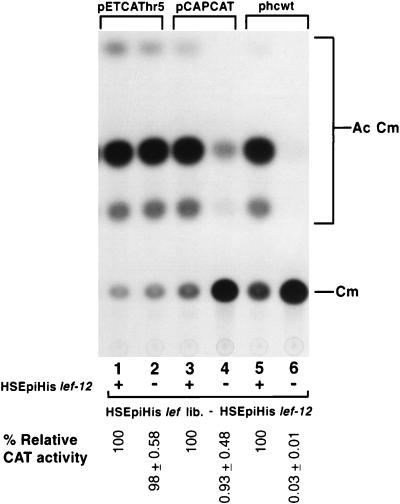
Since HSEpiHis lef-12 was important for expression from the late reporter plasmid, pCAPCAT, in combination with the HSEpiHis lef library in SF-21 cells, it was of interest to determine whether HSEpiHis lef-12 had an effect on early and very late gene expression. To test this, HSEpiHis lef-12 was cotransfected with the HSEpiHis lef library containing all of the lef genes except HSEpiHis lef-12 along with reporter plasmids containing the CAT gene under early (pETCAThr5) or very late (phcwt) promoter control (Fig. 5). Expression of pETCAThr5 was unaffected by replacement of HSEpiHis lef-12 by its frameshifted version (Fig. 6; compare lanes 1 and 2), indicating that lef-12 does not stimulate expression from this early promoter. In contrast, substitution of the frameshifted HSEpiHis lef-12 resulted in dramatic decreases in CAT expression from both pCAPCAT and phcwt (compare lanes 3 and 4 and lanes 5 and 6, respectively), indicating the involvement of a functional HSEpiHis lef-12 gene product in both late and very late gene expression.
FIG. 5.
Effect of lef-12 on early, late, and very late gene expression in SF-21 cells. The HSEpiHis lef library minus HSEpiHis lef-12 was cotransfected with reporter plasmids containing the CAT gene under early (pETCAThr5) (lanes 1 and 2), late (pCAPCAT) (lanes 3 and 4), and very late (phcwt) (lanes 5 and 6) promoter control in the presence of HSEpiHis lef-12 (+) (lanes 1, 3, and 5) or its frameshifted version (−) (lanes 2, 4, and 6). HSEpiHis vlf-1 was added in lanes 5 and 6. The CAT activities shown below the lanes are relative to those of each of the reporter plasmids cotransfected with the HSEpiHis lef library and HSEpiHis lef-12. The acetylated chloramphenicol products (Ac Cm) and unacetylated substrate (Cm) are indicated on the right.
FIG. 6.
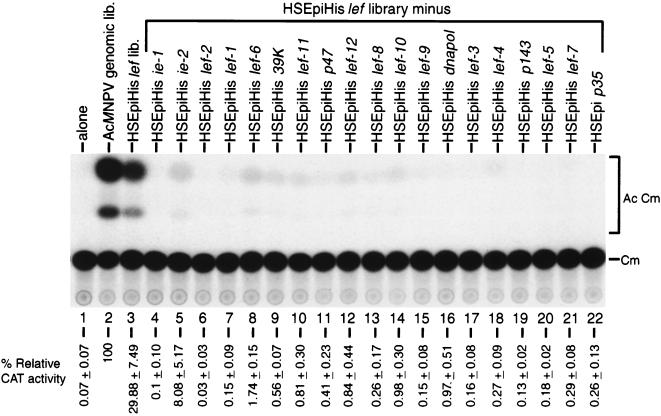
Contribution of each HSEpiHis lef to late gene expression. Transient expression assays showing the levels of CAT activity from SF-21 cells transfected with the late reporter plasmid pCAPCAT alone (lane 1) or cotransfected with pCAPCAT and the AcMNPV genomic library (lane 2), the HSEpiHis lef library (lane 3), or the HSEpiHis lef library lacking one of the 19 HSEpiHis lef genes (indicated above the lanes) (lanes 4 through 22). Cotransfections in lanes 4 through 22 each contained the empty vector pHSEpiHisVI+ to equal the total amount of DNA in lane 3. The acetylated chloramphenicol products (Ac Cm) and unacetylated substrate (Cm) are indicated on the right. Relative CAT activities are shown below each lane.
Effect of omitting each EpiHis lef library clone on expression from a late promoter in SF-21 cells.
To determine the specific contribution of each HSEpiHis lef to late gene expression from the reporter plasmid pCAPCAT, each lef was removed individually from cotransfections which included all other members of the HSEpiHis lef library and replaced with the control vector plasmid pHSEpiHisVI+ (Fig. 6). Removal of HSEpiHis ie-2 resulted in only a 3.7-fold decrease in relative CAT activity (Fig. 6, lane 5 versus lane 3), while individual removal of the other HSEpiHis lef genes, such as HSEpiHis lef-12 (lane 12), resulted in a 35-fold or greater decrease in relative CAT activity (lanes 4 and 6 through 22).
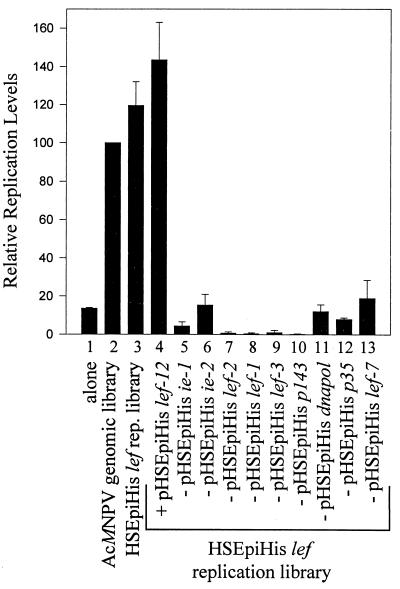
lef-12 does not play a role in DNA replication.
In previous experiments which identified late expression factors with roles in DNA replication, it was established that p47* was not required in hr-dependent plasmid replication assays. Since lef-12 was also provided by p47*, it is likely that like p47, lef-12 is not required for DNA replication. However, it was of interest to investigate whether lef-12 had any effect on DNA replication in the context of the pHSEpiHis lef replication library. In transient DNA replication assays, the pHSEpiHis lef replication library gave levels of replication approximately 80% of those seen with the AcMNPV genomic library (Fig. 7, lanes 2 and 3). When pHSEpiHis lef-12 was added to the pHSEpiHis replication lef library, there was no significant difference in the levels of plasmid DNA replication (lane 4). To determine the specific contribution of each HSEpiHis lef on transient DNA replication, we removed each HSEpiHis lef individually from the assay system. Removal of HSEpiHis ie-1, lef-2, lef-1, lef-3, p143, and p35 (lanes 5 and 7 through 10) strongly reduced replication levels, indicating a role in viral DNA replication, whereas removal of pHSEpiHis ie-2, pHSEpiHis dnapol, and pHSEpiHis lef-7 (lanes 6, 11, and 13) decreased transient replication levels partially, supporting the view that these lef genes are stimulatory for DNA replication under these transient assay conditions (28).
FIG. 7.
Contribution of pHSEpiHis lef-12 and each HSEpiHis replication lef to plasmid DNA replication. SF-21 cells were cotransfected with pCAPCAT (lane 1), pCAPCAT and the complete AcMNPV genomic library (lane 2), pCAPCAT and the HSEpiHis lef replication library (lane 3), or pCAPCAT and the HSEpiHis lef replication library with the addition of pHSEpiHis lef-12 or lacking one of the nine lef genes involved in plasmid replication (lanes 4 to 13). Levels of replicated plasmid were quantitated relative to those observed in the presence of the AcMNPV genomic library. The results shown are representative of two independent experiments.
DISCUSSION
We have constructed a set of 18 plasmids in which each previously identified lef ORF is expressed as an epitope-tagged fusion protein from the D. melanogaster hsp70 promoter. Failure of this library to support late gene expression in transient expression assays in SF-21 cells led to the identification of a 19th lef, lef-12 (ORF41), located adjacent to p47. The set of 19 plasmids, designated the HSEpiHis lef library, supports gene expression from a late viral promoter in transient assays.
lef-12 potentially encodes a 181-amino-acid polypeptide with a predicted molecular mass of 21,058 Da with no significant similarities to other genes in the protein or nucleic acid databases. lef-12 is predicted to be expressed as an early gene, since a CAGT motif is found at an appropriate distance (52 bp) upstream from the lef-12 initiation codon, and the nearest TAAG sequence, a characteristic promoter element of late and very late genes, is 711 bp upstream from the lef-12 initiation codon.
In the context of the HSEpiHis lef library in SF-21 cells, lef-12 is necessary for expression of a reporter gene under control of the late capsid or very late polyhedrin promoters but does not affect the early ETL promoter. Although lef-12 activates late gene expression at least 10-fold in the context of the HSEpiHis lef library in SF-21 cells, it exerts only a 3-fold effect in TN-368 cells in this context. Whether this gene is essential to AcMNPV infection remains to be determined.
The difference in the relative level of stimulation by lef-12 observed in the two contexts, the AcMNPV genomic library or the HSEpiHis lef library, suggests that another AcMNPV gene may be functionally redundant with lef-12, that lef-12 may negatively affect transcription from another lef promoter or that the relative levels of expression of the lef genes may influence gene expression in these transient assays. As shown in Fig. 3, the relative levels of expression of members of the HSEpiHis lef library varied widely and probably do not reflect the levels of lef expression found in an AcMNPV infection or in transient assays involving the AcMNPV genomic library.
Clues to why lef-12 was not identified in earlier experiments (51) may be obtained from the failure of a pair of plasmid subclones, one containing an intact p47 and the other containing an intact lef-12, to substitute for plasmid p47* in the AcMNPV genomic library. The presence of both plasmids should have provided intact copies of both p47 and lef-12 and thus restored activity, but they did not. The reason these plasmids were unable to supply the functions of both of these genes is unknown. However, since each plasmid also contained frameshifted versions of either p47 or lef-12, it is possible that one or both of the truncated lef-12 or p47 gene products exerted a dominant negative effect. The effect seemed to be limited to the context of SF-21 cells transfected with the AcMNPV genomic library since transient assays using the HSEpiHis lef library show clearly that lef-12 is involved in baculovirus late gene expression. The fact that lef-12 exerts little or no effect on transient late gene expression in the context of the genomic library is an additional reason why lef-12 was not identified in the initial study of lef genes within p47* since these studies focused on finding lef genes within the context of the genomic library (51).
Through the constitutive expression of each LEF, we have determined that each LEF plays an independent role in expression of reporter genes under the transcriptional control of the late capsid promoter. Therefore, IE-1 and IE-2 appear to have roles in AcMNPV late gene expression, in addition to their roles in transactivation of other lef genes. IE-1 and IE-2 influence the steady-state levels of plasmid DNA in a transient plasmid DNA replication assay (20, 28). Transient late gene expression and AcMNPV late gene expression are both dependent on DNA replication, and therefore IE-1 and IE-2 may exert their apparent transregulatory effects through DNA replication. In vitro transcription experiments with nuclear extracts containing subsets of the HSEpiHis LEFs may uncouple the dependence of baculovirus transcription on DNA replication and help to resolve the roles of IE-1 and IE-2 in DNA replication and/or late gene transcription.
lef-12 has a role in late gene transcription and not virus DNA replication. p47* was not necessary for replication in transient assays using the lef library (28), and the addition of pHSEpiHis lef-12 had no impact in assays using the HSEpiHis lef replication library. Therefore, like p47, lef-12 is a transcription lef. lef-12 is the only transcription-specific lef which demonstrates cell line specificity.
ACKNOWLEDGMENTS
We thank Jason Todd, who constructed pHSEpiHisVI+, pHSEpiHis lef-4, and pHSEpiHis lef-6. We also thank Somasekar Seshagiri, who constructed pHSEpip35VI+. Thanks go to Jeanne McLachlin for critical reading of the manuscript and for sequencing the ends of the genomic clones and to Domagoj Vucic, Song Yang, and William Kaiser for assistance with figures.
This work was supported in part by Public Health Service grant AI23719 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 2.Barrett J W, Lauzon H A, Mercuri P S, Krell P J, Sohi S S, Arif B M. The putative LEF-1 proteins from two distinct Choristoneura fumiferana multiple nucleopolyhedroviruses share domain homology to eukaryotic primases. Virus Genes. 1996;13:229–237. doi: 10.1007/BF00366983. [DOI] [PubMed] [Google Scholar]
- 3.Bump N J, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Licari P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 4.Carson D D, Guarino L A, Summers M D. Functional mapping of an AcNPV immediately early gene which augments expression of the ie-1 trans-activated 39k gene. Virology. 1988;162:444–451. doi: 10.1016/0042-6822(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 5.Carson D D, Summers M D, Guarino L A. Molecular analysis of a baculovirus regulatory gene. Virology. 1991;182:279–286. doi: 10.1016/0042-6822(91)90671-w. [DOI] [PubMed] [Google Scholar]
- 6.Carstens E B, Lu A L, Chan H. Sequence, transcriptional mapping, and overexpression of p47, a baculovirus gene regulating late gene expression. J Virol. 1993;67:2513–2520. doi: 10.1128/jvi.67.5.2513-2520.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C J, Thiem S M. Differential infectivity of two Autographa californica nucleopolyhedrovirus mutants on three permissive cell lines is the result of lef-7 deletion. Virology. 1997;227:88–95. doi: 10.1006/viro.1996.8341. [DOI] [PubMed] [Google Scholar]
- 8.Choi J, Guarino L A. Expression of the IE1 transactivator of Autographa californica nuclear polyhedrosis virus during viral infection. Virology. 1995;209:99–107. doi: 10.1006/viro.1995.1234. [DOI] [PubMed] [Google Scholar]
- 9.Clem R J, Fechheimer M, Miller L K. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- 10.Clem R J, Miller L K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans J T, Leisy D J, Rohrmann G F. Characterization of the interaction between the baculovirus replication factors LEF-1 and LEF-2. J Virol. 1997;71:3114–3119. doi: 10.1128/jvi.71.4.3114-3119.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field J, Nikawa J, Broek D, MacDonald B, Rodgers L, Wilson I A, Lerner R A, Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988;8:2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarino L A, Dong W. Expression of an enhancer-binding protein in insect cells transfected with the Autographa californica nuclear polyhedrosis virus IE1 gene. J Virol. 1991;65:3676–3680. doi: 10.1128/jvi.65.7.3676-3680.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino L A, Summers M D. Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J Virol. 1986;57:563–571. doi: 10.1128/jvi.57.2.563-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarino L A, Summers M D. Nucleotide sequence and temporal expression of a baculovirus regulatory gene. J Virol. 1987;61:2091–2099. doi: 10.1128/jvi.61.7.2091-2099.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hang X, Dong W, Guarino L A. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J Virol. 1995;69:3924–3928. doi: 10.1128/jvi.69.6.3924-3928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey A J, Bidwai A P, Miller L K. Doom, a product of the Drosophila mod(mdg4) gene, induces apoptosis and binds to baculovirus inhibitor-of-apoptosis proteins. Mol Cell Biol. 1997;17:2835–2843. doi: 10.1128/mcb.17.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hink W F. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970;226:466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- 20.Kool M, Ahrens C, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;91:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kool M, van den Berg P M M M, Tramper J, Goldbach R W, Vlak J M. Location of two putative origins of DNA replication of Autographa californica nuclear polyhedrosis virus. Virology. 1993;192:94–101. doi: 10.1006/viro.1993.1011. [DOI] [PubMed] [Google Scholar]
- 22.Leisy D J, Rohrmann G F. Characterization of the replication of plasmids containing hr in baculovirus-infected Spodoptera frugiperda cells. Virology. 1993;196:722–730. doi: 10.1006/viro.1993.1529. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Passarelli A L, Miller L K. Identification, sequence, and transcriptional mapping of lef-3, a baculovirus gene involved in late and very late gene expression. J Virol. 1993;67:5260–5268. doi: 10.1128/jvi.67.9.5260-5268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu A, Carstens E B. Immediate-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology. 1993;195:710–718. doi: 10.1006/viro.1993.1422. [DOI] [PubMed] [Google Scholar]
- 25.Lu A, Krell P J, Vlak J M, Rohrmann G F. Baculovirus DNA replication. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Press; 1997. pp. 171–192. [Google Scholar]
- 26.Lu A, Miller L K. Differential requirements for baculovirus late expression factor genes in two cell lines. J Virol. 1995;69:6265–6272. doi: 10.1128/jvi.69.10.6265-6272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu A, Miller L K. Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis. J Virol. 1994;68:6710–6718. doi: 10.1128/jvi.68.10.6710-6718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu A, Miller L K. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J Virol. 1995;69:975–982. doi: 10.1128/jvi.69.2.975-982.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu A, Miller L K. Species-specific effects of the hcf-1 gene on baculovirus virulence. J Virol. 1996;70:5123–5130. doi: 10.1128/jvi.70.8.5123-5130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLachlin J R, Miller L K. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J Virol. 1994;68:7746–7756. doi: 10.1128/jvi.68.12.7746-7756.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merrington C L, Kitts P A, King L A, Possee R D. An Autographa californica nucleopolyhedrovirus lef-2 mutant: consequences for DNA replication and very late gene expression. Virology. 1996;217:338–348. doi: 10.1006/viro.1996.0121. [DOI] [PubMed] [Google Scholar]
- 32.Morris T D, Miller L K. Promoter influence of baculovirus-mediated gene expression in permissive and nonpermissive insect cell lines. J Virol. 1992;66:7397–7405. doi: 10.1128/jvi.66.12.7397-7405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris T D, Todd J W, Fisher B, Miller L K. Identification of lef-7: a baculovirus gene affecting late gene expression. Virology. 1994;200:360–369. doi: 10.1006/viro.1994.1200. [DOI] [PubMed] [Google Scholar]
- 34.Nissen M S, Friesen P D. Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J Virol. 1989;63:493–503. doi: 10.1128/jvi.63.2.493-503.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohresser M, Morin N, Cerutti M, Delsert C. Temporal regulation of a complex and unconventional promoter by viral products. J Virol. 1994;68:2589–2597. doi: 10.1128/jvi.68.4.2589-2597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman and Company; 1992. [Google Scholar]
- 37.Partington S, Yu H, Lu A, Carstens E B. Isolation of temperature sensitive mutants of Autographa californica nuclear polyhedrosis virus: phenotype characterization of baculovirus mutants defective in very late gene expression. Virology. 1990;175:91–102. doi: 10.1016/0042-6822(90)90189-x. [DOI] [PubMed] [Google Scholar]
- 38.Passarelli A L, Miller L K. Identification and characterization of lef-1, a baculovirus gene involved in late and very late gene expression. J Virol. 1993;67:3481–3488. doi: 10.1128/jvi.67.6.3481-3488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passarelli A L, Miller L K. Identification and transcriptional regulation of the baculovirus lef-6 gene. J Virol. 1994;68:4458–4467. doi: 10.1128/jvi.68.7.4458-4467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Passarelli A L, Miller L K. Identification of genes encoding late expression factors located between 56.0 and 65.4 map units of the Autographa californica nuclear polyhedrosis virus genome. Virology. 1993;197:704–714. doi: 10.1006/viro.1993.1646. [DOI] [PubMed] [Google Scholar]
- 41.Passarelli A L, Miller L K. Three baculovirus genes involved in late and very late gene expression: ie-1, ie-n, and lef-2. J Virol. 1993;67:2149–2158. doi: 10.1128/jvi.67.4.2149-2158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passarelli A L, Todd J W, Miller L K. A baculovirus gene involved in late gene expression predicts a large polypeptide with a conserved motif of RNA polymerases. J Virol. 1994;68:4673–4678. doi: 10.1128/jvi.68.7.4673-4678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson M, Bjornson R, Pearson G, Rohrmann G. The Autographa californica baculovirus genome: evidence for multiple replication origins. Science. 1992;257:1382–1384. doi: 10.1126/science.1529337. [DOI] [PubMed] [Google Scholar]
- 44.Prikhod’ko E A, Miller L K. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J Virol. 1998;72:684–692. doi: 10.1128/jvi.72.1.684-692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rankin C, Ooi B G, Miller L K. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene. 1988;70:39–50. doi: 10.1016/0378-1119(88)90102-3. [DOI] [PubMed] [Google Scholar]
- 46.Rice W C, Miller L K. Baculovirus transcription in the presence of inhibitors and in nonpermissive Drosophila cells. Virus Res. 1986;6:155–172. doi: 10.1016/0168-1702(86)90047-x. [DOI] [PubMed] [Google Scholar]
- 47.Rodems S M, Pullen S S, Friesen P D. DNA-dependent transregulation by IE1 of Autographa californica nuclear polyhedrosis virus: IE1 domains required for transactivation and DNA binding. J Virol. 1997;71:9270–9277. doi: 10.1128/jvi.71.12.9270-9277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seshagiri S, Miller L K. Baculovirus inhibitors of apoptosis (IAPs) block activation of Sf-caspase-1. Proc Natl Acad Sci USA. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thiem S M, Miller L K. Identification, sequence, and transcriptional mapping of the major capsid protein gene of the baculovirus Autographa californica nuclear polyhedrosis virus. J Virol. 1989;63:2008–2018. doi: 10.1128/jvi.63.5.2008-2018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Todd J W, Passarelli A L, Lu A, Miller L K. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J Virol. 1996;70:2307–2317. doi: 10.1128/jvi.70.4.2307-2317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Todd J W, Passarelli A L, Miller L K. Eighteen baculovirus genes, including lef-11, p35, 39K, and p47, support late gene expression. J Virol. 1995;69:968–974. doi: 10.1128/jvi.69.2.968-974.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torok I, Karch F. Nucleotide sequences of heat shock activated genes in Drosophila melanogaster. I. Sequences in the regions of the 5′ and 3′ ends of the hsp 70 gene in the hybrid plasmid 56H8. Nucleic Acids Res. 1980;8:3105–3123. doi: 10.1093/nar/8.14.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaughn J L, Goodwin R H, Tompkins G J, McCawley P. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera: Noctuidae) In Vitro. 1977;13:213–217. doi: 10.1007/BF02615077. [DOI] [PubMed] [Google Scholar]