Machine learning tool TEEBoT predicts an individual’s tissue-specific gene expression from his/her blood transcriptome.
Abstract
Complex diseases are mediated via transcriptional dysregulation in multiple tissues. Thus, knowing an individual’s tissue-specific gene expression can provide critical information about her health. Unfortunately, for most tissues, the transcriptome cannot be obtained without invasive procedures. Could we, however, infer an individual’s tissue-specific expression from her whole blood transcriptome? Here, we rigorously address this question. We find that an individual’s whole blood transcriptome can significantly predict tissue-specific expression levels for ~60% of the genes on average across 32 tissues, with up to 81% of the genes in skeletal muscle. The tissue-specific expression inferred from the blood transcriptome is almost as good as the actual measured tissue expression in predicting disease state for six different complex disorders, including hypertension and type 2 diabetes, substantially surpassing the blood transcriptome. The code for tissue-specific gene expression prediction, TEEBoT, is provided, enabling others to study its potential translational value in other indications.
INTRODUCTION
Most common complex (non-Mendelian) diseases involve dysfunction in multiple tissues organs. For instance, hypertension, which is characterized by elevated arterial pressure, involves metabolic changes in the heart, blood vessels, brain, kidney, etc. (1). Furthermore, the tissue and organ dysfunction is primarily driven by and reflected in transcriptional changes in various tissues and organs (2). Transcriptional variance mediates, in large part, causal links between genotype and complex traits (3). Thus, the knowledge of tissue-specific gene expression (TSGE) profile can lead to a better understanding of diseases etiology, enabling patient subtyping and assessing drug efficacy (4, 5). However, except for easily accessible tissues such as the blood, muscle, skin, etc. and, in some cases, biopsies, organ and TSGE profiles cannot be readily obtained, presenting a challenge for transcription-based investigation of complex diseases. This limitation naturally gives rise to two important research questions: (i) To what extent can we predict an individual’s TSGE based solely on his/her whole blood gene expression (WBGE), and (ii) can the predicted tissue-specific expression reflect disease states better than what can be gleaned on those directly from the WBGE?
Cross-individual variance in TSGE can be explained, to some extent, by the genotypic variance, forming the basis of expression quantitative trait loci (eQTL). A widely used tool, PrediXcan, predicts the TSGE of a gene based on the gene’s eQTL single-nucleotide polymorphisms (or eSNPs) (6). However, a priori, such a tool has limited scope, as it can only predict expression of a minority of genes (about 10% genes on average across tissue) that have significant tissue-specific eSNPs (6). Furthermore, the previous prediction models based on blood expression [e.g., (7) and (8)] do not use the expression of other genes to predict the expression of a given gene, missing on the possibility of exploiting potentially shared regulatory programs between tissues. In contrast, similar to a very recent work (9), here, we use an individual’s WBGE, including the whole blood splicing (WBSp) profile, to predict the expression of each gene in a particular tissue of the individual. Notably, however, in contrast to the previous work, we control for demographic confounders via a log-likelihood ratio (LLR) test.
We proceed by building a linear model based on Genotype-Tissue Expression (GTEx) data (10) for 32 primary tissues having at least 65 samples each. We find that an individual’s WBGE and splicing profile significantly inform tissue-specific expression levels (above and beyond various demographic variable) for substantive fractions of genes, with a mean of 59% of genes across 32 tissues, up to 81% for muscle-skeletal tissue, based on likelihood ratio test with false discovery rate (FDR) threshold of 5%. The splicing profile contributes further beyond the gene expression profile, and for the subset of genes having eSNPs, genotype makes further significant contributions. We find that the genes with highly predictable expression are not biased toward housekeeping genes, proportionally representing tissue-specific genes. Moreover, these genes tend to have a greater number of protein interaction partners, which may suggest a contribution by shared gene networks toward expression predictability. Last, in many cases, the predicted tissue-specific expression can be used as a surrogate for actual expression in predicting disease state for several complex diseases far better than using the whole blood expression. Overall, our work establishes the utility and limits of whole blood transcriptome (WBT) in estimating gene expression in other tissues, leading a basis for future translationally motivated applications. We provide our software pipeline for predicting TSGE, named TEEBoT (Tissue Expression Estimation using Blood Transcriptome), in a user friendly and publicly available form.
RESULTS
Overview of TEEBoT: A pipeline for TSGE prediction
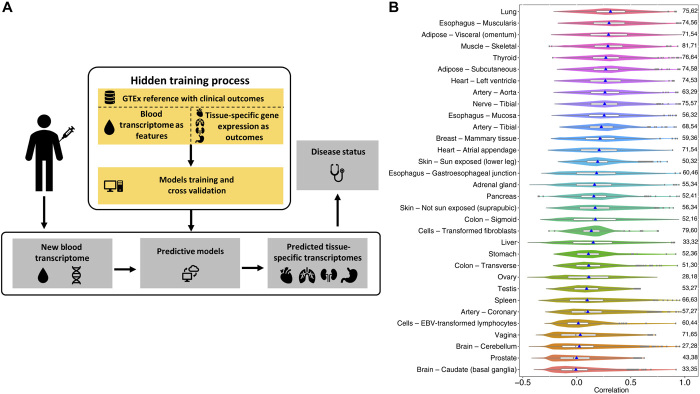
Figure 1A illustrates the overall motivation and TEEBoT pipeline. Our goal is to assess the predictability of TSGE using all available and easily accessible information about the individual, which includes WBT, his/her genotype, as well as basic demographic information on age, gender, and race. Normalized expression data were obtained from GTEx V6 (11) for 32 primary tissues (60% of all tissues) for which both the gene expression and WBT were available for at least 65 individuals (fig. S1).
Fig. 1. Overall approach and its performance.
(A) Overall approach. We train a generalized linear model to predict tissue-specific gene expression given the WBT of an individual, which can then be used to predict disease state. For CV, the model is trained on a subset (training set) of samples and tested on the remaining (test) samples in multiple train-test splits. “New blood transcriptome” refers to the transcriptome obtained from a new patient. (B) Prediction accuracy [in terms of cross-validation PCC (Pearson correlation coefficient)] of gene expression in target tissues from the blood expression using model M2 [WBGE + WBSp + CF (confounding factors)]. Only the genes with LLR test (M2 ~ CF, FDR ≤ 0.05) are included in this plot. The blue points mark the mean values, and the fractions of the genes with significant contribution from transcriptome toward prediction over CF (LLR test FDR ≤ 0.05) are indicated on the right side of each violin plot.
For each tissue and for each gene, we have fit three nested regression models to estimate TSGE: The prime model (M2; see Methods), whose results are described in the main text, is based on WBGE, WBSp information, and three demographic “confounding” factors (CFs)—age, race, and sex. To reduce the dimensionality of the modeling task, instead of using the expression (respectively, splicing) levels of all genes in blood, we estimated the principal components (PCs) using WBGE (respectively, WBSp) across all individuals and used the sample-specific scores of the top 10 PCs (top 20 PCs for WBSp) explaining 99% of variance as features. The whole-genome tissue-specific splicing profiles, which comprise the percent spliced in (PSI) values for annotated local splicing events in the genome, were obtained from (12); we note that, although we use the splicing profile as features, we only predict the overall gene expression and not the expression of specific isoforms. To assess the value of using splicing information, we additionally built and tested our base model, “WBGE + CF” model (M1), which uses only the WBGE PCs and CF variables. Last, to estimate the contribution of SNPs, we fit a third “WBGE + WBSp + SNP + CF” model (M3); although this model is most inclusive, it covers only a small fraction of about 10% of the genes, those having at least one eSNPs, as reported previously by the GTEx consortium (11). For those genes, we used the top five PCs of the genotype profile of eSNPs detected in a cross-validation (CV) manner to avoid overfitting. Below, we present the results obtained using our prime model M2. While results based on models M1 and M3 are mentioned briefly in context as appropriate, their details are provided in Supplementary Results for brevity and focus.
The predictive power of WBSp and expression information (the M2 model)
For each of the 17,031 genes, in each of the 32 tissues, we fit the regression model M2 and estimate the CV accuracies using a Pearson correlation coefficient (PCC) between the predicted and observed expression across individuals. First, as baseline, we assessed the contribution of WBT (i.e., WBGE and WBSp) over the demographic CFs via a LLR test and found that, on average, across tissues, for 59% of genes, WBT makes a significant contribution toward TSGE prediction, with a maximum of 81% of the genes in the muscle-skeletal tissue; the fraction of these predicted genes and their prediction accuracies are shown in Fig. 1B. Qualifying the LLR test does not necessarily imply that a gene’s expression is predicted with high accuracy. Figure S2 shows the corresponding plots for all genes, and table S1 shows the number of genes with accuracy above various thresholds. For instance, on average, for ~3000 (18%) of the genes, WBT makes significant contribution toward their TSGE prediction (LLR FDR ≤ 0.05) and they have a CV PCC ≥ 0.3, up to 6763 genes in the muscle. Directly comparing with results in (9) the fraction of genes with prediction accuracy ρ > 0.3, we find that in 14 of the 25 common tissues, our method detects a greater fraction of genes.
A similar assessment of the base model M1 (without WBSp) is provided in results S1 (also fig. S3 and table S2), and a direct comparison of model M2 with the model M1 is provided in results S2, clearly establishing the contribution of WBSp in predicting TSGE above and beyond WBGE alone; for instance, on average, for 43.2% of the genes, WBSp makes a significant additional contribution (likelihood ratio test FDR ≤ 0.05), up to 70.7% for muscle-skeletal tissue.
While SNPs are expected to contribute to TSGE prediction, as mentioned earlier, SNP-based prediction is applicable to ~10% of the genes that have a significant eSNP. For the subset of these genes, we have quantified the accuracy of the model M3, which additionally includes eSNPs. As expected, for ~60% of the genes, on average, across tissues (this corresponds to ~6% of all genes), eSNPs make significant additional contributions (results S3 and figs. S4 and S5). We have also compared model M2 with an SNP-only model M4 that we have constructed [comparable to a previous tool PrediXcan (6)] and found that overall WBT is a better predictor of TSGE than eSNPs alone (results S4 and table S3). We also found that genes that have eSNPs exhibit greater predictability by M2, although M2 does not include SNPs (results S5 and fig. S6).
We further assessed the generalizability of our model by training the M2 model on samples from GTEx V6 and testing its accuracy on samples exclusive to GTEx V8. Figure S7 compares the CV accuracy in GTEx V6 mentioned above with the accuracy in the proxy independent samples. In 13 of 31 tissues, the accuracy in the proxy independent samples is greater. Furthermore, their prediction performance concordance is high (PCC: min = 0.21, median = 0.55, max = 0.75) based on Pearson correlation between the prediction accuracy of the two tests across genes in each tissue.
Characteristics of genes whose TSGE is predictable by WBT (model M2)
We investigated the distinctive properties of tissue-specific predictable genes (TSPGs) by the M2 model in terms of their expression breadth, evolutionary conservation, and network connectivity. In each tissue, we identified TSPG as the genes for which WBT contributed significantly relative to CF (LLR FDR ≤ 0.05) and were among the top 25% most predictable based on CV PCC. First and quite notably, we observe that the TSPGs were quite different in each tissue (fig. S8), with an average Jaccard index of 0.13 across all tissue pairs.
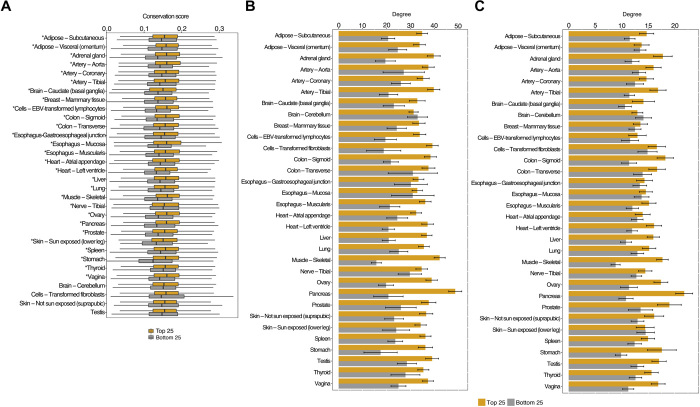
We next identified the enriched Gene Ontology (GO) biological processes in each tissue for the highly predictable genes (LLR FDR ≤ 0.05, CV PCC ≥ 0.5) using the Database for Annotation, Visualization and Integrated Discovery (13). The TreeMap view using REVIGO (14) for the 17 tissues with at least five significantly enriched terms (FDR ≤ 0.05) is provided in data file S1, and a combined view of GO terms in all tissues is provided in data file S2. By and large, TSPGs are enriched for numerous fundamental cellular processes, including metabolic processes, RNA processing, translation, transcription, etc. But notably, in a few cases, there is an enrichment for highly tissue-specific or tissue-relevant processes, such as “cardiac muscle cell action potential” in the heart and “cell morphogenesis involved in neuron differentiation” in the nerve. We additionally assessed whether the broadly expressed housekeeping genes (see Methods) are overrepresented among the TSPG, relative to genes with tissue-specific expression. As shown in fig. S9, we did not observe a substantive bias, attesting to broad utility of imputing TSGE. Looking specifically at the predictability of transcription factor (TF), overall, in 19 of 32 tissues, TF was significantly more predictable (Wilcoxon test, P ≤ 0.05) than other genes; the opposite is true in only four cases. Table S4 lists highly predictable (PCC ≥ 0.5) TFs in all tissues. We found that, in a vast majority of tissues, the TSPGs are evolutionarily more conserved (Fig. 2A).
Fig. 2. Distinguishing features of genes with predictable TSGE.
(A) More predictable genes are more conserved. We used the 46 mammalian species PhastCons score downloaded from UCSC (University of California Santa Cruz) genome browser. (The tissues with asterisks have significantly more conservation scores for the top 25 percentile predictable genes than the bottom 25 percentile ones). The bar plots (with means ± 95% confidence interval) shows comparison of the overall degree and (B) the degree of housekeeping genes (C) of the top 25 and bottom 25 percentile predictable genes across all the tissues.
Next, we assessed whether the predictability of TSPG may be related to their interactions with other genes, which tend to be functionally related and have similar expression profile (15, 16). We therefore compared the degree distribution of TSPG in a protein interaction network (PIN) with the background (see Methods). We also assessed whether broadly expressed housekeeping genes, by virtue of being expressed both in whole blood and the target tissue of interest, may better inform the TSGE. We therefore obtained the overall degree in the PIN and the degree relative only to housekeeping genes. Figure 2 (B and C) shows that TSPGs have much greater connectivity, both overall, and relative to housekeeping genes. To further probe the potential mechanism underlying this observation, for each of the most highly predictable genes g in a tissue (PCC ≥ 0.7), we tested whether g preferentially interacts with those genes whose expression values are most predictive of g’s TSGE (see Methods). We tested this hypothesis in each tissue independently using a one-sided paired Wilcoxon test across genes comparing interactions with predictive genes and the rest using the fraction of genes in either class that interact with a given gene. We performed this test only for the tissues with at least five genes with nonzero interactions with the predictive features. As shown in table S5, in 11 of the 12 tissues in which we could test this hypothesis, it is supported (P ≤ 0.05). We also noticed that paralogous gene pairs have a slightly greater than expected tendency to be among the highly predictable genes; while the background probability that a random pair of genes are paralogs is ~1%, among the top 1000 most predictable genes in each tissue, this probability is, on average, ~1.4%. However, paralogy explain a very small fraction of predictable genes.
For several genes, their TSGE was highly predictable (PCC ≥ 0.7) in multiple tissues. We investigated whether TSGE prediction model is tissue specific by assessing whether the same or different WBT gene features were used in different tissues to predict the gene’s expression. For the 340 genes that are very highly predictable in multiple tissues, we estimated the overlap between the top 100 most robustly predictive features (see Methods) for a given gene in two different tissues. The mean overlap between two sets of 100 features was only 8, strongly suggesting a tissue-specific model. To further probe the mechanism underlying the model’s tissue specificity, we assessed whether the tissue-specific predictive features (genes) exhibit a tissue-biased expression. Consider a gene g that is highly predictable in, say, two tissues T1 and T2, respectively, by feature sets F1 and F2. We tested whether a predictive gene in F1 has a higher expression in T1 compared to T2. For each of the 340 cases above and for each tissue-specific predictive gene feature, we estimated the fold difference of its expression in T1 relative to T2. We found a fold difference ≥ 1.5 in 66%, ≥2 in 56%, and ≥5 in 37% of the cases, suggesting that tissue-specific prediction uses distinct features, particularly, those with higher expression in the specific tissue.
We ascertained that predictability of a particular gene g is minimally dependent on the expression of gene g in blood. We computed gene expression PCs in the blood with and without a gene and found that the two PCs are practically identical (cross-sample Pearson correlation > 0.99) for all genes in a random sample of 100 genes.
Utility of WBT predicted tissue-specific expression in predicting complex diseases
Last, we assessed the extent to which the predicted TSGE can reveal tissue-specific disease-associated genes (DGs) and predict disease states. For each disease annotated in GTEx and for each tissue, we considered the number of samples available in the tissue that were annotated as positive for the disease and those that were annotated as negative. We retained the disease-tissue pairs, having at least 25 cases (positive for the disease) and 25 control (negative for the disease) samples in the particular tissue. This resulted in 83 disease-tissue pairs, involving five diseases (MHHTN-hypertension, MHT2D–type 2 diabetes, MHHRTATT–acute myocardial infarction, MMHRTDIS–ischemic heart disease, and MHCOPD–chronic respiratory disease) across 30 tissues.
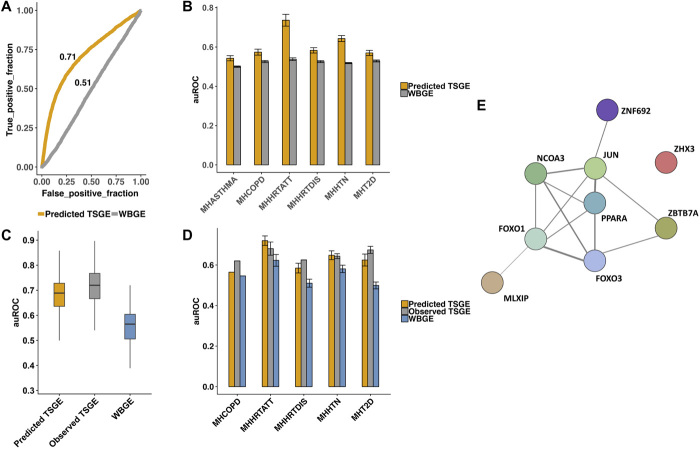
We first assessed the extent to which DGs, ascertained based on observed TSGE (see below), can be identified on the basis of the predicted TSGE. For each of the 83 disease-tissue pairs, we identified a reference set of DGs, whose tissue-specific expression was significantly different between cases and control individuals (Wilcoxon FDR ≤ 0.2). We then quantified the accuracy with which the predicted TSGE could distinguish DGs from the rest of the genes. As shown in table S6, on average, across 83 cases, predicted TSGE could distinguish DGs from the other genes with an area under the receiver operating characteristic curve (auROC) of 0.6 (with 19 cases having >0.7 of auROC). In contrast, WBGE failed to predict DGs (average auROC of 0.52 with zero cases having >0.7 of auROC). The result for hypertension–artery tibial pair is shown in Fig. 3A, and disease-wise summary across all tissues is shown in Fig. 3B.
Fig. 3. Predicting disease genes and disease state using predict TSGE.
(A) auROC (area under the receiver operating characteristic curve) for prediction of genes whose observed artery tibial expression is associated with hypertension based on predicted TSGE in artery tibial and WBGE. (B) Generalization of (A) to all disease-tissue pairs: disease-wise cross-tissue summary of auROCs. (C) Predicting hypertension state based on observed and predicted TSGE in artery tibial, as well as WBGE. (D) Generalization of (C) to all disease-tissue pairs: disease-wise cross-tissue summary of auROCs. (E) Functional connection among the nine TFs that are highly predictable in artery tibial and whose predicted TSGE are highly predictive of hypertension status (see text for details).
Next, we assessed the extent to which the TSGE can predict the disease state. For this, in each disease-tissue pair, we used the genes that were highly predictable in the tissue from WBGE (LLR FDR ≤ 0.05 and PCC ≥ 0.3) as features for building the pertaining disease/control predictors. We then compared the prediction accuracy when using their (i) actual TSGE, (ii) predicted TSGE, and (iii) WBGE (see Methods). Of the 83 disease-tissue pairs above, we focus on the 23 cases where the baseline CV prediction accuracy auROC based on the actual TSGE was at least 0.6. Analyses of these 23 cases are shown in fig. S10 and table S7. Results for one specific case of hypertension–artery tibial are shown in Fig. 3C, and disease-wise summary across tissues are shown in Fig. 3D. These results shown that (i) the accuracies using the predicted TSGE is comparable to those using observed TSGE (average fractional difference = 0.3%), (ii) predicted TSGE performs substantially better than WBGE (average fractional difference = 12%), and (iii) WBGE performance is modest (average auROC = 0.57). Overall, these results suggest that predicted TSGE can provide insights into tissue-specific disease-linked genes, can predict disease-state, comparable to observed TSGE, and is superior to WBGE.
We illustrate the above results for the specific case of hypertension–artery tibial pair (Fig. 3, A and C). There are 108 genes (i) whose gene expression in artery tibial are highly predictable using WBT (PCC ≥ 0.5) and (ii) whose predicted TSGE was differential between hypertensive individuals relative to control group (P ≤ 0.05).These genes are enriched for two major functional categories, various acid metabolism including carboxylic acid and various ion and carboxylic acid transports, both of which have functional links with hypertension (17, 18). The 108 genes include nine TFs shown in Fig. 3E, eight of which are functionally related based on a diverse array of evidence according to the Search Tool for the Retrieval of Interacting Genes/Proteins database (19). PPAR (peroxisome proliferator–activated receptor)–α and NCOA3 (also known as SRC3) form the two hubs. PPAR-α, by virtue of its involvement in control of vascular tone, has been suggested as an important target for hypertension (20). SRC3 is also known to regulate smooth muscle cell transcription, thus regulating hypertension (21). Other TFs also have links to hypertension. FOXO1 is involved in vascular homeostasis (22), and FOXO3 variants have been linked to blood pressure (23). ZNF692 (also known as AREBP) is linked by Genome-Wide Association Study to systolic blood pressure in gene cards database. ZHX3 (24) and MLXIP (25) have been linked to coronary artery disease and hypertension. JUN (also known as AP-1) was associated with arterial stiffness in elderly hypertensive patients (26). Overall, this example illustrates the potential clinical value of WBT-based TSGE prediction.
DISCUSSION
Charting TSGE profiles in individuals is important for understanding complex diseases, a realization that has been one of the prime motivations of the GTEx consortium (27). Here, we used the availability of genotypes and tissue-wise gene expression profiles in dozens of tissues across hundreds of individuals in the GTEx database to build models that predict TSGE profiles from the blood, which is by far the most readily available tissue. This particular scenario has not been comprehensively evaluated previously. Furthermore, we show that the global splicing profile in the blood significantly contributes to the predictability of TSGE in other tissues of the same individual. While in this work we predict gene expression, it will be an important future goal to further assess the possibility of predicting isoform expression in the target tissue.
Our results show that the more predictable genes have a greater connectivity to other genes in a protein-protein interaction network. However, these highly predictable genes are not particularly biased toward broadly expressed housekeeping genes and proportionally represent genes with tissue-restricted expression, attesting to a broad utility of the approach.
The predictability across tissues and genes within a tissue are variable. With regards to intergene variation in predictability, we observed that genes with greater number of protein interactions tend to be predicted better, suggesting that gene-gene interaction–based regulatory networks may play a role in this phenomenon. This is also consistent with the tendency of predictable genes to be involved in fundamental cellular processes and being evolutionary more conserved. Greater connectivity is expected of regulatory protein, such as TFs, and consistently, TFs exhibit greater predictability. The intertissue variability is harder to assess. There is certainly a sample size effect where tissues with higher sample size have higher CV accuracy (Spearman correlation = 0.71). We speculate that a greater complexity and heterogeneity in cellular composition of a tissue could adversely affect the prediction accuracy. Likewise, it is possible that a greater immune infiltration in the normal tissue could favorably affect the prediction accuracy (because our model is based on WBT). These possibilities, however, are challenging to assess because of the lack of relevant experimental data.
We find that the predicted TSGE using model M2, which learns from WBT alone, performs far better than the source WBT in predicting disease state. That is, the global expression and splicing profile in blood captures clinically relevant information indirectly via predicted TSGE in other tissues better than when it is used directly as a surrogate for TSGE. We note that among the top predictable disease-tissue pairs, some of the revealed tissues are relevant to the phenotype, e.g., involvement of the nerve, artery, muscle, and heart in heart attack and hypertension. There are others that seem counterintuitive at first but reveal established connection in the literature, e.g., parallels between lung conditions and heart attack. However, we note that there are yet others that are hard to interpret, such as association between expression in the skin and transformed fibroblasts, with hypertension. These analyses, by their nature, are correlative, and because of global associations in gene expression across tissues, it is hard to ascribe causality.
Transcriptome-based prognostic markers are starting to be developed for complex diseases (28–30), including cancer (31). While blood transcriptome can help detect biomarkers in some cases, as we have shown, accurate models to predict tissue-specific expression based on the blood transcriptome can be more effective in this regard. In the future, investigations into our ability to predict the transcriptome of the complex ecosystem of a tumor would further extend the utility of this approach. Together, our results provide a comprehensive and positive response to the two research questions we have set to study: It charts the extent to which human tissue expression can be predicted from blood transcriptomics in 25 human tissues, and based on the latter, it lays a basis for the future utilization of blood expression data for building predictive models of complex disorders.
METHODS
Linear models for gene expression predictability
We used three different linear regression models to predict a gene’s expression in a tissue (other than whole blood)
where Ygj is the expression of gth gene in the target tissue in the jth sample and PCjk(WBGE), PCjk(WBSp), and PCjk(eSNP) denote the value of kth PC of WBGE, WBSp, and eSNPs for jth sample, respectively. Agej, Sexj, and Racej denote the age, sex, and race of the jth sample, respectively. egj denotes the error term for the gth gene in the jth sample. Note that, instead of using all the genes’ expression and splicing in WB, we use a reduced PC representation to prevent overfitting while still capturing the variability. Specifically, we use the top 10 PCs based on the WBGE and 20 PCs for WBSp across the GTEx individuals as representative WBC transcriptomic features, capturing 99% variance. To avoid overfitting, the eSNPs used in model M3 are determined in each of the CV step and then the top five PCs of these eSNPs are used for prediction; we detect eSNPs only from the training samples, and we used PCs of the detected eSNPs to capture the ancestry, as is conventional in the standard eQTLs studies. eSNPs that are present within the 1-Mb region of the corresponding gene were used in the model. LASSO package from R is used to build the regression model, and results are computed for fivefold CV with 25 independent iterations.
To assess the contribution of SNPs without the WBT in the prediction of gene expression, we build model M4
Only the eSNPs within the 1-Mb region of the gene “g,” identified in the training set alone, are considered. Apart from these models, we also implemented a baseline model M0 based only on the confounders: age, sex, and race.
LLR test to identify genes whose expression is informed by various model features
For each of the three models (M1, M2, and M3), we assess for each gene whether its expression predictability has significant contribution from WBGE, (WBGE+WBSp), and (WBGE+WBSp+eSNPs), respectively, above and beyond age, race, and sex. For each gene, we compare the model (M1, M2, or M3) with the null model M0 using the LLR test using R package “lmtest.” The P value indicates the significance of the contribution by the additional features. We apply FDR ≤ 0.05 to select the genes, henceforth called the “significant gene” with respect to a particular model. With regard to finding the significant genes that has significant contribution from eSNPs above and beyond WBT, we compare model M4 with M3 using the LLR test as above.
Characterizing predictable genes
Housekeeping genes
Housekeeping genes (HK) (3791) were obtained in (32), of which 3342 genes were common to the GTEx gene sets and were considered.
Tissue specificity
For estimating tissue specificity of each gene, we use GTEx data version 6. For each of the gene in a target tissue, we calculate its tissue specificity as log2 of ratio of the mean gene expression in target tissue to the mean gene expression in rest of the tissues and consider the genes with their tissue specificity among the top 25 percentile.
Connectivity
The PIN is obtained from (33). From this network, we extract the degrees of connectivity of the top and bottom 25 percentile predictable genes, which are considered as foreground and control for comparison of their degrees. Later, we perform a similar comparison by considering only the connectivity with the housekeeping genes.
Identifying the most predictive features of a gene
For a given gene in a particular tissue, we find the list of genes whose expression in whole blood contribute significantly toward its expression prediction. We consider the top five PCs (most frequently appearing across independent tuns) of blood gene expression that contribute to the prediction, and for each PC, we identify the top 20 genes that are most correlated with the corresponding PC. Overall, this yields 100 genes (across five PCs), denoted as S(g), contributing significantly toward g’s TSGE prediction.
Analysis of disease prediction
In addition, we estimate disease predictability specific to tissue, taking into account all the genes whose expression are significantly predictable (FDR LLR ≤ 0.05 and predictability score ≥ 0.3). To do so, we build a LASSO model and estimate auROC in a CV fashion.
Code availability
TEEBoT is available on Github (https://github.com/mbasugit/Imputation).
Acknowledgments
We are grateful to the help from F. Schischlik, D. Wu, S. Patkar, and A. Singh. This work used the computational resources of the NIH HPC Biowulf cluster. Funding: This work is supported in part by the Intramural Research Program of the NIH, National Cancer Institute. S.H. is funded in part by NSF award 1564785. Author contributions: E.R. and S.H. designed and supervised this project. M.B. and K.W. performed the analyses. M.B., K.W., and S.H. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Our model is based on the GTEx database, and while the processed GTEx data are freely available from their portal, the protected data (imputed SNPs and demographic information) are controlled and require individual laboratories to request for access. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabd6991/DC1
REFERENCES AND NOTES
- 1.Dai X., Hua L., Chen Y., Wang J., Li J., Wu F., Zhang Y., Su J., Wu Z., Liang C., Mechanisms in hypertension and target organ damage: Is the role of the thymus key? (Review). Int. J. Mol. Med. 42, 3–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cookson W., Liang L., Abecasis G., Moffatt M., Lathrop M., Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10, 184–194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nica A. C., Montgomery S. B., Dimas A. S., Stranger B. E., Beazley C., Barroso I., Dermitzakis E. T., Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLOS Genet. 6, e1000895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu M., Sharmin M., Das A., Nair N. U., Wang K., Lee J. S., Chang Y.-P. C., Ruppin E., Hannenhalli S., Prediction and subtyping of hypertension from pan-tissue transcriptomic and genetic analyses. Genetics 207, 1121–1134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant E. P., Pickard M. D., Briskin M. J., Gutierrez-Ramos J.-C., Gene expression profiles: Creating new perspectives in arthritis research. Arthritis Rheum. 46, 874–884 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Gamazon E. R., Wheeler H. E., Shah K. P., Mozaffari S. V., Aquino-Michaels K., Carroll R. J., Eyler A. E., Denny J. C.; GTEx Consortium, Nicolae D. L., Cox N. J., Im H. K., A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J., Gamazon E. R., Pierce B. L., Stranger B. E., Im H. K., Gibbons R. D., Cox N. J., Nicolae D. L., Chen L. S., Imputing gene expression in uncollected tissues within and beyond GTEx. Am. J. Hum. Genet. 98, 697–708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halloran J. W., Zhu D., Qian D. C., Byun J., Gorlova O. Y., Amos C. I., Gorlov I. P., Prediction of the gene expression in normal lung tissue by the gene expression in blood. BMC Med. Genomics 8, 77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W., Liu X., Leng F., Li W., Blood-based multi-tissue gene expression inference with Bayesian ridge regression. Bioinformatics 36, 3788–3794 (2020). [DOI] [PubMed] [Google Scholar]
- 10.GTEx Consortium , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., Foster B., Moser M., Karasik E., Gillard B., Ramsey K., Sullivan S., Bridge J., Magazine H., Syron J., Fleming J., Siminoff L., Traino H., Mosavel M., Barker L., Jewell S., Rohrer D., Maxim D., Filkins D., Harbach P., Cortadillo E., Berghuis B., Turner L., Hudson E., Feenstra K., Sobin L., Robb J., Branton P., Korzeniewski G., Shive C., Tabor D., Qi L., Groch K., Nampally S., Buia S., Zimmerman A., Smith A., Burges R., Robinson K., Valentino K., Bradbury D., Cosentino M., Diaz-Mayoral N., Kennedy M., Engel T., Williams P., Erickson K., Ardlie K., Winckler W., Getz G., De Luca D., MacArthur D., Kellis M., Thomson A., Young T., Gelfand E., Donovan M., Meng Y., Grant G., Mash D., Marcus Y., Basile M., Liu J., Zhu J., Tu Z., Cox N. J., Nicolae D. L., Gamazon E. R., Im H. K., Konkashbaev A., Pritchard J., Stevens M., Flutre T., Wen X., Dermitzakis E. T., Lappalainen T., Guigo R., Monlong J., Sammeth M., Koller D., Battle A., Mostafavi S., Carthy M. M., Rivas M., Maller J., Rusyn I., Nobel A., Wright F., Shabalin A., Feolo M., Sharopova N., Sturcke A., Paschal J., Anderson J. M., Wilder E. L., Derr L. K., Green E. D., Struewing J. P., Temple G., Volpi S., Boyer J. T., Thomson E. J., Guyer M. S., Ng C., Abdallah A., Colantuoni D., Insel T. R., Koester S. E., Little A. R., Bender P. K., Lehner T., Yao Y., Compton C. C., Vaught J. B., Sawyer S., Lockhart N. C., Demchok J., Moore H. F., The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Wu D., Zhang H., Das A., Basu M., Malin J., Cao K., Hannenhalli S., Comprehensive map of age-associated splicing changes across human tissues and their contributions to age-associated diseases. Sci. Rep. 8, 10929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang D. W., Sherman B. T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M. W., Lane H. C., Lempicki R. A., DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169–W175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supek F., Bošnjak M., Škunca N., Šmuc T., Revigo summarizes and visualizes long lists of gene ontology terms. PLOS ONE 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver S., Guilt-by-association goes global. Nature 403, 601–602 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Yu H., Braun P., Yıldırım M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J.-F., Dricot A., Vazquez A., Murray R. R., Simon C., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A.-S., Motyl A., Hudson M. E., Park J., Xin X., Cusick M. E., Moore T., Boone C., Snyder M., Roth F. P., Barabási A.-L., Tavernier J., Hill D. E., Vidal M., High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ives H. E., Ion transport defects and hypertension where is the link? Hypertension 14, 590–597 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Das U. N., Essential fatty acids and their metabolites in the context of hypertension. Hypertens. Res. 33, 782–785 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L. J., STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Usuda D., Peroxisome proliferator-activated receptors for hypertension. World J. Cardiol. 6, 744–754 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H. J., Haque Z., Lu Q., Li L., Karas R., Mendelsohn M., Steroid receptor coactivator 3 is a coactivator for myocardin, the regulator of smooth muscle transcription and differentiation. Proc. Natl. Acad. Sci. U.S.A. 104, 4065–4070 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dharaneeswaran H., Abid R., Yuan L., Dupuis D., Beeler D., Spokes K. C., Janes L., Sciuto T., Kang P. M., Jaminet S.-C. S., Dvorak A., Grant M. A., Regan E. R., Aird W. C., FOXO1-mediated activation of akt plays a critical role in vascular homeostasis. Circ. Res. 115, 238–251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris B. J., Chen R., Donlon T. A., Evans D. S., Tranah G. J., Parimi N., Ehret G. B., Newton-Cheh C., Seto T., Willcox D. C., Masaki K. H., Kamide K., Ryuno H., Oguro R., Nakama C., Kabayama M., Yamamoto K., Sugimoto K., Ikebe K., Masui Y., Arai Y., Ishizaki T., Gondo Y., Rakugi H., Willcox B. J., Association analysis of FOXO3 longevity variants with blood pressure and essential hypertension. Am. J. Hypertens. 29, 1292–1300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamada Y., Yasukochi Y., Kato K., Oguri M., Horibe H., Fujimaki T., Takeuchi I., Sakuma J., Identification of 26 novel loci that confer susceptibility to early-onset coronary artery disease in a Japanese population. Biomed. Rep. 9, 383–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alobeidy B. F., Li C., Alzobair A. A., Liu T., Zhao J., Fang Y., Zheng F., The association study between twenty one polymorphisms in seven candidate genes and coronary heart diseases in chinese han population. PLOS ONE 8, e66976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q., Han L., Du Q., Zhang M., Zhou S., Shen X., The association between oxidative stress, activator protein-1, inflammatory, total antioxidant status and artery stiffness and the efficacy of olmesartan in elderly patients with mild-to-moderate essential hypertension. Clin. Exp. Hypertens. 38, 365–369 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Ongen H., Brown A. A., Delaneau O., Panousis N. I., Nica A. C.; GTEx Consortium, Dermitzakis E. T., Estimating the causal tissues for complex traits and diseases. Nat. Genet. 49, 1676–1683 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Redei E. E., Andrus B. M., Kwasny M. J., Seok J., Cai X., Ho J., Mohr D. C., Blood transcriptomic biomarkers in adult primary care patients with major depressive disorder undergoing cognitive behavioral therapy. Transl. Psychiatry 4, e442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampson D., Yager T. D., Fox B., Shallcross L., Hugh L. M., Seldon T., Rapisarda A., Hendriks R. A., Brandon R. B., Navalkar K., Simpson N., Stafford S., Gil E., Venturini C., Tsaliki E., Roe J., Chain B., Noursadeghi M., Blood transcriptomic discrimination of bacterial and viral infections in the emergency department: A multi-cohort observational validation study. BMC Med. 18, 185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heidecker B., Kittleson M. M., Kasper E. K., Wittstein I. S., Champion H. C., Russell S. D., Hruban R. H., Rodriguez E. R., Baughman K. L., Hare J. M., Transcriptomic biomarkers for the accurate diagnosis of myocarditis. Circulation 123, 1174–1184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Y., Zhao B., Liu Z., Ren X., Zhao W., Li Z., You L., Zhao Y., Molecular subtyping of pancreatic cancer: Translating genomics and transcriptomics into the clinic. J. Cancer 8, 513–522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisenberg E., Levanon E. Y., Human housekeeping genes, revisited. Trends Genet. 29, 569–574 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Schaefer M. H., Fontaine J.-F., Vinayagam A., Porras P., Wanker E. E., Andrade-Navarro M. A., HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLOS ONE 7, e31826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GTEx Consortium , The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo X., Lin W., Bao J., Cai Q., Pan X., Bai M., Yuan Y., Shi J., Sun Y., Han M.-R., Wang J., Liu Q., Wen W., Li B., Long J., Chen J., Zheng W., A comprehensive cis-eQTL analysis revealed target genes in breast cancer susceptibility loci identified in genome-wide association studies. Am. J. Hum. Genet. 102, 890–903 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabd6991/DC1