Abstract
Over the past century, early advances in understanding the identity of the chemicals that collectively form a living plant have led scientists to deeper investigations exploring where these molecules localize, how they are made, and why they are synthesized in the first place. Many small molecules are specific to the plant kingdom and have been termed plant secondary metabolites, despite the fact that they can play primary and essential roles in plant structure, development, and response to the environment. The past 100 yr have witnessed elucidation of the structure, function, localization, and biosynthesis of selected plant secondary metabolites. Nevertheless, many mysteries remain about the vast diversity of chemicals produced by plants and their roles in plant biology. From early work characterizing unpurified plant extracts, to modern integration of ‘omics technology to discover genes in metabolite biosynthesis and perception, research in plant (bio)chemistry has produced knowledge with substantial benefits for society, including human medicine and agricultural biotechnology. Here, we review the history of this work and offer suggestions for future areas of exploration. We also highlight some of the recently developed technologies that are leading to ongoing research advances.
The past century has led to remarkable advances in illuminating the chemical repertoire, biosynthetic pathways, localization patterns, and diverse functions of plant secondary metabolites.
Introduction
A hundred years ago, scientists were trying to understand the fundamental question, “What makes a plant?” Since then, we have learned a great deal about the chemical constituents of plants and their functions. But, there is still much exploration needed to answer this question; tens of thousands of plant chemicals (known as secondary metabolites, phytochemicals, or natural products) remain to be identified, and many mysteries remain even in some of the most well-studied organisms and pathways.
The term “secondary metabolite” suffers from the implication that the chemical in question is of secondary importance, and the term “specialized metabolite” is sometimes preferred. Even this may have negative connotations, as specialization can be taken to mean “not of general interest,” as seen by the relative numbers of papers on plant secondary metabolism compared with, say, plant development, in high-impact general interest journals over the years. This situation is changing, and here, we hope to further explode the myth by pointing out the major advances that the study of plant secondary metabolism has made in the areas of phylogenetics, genome evolution, basic biochemistry, plant defense, ecology, and plant development. Important principles of biology have indeed been learned by the study of plant secondary metabolism (some are highlighted in Fig. 1).
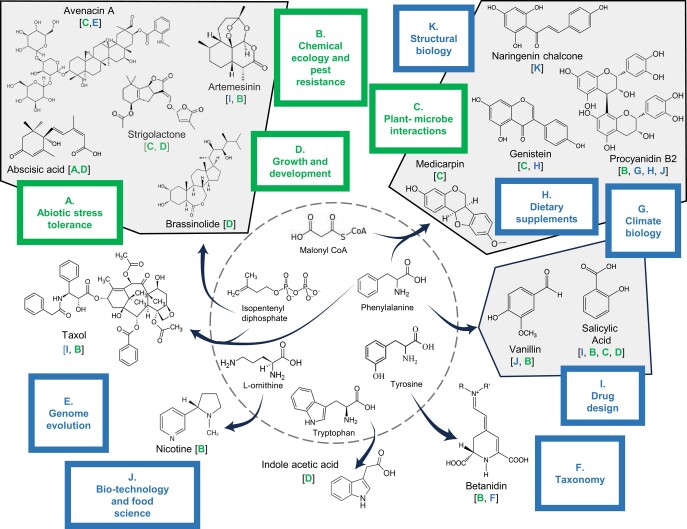
Figure 1.
Structures of a selection of plant secondary metabolites, showing their biosynthetic origins, in planta functions, and uses or importance in plant biology. The bracket under each compound indicates the in planta function (green letter referring to a green box) and potential use or area of biology that the study of the compound has impacted (blue letter referring to a blue box). Arrows link precursors from primary metabolism (central circle) to the end products. Salicylic acid can be derived from phenylalanine or isochorismate (shikimate pathway intermediate).
The difference between our current approach to plant secondary metabolism and the work done a century ago is largely a question of techniques and ways of thinking. We therefore also highlight the major technological and intellectual innovations over the past 100 yr. We discuss the examples of particular interest and offer our predictions for the future of the field. This is a vast area, and our selection of examples cannot encompass all of the important work in this topic. Therefore, we invite interested readers to continue to explore this broad and exciting topic beyond this review.
The era of chemistry and function
A key driver of early phytochemistry was understanding the compounds that gave medicinal plants their healing abilities. As a classic review of phytochemistry states, “The desire to discover the healing virtues of plants or again, to extract deadly poisons from them, is one of the oldest endeavors of mankind” (Burrell 1937). By the early 20th century, organic compounds from plants were generally classified by whether they were commonly found in many plant species (today, many of these compounds could be described as primary metabolites) or whether they were specific to certain plants. Compounds identified as specific to medicinal or poisonous plants were treated with particular interest, and chemical characterization of these types of compounds led to some of the first plant-derived medicinal compounds: aspirin (salicylic acid), morphine, digitoxin, and quinine (Leroux 1830; Fisch 1985; Schmitz 1985; Achan et al. 2011). As characterization of plant metabolites has improved, subsequent research has shown that many “specific” compounds were in fact much more broadly distributed in plants than previously realized. Salicylic acid is an excellent example of this—originally identified as the medicinal component in willow (Salix spp) bark, we know now that salicylic acid is an important defense hormone found across the plant kingdom (Peng et al. 2021).
Early attempts to identify the medicinal components of plants led to the discovery and characterization of many secondary metabolites, including alkaloids, glucosides, anthocyanins, carotenoids, and terpenes (Burrell 1937). Bioactivity assays were frequently used to determine which plants made compounds with “desirable” effects. These assays would apply plant extracts to tissues, animals, or microbes of interest. If favorable biological changes were induced by the extracts, then natural product chemistry techniques would be used to isolate the causative plant metabolite. These assays were frequently integrated with chemical synthesis to identify and refine compounds of interest. For instance, the discovery of aspirin involved treating feverish patients with extracts of willow bark, which had high levels of salicin, a compound convertible to salicylic acid (Montinari et al. 2019; Mahdi et al. 2006). Chemical structure confirmation was achieved by comparing the natural compound with synthesized salicylic acid. Further synthetic modifications resulted in acetylsalicylic acid, the water-soluble compound used in the commercial formulation for aspirin. Over several decades, synthetic analogs of acetylsalicylic acid were generated to create a number of other nonsteroidal anti-inflammatory drugs, including ibuprofen. This integrated natural product and synthetic chemistry approach to medicine has served as a template for the discovery and creation of a wide range of drugs ranging from chemotherapeutics to antibiotics.
Studying plant chemistry in the context of animals and insects became an expanding theme that gave rise to important discoveries in understanding the functions of plant metabolites. Starting in the 1960s, the work of Tony Swain, Jeffrey Harborne, and later Thomas Hartmann and Tom Mabry, among others, led to the rebirth of chemical ecology (Hartmann 2008) and chemotaxonomy (Bate-Smith 1962). Swain, along with Bate-Smith, Haslam, and White, coined the term “polyphenol,” and the technique of paper chromatography allowed polyphenols to be used as markers in broad taxonomic studies (Swain 1985). By the mid-20th century, explorations in the field of chemical ecology were leading to the discovery of roles of plant metabolites in regulating insect behavior and biology (Dethier 1941). The 1940s to 1960s also saw the birth of our understanding of inducible (phytoalexin) as opposed to static (phytoanticipin) antimicrobial chemical defenses in plants (Cruickshank and Perrin 1961; Tomiyama et al. 1968; VanEtten et al. 1994), followed by appreciation of the role of compounds such as flavonoids and (later) sinapate esters as protectants against abiotic stress (UV light) (Markham and Mabry 1975; Li et al. 1993; Sheahan 1996).
The desire to understand what controls plant growth and development led to the identification of other groups of bioactive plant secondary metabolites. Although compounds now recognized as plant hormones are believed to be distributed across the plant kingdom, several are structurally related to other secondary metabolites with more specialized distributions and different functions. Thus, auxin is in the same indole family as the Arabidopsis (Arabidopsis thaliana) phytoalexin camalexin, gibberellins are diterpenes, abscisic acid is derived from carotenoids, and brassinosteroids are triterpenes. Certain flavonoids function as inhibitors of auxin transport (Jacobs and Rubery 1988), and a coniferyl alcohol dimer (lignan) was proposed to act as a cytokinin, although this has recently been challenged (Witvrouw et al. 2023). Here, we consider “plant hormones” only from the perspective of where their study provides broader insight into secondary metabolism or has led to the development of innovative techniques for studying plant metabolism.
Figure 1 summarizes the biological activities of a selection of well-studied plant secondary metabolites and also indicates their usefulness to humans and, where appropriate, to the study of biology in general.
The era of biosynthesis
Model systems for the study of plant secondary metabolism
By their nature and with some exceptions, most plant secondary metabolites are not widely present throughout the plant kingdom. There are also limits to distribution at the level of chemical classes, with, for example, betalains replacing anthocyanins in the Caryophyllales and isoflavonoids being generally, but not exclusively, restricted to the Leguminosae. Multiple model species have therefore been developed to study plant secondary metabolism, especially at the molecular genetic level. Early studies prior to the application of genetic approaches targeted species irrespective of their genetic complexity or tractability, but work soon began to focus on model species with advanced genetic features such as easy transformability and availability of mutant collections. Unfortunately, Arabidopsis, although possessing many of the conserved secondary metabolic pathways such as phenylpropanoid and triterpene, is not always a good model for studying other aspects of metabolism/ecology. For example, although our understanding of the biosynthesis of flavonoid-derived condensed tannins was accelerated through study of the set of Arabidopsis transparent testa mutants (Appelhagen et al. 2014), Arabidopsis has since been shown to lack several of the enzymatic steps involved in tannin biosynthesis in other species (Yu et al. 2023). Madagascar periwinkle (Catharanthus roseus) has been and continues to be widely studied as a model plant for alkaloid biosynthesis (Kulagina et al. 2022), and the wild tobacco Nicotiana alata, with facile transformation and the existence of distinct field populations, has been an important model for studies in chemical ecology (Kumar et al. 2014). Barrel medic (Medicago truncatula), with highly developed genetic resources (Tadege et al. 2008), has served as one model for (iso)flavonoid and triterpene biosynthesis (Liu et al. 2018; Ribeiro et al. 2020). With ever-increasing improvements in genomic/transcriptomic resources (One Thousand Plant Transcriptomes Initiative 2019) and synthetic biology and rapid transient expression technologies (Owen et al. 2017), plant transformation efficiency is now the major hurdle to investigations of secondary metabolism across the broad diversity of plant species.
Unraveling the biosynthesis of secondary metabolites
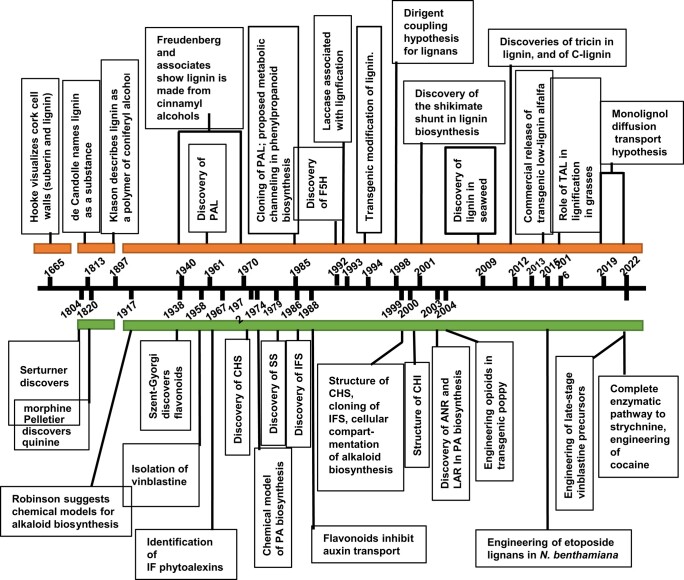
Phase 1: Chemistry informs biochemistry. Early work on the biosynthesis of plant secondary metabolites relied heavily on the principles of organic chemistry. The pioneering work of Robinson was among the first to contemplate a role for enzymes in the assembly of complex plant metabolites (Robinson 1917), leading to a Nobel Prize in Chemistry in 1947. Chemical synthesis or semisynthesis was used as a guide to how these molecules might be assembled in the plant under enzymatic control. For example, the chemical assembly of condensed tannins through nucleophilic addition via a carbocation intermediate was proposed as the basis for their biosynthesis from flavonoid precursors (Haslam 1974; Roux and Ferreira 1982). Biosynthetic pathways were also interrogated through labeling experiments, usually utilizing 14C-labeled precursors which had to be custom synthesized. Examples include pioneering studies on the biosynthesis of lignin from L-phenylalanine and coniferyl alcohol (Freudenberg 1965), of vanillin from ferulic acid (Zenk 1965) and of benzylisoquinoline alkaloids from tyrosine (Rueffer and Zenk 1987). At the same time, in vitro assays were being developed to examine whether crude extracts from plants might contain predicted enzyme activities for involvement in the synthesis of a range of different secondary metabolites (e.g. Koukol and Conn 1961; Britsch and Grisebach 1986; Kochs and Grisebach 1986, e.g. in the phenylpropanoid/flavonoid pathway), and pathways were “assembled” based on the ordering of such activities. Figure 1 gives a brief indication of the biosynthetic origins (precursors from primary metabolism) of a selection of secondary metabolites, and Fig. 2 provides a historical timeline for the identification and characterization of selected phenylpropanoids, flavonoids, alkaloids, and their biosynthetic pathways.
Figure 2.
Historical timeline for research on lignin, lignans, flavonoids, and alkaloids, with a focus on phenolic pathways. ANR, anthocyanidin reductase; CHI, chalcone isomerase; CHS, chalcone synthase; F5H, ferulate:coniferaldehyde 5-hydroxylase; IF, isoflavonoid; IFS, isoflavone synthase; LAR, leucoanthocyanidin reductase; PA, proanthocyanidin (condensed tannin); PAL, L-phenylalanine ammonia-lyase; SS, strictosidine synthase; TAL, L-tyrosine ammonia-lyase. The enzymes are referenced further in the text.
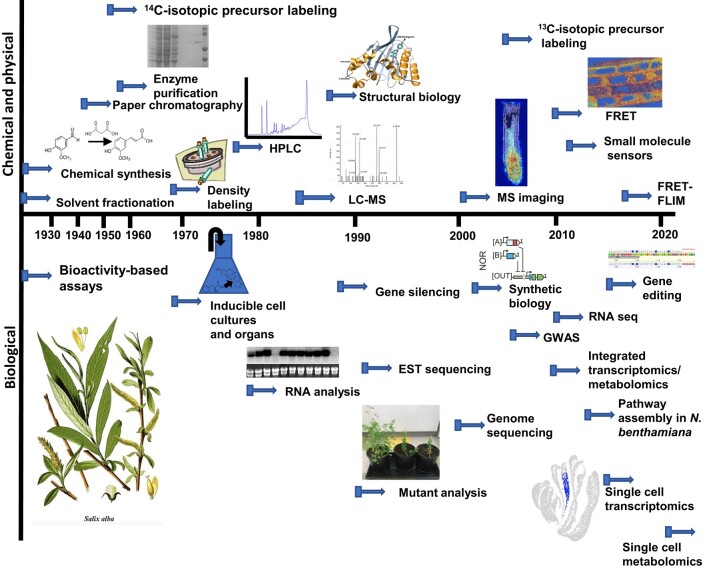
Phase 2: Molecular biology enters the race. Large strides in our understanding of the molecular basis of plant secondary metabolism were made in the 1980s following the development of inducible systems such as cell cultures or biotically/abiotically stressed tissues (Hadwiger and Schwochau 1971; Kreuzaler and Hahlbrock 1973; Lawton et al. 1980; Zenk 1991) to which the tools of the emerging discipline of molecular biology could be applied. A comparison of the timelines for discoveries (Fig. 2) and for the adoption of techniques to the study of plant metabolism (Fig. 3) shows the enormous impact of molecular biology on the field. Once a reliable assay had been developed for an enzyme that was induced in parallel with an associated metabolite, it was then possible to purify the enzyme to homogeneity, sequence peptides derived from the enzyme protein, design oligonucleotide probes corresponding to the protein sequence, and screen cDNA or genomic libraries to identify the corresponding gene and provide DNA probes to enable the detection of transcripts on RNA blots (e.g. Kreuzaler et al. 1983; Edwards et al. 1985). In some cases, plant genes were cloned based on their homology to conserved regions in genes from mammalian species (e.g. Learned and Fink 1989).
Figure 3.
Timeline for the development of techniques that have driven research on plant secondary metabolites. Pictures are from: MS imaging (Zhang et al. 2023); FRET (Herud-Sikimić et al. 2021); mutant analysis (Adiji et al. 2021); synthetic biology (Brophy et al. 2022); and single-cell transcriptomics (Nolan et al. 2023). The picture bottom left is a vintage lithograph showing the white willow, Salix alba (original book source: Prof. Dr Otto Wilhelm Thomé, Flora von Deutschland, Österreich und der Schweiz 1885, Gera (Germany)—public domain, https://commons.wikimedia.org/w/index.php?curid=2358667). The figure from Brophy et al. (2022) is reprinted with permission from AAAS. Other images are our own or licensed under a Creative Commons Attribution License (CC BY, https://creativecommons.org/licenses/by/4.0/) by Zhang et al. (2023), Herud-Sikimić et al. (2021), and Nolan et al. (2023). EST, expressed sequence tag; FLIM, fluorescence lifetime imaging microscopy.
Experiments, in which a particular metabolite or sets of metabolites were induced as a result of application of a chemical, elicitor molecule, or a light stimulus, were then designed to address the question of whether the induction involved de novo gene expression (Hahlbrock and Grisebach 1979; Hahlbrock and Scheel 1989). Earlier studies on environmentally controlled or elicitor-induced plant metabolism had relied on cumbersome techniques such as density labeling with deuterated water to determine whether enzymes were being synthesized de novo (Filner and Varner 1967; Lawton et al. 1980). Now, antibodies could be used as reagents for in vitro translation assays to determine whether translatable mRNA activities were being increased as a result of the chemical, environmental, or developmental stimulus (e.g. Kreuzaler et al. 1983), and nuclear run-off experiments could address control at the level of transcription itself (Somssich et al. 1989). The cDNAs could also be used as probes for in situ hybridization experiments to address the cellular localization of the transcripts and, by extension, the pathway (Schmelzer et al. 1988; St Pierre et al. 1999). Together, this work led to the appreciation that plant secondary metabolism was under complex spatial and temporal control, often regulated primarily at the level of gene expression (Hahlbrock and Scheel 1989). For example, transcriptional regulation at the levels of L-phenylalanine ammonia-lyase (PAL) (Edwards et al. 1985), chalcone synthase (CHS) (Bell et al. 1984), and 3-hydroxymethylglutaryl coenzyme A reductase (HMGR) (Choi et al. 1992) were proposed as contributing to control of phenylpropanoid, flavonoid, and terpenoid biosynthesis, respectively. It also became apparent, by comparison of levels of transcripts and their corresponding proteins, that posttranslational control could be important, particularly for key entry-point enzymes into secondary metabolism such as HMGR (Korth et al. 2000).
Phase 3: “Omics” changes the game. Between the mid-1980s and the mid-1990s, many of the key enzymes in the biosynthesis of different classes of plant secondary metabolite had been identified at the molecular level. However, further progress was often impeded by uncertainty about and unavailability of potential intermediates for later pathway reactions. The advent of RNA and genome sequencing in the early 2000s made available a powerful resource for studies on plant secondary metabolism (Kellner et al. 2015; Wurtzel and Kutchan 2016). It was possible to obtain the total complement of all classes of enzymes expressed within an organism, a particular tissue, or in response to a particular biotic or abiotic stimulus, providing a set of candidate genes that fitted predicted expression patterns to test for involvement of their encoded proteins in a “missing reaction” within a biosynthetic pathway. It became clear that specific classes of enzymes had been recruited, through evolution, for the purpose of elaborating plant secondary metabolites (Modolo et al. 2009). These include cytochrome P450 enzymes or 2-oxoglutarate–dependent dioxygenases for hydroxylation reactions, uridine diphosphate glycosyltransferases for addition of sugar moieties, BAHD-family acyltransferases for acyl group transfer, etc. The gene candidates could be selected and expressed heterologously in a microbe (generally Escherichia coli, but with yeast [usually Saccharomyces cerevisiae] a favored host for membrane-bound enzymes such as the P450s) or transiently expressed in a plant host such as N. benthamiana (see below for subsequent developments of this technology) to determine the catalytic activity of the encoded enzyme. At first, recombinant enzymes were tested against a range of predicted substrates, providing tentative in vitro functions, the physiological relevance of which could then be determined by subsequent genetic analysis. This approach was, however, still cumbersome for the elucidation of multistep pathways and still severely limited by the availability of pathway intermediates to test as substrates.
The development of robust metabolomic technologies allowed the interrogation of metabolite/transcript cooccurrence for predicting the involvement of genes in secondary metabolite biosynthesis (Saito and Matsuda 2010). Subsequently, coupling genome mining and metabolomics with multigene transient expression systems in N. benthamiana has provided an efficient route to deciphering multistep pathways using a gain-of-function approach. Excellent examples are the discovery of the pathways to the etoposide lignans in mayapple (Podophyllum peltatum; Lau and Sattely 2015; Box 1), colchicine in Gloriosa superba (Nett et al. 2020), strychnine in Strychnos nux-vomica (Hong et al. 2022), momilactone diterpenes in rice (Oryza sativa) (De La Peña and Sattely 2021), limonoid triterpenes in Citrus and Meliaceae species (De La Peña et al. 2023), QS saponins (vaccine adjuvants) in soap bark tree (Quillaja saponaria) (Reed et al. 2023), and the late stages of vinblastine synthesis in C. roseus (Grzech et al. 2023). In these studies, transcriptome data were generated to provide a catalog of differentially expressed transcripts in tissues producing the end products and candidates were selected based on expression profile and enzyme class predicted from a chemically logical theoretical pathway and single and multigene expression through leaf infiltration of expression constructs was used to build the pathway step-by-step in N. benthamiana. LC-MS–based metabolite profiling was used to demonstrate the sequential appearance and disappearance of pathway intermediates as consecutive enzymes were sequentially coexpressed. Because flux from primary metabolism, or earlier branches of secondary metabolism, may not be specifically channeled into the engineered pathway, yields of end products could be quite low. However, it has often been possible to increase flux into the engineered pathway. For example, activation of the general phenylpropanoid pathway through coexpression of a MYB transcription factor massively increased etoposide formation, to levels higher than in the plant of origin, in N. benthamiana coinfiltrated with the structural genes for the pathway (Kim et al. 2022), confirming the utility of N. benthamiana as a chassis for commercial production of high value plant metabolites. In the case of terpene production, coexpression of a feedback insensitive form of HMG-CoA reductase increased the yields of triterpenes by 4- to 10-fold (Reed et al. 2017), and momilactone production could be enhanced by rerouting diterpene biosynthesis from the chloroplastic to the cytosolic, high-flux mevalonate pathway (De La Peña and Sattely 2021).
BOX 1.
Case study: Discovery of the etoposide pathway
The paper published by Lau and Sattely (2015) is a notable example of coupling transcriptomics, chemical intuition, and metabolite analysis to unravel a complex biosynthetic pathway. The authors were searching for the genes that synthesize etoposide, a chemotherapeutic agent produced naturally by Mayapple. Four genes in this pathway had been previously discovered, over a span of about 17 yr. To further unravel the remaining biosynthetic genes, Lau and Sattely (2015) identified biosynthetic gene candidates by searching for enzymes that were coexpressed in the same tissues and environmental conditions as the known genes in the pathway. Then, they heterologously expressed these genes in N. benthamiana and measured the biosynthetic products. This led to the identification of 6 genes that, when combined, produced etoposide aglycone, the immediate precursor to etoposide. More recently, the Sattely lab has demonstrated a semi-semisynthetic approach to producing etoposide precursors at a milligram scale in N. benthamiana. Overall, this approach demonstrates a compelling example of how recently developed ideas and technology can rapidly lead to substantial discoveries in complex plant metabolic pathways.
Genetic loss of function provides critical information on the involvement of a gene and its encoded protein in a particular biological function. Candidate genes identified by the approaches above can be functionally characterized by isolation of mutants and impacts on secondary metabolism determined by metabolomics or targeted metabolite profiling (e.g. Adiji et al. 2021 as one of many examples). The advent of widely applicable gene editing technologies removes the requirement for available mutant populations, and virus-induced gene silencing (VIGS) has been and continues to be developed as a rapid approach to gene knock-down. VIGS applies recombinant viruses to transiently downregulate the expression of targeted genes. This technology has found use in identifying genes involved in the biosynthesis of several types of secondary metabolites, including monoterpene indole alkaloids (MIAs) in opium poppy (Papaver somniferum) (Wijekoon and Facchini 2012; Salim et al. 2013; Chen et al. 2020), bisindole alkaloids in C. roseus (Liscombe and O’Connor 2011), and acyl sugars in Solanum and Nicotiana species (Slocombe et al. 2008).
Random assembly of secondary metabolite-derived polymers
Polymers of secondary metabolites in the form of lignin (derived from hydroxycinnamyl alcohols) and condensed tannins (derived from flavonoids) constitute a substantial proportion of the fixed carbon on the planet. Other plant polymers derived wholly or in part from secondary metabolites include suberin (fatty acids, hydroxycinnamic acids associated with lignin), melanin (phenolics), phlobaphenes (flavonoids), and sporopollenin (phenolics, fatty acids, flavonoids). With the exception of the highly inert polymer sporopollenin, the structure of which was only recently discovered (Li et al. 2019), the biosynthesis of the building blocks of these polymers has been intensively studied and reviewed elsewhere (Dixon and Barros 2019; Yu et al. 2023). However, in contrast to other plant polymers such as polysaccharides and proteins, the assembly of the monomeric units into polymers requires less precise enzymatic control. In the case of melanin, the polymer results from the oxidation of vacuolar phenolics and possibly other molecules after rupture of the chloroplast membrane releases polyphenol oxidase (Glagoleva et al. 2020). After many years of debate, it now appears that the monomers that constitute the chains of condensed tannins are linked nonenzymatically via nucleophilic attack between an activated flavan-3-ol or leucoanthocyanidin carbocation extension unit and a flavan-3-ol starter unit (Liu et al. 2016; Jun et al. 2021); genetic analysis has failed to demonstrate any type of polymerase that would yield the observed linkage patterns but rather reveals proteins that appear to control the spatial distribution of starter and extension units (Yu et al. 2023).
In the case of lignin, it has been known for many years that the monolignols (hydroxycinnamyl alcohols) are polymerized via a free radical reaction initiated by the action of laccases and peroxidases. However, the large size of the laccase and peroxidase gene families in plants has hampered genetic analysis of this process, and there has been much debate as to the extent to which the polymerization is purely chemical or requires some type of template (Ralph et al. 2008). As in many things, the truth probably lies somewhere in the middle; the discovery of C-lignin composed only of catechyl alcohol units (Chen et al. 2012) and evidence for the incorporation of structurally diverse nonconventional units into the lignin of grasses (e.g. the flavonoid tricin), and other species (e.g. stilbenes and polyamines) argues against strict proteinaceous control of coupling (Ralph et al. 2019), whereas a picture is now emerging of precise spatial control of the initiation of macromolecular lignin assembly in the Casparian strip of the root involving dirigent proteins, previously implicated in stereoselective lignan coupling (Davin and Lewis 2000; Hosmani et al. 2013; Wang et al. 2022).
The above examples highlight the importance of cell biology in the accumulation of secondary metabolite-derived polymers and the need for unbiased genetic analysis in elucidating the complete biosynthetic machinery.
Biological insights derived from studies on plant secondary metabolism
Secondary metabolite synthesis is under complex spatial control
Secondary metabolites are seldom found throughout all plant tissues but rather are deposited in specific tissues or even specialized organs. Glandular trichomes are common sites of storage of metabolites required on the outer surface of the plant and can be isolated in pure form, facilitating the elucidation of pathways involved in the biosynthesis of their contents through RNA sequencing. A classic example is the interrogation of the peppermint (Mentha x piperita) glandular trichome transcriptome for genes involved in the biosynthesis of menthol and related monoterpenes (Lange et al. 2000). In some cases, secondary metabolites are localized to the leaf epidermis, and transcriptome analysis of the C. roseus leaf “epidermone” revealed the presence of transcripts for the complete MIA pathway, including a transporter for leaf surface secretion (Murata et al. 2008; Yu and De Luca 2013).
However, not all secondary metabolites are synthesized in the organs in which they ultimately accumulate. A well know example is nicotine in Nicotiana species, which accumulates in leaves but is synthesized in roots (Zenkner et al. 2019), its transport being facilitated by a purine uptake permease (Kato et al. 2015). Application of single cell metabolomics analysis revealed that, although most MIA precursors are indeed found in the epidermal cells of C. roseus, the major MIAs vindoline and serpentine localize to idioblast cells, and their precursor strictosidine accumulates in both cell types (Yamamoto et al. 2019). The biosynthesis of benzyisoquinoline alkaloid biosynthesis in the opium poppy is equally complex: the initial steps of biosynthesis of several of the alkaloids appear to occur in companion cells and sieve elements, with final accumulation of the alkaloids in laticifers (reviewed in Beaudoin and Facchini 2014). Surprisingly, however, the pathway gene transcripts are localized to companion cells, whereas the proteins themselves are found in both sieve elements and laticifers, and shotgun proteomics has confirmed the presence of enzymes specific for the final steps of morphine biosynthesis to be present in laticifers (Onoyovwe et al. 2013). MS imaging techniques (Fig. 3) hold promise for higher resolution understanding of the spatial distributions of the precursors and end products of plant secondary metabolism (Dong and Aharoni 2022). This understanding is assisted by our increasing knowledge of transporters for secondary metabolites (Gani et al. 2021), although in some cases (e.g. monolignol transport) genetic analysis has failed to reveal transport mechanisms, and passive diffusion is becoming increasingly accepted (Perkins et al. 2022). At the same time, the release of floral volatiles to the atmosphere, long believed to occur via passive diffusion, has recently been shown to be mediated by nonspecific lipid transfer proteins (Liao et al. 2023).
Metabolic channeling provides an additional layer of pathway regulation
Metabolic channeling is the passing of intermediates of enzymatic reactions from one enzyme to the next without equilibration into the cellular milieu (cytosol). It provides a means to regulate flux into bifurcating pathways, to enhance catalytic activity, and to protect highly reactive intermediates from undesirable chemical conversion or from causing cellular toxicity (Winkel-Shirley 1999). Although first postulated in mammalian metabolism, many of the best examples of metabolic channeling have been demonstrated, or at least proposed, in plants. Unfortunately, metabolic channeling can be used as a convenient catch-all interpretation when metabolic data do not fit the current paradigm, in some cases perhaps because the model is incorrect. Data supporting metabolic channeling require careful interpretation and usually confirmation via multiple approaches.
Metabolic channeling has been proposed to operate in the synthesis of many types of plant secondary metabolites, including terpenoids, alkaloids, phenylpropanoids, flavonoids, and cyanogenic glycosides (Gutensohn et al. 2022; Jørgensen et al. 2005). Techniques for determining protein–protein interactions have been widely used in these studies. For example, bimolecular fluorescence complementation (BiFC) and coimmunoprecipitation approaches revealed that soybean (Glycine max) isoflavone synthase interacted with CHS, chalcone isomerase (CHI), and cinnamate 4-hydroxylase (C4H), suggesting the formation of an isoflavone biosynthesis metabolon (Dastmalchi et al. 2016). But metabolic channeling is not necessarily conserved in the same pathway between different species. For example, snapdragon (Antirrhinum majus) flavone synthase II (FNS II) was shown to interact with CHS, CHI, and dihydroflavanol reductase (DFR) by yeast 2-hybrid and BiFC assays (Fujino et al. 2018). In contrast, using similar methods, no interaction between FNS II and CHS could be verified in torenia (Torenia hybrida), where FNS II interacted instead with flavonoid 3-hydroxylase (F3H) (Fujino et al. 2018). In a further example from the flavonoid pathway, Förster resonance energy transfer (FRET) detected by fluorescence lifetime imaging microscopy (FLIM) (FRET-FLIM) provided in vivo evidence that DFR competed with flavonol synthase (FLS) in physical binding to CHS in Arabidopsis; this suggests that channeling may operate to determine the direction of flux into flavonols and anthocyanins/proanthocyanidins (Crosby et al. 2011). Metabolic channeling is likely a dynamic process that is potentially dependent upon physiological conditions, an aspect that requires further study.
It was proposed that the membrane-associated cytochrome P450 enzymes C4H, flavanone 3′-hydroxylase (F3′H), and FNS II act to anchor the phenylpropanoid/flavonoid enzyme complex to the cytosolic side of the endoplasmic reticulum (Winkel-Shirley 1999, 2001; Fujino et al. 2018), and C4H, coumaroyl shikimate 3′-hydroxylase (C3′H), and ferulate/coniferaldehyde 5-hydroxylase (F5H) P450s were assumed to operate similarly in the pathway to monolignols. Direct evidence of weak PAL/C4H interactions came from FRET studies in N. tabacum (Achnine et al. 2004), and FLIM revealed the consecutive P450s C4H and C3′H to be colocalized on the endoplasmic reticulum (ER) membrane and restricted in their lateral diffusion by association with other ER proteins (Bassard et al. 2012). The operationally soluble enzymes hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase and 4-coumarate:CoA ligase were partially associated with the ER when the 2 P450s were coexpressed. It was subsequently shown that the 3 P450 enzymes of the monolignol pathway, although in spatial proximity to one another on the ER membrane, do not interact directly but rather through the intermediacy of 2 members of the membrane sterol binding protein family, which themselves can form multimers to allow organization of P450 clusters with resultant stabilization of the P450 enzymes (Gou et al. 2018).
Although most examples of metabolic channeling in plant secondary metabolism have been proposed to involve complexes anchored to the ER, compartmentation between cytosol and organelles can also occur. During the biosynthesis of MIA alkaloids in C. roseus, strictosidine is formed from secologanin and tyrosine by a Pictet–Spengler reaction catalyzed by strictosidine synthase (Treimer and Zenk 1979); many plants contain putative orthologs of strictosidine synthase with functions yet to be determined (Kibble et al. 2010). Strictosidine is then deglycosylated by an enzyme, strictosidine β-glucosidase (SDG), that multimerizes to form a shield protecting the reactive aglycone, which spontaneously reorganizes to yield, among other products, cathenamine and epi-cathenamine, which are downstream intermediates for the formation of MIAs. The enzymes that initiate further conversion of these products may be localized to the nucleus, where they may physically interact with the SDG multimers, forming a nuclear metabolon that is thought to further protect reactive intermediates until more stable compounds are generated for subsequent oxidation by cytochrome P450 enzymes localized on the ER (Stavrinides et al. 2015). Nuclear localization of secondary metabolism warrants further study.
Plant secondary metabolism evolves through a variety of mechanisms
Because many secondary metabolites are involved in the interactions of plants with their environment, their appearance in plant lineages is intimately associated with plant microbe/pest/predator coevolution. In some cases, the same metabolites are made by both the plant and its potential pest/predator. Aside from ecological considerations, this situation is of interest for the light it sheds on the evolutionary flexibility of secondary metabolite biosynthesis. A remarkable example concerns the iridoid sex pheromones, which are synthesized in the hind legs of female pea aphids (Acyrthosiphon pisum) by the same sequence of reactions as the iridoids in plants, although 6 enzymes of the insect pathway are unrelated to their plant counterparts (Köllner et al. 2022). The same situation of common intermediates but involvement of different classes of enzymes is seen in the case of the diterpene gibberellins of plants, fungi, and bacteria (Salazar-Cerezo et al. 2018), highlighting the operation of cross-kingdom convergent evolution of secondary biosynthetic pathways. At the same time, phylogenetic studies have revealed that some enzymes of plant secondary metabolism have arisen through horizontal gene transfer, such as the origin of PAL from soil bacteria and fungi during symbioses early in land colonization (Emiliani et al. 2009). A number of enzymes of secondary metabolism have also been coopted during evolution from enzymes of primary metabolism (Sonawane et al. 2020; Lou et al. 2022), an understanding of which can inform rational metabolic engineering.
Some enzymes of secondary metabolism, for example, the terpene synthases, are remarkably promiscuous, based on the contours and dynamics, rather than catalytic residues or mechanism, of their active sites that direct the folding of the tertiary carbocation toward a large range of potential products (Greenhagen et al. 2006; Jia et al. 2019). Subsequent modification of terpene structures largely includes hydroxylation reactions, most of which are catalyzed by cytochrome P450 enzymes, and kingdom-wide phylogenetic analysis has suggested that the CYP716 family has evolved specifically toward triterpenoid biosynthesis in eudicots, with neofunctionalization of the CYP716 family coinciding with the emergence of oxidosqualene cyclases with specific roles in secondary metabolism (Miettinen et al. 2017).
In contrast to the situation in fungi, where secondary metabolic genes are usually clustered together in operons, it was thought, until the late 1990s, that the genes involved in plant secondary metabolism were randomly distributed throughout the genome. This appears true in many cases, including, for example, the genes of general phenylpropanoid metabolism and the lignin and anthocyanin branches. However, the picture changed in 1997, when it was shown that all 5 structural genes (a tryptophan synthase homolog and 4 cytochrome P450s) for the biosynthesis of anti-insect benzoxazinones in grasses were linked in an operon-like structure (Frey et al. 1997). Following the subsequent discovery of the genomic organization of the avenacin pathway in oat (Avena sativa), the momilactone pathway in rice, and the thalianol pathway in Arabidopsis (Qi et al. 2004; Wilderman et al. 2004; Field and Osbourn 2008), diterpene and triterpene biosynthesis emerged as a focus area for studies on metabolic diversification through independent evolution of gene clusters in plants. Gene clusters have since been discovered in several other areas of plant secondary metabolism (Bharadwaj et al. 2021), including the biosynthesis of the surface β-diketone waxes in wheat (Triticum aestivum) and barley (Hordeum vulgare) where the cluster contains a polyketide synthase, a cytochrome, P450 and a hydrolase/carboxylase, functions that were previously considered to be part of a multifunctional protein (Hen-Avivi et al. 2016)
Clustering of genes allows for more highly coordinated coregulation, as well as opportunities for regulation via targeted chromatin remodeling (Osbourn 2010). Analysis of the structure of the thalianol cluster in a wide range of A. thaliana accessions and related Arabidopsis species showed that the cluster is largely fixed, with a low frequency of deleterious haplotypes, and that chromosomal inversion can shuttle distant genes into the cluster to enable cluster compaction (Liu et al. 2020). Cross-cluster effects may also play a part in the maintenance of biosynthetic gene clusters. For example, in addition to the momilactone cluster on chromosome 4, rice also contains a gene cluster on chromosome 2 for synthesis of related phytocassane diterpenoids, which contains 2 genes encoding related cytochrome P450 enzymes (CYP76M7/8). CRISPR-Cas9 editing to remove the cluster on chromosome 2 resulted in phytotoxicity due to apparent buildup of momilactone pathway intermediates requiring CYP76M7/8 for further conversion (Li et al. 2022).
Plants also contain examples of highly tandemly duplicated genes, the best known being the leucine-rich repeat disease resistance genes in many plants (van Wersch and Li 2019). The tandem arrangement may provide a “reservoir” for generation of gene diversity through site-directed recombination or ensure cosegregation of genes that are most effective when expressed in concert. In the area of secondary metabolism, the ∼100 Arabidopsis small molecule uridine diphosphate glycosyl transferase (UGT) genes are scattered throughout the genome but cluster into groups of up to 7 genes. The over 300 M. truncatula UGT genes are distributed similarly, but the number of genes in each array is greater, with one locus on chromosome 6 containing 27 tandemly arrayed UGT genes and CYP genes often cocluster with the UGT genes (Young et al. 2011). It has yet to be shown whether this particular organization provides a reservoir for evolutionary adaptation of UGT functions.
It is now becoming apparent that plant secondary metabolism has evolved to include built-in redundancies. This likely provides both flexibility and protection against fluctuations in precursor flux. Examples include dual pathways for O-methylation (at the free acid and CoA ester levels) and 3′-hydroxylation (direct or via the “shikimate shunt”) in monolignol biosynthesis (Ye et al. 1994; Dixon and Barros 2019; Vanholme et al. 2019) and parallel pathways to epicatechin as a starter or extension unit in condensed tannin biosynthesis (reviewed in Yu et al. 2023). Such phenomena complicate the genetic analysis of pathway structure.
Manipulation of secondary metabolism provides a paradigm for plant ecology
The field of plant chemical ecology was developed from the discoveries in the 1940s to 1980s that characterized secondary metabolites and their biological activities against insect predators and microbial pathogens. The subsequent discoveries of pathway genes have now allowed ecological predictions to be tested in natural environments. A good example is the work of Baldwin and associates with the wild tobacco N. alata in habitat in the Great Basin Desert in Utah, which established that release of volatiles from insect larval-attacked leaves can attract predatory insects in nature (Kessler and Baldwin 2001), and that adjacent plants can perceive volatile signals from attacked plants to induce preemptive defense pathways (Baldwin et al. 2006). Planting transgenically modified N. alata with strongly reduced nicotine levels in its natural environment confirmed that nicotine does indeed protect the plant from larval feeding (Steppuhn et al. 2004) and demonstrated that insect predators (wolf spiders) prefer larvae that have fed on plants lacking nicotine (Kumar et al. 2014). Furthermore, use of plant-mediated RNAi to downregulate an insect cytochrome P450 implicated in the exhalation of digested nicotine from the spiracles resulted in increased spider attack, showing that larvae themselves utilize nicotine as a defense (Kumar et al. 2014).
The optimal defense theory (McKey 1979) states that the evolution and allocation of defenses is based on both the fitness value and vulnerability of individual plant parts to simultaneously optimize growth and defense. This theory was recently tested for the glucosinolate defense compounds of A. thaliana, which naturally exhibit a gradient of distribution from the youngest (highest) to the oldest leaves, where the compounds are synthesized in the lower mature leaves and transported to the younger leaves, which are the preferred tissues for herbivory. Modifying this gradient by knock-out of 2 glucosinolate transporter genes altered the feeding preference of larvae of the generalist insect herbivore African cotton leafworm (Spodoptera littoralis) in a way consistent with the tenets of the optimal defense theory (Hunziker et al. 2021). However, glucosinolates appear to have functions beyond protection against herbivory and pathogen attack. Levels of aliphatic glucosinolates in Arabidopsis are regulated by a transcriptional cascade involving auxin-sensitive AUXIN/INDOLE-3-ACETIC ACID-INDUCIBLE (Aux/IAA) repressors, with elevated glucosinolate levels being necessary for correct stomatal regulation under drought (Salehin et al. 2019).
It can be expected that the ecological functions of more and more secondary metabolites will be examined through field studies with genetically modified plants in the coming years.
Cutting-edge technologies are paving the road to discoveries in secondary metabolism research
Emerging technologies are enabling understandings of metabolites and their functional roles in plants. Here, we highlight recently developed or improved technical approaches and how these have influenced the field of plant metabolic biology.
Genome-wide association studies allow unbiased analysis of gene function
Genome-wide association studies (GWAS) are well known in the biomedical field but have now become a powerful tool in many other areas of biology, including plants (Tibbs Cortes et al. 2021). Linking traits to genotypes across populations allows an unbiased approach to discovery of genes impacting important traits, and this approach is expected to have broad impacts for plant secondary metabolism in the future. As one example, in a GWAS analysis of more than 1,000 individual lines of black cottonwood (Populus trichocarpa), some single nucleotide polymorphisms (SNPs) associated with lignin content across the Populus species range and in plants grown under different environmental conditions resided in a 5-enolpyruvylshikimate-3-phosphate synthase-like (EPSP) gene (Xie et al. 2018). EPSP synthase is an enzyme of the shikimate pathway that provides precursors for phenylpropanoid biosynthesis, but the rare alleles were not associated with enzymatic properties but rather structural changes that converted the enzyme to a transcription factor that acted as a repressor of lignin biosynthesis (Xie et al. 2018). An approach in which untargeted, qualitative metabolic traits are subjected to GWAS analysis (QT-GWAS) appears promising in uncovering gene metabolite relations, revealing enzymes of chroman, nucleotide, and neolignan biosynthesis in Arabidopsis (Brouckaert et al. 2023).
To determine the genetic basis for production of defensive terpene metabolites secreted by maize roots, a combination of metabolite-based QTL mapping, GWAS analysis, and use of near-isogenic lines was applied, leading to the identification of a terpene cyclase involved in the synthesis of the nonvolatile antibiotic β-costic acid (Ding et al. 2017). Such approaches provide powerful tools for molecular breeding to enhance biotic stress tolerance.
Single-cell transcriptomics is enabling deep characterization of phytohormone signaling leading to the identification of regulators
Another rapidly evolving method that is advancing our understanding of gene function is single-cell sequencing. The recent explosion of use of single-cell transcriptomics has led to the extensive generation of data on thousands of genes underlying plant development and stress response. Several of these studies have already made progress deepening our understanding of small molecule biosynthesis and signaling. For example, single-cell transcriptomic studies during phosphate deficiency revealed cytokinin production in the central vasculature, which has a cascading effect to promote root hair development in the epidermis (Wendrich et al. 2020). In another example, single-cell RNA sequencing of developing Arabidopsis lateral roots revealed camalexin, an indole phytoalexin, as a regulator of lateral root formation (Serrano-Ron et al. 2021). Other studies have found additional roles for auxin, cytokinin, and gibberellin in various developmental processes, including root development (Torii et al. 2020), leaf vein development (Liu et al. 2022), and tissue regeneration (Efroni et al. 2016). In addition, single-cell transcriptomics has been used to analyze genes that respond to phytohormone stimulus. For example, single-cell analysis in brassinosteroid-blind mutants revealed non-cell-autonomous functions of brassinosteroid signaling (Graeff et al. 2021). Another single-cell study of the brassinosteroid response in roots identified responsive transcription factors expressed with high specificity in the elongating cortex (Nolan et al. 2023). In the future, it will be important to delineate the transcriptomic responses to abiotic and biotic stresses at the single cell level and explore methods capable of combining single-cell transcriptomics with metabolomics measurements to address the involvement of unidentified regulatory natural products.
Technological advances to visualize localization patterns of small molecules
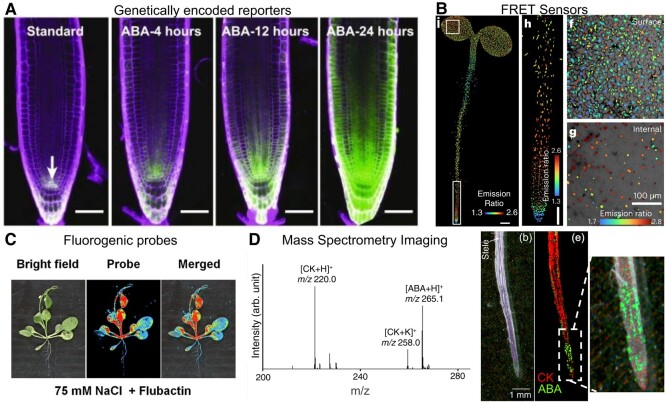
Key advances in small molecule biology have been enabled by the development of strategies to visualize phytochemicals across space and time using chemical and genetic-based approaches. The most common technique for visualizing metabolite activity is to design and measure genetically encoded reporters. This technique has been mostly applied to plant hormones in the context of development. Promoter fusions, which typically rely on synthetic promoters that are activated by phytohormone-specific transcription factors, can be used to measure the activities of auxin (Ulmasov et al. 1997), ABA (Wu et al. 2018), brassinosteroids (Chaiwanon and Wang 2015), and cytokinins (Liu and Müller 2017). For example, DR5 is a synthetic promoter element that contains 7 repeats of an auxin-response element (Ulmasov et al. 1997). DR5 has been used extensively to drive various reporters, including GUS, GFP, and luciferase (LUC) to quantify auxin-regulated processes. 6xABRE is another synthetic promoter used to visualize the transcriptional response to ABA (Wu et al. 2018; Fig. 4A). However, it is advised to use multiple strategies to test conclusions using synthetic promoter:reporter lines, as it has been shown that these lines do not always reflect known phytohormone concentrations (Chandler and Werr 2015). A comparison of multiple approaches for the visualization of ABA in plant roots and shoots is given in Fig. 4. Another genetic approach to visualizing phytohormones uses signal-induced degradation to indirectly measure phytohormone levels. This approach is exemplified by R2D2 (ratiometric version of 2 DIIs), which leverages an interaction between auxin, an Aux/IAA DII domain, and a ubiquitin ligase to measure auxin levels (Liao et al. 2015). In this system, the DII domain is fused to the reporter n3xVenus and the mDII domain, which is not degraded by auxin, is fused to nTdTomato. Changes in auxin levels are determined by measuring the ratio of yellow to red fluorescence. FRET sensors are another way to measure small molecule activity. For instance, a FRET reporter for auxin that utilizes an engineered tryptophan repressor protein-binding pocket allows for transient and reversible detection of IAA (Herud-Sikimić et al. 2021). A FRET reporter for ABA relies on the signal-induced interaction between covalently linked ABA receptor domains (such as pyrabactin resistance 1 and PP2C-type phosphatase domains) to measure changes in ABA localization and concentration (Jones et al. 2014; Waadt et al. 2014; Rowe et al. 2023; Fig. 4B).
Figure 4.
Examples of techniques used to visualize the localization of secondary metabolites. Each technology shown here is designed to image abscisic acid (ABA). A) A genetically encoded reporter (Wu et al. 2018); B) a FRET sensor (Rowe et al. 2023); C) a fluorogenic, small molecule-based probe (Yang et al. 2022), and this image was reprinted with permission from Yang et al. (2022) copyright 2022 American Chemical Society; and D) an MALDI MS imaging method for imaging phytohormones cytokinin (CK) and ABA (Shiono et al. 2017), and this image was reprinted with permission from Shiono et al. (2017), copyright 2017 American Chemical Society. Images are licensed under a Creative Commons Attribution License (CC BY, https://creativecommons.org/licenses/by/4.0/) by Wu et al. (2018) and Rowe et al. (2023).
Currently, there are few examples of biosensors for secondary metabolites beyond the classical hormones, which reflects the general lack of detailed information required to generate these types of reporters. Designing receptor-based biosensors requires atomic scale resolution, which is lacking for most secondary metabolite receptors, if known (Nemhauser and Torii 2016). Likewise, promoter-based biosensors require the identification of highly specific sequences responsive only to the metabolite of interest. Further elucidation of these interactions will expand the use of biosensors and lead to understanding of the localization patterns of secondary metabolites and their potential functions as signal molecules. Global protein–metabolite interactome studies could provide a basis for technology development in this space (Luzarowski et al. 2021).
Direct imaging of metabolites can occasionally be performed if the chemical of interest already has natural fluorescence properties. For example, phenolic compounds such as hydroxycinnamic acids, coumarins, stilbenes, and styrylpyrones are often auto-fluorescent under the proper excitation wavelength (Hutzler et al. 1998). Phenylcoumarans and stilbenes are major chromophore monomers in lignin and have enabled nondestructive imaging of lignin (Maceda and Terrazas 2022). The fluorescence of lignin can be further amplified by dyes or by feeding monolignols with small fluorescent tags (Tobimatsu et al. 2013). Flavonoids are other phenolic metabolites that naturally possess low levels of fluorescence, which can be enhanced with dye conjugates to reveal localization and intensity, as well as uncover their regulatory interactions with signaling pathways and phytohormones (Lewis et al. 2011; Muhlemann et al. 2018). However, the vast majority of metabolites do not have inherent fluorescence. To address this, bioactive compounds with fluorescent tags have been developed to track brassinosteroids (Irani et al. 2012), auxins (Pařízková et al. 2021; Hayashi et al. 2014), and jasmonates (Liu and Sang 2013). While such bioactive fluorescent tags have provided information on the spatiotemporal dynamics of these molecules, they typically cannot be utilized to determine activity (i.e. protein-binding or participation in enzymatic reactions). One strategy for enabling measurements of metabolite activity is the design of synthetic protein-responsive small molecule reporters that change fluorescence properties upon protein interactions. For example, the computationally optimized synthetic compound, flubactin, increases in fluorescence 8-fold upon binding to the ABA pyrabactin resistance 1 receptor (Yang et al. 2022; Fig. 4C). Protein-responsive fluorescent reporters have also been developed for strigolactone (de Saint Germain et al. 2022) and retinaldehyde (Dickinson et al. 2021). These reporters enable access to a key element of secondary metabolite function, interactions with signaling proteins. However, an obvious limitation of pro-fluorescent and fluorescent reporters is that they can only be designed where the identity of a secondary metabolite is known, and the vast majority of secondary metabolites in plants remain unknown.
In order to explore the “black box” of secondary metabolism, chemical imaging technologies have provided avenues for direct measurements of metabolites in situ and in vivo. For example, desorption electrospray ionization and matrix assisted laser desorption/ionization (MALDI) MS imaging has been applied to map the spatial profiles of primary metabolites (Zhang et al. 2023), glucosinolates (Sarsby et al. 2012), lipids (Sarabia et al. 2018), alkaloids (Conceição et al. 2021), and phytohormones (Shiono et al. 2017; Fig. 4D) across plant tissue sections of interest. This work has revealed localization patterns of known metabolites as well as mass to charge signatures of many unknown compounds. MS imaging is capable of measuring relative levels of mass to charge ratios of tens to hundreds of small molecules with spatial resolution ranging from 5 to 250 μm. However, a limitation of this technology is that it cannot perform dynamic measurements. Two chemical imaging techniques that are nondestructive and capable of measuring dynamic information are Raman and NMR microscopy. Raman spectroscopy is a selective technique that measures vibrational modes of molecules. When applied as a microscopic method, it has been used to measure different classes of secondary metabolites (Yang et al. 2017) and cell wall polymers (Gierlinger and Schwanninger 2006) with high spatial resolution (1 to 10 μm). NMR microscopy is related to magnetic resonance imaging (MRI), which has been widely used for whole body imaging in human medical applications. In plants, NMR microscopy is ideally suited to measure water flow, but can also be applied to track C13 labeled plant metabolites (Köckenberger et al. 2004).
Advances in plant synthetic biology are deepening our understanding of how to produce and utilize secondary metabolites
Due to the ability of plants to synthesize and store large amounts of metabolites of interest for human health and society, both primary and secondary metabolism are now major targets of plant synthetic biology efforts (Maeda 2019), and these advances have led to engineering of secondary metabolite pathways with applications in medicine, agriculture, and renewable fuels in sustainable ways (Goold et al. 2018; Liu et al. 2023). This research has also been critical for generating deeper understandings of metabolite biosynthesis and regulation. For example, elucidating the biological pathway for colchicine biosynthesis generated understanding of the tissue specificity of alkaloid biosynthesis in medicinal plants in addition to providing engineering strategies for the production of this medicinal compound (Nett et al. 2020). In spite of this and the other examples given earlier, the discovery of natural product biosynthesis pathways still remains a major obstacle for achieving the full potential for engineering secondary metabolites in plant and microbes (Anarat-Cappillino et al. 2014). Advances will rely not only on discovery of pathway genes but also on deployment of tunable synthetic regulatory circuits (Nemhauser and Torii 2016).
Another branch of synthetic biology research is to utilize metabolic pathways to understand functions or to engineer new traits. As previously mentioned, the development of genetically encoded biosensors is a growing area of plant biology research. This includes the development of small molecule reporters to study the localization of signaling components and the creation of plant “sentinels” that produce dramatic phenotypic changes in response to external chemical cues (Jez et al. 2016; Uslu and Grossmann 2016). One of the earliest synthetic biology examples in plants was the engineering of plant metabolism for phytoremediation of toxins (Bizily et al. 2000). An exciting recently developed approach is the synthetic transfer of features of plant secondary metabolite regulation into heterologous systems for precise control of regulatory processes. For example, the auxin-inducible degron system, used to rapidly degrade proteins in organisms that do not endogenously synthesize auxin, has already made important contributions to biological research (Nishimura et al. 2009). Another current theme in synthetic biology is to generate strategies for precise tuning of secondary metabolite localization to improve plant growth or stress response. For example, researchers have rewired phytohormone pathways to control plant development and architecture (Brophy et al. 2022). Finally, there is potential in the future for engineering plant–microbial interactions, which often involve metabolite-based communication, for improving plant growth and stress responses (Ke et al. 2021).
Conclusions
The past century has seen enormous advances in our understanding of the broad repertoire of chemicals elaborated by plants, their localizations, functions, and biosynthetic pathways. These developments have in many ways tracked the technological developments in analytical chemistry and molecular and cellular biology. There is a long history of the use of plant secondary metabolites in human health and society. Continuing the discovery of new molecules and pathways will lead to further societal benefits. At the same time, the study of plant secondary metabolism itself has led to a number of conceptual advances in biology and has raised a number of questions that, even if recognized, could not have been addressed by early workers in the field (see “OUTSTANDING QUESTIONS”). We are entering an exciting period where advances in gene discovery and synthetic biology are opening up possibilities for the sustainable production of a wide range of plant secondary metabolites as replacements for traditional medicines, food ingredients, and fuels. To further our understanding of the functions of secondary metabolites in plant ecosystems, particularly under climate change, it will be necessary to manipulate these compounds in a wide variety of species beyond crops. Recent advances combining VIGS with CRISPR Cas9-based gene editing (e.g. Ellison et al. 2020) promise solutions to the need for species-independent and genotype-independent genetic transformation methods for rapid pathway manipulation, thereby allowing scientists to expand the legacy of the past century's insights into plant secondary metabolism.
ADVANCES.
High-throughput gene sequencing, MS, and AI technologies are helping to assemble the complement of secondary metabolites across the plant kingdom.
Studies in plant–insect ecology highlight the evolutionary flexibility of secondary metabolite biosynthesis and the roles of secondary metabolites in multitrophic interactions.
Coupling genome mining and metabolomics with multigene transient expression systems provide a pipeline for gene discovery in complex pathways.
The discovery of plant metabolite gene clusters is a major advance in understanding the biosynthesis and evolution of secondary metabolites and plant genome evolution.
Single-cell “omics” and small molecule biosensors hold future promise in determining the roles of specialized metabolites as signal compounds.
OUTSTANDING QUESTIONS.
What are the chemical identities of the myriad of specialized metabolites that have remained uncharacterized?
What are the genes that synthesize and respond to specialized metabolites, and can we improve the speed and reliability of plant transformation and VIGS to enable their study in any species of interest?
What are the roles of secondary metabolites in signaling, and can this be addressed in the future through proteome:metabolome-level interaction mapping?
How do we improve our ability to measure and engineer these metabolites with high spatial precision?
What translational applications for plant specialized metabolites will be developed in the future?
Acknowledgments
A.J.D. would like to thank Philip Benfey for many years of exemplary mentorship and guidance and for seeing the potential for understanding the chemistry underlying developmental plant biology. R.A.D. thanks Vernon Butt, Derek Bendall, and Chris Lamb for inspiration during the early part of his career.
Contributor Information
Richard A Dixon, BioDiscovery Institute and Department of Biological Sciences, University of North Texas, Denton, TX 76203, USA.
Alexandra Jazz Dickinson, Department of Cell and Developmental Biology, University of California at San Diego, La Jolla, CA 92093, USA.
Funding
Work in the authors’ laboratories was funded by the Center for Bioenergy Innovation, a U.S. Department of Energy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science (to R.A.D.); the National Science Foundation Division of Integrated Organismal Systems (award number 1456286 to R.A.D.); the National Science Foundation Early-Concept Grants for Exploratory Research program (collaborative EAGER 2028649 to A.J.D.); the National Institutes of Health (NIGMS 1R35GM147216 to A.J.D.); and the Hellman Foundation (to A.J.D.).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, Rosenthal PJ, D'Alessandro U. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar J. 2011:10(1):144. 10.1186/1475-2875-10-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell. 2004:16(11):3098–3109. 10.1105/tpc.104.024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adiji O, Docampo-Palacios ML, Alvarez-Hernandez A, Pasinetti GM, Wang X, Dixon RA. UGT84F9 Is the major flavonoid UDP-glucuronosyltransferase in Medicago truncatula. Plant Physiol. 2021:185(4):1617–1637. 10.1093/plphys/kiab016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anarat-Cappillino G, Sattely ES. The chemical logic of plant natural product biosynthesis. Curr Opin Plant Biol. 2014:19:51–58. 10.1016/j.pbi.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen I, Thiedig K, Nordholt N, Schmidt N, Huep G, Sagasser M, Weisshaar B. Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta. 2014:240(5):955–970. 10.1007/s00425-014-2088-0 [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Halitschke R, Paschold A, von Dahl CC, Preston CA. Volatile signaling in plant-plant interactions: “talking trees” in the genomics era. Science. 2006:311(5762):812–815. 10.1126/science.1118446 [DOI] [PubMed] [Google Scholar]
- Bassard JE, Richert L, Geerinck J, Renault H, Duval F, Ullmann P, Schmitt M, Meyer E, Mutterer J, Boerjan W, et al. Protein-protein and protein-membrane associations in the lignin pathway. Plant Cell. 2012:24(11):4465–4482. 10.1105/tpc.112.102566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate-Smith EC. The phenolic constituents of plants and their taxonomic significance. I. Dicotyledons. Bot J Linnean Soc. 1962:58(371):95–173. 10.1111/j.1095-8339.1962.tb00890.x [DOI] [Google Scholar]
- Beaudoin GA, Facchini PJ. Benzylisoquinoline alkaloid biosynthesis in opium poppy. Planta. 2014:240(1):19–32. 10.1007/s00425-014-2056-8 [DOI] [PubMed] [Google Scholar]
- Bell JN, Dixon RA, Bailey JA, Rowell PM, Lamb CJ. Differential induction of chalcone synthase mRNA activity at the onset of phytoalexin accumulation in compatible and incompatible plant-pathogen interactions. Proc Natl Acad Sci U S A. 1984:81(11):3384–3388. 10.1073/pnas.81.11.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj R, Kumar SR, Sharma A, Sathishkumar R. Plant metabolic gene clusters: evolution, organization, and their applications in synthetic biology. Front Plant Sci. 2021:12:697318. 10.3389/fpls.2021.697318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizily SP, Rugh CL, Meagher RB. Phytodetoxification of hazardous organomercurials by genetically engineered plants. Nat Biotechnol. 2000:18(2):213–217. 10.1038/72678 [DOI] [PubMed] [Google Scholar]
- Britsch L, Grisebach H. Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur J Biochem. 1986:156(3):569–577. 10.1111/j.1432-1033.1986.tb09616.x [DOI] [PubMed] [Google Scholar]
- Brophy JAN, Magallon KJ, Duan L, Zhong V, Ramachandran P, Kniazev K, Dinneny JR. Synthetic genetic circuits as a means of reprogramming plant roots. Science. 2022:377(6607):747–751. 10.1126/science.abo4326 [DOI] [PubMed] [Google Scholar]
- Brouckaert M, Peng M, Höfer R, Houari IE, Darrah C, Storme V, Saeys Y, Vanholme R, Goeminne G, Timokhin VI, et al. QT-GWAS: a novel method for unveiling biosynthetic loci affecting qualitative metabolic traits. Mol Plant. 2023:16(7):1212–1227. 10.1016/j.molp.2023.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RC. Phytochemistry. What it is and how it has developed? J Chem Educ. 1937:14(11):520. 10.1021/ed014p520 [DOI] [Google Scholar]
- Chaiwanon J, Wang ZY. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol. 2015:25(8):1031–1042. 10.1016/j.cub.2015.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW, Werr W. Cytokinin–auxin crosstalk in cell type specification. Trends Plant Sci. 2015:20(5):291–300. 10.1016/j.tplants.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J. A polymer of caffeyl alcohol in plant seeds. Proc Natl Acad Sci U S A. 2012:109(5):1772–1177. 10.1073/pnas.1120992109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Chen X, Hagel JM, Facchini PJ. Virus-induced gene silencing to investigate alkaloid biosynthesis in Opium Poppy. Meth Mol Biol. 2020:2172:75–92. 10.1007/978-1-0716-0751-0_7 [DOI] [PubMed] [Google Scholar]
- Choi D, Ward BL, Bostock RM. Differential induction and suppression of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes in response to Phytophthora infestans and to its elicitor arachidonic acid. Plant Cell. 1992:4(10):1333–1344. 10.1105/tpc.4.10.1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição RS, Perez CJ, Branco A, Botura MB, Ifa DR. Identification of Sassafras albidum alkaloids by high-performance thin-layer chromatography tandem mass spectrometry and mapping by desorption electrospray ionization mass spectrometry imaging. J Mass Spectrom. 2021:56(1):e4674. 10.1002/jms.4674 [DOI] [PubMed] [Google Scholar]
- Crosby KC, Pietraszewska-Bogiel A, Gadella TWJ, Winkel BSJ. Förster resonance energy transfer demonstrates a flavonoid metabolon in living plant cells that displays competitive interactions between enzymes. FEBS Lett. 2011:585(14):2193–2198. 10.1016/j.febslet.2011.05.066 [DOI] [PubMed] [Google Scholar]
- Cruickshank IAM, Perrin DR. Studies on phytoalexins. III. The isolation, assay, and general properties of a phytoalexin from Pisum sativum L. Australian J Biol Sci. 1961:14(3):336–348. 10.1071/BI9610336 [DOI] [Google Scholar]
- Dastmalchi M, Bernards MA, Dhaubhadel S. Twin anchors of the soybean isoflavonoid metabolon: evidence for tethering of the complex to the endoplasmic reticulum by IFS and C4H. Plant J. 2016:85(6):689–706. 10.1111/tpj.13137 [DOI] [PubMed] [Google Scholar]
- Davin LB, Lewis NG. Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 2000:123(2):453–462. 10.1104/pp.123.2.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Peña R, Hodgson H, Liu JCT, Stephenson MJ, Martin AC, Owen C, Harkess A, Leebens-Mack J, Jimenez LE, Osbourn A, et al. Complex scaffold remodeling in plant triterpene biosynthesis. Science. 2023:379(6630):361–368. 10.1126/science.adf1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Peña R, Sattely ES. Re-routing plant terpene biosynthesis enables momilactone pathway elucidation. Nat Chem Biol. 2021:17(2):205–212. 10.1038/s41589-020-00669-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Schouveiler P, Pillot JP, Singh AV, Chevalier A, Daignan Fornier S, Guillory A, Bonhomme S, Rameau C, et al. Expansion of the strigolactone profluorescent probes repertory: the right probe for the right application. Front Plant Sci. 2022:13:887347. 10.3389/fpls.2022.887347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethier VG. Chemical factors determining the choice of food plants by Papilio larvae. Am Nat. 1941:75(756):61–73. 10.1086/280929 [DOI] [Google Scholar]
- Dickinson AJ, Zhang J, Luciano M, Wachsman G, Sandoval E, Schnermann M, Dinneny JR, Benfey PN. A plant lipocalin promotes retinal-mediated oscillatory lateral root initiation. Science. 2021:373(6562):1532–1536. 10.1126/science.abf7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Huffaker A, Köllner TG, Weckwerth P, Robert CAM, Pencer JL, Lipka AE, Schmelz EA. Selinene volatiles are essential precursors for maize defense promoting fungal pathogen resistance. Plant Physiol. 2017:175(3):1455–1468. 10.1104/pp.17.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Barros J. Lignin biosynthesis: old roads revisited and new roads explored. Open Biol. 2019:9(12):190215. 10.1098/rsob.190215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aharoni A. Image to insight: exploring natural products through mass spectrometry imaging. Nat Prod Rep. 2022:39(7):1510–1530. 10.1039/D2NP00011C [DOI] [PubMed] [Google Scholar]
- Edwards K, Cramer CL, Bolwell GP, Dixon RA, Schuch W, Lamb CJ. Rapid transient induction of phenylalanine ammonia-lyase mRNA in elicitor-treated bean cells. Proc Natl Acad Sci U S A. 1985:82(20):6731–6673. 10.1073/pnas.82.20.6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell. 2016:165(7):1721–1733. 10.1016/j.cell.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison EE, Nagalakshmi U, Gamo ME, Huang P-j, Dinesh Kumar S, Voytas D. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants. 2020:6(6):620–624. 10.1038/s41477-020-0670-y [DOI] [PubMed] [Google Scholar]
- Emiliani G, Fondi M, Fani R, Gribaldo S. A horizontal gene transfer at the origin of phenylpropanoid metabolism: a key adaptation of plants to land. Biol Direct. 2009:4(1):7. 10.1186/1745-6150-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE. Metabolic diversification-independent assembly of operon-like gene clusters in different plants. Science. 2008:320(5875):543–547. 10.1126/science.1154990 [DOI] [PubMed] [Google Scholar]
- Filner P, Varner JE. A test for de novo synthesis of enzymes: density labeling with H2O18 of barley alpha-amylase induced by gibberellic acid. Proc Natl Acad Sci U S A. 1967:58(4):1520–1526. 10.1073/pnas.58.4.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch C. William withering: an account of foxglove and some of its medical uses. 1785–1985. J Am Coll Cardiol. 1985:5(5):1A–2A. 10.1016/S0735-1097(85)80456-3 [DOI] [PubMed] [Google Scholar]
- Freudenberg K. Lignin: its constitution and formation from p-hydroxycinnamyl alcohols: lignin is duplicated by dehydrogenation of these alcohols; intermediates explain formation and structure. Science. 1965:148(3670):595–600. 10.1126/science.148.3670.595 [DOI] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997:277(5326):696–699. 10.1126/science.277.5326.696 [DOI] [PubMed] [Google Scholar]
- Fujino N, Tenma N, Waki T, Ito K, Komatsuzaki Y, Sugiyama K, Yamazaki T, Yoshida S, Hatayama M, Yamashita S, et al. Physical interactions among flavonoid enzymes in snapdragon and torenia reveal the diversity in the flavonoid metabolon organization of different plant species. Plant J. 2018:94(2):372–392. 10.1111/tpj.13864 [DOI] [PubMed] [Google Scholar]
- Gani U, Vishwakarma RA, Misra P. Membrane transporters: the key drivers of transport of secondary metabolites in plants. Plant Cell Rep. 2021:40(1):1–18. 10.1007/s00299-020-02599-9 [DOI] [PubMed] [Google Scholar]
- Gierlinger N, Schwanninger M. Chemical imaging of poplar wood cell walls by confocal Raman microscopy. Plant Physiol. 2006:140(4):1246–1254. 10.1104/pp.105.066993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glagoleva AY, Shoeva OY, Khlestkina EK. Melanin pigment in plants: current knowledge and future perspectives. Front Plant Sci. 2020:11:770. 10.3389/fpls.2020.00770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold HD, Wright P, Hailstones D. Emerging opportunities for synthetic biology in agriculture. Genes (Basel). 2018:9(7):341. 10.3390/genes9070341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Ran X, Martin DW, Liu CJ. The scaffold proteins of lignin biosynthetic cytochrome P450 enzymes. Nat Plants. 2018:4(5):299–310. 10.1038/s41477-018-0142-9 [DOI] [PubMed] [Google Scholar]
- Graeff M, Rana S, Wendrich JR, Dorier J, Eekhout T, Aliaga Fandino AC, Guex N, Bassel GW, De Rybel B, Hardtke CS. A single-cell morpho-transcriptomic map of brassinosteroid action in the Arabidopsis root. Mol Plant. 2021:14(12):1985–1999. 10.1016/j.molp.2021.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhagen BT, O'Maille PE, Noel JP, Chappell J. Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc Natl Acad Sci U S A. 2006:103(26):9826–9831. 10.1073/pnas.0601605103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzech D, Hong B, Caputi L, Sonawane PD, O’Connor SE. Engineering the biosynthesis of late-stage vinblastine precursors precondylocarpine acetate, catharanthine, tabersonine in Nicotiana benthamiana. ACS Synth Biol. 2023:12(1):27–34. 10.1021/acssynbio.2c00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutensohn M, Hartzell E, Dudareva N. Another level of complexity: the role of metabolic channeling and metabolons in plant terpenoid metabolism. Front Plant Sci. 2022:13:954083. 10.3389/fpls.2022.954083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger LA, Schwochau ME. Ultraviolet light-induced formation of pisatin and phenylalanine ammonia lyase. Plant Physiol. 1971:47(4):588–590. 10.1104/pp.47.4.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahlbrock K, Grisebach H. Enzymic controls in the biosynthesis of lignin and flavonoids. Annu Rev Plant Biol. 1979:30(1):105–130. 10.1146/annurev.pp.30.060179.000541 [DOI] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev Plant Physiol Plant Mol Biol. 1989:40(1):347–369. 10.1146/annurev.pp.40.060189.002023 [DOI] [Google Scholar]
- Hartmann T. The lost origin of chemical ecology in the late 19th century. Proc Natl Acad Sci U S A. 2008:105(12):4541–4546. 10.1073/pnas.0709231105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam E. Biogenetically patterned synthesis of procyanidins. J Chem Soc Chem Commun. 1974:1974(15):594–595. 10.1039/c39740000594 [DOI] [Google Scholar]
- Hayashi K, Nakamura S, Fukunaga S, Nishimura T, Jenness MK, Murphy AS, Motose H, Nozaki H, Furutani M, Aoyama T. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc Natl Acad Sci U S A. 2014:111(31):11557–11562. 10.1073/pnas.1408960111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hen-Avivi S, Savin O, Racovita RC, Lee W-S, Adamski NM, Malitsky S, Almekias-Siegl E, Levy M, Vautrin S, Bergès H, et al. A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell. 2016:28(6):1440–1460. 10.1105/tpc.16.00197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herud-Sikimić O, Stiel AC, Kolb M, Shanmugaratnam S, Berendzen KW, Feldhaus C, Höcker B, Jürgens G. A biosensor for the direct visualization of auxin. Nature. 2021:592(7856):768–772. 10.1038/s41586-021-03425-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Grzech D, Caputi L, Sonawane P, Rodriguez Lopez CE, Kamileen MO, Hernandez Lozada NJ, Grabe V, O’Connor S. Biosynthesis of strychnine. Nature. 2022:607(7919):617–622. 10.1038/s41586-022-04950-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani PS, Kamiya T, Danku J, Naseer S, Geldner N, Guerinot ML, Salt DE. Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based casparian strip in the root. Proc Natl Acad Sci U S A. 2013:110(35):14498–11503. 10.1073/pnas.1308412110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker P, Lambertz SK, Weber K, Crocoll C, Halkier BA, Schulz A. Herbivore feeding preference corroborates optimal defense theory for specialized metabolites within plants. Proc Natl Acad Sci U S A. 2021:118(47):e2111977118. 10.1073/pnas.2111977118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzler P, Fischbach R, Heller W, Jungblut T, Reuber S, Schmitz R, Veit M, Weissenbock G, Schnitzler J. Tissue localization of phenolic compounds in plants by confocal laser scanning microscopy. J Exp Bot. 1998:49(323):953–965. 10.1093/jxb/49.323.953 [DOI] [Google Scholar]
- Irani NG, Di Rubbo S, Mylle E, Van den Begin J, Schneider-Pizoń J, Hniliková J, Šíša M, Buyst D, Vilarrasa-Blasi J, Szatmári AM, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol. 2012:8(6):583–589. 10.1038/nchembio.958 [DOI] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988:241(4863):346–349. 10.1126/science.241.4863.346 [DOI] [PubMed] [Google Scholar]
- Jez JM, Lee SG, Sherp AM. The next green movement: plant biology for the environment and sustainability. Science. 2016:353(6305):1241–1244. 10.1126/science.aag1698 [DOI] [PubMed] [Google Scholar]
- Jia M, Mishra SK, Tufts S, Jernigan RL, Peters RJ. Combinatorial biosynthesis and the basis for substrate promiscuity in class I diterpene synthases. Metab Eng. 2019:55:44–58. 10.1016/j.ymben.2019.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Danielson JA, Manojkumar SN, Lanquar V, Grossmann G, Frommer WB. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife. 2014:3:e01741. 10.7554/eLife.01741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Møller BL. Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol. 2005:8(3):280–291. 10.1016/j.pbi.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Jun JH, Lu N, Docampo-Palacios M, Wang X, Dixon RA. Dual activity of anthocyanidin reductase supports the dominant plant proanthocyanidin extension unit pathway. Sci Adv. 2021:7(20):eabg4682. 10.1126/sciadv.abg4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Shitan N, Shoji T, Hashimoto T. Tobacco NUP1 transports both tobacco alkaloids and vitamin B6. Phytochemistry. 2015:113:33–40. 10.1016/j.phytochem.2014.05.011 [DOI] [PubMed] [Google Scholar]
- Ke J, Wang B, Yoshikuni Y. Microbiome engineering: synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021:39(3):244–261. 10.1016/j.tibtech.2020.07.008 [DOI] [PubMed] [Google Scholar]
- Kellner F, Kim J, Clavijo BJ, Hamilton JP, Childs KL, Vaillancourt B, Cepela J, Habermann M, Steuernagel B, Clissold L, et al. Genome-guided investigation of plant natural product biosynthesis. Plant J. 2015:82(4):680–692. 10.1111/tpj.12827 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Defensive function of herbivore-induced plant volatile emissions in nature. Science. 2001:291(5511):2141–2144. 10.1126/science.291.5511.2141 [DOI] [PubMed] [Google Scholar]
- Kibble NAJ, Sohani MM, Shirley N, Byrt C, Roessner U, Bacic A, Schmidt O, Schultz CJ. Phylogenetic analysis and functional characterisation of strictosidine synthase-like genes in Arabidopsis thaliana. Funct Plant Biol. 2010:36(12):1098–1109. 10.1071/FP09104 [DOI] [PubMed] [Google Scholar]
- Kim SS, Wengier DL, Ragland CJ, Sattely ES. Transcriptional reactivation of lignin biosynthesis for the heterologous production of etoposide aglycone in Nicotiana benthamiana. ACS Synth Biol. 2022:11(10):3379–3387. 10.1021/acssynbio.2c00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Grisebach H. Enzymic synthesis of isoflavones. Eur J Biochem. 1986:155(2):311–318. 10.1111/j.1432-1033.1986.tb09492.x [DOI] [PubMed] [Google Scholar]
- Köckenberger W, De Panfilis C, Santoro D, Dahiya P, Rawsthorne S. High resolution NMR microscopy of plants and fungi. J Microsc. 2004:214(2):182–189. 10.1111/j.0022-2720.2004.01351.x [DOI] [PubMed] [Google Scholar]
- Köllner TG, David A, Luck K, Beran F, Kunert G, Zhou J-J, Caputi L, O’Connor SE. Biosynthesis of iridoid sex pheromones in aphids. Proc Natl Acad Sci U S A. 2022:119(42):e2211254119. 10.1073/pnas.2211254119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth KL, Jaggard DAW, Dixon RA. Developmental and light-regulated post-translational control of 3-hydroxy-3-methylglutaryl-CoA reductase levels in potato. Plant J. 2000:23(4):507–516. 10.1046/j.1365-313x.2000.00821.x [DOI] [PubMed] [Google Scholar]
- Koukol J, Conn EE. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J Biol Chem. 1961:236(10):2692–2698. 10.1016/S0021-9258(19)61721-7 [DOI] [PubMed] [Google Scholar]
- Kreuzaler F, Hahlbrock K. Flavonoid glycosides from illuminated cell suspension cultures of Petroselinum hortense. Phytochemistry. 1973:12(5):1149–1152. 10.1016/0031-9422(73)85031-9 [DOI] [Google Scholar]
- Kreuzaler F, Ragg H, Fautz E, Kuhn DN, Hahlbrock K. UV induction of chalcone synthase mRNA in cell suspension cultures of Petroselinum hortense. Proc Natl Acad Sci U S A. 1983:80(9):2591–2593. 10.1073/pnas.80.9.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulagina N, Méteignier L-V, Papon N, O'Connor SE, Courdavault V. More than a Catharanthus plant: a multicellular and pluri-organelle alkaloid-producing factory. Curr Opin Plant Biol. 2022:67:102200. 10.1016/j.pbi.2022.102200 [DOI] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Steppuhn BI. Natural history-driven, plant-mediated RNAi-based study reveals CYP6B46's role in a nicotine-mediated antipredator herbivore defense. Proc Natl Acad Sci U S A. 2014:111(4):1245–1252. 10.1073/pnas.1314848111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R. Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc Natl Acad Sci U S A. 2000:97(6):2934–2939. 10.1073/pnas.97.6.2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau W, Sattely ES. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015:349(6253):1224–1228. 10.1126/science.aac7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton MA, Dixon RA, Lamb CJ. Elicitor modulation of L-phenylalanine ammonia-lyase in French bean cell suspension cultures. Biochim Biophys Acta. 1980:633(2):162–175. 10.1016/0304-4165(80)90402-X [DOI] [PubMed] [Google Scholar]
- Learned RM, Fink GR. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Arabidopsis thaliana is structurally distinct from the yeast and animal enzymes. Proc Natl Acad Sci U S A. 1989:86(8):2779–2783. 10.1073/pnas.86.8.2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux H. Discovery of salicine. J Chim Med. 1830:6:340–432. [Google Scholar]
- Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BS, Muday GK. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 2011:156(1):144–164. 10.1104/pp.111.172502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FS, Phyo P, Jacobowitz J, Hong M, Weng JK. The molecular structure of plant sporopollenin. Nat Plants. 2019:5(1):41–46. 10.1038/s41477-018-0330-7 [DOI] [PubMed] [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993:5(2):171–179. 10.2307/3869583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhang J, Li Z, Peters RJ, Yang B. Dissecting the labdane-related diterpenoid biosynthetic gene clusters in rice reveals directional cross-cluster phytotoxicity. New Phytol. 2022:233(2):878–889. 10.1111/nph.17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao P, Maoz I, Shih ML, Lee JH, Huang XQ, Morgan JA, Dudareva N. Emission of floral volatiles is facilitated by cell-wall non-specific lipid transfer proteins. Nat Commun. 2023:14(1):330. 10.1038/s41467-023-36027-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods. 2015:12(3):207–210. 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe DK, O'Connor SE. A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry. 2011:72(16):1969–1977. 10.1016/j.phytochem.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Cheema J, Vigouroux M, Hill L, Reed J, Paajanen P, Yant L, Osbourn A. Formation and diversification of a paradigm biosynthetic gene cluster in plants. Nat Commun. 2020:11(1):5354. 10.1038/s41467-020-19153-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ha CM, Dixon RA. Functional genomics in the study of metabolic pathways in Medicago truncatula: an overview. Methods Mol Biol. 2018:1822:315–337. 10.1007/978-1-4939-8633-0_20 [DOI] [PubMed] [Google Scholar]
- Liu C, Wang X, Shulaev V, Dixon RA. A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat Plants. 2016:2(12):16182. 10.1038/nplants.2016.182 [DOI] [PubMed] [Google Scholar]
- Liu J, Müller B. Imaging TCSn::GFP, a synthetic cytokinin reporter, in Arabidopsis thaliana. Methods Mol Biol. 2017:1497:81–90. 10.1007/978-1-4939-6469-7_9 [DOI] [PubMed] [Google Scholar]
- Liu S, Sang R. Bioactive fluorescent jasmonate designed by molecular modeling and its migration in tomato visualized by fluorescent molecular imaging. Tetrahedron. 2013:69(2):844–848. 10.1016/j.tet.2012.10.105 [DOI] [Google Scholar]
- Liu X, Zhang P, Zhao Q, Huang AC. Making small molecules in plants: a chassis for synthetic biology-based production of plant natural products. J Integr Plant Biol. 2023:65(2):417–443. 10.1111/jipb.13330 [DOI] [PubMed] [Google Scholar]
- Liu Z, Wang J, Zhou Y, Zhang Y, Qin A, Yu X, Zhao Z, Wu R, Guo C, Bawa G, et al. Identification of novel regulators required for early development of vein pattern in the cotyledons by single-cell RNA-sequencing. Plant J. 2022:110(1):7–22. 10.1111/tpj.15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y-R, Pichersky E, Last RL. Deep roots and many branches: origins of plant-specialized metabolic enzymes in general metabolism. Curr Opin Plant Biol. 2022:66:102192. 10.1016/j.pbi.2022.102192 [DOI] [PubMed] [Google Scholar]
- Luzarowski M, Vicente R, Kiselev A, Wagner M, Schlossarek D, Erban A, de Souza LP, Childs D, Wojciechowska I, Luzarowska U, et al. Global mapping of protein-metabolite interactions in Saccharomyces cerevisiae reveals that Ser-Leu dipeptide regulates phosphoglycerate kinase activity. Commun Biol. 2021:4(1):181. 10.1038/s42003-021-01684-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceda A, Terrazas T. Fluorescence microscopy methods for the analysis and characterization of lignin. Polymers (Basel). 2022:14(5):961. 10.3390/polym14050961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda HA. Harnessing evolutionary diversification of primary metabolism for plant synthetic biology. J Biol Chem. 2019:294(45):16549–16566. 10.1074/jbc.REV119.006132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi JG, Mahdi AJ, Bowen ID. The historical analysis of aspirin discovery, its relation to the willow tree and antiproliferative and anticancer potential. Cell Prolif. 2006:39(2):147–155. 10.1111/j.1365-2184.2006.00377.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham KR, Mabry TJ. Ultraviolet-visible and proton magnetic resonance spectroscopy of flavonoids. In: Harborne HB, Mabry TJ, Mabry H, editors. The flavonoids. New York (NY): Academic Press; 1975. p. 45–77. [Google Scholar]
- McKey D. The distribution of secondary compounds within plants. In: Rosenthal GA, Janzen DH, editors. Herbivores: their interaction with secondary plant metabolites. New York (NY): Academic Press; 1979. p. 55–134. [Google Scholar]
- Miettinen K, Pollier J, Buyst D, Arendt P, Csuk R, Sommerwerk S, Moses T, Mertens J, Sonawane PD, Pauwels L, et al. The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat Commun. 2017:8(1):14153. 10.1038/ncomms14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolo LV, Reichert AI, Dixon RA. Introduction to the different classes of biosynthetic enzymes. In: Osbourn A, Lanzotti V, editors. Plant-derived natural products, synthesis, function and application. New York (NY): Springer; 2009. p. 143–163. [Google Scholar]
- Montinari MR, Minelli S, De Caterina R. The first 3500 years of aspirin history from its roots—a concise summary. Vascul Pharmacol. 2019:113:1–8. 10.1016/j.vph.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Muhlemann JK, Younts TLB, Muday GK. Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proc Natl Acad Sci U S A. 2018:115(47):11188–11197. 10.1073/pnas.1811492115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata J, Roepke J, Gordon H, De Luca V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell. 2008:20(3):524–542. 10.1105/tpc.107.056630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Torii KU. Plant synthetic biology for molecular engineering of signalling and development. Nat Plants. 2016:2(3):16010. 10.1038/nplants.2016.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett RS, Lau W, Sattely ES. Discovery and engineering of colchicine alkaloid biosynthesis. Nature. 2020:584(7819):148–153. 10.1038/s41586-020-2546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Meth. 2009:6(12):917–292. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Nolan TM, Vukašinović N, Hsu CW, Zhang J, Vanhoutte I, Shahan R, Taylor IW, Greenstreet L, Heitz M, Afanassiev A, et al. Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science. 2023:379(6639):eadf4721. 10.1126/science.adf4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative . One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019:574(7780):679–685. 10.1038/s41586-019-1693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoyovwe A, Hagel JM, Chen X, Khan MF, Schriemer D, Facchini PJ. Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers. Plant Cell. 2013:25(10):4110–4122. 10.1105/tpc.113.115113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. Gene clusters for secondary metabolic pathways: an emerging theme in plant biology. Plant Physiol. 2010:154(2):531–535. 10.1104/pp.110.161315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C, Patron NJ, Huang A, Osbourn A. Harnessing plant metabolic diversity. Curr Opin Chem Biol. 2017:40:24–30. 10.1016/j.cbpa.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pařízková B, Žukauskaitė A, Vain T, Grones P, Raggi S, Kubeš MF, Kieffer M, Doyle SM, Strnad M, Kepinski S, et al. New fluorescent auxin probes visualise tissue-specific and subcellular distributions of auxin in Arabidopsis. New Phytol. 2021:230(2):535–549. 10.1111/nph.17183 [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang J, Li X, Zhang Y. Salicylic acid: biosynthesis and signaling. Annu Rev Plant Biol. 2021:72(1):761–791. 10.1146/annurev-arplant-081320-092855 [DOI] [PubMed] [Google Scholar]
- Perkins ML, Schuetz M, Unda F, Chen KT, Bally MB, Kulkarni JA, Yan Y, Pico J, Castellarin SD, Mansfield SD, et al. Monolignol export by diffusion down a polymerization-induced concentration gradient. Plant Cell. 2022:34(5):2080–2095. 10.1093/plcell/koac051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci U S A. 2004:101(21):8233–8238. 10.1073/pnas.0401301101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Brunow G, Harris P, Dixon RA, Boerjan W. Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? Rec Adv Polyphenols Res. 2008:1:36–66. 10.1002/9781444302400.ch2 [DOI] [Google Scholar]
- Ralph J, Lapierre C, Boerjan W. Lignin structure and its engineering. Curr Opin Biotechnol. 2019:56:240–249. 10.1016/j.copbio.2019.02.019 [DOI] [PubMed] [Google Scholar]
- Reed J, Orme A, El-Demerdash A, Owen C, Martin LBB, Misra RC, Kikuchi S, Rejzek M, Martin AC, Harkess A, et al. Elucidation of the pathway for biosynthesis of saponin adjuvants from the soapbark tree. Science. 2023:379(6638):1252–1264. 10.1126/science.adf3727 [DOI] [PubMed] [Google Scholar]
- Reed J, Stephenson MJ, Miettinen K, Brouwer B, Leveau A, Brett P, Goss RJM, Goossens A, O’Connell MA, Osbourn A. A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metabolic Eng. 2017:42:185–193. 10.1016/j.ymben.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro B, Lacchini E, Bicalho KU, Mertens J, Arendt P, Vanden Bossche R, Calegario G, Gryffroy L, Ceulemans E, Buitink J, et al. A seed-specific regulator of triterpene saponin biosynthesis in Medicago truncatula. Plant Cell. 2020:32(6):2020–2042. 10.1105/tpc.19.00609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. A theory of the mechanism of the phytochemical synthesis of certain alkaloids. J Chem Soc Trans. 1917:111(0):876–899. 10.1039/CT9171100876 [DOI] [Google Scholar]
- Roux D, Ferreira D. Structure and function in the biomimetic synthesis of linear, angular and branched condensed tannins. Pure Appl Chem. 1982:54(12):2465–2478. 10.1351/pac198254122465 [DOI] [Google Scholar]
- Rowe J, Grangé-Guermente M, Exposito-Rodriguez M, Wimalasekera R, Lenz MO, Shetty KN, Cutler SR, Jones AM. Next-generation ABACUS biosensors reveal cellular ABA dynamics driving root growth at low aerial humidity. Nat Plants. 2023:9(7):1103–1115. 10.1038/s41477-023-01447-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueffer M, Zenk MH. Distant precursors of benzylisoquinoline alkaloids and their enzymatic formation. Z Naturforsch. 1987:42(4):319–332. 10.1515/znc-1987-0402 [DOI] [Google Scholar]
- Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol. 2010:61(1):463–489. 10.1146/annurev.arplant.043008.092035 [DOI] [PubMed] [Google Scholar]
- Salazar-Cerezo S, Martínez-Montiel N, García-Sánchez J, Pérez-y-Terrón R, Martínez-Contreras RD. Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria. Microbiol Res. 2018:208:85–98. 10.1016/j.micres.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Salehin M, Li B, Tang M, Katz E, Song L, Ecker JR, Kliebenstein DJ, Estelle M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat Commun. 2019:10(1):4021. 10.1038/s41467-019-12002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim V, Yu F, Altarejos J, De Luca V. Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant J. 2013:76(5):754–765. 10.1111/tpj.12330 [DOI] [PubMed] [Google Scholar]
- Sarabia LD, Boughton BA, Rupasinghe T, van de Meene AML, Callahan DL, Hill CB, Roessner U. High-mass-resolution MALDI mass spectrometry imaging reveals detailed spatial distribution of metabolites and lipids in roots of barley seedlings in response to salinity stress. Metabolomics. 2018:14(5):63. 10.1007/s11306-018-1359-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarsby J, Towers MW, Stain C, Cramer R, Koroleva OA. Mass spectrometry imaging of glucosinolates in Arabidopsis flowers and siliques. Phytochemistry. 2012:77:110–118. 10.1016/j.phytochem.2012.01.026 [DOI] [PubMed] [Google Scholar]
- Schmelzer E, Jahnen W, Hahlbrock K. In situ localization of light-induced chalcone synthase mRNA, chalcone synthase, and flavonoid end products in epidermal cells of parsley leaves. Proc Natl Acad Sci U S A. 1988:85(9):2989–2993. 10.1073/pnas.85.9.2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. Friedrich Wilhelm Serturner and the discovery of morphine. Pharm Hist. 1985:27:61–74. PMID: 11611724. [PubMed] [Google Scholar]
- Serrano-Ron L, Perez-Garcia P, Sanchez-Corrionero A, Gude I, Cabrera J, Ip PL, Birnbaum KD, Moreno-Risueno MA. Reconstruction of lateral root formation through single-cell RNA sequencing reveals order of tissue initiation. Mol Plant. 2021:14(8):1362–1378. 10.1016/j.molp.2021.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan JJ. Sinapate esters provide greater UV-B attenuation than flavonoids in Arabidopsis thaliana (Brassicaceae). Am J Botany. 1996:83(6):679–686. 10.1002/j.1537-2197.1996.tb12757.x [DOI] [Google Scholar]
- Shiono K, Hashizaki R, Nakanishi T, Sakai T, Yamamoto T, Ogata K, Harada KI, Ohtani H, Katano H, Taira S. Multi-imaging of cytokinin and abscisic acid on the roots of rice (Oryza sativa) using matrix-assisted laser desorption/ionization mass spectrometry. J Agric Food Chem. 2017:65(35):7624–7628. 10.1021/acs.jafc.7b02255 [DOI] [PubMed] [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, Besser K, Welsby NA, Harper A, Aziz N, Li Y, Larson TR, Giovannoni J, et al. Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol. 2008:148(4):1830–1846. 10.1104/pp.108.129510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich IE, Bollmann J, Hahlbrock K, Kombrink E, Schulz W. Differential early activation of defense-related genes in elicitor-treated parsley cells. Plant Mol Biol. 1989:12(2):227–234. 10.1007/BF00020507 [DOI] [PubMed] [Google Scholar]
- Sonawane PD, Jozwiak A, Panda S, Aharoni A. ‘Hijacking’ core metabolism: a new panache for the evolution of steroidal glycoalkaloids structural diversity. Curr Opin Plant Biol. 2020:55:118–128. 10.1016/j.pbi.2020.03.008 [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Vazquez-Flota F, De Luca V. Multicellular compartmentation of Catharanthus roseus alkaloid biosynthesis predicts intercellular translocation of a pathway intermediate. Plant Cell. 1999:11(5):887–900. 10.1105/tpc.11.5.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides A, Tatsis EC, Foureau E, Caputi L, Kellner F, Courdavault V, O'Connor SE. Unlocking the diversity of alkaloids in Catharanthus roseus: nuclear localization suggests metabolic channeling in secondary metabolism. Chem Biol. 2015:22(3):336–341. 10.1016/j.chembiol.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A, Gase K, Krock B, Halitschke R, Baldwin IT. Nicotine's defensive function in nature. PLoS Biol. 2004:2(8):e217. 10.1371/journal.pbio.0020217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain T. Plant phenolics: past and future. Annu Proc Phytochem Soc Europe. 1985:25:453–468. [Google Scholar]
- Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008:54(2):335–347. 10.1111/j.1365-313X.2008.03418.x [DOI] [PubMed] [Google Scholar]
- Tibbs Cortes L, Zhang Z, Yu J. Status and prospects of genome-wide association studies in plants. Plant Genome. 2021:14(1):e20077. 10.1002/tpg2.20077 [DOI] [PubMed] [Google Scholar]
- Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W, et al. Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 2013:76(3):357–366. 10.1111/tpj.12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyama K, Sakuma T, Ishizaka N, Sato N, Katsui N, Tagasugi M, Masamune T. A new antifungal substance isolated from resistant potato tuber tissue. Phytopathol. 1968:58:115–116. [Google Scholar]
- Torii K, Kubota A, Araki T, Endo M. Time-series single-cell RNA-seq data reveal auxin fluctuation during endocycle. Plant Cell Physiol. 2020:61(2):243–254. 10.1093/pcp/pcz228 [DOI] [PubMed] [Google Scholar]
- Treimer JF, Zenk MH. Purification and properties of strictosidine synthase, the key enzyme in indole alkaloid formation. Eur J Biochem. 1979:101(1):225–233. 10.1111/j.1432-1033.1979.tb04235.x [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997:9(11):1963–1971. 10.1105/tpc.9.11.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uslu VV, Grossmann G. The biosensor toolbox for plant developmental biology. Curr Opin Plant Biol. 2016:29:138–147. 10.1016/j.pbi.2015.12.001 [DOI] [PubMed] [Google Scholar]
- VanEtten HD, Mansfield JW, Bailey JA, Farmer EE. Two classes of plant antibiotics: phytoalexins versus “phytoanticipins”. Plant Cell. 1994:6(9):1191–1192. 10.2307/3869817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, De Meester B, Ralph J, Boerjan W. Lignin biosynthesis and its integration into metabolism. Curr Opin Biotechnol. 2019:56:230–239. 10.1016/j.copbio.2019.02.018 [DOI] [PubMed] [Google Scholar]
- van Wersch S, Li X. Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci. 2019:24(8):688–699. 10.1016/j.tplants.2019.05.005 [DOI] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, Schroeder JI. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife. 2014:3:e01739. 10.7554/eLife.01739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cao Y, Liang X, Zhuang J, Wang X, Qin F, Jiang C. A dirigent family protein confers variation of casparian strip thickness and salt tolerance in maize. Nat Commun. 2022:13(1):2222. 10.1038/s41467-022-29809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich JR, Yang B, Vandamme N, Verstaen K, Smet W, Van de Velde C, Minne M, Wybouw B, Mor E, Arents HE, et al. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science. 2020:370(6518):eaay4970. 10.1126/science.aay4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijekoon CP, Facchini PJ. Systematic knockdown of morphine pathway enzymes in opium poppy using virus-induced gene silencing. Plant J. 2012:69(6):1052–1063. 10.1111/j.1365-313X.2011.04855.x [DOI] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-imara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004:135(4):2098–2105. 10.1104/pp.104.045971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant. 1999:107(1):142–149. 10.1034/j.1399-3054.1999.100119.x [DOI] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001:126(2):485–493. 10.1104/pp.126.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvrouw K, Kim H, Vanholme R, Goeminne G, Ralph J, Boerjan W, Vanholme B. Reassessing the claimed cytokinin-substituting activity of dehydrodiconiferyl alcohol glucoside. Proc Natl Acad Sci U S A. 2023:120(9):e2123301120. 10.1073/pnas.2123301120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Duan L, Pruneda-Paz JL, Oh DH, Pound M, Kay S, Dinneny JR. The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA-mediated transcriptional regulation. Plant Physiol. 2018:177(4):1650–1665. 10.1104/pp.18.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzel E, Kutchan T. Plant metabolism, the diverse chemistry set of the future. Science. 2016:353(6305):1232–1236. 10.1126/science.aad2062 [DOI] [PubMed] [Google Scholar]
- Xie M, Muchero W, Bryan AC, Yee K, Guo HB, Zhang J, Tschaplinski TJ, Singan VR, Lindquist E, Payyavula RS, et al. A 5-enolpyruvylshikimate 3-phosphate synthase functions as a transcriptional repressor in Populus. Plant Cell. 2018:30(7):1645–1660. 10.1105/tpc.18.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Takahashi K, Caputi L, Mizuno H, Rodriguez-Lopez CE, Iwasaki T, Ishizaki K, Fukaki H, Ohnishi M, Yamazaki M, et al. The complexity of intercellular localisation of alkaloids revealed by single-cell metabolomics. New Phytol. 2019:224(2):848–859. 10.1111/nph.16138 [DOI] [PubMed] [Google Scholar]
- Yang JF, Chen WJ, Zhou LM, Hewage KAH, Fu YX, Chen MX, He B, Pei RJ, Song K, Zhang JH, et al. Real-time fluorescence imaging of the abscisic acid receptor allows nondestructive visualization of plant stress. ACS Appl Mater Interfaces. 2022:14(25):28489–28500. 10.1021/acsami.2c02156 [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang X, Zhao C, Tian G, Zhang H, Xiao H, He L, Zheng J. Chemical mapping of essential oils, flavonoids and carotenoids in citrus peels by Raman microscopy. J Food Sci. 2017:82(12):2840–2846. 10.1111/1750-3841.13952 [DOI] [PubMed] [Google Scholar]
- Ye Z-H, Kneusel RE, Matern U, Varner JE. An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell. 1994:6(10):1427–1439. 10.1105/tpc.6.10.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011:480(7378):520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, De Luca V. ATP-binding cassette transporter controls leaf surface secretion of anticancer drug components in Catharanthus roseus. Proc Natl Acad Sci U S A. 2013:110(39):15830–15835. 10.1073/pnas.1307504110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Song Y, Lin J, Dixon RA. The complexities of proanthocyanidin biosynthesis and its regulation in plants. Plant Commun. 2023:4(2):100498. 10.1016/j.xplc.2022.100498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenk MH. Biosynthese von vanillin in Vanilla planifolia. Andr Z Pflanzenphysiol. 1965:53:404–414. [Google Scholar]
- Zenk MH. Chasing the enzymes of secondary metabolism: plant cell cultures as a pot of gold. Phytochemistry. 1991:30(12):3861–3863. 10.1016/0031-9422(91)83424-J [DOI] [Google Scholar]
- Zenkner FF, Margis-Pinheiro M, Cagliari A. Nicotine biosynthesis in Nicotiana: a metabolic overview. Tobacco Sci. 2019:56(1):1–9. 10.3381/18-063 [DOI] [Google Scholar]
- Zhang T, Noll SE, Peng JT, Klair A, Tripka A, Stutzman N, Cheng C, Zare RN, Dickinson AJ. Chemical imaging reveals diverse functions of tricarboxylic acid metabolites in root growth and development. Nat Commun. 2023:14(1):2567. 10.1038/s41467-023-38150-z [DOI] [PMC free article] [PubMed] [Google Scholar]