ABSTRACT
The recruitment of ATG12–ATG5-ATG16L1 complex to phagophore mediated by the specific interaction between ATG16L1 and WIPI2, is pivotal to the formation of autophagosomes during macroautophagy. Recently, we reported that ATG16L1 contains two distinct WIPI2-binding sites, the previously reported WIPI2-binding site (WBS1), and the newly identified site (WBS2). By determining the crystal structures of WIPI2 with ATG16L1 WBS1 and WBS2 respectively, we uncovered that, unlike ATG16L1 WBS1, ATG16L1 WBS2 and its binding mechanism to WIPI2 are conserved from yeast to mammals. Using cell-based functional assays, we further demonstrated that the integrity of two WIPI2-binding sites of ATG16L1 is essential for normal autophagic flux. In summary, our study provided mechanistic insights into the interaction of two key autophagic proteins, ATG16L1 and WIPI2, and revealed a dual-binding-site mode adopted by ATG16L1 to associate with WIPI2.
Abbreviations: ATG: autophagy-related protein; CCD: coiled-coil domain; ITC: isothermal titration calorimetry; PI3KC3-C1: class III phosphatidylinositol 3-kinase complex I; PtdIns3P: phosphatidylinositol-3-phosphate; ULK: Unc-51-like kinase; WBS: WIPI2-binding site; WIPI: WD repeat domain phosphoinositide-interacting protein.
KEYWORDS: Macroautophagy, WIPI2, ATG16L1, autophagosome, autophagy
Macroautophagy (hereafter autophagy) is a highly conserved cellular degradation process involving the formation of double-membrane autophagosomes that sequester intracellular substrates destined to lysosomal turnover, including protein aggregates, and damaged organelles. The coordinated process of autophagy involves a set of core autophagy-related (ATG) proteins. In starvation-induced autophagy, the Unc-51-like kinase (ULK) complex can be activated by several signaling pathways and is targeted to the autophagosome initiation site, which in turn leads to the recruitment and phosphorylation of multiple downstream ATG proteins, including the autophagy-specific class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1). Subsequently, the recruited PI3KC3-C1 generates substantial phosphatidylinositol-3-phosphate (PtdIns3P) to recruit effectors such as the WD repeat domain phosphoinositide-interacting protein (WIPI) family proteins (WIPI1 to WIPI4 in mammals). WIPI2 recruits and allosterically activates the ATG12–ATG5-ATG16L1 complex to catalyze conjugation of ATG8 family proteins to the phosphatidylethanolamine present in autophagosomal phagophore membrane, which promotes the elongation and closure of the phagophore. Therefore, the direct interaction between WIPI2 and ATG16L1 is considered to functionally link between PtdIns3P production and the lipidation of ATG8 family proteins, two essential steps in autophagosome formation. However, the detailed molecular mechanism governing the interaction of ATG16L1 with WIPI2 is not fully elucidated.
ATG16L1 contains an N-terminal ATG5-binding domain for associating with the ATG12–ATG5 conjugate and assembles into the ATG12–ATG5-ATG16L1 complex. This domain is followed by a dimeric coiled-coil domain (CCD) and a C-terminal WD40 domain (Figure 1A). WIPI2 consists of an N-terminal WD40 domain that acquires a 7-bladed β-propeller and a C-terminal unstructured region (Figure 1A). The WD40 domain of WIPI2 interacts with the ATG12–ATG5-ATG16L1 complex and the phagophore membrane by recognizing PtdIns3P through its FRRG motif. The site of ATG16L1 that interacts with WIPI2 was mapped to a region adjacent to ATG16L1 CCD about ten years ago. However, the removal of this WIPI2-binding site just reduces but not completely abolishes the phagophore localization of ATG16L1, implying the existence of uncharacterized alternative and potentially redundant mechanism for ATG16L1 recruitment to the phagophore.
Figure 1.
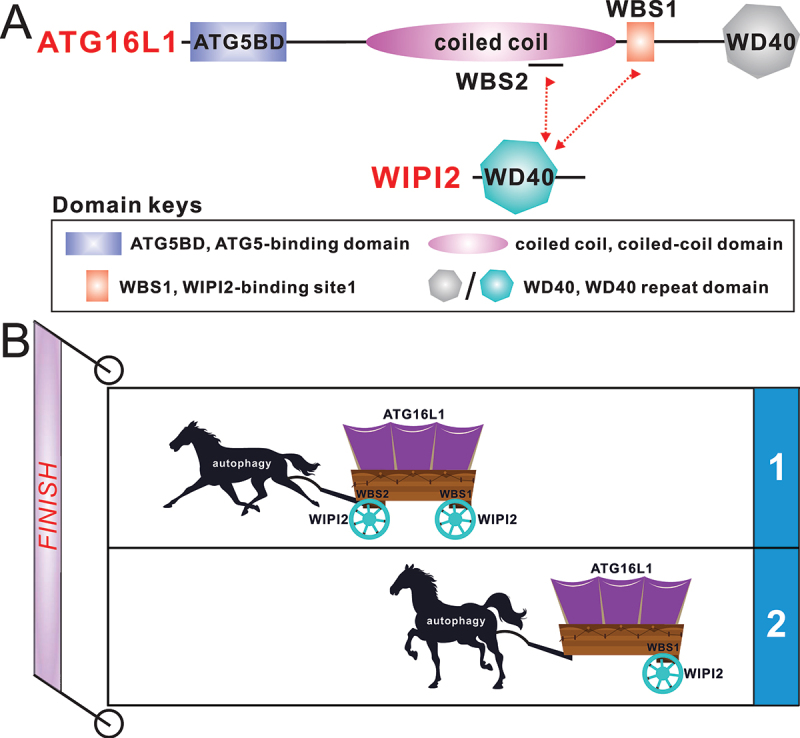
(A) the schematic diagram showing the domain organizations of ATG16L1 and WIPI2. In this drawing, the interactions of ATG16L1 with WIPI2 are highlighted and indicated by two-way arrows. (B) a cartoon model depicting that ATG16L1 adopts a dual binding mode to interact with WIPI2, which can drive mammalian autophagy process more effectively than through single binding modes. The templates of horse and carriage both come from “https://all-free-download.Com/”. WBS, WIPI2-binding site.
In our recently published study [1], we systemically characterized the interaction between ATG16L1 and WIPI2. We firstly conducted quantitative isothermal titration calorimetry (ITC)-based analyses with purified relevant ATG16L1 fragments and the core WD40 domain of WIPI2 (hereafter WIPI2Δ), and revealed that ATG16L1 has two distinct WIPI2-binding sites, i.e., the previously reported WIPI2-binding site 1 (WBS1) and a newly identified site, WBS2, which localizes within the CCD region of ATG16L1. We subsequently elucidated the detailed binding mechanism and uncovered the key binding determinants for the interaction of WIPI2 with ATG16L1 by solving the two high-resolution crystal structures of WIPI2Δ in complex with ATG16L1 WBS1 and WBS2, respectively. Based on the two structures together with relevant biochemical analyses using WIPI2Δ, we revealed that ATG16L1 WBS1 and WBS2 are competitive in binding to the same WIPI2 molecule, but they can simultaneously bind to different WIPI2 molecules.
Strikingly, our detailed structure and sequence alignment analyses showed that WBS1 of ATG16L1 is only present in mammals, whereas ATG16L1 WBS2 and its binding mode with WIPI2 are well conserved within both yeast and mammals, which implies the potential significance of ATG16L1 WBS2 in autophagy. To uncover the functional relevance of ATG16L1 WBS1 and WBS2, we generated an atg16l1−/− knockout HeLa cell line, and back-transfected with plasmids to stably express the wild-type ATG16L1 or distinct ATG16L1 mutants, such as the D164R mutant, which disrupts WBS2, the L224Q mutant, which abolishes WBS1, or the DRLQ mutant, which simultaneously disrupts both WBS1 and WBS2. By evaluating the extent of SQSTM1/p62 degradation and LC3B lipid modification levels, we demonstrated that ATG16L1 WBS2 is indispensable for the normal progression of bulk autophagy, while ATG16L1 WBS1 facilitates further the autophagic flux.
Interestingly, although the dimeric ATG16L1(124–247) fragment, which contains intact WBS1 and WBS2 of ATG16L1, forms a stable 2:2 stoichiometric complex with WIPI2 in solution, our structural modeling analysis showed that the dimeric ATG16L1 fragment can theoretically bind four WIPI2 molecules through its WBS1 and WBS2. Compared with the previously proposed single-binding-site mode, the dual-binding-site mode of ATG16L1/WIPI2 interaction uncovered in our study provides a more efficient way to recruit the ATG12–ATG5-ATG16L1 complex to phagophore membrane decorated with WIPI2, thereby effectively driving the autophagy process (Figure 1B).
All in all, through systematically biochemical, structural and cell-based functional studies, we uncovered the molecular mechanism by which ATG16L1 adopts a dual-binding-site mode to interact with WIPI2 during mammalian autophagy.
Funding Statement
The work was supported by grants from NSFC (92253301, 32071219), STCSM (20×D1425200), and CAS (JCTD-2022-10).
Disclosure statement
No potential conflict of interest was reported by the authors.
Reference
- [1].Gong X, Wang Y, Tang Y, et al. ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy. Sci Adv. 2023;9:eadf0824. [DOI] [PMC free article] [PubMed] [Google Scholar]


