Abstract
Autocrine signaling pathways regulated by RAPID ALKALINIZATION FACTORs (RALFs) control cell wall integrity during pollen tube germination and growth in Arabidopsis (Arabidopsis thaliana). To investigate the role of pollen-specific RALFs in another plant species, we combined gene expression data with phylogenetic and biochemical studies to identify candidate orthologs in maize (Zea mays). We show that Clade IB ZmRALF2/3 mutations, but not Clade III ZmRALF1/5 mutations, cause cell wall instability in the sub-apical region of the growing pollen tube. ZmRALF2/3 are mainly located in the cell wall and are partially able to complement the pollen germination defect of their Arabidopsis orthologs AtRALF4/19. Mutations in ZmRALF2/3 compromise pectin distribution patterns leading to altered cell wall organization and thickness culminating in pollen tube burst. Clade IB, but not Clade III ZmRALFs, strongly interact as ligands with the pollen-specific Catharanthus roseus RLK1-like (CrRLK1L) receptor kinases Z. mays FERONIA-like (ZmFERL) 4/7/9, LORELEI-like glycosylphosphatidylinositol-anchor (LLG) proteins Z. mays LLG 1 and 2 (ZmLLG1/2), and Z. mays pollen extension-like (PEX) cell wall proteins ZmPEX2/4. Notably, ZmFERL4 outcompetes ZmLLG2 and ZmPEX2 outcompetes ZmFERL4 for ZmRALF2 binding. Based on these data, we suggest that Clade IB RALFs act in a dual role as cell wall components and extracellular sensors to regulate cell wall integrity and thickness during pollen tube growth in maize and probably other plants.
Pollen-specific RAPID ALKALINIZATION FACTORs interact with receptor kinases, GPI-anchored proteins, and cell wall proteins to regulate cell wall integrity during pollen tube growth in maize.
IN A NUTSHELL.
Background: As sperm cell transporting vehicles, pollen tubes of flowering plants grow through the transmitting tissues of the style to deliver immobile sperm cells for double fertilization and seed production. In maize (Zea mays), pollen tubes grow up to 1 cm/h through elongated styles (silks) before their sperm cell cargo is released inside ovules. In the model plant Arabidopsis (Arabidopsis thaliana), it has been shown that pollen tube secreted RAPID ALKALINIZATION FACTOR (RALF) proteins interact with receptor complexes and cell wall proteins to regulate pollen tube integrity during pollen germination and tube growth. Whether the identified mechanisms exist in other flowering plants was not known.
Question: Are the mechanisms that regulate pollen tube integrity evolutionarily conserved among flowering plants? We addressed this question by using maize, as a grass and crop species that has sexual organs with very different morphology from those of Arabidopsis.
Findings: Based on phylogenetic analysis, RALF proteins from maize and Arabidopsis can be classified into different clades. We show that maize Clade IB ZmRALFs are mainly located in the pollen tube cell wall and are required for proper pollen tube growth. ZmRALFs from Clade IB, but not ZmRALFs from Clade III, strongly interact as ligands with cell surface receptor-like kinases, co-receptors, and cell wall proteins from both maize and Arabidopsis. The strongest binding affinity was observed with LRX/PEX cell wall proteins. Misregulation of Clade IB RALFs affected pectin patterning of the pollen tube cell wall. In summary, our findings indicate that Clade IB RALF signaling mechanisms are partly conserved between both species and that RALF proteins play a dual role as cell wall components and extracellular sensors in regulating cell wall integrity.
Next steps: It is now important to identify and study components of RALF downstream signaling and to establish the dual role of RALF interaction/competition between cell wall proteins and cell surface receptors. Moreover, the function of pollen tube-specific RALFs of Clade III that are not located in the cell wall remains to be elucidated to better unravel the multiple roles of this protein family during pollen tube growth.
Introduction
Flowering plants (angiosperms) have evolved pollen tubes that carry their passive and immobile sperm cell cargo deep into the maternal reproductive tissues toward the embryo sac for double fertilization (Dresselhaus et al. 2016). During their journey from the stigma through the transmitting tract of the style toward ovules, pollen tubes secrete many small proteins for communication with various maternal tissues (Bircheneder and Dresselhaus 2016; Ge et al. 2019a; Johnson et al. 2019; Kim et al. 2021; Ogawa and Kessler 2023). These include RAPID ALKALINIZATION FACTORs (RALFs) that regulate hydration as well as cell wall integrity during pollen germination, tube growth, and reception in the model plant Arabidopsis (Arabidopsis thaliana) (Ge et al. 2017; Mecchia et al. 2017; Liu et al. 2021; Gao et al. 2022; Zhong et al. 2022). First discovered in tobacco (Nicotiana tabacum) leaves (Pearce et al. 2001), RALF genes were found to be universally distributed among land plants with 1 to 3 family members in mosses and up to 37 members in eudicots like Arabidopsis (Campbell and Turner 2017; Abarca et al. 2021). Functional studies have been mainly performed in Arabidopsis: it was shown, for example, that AtRALF1 overexpression or application of the peptide inhibited rapid root growth (Matos et al. 2008; Li et al. 2022), while downregulation resulted in increased root length (Bergonci et al. 2014). AtRALF34 was reported as part of a signaling network that regulates lateral root initiation (Gonneau et al. 2018). Leaf-expressed AtRALF23 inhibits immune responses (Stegmann et al. 2017; Gronnier et al. 2022) and induces stomatal closure (Yu et al. 2018). RALFs were also reported to play important roles in reproduction: initially, it was shown in tomato (Solanum lycopersicum) that in vitro application of pollen-specific SlPRALF inhibited pollen germination and that SlPRALF interacts with pollen-specific leucine-rich repeat (LRR)/extensin-like chimaera proteins (LRXs) (Covey et al. 2010). In Arabidopsis, it was later shown that AtRALF23/33 regulates the hydration of pollen grains at the stigma (Liu et al. 2021), while antagonistic AtRALF1/22/23/33 generated by the stigma (sRALFs) and compatible pollen-derived AtRALF10/11/12/13/25/26/30 (pRALFs) regulate pollen tube penetration at the stigma (Lan et al. 2023). AtRALF4/19 redundantly regulates pollen germination and tube growth. It was further shown that this regulation occurs via binding to LRX cell wall proteins (Mecchia et al. 2017; Wang et al. 2018), as well as via an autocrine signaling pathway by binding to pollen-specific Catharanthus roseus receptor-like kinase 1-like (CrRLK1Ls) family members ANXUR 1 (ANX1), ANX2, BUDDA's PAPER SEAL 1 (BUPS1), and BUPS2 (Ge et al. 2017). It was further reported that glycosylphosphatidylinositol (GPI)-anchor proteins LLG2 and LLG3 serve as co-receptors in BUPS/ANX-RALF signaling to regulate integrity during pollen tube germination and growth in Arabidopsis (Feng et al. 2019; Ge et al. 2019c). AtRALF6/7/16/36/37 were shown to establish a polytube block and to control pollen tube reception and rupture inside targeted embryo sacs via CrRLK1L family members FERONIA 1 (FER1), ANJEA (ANJ), and HERCULES RECEPTOR KINASE 1 (HERK1) (Zhong et al. 2022). Notably, another report suggests that AtRALF4/19 act via the FER-LRE module to induce pollen tube rupture and sperm release (Gao et al. 2022).
Structural studies elucidated the interactions using AtRALF23 as an example that binds to a surface groove of LLG2 with a conserved N-terminal recognition motif and thereby mediates interaction with FER forming a heterotrimeric complex (Xiao et al. 2019). Crystal structures of LRX2 and LRX8 binding with AtRALF4 revealed a dimeric arrangement of LRX proteins of which each monomer directly interacts with its N-terminal LRR domain to one RALF peptide (Moussu et al. 2020).
Compared with Arabidopsis, little is known about RALF-LLG-CrRLK1L and RALF-LRX signaling pathways in other plant species. Only the LRX proteins PEX1/2 (Rubinstein et al. 1995a, b) that were reported to affect pollen tube growth in maize (Zea mays), and OsRALF17 and OsRALF19, the rice (Oryza sativa) homologs of Arabidopsis RALF4/19, were shown to be involved in pollen hydration, germination, and pollen tube elongation (Kim et al. 2023). Except for these two reports, almost nothing is known to our knowledge about other components of these pathways during reproduction in grasses like maize, a species that probably generates the fastest-growing pollen tubes (Barnabas and Fridvalszky 1984). Pollen hydration and germination occur in maize within 5 min after landing on papilla silk hair structures (Bedinger 1992), before pollen tubes penetrate the silk and enter the transmitting tract where they grow up to 30 cm at a speed of more than 10 mm/h toward the surface of the ovule. They are further guided toward the micropylar region of the ovule, grow inside, and release their sperm cell cargo into the receptive synergid cell (Zhou et al. 2017). It was speculated that maize pollen tubes secrete various small proteins along their journey to trigger, for example, papilla hair cell recognition and penetration, nutrient release and cell wall softening during transmitting tract growth, ovule penetration as well as synergid cell death, and pollen tube rupture (Dresselhaus et al. 2011).
To study the role of pollen-specific RALF proteins in maize, we searched the genome of the inbred line B73 for members of the RALF gene family and combined expression and phylogenetic studies with biochemical interaction studies to elucidate similarities and differences to the above-reported findings in Arabidopsis. Three RALF clades were identified. Pollen-expressed Clade IB and Clade III ZmRALFs were further compared throughout this report. ZmRALFs levels were downregulated by RNAi to investigate milder phenotypes and full knockouts were developed using CRISPR/Cas9. Pollen germination and tube growth, as well as cell wall composition and thickness, were analyzed in the mutants. Binding affinities were compared between ZmRALF interaction partners, and competition studies were included to investigate the consequences of ZmRALF signaling dynamics and competition for the mobile ligand.
Results
Pollen-expressed maize RALFs belong to Clade IB or Clade III
RNA-seq data were generated from pollen grains, pollen tubes, and other reproductive and vegetative tissues of the maize inbred line B73 and deposited in the CoNekT online database (https://evorepro.sbs.ntu.edu.sg). Among the most strongly and specifically expressed genes in pollen grains and pollen tubes, we identified three RALF genes with transcript per million (TPM) expression values >7,000 (Supplementary Fig. S1). Sequences of these 3 genes, named ZmRALF1/2/3, were used as a query for various genome-wide BLAST searches to identify all members of the RALF gene family in maize. In total, 24 RALF-like genes were identified of which 9 are expressed in pollen grains and tubes. ZmRALF1/2/3/5 each shows a strong (>2,500 TPM) and pollen-specific expression pattern, while ZmRALF4/6/7/8 each shows moderate (50 to 400 TPM) expression levels. ZmRALF9 is lowly expressed in pollen but strongly expressed in roots. Other members like ZmRALF12 are broadly expressed in vegetative tissues like the stigma, root, leaf, and seed (Supplementary Fig. S1).
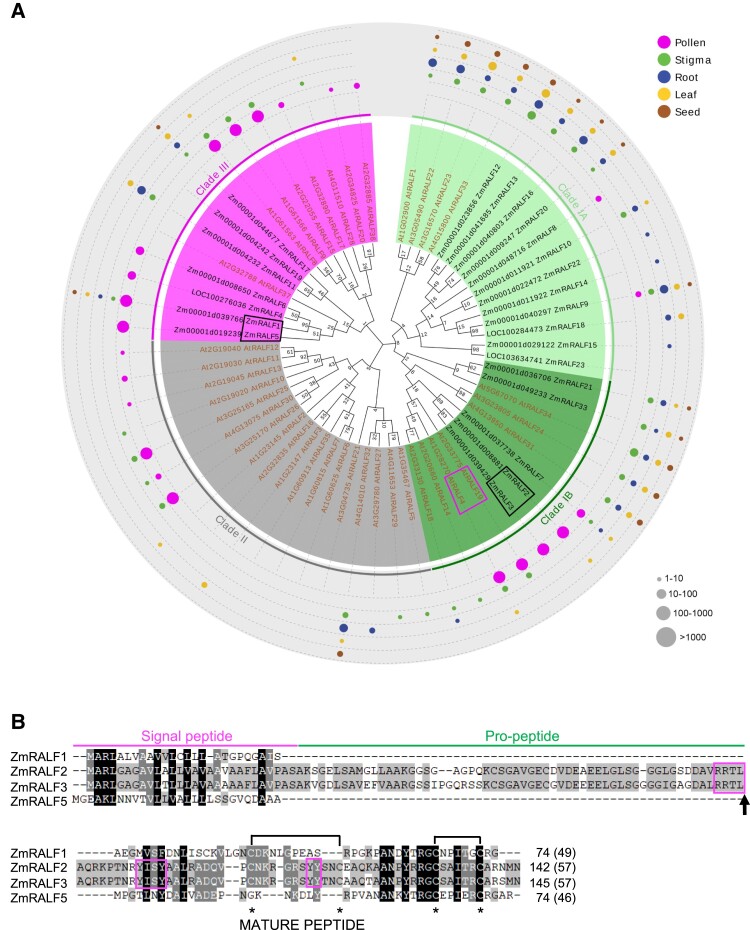
The RALF family in Arabidopsis is expanded and contains 37 members (Campbell and Turner 2017; Abarca et al. 2021). Phylogenetic analysis was done by using and comparing the 24 maize and 37 Arabidopsis RALFs. The generated tree was combined with expression data from 5 tissues including pollen (Fig. 1A). Predicted signal peptides and pro-peptides were removed and only mature protein sequences were used for comparison. Seventeen out of 24 maize RALFs belong to Clade IA and Clade IB, while the remaining 7 maize RALFs belong to Clade III. While Clade IA contains mainly vegetatively expressed RALFs from both species, Clade IB contains pollen-expressed AtRALF4/19 and ZmRALF2/3 representing their closest maize homologs that are also strongly expressed in pollen. Another maize homolog of AtRALF4/19 from Clade IB, ZmRALF7, only shows moderate expression in pollen (Fig. 1A). Clade II contains only Arabidopsis RALFs, of which a few are expressed in pollen, while Clade III contains members from both plant species, of which several genes show expression in pollen, with ZmRALF1/5 belonging to this clade. The structural differences between the mature RALF proteins are visualized in Supplementary Fig. S2. Clade IA and Clade IB RALFs contain typical RALF structures (Campbell and Turner 2017) including YISY and YY interaction domains and 4 conserved cysteines. Clade IA RALFs show strong N-terminal variation, while Clade IB RALFs are less conserved at their C-termini. The YISY box, but not the YY motif, is still recognizable in Clade II RALFs. Both motifs disappeared in Clade III RALFs together with the first 2 conserved cysteines.
Figure 1.
Phylogenetic and expression analysis of maize and Arabidopsis RALF protein families. A) Phylogram using predicted mature maize (24 genes) and Arabidopsis (37 genes) peptide sequences. Arabidopsis AtRALFs are indicated in brown and maize ZmRALFs in black. RALFs being investigated in more detail in this study are highlighted using rectangles. Clades IA and Clade IB indicate RALFs with conserved RXXL/RXLX S1P protease cleavage sites and YISY as well as YY domains, which are lacking in Clade II and Clade III RALFs. Colored dots show expression patterns of RALF genes in the 5 selected tissues indicated. Dot size correlates to expression level determined by TPM values as indicated. See Supplementary Fig. S1 for details about expression values and Supplementary Fig. S2 for amino acid conservation in Clades I to III RALFs. The values at the nodes represent percent of bootstrap confidence level. B) Amino acid alignment of ZmRALF1/2/3/5 proteins. RRTL S1P protease cleavage site (arrow) and YISY as well as YY interaction domains of Clade I RALFs are boxed in magenta. Cysteine residues are marked with asterisks. Intramolecular disulfide bridges are indicated. Length of predicted mature peptides is indicated in brackets behind full length amino acid numbers.
Protein sequence alignment of the 4 most strongly expressed RALFs from maize pollen shows that ZmRALF2/3 contains typical RALF structures (Fig. 1B; Supplementary Fig. S2). These include a RXXL/RXLX (X represents any amino acid) motif for cleavage by SITE-1 PROTEASE (S1P) to remove the pro-peptide and to generate the mature peptide (Matos et al. 2008; Stegmann et al. 2017), 4 conserved cysteine residues to fold a proper structure, a YISY motif as part of an N-terminal α-helix for interaction with LLG co-receptors as well as the YY double tyrosine amino acid motif and partly conserved C-terminal region for likely interaction with CrRLK1L receptor kinases (Xiao et al. 2019). Based on structural studies using their close homolog AtRALF4 (Moussu et al. 2020), it is further likely that ZmRALF2/3 can interact with LRX/PEX cell wall proteins. In contrast, ZmRALF1, which is encoded by the most highly expressed RALF gene in maize pollen, and ZmRALF5 lacks those motifs, suggesting that these RALFs may have different functions during maize pollen germination and growth compared with RALFs containing S1P, YISY, and YY motifs, respectively. Such genes were previously identified as RALF-like genes (Campbell and Turner 2017), although they still fit the model of the RALF family (Silverstein et al. 2007). Thus, we also name them ZmRALFs throughout the article.
Downregulation of pollen-specific RALFs leads to pollen tube burst and male gamete transmission reduction
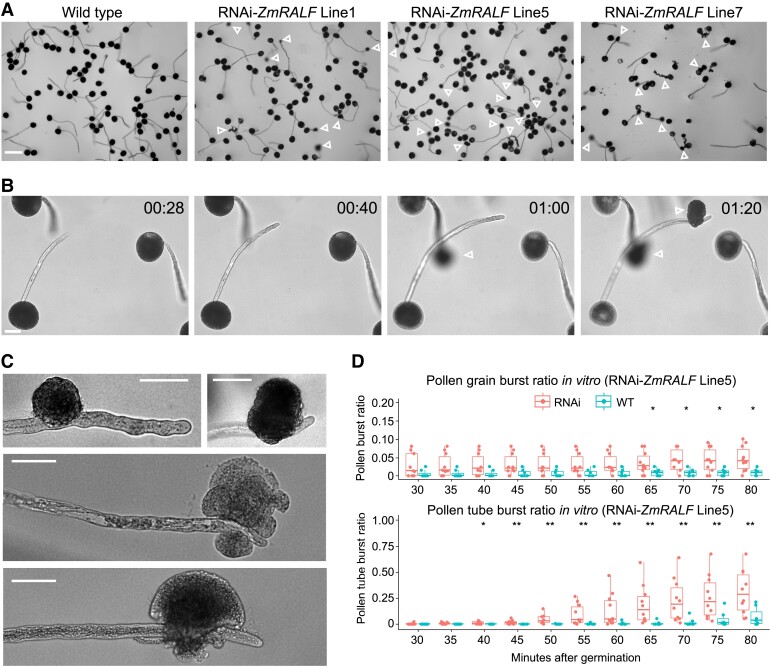
To investigate the function of pollen-specific RALFs in maize, an RNAi-ZmRALFs gene silencing construct was generated based on ZmRALF1/2/3-specific sequences (see Materials and methods for details) and transformed into maize. As shown in Supplementary Fig. S3, ZmRALF1/2/3 were downregulated to different levels in independent RNAi-ZmRALFs mutant lines. Line1, Line5, and Line7 showed a reduction of transcript levels to about 50% compared with that of corresponding wild-type (WT) plants and were thus selected for further phenotypic analyses. Pollen germination and growth were compared in vitro between RNAi-ZmRALFs and regenerated plants lacking the transgene (used as a WT control). On average, the pollen germination ratio was very similar in RNAi-ZmRALF lines (65.71%) and WT plants (65.68%). Following pollen tube growth, pollen tubes from RNAi-ZmRALF mutant lines started to burst, while a burst of WT pollen tubes was initially rare (Fig. 2A). A detailed analysis showed that bursts of pollen tubes occurred exclusively at the sub-apical region but never at the very tip (Fig. 2, B and C). Some pollen tubes even continued their growth despite bursts and loss of substantial cytoplasmic contents (Fig. 2C). At 80 min after germination in vitro, more than 30% of pollen tubes from RNAi-ZmRALF lines had burst, while only 7.2% of WT pollen tubes had burst under identical conditions (Fig. 2D).
Figure 2.
Pollen tubes of RNAi-ZmRALF mutant maize lines burst in vitro. A) Germination and growth of pollen from WT and 3 RNAi-ZmRALF lines after 1 h. Open white arrowheads point toward burst pollen tubes. B) Time-lapse imaging of pollen from RNAi-ZmRALF mutant Line5 at indicated time points. One pollen tube burst at 60 and the second one after 80 min. Open white arrowheads point toward burst pollen tubes. C) Examples of burst pollen tubes. Note that all pollen tubes burst at the shank but never at the apex of the tube. D) Burst ratio of pollen grains and growing pollen tubes from RNAi-ZmRALF mutant Line5 was determined over time. Un-germinated pollen grains (top) and germinated tubes (bottom) were recorded. Whiskers extend to min and max, box boundaries represent the 25th percentile (lower) and 75th percentile (upper), the lines inside boxes represent medians, and dots represent the individual data for each documented pollen germination image. For each pollen germination time point, >200 pollen grains were calculated. Asterisks indicate a significant difference between RNAi-ZmRALF line and the WT (Student’s t-test; *P < 0.05, **P < 0.01). Scale bars are 200 μm in A) and 50 μm in B and C).
To further explore whether the reduction of ZmRALF expression levels in mutant pollen has an effect in vivo, we determined the male and female transmission efficiency of RNAi-ZmRALF mutant lines (Table 1). RNAi-ZmRALF mutant lines were either crossed as the maternal or paternal parent with WT plants (inbred line B73). Because transgenic lines often contain multiple integrations, we calculated an expected segregation ratio of ≥1:1 when RNAi-ZmRALF lines were crossed with WT plants and ≥3:1 in self-crosses of RNAi-ZmRALF mutant lines. As shown in Table 1, when Line1 and Line5 were used as the pollen donor, transmission efficiencies of 0.72 and 0.24, respectively, were obtained. This is much lower than the expected number of ≥1. Similarly, transmission efficiencies of 0.69 and 0.44, respectively, were observed during self-crossings, which are much lower than the expected number of ≥3. In contrast, when we used the same lines as the female parent, transmission efficiencies of 0.94 and 1.0, respectively, were obtained, which are very close to the expected number of ≥1. These genetic studies demonstrate that the reduction of ZmRALF levels in pollen affects male, but not female gamete transmission.
Table 1.
Transmission efficiency of RNAi-ZmRALFs mutant maize lines
| RNAi-RALF mutant | Female | × | Male | Numbers of plants | Segregation expected | Segregation observed | ||
|---|---|---|---|---|---|---|---|---|
| RNAi | WT | total | ||||||
| Line1 | RNAi-ZmRALFs | × | RNAi-ZmRALFs | 59 | 85 | 144 | ≥3:1 | 0.69:1* |
| Line1 | RNAi-ZmRALFs | × | B73 | 31 | 33 | 64 | ≥1:1 | 0.94:1 |
| Line1 | B73 | × | RNAi-ZmRALFs | 33 | 46 | 79 | ≥1:1 | 0.72:1* |
| Line5 | RNAi-ZmRALFs | × | RNAi-ZmRALFs | 54 | 123 | 177 | ≥3:1 | 0.44:1* |
| Line5 | RNAi-ZmRALFs | × | B73 | 39 | 39 | 78 | ≥1:1 | 1:1 |
| Line5 | B73 | × | RNAi-ZmRALFs | 19 | 79 | 98 | ≥1:1 | 0.24:1* |
Mutant lines were crossed with WT (inbred lines B73) plants as indicated. A transmission efficiency of ≥1:1 and ≥3:1, respectively, is expected as lines may contain multiple transgene integrations. RNAi transgenes are normally transmitted (100%) via the female gametophyte, but <20% by the male gametophyte (marked by asterisks).
Only Clade IB ZmRALFs locate to the cell wall in vitro
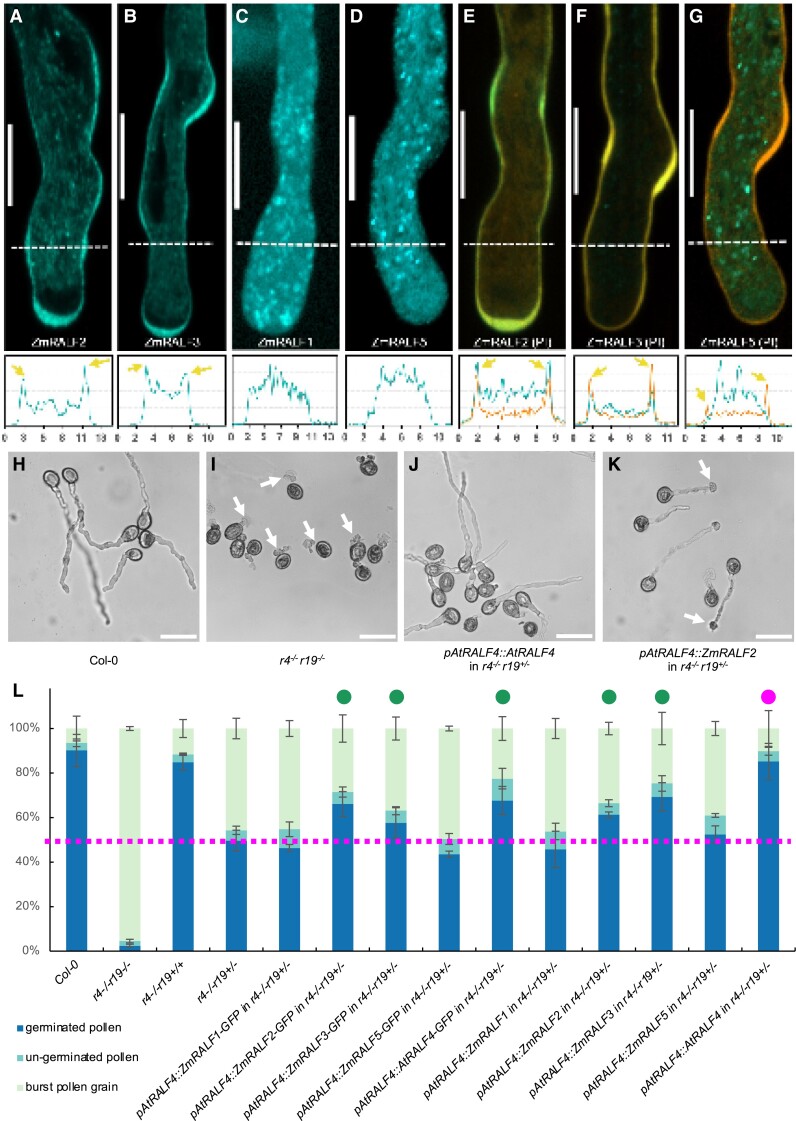
Previous reports showed that GFP-tagged NtRALF was detected first in the ER and later in the cell wall of epidermal cells (Escobar et al. 2003), which is similar to pollen-expressed AtRALF4/19 that were mainly observed in the apoplast (Ge et al. 2017) and cell wall (Mecchia et al. 2017). Thus, to explore the subcellular localization of Clade IB and Clade III ZmRALFs, transgenic Arabidopsis plants expressing the 4 ZmRALFs used in this study were generated. Around 2 kb of genomic sequence upstream of the ORF of AtRALF4 was taken as a promoter and eGFP was cloned C-terminally to ZmRALF-ORFs. Arabidopsis pollen tubes from at least 3 independent transgenic lines were observed by using propidium iodide (PI) for the counter staining of the cell wall. As shown in Fig. 3, A to G, ZmRALF2/3-eGFP-expressing pollen tubes display a slight overexpression phenotype indicated by their bumpy growth behavior. Their eGFP fusion proteins are visible in the ER, in small cytoplasmic vesicles, and finally accumulate at the pollen tube tip and the cell wall showing co-localization with PI. Thick cell wall regions accumulate larger quantities of ZmRALF2/3-eGFP during transient growth retardation. In contrast, ZmRALF1/5-eGFP could be observed in the ER and larger cytoplasmic vesicles or granules, but never in the very tip of the pollen tube or cell wall, indicating that they are not secreted to the apoplast in vitro.
Figure 3.
Localization and complementation studies of maize Clade IB and Clade III RALFs in Arabidopsis pollen tubes. A to G) Around 2 kb upstream of the AtRALF4 gene was used as a promoter to express maize RALF proteins fused C-terminally to eGFP in Arabidopsis pollen tubes. Top row shows images of indicated ZmRALFs with and without PI counter staining, respectively. Bottom row shows correlated relative green and/or red fluorescence intensity each 10 μm from the pollen tube apex (illustrated by white dashed lines). The x-axis values represent the distance in μm from the first dash at the left along the dashed line and the y-axis shows relative fluorescence intensity. Arrowheads indicate fluorescence signal enrichment in the cell wall. Scale bars are 10 μm. H) In vitro growth of WT pollen tubes. I) Burst pollen tubes of AtRALF4/19 double homozygous mutants (r4−/− r19−/−) immediately after germination. Arrows point toward burst pollen tubes. J)AtRALF4 can fully restore in vitro germination and growth of r4−/− r19+/− mutant pollen. Scale bars are 50 μm in H to K). K) Pollen tubes from a partially complemented ZmRALF2 transgenic plant showed partial rescue of germination and a late bursting phenotype in the r4−/− r19+/− mutant background. Arrows point toward burst pollen tubes. L) Quantification of in vitro pollen germination and burst ratio in transgenic lines indicated. Magenta dotted line indicates germination ratio of 50%. Green dots represent transgenic lines that partially complemented the mutant r4−/− r19+/− phenotype. The magenta dot indicates AtRALF4 that fully rescued the pollen grain burst phenotype of the AtRALF4/19 double mutant. The data are presented as the mean value ± Sds, n > 200.
Clade IB ZmRALFs partially complement Arabidopsis ralf4/19 mutants
Although ZmRALF2/3 belong to the same clade as AtRALF4/19, it was unclear whether they are able to functionally complement each other in planta. Therefore, the above RALFs from maize and AtRALF4 from Arabidopsis as a control were expressed with and without eGFP fusions in the ralf4/19 mutant background. As shown in Fig. 3H, WT Col-0 pollen germinated well in vitro on a solid germination medium with a germination ratio of around 90% and only a small portion of pollen grains burst (Fig. 3L). As reported before (Ge et al. 2017; Mecchia et al. 2017), almost all pollen grains of the double homozygous ralf4/19 mutant burst immediately after germination (Fig. 3, I and L). Due to gene redundancy, the pollen germination of single mutants behaved similar to that of the WT (Fig. 3L), and around half of the pollen grains from the heterozygous ralf4−/−19+/− mutant burst, while the other half germinated normally (Fig. 3L). Full complementation was observed using AtRALF4 (Fig. 3, J and L), while the addition of eGFP reduced the complementation ratio (Fig. 3L). ZmRALF2/3 can partially restore the germination burst phenotype, but pollen tube burst was observed later during further growth (Fig. 3K). The eGFP-tag had no influence on ZmRALF2/3 complementation of immediate pollen tube burst (Fig. 3L). In contrast, Clade III ZmRALF1/5 could not rescue the pollen germination burst phenotype (Fig. 3L). In conclusion, although ZmRALF2/3 and AtRALF4/19 are the closest homologs (Fig. 1A), ZmRALF2/3 can only partially complement the ralf4/19 mutant. Together, with the above, findings showing that ZmRALF mutants display a pollen tube growth but not a pollen germination defect in maize, these data indicate functional diversification of pollen-specific Clade IB RALF genes in different plant species.
Pollen tube integrity is regulated by ZmRALF2/3
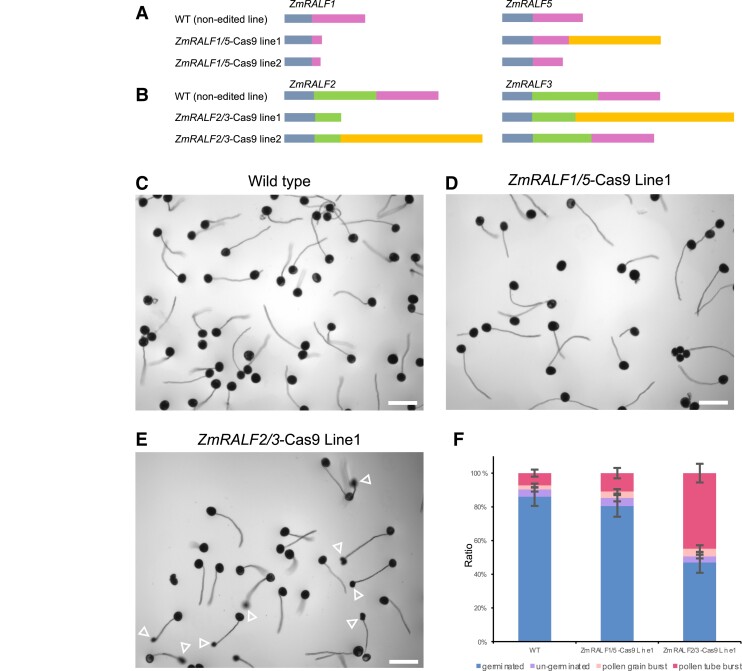
Based on phylogenetic analyses, subcellular localization patterns and complementation results, we assumed that Clade IB and Clade III ZmRALFs possess distinct functions in pollen germination and pollen tube growth. To investigate the function of the 2 clades independently, CRISPR-Cas9 editing mutants were generated. After maize transformation, crossing and genome editing pattern analysis, Cas9-free homozygous mutant lines were used for in vitro pollen germination tests. As shown in Fig. 4, A and B, 2 independent transgenic events from each transformation with different genome editing patterns were selected for further phenotypic analysis. In ZmRALF1/5-Cas9 line1, a 1 bp deletion in ZmRALF1 caused a premature stop codon generating a truncated peptide of 34 aa (instead of 74 aa still including a signal peptide of 25 aa). A 2 bp insertion in ZmRALF5 caused a frame shift mutation generating a non-sense peptide sequence of 146 aa instead of 74 aa (including a signal peptide of 28 aa). In ZmRALF1/5-Cas9 line2, a 2 bp deletion in ZmRALF1 caused a premature stop codon generating a truncated peptide of 34 aa, and a 1 bp insertion in ZmRALF5 generated a shortened peptide of 69 aa. In ZmRALF2/3-Cas9 line1, a 1 bp insertion in ZmRALF2 caused a premature stop codon generating a truncated peptide of 52 aa (instead of 142 aa) excluding the predicted mature peptide sequence and a 1 bp insertion in ZmRALF3 caused a frameshift mutation generating a 203 aa non-sense peptide lacking the predicted mature peptide sequence. In ZmRALF2/3-Cas9 line2, a 1 bp insertion in ZmRALF2 caused a frame shift mutation generating a 183 aa non-sense peptide lacking the predicted mature peptide sequence. A 21 bp deletion in ZmRALF3 caused the lack of the 8 aa SSIPGQRS sequence in the pro-peptide that was replaced by G. Consequently, the mature peptide sequence was kept leading to a single ZmRALF2 mutation.
Figure 4.
Editing pattern and phenotypic analysis of ZmRALF1/5-Cas9 and ZmRALF2/3-Cas9 mutant lines. A) Genome editing pattern of ZmRALF1 and ZmRALF5 in mutant lines indicated. Blue-gray boxes show N-terminal signal peptides and pink boxes predicted mature peptides. Orange box indicates nonsense sequence. B) Genome editing pattern of ZmRALF2 and ZmRALF3 in mutant lines indicated. Color code of boxes as in A). Green box additionally shows the cleaved pro-peptide sequence (see also Fig. 1B; Supplementary Fig. S2A). C to E) In vitro pollen germination of WT (non-edited transgenic line) in C), ZmRALF1/5-Cas9 Line1 D), and ZmRALF2/3-Cas9 Line1 E), respectively. Pollen grains were germinated on PGM for 60 min. Arrowheads in E) point toward burst pollen tubes. Scale bars are 200 μm. F) Quantification of pollen burst ratios in WT (non-edited transgenic line), ZmRALF1/5-Cas9 Line1 and ZmRALF2/3-Cas9 Line1, respectively. n > 200 in every plant from each line. The data are presented as the mean value ± Sds from 3 independent pollen germination experiments.
During in vitro pollen germination tests, ZmRALF1/5-Cas9 lines behaved similarly to WT plants, while pollen tubes from ZmRALF2/3-Cas9 lines burst during growth (Fig. 4, C to F), which is very similar to ZmRALFs-RNAi lines reported above. This result clearly indicates that during in vitro pollen tube growth, ZmRALF2/3 from Clade IB contribute to the regulation of pollen tube integrity maintenance while ZmRALF1/5 from Clade III have no obvious impact suggesting that ZmRALFs from Clade IB and Clade III have distinct functions.
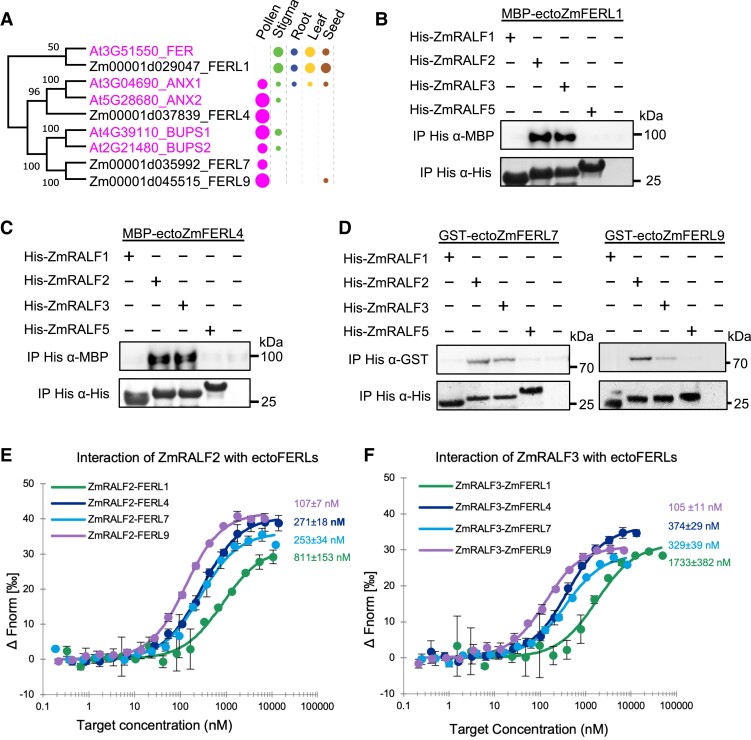
Pollen-specific CrRLK1Ls ZmFERL4/7/9 interact with Clade IB RALFs
In Arabidopsis, it was shown that AtRALFs interact with C. roseus receptor-like kinases (CrRLK1Ls). The most prominent member of the family, FER, which is not expressed in pollen, interacts with root- and stigma-expressed Clade IA AtRALF1/23/33 (Haruta et al. 2014; Liu et al. 2021) as well as pollen- and ovule-expressed Clade IB AtRALF4/19/34 (Gao et al. 2022). Further CrRLK1L members including THESEUS1 and pollen-specific ANX1/2 and BUPS1/2 were reported as Clade IB AtRALF4/19/34 receptors (Ge et al. 2017; Mecchia et al. 2017; Gonneau et al. 2018). Notably, a recent report showed that Clade II AtRALF6/7/16 and Clade III AtRALF36/37 act via FER, ANJ, and HERK1 receptor kinases (Zhong et al. 2022). The single CrRLK1L gene of the liverwort Marchantia polymorpha, MpFERONIA (MpFER), is required for cell expansion and maintenance of cellular integrity pointing toward an overall conserved function (Mecchia et al. 2022). To explore the CrRLK1L family of maize, the above proteins from Arabidopsis were used as a query in a BLAST search of the maize genome. We identified 17 CrRLK1L proteins in maize (Supplementary Fig. S4), which is identical to the number of CrRLK1L proteins in Arabidopsis. According to its founding member, we named all maize CrRLK1L proteins FER-Like (FERL). ZmFERL1/2/3 genes show a broad expression in vegetative tissues and likely encode the putative FER orthologs. Three ZmFERLs are strongly and specifically expressed in pollen. According to its sequence identity and expression pattern, ZmFERL4 appears to be the single putative ortholog of Arabidopsis ANX1/2 in pollen and pollen tubes, while ZmFERL7/9 showed the highest sequence similarity to BUPS1/2. It should be noted that the kinase domain of ZmFERL7 is shortened compared with the homologs from Arabidopsis (Supplementary Fig. S5) and it thus may not be functional. In summary, it seems that the CrRLK1L family is highly conserved in maize and Arabidopsis, and all Arabidopsis CrRLK1L members have close homologs in maize. However, MDS1-4 homologs are lacking in maize and ZmFERL16/17, in contrast to their closest homologs, are missing a kinase domain indicating variation and differences in CrRLK1L signaling in both species.
The phylogenetic relationship and expression of pollen-specific ZmFERLs are summarized in Fig. 5A. To investigate whether N-terminal-tagged ectodomains of pollen-specific ZmFERL4/7/9 as well as non-pollen expressed ZmFERL1 can interact with pollen-expressed Clade IB and Clade III ZmRALFs, we first purified recombinant proteins generated in Escherichia coli and assayed for their quality. As shown in Supplementary Fig. S6A, dynamic light scattering (DLS) analysis demonstrated the distribution of the hydrodynamic radius of purified proteins indicating that the purified protein solution contains highly homogeneous particles implying uniform and thus proper folding. In addition, nano-differential scanning fluorimetry (nanoDSF) assays revealed that recombinant ectoFERLs are folded and unfolded at around 50 °C indicating that they could be used for further studies. We also found that they do not aggregate with increasing temperature (Supplementary Fig. S6, B and C). Afterwards, pull-down assays were performed with C-terminal His-tagged mature ZmRALF1/2/3/5 peptides. As shown in Fig. 5, B to D, Clade IB ZmRALF2/3, but not Clade III ZmRALF1/5, significantly interact with all ZmFERLs being tested. To further uncover the preference of Clade IB ZmRALFs binding toward different ZmFERLs, microscale thermophoresis (MST) assays were performed. While ZmFERL1 showed the lowest binding affinity (Kd of 810 nm with ZmRALF2 and 1,733 nm with ZmRALF3), ZmFERL9 showed the strongest affinity (Kd of 104 to 107 nm with both ZmRALFs), while ZmFERL4/7 had slightly weaker interactions with both ZmRALFs with Kds between 253 and 374 nm (Fig. 5, E and F). In conclusion, the binding affinity between ZmRALF2/3 and pollen-specific ZmFERLs was much higher compared with that of the non-pollen expressed FER homolog ZmFERL1. Binding affinities of Clade III ZmRALF1 with ZmFERL7 and ZmFERL9 (about 3,530 and 1,900 nm, respectively) were substantially weaker compared with the affinity of RALF2 to ZmFERL7 and ZmFERL9 (615 and 545 nm, respectively) measured in parallel (Supplementary Fig. S7). Next, we tested whether ZmFERLs can also interact with Clade IB RALFs from Arabidopsis and vice versa. As shown in Supplementary Fig. S8, A and B, AtRALF4 interacted with all 4 maize ZmFERLs being tested, while AtRALF19 showed only a strong interaction with ZmFERL1. Similarly, ZmRALF2 interacted more strongly with ANX1/2 and BUPS1/2 compared with ZmRALF3, and Clade III ZmRALF1/5 did not interact with Arabidopsis CrRLK1Ls (Supplementary Fig. S8C). A very weak interaction was observed between ZmRALF1/5 and BUPS2. These findings indicate binding preferences between Clade IB RALFs and CrRLK1Ls from maize and Arabidopsis.
Figure 5.
Clade IB, but not Clade III pollen tube-expressed RALFs, interacts strongly with CrRLK1L receptor kinases. A) Phylogenetic analysis of ectodomains of selected Arabidopsis and maize FERONIA-like (FERL) receptor kinases. Expression pattern and relative transcript levels are indicated by dots (see Fig. 1A for explanations). Supplementary Figs. S4 and S5 list all maize and Arabidopsis CrRLK1Ls and shows detailed expression values and protein structures. The values at the nodes represent percent of bootstrap confidence level. B to D) Pull-down assays using MBP-tagged ectodomains of the FER homolog ZmFERL1 B), the ANX1/2 homolog ZmFERL4 C) as well as GST-tagged ectodomains of the BUPS1/2 homologs ZmFERL7/9 D) each in combination with His-tagged ZmRALF1/2/3/5 as indicated. Only ZmRALF2/3 interact significantly with CrRLK1Ls. Input images can be found in Supplementary Fig. S15. E and F) MST binding affinity assays using ZmRALF2/3 in combination with ZmFERL1/4/7/9. Kd values are indicated. Notably, the lowest binding affinity occurs with the FER homolog ZmFERL1, which is not expressed in pollen. See also Supplementary Fig. S7 for MST comparisons with Clade III ZmRALF1 showing very weak interaction. The data are presented as the mean value ± Sds from 3 technical replicates, and similar results can be detected with 3 independent experiments.
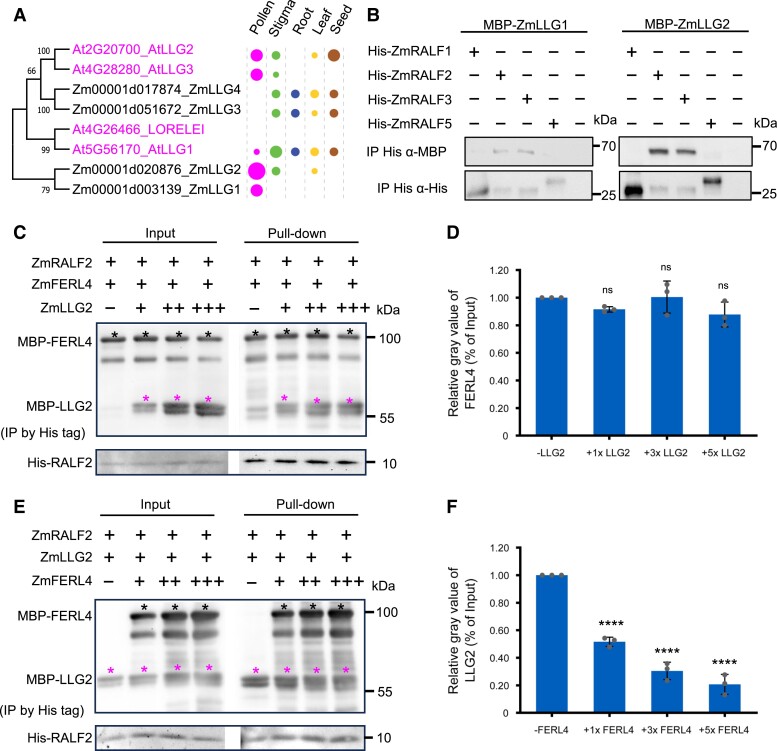
Clade IB RALFs interact with LORELEI-like GPI-anchored proteins
It was further shown in Arabidopsis that pollen expressed LORELEI-like GPI-anchored protein 2 (AtLLG2) and AtLLG3 act as co-receptors in RALF-CrRLK1L interaction complexes (Ge et al. 2019c). Structural analysis revealed that the N-terminal domain of mature Clade IB RALFs functions as a “glue” to generate a FER-RALF23-LLG2 receptor complex in Arabidopsis (Xiao et al. 2019). In maize, we identified 4 LORELEI-like GPI-anchored proteins named as ZmLLG1-4 (Supplementary Fig. S9). ZmLLG1/2 displays a pollen-specific expression pattern (Fig. 6A), which is similar to LLG2/3 in Arabidopsis (Ge et al. 2019c). Notably, ZmLLG3/4 expressed in vegetative tissues is slightly more similar to LLG2/3 compared with ZmLLG1/2. Both Clade IB ZmRALF2/3 interacted with ZmLLG1/2, while interaction with Clade III ZmRALF1/5 was not observed (Fig. 6B). Sequence alignment revealed conservation among LLGs from Arabidopsis and maize, respectively (Supplementary Fig. S9). The region covering 8 cysteines is highly conserved including an Asn-Asp (ND) domain and 2 glycines between 7th and 8th cysteines that were reported to represent the core responsible for RALF binding in Arabidopsis (Xiao et al. 2019). We thus assume that all maize ZmLLGs can also interact with ZmRALFs in a similar manner. Notably, ZmLLG1 lacks the C-terminal omega-side and GAS-domain for GPI-anchor addition indicating that it is likely not located at the surface of the plasma membrane. We thus suppose that ZmLLG2, which interacted stronger with Clade IB ZmRALFs compared with ZmLLG1, and whose gene shows also a stronger expression level, represents the major maize homolog of pollen-expressed Arabidopsis LLGs.
Figure 6.
Binding of Clade IB maize RALFs with LORELEI-like-GPI-anchored (LLG) co-receptors can be outcompeted by FERL receptors, but not vice versa. A) Phylogenetic tree of LLG proteins from maize and Arabidopsis. Expression pattern and relative transcript levels are indicated by dots (see Fig. 1A for explanations). The values at the nodes represent percent of bootstrap confidence level. B) Pull-down assay using MBP-tagged LLGs and His-tagged RALFs as indicated. Input images can be found in Supplementary Fig. S15. C) Competitive pull-down assays of RALF2 with FERL4 by LLG2. LLG2 at relative concentrations of 1×, 3×, and 5× were added to a solution containing RALF2 and FERL4. Black asterisks indicate protein bands of FERL4 and magenta asterisks indicate LLG2 protein bands. D) Quantification of relative gray values of FERL4 bands in pull-down assays relative to the input. Bar plot shows mean value and standard deviation. Experiments were repeated 3 times. Individual data points are indicated. ns, Not significant. Unpaired t-test, compared with the −LLG2 control. E) Competitive pull-down assays of RALF2 with LLG2 by FERL4. FERL4 at concentrations of 1×, 3×, and 5× was added to a solution containing RALF2 and LLG2. Black asterisks indicate protein bands of FERL4 and magenta asterisks indicate LLG2 protein bands. F) Quantification of relative gray values of LLG2 bands in pull-down relative to the input. Bar plot shows the mean value and standard deviation. Experiments were repeated 3 times. Individual data points are indicated. ****P < 0.0001. Unpaired t-test, compared with the −LLG2 control.
Considering that ZmLLGs may act as co-receptors of ZmRALF-FERL complexes, we examined the competitive binding of ZmLLG2 with ZmFERL4 for ZmRALF2. Results showed that ZmLLG2 could not compete for the binding of ZmFERL4 for ZmRALF2 (Fig. 6, C and D). However, ZmFERL4 outcompetes ZmLLG2 binding to ZmRALF2 (Fig. 6, E and F). This finding indicated that ZmLLGs may play an auxiliary role in the binding process of ZmFERL4 and ZmRALF2.
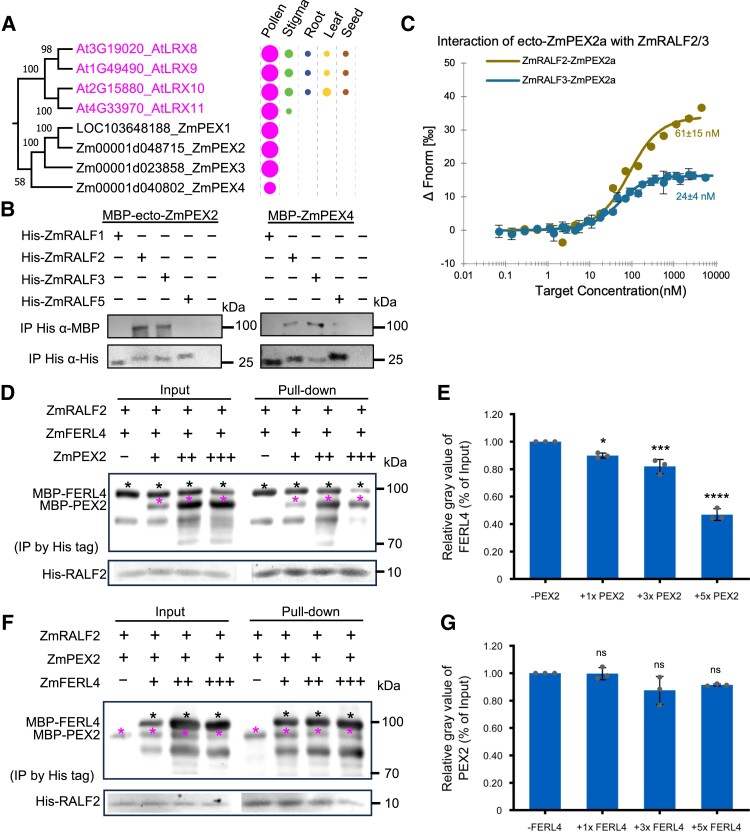
Clade IB ZmRALFs interact with PEX cell wall proteins
After elucidating that the ZmFERL4/7/9-ZmLLG1/2 receptor complex is likely involved in ZmRALF2/3-regulated pollen tube growth, we next aimed to study whether ZmRALFs can also interact with ZmPEX LRR-extensin-like proteins. Previous reports showed that the homologs of ZmPEX cell wall proteins (named LRX in Arabidopsis) directly interact with RALFs and sustain pollen germination and tube growth via the CrRLK1L pathway (Mecchia et al. 2017; Moussu et al. 2020). Until now, it was only known that in maize ZmPEXs localize toward the intine of mature pollen grains and the callosic sheath of pollen tubes (Rubinstein et al. 1995a, b). Their homologs in Arabidopsis were recently shown to interact with AtRALF4/19 and stabilize the pollen tube cell wall during germination by an unknown mechanism (Mecchia et al. 2017; Moussu et al. 2020). We used ZmPEX2 as a query and identified a total of 19 PEX/LRX genes in the maize genome and confirmed 11 LRX genes previously identified in Arabidopsis (Supplementary Fig. S10). This family is thus larger in maize compared with Arabidopsis. Similar to the 4 Arabidopsis homologs, AtLRX8-11, the 4 ZmPEX1-4 genes are specifically expressed in pollen (Fig. 7A) with ZmPEX3 belonging to the group of the most strongly expressed genes in pollen. In addition to their N-terminal LRR domain, PEX/LRX proteins contain a conserved cysteine-rich motif to enhance homodimer formation (Moussu et al. 2020) and a proline-rich extension domain with multiple SPPPP repeats that extend to other cell wall components for anchoring (Herger et al. 2019). A sequence alignment of the 8 PEX and LRX proteins expressed in pollen of maize and Arabidopsis, respectively, is shown in Supplementary Fig. S11. The N-terminal N-D domain, LRR domain, and 11 cysteines are highly conserved, while the proline-rich C-terminal region shows variation between SPPPP repeats. ZmPEX1-3 are slightly larger compared with their 4 Arabidopsis homologs. ZmPEX4 lacks the extensin-like domain and thus is strictly speaking not a PEX/LRX protein despite its high sequence identity of about 74% at the N-terminal part to the other PEX/LRX proteins.
Figure 7.
Cell wall-localized maize PEX proteins physically interact via their LRR-domain with Clade IB RALFs and outcompete RALFs from FERL receptor complexes. A) Phylogenetic analysis of pollen-specific chimeric LRR-extension-like proteins in maize and Arabidopsis. The signal peptide sequence and conserved proline-rich extensin domain were removed before alignment. Expression pattern and relative transcript levels are shown as explained in Fig. 1A. The complete phylogenetic tree and detailed expression data including all LRR-extension-like proteins in maize and Arabidopsis as well as a protein alignment are shown in Supplementary Figs. S10 and S11. The values at the nodes represent percent of bootstrap confidence level. B) Pull-down experiment of MBP-tagged N-terminal LRR domain containing region of ZmPEX2 (ZmPEX2a) and full length ZmPEX4. C) MST binding affinity assay showing that ZmRALF2/3 have strong binding affinity with ZmPEX2a: ZmRALF2, Kd = 60 nm; ZmRALF3, Kd = 24 nm. D) Competitive pull-down assays of RALF2 with FERL4 by PEX2. PEX2 at concentrations of 1×, 3×, and 5× were added to a solution containing RALF2 and FERL4. Black asterisks indicate protein bands of FERL4 and magenta asterisks indicate PEX2 protein bands. The data are presented as the mean value ± Sds from 3 technical replicates, and similar results can be detected with 3 independent experiments. E) Quantification of relative gray values of FERL4 bands in pull-down relative to the input. Bar plot shows mean value and standard deviation. Experiments were repeated 3 times. Individual data points are indicated. *P < 0.05, ***P < 0.001, ****P < 0.0001. Unpaired t-test, compared with the −PEX2 control. F) Competitive pull-down assays of RALF2 with PEX2 by FERL4. FERL4 at concentrations of 1×, 3×, and 5× were added to a solution containing RALF2 and PEX2. Black asterisks indicate protein bands of FERL4 and magenta asterisks indicate PEX2 protein bands. G) Quantification of relative gray values of PEX2 bands in pull-downs relative to the input. Bar plot shows mean value and standard deviation. Experiments were repeated 3 times. Individual data points are indicated. ns, Not significant. Unpaired t-test, compared with the −FERL4 control.
Purified recombinant PEX2 protein was generated in E. coli and assayed for its quality. As shown in Supplementary Fig. S6A, DLS analysis and nanoDSF assays revealed that recombinant PEX2 is folded and unfolds at around 50 °C. Aggregation was not observed by increasing the temperature (Supplementary Fig. S6, B and C). Afterwards, pull-down experiments were carried out with Clade IB and Clade III ZmRALFs with the N-terminal MBP-tagged LRR domain of ZmPEX2 and full-length ZmPEX4 (Fig. 7B). Only ZmRALF2/3 interacted with both ZmPEX proteins. MST measurements showed that ZmRALF2 binds to the LRR-domain of ZmPEX2 with a Kd of 60 nm, while the binding affinity between ZmRALF3 and ZmPEX2 is even higher with a Kd of 24 nm (Fig. 7C). Similar to the interaction with maize ZmPEX2 and ZmPEX4, Clade IB ZmRALFs can also interact with pollen-expressed AtLRX9-11 from Arabidopsis (Supplementary Fig. S12). Subsequent competitive binding analysis of ZmPEX2 with ZmFERL4 for ZmRALF2 showed that ZmPEX2 could outcompete ZmFERL4 for binding of the ligand ZmRALF2 (Fig. 7, D and E), while ZmFERL4 could not outcompete ZmPEX2 for ZmRALF2 binding (Fig. 7, F and G). This indicates the preferential binding of ZmPEX2 to ZmRALF2. In summary, binding of Clade IB ZmRALFs to cell wall ZmPEXs is preferred compared with binding with pollen-expressed cell surface receptors ZmFERL4/7/9 (Fig. 5, E, and F).
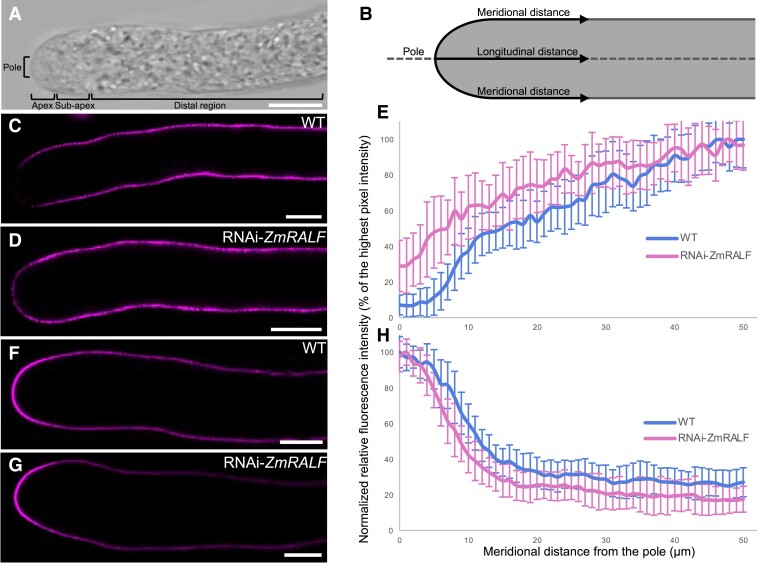
Clade IB ZmRALFs regulate pectin distribution and wall thickness in growing pollen tubes
Proper biochemical composition and spatial distribution of cell wall material are critical for maintaining pollen tube growth as mutants defective in the synthesis of cell wall components exhibit pollen tube growth phenotypes such as slow or wavy growth, growth arrest, and burst (Jiang et al. 2005; Nishikawa et al. 2005; Wang et al. 2011; Chebli et al. 2012; Oelmüller et al. 2023). To investigate cell wall material distribution in the maize pollen tube, we first defined the different regions of growing pollen tubes. As shown in Fig. 8, A and B, the first ∼3 to 4 μm of a growing pollen tube was defined as the “apex,” which is characterized by the clear zone extending from the outermost tip “pole.” The next ∼5 to 10 μm is considered as the “sub-apex” region, and the “distal region” begins from ∼10 μm from the tip.
Figure 8.
Pectin distribution in the pollen tube cell wall is altered in RNAi-ZmRALF mutants. A) Bright field image of a growing maize pollen tube showing various zones. The major growing zone is located at the pole and apex. The apex of the pollen tube is ∼3 to 4 μm, while the sub-apical region is ∼5 to 10 μm in length. Approximately 10 μm from the pole begins the distal region of the pollen tube. B) Schematic figure of a maize pollen tube. Fluorescent intensity in C), D), F), and G) was measured along meridional distances. C and D) Representative images of JIM5 labeled WT and RNAi-ZmRALF mutant pollen tubes, respectively. JIM5 monoclonal antibody labels de-esterified pectin/homogalacuronan. E) Fluorescent intensity quantification of JIM5 labeled WT and RNAi-ZmRALF mutant pollen tubes. n = 26 for both WT and RNAi-ZmRALF pollen tubes. The data are presented as the mean value ± Sds. F and G) Representative images of LM20 labeled WT and RNAi-ZmRALF mutant pollen tubes, respectively. LM20 monoclonal antibody labels esterified pectin/homogalacuronan. H) Fluorescent intensity quantification of LM20 labeled WT and RNAi-ZmRALF mutant pollen tubes. For WT pollen tubes, n = 19 and for RNAi-ZmRALF pollen tubes, n = 21. The data are presented as the mean value ± Sds. Scale bars = 10 μm.
Pectins, which are also known as pectic polysaccharides, are the main and most important component of the pollen tube cell wall. Pectins are a heterogenous group of galacturonyl polymers containing galacturonic acid in the backbone that can be esterified or de-esterified at their carboxylic acid residues by methyl groups. Both forms can be labeled with JIM5 and LM20 antibodies that specifically recognize pectin molecules with low and high degrees of methyl esterification, respectively (Knox et al. 1990; Clausen et al. 2003; Verhertbruggen et al. 2009). As shown in Fig. 8, C and E, the amount of de-esterified pectin in WT maize pollen tubes is almost absent at the first 4 to 5 μm from the very tip of growing pollen tubes, then it slightly increases and reaches a plateau at around 40 μm along the meridional distance. In contrast, in RNAi-ZmRALFs mutant pollen tubes, de-esterified pectin can be detected from the very tip of the pollen tube and reaches similar levels to WT pollen from about 35 μm from the tip (Fig. 8, D and E). The amount of esterified pectin is similar in the apex of WT and RNAi-ZmRALFs mutant pollen tubes but decreases faster in the sub-apical region and remains lower in the distal region (Fig. 8, F to H).
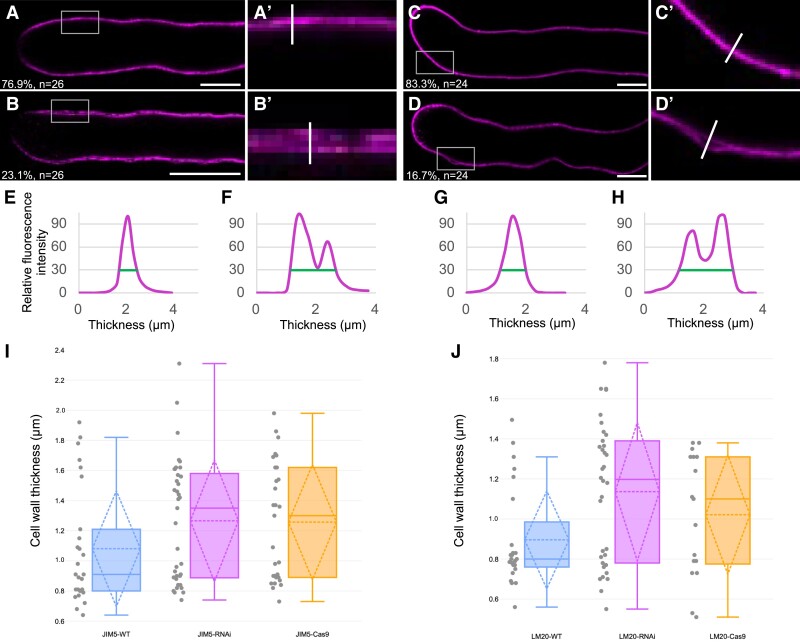
Although different distribution patterns of de-esterified pectin and esterified pectin can be observed along mutant pollen tubes, it was unclear whether this distribution pattern may also affect the wall stiffness and/or thickness. Therefore, we optimized and finally increased the concentration of the fixative to facilitate its penetration into in vitro grown pollen tubes and to enhance the fixation rate. Significant differences in cell wall structure and thickness were detected in mutant pollen tubes compared with those of WT plants. As shown in Fig. 9, A and B, both esterified and de-esterified pectin-labeled WT pollen tubes can be classified into 2 categories, (i) pollen tubes with normal and linear cell wall structure (Fig. 9, A and C) and (ii) pollen tube with split cell wall structure (Fig. 9, B and D), which caused increased cell wall thickness (Fig. 9, E to H). Although splitting of cell walls can also be observed in WT pollen, the frequency increased from 23.1% to 59.5% in RNAi-ZmRALF and to 55.2% in ZmRALF2/3-Cas9 mutant pollen tubes using the JIM5 antibody, while the observed frequency increased from 16.7% to 47.1% and 52.9%, respectively, by using the LM20 antibody (Fig. 9, A to D; Supplementary Fig. S13). Moreover, the cell wall thickness in the sub-apex increased by >30% in RNAi-ZmRALF and ZmRALF2/3-Cas9 mutant pollen from about 0.8 to 0.95 μm in WT pollen tubes to about 1.15 to 1.5 μm depending on the usage of the LM20 and JIM5 antibody, respectively (Fig. 9, I and J). A similar cell wall thickness increase could be observed in crystalline cellulose staining by using the CBM3a antibody (Supplementary Fig. S14).
Figure 9.
Cell wall organization pattern is altered in ZmRALF mutants. A and B) Representative images of JIM5 labeled WT pollen tubes. Cell walls either showed a thin homogenous line or were thicker and dilated showing partially 2 lines. The ratio of each category and the number of pollen tubes that were analyzed are indicated. A′ and B′) Enlargement of gray boxes in A and B), respectively. Scale bars = 10 μm. See Supplementary Fig. S13, A to D for ZmRALF mutant lines. C and D) Representative images of LM20 labeled WT pollen tubes. The ratio of each category like in A and B) and the number of pollen tubes that were analyzed are indicated. C′ and D′) Enlargement of gray boxes in C and D), respectively. Scale bars = 10 μm. See Supplementary Fig. S13, E to H for ZmRALF mutant lines. E to H) Quantification of relative fluorescence intensity of white lines shown in A′ to D′). Thickness of cell walls was measured with the criteria that relative fluorescence intensity was 30% (green lines). I and J) Box plot analysis of pollen tube cell wall thickness WT and ZmRALFs mutant lines after labeling with JIM5 I) and LM20 J), respectively. The thickness was defined with the same criteria as in E to H). Whiskers extend to min and max, box boundaries represent the 25th percentile (lower) and 75th percentile (upper), the lines inside boxes represent medians, Sd is displayed as a dashed rhombus, along with the mean and median values, and dots represent the individual data for each documented pollen tube.
Discussion
Mobile and secreted RALF ligands and their CrRLK1L receptors evolved in bryophytes. There exists, for example, a single CrRLK1L and 3 RALF genes in M. polymorpha (Galindo-Trigo et al. 2016; Mecchia et al. 2017). A strong amplification of RALF genes was reported in angiosperms. While the basal angiosperm Amborella trichopoda contains 9 genes, >30 genes were reported in various eudicot and monocot families (Campbell and Turner 2017; Abarca et al. 2021). So far, research on RALF functions focused on eudicots and was especially conducted in Arabidopsis. It has been shown, for example, that many RALFs are active in pollen functions and regulate among others pollen hydration and penetration in stigmatic papilla cells, pollen tube growth and cell wall integrity, pollen tube exit from the transmitting tract, as well as pollen tube perception in the egg apparatus (e.g. Ge et al. 2017, 2019c; Mecchia et al. 2017; Zhong et al. 2022; Gao et al. 2023; Lan et al. 2023). However, it remained unclear to what extent obtained knowledge can be transferred to other plant species and whether these proteins were co-opted for pollen function before the split of monocots and dicots. In monocots, one study reported that in the grass Saccharum officinarum (sugarcane), SacRALF1 is involved in the regulation of tissue expansion (Mingossi et al. 2010) and OsRALF17 and OsRALF19 were recently shown to be involved in pollen hydration, germination, and pollen tube elongation (Kim et al. 2023). By comparing RALFs from the eudicot model Arabidopsis and the grass model maize, we distinguished 3 clades. Our nomenclature differs from a previous phylogenetic study comparing 795 identified RALFs (Campbell and Turner 2017). Four major clades we reported in that study with Clades I to III RALFs containing an RRXL protease cleavage site and the YISY motif for receptor interaction, while Clade IV contains all other RALFs. In the meantime, we have learned that the YISYxxLRRN domain mediates interaction with the LLG co-receptor forming a heterotrimeric complex with the CrRLK1L member FER (Xiao et al. 2019). Moreover, 4 tyrosine residues are involved in LRX-binding including the 2 Ys of the YISY motif and 2 additional ones in the YY motif in the middle of mature RALP peptides (Moussu et al. 2020). Notably, in the modified nomenclature suggested in this report, closely related Clade IA and Clade IB contain all RALFs harboring the above-described motifs, while these motifs are highly degenerated and lacking in Clade II and Clade III RALFs, respectively. Moreover, Clade IB RALFs that are more conserved after the cleavage site contain reproductive and pollen-specific RALFs (Ge et al. 2017; Mecchia et al. 2017; Gao et al. 2022) including RALFs of basal land plants like mosses, and thus appear to represent the most original clade. Clade IA, which is more variable after the cleavage site, contains mainly vegetatively and sporophytically expressed RALFs in the 2 analyzed species, including well-described RALFs like AtRALF1, which may have acquired specific sub-functions during land plant evolution, like regulation of root and root hair development (Haruta et al. 2014; Zhu et al. 2020) and AtRALF23/33 playing roles in papilla hair cell functions (Liu et al. 2021). Arabidopsis Clade II RALFs that do not exist in maize, and which still contain a degenerated YISY-motif, and even some members of Clade III were recently shown to interact with CrRLK1L receptor kinases in vitro (Zhong et al. 2022). However, binding affinities were lower compared with those measured for ZmRALF2/3 in this report. In contrast to the above findings, maize Clade III ZmRALF1/5 do interact with CrRLK1Ls with very low (almost unspecific) binding and likely acquired novel functions during evolution and the pollen tube journey through the specified transmitting tract tissue of the grasses (Zhou et al. 2017). It will now be important to study the role of Clade III ZmRALFs but also to elucidate whether Clade II and Clade III RALFs can still interact with LLG co-receptors and LRX/PEX cell wall proteins.
Quantitative biochemical assays revealed that Clade IB AtRALF4 from Arabidopsis pollen binds LLGs and LRX cell-wall modules with drastically different binding affinities, and with distinct and mutually exclusive binding modes (Moussu et al. 2020). The binding of AtRALF4 with the cell wall protein AtLRX8 could be very strong with a Kd of 3.5 nm (Moussu et al. 2020), while the strongest interaction with receptor kinases ANXs/BUPSs was reported as 310 nm (Ge et al. 2017). This drastic difference in binding affinity could be partially explained by optimized protein folding and purification systems in the first report, but also by expressing recombinant proteins in different heterologous systems that may affect the formation, for example, of disulfide bonds and glycosylation. However, it points toward the same finding in the present study that ZmRALF2/3 binds on average much stronger to ZmPEXs compared with pollen-specific ZmFERLs, and even much weaker to ZmFERL1 which is not expressed in pollen.
Taking also into consideration that the binding of LRX/PEX cell wall proteins and LGG-CrRLK1L receptor complexes to the RALF motif YISY motif is mutually exclusive, we propose dosage-dependent RALF signaling: we think that during pollen tube tip growth, RALFs possess a dual role and serve as sensor and cell wall component secreted together with cell wall material at the pollen tube apex. While the cell wall is initially thin and fragile at the apex-subapical region, finely balanced RALF signaling is required to integrate cell wall thickness and stiffness with pollen tube growth speed and exocytosis. This allows the cell to reduce the pollen tube growth speed to secrete, for example, additional and sufficient cell wall material components and to provide time to regulate the stiffness of the wall. The thicker the cell wall, the more RALF is bound to LRX/PEX cell wall proteins due to their higher binding affinity. This hypothesis is supported by recent findings showing that the LRX8-RALF4 complex in Arabidopsis interacts with de-methyl esterified pectins that are generated at the subapical region of the growing pollen tube. It was further shown that the LRX8-RALF4-pectin interaction patterns cell wall's polymers into a reticulated network essential for cell wall integrity (Moussu et al. 2023). Depletion of the RALF cell wall component and sensor in RNAi and CRISPR/Cas mutants leads to loss of cell wall thickness and stiffness (control) culminating in pollen tube burst in the sub-apical region. We note that (i) the described RALF interactions are also pH-dependent (Moussu et al. 2020) and (ii) its induced signaling modifies the redox status of the cell wall by RALF-induced generation of reactive oxygen species (ROS), which stimulates pollen hydration and tube growth while reducing the pollen burst rate likely by increasing cell wall stiffness (Feng et al. 2019; Liu et al. 2021; Zhou and Dresselhaus 2023).
With the exception of the above-described regulation of ROS levels and recently reported Ca2+ signaling (Gao et al. 2023), little is known about RALF-induced downstream signaling processes associated with pollen/pollen tube functions. In roots, it has been shown that AtRALF1-FER interaction causes phosphorylation and thus the inhibition of plasma membrane H+-ATPase2 explaining extracellular alkalinization (Haruta et al. 2014). AtRALF1 also enhances the interaction of FER with the receptor-like cytoplasmic kinase RIPK, which mutually phosphorylates each other to regulate cell growth in roots (Du et al. 2016). AtRALF1 treatment promotes direct phosphorylation of ERBB3 binding protein 1 (EBP1) by FER leading to its accumulation in the nucleus and inhibition of RALF peptide responses (Li et al. 2018). It was further shown that AtRALF1 promotes FER-mediated phosphorylation of the elongation factor eIF4E1. Phosphorylated eIF4E1 regulates the synthesis of root hair proteins including ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4), which is required for root hair growth (Zhu et al. 2020). It will now be important to identify downstream signaling components to further the hypothesis that RALFs serve as sensors and activators of cell wall thickness and stiffness regulation during growth. This is technically challenging in species like maize, taking into consideration that also in Arabidopsis only a few downstream targets like the receptor-like cytoplasmic kinase named MARIS have been identified so far (Boisson-Dernier et al. 2015).
Together with callose, pectins are the main components of the pollen tube cell wall, and their distribution pattern is of vital importance for maintaining proper pollen tube growth. In growing pollen tubes, high methyl-esterified (soft) pectins are deposited at the tip of the pollen tube via exocytosis, and de-esterification takes place in the sub-apical region by the activity of pectin methylesterases (PMEs). The activity of PMEs in the very apical region is prevented by PME inhibitors (PMEIs) that are secreted within the same secretory vesicles (Scholz et al. 2020). Recently, it was reported that PMEs can abolish RALF1 perception during seedling root growth, and the application of free demethylated oligogalacturonides also reduces RALF1 activity. It was therefore proposed that demethylated and thus negatively charged pectin may act as a signaling scaffold for positively charged RALF peptides to modulate its output signaling (Rößling et al. 2023). In root hairs, dual roles of RALF22 on the cell wall structure and signaling were reported. On one hand, RALF22 induces the formation of a complex with LLG1 and FER to trigger adaptive cellular responses, while on the other hand, RALF22 directs the compaction of charged pectin polymers into periodic circumferential rings at the tip of root hairs together with LRX1 cell wall protein to maintain cell wall assembly and expansion (Schoenaers et al. 2023). Using immunolabeling of maize pollen tubes, we discovered that the spatial distribution of pectins is substantially altered under low RALF levels, and cell wall morphology is compromised ultimately leading to the bursting of pollen tubes. It remained unclear whether reduced RALF levels led, for example, to faster endocytosis of PMEIs as indicated by higher levels of de-esterified pectins in the apex, to reduced ROS levels required to increase the stiffness of the wall in the sub-apical region or to reduced dimerization of ZmPEXs that likely contribute to the stabilization of the cell wall. It is also unclear why pollen germination is not affected in the different ZmRALF2/3 mutants in maize and whether reduced RALF levels affect the Ca2+-gradient in the pollen tube apex that was reported to be dependent on RALF signaling (Gao et al. 2023). We assume that the latter is unlikely, otherwise, pollen tubes would not be able to germinate and grow larger distances before bursting.
In conclusion, we have reported that the autocrine pollen tube signaling pathway regulating cell wall integrity during pollen tube growth involving Clade IB RALFs interacting with CrRLK1L-LLG receptor complexes and with LRX/PEX cell wall proteins is partly conserved between Arabidopsis and maize, and thus likely also in other angiosperms. This is supported by a recent study in rice showing that the homologs of Arabidopsis RALF4/19 are also involved in pollen germination and tube growth (Kim et al. 2023).
In contrast to Arabidopsis and rice, mutation of the 3 most highly expressed pollen RALF homologs in maize has no obvious effect on pollen germination, which might be associated with “high-speed growth” in this species. We further suggest that RALF signaling occurs in a dosage-dependent manner integrating RALF interaction with receptor complexes and cell wall proteins, respectively, indicating that Clade IB RALFs serve as sensors of cell wall integrity and thickness during pollen tube growth. The idea that RALFs somehow serve as sensors for cell wall integrity is not new and has been proposed before (Gonneau et al. 2018; Ge et al. 2019b), but here we provide a mechanistic explanation for the burst phenotype that indirectly links the cell wall to cell surface receptors via different RALF binding affinities. Moreover, we link the regulation of pollen cell wall structure via the distribution pattern of soft methyl-esterified versus stiffer de-esterified pectins to RALF signaling in the sub-apical region of the growing pollen tube. So far, it has only been described that FER signaling is crucial for maintaining de-esterified pectin at the filiform apparatus of the embryo sac in Arabidopsis (Duan et al. 2020). In summary, the comparison of RALF signaling in slow germinating and growing pollen tubes in a eudicot species (Arabidopsis) with that in a fast germinating and growing monocot and grass species (maize) provides important knowledge about similarities and differences among the 2 species and leads to a large number of suggestions for future research directions. It is now important to better elucidate downstream signaling processes and to figure out functions of more recently evolved Clade III RALFs like ZmRALF1/5 that do not regulate cell wall integrity during pollen tube growth.
Materials and methods
Plant materials and transgene transmission studies
All maize (Z. mays) plants were grown under the same conditions in a greenhouse at 26 °C under illumination of 400 μm s−1 m−2 (measured 70 cm from the soil surface) using 400 W SON-T Agro and HPI-T Plus lamps with 16 h light and 8 h darkness at 21 °C. Air humidity was kept constant at about 60%. Pollen used for in vitro germination assays was collected as follows: old pollen grains were removed in the early morning by shaking and newly shed pollen grains were collected 2 h later and tested for successful germination in vitro. Only pollen germinating with >80% frequency was used for further analysis. Sampling was repeated on at least 3 different days. For transgene transmission efficiency studies, ears were covered with white paper bags before silk emergence to avoid pollen contamination. Transgenic plants were identified by PCR and DNA agarose gel electrophoresis using RNAi-ZmRALF-specific primers (see details below and Supplementary Table S1 for primer sequences). Transgenic/mutant lines were crossed with inbred line B73, which was used as a WT control, and transmission of transgenes was detected as described above. Arabidopsis (A. thaliana) ecotype Columbia-0 (Col-0) seeds were sown on soil and kept for 2 to 3 d at 4 °C in the dark for stratification. Seedlings were then grown under long-day conditions at 21 °C for 16 h of light under illumination of 150 μm s−1 m−2 and 8 h of darkness at 18 °C at 60% air humidity.
Gene identification
To identify the RALF gene family in maize (ZmRALFs), searches with 2 databases were performed. ZmRALF1/2/3 mature peptides were used as queries to run BLASTP in the non-redundant protein sequences (nr) of Z. mays (taxi:4577) at NCBI (https://blast.ncbi.nlm.nih.gov). To avoid missing genes that were not annotated, the same queries were used to run TBLASTN using the Gramene database (http://ensembl.gramene.org/). The BLOSUM62 matrix was used in both searches. Output sequences were manually selected taking into consideration a query cover >50% and an E-score <0.05. Selected peptides were further confirmed according to the cysteine residues arrange model (Silverstein et al. 2007). After RALF peptide identification, corresponding locus tags (B73 RefGen_v4) and gene symbols were extracted from protein introduction pages. Similar procedures were performed to obtain ZmCrRLK1L, ZmLLG, and ZmLRX/ZmPEX gene families in maize. Arabidopsis FER, ANXs, BUPSs, and LORELEI proteins were used as queries to BLAST the maize genome. Because of the highly variable proline-rich extension-like domain in LRX proteins (Herger et al. 2019), only ectodomains of ZmPEX1 and ZmPEX2 were used to identify the whole ZmLRX/ZmPEX family in the maize genome. Outputs were manually checked according to structure features (Boisson-Dernier et al. 2011; Liu et al. 2016; Herger et al. 2019).
Plasmid construction and plant transformation
Considering that 9 RALF genes are expressed in mature maize pollen (Supplementary Fig. S1), an RNAi construct was generated to downregulate the whole gene family. Gene-specific transcribed regions of ZmRALF1/2/3 together generating about 90% of RALF transcripts in maize pollen were cloned after DNA amplification from genomic DNA in one vector and the RNAi-ZmRALF vector was then generated as follows: 604 bp of ZmRALF1/2/3 including 190 bp of ZmRALF1, 212 bp of ZmRALF2, and 202 bp of ZmRALF3 that was synthesized by Thermo Fisher Scientific. For primer sequences see Supplementary Table S1. After digestion with the respective restriction enzymes, fragments were cloned into the corresponding splicing sites of the pUbi-iF2 vector (DNA Cloning Service) that contains the maize Ubi promoter. Corresponding vector regions were always sequenced. To generate CRISPR-Cas9 mutant maize plants, guide RNAs (gRNAs) were designed with Breaking-Cas (Oliveros et al. 2016) and CRISPR-Pv2.0 (Lei et al. 2014) online platforms. Corresponding sequences are included in Supplementary Table S1. The integration of gRNAs to pGW-Cas9 was carried out as previously reported (Char et al. 2017). Transformation constructs were verified by sequencing and restriction enzyme digestion. The maize inbred line HiAB hybrid was used for stable maize transformation. For ZmRALF sub-localization and complementation studies in Arabidopsis, 2,286 bp upstream of the ATG start codon of the Arabidopsis AtRALF4 gene was used as the endogenous promoter, followed with ORFs of ZmRALF genes cloned from maize pollen cDNA. Firstly, the AtRALF4 promoter region was cloned from Arabidopsis genomic DNA and the promoter was integrated into pB7FWG2.0 or pB2GW7 by restriction enzyme digestion and T4 DNA ligation. Next, ZmRALF fragments were transferred individually by LR reaction using Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific). Constructs containing an eGFP sequence fused C-terminally to ZmRALFs were cloned into pB7FWG2.0 and those lacking a tag into pB2GW7, respectively. Finally, gene constructs were transformed into Arabidopsis ecotype Col-0.
To generate recombinant proteins in E. coli, predicted mature peptide regions of ZmRALF1/2/3/5 genes were cloned into plasmid pET53-DEST (NovaPro) for subsequent MST and competition pull-down experiments, and were additionally cloned into pET32b (Novagen) for pull-down experiments using gene-specific primers as indicated (Supplementary Table S1). In the pET53 expression vector, an N-terminal 6×His tag is combined with ZmRALFs to form the recombinant protein, which is insoluble and expressed in inclusion bodies and requires denaturation, purification, and oxidization. In the pET32 expression vector, N-terminal His and TrxA tags were used as they were reported to promote protein solubility (Costa et al. 2014). All receptor-like kinases in this study were cloned individually into the pMAL-p2p plasmid (provided by Kamila Kalinowska) with an N-terminal MBP tag, including the ectodomains of ZmFERL1/4/7/9 and ZmPEX2/4, as well as whole proteins without a signal peptide of ZmLLG1/2 and AtLRX9/10/11. Ectodomains of ZmFERL7/9 were cloned into pGEX-6P1 (GE Healthcare) with an N-terminal GST tag. Constructs were transformed into E. coli (strain BL21).
RNA extraction, RT-qPCR and gene expression analysis
The Trizol Plus RNA purification kit (Thermo Fisher Scientific) was used for total RNA extraction from maize pollen. Purified RNA was reverse transcribed to cDNA using RevertAid H minus reverse transcriptase (Thermo Fisher Scientific). ZmRALF gene-specific qPCR primers (Supplementary Table S1) were designed and tested in standard PCR reactions. In general, the assay involved 2 biological replicates, each represented by 2 cDNA pools that were used as templates for 3 technical replicate reactions. Normalization was based on 2 internal maize reference genes, encoding the maize membrane protein PB1A10.07c (reference gene 1, Zm00001d018359) and cullin (reference gene 2, Zm00001d024855), which were previously reported to be stably expressed (Manoli et al. 2012). The resulting calibrated normalized relative quantities were exported into Excel sheets and the ΔΔCT method (Livak and Schmittgen 2001) was used for further analysis and calculation of fold changes. To compare tissue-specific gene expression patterns of maize and Arabidopsis genes described in this study, mRNA expression levels in the pollen, stigma, root, leaf, and seed of maize were extracted from the CoNekT online database (https://evorepro.sbs.ntu.edu.sg/; Julca et al. 2021). Arabidopsis pollen mRNA expression levels were extracted from Loraine et al. (2013) and expression levels in other tissues were obtained from Genevestigator (www.genevestigator.com). Please note that the provided gene expression values of maize and Arabidopsis genes in Supplementary Figs. S4 and S10 are not comparable.
Protein sequences and phylogenetics analysis
Protein sequences were downloaded from the Universal Protein Resource (UniProt). The MUSCLE method was used for multiple sequence alignment (Edgar 2004). The phylogenetic tree was constructed based on the neighbor-joining statistical method, 1,000 bootstrap, and Dayhoff model. The phylogenetic trees were further optimized on the website of Evolview (Subramanian et al. 2019). For alignment of RALFs, predicted signal peptide and propeptide (if they existed) were cut off and only predicted mature RALF sequences were used. Similarly, for the alignment of CrRLK1Ls, only the ectodomains were used for analysis. Proteins without signal peptides were used to align LORELEI-like GPI-anchored proteins. Sequence alignment results were shown by using Jalview (Waterhouse et al. 2009) and colored by Clustal_X (Thompson et al. 1997). Alignments and machine-readable tree files are provided in Supplementary File 1.
Protein purification and pull-down assays
Purification of soluble TrxA-His-RALF recombinant proteins and MBP/GST tagged proteins generated in E. coli was modified from the protein purification protocol of the QIAexpressionist (Qiagen) as follows: 2 mL of E. coli containing the corresponding protein expression plasmid was added to 1 L LB-medium with the corresponding antibiotic and incubated in a 37 °C shaker at 200 rpm. Once an OD600 of 0.5 to 0.7 was reached, isopropyl β-D-1-thiogalactopyranoside (IPTG, SERVA) was added (1 mm IPTG was used for His-tagged proteins and 0.1 to 0.5 mm IPTG for GST/MBP-tagged proteins) and further incubated in a 30 °C shaker at 200 rpm for another 3 to 4 h. Afterwards, bacteria were pelleted by centrifugation. The pellet was resuspended in 40 mL chilled lysis buffer (His lysis buffer: 50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0; GST/MBP lysis buffer: 50 mm Tris pH 7.5, 150 mm NaCl, 0.05% CA630 from Sigma-Aldrich) containing protease inhibitor, 0.5 mg/mL lysozyme, and 1 mm PMSF. Cells were shattered by repeated sonication until samples became transparent. Lysates were collected by centrifugation at 4 °C for 20 min at 16,000 × g. Supernatants were separated from lysates and incubated with affinity beads at 4 °C overnight (His beads: Ni-NTA from QIAGEN; GST beads: glutathione-agarose beads from ROTH; MBP beads: amylose beads from NEB). Protein-binding beads were collected by centrifugation and washed several times with washing buffer (His washing buffer: 50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole, pH 8.0; GST/MBP washing buffer: 100 mm Tris at pH 8.0) to remove unspecific binding proteins. Proteins were eluted from beads by incubating with elution buffer at 4 °C for 2 to 4 h (His elution buffer: 50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole, adjust pH to 8.0; GST elution buffer: 15 mg/mL reduced glutathione in 100 mm Tris; MBP elution buffer: 25 mg/mL maltose in 100 mm Tris). Proteins were further desalted with PD 10 desalting columns from GE-Healthcare. FLAG-tagged ectodomains of Arabidopsis ANX1/2 and BUPS1/2 proteins were obtained from a previous study (Ge et al. 2017).
Purification of insoluble His-RALF recombinant proteins generated in E. coli was performed under denaturing conditions according to a previous report (Okuda et al. 2013) with some modifications. Cell cultures with an OD600 of 0.6 to 0.8 were induced in 800 mL LB medium containing 2% w/v glucose by 1 mm IPTG at 37 °C for 26 h. Cells were harvested by centrifugation. Subsequently, pellets were homogenized in 40 mL suspension buffer (1 mm EDTA, 50 mm NaCl, 50 mm Tris at pH 8.0) containing protease inhibitor, 1 mg/mL lysozyme, and 2.5 μg/mL DNase I for 1 h on ice and disrupted by sonication. Inclusion bodies were obtained from sonicated extracts by centrifugation. Afterwards, inclusion bodies were solubilized by incubation in a denaturating solution (6 m guanidine hydrochloride, 1 mm β-mercaptoethanol, 5 mm imidazole, 500 mm NaCl, 50 mm Tris at pH 8.0) for 1 h at room temperature. Solubilized His-RALF proteins were centrifuged and ultra-filtrated to remove precipitates. Further, protein renaturation and refolding were carried out on HisTrap FF columns (GE Healthcare) with a linear gradient of 6 to 0 m urea buffer containing 1 mm β-mercaptoethanol, 5 mm imidazole, 500 mm NaCl, and 50 mm Tris (pH 8.0) using an ÄKTA system (GE Healthcare). The first refolded His-RALF protein was eluted from the column with a solution containing 1 mm β-mercaptoethanol, 500 mm imidazole, 500 mm NaCl, and 50 mm Tris (pH 8.0). Eluted protein solutions were dialyzed for 3 d at 4 °C in redox buffer containing 100 mM l-arginine ethyl ester dihydrochloride (Sigma-Aldrich), 1 mm oxidized glutathione (Wako), and 10 mm reduced glutathione (Wako), and 50 mm Tris (pH 8.0), to refold them for a second time. In the last step, refolded proteins were dialyzed for 3 h at 4 °C in PBS buffer (pH 7.4) and concentrated by ultrafiltration (Amicon Ultra Ultracel-3K; Millipore).
In vitro binding or pull-down assays were carried out as described (Ge et al. 2017). ZmRALFs and AtRALFs were generated containing a His-tag and MBP/GST-tags were used for ZmFERLs, AtANXs, AtBUPSs, ZmLLGs, ZmPEXs, and AtLRXs. Briefly, samples were mixed in 500 µL binding buffer (20 mm Tris, pH 7.5, 1% CA630) and rotated at 4 °C for 2 h before addition of His-beads (Ni-NTA, QIAGEN). After continuous rotation for another 2 h, beads were pelleted and washed 5 times. Samples were boiled in an SDS-loading buffer and analyzed by SDS-PAGE and Western blot. For antibodies, we used a His antibody (6×-His Tag Monoclonal Antibody, 1:5,000 dilution, catalog no. MA1-21315, Thermo Fisher Scientific), an MBP antibody (Anti-MBP Monoclonal Antibody, HRP conjugated, 1:2,000 dilution, catalog no. E8038S, NEB), a GST antibody (GST Tag Monoclonal Antibody, 1:5,000 dilution, catalog no. 13-6700, Thermo Fisher Scientific) and a FLAG antibody (anti-Flag-HRP, 1:1,000 dilution, catalog no. A8592, Sigma-Aldrich). Competition pull-down studies were consistent with those described above, with 3 different proteins mixed in proportions. For interaction studies of biotinylated AtRALF4/19 with GST-tagged ZmFERL7/9 and MBP-tagged ZmFERL1/4, the procedure was the same as described above except that His-beads were substituted with streptavidin magnetic particles (Spherotech).
In vitro pollen germination
Fresh maize pollen was germinated on the surface of a solid pollen germination medium (PGM) in Petri dishes. First, PGM [10% w/v sucrose (Roth), 0.005% w/v H3BO3 (Sigma-Aldrich), 10 mm CaCl2 (Sigma-Aldrich), 0.05 mm KH2PO4 (Merck), 6% w/v PEG 4000 (Merck-Schuchardt), pH 5.]) was prepared as described (Schreiber et al. 2004). After all components were mixed, an equal volume of heated 0.6% w/v NuSieve GTG Agarose (Lonza) was added. The mixture was poured into 3 cm Petri dishes, which were ready for usage after cooling down to room temperature. Fresh pollen was obtained as described above and spread directly onto Petri dishes. Pollen germination and growth status were observed and photographed by using a Nikon microscope (Nikon Eclipse TE2000-S) with a 4× objective (Plan Fluor DL 4×/0.13, PHL). Pollen germination status was divided into 4 categories: (i) germinated pollen growing normally, (ii) un-germinated pollen, (iii) burst pollen at germination, and (iv) burst pollen tube. Calculations and statistics were made using the ggpubr package in R (https://CRAN.R-project.org/package=ggpubr). In vitro germination of Arabidopsis pollen was performed as follows: mature pollen was germinated on the surface of solid PGM [18% w/v sucrose, 0.01% w/v H3BO3, 5 mm CaCl2, 5 mm KCl, 1 mm MgSO4 (Merck), pH 7.5; 1.5% NuSieve GTG agarose] in Petri dishes that were placed in humid boxes and incubated at room temperature for 4 h. Solid PGM-containing pollen tubes were cut out and placed on a microscope slide with 150 μL liquid PGM solution (lacking agarose). To visualize the cell wall, PI (Sigma-Aldrich) staining was carried out. About 10 mm PI in PGM solution werasapplied to the surface of germinated pollen tubes and incubated for 5 min. Fluorescence images were observed and collected using a ZEISS LSM 980 Airyscan2 Confocal Laser Scanning microscope with a 63× (Plan-Apochromat 63×/1.40 Oil DIC M27) objective. The fluorescence of eGFP/PI was detected with excitation at 488 nm/561 nm and emission between 505 and 550 nm/580 and 650 nm, master gain between 800 and 900 V, Pinhole was set always at 1 AU.
MST assays
To test the binding affinity of maize RALF peptides with the corresponding interactors, MST assays were carried out by using a Monolish NT.115 (NanoTemper Technologies). His-tagged RALF proteins were labeled with red dyes with the His-Tag labeling kit RED-tris-NTA 2nd Generation (NanoTemper Technologies) according to the manufacturers’ recommendation. His-tagged RALF proteins were diluted to 200 nm in PBST buffer (137 mm NaCl, 2.5 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, 0.05% Tween-20 at pH 7.4). A volume of 100 μL protein solution (200 nm) was mixed with 100 μL dye solution (100 nm) and incubated at room temperature for 30 min. After centrifugation, 20 nm dye-labeled proteins were mixed with a serial dilution of interactors (ZmFERL1/4/7/9, ZmPEX2/4). Samples were loaded into glass capillaries (NT.115 Standard Treated Capillaries) and measured at 40% MST power and 10% LED power. Recorded data were analyzed with MO. Affinity Analysis Software v2.3 (NanoTemper Technologies).
DLS and nanoDSF assay
DLS was performed using Prometheus Panta according to instructions for the instrument. NanoDSF was performed using Prometheus NT.48 equipped with back-reflection mode, according to instructions for the instrument. Samples were loaded in Prometheus Series capillaries and exposed at thermal stress from 20 °C to 95 °C by thermal ramping rate of 1 °C/min.
Immunohistochemistry
For fluorescence labeling of maize pollen tube cell wall epitopes, fresh pollen grains were collected and germinated in liquid PGM within ibiTreat μ-Slide VI 0.4 channels (Ibidi). Forty-five minutes after germination, liquid PGM was replaced with fixative solution [100 mm PIPES buffer, pH 6.9, 4 mm MgSO4, 4 mm EGTA, 10% (w/v) sucrose, and 5% (w/v) formaldehyde] and pollen tubes were fixed at room temperature for 90 min. After washing with PBS buffer (137 mm NaCl, 2.5 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4 at pH 7.4), pollen tubes were incubated with PBS buffer containing 3.5% (w/v) BSA at room temperature for 30 min and then washed with PBS buffer. All antibodies were diluted in PBS buffer with 3.5% (w/v) BSA. Incubation was done at 4 °C overnight for the primary antibody and 3 h at 30 °C for the secondary antibody. De-esterified and esterified pectins were labeled with JIM5 (1:50 dilution, catalog no. ELD004, Kerafast) and LM20 (1:50 dilution, catalog no. ELD003, Kerafast) antibodies, respectively, followed by Alexa Fluor 594 anti-rat IgG (1:100 dilution, catalog no. A-11007, Thermo Fisher Scientific) staining. Crystalline cellulose was labeled with CBM3a (1:200 dilution; catalog no. CZ00411, Nzytech, provided by Paul Knox, University of Leeds, UK), followed by an anti-poly-His antibody (1:12 dilution; catalog no. H1029, Sigma-Aldrich) and subsequent incubation with Alexa Fluor 594 anti-rat IgG (1:100 dilution, catalog no. A-11007, Thermo Fisher Scientific). Negative controls were performed by omitting primary or secondary antibody incubation. Pollen tubes were finally kept in ROTIMount FluorCare (Carl Roth) and fluorescence images were collected using a ZEISS LSM 980 Airyscan2 Confocal Laser Scanning microscope with a 63× (Plan-Apochromat 63×/1.40 Oil DIC M27) objective. The fluorescence of Alexa Fluor 594 anti-rat IgG was detected with excitation at 561 nm and emission between 600 and 680 nm, master gain between 800 and 900 V, pinhole was set always at 1 AU.
Statistical analysis
Statistical analyses were conducted as described in the text and figure legends. Statistical data are provided in Supplementary Data Set 2.
Accession numbers
Accession numbers of experimentally analyzed genes in this study are as follows: ZmRALF1 (Zm00001d039766), ZmRALF2 (Zm00001d008881), ZmRALF3 (Zm00001d039429), ZmRALF5 (Zm00001d019239), ZmFERL1 (Zm00001d029047), ZmFERL4 (Zm00001d037839), ZmFERL7 (Zm00001d035992), ZmFERL9 (Zm00001d045515), ZmLLG1 (Zm00001d003139), ZmLLG2 (Zm00001d020876), ZmLLG3 (Zm00001d051672), ZmLLG4 (Zm00001d017874), ZmPEX1 (LOC103648188), ZmPEX2 (Zm00001d048715), ZmPEX3 (Zm00001d023858), ZmPEX4 (Zm00001d040802), AtRALF4 (At1G28270), AtRALF19 (At2G33775), ANX1 (At3G04690), ANX2 (At5G28680), BUPS1 (At4G39110), BUPS2 (At2G21480), AtLRX9 (At1G49490), AtLRX10 (At2G15880), and AtLRX11 (At4G33970). See Supplemental Data Set 1 for all genes.
Supplementary Material
Acknowledgments
We thank Kamila Kalinowska (University of Regensburg) for providing the pMAL-p2p vector and Armin Hildebrand for plant care.
Contributor Information
Liang-Zi Zhou, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany; National Key Laboratory of Wheat Improvement, College of Life Sciences, Shandong Agricultural University, Tai’an, Shandong 271018, China.
Lele Wang, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany.
Xia Chen, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany.
Zengxiang Ge, Peking-Tsinghua Center for Life Sciences at College of Life Sciences, Peking University, Beijing 100871, China.
Julia Mergner, Chair of Proteomics and Bioanalytics, Technical University of Munich (TUM), 85354 Freising, Germany.
Xingli Li, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany.
Bernhard Küster, Chair of Proteomics and Bioanalytics, Technical University of Munich (TUM), 85354 Freising, Germany; Bavarian Center for Biomolecular Mass Spectrometry (BayBioMS), Technical University of Munich (TUM), 85354 Freising, Germany.
Gernot Längst, Biochemistry Center Regensburg, University of Regensburg, 93053 Regensburg, Germany.
Li-Jia Qu, Peking-Tsinghua Center for Life Sciences at College of Life Sciences, Peking University, Beijing 100871, China.
Thomas Dresselhaus, Cell Biology and Plant Biochemistry, University of Regensburg, 93053 Regensburg, Germany.
Author contributions
T.D. designed the project and L.-Z.Z. together with L.W. and X.C. performed most of the experiments. T.D., L.-Z.Z., G.L., and L.-J.Q. supervised the study and contributed to the analyses and data interpretation. Z.G., X.L., J.M., and B.K. contributed with material and data analyses. L.-Z.Z., L.W., X.C., and T.D. wrote the article with input from all authors.
Supplementary data
The following materials are available in the online version of this article.
Supplementary Figure S1. Expression pattern of 24 maize RALF genes in indicated tissues.
Supplementary Figure S2. RALF protein sequences from maize and Arabidopsis diverge across 3 major phylogenetic clades.
Supplementary Figure S3. Relative expression levels of 3 ZmRALFs in pollen tubes of WT compared with in those of 6 RNAi-ZmRALFs mutant lines.
Supplementary Figure S4. Phylogenetic and expression analysis of 17 maize CrRLKs1L receptor kinases to identify putative orthologs among 17 Arabidopsis CrRLK1Ls.
Supplementary Figure S5. Phylogenetic tree of 34 CrRLK1L proteins from maize and Arabidopsis showing their domain organization and expression pattern patterns.
Supplementary Figure S6. Quality controls of recombinant proteins used in this study by DLS and nanoDSF technology.
Supplementary Figure S7. Clade IB ZmRALF2 binds more strongly to CrRLK1L receptor kinases compared with Clade III ZmRALF1.
Supplementary Figure S8. Maize CrRLK1L receptor kinases can interact with Clade IB Arabidopsis RALFs and vice versa.
Supplementary Figure S9. Alignment of maize and Arabidopsis LLGs.
Supplementary Figure S10. Phylogenetic analysis and expression pattern of PEX/LRX proteins and genes from maize and Arabidopsis.
Supplementary Figure S11. Sequence alignment of pollen-expressed LRR-extensin-like cell wall proteins named PEX in maize and LRX in Arabidopsis.
Supplementary Figure S12. Maize Clade IB RALFs can interact with Arabidopsis LRX proteins.
Supplementary Figure S13. Disturbance of cell wall organization in ZmRALF mutants.
Supplementary Figure S14. Cell wall organization and thickness are defective in ZmRALF mutants.
Supplementary Figure S15. Images of input controls of pulldown experiments.
Supplementary Table S1. List of PCR primers used in this study.
Supplementary Data Set 1. Gene ID and protein information of all maize ZmRALFs, ZmFERLs, ZmPEX/LRXs, and ZmLLGs.
Supplementary Data Set 2. Statistical data.
Supplementary File 1. Alignments and machine-readable tree files for phylogenetic analysis.
Funding
The German Research Foundation (DFG) is acknowledged for financial support via SFB 924 (to T.D. and B.K.), SFB 960 (to G.L.) and research unit FOR 5098 ICIPS (to T.D.), and the China Scholarship Council (CSC) for a fellowship to L.W.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Abarca A, Franck CM, Zipfel C. Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 2021:187(2):996–1010. 10.1093/plphys/kiab308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabas B, Fridvalszky L. Adhesion and germination of differently treated maize pollen grains on the stigma. Acta Bot Hung. 1984:30(3-4):329–332. [Google Scholar]
- Bedinger P. The remarkable biology of pollen. Plant Cell. 1992:4(8):879–887. 10.1105/tpc.4.8.879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonci T, Ribeiro B, Ceciliato PH, Guerrero-Abad JC, Silva-Filho MC, Moura DS. Arabidopsis thaliana RALF1 opposes brassinosteroid effects on root cell elongation and lateral root formation. J Exp Bot. 2014:65(8):2219–2230. 10.1093/jxb/eru099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bircheneder S, Dresselhaus T. Why cellular communication during plant reproduction is particularly mediated by CRP signalling. J Exp Bot. 2016:67(16):4849–4861. 10.1093/jxb/erw271 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Franck CM, Lituiev DS, Grossniklaus U. Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc Natl Acad Sci U S A. 2015:112(39):12211–12216. 10.1073/pnas.1512375112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Kessler SA, Grossniklaus U. The walls have ears: the role of plant CrRLK1Ls in sensing and transducing extracellular signals. J Exp Bot. 2011:62(5):1581–1591. 10.1093/jxb/erq445 [DOI] [PubMed] [Google Scholar]
- Campbell L, Turner SR. A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front Plant Sci. 2017:8:37. 10.3389/fpls.2017.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding MH, Becraft PW, Meyers BC, Walbot V, Wang K, et al. . An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol J. 2017:15(2):257–268. 10.1111/pbi.12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli Y, Kaneda M, Zerzour R, Geitmann A. The cell wall of the Arabidopsis pollen tube–spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol. 2012:160(4):1940–1955. 10.1104/pp.112.199729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen MH, Willats WG, Knox JP. Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr Res. 2003:338(17):1797–1800. 10.1016/S0008-6215(03)00272-6 [DOI] [PubMed] [Google Scholar]
- Costa S, Almeida A, Castro A, Domingues L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol. 2014:5:63. 10.3389/fmicb.2014.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey PA, Subbaiah CC, Parsons RL, Pearce G, Lay FT, Anderson MA, Ryan CA, Bedinger PA. A pollen-specific RALF from tomato that regulates pollen tube elongation. Plant Physiol. 2010:153(2):703–715. 10.1104/pp.110.155457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Lausser A, Marton ML. Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann Bot. 2011:108(4):727–737. 10.1093/aob/mcr017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresselhaus T, Sprunck S, Wessel GM. Fertilization mechanisms in flowering plants. Curr Biol. 2016:26(3):R125–R139. 10.1016/j.cub.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Li X, Chen J, Chen W, Li B, Li C, Wang L, Li J, Zhao X, Lin J, et al. . Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc Natl Acad Sci U S A. 2016:113(51):E8326–E8334. 10.1073/pnas.1609626113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Liu MJ, Kita D, Jordan SS, Yeh FJ, Yvon R, Carpenter H, Federico AN, Garcia-Valencia LE, Eyles SJ, et al. . FERONIA controls pectin- and nitric oxide-mediated male-female interaction. Nature. 2020:579(7800):561–566. 10.1038/s41586-020-2106-2 [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004:5(1):113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar NM, Haupt S, Thow G, Boevink P, Chapman S, Oparka K. High-throughput viral expression of cDNA-green fluorescent protein fusions reveals novel subcellular addresses and identifies unique proteins that interact with plasmodesmata. Plant Cell. 2003:15(7):1507–1523. 10.1105/tpc.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Liu C, Fu R, Zhang M, Li H, Shen L, Wei Q, Sun X, Xu L, Ni B, et al. . LORELEI-LIKE GPI-ANCHORED PROTEINS 2/3 regulate pollen tube growth as chaperones and coreceptors for ANXUR/BUPS receptor kinases in Arabidopsis. Mol Plant. 2019:12(12):1612–1623. 10.1016/j.molp.2019.09.004 [DOI] [PubMed] [Google Scholar]
- Galindo-Trigo S, Gray JE, Smith LM. Conserved roles of CrRLK1L receptor-like kinases in cell expansion and reproduction from algae to angiosperms. Front Plant Sci. 2016:7:1269. 10.3389/fpls.2016.01269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Wang C, Xi Y, Shao Q, Hou C, Li L, Luan S. RALF signaling pathway activates MLO calcium channels to maintain pollen tube integrity. Cell Res. 2023:33(1):71–79. 10.1038/s41422-022-00754-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Wang C, Xi Y, Shao Q, Li L, Luan S. A receptor-channel trio conducts Ca(2+) signalling for pollen tube reception. Nature. 2022:607(7919):534–539. 10.1038/s41586-022-04923-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, Garcia-Valencia LE, Zhong S, et al. . Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science. 2017:358(6370):1596–1600. 10.1126/science.aao3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Cheung AY, Qu LJ. Pollen tube integrity regulation in flowering plants: insights from molecular assemblies on the pollen tube surface. New Phytol. 2019a:222(2):687–693. 10.1111/nph.15645 [DOI] [PubMed] [Google Scholar]
- Ge Z, Dresselhaus T, Qu LJ. How CrRLK1L receptor complexes perceive RALF signals. Trends Plant Sci. 2019b:24(11):978–981. 10.1016/j.tplants.2019.09.002 [DOI] [PubMed] [Google Scholar]
- Ge Z, Zhao Y, Liu MC, Zhou LZ, Wang L, Zhong S, Hou S, Jiang J, Liu T, Huang Q, et al. . LLG2/3 are co-receptors in BUPS/ANX-RALF signaling to regulate Arabidopsis pollen tube integrity. Curr Biol. 2019c:29(19):3256–3265 e3255. 10.1016/j.cub.2019.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonneau M, Desprez T, Martin M, Doblas VG, Bacete L, Miart F, Sormani R, Hematy K, Renou J, Landrein B, et al. . Receptor kinase THESEUS1 is a rapid alkalinization factor 34 receptor in Arabidopsis. Curr Biol. 2018:28(15):2452–2458.e4. 10.1016/j.cub.2018.05.075 [DOI] [PubMed] [Google Scholar]
- Gronnier J, Franck CM, Stegmann M, DeFalco TA, Abarca A, von Arx M, Dünser K, Lin W, Yang Z, Kleine-Vehn J, et al. . Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. Elife. 2022:11:e74162. 10.7554/eLife.74162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science. 2014:343(6169):408–411. 10.1126/science.1244454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herger A, Dunser K, Kleine-Vehn J, Ringli C. Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr Biol. 2019:29(17):R851–R858. 10.1016/j.cub.2019.07.039 [DOI] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell. 2005:17(2):584–596. 10.1105/tpc.104.027631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Harper JF, Palanivelu R. A fruitful journey: pollen tube navigation from germination to fertilization. Annu Rev Plant Biol. 2019:70(1):809–837. 10.1146/annurev-arplant-050718-100133 [DOI] [PubMed] [Google Scholar]
- Julca I, Ferrari C, Flores-Tornero M, Proost S, Lindner AC, Hackenberg D, Steinbachová L, Michaelidis C, Gomes Pereira S, Misra CS, et al. . Comparative transcriptomic analysis reveals conserved programmes underpinning organogenesis and reproduction in land plants. Nat Plants. 2021:7(8):1143–1159. 10.1038/s41477-021-00958-2 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kim JH, Hong WJ, Kim EY, Kim MH, Lee SK, Min CW, Kim ST, Park SK, Jung KH, et al. . Rice pollen-specific OsRALF17 and OsRALF19 are essential for pollen tube growth. J Integr Plant Biol. 2023:65(9):2218–2236. 10.1111/jipb.13508 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Jeon BW, Oh E, Seo PJ, Kim J. Peptide signaling during plant reproduction. Trends Plant Sci. 2021:26(8):822–835. 10.1016/j.tplants.2021.02.008 [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta. 1990:181(4):512–521. 10.1007/BF00193004 [DOI] [PubMed] [Google Scholar]
- Lan Z, Song Z, Wang Z, Li L, Liu Y, Zhi S, Wang R, Wang J, Li Q, Bleckmann A, et al. . Antagonistic RALF peptides control an intergeneric hybridization barrier on Brassicaceae stigmas. Cell. 2023:186(22):4773–4787.e12. 10.1016/j.cell.2023.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014:7(9):1494–1496. 10.1093/mp/ssu044 [DOI] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, Li X, Li X, Zhu S, Wang L, Wang Y, Liao H, Luan S, et al. . EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol. 2018:16(10):e2006340. 10.1371/journal.pbio.2006340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chen H, Alotaibi SS, Pěnčík A, Adamowski M, Novák O, Friml J. RALF1 peptide triggers biphasic root growth inhibition upstream of auxin biosynthesis. Proc Natl Acad Sci U S A. 2022:119(31):e2121058119. 10.1073/pnas.2121058119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Shen L, Xiao Y, Vyshedsky D, Peng C, Sun X, Liu Z, Cheng L, Zhang H, Han Z, et al. . Pollen PCP-B peptides unlock a stigma peptide-receptor kinase gating mechanism for pollination. Science. 2021:372(6538):171–175. 10.1126/science.abc6107 [DOI] [PubMed] [Google Scholar]
- Liu X, Castro C, Wang Y, Noble J, Ponvert N, Bundy M, Hoel C, Shpak E, Palanivelu R. The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell. 2016:28(5):1035–1052. 10.1105/tpc.15.00703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Loraine AE, McCormick S, Estrada A, Patel K, Qin P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013:162(2):1092–1109. 10.1104/pp.112.211441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A. Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol. 2012:169(8):807–815. 10.1016/j.jplph.2012.01.019 [DOI] [PubMed] [Google Scholar]
- Matos JL, Fiori CS, Silva-Filho MC, Moura DS. A conserved dibasic site is essential for correct processing of the peptide hormone AtRALF1 in Arabidopsis thaliana. FEBS Lett. 2008:582(23–24):3343–3347. 10.1016/j.febslet.2008.08.025 [DOI] [PubMed] [Google Scholar]
- Mecchia MA, Rövekamp M, Giraldo-Fonseca A, Meier D, Gadient P, Vogler H, Limacher D, Bowman JL, Grossniklaus U. The single Marchantia polymorpha FERONIA homolog reveals an ancestral role in regulating cellular expansion and integrity. Development. 2022:149(19):dev200580. 10.1242/dev.200580 [DOI] [PubMed] [Google Scholar]
- Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martinez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, et al. . RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science. 2017:358(6370):1600–1603. 10.1126/science.aao5467 [DOI] [PubMed] [Google Scholar]
- Mingossi FB, Matos JL, Rizzato AP, Medeiros AH, Falco MC, Silva-Filho MC, Moura DS. SacRALF1, a peptide signal from the grass sugarcane (Saccharum spp.), is potentially involved in the regulation of tissue expansion. Plant Mol Biol. 2010:73(3):271–281. 10.1007/s11103-010-9613-8 [DOI] [PubMed] [Google Scholar]
- Moussu S, Broyart C, Santos-Fernandez G, Augustin S, Wehrle S, Grossniklaus U, Santiago J. Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc Natl Acad Sci U S A. 2020:117(13):7494–7503. 10.1073/pnas.2000100117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussu S, Lee HK, Haas KT, Broyart C, Rathgeb U, De Bellis D, Levasseur T, Schoenaers S, Fernandez GS, Grossniklaus U, et al. . Plant cell wall patterning and expansion mediated by protein-peptide-polysaccharide interaction. Science. 2023:382(6671):719–725. 10.1126/science.adi4720 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D. Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 2005:5(1):22. 10.1186/1471-2229-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmüller R, Tseng YH, Gandhi A. Signals and their perception for remodelling, adjustment and repair of the plant cell wall. Int J Mol Sci. 2023:24(8):7417. 10.3390/ijms24087417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa ST, Kessler SA. Update on signaling pathways regulating polarized intercellular communication in Arabidopsis reproduction. Plant Physiol. 2023:193(3):1732–1744. 10.1093/plphys/kiad414 [DOI] [PubMed] [Google Scholar]
- Okuda S, Suzuki T, Kanaoka MM, Mori H, Sasaki N, Higashiyama T. Acquisition of LURE-binding activity at the pollen tube tip of Torenia fournieri. Mol Plant. 2013:6(4):1074–1090. 10.1093/mp/sst050 [DOI] [PubMed] [Google Scholar]
- Oliveros JC, Franch M, Tabas-Madrid D, San-Leon D, Montoliu L, Cubas P, Pazos F. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016:44(W1):W267–W271. 10.1093/nar/gkw407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr. RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci U S A. 2001:98(22):12843–12847. 10.1073/pnas.201416998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rößling A-K, Dünser K, Kalmbach L, Barbez E, Kleine-Vehn J. Pectin methylesterase activity is required for RALF1 peptide signalling output. bioRxiv 553913. 10.1101/2023.08.18.553913, 19 August 2023, preprint: not peer reviewed. [DOI]
- Rubinstein AL, Broadwater AH, Lowrey KB, Bedinger PA. Pex1, a pollen-specific gene with an extensin-like domain. Proc Natl Acad Sci U S A. 1995a:92(8):3086–3090. 10.1073/pnas.92.8.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AL, Marquez J, Suarez-Cervera M, Bedinger PA. Extensin-like glycoproteins in the maize pollen tube wall. Plant Cell. 1995b:7(12):2211–2225. 10.2307/3870163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenaers S, Lee HK, Gonneau M, Faucher E, Levasseur T, Akary E, Claeijs N, Moussu S, Broyart C, Balcerowicz D, et al. A pectin-binding peptide with a structural and signaling role in the assembly of the plant cell wall. bioRxiv 554416. 10.1101/2023.08.23.554416, 23 August 2023, preprint: not peer reviewed. [DOI]
- Scholz P, Anstatt J, Krawczyk HE, Ischebeck T. Signalling pinpointed to the tip: the complex regulatory network that allows pollen tube growth. Plants (Basel). 2020:9(9):1098. 10.3390/plants9091098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber DN, Bantin J, Dresselhaus T. The MADS box transcription factor ZmMADS2 is required for anther and pollen maturation in maize and accumulates in apoptotic bodies during anther dehiscence. Plant Physiol. 2004:134(3):1069–1079. 10.1104/pp.103.030577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein KA, Moskal WA Jr, Wu HC, Underwood BA, Graham MA, Town CD, VandenBosch KA. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007:51(2):262–280. 10.1111/j.1365-313X.2007.03136.x [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science. 2017:355(6322):287–289. 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- Subramanian B, Gao S, Lercher MJ, Hu S, Chen W-H. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019:47(W1):W270–W275. 10.1093/nar/gkz357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997:25(24):4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr Res. 2009:344(14):1858–1862. 10.1016/j.carres.2008.11.010 [DOI] [PubMed] [Google Scholar]
- Wang W, Wang L, Chen C, Xiong G, Tan XY, Yang KZ, Wang ZC, Zhou Y, Ye D, Chen LQ. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J Exp Bot. 2011:62(14):5161–5177. 10.1093/jxb/err221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang K, Yin G, Liu X, Liu M, Cao N, Duan Y, Gao H, Wang W, Ge W, et al. . Pollen-expressed leucine-rich repeat extensins are essential for pollen germination and growth. Plant Physiol. 2018:176(3):1993–2006. 10.1104/pp.17.01241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009:25(9):1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Stegmann M, Han Z, DeFalco TA, Parys K, Xu L, Belkhadir Y, Zipfel C, Chai J. Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature. 2019:572(7768):270–274. 10.1038/s41586-019-1409-7 [DOI] [PubMed] [Google Scholar]
- Yu Y, Chakravorty D, Assmann SM. The G protein beta-subunit, AGB1, interacts with FERONIA in RALF1-regulated stomatal movement. Plant Physiol. 2018:176(3):2426–2440. 10.1104/pp.17.01277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li L, Wang Z, Ge Z, Li Q, Bleckmann A, Wang J, Song Z, Shi Y, Liu T, et al. . RALF peptide signaling controls the polytubey block in Arabidopsis. Science. 2022:375(6578):290–296. 10.1126/science.abl4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LZ, Dresselhaus T. Multiple roles of ROS in flowering plant reproduction. Adv Bot Res. 2023:105:139–177. 10.1016/bs.abr.2022.10.002 [DOI] [Google Scholar]
- Zhou L-Z, Juranic M, Dresselhaus T. Germline development and fertilization mechanisms in maize. Mol Plant. 2017:10(3):389–401. 10.1016/j.molp.2017.01.012 [DOI] [PubMed] [Google Scholar]
- Zhu S, Martínez Pacheco J, Estevez JM, Yu F. Autocrine regulation of root hair size by the RALF-FERONIA-RSL4 signaling pathway. New Phytol. 2020:227(1):45–49. 10.1111/nph.16497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.