Abstract
Gfi-1 is a cellular proto-oncogene that was identified as a target of provirus integration in T-cell lymphoma lines selected for interleukin-2 (IL-2) independence in culture and in primary retrovirus-induced lymphomas. Gfi-1 encodes a zinc finger protein that functions as a transcriptional repressor. Here we show that Gfi-1B, a Gfi-1 related gene expressed in bone marrow and spleen, also encodes a transcriptional repressor. IL-6-induced G1 arrest and differentiation of the myelomonocytic cell line M1 were linked to the downregulation of Gfi-1B and the parallel induction of the cyclin-dependent kinase inhibitor p21WAF1. Experiments addressing the potential mechanism of the apparent coordinate regulation of these genes revealed that Gfi-1B represses p21WAF1 directly by binding to a high-affinity site at −1518 to −1530 in the p21WAF1 promoter. Forced expression of Gfi-1B, but not of Gfi-1B deletion mutants lacking the repressor domain, blocked the IL-6-mediated induction of p21WAF1 and inhibited G1 arrest and differentiation. We conclude that Gfi-1B is a direct repressor of the p21WAF1 promoter, the first such repressor identified to date, and that sustained expression of Gfi-1B blocks IL-6-induced G1 arrest and differentiation of M1 cells perhaps because it prevents p21WAF1 induction by IL-6.
Hematopoiesis is a process that takes place in the bone marrow throughout the life of an individual. During this process a small number of hematopoietic stem cells respond to microenvironmental cues to either divide and self-renew or differentiate into hematopoietic progenitors committed to specic hematopoietic lineages. The committed hematopoietic progenitors, in turn, also undergo self-renewal or terminal differentiation. The maintenance of the hematopoietic stem cells and their selection, commitment, and maturation along different hematopoietic lineages depend on cell-to-cell and cell-to-stroma interactions, secreted cytokines, and intracellular signaling molecules (45, 46). The molecular mechanisms involved in regulating hematopoietic cell commitment and differentiation can be addressed in differentiating hematopoietic tissues in intact animals (64) and in cell lines that can be induced to differentiate (35, 38). With both systems, a number of molecules, including growth factors, receptors, and transcription factors, have been identified and shown to contribute to hematopoiesis in a hierarchical order (10, 29, 42, 65).
The myelomonocytic cell line M1 undergoes G1 arrest and differentiation following exposure to interleukin-6 (IL-6) or leukemia inhibitory factor (15, 50). During this process the expression of several signaling molecules is altered. c-myb is downregulated within 3 h from the start of the exposure to IL-6 or leukemia inhibitory factor (26). This is followed by the downregulation of c-myc (25). Overexpression of either c-myb or c-myc inhibits IL-6-induced differentiation of M1 cells (25, 26, 49), suggesting that the downregulation of these molecules is required for differentiation. Another molecule whose expression changes during differentiation of the M1 cells is the cyclin-dependent kinase inhibitor p21WAF1, which is induced following exposure to IL-6 (57). One factor known to induce p21WAF1 is p53 (12). Since M1 cells, however, do not express p53 (62), the induction of p21WAF1 in these cells is p53 independent. Evidence presented in this report indicates that the p53-independent induction of p21WAF1 in differentiating M1 cells depends on the downregulation of the Gfi-1 homolog Gfi-1B, which functions as a direct repressor of p21WAF1.
Gfi-1 is a cellular proto-oncogene that was identified as a target of provirus integration in retrovirus-induced T-cell-lymphoma lines selected for IL-2 independence in culture and in primary retrovirus-induced lymphomas (16, 47, 48). Our early studies have shown that Gfi-1 encodes a transcriptional repressor (66) whose repressor function depends on a novel repressor domain SNAG (19). Forced expression of Gfi-1 inhibits apoptosis of primary thymocytes and immortalized T cells (20) and abrogates G1 arrest induced by IL-2 withdrawal from IL-2-dependent T-cell lines (16, 20). Gfi-1B was cloned by low-stringency hybridization to a probe derived from the Gfi-1 zinc finger region. Interestingly, Gfi-1B was also found to be a target of provirus integration in a subset of B-cell lymphomas induced by Moloney murine leukemia virus (Mo-MuLV) in Eμ-myc transgenic, pim-1/pim-2 knockout mice (39a).
Here we show that Gfi-1B also encodes a SNAG domain containing a transcriptional repressor and that the p21WAF1 promoter is one of its direct targets. IL-6 treatment of M1 cells results in the downregulation of Gfi-1B and the parallel induction of p21WAF1. The induction of p21WAF1 is a direct consequence of the downregulation of Gfi-1B. Inhibition of the induction of p21WAF1 by the forced expression of Gfi-1B inhibits cell cycle arrest and differentiation of the M1 cells, suggesting that these processes depend on p21WAF1 expression. Although p21WAF1 expression can be induced by a multitude of transcriptional activators (5, 8, 21, 36, 37, 43, 44, 54–56, 58, 64), Gfi-1B appears to be its only repressor known to date.
(This work was carried out to partially fulfill the Ph.D. thesis requirements of B.T. at the University of Pennsylvania School of Medicine.)
MATERIALS AND METHODS
Isolation and sequencing of Gfi-1B cDNA.
A 2.9-kb EcoRI fragment from rat genomic DNA hybridizing to a DNA probe from the zinc finger region of the rat Gfi-1 gene (40% formamide, 6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] at 37°C) was cloned into the lambda phage vector lambda ZapII (Stratagene). A 400-bp, repeat-free PstI fragment isolated from this clone was hybridized to RNA from different mouse tissues and was shown to be expressed primarily in spleen and bone marrow. Screening a murine spleen cDNA library (Stratagene) with the same probe identified seven cDNA clones representing a novel Gfi-1-related gene which was named Gfi-1B. The pBluescript(SK−)/Gfi-1B plasmids generated by in vivo excision (according to the manufacturer’s protocol) were subjected to sequence analysis (Sequenase kit; U.S. Biochemical Corp.) with vector- and Gfi-1B-specific oligonucleotide primers. The sequences obtained were analyzed with the GCG sequence analysis software package (Genetics Computer Group, Madison, Wis.).
Northern and Western blotting.
RNA was isolated from different mouse tissues or cultured cells by the method of Chomczinski and Sacchi (9). Total RNA (15 μg) was electrophoresed in 1% formaldehyde agarose gel. Following transfer to Hybond-N nylon membranes (Amersham), the RNA was hybridized to DNA probes as described in the Results section.
M1 cells before and after exposure to IL-6 were lysed in radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 7.2], 150 mM NaCl, 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 1 mM EDTA), and the resulting lysates were electrophoresed by SDS-10% polyacrylamide gel electrophoresis. Western blots of these lysates were subsequently probed with antibodies to Gfi-1B (1:1,000 dilution) or p21WAF1 (1:250 dilution) [p21(C19); Santa Cruz Biotechnology]. The Gfi-1B antibody was raised as follows. The amino-terminal region of Gfi-1B (amino acids 1 to 158) was tagged at its carboxy terminus with a 6-amino-acid histidine tag and was expressed in Escherichia coli. Protein purified by affinity chromatography with a Nickel column was mixed with Freund adjuvant and injected into rabbits (0.5 mg/rabbit). Antibody-positive sera were collected after three sequential monthly injections.
Bacterial GST-Gfi-1B fusion protein and Gfi-1B DNA binding: random oligonucleotide selection.
To express the Gfi-1B zinc finger domain in E. coli, we amplified the associated DNA sequence by PCR. BamHI and EcoRI restriction sites were introduced at the ends of the amplified DNA, which was then digested with BamHI and EcoRI and subcloned in frame with glutathione S-transferase (GST) in the pGEX vector (Pharmacia).
Bacterial cells transformed with the resulting expression construct pGEX-Gfi-1B were grown to log phase and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (Sigma). At 2 h following induction, the bacteria were pelleted and sonicated in ice-cold phosphate-buffered saline (PBS) containing the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (Sigma) (1 mM). The lysates were then clarified by centrifugation at 15,000 × g and mixed with glutathione-linked agarose beads (Sigma).
To identify the DNA-binding consensus for Gfi-1B, a random double-stranded oligonucleotide pool was generated as described previously (66). A 1-μg portion of the mixture of double-stranded oligonucleotides containing an 18-bp random sequence at the center was incubated with 5 μg of the GST-Gfi-1B fusion protein attached to glutathione-linked agarose beads. Incubation was carried out at 25°C in binding buffer (25 mM HEPES [pH 7.5], 100 mM KCl, 0.1 mM ZnSO4, 10 mM MgCl2, 0.1% Nonidet P-40, 1 mM dithiothreitol [DTT], 5% glycerol, poly[dI-dC] [0.2 mg/ml], bovine serum albumin [0.2 mg/ml]) for 30 min. The beads were then centrifuged and washed three times with binding buffer and then boiled for 5 min in water. The oligonucleotide mixture eluted by boiling was used as a template for PCR amplification. After four rounds of selection and amplification, the PCR products were cloned into pBluescript. Plasmid DNA from 80 individual E. coli transformants was sequenced to determine the binding consensus for Gfi-1B.
Cell lines and cultures.
M1 cells (TIB 192) and NIH 3T3 cells (TIB 163) were obtained from the American Type Culture Collection (Rockville, Md.), while the 10-1 cells (23) were provided by J. Sherley (Fox Chase Cancer Center). 10-1 cells and NIH 3T3 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin, 1% streptomycin, and 1% kanamycin, while M1 cells were grown in RPMI supplemented with 10% endotoxin-free fetal bovine serum (Gibco BRL) and 1% penicillin, 1% streptomycin, and 1% kanamycin. M1 cells were seeded at 2 × 105 cells per ml prior to exposure to murine recombinant IL-6 (50 ng/ml) (R&D Systems).
Expression and reporter constructs.
To generate pCMV5/Gfi-1B, the 1.7-kb full-length Gfi-1B cDNA was released from pBluescript(SK−) by double digestion with EcoRI and SalI and then subcloned between EcoRI and SalI in the CMV5 expression vector (1) or between EcoRI and XhoI in the expression vector pcDNA3 (Invitrogen).
To generate the VP16/Gfi-1B chimera, the sequence encoding the acidic activation domain of VP16 (60), including the Kozak consensus, was amplified by PCR. EcoRI and BamHI restriction sites were introduced via the PCR reaction at the 5′ and 3′ ends of the amplified sequence. The amplified DNA was then digested with EcoRI and BamHI. The sequence encoding the zinc finger domain of Gfi-1B was also amplified and digested with EcoRI and BamHI. The two PCR-amplified DNA fragments were ligated together at the BamHI site, and the resulting hybrid DNA fragment was cloned into the EcoRI site of the expression vector pCMV5.
To generate the ΔSNAG mutant (which lacks the first 20 amino acids of Gfi-1B), an oligonucleotide homologous to the zinc finger region 3′ of the HindIII site at nucleotide 765 and another oligonucleotide containing an EcoRI restriction site, the Gfi-1B ATG, and 15 nucleotides encoding amino acids 21 to 25 of Gfi-1B were used in an amplification reaction with the Gfi-1B cDNA as template. The product of the reaction was digested with EcoRI and HindIII and ligated into a pDNA3-based construct containing the zinc finger region of the Gfi-1B cDNA from the HindIII site to the unique BamHI site at nucleotide 1390 in the 3′-untranslated region of the gene. The delta amino terminus (ΔNter) construct was made in a similar manner with the same first oligonucleotide (in the zinc finger region) and another oligonucleotide containing an EcoRI restriction site, the Gfi-1B ATG, and 15 nucleotides encoding amino acids 164 to 168. The product of the amplification reaction was digested with EcoRI and HindIII and ligated into a pDNA3-based construct containing the HindIII-to-BamHI fragment of Gfi-1B. All clones were sequenced to ensure that the final product contained no additional mutations.
Transfections.
NIH 3T3 cells were transiently transfected using Lipofectamine reagent (Gibco BRL) as suggested by the manufacturer. About 1.8 × 105 cells were seeded in 35-mm-diameter petri dishes 16 h prior to transfection. Reporter plasmids (1.8 μg) and 100 ng of pCMV5 empty vector, pCMV5/Gfi-1B, or pCMV5/VP16/Gfi-1B were cotransfected.
10-1 cells were transfected by the calcium phosphate precipitation method (17). About 3.5 × 105 cells were seeded in 60-mm-diameter petri dishes prior to transfection. Portions (1.5 μg) of different p21WAF1 promoter constructs and 3 μg of pCMV5/Gfi-1B or 0.25 μg of p21WAF1 promoter constructs and 4.25 μg of pCMV5/VP16/Gfi-1B were cotransfected.
Stable transfections were performed by electroporation (19). pcDNA3 empty vector and pcDNA3/Gfi-1B were prepared with Qiagen columns (QIAGEN, Inc.). The plasmid DNAs were gel purified following linearization by EcoRI digestion. M1 cells were seeded 16 h prior to transfection. Electroporation of 107 M1 cells, suspended in 250 μl of medium (without antibiotics), was carried out in 0.45-μm cuvettes (Bio-Rad) with 20 μg of DNA. Samples were pulsed at 960 μF and 260 mV and placed in culture for 48 h before selection. The cells were then diluted and seeded at 5 × 104 cells per ml in growth medium containing 400 μg of geneticin per ml (G418 sulfate; Gibco BRL) and divided into aliquots in 24-well plates (5 × 104 cells/well). After 3 to 4 weeks, cultures from wells containing surviving cells were expanded and subcloned in agar in the presence of G418. The Gfi-1B/ΔSNAG and Gfi-1B/ΔNter transfectants were maintained as mass cultures. Fully selected transfectants were maintained in G418 at 200 μg/ml.
Chloramphenicol acetyltransferase (CAT) assays.
NIH 3T3 or 10-1 cells were cotransfected with Gfi-1B, VP16/Gfi-1B, and CAT reporter constructs as described above. Cell lysates were collected 40 h later by four consecutive freeze-thaw cycles and were normalized for transfection efficiency by a β-galactosidase assay (6). Lysates were analyzed for CAT activity by thin-layer chromatography (61).
In vitro translation, preparation of M1 nuclear extracts, and electrophoretic mobility shift assays (EMSAs).
Gfi-1B was in vitro transcribed and translated by the TNT T7-coupled reticulocyte lysate system (Promega). Nuclear extracts were prepared from untreated and IL-6-treated M1 cells collected at 36 h after the start of the IL-6 treatment by the protocol of Kramer and Keller (30). Briefly, 1 × 108 to 2 × 108 cells were collected in 25 ml of cold PBS and pelleted. After being washed with 1 ml of buffer A [10 mM HEPES-KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT], the cells were resuspended in 0.5 ml of the same buffer and they were forced several times through a 25-gauge-by-0.625-inch hypodermic needle. Collected cell nuclei were resuspended in 0.2 ml of buffer C (20 mM HEPES-KOH [pH 7.9], 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, pepsin [1 μg/ml]). The nuclear extracts were dialyzed against 400 ml of buffer D [20 mM HEPES-KOH (pH 7.9), 20% glycerol, 0.1 M NaCl, 0.2 mM EDTA, 0.5 mM DTT]. The protein concentration of the nuclear extracts was determined by the Bradford assay (Bio-Rad).
EMSAs were done with a double-stranded oligonucleotide probe containing the Gfi-1B binding site (5′-CCGAAGTACCGTGATTTCAGGCATGCAC-3′, annealed to its complementary strand). A mutant oligonucleotide (AATC to GGTC) was also generated. The wild-type double-stranded oligonucleotides were end labeled with [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). One-tenth of the Gfi-1B translated in vitro from RNA derived from 1 μg of pcDNA3/Gfi-1B or 12 μg of M1 nuclear extracts was incubated with 104 cpm of the end-labeled oligonucleotides (20 min at room temperature) in a 20-μl final volume in binding buffer [12 mM HEPES (pH 7.6), 12% glycerol, 4 mM Tris, 60 mM KCl, 1 mM EDTA, 1 mM DTT, poly(dI-dC) (2 μg)]. To compete the protein binding, excess wild-type or mutant double-stranded oligonucleotides were incubated with the in vitro-translated product (100- to 200-fold) or the M1 nuclear extracts (100-fold) for 15 min prior to the addition of the labeled probe. Samples were then electrophoresed at 25 mA in nondenaturing polyacrylamide gels at 4°C for 3 h in 0.5× TBE buffer (45 mM Tris-borate–1 mM EDTA [pH 8.0]) (66).
Cell cycle analysis.
Parental M1, M1/vector, and M1/Gfi-1B cells were seeded at 2 × 105 cells per ml. After IL-6 treatment (50 ng/ml), the cells were harvested at sequential time points as indicated below. Harvested cells were washed with PBS and resuspended in fluorescence-activated cell sorter buffer (3.4 mM sodium citrate, 10 mM NaCl, 0.1% Nonidet P-40, 75 μM ethidium bromide) (51). The DNA content of these cells was then determined by flow cytometry. The data were analyzed with the CellQuest package (Becton Dickinson).
M1 cell attachment to the surface of the petri dish.
Parental M1, M1/vector, and M1/Gfi-1B cells were seeded at 2 × 105 cells per ml, and they were treated with IL-6 (50 ng/ml). After 48 h, the medium and the suspension cells were removed, the dishes were washed twice with PBS, and the cells were photographed with an inverted microscope (Nikon).
Mac-1 expression.
Parental M1, M1/vector, and M1/Gfi-1B cells were collected at 96 h following IL-6 treatment. Expression of Mac-1 was then determined by staining the cells with the anti-mouse CD11b monoclonal antibody M1/70 (Pharmigen), which is directed against the Mac-1 αm chain. This chain is known to be induced during differentiation along the macrophage lineage (4). Stained cells were analyzed by flow cytometry with a fluorescence-activated cell sorter and the CellQuest software package (Becton Dickinson).
Nucleotide sequence accession number.
The cDNA sequence for Gfi-1B has been submitted to GenBank under accession no. AF017275.
RESULTS
Gfi-1B encodes a Gfi-1-related zinc finger protein that is expressed in bone marrow and spleen.
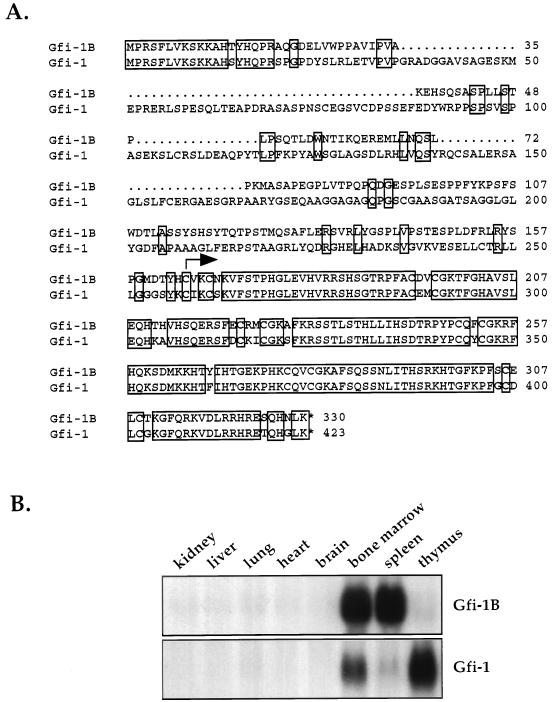
With a DNA probe derived from the zinc finger region of Gfi-1 (16) and low-stringency hybridization, we cloned three novel zinc finger protein-encoding genes from rat DNA. A full-length cDNA clone of one of these genes, Gfi-1B, isolated from a murine spleen cDNA library (Stratagene), encodes a 330-amino-acid zinc finger protein which is 97% identical to Gfi-1 in the zinc finger region. The homology between the two proteins, amino-terminal to their zinc finger domains, is restricted to a 20-amino-acid peptide which defines the Gfi-1 repressor domain SNAG (19) (Fig. 1A). SNAG is shared by Gfi-1 and Gfi-1B, the members of the Snail/Slug family of proteins found in vertebrates (19), the zinc finger protein IA-1, which is overexpressed in neuroendocrine neoplasms (32), and the homeobox protein Gsh-1, which is involved in pituitary development (34).
FIG. 1.
Gfi-1B encodes a Gfi-1-related zinc finger protein that is expressed in bone marrow and spleen. (A) Predicted amino acid sequence of Gfi-1B and comparison with Gfi-1. The two proteins exhibit a high degree of homology in the SNAG repressor domain (amino acids 1 to 19) and the zinc finger domain (amino acids 164 to 329). The arrow indicates the start of the zinc finger region. (B) Tissue distribution of Gfi-1B and Gfi-1. Gfi-1B is expressed in bone marrow and spleen, whereas Gfi-1 is expressed primarily in bone marrow and thymus.
Gfi-1 is expressed in bone marrow, thymus, and to a lesser extent in spleen (Fig. 1B), as well as in testes (16). Hybridization of a Gfi-1B cDNA probe derived from the unique non-zinc finger region of the gene to normal RNA from different mouse tissues revealed that Gfi-1B is expressed primarily in bone marrow and spleen as a single 1.8-kb RNA (Fig. 1B). The two genes differ with regard to their chromosomal locations. Thus, in mice Gfi-1 maps to chromosome 5 (3), whereas Gfi-1B maps to chromosome 2 near B-myc (59a). Similarly, in humans Gfi-1 maps to chromosome 1p22 (3), whereas Gfi-1B maps within a sequenced cosmid (GenBank accession number AC000393) derived from human chromosome 9q34.
Gfi-1B is a sequence-specific DNA-binding protein that functions as a transcriptional repressor.
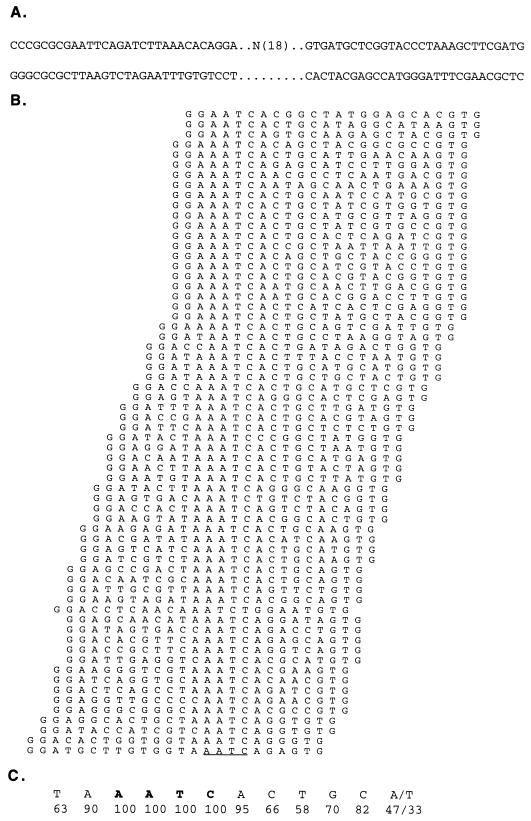
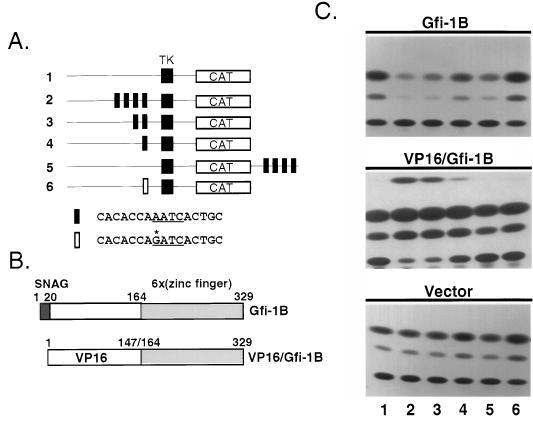
The sequence homologies between Gfi-1B and Gfi-1 suggested that the latter may also be a sequence-specific DNA-binding protein and that it may also function as a transcriptional repressor. To address the first hypothesis, we used the random oligonucleotide binding and selection strategy (66). The oligonucleotide library selected after four rounds of Gfi-1B binding was cloned into pBluescript, and the isolated clones were sequenced. Of 80 sequenced clones, all contained at least one AATC motif. Aligning the sequences of 64 clones with one or two AATC motifs revealed that the Gfi-1B DNA-binding consensus sequence (Fig. 2 and Table 1) is virtually identical to the DNA-binding consensus sequence of Gfi-1 (66). To examine whether Gfi-1B also functions as a transcriptional repressor, NIH 3T3 cells, which express neither Gfi-1 nor Gfi-1B (data not shown), were cotransfected with a CMV5/Gfi-1B expression construct and a CAT reporter construct. The CAT gene in the latter construct was under the control of a minimal thymidine kinase promoter from herpes simplex virus with one to four concatamerized Gfi-1/Gfi-1B binding sites upstream or four sites downstream of the reporter (19). Alternatively, this construct contained a single mutant site (GATC as opposed to AATC) upstream of the reporter (Fig. 3A). The results (Fig. 3C, upper panel) showed that Gfi-1B represses all constructs containing wild-type Gfi-1B binding sites but has no effect on the construct containing the mutant site. The strength of the repression was proportional to the number of Gfi-1B binding sites in the reporter construct. Cotransfection of the same reporter constructs with an expression construct of a VP16/Gfi-1B chimera (Fig. 3B) revealed that the chimera activates the reporters that are repressed by wild-type Gfi-1B (Fig. 3C, middle panel). These results collectively indicate that Gfi-1B is an additive, distance-independent, active transcriptional repressor which functions by binding to the same DNA sequence as Gfi-1.
FIG. 2.
Definition of the Gfi-1B binding site. (A) Double-stranded DNA oligonucleotides used for Gfi-1B binding and PCR amplification. The original oligonucleotide pool contained an 18-bp random sequence flanked by fixed sequences that were used for PCR amplification. (B) Alignment of 64 sequences of the selected oligonucleotides containing one AATC motif (underlined). (C) Deduced Gfi-1B binding site. The numbers show the frequencies of individual bases at each position.
TABLE 1.
Derivation of the Gfi-1B consensus binding site
| Totala | No. of oligonucleotides withb:
|
% of oligonucleotides withc:
|
Consensus binding site 5′ to 3′ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | C | G | T | A | C | G | T | ||
| 23 | 5 | 7 | 5 | 6 | 22 | 30 | 22 | 26 | |
| 27 | 10 | 7 | 5 | 5 | 37 | 26 | 19 | 19 | |
| 32 | 10 | 6 | 11 | 5 | 31 | 19 | 34 | 16 | |
| 35 | 11 | 6 | 9 | 9 | 31 | 17 | 26 | 26 | |
| 37 | 8 | 9 | 11 | 9 | 22 | 24 | 30 | 24 | |
| 40 | 13 | 9 | 10 | 8 | 33 | 23 | 25 | 20 | |
| 43 | 2 | 13 | 1 | 27 | 5 | 30 | 2 | 63 | T |
| 61 | 55 | 5 | 0 | 1 | 90 | 8 | 0 | 2 | A |
| 64 | 0 | 0 | 0 | 64 | 100 | 0 | 0 | 0 | A |
| 64 | 64 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | A |
| 64 | 0 | 0 | 0 | 64 | 0 | 0 | 0 | 100 | T |
| 64 | 0 | 64 | 0 | 0 | 0 | 100 | 0 | 0 | C |
| 64 | 61 | 1 | 0 | 2 | 95 | 2 | 0 | 3 | A |
| 64 | 4 | 42 | 18 | 0 | 6 | 66 | 28 | 0 | C |
| 64 | 10 | 2 | 15 | 37 | 16 | 3 | 23 | 58 | T |
| 64 | 6 | 6 | 45 | 7 | 9 | 9 | 70 | 11 | G |
| 60 | 5 | 49 | 1 | 5 | 8 | 82 | 2 | 8 | C |
| 57 | 27 | 4 | 7 | 19 | 47 | 7 | 12 | 33 | A/T |
| 51 | 18 | 12 | 7 | 14 | 35 | 24 | 14 | 27 | |
| 45 | 10 | 9 | 15 | 11 | 22 | 20 | 33 | 24 | |
| 40 | 7 | 12 | 13 | 8 | 18 | 30 | 33 | 20 | |
| 36 | 9 | 5 | 13 | 11 | 25 | 14 | 36 | 31 | |
| 29 | 9 | 8 | 5 | 7 | 31 | 28 | 17 | 24 | |
| 23 | 6 | 4 | 7 | 6 | 26 | 17 | 30 | 26 | |
Total number of selected oligonucleotides at a given position.
Number of selected oligonucleotides that carried that base at a given position.
Percentage of selected oligonucleotides that carried that base at a given position. When the frequency of selection of a base at a given position was statistically significant to a P value of <0.05, the base is identified by a capital letter in the binding site column. Bases selected at a frequency of 100% are in boldface.
FIG. 3.
Gfi-1B represses whereas a VP16/Gfi-1B chimera activates promoters containing a Gfi-1B binding site(s). (A) Schematic diagram of six different CAT reporter constructs. CAT expression by these constructs is under the control of a minimal herpes simplex virus thymidine kinase promoter (construct 1) with one (construct 4), two (construct 3) or four (construct 2) Gfi-1B binding sites placed upstream or four sites placed downstream (construct 5) of the CAT gene. Alternatively, one mutant site with a GATC instead of an AATC core motif was placed upstream of the thymidine kinase promoter (construct 6). (B) Schematic diagram of Gfi-1B and a VP16/Gfi-1B chimera. In this chimera the Gfi-1B amino-terminal region (amino acids 1 to 164) was replaced by the acidic activation domain of VP16 (amino acids 1 to 147). (C) Cotransfection of the six CAT reporter constructs (1.8 μg) shown in panel A with a CMV5/Gfi-1B construct (100 ng) into NIH 3T3 cells results in repression of CAT gene expression (top panel, lanes 2 to 5; the lane number corresponds to the number of the reporter construct used in that lane, as shown in panel A), while cotransfection of the same reporter constructs with the CMV5 empty vector (100 ng) does not (bottom panel). Cotransfection of the reporter constructs with a CMV5 expression construct of the VP16/Gfi-1B chimera (100 ng) shown in panel B induces CAT expression from the same constructs that were repressed by Gfi-1B (middle panel). Mutation of the AATC motif to GATC abrogates both the repression and the induction of CAT expression (lane 6).
G1 arrest and differentiation induced by exposure of the myelomonocytic cell line M1 to IL-6 depend on the IL-6-induced downregulation of Gfi-1B.
Both Gfi-1B and Gfi-1 are expressed in the bone marrow. However, the two genes are differentially expressed in the cells that leave the bone marrow to populate the thymus and the spleen. Thus, thymocytes express mainly Gfi-1, while splenocytes express almost exclusively Gfi-1B (Fig. 1B). Additional hematopoietic-lineage-specific expression studies showed that Gfi-1B is expressed in early hematopoietic cells and that its expression declines with differentiation (44a).
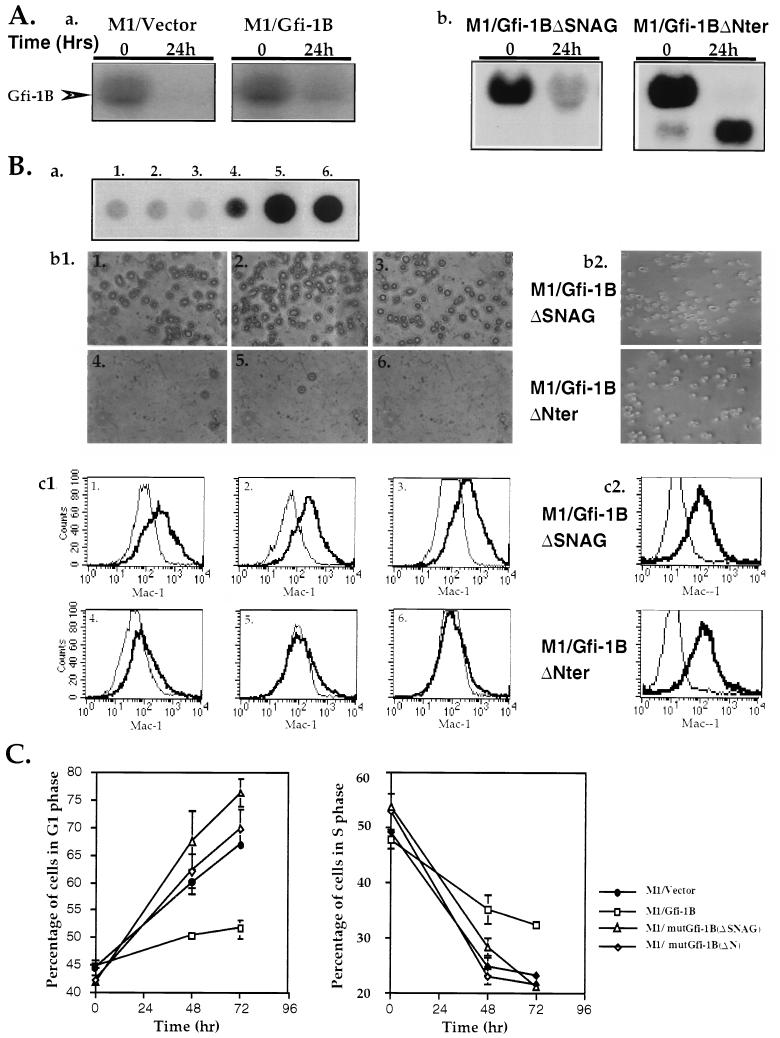
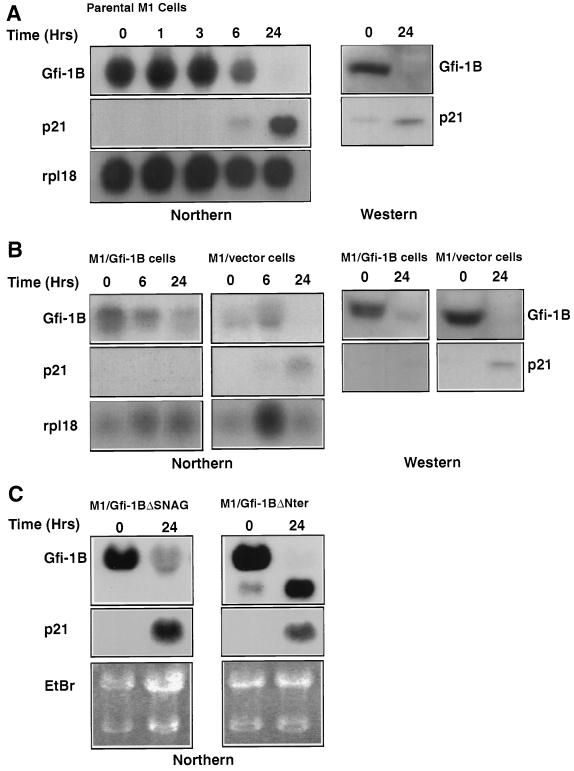
This pattern of expression suggested that Gfi-1B may be involved in the differentiation of hematopoietic cells. To test this hypothesis we first examined whether IL-6-induced differentiation of the myelomonocytic cell line M1 is associated with changes in Gfi-1B expression. The results showed that growing M1 cells express substantial levels of Gfi-1B and that Gfi-1B expression declines to undetectable levels within 24 h, following exposure to IL-6 (Fig. 4Aa, left panel). The downregulation of Gfi-1B could either be the cause or the result of differentiation. To distinguish between these possibilities, we examined whether forced expression of Gfi-1B inhibits IL-6-induced differentiation. Gfi-1B was stably transfected into M1 cells by using a pcDNA3 expression construct. Of 20 transfected M1 cell clones, 3 overexpress Gfi-1B as determined by the sustained expression of the gene following exposure of the clones to IL-6. Fig. 4Aa (right panel) shows the expression of Gfi-1B in one of these clones before and 24 h after the start of IL-6 treatment. Fig. 4Ba shows a dot blot analysis of RNA isolated from wild-type and vector-transfected M1 clones (blots 1 and 2, respectively) and four Gfi-1B-transfected clones at 24 h after exposure to IL-6. Of the latter clones, one (blot 3) does not express Gfi-1B constitutively, whereas the other three (blots 4, 5, and 6) do. The same cell clones were then scored for IL-6-induced differentiation, which was measured by their relative attachment to the surface of the petri dish at 48 h and by the relative expression of Mac-1, a marker of mature macrophages, granulocytes, and natural killer cells (2) at 96 h following exposure to IL-6. Mac-1 expression was determined by immunofluorescence staining and flow cytometry (4). The results revealed that forced expression of Gfi-1B prevented both cell attachment (Fig. 4Bb1) and Mac-1 induction (Fig. 4Bc1). Clone 3, however, which had been transfected with the Gfi-1B construct but did not overexpress Gfi-1B, underwent differentiation in a way similar to that of the parental and vector-transfected cells. Identical results were obtained with three mass cultures of M1 cells infected with the MSCV retrovirus vector (24) or MSCV-based Gfi-1B expression constructs (data not shown).
IL-6-induced differentiation of M1 cells has been linked to the partial G1 arrest which occurs at approximately 48 h following exposure to IL-6 (30). Since forced expression of Gfi-1B blocks differentiation, we examined whether it also inhibits IL-6-induced G1 arrest. To this end, five of the M1 cell clones tested for differentiation (clones 1, 2, 4, 5, and 6) were examined for their cell cycle distribution before and at 48 and 72 h following exposure to IL-6. Cell cycle distribution was determined by ethidium bromide staining and flow cytometry (51). The results (Fig. 4C) showed that forced expression of Gfi-1B indeed impairs the partial G1 arrest induced by IL-6 without changing cell viability.
FIG. 4.
Forced expression of Gfi-1B inhibits IL-6-induced G1 arrest and differentiation of the murine myeloblastic leukemia cell line M1. (Aa) Northern blot of total-cell RNA derived from clones of M1 cells transfected with the pcDNA3 vector (M1/vector) or a pcDNA3 expression construct of Gfi-1B (M1/Gfi-1B) probed with a Gfi-1B cDNA probe. Cells were harvested before (time 0) and 24 h after treatment with IL-6. (Ab) Northern blot of total-cell RNA derived from M1 cells stably transfected with pcDNA3-based expression constructs of Gfi-1BΔSNAG or Gfi-1BΔNter probed with a Gfi-1B cDNA probe. Cells were also harvested before (time 0) and 24 h after treatment with IL-6. (Ba) Gfi-1B expression in six M1 cell clones 24 h after exposure to IL-6. Clone 1 was untransfected parental M1, clone 2 was transfected with pcDNA3, and clones 3 to 6 were transfected with the pcDNA3/Gfi-1B construct. (Bb1 and Bc1) Constitutive expression of Gfi-1B blocks IL-6-induced differentiation of M1 cells. Differentiation was assessed by measuring the relative cell attachment to the surface of the petri dish at 48 h after IL-6 treatment (b1) and Mac-1 expression at 96 h after IL-6 treatment (c1). The superimposed graphs in panel Bc depict fluorescent intensity versus cell number in untreated and IL-6-treated cell cultures. Graphs 1 to 6 in both panels Bb1 and Bc1 correspond to the six M1 clones analyzed for Gfi-1B expression after the IL-6 treatment shown in panel Ba. (Bb2 and Bc2) M1 cells stably transfected with Gfi-1BΔSNAG or Gfi-1BΔNter do not differentiate in response to IL-6 as determined by cell attachment to plastic (Bb2) and by Mac1 expression (Bc2). Identical results were obtained from four independent cultures of M1 cells expressing these mutants. (C) Constitutive expression of Gfi-1B in M1 cells inhibits IL-6-induced G1 arrest. M1/vector, M1/Gfi-1B, M1/mutGfi-1B(ΔSNAG), and M1/mutGfi-1B(ΔN) cells were examined for cell cycle distribution at 0, 48, and 72 h after IL-6 treatment. Each percentage of cells in G1 or S represents an average of three independent experiments.
To determine whether the inhibition of the IL-6-induced differentiation of M1 cells that stably overexpress Gfi-1B depends on the ability of Gfi-1B to repress transcription, we stably transfected M1 cells with pcDNA3-based expression constructs of Gfi-1BΔSNAG and Gfi-1BΔNter. These Gfi-1B mutants harbor deletions of the SNAG domain (amino acids 2 to 20) or the entire amino-terminal region (amino acids 2 to 163), respectively, and lack transcriptional repression activity (59). The transfected cells were then treated with IL-6, and they were examined at the indicated time points for Gfi-1B expression, differentiation, and cell cycle distribution. Expression of Gfi-1B was determined by Northern blotting, differentiation was determined by cell attachment to plastic, and Mac-1 expression and cell cycle distribution were determined by flow cytometry as described above. The results (Fig. 4) revealed that Gfi-1BΔSNAG and Gfi-1BΔNter fail to inhibit the IL-6-induced G1 arrest and differentiation of M1 cells. Therefore, the effects of Gfi-1B on IL-6-induced cell cycle arrest and differentiation of the M1 cells depend on its ability to repress transcription.
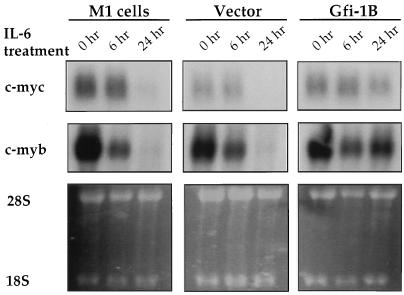
Epistatic relationship of the downregulation of Gfi-1B, c-myc, and c-myb during IL-6-induced differentiation of M1 cells.
At least two other oncogenes, c-myb and c-myc, are known to be downregulated during IL-6-induced differentiation of the M1 cells and their downregulation is necessary for differentiation (25, 26). To map the action of Gfi-1B epistatically relative to c-myb and c-myc, parental as well as vector or Gfi-1B-transfected M1 cells were examined for c-myb or c-myc expression following exposure to IL-6. The results confirmed that both c-myb and c-myc are downregulated during differentiation and that the downregulation of c-myb precedes that of c-myc (Fig. 5). Forced expression of Gfi-1B inhibited the downregulation of c-myc (Fig. 5, upper panel), suggesting that Gfi-1B functions upstream of c-myc. On the other hand, forced expression of Gfi-1B had no effect on the initial drop of c-myb expression occurring within 3 h (26) but blocked the complete repression of c-myb occurring between 6 and 24 h following exposure to IL-6 (Fig. 5, middle panel). This suggests that the downregulation of c-myb during M1 cell differentiation is a complex process accomplished in two steps that are under the control of different regulatory processes. Gfi-1B downregulation precedes the second but not the first wave of c-myb downregulation. Although these data show that Gfi-1B contributes to the regulation of c-myc and c-myb in the context of myelomonocytic cell differentiation, they do not imply that its role in the regulation of these genes is direct.
FIG. 5.
Epistatic relationship of Gfi-1B, c-myc, and c-myb during IL-6-induced differentiation of M1 cells. Northern blots of total RNA from parental M1 cells and pcDNA3 and pcDNA3/Gfi-1B-transfected M1 cells were hybridized to c-myc and c-myb probes. Cells were harvested at 0, 6, and 24 h following exposure to IL-6. Ethidium bromide staining of the RNA is presented as a measure of loading.
IL-6 treatment of M1 cells downregulates Gfi-1B and upregulates p21WAF1 with similar kinetics: forced expression of Gfi-1B inhibits IL-6-mediated induction of p21WAF1.
Progression through the cell cycle depends on the activity of a set of kinases, the cyclin-dependent kinases, which are regulated by binding to their regulatory subunits, the cyclins (18, 41, 52), and by phosphorylation (33). The cyclin–cyclin-dependent kinase active complexes are also regulated by cyclin-dependent kinase inhibitors, which bind either to the cyclin-dependent kinase or the cyclin component of the complexes or both (22, 53). Upregulation of cyclin-dependent kinase inhibitors induces cell cycle arrest (14). Given that the expression of Gfi-1B prevents G1 arrest, the cyclin-dependent kinase (cdk) inhibitors are good candidate targets for Gfi-1B repression. To address this hypothesis, we probed Northern blots of total-cell RNA from untreated and IL-6-treated M1 cells with Gfi-1B, p27KIP1, p16INK4A, and p21WAF1 probes. Northern blots of total-cell RNA derived from M1/Gfi-1B cell lines were probed in parallel. The results showed that the downregulation of Gfi-1B in IL-6-treated M1 cells is associated with the upregulation of p21WAF1 and that the kinetics of the downregulation of Gfi-1B and the upregulation of p21WAF1 are similar (Fig. 6A, left panels). However, the upregulation of p21WAF1 was abrogated in M1 cells transfected with pcDNA3/Gfi-1B and expressing Gfi-1B constitutively. This is shown in Fig. 6B, which presents a repeat of the experiment shown in Fig. 6A with pcDNA3- or pcDNA3/Gfi-1B-transfected cells. The changes in the expression of Gfi-1B and p21WAF1 at the RNA level paralleled similar changes at the protein level (Fig. 6A and B, right panels).
FIG. 6.
Induction of p21WAF1 during IL-6-induced differentiation of M1 cells depends on the downregulation of Gfi-1B. (A, left) Northern blots of total-cell RNA from parental M1 cells harvested at the indicated time points following IL-6 treatment. The blot was hybridized to a probe derived from the unique non-zinc finger region of Gfi-1B (upper panels), a p21WAF1 probe derived from the coding region of the gene (middle panel), and the ribosomal protein rpl18 (lower panel). (A, right) Western blots of lysates from the same cells, harvested at 0 and 24 h after treatment with IL-6 and probed with the anti-Gfi-1B antiserum (upper panel) or with the anti-p21 antibody (lower panel). (B, left) Northern blots of M1 cells stably transfected with pcDNA3 or pcDNA3/Gfi-1B and harvested at the indicated time points following treatment with IL-6. The blots were hybridized to the same Gfi-1B probe (upper panels), p21WAF1 probe (middle panels), or ribosomal protein probe (lower panels) listed for panel A. (B, right) Western blots of lysates from the cells shown on the left side of panel B probed with anti-Gfi-1B (upper panel) or anti-p21WAF1 (lower panel) antibodies. (C) Forced expression of Gfi-1BΔSNAG or Gfi-1BΔNter constructs in M1 cells fails to inhibit IL-6-mediated induction of p21WAF1. Northern blots of M1 cells stably transfected with pcDNA3-based expression constructs of Gfi-1BΔSNAG or Gfi-1BΔNter and harvested at the indicated time points following treatment with IL-6 are shown. The blots were probed with Gfi-1B (upper panels) or p21WAF1 (middle panels) probes. RNA loading was determined by staining the gel with ethidium bromide (lower panel).
Gfi-1BΔSNAG and Gfi-1BΔNter, two Gfi-1B mutants defective in transcriptional repression function, failed to inhibit IL-6-induced G1 arrest and differentiation of M1 cells (Fig. 4). To determine whether these mutants were also defective in preventing induction of p21WAF1, we examined the expression of Gfi-1B and p21WAF1 in Gfi-1BΔSNAG- and Gfi-1BΔNter-transfected M1 cells before and after IL-6 treatment. The results (Fig. 6C) revealed that overexpression of the mutants does not inhibit p21WAF1 induction. These results confirmed that the downregulation of Gfi-1B and the induction of p21WAF1 are linked and suggested that Gfi-1B may be a physiological direct repressor of p21WAF1. Of the other two cyclin-dependent kinase inhibitors whose expression was examined in these cells, p16INK4A was undetectable and p27KIP1 exhibited a pattern of expression that did not correlate with that of Gfi-1B (data not shown).
Gfi-1B represses the p21WAF1 promoter.
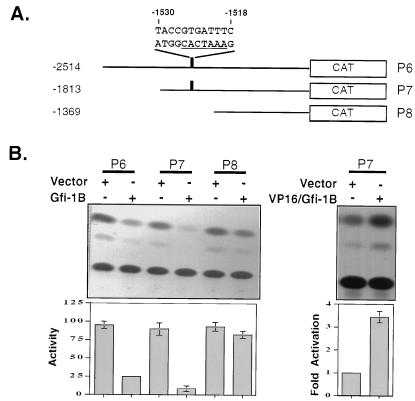
To determine whether the downregulation of p21WAF1 by Gfi-1B is due to direct repression of the p21WAF1 promoter, we cotransfected the p53-deficient mouse embryonic fibroblast cell line 10-1 (23) with a Gfi-1B expression construct and three progressively deleted p21WAF1 promoter-CAT reporter constructs: P6 starting at −2514, P7 starting at −1813, and P8 starting at −1369 from the transcription start site (Fig. 7A) (13). The results showed that Gfi-1B represses the P6 and P7 p21WAF1 promoter constructs but fails to repress the P8 promoter construct (Fig. 7B, left panel), suggesting that the promoter region responsible for the repression maps at −2514 to −1369. Cotransfection of the P7 p21WAF1 reporter construct with the VP16/Gfi-1B chimera (Fig. 3) showed that the P7 promoter which is repressed by Gfi-1B is activated by the fusion protein (Fig. 7B, right panel). These results support the hypothesis that Gfi-1B is a direct repressor of the p21WAF1 promoter.
FIG. 7.
Gfi-1B is a direct repressor of the p21WAF1 promoter. (A) Schematic diagram of sequentially deleted p21WAF1 promoter-CAT reporter constructs. The location and sequence of a putative Gfi-1B binding site at −1530 to −1518 are marked. (B) Gfi-1B represses whereas the VP16/Gfi-1B chimera activates the p21WAF1 promoter. The p53-deficient mouse embryonic fibroblast cell line 10-1 was cotransfected as indicated with the reporter constructs shown in panel A and Gfi-1B (left panels) or the VP16/Gfi-1B chimera (right panels). Repression and activation were quantitatively measured with a phosphorimager. Repression measurements were based on three independent experiments. Activation measurements were based on two independent experiments.
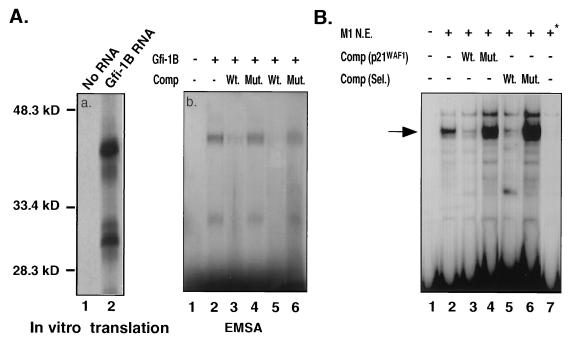
Computer-assisted analysis of the nucleotide sequence of the p21WAF1 promoter revealed a putative high-affinity Gfi-1B binding site between −1518 and −1530 (Fig. 7A). To confirm that Gfi-1B binds this site, we carried out EMSAs with a synthetic double-stranded oligonucleotide corresponding to this site as a probe and in vitro-translated Gfi-1B protein. Figure 8Aa, lane 2, shows an autoradiogram of “in vitro”-translated Gfi-1B labeled with [35S]methionine. The smaller-size bands are likely to represent the products of alternative translational initiation. Figure 8Ab, lane 2, shows that Gfi-1B binds to and shifts the oligonucleotide corresponding to the putative Gfi-1B binding site in the p21WAF1 promoter. The binding is specific in that it is competed by excess unlabeled wild-type but not mutant (AATC to GGTC) oligonucleotide (lanes 3 and 5 and lanes 4 and 6, respectively). The faster-migrating band could be due to complexes of the oligonucleotide with the smaller-size Gfi-1B protein products observed in Fig. 8Aa. EMSAs were also carried out with the same oligonucleotide derived from the sequence of the p21WAF1 promoter and nuclear extracts from IL-6-treated (36 h) and untreated M1 cells (Fig. 8B). The results showed that nuclear extracts from untreated (lane 2) M1 cells also bind to the probe, giving rise to one major shifted band which is competed by excess unlabeled wild-type oligonucleotide (lane 3) but not by its nonbinding mutant (lane 4) (Fig. 8B). The shift was not detected in nuclear extracts, however, from IL-6-treated (36 h) M1 cells (lane 7) in which Gfi-1B is no longer expressed (Fig. 8). An oligonucleotide derived from the Gfi-1B binding site selection experiment and a nonbinding mutant of the same oligonucleotide were also used for competition experiments, and the results were identical (Fig. 8B, lanes 5 and 6).
FIG. 8.
Gfi-1B binds the p21WAF1 promoter directly. (Aa) [35S]methionine-labeled, in vitro-translated Gfi-1B was electrophoresed in 10% SDS-PAGE. Lanes: 1, products of in vitro translation carried out in the absence of exogenously added RNA; 2, in vitro translation in the presence of Gfi-1B RNA. The lower band is likely to represent a Gfi-1B product translated from an internal AUG. (Ab) A 28-bp double-stranded oligonucleotide containing the putative Gfi-1B binding site of the p21WAF1 promoter was 32P labeled and used as a probe in an EMSA. Lane 1 contains the oligonucleotide probe and reticulocyte lysate only (negative control). Comp, oligonucleotide competition; Wt and Mut, wild-type and mutant oligonucleotide competitors, respectively. In lanes 3 and 4, the oligonucleotide competitors were added at a 100-fold excess, whereas in lanes 5 and 6 they were added at a 200-fold excess. (B) The same double-stranded oligonucleotide probe used in panel A was incubated with nuclear extracts from untreated (lanes 2 to 6) or IL-6-treated (lane 7) M1 cells. The major shifted band, indicated by an arrow, was competed by excess wild-type oligonucleotide competitors comp (p21WAF1) derived from the p21WAF1 promoter, and the Comp (Sel.) band was derived via the oligonucleotide selection experiment (lanes 3 and 5). Nonbinding mutants of the same oligonucleotides did not compete (lanes 4 and 6).
DISCUSSION
The data presented in this report show that the Gfi-1-related gene Gfi-1B encodes a 329-amino acid protein which is homologous to Gfi-1 in the zinc finger and SNAG repressor domains but which diverges completely from Gfi-1 in the region between the two domains. As expected from these findings, the Gfi-1B protein binds the same DNA sequence as Gfi-1 (66) and functions also as a transcriptional repressor (19, 66). One of the targets of the Gfi-1B repressor is the cyclin-dependent kinase inhibitor p21WAF1.
During IL-6-induced differentiation of the myelomonocytic cell line M1 along the macrophage lineage, Gfi-1B is downregulated to undetectable levels, whereas p21WAF1 is induced. In parallel with these phenomena the M1 cells undergo partial G1 arrest at 48 h from the start of the exposure to IL-6 and they differentiate along the macrophage lineage. Forced expression of Gfi-1B in M1 cells blocks the IL-6-mediated induction of p21WAF1 and inhibits G1 arrest and differentiation. However, Gfi-1B mutants lacking the SNAG domain or the entire amino-terminal region of Gfi-1B fail to inhibit both the induction of p21WAF1 and the cell cycle arrest and differentiation of these cells. These data suggest that IL-6 downregulates Gfi-1B, whose expression in proliferating cells maintains p21WAF1 in a repressed state. Downregulation of Gfi-1B derepresses the p21WAF1 promoter and contributes to the induction of p21WAF1. Expression of p21WAF1 is likely to contribute to the induction of G1 arrest and differentiation in IL-6-treated M1 cells.
If the inhibitory effect of Gfi-1B on the differentiation of M1 cells is due to the repression of p21WAF1, M1 cells defective in p21WAF1 should fail to differentiate in response to IL-6. Preliminary data show that M1 cells overexpressing antisense p21WAF1 indeed failed to undergo G1 arrest and differentiation despite the fact that they downregulated Gfi-1B normally in response to IL-6 (data not shown). These data suggest that p21WAF1 induction may play an important regulatory role in hematopoietic cell differentiation, and they are in agreement with earlier data showing that mice with homozygous inactivation of p21WAF1 exhibit abnormalities in the differentiation of other differentiating cell types such as keratinocytes (40).
M1 cells have been extensively studied as a model of myeloid cell differentiation. Studies with this model system have shown that the differentiation process is marked by changes in the expression of a series of genes (35). Some of these genes are likely to play a regulatory role, whereas others may be important for the expression of the differentiated cell phenotype (2). Two of the regulatory genes, namely c-myb (49) and c-myc (25), behave in a way similar to Gfi-1B during differentiation in that they are both downregulated. Moreover, downregulation of these transcriptional activators is obligatory for differentiation (49) and neither is known to regulate the p21WAF1. Of these, c-myb is downregulated within the first 3 h, while c-myc is downregulated beyond the 6-h time point following exposure to IL-6. As expected from these findings, forced expression of c-myb blocks the downregulation of c-myc and abolishes differentiation (49). Overexpression of c-myc, on the other hand, does not affect c-myb expression and blocks differentiation at an intermediate state (25). The data in this report showed that overexpression of Gfi-1B blocks the downregulation of c-myc and therefore suggest that the downregulation of Gfi-1B not only precedes but is also required for the downregulation of c-myc. The same data revealed unexpectedly that the downregulation of c-myb is a more complex process that takes place in two differentially regulated waves. The first wave, which occurs within the first 3 h following exposure to IL-6, precedes and is independent of the downregulation of Gfi-1B. However, the second wave of c-myb downregulation, which occurs more than 6 h following exposure to IL-6, is blocked by the overexpression of Gfi-1B. These data provide a temporal framework that is required to further explore the potential physiological relationships between these three transcription factors that are sequentially involved in regulating myeloid cell differentiation.
The regulation of the cyclin-dependent kinase inhibitor p21WAF1 has been the subject of intense study. Earlier studies had shown that it is induced by the tumor suppressor gene p53 (12) and that the induction of p21WAF1 by p53 may be sufficient to explain at least some of the p53-elicited biological effects. However, subsequent studies showed that p21WAF1 may also be induced by p53-independent mechanisms (11, 27, 28, 39). Transcriptional regulators other than p53 that contribute to the induction of the p21WAF1 promoter include MyoD and the adapter protein p300 (21, 44, 54), the receptor for vitamin D3 (36), the retinoic acid receptor (37), C/EBPα (58), STAT1 (8), SP1 (5), SP3 (43), AP2 (64), homeobox protein Gax (55), and BRCA1 (56). All the factors regulating p21WAF1 expression known to date, however, do so by inducing the activation of its promoter. The data in this report identify Gfi-1B as a repressor of this promoter, the only such repressor yet known, and demonstrate that p21WAF1 expression is regulated by both positive and negative regulators of transcription. The expression of p21WAF1 therefore may depend on the balance between transcriptional inducers and repressors. Since p53 is one of the main inducers of p21WAF1, Gfi-1B is likely to compete with p53-elicited biological effects such as cell cycle arrest and apoptosis. The data presented in this report showed that Gfi-1B indeed promotes cell cycle progression and therefore support this hypothesis. We therefore suggest that mutations that inactivate the p53 gene, which indeed are the most commonly observed mutations in human cancer (7), and mutations that induce the expression or enhance the activity of Gfi-1B and perhaps Gfi-1 may exert overlapping biological effects and may act synergistically in oncogenesis.
The preceding data showed that Gfi-1B is involved in myelomonocytic cell differentiation. However, the findings showing that Gfi-1B is a direct repressor of p21WAF1 and a potential competitor of p53 suggested that Gfi-1B may also play a role in oncogenesis. This was confirmed recently by experiments showing that Gfi-1B like Gfi-1 is also a target for provirus integration in retrovirus-induced rodent lymphomas. Specifically, Gfi-1B was shown to be involved in Mo-MuLV-induced B-cell lymphomas in Eμ-myc transgenic, pim-1/pim-2 knockout mice (39a). Gfi-1B involvement appears to be specific for these tumors because analysis of 217 DNA samples from 105 Mo-MuLV-induced rat T-cell lymphomas revealed no Gfi-1B involvement (data not shown). The reason for this specificity is that Gfi-1B, like Gfi-1, may cooperate with c-myc (42, 47), although perhaps only in the absence of pim activity.
In summary, the data in this report showed that Gfi-1B, the second known member of the Gfi-1 family of zinc finger proteins, is also a SNAG-domain-containing transcriptional repressor. The Gfi-1B repressor is involved in regulating the process of hematopoietic cell differentiation. In addition, it is involved in lymphoid cell oncogenesis. The differentiation controlling function and perhaps the oncogenic function of Gfi-1B may depend at least partially on the ability of the protein to repress the expression of the cyclin-dependent kinase inhibitor p21WAF1. Although the expression of p21WAF1 can be induced by a multitude of transcriptional activators (5, 8, 21, 36, 37, 43, 44, 54–56, 58, 64), Gfi-1B appears to be its only repressor known to date.
ACKNOWLEDGMENTS
We thank H. Mikkers and A. Berns for communicating their data prior to publication. We also thank M. Flubacher (Fox Chase Cancer Center [FCCC]) for providing Northern blots for the initial examination of Gfi-1B expression, K. Stamatakis for help with the generation of the Gfi-1BΔSNAG and Gfi-1BΔNter constructs, J. Sherley and Y. Liu (FCCC) for the 10-1 cell line, B. Calabretta and T. Skorski (Kimmel Cancer Center) for the c-myc and c-myb probes, and P. Bateman for secretarial assistance.
This work was supported by the Public Health Service grant RO1 CA-56110. Additional support was provided by Public Health Service grant CA-06927 and by an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center. W.S.E.-D. is an Assistant Investigator of the Howard Hughes Medical Institute. T.-Y.Y. was supported by an L. Greenwald Postdoctoral Fellowship (FCCC). B.T. was a graduate student in the Cell and Molecular Biology program of the University of Pennsylvania School of Medicine.
REFERENCES
- 1.Anderson S, Davis D H, Dahlbäck H, Jörnvall H, Russell D W. Cloning, structure and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 2.Arnaout M A. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood. 1990;75:1037–1050. [PubMed] [Google Scholar]
- 3.Bell D W, Taguchi T, Jenkins N A, Gilbert D J, Copeland N G, Gilks C B, Zweidler-McKay P, Grimes H L, Tsichlis P N, Testa J R. Chromosomal localization of a gene, GFI1, encoding a novel zinc finger protein reveals a new syntenic region between man and rodents. Cytogenet Cell Genet. 1995;70:263–267. doi: 10.1159/000134048. [DOI] [PubMed] [Google Scholar]
- 4.Bies J, Hoffman B, Amanullah A, Giese T, Wolff L. B-Myb prevents growth arrest associated with terminal differentiation of monocytic cells. Oncogene. 1996;12:355–363. [PubMed] [Google Scholar]
- 5.Biggs J R, Kudlow J E, Kraft A S. The role of the transcription factor Sp1 in regulating the expression of the WAF1/CIP1 gene in U937 leukemic cells. J Biol Chem. 1996;271:901–906. doi: 10.1074/jbc.271.2.901. [DOI] [PubMed] [Google Scholar]
- 6.Bignon C, Daniel D, Dijiane J. β-Galactosidase and chloramphenicol acetyltransferase assays in 96-well plates. BioTechniques. 1993;15:243–245. [PubMed] [Google Scholar]
- 7.Bosari S, Viale G. The clinical significance of p53 aberrations in human tumors. Virchows Arch. 1995;427:229–241. doi: 10.1007/BF00203389. [DOI] [PubMed] [Google Scholar]
- 8.Chin Y E, Kitagawa M, Su W C, You Z H, Iwamoto Y, Fu X Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Collins S J, Tsai S, Bernstein I. Retinoic acid receptors in hematopoiesis. Curr Top Microbiol Immunol. 1996;211:7–15. doi: 10.1007/978-3-642-85232-9_2. [DOI] [PubMed] [Google Scholar]
- 11.Elbendary A, Berchuck A, Davis P, Havrilesky L, Bast R C, Jr, Iglehart J D, Marks J R. Transforming growth factor beta 1 can induce CIP1/WAF1 expression independent of the p53 pathway in ovarian cancer cells. Cell Growth Differ. 1994;5:1301–1307. [PubMed] [Google Scholar]
- 12.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 13.El-Deiry W S, Tokino T, Waldman T, Oliner J D, Velculescu V E, Burrell M, Hill D E, Healy E, Rees J L, Hamilton S R, Kinzler K W, Vogelstein B. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 14.Elledge S J, Winston J, Harper J W. A question of balance: the role of cyclin-kinase inhibitors in development and tumorigenesis. Trends Cell Biol. 1996;6:388–392. doi: 10.1016/0962-8924(96)10030-1. [DOI] [PubMed] [Google Scholar]
- 15.Gearing D P, Gough N M, King J A, Hilton D J, Nicola N A, Simpson R J, Nice E C, Kelso A, Metcalf D. Molecular cloning and expression of cDNA encoding a murine myeloid leukemia inhibitory factor (LIF) EMBO J. 1987;6:3995–4002. doi: 10.1002/j.1460-2075.1987.tb02742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilks C B, Bear S E, Grimes H L, Tsichlis P N. Progression of interleukin-2 (IL-2)-dependent rat T cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graña X, Reddy E P. Cell cycle control in mammalian cells: role of cyclins, cyclin-dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs) Oncogene. 1995;11:211–219. [PubMed] [Google Scholar]
- 19.Grimes H L, Chan T O, Zweidler-McKay P A, Tong B, Tsichlis P N. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol. 1996;16:6263–6272. doi: 10.1128/mcb.16.11.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimes H L, Gilks C B, Chan T O, Porter S, Tsichlis P N. The Gfi-1 proto-oncoprotein represses Bax expression and inhibits T-cell death. Proc Natl Acad Sci USA. 1996;93:14569–14573. doi: 10.1073/pnas.93.25.14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 22.Harper J W, Elledge S J. Cdk inhibitors in development and cancer. Curr Opin Genet Dev. 1996;6:56–64. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 23.Harvey D M, Levine A J. p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev. 1991;5:2375–2385. doi: 10.1101/gad.5.12b.2375. [DOI] [PubMed] [Google Scholar]
- 24.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:1–11. [PubMed] [Google Scholar]
- 25.Hoffman-Liebermann B, Liebermann D A. Interleukin-6- and leukemia inhibitory factor-induced terminal differentiation of myeloid leukemia cells is blocked at an intermediate stage by constitutive c-myc. Mol Cell Biol. 1991;11:2375–2381. doi: 10.1128/mcb.11.5.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman-Liebermann B, Liebermann D A. Suppression of c-myc and c-myb is tightly linked to terminal differentiation induced by IL6 or LIF and not growth inhibition in myeloid leukemia cells. Oncogene. 1991;6:903–909. [PubMed] [Google Scholar]
- 27.Jiang H, Lin J, Su Z Z, Collart F R, Huberman E, Fisher P B. Induction of differentiation in human promyelocytic HL-60 leukemia cells activates p21, WAF1/CIP1, expression in the absence of p53. Oncogene. 1994;9:3397–3406. [PubMed] [Google Scholar]
- 28.Johnson M, Dimitrov D, Vojta P J, Barrett J C, Noda A, Pereira-Smith O M, Smith J R. Evidence for a p53-independent pathway for upregulation of SDI1/CIP1/WAF1/p21 RNA in human cells. Mol Carcinog. 1994;11:59–64. doi: 10.1002/mc.2940110202. [DOI] [PubMed] [Google Scholar]
- 29.Kelley K W, Arkins S, Minshall C, Liu Q, Dantzer R. Growth hormone, growth factors and hematopoiesis. Horm Res. 1996;45:38–45. doi: 10.1159/000184757. [DOI] [PubMed] [Google Scholar]
- 30.Kramer A, Keller W. Preparation and fractionation of mammalian extracts active in pre-mRNA splicing. Methods Enzymol. 1990;181:3–19. doi: 10.1016/0076-6879(90)81107-6. [DOI] [PubMed] [Google Scholar]
- 31.Krystosek A, Sachs L. Control of lysozyme induction in the differentiation of myeloid leukemic cells. Cell. 1976;9:675–684. doi: 10.1016/0092-8674(76)90131-8. [DOI] [PubMed] [Google Scholar]
- 32.Lan M S, Russell E K, Lu J, Johnson B E, Notkins A L. IA-1, a new marker for neuroendocrine differentiation in human lung cancer cell lines. Cancer Res. 1993;53:4169–4171. [PubMed] [Google Scholar]
- 33.Lew D J, Kornbluth S. Regulatory roles of cyclin-dependent kinase phosphorylation in cell cycle control. Curr Opin Cell Biol. 1996;8:795–804. doi: 10.1016/s0955-0674(96)80080-9. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Zeitler P S, Valerius M T, Small K, Potter S S. Gsh-1, an orphan Hox gene, is required for normal pituitary development. EMBO J. 1996;15:714–724. [PMC free article] [PubMed] [Google Scholar]
- 35.Liebermann D A, Hoffman B. Differentiation primary response genes and proto-oncogenes as positive and negative regulators of terminal hematopoietic cell differentiation. Stem Cells. 1994;12:352–369. doi: 10.1002/stem.5530120402. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Lee M H, Cohen M, Bommakanti M, Freedman L P. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 37.Liu M, Iavarone A, Freedman L P. Transcriptional activation of the human p21 (WAF1/CIP1) gene by retinoic acid receptor. Correlation with retinoid induction of U937 cell differentiation. J Biol Chem. 1996;271:31723–31728. doi: 10.1074/jbc.271.49.31723. [DOI] [PubMed] [Google Scholar]
- 38.Meier R W, Chen T, Mathews S, Niklaus G, Tobler A. The differentiation pathway of HL60 cells is a model system for studying the specific regulation of some myeloid genes. Cell Growth Differ. 1992;3:663–669. [PubMed] [Google Scholar]
- 39.Michieli P, Chedid M, Lin D, Pierce J H, Mercer W E, Givol D. Induction of WAF1/CIP1 by a p53-independent pathway. Cancer Res. 1994;54:3391–3395. [PubMed] [Google Scholar]
- 39a.Mikkers, H., and A. Berns. Personal communication.
- 40.Missero C, Di Cunto F, Kiyokawa H, Koff A, Dotto G P. The absence of p21Cip1/WAF1 alters keratinocyte growth and differentiation and promotes ras-tumor progression. Genes Dev. 1996;10:3065–3075. doi: 10.1101/gad.10.23.3065. [DOI] [PubMed] [Google Scholar]
- 41.Morgan D O. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 42.Orkin S H. Development of hematopoietic system. Curr Opin Genet Dev. 1996;6:597–602. doi: 10.1016/s0959-437x(96)80089-x. [DOI] [PubMed] [Google Scholar]
- 43.Prowse D M, Bolgan L, Molnar A, Dotto G P. Involvement of the Sp3 transcription factor in induction of p21Cip1/WAF1 in keratinocyte differentiation. J Biol Chem. 1997;272:1308–1314. doi: 10.1074/jbc.272.2.1308. [DOI] [PubMed] [Google Scholar]
- 44.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Ryan, K., I. Hayes, R. J. B. Nibbs, J. W. Baird, B. Tong, P. N. Tsichlis, J. D. Ansell, N. Hole, and G. J. Graham. Submitted for publication.
- 45.Sachs L. The control of growth and differentiation in normal and leukemic blood cells. Cancer. 1990;65:2196–2206. doi: 10.1002/1097-0142(19900515)65:10<2196::aid-cncr2820651006>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 46.Sachs L. The control of hematopoiesis and leukemia: from basic biology to the clinic. Proc Natl Acad Sci USA. 1996;93:4742–4749. doi: 10.1073/pnas.93.10.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheijen B, Jonkers J, Acton D, Berns A. Characterization of pal-1, a common proviral insertion site in murine leukemia virus-induced lymphomas of c-myc and Pim-1 transgenic mice. J Virol. 1997;71:9–16. doi: 10.1128/jvi.71.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt T, Zornig M, Beneke R, Moroy T. MoMuLV proviral integrations identified by Sup-F selection in tumors from infected myc/pim bitransgenic mice correlate with activation of the gfi-1 gene. Nucleic Acids Res. 1996;24:2528–2534. doi: 10.1093/nar/24.13.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selvakumaran M, Liebermann D A, Hoffman-Liebermann B. Deregulated c-myb disrupts interleukin-6- or leukemia inhibitory factor-induced myeloid differentiation prior to c-myc: role in leukemogenesis. Mol Cell Biol. 1992;12:2493–2500. doi: 10.1128/mcb.12.6.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shabo Y, Lotem J, Rubinstein M, Revel M, Clark S C, Wolf S F, Kamen R, Sachs L. The myeloid blood cell differentiation-inducing protein MGI-2A is interleukin-6. Blood. 1988;72:2070–2073. [PubMed] [Google Scholar]
- 51.Sherley J L, Kelly T J. Regulation of human thymidine kinase during the cell cycle. J Biol Chem. 1988;263:8350–8358. [PubMed] [Google Scholar]
- 52.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 53.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 54.Skapek S X, Rhee J, Spicer D B, Lassar A B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 55.Smith R C, Branellec D, Gorski D H, Guo K, Perlman H, Dedieu J-F, Pastore C, Mahfoudi A, Denèfle P, Isner J M, Walsh K. p21CIP1-mediated inhibition of cell proliferation by overexpression of the gax homeodomain gene. Genes Dev. 1997;11:1674–1689. doi: 10.1101/gad.11.13.1674. [DOI] [PubMed] [Google Scholar]
- 56.Somasundaram K, Zhang H, Zeng Y-X, Houvras Y, Peng Y, Zhang H, Wu G S, Licht J D, Weber B L, El-Deiry W S. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CIP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 57.Steinman R A, Hoffman B, Iro A, Guillouf C, Liebermann D A, el-Houseini M E. Induction of p21 (WAF-1/CIP1) during differentiation. Oncogene. 1994;9:3389–3396. [PubMed] [Google Scholar]
- 58.Timchenko N A, Wilde M, Nakanishi M, Smith J R, Darlington G J. CCAAT/enhancer-binding protein α (C/EBP α) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 59.Tong B. Gfi-1B represses p21WAF1 expression and inhibits myeloid cell differentiation. Ph.D. thesis. Philadelphia: University of Pennsylvania; 1997. [Google Scholar]
- 59a.Tong, B., N. Copeland, and N. Jenkins. Unpublished data.
- 60.Vasavada H A, Ganguly S, Germino F J, Wang Z X, Weissman S M. A contingent replication assay for the detection of protein-protein interactions in animal cells. Proc Natl Acad Sci USA. 1991;88:10686–10690. doi: 10.1073/pnas.88.23.10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weeks D L, Jones N C. E1A control of gene expression is mediated by sequences 5′ to the transcriptional starts of the early viral genes. Mol Cell Biol. 1983;3:1222–1234. doi: 10.1128/mcb.3.7.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Wild-type p53 induces apoptosis of myeloid leukemic cells that is inhibited by interleukin-6. Nature. 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 63.Zanjani E D, Almeida-Porada G, Flake A W. The human/sheep xenograft model: a large animal model of human hematopoiesis. Int J Hematol. 1996;63:179–192. doi: 10.1016/0925-5710(96)00445-8. [DOI] [PubMed] [Google Scholar]
- 64.Zeng Y X, Somasundaram K, El-Deiry W S. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;15:78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 65.Zhang D E, Hohaus S, Voso M T, Chen H M, Smith L T, Hetherington C J, Tenen D G. Function of PU.1 (Spi-1), C/EBP, and AML1 in early myelopoiesis: regulation of multiple myeloid CSF receptor promoters. Curr Top Microbiol Immunol. 1996;211:137–147. doi: 10.1007/978-3-642-85232-9_14. [DOI] [PubMed] [Google Scholar]
- 66.Zweidler-McKay P A, Grimes H L, Flubacher M M, Tsichlis P N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]