Abstract
The ability of the IκBα protein to sequester dimeric NF-κB/Rel proteins in the cytoplasm provides an effective mechanism for regulating the potent transcriptional activation properties of NF-κB/Rel family members. IκBα can also act in the nucleus as a postinduction repressor of NF-κB/Rel proteins. The mechanism by which IκBα enters the nucleus is not known, as IκBα lacks a discernible classical nuclear localization sequence (NLS). We now report that nuclear localization of IκBα is mediated by a novel nuclear import sequence within the second ankyrin repeat. Deletion of the second ankyrin repeat or alanine substitution of hydrophobic residues within the second ankyrin repeat disrupts nuclear localization of IκBα. Furthermore, a region encompassing the second ankyrin repeat of IκBα is able to function as a discrete nuclear import sequence. The presence of a discrete nuclear import sequence in IκBα suggests that cytoplasmic sequestration of the NF-κB/Rel–IκBα complex is a consequence of the mutual masking of the NLS within NF-κB/Rel proteins and the import sequence within IκBα. Nuclear import may be a conserved property of ankyrin repeat domains (ARDs), as the ARDs from two other ARD-containing proteins, 53BP2 and GABPβ, are also able to function as nuclear import sequences. We propose that the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences.
Directional transport of proteins through the nuclear pore complex provides a powerful regulatory mechanism for controlling gene expression, as illustrated by the NF-κB/Rel family of transcription factors (for reviews, see references 3, 5, and 30). Association of the inhibitor of κBα (IκBα) protein with dimeric NF-κB/Rel complexes containing either c-Rel or p65 (RelA) results in the sequestration of the Rel dimer in the cytoplasm, through masking of the nuclear localization sequences (NLSs) within Rel proteins (4, 20, 26, 41, 65, 77). In response to a variety of extracellular stimuli, including proinflammatory cytokines, viral infection, bacterial lipopolysaccharide, phorbol esters, oxidants, and UV light, IκBα becomes inducibly phosphorylated at serine residues 32 and 36 (9, 10, 15, 72). The recently identified protein kinase complex, IKK (IκB kinase), phosphorylates IκBα at these N-terminal serine residues and targets IκBα for ubiquitin-dependent degradation by the 26S proteasome (16, 48, 60, 63, 76). Degradation of IκBα enables the free Rel dimer to translocate to the nucleus and activate κB-dependent gene expression. One of the target genes of Rel proteins is the IκBα gene itself, resulting in the rapid induction of newly synthesized IκBα protein (1, 42, 45, 70).
Several lines of evidence have led to the suggestion that newly synthesized IκBα can function in the nucleus as a postinduction repressor of κB-dependent gene expression. First, ectopically overexpressed IκBα is readily detected in the nucleus, consistent with the suggestion that IκBα has a nuclear function (13, 50, 77). Second, following cytokine stimulation of cells, a significant fraction of newly synthesized endogenous IκBα appears transiently in the nucleus (1). Nuclear expression of IκBα correlates with inhibition of NF-κB-dependent transcription and disappearance of NF-κB from the nucleus (1). Third, brief stimulation of wild-type fibroblasts with tumor necrosis factor alpha (TNF-α) results in a transient activation of nuclear NF-κB (6). In contrast, brief stimulation of IκBα null-mutant fibroblasts with TNF-α results in a sustained level of nuclear NF-κB (6). Finally, IκBα can inhibit NF-κB-dependent transcription in the nucleus in vivo and can remove Rel proteins from functional preinitiation complexes in vitro (73). Taken together, these results are consistent with a model in which newly synthesized IκBα proteins can enter the nucleus, displace dimeric Rel proteins from DNA, and export Rel proteins from the nucleus to the cytoplasm. Implicit in this model is the ability of both Rel and IκBα proteins to enter the nucleus. In contrast, this model postulates that the Rel-IκBα complex is exported from the nucleus and is efficiently retained in the cytoplasm.
Nuclear import of Rel proteins is accomplished by virtue of an NLS located at the C-terminus of the Rel homology domain. The Rel-derived NLS is characterized by a short stretch of basic amino acids that resembles a classical NLS typified by the NLS of the simian virus 40 (SV40) large T protein (4, 26, 29, 31, 43, 64, 77). Nuclear import of proteins bearing such classical NLSs is accomplished by a soluble heterodimeric protein complex consisting of a 60-kDa protein, importin-α, and a 90-kDa protein, importin-β (12, 19, 33, 40, 51, 52, 59, 74). Importin-α binds to NLS-containing proteins and, through interaction with importin-β, mediates the docking of the NLS-containing protein to nucleoporins and the subsequent translocation of the NLS-containing protein to the nucleus (12, 19, 33–35, 37, 40, 51, 52, 59, 61, 74). Although a direct involvement of the importin-α–importin-β (importin-α–β) receptor in the nuclear import of Rel proteins has not been demonstrated, the presence of a classical NLS within Rel proteins suggests that nuclear import of Rel proteins is mediated by an importin-α–β-dependent pathway.
Similar to nuclear import, nuclear protein export is also a sequence-dependent receptor-mediated process. One class of nuclear export sequences (NESs) is characterized by a cluster of five leucine or isoleucine residues, each separated by one or two amino acid residues (for reviews, see references 36 and 54). NESs from several proteins, including the Rev protein of human immunodeficiency virus and the protein inhibitor of cyclic AMP-dependent protein kinase (21, 75), have been described. IκBα contains a sequence located between the ankyrin repeat domain (ARD) and the C-terminal PEST domain which resembles the previously described NESs in Rev and protein kinase inhibitor. The IκBα-derived NES can functionally substitute for the NES in Rev and is required for IκBα-mediated nuclear export of NF-κB (2, 24). IκBα-mediated nuclear export of Rel proteins is likely mediated by exportin 1 (CRM1), a recently identified importin-β-related protein that mediates the nuclear export of NES-containing proteins (22, 25, 56, 69).
The mechanism by which IκBα is able to localize to the nucleus is not known. IκBα does not contain a region of basic residues that resembles previously characterized NLSs. Thus, it has been proposed that the small size of IκBα might allow passive, NLS-independent accumulation of IκBα in the nucleus (1, 77). We now demonstrate that nuclear localization of IκBα is mediated by a novel nuclear import sequence within the second ankyrin repeat of IκBα. A region encompassing the second ankyrin repeat from IκBα can functionally substitute for the classical NLS in nucleoplasmin. ARDs from other proteins, including 53BP2 and GABPβ, are also able to function as nuclear import sequences. We propose that the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences.
MATERIALS AND METHODS
Construction of recombinant DNA molecules.
The construction of recombinant DNA molecules was performed according to standard techniques (66). Mutant p40 and MAD3 cDNAs were generated from the respective cDNAs encoding either the wild-type or the epitope-tagged proteins from phagemid single-strand DNA (66). The presence of each mutation within the respective cDNAs was confirmed by nucleotide sequence analysis. Typically, two independent isolates of each mutant p40 or MAD3 gene were separately subcloned into expression vectors and independently assayed for function. In no cases were any differences found between independent isolates of the same mutation. The p40 and MAD3 genes were expressed in chicken embryo fibroblasts (CEF) by using a spleen necrosis virus (SNV)-driven retroviral vector derived from pJD214 (17) and in COS-1 cells by using either the SNV-driven vector or a cytomegalovirus (CMV)-derived vector (9). The epitope-tagged p40 protein (LBD-p40) contains a C-terminal 18-amino-acid peptide derived from the ligand binding domain (LBD) of the platelet-derived growth factor. The LBD epitope tag consists of the sequence EVIVVPHSLPFML. A plasmid containing a segment of DNA encoding the LBD epitope tag and affinity-purified antipeptide sera against the LBD epitope tag were provided by Dan Donoghue (University of California). The epitope-tagged MAD3 protein (myc-MAD3) contains a C-terminal 11-amino-acid peptide derived from the c-Myc protein. The c-Myc epitope tag consists of the sequence MEQKLISEEDL. A modified pcDNA1 plasmid (Invitrogen) containing a segment of DNA encoding the myc epitope tag was provided by Gideon Dreyfuss (University of Pennsylvania). The epitope tags did not significantly alter the localization or the biochemical properties of the respective IκBα proteins.
The full-length c-Rel gene (11) was subcloned into pCMV4 as an XbaI fragment. The CMV-derived expression vectors for p65 (RelA) and IκBβ were obtained from Dean Ballard (Vanderbilt University). The cDNA encoding 53BP2 was obtained from Louie Naumovski (Stanford University). The cDNA encoding GABPβ was obtained from Mark Martin (University of Missouri). The cDNA encoding Notch1 was obtained from Anthony Capobianco (University of California). The cDNAs encoding myc-tagged nucleoplasmin core (NPc) and myc-tagged NPc-M9 were obtained from Gideon Dreyfuss (University of Pennsylvania). The cDNA encoding chicken muscle pyruvate kinase (PK) was obtained from Dan Donoghue. To facilitate construction of the NPc fusion proteins, a PmlI restriction site and a termination codon were introduced after the nucleoplasmin open reading frame. All NPc fusion genes were constructed by the insertion of blunt-ended fragments into the PmlI restriction site. Some of the inserts were obtained by PCR prior to cloning into the NPc expression vector, while other inserts were isolated out of their respective cDNA clones by the use of specific restriction enzyme sites. The complete nucleotide sequence of the PCR-derived inserts was determined to confirm faithful amplification of the cDNA. For construction of the myc-tagged PK fusion proteins, PCR amplification of a cDNA encoding PK was used to introduce a BamHI site upstream of codon 17 and a PmlI site downstream of codon 524. This fragment was subcloned into the appropriate myc-tagged expression vectors. Details of all plasmid constructions and primer sequences are available upon request.
Cell culture and transfection.
CEF were obtained from Spafas and grown in M199 containing 10% tryptose phosphate and 10% fetal calf serum (FCS). DNA transfections into CEF were performed with calcium phosphate coprecipitates as previously described (65). The biochemical properties of the Rel or p40 proteins were typically analyzed 4 to 5 days after transfection of CEF with the appropriate plasmids. Monkey COS-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FCS. Wild-type (WT+/+) and mutant 3T3 fibroblasts, and primary mouse embryo fibroblasts (MEF) lacking both the c-Rel and the p65 genes (c-Rel/p65−/−) were prepared in David Baltimore’s laboratory (California Institute of Technology) and were grown in DMEM containing 10% donor calf serum (7, 67). Transfections into COS-1 cells were performed on 35-mm-diameter plates by using Lipofectamine with a total of 2 μg of plasmid DNA in accordance with the directions from the manufacturer (GIBCO BRL). Transfections into 3T3 cells and into MEF were performed on 35-mm-diameter plates by using LipofectaminePLUS with a total of 1 μg of plasmid DNA in accordance with the directions from the manufacturer (GIBCO BRL). The cellular localization and the biochemical properties of the ectopically expressed proteins were typically analyzed 36 to 48 h after transfection of the COS-1 cells, 3T3 fibroblasts, or MEF with the appropriate plasmids.
Antibodies.
The following primary antibodies for detection of the respective ectopically expressed proteins were used: rabbit polyclonal anti-p40 (R1807), rabbit polyclonal anti-MAD3 (Santa Cruz Biotechnology), rabbit affinity-purified anti-LBD (Dan Donoghue), mouse monoclonal anti-myc (Sigma), rabbit polyclonal anti-β-galactosidase (Chemicon International), rabbit polyclonal anti-Rel (28), mouse monoclonal anti-c-Rel (HY87) (Henry R. Bose, Jr., University of Texas), rabbit polyclonal anti-p65 (Santa Cruz Biotechnology), and mouse monoclonal anti-p65 (Boehringer Mannheim). The appropriate anti-rabbit or anti-mouse fluorescein isothiocyanate-conjugated secondary antibody (Jackson Labs) or anti-rabbit Cy5-conjugated secondary antibody (Jackson Labs) was used for detection of the ectopically expressed proteins by indirect immunofluorescence. The appropriate anti-rabbit (Amersham) or anti-mouse (New England Biolabs) immunoglobulin G (IgG) conjugated to horseradish peroxidase was used in conjunction with the enhanced chemiluminescence system (ECL; Amersham) for detection of the ectopically expressed proteins by immunoblot analysis.
Indirect immunofluorescence.
Indirect immunofluorescence assays using CEF, COS-1 cells, 3T3 fibroblasts, or MEF were conducted on coverslips with the appropriate antisera as previously described (28). The coverslips were mounted onto glass slides with Mowiol containing 2.5% DABCO (Sigma).
Biochemical experiments.
Cell lysates for the coimmunoprecipitation analysis were prepared in ELB (50 mM Tris-HCl [pH 7.9], 250 mM NaCl, 0.1% Triton X-100, 5 mM EDTA, and 1 mM dithiothreitol). Protease inhibitors and phosphatase inhibitors were routinely included in the lysis buffers. The protease inhibitors used were 1 mM phenylmethylsulfonyl fluoride; antipain, aprotinin, leupeptin, and soybean trypsin-chymotrypsin inhibitor (5 μg/ml each); and pepstatin (0.5 μg/ml). The phosphatase inhibitors used were 0.4 mM sodium orthovanadate and 1 mM sodium fluoride. Equivalent aliquots of ELB cell lysates were used for coimmunoprecipitation analysis. Immunoprecipitation of LBD-tagged p40 proteins was performed with 3 μl of affinity-purified anti-LBD serum per sample. The immunoprecipitations were conducted in antibody excess to ensure quantitative precipitation of the LBD-tagged p40 proteins. DNA-binding of Rel proteins was determined by electrophoretic mobility shift assay as previously described (65).
IκBα localization and expression following TNF-α and CHX treatment.
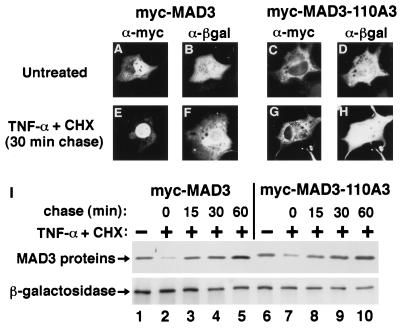
COS-1 cells (on 35-mm-diameter plates) were cotransfected with 0.5 μg of a CMV-derived β-galactosidase expression vector and 1.5 μg of either the myc-MAD3 or the myc-MAD3-110A3 expression vector. At 36 h posttransfection, the transfected COS-1 cells were either refed with complete medium (DMEM containing 10% FCS) or were cultured in complete medium containing cycloheximide (CHX) (100 μg/ml; Sigma) and TNF-α (10 ng/ml; Chemicon International) for 4 h. The TNF-α and CHX were subsequently removed, and the transfected cells were washed three times with DMEM and refed with complete medium. At 0, 15, 30, or 60 min following removal of the TNF-α and CHX, the chase samples were fixed for double-label indirect immunofluorescence. ELB cell lysates of the untreated and the TNF-α- and CHX-treated samples were collected in parallel for determination of the protein levels of the ectopically expressed proteins.
RESULTS
Nuclear localization of IκBα requires the integrity of a hydrophobic cluster of amino acids located within the second ankyrin repeat.
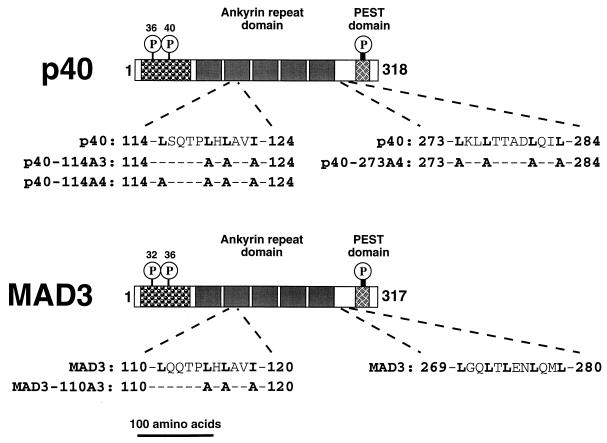
Both the mammalian (MAD3) and the avian (p40) IκBα proteins contain two clusters of hydrophobic residues that resemble previously described NESs (14, 21, 24, 39, 75). One cluster of hydrophobic residues (amino acids 114 to 124 in p40 [Fig. 1]) is located within the second ankyrin repeat, while a C-terminal cluster of hydrophobic residues (amino acids 273 to 284 in p40 [Fig. 1]) is located between the ARD and the acidic and serine-rich PEST domain. The hydrophobic cluster within the second ankyrin repeat is highly conserved among other IκB proteins, while the C-terminal hydrophobic amino acids are unique to the IκBα proteins (14, 27, 39, 46, 47, 55, 71). The C-terminal cluster of hydrophobic amino acids of the mammalian IκBα protein is required for nuclear export of NF-κB following coinjection of in vitro-synthesized IκBα and NF-κB into Xenopus oocyte nuclei (2). However, the role of these NES-like sequences in the distribution of IκBα between the nucleus and the cytoplasm has not been established.
FIG. 1.
Domain organization of IκBα. The avian (p40) and mammalian (MAD3) IκBα proteins are represented by long rectangular boxes. The numbers to the left of each box indicate the first amino acid of each protein, and the numbers to the right of each box indicate the total number of amino acids in each protein. The IκBα proteins contain an N-terminal regulatory domain, a central domain containing five ankyrin repeats, and a C-terminal acidic and serine-rich (PEST) domain. The sites of N-terminal cytokine-inducible serine phosphorylation and the sites of constitutive serine phosphorylation within the C-terminal PEST domain of IκBα are indicated by the circled P’s. The amino acid sequences of two clusters of hydrophobic residues are indicated in the single-letter code below the rectangle representing each IκBα protein. The residues relevant to the present work are in boldface type, and the mutations introduced into the IκBα proteins are indicated. The scale of this line drawing is indicated by the length of the bar at the bottom of the figure.
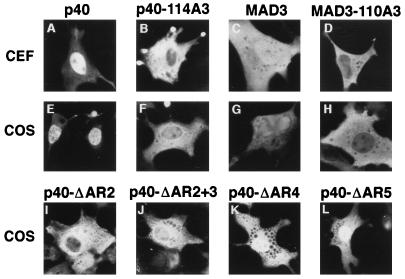
Mutant IκBα proteins containing amino acid substitutions within either the region between residues 114 and 124 or the region between residues 273 and 284 were constructed (Fig. 1). Expression vectors coding for the wild-type and mutant IκBα proteins were transfected into CEF and into COS-1 cells. The cellular distribution of the IκBα proteins was determined by indirect immunofluorescence. As previously reported (13, 50, 77), the wild-type p40 protein was predominantly nuclear (Fig. 2A and E; Table 1) while the wild-type MAD3 protein was distributed throughout both the nucleus and the cytoplasm (Fig. 2C and G; Table 1) in both CEF and COS-1 cells. Alanine substitution of leucine residues 119 and 121 and of isoleucine residue 124 in the second ankyrin repeat of p40 (p40-114A3) or alanine substitution of the corresponding hydrophobic residues in MAD3 (MAD3-110A3 [Fig. 1]) significantly reduced nuclear accumulation of the IκBα proteins (Fig. 2B, D, F, and H; Table 1). Fusion of the classical NLS derived from the SV40 large T protein onto the cytoplasmic p40-114A3 protein (p40-114A3-NLS) restored the nuclear localization of the mutant p40-114A3 protein (Table 1).
FIG. 2.
Cellular localization of wild-type and mutant IκBα proteins. CEF were transfected with SNV-derived retroviral vectors (A to D) and COS-1 cells were transfected with CMV-derived expression vectors (E to H) encoding either wild-type p40 (A and E), p40-114A3 (B and F), MAD3 (C and G), or MAD3-110A3 (D and H), or COS-1 cells were transfected with SNV-derived expression vectors encoding either p40-ΔAR2 (I), p40-ΔAR2+3 (J), p40-ΔAR4 (K), or p40-ΔAR5 (L). The p40-114A3 protein contains alanine substitutions for leucine 119, leucine 121, and isoleucine 124 in p40. The MAD3-110A3 protein contains alanine substitutions for leucine 115, leucine 117, and isoleucine 120 in MAD3. The p40-ΔAR2 protein contains a deletion of amino acids 98 to 142, encompassing the second ankyrin repeat in p40. The p40-ΔAR2+3 protein contains a deletion of amino acids 117 to 188, encompassing the second and third ankyrin repeats in p40. The p40-ΔAR4 protein contains a deletion of amino acids 189 to 222, encompassing the fourth ankyrin repeat in p40. The p40-ΔAR5 protein contains a deletion of amino acids 223 to 256, encompassing the fifth ankyrin repeat in p40. The cellular localization of the proteins in transfected cells was determined by indirect immunofluorescence with anti-p40 or anti-MAD3 serum. The cells shown are representative of more than 200 cells that were positive for the expression of the indicated proteins (see Table 1 for quantitation).
TABLE 1.
Localization of IκBα proteins in CEF and in COS-1 cells
| Vector (cell line) and IκBα proteina | Nb | N/Cc | Cd |
|---|---|---|---|
| SNV LTR (CEF) | |||
| p40 | 55 | 44 | 1 |
| p40-114A3 | 6 | 22 | 72 |
| MAD3 | 13 | 85 | 2 |
| MAD3-110A3 | 1 | 22 | 77 |
| SNV LTR (COS-1) | |||
| p40 | 30 | 60 | 10 |
| p40-114A3 | 2 | 22 | 76 |
| p40-114A4 | 1 | 17 | 82 |
| p40-273A4 | 22 | 76 | 2 |
| p40-ΔAR2 | <1 | 24 | 76 |
| p40-ΔAR2+3 | <1 | 16 | 84 |
| p40-ΔAR4 | 2 | 71 | 27 |
| p40-ΔAR5 | 13 | 78 | 9 |
| CMV (COS-1) | |||
| p40 | 52 | 47 | 1 |
| p40-114A3 | 9 | 68 | 23 |
| p40-114A4 | 11 | 63 | 26 |
| p40-273A4 | 38 | 61 | 1 |
| p40-NLS | 96 | 4 | <1 |
| p40-114A3-NLS | 58 | 28 | 14 |
| MAD3 | 22 | 65 | 13 |
| MAD3-110A3 | 4 | 5 | 91 |
The avian (p40) or mammalian (MAD3) wild-type and mutant IκBα proteins were expressed in CEF by using an SNV-derived retroviral vector. The IκBα proteins were also expressed in COS-1 cells by using either the SNV-derived vector or a CMV-derived expression vector. The cellular localization of the indicated IκBα protein was determined by indirect immunofluorescence. A total of 200 cells that were positive for expression of the respective IκBα protein were scored. LTR, long terminal repeat.
The percentage of cells that displayed predominantly nuclear staining is expressed relative to the total number of cells that displayed staining for the respective IκBα protein.
The percentage of cells that displayed predominantly whole-cell staining is expressed relative to the total number of cells that displayed staining for the respective IκBα protein.
The percentage of cells that displayed predominantly cytoplasmic staining is expressed relative to the total number of cells that displayed staining for the respective IκBα protein.
Similar to that in the mutant p40-114A3 protein, alanine substitution of four hydrophobic residues within the p40 region between residues 114 and 124 (p40-114A4 [Fig. 1]) resulted in a mutant p40 protein that was predominantly cytoplasmic in COS-1 cells (Table 1). In contrast, neither deletion of the C-terminal 51 amino acids including the region between residues 273 and 284 (p40-Δ267) nor alanine substitution of four leucine residues within the region between residues 273 and 284 of p40 (p40-273A4 [Fig. 1]) significantly altered the cellular distribution of p40 in COS-1 cells (Table 1 and data not shown).
As alanine substitution of hydrophobic residues within the second ankyrin repeat of IκBα disrupted nuclear localization of IκBα, we asked whether deletion of the second ankyrin repeat would similarly disrupt nuclear localization of IκBα. Deletion of the second ankyrin repeat of p40 (p40-ΔAR2) or deletion of both the second and the third ankyrin repeats of p40 (p40-ΔAR2+3) resulted in mutant p40 proteins that were predominantly cytoplasmic in COS-1 cells (Fig. 2I and J; Table 1). In contrast, deletion of the fourth (p40-ΔAR4) or the fifth (p40-ΔAR5) ankyrin repeats of p40 had only a modest effect on the relocalization of p40 from the nucleus to the cytoplasm in COS-1 cells (Fig. 2K and L; Table 1).
The wild-type and mutant IκBα proteins were expressed at equivalent levels, as determined by anti-p40 or anti-MAD3 immunoblot analysis (data not shown, but see Fig. 9). Furthermore, the turnover rates of the wild-type IκBα and the mutant IκBα-A3 proteins were equivalent, as determined by pulse-chase analysis in CEF and in COS-1 cells (data not shown). Thus, the inability of the IκBα-A3 proteins to accumulate in the nucleus is not due to increased turnover of these mutant proteins. Rather, our results indicate that nuclear localization of IκBα is sequence dependent and requires the integrity of hydrophobic residues within the second ankyrin repeat. In contrast to previous suggestions (1, 77), nuclear localization of IκBα does not occur by passive diffusion through the nuclear pore.
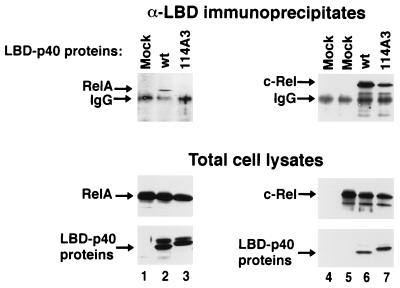
FIG. 9.
Coimmunoprecipitation analysis of wild-type and mutant IκBα proteins. COS-1 cells were mock transfected (lane 4), transfected with a CMV-derived expression vector encoding p65 (RelA) (lanes 1 to 3), or transfected with a CMV-derived expression vector encoding c-Rel (lanes 5 to 7). CMV-derived expression vectors encoding either the wild-type LBD-p40 (lanes 2 and 6) or the LBD-p40-114A3 protein (lanes 3 and 7) were included in some transfections. Cell lysates were subjected to immunoprecipitation with affinity-purified anti-LBD rabbit IgG followed by immunoblot analysis with either anti-p65 mouse IgG (top panels, lanes 1 to 3), or anti-c-Rel mouse IgG (top panels, lanes 4 to 7). Cell lysates were also subjected to direct immunoblot analysis with anti-p65 rabbit serum (middle panels, lanes 1 to 3), with anti-c-Rel rabbit serum (middle panels, lanes 4 to 7), or with anti-p40 rabbit serum (bottom panels, lanes 1 to 7). The p65 (RelA), c-Rel, and p40 proteins are indicated by arrows. Only the relevant portions of each immunoblot are shown.
Nuclear localization of IκBα is independent of p50, p52, p65 (RelA), or c-Rel.
To determine whether nuclear localization of wild-type IκBα protein is dependent on the presence of endogenous p50, p52, p65 (RelA), or c-Rel, we determined the cellular distribution of wild-type and mutant MAD3 proteins in fibroblasts which lack these Rel proteins. The ectopically expressed wild-type MAD3 protein was distributed throughout both the nucleus and the cytoplasm in WT+/+ and in fibroblasts lacking both copies of the NFκB1, p65 (RelA), NFκB1 and NFκB2, NFκB1 and p65 genes, and c-Rel and p65 (RelA) (p50−/−, p65−/−, p50/p52−/−, p50/p65−/−, and c-Rel/p65−/−, respectively) (Fig. 3A to F, respectively) (quantified in Table 2). Furthermore, the mutant MAD3-110A3 protein remained predominantly cytoplasmic when ectopically expressed in these cell types (Table 2). Therefore, the nuclear localization of ectopically expressed IκBα does not require expression of these endogenous Rel proteins.
FIG. 3.
Cellular distribution of IκBα is independent of the p50, p52, p65 (RelA), and c-Rel proteins. WT+/+ (A), p50−/− (B), p65−/− (C), p50/p52−/− (D), and p50/p65−/− (E) mouse 3T3 fibroblasts or c-Rel/p65−/− primary MEF (F) were transfected with CMV-derived expression vectors coding for wild-type MAD3. The cellular localization of the ectopically expressed MAD3 protein was determined by indirect immunofluorescence with anti-MAD3 serum. The results shown are representative of at least 100 cells that were positive for expression of the wild-type MAD3 protein (see Table 2 for quantitation).
TABLE 2.
Localization of IκBα proteins
| Cell line and IκBα proteina | Nb | N/Cc | Cd |
|---|---|---|---|
| WT+/+ 3T3 | |||
| MAD3 | 1 | 70 | 29 |
| MAD3-110A3 | 1 | 35 | 64 |
| p50−/− 3T3 | |||
| MAD3 | 1 | 77 | 22 |
| MAD3-110A3 | 2 | 17 | 81 |
| p65−/− 3T3 | |||
| MAD3 | 1 | 80 | 19 |
| MAD3-110A3 | 1 | 21 | 78 |
| p50/p52−/− 3T3 | |||
| MAD3 | 3 | 86 | 11 |
| MAD3-110A3 | 1 | 15 | 84 |
| p50/p65−/− 3T3 | |||
| MAD3 | 2 | 82 | 16 |
| MAD3-110A3 | 8 | 27 | 65 |
| c-Rel/p65−/− MEF | |||
| MAD3 | 3 | 86 | 11 |
| MAD3-110A3 | 2 | 34 | 64 |
Wild-type (MAD3) and mutant (MAD3-110A3) mammalian IκBα proteins were expressed in the indicated 3T3 mouse fibroblasts or MEF. The cellular localization of the ectopically expressed MAD3 proteins was determined by indirect immunofluorescence. At least 100 cells that were positive for expression of the respective MAD3 protein were examined.
The percentage of cells that displayed predominantly nuclear staining is expressed relative to the total number of cells that displayed staining for the respective MAD3 protein.
The percentage of cells that displayed predominantly whole-cell staining is expressed relative to the total number of cells that displayed staining for the respective MAD3 protein.
The percentage of cells that displayed predominantly cytoplasmic staining is expressed relative to the total number of cells that displayed staining for the respective MAD3 protein.
Nuclear localization of newly synthesized IκBα requires the integrity of hydrophobic residues within the second ankyrin repeat.
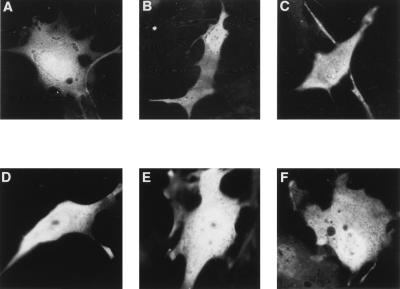
It has previously been demonstrated that upon cytokine stimulation of cells, a significant fraction of newly synthesized endogenous IκBα appears transiently in the nucleus (1). To determine whether nuclear localization of newly synthesized IκBα requires the integrity of the second ankyrin repeat, COS-1 cells ectopically expressing either a wild-type epitope-tagged MAD3 protein (myc-MAD3) or a mutant epitope-tagged MAD3-110A3 protein (myc-MAD3-110A3) were treated with TNF-α for 4 h in the presence of CHX and the cellular distribution and expression levels of the newly synthesized MAD3 proteins were analyzed at successive time points following removal of the TNF-α and CHX (Fig. 4; Table 3). An expression vector coding for β-galactosidase was cotransfected with the respective MAD3 expression vectors to control for transfection efficiency. TNF-α treatment of COS-1 cells expressing either wild-type myc-MAD3 or mutant myc-MAD3-110A3 resulted in a significant decline in the abundance of both myc-MAD3 (Fig. 4, compare lanes 1 and 2) and myc-MAD3-110A3 (Fig. 4, compare lanes 6 and 7). Within 30 min following removal of the TNF-α and CHX, the expression levels of myc-MAD3 (Fig. 4, compare lanes 1 and 4) and myc-MAD3-110A3 (Fig. 4, compare lanes 6 and 9) were restored to nearly untreated levels as a result of new synthesis of the respective IκBα proteins. The newly synthesized myc-MAD3 could readily be detected in the nucleus of COS-1 cells following removal of the TNF-α and CHX (Fig. 4E; Table 3). In contrast, the newly synthesized mutant myc-MAD3-110A3 protein did not accumulate in the nucleus but rather was detected primarily in the cytoplasm of COS-1 cells following removal of the TNF-α and CHX (Fig. 4G; Table 3). Therefore, nuclear localization of newly synthesized IκBα following cytokine stimulation requires the integrity of hydrophobic residues within the second ankyrin repeat.
FIG. 4.
Nuclear localization of newly synthesized IκBα requires the integrity of the second ankyrin repeat. (A to H) COS-1 cells were cotransfected with CMV-derived expression vectors encoding β-galactosidase and either epitope-tagged MAD3 (myc-MAD3) or epitope-tagged MAD3-110A3 (myc-MAD3-110A3). At 36 h posttransfection, the transfected cells were either refed with complete medium (A to D), or were cultured in complete medium containing TNF-α (10 ng/ml) and CHX (100 μg/ml) for 4 h. The TNF-α and CHX were subsequently removed, and the transfected cells were chased in complete medium for 0, 15, 30, or 60 min. The localization of the β-galactosidase protein (B, D, F, and H) and either the myc-MAD3 (A and E) or the myc-MAD3-110A3 (C and G) protein was determined by anti-β-galactosidase (α-βgal) and anti-myc (α-myc) double-label indirect immunofluorescence, as indicated. The cellular localization of the ectopically expressed proteins at 30 min posttreatment is shown (E to H). The cells shown are representative of more than 25 cells that were positive for expression of both β-galactosidase and the respective myc-MAD3 protein (see Table 3 for quantitation). (I) For determination of the protein levels of the ectopically expressed proteins, cell lysates of the untreated and the TNF-α- and CHX-treated samples were collected in parallel. Equivalent amounts of each cell lysate were subjected to immunoblot analysis, and the levels of the ectopically expressed proteins from untreated samples (lanes 1 and 6) or from samples treated with TNF-α and CHX for 4 h and subsequently chased in complete medium for 0 (lanes 2 and 7), 15 (lanes 3 and 8), 30 (lanes 4 and 9), or 60 (lanes 5 and 10) min were determined by anti-MAD3 and anti-β-galactosidase immunoblots, as indicated. The myc-MAD3 and β-galactosidase proteins are indicated by arrows. Only the relevant portions of each immunoblot are shown.
TABLE 3.
Localization of IκBα proteins in β-galactosidase-positive cells following TNF-α and CHX treatment
| IκBα protein and treatment or time posttreatment (min)a | Nb | N/Cc | Cd | No MAD3e |
|---|---|---|---|---|
| myc-MAD3 | ||||
| No treatment | 28 | 40 | 4 | 28 |
| 0 | 8 | 8 | <4 | 84 |
| 15 | 28 | 20 | <4 | 52 |
| 30 | 36 | 24 | 4 | 36 |
| 60 | 16 | 52 | 4 | 28 |
| myc-MAD3-110A3 | ||||
| No treatment | <4 | 4 | 84 | 12 |
| 0 | 4 | 4 | 12 | 80 |
| 15 | 4 | 4 | 44 | 48 |
| 30 | <4 | 4 | 68 | 28 |
| 60 | <4 | 8 | 72 | 20 |
COS-1 cells were cotransfected with CMV-derived expression vectors encoding β-galactosidase and either epitope-tagged myc-MAD3 or epitope-tagged myc-MAD3-110A3. At 36 h posttransfection, the transfected cells were either refed with complete medium (No treatment) or were cultured in complete medium containing TNF-α (10 ng/ml) and CHX (100 μg/ml) for 4 h. Following the 4-h treatment with TNF-α and CHX, the transfected cells were washed and chased in complete medium for either 0, 15, 30, or 60 min to allow protein synthesis. The cellular localization of the ectopically expressed proteins was determined by double-label indirect immunofluorescence using anti-myc IgG and anti-β-galactosidase sera. At least 25 cells that were positive for coexpression of the respective myc-MAD3 and the β-galactosidase proteins were examined.
The percentage of doubly labeled cells that displayed predominantly nuclear staining of the respective myc-MAD3 protein is expressed relative to the total number of cells that displayed staining for β-galactosidase.
The percentage of doubly labeled cells that displayed predominantly whole-cell staining of the respective myc-MAD3 protein is expressed relative to the total number of cells that displayed staining for β-galactosidase.
The percentage of doubly labeled cells that displayed predominantly cytoplasmic staining of the respective myc-MAD3 protein is expressed relative to the total number of cells that displayed staining for β-galactosidase.
The percentage of β-galactosidase cells in which staining of the respective myc-MAD3 protein could not be detected is expressed relative to the total number of cells that displayed staining for β-galactosidase.
The second ankyrin repeat in IκBα functions as a discrete nuclear import sequence.
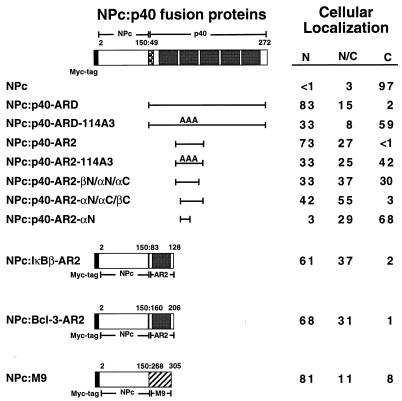
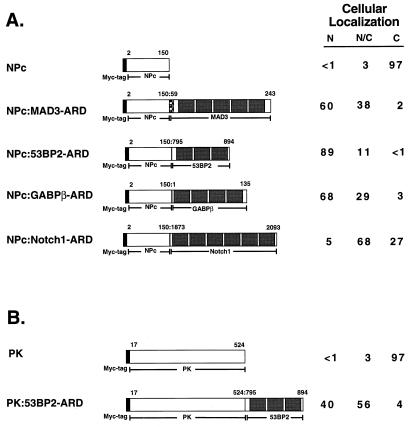
To determine if IκBα contains a nuclear import sequence that can functionally substitute for a classical NLS, the IκBα ARD was fused onto NPc, and the cellular distribution of the NPc-ARD fusion protein was determined by indirect immunofluorescence in COS-1 cells. Nucleoplasmin is normally a nuclear protein (62), and deletion of its C-terminal bipartite NLS prevents the nuclear localization of NPc (Fig. 5 and 6A) (49). Fusion of the p40 ARD onto NPc (NPc–p40-ARD) relocalized NPc to the nucleus (Fig. 5 and 6B). In contrast, the p40 ARD containing the A3 mutation (NPc–p40-ARD-114A3) was not able to efficiently relocalize NPc to the nucleus (Fig. 5 and 6C). Similar to the intact ARD of p40, fusion of the second ankyrin repeat of p40 onto NPc (NPc–p40-AR2) also efficiently relocalized NPc to the nucleus (Fig. 5 and 6D), while the A3 mutation (NPc–p40-AR2-114A3) markedly reduced the ability of the second ankyrin repeat to relocalize NPc to the nucleus (Fig. 5 and 6E). The second ankyrin repeat of p40 was as efficient as the M9 nuclear import sequence for relocalization of NPc to the nucleus (Fig. 5 and 6F). Thus, the second ankyrin repeat of IκBα contains a nuclear import signal that can functionally substitute for the NLS in nucleoplasmin.
FIG. 5.
Nuclear import function of the second ankyrin repeat from IκB proteins. Fusion proteins between NPc and either the avian IκBα protein (p40), the mammalian IκBβ protein, the mammalian Bcl-3 protein, or the M9 nuclear import signal from hnRNP A1 are indicated (colons show fusions). The NPc protein comprises amino acids 2 through 150 of nucleoplasmin and contains an N-terminal epitope tag derived from the c-Myc protein. The ARD from p40, the second ankyrin repeat from p40 (p40-AR2), the second ankyrin repeat from IκBβ (IκBβ-AR2), the second ankyrin repeat from Bcl-3 (Bcl-3-AR2), or the M9 nuclear import signal was fused onto the C terminus of NPc. The following NPc-p40 fusion proteins were constructed with the indicated p40-derived amino acids (in parentheses): p40-ARD (49 to 272), p40-AR2 (103 to 149), p40-AR2-βN/αN/αC (103 to 138), p40-AR2-αN/αC/βC (117 to 149), and p40-AR2-αN (113 to 130). The IκBβ-derived amino acids used to construct the NPc–IκBβ-AR2 fusion protein were 83 to 128. The Bcl-3-derived amino acids used to construct the NPc–Bcl-3-AR2 fusion protein were 160 to 206. The hnRNP A1-derived amino acids used to construct the NPc-M9 fusion protein were 268 to 305. The cellular localization of each fusion protein was determined in COS-1 cells with anti-myc IgG. Cells that were positive for expression of the indicated fusion proteins were classified as having predominantly nuclear staining (N), staining that was distributed equally between the nucleus and the cytoplasm (N/C), or staining that was predominantly cytoplasmic (C). At least 200 cells that were positive for expression of each fusion protein were scored, and the percentage of cells in each category is given.
FIG. 6.
Nuclear localization of NPc upon fusion of the second ankyrin repeat of IκBα. COS-1 cells were transfected with CMV-based expression vectors encoding NPc (A), NPc–p40-ARD (B), NPc–p40-ARD-114A3 (C), NPc–p40-AR2 (D), NPc–p40-AR2-114A3 (E), or NPc-M9 (F). The NPc fusion proteins contain an N-terminal epitope tag derived from the c-Myc protein. The cellular localization of the NPc proteins in transfected cells was determined by indirect immunofluorescence with anti-myc IgG. The cells shown are representative of more than 200 cells that were positive for the expression of the indicated proteins.
The hydrophobic cluster within the second ankyrin repeat is highly conserved among other IκB family members (14, 27, 39, 46, 47, 55, 71). As a nuclear function has previously been proposed for several other IκB family members, including IκBβ and Bcl-3 (8, 23, 55, 57, 73), we asked whether the second ankyrin repeat of either IκBβ or of Bcl-3 would be able to functionally substitute for the classical NLS in nucleoplasmin. Fusion of the second ankyrin repeat from either IκBβ or from Bcl-3 onto NPc efficiently relocalized NPc to the nucleus (Fig. 5). Thus, nuclear import is a conserved property of the second ankyrin repeat from these IκB proteins.
The recently described crystal structure of the ARD-containing protein 53BP2 reveals that an individual ankyrin repeat consists of a short N-terminal β-hairpin and N- and C-terminal α-helices that pack in an antiparallel fashion (32). The β-hairpin which initiates the adjacent ankyrin repeat provides critical amino acid interactions that stabilize the previous ankyrin repeat (32). To define the minimal structural requirements for the nuclear import function of the second ankyrin repeat, further truncations of the second ankyrin repeat were fused onto NPc, and their ability to relocalize NPc to the nucleus was determined. Deletion of either the N-terminal (NPc–p40-AR2-αN/αC/βC) or the C-terminal (NPc–p40-AR2-βN/αN/αC) β-hairpin reduced the ability of the second ankyrin repeat to relocalize NPc to the nucleus (Fig. 5). A sequence encompassing the N-terminal α-helix of the second ankyrin repeat (NPc–p40-AR2-αN) was unable to function as a nuclear import signal when fused onto NPc (Fig. 5). Thus, an extended ankyrin repeat region which includes an N-terminal β-hairpin, an N-terminal α-helix, a C-terminal α-helix, and an adjacent C-terminal β-hairpin constitutes a fully functional nuclear import sequence.
The ability of NPc to form oligomers is likely a critical factor in its ability to remain in the cytoplasm in the absence of a nuclear import signal (49). Expression of the various NPc–IκBα-ARD and the NPc–IκB-AR2 fusion proteins was confirmed by immunoblot analysis (data not shown). Fusion of the IκBα ARD or the second ankyrin repeat from IκB proteins onto NPc did not disrupt NPc oligomer formation (data not shown).
The ARDs of diverse proteins contain a functional nuclear import signal.
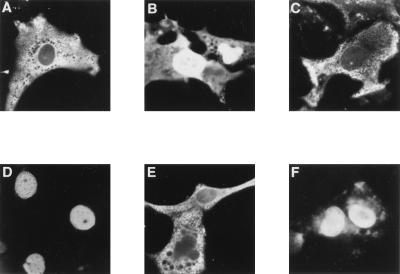
To determine whether nuclear import is a common property of ARDs, the ARDs from several other ARD-containing proteins were fused onto NPc and their cellular distribution was determined by indirect immunofluorescence in COS-1 cells. Fusion of the first, second, and third ankyrin repeats from the p53-associated protein, 53BP2 (53), or fusion of the ARD from GABPβ (44), a transcription factor which belongs to the Ets family of proteins, onto NPc efficiently relocalized NPc to the nucleus (Fig. 7A and 8B and C). In contrast, fusion of the ARD from the Notch1 protein (18, 38), a transmembrane receptor protein, onto NPc did not efficiently relocalize NPc to the nucleus (Fig. 7A and 8D). The localization of the NPc-ARD fusion proteins was also examined in WT+/+, p50−/−, and p65−/− 3T3 fibroblasts. While the NPc protein remained predominantly cytoplasmic, fusion of the ARDs from MAD3, 53BP2, and GABPβ onto NPc markedly relocalized NPc to the nucleus in all three of these cell types (Table 4). Expression of the various NPc-ARD fusion proteins was confirmed by immunoblot analysis (data not shown). Fusion of the ARDs onto NPc did not disrupt NPc oligomer formation (data not shown). Thus, the nuclear import function of ARDs derived from several ARD-containing proteins are independent of either p50 or p65.
FIG. 7.
Nuclear import function of ankyrin repeat domains. (A) The structures of fusion proteins between NPc and the ARDs from the mammalian IκBα protein (MAD3), 53BP2, GABPβ, and Notch1 are indicated. The NPc protein comprises amino acids 2 through 150 of nucleoplasmin and contains an N-terminal epitope tag derived from the c-Myc protein. The ARDs from the indicated proteins were fused onto the C terminus of NPc. The amino acid residues from each protein that were used to construct each of the NPc-ARD fusion proteins are indicated. The cellular localization of the NPc-ARD fusion proteins was determined in COS-1 cells using anti-myc IgG. (B) The structure of a fusion protein between PK and the ARD from 53BP2 is indicated. Amino acids 17 through 524 of PK were used to construct the fusion protein, which also contains an N-terminal epitope tag derived from the c-Myc protein. The amino acid residues that were used to construct the PK–53BP2-ARD fusion protein are indicated. The cellular localization of the PK–53BP2-ARD fusion protein was determined in COS-1 cells using anti-myc IgG. For both panels, cells that were positive for expression of the indicated fusion proteins were classified as having predominantly nuclear staining (N), staining that was distributed equally between the nucleus and the cytoplasm (N/C), or staining that was predominantly cytoplasmic (C). At least 200 cells that were positive for expression of each fusion protein were scored, and the percent of cells in each category is given. Colons show fusions.
FIG. 8.
Nuclear localization of NPc or PK upon fusion of ARDs. COS-1 cells were transfected with CMV-based expression vectors encoding NPc–MAD3-ARD (A), NPc–53BP2-ARD (B), NPc–GABPβ-ARD (C), NPc–Notch1-ARD (D), PK (E), or PK–53BP2-ARD (F). The NPc and PK fusion proteins contain an N-terminal epitope tag derived from the c-Myc protein. The cellular localization of the NPc and PK fusion proteins in transfected cells was determined by indirect immunofluorescence with anti-myc IgG. The cells shown are representative of more than 200 cells that were positive for the expression of the indicated proteins.
TABLE 4.
Localization of NPc-ARD fusion proteins
| Cell line and protein or fusion proteina | Nb | N/Cc | Cd |
|---|---|---|---|
| WT+/+ 3T3 | |||
| NPc | <1 | 10 | 90 |
| NPc–MAD3-ARD | 27 | 65 | 8 |
| NPc–53BP2-ARD | 40 | 53 | 7 |
| NPc–GABPβ-ARD | 40 | 46 | 14 |
| NPc–Notch1-ARD | 19 | 60 | 21 |
| p50−/− 3T3 | |||
| NPc | <1 | 9 | 91 |
| NPc–MAD3-ARD | 48 | 47 | 5 |
| NPc–53BP2-ARD | 46 | 51 | 3 |
| NPc–GABPβ-ARD | 53 | 37 | 10 |
| NPc–Notch1-ARD | 12 | 53 | 35 |
| p65−/− 3T3 | |||
| NPc | 1 | 4 | 95 |
| NPc–MAD3-ARD | 58 | 39 | 3 |
| NPc–53BP2-ARD | 32 | 60 | 8 |
| NPc–GABPβ-ARD | 41 | 43 | 16 |
| NPc–Notch1-ARD | 7 | 36 | 57 |
NPc or NPc-ARD fusion proteins containing an N-terminal c-Myc-derived epitope tag were expressed in the indicated 3T3 mouse fibroblasts. The cellular localization of each fusion protein was determined by using anti-myc IgG. At least 100 cells that were positive for expression of each fusion protein were scored.
The percentage of cells that displayed predominantly nuclear staining is expressed relative to the total number of cells that displayed staining for the respective NPc fusion protein.
The percentage of cells that displayed predominantly whole-cell staining is expressed relative to the total number of cells that displayed staining for the respective NPc fusion protein.
The percentage of cells that displayed predominantly cytoplasmic staining is expressed relative to the total number of cells that displayed staining for the respective NPc fusion protein.
PK is a protein of cytoplasmic origin, and fusion of a classical NLS onto PK localizes PK to the nucleus (43, 49). To determine whether an ARD generally directs nuclear localization, the ARD of 53BP2 was fused onto PK and the cellular localization of the PK–53BP2-ARD fusion protein was determined. As expected, PK was predominantly cytoplasmic when ectopically expressed in COS-1 cells (Fig. 7B and 8E). Fusion of the 53BP2 ARD onto PK significantly localized PK from the cytoplasm to the nucleus (Fig. 7B and 8F). Expression of the PK-53BP2 protein was confirmed by immunoblot analysis (data not shown). Thus, the 53BP2 ARD can functionally substitute for a classical NLS from a normally nuclear protein and also specify the nuclear import of a normally cytoplasmic protein.
Hydrophobic residues within the nuclear import sequence of IκBα are required for association with p65 (RelA) but not c-Rel.
The identification of a discrete nuclear import sequence within IκBα indicates that nuclear localization of both IκBα and Rel proteins are dependent upon cis-acting sequences within the respective proteins. However, both the p65-IκBα and the c-Rel–IκBα complexes are sequestered in the cytoplasm (4, 20, 26, 65, 77). Cytoplasmic sequestration of these Rel-IκBα complexes indicates that both the Rel NLS and the IκBα nuclear import sequence are functionally inactive in the Rel-IκBα complex. The p65 NLS has previously been shown to be masked in the context of the p65-IκBα complex (4, 26, 77). However, the mechanism by which the IκBα nuclear import sequence is functionally inactivated within either the p65-IκBα or the c-Rel–IκBα complex is not known.
We first examined the ability of the wild-type p40 and the p40-114A3 proteins to associate with p65 by coimmunoprecipitation analysis (Fig. 9). COS-1 cells were cotransfected with CMV-driven expression vectors encoding p65 and either a wild-type epitope-tagged p40 protein (p40-LBD) or a mutant epitope-tagged p40-114A3 protein (LBD-p40-114A3). The p65 protein was not detected in anti-LBD immunoprecipitates when singly transfected into COS-1 cells (Fig. 9, upper panel, lane 1) but was readily detected from COS-1 cells cotransfected with wild-type LBD-p40 (Fig. 9, upper panel, lane 2). Furthermore, p65 was not detected in anti-LBD immunoprecipitates when cotransfected with the LBD-p40-114A3 protein (Fig. 9, upper panel, lane 3).
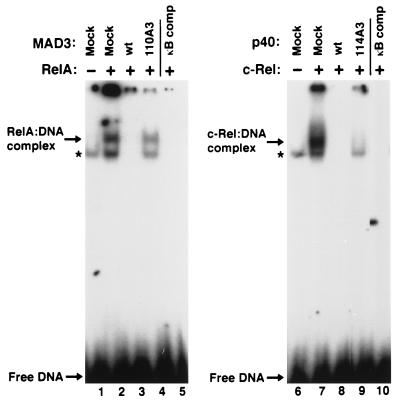
As an independent measure of the ability of wild-type or mutant IκBα proteins to associate with p65, we examined the ability of wild-type and mutant IκBα proteins to inhibit the DNA-binding activity of p65. Coexpression of MAD3 with p65 inhibited DNA-binding by p65 (Fig. 10, lane 3). In contrast, the mutant MAD3-110A3 protein was markedly reduced in its ability to inhibit DNA-binding by p65 or the endogenous DNA-binding activity from COS-1 cells (Fig. 10, compare lanes 3 and 4). Similarly, wild-type p40 inhibited DNA binding by p65, while the mutant p40-114A3 protein was markedly reduced in its ability to inhibit DNA binding by p65 in COS-1 cells (data not shown). The steady-state levels of p65 and of the IκBα proteins within the respective cell lysates were approximately equivalent, as determined by immunoblot analysis (data not shown). The IκBα-A3 proteins were also markedly deficient for inhibition of NF-κB-dependent luciferase gene expression relative to the wild-type IκBα proteins in cells treated with TNF-α or cotransfected with an expression vector encoding the Tax protein of human T-cell leukemia virus type 1 (data not shown). Taken together, these results show that the mutant IκBα-A3 proteins are markedly reduced in their ability to associate with or to inhibit the DNA binding of p65. Our results suggest that hydrophobic residues within the second ankyrin repeat of IκBα participate in critical amino acid contacts between IκBα and p65.
FIG. 10.
Inhibitory properties of wild-type and mutant IκBα proteins. COS-1 cells were mock transfected (lanes 1 and 6) or were transfected with CMV-derived expression vectors encoding either p65 (RelA) (lanes 2 to 5) or c-Rel (lanes 7 to 10). CMV-derived expression vectors encoding wild-type MAD3 (lane 3), mutant MAD3-110A3 (lane 4), wild-type p40 (lane 8), or mutant p40-114A3 (lane 9) were included in some transfections. Cell lysates were analyzed for proteins that bound to a 32P-labeled oligonucleotide containing a palindromic κB site. A 100-fold excess of the unlabeled palindromic κB oligonucleotide was included in some DNA-binding reaction mixtures (lanes 5 and 10). The DNA-binding reaction mixtures were electrophoresed through a 5% nondenaturing polyacrylamide gel. The positions of the respective Rel-DNA complexes and unbound DNA are indicated by arrows. The endogenous Rel DNA-binding activity in COS-1 cells is denoted by an asterisk.
In contrast to p65, c-Rel was readily detected in α-LBD immunoprecipitates from COS-1 cells transfected with the mutant LBD-p40-114A3 protein (Fig. 9, upper panel, lane 7). The c-Rel protein was also able to associate with the p40-114A3 protein in the Saccharomyces cerevisiae two-hybrid system (data not shown). Furthermore, coexpression of either wild-type p40 (Fig. 10, lane 8) or p40-114A3 (Fig. 10, lane 9) with c-Rel inhibited DNA binding by c-Rel. Similar to the MAD3-110A3 protein, the p40-114A3 protein was markedly reduced in its ability to inhibit the endogenous DNA-binding activity in COS-1 cells (Fig. 10, compare lanes 8 and 9). The steady-state levels of c-Rel and of the p40 proteins within the respective cell lysates were approximately equivalent, as determined by immunoblot analysis (data not shown). Taken together, these results show that hydrophobic residues within the second ankyrin repeat of IκBα are not critically required for association of IκBα with c-Rel.
DISCUSSION
The IκBα protein normally sequesters dimeric Rel proteins in the cytoplasm in unstimulated cells (4, 20, 26, 65, 77). Upon cellular stimulation, IκBα becomes inducibly phosphorylated and degraded, enabling the dimeric Rel complex to translocate to the nucleus (9, 10, 15, 16, 48, 60, 63, 72, 76). In activated cells, the cellular pool of IκBα is rapidly replenished and newly synthesized IκBα enters the nucleus and exports the dimeric Rel complex to the cytoplasm (1, 2, 42, 45, 70). The small size of IκBα and the absence of a discernible classical NLS in IκBα have led to the previous suggestion that IκBα might enter the nucleus by passive diffusion (1, 77). Our results now demonstrate that nuclear localization of newly synthesized IκBα does not occur by passive diffusion. Rather, nuclear localization of IκBα is specified by a novel nuclear import sequence within the second ankyrin repeat.
The second ankyrin repeat of IκBα is the predominant nuclear import sequence in the context of the full-length IκBα protein, as either alanine substitution of hydrophobic residues within the second ankyrin repeat or deletion of the second ankyrin repeat markedly relocalizes IκBα to the cytoplasm. However, the other ankyrin repeats within the ARD of IκBα might also contribute to the nuclear localization of the full-length IκBα protein. In particular, neither mutations within the second ankyrin repeat nor deletion of the second ankyrin repeat fully restricts IκBα to the cytoplasm. Furthermore, deletion of the fourth or the fifth ankyrin repeat of IκBα partially relocalizes IκBα to the cytoplasm. Finally, similar to fusion of the second ankyrin repeat, fusion of the third, fourth, or fifth ankyrin repeats of IκBα onto NPc also relocalizes NPc from the cytoplasm to the nucleus (64a). Therefore, although the second ankyrin repeat of IκBα is the predominant nuclear import sequence, the other ankyrin repeats may also contribute to the nuclear localization of the full-length IκBα protein.
The hydrophobic residues within the second ankyrin repeat are highly conserved among other IκB family members (14, 27, 39, 46, 47, 55, 71). The ability of the second ankyrin repeat from IκBα, IκBβ, or Bcl-3 to each specify nuclear import of NPc suggests that nuclear localization of these IκB family members might be mediated by a nuclear import function encoded within their second ankyrin repeat.
To further define the sequence requirements for the nuclear import function of the second ankyrin repeat of IκBα, and by analogy with the 53BP2 crystal structure (32), we initially chose an extended ankyrin repeat region that would encompass the N-terminal β-hairpin and the two α-helices of the second ankyrin repeat, and the N-terminal β-hairpin of the third ankyrin repeat. Deletion of either β-hairpin reduced the nuclear import function of this extended ankyrin repeat region. Furthermore, an 18-amino-acid sequence encompassing just the N-terminal α-helix was not sufficient to mediate nuclear import. Our results suggest that a fully functional ankyrin repeat-derived nuclear import sequence is comprised of an N-terminal β-hairpin, two α-helices, and the β-hairpin of the adjacent ankyrin repeat. The 53BP2 crystal structure shows that amino acid interactions between adjacent β-hairpins, and between a conserved histidine residue within the N-terminal α-helix and the backbone of the β-hairpin from the adjacent ankyrin repeat provide interactions necessary for the stability of an ankyrin repeat (32). Our results are consistent with the notion that the ability of the second ankyrin repeat to function as a nuclear import sequence critically requires amino acid interactions that maintain the structural integrity of that ankyrin repeat.
The ability of the ARDs from other ARD-containing proteins, such as 53BP2 and GABPβ, to functionally substitute for a classical NLS suggests that nuclear import is not a unique property of the IκBα ARD. However, nuclear import is not a general property of all ARDs, as the ARD from Notch1 is a poor nuclear import sequence. Thus, the ability of ARDs to specify nuclear import is not equivalent among all ARD-containing proteins. The precise structural determinants of individual ankyrin repeats within an intact ARD which are necessary for nuclear import function will need to be identified to understand how specific ARDs specify nuclear import.
At least two mechanisms can be envisioned to understand how the ARD mediates nuclear localization of IκBα. One possibility is that nuclear import of IκBα occurs via a piggyback mechanism, in which the IκBα-derived nuclear import sequence mediates association with another protein that contains a classical NLS, and subsequent nuclear import of IκBα occurs via the importin-α–β-mediated import pathway. If nuclear import of IκBα is accomplished via such a piggyback mechanism, our results strongly suggest that members of the Rel transcription factor family are not the carrier protein. First, significant nuclear accumulation of ectopically expressed IκBα is observed in p50−/−, p65−/−, p50/52−/−, p50/p65−/−, and c-Rel/p65−/− fibroblasts. Second, the ability of ARDs from other ARD-containing proteins to functionally substitute for a classical NLS reveals that nuclear import is not a property restricted to the ARD of IκBα. Third, the isolated ARDs from IκBα, GABPβ, and 53BP2 are able to function as discrete nuclear import sequences in 3T3 fibroblasts that lack either p50 or p65. Taken together, these results are not consistent with a model in which the nuclear import function of the ARD of IκBα is mediated via a piggyback interaction with Rel proteins. However, it remains possible that a non-Rel-related protein mediates nuclear import of IκBα through the importin-α–β-mediated import pathway.
An alternative model is that nuclear import of IκBα occurs via an importin-α–β-independent receptor-mediated pathway (for reviews, see references 36 and 54). A precedent for a receptor-mediated nuclear import pathway that is independent of the importin-α–β receptor has recently been established for the nuclear import of hnRNP A1 (58). Nuclear import of hnRNP A1 is mediated through interaction of the M9 transport sequence in hnRNP A1 with a novel importin-β-related receptor termed transportin (58, 68). As the individual ankyrin repeats within the ARD of IκBα bear no sequence resemblance to either classical NLSs or to the M9 motif, nuclear import of ARDs would likely be mediated by an as yet unidentified receptor. The identification of proteins that interact with the wild-type but not mutant ARD of IκBα should provide additional insight into how nuclear import of ARD-containing proteins is accomplished.
Our identification of a discrete nuclear import sequence within IκBα indicates that nuclear localization of both IκBα and Rel proteins is dependent upon cis-acting sequences within the respective proteins. However, the Rel-IκBα complex is sequestered in the cytoplasm (4, 20, 26, 65, 77). The p65 (RelA) NLS is required for association with IκBα and is masked within the p65-IκBα complex, consistent with the suggestion that the p65 NLS participates in direct amino acid contacts with IκBα (4, 26, 77). We now provide evidence that hydrophobic residues within the IκBα nuclear import sequence are required for association with p65. Our results suggest that amino acids within the IκBα nuclear import sequence participate in critical contacts with p65-derived amino acids. In contrast to the p65-IκBα complex, hydrophobic residues within the IκBα nuclear import sequence are not required for association with c-Rel. However, the relatively large size of the IκBα nuclear import sequence suggests that other amino acids within the IκBα nuclear import sequence may participate in direct amino acid contacts with c-Rel. Alternatively, the IκBα nuclear import sequence, though not directly involved in critical contacts with c-Rel, may be buried within the c-Rel–IκBα complex. Despite differences in the nature of amino acid contacts within the respective p65-IκBα and the c-Rel–IκBα complexes, it is clear that mutual masking of the NLS on Rel proteins and the IκBα nuclear import sequence would provide an efficient mechanism for cytoplasmic sequestration of the Rel-IκBα complexes.
ACKNOWLEDGMENTS
We thank Andrew Chappell for technical assistance and Dirk Görlich, J. Alan Diehl, and members of the Hannink laboratory for stimulating conversations. We thank Fernando Arenzana-Seisdedos, Dean Ballard, David Baltimore, Henry R. Bose, Anthony Capobianco, Dan Donoghue, Gideon Dreyfuss, Mark Martin, Gary Nabel, and Louie Naumovski for providing various reagents.
This work was supported by Public Health Service grant CA-55027 from the National Cancer Institute, by USDA NRICGP award 95-04073, by University of Missouri Research Board grant RB97-175, and by the University of Missouri Molecular Biology Program.
REFERENCES
- 1.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 5.Beg A A, Baldwin A S., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB in the immune system. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 6.Beg A A, Sha W C, Bronson R T, Baltimore D. Constitutive NF-κB activation, enhanced granulopoiesis, and neonatal lethality in IκBα deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 7.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature (London) 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 8.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 directly transactivates through kappa-B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 9.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 11.Capobianco A J, Simmons D L, Gilmore T D. Cloning and expression of a chicken c-Rel cDNA: unlike p59v-rel, p68c-rel is a cytoplasmic protein in chicken embryo fibroblasts. Oncogene. 1990;5:257–266. [PubMed] [Google Scholar]
- 12.Chi N, Adam E, Adam S. Sequence and characterization of cytoplasmic nuclear import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cressman D E, Taub R. IκBα can localize in the nucleus but shows no direct transactivation potential. Oncogene. 1993;8:2567–2573. [PubMed] [Google Scholar]
- 14.Davis N, Ghosh S, Simmons D L, Tempst P, Liou H C, Baltimore D, Bose H R., Jr Rel-associated pp40: an inhibitor of the Rel family of transcription factors. Science. 1991;253:1268–1271. doi: 10.1126/science.1891714. [DOI] [PubMed] [Google Scholar]
- 15.DiDonato J A, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature (London) 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty J P, Temin H M. High mutation rate of a spleen necrosis virus-based retrovirus vector. Mol Cell Biol. 1986;6:4387–4395. doi: 10.1128/mcb.6.12.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 19.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 20.Ernst M K, Dunn L L, Rice N R. The PEST-like sequence of IκBα is responsible for inhibition of DNA binding but not for cytoplasmic retention of c-Rel or RelA homodimers. Mol Cell Biol. 1995;15:872–883. doi: 10.1128/mcb.15.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–484. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 22.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 23.Franzoso G, Bours V, Park S, Tomita-Yamaguchi M, Kelly K, Siebenlist U. The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-kappa B-mediated inhibition. Nature (London) 1992;359:339–342. doi: 10.1038/359339a0. [DOI] [PubMed] [Google Scholar]
- 24.Fritz C C, Green M R. HIV Rev uses a conserved cellular protein export pathway for nucleocytoplasmic transport of viral RNAs. Curr Biol. 1996;6:848–854. doi: 10.1016/s0960-9822(02)00608-5. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 26.Ganchi P A, Sun S C, Greene W C, Ballard D W. IκBα/MAD-3 masks the nuclear localization signal of NF-κB p65 and requires the transactivation domain to inhibit NF-κB p65 DNA binding. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisler R, Bergmann A, Hiromi Y, Nusslein-Volhard C. cactus, a gene involved in dorsoventral pattern formation of Drosophila, is related to the IκB gene family of vertebrates. Cell. 1992;71:613–621. doi: 10.1016/0092-8674(92)90595-4. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore T D, Temin H M. Different localization of the product of the v-rel oncogene in chicken fibroblasts and spleen cells correlates with transformation by REV-T. Cell. 1986;44:791–800. doi: 10.1016/0092-8674(86)90845-7. [DOI] [PubMed] [Google Scholar]
- 29.Gilmore T D, Temin H M. v-Rel oncoproteins in the nucleus and in the cytoplasm transform chicken spleen cells. J Virol. 1988;62:703–714. doi: 10.1128/jvi.62.3.703-714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilmore T D, Koedood M, Piffat K A, White D W. Rel/NF-κB/IκB proteins and cancer. Oncogene. 1996;13:1367–1378. [PubMed] [Google Scholar]
- 31.Goldfarb D S, Gariepy J, Schoolnik G, Kornberg R D. Synthetic peptides as nuclear localization signals. Nature (London) 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- 32.Gorina S, Pavletich N P. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 33.Görlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 34.Görlich D, Kostka S, Kraft R, Dingwall C, Laskey R, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 35.Görlich D, Vogel G, Mills A, Hartmann E, Laskey R. Distinct functions for the two importin subunits in nuclear protein import. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 36.Görlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 37.Görlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 38.Guan E, Wang J, Laborda J, Norcross M, Baeuerle P A, Hoffman T. T cell leukemia-associated Notch/Translocation-associated Notch homologue has IκB-like activity and physically interacts with Nuclear Factor-κB proteins in T cells. J Exp Med. 1996;183:2025–2032. doi: 10.1084/jem.183.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haskill S, Beg A A, Thompkins S M, Morris J S, Yurochko A D, Sampson-Johannes A, Mondal K, Ralph P, Baldwin A S., Jr Characterization of an immediate-early gene induced in adherent monocytes that encodes IκB-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 40.Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue J I, Kerr L D, Rashid D, Davis N, Bose H R, Jr, Verma I M. Direct association of pp40/IκB-β with Rel/NF-κB transcription factors: Role of ankyrin repeats in the inhibition of DNA binding activity. Proc Natl Acad Sci USA. 1992;89:4333–4337. doi: 10.1073/pnas.89.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito C Y, Kazantsev A G, Baldwin A S., Jr Three NF-κB sites in the IκBα promoter are required for induction of gene expression by TNF-α. Nucleic Acids Res. 1994;22:3787–3792. doi: 10.1093/nar/22.18.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 44.La Marco K, Thompson C C, Byers B P, Walton E M, McKnight S L. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- 45.Le Bail O, Schmidt-Ulrich R, Israel A. Promoter analysis of the gene encoding the IκBα/MAD-3 inhibitor of NF-κB: positive regulation by members of the Rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Nabel G J. A new member of the IκB protein family, IκBɛ, inhibits RelA (p65)-mediated NF-κB transcription. Mol Cell Biol. 1997;17:6184–6190. doi: 10.1128/mcb.17.10.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liou H C, Nolan G P, Ghosh S, Fujita T, Baltimore D. The NF-kappa B precursor, p105, contains an internal I kappa B-like inhibitor that preferentially inhibits p50. EMBO J. 1992;11:3003–3009. doi: 10.1002/j.1460-2075.1992.tb05370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 49.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 50.Morin P J, Gilmore T D. The C-terminus of the NF-κB p50 precursor and an IκB isoform contain transcription activation domains. Nucleic Acids Res. 1992;20:2453–2458. doi: 10.1093/nar/20.10.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at the nuclear pore complex. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 bind nuclear localization signal and β interacts with peptide repeat containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naumovski L, Cleary M L. The p53-binding protein 53BP2 also interacts with Bcl2 and impedes cell cycle progression at G2/M. Mol Cell Biol. 1996;16:3884–3892. doi: 10.1128/mcb.16.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 55.Ohno H, Takimoto G, McKeithan T W. The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell. 1990;60:991–997. doi: 10.1016/0092-8674(90)90347-h. [DOI] [PubMed] [Google Scholar]
- 56.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 57.Phillips R J, Ghosh S. Regulation of IκBβ in WEHI 231 mature B cells. Mol Cell Biol. 1997;17:4390–4396. doi: 10.1128/mcb.17.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 59.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear-protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 61.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 62.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1988;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez M S, Wright J, Thompson J, Thomas D, Baleux F, Virelizier J-L, Hay R T, Arenzana-Seisdedos F. Identification of lysine residues required for signal-induced ubiquitination and degradation of IκBα in vivo. Oncogene. 1996;12:2425–2435. [PubMed] [Google Scholar]
- 64.Rottjakob E M, Sachdev S, Leanna C A, McKinsey T A, Hannink M. PEST-dependent cytoplasmic retention of v-Rel by IκBα: evidence that IκBα regulates the cellular localization of c-Rel and v-Rel by distinct mechanisms. J Virol. 1995;70:3176–3188. doi: 10.1128/jvi.70.5.3176-3188.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64a.Sachdev, S., and M. Hannink. Unpublished data.
- 65.Sachdev S, Rottjakob E M, Diehl J A, Hannink M. IκBα mediated inhibition of nuclear transport and DNA-binding by Rel proteins are separable functions: phosphorylation of C-terminal serine residues of IκBα is specifically required for inhibition of DNA-binding. Oncogene. 1995;11:811–823. [PubMed] [Google Scholar]
- 66.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 67.Sha W C, Liou H C, Tuomanen E I, Baltimore D. Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 68.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 70.Sun S-C, Ganchi P A, Ballard D W, Greene W C. NF-κB controls expression of inhibitor κB-α: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 71.Thompson J E, Phillips R J, Erdjument-Bromage H, Tempst P, Ghosh S. IκBβ regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 72.Traenckner E B, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tran K, Merika M, Thanos D. Distinct functional properties of IκBα and IκBβ. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1α as a functional receptor for nuclear localization signals. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 75.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–474. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 76.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 77.Zabel U, Henkel T, Silva M S, Baeuerle P A. Nuclear uptake control of NF-κB by MAD-3, an IκB protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]