Abstract
Background
Intensive care research has predominantly relied on conventional methods like randomized controlled trials. However, the increasing popularity of open-access, free databases in the past decade has opened new avenues for research, offering fresh insights. Leveraging machine learning (ML) techniques enables the analysis of trends in a vast number of studies.
Objective
This study aims to conduct a comprehensive bibliometric analysis using ML to compare trends and research topics in traditional intensive care unit (ICU) studies and those done with open-access databases (OADs).
Methods
We used ML for the analysis of publications in the Web of Science database in this study. Articles were categorized into “OAD” and “traditional intensive care” (TIC) studies. OAD studies were included in the Medical Information Mart for Intensive Care (MIMIC), eICU Collaborative Research Database (eICU-CRD), Amsterdam University Medical Centers Database (AmsterdamUMCdb), High Time Resolution ICU Dataset (HiRID), and Pediatric Intensive Care database. TIC studies included all other intensive care studies. Uniform manifold approximation and projection was used to visualize the corpus distribution. The BERTopic technique was used to generate 30 topic-unique identification numbers and to categorize topics into 22 topic families.
Results
A total of 227,893 records were extracted. After exclusions, 145,426 articles were identified as TIC and 1301 articles as OAD studies. TIC studies experienced exponential growth over the last 2 decades, culminating in a peak of 16,378 articles in 2021, while OAD studies demonstrated a consistent upsurge since 2018. Sepsis, ventilation-related research, and pediatric intensive care were the most frequently discussed topics. TIC studies exhibited broader coverage than OAD studies, suggesting a more extensive research scope.
Conclusions
This study analyzed ICU research, providing valuable insights from a large number of publications. OAD studies complement TIC studies, focusing on predictive modeling, while TIC studies capture essential qualitative information. Integrating both approaches in a complementary manner is the future direction for ICU research. Additionally, natural language processing techniques offer a transformative alternative for literature review and bibliometric analysis.
Keywords: BERTopic, critical care, eICU, machine learning, MIMIC, Medical Information Mart for Intensive Care, natural language processing
Introduction
The start of critical care as a medical subspecialty can be traced back to a polio epidemic during which a substantial number of patients needed prolonged mechanical ventilation [1]. Over time, the field of critical care has experienced significant growth and continual evolution. Research in this field has played a pivotal role in unraveling the complexities of numerous diseases and treatment modalities, driving substantial advancements in clinical practice over the past decades [2]. Groundbreaking studies have investigated critical areas such as sepsis, mechanical ventilation, acute lung and kidney injuries, intensive care unit (ICU) delirium, and sedation in critically ill patients [3].
These research studies have often been conducted in traditional ways such as prospective and randomized controlled trials [4], cohort and observational studies, clinical trials [5], and clinical and translational research [6]. These traditional methods have revolutionized patient care and improved outcomes significantly. For instance, the implementation of protocol-driven, goal-directed management of sepsis and appropriate fluid therapy has led to remarkable reductions in mortality rates [7,8], and these findings have been integral in developing evidence-based practice guidelines that are now the gold standard [9,10].
Despite their undeniable merits, traditional research methods in intensive care also come with several limitations [11]. Clinical trials are known for their high costs [12], stringent standardization requirements, and ethical oversight [13]. Data collection can be laborious, prone to human errors, and constrained in terms of quantity and granularity [14]. Moreover, obtaining patient consent for most randomized controlled trials in the ICU poses challenges [15], necessitating alternative consent models. These limitations have become increasingly apparent as medical complexity continues to grow exponentially [16].
The advent of electronic health records (EHRs) has heralded a new era in clinical research by facilitating the digitization of health care systems [17]. In this era of data science, a more integrated approach can be adopted, using machine learning (ML) algorithms to tackle the complexity of critical illness [18]. Open-access databases (OADs), such as the Medical Information Mart for Intensive Care (MIMIC) database [19] and the Philips eICU Collaborative Research Database (eICU-CRD) [20], have played a transformative role by enabling free data sharing.
The concept of free and open databases plays a pivotal role in promoting data sharing and advancing medical knowledge in accordance with the findable, accessible, interoperable, and reusable (FAIR) guiding principle. The FAIR principles, which emphasize that data should be findable, accessible, interoperable, and reusable, are essential for fostering a collaborative and transparent scientific research environment [21,22]. By removing barriers to access, free, and open databases allow researchers, regardless of their affiliations or resources, to contribute to and benefit from the collective pool of information. Accessibility fosters inclusivity and diversity in research, promoting a broader range of perspectives and approaches to medical challenges. This democratization of knowledge leads to a more equitable distribution of information. Researchers can now leverage these vast repositories of information for ML and artificial intelligence studies, marking a departure from traditional intensive care (TIC) research approaches.
Conducting a literature review [23] to investigate the disparities between traditional ICU research and studies based on open-access data sets holds significant importance as it provides a comprehensive understanding of the strengths and limitations of the latter. However, conventional methods of literature reviews and bibliometric analysis have their limitations, especially when dealing with large-scale literature due to computational complexity and the labor-intensive nature of manual interpretations [24-26]. To address these challenges, natural language processing (NLP) offers a promising avenue, while topic modeling techniques can be used to extract various topic themes from extensive data sets [27,28].
Built on the foundations of bidirectional encoder representations from transformers (BERT), BERTopic introduces a novel approach to topic modeling [29,30]. Unlike traditional unsupervised models like latent Dirichlet allocation, which rely on “bag-of-words” model [31], BERTopic overcomes the problem of semantic information loss, significantly enhancing the accuracy of generated topics, and providing more interpretable compositions for each topic, which greatly facilitates the classification of topics.
With the aid of BERTopic, this study aims to shed light on the disparities and commonalities between studies conducted through OADs and TIC research. By analyzing the overall trends and patterns in these 2 groups, we seek to identify knowledge gaps and explore avenues for complementary contributions between these research approaches.
Methods
Data
Data Filtering
We performed an ML-based analysis of research abstracts in the Web of Science (WoS) database to automatically categorize the research papers to conduct this literature mapping analysis. There was no limit to the year of publication of the articles. The search query consisted of the following keyword to identify all the studies that were published under the umbrella of intensive care: (“ICU” OR “intensive care”). The search terms were deliberately left to be broad to cover broad spectrums of journals in the field.
The inclusion criteria were as follows: (1) written in English, (2) articles that had keywords related to intensive care, (3) articles that had the article type of “article” or “review.” We excluded articles with incomplete data fields (eg, title, abstract, publication year, and paper citation). The articles included were then further processed to identify if they were studies using OADs. These articles were labeled as “open-access database,” while the rest of the articles extracted were labeled as “traditional intensive care.”
The search used for this study was performed on January 18, 2023, from WoS. This generated 227,893 search results, which were subsequently reselected using Python. An advanced search from PubMed was done based on the broad search terms of ICU studies used from previous Cochrane ICU literature review [32] to ensure the accuracy of the results. The numbers corroborated with a discrepancy of 4.9% (227,893 WoS keyword search vs 239,748 PubMed ICU keyword search).
Selection Criteria for OADs
A title search using keywords from all currently existing OADs was conducted to identify OAD studies. These include (1) MIMIC [19], (2) eICU-CRD [20], (3) Amsterdam University Medical Centers Database (AmsterdamUMCdb) [33], (4) High Time Resolution ICU Dataset (HiRID) [34], and (5) Pediatric Intensive Care database [35]. We avoided including only keywords in the search and restricted the search years by the year that the OAD was made publicly available to reduce the inadvertent inclusion of incorrect articles due to keywords. For instance, the search term for OADs published with the MIMIC database included title keyword search with the terms (“MIMIC-IV” OR “MIMIC-III” OR “MIMIC-II” OR “MIMIC Dataset” OR “medical information mart for intensive care” OR “MIMIC IV” OR “MIMIC III” OR “MIMIC II”) in studies that were published after 2003. The title keyword search for the searches and the year of cutoff for each OAD are presented in Multimedia Appendix 1.
Furthermore, to ensure the accuracy of the supervised keyword classification, a manual review of the classification by 2 critical care physicians was done for 100 articles from each category that were randomly selected. The review was done independently with the physicians labeling the extract publications into OAD and TICs. An accuracy of 99% was achieved on independent reviews, and full agreement was achieved after discussion on the discrepancy. The final results were matched with the supervised keyword classification.
We performed a bibliometric analysis by directly extracting publication details from the WoS database using Python (Python Software Foundation). The analysis involved assessing the number of articles published per year, calculating total citation counts, and identifying the top journals that published intensive care-related articles. Comprehensive results are presented in Multimedia Appendix 2.
Data Analysis
Uniform Manifold Approximation and Projection
Uniform manifold approximation and projection (UMAP) is a manifold learning technique for dimension reduction, which can identify key structures in high-dimensional data space and map them to low-dimensional space to accomplish dimensionality reduction. Compared to other dimensionality reduction algorithms, such as principal component analysis [36], UMAP can retain more global features [37]. In this paper, we constructed a corpus consisting of abstract words from all studies. However, due to the massive size of the corpus, visualizing and analyzing the high-dimensional data to explore the differences in the vocabulary patterns between the OAD and TIC studies is a challenge. The UMAP package in Python, which implements the UMAP algorithm, was used to project the high-dimensional corpus to 4 dimensions. By cross plotting each dimension, we were able to investigate underlying differences in corpus distribution between OAD and TIC studies.
BERTopic
Topic modeling can help us explore the similarities and differences between research topics in OAD and TIC studies. Unlike conventional topic modeling models, BERTopic uses the BERT framework for embeddings, enabling a deeper understanding of semantic relationships [30]. The BERTopic model was implemented by the BERTopic package in Python and divided 146,727 studies into 30 topic IDs. We also performed latent Dirichlet allocation topic modeling through Python’s LdaModel package for comparison. Through the review of topic keywords by 2 critical care physicians, BERTopic exhibited superior accuracy and sophistication in topic identification, with enhanced interpretability and scientific rigor.
Consequently, the BERTopic model was used for the final analysis. Each of these topics was given a corresponding clinical research category. The overlapping categories were merged into topic families for easier comparisons. By using these advanced techniques, we were able to uncover hidden patterns and relationships within the literature and provide insights into the current state of intensive care research.
Results
A total of 227,893 records were identified from the WoS database on January 18, 2023, of which 195,463 full records were subsequently processed. Records were excluded if they are not “article” or “review” or if they do not contain keywords related to intensive care. After exclusions, 145,426 articles were identified as TIC studies and 1301 articles were categorized as OAD (Figure 1).
Figure 1.
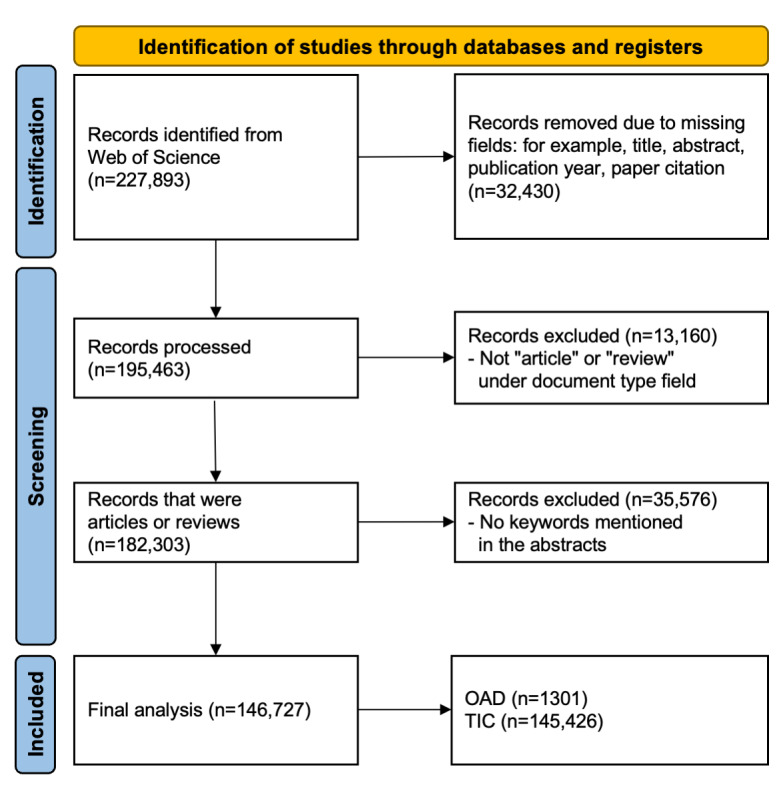
PRISMA (Preferred Reporting Items for Systematic Review and Meta-Analysis) diagram of the study. The final studies were divided into open-access database (OAD) and traditional intensive care (TIC) studies.
We examined the number of articles published per year to analyze the trends in TIC and OAD studies (Figure 2). Over the past 2 decades, TIC studies have experienced exponential growth, culminating in a peak of 16,378 articles in 2021. A subsequent decline in the number of publications occurred in 2022, likely attributable to delayed indexing within the WoS database and a reduction in COVID-19–related studies as the pandemic stabilized [38]. In contrast, the first OAD study emerged in 2003, with its popularity experiencing a consistent upsurge since 2018. Nonetheless, the number of OAD publications remains markedly lower in comparison to TIC publications.
Figure 2.

Number of publications of open-access database (OAD) and traditional intensive care (TIC) studies by year. The first study in the OAD category started in the year 2003.
The OAD studies were published most frequently in new open-access journals such as Frontiers in Medicine, Frontiers in Cardiovascular Medicine, and Scientific Reports while the TIC studies were published most frequently in established journals like Critical Care Medicine, Intensive Care Medicine, and Critical Care (Multimedia Appendix 2). Further analysis of keywords from the abstracts showed 2.4% (3492/145,426) TIC studies were meta-analyses or systematic reviews, while only 0.08% (1/1301) OAD study was in this category. There were 5.61% (73/1301) OAD studies, and 7.43% (10,799/145,426) TIC studies that had the keyword of “cost.” Examples of the data fields that are available within OADs such as MIMIC and eICU-CRD are listed in Textbox 1. Some information fields such as end-of-life goals and values and health care provider psychology are not available within the current EHRs extracted for OADs.
Examples of information available in the current open-access databases (OADs) and examples of information not available in OADs.
Examples of information that is available in current OADs
Patient information: demographics and social set-up
Hospital context: admission time and discharge time, intensive care unit (ICU) and hospital admissions, and pre-ICU admission
Diagnosis: physician-curated ICU diagnosis and data-driven phenotypes
Intervention: medications, procedures, and organ support
Diagnostics: blood test, microbiology, and scans
Clinical texts: clinical notes and diagnostic reports
Physiological monitoring: basic monitoring and waveforms
Examples of information that is not readily available in current OADs
Patient information: family set up and visiting, financial information, and special populations
Hospital context: post-ICU discharge details, delayed admission or discharge, and health personnel psychology
Diagnosis: pre-ICU history and diagnosis requiring clinical symptoms
Intervention: indications for interventions, complications, and intraoperative and postoperative
Diagnostics: pathology photographs, imaging, and molecular or genetic studies
Clinical texts: patient narratives, end-of-life goals and patient value, and health personnel behavior
Physiological monitoring: advanced monitoring
The UMAP algorithm was used to project the high-dimension corpus to 4 dimensions and allowed exploration of the vocabulary patterns between the OAD and TIC studies (Figure 3). The projection values are represented by the x-axis, while the densities are represented by the y-axis. The presence of considerable overlap between TIC studies and OAD studies suggests that they share a substantial number of common terminologies, which may correspond to similar research topics. Nonetheless, TIC studies exhibit a more extensive coverage than OAD studies, which may stem from broader research scope and extended research duration.
Figure 3.

Corpus distribution of open-access database and traditional intensive care studies along dimensions 1-4. OAD: open-access database; TIC: traditional intensive care.
Subsequently, the BERTopic model was then used to generate 30 topic IDs (Figure 4). The internal commonalities of each topic ID were reviewed by critical care physicians and assigned a specific subtopic in intensive care research. The model was able to automatically classify the topics with high interpretability and the topic components were interpreted with ease. For instance, components in topic ID 5 consist of, in decreasing order of weightage: “learning,” “model,” “machine,” “machine learning,” “models,” “data,” “prediction,” and “performance.” This topic was consequently labeled “predictive model” (topic ID 5 in Multimedia Appendix 3).
Figure 4.

The ratio of open-access database and traditional intensive care studies within each topic was identified by BERTopic.
The overall topic distribution in TIC studies was more uniform, while the OAD studies tended to be concentrated on several topics including topic ID 2 (kidney injury), 5 (predictive model), and 13 (sepsis). Some topics that were missing in OAD studies included 6 (pediatrics care), 21 (viral infections), 23 (health personnel and psychology), and 28 (nutrition and rehabilitation).
The similarity matrix shows that there was little overlap between the topics (Multimedia Appendix 4). To facilitate the interpretability of the categories, the overlapping topic IDs were merged to form the final 22 topic families (Multimedia Appendix 3).
Topics such as “healthcare associated infection,” “thoracic surgeries,” and “pregnancy related” research were among the more frequently discussed 15 topics in TIC studies but have limited publications in OAD studies. The topics of “predictive model,” “obesity,” and “fungal infections” were popular in OAD studies but not the TIC studies. Overall, the topic distributions of the TIC studies were distributed more evenly with the topic family of sepsis accounting for a quarter of the studies, while publications in the OAD studies were heavily skewed toward the predictive model (>40%) and sepsis (>30%; Figure 5).
Figure 5.

The top 15 topic families represented in open-access database and traditional intensive care studies.
Discussion
Principal Results
This study conducted a comprehensive review and bibliometric analysis of OAD and TIC studies. NLP was used to facilitate this large-scale literature review. Studies using OADs mainly concentrated on a few topics, such as predictive modeling, while TIC studies covered a wider range of topics with a more balanced distribution.
Advantages of OAD Studies
OAD studies offer several advantages that have contributed to their increasing popularity in intensive care research. The granularity of data and easy access to large-cohort databases, such as MIMIC [39], has enabled researchers to perform predictive modeling and conduct various secondary analyses efficiently [40,41]. This accessibility has provided valuable opportunities for exploring specific aspects of patient care, evident in studies investigating phenomena like “weekend effects” and circadian rhythms in ICU patients before discharge [42-46]. The vast amount of longitudinal and time series data available in OADs has also facilitated the implementation of complex ML and deep learning methods [47].
Limitations of OAD Studies
However, it is crucial to acknowledge the retrospective nature of OAD data, which inherently limits the assessment of confounding factors and the ability to draw strong causal conclusions. The observational design of OAD studies may result in lower-quality evidence according to the GRADE (Grading of Recommendations, Assessment, Development, and Evaluations) framework [48,49], and thus, the research from OAD studies has yet to be fully integrated into existing evidence-based guidelines, as exemplified by the omission of OAD studies in the 2021 sepsis guidelines [50]. Nevertheless, OADs remain a valuable resource for supplementing and complementing TIC studies, providing unique insights and enhanced predictive scores for intensive care settings.
Furthermore, approximately 50% of the studies using OADs published focused on predictive modeling. The increased usage of ML methods in predictive modeling has not been without critique. Some medical prediction problems inherently possess linear characteristics, and the selection of features may predominantly focus on already known strong predictors, leading to limited improvements in prediction accuracy with ML [51]. Additionally, interstudy heterogeneity poses a challenge in comparing results obtained from different ML models applied to the same data sets [52]. The ethical implications of relying solely on ML models to make high-risk health care decisions instead of involving clinical expertise are also relevant considerations [51,53].
While OADs provide comprehensive patient data, there are certain limitations in their ability to capture specific information essential for certain critical care research areas. Notably, data fields related to qualitative aspects such as ethics and end-of-life care [54,55], and health care personnel psychology [56] may be challenging, if not impossible, to obtain through OADs generated from EHRs. Consequently, TIC studies have played a pivotal role in addressing these limitations by capturing critical information that is integral to understanding ethical considerations, patient experiences, and health care provider psychology in intensive care [57,58].
Synergy Between OAD and TIC Studies
The synergy between OAD and TIC studies is a promising approach to enhance the comprehensiveness and robustness of intensive care research. OADs, with their large cohort sizes, can serve as external validation cohorts for ML models developed from TIC studies, potentially reducing the sample sizes required for prospective research. Furthermore, OAD studies can corroborate the results of TIC studies, benefiting from larger sample sizes and real-world data, thus providing more practical insights for implementation in intensive care settings [43]. The integration of OAD and TIC studies presents an opportunity to bridge the gaps in data availability and research methodologies, ultimately enriching the understanding and practice of critical care medicine.
Potential Impact of NLP
The usage of large language models such as BERTopic has proven to be a valuable tool for large-scale literature review and topic extraction [58]. This approach has enabled accurate, reliable, and granular topic generation, offering clinicians a more effective means of interpreting data compared to traditional bag-of-words models [59]. The potential of NLP to analyze scientific articles and identify trends and knowledge gaps holds promise for shaping the future of research in critical care medicine. As the volume of publications in critical care continues to grow and large language modeling continues to advance in health care [60], AI technology will be crucial in efficiently identifying and predicting emerging trends.
Future Directions
Future research in the field of critical care can explore novel applications of ML beyond predictive modeling. For instance, using ML to study patterns in how papers are cited, shared, and discussed on the web could help predict their potential impact on the scientific community. This analysis can aid in identifying highly influential papers and understanding the factors that contribute to their recognition. Additionally, investigations into methods for enhancing the interpretability and transparency of ML algorithms in critical care research would further facilitate the ethical and responsible use of AI technologies.
Strengths and Limitations
The study’s application of NLP-driven in analyzing scientific articles and identifying trends highlights the potential impact of AI technologies in streamlining literature reviews and identifying emerging trends more efficiently.
Another notable strength of this study is the usage of the WoS database, the world’s oldest and most extensively used repository of research publications and citations, encompassing approximately 34,000 journals [61]. The comprehensiveness of this database provides a robust representation of the literature in the field of intensive care research. Nevertheless, it is essential to acknowledge that some articles published in nonindexed journals might not have been captured, and future studies could benefit from considering additional databases to supplement our findings.
One other limitation lies in the classification of OAD and TIC studies, which may be subject to variations in the interpretation of keywords. However, we optimized the keyword combinations during the search process in the WoS database and implemented Python filtering techniques, resulting in a relatively high level of accuracy in our classifications. The number of studies was further corroborated with a manual search on PubMed and a review of the classifications of the studies was done by critical care physicians.
Although there were no specific language restrictions, the nature of the search term being in English inadvertently excluded valuable contributions from non-English research. This may potentially limit the generalizability of our findings to a broader international audience. In future investigations, the inclusion of articles from various languages could offer a more comprehensive and diverse perspective on intensive care research.
Conclusions
This study has provided valuable insights into the expanding landscape of intensive care research through a comprehensive bibliometric analysis of a large number of publications by leveraging NLP technologies. While OAD studies have demonstrated significant promise, it is essential to view them as a complementary approach rather than a replacement for TIC studies. The unique strength of TIC studies lies in their ability to capture crucial qualitative information, which is essential for comprehensive and ethical decision-making. The integration of both OAD and TIC studies offers a synergistic approach to enriching our understanding of critical care medicine and advancing patient care outcomes. As NLP technology continues to advance, it holds the potential to offer a feasible and transformative alternative for literature review and bibliometric analysis.
Acknowledgments
We thank Dr Nicholas Brian Shannon for assistance with the manual review of the supervised keyword classification. This work was supported by the Duke-NUS Signature Research Programme, funded by the Ministry of Health, Singapore.
Abbreviations
- AmsterdamUMCdb
Amsterdam University Medical Centers Database
- BERT
bidirectional encoder representations from transformers
- EHR
electronic health record
- eICU-CRD
eICU Collaborative Research Database
- FAIR
findable, accessible, interoperable, and reusable
- GRADE
Grading of Recommendations, Assessment, Development, and Evaluations
- HiRID
High Time Resolution ICU Dataset
- ICU
intensive care unit
- MIMIC
Medical Information Mart for Intensive Care
- ML
machine learning
- NLP
natural language processing
- OAD
open-access database
- TIC
traditional intensive care
- UMAP
uniform manifold approximation and projection
- WoS
Web of Science
Search terms for open-access database (OAD) studies with the cutoff by the years of publications.
Top 20 journals ranked by total citation in which the open-access database and traditional intensive care studies were published. The average citation per article was obtained with the total citation/total number of articles. The citation counts were obtained from Web of Science.
Topic ID and topic family and the components and weightage in each of the categories.
Similarity matrix of 30 topics.
Data Availability
The data sets generated during and/or analyzed during this study are available from the corresponding author on reasonable request. The complete set of code used in this study is readily available for download on GitHub [62].
Footnotes
Authors' Contributions: YK and NL played key roles in the conceptualization of the project. RY was responsible for formalizing the methodology and conducting data curation with the advisory of YK. YK contributed to the validation of the data, ensuring its relevance to the research objectives. RY took the lead in visualizing the data. Both YK and RY drafted the original manuscript. NL served as the project supervisor, overseeing the implementation, and providing valuable input in the writing, review, and editing phases.
Conflicts of Interest: None declared.
References
- 1.Kelly FE, Fong K, Hirsch N, Nolan JP. Intensive care medicine is 60 years old: the history and future of the intensive care unit. Clin Med (Lond) 2014;14(4):376–379. doi: 10.7861/clinmedicine.14-4-376. https://linkinghub.elsevier.com/retrieve/pii/14/4/376 .14/4/376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook D, Brower R, Cooper J, Brochard L, Vincent JL. Multicenter clinical research in adult critical care. Crit Care Med. 2002;30(7):1636–1643. doi: 10.1097/00003246-200207000-00039. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg AL, Tripathi RS, Blum J. The most influential articles in critical care medicine. J Crit Care. 2010;25(1):157–170. doi: 10.1016/j.jcrc.2008.12.010. https://europepmc.org/abstract/MED/19327953 .S0883-9441(08)00265-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Granholm A, Alhazzani W, Derde LPG, Angus DC, Zampieri FG, Hammond NE, Sweeney RM, Myatra SN, Azoulay E, Rowan K, Young PJ, Perner A, Møller MH. Randomised clinical trials in critical care: past, present and future. Intensive Care Med. 2022;48(2):164–178. doi: 10.1007/s00134-021-06587-9. https://europepmc.org/abstract/MED/34853905 .10.1007/s00134-021-06587-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markey KA, Ottridge R, Mitchell JL, Rick C, Woolley R, Ives N, Nightingale P, Sinclair AJ. Assessing the efficacy and safety of an 11β-hydroxysteroid dehydrogenase type 1 inhibitor (AZD4017) in the idiopathic intracranial hypertension drug trial, IIH:DT: clinical methods and design for a phase II randomized controlled trial. JMIR Res Protoc. 2017;6(9):e181. doi: 10.2196/resprot.7806. https://www.researchprotocols.org/2017/9/e181/ v6i9e181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdonk F, Feyaerts D, Badenes R, Bastarache JA, Bouglé A, Ely W, Gaudilliere B, Howard C, Kotfis K, Lautrette A, Le Dorze M, Mankidy BJ, Matthay MA, Morgan CK, Mazeraud A, Patel BV, Pattnaik R, Reuter J, Schultz MJ, Sharshar T, Shrestha GS, Verdonk C, Ware LB, Pirracchio R, Jabaudon M. Upcoming and urgent challenges in critical care research based on COVID-19 pandemic experience. Anaesth Crit Care Pain Med. 2022;41(5):101121. doi: 10.1016/j.accpm.2022.101121. https://linkinghub.elsevier.com/retrieve/pii/S2352-5568(22)00102-3 .S2352-5568(22)00102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurnani PK, Patel GP, Crank CW, Vais D, Lateef O, Akimov S, Balk R, Simon D. Impact of the implementation of a sepsis protocol for the management of fluid-refractory septic shock: a single-center, before-and-after study. Clin Ther. 2010;32(7):1285–1293. doi: 10.1016/j.clinthera.2010.07.003.S0149-2918(10)00230-4 [DOI] [PubMed] [Google Scholar]
- 8.Wang JL, Chin CS, Chang MC, Yi CY, Shih SJ, Hsu JY, Wu CL. Key process indicators of mortality in the implementation of protocol-driven therapy for severe sepsis. J Formos Med Assoc. 2009;108(10):778–787. doi: 10.1016/S0929-6646(09)60405-8. https://linkinghub.elsevier.com/retrieve/pii/S0929-6646(09)60405-8 .S0929-6646(09)60405-8 [DOI] [PubMed] [Google Scholar]
- 9.Levy MM, Evans LE, Rhodes A. The surviving sepsis campaign bundle: 2018 update. Crit Care Med. 2018;46(6):997–1000. doi: 10.1097/CCM.0000000000003119. https://journals.lww.com/ccmjournal/fulltext/2018/06000/the_surviving_sepsis_campaign_bundle__2018_update.21.aspx .00003246-201806000-00021 [DOI] [PubMed] [Google Scholar]
- 10.Fan E, Del Sorbo L, Goligher EC, Hodgson CL, Munshi L, Walkey AJ, Adhikari NKJ, Amato MBP, Branson R, Brower RG, Ferguson ND, Gajic O, Gattinoni L, Hess D, Mancebo J, Meade MO, McAuley DF, Pesenti A, Ranieri VM, Rubenfeld GD, Rubin E, Seckel M, Slutsky AS, Talmor D, Thompson BT, Wunsch H, Uleryk E, Brozek J, Brochard LJ. An official American Thoracic Society/European Society of Intensive Care Medicine/Society of Critical Care Medicine clinical practice guideline: mechanical ventilation in adult patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2017;195(9):1253–1263. doi: 10.1164/rccm.201703-0548ST. https://www.atsjournals.org/doi/10.1164/rccm.201703-0548ST . [DOI] [PubMed] [Google Scholar]
- 11.Goldfrad C, Vella K, Bion JF, Rowan KM, Black NA. Research priorities in critical care medicine in the UK. Intensive Care Med. 2000;26(10):1480–1488. doi: 10.1007/s001340000628. [DOI] [PubMed] [Google Scholar]
- 12.Moore TJ, Heyward J, Anderson G, Alexander GC. Variation in the estimated costs of pivotal clinical benefit trials supporting the US approval of new therapeutic agents, 2015-2017: a cross-sectional study. BMJ Open. 2020;10(6):e038863. doi: 10.1136/bmjopen-2020-038863. https://bmjopen.bmj.com/lookup/pmidlookup?view=long&pmid=32532786 .bmjopen-2020-038863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umscheid CA, Margolis DJ, Grossman CE. Key concepts of clinical trials: a narrative review. Postgrad Med. 2011;123(5):194–204. doi: 10.3810/pgm.2011.09.2475. https://europepmc.org/abstract/MED/21904102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maré IA, Kramer B, Hazelhurst S, Nhlapho MD, Zent R, Harris PA, Klipin M. Electronic data capture system (REDCap) for health care research and training in a resource-constrained environment: technology adoption case study. JMIR Med Inform. 2022;10(8):e33402. doi: 10.2196/33402. https://medinform.jmir.org/2022/8/e33402/ v10i8e33402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Hearn K, Gibson J, Krewulak K, Porteous R, Saigle V, Sampson M, Tsampalieros A, Barrowman N, Cameron S. Consent models in Canadian critical care randomized controlled trials: a scoping review. Can J Anaesth. 2022;69(4):513–526. doi: 10.1007/s12630-021-02176-y. https://link.springer.com/article/10.1007/s12630-021-02176-y .10.1007/s12630-021-02176-y [DOI] [PubMed] [Google Scholar]
- 16.Ghassemi M, Celi LA, Stone DJ. State of the art review: the data revolution in critical care. Crit Care. 2015;19(1):118. doi: 10.1186/s13054-015-0801-4. https://ccforum.biomedcentral.com/articles/10.1186/s13054-015-0801-4 .10.1186/s13054-015-0801-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bates DW, Saria S, Ohno-Machado L, Shah A, Escobar G. Big data in health care: using analytics to identify and manage high-risk and high-cost patients. Health Aff (Millwood) 2014;33(7):1123–1131. doi: 10.1377/hlthaff.2014.0041.33/7/1123 [DOI] [PubMed] [Google Scholar]
- 18.Mlodzinski E, Wardi G, Viglione C, Nemati S, Crotty Alexander L, Malhotra A. Assessing barriers to implementation of machine learning and artificial intelligence-based tools in critical care: web-based survey study. JMIR Perioper Med. 2023;6:e41056. doi: 10.2196/41056. https://periop.jmir.org/2023//e41056/ v6i1e41056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson AEW, Bulgarelli L, Shen L, Gayles A, Shammout A, Horng S, Pollard TJ, Hao S, Moody B, Gow B, Lehman LWH, Celi LA, Mark RG. MIMIC-IV, a freely accessible electronic health record dataset. Sci Data. 2023;10(1):1. doi: 10.1038/s41597-022-01899-x. doi: 10.1038/s41597-022-01899-x.10.1038/s41597-022-01899-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O. The eICU collaborative research database, a freely available multi-center database for critical care research. Sci Data. 2018;5:180178. doi: 10.1038/sdata.2018.178. doi: 10.1038/sdata.2018.178.sdata2018178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inau ET, Sack J, Waltemath D, Zeleke AA. Initiatives, concepts, and implementation practices of FAIR (findable, accessible, interoperable, and reusable) data principles in health data stewardship practice: protocol for a scoping review. JMIR Res Protoc. 2021;10(2):e22505. doi: 10.2196/22505. https://www.researchprotocols.org/2021/2/e22505/ v10i2e22505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, 't Hoen PAC, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. doi: 10.1038/sdata.2016.18.sdata201618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahadoran Z, Mirmiran P, Kashfi K, Ghasemi A. Importance of systematic reviews and meta-analyses of animal studies: challenges for animal-to-human translation. J Am Assoc Lab Anim Sci. 2020;59(5):469–477. doi: 10.30802/AALAS-JAALAS-19-000139. https://europepmc.org/abstract/MED/32727637 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haidich AB. Meta-analysis in medical research. Hippokratia. 2010;14(Suppl 1):29–37. https://europepmc.org/abstract/MED/21487488 . [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson DF, Walker CK. A descriptive and historical review of bibliometrics with applications to medical sciences. Pharmacotherapy. 2015;35(6):551–559. doi: 10.1002/phar.1586. [DOI] [PubMed] [Google Scholar]
- 26.Donthu N, Kumar S, Mukherjee D, Pandey N, Lim WM. How to conduct a bibliometric analysis: an overview and guidelines. J Bus Res. 2021;133:285–296. doi: 10.1016/j.jbusres.2021.04.070. https://www.sciencedirect.com/science/article/pii/S0148296321003155?via%3Dihub . [DOI] [Google Scholar]
- 27.Zhao W, Chen JJ, Perkins R, Liu Z, Ge W, Ding Y, Zou W. A heuristic approach to determine an appropriate number of topics in topic modeling. BMC Bioinformatics. 2015;16(Suppl 13):S8. doi: 10.1186/1471-2105-16-S13-S8. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-16-S13-S8 .1471-2105-16-S13-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doanvo A, Qian X, Ramjee D, Piontkivska H, Desai A, Majumder M. Machine learning maps research needs in COVID-19 literature. Patterns (N Y) 2020;1(9):100123. doi: 10.1016/j.patter.2020.100123. https://linkinghub.elsevier.com/retrieve/pii/S2666-3899(20)30164-1 .S2666-3899(20)30164-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. ArXiv. Preprint posted online on May 24 2019. 2018 doi: 10.48550/arXiv.1810.04805. [DOI] [Google Scholar]
- 30.Grootendorst M. BERTopic: neural topic modeling with a class-based TF-IDF procedure. ArXiv. Preprint posted online on March 11 2022. 2022 doi: 10.48550/arXiv.2203.05794. [DOI] [Google Scholar]
- 31.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. J Mach Learn Res. 2003;3:993–1022. https://www.jmlr.org/papers/volume3/blei03a/blei03a.pdf?ref=http://githubhelp.com . [Google Scholar]
- 32.Delaney A, Bagshaw SM, Ferland A, Manns B, Laupland KB, Doig CJ. A systematic evaluation of the quality of meta-analyses in the critical care literature. Crit Care. 2005;9(5):R575–R582. doi: 10.1186/cc3803. https://ccforum.biomedcentral.com/articles/10.1186/cc3803 .cc3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoral PJ, Peppink JM, Driessen RH, Sijbrands EJG, Kompanje EJO, Kaplan L, Bailey H, Kesecioglu J, Cecconi M, Churpek M, Clermont G, van der Schaar M, Ercole A, Girbes ARJ, Elbers PWG. Sharing ICU patient data responsibly under the Society of Critical Care Medicine/European Society of Intensive Care Medicine joint data science collaboration: the Amsterdam university medical centers database (AmsterdamUMCdb) example. Crit Care Med. 2021;49(6):e563–e577. doi: 10.1097/CCM.0000000000004916. https://europepmc.org/abstract/MED/33625129 .00003246-202106000-00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faltys M, Zimmermann M, Lyu X, Hüser M, Hyland S, Rätsch G, Merz T. HiRID, a high time-resolution ICU dataset. PhysioNet. 2021. [2024-04-02]. https://physionet.org/content/hirid/1.1.1/
- 35.Zeng X, Yu G, Lu Y, Tan L, Wu X, Shi S, Duan H, Shu Q, Li H. PIC, a paediatric-specific intensive care database. Sci Data. 2020;7(1):14. doi: 10.1038/s41597-020-0355-4. doi: 10.1038/s41597-020-0355-4.10.1038/s41597-020-0355-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maćkiewicz A, Ratajczak W. Principal components analysis (PCA) Comput Geosci. 1993;19(3):303–342. doi: 10.1016/0098-3004(93)90090-R. [DOI] [Google Scholar]
- 37.McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. ArXiv. Preprint posted online on September 18 2020. 2018 doi: 10.48550/arXiv.1802.03426. [DOI] [Google Scholar]
- 38.Murray CJL. COVID-19 will continue but the end of the pandemic is near. Lancet. 2022;399(10323):417–419. doi: 10.1016/S0140-6736(22)00100-3. https://europepmc.org/abstract/MED/35065006 .S0140-6736(22)00100-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mark R. Secondary Analysis of Electronic Health Records. Cham, Switzerland: Springer International Publishing; 2016. The story of MIMIC; pp. 43–49. [Google Scholar]
- 40.Alghatani K, Ammar N, Rezgui A, Shaban-Nejad A. Predicting intensive care unit length of stay and mortality using patient vital signs: machine learning model development and validation. JMIR Med Inform. 2021;9(5):e21347. doi: 10.2196/21347. https://medinform.jmir.org/2021/5/e21347/ v9i5e21347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu D, Zheng M, Sepulveda NA. Using artificial neural network condensation to facilitate adaptation of machine learning in medical settings by reducing computational burden: model design and evaluation study. JMIR Form Res. 2021;5(12):e20767. doi: 10.2196/20767. https://formative.jmir.org/2021/12/e20767/ v5i12e20767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Ho KM, Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Crit Care. 2019;23(1):112. doi: 10.1186/s13054-019-2411-z. https://ccforum.biomedcentral.com/articles/10.1186/s13054-019-2411-z .10.1186/s13054-019-2411-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer A, Zverinski D, Pfahringer B, Kempfert J, Kuehne T, Sündermann SH, Stamm C, Hofmann T, Falk V, Eickhoff C. Machine learning for real-time prediction of complications in critical care: a retrospective study. Lancet Respir Med. 2018;6(12):905–914. doi: 10.1016/S2213-2600(18)30300-X.S2213-2600(18)30300-X [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Zhu Z, Zhao C, Guo Y, Chen D, Wei Y, Jin J. Central venous pressure measurement is associated with improved outcomes in septic patients: an analysis of the MIMIC-III database. Crit Care. 2020;24(1):433. doi: 10.1186/s13054-020-03109-9. https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-03109-9 .10.1186/s13054-020-03109-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faust L, Feldman K, Chawla NV. Examining the weekend effect across ICU performance metrics. Crit Care. 2019;23(1):207. doi: 10.1186/s13054-019-2479-5. https://ccforum.biomedcentral.com/articles/10.1186/s13054-019-2479-5 .10.1186/s13054-019-2479-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson S, Villarroel M, Harford M, Finnegan E, Jorge J, Young D, Watkinson P, Tarassenko L. Vital-sign circadian rhythms in patients prior to discharge from an ICU: a retrospective observational analysis of routinely recorded physiological data. Crit Care. 2020;24(1):181. doi: 10.1186/s13054-020-02861-2. https://ccforum.biomedcentral.com/articles/10.1186/s13054-020-02861-2 .10.1186/s13054-020-02861-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie F, Yuan H, Ning Y, Ong MEH, Feng M, Hsu W, Chakraborty B, Liu N. Deep learning for temporal data representation in electronic health records: a systematic review of challenges and methodologies. J Biomed Inform. 2022;126:103980. doi: 10.1016/j.jbi.2021.103980. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(21)00309-9 .S1532-0464(21)00309-9 [DOI] [PubMed] [Google Scholar]
- 48.Guyatt GH, Oxman AD, Kunz R, Vist GE, Falck-Ytter Y, Schünemann HJ, GRADE Working Group What is "quality of evidence" and why is it important to clinicians? BMJ. 2008;336(7651):995–998. doi: 10.1136/bmj.39490.551019.BE. https://europepmc.org/abstract/MED/18456631 .336/7651/995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck-Ytter Y, Alonso-Coello P, Schünemann HJ. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336(7650):924–926. doi: 10.1136/bmj.39489.470347.AD. https://europepmc.org/abstract/MED/18436948 .336/7650/924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, Machado FR, Mcintyre L, Ostermann M, Prescott HC, Schorr C, Simpson S, Wiersinga WJ, Alshamsi F, Angus DC, Arabi Y, Azevedo L, Beale R, Beilman G, Belley-Cote E, Burry L, Cecconi M, Centofanti J, Coz Yataco A, De Waele J, Dellinger RP, Doi K, Du B, Estenssoro E, Ferrer R, Gomersall C, Hodgson C, Hylander Møller M, Iwashyna T, Jacob S, Kleinpell R, Klompas M, Koh Y, Kumar A, Kwizera A, Lobo S, Masur H, McGloughlin S, Mehta S, Mehta Y, Mer M, Nunnally M, Oczkowski S, Osborn T, Papathanassoglou E, Perner A, Puskarich M, Roberts J, Schweickert W, Seckel M, Sevransky J, Sprung CL, Welte T, Zimmerman J, Levy M. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit Care Med. 2021;49(11):e1063–e1143. doi: 10.1097/CCM.0000000000005337. https://journals.lww.com/ccmjournal/fulltext/2021/11000/surviving_sepsis_campaign__international.21.aspx .00003246-202111000-00021 [DOI] [PubMed] [Google Scholar]
- 51.Volovici V, Syn NL, Ercole A, Zhao JJ, Liu N. Steps to avoid overuse and misuse of machine learning in clinical research. Nat Med. 2022;28(10):1996–1999. doi: 10.1038/s41591-022-01961-6. https://www.nature.com/articles/s41591-022-01961-6 .10.1038/s41591-022-01961-6 [DOI] [PubMed] [Google Scholar]
- 52.Fleuren LM, Klausch TLT, Zwager CL, Schoonmade LJ, Guo T, Roggeveen LF, Swart EL, Girbes ARJ, Thoral P, Ercole A, Hoogendoorn M, Elbers PWG. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46(3):383–400. doi: 10.1007/s00134-019-05872-y. https://europepmc.org/abstract/MED/31965266 .10.1007/s00134-019-05872-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoon CH, Torrance R, Scheinerman N. Machine learning in medicine: should the pursuit of enhanced interpretability be abandoned? J Med Ethics. 2022;48(9):581–585. doi: 10.1136/medethics-2020-107102. http://jme.bmj.com/lookup/pmidlookup?view=long&pmid=34006600 .medethics-2020-107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillett GR. Intensive care unit research ethics and trials on unconscious patients. Anaesth Intensive Care. 2015;43(3):309–312. doi: 10.1177/0310057X1504300304.20140695 [DOI] [PubMed] [Google Scholar]
- 55.Aulisio MP, Chaitin E, Arnold RM. Ethics and palliative care consultation in the intensive care unit. Crit Care Clin. 2004;20(3):505–523, x-xi. doi: 10.1016/j.ccc.2004.03.006.S0749070404000090 [DOI] [PubMed] [Google Scholar]
- 56.Raudenská J, Steinerová V, Javůrková A, Urits I, Kaye AD, Viswanath O, Varrassi G. Occupational burnout syndrome and post-traumatic stress among healthcare professionals during the novel coronavirus disease 2019 (COVID-19) pandemic. Best Pract Res Clin Anaesthesiol. 2020;34(3):553–560. doi: 10.1016/j.bpa.2020.07.008. https://europepmc.org/abstract/MED/33004166 .S1521-6896(20)30063-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson JE, Jones C, Bienvenu OJ. Family response to critical illness: postintensive care syndrome-family. Crit Care Med. 2012;40(2):618–624. doi: 10.1097/CCM.0b013e318236ebf9. [DOI] [PubMed] [Google Scholar]
- 58.White DB, Angus DC, Shields AM, Buddadhumaruk P, Pidro C, Paner C, Chaitin E, Chang CCH, Pike F, Weissfeld L, Kahn JM, Darby JM, Kowinsky A, Martin S, Arnold RM. A randomized trial of a family-support intervention in intensive care units. N Engl J Med. 2018;378(25):2365–2375. doi: 10.1056/NEJMoa1802637. https://www.nejm.org/doi/10.1056/NEJMoa1802637 . [DOI] [PubMed] [Google Scholar]
- 59.Popoff B, Occhiali É, Grangé S, Bergis A, Carpentier D, Tamion F, Veber B, Clavier T. Trends in major intensive care medicine journals: a machine learning approach. J Crit Care. 2022;72:154163. doi: 10.1016/j.jcrc.2022.154163.S0883-9441(22)00192-7 [DOI] [PubMed] [Google Scholar]
- 60.Yang R, Tan TF, Lu W, Thirunavukarasu AJ, Ting DSW, Liu N. Large language models in health care: development, applications, and challenges. Health Care Sci. 2023;2(4):255–263. doi: 10.1002/hcs2.61. https://onlinelibrary.wiley.com/doi/10.1002/hcs2.61 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Birkle C, Pendlebury DA, Schnell J, Adams J. Web of science as a data source for research on scientific and scholarly activity. Quant Sci Stud. 2020;1(1):363–376. doi: 10.1162/qss_a_00018. https://direct.mit.edu/qss/article/1/1/363/15569/Web-of-Science-as-a-data-source-for-research-on . [DOI] [Google Scholar]
- 62.GitHub. https://github.com/YangRui525/Comparing-OAD-and-TIC-Studies .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Search terms for open-access database (OAD) studies with the cutoff by the years of publications.
Top 20 journals ranked by total citation in which the open-access database and traditional intensive care studies were published. The average citation per article was obtained with the total citation/total number of articles. The citation counts were obtained from Web of Science.
Topic ID and topic family and the components and weightage in each of the categories.
Similarity matrix of 30 topics.
Data Availability Statement
The data sets generated during and/or analyzed during this study are available from the corresponding author on reasonable request. The complete set of code used in this study is readily available for download on GitHub [62].


