Abstract
Despite recent advances in the development of scaffold-based three-dimensional (3D) cell models, challenges persist in imaging and monitoring cell behavior within these complex structures due to their heterogeneous cell distribution and geometries. Incorporating sensors into 3D scaffolds provides a potential solution for real-time, in situ sensing and imaging of biological processes such as cell growth and disease development. We introduce a 3D printed hydrogel-based scaffold capable of supporting both surface-enhanced Raman scattering (SERS) biosensing and imaging of 3D breast cancer cell models. The scaffold incorporates plasmonic nanoparticles and SERS tags, for sensing and imaging, respectively. We demonstrate the scaffold’s adaptability and modularity in supporting breast cancer spheroids, thereby enabling spatial and temporal monitoring of tumor evolution.
Keywords: SERS, biosensing, bioimaging, 3D printing, tumor microenvironment, 3D cell models
Introduction
The transition from traditional in vitro two-dimensional (2D) cell cultures to more complex three-dimensional (3D) configurations is crucial in cancer research, toward reproducing biological processes under more realistic conditions, thereby facilitating the translation of in vitro results to the clinic.1 Traditional 2D approaches fail to replicate the complexity of the cancer microenvironment, whereas animal models, besides ethical concerns, show divergences in terms of immune system and cell genotype.2 In contrast, 3D cancer models promote cell–cell interactions and cell–extracellular matrix (ECM) interactions, while supporting cell growth for different cell types.1 Spheroids, or tumoroids in particular, offer a high degree of flexibility in recreating various aspects of the tumor microenvironment (TME), including heterotypic cell mixtures, cellular morphology, motility, growth, drug resistance, and metastatic behavior.3,4 When supported by 3D printed scaffolds, cells can also adhere, proliferate, differentiate, migrate, and invade, better recreating the real scenario.5−8
Despite the development of numerous 3D cell models and scaffold/cell combinations, imaging and monitoring cell behavior within such complex structures remains challenging.9,10 Incorporation of sensors into scaffolds designed to support 3D cell growth may offer the possibility of in situ sensing and imaging relevant biological processes, thereby providing real-time information on different aspects such as cell growth, disease development, or response to drug treatments. The versatility of 3D printing techniques, especially regarding ink composition, makes it particularly interesting to control the spatial distribution of nanoparticle (NP)-based sensors and labels into ink formulations.11−14 Among the various printing techniques available, extrusion-based 3D printing is suitable for biomedical applications because hydrogels, highly biocompatible polymer materials sharing characteristics of living tissues, can be readily printed with sufficient spatial resolution. Additionally, ink development can leverage the well-studied hydrogel chemistry to tune the composition of the inks, in terms of chemical composition, crosslinking strategies, rheological properties, and compatibility with nano- and biomaterials.15,16
We and others have demonstrated the incorporation of plasmonic NPs within 3D scaffolds for surface-enhanced Raman scattering (SERS)-based sensing and imaging of model molecules and cells, respectively.17−23 Whereas bare ligand-free anisotropic plasmonic NPs enable label-free sensing of cell-secreted metabolites, NPs labeled with Raman-active molecules (Raman reporters) on their surface, also known as SERS tags, allow for SERS imaging with a well-defined Raman fingerprint. Efforts have focused on synthesizing biocompatible and stable SERS tags that can be effectively internalized by different cell types, to provide a multiplex imaging platform for biological systems (simultaneous detection of different SERS tags with a single illumination laser beam and narrow Raman peaks).24,25 Compared to other optical imaging techniques, such as fluorescence microscopy, SERS additionally offers various advantages including negligible photobleaching during imaging sessions (avoiding well-known loss of fluorescence signal overtime) and the possibility to irradiate samples within the near-infrared (NIR) biological transparency windows, thereby enhancing light penetration into living tissues.26 Indeed, this latter feature can also be exploited toward the use of NIR-luminescent NPs as nanothermometers. For example, rare earth (Nd3+ in particular)-based fluorescent nanothermometers display temperature dependent photoluminescence (PL) within biological windows, and have thus become highly attractive for measuring local temperature in cellular environments.27,28 However, SERS sensing and imaging, as well as NIR-nanothermometry, still face significant challenges such as the need for optically transparent materials and improved data analysis methods suitable for 3D models, in which background noise often hinders signal quality.19,26
Although our previous studies provided the foundation for scaffold-based imaging, various hurdles were identified including poor resolution and material transparency, which limited their application to more complex biological models. We present herein a different scaffold formulation that allows in situ SERS biosensing and bioimaging of human breast cancer 3D tumoroids, over extended periods of time and in a noninvasive manner. The developed ink formulation, based on polyethylene glycol diacrylate (PEGDA), offers excellent printing resolution in 3D, along with chemical compatibility with inorganic NPs, including bare (ligand-free) metal NPs and SERS tags, for sensing and imaging purposes, respectively. Additionally, we present proof-of-concept results on the simultaneous incorporation of photoluminescent nanothermometers, as a means to measure temperature with high spatial resolution and accuracy along scaffold fibers. To illustrate the practical utility of these versatile scaffolds, we integrated as the cellular environment an optimized extracellular matrix, where human breast cancer and stromal cells could be grown into aggregates resembling heterogeneous tumoroids. Our findings support the use of these multifunctional scaffolds as a platform to study complex cell cultures in realistic environments such as tumor models, via noninvasive and noncytotoxic techniques.
Results and Discussion
Scaffold Fabrication and Characterization
3D plasmonic scaffolds were fabricated by extrusion printing of a PEGDA-based formulation comprising PEGDA, κ-carrageenan, fumed silica, and gold nanorods (AuNRs). From our previous benchmarking of AuNRs vs. gold nanostars (AuNSs), with different surface ligands, bare ligand-free AuNRs were identified as the best performing SERS enhancers within hydrogel scaffolds.17 In the present work, AuNRs labeled with 2-naphthalenethiol (2NAT) were employed as SERS tags for scaffold imaging (AuNR@2NAT). The main localized surface plasmon resonance (LSPR) of the synthesized AuNRs was tuned to match the 785 nm excitation wavelength of the laser employed for SERS studies, and TEM images confirmed the expected morphology of the NPs (Figure S1A,B). In the case of AuNR@2NAT, the SERS spectrum of the NP dispersion in water additionally matched the characteristic molecular fingerprint of 2NAT and the NP concentration was tuned to maximize the SERS signal within the scaffold (Figure S1C).
The choice of PEGDA for the ink formulation was based on the rapid photo-crosslinking of this biocompatible polymer, which ensures shape retention and fidelity of the printed constructs. Fumed silica and κ-carrageenan were added to improve printability and elasticity, respectively. Rheological characterization of the ink confirmed the shear-thinning behavior of the formulation, which facilitates 3D printing and ensures recovery of the viscoelastic properties after extrusion (Figure S2A,B). Swelling and biocompatibility tests of the printed scaffolds (Figure S2C,D) demonstrated high stability in cell media and no cytotoxic effects, respectively. The amount of photoinitiator was optimized to maximize viability of the MCF7 cell line in contact with the scaffold (Figure S2D). The long-term stability of the scaffold was also verified using reflection confocal microscopy (Figure S3).
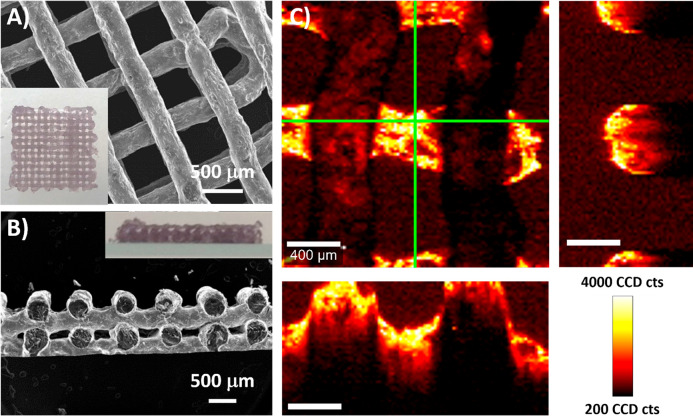
We thus printed 3D scaffolds (Figure 1A) in the form of multilayer grids, with an interlinear distance of 800 μm and a varying number of layers (from 2 to 6), using a RegenHu Discovery bioprinter equipped with an integrated UV lamp source (365 nm). Thanks to the rapid photo-crosslinking of each layer, well-defined round-shaped fibers were obtained and no collapse between layers was observed (Figure 1A,B). The printed fibers had a diameter of 292 ± 27 μm and a window width of 486 ± 32 μm. Compared to our previously published works, an improved definition of the cross-section was achieved with this formulation, comprising well-defined windows between fibers in all directions of the scaffold (XYZ).17,18 The high definition of the scaffold, as well as the uniform distribution of AuNRs within the fibers, were confirmed by SERS mapping of the characteristic signal of a model molecule, 4-mercaptobenzoic acid (MBA) (Figure 1C). Three-dimensional reconstruction of the MBA SERS signal revealed the grid-like and layered structure of the scaffold, with the different layers being perfectly distinguished from each other. When scanning in the Z direction, the MBA SERS signal could be recorded throughout the different layers of the scaffold, down to 1000 μm in depth (Figure S4).
Figure 1.
(A) SEM image showing a top view 3D-printed scaffold (1 cm × 1 cm). A top-view photograph is shown as an inset. (B) SEM image showing a cross-section of the scaffold. A side-view photograph is shown as an inset. (C) SERS maps based on the intensity of the MBA peak at 1084 cm–1, showing details of the XY plane (focused on the second layer of the scaffold) and corresponding XZ and YZ planes, defined by green lines (scale bars: 400 μm).
3D Cell Culture and SERS Imaging
We next studied the use the PEGDA scaffolds to support the growth of cancer cells. We have previously shown that similar scaffolds offer a suitable substrate for HeLa epithelial cell growth and that the grid-like 3D printing design provides imaging windows that allow cell monitoring.17 The improved spatial resolution of the present PEGDA scaffolds in the Z-axis was expected to improve the 3D distribution of cells, which would be located in and around the scaffold fibers, in a three-dimensional fashion. To test this hypothesis, we first coated the scaffold fibers with a mixture of fibronectin and collagen, two major components of the ECM, known to aid cell adhesion, followed by immersion of the modified scaffold in a suspension of MCF7 breast cancer epithelial cells (see scheme in Figure S5). Immunofluorescence staining was used to confirm the successful adhesion of both biomolecules onto the scaffold (Figure S6). MCF7 cells, previously transfected to express green fluorescent protein (GFP), were observed to adhere onto the coated scaffold surface, with lamellipodia and filopodia developing over time, both being features of cell spreading and key to migration (Figure 2A,B, Figures S7–S9). The addition of fibronectin and collagen, as well as the experimental setup involving rotation of the scaffold inside the cell suspension (see Experimental Section for more detail), were key to achieve efficient cell adherence; in trials lacking both ECM components or under static conditions, cells were observed to pass straight through the voids in between the fibers, all the way to the underlying (bottom) surface (Figure S10), as also observed in previous work involving scaffolds with imaging windows.28
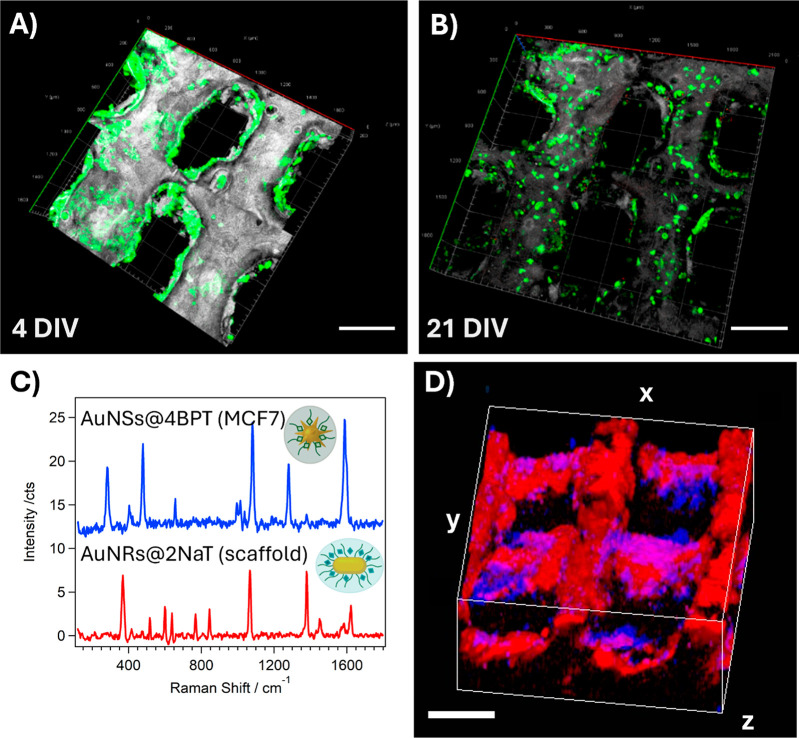
Figure 2.
(A,B) 3D confocal fluorescence microscopy reconstructions of (GFP-expressing) MCF7 cells cultured on scaffolds coated with fibronectin and collagen, after 4 (A) and 21 (B) days in vitro (DIV) (Scale bars: 500 μm). (C) SERS spectra of AuNR@2NAT in the scaffold and AuNS@4BPT in labeled MCF7 cells. (D) 3D SERS reconstruction of MCF7 cells labeled with AuNS@4BPT (blue) on a scaffold labeled with AuNR@2NAT (red) after 5 DIV. The cube dimensions are 2 × 2 × 1 mm3 (scale bars: 500 μm).
We aimed, as a major target, at the implementation of SERS as a dual imaging/sensing tool, to monitor live 3D tumor models over extended periods of time. We thus explored the possibility of monitoring the growth of MCF7 cells in 3D PEGDA scaffolds by SERS. For that purpose, the scaffolds were labeled with AuNR@2NAT, whereas MCF7 cells were labeled with 4-bisphenylthiol-encoded AuNSs (AuNS@4BPT) (see Figure S1D–H for SERS tag characterization). It is important to highlight the wide range of available SERS tags.29 The criteria guiding the signal assignment in each case were devised to optimize the SERS signal while maximizing multiplexing. Cells were incorporated into the scaffold again, after fibronectin and collagen coating, as previously described (see scheme in Figure S5). Characteristic spectra for both SERS tags were identified (Figure 2C) and the recorded SERS signals were used (Figure 2D) for simultaneous 3D reconstruction of the labeled scaffold (red) and the cells (blue). A homogeneous distribution of MCF7 cells was observed all over the scaffold, further demonstrating its biocompatible nature and suitability to support cell growth. It should be stressed that, the specific SERS fingerprint for each tag allowed a clear distinction of the scaffold and cells, upon simultaneous 3D SERS imaging under the same 785 nm excitation laser.
In these initial assays, we observed that MCF7 cells grow and migrate on the scaffolds for up to 21 days in vitro (DIV), which is particularly long for an in vitro 3D model (Figure 2B). After ca. 2 weeks, the number of cells adhered to the scaffolds was found to decrease, possibly due to the delicate nature of cell adhesion onto the scaffold. The observation that cells were not migrating to the underlying substrate (Figure S11) and the absence of cell cytotoxicity might also suggest, either a breakdown of the ECM components, leading to degradation of the local microenvironment, or that MCF7 cells require certain cues to chemotactically support their growth.30,31 To design a more realistic model, where multiple cell types can grow and form aggregates, we introduced various changes to our model, namely the use of a complex ECM such as Matrigel, in which cells would be suspended, and the incorporation of fibroblasts, which are known to contribute to ECM production. SERS tag-labeled MCF7 (expressing red fluorescent protein, RFP) and Human Dermal Fibroblasts (HDF, expressing GFP) were combined in a 1:2 ratio and embedded in Matrigel, which was subsequently introduced into the scaffold (Figure S12). Via confocal and SERS imaging, we were able to identify both cell populations with high resolution in a relatively large volume (over 500 μm3) (Figure S13). Although both techniques allowed us to identify cell aggregates after 5 DIV, the lack of extended fibroblast morphology called for further optimization of the ECM composition and concentration. We thus selected a combination of Matrigel at lower concentration (1.5 mg/mL instead of 3.7 mg/mL) with collagen type I. Spheroid growth was also improved toward SERS monitoring of biological processes at different stages and over longer periods of time. We explored the incorporation of preformed spheroids, which would act as nucleation points for subsequent expansion and tumoroid growth, either stimulated by the release of chemotactic factors, or by remodeling of the local ECM microenvironment.32,33 Indeed, using reflectance confocal microscopy to monitor collagen organization, we observed changes in the pattern of ECM deposition around spheroids, when compared with control experiments without cells or spheroids (Figure S14).34
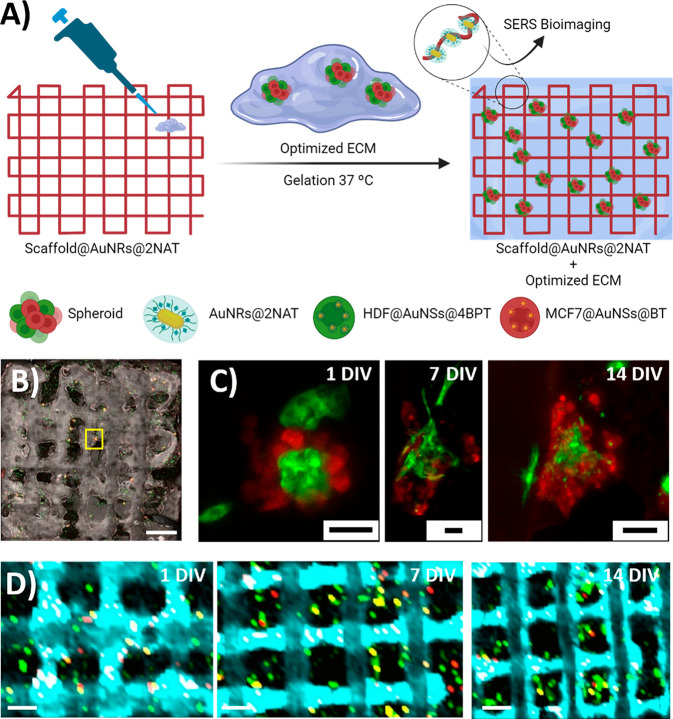
A combination of free cells and spheroids, composed of MCF7 and HDF cells, previously transfected and labeled with SERS tags, were combined with the optimized ECM solution and pipetted onto the scaffold (Figure 3A). We then used fluorescence microscopy to observe the reorganization of cell distribution and clustering of cells around preformed spheroids, during the first week (Figure 3B and Figure S15). At the later time points, we could identify extended fibroblast morphology, proliferation of MCF7 cells, and progressive filling of imaging windows. In some cases, the original spheroids developed into giant tumoroids measuring nearly 500 μm × 500 μm, in the XY plane (Figures S15, S16). Control samples, produced in the absence of any scaffold, showed similar changes in cell distribution at early time points, but signs of poor cell proliferation and lack of 3D model growth after 10 DIV (Figure S17). It was thus observed that, the presence of the PEGDA scaffold not only aided as a guide to repeatedly locate the same spot for imaging, but also appeared to promote spheroid growth. Although the scaffold itself does not comprise any immunostimulatory components and thus should not enhance cell proliferation by any molecular stimulus, we hypothesize that the porous and biocompatible nature of the 3D structure helps retain the original ECM, as well as the subsequent cell-generated ECM, in turn enabling suspended 3D cell growth and closely resembling the conditions found in a natural environment. Indeed, a study of the cell cytokines secreted in these 3D models (Figure S18) showed a time-dependent increase in the presence of molecules such as IL-6 (known to play a significant role in cancer cell proliferation and metastasis),30 IL-8 (involved in stimulating CAFs),35 Macrophage Migration Inhibitory Factor (MIF) (pro-oncogenic factor in the TME),36 and SERPIN E1 (associated with poor breast cancer prognosis).37 The presence of these cytokines suggests that these models are of biological relevance to study the breast TME.
Figure 3.
Monitoring of spheroidal growth (MCF7 in red, HDF in green) in a PEGDA scaffold over time. (A) Schematic view of the preparation of the scaffold containing AuNR@2NAT, optimized ECM, and SERS-labeled MCF7 and HDF cells and spheroids. (B) MIP from Z-stack confocal fluorescence imaging of spheroid growth after 1 DIV. Scale bar: 1000 μm. (C) Confocal microscopy images at different times of the spheroid found in the yellow square in (B). Scale bars: 50 μm. (D) Merged XY SERS maps showing spheroid growth within a scaffold labeled with AuNR@2NAT (cyan), MCF7 cells labeled with AuNS@BT (red), and HDF cells labeled with AuNS@4BPT (green). Scale bars: 500 μm.
To perform live SERS imaging within the same model configuration, the scaffold was labeled with AuNR@2NAT SERS tags, whereas MCF7 and HDF cells within the spheroids were labeled with benzenethiol decorated AuNSs (AuNS@BT) and AuNS@4BPT, respectively. The maps in Figure 3D demonstrate a clear differentiation of all three SERS tags over the studied time points. The laser power used for SERS measurements was adjusted to avoid excessive plasmonic heating (and prevent cell death or possible denaturation of ECM components) while ensuring a meaningful SERS signal from all SERS tag-labeled cells.26 SERS imaging allowed us to precisely locate tumoroids within the scaffold over long periods of time (cell cultures were extended up to 18 DIV), and even to acquire a 3D reconstruction of cell distribution within a scaffold window, down to 600 μm in depth (Video S1). It should be mentioned here that extensive data analysis is required when working with multiple SERS tags in 3D models (Figure S19). Indeed, the significance of data analysis for SERS measurements is emphasized by the growing body of research that utilizes this powerful tool to unravel the intricacies of the collected data.24,38−41 Using complementary confocal fluorescence and SERS imaging, we were able to image the same area with high resolution, correctly distinguishing the two labeled cell types due to their distinct fluorescence and SERS signals (Figure 4). SERS maps taken at equidistant heights (different Z planes), with a step size of 20 μm within the same region, allowed us to clearly distinguish the cells with respect to the scaffold (Figure S20). These findings hold promise for advancing SERS performance in 3D cell models, offering distinct advantages that address well-known limitations faced by fluorescence imaging. The multiplexing ability of SERS proves particularly beneficial in complex biological systems. Additionally, the improved photostability and absence of photobleaching contribute to ensuring the stability of the sample during the measurements. Furthermore, the high penetration depth of SERS in biological tissues presents a notable advantage over conventional fluorescence imaging techniques.
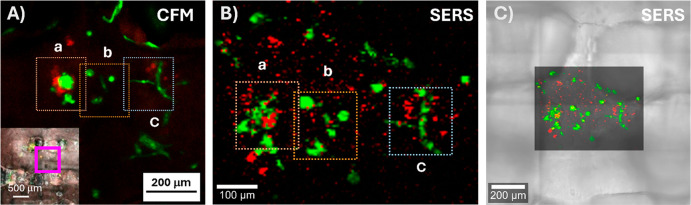
Figure 4.
Equivalent fluorescence and SERS imaging of cell aggregates or spheroids (RFP-expressing MCF7 in red, GFP-expressing HDF in green) inside the scaffold. (A) Confocal fluorescence image showing cell organization in the area marked with a purple frame (inset) after 4 DIV. (B) High-resolution SERS map of the same region after 4 DIV. (C) High-resolution SERS map of the same region overlapping the optical image from the scaffold. SERS images show HDF cells labeled with AuNS@4BPT (in green) and MCF7 cells with AuNS@BT (in red). Dotted square frames in A and B correspond to the same regions in the sample.
4D Tumor Monitoring
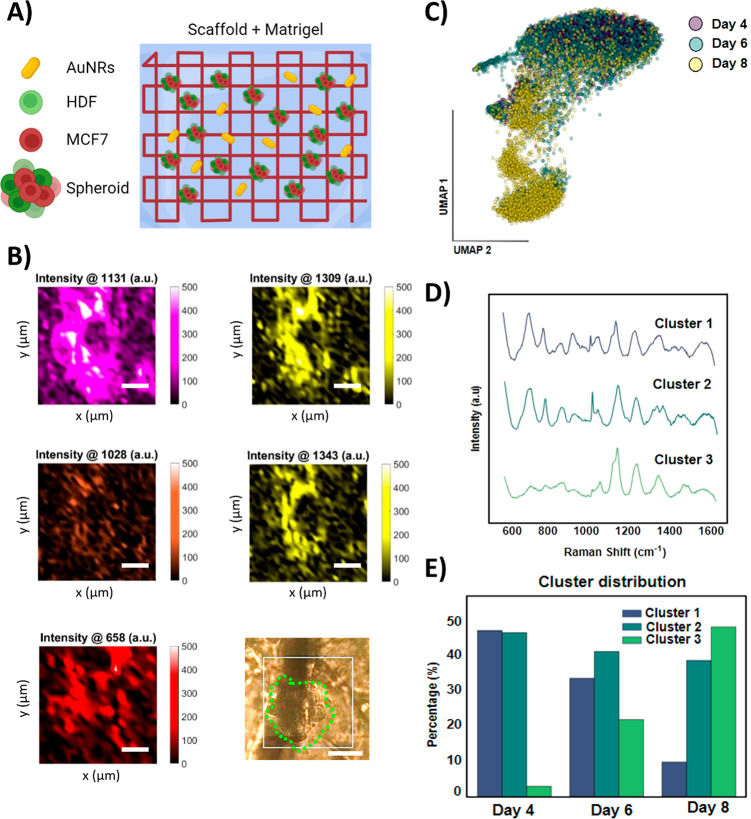
We thus proceeded to evaluate the SERS sensing performance of the composite scaffold in the presence of cells, to retrieve biological information from our 3D models. In contrast to SERS imaging, SERS sensing is generally based on significantly weaker signals, arising from molecules adsorbed on unlabeled AuNRs as SERS enhancers. With the aim of detecting cell-derived metabolites, cells were placed near the scaffold by using the originally described fibronectin/collagen-coating procedure (as opposed to embedding cells in the ECM). SERS maps were then collected at different time points from an extensive region (2 mm × 2 mm) of the scaffold, using a 40× immersion objective. After processing the collected data by Principal Component Analysis (PCA), we initially identified the principal component in the spectra as the background of our PEGDA scaffold. After 3 days of incubation, an intense signal was detected around 1130 cm–1 (Figure S21A,B), which we attribute to cell-induced changes in the proteins used to coat the scaffold.42 However, to effectively detect similar biomolecules and ECM remodeling induced by the preformed spheroids, AuNRs were dispersed in the matrix surrounding the cells, thus in closer proximity to cells and secreted metabolites. We thus printed AuNR-free scaffolds and embedded the cells and spheroids in the optimized ECM, in which AuNRs were also included (Figure 5A). Recognizing a risk of potential endocytosis of ECM-embedded AuNRs by cells, we employed TEM imaging to discard this possibility. The whole sample (scaffold with cells embedded in optimized ECM with AuNRs) was resin embedded and sections imaged to identify the location of AuNRs. As shown in Figure S22, whereas AuNSs could be clearly identified inside cells, AuNRs were only found extracellularly in material resembling Matrigel or other ECMs.
Figure 5.
(A) Scheme of the scaffold with optimized ECM-containing spheroids made of MCF7 and HDF cells, together with ligand-free AuNRs. (B) SERS peak intensity maps (150 μm × 150 μm) for those peaks identified in PCA analysis around a cell spheroid (green dashed line in the optical image) after 4 DIV (Scale bars = 50 μm). (C) Uniform Manifold Approximation and Projection (UMAP) of the whole spectra collected over the course of the cell culture experiment at 4, 6, and 8 DIV (data points are color-coded by the DIV, as labeled). (D) SERS spectra of the different clusters identified in (C). (E) Percentage of the distribution of spectra at different DIV across the three clusters.
For live SERS sensing, spectra were recorded from small regions of the ECM matrix surrounding the spheroids, using a 40× water immersion objective with a step size of 5 μm in a 150 μm × 150 μm area. Similar measurements were repeated after 4, 6, and 8 DIV. To identify the main sources of SERS signal variability across the imaged sample, PCA was again carried out. In this case, three main components were determined for samples displaying peaks at 658, 1028, 1131, 1309, and 1343 cm–1 (Figure S21C). The peak at 1131 cm–1 (also observed in the previous scaffold configuration) was associated with an increase in the amount of proteins from degraded or remodeled ECM, as proven in control degradation experiments of the matrix in which the peak at 1131 cm–1 remained but those at 1003 and 1032 cm–1 (usually associated with phenylalanine residues) were not identified, suggesting structural changes in the amino acids of collagen (Figure S21D).43,44Figure 5B shows representative SERS maps of a spheroid and its surrounding area, highlighting the predominant Raman peaks. Comparing the SERS maps to the optical image obtained from Raman microscopy, it can be clearly observed that the maximum intensity of the peaks at 1131, 1309, and 1343 cm–1 is located in the perispheroid region (outside the spheroid). Therefore, we observed a spatial diffusive pattern that could be connected to the metabolic activity of the spheroids or to ECM remodeling.45 According to previous literature reports, perispheroidal or similar pericellularly located signals can be associated with C–N stretching and CH2 and CH3 twisting of proteins, whereas signals located at 1028 and 658 cm–1 may be associated with cell-secreted metabolites, such as phenylalanine and histidine, respectively.42 These findings, in addition to fluorescence and reflection imaging studies of cell growth and ECM remodeling, respectively, suggest that breast cancer spheroids grown in scaffolds play an active role in the reorganization of their ECM, and that these changes can be observed using noninvasive techniques such as SERS.46 Although these techniques benefit from the ability to spatially resolve biological and physical changes, processing of large data sets to observe trends is complicated. We thus employed Uniform Manifold Approximation and Projection (UMAP) visualization to analyze the bulk data collected over time, specifically focusing on the spectra recorded at 4, 6, and 8 DIV, comprising a total of 13,417 spectra. As depicted in Figure 5C, the distribution of data points suggests that certain vibrational changes are related to progression over time. Notably, dimensional reduction using UMAP revealed that temporal variability predominates over spatial heterogeneity. Hence, spectra from the same time point share more similarities with each other and are typically closer neighbors in the UMAP space, compared to spectra from different time points. Furthermore, the k-means analysis (an unsupervised algorithm for clustering similar data points together) identified a significant distribution of time points within each of the three created clusters, as well as a distinctive spectral fingerprint for each cluster centroid, which represents the average spectra for each cluster (Figure 5C,D). Consequently, we could observe in Figure 5D that most spectra at 4 DIV are grouped within cluster 1, in contrast with the higher abundance of points from 8 DIV in cluster 3. This profile, along with the more balanced cluster distribution for 6 DIV, supports the notion that SERS spectra can reflect changes over time within the cellular system. Specifically, these changes suggest an increase in collagen and proteoglycan deposition at earlier stages in the 3D model, potentially reflecting remodeling of the TME.
Detecting changes in the ECM poses a considerable challenge, given its intricate composition, which encompasses various secondary structures, alignments, and crosslinking states.47 Recent findings indicate that cancer cells instigate alterations in collagen well before metastasis, resulting in fluctuations in collagen crosslinking and the formation of a fibrotic microenvironment.48 SERS represents a promising tool for tracking ECM modifications, including increased deposition and variations in composition, both of which are associated with increasing risk of breast carcinoma. Notably, the Raman shifts observed around 830 and 1020 cm–1 within the spectral clusters can be linked to collagen density and the presence of proteoglycans, respectively.48 These observed spectral profiles may potentially reflect the remodeling of ECM barriers, facilitating cell migration and the creation of a premetastatic niche. The surveillance of changes in ECM structure and composition is of relevance to advance our understanding of cancer progression and developing effective treatment strategies.
Finally, as proof of concept, we additionally endeavored to monitor temperature using LaOCl:Nd3+ nanothermometers28 (Figures S23–S25) within our model configuration incorporating the optimized ECM, as depicted in Figure S26. We initially observed that the addition of components (AuNPs, cells, scaffold) in the optical light path affected the reproducibility of our measurements. Specifically, when measurements were conducted focused on fibers at the top layer of the scaffold, sufficient precision and accuracy was achieved regardless of the presence of optimized ECM and/or cells and spheroids. However, when measurements were conducted on lower fibers, calibration curves were less accurate, and the measured temperature deviated further from the chamber temperature. As such, the varying volumes of ECM at different locations within the scaffold had a pronounced impact on the precision and accuracy of the measurements. This variability also poses problems in generating 3D temperature maps, because single point calibrations are required at each location, significantly increasing the time required to conduct a measurement. To enhance the feasibility of this approach for local temperature monitoring, aspects such as nanothermometer sensitivity, ECM composition and geometry, and Raman microscope setup all require optimization, something which we will focus on in future work.
Conclusions
The present study showcases the versatility of formulation and configuration of 3D cell culture scaffolds, toward the integration of plasmonic nanoparticles for SERS biosensing and bioimaging. Hydrogel-based scaffolds were prepared, comprising AuNRs for biosensing of breast cancer cell-secreted metabolites, and SERS-labeled NPs for 3D bioimaging. Fibronectin and collagen-coated scaffolds not only enabled monitoring of MCF7 cell growth and distribution, but also facilitated in situ monitoring of the cell microenvironment. More realistic tumor models were accomplished by incorporating spheroids comprising both MCF7 and HDF cells embedded in a Matrigel-based ECM matrix to the scaffold, and such models were monitored for up to 21 days in vitro. The implementation of multiplex live SERS imaging demonstrated a high spatial resolution, allowing correlation of morphological cell differences and scaffold structure with confocal fluorescence microscopy. Finally, live SERS biosensing was performed within the same 3D configuration, using PCA and UMAP analysis to reveal spatial and temporal profiles of cell-secreted molecules. These results, in combination with fluorescence and reflection microscopy, as well as cytokine analysis, suggest that breast tumoroids are capable of physically and biologically changing their ECM in a trend associated with cancer cell TME plasticity. Employing bulk data spectral analytical algorithms an unbiased approach can be applied to monitor the evolution of spheroids within inherently challenging environments. In addition to SERS biosensing within a 3D configuration, we also explored the integration of nanothermometers into the scaffold formulation, as a proof of concept to monitor temperature fluctuations. The use of nanothermometers embedded in the scaffolds allowed us to determine temperatures in the microscale, achieving a higher spatial resolution than other methods such as thermocouples. Unfortunately, we encountered difficulties, namely below-par precision and accuracy, when temperature measurements were performed in the presence of ECM. Although further analysis is required to gain a comprehensive understanding of how to improve these conditions in live cellular 3D scaffold configurations, this approach serves as a proof of concept for local temperature monitoring within biological environments. Future efforts will be also dedicated to improving the transparency of the materials and the diffusion of metabolites into the hydrogel matrix, thereby reducing background signals derived from the polymeric backbone and improving SERS detection of cell-secreted molecules, respectively. We expect that these changes will play an important role in the enhancement of biomarker SERS signals, in addition to facilitating in situ and non-invasive temperature measurements. In conclusion, the work presented demonstrates the performance of 3D printed plasmonic scaffolds as versatile platforms for SERS biosensing and bioimaging to monitor the growth of tumor models in a 3D context.
Experimental Section
Materials
Hexadecyltrimethylammonium bromide (CTAB, ≥99.0%), 5-bromosalicylic acid (5 BrSA, technical grade, 90%), hydrogen tetrachloroaurate trihydrate (HAuCl4·3H2O, ≥99.9%), silver nitrate (AgNO3, ≥99.9%), l-ascorbic acid (AA, ≥99%), sodium borohydride (NaBH4, 99%), poly(ethylene glycol) diacrylate (PEGDA, Mn 700), fumed silica (0.2–0.3 μm average particle size), (2-hydroxy-4′-(2-hydroxyethoxy)2-methylpropiophenone) (98%), O-[2-(3-mercaptopropionylamino)ethyl]-O′-methylpolyethylene glycol (PEG, MW 5000 g/mol), biphenyl-4-thiol (4BPT, 97%), 4-methylbenzenethiol (MBT, 98%), 2-naphthalenethiol (2NAT, 99%), benzenethiol (BT, 98%), 4-mercaptobenzoic acid (MBA, 90%), poly(isobutylene-alt-maleic anhydride) (average MW ∼6000 g/mol), poly-l-arginine hydrochloride (PA, Aldrich no. 26982–20–7 >70 000 Da), dodecylamine (98%), tetrahydrofuran (THF, 99.85%, extra dry), chloroform (CHCl3, ≥99.8%), sodium hydroxide (NaOH, >97%) and oleylamine (OAm, 70%) were all purchased from Sigma-Aldrich. κ-Carrageenan was purchased from Fisher. LaCl3·7H2O, 99.99%, NdCl3 anhydrous, 99.9%, stored under Ar and oleic acid (OA, 90%), purchased from Alfa Aesar. Ethanol (absolute), toluene (extrapure), and chloroform (ACS basics, stabilized with ethanol) purchased from Scharlau. The amphiphilic polymer dodecylamine grafted (75%) poly(isobutylene-alt-maleic anhydride) (PMA) was synthesized following already published procedures.49,50 Type I collagen and Matrigel were purchased from Corning. Cell Tracker Deep Red, DMEM, fetal bovine serum (FBS), and penicillin–streptomycin (PS) were purchased from Invitrogen. FBS and PS were used at 10% and 1% respectively to prepare complete DMEM (cDMEM). The MTT kit was purchased from Roche. All chemicals were used as received. Milli-Q water was used in all experiments. All glassware used for AuNRs synthesis was washed with aqua regia, rinsed with water and dried before use.
Nanoparticle Synthesis
AuNRs were prepared following a well-established procedure.51,52 First, seeds were prepared by adding 25 μL of a 50 mM HAuCl4 solution to 4.7 mL of 0.1 M CTAB solution at 30 °C and the mixture was stirred for 5 min. Then, 300 μL of a freshly prepared 10 mM NaBH4 solution was injected under vigorous stirring and the solution was kept for 30 min at room temperature to consume excess borohydride. For AuNR growth, 45 mg of 5-BrSA was added to 25 mL of 0.1 M CTAB and, after complete dissolution, 480 μL of 0.01 m AgNO3 was added. The solution was stirred for 15 min at room temperature. To perform the prereduction of Au (III) to Au (I), 500 μL of 50 mM HAuCl4 solution was added to the mixture. Prereduction was monitored by UV–vis spectroscopy until the value of the absorbance at 396 nm was 0.8–0.85, to obtain AuNRs with a longitudinal plasmon band around 780 nm.52 At this time, 130 μL of a 100 mM AA solution was added. After 30 s, 80 μL of seed solution was added under vigorous stirring. The mixture was then left undisturbed for 4 h. The resulting solution was centrifuged at 8000 rpm for 30 min, to purify the AuNRs. The precipitate was collected and AuNRs were redispersed in a 1 mM CTAB solution.
AuNSs were prepared following a previously reported procedure.29 Gold seeds were synthesized by the addition of 25 mL of 34 mM citrate solution to 500 mL of a boiling 0.5 × 10–3 M HAuCl4 solution under vigorous stirring. After 15 min, the solution was cooled and stored at 4 °C until further use. For AuNS growth, 50 μL of seed solution was added to 10 mL 0.25 mM HAuCl4 and 10 μL HCl 1 M at room temperature under moderate stirring. Immediately after, 100 μL of 3 mM AgNO3 and 50 μL of 100 mM ascorbic acid were added simultaneously. When the solution turned from green to blue, 150 μL of 0.1 mM PEG-SH were added, and the solution was stirred for 15 min. For AuNS purification, the solution was centrifuged at 2000 rpm for 10 min and the precipitate redispersed in water.
SERS Tags
AuNSs and AuNRs coated with 4BPT, 2NAT, MBT, or BT were synthesized and characterized as previously reported.25 The final concentration of the SERS tags was [Au0] = 0.5 mM, corresponding to 1.9 × 1010 NP/mL. We employed 2NAT-coated AuNRs as SERS tags for SERS labeling of the scaffold. For cell labeling, we employed either 4BPT, MBT, or BT-coated AuNSs for MCF7 cells, whereas HDF cells were labeled with 4BPT-labeled AuNSs. Although all SERS tags would be appropriate, higher SERS signal intensities were obtained when combining HDF with AuNS@4BPT. SERS tags were coated with (positively charged) poly-l-arginine hydrochloride (PA) to enhance cell uptake, as reported.29
Synthesis of Nanothermometers
Core–shell LaOCl3:Nd3+@LaOCl3 nanothermometers were prepared following a recently reported protocol.28 Briefly, for a final amount of 1 mmol of LaOCl/Nd3+NPs, 0.98 mmol of LaCl3·7H2O and 0.02 mmol of anhydrous NdCl3 were mixed in methanol. The mixture was stirred in OAm at 80 °C under a vacuum for 1 h. Afterward, the temperature was quickly raised to 210 °C and the reaction left under reflux in argon atmosphere for 1 h. Then, 4 mL of oleic acid was added to quench the reaction and when the temperature dropped to 180 °C, a few seconds after the addition, the mixture was cooled under an air stream. The NPs were then collected using toluene and centrifugation at 120 g for 15 min. The NPs were washed using a mixture of 5 mL:2 mL toluene:ethanol and centrifuged at 120g for 10 min. The same synthetic and purification procedure described above was used to grow an inert LaOCl shell on the NPs. In this case 0.5 mmol of LaCl3.7H2O was mixed with previously prepared 1 mmol of core nanoparticles. The final core–shell NPs were dispersed in 10 mL CHCl3 to achieve a concentration of 30 mM of the core–shell nanothermometers (confirmed by ICP-MS). To make nanothermometers water-soluble, PMA coating was performed following a previously reported procedure.28
Nanoparticle Characterization
TEM images were collected using a transmission electron microscope JEOL JEM-1400PLUS operating at 120 kV. A drop of the dispersion was deposited on a carbon-coated copper grid and dried under ambient conditions. UV–vis–NIR optical extinction spectra were recorded using an Agilent 8453 UV–vis diode array spectrophotometer.
Ink Preparation
Prior to the incorporation of gold NPs to the ink, the AuNR solution was gently washed to remove excess CTAB. AuNRs were first centrifuged at 8000 rpm for 10 min and redispersed in 1 mM CTAB. Then, a second centrifugation step was performed and AuNRs were redispersed in Milli-Q water. The concentration of gold for the ink was adjusted to [Au0] = 1 mM. Then, 500 mg of PEGDA was dissolved in 1 mL of AuNRs solution. Twenty mg of κ-carrageenan, 150 mg of fumed silica and 5 mg of 2-hydroxy-4′-(2-hydroxyethoxy) 2-methylpropiophenone (Irgacure 2959) were also added and the ink was thoroughly mixed using a Thinky Mixer at 3500 rpm for 1 min. For inks containing AuNRs labeled with Raman reporters, the same procedure was followed. Samples were stored at 4 °C before usage.
3D Printing of Scaffolds
A multiheaded 3D Discovery bioprinter (RegenHU, Switzerland) was used to print the scaffolds. A high precision plunger dispenser was used at a constant volume flow rate of 1.5 μL/s using a stainless-steel needle with an inner diameter of 0.41 mm, at 10 mm/s printing speed. The G-code for square scaffolds was produced using BIOCAD software (RegenHU, Switzerland) with 0.8 mm spacing. In situ UV-crosslinking was performed after the deposition of each layer using the light curing cartridge at 365 nm (500 mW).
Scaffold Characterization
Rheological properties were studied using a MCR 302 rheometer (Anton Paar, Spain). All tests were carried out in triplicate at 25 °C, using 25 mm parallel plate geometry and a solvent trap to prevent water evaporation. First, the ink was subjected to an amplitude sweep to determine the linear viscoelastic region, where both the storage (G′) and loss (G′’) moduli are parallel and independent of the applied amplitude. Frequency sweeps were carried out from 0.1 to 100 rad s–1 at a fixed strain determined from the amplitude sweeps. Shear stress and viscosity were measured for shear rates from 0.01 to 1500 s–1, gap = 1 mm. Oscillatory-rotational-oscillatory tests were performed to characterize the materials recovery behavior by monitoring the material at rest for 120 s, followed by a shear rate of 10 s–1 for another 120 s and finally monitor materials at rest for another 300 s.
The swelling capacity and stability of the as-prepared hydrogels was studied by a general gravimetric method. Samples (n = 3) were incubated at 37 °C in cell culture media (cDMEM) and at selected time intervals the swollen hydrogels were removed, the excess of liquid absorbed with filter paper and the scaffold weighed. The swelling ratio (SR) was calculated using eq 1:
| 1 |
where Ws is the weight of the swollen sample and Wd is the weight of the dried hydrogel sample.
Scanning Electron Microscopy (SEM) was performed using a JEOL JSM-6490LV operating at an accelerated voltage of 15 kV and at a working distance of 15 mm.
SERS Measurements
For immersion measurements, the scaffolds were placed on a quartz slide (24 × 60 mm) and a custom-made 3D printed holder (Figure S27) was placed around the scaffold using two-component silicon dentist glue (Proclinic Products), thereby providing a water-tight seal around the scaffold. cDMEM or water was then added to avoid scaffold drying. Different scaffold configurations were employed in this study. 3D SERS imaging of the scaffolds was performed by labeling the scaffold with AuNR@2NAT and coated with MCF7 cells labeled with AuNS@4BPT. A cell coculture was also imaged by SERS using MCF7 cells labeled with AuNS@MBT and HDF cells labeled with AuNS@4BPT. The scaffold was labeled with AuNR@2NAT. Finally, scaffolds labeled with AuNS@2NAT and combined with optimized ECM containing spheroids of MCF7 cells labeled with AuNS@BT, and HDF labeled with AuNS@4BPT, were also imaged by SERS.
Live-SERS imaging and sensing was performed using a confocal Raman microscope (inVia Reflex, Renishaw, U.K.) equipped with 1024 × 512 CDD detectors, using a 785 nm laser excitation source (maximum output 270 mW) and a 1200 l/mm diffraction grating. SERS maps were recorded using a 40× immersion objective (numerical aperture, NA = 0.8; Nikon, Japan) at 50% laser power (36 mW at surface) and 1 s integration time. For SERS imaging, maps of the selected areas were acquired with a resolution of 70 μm in X and Y. For Z measurements, the map of one selected area was acquired with a resolution of 50 μm in X and Y, and 100 μm in Z. SERS data were first analyzed using the WiRE4.4 software (Renishaw, U.K.) to correct the baseline in the spectra and eliminate cosmic rays. Figure S19 shows the difference in the resulting SERS maps, considering the peak intensity of the characteristic tags (1380, 1280, and 1000 cm–1 for 2NAT, 4BPT, and BT, respectively) versus the ones performed considering reference spectra for each tag. As can be seen for 2NAT, both methods revealed the same results because this signal was the most intense one of the three, due to its higher abundance and more homogeneous distribution within the scaffold. In the case of 4BPT and BT, the use of PCA analysis avoids misassignment of the signals because the background of the scaffold grid can be avoided. Therefore, SERS mappings were analyzed by PCA in Matlab, which considers the full spectrum to assign the typical fingerprints of SERS tags.23
For SERS sensing of metabolites, scaffolds were swollen in a 1 mM MBA for 30 min. SERS data were analyzed using True Component Analysis (TCA) provided by the software FIVE (WITec GmbH, Germany). For SERS sensing with live spheroids, maps from selected areas (100 μm × 100 μm in X and Y) were acquired with a resolution of 5 μm in X and Y, from 3 different areas of each sample. SERS data were first analyzed using the WiRE4.4 software and analyzed using PCA (PCA function in Matlab). To explore the temporal variability and spectral similarity in the collected data, we employed the UMAP technique. UMAP is a nonlinear dimensionality reduction method that can effectively capture the underlying structure of high-dimensional data.
For UMAP visualization, we used the Python UMAP-learn package. We applied the UMAP algorithm with a specified parameter of n_neighbors = 50. The transformed data containing 13417 spectra were then plotted in a 2D space, where each point represents a single spectrum. Data points in the UMAP plot were color-coded according to the days in vitro (DIV), when the spectra were recorded, with distinct colors for 4, 6, and 8 DIV, as well as other time points. This approach enabled us to visually assess the similarity of spectra within the same time point and to evaluate the extent of temporal variability across the data set.
K-Means Clustering
Upon PCA transformation, we employed k-means clustering to identify patterns within the data and to explore their temporal distribution. First, we normalized the data using the StandardScaler from the sklearn.preprocessing module, which standardizes the data by removing the mean and scaling it to unit variance. Next, we applied PCA to reduce the dimensionality of the normalized data. We retained the first two principal components for subsequent analysis. We then used the KMeans class from the sklearn.cluster module to perform k-means clustering with three clusters (n clusters = 3). We initialized the algorithm with a random state of 42 to ensure reproducibility. We assigned cluster labels to each spectrum and computed the average spectrum for each cluster by grouping the data by the assigned cluster labels. We plotted the average spectrum for each cluster to visualize the spectral fingerprints of the cluster centroids. This clustering approach allowed us to assess the distribution of spectra at different DIV across the three clusters and to observe temporal changes in the spectral data.
For high-resolution SERS sensing and imaging, measurements were performed with a confocal Raman microscope (Alpha300R, WITec GmbH, Germany) coupled through an optic multifiber to the spectrometer (UHTS 400S-NIR, WITec GmbH, Germany) equipped with a 300 g/mm grating and a back-illuminated deep depletion CCD detector (DU401, Andor, U.K.). A 785 nm laser excitation source (maximum output 79 mW) with spot size close to diffraction limit and a 20× water immersion dip-in objective (Achroplan, NA = 0.5, Zeiss, Germany) were used for all measurements.
For scaffolds with MCF7 cells, spectra were recorded at spectral center of 1600 cm–1, integration time of 0.05 s and laser power of 10 mW over a volume of 2000 × 2000 × 1000 μm3 with a step size of 13.33 μm in (XY) and 50 μm (Z). For the PEGDA scaffold filled with Matrigel containing embedded MCF7 and HDF, SERS measurements were performed by recording the spectral center of 1500 cm–1. A volumetric region of 650 μm × 500 μm × 350 μm was imaged with 0.05 s integration time, at 10 mW laser power and step size of 5 μm in (XY) and 20 μm (Z). In both cases, spectra were analyzed using cosmic ray removal (CRR), background subtraction (Sub, shape) before applying the TCA tool, all implemented in the equipment software FIVE to identify the known fingerprint of SERS tags.
Temperature Measurements
For calibration of the scaffold with nanothermometers, a temperature and environmental control cell, Linkam THMS600, was used. The samples were placed in a quartz (optical quality) Petri dish of 22 mm in diameter and max. volume of 700 μL, which was filled with optiMEM, and the scaffold. Photoluminescence measurements were acquired with the same confocal Raman microscope (Alpha300R, WITec GmbH, Germany) as the 3D SERS measurements, but using the 532 nm diode laser excitation source and a 20× air objective (NA = 0.4, Nikon, Japan). The resulting photoluminescence was collected in the NIR spectrometer (UHTS 400S-NIR, WITec GmbH, Germany) equipped with the 300 g/mm grating and collected in the DU401 CCD in the 800 to 1000 nm wavelength range. The integration time was set to 0.1 s and averaged over 10 accumulations. Different measurements were performed using line and point acquisition in the fixed 800 to 1000 nm wavelength range.
To acquire the calibration curve, NIR photoluminescence spectra were recorded at different temperatures between 5 and 70 °C, allowing the temperature to stabilize for at least 5 min before each measurement. Each measurement comprised 20 spectra along a 300 μm line, collected using 532 nm irradiation at 40 mW power using 10 accumulations at 0.1 s. Afterward, a background removal filter and averaging of the 20 spectra was calculated to all the measurements prior to integration of the whole bands centered at 820 and 900 nm.
Scaffold Preparation for Cell Studies
To sterilize scaffolds for cell studies, scaffolds were exposed to ethanol (50% v/v in sterile water) for 10 min, followed by 90% (v/v) ethanol for another 10 min. Following this, three washes in nanopure sterile water were undertaken. Scaffolds were then placed under a UV germicidal lamp for 3 min. To sterilize the holders and substrate (glass or quartz, slide or coverslip), these were all washed first with detergent, nanopure sterile water, dried and then placed under a UV lamp for 3 min. To reuse holders and substrates, a washing step with Tergazyme (1% w/v, 1 h) was incorporated to remove any cellular-derived biomolecules.
Cell Culture and Lentivirus Transformation
MCF7 (HTB-22, ATCC) and HDF (HDFa, ThermoFisher) cells were cultured in DMEM supplemented with 10% (v/v) FBS and 1% (v/v) PS. Passages were conducted using trypsin-EDTA. Cells were routinely checked for mycoplasma and confluence was controlled in subpassages. For GFP and RPF expression, cells were transfected with GFP- (pLenti CMV GFP Hygro (656–4, Addgene) or RFP (pLenti CMV RFP Hygro (LVP1226, AMSBIO) according to the manufacturer’s instructions.
3D Cell Culture Conditions
SERS Tag Incubation and Cell Preparation
MCF7 and HDF cells were seeded in 24-well plates at seeding densities of 5 × 104 and 3 × 104 cells/well/500 μL, respectively. Once adhered, SERS tags were added (350 μL/well) at a final concentration of 25 μM [Au0]. After overnight incubation, nonuptaken SERS tags were removed via washing and cells detached via trypsinization. Cells were adjusted to 3.84 × 104 cells/mL and mixed in a 1:2 ratio MCF7:HDF cells for spheroid formation.
Spheroid Formation
To form spheroids, μ-mold inserts were used (Microtissues, Sigma). In brief, low-melting weight agarose (2% w/v) was dissolved in sterile saline solution and 330 μL pipetted into sterile autoclaved μ-molds. The #24–96 series (400 μm individual spheroid mold diameter) was used, resulting in ca. 96 spheroids/well of a 24-well plate. Once hardened, agarose μ-molds were removed, placed in a 24-well plate, and covered with sterile PBS. Between 12 and 16 μ-molds were made per experiment. The 24-plate was stored at 37 °C in a humidified incubator until use (maximum 1–2 days prior to cell seeding). PBS solution was removed from the agarose μ-molds, followed by two washes with cDMEM. Care was taken to not damage the agarose μ-mold. The remaining cDMEM was removed and 75 μL of MCF7:HDF cell solution (equivalent to 2880 cells/ μ-mold or 30 cells/spheroid) added to the agarose μ-mold; without disturbing the 24-well plate, the cells were left to sediment in their μ-wells (ca. 30 min) prior to addition of cDMEM (600 μL/well). Spheroids were transferred to the incubator and left for 48 h for spheroid formation to occur. Occasionally, spheroids were observed under an upright microscope and images taken to document spheroid growth (see Figure S28). Meanwhile, the process of cell seeding and exposure to NPs was repeated to achieve a suspension of cells, not aimed for spheroid formation, to be combined with the spheroids once formed. The timings were coordinated so that spheroids and cell suspension were ready simultaneously. After overnight exposure to SERS tags, nonuptaken SERS tags were removed via washing and cells detached via trypsinization. Cells were adjusted to 8 × 105 cells/mL and mixed in a 1:2 ratio, MCF7:HDF cells. The volume of cells and number of spheroids required for the preparation of the scaffolds was calculated based on the seeding density of 4 × 104 cells, plus the spheroids obtained from 8 μ-molds per scaffold. For example, for the preparation of two scaffolds, 100 μL of the cell suspension at 8 × 105 cells/mL (equivalent to 8 × 104 cells) was centrifuged with spheroids collected from 16 μ-molds (theoretically equivalent to 1500 spheroids although optical observations confirm the successful formation of ca. 50%) to concentrate the sample, finally resuspending the pellet in 100 μL optimized ECM matrix (see below).
Optimized ECM Formulation
In initial studies we used Matrigel alone (7.5 mg/mL), but the final formulation was composed of Matrigel and collagen (1.5 and 0.8 mg/mL, respectively), incorporating cells, spheroids, and SERS sensing AuNRs (0.5 mM) where necessary. The order in which the components were mixed was crucial to avoid aggregation of AuNRs and ECM components; working on ice, first the AuNRs (previously washed of excess surfactant, as described for the ink preparation and adjusted to at least 5 mM) were mixed with concentrated Matrigel, followed by the addition of a concentrated cell suspension and any extra media required to adjust the final component concentrations, and finally collagen. For imaging applications with SERS labeled cells, AuNRs were omitted from the sensing formulation. For control degradation experiments, the nonenzymatic proprietary cell recovery solution of Corning was employed.
Coated Scaffold 3D Model Fabrication
Pre-cleaned scaffolds were immersed in a solution of fibronectin (50 μg/mL) and collagen type I (25 μg/mL) for 1 h at 37 °C under rotation. The scaffolds were rinsed with warmed medium to remove any unbound proteins and immersed in a solution of cells (1 × 106 cells/mL) for 2 h with rotation at 37 °C. The cell and protein coated scaffold was subsequently placed in the center of the substrate under sterile conditions with minimal transfer of liquid to avoid wetting the surrounding substrate area. An in-house 3D printed holder was placed around the scaffold using two-component glue, as previously described for SERS measurements. cDMEM was then added to avoid drying of the scaffold. To verify the presence of fibronectin and collagen, immunostaining was used. Scaffolds were first blocked using 1% bovine serum albumin in PBS buffer for 1 h. Primary antibodies antifibronectin (Sigma) (1/400) and anticollagen type I (COL-1; Abcam) (1/2000) were added for 1 h, followed by secondary matched antibodies (both Abcam, 1/1000) for 1 h.
ECM 3D Model Fabrication
Pre-cleaned scaffolds were placed at the center of the substrate under sterile conditions, with minimal transfer of liquid to avoid wetting the surrounding substrate area. An in-house 3D printed holder was placed around the scaffold using two-component glue, as previously described for SERS measurements. cDMEM was then added to avoid drying of the scaffold. Then, 50 μL of optimized ECM containing single cells and spheroids (see above) was added to the scaffold and allowed to infiltrate for 5 min, before carefully transferring the scaffold to the incubator in a humidified chamber. After ca. 40 min, cDMEM was added and the sample returned to the incubator. Samples without scaffolds were also made using the same cells/spheroid mixture, simply pipetting 50 μL of the solution into a μ-angiogenesis slide (Ibidi, Germany).
Confocal Microscopy Imaging
A Zeiss 880 LSM inverted confocal microscope was used for fluorescence and reflection imaging. Objectives included Plan-Apochromat 10 × /0.45 NA, Plan-Apochromat 20×/0.8 NA, and EC Plan-Neofluor 40×/1.30 Oil DIC, and excitation lines included 405, 488, 561, and 633 nm, suitable for DAPI and AF405, GFP, RFP, and AF633 imaging. The excitation lines 488 and 633 nm were also used for reflection imaging of collagen and the scaffold, respectively. In most cases, images correspond to MIPs of z-stacks, to obtain the best resolution and 3D rendering. Efforts were made to keep the pinhole to a minimum, thus maximizing resolution of the used objective.
Electron Microscopy Preparation
To view the cell distribution in the scaffolds via TEM, scaffolds were either processed as a whole or dissected into quarters. Samples were processed in Eppendorf tubes. Samples were fixed in a solution of 2.5% glutaraldehyde and 2% formaldehyde in Sorensen’s buffer. Samples were left at 4 °C for 4 h, following which the fixative was replaced with Sorensen’s buffer alone and samples stored at 4 °C overnight. The following day, samples were further fixed and stained with OsO4 (1% in nanopure water) on ice for 1 h. The samples were washed three times with nanopure water, followed by an ethanol dehydration series starting at 30% and ending with three 100% ethanol washes, 30 min each. TEM samples were transferred to a glass coverslip substrate and embedded in Spurr’s resin. Once hardened, samples were immersed in liquid nitrogen to encourage separation of the sample from the underlying substrate. Samples were trimmed and cut at 100 nm for imaging, using 50- or 100-square grids in a JEOL JEM-1400PLUS operating at 120 kV.
Immunological Analysis
To determine the cytokine profile of 3D models, supernatants from 3D cell models, composed of cells and spheroids embedded in optimized ECM, were collected at different time points and stored at −20 °C. We first conducted a membrane-based sandwich immunoassay array (RnD, Proteome Profiler Human Cytokine Array Kit), capable of detecting the presence of 36 different cytokines from selected few samples, to determine which cytokines were of most interest for subsequent ELISA cytokine detection. IL-6, IL-8, MIF, and SERPIN E1 all gave positive results, and we thus screened all supernatants to determine the concentration of these cytokines using a sandwich ELISA setup (RnD, DuoSet kits). Both the membrane-based immunoassay and the ELISA were conducted following the instructions provided with each kit.
All drawings were created with Biorender License Agreement Number No. UP260UQAI3.
Acknowledgments
Financial support is acknowledged from the European Research Council (ERC Advanced Grant 787510, 4DbioSERS). C.G.A acknowledges a Juan de la Cierva Incorporación fellowship (IJC2019-040827-I).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.4c00543.
Video S1: SERS maps of the different components of a scaffold containing preformed spheroids of MCF7 and HDF cells at different heights of the scaffold in an imaging hole filled with optimized ECM (MP4)
Additional details on the fabrication and characterization of nanoparticles and scaffolds, 3D cell growth and imaging data as well as experimental setups and supporting results (PDF)
Author Present Address
# Institut Pasteur, Université Paris Cité, 25–28 Rue du Dr. Roux, 75015 Paris, France
Author Present Address
∇ Biobide Spain (BBD BIOPHENIX, S.L.U.), Paseo Mikeletegi 56, Bajo, Donostia-San Sebastian 20009, Spain
Author Present Address
⊗ CIC nanoGUNE, Basque Research and Technology Alliance (BRTA), 20018 Donostia-San Sebastián, Spain
Author Contributions
¶ C.G.-A. and M.H.-L. contributed equally. C.G.-A. and M.H.-L. conceived the idea, designed the experiments, and wrote the manuscript, under the supervision of L.M.L.-M.; C.G.-A. carried out the fabrication and characterization of the scaffold, nanoparticle synthesis and SERS sensing and imaging. M.H.-L. performed 3D cell culture studies, confocal microscopy imaging and immunological analysis. E.L. and J.L. performed high resolution SERS imaging and data analysis. C.R.-L. performed nanothermometer synthesis and temperature measurements. P.P. and D.J.d.A. synthesized and characterized the SERS tags. B.M. performed initial cell cultures, lentivirus transformation and confocal microscopy analysis. J.P. performed machine learning analysis of SERS sensing data. All authors participated in the discussion of results and have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Meng F.; Meyer C. M.; Joung D.; Vallera D. A.; McAlpine M. C.; Panoskaltsis-Mortari A. 3D Bioprinted In Vitro Metastatic Models via Reconstruction of Tumor Microenvironments. Adv. Mater. 2019, 31, 1806899. 10.1002/adma.201806899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H.; Esworthy T.; Zhou X.; Hann S. Y.; Glazer R. I.; Li R.; Zhang L. G. Engineering a Novel 3D Printed Vascularized Tissue Model for Investigating Breast Cancer Metastasis to Bone. Adv. Healthc. Mater. 2020, 9, e1900924 10.1002/adhm.201900924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal G.; Li W. A Versatile 3D Tissue Matrix Scaffold System for Tumor Modeling and Drug Screening. Sci. Adv. 2017, 3, e1700764 10.1126/sciadv.1700764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K. M.; Cukierman E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell 2007, 130, 601–610. 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Rijal G.; Li W. 3D Scaffolds in Breast Cancer Research. Biomaterials 2016, 81, 135–156. 10.1016/j.biomaterials.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Augustine R.; Kalva S. N.; Ahmad R.; Zahid A. A.; Hasan S.; Nayeem A.; McClements L.; Hasan A. 3D Bioprinted Cancer Models: Revolutionizing Personalized Cancer Therapy. Transl. Oncol. 2021, 14, 101015. 10.1016/j.tranon.2021.101015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M.; Lemma E. D.; Bertels S.; Blasco E.; Barner-Kowollik C.; Wegener M.; Bastmeyer M. 3D Scaffolds to Study Basic Cell Biology. Adv. Mater. 2019, 31, 1808110. 10.1002/adma.201808110. [DOI] [PubMed] [Google Scholar]
- Do A. V.; Khorsand B.; Geary S. M.; Salem A. K. 3D Printing of Scaffolds for Tissue Regeneration Applications. Adv. Healthc. Mater. 2015, 4, 1742–1762. 10.1002/adhm.201500168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Aristizabal P.; Perumal G.; García-Astrain C.; Liz-Marzán L. M.; Izeta A. Trends in Tissue Bioprinting, Cell-Laden Bioink Formulation, and Cell Tracking. ACS Omega 2022, 7, 16236. 10.1021/acsomega.2c01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel C. S.; Holme M. N.; Gopal S.; Stevens M. M. Advances in High-Resolution Microscopy for the Study of Intracellular Interactions with Biomaterials. Biomaterials 2020, 226, 119406. 10.1016/j.biomaterials.2019.119406. [DOI] [PubMed] [Google Scholar]
- Chizari K.; Daoud M. A.; Ravindran A. R.; Therriault D. 3D Printing of Highly Conductive Nanocomposites for the Functional Optimization of Liquid Sensors. Small 2016, 12, 6076–6082. 10.1002/smll.201601695. [DOI] [PubMed] [Google Scholar]
- Deng Z.; Hu T.; Lei Q.; He J.; Ma P. X.; Guo B. Stimuli-Responsive Conductive Nanocomposite Hydrogels with High Stretchability, Self-Healing, Adhesiveness, and 3D Printability for Human Motion Sensing. ACS Appl. Mater. Interfaces 2019, 11, 6796–6808. 10.1021/acsami.8b20178. [DOI] [PubMed] [Google Scholar]
- Guo S. Z.; Yang X.; Heuzey M. C.; Therriault D. 3D Printing of a Multifunctional Nanocomposite Helical Liquid Sensor. Nanoscale 2015, 7, 6451–6456. 10.1039/C5NR00278H. [DOI] [PubMed] [Google Scholar]
- Trampe E.; Koren K.; Akkineni A. R.; Senwitz C.; Krujatz F.; Lode A.; Gelinsky M.; Kühl M. Functionalized Bioink with Optical Sensor Nanoparticles for O2 Imaging in 3D-Bioprinted Constructs. Adv. Funct. Mater. 2018, 28, 1804411. 10.1002/adfm.201804411. [DOI] [Google Scholar]
- GhavamiNejad A.; Ashammakhi N.; Wu X. Y.; Khademhosseini A. Crosslinking Strategies for 3D Bioprinting of Polymeric Hydrogels. Small 2020, 16, e2002931 10.1002/smll.202002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam R. Y.; Smith L. J.; Shoichet M. S. Engineering Cellular Microenvironments with Photo- and Enzymatically Responsive Hydrogels: Toward Biomimetic 3D Cell Culture Models. Acc. Chem. Res. 2017, 50, 703–713. 10.1021/acs.accounts.6b00543. [DOI] [PubMed] [Google Scholar]
- García-Astrain C.; Lenzi E.; Jimenez de Aberasturi D.; Henriksen-Lacey M.; Binelli M. R.; Liz-Marzán L. M. 3D-Printed Biocompatible Scaffolds with Built-In Nanoplasmonic Sensors. Adv. Funct. Mater. 2020, 30, 2005407. 10.1002/adfm.202005407. [DOI] [Google Scholar]
- Plou J.; Molina-Martínez B.; García-Astrain C.; Langer J.; García I.; Ercilla A.; Perumal G.; Carracedo A.; Liz-Marzán L. M. Nanocomposite Scaffolds for Monitoring of Drug Diffusion in Three-Dimensional Cell Environments by Surface-Enhanced Raman Spectroscopy. Nano Lett. 2021, 21, 8785–8793. 10.1021/acs.nanolett.1c03070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi E.; Jimenez De Aberasturi D.; Henriksen-Lacey M.; Piñeiro P.; Muniz A. J.; Lahann J.; Liz-Marzán L. M. SERS and Fluorescence-Active Multimodal Tessellated Scaffolds for Three-Dimensional Bioimaging. ACS Appl. Mater. Interfaces 2022, 14, 20708–20719. 10.1021/acsami.2c02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman S. E.; McCracken J. M.; Miller L. A.; Jayalath S.; Nuzzo R. G. Biocompliant Composite Au/PHEMA Plasmonic Scaffolds for 3D Cell Culture and Noninvasive Sensing of Cellular Metabolites. Adv. Healthc. Mater. 2021, 10, 2001040. 10.1002/adhm.202001040. [DOI] [PubMed] [Google Scholar]
- Almohammed S.; Alruwaili M.; Reynaud E. G.; Redmond G.; Rice J. H.; Rodriguez B. J. 3D-Printed Peptide-Hydrogel Nanoparticle Composites for Surface-Enhanced Raman Spectroscopy Sensing. ACS Appl. Nano Mater. 2019, 2, 5029–5034. 10.1021/acsanm.9b00940. [DOI] [Google Scholar]
- Jaitpal S.; Chavva S. R.; Mabbott S. 3D Printed SERS-Active Thin-Film Substrates Used to Quantify Levels of the Genotoxic Isothiazolinone. ACS Omega 2022, 7, 2850–2860. 10.1021/acsomega.1c05707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.; Vikesland P. J. SERS-Active Printable Hydrogel for 3D Cell Culture and Imaging. Anal. Chem. 2023, 95, 18055–18064. 10.1021/acs.analchem.3c02641. [DOI] [PubMed] [Google Scholar]
- Zhuo X.; Henriksen-Lacey M.; Jimenez de Aberasturi D.; Sanchez-Iglesias A.; Liz-Marzán L. M. Shielded Silver Nanorods for Bioapplications. Chem. Mater. 2020, 32, 5879–5889. 10.1021/acs.chemmater.0c01995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez de Aberasturi D.; Henriksen-Lacey M.; Litti L.; Langer J.; Liz-Marzán L. M. Using SERS Tags to Image the Three-Dimensional Structure of Complex Cell Models. Adv. Funct. Mater. 2020, 30, 1909655. 10.1002/adfm.201909655. [DOI] [Google Scholar]
- Lenzi E.; Henriksen-Lacey M.; Molina B.; Langer J.; De Albuquerque C. D. L.; Jimenez De Aberasturi D.; Liz-Marzán L. M. Combination of Live Cell Surface-Enhanced Raman Scattering Imaging with Chemometrics to Study Intracellular Nanoparticle Dynamics. ACS Sens. 2022, 7, 1747–1756. 10.1021/acssensors.2c00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla M.; Zhang Y.; Liz-Marzán L. M. Subtissue Plasmonic Heating Monitored with CaF2:Nd3+,Y3+ Nanothermometers in the Second Biological Window. Chem. Mater. 2018, 30, 2819–2828. 10.1021/acs.chemmater.8b00806. [DOI] [Google Scholar]
- Renero-Lecuna C.; Herrero A.; Jimenez De Aberasturi D.; Martínez-Flórez M.; Valiente R.; Mychinko M.; Bals S.; Liz-Marzán L. M. Doped Lanthanum Oxychloride Nanocrystals as Nanothermometers. J. Phys. Chem. C 2021, 125, 19887–19896. 10.1021/acs.jpcc.1c05828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez De Aberasturi D.; Serrano-Montes A. B.; Langer J.; Henriksen-Lacey M.; Parak W. J.; Liz-Marzán L. M. Surface Enhanced Raman Scattering Encoded Gold Nanostars for Multiplexed Cell Discrimination. Chem. Mater. 2016, 28, 6779–6790. 10.1021/acs.chemmater.6b03349. [DOI] [Google Scholar]
- Kalluri R. The Biology and Function of Fibroblasts in Cancer. Nat. Rev. Cancer 2016, 16, 582–598. 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- Spaw M.; Anant S.; Thomas S. M. Stromal Contributions to the Carcinogenic Process. Mol. Carcinog. 2017, 56, 1199–1213. 10.1002/mc.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubba F.; Pouchol C.; Ferrand N.; Vidal G.; Almeida L.; Perthame B.; Sabbah M. A Chemotaxis-Based Explanation of Spheroid Formation in 3D Cultures of Breast Cancer Cells. J. Theor. Biol. 2019, 479, 73–80. 10.1016/j.jtbi.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Girigoswami K.; Saini D.; Girigoswami A. Extracellular Matrix Remodeling and Development of Cancer. Stem Cell Rev. Rep. 2021, 17, 739–747. 10.1007/s12015-020-10070-1. [DOI] [PubMed] [Google Scholar]
- Nerger B. A.; Brun P. T.; Nelson C. M. Microextrusion Printing Cell-Laden Networks of Type I Collagen with Patterned Fiber Alignment and Geometry. Soft Matter 2019, 15, 5728–5738. 10.1039/C8SM02605J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.; Chen J.; Yao H.; Liu J.; Yu S.; Lao L.; Wang M.; Luo M.; Xing Y.; Chen F.; Huang D.; Zhao J.; Yang L.; Liao D.; Su F.; Li M.; Liu Q.; Song E. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856. 10.1016/j.cell.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Verjans E.; Noetzel E.; Bektas N.; Schütz A. K.; Lue H.; Lennartz B.; Hartmann A.; Dahl E.; Bernhagen J. Dual Role of Macrophage Migration Inhibitory Factor (MIF) in Human Breast Cancer. BMC Cancer 2009, 9, 1–18. 10.1186/1471-2407-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi I.; Petersen R. M.; Thompson E. W.; Roberts-Thomson S. J.; Monteith G. R. Hypoxia-Induced Reactive Oxygen Species Mediate N-Cadherin and SERPINE1 Expression, EGFR Signalling and Motility in MDA-MB-468 Breast Cancer Cells. Sci. Rep. 2017, 7, 15140. 10.1038/s41598-017-15474-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschulat A.; Drescher D.; Kneipp J. Surface-Enhanced Raman Scattering Hybrid Nanoprobe Multiplexing and Imaging in Biological Systems. ACS Nano 2010, 4, 3259–3269. 10.1021/nn100280z. [DOI] [PubMed] [Google Scholar]
- Huefner A.; Kuan W. L.; Barker R. A.; Mahajan S. Intracellular SERS Nanoprobes for Distinction of Different Neuronal Cell Types. Nano Lett. 2013, 13, 2463–2470. 10.1021/nl400448n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventi A. A.; Billimoria K.; Bartczak D.; Laing S.; Goenaga-Infante H.; Faulds K.; Graham D. New Model for Quantifying the Nanoparticle Concentration Using SERS Supported by Multimodal Mass Spectrometry. Anal. Chem. 2023, 95, 2757–2764. 10.1021/acs.analchem.2c03779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAughtrie S.; Lau K.; Faulds K.; Graham D. 3D Optical Imaging of Multiple SERS Nanotags in Cells. Chem. Sci. 2013, 4, 3566–3572. 10.1039/c3sc51437d. [DOI] [Google Scholar]
- Kneipp J.; Kneipp H.; Wittig B.; Kneipp K. Novel Optical Nanosensors for Probing and Imaging Live Cells. Nanomedicine 2010, 6, 214–226. 10.1016/j.nano.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Milewska A.; Sigurjonsson O. E.; Leosson K. SERS Imaging of Mesenchymal Stromal Cell Differentiation. ACS Appl. Bio Mater. 2021, 4, 4999–5007. 10.1021/acsabm.1c00286. [DOI] [PubMed] [Google Scholar]
- Rabotyagova O. S.; Cebe P.; Kaplan D. L. Collagen Structural Hierarchy and Susceptibility to Degradation by Ultraviolet Radiation. Mater. Sci. Eng., C 2008, 28, 1420–1429. 10.1016/j.msec.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameni M.; Dosescu J.; Moin K.; Sloane B. F. Functional Imaging of Proteolysis: Stromal and Inflammatory Cells Increase Tumor Proteolysis. Mol. Imaging 2003, 2, 159–175. 10.1162/153535003322556903. [DOI] [PubMed] [Google Scholar]
- Nguyen A. K.; Molley T. G.; Kardia E.; Ganda S.; Chakraborty S.; Wong S. L.; Ruan J.; Yee B. E.; Mata J.; Vijayan A.; Kumar N.; Tilley R. D.; Waters S. A.; Kilian K. A. Hierarchical Assembly of Tryptophan Zipper Peptides into Stress-Relaxing Bioactive Hydrogels. Nat. Commun. 2023, 14, 1–13. 10.1038/s41467-023-41907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholt M. S.; Serio A.; Albro M. B. Raman Spectroscopy: Guiding Light for the Extracellular Matrix. Front. Bioeng. Biotechnol. 2019, 7, 303. 10.3389/fbioe.2019.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paidi S. K.; Rizwan A.; Zheng C.; Cheng M.; Glunde K.; Barman I. Label-Free Raman Spectroscopy Detects Stromal Adaptations in Pre-Metastatic Lungs Primed by Breast Cancer. Cancer Res. 2017, 77, 247. 10.1158/0008-5472.CAN-16-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreyling W. G.; Abdelmonem A. M.; Ali Z.; Alves F.; Geiser M.; Haberl N.; Hartmann R.; Hirn S.; De Aberasturi D. J.; Kantner K.; Khadem-Saba G.; Montenegro J. M.; Rejman J.; Rojo T.; De Larramendi I. R.; Ufartes R.; Wenk A.; Parak W. J. In Vivo Integrity of Polymer-Coated Gold Nanoparticles. Nat. Nanotechnol. 2015, 10, 619–623. 10.1038/nnano.2015.111. [DOI] [PubMed] [Google Scholar]
- Castellanos-Rubio I.; Rodrigo I.; Olazagoitia-Garmendia A.; Arriortua O.; Gil De Muro I.; Garitaonandia J. S.; Bilbao J. R.; Fdez-Gubieda M. L.; Plazaola F.; Orue I.; Castellanos-Rubio A.; Insausti M. Highly Reproducible Hyperthermia Response in Water, Agar, and Cellular Environment by Discretely PEGylated Magnetite Nanoparticles. ACS Appl. Mater. Interfaces 2020, 12, 27917–27929. 10.1021/acsami.0c03222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarabelli L.; Grzelczak M.; Liz-Marzán L. M. Tuning Gold Nanorod Synthesis through Prereduction with Salicylic Acid. Chem. Mater. 2013, 25, 4232–4238. 10.1021/cm402177b. [DOI] [Google Scholar]
- Scarabelli L.; Sánchez-Iglesias A.; Pérez-Juste J.; Liz-Marzán L. M. A “Tips and Tricks” Practical Guide to the Synthesis of Gold Nanorods. J. Phys. Chem. Lett. 2015, 6, 4270–4279. 10.1021/acs.jpclett.5b02123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.