Abstract
Recently, it was found that if either the TATA binding protein or RNA polymerase II holoenzyme is artificially tethered to a promoter, transcription is activated. This finding provided presumptive evidence that upstream activating proteins function by recruiting components of the preinitiation complex (PIC) to the promoter. To date, however, there have been no studies demonstrating that upstream factors actually recruit components of the PIC to the promoter in vivo. Therefore, we have studied the mechanism of action of two disparate transactivating domains. We present a series of in vivo functional assays that demonstrate that each of these proteins targets different components of the PIC for recruitment. We show that, by targeting different components of the PIC for recruitment, these activating domains can cooperate with each other to activate transcription synergistically and that, even within one protein, two different activating subdomains can activate transcription synergistically by cooperating to recruit different components of the PIC. Finally, considering our work together with previous studies, we propose that certain transcription factors both recruit components of the PIC and facilitate steps in transcriptional activation that occur subsequent to recruitment.
When examined in vitro, promoter-specific transactivating factors bind to several proteins in the preinitiation complex (PIC), and there is much evidence that these interactions are essential for transactivation to occur (8, 10, 18, 27, 33). However, the immediate functional consequence of the interaction between transactivating factors and the PIC has yet to be studied in an in vivo system. Therefore, it has been unclear whether upstream factors activate transcription in vivo by modifying proteins that are already assembled in the PIC or whether they do so by recruiting proteins into the assembling PIC. Furthermore, the specific steps in the formation of an active PIC that are regulated by upstream activating proteins have not been identified.
Several recent studies have demonstrated that artificial recruitment of either of two components of the PIC to the promoter in vivo is sufficient to activate transcription. Specifically, it has been shown that artificial recruitment of TATA binding protein (TBP) or the RNA polymerase II holoenzyme activates transcription (1, 5, 7, 13, 32). These findings suggested that promoter-bound factors might stimulate transcription if they recruit either TBP or the holoenzyme into an assembling PIC. It has yet to be demonstrated, however, that promoter-specific transactivating factors actually do recruit components of the PIC in an in vivo system, and if upstream activators do indeed recruit components of the PIC, it is not known whether each has a generalized recruiting function or whether different transactivating proteins recruit different components of the PIC. Additionally, the significance of the finding that recruitment of either TBP or the holoenzyme is sufficient to activate transcription is unknown: i.e., would recruitment of one of these by an upstream activator preclude further activation by the other, or would simultaneous recruitment of TBP and the holoenzyme result in additive or even synergistic transcriptional activity?
We present a series of functional assays that were designed to address these issues. The results of each are consistent with several possible interpretations; however, when the results of all of the assays are considered together they strongly suggest that in vivo (i) upstream activators can recruit at least two components of the PIC in a stepwise manner, one of which is TBP; (ii) disparate activating domains target different components of the PIC for recruitment; (iii) activating proteins on the same promoter that recruit different components of the PIC activate transcription synergistically; and (iv) separate activating subdomains within one protein that recruit different components of the PIC activate transcription synergistically. We also discuss the likelihood that some transcription factors activate transcription by recruitment and then through further interaction with certain proteins after they are assembled in the PIC.
MATERIALS AND METHODS
Tissue culture and transfections.
C33a cells were grown and transfected on 60-mm-diameter plates as described elsewhere (31). For each plate, 0.4 μg of the reporter plasmid and 0.1 μg of each expression vector were transfected, unless otherwise indicated. Empty vector was used to bring total DNA to 5 μg for each transfection. Cells were collected 36 h after transfection, and chloramphenicol acetyltransferase (CAT) activity was determined as described elsewhere (31). Each experiment was performed a minimum of three times, and the results of representative experiments are presented.
Plasmids.
pGal4-VP16Δ456 (15), pGal4-Sp1 (25), pCGNTBP (30), pCGNTBPAS (30), and pG2E1B-CAT and pE1B-CAT (16) have all been described previously. To construct pGal4-TBP, p6His-T-hTBP (a gift from R. G. Roeder) was digested with NdeI, blunted, and then digested with BamHI to obtain the cDNA for hTBP. This was cloned between the SmaI and BamHI sites of PM1 (23). To construct pLexA-VP16Δ456, pGal4-VP16Δ456 was digested with BamHI, blunted, and then digested with EcoRI, and the resultant fragment was cloned between the SmaI and EcoRI sites of pBXL1 (a gift from P. Broad). To construct pG2L2E1B-CAT, pL6EC (a gift from P. Broad) was digested with XhoI to obtain two LexA binding sites. These were cloned by blunt end ligation into the XbaI site of pG2E1B-CAT. To construct LexA-TBP, Gal4-TBP was digested with EcoRI and the resultant fragment was cloned into the EcoRI site of pBXL1. pG2-CAT was constructed by digesting pG2E1B-CAT with XbaI and BamHI to remove the TATA box and then closed by blunt end ligation. pL2E1B-CAT was constructed by cloning the two LexA sites obtained by XhoI digestion from pL6EC into the XhoI site of pE1B-CAT. pG2RS-CAT was constructed by cloning the sequence 5′-CTAGAGGGTGTAAAGTACT-3′ between the BamHI and XbaI sites of pG2E1B-CAT. Gal4-(Sp1-VP16Δ456) was constructed by digesting pGal4-Sp1 with SalI and RsaI to obtain the coding sequence for the Sp1 activating domain. For cloning purposes, oligonucleotides were used to add the sequence 5′-GAATTCCCGGG-3′ at the SalI site and the sequence 5′-ACTCTCAGGACAGGGTACCGAATTC-3′ at the RsaI site. The Sp1 sequence is in boldface. EcoRI sites are underlined, and these were used to clone the Sp1 activating domain in frame in the EcoRI site in Gal4-VP16Δ456. pCGNTBPAS was used to express TBPRS.
Primer extension assays.
Transfections were performed in exactly the same manner as they were performed for the associated CAT assays, and total RNA was isolated 36 h after transfection with RNeasy (Qiagen). The Primer Extension System (Promega) was used according to the manufacturer’s recommendations to assay for CAT transcripts. Twenty micrograms of RNA was used for each assay, except that 40 μg was used for assessing the CAT transcript induced by Gal4-Sp1 and the combination of Gal4-Sp1 and TBP. In addition, primer extensions were done in parallel for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with 20 μg of the RNA from each sample. The sequence 5′-GGGATATATCAACGGTGG-3′ was used for the CAT primer, and the sequence 5′-GACCTTCACCTTCCCCAT-3′ was used for the GAPDH primer. Each assay was performed with RNA prepared from two different transfections, and the results were similar each time.
Immunoblotting.
pGal4-TBP (3 μg), pGal4-VP16Δ456 (4 μg), pGal4-Sp1 (1 μg), PM1 (4 μg), pLexA-VP16Δ456 (1 μg), and pBLX1 (1 μg) were cotransfected with the quantity of expression vector indicated into C33a cells as outlined above. Thirty-six hours after transfection, immunoblotting was performed on cell lysates with either anti-Gal4 polyclonal antiserum (Santa Cruz) or anti-LexA polyclonal antiserum (a gift from R. Brent, Massachusetts General Hospital) as described previously (31). Each immunoblot assay was performed twice with protein lysates prepared from two different transfections, and the results were similar each time.
RESULTS
Upstream activators differ in their abilities to recruit TBP to the promoter in vivo.
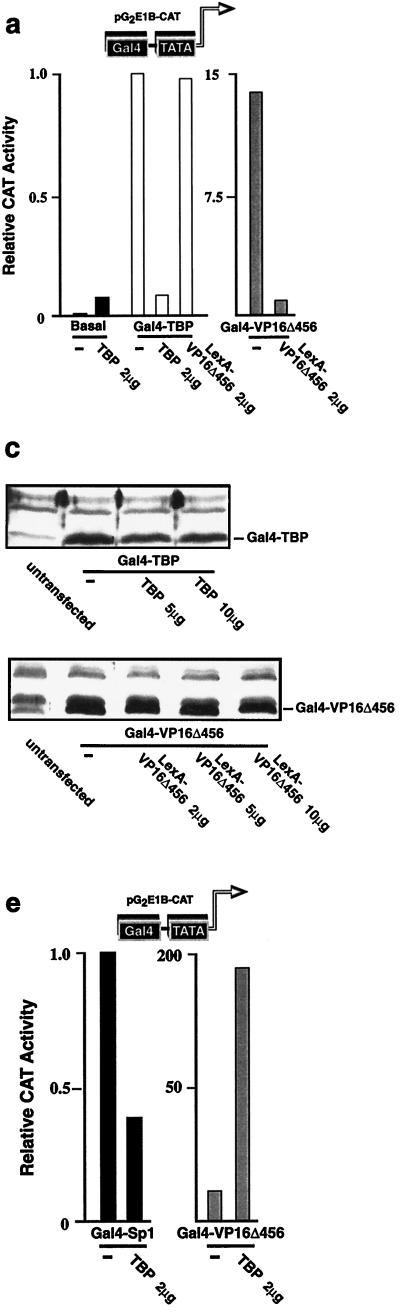
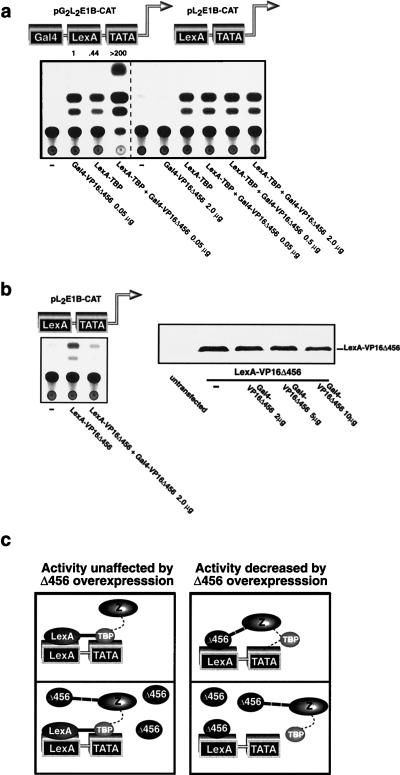
Transcriptional activators differ in their responses to overexpression of TBP (22). This most likely reflects differences in the mechanisms by which activators function. Therefore, we reasoned that overexpression of TBP might serve as a tool for dissecting the mechanism of activator function in vivo. We first examined the effect of TBP overexpression on the activity of promoter-bound TBP. When TBP is artificially tethered to a promoter by the DNA-binding domain of the yeast transcription factor Gal4 (Gal4), as a Gal4-TBP fusion protein, transcription is activated (32) (Fig. 1a). Overexpression of TBP disrupted activation by Gal4-TBP (Fig. 1a). This effect was specific since expression of the transcriptional activator LexA-VP16Δ456 (a chimeric protein in which a transactivation domain corresponding to VP16 amino acids 413 to 456 [20] is fused to the DNA-binding domain of the bacterial transcription factor LexA) failed to inhibit the activity of Gal4-TBP, even though it completely inhibited transactivation by Gal4-VP16Δ456 (Fig. 1a), and in contrast to its inhibitory effect on Gal4-TBP activity, overexpression of TBP activated the basal promoter (Fig. 1a) and potentiated activation by Gal4-VP16Δ456 (see below and Fig. 1e). The effect of overexpression of TBP on the activity of Gal4-TBP was at the mRNA level (Fig. 1b), and the effects of overexpression of TBP and LexA-VP16Δ456 were not due to a reduction in expression of the Gal4-TBP or Gal4-VP16Δ456 protein (Fig. 1c). One possible model to explain the mechanism by which overexpression of TBP inhibits Gal4-TBP activity is depicted in Fig. 1d: when Gal4-TBP is bound to a promoter, the TBP moiety weakly nucleates the assembly of the remaining components (Z) that are necessary to form an active PIC (Fig. 1d, upper panel); when TBP is overexpressed, it competes with the promoter-bound Gal4-TBP for binding to a protein(s) that is a component of the assembling PIC, and so it inhibits transcription by blocking the assembly of an active PIC (Fig. 1d, lower panel). In this model, the effect of overexpression of TBP would be analogous to the proposed mechanism for transcriptional squelching (9). Overexpression of TBP also inhibited transactivation by Sp1 (Fig. 1e); in contrast, however, TBP overexpression enhanced activation by Gal4-VP16Δ456 (Fig. 1e). The effect of overexpression of TBP on the activity of Gal4-Sp1 and Gal4-VP16Δ456 was at the mRNA level (Fig. 1f), and the effect of overexpression of TBP was not due to an alteration in the levels of Gal4-Sp1 and Gal4-VP16Δ456 proteins (Fig. 1g). One possible model to explain these findings is presented in Fig. 1h: Sp1 efficiently recruits TBP to the promoter (either directly or through an adapter, i.e., a coactivator or TBP-associated factor [TAF], and the resulting Sp1-TBP complex acts in a manner similar to that of Gal4-TBP (as described above) in respect to its potential to nucleate an active PIC (Fig. 1h, left upper panel), which is disrupted by TBP overexpression (Fig. 1h, left lower panel); VP16Δ456 efficiently recruits components of the PIC that assemble downstream of TBP, but TBP only transiently or weakly associates with this complex, either stochastically or through a weak interaction with the TATA box, other components of the PIC, or VP16Δ456 itself, to complete the formation of an active PIC (Fig. 1h, right upper panel); however, if TBP is overexpressed, the interaction of the assembling PIC and TBP is driven by the increased concentration of TBP such that formation of a complete PIC is favored and transcription is augmented (Fig. 1h, right lower panel). We sought further evidence in support of these models.
FIG. 1.
Activators target different components of the PIC for recruitment. (a) Overexpression of TBP inhibits the activity of promoter-bound TBP. When TBP is overexpressed with a reporter that contains Gal4 binding sites and a TATA box, transcription is activated. However, activation of the same promoter by Gal4-TBP is inhibited by overexpression of TBP. Gal4-TBP activity is unaffected by overexpression of LexA-VP16Δ456, even though LexA-VP16Δ456 squelches Gal4-VP16Δ456. (b) Cells were transfected in exactly the same manner as for panel a, and primer extensions for CAT and GAPDH mRNA were performed in parallel for each sample. The extension products were of the expected sizes. Numbers at left show sizes of DNA standards. (c) Overexpression of TBP does not affect the expression of Gal4-TBP protein, and expression of LexA-VP16Δ456 does not affect the expression of Gal4-VP16Δ456 protein when assessed by immunoblotting. (d) One possible model for the mechanism of inhibition of Gal4-TBP activity by TBP overexpression (see text for details). (e) Overexpression of TBP inhibits the activity of Gal4-Sp1 but potentiates the activity of Gal4-VP16Δ456. (f) Cells were transfected in exactly the same manner as for panel e, and primer extensions for CAT and GAPDH mRNA were performed in parallel for each sample. The extension products were of the expected sizes. The contrast of the image was increased to facilitate the visualization of the CAT mRNA in the cells transfected with the Gal4-Sp1 expression vector and the cells cotransfected with the Gal4-Sp1 and TBP expression vector. Numbers at left show sizes of DNA standards. (g) Overexpression of TBP does not affect the expression of the Gal4-Sp1 or Gal4-VP16Δ456 proteins when assessed by immunoblotting. (h) One possible model for the mechanisms of transcriptional inhibition and potentiation by TBP overexpression (see text for details).
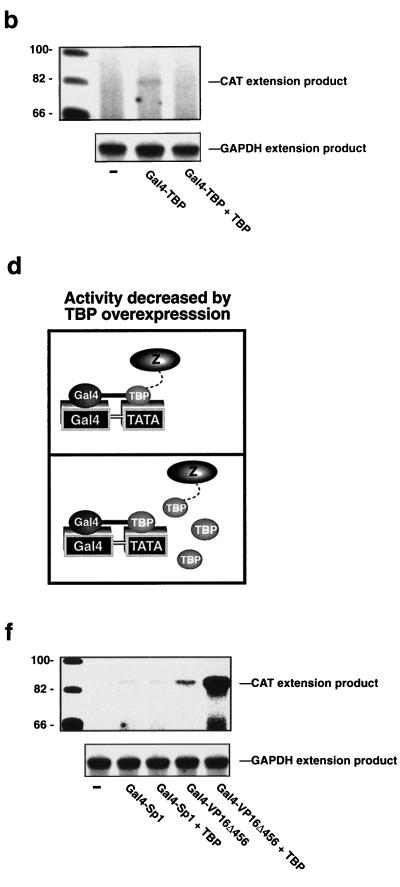
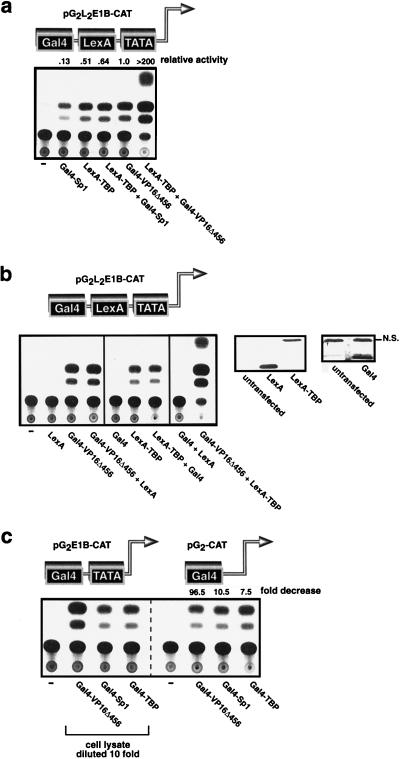
To simplify the examination of the mechanism by which transactivating proteins function, we eliminated a variable in the assembly of the PIC by “clamping” TBP to the promoter by using the LexA DNA-binding domain. We reasoned that a transcriptional activator that functions solely by recruiting TBP would have no effect on promoter activity if TBP was stably fixed on the promoter by LexA (as a LexA-TBP fusion protein [5]); however, an activator that normally functions at a step after TBP recruitment in the assembly of a functional PIC would act in concert with LexA-TBP and further stimulate transcription. If either Gal4-Sp1 or LexA-TBP was brought to a promoter that contains both Gal4 and LexA operators upstream from a TATA box, transcription was activated (Fig. 2a). However, when both were expressed together, the Gal4-Sp1 was inert as evidenced by the fact that the resulting promoter activity was no higher than that induced by LexA-TBP alone (Fig. 2a). The lack of further activation by Gal4-Sp1 was not simply due to the intrinsically low level of activity of Sp1, since the combinations of Gal4-Sp1 and LexA-VP16Δ456 (data not shown) and LexA-Sp1 and Gal4-VP16Δ456 (below and Fig. 4a) activated transcription synergistically. In contrast to the lack of cooperativity between Gal4-Sp1 and LexA-TBP, Gal4-VP16Δ456 cooperated with LexA-TBP to activate transcription synergistically (Fig. 2a). This effect was dependent upon the transactivation domains of the chimeric proteins (Fig. 2b). One possible explanation for these findings is that Sp1 normally activates transcription by recruiting TBP to the promoter; therefore, Sp1 has no additional effect if TBP is already bound to the promoter (in this case by LexA). Conversely, VP16Δ456 does act cooperatively with LexA-TBP, so it is conceivable that VP16Δ456 normally functions in the formation of an active PIC at a step that occurs downstream of TBP recruitment. Although these results are consistent with several possible mechanisms, when considered in the context of the results presented in Fig. 1a and e they provide further evidence for the models presented in Fig. 1d and h in which Sp1 recruits TBP and VP16Δ456 recruits another component(s) into the PIC. Further evidence for these models is provided by the experiments described below.
FIG. 2.
Sp1 recruits TBP, and VP16Δ456 targets a step that is downstream of TBP recruitment to activate transcription. (a) Gal4-Sp1 is inert if TBP is tethered to the promoter by LexA, but Gal4-VP16Δ456 cooperates with LexA-TBP to activate transcription synergistically. (b) Synergistic activation induced by the combination of Gal4-VP16Δ456 and LexA-TBP is dependent upon the VP16Δ456 and TBP moieties in these chimeric proteins. This is evidenced by the fact that the Gal4 and LexA DNA-binding domains are inactive in this assay (left panel) even though the Gal4 and LexA DNA-binding domains are readily expressed in transfection assays when assessed by immunoblotting (right panels). NS, nonspecific bond. (c) Gal4-Sp1 and Gal4-TBP are less dependent upon the TATA box than Gal4-VP16Δ456 for their transactivation function. “fold decrease” represents the ratio of the activity of each activator in the presence to that in the absence of the TATA box. The cell lysates for the assays depicted in the left panel were diluted 10-fold to facilitate the comparison between the two reporter constructs.
FIG. 4.
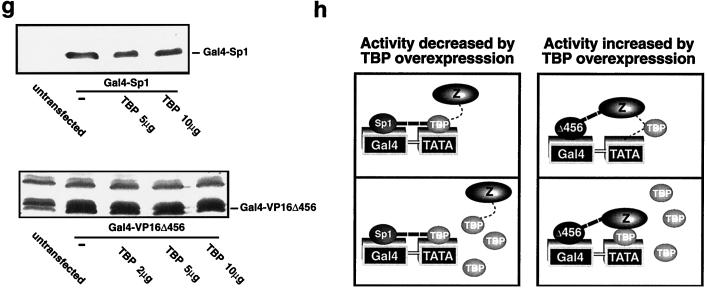
VP16Δ456 recruits a component(s) that is assembled into the PIC downstream of TBP to activate transcription. (a) When Gal4-VP16Δ456 was tethered to the promoter, it potentiated the activity of promoter-bound LexA-TBP in a synergistic manner (left panel). Gal4-VP16Δ456 does not cooperate with LexA-TBP unless it is tethered to the promoter, even when high concentrations of the Gal4-VP16Δ456 expression vector are transfected (right panel). (b) Even when Gal4-VP16Δ456 is not tethered to the promoter, it can still interact with its transactivation target(s), as indicated by the fact that it squelches the activity of LexA-VP16Δ456 (left panel) but not that of LexA-TBP (a). Overexpression of Gal4-VP16Δ456 does not affect cellular levels of LexA-VP16Δ456 as assessed by immunoblotting (right panel). (c) A model for the interactions suggested by the findings that Gal4-VP16Δ456 squelches the activity of promoter-bound LexA-VP16Δ456 but not promoter-bound LexA-TBP.
We next examined the contribution of TATA box function to the assembly of an active PIC by Sp1, VP16Δ456, and TBP. The TATA box has a specific affinity for TBP, and so it is likely that it participates in the assembly of the PIC by contributing to the stabilization of TBP on the promoter. Therefore, it might be expected that deletion of the TATA box would have less of an effect on the activity of a transactivating protein that efficiently recruits TBP to the promoter than on that of a transactivating protein that cannot recruit or can only weakly recruit TBP. In accord with this prediction and the models we have presented for activation, we found that Gal4-Sp1 and Gal4-TBP were markedly less dependent upon the TATA box than Gal4-VP16Δ456. Deletion of the TATA box resulted in a 96.5-fold decrease of the activity of Gal4-VP16Δ456 but only 10.5- and 7.5-fold decreases of the activity of Gal4-Sp1 and Gal4-TBP, respectively (Fig. 2c). Indeed, Gal4-VP16Δ456 was the strongest activator of the three when the TATA box was present, but it had slightly less activity than Gal4-Sp1 and less than half the activity of Gal4-TBP when the TATA box was deleted (Fig. 2c). The simplest explanation for these findings is that Gal4-VP16Δ456 activates transcription by targeting a step after TBP recruitment, and so it is more dependent upon the TATA box to stabilize TBP at the promoter than is either Gal4-Sp1 or Gal4-TBP. In accord with this hypothesis is the finding that, if the TATA box is mutated, the activity of LexA-Gal11, an artificial construct that activates transcription in Saccharomyces cerevisiae by recruiting only the RNA polymerase II holoenzyme, is attenuated to a greater degree than that of an activator that contacts both TBP and TFIIB (1a). Thus, these findings are consistent with and therefore provide further corroborative evidence for the models presented in Fig. 1d and h in which Sp1 recruits TBP to the promoter and VP16Δ456 recruits a component that assembles into the PIC after TBP.
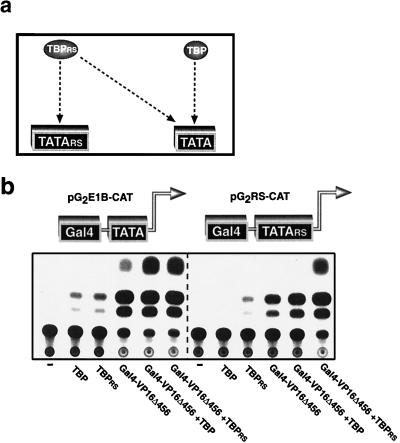
Finally, to confirm that VP16Δ456 transactivation truly is limited by the amount of TATA-bound TBP, we performed an experiment in which it was possible to regulate the TBP-TATA interaction. We took advantage of the relaxed-specificity TBP (TBPRS) developed by Strubin and Struhl (28). This TBP derivative contains mutations on its DNA binding surface that allow it to recognize both the canonical TATA box sequence and the altered TATA box sequence TGTAAA (TATARS), whereas wild-type TBP recognizes only the canonical TATA box (28) (Fig. 3a). As has been demonstrated previously, overexpression of either TBP or TBPRS increased the basal activity of a promoter that contains a canonical TATA box (Fig. 3b, left panel), while only TBPRS increased activity when the TATA box was replaced with TATARS (Fig. 3b, right panel). As would be expected, overexpression of either TBP or TBPRS stimulated transactivation by Gal4-VP16Δ456 to approximately the same extent when assessed with a reporter gene that contains a canonical TATA box in its promoter (Fig. 3b, left panel). In marked contrast, however, as would be predicted if the model presented in Fig. 1h is correct, TBPRS was considerably more effective than wild-type TBP in augmenting Gal4-VP16Δ456 transactivation when the canonical TATA box was replaced with TATARS (Fig. 3b, right panel). These findings indicate that overexpressed TBP must interact with the TATA box to effectively augment transactivation by Gal4-VP16Δ456. Therefore, it is most likely that overexpression of TBP enhances activation by VP16Δ456 because the increased concentration of TBP drives the formation of an active PIC as depicted in the model in Fig. 1h.
FIG. 3.
Overexpressed TBP must be incorporated into the PIC to cooperate with VP16Δ456. (a) TBPRS binds to both a canonical TATA box and a mutant TATA box (TATARS); TBP binds only a canonical TATA box. (b) Overexpression of either TBPRS or TBP increases the activity of a basal promoter that contains a canonical TATA box (left panel); only TBPRS increases the activity of the basal promoter when the TATA box is replaced by TATARS (right panel). Both TBPRS and TBP cooperate with VP16Δ456 with equal effectiveness to activate a promoter that contains a canonical TATA box (left panel); TBPRS cooperates with VP16Δ456 much more effectively than does TBP to activate a promoter that contains TATARS (right panel).
An upstream activator can recruit a component that assembles in the PIC downstream of TBP.
The results presented thus far are consistent with either of two mechanisms for transactivation by VP16Δ456: (i) VP16Δ456 stimulates transcription by recruiting proteins into the assembling PIC, or (ii) VP16Δ456 stimulates transcription by modifying and thereby activating a protein(s) that is already assembled in the PIC. To distinguish between these two possibilities, we sought to determine if it is necessary for VP16Δ456 to be tethered to the promoter to further activate promoter-bound TBP. We reasoned that, if VP16Δ456 functions solely by interacting with proteins that are already assembled in the PIC and it is present at a sufficient concentration in proximity to the PIC, it would not be necessary for VP16Δ456 to be promoter bound to activate transcription. However, if VP16Δ456 normally functions as a tether to recruit proteins to the promoter and stabilize them in the PIC, VP16Δ456 itself would have to be stably bound to the promoter to function as a transcriptional activator. When Gal4-VP16Δ456 was brought to the promoter, it potentiated the activity of promoter-bound LexA-TBP in a synergistic manner (Fig. 4a, left panel), but Gal4-VP16Δ456 that was not bound to the promoter had no effect whatsoever (activating or squelching) on the activity of promoter-bound LexA-TBP (Fig. 4a, right panel). This was true even at much higher concentrations than were necessary for synergy when Gal4-VP16Δ456 was promoter bound (Fig. 4a, right panel), suggesting that Gal4-VP16Δ456 must be bound to the promoter to recruit a component(s) into the PIC to activate transcription. Alternatively, it was conceivable that the local concentration of VP16Δ456 is high enough to interact with proteins in the PIC only if it is tethered near the PIC on the promoter, so we examined this possibility. When 2 μg of Gal4-VP16Δ456 was transfected, it squelched the activity of LexA-VP16Δ456 (Fig. 4b, left panel); however, the same concentration of Gal4-VP16Δ456 neither squelched nor activated the activity of LexA-TBP when there was no Gal4 binding site on the promoter (Fig. 4a, right panel; also see analogous results in Fig. 1a). The fact that overexpressed Gal4-VP16Δ456 squelches activation by promoter-bound LexA-VP16Δ456 but not LexA-TBP-mediated activation strongly suggests that Gal4-VP16Δ456 blocks transactivation by LexA-VP16Δ456 because it specifically competes with the promoter-bound LexA-VP16Δ456 for binding to its target in the PIC and thereby interferes with the ability of LexA-VP16Δ456 to recruit its target protein(s) (see model in Fig. 4c). Therefore, it apparently is not sufficient for VP16Δ456 to simply bind to its target(s) to activate transcription. Thus, the results presented above are most consistent with a mechanism in which VP16Δ456 activates transcription at least in part by recruiting components of the PIC to the promoter. It is known that VP16 also activates transcription at a postinitiation step. Our data, however, suggests that recruitment is a requisite first step for transcriptional activation by VP16Δ456.
Upstream activating domains that recruit different components of the PIC can cooperate to activate transcription synergistically.
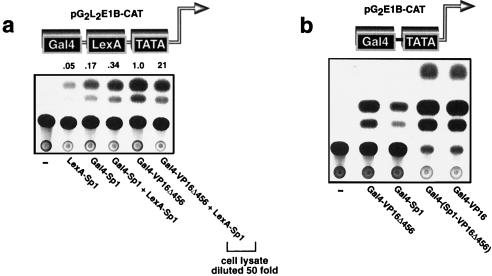
It has been proposed that synergistic transcriptional activation might occur when two components of the PIC, such as TBP and the RNA polymerase holoenzyme, are recruited by different activators on the promoter (26). Since our findings suggest that Sp1 and VP16Δ456 recruit different components of the PIC, we sought to determine if they would activate transcription synergistically when bound to the same promoter. We found that when LexA-Sp1 was brought to a promoter with Gal4-Sp1, transcriptional activity was increased about twofold over the level observed when only Gal4-Sp1 was bound to the same promoter (Fig. 5a). However, when LexA-Sp1 was brought to a promoter with Gal4-VP16Δ456, transcriptional activity increased 21-fold over that induced by Gal4-VP16Δ456 alone and 420-fold over the level induced by LexA-Sp1 alone (Fig. 5a). These findings are consistent with a mechanism in which Sp1 and VP16Δ456 recruit different proteins into the PIC and thereby cooperate when bound to the same promoter to activate transcription synergistically.
FIG. 5.
Transcriptional synergy. (a) Sp1 cooperates with VP16Δ456 but not with itself to activate transcription synergistically. Relative promoter activity is indicated above each assay. The cell lysate from one assay was diluted as indicated to facilitate quantification of activity. (b) Activating subdomains within one protein can cooperate to activate transcription synergistically. A single chimeric protein, encoded by Gal4-(Sp1-VP16Δ456), containing the activation domains of Sp1 and VP16Δ456 fused to the Gal4 DNA-binding domain is a strong transcriptional activator.
Generally, several activators must act in concert on a eukaryotic promoter for maximal transcriptional activity, and yet some, such as VP16, can act alone to strongly activate promoters (reviewed in reference 19). It is possible that the transactivation domains of some activators contain subdomains that independently target different components of the PIC for recruitment and that these subdomains cooperate with each other to activate transcription synergistically. To test this possibility, we constructed a vector that expresses a single chimeric protein that contains both the Sp1 activation domain and VP16Δ456 fused to the Gal4 DNA-binding domain [Gal4-(Sp1-VP16Δ456)]. We reasoned that if the Sp1 and VP16Δ456 domains in this chimeric protein could act independently to recruit their normal targets to the PIC, they would cooperate and activate transcription strongly. The transcriptional activity induced by Gal4-(Sp1-VP16Δ456) was at least 200-fold greater than the activity induced by either Gal4-Sp1 or Gal4-VP16Δ456 (Fig. 5b) and equivalent to that of the strong transcriptional activator Gal4-VP16 (which contains the full-length VP16 activating domain fused to Gal4 [24]), suggesting that some transcription factors may activate transcription efficiently because they contain subdomains that target different components of the PIC for recruitment. Indeed, VP16 has been shown to bind to several different proteins in the PIC, including TBP, either directly (27) or indirectly through a TAF (10, 14), and proteins that assemble into the PIC after TBP (17, 18, 21, 33).
DISCUSSION
It has recently been demonstrated that artificial recruitment of either of two components of the PIC, TBP and the RNA polymerase holoenzyme, is sufficient to activate transcription (1, 5, 7, 13, 32). This finding suggested that upstream transcriptional activators might function at least in part by recruiting these proteins into an assembling PIC. Here we have used a series of functional assays to evaluate whether upstream activators do indeed recruit components of the PIC. The results of these assays provide evidence that Sp1 recruits TBP to the promoter and that VP16Δ456 functions to activate transcription at least in part by recruiting a component(s) that assembles into the PIC downstream of TBP.
First, we demonstrated that overexpression of TBP reduces the activity of promoter-bound TBP and Sp1 but potentiates the activity of VP16Δ456. This suggests that endogenous TBP levels are not limiting for activation by Sp1 but that they are limiting for activation by VP16Δ456. One possible explanation for this finding is that Sp1 efficiently recruits TBP to the promoter but VP16Δ456 cannot. Therefore, an increase in the cellular concentration of TBP would only squelch activation by Sp1, but it could facilitate activation by VP16Δ456 (Fig. 1h). We then examined the activity of both Sp1 and VP16Δ456 when TBP was constitutively bound to the promoter by the LexA DNA-binding domain. We found that, while Sp1 had no further effect on promoter activity if TBP was bound in this manner, VP16Δ456 potentiated the activity of the promoter-bound TBP synergistically. These findings suggest that activation by Sp1 is solely a result of TBP recruitment and that VP16Δ456 activation results from a step(s) downstream of TBP recruitment. Next, reasoning that the TATA box serves to stabilize TBP on the promoter, we examined the effect of deletion of the TATA box on the activity of promoter-bound TBP, Sp1, and VP16Δ456. Whereas Gal4-VP16Δ456 was the strongest activator when a TATA box was present, it became the weakest activator when the TATA box was deleted—its activity falling to less than half that of Gal4-TBP in the absence of the TATA box. These results suggest that Gal4-VP16Δ456 is more dependent upon the TATA box for TBP recruitment than is either Gal4-TBP or Gal4-Sp1 and that VP16Δ456 activates transcription by targeting a step that occurs downstream of TBP recruitment. This was further demonstrated by the finding that overexpressed TBP enhances VP16Δ456 transactivation only when the TBP is bound to the TATA box (Fig. 3b). We then found that, when VP16Δ456 was expressed at a level at which it readily squelched itself, it had no effect whatsoever on the activity of promoter-bound TBP (Fig. 1a and 4a and b). This suggests that when expressed at this level VP16Δ456 self-squelches because it specifically interacts with its normal target protein(s) in the PIC and thereby interferes with the ability of promoter-bound VP16Δ456 to recruit and/or stabilize its target protein(s) in the PIC. Conversely, that this level of Gal4-VP16Δ456 does not potentiate the activity of promoter-bound TBP, even though it can squelch promoter-bound LexA-VP16Δ456, suggests that VP16Δ456 must be tethered to the promoter to recruit a protein(s) into the PIC to activate transcription (Fig. 4c). Finally, consistent with the hypothesis that Sp1 and VP16Δ456 recruit different components of the PIC was the finding that, when they were both brought to the same promoter, either as separate proteins or together in one chimeric protein, they activated transcription synergistically.
The protein interactions suggested by our findings have been shown to occur either in vitro or in yeast two-hybrid systems. However, the mechanism by which these interactions result in transactivation was not addressed. Specifically, it has been demonstrated that Sp1 interacts with a protein that is tightly associated with TBP in the TFIID complex, hTAFII130, and its Drosophila homolog, dTAFII110 (6, 8, 12, 29), and that VP16Δ456 can bind to the mediator subcomplex in a yeast RNA polymerase II holoenzyme (11). Our data advances these findings by providing evidence that these interactions result in recruitment and that this is, at least in part, the mechanism by which Sp1 and VP16Δ456 activate transcription. It is notable that VP16 has also been shown to interact with TAFII40 (10), suggesting that VP16 should also recruit TFIID to the promoter. However, it was shown that the C-terminal 39 amino acids of VP16 are necessary for this interaction (10). These amino acids are deleted from VP16Δ456; hence, VP16Δ456 is deficient in this function. It is possible that the Sp1-VP16Δ456 construct we studied was as efficient an activator as intact VP16 because the Sp1 portion of the chimeric protein compensated for the loss of this function by binding to hTAFII130.
Several models have been proposed for the mechanism by which transactivating proteins function together in a synergistic fashion. Indeed, it is likely that there are several mechanisms by which they can act in a synergistic fashion. We have provided evidence that some transcription factors may cooperate to activate transcription synergistically when they simultaneously but independently recruit different components of the PIC. Carey and coworkers originally proposed a model for synergy in which multiple activator molecules simultaneously contact a single target in the PIC and thereby stabilize it at the promoter (4). In this model, an arithmetic increase in the number of activating molecules on the promoter should lead to an exponential increase in stability, which would result in a synergistic increase in transcription. Alternately, a kinetic model in which multiple activator molecules contact a target in a sequential manner has been proposed (3). Increasing the number of activator molecules on the promoter would increase the frequency of activator-target functional contacts. Once a threshold frequency of activator-target interactions is surpassed, transcription would occur. Thus, increasing the number of activators would result in synergy once the threshold is exceeded.
Several transcription factors have been shown to activate transcription by targeting steps that occur postinitiation. Indeed, VP16 has been shown to facilitate elongation, and this is thought to be a result of the interaction of VP16 with TFIIH (2, 34). Therefore, considering these findings together with our results, it is likely that VP16 and other transcription factors function as transcriptional activators both by recruiting components of the PIC and by activating a postinitiation function of the assembled PIC.
ACKNOWLEDGMENTS
We thank J. T. Lis for critical review of the manuscript. We thank A. Berk for pGal4-VP16Δ456, W. Schaffner for pGal4-Sp1, W. Herr for pCGNTBP and pCGNTBPAS, M. R. Green for pG2E1B-CAT and pE1B-CAT, R. G. Roeder for p6His-T-hTBP, I. Sadowski for PM1, and P. Broad for pBXL1 and pL6EC.
This work was supported by a grant from the NIH.
REFERENCES
- 1.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 1a.Barberis, A., and M. Ptashne. Personal communication.
- 2.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carey M, Kolman J, Katz D A, Gradoville L, Barberis L, Miller G. Transcriptional synergy by the Epstein-Barr virus transactivator ZEBRA. J Virol. 1992;66:4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey M, Lin Y S, Green M R, Ptashne M. A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature. 1990;345:361–364. doi: 10.1038/345361a0. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 7.Farrell S, Simkovich N, Wu Y, Barberis A, Ptashne M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996;10:2359–2367. doi: 10.1101/gad.10.18.2359. [DOI] [PubMed] [Google Scholar]
- 8.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 10.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 11.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 12.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 13.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 14.Klemm R D, Goodrich J A, Zhou S, Tjian R. Molecular cloning and expression of the 32-kDa subunit of human TFIID reveals interactions with VP16 and TFIIB that mediate transcriptional activation. Proc Natl Acad Sci USA. 1995;92:5788–5792. doi: 10.1073/pnas.92.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi N, Boyer T G, Berk A J. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 17.Lin Y S, Green M R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991;64:971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 19.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 20.Regier J L, Shen F, Triezenberg S J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts S G, Ha I, Maldonado E, Reinberg D, Green M R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993;363:741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- 22.Sadovsky Y, Webb P, Lopez G, Baxter J D, Fitzpatrick P M, Gizang-Ginsberg E, Cavailles V, Parker M G, Kushner P J. Transcriptional activators differ in their responses to overexpression of TATA-box-binding protein. Mol Cell Biol. 1995;15:1554–1563. doi: 10.1128/mcb.15.3.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 24.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 25.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stargell L A, Struhl K. Mechanisms of transcriptional activation in vivo—two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 27.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 28.Strubin M, Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992;68:721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- 29.Tanese N, Saluja D, Vassallo M F, Chen J L, Admon A. Molecular cloning and analysis of two subunits of the human TFIID complex: hTAFII130 and hTAFII100. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tansey W P, Ruppert S, Tjian R, Herr W. Multiple regions of TBP participate in the response to transcriptional activators in vivo. Genes Dev. 1994;8:2756–2769. doi: 10.1101/gad.8.22.2756. [DOI] [PubMed] [Google Scholar]
- 31.Weintraub S J, Chow K, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 32.Xiao H, Friesen J D, Lis J T. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]