Abstract
Background Diffuse midline glioma (DMG) is a devastating pediatric brain tumor unresponsive to hundreds of clinical trials. Approximately 80% of DMGs harbor H3K27M oncohistones, which reprogram the epigenome to increase the metabolic profile of the tumor cells.
Methods We have previously shown preclinical efficacy of targeting both oxidative phosphorylation and glycolysis through treatment with ONC201, which activates the mitochondrial protease ClpP, and paxalisib, which inhibits PI3K/mTOR, respectively.
Results ONC201 and paxalisib combination treatment aimed at inducing metabolic distress led to the design of the first DMG-specific platform trial PNOC022 (NCT05009992).
Conclusions Here, we expand on the PNOC022 rationale and discuss various considerations, including liquid biome, microbiome, and genomic biomarkers, quality-of-life endpoints, and novel imaging modalities, such that we offer direction on future clinical trials in DMG.
Keywords: pediatric neuro-oncology, clinical trials, diffuse midline glioma, ONC201, paxalisib
Background
Diffuse midline glioma (DMG) is the most lethal childhood cancer, with a median survival of only 10 months.1 There are 200–300 cases of DMG in the United States annually, each arising in midline structures, such as the spinal cord, pons, and thalamus.2,3 Approximately 80% of DMGs harbor somatic missense mutations, which substitute methionine for lysine at position 27 of histone H3.1 or H3.3, known together as the H3K27M mutations.4–7 These H3K27M mutations drive global epigenetic aberrancies including genomic hypomethylation, decreased H3K27 trimethylation (H3K27me3), and increased H3K27 acetylation (H3K27ac).8–13
H3K27M-mutant DMGs depend on methionine metabolism, with restriction of this amino acid increasing the survival of in vivo models.14 To exploit tumor dependency on high metabolic activity, we performed a preclinical investigation of the mitochondrial targeting drugs ONC201 and ONC206, demonstrating that they decrease DMG cell viability in vitro and increase survival of in vivo models.15 ClpP activation is achieved by a number of drugs including ADEPs, D9, TR, and ONC drugs. ClpP targeting was first achieved as an anti-bacterial strategy using the natural products acyldepsipeptide antibodies (ADEPs), which showed efficacy in ClpP activation.16 D9 is a small molecule that mimics the natural structure of ClpX, the ClpP chaperon.17 D9 has been used as a potent ClpP agonist to better understand the function of this mitochondrial protease.17 TR compounds are a novel series of imipridone that have shown stringent efficacy in ClpP targeting with some reported to be an order of magnetite more effective than ONC compounds in inducing breast tumor cell kill.18 ONC201, the original imipridone drug, was reported to induce TRAIL apoptotic pathway by inhibiting AkT and ERK phosphorylation of Foxo3a. However, subsequent studies showed ONC201 as ClpP agonist.16–21 Caseinolytic peptidase proteolytic subunit (ClpP) protein localizes to the inner mitochondrial membrane and hydrolyses excess or misfolded proteins.20,22 ClpP is a key regulator of the mitochondrial stress response, and a target for disruption of mitochondrial metabolic pathways in cancer. ONC201 efficacy has been studied in a number of cancers including glioblastoma, hematological malignancies, and prostate cancer.23–26 Indeed, initial studies in glioblastoma, showed ONC201 efficacy in a patient with K3K27M mutation.26 To validate this, we used preclinical models of DMG. Our in vitro and in vivo studies showed that ONC201 and ONC206 allosterically bind to and increase the mitochondrial protease ClpP activity.27 Accordingly, treatment in DMG drives mitochondrial degradation, in turn causing an integrated stress response (ISR) and oxidative phosphorylation (OXPHOS) impairment; interestingly, astrocytic lineage differentiation of tumor cells is also observed.15 These effects of ONC201 combined with epigenetic downregulation of cell cycle-related pathways correlate with improved survival beyond what has been previously seen in this patient population. Specifically, Venneti et al. demonstrated that ONC201 correlated with prolonged overall survival of 21.7 months in patients with newly diagnosed DMG treated after radiation but before disease progression and 9.3 months in patients with progressive disease.28 While ONC206 may have similar properties, the drug remains in early phase, dose escalation studies. This is in contrast to the much larger experience with ONC201 and defined recommended dosing. Thus, we aimed to build on these promising findings with single-agent ONC201 through the addition of novel agents on an ONC201 backbone therapy.
However, H3K27M-mutant DMGs are notable for enhanced glycolysis, potentially offering a compensatory mechanism for tumor energetics following diminished OXPHOS activity.29 This metabolic shift is likely mediated by overactivity of the PI3K/mTOR pathway, which indirectly drives glycolysis.30–32 The PI3K/mTOR signaling pathway controls multiple cellular processes including metabolism, motility, proliferation, growth, and survival.33,34 For example, the highly conserved AMPK protein, is a key energy sensor and homeostasis regulator such as glucose metabolism.35 AMPK promotes glucose uptake by activating PI3K emphasizing the role of PI3K/mTOR in cellular energy metabolism.36 Furthermore, mTOR inhibition leads to suppression of glutathione production in multiple pediatric brain tumors.37 Because glutathione is critically important to mitochondrial function, suppression of glutathione production may increase the efficacy of mitochondrial targeting drugs such as ONC201. The PI3K/mTOR pathway is also activated as a response to ROS accumulation.38 Therefore, we combined ONC201 with GDC-0084 (paxalisib), a clinically available blood-brain barrier (BBB) penetrative PI3K/mTOR inhibitor with promise in atypical rhabdoid teratoid tumor, glioblastoma, and pediatric high-grade glioma (pHGG).39–42 This allowed us to investigate the dual inhibition of OXPHOS and glycolysis. We showed that ONC201 plus paxalisib synergizes in altering OXPHOS and glycolysis to induce metabolic distress.43 This led to the design of the first DMG-specific platform trial PNOC022 (NCT05009992).
Here, we report the PNOC022 rationale and design, review the motivating liquid biome and microbiome work leading to the clinical trial, explore genomic biomarkers of response to the treatment, discuss quality-of-life endpoints, consider the integration of novel imaging modalities to monitor and guide treatment across clinical course, and offer direction for the next generation of clinical trials in DMG.
Challenges in Clinical Trial Design for DMGs
The rapid pace of recent discoveries in the field of DMG has invigorated laboratory and translational research efforts. However, the ultimate objective to develop a therapeutic trial that improves outcomes has yet to be fulfilled.
Unique to DMG, the scientific community is engaged in an ongoing debate regarding the choice of endpoints that will most effectively define success or futility of the hypothesis being tested in clinical settings. Numerous challenges further complicate this endeavor, with the rarity of the disease being a significant obstacle. The utilization of imprecise model systems during preliminary laboratory-based investigations and the prioritization of therapies and relevant biomarkers to evaluate in homogeneous patient cohorts pose further difficulties. These considerations must be balanced with the limited resources available for conducting trials. While the ultimate goal is achieving a cure, there are known confounding factors when studying endpoints, including OS endpoints affected by multiple sequential therapies and incomplete knowledge of subtype-specific natural history. More cost-effective, single-arm phase 2 studies require consensus-validated endpoints that use patient-level data from historically and contemporarily treated populations, ensuring uniform characterization. Alternatively, concurrently controlled randomized studies can be employed. Unfortunately, the current “standard of care” for DMG is far from curative, with any meaningful responses primarily attributable to focal radiotherapy. The acquisition of pharmacodynamic biomarkers introduces more challenges, necessitating multiple tumor tissue samples or less invasive, nontumor liquid biopsies for longitudinal assessments using validated assays. Given the complexity and challenges posed by DMG, well-conducted clinical research is crucial.
The Food and Drug Administration (FDA) considers concurrently controlled randomized studies as the “gold standard” for determining efficacy. Disease stakeholders for DMG, including patients/families and researchers, would prefer nonrandomized studies until a reasonable standard of care is established. An alternative to concurrently controlled trials is utilizing what the FDA calls “external controls.” While not always consisting of contemporaneously treated patients, external controls can provide a control arm for confirming endpoints. Traditional historical controls commonly used in phase 2 trials rely on aggregated results and rarely include sufficient individual patient-level data. Thus, these options lack the comprehensiveness or contemporaneity required to serve as adequate external controls. Recent guidance has been published to identify patients who would be reliable for this purpose, with comprehensive longitudinal clinical, biological, and treatment data at the patient-level providing the minimum required information. The intent of such controls is to assume the role of the concurrent treatment control group, theoretically adhering to all eligibility requirements and follow-up assessments as planned for the intervention. The evident advantage would be a significant reduction in the required sample size for determining trial endpoints while minimizing children’s exposure to a control arm with an inadequate standard of care. Substantial resources are necessary to develop the enriched cohorts required for clinical trials and regulatory application of external controls. However, the potential benefits include addressing sample size and concurrent control issues and overcoming specific clinical research challenges present in DMG.
Within PNOC022, we aimed to tackle some of these challenges through specific strategies. First, we only advanced combination therapies into the clinical setting after thorough assessments in multiple model systems and laboratories, ensuring a higher confidence level in their potential efficacy. Second, we molecularly characterize each patient’s tumor to assess outcomes within a subtype-specific context. Additionally, we have incorporated the collection of CSF as a potential surrogate for on-treatment biopsies. Furthermore, our study integrates various correlative studies, including microbiome investigations and applies contemporary disease-specific, pediatric radiographic assessment criteria through the use of Response Assessment in Pediatric Neuro-Oncology (RAPNO) specific to DMG.
Despite these advancements, endpoints remain a continued challenge in DMG, even with the application of RAPNO guidelines.
Disease Cohorts and Rationale
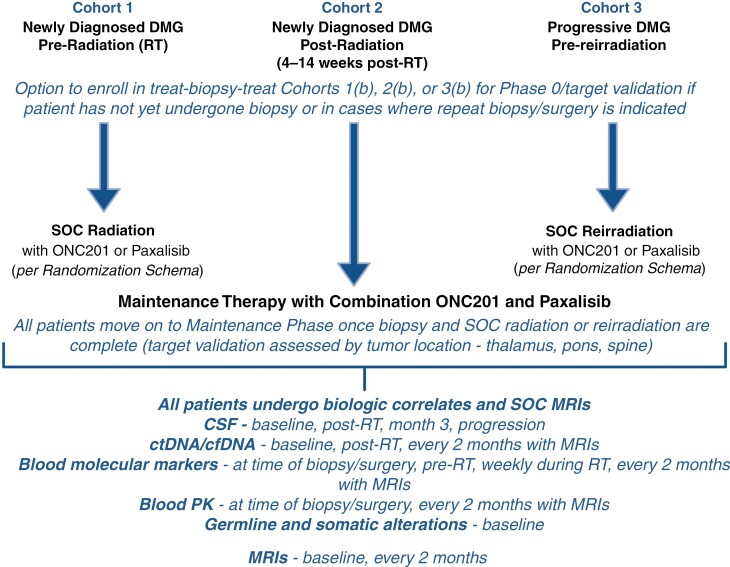
PNOC022 encompasses a multi-cohort, multi-arm adaptive trial design. At the outset of trial development, the goal was to create therapy options for patients at all stages of the disease, from initial diagnosis to disease progression. Investigators wanted to (a) explore the potential efficacy of the therapeutic approach at each stage of the disease, (b) address questions of drug penetration into the tumor with consideration of pre and postradiation effects, and (c) investigate biomarkers of response/resistance within the tumor tissue. To this end, the trial includes 3 cohorts—newly diagnosed, preradiation (cohort 1); newly diagnosed, postradiation (cohort 2); and recurrent (cohort 3; Figure 1). Each cohort has presurgical and postsurgical arms to allow a treat-biopsy-treat approach for patients who have not yet undergone standard-of-care biopsy/resection and are enrolled before surgery. Assignment into each combination arm is determined through a priori randomization at study entry.
Figure 1.
Schematic of PNOC022. Each cohort (1, 2, 3) is represented at the top of the figure with each phase of treatment within the respective cohort outlined as the figure moves from top to bottom. Description and frequency of collected biologic and imaging correlates on PNOC022 is detailed at the bottom of the figure. CSF, cerebrospinal fluid; ctDNA, circulating tumor DNA; DMG, diffuse midline glioma; MRI, magnetic resonance imaging; PK, pharmacokinetics; RT, radiation therapy. Created with Biorender.com.
The presurgical target validation arms require dosing at a specific time point before surgery. This design facilitates the investigation of the pharmacokinetics of drug penetration into the tumor and the pharmacodynamic effects of the drug on tissue. Further in the target validation arms and as means to assess differential drug penetration, we aim to enroll at least five patients each with disease at the following anatomic locations—thalamic, pons, and spine. Within cohorts 1 and 3, investigated drugs are also given concomitantly with upfront or reirradiation. This design allows both safety and efficacy assessment of combination therapy with radiation. After radiation for cohorts 1 and 3 and at the time of study entry for cohort 2, patients begin maintenance therapy with combination therapy. This design allows both safety and efficacy assessment of combination therapy with radiation and exploration of the added impact of combinatorial strategies. The current target validation design investigates posttreatment tumor tissue after ONC201 is given on day −1 or days −2 and −1 and paxalisib on day −1. For cohorts 1 and 3 patients are then randomized to receive either ONC201 or paxalisib with radiation therapy. Within the maintenance phase for cohort 1 and 3 as well as at study entry for cohort 2, all patients receive the combination therapy of ONC201 and paxalisib. An adaptive trial design allows the incorporation of new combination strategies throughout the life of the trial. By offering a flexible and multi-arm/multi-cohort design, PNOC022 aims to comprehensively address toxicity, efficacy, and biomarker identification for patients with DMG, regardless of disease stage, and across a wide-ranging combination of therapies.
Statistical Design (Platform Design and Endpoints)
The trial contains 3 cohorts based on the stage of the disease and 2–3 phases within each cohort: target validation, upfront or reirradiation, and maintenance. Within each disease phase cohort, patients are randomized to one of the study arms or assigned a study arm in the setting of prior exposure to treatment on an alternate arm. The adaptive platform design allows new treatment combination arms to be added and available for additional randomization options during the trial and also provides a mechanism that patients can switch to new cohorts and arms as their disease progresses.
The trial uses the novel Bayesian drug combination platform trial design with adaptive shrinkage (ComPAS) for all cohorts.44 ComPAS allows for dropping ineffective drug combinations and adding new combinations to the ongoing clinical pipeline adaptively based on the accumulating trial data. The treatment maintenance combinations are initially formed with ONC201 and paxalisib, with ONC201 as the backbone therapy and paxalisib as the novel agent. For the target validation and radiation phases, assigned patients will receive only single agent, monotherapy according to the study arm to which they were randomized at the study entry. The novel agents and ONC201 combinations will then be given during the maintenance phase. Each combination will undergo 2 interim analyses, at which point, a decision will be made to stop the arm early for futility, graduate the arm for superiority, or continue the arm to the next interim or final stage. Patients will also be stratified within each arm according to known molecular and clinical prognostic markers of H3 status, TP53 wild-type versus mutant, and age at diagnosis. The ComPAS design decisions for interim and final analyses will be made in comparison to the appropriate historical control using a Bayesian hierarchical model. The advantage of this model is the ability to borrow information across arms. There are 2 settings where borrowing may be advantageous: first, if efficacy (either highly efficacious or not-efficacious) is similar at the interim analysis in the same 2 arms in Cohorts 1 and 2; second, if efficacy is identical for 2 arms that only differ by the number of days the drug was given before the target validation phase (ie, 1 or 2 days).
For cohorts 1 and 2, the primary objective is to assess the efficacy of the combination therapies based on median progression-free survival at 6 months (PFS6). PFS6 is defined as the percentage of patients alive and free from progression at 6 months (26 weeks). The historical control PFS6 rate is 39% based on an ONC201 monotherapy trial (NCT03416530).45 The maximum sample size is 33, and the interim analyses will occur when 11 and 22 patients have been enrolled. For cohort 3, the primary objective is to assess the efficacy of the combination therapies based on median overall survival at 7 months (OS7) based on patient-level data from published data.46–51 OS7 is defined as the percentage of patients alive at 7 months (30 weeks). The historical control OS7 rate is 50% based on published literature. The maximum sample is 42; the interim analyses will occur when 14 and 28 patients have been enrolled.
Correlative Studies of PNOC022
Cell-Free Tumor DNA (cf-tDNA)
Over the past decade, liquid biopsy technologies have emerged to monitor the treatment response of various solid tumors on a clinical and molecular level. These technologies involve assessing plasma and cerebrospinal fluid (CSF) for extracted cell-free tumor DNA (cf-tDNA). Recent studies have shown that these liquid biopsy techniques can effectively detect patient-specific tumor variants in both DMG and nonDMG tumors and even correlate or predict tumor growth and response to treatment.52–56 We and others have shown the utility of digital droplet PCR (ddPCR) to measure circulating tumor DNA (ctDNA) in H3K27M DMGs.53,57–61
Probing for H3F3A (H3.3) K27M mutations, studies showed higher detectable levels of ctDNA in samples of cf-tDNA derived from CSF compared to serum or plasma, as further described below.53,57–60 However, the clinical utility of probing H3.3K27M cf-tDNA for therapeutic monitoring of response and resistance in ongoing and prospective trials was unclear. Early clues of the feasibility of clinical assessment of longitudinal ctDNA in DMGs came from a pilot precision trial utilizing upfront biopsies for designing a biopsy-informed therapy (NCT02274987).62 This trial reported the detection and monitoring of H3F3A and HIST1H3B (H3.1) wild-type (WT) and mutant alleles and assessment of mutation allele frequency (MAF) in 11/13 (85%) of subjects at diagnosis, (6/6; 100%) at postradiation, (7/7; 100%) at treatment, (5/7; 71%) at progression, and (5/5; 100%) at the end of the study. However, the MAF positive detection rate remained very low, hindering further clinical interpretations.
While monitoring MAF is a powerful approach for detecting clinically relevant histone and driver mutations in the pHGG liquid biome, this approach is suitable only for monitoring a limited number of small nucleotide variants (SNVs). It is, therefore, insufficient to detect larger-scale genomic alterations. Capturing intra-tumoral mutational heterogeneity requires comprehensive mutation profiling in ctDNA, including detection of insertions/deletions as well as focal and broad copy number variations (CNVs). To address this unmet need, a targeted, hybrid capture-based next-generation sequencing platform covering a panel of 523 cancer-associated genes (TSO500ctDNATM), encompassing all major prognostic and driver mutations associated with pHGG, was employed.62,63 This approach successfully detected tumor-associated gene CNVs in DMG patient CSF, providing an opportunity for longitudinal gene CNV monitoring to understand the genomic drivers of pHGG evolution and disease progression.61
To further establish the clinical utility of ctDNA profiling, investigators in the ONC014 phase 1 trial of ONC201 in children and young adults (NCT03416530) conducted an arm with 24 patients with H3K27M-mutant DMG enrolled after radiation to undergo serial CSF collection.45 Patients underwent serial lumbar puncture (LP) for cf-tDNA analysis at 0, 2, and 6 months on therapy, while patients enrolled at the University of Michigan underwent monthly serial plasma collection. ddPCR analysis of cf-tDNA samples was performed, and variant allele fraction (VAF) was compared to radiographic measured by maximal 2D tumor area on magnetic resonance imaging (MRI). Patients were screened before LP to confirm clinical safety, with particular concern given to signs of increased intracranial pressure or rapidly changing neurologic exams. No adverse outcomes related to the procedure were observed.
Importantly, change in H3.3K27M VAF over time (“VAF delta”) correlated with prolonged PFS in both CSF and plasma samples, predicting progression and sustained response and possible differentiation of pseudo-progression and pseudo-response.64 Indeed, VAF “spikes” (an increase of at least 25%) preceded tumor progression in many cases.64
These results confirmed the feasibility and utility of serial cf-tDNA in both plasma and CSF of DMG patients to supplement radiographic monitoring. CSF allows for assessing other tumor biomarkers that may be enriched in spinal fluid, such as cell-free DNA methylation sequencing, exosomal RNA, and mitochondrial DNA (mtDNA). However, CSF collection carries more procedural risk than a blood draw, thus encouraging further research to optimize plasma diagnostics for DMG to complement or potentially replace CSF diagnostics. Multiple academic institutions and biotechnology companies are optimizing these methods and transitioning these tests to a CLIA-certified setting will allow for improved decision-making in the clinical management of DMG patients and potentially broader glioma patient populations.
Microbiome
The gut microbiome is vital in various diseases, including central nervous system diseases and cancer.65–68 For adult HGG, there is emerging evidence that the gut microbiome plays a role in pathogenesis.69 Metabolites, such as tryptophan, are strongly influenced by the gut microbiome and can modify glioma’s microenvironment by directly affecting T cells, dendritic cells, tumor-associated macrophages, and antigen-presenting cells.70 Recent data on pediatric DMGs and the microbiome revealed a potential role of the microbiome on PFS and OS. Specifically, the Firmicutes/Bacteroidetes ratio, which serves as a parameter of normal intestinal homeostasis, was observed to be unfavorable at diagnosis, and some components, such as Flavobacteriaceae and Bacillales, were associated with a higher risk of disease progression and death.71 To better understand and elucidate the underlying mechanisms of these observations, it is vital to gain insight into the microbial composition of trial patients with DMG, as is currently being performed in the PNOC22 clinical trials.
Predictive Genomic Biomarkers of ONC201 in DMG
DMGs harbor complex genomes with frequent somatic inactivation of TP53, chromosomal instability (eg, loss of 10q), and high-level amplification of oncogenes (eg, PDGFRA, MET, EGFR).72 We and others have demonstrated that ONC201 binds and activates ClpP, that CLPP depletion abrogates ONC201 sensitivity, and that CLPP expression levels predict ONC201 sensitivity in DMG and other cancer types.15,22,43,73CLPP is located on 19p13.3, a region also susceptible to TP53-associated genomic instability in DMGs and thus potentially predictive of ONC201 sensitivity. Notably, protein-coding genes are dosage sensitive in cancer, and CLPP CNVs also explain CLPP expression differences between cancer cell lines (https://depmap.org/portal/gene/CLPP). Joint DMG whole genome and RNA sequencing in PNOC022 will inform whether high CLPP copy number and expression levels predict ONC201 sensitivity in DMG patients.
Studies in glioblastoma (GBM) and breast cancer provided evidence that ONC201 and ONC206 drive rapid depletion of mtDNA copy number, an effect not seen with other metabolic drugs such as metformin.74,75 Large-scale analysis of whole cancer genomes revealed an order of magnitude difference in mtDNA copy number between GBMs (~70 to 800 copies per cell).76 So far, however, little is known to what extent mtDNA copy number varies between H3K27M-mutant and H3WT DMGs and whether it will modulate ONC201 sensitivity. We will address this question in PNOC022 based on the quantification of mtDNA copy number levels from DMG whole genomes using pipelines developed within the Pan-Cancer Analysis of Whole Genomes Consortium.76
Oncogenic signaling pathways have also been nominated to confer resistance to ONC201. In 2021, He et al. observed that high expression levels of EGFR, the constitutively active form of EGFR (EGFRvIII), and EGFR staining in GBM are determinants of poor preclinical and clinical response to ONC201.77 A recent independent effort to identify combinatorial biomarkers using transcriptomics and proteomics in a large panel of cancer cell lines has also highlighted ClpP and EGFR as predictors of ONC201 and ONC206 sensitivity.78 A similar ONC201 resistance mechanism is also studied in H3K27M-mutant DMGs that harbor oncogenic EGFR mutations, high-level EGFR amplifications, or high EGFR expression levels and will be addressed based on RNA- and whole genome sequencing in PNOC022.79
H3K27M-mutant DMGs are likely stalled in a cellular state between neural stem cells and oligodendrocyte precursor cells (OPC) with a subpopulation of DMGs being defined by high levels of EGFR expression.56–58 This subpopulation resembles a developmental EGFR-high intermediate progenitor cell state termed Pre-OPC80–85 and we observed that ONC201 drives the differentiation of OPC-like DMG cells toward other cell states which raises the possibility of a nongenetic and cell-intrinsic ONC201-resistant subpopulation of Pre-OPC-like DMG cells, thus necessitating combination therapies such as ONC201 and paxalisib.15
Quality of Life and Patient-Reported Outcomes
Along with toxicity, efficacy, and biomarker assessments, PNOC022 aims to collect details on patient experience throughout participation in the trial. Patient-reported outcomes (PROs) are reports of the status of a patient’s health condition that come directly from the patient and/or proxy (in patients less than 18 years of age) without interpretation of the response by a clinician.86 PROs are increasingly acknowledged as an essential source of information about patient and/or proxy experience during cancer treatment. These assessments allow patients/proxies to report on side effects and overall quality-of-life during and after treatment; as such, they are being utilized as direct measures of outcomes in clinical trials.87–95 PROs may provide insight into how patients/proxies perceive the care they have received, and can be administered longitudinally throughout the clinical course. Consensus as to which measures to include within clinical trials is beginning to come into focus for various patient populations. For example, a recent publication from the National Clinical Trials Network (NCTN) Adolescent and Young Adult (AYA) PRO Task Force put forth a recommended core battery of PRO measures to include in clinical trials targeting the AYA population. This battery has inherent flexibility, allowing the PROs to shift based on the trial population and specific study objectives. There are few prospective measures of patient/proxy perception of and satisfaction with participation in a clinical trial, with almost no prospective data within the pediatric cancer patient population.93–95 Within PNOC022, we hypothesized that there will be at least some perceived benefit from participation in PNOC022 and that the perceived benefit, or lack thereof, of study participation will be variable over time, depending on the intensity of the phase of therapy at the time of questionnaire administration.
To understand how the patient and/or proxy perceives trial participation, we created a four-question PRO (“DMG-ACT PRO”). The DMG-ACT PRO is administered at postresection/biopsy; end of radiation; beginning of cycles 3, 6, 12, 18, and 24; and end of treatment. The questions offer insight into the overall satisfaction of study participation, the likelihood of recommending DMG-ACT or a similar study to a future patient, and the retrospective likelihood of participating again in this study. The final question centers around why the patient/proxy answered as they did about the retrospective likelihood of participating in the study with 4 prewritten choices and an option for free text response.
Consideration of Novel Imaging Assessment Integration
Standard contrast-enhanced MRI is the primary clinical imaging modality to determine treatment response.96 However, the conventional MRI technique has its limitations. For example, differentiation between pseudo-progression or tumor progression/recurrence can be difficult. In DMGs, pseudo-progression is commonly seen and has been described in up to 50% of patients.97,98 This can cause a clinical dilemma with the risk of early termination of treatment in a clinical trial. Therefore, further optimization of imaging techniques is needed and advanced MRI techniques and positron emission tomography (PET) may be helpful.
Advanced MRI Techniques
Advanced MRI methods that are now available within standard MR sequencing options, including MR perfusion (MRP) and MR spectroscopy (MRS), have emerged. MRP is used for imaging the vascularity of the brain and the tumor, with different parameters such as arterial-spin labeling (ASL) and cerebral blood flow (CBF). In DMG, the prognostic value of MRP has been demonstrated.99 For instance, increased ASL-CBF can be observed in pseudo-progression compared to actual progression.97 Further, a difference in perfusion values between tumors with H3.1K27M and H3.3K27M mutations was found in pediatric DMGs, suggesting that this imaging modality may offer prognostic and therapeutic insights.97 MRS measures biochemical changes in the brain, and the most commonly used technique is proton (1H; hydrogen) spectroscopy. Cellular metabolites are measured, such as choline (Cho), creatine, lactate, lipids, and myoinositol. The biochemical profiles vary by brain regions and with brain maturation. Proton MRS can help differentiate types of brain tumors, especially those found in the infratentorial compartment, by looking at ratios of N-acetyl aspartate (NAA), creatine, Cho, and lactate.100
In one prospective study of 36 patients with pontine DMGs after radiation therapy, it was found that patients with higher Cho: NAA values were at greater mortality risk. Additionally, an increase in Cho: NAA inversely correlated with survival throughout the disease course.101 Further efforts are needed to determine what metabolic measures on 1H-MRS at diagnosis are the best survival or treatment response predictors.100 Indeed, although the diagnostic value of MRS in differentiating between tumor recurrence or radiation necrosis has been shown in a systematic review of 28 studies covering adult brain tumors, similar work is currently absent for DMGs.102
PET/18F-FDG
PET visualizes metabolic features, which can assist in determining the most active tumor location, thereby guiding a biopsy’s target tissue and discriminating between pseudo-progression and tumor progression. Ideally, PET and MRI scans are performed in a single session on a hybrid PET/MRI system in pediatric patients, minimizing the diagnostic burden for the patient.
18F-fluorodeoxyglucose (18F-FDG) was one of the first radiotracers used for PET imaging. Unfortunately, however, the normal brain has a physiologic intense 18F-FDG uptake, which limits the sensitivity for detecting brain lesions. This contributes to limited 18F-FDG activity in DMG and, subsequently, contradictory results regarding the correlation between PET parameters and survival in this disease.99
In recent years, radiolabeled amino acids such as 11C-methyl-l-methionine (11C-MET), 18F-fluoroethyl-l-tyrosine (18F-FET), and 18F-fluoro-l-dihydroxyphenylalanine (18F-DOPA) have been widely introduced for brain tumor imaging and are some of the most commonly applied radiotracers.103 Importantly, these tracers have a minimal uptake in normal brain compared to 18F-FDG, each capable of crossing the BBB. Their uptake depends on the expression of the amino acid transporter, L-type amino acid transporter 1.99
11C-MET is the oldest amino acid tracer used in brain tumor imaging. Few studies have been performed in DMG patients but have shown that 11C-MET-PET can visualize the tumors in most cases.104,105 Although 11C-MET-PET has proven its clinical value, 18F-FET-PET has logistic advantages over 11C-MET-PET because of its longer half-life, allowing for dynamic scanning with the evaluation of time-activity-curves (TACs) and independence from a cyclotron.
18F-DOPA has been studied in children with DMGs, with 18F-DOPA-PET capable of differentiating between H3K27M-mutant and H3WT DMGs and predicting outcomes.106,107 A recent retrospective study in 15 pediatric brain tumor patients, of which 5 had confirmed H3K27-altered DMG and 2 had a clinical/radiologic diagnosis of pontine DMG, analyzed the added value of dynamic 18F-DOPA-PET and computed tomography parameters.108 In this study, the TAC dynamic PET parameter identified patients at higher risk of disease progression and death.
In adults with gliomas, 18F-FET-PET is widely used, and evidence-based guidelines are described by the EANM/EANO/RANO working group.109 In 2015, Dunkl et al. described the first cohort of pediatric patients with brain tumors who received dynamic 18F-FET-PET for further diagnostic scanning because of complex initial imaging findings on MRI.110 They found that this approach may be helpful for the identification of newly diagnosed brain lesions suggestive of glioma and in the diagnosis of tumor progression or recurrence.
A case report from an adult with an H3K27M-mutant DMG showed high 18F-FET uptake and a short time to peak.111 Specific to the pediatric population, the diagnostic accuracy and clinical impact of 18F-FET have been reported in a prospective study with 169 scans in 97 children and adolescents comprising a variety of high- and low-grade tumors, including six children with H3K27M-mutant DMG.112 Indeed, 18F-FET-PET showed significantly higher accuracy for detecting tumors in both untreated and treated lesions than MRI and altered the treatment plan in 33% of patients with a clinical indication for additional imaging.
Advanced MRI and PET imaging can offer additional value in managing DMG, including use at diagnosis to define the best target for biopsy and assistance in radiation planning. During treatment, it may correlate with survival or treatment response and discriminate between treatment-related effects versus tumor progression. However, a larger, homogenous cohort of DMG patients needs to be studied with sequential scan time points to prove further the additional value of metabolic imaging approaches, such as 18F-FET-PET, and determine how these can be integrated into clinical care. By incorporating 18F-FET-PET imaging into PNOC022, we will be able to address some of these questions and also assess utility within a clinical trial setting for children with DMGs.
Conclusions
DMG remains a deadly disease that deserves thoughtful, multidisciplinary, and multifaceted clinical trial design in the quest to find a cure. PNOC022 provides an adaptive platform that allows near real-time incorporation of new treatment arms based on an evolving understanding of potential disease vulnerabilities. The trial also incorporates collection of novel pharmacokinetic, molecular, genomic, and imaging biomarkers that will inform on tumors’ mechanisms of response and resistance. Each piece of the trial has been developed with expertise from basic scientists to clinical researchers to pharmacologists to imaging specialists. Further, the PNOC mission includes trial access for as many children and young adults as feasible, regardless of geographic location. To that end, PNOC022 is currently open at 31 sites across 5 countries, including several sites in Australia/New Zealand and Israel. Through the design and collaboration of PNOC022, we will greatly augment our understanding of DMG with the ultimate goal of advancing more effective therapy combinations in rapid pace.
Acknowledgments
This article appears as part of the supplement “H3 K27M-mutant Glioma: Disease State Overview,” sponsored by Chimerix, Inc. We thank Truman Knowles for his support in the editing process.
Contributor Information
Sabine Mueller, Department of Neurology, Neurosurgery and Pediatrics, University of California, San Francisco, California, USA.
Cassie Kline, Division of Oncology, Department of Pediatrics, Children’s Hospital of Philadelphia, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Andrea Franson, Department of Pediatrics, University of Michigan, Ann Arbor, Michigan, USA.
Jasper van der Lugt, Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Michael Prados, Department of Neurosurgery and Pediatrics, University of California, San Francisco, San Francisco, California, USA.
Sebastian M Waszak, Department of Neurology, University of California, San Francisco, San Francisco, California, USA; Laboratory of Computational Neuro-Oncology, Swiss Institute for Experimental Cancer Research, School of Life Sciences, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland.
Sabine L A Plasschaert, Princess Máxima Center for Pediatric Oncology, Utrecht, The Netherlands.
Annette M Molinaro, Division of Biomedical Statistics and Informatics, Department of Neurosurgery, University of California, San Francisco, San Francisco, California, USA.
Carl Koschmann, Department of Pediatrics, University of Michigan, Ann Arbor, Michigan, USA.
Javad Nazarian, Research Center for Genetic Medicine, Children’s National Health System, Washington, District of Columbia, USA; Brain Tumor Institute, Children’s National Health System, Washington, District of Columbia, USA; DMG Research Center, Department of Pediatrics, University Children’s Hospital, University of Zurich, Zürich, Switzerland.
Funding
C.K. is supported by NIH/NINDS [grant numbers R01-NS124607 and R01-NS119231] and C.K., S.M.W., S.M., and J.N. by the Department of Defense [grant number CA201129P1]. C.Kl. is supported by a Bristol Myers Squibb Foundation Robert A. Winn Diversity in Clinical Trials Career Development Award. A.F. is supported by the Rogel Cancer Center and Hyundai Hope on Wheels. S.M.W. is supported by the École Polytechnique Fédérale de Lausanne. PNOC022 is supported by the PNOC Foundation, Storm the Heavens, Chad Tough Defeat DIPG. Kazia and Chimerix provide the study drugs free of charge.
Conflict of interest statement
None declared.
Supplement sponsorship
This article appears as part of the supplement “H3 K27M-mutant Glioma: Disease State Overview,” sponsored by Chimerix, Inc.
References
- 1. Jansen MH, Veldhuijzen van Zanten SE, Sanchez Aliaga E, et al. Survival prediction model of children with diffuse intrinsic pontine glioma based on clinical and radiological criteria. Neuro-oncology. 2015;17(1):160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Findlay IJ, De Iuliis GN, Duchatel RJ, et al. Pharmaco-proteogenomic profiling of pediatric diffuse midline glioma to inform future treatment strategies. Oncogene. 2022;41(4):461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warren KE. Diffuse intrinsic pontine glioma: poised for progress. Front Oncol. 2012;2:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 5. Wu G, Broniscer A, McEachron TA, et al.. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buczkowicz P, Hoeman C, Rakopoulos P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet. 2014;46(5):451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol (Berl). 2012;124(3):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bender S, Tang Y, Lindroth AM, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. [DOI] [PubMed] [Google Scholar]
- 9. Pasini D, Malatesta M, Jung HR, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of polycomb group target genes. Nucleic Acids Res. 2010;38(15):4958–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piunti A, Hashizume R, Morgan MA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017;23(4):493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larson JD, Kasper LH, Paugh BS, et al. Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell. 2019;35(1):140–155.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jain SU, Rashoff AQ, Krabbenhoft SD, et al. H3 K27M and EZHIP impede H3K27-methylation spreading by inhibiting allosterically stimulated PRC2. Mol Cell. 2020;80(4):726–735.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krug B, De Jay N, Harutyunyan AS, et al. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M gliomas. Cancer Cell. 2019;35(5):782–797.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Golbourn BJ, Halbert ME, Halligan K, et al. Loss of MAT2A compromises methionine metabolism and represents a vulnerability in H3K27M mutant glioma by modulating the epigenome. Nat Cancer. 2022;3(5):629–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Przystal JM, Cosentino CC, Yadavilli S, et al. Imipridones affect tumor bioenergetics and promote cell lineage differentiation in diffuse midline gliomas. Neuro-Oncol. Published online February 14, 2022;24(9):noac041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cole A, Wang Z, Coyaud E, et al. Inhibition of the mitochondrial protease ClpP as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 2015;27(6):864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conlon BP, Nakayasu ES, Fleck LE, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503(7476):365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Sagarra MR, Mayo I, Marco S, et al. Mitochondrial localization and oligomeric structure of HClpP, the human homologue of E. coli ClpP. J Mol Biol. 1999;292(4):819–825. [DOI] [PubMed] [Google Scholar]
- 19. Graves PR, Aponte-Collazo LJ, Fennell EMJ, et al. Mitochondrial protease ClpP is a target for the anticancer compounds ONC201 and related analogues. ACS Chem Biol. 2019;14(5):1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishizawa J, Zarabi SF, Davis RE, et al. Mitochondrial ClpP-mediated proteolysis induces selective cancer cell lethality. Cancer Cell. 2019;35(5):721–737.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacques S, Van Der Sloot AM, Huard C C, et al. Imipridone anticancer compounds ectopically activate the ClpP Protease and represent a new scaffold for antibiotic development. Genetics. 2020;214(4):1103–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bonner ER, Waszak SM, Grotzer MA, Mueller S, Nazarian J.. Mechanisms of imipridones in targeting mitochondrial metabolism in cancer cells. Neuro-oncology. 2021;23(4):542–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chari A, Barlogie B.. Imipridone ONC201: combination therapy in hematologic malignancies. Cell Cycle. 2018;17(16):1947–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edwards H, Ge Y.. ONC201 shows promise in AML treatment. Cell Cycle. 2018;17(3):277–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lev A, Lulla AR, Ross BC, et al. ONC201 targets AR and AR-V7 signaling, reduces PSA, and synergizes with everolimus in prostate cancer. Mol Cancer Res. 2018;16(5):754–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arrillaga-Romany I, Odia Y, Prabhu VV, et al. Biological activity of weekly ONC201 in adult recurrent glioblastoma patients. Neuro-oncology. 2020;22(1):94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prabhu VV, Morrow S, Rahman Kawakibi A, et al. ONC201 and imipridones: anti-cancer compounds with clinical efficacy. Neoplasia. 2020;22(12):725–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Venneti S, Kawakibi AR, Ji S, et al. Clinical efficacy of ONC201 in H3K27M-mutant diffuse midline gliomas is driven by disruption of integrated metabolic and epigenetic pathways. Cancer Discov. Published online August 16, 2023;13:OF1–OF24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung C, Sweha SR, Pratt D, et al. Integrated metabolic and epigenomic reprograming by H3K27M mutations in diffuse intrinsic pontine gliomas. Cancer Cell. 2020;38(3):334–349.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magaway C, Kim E, Jacinto E.. Targeting mTOR and metabolism in cancer: lessons and innovations. Cells. 2019;8(12):1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Georgescu MM, Islam MZ, Li Y, et al. Global activation of oncogenic pathways underlies therapy resistance in diffuse midline glioma. Acta Neuropathol Commun. 2020;8(1):111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paugh BS, Broniscer A, Qu C, et al. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29(30):3999–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makinoshima H, Takita M, Saruwatari K, et al. Signaling through the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) axis is responsible for aerobic glycolysis mediated by glucose transporter in epidermal growth factor receptor (EGFR)-mutated lung adenocarcinoma. J Biol Chem. 2015;290(28):17495–17504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ribback S, Cigliano A, Kroeger N, et al. PI3K/AKT/mTOR pathway plays a major pathogenetic role in glycogen accumulation and tumor development in renal distal tubules of rats and men. Oncotarget. 2015;6(15):13036–13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X, Lin Y, Kemper T, et al. AMPK and Akt/mTOR signalling pathways participate in glucose-mediated regulation of hepatitis B virus replication and cellular autophagy. Cell Microbiol. 2020;22(2):e13131. [DOI] [PubMed] [Google Scholar]
- 36. Aldonza MBD, Hong JY, Bae SY, et al. Suppression of MAPK signaling and reversal of mTOR-dependent MDR1-associated multidrug resistance by 21α-methylmelianodiol in lung cancer cells. PLoS One. 2015;10(6):e0127841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poore B, Yuan M, Arnold A, et al. Inhibition of mTORC1 in pediatric low-grade glioma depletes glutathione and therapeutically synergizes with carboplatin. Neuro-oncology. 2019;21(2):252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montes DK, Brenet M, Muñoz VC, et al. Vasopressin activates Akt/mTOR pathway in smooth muscle cells cultured in high glucose concentration. Biochem Biophys Res Commun. 2013;441(4):923–928. [DOI] [PubMed] [Google Scholar]
- 39. Heffron TP, Ndubaku CO, Salphati L, et al. Discovery of clinical development candidate GDC-0084, a brain penetrant inhibitor of PI3K and mTOR. ACS Med Chem Lett. 2016;7(4):351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He C, Xu K, Zhu X, et al. Patient-derived models recapitulate heterogeneity of molecular signatures and drug response in pediatric high-grade glioma. Nat Commun. 2021;12(1):4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wen PY, de Groot JF, Battiste J, et al. Paxalisib in patients with newly diagnosed glioblastoma with unmethylated MGMT promoter status: final phase 2 study results. J Clin Oncol. 2022;40(16_suppl):2047–2047. [Google Scholar]
- 42. Wen PY, de Groot J, Battiste JD, et al. Abstract LB125: pharmacokinetics of paxalisib in phase 2 clinical study in glioblastoma (GBM) with unmethylated O6-methylguanine-methyltransferase (MGMT) promotor status. Cancer Res. 2021;81(13_Supplement):LB125–LB125. [Google Scholar]
- 43. Jackson ER, Duchatel RJ, Staudt DE, et al. ONC201 in combination with paxalisib for the treatment of H3K27-altered diffuse midline glioma. Cancer Res. Published online May 5, 2023;83(14):2421–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang R, Shen J, Yuan Y.. ComPAS. A Bayesian drug combination platform trial design with adaptive shrinkage. Stat Med. 2019;38(7):1120–1134. [DOI] [PubMed] [Google Scholar]
- 45. Gardner SL, Allen JC, Zaky WT, et al. ONC201 in previously-irradiated pediatric H3 K27M-mutant glioma. J Clin Oncol. 2019;37(15_suppl):10046–10046. [Google Scholar]
- 46. Lassaletta A, Strother D, Laperriere N, et al. Reirradiation in patients with diffuse intrinsic pontine gliomas: the Canadian experience. Pediatr Blood Cancer. 2018;65(6):e26988. [DOI] [PubMed] [Google Scholar]
- 47. Morales La Madrid A, Santa-María V, Cruz Martinez O, et al. Second re-irradiation for DIPG progression, re-considering “old strategies” with new approaches. Childs Nerv Syst. 2017;33(5):849–852. [DOI] [PubMed] [Google Scholar]
- 48. Tsang DS, Oliveira C, Bouffet E, et al. Repeat irradiation for children with supratentorial high-grade glioma. Pediatr Blood Cancer. 2019;66(9):e27881. [DOI] [PubMed] [Google Scholar]
- 49. Kline C, Liu SJ, Duriseti S, et al. Reirradiation and PD-1 inhibition with nivolumab for the treatment of recurrent diffuse intrinsic pontine glioma: a single-institution experience. J Neurooncol. 2018;140(3):629–638. [DOI] [PubMed] [Google Scholar]
- 50. Müller K, Scheithauer H, Pietschmann S, et al. Reirradiation as part of a salvage treatment approach for progressive non-pontine pediatric high-grade gliomas: preliminary experiences from the German HIT-HGG study group. Radiat Oncol. 2014;9(1):177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Freese C, Takiar V, Fouladi M, et al. Radiation and subsequent reirradiation outcomes in the treatment of diffuse intrinsic pontine glioma and a systematic review of the reirradiation literature. Pract Radiat Oncol. 2017;7(2):86–92. [DOI] [PubMed] [Google Scholar]
- 52. Wang Y, Springer S, Zhang M, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci USA. 2015;112(31):9704–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bruzek AK, Ravi K, Muruganand A, et al. Electronic DNA analysis of CSF cell-free tumor DNA to quantify multi-gene molecular response in pediatric high-grade glioma. Clin Cancer Res. 2020;26(23):6266–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu APY, Smith KS, Kumar R, et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39(11):1519–1530.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang TY, Piunti A, Lulla RR, et al. Detection of histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol Commun. 2017;5(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Izquierdo E, Proszek P, Pericoli G, et al. Droplet digital PCR-based detection of circulating tumor DNA from pediatric high grade and diffuse midline glioma patients. Neuro-Oncol Adv. 2021;3(1):vdab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stallard S, Savelieff MG, Wierzbicki K, et al. CSF H3F3A K27M circulating tumor DNA copy number quantifies tumor growth and in vitro treatment response. Acta Neuropathol Commun. 2018;6(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Panditharatna E, Kilburn LB, Aboian MS, et al. Clinically relevant and minimally invasive tumor surveillance of pediatric diffuse midline gliomas using patient-derived liquid biopsy. Clin Cancer Res. 2018;24(23):5850–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li D, Bonner ER, Wierzbicki K, et al. Standardization of the liquid biopsy for pediatric diffuse midline glioma using ddPCR. Sci Rep. 2021;11(1):5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bonner ER, Harrington R, Eze A, et al. Circulating tumor DNA sequencing provides comprehensive mutation profiling for pediatric central nervous system tumors. npj Precis Oncol. 2022;6(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mueller S, Jain P, Liang WS, et al. A pilot precision medicine trial for children with diffuse intrinsic pontine glioma—PNOC003: a report from the pacific pediatric neuro-oncology consortium. Int J Cancer. Published online April 3, 2019;145(7):1889–1901. [DOI] [PubMed] [Google Scholar]
- 63. Nikbakht H, Panditharatna E, Mikael LG, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun. 2016;7(1):11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cantor E, Wierzbicki K, Tarapore RS, et al. Serial H3K27M cell-free tumor DNA (cf-tDNA) tracking predicts ONC201 treatment response and progression in diffuse midline glioma. Neuro-oncology. 2022;24(8):1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sepich-Poore GD, Zitvogel L, Straussman R, et al. The microbiome and human cancer. Science. 2021;371(6536):eabc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li J, Zhao S, Lee M, et al. Reliable tumor detection by whole-genome methylation sequencing of cell-free DNA in cerebrospinal fluid of pediatric medulloblastoma. Sci Adv. 2020;6(42):eabb5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Welton JL, Loveless S, Stone T, et al. Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J Extracell Vesicles. 2017;6(1):1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Peng Y, Zheng D, Zhang X, et al. Cell-free mitochondrial DNA in the CSF: a potential prognostic biomarker of anti-NMDAR encephalitis. Front Immunol. 2019;10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liang J, Li T, Zhao J, Wang C, Sun H.. Current understanding of the human microbiome in glioma. Front Oncol. 2022;12:781741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. De Cecco L, Biassoni V, Schiavello E, et al. DIPG-36. The brain–gut–microbiota axis to predict outcome in pediatric diffuse intrinsic pontine glioma. Neuro-Oncology. 2022;24(Supplement_1):i26–i26. [Google Scholar]
- 72. Kline C, Jain P, Kilburn L, et al. Upfront biology-guided therapy in diffuse intrinsic pontine glioma: therapeutic, molecular, and biomarker outcomes from PNOC003. Clin Cancer Res. 2022;28(18):3965–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fennell EMJ, Aponte-Collazo LJ, Pathmasiri W, et al. Multi-omics analyses reveal ClpP activators disrupt essential mitochondrial pathways in triple-negative breast cancer. Front Pharmacol. 2023;14:1136317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ishida CT, Zhang Y, Bianchetti E, et al. Metabolic reprogramming by Dual AKT/ERK Inhibition through imipridones elicits unique vulnerabilities in glioblastoma. Clin Cancer Res 2018;24(21):5392–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Greer YE, Porat-Shliom N, Nagashima K, et al. ONC201 kills breast cancer cells in vitro by targeting mitochondria. Oncotarget. 2018;9(26):18454–18479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yuan Y, Ju YS, Kim Y, et al. ; PCAWG Consortium. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet. 2020;52(3):342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. He Y, Li J, Koga T, et al. Epidermal growth factor receptor as a molecular determinant of glioblastoma response to dopamine receptor D2 inhibitors. Neuro-oncology. 2021;23(3):400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Morrow S, Nath K, Zhang Y, et al. Abstract 393: predictive biomarker evaluation and molecular differentiation for imipridones ONC201 and ONC206. Cancer Res. 2021;81(13_Supplement):393–393. [Google Scholar]
- 79. Kawakibi AR, Tarapore R, Gardner S, et al. CTNI-61. Clinical efficacy and predictive biomarkers of ONC201 in H3K27M-mutant diffuse midline glioma. Neuro-Oncology. 2022;24(Supplement_7):vii86–vii87. [Google Scholar]
- 80. Liu I, Jiang L, Samuelsson ER, et al. The landscape of tumor cell states and spatial organization in H3-K27M mutant diffuse midline glioma across age and location. Nat Genet. 2022;54(12):1881–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huang W, Bhaduri A, Velmeshev D, et al. Origins and proliferative states of human oligodendrocyte precursor cells. Cell. 2020;182(3):594–608.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Trevino AE, Müller F, Andersen J, et al. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell. 2021;184(19):5053–5069.e23. [DOI] [PubMed] [Google Scholar]
- 83. Andersen J, Thom N, Shadrach JL, et al. Single-cell transcriptomic landscape of the developing human spinal cord. Nat Neurosci. 2023;26(5):902–914. [DOI] [PubMed] [Google Scholar]
- 84. Filbin MG, Tirosh I, Hovestadt V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science. 2018;360(6386):331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jessa S, Blanchet-Cohen A, Krug B, et al. Stalled developmental programs at the root of pediatric brain tumors. Nat Genet. 2019;51(12):1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Thorlund K, Dron L, Park JJ, Mills EJ.. Synthetic and external controls in clinical Trials – A primer for researchers. Clin Epidemiol. 2020;Volume 12:457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Coens C, Pe M, Dueck AC, et al. ; Setting International Standards in Analyzing Patient-Reported Outcomes and Quality of Life Endpoints Data Consortium. International standards for the analysis of quality-of-life and patient-reported outcome endpoints in cancer randomised controlled trials: recommendations of the SISAQOL Consortium. Lancet Oncol. 2020;21(2):e83–e96. [DOI] [PubMed] [Google Scholar]
- 88. Basch E. Missing patients’ symptoms in cancer care delivery—the importance of patient-reported outcomes. JAMA Oncol. 2016;2(4):433–434. [DOI] [PubMed] [Google Scholar]
- 89. Leahy AB, Steineck A.. Patient-reported outcomes in pediatric oncology: the patient voice as a gold standard. JAMA Pediatr. 2020;174(11):e202868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Reeve BB, Withycombe JS, Baker JN, et al. The first step to integrating the child’s voice in adverse event reporting in oncology trials: a content validation study among pediatric oncology clinicians. Pediatr Blood Cancer. 2013;60(7):1231–1236. [DOI] [PubMed] [Google Scholar]
- 91. Leahy AB, Feudtner C, Basch E.. Symptom monitoring in pediatric oncology using patient-reported outcomes: why, how, and where next. The Patient. 2018;11(2):147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dobrozsi S, Yan K, Hoffmann R, Panepinto J.. Patient-reported health status during pediatric cancer treatment. Pediatr Blood Cancer. 2017;64(4):e26295. [DOI] [PubMed] [Google Scholar]
- 93. Roth ME, Parsons SK, Ganz PA, et al. Inclusion of a core patient-reported outcomes battery in adolescent and young adult cancer clinical trials. J Natl Cancer Inst. 2023;115(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cheng JD, Hitt J, Koczwara B, et al. Impact of quality of life on patient expectations regarding phase I clinical trials. J. Clin. Oncol 2000;18(2):421–428. [DOI] [PubMed] [Google Scholar]
- 95. DasMahapatra P, Raja P, Gilbert J, Wicks P.. Clinical trials from the patient perspective: survey in an online patient community. BMC Health Serv Res. 2017;17(1):166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cooney TM, Cohen KJ, Guimaraes CV, et al. Response assessment in diffuse intrinsic pontine glioma: recommendations from the response assessment in pediatric neuro-oncology (RAPNO) working group. Lancet Oncol. 2020;21(6):e330–e336. [DOI] [PubMed] [Google Scholar]
- 97. Calmon R, Puget S, Varlet P, et al. Cerebral blood flow changes after radiation therapy identifies pseudoprogression in diffuse intrinsic pontine gliomas. Neuro-oncology. 2018;20(7):994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Carceller F, Fowkes LA, Khabra K, et al. Pseudoprogression in children, adolescents and young adults with non-brainstem high grade glioma and diffuse intrinsic pontine glioma. J Neurooncol. 2016;129(1):109–121. [DOI] [PubMed] [Google Scholar]
- 99. Lovibond S, Gewirtz AN, Pasquini L, Krebs S, Graham MS.. The promise of metabolic imaging in diffuse midline glioma. Neoplasia. 2023;39:100896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Blüml S, Saunders A, Tamrazi B.. Proton MR spectroscopy of pediatric brain disorders. Diagnostics (Basel, Switzerland). 2022;12(6):1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Steffen-Smith EA, Shih JH, Hipp SJ, Bent R, Warren KE.. Proton magnetic resonance spectroscopy predicts survival in children with diffuse intrinsic pontine glioma. J Neurooncol. 2011;105(2):365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Smith EJ, Naik A, Shaffer A, et al. Differentiating radiation necrosis from tumor recurrence: a systematic review and diagnostic meta-analysis comparing imaging modalities. J Neurooncol. 2023;162(1):15–23. [DOI] [PubMed] [Google Scholar]
- 103. Verger A, Kas A, Darcourt J, Guedj E.. PET Imaging in neuro-oncology: an update and overview of a rapidly growing area. Cancers. 2022;14(5):1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Rosenfeld A, Etzl M, Bandy D, et al. Use of positron emission tomography in the evaluation of diffuse intrinsic brainstem gliomas in children. J Pediatr Hematol Oncol. 2011;33(5):369–373. [DOI] [PubMed] [Google Scholar]
- 105. Tinkle CL, Duncan EC, Doubrovin M, et al. Evaluation of 11 C-Methionine PET and anatomic MRI associations in diffuse intrinsic pontine glioma. J Nucl Med. 2019;60(3):312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Piccardo A, Tortora D, Mascelli S, et al. Advanced MR imaging and 18F-DOPA PET characteristics of H3K27M-mutant and wild-type pediatric diffuse midline gliomas. Eur J Nucl Med Mol Imaging. 2019;46(8):1685–1694. [DOI] [PubMed] [Google Scholar]
- 107. Morana G, Tortora D, Bottoni G, et al. Correlation of multimodal 18 F-DOPA PET and conventional MRI with treatment response and survival in children with diffuse intrinsic pontine gliomas. Theranostics. 2020;10(26):11881–11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fiz F, Bini F, Gabriele E, et al. Role of dynamic parameters of 18F-DOPA PET/CT in pediatric gliomas. Clin Nucl Med. 2022;47(6):517–524. [DOI] [PubMed] [Google Scholar]
- 109. Albert NL, Weller M, Suchorska B, et al. Response assessment in neuro-oncology working group and European association for neuro-oncology recommendations for the clinical use of PET imaging in gliomas. Neuro-oncology. 2016;18(9):1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Dunkl V, Cleff C, Stoffels G, et al. The usefulness of dynamic O-(2-18F-fluoroethyl)-l-tyrosine PET in the clinical evaluation of brain tumors in children and adolescents. J Nucl Med. 2015;56(1):88–92. [DOI] [PubMed] [Google Scholar]
- 111. Vettermann FJ, Unterrainer M, Ruf V, et al. Dual PET imaging of an H3K27M-mutant glioma with 18F-GE-180 and 18F-FET PET. Clin Nucl Med. 2020;45(12):992–993. [DOI] [PubMed] [Google Scholar]
- 112. Marner L, Lundemann M, Sehested A, et al. Diagnostic accuracy and clinical impact of [18F]FET PET in childhood CNS tumors. Neuro-oncology. 2021;23(12):2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]