Significance
Congenital scoliosis (CS) is the most common congenital spinal disorder caused by congenital vertebral malformations (CVMs), influenced by genetic and environmental factors and exhibiting diverse clinical presentations. Here, we identified the critical roles of Vangl1 and Vangl2, two core components in the Wnt/planar cell polarity (Wnt/PCP) signaling pathway, in vertebral development and in predisposition to CVMs in CS patients. We found that in Vangl mutant mouse models, the CVMs present in a Vangl gene dose- and gestational hypoxia-dependent manner. Our studies reveal a complex etiology of CS and its association with Wnt/PCP signaling.
Keywords: congenital vertebral malformation, congenital scoliosis, somite, VANGL1/2, planar cell polarity (PCP)
Abstract
Congenital scoliosis (CS), affecting approximately 0.5 to 1 in 1,000 live births, is commonly caused by congenital vertebral malformations (CVMs) arising from aberrant somitogenesis or somite differentiation. While Wnt/ß-catenin signaling has been implicated in somite development, the function of Wnt/planar cell polarity (Wnt/PCP) signaling in this process remains unclear. Here, we investigated the role of Vangl1 and Vangl2 in vertebral development and found that their deletion causes vertebral anomalies resembling human CVMs. Analysis of exome sequencing data from multiethnic CS patients revealed a number of rare and deleterious variants in VANGL1 and VANGL2, many of which exhibited loss-of-function and dominant-negative effects. Zebrafish models confirmed the pathogenicity of these variants. Furthermore, we found that Vangl1 knock-in (p.R258H) mice exhibited vertebral malformations in a Vangl gene dose- and environment-dependent manner. Our findings highlight critical roles for PCP signaling in vertebral development and predisposition to CVMs in CS patients, providing insights into the molecular mechanisms underlying this disorder.
Vertebrae formation depends on somitogenesis and somite differentiation, and the failure of these developmental processes gives rise to congenital vertebral malformations (CVMs), which in turn can clinically manifest as congenital scoliosis (CS), congenital kyphosis, or kyphoscoliosis (1). During somitogenesis, the periodic formation and segmentation of symmetrical somites from the presomitic mesoderm (PSM) rely on the clock and wavefront signaling system regulated by the Wnt/ß-catenin, Notch, and fibroblast growth factor (Fgf) signaling pathways (2). Perturbations of these signaling pathways can disrupt somite development in both mice and humans, leading to CVMs (1). For example, knockout of Fgf4 leads to a range of vertebral defects and Wnt3a mutant mice exhibit axial truncation due to impaired Fgf and Wnt/ß-catenin signaling (3, 4), respectively. Wnt5a mutant mice also exhibit truncation of the posterior body axis (5). In humans, mutations in WNT5A, its receptor ROR2, or other core components of the Wnt signaling pathway (e.g., DVL1 and DVL3) cause Robinow syndrome, a disease characterized by multiple bone abnormalities. Some patients also have genital abnormalities, abnormal facial features, and spinal defects (6–10). It is well known that Wnt5a can elicit signaling responses to activate or inhibit canonical Wnt/ß-catenin signaling in a context-dependent manner (11) or a ß-catenin-independent manner to regulate polarized cell behaviors through the planar cell polarity (PCP) signaling pathway (12, 13). However, whether PCP signaling is directly involved in somitogenesis or somite differentiation and associated with CVMs remains unclear.
PCP refers to the coordinated orientation and alignment of cells within the plane of a tissue and is established by the asymmetrical subcellular localization of six core components, Frizzled (Fz), Van Gogh (Vang), Flamingo (Fmi), Dishevelled (Dsh), Prickle (Pk), and Diego (Dgo) (14). In vertebrates, PCP controls many fundamental cellular and developmental processes, such as collective cell movement and convergent extension (CE), which coordinate the elongation and narrowing of the anterior–posterior (A–P) body axis (15). Disruption of PCP signaling has been associated with severe defects in humans and mice, the most common of which are neural tube defects (NTDs) ranging from spina bifida in humans to craniorachischisis in mice (14, 16). Although skeletal defects have been observed in PCP mutant mice (e.g., Prickle1, Dvl2, Fat4, and Dchs1 mutants) (17–20), there is no clear evidence that the disturbance of PCP components is involved in human vertebral malformations.
Vangl1 and Vangl2, the vertebrate homologs of Drosophila Vang, are two highly conserved four-pass transmembrane core proteins that are mainly dedicated to PCP signaling and required for various embryonic developmental processes (21–23). Vangl2 Loop-tail (Lp) mutant mice showed skeletal anomalies, including shortened limbs and misshapen and fused vertebrae and ribs (22, 24, 25). In humans, VANGL1 and VANGL2 mutations have been reported in association with NTDs (26, 27). Interestingly, CVMs are concomitant with NTDs in a significant fraction of patients (28, 29). However, the mechanism underlying their clinical association is unknown, and PCP signaling has not been implicated in the pathogenesis of CVMs.
Here, we found that Vangl1- and Vangl2-deficient mice exhibited severe CVMs in a dose-dependent manner as a result of defects in somite development, resembling the spinal anomalies of CS patients. We subsequently analyzed a multicenter and multiethnic cohort of CS patients and identified many rare and deleterious VANGL1 and VANGL2 variants. The variants’ pathogenicity was confirmed by in vitro and in vivo functional analyses in cultured cells, zebrafish, and genetically modified mouse models. We found that both polygenic and environmental factors significantly enhanced the penetrance of CVMs in Vangl1 mutant mice. Collectively, our findings revealed a critical role for PCP signaling in vertebral development and genetic susceptibility to the development of CVMs in CS patients.
Results
Somite Development Defects and Vertebral Malformations Caused by Loss of Vangl1 and Vangl2.
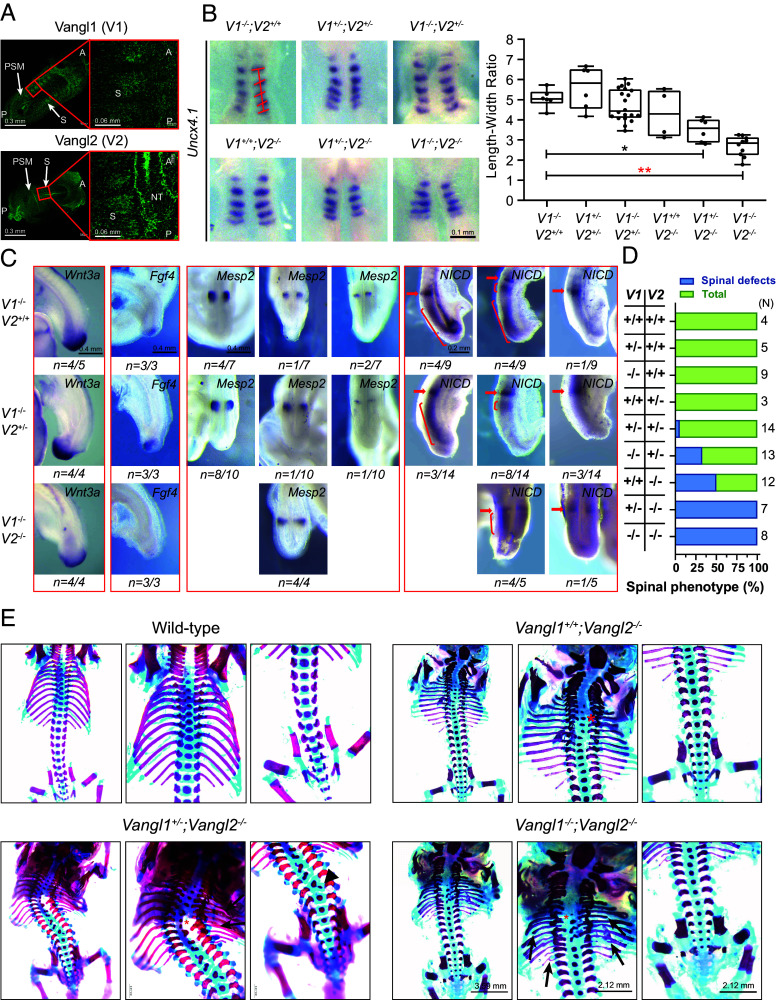
Although the function of Vangl1 and Vangl2 in NT development has been well studied, their roles in somite and vertebral development remain unclear. Here, we first examined the expression patterns of Vangl1 and Vangl2 in wild-type embryos at embryonic stage E8.5 by whole-mount immunofluorescence staining. We found that Vangl1 and Vangl2 are both ubiquitously expressed in PSM, somite, and NT with Vangl2 being enriched in NT (Fig. 1A), suggesting a potential role for Vangl genes in somitogenesis and/or somite differentiation. To investigate somitogenesis in Vangl1 and Vangl2 mutants, we performed whole-mount in situ hybridization using a variety of somitogenesis markers. The staining of caudal somite marker Uncx4.1 showed significantly shortened and widened somite-forming regions in Vangl1−/−;Vangl2−/− double null embryos at E8.5 (Fig. 1B). Because Vangl1−/− mutant mice are morphologically normal (21, 22) and have no defects in somite formation (Fig. 1B), they were used as control embryos in subsequent analyses. Wnt3a, Fgf4, and Fgf8 that are expressed in the PSM and tailbud are required for paraxial mesoderm development and somitogenesis. Fgf4 and Fgf8 together are essential for the proper expressions of segmentation clock genes required for somite formation (3, 30–32). Fgf4 mutant mice have multiple vertebral defects resembling CVMs (4). Notably, the expression levels of Wnt3a, Fgf4, and Fgf8 were markedly reduced in the Vangl1/2 double homozygous null mutant (Fig. 1C and SI Appendix, Fig. S1). Mesp2 is a Notch signaling downstream target that regulates the segmentation of paraxial mesoderm by defining the future somite boundary position (33). Importantly, Mesp2 is dynamically expressed and serves as a cyclic factor regulating the oscillation of Notch activity, thereby initiating a segmentation program in the anterior PSM (34). Mesp2 showed a clear oscillatory pattern in control embryos, whereas its dynamic expression is significantly disturbed in the Vangl1/2 double mutant, where only one pattern was exhibited (Fig. 1C). Formation of Notch1 intracellular domain (NICD) is a key marker for Notch signaling pathway activation and Hes7 and Lfng are critical oscillating Notch pathway genes that control somitogenesis (4, 35). We found that the oscillating patterns of NICD were remarkably compromised, with most Vangl1−/− embryos displaying both a sharp and long band in the PSM, while in contrast, a sharp and shorter band was dominantly detected with the loss of both Vangl1 and Vangl2 (Fig. 1C). The oscillating patterns of Hes7 and Lfng remained largely unaffected in Vangl mutants, but their expressions were more restricted to the caudal regions (SI Appendix, Fig. S1). The expression regions of Notch ligands Dll1 and Dll3 were also more limited with the loss of Vangl1 and Vangl2 (SI Appendix, Fig. S1). Moreover, we examined the expressions of T-box transcription factor T (Tbxt, also called brachyury or T) and T-box transcription factor 6 (Tbx6), which are expressed in the primitive streak, tail bud, and PSM and essential for the specification of the posterior paraxial mesoderm (36, 37). We found that the expression of Tbx6 was also restricted to the caudal region in Vangl1−/−;Vangl2−/− double-mutant embryos (SI Appendix, Fig. S1). We speculated that the expression pattern changes of somitogenesis genes may result from an alteration in PSM morphology due to loss of PCP. Therefore, we measured PSM length and width at the 23 to 25 somite stage in E9.5 embryos. Interestingly, there is no significant difference in PSM length among the different genotypes, whereas the width of the PSM in Vangl1/2 double null embryos is significantly wider (SI Appendix, Fig. S2). However, this is largely because the NT is not closed rather than a wider PSM in the mutants. Together, these results demonstrate an important role of Vangl genes in the formation of mouse somites. Although the underlying mechanism is unclear, Vangl deficiency has a significant impact on key somitogenesis gene expression and the oscillating patterns of Notch signaling, leading to aberrant somite development.
Fig. 1.
Somite development defects and vertebral malformations in Vangl1 and Vangl2 mutant mouse embryos. (A) Vangl1 and Vangl2 expression in the PSM, NT, and somites (S) of E8.5 wild-type embryos detected by immunofluorescence of endogenous Vangl proteins. A, anterior; P, posterior. (B) Whole-mount in situ hybridizations of Uncx4.1 in 5 to 6 somites stage of mouse embryos. ImageJ quantified the width and length of somites. The width was calculated on the basis of the average of each somite and the length was determined by the Uncx4.1 positive region along the A–P body axis. The significance of the differences between the groups was calculated by a one-way ANOVA test, F = 16.18, *P = 0.0038, **P < 0.0001. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values. (C) Whole-mount in situ hybridizations of Wnt3a, Fgf4, and Mesp2 and whole-mount immunohistochemistry of NICD in 23 to 25 somite stage of Vangl mutant mice at E9.5. The red arrow and bracket denote a variable cyclic pattern. (D) Penetrance of spinal phenotypes (rib and vertebrae) of Vangl mutant mice. The number of analyzed animals for each genotype is listed on the right side of the chart. (E) Spinal phenotypes of E17.5 to E18.5 Vangl1 and Vangl2 mutant mice. Red asterisks indicate vertebral fusion, vertebral misalignment, and delayed vertebral ossification. Arrowheads point to hemivertebrae, and arrows point to rib fusion, rib shortening/missing, and rib bifurcation. V1 denotes Vangl1, and V2 represents Vangl2.
Because Notch signaling dysregulation and defects in somitogenesis are strongly associated with vertebral malformations, we analyzed the axial skeletons of Vangl mutants at E18.5. The skeletons exhibited phenotypes including hemivertebrae, vertebral fusion, vertebral misalignment, delayed vertebral ossification, rib fusion, rib shortening/missing, and rib bifurcation. The severity and penetrance of the anomalies increased with the reduction in dosage of the Vangl genes (Fig. 1 D and E and SI Appendix, Fig. S3). The mutant mice exhibited remarkable curvature of the spine in a pattern reminiscent of CS patients. These findings indicate that Vangl genes are required for the normal development of the axial skeleton and may be candidates underlying CS susceptibility.
Identification of Deleterious VANGL1 and VANGL2 Variants in CS Patients.
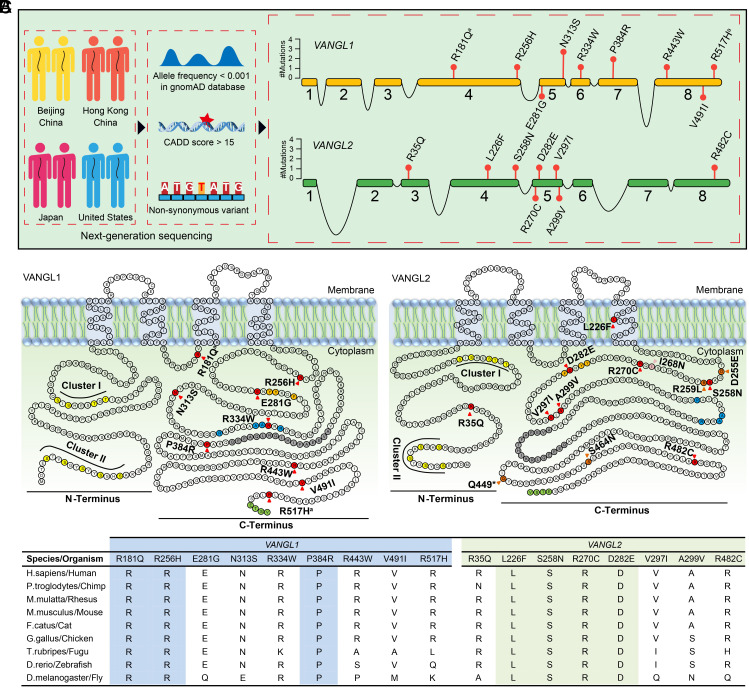
Because Vangl mutant mice displayed CVMs, we analyzed VANGL1 and VANGL2 rare variant alleles in the multicenter and multiethnic sequencing data of 782 subjects with CS (Fig. 2A). Nine VANGL1 and eight VANGL2 rare nonsynonymous missense variants were identified in 19 patients (Table 1 and Fig. 2 A and B). One VANGL1 variant (p.E281G) and two VANGL2 variants (p.L226F and p.A299V) were absent from public databases, including Exome Aggregation Consortium, Genome Aggregation Database (gnomAD), and approximately 8,000 in-house sequenced whole-exome and whole-genome datasets (Table 1). Among the above variants, the VANGL1 p.E281G and VANGL2 p.L226F were deemed likely to be gene-disrupting by Polyphen2, Sorting Intolerant from Tolerant (SIFT), Rare Exome Variant Ensemble Learner (REVEL), and Combined Annotation Dependent Depletion (CADD) prediction tools. One VANGL2 variant (p.R270C) and three VANGL1 variants (p.R181Q, p.R256H, and p.R443W) were predicted to be highly damaging (REVEL > 0.8 and CADD > 20), and their encoded amino acids were highly conserved (Fig. 2C). Intriguingly, the VANGL1 variants (p.R181Q and p.R256H) were identified on the same allele in the same patient and were not detected in their healthy mother (SI Appendix, Fig. S4). Unfortunately, we could not validate whether the two variants (p.R181Q and p.R256H) were de novo because the patient’s biological father had passed away. Additionally, three unrelated subjects shared the same missense VANGL1 variant (p.N313S) with extremely low frequency in gnomAD (0.008% in the general population vs. 0.384% in the cohort), indicating an enrichment in our cohort. The VANGL1 variant (p.P384R), predicted to be damaging and highly conserved, recurrent in two unrelated subjects. The rest of the VANGL1 variants (p.R334W, p.V491I, and p.R517H) and VANGL2 variants (p.R35Q, p.S258N, p.D282E, p.V297I, and p.R482C) were also rare in the gnomAD database and predicted to be deleterious by at least one of the four prediction methods (Table 1).
Fig. 2.
Identification of VANGL1 and VANGL2 variants in CS patients. (A) Brief workflow of the identification of VANGL1 and VANGL2 variants in a multicenter CS cohort (Left) and the location of the identified variants (Right). The numbered orange and green boxes indicate the coding exons of VANGL1 and VANGL2. (B) Amino acid residues of VANGL1 and VANGL2 defining sequence landmarks and signature motifs are depicted in different colors, including rare variants identified in our multicenter cohort (red), looptail variants (dark orange), Curly Bob variant (pink), two highly conserved phosphorylation clusters (yellow, cluster I and cluster II), trans-Golgi network sorting motif (orange), p97/VCP-interacting motif (blue), coiled-coil domain (gray), and PDZ-binding motif (light green). aThe same variant was found in reported NTDs patients. (C) Conservation and multiple alignments of VANGL1 and VANGL2 variants. Highlighted variants are conserved among all listed species.
Table 1.
Genotype and mutational features of subjects with VANGL1 and VANGL2 variants
| In vitro functional test | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Ethnicity | Gene | Nucleotide change | Protein change | gnomAD Exome ALL | gnomAD Exome EAS | SIFT | PolyPhen-2 | REVEL | CADD | Gerp | WB | IF (vs. WT) | DN (VANGL2) |
| HK0001* | Southern Han | VANGL1 | c. 542G>A +c.767G>A | p.Arg181Gln (p.R181Q) | 0.00001663 | 0 | D | D | 0.795 | 28 | 5.52 | * | ** | ** |
| +p.Arg256His (p.R256H) | 0.0000183 | 0 | D | D | 0.875 | 32 | 5.73 | |||||||
| SCO2003P0130 | Northern Han | VANGL1 | c.842A>G | p.Glu281Gly (p.E281G) | 0 | 0 | D | D | 0.94 | 29.4 | 5.66 | * | ** | ** |
| SCO2003P0531 | Northern Han | VANGL1 | c.938A>G | p.Asn313Ser (p.N313S) | 0.00008226 | 0 | T | B | 0.214 | 16.5 | 3.35 | ns | * | * |
| SCO2003P1220 | ||||||||||||||
| SCO2003P1319 | ||||||||||||||
| SCO2003P1232 | Southern Han | VANGL1 | c.1000C>T | p.Arg334Trp (p.R334W) | 0.000009142 | 0 | D | B | 0.558 | 23 | 3.64 | ns | * | * |
| SCO1906P0098 | Northern Han | VANGL1 | c.1151C>G | p.Pro384Arg (p.P384R) | 0.00001829 | 0 | D | D | 0.797 | 28 | 5.44 | ns | * | * |
| SCO2003P2246 | ||||||||||||||
| SCO1908P0074 | Northern Han | VANGL1 | c.1327C>T | p.Arg443Trp (p.R443W) | 0.00004989 | 0.0001004 | D | D | 0.609 | 31 | 4.95 | * | ** | * |
| SCO2003P1178 | Northern Han | VANGL1 | c.1471G>A | p.Val491Ile (p.V491I) | 0.00003326 | 0 | T | B | 0.211 | 19 | 3.41 | ns | * | * |
| SCO2003P0196 | Northern Han | VANGL1 | c.1550G>A | p.Arg517His (p.R517H) | 0.00006399 | 0.0001105 | T | D | 0.467 | 25.3 | 5.4 | ns | * | * |
| SCO1908P0163 | Southern Han | VANGL2 | c.104G>A | p.Arg35Gln (p.R35Q) | 0.000009141 | 0 | T | D | 0.371 | 24.6 | 4.65 | ns | * | * |
| SCO2003P0613 | Southern Han | VANGL2 | c.676C>T | p.Leu226Phe (p.L226F) | 0 | 0 | D | D | 0.781 | 24 | 1.27 | * | ** | ** |
| US0001† | Turkish | VANGL2 | c.773G>A | p.Ser258Asn (p.S258N) | 0.00002022 | 0 | T | B | 0.467 | 24 | 5.26 | ns | * | * |
| US0002‡ | Hispanic | VANGL2 | c.808C>T | p.Arg270Cys (p.R270C) | 0.00006399 | 0 | D | D | 0.889 | 29.5 | 3.99 | * | * | * |
| JP0001 | Japanese | VANGL2 | c.846C>A | p.Asp282Glu (p.D282E) | 0.00000914 | 0 | T | D | 0.597 | 22.7 | 2.99 | * | * | * |
| SCO2003P0386 | Northern Han | VANGL2 | c.889G>A | p.Val297Ile (p.V297I) | 0.00004989 | 0 | T | B | 0.255 | 17.3 | 3.99 | ns | ** | * |
| SCO2003P0525 | Northern Han | VANGL2 | c.896C>T | p.Ala299Val (p.A299V) | 0 | 0 | T | B | 0.419 | 23.1 | 4.91 | ns | * | * |
| SCO2105P3029 | Northern Han | VANGL2 | c.1444C>T | p.Arg482Cys (p.R482C) | 0.00008227 | 0 | T | D | 0.4 | 25.4 | 4.17 | ns | * | * |
Transcripts: NM_138959.2 (VANGL1) and NM_020335.2 (VANGL2). Abbreviations: CADD, Combined Annotation Dependent Depletion; Gerp, genomic evolutionary rate profiling; WB, western blot; IF, immunofluorescence; DN, dominant negative effect on wild-type VANGL2; NA, not available; T, tolerance; B, benign; D, deleterious; ns, not significant; *P < 0.05; **P < 0.05; and significant decrease in membrane to total ratio of VANGL1 and VANGL2 in IF, calculated by the one-way ANOVA test with mean difference greater than 0.2.
*Both variants were confirmed on the same allele, and the inheritance was unknown because the father had passed away.
†The patient was diagnosed with hemifacial microsomia, Goldenhar syndrome (also known as oculo-auriculo-vertebral spectrum disorder) with hemivertebrae.
‡The patient was diagnosed with VACTERL (Vertebral abnormalities, anal atresia, cardiac defects, tracheal anomalies, esophageal atresia, renal and radial abnormalities, and limb abnormalities) syndrome.
Except for VANGL1 p.R181Q and VANGL2 p.R35Q and p.L226F, all the variants were located in the carboxyl-terminal cytosolic region, which contains many critical functional domains and motifs (23) (Fig. 2B). Notably, VANGL1 p.E281G and VANGL2 p.D282E were adjacent to the trans-Golgi network (TGN) sorting motif (YYXXF) required for VANGL trafficking from the Golgi to the cell surface (38). Two VANGL2 variants (p.S258N and p.R270C) were in close proximity to the pathogenic mouse mutations Lp p.R259L and p.D255E and Curly Bob p.I268N, respectively (39–41) (Fig. 2B).
All probands carrying rare VANGL variants were affected by various severities of spinal deformities, ranging from one vertebral malformation (single-segmented hemivertebrae) to extensive fusion of vertebrae, with maximum Cobb angles from 21.7 to 135 degrees (SI Appendix, Fig. S5A and Table S1). Six patients had accompanying rib anomalies. Interestingly, compared to the cohort without rare VANGL variants, CS patients with rare VANGL1 or VANGL2 variants had an unexpectedly high ratio of intraspinal abnormalities (28.0% vs. 80.0%, P < 0.001), including syringomyelia, diastematomyelia, tethered spinal cord, spinal bifida, dermal sinus, etc. (SI Appendix, Fig. S5B and Table S2).
Reduced Expression and Compromised Membrane Localization Caused by VANGL1 and VANGL2 Variants.
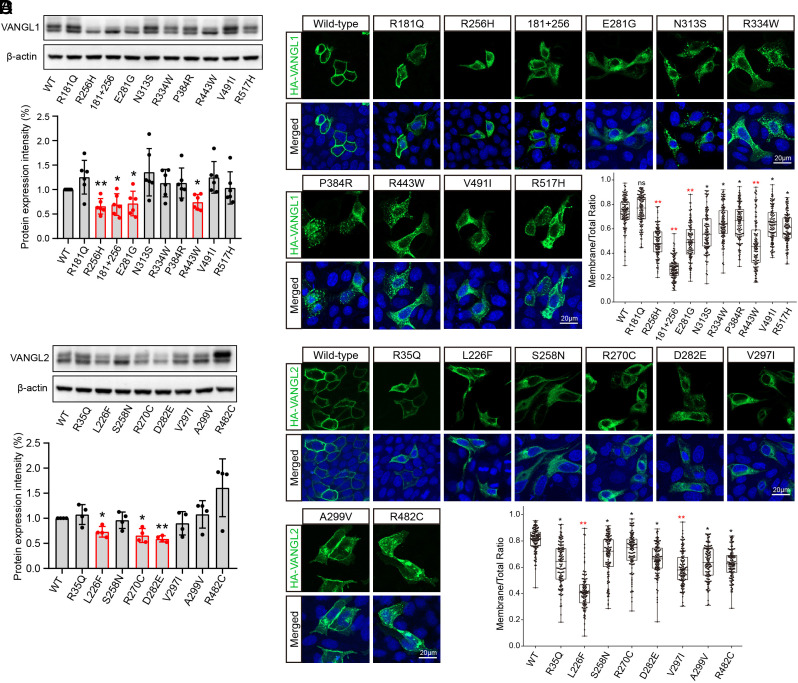
Vangl proteins are phosphorylated and transported from the endoplasmic reticulum (ER) to the plasma membrane, where they function as key components of PCP signaling (42, 43). Phosphorylation of Vangl is required for establishing PCP in vivo (42, 44–46). To interrogate the function of CS-associated VANGL1 and VANGL2 variants, we first analyzed VANGL mutants by immunoblotting in HEK293T and MDCK cells (Fig. 3 A and B and SI Appendix, Fig. S6). VANGL protein appears as two bands on the electrophoresis gel; the higher band is basally phosphorylated VANGL, and the lower band is unphosphorylated VANGL (42, 44, 47). Previously, we have shown that the Lp mutations (Vangl2 p.D255E and p.S464N) that cause looptail or the craniorachischisis phenotype in mice lead to a markedly reduced total protein level and loss of phosphorylation (40, 44). The Lp mutant Vangl2 was trapped in the ER and subjected to ER-associated proteasomal degradation (43, 48, 49). In our current study, some VANGL1 (p.R256H, p.R181Q+R256H, p.E281G, and p.R443W) and VANGL2 (p.L226F, p.R270C, and p.D282E) variants also showed significant decreases in expression levels (P < 0.05, Fig. 3 A and B and SI Appendix, Fig. S6). The phosphorylation of VANGL1 variants (p.R256H, p.R181Q+R256H, p.E281G, and p.R443W) and the VANGL2 variant (p.D282E) was nearly abolished (Fig. 3 A and B and SI Appendix, Fig. S6).
Fig. 3.
Effects of VANGL1 and VANGL2 variants on expression and localization. (A and B) Protein expression of VANGL1 and VANGL2 mutants in HEK293T cells. Band intensities of western blot gels were quantitated using ImageJ and normalized with β-actin. Each experiment was repeated at least three times. The significance of the differences was calculated by a one-way ANOVA test, F = 8.77 (A), 10.67 (B). Error bars show the mean ± SD. The panels labeled in red show a significant difference, compared with the corresponding wild-type group, *P < 0.05, **P < 0.01. (C and D) Intracellular localization of wild-type and mutant VANGL1 and VANGL2 proteins. The HA-tagged VANGL1/2 proteins were stained in green, and nuclei were visualized by DAPI in blue. Membrane localization ratios of wild-type and mutant VANGL1 and VANGL2 were analyzed by ImageJ in more than 100 cells for each group. The significance of the differences was calculated by a one-way ANOVA test, F = 114.50 (C), 69.72 (D). *P < 0.05, **P < 0.01, and the mean difference between wild-type and labeled groups exceeded 0.2. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values.
Localization on the plasma membrane is required for VANGL1 and VANGL2 to establish and maintain normal PCP signaling (23). Mouse Lp mutations and VANGL variants associated with NTDs in humans cause defective Vangl membrane targeting (49). In our study, wild-type VANGL1 and VANGL2 were primarily targeted to the plasma membrane with a typical “mesh-like” staining pattern, whereas most of the VANGL1 and VANGL2 CS-associated variants, except for the VANGL1 p.R181Q variant, were not well localized to the plasma membrane (Fig. 3 C and D). For the most part, the VANGL variants accumulated intracellularly, illustrated by strong colocalization with the ER (SI Appendix, Fig. S7). VANGL1 (p.R256H, p.R181Q+R256H, p.E281G, and p.R443W) and VANGL2 (p.L226F and p.V297I) variants displayed severely affected or completely abolished membrane localization (red asterisks in quantitative data of Fig. 3 C and D and SI Appendix, Fig. S8). Overall, these findings demonstrate partial or complete loss-of-function effects caused by VANGL1 and VANGL2 variants identified in CS patients.
VANGL1 and VANGL2 Variants Display Dominant-Negative Effects.
VANGL1 and VANGL2 are paralogs with 72% sequence identity and a similar structure. VANGL proteins can interact with each other and potentially assemble as dimers or oligomers (23, 50). Because in our studies, all identified VANGL variants are heterozygous and Vangl heterozygous null animals are morphologically normal, we sought to determine the effect of mutant VANGL on wild-type VANGL by coexpressing them in a 1:1 ratio to examine their subcellular localization pattern (Fig. 4 A and B). When wild-type VANGL1 or VANGL2 were cotransfected (HA-tagged), wild-type VANGL2 (FLAG-tagged) mainly localized to the plasma membrane, as expected. However, the cotransfection of mutant VANGL1 or VANGL2 impaired the transportation of wild-type VANGL2 to the cell surface. Wild-type VANGL2 was colocalized with mutant VANGL proteins intracellularly (Fig. 4 A and B). The quantitative analysis of numerous cells revealed the dominant-negative effects of all identified VANGL variants, which were very significant in three (VANGL1 p.R181Q+R256H, p.E281G, and VANGL2 p.L226F; red asterisks in the statistical data shown in Fig. 4 A and B). These results suggest that mutant VANGL also interferes with the normal transportation of wild-type VANGL, leading to the mislocalization of both mutant and wild-type VANGL, disrupting PCP signaling.
Fig. 4.
VANGL1 and VANGL2 variants exhibit dominant-negative effects. (A and B) Colocalization of HA-tagged wild-type and mutant VANGL1/2 (red) with Flag-tagged wild-type VANGL2 (green) in MDCK cells. Membrane localization ratios of wild-type and mutant VANGL1 and VANGL2 were analyzed by ImageJ in more than 100 cells for each group. The significance of the differences was calculated by a one-way ANOVA test, F = 49.04 (A), 17.24 (B). *P < 0.05, **P < 0.01, and the mean difference between wild-type and labeled groups exceeded 0.2. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values.
Mutant VANGL1 and VANGL2 Fail to Rescue CE Defects in Zebrafish Embryos.
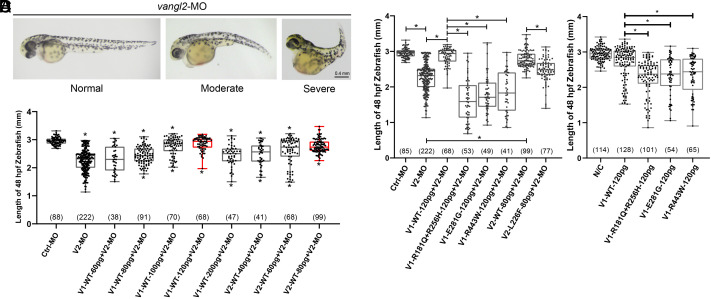
Knockdown or disruption of vangl2 in zebrafish, which causes CE movement defects and shortened A–P body axis, is a well-established in vivo model for testing Vangl function (51). We employed this system to assess the function of VANGL variants. Compared to the control-morpholino (MO) group, zebrafish embryos injected with vangl2-MO and raised until 2 d postfertilization exhibited a notable decrease in body length of the A–P axis (P < 0.05, Fig. 5 A and B). To test whether VANGL1 or VANGL2 could rescue the CE defects, we coinjected zebrafish embryos with vangl2-MO and either wild-type VANGL1 or wild-type VANGL2 mRNA at different dosages. We compared the body lengths of zebrafish to identify the optimal dosages for rescuing CE defects. Zebrafish injected with 120 pg VANGL1 or 80 pg VANGL2 showed no significant difference in body length, compared to those in the control-MO group (highlighted in red in Fig. 5B). Using these optimal dosages, we coinjected either VANGL1 or VANGL2 mutant mRNA with vangl2-MO to evaluate the pathogenicity of VANGL mutants. We selected three VANGL1 variants (p.R181Q+R256H, p.E281G, and p.R443W) and one VANGL2 variant (p.L226F) that exhibited the most deleterious functional effects on VANGL functions in the cellular assays (Figs. 3 and 4). Consistently, each of the mutant mRNAs failed to rescue the CE defects in zebrafish (P < 0.05) (Fig. 5C). Of the variants tested, VANGL1 p.R181Q+R256H seemed to be the most deleterious, and the phenotype was even more severe than that in the vangl2-MO group (Fig. 5 C and D). Furthermore, we directly investigated the dominant negative effect of the VANGL1 variants (p.R181Q+R256H, p.E281G, and p.R443W) in zebrafish embryos by evaluating their impact when injected alone (without vangl2-MO). Zebrafish embryos injected with VANGL1 mutant mRNA (p.R181Q+R256H, p.E281G, and p.R443W) alone exhibited significant CE defects compared to the group injected with wild-type one (Fig. 5D). Overall, these in vivo results confirm the loss-of-function status of the VANGL1 and VANGL2 variants and some VANGL variants also show dominant-negative effects.
Fig. 5.
Mutant VANGL1 and VANGL2 fail to rescue CE defects in zebrafish embryos. (A) Illustrations of normal, moderate, and severe defects in CE in zebrafish embryos injected with vangl2-MO (morpholino). (B) The optimal dosage of human VANGL1 and VANGL2 wild-type mRNA to rescue CE defects in zebrafish embryos. The reduced body length of zebrafish embryos caused by 8 ng vangl2-MO was fully rescued by 120 pg human VANGL1 or 80 pg human VANGL2 wild-type mRNA. The significance of the differences was calculated by a one-way ANOVA test, F = 47.60. The Top and Bottom asterisks indicate significant differences compared to the control-MO group and the vangl2-MO group, respectively. (C) The comparison of body length in zebrafish embryos between control-MO (Ctrl-MO), vangl2-MO (V2-MO), and rescue groups. The significance of the differences was calculated by a one-way ANOVA test, F = 117.60. (D) The comparison of body length in zebrafish embryos after mRNA injection between blank control, human VANGL1 wild-type, and human VANGL1 mutant groups. The statistical significance of differences between the groups was calculated by the one-way ANOVA test, F = 41.83. *P < 0.05. Box plots show the center line as the median, box limits as the upper and lower quartiles, and whiskers as the minimum to maximum values. The number in parentheses indicates the number of analyzed zebrafish embryos. V1 and V2 denote Vangl1 and Vangl2, respectively.
Characterization of a Vangl1 Knock-In Mouse Model.
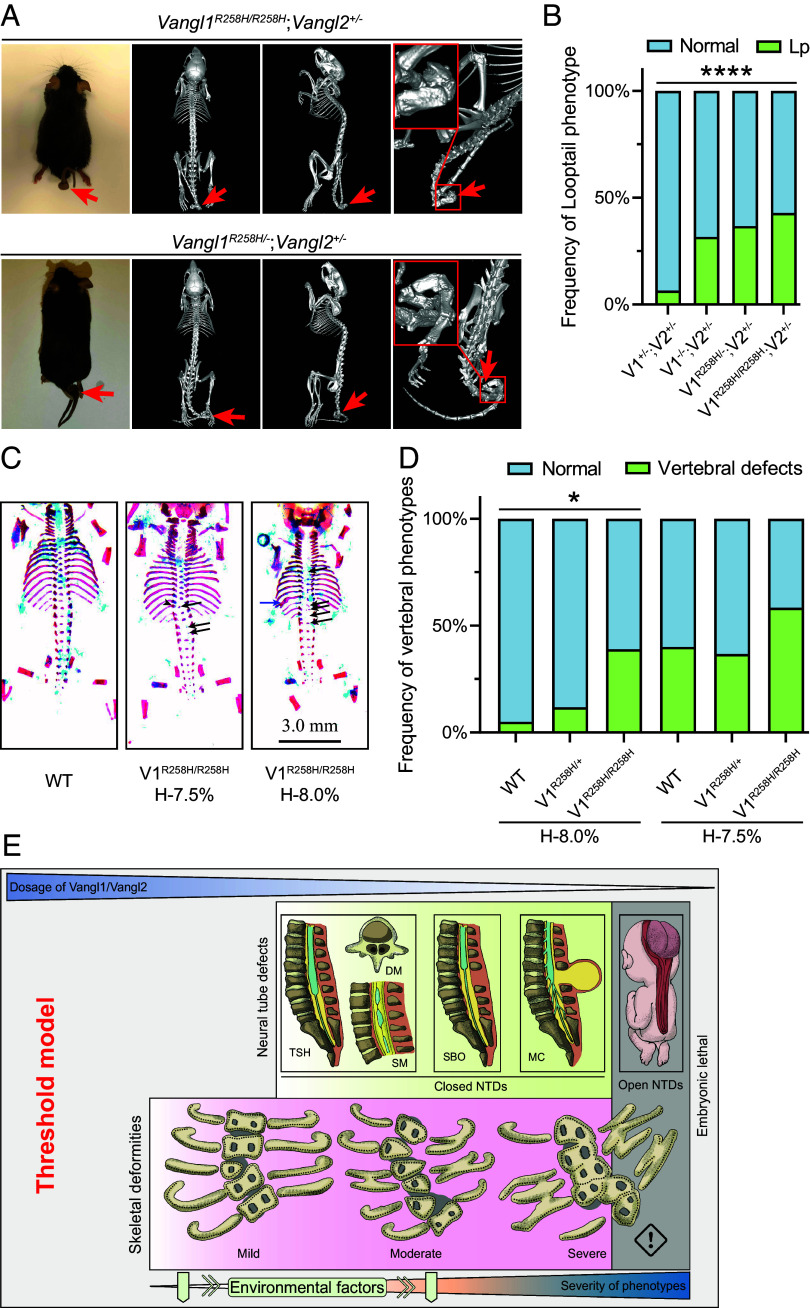
Our experiments showed that while the VANGL1 p.R181Q+R256H or p.R256H variants severely impaired VANGL function (Table 1 and Figs. 2–4), the p.R181Q variant alone had no significant influence (Fig. 3 A and C), indicating that the deleterious effect was mainly caused by the p.R256H variant. To further understand the functional impact of the VANGL1 p.R256H variant on CVMs, we generated a Vangl1-R258H knock-in mouse model, the equivalent of the human VANGL1 p.R256H substitution. Both Vangl1R258H/+ and Vangl1R258H/R258H mice were viable and fertile, but surprisingly, no skeletal abnormalities were observed (data not shown). Because axial skeletal development is sensitive to the overall gene dosage of Vangl1 and Vangl2 in mice (Fig. 1), we crossed Vangl1R258H mutant mice with Vangl2+/− or Vangl1+/−;Vangl2+/−mice, which are nearly indistinguishable from wild-type mice. Only a small fraction of Vangl1+/−;Vangl2+/− mice showed the looptail phenotype (5/76, 6.58%) (Fig. 6 A and B). Intriguingly, with the reduction of Vangl gene dosage, a higher penetrance of looptail was observed in Vangl1R258H/R258H;Vangl2+/− (30/70, 42.86%) and Vangl1R258H/−;Vangl2+/− (33/90, 36.67%) mice in comparison to Vangl1−/−;Vangl2+/−(31/98, 31.63%) and Vangl1+/−;Vangl2+/−(5/76, 6.58%) mice (Fig. 6 A and B). Vaginal atresia was also detected in some Vangl1R258H/−;Vangl2+/− and Vangl1R258H/R258H;Vangl2+/− mice (data not shown). These results suggest that the missense Vangl1 p.R258H mutation not only impairs its own function but also has a dominant-negative effect on wild-type Vangl in the development of the mouse axial skeleton. We performed microcomputed tomography (micro-CT) scanning of adult Vangl1R258H/−;Vangl2+/− and Vangl1R258H/R258H;Vangl2+/− mice and found that hemivertebrae or vertebral malformations were mainly restricted to the caudal vertebrae in the curly tail (Fig. 6 A and B).
Fig. 6.
Vangl1-R258H knock-in mice display vertebral defects in a Vangl gene dose- and hypoxia-dependent manner. (A) Phenotype of the 2-mo-old Vangl1R258H/R258H;Vangl2+/− and Vangl1R258H/−;Vangl2+/− mice by MicroCT. Looptail was observed, and vertebral malformations are indicated by red arrows. (B) Penetrance of looptail (Lp) in various Vangl1-R258H mutant mice. The significance of the differences between the groups was calculated by a chi-squared test, χ2 = 27.56, P < 0.0001. (C) Dorsal view of skeletal preparation of E17.5 Vangl1R258H/R258H embryos with hypoxia. Black arrows indicate vertebral ossification abnormalities; blue arrows indicate fused rib; arrowheads indicate hemivertebrae. (D) The penetrance of vertebral defects in wild-type, Vangl1R258H/+, and Vangl1R258H/R258H mouse embryos induced by mild hypoxia. The significance of the differences between the groups was calculated by the chi-squared test, χ2 = 8.00 (H-8.0%), 2.77 (H-7.5%). *P < 0.05. V1 and V2 denote Vangl1 and Vangl2, respectively. H-8.0% and H-7.5% denote hypoxia at 8.0% and 7.5% oxygen concentration, respectively. (E) Threshold model of Vangl gene dosage in birth defects. The decrease in Vangl1 and Vangl2 gene dosage and the addition of environmental factors increase the penetrance and severity of skeletal and neurologic phenotypes. The threshold of Vangl gene dosage for vertebral defects and NT defects is different, with the former more sensitive to Vangl dosage. TSH, tethered spinal cord; DM, diastematomyelia; SM, syringomyelia; SBO, spinal bifida occulta; MC, meningocele; NTDs, neural tube defects.
Perinatal Hypoxia Increases the Penetrance of Vertebral Defects in a Vangl1 Knock-In Mouse Model.
In addition to genetic factors, environmental risk factors, such as hypoxia and vitamin deficiency, are associated with the occurrence of CVMs (52). In particular, there is solid evidence that gestational hypoxia in pregnant mice gives rise to rib and vertebra malformations in newborns (53). Another study of mouse embryos revealed that haploinsufficiency of Notch signaling pathway genes (e.g., Mesp2), combined with short-term gestational hypoxia, significantly increases the penetrance and severity of CVMs, demonstrating that an environmental factor coupled with a genetic deficit plays an important role in the etiology of CS (54). Therefore, we evaluated possible gene–environment interactions using the Vangl1R258H CS-susceptible mouse model. Pregnant mice carrying E9.5 wild-type, Vangl1R258H/+, or Vangl1R258H/R258H embryos were exposed to 7.5% or 8.0% short-term hypoxia. A sharp threshold at 8.0% was observed. The lower oxygen concentration (7.5%) induced obvious CVMs in wild-type embryos, whereas the effect of 8.0% oxygen on wild-type embryos was minimal (Fig. 6 C and D). Compared with wild-type embryos, Vangl1R258H/R258H embryos exhibited severe vertebral defects and a high prevalence of spinal deformities after hypoxia exposure (Fig. 6 C and D). This result provides evidence for a VANGL–environment interaction in the etiology of CS.
Discussion
In this study, we identified the critical roles of two core PCP signaling genes, Vangl1 and Vangl2, in vertebral development and predisposition to human CVMs. Vangl-mutant mice showed defects in somitogenesis, possibly via the disturbance of Notch signaling, and exhibited multiple spine and rib anomalies, including hemivertebrae, the most common cause of CS. These findings are supported by our genetic studies in patients with CS, in whom we identified a number of rare variants in VANGL1 and VANGL2 with confirmed loss-of-function and dominant-negative effects. We further addressed the in vivo functional significance of the most deleterious variants using both zebrafish and mouse models. The Vangl1-R258H knock-in mice developed vertebral malformations in a Vangl gene dose- and perinatal hypoxia–dependent manner. Collectively, our results demonstrate the importance of PCP signaling in somite development and the pathogenesis of CVMs.
The development of somite includes somite formation, commonly referred to as somitogenesis, and somite differentiation (2, 55, 56). During somitogenesis, the PSM progressively segments into bilaterally symmetrical epithelial somites in an A–P direction, controlled by the segmentation clock and determination front. Somite differentiation refers to the maturation and differentiation of somites into the sclerotome, dermatome, and myotome, which eventually develop into various tissues including vertebrae (2, 55, 56). Some proteins specifically influence one of these two processes, while others have an impact on both. Fat4 and Dchs1 mutant mice exhibit malformed vertebrae that are split and fused. While somite formation occurs normally in these mutants, Fat4 and Dchs1 are required specifically for the proliferation of sclerotome cells as they differentiate into vertebrae. The loss of Fat4/Dchs1 results in decreased sclerotome proliferation and defective vertebral morphogenesis (20). The expression pattern of endogenous Vangl proteins, the defects in somite formation, and the changes of the key somitogenesis markers in Vangl mutant mice together support an important role of Vangl genes in the regulation of somitogenesis. Although the expressions of Vangl1 and Vangl2 in somites and complex vertebral malformations in Vangl mutant mice also suggest a possible role of Vangl genes in somite differentiation, it is difficult to distinguish their specific roles in somitogenesis and somite differentiation. Precise genetic tests (e.g., conditional deletion of Vangl1 and Vangl2 specifically in formed somites) are required to address this question.
Abnormal somite development during embryogenesis causes CVMs, which in turn cause CS, a spinal curvature disorder with unclear genetic etiology resulting from developmental failure of vertebral formation, segmentation, or both (1, 57). CS has an incidence of approximately 0.5 to 1 case per 1,000 live births and often progresses rapidly, leading to back pain, respiratory and psychologic distress, pulmonary and cardiac dysfunction, and disability (58). Genetic factors contribute to the development of CS, and different inheritance modes have been proposed (57–61). Previous studies have revealed pathogenic genes and copy number variants associated with CVMs, such as TBX6 (MIM #602427), TBXT (MIM #601397), PAX1 (MIM #167411), and DLL1 (MIM #606582) (58, 62–66). Considering the low recurrence rate of CS in descendants, the recessive or compound heterozygous model is preferred. We have proposed a compound heterozygous model of CVMs caused by biallelic variants of TBX6 with one rare loss-of-function TBX6 variant and one common hypomorphic allele in trans (58, 59, 62, 67, 68). This compound inheritance gene dosage model explains up to 11% of CS cases in different populations (58, 62, 69, 70). However, the VANGL1 and VANGL2 variants detected in this study are all heterozygous, and the inheritance pattern is unknown due to a lack of parental genetic information. Although we validated the deleterious effects of these variants, we found that the mutation alone (VANGL1-R256H/Vangl1-R258H) was not sufficient to cause CVMs in mice, suggesting the existence of additional risk factors or a difference in gene dosage sensitivity between humans and mice.
Digenic or oligogenic models have been proposed for many diseases (71, 72). For example, the digenic combination of SMAD6 and BMP2 variants has been reported in nonsyndromic midline craniosynostosis (71). An oligogenic effect of MKL2, MYH7, and NKX2-5 variants has been demonstrated in cardiac anomalies (72). The digenic variants of VANGL2, CELSR1, SCRIB, DVL3, and PTK7 may also be responsible for NTDs (73, 74). Recently, we evaluated the mutation spectrum of TBX6-mediated genes and proposed an oligogenic model of CS (60). In our current study, the oligogenic model may represent additional risk variants in other genes involved in somite development. Moreover, patients may have somatic mutations in addition to the germline variants. This phenomenon has been observed in NTDs (75–77). For example, somatic mutations in CELSR1, FZD6, and VANGL1 in NT lesions are associated with NTDs (75). In mice, mosaic deletion of Vangl2 in as few as 16% of neuroepithelial cells is sufficient to cause NTDs (77). These findings support the hypothesis that CS may be induced by a combined effect of germline susceptibility variants and somatic mutations, explaining the low familial recurrence rate and phenotypic variation in CS patients. Validation of this model requires future genetic studies on malformed vertebral tissues.
Various environmental factors can induce CS (52, 54, 57). In the Vangl1R258H CS-susceptible mouse model, short-term hypoxia exposure significantly increased the severity and penetrance of vertebral defects. Sparrow et al. demonstrated that gestational hypoxia can disrupt the segmentation process during somitogenesis by interfering with the periodic expression of Notch1 and blocking Fgf signaling (54). Given the compromised Notch signaling activity in Vangl mutant mice, the concomitant dysregulation of Notch signaling caused by genetic variants and the additional effect of hypoxia on Notch, Fgf signaling or both may explain the increasing severity and penetrance of vertebral defects. Various degrees of dysregulation of somitogenesis at different time points can trigger spinal phenotypes ranging from tail to thoracic or lumbar region. Vangl1R258H mutant mice with additional loss of the Vangl2 allele displayed hemivertebrae restricted to the tail; however, hypoxia-treated Vangl1R258H mutant mice developed thoracic and lumbar hemivertebrae. The severe dysregulation of somitogenesis will easily influence the early-formed somites that give rise to the structures at the thoracic and lumbar spine level, whereas the mild dysregulation of somitogenesis may only manifest in the caudal region because the effects may need to be accumulated and intensified over multiple segmentation cycles to reach a critical threshold. Environmental factors such as hypoxia can enhance somitogenesis defects and thus lead to CVMs in more rostral regions. All these effects could be a combined result of genetic variants and gestational hypoxia.
CS can manifest as an isolated spinal defect or multisystem malformations complicated with other anomalies (58, 78, 79). Liu et al. found that over 20.0% of CS patients have intraspinal malformations, of which the most common type is diastematomyelia (78). In our cohort, 29.1% of the CS patients had intraspinal deformities; the prevalence in those with VANGL1 and VANGL2 variants was substantially higher (80.0%), suggesting a strong correlation between CVMs, intraspinal defects, and VANGL. Interestingly, compared to previously reported NTD patients carrying VANGL variants (26–29), the NTD phenotypes of our CS patients with rare VANGL variants were mild. These patients mainly demonstrated syringomyelia, diastematomyelia, and tethered spinal cord, indicating a phenotypic spectrum associated with VANGL variants. Although there was a clear clinical association between NTDs and CVMs, the underlying pathogenesis was unclear. Somite development and neurulation are closely connected during embryonic development and share some critical signaling pathways. Signals from the NT and notochord can also contribute to the formation and differentiation of somites (80). For example, the transformation of ventromedial somites into the sclerotome requires Sonic hedgehog (Shh) and Noggin signaling molecules secreted by the NT and ventral notochord (81, 82). The development of somites relies on the integrity of the NT. Removal of the NT during embryogenesis causes failure of segmentation or hypoplasia of the vertebrae (83). From this perspective, it is also possible that CVMs observed in Vangl mutant mice are a secondary effect from failed NT closure due to disruption of PCP signaling. However, based on the data we have in mouse models (a higher level of Vangl deficiency is required for developing NTDs than CVMs) and human patients (CVMs are more pronounced than NTDs in VANGL variant carriers), we tend to believe that Vangl genes play independent roles in both somite development and NT closure. It would be interesting to investigate whether other PCP components are also involved in the pathogenesis of CS comorbid with NTDs.
Vangl are tetraspan transmembrane proteins with cytoplasmic N- and C-terminals and contain many functional domains and motifs. We have previously identified two clusters of highly conserved phosphorylation sites in Vangl N-terminals required for PCP in zebrafish gastrulation and mouse development (42, 44). Interestingly, some variants, particularly those in VANGL1 (p.R256H, p.R181Q+R256H, p.E281G, and p.R443W), disrupted VANGL phosphorylation and showed poor membrane localization. The long C-terminal region harbors multiple signal motifs and interaction domains, including a TGN sorting motif, a p97/valosin-containing protein (VCP)-interacting motif, a coiled-coil domain, and a PDZ-binding motif at the end of the C-terminals (23, 43). The VANGL1 p.E281G and VANGL2 p.D282E variants are adjacent to the TGN sorting motif, which affected their intracellular trafficking. The VANGL1 p.R256H and VANGL2 p.S258N and p.R270C variants are located in the same region as the well-known Vangl2 Lp and Curly Bob mutations. Similar to the Vangl2 Lp mutant, these variants caused ER retention of Vangl proteins. Because the heterozygous Lp mutation can cause looptail or looptail with spina bifida in mice, our findings support a potential causal effect of VANGL risk variants in causing CS and/or NTD in humans. To further understand the impact of VANGL variants, we analyzed the three-dimensional VANGL structures predicted by AlphaFold (SI Appendix, Fig. S9, https://alphafold.ebi.ac.uk) (84). Most of the variants were located in the α-helix or the β-sheet, and three VANGL1 variants (p.R256H, p.E281G, and p.V491I) and four VANGL2 variants (p.S258N, p.R270C, p.D282E, and p.R482C) were located within the core structure of the protein, which is also the location of Lp and Curly Bob mutations. These findings support the functional importance of the identified VANGL variants.
Vangl are mainly dedicated to PCP signaling but some studies also suggested non-PCP roles of Vangl genes (85–87). Although it is most likely that the CVMs caused by Vangl deficiency is due to the defective PCP signaling, it is also possible that their non-PCP functions contribute to the phenotype. VANGL1 has been reported to be associated with adolescent idiopathic scoliosis (AIS) (88). However, AIS and CS are distinct spinal disorders with different clinical presentations and underlying causes. AIS typically manifests during pubertal growth and has no CVMs. Notably, another AIS genetic study failed to identify an association between VANGL1 and AIS (89).
Overall, we have revealed the important function of core PCP genes Vangl1 and Vangl2 in somite and vertebral development and their predisposing role in the development of CVMs. Our data also suggest a Vangl gene–environment interaction model that regulates the formation of the axial skeleton. The penetrance and severity of CVMs and NTDs increase as the Vangl gene dosage reduces (Fig. 6E). Other genetic variants, either germline or somatic, and environmental factors may lower the threshold for developing isolated or concomitant phenotypes.
Materials and Methods
Study Approval.
The present multicenter study was conducted in accordance with the tenets of the Declaration of Helsinki. In mainland China, ethical approval was acquired from the Institutional Review Board of Peking Union Medical College Hospital (PUMCH, Approval No. JS-2364). In Hong Kong, ethics approval was acquired from the Institutional Review Board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU/HA HKW IRB Ref# UW 15-216). In the United States, ethical approval was acquired from the Baylor College of Medicine IRB (H-29697). In Japan, ethical approval was acquired from the Institutional Review Board of the RIKEN Yokohama Institute (Approval No. 17-16-40). Written informed consent was obtained from affected individuals and the participating parents and siblings. All zebrafish and mouse experiments were conducted in compliance with the Guidelines from The Committee on Use of Laboratory Animals for Teaching and Research of The University of Hong Kong.
Recruitment of Participants, Sequencing, and Variant Detection.
Patients diagnosed with CS were included, and those with a previous molecular diagnosis [for example, TBX6-associated CS (TACS) (58, 67)] were excluded. Patients with VANGL1 or VANGL2 variants were identified in multicenter and multiethnic CS cohorts from mainland China (708 cases), the Hong Kong Special Administrative Region of China (67 cases), Japan (1 case), and the United States (6 cases) under the framework of the DISCO (Deciphering disorders Involving Scoliosis and COmorbidities, http://www.discostudy.org/) study. Genomic DNA was extracted from peripheral venous blood samples. Whole-exome sequencing was performed independently at different centers. The sequencing data were annotated using the Peking Union Medical college hospital Pipeline (PUMP) and HKU Pipeline as previously described (59, 61, 62, 66, 68, 90). Whole-exome data of Japanese patients were also addressed and filtered based on previous studies (91, 92). Whole-exome sequencing data generated in the United States were analyzed as previously described using the Baylor College of Medicine Human Genome Sequencing Center-designed core capture reagent 29 (52 Mb, Nimblegen) and an Illumina HiSeq2000. Data were processed using the local Mercury pipeline, with alignment using BWA-aln, variant calling by Atlas2, and annotation using an in-house developed Cassandra pipeline (93). The following criteria further filtered ultrarare variants of VANGL1 and VANGL2: 1) predicted to alter amino acid sequence; 2) allele frequency < 0.001 in gnomAD (https://gnomad.broadinstitute.org/); and 3) CADD score > 15.
Mouse Experiments.
All mice used in this study were maintained on C57BL/6 background. If all genotypes are available, embryos from same littermate were used for each experiment, but if one experiment needs multiple embryos with same genotype, embryos from different mating pairs were grouped for studies and N number was provided. Generation of the Vangl1 knock-in mouse model, skeletal preparation, whole-mount immunofluorescence staining, whole-mount immunofluorescence staining, whole-mount immunohistochemistry, micro-CT analyses, and hypoxia treatment were elucidated in SI Appendix.
Zebrafish Microinjection.
The AB zebrafish strain was used for all the microinjections. Defined MOs and mRNAs were injected into one-cell stage embryos passing through the yolk. Injection and measuring methods were described in SI Appendix.
Sanger Sequencing.
Primers to amplify the corresponding exons of VANGL1 and VANGL2 (GenBank: NM_138959.2 and NM_020335.2, respectively) were designed with Primer-BLAST (94), and an appropriate amount of genomic DNA was used for PCR. A DNA Analyzer (Applied Biosystems) was used to confirm the candidate variants, and segregation analyses were performed using family member data (if available).
Cellular Experiments.
Human full-length VANGL1 and VANGL2 cDNAs were subcloned into pcDNA3.1 vector with a hemagglutinin (HA) epitope or FLAG tag (DYKDDDDK) inserted into the N terminus. HEK293T and MDCK cells were subjected to transfection for subsequent cellular assays. Detailed protocols of plasmid construction, immunoblotting, and immunofluorescence were incorporated within SI Appendix.
Statistical Analysis.
One-way ANOVA was conducted to compare differences in immunoblotting, membrane localization density, and body length of zebrafish embryos. A chi-squared test was used to calculate the penetrance of vertebral defects in mouse embryos. All statistical tests are declared in the figure legends. The significance threshold was set at P < 0.05. Continuous variables were expressed as the mean ± SD, as indicated in the figure legends. SPSS 24.0 (Chicago) and GraphPad Prism 8 (CA) were used for data analyses.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (DOCX)
Acknowledgments
We would like to thank all patients and their family members for participating in this study. We thank Jing Guo, The University of Hong Kong, for assistance with confocal microscopy, and Jacky Hung, The University of Hong Kong, for assistance with zebrafish breeding and maintenance. We thank GeneSeeq Inc. and Novogene Inc. for exome sequencing technical support. We thank Beijing Ekitech Co., Ltd. for support in bioinformatic analyses and data management. This research was funded in part by the National Key Research and Development Program of China (2022YFC2703102, 2023YFC2507700, and 2023YFC2507701), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-051 to T.J.Z. and N.W., 2021-I2M-1-052 to Z.W., and 2023-I2M-C&T-A-003 to T.J.Z.), National Natural Science Foundation of China (82072391 to N.W., 81930068 and 81772299 to Z.W., 82172525 to G.Q., 82172382 to T.J.Z., and 82372366 to S.W.), National High Level Hospital Clinical Research Funding (2022-PUMCH-D-004 to T.J.Z. and N.W.and 2022-PUMCH-C-033 to N.W.), Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2019PT320025 to N.W.), China Postdoctoral Science Foundation (No. 2019M663272 to Y. Ye), National Natural Science Foundation of China Incubation Project of Guangdong Provincial People's Hospital (No. KY0120220040 to Y. Ye), Guangdong Basic and Applied Basic Research Foundation (No. 2022A1515111091 to Y. Ye), Guangzhou Municipal Science and Technology Project (No. 2023A04J0500 to Y. Ye), the Chinese University of Hong Kong start-up and direct grants (2022.085 to B.G.), Lo Kwee Seong Foundation (to B.G.), the Health and Medical Research Fund (No. 06171406 to B.G.), Innovation Technology Commission Fund (Health@InnoHK to B.G. at Center for Translational Stem Cell Biology), General Research Fund (No. 17118120 and No. 17101422 to B.G. and No. 17114519 to Y.-Q.S.), the Tam Sai Kit Endowment Fund (to K.D.K.L.), US National Institutes of Health/National Human Genome Research Institute/National Heart, Lung, and Blood Institute UM1 HG006542 to the Baylor-Hopkins Center for Mendelian Genomics, and US National Institutes of Health/National Human Genome Research Institute U01 HG011758 to the Baylor College of Medicine Genomic Research to Elucidate the Genetics of Rare (BCM-GREGoR) programs.
Author contributions
X.F., Y. Ye, J.Z., J.P.Y.C., T.J.Z., B.G., and N.W. designed research; X.F., Y. Ye, and J.Z. performed research, contributed new reagents/analytic tools and analyzed data; Y.Ye, Y.Z., S.Z., N.O., Z.Z., Y.N., Y. Yonezawa, G.L., M.L., X. Li, P.W.H.C., K.X., K.T., S.W., V.N.T.C., Y.-Q.S., Y. Yang, K.D.K.L., G.Q., K.W., Z.W., J.E.P., S.I., J.P.Y.C., T.J.Z., N.W., D.d.I.S.a.C.s.g. and J.E.O.S.R.G. recruitment of participants, sequencing, gathered clinical histories, performed clinical examinations, and collected DNA and tissue samples for the study; J.P.Y.C. and N.W. led the clinical work in Hong Kong and Beijing, respectively. N.O., Y. Yonezawa, K.T., T.K., K.W., and S.I. led the clinical work in Japan. J.E.P. and J.R.L. led the clinical work in USA; N.W. and T.J.Z. led the genetic studies; B.G. led the functional studies; K.S.L., P.S.L., and C.Y.H.L. generated the Vangl1 knock-in mice; J.C.W.M., S.Z., J.X., V.N.T.C., Y.-Q.S., X. Lin, and X.W. assisted in genetic or functional studies; N.W., B.G., T.J.Z., and J.P.Y.C. jointly supervised the whole project; and X.F., Y. Ye, J.Z., B.G., and N.W. wrote the paper.
Competing interests
J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Genetics Center, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical genomic sequencing offered in the Baylor Genetics Laboratory (http://bmgl.com).
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Jason Pui Yin Cheung, Email: cheungjp@hku.hk.
Terry Jianguo Zhang, Email: zhangjianguo@pumch.cn.
Bo Gao, Email: bogao@cuhk.edu.hk.
Nan Wu, Email: dr.wunan@pumch.cn.
Collaborators: Guixing Qiu, Nan Wu, Jianguo Zhang, Zhihong Wu, Shengru Wang, Sen Liu, Ziquan Li, Yang Yang, Zhengye Zhao, Guilin Chen, Guozhuang Li, Yuanpeng Zhu, Jihao Cai, Di Liu, Kexin Xu, Jianle Yang, Aoran Maheshati, Qing Li, Jingyi Xie, Xiangjie Yin, Jie Wang, Zihua Li, Zhifa Zheng, Kun Fang, Xiangyu Nie, Xi Cheng, Wen Wen, Xinyu Yang, Yuanqiang Zhang, Lian Liu, Lianlei Wang, Na Chen, Jiachen Lin, Mao Lin, Lina Zhao, Fei Liu, Yuchen Niu, Qing Liu, Guangxi Gao, Shuai Li, Yueyan Bai, Sen Zhao, Yongyu Ye, Hengqiang Zhao, Zefu Chen, Jiaqi Liu, Zihui Yan, Chenxi Yu, Jiashen Shao, Nao Otomo, Yoshiro Yonezawa, Kazuki Takeda, Yoji Ogura, Noriaki Kawakami, Toshiaki Koatani, Teppei Suzuki, Koki Uno, Hideki Sudo, Satoshi Inami, Hiroshi Taneichi, Hideki Shigematsu, Kei Watanabe, Ryo Sugawara, Yuki Taniguchi, Shohei Minami, Masaya Nakamura, Morio Matsumoto, Shiro Ikegawa, and Kota Watanabe
Data, Materials, and Software Availability
All data supporting the findings of this study are available within the paper and its SI Appendix. The raw genetic sequencing data for CS patients have been deposited in the Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/gsa-human/) (95) under accession number HRA006007. All raw sequencing data deposited in GSA are under restricted access and only academic use would be approved.
Supporting Information
References
- 1.Pourquie O., Vertebrate segmentation: From cyclic gene networks to scoliosis. Cell 145, 650–663 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz-Cuadros M., et al. , In vitro characterization of the human segmentation clock. Nature 580, 113–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takada S., et al. , Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8, 174–189 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Anderson M. J., Magidson V., Kageyama R., Lewandoski M., Fgf4 maintains Hes7 levels critical for normal somite segmentation clock function. Elife 9, e55608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaguchi T. P., Bradley A., McMahon A. P., Jones S., A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development 126, 1211–1223 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Zhang C., et al. , Novel pathogenic variants and quantitative phenotypic analyses of Robinow syndrome: WNT signaling perturbation and phenotypic variability. HGG Adv. 3, 100074 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roifman M., et al. , De novo WNT5A-associated autosomal dominant Robinow syndrome suggests specificity of genotype and phenotype. Clin. Genet. 87, 34–41 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Bunn K. J., et al. , Mutations in DVL1 cause an osteosclerotic form of Robinow syndrome. Am. J. Hum. Genet. 96, 623–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J. J., et al. , DVL3 alleles resulting in a -1 frameshift of the last exon mediate autosomal-dominant Robinow syndrome. Am. J. Hum. Genet. 98, 553–561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White J. J., et al. , WNT signaling perturbations underlie the genetic heterogeneity of Robinow syndrome. Am. J. Hum. Genet. 102, 27–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikels A. J., Nusse R., Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 4, e115 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao B., Wnt regulation of planar cell polarity (PCP). Curr. Top. Dev. Biol. 101, 263–295 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Yang Y., Mlodzik M., Wnt-Frizzled/planar cell polarity signaling: Cellular orientation by facing the wind (Wnt). Annu. Rev. Cell Dev. Biol. 31, 623–646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler M. T., Wallingford J. B., Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 18, 375–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray R. S., Roszko I., Solnica-Krezel L., Planar cell polarity: Coordinating morphogenetic cell behaviors with embryonic polarity. Dev. Cell 21, 120–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallingford J. B., Niswander L. A., Shaw G. M., Finnell R. H., The continuing challenge of understanding, preventing, and treating neural tube defects. Science 339, 1222002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamblet N. S., et al. , Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development 129, 5827–5838 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Yang T., Bassuk A. G., Fritzsch B., Prickle1 stunts limb growth through alteration of cell polarity and gene expression. Dev. Dyn. 242, 1293–1306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C., et al. , Null and hypomorph Prickle1 alleles in mice phenocopy human Robinow syndrome and disrupt signaling downstream of Wnt5a. Biol. Open 3, 861–870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuta A., et al. , Fat4-Dchs1 signalling controls cell proliferation in developing vertebrae. Development 143, 2367–2375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torban E., et al. , Genetic interaction between members of the Vangl family causes neural tube defects in mice. Proc. Natl. Acad. Sci. U.S.A. 105, 3449–3454 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song H., et al. , Planar cell polarity breaks bilateral symmetry by controlling ciliary positioning. Nature 466, 378–382 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailly E., Walton A., Borg J. P., The planar cell polarity Vangl2 protein: From genetics to cellular and molecular functions. Semin. Cell Dev. Biol. 81, 62–70 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Stein K. F., Mackensen J. A., Abnormal development of the thoracic skeleton in mice homozygous for the gene for looped-tail. Am. J. Anat. 100, 205–223 (1957). [DOI] [PubMed] [Google Scholar]
- 25.Greene N. D., Gerrelli D., Van Straaten H. W., Copp A. J., Abnormalities of floor plate, notochord and somite differentiation in the loop-tail (Lp) mouse: A model of severe neural tube defects. Mech. Dev. 73, 59–72 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Kibar Z., et al. , Mutations in VANGL1 associated with neural-tube defects. N. Engl. J. Med. 356, 1432–1437 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Lei Y. P., et al. , VANGL2 mutations in human cranial neural-tube defects. N. Engl. J. Med. 362, 2232–2235 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Kibar Z., et al. , Contribution of VANGL2 mutations to isolated neural tube defects. Clin. Genet. 80, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kibar Z., et al. , Novel mutations in VANGL1 in neural tube defects. Hum. Mutat. 30, E706–E715 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crossley P. H., Martin G. R., The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439–451 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Naiche L. A., Holder N., Lewandoski M., FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 4018–4023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boulet A. M., Capecchi M. R., Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 371, 235–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saga Y., Hata N., Koseki H., Taketo M. M., Mesp2: A novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev. 11, 1827–1839 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Morimoto M., Takahashi Y., Endo M., Saga Y., The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature 435, 354–359 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Bessho Y., Hirata H., Masamizu Y., Kageyama R., Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451–1456 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson V., Manson L., Skarnes W. C., Beddington R. S., The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development 121, 877–886 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Chapman D. L., Agulnik I., Hancock S., Silver L. M., Papaioannou V. E., Tbx6, a mouse T-Box gene implicated in paraxial mesoderm formation at gastrulation. Dev. Biol. 180, 534–542 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Guo Y., Zanetti G., Schekman R., A novel GTP-binding protein-adaptor protein complex responsible for export of Vangl2 from the trans Golgi network. Elife 2, e00160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guyot M. C., et al. , A novel hypomorphic Looptail allele at the planar cell polarity Vangl2 gene. Dev. Dyn. 240, 839–849 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kibar Z., et al. , Ltap, a mammalian homolog of Drosophila Strabismus/Van Gogh, is altered in the mouse neural tube mutant Loop-tail. Nat. Genet. 28, 251–255 (2001). [DOI] [PubMed] [Google Scholar]
- 41.El-Hassan A. R., et al. , Identification and characterization of a novel chemically induced allele at the planar cell polarity gene Vangl2. Mamm. Genome 29, 229–244 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Yang W., et al. , Wnt-induced Vangl2 phosphorylation is dose-dependently required for planar cell polarity in mammalian development. Cell Res. 27, 1466–1484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng D., et al. , Regulation of Wnt/PCP signaling through p97/VCP-KBTBD7-mediated Vangl ubiquitination and endoplasmic reticulum-associated degradation. Sci. Adv. 7, eabg2099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao B., et al. , Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev. Cell 20, 163–176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strutt H., Gamage J., Strutt D., Reciprocal action of Casein Kinase I epsilon on core planar polarity proteins regulates clustering and asymmetric localisation. Elife 8, e45107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly L. K., Wu J., Yanfeng W. A., Mlodzik M., Frizzled-induced Van Gogh phosphorylation by CK1 epsilon promotes asymmetric localization of core PCP factors in Drosophila. Cell Rep. 16, 344–356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng D., He Z., Gao B., Analysis of the ubiquitination and phosphorylation of Vangl proteins. Bio. Protoc. 12, e4533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iliescu A., Gravel M., Horth C., Kibar Z., Gros P., Loss of membrane targeting of Vangl proteins causes neural tube defects. Biochemistry 50, 795–804 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Iliescu A., Gravel M., Horth C., Gros P., Independent mutations at Arg181 and Arg274 of Vangl proteins that are associated with neural tube defects in humans decrease protein stability and impair membrane targeting. Biochemistry 53, 5356–5364 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Belotti E., et al. , Molecular characterisation of endogenous Vangl2/Vangl1 heteromeric protein complexes. PLoS One 7, e46213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jessen J. R., et al. , Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 4, 610–615 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., Yu X., Shen J., Environmental aspects of congenital scoliosis. Environ. Sci. Pollut. Res. Int. 22, 5751–5755 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Ingalls T. H., Curley F. J., Principles governing the genesis of congenital malformations induced in mice by hypoxia. N. Engl. J. Med. 257, 1121–1127 (1957). [DOI] [PubMed] [Google Scholar]
- 54.Sparrow D. B., et al. , A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 149, 295–306 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Maroto M., Bone R. A., Dale J. K., Somitogenesis. Development 139, 2453–2456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hubaud A., Pourquie O., Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709–721 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Giampietro P. F., et al. , Clinical, genetic and environmental factors associated with congenital vertebral malformations. Mol. Syndromol. 4, 94–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu N., et al. , TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 372, 341–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W., et al. , TBX6 missense variants expand the mutational spectrum in a non-Mendelian inheritance disease. Hum. Mutat. 41, 182–195 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y., et al. , Mutational burden and potential oligogenic model of TBX6-mediated genes in congenital scoliosis. Mol. Genet. Genomic Med. 8, e1453 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao S., et al. , Unraveling the genetic architecture of congenital vertebral malformation with reference to the developing spine. Nat. Commun. 15, 1125 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng X., et al. , Genetic variants of TBX6 and TBXT identified in patients with congenital scoliosis in Southern China. J. Orthop. Res. 39, 971–988 (2021). [DOI] [PubMed] [Google Scholar]
- 63.Ghebranious N., et al. , A missense T (Brachyury) mutation contributes to vertebral malformations. J. Bone Miner. Res. 23, 1576–1583 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Barhoumi T., et al. , Delta like-1 gene mutation: A novel cause of congenital vertebral malformation. Front. Genet. 10, 534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giampietro P. F., et al. , An analysis of PAX1 in the development of vertebral malformations. Clin. Genet. 68, 448–453 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Lai W., et al. , Identification of copy number variants in a Southern Chinese cohort of patients with congenital scoliosis. Genes (Basel) 12, 1213 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J., et al. , TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: Further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet. Med. 21, 1548–1558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao S., et al. , Diagnostic yield and clinical impact of exome sequencing in early-onset scoliosis (EOS). J. Med. Genet. 58, 41–47 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lefebvre M., et al. , Autosomal recessive variations of TBX6, from congenital scoliosis to spondylocostal dysostosis. Clin. Genet. 91, 908–912 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Takeda K., et al. , Compound heterozygosity for null mutations and a common hypomorphic risk haplotype in TBX6 causes congenital scoliosis. Hum. Mutat. 38, 317–323 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Timberlake A. T., et al. , Two locus inheritance of non-syndromic midline craniosynostosis via rare SMAD6 and common BMP2 alleles. Elife 5, e20125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gifford C. A., et al. , Oligogenic inheritance of a human heart disease involving a genetic modifier. Science 364, 865–870 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L., et al. , Digenic variants of planar cell polarity genes in human neural tube defect patients. Mol. Genet. Metab. 124, 94–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murdoch J. N., et al. , Genetic interactions between planar cell polarity genes cause diverse neural tube defects in mice. Dis. Model Mech. 7, 1153–1163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tian T., et al. , Somatic mutations in planar cell polarity genes in neural tissue from human fetuses with neural tube defects. Hum. Genet. 139, 1299–1314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.D’Gama A. M., Walsh C. A., Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci. 21, 1504–1514 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Galea G. L., et al. , Cell non-autonomy amplifies disruption of neurulation by mosaic Vangl2 deletion in mice. Nat. Commun. 12, 1159 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y. T., et al. , A retrospective study of congenital scoliosis and associated cardiac and intraspinal abnormities in a Chinese population. Eur. Spine J. 20, 2111–2114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu N., et al. , Retrospective analysis of associated anomalies in 636 patients with operatively treated congenital scoliosis. J. Bone Joint Surg. Am. 105, 537–548 (2023). [DOI] [PubMed] [Google Scholar]
- 80.Ye Y., et al. , Exploring the association between congenital vertebral malformations and neural tube defects. J. Med. Genet. 60, 1146–1152 (2023). [DOI] [PubMed] [Google Scholar]
- 81.Marcelle C., Stark M. R., Bronner-Fraser M., Coordinate actions of BMPs, Wnts, Shh and Noggin mediate patterning of the dorsal somite. Development 124, 3955–3963 (1997). [DOI] [PubMed] [Google Scholar]
- 82.Wagner J., Schmidt C., Nikowits W. Jr., Christ B., Compartmentalization of the somite and myogenesis in chick embryos are influenced by Wnt expression. Dev. Biol. 228, 86–94 (2000). [DOI] [PubMed] [Google Scholar]
- 83.Colbjorn Larsen K., Fuchtbauer E. M., Brand-Saberi B., The neural tube is required to maintain primary segmentation in the sclerotome. Cells Tissues Organs. 182, 12–21 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Tunyasuvunakool K., et al. , Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Glasco D. M., et al. , The mouse Wnt/PCP protein Vangl2 is necessary for migration of facial branchiomotor neurons, and functions independently of Dishevelled. Dev. Biol. 369, 211–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sittaramane V., et al. , The cell adhesion molecule Tag1, transmembrane protein Stbm/Vangl2, and Lamininalpha1 exhibit genetic interactions during migration of facial branchiomotor neurons in zebrafish. Dev. Biol. 325, 363–373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gong Y., et al. , Vangl2 limits chaperone-mediated autophagy to balance osteogenic differentiation in mesenchymal stem cells. Dev. Cell 56, 2103–2120.e9 (2021), 10.1016/j.devcel.2021.06.011. [DOI] [PubMed] [Google Scholar]
- 88.Andersen M. R., et al. , Mutation of the planar cell polarity gene VANGL1 in adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 42, E702–E707 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Xu L., et al. , VANGL1 is not associated with the susceptibility of adolescent idiopathic scoliosis in the Chinese population. Spine (Phila Pa 1976) 43, E580–E584 (2018). [DOI] [PubMed] [Google Scholar]
- 90.Wang K., et al. , Perturbations of BMP/TGF-beta and VEGF/VEGFR signalling pathways in non-syndromic sporadic brain arteriovenous malformations (BAVM). J. Med. Genet. 55, 675–684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Otomo N., et al. , Bi-allelic loss of function variants of TBX6 causes a spectrum of malformation of spine and rib including congenital scoliosis and spondylocostal dysostosis. J. Med. Genet. 56, 622–628 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Guo L., et al. , Identification of biallelic EXTL3 mutations in a novel type of spondylo-epi-metaphyseal dysplasia. J. Hum. Genet. 62, 797–801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bainbridge M. N., et al. , Whole-genome sequencing for optimized patient management. Sci. Transl. Med. 3, 87re83 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ye J., et al. , Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.CNCB-NGDC Members and Partners, Database resources of the national genomics data center, China national center for bioinformation in 2023. Nucleic Acids Res. 51, D18–D28 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (DOCX)
Data Availability Statement
All data supporting the findings of this study are available within the paper and its SI Appendix. The raw genetic sequencing data for CS patients have been deposited in the Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/gsa-human/) (95) under accession number HRA006007. All raw sequencing data deposited in GSA are under restricted access and only academic use would be approved.