SUMMARY
Although gut and lymph node (LN) memory CD4 T cells represent major HIV and simian immunodeficiency virus (SIV) tissue reservoirs, the study of the role of dendritic cells (DCs) in HIV persistence has long been limited to the blood due to difficulties to access lymphoid tissue samples. In this study, we show that LN migratory and resident DC subpopulations harbor distinct phenotypic and transcriptomic profiles. Interestingly, both LN DC subpopulations contain HIV intact provirus and inducible replication-competent HIV despite the expression of the antiviral restriction factor SAMHD1. Notably, LN DC subpopulations isolated from HIV-infected individuals treated for up to 14 years are transcriptionally silent but harbor replication-competent virus that can be induced upon TLR7/8 stimulation. Taken together, these results uncover a potential important contribution of LN DCs to HIV infection in the presence of ART.
Graphical abstract
In brief
Although DCs play a major role during HIV dissemination, their implications in immunopathogenesis and persistence has long-time been neglected. Banga et al. demonstrate that lymph node DCs containing inducible replication-competent virus can be detected after years of suppressive ART, opening new avenues for developing therapeutic interventions for HIV cure.
INTRODUCTION
Pioneering studies have demonstrated that HIV persistence under anti-retroviral therapy (ART) can mainly be attributed to the ability of HIV to establish a stable viral reservoir within CD4 T cells endowed with a long half-life and capacity to proliferate.1–9
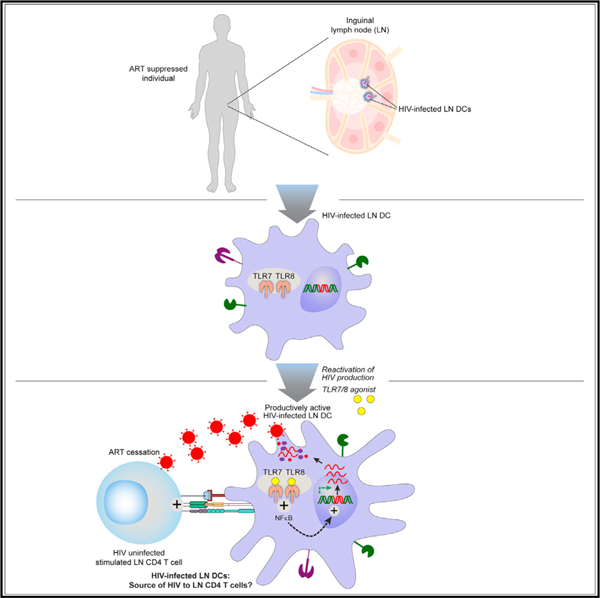
Following ART interruption, the actual model proposes that virus replication might be induced from latently infected CD4 T cells following T cell receptor (TCR) or micro-environmental driven stimulations and/or following stochastic events.10–13 Interestingly, to identify the cellular source of the rebounding virus, a pioneer study performed in situ hybridization of lymph node (LN) and Gut-associated lymphoid tissue (GALT) biopsies collected at the time of viral rebound from otherwise fully suppressed individuals.11 The authors underscored the presence of HIV RNA in GALT and LN germinal center (GC) area at the time of viral rebound and proposed that HIV production in the B cell follicles may precede viral dissemination.11 However, the immuno-virological mechanisms associated with these observations remain elusive but might be associated with an increased susceptibility of T follicular helper (Tfh) cells to HIV infection,14 the clonal expansion of HIV-infected Tfh cells,15 and/or a limited elimination of HIV-infected Tfh cells in GC areas.16 However, because the Tfh cell differentiation is a multi-stage process requiring long-lasting interactions with LN dendritic cells (DCs) and pre-GC B cells in a specific cytokine/chemokine microenvironment, we postulated that in the event of HIV infection of LN DCs, HIV-infected LN DCs may not only stimulate but also efficiently transmit HIV infection to antigen-specific CD4 T cells.
LNs are unique microenvironments that harbor heterogeneous myeloid DC subsets called conventional DCs (cDCs) that are classified on the basis of their phenotype and functions.17,18 In tissues, cDCs are continuously replenished from a population of hematopoietic precursor DCs (pre-cDCs) that originate from bone marrow and circulate in the blood before reaching tissues where they can differentiate into mature DCs through the process called cDCpoiesis.19,20
LN myeloid DCs can be identified on the basis of their original tissue location, i.e., ‘‘resident DCs’’ and ‘‘migratory DCs.’’ In particular, LN resident DCs differentiate in, and spend their entire lives within LN tissues.21 On the other hand, migratory DCs can migrate from peripheral tissues (e.g., from the genital mucosa to the inguinal draining LNs) bearing antigens.22 In the process of maturation, these DCs upregulate the expression of co-stimulatory, adhesion and MHC molecules and the chemokine receptor CCR7, allowing them to migrate to local draining LNs.22
In contrast to the well-studied CD4 T cells and macrophages,23–29 the data regarding the potential role of DCs, in general and as HIV reservoirs, are rare. The investigations were mainly hindered due to the low frequency of DCs, scarce availability of tissue DCs, and therefore the high reliance on experiments performed either on blood DCs or on in-vitro-derived DCs. These studies revealed that although in-vitro-derived DCs were permissive to various HIV strains,30 de novo virus production occurred only intermittently, mainly due to reverse transcription blocks mediated by restriction factors such as SAM-domain- and HD-domain-containing protein (SAMHD1).31–33 However, the conclusions proposed on the basis of in vitro generated DCs might not totally reflect the complexity of DCs in vivo.22 More importantly, recent reports suggested that DC lifespan and survival are also highly dependent on their anatomical locations and local microenvironment19,34 and therefore could be much longer than previously assumed, making HIV-infected LN DCs potential candidates for harboring replication-competent viruses over time.
Given the enhanced T cell scanning activity of mature LN DCs and their pivotal role in inducing T cell immune responses, we hypothesized that in the event of infection of LN DCs with replication-competent HIV, DCs could act as a yet underestimated cellular source of HIV. In this context, we have therefore evaluated the potential presence of HIV-infected LN-derived myeloid DCs during HIV infection and under suppressive ART.
RESULTS
Characterization of LN myeloid DC transcriptomic profile by scRNA-seq
Due to the paucity of data regarding human myeloid LN DC subpopulations, we FACS sorted the two identified LN myeloid DC subpopulations, i.e., LN migratory (CD3−CD20−HLA-DR+CD45+CD11c+CCR7+) and resident (CD3−CD20−HLA-DR+CD45+CD11c+CCR7−) DCs from inguinal LNs (Figure S1A) isolated from ART-treated HIV-infected individuals (N = 3) and performed a comprehensive transcriptomic characterization by single-cell RNA sequencing (scRNA-seq).
Of note, the purity of sorting experiments was ≥98% for each sorted cell subset and for all experiments performed in the present manuscript (average purity for LN resident DCs: 99.44%; migratory DCs: 99.16%; LN CD4 T cells: 98.9%; Figure S1C; Table 1). Notably, none of the sorted LN DC subpopulations expressed either CD3 or CD20 transcripts. When present, the ‘‘contaminating’’ cells were DCs harboring either reduced CD45 and/or CCR7 expression (Figure S1C). In addition, to further demonstrate the absence of CD4 T cells within the sorted LN DC subpopulations, a quantification of T cell receptor excision circles (TRECs) was performed on sorted LN DC subpopulations as previously described (Figure S1E).35 The cumulative data demonstrated that although TREC copies were consistently detected within CD4 T cells, ranging from 500,000 to 50 CD4 T cells (Figure S1D), TRECs were not detected within 40,000 sorted LN resident or migratory DCs (Figure S1E).
Table 1.
Cohort clinical characteristics
| Patient ID | Age | Sex | CD4 cell count (cells/μL) | VL (copies/mL) | Time on suppressive ART (years) | Current ART treatment regimena | Assay | No. of LN Mononuclear cells collected (million) | Purity LN resident DCs (%) | Purity LN migratory DCs (%) | Purity LN CD4/PD-1+ or PD-1−CD4 T cells |
|---|---|---|---|---|---|---|---|---|---|---|---|
| #118 | 28 | M | 538 | 1.40E + 4 | viremic | N/A | CA, TILDA, viral production | 360 | 99 | 99 | 100 |
| #128 | 30 | M | 158 | 2.50E + 4 | viremic | N/A | viral production | 150 | 99.8 | 98.1 | 99 |
| #148 | 37 | M | 717 | 2.00E + 4 | viremic | N/A | MC | 120 | N/A | N/A | N/A |
| #149 | 38 | M | 280 | 1.80E + 4 | viremic | N/A | MC, TILDA, viral production | 150 | 100 | 98.1 | 99 |
| #157 | 22 | M | 507 | 1.90E + 3 | viremic | N/A | MC, INT DNA, CA | 100 | 99 | 100 | 98 |
| #158 | 28 | M | 279 | 3.10E + 5 | viremic | N/A | MC, INT DNA, CA | 90 | 99.1 | 100 | 100 |
| #1001 | 27 | M | 655 | 3.22E + 4 | viremic | N/A | INT DNA | N/D | 98.6 | 100 | 100 |
| #1003 | 30 | M | 913 | 6.96E + 4 | viremic | N/A | MC | N/D | N/A | N/A | N/A |
| #1008 | 23 | M | 566 | 4.94E + 5 | viremic | N/A | MC, CA, TILDA | N/D | 98.2 | 98.4 | 99.4 |
| #1009 | 45 | M | 894 | 1.79E + 4 | viremic | N/A | MC | N/D | N/A | N/A | N/A |
| #1010 | 35 | M | 803 | 1.44E + 4 | viremic | N/A | CA, TILDA, viral production | N/D | 100 | 98.5 | 100 |
| #1013 | 50 | F | 428 | 2.15E + 5 | viremic | N/A | INT DNA, viral production | N/D | 99.8 | 100 | 99.3 |
| #1014 | 29 | M | 381 | 4.16E + 5 | viremic | N/A | MC, viral production | N/D | 99.3 | 99.1 | 100 |
| #1017 | 37 | F | 498 | 1.74E + 4 | viremic | N/A | INT DNA | N/D | 99 | 99.5 | 98.1 |
| #1019 | 31 | M | 855 | 1.79E + 4 | viremic | N/A | HIV sequencing | N/D | 98 | 100 | 98 |
| #1024 | 31 | M | 546 | 4.63E + 4 | viremic | N/A | HIV sequencing | N/D | 99 | 100 | 98.1 |
| #1025 | 29 | M | 637 | 1.21E + 5 | viremic | N/A | MC, HIV sequencing | N/D | 99.1 | 98 | 98.3 |
| #1027 | 31 | F | 521 | 6.08E + 4 | viremic | N/A | MC | N/D | N/A | N/A | N/A |
| #161 | 56 | M | 677 | <50 | 5 | TDF, FTC, DRV/r | MC, INT DNA, CA, TILDA, VOA | 80 | 99.4 | 99.1 | 98.9 |
| #136 | 44 | M | 1,326 | <50 | 10 | 3TC, ABC, RAL | scRNA-seq, MC, VOA, DC-T cell stimulation | 150 | 98.5 | 98 | 98.1 |
| #109 | 44 | M | 625 | <50 | 0.5 | FTC, TDF | MC, INT DNA, TILDA, VOA, RAM | 60 | 100 | 100 | 100 |
| #082 | 52 | M | 955 | <50 | 2.2 | 3TC/AZT, IDV | scRNA-seq, INT DNA, CA RNA, TILDA | 80 | 100 | 98.2 | 98 |
| #142 | 37 | F | 272 | <50 | 14 | FTC, TDF, DRV/r | MC, VOA, DC-T cell stimulation | 90 | 100 | 98.3 | 98.5 |
| #122 | 35 | M | 517 | <50 | 19 | DTG, ABC, 3TC | MC, INT DNA, VOA | 85 | 100 | 100 | 98 |
| #078 | 39 | M | 728 | <50 | 4.6 | FTC, TDF, EFV | MC | 60 | N/A | N/A | N/A |
| #058–2 | 48 | F | 666 | <50 | 12 | 3TC/AZT, EFV | MC | 40 | N/A | N/A | N/A |
| #075 | 44 | M | 417 | <50 | 3.2 | FTC, TDF, EFV | MC | 90 | N/A | N/A | N/A |
| #103 | 41 | M | 318 | <50 | 1.1 | FTC, TDF, EFV | MC, VOA | 100 | 100 | 100 | 100 |
| #155 | 43 | F | 852 | <50 | 1 | NVP, 3TC, FTC | MC | 10 | N/A | N/A | N/A |
| #139 | 35 | M | 571 | <50 | 7.5 | FTC, TDF, EFV | MC | 30 | N/A | N/A | N/A |
| #163 | 34 | F | 699 | <50 | 17 | FTC, TDF, RPV | MC | 10 | N/A | N/A | N/A |
| #159 | 45 | M | 336 | <50 | 6 | EVG, FTC, TDF | VOA | 40 | 100 | 100 | 100 |
| #104 | 41 | F | 448 | <50 | 4 | FTC, TDF, RPV | VOA, RAM | 50 | 99.2 | 98 | 98 |
| #133 | 41 | M | 709 | <50 | 8 | FTC, TDF, EVG | VOA, RAM | 60 | 100 | 100 | 100 |
| #011–2 | 54 | M | 954 | <50 | 12 | ATV/r, FTC, TDF | scRNA-seq, CA | 120 | 100 | 100 | 98 |
| #R003 | 33 | M | 902 | <50 | 2.9 | FTC, TDF, RPV | QVOA | 20 | QVOA pool: | QVOA pool: | QVOA pool: |
| #045 | 41 | M | 442 | <50 | 7.6 | FTC, TDF, ETR | QVOA | 78 | 100 | 98.1 | 99 |
| #105 | 46 | F | 712 | <50 | 7.9 | DRV/r, RAL, MVC | QVOA | 10 | |||
| #099 | 46 | M | 437 | <50 | 3.5 | FTC, TDF, RTV, ATV | QVOA | 27 | |||
| #094 | 46 | M | 440 | <50 | 6.6 | FTC, TDF, EVG | QVOA | 68 | |||
| #093 | 41 | F | 438 | <50 | 4.7 | ABC, DTG, 3TC | QVOA | 17.5 | |||
| #169 | 30 | M | 336 | <50 | 4 | RPV/DTG | DC-T cell stimulation | 40 | 100 | 99.1 | 99 |
Abbreviations: VL, viral load at sampling date; FTC, emtricitabine; TDF, tenofovir disoproxil fumarate; RTV, ritonavir; ATV, atazanavir; ATV/r, atazanavir boosted with ritonavir; 3TC, lamivudine; ABC, abacavir; EFV, efavirenz; EVG, elvitegravir; RAL, raltegravir; DTG, dolutegrvir; AZT, zidovudine; DRV/r, darunavir boosted with ritonavir; IDV, idinavir; NVP, nevirapine; RPV, rilpivirine; ETR, etravirine; MVC, maraviroc; scRNA-seq, single-cell RNA sequencing; MC, mass cytometry; INT DNA, integrated DNA; CA, cell-associated HIV RNA; TILDA, tat-rev limiting dilution assay; VOA, viral outgrowth assay; QVOA, quantitative viral outgrowth assay; RAM, resistance associated mutation; N/A, not applicable; N/D, not determined. Time on suppressive ART refers to the time that the individual maintained continuous suppression of viremia to below the limit of detection up until the sampling date.
We first assessed the differentially expressed gene signatures between the two sorted LN DC subpopulations as previously described (Figure 1).36,37 The representative example and cumulative data obtained from a mean of 3,239 resident and 2,344 migratory DCs per subject analyzed revealed a clear segregation between these two subsets on the basis of expression of >300 genes (Figures 1A and 1B; Tables S1 and S2). In particular, LN migratory DCs expressed significantly higher levels of transcripts encoding for the lineage defining transcription factor IRF4, the chemokine receptor CCR7, the interleukin (IL)-7 cytokine receptor IL-7R, and the anti-apoptotic/survival factor BIRC3, compared with LN resident DCs (p < 0.05) (Figures 1B–1E and 1G). In contrast, LN resident DCs expressed significantly higher levels of transcripts encoding for the transcription factor IRF8 and the anti-viral restriction factor SAMHD1 compared with migratory DCs (p < 0.05) (Figures 1B, 1F, and 1H), supporting previous features and specialized functions delineated to LN resident and migratory DC subpopulations in vivo.22
Figure 1. Characterization of LN resident and migratory DCs.
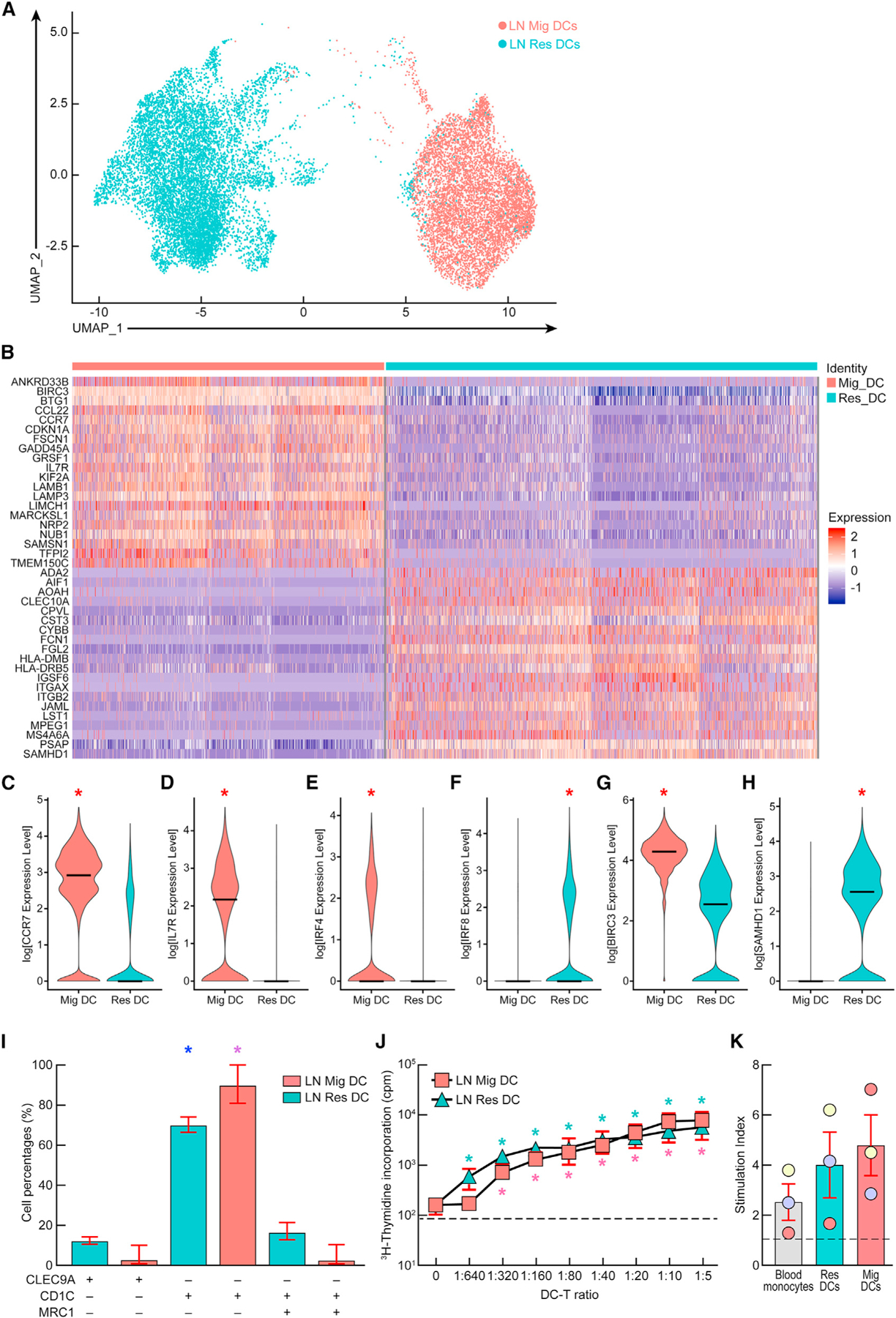
(A) Ex vivo transcriptomic uniform manifold approximation and projection (UMAP) profile of LN resident and migratory DCs from aviremic ART-treated HIV-infected individuals (N = 3).
(B) Heatmap representing the top 20 differentially expressed genes within LN resident and migratory DCs.
(C–H) Violin plots showing the normalized expression level of differentially expressed genes between LN resident and migratory DCs.
(I) Percentages of cDC1, cDC2, and DC3-like cells within LN resident and migratory DCs of HIV-infected individuals (N = 3).
(J and K) (J) Stimulatory potential of LN resident or migratory DCs as assessed by mixed leukocyte reaction (MLR) assay or using HIV-specific stimulation (K). Individuals are color coded (K).
Bars correspond to mean (C–I and K). Red bars correspond to mean ± SEM (I–K). Dashed lines represent the cutoff for positivity (J and K). Red stars indicate statistical significance (*p < 0.05) (C–H). Statistical significance of cDC2-type LN resident (blue stars) or migratory (purple stars) DCs compared with cDC1 and DC3-type LN DCs (*p < 0.05) (I). Statistical significance of LN resident (green stars) or migratory (pink stars) DCs:CD4 T cell MLR cultures at various DC:T cell ratios compared with CD4 T cells alone (*p < 0.05) (J). Statistical significance was assessed with the non-parametric Wilcoxon rank sum test implemented in the ‘‘FindMarkers’’ function of the Seurat package (C–H) or using one-way ANOVA followed by either multiple comparison test (I) or Wilcoxon matched-pairs two-tailed signed rank test (J and K). ‘‘Res DC’’ corresponds to LN resident DCs and ‘‘Mig DC’’ corresponds to LN migratory DCs.
In addition to their migratory potential, DCs are also classified into distinct subsets based on their phenotypic characteristics and functional abilities to prime T cells. We therefore investigated the distribution of cDC1, cDC2, and DC3 subsets within migratory and resident DCs based on the expression of distinct transcripts. Briefly, cDC1 were identified on the basis of the expression of CLEC9A transcript, cDC2 were identified on the basis of the expression of CD1c transcript in the absence of MRC1 transcript, and DC3 were identified on the basis of the co-expression of CD1c and MRC1 transcripts.17,18 The cumulative data revealed that both LN resident and migratory DCs were enriched in cells expressing markers of cDC2 cells (mean: 74% and 94%, respectively), whereas much smaller fractions of cells harbored markers associated with cDC1 or DC3 cells (resident DCs: mean cDC1, 10% and mean DC3, 14%; migratory DCs: mean cDC1, 2.5% and mean DC3, 3.4%) (p < 0.05) (Figure 1I).
We next investigated the co-stimulatory potential of LN resident and migratory DCs. The cumulative data indicated that both LN resident and migratory DCs were able to induce a dose-dependent CD4 T cell proliferation by mixed leukocyte reaction (MLR), suggesting that both LN DC subpopulations were endowed with a stimulatory potential (p < 0.05) (Figure 1J). Of note, no notable differences, however, were observed between LN resident and migratory DCs (p > 0.05) (Figure 1J). In addition, both LN DC subpopulations were able to induce HIV-specific memory CD8 T cell proliferation in an autologous assay, albeit at low levels, which was not significantly different from blood monocytes (p > 0.05) (Figure 1K; Table S3). Notably, no significant differences were observed between LN resident and migratory DCs (p > 0.05) (Figure 1K). Taken together, the data indicated that both LN resident and migratory DCs isolated from ART-treated HIV-infected individuals were able to stimulate HIV-specific memory CD8 T cells in vitro.
LN DC phenotypic characterization by MC
We subsequently assessed the protein expression of markers that were differentially expressed between the two LN DC subpopulations (as identified in Figures 1A–1H) by mass cytometry (MC) (gating strategy represented in Figure S2).
The representative example and the cumulative protein expression profiles confirmed that the two LN DC subpopulations clustered away from naive CD4 T cells and from each other (Figures 2A and S3A). In particular, LN migratory DCs expressed increased protein expression level of BIRC3 (Figures 2B and 2C), PD-L1, PD-L2, CD155, CD127, CD40, CD86, and IRF4 compared with LN resident DCs (p < 0.05) (Figure S3). In contrast, and consistent with the scRNA-seq data, LN resident DCs expressed increased protein expression level of SAMHD1 compared with migratory DCs (p < 0.05) (Figures 2B, 2C, and S3). These data revealed some specific characteristics of LN DCs directly isolated ex vivo from LN tissues.
Figure 2. Phenotypic profile of LN resident and migratory DCs by mass cytometry.

(A) t-SNE plots depicting the surface expression patterns of CD4, CCR5, CXCR4, DC SIGN, CD169, and CD163 directly ex vivo on naive CD4 T cells, LN migratory and LN resident DCs from a treated HIV-infected individual (#161).
(B) Representative mass cytometry profile of BIRC3 and SAMHD1 expression levels in naive CD4 T cells, LN Tfh cells, LN resident, and LN migratory DCs isolated directly ex vivo from a representative HIV-uninfected, viremic (#1009), and aviremic ART-treated HIV-infected individual (#163).
(C) Frequencies of LN resident and LN migratory DCs expressing CD4, CCR5, CXCR4, DC-SIGN, CD169, and CD163 or mean signal intensity (MSI) of SAMHD1 and BIRC3 in naive CD4 T cells, LN resident, and LN migratory DCs isolated directly ex vivo from HIV-uninfected (N = 4), viremic (N = 6) and ART-treated HIV-infected individuals (N = 7).
Red bars correspond to mean ± SEM (C). Red stars indicate statistical significance (*p < 0.05) for intra-group comparisons, i.e., LN migratory DCs versus LN resident DCs (C). Green stars indicate statistical significance (*p < 0.05) for intra-group comparisons of LN migratory DCs, LN resident DCs, and naive CD4 T cells (C). Statistical significance was obtained using one-way ANOVA (Kruskal-Wallis test) followed by Wilcoxon matched-pairs two-tailed signed rank test.
Interestingly, both LN DC subpopulations expressed high levels of the HIV receptor CD4 and co-receptors CXCR4, CCR5, CD163, CD169, and DC-SIGN (Figures 2A and 2C), raising the potential susceptibility to HIV infection.
LN DCs are susceptible to HIV infection in vitro
We next evaluated the susceptibility of LN DC subpopulations isolated directly ex vivo from LNs of HIV-uninfected individuals to R5-tropic and X4-tropic HIV infection in vitro. The representative example and cumulative data indicated that both DC subsets were susceptible to CCR5-tropic and X4-tropic HIV infection in vitro (Figures 3A–3C). Interestingly, the proportions of both CCR5-enhanced green fluorescent protein (EGFP) and CXCR4-EGFP transduced LN migratory DCs were significantly higher compared with LN resident DCs (p < 0.05; mean LN migratory CCR5-EGFP+: 5% versus 3%; and mean LN migratory CXCR4-EGFP+: 8.2% versus 4.5%) (Figures 3A–3C).
Figure 3. LN resident and migratory DCs are susceptible to HIV infection in vitro.
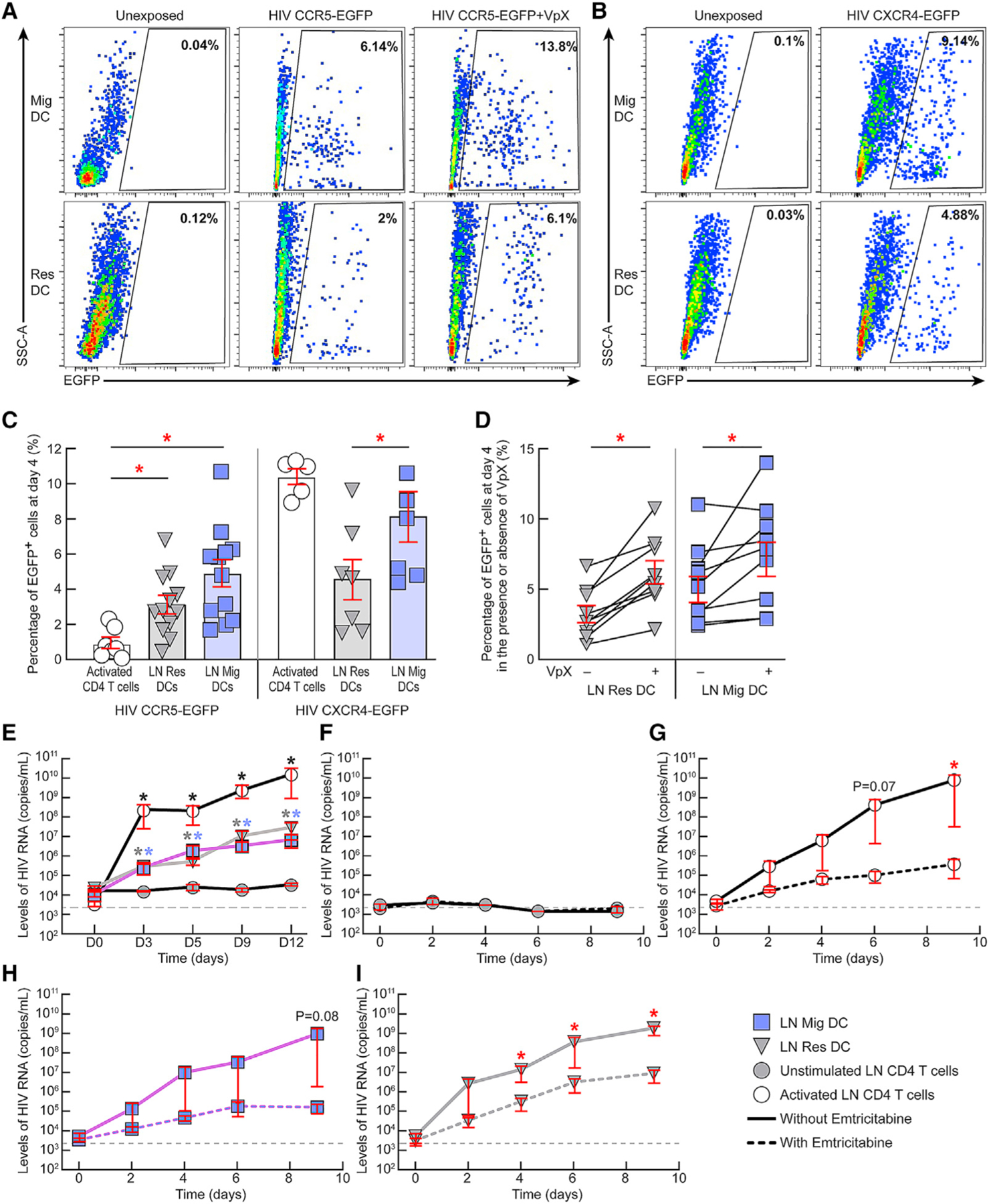
(A and B) Representative flow cytometry profiles of LN resident and migratory DCs isolated from a representative HIV-uninfected individual showing their relative susceptibility to (A) CCR5-tropic HIV-derived vector encoding for EGFP in the presence or absence of SAMHD1-modulating factor, i.e., Vpx at day 4 post-exposure or (B) to CXCR4-tropic HIV-derived vector encoding for EGFP.
(C) Percentages of EGFP+ LN DCs at day 4 post-exposure to either CCR5-tropic (N = 12) or CXCR4-tropic (N = 7) HIV-derived vectors. Activated LN CD4 T cells were added as controls (N = 5).
(D) Percentage of EGFP+ LN DCs at day 4 post-exposure to CCR5-tropic HIV-derived vector encoding for EGFP in the presence or absence of Vpx (N = 9).
(E) Levels of HIV RNA (copies/mL) in culture supernatants of unstimulated LN CD4 T cells, activated LN CD4 T cells, LN migratory DCs, and LN resident DCs of HIV-uninfected individuals exposed to HIV Ba-L (N = 3).
(F–I) Levels of HIV RNA (copies/mL) in culture supernatants of unstimulated LN CD4 T cells (F), activated LN CD4 T cells (G), LN migratory DCs (H), and LN resident DCs (I) of HIV-uninfected individuals exposed to HIV Ba-L in the presence or absence of emtricitabine (N = 3).
Red bars correspond to mean ± SEM (C–I). Red stars indicate statistical significance (*p < 0.05) of intra-group comparisons (C and D) or of conditions cultured without emtricitabine compared with conditions with emtricitabine (G and I). Black, gray, and violet stars indicate statistical significance (*p < 0.05) of activated CD4 T cells, LN resident, and migratory DCs, respectively, compared with the levels of unstimulated CD4 T cells at the corresponding time points (E). Wells with detectable HIV-1 RNA (≥2,000 HIV-1 RNA copies/mL) were referred to as HIV-1 RNA-positive wells (E–I). Gray dashed lines (E–I) represent the limit of detection. Statistical significance (p values) was obtained using one-way ANOVA (Kruskal-Wallis test) followed by Wilcoxon matched-pairs two-tailed signed rank test (C–E) or following a one-tailed ratio paired t test (F–I). “Res DC” corresponds to LN resident DCs and “Mig DC” corresponds to LN migratory DCs.
Because high levels of the anti-viral restriction factor SAMHD1 were detected within LN resident DCs, we assessed whether the complementation of HIV-2 SAMHD1 counteracting protein, i.e., Vpx, could influence the susceptibility of DC transduction with CCR5-tropic HIV-derived vector. The representative examples and cumulative data indicated that the presence of Vpx at the time of exposure to CCR5-tropic HIV-derived vector significantly increased the susceptibility of both LN resident and migratory DC transduction (p < 0.05; LN resident DCs from mean 3% to 6.4%; LN migratory DCs from mean 5% to 7.1%) (Figures 3A and 3D).
We further evaluated whether LN DC subpopulations isolated directly ex vivo from LNs of HIV-uninfected individuals and exposed to CCR5-tropic replication-competent HIV-1 labderived variant (HIV Ba-L) were productively infected by HIV in vitro (N = 3). The cumulative data indicated that HIV-RNA levels were significantly increased in days 3, 5, 9, and 12 culture supernatants of both LN resident and migratory DCs, compared with unstimulated CD4 T cells at the corresponding time points (p < 0.05) (Figure 3E). To further evaluate whether LN DC subpopulations supported active cycles of de novo replication of CCR5-tropic HIV in vitro, LN DC subpopulations isolated from 3 HIV-uninfected individuals were exposed to HIV Ba-L in the presence or absence of reverse transcriptase inhibitor, i.e., emtricitabine (FTC). In parallel, unstimulated and activated sorted CD4 T cells from the same individuals were used as controls (Figures 3F–3I). The cumulative data indicated that HIV-RNA levels significantly increased at days 4, 6, and 9 in culture supernatants of LN resident DCs (Figure 3I) and at day 9 for activated LN CD4 T cells cultured in the absence of FTC compared with those cultured in the presence of FTC (p < 0.05) (Figure 3G). Notably, HIV-RNA levels at day 9 reached border-line significance for LN migratory DCs cultured in the absence of FTC compared with conditions with FTC (p = 0.08) (Figure 3H). Taken together, these data demonstrated that LN DC subpopulations were productively infected and supported active cycles of de novo replication of R5 tropic HIV in vitro.
Ex-vivo-isolated LN DCs of viremic HIV-infected individuals harbor genome-intact HIV
We next assessed the relative distribution of defective and intact HIV-1 sequences among LN DC subpopulations. For this purpose, we FACS sorted LN migratory and resident DCs from three chronic viremic HIV-infected individuals (N = 3) and concurrently analyzed all proviral HIV-1 sequences at single-genome level by near full-genome amplification using full-length individual proviral sequencing (FLIP-seq) and their respective chromosomal integration sites by matched integration site and proviral sequencing (MIP-seq) as previously described.38 Of note, PD-1+/Tfh and LN PD-1−/non-Tfh CD4 T cells were sorted in parallel from the same individuals as internal controls (Figure 4).
Figure 4. LN resident and migratory DCs are enriched with genome-intact HIV DNA.

(A) Maximum-likelihood phylogenetic trees of all HIV-1 sequences obtained from LN resident and migratory DCs of the 3 chronic viremic HIV-infected individuals (#1024, #1025, and #1019; 8–9 kb amplicons) as assessed by FLIP-seq and MIP-seq assays. Sequences from LN PD-1+/Tfh and LN PD-1−/non-Tfh cells are also depicted. Chromosomal integration site coordinates for the respective sequences are indicated.
(B) Pie charts reflecting the relative proportion of intact and defective HIV-1 sequences.
(C and D) Frequencies of cells harboring either intact (C) or defective (D) HIV-1 sequences (cells/million) as assessed by both FLIP-seq and MIP-seq assays (2 assessments per individual). HIV-infected individuals are color coded (C and D). LN cell populations are color coded (C and D). Histograms represent minimum to maximum of the range and the line corresponds to the median (C and D). Red bars correspond to 95% confidence interval. “Res DC” corresponds to LN resident DCs and “Mig DC” corresponds to LN migratory DCs.
Notably, consistent with previous studies, PD-1+/Tfh cells harbored the highest proportion of HIV-infected cells harboring at least one copy of a viral sequence as assessed by LTR gag count by droplet digital PCR (ddPCR) (average of 11.16%),39 whereas LN migratory and resident DCs harbored on an average 0.4% and 0.21% of HIV-infected cells, respectively (Table S4).
Interestingly, we found that, within the LN DCs, although the proviral landscape consisted of some defective proviruses with large deletions and hypermutations, a large number of individual proviral DNA sequences isolated from LN DCs, particularly LN migratory DCs were genome intact (55% genome intact within LN migratory DCs [n = 16,000 cells] compared with 40% in LN resident DCs [n = 36,400 cells], 16% in LN PD-1+/Tfh cells [n = 4,000 cells] and none within LN PD-1−/non-Tfh cells [n = 15,770 cells]) (Figures 4A–4D; Tables S4 and S5). Of note, consistent with previous studies, Tfh cells harbored the highest frequency of intact proviruses per million cells14,39,40 (Figure 4C). Notably, we detected on an average 120 LN migratory DCs/million and 57 LN resident DCs/million containing intact proviral sequences (Figure 4C).
Interestingly, the maximum-likelihood phylogenetic trees showed that the proviral sequences from LN PD-1+/Tfh cells and LN DC subpopulations were frequently intermingled. Moreover, sequences from LN DC subpopulations were frequently in immediate phylogenetic proximity to sequences from LN PD-1+/Tfh cells (Figure 4A). Together, these data did not suggest phylogenetic compartmentalization between sequences from LN DC subpopulations and LN CD4 T cells in viremic individuals.
Taken together, these results confirmed the presence of LN DCs infected with intact replication-competent HIV in vivo.
Ex-vivo-isolated LN DCs of viremic HIV-infected individuals are transcriptionally active and produce HIV upon TLR7/8 stimulation
We next performed a comprehensive virological assessment of purified LN migratory and resident DCs isolated directly ex vivo from viremic HIV-infected individuals (N = 5) (Figure 5). Notably, because LN PD-1high/Tfh are known to be enriched in HIV-infected cells compared with LN PD-1−/non-Tfh CD4 T cells,14 both populations were also sorted from the same individuals and used as internal controls (Figure S1B).
Figure 5. LN DCs of viremic individuals are HIV-infected, transcriptionally active, and produce HIV upon TLR7/8 in vitro stimulation.

(A–F) LN resident and migratory DCs were isolated from untreated viremic HIV-infected individuals.
(A) Levels of integrated HIV DNA (levels/million cells) within LN resident and migratory DCs (N = 5).
(B) Levels of cell-associated unspliced HIV gag RNA within LN resident and migratory DCs (N = 5).
(C and D) (C) Frequencies of cells containing unspliced HIV gag RNA (cells/million) (C) and multi-spliced tat-rev RNA (cells/million) (D) in LN resident and migratory DCs (N = 5).
(E and F) (E) Levels of HIV production in LN resident and migratory DC culture supernatants at day 7 post-stimulation with TLR7/8 agonist as assessed by HIV-RNA levels (copies/mL) (E) or HIV P24 levels (pg/mL) (F) (N = 6).
Autologous LN PD-1+/Tfh and LN PD-1−/non-Tfh cells were used as controls (A–F). LN PD-1+/Tfh and LN PD-1−/non-Tfh cells were stimulated or not with anti-CD3/anti-CD28 mAbs for 3 days (E and F). Dashed line represents the limit of detection (A–F). HIV-infected individuals are color coded (A–D). Red bars correspond to mean ± SEM (A–F). Red stars indicate statistical significance (*p < 0.05). Statistical significance (p values) was obtained using one-way ANOVA (Kruskal-Wallis test) followed by Wilcoxon matched-pairs one-tailed signed rank test (E and F). ‘‘Res DC’’ corresponds to LN resident DCs and ‘‘Mig DC’’ corresponds to LN migratory DCs.
The results confirmed that LN PD-1+/Tfh cells harbored the highest frequency of cells containing integrated HIV DNA (11,800 cells/million) compared with both LN DC subpopulations and LN PD-1−/non-Tfh CD4 T cells (Figure 5A).14 Interestingly, both LN migratory and resident DCs contained integrated HIV DNA (1,875 cells/million in LN migratory DCs and 620 cells/million in resident DCs) (Figure 5A). In addition, both LN DC subpopulations harbored cells containing HIV unspliced gag RNA and ms tat-rev HIV RNA at levels and frequencies comparable to those detected in LN PD-1+/Tfh cells (p > 0.05; Figures 5B–5D).
We next evaluated the capacity of ex-vivo-isolated LN DC subpopulations to support active HIV RNA and/or HIV P24 production in vitro upon Toll-like receptor (TLR)7/8 stimulation.41,42 For this purpose, ex-vivo-isolated sorted LN DC subpopulations were exposed or not to TLR7/8 agonist (R848),41,42 whereas autologous LN PD-1+/Tfh and LN PD-1−/non-Tfh cells were stimulated or not with anti-CD3/anti-CD28 mAbs in vitro as previously described.24,43 Notably, in these experimental settings, no target cells were added to the cell culture. HIV production was assessed as levels of HIV RNA and HIV P24 in culture supernatants at day 7.
The cumulative data indicated that HIV production was detected in the absence of stimulation in the culture supernatants of CD4 T cells and both LN DC subpopulations (Figures 5E and 5F). However, HIV-RNA levels were significantly increased in culture supernatants of stimulated conditions compared with unstimulated conditions for both LN resident and migratory DC subpopulations (p < 0.05) (Figure 5E; 20-fold higher for LN resident DCs; 860-fold increase for LN migratory DCs). HIV P24 levels were significantly increased in culture supernatants of stimulated LN migratory DCs compared with unstimulated conditions (p < 0.05; Figure 5F; 6.4-fold increase for LN migratory DCs), whereas no significant increase was detected in stimulated LN resident DC cultures (p > 0.05; Figure 5F; 1.25-fold higher for LN resident DCs). These data showed that HIV production from LN DC subpopulations was inducible upon TLR7/8 stimulation in vitro.
Taken together, the virological assessments performed in LN DCs isolated directly ex vivo from viremic HIV-infected individuals demonstrated that LN DC subpopulations were infected by HIV in vivo, were transcriptionally active, and could support active viral production upon TLR7/8 stimulation in vitro.
HIV-infected LN DCs containing replication-competent HIV are still detectable after 14 years of suppressive therapy
We next assessed the presence of HIV-infected DCs in ART-treated HIV-infected individuals. For this purpose, we FACS sorted LN resident and migratory DCs and total LN CD4 T cells from ART-treated aviremic HIV-infected individuals and performed a comprehensive virological assessment (Figure 6). Of note, due to the limited number of DCs obtained, assays were performed on distinct donors when mentioned (Table 1).
Figure 6. HIV-infected LN DCs containing replication-competent and infectious HIV are still detectable despite years of ART.

(A–N) LN resident and migratory DCs were isolated from treated aviremic HIV-infected individuals.
(A) Frequencies of LN DCs harboring integrated HIV DNA (cells per million) directly ex vivo (N = 4).
(B) Frequencies of LN DCs harboring cell-associated unspliced HIV gag RNA or multi-spliced tat-rev HIV RNA (cells per million) directly ex vivo (N = 3).
(C) Levels of HIV RNA (copies/mL) produced in culture supernatants of unstimulated VOA conditions (N = 3).
(D) Levels of HIV RNA (copies/mL) produced in culture supernatants of stimulated VOA conditions (N = 9).
(E) Proportion of HIV-infected individuals with detectable HIV RNA (≥20 HIV RNA copies/mL) in stimulated VOA conditions at day 14 (N = 9).
(F) Levels of HIV RNA (copies/mL) produced in the culture supernatants at day 14 in the stimulated VOA conditions (N = 9).
(G) Correlation between duration of ART (in years) and levels of HIV RNA (copies/mL) produced in the stimulated VOA culture supernatants at day 14 (N = 9).
(H–J) (H) Estimated decay slopes for HIV-RNA levels in stimulated VOA culture supernatants of LN CD4 T cells (H), LN migratory DCs (I), or LN resident DCs (J) with the duration of suppressive therapy.
(K) Proportion of HIV-infected individuals with detectable HIV-1 RNA (≥20 HIV RNA copies/mL) in LN cell populations at day 14 of the in vitro HIV-1 infection assay (N = 9).
(L) Levels of HIV RNA (copies/mL) in culture supernatants at day 14 in the in vitro infection assay (N = 9).
(M) Schematic representation of the QVOA preformed on LN resident and migratory DCs. HIV-infected individuals were considered as replicates at 40,000 cell concentration (N = 9).
(N) Estimated mean frequencies of LN resident and migratory DCs harboring inducible replication-competent virus/RNA units per million (RUPM) cells.
Dashed line represents the limit of detection (A–D), (F), and (L). Undetectable values were arbitrarily defined as 10 HIV RNA copies/mL (C), (D), (F–J), and (L). HIV-infected individuals were color coded (A), (B), (F), and (L). Histograms correspond to the estimated mean (E), (F), (K), (L), and (N). Red bars correspond to mean ± SEM (D), (F), and (L) or to lower and upper confidence interval at 0.95 (N). Red stars indicate statistical significance (*p < 0.05) (D–F) and (K). Statistical significance (p values) was obtained using one-way ANOVA (Kruskal-Wallis test) followed by Wilcoxon matched-pairs one-tailed signed rank test (D) and (F) or using Spearman rank test for correlations (G) or using chi-square test for proportion of positive individuals (E) and (K). ‘‘Estimate’’ refers to the decay slopes for LN CD4 T cells (H), LN migratory DCs (I), or LN resident DCs (J). Red color wells correspond to HIV-RNA-negative wells, whereas green wells correspond to HIV-RNA-positive wells (M). ‘‘Res DC’’ corresponds to LN resident DCs and ‘‘Mig DC’’ corresponds to LN migratory DCs.
The results showed that integrated HIV DNA was detected within LN CD4 T cells of all patients tested, with a mean integrated HIV DNA copy number 1,873/million cells, which is consistent with previous findings (Figure 6A).44 Interestingly, HIV-integrated DNA was detected in both LN migratory and resident DCs despite suppressive ART with a frequency of about 125 cells/million cells in LN resident DCs and about 300 cells/million cells in migratory DCs (Figure 6A).
As previously mentioned, we did not observe any contamination of the sorted DC subpopulations by CD4 T cells. Nevertheless, we estimated the theoretical consequences of such type of contamination on the frequency of resident or migratory DCs containing integrated HIV DNA, considering the unlikely event that all non-pure cells were CD4 T cells. Given the average purity of the DC subpopulations, i.e., 99.44% for LN resident DCs and 99.16% for migratory DCs, the 40,000 sorted LN resident or migratory DCs for the quantification of cells harboring integrated HIV DNA would therefore be contaminated with 224 and 336 ‘‘non-pure cells,’’ respectively. Based on the average frequency of HIV-infected CD4 T cells of treated HIV individuals, i.e., 1,873 cells/million, the potential number of sorted HIV-infected CD4 T cells would be of 0.42 and 0.63 cells among sorted LN resident or migratory DCs, respectively. Consequently, we can confidently state that if such a theoretical contamination were present, it would have limited consequences on the observations made.
In contrast to the data obtained in viremic HIV-infected individuals, unspliced gag or ms tat-rev HIV RNA was not detected in both LN migratory and resident DCs isolated directly ex vivo from ART-treated individuals (Figure 6B). Based on these observations, we evaluated the capacity of ex-vivo-isolated LN DC subpopulations from ART-treated individuals to de novo produce replication-competent viruses in vitro. To address this issue, we performed a viral outgrowth assay (VOA) on sorted LN migratory and resident DCs from ART-treated individuals. Briefly, LN DC subpopulations were either stimulated with TLR7/8 agonist, pulsed with SEB to favor virological synapse formation, and co-cultured with allogeneic pre-stimulated blood CD4 T cells isolated from uninfected donors (N = 9; referred to as ‘‘stimulated VOA’’) or, alternatively, left unstimulated and co-cultured with allogeneic pre-stimulated blood CD4 T cells isolated from HIV-uninfected donors (N = 3; referred to as ‘‘unstimulated VOA’’). To fully appreciate the dynamic of HIV infection within these cell populations, we included individuals fully suppressed for 6 months to 19 years (Table 1). As internal controls, LN CD4 T cells were stimulated with anti-CD3/anti-CD28 mAbs or TLR7/8 agonist (Figure S4). HIV RNA levels were assessed in the VOA culture supernatants at days 0, 5, and 14 as previously described.40,44
Importantly, and consistent with previous studies, HIV RNA was not detected in culture supernatants of CD4 T cells exposed to TLR7/8 agonist,45 whereas anti-CD3/CD28 mAbs stimulation of CD4 T cells consistently reactivated HIV production in all individuals tested (p < 0.05) (N = 5; Figure S4).
The cumulative data of levels of HIV RNA detected in the culture supernatants of DCs at various time points indicated that in the absence of DC stimulation, neither of the LN DCs were able to transmit HIV infection to activated CD4 T cells in vitro (p > 0.05) (Figure 6C). However, upon stimulation, HIV-1 RNA levels significantly increased in the culture supernatants of LN migratory DCs at days 5 and 14 compared with levels at day 0 and at day 14 for LN CD4 T cells compared with levels at day 0 (p < 0.05) (Figure 6D). These data suggested that in TLR7/8 stimulated conditions, the HIV RNA detected in the VOA culture supernatants likely resulted from viruses produced de novo from infected DCs and not from HIV virions adherent to LN DC processes.
Furthermore, the cumulative data of the TLR7/8 stimulated conditions at day 14 showed that inducible HIV RNA was more frequently detected within LN CD4 T cells (88% of the individuals tested) and in LN migratory DCs (77% of the individuals tested) compared with LN resident DCs (33% of the individuals tested) (Figure 6E). Moreover, the HIV-RNA levels were significantly higher in the culture supernatants of LN migratory DC and LN CD4 T cell supernatants compared with LN resident DCs (p < 0.05) (Figure 6F). Notably, no significant differences were observed between the HIV-RNA levels detected in the culture supernatants of LN migratory DC and CD4 T cells (p > 0.05) (Figure 6F). Interestingly, the virions produced in the culture supernatants of LN CD4 T cells, LN resident DCs and migratory DCs of 3 ART-treated HIV-infected individuals tested revealed a strong homology between HIV sequences obtained from LN CD4 T cells and LN migratory DCs (100%) and between LN CD4 T cells and LN resident DCs (99%) (Table S6) and did not harbor any drug resistance mutations in any of the individuals tested (Table S7).
We next determined the relationship between the duration of treatment and the levels of HIV RNA detected at day 14 in the VOA culture supernatants. Interestingly, a significant inverse correlation was observed between the duration of treatment and the levels of HIV RNA in the culture supernatants of LN migratory DCs (r = −0.87, p < 0.05) or with LN CD4 T cells (r = −0.67, p = 0.05) (Figure 6G). Of note, HIV RNA was consistently detected in culture supernatants of LN migratory DCs from HIV-infected individuals who were treated for up to 14 years and detectable in culture supernatant of LN resident DCs in one individual treated for up to 19 years. A logarithmic regression model with first order kinetics was applied to best fit HIV-RNA levels in LN migratory and resident DCs and LN CD4 T cells with duration of ART as previously described.46 Based on this analysis, the decay slopes indicated a decay rate of about −0.5 within LN migratory DCs, −0.3 within LN CD4 T cells, and 0.01 within LN resident DCs (Figures 6H–6J).
Additionally, to assess whether the produced virions were, indeed, infectious, we performed an in vitro HIV infection assay as previously described.40,44 The cumulative data showed the presence of HIV RNA in 44% of the culture supernatants of LN migratory DCs (Figure 6K), indicating that the virions produced from LN migratory DCs were frequently infectious despite years of ART. Infectious viruses were detected in stimulated culture supernatants of LN migratory DCs and CD4 T cells isolated from patients treated for up to 5 years (Figure 6L).
Finally, to estimate the frequencies of HIV-infected LN DCs containing inducible replication-competent virus, LN resident and migratory DCs were sorted from 6 aviremic ART-treated HIV-infected individuals (mean duration of suppressive therapy: 5.65 years) and subjected to a quantitative VOA (QVOA) with single-replicate-cell dilutions ranging from 3.75 × 105 to 4 × 103 cells/wells (Figure 6M). HIV RNA was assessed in the culture supernatants at day 14 as previously described.40 Of note, the data generated from the 9 individuals using the conventional VOA (4 × 104 cells; Figures 6D–6F) were also considered in the quantification wherein each individual was considered as a replicate (Figure 6M). Notably, we estimate that between 1.2 and 112 HIV-infected LN migratory DCs and between 0.5 and 47 LN resident DCs would contain HIV proviral DNA within the VOAs (Figures 6D–6F and 6K–6N). The frequencies of HIV-infected LN DCs containing inducible replication-competent virus were estimated by using the extreme limiting dilution assay as previously described (Figures 6M and 6N).40 These analyses provided the frequencies of cells harboring inducible replication-competent viruses as the average RNA units per million (RUPM) cells for each LN DC subpopulation in the 15 individuals studied.
The results indicated that the average RUPM frequencies of LN resident and migratory DCs harboring inducible replication-competent virus post-TLR7/8 agonist exposure reached 8 and 21 cells/million cells, respectively (Figure 6N). Taken together, the data demonstrated that LN resident and migratory DCs containing inducible replication-competent virus are detectable despite years of suppressive antiretroviral therapy and may therefore represent a yet untapped source of infectious HIV-1 in LN tissues in vivo.
DISCUSSION
The primary function of DCs is antigen presentation to T cells. Therefore, DCs may play a fundamental role in the reactivation of HIV replication following the stimulation of antigen-specific HIV-infected CD4 T cells after ART interruption. In this study, we postulated that LN DCs might be infected with inducible replication-competent HIV in vivo and may thus have an additional role in serving as source of HIV virions to infect uninfected CD4 T cells and amplifying viral replication. We therefore investigated the potential presence of HIV-infected LN DCs during HIV infection and under ART.
This study focused on a comprehensive transcriptomic combined with the phenotypic characterization of human LN resident and migratory DCs.22,47 We found >300 differentially expressed genes segregated LN resident from migratory DCs within LN tissues. Their functions were defined by divergent repertoires of integrins, cytokines/chemokine ligand, and receptor molecules tailored to counter various pathogen breaches, orchestrated by a set of unique transcription factors.
Moreover, we showed that LN DCs express the HIV receptor CD4 and co-receptors CCR5 and CXCR4, viral capture receptors, such as DC-SIGN and CD169 (Siglec-1)48–50 and were susceptible to HIV infection in vitro despite the detectable expression of SAMHD1. Furthermore, both LN DC subpopulations supported active cycles of de novo replication of CCR5-tropic HIV in vitro.
Using a comprehensive virological assessment of HIV-infected cells encompassing frequencies of cells integrated HIV-1 DNA, frequencies and copy number of cells with HIV-1 RNA, we observed a reduced proportion of latently infected DCs compared with Tfh cells in viremic HIV-infected individuals. Indeed, consistent with this observation, LN migratory and resident DCs isolated from viremic HIV-infected individuals harbored 55% and 40% genome-intact proviruses, respectively, that were integrated within active host transcription units. In addition, APOBEC3-mediated hypermutations within HIV genomes in LN DCs were infrequently detected. Interestingly, HIV-infected resident and migratory DCs were transcriptionally active directly ex vivo and could produce HIV virions in vitro upon TLR7/8 stimulation.
We next showed that LN DCs harboring integrated HIV DNA could still be detected in all ART-treated HIV-infected individuals analyzed. Although, we did not detect HIV transcriptional activity within LN DCs isolated directly ex vivo from ART-treated HIV-infected individuals, replication-competent virus was retrievable from LN CD4 T cells and LN migratory DCs despite years of suppressive ART (up to 14 years), in the absence of any detectable viral blip.
DCs are known for their endo-phagocytic capacities.51 These properties could influence the quantification of infected cells by qPCR in the event that a DC had endo-phagocytosed an HIV-infected CD4 T cell. However, this phenomenon would probably not influence the results generated by VOA because TLR7/8 stimulation acts on the genomic DNA and not on the potential endo-phagocytosed material. In addition, LN DCs subpopulations were stimulated with TLR7/8 agonist in this study. Although TLR7 and TLR8 are largely expressed by myeloid and plasmacytoid DCs (pDCs), B cells, monocytes, and macrophages, these receptors are poorly expressed by CD4 T cells.52 Consistent with TLR7 and/or TLR8 expression, TLR7/8 agonist did not induce direct HIV production from isolated CD4 T cells in vitro, whereas HIV RNA was consistently detected in TLR7/8-stimulated DC culture supernatants. Taken together, this body of evidence supports the fact that the viral RNA detected in TLR7/8-stimulated DC VOA culture supernatants originated from HIV-infected DCs.
Notably, SAMHD1 protein levels were significantly higher in LN resident DCs compared with migratory DCs of viremic HIV individuals. However, despite differences in SAMHD1 protein levels, both LN DC subpopulations harbored similar frequencies of intact virus in viremic HIV-infected individuals and similar levels of HIV RNA produced in DC culture supernatants, suggesting that SAMHD1 levels are probably not associated with the distinct HIV induction capacity of LN DCs in treated conditions. However, whether or not the distinct inducibility of HIV production between infected LN DCs and infected CD4 T cells depends on the mode of stimulation and/or other factors such as distinct proviral landscapes remains to be further investigated.
The decay slopes of the inducible reservoir indicated a decay rate of −0.5 within LN migratory DCs, −0.3 within LN CD4 T cells, and 0.01 within LN resident DCs. Notably, such decay curve may on one hand underestimate the inducible reservoir because the inducible reservoir as assessed by VOAs rely on the inducibility of the virus post-stimulation and on the other hand, overestimate the inducible reservoir because they do not factor in the contribution of the cellular and humoral immune responses that may influence the decay rates in vivo. Therefore, the contribution of distinct LN cell populations to viral rebound kinetics needs to be evaluated in future analytical treatment interruption studies.
Taken together, the virological findings indicate that LN DCs probably represent a yet underestimated cellular source of replication-competent virus, functionally capable of resuming active viral production upon stimulation. Additional investigations would be however required to fully delineate the role of DCs in long-term HIV persistence.
In this context, most clinical trials have evaluated the impact of latency reversing agents (LRAs) on mainly T cell reservoirs. Of note, the efficiency of histone deacetylase inhibitors (HDACis) to reactivate HIV replication from myeloid cells has been evaluated in few cell line models and has shown to alter activation states of myeloid cells under certain experimental conditions.53,54 However, whether or not HDACis may efficiently reactivate HIV replication from HIV-infected LN DCs needs to be further evaluated. In addition, it can be postulated that the effects of some of these LRAs on HIV-infected cells can be both direct and indirect (e.g., through cytokine secretion). Indeed, two independent rhesus macaque studies noted the ability of TLR7 agonist treatment to induce potent immune responses and transiently reactivate HIV viremia by a mechanism involving immune activation of pDCs and natural killer (NK) cells and the secretion of soluble factors in an interferon (IFN)-α-dependent manner.55 In addition, it has been proposed that tumor necrosis factor alpha (TNF-α), which is secreted by myeloid DCs subsequent to TLR8 triggering, may act in a paracrine manner in the activation of HIV from neighboring latently infected CD4 T cells, which do not express TLR8, thus revealing a potential indirect mechanism by which TLR8 triggering can promote viral replication.56 Furthermore, the presence of type I IFN (direct) and IL-15 (indirect) has been shown to induce DC activation in several cancer models.57,58 However, whether or not either of these stimuli may also act as a LRA and result in the reactivation of HIV replication from infected DCs remains to be evaluated in future studies.
The potential mechanisms associated with the detection of HIV-infected DCs despite ART remain to be elucidated but could be attributed to four main non-mutually exclusive phenomena: (1) the presence of low-level ongoing viral replication, (2) a long half-life of DCs, and/or proliferation of (3) pre-cDCs, or (4) mature DCs.
Although it is difficult to formally rule out the possibility of infrequent replication events either in reproductive organs and/or in LN tissues, no detectable evidence of HIV molecular evolution, indicative of replication, has been demonstrated till date during continuous therapy.59–62
The half-life of human LN DCs and the potential influence of DC maturation/activation on their lifespan remain unknown. However, the studies performed in mice indicated that the half-life of myeloid DCs (cDCs) in lymphoid and non-lymphoid tissues was estimated to range from days to weeks (between 2 and 14 days) before being replenished by newly produced DCs from pre-DCs through a process called DCpoeisis that involves pre-DC proliferation and differentiation.19 Interestingly, recent evidences highlighted that pre-DCs are susceptible to HIV infection of CCR5 and CXCR4-tropic strains mediated through SIGLEC-1-dependant capture mechanisms.49 Whether or not the persistence of HIV-infected DCs despite suppressive ART is associated with the infection and the proliferation of HIV-infected pre-cDCs remains, however, to be established. Alternately, LN DCs may survive longer than previously anticipated in the LN tissues potentially through the expression of anti-apoptotic/cell survival molecules, such as BIRC3 and/or BIRC5, as previously proposed for CD4 T cells, but it remains to be formally demonstrated.9
Taken together, this study demonstrates that LN DCs harboring inducible replication-competent HIV are detectable despite years of suppressive ART. These observations may have major implications in the design of immunological strategies aiming to achieve a functional cure of HIV infection.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthieu Perreau (matthieu.perreau@chuv.ch).
Materials availability
This study did not generate unique reagents.
Data and code availability
RNA-Seq data was deposited to Gene Expression Omnibus (GEO) with the following accession numbers GEO: GSE196066, which can also be found in the key resources table.
Near-full length HIV proviral sequences obtained from LN DC subpopulations and CD4 T cells were deposited to GenBank with the following accession numbers: OM795243-291, which can also be found in the key resources table.
Protease/reverse transcriptase and Integrase sequences from HIV virion-associated RNA were deposited in the Sequence Read Archive (SRA) database with the following accession number: SMN35564308-315, which can also be found in the key resources table.
Patient derived HIV proviral sequences: Due to study participant confidentiality concerns, viral sequencing data cannot be publicly released, but will be made available to investigators upon reasonable request and after signing a data transfer agreement.
This paper does not report original code.
Publicly available software and code used in this study are listed in the key resources table. Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| LNMC samples from study participants living with HIV | This study, Centre hospitalier universitaire vaudois (CHUV), Lausanne Switzerland | N/A |
|
| ||
| Flow Cytometry | ||
|
| ||
| APC-H7-conjugated mouse anti-human anti-HLA-DR (G46-6) | Becton Dickinson | 561358 |
| APC conjugated mouse anti-human anti-CD4 (RPA-T4) | Becton Dickinson | 555349 |
| ECD-conjugated mouse anti-human anti-CD3 (UCHT1) | Beckman Coulter | A07748 |
| PE-Cy7-conjugated mouse anti-human anti-CD11c (B-ly6) | Becton Dickinson | 561356 |
| PerCP-Cy5.5 conjugated mouse anti-human anti-CD45 (H-I30) | Becton Dickinson | 332784 |
| Alexa Fluor 700 conjugated mouse anti-human anti-CCR7 (150503) | Becton Dickinson | 561143 |
| FITC-conjugated mouse anti-human anti-CD20 (clone 2H7). | Becton Dickinson | 555622 |
| BV421 conjugated mouse anti-human CD279 (EH12) | Becton Dickinson | 565935 |
| PerCP-Cy5.5 conjugated mouse anti-human anti-CD8 (SK1) | Becton Dickinson | 341050 |
| PE-CF594-conjugated mouse anti-human anti-CD45RA (HI100) | Becton Dickinson | 562298 |
|
| ||
| Mass Cytometry | ||
|
| ||
| Ultra-LEAF™ Purified anti-human CD8a (RPA-T8) | Biolegend | 301073 |
| Ultra-LEAF™ Purified anti-human CD4 (RPA-T4) | Biolegend | 300570 |
| Purified anti-human CD45 Antibody (HI30) | Biolegend | 304002 |
| Purified anti-human CD19 (HIB19) | Biolegend | 302202 |
| Purified anti-human/mouse/rat CD278 (ICOS) (C398.4A) | Biolegend | 313502 |
| Purified IRF4 (IRF4.3E4) | Biolegend | 646402 |
| Purified anti-human CCR5 (J418F1) | Biolegend | 359102 |
| SAMHD1 antibody (AA 426-657) (FITC) | Antibodies-online.com | ABIN7177151 |
| Purified anti-human CD163 (GHI/61) | Biolegend | 3145010B |
| Purified anti-human IgD (IA6-2) | Biolegend | 348202 |
| Purified anti-human BIRC3 (C-IAP2) | Biolegend | 667702 |
| Anti-Human CD274/PD-L1 (29E.2A3)-148Nd | Fluidigm | 3148017B |
| Anti-Human CD127/IL-7Ra (A019D5)-149Sm | Fluidigm | 3149011B |
| Anti-Human CD134/OX40 (ACT35)-150Nd | Fluidigm | 3150023B |
| Purified anti-human CD123 (6H6) | Biolegend | 306002 |
| Purified anti-human CD21 (Bu32) | Biolegend | 354902 |
| Purified anti-human DC-SIGN (9E9A8) | Biolegend | 330102 |
| Anti-Human CD27 (L128)-155Gd | Fluidigm | 3155001B |
| Anti-Human CD86/B7.2 (IT2.2)-156Gd | Fluidigm | 3156008B |
| Anti-Human CD169 (7-239)-158Gd | Fluidigm | 3158027B |
| Anti-Human CD197/CCR7 (G043H7)-159Tb | Fluidigm | 3159003A |
| Anti-Human CD14 (M5E2)-160Gd | Fluidigm | 3160001B |
| Purified anti-human CD1c (L161) | Biolegend | 331502 |
| Anti-Human CD11c (Bu15)-162Dy | Fluidigm | 3162005B |
| Purified anti-human CD85j (GHI/75) | Biolegend | 333702 |
| APC-conjugated anti-IRF8 (V3GYWCH) | Invitrogen | 17-9852-82 |
| Anti-APC (APC003)-163Dy | Fluidigm | 3163001B |
| Anti-FITC (FIT-22)-144Nd | Fluidigm | 3144006B |
| Anti-Human CD185/CXCR5 (RF8B2)-164 | Fluidigm | 3164029B |
| Anti-Human CD40 (5C3)-165Ho | Fluidigm | 3165005B |
| Purified anti-human CD155 (PVR) (SKII.4) | Biolegend | 337602 |
| Purified anti-human CD38 (HIT2) | Biolegend | 303502 |
| Anti-Human CD45RA (HI100)-169Tm | Fluidigm | 3169008B |
| Anti-Human CD3 (UCHT1)-170Er | Fluidigm | 3170001B |
| Anti-Human CD273/PDL2 (24F.10C12)-172Yb | Fluidigm | 3172014B |
| Anti-Human CD184/CXCR4 (12G5)-173Yb | Fluidigm | 3173001B |
| Purified anti-human HLA-DR (L243) | Biolegend | 307602 |
| Purified anti-human PD-1(EH12.2H7) | Biolegend | 329925 |
| Anti-Human CD11b/Mac-1 (ICRF44)-209Bi | Fluidigm | 3209003B |
| Purified NA/LE Mouse Anti-Human CD3 | Becton Dickinson | 555329 |
| Purified Mouse Anti-Human CD28 | Becton Dickinson | 555726 |
|
| ||
| Virus strainss | ||
|
| ||
| Human Immunodeficiency Virus Type 1 (HIV-1) BaL | HIV reagent program | 510 |
| Human Immunodeficiency Virus Type 1 (HIV-1)-CCR5 EGFP | Gift From Nicola Manel | Ref. 68; Manel et al.63 |
| Human Immunodeficiency Virus Type 1 (HIV-1)-CXCR4 EGFP | Gift From Nicola Manel | Ref. 68; Manel et al.63 |
| VpX Lentivirus | Gift from Fabio Condotti | Bobadilla et al. (2013).64 Gene Therapy |
|
| ||
| Reagents, chemicals, peptides, recombinant proteins | ||
|
| ||
| HIV peptides | JPT Peptide Technology | Customized |
| TLR-7/8 agonist | InvivoGen | CL264 |
| SEB | Sigma-Aldrich | S4881 |
| Human IL-2 IS, research grade | Miltenyi | 130-097-743 |
| Emtricitabine | NIH Reagents Program | HRP-10071 |
| Thymidine, [Methyl-3H], in 2% ethanol, 1 mCi | perkinelmer | NET027W001MC |
|
| ||
| Critical commercial assays | ||
|
| ||
| Single cell RNAseq (Seq-well; refer to Gierahn et al.36) | ||
| polydimethylsiloxane (PDMS) | ChemGenes Corporation, Wilmington, MA, USA | #MACOSKO201110 |
| polycarbonate membrane | Sterlitech Corporation, Auburn, WA, USA | #PCT00162X22100 |
| barcoded with mRNA capturing beads | ChemGenes Corporation, Wilmington, MA, USA). | #MACOSKO201110 |
| cDNA library preparation - Nextera XT DNA Library Preparation Kit) | Illumina, Inc., San Diego, CA, USA | #FC-131-1024 |
| NGS HIV resistance testing (Sentosa SQ HIV Genotyping Assay) | Vela Diagnostics, Hamburg | |
| DNeasy Blood and Tissue Kit | Qiagen | 69504 |
| MIPseq protocol | Refer to Einkauf et al.38 | |
| EasySep Human CD8+ Tcell IsolationKit | Stemcell | 17953 |
| EasySep™ Human CD4+ T Cell Isolation Kit | Stem Cell | 17952 |
| Human IFNγ ELISpotPRO assay KIT | MABTECH | 3420-2APW-2 |
| Cobas 6800 | Cobas HIV-1; Roche Diagnostics AG | KIT COBAS 6800/8800 HIV 96T IVD; Référence : 7000995190 |
| HIV-1 P24 Electrochemiluminiscence assay | Elecsys HIV Duo de Roche | 08 836 973 190 (300 tests) |
| IFNγ ELISpot assay KIT | MABTECH | 3420-2APW-2 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Invitrogen | L34957 |
|
| ||
| Software and algorithms | ||
|
| ||
| STAR aligner software | ENCODE, version 2.7 | https://www.encodeproject.org/software/star/ |
| R package | R Core Team and R Foundation for Statistical Computing, version 4.0.2 | https://www.r-project.org |
| Seurat package for R (v.4.0.0) | ||
| DropEst (version 0.8.5) | ||
| SmartGene ASP-IDNS®−5 pipeline for NGS data using the ‘‘Provirus Pipeline’’ version 2.0.5_HIV1_v1.4 | ||
| ANRS algorithm | http://www.hivfrenchresistance.org | |
| MIPseq data analysis | Refer to Einkauf et al.38 | N/A |
| FlowJo 10 | FlowJo, LLC | https://www.flowjo.com |
| Graphpad Prism 9 | Graphpad | https://graphpad.com |
| Cytobank software package | Cytobank | https://premium.cytobank.org/cytobank/ |
| Prism | Graphpad, version 8.2.1 | https://www.graphpad.com/scientific-software/prism |
|
| ||
| Sequence | ||
|
| ||
| RNAseq sequences for LN DCs | Gene Expression Omnibus (GEO) | GSE196066 |
| Near-full length proviral sequences obtained from LN DC subpopulations and CD4 T cells | GenBank | OM795243-OM795291. |
| Protease/reverse transcriptase and Integrase sequences from HIV virion-associated RNA | Sequence Read Archive (SRA) database | SMN35564308-315 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Study participants
Forty two HIV-1 infected adult volunteers and fourteen HIV-uninfected individuals were enrolled in the present study. HIV-1-infected study participants were recruited at the Centre hospitalier universitaire vaudois (CHUV) Hospital, Lausanne, Switzerland. Clinical characteristics of the enrolled individuals are depicted in Table 1. No statistical method was used to predetermine sample size. The influence of gender was not measured in this study. Treated individuals received ART for 6 months to 19 years (Table 1). No exclusion criteria was implemented except with regards to the isolation of inducible replication competent virus from LN DCs, which were exclusively performed on cells isolated from treated aviremic HIV-1 infected individuals with undetectable viremia (HIV RNA levels <50 copies per ml of plasma) for ≥6 months.
Study approval
Cell samples collected from HIV-uninfected and HIV-infected individuals were collected in a prospective study approved by the La Commission cantonale d’éthique de la recherche sur l’être humain (CER-VD) (#2018–01932) and all subjects provided a written informed consent.
METHOD DETAILS
Cell isolation
Inguinal lymph node biopsies were collected and lymph node mononuclear cells were isolated by mechanical disruption as previously described.65 Lymph node mononuclear cells were cryopreserved in liquid nitrogen for long-term storage. Total LNMCs isolated for each individual are shown in Table 1.
Cell culture
Cells were cultured in RPMI (Gibco; Life Technologies) containing 10% heat-inactivated FBS (Institut de Biotechnologies Jacques Boy), 100 IU/ml penicillin and 100 μg/ml streptomycin (Bio Concept).
Antibodies
The following antibodies were used for sorting experiments: APC-H7-conjugated anti-HLA-DR (clone G46-6), APC conjugated anti-CD4 (clone RPA-T4), ECD-conjugated anti-CD3 (clone UCHT1), PE-Cy7-conjugated anti-CD11c (clone B-ly6), PerCP-Cy5.5 conjugated anti-CD45 (H-I30), Alexa Fluor 700 conjugated anti-CCR7 (150503), FITC-conjugated anti-CD20 (clone 2H7) and BV421 conjugated anti-PD-1 (EH12). All antibodies including purified coating anti-CD3 (clone UCHT1) and anti-CD28 (clone CD28.2) mAbs were purchased from BD (Becton, Dickinson; CA, USA). The following antibodies were used for mass cytometry experiments: 113In-conjugated anti-CD8 (RPA-T8), 115In-conjugated anti-CD4 (RPA-T4), 141Pr-conjugated anti-CD45 (HI30), 142nd-conjugated anti-CD19 (HIB19), 143Nd-conjugated anti-ICOS (C398.4A) or anti-IRF4 (IRF4.3E4), 144Nd-conjugated anti-CCR5 (J418F1) or anti-FITC to detect FITC-conjugated anti-SAMHD1 (AA256-370); 145Nd-conjugated anti-CD163 (GHI/61), 146Nd-conjugated anti-IgD (IA6-2) or anti-BIRC3 (C-IAP2), 148Sm-conjugated anti-PD-L1 (29E.2A3), 149Sm-conjugated anti-CD127 (A019D5), 150Nd-conjugated anti-OX40 (ACT-35), 151Eu-conjugated anti-CD123 (6H6), 152Sm-conjugated anti-CD21 (BL13), 153Eu-conjugated anti-OX-40L (11C3.1), 154Sm-conjugated anti-DC-SIGN (9E9A8), 155Gd-conjugated anti-CD27 (L128), 156Gd-conjugated anti-CD86 (IT2.2), 158Gd-conjugated anti-CD169 (7–239), 159Tb-conjugated anti-CCR7 (G043H7), 160Gd-conjugated anti-CD14 (M5E2), 161Dy-conjugated anti-CD1c (L161), 162Dy-conjugated anti-CD11c (Bu15), 163Dy-conjugated anti-CD85j (GHI/75) or anti-APC to detect APC-conjugated anti-IRF8 (7G11A45), 164Dy-conjugated anti-CXCR5 (51505), 165Ho-conjugated anti-CD40 (5C3), 166Er-conjugated anti-CD155 (SKII.4), 167Er-conjugated anti-CD38 (HIT2), 168Er conjugated IgG1 isotype control, 169Yb-conjugated anti-CD45RA (HI100), 170Er-conjugated anti-CD3 (UCHT1), 172Yb-conjugated anti-PD-L2 (24F.10C12), 173Yb-conjugated anti-CXCR4 (12G5), 174Yb-conjugated anti-HLA-DR (L243), 175Lu-conjugated anti-PD-1(EH12.2H7); 209Bi-conjugated anti-CD11b (ICRF44) and 191Ir was used to label DNA. Purified antibodies were either purchased from Biolegend and metal-conjugated or were purchased from Fluidigm/DVS.
Sorting of LN DC sub-populations
Cryopreserved lymph node mononuclear cells were thawed and then stained with Aqua LIVE/DEAD stain kit (4°C; 15 min) and then with a cocktail of anti-CD3 ECD, anti-CD4 APC, anti-CD45 PerCp-Cy5.5, anti-PD-1 PB, anti-HLA-DR APC-H7, anti-CD11c PeCy7 and anti-CCR7 AF700 antibodies (4°C; 25 min). Viable LN migratory and LN resident DC subpopulations were sorted using FACSAria (Beckton & Dickinson). In some experiments, LN PD-1+/Tfh or PD-1− and/or total LN CD4 T cells were also sorted. In all sorting experiments the grade of purity of the sorted cell populations was R 98%.
Mass cytometry
Freshly isolated inguinal lymph node mononuclear cells from healthy HIV uninfected, viremic and ART treated HIV-infected individuals were resuspended (106 cells/ml) in complete RPMI medium, barcoded with anti-CD45 metal-conjugated antibodies and incubated (20 min; 4 °C). Cells were washed, fixed with 2.4% PFA (10 min; room temperature), pooled and then incubated with metal-conjugated antibodies directed against a panel of 38 surface parameters including lineage markers for T-cell, B-cell and DCs (20 min; 4 °C). Cells were washed, fixed with 2.4% PFA (10 min; room temperature). In some experiments, after surface staining, cells were permeabilized for 1 h at 4°C with Foxp3 Fixation/Permeabilization kit (eBioscience). Cells were washed and stained with APC-conjugated anti-IRF8, 143Nd conjugated-anti-IRF4, FITC-conjugated anti-SAMHD1 and 146Nd conjugated anti-BIRC3 (20 min; 4 °C). FMO controls with no intracellular antibodies was consistently performed. Cells were washed, incubated with 144Nd-conjugated anti-FITC and with 152Sm-conjugated anti-APC mAbs (20 min; 4 °C) and fixed with 2.4% PFA (10 min; room temperature). Total cells were identified by DNA intercalation (1μM Cell-ID Intercalator, Fluidigm/DVS Science) in 2% PFA at 4 °C overnight. Labeled samples were acquired on a CyTOF1 instrument that was upgraded to CyTOF2 (Fluidigm) using a flow rate of 0.045 ml/min. Data were analyzed using Fluidigm Cytobank software package (Cytobank, Mountain View, CA) or using FlowJo version 10.7.1. At least 300,000 events were acquired for each sample.
Single cell RNAseq
LN resident and LN migratory DCs were FACS sorted (N = 20,000 cells per population) on the basis of CCR7 expression from three ART treated HIV-infected individuals (N=3) and subjected to single cell RNAseq approach i.e Seq-Well as previously described.36,66 Briefly, sorted DCs were loaded onto a functionalized-polydimethylsiloxane (PDMS) array pre-loaded with uniquely barcoded with mRNA capturing beads (#MACOSKO201110, ChemGenes Corporation, Wilmington, MA, USA). Cell loading was followed by the reversible sealing of the array using a functionalized semipermeable polycarbonate membrane (#PCT00162X22100, Sterlitech Corporation, Auburn, WA, USA). Cell lysis through the semi-permeable membrane was followed by the hybridization of the released cellular mRNA to the bead-bound poly(dT) oligonucleotides that also contain a universal ‘PCR handle’ primer sequence, a cell barcode and a unique molecular identifier (UMI). Subsequently, the beads were removed from the array and pooled for reverse transcription, Exonuclease I treatment, second-strand synthesis, and whole transcriptome amplification (WTA). For each array sample, 1 ng of purified WTA product was used for cDNA library preparation (Nextera XT DNA Library Preparation Kit, #FC-131-1024, Illumina, Inc., San Diego, CA, USA), using 1 μL of Custom P5 hybrid oligo and 1 μL of Nextera N700 Index oligo. Paired-end sequencing was performed on Illumina NextSeq 500 sequencer with an Illumina NextSeq 500/550 High Output Kit v2.5 (75 Cycles) at a final concentration of 2.6 pM. Four Seq-Well arrays were sequenced per NextSeq500 sequencing run. Raw sequencing reads were converted from bcl files to FASTQs using bcl2fastq and demultiplexed according to the Nextera N700 indices that corresponded to individual samples. For each sample, droptag (version 0.8.5) was used to generate tagged fastq files for alignment. STAR (version 2.7.0) was used for alignments against the human reference genome hg38 (ENCODE release27, primary assembly)67 and DropEst (version 0.8.5) was subsequently used to generate the count matrices.68 Count matrices were pre-filtered on the basis of transcript detection (≥750 and % 4000 UMIs per cell) for inclusion in downstream analyses. For further processing, the counts matrices were imported into ≤ (v.4.0.2) and subsequently analyzed using the Seurat package69 for R (v.4.0.0). The count matrix was filtered to retain cells that had less than 20% of mitochondrial gene counts. Genes not detected in at least 5 cells were removed. The counts were log-normalized, integrated and scaled using the NormalizeData, IntegrateData and ScaleData functions implemented in the Seurat package. Most variable genes detection and principal component (PC) reduction were performed using the FindVariableFeatures and RunPCA functions using default parameters. Cell clustering of all cells was performed using the shared nearest neighbor modularity optimization–based algorithm implemented in the FindNeighbors and FindClusters functions, using 20 PCs and a 1.5 resolution. Uniform manifold approximation and projection (UMAP) dimensionality reduction was performed using the RunUMAP function of the Seurat package with parameters reduction = ‘‘pca’’ and 20 PCs. Genes differentially expressed between LN resident and LN migratory DC sub-populations were determined using the FindMarkers function, which performs a Wilcoxon Rank Sum test on genes with over 0.25-fold difference, on log-scale, between the two clusters, with parameters min.pct = 0.25 and only.pos = TRUE. In addition, LN resident and migratory DC were scored as cDC1/2 and DC3 subsets based on CLEC9A-expression (cDC1); CD1c+ MRC1− (cDC2) and CD1c+MRC1+ (DC3).17,18
Data Availability Statement
Raw and processed RNAseq data are available on the gene expression omnibus (GEO) in GSE196066.
IFNγ ELISpot assay
Cryo-preserved blood mononuclear cells were rested for 6 h at 37°C, and then 200,000 cells were stimulated or not with HIV immunodominant peptides (1 μg of each single peptide) in 100 μl of complete media (RPMI plus 10% FBS) in tripli#e conditions. 200 ng/ml of staphylococcal enterotoxin B (SEB) was used as a positive control on 50,000 cells. Results were expressed as the mean number of SFU/106 cells. Responses exceeding 55 SFU/106 and with a 5-fold increase as compared to unstimulated conditions were scored as positive.
DC-T cell stimulation capacity
The capacity of LN DC subpopulations to stimulate T cell activation was assessed using two strategies; 1) via a mixed leukocyte reaction (MLR) wherein sorted viable LN DC sub-populations (eight, two-fold serial dilutions from 2.104-156 cells) from HIV uninfected individuals (N=3) were mixed with CD4 T cells from allogeneic donors (105 cells) for 5 days at 37°C and 5% CO2 in 96-well U-bottom plates in complete RPMI media in a total volume of 120μl per well. In parallel, unstimulated CD4 T cells and CD4 T cells stimulated with anti-CD3/anti-CD28 mAbs (eight, two-fold serial dilutions from 50 μg/mL-0.39 μg/mL) were used as controls. 1 μCi/well of [3H]-thymidine was added at day 4 to the cultures and proliferation of T cells was assessed by [3H]-Thymidine incorporation after 18 hours with a scintillation counter. The results were plotted as [3H]-Thymidine incorporation in counts per min (cpm); and 2) sorted viable LN DC sub-populations (2.104 cells) from ART treated HIV-infected individuals (N=3) were pulsed with HIV peptides and co-cultured with sorted autologous memory CD8 T cells in a 1:10 ratio for 5 days at 37°C and 5% CO2 in 96-well U-bottom plates in complete RPMI media in a total volume of 200μl per well. As control, autologous memory CD8 T cells were cultured alone or co-cultured with blood monocytes pulsed with HIV peptides. 1 μCi/well of [3H]-thymidine was added at day 4 to the cultures and proliferation of HIV-specific memory CD8 T cells was assessed by [3H]-Thymidine incorporation after 18 hours with a scintillation counter. The results were plotted as stiulation index (each response compared to CD8 T cells cultured alone).
Preparation of high-titer purified HIV-1 virus stocks
The CCR5-tropic and CXCR4-tropic HIV-derived vector encoding for EGFP (HIVR5GFP and HIVX4GFP) and were obtained as a kind gift from Nicola Manel.63 The proviral plasmid HIV R5GFP and X4GFP were derived from NL4-3 with Ba-L env, ∆nef, and GFP in nef.63 Both constructs were amplified as previously described.70 Briefly, viral particles were prepared by transfection of 293 T cells. For this purpose, 10–12 million 293 T cells were seeded per 15cm tissue culture dish in DMEM media with 10% FBS and Penicillin-Streptomicin, Gentamicin (50 μg/ml, GIBCO). The next day, cells were transfected with 70 μg of plasmid DNA, through Calcium phosphate transfection (Takara) as previously described.70 The next day, media was replaced with DMEM with 10% FCS. After 2 days, media was filtered at 0.45 μM, collected into 38.5mL ultraclear centrifuge tubes (Herolab 253050), centrifuged at 50,000 g for 2 hours at 16°C. Virus pellet was re-suspended in 1 mL PBS, aliquoted and snap frozen in dry ice and then at −80°C for later use. Viral titers were determined by the assessment of HIV P24 antigen by ECL COBAS HIV-1 Ag (Roche; Switzerland), HIV RNA by COBAS HIV-1 Test (Roche; Switzerland; copies/mL) and the percentage of EGFP+ CD4 T cells with increasing virus doses at day 4 post-transduction by flow cytometry. Endotoxin levels were below the detection limit (Limulus amebocyte lysate assay; Sigma).
Susceptibility of LN DCs to HIV infection
Sorted viable LN DC sub-populations (5.104) from HIV uninfected individuals were pelleted and resuspended either in complete RPMI media or in dilutions of CCR5-tropic (N=12) or CXCR4-tropic (N=7) HIV-derived vector encoding for EGFP (CCR5-tropic: 43.6pg of P24; CXCR4-tropic: 50pg of P24) in the presence or absence of HIV-2 protein i.e. Vpx-containing lentivectors with the indicated dose in a final volume of 50μl (individuals performed for Vpx N=9). Of note, activated purified CD4 T cells from the same individuals were used as internal controls. 24 hours post-exposure, cultures were supplemented with fresh complete 10% RPMI media. Finally, cells were washed with PBS, stained for live cells, fixed and assessed for GFP expression by flow cytometry on day 4.
Integrated HIV DNA
LN DC sub-populations were FACS sorted from viremic (2.104-10.104 cells; N=5) or ART-treated HIV-infected individuals (40–65.103 cells; N=4). In parallel, autologous LN PD-1−/non-Tfh and Tfh cells were sorted from viremic HIV-infected individuals and total LN CD4 T cells were sorted from ART treated HIV-infected individuals as internal controls. Cell populations were lysed using lysis buffer (10 mM Tris-HCl, pH 8.0, 50 nM KCl, 400 μg/ml proteinase K; Invitrogen) and integrated HIV DNA and CD3 gene copy numbers were determined using a cross-clade ultrasensitive nested Alu PCR, as previously described.71 The frequency of cells harboring integrated HIV DNA per million cells was calculated as previously described.71
Quantification of cell-associated HIV RNA
Cell-associated HIV RNA from individual samples was extracted from sorted LN DC subpopulations for the quantification of total cell-associated gag HIV RNA from viremic HIV-infected individuals (2.104 cells; N=5). LN Tfh, LN PD-1−/non-Tfh and total CD4 T cells sorted from the same individuals were used as internal controls. Quantification of total cell-associated gag HIV-RNA was performed as previously described.40 For the estimation of frequencies of cells containing gag and tat-rev HIV-RNA, a limiting dilution assay was performed.72 Briefly, cell-associated HIV RNA was extracted from different cell concentrations (2-fold limiting dilutions of 103, 500, 250, 175, 87 and 43 cells and 10 replicates per condition; total 2.104 cells) of sorted LN DC subpopulations from viremic HIV-infected individuals (N=5). On the other hand, cell-associated HIV RNA was extracted from different cell concentrations (2-fold limiting dilutions of 1.103, 5.102, 250, 100, 50 cells and 20 replicates per condition; total 4.104 cells) of sorted LN DC subpopulations from aviremic ART treated HIV-infected individuals (N=3). In parallel, autologous LN PD-1−/non-Tfh and Tfh cells were sorted from viremic HIV-infected individuals and total LN CD4 T cells were sorted from ART treated HIV-infected individuals as internal controls. Extracted cell-associated HIV-1 RNA was subjected to DNase treatment (all from RNAqueous-4PCR Kit, Ambion). RNA standard curves were generated after isolation and quantification of viral RNA from supernatant of ACH2 cultures.73 One-step cDNA synthesis and pre-amplification were performed by using the following primers for gag: ULF1: 5′-ATGCCACGTAAGCGAAACTCTGGGTCTCTCTDGTTAGAC-3′; UR1: 5′-CCATCTCTCTCCTTCTAGC-3′ and for tat-rev: tat1.4 (outer nested): 5-TGG CAG GAA GAA GCG GAG A −3; reverse: 5- GGATCT GTC TCT GTC TCT CTC TCC ACC −3 as previously described.72 Real-time PCR was performed using a Roche Light Cycler 480II with the following primers for gag: LambdaT: 5′-ATGCCACGTAAGCGAAACT-3′; UR2: 5′-CTGAGGGATCTCTAGTTACC-3′; and probes: 56-FAM 5′-CACTCAAGG/ZEN/CAAGCTTTATTGAGGC-3′ IABkFQ23.71 And for tat/rev: tat2 (inner nested):5- ACA GTC AGA CTC ATC AAGTTT CTC TAT CAA AGC A −3; reverse: 5- GGATCT GTC TCT GTC TCT CTC TCC ACC −3; MS-HIV-FAMZEN; 5- /56-FAM/TTC CTT CGG /ZEN/GCC TGT CGG GTC CC/3IABkFQ/ −3. The results were expressed in cells expressing HIV gag RNA or tat-rev RNA per million cells.
LN DC HIV production
HIV production from LN DCs was assessed using two distinct strategies: first, sorted viable LN DC subpopulations (4.104) from HIV uninfected individuals (N=3) were exposed or not to HIV lab-derived variant i.e. HIV Ba-L (NIH AIDS reagent program; #510) at 10pg P24 in a final volume of 50μl (N=3). In parallel, total CD4 T cells (4.104) were sorted as internal controls. CD4 T cells were either exposed to the same concentration of HIV Ba-L directly ex vivo or after activation with anti-CD3/anti-CD28 mAbs for 3 days. 24 hours post-exposure to HIV Ba-L, cultures were washed thrice with complete RPMI media and resuspended in a final volume of 100μl in fresh complete 10% RPMI media. HIV production was assessed in the culture supernatants by the measurement of levels of HIV RNA by COBAS HIV-1 Test (Roche; Switzerland; copies/mL) following a 1/100 medium dilution in basement matrix buffer (Ruwag Handels AG) of all culture supernatants on day 0, 3, 5, 9 and 12 as previously described.40 Wells with detectable HIV-1 RNA (≥ 2000 HIV-1 RNA copies/ml) were referred to as HIV-1 RNA-positive wells. As a second strategy: sorted viable LN DC subpopulations (5.104) from viremic HIV-infected individuals (N=6) were exposed or not to TLR-7/8 agonist (1μg/mL). In parallel, autologous LN PD-1+/Tfh and LN PD-1−/non-Tfh cells (5.104) were sorted as internal controls and exposed or not to coated anti-CD3/anti-CD28 mAbs stimulation as previously described.24,43 Notably, in this experimental settings - no target cells were added to the cell culture. HIV production was assessed by the measurement of levels of HIV RNA by COBAS HIV-1 Test (Roche; Switzerland; copies/mL) following 1/50 medium dilution in basement matrix buffer; or by HIV P24 (Electrochemiluminiscence assay; pg/mL) detection in DC culture supernatants on day 7 as previously described.40 Wells with detectable HIV-1 RNA (≥ 1000 HIV-1 RNA copies/ml) were referred to as HIV-1 RNA-positive wells. Wells with detectable HIV-1 P24 (≥ 1000 pg/mL) were referred to as HIV-1 P24-positive wells.
LN DC HIV replication
Sorted viable LN DC subpopulations (4.104) isolated from HIV-uninfected individuals (N=3) were exposed to CCR5-tropic replication competent HIV-1 lab-derived variant i.e. HIV Ba-L (#510 from NIH AIDS reagent program) at 10pg P24 for 36 hours in a final volume of 100μl. In parallel, total CD4 T cells (4.104) were sorted as internal controls. CD4 T cells were exposed to the same concentration of HIV Ba-L directly ex vivo or after activation with anti-CD3/anti-CD28 mAbs for 3 days. 36 hours post-exposure to HIV Ba-L, LN DC subpopulations and CD4 T cells were washed thrice with complete RPMI media, and cells were cultured for additional 9 days in the presence or absence of reverse transcriptase inhibitor i.e Emtricitabine (75nM). HIV production was assessed in the culture supernatants by the measurement of levels of HIV RNA by COBAS HIV-1 Test (Roche; Switzerland; copies/mL) following a 1/100 medium dilution in basement matrix buffer (Ruwag Handels AG) of all culture supernatants on day 0, 2, 4, 6 and 9 as previously described.40 Wells with detectable HIV-1 RNA (≥ 2000 HIV-1 RNA copies/ml) were referred to as HIV-1 RNA-positive wells.
LN DC Viral outgrowth assay (VOA)
Viable LN DC subpopulations (4.104) were sorted from ART treated HIV-infected individuals (N=9). In some conditions, LN DC subpopulations were left unstimulated, and co-cultured with allogeneic pre-stimulated blood CD4 T cells isolated from healthy donors (‘‘unstimulated settings’’). Alternatively, sorted LN DC subpopulations were pulsed with SEB (250ng/mL) in order to mimic the infection spread through a virological synapse, and co-cultured with allogeneic pre-stimulated blood CD4 T cells isolated from healthy donors in the presence of TLR-7/8 agonist (R848; 1μg/mL) (‘‘stimulated settings’’). Total LN CD4 T cells stimulated with anti-CD3/CD28 mAbs for 3 days were used as internal controls in both unstimulated and stimulated VOA settings. IL-2 (10 units/mL) was added to all cultures. Pre-stimulated blood CD4 T cells were re-added to all cultures at day 4, 7 and 11 in both unstimulated and stimulated VOA settings. HIV production was assessed in the undiluted culture supernatants of the DC VOA by the measurement of levels of HIV RNA by COBAS HIV-1 Test (Roche; Switzerland; copies/mL) at days 0, 5 and 14 in both unstimulated and stimulated VOA settings as previously described.40 Wells with detectable HIV-1 RNA (≥20 HIV-1 RNA copies/ml) were referred to as HIV-1 RNA-positive wells. with detectable HIV-1 RNA (≥20 HIV-1 RNA copies/ml) were referred to as HIV-1 RNA-positive wells. In some experiments, purified LN total CD4 T cells (106) were stimulated either with TLR7/8 agonist (R848; 1μg/mL) or with anti-CD3/CD28 mAbs for 3 days (N=5) and HIV production was assessed in 1/10 diluted culture supernatants by the measurement of levels of HIV RNA as described above at days 0, 5 and 14. Wells with detectable HIV-1 RNA (≥200 HIV-1 RNA copies/ml) were referred to as HIV-1 RNA-positive wells. For the estimation of frequencies of HIV-infected LN DCs, a quantitative viral outgrowth assay (Q-VOA) was performed using viable sorted LN DC subpopulations at single-replicate cell dilutions ranging from 3.75.105 to 4.103 cells per well from aviremic ART treated HIV-infected individuals (N=6) using the ‘‘stimulated settings’’. Of note, the data generated from the 9 individuals using the ‘‘stimulated settings’’ of conventional VOA (4.104 cells) were also considered in the quantification wherein each individual was considered as a replicate. The frequencies of HIV-infected cells at the cohort level (N=15) were estimated in LN resident and migratory DCs using extreme limiting dilution assay (ELDA) as previously described and expressed as RNA unit per million or RUPM frequencies.40
Decay slope estimation
A logarithmic regression model with first order kinetics was applied to best fit HIV RNA levels in LN Dc subpopulations and in LN CD4 T cells with duration of ART as previously described.46
In vitro HIV-1 infection assay
CD4 T cells from HIV- uninfected individuals were activated for 48 h with anti-CD3 and anti-CD28 mAbs in complete RPMI medium supplemented with IL-2 (10 units/ml). Activated CD4 T cells (106 cells/ml) were washed and exposed (6 h, 37 °C, 5% CO2) to 100 μl–1 mL culture supernatants collected at day 14 of the LN DC viral outgrowth assay (N=9). Following a 6-h exposure, cells were washed twice with complete medium and cultured for 14 additional days in complete RPMI medium. The presence of infectious virus was assessed by the measurement of HIV RNA (COBAS HIV-1 Test; Roche; Switzerland; copies/mL) in the culture supernatants at days 0, 5 and 14 after the inoculations as previously described.40
Screening for drug assicated resistance mutations
Viral outgrowth assay (VOA) culture supernatants of LN resident and migratory DCs from 3 ART treated fully suppressed HIV-infected individuals (Table 1), were subjected to Vela NGS HIV resistance testing as previously described.74 Briefly, following HIV RNA extraction from the VOA culture supernatants, protease/reverse transcriptase (PR/RT) and integrase (IN) coding regions were amplified (1500 bp-length PR/RT amplicon includes PR codons 1–99 and RT codons 1–411, while the IN amplicon covers codons 1–289 (~1000 bp) and sequenced using the ViroKey SQ FLEX HIV 1 Genotyping Assay (Vela Diagnostics, Hamburg, Germany) running on the Vela platform as previously described.74 The library was sequenced on the Sentosa SQ 301 Sequencer (Ion Torrent PGM). NGS data analysis was performed by extracting FASTQ files from the Vela System and transferring to the SmartGene ASP-IDNS®−5 pipeline for NGS data using Advanced Sequencing Platform (ASP) (IDNS® 5 v3.12.0), as previously described.74 The resistance associated mutations in the three HIV-1 regions were identified using the ASI Drug Resistance Algorithm ANRS 2022 v.33 (2019, v.30, available at http://www.hivfrenchresistance.org).
HIV virion-associated RNA sequences accession Number(s)
Sequences were submitted to Sequence Read Archive (SRA) database under accession numbers: SMN35564308-315.
Whole genome amplification
Genomic DNA from individual samples was extracted from ≈3 ×105 sorted LN DC subpopulations, PD-1+/Tfh and LN PD-1−/non-Tfh cells using a QIAGEN DNeasy Blood & Tissue or FFPE Tissue kit (N=3). Genomic DNA diluted to single genome levels based on Poisson distribution statistics and droplet digital PCR (ddPCR) was subjected to multiple displacement amplification (MDA) with phi29 polymerase (QIAGEN REPLI-g Single Cell Kit, catalog 150345), as per the manufacturer’s protocol and as previously described.38 Following this, DNA from each well was split and separately subjected to HIV sequencing and integration site analysis.
HIV near-full-genome sequencing
DNA resulting from whole-genome amplification reactions was subjected to HIV-1 near-full-genome amplification using a 1-amplicon and/or non-multiplexed 5-amplicon approach as previously described.38,75 PCR products were visualized by agarose gel electrophoresis. All near-full-length and/or 5-amplicon positive amplicons, and selected sequences with major deletions (<8kb in gel size or <5 amplicons positive) were subjected to Illumina MiSeq sequencing at the MGH DNA Core facility with a median of 1000 reads per base. Resulting short reads were de novo assembled using Ultracycler v1.0 and aligned to HXB2 to identify large deleterious deletions (fragment size <8kb of the amplicon aligned to HXB2), out-of-frame indels, premature/lethal stop codons, internal inversions, or packaging signal deletions, using either an automated in-house pipeline written in R scripting language76 or using the Los Alamos HIV Sequence Database Hypermut 2.0 program for APOBEC-3G/3F–associated hypermutations.77 Viral sequences (>8kb) that lacked all the above listed mutations were classified as ‘‘genome-intact.’’ Multiple sequence alignments were performed using MUSCLE.78 Phylogenetic distances between sequences were examined using Clustal X–generated neighbor-joining algorithms.79 Viral sequences were considered clonal if they had completely identical consensus sequences with the same integration site; single nucleotide variations in primer binding sites were not considered for clonality analysis.
Integration site analysis
Integration sites associated with each viral sequence were obtained using 1) integration site loop amplification (ISLA)80; 2) ligation-mediated PCR (LM-PCR; Lenti-X Integration Site Analysis Kit; Clontech, catalog 631263)81 and/or 3) nonrestrictive linear amplification–mediated PCR (nrLAM-PCR)82 as previously described.38 Amplified near-full-length viral sequences and viral-host junctions were analyzed by Illumina MiSeq next-generation sequencing. MiSeq paired-end FASTQ files were demultiplexed and analyzed using bioinformatics approach based on either the alignment of small reads (142 bp) simultaneously to human reference genome GRCh38 and HIV-1 reference genome HXB2 using BWA-MEM83 or alternatively, small reads were de novo assembled using Ultracycler v1.0, generating one consensus sequence from each contig, which were then mapped to HXB2 using R library Biostrings local pairwise alignment to identify terminal nucleotides of the viral genome. Then, the HIV-1 part of the contig consensus was trimmed, and the remaining portion (>20 bp) was submitted to Web-based BLAT as previously described.38,84 The final list of integration sites and corresponding chromosomal annotations was obtained using Ensembl (v86, http://www.ensembl.org). Pseudogenes were determined using UCSC Genome Browser (http://www.genome.ucsc.edu/) and GENCODE (v28, https://www.gencodegenes.org/). Repetitive genomic sequences harboring HIV-1 integration sites were identified using RepeatMasker (http://www.repeatmasker.org/).
Proviral sequence accession Number(s)
Sequences were submitted to GenBank under accession numbers: Sequences were submitted to GenBank under accession numbers OM795243-OM795291.
TREC analysis
Quantification of TREC copies was performed in sorted viable LN resident and migratory DC subpopulations (4.104) from HIV uninfected individuals (N=3) as previously described.35 In addition, ten-fold serial dilutions of viable CD4 T cells ranging from 5.105 to 1 cell, were used as controls. Briefly, sorted cells were re-suspended in 2μl of complete RPMI media and directly spotted onto anonymized Guthrie cards (903; GE Healthcare). The Guthrie cards were air dried for 24 hours at room temperature until further analysis. DNA was extracted from each entire spot by punching out 3.2 mm circles, eluted into Generation DNA Elution Solution (QIAGEN) and subjected to real-time quantitative PCR (RT-qPCR) for TRECs as previously described.35 β-actin (ACTB) was used as a house-keeping gene. The data were plotted as TREC copies detected/sample.
QUANTIFICATION AND STATISTICAL ANALYSES
Statistics
Statistical significance (P values) was obtained using one-way ANOVA (Kruskal-Wallis test) followed by Mann-Whitney test, Wilcoxon Matched-pairs two-tailed Signed Rank test or ratio paired-t test. Spearman rank test was used for correlations. The analyses of multiple comparisons were taken into account for the calculation of statistical significance. P values less than 0.05 were considered significant. All P values for scRNAseq are presented as adjusted P values based on bonferroni correction. Transcripts with adjusted P values less than 0.05, log2 fold change higher than 0.25 and expressed in at least 25% of the cells were considered as significant. Analyses were performed using Prism (GraphPad).
Supplementary Material
Highlights.
LN DCs support active cycles of de novo HIV replication
LN DCs harbor intact HIV proviruses within active host transcription units
LN DCs harbor inducible replication-competent HIV despite years of suppressive ART
ACKNOWLEDGMENTS
We are grateful to the following people and facilities: Michael Moulin, Manon Geiser, Rachel Mamin, Thibaut Decaillon, Manuela Lavelli, Philippe Kiehl, Claudia Lima De Pavia Campos, Silvia Sabatino, and Gonzalo Tapia for technical assistance; Prof. Alexandra Calmy and Dr. Hélène Buvelot, Division of Infectious Diseases, University Hospital Geneva, University of Geneva, Switzerland, for patient recruitment; Nicola Manel and Helena Izquierdo-Fernandez for plasmids; Bastien Mangeat and the Gene Expression Core Facility team (EPFL, Lausanne, Switzerland), Lausanne Genomic Technologies Facility, University of Lausanne, Switzerland; Marion Leleu at the BioInformatics Competence Center, University of Lausanne, Switzerland; Aaron Weddle and John Weddle for their assistance with the figures. This work was supported by NIH grants AI117841, AI120008, AI130005, DK120387, AI152979, and AI155233 to M.L., Swiss National Science Foundation grant 320030_200912 and Freedom Forever to M.P., and an educational grant from FONDATION MACHAON to R.B.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2023.08.020.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. (1999). Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5, 512–517. [DOI] [PubMed] [Google Scholar]
- 2.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, and Siliciano RF (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9, 727–728. [DOI] [PubMed] [Google Scholar]
- 3.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, et al. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo HH, Hua S, Chen HR, Ouyang Z, et al. (2017). Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest. 127, 2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McManus WR, Bale MJ, Spindler J, Wiegand A, Musick A, Patro SC, Sobolewski MD, Musick VK, Anderson EM, Cyktor JC, et al. (2019). HIV-1 in lymph nodes is maintained by cellular proliferation during antiretroviral therapy. J. Clin. Invest. 129, 4629–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, et al. (2018). Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc. Natl. Acad. Sci. USA 115, E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonetti FR, Zhang H, Soroosh GP, Duan J, Rhodehouse K, Hill AL, Beg SA, McCormick K, Raymond HE, Nobles CL, et al. (2021). Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J. Clin. Invest. 131, e145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, et al. (2017). Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13, e1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo HH, Ahmad R, Lee GQ, Gao C, Chen HR, Ouyang Z, Szucs MJ, Kim D, Tsibris A, Chun TW, et al. (2018). Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4+ T cells. Immunity 48, 1183–1194.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagne M, Michaels D, Schiralli Lester GM, Gummuluru S, Wong WW, and Henderson AJ (2019). Strength of T cell signaling regulates HIV-1 replication and establishment of latency. PLoS Pathog. 15, e1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, Khoruts A, Estes JD, Anderson J, Callisto SP, et al. (2015). Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc. Natl. Acad. Sci. USA 112, E1126–E1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banga R, Munoz O, and Perreau M (2021). HIV persistence in lymph nodes. Curr. Opin. HIV AIDS 16, 209–214. [DOI] [PubMed] [Google Scholar]
- 13.Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong MAWP, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, et al. (2021). Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 27, 2085–2098. [DOI] [PubMed] [Google Scholar]
- 14.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, and Pantaleo G (2013). Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J. Exp. Med. 210, 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkenschlager J, Finkin S, Ramos V, Kraft J, Cipolla M, Nowosad CR, Hartweger H, Zhang W, Olinares PDB, Gazumyan A, et al. (2021). Dynamic regulation of TFH selection during the germinal centre reaction. Nature 591, 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, et al. (2015). B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat. Med. 21, 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginhoux F, Guilliams M, and Merad M (2022). Expanding dendritic cell nomenclature in the single-cell era. Nat. Rev. Immunol. 22, 67–68. [DOI] [PubMed] [Google Scholar]
- 18.Kvedaraite E, and Ginhoux F (2022). Human dendritic cells in cancer. Sci. Immunol. 7, eabm9409. [DOI] [PubMed] [Google Scholar]
- 19.Cabeza-Cabrerizo M, van Blijswijk J, Wienert S, Heim D, Jenkins RP, Chakravarty P, Rogers N, Frederico B, Acton S, Beerling E, et al. (2019). Tissue clonality of dendritic cell subsets and emergency DCpoiesis revealed by multicolor fate mapping of DC progenitors. Sci. Immunol. 4, eaaw1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabeza-Cabrerizo M, Cardoso A, Minutti CM, Pereira da Costa M, and Reis e Sousa C (2021). Dendritic cells revisited. Annu. Rev. Immunol. 39, 131–166. [DOI] [PubMed] [Google Scholar]
- 21.Chen K, Wang JM, Yuan R, Yi X, Li L, Gong W, Yang T, Li L, and Su S (2016). Tissue-resident dendritic cells and diseases involving dendritic cell malfunction. Int. Immunopharmacol. 34, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segura E, Valladeau-Guilemond J, Donnadieu MH, Sastre-Garau X, Soumelis V, and Amigorena S (2012). Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 209, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton KL, Collins DR, Lengieza J, Ghebremichael M, Dotiwala F, Lieberman J, and Walker BD (2018). Resistance of HIV-infected macrophages to CD8+ T lymphocyte–mediated killing drives activation of the immune system. Nat. Immunol. 19, 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganor Y, Real F, Sennepin A, Dutertre CA, Prevedel L, Xu L, Tudor D, Charmeteau B, Couedel-Courteille A, Marion S, et al. (2019). HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 4, 633–644. [DOI] [PubMed] [Google Scholar]
- 25.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, et al. (2016). Macrophages sustain HIV replication in vivo independently of T cells. J. Clin. Invest. 126, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko A, Kang G, Hattler JB, Galadima HI, Zhang J, Li Q, and Kim W (2019). Macrophages but not astrocytes harbor HIV DNA in the brains of HIV-1-infected aviremic individuals on suppressive antiretroviral therapy. J. Neuroimmune Pharmacol. 14, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Kang G, Duan L, Lu W, Katze MG, Lewis MG, Haase AT, and Li Q (2015). SIV infection of lung macrophages. PLoS One 10, e0125500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plitnik T, Sharkey ME, Mahboubi B, Kim B, and Stevenson M (2018). Incomplete suppression of HIV-1 by SAMHD1 permits efficient macrophage infection. Pathog. Immun. 3, 197–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zalar A, Figueroa MI, Ruibal-Ares B, Baré P, Cahn P, de Bracco MM, and Belmonte L (2010). Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antiviral Res. 87, 269–271. [DOI] [PubMed] [Google Scholar]
- 30.Pion M, Granelli-Piperno A, Mangeat B, Stalder R, Correa R, Steinman RM, and Piguet V (2006). APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J. Exp. Med. 203, 2887–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, et al. (2012). SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cribier A, Descours B, Valadão ALC, Laguette N, and Benkirane M.J.C.r. (2013). Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043. [DOI] [PubMed] [Google Scholar]
- 33.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Ségéral E, Yatim A, Emiliani S, Schwartz O, and Benkirane M (2011). SAMHD1 is the dendritic-and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang WLW, Baumgarth N, Eberhardt MK, Lee CY, Baron CA, Gregg JP, and Barry PA (2007). Exposure of myeloid dendritic cells to exogenous or endogenous IL-10 during maturation determines their longevity. J. Immunol. 178, 7794–7804. [DOI] [PubMed] [Google Scholar]
- 35.Borte S, von Döbeln U, Fasth A, Wang N, Janzi M, Winiarski J, Sack U, Pan-Hammarström Q, Borte M, and Hammarström L (2012). Neonatal screening for severe primary immunodeficiency diseases using high-throughput triplex real-time PCR. Blood 119, 2552–2555. [DOI] [PubMed] [Google Scholar]
- 36.Gierahn TM, Wadsworth MH, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, and Shalek AK (2017). Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kazer SW, Aicher TP, Muema DM, Carroll SL, Ordovas-Montanes J, Miao VN, Tu AA, Ziegler CGK, Nyquist SK, Wong EB, et al. (2020). Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat. Med. 26, 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, et al. (2019). Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 129, 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo H-H, et al. (2020). Blood and lymph node dissemination of clonal genome-intact HIV-1 DNA sequences during suppressive antiretroviral therapy. J. Infect. Dis. 222, 655–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, and Perreau M (2016). PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med. 22, 754–761. [DOI] [PubMed] [Google Scholar]
- 41.Bertram KM, Botting RA, Baharlou H, Rhodes JW, Rana H, Graham JD, Patrick E, Fletcher J, Plasto TM, Truong NR, et al. (2019). Identification of HIV transmitting CD11c+ human epidermal dendritic cells. Nat. Commun. 10, 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smed-Sörensen A, Loré K, Vasudevan J, Louder MK, Andersson J, Mascola JR, Spetz AL, and Koup RA (2005). Differential susceptibility to human immunodeficiency virus Type 1 infection of myeloid and plasmacytoid dendritic cells. J. Virol. 79, 8861–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banga R, Rebecchini C, Procopio FA, Noto A, Munoz O, Ioannidou K, Fenwick C, Ohmiti K, Cavassini M, Corpataux JM, et al. (2019). Lymph node migratory dendritic cells modulate HIV-1 transcription through PD-1 engagement. PLoS Pathog. 15, e1007918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Banga R, Procopio FA, Ruggiero A, Noto A, Ohmiti K, Cavassini M, Corpataux JM, Paxton WA, Pollakis G, and Perreau M (2018). Blood CXCR3+ CD4 T cells are enriched in inducible replication competent HIV in aviremic antiretroviral therapy-treated individuals. Front. Immunol. 9, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Wang Z, Hou Y, Liu X, Hong J, Shi X, Huang X, Zhang T, Liao X, and Zhang L (2023). Novel TLR7/8 agonists promote activation of HIV-1 latent reservoirs and human T and NK cells. Front. Microbiol. 14, 1033448. 10.3389/fmicb.2023.1033448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chun TW, Justement JS, Moir S, Hallahan CW, Maenza J, Mullins JI, Collier AC, Corey L, and Fauci AS (2007). Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195, 1762–1764. [DOI] [PubMed] [Google Scholar]
- 47.Eisenbarth SC (2019). Dendritic cell subsets in T cell programming: location dictates function. Nat. Rev. Immunol. 19, 89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, et al. (2000). DC-SIGN, a dendritic cell–specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100, 587–597. [DOI] [PubMed] [Google Scholar]
- 49.Ruffin N, Gea-Mallorquí E, Brouiller F, Jouve M, Silvin A, See P, Dutertre CA, Ginhoux F, and Benaroch P (2019). Constitutive Siglec-1 expression confers susceptibility to HIV-1 infection of human dendritic cell precursors. Proc. Natl. Acad. Sci. USA 116, 21685–21693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izquierdo-Useros N, Lorizate M, McLaren PJ, Telenti A, Kräusslich HG, and Martinez-Picado J (2014). HIV-1 capture and transmission by dendritic cells: the role of viral glycolipids and the cellular receptor Siglec-1. PLoS Pathog. 10, e1004146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Veenhuis RT, Abreu CM, Costa PAG, Ferreira EA, Ratliff J, Pohlenz L, Shirk EN, Rubin LH, Blankson JN, Gama L, et al. (2023). Monocyte-derived macrophages contain persistent latent HIV reservoirs. Nat. Microbiol. 8, 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito T, Amakawa R, Kaisho T, Hemmi H, Tajima K, Uehira K, Ozaki Y, Tomizawa H, Akira S, and Fukuhara S (2002). Interferon-alpha and interleukin-12 are induced differentially by toll-like receptor 7 ligands in human blood dendritic cell subsets. J. Exp. Med. 195, 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chateauvieux S, Eifes S, Morceau F, Grigorakaki C, Schnekenburger M, Henry E, Dicato M, and Diederich M (2011). Valproic acid perturbs hematopoietic homeostasis by inhibition of erythroid differentiation and activation of the myelo-monocytic pathway. Biochem. Pharmacol. 81, 498–509. [DOI] [PubMed] [Google Scholar]
- 54.Kula A, Delacourt N, Bouchat S, Darcis G, Avettand-Fenoel V, Verdikt R, Corazza F, Necsoi C, Vanhulle C, Bendoumou M, et al. (2019). Heterogeneous HIV-1 reactivation patterns of disulfiram and combined disulfiram+Romidepsin treatments. J. Acquir. Immune Defic. Syndr. 80, 605–613. [DOI] [PubMed] [Google Scholar]
- 55.Tsai A, Irrinki A, Kaur J, Cihlar T, Kukolj G, Sloan DD, and Murry JP (2017). Toll-like receptor 7 agonist GS-9620 induces HIV expression and HIV-specific immunity in cells from HIV-infected individuals on suppressive antiretroviral therapy. J. Virol. 91. e02166–e02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlaepfer E, and Speck RF (2011). TLR8 activates HIV from latently infected cells of myeloid-monocytic origin directly via the MAPK pathway and from latently infected CD4+ T cells indirectly via TNF-a. J. Immunol. 186, 4314–4324. [DOI] [PubMed] [Google Scholar]
- 57.Chu TH, Vo MC, Lakshmi TJ, Ahn SY, Kim M, Song GY, Yang DH, Ahn JS, Kim HJ, Jung SH, et al. (2022). Novel IL-15 dendritic cells have a potent immunomodulatory effect in immunotherapy of multiple myeloma. Transl. Oncol. 20, 101413. 10.1016/j.tranon.2022.101413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, and Tough DF (2002). Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood 99, 3263–3271. [DOI] [PubMed] [Google Scholar]
- 59.Bozzi G, Simonetti FR, Watters SA, Anderson EM, Gouzoulis M, Kearney MF, Rote P, Lange C, Shao W, Gorelick R, et al. (2019). No evidence of ongoing HIV replication or compartmentalization in tissues during combination antiretroviral therapy: implications for HIV eradication. Sci. Adv. 5, eaav2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kearney MF, Wiegand A, Shao W, McManus WR, Bale MJ, Luke B, Maldarelli F, Mellors JW, and Coffin JM (2017). Ongoing HIV replication during ART reconsidered. Open Forum Infect. Dis. 4, ofx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Pond SLK, Chung YS, Penugonda S, Chipman J, Fletcher CV, et al. (2016). Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, et al. (2014). Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. USA 111, 2307–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manel N, Hogstad B, Wang Y, Levy DE, Unutmaz D, and Littman DR (2010). A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature 467, 214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bobadilla S, Sunseri N, and Landau NR (2013). Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther 20, 514–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pantaleo G, Graziosi C, Demarest JF, Butini L, Montroni M, Fox CH, Orenstein JM, Kotler DP, and Fauci AS (1993). HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 362, 355–358. [DOI] [PubMed] [Google Scholar]
- 66.Hughes TK, Wadsworth MH, Gierahn TM, Do T, Weiss D, Andrade PR, Ma F, de Andrade Silva BJ, Shao S, Tsoi LC, et al. (2020). Second-strand synthesis-based massively parallel scRNA-seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity 53, 878–894.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petukhov V, Guo J, Baryawno N, Severe N, Scadden DT, Samsonova MG, and Kharchenko PV (2018). dropEst: pipeline for accurate estimation of molecular counts in droplet-based single-cell RNA-seq experiments. Genome Biol. 19, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barde I, Salmon P, and Trono D (2010). Production and titration of lentiviral vectors. Curr. Protoc. Neurosci. Chapter 4. Unit 4.21. [DOI] [PubMed] [Google Scholar]
- 71.Vandergeeten C, Fromentin R, Merlini E, Lawani MB, DaFonseca S, Bakeman W, McNulty A, Ramgopal M, Michael N, Kim JH, et al. (2014). Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 88, 12385–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin A, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, et al. (2015). A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBiomedicine 2, 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandergeeten C, Fromentin R, DaFonseca S, Lawani MB, Sereti I, Lederman MM, Ramgopal M, Routy J, Sékaly R, and Chomont N (2013). Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood 121, 4321–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bonifacio MA, Genchi C, Lagioia A, Talamo V, Volpe A, and Mariggiò MA (2022). Analytical assessment of the vela diagnostics NGS assay for HIV genotyping and resistance testing: the Apulian experience. Int. J. Mol. Sci. 23, 2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee GQ, Bangsberg DR, Mo T, Lachowski C, Brumme CJ, Zhang W, Lima VD, Boum Y, Mwebesa BB, Muzoora C, et al. (2017). Prevalence and clinical impacts of HIV-1 intersubtype recombinants in Uganda revealed by near-full-genome population and deep sequencing approaches. AIDS 31, 2345–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R Core Team. (2013). A Language and Environment for Statistical Computing (R Foundation for Statistical Computing). [Google Scholar]
- 77.Rose PP, and Korber BT (2000). Detecting hypermutations in viral sequences with an emphasis on G –> A hypermutation. Bioinformatics 16, 400–401. [DOI] [PubMed] [Google Scholar]
- 78.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- 80.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, and Frenkel LM (2014). HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Serrao E, Cherepanov P, and Engelman AN (2016). Amplification, next-generation sequencing, and genomic DNA mapping of retroviral integration sites. J. Vis. Exp. 109, e53840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W, Wolf S, Glimm H, Schmidt M, and von Kalle C (2010). Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 5, 1379–1395. [DOI] [PubMed] [Google Scholar]
- 83.Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kent WJ (2002). BLAT–the BLAST-like alignment tool. Genome Res. 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data was deposited to Gene Expression Omnibus (GEO) with the following accession numbers GEO: GSE196066, which can also be found in the key resources table.
Near-full length HIV proviral sequences obtained from LN DC subpopulations and CD4 T cells were deposited to GenBank with the following accession numbers: OM795243-291, which can also be found in the key resources table.
Protease/reverse transcriptase and Integrase sequences from HIV virion-associated RNA were deposited in the Sequence Read Archive (SRA) database with the following accession number: SMN35564308-315, which can also be found in the key resources table.
Patient derived HIV proviral sequences: Due to study participant confidentiality concerns, viral sequencing data cannot be publicly released, but will be made available to investigators upon reasonable request and after signing a data transfer agreement.
This paper does not report original code.
Publicly available software and code used in this study are listed in the key resources table. Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| LNMC samples from study participants living with HIV | This study, Centre hospitalier universitaire vaudois (CHUV), Lausanne Switzerland | N/A |
|
| ||
| Flow Cytometry | ||
|
| ||
| APC-H7-conjugated mouse anti-human anti-HLA-DR (G46-6) | Becton Dickinson | 561358 |
| APC conjugated mouse anti-human anti-CD4 (RPA-T4) | Becton Dickinson | 555349 |
| ECD-conjugated mouse anti-human anti-CD3 (UCHT1) | Beckman Coulter | A07748 |
| PE-Cy7-conjugated mouse anti-human anti-CD11c (B-ly6) | Becton Dickinson | 561356 |
| PerCP-Cy5.5 conjugated mouse anti-human anti-CD45 (H-I30) | Becton Dickinson | 332784 |
| Alexa Fluor 700 conjugated mouse anti-human anti-CCR7 (150503) | Becton Dickinson | 561143 |
| FITC-conjugated mouse anti-human anti-CD20 (clone 2H7). | Becton Dickinson | 555622 |
| BV421 conjugated mouse anti-human CD279 (EH12) | Becton Dickinson | 565935 |
| PerCP-Cy5.5 conjugated mouse anti-human anti-CD8 (SK1) | Becton Dickinson | 341050 |
| PE-CF594-conjugated mouse anti-human anti-CD45RA (HI100) | Becton Dickinson | 562298 |
|
| ||
| Mass Cytometry | ||
|
| ||
| Ultra-LEAF™ Purified anti-human CD8a (RPA-T8) | Biolegend | 301073 |
| Ultra-LEAF™ Purified anti-human CD4 (RPA-T4) | Biolegend | 300570 |
| Purified anti-human CD45 Antibody (HI30) | Biolegend | 304002 |
| Purified anti-human CD19 (HIB19) | Biolegend | 302202 |
| Purified anti-human/mouse/rat CD278 (ICOS) (C398.4A) | Biolegend | 313502 |
| Purified IRF4 (IRF4.3E4) | Biolegend | 646402 |
| Purified anti-human CCR5 (J418F1) | Biolegend | 359102 |
| SAMHD1 antibody (AA 426-657) (FITC) | Antibodies-online.com | ABIN7177151 |
| Purified anti-human CD163 (GHI/61) | Biolegend | 3145010B |
| Purified anti-human IgD (IA6-2) | Biolegend | 348202 |
| Purified anti-human BIRC3 (C-IAP2) | Biolegend | 667702 |
| Anti-Human CD274/PD-L1 (29E.2A3)-148Nd | Fluidigm | 3148017B |
| Anti-Human CD127/IL-7Ra (A019D5)-149Sm | Fluidigm | 3149011B |
| Anti-Human CD134/OX40 (ACT35)-150Nd | Fluidigm | 3150023B |
| Purified anti-human CD123 (6H6) | Biolegend | 306002 |
| Purified anti-human CD21 (Bu32) | Biolegend | 354902 |
| Purified anti-human DC-SIGN (9E9A8) | Biolegend | 330102 |
| Anti-Human CD27 (L128)-155Gd | Fluidigm | 3155001B |
| Anti-Human CD86/B7.2 (IT2.2)-156Gd | Fluidigm | 3156008B |
| Anti-Human CD169 (7-239)-158Gd | Fluidigm | 3158027B |
| Anti-Human CD197/CCR7 (G043H7)-159Tb | Fluidigm | 3159003A |
| Anti-Human CD14 (M5E2)-160Gd | Fluidigm | 3160001B |
| Purified anti-human CD1c (L161) | Biolegend | 331502 |
| Anti-Human CD11c (Bu15)-162Dy | Fluidigm | 3162005B |
| Purified anti-human CD85j (GHI/75) | Biolegend | 333702 |
| APC-conjugated anti-IRF8 (V3GYWCH) | Invitrogen | 17-9852-82 |
| Anti-APC (APC003)-163Dy | Fluidigm | 3163001B |
| Anti-FITC (FIT-22)-144Nd | Fluidigm | 3144006B |
| Anti-Human CD185/CXCR5 (RF8B2)-164 | Fluidigm | 3164029B |
| Anti-Human CD40 (5C3)-165Ho | Fluidigm | 3165005B |
| Purified anti-human CD155 (PVR) (SKII.4) | Biolegend | 337602 |
| Purified anti-human CD38 (HIT2) | Biolegend | 303502 |
| Anti-Human CD45RA (HI100)-169Tm | Fluidigm | 3169008B |
| Anti-Human CD3 (UCHT1)-170Er | Fluidigm | 3170001B |
| Anti-Human CD273/PDL2 (24F.10C12)-172Yb | Fluidigm | 3172014B |
| Anti-Human CD184/CXCR4 (12G5)-173Yb | Fluidigm | 3173001B |
| Purified anti-human HLA-DR (L243) | Biolegend | 307602 |
| Purified anti-human PD-1(EH12.2H7) | Biolegend | 329925 |
| Anti-Human CD11b/Mac-1 (ICRF44)-209Bi | Fluidigm | 3209003B |
| Purified NA/LE Mouse Anti-Human CD3 | Becton Dickinson | 555329 |
| Purified Mouse Anti-Human CD28 | Becton Dickinson | 555726 |
|
| ||
| Virus strainss | ||
|
| ||
| Human Immunodeficiency Virus Type 1 (HIV-1) BaL | HIV reagent program | 510 |
| Human Immunodeficiency Virus Type 1 (HIV-1)-CCR5 EGFP | Gift From Nicola Manel | Ref. 68; Manel et al.63 |
| Human Immunodeficiency Virus Type 1 (HIV-1)-CXCR4 EGFP | Gift From Nicola Manel | Ref. 68; Manel et al.63 |
| VpX Lentivirus | Gift from Fabio Condotti | Bobadilla et al. (2013).64 Gene Therapy |
|
| ||
| Reagents, chemicals, peptides, recombinant proteins | ||
|
| ||
| HIV peptides | JPT Peptide Technology | Customized |
| TLR-7/8 agonist | InvivoGen | CL264 |
| SEB | Sigma-Aldrich | S4881 |
| Human IL-2 IS, research grade | Miltenyi | 130-097-743 |
| Emtricitabine | NIH Reagents Program | HRP-10071 |
| Thymidine, [Methyl-3H], in 2% ethanol, 1 mCi | perkinelmer | NET027W001MC |
|
| ||
| Critical commercial assays | ||
|
| ||
| Single cell RNAseq (Seq-well; refer to Gierahn et al.36) | ||
| polydimethylsiloxane (PDMS) | ChemGenes Corporation, Wilmington, MA, USA | #MACOSKO201110 |
| polycarbonate membrane | Sterlitech Corporation, Auburn, WA, USA | #PCT00162X22100 |
| barcoded with mRNA capturing beads | ChemGenes Corporation, Wilmington, MA, USA). | #MACOSKO201110 |
| cDNA library preparation - Nextera XT DNA Library Preparation Kit) | Illumina, Inc., San Diego, CA, USA | #FC-131-1024 |
| NGS HIV resistance testing (Sentosa SQ HIV Genotyping Assay) | Vela Diagnostics, Hamburg | |
| DNeasy Blood and Tissue Kit | Qiagen | 69504 |
| MIPseq protocol | Refer to Einkauf et al.38 | |
| EasySep Human CD8+ Tcell IsolationKit | Stemcell | 17953 |
| EasySep™ Human CD4+ T Cell Isolation Kit | Stem Cell | 17952 |
| Human IFNγ ELISpotPRO assay KIT | MABTECH | 3420-2APW-2 |
| Cobas 6800 | Cobas HIV-1; Roche Diagnostics AG | KIT COBAS 6800/8800 HIV 96T IVD; Référence : 7000995190 |
| HIV-1 P24 Electrochemiluminiscence assay | Elecsys HIV Duo de Roche | 08 836 973 190 (300 tests) |
| IFNγ ELISpot assay KIT | MABTECH | 3420-2APW-2 |
| LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit | Invitrogen | L34957 |
|
| ||
| Software and algorithms | ||
|
| ||
| STAR aligner software | ENCODE, version 2.7 | https://www.encodeproject.org/software/star/ |
| R package | R Core Team and R Foundation for Statistical Computing, version 4.0.2 | https://www.r-project.org |
| Seurat package for R (v.4.0.0) | ||
| DropEst (version 0.8.5) | ||
| SmartGene ASP-IDNS®−5 pipeline for NGS data using the ‘‘Provirus Pipeline’’ version 2.0.5_HIV1_v1.4 | ||
| ANRS algorithm | http://www.hivfrenchresistance.org | |
| MIPseq data analysis | Refer to Einkauf et al.38 | N/A |
| FlowJo 10 | FlowJo, LLC | https://www.flowjo.com |
| Graphpad Prism 9 | Graphpad | https://graphpad.com |
| Cytobank software package | Cytobank | https://premium.cytobank.org/cytobank/ |
| Prism | Graphpad, version 8.2.1 | https://www.graphpad.com/scientific-software/prism |
|
| ||
| Sequence | ||
|
| ||
| RNAseq sequences for LN DCs | Gene Expression Omnibus (GEO) | GSE196066 |
| Near-full length proviral sequences obtained from LN DC subpopulations and CD4 T cells | GenBank | OM795243-OM795291. |
| Protease/reverse transcriptase and Integrase sequences from HIV virion-associated RNA | Sequence Read Archive (SRA) database | SMN35564308-315 |
Raw and processed RNAseq data are available on the gene expression omnibus (GEO) in GSE196066.


