Abstract
Malignant peripheral nerve sheath tumors (MPNSTs) are chemotherapy resistant sarcomas that are a leading cause of death in neurofibromatosis type 1 (NF1). Although NF1-related MPNSTs derive from neural crest cell origin, they also exhibit intratumoral heterogeneity. TP53 mutations are associated with significantly decreased survival in MPNSTs, however the mechanisms underlying TP53-mediated therapy responses are unclear in the context of NF1-deficiency. We evaluated the role of two commonly altered genes, MET and TP53, in kinome reprograming and cellular differentiation in preclinical MPNST mouse models. We previously showed that MET amplification occurs early in human MPNST progression and that Trp53 loss abrogated MET-addiction resulting in MET inhibitor resistance. Here we demonstrate a novel mechanism of therapy resistance whereby p53 alters MET stability, localization, and downstream signaling leading to kinome reprogramming and lineage plasticity. Trp53 loss also resulted in a shift from RAS/ERK to AKT signaling and enhanced sensitivity to MEK and mTOR inhibition. In response to MET, MEK and mTOR inhibition, we observed broad and heterogeneous activation of key differentiation genes in Trp53-deficient lines suggesting Trp53 loss also impacts lineage plasticity in MPNSTs. These results demonstrate the mechanisms by which p53 loss alters MET dependency and therapy resistance in MPNSTS through kinome reprogramming and phenotypic flexibility.
Subject terms: Sarcoma, Oncogenes, Growth factor signalling, Differentiation, Genetic markers
Introduction
Malignant peripheral nerve sheath tumors (MPNSTs) are aggressive, chemoresistant sarcomas arising from Schwann cells that are the leading cause of death in patients with Neurofibromatosis Type 1 (NF1) [1]. NF1 is an autosomal dominant tumor predisposition syndrome caused by inactivating mutations in the NF1 gene [2–4]. NF1 is a tumor suppressor gene that encodes neurofibromin, a critical negative regulator of RAS [5]. NF1-related MPNSTs exhibit deregulated RAS signaling caused by loss of heterozygosity of NF1 along with additional tumor suppressor loss (TP53, CDKN2A, SUZ12, PTEN) and receptor tyrosine kinase (RTK) amplification (MET, EGFR, PDGFR) [6–12]. As such, targeted therapies against RTKs and RAS effectors including MEK have been proposed as a treatment option for MPNSTs. Even with promising preclinical results, clinical trials featuring tyrosine kinase inhibitors have not been successful to date [13–16]. Because of their aggressive clinical behavior, the 5 year survival rate remains only at 10–50% [17–20].
Although histologic and genomic MPNST subtypes have been described, these categories are not therapeutically relevant. The MPNST chemotherapy regimen has remained largely unchanged since the incorporation of doxorubicin in the 1980’s. The lack of actionable MPNST subtypes remains a major barrier to effective treatment, particularly given the vast differences in kinase signaling between histologically identical tumors [14, 21–25]. Additionally, MPNSTs are known to exhibit divergent states of differentiation leading to intratumoral heterogeneity. For example, MPNSTs can contain cellular regions comprised of malignant muscle, bone, fat, nerve and cartilage cells [26]. It has long been suspected that the differentiation states of various MPNST histologic subtypes contribute to therapy resistance, but confirmatory data is lacking. Thus the identification of predictive biomarkers for MPNSTs continues to be an area of intense study [27]. 25–60% of MPNSTs are p53-deficient [9, 10, 28], which is associated with significantly diminished survival [28–30] and poor response to neo-adjuvant chemotherapy [31]. Additionally, MET [32] and PI3K/AKT/mTOR pathway [33] activation are both associated with poor prognosis, and animal modeling indicates that genetic activation of these pathways in the context of NF1 loss drives MPNST growth and is targetable therapeutically [34, 35]. MET or its ligand, HGF, are amplified in 25–50% of MPNSTs [7, 8], which results in activation of downstream effectors including RAS, PI3K, and STAT3. PI3K may also be activated in MPNST via loss of PTEN, its major upstream negative regulator [36], or via amplification of other RTKs including EGFR [37]. Collectively, the PI3K/AKT/mTOR pathway is activated in 50% of MPNSTs [33]. Crosstalk between p53 and PI3K/AKT/mTOR occurs in a cell type and stress-dependent manner, in which positive or negative regulation of either pathway is highly contextual and may be reciprocal [38]. Defining the role of these pathways in therapeutic response is critical to predicting effective targeted therapies and predictive biomarkers for future MPNST trials.
Consistent with these clinical observations, we previously found that a Trp53-deficient mouse model of NF1-related MPNST exhibited faster tumor growth and was more resistant to chemotherapy compared to Trp53-intact models. Interestingly, the Trp53-deficient model was also less sensitive to single agent and combination MEK and MET inhibition despite sustained repression of ERK phosphorylation. Trp53-deficient tumors also exhibited unusually high AKT activation both at baseline and in response to targeted therapy [25, 34]. In this study, we aimed to define the role of p53 in regulating kinase signaling, targeted therapy response, and cellular differentiation in NF1-related MPNSTs. Transcriptomic and phospho-proteomic analysis revealed multiple mechanisms of resistance, including deregulation of MET stability and localization, deregulated PI3K/AKT/mTOR signaling, and altered lineage plasticity. In contrast to these results, we found that p53 loss actually increased sensitivity to mTOR inhibition, which was associated with broad and persistent kinome activation. Excitingly, combined mTOR and MEK inhibition reversed clonal selection for p53-deficiency and was the most effective drug combination in all models, regardless of p53 status. Lineage plasticity, as defined by transcriptional profiling, was closely linked to tyrosine kinase inhibitor resistance. Collectively, these data suggest that p53 acts as a master regulator of tyrosine kinase signaling and mediates oncogene-addiction and cell fate in MPNSTs.
Results
p53-deficiency is associated with MET inhibitor resistance in MPNSTs
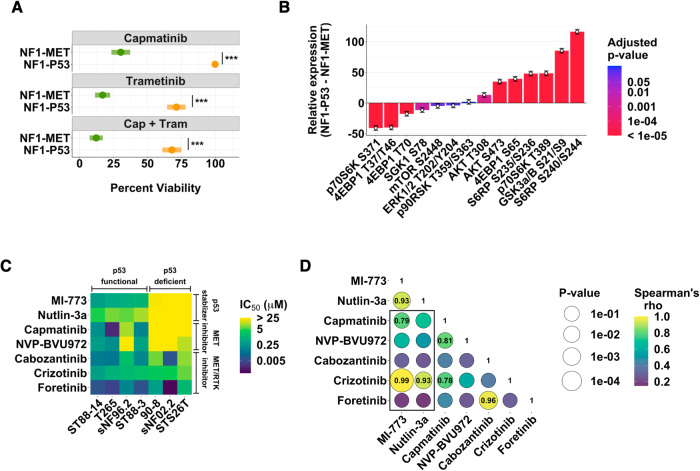
Previously, we compared response to MET and MEK inhibition in tumor xenografts derived from MPNSTs of genetically engineered mouse models of NF1, including a Met-amplified, Trp53-wildtype model (NF1-MET; genotype: Nf1fl/ko;lox-stop-loxMETtg/+;Plp-creERTtg/+) and a Trp53 deficient model (NF1-P53; genotype: Nf1ko/+;p53R172H;Plp-creERTtg/+). NF1-P53 MPNSTs were less sensitive to both MET and MEK inhibition in vivo [34]. To further investigate the molecular mechanisms of drug resistance, we generated tumor cell line isolates from these murine MPNSTs. Cell viability analysis confirmed that the MPNST cell lines maintained their drug sensitivity phenotypes in vitro, as the NF1-P53 cell line was resistant to single agent MET (capmatinib) and MEK (trametinib) inhibition as well as combination therapy (Fig. 1A). A targeted analysis of the RAS/ERK and PI3K/AKT pathways with reverse phase protein arrays (RPPA) was used to identify both immediate (2 h) and adaptive (48 h) kinome responses to MET and MEK inhibition. After controlling for drug treatment and exposure time, several PI3K effectors (including AKT and S6) were significantly activated in the NF1-P53 cells, suggesting PI3K/AKT pathway activation may promote resistance to MET and MEK inhibition (Fig. 1B, Supplementary Fig. 2).
Fig. 1. p53 deficiency is associated with MET inhibitor resistance in MPNSTs.
A Percent viability of NF1-MET and NF1-P53 cells after 72 h of capmatinib (100 nM), trametinib (40 nM) or combination (capmatinib 100 nM, trametinib 40 nM) treatment. B Change in phospho-site activation of MET effectors in NF1-P53 cells relative to NF1-MET cells upon capmatinib (100 nM), trametinib (100 nM), or combination (capmatinib 100 nM, trametinib 100 nM) treatment for 2 and 48 h. See also Supplementary Fig. 2. C IC50 of p53 stabilizing drugs and MET inhibitors against a panel of human MPNST cell lines. D Spearman’s correlations (color) and significance (size) between the IC50 of the drugs in (C). The rho value of correlations with a p-value < 0.5 are indicated in their respective bubble. The black box indicates the correlations between p53 stabilizing drugs and MET inhibitors. ***p < 0.001.
To determine if p53 also regulates sensitivity to MET inhibition in human MPNSTs, we screened a panel of 6 NF1-related and 1 sporadic (STS26T) MPNST cell lines for functional p53 based on sensitivity to the p53 stabilizing drugs, MI-773 and nutlin-3a. These p53 stabilizers inhibit MDM2-mediated p53 degradation and are selectively active in p53 wild-type cells [39]. Based on the IC50 of these drugs, we classified four cell lines as being “p53-functional” and three cell lines as being “p53-deficient” (Fig. 1C, Supplementary Fig. 3A), which was consistent with a recent genomic analysis of MPNST cell lines [40]. In addition, we screened the cell lines for sensitivity to of a panel of MET inhibitors. All but one of the p53-intact cell lines were sensitive to MET inhibition while, the p53-deficient cells were profoundly resistant to MET inhibition (Fig. 1C, Supplementary Fig. 3A). The IC50 between several of the p53 stabilizers and MET inhibitors significantly correlated, similar to drugs within the same class (Fig. 1D, Supplementary Fig. 3B), suggesting a critical role for p53 in regulating MET-dependency.
p53 loss drives MET inhibitor resistance in MPNSTs
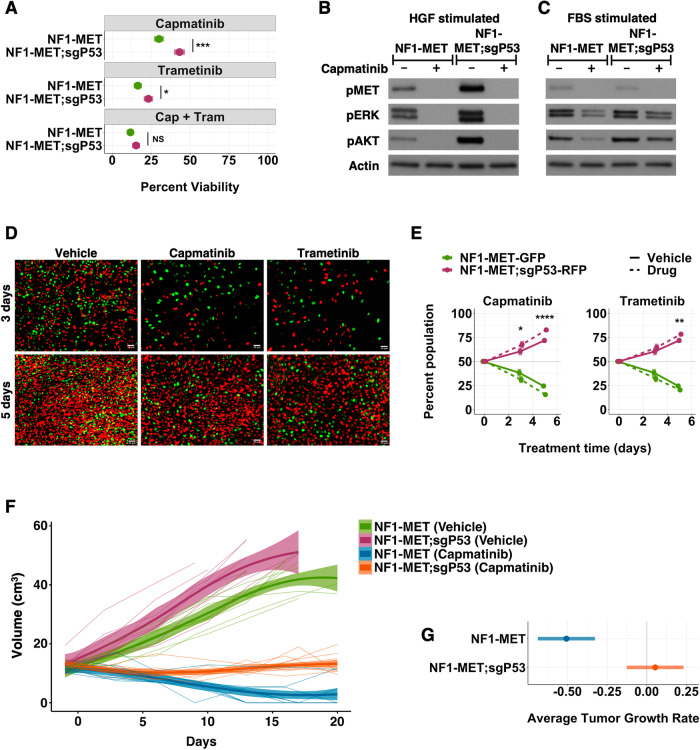
To evaluate the impact of p53 loss-of-function in the context of MET-addiction, we used CRISPR-Cas9-mediated knockout of p53 in murine NF1-MET cells to create the NF1-MET;sgP53 line. p53 protein levels as well as the p53 target gene, p21, were diminished in NF1-MET;sgP53 cells (Supplementary Fig. 4A, B). Cell viability analysis demonstrated that NF1-MET;sgP53 cells were significantly less sensitive to MET inhibition than NF1-MET cells; however, combined MET-MEK inhibition restored drug sensitivity (Fig. 2A). Interestingly, the magnitude of resistance was less in the isogenic cells compared to the NF1-MET and NF1-P53 cells (Supplemental Fig. 4C), suggesting the possibility of additional genetic modifiers in the NF1-P53 cell line. CDKN2A loss is a key step in pre-MPNST transformation [41] and contributes to drug resistance in other tumor types [42, 43]. We evaluated the NF1-MET and NF1-P53 cell lines for spontaneous Cdkn2a loss, however Cdkn2a was deleted only in the NF1-MET cell line (Supplemental Fig. 4D), indicating Cdkn2a loss does not contribute to increased drug resistance in the NF1-P53 cell line. We next evaluated the impact of p53 on HGF-induced MET signaling and observed augmented MET signaling in NF1-MET;sgP53 cells, yet capmatinib inhibited HGF-dependent ERK and AKT activation regardless of p53 status (Fig. 2B). Stimulation with FBS (which contains other growth factors as well as HGF) also induced pERK and pAKT. Interestingly, the effect of capmatinib was quite modest in NF1-MET;sgP53 cells, suggesting capmatinib resistance may be partially mediated by parallel pathway activation that converges on ERK and AKT (Fig. 2C).
Fig. 2. p53 loss drives MET inhibitor resistance in MPNSTs.
A Percent viability of NF1-MET and NF1-MET;sgP53 cells after 72 h of capmatinib (100 nM), trametinib (40 nM) or combination (capmatinib 100 nM, trametinib 40 nM) treatment. B, C Western blot of NF1-MET and NF1-MET;sgP53 cells treated with capmatinib (100 nM) for 2 h and stimulated with HGF (B) or 10% FBS (C) for 15 min. Images (D) and flow cytometry analysis (E) of GFP labeled NF1-MET and RFP labeled NF1-MET;sgP53 cells after 3 and 5 days of treatment with vehicle (DMSO), capmatinib (100 nM), or trametinib (40 nM). F Individual tumor growth curves, LOESS curves, and 95% ribbons for vehicle or capmatinib (30 mg/kg BID) treated NF1-MET and NF1-MET;sgP53 xenografts. G Pairwise comparison of growth trend estimates in capmatinib treated NF1-MET (p-value = 0.0083) and NF1-MET;sgP53 (p-value = 0.089) tumors to 0 (grey line). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Resistant clonal populations within heterogeneous MPNSTs may explain the clinical failures of targeted kinase inhibitors in MPNSTs [44]. To determine if p53 is a key driver of clonal selection and drug resistance in MPNSTs, we performed a clonal competition assay using labeled isogenic NF1-MET-GFP and NF1-MET;sgP53-RFP cell lines. Notably, when cultured separately, NF1-MET;sgP53 cells do not have a significant proliferative advantage compared to NF1-MET cells (Supplementary Fig. 4E). In contrast, when cocultured NF1-MET;sgP53-RFP cells had a strong growth advantage, which was significantly enhanced by both MET or MEK inhibition (Fig. 2D, E). After 5 days of capmatinib treatment, NF1-MET;sgP53-RFP cells comprised 83% of the culture (Fig. 2D, E), indicating that p53 loss drives strong clonal selection with MET inhibition.
To evaluate p53 loss and capmatinib sensitivity in vivo, we treated orthotopic MPNST xenografts with capmatinib. Tumor growth rate was significantly increased in the NF1-MET;sgP53 tumors compared to the parental tumors (p-value = 4.7e-4) (Fig. 2F; Supplementary Fig. 4F, G). Capmatinib strongly inhibited tumor growth in both the NF1-MET and NF1-MET;sgP53 models (Fig. 2F; Supplementary Fig. 4F), however only the NF1-MET tumors regressed on treatment (p-value = 0.0083), while the NF-MET;sgP53 tumors remained stable throughout treatment, with a subset of tumors actually showing increased growth (Fig. 2F, G; Supplementary Fig. 4F). These data confirm our in vitro findings and further demonstrate that p53 loss promotes MET inhibitor resistance.
Met inhibition induces p53-dependent lineage plasticity
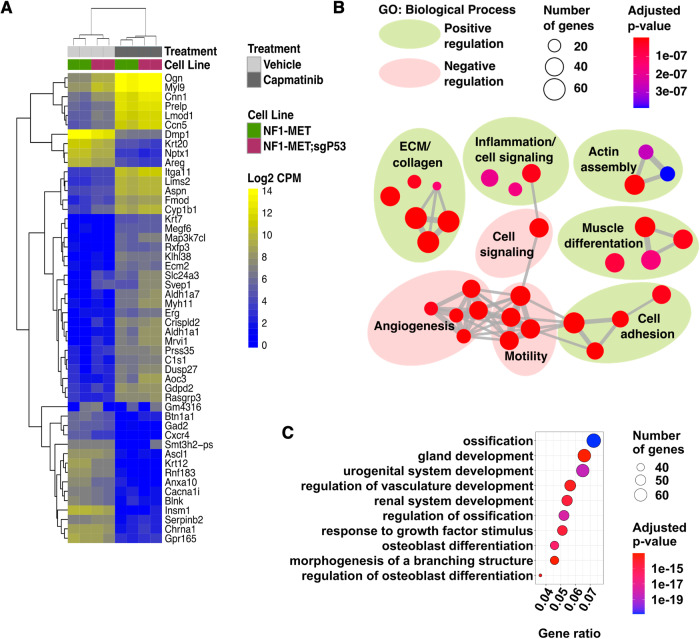
To understand how loss of p53 drives resistance we used RNA-seq to examine capmatinib-induced transcriptional changes in NF1-MET and NF1-MET;sgP53 cells. Unsupervised hierarchical clustering identified strong clustering by treatment followed by genotype, with p53 status modifying expression of some gene subsets (Fig. 3A). GO term enrichment analysis of genes upregulated by MET inhibition identified biological processes related to positive regulation of actin organization, cell adhesion, collagen deposition/ossification, and muscle differentiation (Fig. 3B, Supplementary Fig. 5A). We next examined genes that were downregulated in capmatinib treated NF1-MET;sgP53 cells and identified biological processes related to bone and kidney development (Fig. 3C, Supplementary Fig. 5B). Together, these data suggest that response to MET inhibition may be partially mediated through the induction of linage plasticity pathways in MPNSTs and that p53 loss disrupts this process to promote drug resistance. As differentiation and cell cycle arrest are coupled, we next examined whether p53 loss altered expression of known p53 target genes that promote cell cycle arrest. Indeed, expression of the cell cycle regulators Cdkn1a and Zmat3 were lost in the NF1-MET;sgP53 cells, while expression of genes involved in senescence or apoptosis were unchanged (Supplementary Fig. 5C).
Fig. 3. Met inhibition induces p53-dependent linage plasticity.
A Unsupervised hierarchical clustering of top 50 differentially expressed genes in NF1-MET and NF1-MET;sgP53 cells treated with capmatinib (100 nM) for 24 h. B GO term enrichment analysis of capmatinib-induced genes. The top 30 most significantly enriched biological process terms (by adjusted p-value) are shown. Connecting grey lines represent relatedness of the pathways, while dot size indicates the number of genes differentially expressed in the pathway. See Supplementary Fig. 5B for individual terms. C Top 10 most significantly enriched biological process GO terms derived from genes that were decreased in capmatinib treated NF1-MET;sgP53 cells. See Supplementary Fig. 5C for expanded top 30 terms.
p53 regulates MET stability and localization
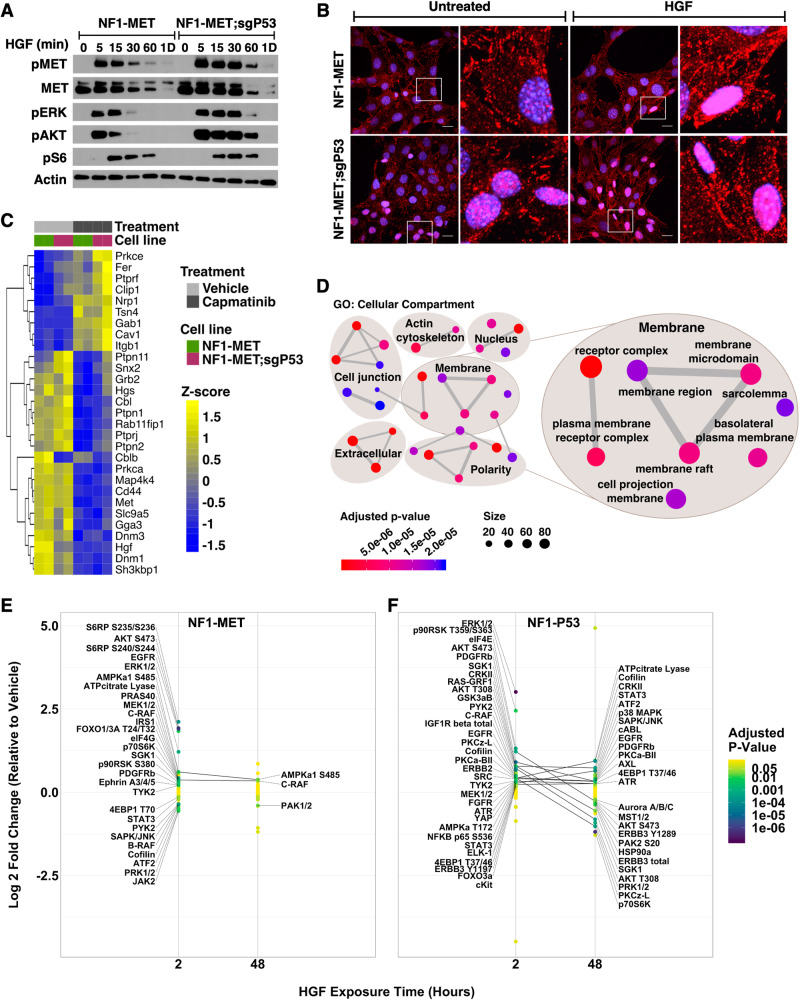
RTK expression, activation, and recycling are tightly regulated processes that ensure RTK regulation in normal physiological conditions. Deregulation of this cycle of RTK activation and recycling is often observed in cancers, such as the MET exon 14 deletions found in lung cancers [45]. To determine whether RTK spaciotemporal regulation promoted the enhanced MET signaling observed in p53-deficient MPNST cells (Figs. 1B & 2B, C), we measured the kinetics of MET activation and turnover. In NF1-MET cells an immediate, and expected, increase in MET activation was observed within 5 min of HGF treatment that quickly diminished along with total MET levels over time (Fig. 4A). In contrast, HGF-treatment induced a drastic pMET increase that remained elevated for 60 min in NF1-MET;sgP53 cells. Both phosphorylated and total MET were persistently elevated in the NF1-MET;sgP53 cells, which corresponded to increased and prolonged activation of both ERK and AKT (Fig. 4A). Activation of the mTOR effector S6 was similar between the two cell lines.
Fig. 4. p53 regulates MET stability, localization, and effector signaling.
A Time course western blot of NF1-MET and NF1-MET;sgP53 cells stimulated with HGF for 5 min to 1 day. B Representative images of phospho-MET localization (red) after 5 min of HGF stimulation. C Expression of genes associated with MET stability and localization after 24 h of vehicle (DMSO) or capmatinib (100 nM) treatment. D GO analysis of Cellular Compartment terms downregulated in the NF1-MET;sgP53 cell line compared to the parental NF1-MET line. See Supplementary Fig. 5D for individual terms. Change in phospho-protein expression in NF1-MET (E) and NF1-P53 (F) cells after HGF treatment. Significantly increased or decreased proteins at each time point are labeled and proteins that are significant at both the 2 and 48 h time points are connected. See Table 1 for phosphosites. Color indicates P-value.
To examine whether p53 loss also alters MET subcellular localization, we performed immunostaining of MET after HGF-treatment. In normal conditions, HGF treatment results in MET localization and activation at the plasma membrane; however MET can also be internalized to the nucleus, where it’s function is incompletely understood [46]. At baseline, pMET was localized to the cytoplasm in the NF1-MET cells, whereas we observed both nuclear and cytoplasmic staining in NF1-MET;sgP53 cells. Interestingly, HGF dramatically increased nuclear MET localization in the p53-deficient NF1-MET;sgP53 cells, while treatment induced nuclear MET localization only in a small percentage of NF1-MET cells (Fig. 4B). In several cancers, nuclear MET is associated with drug resistance and poor prognosis [47–51]. These results suggest that p53 loss induces nuclear MET localization, promoting tumor aggressive phenotypes in MPNST cells.
To understand how loss of p53 promotes increased stability and nuclear localization of MET, we used RNA-seq to examine the expression of genes involved in MET activation and turnover. Capmatinib treatment induced sweeping compensatory expression changes in both cell lines (Fig. 4C). In NF1-MET;sgP53 cells, expression of genes critical for MET degradation, Sh3kpb1 and Cblb, [52–54] were significantly downregulated compared to the parental cell line. Conversely, Prkce expression, which is required for nuclear MET translocation [55], was significantly upregulated in the NF1-MET;sgP53 cells in the presence of capmatinib. GO cellular compartment enrichment analysis revealed that p53 loss promoted significant downregulation in plasma membrane and receptor organization pathways (Fig. 4D, Supplementary Fig. 5D).
MET and the RTK EGFR share much of the same recycling machinery [54, 56], and EGFR is also frequently amplified in MPNST [37]. To determine if p53 loss also enhanced EGFR activation, we treated the NF1-MET and NF1-MET;sgP53 cells with EGF. Strikingly, EGF induced much stronger EGFR phosphorylation in the NF1-MET;sgP53 cells than the parental cell line (Supplementary Fig. 6), similar to the increased MET activation upon HGF treatment. This corresponded with increased and prolonged activation of pERK, pAKT, and pS6. These results suggest an important role for p53 in regulating both MET and EGFR signaling in MPNSTs and may partially explain resistance to EGFR inhibition in MPNSTs [57].
To more broadly examine the effect of p53 expression on MET signaling, we used RPPA to evaluate the response of 98 protein phosphosites to short and extended HGF stimulation. After 2 h of HGF treatment, numerous phosphosites were significantly upregulated in both the NF1-MET and NF1-P53 cell lines, yet the number of upregulated sites as well as the magnitude of change was higher in the p53-deficient cells (Fig. 4E, F, Table 1). Additionally, phosphorylation of several proteins, including STAT3, JAK2, and B-RAF, were significantly decreased in the NF1-MET cell line after just 2 h HGF treatment, (Fig. 4E, Table 1) suggesting that rapid negative feedback signaling in response to MET activation is p53-dependent. Remarkably, after 48 h of HGF treatment, only 3/98 phospho-sites were significantly changed in the NF1-MET cells compared to 27/98 sites in the NF1-P53 cells (Fig. 4E, F, Table 1). Notably, STAT3 Y705 phosphorylation, which is sustained by perinuclear MET [58], was among the most significantly increased phosphosites in the p53-deficient cells after 48 h of HGF exposure (Fig. 4F, Table 1). Collectively, these data suggest that p53 loss moderates MET addiction by modulating the location, timing, and magnitude of MET effector signaling.
Table 1.
Reverse phase protein array fold change in expression relative to vehicle.
| Cell.line | Time | gene | logFC | t | SE | adj.P.Val |
|---|---|---|---|---|---|---|
| NF1-MET | 2 | AKT S473 | 1.922302123 | 13.17752484 | 0.145877329 | 8.98E-07 |
| NF1-MET | 2 | S6RP S240/S244 | 1.901331348 | 12.18886476 | 0.155989207 | 1.13E-06 |
| NF1-MET | 2 | S6RP S235/S236 | 2.113685262 | 7.427556054 | 0.284573451 | 0.000191344 |
| NF1-MET | 2 | SAPK/JNK T183/Y185 | −0.357780978 | −7.176927867 | 0.04985155 | 0.00020508 |
| NF1-MET | 2 | PRAS40 T246 | 0.427694519 | 6.446338782 | 0.066346888 | 0.000486589 |
| NF1-MET | 2 | ERK1/2 T202/Y204 | 1.211520394 | 6.189667411 | 0.195732713 | 0.0006043 |
| NF1-MET | 2 | p90RSK S380 | 0.279372169 | 5.196687333 | 0.053759665 | 0.002642982 |
| NF1-MET | 2 | IRS1 S612 | 0.351537864 | 5.113142028 | 0.068751829 | 0.002654418 |
| NF1-MET | 2 | Cofilin S3 | −0.386870886 | −5.04811324 | 0.076636729 | 0.002654418 |
| NF1-MET | 2 | B-RAF S445 | −0.373076735 | −4.73781656 | 0.078744445 | 0.004054089 |
| NF1-MET | 2 | FOXO1 T24/FOXO3a T32 | 0.350437141 | 4.692337306 | 0.074682854 | 0.004054089 |
| NF1-MET | 2 | p70S6K S371 | 0.34623766 | 4.503365928 | 0.076884194 | 0.00497755 |
| NF1-MET | 2 | TYK2 Y1054/Y1055 | 0.207025297 | 4.48444698 | 0.046165179 | 0.00497755 |
| NF1-MET | 2 | EGFR Y1068 | 1.833693254 | 4.169044594 | 0.439835366 | 0.008210613 |
| NF1-MET | 2 | AMPKa1 S485 | 0.60751555 | 4.096781071 | 0.148290948 | 0.008754974 |
| NF1-MET | 2 | MEK1/2 S217/S221 | 0.382514877 | 4.021501455 | 0.095117428 | 0.009434676 |
| NF1-MET | 2 | JAK2 Y1007 | −0.563544454 | −3.981228378 | 0.141550396 | 0.009459995 |
| NF1-MET | 2 | ATF2 T71 | −0.493146086 | −3.956642856 | 0.124637503 | 0.009459995 |
| NF1-MET | 2 | C-RAF S338 | 0.366910981 | 3.629966153 | 0.101078348 | 0.016447315 |
| NF1-MET | 2 | elF4G S1108 | 0.347419148 | 3.590275085 | 0.096766721 | 0.016447315 |
| NF1-MET | 2 | PRK1 T774/PRK2 T816 | −0.554399931 | −3.579608311 | 0.154877261 | 0.016447315 |
| NF1-MET | 2 | SGK1 S78 | 0.328678952 | 3.158633033 | 0.10405734 | 0.033940163 |
| NF1-MET | 2 | Ephrin A3 Y799/A4 Y799/A5 Y833 | 0.211585037 | 3.150067002 | 0.067168424 | 0.033940163 |
| NF1-MET | 2 | 4EBP1 T70 | −0.141851826 | −3.118418469 | 0.045488387 | 0.03454584 |
| NF1-MET | 2 | PDGFRb Y751 | 0.224593878 | 3.041081528 | 0.073853291 | 0.038423831 |
| NF1-MET | 2 | PYK2 Y402 | −0.357038387 | −2.963657163 | 0.12047223 | 0.042611794 |
| NF1-MET | 2 | ATPcitrate Lyase S454 | 0.578983064 | 2.92971822 | 0.197624147 | 0.042611794 |
| NF1-MET | 2 | PKCa-BII T638/T641 | −0.268588053 | −2.92711569 | 0.091758605 | 0.042611794 |
| NF1-MET | 2 | STAT3 Y705 | −0.199537985 | −2.901091404 | 0.068780316 | 0.043228251 |
| NF1-MET | 2 | p90RSK T359/S363 | 0.299326773 | 2.790135479 | 0.107280372 | 0.051579862 |
| NF1-MET | 2 | NFKB p65 S536 | 0.384217692 | 2.734680663 | 0.140498193 | 0.055441102 |
| NF1-MET | 2 | AKT T308 | −0.140812972 | −2.609527129 | 0.053961107 | 0.068011589 |
| NF1-MET | 2 | Paxillin Y118 | −0.425764858 | −2.462668149 | 0.172887629 | 0.086838723 |
| NF1-MET | 2 | Chk1 S345 | −0.385540662 | −2.430346807 | 0.158636068 | 0.089515615 |
| NF1-MET | 2 | AXL Y702 | −0.224327571 | −2.361084704 | 0.095010387 | 0.098886672 |
| NF1-MET | 2 | PAK1 S199/S204-PAK2 S192/S197 | 0.288540675 | 2.336184697 | 0.123509359 | 0.100219981 |
| NF1-MET | 2 | cABL Y245 | 0.256465918 | 2.323768124 | 0.110366398 | 0.100219981 |
| NF1-MET | 2 | FRS2a Y436 | 0.130796537 | 2.147269734 | 0.060912951 | 0.134777023 |
| NF1-MET | 2 | FOXO1 S256 | −0.159413387 | −2.108642238 | 0.075600016 | 0.137625892 |
| NF1-MET | 2 | Raf S259 | −0.354879646 | −2.107342191 | 0.168401529 | 0.137625892 |
| NF1-MET | 2 | AMPKb1 S108 | 0.227215981 | 2.077323391 | 0.109379205 | 0.141727924 |
| NF1-MET | 2 | Acetyl CoAC S79 | 0.194756604 | 1.987884072 | 0.097971812 | 0.162333371 |
| NF1-MET | 2 | eNOS S1177 | −0.132242743 | −1.90489787 | 0.069422485 | 0.183577551 |
| NF1-MET | 2 | elF4E S209 | 0.20749804 | 1.888237211 | 0.109889816 | 0.184719755 |
| NF1-MET | 2 | LIMK1 T508/LIMK2 T505 | 0.502971608 | 1.874645387 | 0.268302267 | 0.184957224 |
| NF1-MET | 2 | ELK-1 S383 | −0.126221239 | −1.848627836 | 0.06827834 | 0.189327013 |
| NF1-MET | 2 | p70S6K T389 | −0.223079564 | −1.830005003 | 0.121901068 | 0.19138652 |
| NF1-MET | 2 | CRKII Y221 | 0.161829588 | 1.71803078 | 0.094194813 | 0.224956683 |
| NF1-MET | 2 | AMPKa T172 | −0.106060366 | −1.711454358 | 0.061970899 | 0.224956683 |
| NF1-MET | 2 | STAT5 Y694 | −0.12552217 | −1.648615013 | 0.076137952 | 0.245083145 |
| NF1-MET | 2 | STAT4 Y693 | 0.188068483 | 1.636372925 | 0.114930087 | 0.245246876 |
| NF1-MET | 2 | PKCz-L T410/T403 | −0.340169334 | −1.613562251 | 0.210818847 | 0.249846724 |
| NF1-MET | 2 | MST1 T183/MST2 T180 | −0.116366722 | −1.55288187 | 0.074935978 | 0.270957325 |
| NF1-MET | 2 | HSP27 S82 | 0.204477521 | 1.433895702 | 0.142602786 | 0.314784611 |
| NF1-MET | 2 | PAK2 S20 | 0.075958983 | 1.427312817 | 0.053218175 | 0.314784611 |
| NF1-MET | 2 | PTEN S380 | 0.09618027 | 1.425906581 | 0.067452013 | 0.314784611 |
| NF1-MET | 2 | Aurora AT288/BT232/CT198 | −0.075020147 | −1.388531212 | 0.05402842 | 0.328084638 |
| NF1-MET | 2 | IGF1R beta total | −0.109648759 | −1.313599187 | 0.083472005 | 0.358184767 |
| NF1-MET | 2 | SUMO2/3 total | −0.088966648 | −1.309940521 | 0.067916555 | 0.358184767 |
| NF1-MET | 2 | PDK1 S241 | 0.119972047 | 1.283834566 | 0.093448214 | 0.36659027 |
| NF1-MET | 2 | EGFR | −0.115202589 | −1.212201788 | 0.095035819 | 0.395688162 |
| NF1-MET | 2 | HSP90a T5/T7 | 0.061561236 | 1.211513227 | 0.050813507 | 0.395688162 |
| NF1-MET | 2 | STAT1 Y701 | 0.091864833 | 1.197848256 | 0.076691544 | 0.397412797 |
| NF1-MET | 2 | p38 MAPK T180/Y182 | −0.090398979 | −1.159134316 | 0.077988356 | 0.414215723 |
| NF1-MET | 2 | MCSFR Y732 | 0.09161124 | 1.127143873 | 0.081277326 | 0.425867883 |
| NF1-MET | 2 | FOXO3a S256 | 0.064443492 | 1.113218684 | 0.057889337 | 0.425867883 |
| NF1-MET | 2 | ERBB2 Y1248 | 0.054571225 | 1.102954623 | 0.049477307 | 0.425867883 |
| NF1-MET | 2 | EGFR Y1148 | 0.057698423 | 1.098267471 | 0.052535857 | 0.425867883 |
| NF1-MET | 2 | MET Y1234/Y1235 | 0.062520874 | 1.084193355 | 0.057665797 | 0.428245587 |
| NF1-MET | 2 | ALK Y1586 | 0.048094682 | 1.007265605 | 0.047747766 | 0.453860046 |
| NF1-MET | 2 | IGF1R Y1135/Y1136-IR Y1150/Y1151 | 0.048104918 | 0.995876617 | 0.048304094 | 0.453860046 |
| NF1-MET | 2 | PKCa-BII T638/T641.1 | 0.100953368 | 0.992699373 | 0.101695811 | 0.453860046 |
| NF1-MET | 2 | ERBB3 total | −0.089390304 | −0.991566511 | 0.090150588 | 0.453860046 |
| NF1-MET | 2 | SEK1/MKK4 S80 | 0.062495857 | 0.984466402 | 0.06348196 | 0.453860046 |
| NF1-MET | 2 | mTOR S2448 | 0.11995107 | 0.983356482 | 0.121981268 | 0.453860046 |
| NF1-MET | 2 | ATR S428 | 0.06010448 | 0.916346836 | 0.065591409 | 0.48923586 |
| NF1-MET | 2 | Insulin Rec beta total | 0.139451477 | 0.907646742 | 0.153640696 | 0.48923586 |
| NF1-MET | 2 | 4EBP1 S65 | −0.071161791 | −0.898915627 | 0.079164039 | 0.48923586 |
| NF1-MET | 2 | 4EBP1 T37/46 | 0.086559049 | 0.837613545 | 0.103340077 | 0.517529735 |
| NF1-MET | 2 | cKit Y719 | −0.047101592 | −0.822027741 | 0.057299273 | 0.517529735 |
| NF1-MET | 2 | EGFR Y1173 | −0.060353872 | −0.821221992 | 0.073492761 | 0.517529735 |
| NF1-MET | 2 | FAK Y576/Y577 | −0.084928722 | −0.817651773 | 0.103869061 | 0.517529735 |
| NF1-MET | 2 | ERBB3 Y1197 | 0.03893665 | 0.745070258 | 0.052259031 | 0.5603899 |
| NF1-MET | 2 | RAS-GRF1 S916 | −0.037999327 | −0.725068367 | 0.052407923 | 0.567581099 |
| NF1-MET | 2 | ERBB3 Y1289 | −0.047112954 | −0.681180319 | 0.069163704 | 0.591686368 |
| NF1-MET | 2 | ATF2 T69/71 | 0.287905702 | 0.553428151 | 0.520222365 | 0.678726405 |
| NF1-MET | 2 | cKit Y719.1 | −0.026938522 | −0.523060932 | 0.05150169 | 0.694087297 |
| NF1-MET | 2 | PTEN total | −0.038462342 | −0.465095131 | 0.082697795 | 0.730972672 |
| NF1-MET | 2 | JAK1 Y1022/Y1023 | −0.02886501 | −0.446166932 | 0.064695539 | 0.737499192 |
| NF1-MET | 2 | ERK 1/2 total | −0.021646902 | −0.430497215 | 0.050283489 | 0.741470992 |
| NF1-MET | 2 | cAbl T735 | 0.015672364 | 0.28602473 | 0.054793736 | 0.840427895 |
| NF1-MET | 2 | PAK1 T423/PAK2 T402 | 0.015374589 | 0.283991317 | 0.054137532 | 0.840427895 |
| NF1-MET | 2 | IGF1R Y1131/IR Y1146 | −0.018281078 | −0.233796222 | 0.078192357 | 0.871708101 |
| NF1-MET | 2 | YAP S127 | 0.020694077 | 0.215920833 | 0.095841039 | 0.876766942 |
| NF1-MET | 2 | FGFR Y653/Y654 | 0.010820267 | 0.192137219 | 0.056315308 | 0.885142626 |
| NF1-MET | 2 | GSK3aB S21/S9 | −0.024481708 | −0.182183964 | 0.134379047 | 0.885142626 |
| NF1-MET | 2 | SRC Family Y416 | −0.017649341 | −0.160619595 | 0.109882865 | 0.892964663 |
| NF1-MET | 2 | SRC Y527 | 0.003733285 | 0.060600902 | 0.061604442 | 0.962342908 |
| NF1-MET | 2 | ERBB4 total | −0.001936876 | −0.017160856 | 0.112865928 | 0.986575511 |
| NF1-P53 | 2 | ERK1/2 T202/Y204 | 3.010364583 | 17.13568196 | 0.175678131 | 5.99E-08 |
| NF1-P53 | 2 | PDGFRb Y751 | 0.904525529 | 10.11412579 | 0.089431904 | 1.28E-05 |
| NF1-P53 | 2 | GSK3aB S21/S9 | 0.669624449 | 8.432233252 | 0.079412467 | 6.15E-05 |
| NF1-P53 | 2 | AKT T308 | 0.71647495 | 6.663274863 | 0.107525949 | 0.000511218 |
| NF1-P53 | 2 | AKT S473 | 1.218304914 | 6.49272911 | 0.187641421 | 0.000526599 |
| NF1-P53 | 2 | elF4E S209 | 1.313683465 | 6.217691262 | 0.211281553 | 0.000665189 |
| NF1-P53 | 2 | IGF1R beta total | 0.489705544 | 5.435102443 | 0.090100518 | 0.001972004 |
| NF1-P53 | 2 | C-RAF S338 | 0.494346801 | 5.203431476 | 0.095004 | 0.002497863 |
| NF1-P53 | 2 | MEK1/2 S217/S221 | 0.340689965 | 5.079591142 | 0.067070352 | 0.002497863 |
| NF1-P53 | 2 | FGFR Y653/Y654 | 0.334713668 | 5.078976972 | 0.065901789 | 0.002497863 |
| NF1-P53 | 2 | RAS-GRF1 S916 | 0.718165835 | 4.879138263 | 0.147191122 | 0.003191807 |
| NF1-P53 | 2 | CRKII Y221 | 0.780558668 | 4.761242432 | 0.163940123 | 0.003585727 |
| NF1-P53 | 2 | TYK2 Y1054/Y1055 | 0.343089769 | 4.350194218 | 0.07886769 | 0.006821601 |
| NF1-P53 | 2 | p90RSK T359/S363 | 2.446636721 | 4.232685332 | 0.578034163 | 0.007818765 |
| NF1-P53 | 2 | YAP S127 | 0.30150881 | 4.078700928 | 0.073922755 | 0.009320997 |
| NF1-P53 | 2 | EGFR Y1068 | 0.436628558 | 4.061619405 | 0.107501101 | 0.009320997 |
| NF1-P53 | 2 | PKCz-L T410/T403 | 0.429872432 | 3.905731493 | 0.110061952 | 0.011659665 |
| NF1-P53 | 2 | Cofilin S3 | 0.419253559 | 3.722958236 | 0.112613017 | 0.015416075 |
| NF1-P53 | 2 | ERBB2 Y1248 | 0.403440708 | 3.682397883 | 0.109559239 | 0.015444474 |
| NF1-P53 | 2 | AMPKa T172 | 0.294335447 | 3.665033139 | 0.080309082 | 0.015444474 |
| NF1-P53 | 2 | SGK1 S78 | 0.78686947 | 3.243479394 | 0.242600422 | 0.032293889 |
| NF1-P53 | 2 | PKCa-BII T638/T641.1 | 0.411619851 | 3.196770414 | 0.128761155 | 0.032500124 |
| NF1-P53 | 2 | FOXO3a S256 | 0.180072557 | 3.191579087 | 0.056421148 | 0.032500124 |
| NF1-P53 | 2 | SRC Y527 | 0.393073974 | 3.129259805 | 0.125612445 | 0.03361802 |
| NF1-P53 | 2 | PYK2 Y402 | 0.611115205 | 3.129124953 | 0.195299074 | 0.03361802 |
| NF1-P53 | 2 | NFKB p65 S536 | 0.285184916 | 3.091711009 | 0.092241777 | 0.03467691 |
| NF1-P53 | 2 | ELK-1 S383 | 0.241889867 | 3.033426738 | 0.079741457 | 0.037253839 |
| NF1-P53 | 2 | STAT3 Y705 | 0.264961472 | 2.993820517 | 0.088502791 | 0.038371634 |
| NF1-P53 | 2 | ATR S428 | 0.333482114 | 2.979610307 | 0.111921386 | 0.038371634 |
| NF1-P53 | 2 | ERBB3 Y1197 | 0.209400955 | 2.921352907 | 0.071679445 | 0.040882897 |
| NF1-P53 | 2 | 4EBP1 T37/46 | 0.232743611 | 2.910281191 | 0.079972895 | 0.040882897 |
| NF1-P53 | 2 | cKit Y719 | 0.17288198 | 2.882589378 | 0.059974543 | 0.041716331 |
| NF1-P53 | 2 | HSP27 S82 | 0.546905289 | 2.625662433 | 0.208292308 | 0.065387908 |
| NF1-P53 | 2 | EGFR Y1148 | 0.300051483 | 2.338387089 | 0.128315575 | 0.107877848 |
| NF1-P53 | 2 | PAK2 S20 | 0.421223599 | 2.311456784 | 0.182232954 | 0.110081135 |
| NF1-P53 | 2 | JAK1 Y1022/Y1023 | 0.226719736 | 2.282915398 | 0.099311493 | 0.112739035 |
| NF1-P53 | 2 | elF4G S1108 | 0.447932511 | 2.190751451 | 0.204465235 | 0.120340855 |
| NF1-P53 | 2 | LIMK1 T508/LIMK2 T505 | −0.445656431 | −2.166426331 | 0.205710402 | 0.120340855 |
| NF1-P53 | 2 | ATF2 T69/71 | −4.483851066 | −2.156060658 | 2.079649777 | 0.120340855 |
| NF1-P53 | 2 | Ephrin A3 Y799/A4 Y799/A5 Y833 | 0.219927822 | 2.155791819 | 0.102017189 | 0.120340855 |
| NF1-P53 | 2 | AXL Y702 | 0.16013891 | 2.144282221 | 0.074681825 | 0.120340855 |
| NF1-P53 | 2 | Chk1 S345 | −0.867735767 | −2.141068475 | 0.405281651 | 0.120340855 |
| NF1-P53 | 2 | EGFR Y1173 | 0.594358949 | 2.125542156 | 0.279626987 | 0.120340855 |
| NF1-P53 | 2 | p90RSK S380 | 0.206042768 | 2.123177847 | 0.097044516 | 0.120340855 |
| NF1-P53 | 2 | eNOS S1177 | 0.349582937 | 2.117582399 | 0.165085872 | 0.120340855 |
| NF1-P53 | 2 | S6RP S235/S236 | −0.438272388 | −2.111031895 | 0.207610501 | 0.120340855 |
| NF1-P53 | 2 | SAPK/JNK T183/Y185 | 0.226261372 | 2.047154124 | 0.110524835 | 0.130212957 |
| NF1-P53 | 2 | JAK2 Y1007 | 0.339950076 | 2.035897464 | 0.166977995 | 0.130212957 |
| NF1-P53 | 2 | SUMO2/3 total | 0.188737227 | 2.031289417 | 0.092914985 | 0.130212957 |
| NF1-P53 | 2 | Raf S259 | 0.350285464 | 1.942261354 | 0.180349294 | 0.149278961 |
| NF1-P53 | 2 | PAK1 T423/PAK2 T402 | 0.112535305 | 1.900196996 | 0.059222968 | 0.15750216 |
| NF1-P53 | 2 | 4EBP1 T70 | 0.225779621 | 1.866683435 | 0.120952282 | 0.162348866 |
| NF1-P53 | 2 | SEK1/MKK4 S80 | 0.270497053 | 1.86055151 | 0.145385415 | 0.162348866 |
| NF1-P53 | 2 | PKCa-BII T638/T641 | 0.311119633 | 1.766824766 | 0.176089695 | 0.187145595 |
| NF1-P53 | 2 | ALK Y1586 | 0.132000358 | 1.730004546 | 0.076300585 | 0.195442004 |
| NF1-P53 | 2 | MET Y1234/Y1235 | 0.136078743 | 1.712320537 | 0.079470368 | 0.195442004 |
| NF1-P53 | 2 | Acetyl CoAC S79 | 0.192622266 | 1.709275461 | 0.112692349 | 0.195442004 |
| NF1-P53 | 2 | cKit Y719.1 | −0.095457855 | −1.55748383 | 0.061289789 | 0.247059458 |
| NF1-P53 | 2 | Insulin Rec beta total | 0.243733375 | 1.522261582 | 0.160112676 | 0.257181237 |
| NF1-P53 | 2 | PTEN total | 0.271065562 | 1.481409013 | 0.182978205 | 0.270097317 |
| NF1-P53 | 2 | ATF2 T71 | −0.170476165 | −1.469068285 | 0.116043731 | 0.270969174 |
| NF1-P53 | 2 | mTOR S2448 | 0.236164957 | 1.413586657 | 0.167067902 | 0.291137456 |
| NF1-P53 | 2 | cAbl T735 | 0.233992294 | 1.383691566 | 0.169107263 | 0.300280774 |
| NF1-P53 | 2 | AMPKb1 S108 | 0.189719744 | 1.351725949 | 0.140353704 | 0.306315814 |
| NF1-P53 | 2 | IRS1 S612 | 0.147356104 | 1.344166693 | 0.109626362 | 0.306315814 |
| NF1-P53 | 2 | PRK1 T774/PRK2 T816 | 0.492186186 | 1.335865163 | 0.368440018 | 0.306315814 |
| NF1-P53 | 2 | AMPKa1 S485 | 0.173551816 | 1.331212781 | 0.130371206 | 0.306315814 |
| NF1-P53 | 2 | PRAS40 T246 | 0.210379849 | 1.315503715 | 0.159923417 | 0.308949708 |
| NF1-P53 | 2 | ERBB3 Y1289 | 0.239761306 | 1.306480341 | 0.183516964 | 0.308949708 |
| NF1-P53 | 2 | 4EBP1 S65 | 0.120729161 | 1.284716269 | 0.093973404 | 0.314814533 |
| NF1-P53 | 2 | STAT1 Y701 | −0.146905554 | −1.261254191 | 0.116475771 | 0.321611473 |
| NF1-P53 | 2 | SRC Family Y416 | −0.172644004 | −1.100542592 | 0.156871715 | 0.401776139 |
| NF1-P53 | 2 | FOXO1 S256 | 0.161319084 | 1.087436909 | 0.148347994 | 0.403766118 |
| NF1-P53 | 2 | B-RAF S445 | 0.086568042 | 1.036768481 | 0.083497949 | 0.427878345 |
| NF1-P53 | 2 | IGF1R Y1135/Y1136-IR Y1150/Y1151 | 0.086052305 | 1.016182652 | 0.084681927 | 0.434472241 |
| NF1-P53 | 2 | STAT4 Y693 | 0.192308579 | 0.967008494 | 0.198869586 | 0.458785274 |
| NF1-P53 | 2 | MCSFR Y732 | 0.136118264 | 0.95601657 | 0.142380654 | 0.459650554 |
| NF1-P53 | 2 | EGFR | 0.096651035 | 0.938840801 | 0.102947203 | 0.463832839 |
| NF1-P53 | 2 | p38 MAPK T180/Y182 | −0.066818407 | −0.922426986 | 0.072437611 | 0.463832839 |
| NF1-P53 | 2 | cABL Y245 | 0.113245687 | 0.920949879 | 0.122966178 | 0.463832839 |
| NF1-P53 | 2 | p70S6K T389 | −0.139200662 | −0.898884989 | 0.154859257 | 0.47177771 |
| NF1-P53 | 2 | ERK 1/2 total | 0.129601234 | 0.777377268 | 0.166716007 | 0.538749885 |
| NF1-P53 | 2 | FRS2a Y436 | 0.101692386 | 0.777330153 | 0.130822644 | 0.538749885 |
| NF1-P53 | 2 | ERBB3 total | 0.157347755 | 0.733829217 | 0.214420128 | 0.561931424 |
| NF1-P53 | 2 | Aurora AT288/BT232/CT198 | 0.052414213 | 0.723617075 | 0.072433633 | 0.561931424 |
| NF1-P53 | 2 | ERBB4 total | −0.085129277 | −0.714727449 | 0.119107329 | 0.561931424 |
| NF1-P53 | 2 | Paxillin Y118 | 0.40248955 | 0.667079652 | 0.603360557 | 0.588378613 |
| NF1-P53 | 2 | HSP90a T5/T7 | 0.127216857 | 0.62179464 | 0.204596259 | 0.613628774 |
| NF1-P53 | 2 | FAK Y576/Y577 | 0.076011002 | 0.453037429 | 0.167780844 | 0.732402293 |
| NF1-P53 | 2 | PAK1 S199/S204-PAK2 S192/S197 | 0.075967466 | 0.413771172 | 0.183597774 | 0.754794122 |
| NF1-P53 | 2 | ATPcitrate Lyase S454 | 0.089061158 | 0.391078908 | 0.227731938 | 0.764192257 |
| NF1-P53 | 2 | S6RP S240/S244 | −0.038460057 | −0.368639052 | 0.10432985 | 0.77335535 |
| NF1-P53 | 2 | PTEN S380 | −0.021371962 | −0.27591531 | 0.077458411 | 0.837988619 |
| NF1-P53 | 2 | IGF1R Y1131/IR Y1146 | 0.043792192 | 0.236068329 | 0.185506424 | 0.860742894 |
| NF1-P53 | 2 | FOXO1 T24/FOXO3a T32 | 0.0214416 | 0.182521197 | 0.117474574 | 0.894284957 |
| NF1-P53 | 2 | PDK1 S241 | −0.009597387 | −0.103406735 | 0.092812011 | 0.948038149 |
| NF1-P53 | 2 | MST1 T183/MST2 T180 | −0.008415726 | −0.0901971 | 0.093303735 | 0.948751765 |
| NF1-P53 | 2 | p70S6K S371 | −0.013011528 | −0.06305636 | 0.20634759 | 0.960437771 |
| NF1-P53 | 2 | STAT5 Y694 | 0.001268889 | 0.015436168 | 0.082202321 | 0.987932164 |
| NF1-MET | 48 | AMPKa1 S485 | 0.37796374 | 4.946710116 | 0.076407093 | 0.018034171 |
| NF1-MET | 48 | PAK1 S199/S204-PAK2 S192/S197 | −0.40007899 | −4.819039561 | 0.083020483 | 0.018034171 |
| NF1-MET | 48 | C-RAF S338 | 0.33997879 | 4.054645186 | 0.083849209 | 0.0477197 |
| NF1-MET | 48 | MET Y1234/Y1235 | −0.138757341 | −3.791211758 | 0.036599734 | 0.058400691 |
| NF1-MET | 48 | cKit Y719 | −0.205340704 | −3.66161783 | 0.056079229 | 0.059572304 |
| NF1-MET | 48 | PTEN S380 | −0.139448487 | −3.091750027 | 0.045103416 | 0.110263618 |
| NF1-MET | 48 | PYK2 Y402 | −0.221909975 | −3.05359782 | 0.072671645 | 0.110263618 |
| NF1-MET | 48 | IGF1R beta total | −1.190846864 | −3.038981056 | 0.391857285 | 0.110263618 |
| NF1-MET | 48 | Paxillin Y118 | −1.072641782 | −3.026197152 | 0.354452049 | 0.110263618 |
| NF1-MET | 48 | ATF2 T69/71 | 0.853900834 | 2.538571829 | 0.336370562 | 0.239282081 |
| NF1-MET | 48 | EGFR Y1068 | 0.568572377 | 2.509580101 | 0.226560761 | 0.239282081 |
| NF1-MET | 48 | HSP27 S82 | −0.414401695 | −2.319399559 | 0.178667661 | 0.312171404 |
| NF1-MET | 48 | B-RAF S445 | −0.141869056 | −2.147974306 | 0.066047836 | 0.3283577 |
| NF1-MET | 48 | STAT4 Y693 | −0.217067811 | −2.146893135 | 0.101107879 | 0.3283577 |
| NF1-MET | 48 | JAK1 Y1022/Y1023 | −0.140156149 | −2.123675315 | 0.065996976 | 0.3283577 |
| NF1-MET | 48 | AXL Y702 | −0.19260434 | −2.060228379 | 0.093486888 | 0.3283577 |
| NF1-MET | 48 | ATF2 T71 | 0.308000079 | 2.041506752 | 0.150868999 | 0.3283577 |
| NF1-MET | 48 | FOXO1 T24/FOXO3a T32 | 0.180279826 | 2.009693407 | 0.089705139 | 0.3283577 |
| NF1-MET | 48 | SGK1 S78 | 0.200769041 | 2.007064195 | 0.1000312 | 0.3283577 |
| NF1-MET | 48 | PRAS40 T246 | 0.126670824 | 1.988108033 | 0.063714256 | 0.3283577 |
| NF1-MET | 48 | ATR S428 | 0.117317044 | 1.982296893 | 0.059182378 | 0.3283577 |
| NF1-MET | 48 | PKCz-L T410/T403 | 0.383182032 | 1.88883495 | 0.202866869 | 0.359392363 |
| NF1-MET | 48 | FRS2a Y436 | −0.136031345 | −1.870908783 | 0.0727087 | 0.359392363 |
| NF1-MET | 48 | EGFR | −0.122447681 | −1.85483888 | 0.066015265 | 0.359392363 |
| NF1-MET | 48 | ERBB3 Y1289 | 0.079814489 | 1.826726654 | 0.043692629 | 0.362257368 |
| NF1-MET | 48 | AKT S473 | −0.179936555 | −1.716076269 | 0.104853472 | 0.416719138 |
| NF1-MET | 48 | LIMK1 T508/LIMK2 T505 | 0.223147981 | 1.688686794 | 0.132142907 | 0.416719138 |
| NF1-MET | 48 | 4EBP1 S65 | −0.102675468 | −1.678317444 | 0.061177621 | 0.416719138 |
| NF1-MET | 48 | cAbl T735 | 0.08577078 | 1.650350497 | 0.051971251 | 0.421685598 |
| NF1-MET | 48 | ERK1/2 T202/Y204 | −0.185725311 | −1.500794006 | 0.123751367 | 0.52142927 |
| NF1-MET | 48 | eNOS S1177 | −0.0786173 | −1.413602386 | 0.055614861 | 0.566481485 |
| NF1-MET | 48 | YAP S127 | −0.119722462 | −1.408627137 | 0.084992301 | 0.566481485 |
| NF1-MET | 48 | MCSFR Y732 | 0.11587647 | 1.352506833 | 0.085675331 | 0.583643249 |
| NF1-MET | 48 | Aurora AT288/BT232/CT198 | 0.079586916 | 1.35007624 | 0.058949942 | 0.583643249 |
| NF1-MET | 48 | ERBB3 Y1197 | 0.077650264 | 1.332247073 | 0.058285183 | 0.583643249 |
| NF1-MET | 48 | FAK Y576/Y577 | −0.131890129 | −1.310767883 | 0.100620507 | 0.586546235 |
| NF1-MET | 48 | ELK-1 S383 | −0.070712657 | −1.20347983 | 0.058756827 | 0.644081422 |
| NF1-MET | 48 | CRKII Y221 | 0.100750104 | 1.20012136 | 0.08394993 | 0.644081422 |
| NF1-MET | 48 | cABL Y245 | 0.074552422 | 1.191318343 | 0.062579765 | 0.644081422 |
| NF1-MET | 48 | GSK3aB S21/S9 | 0.096144411 | 1.178779481 | 0.081562678 | 0.644081422 |
| NF1-MET | 48 | ERK 1/2 total | −0.045697609 | −1.129934619 | 0.040442701 | 0.674948012 |
| NF1-MET | 48 | Ephrin A3 Y799/A4 Y799/A5 Y833 | 0.060344409 | 1.079496162 | 0.055900531 | 0.695618534 |
| NF1-MET | 48 | RAS-GRF1 S916 | 0.063022824 | 1.050403438 | 0.059998684 | 0.695618534 |
| NF1-MET | 48 | AKT T308 | −0.070924821 | −1.048968893 | 0.067613846 | 0.695618534 |
| NF1-MET | 48 | Chk1 S345 | 0.16047593 | 1.024497505 | 0.156638673 | 0.695618534 |
| NF1-MET | 48 | PDK1 S241 | 0.109884181 | 1.020968175 | 0.10762743 | 0.695618534 |
| NF1-MET | 48 | p90RSK S380 | 0.073311803 | 1.003218825 | 0.073076582 | 0.695618534 |
| NF1-MET | 48 | mTOR S2448 | −0.102220137 | −0.997490811 | 0.102477272 | 0.695618534 |
| NF1-MET | 48 | PRK1 T774/PRK2 T816 | 0.22716092 | 0.980105562 | 0.231771891 | 0.697952328 |
| NF1-MET | 48 | p38 MAPK T180/Y182 | 0.073336447 | 0.908135058 | 0.080755 | 0.75402912 |
| NF1-MET | 48 | p70S6K S371 | 0.079345336 | 0.889890787 | 0.089163005 | 0.757397037 |
| NF1-MET | 48 | Acetyl CoAC S79 | 0.043279678 | 0.833547946 | 0.051922242 | 0.789218168 |
| NF1-MET | 48 | ERBB3 total | 0.078842399 | 0.82899535 | 0.095105961 | 0.789218168 |
| NF1-MET | 48 | cKit Y719.1 | −0.04353576 | −0.810352496 | 0.053724472 | 0.793382834 |
| NF1-MET | 48 | Insulin Rec beta total | 0.076941321 | 0.75785152 | 0.101525588 | 0.822495297 |
| NF1-MET | 48 | JAK2 Y1007 | 0.061968239 | 0.743313017 | 0.083367622 | 0.822495297 |
| NF1-MET | 48 | PAK2 S20 | −0.041619237 | −0.732194507 | 0.056841777 | 0.822495297 |
| NF1-MET | 48 | elF4E S209 | 0.061350183 | 0.724686088 | 0.084657597 | 0.822495297 |
| NF1-MET | 48 | IGF1R Y1131/IR Y1146 | −0.065404199 | −0.697273547 | 0.093799914 | 0.835006176 |
| NF1-MET | 48 | ERBB4 total | −0.055801468 | −0.660584693 | 0.084472845 | 0.835006176 |
| NF1-MET | 48 | STAT1 Y701 | −0.090622465 | −0.659315913 | 0.137449231 | 0.835006176 |
| NF1-MET | 48 | PDGFRb Y751 | 0.040637743 | 0.657080039 | 0.061845956 | 0.835006176 |
| NF1-MET | 48 | PAK1 T423/PAK2 T402 | 0.030554298 | 0.607107384 | 0.050327666 | 0.860421265 |
| NF1-MET | 48 | IRS1 S612 | 0.048608617 | 0.593295138 | 0.08192991 | 0.860421265 |
| NF1-MET | 48 | Cofilin S3 | 0.033663051 | 0.591051049 | 0.056954558 | 0.860421265 |
| NF1-MET | 48 | p70S6K T389 | −0.053843581 | −0.57520891 | 0.093607001 | 0.862894592 |
| NF1-MET | 48 | STAT5 Y694 | −0.040637038 | −0.486271077 | 0.083568693 | 0.877044712 |
| NF1-MET | 48 | FOXO1 S256 | 0.020685928 | 0.480263471 | 0.04307204 | 0.877044712 |
| NF1-MET | 48 | p90RSK T359/S363 | 0.085342308 | 0.458611917 | 0.186088291 | 0.877044712 |
| NF1-MET | 48 | elF4G S1108 | 0.028573198 | 0.451725765 | 0.063253417 | 0.877044712 |
| NF1-MET | 48 | ERBB2 Y1248 | 0.022343103 | 0.450190976 | 0.049630277 | 0.877044712 |
| NF1-MET | 48 | 4EBP1 T37/46 | −0.031266332 | −0.428262034 | 0.073007481 | 0.877044712 |
| NF1-MET | 48 | FGFR Y653/Y654 | 0.024775964 | 0.413252934 | 0.05995351 | 0.877044712 |
| NF1-MET | 48 | S6RP S240/S244 | 0.087083602 | 0.412707785 | 0.211005474 | 0.877044712 |
| NF1-MET | 48 | PTEN total | 0.033659637 | 0.408980929 | 0.082301239 | 0.877044712 |
| NF1-MET | 48 | MEK1/2 S217/S221 | 0.025026013 | 0.406845664 | 0.0615123 | 0.877044712 |
| NF1-MET | 48 | SEK1/MKK4 S80 | 0.031709205 | 0.376866596 | 0.08413907 | 0.877044712 |
| NF1-MET | 48 | EGFR Y1148 | −0.042305995 | −0.368430913 | 0.114827484 | 0.877044712 |
| NF1-MET | 48 | SAPK/JNK T183/Y185 | −0.025225966 | −0.359730068 | 0.070124708 | 0.877044712 |
| NF1-MET | 48 | SUMO2/3 total | 0.021384676 | 0.358908705 | 0.059582494 | 0.877044712 |
| NF1-MET | 48 | PKCa-BII T638/T641.1 | 0.032736948 | 0.358506867 | 0.091314704 | 0.877044712 |
| NF1-MET | 48 | EGFR Y1173 | −0.018738898 | −0.34580907 | 0.054188568 | 0.877044712 |
| NF1-MET | 48 | Raf S259 | −0.032928323 | −0.345649482 | 0.095265073 | 0.877044712 |
| NF1-MET | 48 | ATPcitrate Lyase S454 | 0.040289704 | 0.322310662 | 0.125002702 | 0.886868641 |
| NF1-MET | 48 | FOXO3a S256 | −0.011309756 | −0.281400429 | 0.04019097 | 0.9119234 |
| NF1-MET | 48 | IGF1R Y1135/Y1136-IR Y1150/Y1151 | −0.018267631 | −0.258864553 | 0.0705683 | 0.920831358 |
| NF1-MET | 48 | S6RP S235/S236 | 0.045152057 | 0.236236347 | 0.191130863 | 0.922691109 |
| NF1-MET | 48 | AMPKa T172 | −0.014678808 | −0.219498568 | 0.066874278 | 0.922691109 |
| NF1-MET | 48 | MST1 T183/MST2 T180 | 0.011988399 | 0.211474763 | 0.056689504 | 0.922691109 |
| NF1-MET | 48 | SRC Y527 | 0.019042718 | 0.207657342 | 0.091702598 | 0.922691109 |
| NF1-MET | 48 | STAT3 Y705 | 0.010631514 | 0.158180199 | 0.067211407 | 0.939348158 |
| NF1-MET | 48 | TYK2 Y1054/Y1055 | 0.011323118 | 0.156856502 | 0.072187748 | 0.939348158 |
| NF1-MET | 48 | HSP90a T5/T7 | 0.006759779 | 0.150928521 | 0.044787949 | 0.939348158 |
| NF1-MET | 48 | NFKB p65 S536 | 0.009859336 | 0.101787287 | 0.096862159 | 0.969489151 |
| NF1-MET | 48 | AMPKb1 S108 | 0.005774302 | 0.083278346 | 0.06933738 | 0.974302478 |
| NF1-MET | 48 | PKCa-BII T638/T641 | −0.003882564 | −0.059201386 | 0.065582313 | 0.974469857 |
| NF1-MET | 48 | 4EBP1 T70 | −0.002416504 | −0.057837829 | 0.041780675 | 0.974469857 |
| NF1-MET | 48 | SRC Family Y416 | −0.006148277 | −0.039082924 | 0.15731363 | 0.979328004 |
| NF1-MET | 48 | ALK Y1586 | −1.96E-06 | −4.63E-05 | 0.04231125 | 0.999963801 |
| NF1-P53 | 48 | p70S6K S371 | −1.188965716 | −14.70413955 | 0.080859251 | 5.37E-07 |
| NF1-P53 | 48 | Cofilin S3 | 0.935505427 | 8.510809545 | 0.109919676 | 0.000104186 |
| NF1-P53 | 48 | STAT3 Y705 | 0.642420785 | 8.15328966 | 0.078792832 | 0.000107972 |
| NF1-P53 | 48 | AKT T308 | −0.815153411 | −7.176297718 | 0.113589687 | 0.00029118 |
| NF1-P53 | 48 | PKCz-L T410/T403 | −1.023016875 | −6.821386732 | 0.14997198 | 0.000381664 |
| NF1-P53 | 48 | CRKII Y221 | 0.730429189 | 6.413123032 | 0.11389602 | 0.000572755 |
| NF1-P53 | 48 | PRK1 T774/PRK2 T816 | −0.916501363 | −5.934462616 | 0.154437128 | 0.001006921 |
| NF1-P53 | 48 | cABL Y245 | 0.35898297 | 5.507073199 | 0.0651858 | 0.001718896 |
| NF1-P53 | 48 | AXL Y702 | 0.323622067 | 5.245046716 | 0.061700512 | 0.002331113 |
| NF1-P53 | 48 | PDGFRb Y751 | 0.324597283 | 4.420434338 | 0.073431083 | 0.008434293 |
| NF1-P53 | 48 | HSP90a T5/T7 | −0.356350306 | −4.275722489 | 0.083342712 | 0.009401693 |
| NF1-P53 | 48 | SAPK/JNK T183/Y185 | 0.399355681 | 4.254391705 | 0.093869044 | 0.009401693 |
| NF1-P53 | 48 | ATPcitrate Lyase S454 | 0.954435669 | 4.009396519 | 0.238049707 | 0.013409643 |
| NF1-P53 | 48 | ERBB3 Y1289 | −0.294289464 | −3.814946532 | 0.077141177 | 0.017668328 |
| NF1-P53 | 48 | SGK1 S78 | −0.551031351 | −3.645740019 | 0.151143896 | 0.021128552 |
| NF1-P53 | 48 | PAK2 S20 | −0.303591378 | −3.629206001 | 0.083652286 | 0.021128552 |
| NF1-P53 | 48 | PKCa-BII T638/T641 | −0.731892773 | −3.609761183 | 0.202753793 | 0.021128552 |
| NF1-P53 | 48 | ATR S428 | 0.254886444 | 3.519007289 | 0.072431349 | 0.022329025 |
| NF1-P53 | 48 | EGFR Y1068 | 0.328859293 | 3.509537826 | 0.093704445 | 0.022329025 |
| NF1-P53 | 48 | Aurora AT288/BT232/CT198 | −0.217101752 | −3.438953087 | 0.063130187 | 0.022329025 |
| NF1-P53 | 48 | p38 MAPK T180/Y182 | 0.437365607 | 3.428799436 | 0.127556486 | 0.022329025 |
| NF1-P53 | 48 | ERBB3 total | −0.382293598 | −3.420964573 | 0.111750236 | 0.022329025 |
| NF1-P53 | 48 | AKT S473 | −0.271305264 | −3.414708506 | 0.079451954 | 0.022329025 |
| NF1-P53 | 48 | ATF2 T71 | 0.523404131 | 3.362957619 | 0.15563804 | 0.023537137 |
| NF1-P53 | 48 | 4EBP1 T37/46 | 0.259673818 | 3.087643825 | 0.084100963 | 0.037339208 |
| NF1-P53 | 48 | PKCa-BII T638/T641.1 | 0.324103867 | 3.069849701 | 0.105576461 | 0.037339208 |
| NF1-P53 | 48 | MST1 T183/MST2 T180 | −0.245253836 | −2.971174467 | 0.082544407 | 0.043163301 |
| NF1-P53 | 48 | ELK-1 S383 | 0.194285926 | 2.686687406 | 0.072314303 | 0.070421213 |
| NF1-P53 | 48 | ATF2 T69/71 | 4.92824451 | 2.666931937 | 1.847907868 | 0.070513448 |
| NF1-P53 | 48 | IGF1R Y1131/IR Y1146 | −0.206288428 | −2.529022478 | 0.081568444 | 0.087817939 |
| NF1-P53 | 48 | S6RP S235/S236 | −1.284256801 | −2.467278564 | 0.520515526 | 0.091819324 |
| NF1-P53 | 48 | PYK2 Y402 | −0.236371173 | −2.463773255 | 0.095938688 | 0.091819324 |
| NF1-P53 | 48 | FRS2a Y436 | 0.195977734 | 2.452512663 | 0.079908959 | 0.091819324 |
| NF1-P53 | 48 | ERK1/2 T202/Y204 | −0.275334191 | −2.376040759 | 0.115879406 | 0.102426463 |
| NF1-P53 | 48 | GSK3aB S21/S9 | 0.195463433 | 2.285629702 | 0.085518417 | 0.117184977 |
| NF1-P53 | 48 | AMPKa1 S485 | 0.502817468 | 2.159942433 | 0.232792069 | 0.140846941 |
| NF1-P53 | 48 | FAK Y576/Y577 | −0.281971411 | −2.152030979 | 0.131025721 | 0.140846941 |
| NF1-P53 | 48 | MCSFR Y732 | −0.289477295 | −2.048184043 | 0.141333634 | 0.164836444 |
| NF1-P53 | 48 | elF4E S209 | −0.259668428 | −1.94110306 | 0.133773643 | 0.191172735 |
| NF1-P53 | 48 | S6RP S240/S244 | −0.634754883 | −1.934064596 | 0.328197354 | 0.191172735 |
| NF1-P53 | 48 | PRAS40 T246 | −0.208445063 | −1.862091972 | 0.111941336 | 0.211181199 |
| NF1-P53 | 48 | ALK Y1586 | −0.086544806 | −1.800294748 | 0.048072576 | 0.226392758 |
| NF1-P53 | 48 | AMPKa T172 | 0.122843009 | 1.793466054 | 0.06849475 | 0.226392758 |
| NF1-P53 | 48 | Raf S259 | −0.158437654 | −1.75036374 | 0.090516988 | 0.237983813 |
| NF1-P53 | 48 | cKit Y719.1 | −0.127652185 | −1.684118643 | 0.07579762 | 0.260009307 |
| NF1-P53 | 48 | PTEN total | −0.63558934 | −1.57251607 | 0.404186228 | 0.305673729 |
| NF1-P53 | 48 | SRC Y527 | 0.152181892 | 1.515055344 | 0.100446425 | 0.315011785 |
| NF1-P53 | 48 | C-RAF S338 | 0.152179573 | 1.499208411 | 0.101506616 | 0.315011785 |
| NF1-P53 | 48 | PAK1 S199/S204-PAK2 S192/S197 | −0.369317218 | −1.494488474 | 0.247119482 | 0.315011785 |
| NF1-P53 | 48 | TYK2 Y1054/Y1055 | 0.078381165 | 1.488168541 | 0.052669548 | 0.315011785 |
| NF1-P53 | 48 | JAK1 Y1022/Y1023 | −0.125996483 | −1.48726241 | 0.084717049 | 0.315011785 |
| NF1-P53 | 48 | ERBB3 Y1197 | −0.117075464 | −1.477708348 | 0.079227721 | 0.315011785 |
| NF1-P53 | 48 | SRC Family Y416 | 0.127634129 | 1.45584991 | 0.08766984 | 0.319492542 |
| NF1-P53 | 48 | FGFR Y653/Y654 | 0.151703263 | 1.436909197 | 0.105576095 | 0.319492542 |
| NF1-P53 | 48 | PDK1 S241 | 0.088973018 | 1.433392793 | 0.062071624 | 0.319492542 |
| NF1-P53 | 48 | SUMO2/3 total | 0.103132847 | 1.407136185 | 0.073292726 | 0.326997325 |
| NF1-P53 | 48 | Ephrin A3 Y799/A4 Y799/A5 Y833 | −0.180092097 | −1.277949833 | 0.140922666 | 0.386887774 |
| NF1-P53 | 48 | B-RAF S445 | 0.100969172 | 1.268238144 | 0.079613732 | 0.386887774 |
| NF1-P53 | 48 | cAbl T735 | −0.125213221 | −1.263601749 | 0.099092314 | 0.386887774 |
| NF1-P53 | 48 | p70S6K T389 | −0.153886111 | −1.24316367 | 0.123785882 | 0.392281236 |
| NF1-P53 | 48 | Chk1 S345 | 0.630391381 | 1.228628023 | 0.513085628 | 0.394308818 |
| NF1-P53 | 48 | p90RSK T359/S363 | 0.13560333 | 1.067531949 | 0.127025079 | 0.490070134 |
| NF1-P53 | 48 | STAT4 Y693 | −0.115635787 | −1.040006984 | 0.11118751 | 0.500880595 |
| NF1-P53 | 48 | Insulin Rec beta total | 0.078408158 | 1.029315133 | 0.076175075 | 0.500880595 |
| NF1-P53 | 48 | ERBB2 Y1248 | −0.09450299 | −0.981403598 | 0.096293707 | 0.52687515 |
| NF1-P53 | 48 | FOXO3a S256 | −0.045673628 | −0.940066001 | 0.048585555 | 0.548815244 |
| NF1-P53 | 48 | MEK1/2 S217/S221 | 0.15845465 | 0.909935188 | 0.174138391 | 0.562851827 |
| NF1-P53 | 48 | MET Y1234/Y1235 | −0.098355252 | −0.882576555 | 0.111441044 | 0.568244527 |
| NF1-P53 | 48 | STAT1 Y701 | −0.3231581 | −0.88045342 | 0.367035998 | 0.568244527 |
| NF1-P53 | 48 | PAK1 T423/PAK2 T402 | −0.050512557 | −0.828956087 | 0.060935142 | 0.598874899 |
| NF1-P53 | 48 | cKit Y719 | 0.052637198 | 0.81258753 | 0.064777266 | 0.60294147 |
| NF1-P53 | 48 | mTOR S2448 | −0.079164867 | −0.770668553 | 0.102722327 | 0.618645183 |
| NF1-P53 | 48 | EGFR Y1148 | 0.103185775 | 0.758926123 | 0.135962872 | 0.618645183 |
| NF1-P53 | 48 | Paxillin Y118 | −0.196136641 | −0.743697607 | 0.263731708 | 0.618645183 |
| NF1-P53 | 48 | PTEN S380 | −0.069940382 | −0.741307325 | 0.094347351 | 0.618645183 |
| NF1-P53 | 48 | JAK2 Y1007 | 0.053117456 | 0.737827661 | 0.071991685 | 0.618645183 |
| NF1-P53 | 48 | 4EBP1 S65 | 0.077175391 | 0.680140851 | 0.113469718 | 0.654984852 |
| NF1-P53 | 48 | IRS1 S612 | 0.063683041 | 0.658489792 | 0.096710749 | 0.66350465 |
| NF1-P53 | 48 | ERK 1/2 total | −0.044954448 | −0.644743217 | 0.069724578 | 0.665841259 |
| NF1-P53 | 48 | AMPKb1 S108 | −0.107233733 | −0.626869931 | 0.171062174 | 0.671449617 |
| NF1-P53 | 48 | Acetyl CoAC S79 | 0.054099695 | 0.595576941 | 0.090835779 | 0.687640734 |
| NF1-P53 | 48 | ERBB4 total | −0.060806728 | −0.520620945 | 0.116796546 | 0.739097423 |
| NF1-P53 | 48 | YAP S127 | 0.029101588 | 0.47467031 | 0.061309055 | 0.76768547 |
| NF1-P53 | 48 | EGFR | −0.056524183 | −0.443436335 | 0.127468542 | 0.784228074 |
| NF1-P53 | 48 | SEK1/MKK4 S80 | −0.068063891 | −0.427633572 | 0.15916405 | 0.787987731 |
| NF1-P53 | 48 | 4EBP1 T70 | 0.037997429 | 0.405895785 | 0.093613756 | 0.796631623 |
| NF1-P53 | 48 | elF4G S1108 | −0.033177858 | −0.384966873 | 0.086183669 | 0.804289504 |
| NF1-P53 | 48 | LIMK1 T508/LIMK2 T505 | −0.035473615 | −0.374082289 | 0.094828374 | 0.804289504 |
| NF1-P53 | 48 | EGFR Y1173 | −0.122134664 | −0.351948796 | 0.347023958 | 0.813169445 |
| NF1-P53 | 48 | eNOS S1177 | −0.019962912 | −0.33435334 | 0.059706033 | 0.818325836 |
| NF1-P53 | 48 | FOXO1 T24/FOXO3a T32 | −0.06477384 | −0.320081991 | 0.202366399 | 0.820782828 |
| NF1-P53 | 48 | HSP27 S82 | 0.669621519 | 0.3086704 | 2.169373926 | 0.820957233 |
| NF1-P53 | 48 | RAS-GRF1 S916 | −0.021866434 | −0.248226928 | 0.088090498 | 0.860332418 |
| NF1-P53 | 48 | IGF1R Y1135/Y1136-IR Y1150/Y1151 | −0.009133196 | −0.180227818 | 0.050675841 | 0.905751821 |
| NF1-P53 | 48 | STAT5 Y694 | 0.013476121 | 0.167466405 | 0.080470592 | 0.906439401 |
| NF1-P53 | 48 | p90RSK S380 | 0.006947483 | 0.118612177 | 0.058573097 | 0.935922833 |
| NF1-P53 | 48 | NFKB p65 S536 | −0.009933543 | −0.081245524 | 0.12226573 | 0.95186899 |
| NF1-P53 | 48 | IGF1R beta total | −0.01082882 | −0.064614954 | 0.167589995 | 0.95186899 |
| NF1-P53 | 48 | FOXO1 S256 | 0.013225355 | 0.061644904 | 0.214540926 | 0.95186899 |
p53 loss drives mTOR Dependency in MPNSTs
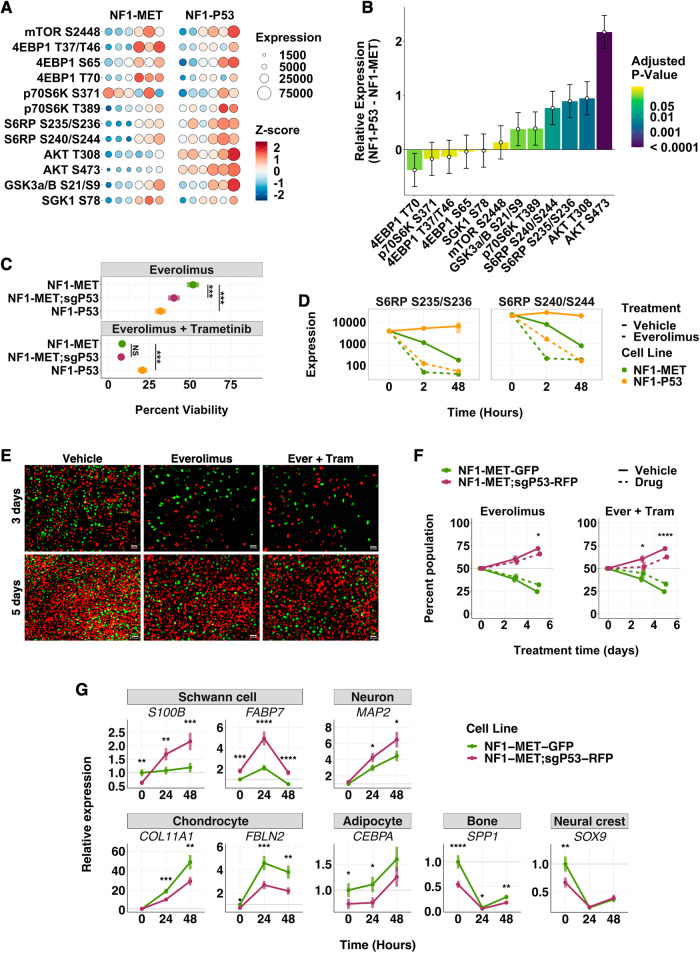
AKT activation was consistently elevated and sustained in p53-deficient MPNST cell lines and is targetable therapeutically, either directly or via its downstream effectors. To determine the scope of AKT/mTOR pathway activation in vivo, we assessed the phosphorylation status of mTORC1 and mTORC2 pathway effectors in MPNST tumorgrafts by RPPA. Globally, phosphorylation was significantly increased in NF1-P53 tumors compared to NF1-MET tumors (p-value = 0.0034) (Fig. 5A), with increased activation of 7/12 phosphosites in NF1-P53 tumors (Fig. 5B). AKT and S6RP phosphorylation were significantly increased in the NF1-P53 tumors (Fig. 5B), suggesting increased dependency on the AKT/mTOR pathway. Regardless of p53 status, treatment with the AKT inhibitor, afuresertib, had no effect on the MPNST cell lines, either as a single agent or in combination with trametinib (Supplementary Fig. 7A, B). However, treatment with the mTOR inhibitor, everolimus, significantly decreased viability in the p53-deficient cell lines compared to the p53-intact cells (Fig. 5C). This enhanced inhibition of the p53-deficient cells was observed even though everolimus strongly inhibited downstream pS6RP regardless of p53 status (Fig. 5D). Further, combination therapy of mTOR (everolimus) and MEK (trametinib) inhibition reversed clonal selection for p53 loss (Fig. 5E, F).
Fig. 5. p53 deficiency induces mTOR dependency.
A mTORC1/mTORC2 phospho-protein expression (size) and z - score (color) of NF1-MET and NF1-P53 tumors (6 animals/group). B Contrast estimates +/− SE of mTORC1/mTORC2 phospho-protein expression in NF1-P53 tumors compared to NF1-MET tumors. Color indicates P-value. C Percent viability of NF1-MET, NF1-MET;sgP53, and NF1-P53 cells after 72 h of everolimus (20 nM) or combination (everolimus 20 nM, trametinib 40 nM) treatment. D Phospho-S6RP expression measured over time by RPPA after vehicle (DMSO) or everolimus (100 nM) treatment. Images (E) and flow cytometry analysis (F) of GFP labeled NF1-MET and RFP labeled NF1-MET;sgP53 cells after 3 and 5 days of treatment with vehicle (DMSO), everolimus (20 nM), or combination everolimus (20 nM) and trametinib (40 nM). G Expression (relative to the housekeeping gene PPIA) of cell fate markers by qRT-PCR upon treatment with combination everolimus (20 nM) and trametinib (40 nM). See also Supplementary Fig. 9E. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
As combination mTOR and MEK inhibition was so effective in inhibiting MPNST cell growth, we next asked whether treatment reversed the nuclear localization of MET leading to global downregulation of MAPK and AKT/mTOR signaling. Unexpectedly, combination everolimus and trametinib actually induced ligand-independent MET localization specifically in the NF1-MET;sgP53 cells compared to the parental line (Supplementary Fig. 8A). Moreover, treatment with everolimus or the dual PI3K/mTOR inhibitor BEZ235 induced stronger compensatory ERK and AKT activation in NF1-P53 cells, consistent with increased MET activation (Supplementary Fig. 8B). These data reinforce the role of p53 in regulating MET localization and effector signaling in response to diverse stimuli.
We next tested whether the excessive AKT activation induced by everolimus resulted in p53-independent oncogene induced apoptosis or senescence in p53 null MPNST cells [59, 60]. Combination everolimus and trametinib treatment did induce apoptosis in the NF1-MET;sgP53 cells compared to the parental cell line, however, the difference was minor (Supplementary Fig. 9A, B). We also did not observe senescence based on expression of senescence-associated secretory phenotype markers (Supplementary Fig. 9C), or by staining with the senescence marker beta-galactosidase after 7 days of drug treatment (Supplementary Fig. 9D).
We evaluated whether differentiation of MPNST cells was impacted by combined everolimus and trametinib treatment. We observed that mTOR and MEK inhibition significantly increased expression of the Schwann cell markers S100B and FABP7 in NF1-MET;sgP53 but not NF1-MET cells (Fig. 5G). We also examined differentiation marker expression of neuron, chondrocyte, adipocyte, bone, kidney, endothelial, and muscle cells, as well as the multipotency marker SOX9, which is expressed in neural crest cells. Combined MEK and mTOR inhibition resulted in increase of the neuronal marker MAP2 in NF1-MET;sgP53 cells, while chondrocyte and adipocyte differentiation were strongly induced in NF1-MET cells (Fig. 5G). Interestingly, both kidney and muscle markers were strongly induced by treatment in both NF1-MET and NF1-MET;sgP53 line (Supplementary Fig. 9E). Consistent with the induction of multiple differentiation pathways upon mTOR and MEK inhibition, the multipotency marker SOX9 was decreased in both cell lines (Fig. 5G). These results suggest that lineage differentiation is altered by mTOR and MEK inhibition, with p53-deficient MPNST cells most sensitive to shifts back towards differentiated Schwann cell states.
Discussion
Resistance to both chemotherapy and targeted kinase inhibition in NF1-related MPNSTs is a daunting clinical challenge. Although MPNSTs harbor complex genomic alterations, copy number gains of RTKs, such as MET, PDGFRα, and EGFR are commonly detected [34, 61]. Moreover, autocrine MET-HGF signaling has been demonstrated to promote acquired resistance to MEK inhibition [61]. Previously, we observed that p53 status impacted the therapeutic response to combined MET and MEK inhibition in Met-amplified MPNST tumorgrafts. Even though it is known that p53 is an independent predictor of poor survival and poor response to neoadjuvant chemotherapy in MPNSTs [28, 31], how p53 function influences MPNST therapeutic response is not fully understood. We examined whether p53 has a ‘non-canonical’ function and discovered a novel role for p53 in modulating kinome adaptations to targeted therapy. A comprehensive transcriptional and phosphoproteomic analysis revealed multiple mechanisms of resistance, including deregulation of MET stability, localization, and effector activation.
We used 3 genetic models of p53-deficient, Met-amplified MPNST cells to examine targeted therapy resistance. The NF1-p53 cell line was generated from a murine MPNST driven by loss of Trp53 and Nf1, with Met amplification occurring spontaneously, while the NF1-MET cell line was driven by amplification of Met in the context of Nf1 loss [34], with spontaneous deletion of Cdkn2a. We further used Crispr-Cas9 to generate a p53-deficient isogenic cell line, NF1-MET;sgP53, from the NF1-MET cells to identify p53-specific regulation of drug response. In general, the p53-deficient cells demonstrated similar phenotypes, although in several experiments drug responses were exaggerated in the NF1-P53 cells compared to the NF1-MET;sgP53 cells. These stronger phenotypes may be caused by additional genomic alterations caused by early loss of p53, compared to our isogenic model in which p53 loss was a late event. Interestingly, Cdkn2a was wildtype in NF1-P53 cells, but deleted in NF1-MET cells. In patients with NF1, CDKN2A is an early loss to promote atypical neurofibroma [41], while additional loss of tumor suppressors (e.g. TP53) and/or amplification of RTKs (e.g. MET or EGFR) is required for malignant transformation to MPNST [8]. Understanding how the stepwise timing and clonal evolution of MPNST dictates drug response is critical for developing new therapies.
p53-deficient MPNSTs exhibited increased baseline MET activation suggesting that p53 loss disinhibits amplified MET signaling, an effect that is exaggerated in response to HGF. To determine if p53 is a key driver of clonal selection and drug resistance in MPNSTs, we performed clonal competition assays and discovered that p53-deficient cells had a significant growth advantage in response to MET or MEK inhibition. RNA-seq analysis revealed that p53 regulates expression of several genes involved in MET localization and receptor turnover, resulting in altered signaling kinetics and effector activation. p53 loss resulted in decreased Cblb expression, an E3 ubiquitin ligase that targets both MET [52, 53] and EGFR [56] for lysosomal degradation. p53 loss also induced Prkce expression, which is required for nuclear MET translocation [55], and increased activation of AKT, which is required for nuclear EGFR translocation [62]. Collectively, these data suggest a model in which p53 loss causes increased MET localization to the nucleus. In breast and ovarian cancers nuclear MET results in altered effector activation leading to increased calcium signaling [63], PARP activation [51], and YAP-dependent transcriptional activation [50] to promote drug resistance and invasion/metastasis. Interestingly, nuclear MET also induces sustained STAT3 activation [58], which occurred in response to HGF in our non-isogenic NF1-P53 cell line. In oncogene-addicted cancers, a positive feedback look that promotes STAT3 activation induces resistance to targeted therapy [64]. As the majority of MPNSTs demonstrate loss of function of p53, our findings may partially explain the unexpected failure of the EGFR inhibitor erlotinib in a previous clinical trial in MPNST [57]. Overall, these data indicate that p53 status should be evaluated as a predictive biomarker of response to RTK inhibition in future MPNST clinical trials.
Even though p53-deficiency promoted resistance to MET and MEK inhibition, we found that p53 loss actually increased sensitivity to mTOR inhibition. Combined MEK and mTOR inhibition is currently being evaluated in MPNST in a phase 2 clinical trial (NCT03433183) [65]. Early reports indicate only 2/21 enrolled patients responded to treatment [66]. It is unknown what genetic factors may have contributed to response in these patients. In genetically engineered mouse models of MPNST mTOR [67] or combined mTOR and MEK inhibition [68] were highly effective in a Nf1/Tp53 inactivated model but relatively less effective in an p53 intact ΔNf1/ ΔPten model [35]. Previously, TP53 loss or mutation has been reported to either decrease [69, 70] or increase [59, 71, 72] sensitivity to mTOR inhibition, depending on the context. Similar to our findings, human rhabdomyosarcoma cells lacking functional p53 are sensitive to rapamycin, while overexpression of p53 induces rapamycin resistance [59]. In these studies mechanistically, in the absence of p53, rapamycin induced sustained activation of the ASK1/JNK signaling cascade resulting in persistent c-Jun hyperphosphorylation and subsequent p53-independent apoptosis [60]. Restoration of p53 expression resulted in rapamycin resistance by inducing cell cycle arrest instead of JNK/c-Jun-mediated apoptosis [59, 60]; however, we found no evidence of substantial apoptosis in the context of mTOR inhibition in MPNST. Rather, mTOR and MEK inhibition induced Schwann cell and neuronal differentiation exclusively in p53 null cells in association with increased treatment sensitivity.
Induction of differentiation using epigenetic therapies has been proposed as a treatment for sarcomas [73], however our data indicate that kinase inhibitors may also be used to induce differentiation and cell cycle arrest in MPNSTs. Additionally, we show p53 loss plays a paradoxical role in differentiation therapy response, by causing resistance to MET inhibition and increased sensitivity to mTOR inhibition. Normal Schwann cells are capable of undergoing dedifferentiation followed by redifferentiation in cases of peripheral nerve injury to support nerve repair [74]. However, these reprogramming pathways are altered in disease states, including NF1, resulting in induction of bone, cartilage, muscle, and adipocyte differentiation pathways [75, 76]. p53 is also a critical regulator of reprogramming, as well as tissue development and mesenchymal stem cell differentiation pathways [77, 78]. This less conventional role of p53 may be particularly important for sarcomas such as MPNST, as additional loss of p53 in the context of NF1-defficiency may further contribute to deregulation of these key differentiation/reprogramming pathways to cause lineage plasticity. As an added layer of complexity, both MET and mTOR also regulate these processes. Upon nerve injury, MET [79] and mTOR [80] are required for the reprogramming of mature Schwann cells into dedifferentiated repair Schwann cells. In addition to this, MET plays a key role in embryonic muscle and nervous system development [81], while mTOR regulates bone development and chondrocyte differentiation [82, 83]. It remains unclear how differential lineage plasticity so precisely directs drug response in the presence or absence of p53 in MPNST. Differentiation therapy has been proposed as a p53-independent therapeutic strategy for p53-mutant carcinomas [84], but our data suggests that in MPNSTs there are also alternative p53-dependent mechanisms guiding differentiation therapy response. Collectively, these data suggest that there are key tissue-specific functions of p53 that contribute to drug response in MPNST and other sarcomas outside of the more classical tumor suppressor functions of p53.
Conclusion
Our data reveals profound kinome signaling plasticity in MPNST cells and a complex interplay between clonal subpopulations that is influenced by p53. Apart from the well-known tumor suppressor role of p53, we demonstrate that p53-deficiency promotes acquired resistance to targeted kinase inhibition by modulating kinome signaling and MET localization. Moreover, p53-deficiency enhances lineage plasticity which also contributes to kinase inhibitor resistance. Understanding how p53 and other commonly altered genes modulate treatment response is critical for the advancement of precision medicine approaches for MPNST patients.
Methods
Cell culture and drugs
Mouse-derived MPNST cell lines described previously [34] were cultured at 37 °C in a humidified atmosphere in 5% CO2 in low pH DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Corning, lot #35070165) and 1% Penicillin Streptomycin (Thermo Fisher Scientific), unless otherwise indicated. Cell lines were verified to be free of Mycoplasma contamination every 6 months by PCR (ATCC). Human-derived MPNST cell lines were cultured as described previously [85]. Capmatinib (Novartis), trametinib (Novartis), everolimus (Selleckchem), and afuresertib (Selleckchem) solutions were prepared in DMSO. For human cell line [86] IC50 experiments, drugs were purchased as 10 mM stock solutions (Selleckchem) and handled as previously described [85]. All cell culture experiments were performed 3 independent times, unless otherwise noted.
Viability and dose response
For cell viability experiments, cells were allowed to adhere and then treated with DMSO or the indicated drug dose. After 72 h, cells were trypsinized (Thermo Fisher Scientific) and trypan blue negative cells were counted using a TC20 Automated Cell Counter (Bio-Rad) to determine percent viability relative to vehicle treatment. To meet the assumption of equal variance, data were log transformed and pairwise comparisons of beta regressions were done using the betareg (v 3.1-3) and emmeans (v 1.4.5) packages and plotted using the ggplot2 (v 3.3.0) package in R (v 3.6.3). Emmeans and ggplot2 were also used for comparison of doubling times. For human MPNST cell lines, the IC50 was determined as previously described [85]. Briefly, cells were allowed to adhere and then treated with DMSO or the indicated serially diluted drug (4.6 nM–10 μM) for 72 h, cell viability was measured by ATPlite Luminescence Assay (PerkinElmer), and IC50 was calculated using GraphPad Prism 7. The rcorr function in the Hmisc (v4.4-1) package was used to generate spearman correlations of IC50. The ggplot2 (v 3.3.0) and ggpubr (v 0.2.5) packages were used to plot an IC50 matrix and correlogram, respectively, using R (v 3.6.3). For dose combination matrices, 2500 cells/well were plated in a 96 well plate, allowed to adhere, and then treated with the indicated serially diluted drug or vehicle for 72 h. Cell viability was measured using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) (Promega) following the manufacture’s protocol. Absorbance was read using Synergy Neo microplate reader (BioTek) and normalized to cell number using a standard curve generated by GraphPad Prism 7. Mean percent inhibitions relative to vehicle controls were plotted in dose response matrices using the synergyfinder package (v 2.0.12) in R (v 3.6.3).
Reverse phase protein array
For cell culture experiments, cells were seeded in a 6-well plate, allowed to adhere overnight, and then treated in replicates of 6 with DMSO or the indicated drug or ligand dose for 2 or 48 h. To harvest samples, the cells were washed thoroughly 3 times with ice cold PBS, and plates were immediately snap frozen on dry ice to preserve the integrity of the phosphoproteome. For mouse tumorgrafts, we analyzed data, where some of this data was used in a previous study [25]. Briefly, immediately following the euthanasia of tumor-bearing mice, 15–25 mg portions of each tumor were transplanted into the flank of NSG-SCID mice using a 10-gauge trochar. When the tumor volume reached approximately 150 mm3, mice were euthanized, and tumors were immediately harvested and snap-frozen in liquid nitrogen within 20 min upon surgical resection to preserve the integrity of the phosphoproteome. Six tumors were assessed for each genotype. All animal experimentation in this study was approved by the Van Andel Institute’s Internal Animal Care and Use Committee (XPA-19-04-001). Specimens were then embedded in an optimal cutting temperature compound (Sakura Finetek, Torrance, CA, USA), cut into 8 μm cryo-sections, mounted on uncharged glass slides, and stored at −80 °C until use. Each slide was fixed in 70% ethanol (Sigma Aldrich, Darmstadt, Germany), washed in deionized water, stained with hematoxylin (Sigma Aldrich, Darmstadt, Germany) and blued in Scott’s Tap Water substitute (Electron Microscopy Sciences), and dehydrated through an ethanol gradient (70%, 95%, and 100%) and xylene (Sigma Aldrich, Darmstadt, Germany). In order to prevent protein degradation, complete protease inhibitor cocktail tablets (Roche Applied Science, Basel, Switzerland) were added to the ethanol, water, hematoxylin, and Scott’s Tap Water substitute [87]. Cells were lysed in a 1:1 solution of 2× Tris-Glycine SDS Sample buffer (Invitrogen Life Technologies, Carlsbad, CA, USA) and Tissue Protein Extraction Reagent (Pierce, Waltham, MA, USA) supplemented with 2.5% of 2-mercaptoethanol (Sigma Aldrich, Darmstadt, Germany). Cell lysates were boiled for 8 min and stored at −80 °C. Reverse phase protein microarray construction and immunostaining was performed as previously described [25] A total of 98 protein sites passed quality control metrics and were used for analysis. Differential expression was performed using R package “limma” [88] and R (v 3.6.0). For differential activation analysis, P-values for delta-delta differences in limma fold changes were generated in R (v 3.6.3) using Wald tests and adjusted using the BH method. Adjusted P-values of less than 0.05 were considered significant. For tumors, the pairwise difference between genotypes in mean centered phospho-site expression (Z score) was assessed using a linear mixed-effects model with a random intercept for each phospho-site using the lme4 (v 1.1-23) and emmeans (v 1.4.5) packages in R with the selected phospho-sites. Pairwise differences between genotypes in log transformed expression were also assessed for each individual phospho-site using emmeans. A comprehensive analysis including all 98 tumorgraft phospho-sites was previously published [25] Balloon plots were created from the limma fold changes (cell lines) or z-scores (tumors) in R (v 3.6.3) using the ggballoonplot function in the ggpubr package (v 0.2.5). Labeled fold change, differential activation, waterfall, and site-specific expression plots were created using ggplot2 (v 3.3.0) in R (v 3.6.3).
Lentiviral transduction
To generate the NF1-MET;sgP53 cell line, the pLentiCRISPRv2 vector (Addgene #52961) [89] was digested with BsmBI and dephosphorylated with CIP (New England BioLabs) following the manufactures instructions and then purified with using Qiaquick Gel Extraction kit (Qiagen). The oligonucleotides 5’-cac cgA GCC AAG TCT GTT ATG TGC A-3’ and 5’-aaa cTG CAC ATA ACA GAC TTG GCT c-3’ were annealed and then ligated into the vector using T4 PNK and T4 ligase (Thermo Fisher Scientific). One Shot competent cells (Thermo Fisher Scientific) were transformed and candidate colonies were verified by diagnostic digestion. Lentivirus was produced by lipofectamine transfection (Thermo Fisher Scientific) of HLA 293 cells kindly provided by Dr. Bart Williams with either the pLentiCRISPRv2;sgP53 plasmid or the pLentiCRISPRv2 plasmid (as an empty vector control), along with the envelop and packaging plasmids pCMV-VSV-G (Addgene #8454) [90] and psPAX2 (Addgene #12260). NF1-MET cells were infected with 500 μL of 0.45 μm filtered virus with polybrene (EMD Millipore), and 48 h post infection transduced cells were selected for by 2 μg/mL puromycin (Thermo Fisher Scientific) treatment. To generate H2B GFP and RFP labeled cells, lentivirus was produced by lipofectamine transfection of Phoenix 293 cells kindly provided by Dr. Bart Williams with either LV-GFP (Addgene #25999) [91] or pHIV-H2BmRFP (Addgene #18982) [92] plasmids, along with pCMV-VSV-G and psPAX2. Filtered lentivirus was concentrated using Lenti-X Concentrator (Takara). NF1-MET;sgEmptyVector and NF1-MET;sgP53 cells were infected with GFP or RFP lentivirus to create NF1-MET-GFP and NF1-MET;sgP53-RFP cell lines, respectively.
Competition assay
For clonal competition assays, single cell suspensions of NF1-MET-GFP and NF1-MET;sgP53-RFP cells were prepared in sorting buffer (HBSS without Ca2+, Mg2+, and Phenol Red (Thermo Fisher Scientific) with 25 mM HEPES, 2 mM EDTA, 2% FBS, and 10 mg/mL DAPI added). 7,500 GFP+ and 7,500 RFP+ single live cells were sorted into each well of a 24 well culture plate containing low pH DMEM with 10% FBS using a MoFlo Astrios cell sorter with Summit v6.3 software (Beckman Coulter). Sorting was performed at 25 psi using a 100um nozzle, with Purify for the abort mode and a drop envelope set to 1–2 drops. Cells were selected using SSC vs FSC, and single cells using both SSC area vs height and area vs width. Live cells (DAPI negative) were identified using the 355-448/59 channel, and GFP and RFP signals were detected using the 488-510/20 and 561-614/20 channels, respectively.
After cells adhered they were treated with DMSO or the indicated drug dose and cocultured for the indicated timepoints. The cocultured cells were analyzed to determine the percentage of single, live cells that were either GFP+ or RFP+ using a CytoFLEX S flow cytometer with CytExpert v2.4 software (Beckman Coulter). Analysis was performed using FlowJo v10.7 software (BD Life Sciences). Single cells were identified using SSC vs FSC, followed by SSC area vs height. Live cells (DAPI negative) were identified using the 405-450/45 channel, and GFP and RFP signals were detected using the 488-525/40 and 561-610/20 channels, respectively. Representative images of the gating strategy are shown in Supplementary Fig. 1. Pairwise comparisons of beta regressions were done using the betareg (v 3.1-3) and emmeans (v 1.4.5) packages and plotted using the ggplot2 (v 3.3.0) package in R (v 3.6.3).
Western blotting
Cells were grown overnight followed by serum starving overnight for a final confluency of 90%, and then treated with drug and/or stimulated with 10% serum or 100 ng/mL HGF or EGF as indicated. Cell lysate collection and immunoblotting were done as previously described [34]. Primary antibodies were purchased from Cell Signaling: p53 (#2524), phospho-MET Y1234/1235 (#3077), MET (#3127), phospho-AKT S473 (#9271), phospho-ERK T202/Y204 (#9101), phospho-S6 S235/236 (#4858), β-Actin (#3700).
RT-qPCR
RNA was isolated using RNeasy Mini Kit and RNase-Free DNase set (Qiagen) following the manufactures protocol. cDNA was synthesized using SuperScript II Reverse Transcriptase with Oligo (dT) 12-18mer Primers (Thermo Fisher Scientific) using Tetrad 2 (BioRad) following the manufactures protocols. qPCR was done on Step One Plus (Applied Biosystems) using Fast Start Universal SYBR Green Master Mix (Roche). Primer sequences are listed in Table 2. Relative expression was calculated using the ddCt method and plotted using GraphPad Prism 7 (Cdkn1a) or using the ggplot2 (v 3.3.0) package in R (v 3.6.3). For statistical analysis, a linear mixed-effects model with random intercepts for experimental and technical replicates was used to summarize relative expression from 4 separate experiments using the lme4 (v 1.1-23) and emmeans (v 1.4.5) packages in R. All p-values were automatically adjusted using the Tukey method.
Table 2.
Primer sequences.
| Gene | Forward | Reverse |
|---|---|---|
| Acta2 | CGAAACCACCTATAACAGCATCA | GCGTTCTGGAGGGGCAAT |
| Cdkn1a | CTTGCACTCTGGTGTCTG | CTTGGAGTGATAGAAATCTGTCA |
| Cebpa | AGGTGCTGGAGTTGACCAGT | CAGCCTAGAGATCCAGCGAC |
| Col11a1 | GACCAGAAGACACACTGAAAGCA | TCCATGCCATCTGAGTAGTCAAGA |
| Fabp7 | CTCTGGGCGTGGGCTTT | TTCCTGACTGATAATCACAGTTGGTT |
| Fbln2 | AGTGGCCGTAAGTATGCTGC | GGAAGCTGGTAGCAAATGAGC |
| Il1B | TGTAATGAAAGACGGCACACC | TCTTCTTTGGGTATTGCTTGG |
| Il10 | CCCTTTGCTATGGTGTCCTT | TGGTTTCTCTTCCCAAGACC |
| Il6 | GAGGATACCACTCCCAACAGACC | AAGTGCATCATCGTTGTTCATACA |
| Map2 | TCTAAAGAACATCCGTCACAGG | GGTGAGCATTGTCAAGTGAGC |
| Mcp1 | GCATCCACGTGTTGGCTCA | CTCCAGCCTACTCATTGGGATCA |
| Ppia | GGCAAATGCTGGACCAAAC | CATTCCTGGACCCAAAACG |
| S100B | CTGGAGAAGGCCATGGTTGC | CTCCAGGAAGTGAGAGAGCT |
| Sox9 | TCCACGAAGGGTCTCTTCTC | AGGAAGCTGGCAGACCAGTA |
| Spp1 | TCTCCTTGCGCCACAGAATG | TCCTTAGACTCACCGCTCTT |
| Synpo | CATCGGACCTTCTTCCTGTG | TCGGAGTCTGTGGGTGAG |
| Tubb5 | ATGCCATGTTCATCGCTTAT | TTGTTCGGTACCTACATTGG |
| Vcam1 | TCGCTCAAATCGGTGACTC | ACAGGCTCCATGGTCAGAAC |
Mouse xenograft models
Six- to eight-week-old female NSG-SCID mice were injected subcutaneously with one million cells of either NF1-MET or NF1-MET;sgP53 (10 mice/cell line). Mice were randomly assigned to each treatment group by tumor growth rate, so that growth rates were matched across treatment groups. The investigator was not blinded. Tumors were measured twice weekly with calipers. Once tumors reached approximately 150 mm3, 5 mice from each cell line group were treated with either 30 mg/kg capmatinib or vehicle (0.5% methylcellulose) twice daily for 15 days or until tumors reach 2500 mm3. A second experiment was performed exactly the same way, for a final total of 10 mice in each cell line and treatment group. All animal experimentation in this study was approved by the Van Andel Institute’s Internal Animal Care and Use Committee (XPA-19-04-001). Sample sizes were selected based on our previously published study of NF1-MET tumorgrafts [34].
A linear mixed-effects model was used to determine if there were significant differences between the results of experiment one and experiment two. The models were stratified by treatment group. Data were square root transformed before being entered into the model. The model used a 3-way interaction for timepoint, cell-line, and experiment and also included a random intercept for tumor ID. A linear mixed-effects model was used for the combination analysis. Data were square root transformed before being entered into the model. This was a linear mixed effects model with a 4-way interaction between timepoint, experiment, treatment group, and cell-line. A random intercept for tumor-id was also included. All statistical analyses were run using R (v. 3.6.0) and all linear regression models were run using the lme4 package. The emtreands function from the emmeans package was used for the contrasts/comparisons that were run on the regression output to obtain growth trend estimates for the average tumor growth rate for each treatment/genotype group. All p-values that were obtained through the emmeans output were automatically multiple-testing corrected via the Tukey method.
Immunofluorescence
Cells were plated at 50,000 cells per well on glass coverslips (Fisherbrand) in a 24 well plate and allowed to attach overnight. The next day cells were serum-starved for 16 h. The third day cells were treated with 100 ng/mL HGF for 5 min at 37 °C, washed three times with phosphate buffered saline + 0.5% tween 20 (PBST), and fixed with 4% formaldehyde in PBS for 15 min. Cells were permeabilized with ice-cold methanol for 3 min at −20 °C. Samples were blocked (5% normal goat serum, 1% BSA, 0.3% Triton X-100 in PBS) for 1 h at room temperature in a humidity chamber. Antibodies against phospho-MET (Y1230/1234/1235; Abcam #5662) were diluted 1:100 in antibody dilution buffer (1% BSA and 0.3% triton-x 100 in PBS) and incubated with the cells overnight at 4 °C in a humidity chamber. Primary antibodies were detected using Alexa Flour secondary antibody goat-against-rabbit 594 (Life Technologies #8889) at 1:500 and incubated for 40 min at room temperature. Cell nuclei were stained with DAPI and samples were mounted with ProLong Gold antifade reagent (Life Technologies). For imaging, at three regions of interest were imaged per sample using a 60x Plan Apo VC oil immersion objective with 1.4 NA on a Nikon A1 plus-RSi laser scanning confocal microscope (Nikon Elements software). Image resolution was 1024 × 1024 of z-slices covering the entirety of cell thickness. PMT levels were set using controls with 403 and 561 solid-state lasers. FIJI was used to generate maximum intensity projection TIFF images.
RNA sequencing
Cells were plated in a 6-well plate, grown in 10% FBS overnight, and then treated in duplicate with 100 nM capmatinib or DMSO. RNA was isolated using RNeasy Mini Kit and RNase-Free DNase set (Qiagen) following the manufactures protocol. Libraries were prepared by the Van Andel Genomics Core from 500 ng of total RNA using the KAPA mRNA Hyperprep kit (v4.17) (Kapa Biosystems, Wilmington, MA USA). RNA was sheared to 300–400 bp. Prior to PCR amplification, cDNA fragments were ligated to IDT for Illumina TruSeq UD Indexed adapters (Illumina Inc, San Diego CA, USA). Quality and quantity of the finished libraries were assessed using a combination of Agilent DNA High Sensitivity chip (Agilent Technologies, Inc.), QuantiFluor® dsDNA System (Promega Corp., Madison, WI, USA), and Kapa Illumina Library Quantification qPCR assays (Kapa Biosystems). Individually indexed libraries were pooled and 50 bp, paired end sequencing was performed on an Illumina NovaSeq6000 sequencer using an S2, 100 bp sequencing kit (Illumina Inc., San Diego, CA, USA) to a minimum raw depth of 41.4 M reads with an average of 47.2 M reads per sample. Base calling was done by Illumina RTA3 and output of NCS was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v1.9.0. Sequencing adapters were trimmed using Trimgalore v0.4.2 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Bases with a quality score less than 20 were also removed from the ends of reads. Trimmed data were quality controlled with FastQC v0.11.7 [93] and then mapped with STAR v2.5.2b to the mm10 genome using the default settings [94]. Raw gene counts (minimum of 30 M counts per sample and a mean of 34.92) generated by STAR were imported into R v3.6.0. Genes with less than 10 counts in a minimum of two samples were immediately removed from all further analysis; this minimizes multiple testing adjustments and removes low-expressed genes unlikely to be biologically meaningful. For all differential expression contrasts using a subset of the data, the count data were further filtered so that, for the samples being used in the contrast, a minimum of two samples have > 0 counts. A quasi-likelihood negative binomial generalized log-linear model was then fit to the filtered count data using the weighted trimmed mean of M-values to normalize for library size and composition biases [95]. P-values were generated using empirical Bayes quasi-likelihood F-tests, and then adjusted using the BH method; adjusted P-values less than 0.05 were considered significant. Gene ontology enrichment analyses were done using the clusterProfiler R package v3.14.3 using the function ‘goseq’ with KEGG ontologies. Heatmaps were generated from library-size normalized counts centered across genes (z-scores) using the pheatmap package v1.0.12.
Apoptosis assay
NF1-MET and NF1-MET;sgP53 cells were plated in a 6-well plate, grown in 10% FBS overnight, and then treated with 20 nM everolimus and 40 nM trametinib or DMSO for the indicated time points. Cells were harvested and stained with Annexin V Alexa Fluor 647 Conjugate (Invitrogen) and DAPI according to the manufactures protocol. Stained samples were acquired on a Beckman Coulter CytoFLEX S running CytExpert v2.4. AnnexinV-A647 and DAPI were detected in the 640-660/20 and 405-450/45 channels respectively. Data were analyzed using FlowJo v10.x. An initial gate was set on a forward vs side scatter plot to include both live and dead cells while excluding small debris and noise. Aggregated cells were removed for analysis using a gate on side scatter area vs side scatter height. The single cells were then analyzed for health on a plot of Annexin V vs DAPI. For statistical analysis, the Annexin V and/or DAPI positive cell percentages from 4 separate experiments were calculated using a beta family generalized linear mixed effect model with a random intercept for the experimental run and a logit link using glmmTMB (v1.1.3) in R. Treatment verses control odds ratios were assessed using emmeans (v1.4.5).
Supplementary information
Acknowledgements
The authors thank the VARI Vivarium and Transgenics Core and the VARI Optical Imaging Core (confocal microscopy facilities), as well as Zach Madaj for advice on statistical analysis, and Drs Alex Zhong and Payton Stevens for their advice on lentiviral production. Drug profiling was done at the High Throughput Screening Core, Perelman School of Medicine, University of Pennsylvania. This study was funded in part by grants from the Neurofibromatosis Research Program of the Department of Defense (W81XWH-21-1-0224 and W81XWH-19-1-0537), the Children’s Tumor Foundation Young Investigator Award (CTF-2018-01-009), the Children’s Tumor Foundation (CTF-2015-05-006 and CTF-2019-05-004), NF Michigan, and the Van Andel Institute.
Author contributions
JLG, LEM, EAT and CJE designed and performed experiments and helped write the manuscript. EW and IB analyzed data and helped write the manuscript. KW, SG, and JMF designed and performed experiments. RTCS, JLS, MaA, JW, and EFP provided technical expertise, performed experiments, and helped write the manuscript. MeA, and TVW performed experiments. CRG and MRS oversaw the project and helped write the manuscript.
Funding
Department of Defense NFRP (W81XWH-21-1-0224) (MRS), Children’s Tumor Foundation Young Investigator Award (CTF-2018-01-009; 10.48105/pc.gr.88576) (JLG), NF Michigan (MRS), Children’s Tumor Foundation (CTF-2015-05-006 and CTF-2019-05-004) (JMF), Department of Defense NFRP (W81XWH-19-1-0537) (JMF).
Data availability
The RNA-seq data generated in this study were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE225747 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225747).
Code availability
R code used in this study is available upon request.
Competing interests
EFP is a paid consultant of and has stock equity in Theralink Technologies, Inc., Perthera Inc., and Ceres Nanosciences Inc. JW has stock ownership in Theralink Technologies LLC.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41388-024-03000-9.
References
- 1.Duong TA, Sbidian E, Valeyrie-Allanore L, Vialette C, Ferkal S, Hadj-Rabia S, et al. Mortality associated with neurofibromatosis 1: a cohort study of 1895 patients in 1980-2006 in France. Orphanet J Rare Dis. 2011;6:18. doi: 10.1186/1750-1172-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huson SM, Compston DA, Harper PS. A genetic study of von Recklinghausen neurofibromatosis in south east Wales. II. Guidelines for genetic counselling. J Med Genet. BMJ Publ Group Ltd. 1989;26:712–21. doi: 10.1136/jmg.26.11.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell Elsevier. 1990;62:187–92. doi: 10.1016/0092-8674(90)90252-A. [DOI] [PubMed] [Google Scholar]
- 4.Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Sci Am Assoc Advancement Sci. 1990;249:181–6. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- 5.DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, et al. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell Elsevier. 1992;69:265–73. doi: 10.1016/0092-8674(92)90407-4. [DOI] [PubMed] [Google Scholar]
- 6.Legius E, Dierick H, Wu R, Hall BK, Marynen P, Cassiman J-J, et al. TP53 mutations are frequent in malignant NFI tumors. Genes Chromosomes Cancer. 1994;10:250–5. doi: 10.1002/gcc.2870100405. [DOI] [PubMed] [Google Scholar]
- 7.Rao UN, Sonmez-Alpan E, Michalopoulos GK. Hepatocyte growth factor and c-MET in benign and malignant peripheral nerve sheath tumors. Hum Pathol. 1997;28:1066–70. doi: 10.1016/S0046-8177(97)90060-5. [DOI] [PubMed] [Google Scholar]
- 8.Mantripragada KK, Spurlock G, Kluwe L, Chuzhanova N, Ferner RE, Frayling IM, et al. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res. 2008;14:1015–24. doi: 10.1158/1078-0432.CCR-07-1305. [DOI] [PubMed] [Google Scholar]
- 9.Verdijk RM, den Bakker MA, Dubbink HJ, Hop WCJ, Dinjens WNM, Kros JM. TP53 mutation analysis of malignant peripheral nerve sheath tumors. J Neuropathol Exp Neurol. 2010;69:16–26. doi: 10.1097/NEN.0b013e3181c55d55. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Deshmukh H, Payton JE, Dunham C, Scheithauer BW, Tihan T, et al. Array-based comparative genomic hybridization identifies CDK4 and FOXM1 alterations as independent predictors of survival in malignant peripheral nerve sheath tumor. Clin Cancer Res. 2011;17:1924–34. doi: 10.1158/1078-0432.CCR-10-1551. [DOI] [PubMed] [Google Scholar]
- 11.De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nat Nat Publ Group. 2014;514:247–51. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 12.Brohl AS, Kahen E, Yoder SJ, Teer JK, Reed DR. The genomic landscape of malignant peripheral nerve sheath tumors: diverse drivers of Ras pathway activation. Sci Rep. Nat Publ Group. 2017;7:14992. doi: 10.1038/s41598-017-15183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutmann DH, Blakeley JO, Korf BR, Packer RJ. Optimizing biologically targeted clinical trials for neurofibromatosis. Expert Opin Investig Drugs. 2013;22:443–62. doi: 10.1517/13543784.2013.772979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin E, Lamba N, Flucke UE, Verhoef C, Coert JH, Versleijen-Jonkers YMH, et al. Non-cytotoxic systemic treatment in malignant peripheral nerve sheath tumors (MPNST): A systematic review from bench to bedside. Crit Rev Oncol Hematol. 2019;138:223–32. doi: 10.1016/j.critrevonc.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Natalie Wu LM, Lu QR. Therapeutic targets for malignant peripheral nerve sheath tumors. Future Neurol Future Med. 2019;14:FNL7. doi: 10.2217/fnl-2018-0026. [DOI] [Google Scholar]
- 16.Staedtke V, Bai R-Y, Blakeley JO. Cancer of the Peripheral Nerve in Neurofibromatosis Type 1. Neurother J Am Soc Exp Neurother. 2017;14:298–306. doi: 10.1007/s13311-017-0518-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans DGR, Baser ME, McGaughran J, Sharif S, Howard E, Moran A. Malignant peripheral nerve sheath tumours in neurofibromatosis 1. J Med Genet. 2002;39:311–4. doi: 10.1136/jmg.39.5.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari A, Bisogno G, Macaluso A, Casanova M, D’Angelo P, Pierani P, et al. Soft-tissue sarcomas in children and adolescents with neurofibromatosis type 1. Cancer. 2007;109:1406–12. doi: 10.1002/cncr.22533. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari A, Miceli R, Rey A, Oberlin O, Orbach D, Brennan B, et al. Non-metastatic unresected paediatric non-rhabdomyosarcoma soft tissue sarcomas: Results of a pooled analysis from United States and European groups. Eur J Cancer Elsevier. 2011;47:724–31. doi: 10.1016/j.ejca.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson KL, Al Sannaa GA, Kivlin CM, Ingram DR, Landers SM, Roland CL, et al. Patterns of recurrence and survival in sporadic, neurofibromatosis type 1-associated, and radiation-associated malignant peripheral nerve sheath tumors (MPNSTs) J Neurosurg. 2017;126:319–29. doi: 10.3171/2015.12.JNS152443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll SL. The challenge of cancer genomics in rare nervous system neoplasms: malignant peripheral nerve sheath tumors as a paradigm for cross-species comparative oncogenomics. Am J Pathol Elsevier. 2016;186:464–77. doi: 10.1016/j.ajpath.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong J, Li B, Lin D, Zhou Q, Huang D. Advances in targeted therapy and immunotherapy for non-small cell lung cancer based on accurate molecular typing. Front Pharmacol. Frontiers; 2019;10. https://www.frontiersin.org/articles/10.3389/fphar.2019.00230/full. [DOI] [PMC free article] [PubMed]
- 23.Farid M, Demicco EG, Garcia R, Ahn L, Merola PR, Cioffi A, et al. Malignant Peripheral Nerve Sheath Tumors. Oncologist. 2014;19:193–201. doi: 10.1634/theoncologist.2013-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seebacher NA, Stacy AE, Porter GM, Merlot AM. Clinical development of targeted and immune based anti-cancer therapies. J Exp Clin Cancer Res. 2019;38:156. doi: 10.1186/s13046-019-1094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grit JL, Pridgeon MG, Essenburg CJ, Wolfrum E, Madaj ZB, Turner L, et al. Kinome profiling of NF1-related MPNSTs in response to kinase inhibition and doxorubicin reveals therapeutic vulnerabilities. Genes. 2020;11:331. doi: 10.3390/genes11030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suresh TN, Harendra Kumar ML, Prasad CSBR, Kalyani R, Borappa K. Malignant peripheral nerve sheath tumor with divergent differentiation. Indian J Pathol Microbiol. 2009;52:74–6. doi: 10.4103/0377-4929.44971. [DOI] [PubMed] [Google Scholar]
- 27.Hanemann CO, Blakeley JO, Nunes FP, Robertson K, Stemmer-Rachamimov A, Mautner V, et al. Current status and recommendations for biomarkers and biobanking in neurofibromatosis. Neurology. 2016;87:S40–8. doi: 10.1212/WNL.0000000000002932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Høland M, Kolberg M, Danielsen SA, Bjerkehagen B, Eilertsen IA, Hektoen M, et al. Inferior survival for patients with malignant peripheral nerve sheath tumors defined by aberrant TP53. Mod Pathol Nat Publ Group. 2018;31:1694–707. doi: 10.1038/s41379-018-0074-y. [DOI] [PubMed] [Google Scholar]
- 29.Brekke HR, Kolberg M, Skotheim RI, Hall KS, Bjerkehagen B, Risberg B, et al. Identification of p53 as a strong predictor of survival for patients with malignant peripheral nerve sheath tumors. Neuro-Oncol. 2009;11:514–28. doi: 10.1215/15228517-2008-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou C, Smith KD, Liu J, Lahat G, Myers S, Wang W-L, et al. Clinical, pathological, and molecular variables predictive of malignant peripheral nerve sheath tumor outcome. Ann Surg. 2009;249:1014–22. doi: 10.1097/SLA.0b013e3181a77e9a. [DOI] [PubMed] [Google Scholar]
- 31.Karpinsky G, Krawczyk MA, Izycka-Swieszewska E, Fatyga A, Budka A, Balwierz W, et al. Tumor expression of survivin, p53, cyclin D1, osteopontin and fibronectin in predicting the response to neo-adjuvant chemotherapy in children with advanced malignant peripheral nerve sheath tumor. J Cancer Res Clin Oncol. 2018;144:519–29. doi: 10.1007/s00432-018-2580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres KE, Zhu Q-S, Bill K, Lopez G, Ghadimi MP, Xie X, et al. Activated MET is a molecular prognosticator and potential therapeutic target for malignant peripheral nerve sheath tumors. Clin Cancer Res J Am Assoc Cancer Res. 2011;17:3943–55. doi: 10.1158/1078-0432.CCR-11-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endo M, Yamamoto H, Setsu N, Kohashi K, Takahashi Y, Ishii T, et al. Prognostic significance of AKT/mTOR and MAPK pathways and antitumor effect of mTOR inhibitor in NF1-related and sporadic malignant peripheral nerve sheath tumors. Clin Cancer Res. 2013;19:450–61. doi: 10.1158/1078-0432.CCR-12-1067. [DOI] [PubMed] [Google Scholar]
- 34.Peacock JD, Pridgeon MG, Tovar EA, Essenburg CJ, Bowman M, Madaj Z, et al. Genomic status of MET potentiates sensitivity to MET and MEK inhibition in NF1-related malignant peripheral nerve sheath tumors. Cancer Res. 2018;78:3672–87. doi: 10.1158/0008-5472.CAN-17-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson AL, Anderson LK, Greeley AD, Keng VW, Rahrmann EP, Halfond AL, et al. Co-targeting the MAPK and PI3K/AKT/mTOR pathways in two genetically engineered mouse models of Schwann cell tumors reduces tumor grade and multiplicity. Oncotarget. 2014;5:1502–14. doi: 10.18632/oncotarget.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradtmöller M, Hartmann C, Zietsch J, Jäschke S, Mautner V-F, Kurtz A, et al. Impaired Pten Expression in Human Malignant Peripheral Nerve Sheath Tumours. PLoS ONE. 2012;7:e47595. doi: 10.1371/journal.pone.0047595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holtkamp N, Malzer E, Zietsch J, Okuducu AF, Mucha J, Mawrin C, et al. EGFR and erbB2 in malignant peripheral nerve sheath tumors and implications for targeted therapy. Neuro-Oncol. 2008;10:946–57. doi: 10.1215/15228517-2008-053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui D, Qu R, Liu D, Xiong X, Liang T, Zhao Y. The Cross Talk Between p53 and mTOR Pathways in Response to Physiological and Genotoxic Stresses. Front Cell Dev Biol. 2021;9. https://www.frontiersin.org/articles/10.3389/fcell.2021.775507. [DOI] [PMC free article] [PubMed]
- 39.Shen H, Maki CG. Pharmacologic activation of p53 by small-molecule MDM2 antagonists. Curr Pharm Des. 2011;17:560–8. doi: 10.2174/138161211795222603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magallón-Lorenz M, Terribas E, Ortega-Bertran S, Creus-Bachiller E, Fernández M, Requena G, et al. Deep genomic analysis of malignant peripheral nerve sheath tumor cell lines challenges current malignant peripheral nerve sheath tumor diagnosis. iScience. 2023;26:106096. doi: 10.1016/j.isci.2023.106096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pemov A, Hansen NF, Sindiri S, Patidar R, Higham CS, Dombi E, et al. Low mutation burden and frequent loss of CDKN2A/B and SMARCA2, but not PRC2, define premalignant neurofibromatosis type 1–associated atypical neurofibromas. Neuro-Oncol. 2019;21:981–92. doi: 10.1093/neuonc/noz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty S, Utter MB, Frias MA, Foster DA. Cancer cells with defective RB and CDKN2A are resistant to the apoptotic effects of rapamycin. Cancer Lett. 2021;522:164–70. doi: 10.1016/j.canlet.2021.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang J, Gu Y, Liu J, Wu R, Fu L, Zhao J, et al. Coexistence of p16/CDKN2A homozygous deletions and activating EGFR mutations in lung adenocarcinoma patients signifies a poor response to EGFR-TKIs. Lung Cancer. 2016;102:101–7. doi: 10.1016/j.lungcan.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Moon C-I, Tompkins W, Wang Y, Godec A, Zhang X, Pipkorn P, et al. Unmasking intra-tumoral heterogeneity and clonal evolution in NF1-MPNST. Genes Multidiscip Digital Publ Inst. 2020;11:499. doi: 10.3390/genes11050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol Publ Int Assoc Study Lung Cancer. 2016;11:1493–502. doi: 10.1016/j.jtho.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Pozner-Moulis S, Pappas DJ, Rimm DL. Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res Am Assoc Cancer Res. 2006;66:7976–82. doi: 10.1158/0008-5472.CAN-05-4335. [DOI] [PubMed] [Google Scholar]
- 47.Pozner-Moulis SA, Pappas D, Rimm DL. Nuclear localization of a fragment of the HGF receptor (Met) Cancer Res Am Assoc Cancer Res. 2006;66:45–45. [Google Scholar]
- 48.Tey SK, Tse EYT, Mao X, Ko FCF, Wong AST, Lo RC-L, et al. Nuclear Met promotes hepatocellular carcinoma tumorigenesis and metastasis by upregulation of TAK1 and activation of NF-κB pathway. Cancer Lett. 2017;411:150–61. doi: 10.1016/j.canlet.2017.09.047. [DOI] [PubMed] [Google Scholar]
- 49.Xie Y, Fan H, Lu W, Yang Q, Nurkesh A, Yeleussizov T, et al. Nuclear MET requires ARF and is inhibited by carbon nanodots through binding to phospho-tyrosine in prostate cancer. Oncogene. 2019;38:2967–83. doi: 10.1038/s41388-018-0608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30:937–45. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- 51.Du Y, Yamaguchi H, Wei Y, Hsu JL, Wang H-L, Hsu Y-H, et al. Blocking c-Met-mediated PARP1 phosphorylation enhances anti-tumor effects of PARP inhibitors. Nat Med. 2016;22:194–201. doi: 10.1038/nm.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peschard P, Ishiyama N, Lin T, Lipkowitz S, Park M. A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. J Biol Chem. 2004;279:29565–71. doi: 10.1074/jbc.M403954200. [DOI] [PubMed] [Google Scholar]
- 53.Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell Elsevier. 2007;27:474–85. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 54.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–90. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 55.Kermorgant S, Zicha D, Parker PJ. PKC controls HGF-dependent c-Met traffic, signalling and cell migration. EMBO J. 2004;23:3721–34. doi: 10.1038/sj.emboj.7600396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pennock S, Wang Z. A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol. 2008;28:3020–37. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albritton KH, Rankin C, Coffin CM, Ratner N, Budd GT, Schuetze SM, et al. Phase II study of erlotinib in metastatic or unresectable malignant peripheral nerve sheath tumors (MPNST) J Clin Oncol Am Soc Clin Oncol. 2006;24:9518–9518. doi: 10.1200/jco.2006.24.18_suppl.9518. [DOI] [Google Scholar]
- 58.Kermorgant S, Parker PJ. Receptor trafficking controls weak signal delivery: a strategy used by c-Met for STAT3 nuclear accumulation. J Cell Biol. 2008;182:855–63. doi: 10.1083/jcb.200806076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang S, Liu LN, Hosoi H, Dilling MB, Shikata T, Houghton PJ. p53/p21CIP1 cooperate in enforcing rapamycin-induced G1 arrest and determine the cellular response to rapamycin. Cancer Res Am Assoc Cancer Res. 2001;61:3373–81. [PubMed] [Google Scholar]
- 60.Huang S, Shu L, Dilling MB, Easton J, Harwood FC, Ichijo H, et al. Sustained activation of the JNK cascade and rapamycin-induced apoptosis are suppressed by p53/p21(Cip1) Mol Cell. 2003;11:1491–501. doi: 10.1016/S1097-2765(03)00180-1. [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Pollard K, Calizo A, Pratilas CA. Activation of Receptor Tyrosine Kinases Mediates Acquired Resistance to MEK Inhibition in Malignant Peripheral Nerve Sheath Tumors. Cancer Res. 2021;81:747–62. doi: 10.1158/0008-5472.CAN-20-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W-C, Chen Y-J, Li L-Y, Wei Y-L, Hsu S-C, Tsai S-L, et al. Nuclear translocation of epidermal growth factor receptor by AKT-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–68. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008;283:4344–51. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee H-J, Zhuang G, Cao Y, Du P, Kim H-J, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 65.Sarcoma Alliance for Research through Collaboration. SARC031: A Phase 2 Trial of the MEK Inhibitor Selumetinib (AZD6244 Hydrogen Sulfate) in Combination With the mTOR Inhibitor Sirolimus for Patients With Unresectable or Metastatic Malignant Peripheral Nerve Sheath Tumors [Internet]. clinicaltrials.gov; 2023 Mar. Report No.: NCT03433183. Available from: https://clinicaltrials.gov/study/NCT03433183.
- 66.Kim A. SARC031: The phase 2 trial of selumetinib and sirolimus for patients with unresectable or metastatic malignant peripheral nerve sheath tumors. Abstr 2023 NF Conf CTF [Internet]. Fairmont Scottsdale Princess Arizona; 2023. page 18. Available from: https://bv-nf.de/wp-content/uploads/2023/08/23_NFConferenceAbstractBook_web_revised.pdf.
- 67.Johannessen CM, Johnson BW, Williams SMG, Chan AW, Reczek EE, Lynch RC, et al. TORC1 Is Essential for NF1-Associated Malignancies. Curr Biol. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 68.Malone CF, Fromm JA, Maertens O, DeRaedt T, Ingraham R, Cichowski K. Defining key signaling nodes and therapeutic biomarkers in NF1-mutant cancers. Cancer Discov Am Assoc Cancer Res. 2014;4:1062–73. doi: 10.1158/2159-8290.CD-14-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kojima K, Shimanuki M, Shikami M, Samudio IJ, Ruvolo V, Corn P, et al. The dual PI3 kinase/mTOR inhibitor PI-103 prevents p53 induction by Mdm2 inhibition but enhances p53-mediated mitochondrial apoptosis in p53 wild-type AML. Leukemia. 2008;22:1728–36. doi: 10.1038/leu.2008.158. [DOI] [PubMed] [Google Scholar]
- 70.Kong N, Tao W, Ling X, Wang J, Xiao Y, Shi S, et al. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci Transl Med. 2019;11:eaaw1565. doi: 10.1126/scitranslmed.aaw1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cordani M, Oppici E, Dando I, Butturini E, Dalla Pozza E, Nadal-Serrano M, et al. Mutant p53 proteins counteract autophagic mechanism sensitizing cancer cells to mTOR inhibition. Mol Oncol. 2016;10:1008–29. doi: 10.1016/j.molonc.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyake N, Chikumi H, Takata M, Nakamoto M, Igishi T, Shimizu E. Rapamycin induces p53-independent apoptosis through the mitochondrial pathway in non-small cell lung cancer cells. Oncol Rep. 2012;28:848–54. doi: 10.3892/or.2012.1855. [DOI] [PubMed] [Google Scholar]
- 73.Luther G, Rames R, Wagner ER, Zhu G, Luo Q, Bi Y, et al. Molecular basis of differentiation therapy for soft tissue sarcomas. Trends Cancer Res. 2010;6:69–90. [PMC free article] [PubMed] [Google Scholar]
- 74.Boerboom A, Dion V, Chariot A, Franzen R. Molecular Mechanisms Involved in Schwann Cell Plasticity. Front Mol Neurosci. 2017;10:38. doi: 10.3389/fnmol.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mazuelas H, Magallón-Lorenz M, Fernández-Rodríguez J, Uriarte-Arrazola I, Richaud-Patin Y, Terribas E, et al. Modeling iPSC-derived human neurofibroma-like tumors in mice uncovers the heterogeneity of Schwann cells within plexiform neurofibromas. Cell Rep. 2022;38:110385. doi: 10.1016/j.celrep.2022.110385. [DOI] [PubMed] [Google Scholar]
- 76.Masaki T, Qu J, Cholewa-Waclaw J, Burr K, Raaum R, Rambukkana A. Reprogramming Adult Schwann Cells to Stem Cell-Like Cells by Leprosy Bacilli Promotes Dissemination of Infection. Cell. 2013;152:51–67. doi: 10.1016/j.cell.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Velletri T, Xie N, Wang Y, Huang Y, Yang Q, Chen X, et al. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis. 2016;7:e2015. doi: 10.1038/cddis.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rivlin N, Koifman G, Rotter V. p53 orchestrates between normal differentiation and cancer. Semin Cancer Biol. 2015;32:10–7. doi: 10.1016/j.semcancer.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 79.Ko KR, Lee J, Lee D, Nho B, Kim S. Hepatocyte Growth Factor (HGF) Promotes Peripheral Nerve Regeneration by Activating Repair Schwann Cells. Sci Rep. Nat Publ Group. 2018;8:8316. doi: 10.1038/s41598-018-26704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Norrmén C, Figlia G, Pfistner P, Pereira JA, Bachofner S, Suter U. mTORC1 Is Transiently Reactivated in Injured Nerves to Promote c-Jun Elevation and Schwann Cell Dedifferentiation. J Neurosci Soc Neurosci. 2018;38:4811–28. doi: 10.1523/JNEUROSCI.3619-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Birchmeier C, Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol Elsevier. 1998;8:404–10. doi: 10.1016/S0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- 82.H’ng CH, Khaladkar A, Rosello-Diez A. Look who’s TORking: mTOR-mediated integration of cell status and external signals during limb development and endochondral bone growth. Front Cell Dev Biol. 2023;11:1153473. doi: 10.3389/fcell.2023.1153473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen J, Long F. mTOR signaling in skeletal development and disease. Bone Res Nat Publ Group. 2018;6:1–6. doi: 10.1038/s41413-017-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Enane FO, Saunthararajah Y, Korc M. Differentiation therapy and the mechanisms that terminate cancer cell proliferation without harming normal cells. Cell Death Dis. 2018;9:912. doi: 10.1038/s41419-018-0919-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J, Grovola MR, Xie H, Coggins GE, Duggan P, Hasan R, et al. Comprehensive pharmacological profiling of neurofibromatosis cell lines. Am J Cancer Res. 2017;7:923–34. [PMC free article] [PubMed] [Google Scholar]
- 86.Miller SJ, Rangwala F, Williams J, Ackerman P, Kong S, Jegga AG, et al. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 2006;66:2584–91. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- 87.Pin E, Federici G, Petricoin EF. Preparation and use of reverse protein microarrays. Curr Protoc Protein Sci. 2014;75:27.7.1–27.7.29. doi: 10.1002/0471140864.ps2707s75. [DOI] [PubMed] [Google Scholar]
- 88.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat Med. 2010;16:821–7. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Welm BE, Dijkgraaf GJP, Bledau AS, Welm AL, Werb Z. Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell. 2008;2:90–102. doi: 10.1016/j.stem.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010 [cited 2020 Oct 5]; Available from: http://www.bioinformatics.babraham.ac.uk/projects/.
- 94.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE225747 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE225747).
R code used in this study is available upon request.