Abstract
In this work, 710 CDSs corresponding to over 290 000 codons equally distributed between Brassica napus, Arabidopsis thaliana, Lycopersicon esculentum, Nicotiana tabacum, Pisum sativum, Glycine max, Oryza sativa, Triticum aestivum, Hordeum vulgare and Zea mays were considered. For each amino acid, synonymous codon choice was determined in the presence of A, G, C or T as the initial nucleotide of the subsequent triplet; data were statistically analysed under the hypothesis of an independent assortment of codons. In 33.4% of cases, a frequency significantly (P = 0.01) different from that expected was recorded. This was mainly due to a pervasive intercodon TpA and CpG deficiency. As a general rule, intercodon TpAs and CpGs were preferably replaced by CpAs and TpGs, respectively. In several instances, codon frequencies were also modified to avoid homotetramer and homotrimer formation, to reduce intercodon ApCs downstream {1,2} GG or AG dinucleotides, as well as to increase GpA or ApG intercodons under certain contexts. Since TpA, CpG and homotetra(tri)mer deficiency directly or indirectly accounted for 77% of significant variation in the codon frequency, it can be concluded that codon usage mirrors precise needs at the DNA structure level. Plant species exhibited a phylogenetically-related adaptation to structural constraints. Codon usage flexibility was reflected in strikingly different arrays of optimum codons for probe design.
INTRODUCTION
DNA sequence analyses have demonstrated that synonymous codons are used differently by organisms and each type of genome has a particular coding strategy (1,2).
In Escherichia coli and yeast, synonymous codon choice patterns are related to the abundance of isoaccepting tRNAs (3,4); moreover, the extent of the bias in codon usage is positively correlated to the level of gene expression (2,5,6). In multicellular organisms, a functional adaptation of tRNA population to codon frequency has also been hypothesised (5). However, interpretation of codon usage patterns in these species is complicated by cell-specific, tissue-specific and developmentally-regulated gene expression (7,8); furthermore, data concerning the composition of tRNA populations in different cell lines are nearly absent. To our knowledge, the only evidence collected for plants concerns zein synthesis in maize (9). The codon usage pattern adopted in this case was found to fit well with the array of most abundant tRNA isoacceptors in the endosperm.
It should be noted that, even for functionally homologous genes, remarkable differences in codon usage exist across species (10,11). In particular, the G+C percentage in the silent codon third nucleotide position (G3+C3) is higher in monocots than in dicots and the difference is especially evident if members of the Gramineae family are considered (73.5 versus 45.0%) (12). Moreover, within monocots, genes can be classified into two groups: those with a narrow codon bias and a high G3+C3 value and those with a broader codon usage and a relatively lower G3+C3 percentage (10,12). This separation resembles that of vertebrate nuclear genes (5,13).
Differences in the codon bias between genes in the same organism have been attributed to the G+C variation throughout the genome (5). This can be due to the dispersion of large (>300 kb) isochores homogeneous for G+C content (14).
A highly stable non-random dinucleotide frequency pattern has been identified in bulk genomic DNA which has been called ‘general design’ (9,15). Closely related species present more similar general designs than unrelated organisms (16); these characteristic dinucleotide frequencies may reflect the response of the genome to evolutionary selection pressures (15). There may be factors, such as base-step conformational tendencies, methylation patterns, DNA replication and repair mechanisms and context-dependent mutation patterns which influence the compositional and structural patterns of a genomic sequence (17,18); dinucleotide relative abundance values constitute a ‘genome signature’ which may reflect the influence of such factors (17,19).
In plants, most dinucleotide frequency studies have been focused on CpG and TpA occurrences (20–23). It has been noted that CpG dinucleotides in coding sequences occurred less frequently than expected on the basis of the G+C content of the sequences; this shortage, however, decreased and vanished as the G+C level increased (24). In fact, the distribution of CpG dinucleotides in eukaryotic genomes revealed two basic patterns: in the first pattern, CpGs were few in number, frequently methylated and scattered along the DNA in both coding and non-coding sequences; in the second, CpGs were at a frequency close to the expected, unmethylated and clustered in DNA segments called CpG islands (25,26).
CpG under-representation is commonly ascribed to the classical methylation→deamination→mutation mechanism: methylation of cytosine in position 5, followed by deamination of 5-methylcytosine produces (when unrepaired) the conversion of CpG to TpG (27). However, this hypothesis cannot account for CpG suppression in animal mitochondria (28) or chloroplast genomes (21) which lack methylase activity. Moreover, in many vertebrate sequences, CpG suppression is not associated with a significant abundance of the dinucleotide TpG, reflecting an irregular distribution of unmethylated CpG regions across the genome (29). Therefore, CpG deficiencies may in some circumstances be due to structural constraints operating at DNA level (30–32). It should be remembered that the CpG dinucleotide exhibits the greatest thermodynamic stacking energy of all dinucleotides (33,34), hence its frequency reduction might facilitate DNA replication and transcription (29).
Besides CpG, the dinucleotide TpA is also under-represented in most life forms (35–37). In the human genome, while CpG frequency is lowest in transcriptionally silent DNA, TpA is most stringently avoided in DNA designed to be expressed as mRNA (36). TpA paucity may reflect UpA instability to nucleolytic cleavage in mRNA and the fact that two out of three stop codons start with TpA (36). Moreover, TpA is less stable energetically than all other dinucleotides (33,34), which would provide flexibility for untwisting and bending of the DNA double helix (16); this may explain why TATA sequences are very easy to unwind through protein interaction and are found, among other regions, at the sites of replication origin (38). In all likelihood, restricted TpA usage may help to avoid inappropriate binding of regulatory factors (29).
In this work we focused on dinucleotide frequencies at intercodon sites (codon position {3,4}; where 4 = 1 of the next codon) in several plant species using large cDNA samples. The main purpose of our study was to determine how and by how much intercodon dinucleotides may affect synonymous codon choice in plants. Few previous studies on plant dinucleotide frequencies have been carried out; in addition, a very limited number of species and/or sequences (17,21,23,37) were considered. The results of these studies were often contradictory and sometimes in sharp contrast with those observed in animal systems. It should be noted that evidence was sometimes collected using intergenic DNA and in no instance was analysis carried out at the amino acid level. Synonymous codon usage pattern may have important implications on the level of gene expression in transformation experiments involving donor and recipient organisms using different dialects (7,10,11). It may also indicate the type and strength of factors which are acting on DNA in the presence of amino acid constraints. Finally, it should be considered when designing degenerate primers or probes such as those deduced from N-terminal sequences of proteins. Working on human sequences, it has been demonstrated that the overall probe–target homology could be increased from 66.6 to >82% when codon usage and intercodon dinucleotide frequencies were taken into account (39). In the present study, the same computational method was applied to provide information about optimum codon choice for probe design in model plants and economically important crop species.
MATERIALS AND METHODS
Plant species and gene sequences
In this work, four monocot and six dicot species were considered (Table 1). All monocots were chosen from the Gramineae family whereas dicots were selected from the Brassicaceae, Solanaceae and Leguminosae (Papilionaceae) families. The choice of species was made on the basis of their importance as crop or model plants, and the relative abundance of complete coding DNA sequences (CDS); 60–80 CDSs per species were extracted from the GenBank database (release 112.0) using the NCBI Entrez retrieval system. Duplicate sequences and alleles of the same gene were avoided in order to minimize gene-specific bias. Mitochondrial and chloroplastic DNAs were also excluded from sampling due to the peculiarity of their G+C content (40). Apart from these constraints, CDSs were chosen completely at random.
Table 1. Plant species and composition of the samples.
| Species | No. of sequences | Amino acid-coding triplets |
|---|---|---|
| Arabidopsis thaliana Heynh. | 71 | 28 880 |
| Brassica napus L. | 80 | 28 152 |
| Lycopersicon esculentum Mill. | 61 | 31 811 |
| Nicotiana tabacum L. | 64 | 26 741 |
| Pisum sativum L. | 72 | 30 656 |
| Glycine max Merr. | 73 | 32 239 |
| Oryza sativa L. | 79 | 29 452 |
| Triticum aestivum L. | 76 | 26 594 |
| Hordeum vulgare L. | 72 | 27 489 |
| Zea mays L. | 62 | 29 745 |
The exact number of sequences retrieved and their equivalence to amino acid coding triplets are reported separately for each plant species in Table 1 (a complete list of sampled sequences is available as Supplementary Material at NAR Online, Table S1).
Sequence analysis
After pooling the CDSs sampled in each species, the codon usage and the intercodon values were calculated using an unpublished computer program written by Prof. F. Fabris (Department of Mathematics and Computer Science, University of Udine, Italy). Codon usage was obtained by dividing the number of times a codon occurred by the total number of codons (termination signals included). Intercodon values were calculated at the single codon level, therefore they were equal to the number of cases in which a given codon was followed by either an A, G, C or T.
Intercodon values were entered in contingency tables with synonymous codons as row variables and A4, G4, C4 or T4 (i.e., the first nucleotide following the codon) as column variables. Once these tables were completed, the total χ2 value was computed; provided that the observed χ2 was highly significant (P ≤ 0.01), tables were then analysed with the ACTUS program (Analysis of Contingency Tables Using Simulation) (41) by simulating 1000 such tables. In the simulated tables, cases were assigned randomly to cells with a probability proportional to row and column frequency and under the hypothesis of independence between row and column variables (null hypothesis). The last nucleotide of a given codon and the first of the following one were considered inter-related when the observed intercodon frequency was 990 times out of 1000 greater (thus defined significantly large value) or lower (significantly small value) than the simulated values. This threshold level can be interpreted as a one-tailed significance equal to 1%. There is one main advantage in using ACTUS; in contrast with standard statistical χ2 tests, this method clearly indicates which cells contain significantly higher or lower cases than predicted by independence (41).
Optimum codon and certainty factor
In order to determine the optimum codon (not necessarily the most frequent) for a generic amino acid in the presence of a different N4 (where N = A, C, G or T), the following procedure was adopted. One codon was arbitrarily chosen as optimum (fractional identity = 1.00) and the fractional identities of all synonymous codons were determined accordingly. Codon frequencies in the case of a given N4 were multiplied by fractional identities, the summated results giving the overall percentage similarity (39). This procedure was repeated for each synonymous triplet considered as an optimum codon and the results compared. The real optimum codon for a given N4 was the triplet showing the highest overall percentage similarity. The calculation is illustrated in Table 2 for Arabidopsis thaliana AGA (a) and CGA (b) raised to optimum codons in the case of A4.
Table 2.
| Synonymous codon | Frequency F (%) | (a) AGA | (b) CGA | |||
|---|---|---|---|---|---|---|
| Fractional identity (I) | Product (F × I) | Fractional identity (I) | Product (F × I) | |||
| AGA | 39.2 | 1.00 | 39.2 | 0.66 | 25.9 | |
| AGG | 26.1 | 0.66 | 17.2 | 0.33 | 8.6 | |
| CGA | 10.0 | 0.66 | 6.6 | 1.00 | 10.0 | |
| CGG | 9.3 | 0.33 | 3.1 | 0.66 | 6.2 | |
| CGC | 5.1 | 0.33 | 1.7 | 0.66 | 3.4 | |
| CGT | 10.4 | 0.33 | 3.4 | 0.66 | 6.9 | |
| Overall % similarity | 71.1 | 60.8 | ||||
For each amino acid and intercodon type, the certainty factor was defined as the overall percentage similarity predicted between the optimum codon and the target sequence.
Cluster analysis
The data for codon frequency in the presence of different N4 were entered in a table with plant species on columns and all possible combinations between codons and N4 on rows. After producing a matrix of Euclidean distances between species, a classification algorithm based on the average linkage between groups was applied. The SPSS package was used to perform cluster analysis.
RESULTS AND DISCUSSION
Codon utilisation profiles in the sampled sequences (see Supplementary Material, Table S2) were always in excellent agreement with those reported in the Codon Usage Database (42); actually, in previous work (18) dinucleotide frequencies (i.e., genome signature) showed substantial invariance across 50 kb contigs sampled throughout the genome. On the basis of sample size, representativity of codon choice patterns and the wide experimental evidence regarding the conditions for a genomic signature consistency, it can be assumed that results achieved in this work may be extended to other CDSs.
For all species, the relative frequency of each codon when the subsequent triplet is headed by A, G, C or T and the results of statistical analysis are reported in Supplementary Material, Table S3 (available at NAR Online).
Intercodon TpA suppression
A pervasive under-representation of T3pA4 was observed in all species (Supplementary Material, Table S3). A significant (P = 0.01) intercodon TpA suppression was always recorded for codons AAT (Asn), GAT (Asp), TAT (Tyr), TTT (Phe), ATT (Ile), GGT (Gly), GCT (Ala), GTT (Val), TCT (Ser) and CTT (Leu). With other TpA intercodons, exceptions to this trend mostly involved Gramineae and residues coded for by six triplets. It should be pointed out that a significant T3pA4 deficiency was also observed in Hordeum vulgare, Zea mays and Lycopersicon esculentum (Supplementary Material, Table S3) where no such reduction was previously found in bulk genomic DNA (see 19 for review).
From a quantitative point of view, direct evidence of a T3pA4 suppression was achieved by examining the relative amounts of triplets used to code Asn, Asp, His, Tyr, Cys and Phe in the presence or absence of A4. These amino acids are all coded for by two triplets ending either with T or C; while the C3/T3 ratio averaged 0.45–1.34 for B4 (where B = G, C or T), a mean 2.1-fold increase of C- over T-ending codons was noted in the presence of A4. Of the above-mentioned amino acids, special interest was devoted to Tyr and Asn as they are coded for by TAT and AAT, respectively. When an A-starting codon follows, a TATA sequence or a motif which could be interpreted as a polyadenylation signal (consensus sequence: 5′-AATAAA-3′) (43) is generated. Within coding regions, the presence of such elements should be disfavoured (17). Surprisingly, the ratios between C- and T-ending codons for Tyr and Asn (2.3 and 2.0, respectively) fell within, or only slightly distanced from, the range observed for other amino acids indicating that both TAT3A4 and AAT3A4 tetranucleotides are not rare in plant CDSs. Even in rice, where the ratio TAC3A4/TAT3A4 was highest (3.61), 20 of 79 sequences (25.3%) contained at least one TATA motif which could be avoided through synonymous codon replacement.
In order to verify whether T3pA4 substitution with other intercodons was random, the codon usage for four-coded amino acids (Thr, Gly, Ala, Val and Pro) was studied in more detail. Statistical analysis indicated that when the following codon was headed by A, T3 was preferably replaced by C3. This phenomenon was quantitatively more evident in dicots than in Gramineae, probably because the latter species already show a remarkable preference for C-ending codons (20). In previous work, CpA dinucleotide frequency was found to be correlated with CpG suppression (21,37) as CpA represents the complementary reverse of TpG (see below). In dicots, the coefficient of linear correlation between T3pA4 reduction and C3pA4 increase was highest for Thr (r = 0.925, P = 0.01), Val (r = 0.957, P = 0.01) and Ala (r = 0.912, P = 0.05); for the remaining two amino acids (Gly and Pro), T3pA4 suppression was significantly correlated with an A3pA4 increase (r = 0.949, P = 0.01 and r = 0.850, P = 0.05, respectively). In no instance were the NCG codons of Thr, Ala and Pro increased as a consequence of NCT3pA4 suppression; indeed, in the Gramineae members, a significant reduction in NCG3pA4 was observed in most cases (Supplementary Material, Table S3) (see next paragraph on CpG deficiency for explanation). A clear trend of T3pA4 substitution with C3pA4 was also observed for Ile, Ser and Leu. A remarkable exception concerned Arg where CGT was partially replaced by AGG.
Intercodon CpG deficiency
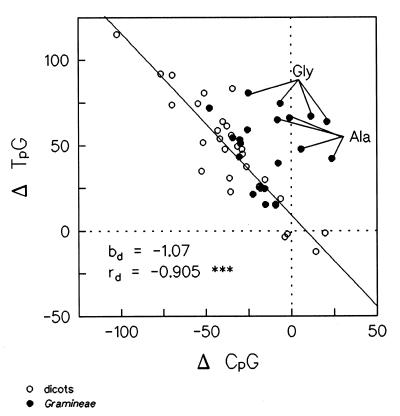
As for T3pA4, the CpG intercodon appeared widely under-represented in both dicots and Gramineae. This phenomenon was particularly evident for amino acids coded for by two or three triplets. In general, C3pG4 deficiency was clearly associated with an over-representation of T3pG4, in accordance with the classical methylation→deamination→mutation scenario causing the conversion of CpG into TpG (44). The effect of the plant species and the type of amino acid upon C3pG4 replacement was evaluated with a correlation analysis taking into account the data relative to the four-coded amino acids. Statistical analysis revealed that in dicots there is a very tight association between C3pG4 reduction and T3pG4 increase (Fig. 1); this pattern was followed by all species, regardless of the amino acid considered. Interestingly, the regression line (Fig. 1) nearly crossed the origin and the confidence limits (P = 0.05) for the regression coefficient (–0.88, –1.26) encompassed the value of –1 (exact replacement of C3pG4 with T3pG4).
Figure 1.
Correlation between C3pG4 depletion and T3pG4 increase in dicots and Gramineae (data relative to the four-coded amino acids: Thr, Val, Pro, Gly and Ala). ***, significant at the probability level, P = 0.001.
Boudraa and Perrin (21) also observed an opposite trend between CpG and TpG frequencies, but very few plant nuclear genes were considered. Work on dicot genomic DNA (17) revealed a strong CpG suppression but none of the dinucleotides which can derive from CpG single-base mutation (CC/GG, TG/CA and AG/CT) was over-represented.
Since CpG deficiency is widespread across amino acids and species, the occurrence of some structural rather than translational constraints can be deduced; actually, a CpG depletion was noted even in silent DNA (36) and this was ascribed to high dinucleotide stacking energy, supercoiling and chromatin packing (29).
In comparison with the observations made in dicots, Gramineae species showed an amino acid-specific behaviour. In particular, for Thr, Val and Pro, cereals and dicots seemed to replace C3pG4 with T3pG4 in a nearly identical manner (Fig. 1). In contrast, Gramineae species clearly departed from the common pattern in the case of Gly and Ala involvement (Fig. 1). In fact, when the data from all species were pooled, the percentage variation explained by regression increased from 36.7 to 73.3%, provided that the latter amino acids were excluded from the analysis. In the absence of Gly and Ala, both the correlation coefficient (r = –0.856, P = 0.01) and the slope of the regression line (–1.00) were not significantly different from those obtained with dicots alone; the values relative to the intercept were also similar (11.16 versus 9.05). The following behaviour of Gramineae in dealing with Gly and Ala codons was observed: (i) compared to other amino acids, C-ending codons were never significantly under-represented; (ii) the observed number of T-ending codons definitely exceeded the expected value, indicating a primary T-choice; (iii) use of codons with terminal nucleotides other than C or T was sometimes restricted (Supplementary Material, Table S3), e.g., in the presence of G4, the GGG codon (Gly) was always avoided to prevent the onset of a G-homotetramer (see below). Restricting the analysis to Gly and Ala (all species), a negative correlation was found between C3pG4 and G3pG4 (r = –0.817, P = 0.01 and r = –0.716, P = 0.05, respectively); however, the values relative to the intercept and the slope of the regression lines clearly indicate that the only effect of a stronger C3pG4 reduction was a lower deficiency of G3pG4.
It was previously noted that, in monocot genomic DNA, CpG frequency was only marginally low to low-normal (17) and that NCG codons were not avoided unlike dicots (12,20). Disaggregating the data at an amino acid level and taking into account the type of following nucleotide led us to discover a more complex situation. A good example can be provided for NCG codon frequency: in monocots NCGs appeared either over-represented, under-represented or at the expected frequency according to the first nucleotide of the following codon; more precisely, no particular influence was determined by G4 or T4 whereas C4 and A4 caused a remarkable NCG increase and decrease, respectively.
Homotetra(tri)mer avoidance
In several instances, intercodon frequencies appeared modified in order to limit the onset of homotetramers. This phenomenon was more relevant for G and C rather than for T and A. To our knowledge, the reasons for avoidance were established only for G-homotetramers; in particular, G runs were found to exert detrimental effects on mRNA stability (45). Interestingly, G- and C-homotetramers were preferably avoided in Gramineae whereas T- and A-tetranucleotides were more frequently omitted in dicots. In a number of cases, homotrimers were also deficient, e.g., AGG3pG4, GCC3pC4, GAA3pA4, ATT3pT4.
Sometimes, homotetra(tri)mer under-representation had a remarkable impact on synonymous codon usage. As previously indicated in Gramineae, GGG3pG4 avoidance was accompanied by a GGC3pG4 frequency which (in contrast to the rule) approached the expected value; the same phenomenon concerned the two binomials [CCC3pC4, CCG3pC4](Pro) and [GCC3pC4, GCG3pC4](Ala) where the second terms were over-represented despite their internal CpG element. In Gramineae, the synonymous WCG (where W = A or T) codon frequency was also significantly enhanced in the presence of C4. Similarly, to prevent the appearance of T-trimers, the ATA codon of Ile was preferentially used; this is surprising not only in view of the internal TpA element, but also considering the less stable codon–anticodon interaction compared to the alternative triplet ATC (3).
A3pC4 reduction and preference for RpR intercodons
It was previously noted that, in eukaryots, dinucleotides of the mixed type, i.e., YpR or RpY (where Y = C or T and R = A or G), are disfavoured (exceptions to this trend are TpG, CpA and sometimes GpC). This fact has been explained in terms of minimal double-helix distortion (30,32). The present study confirmed that, of YpR intercodons, C3pG4 and T3pA4 are clearly suppressed but another two YpR intercodon types, namely T3pG4 and C3pA4, are commonly used in substitution of the former. With regards to RpY intercodons, A3pC4s appeared frequently avoided in both dicots and Gramineae, particularly with a GGA (Gly) or AGA (Arg) codon. In contrast, A3pT4, G3pC4 and G3pT4 (i.e., other RpY intercodons) were only infrequently diminished in dicots, whereas in the Gramineae species the results were even less consistent.
As to YpY intercodons, no sign of a clear preference could be traced in this work. In contrast, the relative abundance of some R3pR4 combinations was fairly evident. Especially in dicots, the intercodons more often over-represented were G3pA4 and A3pG4; interestingly, A3pG4s were favoured when the second codon position was occupied by an R and disfavoured in the presence of Y2 (Supplementary Material, Table S3).
Optimum codon choice and certainty factor
Synthetic probes deduced from amino acid sequence data are increasingly used in plant molecular biology. On the basis of pervasive C3pG4, T3pA4 and homotetra(tri)mer depletion, a remarkable reduction of wrong choices in degenerate positions could be expected. However, it should be considered that intercodon over- and under-representation have to be interpreted in relative terms and that codons giving rise to disfavoured intercodons are not necessarily used less frequently than others. Secondly, when dealing with six-coded amino acids, the overall homology of a codon with respect to all possible substitutes should not be overlooked.
For all plant species, optimum codons and certainty factors are available at NAR Online (Supplementary Material, Table S4). From these data, it can be deduced that: (i) the strength of the structural constraints follows the order: homotetramer > T3pA4 > C3pG4 > homotrimer; (ii) while homotetramer avoidance is a rule applied in all optimum codons but one, plant species sharply differ in their adaptation to TpA and CpG constraints. Actually, some optimum codons giving rise to T3pA4s can occur in dicots but never in Gramineae, which, on the other hand, tend to use optimum codons leading to the formation of C3pG4s. Furthermore, homotrimers formed in dicots with the sharing of an optimum codon always involved T or A, whereas in Gramineae homotrimers were almost invariably of the C- or G-type.
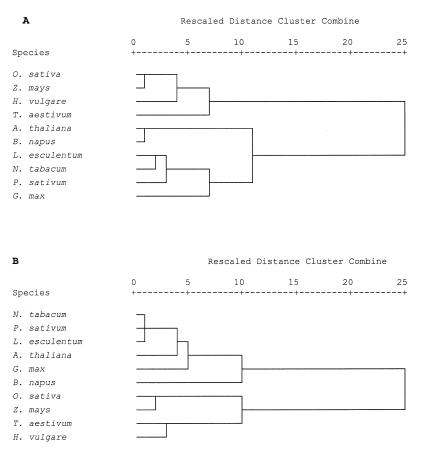
Another relevant issue concerns the flexibility of codon usage in relation to the different intercodons generated. With regards to this factor, plant species appeared to behave in a phylogenetically-related manner. In fact, when cluster analysis was carried out on the data in Table S3 (Supplementary Material), similarity levels were highest between species of the same family (Fig. 2A). Not only were dicots clearly separated from Gramineae but, in agreement with previous findings on G+C content in Brassicaceae (12), A.thaliana and Brassica napus clustered separately from other dicots. This evidence suggests that the frequencies of tetranucleotides, each composed by a codon and the initial of the following one, can not only be used for good probe design, but also for classification purposes and evolutionary studies. These elements all share the advantages characteristic of dinucleotide frequencies: (i) they constitute a genome signature reflecting base-step stacking capacities, duplex curvature and other higher order DNA structural features (29); (ii) they can be easily calculated using the entire available genome sequences without any prior alignment, since they are unaffected by gaps or sequence rearrangements (18); (iii) they allow the tracing of phylogenetic relationships without direct comparison of gene sequences (16). In addition, tetranucleotides (NNN3pN4) are more informative because they also consider the structural constraints connected with homotetramer and homotrimer formation, ApC depletion and RpR preference in different codon contexts and, overall, the amino acid constraint. The utility of this approach was confirmed by comparing clusters obtained from NNN3pN4 and intercodon dinucleotides frequency data. The latter allowed the discrimination of Gramineae but dicot species belonging to the same family were often sorted in different cluster regions (Fig. 2B).
Figure 2.
Dendrogram using average linkage between groups of NNN3pN4 frequency data (A) and intercodon dinucleotide frequency data (B).
Codon usage flexibility also influenced the number of optimum codons recorded in the different situations (Table 3). Maximum values (35 or more) were observed for the two Brassicaceae members and were determined by the addition of some C- or G-ending triplets to the dicot array of optimum codons. Within Gramineae, a striking difference was noted between Triticeae and the Oryza sativa, Zea mays couple. While in the latter species optimum codons were 34 and 29, respectively, in Triticeae (Triticum aestivum and Hordeum vulgare) their number was remarkably lower. The reason for this was the considerable preference for C-ending triplets, regardless of codon neighbourhood. It should be noted that the low flexibility of Triticeae had no adverse effect on certainty factors which were even slightly higher than those found in other species.
Table 3. Number of C3pG4, T3pA4, homotetramers and homotrimers generated by optimum codons in the 10 species considered.
| A.thaliana | B.napus | L.escultenum | N.tabacum | P.sativum | G.max | O.sativa | Z.mays | T.aestivum | H.vulgare | |
|---|---|---|---|---|---|---|---|---|---|---|
| No. optimum codons | 35 | 37 | 29 | 31 | 33 | 34 | 34 | 29 | 25 | 22 |
| No. optimum codons producing: | ||||||||||
| Homotetramer | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| TpA intercodon | 1 | 1 | 3 | 4 | 2 | 0 | 0 | 0 | 0 | 0 |
| CpG intercodon | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 4 | 11 | 11 |
| Homotrimer | 3 | 1 | 6 | 7 | 6 | 3 | 5 | 3 | 2 | 3 |
CONCLUSIONS
It was reported that for large collections of genes (50 or more), the codon signature, defined as the dinucleotide relative abundance at codon positions {1,2}, {2,3} and {3,4} (where 4 = 1 of the next codon), largely adheres to the genome signature (46). In this work, intercodon sequence analysis was carried out at an amino acid level; using this approach, it was possible to ascertain that some intercodon dinucleotide frequencies are significantly shifted from genome signature data (see 19 for review). All the evidence collected consistently indicates that structural constraints determine a non-randomness of codon neighbourhood. This conclusion agrees with the results obtained by Santibánez-Koref and Reich (47) in mammalian CDSs. Since gene sequences were collected randomly, the influence of other factors, namely translational efficiency, cannot be excluded. Interestingly, in the third nucleotide position, disfavoured C and A were preferably replaced by T and G, respectively, and vice versa. Hence, it can be hypothesised that rules affecting nucleotide replacement could have determined nucleotide assortment in the degenerate positions of the genetic code.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. F. Fabris (Department of Mathematics and Computer Science, University of Udine, Italy) for providing a computer program, and Dr P. Ganis (Department of Biology, University of Trieste, Italy) for help with the statistical analyses. This work was partly funded by the European Community in the frame of the INCO-Copernicus project, contract IC15CT961011.
REFERENCES
- 1.Grantham C., Gautier,C., Gouy,M., Mercier,R. and Pavé,A. (1980) Nucleic Acids Res., 8, r49–r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grantham C., Gautier,C., Gouy,M., Jacobzone,M. and Mercier,R. (1981) Nucleic Acids Res., 9, r43–r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikemura T. (1982) J. Mol. Biol., 158, 573–597. [DOI] [PubMed] [Google Scholar]
- 4.Ikemura T. (1981) J. Mol. Biol., 146, 1–21. [DOI] [PubMed] [Google Scholar]
- 5.Ikemura T. (1985) Mol. Biol. Evol., 2, 13–34. [DOI] [PubMed] [Google Scholar]
- 6.Hoekema A., Kastelein,R.A., Vasser,M. and de Boer,H.A. (1987) Mol. Cell. Biol., 7, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fennoy S.L. and Bailey-Serres,J. (1993) Nucleic Acids Res., 21, 5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiapello H., Lisacek,F., Caboche,M. and Hénaut,A. (1998) Gene, 209, GC1–GC38. [DOI] [PubMed] [Google Scholar]
- 9.Viotti A., Balducci,C. and Weil,J.H. (1978) Biochim. Biophys. Acta, 517, 125–132. [DOI] [PubMed] [Google Scholar]
- 10.Brinkmann H., Martinez,P., Quigley,F., Martin,W. and Cerff,R. (1987) J. Mol. Evol., 26, 320–328. [DOI] [PubMed] [Google Scholar]
- 11.Niesbach-Klösgen U., Barzen,E., Bernhardt,J., Rohde,W., Schwarz-Sommer,Z., Reif,H.J., Wienand,U. and Saedler,H. (1987) J. Mol. Evol., 26, 213–225. [Google Scholar]
- 12.Campbell W.H. and Gowri,G. (1990) Plant Physiol., 92, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aota S. and Ikemura,T. (1986) Nucleic Acids Res., 14, 6345–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernardi G., Olofsson,B., Filipski,J., Zerial,M., Salinas,J., Cuny,G., Meunier-Rotival,M. and Rodier,F. (1985) Science, 228, 953–958. [DOI] [PubMed] [Google Scholar]
- 15.Russell G.J., Walker,P.M.B., Elton,R.A. and Subak-Sharpe,J.H. (1976) J. Mol. Biol., 108, 1–23. [DOI] [PubMed] [Google Scholar]
- 16.Karlin S. and Ladunga,I. (1994) Proc. Natl Acad. Sci. USA, 91, 12832–12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlin S. and Mrázek,J. (1997) Proc. Natl Acad. Sci. USA, 94, 10227–10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlin S., Mrázek,J. and Campbell,A.M. (1997) J. Bacteriol., 179, 3899–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlin S., Campbell,A.M. and Mrázek,J. (1998) Annu. Rev. Genet., 32, 185–225. [DOI] [PubMed] [Google Scholar]
- 20.Murray E.E., Lotzer,J. and Eberle,M. (1989) Nucleic Acids Res., 17, 477–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boudraa M. and Perrin,P. (1987) Nucleic Acids Res., 15, 5729–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montero L.M., Filipski,J., Gil,P., Capel,J., Martínez-Zapater,J.M. and Salinas,J. (1992) Nucleic Acids Res., 20, 3207–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver J.L., Marín,A. and Martínez-Zapater,J.M. (1990) Nucleic Acids Res., 18, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montero L.M., Salinas,J., Matassi,G. and Bernardi,G. (1990) Nucleic Acids Res., 18, 1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antequera F. and Bird,A.P. (1988) EMBO J., 7, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tazi J. and Bird,A. (1990) Cell, 60, 909–920. [DOI] [PubMed] [Google Scholar]
- 27.Bird A.P. (1980) Nucleic Acids Res., 8, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardon L.R., Burge,C., Clayton,D.A. and Karlin,S. (1994) Proc. Natl Acad. Sci. USA, 91, 3799–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlin S. and Burge,C. (1995) Trends Genet., 11, 283–290. [DOI] [PubMed] [Google Scholar]
- 30.Nussinov R. (1984) Nucleic Acids Res., 12, 1749–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lennon G.G. and Fraser,N.W. (1983) J. Mol. Evol., 19, 286–288. [DOI] [PubMed] [Google Scholar]
- 32.Nussinov R. (1984) J. Mol. Evol., 20, 111–119. [DOI] [PubMed] [Google Scholar]
- 33.Breslauer K.J., Frank,R., Blöcker,H. and Marky,L.A. (1986) Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delcourt S.G. and Blake,R.D. (1991) J. Biol. Chem., 23, 15160–15169. [PubMed] [Google Scholar]
- 35.Nussinov R. (1981) J. Biol. Chem., 256, 8458–8462. [PubMed] [Google Scholar]
- 36.Beutler E., Gelbart,T., Han,J., Koziol,J.A. and Beutler,B. (1989) Proc. Natl Acad. Sci. USA, 86, 192–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burge C., Campbell,A.M. and Karlin,S. (1992) Proc. Natl Acad. Sci. USA, 89, 1358–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter C.A. (1993) J. Mol. Biol., 230, 1025–1054. [DOI] [PubMed] [Google Scholar]
- 39.Lathe R. (1985) J. Mol. Biol., 183, 1–12. [DOI] [PubMed] [Google Scholar]
- 40.Boudraa M. (1987) Génét. Sél. Evol., 19, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estabrook C.B. and Estabrook,G.F. (1989) Historical Methods, 22, 5–8. [Google Scholar]
- 42.Nakamura Y., Gojobori,T. and Ikemura,T. (1999) Nucleic Acids Res., 27, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallie D.R. (1996) In Owen,M.R.L. and Pen,J. (eds), Transgenic Plants: a Production System for Industrial and Pharmaceutical Proteins. John Wiley & Sons Ltd, Chichester, UK, pp. 49–74.
- 44.Bird A.P. (1980) Nucleic Acids Res., 8, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williamson J.R. (1994) Annu. Rev. Biophys. Biomol. Struct., 23, 703–730. [DOI] [PubMed] [Google Scholar]
- 46.Karlin S. and Mrázek,J. (1996) J. Mol. Biol., 262, 459–472. [DOI] [PubMed] [Google Scholar]
- 47.Santibánez-Koref M. and Reich,J.G. (1986) Biomed. Biochim. Acta, 45, 737–748. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.