Abstract
Amplification of genes involved in signal transduction and cell cycle control occurs in a significant fraction of human cancers. Loss of p53 function has been proposed to enable cells with gene amplification to arise spontaneously during growth in vitro. However, this conclusion derives from studies employing the UMP synthesis inhibitor N-phosphonacetyl-l-aspartate (PALA), which, in addition to selecting for cells containing extra copies of the CAD locus, enables p53-deficient cells to enter S phase and acquire the DNA breaks that initiate the amplification process. Thus, it has not been possible to determine if gene amplification occurs spontaneously or results from the inductive effects of the selective agent. The studies reported here assess whether p53 deficiency leads to spontaneous genetic instability by comparing cell cycle responses and amplification frequencies of the human fibrosarcoma cell line HT1080 when treated with PALA or with methotrexate, an antifolate that, under the conditions used, should not generate DNA breaks. p53-deficient HT1080 cells generated PALA-resistant variants containing amplified CAD genes at a frequency of >10−5. By contrast, methotrexate selection did not result in resistant cells at a detectable frequency (<10−9). However, growth of HT1080 cells under conditions that induced DNA breakage prior to selection generated methotrexate-resistant clones containing amplified dihydrofolate reductase sequences at a high frequency. These data demonstrate that, under standard growth conditions, p53 loss is not sufficient to enable cells to produce the DNA breaks that initiate amplification. We propose that p53-deficient cells must proceed through S phase under conditions that induce DNA breakage for genetic instability to occur.
It was proposed over 80 years ago that the altered growth characteristics of cancer cells are due to the gain or loss of genetic material (8). Since that time, changes in the copy numbers of genes responsible for controlling cell growth, such as the mutation or loss of tumor suppressor genes or the amplification of various oncogenes (27, 33, 63, 66), have been identified in nearly all human cancers. Normal, diploid cells do not undergo such genetic alterations due to the combined effects of DNA damage repair mechanisms and cell cycle checkpoints. Checkpoints minimize the proliferation of cells with genetic abnormalities either by arresting the cell cycle to allow adequate time for repair or by removing cells containing damaged genomes from the cycling population through induction of premature senescence or apoptosis (for reviews, see references 29, 41 and 56).
Gene amplification, a process by which subchromosomal portions of the genome increase in copy number, has been observed frequently in many human cancers (7) but not in normal cells (67, 73, 82, 85). This implies that normal cells possess control mechanisms which are inactivated during tumor cell initiation and/or progression to allow such genetic anomalies to arise. In vitro, amplification can allow cells to become resistant to antimetabolites such as N-phosphonacetyl-l-aspartate (PALA), an inhibitor of the CAD enzyme complex, which is responsible for catalyzing the first three steps of de novo pyrimidine biosynthesis (71). The amplification process can be initiated by chromosome breakage (13, 28, 39, 48, 67, 69, 78, 80, 81) and can lead to the formation of a variety of chromosomal structures, including extrachromosomal elements (double minute chromosomes), expanded chromosomal regions (homogeneously staining regions), and dicentric chromosomes. Given the importance of chromosome breakage in initiating gene amplification, studies characterizing the cellular mechanisms that regulate the amplification process have focused on cell cycle pathways which either limit the propagation of cells containing broken chromosomes or prevent such structural chromosomal changes from occurring.
The p53 protein has been identified as a vital transcriptional regulator in the pathway required for cell cycle arrest or apoptosis in response to DNA breakage (22, 31, 36, 37). p53 also mediates a cell cycle arrest, independent of DNA damage, in response to reduced ribonucleotide levels such as can be produced by PALA treatment (45). Consistent with its role in the breakage-mediated cell cycle arrest pathway, loss of p53 function is required for gene amplification to occur in human cells in response to selection with the antimetabolite PALA (47, 79, 85). The capacity of p53-deficient cells to generate PALA-resistant variants can be explained by the role of p53 in two cell cycle checkpoints. First, p53-deficient cells treated with PALA enter into S phase with inadequate ribonucleoside triphosphate (rNTP) pools, resulting in induction of double-strand DNA breaks (DSBs), while similarly treated, wild-type p53-containing cells undergo a G0/G1 cell cycle arrest and do not undergo breakage (6, 23, 57). Second, loss of p53 allows cells containing broken chromosomes to continue to proliferate, allowing cycles of DNA breakage and rejoining to occur, resulting in the generation of rare variants containing amplified sequences. The contribution of these two p53-mediated checkpoints activated by PALA treatment helps explain why the frequency of gene amplification in normal cells is at least 4 orders of magnitude lower than that in cells with a defective p53 pathway (47, 73, 82, 85).
Many studies characterizing the role of p53 in the control of genetic stability have used gene amplification as the model and PALA as the selective agent. Luria-Delbruck fluctuation analyses have been interpreted to indicate that amplification is a spontaneous process and that application of the selective agent enables detection of rare variants with an increase in the copy number of an appropriate genetic locus (35, 74). This model predicts that as p53-deficient cells proceed through S phase under standard growth conditions, they undergo DNA breakage throughout the genome at a detectable frequency. As such cells lack an arrest response triggered by DNA breakage, variants with local increases in random genomic regions should accumulate, so long as such increases do not result in a growth disadvantage. Cells with amplification of loci that enable survival during a particular growth challenge will then emerge as resistant variants. However, treatment of p53-deficient cells with PALA not only selects for those that have undergone CAD gene amplification but, as described above, can also cause the DSBs that initiate the process. Thus, previous studies have not been able to determine if treatment with PALA merely selects for preexisting variants in the population or if PALA induces the amplification process. The latter would indicate that p53 loss may create a genetic background permissive for amplification but that appropriate growth challenges must be present for genome destabilization to occur.
Determination of whether loss of p53 function is sufficient for the induction of genetic instability requires an experimental system that allows detection of preexisting variants which have already undergone amplification but does not induce the DSBs that would initiate the process in a p53-deficient cell line. Methotrexate (MTX), an antifolate which inhibits dihydrofolate reductase (DHFR), can be used to select for variants which resist its toxic effects through amplification of the DHFR locus (see reference 61 for a review). MTX causes a p53-independent growth arrest in normal human fibroblasts (45) and, as shown here, in the p53-deficient human fibrosarcoma cell line HT1080. We reasoned that if HT1080 cells acquire copy number changes during unchallenged growth due to their p53 deficiency, they should generate variants with amplified DHFR genes that should grow in the presence of MTX. We show that no such variants are generated in a population exceeding 109 cells. However, passage of HT1080 cells under conditions that induce chromosome breakage generated numerous cells that resist MTX due to DHFR gene amplification. These data demonstrate that initiation of the amplification process under nonchallenging growth conditions occurs with immeasurably low probability in this p53-deficient cell line. The data further reveal that amplification is largely an induced event that requires entry into and progression through S phase under conditions that increase the probability of DNA breakage. The relevance of these observations to genetic instability and tumor progression in vivo is discussed.
MATERIALS AND METHODS
Cell culture.
HT1080, a human fibrosarcoma tumor cell line, and WS1, a normal diploid human embryonic skin cell strain, were obtained from the American Type Culture Collection (Rockville, Md.). Cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% dialyzed fetal calf serum (Sigma) and nonessential amino acids (Life Technologies Inc., Gaithersburg, Md.). PALA was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute (Bethesda, Md.); MTX, VP16, and 1-β-d-arabinofuranosylcytosine (araC) were obtained from Sigma. The same lot of each drug was used throughout all experiments. Cell doubling times were approximately 20 to 24 h for HT1080 and 24 to 28 h for WS1. Cells were repeatedly analyzed and found to be free of mycoplasma contamination by in situ staining with Hoescht dye (11).
Cell cycle analysis.
Cells were synchronized by growth to confluence and held for 72 h in medium containing 0.1% serum. PALA-treated cells had 100 μM PALA added to the low-serum medium for the last 24 h. Cells were removed from the plates and were (i) treated with 4 Gy of gamma radiation (approximately 3.8 Gy/min at a distance of 40 cm) from a 60Co source (Gammabeam 150-C) and plated 1:4, (ii) passed 1:4 into medium containing drugs at various concentrations, or (iii) passed 1:4 into normal growth medium. Pulse-labeled cells were grown for 72 h, at which point they were treated with 10 μM bromodeoxyuridine (BrdU) for 1 h. Cells continuously labeled with BrdU were prepared identically, but 65 μM BrdU was present during the entire 72-h treatment or postirradiation period (45). At no time did the treated cells approach confluence. After treatment, cells were removed from the plates by using trypsin and were fixed with 70% ice-cold ethanol. Fixed cells were permeabilized, reacted with fluorescein isothiocyanate (FITC)-conjugated anti-BrdU antibody (Pharmingen), and counterstained with propidium iodide as previously described (45). Labeled cells were analyzed on a Becton-Dickinson FACScan, using Cell Quest software.
Selection for cells containing amplified DNA.
HT1080 or WS1 cells (1 × 104 to 5 × 105) were plated on 10-cm-diameter plates and allowed to recover for 24 h. The plates were washed with phosphate-buffered saline (PBS), and medium containing various concentrations of MTX or PALA was added. Media were changed every 3 days either until resistant colonies of >50 cells arose (14 to 21 days) or for 30 days, at which point selection was stopped. Colonies of >50 cells were isolated and expanded for later analysis. Plates were stained with crystal violet, and the number of resistant colonies was recorded. All experiments were done at least in duplicate.
For analysis of mechanisms of PALA resistance in HT1080 cells, cells were grown in slide chambers (Nunc) or on glass slides. After selection as described above, slides containing resistant colonies were treated with methanol-acetic acid (3:1) to fix the cells and were subjected to fluorescent in situ hybridization (FISH) as detailed below.
Cell photography.
HT1080 cells (2 × 105) were plated on 15-cm-diameter plates and allowed to grow for 24 h in normal medium. The cells were then grown in normal medium or medium containing either 54 μM PALA or 50 nM MTX. Photographs of cells on the same portion of the plate were taken every 24 h over a 10-day period with Kodak TMAX 400 black-and-white print film and a Nikon Diaphot microscope (magnification, × 40). Negatives were scanned and pictures were assembled by using Adobe Photoshop 3.0.5 (Adobe Software).
Pretreatment with DNA-damaging agents.
Cells (5 × 105) were plated on 15-cm plates-diameter and allowed to grow for 24 h. DNA-damaging agents were added to the medium at various concentrations and for various times. The plates were rinsed twice with PBS, and the cells were allowed to recover in normal medium for 1 to 4 days. Treated cells were split 1:4 and allowed to recover in normal medium for 12 h. Cells were selected in either 50 or 100 nM MTX; selective medium was replaced every 3 days until colonies formed (14 to 21 days). Pools of resistant cells and independent clones from separate plates were picked, transferred to 12-well dishes, and expanded up to 10-cm-diameter plates. Cells in duplicate plates were fixed and stained with crystal violet, and the number of resistant colonies was counted.
FISH.
FISH was done as described previously (81). Briefly, cells were allowed to grow to 50% confluence and treated with colcemid (0.1 μg/ml) for 30 min. Cells were harvested and resuspended in 75 mM KCl for 25 min. After centrifugation, cells were fixed in multiple washes of methanol-acetic acid (3:1) and dropped onto slides. Chromosome spreads were dried for 24 h, treated with 100 μg of RNase A (Sigma) per ml in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 h and dehydrated in an ethanol series (70, 80, and 100%). The chromosome spreads were denatured in 70% formamide–2× SSC at 72°C for 2 min and dehydrated in an ice-cold ethanol series. Chromosome spreads were hybridized with either a human DHFR (phage lambda J1, a gift from G. Attardi [84]) or CAD (cosmid HuCad69, a gift from O. Chernova and G. Stark) probe labeled with biotin-11-dUTP by random priming with the BioPrime Labeling System (Life Technologies), according to the manufacturer’s protocol. Twenty five nanograms of biotinylated DHFR or CAD probe was mixed with 2 μg of human Cot1 DNA (Life Technologies) in 12% dextran sulfate–2× SSC–60% formamide for 12 h at 37°C in a humidified chamber. Hybridizations were washed sequentially in 4× and 2× SSC at 45°C and in 0.1× SSC at 65°C. Probes were visualized by using FITC conjugated avidin. In some cases, the signal was amplified by using an intermediate biotinylated antiavidin treatment. Metaphase spreads were visualized by using a triple band-pass filter (Chroma) on a Zeiss standard WL epifluorescence microscope with a Neofluor oil immersion objective (63× or 100×). Photographs were taken with Ektachrome 400 ASA color slide film (Kodak).
Immunoblot analysis.
Protein immunoblot analyses were done as previously described (5). Briefly, 106 cells were serum starved for 48 h and released into medium containing the specified drugs. After treatment, the cells were washed in PBS and lysed in a sodium dodecyl sulfate (SDS) lysis buffer containing protease inhibitors. Protein concentrations were determined by a modified Lowry assay (Bio-Rad), and cell lysates were aliquoted and frozen in dry ice-ethanol. Twenty micrograms of each sample was run on sodium dodecyl sulfate (SDS)-polyacrylamide gels (6.5 or 10% polyacrylamide) and blotted onto supported nitrocellulose membranes (Schleicher & Schuell). Blots were probed with primary antibodies to p53 (Ab-6; Santa Cruz Biotechnology), pRb (3C8; Canji, Inc., San Diego, Calif.), or β-actin (Sigma). Proteins were visualized by using horseradish peroxidase-conjugated secondary antibodies (Amersham) and a chemiluminescence detection system (Dupont NEN). Exposed autoradiograms were scanned, and relative amounts of signal were quantified by using NIH Image (National Institutes of Health).
RESULTS
Effects of nucleotide depletion on cell cycle progression in HT1080 cells.
Loss of normal p53 function, or disruption of other elements in the signal transduction pathways in which p53 participates, is required for cells to undergo PALA-selected gene amplification (47, 79, 85). Therefore, we determined the integrity of p53-mediated cell cycle arrest responses in HT1080 cells after treatment with ionizing radiation or PALA. The human normal diploid fibroblast (NDF) strain WS1, which expresses wild-type p53, provided a normal-cell control. Cells were synchronized by serum starvation and then either irradiated or released into medium containing PALA. Cells were pulse-labeled with BrdU 72 h later, allowing determination of the percentage of cells that could enter S phase and continue to proliferate after these treatments.
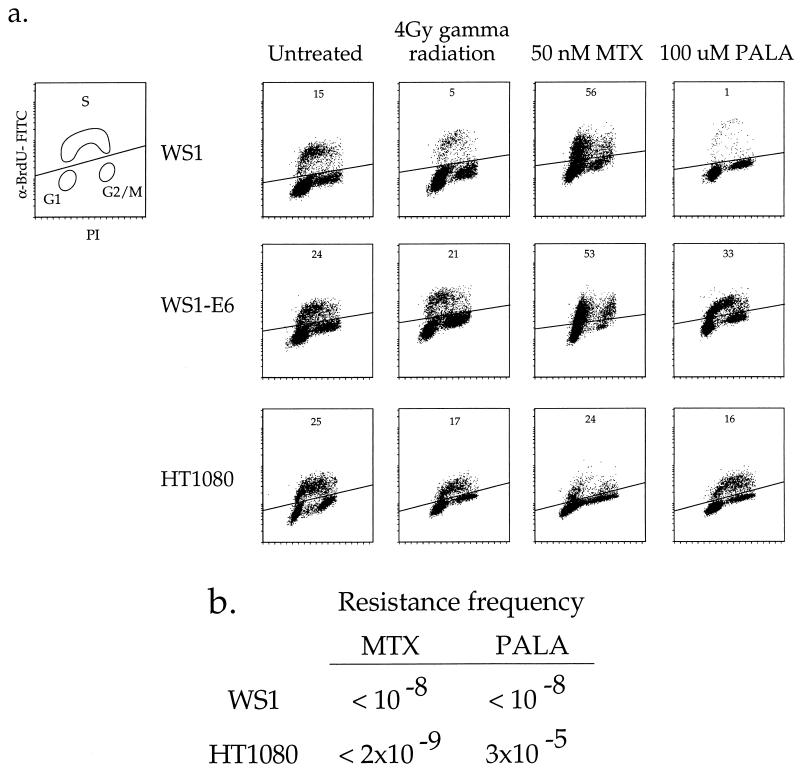
The cell cycle analyses shown in Fig. 1a demonstrate that HT1080 cells do not arrest subsequent to challenge with PALA or gamma radiation, indicating that they have defective p53-mediated ribonucleotide and DNA damage responses. Treatment of WS1 cells with 4 Gy of gamma radiation or 100 μM PALA resulted in significant reductions (67 and 93%, respectively) in the number of BrdU-positive cells compared to the untreated control. In contrast, WS1-E6 cells (in which the p53 protein is degraded by an ubiquitin-dependent pathway [60, 77]) and HT1080 cells display no or only small reductions in the percentage of BrdU-positive cells 72 h after similar radiation and PALA treatments. Continuous labeling of HT1080 cells with BrdU, which allows determination of the extent of progression through multiple cell cycles (45), also showed that HT1080 cells failed to arrest in the first or subsequent cell cycles after treatment with either radiation or PALA (data not shown). These cell growth characteristics are consistent with the presence of a defective p53-dependent signal transduction pathway.
FIG. 1.
Amplification potential and cell cycle response of HT1080 and WS1 cells to radiation and antimetabolite treatments. (a) HT1080 and WS1 cells were synchronized by confluence and serum starvation for 72 h and treated as follows: (i) released into normal growth medium, (ii) irradiated with 4 Gy of gamma radiation and released into growth medium, (iii) released into growth medium containing 100 μM PALA, or (iv) released into medium containing 50 nM MTX. All treatments lasted for 72 h, at which point the cells were treated with 10 μM BrdU for 30 min. The y axis (anti-BrdU–FITC [α-BrdU-FITC]) is on a log scale, the x axis (propidium iodide [PI] level) is on a linear scale, and the diagonal line separates BrdU-positive cells (above the line) from BrdU-negative cells (below the line). The diagram in the upper left shows the positions of cells in different parts of the cell cycle. The numbers at the tops of the plots indicate the percentages of cells that are BrdU positive after each treatment. (b) Amplification frequencies were determined by selecting 3 × 104 to 5 × 106 cells per 15-cm-diameter plate with either PALA (9 times the LD50) or MTX (2.5 to 9 times the LD50). Selections were applied for 2 to 4 weeks, after which resistant colonies were scored by staining with crystal violet. The resistance frequency was determined by dividing the number of resistant colonies by the total number of cells selected.
We next characterized the cell cycle responses of HT1080 and WS1 cells to MTX. Cells were treated with 50 nM MTX (approximately 2.5 times the 50% lethal dose [LD50]) for 72 h and then exposed to a 1-h pulse of BrdU. Treatment of HT1080 or WS1 cells with 50 nM MTX stopped cell cycle progression, as demonstrated by a lack of BrdU-positive cells throughout S phase (Fig. 1a). The discrete accumulation of 2N (G1) and 4N (G2/M or tetraploid G1) populations represents cells that can incorporate BrdU during the pulse-labeling due to the ability of BrdU to transiently overcome the MTX-induced thymidine pool depletion. However, the lack of BrdU-positive cells throughout S phase (compared with untreated controls) indicates that MTX-treated HT1080 and WS1 cells fail to progress through the cell cycle. The facts that the WS1 cells accumulate primarily in G1 and that the HT1080 cells are found in both G1 and G2/M reflect the inefficient synchronization of the HT1080 cells by serum starvation; tumor cell lines rarely synchronize as well as primary strains in response to low serum concentrations (unpublished observations). Similar results were obtained when cells were treated with MTX concentrations of from 50 to 500 nM (2.5 to 25 times the LD50) (data not shown). The failure of HT1080 cells to grow in the presence of MTX is consistent with previous findings showing that MTX induces a p53-independent cell cycle arrest in NDFs (45).
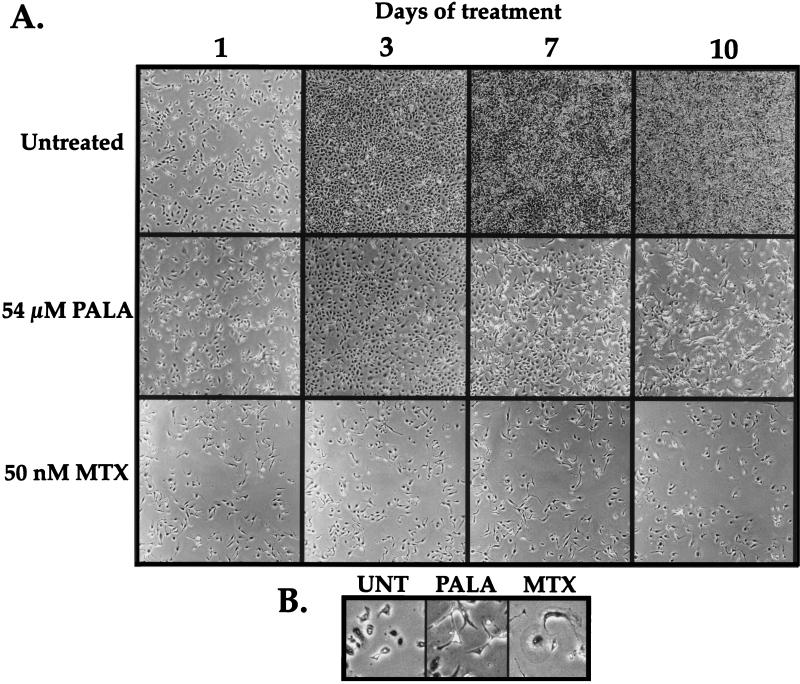
Photographic analysis of cells during normal growth and during treatment with MTX or PALA supports the conclusion that these drugs elicit distinctly different growth responses in HT1080 cells. Untreated cells quickly became confluent, whereas the MTX-treated cells failed to proliferate during this period (Fig. 2A). By contrast, PALA-treated cells initially increased in number, but continued PALA treatment resulted in fewer viable cells, due to cell death induced by S-phase progression in the presence of limiting nucleotide pools. These observations are consistent with the finding of an aberrant p53-mediated arrest response in HT1080 cells.
FIG. 2.
Growth potential and morphology of HT1080 cells treated with antimetabolites. (A) Photographs of HT1080 cells grown under normal conditions or treated with 54 μM PALA or 50 nM MTX. The same field of cells for each treatment was photographed every 24 h for 10 days. (b) Magnified view of cells pictured in panel A, showing differences in cell morphology after the different treatments. Untreated cells at day 1 (UNT) are shown for comparison, as these cells were at the same approximate density as the pictured PALA- and MTX-treated cells at day 10.
Gene amplification is dependent upon selective conditions in HT1080 cells.
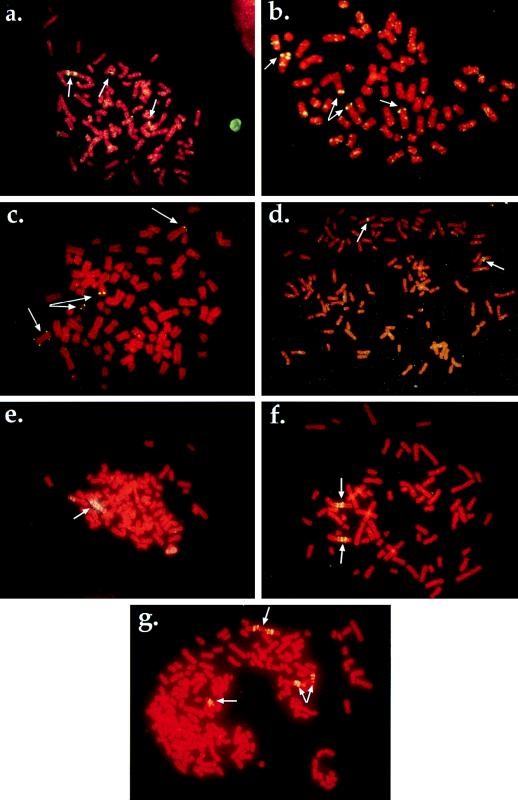
Current models predict that DNA amplification should occur at low rates in the absence of selective pressure in p53-deficient cells (for example, see references 57 and 74). Given the absence of p53-mediated arrest responses in HT1080 cells, they should be able to undergo DNA amplification. To test this hypothesis, we subjected HT1080 cells to PALA or MTX selection and determined the frequencies of drug resistance. Parallel selections with WS1 cells were done to provide rates for normal cells with an intact p53 pathway. HT1080 cells gave rise to PALAr colonies at a frequency of ∼3 × 10−5 (Fig. 1b). FISH analyses of populations and clones of PALAr cells indicated that resistance was due to CAD gene amplification as well as other mechanisms (see Fig. 4a to c). A separate analysis of multiple, independently isolated PALAr HT1080 clones revealed that a significant fraction (36%) were PALAr due to gene amplification, while clones displaying additional copies of a CAD-containing chromosome (38%), isochromosome 2p formation (8%), and other, non-copy-number mediated mechanisms (18%) were also found. The finding that human cells can acquire PALA resistance by mechanisms other than amplification is consistent with previous studies (68). Importantly, the generation of clones containing amplified CAD sequences indicates that HT1080 cells are capable of undergoing the breakage events that initiate and propagate the amplification process.
FIG. 4.
Mechanisms of resistance to PALA and MTX in HT1080 cells. (a) Amplification of the CAD locus in PALAr HT1080 cells. Arrows indicate localization of the CAD locus to two unamplified chromosomal sites and one amplified region. Metaphase was produced from a population of PALAr HT1080 but is representative of the amplification observed in a majority of the cells in the population. (b) Resistance to PALA mediated by aneuploidy. Arrows indicate the presence of four copies of a chromosome containing CAD sequences. (c) Amplification in HT1080 due to chromosome breakage. Arrows indicate that this PALAr clone contains two apparently normal chromosomes 2 containing CAD sequences. Also present are a copy of an isochromosome 2p and a small chromosomal fragment bearing CAD sequences. (d) Untreated HT1080 cell metaphase. Arrows indicate the localization of the DHFR probe to two single chromosomal sites. (e) Localization of the DHFR probe to an expanded chromosomal array. Metaphase was derived from a pool of cells treated with araC for 4 h and resistant to 50 nM MTX. Similar structures were observed in all pools examined (six of six). (f) Multiple copies of the DHFR locus on two different chromosomes in a clone derived from cells treated with 10 μM VP16 and resistant to 100 nM MTX. This metaphase is representative of amplification observed in other independent clones derived from different DNA breakage treatments (six of seven examined). (g) Evidence of ongoing genetic instability. The same cells shown in panel f were subjected to two rounds of stepwise selection, first at 200 nM MTX and then at 400 nM MTX. The metaphase shown was derived from a pool of cells resistant to 400 nM MTX. Arrows indicate regions of amplification, with one site (top) consisting of an inverted duplication that was not present in the initial clone (see panel f). This figure was compiled by using Adobe Photoshop 3.0.5 (Adobe Systems Inc.).
In contrast to the results obtained with PALA, selection of HT1080 cells with MTX generated no resistant colonies from >5 × 108 cells selected. We varied both the MTX concentration (2.5 to 9 times the LD50) and selection density (200 to 6,000 cell/cm2) to ensure that the lack of MTXr HT1080 cells was not due to the stringency of the selection or to the density of the selected cells; however, neither modification of the selective protocol yielded resistant variants. Instead, treatment of HT1080 cells with MTX induced a growth arrest and morphologic changes distinct from those seen in PALA-treated cells, including cellular enlargement and the development of pronounced cytoplasmic extensions (Fig. 2B). MTX-treated HT1080 cells remained growth arrested or died slowly, as 10 to 20% of the cells released from the plate after 4 weeks of drug exposure. These data indicate that neither DHFR gene amplification nor other mutational mechanisms which produce MTX resistance (24, 25, 61) occurred during unchallenged growth at measurable frequencies in these cells. Selection of WS1 cells with either PALA or MTX also failed to generate resistant colonies at a measurable frequency (Fig. 1b), consistent with previous analyses demonstrating a barrier to gene amplification or other mechanisms of resistance in NDFs (73, 82).
HT1080 cells undergo DHFR gene amplification after exposure to conditions that cause DNA breakage.
The data presented above and elsewhere (47, 57, 85) are not consistent with the proposal that p53-deficient cells acquire drug resistance during unperturbed growth. Rather, they are more consistent with the hypothesis that the acquisition of drug resistance requires inappropriate cell cycle progression in the presence of the selective agent. It is known that failure to arrest in the presence of PALA can result in the formation of DSBs (6, 23, 54), which should stimulate the generation of amplified sequences. It is likely that the inability of HT1080 cells to progress through S phase when treated with MTX prevents the formation of DSBs that could initiate the amplification process. If treatment with MTX prevents initiation of amplification, we reasoned it might be possible to overcome this barrier by using alternative breakage strategies prior to selecting for MTX resistance.
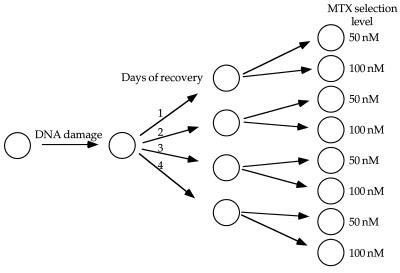
We tested the ability of diverse DNA breakage treatments to initiate the amplification process at the DHFR locus in HT1080 cells. VP16, a topoisomerase II inhibitor; araC, a DNA replication chain terminator; and PALA have all been shown to generate DSBs in cycling cells (12, 20, 38, 55). HT1080 and WS1 cells were treated with these DNA-breakage agents as indicated in Table 1 and Materials and Methods. A recovery period in nonselective medium of 1 to 4 days was utilized to allow cells to accumulate additional copies of the DHFR locus. Cells were then selected with MTX at 2.5 and 5 times the LD50 (50 and 100 nM, respectively) (Fig. 3).
TABLE 1.
MTXr colonies are generated by the DNA damage-recovery-selection strategya
| Cells | Treatment | Viability (%) | MTX concn (nM) | MTXr colonies/plate after the following days of recovery before selection:
|
||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||||
| HT1080 | Untreated | 100 | 50 | 0 | 0 | 0 | 0 | 0 |
| 100 | 0 | 0 | 0 | 0 | 0 | |||
| PALA, 54 μM, 36 h | 36 | 50 | 0 | 3 | 11 | 12 | 15 | |
| 100 | 0 | 0 | 0 | 0 | 0 | |||
| VP16, 2 μM, 1 h | 47 | 50 | 0 | 2 | 4 | >50 | NDb | |
| 100 | 0 | 0 | 0 | 0 | ND | |||
| VP16, 10 μM, 1 h | 13 | 50 | 0 | 1 | 1 | 7 | >50 | |
| 100 | 0 | 0 | 0 | 1 | 0 | |||
| araC, 10 μM, 4 h | 52 | 50 | 0 | 0 | 2 | 15 | ND | |
| 100 | 0 | 0 | 0 | 0 | ND | |||
| araC, 10 μM, 16 h | 17 | 50 | 0 | 0 | 20 | 33 | >50 | |
| 100 | 0 | 0 | 0 | 0 | 0 | |||
| WS1 | All | 7–100 | 50 | 0 | 0 | 0 | 0 | 0 |
| 100 | 0 | 0 | 0 | 0 | 0 | |||
HT1080 and WS1 cells were treated as indicated in Fig. 3. Cell viability was measured, by trypan blue exclusion, as the percentage of cells alive 72 h after treatment compared to that for the untreated control.
ND, not determined.
FIG. 3.
Strategy for the establishment of MTXr HT1080 cells. Cells were plated at 5 × 105 per 15-cm-diameter plate and either left untreated or treated with one of the following: 54 μM PALA for 48 h, 2 μM VP16 for 1 h, 10 μM VP16 for 1 h, 10 μM araC for 4 h, or 10 μM araC for 16 h. After treatment, plates were split 1:4 and cells were allowed to recover in normal medium for 1 to 4 days. After recovery, cells were split 1:2 and selected with either 50 or 100 nM MTX. The selective medium was changed every 3 days.
Pretreatment of HT1080 cells with DNA-damaging agents allowed the recovery of resistant colonies under all treatment conditions, whereas the untreated HT1080 cells, and WS1 cells after all treatments, failed to give rise to resistant colonies (Table 1). Omitting the recovery period in nonselective medium (shown as 0 days of recovery in Table 1) or treating the cells simultaneously with both a DNA-damaging agent and MTX (data not shown) resulted in the MTX-induced growth arrest described earlier and no resistant colonies.
Resistance to MTX can be achieved through mechanisms other than amplification of the DHFR locus, such as mutations affecting drug transport or affinity (24, 25, 61). We performed FISH with a human DHFR probe on metaphase spreads obtained from six independently isolated pools and seven clones of MTXr HT1080 cells to determine the fractions of pools and clones containing amplified DHFR sequences. All six pools and the majority of the clones examined (six of seven) contained amplified DHFR sequences arranged in expanded chromosomal arrays (Fig. 4d to f). Amplification mediated by extrachromosomal elements, such as double minute chromosomes, was not observed.
MTXr HT1080 cells still maintain an MTX-induced growth arrest response.
There were two possible explanations for the ability to generate HT1080 cells containing amplified DHFR genes after treatment with DNA breakage agents. As hypothesized, the induced DNA breakage may have initiated the formation of amplicons containing extra DHFR genes, allowing the cells to survive the MTX selection. Alternatively, treatment with the DNA-damaging agents may have caused mutations that abrogated the MTX-induced growth arrest, allowing the cells to progress through S phase during MTX challenge, which would induce chromosome breakage, as occurs during PALA selection. These possibilities were distinguished by determining if the HT1080 variants containing amplified DHFR genes still underwent a growth arrest in response to higher doses of MTX. We subjected MTXr HT1080 cells to drug concentrations either one or five times the original selective level and monitored their ability to progress through the cell cycle as measured by BrdU incorporation. As expected, all resistant cells cycled when challenged with the MTX concentration used for selection (either 50 or 100 nM MTX, depending on the clone analyzed). However, elevating the concentration to five times the selective level (250 or 500 nM MTX, respectively) resulted in a growth arrest (data not shown). The arrested cells exhibited the same morphologic changes as observed in the MTX-sensitive parental cells. In contrast, PALAr HT1080 cells containing amplified CAD sequences were able to progress through the cell cycle even in the presence of PALA concentrations exceeding five times the level used for selection. These results indicate that the MTX-induced growth arrest response is still intact in the MTXr HT1080 clones and that the generation of variants containing amplified DHFR genes resulted from the clastogenic effects of the drug treatments employed prior to MTX selection.
While increasing the selective concentration of MTX fivefold in the MTXr HT1080 cells caused a growth arrest, a more modest twofold increase in the selective level allowed the isolation of clones resistant to higher MTX concentrations. Clones resistant to two or four times the original selective concentrations were obtained by using stepwise increases in drug concentration. Clones resistant to higher MTX concentrations were isolated and analyzed by FISH to determine the mechanism of resistance. We detected chromosomes containing larger and more complex amplicon structures than those found in cells resistant to lower drug concentrations (Fig. 4f and g). These complex amplicon structures were observed in metaphase spreads from multiple independent clones and are indicative of a mechanism involving chromosome breaks induced by bridge-breakage-fusion cycles (50). The observed increase in amplicon complexity indicates that the initiating event in the amplification process produced genetic instability that was manifested for many succeeding cell generations.
Response of cell cycle control proteins to different growth conditions.
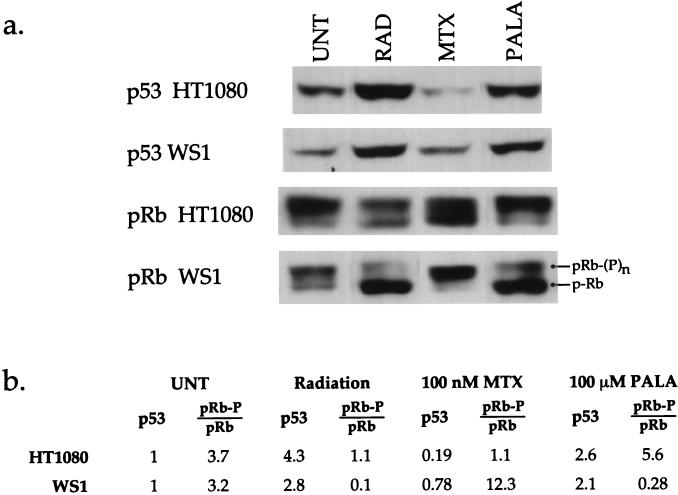
HT1080 cells clearly exhibit divergent responses to antimetabolites, as demonstrated by their growth during PALA treatment and growth arrest with MTX. The former response is typical of a tumor cell with defective p53 cell cycle control pathways, while the latter indicates that some growth controls must still be intact in these cells. This finding raised the question of how G1 cell cycle control proteins would respond following each treatment. We therefore examined the expression patterns of p53 and pRb in HT1080 and WS1 cells by immunoblot analysis after radiation, PALA, or MTX treatment.
HT1080 and WS1 cells were synchronized by serum depletion and then exposed to 4 Gy of gamma radiation or were treated with 100 μM PALA or 100 nM MTX for 72 h. Cell extracts were prepared, and expression levels of p53 and pRb were determined after separation by SDS-polyacrylamide gel electrophoresis and transfer to nitrocellulose. Analysis with a β-actin antibody verified that equal amounts of protein were present in each sample (data not shown). Results are shown for treatment with 100 nM MTX, but equivalent results were obtained after treatment with 50 nM MTX. The p53 levels in WS1 and HT1080 underwent similar increases after treatment with gamma irradiation or PALA. By contrast, MTX treatment resulted in either no change or a slight reduction in p53 levels. The responses of pRb to PALA and radiation correlate well with the observed cell cycle response. WS1 cells, which undergo an arrest after these treatments, exhibited the predominantly hypophosphorylated (nonproliferative) form of pRb, while HT1080 cells, which fail to arrest with PALA, had substantial levels of hyperphosphorylated (proliferative) pRb (Fig. 5). However, WS1 and HT1080 cells show significant differences in pRb expression after MTX treatment. The ratio of hyper- to hypophosphorylated pRb in WS1 cells increases approximately fourfold after treatment with MTX, compared to that in the untreated controls. This trend is reversed in HT1080 cells, with the ratio of hyper- to hypophosphorylated pRb being reduced to almost 1:1. Thus, while HT1080 and WS1 cells have very similar growth arrest responses to MTX treatment, they display very different pRb protein responses. The mechanism by which MTX treatment elicits different pRb phosphorylation patterns yet still induces a growth arrest remains to be elucidated. However, these data emphasize that predictions concerning genetic stability are difficult to make based solely upon changes in expression of cell cycle proteins, a finding consistent with reports for other tumor cell lines (42, 46, 53).
FIG. 5.
Effects of radiation, PALA, and MTX on p53 and pRb expression in HT1080 and WS1 cells. (a) Immunoblots indicating protein levels of p53 and pRb in HT1080 and WS1 cells. All p53 blots are from a single exposure of equivalent amounts of sample run on the same gel, as verified by probing with β-actin (data not shown). pRb blots represent equivalent amounts of sample run on the same gel; however, exposure times varied, due to different absolute amounts of pRb protein. Images which allowed proper quantification of the pRb ratios were selected. Since the ratio of the hyperphosphorylated to hypophosphorylated forms of the protein within each sample was calculated, use of different exposures does not affect the calculated ratio value. UNT, untreated; RAD, radiation. (b) Relative expression levels of p53 and ratios of hyper- to hypophosphorylated pRb after treatments. The p53 levels indicated are corrected for loading differences by comparison with expression of an internal standard (β-actin) (data not shown), and values given are relative to those for the untreated control. The ratios given for pRb represent the ratio of the upper, hyperphosphorylated pRb band to the lower, hypophosphorylated pRb band. This figure was compiled by using Canvas 3.5.4 (Deneba Software, Miami, Fla.).
DISCUSSION
This study addresses the question of whether p53 deficiency is sufficient to generate genomic instability or whether cell cycle progression under suboptimal growth conditions is additionally required. We approached this problem by comparing the growth responses of HT1080 cells to two antiproliferative agents, PALA and MTX, that affect different facets of nucleotide biosynthesis. The data indicate that gene amplification in HT1080 cells is largely an induced rather than a spontaneous process, but they also provide a way of reconciling previous studies suggesting that some cell lines can undergo amplification spontaneously.
HT1080 cells have been used in a number of studies investigating amplification and cell cycle control, but discrepancies concerning the functional status of the p53 pathway in this cell line have emerged (e.g., compare references 10 and 40). It is possible that multiple variants of HT1080 currently exist. We obtained a fresh culture of HT1080 cells from the American Type Culture Collection for these experiments and employed several molecular analyses to characterize the p53 protein. Single-strand conformational polymorphism analysis of p53 exons 5 to 9 failed to reveal any mutations, and treatment with gamma radiation induced expression of both p53 and p21 (data not shown), demonstrating the existence of a wild-type gene and protein in the HT1080 cells employed here. Furthermore, HT1080 does not rereplicate its genome when treated for extended time periods with the mitotic inhibitor colcemid, consistent with the presence of wild-type p53 (references 15 and 21 and unpublished observations). However, the absence of a G1 arrest response after treatment with gamma radiation or PALA and the emergence of variants with CAD gene amplification in response to PALA selection unequivocally demonstrate that downstream components of the p53-mediated DNA breakage and rNTP arrest pathways are not functioning properly.
The ability of MTX to induce a cell cycle arrest in HT1080 cells contrasts with the cell cycle progression observed with PALA and most likely derives from the different metabolic effects of these drugs. PALA inhibits the first steps of de novo pyrimidine biosynthesis (71), reducing levels of all pyrimidine ribo- and deoxyribonucleotides (45, 52), and results in p53 activation (45). By contrast, MTX should primarily inhibit thymidylate biosynthesis and reduce levels of thymidine-based nucleotides at the concentrations employed here (51). It is reasonable to propose that pRb is an important component of the MTX-induced growth arrest, since pRb sequesters the transcriptional activator E2F-1, which is required for regulating the expression of genes necessary for progression through S phase, including DHFR and thymidylate synthase genes (16). Accordingly, pRb+/+ and pRb+/− mouse embryonic fibroblasts (MEFs) express lower levels of DHFR mRNA and protein than isogenic pRb−/− MEFs (2). Similar differences in DHFR expression were obtained for human cells containing either wild-type or defective pRb protein (44). More importantly, MTX induced a G1/S arrest in Rb+/+ MEFs, while Rb−/− MEFs progressed into S phase (2). HT1080 cells have two normal copies of the Rb gene (3). The presence of wild-type pRb may result in reduced expression of DHFR and a reduced ability to overcome the depletion of thymidine-based nucleotides caused by the MTX treatment, ultimately leading to a growth arrest. We would predict that abrogation of pRb function, through binding of human papillomavirus type 16 E7 protein, would eliminate the MTX-induced arrest in HT1080 cells. However, despite numerous attempts, we have been unable to obtain HT1080 cells expressing sufficient E7 to abrogate pRb function (unpublished observations).
We have examined a number of other tumor cell lines in search of those with an aberrant p53 pathway and normal MTX-induced growth arrest, but we have been unsuccessful. Instead, the majority of the tumor cell lines examined displayed defects in multiple aspects of cell cycle control (e.g., failure to arrest in response to both PALA and MTX and continued replication in the presence of colcemid, etc.) (data not shown). This finding is not surprising, since most tumor cell lines have defects in the cell cycle control pathways involving p53 and pRb (see reference 65 for a review). As well, many cell lines contain activated oncogenes whose functions can lead to genomic rearrangements (17, 18, 19, 26, 32). Thus, cell lines such as HT1080 that maintain some aspects of normal checkpoint control and a relatively stable genome appear to be exceptions to the rule. HT1080 may be representative of a rare class of tumor cell that is capable of maintaining a minimally altered genome under normal growth conditions (58) but has a genetic background that allows for the generation of genetic abnormalities in response to growth challenges. As such, it provides a valuable resource for examining factors which initiate the process of genetic instability.
Experiments showing that gene amplification requires abrogation of p53 function were done prior to the finding that PALA can cause a damage-independent cell cycle arrest due to rNTP depletion (45). By utilizing a selective agent other than PALA, we were able to determine that the failure to stop cell cycle progression under suboptimal growth conditions was an important step in the generation of amplified sequences. PALA and MTX can cause DSBs, but only when the treated cells progress through the cell cycle in the presence of the drug (6, 23, 43, 55). In cells lacking the MTX-induced growth arrest, MTX functions both to initiate the amplification process by generating DSBs and to select for those cells with increased DHFR copy number, analogous to the action of PALA in p53-deficient cells. The fact that HT1080 cells have an intact MTX-induced growth arrest allowed us to differentiate between cells that had undergone amplification prior to selection and those that required an inductive event to start the process. The results presented here indicate that simply loss of the p53-mediated breakage sensor is insufficient to allow gene amplification to occur. Rather, cells need to have lost the ability to halt progression through the cell cycle under growth conditions that result in the generation of chromosome breaks.
Our data reveal that a consequence of inducing the amplification process is genomic instability in subsequent cell divisions. Continuing genetic instability has been reported to result from other treatments that induce DNA breakage, such as X irradiation (49) or chromosome destabilization induced by integration of extrachromosomal elements (59), and is consistent with a mechanism of amplification via bridge-breakage-fusion cycles (14, 48, 59, 69). As shown here, a treatment as short as 60 min with a DNA breakage agent (e.g., VP16) was sufficient to produce the unstable chromosomal intermediates that enabled copy number alterations to continue for many cell cycles. This single initiating event produced cells resistant to increasing concentrations of MTX, despite the presence of an intact growth arrest pathway. The perpetuation of genetic instability may have therapeutic implications, as tumor progression and/or resistance to different drugs could arise many cell generations after a single course of therapy with a clastogenic agent.
The results presented here demonstrate that amplification of the DHFR locus, and probably other loci as well, does not occur spontaneously at detectable frequencies in HT1080 cells under nonperturbing growing conditions. DNA amplification can occur by a variety of mechanisms (see reference 81 for a review and references), but whether the molecular substrates that ultimately form amplicons arise spontaneously during unchallenged cell growth or whether an inductive stimulus must first be applied has been heavily debated (4, 35, 57, 62, 74, 76). Luria-Delbruck fluctuation analyses (74) and the detection of gene amplification by flow sorting (35) have been cited as evidence that some cell lines generate amplicons prior to application of a selective agent. In contrast, equally compelling studies show that the antimetabolites used to select for amplified sequences, such as PALA, MTX, and hydroxyurea (9, 30, 57, 72, 75), or clastogenic agents, such as gamma radiation and topoisomerase inhibitors (this report and references 20 and 64), can increase baseline amplification frequencies by up to 104-fold. These studies are consistent with models for amplification in which the induction of DSBs is required for amplicon production (80, 81).
The data from this study suggest a way of reconciling these two apparently contradictory views of amplicon genesis. The spontaneous amplification inferred in some studies may reflect the consequences of earlier events that destabilized the genome, such as those discussed above. Alternatively, mutations that compromise DNA repair or cause DNA breakage due to inappropriate entry into or progression through S phase could lead to a high intrinsic rate of structural chromosome changes. For example, expression of an activated Ha-ras oncogene can lead to inappropriate entry into S phase (70) and chromosomal alterations (18, 19), and overexpression of Mos and activated mitogen-activated protein kinase in p53-deficient cells greatly enhances genomic instability (26). Inactivation of the Rb gene or overexpression of the E2F transcription factor can promote entry into S phase under conditions that could lead to chromosome breakage (1, 34, 83). Thus, cells which undergo frequent chromosomal alterations may be able to generate variants containing amplified DNA in the absence of a selective agent, whereas cells incapable of such spontaneous alterations would require an exposure to an inductive agent for gene amplification to occur.
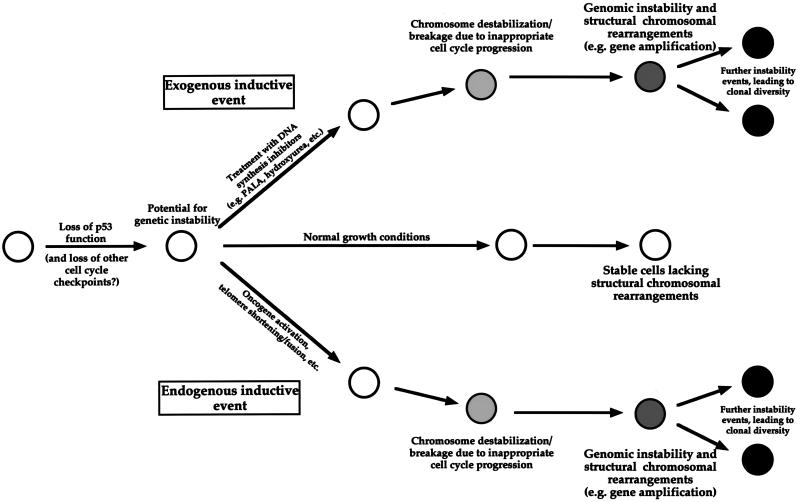
A simplified model, consistent with the data presented here and in the studies cited above, is shown in Fig. 6. Cells that have lost function of the p53 pathway, and possibly other cell cycle checkpoints, have the potential for undergoing structural chromosomal alterations or ploidy changes. Under nonselective growth conditions, such cells rarely, if ever, undergo gross structural chromosomal rearrangements. However, abnormal growth conditions, produced by exogenous agents (such as antimetabolites) or by defects in endogenous growth-regulatory pathways (e.g., the activation of oncogenes), can induce genome destabilization. Lacking the appropriate DNA damage recognition and/or repair checkpoints, cells subjected to the stresses created by such conditions should enter and continue through the cell cycle and undergo chromosome breakage and/or nondisjunction at an increased frequency. These alterations can subsequently generate additional manifestations of genetic instability, including gene amplification, translocations, and deletions, etc. An implication of this model is that the frequent oncogene activation in p53-deficient tumor cells may serve to both induce S-phase entry and produce the genomic destabilization that drives neoplastic progression.
FIG. 6.
Genetic instability and gene amplification depend on the cellular response to aberrant growth conditions.
ACKNOWLEDGMENTS
We thank members of the Wahl lab and Walter Eckhart for insightful discussions and comments during the preparation of the manuscript and Steve Linke for technical assistance.
This work was supported by the National Institutes of Health and the G. Harold and Leila Y. Mathers Charitable Foundation. T.G.P. was supported, in part, by a Public Health Service Genome Training Grant and the H. A. and Mary K. Chapman Charitable Trust.
REFERENCES
- 1.Almasan A, Linke S P, Paulson T G, Huang L, Wahl G M. Genetic instability as a consequence of inappropriate entry into and progression through S-phase. Cancer Metas Rev. 1995;14:59–73. doi: 10.1007/BF00690212. [DOI] [PubMed] [Google Scholar]
- 2.Almasan A, Yin Y, Kelly R, Lee E, Bradley A, Li W, Bertino J, Wahl G. pRb deficiency leads to inappropriate entry into S-phase, activation of E2F responsive genes and apoptosis. Proc Natl Acad Sci USA. 1995;92:5436–5440. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson M J, Fasching C L, Xu H J, Benedict W F, Stanbridge E J. Chromosome 13 transfer provides evidence for regulation of RB1 protein expression. Genes Chromosomes Cancer. 1994;9:251–260. doi: 10.1002/gcc.2870090405. [DOI] [PubMed] [Google Scholar]
- 4.Andrulis I L, Argonza R, Cairney A E. Molecular and genetic characterization of human cell lines resistant to l-asparaginase and albizziin. Somat Cell Mol Genet. 1990;16:59–65. doi: 10.1007/BF01650480. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. One-dimensional gel electrophoresis of proteins, 10.2. In: Jannsen K, editor. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1994. [Google Scholar]
- 6.Ayusawa D, Shimizu K, Koyama H, Takeishi K, Seno T. Accumulation of DNA strand breaks during thymineless death in thymidylate synthase-negative mutants of mouse FM3A cells. J Biol Chem. 1983;258:12448–12454. [PubMed] [Google Scholar]
- 7.Benner S E, Wahl G M, Von H D. Double minute chromosomes and homogeneously staining regions in tumors taken directly from patients versus in human tumor cell lines. Anticancer Drugs. 1991;2:11–25. doi: 10.1097/00001813-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Boveri T (trans.) The origin of malignant tumours. Baltimore, Md: Williams and Wilkins; 1929. [Google Scholar]
- 9.Brown P C, Tlsty T D, Schimke R T. Enhancement of methotrexate resistance and dihydrofolate reductase gene amplification by treatment of mouse 3T6 cells with hydroxyurea. Mol Cell Biol. 1983;3:1097–1107. doi: 10.1128/mcb.3.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Hall I, Lansing T, Gilmer T, Kastan M. Separate pathways for p53 induction by ionizing radiation and N-(Phosphonoacetyl)-l aspartate. Cancer Res. 1996;56:3659–3662. [PubMed] [Google Scholar]
- 11.Chen T R. In situ detection of mycoplasma contamination in cell cultures by fluorescent Hoechst 33258 stain. Exp Cell Res. 1977;104:255–262. doi: 10.1016/0014-4827(77)90089-1. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Goz B, Kirkman L. An analysis of vincristine-resistance in BHK cells pretreated with 1-beta-d-arabinofuranosylcytosine. Anticancer Res. 1993;13:249–255. [PubMed] [Google Scholar]
- 13.Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89:215–225. doi: 10.1016/s0092-8674(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 14.Cowell J K, Miller O J. Occurrence and evolution of homogeneously staining regions may be due to breakage-fusion-bridge cycles following telomere loss. Chromosoma. 1983;88:216–221. doi: 10.1007/BF00285623. [DOI] [PubMed] [Google Scholar]
- 15.Cross S M, Sanchez C A, Morgan C A, Schimke M K, Ramel S, Idzerda R L, Raskind W H, Reid B J. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–6. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 16.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denko N, Stringer J, Wani M, Stambrook P. Mitotic and post mitotic consequences of genomic instability induced by oncogenic Ha-ras. Somat Cell Mol Genet. 1995;21:241–253. doi: 10.1007/BF02255779. [DOI] [PubMed] [Google Scholar]
- 18.Denko N C, Giaccia A J, Stringer J R, Stambrook P J. The human Ha-ras oncogene induces genomic instability in murine fibroblasts within one cell cycle. Proc Natl Acad Sci USA. 1994;91:5124–5128. doi: 10.1073/pnas.91.11.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vries J E, Kornips F H, Marx P, Bosman F T, Geraedts J P, ten Kate J. Transfected c-Ha-ras oncogene enhances karyotypic instability and integrates predominantly in aberrant chromosomes. Cancer Genet Cytogenet. 1993;67:35–43. doi: 10.1016/0165-4608(93)90041-j. [DOI] [PubMed] [Google Scholar]
- 20.Di Leonardo A, Cavolina P, Maddalena A. DNA topoisomerase II inhibition and gene amplification in V79/B7 cells. Mutat Res. 1993;301:177–182. doi: 10.1016/0165-7992(93)90075-7. [DOI] [PubMed] [Google Scholar]
- 21.Di Leonardo A, Khan S H, Linke S P, Greco V, Seidita G, Wahl G M. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 22.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 23.Di Leonardo A, Linke S P, Yin Y, Wahl G M. Cell cycle regulation of gene amplification. Cold Spring Harbor Symp Quant Biol. 1993;58:655–667. doi: 10.1101/sqb.1993.058.01.073. [DOI] [PubMed] [Google Scholar]
- 24.Flintoff W F, Davidson S V, Siminovitch L. Isolation and partial characterization of three methotrexate-resistant phenotypes from Chinese hamster ovary cells. Somat Cell Genet. 1976;2:245–261. doi: 10.1007/BF01538963. [DOI] [PubMed] [Google Scholar]
- 25.Frei E, Rosowsky A, Wright J E, Cucchi C A, Lippke J A, Ervin T J, Jolivet J, Haseltine W A. Development of methotrexate resistance in a human squamous cell carcinoma of the head and neck in culture. Proc Natl Acad Sci USA. 1984;81:2873–2877. doi: 10.1073/pnas.81.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukasawa K, Vande Woude G F. Synergy between the Mos/mitogen-activated protein kinase pathway and loss of p53 function in transformation and chromosome instability. Mol Cell Biol. 1997;17:506–518. doi: 10.1128/mcb.17.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 28.Hahn P J. Molecular biology of double-minute chromosomes. Bioessays. 1993;15:477–484. doi: 10.1002/bies.950150707. [DOI] [PubMed] [Google Scholar]
- 29.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 30.Hill A, Schimke R. Increased gene amplification in L5178Y mouse lymphoma cells with hydroxyurea-induced chromosomal aberrations. Cancer Res. 1985;45:5050–5057. [PubMed] [Google Scholar]
- 31.Huang L-C, Clarkin K C, Wahl G M. Sensitivity and selectivity of the p53-mediated DNA damage sensor. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hundley J E, Koester S K, Troyer D A, Hilsenbeck S G, Subler M A, Windle J J. Increased tumor proliferation and genomic instability without decreased apoptosis in MMTV-ras mice deficient in p53. Mol Cell Biol. 1997;17:723–731. doi: 10.1128/mcb.17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson B E, Battey J, Linnoila I, Becker K L, Makuch R W, Snider R H, Carney D N, Minna J D. Changes in the phenotype of human small cell lung cancer cell lines after transfection and expression of the c-myc proto-oncogene. J Clin Invest. 1986;78:525–32. doi: 10.1172/JCI112604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 35.Johnston R, Beverley S, Schimke R. Rapid spontaneous dihydrofolate reductase gene amplification shown by fluorescence-activated cell sorting. Proc Natl Acad Sci USA. 1983;80:3711–3715. doi: 10.1073/pnas.80.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 37.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kufe, D. W., J. D. Griffin, and D. R. Spriggs. 1985. Cellular and clinical pharmacology of low-dose ara-C. Semin. Oncol. 2(Suppl. 3):200–207. [PubMed]
- 39.Kuo M T, Vyas R C, Jiang L X, Hittelman W N. Chromosome breakage at a major fragile site associated with P-glycoprotein gene amplification in multidrug-resistant CHO cells. Mol Cell Biol. 1994;14:5202–5211. doi: 10.1128/mcb.14.8.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labrecque S, Matlashewski G J. Viability of wild type p53-containing and p53-deficient tumor cells following anticancer treatment: the use of human papillomavirus E6 to target p53. Oncogene. 1995;11:387–392. [PubMed] [Google Scholar]
- 41.Lee J M, Bernstein A. Apoptosis, cancer and the p53 tumour suppressor gene. Cancer Metastasis Rev. 1995;14:149–161. doi: 10.1007/BF00665797. [DOI] [PubMed] [Google Scholar]
- 42.Li C Y, Nagasawa H, Dahlberg W K, Little J B. Diminished capacity for p53 in mediating a radiation-induced G1 arrest in established human tumor cell lines. Oncogene. 1995;11:1885–1892. [PubMed] [Google Scholar]
- 43.Li J C, Kaminskas E. Accumulation of DNA strand breaks and methotrexate cytotoxicity. Proc Natl Acad Sci USA. 1984;81:5694–5698. doi: 10.1073/pnas.81.18.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W, Fan J, Hochhauser D, Banerjee D, Zielinski Z, Almasan A, Yin Y, Kelly R, Wahl G M, Bertino J R. Lack of functional retinoblastoma protein mediates increased resistance to antimetabolites in human sarcoma cell lines. Proc Natl Acad Sci USA. 1995;92:10436–10440. doi: 10.1073/pnas.92.22.10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linke S P, Clarkin K C, Di L A, Tsou A, Wahl G M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 46.Little J B, Nagasawa H, Keng P C, Yu Y, Li C Y. Absence of radiation-induced G1 arrest in two closely related human lymphoblast cell lines that differ in p53 status. J Biol Chem. 1995;270:11033–11036. doi: 10.1074/jbc.270.19.11033. [DOI] [PubMed] [Google Scholar]
- 47.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tisty T D. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 48.Ma C, Martin S, Trask B, Hamlin J L. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 49.Marder B A, Morgan W F. Delayed chromosomal instability induced by DNA damage. Mol Cell Biol. 1993;13:6667–6677. doi: 10.1128/mcb.13.11.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 51.Moran R G, Mulkins M, Heidelberger C. Role of thymidylate synthetase activity in development of methotrexate cytotoxicity. Proc Natl Acad Sci USA. 1979;76:5924–5928. doi: 10.1073/pnas.76.11.5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moyer J D, Smith P A, Levy E J, Handschumacher R E. Kinetics of N-(phosphonacetyl)-l-aspartate and pyrazofurin depletion of pyrimidine ribonucleotide and deoxyribonucleotide pools and their relationship to nucleic acid synthesis in intact and permeabilized cells. Cancer Res. 1982;42:4525–4531. [PubMed] [Google Scholar]
- 53.Nagasawa H, Li C Y, Maki C G, Imrich A C, Little J B. Relationship between radiation-induced G1 phase arrest and p53 function in human tumor cells. Cancer Res. 1995;55:1842–1846. [PubMed] [Google Scholar]
- 54.Nelson W G, Kastan M B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson W G, Kastan M B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 57.Poupon M F, Smith K A, Chernova O B, Gilbert C, Stark G R. Inefficient growth arrest in response to dNTP starvation stimulates gene amplification through bridge-breakage-fusion cycles. Mol Biol Cell. 1996;7:345–354. doi: 10.1091/mbc.7.3.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasheed S. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 59.Ruiz J C, Wahl G M. Chromosomal destabilization during gene amplification. Mol Cell Biol. 1990;10:3056–3066. doi: 10.1128/mcb.10.6.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 61.Schimke R T. Gene amplification in cultured cells. J Biol Chem. 1988;263:5989–5992. [PubMed] [Google Scholar]
- 62.Schimke R T. Gene amplification; what are we learning? Mutat Res. 1992;276:145–149. doi: 10.1016/0165-1110(92)90004-s. [DOI] [PubMed] [Google Scholar]
- 63.Seeger R C, Brodeur G M, Sather H, Dalton A, Siegel S E, Wong K Y, Hammond D. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 64.Sharma R C, Schimke R T. Enhancement of the frequency of methotrexate resistance by G-radiation in Chinese hamster ovary and mouse 3T6 cells. Cancer Res. 1989;49:3861–3866. [PubMed] [Google Scholar]
- 65.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 66.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A, Press M. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 67.Smith K A, Agarwal M L, Chernov M V, Chernova O B, Deguchi Y, Ishizaka Y, Patterson T E, Poupon M F, Stark G R. Regulation and mechanisms of gene amplification. Philos Trans R Soc London B. 1995;347:49–56. doi: 10.1098/rstb.1995.0008. [DOI] [PubMed] [Google Scholar]
- 68.Smith K A, Chernova O B, Groves R P, Stark M B, Martinez J L, Davidson J N, Trent J M, Patterson T E, Agarwal A, Duncan P, Agarwal M L, Stark G R. Multiple mechanisms of N-phosphonacetyl-l-aspartate resistance in human cell lines: carbamyl-P synthetase/aspartate transcarbamylase/dihydro-orotase gene amplification is frequent only when chromosome 2 is rearranged. Proc Natl Acad Sci USA. 1997;94:1816–1821. doi: 10.1073/pnas.94.5.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith K A, Stark M B, Gorman P A, Stark G R. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc Natl Acad Sci USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan, N. F., R. W. Sweet, M. Rosenberg, and J. R. Feramisco. 1986. Microinjection of the ras oncogene protein into nonestablished rat embryo fibroblasts. Cancer Res. 6427–6432 [PubMed]
- 71.Swyryd E A, Seaver S S, Stark G R. N-(Phosphonacetyl)-l-aspartate, a potent transition state analog inhibitor of aspartate transcarbamylase, blocks proliferation of mammalian cells in culture. J Biol Chem. 1974;249:6945–6950. [PubMed] [Google Scholar]
- 72.Tlsty T, Brown P C, Johnston R, Schimke R T. Enhanced frequency of generation of methotrexate resistance and gene amplification in cultured mouse and hamster cell lines. In: Schimke R T, editor. Gene amplification. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 231–238. [Google Scholar]
- 73.Tlsty T D. Normal diploid human and rodent cells lack a detectable frequency of gene amplification. Proc Natl Acad Sci USA. 1990;87:3132–3136. doi: 10.1073/pnas.87.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tlsty T D, Margolin B H, Lum K. Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria-Delbruck fluctuation analysis. Proc Natl Acad Sci USA. 1989;86:9441–9445. doi: 10.1073/pnas.86.23.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varshavsky A. Phorbol ester dramatically increases incidence of methotrexate-resistant mouse cells: possible mechanisms and relevance to tumor promotion. Cell. 1981;25:561–572. doi: 10.1016/0092-8674(81)90074-x. [DOI] [PubMed] [Google Scholar]
- 76.Wahl G, Gaudray P, Carroll S, Proctor N. Is gene amplification inducible? In: Dulbecco R, De Vita V, Santi L, Zardi L, editors. Cancer frontiers, proceedings of the Second International Conference on Progress in Cancer Research. Totowa, N.J: Humana Press; 1986. pp. 71–98. [Google Scholar]
- 77.Werness B A, Levine A J, Howley P M. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 78.Wettergren Y, Kullberg A, Levan G. Drug-specific rearrangements of chromosome 12 in hydroxyurea-resistant mouse SEWA cells: support for chromosomal breakage model of gene amplification. Somat Cell Mol Genet. 1994;20:267–285. doi: 10.1007/BF02254717. [DOI] [PubMed] [Google Scholar]
- 79.White A E, Livanos E M, Tlsty T D. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 80.Windle B, Draper B W, Yin Y X, O’Gorman S, Wahl G M. A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev. 1991;5:160–174. doi: 10.1101/gad.5.2.160. [DOI] [PubMed] [Google Scholar]
- 81.Windle B E, Wahl G M. Molecular dissection of mammalian gene amplification: new mechanistic insights revealed by analyses of very early events. Mutat Res. 1992;276:199–224. doi: 10.1016/0165-1110(92)90009-x. [DOI] [PubMed] [Google Scholar]
- 82.Wright J A, Smith H S, Watt F M, Hancock M C, Hudson D L, Stark G R. DNA amplification is rare in normal human cells. Proc Natl Acad Sci USA. 1990;87:1791–1795. doi: 10.1073/pnas.87.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang J K, Masters J N, Attardi G. Human dihydrofolate reductase gene organization. Extensive conservation of the G + C-rich 5′ non-coding sequence and strong intron size divergence from homologous mammalian genes. J Mol Biol. 1984;176:169–187. doi: 10.1016/0022-2836(84)90419-4. [DOI] [PubMed] [Google Scholar]
- 85.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]