Abstract
The lateral septum (LS) is a midline, subcortical structure, which regulates social behaviors that are frequently impaired in neurodevelopmental disorders including schizophrenia and autism spectrum disorder. Mouse studies have identified neuronal populations within the LS that express a variety of molecular markers, including vasopressin receptor, oxytocin receptor, and corticotropin releasing hormone receptor, that control specific facets of social behavior. Despite its critical role in the regulation of social behavior and notable gene expression patterns, comprehensive molecular profiling of the human LS has not been performed. Here, we conducted single nucleus RNA-sequencing (snRNA-seq) to generate the first transcriptomic profiles of the human LS using postmortem human brain tissue samples from 3 neurotypical donors. Our analysis identified 4 transcriptionally distinct neuronal cell types within the human LS that are enriched for TRPC4, the gene encoding Trp-related protein 4. Differential expression analysis revealed a distinct LS neuronal cell type that is enriched for OPRM1, the gene encoding the μ-opioid receptor. Leveraging recently collected mouse LS snRNA-seq datasets, we also conducted a cross-species analysis. Our results demonstrate that TRPC4 enrichment in the LS is highly conserved between human and mouse, while FREM2, which encodes FRAS1 related extracellular matrix protein 2, is enriched only in the human LS. Together, these results highlight transcriptional heterogeneity of the human LS, and identify robust marker genes for the human LS.
INTRODUCTION
The lateral septum (LS) is a subcortical brain region located along the medial boundaries of the lateral ventricle that controls several aspects of social behavior. Human fMRI studies and clinical case reports demonstrated LS involvement in social cognitive processes such as moral sentiments [1–3], social attachment [4], and sexual behaviors [5], while functional studies in rodents demonstrated that molecularly-defined subpopulations of LS neurons critically regulate social behaviors [6–11] and mediate stress-induced social deficits [12]. To more comprehensively characterize the molecular diversity of these LS cell types, recent single nucleus RNA-sequencing (snRNA-seq) studies in mice identified dozens of unique neuronal subtypes that are both transcriptionally and spatially distinct [13–16]. However, no studies to date have investigated the molecular and transcriptional heterogeneity of the human LS. Understanding the molecular composition of the LS at single nucleus resolution is essential for understanding the neural substrates that control human social behaviors, many of which are impaired in neurodevelopmental and neuropsychiatric disorders, including autism spectrum disorder (ASD) [17–19], schizophrenia [20], and bipolar disorder (BPD) [21,22].
To understand the molecular diversity of the human LS, we generated snRNA-seq data from postmortem human LS tissue (N=3 brain donors). We identified 22 transcriptionally distinct cell clusters with 4 neuronal subtypes originating from the LS. These neuronal clusters exhibited high expression of transient receptor potential cation channel subfamily C member 4 (TRPC4), which has also been identified as an LS marker in mouse snRNA-seq studies [13], suggesting potential conservation of transcriptional markers across species. Cross-species analysis confirmed TRPC4 as a transcriptional marker of the LS in humans and mice, and identified FRAS1 related extracellular matrix 2 (FREM2) as a human-specific marker. Single molecule in situ hybridization (smFISH) studies confirmed FREM2 expression in the LS of an independent donor. This dataset is a resource for understanding molecular heterogeneity within the human LS and highlights additional avenues of research within the field, particularly in understanding the spatial topography of transcriptionally distinct LS neuronal subtypes.
MATERIALS AND METHODS
Postmortem Human Tissue Samples
Postmortem human brain tissue from adult male donors (N=4) of Caucasian, African, or Hispanic ancestry spanning ages 47–62 years were obtained at the time of autopsy following informed consent from legal next of kin, through the Maryland Department of Health IRB protocol #12–24 and from the Department of Pathology at Western Michigan University Homer Stryker MD School of Medicine, under the WCG protocol #20111080. Details regarding tissue acquisition, processing, dissection, clinical characterization, diagnosis, neuropathological examination, RNA extraction, and quality control (QC) measures have been previously published [23]. Using a standardized protocol, all donors were subjected to clinical characterization and diagnosis. Macro- and microscopic neuropathological examinations were performed, and subjects with evidence of obvious neuropathology were excluded. All donors were negative for illicit drugs of abuse. Demographics for donors included in the study are listed in Table S1. To dissect the LS, fresh frozen coronal brain slabs with clearly visible caudate nucleus, putamen, internal capsule, anterior commissure, corpus callosum, fornix, and, anterior nucleus accumbens (MNI: 0.68) were selected. Using a hand-held dental drill, approximately 10 X 20 mm tissue blocks were dissected encompassing the corpus callosum as the dorsal landmark, anterior commissure as the ventral landmark, and fornix as the medial landmark. Tissue blocks were stored in sealed cryogenic bags at −80°C until cryosectioning.
Tissue Processing for Anatomical Validation
Fresh frozen tissue blocks were placed inside the cryostat (Leica CM3050s) at −15°C for 30 minutes prior to cryosectioning. Tissue blocks were mounted on a round chuck with Optimal Temperature Compound (TissueTek Sakura, Cat #4583) for an additional 10–15 minutes. For each donor (N=4), a series of 30 micron tissue sections were discarded until a flat surface was reached. The next 6–8 consecutive 10 microns tissue sections were collected on microscope slides (VWR SuperFrost Microscope Slides, Cat #48311703). These slides were stored in slide boxes inside cryogenic bags at −80°C until ready for single molecule fluorescent in situ hybridization (smFISH) experiments.
Single Molecule Fluorescent in situ Hybridization (smFISH) for Anatomical Validation and Co-Expression Visualization
RNAScope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics [ACD], Cat #323100) and 4-Plex Ancillary Kit (ACD, Cat #323120) were used for smFISH. Tissue sections on microscope slides were fixed in 10% neutral buffered formalin solution (Sigma-Aldrich, Cat #HT501128) for 30 minutes at RT then underwent ethanol dehydration steps with 50%, 70%, and 100% ethanol solutions. Following dehydration, tissue sections underwent permeabilization treatments with hydrogen peroxide for 10 minutes at RT and protease IV for 30 minutes. Tissue sections on microscope slides were incubated for 2 hours at 40°C with commercially available probes. The slides were stored in 4x saline sodium citrate (Quality Biological, Cat #351–003–101) at 4°C for up to 72 hours. Probes were amplified and fluorescently labeled with fluorescent opal dye (Akoya Biosciences [AB]) diluted in TSA Buffer (ACD, Cat #322809) at a 1:500 ratio. For anatomical validation, the following probe and fluorescent opal dye combinations were used: Opal 520 (AB, Cat #FP1487001KT) for MBP (ACD, Cat #411051-C4), Opal 570 (AB, Cat #FP1488001KT) for ELAVL2 (ACD, Cat #540301-C3), Opal 620 (AB, Cat #FP1495001KT) for PPP1R1B (ACD, Cat #477021), and Opal 690 (AB, Cat #FP1497001KT) for TRPC4 (ACD, Cat #1226941-C2). For visualization of OPRM1 expression in the LS, the following probe and fluorescent opal dye combinations were used: Opal 520 (AB, Cat #FP1487001KT) for MBP (ACD, Cat #411051), Opal 570 (AB, Cat #FP1488001KT) for OPRM1 (ACD, Cat #410681-C4), Opal 620 (AB, Cat #FP1495001KT) for GAD1 (ACD, Cat #404031-C3), and Opal 690 (AB, Cat #FP1497001KT) for TRPC4 (ACD, Cat #1226941-C2). For visualization of FREM2 expression in the LS, the following probe and fluorescent opal dye combinations were used: Opal 520 (AB, Cat #FP1487001KT) for FREM2 (ACD, Cat #482841), Opal 570 (AB, Cat #FP1488001KT) for ELAVL2 (ACD, Cat #540301-C3), Opal 620 (AB, Cat #FP1495001KT) for PPP1R1B (ACD, Cat #477021-C4), and Opal 690 (AB, Cat #FP1497001KT) for TRPC4 (ACD, Cat #1226941-C2). After a brief wash step, DAPI staining was applied on each slide for 20 seconds then coverslipped with microscope slide cover glass (Fisher Scientific, Cat #22–050-232) and 80uL Fluoromount-G (ThermoFisher, Cat #00-4958-02). Slides were allowed to dry at RT for 24 hours prior to imaging.
Imaging for Anatomical Validation and Co-Expression Visualization
For anatomical validation, the imaging protocol from Ramnauth et al, 2023 [24] was implemented. Briefly, images were acquired using AX Nikon Ti2-E confocal fluorescence microscope equipped with NIS-Elements (v5.42.02) with 2x magnification (Nikon PLAN APO λ D 2x/0.1 objective) with a pinhole of 1.0 AU and the following laser power (LP) and gain (G) settings: DAPI: 27.00LP/18.00G, Opal 520: 28.29LP/25.00G, Opal 570: 25.00LP/20.00G, Opal 620: 25.00LP/20.00G, Opal 690: 25.00LP/25.00G, Lipofuscin: 28.29LP/25.00G. For co-expression validations, the following LP and G settings were used: 20x magnification (Nikon PLAN APO λ 20x/0.80 objective) with a pinhole of 1.0 AU and the following LP/G settings: DAPI: 5.00LP/2.00G, Opal 520: 5.00LP/1.00G, Opal 570: 5.00LP/3.00G, Opal 620: 5.00LP/3.00G, Opal 690: 5.00LP/5.00G, Lipofuscin: 7.00LP/5.00G.
snRNA-seq Data Generation
After anatomical validation, the 3 out of 4 fresh frozen tissue block samples containing the broadest extent of the LS were selected for snRNA-seq data generation. For each donor (N=3), 1–2 sections of 10 micron tissue were discarded to reach a flat surface, then the adjacent 3 sections of 10 micron tissue were collected on microscope slides (VWR SuperFrost Microscope Slides, Cat #48311703). Slides were stored in a slide box in −80°C. Prior to sample collection, each block was scored using a Single Edge Razor Blade (ULINE, Cat #H-595B) to minimize contamination from neighboring brain regions. The adjacent 3–5 sections of 100 micron tissue sections were collected in a pre-chilled 2mL microcentrifuge tube (Eppendorf Protein LoBind Tube, Cat #22431102) to total a volume of approximately 40 mg for each donor. Collected samples were stored in −80°C until ready for nuclei sorting. The next 3 sections of 10 micron tissue sections were collected on microscope slides then stored in a slide box in −80°C until ready for validation experiments using smFISH.
Nuclei from each donors9 tissue samples were extracted as previously described [13]. Nuclei EZ Lysis Buffer (MilliporeSigna, Cat #NUC101) was chilled on wet ice then added to each 20mL PCR tube containing frozen tissue. Each tissue sample was homogenized using separate pairs of glass douncers and pestles per donor then filtered through 70μm mesh filter on separate 50 mL conical tubes. Filtrate was centrifuged at 500 rcf for 5 minutes at 4°C. Resulting pellets were washed with a total volume of 3 mL EZ Lysis Buffer. Final pellet was washed 3 times with wash buffer solution (1x PBS, 1% BSA, and 0.2U/uL RNAse Inhibitor). Nuclei underwent a 30 minute incubation on wet ice with Alexa Fluor 488-conjugated anti-NeuN-AF488 (Millipore Sigma, #MAB377X) diluted in dilution wash buffer at a 1:1000 ratio. Nuclei were labeled with propidium iodide (PI) diluted in a staining buffer solution (1x PBS, 3% BSA, and 0.2U/uL RNase inhibitor) at a 1:500 ratio. Samples were filtered through 35μm filter top FACS tubes then underwent fluorescent activated nuclear sorting (FANS) on Bio-Rad S3e Cell Sorter at the Lieber Institute for Brain Development. Gating criteria were set to select for whole, singlet nuclei by forward/side scatter then G0G1 nuclei by PI fluorescence and neuronal nuclei by Alexa Fluor 488 fluorescence. For each donor, approximately 9000 nuclei were collected with 2:1 enrichment for neuronal nuclei into tubes each containing 23.1uL solution from 10x Genomics Single Cell 39 Reagent kit (without reverse transcriptase). Reverse Transcription enzyme and water were added. cDNA and libraries were prepared according to the instructions provided by the manufacturer (10x Genomics Chromium Next GEM Single Cell 3’ Reagent Kits v3.1 Dual Index, CG000315 RevE) then sequenced on the Illumina NovaSeq6000 at the Johns Hopkins University Single Cell & Transcriptomics Core.
snRNA-seq Raw Data Processing
FASTQ files for snRNA-seq sample libraries from 3 donors were aligned to the human genome (GRCh38/Hg38, Ensembl release 98), using 10x Genomics9 software [25], cellranger count (version 7.2.0). Raw feature-barcode files were analyzed in R v4.3.1, using the Bioconductor suite [26,27] of single-cell analytical packages version 3.17. Droplets containing nuclei were identified using the emptyDrops() function from the DropletUtils [28],[29] package v1.20.0 using a data-driven lower threshold. Next, high quality nuclei were identified as those with less than 5% of reads mapping to the mitochondrial genome. Furthermore, following mitochondrial mapping rate thresholding, sample specific median absolute deviation (MAD) thresholds were identified for the number of UMI counts and detected genes per cell. The median value of the number of UMI counts and detected genes per cell for one particular sample (“Sample 1”) was much higher than the other two samples, and thus using an adaptive threshold for both UMI counts and detected features would have removed several hundred high quality nuclei. Therefore, only a UMI count threshold and mitochondrial mapping rate was used to remove low quality quality nuclei from Sample 1. In addition to thresholds for mitochondrial rate mapping, total UMI counts, and detected features, preliminary dimensionality reduction and clustering identified a cluster of low quality neuronal nuclei. Particularly, nuclei within this cluster exhibited expression of neurons, such as GAD1, SYT1, and SNAP25, but had abnormally low number of UMIs and detected genes. These nuclei were removed from further analysis. Following removal of low quality nuclei, sample specific doublet scores were calculated using the computeDoubletDensity() from the scDblFinder [30] v1.14.0 to identify any doublet driven clusters. These QC measures resulted in a final total of 9,225 nuclei with an average UMI count of ~23,027 per nucleus and an average number of detected features per nucleus of ~5,067.
Feature Selection, Dimensionality Reduction, and Clustering
Feature selection was conducted by first calculating binomial deviance residuals. The top 2000 highly deviant genes (HDGs) were identified by ranking each gene according to its total deviance. Principal component analysis (PCA) was then performed within the 2000 HDG feature space [31]. To avoid batch effects driven by donor/sample, we performed batch correction with mutual nearest neighbors within the reduced principal component space using the reducedMNN() function within the batchelor [32] package v1.16.0. Graph based clustering was then performed within the mutual nearest neighbors, PC-reduced space using k=20 nearest neighbors and the Louvain method for community detection [33], yielding 24 preliminary clusters. To visually inspect clusters, we generated a t-distributed Stochastic Neighbor Embedding (t-SNE) [34] using the top 50 mutual nearest neighbors-corrected PCs. Cluster identity was determined using previously identified transcriptional markers of cell types within the striatum [35–37], medial septum [14], and lateral septum [38]. Initial clustering identified 3 oligodendrocyte populations that were merged to generate a single oligodendrocyte cluster. Log2-normalized counts were generated using the multiBatchNorm()function provided by the batchelor package [32] v1.16.0 with the donor or sample representing the batch.
Identification of Cluster Specific Marker Genes
Cluster specific marker genes were identified using an approach previously described with minor modifications. Briefly, log2-normalized counts were used to calculate two sets of statistics. The first set implemented the findMarkers() function provided by the scran [39] package v1.28.2 to perform pairwise t-tests and identify differences between each cluster. The second set utilized the findMarkers_1vAll() function provided by the DeconvoBuddies [40] package v0.99.0 to perform cluster-versus-all-other-nuclei testing. t-statistics were calculated for each gene per cluster as previously described [36].
Comparison of Transcriptionally Distinct Cell Populations Across Human and Mice
To begin comparing transcriptomic profiles of transcriptionally distinct cell populations across human and mice, t-statistics were calculated per gene per cluster as previously described [36]. Shared homologs were then identified using JAX lab homology report (http://www.informatics.jax.org/downloads/reports/HOM_AllOrganism.rpt) and the “DB.Class.Key” identifier. For initial cross-species analysis focused on the correlation of every human and mouse cluster, we used the 885 shared homologs of the top 100 marker genes for each human or mouse cluster. All 16,562 shared homologs between human and mouse were used for the identification of conserved and divergent marker genes of LS neurons. Pearson9s correlation coefficient was then calculated for t-statistics of shared homologs using the cor() function provided by base R.
Human LS cell types were compared to the 10x genomics whole mouse brain taxonomy using MapMyCells [41], an interactive online tool provided by the NIH9s Brain Initiative Cell Census Network and the Allen Institute. First, human-mouse orthologs were identified using the conversion table provided by Allen Institute’s GeneOrthology package. Raw count matrices including all orthologs were uploaded to MapMyCells and the hierarchical correlation mapping algorithm was used.
RESULTS
Characterization of transcriptionally distinct cell types within the human lateral septum
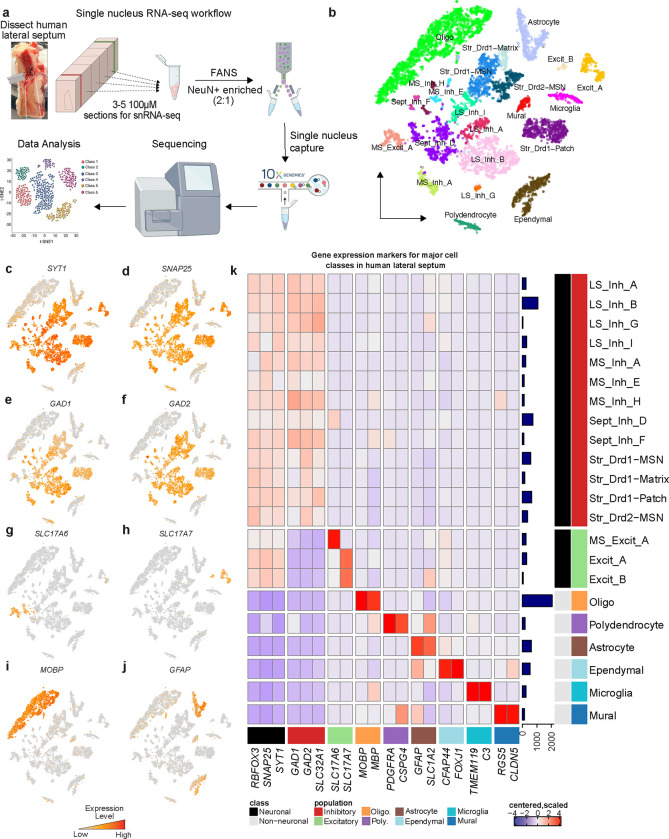
The LS is medial and dorsal to the medial septum (MS) and lateral to the caudate nucleus (CN). These regions are transcriptionally and functionally distinct from the LS and vary along the anterior-posterior axis. To ensure that dissections were enriched for LS rather than the neighboring regions, we performed RNAscope smFISH with probes for TRPC4 to identify the LS, ELAV like RNA binding protein 2 (ELAVL2) to identify the medial septum, protein phosphatase 1 regulatory inhibitor subunit 1B (PPP1R1B) to identify the striatum, and myelin basic protein (MBP) to identify surrounding white matter (Fig. S1), based on regional markers identified in the mouse [36,38]. Following anatomical validation, we captured single nuclei from the LS of 3 human brain donors using the 10x Genomics Chromium platform (Fig. 1a). Following quality control (see STAR Methods), 9,225 nuclei were divided into 22 transcriptionally distinct cell types (Fig. 1b, Table S2, Fig. S2). These clusters included 16 neuronal cell types that exhibit high expression of pan-neuronal marker genes including synaptotagmin 1 (SYT1), synaptosome associated protein 25 (SNAP25), and RNA binding fox-1 homolog 3 (RBFOX3/NeuN) (Fig. 1c,d,k). We also identified 6 non-neuronal clusters including oligodendrocytes, marked by myelin associated oligodendrocyte basic protein (MOBP) expression (Fig. 1e,k) and astrocytes, marked by glial fibrillary acidic protein (GFAP) expression (Fig. 1f,k). The LS is predominantly inhibitory, containing GABAergic neurons that are marked by glutamate decarboxylase 1 (GAD1) and glutamate decarboxylase 2 (GAD2), genes encoding enzymes critical for the synthesis of gamma-aminobutyric acid (GABA) (Fig. 1g,h,k). Several distinct clusters exhibited expression of both solute carrier family 17 member 6 (SLC17A6/VGLUT2) and solute carrier family 17 member 7 (SLC17A7/VGLUT1) (Fig. 1i–k). The MS contains glutamatergic neurons suggesting that these cells may not have originated from the LS, but rather a neighboring brain region.
Figure 1.
snRNA-seq of human lateral septum identifies 22 transcriptionally distinct cell populations. a, Description of workflow to dissect human lateral septum, dissociate nuclei, and perform droplet based single nucleus capture. b, t-distributed Stochastic Neighbor Embedding of 9,225 nuclei from human lateral septum colored by cluster identitiy. Feature plots demonstrating distribution of expression across cell types for c, SYT1, d, SNAP25, e, GAD1, f, GAD2, g, SLC17A6, h, SLC17A7, i, MOBP, and j, GFAP . k, Heatmap of expression values for general cell class marker genes used to determine cluster identity. Color of square corresponds to centered and scaled expression values (logcounts).
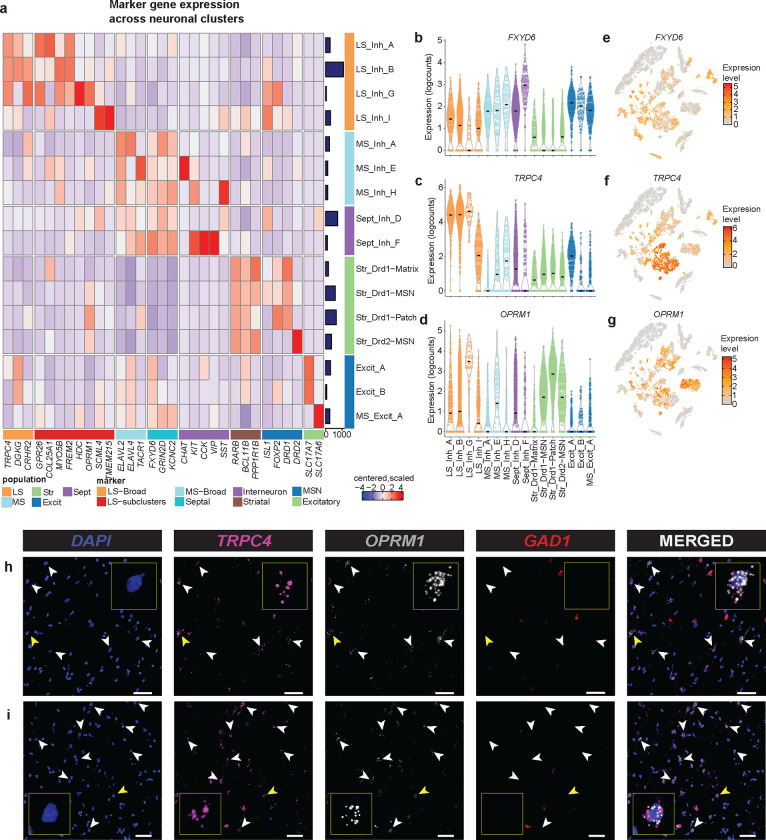
To determine the brain region of origin for each cluster, we used established marker genes (Fig. 2, Fig. S3). First, clusters representing septal nuclei were identified by expression of FXYD domain containing ion transport regulator (FXYD6; Fig. 2a,b,e), a gene that is abundantly expressed in mouse in both MS and LS [14]. Although this gene is detected in all neuronal clusters, it is most highly expressed in nuclei derived from the MS and LS (Fig. 2a,b,e). To delineate between the LS and MS, we queried expression of TRPC4, diacylglycerol kinase gamma (DGKG), and corticotropin releasing hormone receptor 2 (CRHR2), LS marker genes that were identified in rodent studies (Fig. 2a,c,f) [8,13,14,42]. Four clusters exhibited relatively high expression of these genes (LS_Inh_A, LS_Inh_B, LS_Inh_G, LS_Inh_I) and were classified as LS neuronal subtypes. MS neuronal subtypes (MS_Inh_A, MS_Inh_E, MS_Inh_H, MS_Excit_A) were identified using ELAVL2 (Fig. 2a). MS_Excit_A exhibits high expression of SLC17A6 (Fig. 2a). SLC17A6 marks a discrete population of excitatory MS neurons that is important for arousal and wakefulness in mouse models [43]. We also used a panel of molecular markers (retinoic acid receptor beta (RARB), BCL11 transcription factor B (BCL11B), PPP1R1B) to identify nuclei originating from the CN, or striatum (Fig. 2b, Fig. S3). Additionally, striatal neurons are both spatially and transcriptionally distinct [35–37,44–46]. Our dataset contains both DRD1-expressing patch and matrix cells as denoted by expression of opioid receptor mu 1 (OPRM1), crystallin mu (CRYM), and semaphorin 5B (SEMA5B; Fig. 2a, Fig. S3).
Figure 2.
Identification of novel marker genes of human lateral septum (LS). a, Heatmap of expression values for neuronal clusters within the human LS, medial septum (MS), and striatum (Str). Violin plots of expression values for b, FXYD6, c, TRPC4, and d, OPRM1. Feature plots demonstrating distribution of expression values for e, FXYD6, f, TRPC4, and g, OPRM1. h, Representative smFISH images demonstrating the coexpression of TRPC4 and OPRM1 transcripts in tissue section collected from the anterior end and i, posterior end of tissue sections collected for snRNA-seq data generation. White bar indicates 50μm.
Identification of transcriptionally distinct neuronal subtypes within the human lateral septum
We next investigated LS neuronal subtype heterogeneity by identifying genes that are preferentially enriched within each subtype using a cluster-versus-all-other-clusters approach (Fig. 2a, Table S3). While this analysis will identify genes that are most abundant within each cluster, it will not specifically identify selective markers. For example, FREM2 is highly expressed within the LS_Inh_B cluster, and is identified as a marker gene for this cluster. However, this gene is widely expressed in most LS clusters originating from the anterior and middle LS (LS_Inh_A, LS_Inh_B, LS_Inh_G), and may represent an additional marker of the human LS (Fig. 2a). G protein-coupled receptor 26 (GPR26), which encodes an orphan GPCR that is important for satiety and feeding behaviors [47], is preferentially enriched within LS_Inh_A, while transmembrane protein 215 (TMEM215) and Scm polycomb group protein like 4 (SCML4) were identified as marker genes for LS_Inh_I. Cells within the LS_Inh_I cluster do not express canonical LS marker genes to the same extent as other LS clusters. However, cells within this cluster are not significantly enriched for any MS or CN markers (Fig. 2a, Fig. S3). Enrichment of LS marker genes and depletion of striatal and MS marker genes led us to classify LS_Inh_I as an LS neuronal subtype.
The μ-opioid receptor is expressed in several transcriptionally distinct neuronal subtypes in rodent LS [16], and regulates opioid-dependent behaviors that model specific aspects of human drug addiction [48,49]. We observe distributed expression of OPRM1 across LS neuronal subtypes with preferential enrichment in the LS_Inh_G cluster (Fig. 2d,g). We validated OPRM1 expression in the human LS using smFISH. We multiplexed probes for TRPC4, GAD1, and OPRM1 in tissue sections originating from the anterior and posterior portions of the LS, and found co-expression of TRPC4 and OPRM1, confirming OPRM1 expression in human LS neurons (Fig. 2h–i). OPRM1-expressing neurons do not appear to be spatially organized as OPRM1 punctae were identified within both anterior and posterior tissue sections. Surprisingly, cells expressing TRPC4 and OPRM1 did not express GAD1 (Fig. 2h–i).
Transcriptional signatures of discrete LS neuronal populations are conserved across mice and humans
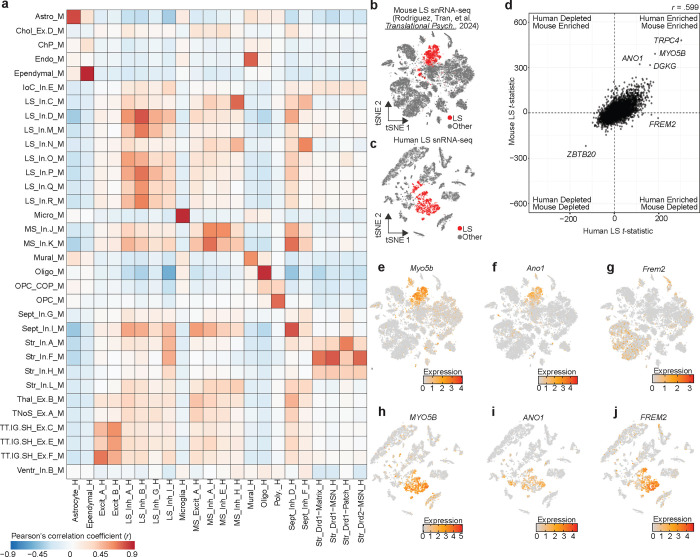
We next investigated conservation of transcriptional signatures of LS cell types across human and mouse using two independent datasets. First, we utilized MapMyCells [41], an online application that allows users to compare their data to high quality, large-scale single cell sequencing datasets. Using the 10x Genomics whole mouse brain taxonomy dataset [41], we calculated the percentage of human cells from each cluster that map onto each broad mouse cell class. Almost 100% of each non-neuronal population map onto the same non-neuronal population in mouse (Fig. S4). A similar trend was observed with neurons originating from the LS_Inh_A, LS_Inh_B, and LS_Inh_G populations mapping to GABAergic LS mouse neurons (Fig. S4a,b). LS_Inh_I, a population of human LS neurons marked by the expression of TRPC4 and the absence of many broad striatal markers, mapped to GABAergic populations in the mouse striatum (Fig. S4a–c). We were particularly interested in identifying LS cell types and marker genes that are similar between human and mouse. Using a recently published transcriptional atlas of the mouse LS [13], we calculated t-statistics for each gene per cluster within each dataset and identified shared homologs for the top marker genes of each cluster. Then, we calculated the pearson9s correlation coefficient (r) by correlating each cluster9s t-statistics across species. Transcriptional signatures for non-neuronal populations such as microglia, oligodendrocytes, and ependymal cells are highly correlated between human and mouse (Fig. 3a). The Excit_A and Excit_B neuronal clusters are also highly correlated with mouse clusters originating from the tenia tecta, indusium griseum, and septohippocampal nucleus (Fig. 3a). This correlation is primarily driven by the presence of SLC17A7 transcripts (Fig. 1k).
Figure 3.
Identification of conserved transcriptional markers for lateral septum (LS) neurons. a, Heatmap of Pearson s correlation coefficients calculated by correlating the t-statistics of the top 100 marker genes from each cluster that has a homologous partner in either species. b-c, tSNE of mouse and human snRNA-seq datasets containing LS neurons. Cells are colored by LS neuronal identity. d, t-statistics were calculated for all LS neurons in mouse or human and correlated to identify conserved transcriptional markers. e-g,Feature plots demonstrating the distribution of expression of Myo5b, Ano1, and Frem2 in mouse snRNA-seq dataset containing the LS. h-j, Feature plots demonstrating distribution of expression of MYO5B, ANO1, and FREM2 in human snRNA-seq dataset containing the LS.
Interestingly, no particular LS neuronal cluster exhibited a high degree of conservation across species. Instead, LS neuronal clusters exhibited broad conservation, (Fig. 3a) suggesting that primary markers of LS neurons are conserved while distinct marker genes for discrete subtypes may exhibit divergence across species. To identify both conserved and divergent marker genes for LS neurons across the human and mouse, we first collapsed all neuronal LS clusters within each species (Fig. 3b,c), and then recalculated t-statistics for all 16,562 shared homologs between human and mouse. Overall, gene enrichment within mouse and human neuronal LS clusters are correlated (r = .599, p < 2.2×10−16, Fig. 3d). As expected, TRPC4 and DGKG were enriched in neuronal LS clusters in both the human and mouse (Fig. 3d). We also identified ANO1 and MYO5B as strong markers of the LS in both humans and mice (Fig. 3d–i). Additionally, this analysis identified that FREM2, a marker gene for LS_Inh_B, was specifically enriched only within neuronal LS clusters from the human, and represents a potentially species-divergent LS marker gene (Fig. 3g–j).
In situ validation of FREM2 as a marker gene for human LS neurons
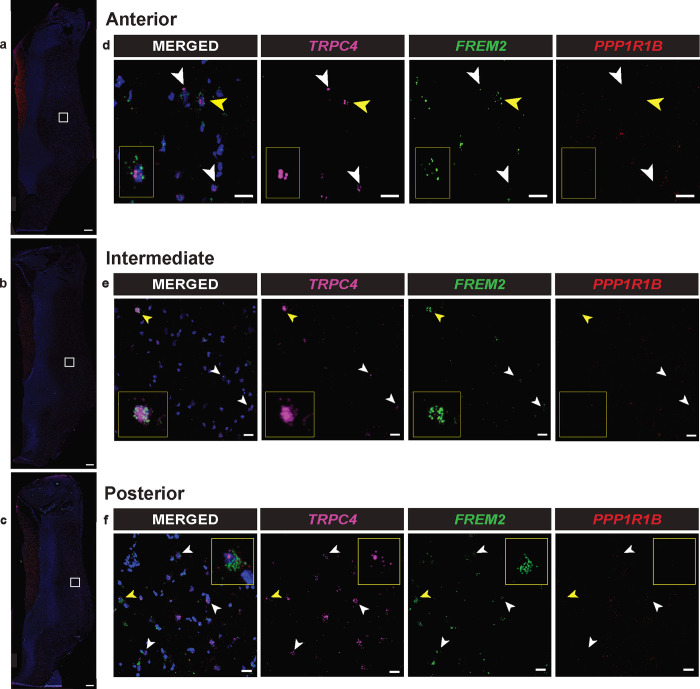
Identification of additional markers of the human LS is important as there are no distinct anatomical landmarks that distinguish the LS from surrounding brain regions. Thus, anatomical validation of tissue sections containing the LS relies on targeted amplification of biomolecules, such as mRNA transcripts, to ensure accurate dissection. Our cross-species analysis identified FREM2 as a selective marker gene for human LS neurons (Fig. 3d,j). We next sought to validate this finding in situ by probing for FREM2. Because gene expression is spatially organized within the LS [42], we chose to perform smFISH with probes for TRPC4, FREM2, and PPP1R1B on sections that span the anterior-posterior axis. PPP1R1B was included as it marks medium spiny neurons (MSNs) within the striatum [36,37]. We hypothesized that FREM2 would localize to the LS and be co-expressed with TRPC4. smFISH experiments were performed on LS tissue from a donor that was not used for sequencing studies, increasing confidence that these results are generalizable across individual donors. FREM2 is highly expressed across the anterior-posterior axis of the LS, confirming it as a transcriptional marker of the LS (Fig. 4a–c). A subset of neurons within the LS do co-express TRPC4 and FREM2 (Fig. 4a–c). However, there are a number of cells within the LS that only express one of these genes. This is expected as TRPC4 was highly expressed in all neuronal clusters originating from the LS (Fig. 2c,f), while FREM2 was primarily enriched in LS_Inh_A and LS_Inh_B (Fig. 3j), but was comparatively lower than TRPC4. The difference in enrichment is further demonstrated by the t-statistic calculations as the t-statistic is higher for TRPC4 than FREM2 (Fig. 3d).
Figure 4.
In situ validation of FREM2 expression in the lateral septum (LS). a, Representative 2x magnification images taken using an anterior (z=0μm), b, an intermediate (z+650μm), and c, a posterior (z+1210μm) tissue section from a postmortem tissue sample containing the LS from the right hemisphere. Notably, the septal regions from the right and left hemispheres are merged in this donor. All three images underwent gamma manipulation in ImageJ to clarify the boundaries of each tissue sample. White bars indicate 1000μm. d, 40x magnification images of the area demarcated by white squares in 2x images from anterior (z=0μm), e, intermediate (z+650μm), and f, posterior (z+1210μm) tissue sections. White bar indicates 20μm.
DISCUSSION
The LS regulates evolutionarily conserved behaviors that are frequently dysregulated in neuropsychiatric and neurodevelopmental disorders. Despite its essential function in regulating social and affective behaviors, the molecular and transcriptional heterogeneity of cell types within the LS has only recently been addressed. snRNA-seq studies of the mouse LS identified dozens of transcriptionally distinct neuronal subtypes that are implicated in social and affective behaviors [13–16]. For example, our recent snRNA-seq study of the mouse LS demonstrated that transcriptionally distinct neuronal subtypes are differentially regulated by TrkB signaling [13], providing insight about cell type-specific function of BDNF-TrkB in the LS in social recognition. Additionally, multimodal studies have identified electrophysiological properties specific to transcriptionally distinct neuronal subtypes across the anterior-posterior extent of the mouse LS [15]. While molecular and cellular properties of LS neurons can be functionally interrogated in rodent models, a major limitation is the ability to relate these findings to analogous populations of neurons within the human brain.
To address this limitation, we used snRNA-seq to generate the first comprehensive molecular profiles of human LS cell types. Our study identified 16 transcriptionally distinct neuronal subtypes, including 4 that are LS specific (Fig. 1b). The human LS is surrounded by two GABAergic brain regions that exhibit high levels of expression of GAD1 and SLC32A1, the gene encoding the vesicular GABA transporter. Thus, accurate identification of human LS populations cannot rely on the presence of GABAergic markers alone. Hence for our dissections, we primarily relied on expression of TRPC4, a gene that was identified as a marker of the mouse LS [13]. smFISH studies confirmed that human LS neurons exhibited high expression of TRPC4 (Fig. S1). The snRNA-seq data also confirmed the presence of this gene within LS (Fig. 2a,c,f). In addition to LS-specific marker genes, we also found that transcriptional markers of the broad septal complex in the mouse, FXYD6 and DGKG, were enriched within neuronal clusters originating from human LS and MS (Fig. 2a,b,e). Interestingly, DGKG was specifically enriched within human LS neuronal clusters (Fig. 2a). The enrichment of mouse LS marker genes in human LS populations suggest that region-specific transcriptional markers may be conserved across species.
To investigate cross-species enrichment of marker genes, we first compared t-statistics on a per cluster basis for human and mouse clusters. Correlation of t-statistics between clusters demonstrated that human and mouse LS neuronal clusters are broadly conserved, without any specific human cluster mapping to one mouse cluster (Fig. 3a). This is in contrast to glial populations which exhibit high correlation coefficients across the two species (Fig. 3a). Broad conservation suggests that marker genes of the entire LS, instead of the markers of discrete neuronal subtypes, are conserved across species. We next correlated LS-specific t-statistics between human and mouse. This analysis found that TRPC4 was highly conserved, confirming the findings from smFISH studies in both human (Fig. S1) and mouse [13]. FREM2 was identified as a human-specific LS marker gene and validated via smFISH results (Fig. 3d,j & Fig. 4a–c). To our knowledge, no other studies have identified FREM2 as a marker gene of the LS.
Our dataset includes 4 transcriptionally distinct LS neuronal subtypes (LS_Inh_A, LS_Inh_B, LS_Inh_G, LS_Inh_I). All neuronal subtypes exhibit expression of TRPC4, DGKG, and FREM2, but are also enriched for cluster-specific genes (Fig. 2a). OPRM1, the gene encoding the μ-opioid receptor, was expressed in multiple LS clusters, but primarily enriched within the LS_Inh_G cluster (Fig. 2a,d). This finding is in agreement with recent snRNA-seq of the mouse LS that found Oprm1 to be expressed in several LS neuronal subtypes [16]. Opioid-related deaths continue to climb, thus identifying the neuronal subtypes that may regulate opioid addiction-related behaviors is critical for understanding how drugs impact the brain and the development of future therapeutics. Corticotropin releasing hormone (CRH) signaling within the LS is critically important for social behaviors in mice [8]. Interestingly, LS_Inh_G is also significantly enriched for CRHR2 (Fig. 2a & Fig. S3), a gene encoding a receptor for CRH signaling. LS_Inh_I was enriched for TRPC4, but also exhibited expression of transcriptional markers of striatal MSNs BCL11B, PPP1R1B, and ISL1 [36,37,46] (Fig. 2a). A large portion of the neurons in LS_Inh_I originates from a single donor in which the tissue punches were taken from a more posterior portion of the LS, suggesting that this cluster may represent a cell type that marks the transition from LS to striatum (Table S2, Fig. S1). The presence of a transcriptionally distinct LS neuronal subtype originating from a specific position along the rostral-caudal axis suggests that some clusters may be both transcriptionally and spatially distinct, similar to observations in mouse. It is important to note that this dataset does not contain neurotensin neurons. Within the mouse, neurotensin neurons regulate social approach behavior [12] and are modulated by opioid withdrawal [16]. However, the absence of neurotensin neurons may be due to technical issues with capture of NTS mRNA. A recently published snRNA-seq study of the mouse LS from our group utilized the same 39 mRNA capture technology used for this study and likewise failed to identify neurotensin neurons [13]. Thus, identification of human neurotensin neurons may require additional studies focused on the amplification of NTS mRNA via smFISH or spatial transcriptomic approaches.
In conclusion, single nucleus resolution allowed us to identify cluster-specific marker genes, such as OPRM1, while also providing the ability to identify marker genes for the broad LS. While snRNA-seq provides unprecedented resolution into the transcriptome of LS neurons, it does not provide information regarding its spatial organization. Mouse studies have identified that the LS is composed of non-overlapping spatial domains that contain transcriptionally distinct LS nuclei [15,42]. Our data suggests that the human LS is also composed of transcriptionally and spatially distinct neuronal cell types. Future studies of the human LS will require spatial transcriptomics to understand how transcriptionally distinct cell types are spatially organized, both within a single tissue section and along the anterior-posterior axis.
Supplementary Material
Supplementary Table 1. Donor demographic information. Demographic information on brain donors including Brain ID, diagnosis information, sex, race, age, and manner of death. An additional column containing the type of experiment (RNAscope vs snRNA-seq) the tissue was used for is also included.
Supplementary Table 2. Sample representation by cluster. snRNA-seq data was generated from three different donors and identified 22 transcriptionally distinct cell types. This table contains the number of nuclei within each cluster by the sample/donor.
Supplementary Table 3. Top 50 marker genes for each cluster. Differential expression testing was performed via 1-versus-all or pairwise testing. This table contains the top 50 marker genes by adjusted p-value for each cluster by each testing method.
ACKNOWLEDGEMENTS
A portion of Figure 1 was created with BioRender.com. We thank the LIBD neuropathology team, particularly James Tooke and Amy Deep-Soboslay, for curation of the brain samples and assistance with tissue dissections. We thank Kristen Maynard for feedback on the manuscript. Finally, we thank the families of Connie and Steve Lieber and Milton and Tamar Maltz for their generous support.
FUNDING
Funding for this project was provided by the Lieber Institute for Brain Development and National Institute of Health award R01MH105592 (KM).
Footnotes
CONFLICT OF INTEREST
The authors do not report any conflict of interest.
DATA AVAILABILITY
Raw sequencing data is publicly available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1093410. All code for analyzing the data is publicly available at https://github.com/LieberInstitute/ls_molecular-profiling. Any additional information required is available from the lead contact upon request.
REFERENCES
- 1.Moll J., Bado P., de Oliveira-Souza R., Bramati I.E., Lima D.O., Paiva F.F., Sato J.R., Tovar-Moll F., and Zahn R. (2012). A neural signature of affiliative emotion in the human septohypothalamic area. J. Neurosci. 32, 12499–12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahn R., de Oliveira-Souza R., and Moll J. (2020). Moral motivation and the basal forebrain. Neurosci. Biobehav. Rev. 108, 207–217. [DOI] [PubMed] [Google Scholar]
- 3.Moll J., Zahn R., de Oliveira-Souza R., Bramati I.E., Krueger F., Tura B., Cavanagh A.L., and Grafman J. (2011). Impairment of prosocial sentiments is associated with frontopolar and septal damage in frontotemporal dementia. Neuroimage 54, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Insel T.R., and Young L.J. (2001). The neurobiology of attachment. Nat. Rev. Neurosci. 2, 129–136. [DOI] [PubMed] [Google Scholar]
- 5.Gorman D.G., and Cummings J.L. (1992). Hypersexuality following septal injury. Arch. Neurol. 49, 308–310. [DOI] [PubMed] [Google Scholar]
- 6.Clemens A.M., Wang H., and Brecht M. (2020). The lateral septum mediates kinship behavior in the rat. Nat. Commun. 11, 3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leroy F., Park J., Asok A., Brann D.H., Meira T., Boyle L.M., Buss E.W., Kandel E.R., and Siegelbaum S.A. (2018). A circuit from hippocampal CA2 to lateral septum disinhibits social aggression. Nature 564, 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de León Reyes N.S., Sierra Díaz P., Nogueira R., Ruiz-Pino A., Nomura Y., de Solis C.A., Schulkin J., Asok A., and Leroy F. (2023). Corticotropin-releasing hormone signaling from prefrontal cortex to lateral septum suppresses interaction with familiar mice. Cell 186, 4152–4171.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azevedo E.P., Tan B., Pomeranz L.E., Ivan V., Fetcho R., Schneeberger M., Doerig K.R., Liston C., Friedman J.M., and Stern S.A. (2020). A limbic circuit selectively links active escape to food suppression. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sailer L.L., Park A.H., Galvez A., and Ophir A.G. (2022). Lateral septum DREADD activation alters male prairie vole prosocial and antisocial behaviors, not partner preferences. Commun. Biol. 5, 1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto M., Brito S.I., Venner A., Pasqualini A.L., Yang T.L., Allen D., Fuller P.M., and Anthony T.E. (2022). Lateral septum modulates cortical state to tune responsivity to threat stimuli. Cell Rep. 41, 111521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L., Durand-de Cuttoli R., Aubry A.V., Burnett C.J., Cathomas F., Parise L.F., Chan K.L., Morel C., Yuan C., Shimo Y., et al. (2023). Social trauma engages lateral septum circuitry to occlude social reward. Nature 613, 696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez L.A., Tran M.N., Garcia-Flores R., Oh S., Phillips R.A., Pattie E.A., Divecha H.R., Kim S.H., Shin J.H., Lee Y.K., et al. (2024). TrkB-dependent regulation of molecular signaling across septal cell types. Transl. Psychiatry 14, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turrero García M., Stegmann S.K., Lacey T.E., Reid C.M., Hrvatin S., Weinreb C., Adam M.A., Nagy M.A., and Harwell C.C. (2021). Transcriptional profiling of sequentially generated septal neuron fates. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid C.M., Ren Y., Xie Y., García M.T., Tran D., Vu S., Liu J., Adam M.A., Hanson S.M., Pisco A.O., et al. (2024). Multimodal classification of neurons in the lateral septum. BioRxiv. [Google Scholar]
- 16.Simon R.C., Fleming W.T., Senthilkumar P., Briones B.A., Ishii K.K., Hjort M.M., Martin M.M., Hashikawa K., Sanders A.D., Golden S.A., et al. (2024). Opioid-driven disruption of the septal complex reveals a role for neurotensin-expressing neurons in withdrawal. BioRxiv. [Google Scholar]
- 17.Frye R.E. (2018). Social skills deficits in autism spectrum disorder: potential biological origins and progress in developing therapeutic agents. CNS Drugs 32, 713–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osterling J., and Dawson G. (1994). Early recognition of children with autism: a study of first birthday home videotapes. J. Autism Dev. Disord. 24, 247–257. [DOI] [PubMed] [Google Scholar]
- 19.Yeung M.K. (2022). A systematic review and meta-analysis of facial emotion recognition in autism spectrum disorder: The specificity of deficits and the role of task characteristics. Neurosci. Biobehav. Rev. 133, 104518. [DOI] [PubMed] [Google Scholar]
- 20.Green M.F., Horan W.P., and Lee J. (2015). Social cognition in schizophrenia. Nat. Rev. Neurosci. 16, 620–631. [DOI] [PubMed] [Google Scholar]
- 21.Depp C.A., Mausbach B.T., Harvey P.D., Bowie C.R., Wolyniec P.S., Thornquist M.H., Luke J.R., McGrath J.A., Pulver A.E., and Patterson T.L. (2010). Social competence and observer-rated social functioning in bipolar disorder. Bipolar Disord. 12, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillissie E.S., Lui L.M.W., Ceban F., Miskowiak K., Gok S., Cao B., Teopiz K.M., Ho R., Lee Y., Rosenblat J.D., et al. (2022). Deficits of social cognition in bipolar disorder: Systematic review and meta-analysis. Bipolar Disord. 24, 137–148. [DOI] [PubMed] [Google Scholar]
- 23.Lipska B.K., Deep-Soboslay A., Weickert C.S., Hyde T.M., Martin C.E., Herman M.M., and Kleinman J.E. (2006). Critical factors in gene expression in postmortem human brain: Focus on studies in schizophrenia. Biol. Psychiatry 60, 650–658. [DOI] [PubMed] [Google Scholar]
- 24.Ramnauth A.D., Tippani M., Divecha H.R., Papariello A.R., Miller R.A., Pattie E.A., Kleinman J.E., Maynard K.R., Collado-Torres L., Hyde T.M., et al. (2023). Spatially-resolved transcriptomics of human dentate gyrus across postnatal lifespan reveals heterogeneity in markers for proliferation, extracellular matrix, and neuroinflammation. BioRxiv. [Google Scholar]
- 25.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amezquita R.A., Lun A.T.L., Becht E., Carey V.J., Carpp L.N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C., et al. (2020). Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lun A.T.L., Riesenfeld S., Andrews T., Dao T.P., Gomes T., participants in the 1st Human Cell Atlas Jamboree, and Marioni J.C. (2019). EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffiths J.A., Richard A.C., Bach K., Lun A.T.L., and Marioni J.C. (2018). Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun. 9, 2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Germain P.-L., Lun A., Garcia Meixide C., Macnair W., and Robinson M.D. (2021). Doublet identification in single-cell sequencing data using scDblFinder. F1000Res. 10, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Townes F.W., Hicks S.C., Aryee M.J., and Irizarry R.A. (2019). Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model. Genome Biol. 20, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haghverdi L., Lun A.T.L., Morgan M.D., and Marioni J.C. (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blondel V.D., Guillaume J.-L., Lambiotte R., and Lefebvre E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008. [Google Scholar]
- 34.Van der Maaten L., and Hinton G. (2008). Visualizing data using t-SNE. Journal of machine learning research 9. [Google Scholar]
- 35.Savell K.E., Tuscher J.J., Zipperly M.E., Duke C.G., Phillips R.A., Bauman A.J., Thukral S., Sultan F.A., Goska N.A., Ianov L., et al. (2020). A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Sci. Adv. 6, eaba4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran M.N., Maynard K.R., Spangler A., Huuki L.A., Montgomery K.D., Sadashivaiah V., Tippani M., Barry B.K., Hancock D.B., Hicks S.C., et al. (2021). Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron 109, 3088–3103.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips R.A., Tuscher J.J., Fitzgerald N.D., Wan E., Zipperly M.E., Duke C.G., Ianov L., and Day J.J. (2023). Distinct subpopulations of D1 medium spiny neurons exhibit unique transcriptional responsiveness to cocaine. Mol. Cell. Neurosci. 125, 103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez L.A., Tran M.N., Garcia-Flores R., Pattie E.A., Divecha H.R., Kim S.H., Shin J.H., Lee Y.K., Montoya C., Jaffe A.E., et al. (2023). TrkB-dependent regulation of molecular signaling across septal cell types. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lun A.T.L., McCarthy D.J., and Marioni J.C. (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. [version 2; peer review: 3 approved, 2 approved with reservations]. F1000Res. 5, 2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huuki-Myers L.A., Maynard K.R., Hicks S.C., Zandi P., Kleinman J.E., Hyde T.M., Goes F.S., and Collado-Torres L. (2023). DeconvoBuddies: a R/Bioconductor package with deconvolution helper functions. [Google Scholar]
- 41.Yao Z., van Velthoven C.T.J., Kunst M., Zhang M., McMillen D., Lee C., Jung W., Goldy J., Abdelhak A., Aitken M., et al. (2023). A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain. Nature 624, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Besnard A., and Leroy F. (2022). Top-down regulation of motivated behaviors via lateral septum sub-circuits. Mol. Psychiatry 27, 3119–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An S., Sun H., Wu M., Xie D., Hu S.-W., Ding H.-L., and Cao J.-L. (2021). Medial septum glutamatergic neurons control wakefulness through a septo-hypothalamic circuit. Curr. Biol. 31, 1379–1392.e4. [DOI] [PubMed] [Google Scholar]
- 44.Märtin A., Calvigioni D., Tzortzi O., Fuzik J., Wärnberg E., and Meletis K. (2019). A spatiomolecular map of the striatum. Cell Rep. 29, 4320–4333.e5. [DOI] [PubMed] [Google Scholar]
- 45.Andraka E., Phillips R.A., Brida K.L., and Day J.J. (2023). Chst9 Marks a Spatially and Transcriptionally Unique Population of Oprm1 -Expressing Neurons in the Nucleus Accumbens. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He J., Kleyman M., Chen J., Alikaya A., Rothenhoefer K.M., Ozturk B.E., Wirthlin M., Bostan A.C., Fish K., Byrne L.C., et al. (2021). Transcriptional and anatomical diversity of medium spiny neurons in the primate striatum. Curr. Biol. 31, 5473–5486.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen D., Liu X., Zhang W., and Shi Y. (2012). Targeted inactivation of GPR26 leads to hyperphagia and adiposity by activating AMPK in the hypothalamus. PLoS ONE 7, e40764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y.-L., Wei S., Liu Q., Gong Q., Zhang Q.-J., Zheng T.-G., Yong Z., Chen F., Lawrence A.J., and Liang J.-H. (2022). Mu-opioid receptors in septum mediate the development of behavioural sensitization to a single morphine exposure in male rats. Addict. Biol. 27, e13066. [DOI] [PubMed] [Google Scholar]
- 49.Le Merrer J., Gavello-Baudy S., Galey D., and Cazala P. (2007). Morphine self-administration into the lateral septum depends on dopaminergic mechanisms: Evidence from pharmacology and Fos neuroimaging. Behav. Brain Res. 180, 203–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Donor demographic information. Demographic information on brain donors including Brain ID, diagnosis information, sex, race, age, and manner of death. An additional column containing the type of experiment (RNAscope vs snRNA-seq) the tissue was used for is also included.
Supplementary Table 2. Sample representation by cluster. snRNA-seq data was generated from three different donors and identified 22 transcriptionally distinct cell types. This table contains the number of nuclei within each cluster by the sample/donor.
Supplementary Table 3. Top 50 marker genes for each cluster. Differential expression testing was performed via 1-versus-all or pairwise testing. This table contains the top 50 marker genes by adjusted p-value for each cluster by each testing method.
Data Availability Statement
Raw sequencing data is publicly available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1093410. All code for analyzing the data is publicly available at https://github.com/LieberInstitute/ls_molecular-profiling. Any additional information required is available from the lead contact upon request.