Abstract
Mitochondria tailor their morphology to execute their specialized functions in different cell types and/or different environments. During spermatogenesis, mitochondria undergo continuous morphological and distributional changes with germ cell development. Deficiencies in these processes lead to mitochondrial dysfunction and abnormal spermatogenesis, thereby causing male infertility. In recent years, mitochondria have attracted considerable attention because of their unique role in the regulation of piRNA biogenesis in male germ cells. In this review, we describe the varied characters of mitochondria and focus on key mitochondrial factors that play pivotal roles in the regulation of spermatogenesis, from primordial germ cells to spermatozoa, especially concerning metabolic shift, stemness and reprogramming, mitochondrial transformation and rearrangement, and mitochondrial defects in human sperm. Further, we discuss the molecular mechanisms underlying these processes.
Keywords: Mitochondria, Spermatogenesis, Male germ cells, Human infertility, Gene mutation
Introduction
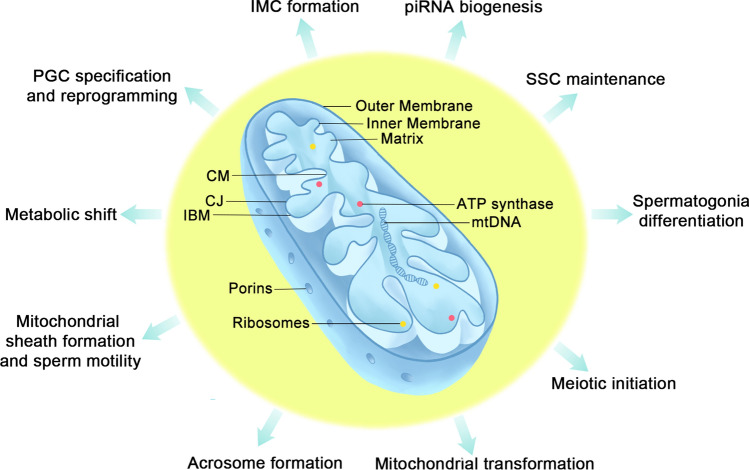
Mammalian mitochondria are multitasked organelles thought to originate from the endosymbiosis of alpha-proteobacterium [1]. Their roles exceed oxidative phosphorylation (OXPHOS) and they have been implicated to participate in many other cellular processes, including apoptosis, autophagy, calcium homeostasis, aging, signaling, stem cell renewal, and immune responses [2]. Similar to their bacterial ancestor, mitochondria comprise two structurally and functionally different membranes: the outer mitochondrial membrane (OMM) and inner mitochondrial membrane (IMM). The space between the OMM and IMM is called the intermembrane space, and the IMM encapsulates matrix compartments. The OMM is the principal platform for mitochondrial signaling, whereas the IMM mainly participates in energy conversion [3] (Fig. 1). Cristae are no longer seen as extended infoldings of the IMM; however, they are connected to the inner boundary membrane (IBM), which is the portion of the IMM parallel to the OMM and is enriched in protein import systems essential for mitochondrial fusion and nucleus-encoded proteins, via pore- or slit-like structures called cristae junctions (CJs) [4].
Fig. 1.
Mitochondrial structure and functions during male germ cell development. The mitochondrial lumen is named matrix, where Krebs cycle, mitochondrial DNA replication and proteins biosynthesis occur. The matrix is encompassed by the inner mitochondrial membrane (IMM) and outer mitochondrial membrane (OMM). The IMM can be divided into three parts: inner boundary membrane (IBM) (the part of the IMM running parallel to the OMM), cristae membranes (CMs), which are deep and pleomorphic invaginations occupying the surface area and houses mitochondrial respiration machinery, and cristae junctions (CJs), which are pore- or slit-like membrane structures connecting IBM and CM. Mitochondrial porins, or voltage-dependent anion-selective channels (VDACs) are major metabolite channels in the outer membrane connecting the transport of molecules and mitochondrial metabolism
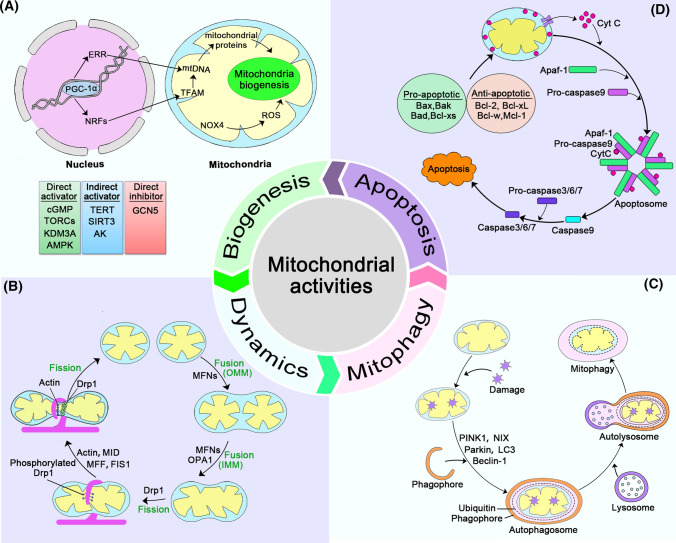
Mitochondrial activities during its life cycle include biogenesis, fusion and fission, cristae remodeling, clearance, and apoptosis (Fig. 2). These processes are vitally important for maintaining proper cellular function, and disruption of any of these events leads to the deterioration of bioenergetics and rapid induction of apoptosis, particularly in energy-craving cells such as myocytes and neurons. Mitochondrial biogenesis can be defined as the generation and development of mitochondria, which is regulated by factors encoded by both the nuclear genome and some mitochondrial elements. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) and nuclear respiration factors (NRFs), especially NRF1, are the main regulators. Some environmental factors, including multiple activators and inhibitors, can adjust the activity of mitochondrial biogenesis by changing the concentration of PGC-1α [5–7]. (Fig. 2A) [8]. Through continuous fusion and fission, mitochondria adjust their morphology, number, distribution, size, and motility. This flexible and adaptable feature of mitochondria, involving their morphology and subcellular distribution is collectively called mitochondrial dynamics (Fig. 2B), which has been summarized in multiple reports [4, 9–12]. The proper balance of mitochondrial dynamics is of great significance, and mutated fusion/fission proteins can lead to various human diseases [10]. Mitochondrial dynamics influence complex signaling pathways and affect gene expression, involving in multiple processes ranging from metabolism, cell cycle, and cell differentiation to cell senescence [11, 13]. Mitochondria first adjust their structure and composition to cope with cellular stress, including antioxidant action, DNA repair, protein folding, and degradation. If the damage is beyond the capacity of these systems, mitochondria will activate border quality control systems, such as mitophagy, to remove the damaged mitochondria [14]. Generally, mitochondrial clearance mainly consists of three pathways: PINK1/Parkin-mediated mitophagy, OMM receptor-mediated autophagy, and lipid-mediated mitophagy. Thereinto, the PINK1/Parkin pathway is the most critical course (Fig. 2C) [14–16].
Fig. 2.
Activities involving the mitochondrion. A Mitochondria biogenesis: PGC-1α is the core hub of mitochondrial biogenesis. Multiple elements can affect this process, including direct activators (the green part), indirect activators (the blue part), and direct inhibitors (the pink part). Through interaction with many transcription factors/proteins, such as estrogen-related receptors and nuclear respiration factors, PGC-1α controls the transcription of key mitochondrial enzymes and mtDNA synthesis. The alteration of mtDNA influences mitochondrial proteins to modify mitochondrial biogenesis. In addition, ROS directly impacts the process, which is regulated by nicotinamide adenine dinucleotide phosphate oxidase 4. B Mitochondrial fusion and fission: during OMM fusion, MFNs on the OMM of two mitochondria recognize each other with independent IMMs and matrix components; for IMM fusion, two IMMs and the matrix are fused, and both MFNs and OPA1 participate in this process. In contrast, fission refers to dividing one mitochondrion into two mitochondria under the cooperation of Drp1, actin, MFF, MID, and FIS1. C Mitochondrial mitophagy: PINK1/Parkin-mediated mitophagy is a predominant process of mitochondrial degeneration. The damaged mitochondria are usually recognized by the autophagosomes under the action of several elements, such as PINK1, Parkin, NIX, Beclin-1, LC3, and ubiquitin, and then the complex is delivered to the lysosome for degradation. D Mitochondrial mediated apoptosis: Upon receiving an apoptotic stimulus, cytochrome c is released from the intermembrane space of the mitochondrion into the cytoplasm and binds to Apaf-1, forming an “apoptosome.” Apoptosome subsequently recruits and facilitates the activation of procaspase-9, which targets other caspases, including caspase3/6/7, to finally activate cell apoptosis
There are two main apoptotic signaling pathways: the extrinsic (also named death receptor) pathway and the other is intrinsic, or mitochondrial, pathway (Fig. 2D). Both pathways converge upon the activation of caspase 3 and caspase 7 [17]. Numerous cellular stressors, such as growth-factor deprivation or DNA damage, can induce the mitochondrial pathway of apoptosis to kill cells. Mitochondrial outer membrane permeabilization (MOMP) is critical for the release of soluble proteins, such as cytochrome c (Cyt c), from the mitochondrial intermembrane space, resulting in cell death. Cyt c then binds to the adaptor molecule apoptotic peptidase activating factor 1 (Apaf-1), to form a complex called the apoptosome, which initiates the activation of the executioner caspases [18]. Importantly, Cyt c-mediated apoptosis is required for spermatogenesis, especially for spermatogonia maintenance [19].
Male germ cell development is a sophisticated process during which mitochondria undergo continuous changes to accommodate the varied energy demands of different cell types. Accumulating evidence has revealed the essential role of mitochondria in spermatogenesis, and the disruption of mitochondrial dynamics and functions disrupts spermatogenesis, thereby leading to male infertility. In this review, we discuss the dynamic changes in mitochondria during spermatogenesis, from primordial germ cells (PGCs) to spermatozoa, in mammals. Specifically, we highlight new insights into mitochondrial protein-mediated regulation of germ cell development, including special germinal granule formation, metabolic shift, mitochondrial transformation, and rearrangement. Recent studies on molecular mechanisms underlying mitochondrial functions during spermatogenesis in mouse models, as well as mitochondrial defects in humans, are discussed.
Dynamic changes in mitochondrial morphology and distribution during male germ cell development
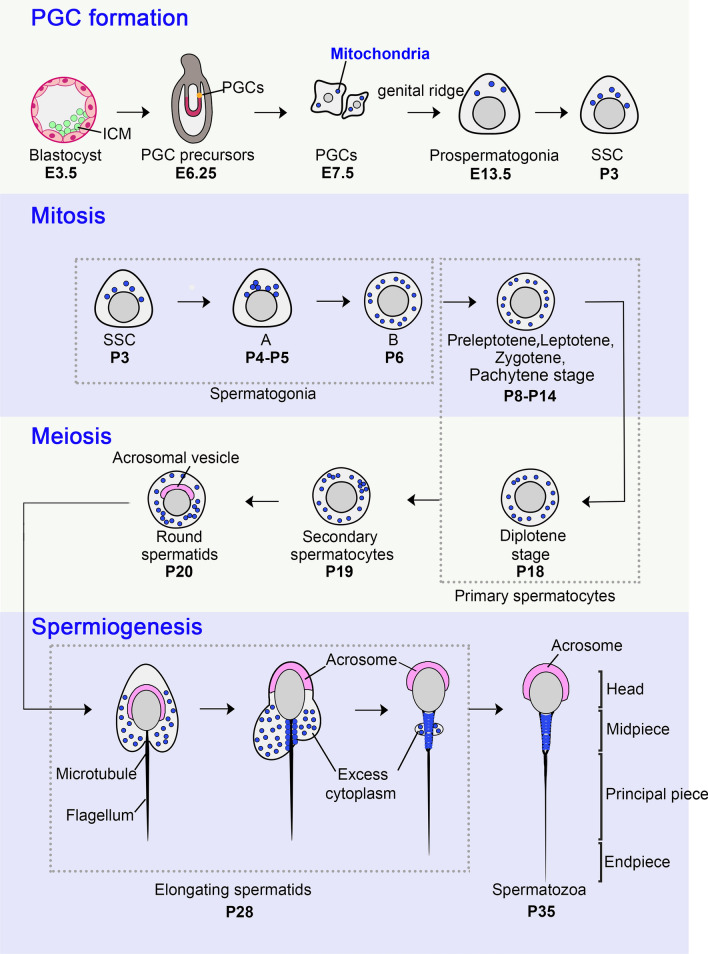
The development of male germ cells is a tightly coordinated and complex process, which initiates from early embryogenesis and forms mature spermatozoa through spermatogonia mitosis, spermatocyte meiosis, and spermiogenesis on postnatal days. Shortly after fertilization in mice, germ cells first develop as primordial germ cells (PGC), which are derived from post-implantation epiblast cells that have the ability to generate all somatic cell and PGC precursors (embryonic day 3.5 [E3.5]–E6.25). PGCs emerge inside the extraembryonic mesoderm at the posterior end of the primitive steak as a cluster at E7.25 and settle in the fetal gonads [20]. Subsequently, they rapidly proliferate with an average doubling time of approximately 16 h and migrate to the genital ridge, where sex determination occurs at E12.5 [21]. If XX, they initiate oogenesis and arrest in meiosis by E14.5 and remain in this state until puberty. If XY, they transition to prospermatogonia (gonocytes). Prospermatogonia continue to proliferate for several days before entering a period of quiescence around E16.5, during which DNA methylation patterns are re-established [22, 23]. Spermatogenesis is initiated shortly after birth and lasts for approximately 35 days in mice. Prospermatogonia re-enter mitosis around postnatal day (P) 1.5–3.5 and develop into spermatogonial stem cells (SSCs), which can self-renew and produce large numbers of spermatogonia [24]. During the first postnatal week, the undifferentiated spermatogonia proliferate by consecutive mitotic divisions and mature into primary spermatocytes, which enter the meiotic process around P12 [25]. After two sequential meiotic divisions, round spermatids, the earliest post-meiotic cells, are produced around P20-P22. Spermatids develop from round cells into elongating cells within 2–3 weeks, turning into highly specialized spermatozoa that are ultimately released into the seminiferous tubule lumen from approximately P35 [25], and undergo sperm maturation in the epididymis (Fig. 3).
Fig. 3.
Cellular distribution of mitochondria during male germ cell development. Shortly after fertilization, blastocyst at embryonic day (E) 3.5 evolves and forms primordial germ cell (PGC) at E7.5. The PGC passes through the genital ridge to produce prospermatogonia (pro-SG) which usually appears at E13.5, finally giving rise to spermatogonial stem cell (SSC) at postnatal day 3 (P3). Spermatogonia (SG) undergoes consecutive mitotic divisions and matures into primary spermatocytes at P8, which enter the meiotic process. Through the first meiotic division, each spermatocyte (Spc I) through leptotene, zygotene, pachytene, and finally diplotene stages, yields a pair of secondary spermatocytes (Spc II) around P19. The final phase is spermiogenesis, during which the round haploid spermatid (RS) goes through dramatic morphological and structural reorganization, forming spermatozoa (Spz) at P35. During this developmental course, the distribution of mitochondria (blue dots) is dynamically changed. In PGC and pro-SG, mitochondria are less and scattered but large. In SSC, mitochondria remain few and are gradually distributed near the nucleus; the number of mitochondria increases, often scattered with some clusters in spermatogonia. Among primary spermatocytes, mitochondria appear like capsules and are still dispersed with some clusters, while in secondary spermatocytes, more clusters appear. In spermatids, including round spermatids and elongated spermatids, and spermatozoa, the mitochondria gradually move to the opposite side of the acrosome, surround the axon, and finally align at the midpiece of spermatozoa
In general, undifferentiated stem cells tend to have reduced mitochondrial function and mainly rely on glycolysis, whereas differentiating cells contain active mitochondria with high membrane potentials and increased oxidative phosphorylation as described in previous reviews [26, 27]. Germ cell development proceeds through mitosis and meiosis, producing several strikingly different cell types, including undifferentiated and differentiating cells. Concomitantly, mitochondria undergo marked dynamic changes to satisfy different energy requirements, including changes in number, morphology, distribution, and energy metabolism (Fig. 3).
In mouse PGCs, mitochondria are sparse but large, roundish-oval-like structures with only a few cristae regularly oriented in a shelf-like fashion, which are thought to be in a naïve state and have limited capacities to produce energy [28–30]. In humans, mitochondrial number decreases approximately 100,000 in zygotes to approximately 10 in PGCs [31] and increases with PGC proliferation. However, male PGCs undergo a metabolic shift from glycolysis to oxidative phosphorylation (OXPHOS) during PGC differentiation. Glycolysis and OXPHOS in these PGCs at E9.5 are maintained at levels similar to those in embryonic stem cells (ESCs); subsequently, glycolysis decreases after E11.5, and OXPHOS is elevated after E9.5. Compared with the metabolic profiles of ESCs and gonadal somatic cells (somas), male PGCs at E13.5 present strikingly increased OXPHOS activity and decreased glycolysis. Notably, glycolysis and OXPHOS levels are essential for the control of PGC reprogramming and specification of pluripotent stem cells (PSCs) into PGCs in culture [32]. Mitochondria in P1 mouse prospermatogonia are relatively undeveloped and round with few and indistinct cristae, whereas at P4, they develop increased numbers of electron-dense cristae [33, 34]. A metabolic shift also occurs during the perinatal quiescence period; the genes associated with OXPHOS transiently increase in expression among quiescent prospermatogonia and then decrease during cell cycle re-entry [24], suggesting that mitochondria play a role in preventing meiotic initiation.
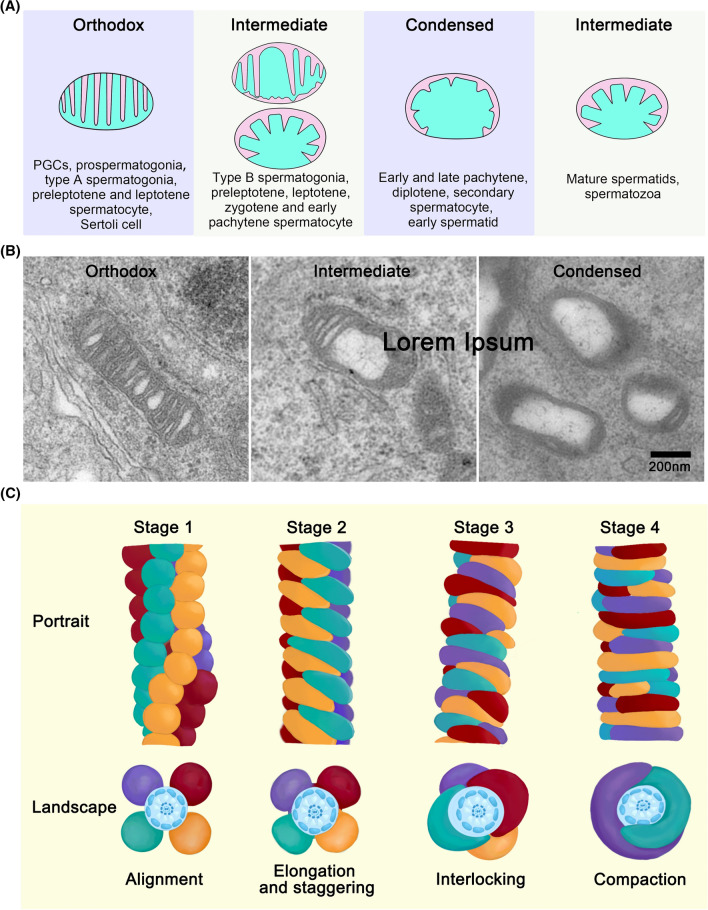
Three types of mitochondrion are observed during spermatogenesis: the orthodox type in Sertoli cells, spermatogonia, preleptotene, and leptotene spermatocytes; intermediate type in zygotene spermatocytes; and condensed type in pachytene spermatocytes, secondary spermatocytes, and early spermatids [35] (Fig. 4A, B). In type A spermatogonia, mitochondria exhibit an orthodox form characteristic of an ovoidal shape and lamellar cristae with an interposed electron-dense matrix. In type B spermatogonia and leptotene spermatocytes, the space between the two lamellar cristae of some mitochondria starts to increase in size, leading to dilatation of the intracristal spaces. Many mitochondria are elongated and localized around the nucleus in zygotene and early pachytene spermatocytes. In late pachytene spermatocytes, mitochondria become small and round, forming aggregates as clusters that are randomly distributed throughout the cytoplasm. Morphologically, mitochondria in late pachytene spermatocytes are regarded as the typical “condensed” type, in which the inner space with its dense matrix is flattened against the outer membrane by considerable expansion of one or more intracristal spaces [35]. Mitochondria maintain a “condensed” shape; however, the aggregates are not observable in a broad spectrum of maturating germ cells, including diplotene and secondary spermatocytes and Golgi and cap phase spermatids, until acrosome phase spermatids.
Fig. 4.
Morphological changes of mitochondria and mitochondrial sheath formation during male germ cell development. A In PGCs, prospermatogonia and type A spermatogonia, there are mainly “orthodox” mitochondria whose typical lamellar cristae are thin and near each other. When the cells evolve into type B spermatogonia, the mitochondrion whose intracristal space is enlarged and expanded are usually called “intermediate” mitochondria, and small vesicles appear gradually. This kind of mitochondrion is also presented among preleptotene, leptotene, zygotene, and some early pachytene stage spermatocytes. During this process, the cristae first retract and form some vesicles and then form typical “intermediate” mitochondria whose partial cristae are compact. As the intracristal space is further increase and dilate, “condensed” mitochondria are formed. Some early pachytene and late pachytene, diplotene, secondary spermatocytes, and early spermatids present “condensed” morphology. Their mitochondrial cristae are abundant, compact, and convoluted, and the cristae gradually evolving into a packed concentric substance flattening to the OMM, and the matrix is large, appearing like an ellipse. However, the mitochondria gradually turn into a typical “intermediate” type in mature spermatids and spermatozoa. B Represent TEM photographs displaying three typical types of mitochondrial morphologies, “orthodox”, “intermediate”, and “condensed” in mice. C Ho’s model shows mitochondrial sheath formation in spermatids. At stage 1, the regular alignment of spherical mitochondria makes four dextral longitudinal arrays. At stage 2, spherical mitochondria gradually assume a crescent-like form, and the elongating two ends are staggered between the mitochondria at the adjacent arrays. Each crescent-like mitochondrion span half of one gyre at the end of stage 2. At stage 3, each pair of mitochondria slide each other through a specific contact point, making a final sinistral double-helical structure. At stage 4, the mitochondrion shape changes from crescent-like to rod-like, making a sinistral double helix through an end-to-end contact, with each mitochondrion spanning three-fifths of one gyre in mature sperm
During spermiogenesis, a greater number of mitochondria in the Golgi and cap phase spermatids are arranged along the plasma membrane. Subsequently, “condensed” mitochondria in the late acrosomal phase and early maturation phase spermatids gradually develop more convoluted cristae. Simultaneously, partial mitochondria begin to move towards the flagellum, whereas the remaining mitochondria continuously aggregate and separate from mature spermatids or testis spermatozoa within the residual bodies [35]. Gradually, the intracristal spaces of flagellar mitochondria are reduced, and the cristae stretch themselves into the interior of the organelles; meanwhile, mitochondria lose their “condensed” appearance. By regularly arranging along with the middle piece of spermatozoa, mitochondria form mitochondrial sheaths that surround the axonemal complex and nine outer dense fibers (ODFs) [35, 36] (Fig. 3).
During mitochondrial sheath development in mammalian spermatozoa, mitochondria gradually change from spheroid to long and rod-like shape, surrounding the axonemal complex and ODFs [36]. Intriguingly, the length of the middle piece and number of gyres in the mitochondrial sheath are uniform and heritable within the same mammalian species [37]. In the mouse spermatid tail, the mitochondrial helical sheath begins to form during late spermiogenesis around step 15. In 1988, Otani et al. first divided mouse mitochondrial sheath formation into four synchronized developmental stages according to observations from cryo-fracturing method with high-resolution scanning electron microscopy (SEM) [38]. Ho et al. revised the model by using new image processing software to reconstruct mature mouse sperm middle piece from ultrathin serial sections during spermiogenesis [39]. In Ho’s model (Fig. 4C), spherical mitochondria regularly arrange in four dextral longitudinal arrays at stage 1, with two adjacent arrays of alternately arranged spherical mitochondria. At stage 2, all spherical mitochondria elongate to a crescent-like form, and the elongating two ends are staggered between the mitochondria at the adjacent arrays. Pairs of mitochondria from opposing arrays contact each other, forming approximately 70 doughnut-like structures. At the end of stage 2, each crescent-like mitochondrion spans half of one gyre. At stage 3, mitochondria continue to elongate to three-fifths of one gyre and slide each other through the contact point in a specific pattern, making a final sinistral double-helical structure. At stage 4, crescent-like mitochondria become rod-like and contact end-to-end, forming a sinistral double helix, with each mitochondria spanning three-fifths of one gyre in mature sperm [39].
Mitochondria in PGCs
PGCs have distinct metabolic properties that require a dynamic balance between glycolysis and OXPHOS during differentiation. OXPHOS and glycolysis activities in PGCs are initially comparable to those in ESCs; however, OXPHOS increases after E9.5, while glycolysis decreases after E11.5, as revealed by single-cell RNA-seq over a developmental time course referred to as a “pseudotime analysis” [32, 40]. Glycolytic inhibitor markedly blocks PGC reprogramming in both feeder cell co-cultured and feeder-free PGCs, suggesting a direct role of glycolysis activity in PGC reprogramming. Furthermore, ATP production by OXPHOS is required for the initial step to induce PGC-like cells [32] from mouse ESCs in the developing embryo [20, 41]), which relies primarily on glycolysis for energy production [42]. These findings suggest that the energy metabolism profile is gradually established during differentiation and that mitochondria play an essential role in PGC specification.
Traditionally, two canonical functions have been ascribed to mitochondria: one is to regulate catabolism through ATP production by OXPHOS, and the other is to support anabolism through the production of tricarboxylic acid (TCA) cycle metabolites such as citrate and oxaloacetate, which generate macromolecules such as lipids and nucleotides. Mitochondria have also gained recognition as signaling organelles that dictate stem cell fate through reactive oxygen species (ROS), metabolites of the TCA cycle, regulation of NAD+/NADH ratios, and pyruvate metabolism [3]. α-Ketoglutarate (αKG), a key intermediate generated in the TCA cycle, is a substrate for a wide range of enzymatic reactions, and as a cofactor in many enzymatic reactions, it has pleiotropic properties involved in several physiological processes besides cellular metabolism. In addition to its significant role in pluripotency acquisition and maintenance of mESCs, αKG is able to specifically enhances PGC fate [40]. PGC-like cells exhibit increased expression of the genes encoding IDH2 (αKG-producing enzyme) and COX7A1 (a central regulator of mitochondrial oxidative metabolism), suggesting of enhanced oxidative metabolism and αKG synthesis. Increased αKG or decreased glycolysis levels favor the retention of mESC self-renewal over exit from naïve pluripotency. Repressing glycolysis or lifting αKG levels makes mESC prefer retention of self-renewal over exit from naïve pluripotency. Notably, αKG safeguards the transient developmental potential for PGC fate by retaining the particular epigenetic state of competent EpiLCs, at least partially including H3K9me2, H3K27me3, and DNMT3b [40]. These findings link the TCA metabolite αKG with the oxidative metabolic state and histone methylation status, ultimately influencing the naïve pluripotency of mESCs and PGC fate.
Most small non-coding RNAs (sncRNAs), including microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), endogenous small interfering RNAs (endo-siRNAs), and promoter-associated small RNAs, are encoded by the nuclear genome and participate in transcriptional or post-transcriptional regulation, mobile silencing, and chromatin remodeling. Additionally, the mitochondrial genome is known to encode thousands of small RNAs, called mitosRNAs, and has been proposed to play important roles in synchronization and nuclear-mitochondrial communication [43, 44]. Next-generation sequencing (NGS) and bioinformatic analysis of the sncRNAs in mouse male E23 PGCs, spermatogonia cells, spermatozoa, oocytes, and zygotes have shown that although mitochondrial sncRNAs account for less than 1% of the total sncRNAs, they are cell type-specific, and all mitochondrial genes encode mitosRNAs. Interestingly, mito-piRNAs are the most abundant among mitosRNAs, accounting for 80–90% mitosRNAs. In PGCs, miRNA accounts for approximately 8% of the mitosRNA population, which is higher than that in other cell types. Furthermore, chromosome distribution analysis revealed an enrichment of mitosRNA on chromosome 2 in all cell types; however, mitosRNAs from PGCs have also shown higher enrichment on chromosomes other than chromosome 2 than on other cell types, indicating the distinct characteristics of PGC mitosRNAs [45]. Notably, mitosRNAs are not only present in PGCs but also in spermatogenic cells, oocytes, and zygotes, suggesting their significant regulatory roles in spermatogenesis, gamete differentiation, and fertilization. Although more data are needed to elucidate the biological function of mitosRNAs, their presence in male germ cells, gametes, and zygotes opens the door to new directions for mitochondrial function investigation, especially the communication between the mitochondrion and nucleus. Even though cellular metabolism is closely correlated with pluripotency and differentiation of stem cells and the abundance of mitosRNA in germ cells, it remains unclear how metabolism and mitosRNA affect cell fate, and the role of mitochondria in PGCs is largely unknown. With single-cell analytics technology [46], in-depth studies on mitochondrial function will continue to enrich our understanding of stem cells and PGCs.
Mitochondria in prospermatogonia
Prospermatogonia are transitory cells of the male germ cell line between PGCs and SSCs. In mammals, mitochondria in prospermatogonia are comparatively undeveloped and appear round with enlarged cristae edges that are few and indistinct. The transition from silent prospermatogonia to active spermatogonia represents the inception of spermatogenesis, and with this transition, the number of mitochondrial cristae increases [34]. In prospermatogonia, mitochondria with enlarged cristae are connected to numerous electron-dense particles of ribonucleoproteins (called “Nuage”) that are usually observed in prospermatogonia, SSCs, and spermatogonia. Nuage is a discrete, dense fibrous cytoplasmic organelle that lacks a surrounding membrane, is often closely adjacent to the germ cell nuclear envelope, and is enriched with RNA and RNA-binding proteins [47]. In particular, a distinct Nuage form, inter-mitochondrial cement (IMC), appears, whose components (derived from the mitochondrion) form the adhesion between mitochondria. In mouse spermatogonia, IMC is observable via electron microscopy at around E15 and continuously exists in primary spermatocytes until it is no longer discernible in spermatids [47].
The Tudor domain-containing 1/Mouse Tudor repeat-1 (Tdrd1/Mtr-1) protein is located in the cytoplasm of fetal prospermatogonia, postnatal spermatogonia, spermatocytes, and round spermatids. It is present in the IMC in spermatogonia, spermatocytes, and chromatoid bodies of round spermatids [48, 49]. Targeted deletion of TDRD1 (Tdrd1tm1/tm1) in mice leads to male infertility, whereas female Tdrd1tm1/tm1 mice are fertile. In Tdrd1tm1/tm1 testes, spermatogenesis cannot proceed further than haploid step 7 round spermatids up. Notably, IMCs are virtually absent in Tdrd1tm1/tm1 pro-spermatogonia and spermatocytes [49]. Likewise, MVH/DDX4 is another Nuage-associated protein essential for spermatogenesis and is present in germ cells from prenatal gonocytes at E10.5 to postnatal spermatogonia until round spermatids [50, 51]. Importantly, MVH/DDX4 acts as the upstream of TDRD1/Mtr-1 to regulate the cellular distribution of TDRD1/Mtr-1. In wild-type spermatogonia, TDRD1/Mtr-1 exhibits a delicate granular appearance that corresponds to the IMC. However, TDRD1/Mtr-1 is diffusely distributed in the cytoplasm or, more notably, exists as a crescent-like aggregate around the nucleus in Mvh1098/1098 spermatocytes [49]. In mice, Mvh1098/1098 gene-targeted mutants are male-sterile (whereas female are fertile) due to an arrest at the prophase of meiosis, and the majority of spermatocytes are lost [50]. IMCs are absent among clustered mitochondria in the remaining Mvh1098/1098 spermatocytes and are observed only in a small set of Mvh1098/1098 fetal spermatogonia [49]. Further, IMC is absent in Tdrd1tm1/tm1 oocytes and is markedly reduced in Mvh1098/1098 oocytes; however, both Tdrd1tm1/tm1 and Mvh1098/1098 females are fertile, suggesting that IMC is not a direct prerequisite for oogenesis and female fertility but is indispensable for germ cell development and male fertility in mice [49].
MITOPLD (Zucchini in Drosophila) belongs to the phospholipase D superfamily conserved among diverse species, from bacteria to humans, which gathers in the OMM based on its mitochondria-targeted signal in the N-terminal region. Two roles have been reported for MITOPLD: one is to facilitate mitochondrial fusion by hydrolyzing cardiolipin (CL) to generate phosphatidic acid (PA), comprising a novel lipid signaling pathway on the mitochondrial surface [52], and the other is to be closely associated with gonocyte differentiation and spermatogenesis through the regulation of the piRNA biogenesis pathway [53, 54]. In mice, MITOPLD is highly expressed in the testes from E16.5 and adult stage, and in growing oocytes. Mitopld−/− mice are grossly normal in appearance but exhibit male infertility, whereas female mice are fertile [53, 54]. Mitopld−/− male mice display mitotic arrest, DNA damage, and piRNA blockage [54]. In E16.5 Mitopld mutant prospermatogonia, the localization of piRNA pathway proteins is disrupted, including MILI, TDRD1, and MIWI2, which show crescent-shaped signals around the nuclei rather than granular signals in the cytoplasm in wild-type prospermatogonia. This diffuse distribution phenomenon is similar to the abnormal localization in Mvh mutant prospermatogonia [53]. In addition, mitochondria accumulate in a particular region neighboring the nucleus in Mitopld−/− prospermatogonia. These results demonstrate that MITOPLD is essential for mitochondrial distribution and nuage formation, and MITOPLD and MVH may play a role in the same context [53].
GASZ is a germ cell-specific protein that is expressed in both male and female germ cells. It localizes to the outer mitochondrial membrane based on its C-terminal mitochondrial targeting sequence and is mainly enriched in the IMC structure in both prospermatocytes and spermatocytes [55]. Similar to other IMC components, Gasz knockout mutants exhibit male infertility due to a zygotene-pachytene block and normal female fertility, which is reminiscent of IMC components such as MILI, MIWI2, TDRD1, and MVH knockout mouse models. The localization and/or protein levels of multiple IMC components and piRNA pathway proteins are disrupted upon GASZ deletion, such as the absence of MILI, diffused cytoplasmic distribution of TDRD1 and MVH in E16.5 prospermatogonia and P0 spermatogonia, and dramatically decreased protein levels of MIWI, TDRD1, MVH, MILI, TDRD6, and TDRD7 in P10 and P14 testes, as well as the absence of IMC in germ cells. Markedly, piRNAs are decreased, and thus retrotransposons are increased upon Gasz deletion, similar to that of MILI and MIWI2 testes [55]. Notably, the mitochondrial localization sequence (MLS) is essential for Gasz to regulate spermatogenesis, as demonstrated by MLS deletion Gasz mice (GaszΔNLS/NLS), exhibiting a phenotype similar to that of global Gasz knockout mice. This further demonstrates the role of GASZ in IMC maintenance, piRNA biogenesis, and mitochondrial dynamics [55]. Since GASZ can interact with multiple critical IMC components and mitochondrially located proteins, such as MIWI, DDX4, TDRD1, MFN1, and MFN2, it probably regulates a protein-RNA complex formation, maintenance, and subsequent spermatogenesis.
Mitochondria in spermatogonia
In mammals, SSCs, progenitors, and differentiating spermatogonia comprise a subset of the heterogeneous spermatogonial population [56], and notable changes in gene expression exist even between closely related cell types. For example, over 1400 genes are differentially expressed during the SSC-to-progenitor transition [57]. Compared to other stem cell types, our understanding remains particularly limited to SSC metabolism, metabolic changes during spermatogonial differentiation, and their influence on cell fate. However, huge data generated by powerful single-cell RNA sequencing (scRNA-seq) from mouse and human testicular cell populations has shed light on gene expression dynamics related to mitochondrial function and metabolism across discrete spermatogonial subsets, as summarized in [58]. Accordingly, SSCs prefer a hypoxic environment, which assists glycolytic metabolism essential for SSCs to maintain regenerative capacity. SSC differentiation occurs with increased OXPHOS activity, which is crucial for sustaining spermatogenesis and male fertility. Inhibition of OXPHOS prevents spermatogonia differentiation by inhibiting the respiratory chain complex or by eliminating mitochondrial membrane potential (MMP) [59]. OXPHOS affects spermatogonia apoptosis and self-renewal by triggering ROS production, as previously described [60]. Therefore, during spermatogonia differentiation, metabolism is likely to shift from glycolysis to aerobic respiration because researchers have found that key glycolytic enzymes HK2, ALDOA, PGK1, PKM, and LDHA are significantly reduced [59], whereas mitochondrial regulatory factors are upregulated [59].
When SSCs were compared with ID4-EGFP undifferentiated progenitor cells, the statistically significant differences in mitofusin protein MFN1/MFN2 expression demonstrated that preparation for mitochondrial fusion is likely to begin after the transition from SSCs to progenitor cells to facilitate the differentiation of spermatogonia [57]. Several studies have elucidated the function of mitofusin protein in regulating spermatogonial differentiation. For example, regardless of the conditional deletion of Mfn1 and Mfn2 alone or simultaneously with the male germline-specific Stra8-Cre driver (beginning at P3 in undifferentiated spermatogonia) [61], the mutant males are infertile [62, 63]. The Mfn1/Mfn2 double mutant testes display a significant reduction in spermatocyte numbers as early as P24 and a dramatic decrease in the number of differentiating spermatogonia; however, no apparent influence on the undifferentiated population in adulthood is exhibited [62, 63]. Interestingly, Mfn1 single deletion showed a much more severe phenotype than that following Mfn2 single deletion; the first round of spermatogenesis is disrupted in Mfn1 single deletion testes but can be completed upon the loss of Mfn2 alone. In addition to the mitochondrial fragmentation, Mfn2 single deletion results in increased mitochondria-endoplasmic reticulum (ER) distance, fragmented ER, and increased thickness of the IMC structure, which are not observed in Mfn1 single knockout testes, indicating the unique roles of Mfn1 and Mfn2 in the regulation of spermatogenesis [62, 63]. Furthermore, Mfn1/Mfn2 double knockout leads to mitochondrial fragmentation and reduced OXPHOS components in undifferentiated spermatogonia, differentiated spermatogonia and spermatocytes. Notably, after long-term mitofusin loss, both Mfn1 and Mfn2 single mutants exhibit severe depletion of differentiating spermatogonia, similar to the level observed in Mfn1/Mfn2 double mutants [63], together with the age-dependent phenotype in Mfn2 single mutants [62], suggesting that both Mfn1 and Mfn2 are essential for the maintenance of differentiating spermatogonia. For the metabolic style, probably unlike glycolysis-preferring SSCs, differentiating spermatogonia demand mitochondrial augmentation and glycolysis-OXPHOS transition to retain functionality [58].
However, a limitation of Stra8-Cre is that it only affects STRA8-expressing cells, whereas a rare population of self-renewing SSCs does not express STRA8 and thus cannot be detected. Interestingly, another group established Ddx4-Cre (commenced expression in gonocytes at E15.5 [64])-mediated conditional knockout of Mfn1 and Mfn2 in mice, referred to as Mfn1f/f; Ddx4-Cre and Mfn2f/f; Ddx4-Cre, respectively [65, 66]. Both Mfn1f/f; Ddx4-Cre and Mfn2f/f; Ddx4-Cre males were infertile, accompanied by meiosis blocks at the zygotene and pachytene stages. Mfn2f/f; Ddx4-Cre mouse testes displayed reduced germ cells early at P14, leaving much fewer spermatocytes at P14, and no spermatids were detected at P35 and later. Notably, Mfn2f/f; Ddx4-Cre testes exhibited significantly decreased spermatogonia differentiation and increased undifferentiated spermatogonia at P14 and P21, similar to the spermatogonia differentiation block in Mfn1f/f; Ddx4-Cre testes [62]. In addition, ROS levels and DNA oxidation were elevated in differentiating spermatogonia and spermatocytes in both Mitofusion mutants, which is probably the direct reason for cell apoptosis. Interestingly, Mfn2f/f; Ddx4-Cre testes harbor a series of distinct defects compared to those of Mfn1f/f; Ddx4-Cre testes, with normal mitochondrial morphology in P0 prospermatogonia and gradual fragmentation from P5 but become relatively normal around P13. Whereas Mfn1f/f; Ddx4-Cre testes showed severely swollen and enlarged mitochondria in P0 prospermatogonia, relatively normal mitochondria at P5, and significantly smaller mitochondria at P8. Furthermore, mitochondrial Ca2+ levels and the number of fragmented ERs are dramatically increased in Mfn2f/f; Ddx4-Cre germ cells, whereas no obvious change was observed in Mfn1f/f; Ddx4-Cre germ cells [65]. In addition, the introduction of Mfn2 could rescue the phenotype of Mfn2f/f; Ddx4-Cre testes, but not Mfn2f/f; Ddx4-Cre testes [65]. Taken together, these data further confirm the non-redundant and distinct roles of Mfn1 and Mfn2 in regulating spermatogenesis via germ cell differentiation, mitochondrial morphology, mitochondria-ER contact membrane, mitochondrial Ca2+ homeostasis, metabolic shift, and DNA oxidation.
SHP2 is a ubiquitously expressed protein tyrosine phosphatase that plays an important role in cytosolic signal transduction to regulate cell growth, cell development, tissue inflammation, and cellular chemotaxis. Apart from its cytoplasm-nuclei shuttle localization associated with the signal transducer, it has also been detected in the mitochondria of rat brains [67, 68]. SHP2 is highly expressed in the nuclei and cytoplasm of Sertoli cells and in the cytoplasm of undifferentiated spermatogonia; however, it exhibits significantly decreased levels in meiotic spermatocytes in mice [69, 70]. Ddx4-Cre-mediated Shp2 deletion results in male infertility with progressive germ cell loss and thus Sertoli-only phenotype in adults due to blocked proliferation of SSCs and undifferentiated spermatogonia; therefore, SSCs fail to self-renew and germ cells cannot be replenished [69]. Additionally, Stra8-Cre-mediated Shp2 deletion causes male infertility, accompanied by accelerated differentiation of spermatogonia, thus disrupting spermatogonial proliferation/differentiation balance [70]. Furthermore, the loss of SHP2 in Sertoli cells causes premature SSC differentiation [71]. However, the limitations of these studies are that they did not detect whether SHP2 colocalizes with mitochondria in the testes and the unclear molecular mechanisms, revealing the need for a clear mechanism to investigate for SHP2 in maintaining SSCs and undifferentiated spermatogonia.
Mitochondria in spermatocytes
Spermatocytes are certain cells produced after mitosis and proliferation of spermatogonia and they finally differentiate into mature sperm cells. They are divided into primary spermatocytes (preleptotene, leptotene, zygotene, and pachytene stages) and secondary spermatocytes, and changes in mitochondrial morphology are described above (Fig. 4A, B). As meiosis begins, mitochondria become more abundant; they aggregate, fuse, and finally promote OXPHOS [63]. The high expression of COXII encoded by the mitochondrial genome in spermatocytes at the pachytene stage indicates a higher energy requirement for meiotic cells [72]. IMC exists in all primary spermatocytes, and several key piRNA pathway proteins located in IMC and/or mitochondria play significant roles in meiosis, such as MIWI, TDRD6, MOV10L1, and TDRKH [73]. Here, we will mainly focus on other proteins and/or factors involved in mitochondrial function during meiosis, including mitochondria-located proteins and mtDNA copy number.
Elongation factor 4 (EF4) is located in the outer mitochondrial membrane and co-stains with mitochondrial ribosomes based on its mitochondrial targeting signal in the N terminus [74]. Although EF4 is a conserved protein detected in almost all known genomes [75], its ablation in organisms including bacteria and higher plants, has produced no obvious phenotype, making its function largely unknown. However, a study on its role in the reproductive system reported its high expression in spermatocytes and round spermatids, whereas it was weakly expressed in Sertoli cells, spermatogonia and elongated spermatids [74]. Knockout (KO) of Ef4 in mice leads to male infertility, with round spermatids ceasing to develop at developmental steps 5–8, caused by mitochondrial dysfunction. Mitochondrial defects are observable as early as in spermatogonia, and mitochondria are completely absent in the IMM of spermatogonia and spermatocytes in Ef4 KO mice. In addition, mitochondria displayed disjointed mitochondrial sheaths formed with irregular size and seriously reduced membrane organization or absent cristae, as well as decreased levels of mtDNA-encoded OXPHOS subunits, in Ef4 KO mice. Furthermore, EF4 deletion results in accelerated mitochondrial translation, causing excessive production of possibly incompletely folded products, accompanied by a reduced mTOR signaling pathway and reduced cytoplasmic translation in the testis. Interestingly, unlike the EF4 deletion testis, EF4 depletion in the heart results in normal mitochondria, increased mTOR signaling pathway and increased cytoplasmic translation, suggesting the distinct role of EF4 in different types of tissues and cells. Taken together, this finding elucidates the cross-talk between EF4-dependent mitochondrial quality control and the mTOR pathway, and when such a cross-talk is uncoupled, developmental defects are expected to occur, such as defects in spermatogenesis [74]. MFN2 is also involved in the mRNA translation during spermatogenesis via its association with the polysome and interacts with the translational regulator MSY2 (exclusively enriched in both the meiotic and post-meiotic germ cells and participates in both transcription and mRNA translation [76, 77]) in spermatogenic cells to regulate the fate of MSY2-bound gamete-specific mRNAs [62].
Another study revealed a new insight into the role of mitochondrial proteins in regulating epigenetic modification during spermatogenesis. Prohibitin (PHB or PHB1) is an evolutionarily conserved IMM protein that participates in both mitochondrial (respiratory chain subunit assembly, biogenesis, and mitophagy) and non-mitochondrial functions [78–80]. PHB is especially highly expressed in spermatocytes, and its loss in spermatocytes using Stra8-Cre leads to male infertility with meiosis blockage at the pachytene stage [81]. Apart from the reduced respiratory complex subunits in PHB mutant spermatocytes, STAG3 shows a significant decrease, a component of meiosis-specific cohesion essential for chromosome pairing and formation of the synaptonemal complex (SC), level is significantly reduced. Meanwhile, incomplete SC formation and impaired crossover have been detected in PHB mutant spermatocytes [81]. Interestingly, to explain how the IMM located in PHB can influence synapsis and double-strand break (DSB) repair occurring in the nucleus, the axis of PHB-JAK2-H3Y14ph has been demonstrated to be a novel mechanism in both the safeguarding of the stabilization of the meiotic STAG3 cohesin complex and regulating heterochromatin formation in spermatocytes [81]. PHB in spermatocytes predominantly affects the JAK/STAT and PI3K/AKT pathways, which can exert regulatory effects on meiotic cells via a non-canonical pathway likely involving dynamic epigenetic changes in histone modifications [81]. Therefore, this study adds a new layer of regulatory complexity of mitochondria in regulating epigenetic modifications and, thus, male fertility.
Mutations in mtDNA are a common cause of mitochondrial diseases, including male infertility, mtDNA point mutations, and deletions that negatively affect semen quality in humans. Mitochondrial transcription factor A (TFAM) wraps mammalian mtDNA, packaged into mitochondrial nucleoids [82], and directly influences the mtDNA copy number. The deletion of TFAM can lead to defects in spermatogenesis; the older the age, the worse the defects. Compared with PolgAmutTfamWT testes, PolgAmutTfamko testes show many severe defects with gradual loss of late-stage spermatocytes and round spermatids, and the absence of sperm in the cauda after spermiogenesis is completed around P25 [83]. However, overexpression of TFAM (PolgAmutTfamOE mice) in the mtDNA mutator PolgAmut/mut mice, which bear an exonuclease-deficient version of the catalytic subunit of the mitochondrial DNA polymerase and develop a premature aging phenotype with reduced male fertility [84, 85], can rescue the fertility phenotype and normalize testis morphology. Furthermore, consistent with morphology, PolgAmutTfamOE mice showed better preserved COX activity and normal mitochondrial cristae remodeling than those in PolgAmutTfamwt mice and PolgAmutTfamko mice [83]. Importantly, reversed global proteomic changes in PolgAmutTfamOE mouse spermatocytes, especially restoring the protein levels of OXPHOS, mitochondrial translation, and mitochondrial RNA regulation. Based on these data, increasing mtDNA copy number would be an elegant way to treat the respiratory chain dysfunction caused by heteroplasmic mutations, providing a potential target for pharmacological agents.
Reactive oxygen species (ROS) or intermediates are produced by the incomplete reduction of oxygen. Lipid hydroperoxides are a type of ROS whose biological functions are still poorly understood. Phospholipid hydroperoxide glutathione peroxidase (PHGPx) is a selenoprotein belonging to the family of glutathione peroxidases. It has been engaged in antioxidative defense and spermatogenesis and comprises almost the entire selenium content of mammalian testes [86]. PHGPx has three different forms in the testes: a cytosolic enzyme, a protein with a mitochondrion-targeting peptide, and a splice variant targeting the nucleus [87–89]. PHGPx is specifically expressed in postmeiotic male germ cells and Leydig cells in mouse testes, and it is conserved from Drosophila to humans and localized in the cytoplasm of round spermatids and the mitochondrial sheath of the spermatozoa midpiece [90]. PHGPx expression is dramatically decreased in patients with oligoasthenozoospermia, a defect associated with a significant decrease in both the number and the motility of spermatozoa [91]. Spermatocyte-specific PHGPx KO mice are infertile, with a significant decrease in the number of spermatozoa, substantial reduction in mitochondrial membrane potential, mitochondrial swelling, and hairpin-like bend at the midpiece of epididymal spermatozoa, leading to a severe reduction in the forward motility of spermatozoa [91].
Through immunoprecipitation (IP)-Mass spectrometric (MS) analysis using an anti-MILI antibody in germline stem (GS) cells, several MILI-binding proteins have been identified, including several well-known piRNA pathway components, such as MOV10L1. Unexpectedly, Glycerol-3-phosphate acyltransferase 2 (GPAT2), an OMM-located protein, interacts with MILI, and knockdown of GPAT2 results in impaired piRNA production in GS cells [92]. In mammals, there are four GPAT isoforms, GPAT1-GPAT4, which differ in their subcellular location, tissue expression pattern, substrate preference, transcriptional regulation, and sensitivity to sulfhydryl group reagents such as N-ethylmaleimide [93]. GPAT1 and GPAT2 are located in the mitochondria, whereas GPAT3 and GPAT4 dwell in the ER. Unlike other GPAT members, which are highly expressed in lipogenic tissues such as the liver and adipose tissue and are essential for the de novo synthesis of triacylglycerol and glycerophospholipids, GPAT2 shows the highest expression in the testis [94]. During mouse spermatogenesis, GPAT2 is expressed from E13.5 to the adult stage, with maximal expression at the pachytene stage [95, 96], and its transcription is controlled via epigenetic mechanisms in combination with retinoic acid [95]. GPAT2-deficient female mice are fertile, while male mice are infertile, with rarely detected pachytene spermatocytes and absence of sperm in the caudal epididymis [96]. Notably, GPAT2-deficient E15.5 prospermatogonia exhibit impaired IMC formation, decreased piRNA production, and DNA methylation of the retrotransposon genes, and thus elevated expression levels of IAP and Line-1 transcripts. Interestingly, the binding of PIWIL4 with piRNA is required for PIWIL4 to translocate from the cytoplasm to the nucleus, and PIWIL4 failed to enter the nucleus in GPAT2-deficient E15.5 prospermatogonia, suggesting that PIWIL4 proteins do not bind to piRNAs under these conditions [96]. Furthermore, the loss of germ cells in GPAT2-deficient mice occurred much earlier, at around D7, than at D14 in MILI-deletion mice. Since spermatogonia commence cell division at approximately D2 after birth, and apoptosis is detected in D3 GPAT2-deficient testes, indicating the role of GPAT2 in inhibiting spermatogonia apoptosis [96]. Although GPAT2 can directly bind to MILI, and GPAT2- and MILI-deficient mice share similar phenotypes, including apoptotic cell death of pachytene spermatocytes and reduction in the DNA methylation of retrotransposons, however, spermatogonia are dramatically decreased in GPAT2-deficient mice but not in MILI-deficient mice, indicating the additional roles of GPAT2 during spermatogenesis [96], including piRNA processing and germ cell apoptosis.
Some proteins are highly expressed during meiosis; however, their roles in spermatogenesis remain unexplored. For instance, in mice, the short Ca2+-binding mitochondrial carrier (SCaMC) is one of the two mitochondrial carriers responsible for transporting ATP across the IMM [97]. SCaMC-1L is preferentially expressed in the testis, and its intracellular localization exhibits dynamic changes during spermiogenesis with no limitation to mitochondrial structures. SCaMC-1L is highly expressed in the IMC of spermatocytes, chromatoid bodies in spermatids, mitochondrial sheaths in the middle pieces of spermatozoa, and other unknown germinal granules [98].
Mitochondria in spermatids and spermatozoa
Haploid spermatids are formed after meiotic division of spermatocytes and undergo a series of morphological transformations, termed spermiogenesis, into mature spermatozoa. After completing a series of metamorphosis processes, spermatids develop into spermatozoa, which are motile male gametes with a round or elongated heads and long posterior flagella. In the early stages of spermatozoa formation, the cytoplasm is relatively evenly surrounded by the nucleus, and the mitochondria move to the opposite side of the acrosome. They gather at the base of the axons and form a mitochondrial sheath in the middle of the spermatid tail [99]. As the cells develop into mature spermatozoa, the cytoplasm is reduced gradually, and finally, only a moon bud-shaped cytoplasmic band between the acrosome and nucleus is left. Mitochondria are observed to be arranged in the periphery of tail microtubules and are pivotal to the metabolism and motility of spermatozoa [100]. They engage in energy generation, respiration balance, calcium modulation, flagellar motility, sperm capacitation, acrosome reactions, and apoptotic pathways. Any alteration in the mitochondrial function described above is linked to a reduction in sperm quality or male infertility. The morphology and distribution of mitochondria dramatically change during spermiogenesis [35]; elongate and cluster during meiosis, a fragment in post-meiotic spermatids arranged longitudinally around the axoneme of the midpiece, and forming a compact mitochondrial sheath in a step-wise and coordinated manner, which fuels sperm motility [39]. Although the molecular mechanisms that underlie mitochondrial functions have mostly been deciphered using cultured cells, the role of mitochondrial activities in spermatids and spermatozoa remains largely unknown. Nevertheless, recent studies have revealed several key factors during spermiogenesis, especially in regulating mitochondrial transformation and rearrangements.
Recently, a study revealed that fully saturated tetrapalmitoyl cardiolipin (TPCL) is not limited to the mitochondrion but is also located in the sperm acrosome and regulates male meiosis during spermatogenesis [101], which challenges the established notion that cardiolipin (CL) is only located in the mitochondrion [102]. In eukaryotes, CL is a phospholipid that carries four fatty acids with a strong preference for unsaturated chains and is specifically located in the mitochondrial membranes. CL comprises 10–20% of the lipid content of the IMM [103] and participates in several mitochondrial functions, including mitochondrial energy production [104], cristae morphology [105], fusion and fission [106–108], apoptosis [109] and mitophagy [110]. The accumulation of monolyso cardiolipin (MLCL), a variant of CL, in the mitochondrion results in Barth syndrome (BTHS) in humans [111]. TPCL, a fully saturated species of CL, was first discovered in the testes of rats [112], and it exists in the testes of mice and humans [101], confirming that its presence is a common phenomenon. Interestingly, TPCL is exclusively expressed in the testis, but not in other tissues, and is more abundant than any other CL species [101]. TPCL emerges with germ cells, reaches the maximal level in spermatids, and partially decreases in mature sperm from caudal epididymis. Except for its location in the mitochondria of the sperm midpiece, it is unexpectedly distinctively expressed in the acrosome, because CL is traditionally present in the mitochondrion [101]. Because TPCL originates from mitochondria, its distinct location in the acrosome implies a novel function of mitochondria in acrosome formation. Furthermore, the deletion of TAZ in mice, a mitochondrial enzyme responsible for the high unsaturation of CL [113], leads to the prevention of TPCL formation and male infertility owing meiosis inhibition with the dramatic loss of round spermatids and the near absence of elongated spermatids. Condensed mitochondria are abundant form in normal spermatocytes and gradually transition to intermediate and orthodox states in spermatids. However, all TAZ-deficient germ cells show a significant increase in orthodox mitochondria and a decrease in condensed mitochondria in spermatocytes, indicating the role of TAZ in regulating the mitochondrial transformation from orthodox to condensed type during early meiosis [101]. Notably, several mitochondrial proteins, including Ant4, Spata18, Suox, Smcp, Gldc, and Slc25a19, are present in the acrosome. ANT4 is located in the IMM, and Ant4-deficient male mice are infertile due to early meiosis arrest [114, 115]. SPATA18 is a mitochondrion-eating protein that regulates the clearance of damaged mitochondria [116, 117]. ANT4, SPATA18, and SUOX also localize to the mitochondrial piece and acrosome of sperm, similar to the expression pattern of TAZ. Interestingly, Ant4-deficient mice display a similar but more severe phenotype than that in TAZ-deletion mice, with a complete lack of TPCL, transformational inhibition of orthodox into condensed mitochondria, and complete loss of spermatids. Furthermore, Ant4, Spata18, and Suox are proved to be present in nascent acrosomal granules, suggesting the translocation of these mitochondrial proteins into the acrosome [101]. These data demonstrate the regulators of mitochondrial transformation from orthodox to condensed form and provokingly reveal a novel function of the mitochondrion in the biogenesis of acrosome, although detailed mechanisms need to be developed.
Several ARM family proteins have been reported to be related to mitochondria [118–120] or spermatogenesis [121, 122]; however, their roles in mitochondrial sheath formation remain largely unexplored until the study of Armadillo repeat-containing 12 (ARMC12) function in knockout mice. ARMC12 is an OMM protein that is evolutionarily conserved among species, enriched in the testis, and begins to express around D20, corresponding to the haploid round spermatid stage. The protein is expressed in steps 10–16 spermatids and localizes in the midpiece of step 16 spermatids [123]. The knockout of Armc12 in mice causes male infertility due to dramatically decreased sperm motility and defects in sperm-zona pellucida binding capacity during in vitro fertilization (IVF) [123]. Wild-type (WT) spermatozoa can pass the oviduct through the utero-tubal junction (UTJ), whereas ARMC12-deficient sperm arrives at the uterus near the UTJ and fails to pass through the UTJ into the oviduct because they cannot keep their head at the apex while swimming [123]. Mitochondria start disorganizing in step 16 spermatids, which cannot coil along the flagellum at the interlocking step, implying the role of ARMC12 in spatiotemporal mitochondrial dynamics during mitochondrial formation [123]. Furthermore, ARMC12 interacts with several mitochondrion-related proteins in male germ cells, including TBC1D21, GK2, MIC60, VDAC2, and VDAC3. TBC1D21 and GK2 show testis-enriched expression in both mice and humans. GK2 is specifically localized to the OMM in the round and elongating spermatids, interacts with PLD6 (MitoPLD) and induces PLD6 and phosphatidic acid (PA)-dependent mitochondrial clustering in cells [124]. Gk2 knockout mice show male infertility with similar mitochondrial defects to those observed in ARMC12-deficient mice, including an improper arrangement of mitochondria at the interlocking step, disorganization of mitochondrial sheath formation, and failure to pass through the UTJ [124, 125]. Similarly, Tbc1d21 knockout male mice are sterile, with an abnormality in spermatozoa from the cauda epididymis. Tbc1d21 knockout spermatids show an abnormal interlocking structure and thus improper mitochondrial sheath formation [123, 126]. Furthermore, in the absence of TBC1D21, the interactions between ARMC12-VDAC2/3 are disrupted, revealing the cooperative role of ARMC12 and TBC1D21 in regulating proper mitochondrial sheath formation [123]. Interestingly, the mRNA levels of TBC1D21 in sperm cells decreased in men with teratozoospermia (n = 13) compared with those in healthy controls (n = 8), suggesting a significant association between TBC1D21 and the quantity of human sperm [126].
Mitochondrial fission is controlled by GTPase dynamin-related protein 1 (Drp1), whose recruitment is mediated by several OMM proteins, including mitochondrial fission factor (MFF), mitochondrial fission protein 1 (FIS1), and mitochondrial dynamics proteins of 49 kDa and 51 kDa (MiID49 and MID51, respectively) [127]. DRP1 deletion in mice is embryonically lethal [128], and no male germ cell-specific knockout mice have been reported. Male germ cell-specific Fis1 knockout mice using Stra8-Cre driver exhibits male infertility, showing multinucleated spermatid giant cells, accumulated aberrant mitochondria in round spermatids and giant cells, and fragmented and dispersed acrosome structures throughout the cytoplasm in the spermatids [129]. This study investigated the essential role of mitophagy in regulating mitochondrial density in spermatids; however, it remains unclear how mitochondria are being cleared and why the acrosome is also being impaired upon FIS1 deletion, leaving a question about the relationship between mitochondrial function and acrosome formation. Furthermore, mice with a gene trap disruption of Mff (Mffgt) die prematurely with a mean life span of 13 weeks caused by severe dilated cardiomyopathy and heart failure, and display decreased fertility in both males and females [130]. Ablation of MFF does not impair the generation of any one cell type, including spermatogonia, spermatocytes, spermatids and spermatozoa; however, sperm from the caudal epididymis often bear morphological abnormalities in the midpiece, presenting as disjointed mitochondrial sheaths with gaps between adjacent organelles, kinks in over 60% of Mffgt sperm, and swollen mitochondria, and thus exhibit poor recruitment of mitochondria to the sperm midpiece and/or cannot properly wrap around the axoneme. Subsequently, Mffgt sperm showed reduced respiratory chain complex IV activity, motility, and fertility [131]. Although more data are needed to elucidate the role of mitochondrial fission in mitochondrial rearrangement, this study suggests that fission is vital for producing relatively small mitochondria that are more easily be packed around the midpiece. After the alignment of mitochondria in the midpiece of spermatozoa, the connection between mitochondria and outer-dense fibers (ODFs) need to be well maintained, and multiple proteins play an essential role in the mitochondria-ODF binding, including GOPC, GPX4 and KLC3, and SPATA19, etc., as reviewed previously [39].
In summary, mitochondria shoulder pivotal missions during germ cell evolution, both in the development and maintenance of germ cells and gametes, as well as in successful fertilization and early embryonic development. Mitochondrial functions are not limited to energy support but also extend to stemness maintenance and reprogramming, metabolic shift, spermatogonia differentiation, Nuage formation, piRNA processing, epigenetic modification, acrosome formation, sperm motility, etc. (Fig. 1). Vital mitochondrial regulators are summarized in Table 1.
Table 1.
Summarization of key mitochondrial regulators during male germ cell development
| Germ cell types | Name | Mitochondrial location | Functions | Phenotypes in mutant mice | References |
|---|---|---|---|---|---|
| Primordial germ cell (PGC) | αKG | Matrix | Mouse ESC self-renewal, differentiation of human ESCs, PGC fate enhancement | Not available | [40] |
| mitosRNA | Matrix | Synchronization and nuclear-mitochondrial communication | Not available | [45] | |
| Prospermatogonia (gonocytes) | TDRD1 | IMC | piRNA biogenesis | Male infertile, female fertile | [48, 49] |
| MITOPLD | IMC, mitochondria | Primary piRNA processing | Male infertile, female fertile | [53, 54] | |
| GASZ | IMC, mitochondria | piRNA metabolism | Male infertile, female fertile | [55, 66] | |
| Spermatogonia | MFN1 | OMM | Mitochondrial fusion | Male infertile | [62, 63, 65, 66] |
| MFN2 | OMM, MAM | Mitochondrial fusion | Male infertile | [62, 63, 65] | |
| SHP2 | Mitochondria | Cell growth and development, tissue inflammation, and cellular chemotaxis | Male infertile | [69, 70] | |
| Spermatocyte | EF4 | OMM | Quality-control in translation | Male infertile | [74] |
| PHB | IMM | Respiratory chain subunits assembly, mitochondrial biogenesis, and mitophagy | Male infertile | [81] | |
| TFAM | Matrix | mtDNA copy number | Male infertile | [83] | |
| PHGPx | Mitochondria | antioxidative defense | Male infertile | [91] | |
| GPAT2 | Mitochondria | de novo synthesis of triacylglycerol and glycerophospholipids | Male infertile, female fertile | [92, 96] | |
| Spermatid and spermatozoon | TAZ | IMM | Tetrapalmitoyl-CL (TPCL) formation | Male infertile | [101] |
| ANT4 | Mitochondria | Exchange ADP and ATP across lipid bilayers | Male infertile | [101, 114, 115] | |
| ARMC12 | OMM | Mitochondrial sheath formation | Male infertile | [123] | |
| GK2 | OMM | Glycerol metabolism, mitochondrial sheath formation | Male infertile | [124, 125] | |
| TBC1D21 | OMM | Mitochondrial sheath formation | Male infertile | [123, 126] | |
| FIS1 | OMM | Mitochondrial fission and mitophagy | Male infertile | [129] | |
| MFF | OMM | Mitochondrial sheath formation | Male sub-infertile | [131] |
αKG, α-ketoglutarate; ESC, embryonic stem cell; TDRD1, Tudor domain-containing 1/Mouse Tudor repeat-1; IMC, inter-mitochondrial cement; MitoPLD, phospholipase D family member 6; OMM, outer mitochondrial membrane; MAM, mitochondrial-associated ER membrane; IMM, inner mitochondrial membrane; MFN, The mitochondrial fusion factor mitofusins; EF4, elongation factor 4; PHB, prohibitin; TFAM, mitochondrial transcription factor A; PHGPx: phospholipid hydroperoxide glutathione peroxidase; GPAT2, glycerol-3-phosphate acyltransferase 2; TAZ, tafazzin; ANT4, ADP/ATP translocase 4; ARMC12, armadillo repeat-containing 12; GK2, glycerin kinase 2; TBC1D21, TBC1 domain family member 21; MFF, mitochondrial fission factor
The role of Sertoli cell-derived factors in regulating mitochondrial function
Sertoli cells are testicular somatic cells that are essential for testis formation and spermatogenesis via direct contact with germ cells and by controlling the environmental milieu within the seminiferous tubules. During meiosis entry, mitochondria undergo a morphological transformation from the orthodox type with rich cristae into the intermediate type with an initial dilatation of intracristal spaces in early spermatocytes, to the condensed type with almost no cristae which are typically in pachytene spermatocytes and early spermatids [35]. Studies have shown that Sertoli cells participate in regulating mitochondrial morphology through important paracrine modulators, one of which is activin signaling [132]. Activins belong to the transforming growth factor-beta (TGF-β) superfamily and were originally isolated for their capacity to stimulate the release of follicle-stimulating hormone (FSH) [133, 134]. Except for their multiple biological functions, including the control of inflammation, fibrosis, developmental biology, and tumorigenesis, activins are well known for their role in regulating reproduction, as reviewed previously [132, 135]. The testis is one of the major sites for activin production; activin A levels begin to increase at approximately E12.5 testis, reaching a peak on the day of birth [136], and in postnatal testis, activin A levels are highest in the first week in mice [137, 138]. In cell culture experiments, activin A secreted by Sertoli cells can act on meiotic germ cells and maintain the physiological “condensed” phenotype of mitochondria in primary spermatocytes in vitro. The number of condensed mitochondria is decreased upon the inactivation of activin A [139]. Furthermore, activin A has been shown to be essential in maintaining normal energy balance mainly via its downstream effects on mitochondrial energy metabolism, since the mutation of activin A-encoding gene Inhba, InhbaBK/BK mice (the sequence encoding the Inhba mature domain is replaced with that of Inhbb, rendering the gene product functionally hypomorphic [140]) exhibits many disproportionately small tissues relative to the total body weight accompanied by increased mitochondrial energy metabolism and upregulated genes associated with enhanced mitochondrial biogenesis and function [141]. Regarding the influence of activin on male fertility, InhbaBK/BK mice bear small testes in size with a 9-day delay in the onset of fertility [140], and the complete loss of activin A causes male infertility due to testicular dysgenesis characterized by reduced Sertoli cell numbers and failure in testicular cord elongation in Inhba−/− mice [136]. However, activin B-deficient Inhbb−/− mice remain fertile[142], indicating the essential role of activin A but not activin B in testicular development. Although the mitochondrial dysfunction has been explored in other tissues in the previous studies [141], the mitochondrial morphology and function in the testis are yet to be discovered, making the mechanism by which activin regulates morphology transition and function during spermatogenesis largely unknown.
Mitochondrial defects and their links to infertility in humans
Infertility affects approximately10–15% of couples globally, and male factors account for approximately 50% of infertile couples overall [143], including reduced spermatozoa numbers (oligozoospermia or azoospermia), increased abnormal spermatozoa with morphological defects (teratozoospermia) and reduced sperm motility (asthenozoospermia). Mitochondrial dysfunction reduces aerobic energy production, which is essential not only for sperm motility but also for hyperactivation, acrosome reaction and oocyte penetration. Considering the vital role of the mitochondria in sperm motility, individuals with asthenozoospermia (low sperm motility) are suitable candidates for mutation screening. The link between primary mitochondrial defects and male infertility was first reported in four-generation men with mitochondrial encephalomyopathy [144], a heterogeneous clinical disorder due to abnormal mitochondrial dysfunction, especially affecting the brain, nervous system, and muscles [145]. Neither the patient nor his four brothers had children, whether or not they had been married. Up to 92% of sperm from the patient were abnormal in the midpieces, characterized by increased matrix, thickening of the membranes (especially the outer membranes), and a disarrangement of cristae [144]. Abnormal mitochondrial function has been shown to be associated with asthenozoospermia/oligoasthenozoospermia, and the mitochondrial abnormalities include a short midpiece, total absence of mitochondrial sheath, defective assembly, and/or clustering of mitochondria. However, the primary mitochondrial sheath defects confined to the midpiece are relatively rare, as previously reviewed [146]. A combination of axonemal abnormalities are much more common, such as a special type of asthenozoospermia called multiple morphological anomalies of sperm flagella (MMAF), which is usually characterized as a combination of flagellar morphological abnormalities (absent, short, bent, coiled, and irregular flagella) and seriously reduced motile ability; however, usually, no primary ciliary dyskinesia (PCD) respiratory features are observed. Spermatozoa from patients with MMAF usually bear developmental defects of the mitochondrial sheath, flagellum elongation and mainly absence of central pairs in the axoneme [146]. The relationship between sperm parameters and mitochondrial defects, such as electron transport chain (ETC), reactive oxygen species (ROS), mtDNA integrity, calcium homoeostasis, and apoptosis pathway, has been summarized in recent literature [100, 147, 148]. Moreover, the sperm flagellum harbors distinct peri-axonemal structures, which are absent in other motile cilia, including the mitochondrial sheath in the midpiece, ODFs in the midpiece and the proximal part of the principal piece, and the fibrous sheath in the principal piece [149]. Interestingly, approximately 20 MMAF-related genes have been identified in the last 5 years, and most of their mutations lead to defects in the mitochondrial sheath in patients with MMAF and/or knockout/mutation transgenic mouse models (Table 2), such as CFAP251, whose mutation leads to mainly mitochondrial sheath defects in men.
Table 2.
Mitochondrial defects caused by gene mutated in MMAF infertile men
| Gene | location | Sperm phenotype in human | Phenotypes in mutant mice | References |
|---|---|---|---|---|
| WDR66 (CFAP251) | Sperm flagellum | Abnormal midpiece length, dysplasia of mitochondrial sheath, with very low mitochondrial density | Not available | [151] |
| AK7 | Sperm flagellum and cilia | Serious disarrangement of the (9 + 2) axonemal structure, incomplete mitochondrial sheath and dysplasia of the fibrous sheath | PCD and MMAF phenotypes, severe in mice than that of human | [152–154] |
| CFAP58 | Sperm flagellum, high in midpiece | Absence and/or disorganization of central pair of microtubules, nine doublets of microtubules, nexin, dynein arms, disarranged mid-piece with shortened mitochondrial sheath, misshapen mitochondrial sheath, fibrous sheath, and outer dense fibers | Typical MMAF phenotypes and infertility | [155–157] |
| FSIP2 | Fibrous sheath in mammalian, centriole in human | Absence of mitochondrial sheath from the midpiece although the outer dense fibers remain present, hypertrophic FS extended up to the sperm neck, variable defects of axoneme | Typical MMAF phenotypes and infertility in mutation knock in mice, no gross anomaly except for sperm tails with increased length in over expression mice | [158–161] |
| DNAH1 | Inner dynein arm | Supernumerary dense fibers with absence of mitochondrion, lack of inner dynein arm, disorganized fibrous sheath and microtubule doublets | Infertile due to defects in sperm motility, but no MMAF phenotypes | [162–164] |
| DNAH2 | Inner dynein arm | Severely disarranged axonemal structures with mitochondrial sheath defection | MMAF phenotypes, characterized with absence or irregular mitochondrial arrangement, misaligned outer dense fiber and microtubule doublets in the midpiece of sperm | [165–167] |
| DNAH6 | Inner dynein arm | Disorganized mitochondrial sheath or fibrous sheath, and a lack of axonemal central pair complex | Not available | [168] |
| DNAH10 | Inner dynein arm | Absence of inner dynein arms, and a dramatic disarrangement of peri-axonemal structures including mitochondrial sheath, outer dense fibers, and fibrous sheath | MMAF and infertility in both knockout and mutant knock in mice | [169] |
| DNAH17 | Outer dynein arm | Distorted axonemal structures with a high frequency of abnormal or fragmented mitochondrial sheath, disrupted 9 + 2 microtubular array | MMAF and infertility | [170–172] |
| CFAP43 | Sperm flagellum | Severe axonemal and peri-axonemal defects affecting the mitochondrial sheath, outer dense fibers, and fibrous sheath | Typical MMAF phenotypes and infertility | [173, 174] |
| CFAP44 | Sperm flagellum | Similar with that of CFAP44 | MMAF [174] or asthenozoospermia without MMAF [173] | [173, 174] |
| CFAP65 | Acrosome and midpiece using N-terminal antibody, whole sperm flagellum using C-terminal antibody | Misshapen heads and multiple tail abnormalities including poorly assembled mitochondrial sheaths and misarranged fibrous sheaths | MMAF, with malformed sperm head with abnormal acrosomes, misassembled mitochondrial sheath, deficient manchette organization, and destroyed 9 + 2 structures | [175–178] |
| CFAP69 | Midpiece of sperm flagellum | Head malformations, in particular thin heads and an abnormal acrosomal region | MMAF, with detached mitochondrial sheath, irregular outer dense fibers, disorganized fibrous sheath, disrupted axoneme | [179] |
| TTC21A | NA | Disorganized axonemal components, hyperplasia of the fibrous sheaths | Asthenoteratospermia and subfertility, mid-pieces could not be formed without being surrounded by mitochondria and without relocation of the annulus | [180] |
| TTC29 | Concentrated at the base of sperm flagella and faintly along flagella | A reduced mitochondrial sheath and abnormal axonemal cross sections with a lack of the central pair | Asthenoteratospermia and subfertility (no MMAF) | [181, 182] |
During spermiogenesis, mitochondria are precisely controlled to align into the midpiece of the flagellum; however, a number of mitochondria are tagged for incorporation into the midpiece, while the majority remain within the spermatid cytoplasm and are eventually destroyed. In addition, the molecular mechanism by which mitochondria are recruited and subsequently attached in the midpiece remains largely unknown. However, mitochondrial sheath defects frequently occur in models that harbor defects in the manchette structure, ODFs, and/or 9 + 2 structures, especially in patients with MMAF, providing promising clues to discover the mechanism of mitochondrial recruitment, attachment, and maintenance in the midpiece during sperm tail formation.
Conclusion and future perspectives
In this review, we describe the dynamic changes in mitochondria during germ cell development, from PGCs to spermatozoa. Beyond discussing the characteristics of mitochondria, we aimed to highlight the new roles of mitochondrion-located regulators during spermatogenesis. Together with recent findings that mitochondria could extend their influence to epigenetic modifications in the nucleus and acrosome formation, these emerging roles imply that the mitochondrion has multifaceted functions in regulating germ cell development.
Male germ cells have distinct metabolic requirements depending on their differentiation state. The burst of scRNA-seq data and related functional studies enabled us to reveal the metabolic patterns in a different types of germ cells. Metabolism plays a crucial role in regulating SSC maintenance and differentiation, and maintenance of glycolysis is imperative to sustain the long-term regenerative capacity of SSCs. Mitochondrial biogenesis and activities are elevated in progenitor and differentiating spermatogonia, suggesting that a switch from glycolysis to OXPHOS occurs in association with spermatogonial differentiation [58]. In contrast, spermatocytes and spermatids require lactate and pyruvate for survival, OXPHOS activity increases during meiotic prophase I, and mitochondrial fusion proteins play pivotal roles during the metabolic shift, as reviewed previously [12]. Therefore, OXPHOS fuels mammalian spermatogenesis and, in particular, meiosis. However, future studies should address this mechanism via the establishment of more models by genetically removing other mitochondrial proteins.
The finding that TPCL and other mitochondrial proteins, can assemble into the acrosome, suggests a novel role for the mitochondria in acrosome formation. Condensed mitochondria probably provide an additional source for the acrosome; even though the contribution of mitochondria to the acrosome mass seems small, it is pivotal because the absence of TPCL leads to the inhibition of acrosome formation and male infertility [101]. This study also raises further questions, such as what signal(s) initiate the translocation of mitochondrial proteins into the acrosome? Mitochondrial proteins undergo post-translational modifications when translocated into acrosomes. What are the functions of these proteins upon incorporation into acrosomes? Furthermore, TPCL ablation causes the inhibition of mitochondrial transformation from the orthodox to condensed type, during which cristae undergo dramatic remodeling. Intriguingly, CL plays an emerging role in cristae remodeling [4]. Mammalian germ cell development is a promising process for investigating this intricate issue because various mitochondria change significantly, especially in their morphology. Additionally, several mitochondrial proteins are localized to mouse chromatoid bodies, including piRNA pathway components, as previously reviewed [73, 150], and COX1, which is encoded by the mitochondrial genome and usually confined inside the mitochondrion, is localized to mouse chromatoid bodies [150]. These data indicate a connection between the mitochondrion and other organelles.
In the future, a key area of research will be to define the association between the mitochondrion and other organelles, especially the ER, nucleus, acrosome, and mitochondrion-associated germinal granules. This will require further understanding of how mitochondria transform and interact with other organelles. As discussed here, the mitochondrion undergoes continuous mitochondrial transformation during germ cell development; it will be challenging but intriguing to address the communication and crosstalk between the mitochondrion and other organelles, thus revealing various functions.
Author contributions
XW, LY, and YW reviewed the literature. LY and XW wrote the manuscript. SY conceived and revised the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31801237 to X.W. and 81971444 to S.Y.), and the Strategic Collaborative Research Program of the Ferring Institute of Reproductive Medicine, Ferring Pharmaceuticals, and the Chinese Academy of Sciences (FIRMSC200502 COV02 to S.Y.).
Declarations
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaoli Wang and Lisha Yin contributed equally.
References
- 1.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467(7318):929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 2.Patananan AN, et al. Modifying the mitochondrial genome. Cell Metab. 2016;23(5):785–796. doi: 10.1016/j.cmet.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarty RP, Chandel NS. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28(3):394–408. doi: 10.1016/j.stem.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kondadi AK, Anand R, Reichert AS. Cristae membrane dynamics - a paradigm change. Trends Cell Biol. 2020;30(12):923–936. doi: 10.1016/j.tcb.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutlu B, Puigserver P. GCN5 acetyltransferase in cellular energetic and metabolic processes. Biochim Biophys Acta Gene Regul Mech. 2021;1864(2):194626. doi: 10.1016/j.bbagrm.2020.194626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin E, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470(7334):359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He C, et al. NOX4 modulates macrophage phenotype and mitochondrial biogenesis in asbestosis. JCI Insight. 2019 doi: 10.1172/jci.insight.126551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma A, et al. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep. 2019;20(12):e48395. doi: 10.15252/embr.201948395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan DC. Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol. 2020;15:235–259. doi: 10.1146/annurev-pathmechdis-012419-032711. [DOI] [PubMed] [Google Scholar]
- 11.Giacomello M, et al. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21(4):204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 12.Varuzhanyan G, Chan DC. Mitochondrial dynamics during spermatogenesis. J Cell Sci. 2020 doi: 10.1242/jcs.235937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24(12):761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Yoo SM, Jung YK. A molecular approach to mitophagy and mitochondrial dynamics. Mol Cells. 2018;41(1):18–26. doi: 10.14348/molcells.2018.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ploumi C, Daskalaki I, Tavernarakis N. Mitochondrial biogenesis and clearance: a balancing act. FEBS J. 2017;284(2):183–195. doi: 10.1111/febs.13820. [DOI] [PubMed] [Google Scholar]
- 16.Onishi M, et al. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3):e104705. doi: 10.15252/embj.2020104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol. 2020;21(2):85–100. doi: 10.1038/s41580-019-0173-8. [DOI] [PubMed] [Google Scholar]
- 18.Dorstyn L, Akey CW, Kumar S. New insights into apoptosome structure and function. Cell Death Differ. 2018;25(7):1194–1208. doi: 10.1038/s41418-017-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honarpour N, et al. Adult apaf-1-deficient mice exhibit male infertility. Dev Biol. 2000;218(2):248–258. doi: 10.1006/dbio.1999.9585. [DOI] [PubMed] [Google Scholar]
- 20.Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110(2):521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 21.Lin YT, Capel B. Cell fate commitment during mammalian sex determination. Curr Opin Genet Dev. 2015;32:144–152. doi: 10.1016/j.gde.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vergouwen RP, et al. Proliferative activity of gonocytes, sertoli cells and interstitial cells during testicular development in mice. J Reprod Fertil. 1991;93(1):233–243. doi: 10.1530/jrf.0.0930233. [DOI] [PubMed] [Google Scholar]
- 24.Law NC, Oatley MJ, Oatley JM. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat Commun. 2019;10(1):2787. doi: 10.1038/s41467-019-10596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nebel BR, Amarose AP, Hacket EM. Calendar of gametogenic development in the prepuberal male mouse. Science. 1961;134(3482):832–833. doi: 10.1126/science.134.3482.832. [DOI] [PubMed] [Google Scholar]
- 26.Chen CT, Hsu SH, Wei YH. Mitochondrial bioenergetic function and metabolic plasticity in stem cell differentiation and cellular reprogramming. Biochim Biophys Acta. 2012;1820(5):571–576. doi: 10.1016/j.bbagen.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Rehman J. Empowering self-renewal and differentiation: the role of mitochondria in stem cells. J Mol Med (Berl) 2010;88(10):981–986. doi: 10.1007/s00109-010-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamboni L, Merchant H. The fine morphology of mouse primordial germ cells in extragonadal locations. Am J Anat. 1973;137(3):299–335. doi: 10.1002/aja.1001370305. [DOI] [PubMed] [Google Scholar]
- 29.Clark JM, Eddy EM. Fine structural observations on the origin and associations of primordial germ cells of the mouse. Dev Biol. 1975;47(1):136–155. doi: 10.1016/0012-1606(75)90269-9. [DOI] [PubMed] [Google Scholar]
- 30.Sathananthan H, Pera M, Trounson A. The fine structure of human embryonic stem cells. Reprod Biomed Online. 2002;4(1):56–61. doi: 10.1016/s1472-6483(10)61916-5. [DOI] [PubMed] [Google Scholar]
- 31.Jansen RP, de Boer K. The bottleneck: mitochondrial imperatives in oogenesis and ovarian follicular fate. Mol Cell Endocrinol. 1998;145(1–2):81–88. doi: 10.1016/s0303-7207(98)00173-7. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi Y, et al. Distinct requirements for energy metabolism in mouse primordial germ cells and their reprogramming to embryonic germ cells. Proc Natl Acad Sci USA. 2017;114(31):8289–8294. doi: 10.1073/pnas.1620915114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baillie AH. The histochemistry and ultrastructure of the genocyte. J Anat. 1964;98:641–645. [PMC free article] [PubMed] [Google Scholar]
- 34.Busada JT, et al. Retinoic acid induces multiple hallmarks of the prospermatogonia-to-spermatogonia transition in the neonatal mouse. Biol Reprod. 2014;90(3):64. doi: 10.1095/biolreprod.113.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Martino C, et al. Morphological, histochemical and biochemical studies on germ cell mitochondria of normal rats. Cell Tissue Res. 1979;196(1):1–22. doi: 10.1007/BF00236345. [DOI] [PubMed] [Google Scholar]
- 36.Fawcett DW. The mammalian spermatozoon. Dev Biol. 1975;44(2):394–436. doi: 10.1016/0012-1606(75)90411-x. [DOI] [PubMed] [Google Scholar]
- 37.Woolley DM, Beatty RA. Inheritance of midpiece length in mouse spermatozoa. Nature. 1967;215(5096):94–95. doi: 10.1038/215094a0. [DOI] [PubMed] [Google Scholar]
- 38.Otani H, et al. Development of mitochondrial helical sheath in the middle piece of the mouse spermatid tail: regular dispositions and synchronized changes. Anat Rec. 1988;222(1):26–33. doi: 10.1002/ar.1092220106. [DOI] [PubMed] [Google Scholar]
- 39.Ho HC, Wey S. Three dimensional rendering of the mitochondrial sheath morphogenesis during mouse spermiogenesis. Microsc Res Tech. 2007;70(8):719–723. doi: 10.1002/jemt.20457. [DOI] [PubMed] [Google Scholar]
- 40.Tischler J, et al. Metabolic regulation of pluripotency and germ cell fate through alpha-ketoglutarate. EMBO J. 2019 doi: 10.15252/embj.201899518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawson KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp. 1994;182:68–84 . doi: 10.1002/9780470514573.ch5. [DOI] [PubMed] [Google Scholar]
- 42.Leese HJ, Barton AM. Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J Reprod Fertil. 1984;72(1):9–13. doi: 10.1530/jrf.0.0720009. [DOI] [PubMed] [Google Scholar]
- 43.Ro S, et al. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res. 2013;23(6):759–774. doi: 10.1038/cr.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vendramin R, Marine JC, Leucci E. Non-coding RNAs: the dark side of nuclear-mitochondrial communication. EMBO J. 2017;36(9):1123–1133. doi: 10.15252/embj.201695546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larriba E, Rial E, Del Mazo J. The landscape of mitochondrial small non-coding RNAs in the PGCs of male mice, spermatogonia, gametes and in zygotes. BMC Genom. 2018;19(1):634. doi: 10.1186/s12864-018-5020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacDonald JA, et al. A nanoscale, multi-parametric flow cytometry-based platform to study mitochondrial heterogeneity and mitochondrial DNA dynamics. Commun Biol. 2019;2:258. doi: 10.1038/s42003-019-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eddy EM. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 48.Chuma S, et al. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech Dev. 2003;120(9):979–990. doi: 10.1016/s0925-4773(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 49.Chuma S, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA. 2006;103(43):15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanaka SS, et al. The mouse homolog of drosophila vasa is required for the development of male germ cells. Genes Dev. 2000;14(7):841–853. [PMC free article] [PubMed] [Google Scholar]
- 51.Onohara Y, et al. Localization of mouse vasa homolog protein in chromatoid body and related nuage structures of mammalian spermatogenic cells during spermatogenesis. Histochem Cell Biol. 2010;133(6):627–639. doi: 10.1007/s00418-010-0699-5. [DOI] [PubMed] [Google Scholar]
- 52.Choi SY, et al. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8(11):1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20(3):364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H, et al. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20(3):376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma L, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5(9):e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Rooij DG. Spermatogonial stem cell renewal in the mouse. I. Normal situation. Cell Tissue Kinet. 1973;6(3):281–287. doi: 10.1111/j.1365-2184.1973.tb01617.x. [DOI] [PubMed] [Google Scholar]
- 57.Helsel AR, et al. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144(4):624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lord T, Nixon B. Metabolic changes accompanying spermatogonial stem cell differentiation. Dev Cell. 2020;52(4):399–411. doi: 10.1016/j.devcel.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 59.Chen W, et al. A bioenergetic shift is required for spermatogonial differentiation. Cell Discov. 2020;6:56. doi: 10.1038/s41421-020-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morimoto H, et al. ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell. 2013;12(6):774–786. doi: 10.1016/j.stem.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 61.Sadate-Ngatchou PI, et al. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46(12):738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, et al. MFN2 interacts with nuage-associated proteins and is essential for male germ cell development by controlling mRNA fate during spermatogenesis. Development. 2021 doi: 10.1242/dev.196295. [DOI] [PubMed] [Google Scholar]
- 63.Varuzhanyan G, et al. Mitochondrial fusion is required for spermatogonial differentiation and meiosis. Elife. 2019 doi: 10.7554/eLife.51601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gallardo T, et al. Generation of a germ cell-specific mouse transgenic Cre line. Vasa-Cre Genesis. 2007;45(6):413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen W, et al. MFN2 plays a distinct role from MFN1 in regulating spermatogonial differentiation. Stem Cell Rep. 2020;14(5):803–817. doi: 10.1016/j.stemcr.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, et al. GASZ and mitofusin-mediated mitochondrial functions are crucial for spermatogenesis. EMBO Rep. 2016;17(2):220–234. doi: 10.15252/embr.201540846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arachiche A, et al. Localization of PTP-1B, SHP-2, and Src exclusively in rat brain mitochondria and functional consequences. J Biol Chem. 2008;283(36):24406–24411. doi: 10.1074/jbc.M709217200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salvi M, et al. Tyrosine phosphatase activity in mitochondria: presence of Shp-2 phosphatase in mitochondria. Cell Mol Life Sci. 2004;61(18):2393–2404. doi: 10.1007/s00018-004-4211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Puri P, et al. The transition from stem cell to progenitor spermatogonia and male fertility requires the SHP2 protein tyrosine phosphatase. Stem Cells. 2014;32(3):741–753. doi: 10.1002/stem.1572. [DOI] [PubMed] [Google Scholar]
- 70.Li Y, et al. The role of tyrosine phosphatase Shp2 in spermatogonial differentiation and spermatocyte meiosis. Asian J Androl. 2020;22(1):79–87. doi: 10.4103/aja.aja_49_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu X, et al. Deletion of the tyrosine phosphatase Shp2 in Sertoli cells causes infertility in mice. Sci Rep. 2015;5:12982. doi: 10.1038/srep12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saunders PT, et al. Mitochondrial cytochrome C oxidase II messenger ribonucleic acid is expressed in pachytene spermatocytes at high levels and in a stage-dependent manner during spermatogenesis in the rat. Biol Reprod. 1993;48(1):57–67. doi: 10.1095/biolreprod48.1.57. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, et al. Mitochondria associated germinal structures in spermatogenesis: piRNA pathway regulation and beyond. Cells. 2020 doi: 10.3390/cells9020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao Y, et al. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nat Struct Mol Biol. 2016;23(5):441–449. doi: 10.1038/nsmb.3206. [DOI] [PubMed] [Google Scholar]
- 75.Margus T, Remm M, Tenson T. Phylogenetic distribution of translational GTPases in bacteria. BMC Genom. 2007;8:15. doi: 10.1186/1471-2164-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu W, et al. Mammalian male and female germ cells express a germ cell-specific Y-Box protein, MSY2. Biol Reprod. 1998;59(5):1266–1274. doi: 10.1095/biolreprod59.5.1266. [DOI] [PubMed] [Google Scholar]
- 77.Yang J, et al. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci USA. 2005;102(16):5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ande SR, et al. Prohibitin in adipose and immune functions. Trends Endocrinol Metab. 2016;27(8):531–541. doi: 10.1016/j.tem.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 79.Ko KS, et al. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology. 2010;52(6):2096–2108. doi: 10.1002/hep.23919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hernando-Rodriguez B, Artal-Sanz M. Mitochondrial quality control mechanisms and the PHB (Prohibitin) complex. Cells. 2018 doi: 10.3390/cells7120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang LF, et al. PHB regulates meiotic recombination via JAK2-mediated histone modifications in spermatogenesis. Nucleic Acids Res. 2020;48(9):4780–4796. doi: 10.1093/nar/gkaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kukat C, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA. 2011;108(33):13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang M, et al. Increased total mtDNA copy number cures male infertility despite unaltered mtDNA mutation load. Cell Metab. 2017;26(2):429–436. doi: 10.1016/j.cmet.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 84.Kujoth GC, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309(5733):481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 85.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 86.Ursini F, Bindoli A. The role of selenium peroxidases in the protection against oxidative damage of membranes. Chem Phys Lipids. 1987;44(2–4):255–276. doi: 10.1016/0009-3084(87)90053-3. [DOI] [PubMed] [Google Scholar]
- 87.Pushpa-Rekha TR, et al. Rat phospholipid-hydroperoxide glutathione peroxidase. cDNA cloning and identification of multiple transcription and translation start sites. J Biol Chem. 1995;270(45):26993–26999. doi: 10.1074/jbc.270.45.26993. [DOI] [PubMed] [Google Scholar]
- 88.Arai M, et al. Import into mitochondria of phospholipid hydroperoxide glutathione peroxidase requires a leader sequence. Biochem Biophys Res Commun. 1996;227(2):433–439. doi: 10.1006/bbrc.1996.1525. [DOI] [PubMed] [Google Scholar]
- 89.Pfeifer H, et al. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15(7):1236–1238. [PubMed] [Google Scholar]
- 90.Nayernia K, et al. Phospholipid hydroperoxide glutathione peroxidase: expression pattern during testicular development in mouse and evolutionary conservation in spermatozoa. Mol Reprod Dev. 2004;67(4):458–464. doi: 10.1002/mrd.20039. [DOI] [PubMed] [Google Scholar]
- 91.Imai H. New strategy of functional analysis of PHGPx knockout mice model using transgenic rescue method and Cre-LoxP system. J Clin Biochem Nutr. 2010;46(1):1–13. doi: 10.3164/jcbn.09-94R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shiromoto Y, et al. GPAT2, a mitochondrial outer membrane protein, in piRNA biogenesis in germline stem cells. RNA. 2013;19(6):803–810. doi: 10.1261/rna.038521.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wendel AA, Lewin TM, Coleman RA. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochim Biophys Acta. 2009;1791(6):501–506. doi: 10.1016/j.bbalip.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang S, et al. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2) Arch Biochem Biophys. 2007;465(2):347–358. doi: 10.1016/j.abb.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Garcia-Fabiani MB, et al. Methylation of the Gpat2 promoter regulates transient expression during mouse spermatogenesis. Biochem J. 2015;471(2):211–220. doi: 10.1042/BJ20150730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shiromoto Y, et al. GPAT2 is required for piRNA biogenesis, transposon silencing, and maintenance of spermatogonia in micedagger. Biol Reprod. 2019;101(1):248–256. doi: 10.1093/biolre/ioz056. [DOI] [PubMed] [Google Scholar]
- 97.Chen XJ. Sal1p, a calcium-dependent carrier protein that suppresses an essential cellular function associated with the Aac2 isoform of ADP/ATP translocase in saccharomyces cerevisiae. Genetics. 2004;167(2):607–617. doi: 10.1534/genetics.103.023655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Amigo I, et al. SCaMC-1Like a member of the mitochondrial carrier (MC) family preferentially expressed in testis and localized in mitochondria and chromatoid body. PLOS ONE. 2012;7(7):e40470. doi: 10.1371/journal.pone.0040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.张适, et al. (1987) 精子发生过程中线粒体的变化 %J 南京医学院学报 2: 81–84
- 100.Vertika S, Singh KK, Rajender S. Mitochondria, spermatogenesis, and male infertility—an update. Mitochondrion. 2020;54:26–40. doi: 10.1016/j.mito.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 101.Ren M, et al. Extramitochondrial cardiolipin suggests a novel function of mitochondria in spermatogenesis. J Cell Biol. 2019;218(5):1491–1502. doi: 10.1083/jcb.201808131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ren M, Phoon CK, Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 103.Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52(4):590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 104.Paradies G, et al. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim Biophys Acta. 2014;1837(4):408–417. doi: 10.1016/j.bbabio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 105.Ikon N, Ryan RO. Cardiolipin and mitochondrial cristae organization. Biochim Biophys Acta Biomembr. 2017;1859(6):1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramachandran R. Mitochondrial dynamics: the dynamin superfamily and execution by collusion. Semin Cell Dev Biol. 2018;76:201–212. doi: 10.1016/j.semcdb.2017.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ban T, et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol. 2017;19(7):856–863. doi: 10.1038/ncb3560. [DOI] [PubMed] [Google Scholar]
- 108.Liu R, Chan DC. OPA1 and cardiolipin team up for mitochondrial fusion. Nat Cell Biol. 2017;19(7):760–762. doi: 10.1038/ncb3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petrosillo G, Ruggiero FM, Paradies G. Role of reactive oxygen species and cardiolipin in the release of cytochrome c from mitochondria. FASEB J. 2003;17(15):2202–2208. doi: 10.1096/fj.03-0012com. [DOI] [PubMed] [Google Scholar]
- 110.Chu CT, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Valianpour F, et al. Monolysocardiolipins accumulate in Barth syndrome but do not lead to enhanced apoptosis. J Lipid Res. 2005;46(6):1182–1195. doi: 10.1194/jlr.M500056-JLR200. [DOI] [PubMed] [Google Scholar]
- 112.Wang HY, Jackson SN, Woods AS. Direct MALDI-MS analysis of cardiolipin from rat organs sections. J Am Soc Mass Spectrom. 2007;18(3):567–577. doi: 10.1016/j.jasms.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vreken P, et al. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279(2):378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 114.Brower JV, et al. Evolutionarily conserved mammalian adenine nucleotide translocase 4 is essential for spermatogenesis. J Biol Chem. 2007;282(40):29658–29666. doi: 10.1074/jbc.M704386200. [DOI] [PubMed] [Google Scholar]
- 115.Brower JV, et al. Adenine nucleotide translocase 4 deficiency leads to early meiotic arrest of murine male germ cells. Reproduction. 2009;138(3):463–470. doi: 10.1530/REP-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kitamura N, et al. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLOS ONE. 2011;6(1):e16060. doi: 10.1371/journal.pone.0016060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsuneki M, et al. Mieap suppresses murine intestinal tumor via its mitochondrial quality control. Sci Rep. 2015;5:12472. doi: 10.1038/srep12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wagner F, et al. Armadillo repeat-containing protein 1 is a dual localization protein associated with mitochondrial intermembrane space bridging complex. PLOS ONE. 2019;14(10):e0218303. doi: 10.1371/journal.pone.0218303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lopez-Domenech G, et al. The Eutherian Armcx genes regulate mitochondrial trafficking in neurons and interact with Miro and Trak2. Nat Commun. 2012;3:814. doi: 10.1038/ncomms1829. [DOI] [PubMed] [Google Scholar]
- 120.Chen Z, et al. Global phosphoproteomic analysis reveals ARMC10 as an AMPK substrate that regulates mitochondrial dynamics. Nat Commun. 2019;10(1):104. doi: 10.1038/s41467-018-08004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng W, Ip YT, Xu Z. Gudu, an armadillo repeat-containing protein, is required for spermatogenesis in drosophila. Gene. 2013;531(2):294–300. doi: 10.1016/j.gene.2013.08.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coutton C, et al. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am J Hum Genet. 2019;104(2):331–340. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shimada K, et al. ARMC12 regulates spatiotemporal mitochondrial dynamics during spermiogenesis and is required for male fertility. Proc Natl Acad Sci USA. 2021 doi: 10.1073/pnas.2018355118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen Y, et al. Glycerol kinase-like proteins cooperate with Pld6 in regulating sperm mitochondrial sheath formation and male fertility. Cell Discov. 2017;3:17030. doi: 10.1038/celldisc.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shimada K, et al. Glycerol kinase 2 is essential for proper arrangement of crescent-like mitochondria to form the mitochondrial sheath during mouse spermatogenesis. J Reprod Dev. 2019;65(2):155–162. doi: 10.1262/jrd.2018-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang YY, et al. Deficiency of the Tbc1d21 gene causes male infertility with morphological abnormalities of the sperm mitochondria and flagellum in mice. PLoS Genet. 2020;16(9):e1009020. doi: 10.1371/journal.pgen.1009020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Loson OC, et al. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wakabayashi J, et al. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186(6):805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Varuzhanyan G, et al. Fis1 ablation in the male germline disrupts mitochondrial morphology and mitophagy, and arrests spermatid maturation. Development. 2021 doi: 10.1242/dev.199686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen H, et al. Titration of mitochondrial fusion rescues Mff-deficient cardiomyopathy. J Cell Biol. 2015;211(4):795–805. doi: 10.1083/jcb.201507035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Varuzhanyan G, et al. Mitochondrial fission factor (Mff) is required for organization of the mitochondrial sheath in spermatids. Biochim Biophys Acta Gen Subj. 2021;1865(5):129845. doi: 10.1016/j.bbagen.2021.129845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wijayarathna R, de Kretser DM. Activins in reproductive biology and beyond. Hum Reprod Update. 2016;22(3):342–357. doi: 10.1093/humupd/dmv058. [DOI] [PubMed] [Google Scholar]
- 133.Vale W, et al. Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature. 1986;321(6072):776–779. doi: 10.1038/321776a0. [DOI] [PubMed] [Google Scholar]
- 134.Ling N, et al. Pituitary FSH is released by a heterodimer of the beta-subunits from the two forms of inhibin. Nature. 1986;321(6072):779–782. doi: 10.1038/321779a0. [DOI] [PubMed] [Google Scholar]
- 135.de Kretser DM, et al. Inhibins, activins and follistatin: actions on the testis. Mol Cell Endocrinol. 2001;180(1–2):87–92. doi: 10.1016/s0303-7207(01)00502-0. [DOI] [PubMed] [Google Scholar]
- 136.Mendis SH, et al. Activin A balances Sertoli and germ cell proliferation in the fetal mouse testis. Biol Reprod. 2011;84(2):379–391. doi: 10.1095/biolreprod.110.086231. [DOI] [PubMed] [Google Scholar]
- 137.Barakat B, et al. Inhibin, activin, follistatin and FSH serum levels and testicular production are highly modulated during the first spermatogenic wave in mice. Reproduction. 2008;136(3):345–359. doi: 10.1530/REP-08-0140. [DOI] [PubMed] [Google Scholar]
- 138.Mithraprabhu S, et al. Activin bioactivity affects germ cell differentiation in the postnatal mouse testis in vivo. Biol Reprod. 2010;82(5):980–990. doi: 10.1095/biolreprod.109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meinhardt A, et al. Activin maintains the condensed type of mitochondria in germ cells. Mol Cell Endocrinol. 2000;168(1–2):111–117. doi: 10.1016/s0303-7207(00)00308-7. [DOI] [PubMed] [Google Scholar]
- 140.Brown CW, et al. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat Genet. 2000;25(4):453–457. doi: 10.1038/78161. [DOI] [PubMed] [Google Scholar]
- 141.Li L, et al. Activin signaling: effects on body composition and mitochondrial energy metabolism. Endocrinology. 2009;150(8):3521–3529. doi: 10.1210/en.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vassalli A, et al. Activin/inhibin beta B subunit gene disruption leads to defects in eyelid development and female reproduction. Genes Dev. 1994;8(4):414–427. doi: 10.1101/gad.8.4.414. [DOI] [PubMed] [Google Scholar]
- 143.Agarwal A, et al. A unique view on male infertility around the globe. Reprod Biol Endocrinol. 2015;13:37. doi: 10.1186/s12958-015-0032-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Folgero T, et al. Mitochondrial disease and reduced sperm motility. Hum Reprod. 1993;8(11):1863–1868. doi: 10.1093/oxfordjournals.humrep.a137950. [DOI] [PubMed] [Google Scholar]
- 145.Kubota Y, Mizoguchi M, Goto Y. Mitochondrial encephalomyopathy and skin disease. Nihon Rinsho. 2002;60(Suppl 4):632–635. [PubMed] [Google Scholar]
- 146.Wang WL, Tu CF, Tan YQ. Insight on multiple morphological abnormalities of sperm flagella in male infertility: what is new? Asian J Androl. 2020;22(3):236–245. doi: 10.4103/aja.aja_53_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Boguenet M, et al. Mitochondria: their role in spermatozoa and in male infertility. Hum Reprod Update. 2021;27(4):697–719. doi: 10.1093/humupd/dmab001. [DOI] [PubMed] [Google Scholar]
- 148.Durairajanayagam D, et al. Causes and consequences of sperm mitochondrial dysfunction. Andrologia. 2021;53(1):e13666. doi: 10.1111/and.13666. [DOI] [PubMed] [Google Scholar]
- 149.Linck RW, Chemes H, Albertini DF. The axoneme: the propulsive engine of spermatozoa and cilia and associated ciliopathies leading to infertility. J Assist Reprod Genet. 2016;33(2):141–156. doi: 10.1007/s10815-016-0652-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Haraguchi CM, et al. Chromatoid bodies: aggresome-like characteristics and degradation sites for organelles of spermiogenic cells. J Histochem Cytochem. 2005;53(4):455–465. doi: 10.1369/jhc.4A6520.2005. [DOI] [PubMed] [Google Scholar]
- 151.Auguste Y, et al. Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am J Hum Genet. 2018;103(3):413–420. doi: 10.1016/j.ajhg.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lores P, et al. Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum Mol Genet. 2018;27(7):1196–1211. doi: 10.1093/hmg/ddy034. [DOI] [PubMed] [Google Scholar]
- 153.Fernandez-Gonzalez A, et al. Mutation of murine adenylate kinase 7 underlies a primary ciliary dyskinesia phenotype. Am J Respir Cell Mol Biol. 2009;40(3):305–313. doi: 10.1165/rcmb.2008-0102OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mata M, et al. New adenylate kinase 7 (AK7) mutation in primary ciliary dyskinesia. Am J Rhinol Allergy. 2012;26(4):260–264. doi: 10.2500/ajra.2012.26.3784. [DOI] [PubMed] [Google Scholar]
- 155.Sha Y, et al. Biallelic mutations of CFAP58 are associated with multiple morphological abnormalities of the sperm flagella. Clin Genet. 2021;99(3):443–448. doi: 10.1111/cge.13898. [DOI] [PubMed] [Google Scholar]
- 156.Li ZZ, 2020. The novel testicular enrichment protein Cfap58 is required for Notch-associated ciliogenesis. Biosci Rep. [DOI] [PMC free article] [PubMed]
- 157.He X, et al. Bi-allelic loss-of-function variants in CFAP58 cause flagellar axoneme and mitochondrial sheath defects and asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2020;107(3):514–526. doi: 10.1016/j.ajhg.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Liu M, et al. Novel mutations in FSIP2 lead to multiple morphological abnormalities of the sperm flagella and poor ICSI prognosis. Gene. 2021;781:145536. doi: 10.1016/j.gene.2021.145536. [DOI] [PubMed] [Google Scholar]
- 159.Liu W, et al. Homozygous loss-of-function mutations in FSIP2 cause male infertility with asthenoteratospermia. J Genet Genom. 2019;46(1):53–56. doi: 10.1016/j.jgg.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 160.Martinez G, et al. Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum Reprod. 2018;33(10):1973–1984. doi: 10.1093/humrep/dey264. [DOI] [PubMed] [Google Scholar]
- 161.Fang X, et al. Hypomorphic and hypermorphic mouse models of Fsip2 indicate its dosage-dependent roles in sperm tail and acrosome formation. Development. 2021 doi: 10.1242/dev.199216. [DOI] [PubMed] [Google Scholar]
- 162.Ben Khelifa M, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2014;94(1):95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Amiri-Yekta A, et al. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum Reprod. 2016;31(12):2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 164.Neesen J, et al. Disruption of an inner arm dynein heavy chain gene results in asthenozoospermia and reduced ciliary beat frequency. Hum Mol Genet. 2001;10(11):1117–1128. doi: 10.1093/hmg/10.11.1117. [DOI] [PubMed] [Google Scholar]
- 165.Hwang JY, et al. Genetic defects in DNAH2 underlie male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. Front Cell Dev Biol. 2021;9:662903. doi: 10.3389/fcell.2021.662903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Y, et al. DNAH2 is a novel candidate gene associated with multiple morphological abnormalities of the sperm flagella. Clin Genet. 2019;95(5):590–600. doi: 10.1111/cge.13525. [DOI] [PubMed] [Google Scholar]
- 167.Gao Y, et al. Novel bi-allelic variants in DNAH2 cause severe asthenoteratozoospermia with multiple morphological abnormalities of the flagella. Reprod Biomed Online. 2021;42(5):963–972. doi: 10.1016/j.rbmo.2021.01.011. [DOI] [PubMed] [Google Scholar]
- 168.Tu C, et al. Identification of DNAH6 mutations in infertile men with multiple morphological abnormalities of the sperm flagella. Sci Rep. 2019;9(1):15864. doi: 10.1038/s41598-019-52436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Tu C, et al. Bi-allelic mutations of DNAH10 cause primary male infertility with asthenoteratozoospermia in humans and mice. Am J Hum Genet. 2021;108(8):1466–1477. doi: 10.1016/j.ajhg.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang B, et al. Novel loss-of-function variants in DNAH17 cause multiple morphological abnormalities of the sperm flagella in humans and mice. Clin Genet. 2021;99(1):176–186. doi: 10.1111/cge.13866. [DOI] [PubMed] [Google Scholar]
- 171.Whitfield M, et al. Mutations in DNAH17, encoding a sperm-specific axonemal outer dynein arm heavy chain, cause isolated male infertility due to asthenozoospermia. Am J Hum Genet. 2019;105(1):198–212. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sha Y, et al. DNAH17 is associated with asthenozoospermia and multiple morphological abnormalities of sperm flagella. Ann Hum Genet. 2020;84(3):271–279. doi: 10.1111/ahg.12369. [DOI] [PubMed] [Google Scholar]
- 173.Coutton C, et al. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun. 2018;9(1):686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tang S, et al. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2017;100(6):854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li W, et al. Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J Med Genet. 2020;57(2):89–95. doi: 10.1136/jmedgenet-2019-106344. [DOI] [PubMed] [Google Scholar]
- 176.Zhang X, et al. A novel homozygous CFAP65 mutation in humans causes male infertility with multiple morphological abnormalities of the sperm flagella. Clin Genet. 2019;96(6):541–548. doi: 10.1111/cge.13644. [DOI] [PubMed] [Google Scholar]
- 177.Wang W, et al. Biallelic mutations in CFAP65 lead to severe asthenoteratospermia due to acrosome hypoplasia and flagellum malformations. J Med Genet. 2019;56(11):750–757. doi: 10.1136/jmedgenet-2019-106031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang W, et al. CFAP65 is required in the acrosome biogenesis and mitochondrial sheath assembly during spermiogenesis. Hum Mol Genet. 2021;30(23):2240–2254. doi: 10.1093/hmg/ddab185. [DOI] [PubMed] [Google Scholar]
- 179.Dong FN, et al. Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am J Hum Genet. 2018;102(4):636–648. doi: 10.1016/j.ajhg.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu W, et al. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet. 2019;104(4):738–748. doi: 10.1016/j.ajhg.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lores P, et al. Mutations in TTC29, encoding an evolutionarily conserved axonemal protein, result in asthenozoospermia and male infertility. Am J Hum Genet. 2019;105(6):1148–1167. doi: 10.1016/j.ajhg.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liu C, et al. Bi-allelic mutations in TTC29 cause male subfertility with asthenoteratospermia in humans and mice. Am J Hum Genet. 2019;105(6):1168–1181. doi: 10.1016/j.ajhg.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Li ZZ, 2020. The novel testicular enrichment protein Cfap58 is required for Notch-associated ciliogenesis. Biosci Rep. [DOI] [PMC free article] [PubMed]