Abstract
Holliday junction resolvases (HJRs) are key enzymes of DNA recombination. A detailed computer analysis of the structural and evolutionary relationships of HJRs and related nucleases suggests that the HJR function has evolved independently from at least four distinct structural folds, namely RNase H, endonuclease, endonuclease VII–colicin E and RusA. The endonuclease fold, whose structural prototypes are the phage λ exonuclease, the very short patch repair nuclease (Vsr) and type II restriction enzymes, is shown to encompass by far a greater diversity of nucleases than previously suspected. This fold unifies archaeal HJRs, repair nucleases such as RecB and Vsr, restriction enzymes and a variety of predicted nucleases whose specific activities remain to be determined. Within the RNase H fold a new family of predicted HJRs, which is nearly ubiquitous in bacteria, was discovered, in addition to the previously characterized RuvC family. The proteins of this family, typified by Escherichia coli YqgF, are likely to function as an alternative to RuvC in most bacteria, but could be the principal HJRs in low-GC Gram-positive bacteria and Aquifex. Endonuclease VII of phage T4 is shown to serve as a structural template for many nucleases, including McrA and other type II restriction enzymes. Together with colicin E7, endonuclease VII defines a distinct metal-dependent nuclease fold. As a result of this analysis, the principal HJRs are now known or confidently predicted for all bacteria and archaea whose genomes have been completely sequenced, with many species encoding multiple potential HJRs. Horizontal gene transfer, lineage-specific gene loss and gene family expansion, and non-orthologous gene displacement seem to have been major forces in the evolution of HJRs and related nucleases. A remarkable case of displacement is seen in the Lyme disease spirochete Borrelia burgdorferi, which does not possess any of the typical HJRs, but instead encodes, in its chromosome and each of the linear plasmids, members of the λ exonuclease family predicted to function as HJRs. The diversity of HJRs and related nucleases in bacteria and archaea contrasts with their near absence in eukaryotes. The few detected eukaryotic representatives of the endonuclease fold and the RNase H fold have probably been acquired from bacteria via horizontal gene transfer. The identity of the principal HJR(s) involved in recombination in eukaryotes remains uncertain; this function could be performed by topoisomerase IB or by a novel, so far undetected, class of enzymes. Likely HJRs and related nucleases were identified in the genomes of numerous bacterial and eukaryotic DNA viruses. Gene flow between viral and cellular genomes has probably played a major role in the evolution of this class of enzymes. This analysis resulted in the prediction of numerous previously unnoticed nucleases, some of which are likely to be new restriction enzymes.
INTRODUCTION
Recombination is one of the fundamental aspects of the biochemistry of DNA that results not only in the repair of damage (mutation), but also in the production of combinatorial variation that provides the substrate for natural selection. Over three decades ago Robin Holliday proposed a basic model for recombination that was observed in fungal meiosis (1). This model invoked pairing between complementary single strands coming from two distinct homologous duplexes, with the formation of heteroduplexes joined by a four-way junction. It has been hypothesized that the resolution of this junction, catalyzed by an enzyme called the resolvase, results in the formation of separated recombinant molecules. While the details of the Holliday model have undergone considerable revision with the accumulation of data, the basic concept of strand exchange followed by resolution of the four-way junction has endured all these developments.
A variety of molecular structures are utilized in recombination, including molecules with single-strand gaps, single-strand overhangs and double-strand breaks (2–4). These substrates are processed via similar mechanisms in all DNA-based life forms, from various recombination events in prokaryotes, their plasmids and phages, to meiotic crossing-over in eukaryotes. Studies on the recombination processes in the bacterium Escherichia coli have revealed a range of participating proteins. These components of recombination systems include RecA, which is involved in homologous strand exchange, helicases, such as RuvAB and RecG, that promote strand migration, the RecBCD helicase-nuclease, in which the nuclease domain of RecB generates single-strand regions at the ends of duplexes, and Holliday junction resolvases (HJRs) such as RuvC and RusA (5–8). RecA is a highly conserved P-loop-containing ATPase, which is represented by functionally equivalent orthologs in the three domains of life, bacteria, archaea and euakryotes (9,10). Otherwise, recombination systems show significant differences between these primary domains, with virtually no universal conservation. The helicases RuvAB and RecG, whose functions in recombination partially overlap, are present in almost all bacteria (10–12). The other recombination-specific enzymes identified in the bacterial model system, however, show notably scattered patterns of phyletic distribution (10–12). In particular, with the increasing number of completely sequenced bacterial and archaeal genomes, this trend of a lack of universal conservation is becoming a clear rule among the nucleases and HJRs involved in recombination.
A number of distinct HJRs have been identified in prokaryotes, including RuvC, which is conserved in the majority of bacteria, E.coli RusA, bacteriophage T4 endonuclease VII (EndoVII) (11) and the recently detected HJR from Pyrococcus furiosus, which is conserved in all archaea whose complete genome sequences are available (13). Orthologs of RuvC are encoded by some bacteriophages infecting lactococci and streptococci and orthologs of RusA exist in the genomes of several phages from lactoccoci, staphylococci, E.coli and Mycobacterium tuberculosis (11). Very few HJRs have been identified in eukaryotes. One of these is the yeast mitochondrial resolvase Cce1 (14,15) and the other one is topoisomerase IB from poxviruses (16). The mosaic phyletic distribution of the distinct resolvases among bacteria suggests that the evolution of this critical function involved multiple non-orthologous gene displacements (17).
To investigate the evolution of the resolvases and their relationships with other nucleases, we performed a detailed analysis of their protein sequences using local alignment searches, pattern searches, sequence profile analysis, secondary structure prediction-based threading and structural modeling. We show that the archaeal HJR belongs to a vast superfamily of bacterial and archaeal nucleases that has not been described previously. The structural fold of the archaeal HJR could be modeled using the structure of bacteriophage λ exonuclease, whereas the fold of another branch of the same superfamily is typified by the structure of the very short patch repair endonuclease (Vsr). In addition, we identified new families of nucleases related to RuvC and T4 EndoVII. Experimentally characterized members of these families participate in different processes in DNA repair, recombination and protection against foreign DNA. As a result of this analysis, we predict the HJRs for all bacteria and archaea whose genomes have been sequenced and for several families of viruses.
SEQUENCE AND STRUCTURE ANALYSIS
Analysis of multiple sequences was handled using the SEALS program package (18). Sequence similarity searches were performed using the gapped BLASTP program (19) and the Non-Redundant (NR) protein database at the National Center for Biotechnology Information (NIH, Bethesda, MD). Additional searches of nucleotide sequences translated in all six frames were performed using the gapped TBLASTN program (19) and the sequences of unfinished genomes provided by The Institute for Genome Research, The Sanger Center and the Pseudomonas Genome project. Pattern searches were carried out using the GREF program of the SEALS package or the PHI-BLAST program (20). Iterative database searches with position-specific weight matrices (PSSMs) were performed using the PSI-BLAST program; a cut-off of 0.01, in terms of the expectation (E) value for inclusion of detected sequences into the PSSM, was used unless otherwise indicated (19). Additional profile searches were carried out using hidden Markov models generated from alignments of protein domains using the HMMS program of the HMMER2 package (21). Multiple alignments of protein sequences were constructed using the CLUSTAL_X program (22) or the MACAW program (23) and adjusted manually on the basis of PSI-BLAST results. Secondary structure prediction and secondary structure-based threading were carried out using the PHD (24,25) and PSI-PRED programs (26). The 3-dimensional structures of proteins were manipulated using the SwissPDB viewer program (27) and ribbon diagrams were constructed using the MOLSCRIPT program (28).
IDENTIFICATION AND CLASSIFICATION OF HJRs AND RELATED NUCLEASES
We found that all known HJRs and homologous nucleases fit into three principal structural folds, with the single exception of RusA, for which no structural cognate was identified. The Hsp70/RNase H fold includes RuvC and its homologs, the endonuclease fold includes a large, diverse set of nucleases such as archaeal HJRs, recB-like nucleases, the λ exonuclease family and the Vsr-like nuclease family, and the EndoVII fold includes a family of nucleases typified by phage T4 EndoVII. Below we describe novel superfamilies of nucleases that we identified within each of these folds, analyze the phyletic distribution and probable evolutionary history for each family and show how the available 3-dimensional structures help in understanding the catalytic functions of these enzymes.
THE RuvC SUPERFAMILY OF HJRs
The archetypal bacterial resolvase RuvC has been crystallized and shown to possess an α–β structure that places it in the RNase H fold (after the SCOP classification of protein structures; 29,30). This fold is characteristic of a vast, diverse assemblage of proteins including numerous nucleases, such as the 3′→5′ exonuclease superfamily, retroposon/retroviral integrases and integrases of diverse DNA transposons, including bacteriophage Mu and RNase H itself (31–33). In addition, the fold includes ATP-utilizing enzymes of the HSP70 superfamily that contain two tandem copies of the RNase H structural unit (34,35). In spite of the low overall sequence similarity, the general conservation of the topology and the presence of a conserved acidic residue required for catalysis at the C-terminus of the first β-strand in this fold, along with a similar mono- or polynucleotide binding mode, suggest that all these enzymes have diverged from a common ancestor. Given the functional diversity of the enzymes in this fold, it seems possible that the nuclease activity has arisen independently on multiple occasions from the ancestral nucleotide-binding core. The 3′→5′ nucleases, the integrases and the RNase Hs have been extensively characterized in terms of their sequence diversity and evolutionary relationships (36,37). In contrast to the wide spread of these enzymes, the RuvC family has been restricted to a set of orthologs from different bacteria and bacteriophages; sequence searches have so far failed to identify statistically significant similarity between RuvC and any other proteins of the RNase H fold.
The RuvC family
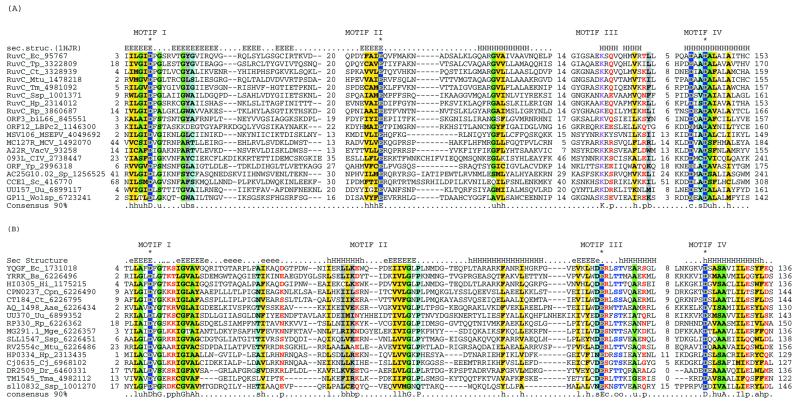
Taking advantage of the diverse set of RuvC sequences that have recently become available as a result of bacterial genome sequencing projects and sensitive profile search methods, we investigated the sequence relationships of the RuvC family. A PSI-BLAST search initiated with the E.coli RuvC sequence detected, after three iterations, not only the orthologous bacterial proteins, but also uncharacterized proteins from Lactococcus and Wolbachia phages, Ureaplasma urealyticum, Yersinia pestis and Melanopus sanguinipes entomopoxvirus. PSI-BLAST searches initiated with the sequences of these newly detected RuvC homologs identified similar proteins in all the poxviruses and Chilo iridiscent virus and also brought the mitochondrial resolvase Cce1 from the yeast Saccharomyces cerevisiae and its Schizosaccharomyces pombe ortholog into the RuvC family. Examination of a multiple alignment using the MACAW program identified five statistically significant motifs (P < 10–8) that are conserved throughout the RuvC family (Fig. 1). A superimposition of these conserved sequence motifs upon the 3-dimensional structure of RuvC (29) suggests that the entire family preserves the core secondary structure elements seen in RuvC, with poorly conserved inserts occurring in the eukaryotic forms (Figs 1 and 2A). The signature motifs of the RuvC family include the acidic residue (typically an aspartate) towards the end of strand 1, a glutamate near the end of conserved strand 4 and two acidic residues (DxxD) associated with the C-terminal α-helix (Figs 1 and 2A). All these residues have been shown to be critical for the resolvase activity of RuvC, with the first, second and last ones forming a spatially juxtaposed acidic triad that could coordinate divalent cations (38).
Figure 1.
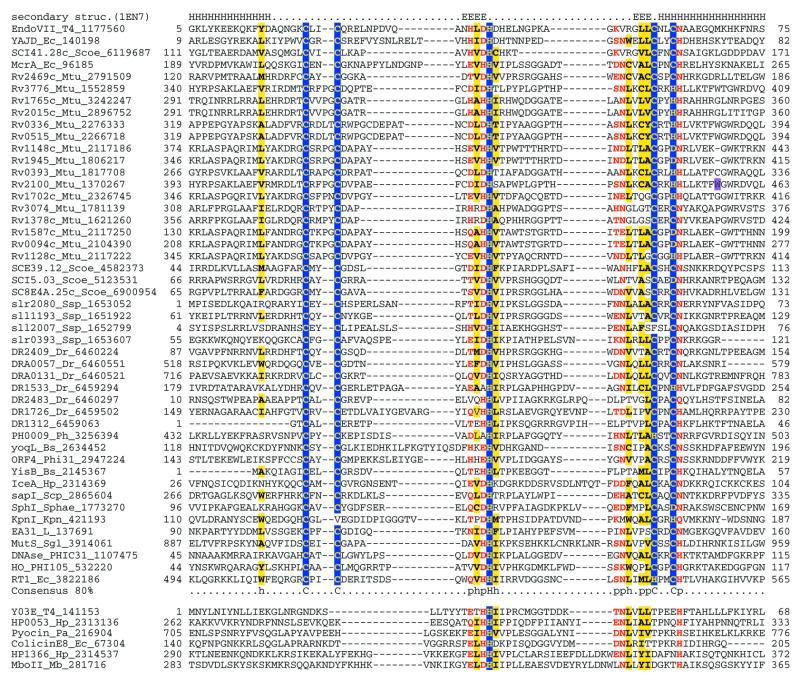
Multiple alignments of the HJRs of the RNase H fold. (A) RuvC family. (B) YqgF family. Each protein is labeled using the gene name followed by the species abbreviation and the GenBank gene identifier. The extent of the domain in each protein is indicated by numbers to the sides of the alignments. The long poorly conserved inserts are replaced by numbers indicating the number of omitted residues. Only Motifs I and IV are aligned between the RuvC and YqgF families. The secondary structure predicted using the PHD program is shown above the alignment with H/h for α-helix and E/e for β-strands (upper case denoting strong prediction and lower case moderate prediction). The shading and coloring are according to the 90% consensus, which is shown underneath the alignment, with the following convention: h, hydrophobic residues (YFWLIVMA); l, aliphatic residues (LIVMA); a, aromatic residues (YFWH), yellow background; p, polar residues (STQNEDRKH), red foreground; s, small residues (SAGTVPNHD), turquoise background; u, tiny residues (GAS), light green background; c, charged residues (KHRED), magenta foreground; b, bulky residues (LIYWFEQRKM), gray background. The residues predicted to form the active site or associated with catalysis are shown in inverse coloring. The species abbreviations are as indicated in Table 1. The following are abbreviations not shown in Table 1: Wolsp, Wolbachia sp.; MSEPV, Melanopus sanguinus entomopox virus; MCV, Molluscum contagiousum virus; VacV, vaccinia virus; biL66 and LBPc2, lactococcal phages biL66 and c2.
Figure 2.
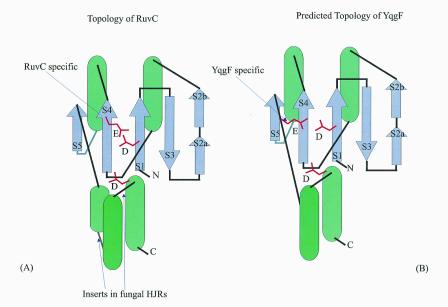
Topological diagrams of the HJRs of the RNase H fold. The α-helices are shown by green cylinders and the β-strands by violet arrows. The strands are numbered in the order of occurrence. Note the distinct positions of conserved glutamates in the RuvC and YqgF families.
These observations indicate that the RuvC family has a much greater spread than previously believed; in particular, Ureaplasma is the first low-GC Gram-positive bacterium, in which a RuvC homolog has been detected. Fungal mitochondrial HJRs have hitherto been considered unrelated to the bacterial HJRs. The unification of Cce1 with the RuvC family suggests that the resolvase function of these proteins is preserved in spite of the considerable sequence divergence and that it can function independently of the RuvAB complex. This relationship is consistent with the ability of S.pombe Cce1 to complement RuvC deficiency in E.coli (39) and the notable biochemical similarities between Cce1 and RuvC, namely a strong preference for four-way junctions and limited sequence specificity in the resolution step (14,40). The Cce1 proteins show significant sequence similarity to the MRS1 protein involved in mitochondrial intron splicing (40). However, the acidic residues critical for the catalytic activity of the RuvC family enzymes (Figs 1 and 2A) are not conserved in this protein (not shown), suggesting that it is an inactive member of this family. It seems likely that MRS1 retains the ability to bind unusual nucleic acid structures and functions in RNA processing in this capacity.
Poxviruses possess hairpin-terminated telomeres that require resolution after replication to produce separated daughter duplexes that are packaged into the virion (41). It has been shown that a viral gene product is involved in this process (42). Viral topoisomerase IB is capable of catalyzing this reaction in vitro, but appears to be required in large quantities and the reaction proceeds slowly (16,43). Thus the newly identified RuvC homologs appear to be strong candidates for the role of the poxvirus telomere resolvase. Experiments based on this prediction have shown that the poxvirus enzyme is indeed capable of resolving Holliday junctions and that the activity critically depends on the conserved acidic residues (44). The presence of a RuvC homolog among the gene products of an iridovirus and many bacteriophages suggests a widespread role in the resolution of branched DNA structures, although, at least in the latter case, a general nuclease function is also possible.
The YqgF family of RuvC homologs
Iterative database searches started with the RuvC protein sequences retrieved, with a moderate statistical significance, a group of proteins typified by E.coli YqgF. Reciprocal searches initiated with the YqgF protein sequences recovered the RuvC family, which supports a homologous relationship between these two protein families. A motif search using the Gibbs sampling procedure as implemented in the PROBE program (45) detects a highly significant motif (P < 10–15) shared by the RuvC and the YqgF families. This motif corresponds to the first two strands of the RuvC proteins and includes the conserved acidic residue occurring at the end of strand 1, which is present in most members of the RNase H fold (33,35). Multiple alignment-based secondary structure prediction for the YqgF family proteins reveals a succession of elements that are compatible with the RNase H fold. Superposition of the multiple alignments of the YqgF and RuvC families with the RuvC 3-dimensional structure indicates that the proximal and distal aspartates of the RuvC catalytic triad are conserved in the YqgF family (Figs 1 and 2). However, the glutamate at the end of strand 4 (Fig. 2A), which is invariant in the RuvC family, is missing in the YqgF family proteins. Instead, they contain a conserved glutamate at the end of strand 5 (Fig. 2B). Given the spatial proximity of the end of strand 5 to the two other conserved acidic residues, functional equivalence of the conserved glutamates in the RuvC and YqgF families appears likely. This strongly suggests that the YqgF family proteins are nucleases with a catalytic mechanism similar to that of RuvC.
The RuvC resolvases are conspicuously absent in the low-GC Gram-positive bacterial lineage, with the exception of Ureaplasma (Table 1). Furthermore, loss of function ruvC mutants of E.coli show a residual HJR activity that cannot be ascribed to the prophage-encoded RusA resolvase (46). With the exception of the spirochetes, the YqgF family is represented in all bacterial lineages, including the mycoplasmas with their highly degenerate genomes. Taken together with the prediction of a RuvC-like enzymatic activity, this suggests that YqgF family proteins could be alternative HJRs whose function partially overlaps with that of RuvC.
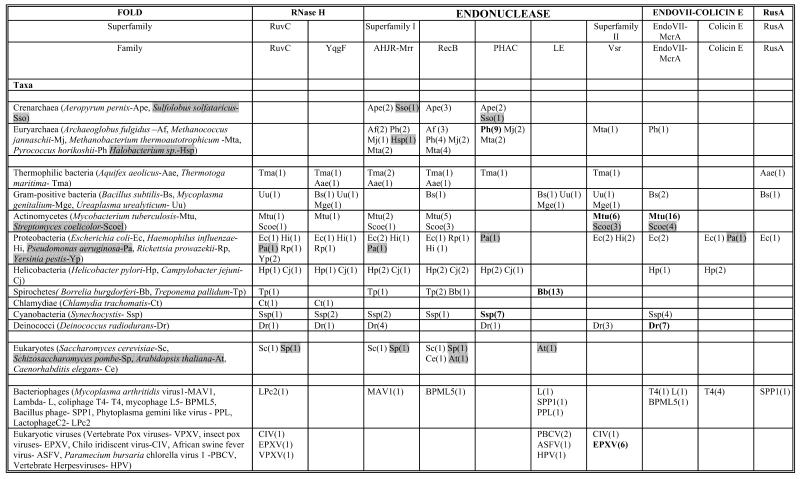
Table 1. Phyletic distribution of HJRs and related nucleasesa.
aOrganisms whose complete genome sequences are not yet available are shaded gray. Major lineage-specific expansions are shown in bold. The numbers in parentheses indicate the number of paralogs within a family that can be detected in a given genome.
Finally, it is of interest that iterative database searches started with the sequences of RuvC and YqgF family proteins produced statistically significant alignments with the HSP70 superfamily of molecular chaperones that contain a duplicated RNase H fold. This suggests a possible sister group relationship between the RuvC-type HJRs and the HSP70 superfamily within the RNase H fold. This hypothesis is also supported by the specific similarity in the conformation of the N-terminal strands of these two protein superfamilies (not shown).
ARCHAEAL HJRs AND THE ENDONUCLEASE FOLD
Fold recognition and phylogenetic affinities of the archaeal HJRs
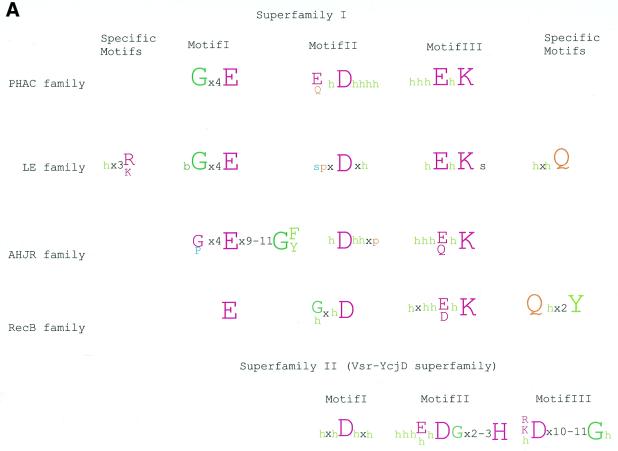
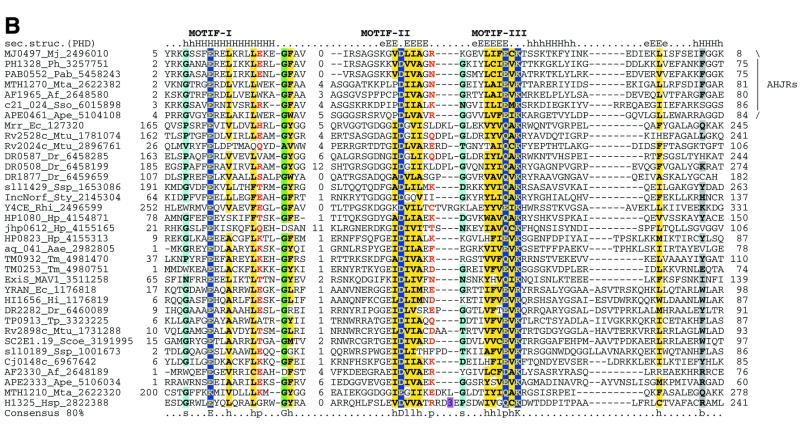
Recently, the archaeal Holliday junction resolvase (hereinafter AHJR) from Pyrococcus furiosus has been cloned and biochemically characterized. The enzyme shows maximal activity on four-way junctions and a weak activity on three-way junctions and looped-out DNA (13). It resolves RecA-generated joined duplexes with the same efficiency as E.coli RuvC and, given its conservation in all sequenced euryarchaeal and crenarchaeal genomes, is likely to be the principal archaeal Holliday junction resolvase. A PSI-BLAST search of the NR database with a PSSM based on an alignment of the archaeal resolvases retrieved a number of uncharacterized proteins from several bacteria as well as the methylated DNA-specific restriction enzyme Mrr (47). All these proteins showed notable conservation of three motifs centered around a constellation of charged residues (Fig. 3A). The N-terminal motif is predicted to form a helix and contains a strictly conserved glutamate. The two distal motifs are predicted to form β-strands and contain another conserved acidic residue (typically aspartate) and a [EDQ]xK signature, respectively. Thus the AHJRs and Mrr-like endonucleases together define a new enzyme family, in which the conserved charged residues could form the active site.
Figure 3.
Multiple alignments of the HJRs and related nucleases of the endonuclease fold. (A) A schematic of the conserved motifs containing the (predicted) catalytic residues. (B) Superfamily I: the AHJR–Mrr family. (C) Superfamily I: the RecB family. (D) Superfamily I: the PHAC family. (E) Superfamily I: the λ exonuclease family. (F) Superfamily II: Vsr homologs. The schematic representation in (A) shows the configuration of the three conserved motifs of superfamilies I and II as well as certain family-specific motifs described in the text. The conserved residues that are present in >25% of the cases are shown by the single letter code in upper case. In other cases the general consensus category for the residues as indicated in the legend to Figure 1 is shown in lower case. The alignment notation is as indicated in the legend to Figure 1. The conserved motifs of each superfamily are indicated above the alignment. All families of superfamily I share three conserved motifs as shown in (A), but because of the absence of extended sequence similarity between the families, the alignment for each family is shown separately (B–F). In (E) only two sequences from B.burgdorferi, BB036 from the chromosome and BBC12 from linear plasmid C, are shown; the remaining plasmid-encoded sequences are nearly identical to these. Blue letters in some of the sequences in the alignment indicate anamolous inserts that have been excised. Additional species abbreviations: Sty, Salmonella typhimurium; Rsph, Rhodopseudomonas spheroides; Hs, Homo sapiens; Ll, Lactococcus lactis; Ban, Bacillus anthracis; Tfo, Thiobacillus ferroxidans; Vic, Vibrio cholerae; Coxb, Coxiella burnetti; Rhi, Rhizobium sp.; LPA118, Listeria phage A118; NPVAC, AcMNPV and NPVOP, nuclear polyhedrosis viruses of Autographa califorinica, Bombyx mori and Orgyia pseudotsugata; EBV, Epstein–Barr virus; KSV, Kaposi sarcoma virus; HSVSA, herpes virus saimiri; HSVEB, equine herpes virus B; VZVD, varicella zoster virus D.
We included all proteins detected in transitive searches with the AHJRs in a PSSM and iteratively searched the NR database to detect more distant relationships. These searches revealed statistically significant similarity between the AHJR–Mrr family and two other protein families, namely the RecB nuclease family (10) and a previously undetected family typified by the C-terminal conserved domain of the PH (Pyrococcus horikoshii)-type ATPases (hereinafter PHAC; 48). All these proteins contain characteristic conserved charged residues associated with the three motifs identified in the AHJR–Mrr family (Fig. 3A). Additionally, in these searches two other proteins families typified by the bacteriophage λ exonuclease and Vsr endonuclease, respectively, showed marginal similarity to the AHJR–Mrr family (E values of 0.3–0.05). The λ exonuclease family shows a conservation pattern that is very similar to that in the AHJR–Mrr–RecB–PHAC proteins and contains readily identifiable counterparts of the three conserved motifs described above. Thus the AHJR–Mrr, RecB-like nuclease, PHAC and λ exonuclease families share a common set of motifs that are predicted to define the nucleolytic active site of these enzymes. Motif analysis using the Gibbs sampling method implemented in the PROBE program (45) confirmed the statistical significance of the three conserved motifs in a set of 130 representatives from each of these four families, with P values below 10–6.
Secondary structure prediction for the AHJR, RecB and PHAC families identified a pattern of strands and helices nearly identical to that seen in the core domain of λ exonuclease (49). Thus all these proteins are predicted to share a common fold with the λ exonuclease (Fig. 4A) and, by inference, with other members of the endonuclease fold (after SCOP; 30), such as EcoRV and PvuII (50). Based on the structure of λ exonuclease, it can be inferred that all these enzymes coordinate a divalent cation (Mg2+) via the conserved aspartate in motif II, the glutamate, glutamine or aspartate in motif III (Fig. 3A and B) and one of the oxygens of the scissile phosphodiester group. The conserved lysine in motif III (Fig. 3A and B) is likely to contact the phosphate of the DNA backbone, as suggested by the presence of an equivalent residue in EcoRV (51).
Figure 4.
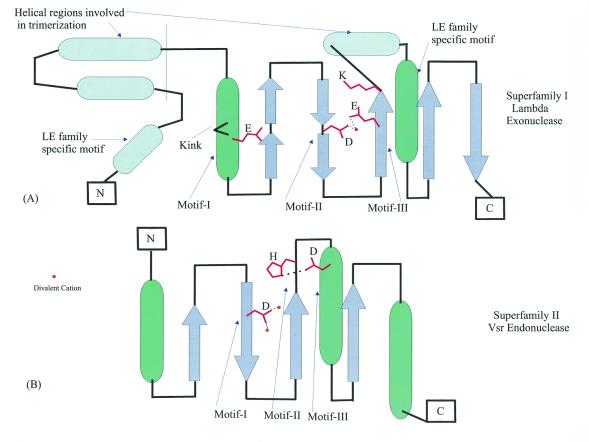
Topological diagrams of the enzymes of the endonuclease fold. (A) λ exonuclease, the structural template for superfamily I. (B) Vsr, the structural template for superfamily II. The positions of conserved motifs indicated in Figure 3 are shown by arrows and the residues involved in catalysis are indicated; the coordinated metal cations are shown by yellow circles.
The Vsr endonuclease family (52), which also showed borderline similarity to AHJRs in iterative database searches, does not contain detectable counterparts to the three motifs that are typical of the four families described above. Recently, the crystal structure of Vsr has been solved and structural comparisons have shown that it is a bona fide member of the endonuclease fold, however, the active site of this nuclease family is distinct from that in the other four families (Figs 3C and 4B; 53,54).
Classification of the endonuclease fold enzymes
Based on the results of exhaustive PSI-BLAST searches and conversed sequence features, the DNases of the endonuclease fold could be classified into distinct groups, namely: superfamily I, which consists of the AHJR family, the RecB family, the PHAC nuclease family and the λ exonuclease (LE) family; superfamily II, which includes the Vsr homologs; classical restriction endonucleases that possess the same fold but show little or no detectable sequence similarity to each other or to superfamilies I and II beyond the principal active residues.
Superfamily I: the AHJR–Mrr family. As a whole, this superfamily is defined by the three motifs containing the (predicted) catalytic residues (Figs 3A and 4). The AHJR–Mrr family can be characterized by an apparent synapomorphy (a shared derived character), namely the G[FY] signature, which resides at the end of the first predicted helix of these proteins (Fig. 3B). Within this family, a number of clusters of orthologs, including the archaeal resolvases proper, can be recognized (see the COG classification at http://www.ncbi.nlm.nih.gov/COG/ ; 55). One of these, typified by E.coli YraN, defines a group of orthologous proteins that are conserved in several bacterial lineages and could function as hitherto unrecognized HJRs or DNA repair enzymes.
The Mrr-like nucleases are found in a number of bacteria, the archaeon Methanobacterium thermoautotrophicum and the yeasts S.cerevisiae and S.pombe, which suggests widespread horizontal mobility. The radioresistant bacterium Deinococcus radiodurans encodes two divergent copies of the Mrr nuclease, while the plasmid from Rhizobium encodes a Mrr protein with a duplication of the nuclease domain. Another orthologous cluster includes members from Helicobacter, Synechocystis, Deinococcus and Thermotoga (DR1877, sll1429, HP1080 and TM0932; Figs 3B and 5), most of which contain hydrophobic signal peptides and membrane-spanning segments, suggesting that they are cell surface-associated nucleases. Helicobacter and Mycobacteria encode proteins in which the AHJR nuclease domain is fused to a superfamily II helicase and an adenine-specific DNA methylase (RvD1-Rv2024c′; Fig. 5). Given that genes coding for similar helicases are present in operons of type III restriction–modification systems (56), it seems likely that in these proteins the AHJR nuclease functions as a restriction enzyme. A single viral member of this family is seen in the genome of the Mycoplasma arthritidis bacteriophage MAV1.
Figure 5.
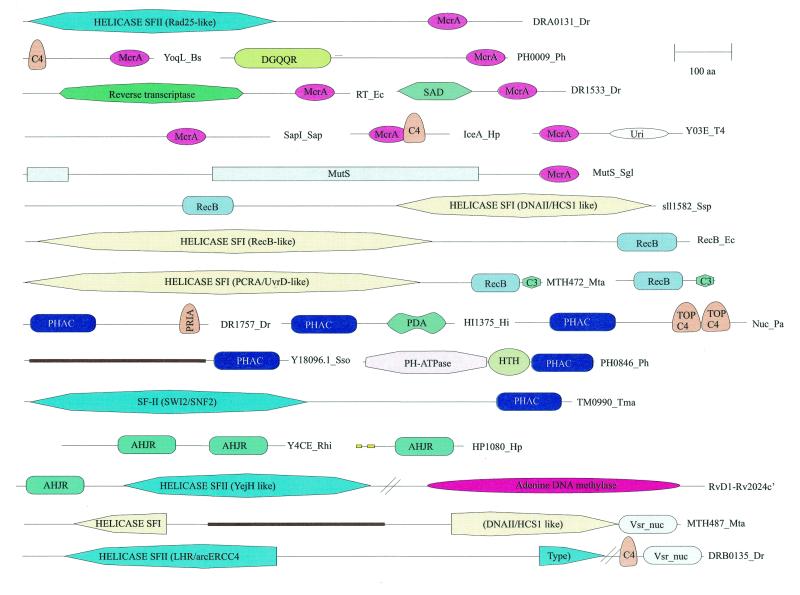
Domain architectures of HJRs and related nucleases. Nuclease domains are indicated by the following abbreviations: McrA, EndoVII fold nuclease domain; RecB, RecB family nuclease of the endonuclease fold; PHAC, PHAC family nuclease of the endonuclease fold; AHJR, nuclease of the archaeal HJR family of the endonuclease fold; Vsr_nuc, nuclease of the Vsr–YcjD superfamily of the endonuclease fold. The proteins are labeled as in the alignment figures. For the helicase domains of superfamilies I and II (SFI and SFII) the closest functionally characterized homologs are indicated. Domain abbreviations: URI, UvrC-Intron nuclease domain; MutS, mismatch repair ATPase; C4, four-cysteine Zn cluster; C3, three three-cysteine Zn cluster; TOP C4, Zn ribbon domain related to that at the C-termini of topoisomerase IA; PriA, predicted Zn-binding domain shared with the PriA helicases; HTH, helix–turn–helix DNA-binding domain; DGQQR, uncharacterized conserved domain found in a diverse set of bacterial and archaeal proteins and designated by its characteristic amino acid signature (L.Aravind and E.V.Koonin, unpublished observations); PDA, PHAC/DGQQR-associated domain (L.Aravind, unpublished observations); SAD, uncharacterized conserved domain associated with the SET domain in several chromatin-associated proteins. A coiled-coil domain inserted into the helicase domain of MTH487 is indicated by a brown bar and signal peptides and transmembrane regions are indicated by yellow bars.
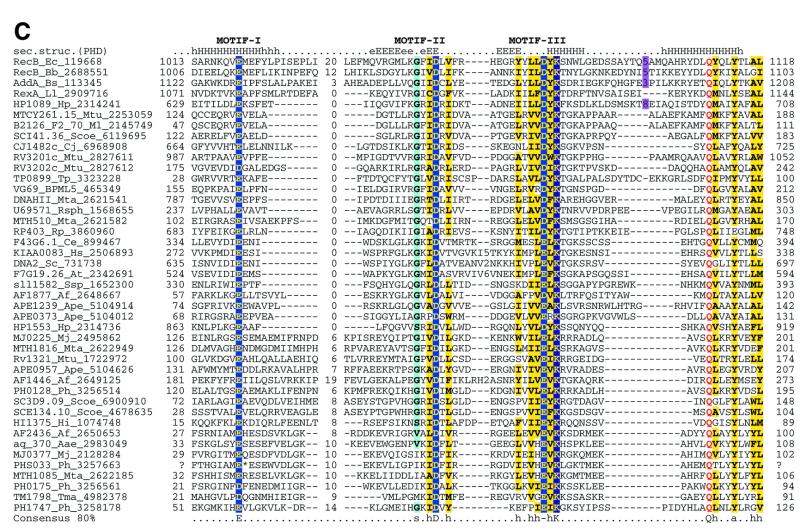
Superfamily I: the RecB family. The RecB family is supported by a synapomorphic motif containing the Qx(3)Y signature, which is located downstream of the three motifs common to the entire superfamily (Fig. 3C) (10). In contrast, motif I, which corresponds to the N-terminal helix, shows considerable divergence within this family (Fig. 3C). The RecB protein, the namesake of this family, consists of a C-terminal nuclease domain fused to a superfamily I helicase domain (57; Fig. 5). As a subunit of the RecBCD recombinase complex, RecB unwinds DNA and preferentially degrades the 3′-strand in an exonucleolytic reaction, only occasionally nicking the 5′-strand (58). Mutagenesis studies on the RecB nuclease domain have shown that the conserved charged residues in motifs II and III are critical for the exonucleolytic activity of RecBCD in both directions, and an active site similar to those of restriction endonucleases has been postulated (57,59). In contrast, another member of this family, the yeast DNA2 protein, combines an N-terminal RecB nuclease domain with a C-terminal superfamily I helicase domain (Fig. 5) and shows 3′-single-strand-specific endonuclease activity (60). Thus different members of the RecB family possess either exonuclease or endonuclease activity.
The RecB nuclease domain shows several apparently independent fusions to superfamily I helicases (Fig. 5), which suggests that this nuclease typically functions in a close association with DNA unwinding. Three of these fusions, namely those in the RecB-like, AddA-like and Rv3201c-like proteins, show a sporadic distribution in the bacterial world, indicating spread by horizontal gene transfer. The DNA2 protein is a eukaryote-specific helicase–nuclease fusion that has been horizontally acquired by cyanobacteria, probably from the plant lineage. Several small RecB family nucleases, typically fused to a C-terminal three-cysteine metal-binding cluster, are common in many archaeal and bacterial genomes, which suggests several still unexplored nuclease functions (10).
Superfamily I: the PHAC family. The PH-type ATPases have been described as a group of proteins that show a specific expansion in the genomes of the pyrococci, although they are also present in one or two copies in some other archaea and bacteria, e.g. Coxiella (61). These proteins contain a distinct ATPase domain followed by a predicted DNA-binding helix–turn–helix domain (48). The C-terminal region of these proteins was identified in the present analysis as a new nuclease family, PHAC, which is related to the AHJR and RecB-like nucleases (Fig. 3D). In addition to the prevalent fusion with PH ATPases, the PHAC domain was detected in combination with other domains (Fig. 5). In particular, some of the archaea and the thermophilic bacterium Thermotoga possess a PHAC domain fused to a SWI2/SNF2-like superfamily II helicase; to our knowledge, fusion of this class of superfamily II helicase domains with a nuclease has not been detected previously. A combination of the PHAC domain with long and short coiled-coil regions, respectively, is seen as a single copy in the crenarchaeon Aeropyrum pernix, and at six copies in Synechocystis. Other unusual domain architectures of the PHAC domain include fusions with a PriA-type Zn finger in Deinococcus and with a topoisomerase C-terminal Zn ribbon domain in Pseudomonas aeruginosa (Fig. 5).
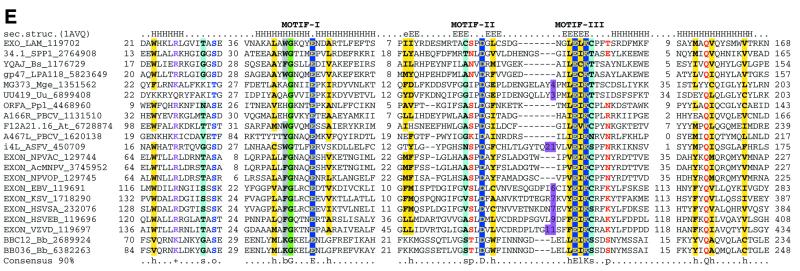
Superfamily I: the LE family. The LE family is typified by λ exonuclease, a toroidal trimeric nuclease that generates single-stranded overhangs involved in the repair and recombination of phage chromosomes (49). Here we describe previously undetected homologs of λ exonuclease encoded in bacterial genomes, namely those of Mycoplasma, Bacillus subtilis and Thiobacillus ferrooxidans (Fig. 3E). These bacterial lineages probably acquired the genes for LE family nucleases from prophages related to the Spiroplasma phage SPP1 or the Listeria phage A118. In addition to the bacterial viruses, members of this family were detected in large eukaryotic cytoplasmic DNA viruses, namely Paramecium bursaria Chlorella virus (PBCV) and African swine fever virus (ASFV), as well as nuclear polyhedrosis viruses and herpesviruses (Fig. 3E). In herpesviruses and nuclear polyhedrosis viruses the respective proteins have been characterized as alkaline exonucleases involved in the processing of viral replication intermediates (62). Interestingly, the plant nuclear genome encodes a nuclease of this family which is closely related to one of the two LE family members from PBCV (Fig. 3E). This protein contains a predicted organellar transit peptide (not shown) and is likely to function in mitochondria or chloroplasts.
Of particular importance is the identification of predicted LE family nucleases in the Lyme disease spirochaete Borrelia burgdorferi (Fig. 3E); an LE family protein is encoded in the linear chromosome of Borrelia and in each of its linear plasmids. No RuvC, RusA or other potential HJRs have hitherto been identified in this organism, as opposed to the other spirochete whose genome has been sequenced, Treponema pallidum, which encodes a RuvC ortholog (63). This is a major puzzle, particularly given the critical role of recombination in Lyme disease pathogenesis (64–66). We propose that in the linear replicons of Borrelia the LE exonuclease family proteins substitute for the classical HJRs. It is likely that they have been acquired from a bacteriophage with a linear genome and were recruited for recombination of linear replicons in Borrelia via the generation of single-stranded overhangs similar to those involved in phage λ recombination (67).
The LE family possesses a clear synapomorphy in the form of an additional motif, N-terminal of the three catalytic motifs of superfamily I, which contains a conserved hydrophobic residue (typically tryptophan) and a basic residue (Fig. 3E). Furthermore, these proteins contain another characteristic C-terminal motif with a conserved glutamine, which could be equivalent to the synapomorphic motif of the RecB family (Fig. 3C and E). Examination of the 3-dimensional structure of λ exonuclease indicates that the N-terminal motif specific to this family forms a helix which is positioned opposite the active site residues (Fig. 4A), suggesting a possible role in binding DNA or distorting DNA structure. The unusual toroidal structure and location of the active site in λ exonuclease suggests that this enzyme encircles the DNA molecule in the process of degradation (50). Trimerization is mediated mainly by two α-helical regions, one of which closely follows the strand containing the catalytic motif III (Fig. 4A) and is conserved in members of the LE family; the other helix is poorly conserved. While these regions are likely to support oligomerization in many of these proteins, the toroidal structure might not necessarily extend to the entire family.
The 3-dimensional structure of λ nuclease has some general implications for the entire superfamily I of the endonuclease fold. The helix corresponding to motif I contains a small residue (glycine or proline) three positions upstream of the conserved glutamate in all members of the LE family, most of the AHJR and PHAC nucleases and some of the RecB family nucleases (Fig. 3B–E). This residue forms a kink in the helix and probably allows some flexibility that enables a DNA sequence structure-dependent conformational change. This is likely to be critical for the function of many members of this family.
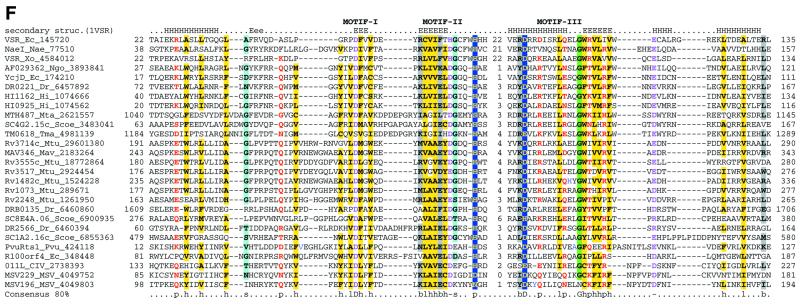
Superfamily II: Vsr-like nucleases. The Vsr nuclease was initially identified as the endonuclease involved in very short patch repair of TG mismatches (52,68). No homologs of the E.coli Vsr nuclease beyond obvious orthologs in certain bacteria have been reported so far, however, the recent solution of the crystal structure of Vsr has shown that it possesses the endonuclease fold (54). Vsr nucleases were detected with low scores in iterative database searches with superfamily I nucleases as queries. In spite of a lack of statistical significance or motif conservation, these alignments are functionally and evolutionarily relevant, given that they match the structural alignments of Vsr with other members of the endonuclease fold, such as λ exonuclease (53). A detailed analysis using iterative sequence searches from different starting points resulted in the detection of a fairly widespread set of Vsr homologs.
Other than Vsr itself, a cluster of orthologous proteins typified by E.coli YcjD is represented in a number of diverse bacteria (Fig. 3F). Mycobacterium tuberculosis encodes five closely related members of the Vsr superfamily, which are probably the result of a recent expansion. Thermotoga and Methanobacterium encode proteins in which the Vsr nuclease domain is fused to superfamily I helicases, whereas in Deinococcus it is fused in a similar orientation to a superfamily II helicase (Fig. 5). There is a similar fusion to a superfamily I helicase in Mycoplasma, but in this case the Vsr nuclease domain is partially disrupted and is likely to be inactive (not shown). Divergent members of the Vsr superfamily were identified in the genomes of two large DNA viruses, namely Chilo iridescent virus and entomopox virus, with a specific expansion of the family in the latter (Fig. 3F and data not shown). Furthermore, the restriction enzyme PvuRts1I, which restricts hydroxymethylcytosine-containing phage DNA (69), was also identified as a divergent member of the Vsr superfamily (Fig. 3F).
A multiple alignment of superfamily II shows three prominent motifs, in which most of the sequence conservation is concentrated (Fig. 3F). Motif I is centered around a conserved aspartate, which is equivalent to the aspartate in motif II of superfamily I (Fig. 3A–E). The crystal structure of Vsr suggests that this aspartate coordinates two divalent cations that are critical for activity (Fig. 4B). Motif II contains the signature bh[DEH] (b is a bulky, typically acidic residue and h is a hydrophobic residue). The charged residue in this signature is equivalent to the basic residue in motif III of superfamily I (Fig. 3A–E), but appears to perform a distinct function because it is not involved in phosphate binding, unlike the corresponding residue in λ exonuclease or the restriction enzymes (53). In contrast, in the Vsr superfamily motif II contains a conserved downstream histidine which, in Vsr itself, is essential for binding the scissile phosphate (53). Motif III encompasses a helix with a conserved negatively charged residue and a tightly associated strand (Figs 3F and 4B). The acidic residue in this helix forms a bond with the conserved histidine and activates it for catalysis (53; Fig. 4B). Thus, within the basic scaffold of the endonuclease fold the Vsr nucleases have evolved an active site and catalytic mechanism that are distinct from those of the superfamily I enzymes.
Restriction endonucleases. Restriction endonucleases are the most divergent members of the endonuclease fold that show very little sequence conservation (70). Only a few closely related proteins that have been disseminated via recent horizontal transfers are easily recognizable. Evolution of restriction enzymes has recently been analyzed in detail (71,72) and here we only briefly discuss their relationships with other nucleases of the endonuclease fold. The crystal structures of several restriction enzymes that show no objectively detectable sequence similarity to each other, namely EcoRV (51), PvuII (73), BamHI (74), EcoRI (75), MutH (71), FokI (76), Cfr10I (77), BglI (78) and MunI (79), have been solved. A comparison of these structures indicates that they are all evolutionarily related and contain a common, ancestral active site comprised of residues equivalent to those that are conserved in motifs II (D at the end of a strand) and III ([EDQ]xK associated with a strand) of superfamily I of the endonuclease fold (50,71; Fig. 3A). Some of these enzymes, e.g. EcoRV, FokI, BglI and MutH, possess an equivalent of the conserved glutamate found in the N-terminal helix of motif I in superfamily I (Fig. 3A), suggesting that this residue was also present in the ancestral endonuclease fold.
Structural comparisons and cleavage specificities have revealed distinct subclasses among the restriction endonucleases. The enzymes that form blunt-ended fragments, such as EcoRV and PvuII, MutH, which cuts only one unmethylated strand of its target site, and BglI, which forms 3′-overhangs, appear to be structural neighbors and form one distinct subclass. MunI, EcoRI and BamHI, which form 5′-overhangs, and FokI, which cleaves non-specifically some distance away from its recognition site, form the second subclass of restriction enzymes (72). These similarities apart, the restriction enzymes show diverse dimerization and DNA-binding modes as well as differences in the details of catalysis (80). One striking example is replacement of the otherwise conserved lysine in the [EQD]xK signature in the BamHI equivalent of motif III. This suggests that selection for the strict site-specificity typical of restriction enzymes has resulted in accommodation of a variety of changes within the framework of the endonuclease fold, with a concomitant erosion of sequence conservation.
The extensive sequence divergence might also have been instigated by a lack of selective forces that, in the case of other nucleases, stem from their functionally critical interactions with multiple, conserved components of the cellular DNA repair machinery. The existence of specific relationships between certain restriction enzymes and other evolutionarily conserved nucleases suggests that restriction enzymes have arisen on multiple occasions from different nuclease lineages. Indeed, some of the restriction enzymes, such as Mrr, are conserved in several lineages and show a distinct, readily recognizable relationship with other conserved nucleases, in this case the AHJR family (see above). These relatively highly conserved enzymes could be evolutionary intermediates in the origin of other highly divergent restriction enzymes. Furthermore, the detection of a relationship between PvuRts1I and Vsr (Fig. 3F) suggests that more divergent and still undetected restriction enzymes could exist that are distantly related to superfamily II, rather than superfamily I, of the endonuclease fold.
THE T4 ENDONUCLEASE VII–COLICIN E FOLD
Unlike RuvC, bacteriophage T4 EndoVII recognizes both Holliday junctions and other perturbations in duplex DNA and shows no sequence specificity (81). Recently, the structure of this protein has been solved and identified as a new fold (81). Sequence searches readily detected EndoVII homologs in mycobacteriophages, such as L5, E.coli, H.pylori (strain J99) and Streptomyces spp. The conservation pattern in these proteins includes two dyads of conserved cysteines and a central conserved histidine and is identical to the pattern seen in the large family of endonucleases typified by the methylcytosine restriction enzyme McrA and certain intron-encoded nucleases (82,83). PSI-BLAST searches (inclusion threshold 0.01) initiated with the sequence of the nuclease domain of McrA revealed statistically significant similarity between T4 EndoVII and its homologs and the McrA-like nuclease family. Additional searches using PSSMs that included both EndoVII-like and McrA-like proteins resulted in the unification of this family with a group of type II restriction enzymes that includes SapI, SphI, KpnI and IceA (Fig. 6). Additionally, the central region, which contains a conserved dyad of a charged residue and an invariant histidine, is shared between the EndoVII–McrA superfamily and colicin E-DNases, pyocin Ap41, type II restriction enzyme MboII and the mobile intron endonucleases of T4 (Fig. 6).
Figure 6.
Multiple alignment of the Holliday junction resolvases and related nucleases of the EndoVII–colicin E fold. The alignment notation is as indicated in the legend to Figure 1. Additional species abbreviations: Sgl, Sarcophyton glaucum; Kpn, Klebsiella pneumoniae; Phi31, Lactococcus phage φ31; PHIC31, Streptomyces phage φC 31; Phi105, Bacillus subtilis phage φ105; Sphae, Streptomyces phaeochromogenes; Scp, Saccharopolyspora sp.; Mb, Moraxella bovis.
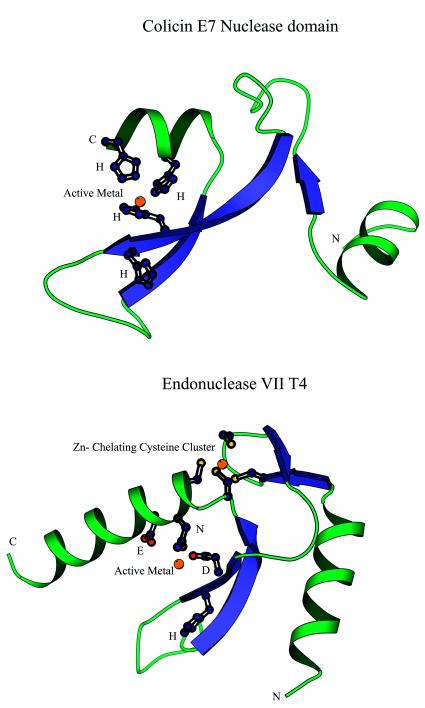
A superposition of the multiple alignment of the EndoVII–McrA superfamily with the 3-dimensional structure of EndoVII suggests a common core structure for all these proteins (Fig. 7A). This structural core encompasses a central hairpin of twisted β-strands, which is flanked on each side by an α-helix, and defines the minimal nuclease domain. At the end of the first α-helix there is a hairpin formed by short β-strands that contain the first conserved cysteine dyad. These cysteines chelate a Zn2+ ion in conjunction with the second cysteine dyad that is located in the beginning of the C-terminal helix (Fig. 7A). Associated with the first strand of the central hairpin is the characteristic signature of this family, namely a dyad of charged residues, in which the first one is either an aspartate or a histidine and the second one is invariably a histidine (Figs 6 and 7A). The first of these residues, along with a polar residue (N,H or Q) in the C-terminal helix that occurs immediately after the last Zn-chelating cysteine, participates in coordination of the active metal. These residues are critical for the catalytic activity of EndoVII (84,85) and from the high degree of conservation across the entire superfamily it can be inferred that they perform the same function in all these nucleases. Chelation of the active metal is augmented by other polar residues in the C-terminal helix, two or three residues downstream of the conserved polar position. The conserved histidine in the first strand faces away from the chelating residues (Fig. 7A) and, in line with the site-directed mutagenesis data (85), we propose that this residue acts as an amphiphile that directs the water to attack the phosphodiester bond in the substrate. DNA binding might result in a conformational change that suitably reorients this histidine for its catalytic function.
Figure 7.
Structures of nucleases of the EndoVII–colicin E fold. (A) Colicin E. (B) EndoVII. The residues involved in chelating the active metal and the stabilizing zinc cluster in EndoVII are shown in the ball-and-stick representation. The orientations of the N-terminal helices differ in the two families. Note the highly conserved histidine shared by these families that faces away from the chelated active metal and is required for catalysis.
The sequences of bacteriocin nucleases, MboII and the mobile T4 endonucleases align well with the EndoVII–McrA superfamily in terms of the residues involved in coordinating the active metal and in catalysis, but lack the two pairs of cysteines that chelate Zn in the latter (Fig. 6). A direct superposition of the structure of E.coli colicin E7 (86) with that of EndoVII based on their sequence alignment shows that the two superfamilies indeed share the basic elements of the structural core as well as the active metal-coordinating and catalytic residues (Fig. 7A and B). However, lack of the Zn-coordinating cysteines in colicin E7 probably results in a different orientation of the N-terminal helix compared to EndoVII. Conservation of the catalytic site and the structural core suggests that although colicin acts as a monomer (86) and EndoVII as a dimer (81), they share a common evolutionary origin and catalytic mechanism. Apparently, the combination of the conserved core with distinct stabilizing α-helical elements has so far obscured this relationship between these two families.
The detection of a number of type II restriction endonucleases in both the EndoVII–McrA and colicin E7 versions of this fold is of interest given the extreme divergence and difficulties in sequence-based identification of restriction enzymes. As discussed above, all type II restriction enzymes whose structures have been determined belong to the endonuclease fold. However, on at least two distinct occasions restriction enzymes appear to have independently emerged from the EndoVII–colicin E fold.
Nucleases of this fold are encoded by several bacteria with a moderate to large genome size (Table 1); some of these could be hitherto unidentified restriction enzymes, whereas others could be involved in DNA repair. The possibility of repair functions is of particular interest in the case of the radioresistant bacterium D.radiodurans, which encodes seven proteins containing the EndoVII–McrA domain, in three of which it is combined with other domains (two of these are shown in Fig. 5). One of these domains is a Rad25-like helicase, which is suggestive of a repair function. Given the role of recombination in DNA repair in Deinococcus (87), it seems possible that EndoVII–McrA-like nucleases recognize and cleave unusual structures left behind after recombination. The most prominent expansion of this domain is seen in the genome of M.tuberculosis (16 members). Most of the mycobacterial EndoVII–McrA-like nucleases contain an unusual four-residue insert between the first pair of cysteines and are highly similar to each other (Fig. 6), which indicates that this family is the result of recent serial duplications.
IMPLICATIONS OF THE PHYLETIC DISTRIBUTION AND POSSIBLE EVOLUTIONARY SCENARIOS FOR HJRs AND RELATED NUCLEASES
Recombination is ubiquitous in DNA-based life and so is Holliday junction resolution. However, the enzymatic machinery for recombination shows little or no conservation between the three primary domains of life, bacteria, archaea and eukaryotes, with the sole exception of the RecA recombinase (9). This is consistent with a major dichotomy of the DNA replication systems between the bacterial and archaeal–eukaryotic lineages (88–90). The phyletic distribution of the HJRs generally follows the same principal division (Table 1). Altogether, HJR activity appears to have evolved within at least four independent structural folds, namely RNase H (the RuvC superfamily), endonuclease, EndoVII–colicin E and RusA. In bacteria the principal ancestral resolvase is clearly one of the RuvC superfamily, whereas in archaea this role belongs to AHJR. Both of these nuclease folds are largely absent in eukaryotes, the only exceptions being the RecB nuclease domain in DNA2, a single LE family member in Arabidopsis, some divergent homologs of Mrr-like nucleases of the AHJR family and fungal mitochondrial RuvC-like resolvases. It appears most likely that these genes have entered the eukaryotic lineage via horizontal transfer from bacteria. The dramatic difference in the abundance of these enzymes between prokaryotes and eukaryotes might have to do with the linear structure of eukaryotic chromosomes and the complex organization of chromatin that could restrict the function of these nucleases. The identity of the eukaryotic HJR(s) remains elusive. Based on experimental data obtained in the vaccinia virus system, topoisomerase IB has been proposed as a candidate for the HJR function in eukaryotes (16), but direct support for this is lacking.
Beyond the principal differences between bacteria, archaea and eukaryotes, there is evidence of numerous horizontal gene transfers, lineage-specific gene losses and non-orthologous gene displacement in the evolution of the HJRs (Table 1). It appears that in bacteria the principal HJR function can be provided by at least four distinct protein families, namely RuvC, YqgF, LE and RusA. The RuvC and YqgF families are each represented by one member in most bacteria. It appears that these families are a product of an ancient duplication, perhaps in the common ancestor of all extant bacteria, with subsequent lineage-specific elimination of RuvC (in low-GC Gram-positive bacteria, Aquifex and Borrelia) or YqgF (in the spirochetes). The case of Borrelia is particularly interesting because in this lineage both RuvC and YqgF have been lost and apparently replaced by LE family nucleases. A correlation between this unusual displacement and the linear structure of both the chromosome and most of the plasmids in Borrelia is obvious and it will be of interest to see whether other bacteria with linear genomes use the same type of HJR. In addition to the major HJRs of the RNase H fold, many bacteria encode one or more proteins of the AHJR–Mrr family. Some of these proteins form clusters of orthologs that are highly conserved in several bacterial lineages (e.g. YraN and its orthologs) and, in principle, could function as alternative HJRs.
In at least three bacteria, E.coli, B.subtilis and A.aeolicus, an additional HJR activity is provided by RusA, an enzyme that appears to be unrelated to other resolvases and is also encoded by many bacteriophages (91). Thus most bacteria seem to encode multiple HJRs, which is compatible with the phenotypes of E.coli ruvC and rusA mutants. The AHJR family is (so far) represented in all archaea, but shows sporadic distribution in bacteria, which suggests that dissemination via horizontal gene transfer and differential gene loss has been important in the evolution of this family. In contrast, no representatives of the RuvC and YqgF families were detected in archaea. Given the generally extensive gene exchange between bacteria and archaea (61), this asymmetry in horizontal dissemination of the HJRs is puzzling. One possible explanation, in line with the general disparity between the replication and recombination machineries in archaea and bacteria, is that there are major mechanistic differences between bacterial and archaeal HJRs which make them non-interchangeable. Under this hypothesis, the bacterial members of the AHJR family enzymes would be predicted to function as general nucleases rather than resolvases.
Identified or predicted nucleases of the AHJR family, the LE family and bacterial RuvC and RusA families are encoded by many bacterial and eukaryotic viruses. There is little doubt that gene flow between cellular and viral genomes has contributed significantly to the observed distribution of these nucleases. In particular, it appears likely that the LE family nuclease in Borrelia was originally acquired from a phage, with subsequent dissemination among the Borrelia replicons. A similar origin appears likely for RuvC of Ureaplasma, which is so far the sole representative of this family in low-GC Gram-positive bacteria. Furthermore, RusA appears to be a typical bacteriophage enzyme and probably has been independently acquired by E.coli, B.subtilis and A.aeolicus from different phages. Endonuclease I of phage T7 functions as an HJR (92), but shows no detectable sequence similarity to any of the known families of nucleases. However, the requirement of certain acidic residues for its activity (93) and secondary structure prediction (not shown) suggest that it could be a divergent version of the endonuclease fold.
T4 EndoVII and the AHJRs belong to large families of DNases, several of which are known or predicted to be involved in DNA repair. This suggests that the ancestral members of the EndoVII and the endonuclease folds might have had a general repair function, from which diverse specificities have been independently derived on multiple occasions during evolution. Several independent fusions between superfamily I and II helicases and nucleases of the endonuclease fold and one fusion of a EndoVII fold nuclease and a superfamily II helicase (Fig. 5) indicate that many of these nucleases function in a close association with helicases that generate their substrates by means of ATP-dependent duplex unwinding. This is supported by the general similarities in the action of the nuclease–helicase proteins, such as RecB (59) and AddA (94) in bacteria and DNA2 in eukaryotes (60). In contrast, no domain fusions have so far been detected for the RuvC and YqgF families. RuvC functions as a non-covalent complex with the hexameric RuvAB helicase whose mode of action is distinct from that of the typically dimeric or monomeric helicases of superfamilies I and II (95).
Unlike the RNase H fold nucleases, enzymes of the endonuclease and the EndoVII folds have undergone prominent, lineage-specific expansions. Examples include the proliferation of EndoVII-like and Vsr-like nucleases in M.tuberculosis and the two distinct expansions of the PHAC family in pyrococci and Synechocystis. In each of these cases the respective paralogs are clearly more closely related to each other than to homologs in other lineages, which suggests a burst of serial tandem duplications followed by dissemination within the given genome. One of the selective forces triggering these expansions is likely to be defense against invading viral or plasmid DNA; alternatively, or in addition, the genes coding for these nucleases might behave as self-propagating, selfish elements. This is consistent with the recent hypothesis that restriction–modification systems are intrinsically mobile elements (96). In this context, the presence of numerous endonuclease and EndoVII fold proteins in bacterial and archaeal genomes provides clues for the evolutionary origins of the diversity of typical restriction endonucleases, which have probably been recruited from these two folds on multiple occasions. The prevalence of DNA parasitism provided a niche for restriction enzymes to evolve from DNA repair endonucleases and to spread across the prokaryotic world. The colicins of the EndoVII fold represent another form of recruitment of nucleases for defense against intra-specific competitors (97). Exploration of the expanded families of predicted nucleases for new restriction activities seems to be promising.
CONCLUSIONS
A survey of the phyletic distribution of HJRs and related nucleases accompanied by a detailed computer analysis of their structural and evolutionary relationships shows that the HJR function has evolved independently from at least four distinct structural folds. In the course of this analysis it became clear that the endonuclease fold includes by far a greater diversity of nucleases than previously suspected and unifies archaeal HJRs, repair nucleases such as RecB and Vsr, restriction enzymes and a large variety of predicted nucleases whose specific activities await experimental investigation. The analysis of this fold involved structure prediction for structurally uncharacterized important enzymes, such as the AHJR and the RecB family nucleases.
The range of RNase H fold HJRs was also expanded beyond the previously characterized RuvC family by the discovery of a second major family of predicted resolvases that are likely to function as an alternative to RuvC in most bacteria, but possibly as the principal HJR in low-GC Gram-positive bacteria and Aquifex. It was shown that EndoVII of phage T4 serves as a structural template for several nucleases, including McrA and other type II restriction enzymes. Furthermore, EndoVII was unified with colicin E7 to define a distinct metal-dependent nuclease fold.
As the result of this analysis, the principal HJRs are now known or confidently predicted for all bacteria and archaea whose genomes have been completely sequenced. Many species encode multiple potential HJRs, which is compatible with the available genetic data. Horizontal gene transfer, lineage-specific gene loss and gene expansion and non-orthologous gene displacement seem to have made major contributions to the evolution of HJRs and related nucleases. The most notable case of displacement is seen in the Lyme disease spirochete B.burgdorferi, which does not have any of the typical HJRs, but instead encodes, in its chromosome and each of the linear plasmids, members of the λ exonuclease family that are predicted to function in recombination. The abundance and diversity of different classes of HJRs and related nucleases in bacteria and archaea stand in sharp contrast to their paucity in eukaryotes. The few enzymes of the endonuclease fold and the RNase H fold that were detected in eukaryotes probably entered the eukaryotic genomes via horizontal transfer from bacteria. The identity of the eukaryotic HJR(s) remains unknown; this function could be performed by topoisomerase IB or by a novel class of enzymes that remains to be discovered.
Different types of likely HJRs and related nucleases were identified in the genomes of diverse bacterial and eukaryotic DNA viruses, in many of which these enzymes have not been detected previously. Gene exchange between viral and cellular genomes probably played a major role in the evolution of this class of enzymes.
This analysis provides a detailed picture of the distribution of HJRs, key enzymes of recombination, in different life forms and offers scenarios for their evolution. On a more practical note, many of the predicted nucleases could turn out to be new restriction enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank B. Moss and T. Senkevich for critical reading of the manuscript and A. Garcia for discussions. K.S.M. is supported by US Department of Energy OBER grant DE-FG02-98ER62583.
REFERENCES
- 1.Holliday R. (1964) Genet. Res., 5, 282–304. [Google Scholar]
- 2.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
- 3.Lindahl T. and West,S.C. (1995) DNA Repair and Recombination. Chapman & Hall, London, UK.
- 4.Smith P.J. and Jones,C. (2000) DNA Recombination and Repair. Oxford University Press, Oxford, UK.
- 5.Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West S.C. (1992) Annu. Rev. Biochem., 61, 603–640. [DOI] [PubMed] [Google Scholar]
- 7.Iwasaki H., Takahagi,M., Shiba,T., Nakata,A. and Shinagawa,H. (1991) EMBO J., 10, 4381–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitby M.C., Vincent,S.D. and Lloyd,R.G. (1994) EMBO J., 13, 5220–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leipe D.D., Aravind,L., Grishin,N.V. and Koonin,E.V. (2000) Genome Res., 10, 5–16. [PubMed] [Google Scholar]
- 10.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharples G.J., Ingleston,S.M. and Lloyd,R.G. (1999) J. Bacteriol., 181, 5543–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen J.A. and Hanawalt,P.C. (1999) Mutat. Res., 435, 171–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komori K., Sakae,S., Shinagawa,H., Morikawa,K. and Ishino,Y. (1999) Proc. Natl Acad. Sci. USA, 96, 8873–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oram M., Keeley,A. and Tsaneva,I. (1998) Nucleic Acids Res., 26, 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White M.F. and Lilley,D.M. (1997) J. Mol. Biol., 266, 122–134. [DOI] [PubMed] [Google Scholar]
- 16.Sekiguchi J., Seeman,N.C. and Shuman,S. (1996) Proc. Natl Acad. Sci. USA, 93, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin E.V., Mushegian,A.R. and Bork,P. (1996) Trends Genet., 12, 334–336. [PubMed] [Google Scholar]
- 18.Walker D.R. and Koonin,E.V. (1997) ISMB, 5, 333–339. [PubMed] [Google Scholar]
- 19.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z., Schaffer,A.A., Miller,W., Madden,T.L., Lipman,D.J., Koonin,E.V. and Altschul,S.F. (1998) Nucleic Acids Res., 26, 3986–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eddy S.R. (1998) Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuler G.D., Altschul,S.F. and Lipman,D.J. (1991) Proteins, 9, 180–190. [DOI] [PubMed] [Google Scholar]
- 24.Rost B. and Sander,C. (1993) J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 25.Rost B., Schneider,R. and Sander,C. (1997) J. Mol. Biol., 270, 471–480. [DOI] [PubMed] [Google Scholar]
- 26.Jones D.T. (1999) J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 27.Guex N. and Peitsch,M.C. (1997) Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 28.Kraulis P.J. (1991) J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 29.Ariyoshi M., Vassylyev,D.G., Iwasaki,H., Nakamura,H., Shinagawa,H. and Morikawa,K. (1994) Cell, 78, 1063–1072. [DOI] [PubMed] [Google Scholar]
- 30.Lo Conte L., Ailey,B., Hubbard,T.J., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice P. and Mizuuchi,K. (1995) Cell, 82, 209–220. [DOI] [PubMed] [Google Scholar]
- 32.Rice P., Craigie,R. and Davies,D.R. (1996) Curr. Opin. Struct. Biol., 6, 76–83. [DOI] [PubMed] [Google Scholar]
- 33.Yang W. and Steitz,T.A. (1995) Structure, 3, 131–134. [DOI] [PubMed] [Google Scholar]
- 34.Bork P., Sander,C. and Valencia,A. (1992) Proc. Natl Acad. Sci. USA, 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aravind L. and Koonin,E.V. (1999) J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 36.Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haren L., Ton-Hoang,B. and Chandler,M. (1999) Annu. Rev. Microbiol., 53, 245–281. [DOI] [PubMed] [Google Scholar]
- 38.Saito A., Iwasaki,H., Ariyoshi,M., Morikawa,K. and Shinagawa,H. (1995) Proc. Natl Acad. Sci. USA, 92, 7470–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitby M.C. and Dixon,J. (1997) J. Mol. Biol., 272, 509–522. [DOI] [PubMed] [Google Scholar]
- 40.White M.F. and Lilley,D.M. (1996) J. Mol. Biol., 257, 330–341. [DOI] [PubMed] [Google Scholar]
- 41.Moss B. (1996) In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincott-Raven, Philadelphia, PA, pp. 2637–2671.
- 42.Stuart D., Ellison,K., Graham,K. and McFadden,G. (1992) J. Virol., 66, 1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palaniyar N., Gerasimopoulos,E. and Evans,D.H. (1999) J. Mol. Biol., 287, 9–20. [DOI] [PubMed] [Google Scholar]
- 44.Garcia A., Aravind,L., Koonin,E.V. and Moss,B. (2000) Proc. Natl Acad. Sci. USA, 97, 8926–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuwald A.F., Liu,J.S., Lipman,D.J. and Lawrence,C.E. (1997) Nucleic Acids Res., 25, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahdi A.A., Sharples,G.J., Mandal,T.N. and Lloyd,R.G. (1996) J. Mol. Biol., 257, 561–573. [DOI] [PubMed] [Google Scholar]
- 47.Waite-Rees P.A., Keating,C.J., Moran,L.S., Slatko,B.E., Hornstra,L.J. and Benner,J.S. (1991) J. Bacteriol., 173, 5207–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aravind L. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kovall R. and Matthews,B.W. (1997) Science, 277, 1824–1827. [DOI] [PubMed] [Google Scholar]
- 50.Kovall R.A. and Matthews,B.W. (1998) Proc. Natl Acad. Sci. USA, 95, 7893–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winkler F.K., Banner,D.W., Oefner,C., Tsernoglou,D., Brown,R.S., Heathman,S.P., Bryan,R.K., Martin,P.D., Petratos,K. and Wilson,K.S. (1993) EMBO J., 12, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hennecke F., Kolmar,H., Brundl,K. and Fritz,H.J. (1991) Nature, 353, 776–778. [DOI] [PubMed] [Google Scholar]
- 53.Tsutakawa S.E., Jingami,H. and Morikawa,K. (1999) Cell, 99, 615–623. [DOI] [PubMed] [Google Scholar]
- 54.Tsutakawa S.E., Muto,T., Kawate,T., Jingami,H., Kunishima,N., Ariyoshi,M., Kohda,D., Nakagawa,M. and Morikawa,K. (1999) Mol. Cell, 3, 621–628. [DOI] [PubMed] [Google Scholar]
- 55.Tatusov R.L., Galperin,M.Y., Natale,D.A. and Koonin,E.V. (2000) Nucleic Acids Res., 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Su P., Im,H., Hsieh,H., Kang,A.S. and Dunn,N.W. (1999) Appl. Environ. Microbiol., 65, 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu M., Souaya,J. and Julin,D.A. (1998) J. Mol. Biol., 283, 797–808. [DOI] [PubMed] [Google Scholar]
- 58.Kuzminov A. (1999) Microbiol. Mol. Biol. Rev., 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J., Chen,R. and Julin,D.A. (2000) J. Biol. Chem., 275, 507–513. [DOI] [PubMed] [Google Scholar]
- 60.Bae S.H., Choi,E., Lee,K.H., Park,J.S., Lee,S.H. and Seo,Y.S. (1998) J. Biol. Chem., 273, 26880–26890. [DOI] [PubMed] [Google Scholar]
- 61.Makarova K.S., Aravind,L., Galperin,M.Y., Grishin,N.V., Tatusov,R.L., Wolf,Y.I. and Koonin,E.V. (1999) Genome Res., 9, 608–628. [PubMed] [Google Scholar]
- 62.Goldstein J.N. and Weller,S.K. (1998) J. Virol., 72, 8772–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian G., Koonin,E.V. and Aravind,L. (2000) Infect. Immun., 68, 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livey I., Gibbs,C.P., Schuster,R. and Dorner,F. (1995) Mol. Microbiol., 18, 257–269. [DOI] [PubMed] [Google Scholar]
- 65.Zhang J.R., Hardham,J.M., Barbour,A.G. and Norris,S.J. (1997) Cell, 89, 275–285. [DOI] [PubMed] [Google Scholar]
- 66.Sung S.Y., McDowell,J.V., Carlyon,J.A. and Marconi,R.T. (2000) Infect. Immun., 68, 1319–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitsis P.G. and Kwagh,J.G. (1999) Nucleic Acids Res., 27, 3057–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sohail A., Lieb,M., Dar,M. and Bhagwat,A.S. (1990) J. Bacteriol., 172, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janosi L., Yonemitsu,H., Hong,H. and Kaji,A. (1994) J. Mol. Biol., 242, 45–61. [DOI] [PubMed] [Google Scholar]
- 70.Roberts R.J. and Macelis,D. (1998) Nucleic Acids Res., 26, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ban C. and Yang,W. (1998) EMBO J., 17, 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bujnicki J.M. (2000) J. Mol. Evol., 50, 39–44. [DOI] [PubMed] [Google Scholar]
- 73.Athanasiadis A., Vlassi,M., Kotsifaki,D., Tucker,P.A., Wilson,K.S. and Kokkinidis,M. (1994) Nature Struct. Biol., 1, 469–475. [DOI] [PubMed] [Google Scholar]
- 74.Newman M., Strzelecka,T., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1995) Science, 269, 656–663. [DOI] [PubMed] [Google Scholar]
- 75.Kim Y.C., Grable,J.C., Love,R., Greene,P.J. and Rosenberg,J.M. (1990) Science, 249, 1307–1309. [DOI] [PubMed] [Google Scholar]
- 76.Wah D.A., Hirsch,J.A., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1997) Nature, 388, 97–100. [DOI] [PubMed] [Google Scholar]
- 77.Bozic D., Grazulis,S., Siksnys,V. and Huber,R. (1996) J. Mol. Biol., 255, 176–186. [DOI] [PubMed] [Google Scholar]
- 78.Newman M., Lunnen,K., Wilson,G., Greci,J., Schildkraut,I. and Phillips,S.E. (1998) EMBO J., 17, 5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deibert M., Grazulis,S., Janulaitis,A., Siksnys,V. and Huber,R. (1999) EMBO J., 18, 5805–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lukacs C.M., Kucera,R., Schildkraut,I. and Aggarwal,A.K. (2000) Nature Struct. Biol., 7, 134–140. [DOI] [PubMed] [Google Scholar]
- 81.Raaijmakers H., Vix,O., Toro,I., Golz,S., Kemper,B. and Suck,D. (1999) EMBO J., 18, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gorbalenya A.E. (1994) Protein Sci., 3, 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shub D.A., Goodrich-Blair,H. and Eddy,S.R. (1994) Trends Biochem. Sci., 19, 402–404. [DOI] [PubMed] [Google Scholar]
- 84.Golz S., Christoph,A., Birkenkamp-Demtroder,K. and Kemper,B. (1997) Eur. J. Biochem., 245, 573–580. [DOI] [PubMed] [Google Scholar]
- 85.Giraud-Panis M.J. and Lilley,D.M. (1996) J. Biol. Chem., 271, 33148–33155. [DOI] [PubMed] [Google Scholar]
- 86.Ko T.P., Liao,C.C., Ku,W.Y., Chak,K.F. and Yuan,H.S. (1999) Struct. Fold Des., 7, 91–102. [DOI] [PubMed] [Google Scholar]
- 87.Battista J.R., Earl,A.M. and Park,M.J. (1999) Trends Microbiol., 7, 362–365. [DOI] [PubMed] [Google Scholar]
- 88.Mushegian A.R. and Koonin,E.V. (1996) Proc. Natl Acad. Sci. USA, 93, 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edgell D.R. and Doolittle,W.F. (1997) Cell, 89, 995–998. [DOI] [PubMed] [Google Scholar]
- 90.Leipe D.D., Aravind,L. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolt E.L., Sharples,G.J. and Lloyd,R.G. (1999) J. Mol. Biol., 286, 403–415. [DOI] [PubMed] [Google Scholar]
- 92.Parkinson M.J. and Lilley,D.M. (1997) J. Mol. Biol., 270, 169–178. [DOI] [PubMed] [Google Scholar]
- 93.Parkinson M.J., Pohler,J.R. and Lilley,D.M. (1999) Nucleic Acids Res., 27, 682–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kooistra J., Haijema,B.J., Hesseling-Meinders,A. and Venema,G. (1997) Mol. Microbiol., 23, 137–149. [DOI] [PubMed] [Google Scholar]
- 95.George H., Kuraoka,I., Nauman,D.A., Kobertz,W.R., Wood,R.D. and West,S.C. (2000) Curr. Biol., 10, 103–106. [DOI] [PubMed] [Google Scholar]
- 96.Kobayashi I., Nobusato,A., Kobayashi-Takahashi,N. and Uchiyama,I. (1999) Curr. Opin. Genet. Dev., 9, 649–656. [DOI] [PubMed] [Google Scholar]
- 97.Kleanthous C., Hemmings,A.M., Moore,G.R. and James,R. (1998) Mol. Microbiol., 28, 227–233. [DOI] [PubMed] [Google Scholar]