Abstract
One of the most intriguing questions facing modern biology concerns how the genome directs the construction of cells, tissues, and whole organisms. It is tempting to suggest that the part of the genome that does not encode proteins contains architectural plans. We are still far from understanding how these plans work at the level of building tissues and the body as a whole. However, the results of recent studies demonstrate that at the cellular level, special non-coding RNAs serve as scaffolds for the construction of various intracellular structures. The term “architectural RNAs” was proposed to designate this subset of non-coding RNAs. In this review, we discuss the role of architectural RNAs in the construction of the cell nucleus and maintenance of the three-dimensional organization of the genome.
Keywords: Non-coding RNA, Nucleus, 3D genome, Liquid condensate, Transcription
Introduction
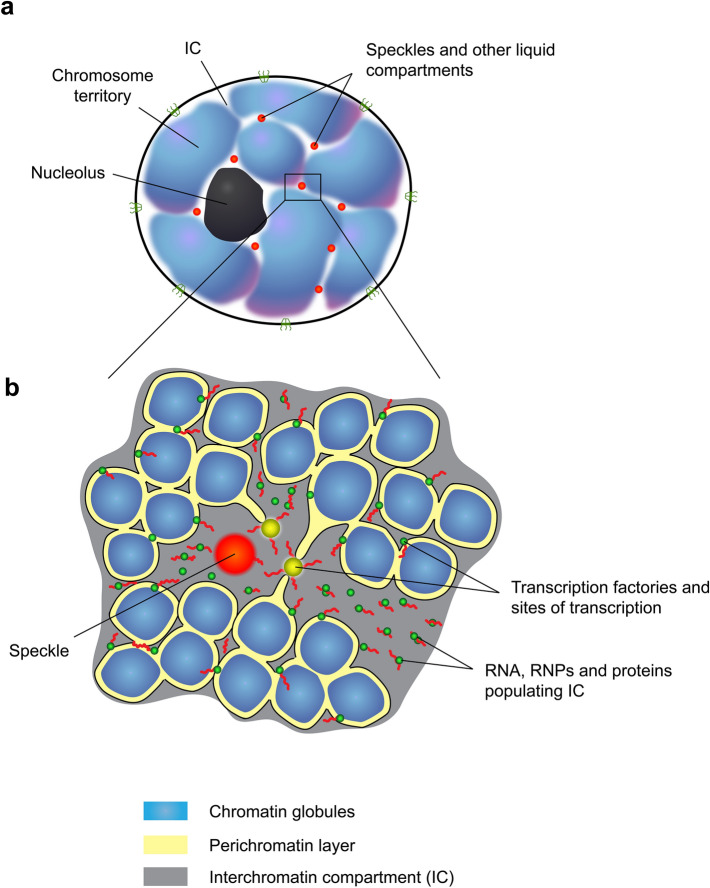
Sequencing of the genomes of humans and a number of other organisms has shown that coding sequences account for only 1.5–2% of the total length of the genome [1]. The functions of the rest of the genome were unclear. It was even suggested that the part of the genome that was proposed to be referred to as junk DNA does not carry any functions at all and is, in a sense, “parasitic”. However, subsequent studies have shown that at least 70% of the entire genome is transcribed [2, 3]. The transcripts of the non-coding part of the genome perform various functions, including regulatory and architectural functions. The idea that non-coding RNAs can participate in maintaining the structure and functional compartmentalization of the cell nucleus arose long ago, when it was demonstrated that inhibition of transcription, or treatment of nuclei with RNase leads to significant changes in nucleus organization. At that time, these observations were interpreted in terms of the participation of RNA in the formation of the nuclear matrix, which was considered to be the structural platform for intranuclear compartmentalization [4–8]. Subsequent studies have demonstrated that a cytoskeleton-like nuclear matrix does not exist [9], which, however, does not exclude the role of RNA in maintaining the structural and functional compartmentalization of the cell nucleus. Modern concepts of the structural and functional compartmentalization of the cell nucleus are largely based on the results of Thomas and Christoph Cremer’s laboratories, which demonstrated that relatively isolated chromosomal territories exist in the interphase nucleus (Fig. 1a). The chromosome territories intermingle with one another and collectively make up the chromatin domain. This domain is spanned by channels that together comprise the interchromatin compartment (IC) [10, 11]. The initial model placed IC mostly between chromosomal territories [11, 12]. With the increase of the resolution of microscopic analysis it becomes clear that IC spans chromosome territories as well. Chromosome territories were found to be composed of chromatin globules and clusters of these globules (chromatin domain clusters) surrounded by IC [13–15]. Transcription is carried out at the border of the chromatin globules and IC in a so-called perichromatin layer (Fig. 1b) [16–18]. Newly synthesized transcripts end up in the IC, which is used for their transport to nuclear pores (Fig. 1b). The same compartment harbors various non-membrane bodies, such as nuclear speckles, paraspeckles, Cajal bodies, PML bodies et cetera [18]. RNA being one of the most represented macromolecules in the IC plays an important role in the formation of this compartment, thus performing an architectural function. In this context, the general properties of RNA are essential, which, for example, contributes to an increase in the mobility of components within macromolecular condensates [19]. Furthermore, a number of non-coding RNAs constitute scaffolds for the assembly of various RNA–protein complexes, providing an increase in the local concentration of certain proteins and the correct positioning of various components of the complex relative to each other. It was these RNAs that were proposed to be termed architectural RNAs. This review examines various aspects of RNA participation in the nuclear architecture functional compartmentalization.
Fig. 1.
Compartmentalization of the cell nucleus. a A schematic of the cell nucleus depicting the existence of chromosome territories and interchromatin compartment (IC) harboring non-membrane nuclear bodies (liquid compartments). b A schematic view of a section of the nucleus showing chromosomal territories composed of chromatin globules. IC surrounds and permeates chromosomal territories. Transcription occurs in a perichromatin layer that lines chromatin globules and is in direct contact with the IC, which contains various nuclear bodies, nascent transcripts, and RNA–protein complexes
Role of RNA in formation of cell nucleus global architecture
Taking into account the fact that a significant portion of primary transcripts is retained in the nucleus [20, 21], one would suppose that they should fulfill some function there. The contribution of that RNA to the nuclear architecture is, perhaps, the first that comes into mind. That is why numerous attempts were made to elucidate the role of RNA in nuclear architecture. In initial experiments, two main strategies were applied. Either transcription was inhibited by drugs, or nuclei were treated with RNase followed by microscopic analysis. Both treatments were reported to cause global reorganization of chromatin within nuclei. More specifically, aggregation of chromatin globules into clumps and repositioning of these clumps toward the nuclear lamina was observed in HeLa cells treated with RNase A [6]. It was also reported that treatment of permeabilized cells with RNase A affected the integrity of centromeric heterochromatin and caused the disintegration of chromocenters (clusters of pericentromeric heterochromatin gathered around nucleoli) [22, 23]. Clearly, treatment of permeabilized cells with RNase is a rather crude experimental approach because the permeabilization itself may contribute to the observed effects. To overcome this problem, in some studies, RNase was delivered to living cells by microinjection [24]. The results essentially confirmed the observations made in permeabilized cells treated with RNase A. The most prominent effect was the aggregation of chromatin clusters at the nuclear periphery [24]. Interestingly, the addition of purified nuclear RNA-to-RNA-depleted nuclei almost completely restored the initial pattern of chromatin distribution within these nuclei [24]. Taking into account the fact that RNA does not contribute to the assembly of nucleosomes, one would consider the participation of RNA in higher levels of chromatin packaging within the cell nucleus. The authors of the above-cited study [24] proposed that a specific fraction of Pol II transcripts termed “chromatin-interlinking RNAs” contribute to the maintenance of the transcriptionally active de-condensed perichromatin layer that is located on the surface of interchromatin channels.
Recently, the contribution of RNA to the global organization of chromatin was addressed by Hi-C analysis of RNase-treated nuclei. It was reported that this treatment did not affect the TAD boundaries and slightly compromised spatial interactions within the inactive chromatin compartment (B-compartment) [25]. The latter may be due to extra-compaction (see above) that prevents long-distant interactions.
At first sight, all of the above-discussed observations do not show what the specific role of RNA is, if any, in maintaining the architecture of the cell nucleus. However, it is worth considering them in the context of the hypothesis that chromatin as a whole and the interchromatin domain constitute two phase-separated compartments within the cell nucleus [26, 27]. Although the existence of the interchromatin compartment is well supported by experimental data [18], the forces ensuring the existence of this compartment (in other words, the forces preventing the collapse of large chromatin masses) remain largely unclear. Taking into account the emerging role of liquid phase separation in biological systems, it is tempting to propose that the IC as a whole represents a phase-separated milieu distinct from the chromatin domain and from nuclear bodies immersed into this milieu. RNA molecules transported along the IC constitute an important architectural component of the IC, providing a scaffold for the assembly of nuclear hnRNP and other RNA–protein complexes [28–30]. Under certain conditions, intrinsically disordered protein − RNA complexes are able to form vesicle-like assemblies. These nucleoprotein vesicles remain highly dynamic and can undergo a reversible vesicle-to-droplet phase transition in response to certain stimuli. [31]. In addition, RNA molecules themselves possess an ability to establish multivalent interactions that can drive the assembly of liquid condensates [32–35]. Recent evidence suggests that the chromatin domain is more solid or gel like [36, 37] and is maintained by polymer–polymer phase separation [38, 39]. In this case, the driving force behind the process is the formation of bridges between nucleosomes located in spatial proximity, which is mediated by chromatin-associated proteins. Bridging-induced collapse of a chromatin fiber can be considered as coil-globule transition. Polymer–polymer phase separation does not require any interactions among bridging proteins. The fluid that surrounds the nucleosomes within collapsed globules does not necessarily phase-separate [38, 39]. Comparing to chromatin domain, the IC is more fluid, and RNA is likely to prevent gelation and enhance fluidity [19, 40]. Chromatin masses are mostly excluded from the IC with the exception of active genes that are located in the perichromatin layer [16–18] and can even loop out into the IC, being attracted to transcription factories (Fig. 1b) [41–43]. The location of active genes at the surface of the IC is likely driven by a high level of histone acetylation [44]. Besides, nascent transcripts that “belong” to the IC [30] constitute an additional anchor of transcribing genes.
To conclude this section, it is necessary to emphasize two important points. First, the above-described organization is highly dynamic. Digestion of nuclear RNA is expected to cause a collapse of the IC. Consequently, the chromatin masses become more aggregated, as was indeed observed in the above-cited studies [6, 24]. Other treatments, such as high-salt extraction, result in the aggregation of the RNA and proteins present in the IC, giving rise to a filament mesh termed an “internal nuclear matrix”, which is in fact a cast of the IC channels [9]. Second, although RNA certainly plays an architectural role in global nuclear organization, this role can hardly be attributed to a particular kind of transcript. Rather, the entire pull of primary transcripts is involved, and the contribution of these transcripts to the organization of the internal nuclear space is temporal and secondary to other more specific functions.
Non-coding RNAs as scaffolds for the assembly of membraneless nuclear bodies and other DNA protein complexes
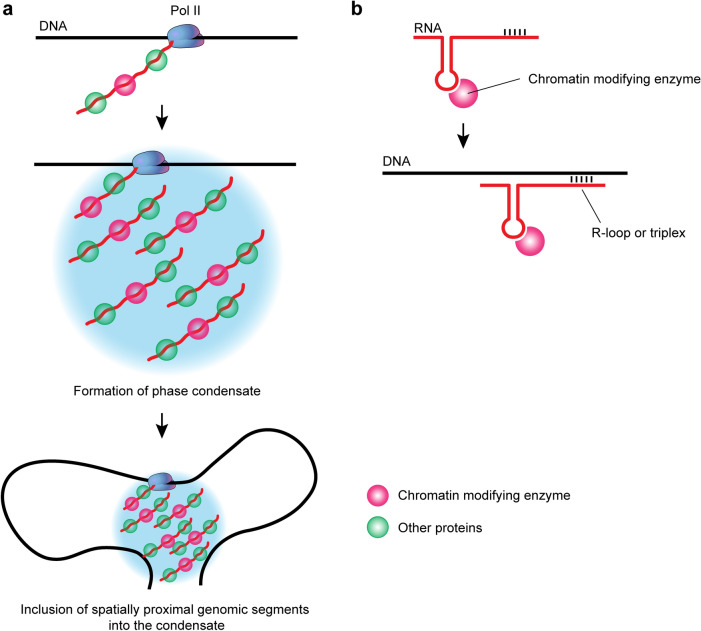
Being a flexible molecule capable of interacting with a number of RNA-binding proteins in a sequence-specific fashion, RNA is an ideal candidate for the scaffold directing the assembly of proteins (RNA–protein complexes). Many non-coding RNAs possess complementary regions of different lengths. Pairing of these regions may either support a specific folding of individual RNAs or result in the association of multiple RNAs in a mesh. Recruiting RNA-binding proteins to this mesh would increase their local concentration to the threshold level necessary to trigger the formation of liquid condensates via interactions of the intrinsically disordered regions present in various RNA-binding proteins (Fig. 2a). In another scenario, folded RNA may ensure the mutual positioning of RNA-binding proteins that is necessary for their assembly in complexes stabilized by specific protein–protein interactions (Fig. 2b). This scaffolding function of RNA has been known since it was demonstrated that both subunits of the translation machine—the ribosome—were assembled on rRNA scaffolds that determine both the shape of ribosomal subunits and mutual positions of individual proteins in the assembly [45–47]. The ribosomal RNAs perfectly fit the definition of architectural RNA that was suggested to designate RNAs that direct the assembly of paraspeckles and several other nuclear bodies [48, 49]. Current studies have demonstrated that non-coding RNAs serve as scaffolds for the assembly and maintaining of various membraneless nuclear compartments (nuclear bodies) [50–52]. Paraspeckles constitute a good example. These are small nuclear bodies that are frequently located close to speckles. Similarly to other nuclear compartments, paraspeckles are involved in the realization of multiple functions, including (i) nuclear retention of mRNA via the adenine to inosine editing process [53], (ii) temporal retention of various regulatory proteins [54, 55], and (iii) microRNA processing [56]. Paraspeckles disappear upon transcription arrest, suggesting that RNA plays a structural function in these nuclear bodies [57]. A specific lncRNA, NEAT1 was found to provide a scaffold for the assembly of paraspeckles [58, 59], reviewed in [60]. This RNA possesses affinity sites for the RNA-binding proteins NONO, SFPQ, RBM14, hnRNP K, FUS, DAZAP1, and hnRNP H3. Each paraspeckle contains approximately 50 NEAT1 RNA molecules held together by RNA–RNA interactions [61]. In addition, the above-mentioned RNA-binding proteins associated with NEAT1 interact with each other and with other proteins, triggering the assembly of a molecular condensate via phase separation [60, 62]. A number of paraspeckle proteins, including FUS and RBM14 that directly interact with NEAT1 RNA, possess intrinsically disordered domains capable of establishing multivalent interactions resulting in the assembly of phase-separated condensates [60]. The association of these proteins with NEAT1 apparently increases their local concentration above a threshold value, enabling the assembly of paraspeckles via phase separation. Besides increasing local concentration, the binding of proteins to specific sites on NEAT1 RNA may ensure the mutual positioning of these proteins, favoring their interaction.
Fig. 2.
RNA as a scaffold for the assembly of phase condensates and protein complexes. a Formation of an RNA–protein condensate on a scaffolding RNA mesh generated by pairing of RNAs containing complementary regions. Increase in the local concentration of RNA-bound proteins above a threshold level leads to phase separation. The formed condensate may retain proteins that are not directly bound to RNA. b Assembly of RNA–protein complexes on a folded RNA scaffold ensuring correct mutual positioning of interacting proteins
In addition to paraspeckles, several other nuclear bodies built on scaffolds of specific lncRNAs were described. Among them are nuclear stress bodies assembled on a scaffold of satellite III lncRNA [63, 64], a perinucleolar compartment assembled on a scaffold of pyrimidine-rich non-coding RNA [65], and Omega speckles built on Drosophila heat shock RNA Omega [66]. Of note, scaffolding RNAs are often transcribed from repetitive sequences and contain repeated motives recognized by RNA-binding proteins. This modularity certainly increases the capabilities of such scaffolds to provide an increase in the local concentration of these RNA-binding proteins [67]. It should be mentioned that conventional bioinformatics tools frequently exclude from the analysis transcripts of repetitive sequences as well as transcripts containing internal repeats. Hence, many architectural RNAs in this category may remain uncharacterized. In some cases, RNA scaffolds enriched in tandem repeats may arise due to the expansion of short repeats in transcribed non-coding DNA sequences, which is associated with the occurrence of a number of neurological and neuromuscular diseases [68–72].
Although nuclear bodies built on the scaffolds of specific RNAs have attracted the most attention of the scientific community, in some cases, any transcripts can play the role of a scaffold. The repair foci constitute a good example. It has been shown that DDR recruits Pol II and other components of the transcription machinery that initiate RNA synthesis. The products of this synthesis—termed damage-induced long non-coding RNA (dilncRNA)—drive the assembly of liquid condensates that contain 53BP and other DDR proteins [73, 74]. Of note, DDR foci are disassembled upon RNase treatment [75, 76], suggesting that they are assembled on an RNA scaffold. Similarly, enhancer RNA (eRNA) appears to serve as a scaffold for the assembly of liquid condensates on superenhancers [77].
It should be mentioned that the exact nature of nuclear bodies scaffolded by RNA is not always well established. Although most of the authors tend to consider nuclear bodies as liquid condensates, some of them do not possess all expected features of liquid condensates [78]. The role of different modes of phase separation in the organization of the cell nucleus has been extensively discussed in several resent reviews [26, 79–81]. For our current discussion, the exact nature of a nuclear body is not of primary interest. We are rather interested in whether this nuclear body is scaffolded by RNA.
Architectural role of lncRNA in higher order chromatin folding
The role of lncRNA in maintaining the higher order chromatin folding may be described by two scenarios: (i) scaffolding of protein complexes mediating histone modification and targeting these complexes to particular areas of the genome and (ii) establishing links between remote genomic elements. As frequently occurs in nature, these scenarios are not mutually exclusive. Nevertheless, below, we shall discuss each of them separately.
Scaffolding of protein complexes mediating histone modification and/or targeting these complexes to particular areas of the genome
Many lncRNAs work in cis via the scaffolding of an assembly of liquid condensates close to the sites of lncRNAs transcription. These condensates may recruit (retain) histone-modifying enzymes due to their affinity to either the scaffolding RNA itself or to some RNA-binding proteins interacting with this RNA (Fig. 3a). This scenario can be exemplified by the MSL complex mediating the hyperactivation of the drosophila male X chromosome. RoX1 and RoX2 RNAs are integral parts of the MSL complex [82]. Their functions are partially redundant and include both scaffolding and targeting of the complex to the affinity sites on the X chromosome. Interestingly, the binding sites for MSL2, one of the key components of the MSL complex, are hidden in RoX1/2 RNAs by stable stem-loop structures that should be remodeled by MLE helicase to trigger the assembly of functional MSL complexes [83, 84]. MSL2 recruits other components of the MSL complex, including MOF histone acetyltransferase, which performs acetylation of histone H4 at lysine K16, causing chromatin decompaction [85]. Of note, RoX and MSL2 form a stable condensate that ensures local trapping of the MSL complex close to the location of RoX transcription, thus ensuring its preferential spreading over the X chromosome [86]. These observations clarify an important question, namely: “Why does lncRNA-directed chromatin remodeling occur in cis close to the site of this lncRNA transcription and regions juxtaposed in the nuclear space to this site?”. A similar mechanism appears to operate during X-chromosome inactivation in mammalian cells. It has long been known that X-chromosome inactivation is directed by lncRNA Xist, which is spread along one of the two X chromosomes in female cells. Xist RNA recruits histone deacetylases and Polycomb complexes [87–89], which repress most of the genes in the inactive copy of the X chromosome. Spreading of Xist over X chromosomes begins from the site of transcription (Xist gene) and several secondary entry sites that are located nearby in the nuclear space [90] and marked by HAS sequences. The question regarding why Xist does not diffuse away from the X chromosome remained obscure until a recent study demonstrated that a complex of Xist RNA with several proteins (PTBP19 MATR3, TDP-43 and CELF11) bound to the affinity sites on this RNA form a condensate that significantly constrained the diffusion of Xist [91].
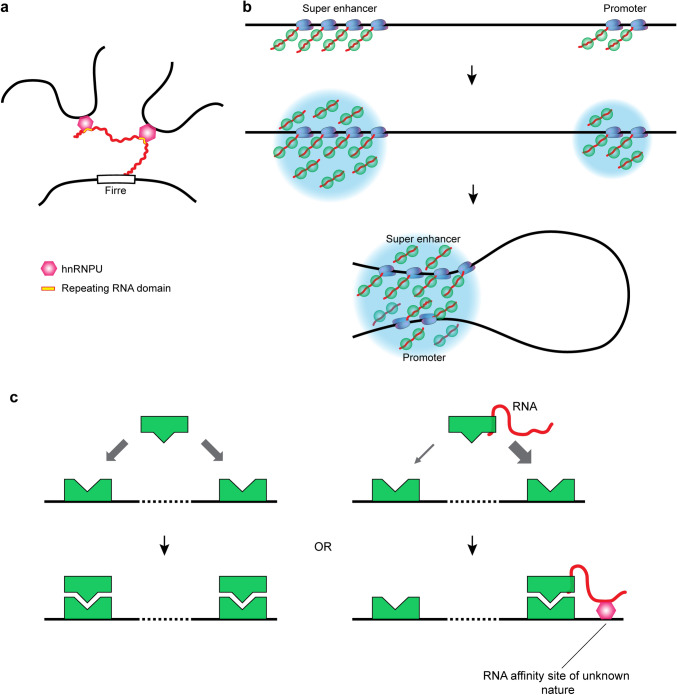
Fig. 3.
Contribution of RNA to targeting chromatin-modifying complexes. a Association of chromatin-modifying enzymes and other proteins with nascent transcripts near the site of transcription may lead to the formation of a phase condensate, which may eventually incorporate remote genomic regions located in spatial proximity to the site of transcription that has nucleated assembly of the liquid condensate. b Delivery of a chromatin-modifying enzyme to a specific genomic site through interaction with RNA that forms RNA–DNA triplex or R-loop with the corresponding genomic site
The expression of specific lncRNAs is typical for imprinted loci. Thus, lncRNA Kcnq1ot1 is paternally expressed antisense to the coding gene Kcnq1 [92]. This lncRNA interacts with H3K9- and H3K27-specific histone methyltransferases, introducing repressive marks into chromatin, and it appears to target these complexes to regions located nearby to the Kcnq1ot1 transcription site [93]. Another lncRNA involved in the establishment of imprinting is Air (Airn) [94]. Recent evidence suggests that Airn mediates the silencing of non-overlapped distant genes via recruiting chromatin-modifying complexes, including PRC2 [95]. Furthermore, the spreading of Airn from the transcription site to the genes to be silenced appears to occur due to a spatial proximity of these regions [95]. The similarity to the mechanism of X-chromosome inactivation directed by Xist RNA seems clear. Of note, both Kcnq1ot1 and Airn act strictly in cis. It is, thus, possible that being complexed with proteins, these lncRNAs also form condensates, which restrict their diffusion.
The scaffolding function of RNA is not limited to the listed examples. Many lncRNAs were found to bind PRC2 and other histone-modifying complexes [96]. The best-studied example is HOTAIR, which serves as a scaffold for polycomb repressive complex 2 (PRC2) and the LSD1/CoREST/REST complex [97]. In contrast to cis-acting lncRNAs, HOTAIR shuffles away from its parental locus (HOXC) and delivers chromatin-repressive complexes to HOXD genes located on another chromosome, thus fulfilling its function in trans [98]. In gastric cancer cells, lncRNA GCAWKR acts as a molecular scaffold of WDR5/KAT2A complexes involved in H3 Lys 4 (H3K4) trimethylation and H3K9 acetylation [99], whereas lncRNA HOXA11-AS provides scaffolding for the chromatin modification factors PRC2, LSD1, and DNMT1 [100].
The mechanisms of attracting RNA complexes with various enzymes to specific sites of the genome are not fully understood. Several studies have demonstrated that the formation of RNA–DNA triplexes and R-loops directs lncRNA-mediated genome targeting of the enzymes modifying epigenetic profiles (Fig. 3b) [101–106].
Establishing links between remote genomic elements
Several mechanisms can be considered by which lncRNAs establish contacts between distant regions of the genome: (i) direct bridging of two or more genomic regions via an RNA molecule, the different parts of which establish contacts with these regions; (ii) assembly of a small liquid compartment (condensate) around an RNA cloud at the site of transcription and the subsequent fusion of this compartment with another compartment of a similar nature assembled at a remote genomic site; (iii) the contribution of lncRNA to the positioning of architectural proteins at specific genomic sites and the modulation of the architectural proteins’ activity (Fig. 4). There are experimental data suggesting that all three scenarios are realized. The first scenario (Fig. 4a) is exemplified by lncRNA Firre, which bears multiple copies of a binding motif for the DNA-associated protein SAFA (also referred to as hnRNPU) and mediates contacts between several genomic regions located on different chromosomes [107]. Similarly, multiple interactions with RNA, which are essential for Polycomb localization to chromatin, may serve to bridge remote chromatin regions together and potentially form Polycomb bodies [108, 109]. Association of genomic regions repressed by Polycomb complexes followed by formation of a Polycomb body (a phase-separated nuclear compartment [110, 111]) is typical for various cells [112, 113]. It is believed that a high local concentration of repressive protein complexes within such compartment provides more stable repression [112]. Whatever is the exact biological function of Polycomb bodies, association of remote genomic fragments within such nuclear compartments shapes the 3D genome.
Fig. 4.
RNA supports 3D genome architecture. a Trans-chromosomal contacts mediated by lncRNA Firre. b A mechanism of RNA-assisted promoter–enhancer communication based on fusing of phase condensates formed around superenhancer and promoter with the assistance of RNA produced from superenhancer and promoter. c Preferential association of a DNA-binding protein with a subset of recognition sites in DNA modulated by association with lncRNA
The other two mechanisms of bridging remote genomic regions with participation of lncRNAs also appear to operate in eukaryotic cells. Most of the enhancers are transcribed, giving rise to short bidirectional transcripts termed enhancer RNA (eRNA) [114, 115]. eRNAs interact with a number of RNA-binding proteins [116–118]. At least at a fraction of the enhancers’ assembly of eRNA complexes with RNA-binding proteins results in the formation of a phase-separated liquid compartment [119]. The fusion of such a compartment with a similar compartment assembled on a target promoter is likely to contribute to establishing enhancer-promoter loops (Fig. 4b). In line with this mechanism, a number of studies have demonstrated that the suppression of eRNA production compromises enhancer-promoter looping [117, 120–122].
Besides the scaffolding assembly of condensates at enhancers, lncRNAs are also likely to modulate the positioning and function of architectural proteins, such as CTCF. CTCF possesses an RNA-binding domain, and the association with RNA was found to be essential for CTCF dimerization [123, 124]. Transcription arrest results in a change of the profile of CTCF binding to genomic DNA and the disruption of at least some of the chromatin loops. Similar effects were observed when mutations were introduced in the DNA sequence encoding Zn fingers involved in RNA binding [124, 125]. These observations suggest that interaction with RNA may ensure preferential CTCF binding to a subset of recognition sites (Fig. 4c). However, it is still not clear whether there is any specificity of CTCF interaction with particular RNAs. Of note, a special group of lncRNAs termed tapRNAs, which are enriched at TAD boundaries and DNA loop anchorage regions have been described. The tapRNA promoters frequently overlap CTCF binding sites, whereas tapRNAs themselves contain specific nucleotide sequence motives that are recognized by Zn fingers [126]. One may speculate that tapRNAs contribute to the positioning and dimerization of CTCF.
The cohesin complex plays an important role in the 3D genome organization [127]. This complex comprises a tripartite ring assembled from SMC1, SMC3, and RAD21 and an additional stromal antigen subunit STAG1 or STAG2. A recent study demonstrated that both STAG1 and STAG2 possess the ability to bind RNA and directly interact with regions containing R loops on dsDNA substrates. Furthermore, in living cells, R loops and SA1/SA2 colocalize at a substantial subset of promoter sites. Taking into account some previous observations [128], the authors propose that, being recruited by promoter and enhancer RNAs, cohesin complexes containing a STAG2 subunit may contribute to the formation of enhancer–promoter loops independently of CTCF [129].
Methods for genome-wide analysis of RNA–DNA interactions
Progress in studying nuclear lncRNAs and their functions in the nuclear architecture and 3D genome organization depends on the ability to identify a genome-wide spectrum of contacts between lncRNAs and genomic DNA. Until recently, only one group of “one-vs-all” biochemical approaches (ChIRP [130], Chart [131], and RAP [90]) had been widely used for addressing this question. These methods are based on the chemical or UV cross-linking of the DNA–protein–RNA complexes in vivo with subsequent purification of the specific complexes that contain a particular RNA with biotinylated complementary oligonucleotides and analysis of the associated DNA fragments via sequencing (Fig. 5a). The application of these methods allowed an extensive study of various lncRNAs, including those involved in the regulation of transcription, particularly in the context of dosage compensation (reviewed in [132]). A drawback of the above-cited methods is that only one RNA can be examined in one experiment. Besides, the experimental design requires that the target RNA be known beforehand (i.e., new RNAs associated with particular genome regions cannot be identified).
Fig. 5.
Methods for studying RNA–chromatin interactions at a genome-wide scale. a “One-vs-all” technologies for mapping sites of chromosomal location for one selected RNA based on the hybridization with complementary biotinylated oligonucleotides. b “All-to-all” technologies based on proximity ligation for generating genome-wide binding profiles for all RNA molecules present in the nucleus (exemplified by Red-C technique). c SPRITE, an “all-to-all” ligation-free technology based on split-pool ligation for identification of multiplex RNA–DNA interactions. d Mapping sites of RNA–chromatin interaction in vivo with RNA–DamID technique
To address these problems, a group of “all-vs-all” methods (MARGI [133], GRID-seq [134], ChAR-seq [135], RADICL-seq [136], and Red-C [137]) has been developed recently, thanks to an increased cost-effectiveness of next-generation sequencing, for the simultaneous identification of the sites of chromatin association for all RNA molecules present in the nucleus. The above-mentioned methods are based on proximity ligation of RNA and DNA in fixed nuclei via a specific bridge adapter, followed by the analysis of the chimeric RNA–DNA molecules by high-throughput sequencing (Fig. 5b). The application of these methods disclosed the global RNA–DNA interactome, the analysis of which allowed researchers not only to support conclusions about the localization of well-known lncRNAs such as MALAT1 [133, 134], RoX [134, 135, 137], and XIST [137] but also to reveal new candidates for the role of architectural RNAs supporting the spatial genome organization. For example, using the Red-C technique, we identified two miRNAs that associate with the repressed chromatin genome wide and may, therefore, contribute to the formation of the heterochromatin nuclear compartment [137]. Notably, the analysis of the RNA–DNA interactome also permitted the features of protein-coding gene transcription to be investigated. Being focused on the contacts between mRNA and its own gene, we showed that introns are spliced out immediately during transcription, thus providing new evidence for a co-transcriptional splicing model [137]. We also demonstrated that the majority of structural genes remain linear in the course of transcription, refuting a popular model suggesting that actively transcribed genes are circularized [138].
Another “all-vs-all” strategy for the identification of RNA–DNA interactions is realized within the framework of SPRITE technology [139], which uses barcodes to label DNA and RNA molecules present in one nucleoprotein complex (Fig. 5c). Using SPRITE, the organization of genomic DNA around two major types of RNA-containing nuclear compartments was revealed—around speckles that contain spliceosomal RNAs (active compartment) and around the nucleolus that contains ribosomal RNA (inactive compartment). The application of a recent modification of SPRITE technology that specifically focuses on RNA–DNA interactions [140] identified hundreds of lncRNAs that form stable nuclear compartments in spatial proximity to their transcriptional loci and regulate genes contained within these loci. In addition, the results of this study support a key role of lncRNAs in the assembly of heterochromatin and various nuclear compartments, including speckles, nucleolus, Cajal bodies, and histone locus bodies [140].
An important advantage of SPRITE over ligation-based approaches is that SPRITE can be used for the analysis of multiplex interactions that occur simultaneously in the cell, whereas ligation-based approaches can only be used to probe pairwise interactions. Another ligation-free method capable of probing multiplex chromatin interactions (GAM [141]) is based on the sequencing of DNA collected from ultrathin cryosections of fixed nuclei. Although this method has been developed for the analysis of DNA–DNA interactions, it potentially could be used for studying RNA–DNA interactions as well.
In the context of this section, RNA immunoprecipitation (RIP) technique is also worth mentioning. In this technique, RNA–protein complexes isolated from fixed cells are subjected to immunoprecipitation with antibodies against a protein of interest followed by the analysis of associated RNA fragments via sequencing. Being coupled with antibodies against chromatin proteins, RIP has long been used for the identification of functional lncRNAs operating on chromatin [142, 143]. The method, however, does not report genomic sites to which chromatin-associated RNAs are recruited and even does not answer a question of whether an RNA is associated with genomic DNA in principle or just bound to a free nucleoplasmic protein.
It should be noted that all of the about-mentioned methods rely on cross-linking, which can potentially introduce artifacts [144]. It would be important to develop technologies allowing an analysis of RNA–DNA interactions in a more native context. A step in this direction was taken in a recent study where a lncRNA of interest (RoX) was tagged with MS2 stem loops and coexpressed with MS2 coat protein (MCP) fused with Dam methyltransferase. Under these conditions, genomic regions occupied by RoX were selectively methylated at GATC sequences, enabling the detection of RoX–genome interactions in vivo with high sensitivity and accuracy [145] (Fig. 5d). New approaches will certainly be of assistance for the in vivo functional characterization of an expanding list of lncRNAs operating in the cell nucleus.
Acknowledgements
This work was done in frame of the Interdisciplinary Scientific and Educational School of Moscow University “Molecular Technologies of the Living Systems and Synthetic Biology”.
Author contributions
Both authors contributed to the literature review and writing of the manuscript.
Funding
This work was supported by grant 075-15-2019-1661 from the Ministry of Science and Higher Education of the Russian Federation.
Declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.International Human Genome Sequencing C Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Encode Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He D, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He DC, Martin T, Penman S. Localization of heterogeneous nuclear ribonucleoprotein in the interphase nuclear matrix core filaments and on perichromosomal filaments at mitosis. Proc Natl Acad Sci U S A. 1991;88:7469–7473. doi: 10.1016/s0309-1651(05)80024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickerson JA, Krochmalnic G, Wan KM, Penman S. Chromatin architecture and nuclear RNA. Proc Natl Acad Sci USA. 1989;86:177–181. doi: 10.1073/pnas.86.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barboro P, D'Arrigo C, Diaspro A, Mormino M, Alberti I, Parodi S, Patrone E, Balbi C. Unraveling the organization of the internal nuclear matrix: RNA-dependent anchoring of NuMA to a lamin scaffold. Exp Cell Res. 2002;279:202–218. doi: 10.1006/excr.2002.5605. [DOI] [PubMed] [Google Scholar]
- 8.Ioudinkova E, Razin SV, Borunova V, de Conto F, Rynditch A, Scherrer K. RNA-dependent nuclear matrix contains a 33 kb globin full domain transcript as well as prosomes but no 26S proteasomes. J Cell Biochem. 2005;94:529–539. doi: 10.1002/jcb.20306. [DOI] [PubMed] [Google Scholar]
- 9.Razin SV, Iarovaia OV, Vassetzky YS. A requiem to the nuclear matrix: from a controversial concept to 3D organization of the nucleus. Chromosoma. 2014;123:217–224. doi: 10.1007/s00412-014-0459-8. [DOI] [PubMed] [Google Scholar]
- 10.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 11.Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathieu U, Jauch A, Emmerich P, Scherthan H, Ried T, Cremer C, Lichter P. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol. 1993;58:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- 12.Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993;1:93–106. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]
- 13.Cremer T, Cremer M, Hubner B, Strickfaden H, Smeets D, Popken J, Sterr M, Markaki Y, Rippe K, Cremer C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589(20 Pt A):2931–2943. doi: 10.1016/j.febslet.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 14.Kirmes I, Szczurek A, Prakash K, Charapitsa I, Heiser C, Musheev M, Schock F, Fornalczyk K, Ma D, Birk U, Cremer C, Reid G. A transient ischemic environment induces reversible compaction of chromatin. Genome Biol. 2015;16:246. doi: 10.1186/s13059-015-0802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szczurek A, Birk U, Knecht H, Dobrucki J, Mai S, Cremer C. Super-resolution binding activated localization microscopy through reversible change of DNA conformation. Nucleus. 2018;9:182–189. doi: 10.1080/19491034.2017.1419846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cremer M, Cremer T. Nuclear compartmentalization, dynamics, and function of regulatory DNA sequences. Genes Chromosomes Cancer. 2018;58:427–436. doi: 10.1002/gcc.22714. [DOI] [PubMed] [Google Scholar]
- 17.Cremer T, Cremer M, Cremer C. The 4D nucleome: genome compartmentalization in an evolutionary context. Biochemistry (Mosc) 2018;83:313–325. doi: 10.1134/S000629791804003X. [DOI] [PubMed] [Google Scholar]
- 18.Cremer T, Cremer M, Hubner B, Silahtaroglu A, Hendzel M, Lanctot C, Strickfaden H, Cremer C. The interchromatin compartment participates in the structural and functional organization of the cell nucleus. BioEssays. 2020;42:e1900132. doi: 10.1002/bies.201900132. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA controls PolyQ protein phase transitions. Mol Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherrer K. Primary transcripts: From the discovery of RNA processing to current concepts of gene expression - Review. Exp Cell Res. 2018;373:1–33. doi: 10.1016/j.yexcr.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Palazzo AF, Lee ES. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet. 2018;9:440. doi: 10.3389/fgene.2018.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 23.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, Choo KH. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caudron-Herger M, Muller-Ott K, Mallm JP, Marth C, Schmidt U, Fejes-Toth K, Rippe K. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus. 2011;2:410–424. doi: 10.4161/nucl.2.5.17736. [DOI] [PubMed] [Google Scholar]
- 25.Barutcu AR, Blencowe BJ, Rinn JL (2019) Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep 20:e48068. 10.15252/embr.201948068 [DOI] [PMC free article] [PubMed]
- 26.Razin SV, Gavrilov AA. The Role of liquid-liquid phase separation in the compartmentalization of cell nucleus and spatial genome organization. Biochemistry (Mosc) 2020;85:643–650. doi: 10.1134/S0006297920060012. [DOI] [PubMed] [Google Scholar]
- 27.Razin SV, Ulianov SV. Divide and rule: phase separation in eukaryotic genome functioning. Cells. 2020 doi: 10.3390/cells9112480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Jove Navarro M, Kashida S, Chouaib R, Souquere S, Pierron G, Weil D, Gueroui Z. RNA is a critical element for the sizing and the composition of phase-separated RNA-protein condensates. Nat Commun. 2019;10:3230. doi: 10.1038/s41467-019-11241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. 2021;22:183–195. doi: 10.1038/s41580-020-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adekunle DA, Hubstenberger A. The multiscale and multiphase organization of the transcriptome. Emerg Top Life Sci. 2020;4:265–280. doi: 10.1042/ETLS20190187. [DOI] [PubMed] [Google Scholar]
- 31.Alshareedah I, Moosa MM, Raju M, Potoyan DA, Banerjee PR. Phase transition of RNA-protein complexes into ordered hollow condensates. Proc Natl Acad Sci U S A. 2020;117:15650–15658. doi: 10.1073/pnas.1922365117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Treeck B, Parker R. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell. 2018;174:791–802. doi: 10.1016/j.cell.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A. 2018;115:2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, Gladfelter AS. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell. 2020;183(1772–1784):e1713. doi: 10.1016/j.cell.2020.11.027. [DOI] [PubMed] [Google Scholar]
- 37.Stephens AD, Liu PZ, Kandula V, Chen H, Almassalha LM, Herman C, Backman V, O'Halloran T, Adam SA, Goldman RD, Banigan EJ, Marko JF. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol Biol Cell. 2019;30:2320–2330. doi: 10.1091/mbc.E19-05-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdel F, Rademacher A, Vlijm R, Tunnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C, Schumacher S, Al Alwash A, Normand C, Herten DP, Engelhardt J, Rippe K. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell. 2020;78:236–249.e7. doi: 10.1016/j.molcel.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erdel F, Rippe K. Formation of chromatin subcompartments by phase separation. Biophys J. 2018;114:2262–2270. doi: 10.1016/j.bpj.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhine K, Vidaurre V, Myong S. RNA Droplets. Annu Rev Biophys. 2020;49:247–265. doi: 10.1146/annurev-biophys-052118-115508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Razin SV, Gavrilov AA, Pichugin A, Lipinski M, Iarovaia OV, Vassetzky YS. Transcription factories in the context of the nuclear and genome organization. Nucleic Acids Res. 2011;39:9085–9092. doi: 10.1093/nar/gkr683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 43.Cook PR, Marenduzzo D. Transcription-driven genome organization: a model for chromosome structure and the regulation of gene expression tested through simulations. Nucleic Acids Res. 2018;46:9895–9906. doi: 10.1093/nar/gky763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gavrilov AA, Shevelyov YY, Ulianov SV, Khrameeva EE, Kos P, Chertovich A, Razin SV. Unraveling the mechanisms of chromatin fibril packaging. Nucleus. 2016;7:319–324. doi: 10.1080/19491034.2016.1190896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agalarov SC, Zheleznyakova EN, Selivanova OM, Zheleznaya LA, Matvienko NI, Vasiliev VD, Spirin AS. In vitro assembly of a ribonucleoprotein particle corresponding to the platform domain of the 30S ribosomal subunit. Proc Natl Acad Sci U S A. 1998;95:999–1003. doi: 10.1073/pnas.95.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Culver GM, Noller HF. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–843. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khaitovich P, Tenson T, Kloss P, Mankin AS. Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry. 1999;38:1780–1788. doi: 10.1021/bi9822473. [DOI] [PubMed] [Google Scholar]
- 48.Chujo T, Hirose T (2017) Nuclear bodies built on architectural long noncoding RNAs: unifying principles of their construction and function. Mol Cells 40:889–896. 10.14348/molcells.2017.0263 [DOI] [PMC free article] [PubMed]
- 49.Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta. 2016;1859:139–146. doi: 10.1016/j.bbagrm.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 50.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 52.Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ. 2012;54:44–54. doi: 10.1111/j.1440-169X.2011.01303.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen LL, Carmichael GG. Nuclear Editing of mRNA 3'-UTRs. Curr Top Microbiol Immunol. 2012;353:111–121. doi: 10.1007/82_2011_149. [DOI] [PubMed] [Google Scholar]
- 54.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, Kai C, Yada T, Suzuki Y, Yamada T, Ozawa T, Kaneki K, Inoue T, Kobayashi M, Kodama T, Wada Y, Sekimizu K, Akimitsu N. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang L, Shao C, Wu QJ, Chen G, Zhou J, Yang B, Li H, Gou LT, Zhang Y, Wang Y, Yeo GW, Zhou Y, Fu XD. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat Struct Mol Biol. 2017;24:816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.e05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43:124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 61.Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell. 2018;70(1038–1053):e1037. doi: 10.1016/j.molcel.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 62.Hirose T, Yamazaki T, Nakagawa S. Molecular anatomy of the architectural NEAT1 noncoding RNA: The domains, interactors, and biogenesis pathway required to build phase-separated nuclear paraspeckles. Wiley Interdisciplin Rev RNA. 2019;10:e1545. doi: 10.1002/wrna.1545. [DOI] [PubMed] [Google Scholar]
- 63.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goenka A, Sengupta S, Pandey R, Parihar R, Mohanta GC, Mukerji M, Ganesh S. Human satellite-III non-coding RNAs modulate heat-shock-induced transcriptional repression. J Cell Sci. 2016;129:3541–3552. doi: 10.1242/jcs.189803. [DOI] [PubMed] [Google Scholar]
- 65.Yap K, Mukhina S, Zhang G, Tan JSC, Ong HS, Makeyev EV. A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol Cell. 2018;72(525–540):e513. doi: 10.1016/j.molcel.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC. Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci. 2000;113(Pt 19):3485–3497. doi: 10.1242/jcs.113.19.3485. [DOI] [PubMed] [Google Scholar]
- 67.Ninomiya K, Hirose T. Short tandem repeat-enriched architectural RNAs in nuclear bodies: functions and associated diseases. Non-coding RNA. 2020 doi: 10.3390/ncrna6010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niimi Y, Takahashi M, Sugawara E, Umeda S, Obayashi M, Sato N, Ishiguro T, Higashi M, Eishi Y, Mizusawa H, Ishikawa K. Abnormal RNA structures (RNA foci) containing a penta-nucleotide repeat (UGGAA)n in the Purkinje cell nucleus is associated with spinocerebellar ataxia type 31 pathogenesis. Neuropathology . 2013;33:600–611. doi: 10.1111/neup.12032. [DOI] [PubMed] [Google Scholar]
- 69.Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, Takahashi M, Matsuura T, Flanigan KM, Iwasaki S, Ishino F, Saito Y, Murayama S, Yoshida M, Hashizume Y, Takahashi Y, Tsuji S, Shimizu N, Toda T, Ishikawa K, Mizusawa H. Spinocerebellar ataxia type 31 is associated with "inserted" penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swinnen B, Robberecht W, Van Den Bosch L (2020) RNA toxicity in non-coding repeat expansion disorders. EMBO J 39:e101112. 10.15252/embj.2018101112 [DOI] [PMC free article] [PubMed]
- 71.Castro AF, Loureiro JR, Bessa J, Silveira I. Antisense transcription across nucleotide repeat expansions in neurodegenerative and neuromuscular diseases: progress and mysteries. Genes. 2020;11:1418. doi: 10.3390/genes11121418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang N, Ashizawa T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr Opin Genet Dev. 2017;44:17–29. doi: 10.1016/j.gde.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michelini F, Pitchiaya S, Vitelli V, Sharma S, Gioia U, Pessina F, Cabrini M, Wang Y, Capozzo I, Iannelli F, Matti V, Francia S, Shivashankar GV, Walter NG, d'Adda di Fagagna F. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat Cell Biol. 2017;19:1400–1411. doi: 10.1038/ncb3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, Cerbino R, Parazzoli D, Rothenberg E, d'Adda di Fagagna F. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019;21:1286–1299. doi: 10.1038/s41556-019-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, Melton DW, Jullien D, Adachi Y. 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci. 2005;118(Pt 9):2043–2055. doi: 10.1242/jcs.02336. [DOI] [PubMed] [Google Scholar]
- 77.Arnold PR, Wells AD, Li XC. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front Cell Develop Biol. 2020;7:377. doi: 10.3389/fcell.2019.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peng A, Weber SC. Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Non-coding RNA. 2019;5:50. doi: 10.3390/ncrna5040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sehgal PB, Westley J, Lerea KM, DiSenso-Browne S, Etlinger JD. Biomolecular condensates in cell biology and virology: Phase-separated membraneless organelles (MLOs) Anal Biochem. 2020;597:113691. doi: 10.1016/j.ab.2020.113691. [DOI] [PubMed] [Google Scholar]
- 80.Lesne A, Baudement MO, Rebouissou C, Forne T. Exploring mammalian genome within phase-separated nuclear bodies: experimental methods and implications for gene expression. Genes. 2019;10:1049. doi: 10.3390/genes10121049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palikyras S, Papantonis A. Modes of phase separation affecting chromatin regulation. Open Biol. 2019;9:190167. doi: 10.1098/rsob.190167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ilik I, Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 2009;6:113–121. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- 83.Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell. 2013;51:174–184. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 84.Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, Toscano S, Wan Y, Spitale RC, Luscombe N, Backofen R, Chang HY, Akhtar A. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51:156–173. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lucchesi JC, Kuroda MI. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 2015;7:a019398. doi: 10.1101/cshperspect.a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Valsecchi CIK, Basilicata MF, Georgiev P, Gaub A, Seyfferth J, Kulkarni T, Panhale A, Semplicio G, Manjunath V, Holz H, Dasmeh P, Akhtar A. RNA nucleation by MSL2 induces selective X chromosome compartmentalization. Nature. 2021;589:137–142. doi: 10.1038/s41586-020-2935-z. [DOI] [PubMed] [Google Scholar]
- 87.Almeida M, Pintacuda G, Masui O, Koseki Y, Gdula M, Cerase A, Brown D, Mould A, Innocent C, Nakayama M, Schermelleh L, Nesterova TB, Koseki H, Brockdorff N. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science. 2017;356:1081–1084. doi: 10.1126/science.aal2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, Brockdorff N. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell. 2017;68(955–969):e910. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Colognori D, Sunwoo H, Kriz AJ, Wang CY, Lee JT. Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol Cell. 2019;74(101–117):e110. doi: 10.1016/j.molcel.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Mancia Leon WR, Damianov A, Chronis C, Papp B, Chen CK, McKee R, Wang XJ, Chau A, Sabri S, Leonhardt H, Zheng S, Guttman M, Black DL, Plath K. A protein assembly mediates Xist localization and gene silencing. Nature. 2020;587:145–151. doi: 10.1038/s41586-020-2703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Gondor A, Grange T, Ohlsson R, Kanduri C. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol. 2004;24:7855–7862. doi: 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 94.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 95.Andergassen D, Muckenhuber M, Bammer PC, Kulinski TM, Theussl HC, Shimizu T, Penninger JM, Pauler FM, Hudson QJ. The Airn lncRNA does not require any DNA elements within its locus to silence distant imprinted genes. PLoS Genet. 2019;15:e1008268. doi: 10.1371/journal.pgen.1008268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma M, Zhang Y, Weng M, Hu Y, Xuan Y, Hu Y, Lv K. lncRNA GCAWKR promotes gastric cancer development by scaffolding the chromatin modification factors WDR5 and KAT2A. Mol Ther. 2018;26:2658–2668. doi: 10.1016/j.ymthe.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, Xie M, Xu L, De W, Wang Z, Wang J. LncRNA HOXA11-as promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 101.Kuo CC, Hanzelmann S, Senturk Cetin N, Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt I, Kurian L, Costa IG. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019;47:e32. doi: 10.1093/nar/gkz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kalwa M, Hanzelmann S, Otto S, Kuo CC, Franzen J, Joussen S, Fernandez-Rebollo E, Rath B, Koch C, Hofmann A, Lee SH, Teschendorff AE, Denecke B, Lin Q, Widschwendter M, Weinhold E, Costa IG, Wagner W. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44:10631–10643. doi: 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Leary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson MJ, Anastasov N. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015;11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 104.Zhao H, Xu Q. Long non-coding RNA DLX6-AS1 mediates proliferation, invasion and apoptosis of endometrial cancer cells by recruiting p300/E2F1 in DLX6 promoter region. J Cell Mol Med. 2020;24:12572–12584. doi: 10.1111/jcmm.15810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blank-Giwojna A, Postepska-Igielska A, Grummt I. lncRNA KHPS1 activates a poised enhancer by triplex-dependent recruitment of epigenomic regulators. Cell Rep. 2019;26(2904–2915):e2904. doi: 10.1016/j.celrep.2019.02.059. [DOI] [PubMed] [Google Scholar]
- 106.Ariel F, Lucero L, Christ A, Mammarella MF, Jegu T, Veluchamy A, Mariappan K, Latrasse D, Blein T, Liu C, Benhamed M, Crespi M. R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol Cell. 2020;77(1055–1065):e1054. doi: 10.1016/j.molcel.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 107.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Long Y, Hwang T, Gooding AR, Goodrich KJ, Rinn JL, Cech TR. RNA is essential for PRC2 chromatin occupancy and function in human pluripotent stem cells. Nat Genet. 2020;52:931–938. doi: 10.1038/s41588-020-0662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wani AH, Boettiger AN, Schorderet P, Ergun A, Munger C, Sadreyev RI, Zhuang X, Kingston RE, Francis NJ. Chromatin topology is coupled to Polycomb group protein subnuclear organization. Nat Commun. 2016;7:10291. doi: 10.1038/ncomms10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Seif E, Kang JJ, Sasseville C, Senkovich O, Kaltashov A, Boulier EL, Kapur I, Kim CA, Francis NJ. Phase separation by the polyhomeotic sterile alpha motif compartmentalizes Polycomb Group proteins and enhances their activity. Nat Commun. 2020;11:5609. doi: 10.1038/s41467-020-19435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, Zhen CY, Ma B, Wang H, Ren X. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem. 2019;294:1451–1463. doi: 10.1074/jbc.RA118.006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pirrotta V, Li HB. A view of nuclear polycomb bodies. Curr Opin Genet Dev. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smigova J, Juda P, Krejci J, Raska I. Structural basis of polycomb bodies. Folia Biol (Praha) 2014;60(Suppl 1):13–20. doi: 10.14712/fb2014060S10013. [DOI] [PubMed] [Google Scholar]
- 114.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, Lauberth SM. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat Struct Mol Biol. 2018;25:687–697. doi: 10.1038/s41594-018-0102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, Wang S, Suter T, Alshareedah I, Gamliel A, Ma Q, Zhang J, Hu Y, Tan Y, Ohgi KA, Jayani RS, Banerjee PR, Aggarwal AK, Rosenfeld MG. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat Struct Mol Biol. 2019;26:193–203. doi: 10.1038/s41594-019-0190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pnueli L, Rudnizky S, Yosefzon Y, Melamed P. RNA transcribed from a distal enhancer is required for activating the chromatin at the promoter of the gonadotropin alpha-subunit gene. Proc Natl Acad Sci U S A. 2015;112:4369–4374. doi: 10.1073/pnas.1414841112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang Y, Su Z, Song X, Liang B, Zeng F, Chang X, Huang D. Enhancer RNA-driven looping enhances the transcription of the long noncoding RNA DHRS4-AS1, a controller of the DHRS4 gene cluster. Sci Rep. 2016;6:20961. doi: 10.1038/srep20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee GS, Chen S, Balk SP, Liu XS, Brown M, Kantoff PW. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saldana-Meyer R, Gonzalez-Buendia E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F, Reinberg D. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014;28:723–734. doi: 10.1101/gad.236869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saldana-Meyer R, Rodriguez-Hernaez J, Escobar T, Nishana M, Jacome-Lopez K, Nora EP, Bruneau BG, Tsirigos A, Furlan-Magaril M, Skok J, Reinberg D. RNA interactions are essential for CTCF-mediated genome organization. Mol Cell. 2019;76(412–422):e415. doi: 10.1016/j.molcel.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldana-Meyer R, Reinberg D, Darzacq X, Tjian R. Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol Cell. 2019;76(395–411):e313. doi: 10.1016/j.molcel.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Amaral PP, Leonardi T, Han N, Vire E, Gascoigne DK, Arias-Carrasco R, Buscher M, Pandolfini L, Zhang A, Pluchino S, Maracaja-Coutinho V, Nakaya HI, Hemberg M, Shiekhattar R, Enright AJ, Kouzarides T. Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018;19:32. doi: 10.1186/s13059-018-1405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kojic A, Cuadrado A, De Koninck M, Gimenez-Llorente D, Rodriguez-Corsino M, Gomez-Lopez G, Le Dily F, Marti-Renom MA, Losada A. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol. 2018;25:496–504. doi: 10.1038/s41594-018-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pan H, Jin M, Ghadiyaram A, Kaur P, Miller HE, Ta HM, Liu M, Fan Y, Mahn C, Gorthi A, You C, Piehler J, Riehn R, Bishop AJR, Tao YJ, Wang H. Cohesin SA1 and SA2 are RNA binding proteins that localize to RNA containing regions on DNA. Nucleic Acids Res. 2020;48:5639–5655. doi: 10.1093/nar/gkaa284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chu C, Spitale RC, Chang HY. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 133.Sridhar B, Rivas-Astroza M, Nguyen TC, Chen W, Yan Z, Cao X, Hebert L, Zhong S. Systematic mapping of RNA-chromatin interactions in vivo. Curr Biol. 2017;27:602–609. doi: 10.1016/j.cub.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li X, Zhou B, Chen L, Gou LT, Li H, Fu XD. GRID-seq reveals the global RNA-chromatin interactome. Nat Biotechnol. 2017;35:940–950. doi: 10.1038/nbt.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bell JC, Jukam D, Teran NA, Risca VI, Smith OK, Johnson WL, Skotheim JM, Greenleaf WJ, Straight AF (2018) Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife 7:e27024. 10.7554/eLife.27024 [DOI] [PMC free article] [PubMed]
- 136.Bonetti A, Agostini F, Suzuki AM, Hashimoto K, Pascarella G, Gimenez J, Roos L, Nash AJ, Ghilotti M, Cameron CJF, Valentine M, Medvedeva YA, Noguchi S, Agirre E, Kashi K, Samudyata LJ, Cazzoli R, Agrawal S, Luscombe NM, Blanchette M, Kasukawa T, Hoon M, Arner E, Lenhard B, Plessy C, Castelo-Branco G, Orlando V, Carninci P. RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat Commun. 2020;11:1018. doi: 10.1038/s41467-020-14337-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gavrilov AA, Zharikova AA, Galitsyna AA, Luzhin AV, Rubanova NM, Golov AK, Petrova NV, Logacheva MD, Kantidze OL, Ulianov SV, Magnitov MD, Mironov AA, Razin SV. Studying RNA-DNA interactome by Red-C identifies noncoding RNAs associated with various chromatin types and reveals transcription dynamics. Nucleic Acids Res. 2020;48:6699–6714. doi: 10.1093/nar/gkaa457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hampsey M, Singh BN, Ansari A, Laine JP, Krishnamurthy S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv Enzyme Regul. 2011;51:118–125. doi: 10.1016/j.advenzreg.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, Trinh V, Aznauryan E, Russell P, Cheng C, Jovanovic M, Chow A, Cai L, McDonel P, Garber M, Guttman M. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174(744–757):e724. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Quinodoz S, Bhat P, Ollikainen N, Jachowicz J, AK B, Chovanec P, Blanco M, Chow A, Markaki Y, Plath K, Guttman M (2020) RNA promotes the formation of spatial compartments in the nucleus. bioRxiv. 10.1101/2020.08.25.267435 [DOI] [PMC free article] [PubMed]
- 141.Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, Fraser J, Dostie J, Game L, Dillon N, Edwards PA, Nicodemi M, Pombo A. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543:519–524. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gilbert C, Svejstrup JQ (2006) RNA immunoprecipitation for determining RNA-protein associations in vivo. Curr Protoc Mol Biol 75:27.4.1–27.4.11. 10.1002/0471142727.mb2704s75 [DOI] [PubMed]
- 143.Hendrickson DG, Kelley DR, Tenen D, Bernstein B, Rinn JL. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gavrilov A, Razin SV, Cavalli G. In vivo formaldehyde cross-linking: it is time for black box analysis. Brief Funct Genomics. 2015;14:163–165. doi: 10.1093/bfgp/elu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cheetham SW, Brand AH. RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites. Nat Struct Mol Biol. 2018;25:109–114. doi: 10.1038/s41594-017-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]