Abstract
The remodeling of the mitochondrial network is a critical process in maintaining cellular homeostasis and is intimately related to mitochondrial function. The interplay between the formation of new mitochondria (biogenesis) and the removal of damaged mitochondria (mitophagy) provide a means for the repopulation of the mitochondrial network. Additionally, mitochondrial fission and fusion serve as a bridge between biogenesis and mitophagy. In recent years, the importance of these processes has been characterised in multiple tissue- and cell-types, and under various conditions. In skeletal muscle, the robust remodeling of the mitochondrial network is observed, particularly after injury where large portions of the tissue/cell structures are damaged. The significance of mitochondrial remodeling in regulating skeletal muscle regeneration has been widely studied, with alterations in mitochondrial remodeling processes leading to incomplete regeneration and impaired skeletal muscle function. Needless to say, important questions related to mitochondrial remodeling and skeletal muscle regeneration still remain unanswered and require further investigation. Therefore, this review will discuss the known molecular mechanisms of mitochondrial network remodeling, as well as integrate these mechanisms and discuss their relevance in myogenesis and regenerating skeletal muscle.
Keywords: Mitochondria, Mitophagy, Biogenesis, Fission, Fusion, Skeletal muscle, Skeletal muscle stem cells, Regeneration
Introduction
Mitochondria are organelles that are vital for energy production, cell survival, and stress regulation. Mitochondria are capable of manipulating both their morphology and function in response to various cellular stimuli. The synthesis of de novo mitochondria, termed biogenesis, is accompanied by increased respiration, metabolic processes, and ATP production, while autophagic degradation of mitochondria (mitophagy) is needed to remove damaged or unnecessary mitochondria (Fig. 1). The balance between biogenesis and mitophagy is important in modulating cell survival and cell fate in various physiological and pathological states [1–8]. The elongation (fusion) and division (fission) of mitochondria serve as a bridge between biogenesis and mitophagy. For instance, mitochondrial fusion prevents mitophagy while fission is an important preceding step needed for mitophagy. Furthermore, mitochondrial fusion can promote the survival of damaged mitochondria through complementation of mitochondrial DNA (mtDNA), whereas fission can remove damaged parts of the mitochondrial network, thereby preventing the entire network from sustaining major damage [3]. In stem cell populations, the switch from a quiescent to an activated state is often (if not always) accompanied by remodeling of the mitochondrial network in order to prime the cell for the increased metabolic demand associated with activation [9]. Mitochondrial turnover is also important in initiating the differentiation program of stem cells to reconstruct damaged tissues. Skeletal muscle is one of the tissues that is highly reliant upon optimal mitochondrial function, given its high metabolic activity. Additionally, skeletal muscle tissue sustains damage due to daily wear and tear, and from numerous disease states, with its repair being intimately dependent upon the turnover of mitochondria within resident skeletal muscle stem cells (satellite cells) [10, 11]. Needless to say, the study of mitochondrial dynamics and turnover in regulating skeletal muscle stem cell function and skeletal muscle tissue regeneration is still in its infancy. However, recent studies have shown the importance of these processes in maintaining skeletal muscle stem cell and skeletal muscle tissue function. Thus, this review will shed light on the molecular mechanisms of mitochondrial dynamics/turnover, their regulation through two major classes of sensors, and their known relevance in skeletal muscle stem cell differentiation and skeletal muscle tissue regeneration.
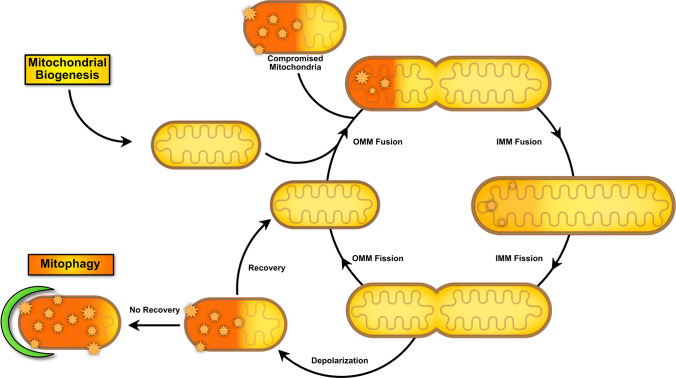
Fig. 1.
Mitochondria life cycle and contribution of dynamic processes in bridging the gap between biogenesis and mitophagy processes. Biogenesis results in the formation of de novo mitochondria that can fuse together to form a reticular network. Healthy mitochondria may also fuse with damaged/compromised mitochondria to allow for complementation of material (substrates, mitochondrial DNA, etc.). In response to numerous stressors ultimately causing depolarization, a portion of the mitochondrial network can split and undergo mitophagy or, in cases where recovery takes place, they can fuse back to the existing network
Mitochondrial biogenesis
Mitochondrial biogenesis is a highly regulated process that is dependent upon the synchronous interaction between mitochondrial and nuclear factors. Over the past two decades, the peroxisome proliferator-activated receptor (PPAR)-gamma coactivator-1 (PPARGC1) family of transcriptional coactivators has emerged as central regulators of mitochondrial biogenesis and metabolism [12]. The PPARGC1 family consists of three members, PPARGC1A, PPARGC1B, and PPARG related coactivator 1 (PPRC1) [13]. These factors interact with transcription factors and nuclear receptors that ultimately enrich the cell with the machinery needed for metabolism [14–16]. PPARGC1A, commonly called PGC1α, has previously been thought to be a necessary mediator of biogenesis [17]; however, growing evidence suggests that there are other factors that are also involved. PPARGC1B, metabolites, and other dietary factors are among the few evolving elements that have been explored to play a role in PPARGC1A-independent mitochondrial biogenesis [18–23]. This section will highlight PPARGC1A-dependent and -independent mechanisms of mitochondrial biogenesis (Fig. 2).
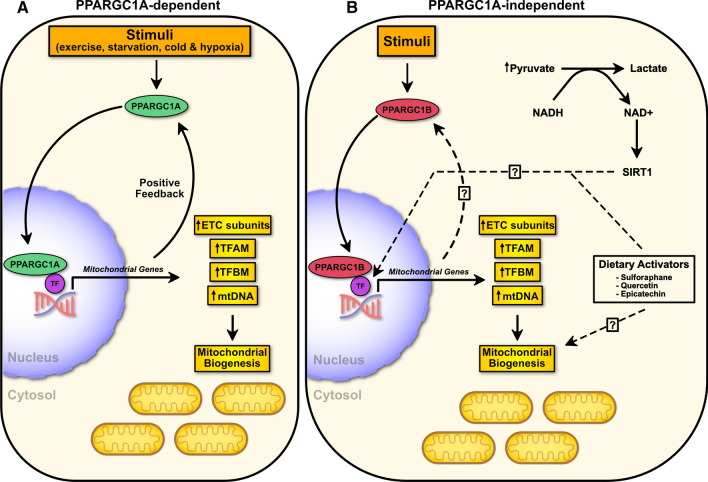
Fig. 2.
Mitochondrial biogenesis overview. a PPARGC1A-dependent mitochondrial biogenesis occurs in response to multiple stimuli leading to the induction of transcriptional coactivator PPARGC1A. This leads to subsequent transcription of mitochondrial biogenesis genes including those of the electron transport chain (ETC), TFAM and TFBM, and mitochondrial DNA (mtDNA). b Mitochondrial biogenesis can also occur independent of PPARGC1A. Transcriptional coactivator PPARGC1B, pyruvate, and dietary activators are among the factors that can induce mitochondrial biogenesis. TF transcription factor
Mechanism of PPARGC1A-dependent Mitochondrial Biogenesis
PPARGC1A is a co-transcriptional regulatory factor that is highly inducible under various physiological conditions including exercise, starvation, cold, and hypoxic stress [15]. These conditions alter energy levels (i.e., increased AMP:ATP ratio) and/or increase stress signaling cascades to trigger mitochondrial biogenesis. To meet the demands of the cell/tissue in these conditions and promote survival, several interconnected pathways are activated that ultimately result in increased levels of PPARGC1A [24, 25]. PPARGC1A can produce an adaptive response by working alongside several transcription factors to promote the transcription of genes involved in metabolism and mitochondrial biogenesis [24, 25].
The canonical transduction of PPARGC1A leads to its binding and coactivation of nuclear respiratory factors (NRFs) and estrogen-related receptors (ESRRs). NRFs are key components in regulating mitochondrial biogenesis, with NRF1 and nuclear factor erythroid 2 like 2 (NFE2L2 or NRF2) being the major targets of PPARGC1A. The significance of these nuclear factors has been established in developmental studies where the knockout of either of these factors results in embryonic lethality [26, 27]. Although homologous, NRF1 and NFE2L2 have slightly different downstream targets. NRF1 forms homodimers on DNA to target a broad range of genes related to oxidative phosphorylation (OXPHOS), mitochondrial membrane transport, detoxification (i.e., glutathione pathway), heme biogenesis, and mitochondrial DNA replication [28]. In contrast, NFE2L2 or GA-binding protein (GABP) seems to be mitochondrial specific with similar targets including mitochondrial transcription factor A (TFAM), mitochondrial transcription factor B (TFBM), and respiratory proteins [29]. The activity of NRF1 has also been shown to be methylation-dependent [30], whereas NFE2L2 requires deacetylated by sirtuin-7 (SIRT7) to form heterotetramers on DNA to enhance transcriptional activity [31].
Compared to NRFs, the physiological function of ESRRs in mitochondrial biogenesis is less studied in its relationship with PPARGC1A. PPARGC1A can target, bind, and activate ESRRA to regulate the expression of genes involved in OXPHOS, fatty acid oxidation, mitochondrial membrane transport, and mitochondrial DNA replication [32, 33]. Interestingly, in skeletal muscle PPARGC1 and ESRR induced regulator, muscle 1 (PERM1) can serve as a positive mediator of mitochondrial biogenesis [34–36]. PERM1 is transcribed by PPARGC1A and ERRA transcription factors and serves as a feed-forward mechanism that elevates the transcription of PPARGC1A, and thus, enhance mitochondrial biogenesis and oxidative capacity [36]. Similar homologues to PERM1 have yet to be identified in other tissues. Nonetheless, the inhibition of ESRRA impairs the ability of PPARGC1A to induce the expression of these genes, and reduces mitochondrial biogenesis as evidenced by a lower mtDNA:nDNA ratio. Interestingly, forced activation of ESRRA was able to induce mitochondrial biogenesis in the absence of PPARGC1A, although not to the same extent [32, 33]. Nonetheless, this suggests that there are likely other mechanisms in place capable of triggering the synthesis of important mitochondrial machinery in the absence of PPARGC1A.
Mechanism of PPARGC1A-independent Mitochondrial Biogenesis
Indeed, PPARGC1A plays a major role in biogenesis; however, the existence of redundant pathways contradicts the idea of a single master regulator. In fact, PPARGC1B has been shown to exhibit similar properties to PPARGC1A and was thought to be an important regulator of mitochondrial biogenesis [34, 35]. Similar to PPARGC1A, PPARGC1B is able to partner with NRF1 and ESRRA to enhance the expression of mitochondrial transcription factors and trigger robust mitochondrial biogenesis [18, 20, 22]. In double knockout models of Ppargc1a and Ppargc1b, skeletal muscle has significantly diminished mitochondrial function, and mtDNA:nDNA content compared to knockout of either alone [36]. It should be noted that the function of PPARGC1B may be tissue-specific since Ppargc1b knockout reduces mitochondrial content in the slow-twitch soleus muscle, and the heart, but not in adipose tissue [37]. Furthermore, Ppargc1b knockout impairs lipid metabolism in the liver, resulting in hepatic steatosis [37]. Thus, PPARGC1B is an important molecule that has a similar function to PPARGC1A and is a chief candidate for further investigation.
Cell studies have identified pyruvate as an important substrate regulating mitochondrial biogenesis in a PPARGC1A-independent manner [21, 23]. Pyruvate treatment is able to increase mitochondrial content as measured by MitoTracker, cytochrome c (CYCS), and electron transport chain (ETC) protein content in PPARGC1A knockdown C2C12 cells and primary myoblasts from Ppargc1a knockout mice [23]. Although the mechanism of pyruvate-induced biogenesis remains unidentified, the authors propose that excess pyruvate can reduce to lactate and produce nicotinamide adenine dinucleotide (NAD+) and SIRT1-mediated biogenesis. A follow-up study demonstrated that pyruvate treatment in C2C12 cells increases PPRC1 and respiratory proteins as a potential compensatory mechanism for the loss of PPARGC1A [21].
Some studies suggest that PPARGC1A is dispensable, particularly when it comes to exercise-induced mitochondrial biogenesis [19, 35]. The emerging roles of NFE2L2, estrogen-related receptor gamma (ESRRG), peroxisome proliferator-activated receptor delta (PPARD), and dietary activators (sulforaphane, quercetin, and epicatechin) have been elegantly highlighted in the context of exercise and skeletal muscle [19]. These factors can interact with members of the PPARGC1 family or with other less defined co-transcriptional regulators to promote the transcription of important mitochondrial genes, including NRF1, TFAM, and OXPHOS-related genes [19].
Mitochondrial fission and fusion
Mitochondria are highly dynamic organelles that continuously undergo cycles of fission and fusion. These processes are fundamental in maintaining mitochondrial morphology and function, and thus, play a central role in maintaining cellular health. On one hand, mitochondrial fission is required during cell division to allow equal distribution of mitochondria in each daughter cell [38]. Mitochondrial fission also enables the efficient removal of damaged mitochondria via selective autophagy (mitophagy) or participates in the induction of cell death [39, 40]. On the other hand, mitochondrial fusion is needed for mtDNA inheritance and helps mitigate stress by mixing contents from partially damaged and healthy mitochondria in a complementary fashion [41, 42]. Mitochondrial fusion is also required during cell differentiation, particularly in stem cell populations, and prevents the autophagic breakdown of mitochondria [9]. Defects in fission/fusion machinery has severe implications and leads to several diseases, particularly in tissues with high energy demands such as the brain, heart, and skeletal muscle. Given the central role of the mitochondria in energy production, metabolism, etc. greater emphasis has been placed on understanding the balance of fission and fusion over the past two decades. This section will underline the mechanism and regulators of mitochondrial fission and fusion (Fig. 3).
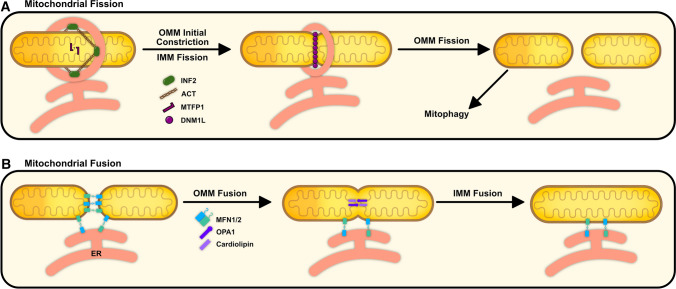
Fig. 3.
Overview of mitochondrial dynamics. a Mitochondrial fission is mediated by ER-mito interplay involving an initial OMM constriction via INF2-mediated ACT polymerization and IMM fission by MTFP1. DNM1L is then recruited to OMM adaptor proteins to complete the final constriction and scission of the mitochondria. b Mitochondrial fusion is mediated by the interaction of MFN1/2 found on the OMM, followed by OPA1 interaction with cardiolipin on the IMM. These events can take place at ER-mito contact sites
Mechanism of mitochondrial fission and fusion
The basis of mitochondrial fission is the constriction and scission of the mitochondrial membrane, resulting in two mitochondria [43, 44]. This mechanism is made possible by a dynamin-like GTPases protein, called dynamin 1-like (DNM1L or DRP1). DNM1L is traditionally found in the cytosol and translocates to the mitochondria in response to several upstream regulators, as explained later. DNM1L migration to the mitochondria is accompanied by its interaction with adaptor proteins found on the outer mitochondrial membrane (OMM). These OMM adaptor proteins include mitochondrial fission 1 (FIS1), mitochondrial fission factor (MFF), and mitochondrial dynamics protein of 49/51 kDa (MID49/51) [43, 44]. For example, the loss of these adaptor proteins, particularly MID49/51, reduce mitochondrial fission, while favouring mitochondrial fusion, and promote resistance to apoptotic stimuli [45]. Nonetheless, once bound to the adaptors, DNM1L oligomerizes around the mitochondria and utilizes GTP hydrolysis to induce a conformational change resulting in the constriction of the OMM and division of the mitochondria [46, 47]. In contrast to the OMM, the constriction of the inner mitochondrial membrane (IMM) has yet to be fully addressed. Overexpression of IMM protein mitochondrial fission process 1 (MTFP1) has been found to elevate mitochondrial fission whereas depletion of MTFP1 results in hyperfusion [48, 49]. The untethering of the mitochondrial membranes may also facilitate IMM fission [50]. One study found that an influx of calcium ions into the mitochondrial matrix can lead to neutralization of tethering by inner membrane mitochondrial protein (IMMT), resulting in a separation of the IMM from the OMM [50]. Taken together, it is well understood that DNM1L mediates fission through its interaction with adaptor proteins found on the OMM, whereas the mechanism of IMM fission has yet to be fully determined.
Mitochondrial fusion is mediated by three major dynamin-like GTPase proteins: mitofusin 1 (MFN1), MFN2, and optic-atrophy 1 (OPA1). The location of these proteins on the mitochondria dictates their function with MFN1/2 being found on the OMM while OPA1 is found on the IMM. MFN1 forms homomultimers or heteromultimers with MFN2 and undergo oligomerization to fuse the OMM [51]. GTP hydrolysis induces a conformational change in MFN1/2 oligomers, pulling and fusing two OMMs [52]. Upon OMM fusion, the long isoform of OPA1 (L-OPA1) mediates the fusion of the IMM through heteromultimer interaction with cardiolipin, once more through the hydrolysis of GTP [53]. The function of OPA1 extends beyond just mitochondrial fusion alone. The enzymatic conversion of L-OPA1 to its short isoform (S-OPA1) via overlapping with the m-AAA protease 1 (OMA1) or YME1-like 1 (YME1L1) peptidase has also been shown to aid in the dissociation of the IMM, favouring fission over fusion [50]. In addition to regulating IMM fusion, OPA1 is a necessary protein in stabilizing mtDNA and maintaining cristae morphogenesis in both long and short configuration [54, 55].
Other important players of mitochondrial dynamics
Indeed, DNM1L, OPA1, MFN1/2 and their related proteins are major mediators of mitochondrial dynamics; however, other mediators have also been shown to play an important role in this process. The endoplasmic reticulum (ER), and lipid molecules are important in priming or enhancing the fission and fusion processes.
ER-mitochondria (ER-mito) contacts have several biological functions including tethering, lipid metabolism, calcium signaling, and in recent years, recognized to mediate mitochondrial dynamics (reviewed elsewhere [56, 57]). ER and mitochondria can form junctions together with the interaction of mitochondrial MFN2 [58]. At these junctions, MFN2 is ubiquitinated at Lys192 by membrane-associated ring-CH type finger 5 (MARCHF5), triggering its oligomerization and providing a platform for ER-mito tethering [58]. These ER-mito sites define the location for not only mitochondrial fission [59–61] but also fusion events [62]. In fact, the majority of mitochondrial fission and fusion events occur at ER-mito contact sites [62]. The constriction of the mitochondria by the ER has been proposed to occur in two sequential phases. First, ER inverted formin 2 (INF2) protein can mediate the initial constriction around the OMM via actin (ACT) polymerization and myosin II (MYH2) mediated contraction [59–61]. This initial constriction is followed by the recruitment of DNM1L and induction of the second constriction to finally divide the mitochondria [59, 60]. A subpopulation of fission proteins (DNM1L, MFF, and FIS1) have also been identified on the ER and were shown to be involved in priming mitochondria for division. Of these proteins, ER-localized MFF was found to function as a platform to allow the recruitment and oligomerization of DNM1L prior to its transfer onto the mitochondria [59]. This mechanism coupled with INF2-mediated ACT polymerization are proposed to be major initiation steps for fission. Furthermore, ER-mito contacts have been shown to play a role in rescuing depolarized mitochondria through a “kiss-and-run” or transient fusion mechanism. This transient fusion event has been proposed to allow the exchange of proteins, metabolites, and ions between healthy and compromised mitochondria [62]. Nonetheless, further investigation is necessary to determine the mechanism(s) involved in sensing and rescuing mitochondria in this manner.
Changes in lipid composition of the OMM/IMM can guide and even enhance mitochondrial fission and is increasingly being studied (reviewed elsewhere [63]). Ceramide, cardiolipin, and phosphatidic acid are among some of the molecules responsible for manipulating DNM1L/OPA1 and thereby regulating mitochondrial dynamics. Ceramide synthesis within cardiomyocytes promotes DNM1L activation, mitochondrial fission, and apoptosis [64]. The inhibition of ceramide synthesis reduced apoptosis and mitochondrial fission, although it is unclear whether the reduction in apoptosis is a direct result of the inhibition of fission [65]. Similarly, cardiolipin can promote mitochondrial fission but also promote fusion depending on its localization. OMM localized cardiolipin enhances DNM1L-mediated fission by enhancing its GTPase activity [66–71]. In contrast, IMM cardiolipin promotes fusion via its interaction with OPA1 [53]. Cardiolipin can be cleaved by mitochondrial-surface phospholipase D (PLD6) to produce phosphatidic acid [72, 73], which has the opposite function on mitochondrial fission. Phosphatidic acid suppresses fission by sequestering DNM1L on the OMM [74, 75], while also promoting fusion events by enhancing MFN1/2 protein interactions [73].
Mitochondrial degradation
Mitochondria undergo dynamic changes to their structure to promote cellular survival as an initial response to cellular stress. In instances of high stress leading to mitochondrial damage and dysfunction, the efficient removal of damaged mitochondria is required to protect against cell death [76]. Several quality control mechanisms, including misfolded protein degradation (AAA protease-mediated) [80–82], vesicular transport of select proteins to the lysosome [79, 80], and autophagic removal of damaged mitochondria (mitophagy) have been identified to help mitigate cellular damage and maintain homeostasis. Of these factors, mitophagy plays the largest role and is present at a crossroad between cell survival and death [1, 3]. Furthermore, the failure of mitophagy has been implicated in various pathological conditions [81–84]. Emerging evidence supports the notion that mitochondria can undergo mitophagy through various mechanisms. These redundant pathways further demonstrate the evolutionary importance of the removal of damaged mitochondria. This section will highlight the various mechanisms of mitophagy (Fig. 4).
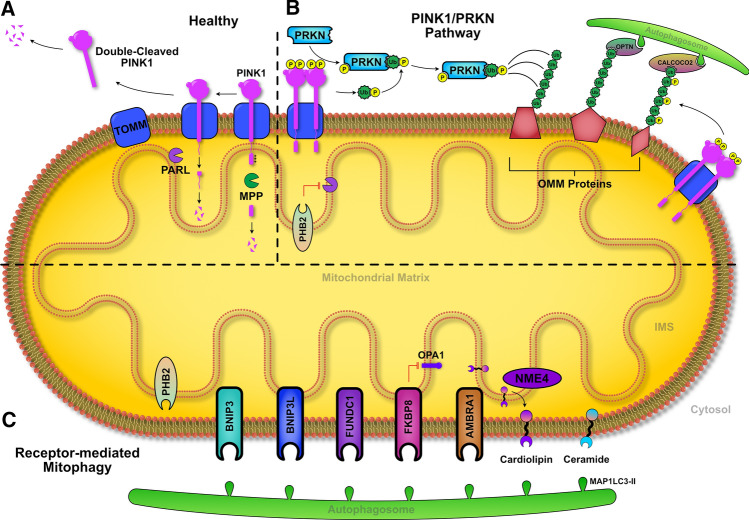
Fig. 4.
Mechanism of mitophagy. a Under normal conditions, PINK1 is partially imported, cleaved by MPP and PARL enzymes, and released into the cytosol to undergo degradation. b Following depolarized, PINK1 can accumulate on the OMM, activating PRKN directly and indirectly (Ub-mediated). The maximally activated PRKN can ubiquitinate OMM proteins, leading to recognition via cargo proteins (OPTN and CALCOCO2), and subsequent degradation through autophagy. c OMM receptor proteins and lipids can also mediate mitophagy independent of PINK1/PRKN through direct interaction with MAP1LC3-II found on the autophagosome
PINK1/PRKN-mediated mitophagy
The most studied mechanism of mitophagy is orchestrated by two proteins: the Ser/Thr kinase, PTEN-induced kinase 1 (PINK1), and the E3-ubitquitin ligase parkin (PRKN) [85, 86]. In mammalian cells, PINK1 serves as a molecular sensor of mitochondrial depolarization [85, 86]. PINK1 targets translocase of outer mitochondrial membrane (TOMM) protein to form a stable complex, enabling its mitochondrial targeting sequence (MTS) and transmembrane (TM) domains to extend into the mitochondrial matrix [85, 86]. In healthy cells, the extension of PINK1 into the matrix enables the breakdown of the MTS domain by mitochondrial processing peptidase (MPP) [87]. Upon this initial cleavage of PINK1, the IMM protein presenilin-associated rhomboid-like (PARL) removes the TM segment, resulting in the dissociation of PINK1 from TOMM [87–91]. The remaining double cleaved PINK1 is broken down within the cytosol via the N-end rule pathway [92] (Fig. 4a). In damaged cells containing compromised (depolarized) mitochondria, PINK1 once again associates with TOMM; however, the MTS domain is unable to extend to the mitochondrial matrix [93, 94]. As a result, the MTS domain is not cleaved by MPP, and concomitantly the TM domain also escapes PARL mediated processing [93, 94], resulting in the accumulation of PINK1 on the OMM. As PINK1 accumulates on the OMM, it becomes highly active after dimerization and autophosphorylation at Ser228 and Ser402 [95], and recruits PRKN to the OMM [94, 96] (Fig. 4b).
PRKN is a unique molecule that is able to autoregulate its own activity. Specifically, the ubiquitin-like (UBL) domain of PRKN ensures a closed conformation of the molecule under resting physiological conditions [97]. PINK1 phosphorylates PRKN at Ser65 to induce a conformation change to an “open” state to initiate the activation and recruitment of PRKN [97–100]. Furthermore, PINK1 accumulation on the OMM leads to the phosphorylation of cytosolic ubiquitin at Ser65 which interacts with phosphorylated PRKN [101–103]. In fact, the addition of a phosphorylated ubiquitin plays an essential role in activating PRKN and inducing mitophagy even in the absence of PINK1-mediated PRKN phosphorylation [100, 103]. The maximally activated PRKN can ubiquitinate multiple proteins on the OMM including MFN1/2, FIS1, TBC1 domain family member 15 (TBC1D15), voltage-dependent anion channels (VDACs), among others [104, 105]. The elongation of ubiquitin chains on the OMM by PRKN, coupled with their phosphorylation by PINK1 at Ser65, provides a feed-forward amplification mechanism that activates and recruits additional PRKN molecules to the OMM [106, 107]. This amplification results in greater recognition of damaged mitochondria and progression of mitophagy [106, 107].
The ubiquitination of OMM proteins allows for their recognition by the autophagic machinery through the interaction of adaptor proteins. Several adaptor proteins have been shown to interact with ubiquitinated OMM proteins including optineurin (OPTN), calcium binding and coiled-coil domain 2 (CALCOCO2), TAX1 binding protein 1 (TAX1BP1), neighbor of BRCA1 gene 1 (NBR1), and sequestosome 1 (SQSTM1). These proteins have a ubiquitin-binding domain and a microtube-associated protein 1 light chain 3 (MAP1LC3) interacting region (LIR) motif that work together to enable the recognition, efficient recruitment of the autophagosome, and degradation of mitochondria [108–110]. Interestingly, OPTN and CALCOCO2 can be recruited in the absence of PRKN, although their contribution may be limited [109]. The contribution of TAX1BP1 is also relatively low, while NBR1 and SQSTM1 have been shown to be dispensable for mitophagy [108, 109].
Mitophagy through multifunctional receptors
Although the PINK1/PRKN pathway of mitophagy is the most studied, other mechanisms exist and the contribution of these PRKN-independent mechanisms may be more important in certain types of cells or tissues. Hypoxia-inducible proteins, OMM spanning proteins, IMM spanning proteins, and lipids are some of the players involved in recruiting the autophagic machinery to induce mitophagy (Fig. 4c).
B-cell lymphoma 2 (BCL2) interacting protein 3 (BNIP3) and sister protein BNIP3-like (BNIP3L or NIX) are situated at the OMM upon depolarization stimuli and have been characterized under hypoxic conditions and for their role in apoptosis [111]. These proteins have also been shown to promote autophagy by competitively replacing BCL2 and BCL2 like 1 (BCL2L1) from Beclin-1 (BECN1), leading to the formation of the autophagosomal machinery, while combined ablation of BNIP3 and BNIP3L significantly reduces hypoxia-induced autophagy [112]. BNIP3 and BNIP3L also contain a LIR motif that can directly interact with MAP1LC3 to induce mitophagy [113–116]. The interaction between MAP1LC3 and BNIP3/BNIP3L is weak under normal growth conditions [114, 117], and is enhanced once BNIP3/BNIP3L are phosphorylated at the LIR motif [117, 118]. Furthermore, BNIP3 has been shown to anchor and inhibit the breakdown of OMM bound PINK1, thus increasing full-length PINK1 accumulation and concomitant PINK1/PRKN-mediated mitophagy [119]. On the other hand, BNIP3L has been shown to improve mitochondrial turnover in Parkinson’s disease patient cells and compensate for lack of functional PINK1/PRKN [120]. Furthermore, BNIP3L does not influence PINK1/PRKN-mediated mitophagy but seems to be an important factor in mediating mitophagy in the absence of PINK1/PRKN.
FUN14 domain-containing 1 (FUNDC1) is another OMM protein that is expressed under hypoxic conditions. FUNDC1 can function independent of PINK1/PRKN and is highly dependent upon its phosphorylation state. Phosphorylation of Tyr18 and Ser13 inhibits the interaction of the FUNDC1 LIR motif with MAP1LC3 [121, 122], whereas Unc-51 like autophagy activating kinase 1 (ULK1)-mediated phosphorylation at Ser17 enhances FUNDC1 and MAP1LC3 association [123]. Under hypoxic conditions, phosphoglycerate mutase 5 (PGAM5) localizes at the mitochondria and dephosphorylates FUNDC1 at Ser13, and thus, promotes mitophagy [121]. Ubiquitination of FUNDC1 by a mitochondrial E3 ligase, MARCHF5, in response to hypoxia, ultimately leads to the degradation of FUNDC1 and reduces mitophagy [124]. Furthermore, hypoxia can induce FUNDC1 accumulation at ER-mito contacts [125]. The association of FUNDC1 with calnexin mediates mitochondrial fission and mitophagy, while the loss of this interaction circumvents mitophagic degradation through hyperfusion [125]. Together, the balance of phosphorylation states, ubiquitination, and regulation of fission provides precise control over FUNDC1-mediated mitophagy under hypoxic conditions.
FK506 binding protein 8 (FKBP8) is among the mitophagy receptors that is involved in apoptotic processes while also regulating cell size through the modulation of the mechanistic target of rapamycin kinase (MTOR) [126]. The precise contribution of FKBP8 to mitophagy is not entirely understood. In overexpression experiments, FKBP8 shows a strong affinity for MAP1LC3A and recruits MAP1LC3A to induce mitophagy in a manner that is independent of PRKN [127]. Interestingly, FKBP8 is not degraded during mitophagy [128]. Instead, FKBP8 can avoid degradation by translocating to the ER [128]. Further, FKBP8 can promote mitochondrial fission and subsequent mitophagy under hypoxic stress in a PRKN-independent manner [129]. Interestingly, FKBP8-induced fission was independent of DNM1L and BNIP3/BNIP3L [129]. Instead, FKBP8 interacts with OPA1 on the IMM, likely inhibiting its partnering with cardiolipin, and thus favouring fission [129]. Nonetheless, further examination of this protein is needed to better elucidate this mechanism.
Autophagy and BECN1 regulator 1 (AMBRA1) is an autophagy protein that has recently been shown to participate in mitophagy similar to other mitophagy receptors. A pool of AMBRA1 can localize on the mitochondria and interact with MAP1LC3 through its LIR motif [130]. In cells overexpressing mitochondria specific AMBRA1, almost all mitochondria are degraded [130]. This occurs in cells that do not express PRKN, and cells with PRKN-knockdown [130, 131]. Interestingly, a small pool of AMBRA1 is capable of binding to BCL2 under normal conditions, and after autophagy induction, AMBRA1 dissociates from BCL2 to bind to BECN1 to progress autophagy [130]. Thus, the multifunctionality of AMBRA1 warrants further study into its role in mitophagy.
Prohibitin 2 (PHB2) is the first IMM receptor to be identified for its role in receptor-mediated mitophagy. Upon proteasomal rupture of the OMM, the LIR motif of PHB2 interacts with MAP1LC3 to induce mitophagy that is independent of PRKN [132]. PHB2 has also been shown to participate in PINK1/PRKN-mediated mitophagy. The depletion of PHB2 on depolarized mitochondria destabilizes PINK1 on the OMM, and thus, prevents recruitment of PRKN [133]. In contrast, the overexpression of PHB2 recruits PRKN and promotes PRKN-mediated mitophagy [133]. In depolarized mitochondria, PHB2 interacts with PARL protease, preventing it from cleaving PGAM5 and PINK1 [133]. The ablation of PHB2 permits PARL-mediated cleavage of PGAM5, which is involved in stabilizing PINK1 on the OMM [133]. Moreover, the lack of PHB2 and full-length PGAM5 destabilizes PINK1 and inhibits mitophagy.
Lipids such as cardiolipin and ceramide have also been implicated as important molecules involved in mitophagy. The translocation of cardiolipin from the IMM to the OMM is mediated by NME/NM23 nucleoside diphosphate kinase 4 (NME4) in response to mitophagic signals [134, 135]. The externalized cardiolipin can freely interact with MAP1LC3 and promote mitophagy [134]. Similarly, ceramide, which is already present on the OMM, can bind MAP1LC3B-II and induce lethal mitophagy [136]. Ablation of the ceramide-binding site on MAP1LC3B prevents endogenous ceramide-mediated mitophagy [136]. The precise mechanism by which ceramide recruits autophagic machinery has yet to be determined.
Interplay between mitochondrial biogenesis, fission/fusion, and mitophagy
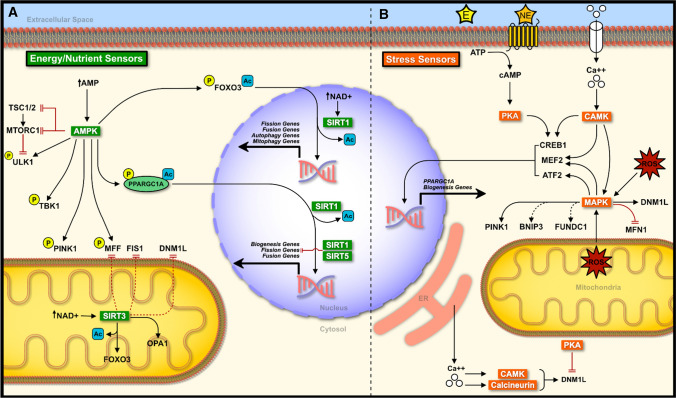
Mitochondria are highly adaptive organelles capable of modulating their morphology and function in response to various cellular stresses. The innate ability of these organelles to adapt is essential for normal cellular function and survival. Furthermore, the de novo formation, dynamic shift in structure, and subsequent removal of damaged mitochondria come together as a major quality control mechanism for this organelle. Alterations in any of these mechanisms can have severe implications on cellular function, potentially leading to disease states [2, 4–8]. Cellular energy/nutrient and stress sensors work together to maintain mitochondrial function through biogenesis, fission/fusion, and mitophagy. There is considerable overlap between energy/nutrient and stress sensors, yet they are still able to maintain a fine-tuned response. This section will discuss major energy/nutrient, and stress sensors in the context of mitochondrial biogenesis, dynamics, and mitophagy (Fig. 5).
Fig. 5.
Interplay between mitochondrial biogenesis, dynamics, and mitophagy. a Under low energy conditions, AMPK phosphorylates multiple targets to promote the turnover of mitochondria through the promotion of mitochondrial biogenesis, fission, and subsequent mitophagy. Low energy conditions can also stimulate the activity of SIRTs resulting in the deacetylation of target proteins and transcription factors. SIRTs promote biogenesis, and mitophagy, while inhibiting fission events. Together AMPK and SIRTs provide a fine-tuned response under low energy conditions. b A number of stress sensors work together to promote mitochondrial homeostasis. PKA, CAMK, calcineurin, and MAPK all promote biogenesis and mitophagy. CAMK, calcineurin, and MAPK also promote fission whereas PKA inhibits fission through the modulation of DNM1L function. Thus, these stress sensors display a similar fine-tuned response to that of AMPK-SIRT. Solid black lines indicate direct activation. Solid red lines indicate direct inhibition. Dotted lines indicate indirect activation (black) or inhibition (red). E epinephrine, NE norepinephrine
Role of energy/nutrient sensors
AMP-activated protein kinase (AMPK) is a major energy sensor in cells that is activated under low energy states, particularly when the ratio of AMP:ATP rises [137, 138]. The activation of AMPK is important in tissues with high energy demands, including the brain, heart, and skeletal muscle. To counter the reduced energy levels within cells, AMPK functions by directly phosphorylating various targets involved in energy metabolism [139–141]. AMPK-mediated phosphorylation of PPARGC1A activates the co-transcription factor, which upregulates genes responsible for ATP production and mitochondrial biogenesis, as discussed earlier. AMPK has also been shown to phosphorylate various epigenetic regulators including DNA methyltransferase 1 (DNMT1), RB binding protein 7 (RBBP7), and histone acetyltransferase 1 (HAT1) in vitro [142]. Phosphorylation of these proteins results in increased acetylation and reduced methylation of histones, ultimately favouring the transcription of PPARGC1A and related biogenesis genes [142]. This epigenetic control may function as a priming step for the accumulation of PPARGC1A prior to its phosphorylation by AMPK.
AMPK is also necessary for mitochondrial fragmentation under starvation conditions [143, 144]. Activated AMPK triggers the phosphorylation of adaptor protein MFF that enhances the recruitment of DNM1L to the OMM, thus, promoting mitochondrial fission [144]. The use of AMPK-mimetic, AICAR, on non-phosphorylatable MFF mutant cells does not lead to mitochondrial fission. Conversely, mutant cells containing constitutively phosphorylated AMPK sites on MFF have greater fragmentation [144]. This landmark study suggests that the phosphorylation of MFF by AMPK is needed for mitochondrial fission. Given that AMPK also activates autophagy, these authors also demonstrated that AMPK-mediated mitochondrial fission preceded mitophagy events [144].
AMPK also phosphorylates several target proteins required for the synthesis of autophagic machinery. AMPK can inactivate the nutrient sensor MTOR complex 1 (MTORC1) directly through the phosphorylation of regulatory-associated protein of MTOR (RAPTOR) [145], or indirectly via phosphorylation of upstream tuberous sclerosis 1/2 (TSC1/2) [146]. Inactivation of MTORC1 and AMPK-mediated phosphorylation of forkhead box O3 (FOXO3) enables its translocation to the nucleus and enhances its transcriptional activity [147]. FOXO3 regulates several autophagy (ULK1, BECN1, MAP1LC3, etc.) and mitophagy (BNIP3 and BNIP3L) factors [148, 149]. AMPK also directly phosphorylates ULK1 at Ser555, enabling it to translocate to the mitochondria and initiate autophagosome formation [150]. Furthermore, AMPK isoform AMPKα2 was found to phosphorylate PINK1 at Ser495 to enhance the recruitment of PRKN and concomitant mitophagy [151]. AMPK-mediated mitophagy can also occur independent of PRKN. For example, phosphorylation of TANK binding kinase 1 (TBK1) at Ser172 via ULK1 can promote recognition and engulfment of damaged mitochondria in a PINK1/PRKN-independent manner [152]. This is possible through TBK1-mediated phosphorylation of autophagy receptors including OPTN, CALCOCO2, and SQSTM1, which serves to enhance their binding capacity [152, 153].
Sirtuins (SIRTs) are a class of nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases that are responsible for a plethora of biological functions [154]. SIRT1 and SIRT3 have been widely investigated with regards to mitochondrial biogenesis. SIRT1 localizes in the nucleus while SIRT3 localizes exclusively in the mitochondria. Similar to AMPK, SIRTs become active during low energy states when the NAD+:NADH ratio shifts toward higher levels of NAD+. Knockdown of SIRT1 or SIRT3 in cells results in reduced fatty acid oxidation/oxidative metabolism, reduced PPARGC1A deacetylation, and elevated reactive oxygen species (ROS) generation with no change in mtDNA:nDNA [155–157]. Sirt1 knockout mice display lower Ppargc1a mRNA [158]; however, it remains unknown whether this leads to reduced mitochondrial content (i.e., mtDNA:nDNA). The overexpression of SIRT1 and SIRT3 is able to increase mitochondrial content, likely through the activation of PPARGC1A [156, 159, 160]. Interestingly, SIRT1 but not SIRT3 overexpression is able to increase mitochondrial oxidative function through increased transcription of PPARGC1A [158, 159].
The role of SIRTs in mitochondrial dynamics is to promote cell survival primarily through the inhibition of mitochondrial fission/fragmentation. The activation of SIRT1 via melatonin or SRT1720 treatment invokes a reduction in DNM1L and MFF levels [161–163]. This is accomplished through the activation of PPARGC1A and inhibition of c-Jun N-terminal kinase (JNK) signaling [161–163]. Interestingly, SIRT1 can prime ACT polymerization around mitochondria and in conjunction with AMPK, can lead to mitochondrial fragmentation [164]. Similarly, SIRT3 promotes mitochondrial fusion. SIRT3 overexpression reduces DNM1L, MFF, and/or FIS1 through the inhibition of suppressor of ras val 2 (SRV2), normalization of AMPK-mediated fission, and suppression of JNK signaling [165–167], while also promoting OPA1-mediated fusion [168]. SIRT3 overexpression may also potentiate the fine tuning of fission/fusion events through the deacetylation and activation of FOXO3, resulting in elevated DNM1L, FIS1, and MFN1 [169]. Sirt3 knockout also leads to inflammation as a result of reduced mitochondrial fusion [170]. Furthermore, SIRT5 can play an auxiliary role in inhibiting mitochondrial fission through the reduction of FIS1 and MID51, which would otherwise lead to fragmentation and mitophagy [171].
Similar to AMPK, SIRTs can promote autophagy and mitophagy under low energy/starvation conditions. Multiple SIRTs can bind and deacetylate FOXO [169, 172–175]. Nicotinamide or resveratrol treatment promote mitophagy through the activation of SIRT1/3, leading to the activation of FOXO3 and subsequent PINK1/PRKN signaling [176, 177]. SIRT1 also impairs PRKN translocation to the mitochondria and delays mitophagy [178]. Furthermore, SIRT3 increases the transcription of genes involved in PINK1/PRKN and BNIP3/BNIP3L-mediated mitophagy through the deacetylation of FOXO3. Sirt3 knockout reduced FOXO3 deacetylation and subsequent Prkn expression in cardiac cells of diabetic animals, whereas overexpression of SIRT3 leads to greater FOXO3 transcriptional activity, greater Prkn expression, and activates mitophagy [175]. Upregulated SIRT3-mediated deacetylation of FOXO3 also promotes receptor-mediated mitophagy through the transcription of Bnip3 and Bnip3l under oxidative stress conditions [169]. In other words, SIRT3 has a crossover role in modulating oxidative stress conditions. This has become apparent given that SIRT3 activates mitogen-activated protein kinase 1/3 (MAPK1/3)-cAMP response element-binding protein 1 (CREB1), leading to elevated BNIP3 and mitophagy [174].
In summary, the energy sensors AMPK and SIRTs play similar roles with respect to mitochondrial turnover such that they promote biogenesis and mitophagy to clear away damaged mitochondria and repopulate the cell with healthy mitochondria. Where these sensors differ is with respect to modulating mitochondrial dynamics. AMPK promotes mitochondrial fission, whereas SIRTs normalize AMPK signaling and may promote mitochondrial fusion [165–167]. It may be possible that these sensors work together to fine-tune the dynamic response of mitochondria under various conditions.
Role of stress sensors
Stress-mediated MAPK signaling plays a part in PPARGC1A activation, particularly in skeletal muscle cells and tissue. Exercise is one of many stressors that can activate MAPK signaling , and thus, increases the expression of Ppargc1a [179]. Exercise-induced activation of MAPK is partially a result of the formation of reactive oxygen species (ROS) and elevated cytosolic calcium ion concentrations [180, 181]. Exercise increases levels of phosphorylated MAPK in the nucleus where it can interact and form a complex with myocyte enhancer factor-2 (MEF2), thereby activating MEF2, and increasing the transcription of Ppargc1a [182, 183]. MAPK can also phosphorylate activating transcription factor 2 (ATF2), another factor known to enhance Ppargc1a transcription [179–181]. The chemical inhibition or dominant-negative mutant of ATF2 significantly reduces Ppargc1a mRNA expression in vitro, while chemical- or exercise-induced activation of MAPK leads to increased PPARGC1A promoter activity [179].
MAPK1/3 have been suggested to be important in mitochondrial dynamics through the manipulation of DNM1L and MFN1 activity. In cancer cells, the phosphorylation of Ser616 on DNM1L by MAPK1/3 increases mitochondrial fission and promotes tumor growth [184], while knockout of Dnm1l inhibits MAPK1/3 mediated mitochondrial fission [184]. In non-cancer cells, Ca2+ influx results in MAPK1/3 activation and subsequent mitochondrial fission through the phosphorylation of Ser616 on DNM1L [185]. Furthermore, MAPK1/3 phosphorylates MFN1 at Thr562 in neuronal cells, inhibiting its function, and thus inhibiting mitochondrial fusion while subsequently increasing fission, and susceptibility to apoptotic stimuli [186]. Taken together, it is possible that under cellular stress the MAPK1/3-mediated DNM1L activation and MFN1 inhibition favours mitochondrial fission and may be an important preceding step in cell death signaling.
MAPK signaling also plays an important role in mitophagy. Early evidence pointed at the localization of MAPK1 at the mitochondria to promote mitophagy in neuronal cells treated with a neurotoxin [187]. MAPK1 and MAPK14 knockdown inhibits mitophagy during starvation and hypoxic conditions while only marginally inhibiting autophagy as a whole [188]. The mechanism of MAPK mediated mitophagy has yet to be fully elucidated; however, there is evidence that MAPK1/3 is an important molecule responsible for the stabilization of PINK1; enabling PINK1/PRKN-mediated mitophagy to occur [189]. Some evidence also suggests that MAPK1/3 signaling can promote PINK1/PRKN-independent mitophagy through the activation of BNIP3 and FUNDC1 [174, 190]; however, more work is needed to fully understand this mechanism.
Calcium signaling is another essential activator of PPARGC1A-dependent mitochondrial biogenesis and mitochondrial fission. Stimulation of skeletal muscle causes the release of stored calcium ions (Ca2+) that activates calcineurin, and calcium/calmodulin-dependent protein kinase IV (CAMK4). Calcineurin interacts with myocyte enhancer factors (MEFs) family of transcription factors that subsequently aid in the transcription of Ppargc1a [191]. PPARGC1A can coactivate MEF transcription in a positive feedback loop, and thus, further increase its own expression [191]. Likewise, the overexpression of active calcineurin in skeletal muscle can increase PPARGC1A protein levels [192]. In contrast, CAMK4 phosphorylates and activates CREB1, a transcription factor that induces Ppargc1a [191, 193]. Thus, CAMK4 may be working synergistically alongside calcineurin to increase Ppargc1a transcription [193]. Together, calcium-mediated mitochondrial biogenesis can work through MEF and CREB1 to ultimately increase Ppargc1a expression [191, 193, 194].
Calcium signaling within cells controls mitochondrial dynamics by triggering calcineurin and CAMK2. In both cases the elevation in cytosolic calcium, which occurs during depolarization events, results in the activation of calcineurin and CAMK2. Activation of calcineurin dephosphorylates DNM1L, promoting its translocation to the mitochondria [195–197]. This was demonstrated by a dominant-negative mutant of calcineurin that effectively blocked mitochondrial fission during depolarization. Calcineurin was found to dephosphorylate DNM1L at Ser637 and Ser656 [195–197]. On the other hand, CAMK2 enhances the activity of DNM1L via Ser616 phosphorylation [198, 199], while inhibition of CAMK2 reduces phosphorylation of DNM1L at Ser616 and inhibits mitochondrial fission following ionizing radiation treatment [198]. Similarly, the chronic downstream activation of CAMK2 by isoproterenol (β-adrenergic receptor agonist) administration increases Ser616 phosphorylation of DNM1L in WT cells but not in dominant-negative DNM1L mutant cells [199]. Further research is needed to identify whether calcineurin-mediated dephosphorylation or CAMK2-mediated phosphorylation events occur to mitochondrial fusion proteins (MFN1/2 and OPA1).
Aside from the role of calcium in mediating mitochondrial fission as a priming step for mitophagy, calcium signaling can also promote mitophagy directly through its interaction with PINK1 and PRKN. Depolarization of the mitochondria promotes intracellular flux of calcium resulting in increased Pink1 mRNA and PINK1 protein expression; potentially as a protective mechanism against increased cytosolic calcium [200]. Another study demonstrated that the OMM protein ras homolog family member T1 (RHOT1) can function as a calcium-sensitive platform for the docking of PRKN during mitophagy [201]. Furthermore, mitochondrial depolarization can recruit calcium-activated CAMK1 to the OMM to promote PINK1/PRKN-mediated mitophagy [202]. To date, the interaction between calcium signaling and OMM mitophagy receptors (BNIP, BNIP3L, FUNDC1, etc.) is not well understood.
Cyclic AMP (cAMP) is another upstream signal that regulates mitochondrial biogenesis. The activation of cAMP can be mediated by various hormonal signals including epinephrine/norepinephrine, glucagon, thyroid hormone, among others [203–208]. It is worth noting that hormonal cAMP signaling can also reduce mitophagy in instances when mitophagy is overactive (i.e., insulin resistance), and thus, it may be important in fine-tuning mitochondrial degradation. cAMP binds and activates protein kinase A (PKA), which then phosphorylates CREB1. Phosphorylated CREB1 can translocate to the nucleus and ultimately promote PPARGC1A, along with other biogenesis promoting proteins [203, 204, 206, 208].
Cytosolic PKA and OMM bound PKA have been established to inhibit DNM1L activity [196, 209, 210]. PKA phosphorylates DNM1L at Ser656 [196] and Ser637 [209], thereby reducing its GTPase activity and preventing its translocation to the mitochondria. This subsequently results in reduced mitochondrial fragmentation, and likely promotes cell survival [196]. Moreover, a balancing act between PKA and calcineurin has been identified, whereby PKA phosphorylates and calcineurin dephosphorylates Ser656 and Ser637 on DNM1L [195–197]. This cycling between phosphorylation states of DNM1L provides fine control over fission activation and inhibition within cells.
cAMP signaling has also been implicated in PINK1/PRKN-mediated and receptor-mediated mitophagy. Under normal conditions, mitochondrial scaffolding protein A-kinase anchoring protein 1 (AKAP1) recruits PKA to the OMM to phosphorylate DNM1L at Ser637 to inhibit its function, and prevent subsequent mitophagy [211]. Following depolarization, PINK1 recruitment to the OMM displaces PKA from AKAP1, enabling DNM1L-mediated fission and mitophagy [211]. Furthermore, CREB1 signaling alongside MAPK1/3 elevates BNIP3 and FUNDC1 to promote mitophagy [174, 190]. Interestingly, activation of PKA results in BNIP3L phosphorylation and dissociates BNIP3L from the OMM, thereby preventing excessive mitophagy, which can lead to insulin resistance [212].
In summary, cellular stress signaling including ROS, Ca2+, and hormones have considerable interplay with respect to their roles in regulating biogenesis, fission/fusion, and mitophagy. Together these cascades formulate a stress response to promote gross mitochondrial remodeling through increased biogenesis, fission, and mitophagy in an attempt to promote cellular function and survival.
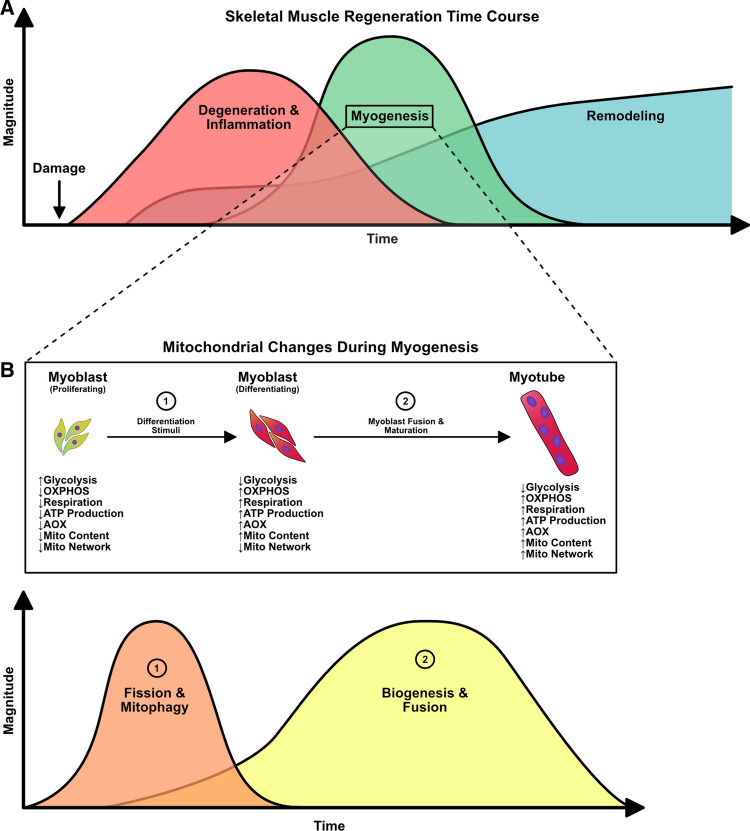
Mitochondrial interplay during skeletal muscle regeneration
Skeletal muscle undergoes continuous cycles of degeneration and regeneration in response to everyday contractile activity or traumatic injury. In order for skeletal muscle to resume normal function, an elaborately coordinated response from different cell types spanning across multiple phases must take place. These phases can be generalized into one of three categories: (1) degeneration/inflammation, (2) myogenesis, and (3) remodeling [213–217]. The first phase occurs immediately following damage and is spearheaded by the infiltration of neutrophils. These cells clear debris through phagocytosis and release chemotactic molecules to attract macrophages, which take over the bulk of the degradation of damaged skeletal muscle tissue [218]. The initial presence of the classical pro-inflammatory phagocytic M1 macrophage and the efficient transition to the alternative anti-inflammatory non-phagocytic M2 macrophage is crucial for the second phase of the regeneration process, myogenesis [216, 218]. Macrophages signal quiescent skeletal muscle stem cells, called satellite cells, triggering their activation into myoblasts. This is accompanied by proliferation, migration, differentiation, and fusion of myoblasts to form de novo skeletal muscle fibres (myofibres) or fusion directly to existing damaged skeletal muscle fibres [213–216]. Throughout these phases, the gross remodeling of the extracellular matrix (ECM) takes place, which involves the removal of old ECM, followed by the deposition of new ECM proteins [214, 219]. These ECM proteins provide the new myofibres with appropriate scaffolding material to mature and resume normal function.
Complete skeletal muscle regeneration can range from 14 to 28 days or more depending on the type/severity of damage and health of the cells involved [220–222]. The multiple steps satellite cells undergo to form myofibres requires extensive intracellular remodeling of the mitochondria. In fact, absence of mitochondrial remodeling has been repeatedly shown to reduce the differentiation capacity of cultured myoblasts [1, 223–228] and diminish the regenerative capacity of skeletal muscle tissue [10, 11, 229, 230]. The mitochondria within skeletal muscle are categorically defined based on their localization within the myofibre. Subsarcolemmal (SS) mitochondrial are located in the periphery of the myofibres and are heavily involved in energy production for transcriptional activities and interact closely with myonuclei [235–237]. On the other hand, intramyofibrillar (IMF) mitochondria are situated at the I-band of the sarcomere and provide energy for muscle contraction [235–237]. Although SS and IMF mitochondria may have divergent physiological roles and locations within muscle, there is evidence that these populations are linked through the network, and that the architecture of individual mitochondria and the configuration of the network are dependent on the metabolic characteristics of the muscle [238–241]. However, the influence of different populations of mitochondria is unknown in regenerating muscle and during myogenesis. Of note, the dynamic nature of these mitochondria vary considerably between mature skeletal muscle tissue and immature myoblasts [242]. Functional assessment of mitochondrial fusion events revealed that mature myofibres display less frequent fusion events compared to myoblasts [242]. The stable contractile structure (i.e., the myosin-actin complex) of mature skeletal muscle provides a physical barrier that partially limits mitochondrial dynamics compared to immature myoblasts [235–237, 242]. Although mitophagy has been shown to change throughout myoblast differentiation [1, 232], the direct impact of the contractile structure on mitophagic processes is unclear. Nonetheless, upon skeletal muscle damage, there is largescale degradation of these contractile structures [243], which may enable greater mitochondrial network remodeling to occur [242]. The nuances of mitochondrial remodeling are far from being completely understood. This section will highlight the current knowledge of mitochondrial involvement and interplay in satellite cell/myoblast function in vitro and during in vivo skeletal muscle regeneration (Fig. 6).
Fig. 6.
Mitochondrial alterations during skeletal muscle regeneration. a Generalized overview of overlapping stages during skeletal muscle regeneration. b Mitochondrial alteration during the “myogenesis” stage, including shift from glycolysis to increased oxidative phosphorylation (OXPHOS) coupled with increased respiration, ATP production, antioxidant (AOX) levels, and mitochondrial content
Mitochondrial fission and mitophagy during myogenesis and skeletal muscle regeneration
Satellite cells and myoblasts are the core of the regeneration process. The differentiation of myoblasts is coupled with metabolic reprogramming that ultimately results in increased OXPHOS and mitochondrial mass to support the newly formed myotubes. One of the features that occurs during the onset of differentiation is mitochondrial fission and mitophagy. The mitochondria present in myoblasts are in an immature state, as indicated by their underdeveloped cristae, low levels of β-oxidation, and low overall respiration [232, 244–246]. Upon differentiation stimuli, myoblasts must generate ATP at a higher rate to support the intracellular remodeling that is accompanied by differentiation. In doing so, differentiated myoblasts shift towards a more oxidative phenotype [225, 226]. To undergo this metabolic shift, the existing mitochondria present in myoblasts must be renewed. In C2C12 myoblasts, differentiation stimuli result in an initial yet dramatic increase in DNM1L levels, autophagy, and mitophagy markers [1, 223, 224, 227, 228]. Both the increase in mitochondrial fission and mitophagy have been shown to be essential for differentiation. The chemical inhibition of DNM1L via mdivi-1 treatment has been shown to reduce DNM1L translocation to the mitochondria, thereby reducing fission, and concomitantly leads to impaired differentiation and negligible myotube formation [223]. Likewise, the knockdown of ATG7 or knockout of Bnip3 in C2C12 myoblasts has been shown to ablate differentiation [1]. In both instances of impaired fission/mitophagy, greater apoptotic activity was noted [1, 223]. Interestingly, ATG7 knockdown and Bnip3 knockout myoblasts display altered DNM1L and OPA1 levels, suggesting a link between autophagy/mitophagy and mitochondrial dynamics [1]. Nonetheless, these results demonstrate that mitochondrial fission and subsequent mitophagy are important processes that enable the progression of myoblast differentiation.
Studies in skeletal muscle tissue have signified the importance of mitochondrial fission in skeletal muscle growth and maintenance of skeletal muscle mass [247–251]. Skeletal muscle tissue-specific knockout of DNM1L results in reduced mitophagy and causes severe muscle wasting and weakness [248]. However, DNM1L deletion in newborn mice results in no alterations in important myogenic regulatory factors (MRFs) such as myogenic differentiation 1 (MYOD1) or myogenin (MYOG) [248]. In contrast, the overexpression of DNM1L and/or FIS1 causes dysfunction of mitochondrial respiration and reduced mtDNA content as a result of excessive mitophagy [247, 249, 250]. The investigation of this fission/mitophagy phenomena has also been noted in the regeneration skeletal muscle tissue. In vivo myoblast-specific knockout of DNM1L does not alter overall satellite cell content or affect the regeneration program following cardiotoxin (CTX)-induced skeletal muscle damage [247], suggesting that DNM1L does not impair satellite cell function during regeneration [247]. Nonetheless, the investigation into other factors has helped characterize the role of fission during skeletal muscle regeneration. Fis1 expression dramatically increased between 3 and 5 days following freeze injury which is at approximately the same time that satellite cells are activated [10]. Fis1 in regenerating skeletal muscle tissue remained slightly above the levels found in undamaged/control tissue for up to 28 days. Consistent with these results, DNM1L is elevated 14 days following cardiotoxin (CTX)-induced skeletal muscle damage [230]. Furthermore, activated ULK1, BNIP3, and MAP1LC3-II were elevated in 14 days post-CTX regenerating skeletal muscle tissue even in animals treated with the autophagy inhibitor, 3-methyladenine (3-MA) [230]. A follow-up study from this same group found an increase in DNM1L, BNIP3, and PINK1/PRKN alongside mitochondrial localization of MAP1LC3B-II in regenerating muscle 7 days post-freeze injury [11]. It would be interesting to investigate whether satellite cells derived from damaged skeletal muscle exhibit a similar response to what is observed in C2C12 myoblasts. Ultimately, this may better elucidate the function of the initial fission/mitophagy processes in satellite cells during skeletal muscle regeneration.
Mitochondrial biogenesis and fusion during myogenesis and skeletal muscle regeneration
As C2C12 myoblasts continue to differentiate and mature into myotubes, so do the mitochondria within the cells. During the differentiation process, myoblasts begin to differentially express MRFs, with MYOD1 and MYOG being two MRFs whose transient expression is required for the progression of myoblast differentiation. In some instances, the elevation of certain MRFs coincides with elevated PPARGC1A, TFAM, cytochrome c oxidase subunit IV (COXIV), and mtDNA [1, 232–234]. Although MYOD1 is expressed early during myoblast differentiation, it has been shown to directly promote the transcription of Ppargc1b or enhance Ppargc1a expression in the presence of activated SIRT1 [158, 235]. The nuclear localization of PPARGC1A, in particular, follows a similar trend to that of MYOG [228]; however, a direct relationship between the two has not been established. The downregulation of Ppargc1a in C2C12 myoblasts increases ROS generation, mitochondrial damage, mitophagy, and results in poor differentiation [232]. Indeed, ROS formation during differentiation may play an important role in promoting mitochondrial biogenesis through MAPK signaling [225]; however, excessive ROS seems to have the opposite effect [232]. This reduction in ROS during the latter half of the differentiation process, as a result of increased antioxidant enzymes [225], may be an important feature for alleviating mitophagy and enabling the repopulation of mitochondria through biogenesis. Moreover, as fission/mitophagy begin to decline and mitochondrial biogenesis machinery increases, so do the levels of fusion protein OPA1 [228]. It is worth noting that the absence of mitophagy during the early stages of myoblast differentiation results in a blunted PPARGC1A response, and decreased mitochondrial protein content [1]. This may emphasize a greater importance of mitophagy in modulating mitochondrial biogenesis. Nonetheless, mitochondrial biogenesis and fusion lead to increased basal and maximal respiration needed for the highly active myotubes [234].
Similar to C2C12 cell culture experiments, MYOD1 and MYOG are elevated at approximately 3 days post-injury in skeletal muscle tissue and are also synchronous with upregulation of mitochondrial biogenesis genes, including Ppargc1b, Pprc1, Nrf1, Nfe2l2, Esrra, and Tfam [10, 229]. In addition to these critical events, Mfn1 and Mfn2 are elevated 3 days following CTX-injury [229], while MFN2 peaks 10 days post-freeze-injury [10]. Although MFN1/2 have well-established roles in maintaining skeletal muscle mass and preventing muscle wasting [44]; the role of MFN1/2 during regeneration has not been explored. In contrast, the role of OPA1 on myogenesis and skeletal muscle regeneration has been characterized. Constitutive skeletal muscle-specific deletion of Opa1 in neonatal mice resulted in a significant reduction of quiescent and activated satellite cells , with no alteration in apoptosis in these cells, which suggests a link between OPA1 and satellite cell self-renewal [255]. Furthermore, treatment of CTX-injured skeletal muscle with a microRNA against Opa1 results in a poor skeletal muscle regenerative response, prolonged inflammation, and reduced regenerating myofibre size [256]. These studies suggest that OPA1 may be an important component in satellite cell self-renewal and regeneration; however, additional studies are required to delineate this relationship. Investigation into OPA1 during skeletal muscle regeneration may provide novel insight into its role in maintaining mitochondrial morphology during the different phases of the regeneration process. Nonetheless, the coupling of mitochondrial biogenesis and fusion likely promotes respiration for the regenerating skeletal muscle; however, this has not been assessed. Furthermore, the inhibition of mitochondrial protein synthesis through the administration of Chloramphenicol or the skeletal muscle-specific knockout of Esrra has been shown to significantly impair regeneration in freeze- and CTX-injury models, respectively [10, 229]. In these instances, regenerating skeletal muscle displayed smaller fibres, reduced mitochondrial density, increased fibrosis, and diminished oxidative enzyme activity [10, 229]. An interesting feature to note is that when AMPK was forcibly activated 3 days post-CTX-injury via AICAR treatment, a significant impairment in the regenerative response was observed. This suggests that early activation of AMPK beyond physiological levels may not be beneficial to mitochondrial biogenesis or regeneration [229]. Instead, it seems to be more beneficial to allow the skeletal muscle tissue to coordinate its regenerative response to damage without the aid of external compounds that would otherwise promote mitochondrial remodeling. Additionally, it is unknown if mitochondrial biogenesis markers continue to increase as MRFs decline during the second half of the regeneration process (i.e., after satellite cell differentiation and formation of de novo myotube/myofibres), or whether mitochondrial respiration is altered in regenerating myofibres.
Concluding remarks
Accumulating evidence demonstrates that mitochondrial dynamics are intimately involved in regulating mitochondrial remodeling processes. The repopulation of mitochondria is an important step in maintaining cellular function and homeostasis in various cell types, under different stress conditions, and in disease states. Furthermore, dysfunction in these processes has been repeatedly shown to impair cellular function, leading to aberrant changes in cell behaviour. In the case of skeletal muscle stem cells, the precise mechanism and role of these processes has yet to be fully elucidated, and greater emphasis is needed in this regard. Nonetheless, the present data highlights the importance of mitochondrial dynamic and turnover processes in facilitating skeletal muscle regeneration. As this field continues to emerge, it may provide valuable information for the development of inhibitor/activator molecules to enhance skeletal muscle regeneration, particularly in diseased states through the targeting of these various mitochondrial mechanisms.
Author contributions
FAR and JQ conceptualized, wrote, and edited the article.
Funding
J.Q. is supported by a Natural Sciences and Engineering Research Council (NSERC) of Canada grant. F.A.R. is a recipient of an Ontario Graduate Scholarship (OGS).
Availability of data and material
Not applicable.
Code availability
Not applicable.
Declaration
Conflict of interest
The authors declare no conflicts of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
The authors consent publication of the current manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baechler BL, Bloemberg D, Quadrilatero J. Mitophagy regulates mitochondrial network signaling, oxidative stress, and apoptosis during myoblast differentiation. Autophagy. 2019;15:1606–1619. doi: 10.1080/15548627.2019.1591672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernardini JP, Lazarou M, Dewson G. Parkin and mitophagy in cancer. Oncogene. 2017;36:1315–1327. doi: 10.1038/onc.2016.302. [DOI] [PubMed] [Google Scholar]
- 3.Bloemberg D, Quadrilatero J. Autophagy, apoptosis, and mitochondria: molecular integration and physiological relevance in skeletal muscle. Am J Physiol Cell Physiol. 2019;317:C111–C130. doi: 10.1152/ajpcell.00261.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rimbaud S, Garnier A, Ventura-Clapier R. Mitochondrial biogenesis in cardiac pathophysiology. Pharmacol Rep. 2009;61:131–138. doi: 10.1016/S1734-1140(09)70015-5. [DOI] [PubMed] [Google Scholar]
- 6.Salem AF, Whitaker-Menezes D, Howell A, et al. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11:4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schumacker PT, Gillespie MN, Nakahira K, et al. Mitochondria in lung biology and pathology: more than just a powerhouse. Am J Physiol Lung Cell Mol Physiol. 2014;306:L962–L974. doi: 10.1152/ajplung.00073.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Li G, Zheng Y, et al. A novel autophagy/mitophagy inhibitor liensinine sensitizes breast cancer cells to chemotherapy through DNM1L-mediated mitochondrial fission. Autophagy. 2015;11:1259–1279. doi: 10.1080/15548627.2015.1056970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu W, Liu Y, Yin H (2019) Mitochondrial dynamics: biogenesis, fission, fusion, and mitophagy in the regulation of stem cell behaviors. In: Stem cells international. https://www.hindawi.com/journals/sci/2019/9757201/. Accessed 5 June 2020 [DOI] [PMC free article] [PubMed]
- 10.Wagatsuma A, Kotake N, Yamada S. Muscle regeneration occurs to coincide with mitochondrial biogenesis. Mol Cell Biochem. 2011;349:139–147. doi: 10.1007/s11010-010-0668-2. [DOI] [PubMed] [Google Scholar]
- 11.Nichenko AS, Southern WM, Tehrani KF, et al. Mitochondrial-specific autophagy linked to mitochondrial dysfunction following traumatic freeze injury in mice. Am J Physiol Cell Physiol. 2020;318:C242–C252. doi: 10.1152/ajpcell.00123.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Investig. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Gleyzer N, Scarpulla RC. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. J Biol Chem. 2011;286:39715–39725. doi: 10.1074/jbc.M111.291575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang H, Ward WF. PGC-1α: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30:145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 16.Sonoda J, Mehl IR, Chong L-W, et al. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci USA. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis1234. Am J Clin Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arany Z, Lebrasseur N, Morris C, et al. The transcriptional coactivator PGC-1beta drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Islam H, Hood DA, Gurd BJ. Looking beyond PGC-1α: emerging regulators of exercise-induced skeletal muscle mitochondrial biogenesis and their activation by dietary compounds. Appl Physiol Nutr Metab. 2020;45:11–23. doi: 10.1139/apnm-2019-0069. [DOI] [PubMed] [Google Scholar]
- 20.Kamei Y, Ohizumi H, Fujitani Y, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci USA. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philp A, Perez-Schindler J, Green C, et al. Pyruvate suppresses PGC1alpha expression and substrate utilization despite increased respiratory chain content in C2C12 myotubes. Am J Physiol Cell Physiol. 2010;299:C240–250. doi: 10.1152/ajpcell.00438.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao D, Liu Y, Liu X, et al. PGC-1 beta-regulated mitochondrial biogenesis and function in myotubes is mediated by NRF-1 and ERR alpha. Mitochondrion. 2010;10:516–527. doi: 10.1016/j.mito.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Wilson L, Yang Q, Szustakowski JD, et al. Pyruvate induces mitochondrial biogenesis by a PGC-1 alpha-independent mechanism. Am J Physiol, Cell Physiol. 2007;292:C1599–1605. doi: 10.1152/ajpcell.00428.2006. [DOI] [PubMed] [Google Scholar]
- 24.Popov DV, Lysenko EA, Kuzmin IV, et al. Regulation of PGC-1α isoform expression in skeletal muscles. Acta Naturae. 2015;7:48–59. doi: 10.32607/20758251-2015-7-1-48-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circ Res. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan JY, Kwong M, Lu R, et al. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ristevski S, O’Leary DA, Thornell AP, et al. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ploumi C, Daskalaki I, Tavernarakis N. Mitochondrial biogenesis and clearance: a balancing act. FEBS J. 2017;284:183–195. doi: 10.1111/febs.13820. [DOI] [PubMed] [Google Scholar]
- 29.Bruni F, Polosa PL, Gadaleta MN, et al. Nuclear respiratory factor 2 induces the expression of many but not all human proteins acting in mitochondrial DNA transcription and replication. J Biol Chem. 2010;285:3939–3948. doi: 10.1074/jbc.M109.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domcke S, Bardet AF, Adrian Ginno P, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 31.Ryu D, Jo YS, Lo Sasso G, et al. A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Schreiber SN, Knutti D, Brogli K, et al. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber SN, Emter R, Hock MB, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci USA. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho Y, Hazen BC, Russell AP, Kralli A. Peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells. J Biol Chem. 2013;288:25207–25218. doi: 10.1074/jbc.M113.489674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho Y, Hazen BC, Gandra PG, et al. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. FASEB J. 2016;30:674–687. doi: 10.1096/fj.15-276360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho Y, Tachibana S, Hazen BC, et al. Perm1 regulates CaMKII activation and shapes skeletal muscle responses to endurance exercise training. Mol Metab. 2019;23:88–97. doi: 10.1016/j.molmet.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J, Wu H, Tarr PT, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 38.Rowe GC, El-Khoury R, Patten IS, et al. PGC-1α is dispensable for exercise-induced mitochondrial biogenesis in skeletal muscle. PLoS ONE. 2012 doi: 10.1371/journal.pone.0041817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zechner C, Lai L, Zechner JF, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lelliott CJ, Medina-Gomez G, Petrovic N, et al. Ablation of PGC-1beta results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamano K, Youle RJ. Coupling mitochondrial and cell division. Nat Cell Biol. 2011;13:1026–1027. doi: 10.1038/ncb2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 44.Chen H, Vermulst M, Wang YE, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos ES, Motori E, Brüser C, et al. Mitochondrial fusion is required for regulation of mitochondrial DNA replication. PLoS Genet. 2019;15:e1008085. doi: 10.1371/journal.pgen.1008085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraus F, Ryan MT. The constriction and scission machineries involved in mitochondrial fission. J Cell Sci. 2017;130:2953–2960. doi: 10.1242/jcs.199562. [DOI] [PubMed] [Google Scholar]
- 47.Ramachandran R. Mitochondrial dynamics: the dynamin superfamily and execution by collusion. Semin Cell Dev Biol. 2018;76:201–212. doi: 10.1016/j.semcdb.2017.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osellame LD, Singh AP, Stroud DA, et al. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci. 2016;129:2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fröhlich C, Grabiger S, Schwefel D, et al. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mears JA, Lackner LL, Fang S, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morita M, Prudent J, Basu K, et al. mTOR controls mitochondrial dynamics and cell survival via MTFP1. Mol Cell. 2017;67:922–935.e5. doi: 10.1016/j.molcel.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 52.Tondera D, Czauderna F, Paulick K, et al. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci. 2005;118:3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- 53.Cho B, Cho HM, Jo Y, et al. Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat Commun. 2017;8:15754. doi: 10.1038/ncomms15754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H, Detmer SA, Ewald AJ, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci. 2004;117:6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 56.Ban T, Ishihara T, Kohno H, et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol. 2017;19:856–863. doi: 10.1038/ncb3560. [DOI] [PubMed] [Google Scholar]
- 57.Anand R, Wai T, Baker MJ, et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919–929. doi: 10.1083/jcb.201308006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H, Smith SB, Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J Biol Chem. 2017;292:7115–7130. doi: 10.1074/jbc.M116.762567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman JR, Lackner LL, West M, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moltedo O, Remondelli P, Amodio G. The mitochondria-endoplasmic reticulum contacts and their critical role in aging and age-associated diseases. Front Cell Dev Biol. 2019 doi: 10.3389/fcell.2019.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugiura A, Nagashima S, Tokuyama T, et al. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 62.Ji W-K, Chakrabarti R, Fan X, et al. Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J Cell Biol. 2017;216:4123–4139. doi: 10.1083/jcb.201610057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abrisch RG, Gumbin SC, Wisniewski BT, et al. Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J Cell Biol. 2020 doi: 10.1083/jcb.201911122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frohman MA. Role of mitochondrial lipids in guiding fission and fusion. J Mol Med (Berl) 2015;93:263–269. doi: 10.1007/s00109-014-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parra V, Eisner V, Chiong M, et al. Changes in mitochondrial dynamics during ceramide-induced cardiomyocyte early apoptosis. Cardiovasc Res. 2008;77:387–397. doi: 10.1093/cvr/cvm029. [DOI] [PubMed] [Google Scholar]
- 68.Kuzmicic J, Parra V, Verdejo HE, et al. Trimetazidine prevents palmitate-induced mitochondrial fission and dysfunction in cultured cardiomyocytes. Biochem Pharmacol. 2014;91:323–336. doi: 10.1016/j.bcp.2014.07.022. [DOI] [PubMed] [Google Scholar]
- 69.Bustillo-Zabalbeitia I, Montessuit S, Raemy E, et al. Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS ONE. 2014;9:e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Francy CA, Alvarez FJD, Zhou L, et al. The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction. J Biol Chem. 2015;290:11692–11703. doi: 10.1074/jbc.M114.610881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Francy CA, Clinton RW, Fröhlich C, et al. Cryo-EM studies of Drp1 reveal cardiolipin interactions that activate the helical oligomer. Sci Rep. 2017;7:10744. doi: 10.1038/s41598-017-11008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Macdonald PJ, Stepanyants N, Mehrotra N, et al. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25:1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stepanyants N, Macdonald PJ, Francy CA, et al. Cardiolipin’s propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell. 2015;26:3104–3116. doi: 10.1091/mbc.E15-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ugarte-Uribe B, Mueller H-M, Otsuki M et al (2014) Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J Biol Chem jbc.M114.575779. 10.1074/jbc.M114.575779 [DOI] [PMC free article] [PubMed]
- 75.Baba T, Kashiwagi Y, Arimitsu N, et al. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J Biol Chem. 2014;289:11497–11511. doi: 10.1074/jbc.M113.531921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi S-Y, Huang P, Jenkins GM, et al. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat Cell Biol. 2006;8:1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 77.Adachi Y, Itoh K, Yamada T, et al. Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol Cell. 2016;63:1034–1043. doi: 10.1016/j.molcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adachi Y, Iijima M, Sesaki H. An unstructured loop that is critical for interactions of the stalk domain of Drp1 with saturated phosphatidic acid. Small GTPases. 2018;9:472–479. doi: 10.1080/21541248.2017.1321614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kubli DA, Gustafsson ÅB. Mitochondria and mitophagy: the Yin and Yang of cell death control. Circ Res. 2012;111:1208–1221. doi: 10.1161/CIRCRESAHA.112.265819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leonhard K, Herrmann JM, Stuart RA, et al. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996;15:4218–4229. doi: 10.1002/j.1460-2075.1996.tb00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leonhard K, Guiard B, Pellecchia G, et al. Membrane protein degradation by AAA proteases in mitochondria: extraction of substrates from either membrane surface. Mol Cell. 2000;5:629–638. doi: 10.1016/s1097-2765(00)80242-7. [DOI] [PubMed] [Google Scholar]
- 82.Karunadharma PP, Basisty N, Chiao YA, et al. Respiratory chain protein turnover rates in mice are highly heterogeneous but strikingly conserved across tissues, ages, and treatments. FASEB J. 2015;29:3582–3592. doi: 10.1096/fj.15-272666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soubannier V, McLelland G-L, Zunino R, et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 84.Soubannier V, Rippstein P, Kaufman BA, et al. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS ONE. 2012;7:e52830. doi: 10.1371/journal.pone.0052830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120:1812–1824. doi: 10.1161/CIRCRESAHA.117.311082. [DOI] [PubMed] [Google Scholar]
- 86.Kerr JS, Adriaanse BA, Greig NH, et al. Mitophagy and Alzheimer’s disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40:151–166. doi: 10.1016/j.tins.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J, Liu W, Li R, Yang H. Mitophagy in Parkinson’s disease: from pathogenesis to treatment. Cells. 2019 doi: 10.3390/cells8070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.VanderVeen BN, Fix DK, Carson JA (2017) Disrupted skeletal muscle mitochondrial dynamics, mitophagy, and biogenesis during cancer cachexia: a role for inflammation. In: Oxidative medicine and cellular longevity. https://www.hindawi.com/journals/omcl/2017/3292087/. Accessed 7 June 2020 [DOI] [PMC free article] [PubMed]
- 89.Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Greene AW, Grenier K, Aguileta MA, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deas E, Plun-Favreau H, Gandhi S, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jin SM, Lazarou M, Wang C, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meissner C, Lorenz H, Weihofen A, et al. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 95.Shi G, Lee JR, Grimes DA, et al. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum Mol Genet. 2011;20:1966–1974. doi: 10.1093/hmg/ddr077. [DOI] [PubMed] [Google Scholar]
- 96.Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lazarou M, Jin SM, Kane LA, Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Okatsu K, Uno M, Koyano F, et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment. J Biol Chem. 2013;288:36372–36384. doi: 10.1074/jbc.M113.509653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aerts L, Craessaerts K, De Strooper B, Morais VA. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J Biol Chem. 2015;290:2798–2811. doi: 10.1074/jbc.M114.620906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okatsu K, Oka T, Iguchi M, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chaugule VK, Burchell L, Barber KR, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iguchi M, Kujuro Y, Okatsu K, et al. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem. 2013;288:22019–22032. doi: 10.1074/jbc.M113.467530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kondapalli C, Kazlauskaite A, Zhang N, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shiba-Fukushima K, Imai Y, Yoshida S, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kane LA, Lazarou M, Fogel AI, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kazlauskaite A, Kondapalli C, Gourlay R, et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 108.Sarraf SA, Raman M, Guarani-Pereira V, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ordureau A, Sarraf SA, Duda DM, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23:886–897. doi: 10.1038/cr.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Heo J-M, Ordureau A, Paulo JA, et al. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamano K, Matsuda N, Tanaka K. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 2016;17:300–316. doi: 10.15252/embr.201541486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hanna RA, Quinsay MN, Orogo AM, et al. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–19104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rikka S, Quinsay MN, Thomas RL, et al. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schwarten M, Mohrlüder J, Ma P, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–698. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 121.Zhu Y, Massen S, Terenzio M, et al. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem. 2013;288:1099–1113. doi: 10.1074/jbc.M112.399345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rogov VV, Stolz A, Ravichandran AC, et al. Structural and functional analysis of the GABARAP interaction motif (GIM) EMBO Rep. 2017;18:1382–1396. doi: 10.15252/embr.201643587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang T, Xue L, Li L, et al. BNIP3 protein suppresses PINK1 kinase proteolytic cleavage to promote mitophagy. J Biol Chem. 2016;291:21616–21629. doi: 10.1074/jbc.M116.733410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koentjoro B, Park J-S, Sue CM. Nix restores mitophagy and mitochondrial function to protect against PINK1/Parkin-related Parkinson’s disease. Sci Rep. 2017;7:44373. doi: 10.1038/srep44373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen G, Han Z, Feng D, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014;54:362–377. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 126.Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 127.Wu W, Tian W, Hu Z, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen Z, Liu L, Cheng Q, et al. Mitochondrial E3 ligase MARCH5 regulates FUNDC1 to fine-tune hypoxic mitophagy. EMBO Rep. 2017;18:495–509. doi: 10.15252/embr.201643309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu W, Lin C, Wu K, et al. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 2016;35:1368–1384. doi: 10.15252/embj.201593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Edlich F, Lücke C. From cell death to viral replication: the diverse functions of the membrane-associated FKBP38. Curr Opin Pharmacol. 2011;11:348–353. doi: 10.1016/j.coph.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 131.Bhujabal Z, Birgisdottir ÅB, Sjøttem E, et al. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017;18:947–961. doi: 10.15252/embr.201643147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saita S, Shirane M, Nakayama KI. Selective escape of proteins from the mitochondria during mitophagy. Nat Commun. 2013;4:1410. doi: 10.1038/ncomms2400. [DOI] [PubMed] [Google Scholar]
- 133.Yoo S-M, Yamashita S, Kim H, et al. FKBP8 LIRL-dependent mitochondrial fragmentation facilitates mitophagy under stress conditions. FASEB J. 2020;34:2944–2957. doi: 10.1096/fj.201901735R. [DOI] [PubMed] [Google Scholar]
- 134.Strappazzon F, Nazio F, Corrado M, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Van Humbeeck C, Cornelissen T, Hofkens H, et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wei Y, Chiang W-C, Sumpter R, et al. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–238.e10. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yan C, Gong L, Chen L, et al. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020;16:419–434. doi: 10.1080/15548627.2019.1628520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kagan VE, Jiang J, Huang Z, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23:1140–1151. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sentelle RD, Senkal CE, Jiang W, et al. Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012;8:831–838. doi: 10.1038/nchembio.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carling D, Mayer FV, Sanders MJ, Gamblin SJ. AMP-activated protein kinase: nature’s energy sensor. Nat Chem Biol. 2011;7:512–518. doi: 10.1038/nchembio.610. [DOI] [PubMed] [Google Scholar]
- 142.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cantó C, Jiang LQ, Deshmukh AS, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jäger S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marin TL, Gongol B, Zhang F, et al. AMPK promotes mitochondrial biogenesis and function by phosphorylating the epigenetic factors DNMT1, RBBP7, and HAT1. Sci Signal. 2017 doi: 10.1126/scisignal.aaf7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ducommun S, Deak M, Sumpton D, et al. Motif affinity and mass spectrometry proteomic approach for the discovery of cellular AMPK targets: identification of mitochondrial fission factor as a new AMPK substrate. Cell Signal. 2015;27:978–988. doi: 10.1016/j.cellsig.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 148.Toyama EQ, Herzig S, Courchet J, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 151.Greer EL, Oskoui PR, Banko MR, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 152.Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 153.Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 154.Tian W, Li W, Chen Y, et al. Phosphorylation of ULK1 by AMPK regulates translocation of ULK1 to mitochondria and mitophagy. FEBS Lett. 2015;589:1847–1854. doi: 10.1016/j.febslet.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 155.Wang B, Nie J, Wu L, et al. AMPKα2 protects against the development of heart failure by enhancing mitophagy via PINK1 phosphorylation. Circ Res. 2018;122:712–729. doi: 10.1161/CIRCRESAHA.117.312317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Seabright AP, Fine NHF, Barlow JP, et al. AMPK activation induces mitophagy and promotes mitochondrial fission while activating TBK1 in a PINK1-Parkin independent manner. FASEB J. 2020;34:6284–6301. doi: 10.1096/fj.201903051R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Richter B, Sliter DA, Herhaus L, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vargas-Ortiz K, Pérez-Vázquez V, Macías-Cervantes MH. Exercise and sirtuins: a way to mitochondrial health in skeletal muscle. Int J Mol Sci. 2019 doi: 10.3390/ijms20112717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gerhart-Hines Z, Dominy JE, Blättler SM, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jing E, Emanuelli B, Hirschey MD, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Amat R, Planavila A, Chen SL, et al. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma Co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J Biol Chem. 2009;284:21872–21880. doi: 10.1074/jbc.M109.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kong X, Wang R, Xue Y, et al. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pfluger PT, Herranz D, Velasco-Miguel S, et al. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ding M, Feng N, Tang D, et al. Melatonin prevents Drp1-mediated mitochondrial fission in diabetic hearts through SIRT1-PGC1α pathway. J Pineal Res. 2018;65:e12491. doi: 10.1111/jpi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li M-Y, Ding J-Q, Tang Q, et al. SIRT1 activation by SRT1720 attenuates bone cancer pain via preventing Drp1-mediated mitochondrial fission. Biochim Biophys Acta Mol Basis Dis. 2019;1865:587–598. doi: 10.1016/j.bbadis.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 167.Qin R, Zhang L, Lin D, et al. Sirt1 inhibits HG-induced endothelial injury: role of Mff-based mitochondrial fission and F-actin homeostasis-mediated cellular migration. Int J Mol Med. 2019;44:89–102. doi: 10.3892/ijmm.2019.4185. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 168.Lovy A, Ahumada-Castro U, Bustos G, et al. Concerted action of AMPK and sirtuin-1 induces mitochondrial fragmentation upon inhibition of Ca2+ transfer to mitochondria. Front Cell Dev Biol. 2020 doi: 10.3389/fcell.2020.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu J, Yan W, Zhao X, et al. Sirt3 attenuates post-infarction cardiac injury via inhibiting mitochondrial fission and normalization of AMPK-Drp1 pathways. Cell Signal. 2019;53:1–13. doi: 10.1016/j.cellsig.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 170.Zhou D, Jiang Y. Sirtuin 3 attenuates neuroinflammation-induced apoptosis in BV-2 microglia. Aging (Albany NY) 2019;11:9075–9089. doi: 10.18632/aging.102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhou J, Shi M, Li M, et al. Sirtuin 3 inhibition induces mitochondrial stress in tongue cancer by targeting mitochondrial fission and the JNK-Fis1 biological axis. Cell Stress Chaperones. 2019;24:369–383. doi: 10.1007/s12192-019-00970-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 172.Wang Q, Xu J, Li X, et al. Sirt3 modulate renal ischemia-reperfusion injury through enhancing mitochondrial fusion and activating the ERK-OPA1 signaling pathway. J Cell Physiol. 2019;234:23495–23506. doi: 10.1002/jcp.28918. [DOI] [PubMed] [Google Scholar]
- 173.Tseng AHH, Shieh S-S, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 174.Tyagi A, Nguyen CU, Chong T, et al. SIRT3 deficiency-induced mitochondrial dysfunction and inflammasome formation in the brain. Sci Rep. 2018;8:17547. doi: 10.1038/s41598-018-35890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guedouari H, Daigle T, Scorrano L, Hebert-Chatelain E. Sirtuin 5 protects mitochondria from fragmentation and degradation during starvation. Biochim Biophys Acta Mol Cell Res. 2017;1864:169–176. doi: 10.1016/j.bbamcr.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 176.Jang S, Kang HT, Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/NADH ratio and SIRT1 protein activation. J Biol Chem. 2012;287:19304–19314. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kobayashi Y, Furukawa-Hibi Y, Chen C, et al. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16:237–243. [PubMed] [Google Scholar]
- 178.Li R, Xin T, Li D, et al. Therapeutic effect of Sirtuin 3 on ameliorating nonalcoholic fatty liver disease: the role of the ERK-CREB pathway and Bnip3-mediated mitophagy. Redox Biol. 2018;18:229–243. doi: 10.1016/j.redox.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yu W, Gao B, Li N, et al. Sirt3 deficiency exacerbates diabetic cardiac dysfunction: role of Foxo3A-Parkin-mediated mitophagy. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1973–1983. doi: 10.1016/j.bbadis.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 180.Das S, Mitrovsky G, Vasanthi HR, Das DK. Antiaging properties of a grape-derived antioxidant are regulated by mitochondrial balance of fusion and fission leading to mitophagy triggered by a signaling network of Sirt1-Sirt3-Foxo3-PINK1-PARKIN. Oxid Med Cell Longev. 2014 doi: 10.1155/2014/345105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 181.Kang HT, Hwang ES. Nicotinamide enhances mitochondria quality through autophagy activation in human cells. Aging Cell. 2009;8:426–438. doi: 10.1111/j.1474-9726.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 182.Di Sante G, Pestell TG, Casimiro MC, et al. Loss of Sirt1 promotes prostatic intraepithelial neoplasia, reduces mitophagy, and delays PARK2 translocation to mitochondria. Am J Pathol. 2015;185:266–279. doi: 10.1016/j.ajpath.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Akimoto T, Pohnert SC, Li P, et al. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 184.Wright DC, Geiger PC, Han D-H, et al. Calcium induces increases in peroxisome proliferator-activated receptor gamma coactivator-1alpha and mitochondrial biogenesis by a pathway leading to p38 mitogen-activated protein kinase activation. J Biol Chem. 2007;282:18793–18799. doi: 10.1074/jbc.M611252200. [DOI] [PubMed] [Google Scholar]
- 185.Zhang Y, Uguccioni G, Ljubicic V, et al. Multiple signaling pathways regulate contractile activity-mediated PGC-1α gene expression and activity in skeletal muscle cells. Physiol Rep. 2014 doi: 10.14814/phy2.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McGee SL, Hargreaves M. Exercise and myocyte enhancer factor 2 regulation in human skeletal muscle. Diabetes. 2004;53:1208–1214. doi: 10.2337/diabetes.53.5.1208. [DOI] [PubMed] [Google Scholar]
- 187.Zhao M, New L, Kravchenko VV, et al. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kashatus JA, Nascimento A, Myers LJ, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yu T, Jhun BS, Yoon Y. High-glucose stimulation increases reactive oxygen species production through the calcium and mitogen-activated protein kinase-mediated activation of mitochondrial fission. Antioxid Redox Signal. 2011;14:425–437. doi: 10.1089/ars.2010.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pyakurel A, Savoia C, Hess D, Scorrano L. Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol Cell. 2015;58:244–254. doi: 10.1016/j.molcel.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hirota Y, Yamashita S, Kurihara Y, et al. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11:332–343. doi: 10.1080/15548627.2015.1023047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Park JH, Ko J, Park YS, et al. Clearance of damaged mitochondria through PINK1 stabilization by JNK and ERK MAPK signaling in chlorpyrifos-treated neuroblastoma cells. Mol Neurobiol. 2017;54:1844–1857. doi: 10.1007/s12035-016-9753-1. [DOI] [PubMed] [Google Scholar]
- 194.Yu W, Xu M, Zhang T, et al. Mst1 promotes cardiac ischemia-reperfusion injury by inhibiting the ERK-CREB pathway and repressing FUNDC1-mediated mitophagy. J Physiol Sci. 2019;69:113–127. doi: 10.1007/s12576-018-0627-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Handschin C, Rhee J, Lin J, et al. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ryder JW, Bassel-Duby R, Olson EN, Zierath JR. Skeletal muscle reprogramming by activation of calcineurin improves insulin action on metabolic pathways. J Biol Chem. 2003;278:44298–44304. doi: 10.1074/jbc.M304510200. [DOI] [PubMed] [Google Scholar]
- 197.Wu H, Kanatous SB, Thurmond FA, et al. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 198.Akimoto T, Ribar TJ, Williams RS, Yan Z. Skeletal muscle adaptation in response to voluntary running in Ca2+/calmodulin-dependent protein kinase IV-deficient mice. Am J Physiol Cell Physiol. 2004;287:C1311–1319. doi: 10.1152/ajpcell.00248.2004. [DOI] [PubMed] [Google Scholar]
- 199.Cereghetti GM, Stangherlin A, Martins de Brito O, et al. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci USA. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yu R, Jin S-B, Lendahl U, et al. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 2019 doi: 10.15252/embj.201899748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bo T, Yamamori T, Suzuki M, et al. Calmodulin-dependent protein kinase II (CaMKII) mediates radiation-induced mitochondrial fission by regulating the phosphorylation of dynamin-related protein 1 (Drp1) at serine 616. Biochem Biophys Res Commun. 2018;495:1601–1607. doi: 10.1016/j.bbrc.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 203.Xu S, Wang P, Zhang H, et al. CaMKII induces permeability transition through Drp1 phosphorylation during chronic β-AR stimulation. Nat Commun. 2016 doi: 10.1038/ncomms13189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gómez-Sánchez R, Gegg ME, Bravo-San Pedro JM, et al. Mitochondrial impairment increases FL-PINK1 levels by calcium-dependent gene expression. Neurobiol Dis. 2014;62:426–440. doi: 10.1016/j.nbd.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Safiulina D, Kuum M, Choubey V, et al. Miro proteins prime mitochondria for Parkin translocation and mitophagy. EMBO J. 2019 doi: 10.15252/embj.201899384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang X, Yuan D, Sun Q, et al. Calcium/calmodulin-dependent protein kinase regulates the PINK1/Parkin and DJ-1 pathways of mitophagy during sepsis. FASEB J. 2017;31:4382–4395. doi: 10.1096/fj.201601096RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Boss O, Bachman E, Vidal-Puig A, et al. Role of the beta(3)-adrenergic receptor and/or a putative beta(4)-adrenergic receptor on the expression of uncoupling proteins and peroxisome proliferator-activated receptor-gamma coactivator-1. Biochem Biophys Res Commun. 1999;261:870–876. doi: 10.1006/bbrc.1999.1145. [DOI] [PubMed] [Google Scholar]
- 208.Gómez-Ambrosi J, Frühbeck G, Martínez JA. Rapid in vivo PGC-1 mRNA upregulation in brown adipose tissue of Wistar rats by a beta(3)-adrenergic agonist and lack of effect of leptin. Mol Cell Endocrinol. 2001;176:85–90. doi: 10.1016/s0303-7207(01)00451-8. [DOI] [PubMed] [Google Scholar]
- 209.Jerums G, Hardy KJ, Eisman JA. The cyclic AMP response to glucagon. Comparison of tissue and plasma cyclic AMP levels in the rabbit. Diabetes. 1977;26:81–88. doi: 10.2337/diab.26.2.81. [DOI] [PubMed] [Google Scholar]
- 210.Miura S, Kai Y, Kamei Y, Ezaki O. Isoform-specific increases in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to beta2-adrenergic receptor activation and exercise. Endocrinology. 2008;149:4527–4533. doi: 10.1210/en.2008-0466. [DOI] [PubMed] [Google Scholar]
- 211.Pfeifle B, Pfeifle R, Faulhaber JD, Ditschuneit H. Thyroid hormone stimulation of lipolysis and cyclic adenosine 3′,5′-monophosphate accumulation in human adipose tissue. Horm Metab Res. 1980;12:711–713. doi: 10.1055/s-2007-999242. [DOI] [PubMed] [Google Scholar]
- 212.Puigserver P, Wu Z, Park CW, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 213.Chang C-R, Blackstone C. Drp1 phosphorylation and mitochondrial regulation. EMBO Rep. 2007;8:1088–1089. doi: 10.1038/sj.embor.7401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Merrill RA, Dagda RK, Dickey AS, et al. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 2011;9:e1000612. doi: 10.1371/journal.pbio.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pryde KR, Smith HL, Chau K-Y, Schapira AHV. PINK1 disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. J Cell Biol. 2016;213:163–171. doi: 10.1083/jcb.201509003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.da Rosa SC, Martens MD, Field JT, et al. Nix induced mitochondrial fission, mitophagy, and myocyte insulin resistance are abrogated by PKA phosphorylation. bioRxiv. 2019 doi: 10.1101/825828. [DOI] [Google Scholar]
- 217.Dumont NA, Bentzinger CF, Sincennes M-C, Rudnicki MA. Satellite cells and skeletal muscle regeneration. Compr Physiol. 2015;5:1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 218.Grounds MD. Towards understanding skeletal muscle regeneration. Pathol Res Pract. 1991;187:1–22. doi: 10.1016/S0344-0338(11)81039-3. [DOI] [PubMed] [Google Scholar]
- 219.Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol. 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- 220.Tidball JG. Mechanisms of muscle injury, repair, and regeneration. Compr Physiol. 2011;1:2029–2062. doi: 10.1002/cphy.c100092. [DOI] [PubMed] [Google Scholar]
- 221.Rahman FA, Krause MP. PAI-1, the plasminogen system, and skeletal muscle. Int J Mol Sci. 2020;21:7066. doi: 10.3390/ijms21197066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tidball JG. Inflammatory processes in muscle injury and repair. Am J Physiol Regul Integr Comp Physiol. 2005;288:R345–R353. doi: 10.1152/ajpregu.00454.2004. [DOI] [PubMed] [Google Scholar]
- 223.Rahman FA, Angus SA, Stokes K, et al. Impaired ECM remodeling and macrophage activity define necrosis and regeneration following damage in aged skeletal muscle. Int J Mol Sci. 2020 doi: 10.3390/ijms21134575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hardy D, Besnard A, Latil M, et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS ONE. 2016;11:e0147198. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Le Moal E, Pialoux V, Juban G, et al. Redox control of skeletal muscle regeneration. Antioxid Redox Signal. 2016;27:276–310. doi: 10.1089/ars.2016.6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- 227.Bloemberg D, Quadrilatero J. Effect of mitochondrial fission inhibition on C2C12 differentiation. Data Brief. 2016;7:634–640. doi: 10.1016/j.dib.2016.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kim B, Kim J-S, Yoon Y, et al. Inhibition of Drp1-dependent mitochondrial division impairs myogenic differentiation. Am J Physiol Regul Integr Comp Physiol. 2013;305:R927–938. doi: 10.1152/ajpregu.00502.2012. [DOI] [PubMed] [Google Scholar]
- 229.Kraft CS, LeMoine CMR, Lyons CN, et al. Control of mitochondrial biogenesis during myogenesis. Am J Physiol Cell Physiol. 2006;290:C1119–1127. doi: 10.1152/ajpcell.00463.2005. [DOI] [PubMed] [Google Scholar]
- 230.Moyes CD, Mathieu-Costello OA, Tsuchiya N, et al. Mitochondrial biogenesis during cellular differentiation. Am J Physiol. 1997;272:C1345–1351. doi: 10.1152/ajpcell.1997.272.4.C1345. [DOI] [PubMed] [Google Scholar]
- 231.Redpath CJ, Khalil MB, Drozdzal G, et al. Mitochondrial hyperfusion during oxidative stress is coupled to a dysregulation in calcium handling within a C2C12 cell model. PLoS ONE. 2013;8:e69165. doi: 10.1371/journal.pone.0069165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sin J, Andres AM, Taylor DJR, et al. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy. 2016;12:369–380. doi: 10.1080/15548627.2015.1115172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.LaBarge S, McDonald M, Smith-Powell L, et al. Estrogen-related receptor-α (ERRα) deficiency in skeletal muscle impairs regeneration in response to injury. FASEB J. 2014;28:1082–1097. doi: 10.1096/fj.13-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Nichenko AS, Southern WM, Atuan M, et al. Mitochondrial maintenance via autophagy contributes to functional skeletal muscle regeneration and remodeling. Am J Physiol Cell Physiol. 2016;311:C190–200. doi: 10.1152/ajpcell.00066.2016. [DOI] [PubMed] [Google Scholar]
- 235.Ainbinder A, Boncompagni S, Protasi F, Dirksen RT. Role of mitofusin-2 in mitochondrial localization and calcium uptake in skeletal muscle. Cell Calcium. 2015;57:14–24. doi: 10.1016/j.ceca.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Koves TR, Noland RC, Bates AL, et al. Subsarcolemmal and intermyofibrillar mitochondria play distinct roles in regulating skeletal muscle fatty acid metabolism. Am J Physiol Cell Physiol. 2005;288:C1074–1082. doi: 10.1152/ajpcell.00391.2004. [DOI] [PubMed] [Google Scholar]
- 237.Cogswell AM, Stevens RJ, Hood DA. Properties of skeletal muscle mitochondria isolated from subsarcolemmal and intermyofibrillar regions. Am J Physiol. 1993;264:C383–389. doi: 10.1152/ajpcell.1993.264.2.C383. [DOI] [PubMed] [Google Scholar]
- 238.Kirkwood SP, Munn EA, Brooks GA. Mitochondrial reticulum in limb skeletal muscle. Am J Physiol Cell Physiol. 1986;251:C395–C402. doi: 10.1152/ajpcell.1986.251.3.C395. [DOI] [PubMed] [Google Scholar]
- 239.Picard M, White K. Turnbull DM (2013) Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J Appl Physiol. 1985;114:161–171. doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Glancy B, Hartnell LM, Malide D, et al. Mitochondrial reticulum for cellular energy distribution in muscle. Nature. 2015;523:617–620. doi: 10.1038/nature14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bleck CKE, Kim Y, Willingham TB, Glancy B. Subcellular connectomic analyses of energy networks in striated muscle. Nat Commun. 2018;9:5111. doi: 10.1038/s41467-018-07676-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Eisner V, Lenaers G, Hajnóczky G. Mitochondrial fusion is frequent in skeletal muscle and supports excitation–contraction coupling. J Cell Biol. 2014;205:179–195. doi: 10.1083/jcb.201312066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Schiaffino S. Losing pieces without disintegrating: contractile protein loss during muscle atrophy. Proc Natl Acad Sci USA. 2017;114:1753–1755. doi: 10.1073/pnas.1700190114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Vertel BM, Fischman DA. Mitochondrial development during myogenesis. Dev Biol. 1977;58:356–371. doi: 10.1016/0012-1606(77)90097-5. [DOI] [PubMed] [Google Scholar]
- 245.Barbieri E, Battistelli M, Casadei L, et al. Morphofunctional and biochemical approaches for studying mitochondrial changes during myoblasts differentiation. J Aging Res. 2011 doi: 10.4061/2011/845379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Robinson MM, Sather BK, Burney ER, et al. Robust intrinsic differences in mitochondrial respiration and H2O2 emission between L6 and C2C12 cells. Am J Physiol Cell Physiol. 2019;317:C339–C347. doi: 10.1152/ajpcell.00343.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Touvier T, De Palma C, Rigamonti E, et al. Muscle-specific Drp1 overexpression impairs skeletal muscle growth via translational attenuation. Cell Death Dis. 2015;6:e1663. doi: 10.1038/cddis.2014.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Favaro G, Romanello V, Varanita T, et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat Commun. 2019;10:2576. doi: 10.1038/s41467-019-10226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Romanello V, Sandri M. Mitochondrial biogenesis and fragmentation as regulators of muscle protein degradation. Curr Hypertens Rep. 2010;12:433–439. doi: 10.1007/s11906-010-0157-8. [DOI] [PubMed] [Google Scholar]
- 250.Romanello V, Guadagnin E, Gomes L, et al. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Romanello V, Sandri M. The connection between the dynamic remodeling of the mitochondrial network and the regulation of muscle mass. Cell Mol Life Sci. 2020 doi: 10.1007/s00018-020-03662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Baldelli S, Aquilano K, Ciriolo MR. PGC-1α buffers ROS-mediated removal of mitochondria during myogenesis. Cell Death Dis. 2014;5:e1515. doi: 10.1038/cddis.2014.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Remels AHV, Langen RCJ, Schrauwen P, et al. Regulation of mitochondrial biogenesis during myogenesis. Mol Cell Endocrinol. 2010;315:113–120. doi: 10.1016/j.mce.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 254.Shintaku J, Peterson JM, Talbert EE, et al. MyoD regulates skeletal muscle oxidative metabolism cooperatively with alternative NF-κB. Cell Rep. 2016;17:514–526. doi: 10.1016/j.celrep.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Tezze C, Romanello V, Desbats MA, et al. Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab. 2017;25:1374–1389.e6. doi: 10.1016/j.cmet.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rodríguez-Nuevo A, Díaz-Ramos A, Noguera E, et al. Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. EMBO J. 2018 doi: 10.15252/embj.201796553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.