Abstract
The bromodomain and extraterminal motif (BET) proteins are critical drug targets for diseases. The precise functions and relationship of BRD2 with other BET proteins remain elusive mechanistically. Here, we used acute protein degradation and quantitative genomic and proteomic approaches to investigate the primary functions of BRD2 in transcription. We report that BRD2 is required for TAF3-mediated Pol II initiation at promoters with low levels of H3K4me3 and for R-loop suppression during Pol II elongation. Single and double depletion revealed that BRD2 and BRD3 function additively, independently, or perhaps antagonistically in Pol II transcription at different promoters. Furthermore, we found that BRD2 regulates the expression of different genes during embryonic body differentiation processes by promoter priming in embryonic stem cells. Therefore, our results suggest complex interconnections between BRD2 and BRD3 at promoters to fine-tune Pol II initiation and elongation for control of cell state.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04349-4.
Keywords: BET proteins, RNA Pol II, Cross-regulation, Embryonic body
Introduction
BET family proteins, including BRD2, BRD3, BRD4, and BRDT, consist of two bromodomains and one extraterminal domain [1]. Increasing numbers of clinically relevant studies have demonstrated that BET protein inhibitors show great therapeutic potential for various pathologies by generally competing for the interactions between bromodomain and acetylated histones, leading to the repression of key genes involved in disease [2–4]. Genomic mapping indicates that BRD2, BRD3, and BRD4 have similar binding profiles at promoter and enhancer regions by recognizing acetylated histones. Previous studies have shown that BRD4 regulates Pol II elongation with p-TEFb [5–7], and BRD2 and BRD3 appear to function in Pol II initiation [8]. However, their molecular effects on initiation factors remain unclear. On the other hand, BRD3 rescues BRD2 deficiency [9], suggesting that BRD2 and BRD3 might function redundantly in cells. However, BRD2-, BRD3-, and BRD4-specific and relationships remain elusive. Investigating unique and coregulated gene targets for individual BRDs would therefore be important for applying BET-specific and pan inhibitors.
BRD2 was demonstrated to regulate gene expression in malignant melanoma, embryonic stem cells, and Th17 cell differentiation [10–12]. Previous protein–protein interaction network analyses have indicated that BRD2 interacts with several proteins, such as E2F1, TBP, SWI/SNF, HDAC11, KAT7, CTCF and topoisomerase I, indicating roles of BRD2 in transcriptional regulation, 3D chromatin organization and DNA repair [13–18]. Previous studies on the molecular mechanisms of BRD2 in Pol II transcription are limited because they primarily used specific gene expression analyses, correlation analyses, BET inhibitor treatment, reporter assays, or aggregate analyses without follow-up mechanistic investigations [11–13]. Although TBP was shown to interact with BRD2 under serum starvation [13], whether they interact under normal conditions and their interdependence for executing effective transcription are unclear. On the other hand, Pol Il was shown to decrease binding after BRD2 depletion [8], but the detailed molecular mechanisms and biological function are also unclear. Thus, the molecular mechanisms of BRD2 in Pol II transcription remain worthy of investigation.
Uncovering the principles for maintaining and establishing cell states is fundamental for understanding development and diseases, so transcriptional regulation during cell lineage commitment has been intensively investigated. Multiple factors, such as LSD1, TAF3, ELL3, and MLL4, have been shown to modulate embryonic stem cell differentiation by controlling distal enhancers [19–22]. BRD2 is essential for early embryo development and proper neural tube maturation, and haploinsufficiency of BRD2 causes epilepsy and severe obesity without causing diabetes during mouse development [23–27]. However, the molecular mechanisms of BRD2-mediated promoter activation during differentiation are not well understood. In particular, understanding BRD2’s effect on cell state control would provide insights into the applications of BET inhibitors in cell reprogramming.
Here, we performed BRD2 ChIA-PET, ChIP-MS, and acute depletion of BRD2 proteins followed by quantitative genomic analyses in murine embryonic stem cells. We found that gene promoters with low-level H3K4me3 modifications required BRD2 to facilitate TAF3-mediated Pol II initiation. Whereas gene promoters with high-level of H3K4me3 modifications, exhibited a reduced requirement for BRD2 to safeguard initiation but needed BRD2 to suppress R-loops for transcriptional elongation. Regarding the roles of other BET proteins (BRD3) in BRD2-mediated transcriptional regulation, we found that BRD2 and BRD3 function additively, independently, or perhaps antagonistically in Pol II transcription at different gene promoters. Furthermore, BRD2 depletion affects the expression of different genes during embryonic stem cell differentiation, likely through promoter priming. Our study reveals a complex regulatory mechanism for BET proteins and provides a foundation for understanding the binding and functional relationships among different BET proteins in development and disease.
Results
BRD2 depletion affects Pol II initiation mediated by TAF3
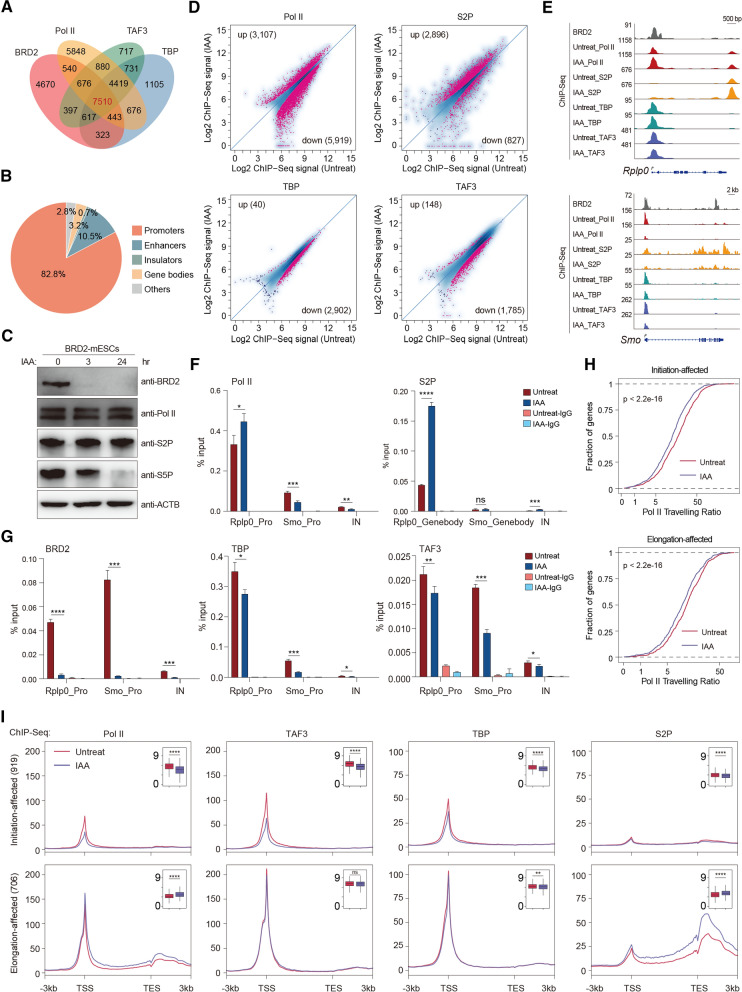
To explore the detailed molecular mechanisms underlying the regulation of Pol II transcription, we first compared the genomic occupancy of BRD2 to that of Pol II and initiation factors, such as TBP and TAF3 (also known as components of TFIID) in mESCs. The overlap analyses for the ChIP-Seq peaks revealed that BRD2, Pol II and initiation factors largely co-occupied gene promoters (Figs. 1A, B), indicating an active role for BRD2 in Pol II regulation. We constructed BRD2-degron mESCs by knocking a mAID-GFP (mini IAA-inducible degradation) tag into the C-terminus of endogenous BRD2 using the CRISPR/Cas9 gene-editing strategies that we previously used [28]. Genotyping, immunofluorescence, and Western blot analyses confirmed that BRD2 protein degron system was efficient (Fig. S1A–D). The Pol II antibody recognized the N-terminal domain (NTD) of RPB1, captured total Pol II, S5P (serine 5 phosphorylated) Pol II antibody recognized initiating Pol II in the promoters, and S2P (serine 2 phosphorylated) Pol II antibody recognized elongating Pol II in the gene bodies. Western blot analyses indicated that BRD2 depletion did not affect the protein levels of total Pol II or S2P Pol II (Fig. 1C), but affect S5P Pol II protein level, consistent with previous report of BRD2 in transcription initiation [8]. To directly investigate the roles of BRD2 in Pol II transcription, we then performed total and S2P Pol II ChIP-Seq before and after BRD2 depletion. The scatter-plot analyses indicated that BRD2 depletion led to both an increase and a decrease in total Pol II occupancy at promoter regions and a preferential increase in S2P Pol II at gene bodies (Fig. 1D, Tables S1–2). These results suggested that BRD2 depletion caused changes in chromatin binding for both total Pol II and elongating Pol II in the genome.
Fig. 1.
BRD2 depletion leads to both Pol II initiation and elongation defects. A Venn diagram displaying the overlapping peaks of the BRD2, Pol II, TAF3, and TBP ChIP-Seq datasets. B The pie chart shows that the overlapping peaks of BRD2, Pol II, TAF3, and TBP ChIP-Seq primarily occupy gene promoters. C Western blot analyses of BRD2, Pol II, S2P Pol II and S5P Pol II protein levels in BRD2 degron mES cells under untreated and IAA-treated conditions. D Scatter plots showing total Pol II (antibody recognized N-terminal of RPB1), S2P (serine 2 phosphorylated RPB1), TBP and TAF3 ChIP-Seq signals at expressed gene promoters and gene bodies (for S2P) before and after BRD2 degradation. Red dots represent the differentially regulated genes identified by DiffBind software (v3.2.2) using DESeq2 as a comparison model (FDR < 0.05). Two replicates were done for each next-generation sequencing experiment. E Genome browser track of BRD2, Pol II, S2P, TBP, and TAF3 ChIP-Seq signals in BRD2 degron mES cells in untreated and 3 h IAA-treated conditions at the Rplp0 and Smo loci. F Pol II ChIP-qPCR analyses of Rplp0 and Smo promoters and intergenic regions in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. Pol II-S2P ChIP-qPCR analyses of the Rplp0 and Smo gene body regions in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. The intergenic region serves as a negative control for ChIP-qPCR. Primers see Table S9. Error bars represent the SD of at least three technical replicates. p values were calculated using Student’s t test (ns: p > 0.05, *p < = 0.05, **p < = 0.01, ***p < = 0.001, ****p < = 0.0001). G BRD2, TBP and TAF3 ChIP-qPCR analyses of Rplp0 and Smo promoters and intergenic regions in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. The qPCR results are represented the same as (F). H The accumulative distribution curve of the Pol ll traveling ratio at initiation and elongation-affected genes upon BRD2 degradation. Initiation-affected genes were defined as genes whose promoters were bound by BRD2, TAF3 and Pol II and showed a decrease in TAF3 signals at promoters identified by DiffBind. Elongation-affected genes were defined as genes whose promoters were bound by BRD2 and Pol II and showed an increase in S2P signals at gene bodies and transcription ending sites identified by DiffBind. Significance was assessed using the Wilcoxon test. I Meta-gene plots of the average Pol II, TAF3, TBP and S2P ChIP-Seq signals at initiation-affected, elongation-affected genes (see methods for more details). Boxplots (insets) displayed the log2 ChIP-Seq signals at ± 100 bp around TSS regions or gene-body regions (Pol II or S2P at elongation-affected genes. Gene body regions were defined as − 1 kb from TSS to transcription termination sites) upon BRD2 depletion. Significance was generated using the ChIP-Seq signals in each gene set under untreat and IAA conditions by the Wilcoxon test (ns p > 0.05, **p < = 0.01, ****p < = 0.0001)
To further decipher the mechanisms of BRD2-mediated transcriptional initiation, we next performed TBP and TAF3 ChIP-Seq after immediate BRD2 depletion. The differential analyses indicated that BRD2 depletion decreased the chromatin binding of TAF3 and TBP at promoters (Fig. 1D). The overlap analysis showed that the majority of genes with decreased TAF3 binding also have decreased Pol II binding, and that genes with increased S2P binding exhibited the most overlap with genes with increased Pol II binding (Fig. S1E, Table S2). Correlation analyses for the ChIP-Seq signal changes consistently indicated that TAF3 and S2P changes were slightly associated with the changes in Pol II ChIP-Seq signals (Fig. S1F). ChIP-qPCR experiments were used to further validate the ChIP-Seq changes in response to BRD2 depletion. Smo and Rplp0 were selected, because Smo encodes the protein smoothened, which plays an important role in Hedgehog signaling [29], and Rplp0 encodes a ribosomal protein, both of which were enriched in functional terms of BRD2 affected genes (Table S2). ChIP-qPCR results revealed that the gene Smo, which showed decreased total Pol II occupancy, also decreased the TAF3 and TBP binding. The gene Rplp0, which showed increased Pol II binding, was associated with higher levels of Pol II occupancy, exhibiting relatively fewer effects on TAF3 and TBP binding, but with increased S2P Pol II binding signals (Figs. 1E–G). Together, these results suggest that BRD2 facilitates TBP and TAF3 binding for many promoters in mESCs.
We next defined initiation- and elongation-affected genes by identifying genes that were co-bound by both Pol II and BRD2, with either decreased TAF3 binding (n = 919) or increased S2P binding (n = 706) (Fig. S1G, Table S3). The initiation-affected genes exhibited decreased Pol II occupancy at the transcription start sties (TSSs) and traveling ratio, consistent with the defects of initiation. The elongation-affected genes were associated with increased Pol II at gene bodies and decreased traveling ratio, also confirming the perturbations in elongation (Fig. 1H, I). Meta-gene analyses demonstrated that initiation-affected genes displayed a decrease in Pol II, TBP, and TAF3 signals at their promoters. Elongation-affected genes contained more total Pol II occupancy at both the promoter and gene body regions but fewer effects on the TBP and TAF3 signals at the promoters after BRD2 depletion (Fig. 1I). Meta-plot and boxplot showed that the effect sizes of TAF3 and TBP are small, however, they are all significantly larger than the effect sizes of genes with unchanged binding (Fig. S1H). In contrast, BRD2 unbound genes did not show obvious binding or changes in these factors (Fig. S1I). Our results collectively suggested that BRD2 functions in Pol II initiation and elongation in mESCs.
BRD2 depletion leads to increased R-loops, potentially underlying the effects of accumulated Pol II occupancy in the gene bodies
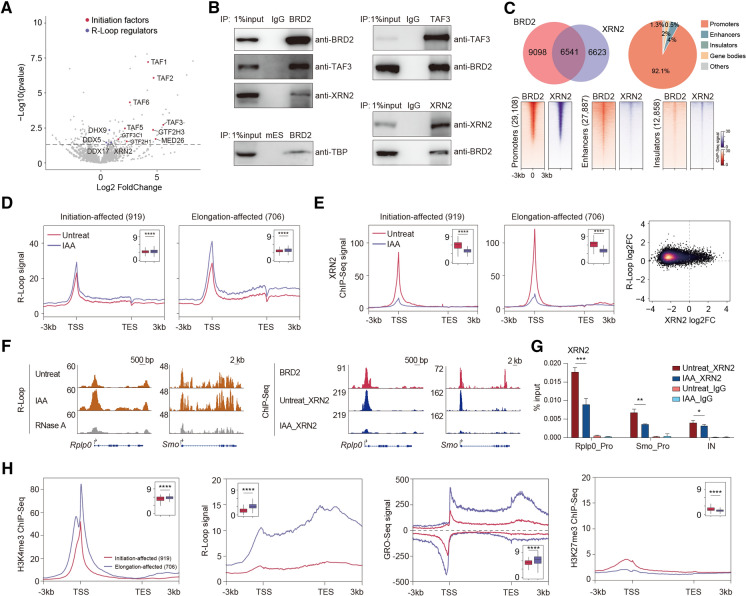
To obtain a deeper understanding of BRD2-mediated Pol II transcriptional regulation, we performed BRD2 ChIP-MS to characterize the BRD2 protein interactome in mESCs, which would provide a resource for pinpointing candidate factors to explain the molecular functions of BRD2. TFIID components, including TAF1, TAF2, and TAF3, were preferentially enriched in the BRD2 ChIP-MS preparations (Fig. 2A, Table S4). The interactions of TBP and TAF3 with BRD2 were independently verified by immunoprecipitation followed by Western blotting experiments (Fig. 2B). In addition, size exclusion chromatography analyses indicated that TAF3 colocalized in the same fractions as BRD2 (Fig. S2A), suggesting that BRD2 interacts with TAF3.
Fig. 2.
BRD2 depletion causes increased R-loop signals potentially underlying the roles of BRD2 in Pol II elongation. A Volcano plot displaying the enriched proteins identified by BRD2 ChIP-MS in mESCs. BRD2 ChIP-MS-enriched proteins were identified by DESeq2 using the PSM (peptide-spectrum match) values in BRD2 and input samples (2 repeats for each sample, p value < 0.05). The enriched initiation factors and R-loop regulators are shown in red and purple, respectively. B Western blot analyses of anti-BRD2, anti-TAF3 and anti-XRN2 immunoprecipitates in isolated chromatin fractions from wild-type mES cells show that BRD2 interacts with TAF3 and XRN2. Western blot analyses of anti-BRD2 immunoprecipitates in isolated chromatin fractions of BRD2 degron mES cells show that BRD2 interacts with TBP. C Pie chart (top left) showing the overlapping ChIP-Seq peaks between BRD2 and XRN2. The pie chart (top right) shows that the overlapping peaks of BRD2, and XRN2 ChIP-Seq occupy promoters. Heatmap plots (bottom) displaying the occupancy of BRD2 and XRN2 at promoters, enhancers, and insulators. D Meta-gene plots (left) of the average R-loop signals at initiation-affected and elongation-affected genes upon BRD2 depletion. Boxplots (insets) show log2 R-loop signals at ± 100 bp around TSSs upon BRD2 depletion. Significance was determined using the Wilcoxon test (****p < = 0.0001). Two replicates were done for each next-generation sequencing experiment. E Meta-gene plots (left) of the average XRN2 ChIP-Seq signals at initiation-affected and elongation-affected genes upon BRD2 degradation. Boxplots (insets) showing log2 XRN2 ChIP-Seq signals at ± 100 bp around TSS regions upon BRD2 depletion. Density scatter plots (right) showing the correlation of binding fold change between R-loop and XRN2 ChIP-Seq signals upon BRD2 depletion. Significance was determined using the Wilcoxon test (****p < = 0.0001). Two replicates were done for each next-generation sequencing experiment. F Left: genome browser track of S9.6 R-loop Cut&Tag signals in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions at the Rplp0 and Smo loci. The RNase A-treated group served as a negative control. Right: genome browser track of XRN2 ChIP-Seq signals in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions at the Rplp0 and Smo loci. G XRN2 ChIP-qPCR analyses of Rplp0 and Smo promoters and intergenic regions in BRD2 degron cells under untreated and 3 h IAA-treated conditions. The intergenic region serves as the negative control for ChIP-qPCR. Primers see Table S9. Error bars represent the SD of at least three technical replicates. p values were calculated using Student’s t test (*: p < = 0.05, **: p < = 0.01). H Meta-gene plots of the average H3K4me3, GRO-Seq, H3K27me3 ChIP-Seq and R-loop signals (public available dataset, see Table S8. The signal distribution is different from our R-loop signals in (D), as they were generated with different library preparation methods for R-loop sequencing) at initiation-affected and elongation-affected genes. Boxplots (insets) showing the log2 ChIP-Seq or GRO-Seq signals at ± 100 bp around TSS regions, or R-loop signals at gene body regions of initiation-affected and elongation-affected genes. Significance was determined using the Wilcoxon test (****p < = 0.0001)
Previous studies have reported that BRD2 suppresses R-loops with TOP1 to inhibit genome instability [18], but the impact of BRD2-mediated R-loop repression on Pol II transcription remains unclear. Interestingly, several proteins involved in R-loop regulation, such as DHX9, DDX5, and XRN2, were detected in the BRD2 ChIP-MS preparation of mESCs (Fig. 2A). These protein interactions were also detected in a published BRD2 IP-MS dataset with a different cell line for investigating the roles of BRD2 in genome stability [18]. Immunoprecipitation followed by Western blotting further validated the interactions between BRD2 and XRN2 (Fig. 2B). Genomic binding analyses also indicated that BRD2 co-occupies gene promoters with XRN2 (Fig. 2C). We then performed R-Loop analyses using a recently published R-Loop CUT&Tag method after BRD2 depletion [30]. Metagene and specific gene examples showed increased R-loop signals in response to BRD2 depletion, and the increase was relatively greater for elongation-affected genes (Fig. 2D), consistent with the prior finding that R-loops may retard Pol II elongation in gene bodies [31, 32], which served as an explanation for the increased Pol II signals in the genes after BRD2 depletion. However, the slight increase in TAF3 and TBP chromatin binding after BRD2 depletion (Fig. 1D) might represent some negative feedback regulations for the transcription elongation suppression that warrant further investigation in the future.
A previous study demonstrated that RNAi knock down both decapping factor and XRN2 lead to increased Pol II occupancy at the gene bodies of ENO1, HSP90AA1, MAT2A, RBM39, etc. [33]. Surprisingly, these genes also showed an increase in Pol II signals at the gene bodies after BRD2 depletion in our mESCs, even though they were performed in different cell lines with distinct perturbations. XRN2 ChIP-Seq was also performed after BRD2 depletion. Meta-gene and specific gene examples revealed that XRN2 decreased chromatin binding after BRD2 depletion, and this decrease was preferentially greater for elongation-affected genes than for initiation-affected genes (Figs. 2E, F). Decreased XRN2 binding was also associated with increased R-loop signals (Fig. 2E). ChIP-qPCR experiments further validated the decreased chromatin binding of XRN2 after BRD2 depletion for both the elongation-affected gene Rplp0 and initiation-affected gene Smo (Fig. 2G). Collectively, our results implicated that BRD2 may function together with R-loop regulators, such as XRN2 and RNA helicases, to suppress R-loops during transcriptional elongation globally. The genome-wide effects of R-loop after BRD2 depletion together with the preferential effects on initiation described above combinatorically contribute to Pol II transcription.
TAF3 directly recognizes H3K4me3-modified histones at promoters, and BRD2 binds acetylated histones in chromatin [34–37]. Therefore, we performed correlation analyses for both initiation-affected and elongation-affected genes. The results showed that initiation-affected genes exhibited a relatively low level of H3K4me3 modifications. In contrast, elongation-affected genes presented a relatively high level of R-loop and GRO-Seq signals (Fig. 2H). These results indicated that the TAF3 binding at promoters containing low levels of H3K4me3 may require BRD2 to enhance its chromatin binding. TAF3 binding at promoters containing high levels of H3K4me3 may not need BRD2 to facilitate its binding, while these genes are highly transcribed and require BRD2 to suppress R-loops for transcription elongation.
BRD2 depletion leads to increased BRD3 binding in a subset of gene promoters
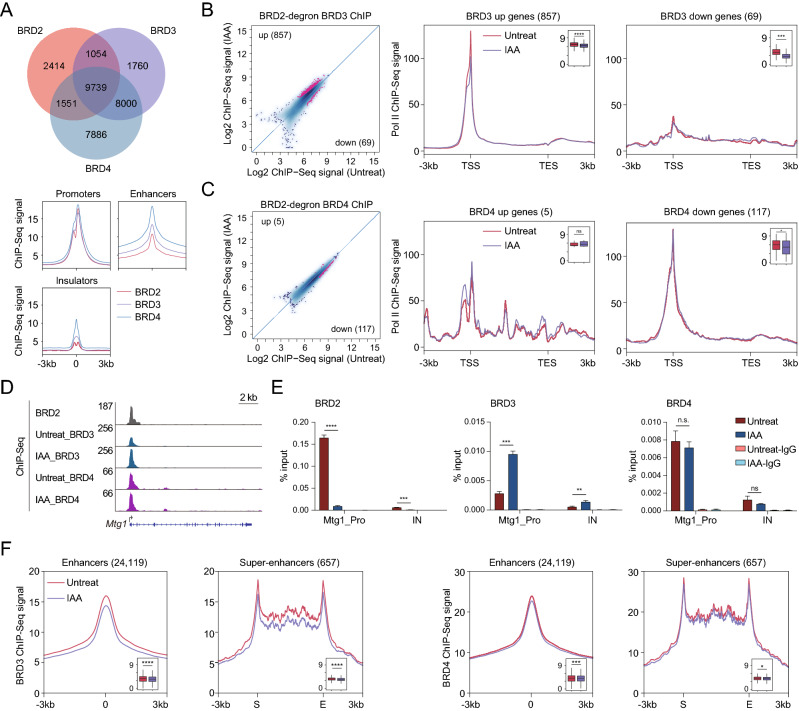
BRD2, BRD3, and BRD4 are bromodomain-containing acetylated histone readers. We hypothesized that other BET proteins might synergistically or antagonistically regulate the molecular functions of BRD2. Genomic localization analyses indicated that BRD2, BRD3, and BRD4 co-occupy the enhancer and promoter regions (Fig. 3A), implicating that BET proteins may interconnect with each other to regulate Pol II [38]. To directly investigate the relationships for chromatin binding among BRD2, BRD3, and BRD4, we performed BRD3 and BRD4 ChIP-Seq after BRD2 depletion. The differential analyses indicated that 857 promoters significantly gained BRD3 binding (BRD3 up genes), and 69 promoters significantly reduced BRD3 binding (BRD3 down genes) after the depletion of BRD2 (Fig. 3B, Table S5). The Pol II occupancy of these genes indicates that genes with decreased BRD3 binding exhibit relatively more changes, and genes with increased BRD3 binding presented relatively fewer changes (Fig. 3B), consistent with a previous report that BRD3 rescues BRD2 deficiency in blood cells [9]. In contrast, the chromatin binding of BRD4 appeared to be affected less than that of BRD3 after BRD2 depletion (Fig. 3C), which is consistent with previous finding that BRD2 RNAi followed BRD4 ChIP-qPCR analyses in Th17 cells [10]. On the other hand, the BRD4 binding-changed genes also seemed not to show obvious Pol II occupancy changes at gene regions after BRD2 depletion (Fig. 3C). ChIP-qPCR experiments validated the increased BRD3 chromatin binding after BRD2 depletion at the Mtg1 gene promoter (Figs. 3D, E). These results suggested that BRD2 competes with BRD3 for binding of subset of gene promoters.
Fig. 3.
BRD2 depletion increases BRD3 binding at a subset of promoters. A Pie chart (top) showing the overlapping peaks among BRD2, BRD3 and BRD4 ChIP-Seq in mESCs. The meta plots (bottom) displaying the occupancy of BRD2, BRD3 and BRD4 at promoters, enhancers and insulators. B Scatter plots (left) showing BRD3 ChIP-Seq signals at expressed gene promoters before and after BRD2 degradation. Red dots represent the differentially regulated genes identified by DiffBind software (v3.2.2) using DESeq2 as a comparison model (FDR < 0.05). Metagene analysis (right) displays the average Pol II ChIP-Seq signals at BRD3 up or down genes (representing genes with up- and downregulated BRD3 ChIP-Seq signals before and after BRD2 depletion). Boxplots (insets) show log2 Pol II ChIP-Seq signals at ± 100 bp around TSS regions upon BRD2 depletion. Significance was determined using the Wilcoxon test (***p < = 0.001; ****p < = 0.0001). Two replicates were done for each next-generation sequencing experiment. C The legend of © is the same as that of (B), but for BRD4. D Genome browser track of BRD3 and BRD4 ChIP-Seq signals in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions at the Mtg1 locus. E BRD2, BRD3 and BRD4 ChIP-qPCR analyses of the Mtg1 promoter and intergenic regions. The intergenic region serves as a negative control for ChIP-qPCR. Primers see Table S9. Error bars represent the SD of at least three technical replicates. p values were calculated using Student’s t test (ns: p > 0.05, **p < = 0.01, ***p < = 0.001, ****p < = 0.0001). F Meta plots of the average BRD3 (left) and BRD4 (right) ChIP-Seq signals at enhancers and super-enhancers before and after BRD2 degradation. Enhancers and super-enhancers were defined using previously published MED1 ChIP-Seq dataset. Boxplots (insets) show log2 BRD3 and BRD4 ChIP-Seq signals at super-enhancer regions and ± 100 bp around the center of enhancer regions. Significance was determined using the Wilcoxon test (*p < = 0.05, *** p < = 0.001, ****p < = 0.0001)
A previous study demonstrated that the BET coactivators BRD3 and BRD4 contained intrinsically disordered regions and formed transcriptional condensates with transcription factors and mediators at super-enhancers for transcription activation [39–41]. Amino acid sequence analyses indicated that BRD2 also contains intrinsically disordered regions (Fig. S2B). However, whether the occupancy of BRD3 and BRD4 at the enhancer regions is dependent on BRD2 is still unclear. We next examined BRD3 and BRD4 chromatin binding in response to BRD2 depletion at enhancer and super-enhancer regions. The meta-analyses showed that BRD2 depletion led to tiny reductions of BRD3 and BRD4 at the enhancer and super-enhancer regions (Fig. 3F). Together, these results suggested that BRD2 depletion increases BRD3 binding at a subset of genes. The interconnections between BRD2 and BRD3 indicated that BRD2 might function in a gene-specific manner with BRD3.
BRD2 and BRD3 depletion reveals both additive and independent effects on Pol II transcription
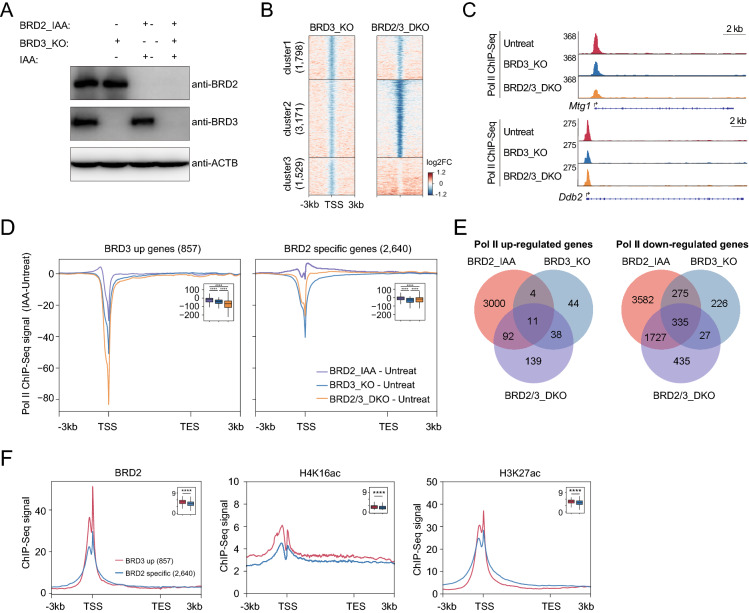
To further examine the functional relationship between BRD2 and BRD3 in Pol II transcription, we knocked out BRD3 in BRD2 degron mES cells. This cell line allows us to investigate the molecular functions of BRD2 and BRD3 both separately and together. Western blot experiments verified BRD3 knockout and BRD2 degradation (Fig. 4A). We performed Pol II ChIP-Seq after depletion of BRD3 or after depletion of both BRD3 and BRD2. To directly investigate the relationships between BRD2 and BRD3 in Pol II transcription, we performed cluster analyses for Pol II ChIP-Seq signals at active gene promoters. The result revealed three gene clusters (Fig. 4B): cluster 1 did not show noticeable differences between BRD3 knock out and BRD2/BRD3 double depletion, indicating little cross regulations of these; cluster 2 exhibited decreased Pol II after BRD3 depletion and much more marked decrease in Pol II after depletion of both BRD2 and BRD3, indicating that BRD2 and BRD3 function additively; cluster 3 showed few decreased Pol II genes after BRD3 depletion and a slightly increased Pol II after BRD2 and BRD3 double depletion, indicating that BRD3 may function independently, or perhaps antagonistically in BRD2-mediated Pol II transcription at these genes.
Fig. 4.
BRD2 functions additively and independently with BRD3 in Pol II transcription. A Western blot validation of BRD3 protein knockout (BRD3_KO) in BRD2 degron mESCs, BRD2 and BRD3 double depletion (BRD2/3_DKO) under 3 h IAA-treated with BRD2 degron mESCs. B Heatmap showing the occupancy changes of Pol II at active gene promoters. The promoters were clustered by the ChIP-Seq signals of untreated, BRD3-depleted and BRD2/3-double depleted cells using k-means. Active gene promoters were downloaded from a previous publication [21]. The color bar indicates the log2FC of ChIP-Seq signals. Two replicates were done for each next-generation sequencing experiment. C Genome browser track of Pol II ChIP-Seq signals in BRD3 protein knockout (BRD3_KO) in BRD2 degron mESCs, BRD2 and BRD3 double depletion (BRD2/3_DKO) conditions at the Mtg1 and Ddb2 loci. D Meta-gene plots of the altered Pol II ChIP-Seq signals at BRD3 up- and BRD2-specific genes upon BRD2 depletion (IAA), BRD3 knockout (KO) or BRD2 and BRD3 double depletion. The altered Pol II ChIP-Seq signals were calculated as Pol II signals in treated samples subtracting Pol II signals in untreated samples. Boxplots (insets) show log2 Pol II ChIP-Seq signals in IAA samples subtracted by signals in untreated samples at ± 100 bp around TSS regions. Significance was determined using the Wilcoxon test (****: p < = 0.0001). E The Venn diagram showed the overlap analyses for the Pol II upregulated genes (left) and downregulated genes (right) after BRD2 depletion (BRD2_IAA), BRD3 depletion (BRD3_KO) and BRD2 and BRD3 double depletion (BRD2/3_DKO). F Meta-gene plots of the BRD2, H4K16ac, H3K27ac ChIP-Seq signals (public available dataset, see Table S8) at BRD3 up- and BRD2-specific genes. Boxplots (insets) show log2 ChIP-Seq signals at ± 100 bp around TSS of BRD3-up and BRD2-specific genes. Significance was determined using the Wilcoxon test (****p < = 0.0001)
We next sought to determine the effects on Pol II transcription considering the relationships between BRD2 and BRD3 in chromatin binding. For the genes with increased BRD3 chromatin binding at their promoters after BRD2 depletion (BRD3 up genes), they showed decreased Pol II binding at the transcription start sites after single depletion of BRD2 or BRD3. Double depletion of BRD2 and BRD3 led to further decrease of Pol II binding at TSSs (Figs. 4C, D). For BRD2-regulated genes that did not show both increase and decrease in BRD3 chromatin binding (BRD2-specific genes), BRD2 depletion slightly increased Pol II signals, BRD3 depletion decreased the Pol II signals at the TSS, and both BRD2 and BRD3 depletion decreased Pol II signals less than BRD3 depletion alone (Figs. 4C, D). These results collectively suggested that BRD2 and BRD3 have both additive and independent functions in Pol II transcription at various gene promoters. In addition, Venn diagram of genes perturbed by a single (BRD2 or BRD3) and double (BRD2 and BRD3) protein showed that some genes were regulated by both BRD2 and BRD3, while others were regulated by BRD2 or BRD3 alone (Fig. 4E), further suggesting the additive and independent roles between BRD2 and BRD3. Moreover, ChIP-Seq signal correlation analyses indicated that the BRD3 up genes tend to have higher abundances of BRD2, BRD3 and histone acetylation occupancy (H4K16ac and H3K27ac) compared with the BRD2 specific genes. In contrast, the BRD4 chromatin binding seemed not to show apparent difference between these two groups of genes (Figs. 4F, S2C). These results implicate that BRD2 and BRD3 might compete with each other’s chromatin binding at high abundance of histone acetylated regions.
BRD2 is dispensable for maintaining promoter-associated chromatin interactions for specific genes in mESCs
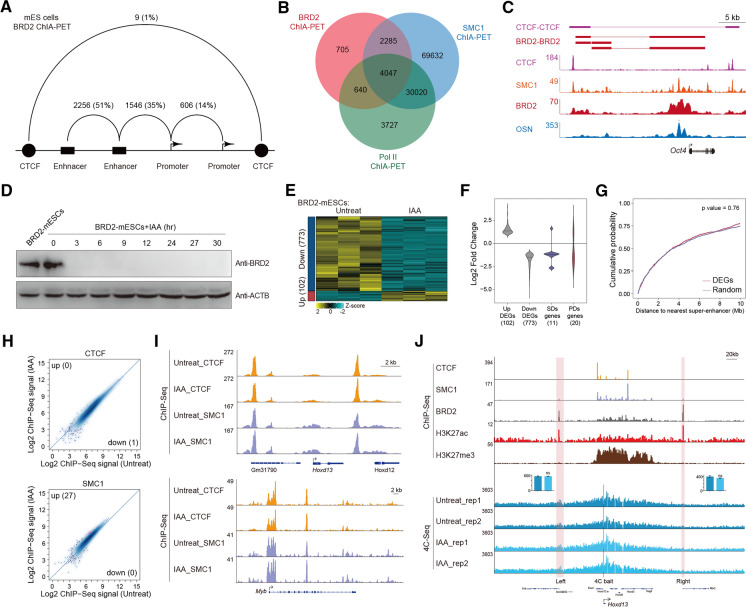
BRD2 is unique compared to BRD3 and BRD4, as BRD2 has been implicated in 3D chromatin organization by interacting with CTCF at the architectural boundaries [17]. To investigate whether BRD2’s functions in 3D genome organization contributed to BRD2-mediated gene expression, we performed BRD2 chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) to capture the BRD2-mediated chromatin interactions in mESCs. The BRD2 ChIA-PET dataset was processed using the previously described pipeline [42], identifying 7,677 high-confidence interactions. Most of these interactions were associated with enhancers and promoters (Fig. 5A). The BRD2 ChIA-PET-identified chromatin interactions were also confirmed by previous Cohesin and Pol II ChIA-PET datasets (Fig. 5B [43, 44]), suggesting the success of the BRD2 ChIA-PET experiment. Interestingly, BRD2 ChIA-PET identified enhancer-promoter interactions for many pluripotency genes in ES cells, such as Oct4 (Fig. 5C), suggesting the potential roles of BRD2 in cell state control.
Fig. 5.
BRD2 is dispensable for specific promoter-associated chromatin interactions. A Summary of the BRD2 ChIA-PET interactions among enhancers, promoters and CTCF binding sites that did not have transcriptional activity. B Venn diagram analyses showing the overlap among ChIA-PET interactions identified by BRD2, RNAPII and Cohesin in mESCs. The Pol II and Cohesin ChIA-PET data were derived from public datasets. C BRD2 ChIA-PET-identified enhancer-promoter interactions for Oct4. ChIA-PET interactions for CTCF-CTCF and BRD2-BRD2 are indicated as purple and red lines, respectively. ChIP-Seq binding profiles for CTCF, SMC1, BRD2, and OCT4/SOX2/NANOG (OSN) are shown at the Oct4 locus. D Western blot analyses of BRD2 protein degradation at the indicated time points after treatment of BRD2-degron-mESCs with IAA. ACTB served as a loading control. E Heatmap of 875 genes with differential expression levels in BRD2-depleted versus untreated mESCs. The normalized RNA-Seq reads were log2-transformed and are displayed in the form of heatmaps for upregulated (red bar) and downregulated (blue bar) groups. Three biological replicates were used for differential analyses in DESeq2 (FDR < 0.05 and |log2FC|> 1). F Violin plot showing the expression fold change of up- and downregulated differentially expressed genes (DEGs) identified in panel E and that of the DEGs in the super-enhancer domains (SDs) and polycomb domains (PDs). G Cumulative distribution of the distance of DEGs to the nearest super-enhancers. Random genes were selected as negative controls. Significance was determined by the Wilcoxon test. H Scatter plots showing CTCF and SMC1 ChIP-Seq signals at CTCF and Cohesin (SMC1) peak sites before and after BRD2 degradation. Red dots represent differentially binding sites identified by DiffBind software (v3.2.2) using DESeq2 as a comparison model (FDR < 0.05). Two replicates were done for each next-generation sequencing experiment. I Genome browser track of CTCF and SMC1 ChIP-Seq signals in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions at the Hoxd13 and Myb loci. J ChIP signals for CTCF, SMC1, BRD2, H3K27ac and H3K27me3 at the Hoxd13 locus (upper). 4C-Seq assay (bottom) analyses for enhancer-promoter interactions at the Hoxd13 locus in BRD2 degron mES cells under untreated and 3 h IAA-treated conditions. The red lines indicate 4C bait sites, and the pink rectangles indicate the 4C prey sites. The bar plots (middle) showing the normalized 4C-Seq signals at the left and right the 4C prey sites, respectively. Error bars represent SD of duplicate reactions. p values were calculated using Student’s t test (ns: p > 0.05)
The majority of BRD2 ChIA-PET interactions localized to enhancers and promoters, indicating that BRD2 occupied mostly active chromatin regions in various cell lineages and may serve as a molecular platform or scaffold for transcriptional regulation. To gain functional insights into the 3D chromatin occupancy of BRD2, multiomics analyses were performed after acute depletion of BRD2. RNA-Seq, CTCF, and Cohesin ChIP-Seq and 4C-Seq were performed after BRD2 depletion to directly investigate the potential roles of BRD2 in the 3D chromatin landscape in mESCs (Fig. 5D). The DESeq2 pipeline was used to identify differentially expressed genes using the RNA-Seq dataset. We found 102 upregulated genes and 773 downregulated genes in response to BRD2 depletion (Fig. 5E, Table S6). Among these differentially expressed genes, 144 had BRD2-mediated ChIA-PET interactions. The Venn diagram showed that there was an enrichment for downregulated genes in the 144 differentially expressed genes-mediated by BRD2 ChIA-PET interactions (Fisher’s exact test p value = 0.01) (Fig. S3B). There were few differentially expressed genes localized in the super-enhancer domains or polycomb domains, which are known to be vulnerable to perturbations in 3D chromatin organization, as previously reported [44] (Fig. 5F). Additionally, these differentially expressed genes did not exhibit obvious differences in the distance to their nearest super-enhancers compared to the randomly selected genes (Fig. 5G). Consistently, the CTCF and Cohesin occupancy genome-wide and at specific loci did not exhibit obvious changes (Figs. 5H, I).
4C-Seq with baits for anti-several genomic loci were performed after BRD2 depletion. Hoxd13 gene expression changed after BRD2 depletion, but its promoter-associated chromatin interactions do not show apparent changes after BRD2 degradation (Fig. 5J). In addition, the enhancer-promoter interaction frequency for the pluripotency gene Sox2 constrained in CTCF-insulated neighborhoods did not obviously change after BRD2 depletion. The interactions between the Sall4 promoter and its proximal or distal enhancer also did not dramatically change after BRD2 depletion (Fig. S3A). However, treatment with the 500 nM BET inhibitor JQ1 for 24 h perturbed these enhancer-promoter interactions, possibly due to secondary effects after long-term transcriptional inhibition by a high concentration of JQ1 or inhibition of other BRDs (Fig. S3A). This evidence collectively suggests that BRD2 is dispensable for maintaining enhancer-promoter interactions for specific genes in mESCs.
BRD2 dynamically regulates key genes in ES cell differentiation
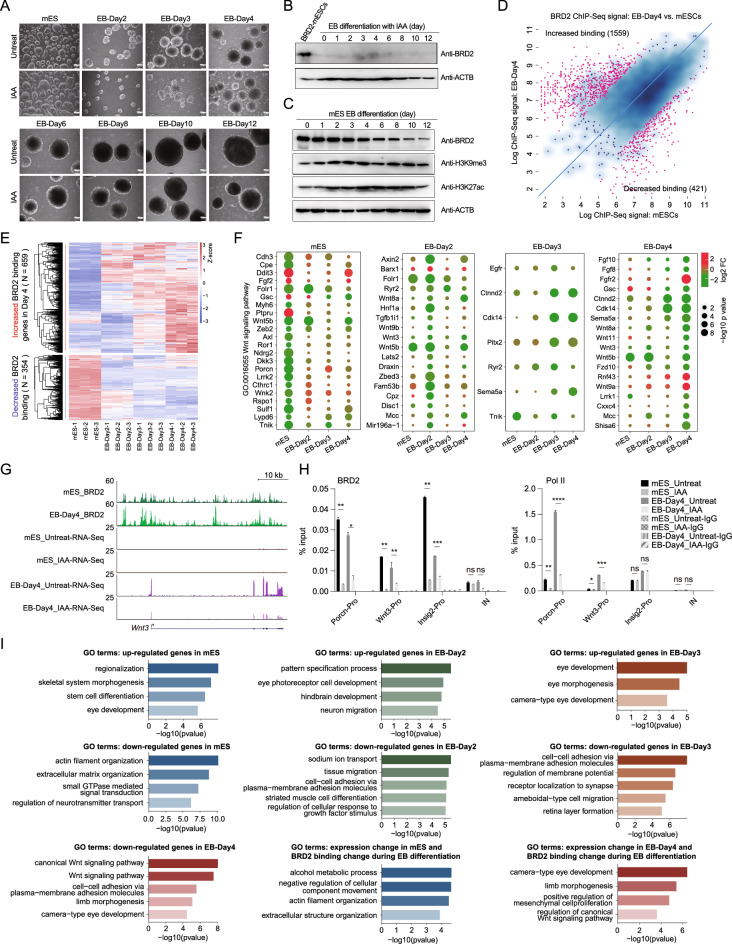
We next sought to investigate the biological functions of BRD2 during ES cell differentiation. mES cells were differentiated into embryoid bodies following a previously published protocol [20, 45], and the cellular morphology and expression of developmental markers confirmed the success of the embryonic body differentiation procedure (Figs. 6A, S4E). The morphology of the differentiated mES cells was similar between the untreated and BRD2-depleted cells (Fig. 6A). BRD2 depletion was efficient during EB differentiation (Fig. 6B). Western blot analyses with ES cell differentiation time course samples indicated that BRD2 protein level, but not H3K9me3 or H3K27ac protein levels, appeared to decrease gradually during embryoid body differentiation (Fig. 6C). We then performed BRD2 ChIP-Seq using untreated and 4 day differentiated (EB-Day-4) mES cells, both of which were not treated with IAA (Table S4). These ChIP-Seq datasets exhibited good correlations between replicates and validated many BRD2 binding sites that have been previously published [11] (Fig. S4A). Differential analyses of the BRD2 ChIP-Seq signals indicated that 421 BRD2 bindings were decreased, while 1559 BRD2 bindings were increased at gene promoters during embryoid body differentiation (Fig. 6D). The ChIP-Seq signals seem to be a little bit of shift to the x-axis. This trend is consistent with the decreased protein level of BRD2 during EB differentiation (Fig. 6C). We also performed BRD2 ChIP-MS on wild-type mESCs and mESCs differentiated into embryoid bodies for 4 days (EB-Day4 cells). The TFIID components, DHX9 and XRN2 were detected in both the wild-type and differentiated conditions. Proteins involved in chromatin remodeling, RNA metabolism, and Pol II regulation were preferentially detected in both mES cells and EB-Day-4 cells (Fig. S4B).
Fig. 6.
BRD2 associates with dynamic gene expression during ES cell differentiation. A Bright field BRD2-mES and embryonic body (EB) time course under untreated and IAA-treated conditions. The scale bar is 100 μm. B Western blot showing the degradation of BRD2 in response to IAA treatment before BRD2-mES differentiation into EBs. C Western blot showing the BRD2, H3K9me3, and H3K27ac protein levels during mESCs differentiation into EBs over time. D Scatter plot showing all detected BRD2 target sites with their differential binding affinities in day-4 EBs compared to mESCs from two replicates of untreated cells. Binding events around promoter regions identified using DiffBind (v3.11), were used for further analysis. If the closest transcription start site (TSS) contained one or more BRD2 peaks within a 2 kb window, then the gene was considered to be regulated for that particular peak. The red dots represent genes where BRD2 exhibited a large change in binding at a later stage of differentiation in day-4 EBs. E BRD2-induced gene expression was correlated with BRD2 binding. Heatmap depicting the expression levels of corresponding genes that exhibited altered BRD2 binding at promoters (increased or decreased). These plotted RNA-Seq data were derived from three biological replicates in untreated cells at four time points: mES, EB day 2, day 3, and day 4. Each row of the heatmap represents one gene. To facilitate the robust detection of gene expression signatures associated with differentiation and modulated by BRD2, we removed the unreliable variations and the nonexpressed genes across all the untreated samples. F Bubble plot showing the changed expression between untreated and BRD2-depleted cells and the statistical significance of representative gene sets related to Wnt signaling at different time points. Significantly affected Wnt signaling-related genes at the specific time points (label on the top of each box) were plotted in each box. The gene expression at other time points were also shown within the same box (labels at the bottom). The color and size of each circle indicate the expression fold change and significance level (− log10 p values) for each selected gene, respectively. G Genome browser track of ChIP-Seq signals of BRD2-mES and BRD2-EB-Day4 and RNA-Seq signals of BRD2-mES and BRD2-EB-Day4 under untreated and IAA-treated conditions at Wnt3 loci. H BRD2 and Pol II ChIP-qPCR analyses of Porcn, Wnt3 and Insig2 (negative control) promoters and intergenic regions (IN). The intergenic region serves as a negative control for ChIP-qPCR. Primers see Table S9. Error bars represent the SD of at least three technical replicates. p values were calculated using Student’s t test (ns: p > 0.05, *p < = 0.05, **p < = 0.01, ***p < = 0.001, ****p < = 0.0001). I GO enrichment analysis for upregulated genes and downregulated genes in mESCs, EB-Day2, EB-Day3 and EB-Day4 cells upon BRD2 depletion. Note: no enriched functions for the upregulated genes in EB-Day4 stage. The GO enrichment for the BRD2 binding changed genes during embryonic body differentiation overlapped with BRD2 affected genes in mESCs, or EB-Day4 cells were shown at the bottom right. GO enrichment analyses were performed by clusterProfiler
To further investigate the biological functions of BRD2 during ES cell differentiation, we differentiated mES cells into embryoid bodies for 2, 3, and 4 days and added IAA to deplete BRD2 proteins 24 h before the cells were collected for RNA-Seq analyses. At least 15.6 million reads were generated for each replicate, and three replicates were used for each condition. These RNA-Seq datasets were of good quality and were highly reproducible (Fig. S4C, Table S6). The differentially expressed genes between differentiated and BRD2-depleted cells 24 h after differentiation were identified. Genes with increased BRD2 binding shows upregulated gene expression, genes with decreased BRD2 binding shows downregulated gene expression (Fig. 6E). The differentially affected gene expression after BRD2 depletion in EB-Day4 cells indicated that the genes were enriched in Wnt signaling (Fig. 6F). ChIP-qPCR was used to validate two genes associated with Wnt signaling. The results showed that BRD2 decreased their binding signals after depletion, and Pol II also decreased the occupancy of the Wnt signaling genes both in embryonic stem cells and embryonic body differentiation after 4 days, and EB-Day4 appeared to show stronger decrease (Figs. 6G, H). Meanwhile, we also showed that the RNA-Seq signals increased at the Wnt signaling-related genes LRRK1 [46] and Shisa6 loci [47] (Fig. S4D), and the BRD2 chromatin binding increased after embryonic body differentiation for 4 days. These genes were downregulated after BRD2 depletion in EB-Day4 cells.
We then performed GO enrichment analysis for differentially expressed genes (DEGs) in mESCs, EB-Day2, EB-Day3, and EB-Day4 cells after BRD2 degradation (Fig. 6I). The results showed that the differentially expressed genes (DEGs) enriched in skeletal system morphogenesis and actin filament organization in mESCs; enriched in eye photoreceptor cell development and sodium ion transport in EB-Day2 cells; enriched in eye development and cell–cell adhesion in EB-Day3 cells; enriched in Wnt signaling pathway in EB-Day4 cells. We noted that genes with decreased expression in embryonic body differentiated cells were enriched in differentiation or development, while the genes with decreased expression in mESCs appeared not to be directly associated with the differentiation or development. In addition, the genes with binding changes during EB differentiation and differentially expressed in mES cells were enriched in the alcohol metabolic process etc. The genes with binding changes during EB differentiation and differentially expressed in EB-Day4 cells were enriched in eye development and limb morphogenesis. Additionally, mRNA expression levels of ES cell markers (Nanog and Sox2), mesoderm markers (T and Wnt3), and endoderm markers (Gata4 and Gata6) were affected by BRD2 depletion during embryoid body differentiation (Fig. S4E). These results suggest that BRD2 facilitates to express key genes related to the embryonic body differentiation process.
BRD2 regulates differential gene expression during ES cell differentiation with primed Pol II regulation at promoters
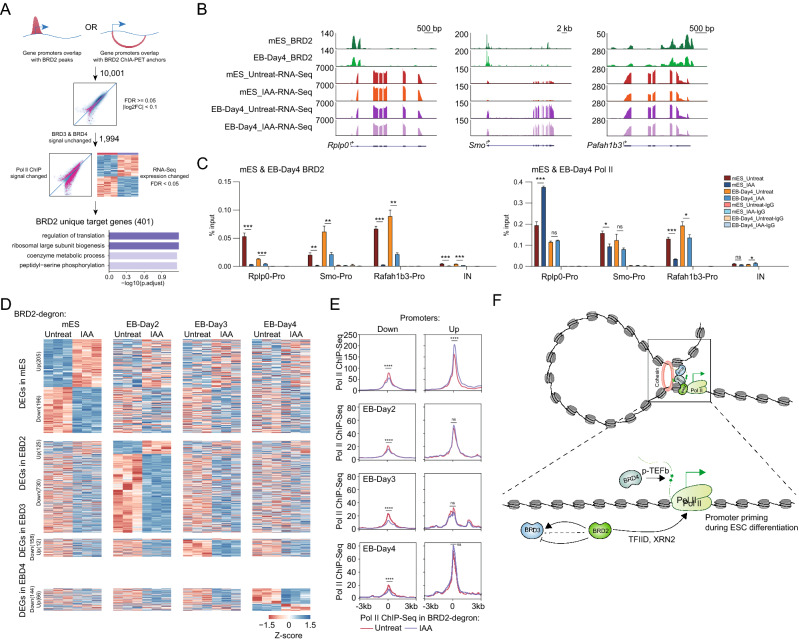
Given the complexity of the BET protein regarding chromatin binding dependence and function in Pol II regulation, we envisioned that it would be crucial to identify the unique targets of BRD2. The BRD2 ChIA-PET and ChIP-Seq datasets identified BRD2-bound genes in the three-dimensional genome. The BRD2 degron followed by Pol II ChIP-Seq and RNA-Seq datasets would help us identify genes that were functionally linked to BRD2. BRD2 depletion followed by BRD3 and BRD4 ChIP-Seq narrowed down the genes solely affected by BRD2. By further intersecting these gene lists, we identified unique BRD2 targets in mESCs (Fig. 7A, Table S7), and expression of these genes exhibited relatively less changes (Tables S6-7), as we used the FDR cutoff for the RNA-Seq gene expression changes for unique target genes to include more genes. GO analyses indicated that the BRD2 unique target genes identified in mESCs were enriched in the regulation of translation and metabolic processes (Fig. 7A). Interestingly, the unique BRD2 target genes in mESCs appeared to become less sensitive to BRD2 depletion during ES cell differentiation (Fig. 7B, Table S6), which were also supported by quantification and statistics analyses of RNA-Seq signals (Fig. S4F). We further validated unique BRD2 targets Rplp0 and Rafah1b3, and gene Smo in ES cells and EB-Day 4 cells using BRD2 and Pol II ChIP-qPCR. Consistent with the gene expression changes, Pol II binding changed in ESCs but little in EB-Day 4 cells (Fig. 7C). We also performed an additional 4-way Venn analysis of the significantly affected genes in mESC and EB-differentiated cells upon BRD2 degradation. There are not many overlaps between the DEGs in mESC and EB-differentiated cells upon BRD2 degradation (Fig. S4G). Thus, we concluded that BRD2 depletion affects the expression of different genes during embryonic body differentiation.
Fig. 7.
BRD2 regulates different gene expression during embryonic body differentiation but exhibits priming regulation of Pol II initiation in ESCs. A Schematic diagram showing the identification of genes regulated solely by BRD2. Genes were first selected if promoters were bound by BRD2 peaks or overlapped with BRD2 ChIA-PET anchors and then filtered by unchanged BRD3 and BRD4 ChIP-Seq signals, as well as changed Pol II and RNA-Seq signals upon BRD2 depletion to obtain a BRD2-uniquely-regulated gene list. GO enrichment terms of BRD2 unique gene targets were identified using the R package clusterProfiler with an adjusted p value cutoff of 0.05. B Genome browser track of ChIP-Seq signals of BRD2-mES and BRD2 EB-Day4 and RNA-Seq signals of BRD2-mES and BRD2 EB-Day4 under untreated and IAA-treated conditions at Rplp0, Smo and Pafah1b3 loci. C BRD2 and Pol II ChIP-qPCR analyses of Rplp0, Smo and Pafah1b3 promoters and intergenic regions in BRD2-mES and BRD2 EB-Day4. The intergenic region serves as the negative control for ChIP-qPCR. Primers see Table S9. Error bars represent the SD of at least three technical replicates. p values were calculated using Student’s t test (ns: p > 0.05, *p < = 0.05, **p < = 0.01, ***p < = 0.001, ****p < = 0.0001). D Heatmap showing expression changes in the differentially expressed genes (DEGs) in mES and EB at 2, 3, and 4 days. For mESCs, heatmap showing expression changes in the 401 unique BRD2 gene targets in response to BRD2 degradation in embryonic bodies differentiated for 2, 3 and 4 days. For differentiated cells, DEGs in that cell type were first identified using DESeq2 (FDR < 0.05, |log2FC|> 1) and then clustered using hierarchical clustering. Expression changes of those DEGs in the rest of the cell types were then plotted following the clustered order. The color bar represents the z score of RNA-Seq expression in each cell type. Three replicates were done for each next-generation sequencing experiment. E Meta plots of Pol II ChIP-Seq signal changes upon BRD2 depletion in mESCs. The promoters for the genes downregulated and upregulated after BRD2 depletion in the mES and EB 2, 3, and 4 day cells are shown as in (D). Significance was determined at ± 100 bp around TSS of the corresponding genes using Wilcoxon test (ns p > 0.05, ****p < = 0.0001). F Model of BRD2-mediated transcription regulation. The model indicates that BRD2, BRD3 and BRD4 co-occupy enhancers and promoters with RNA polymerase II. BRD2 facilitates Pol II initiation via TFIID and suppresses R-loop formation through XRN2 in mESCs. BRD2 primes the gene activation during ES cell differentiation. BRD3 functions additively, independently, or perhaps antagonistically with BRD2 in Pol II transcription. BRD4 mainly promotes Pol II elongation through p-TEFb
To gain further insights into BRD2-regulated gene expression in different cell states, we identified differentially affected genes after BRD2 depletion during ES cell differentiation using a stringent cutoff. The results showed that BRD2-affected genes in various differentiation stages also appeared to be perturbed in ESCs but at a lower magnitude (Fig. 7D). Interestingly, we also observed consistent Pol II occupancy decreases at the promoters of downregulated genes in ESCs, but less significant increase for the upregulated genes (Fig. 7E). This is probably because that the upregulated genes usually express higher level and they may be regulated through multiple mechanisms besides of BRD2-mediated promoter priming. Our results collectively suggest that BRD2 facilitates differentiated cell state control with primed roles in the enhancement of Pol II initiation at promoters in embryonic stem cells.
Discussion
Previous study has showed the initiation and elongation regulations of BRD2 [8], but the detailed molecular mechanism is unclear. Our work revealed that BRD2 facilitates TAF3 binding to genes containing low levels of H3K4me3 for Pol II initiation and connects XRN2 to genes containing high levels of H3K4me3 for suppression of R-loops. Previous study showed the complex relationship among BRD2, BRD3 and BRD4 [48, 49], we provide evidence to show that BRD2 and BRD3 function additively, independently, or perhaps antagonistically for Pol II transcription. We reported that BRD2 is dispensable for specific enhancer-promoter interactions and important for ES cell differentiation, which are also new (Fig. 7F). These findings were observed in mESCs. However, we believe that some of the effects may be generalizable. Our work also identified a unique regulatory circuit for BRD2 and revealed that BRD2 regulates specific gene expression during embryonic body differentiation, but BRD2 has already primed the initiation regulation of these gene promoters in embryonic stem cells.
BRD2/3/4 are known to recognize acetylated histones to regulate transcription [37, 50]. BRD4 has been documented to regulate Pol II pause release, and BRD2 and BRD3 are implicated in redundant function of Pol II initiation. Amino acid sequence conservation analyses for various bromodomain proteins revealed that BRD2 and BRD3 were clustered together with Bdf1 and Bdf2 in yeast and C-terminal regions of TAF1 for their bromodomains compared to other bromodomain factors (Fig. S5A). Conservation analyses for BRD2, BRD3 and BRD4 were performed, and the results showed that BRD2/3/4 bromodomains are conserved among themselves in humans, and BRD2 bromodomain is also conserved across species (Fig. S5B). Our chromatin binding and functional analyses between BRD2 and BRD3 revealed that they regulate Pol II transcription both additively and independently, which has been observed in previous studies with different biological systems [48, 49]. The molecular basis for BRD2- and BRD3-independent and additive functions at different gene promoters needs further investigation. We envisioned that this could be dependent on the levels of local histone acetylation, adjacent binding of other acetylation reader proteins, or co-occupied transcription factors. On the other hand, BRD2 contains intrinsically disordered regions, and may form phase separation-mediated condensates. These biological condensates may concentrate many transcription factors, chromatin regulators and cofactors, which may interfere BRD3 binding after BRD2 depletion. Besides, we also realized that the ChIP-Seq was performed after BRD2 depletion for 3 h, which may not be long enough to have more dramatic effects on BRD3 and BRD4. Several previous studies suggested that BRD2 and BRD4 recognized similar histone acetylation with variable affinities [36, 51]. However, the relationship between the sequence differences and their differential affinity to recognize various histone acetylation is still unclear. Importantly, characterizing the complex relationship between BRD2 and BRD3 would raise broad interest on the molecular basis for the specificity of different BET proteins and put forward higher requirements for the design of inhibitors for specific BET proteins.
Previous studies revealed that BRD2 regulates transcription in melanoma in an H2A.Z.2-dependent manner, and BRD2 regulates developmental poised gene transcription through H2A.Z.1 ubiquitylation [11, 12]. In addition, BRD2 has been implicated in regulating genome stability and CTCF insulator boundaries [17, 18]. Our BRD2 ChIP-Seq dataset, the ChIA-PET dataset, and other datasets generated from different laboratories and cell lines show that BRD2 is predominantly enriched at active promoters and enhancers, even though BRD2 occupies CTCF insulators in the specific cell type [11, 12, 17, 52–54]. BRD2 does not contribute to enhancer-promoter interactions for specific cell identity genes in mES cells, suggesting that coactivators are the accessory factors for executing particular activities in the pre-established chromatin structures.
Using luciferase reporters, previous reports have shown that serum stimulation induces the formation of the BRD2-E2F1-TBP complex for transcriptional activation [13, 16]. Recently, acute depletion of BRD2 followed by Pol II ChIP-Seq analyses showed that BRD2 depletion led to decreased Pol II signals at the transcription start sites and increased Pol II signals for a subset of genes [8]. However, the detailed molecular mechanisms underlying BRD2-mediated Pol II transcription are still unclear. In this study, we showed that BRD2 interacts with TBP and TAF3 and that BRD2 depletion decreases Pol II ChIP signals at transcription start sites and decreases chromatin binding of TBP and TAF3. These results collectively suggest that BRD2 is required for Pol II initiation at many gene promoters, which is consistent with the widespread roles of TFIID in Pol II transcription [55]. BRD2 inhibition increased R-loop signals, which caused DNA double strand breaks and genome instability [18]. We showed that BRD2 depletion increased R-loop signals as well as total and S2P Pol II ChIP signals at gene bodies, and decreased the chromatin binding of XRN2. These results indicate that BRD2 plays a role in Pol II elongation, likely through suppression of R-loops. Whether increased R-loop signals are the cause or consequence of Pol II elongation defects needs additional investigation. It would be informative to examine additional RNA helicases (i.e., Dhx9) for its relationship with BRD2. BRD2 was shown to be enriched in H2A.Z regions, and this histone variant was recently reported to regulate both Pol II initiation and elongation, and nucleosome unwrapping [11, 56, 57]. Thus, we also could not fully rule out the possibility of a direct role of BRD2 in transcription elongation.
Development involves controlling normal cell states, and diseases typically arise due to dysregulation of cell states, so understanding the control of cell states in the transcription level is essential for understanding development and disease [58–61]. Multiple mechanisms have been proposed for controlling cell states. For example, ELL3 and MLL4 prime the enhancer for gene activation during differentiation through the super elongation complex (SEC) or P300 [20, 22, 62, 63], TAF3 controls lineage-specific gene activation by DNA looping with CTCF [19], and LSD1 silences the active enhancer with the NuRD complex during ES cell differentiation [21]. In this study, we found that BRD2 interacts with the TFIID complex and appears to recruit RNA enzymes to suppress R-loops to control genes during cell differentiation. It would be interesting to perform single-cell gene expression analyses after BRD2 depletion to gain detailed information on cell lineages that were responsive to BRD2 depletion. The genes that responded to BRD2 depletion in differentiated cells also exhibited a Pol II response at their promoters after BRD2 depletion. This is unlikely to be caused by insufficient differentiation of mESCs, as the differentiation procedure was performed in the absence of 2i inhibitors and LIF, and the cellular morphology and expression of developmental markers also confirmed our EB differentiation worked. The finding that BRD2 primes the gene promoter’s in ES cells, while these genes would be regulated by BRD2 during embryonic stem cell differentiation, provides a foundation for understanding BET proteins in cell state control.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the members of the Ji laboratory for engaging in helpful discussions. We thank Dr. Richard Young for assistance during the beginning of the studies. We thank Dr. Mario Garcia-Dominguez for the BRD2 wild-type and mutant constructs. We thank the National Center for Protein Sciences at Peking University in Beijing, China, for assistance with high-resolution fluorescence imaging and LYD for help with flow cytometry sorting, DL for help with mass spectrometry detection, and CYS and HXL for help with cell imaging. We also thank Eric Spooner at the Whitehead Proteomics Core for mass spectrometry and Tom Volkert at the Whitehead Genome Technology Core for sequencing.
Author contributions
XJ conceived and supervised the project. CLW generated the degron mES cells and performed BRD2 degradation-related Western blots, 4C-Seq, ChIP-Seq/qPCR, RNA-Seq, differentiation-related analyses, and mass spectrometry analyses. QQX performed most of the analyses, including ChIP-Seq, RNA-Seq-related analyses and 4C-Seq, R-Loop, and conservation analyses. JH initially performed BRD2 ChIP-Seq and expression analyses during the embryonic body differentiation process. XHZ and LC generated the R-loop sequencing data. BA and DD helped with the bioinformatic analyses at the initial stage of the project. XJ performed BRD2 ChIA-PET experiments. All authors contributed to the data analyses and data interpretation. XJ wrote the manuscript with input from JH, CLW, QQX and help from the other authors.
Funding
This work was supported by funds from the Ministry of Science and Technology of China and the National Natural Science Foundation of China (Grants 2017YFA0506600, 31871309 and 32170569, respectively), the Qidong-SLS Innovation Fund, and grants from the Peking-Tsinghua Center for Life Sciences and the Key Laboratory of Cell Proliferation and Differentiation of the Ministry of Education at Peking University School of Life Sciences to X. J. In addition, J.H. is supported in part by the China Postdoctoral Science Foundation (2017M610700). L.C. is supported by Ministry of Science and Technology of China and the National Natural Science Foundation of China (Grants 2021YFA1100500, 32171289).
Data availability
All sequencing datasets generated in this study, including ChIA-PET, ChIP-Seq, RNA-Seq, and 4C-Seq data, have been deposited in GEO with accession GSE160557. Mass spectrometry data can be found in the PRIDE database: PXD030774.
Declarations
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chenlu Wang and Qiqin Xu contributed equally to this work.
Contributor Information
Jie Huang, Email: hjanime@163.com.
Xiong Ji, Email: xiongji@pku.edu.cn.
References
- 1.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell. 2017;67(5–18):e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 7.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 8.Zheng B, Aoi Y, Shah AP, Iwanaszko M, Das S, Rendleman EJ, Zha D, Khan N, Smith ER, Shilatifard A. Acute perturbation strategies in interrogating RNA polymerase II elongation factor function in gene expression. Genes Dev. 2021;35:273–285. doi: 10.1101/gad.346106.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stonestrom AJ, Hsu SC, Jahn KS, Huang P, Keller CA, Giardine BM, Kadauke S, Campbell AE, Evans P, Hardison RC, Blobel GA. Functions of BET proteins in erythroid gene expression. Blood. 2015;125:2825–2834. doi: 10.1182/blood-2014-10-607309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung KL, Zhang F, Jaganathan A, Sharma R, Zhang Q, Konuma T, Shen T, Lee JY, Ren C, Chen CH, et al. Distinct roles of Brd2 and Brd4 in potentiating the transcriptional program for Th17 Cell differentiation. Mol Cell. 2017;65:1068–1080.e1065. doi: 10.1016/j.molcel.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surface LE, Fields PA, Subramanian V, Behmer R, Udeshi N, Peach SE, Carr SA, Jaffe JD, Boyer LA. H2A.Z1 monoubiquitylation antagonizes BRD2 to maintain poised chromatin in ESCs. Cell Rep. 2016;14:1142–1155. doi: 10.1016/j.celrep.2015.12.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vardabasso C, Gaspar-Maia A, Hasson D, Punzeler S, Valle-Garcia D, Straub T, Keilhauer EC, Strub T, Dong J, Panda T, et al. Histone Variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol Cell. 2015;59:75–88. doi: 10.1016/j.molcel.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng J, Dong W, Chen L, Zou T, Qi Y, Liu Y. Brd2 is a TBP-associated protein and recruits TBP into E2F–1 transcriptional complex in response to serum stimulation. Mol Cell Biochem. 2007;294:45–54. doi: 10.1007/s11010-006-9223-6. [DOI] [PubMed] [Google Scholar]
- 14.Bagchi RA, Ferguson BS, Stratton MS, Hu T, Cavasin MA, Sun L, Lin YH, Liu D, Londono P, Song K, et al. HDAC11 suppresses the thermogenic program of adipose tissue via BRD2. JCI Insight. 2018 doi: 10.1172/jci.insight.120159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izumikawa K, Ishikawa H, Yoshikawa H, Fujiyama S, Watanabe A, Aburatani H, Tachikawa H, Hayano T, Miura Y, Isobe T, et al. LYAR potentiates rRNA synthesis by recruiting BRD2/4 and the MYST-type acetyltransferase KAT7 to rDNA. Nucleic Acids Res. 2019;47:10357–10372. doi: 10.1093/nar/gkz747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SC, Gilgenast TG, Bartman CR, Edwards CR, Stonestrom AJ, Huang P, Emerson DJ, Evans P, Werner MT, Keller CA, et al. The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol Cell. 2017;66(102–116):e107. doi: 10.1016/j.molcel.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, Refvik ST, Chiang CM, Xhemalce B, Paull TT, et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019;33:1751–1774. doi: 10.1101/gad.331231.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Lee JE, Lai B, Macfarlan TS, Xu S, Zhuang L, Liu C, Peng W, Ge K. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc Natl Acad Sci USA. 2016;113:11871–11876. doi: 10.1073/pnas.1606857113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013;152:144–156. doi: 10.1016/j.cell.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pal DK, Evgrafov OV, Tabares P, Zhang F, Durner M, Greenberg DA. BRD2 (RING3) is a probable major susceptibility gene for common juvenile myoclonic epilepsy. Am J Hum Genet. 2003;73:261–270. doi: 10.1086/377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velíšek L, Shang E, Velíšková J, Chachua T, Macchiarulo S, Maglakelidze G, Wolgemuth DJ, Greenberg DA. GABAergic neuron deficit as an idiopathic generalized epilepsy mechanism: the role of BRD2 haploinsufficiency in juvenile myoclonic epilepsy. PLoS ONE. 2011;6:e23656. doi: 10.1371/journal.pone.0023656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyuris A, Donovan DJ, Seymour KA, Lovasco LA, Smilowitz NR, Halperin AL, Klysik JE, Freiman RN. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim Biophys Acta. 2009;1789:413–421. doi: 10.1016/j.bbagrm.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2009;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Huang J, Lun K, Li B, Zheng H, Li Y, Zhou R, Duan W, Wang C, Feng Y, et al. Genome-wide analyses of chromatin interactions after the loss of Pol I, Pol II, and Pol III. Genome Biol. 2020;21:158. doi: 10.1186/s13059-020-02067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Liu Z, Jia J. Mechanisms of smoothened regulation in hedgehog signaling. Cells. 2021;10:2138. doi: 10.3390/cells10082138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng ZY, He TT, Gao XM, Zhao Y, Wang J. ZBTB transcription factors: key regulators of the development, differentiation and effector function of T cells. Front Immunol. 2021;12:713294. doi: 10.3389/fimmu.2021.713294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards DS, Maganti R, Tanksley JP, Luo J, Park JJH, Balkanska-Sinclair E, Ling J, Floyd SR. BRD4 prevents R-loop formation and transcription-replication conflicts by ensuring efficient transcription elongation. Cell Rep. 2020;32:108166. doi: 10.1016/j.celrep.2020.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty P, Huang JTJ, Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat Commun. 2018;9:4346. doi: 10.1038/s41467-018-06677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley DL. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021–1036. doi: 10.1016/j.cell.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Lambert JP, Picaud S, Fujisawa T, Hou H, Savitsky P, Uusküla-Reimand L, Gupta GD, Abdouni H, Lin ZY, Tucholska M, et al. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol Cell. 2019;73:621–638.e617. doi: 10.1016/j.molcel.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donczew R, Hahn S. BET family members Bdf1/2 modulate global transcription initiation and elongation in Saccharomyces cerevisiae. Elife. 2021 doi: 10.7554/eLife.69619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boija A, Klein IA, Young RA. Biomolecular condensates and cancer. Cancer Cell. 2021;39:174–192. doi: 10.1016/j.ccell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018 doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daneshvar K, Ardehali MB, Klein IA, Hsieh FK, Kratkiewicz AJ, Mahpour A, Cancelliere SOL, Zhou C, Cook BM, Li W, et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat Cell Biol. 2020;22:1211–1222. doi: 10.1038/s41556-020-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171(1573–1588):e1528. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schujiers J, Lee TI, Zhao K, Young RA. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA. 2012;109:15324–15329. doi: 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berwick DC, Harvey K. LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Hum Mol Genet. 2012;21:4966–4979. doi: 10.1093/hmg/dds342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokue M, Ikami K, Mizuno S, Takagi C, Miyagi A, Takada R, Noda C, Kitadate Y, Hara K, Mizuguchi H, et al. SHISA6 confers resistance to differentiation-promoting Wnt/β-catenin signaling in mouse spermatogenic stem cells. Stem Cell Reports. 2017;8:561–575. doi: 10.1016/j.stemcr.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrieu GP, Denis GV. BET proteins exhibit transcriptional and functional opposition in the epithelial-to-mesenchymal transition. Mol Cancer Res. 2018;16:580–586. doi: 10.1158/1541-7786.MCR-17-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Branigan GL, Olsen KS, Burda I, Haemmerle MW, Ho J, Venuto A, D’Antonio ND, Briggs IE, DiBenedetto AJ. Zebrafish paralogs brd2a and brd2b are needed for proper circulatory, excretory and central nervous system formation and act as genetic antagonists during development. J Dev Biol. 2021 doi: 10.3390/jdb9040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 51.Slaughter MJ, Shanle EK, Khan A, Chua KF, Hong T, Boxer LD, Allis CD, Josefowicz SZ, Garcia BA, Rothbart SB, et al. HDAC inhibition results in widespread alteration of the histone acetylation landscape and BRD4 targeting to gene bodies. Cell Rep. 2021;34:108638. doi: 10.1016/j.celrep.2020.108638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LeRoy G, Chepelev I, DiMaggio PA, Blanco MA, Zee BM, Zhao K, Garcia BA. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome Biol. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders L, Guenther MG, Qi J, Fan ZP, Marineau JJ, Rahl PB, Loven J, Sigova AA, Smith WB, Lee TI, et al. Genome-wide localization of small molecules. Nat Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warfield L, Ramachandran S, Baptista T, Devys D, Tora L, Hahn S. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol Cell. 2017;68(118–129):e115. doi: 10.1016/j.molcel.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mylonas C, Lee C, Auld AL, Cisse II, Boyer LA. A dual role for H2A.Z.1 in modulating the dynamics of RNA polymerase II initiation and elongation. Nat Struct Mol Biol. 2021;28:435–442. doi: 10.1038/s41594-021-00589-3. [DOI] [PubMed] [Google Scholar]
- 57.Wen Z, Zhang L, Ruan H, Li G. Histone variant H2A.Z regulates nucleosome unwrapping and CTCF binding in mouse ES cells. Nucleic Acids Res. 2020;48:5939–5952. doi: 10.1093/nar/gkaa360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bi M, Zhang Z, Jiang YZ, Xue P, Wang H, Lai Z, Fu X, De Angelis C, Gong Y, Gao Z, et al. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat Cell Biol. 2020;22:701–715. doi: 10.1038/s41556-020-0514-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu C, Li L, Zhang Z, Bi M, Wang H, Su W, Hernandez K, Liu P, Chen J, Chen M, et al. A non-canonical role of YAP/TEAD is required for activation of estrogen-regulated enhancers in breast cancer. Mol Cell. 2019;75(791–806):e798. doi: 10.1016/j.molcel.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jang Y, Park YK, Lee JE, Wan D, Tran N, Gavrilova O, Ge K. MED1 is a lipogenesis coactivator required for postnatal adipose expansion. Genes Dev. 2021;35:713–728. doi: 10.1101/gad.347583.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Y, Zhang L, Xiong C, Chen J, Wang L, Wen Z, Yu J, Chen P, Xu Y, Jin J, et al. HIRA complex presets transcriptional potential through coordinating depositions of the histone variants H3.3 and H2A.Z on the poised genes in mESCs. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park YK, Lee JE, Yan Z, McKernan K, O'Haren T, Wang W, Peng W, Ge K. Interplay of BAF and MLL4 promotes cell type-specific enhancer activation. Nat Commun. 2021;12:1630. doi: 10.1038/s41467-021-21893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 2017;45:6388–6403. doi: 10.1093/nar/gkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing datasets generated in this study, including ChIA-PET, ChIP-Seq, RNA-Seq, and 4C-Seq data, have been deposited in GEO with accession GSE160557. Mass spectrometry data can be found in the PRIDE database: PXD030774.