Abstract
Background and aims
Recent evidences highlight a role of the mitochondria calcium homeostasis in the development of colorectal cancer (CRC). To overcome treatment resistance, we aimed to evaluate the role of the mitochondrial sodium-calcium–lithium exchanger (NCLX) and its targeting in CRC. We also identified curcumin as a new inhibitor of NCLX.
Methods
We examined whether curcumin and pharmacological compounds induced the inhibition of NCLX-mediated mitochondrial calcium (mtCa2+) extrusion, the role of redox metabolism in this process. We evaluated their anti-tumorigenic activity in vitro and in a xenograft mouse model. We analyzed NCLX expression and associations with survival in The Cancer Genome Atlas (TCGA) dataset and in tissue microarrays from 381 patients with microsatellite instability (MSI)-driven CRC.
Results
In vitro, curcumin exerted strong anti-tumoral activity through its action on NCLX with mtCa2+ and reactive oxygen species overload associated with a mitochondrial membrane depolarization, leading to reduced ATP production and apoptosis. NCLX inhibition with pharmacological and molecular approaches reproduced the effects of curcumin. NCLX inhibitors decreased CRC tumor growth in vivo. Both transcriptomic analysis of TCGA dataset and immunohistochemical analysis of tissue microarrays demonstrated that higher NCLX expression was associated with MSI status, and for the first time, NCLX expression was significantly associated with recurrence-free survival.
Conclusions
Our findings highlight a novel anti-tumoral mechanism of curcumin through its action on NCLX and mitochondria calcium overload that could benefit for therapeutic schedule of patients with MSI CRC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04311-4.
Keywords: Calcium signaling, Mitochondria, NCLX, Curcumin, Colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and remains a major public health issue. CRC development is a multi-step process involving distinct stages: initiation, promotion and progression that ultimately generates phenotypically altered transformed malignant cells. Understanding the mechanisms that drive tumor hallmarks is critical for the prevention and treatment of CRC progression using drugs acting on these identified mechanisms. Based on the current state of knowledge, CRC develops mainly through a progressive accumulation of genetic alterations by three major mechanisms. The most common is chromosomal instability (CIN) in 75% of CRCs; the second most common is epigenetic modification of DNA methylation, also called the CpG island methylator phenotype (CIMP), in 20% of CRCs; the third most common is microsatellite instability (MSI) or deficiency of DNA-mismatch repair system (dMMR), which occurs in approximately 15% of CRCs [1]. MSI CRC cell lines are more resistant to 5-fluorouracil (5-FU) compared with microsatellite stable (MSS) cell lines [2]. Indeed, determination of the dMMR/MSI status in CRC has prognostic and therapeutic implications [3]. In addition, MSI is associated with a better prognosis in early stage CRC but a worse prognosis at advanced stages [4].
Several preclinical studies support the anti-tumoral properties of curcumin against CRC [5, 6]. Curcumin, also known as turmeric or diferuloylmethane, is a natural yellow-pigmented polyphenol extracted from the plant Curcuma longa. This compound is a highly pleiotropic molecule with numerous targets and mechanisms of action and modulates several pathways [5]. Curcumin is viewed as a potential CRC chemopreventive agent because of its effect on multiple tumor progression pathways [7]. Chemoprevention is a promising strategy for reducing CRC incidence. In CRC, the MMR system strongly influences the sensitivity of cells to curcumin. Specifically, the MMR system modulates curcumin sensitivity through induction of DNA-strand breaks and activation of the G2/M checkpoint [8]. In tumor cells, curcumin participates in mitochondria-dependent apoptotic processes [9]. Curcumin affects mitochondria-associated proteins, such as Bax, Bcl-2, and Bcl-xL, and reactive oxygen species (ROS) production [10]. Curcumin increases rat liver mitochondrial membrane permeability, resulting in swelling, loss of membrane potential, and inhibition of adenosine triphosphate (ATP) synthesis [11]. Recently, intracellular distribution of curcumin has been correlated with mitochondrial staining [12]. Notably, Jelinek and colleagues demonstrated a preferred association of curcumin with cardiolipins, lipids mainly localized in the inner mitochondrial membrane [12]. Despite an abundant literature concerning curcumin and its effects, the molecular mechanisms of action remain poorly described, in particular the link between mitochondrial redox status and mitochondria-associated redox and calcium (Ca2+) signaling mechanisms.
The interplay between Ca2+ and ROS signaling pathways is well established at several subcellular locations and has been implicated in many essential biological functions [13, 14]. Mitochondrial permeability transition pore (mPTP) opening is triggered by different pathological conditions such as Ca2+ overload and oxidative stress [15]. This opening leads to reduced ATP production and allows the release of pro-apoptotic mitochondrial components, namely apoptosis-inducing factor and cytochrome c. Another hallmark of mPTP-induced apoptosis is the loss of mitochondrial membrane potential. Several proteins participate in mitochondrial Ca2+ (mtCa2+) uptake, the mitochondrial calcium uniporter (MCU) and its known regulators, which currently include MICU1, MICU2, EMRE, MCUR1, and MCUb. The mitochondrial Na+/Ca2+/Li+ exchanger (NCLX) mostly mediates mtCa2+ extrusion [16]. Of note, NCLX has been underexplored as a potential therapeutic target pushing cancer cells to apoptosis [17, 18]. In CRC, we have recently shown that downregulation of NCLX results in mtCa2+ overload, increased mitochondrial ROS (mtROS), mitochondrial depolarisation and tumor shrinkage [19, 20]. The benzodiazepine-derived compound CGP37157, an inhibitor of NCLX, promotes mitochondrial damage and induces apoptosis [21].
In the present study, we examined whether curcumin induces a modification of mtCa2+ flux in association with redox metabolism in MSI CRC. Our results indicated that curcumin has a strong anti-tumoral effect through inhibiting NCLX: inhibition of G2/M cell cycle progression, mtCa2+ and ROS overload, mitochondrial membrane depolarization and activation of apoptosis by mPTP opening in MSI CRC cells. In a xenograft mouse model of CRC, NCLX inhibitors decreased CRC growth. In addition, mid-infrared (MIR) spectroscopy coupled with metabolomics gave useful metabolic signatures in terms of circulating biomarkers. Moreover, analysis of CRC in The Cancer Genome Atlas (TCGA) dataset showed that higher NCLX expression was associated with MSI status, and the expression of NCLX in tissue microarrays (TMA) of 302 samples was significantly associated with recurrence-free survival (RFS) in patients with MSI CRC.
Materials and methods
Study design, patient characteristics and survival analysis
From 2003 to 2015, all patients with MSI CRC classified by molecular testing from the cancer biology departments of Poitiers and Tours University Hospitals were included in the multi-center prospective COLOMIN cohort (Cohorte nationale des cancers colorectaux avec instabilité microsatellitaire). This study was designed in accordance with legal requirements and the Declaration of Helsinki, and was approved by the ethics committee of CHU Poitiers and CHRU Tours, France (Comité de protection des personnes Ouest III, no. DC-2008-565 and no. 2018-039).
The main patient characteristics (gender and age) and tumor characteristics (tumor site, Tumor Node Metastasis [TNM] stage, VELIPI criteria [vascular emboli, lymphatic invasion, or perineural invasion], tumor grade, tumor perforation, and initial bowel obstruction) were collected. Whether a case was germline (Lynch syndrome) or sporadic dMMR/MSI was determined as described previously [4]. Information regarding disease recurrences and deaths were collected.
Tumor DNA was extracted from all tumors using the KAPA Express Extract Kit (Roche, Basel, Switzerland). The MSI phenotype was assessed by analyzing microsatellite loci comprising five mononucleotide markers: BAT-25, BAT-26, NR21, NR24 and NR27 (Pentaplex panel) (4). MSI was defined by the presence of instability affecting at least three of the five markers. In the case of one or two markers with instability, a comparative analysis of normal colon and tumor DNA was performed.
RFS was defined as the time from the date of primary tumor curative surgery until the date of recurrence. Patients without recurrence were censored at the last follow-up. Kaplan–Meier survival curves were generated and groups were compared using the log-rank test. P < 0.05 was considered statistically significant. Survival analyses were performed in the Statview environment.
Tissue microarray and immunohistochemistry
Tissue microarray (TMA) construction: TMAs were constructed using formalin-fixed paraffin-embedded tissue samples. For each case, nine cores of 0.6-mm diameter (three in the tumor center, three in the invasive front and three in non-tumor tissue) were transferred from the selected areas to the recipient block, using a TMA workstation (Manual Tissue Arrayer MTA Booster, Alphelys, France). Serial 3-µm sections of the TMA blocks were used for immunohistochemistry. Every tenth section was stained with haematoxylin and eosin to check that the cores adequately represented diagnostic areas.
Immunohistochemistry: Slides were deparaffinised, rehydrated and heated in citrate buffer (pH 6) for antigen retrieval. After blocking endogenous peroxidase with 3% hydrogen peroxide, the primary antibodies were incubated. After incubation with the primary antibody NCLX (HPA040668, Sigma-Aldrich, Saint-Quentin-Fallavier, France) diluted 1:50 overnight at 4 °C or Ki67 (DakoCytomation, Glostrup, Denmark) incubated 30 min at dilution 1:50, immunohistochemistry was performed using the streptavidin–biotin–peroxidase method with diaminobenzidine as the chromogen (Kit LSAB, Dakocytomation, Glostrup, Denmark). Negative controls were obtained after omission of the primary antibody or incubation with an irrelevant antibody.
A semi-quantitative score was assigned based on the intensity of NCLX staining: 0 (no staining), + (moderate staining) and ++ (strong staining). Ki67-positive cells were expressed as a percentage of total cancer cells.
Cell culture and treatments
HCT116 (RRID:CVCL_0291), LoVo (RRID:CVCL_0399) and HT-29 (RRID:CVCL_0320) SW48 (RRID:CVCL_1724) cell lines were purchased from ATCC (Manassas, VA, USA) and cultured in McCoy's medium (Gibco, Thermo Fisher, Illkirch, France) supplemented with 10% fetal bovine serum (FBS, Eurobio, Les Ulis, France), without antibiotics. NCM356 cells (Incell Corporation, LLC, San Antonio, TX, USA) were cultured in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) (Sigma-Aldrich) supplemented with 10% FBS and antibiotics. The cell lines were cultivated at 37 °C in a humidified incubator with 5% CO2. The cell lines were maintained and Mycoplasma absence was tested regularly using the PlasmoTest kit (HEK blue, Invivogen, Toulouse, France). HCT116 NCLX KO cells are the same cells described in Pathak et al., 2020 [19]. NCLX KO cells were generated using a guide RNA (g1) which resulted in a single cut at nucleotide 150 in exon one causing a frameshift mutation and introduction of a stop codon at position 180 in the NCLX open reading frame.
Transcriptomic analysis
The Cancer Genome Atlas (TCGA) dataset of patients with CRC was downloaded from cbioportal47 and analyzed as described previously [48, 49]. Briefly, clinical and transcriptomic data were acquired and processed using, respectively, the TCGABiolinks and edgeR packages in the R environment. Previously published classifications of the TCGA dataset of patients with CRC were obtained from Guinney et al. [28] and Liu et al. [29].
Gene Set Enrichment Analysis (GSEA) was performed on patients with MSI-h CRC from the TCGA transcriptomic dataset using the GSEA JAVA application from the Broad Institute with default parameters (http://software.broadinstitute.org/gsea).
Compounds
Curcumin (10 µM) was purchased from TOCRIS (Bristol, UK); CGP37157 (10 µM) was purchased from Sigma-Aldrich (Merck, Darmstadt, Germany); ITH12575 (10 µM) was purchased from Sigma-Aldrich.
Transfection
Briefly, 2.5 × 105 cells/well were plated in 6-well plates in McCoy’s medium supplemented with 10% FBS without antibiotics. The cells were incubated with a mix of small interfering RNA (siRNA) and lipofectamine in medium without serum for 6 h. After incubation, an equal volume of medium with serum was added to each well. The siRNA sequences directed against NCLX were purchased from Dharmacon (L-007332-00-0005) (La Fayette, CO, USA). Measurement of NCLX WT and NCLX S468T on cell viability in HCT116 cells was achieved by transient over-expression of NCLX WT and NCLX S468T [50] as previously described [50]. Briefly, 1 × 106 cells were incubated with a solution containing 1.5 μg of NCLX WT or NCLX S468Tand 3 μl Lipofectamine 2000 for 3 h. One day later, transfected cells were transferred and treated with 10 µM curcumin for 24 h in 24-well plates.
Reverse transcription and real-time polymerase chain reaction
Total RNA was collected using the Nucleospin RNA Kit (Macherey–Nagel, Hoerdt, France) and transcribed into complementary DNA (cDNA) with the GoScript Reverse Transcription System (Promega, Madison, WI, USA). cDNA was then amplified with the SYBR Green Master kit (Roche, Mannheim, Germany) using a Light Cycler 480 apparatus. PCR was performed in 40 cycles of 15 s at 95 °C and 45 s at 60 °C. The average ΔCt value was calculated for each cell line with respect to the housekeeping gene HPRT1. The primers sequences used are:
HPRT1: forward 5′ CAT-TAT-GCT-GAG-GAT-TTG-GAA-AGG 3′ reverse 5′ CTT-GAG-CAC-ACA-GAG-GGC-TAC-A 3′;
NCLX: forward 5′ GCC-AGC-ATT-TGT-GTC-CAT-TT 3′, reverse 5′AAT-TCG-TCT-CGG-CCA-CTT-AC 3′;
MCU: forward 5′ CGC-CAG-GAA-TAT-GTT-TAT-CCA 3′, reverse 5′ CTT-GTA-ATG-GGT-CTC-TCA-GTC-TGT-T 3′;
MICU1: forward 5′GAG-GCA-GCT-CAA-GAA-GCA-CT 3′, reverse 5′ CAA-ACA-CCA-CAT-CAC-ACA-CG 3′;
MICU2: forward 5′ GGC-AGT-TTT-ACA-GTC-TCC-GC 3′, reverse 5′ AAG-AGG-AAG-TCT-CGT-GGT-GTC 3′;
STIM1: forward 5′ GCC-CTC-AAC-ATA-GAC-CCC-AG 3′, reverse 5′ TCC-ATG-TCA-TCC-ACG-TCG-TCA 3′;
STIM2 forward 5′ TTG-GAC-CCT-TGA-AGA-CAC-TCT 3′, reverse 5′ CCA-GTT-ATG-AGG-TGG-GCG-TG 3′;
Orai1 5′ AGG-TGA-TGA-GCC-TCA-ACG-AG 3′, reverse 5′ CTG-ATC-ATG-AGC-GCA-AAC-AG 3′;
TRPC1 5′ TTA-CTT-GCA-CAA-GCC-CGG-AA 3′, reverse 5′ CTG-CTG-GCA-GTT-AGA-CTG-GG 3′.
Semi-quantitative real-time polymerase chain reaction analyses (redox profiling)
Semi-quantitative real-time polymerase chain reaction (RT-qPCR) analyses were performed as described previously by Kouzi et al. [51]. Briefly, HCT116 cell line was treated for 24 h with 10 µM curcumin, and RNA was isolated using the Maxwell® 16 IVD system and the Maxwell Simply RNA Tissue Kit according to the manufacturer’s instructions (Promega). RNA was reverse transcribed using the SuperScript® VILO cDNA Synthesis Kit (Invitrogen, Waltham, MA, USA) and RT-qPCR was performed using the Universal Probe Library System (Roche, Penzberg, Germany). cDNA, primers, probes and TaqMan master mix were mixed, and PCR was run in a LightCycler® 480 (Roche). The mean cycle threshold (Ct) of the human ACTB and RPL13A genes were used as endogenous controls to normalize the expression of target genes. Each reaction was performed in triplicate. Relative quantification (RQ) gene expression was evaluated by the 2−ΔΔCt method [52]. The list of primers is provided in Supplementary Table S1.
Flow cytometry analysis
Cell cycle
Cells were harvested using trypsin-ethylenediaminetetraacetic acid (EDTA, Thermo Fisher Scientific, San Jose, CA, USA) 24 h after treatments, washed in 1X phosphate-buffered saline (PBS), fixed in cold 70% ethanol and incubated at − 20 °C for at least 2 h. Subsequently, the samples were resuspended in PBS with RNase and stained with 0.025 mg/mL propidium iodide (PI) for 30 min in the dark at room temperature. The DNA content of stained cells was analyzed using a Gallios flow cytometer (Beckman Coulter, Villepinte, France). For each sample, a minimum of 5 × 104 cells were evaluated. Analyses were done using the Kaluza 1.3 software (Beckman Coulter).
Apoptosis
Cells were harvested by Accutase (Thermo Fisher Scientific, San Jose, CA, USA), washed with 1X PBS and then resuspended in annexin-binding buffer following the manufacturer’s instructions (Annexin-PI kit, Thermo Fisher Scientific, San Jose, CA, USA) and analyzed by flow cytometry as described above.
Spheroid cell viability test
The spheroid cell viability test was performed according to the manufacturer’s conditions (Cultrex proliferation—cell viability 3510096K, Bio-Techne, Abingdon, UK). The cells were cultured in extracellular matrix supplemented with Epidermal Growth Factor (EGF) in 96-well plates with round bottoms for 48 h and then treated with 10 µM curcumin for 72 h. To capture the entire spheroid, image fields starting at the center of the well were collected using a 20 × objective lens (Nikon, Champigny-sur-Marne, France).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells were cultured and treated with 1–10 µM curcumin for 24, 48 or 72 h. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg/ml) was added to each well prior to incubation. The cells were incubated at 37 °C for 45 min. Afterward, the supernatant was removed and dimethyl sulphoxide (DMSO) was added to dissolve formazan crystals and absorbance was read at 570 nm using a Mithras LB 940 Multimode Microplate Reader (Berthold, Bad Wildbad, Germany).
Sulforhodamine B (SRB) assay
Cells were cultured and treated with 1–10 µM curcumin for 24, 48, or 72 h.
Cell viability, survival in HBSS and short-term toxicity were evaluated using standard sulforhodamine B (SRB) method after 72 h, 48 h and 24 h treatments, respectively. Briefly, cells were fixed with 50% trichloroacetic acid for 1 h at 4 °C and stained for 15 min with 0.4% SRB solution. Cells were washed 3 times with 1% acetic acid and dye was dissolved with 10 mM trisbase solution over 10 min. Absorbance at 540 nm was read using BioTek Spectrophotometer.
NanoLuc activity assay
The HIF1α NanoLuc protein-reporter HCT116 and HCT116 NFE2L2-C-terminal Luciferase cell lines were purchased from Horizon Discovery (Cambridge, UK) [53]. Cells were seeded in 96-well plates at 3 × 104 cells/well and incubated for 24 h in normoxic or hypoxic conditions (1% O2) with or without 10 µM curcumin or 10 µM CGP37157. Luciferase activity was measured using the Nano-Glo® Reagent (Nano-Glo® Luciferase Assay, Promega) following the supplier’s instruction. Luminescence intensity values were measured by FlexStation-3 (Molecular Devices, San José, CA, USA).
Store-operated calcium-entry measurement by Fura-2 AM
Intracellular Ca2+ imaging was performed as previously described [22, 23]. Cells were plated in 96-well plates at 2 × 104 cells/well 24 h before the experiment. Adherent cells were for loaded with the ratiometric dye Fura2-acetoxymethyl ester (AM; 5 μM) for 45 min at 37 °C and then washed with PBS supplemented with Ca2+. During the experiment, the cells were incubated with Ca2+-free physiologic saline solution (PSS) solution and treated with 2 µM thapsigargin (TG, T7458, Life Technologies, Thermo Fisher) to deplete intracellular store of Ca2+. Ca2+ entry was stimulated by injection of 2 mM of CaCl2. Fluorescence emission was measured at 510 nm using the FlexStation-3 (Molecular Devices) with excitation at 340 and 380 nm. The maximum fluorescence (peak of Ca2+ influx [F340/F380]) was measured and compared with the normal condition. NCLX was inhibited by 10 µM CGP37157 or 10 µM curcumin.
Mitochondrial Ca2+ measurements
Intracellular mitochondria Ca2+ imaging was performed as previously described [19]. To measure mtCa2+ using the Rhod-2 AM dye (543 nm/580–650 nm), the cells were cultured at 60–80% confluency in 96-well plates. The cells were washed with medium without FBS and antibiotic–antimycotic agents. Then, the cells were incubated in medium containing 3 µM Rhod-2 AM (without FBS and antibiotic–antimycotic agents) at 37 °C for 45 min. The cells were washed and kept in PBS (HEPES-buffered saline solution, 140 mM NaCl, 1.13 mM MgCl2, 4.7 mM KCl, 2 mM CaCl2, 10 mM D-glucose, and 10 mM HEPES, adjusted to pH 7.4 with NaOH) containing 2 mM CaCl2 for imaging. The cells were stimulated with 100 µM adenosine triphosphate (ATP) or 10 µM histamine in PBS containing 2 mM CaCl2. The timelapse images were acquired using the FlexStation-3 (Molecular Devices). To measure mtCa2+ using the mt-Cepia, cultured cells were transfected using Lipofectamine 2000 (Invitrogen) with 1,5 µg pCMV CEPIA3mt 2 days before imaging. 24 h before the experiments, transfected cells were plated in 96-well plates. The cells were stimulated with 100 µM adenosine triphosphate (ATP) in PBS containing 2 mM CaCl2. The timelapse images were acquired using the FlexStation-3 (Molecular Devices). N-methyl-d-glucamine (NMDG) was purchased from Sigma-Aldrich. PCMV CEPIA3mt was a gift from Masamitsu Lino (Addgene plasmid # 58219; http://n2t.net/addgene:58219; RRID:Addgene_58219).
Calcium Green-5N (Invitrogen) on permeabilized cells beforehand with digitonin was used to measure mitochondrial Ca2+ uptake and release experiments.
HCT116 permeabilized cells suspended in intracellular Na+-free buffer (130 mM KCl, 10 mM Tris-MOPS (pH 7.4), 10 μM EGTA-Tris, 1 mM KPi, 5 mM malate, 5 mM glutamic acid, 1 μM Ca2+ green-5N). At the starting point, Curcumin or CGP37157 was added following by one pulse of 10 µM Ca2+ at 2 min. Next, 0.5 μM Rutenium Red (RR), an inhibitor of MCU, was added at 6 min to block Ca2+ uptake. Next, the Na+-induced Ca2+ release was initiated by the addition of a Na+ pulse (10 mM at 7 min). Ca2+ uptake/release measurements were performed at 25 °C on a F2710 spectrofluorometer (Hitachi) with the following parameters, λex = 505 nm, λem = 535 nm, slit width: 2.5 nm.
Mitochondrial and cytoplasmic reactive oxygen species measurements
Cells were cultured in 6-well plates at 70–90% confluency. To measure mitochondrial reactive oxygen species (mtROS), 1 × 106 cells were stained with 5 µM MitoSOX (Molecular Probes, Thermo Fisher) in PBS at 37 °C for 40 min [19]. The intensity of staining was measured using a Gallios flow cytometer (Beckman Coulter) and analyzed with the FlowJo software (Tree Star, Ashland, OR, USA).
Cytoplasmic ROS production was measured using the 2′,7′-dichlorofluorescein diacetate (DCFDA) assay according to the manufacturer’s protocol (Molecular Probes, Thermo Fisher). Cells were seeded at 2 × 103 cells/well of 96-well plates 24 h before the experiment. The cells were washed with PSS containing 2 mM Ca2+ and then treated with PSS containing 2 mM Ca2+ and 10 µM DCFDA with or without curcumin or CGP37157. The fluorescence emission was measured at 520 nm using the FlexStation-3 (excitation: 500 nm). The fluorescence rate is reported as relative fluorescence units (RFU) normalised to time 0.
Measurement of mitochondrial permeability transition pore opening
Direct assessment of mitochondrial permeability transition pore (mPTP) opening in HCT116 cells was performed by loading cells with calcein-AM (Sigma-Aldrich) and CoCl2, resulting in mitochondrial localisation of calcein fluorescence [54]. Specifically, the cells were loaded with 1 μM calcein-AM for 30 min at 37 °C in 1 mL (PSS 2 mM Ca2+, pH 7.4) with 1 mM CoCl2. Subsequently, they were incubated with medium free of calcein-AM and CoCl2 and then incubated in reoxygenation medium. mPTP opening was determined from the reduction in mitochondrial calcein signal (slope value) after injection of ionomycin (5 μM). Fluorescence emission was measured at 515 nm using the FlexStation-3. Calcium Green-5 N (Invitrogen) on permeabilized cells beforehand with digitonin was used to measure the Ca2+ retention capacity that reflects mitochondrial function. Ca2+ uptake was followed by measuring extra-mitochondrial Calcium Green-5 N until mPTP opening was achieved [55]. Fluorescence emission was measured at 530 nm.
Measurement of mitochondrial potential
Cells were cultured in 96-well plates at 50–60% confluency. The next day, cells were loaded with Tetramethyl rhodamine (TMRE) dye. The cells were stained with 100 nM TMRE dye in complete growth media and kept at 37 °C in 5% CO2 for 20–30 min. CCCP (50 µM) was used as a positive control [56]. The intensity of staining was measured and analyzed on FlexStation-3 (Molecular Devices).
Patch clamp recording
Traditional patch-clamp electrophysiological recordings were carried out using an Axopatch 200B and Digidata 1440A (Molecular Devices, San José, CA, USA) as previously published [23]. Pipettes were pulled from borosilicate glass capillaries (World Precision Instruments, Sarasota, FL,USA) with a P-1000 Flaming/Brown micropipette puller (Sutter Instrument Company, Novato, CA, USA) and polished using DMF1000 (World Precision Instruments). The resistance of filled pipettes was 2–4 MΩ. Under the whole-cell configuration, only cells with < 8 MΩ series resistance and tight seals (> 16 GΩ) were chosen for recordings. The cells were maintained at a + 30 mV holding potential during experiments. The Clampfit 10.1 software was used for data analysis. The solutions used for this procedure were:
Bath solution: 115 mM sodium methanesulfonate, 10 mM CsCl, 1.2 mM MgSO4, 10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 20 mM CaCl2 and 10 mM glucose (pH adjusted to 7.4 with NaOH);
Pipette solution: 115 mM caesium methanesulfonate, 10 mM caesium BAPTA (Tetrasodium 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate), 5 mM CaCl2, 8 mM MgCl2 and 10 mM HEPES (pH adjusted to 7.2 with CsOH). Based on the Maxchelator software (http://maxchelator.stanford.edu/), the calculated free Ca2+ was 150 nM.
Calcium release-activated channel (CRAC) currents were recorded in the ORAI1 knockout (KO) HEK293 cells expressing 4 µg EYFP-STIM1 along with 1 µg Orai1-CFP. The transfected cells were incubated with or without 10 µM curcumin for 20 min before patch-clamp recording.
Identifying mitochondria–endoplasmic reticulum contact sites using bioluminescence resonance energy transfer
Bioluminescence resonance energy transfer (BRET) assays were carried out as described previously [26]. Briefly, 5–10 × 103 HCT116 cells containing the BRET-based biosensor named MERLIN (Mitochondria-ER Length Indicative Nanosensor) were seeded into a white 96-well plate (Greiner Bio-One, Kremsmünster, Austria) for 48 h. Next, cells were incubated with CGDP for 3 h or transfected with control/NCLX small interfering RNA (siRNA) (30–50 nM) for 48 h. Then, cells were incubated with 5 μM coelenterazine h (Promega, Madison, WI, USA) in phosphate-buffered saline (PBS) for 5 min in the dark. Subsequently, BRET1 measurements were carried out in an INFINITE M PLEX plate reader (Tecan, Switzerland) at room temperature, measuring Renilla luciferase 8 (Rluc8) and mVenus emissions. In every experiment, Rluc8-L1-B33C (donor of the BRET pair) alone was transfected in HCT-116 cells 16 h prior to BRET measurement. The BRET signal was calculated as the acceptor emission relative to the donor emission and corrected by subtracting the background acceptor/donor ratio value detected when RLuc8-L1-B33C is expressed alone.
SeaHorse analysis
The cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) data were obtained using a Seahorse™ XF96 Flux Analyzer from Seahorse Bioscience (Agilent Technologies, Santa Clara, CA, USA) as we described previously [57]. The experiments were performed according to the manufacturer’s instructions. Briefly, HCT116 cells were seeded in XF96 cell culture plates at 2 × 104 cells/well, and then, cells were treated with 10 µM curcumin or CPG37157 for 12 h. On the day of analysis, the culture medium was replaced with XF Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, San Jose, CA, USA) supplemented with 2 mM glutamine and lacking bicarbonate (pH 7.4). The cells were then incubated at 37 °C in a non-CO2 incubator for 1 h and measurements were performed as described in the relevant figure legends. Sequential injection of 10 mM glucose, 1 µM oligomycin, 100 µM dinitrophenol (DNP) and 0.5 µM rotenone/antimycin A permitted the determination of the main respiratory and glycolytic parameters. Finally, the data were normalised to the amount of DNA present in the cells and assayed using Cyquant® Cell Proliferation Assay kit (Thermo Fisher Scientific, San Jose, CA, USA). The data were acquired with the Seahorse Wave Controller and analyzed with the Seahorse Wave Desktop Software.
OmniLog analysis
Metabolic profiling was studied using the OmniLog® Phenotype Microarray™ system (Biolog, Hayward, CA, USA) to evaluate the cell’s ability to metabolise 367 different carbon and energy substrates [51]. According to the manufacturer’s instructions, HCT116 cells were cultured at 2 × 104 cells/well for 24 h in the presence of 10 µM curcumin in PM-M1, PM-M2, PM-M3 and PM-M4 plates (Biolog) in a phenol red-free RPMI-1640-based medium depleted of carbon energy sources (IFM1 medium, Biolog) supplemented with 0.3 mM glutamine, 5% fetal calf serum, 100 U/mL penicillin and 100 μg/mL streptomycin. Following the incubation, 10 µL Redox Dye Mix MB (Biolog, Hayward, CA, USA) was added. Kinetically, tetrazolium reduction was measured at 37 °C over 6 h with the OmniLog® automated incubator-reader. Data are acquired with the Data Collection 2.4 software and analyzed with the PM-M Kinetic and PM-M Parametric software programs. The area under the curve (AUC) of each metabolite consumption was determined using the opm package in the R environment after correction of the background signal with the negative control wells.
Subcutaneous colorectal cancer xenograft
CRC xenografts were established by injecting 5 × 106 tumor cells per mouse. Swiss nude mice were randomized regarding their tumoral volume into treatment groups (n = 8–9/group) after the mean tumor volume reached ∼200 mm3. Mice were given saline with vehicle (DMSO/EtOH) MWF, curcumin at 150 mg/kg per dose (MWF) or CGP37157 at 150 mg/kg (2 times a week). Curcumin, ITH12575 and CGP37157 solutions were prepared at 10 mM in a DMSO/ethanol mixture (60/40). A dilution in a saline solution was carried out to obtain the final desired concentration. All treatments were administered intraperitoneally (IP). Tumors were measured twice each week with calipers and tumor volumes were calculated using the formula (4/3*π* (length/2)*((width/2)*(width/2)). All mice were assessed weekly using whole-body bioluminescent (BLI) imaging to quantify relative amounts of tumor burden. Each mouse was given 150 mg/kg body-weight of D-luciferin potassium salt (Interchim) and anesthetized with ketamine/xylazine. BLI images were acquired 3–5 min after injection and collected in real time until a saturation plateau was reached in the lateral, ventral and dorsal positions. Photons count emitted by the tumor was performed by a photon imager system. Regions of interest were drawn around the tumor and metastases. Tumor burden was quantified as the relative amount of photons produced from the luciferase activity in cells and expressed in cpm using Photovision + software (version 1.3; Biospace Lab). Authorisation (Apafis 19933) was given by the regional ethical committee CEEA—003 (Campus CNRS, Orléans) and MESRI (French Ministry of Higher Education, Research and Innovation).
Mathematical models of tumor growth
The change in tumor volume over time was described using a generalized logistic model as previously published [58]. This model included three parameters: first-order growth rate constant (kgrowth), maximum tumor volume (Vmax) and the power coefficient (γ). These parameters were estimated using nonlinear mixed-effects modeling. This approach aims at quantifying the inter-subject distribution of each model parameter in the population, by estimating their mean and inter-subject standard deviation, as well as quantifying the association of factors of variability (referred to as covariates) and the inter-subject distribution of model parameters. We tested the influence of compounds (CGP37157, ITH12575 and curcumin) as covariates of Vmax, because its inter-subject standard deviation was the only one that could be estimated. This influence was tested using the likelihood ratio test (LRT). This analysis was performed using MonolixSuite2019 (Lixoft®, Antony, France).
Mid-infrared spectroscopy
Total blood collected from the xenograft mouse model was analyzed at D0 (as the beginning of the experiment) and D40 (as 40 days after treatment began) by MIR spectroscopy using a Lumos microspectrophotometer (Bruker, Billerica, MA, USA). Acquisitions were made in transmission mode on a ZnSe multi-plate. Spectrum pre-treatments with smoothed second derivatives and vector normalization were operated. Statistical treatments were performed in the R environment. Statistical models were built with two goals in mind. The first objective was to estimate a decision rule that takes a spectrum as input and returns the probability that the sample belongs to a given group. The second objective was to return a signature of the treatment: a set of wavelengths that characterize the difference between the two compared groups.
The Lasso penalized logistic regression was used, because it allows one to deal with both of the aforementioned goals at once; in particular, it is a powerful method for variable selection [59]. This method is implemented in the glmnet package in the R environment. A k-fold cross-validation function was run to select the penalisation constant that controls the sparsity of the estimator.
Successively, the day and the treatment variable were entered as the dependent variable, Y, in the logistic regression model and coded as 1 for the target group and 0 for the others.
To assess predictive performance, the dataset is randomly divided into a training set and a validation set. If the total effect is large enough, Monte Carlo cross-validation is used with 30 repetitions to obtain 30 sets of misclassification errors [60]. When the total number of samples was smaller than 20, the leave-one-out approaches performed and leads to one estimated error per observation [61].
We fitted the logistic LASSO regression using the training dataset only and predicted the groups of the test data using the fitted models. Then, classification errors were computed for the test dataset. The cut-off value of the probability for classification, which was needed for calculating the test misclassification errors, was set at 0.5.
The MIR spectral variable assignment (cm−1) was as follows: 3320, λCH stretching in lipids; 3220, λOH carboxyl groups; 3100–3050, amide B band of proteins; 2813, lipid band tail (aliphatic chains λCH2/CH3); 1750, λC=O esterified lipids (cholesterol, triglycerides and phospholipids); 1600–1680, amide I band of proteins (sensitive to secondary structure); 1520–1580, amide II band of proteins; 1450, fat deformation λCH; 1380, lipid λCH3 bending; 1350, fat deformation CH3; 980, alcohol function CO; and 900, aromatic rings.
Metabolomic analysis
Liquid chromatography–high-resolution mass spectrometry (LC–HRMS) was performed after an extraction with 400 µL of methanol from 25 µL of plasma. An Ultimate WPS-3000 UPLC system (Dionex, Germany) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) and operated in positive and negative electrospray ionization modes (ESI + and ESI-, respectively; separate analysis for each ionization mode) was used for this analysis. Liquid chromatography was performed using a Phenomenex Kinetex 1.7 µm XB—C18 column (150 mm × 2.10 mm) maintained at 55 °C. Two mobile phase gradients were used. The gradient was maintained at a flow rate of 0.4 mL/min over a runtime of 20 min. Two different columns were used to increase the metabolic coverage. Accordingly, a hydrophilic interaction liquid chromatography (HILIC) column (150 mm × 2.10 mm, 100 Å) was also used. During the full-scan acquisition, which ranged from 58 to 870 m/z, the instrument operated at 70,000 resolution (m/z = 200).
As required for all biological analyses, the pre-analytical and analytical steps of the experiment were validated based on the findings of quality control (QC) samples (mix of all the samples analyzed). Coefficients of variation (CV% = [standard deviation/mean] × 100) were calculated from all metabolite data. If a metabolite had a CV > 30% in QC samples, it was excluded from the final dataset. QC samples were analyzed at the beginning of the run, every 13 samples and at the end of the run.
The samples were subjected to a targeted analysis, based on a library of standard compounds (Mass Spectroscopy Metabolite Library [MSML®] of standards, IROA Technologies, Ann Arbor, MI, USA). The following criteria were followed to identify the metabolites: (1) retention time of the detected metabolite within ± 20 s of the standard reference; (2) exact measured molecular mass of the metabolite within a range of 10 ppm around the known molecular mass of the reference compound; and (3) correspondence between isotopic ratios of the metabolite and the standard reference [62, 63]. The signal value was calculated using the Xcalibur® software (Thermo Fisher Scientific, San Jose, CA, USA) by integrating the chromatographic peak area corresponding to the selected metabolite.
Univariate analysis
The univariate analysis of metabolite levels between groups was realized using the non-parametric Mann–Whitney test in the XLSTAT software (Addinsoft, Paris, France). P < 0.05 was considered significant.
Multivariate analysis
Multivariate analyses were performed as described previously [62, 63] using the SIMCA P + 13.0 software (Umetrics, Umea, Sweden), which included a principal component analysis (PCA) and an orthogonal partial least-squares discriminant analysis (OPLS-DA). The data analysis was preceded by unit variance scaling. The quality of the model was described by the cumulative modeled variation in the X matrix R2 X, the cumulative modeled variation in the Y matrix R2 Y and the cross-validated predictive ability Q2 values.
To evaluate the significance of the created model, cross-validation analysis of variance (CV-ANOVA) was applied. The discriminant metabolites named variable importance in projection (VIP) obtained from OPLS-DA were all considered as responsible for the differences between samples.
Pathway analysis
Metabolites of VIP obtained from the final OPLS-DA model were introduced in the pathway analysis module in the MetaboAnalyst platform.
Quantification and statistical analysis
The data are presented as the mean ± standard deviation (SD) or standard error of the mean (SEM) and were analyzed using GraphPad 7 (GraphPad Software, La Jolla, CA, USA). In the box plot, the box represents the 25th–75th interquartile range, the midline in the box represents the median and the solid square box represents the mean. To test single variables between two groups, a paired Student’s t test or Mann–Whitney test was performed. One-way ANOVA followed by post hoc Tukey’s test was used for multiple comparisons. P < 0.05 was considered to be significant.
Results
Curcumin inhibited proliferation and induced cell cycle arrest of colorectal cancer cells
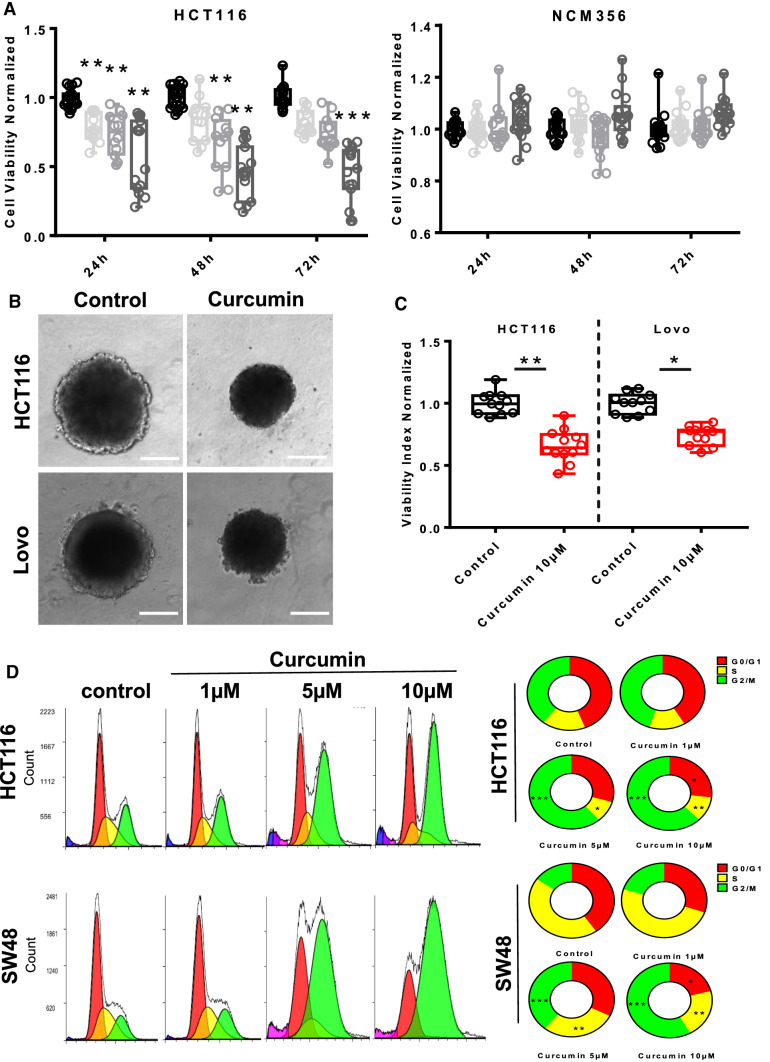
We investigated the role of curcumin in regulating the biology of CRC cells. Curcumin (0–10 µM) dose-dependently reduced the viability of these cells based on the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (Fig. 1A) and the Sulforhodamine B (SRB) assay (Supplementary Fig. 1A). Moreover, 10 µM curcumin decreased the proliferation of HCT116 cells, a CRC cell line, for up to 72 h (Fig. 1A, left). This pattern was identical in two other MSI cell lines, Lovo and SW48 cells (Supplementary Fig. 1B). Interestingly, curcumin had no effect on the proliferation of a non-tumorigenic epithelial colorectal cell line, NCM356 (Fig. 1A right). We next examined whether curcumin affected the proliferation of CRC cells by using the spheroid three-dimensional (3D) assay. There was a significant reduction in HCT116 and Lovo multicellular spheroid volume with respect to the control group after administration of 10 µM curcumin for 48 h (Fig. 1B) confirmed by the measure of the cell viability index (Fig. 1C). Cell-cycle analysis revealed that curcumin induced a large, dose-dependent increase in the percentage of HCT116 and SW48 cells in the G2/M phase (Fig. 1D), suggesting that curcumin can reduce cell proliferation by preventing mitosis.
Fig. 1.
Curcumin exerts anti-proliferative effects on human CRC cells by inducing G2/M cell cycle arrest. A NCM356 and HCT116 were incubated with various concentrations of curcumin (0–10 µM) for 24, 48 or 72 h, and cell viability was measured using the MTT or SRB cell viability assay (supplementary data Fig. 1). (n = 3–5 independent experiments; ANOVA followed by Tukey’s post hoc multiple comparisons, **P < 0.01 and ***P < 0.001). B Lovo and HCT116 3D spheroids were exposed to 10 µM curcumin for 72 h. The area of spheroids after incubation with 10 µM curcumin (72 h) was measured using ImageJ software. C The cell viability index was evaluated by measuring resofulvin reduction in HCT116 and Lovo cells (n = 5 independent experiments; Mann–Whitney test, *P < 0.05, **P < 0.01). D Curcumin triggered G2/M cell cycle arrest in CRC cells (n = 5 independent experiments; ANOVA followed by Tukey’ post hoc multiple comparisons, **P < 0.01 and ***P < 0.001)
Curcumin inhibited mitochondrial Ca2+ extrusion and induced reactive oxygen species production with mitochondrial membrane potential depolarization
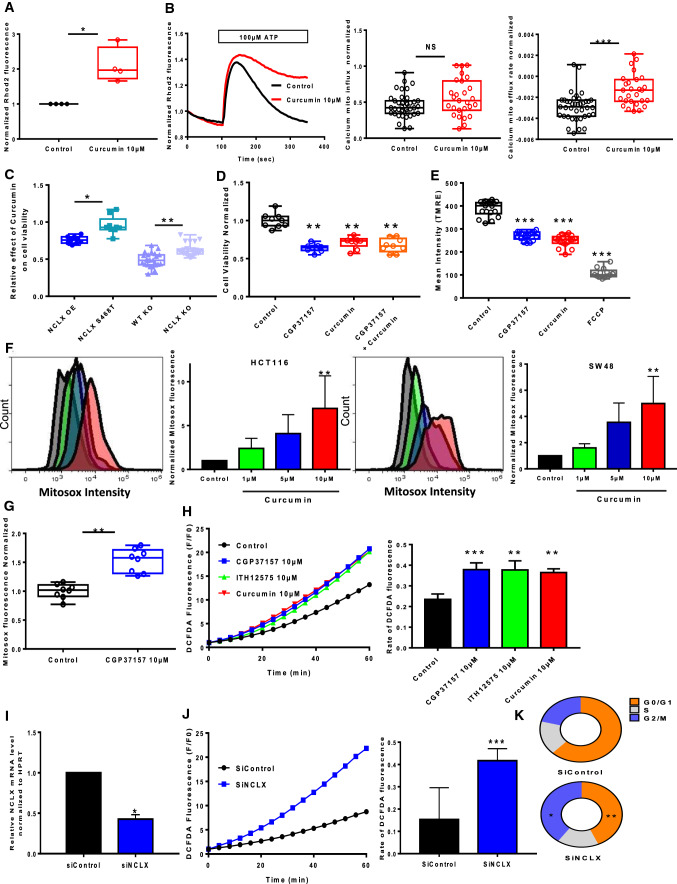
Cell proliferation is controlled by intracellular Ca2+ and ROS, with mitochondria acting as a Ca2+ buffer and ROS generators [14]. We measured mtCa2+ in HCT116 and SW48 cells by loading the cells with Rhod-2AM, an mtCa2+-sensitive dye. Curcumin treatment for 1 h induced a sharp increase in the mtCa2+ concentration under basal conditions (Fig. 2A, Supplementary Fig. 2A). Cells were then stimulated with ATP, a purinergic G protein-coupled receptor (P2Y) agonist that is coupled to phospholipase Cβ activation and subsequent inositol-1,4,5-trisphosphate (IP3) production. IP3 binds to IP3 receptors (IP3R) and then mediates Ca2+ transfer from the endoplasmic reticulum (ER) to the mitochondria. Curcumin had no effect or moderate effect on mtCa2+ uptake, but inhibited mtCa2+ efflux after stimulation with 100 µM ATP on HCT116 cells or with 10 µM histamine on SW48 (Fig. 2B, Supplementary Fig. 2B). Recently, we have shown that NCLX is the most important actor of mtCa2+ extrusion in CRC cells and that inhibiting NCLX (SLC8B1) expression and function prevents mtCa2+ extrusion without affecting mtCa2+ uptake (Supplementary Fig. 2C, D)[19]. To confirm these effects on NCLX, we performed mtCa2+ uptake and release experiments Na+ dependent on permeabilized HCT116 cells (Supplementary Fig. 2E, F). Permeabilized HCT116 cells suspended in intracellular Na+-free buffer. As expected, a slow Ca2+ extrusion was observed after Na+ injection in CGP37157 and curcumin-treated cells without additive effect. The knockdown of either NCLX function (through the over-expression of the S486T mutant, described as a dominant-negative) or expression significantly reduced the effect of curcumin on cell viability (Fig. 2C). Interestingly, the pharmacological inhibition of NCLX, using CGP37157, in addition to curcumin during 24 h did not reduce the cell viability when compared to either curcumin or CGP37157 alone, suggesting that both compounds are acting on the same mechanisms or the same target (Fig. 2D). Altogether, these suggest that curcumin effect on cell viability is mediated by an effect on the NCLX-dependent mtCa2+ efflux. Through the modulation of Na+ influx and Ca2+ efflux, NCLX directly acts on the mitochondrial potential [19]. Here, we observed that both curcumin and CGP37157 induce a depolarization in HCT116 as visualized through the measure of the fluorescence of the mitochondrial potential reporter dye tetramethylrhodamine methyl ester (TMRE). Interestingly, the addition of curcumin and CGP37157 during 1 h does not produce additional depolarization, arguing that both drugs modulate mitochondrial potential through NCLX (Fig. 2E with TMRE).
Fig. 2.
mtCa2+ efflux and ROS production are altered by treatment with curcumin and NCLX targeting in CRC cells. A Global mtCa2+ was evaluated with Rhod2 fluorescence in HCT116 cells treated or not with curcumin (n = 4 independent experiments; Mann–Whitney test, *P < 0.05). B mtCa2+ responses following ATP stimulation (100 μM) were measured in HCT116 cells treated or not with curcumin. The average mtCa2+ influx (peak) (left) and the rates of mtCa2+ efflux (right) are shown (n = 5 independent experiments with 27–39 measurements; Mann–Whitney test, ***P < 0.001). C The knockdown of either NCLX function (through the over-expression of the S486T mutant, described a dominant-negative) or expression significantly reduced the effect of curcumin on cell viability (n = 4 independent experiments of 9 measurements per group for overexpressed NCLX or NCLX S468T *P < 0.05, n = 5 independent experiments of 24 measurements per group for WT KO or NCLX KO cells, **P < 0.01). D CGP37157 and curcumin had no additive effect on cell viability (MTT assay). The results are expressed as the mean ± SD of 3–5 independent experiments (ANOVA followed by Tukey’s post hoc multiple comparisons, *P < 0.05, **P < 0.01). E Mitochondrial membrane potential using the dye TMRE in HCT116 cells treated with 10 μM CGP37157 or 10 μM curcumin or 50 µM FCCP (n = 6, Mann–Whitney test, ***P < 0.01). F HCT116 and SW48 cells were pre-treated with curcumin (1, 5 or 10 μM) for 2 h. The cells were stained with MitoSOX for 40 min. The bar graph presents a quantitative analysis of mtROS. The data represent the mean ± SD (n = 4 independent experiments, **P < 0.01, ANOVA followed by Tukey’s post hoc multiple comparisons). G Effect of CGP37157 pre-treatment on the mtROS level in HCT116. The data are shown as the mean ± SEM of at least three separate experiment with eight measurements each (n = 8, Mann–Whitney test, **P < 0.01). H The effects of curcumin and NCLX blockers (CGP37157 or ITH12575) on cytosolic ROS responses in HCT116 cells were analyzed. DCFDA fluorescent traces are shown as the normalised (F/F0) mean. The histograms show the slope of cytosolic ROS production as a function of time (n = 5 independent experiments with 31–56 measurements per group **P < 0.01, ***P < 0.001; ANOVA followed by Tukey’s post hoc multiple comparisons). I RT-qPCR data showing SLC8B1 mRNA relative to HPRT in HCT116 cells transfected with siRNA against NCLX (n = 4 independent experiments, Mann–Whitney test, *p < 0.05). J Effect of siRNA-mediated NCLX inhibition on cytosolic ROS production evaluated in HCT116 (n = 4 independent experiments with 16–32 measurements per group, Mann–Whiney test, **P < 0.01). K Effect of siRNA against NCLX on cell cycle of HCT116. The results are representative of five independent experiments n = 5 independent experiments; ANOVA followed by Tukey’ post hoc multiple comparisons, **P < 0.01 and ***P < 0.001)
A further consequence of mtCa2+ overload is the generation of mtROS. Using mitoSOX dye to monitor the generation of mtROS, we observed that curcumin induces a dose-dependent increase of mtROS in HCT-116 and SW48 cell lines (Fig. 2F and Supplementary Fig. 2C). 5-FU treatment and pharmacological inhibition of NCLX with CGP37157 also increased mtROS in a same range as curcumin (Fig. 2F and Supplementary Fig. 3A).
All treatments for 1 h including curcumin, pharmacological inhibitors of NCLX (CGP37157 or ITH12575) and downregulation of NCLX with siRNA not only increased mtROS but also enhanced cytosolic ROS demonstrated by the measure of DCFDA (Fig. 2G–J). Here, we observed that the silencing of NCLX expression can also impair cell cycle similarly to curcumin (Fig. 2K). Overall, our results indicate that curcumin may regulate mtCa2+ efflux and ROS production through the inhibition of NCLX.
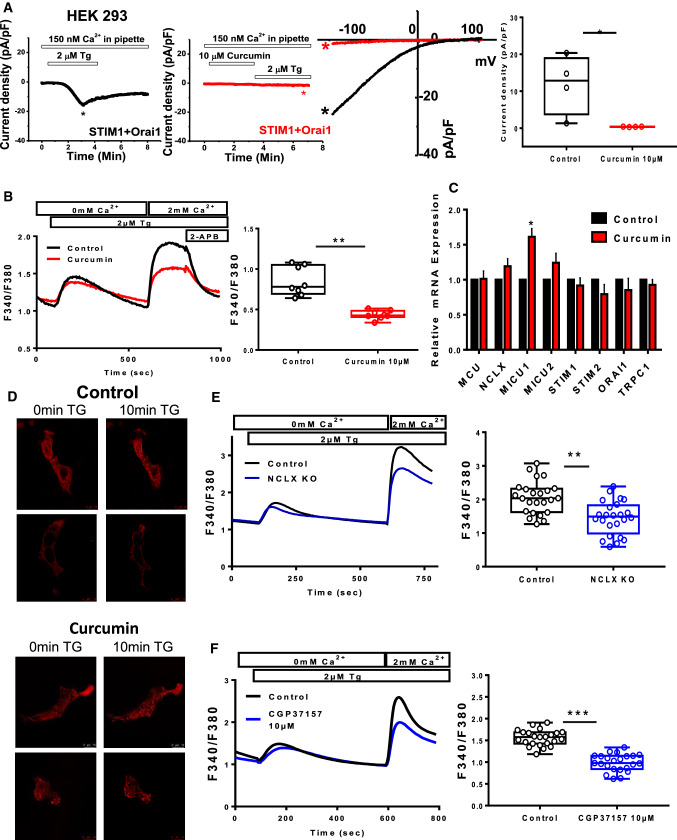
Curcumin and NCLX inhibition decreased store-operated calcium entry
In non-excitable cells, the intracellular Ca2+ concentration is controlled by store-operated calcium entry (SOCE) through CRAC, which are encoded by Orai proteins located at the plasma membrane of CRC cells. This influx is important for several cellular processes, including proliferation, migration and invasion [22]. NCLX inhibition through siRNA knockdown in CRC cells and in HEK293 cells leads to the inhibition of plasma membrane Orai1 channels and reduces SOCE [19, 23]. Here, using the whole-cell patch-clamp technique, we observed that curcumin abolished the Orai1-mediated ICRAC currents induced by Thapsigargin (Tg) in an heterologous expression system co-expressing STIM1 and Orai-1 (Fig. 3A). In addition, curcumin pre-treatment for 1 h reduced SOCE in HCT116 cells (Fig. 3B). To identify the mechanism involved in regulation of SOCE, the messenger RNA (mRNA) levels of the Ca2+ handling proteins MCU, NCLX, MICU1, MICU2, STIM1, STIM2, Orai1, and TRPC1, only MICU1 was significantly increased after 24 h treatment with curcumin, suggesting that its effect is not mediated by transcriptional mechanisms (Fig. 3C). To determine whether curcumin-dependent changes in SOCE and CRAC are mediated by a change in STIM1 oligomerisation, we monitored the formation of STIM1‐RFP puncta in HCT116 cells after Tg-induced ER Ca2+ depletion. Indeed, oligomerisation of STIM1 coupled with ER Ca2+ depletion is necessary for CRAC activation. Numerous puncta were formed following Tg stimulation and curcumin had no significant effect (Fig. 3D). Interestingly, this reduced Ca2+ influx induced by curcumin was also observed in NCLX KO cells (Fig. 3E) or after CGP37157 treatment (Fig. 3F). As expected, molecular silencing and pharmacological inhibition of NCLX reduced Ca2+ influx in a similar manner than curcumin.
Fig. 3.
NCLX inhibition and curcumin treatment decrease SOCE in CRC cells. A Electrophysiological recordings were performed on HEK293 ORAI1 KO cells transfected with ORAI1-CFP and STIM1-YFP. Representative time courses of whole‐cell CRAC currents activated by dialysis of 20 mM BAPTA through the patch pipette and taken at − 100 mV from control cells (black trace) or cells pre-treated with 10 µM curcumin (red trace) are shown. Representative I–V relationships are taken from traces in (A) which are indicated by color‐coded asterisks. Statistical analysis of Ca2+ CRAC currents measured at − 100 mV is shown (n = 4 independent experiments; Mann–Whitney test, *P < 0.05). B Representative traces of SOCE in HCT116 cells pre-treated with curcumin (10 µM for 1 h). The data show the effects of curcumin on SOCE in HCT116 (n = 5 independent experiments; Mann–Whitney test, **P < 0.01). C Relative mRNA levels of MCU, NCLX, MICU1, MICU2, STIM1, STIM2, ORAI1, and TRPC1 genes in HCT116 cells treated or not with 10 µM curcumin for 24 h (n = 5 independent experiments; Mann–Whitney test, *P < 0.05). D/ STIM1-dependent puncta formation a. Representative fluorescent images of HCT116 cells co-expressing STIM1 Cherry (red) showing puncta after treatment with TG (2 µM for 10 min) and pre-treated or not with curcumin for 30 min. E Representative cytosolic Ca2+ measurements in HCT116 WT and NCLX KO cells measured with Fura-2 AM in response to 2 µM TG applied first in nominally Ca2+-free external solution and subsequently with 2 mM external Ca2+. The summary data are presented as the mean ± SD (n = 5 independent experiments, 24 cells for each group; Mann–Whitney test, **P < 0.01). F SOCE was blocked after preincubation with CGP37157 (10 μM for 1 h) (right graph). Ca2+ influx in control and CGP37157-treated cells is shown. The summary data are presented as the mean ± SEM (n = 4 independent experiments, 23 cells for each group; Mann–Whitney test, ***P < 0.001)
Curcumin and NCLX inhibition altered colorectal cancer metabolism
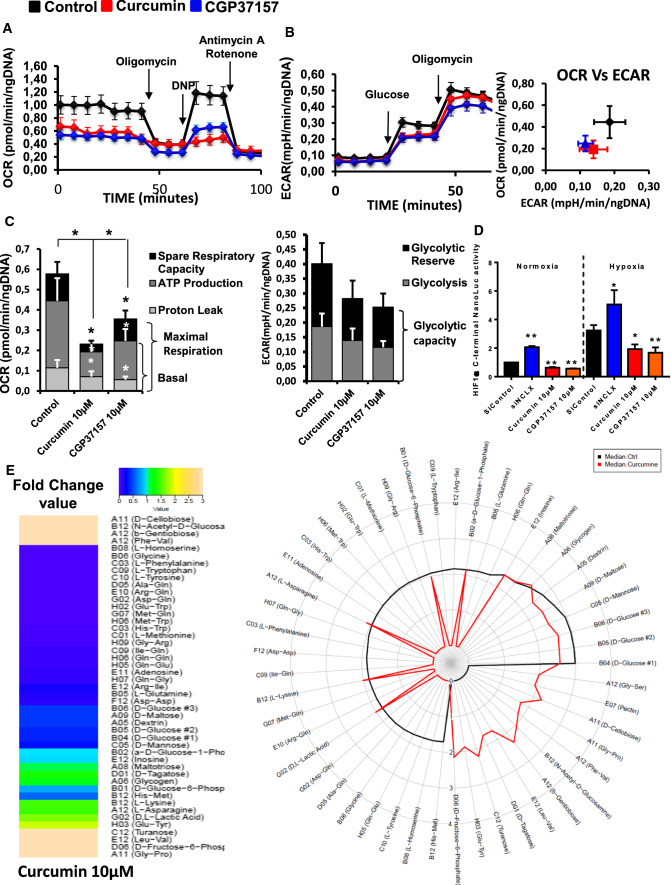
Cancer cell proliferation and mitosis are highly dependent of the cellular energy produced by mitochondria through mitochondrial oxidative phosphorylation [24]. mtCa2+ and mtROS are key players in the control of mitochondrial metabolism and ATP production. Because mtCa2+ regulates three TCA cycle dehydrogenases, we hypothesized that by targeting NCLX, curcumin or CGP37157 affects mtROS and mtCa2+ to limit cellular bioenergetics. To test this hypothesis, we determined the effects of these compounds on mitochondrial respiration (OXPHOS) and aerobic glycolysis (Fig. 4A–C). Both curcumin and CGP37157 treatment reduced the intact cellular oxygen consumption rate (OCR) in HCT116 cells; this change was associated with a reduction in basal respiration, maximal respiration, proton leak and ATP production (Fig. 4C, left). By contrast, curcumin and CGP37157 had no significant effects on glycolysis, glycolytic capacity and glycolytic reserve (Fig. 4C, right). Hypoxia significantly increased the expression of HIF1α compared with normoxia. Under both conditions, NCLX inhibition by curcumin or CGP37157 significantly reduced the activity of HIF1α (Fig. 4D). Surprisingly, we found an opposite effect in cells transfected with siRNA against NCLX. In this condition, the reduction in NCLX increased HIF1α in both normoxic and hypoxic conditions (Fig. 4D). Pathak et al. [19] showed that inhibition of NCLX expression with either short hairpin (shRNA) or CRISPR/Cas9 knockout enhances HIF1a activation and increases the ECAR of HCT116 cells with an increase in glycolysis. Altogether, these data suggest that curcumin may affect cancer cell viability by its action on NCLX and impairing mitochondrial respiration.
Fig. 4.
Targeting NCLX-dependent signaling elicits rapid and dynamic changes in CRC cell metabolism. A Energy metabolism in HCT116 cells pre-treated with curcumin and NCLX blocker (CGP37157). After four measurements of baseline OCR, glucose, oligomycin, DNP, and the mixture of antimycin A and rotenone were injected sequentially, with measurements of OCR recorded after each injection. ATP-linked OCR and OCR due to proton leak can be calculated using basal and oligomycin-sensitive rates. Injection of DNP is used to determine the maximal respiratory capacity. Injection of antimycin A and rotenone allows for the measurement of OCR independent of complex IV. B Cellular aerobic glycolysis was evaluated by measures of the ECAR from HCT116 cells treated with curcumin or CGP37157 (10 µM) following sequential injection (arrow) of glucose (10 mM) and then oligomycin (1 μmol/L) (n = 5 independent experiments; Mann–Whitney test, ***P < 0.001). C The spare respiratory capacity (mitochondrial reserve capacity) was calculated by subtracting the basal from the maximal respiration. The bio-energetic profiles of HCT116 with or without 10 µM curcumin or 10 µM CGP37157 were measured using sequential injection of glucose, oligomycin (1 μmol/L), DNP (100 μmol/L) and the mixture of antimycin A (0.5 μmol/L) and rotenone (0.5 μmol/L). The mitochondrial reserve capacity of untreated control and curcumin- or CGP37157-treated HCT116 cells are shown. The results are presented as the mean ± SD of 5 independent experiments of 3–5 measurements (Mann–Whitney test, *P < 0.05, ***P < 0.001). D HCT116 HIF1α-C-terminal Luc cells incubated under normoxic or hypoxic conditions for 48 h. Four hours before the end of incubation, the cells were treated with CGP37157 or curcumin. The values represent the mean ± SD of 3–6 independent experiments (ANOVA followed by Tukey’s post hoc multiple comparisons, *P < 0.05 **P < 0.01). E The effect of curcumin on the ability of HCT116 cells to metabolise 367 substrates was measured using the OmniLog® Analyzer. The heatmap shows the AUC fold-change data induced by curcumin treatment. The radial plot shows the AUC of metabolized substrates (49 substrates with positives kinetic curves with AUC > 50 in at least one condition) on a logarithmic scale
Previous transcriptomic analysis showed that glycolysis-related genes are upregulated in HCT116 NCLX KO cells [19]. To measure the impact of curcumin on the remodeling of the metabolism, we performed real-time quantitative PCR on a set of 76 genes associated with metabolism. Confirming the bio-energetic results, curcumin treatment had a moderate effect on glycolysis-related genes or antioxidant genes (Supplementary Fig. 4). In terms of energy metabolism, there was a significant decrease in the expression of Glut1 (SLC2A1) and Glut4 (SLC2A4) transporters, as well as hexokinase 2, associated with an increase in glucokinase regulator (GCKR) expression and a decrease in PFKFB1 and B4 (glycolysis activator) expression. These changes favor an overall decrease in carbohydrate metabolism. We also observed a decrease in MCT2 (lactate import) and lactate dehydrogenase A transcripts. Transcript quantification showed an increase in MCT4 (lactate export) and lactate dehydrogenase C associated with the decrease in PDHB (pyruvate dehydrogenase E1 complex or PDC-E1) and a drop in PDP1 (PDC activator) as well as PDC inhibitors (Supplementary Fig. 4). Taken together, these results strongly suggest that curcumin decreases the use of pyruvate for oxidative phosphorylation and increases the lactate efflux. However, in the context of complexes such as PDC, we cannot make assumptions about enzymatic activities. With regard to oxidative metabolism, curcumin induced a decrease in GLRX5 and PRDX3 expression and an increase in the GPX family of enzymes (Supplementary Fig. 4). However, these genes were poorly expressed, suggesting that they would have a moderate effect on ROS levels. Finally, curcumin increased the expression of nuclear factor (erythroid-derived 2)-like 2 (NRF2) target genes (Supplementary Fig. 4). This phenomenon could explain the presence of oxidative stress. NRF2 expression was enhanced following curcumin treatment in normoxic conditions and following curcumin or siNCLX treatment in hypoxic conditions (Supplementary Fig. 3B). However, these treatments did not affect the activity of NRF2 induced by its nuclear translocation (Supplementary Fig. 3C).
To better characterize the metabolic effects of curcumin, we used the OmniLog® Phenotype MicroArray, which allows to generate a map of the metabolism phenotype of cancer cells. Treating HCT116 cells with curcumin blocked the metabolism of l-glutamine, l-tryptophan, l-methionine, adenosine, l-phenylalanine, glycine, l-tyrosine, l-homoserine and different dipeptides (Fig. 4E). By contrast, b-gentiobiose, d-cellobiose, turanose, d-tagatose, d-fructose-6-phosphate, N-acetyl-d-glucosamine pectin and different dipeptides were only used by HCT116 cells treated with curcumin (Fig. 4E). In addition, 14 substrates were found in both curcumin-treated and non-treated HCT116 cells. Finally, curcumin induced a decrease in the use of major substrates, both in terms of biological importance and level of use, such as d-glucose (− 60%), d-mannose (− 69%), d-maltose (− 58%), dextrin (− 55%) and d-glucose-6-phosphate (− 20%), whereas there was an increase in the consumption of substrates such as glycogen (+ 21%), l-asparagine (+ 46%), l-lysine (+ 48%) and d- and l-lactic acid (+ 68%).
Taken together, these results suggest that NCLX inhibition with curcumin suppressed mitochondrial respiration, amino acid metabolism and HIF1α-dependent reduction in glycolytic substrate consumption.
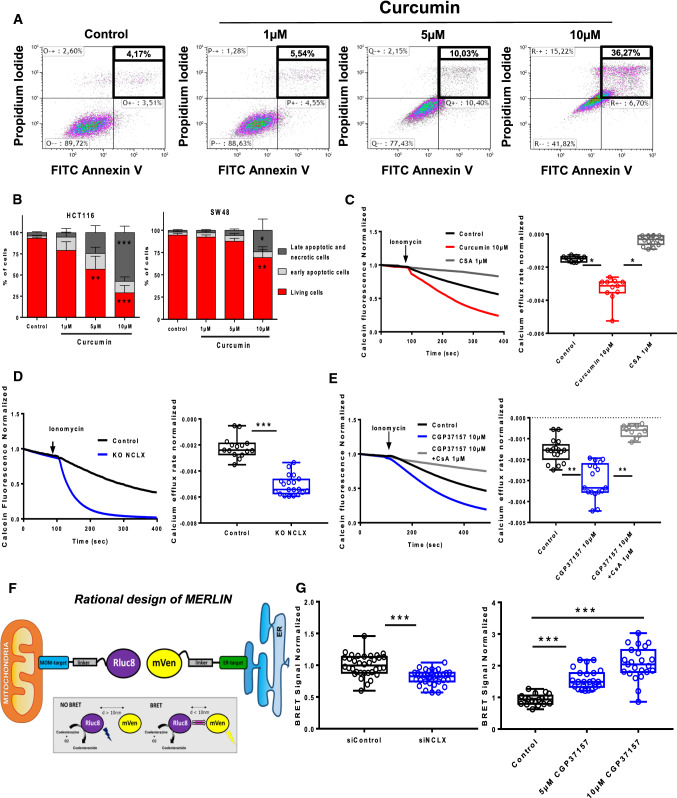
NCLX inhibitors induced apoptosis via mitochondrial permeability transition pore sensitization and modification of endoplasmic reticulum–mitochondria contact sites
High ROS and Ca2+ levels in mitochondria with mitochondria membrane depolarization can trigger cell death following the opening of mPTP [14]. Apoptosis evaluated by flow cytometry indicated that curcumin dose-dependently increased apoptosis in HCT116 and SW48 cells (Fig. 5A and B). To determine whether NCLX plays a role in Ca2+-induced mPTP opening, HCT116 cells were loaded with calcein-AM and cobalt was added to quench the cytosolic fluorescence of calcein. HCT116 cells were pre-treated for 1 h with either 10 µM of curcumin or 1 µM of cyclosporine A (CsA), an mPTP blocker, and exposed to ionomycin to increase mtCa2+ (Fig. 5C). Remarkably, ionomycin induced more notable calcein fluorescence quenching in HCT116 cells pre-treated with curcumin or CGP37157 for 1 h compared to control (DMSO) (Fig. 5C, E). In addition, ionomycin induced greater quenching of calcein fluorescence in HCT116 NCLX KO cells compared with HCT116 WT cells (Fig. 5D). These effects were fully reversed by pre-treatment with CsA, indicating that they are mediated by mPTP (Fig. 5E). Using Calcium Green-5N, we showed that there was greater mPTP activation in siNCLX-transfected HCT116 cells (Supplementary Fig. 3D). Taken together, either NCLX inhibition or curcumin can sensitise CRC cells to mPTP opening and consequently promote cancer cell death.
Fig. 5.
Treatment with curcumin or NCLX inhibitors induces Ca2+-dependent apoptosis by sensitizing mPTP activation. A HCT116 and SW48 were treated with 0–10 µM curcumin for 24 h. The dots indicate the number of Annexin V-/PI- cells (bottom left field, indicates live cells), Annexin V+/PI– cells (bottom right field, indicates early apoptotic cells), Annexin V+/PI+ cells (top right field, indicates late apoptotic cells), and Annexin V−/PI+ cells (top left field, indicates necrotic cells). B The early stage apoptotic cells are shown in grey and the late-stage apoptotic and necrotic cells are shown in black. The results are presented as the mean ± SD of 4–5 independent experiments (*P < 0.05, **P < 0.01 and ***P < 0.005 compared with their respective control; ns: non-significant). C For the mPTP opening experiment, HCT116 cells were loaded with calcein-AM and CoCl2. mPTP opening was monitored by the decrease in calcein fluorescence induced by 5 µM ionomycin in HCT116 cells pre-treated for 1 h with DMSO (black line), 10 µM curcumin (red line) or 1 μM CsA (gray line). mPTP opening occurs after the addition of 5 μM ionomycin (4–5 independent experiments; One-way ANOVA with Dunnett’s multiple comparisons test, *P < 0.05, **P < 0.01 versus DMSO). D Depletion of NCLX expression increased mPTP opening activation in HCT116 cells. Measurement of calcein fluorescence in HCT116 WT and NCLX KO cells after activation of mPTP opening induced by 5 μM ionomycin. The kinetics of mPTP opening was increased when the expression of NCLX was downregulated (5 independent experiments Mann–Whitney test, *P < 0.05). E The CsA pre-treatment prevented the mPTP sensitization induced by CGP37157 treatment. Changes in mitochondrial calcein fluorescence intensities induced by 5 µM ionomycin were measured in HCT116 cells pre-treated for 1 h with 10 µM CGP37157 or = 10 µM CGP37157 and 1 µM CsA (one-way ANOVA with Dunnett’s multiple comparisons test, *P < 0.05, **P < 0.01 and ***P < 0.001 versus DMSO). F Schematic representation of MERLIN based on BRET. The BRET donor was MOM-targeted Rluc8 and the BRET acceptor was mVenus in the ER. In both BRET partners, the linker region is an alpha helix formed by the A(EAAAK)4A motif. BRET occurs upon MOM-Rluc8 stimulation by coelenterazine h and BRET to ER-mVenus. G Targeting NCLX modified the contacts between the ER and mitochondria. Changes in BRET signals in HCT116 cells co-expressing the BRET pair MERLIN, sCal-L1-RLuc, and mVen-L1-B33C after knockdown of NCLX, siRNA control transfection or treatment with CGP37157. The results are based on 3–4 replicates with 18–22 measurements per group. The error bars indicate SD (***P < 0.001, Mann–Whitney test or one-way ANOVA with Dunnett’s multiple comparisons test)
It has increasingly been recognized that the ER and mitochondria communicate at specific membrane contact sites called mitochondria-associated ER membranes (MAM) [25]. Perturbation of MAM function and/or structure elevates oxidative stress, irreparably damages mitochondria and reduces mitochondrial respiration. These effects have an impact on mitochondrial ATP production and apoptosis. Thus, we investigated the role of NCLX on ER–mitochondria contact sites using MERLIN, a newly developed BRET-based proximity biosensor, expressed in HCT116 cells (Fig. 5F) [26]. The MERLIN biosensor allows the detection of changes in distance in the nanometre range between the ER and mitochondria, by measuring the energy transfer between the mitochondrial outer membrane (MOM)-targeted Rluc8 (BRET donor) and the ER-targeted mVenus (BRET acceptor). Silencing NCLX substantially decreased the BRET signal (Fig. 5G), indicating a decrease in contact points between the ER and mitochondria (Fig. 5G, left). However, pharmacological inhibition of NCLX after 3 h pre-treatment strongly increased the BRET signal in a dose-dependent manner, suggesting more intimate contact between the ER and mitochondria under these conditions (Fig. 5G, right). This apparent discrepancy could be explained by temporal differences and/or modes of action between the siRNA approach and CGP37157 treatment. Increased interaction between the mitochondrial network and ER during early ER stress conditions has already been described [27] Taken together, these data suggest that inhibition of NCLX can contribute to apoptosis with complete sensitization of mPTP and to modified architecture and spatiotemporal regulation of ER–mitochondria contact sites.
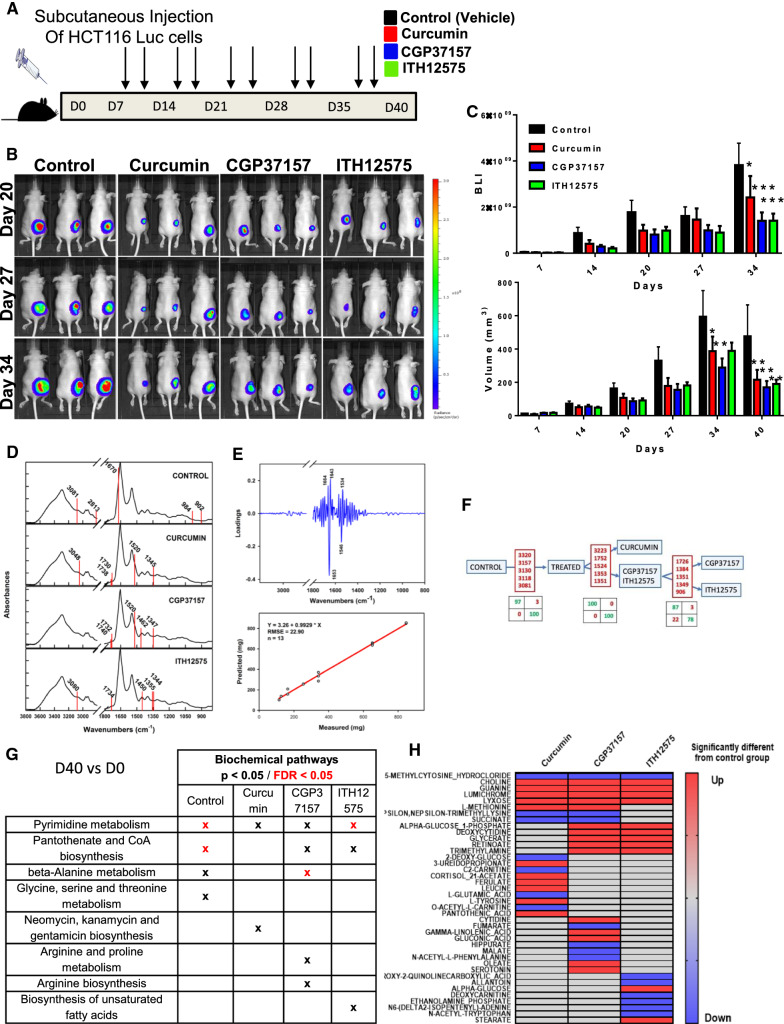
NCLX inhibitors decreased colorectal cancer growth in vivo
The effects of curcumin, CGP37157 and ITH12575 were tested in a CRC-xenografted Swiss nude mouse model (schematic diagram, Fig. 6A). As shown in Fig. 6B (left panel), CRC xenografts that were treated with vehicle (DMSO, Control Group), curcumin, CGP37157 or ITH12575 showed a significant decrease of primary tumor growth at day 40 (tumoral volume or tumoral bioluminescence) compared with control mice. Based on mathematical modeling of tumor growth (Supplementary Fig. 5A, right), the estimated mean (inter-subject standard deviation) model parameters were kgrowth = 0.46 day−1 (–), Vmax = 336 mm3 (0.69) and γ = 0.34. More precisely, tumor volume doubled in 1.5 days and reached a maximum average value of 336 mm3 in the control group. Vmax was significantly decreased by twofold in the presence of either CGP37157 (P = 0.033) or curcumin (P = 0.040). In the presence of ITH12575, there was a non-significant trend for decreased Vmax (P = 0.081). At the end of the study, the impact of treatments on the proliferation of HCT116 tumor xenografts was detected through Ki67 staining (Supplementary Fig. 5B). The results revealed that Ki67-positive cells were significantly decreased in curcumin, CGP37157 and ITH12575 groups (Supplementary Fig. 5B). Moreover, the expression of NCLX in all groups was analyzed by immunohistochemistry. Surprisingly and interestingly, it appears that repeated treatments during 40 days reduced NCLX expression in the tumors and more significantly in ITH12575 group (Supplementary Fig. 5C).
Fig. 6.
Curcumin and NCLX blockers reduced colorectal tumor growth in vivo. A HCT116-Luc cells were implanted subcutaneously into nude mice: control, curcumin-treated, CGP37157-treated, and ITH12575-treated groups (n = 8–9/group). B Representative bioluminescent images of CRC tumors in control, curcumin, CGP37157 and ITH12575 groups at day 20–27 and 34. C BLI value of tumors (expressed in c.p.m) as a function of time recorded in the whole body of mice. Volume of tumors (expressed in mm3) as a function of time recorded in the whole body of mice (Mean ± sem, two-way ANOVA with multiple comparisons test, *P < 0.05, **P < 0.01 and ***P < 0.001 versus control group). D Spectral wavelengths allowing discrimination among the groups superimposed with the mean group spectrum. E Tumor size prediction based on partial least square (PLS) regression showing the loadings (i.e., the correlated spectral variables and the regression model). F The dichotomous model to identify groups from plasma serum with, for each step, the relevant discriminant variables and the consequent confusion matrixes. G Metabolic pathways associated with metabolite sets that were modified between D0 and D40 for each group based on analysis of whole blood samples. H Heatmap analysis of metabolite levels identified at D40 showing the difference between the treated groups and the control group
Metabolic mid-infrared signatures predicted tumor size and were specific for each NCLX blocker
Because we found that curcumin and NCLX inhibitors are regulators of cancer cell metabolism, we aimed to identify some circulating biomarkers of these effects. Using MIR spectroscopy of dried plasma and serum from mice that had been xenografted with CRC cells, we looked for a specific tumor metabolic signature related to NCLX inhibitors and curcumin to gain insights into the similarities between the mechanisms of action of curcumin, CGP37157 and ITH12575 on NCLX.
The analysis was performed in control, curcumin-treated, CGP37157-treated or ITH12575-treated mice. Supplementary Fig. 5D presents spectral signatures between D0 and D40. The five most discriminating variables between the two groups or two subsets were identified as shown in Fig. 6D: control, 3081, 2813, 1670, 984 and 902 cm−1; curcumin, 3048, 1738, 1730, 1520 and 1345 cm−1; CGP37157: 1740, 1732, 1520, 1462 and 1347 cm−1; and ITH12575, 3080, 1734, 1450, 1355 and 1344 cm−1. The signature was mainly linked to protein absorption bands at 1664, 1653, 1643, 1546 and 1534 cm−1. These bands are assigned to the amide vibrations of proteins, suggesting drastic changes in plasma/serum proteins. Interestingly, this signature was correlated with tumor size in the control group and was lost in the treated groups, therefore confirming a ‘treatment’ effect on tumor size (Fig. 6E). To investigate whether curcumin treatment or NCLX blockers yield similar MIR signatures, we performed sequential predictive tests to determine the spectral signatures of mice at D40 according to a dichotomous decision tree (Fig. 6F). Matrices easily discriminated the control group from the treated groups. Moreover, this method allowed us to discriminate the curcumin-treated group from the two other treatments (CGP37157 or ITH12575). The analysis between the CGP37157-treated and ITH12575-treated groups was trickier, which was not surprising given that ITH12575 is a derivative of CGP37157.
In summary, the identified MIR signatures support major metabolic alterations in the mechanisms that underlie the anti-tumoral efficacy of curcumin and other NCLX inhibitors.
High-resolution mass spectrometry full-scan analysis revealed a role for NCLX in the control of protein and amino acid synthesis
To focus on the metabolites that are modified following NCLX inhibition during tumor growth, we used LC-HRMS full-scan analysis of serum extracts from our experimental mouse model and compared metabolomic modulation of each group (between D0 and D40) (Fig. 6G). The variable importance in projection (VIP) list and biochemical pathways allowed us to link or to discriminate each group with a panel of particular metabolites and pathways. For all groups, we observed a change in the metabolism of pyrimidines. Pantothenate and CoA biosynthesis pathways are affected for the control/CGP37157-treated and ITH12575-treated groups. Beta alanine metabolism is also modified under control and CGP37157 conditions. There are also specific modifications such as glycine–serine–threonine metabolism for the control group or arginine metabolism for the CGP37157-treated group. Finally, the biosynthetic pathway of unsaturated fatty acids for the ITH12575-treated group is modified. Taken together, these results suggest an important role of pyrimidine metabolism, pantothenate and CoA biosynthesis, beta alanine metabolism and glycine–serine–threonine metabolism in tumor development and that NCLX inhibitors limit the activation of these pathways. These data suggest metabolic differences over time in each group prior to implantation of tumor cells and during tumor development (Fig. 6G).
To further characterize the metabolic modifications linked to NCLX through the three treatments, we compared the metabolites analyzed in the treated groups to the metabolites in the control group. We found a common base of metabolite modification in our three treated group: choline, guanine, lumichrome, lyxose were upregulated (red color) and methylcytosine was downregulated (blue color) (Fig. 6H). These results suggest a possible anti-tumoral effect by these pathways. We also found metabolites modification such as methionine, nepsilon, succinate or alpha glucose 1-phophate, glycerate, retinoate, deoxycytidine, and trimethylamine that were only present in two groups: CGP/curcumin or CGP/ITH12575 (Fig. 6H). We also found some metabolites found in only one group (Fig. 6H). The combination of these results suggests an important role for NCLX in the control of protein and amino acid synthesis and methylation through the regulation of choline and possibly methionine metabolism (variation common to all 3 groups of inhibitors).
Transcriptomic and tissue microarray analysis highlighted the association between NCLX expression and the MSI status and survival of patients with colorectal cancer
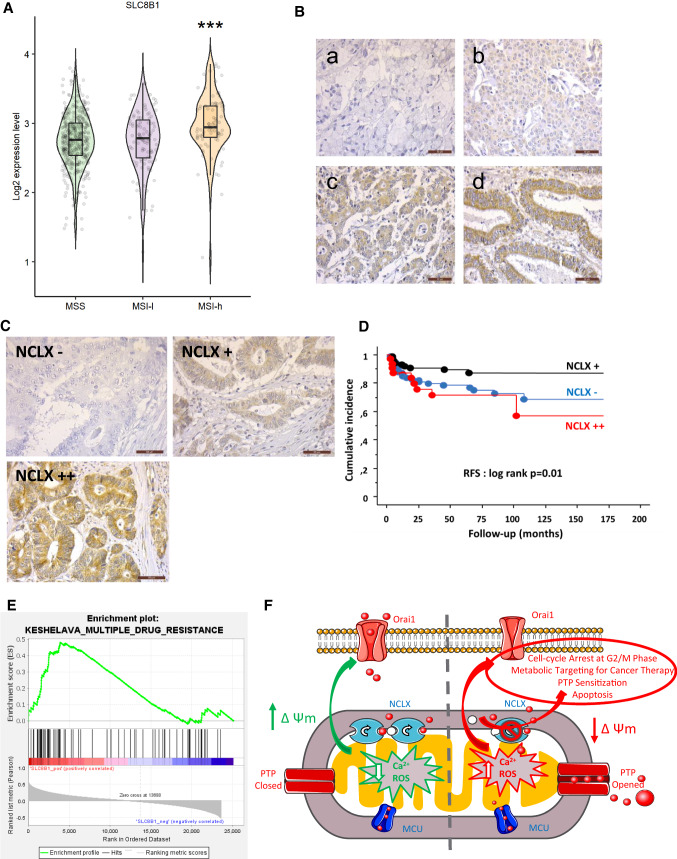
Analysis of patients with CRC in TCGA dataset showed that higher SLC8B1 (NCLX) mRNA expression was associated with MSI status (Fig. 7A). In all tumor stages, SLC8B1 expression tended to be higher in CRC tumors with high MSI (MSI-H) compared to tumors with MSS and low MSI (MSI-L). The difference observed was significant for stage II (Supplementary Fig. 6A). Previous investigations have proposed different classifications of patients with CRC [28, 29]. In the classification proposed by Guinney et al. [28], SLC8B1 expression was higher in CMS1 molecular subtype defined by a worst relapse survival and an MSI status [28] (Supplementary Fig. 6B). In the classification of gastrointestinal cancers proposed by Liu et al. 2018 [29], SLC8B1 mRNA expression was significantly higher in MSI and genome stable (GS) molecular subtypes (Supplementary Fig. 6C). Altogether, these data indicate that SLC8B1 expression is strongly associated to the MSI status of patients with CRC.
Fig. 7.
NCLX expression and clinical parameters of a cohort of patients with MSI CRC. A Boxplots showing the mRNA expression profile of NCLX in primary CRC tissues from the TCGA dataset according to their MSI/MSS status (MSS, n = 423; MSI-Low [MSI-L], n = 100; MSI-High [MSI-H], n = 81). B/ Representative NCLX expression during the differentiation of the colonic epithelium (COLOMIN cohort). C Representative NCLX immunostainings of three groups of patients with CRC classified according to their NCLX expression level (COLOMIN cohort). D Kaplan–Meier plots (cumulative incidence in function of follow-up of patients with CRC) showing RFS of the COLOMIN cohort (P = 0.01 by the log-rank test). E GSEA of the KESHELAVA multiple drug resistance signature in primary MSI CRC tissues from the TCGA dataset. F Schematic diagram of the main findings of the study
NCLX protein expression was next analyzed on TMA-based collection of 302 patients with MSI CRC (COLOMIN cohort). We evaluated the NCLX expression with regard to tumor stages. Most patients at advanced stages had lower NCLX protein expression. Of note, loss of NCLX expression was associated with tumor grade in poorly differentiated cancers (Fig. 7B, a–d; P < 0.0001). Subsequent analysis revealed that the NCLX expression level was inversely associated with perineural invasion (P < 0.0001) and vascular emboli (P < 0.02), suggesting a protective role of this exchanger in CRC. We found that NCLX was expressed in 237 of 261 normal tissue samples (91%), with granular cytoplasmic staining (Fig. 7C). In the cancerous tissues analyzed with TMA, NCLX remained expressed in 183 of 321 samples (57%), with either moderate-intensity (Fig. 7C, panel NCLX +) or high-intensity (Fig. 7C, panel NCLX++) staining; in 138 cancer tissue samples, NCLX expression was lost (Fig. 7C, panel NCLX−).
Based on Cox univariate analysis, NCLX expression was associated with RFS (Fig. 7D). Tumors with loss of NCLX expression (Fig. 7C, panel NCLX-) recurred more than those that retained NCLX expression (Fig. 7C, panel NCLX+). Surprisingly, tumors with increased NCLX expression (Fig. 7C, panel NCLX++) had an even worse prognosis than the patients with retained or lost NCLX expression. This finding suggests that NCLX is normally expressed normally in non-cancerous epithelial cells and its loss of expression is associated with an aggressive progression of epithelial tumors. At the same time, however, over-expression of this exchanger can lead to a worsened evolution of the disease. In line with these data, GSEA of the transcriptomic CRC dataset from TCGA revealed a positive correlation between NCLX expression and the multiple drug resistance signature in patients with MSI (Fig. 7E). Our main results are summarized in a schematic diagram (Fig. 7F).
Discussion
Understanding the mechanisms of curcumin-mediated apoptosis and subsequent cell cycle arrest has important implications for anti-cancer therapy [30]. Over the last few years, the mechanisms of Ca2+ signaling have been greatly improved, especially how its alterations in cancer participate in modifying key processes such as proliferation, metabolism and sensitivity to cell death [18]. Recent evidences highlighted that mtCa2+ is closely associated with the hallmarks of cancer, and MCU and NCLX play key roles in CRC carcinogenesis [19, 31]. The main results of the present study are the demonstration of the mtCa2+-dependent effects of curcumin on NCLX and the potential anti-tumoral effect of NCLX blockers in CRC. Ca2+ homeostasis directly regulates the expression and activation of cell cycle protein complexes [32]. NCLX silencing or curcumin treatment led to an accumulation of cells at the G2/M checkpoint.
Curcumin induced a high mtCa2+ level by blocking mtCa2+ efflux through NCLX. Our data showed that pharmacological inhibition of NCLX in CRC cells caused mtCa2+ overload and an increase in mitochondrial and cytosolic ROS production associated with a mitochondrial membrane depolarization. Kostic et al. demonstrated that mild fluctuations in ΔΨm, which do not affect Ca2+ influx, are sufficient to strongly regulate NCLX [33]. Furthermore, complete mitochondrial depolarization and de-energization lead to reversal of NCLX activity preceding toxic mtCa2+ accumulation [34]. Our results do not allow us to show whether curcumin directly blocks NCLX or whether the action is governed by an effect on the mitochondria membrane potential. However, the study from Mustapha et al. showed that the increase of mitochondrial calcium is nearly occurring in parallel with curcumin accumulation, and precedes ROS production and the ΔΨm decreasing, suggesting a direct impact of curcumin on NCLX in the first instance [35, 36].
Intrinsic apoptosis involves mtCa2+ and mtROS overload and mitochondria membrane depolarization leading to mPTP opening and release of cytochrome c [15]. Curcumin treatment induced apoptosis in a dose-dependent manner. We found that curcumin treatment can lead to a sustain increase of the rate of mPTP activation in CRC cells. Consistently, NCLX KO cells and cells treated with CGP37157 had greater mPTP activation. These effects were reversed by pre-treatment with a PTP inhibitor (CsA). In the same vein as our findings, curcumin induced mtCa2+ overload selectively in malignant breast cancer cells. Of note, the authors found that CGP-37157 and bortezomib co-treatment activates paraptotic signals in cancer cells, similar to curcumin [36]. In addition, it was reported that blockade of MCU by ruthenium red (RR) or Ru360 prevents the increase of mtCa2+ by curcumin, and inhibits the apoptosis induced by curcumin in breast cancer and leukemia cells [36, 37].
In non-excitable cells, including most cancer cells, store-operated Ca2+ entry (SOCE) represents the major and calcium-entry pathways.
Mitochondria control SOCE entry through NCLX and redox signals. Silencing of NCLX induced Orai1 inactivation through oxidation of a critical cysteine (Cys195) in the third transmembrane helix of Orai1 [23]. In agreement with this finding, the replacement of the reactive cysteine residue with serine at a specific Orai1 (C195S) site significantly altered the sensitivity of ICRAC to curcumin, suggesting that the Cys195 is the target residue for the inhibitory action of this compound through ROS signaling [38]. As we previously showed in CRC cell lines with NCLX KO [19], the pharmacological blockade of NCLX by curcumin or CGP37157 decreases SOCE in CRC cells. We also confirmed the effect of curcumin treatment on Orai1-dependent CRAC current.
The fine regulation of Ca2+ in MAM is paramount for the management of metabolism and cell death [39]. Cancer cells need a constitutive transfer of Ca2+ from the ER to mitochondria for their survival [40, 41]. Our data showed that NCLX targeting disrupted MAM contacts. Remodeling of cancer cell metabolism according to nutrient availabilities is a hallmark of cancer [39]. mtCa2+ uptake plays an essential role in the control of aerobic metabolism. In physiological condition, mitochondrial matrix Ca2+ regulates the activity of TCA cycle dehydrogenases [42]. The inhibition of mtCa2+ uptake induced a bio-energetic crisis and metabolic reprogramming. However, the effect of mtCa2+ efflux has been poorly described. Recently, Assali et al. highlighted a key pathway through which mtCa2+ efflux permits a robust activation of respiration in response to profound mitochondrial respiratory uncoupling, while preventing the activation of cell death pathways [43].
In our previous study, we reported that NCLX loss in CRC cells leads to reduced oxygen consumption, increased glycolysis and increased transcription of major glycolytic enzymes and transporters (19). This increased transcription of glycolytic enzymes is under the control of HIF1α. This transcriptional factor is upregulated in response to increased mtROS resulting from loss of NCLX and subsequent increase in mtCa2+. In the present study, we demonstrated that curcumin and NCLX inhibition suppressed mitochondrial respiration and amino acid metabolism without increasing the glycolysis process. This latter finding is in contrast to the effect observed by Pathak et al., suggesting either an adaptation (metabolic reprogramming) of siNCLX-treated cells and NCLX KO cells, or an off-target effect of curcumin or CGP37157. At the molecular level, the differential effect of NCLX KO and NCLX inhibitors on glycolysis can be explained by the expression of HIF1a. We showed that unlike NCLX KO cells, NCLX inhibitors reduced HIF1a expression which is a strong regulator of glycolysis-related genes. Additional experiments should be carried out to refine the understanding of the mechanism of action of NCLX blockers.
In vivo, we demonstrated that NCLX inhibitors reduced the tumor size of NMRI nude mice grafted with HCT116 cells. The effect of curcumin on tumor growth may be explained by its effect on cell proliferation or by induction of apoptosis (see Fig. 1) and by its anti-angiogenic potency as observed in Supplementary Fig. 7 (showing a completely inhibited endothelial tube formation). We thus reasoned that cancer cells with or without NCLX blockers could affect the global metabolism of mice. Interestingly, using MIR spectroscopy on whole blood from xenografted mice, we found a five-element signature in the control group that correlated with tumor size. This correlation was lost in the treated groups, suggesting the possibility to follow and to model tumor growth with minimally invasive peripheral biomarkers. Moreover, spectral analysis at D40 allowed us to isolate a five-element signature specific for each group, thereby allowing discrimination of the experimental groups. Intriguingly, CGP37157 and curcumin caused identical spectral modifications at 1730–1740, 1520 and 1345–1347 cm−1, suggesting a similar global effect in these particular subgroups. To further support our findings, we used metabolomics to identify shared pathways and metabolites among treatments compared with the control group; this analysis produced a similar pattern as seen with MIR spectroscopy. In all groups, we observed a change in pyrimidine metabolism. It is well known that cancer cells alter their metabolism to increase the flux of de novo pyrimidine synthesis to support a high proliferative phenotype [44]. Interestingly, NCLX inhibitors appeared to limit the function of pyrimidine metabolism. More specifically, metabolites of methylcytosine, choline and guanine were particularly affected by the blockade of NCLX. These results provide evidence that NCLX is a key ‘crossroad’ factor in the control of protein and amino acid synthesis and methylation. Choline modulates epigenetic marking of genes, and choline deficiency is correlated with the silencing of several tumor suppressor genes responsible for DNA repair (hMLH1) and cell cycle regulation (p15 and p16) [45].
In the future, we aim to distinguish unique metabolic profiles or easy-to-implement signatures that would be useful as minimally invasive biomarkers of tumoral NCLX expression in a clinical context.
In the first randomized controlled trial using curcumin in combination with FOLFOX chemotherapy for patients with metastatic CRC, researchers showed that this combination is a safe and tolerable treatment that could benefit patients with longer overall survival, but the number of patients was too low to make definitive conclusion [46]. Our TCGA analysis showed that SLC8B1 mRNA levels are significantly higher in CRC tumors with high MSI (MSI-H) compared to tumors with MSS and low MSI (MSI-L). In the classification proposed by Guinney et al. [28], SLC8B1 expression is higher in CMS1 molecular subtype and associated with a worst relapse survival and a MSI status suggesting that mtCa2+ extrusion through NCLX function has a critical role in MSI CRC. In our cohort of patients with MSI CRC, NCLX expression decreased in the advanced CRC stages. NCLX expression was associated with poor tumor differentiation but lower perineural sheathing and/or vascular emboli. Loss of NCLX expression was associated with worse RFS in Cox univariate analysis. Interestingly, we found a subgroup of patients with MSI CRC (13–15%) with increased NCLX expression; they had an even worse prognosis than patients who had maintained or lost NCLX expression. Additional investigations will be necessary to understand the origin of these different subgroups with increased or lost NCLX expression. Moreover, CGP37157 has been reported to prevent neurotoxicity in response to the chemotherapeutic agent salinomycin. This protective effect was not observed when head and neck squamous cell carcinoma cells were co-treated with salinomycin and CGP37157 [47]. Prospective trials need to evaluate the combination of chemotherapy plus curcumin in patients with NCLX-expressing MSI CRC to improve the efficacy and decrease the adverse events of standard treatment.
In summary, our findings provide very strong evidence that curcumin and NCLX blockers inhibit CRC cell growth by enhancing cellular oxidative stress and mtCa2+ overload associated with a mitochondria membrane depolarization, thereby leading to mPTP-dependent cell apoptosis. Our works also show that reducing mitochondrial respiration, HIF1α and MAM perturbations contribute to these mechanisms. These findings support a novel anti-tumoral mechanism for NCLX, particularly in patients with MSI CRC, a clinical finding that has never been described before to our knowledge.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr Virginie André, Julien Sobilo, and Isabelle Domingo for technical assistance. Inserm UMR 1069 is leader of Cancéropole Grand-Ouest 3MC network (Marine Molecules, Metabolism and Cancer), member of Région Centre—Val de Loire thematic research consortium RTR MOTIVHEALTH (Molecular and Technological Innovation for Health) and member of CNRS research group APPICOM (Integrative Approach for a multiscale functional analysis of membrane proteins). We acknowledge Inserm, Université de Tours and Région Centre-Val de Loire for their financial supports.
Author contributions
Study concept and design: MG, WR, and TL. Acquisition of data: MG, SI, JB, AR, TP, XZ, JD, DT, RG, HFR, AL, SL, RC, MO, OS, OH, GFH, DTo, and TL. Analysis and interpretation of data: MG, SI, JB, AR, TP, XZ, DC, JD, DT, VM, HFR, AL, SL, ALP, JFD, PGF, AG, RC, FG, AJGS, PE, OS, OH, GFH, CV, DTo, MT, WR, and TL. Drafting of the manuscript: MG, SI, JB, PE, OS, CV, DTo, MT, WR, and TL. Critical revision of the manuscript for important intellectual content: All. Statistical analysis: MG, SI, JB, VM, PE, DTo, WR, and TL. Obtained funding: MG, AG, DTo, WR, and TL. Study supervision: MG, WR, and TL.
Funding
This project was partly supported by grants on behalf of the following french department committees of Ligue Contre le Cancer Grand-Ouest: 16 (Charente), 37 (Indre-et-Loire), 49 (Maine-et-Loire), 72 (Sarthe) and 85 (Vendée). Maxime Guéguinou is a recipient of a 3-years postdoctoral grant from Région Centre—Val de Loire (Opticoregumine project). Sajida Ibrahim was partly funded by ARFMAD (Association Recherche et Formation dans les Maladies de l’Appareil Digestif). Alison Robert is a recipient of a 3-years doctoral grant from Inserm/ Région Centre – Val de Loire. David Crottès is a recipient of SMART LOIRE VALLEY fellowship from LE STUDIUM Loire Valley Institute for Advanced Studies. COLOMIN cohort was supported in part by a grant from the association “Sport et Collection” and “Ligue Contre le Cancer, Comités départementaux de la Vienne, Charente et Charente-Maritime” (David Tougeron).
Availability of data and materials
Data will be made available on reasonable request.
Declarations
Conflict of interest
No competing interests were disclosed by authors.
Ethics approval and consent to participate
COLOMIN study was designed in accordance with legal requirements and the Declaration of Helsinki, and was approved by the ethics committee of CHU Poitiers and CHRU Tours, France (Comité de protection des personnes Ouest III, no. DC-2008-565 and no. 2018-039).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
William Raoul and Thierry Lecomte (senior authors) have contributed equally to this work.
Contributor Information
Maxime Guéguinou, Email: maxime.gueguinou@univ-tours.fr.
Thierry Lecomte, Email: thierry.lecomte@univ-tours.fr.
References
- 1.Evrard C, Tachon G, Randrian V, Karayan-Tapon L, Tougeron D. Microsatellite instability: diagnosis, heterogeneity, discordance, and clinical impact in colorectal cancer. Cancers (Basel) 2019;11(10):1567. doi: 10.3390/cancers11101567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jo W-S, Carethers JM. Chemotherapeutic implications in microsatellite unstable colorectal cancer. Cancer Biomark. 2006;2(1–2):51–60. doi: 10.3233/CBM-2006-21-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tougeron D, et al. Predictors of disease-free survival in colorectal cancer with microsatellite instability: an AGEO multicentre study. Eur J Cancer. 2015;51(8):925–934. doi: 10.1016/j.ejca.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Tougeron D, et al. Prognosis and chemosensitivity of deficient MMR phenotype in patients with metastatic colorectal cancer: an AGEO retrospective multicenter study. Int J Cancer. 2020;147(1):285–296. doi: 10.1002/ijc.32879. [DOI] [PubMed] [Google Scholar]
- 5.Allegra A, Innao V, Russo S, Gerace D, Alonci A, Musolino C. Anticancer activity of curcumin and its analogues: preclinical and clinical studies. Cancer Invest. 2017;35(1):1–22. doi: 10.1080/07357907.2016.1247166. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz de Porras V, Layos L, Martínez-Balibrea E. Curcumin: a therapeutic strategy for colorectal cancer? Semin Cancer Biol. 2020;73:321–330. doi: 10.1016/j.semcancer.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Katona BW, Weiss JM. Chemoprevention of colorectal cancer. Gastroenterology. 2020;158(2):368–388. doi: 10.1053/j.gastro.2019.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Z, Jin S, Yalowich JC, Brown KD, Rajasekaran B. The mismatch repair system modulates curcumin sensitivity through induction of DNA strand breaks and activation of G 2 M checkpoint. Mol Cancer Ther. 2010;9(3):558–568. doi: 10.1158/1535-7163.MCT-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giordano A, Tommonaro G. Curcumin and cancer. Nutrients. 2019;11(10):2376. doi: 10.3390/nu11102376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Q, et al. Curcumin induces the apoptosis of A549 cells via oxidative stress and MAPK signaling pathways. Int J Mol Med. 2015;36(4):1118–1126. doi: 10.3892/ijmm.2015.2327. [DOI] [PubMed] [Google Scholar]
- 11.Morin D, Barthélémy S, Zini R, Labidalle S, Tillement J-P. Curcumin induces the mitochondrial permeability transition pore mediated by membrane protein thiol oxidation. FEBS Lett. 2001;495(1–2):131–136. doi: 10.1016/S0014-5793(01)02376-6. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Zichri S, et al. Cardiolipin mediates curcumin interactions with mitochondrial membranes. Biochim Biophys Acta Biomembr. 2019;1861(1):75–82. doi: 10.1016/j.bbamem.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Hempel N, Trebak M. Crosstalk between calcium and reactive oxygen species signaling in cancer. Cell Calcium. 2017;63:70–96. doi: 10.1016/j.ceca.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Görlach A, Bertram K, Hudecova S, Krizanova O. Calcium and ROS: a mutual interplay. Redox Biol. 2015;6:260–271. doi: 10.1016/j.redox.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasola A, Bernardi P. Reprint of ‘The mitochondrial permeability transition pore and its adaptive responses in tumor cells’. Cell Calcium. 2015;58(1):18–26. doi: 10.1016/j.ceca.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Pathak T, Trebak M. Mitochondrial Ca2+ signaling. Pharmacol Ther. 2018;192:112–123. doi: 10.1016/j.pharmthera.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero-Garcia S, Prado-Garcia H. Mitochondrial calcium: transport and modulation of cellular processes in homeostasis and cancer. Int J Oncol. 2019;54(4):1155–1167. doi: 10.3892/ijo.2019.4696. [DOI] [PubMed] [Google Scholar]
- 18.Marchi S, Giorgi C, Galluzzi L, Pinton P. Ca2+ fluxes and cancer. Mol Cell. 2020;78(6):1055–1069. doi: 10.1016/j.molcel.2020.04.017. [DOI] [PubMed] [Google Scholar]
- 19.Pathak T, et al. Dichotomous role of the human mitochondrial Na+/Ca2+/Li+ exchanger NCLX in colorectal cancer growth and metastasis. Elife. 2020;9:e59686. doi: 10.7554/eLife.59686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guéguinou M, et al. L′échangeur ionique NCLX—un rôle ambivalent dans la chronologie du cancer colorectal. Med Sci. 2021;37(2):124–126. doi: 10.1051/medsci/2020274. [DOI] [PubMed] [Google Scholar]
- 21.Montero M, et al. Chromaffin-cell stimulation triggers fast millimolar mitochondrial Ca2+ transients that modulate secretion. Nat Cell Biol. 2000;2(2):57–61. doi: 10.1038/35000001. [DOI] [PubMed] [Google Scholar]
- 22.Guéguinou M, et al. SK3/TRPC1/Orai1 complex regulates SOCE-dependent colon cancer cell migration: a novel opportunity to modulate anti-EGFR mAb action by the alkyl-lipid Ohmline. Oncotarget. 2016;7(24):36168. doi: 10.18632/oncotarget.8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben‐Kasus Nissim T et al (2017) Mitochondria control store‐operated Ca2+ entry through Na+ and redox signals. EMBO J [DOI] [PMC free article] [PubMed]
- 24.Grasso D, Zampieri LX, Capelôa T, Van De Velde JA, Sonveaux P. Mitochondria in cancer. Cell Stress. 2020;4(6):114–146. doi: 10.15698/cst2020.06.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Missiroli S, et al. Mitochondria-associated membranes (MAMs) and inflammation. Cell Death Dis. 2018;9(3):329. doi: 10.1038/s41419-017-0027-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hertlein V, et al. MERLIN: a novel BRET-based proximity biosensor for studying mitochondria–ER contact sites. Life Sci. Alliance. 2020;3(1):e201900600. doi: 10.26508/lsa.201900600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bravo R, et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J Cell Sci. 2011;124(Pt 13):2143–2152. doi: 10.1242/jcs.080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guinney J, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33(4):721–735.e8. doi: 10.1016/j.ccell.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng W, Goel A (2020) Curcumin and colorectal cancer: an update and current perspective on this natural medicine. Semin Cancer Biol [DOI] [PMC free article] [PubMed]
- 31.Vultur A, Gibhardt CS, Stanisz H, Bogeski I. The role of the mitochondrial calcium uniporter (MCU) complex in cancer. Pflügers Arch - Eur J Physiol. 2018;470(8):1149–1163. doi: 10.1007/s00424-018-2162-8. [DOI] [PubMed] [Google Scholar]
- 32.Prevarskaya N, Skryma R, Shuba Y. Ion channels in cancer: are cancer hallmarks oncochannelopathies? Physiol Rev. 2018;98(2):559–621. doi: 10.1152/physrev.00044.2016. [DOI] [PubMed] [Google Scholar]
- 33.Kostic M, Katoshevski T, Sekler I. Allosteric regulation of NCLX by mitochondrial membrane potential links the metabolic state and Ca2+ signaling in mitochondria. Cell Rep. 2018;25(12):3465–3475.e4. doi: 10.1016/j.celrep.2018.11.084. [DOI] [PubMed] [Google Scholar]
- 34.Samanta K, Mirams GR, Parekh AB. Sequential forward and reverse transport of the Na+ Ca2+ exchanger generates Ca2+ oscillations within mitochondria. Nat Commun. 2018;9(1):156. doi: 10.1038/s41467-017-02638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moustapha A, et al. Curcumin induces crosstalk between autophagy and apoptosis mediated by calcium release from the endoplasmic reticulum, lysosomal destabilization and mitochondrial events. Cell Death Discov. 2015;1(1):15017. doi: 10.1038/cddiscovery.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon MJ, Kim EH, Kwon TK, Park SA, Choi KS. Simultaneous mitochondrial Ca2+ overload and proteasomal inhibition are responsible for the induction of paraptosis in malignant breast cancer cells. Cancer Lett. 2012;324(2):197–209. doi: 10.1016/j.canlet.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Bae JH, Park J-W, Kwon TK. Ruthenium red, inhibitor of mitochondrial Ca2+ uniporter, inhibits curcumin-induced apoptosis via the prevention of intracellular Ca2+ depletion and cytochrome c release. Biochem Biophys Res Commun. 2003;303(4):1073–1079. doi: 10.1016/S0006-291X(03)00479-0. [DOI] [PubMed] [Google Scholar]
- 38.Shin DH, Nam JH, Lee ES, Zhang Y, Kim SJ. Inhibition of Ca2+ release-activated Ca2+ channel (CRAC) by curcumin and caffeic acid phenethyl ester (CAPE) via electrophilic addition to a cysteine residue of Orai1. Biochem Biophys Res Commun. 2012;428(1):56–61. doi: 10.1016/j.bbrc.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanova H, Kerkhofs M, La Rovere RM, Bultynck G. Endoplasmic reticulum-mitochondrial Ca2+ fluxes underlying cancer cell survival. Front Oncol. 2017;7:70. doi: 10.3389/fonc.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cárdenas C, et al. Selective vulnerability of cancer cells by inhibition of Ca(2+) transfer from endoplasmic reticulum to mitochondria. Cell Rep. 2016;14(10):2313–2324. doi: 10.1016/j.celrep.2016.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gherardi G, Monticelli H, Rizzuto R, Mammucari C. The mitochondrial Ca(2+) uptake and the fine-tuning of aerobic metabolism. Front Physiol. 2020;11:554904. doi: 10.3389/fphys.2020.554904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Assali EA, et al. NCLX prevents cell death during adrenergic activation of the brown adipose tissue. Nat Commun. 2020;11(1):3347. doi: 10.1038/s41467-020-16572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, Cui J, Ma H, Lu W, Huang J (2021) Targeting pyrimidine metabolism in the era of precision cancer medicine. Front Oncol 11 [DOI] [PMC free article] [PubMed]
- 45.Zeisel SH. Dietary choline deficiency causes DNA strand breaks and alters epigenetic marks on DNA and histones. Mutat Res. 2012;733(1–2):34–38. doi: 10.1016/j.mrfmmm.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howells LM, et al. Curcumin combined with FOLFOX chemotherapy is safe and tolerable in patients with metastatic colorectal cancer in a randomized phase IIa trial. J Nutr. 2019;149(7):1133–1139. doi: 10.1093/jn/nxz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scherzed A, et al. Effects of salinomycin and CGP37157 on head and neck squamous cell carcinoma cell lines in vitro. Mol Med Rep. 2015;12(3):4455–4461. doi: 10.3892/mmr.2015.3981. [DOI] [PubMed] [Google Scholar]
- 48.Grossman RL, et al. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ibrahim S, et al. Expression profiling of calcium channels and calcium-activated potassium channels in colorectal cancer. Cancers (Basel) 2019;11(4):561. doi: 10.3390/cancers11040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palty R, et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci U S A. 2010;107(1):436–441. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kouzi F, et al. Disruption of gap junctions attenuates acute myeloid leukemia chemoresistance induced by bone marrow mesenchymal stromal cells. Oncogene. 2020;39(6):1198–1212. doi: 10.1038/s41388-019-1069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Khuc T, Hsu C-WA, Sakamuru S, Xia M. Using β-lactamase and NanoLuc luciferase reporter gene assays to identify inhibitors of the HIF-1 signaling pathway. Methods Mol Biol. 2016;1473:23–31. doi: 10.1007/978-1-4939-6346-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nemani N, et al. MIRO-1 Determines mitochondrial shape transition upon GPCR activation and Ca2+ stress. Cell Rep. 2018;23(4):1005–1019. doi: 10.1016/j.celrep.2018.03.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan X, et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat Cell Biol. 2013;15(12):1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones E, et al. A threshold of transmembrane potential is required for mitochondrial dynamic balance mediated by DRP1 and OMA1. Cell Mol Life Sci. 2017;74(7):1347–1363. doi: 10.1007/s00018-016-2421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picou F, et al. n-3 Polyunsaturated fatty acids induce acute myeloid leukemia cell death associated with mitochondrial glycolytic switch and Nrf2 pathway activation. Pharmacol Res. 2018;136:45–55. doi: 10.1016/j.phrs.2018.08.015. [DOI] [PubMed] [Google Scholar]
- 58.Guéguinou M, et al. Synthetic alkyl-ether-lipid promotes TRPV2 channel trafficking trough PI3K/Akt-girdin axis in cancer cells and increases mammary tumour volume. Cell Calcium. 2021;97:102435. doi: 10.1016/j.ceca.2021.102435. [DOI] [PubMed] [Google Scholar]
- 59.Kim SM, Kim Y, Jeong K, Jeong H, Kim J. Logistic LASSO regression for the diagnosis of breast cancer using clinical demographic data and the BI-RADS lexicon for ultrasonography. Ultrasonography (Seoul, Korea) 2018;37(1):36–42. doi: 10.14366/usg.16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Q-S, Liang Y-Z, Du Y-P. Monte Carlo cross-validation for selecting a model and estimating the prediction error in multivariate calibration. J Chemom. 2004;18(2):112–120. doi: 10.1002/cem.858. [DOI] [Google Scholar]
- 61.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21(15):3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 62.Diémé B, et al. Metabolomics study of urine in autism spectrum disorders using a multiplatform analytical methodology. J Proteome Res. 2015;14(12):5273–5282. doi: 10.1021/acs.jproteome.5b00699. [DOI] [PubMed] [Google Scholar]
- 63.Mavel S, et al. 1H–13C NMR-based urine metabolic profiling in autism spectrum disorders. Talanta. 2013;114:95–102. doi: 10.1016/j.talanta.2013.03.064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on reasonable request.