Abstract
The segregation and maintenance of a dedicated germline in multicellular organisms is essential for species propagation in the sexually reproducing metazoan kingdom. The germline is distinct from somatic cells in that it is ultimately dedicated to acquiring the “totipotency” and to regenerating the offspring after fertilization. The most striking feature of germ cells lies in the presence of characteristic membraneless germ granules that have recently proven to behave like liquid droplets resulting from liquid–liquid phase separation (LLPS). Vasa/Ddx4, a faithful DEAD-box family germline marker highly conserved across metazoan species, harbors canonical DEAD-box motifs and typical intrinsically disordered sequences at both the N-terminus and C-terminus. This feature enables it to serve as a primary driving force behind germ granule formation and helicase-mediated RNA metabolism (e.g., piRNA biogenesis). Genetic ablation of Vasa/Ddx4 or the catalytic-dead mutations abolishing its helicase activity led to sexually dimorphic germline defects resulting in either male or female sterility among diverse species. While recent efforts have discovered pivotal functions of Vasa/Ddx4 in somatic cells, especially in multipotent stem cells, we herein summarize the helicase-dependent and -independent functions of Vasa/Ddx4 in the germline, and discuss recent findings of Vasa/Ddx4-mediated phase separation, germ granule formation and piRNA-dependent retrotransposon control essential for germline development.
Keywords: DEAD-box helicase, Germline, Transposable elements (TE), Phase separation, Spermatogenesis, Spermiogenesis, piRNA
Introduction
In multicellular metazoa, the specification and maintenance of the germline are of paramount importance for the propagation of species. Upon reunion of the parental genomes via fertilization, the germline can regenerate a whole new body with all types of cells and organs, and therefore the germline is considered as ultimate totipotent stem cell [1]. In most species, the capability of germline specification and development is achieved through the highly conserved “germ granules”—the electron-dense, membraneless ribonucleoprotein (RNP) particles, which were given with different names as initially discovered, such as the P bodies in worms, the pole plasm in flies, or nuage (e.g., intermitochondrial cement, chromatoid body) in mammals [2, 3]. Germ granules were initially posited by Weismann, et al., who speculated that the “dark material”, germ cell determinant, is present to carry the hereditary information from generation to generation and, when destroyed, is unable to initiate the germline development [3–5]. After that, mounting evidence has revealed the morphological character and RNA/protein contents of these granules in orchestrating the germline development and maintenance in a wide range of metazoan species. In worms, P granules are distributed uniformly in the cytoplasm of the fertilized, unpolarized one-cell embryo. After symmetry breaking, the P granules become visualized only in the posterior half of the embryo, followed by the embryo dividing that gives rise to a progenitor germ cell containing P granules as well as a somatic, non-P granule-containing daughter cell [6–8]. Previous evidence proposed two successive processes underlying the establishment of the germ granules: (i) the initial assembly of P granules when migrating to the posterior end by cytoplasmic gradient flow; (ii) followed by the degradation or disassembly of anterior P-granule contents [9–11]. Nevertheless, the physical nature as for how the granules are dynamically assembled and disassembled in response to environmental stimuli signaling and developmental cues has only been uncovered until recently by Hyman’s lab [8]. They found that the P granules in Caenorhabditis elegans behave like liquid droplets, which exhibit a highly dynamic morphological transition between soluble form and condensed form via a process known as phase transition (or phase separation). It is referred to as the demixing of contents into two distinct liquid phases in a homogeneous liquid system, and has been well studied in polymer chemistry. This study, perhaps for the first time, attributed the physical nature of liquid droplets to the formation of a non-membrane subcellular structure (germ granules) in cells, thereby linking the phase separation to cell biology in biomedical research [8, 12]. Since then, the field has boomed by intensive studies revealing that many membraneless, nuclear organelles, such as nucleoli, Cajal bodies, and cytoplasmic bodies, e.g., stress granules and Processing bodies (P bodies), hold similar biophysical features as seen in liquid droplets [11, 13, 14]. They are structurally dynamic, size-dependent, labile to shifts in pH, temperature, or ionic strength, and subject to the regulation of post-translational modifications (e.g., arginine methylation). These organelles are characterized by liquid-like behaviors, e.g., dripping, fusion and wetting, and are assembled as a result of liquid–liquid demixing into two-phase systems, also known as liquid–liquid phase separation (LLPS) or coacervation [15–18]. As a complement to the traditional membrane-encapsulated organelles, these membraneless particles are extremely enriched in proteins and nucleic acids with a high density, and thus were recently given the name “biomolecular condensates”. It is worth noting that LLPS significantly impacts the distribution of protein components, with proteins being significantly enriched by 10–300-fold in the coacervate compared to those in the dilute phase [13, 16].
Recent advances have revealed one striking feature that characterizes protein constituents in the membraneless organelles—the presence of abundant intrinsically disordered proteins (IDPs) or hybrid proteins with both ordered domains and intrinsically disordered protein regions (IDPRs) [18, 19]. They prove to be crucial constituents for phase separation. Perhaps one of the best-characterized IDPs is the Vasa (Vas), also known as Mvh (mouse Vasa homolog) or Ddx4 (DEAD-box polypeptide 4) (hereafter we use Ddx4 in mammals), which has been widely studied as a highly conserved, germ cell-specific biomarker necessary for germline specification and maintenance (Fig. 1A) [20–25]. Vasa was first identified as a germline factor following a large-scale genetic screen for maternal-effect recessive lethal mutations in the widely employed genetic model organism—Drosophila over 30 years ago, and was subsequently cloned and shown to be an essential component of polar granule in Drosophila [25, 26]. Thereafter, Vasa homologs have been intensively characterized as a highly conserved germline marker in a myriad of metazoan species. Those extensive studies have illuminated the multifaceted functions of Vasa related to many aspects of RNA metabolism-dependent processes, such as piRNA processing and ribonucleoprotein (RNP)-regulated granule formation, in germline development. Interestingly, recent findings suggest that Vasa/Ddx4 is also expressed in other tissues beyond the germline, such as the stem cells (e.g., neoblasts in planarian) and tumors. In turn, this might justify the fact that germ cells are deemed as ultimate totipotent stem cells, and germline-specific transcriptional machinery is activated in tumor cells (so-called cancer-testis genes) [23, 27, 28]. While extensive studies have been performed and excellent reviews have discussed the versatile roles of Vasa/Ddx4 in the germline and in other tissues among diverse species [22, 23], the exact functions of Vasa/Ddx4 are still far from clear. In this review, we mainly summarize and assess the roles of Vasa/Ddx4 by reconciling its structural domains, including the unstructured tails and the helicase motifs, with phase separation, granule formation and piRNA biogenesis throughout the germline development.
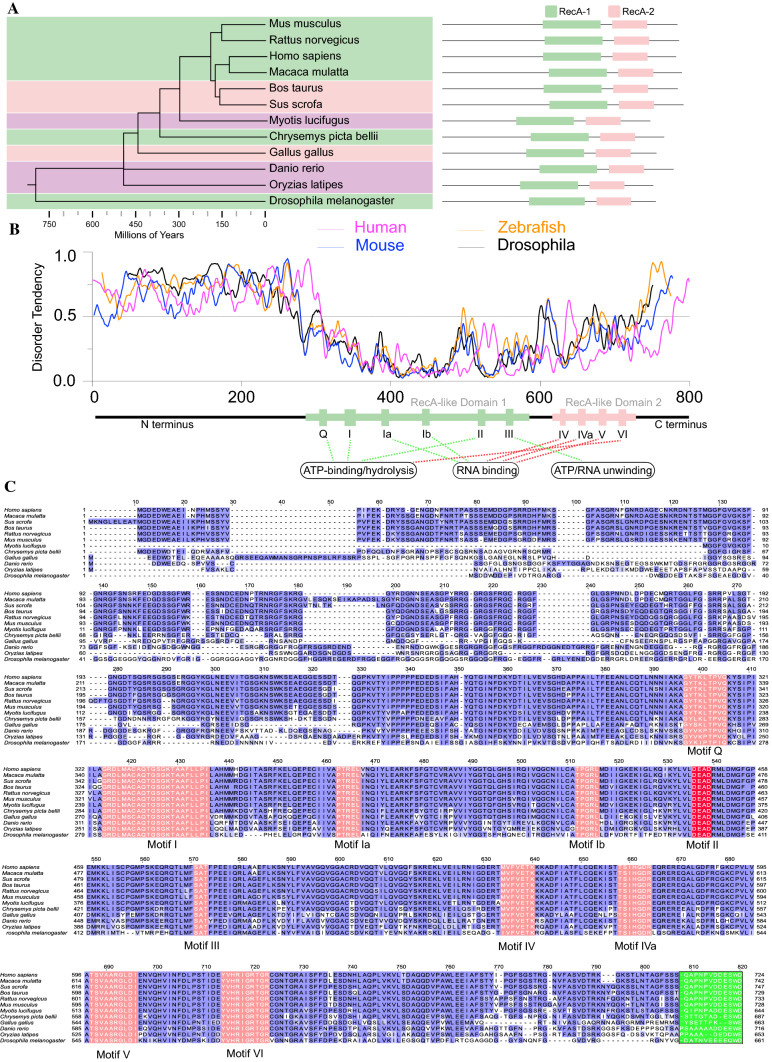
Fig. 1.
Vasa orthologs are evolutionarily conserved and harbor intrinsically disordered regions at both the N-terminus and C-terminus. A The phylogenetic tree of Vasa orthologs from 12 animal species with schematic domain composition illustrated on the right side. Estimated divergence times are obtained from the TimeTree database [102]. B In silico prediction of protein disorder tendency for sequences of Vasa orthologs from mouse (blue), human (red), Drosophila melanogaster (black), and Zebrafish (yellow). Residues above the dotted line are predicted to be disordered by IUPred2A [103]. Conservative regions (DEAD-box helicase domain) are indicated by colored rectangles below. C Multi-sequence alignment of amino acids for Vasa orthologs across 12 metazoan species using Jalview [104]. Amino acids highlighted in red color represent conserved motifs in DEAD-box helicase family (Motifs I–VI) with names labeled, respectively, below. The green color highlights the divergent acidic C-terminal sequence across 12 species
Vasa/Ddx4 is highly conserved across metazoan species in the DEAD-box helicase family
Vasa orthologs are highly evolutionarily conserved and are present in most metazoan species (Fig. 1A, C). Vasa or its mammalian ortholog, Ddx4, harbors consensus DEAD-box helicase motifs, and thus belongs to the DEAD-box family. The DEAD-box protein family was initially defined by the identification of a handful of conserved sequence motifs when comparing a number of evolutionarily conserved RNA metabolism-related proteins, including Vasa, eIF4a, p68, and PL10 et al. [29, 30]. Alignment of amino acid sequences typically revealed at least nine conservative sequence motifs, including motifs Q, I, Ia, Ib, II (DEAD), III, IV, IVa, V and VI, that exhibit minuscule phylogenetic variation across metazoan species as illustrated in Fig. 1C. In general, the helicase core of the DEAD-box family protein is comprised of two recombinase A (RecA)-like domains—the N-terminal domain (NTD) and the C-terminal domain (CTD)—which closely interact with each other to render the bound single- or double-stranded RNA sharply bent through ATP binding and hydrolysis, leading to the disruption of the RNA duplex. As such, the DEAD-box helicase family members can alter (melt) the local secondary RNA structures of the bound substrate RNAs in an ATP-dependent manner. This process is required in a broad range of RNA metabolic processes [31]. Motif II, also known as the Walker B motif, encompasses the canonical core Asp-Glu-Ala-Asp (D-E-A-D) residues that endorse the DEAD name to this family. Motif Q, Motif I (Walker A motif), Motif II and Motif VI are involved in ATP binding and hydrolysis. In comparison, Motifs Ia, Ib, IV and V are composed of consensus residues that interact with the sugar-phosphate backbone of substrate RNAs, as revealed by the crystal structure of Vasa (Fig. 1B, C) [32, 33]. The remaining motifs coordinate the domain interplay that connects RNA unwinding to ATP binding and hydrolysis. It is, however, worth noting that these motifs are not individually dedicated to one function, but often participate in other interactions accompanied by the conformational change of the Vasa/RNA complex [31, 33].
Vasa/Ddx4 is the founding member of the DEAD-box family that hosts the largest number of nucleic acid helicases encoded in metazoa. It harbors all the typical helicase motifs which can change the susceptibility of substrate RNAs to nuclease digestion. However, as is commonly observed for most other DEAD-box family proteins, Vasa/Ddx4 has divergent tails at both N-terminus and C-terminus flanking the canonical helicase domain. The amino acid sequences at both ends significantly vary from Drosophila to higher vertebrates (Fig. 1C). Noteworthily, the sequence diversity at the N-terminus is even present within a closely related species, such as between D. melanogaster and D. simulans [34]. Growing evidence shows that the tail sequences are the intrinsically disordered regions (IDRs), which drive phase separation and endow the Vasa/Ddx4 protein with versatile functions fundamental to germline development as discussed below [29, 30, 35].
Ddx4-mediated nuage formation resulting from phase separation: the tails tell the tale
As aforementioned, while the two tandem RecA-like helicase domains (homologous to the bacterial RecA domain) are highly conserved among members of the DEAD-box family, they typically harbor the variable low-complexity amino acids at their tails, namely intrinsically disordered regions (IDRs) (Fig. 1B, C), with longer N-terminus and shorter C-terminus. IDRs were originally defined by the presence of a disordered stretch of at least 30 amino acids [36], which are often highly enriched in disorder-promoting residues, such as Arg, Gly, Lys, Ser and Pro, but depleted of order-promoting residues, including Leu, Trp, Tyr, Cys, Asn, IIe and Val. Although it has been long known that the function of a protein is dependent on its primary sequence-based 3D structure, growing evidence reveals that abundant proteins play roles without a stable structure in vivo [37]. For instance, the intrinsically disordered proteins (IDPs) significantly differ from those structured proteins or domains in their IDR sequences in many aspects, e.g., low complexity, repetitive sequences (RG, YG, QN et al.), hydrophobicity, charge, and flexibility [38]. IDPs are highly enriched in biomolecular condensates and proven to initiate phase transition both in vitro and in cellulo [18, 38]. While IDPs usually lack tertiary residual contacts, they can bind to a vast number of protein or nucleic acids partners, through short sequence motifs and low-complexity sequences. In particular, these IDPs-interacting partners could significantly differ under different cellular settings.
The IDR sequences have been intensively explored to serve as the primary driving force for the LLPS, a phenomenon of demixing of a liquid phase with another, resulting in the formation of biomolecular condensates, also known as the liquid droplets or membraneless organelles. They are typically micron-sized coacervates, predominantly consisting of ribonucleoprotein (RNP) assemblies that exclude the bulk aqueous phase. These liquid droplets have been extensively studied in different settings, such as the P-bodies, stress granules, Cajal bodies, nucleoli, and so on [11, 39]. It is believed that these membraneless RNP granules, like membrane-encapsulated organelles, provide an alternative local solvent milieu that allows specialized biochemical reactions, but in the meantime, harbor additional advantage that permits an efficient exchange of constituents with surrounding nucleoplasm or cytoplasm. In addition, the absence of encapsulated membrane enables them to quickly respond to external environmental stimuli via rapid assembly and disassembly [11, 14, 15, 40]. On the one hand, the low-complexity sequence in the IDPs is necessary and sufficient to drive phase transition for assembly of liquid droplets independent of the RNA molecules; Proteomic analyses revealed abundant RNA-binding proteins with low-complexity sequences, such as FUS, NANOS, Pumilio et al. enriched in the RNP granules, irrespective of the cell types or tissue origins, possibly implying synergistic enhancement during phase separation among the droplets-contained IDPs [19, 38, 40].
Germ granules are such a typical form of liquid droplets that differ with respect to morphology and RNP contents among diverse species or at varied developmental stages in germline as aforementioned. Ddx4 probably exemplified one of the best-characterized IDPs driving the assembly of germ granules. Nott et al. validated that Ddx4YFP, wherein the conserved DEAD-box cassette was substituted by the YFP sequence, inherently condensed to form liquid-like organelles in the nuclei of HeLa cells [41]. These liquid-like droplets display rapid assembly and disassembly in response to changing surrounding milieus, such as the temperature, salt concentration or tonicity. Furthermore, the disordered sequences of Ddx4 could reversibly form granular structures in vitro, resembling those formed within cells, and the FRAP experiments further demonstrated the similar dynamic internal architecture, indicating that the N-terminal intrinsically disordered region of Ddx4 is the sequence determinant that is sufficient and indispensable to drive the phase separation [41]. Further experiments showed that the droplet stability is primarily maintained by the electrostatic interactions among the interior protein molecules, but could be attenuated by the post-translational methylations via the protein arginine methyltransferase family, e.g., asymmetric dimethylarginine (ADMA) in the six predicted methylation sites at N-terminus of Ddx4 [35, 41]. In agreement with this, a noteworthy feature in the disordered N-terminal tail of Ddx4 lies in that the charged residues are distributed into clustered blocks of net positive and negative charge, consisting of 8–10 residues in length for each block along with pair repeats of FG or RG held in close vicinity [35, 38, 41]. When these residues were swapped to scramble the blocks while simultaneously maintaining the same net charge overall, the newly assembled protein failed to form organelle-like structures in vitro or in cells, indicative of an important role of charge patterning in droplet assembly. Intriguingly, the similar occurrences of short, repeated peptide motifs, such as RG/YG, FG and QN, have been observed to facilitate the assembly of FUS proteins, nuclear pore complex, and P bodies, respectively, suggesting that multivalent interactions, likely through cation-pi interactions, could drive the self-assembly of disordered sequences [6, 18, 42].
Sexually dimorphic functions of Vasa/Ddx4 in metazoa: Vasa/Ddx4 knockout led to gender-specific sterility phenotype in Drosophila and mice
Vas deletion causes female sterility and defective embryonic patterning in Drosophila. In Drosophila, Vas is a maternally derived factor that localizes into two functionally distinct compartments—the pole plasm in the oocytes and the nuage in nurse cells—inside the egg chamber (Fig. 2A). The active transcription and protein translation in the nurse cells produce enough RNAs and proteins, which are subsequently transported to the neighboring oocytes and are essential for oocyte development and embryonic patterning. In the fertilized oocyte, it has been known for decades that the short form of Oskar, short Oskar, is transported via the cytoskeletal microtubules to the posterior tip of the oocyte, and assembles a functional subcellular structure, namely pole plasm, which determines the embryonic anterior–posterior axis and the specification of future primordial germ cells (PGC) [43]. Vasa is specifically recruited to the polar plasm in part through interaction with the C-terminal extended LOTUS domain (eLOTUS) of short Oskar. However, the fact that the long Oskar, which has both LOTUS and OSK domains as in short Oskar, prevents the interaction between Oskar and Vasa, suggest that the N-terminal extension of the long Oskar regulates the interaction between Vasa and Oskar [44]. Noteworthily, the LOTUS domain is highly conserved from bacteria to vertebrates, and was originally defined by its presence in two members of the TUDOR protein family—TDRD5 and TDRD7—in mammals (Table 2) [44, 45]. In the nurse cells of Drosophila, their orthologs Tejas and Tapas are critical for RNA-independent Vasa localization to the nuage and are involved in piRNA-mediated retrotransposon silencing pathway, although the Tudor domains themselves recognize and interact with arginine-methylated, RGG-enriched motifs in the N-terminal tail of Vasa [46] (Table 2). These evidence together suggest that Vasa serves as an upstream regulator in the assemblies of germ plasm in oocytes and nuage in nurse cells.
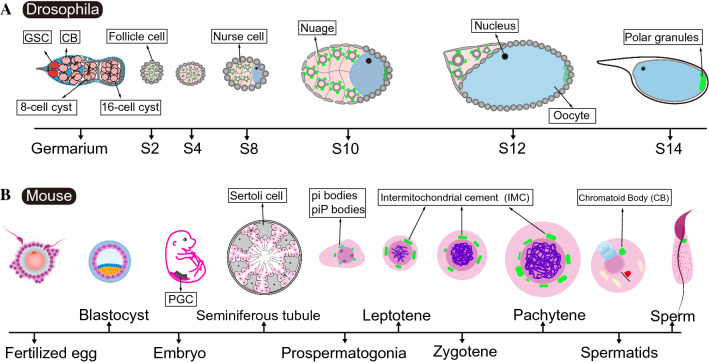
Fig. 2.
Schematic diagrams of germline development and dynamic distribution of nuage in Drosophila and mice. A Anatomy of the ovaries in a single ovariole illustrating nuage formation during Drosophila oogenesis. The Drosophila ovaries comprise 16–20 tubular structures, termed ovarioles, which are arranged as a production line-like assembly of differentiating egg chambers to produce mature eggs (oogenesis). The functional unit of the Drosophila ovary is termed the ovarian follicle, or egg chamber, originating from the germarium that is localized to the anterior tip in each ovariole. Germline stem cells (GSCs) (red) undergo asymmetric cell division, giving rise to one daughter stem cell that remains in contact with the Cap cells, and the other daughter cell destined to differentiate into Cystoblast (CB). Following four rounds of successively synchronous mitotic cell divisions, 16 interconnected cystocytes (light red) are produced, and subsequently bud off the germarium once they are surrounded by somatic follicle cells. During the development of each egg chamber, one of the 16 cystocytes is committed to meiosis and divides into the oocyte at the posterior end, whereas the remaining cells develop into polyploid nurse cells, which are interconnected through the intercellular bridges. Vasa-enriching nuage in nurse cells and Vasa-containing polar granules in oocyte are highlighted in green color. S, stage; B schematic diagram of nuage distribution (green color) in the male mouse germline. Primordial germ cells (PGCs) are induced in a subpopulation of the epiblast in the early embryos starting on embryonic day 6.5 (E6.5). Ddx4 is detectable in PGCs from E10 onwards. Pi-bodies and piP-bodies are present in prospermatogonia in the perinatal testis. Intermitochondrial cement (IMC) is present in the spermatocytes. The chromatoid body (CB) is assembled in haploid spermatids. All granules (green) are localized around the nuclear membrane within the cytoplasm
Table 2.
Representative Vasa/DDX4-interacting partners and their interacting domains
| Species | Protein | Interacting domains | KO phenotype of the interactor | Refs |
|---|---|---|---|---|
| Drosophila | Oskar | Oskar(eLOTUS) vs Vasa(CTD) | No pole plasm, failure of embryonic patterning | [43, 44, 97] |
| Drosophila | eIF5B | eIF5B vs Vasa (C-terminal 595–621) | Vas-eIF5B interaction is required for grk translation and oogenesis | [98] |
| Drosophila | Tapas (Tdrd7 ortholog) | Tapas vs Vasa | Reduced piRNAs, increased TEs, defective piRNA signaling | [99] |
| Drosophila | mei-P26* (mRNA) | 3ʹUTR (U-rich) of mei-P26 mRNA vs Vasa | Defective germline stem cell differentiation in both sexes | [82, 100] |
| Mouse | RanBPM/Ranbp9 | RanBPM(IUS-SPRY) vs Ddx4(N-terminus) | Abolished germline formation in both sexes. Impaired alternative splicing and gene expression. No transposon derepression | [32, 84, 91] |
| Mouse | Mili | Copurify with Ddx4, likely via N-terminus(Ddx4) | Meiotic arrest at zygotene; TEs activation | [50, 51, 68] |
| Mouse | Miwi | Copurify with Ddx4, likely via N-terminus(Ddx4) | Spermiogenic arrest in round spermatids (step 4) | [51, 68, 74] |
| Mouse | Tdrd1 | Copurify with Ddx4, independent of N-terminal arginine methylation | Meiotic arrest at zygotene; derepression of L1 transposons | [64, 101] |
| Mouse | Tdrd6 | Copurify with Ddx4, independent of C-terminus | Spermiogenic arrest in round spermatids. Dispersed CB architecture with mislocalized CB components (Ddx4,Miwi,Mael) | [64, 70] |
| Mouse | Mael | Costa reported the co-purification with Ddx4, but Julio et al. reported no interaction | Zygotene arrest and LINE1 derepression | [57, 60, 73] |
| Mouse | Tdrd5 | LOTUS domain | Zygotene arrest (severe) and spermiogenic arrest (mild); Reduced pachytene piRNA biogenesis | [45] |
| Mouse | Tdrd7 | LOTUS domain | Spermiogenic arrest and age-related cataract; LINE1 derepression | [69] |
For a full list of Vasa interactome, readers are encouraged to refer to the FlyBase [96]
*Note: mei-P26 is an mRNA transcript, but not its translated protein
The first complete Vas-null Drosophila model, namely VasPH165, wherein a 7343-bp deletion removed the entire coding region of Vas, was established in 1998 by Paul Lasko’s laboratory [47] (Table 1). In the female Vas-null Drosophila, a series of defects were observed in both the nurse cells and the oocytes, such as reduced numbers of germline stem cells and developing cysts in the germarium, the tumorous or degenerated egg chambers starting from stage 6, the absence of the oocytes, or undifferentiated oocytes with diffuse nucleoli. This phenomenon could be linked to the mis-regulated localization and/or translation of a handful of oocyte-specific mRNAs, suggesting that Vasa is a master coordinator in regulating Drosophila female germline development [26, 43, 47]. Interestingly, Vas-null males do not exhibit any defects in the male germline, implying a sex-specific role of Vasa in germline development. Thereafter, extensive studies have been designed trying to decipher the specific function of different segments of Vasa protein in Drosophila oogenesis, including the rapidly evolving N-terminal tail, the central RecA-like helicase core, and the acidic carboxyl tail. In general, as summarized in Table 1, the diverse phenotypes in individual Vasa mutants are closely correlated with the extent to which the Vasa mutations caused the deleterious effects. For instance, deletion of the N-terminal tail region (Vas∆3–200) led to defects in Grk translation, Vasa localization as well as transposon control. Mutations in the central helicase domain, even with a single residue substitution, caused a spectrum of phenotypes, including defective oogenesis owing to aberrant Vasa localization and transposon derepression [47–49]. The tail sequences of Vasa protein at both ends are highly divergent across phyla (Fig. 1C). Intriguingly, exclusive deletion of the extreme C-terminal end with seven acidic residues (Fig. 1C) abrogated the pole cell specification and translational activation of Grk, and derepressed the transposons, suggesting an indispensable role of the tail sequence in the germline [48, 49].
Table 1.
Phenotypes with mutant Vasa homologs in Drosophila and in mice
| Species | Vasa variants | Phenotype | Refs |
|---|---|---|---|
| Drosophila | VasPH165 (Vasnull) | Null allele due to entire 7343 bp deletion of coding region of vas. Vasnull ovaries have tumorous germline cysts, disrupted oocyte differentiation, fewer mature eggs | [47] |
| Vas1 | Hypomorphic allele generated by EMS with no mutation in coding region of Vas, but its protein is not expressed after germarium stage. Vas1 females lack polar plasm, PGC and a few posterior segments, and are inviable | [24] | |
| VasLYG2 | P-element-induced hypomorphic vas allele due to residual 2% protein level of wildtype vas; oogenesis is less compromised with numerous laid eggs | [76] | |
| GFP-VasaF504E | Reduced enrichment of GFP-Vasa-F504E to the nuage of nurse cells and ubiquitous distribution in oocyte cytoplasm rescued in the vasaPD/D1 | [44] | |
| VasW660E | VasW660E homozygous females have lower number of embryos with pole cells, disrupted embryonic patterning, mislocalization of vas in nuage and pole plasm, reduced function in piRNA biogenesis | [49] | |
| VasW660F | Less effect on germ cell formation and embryonic development compared to VasW660E | [49] | |
| Vasc.bele | Chimeric Vas with last 7 aa substituted by the C-terminal 7 aa from Vas somatic paralog Bel (DDX3). Vasc.bele exhibits granular localization in the nuage as opposed to a smooth perinuclear ring in wt, but maintains its localization to pole plasm till stage 14 with diminished eggs laid | [49] | |
| Vas∆3–200 or Vas∆636–646 or Vas∆655–661 | Female mutants still can produce more than half number of eggs as in wildtype. Translational failure of Grk. Abnormal Vasa localization. Vas∆3–200 and Vas∆655–661 is critical for transposon control | [48] | |
| Walker A (Motif I): (VasG294A, VasK295N, VasT296V) | Female oogenesis was partially rescued albeit with 2 ~ threefold reduction in the egg number. Translational failure of Grk. Abnormal Vasa localization. transposon control | [48, 75] | |
| VasE400A | Female oogenesis was partially rescued albeit with fivefold reduction of the egg. Translational failure of Grk. Abnormal Vasa localization | [48, 75] | |
| VasD554A | Failure to restore female oogenesis. Translational failure of Grk. Abnormal Vasa localization | [48] | |
| Vas∆636–646 or Vas∆655–661 | Translational failure of Grk. Abnormal Vasa localization. Vas∆655–661 is especially critical for transposon control | [48] | |
| VasHE(R170S), VasQ6(V465M) (aka “Vas6”), VasPW(S518F), VasAS(H520Y), VasD5(G552E), Vas3F(G587E) | Failure to localize either to the perinuclear region of the nurse cells or to the pole plasm. Disrupted formation of perinuclear granule particles | [58] | |
| Vas011(I256N)(aka “Vas4”), Vas4C(P268S), Vas014(I271M) (aka “Vas5”) | Vas011(I256N) localized to pole plasm at a reduced level. Vas4C(P268S) failed to localize to the pole plasm; Vas014(I271M) localized normally to both the perinuclear region and pole plasm; | [58] | |
| Mouse | Ddx4null (Exons 9/10 KO) | Impaired meiotic initiation.Male germline ceased differentiation by the zygotene stage and underwent apoptosis. Normal female fertility | [21] |
| Ddx4+/E446Q (DEAD→EQAD) | Male-specific infertility (dominant-negative effect). No derepression of TEs. Spermiogenic arrest | [51] | |
| Ddx4−/E446Q (DEAD→EQAD) | Phenocopied Mvh-null male testes. TEs (IAP and LINE1) activation. Incomplete meiosis. Female germline development is normal | [51] |
Ddx4 knockout led to male infertility. In the mouse germline, the expression of Ddx4 is detectable starting at embryonic day (E) 10.5–11.5 gonads, just right before the sex is determined [20, 21]. From then on, its protein expression is maintained in both sexes at high levels until post-meiotic spermatids in males, and the primary oocytes in the female mice. In Ddx4 knockout mice, wherein the exons 9 and 10 were deleted causing a null mutation, the proliferative activity of PGC was markedly reduced [21, 50]. After birth, the Ddx4-null spermatogonia continued differentiation and meiotic division, but failed to bypass the zygotene stage, and instead underwent apoptosis. However, in contrast to the Vasa-null Drosophila, the female Ddx4-null mice are completely fertile without any morphological defects in the oocytes [21]. Pillai’s group generated a point-mutation knockin mouse model, Ddx4KI, where the consensus ATPase motif (DEAD→DQAD) was mutated, thereby abolishing the RNA helicase activity. Interestingly, the Ddx4+/KI mice displayed male-specific sterility though both the wildtype and mutant Ddx4DQAD proteins are co-expressed, suggesting a dominant-negative effect for this ATPase-deficient mutant [51]. The spermatogenesis took place efficiently but uniformly arrested at the stage of round spermatids, although it is currently not clear to what extent the function of haploid spermatids was impaired in the Ddx4+/KI mice. However, in contrast to Ddx4-null males, no adverse effect on the piRNA biogenesis pathway was observed in the Ddx4+/KI testis, which was corroborated by the repression of TEs as well as the normal association of Mili- and Miwi-bound piRNA profiles. In comparison, the single mutation in the DEAD motif abolished the Ddx4 expression or destabilized the Ddx4 protein in the Ddx4−/KI mice since Ddx4 is not detected in the Ddx4−/KI mice, leading to similar phenotypes between Ddx4−/KI mice and Ddx4-null mice. In addition, the retrotransposons were derepressed only in the Ddx4−/KI mice but not in the Ddx4+/KI mice, reminiscent of the helicase-independent functions of Ddx4 in male germline differentiation [51].
Ddx4-containing nuage formation and piRNA-mediated retrotransposon control: one size fits all
DDX4 is a master coordinator of nuage organization in the germline. The RNP aggregates are commonly observed in the cytoplasm of the germ cells throughout their life cycle, and thus were given the name “germ granules”, or “nuage”, due to their cloud-like, amorphous and membraneless morphology under transmission electron microscopy (TEM) [52–54] (Fig. 2). In the fetal or perinatal germline in male mice, the perinuclear organelles are vital for the biogenesis of transposable elements (TEs)-derived piwi-interacting RNAs (piRNAs). In general, following transcription by RNA polymerase II and nuclear export to the cytoplasm, there are two proposed processing pathways for piRNA maturation—the primary processing pathway is to cleave the presumably single-stranded precursor piRNA transcripts into thousands of non-overlapping/phased fragments with a strong 5ʹ uridine preference (Mili-bound sense primary piRNAs), while the secondary processing pathway would amplify the piRNA pool of Mili- and Miwi2-bound piRNAs through a so-called “Ping-pong” cycle mechanism [45, 51, 55, 56]. While many details remain elusive concerning what enzymes and how their cofactors are responsible for the primary piRNA processing, it is quite clear that the latter pathway—secondary processing pathway—necessitates the functional integrity of Ddx4-enriching perinuclear nuage, wherein a vast number of PIWI- and Tudor-family protein members dwell in and interplay, as is deduced from the genetically engineered mouse models over the past decades [46, 54, 56, 57]. For instance, genetic deletion of Ddx4 or mutation of the catalytic residue in the DEAD motif led to the altered ultrastructural appearance and the disrupted function of the germ granules as described below [21, 51, 54]. In Drosophila, a single mutation of the amino acid within the N-terminus (e.g., VasHE(R170S)) or in the helicase domain (e.g., VasAS(H520Y)) abrogated the structural integrity of the perinuclear granule particles [58]. While the helicase domain of Vasa/Ddx4 is absolutely essential for executing the substrate RNA unwinding, the disordered tails could maintain the structural architecture of granules alone without the helicase domain, presumably through their interaction with other partners (Table 2) [58]. These genetic evidence suggest that the full-sized Vasa/Ddx4 with both tails and helicase domain is proficient in coordinating the organization and in preserving the functional integrity of the germ granules in both mice and Drosophila. However, it is noteworthy that the secondary “Ping-pong” pathway slightly differs between mice and Drosophila in that different players and cofactors are involved as detailed below.
Pi-bodies and piP-bodies in fetal perinatal germline. In mouse fetal prospermatogonia, there are at least two proposed types of germ granules related to piRNA biogenesis by electron microscopy and immunofluorescence co-staining (Fig. 2B). The first type is called pi-bodies, which comprise Mili/Tdrd1/Gasz/Mov10l1 responsible for processing of the TEs-derived, newly transcribed, precursor long piRNA transcripts; The second type of germ granule shares partial common components of Processing body (P body), and were, thus, named piP-bodies, which harbor Tdrd9/Miwi2/Mael components in addition to the universal P body members, including Ddx6 and Gw182 [54, 59–62]. Both types of granules are most often localized in close vicinity, whereby the processed secondary piRNA can circularize between two types of granules for the efficient production of the mature piRNAs in a feed-forward way, a mechanism for piRNA biogenesis known as the “Ping-pong” cycle. Intriguingly, the Ddx4, as a core piRNA biogenesis factor, was shared by both types of granules. At the molecular level, Mili binds the shorter piRNAs (~ 26nt) while Miwi2 associates with TEs-targeting antisense longer piRNAs (~ 29nt) population [56, 63]. Ddx4 co-localizes and associates with members in both types of nuage in the testis; however, it might not directly interact with piRNAs in the fetal germline as non-crosslinked immunoprecipitation failed to recapitulate the piRNAs profile in the fetal testis lysate. In contrast, its direct binding to pachytene piRNAs was detected in adult testes [50, 64]. Therefore, it is tempting to postulate that Ddx4 might enhance the shuttling of the substrate RNAs between the pi-bodies and piP-bodies by remodeling their secondary structures, thereby facilitating the “Ping-pong” amplification-mediated biogenesis of TE-derived piRNAs [54, 62]. In agreement with this idea, genetic mouse models with deletion of these components remarkably phenocopied each other, as evidenced by the meiotic arrest and aberrant activation of TEs due to the defective de novo DNA methylation.
Intermitochondrial cement (IMC) in spermatocytes. In spermatocytes, the germ granules assemble into highly electron-dense particles with sizes ranging between 60 and 90 nm, or as clusters of 60–90 nm strands, but most often appear as a cementing material connecting the neighboring mitochondria; thus, being initially called “intermitochondrial cement (IMC)”, by Eddy et al. [52, 53] (Fig. 2B). Interestingly, while different germline marker proteins, such as Mael and Ddx25, dynamically associate with the granules at varied stages, Ddx4 is most likely a constitutive member who always stays with and orchestrates the particles [60, 65]. The localization of these particles is highly dynamic within the cytoplasm; however, they are in close contact with the nuclear membrane and the mitochondria, implying that an ATP-dependent RNA-unfolding function presumably enforced by RNA helicase, e.g., Ddx4, is crucial for the functional integrity of nuages [65]. Furthermore, the similar localization pattern and constituents for the granules shared by the fetal spermatogonia and the meiotic spermatocytes suggest that IMC is a continuation of the material descendants from the prospermatogonia. Noteworthily, histological immuno-electron microscopy examination showed that IMC particles are even more abundant in the late stages of spermatocytes, i.e., pachytene spermatocytes. Since the nuage in the prospermatogonia, i.e., pi-bodies and piP-bodies, are the sites for processing of the precursor piRNAs from the TEs via “Ping-pong cycle”, along with the fact that the majority of pachytene piRNAs are derived from discrete piRNA clusters, it is highly likely that the major role of IMC in spermatocytes is to execute similar functions in silencing TEs as in prospermatogonia.
Chromatoid body (CB) in round spermatids. Evolutionarily, IMC in the spermatocytes emanates from the pi-bodies present in the perinatal prospermatogonia as validated by immune-electron microscopy. Some IMC granules might depart from mitochondria, and aggregate further with the isolated clusters of strands into a single discernible electron-dense granular “dot”, giving rise to the chromatoid body (CB) in post-meiotic spermatids (Fig. 2B). Previous studies by immunofluorescence staining and immunoprecipitation have revealed abundant protein components in CB, including the members of piRNA pathway, such as Ddx4, Mael, PIWI family (Miwi, Mili) [57, 60, 66–68], Tudor family (Tdrd1, Tdrd6, Tdrd7) et al. [45, 69, 70]. CB also harbors protein constituents commonly related to RNA processing or metabolism, e.g., Upf1 and Upf2, which are involved in Non-sense-mediated mRNA Decay (NMD) (Table 2) [69]. The convergence of the Tudor-piRNA pathway and NMD pathway in the CB structure reinforces the functional significance of CB in piRNA and mRNA metabolism. However, unlike the piRNAs in the fetal gonocytes, of which the majority are transcribed from large discrete piRNA clusters from TE loci in the genome, ~ 80% of piRNAs in the pachytene spermatocytes and the spermatids stem from the top 50 unannotated intergenic genomic regions termed as “pachytene piRNA clusters”, whose functions are not yet clearly understood [71]. On the other hand, as discussed above, germ cell differentiation can proceed to round spermatids in the Ddx4+/KI testis, whereas meiotic arrest at the zygotene stage occurred in the Ddx4−/KI spermatocytes. The finding that the loss of repeat piRNAs and the presence of MILI-sliced 51-mer pre-piRNA intermediates in the Ddx4−/KI spermatocytes is reminiscent of the functional integrity of the pi-bodies where Ddx4 dwells in together with Mili-Tdrd1, rather than piP-bodies. In the CB of Ddx4+/KI round spermatids, the catalytic-dead Ddx4 remained complexed with the WT Ddx4 and other well-known components (Miwi, Mili and Tdrd1) in the CB. By comparison, immunoprecipitation uncovered that those components alongside the 24–30nt piRNAs are more enriched, implicating that catalytic-dead Ddx4 lost the dynamic remodeling activity of its RNA substrates in vivo, and presumably entrapped the piRNA machinery, leading to the destructive disengagement of its substrates [51, 72]. In addition, a short isoform of Tdrd6 is vastly expressed and physically interacts with Ddx4 in round spermatids, and its deletion led to the diffuse organization of CB with mislocalized Ddx4 and Miwi, whose intensive “singular dot” appearance disappeared [50, 70]. The phenotypes of Tdrd6−/− and Ddx4+/KI testes resemble each other with post-meiotic arrest at the stage of round spermatids. Strikingly, the retrotransposons for LINE1 and IAP are not significantly activated in either Tdrd6−/− or Ddx4+/KI testes, suggesting the essential functions of Ddx4 and CB in regulating the haploid germ cell development independent of TE repression [51, 70, 72, 73]. Another predominant resident of CB is Miwi, which frequently interacts with Ddx4 and, when deleted, showed the spermiogenic arrest at step 4 of round spermatids (Table 2). The RNase activity of Miwi (slicer) conferred by the consensus “DDH” motif is critical to silence the LINE1 transposons independent of “Ping-pong cycle” [74]. The CBs in the round spermatids from either Miwi-null or Miwi slicer-deficient (Miwi+/ADH) testis were fragmented with the LINE1 protein—L1ORF1p—being dramatically upregulated. However, the localizations of most CB constituents appeared not to be affected. Together, these evidences suggest that the Ddx4 abundantly residing in the CB might coordinate and remodel multiple players for both transposon repression and piRNA-independent RNA re-structuring essential for CB-orchestrated haploid spermatid development [56, 74].
Vasa ablation caused nuage loss and derepression of retrotransposons in Drosophila. In Drosophila, the post-meiotic pro-oocyte cell develops into the oocyte with progressive loss of the perinuclear germ granules, whereas the other 15 supporting cells (nurse cells) retain the nuage within the egg chamber (Fig. 2A). Meanwhile, this process is accompanied by the condensing of nuclear karyosomes and the cessation of transcriptional machinery [1, 75, 76]. Thus, in contrast to mammals, the nuage is present in the nurse cells, but not in the germline or soma (i.e., follicle cells), in the female Drosophila (Fig. 2A). Without a functional nuage machinery, it has been found that the follicle cells only encompass the primary piRNA processing pathway, which takes place in another specialized subcellular compartment, namely Yb body [77]. Unlike the majority of pachytene piRNAs generated from large single-stranded piRNA clusters in mice, Drosophila piRNAs are produced from either uni-strand or dual-strand piRNA clusters, such as the 42AB cluster locus in the pericentromeric region of Chr2R. It has been found that the piRNA precursors, once transcribed by RNA Polymerase II, are bound by UAP56, which is localized in the close vicinity of the inner side of the nuclear pores [78]. In the meantime, the perinuclear nuage-enriched Vasa dynamically interacts with Uap56 across the nuclear pores, which facilitates the transport of piRNA precursors from the nuclei to the cytosol for processing into mature piRNAs subsequently. Depletion of Vasa elicited the disassembly and loss of perinuclear granules, and abolished the transposon silencing, but did not impact the expression of protein-coding genes, suggesting a central role of Vasa in nuage-guided transposon control.
The other interesting finding in Drosophila is that there appears to be a tight coupling of nuage to mitochondria during primary piRNA biogenesis. This idea was supported by the colocalization of multiple effector proteins on the surface of mitochondria, such as the Zuc, Armi and Papi [79]. Consistently, their homologs in mice, including MitoPLD, Mov10l1 and Tdrkh, are all present on the outer membrane surface of the mitochondria as well as in the IMC, in mouse germline [59, 62, 80]. However, electron microscopy validated that the perinuclear nuages are not physically close to mitochondria. Intriguingly, genetic deletion of these genes caused similar defects in the primary piRNA processing pathway in both Drosophila and mice, implicating that the mitochondria are indeed pivotal for primary piRNA processing. Then the next question becomes why the mitochondria are so crucial for piRNA biogenesis? Does the mitochondrial membrane surface provide an anchoring site, thereby increasing the local concentration of machinery players for primary piRNA effector protein assembly? This hypothesis seems to be unlikely possible since other similar membrane structures, e.g., endoplasmic reticulum, are also present in each cell. As detailed above, the efficient piRNA processing and movement involve quick RNP remodeling of the nuage, thereby necessitating a large consumption of the ATP energy, as required by the ATPase Vasa/Ddx4 and Armi/Mov10l1 [72]. In this sense, it seems to be plausible to explain why nuages retain close physical contact with the mitochondria, the source of a powerhouse in cells.
Hypothesis: Vasa/Ddx4 fine-tunes the target RNA/protein repertoire in germ granules independent of its sequence specificity
The feature of RNA helicases is their ability to unwind RNA duplexes. We can categorize the RNA helicases into two types based on the catalytic mechanism on the substrates. The first type guides the unwinding of duplex RNA in a canonical ATP-dependent manner by binding and translocating in a 5’-3’ direction along the RNA strand, such as Mov10l1 and Staufen, known as processive RNA helicases [43, 59, 81]. The second type relies on ATP hydrolysis to resolve or melt local secondary RNA structure in a non-processive way to facilitate RNA strands separation or dissociation among RNA/RNA or RNA/protein complexes, including the DEAD-box protein family. As mentioned above, similar to other DEAD-box proteins, the crystal structure revealed that the helicase core region of Vasa tightly folded by the NTD and CTD domains interacts with the backbone phosphate-ribose rather than the base moieties, implying that the RecA-like domains do not bind target RNAs in a sequence-specific manner, but execute its helicase function independent of the RNA sequence, to resolve local short RNA duplexes in the vicinity non-processively [33, 72]. In support of this hypothesis, among 221 candidate mRNAs bound to Vasa identified by Liu et al. in Drosophila embryos, only 24 mRNAs were present along with Vasa in pole cells, and there appeared to be no consensus sequence motifs shared by those mRNAs [82]. Furthermore, the disordered C. elegans protein MEG-3 preferentially binds to ~ 500 mRNAs in a sequence-independent manner with low ribosome coverage within the P granules in vivo, as revealed by individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) [83]. Indeed, the RNP constituents are highly condensed in the coacervate, therefore, there is a peak demand for ATP-dependent unwinding of the RNA duplex to facilitate the timely communication and interaction between RNA-RNA and RNA-proteins. In this regard, the DEAD-box helicases, including Vasa/Ddx4, appear to be the appropriate and global regulators involved in germ granules resulting from phase separation [30, 35].
At the protein level, Vasa/Ddx4 has been shown to interact with other components physically within germ granules, including Oskar and eIF5B in Drosophila [44, 82], and Ranbp9, Mili and Miwi et al. in mice [50, 56, 68, 84] (Table 2). In most cases, this interaction is likely mediated through the IDRs of Vasa/Ddx4, or occasionally via the full-length residues. Since the IDRs exhibit no stable tertiary structure, it is, thus, tempting to posit that Vasa/Ddx4 has a great capacity to facilitate the unwinding/processing of its diverse RNA substrates through repeated, weak, and transient interactions. Therefore, building a proteomic landscape of molecular players in the granules will unambiguously assist in understanding the mechanisms underlying germline development. Two features characterize the protein architecture in the proteinaceous RNP granules. One feature is typified by RNA-binding domains, such as RNA recognition motif (RRM), K-homology (KH) domain, double-stranded RNA-binding domain (dsRBD), et al. The other is the enrichment of low-complexity sequence capable of driving phase transition. This capability endows the RNA-binding protein with the ability to enrich its RNA targets specifically in the RNP granules [14, 18]. In agreement with this hypothesis, high throughput sequencing revealed, for example, that mRNAs with the Pumilio-bound Nanos response elements are especially retained in the Pumilio-containing RNP granules [85]. The core members of the NMD pathway, including ATPase Upf1 and Upf2, are remarkably concentrated in the CB of round spermatids, and a body of evidences showed that CB is an RNA-processing center that enables highly specialized mRNA decay or selective degradation of longer 3’UTR transcripts restricted to a relatively isolated microreactor organelle [67, 86, 87]. It is, thus, conceivable that Ddx4, which is highly condensed in the CB, must be efficient in charge of timely unwinding and sorting out substrate RNA molecules.
Post-translational regulation of Ddx4 protein
Germline proteins are frequently subject to a combination of post-translational modifications in vivo, and intriguingly, these modifications, in turn, fine-tune the Ddx4 functions by modulating the intrinsic protein–protein or protein-RNA interaction networks throughout germline development. As aforementioned, there are at least six putative consensus arginine methylation sites (RGG), part of which were validated through immunoprecipitation and mass spectrometry analysis at the N-terminus of endogenous Ddx4 protein in mouse testes [64]. However, upon treatment of methyltransferase inhibitor (MTA), there are no apparent changes in the binding intensities between Ddx4 and Tdrd1 or Tdrd6, indicating that the interactions are unlikely mediated through the Tudor-Rme2 module as commonly observed between Tudor-PIWI interactions [64]. In turn, the methyl marks attenuate the electrostatic interactions, which play a critical function in the assembly of RNP granules as detailed above. In addition, Ddx4 is also acetylated at Lys405, which is localized in the DEAD-box domain, by the acetyltransferase Hat1 and its cofactor p46 in mouse testis [88]. The acetylation of Ddx4 is developmentally regulated that parallels the dynamic colocalization of Hat1 and p46 in the CB. Once acetylated, the Ddx4-bound target, eIF4B mRNAs, are selectively released from the Ddx4-containing RNP complex, followed by the elevated eIF4B protein translation, suggesting a key role of Ddx4 in regulating the germline-specific protein translation/activation [88].
Perspective
The establishment of a germline lineage is of paramount importance in sexually reproducing metazoan species. Currently, there are at least two general mechanisms exploited to specify the germline in the animal kingdom: (i) preformation—as seen in worms and Drosophila, wherein germ granules are maternally partitioned and loaded to specify the PGCs; (ii) inductive, also termed epigenetic as seen in mice, wherein extrinsic signaling factors, e.g., BMP4, induce the cell–cell interactions to stimulate differentiation into PGCs [1]. Irrespective of what mechanisms employed for PGC specification, germ cells host a myriad of highly conserved molecular determinants, such as Dazl, Vasa and Gcna. In mice, Dazl is activated much earlier than Ddx4 in PGCs, and genome-wide CLIP (crosslinking and immunoprecipitation) profiling revealed that Dazl prefers to recognize and bind mRNA 3ʹUTR containing a UGUU motif, thereby promoting their protein translation [89]. Gcna is an ancient germline marker that also harbors the IDR at the N-terminus, and when deleted, caused male sterility resembling the Ddx4 knockout phenotype [90]. Since Ddx4 is a constitutive component of the germ granules, Kotaja’s laboratory performed the immunoprecipitation combined with crosslinking with a Ddx4-specific antibody, and identified a multitude of protein constituents in the CB structure, of which many have proven to be required for male germline development [66, 67, 84, 86, 87, 91]. Meanwhile, they have also verified that abundant piRNAs, mRNAs and uncharacterized long non-coding RNAs are present in the CB [66, 67]. Nevertheless, since the nuage structures frequently change their morphology and their protein/RNA residents, it is, thus, meaningful to track the dynamic compositions that accommodate the specific needs for germline development [6, 83]. What is more, it has been shown that the Ddx4 antibody-captured cells were not bona fide germ cells when using the ovary sample for immunolabeling for FACS sorting [92–95]. Therefore, it is necessary to exploit an antibody against a common tag, such as HA or Flag, to specifically labels the Ddx4 protein in situ to purify the highly morphologically heterogeneous population of nuages from various stages of germline. In contrast to Dazl, which hosts the well-known RNA-binding domain (RRM) along with the protein–protein interaction domain (DAZ), Ddx4 only harbors the DEAD-box helicase domain flanked by the unstructured IDPR tails at both ends. We, thus, can envision that Ddx4 might not bind specific target RNAs directly. Rather, it works as a core coordinator that facilitates the timely unfolding of duplex RNAs, and proper interactions among RNA/protein constituents within the over-crowded milieu of germ granules through its inherent helicase activity [82]. Further deciphering the dynamic interaction landscape of the RNA and protein components within the germ granules throughout germline development will undoubtedly shed light on our understanding of the mechanisms underlying germline specification and maintenance.
Author contributions
CX generated all figures with help from Muhammad Azhar. JB wrote and revised the draft with help from all authors.
Funding
This work was supported by grants from the Ministry of Science and Technology of China (2019YFA0802600), National Natural Science Foundation of China (31970793, 32170856), the Fundamental Research Funds for the Central Universities” (WK2070000156) and Startup funding (KY9100000001) from USTC.
Availability of data and materials
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors approve the publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Strome S, Updike D. Specifying and protecting germ cell fate. Nat Rev Mol Cell Biol. 2015;16(7):406–416. doi: 10.1038/nrm4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mukherjee N, Mukherjee C. Germ cell ribonucleoprotein granules in different clades of life: from insects to mammals. Wiley Interdiscip Rev RNA. 2021;12:1642. doi: 10.1002/wrna.1642. [DOI] [PubMed] [Google Scholar]
- 3.Gleason RJ, Anand A, Kai T, Chen X. Protecting and diversifying the germline. Genetics. 2018;208(2):435–471. doi: 10.1534/genetics.117.300208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weismann A. Essays upon Heredity and Kindred Biological Problems. 2. Clarendon press; 1891. Continuity of the germ-plasm. [Google Scholar]
- 5.Hegner RW. Effects of removing the germ-cell determinants from the eggs of some chrysomelid beetles. Biol Bull. 1908;16:19–26. doi: 10.2307/1536121. [DOI] [Google Scholar]
- 6.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21(3):403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development. 2010;137(8):1305–1314. doi: 10.1242/dev.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 9.DeRenzo C, Reese KJ, Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. 2003;424(6949):685–689. doi: 10.1038/nature01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hird SN, Paulsen JE, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122(4):1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- 11.Marnik EA, Updike DL. Membraneless organelles: P granules in Caenorhabditis elegans. Traffic. 2019;20(6):373–379. doi: 10.1111/tra.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallo CM, Wang JT, Motegi F, Seydoux G. Cytoplasmic partitioning of P granule components is not required to specify the germline in C elegans. Science. 2010;330(6011):1685–1689. doi: 10.1126/science.1193697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203(6):875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149(4):768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Guzikowski AR, Chen YS, Zid BM. Stress-induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip Rev RNA. 2019;10(3):e1524. doi: 10.1002/wrna.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Goff L, Lecuit T. Developmental biology. Phase transition in a cell. Science. 2009;324(5935):1654–1655. doi: 10.1126/science.1176523. [DOI] [PubMed] [Google Scholar]
- 18.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015;589(1):15–22. doi: 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci USA. 1994;91(25):12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14(7):841–853. doi: 10.1101/gad.14.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lasko P. The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim Biophys Acta. 2013;1829(8):810–816. doi: 10.1016/j.bbagrm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Gustafson EA, Wessel GM. Vasa genes: emerging roles in the germ line and in multipotent cells. BioEssays. 2010;32(7):626–637. doi: 10.1002/bies.201000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schupbach T, Wieschaus E. Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. Rouxs Arch Dev Biol. 1986;195(5):302–317. doi: 10.1007/BF00376063. [DOI] [PubMed] [Google Scholar]
- 25.Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335(6191):611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- 26.Hay B, Jan LY, Jan YN. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell. 1988;55(4):577–587. doi: 10.1016/0092-8674(88)90216-4. [DOI] [PubMed] [Google Scholar]
- 27.Renault AD. vasa is expressed in somatic cells of the embryonic gonad in a sex-specific manner in Drosophila melanogaster. Biol Open. 2012;1(10):1043–1048. doi: 10.1242/bio.20121909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon J, Wessel GM, Yajima M. An unregulated regulator: Vasa expression in the development of somatic cells and in tumorigenesis. Dev Biol. 2016;415(1):24–32. doi: 10.1016/j.ydbio.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linder P. Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34(15):4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5(3):232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 31.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390(12):1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 32.Hong SK, Kim KH, Song EJ, Kim EE. Structural basis for the interaction between the IUS-SPRY domain of RanBPM and DDX-4 in germ cell development. J Mol Biol. 2016;428(21):4330–4344. doi: 10.1016/j.jmb.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 33.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125(2):287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 34.Dehghani M, Lasko P. Multiple functions of the DEAD-box helicase vasa in drosophila oogenesis. Results Probl Cell Differ. 2017;63:127–147. doi: 10.1007/978-3-319-60855-6_6. [DOI] [PubMed] [Google Scholar]
- 35.Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, Fontoura BMA, Weis K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature. 2019;573(7772):144–148. doi: 10.1038/s41586-019-1502-y. [DOI] [PubMed] [Google Scholar]
- 36.Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37(12):509–516. doi: 10.1016/j.tibs.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Das S, Lin YH, Vernon RM, Forman-Kay JD, Chan HS. Comparative roles of charge, pi, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc Natl Acad Sci USA. 2020;117(46):28795–28805. doi: 10.1073/pnas.2008122117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong PA, Vernon RM, Forman-Kay JD. RGG/RG Motif regions in RNA binding and phase separation. J Mol Biol. 2018;430(23):4650–4665. doi: 10.1016/j.jmb.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Seydoux G. The P. Granules of C. elegans: a genetic model for the study of RNA-protein condensates. J Mol Biol. 2018;430(23):4702–4710. doi: 10.1016/j.jmb.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tauber D, Tauber G, Parker R. mechanisms and regulation of RNA condensation in RNP granule formation. Trends Biochem Sci. 2020;45(9):764–778. doi: 10.1016/j.tibs.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57(5):936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9(4):e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Breitwieser W, Markussen FH, Horstmann H, Ephrussi A. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10(17):2179–2188. doi: 10.1101/gad.10.17.2179. [DOI] [PubMed] [Google Scholar]
- 44.Jeske M, Muller CW, Ephrussi A. The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 2017;31(9):939–952. doi: 10.1101/gad.297051.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ding D, Liu J, Midic U, Wu Y, Dong K, Melnick A, Latham KE, Chen C. TDRD5 binds piRNA precursors and selectively enhances pachytene piRNA processing in mice. Nat Commun. 2018;9(1):127. doi: 10.1038/s41467-017-02622-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20(8):724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 47.Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125(9):1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- 48.Dehghani M, Lasko P. In vivo mapping of the functional regions of the DEAD-box helicase Vasa. Biol Open. 2015;4(4):450–462. doi: 10.1242/bio.201410579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dehghani M, Lasko P. C-terminal residues specific to Vasa among DEAD-box helicases are required for its functions in piRNA biogenesis and embryonic patterning. Dev Genes Evol. 2016;226(6):401–412. doi: 10.1007/s00427-016-0560-5. [DOI] [PubMed] [Google Scholar]
- 50.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24(9):887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wenda JM, Homolka D, Yang Z, Spinelli P, Sachidanandam R, Pandey RR, Pillai RS. Distinct roles of RNA helicases MVH and TDRD9 in PIWI slicing-triggered mammalian piRNA biogenesis and function. Dev Cell. 2017;41(6):623–637 e629. doi: 10.1016/j.devcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burgos MH, Fawcett DW. Studies on the fine structure of the mammalian testis. I. Differentiation of the spermatids in the cat (Felis domestica) J Biophys Biochem Cytol. 1955;1(4):287–300. doi: 10.1083/jcb.1.4.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eddy EM. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat Rec. 1974;178(4):731–757. doi: 10.1002/ar.1091780406. [DOI] [PubMed] [Google Scholar]
- 54.Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5(12):e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 56.Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19(8):773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 2006;15(15):2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- 58.Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120(5):1201–1211. doi: 10.1242/dev.120.5.1201. [DOI] [PubMed] [Google Scholar]
- 59.Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, Wang PJ. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29(6):617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15(2):285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17(6):775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 62.van der Heijden GW, Castaneda J, Bortvin A. Bodies of evidence—compartmentalization of the piRNA pathway in mouse fetal prospermatogonia. Curr Opin Cell Biol. 2010;22(6):752–757. doi: 10.1016/j.ceb.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 63.Ding D, Liu J, Dong K, Melnick AF, Latham KE, Chen C. Mitochondrial membrane-based initial separation of MIWI and MILI functions during pachytene piRNA biogenesis. Nucleic Acids Res. 2019;47(5):2594–2608. doi: 10.1093/nar/gky1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirino Y, Vourekas A, Kim N, de Lima AF, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010;285(11):8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yokota S. Nuage proteins: their localization in subcellular structures of spermatogenic cells as revealed by immunoelectron microscopy. Histochem Cell Biol. 2012;138(1):1–11. doi: 10.1007/s00418-012-0962-z. [DOI] [PubMed] [Google Scholar]
- 66.Meikar O, Da Ros M, Liljenback H, Toppari J, Kotaja N. Accumulation of piRNAs in the chromatoid bodies purified by a novel isolation protocol. Exp Cell Res. 2010;316(9):1567–1575. doi: 10.1016/j.yexcr.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 67.Meikar O, Vagin VV, Chalmel F, Sostar K, Lardenois A, Hammell M, Jin Y, Da Ros M, Wasik KA, Toppari J, et al. An atlas of chromatoid body components. RNA. 2014;20(4):483–495. doi: 10.1261/rna.043729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131(4):839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka T, Hosokawa M, Vagin VV, Reuter M, Hayashi E, Mochizuki AL, Kitamura K, Yamanaka H, Kondoh G, Okawa K, et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc Natl Acad Sci USA. 2011;108(26):10579–10584. doi: 10.1073/pnas.1015447108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vasileva A, Tiedau D, Firooznia A, Muller-Reichert T, Jessberger R. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol. 2009;19(8):630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, et al. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 2013;50(1):67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiol J, Spinelli P, Laussmann MA, Homolka D, Yang Z, Cora E, Coute Y, Conn S, Kadlec J, Sachidanandam R, et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell. 2014;157(7):1698–1711. doi: 10.1016/j.cell.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 73.Castaneda J, Genzor P, van der Heijden GW, Sarkeshik A, Yates JR, 3rd, Ingolia NT, Bortvin A. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J. 2014;33(18):1999–2019. doi: 10.15252/embj.201386855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480(7376):264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 75.Durdevic Z, Ephrussi A. Germ cell lineage homeostasis in drosophila requires the vasa RNA helicase. Genetics. 2019;213(3):911–922. doi: 10.1534/genetics.119.302558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rittenhouse KR, Berg CA. Mutations in the Drosophila gene bullwinkle cause the formation of abnormal eggshell structures and bicaudal embryos. Development. 1995;121(9):3023–3033. doi: 10.1242/dev.121.9.3023. [DOI] [PubMed] [Google Scholar]
- 77.Dennis C, Brasset E, Sarkar A, Vaury C. Export of piRNA precursors by EJC triggers assembly of cytoplasmic Yb-body in Drosophila. Nat Commun. 2016;7:13739. doi: 10.1038/ncomms13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, Vreven T, Meignin C, Davis I, Zamore PD, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151(4):871–884. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ge DT, Wang W, Tipping C, Gainetdinov I, Weng Z, Zamore PD. The RNA-binding ATPase, armitage, couples piRNA amplification in nuage to phased piRNA production on mitochondria. Mol Cell. 2019;74(5):982–995986. doi: 10.1016/j.molcel.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20(3):364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gowravaram M, Schwarz J, Khilji SK, Urlaub H, Chakrabarti S. Insights into the assembly and architecture of a Staufen-mediated mRNA decay (SMD)-competent mRNP. Nat Commun. 2019;10(1):5054. doi: 10.1038/s41467-019-13080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu N, Han H, Lasko P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3' UTR. Genes Dev. 2009;23(23):2742–2752. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee CS, Putnam A, Lu T, He S, Ouyang JPT, Seydoux G. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife. 2020;9:e52896. doi: 10.7554/eLife.52896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bao J, Tang C, Li J, Zhang Y, Bhetwal BP, Zheng H, Yan W. RAN-binding protein 9 is involved in alternative splicing and is critical for male germ cell development and male fertility. PLoS Genet. 2014;10(12):e1004825. doi: 10.1371/journal.pgen.1004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goldstrohm AC, Hall TMT, McKenney KM. Post-transcriptional regulatory functions of mammalian pumilio proteins. Trends Genet. 2018;34(12):972–990. doi: 10.1016/j.tig.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bao J, Vitting-Seerup K, Waage J, Tang C, Ge Y, Porse BT, Yan W. UPF2-dependent nonsense-mediated mRNA decay pathway is essential for spermatogenesis by selectively eliminating longer 3'UTR transcripts. PLoS Genet. 2016;12(5):e1005863. doi: 10.1371/journal.pgen.1005863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lehtiniemi T, Kotaja N. Germ granule-mediated RNA regulation in male germ cells. Reproduction. 2018;155(2):R77–R91. doi: 10.1530/REP-17-0356. [DOI] [PubMed] [Google Scholar]
- 88.Nagamori I, Cruickshank VA, Sassone-Corsi P. Regulation of an RNA granule during spermatogenesis: acetylation of MVH in the chromatoid body of germ cells. J Cell Sci. 2011;124(Pt 24):4346–4355. doi: 10.1242/jcs.096461. [DOI] [PubMed] [Google Scholar]
- 89.Li H, Liang Z, Yang J, Wang D, Wang H, Zhu M, Geng B, Xu EY. DAZL is a master translational regulator of murine spermatogenesis. Natl Sci Rev. 2019;6(3):455–468. doi: 10.1093/nsr/nwy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carmell MA, Dokshin GA, Skaletsky H, Hu YC, van Wolfswinkel JC, Igarashi KJ, Bellott DW, Nefedov M, Reddien PW, Enders GC, et al. A widely employed germ cell marker is an ancient disordered protein with reproductive functions in diverse eukaryotes. Elife. 2016;5:e19993. doi: 10.7554/eLife.19993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Puverel S, Barrick C, Dolci S, Coppola V, Tessarollo L. RanBPM is essential for mouse spermatogenesis and oogenesis. Development. 2011;138(12):2511–2521. doi: 10.1242/dev.062505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang H, Zheng W, Shen Y, Adhikari D, Ueno H, Liu K. Experimental evidence showing that no mitotically active female germline progenitors exist in postnatal mouse ovaries. Proc Natl Acad Sci USA. 2012;109(31):12580–12585. doi: 10.1073/pnas.1206600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagner M, Yoshihara M, Douagi I, Damdimopoulos A, Panula S, Petropoulos S, Lu H, Pettersson K, Palm K, Katayama S, et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat Commun. 2020;11(1):1147. doi: 10.1038/s41467-020-14936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Clarkson YL, Weatherall E, Waterfall M, McLaughlin M, Lu H, Skehel PA, Anderson RA, Telfer EE. Extracellular localisation of the C-terminus of DDX4 confirmed by immunocytochemistry and fluorescence-activated cell sorting. Cells. 2019;8(6):578. doi: 10.3390/cells8060578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Navaroli DM, Tilly JL, Woods DC. Isolation of mammalian oogonial stem cells by antibody-based fluorescence-activated cell sorting. Methods Mol Biol. 2016;1457:253–268. doi: 10.1007/978-1-4939-3795-0_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Larkin A, Marygold SJ, Antonazzo G, Attrill H, Dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, et al. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021;49(D1):D899–D907. doi: 10.1093/nar/gkaa1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jeske M, Bordi M, Glatt S, Muller S, Rybin V, Muller CW, Ephrussi A. The crystal structure of the drosophila germline inducer Oskar identifies two domains with distinct vasa helicase- and RNA-binding activities. Cell Rep. 2015;12(4):587–598. doi: 10.1016/j.celrep.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 98.Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131(17):4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 99.Patil VS, Anand A, Chakrabarti A, Kai T. The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in the piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol. 2014;12:61. doi: 10.1186/s12915-014-0061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Page SL, McKim KS, Deneen B, Van Hook TL, Hawley RS. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics. 2000;155(4):1757–1772. doi: 10.1093/genetics/155.4.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16(6):639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 102.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017;34(7):1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- 103.Meszaros B, Erdos G, Dosztanyi Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46(W1):W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.